Abstract
The [URE3] nonchromosomal genetic element is an infectious form (prion) of the Ure2 protein, apparently a self-propagating amyloidosis. We find that an insertion mutation or deletion of HSP104 results in inability to propagate the [URE3] prion. Our results indicate that Hsp104 is a common factor in the maintenance of two independent yeast prions. However, overproduction of Hsp104 does not affect the stability of [URE3], in contrast to what is found for the [PSI+] prion, which is known to be cured by either overproduction or deficiency of Hsp104. Like Hsp104, the Hsp40 class chaperone Ydj1p, with the Hsp70 class Ssa1p, can renature proteins. We find that overproduction of Ydj1p results in a gradual complete loss of [URE3]. The involvement of protein chaperones in the propagation of [URE3] indicates a role for protein conformation in inheritance.
An infectious protein (prion) can, in principle, arise by many mechanisms, including production of a self-modifying protein or a protein that positively regulates its own production (1, 26, 44). However, the known and suspected cases of prions all appear to involve self-propagating protein aggregation (reviewed in references 33, 45, 55, and 57 to 59).
Like yeast viruses, an infectious protein in yeast is expected to be transmitted as a nonchromosomal genetic element. The nonchromosomal genetic elements of Saccharomyces cerevisiae, [URE3] and [PSI+], were identified about 30 years ago (15, 31), but their molecular basis was long obscure. Three unusual genetic properties, each expected of a prion but not of a nucleic acid replicon, were used to identify [URE3] and [PSI+] as prions of the Ure2 and Sup35 proteins, respectively (56). First, if a prion can be cured, it should arise again in the cured strain at some low frequency because the normal form of the protein is still present. Second, overproduction of the normal form of the protein should increase the frequency with which the prion form arises, simply because there is more of the protein present to undergo the prion change. Third, mutants in the gene for the protein should be unable to propagate the prion but the phenotype of these mutants and that produced by the presence of the prion should be the same, because each of these conditions results in a deficiency of the normal form of the protein. [URE3] and [PSI+] have each of these properties as prions of Ure2p and Sup35p, respectively (56). Confirming these assignments are experiments showing that [URE3] truly arises de novo (37), that it is the Sup35 or Ure2 protein (and not the mRNA or the gene) whose overproduction gives rise to [PSI+] or [URE3] (18, 37), and that a regulatory loop is not the basis of the [URE3] phenomenon (37).
Ure2p is a regulator of nitrogen catabolism involved in allowing yeast to use the best nitrogen source available and to turn off the genes involved in the assimilation of poor nitrogen sources (13, 19, 36). A cascade of regulatory factors has been found, with ammonia (a good nitrogen source) inhibiting Mks1p, which in turn inhibits Ure2p, which blocks positive transcription factor Gln3p (14, 21). Gln3p activates transcription of (among others) the allanoate permease gene, DAL5 (47). Because of the chemical resemblance of ureidosuccinate (USA), an intermediate in uracil biosynthesis, to allantoate, a poor nitrogen source, Dal5p can take up USA (54). Thus, the inability of cells blocked in uracil biosynthesis before USA to utilize USA in place of uracil can be used as a measure of Ure2p activity. Schematically, the pathway is NH3 ⊣ Mks1p ⊣ Ure2p ⊣ Gln3p → DAL5 → USA uptake. Either ure2 mutants or cells carrying the [URE3] prion can use USA in place of uracil in spite of the presence of ammonia in the medium (31).
Sup35p and Sup45p are the subunits of the translation termination factor (52, 60). sup35 mutants allow weak nonsense suppressor tRNAs to be more efficient because they have weakened competition by the normal translation termination factors. The presence of the [PSI+] prion confers a phenotype similar to that produced by many recessive sup35 mutations.
Biochemical and cell biological studies indicate that the prion mechanism for [URE3] and [PSI] is amyloid formation (20, 24, 28, 38, 41–43, 53). Ure2p is protease resistant in extracts of [URE3] strains but not in wild-type cells (38), and Ure2p is aggregated specifically in [URE3] cells (20). The Ure2p prion domain readily forms amyloid in vitro and promotes amyloid formation by the native soluble full-length molecule under conditions in which the latter would otherwise be stable (53). Moreover, the pattern of protease-resistant fragments of the in vitro amyloid form of Ure2p closely matches that found for Ure2p in extracts of [URE3] strains (53). Fragments of Ure2p that are unable to assume the prion form in vivo do not form amyloid in vitro (53).
Amyloid is a linear filamentous protein form which is high in β-sheet structure, shows yellow-green birefringence on staining with Congo red, and has a characteristic protease resistance. Over 20 proteins have been shown capable of forming amyloid, and amyloid formation is a key feature of a variety of common diseases, including type II diabetes mellitus (amylin), multiple myeloma (immunoglobulin light chains), Alzheimer's disease (Aβ peptide), Parkinson's disease (α-synuclein), and the transmissible spongiform encephalopathies (PrP). Although determination of the structure of amyloid has been slowed by its irregular form, it is clearly a kind of linear crystal, with some regular features revealed by X-ray fiber diffraction studies, solid-state nuclear magnetic resonance, and cryoelectron microscopy (reviewed in reference 29).
The propagation of [PSI+] requires Hsp104, and [PSI+] is cured by excess Hsp104 (9, 10). This result at once provided key support for the prion model for [PSI+] and indicated that the mechanism of [PSI+] formation involves a conformational change of Sup35p, not a covalent change. Hsp104 is unusual among chaperones in that it does not prevent denaturation but rather promotes renaturation of previously denatured or aggregated proteins (40). It cooperates with Hsp40 (Ydj1p) and Hsp70s in this process (25). The Ssa group of Hsp70 proteins are also essential for [PSI+] propagation (27). Ydj1p is an Hsp40 that is involved in multiple functions, including import of proteins into mitochondria, secretion of mating pheromones, and regulation of the activity of the cytoplasmic Hsp70s (2, 4, 7, 34, 39). In cooperation with the Hsp70 protein Ssa1p, Ydj1p can induce the refolding of denatured protein in vitro even without Hsp104 (34).
Mks1p, a component of the signal transduction cascade regulating nitrogen catabolism (21), is necessary for the de novo generation of [URE3] but not for its propagation (22). We now find that [URE3] requires Hsp104 for its propagation but that, unlike what is found for [PSI+], overexpression of Hsp104 does not cure [URE3]. However, overexpression of Ydj1p results in the gradual loss of [URE3].
MATERIALS AND METHODS
Strains and media.
Strains used are shown in Table 1. Strain 11514 (MATα his3 leu2 lys2 ura3 hsp104::G418; Research Genetics) was crossed with strain HM6B (MATa ura2 can1 trp1 kar1), and URA3 ura2 hsp104::G418 meiotic segregants were chosen for further study. Standard yeast media have been described previously (50). Galactose media included 2% raffinose. USA medium was synthetic dextrose (SD) containing required supplements and 100 μg of sodium USA/ml in place of uracil. Uracil-negative dropout medium was generally not satisfactory for the [URE3] test. Transformation was carried out by a variant of the lithium acetate method (23).
TABLE 1.
Strains of S. cerevisiae
| Strain | Genotype | Source |
|---|---|---|
| 1065 | MATa/MATα ura2/ura2 [ure-o] | F. Lacroute |
| 3310 | MATa kar1 arg1 [URE3-1] | Cytoduction from MA116-8A (M. Aigle) |
| 3310S | MATa kar1 arg1 [ure-o] | Spontaneous [ure-o] from 3310 |
| 4645-8C | MATα kar1 his− ade(2,5) [ure-o] | This work |
| 4645-8CU | MATα kar1 his− ade(2,5) [URE3-1] | Cytoduction from 3310 |
| HM6B | MATa ura2 trp1 can1 kar1 | This work |
| YHE835 | MATα ura2 leu2 his3 trp1 [URE3-1] (Σ1278b background) | This work |
| YHE748 | MATa ura2 leu2 his3 [URE3-1] (Σ1278b) | This work |
| YHE64 | MATα ura2 leu2 trp1 [URE3-2] | 20 |
| 4706-1A | MATα ura2 leu2 his3 trp1 [URE3-1] | P86 (YHE835) × YHE748 |
| 4706-1B | MATa ura2 leu2 his3 [URE3-1] | P86 (YHE835) × YHE748 |
| 4706-1C | MATa ura2 leu2 his3 trp1 P86::LEU2 | P86 (YHE835) × YHE748 |
| 4706-1D | MATα ura2 leu2 his3 P86::LEU2 | P86 (YHE835) × YHE748 |
| 11514 | MATα ura3 his3 leu2 lys2 hsp104::G418 | Research Genetics |
| 4751-11A | MATa ura2 his3 lys2 trp1 can1 kar1 hsp104::G418 | This work |
| 3687 | MATa kar1 ura2 leu2 his− K+ [URE3-2] | 56 |
| 3686 | MATα trp1 ura2 leu2 [ure-o] | 56 |
| 4131 | MATα kar1-1 ura2 leu2 his3 ade2 ure2 | This work |
| DM628-3Aa+ | MATa ura3 leu2 trp1 ade2-1 SUQ5 [PSI+] | D. C. Masison |
Cytoduction.
Cytoplasm can be transferred from one strain (the donor) to another (the recipient) using the kar1 mutant that fails to undergo karyogamy (12). Cytoduction was conducted as described previously (37) using the [URE3] MATa and MATα donor strains 3310 and 4645-8CU, respectively.
Tests for [URE3].
A ura2 strain blocked in USA synthesis can use USA in place of uracil to grow if it carries [URE3]. This is the growth test. In the presence of excess USA, a [URE3] strain takes up more USA than it needs, converts it to uracil, and excretes the excess uracil. This secreted uracil can be detected by the feeding of a lawn of a Ura− strain. In this feeding test, one uses minimal medium without uracil that includes 100 μg USA/ml and a seeded lawn (1.0 ml of a suspension at 0.1 units of optical density at 550 nm) of a diploid ura2/ura2 strain (1065; Table 1). Strains to be tested are streaked on the surface, and plates are observed after 48 h at 30°C. The feeding test can also be used to determine the [URE3] state of strains that do not have a ura2 mutation.
Plasmids.
Plasmids pH218, pH219, pH220, and pH221 were constructed by insertion of the NotI-SalI fragment carrying HSP104 from pFL44L-HSP104 (kindly provided by Magdalena Boguta, Warsaw, Poland) into pRS313, pRS314, pRS423, and pRS424 (11, 51), respectively. pH316 has the GAL1 promoter replacing the ADH1 promoter in CEN LEU2 plasmid pH124 (20). p901 was constructed by PCR amplification of the YDJ1 open reading frame from lambda PM-2784 (ATCC 70080) (48) using oligonucleotides RW43 (5′ ATATACCTC TATACTTTAACGTCAAGGAGAAAAAACCCCGGATCCATGGTTAAAG AAACTAAGTTTTACG 3′) and RW44 (5′ ACCTCTGGCGAAGAAGTCCA AAGCTTCAGCTGCTGCAGGCTCGAGTCATTGAGATGCACATTGAAC ACC 3′) and cotransformation of the PCR product and BamHI-digested pH316 into strain 3687 (35). pA was constructed by PCR amplification of the HSP104 open reading frame from pH218 using oligonucleotides GAL>HSP104 (5′ ATA TACCTCTATACTTTAACGTCAAGGAGAAAAAACCCCGGATCCATGA ACGACCAAACGCAATTTAC 3′) and HSP104>MCS (5′ ACCTCTGGCGA AGAAGTCCAAAGCTTCAGCTGCTGCAGGCTCGAGTTATTAATCTAG GTCATCATCAATTTCC 3′) and cotransformation of the PCR product and BamHI- and XhoI-digested pH316 into strain YHE835. Plasmids isolated from Leu+ transformants by transformation of Escherichia coli were examined for inserts and sequenced.
Identification of the genomic site of transposon insertion.
Chromosomal DNA was isolated from the mutant, treated with EcoRI, and ligated (to circularize it), and the site of integration was amplified by PCR using primers within the LacZ gene (cLacZ67 [5′ TGTGCTGCAAGGCGATTAAGTTG 3′] and LacZ2946 [5′ ATGGCTGAATATCGACGGTTTCC 3′]). This fragment was then sequenced. A second method to accomplish the same goal was use of the “rescue plasmid” pRSQ2 (6). Since the rescue plasmid uses the URA3 gene and our strain carried ura2, we made our strain URA2+ by transforming it with a URA2 gene DNA fragment and then made it ura3 using 5 fluoro-orotic acid selection (5). By either method, the same fragment of chromosomal DNA was detected and, as expected, the insert was in the same gene (see Results).
Western blotting.
Strain YHE835 transformed with pH316 (CEN LEU2 GAL1 promoter) or pA (CEN LEU2 GAL1-HSP104) was streaked for single colonies and grown at 23 or 30°C on plates containing SD plus uracil, His, and Trp or with galactose in place of glucose for 4 to 5 days. After the presence of [URE3] was confirmed by the growth test, some of the single colonies were transferred to 20 ml of the respective media and were grown at 23°C for 12 h to an A550 of 0.5 to 0.8. Cells were collected, suspended in 200 μl of 50 mM Tris-HCl (pH 7.5)–100 mM NaCl–1 mM phenylmethylsulfonyl fluoride–0.001% aprotinin–0.001% pepstatin, and lysed with glass beads. Extracts were centrifuged for 10 min at 15,000 × g at 4°C, and the supernatant fraction was retained. Each extract included about 3 μg of protein per μl. Sample buffer (50 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 10% glycerol, 0.7 M 2-mercaptoethanol, 0.025% bromphenol blue) was added, and samples were boiled for 2 min. Samples (20 μg of protein per lane) were analyzed by electrophoresis on SDS–10% polyacrylamide gel electrophoresis gel, transferred to Immobilon-P membranes (Millipore), and probed with a polyclonal antibody to Hsp104 (Stress Gen Biotechnologies Corp.). Proteins of hsp104::G418 strain 4751-11A were also extracted. Detection was with an alkaline phosphatase-based chemiluminescence detection assay.
RESULTS
Mutant isolation.
To determine what genes are necessary for propagation of the [URE3] prion, we induced mutations in the stably [URE3] haploid strain YHE835, which has the Σ1278b background (3), because [URE3] diploids with this background produced meiotic spore clones with a high proportion of [URE3] segregants, unlike many other strains. We transformed YHE835 [URE3] with a bank of yeast DNA mutagenized with a Tn3-based lacZ LEU2 transposon (6). Since [URE3] makes cells grow slowly, the primary screen was to choose transformant colonies of normal size. Among these, colonies which could not grow on USA but which could grow on uracil were selected. To confirm that these candidates could not maintain [URE3], cytoplasm was transferred by cytoduction from strain 3310 (kar1 [URE3]) and candidates whose cytoductants were all USA− were chosen. These cytoductants were then used as donors to wild-type strain 3310S [ure-o] [rho0] and those which did not transfer [URE3] (implying that they had lost it) were examined further.
Candidate mutants were then mated with a [URE3] strain also having the Σ1278b background (YHE748), and tetrad analysis was carried out. One mutant (P86) for which Leu2+ and USA− each showed 2:2 segregation and generally cosegregated was found. Several spore clones were Leu negative but became USA−. We suspected that these spore clones had spontaneously lost [URE3] and were in fact able to maintain it, as we confirmed by cytoduction experiments using the [URE3] donor strains 3310 and 4645-8CU. The combined results of the cytoduction experiments and the original tetrad data showed that Leu2+ and USA− showed clear 2:2 segregation and cosegregated perfectly in all 12 tetrads except for a single spore clone carrying the mutation which grew poorly on USA. The diploids formed between mutant P86 and the [URE3] strain YHE748 were all stably [URE3], indicating that the P86 mutation is recessive.
To test whether the P86 mutant results in loss of [URE3] rather than, for example, loss of ability to convert USA to uracil, cytoplasm from two USA− spore clones and one USA+ spore clone was transferred to the [ure-o] [rho0] strains 3310S and 4645-8C. Indeed, cytoplasm from the USA− spore clones could not donate [URE3], while the USA+ spore clone did, showing that the mutants specifically lose [URE3]. Diploids formed between mutant segregants and the wild-type [URE3] strains 3310 and 4645-8CU were nearly all slow growing and positive for uracil feeding on USA plates, indicating that the mutation is recessive.
The transposon insertion is in HSP104.
Mutant chromosomal DNA was digested with EcoRI and circularized by ligation, and the site of integration was amplified by PCR using primers in lacZ. The site of integration was also defined using rescue plasmid pRSQ2 (6). By both methods, the site of integration was found to be immediately downstream of bp 1677 in the center of the HSP104 open reading frame.
Complementation tests of P86 mutants with HSP104.
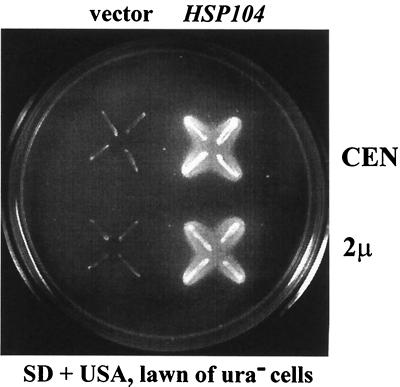
To confirm that the mutation producing loss of [URE3] was the insertion in HSP104 and not a linked mutation, HSP104 on single-copy or high-copy-number plasmids was introduced into the [rho0] P86 mutant strains 4706-1C and 4706-1D (Table 1). The transformants were used as cytoduction recipients from the [URE3] strains 3310 and 4645-8CU, and the cytoductants were subjected to growth and feeding tests. Cells with the control plasmid could not maintain the [URE3] prion, but in cells transformed with the plasmids carrying HSP104, whether on single-copy or multicopy vectors, the mutation was complemented, and on USA the feeding phenomenon was observed (Fig. 1, Table 2). Furthermore, these cytoductants of the P86 mutant complemented with the HSP104 gene could transfer [URE3] by cytoduction to 3310S [ure-o] [rho0] or 4645-8C [ure-o] [rho0] (Table 2). For strain 4706-1D, transformants with a high-copy-number plasmid (pH220) showed more-efficient [URE3] transmission to the recipient than transformants with a low-copy-number plasmid (pH218) (Table 2). When the HSP104-carrying plasmid was lost from the mutant, none of the cells could grow on USA and [URE3] was no longer transferred by cytoduction to the [ure-o] strains (data not shown).
FIG. 1.
The uracil feeding test shows that the P86 mutant is complemented by the HSP104 gene. HSP104 on a single-copy (pH218; CEN HIS3 HSP104) or high-copy-number (pH220; 2μm HIS3 HSP104) plasmid was introduced into ρO P86 mutant strain 4706-1D (MATα ura2 his3 P86::LEU2). The transformants were used as cytoduction recipients from [URE3] strain 3310, and four cytoductants of each were examined by the feeding test on minimal medium without uracil that includes USA and a seeded lawn of diploid ura2 strain 1065. After incubation for 48 h at 30°C, the cells transformed with the plasmids carrying HSP104 grew on USA and showed the feeding phenomenon. Cells with the control vectors (pRS313 [CEN HIS3] or pRS423 [2μm HIS3]) could not grow on USA and could not feed the lawn.
TABLE 2.
Complementation of the P86 mutation by HSP104a
| [URE3-1] donor | Recipient P86 strain | Plasmid | Growth on USA | Feeding on USAb by:
|
|
|---|---|---|---|---|---|
| First cytoductants | Second cytoductants | ||||
| 4645-8CU | 4706-1CρO | CEN vector | − | 0/12 | |
| 4645-8CU | 2μm vector | − | 0/12 | ||
| 4645-8CU | CEN HSP104 | + | 12/12 | 2/2 | |
| 4645-8CU | 2μm HSP104 | + | 12/12 | 2/2 | |
| 3310 | 4706-1DρO | CEN vector | − | 0/12 | |
| 3310 | 2μm vector | − | 0/12 | ||
| 3310 | CEN HSP104 | + | 12/12 | 14/24 | |
| 3310 | 2μm HSP104 | + | 12/12 | 24/24 | |
P86 strains 4706-1CρO and 4706-1DρO were transformed with the indicated TRP1 and HIS3 plasmids, respectively, and the transformants were used as cytoduction recipients from the [URE3] strains 3310 and 4645-8CU. These first cytoductants were tested for growth and feeding on USA and then used as donors in a second cytoduction to wild type [ure-o] recipients (strains 3310S and 4645-8C). The resulting second cytoductants were then tested for feeding on USA. Plasmids used were pH218 (CEN HIS3 HSP104), pH219 (CEN TRP1 HSP104), pRS423 (2μm HIS3), pRS424 (2μm TRP1), pRS313 (CEN HIS3), pRS314 (CEN TRP1), pH220 (2 μm HIS3 HSP104), and pH221 (2μm TRP1 HSP104).
Values are numbers of colonies showing feeding/total number of colonies tested.
hsp104 deletion mutants cannot maintain [URE3].
Transferring cytoplasm from [URE3] strains into an hsp104::G418 null mutant recipient produced cytoductants that were uniformly unable to grow on USA or to feed uracil to a Ura− lawn (the feeding test). When mutant cells were transformed with a plasmid carrying HSP104, whether on single-copy or multicopy vectors, and used as cytoduction recipients, cytoductants were able to grow on USA and the feeding phenomenon was observed (Table 3). As with the P86 mutants, using the hsp104::G418 cytoductants (from the [URE3-1] donor) as donors in cytoduction into a wild-type [ure-o] recipient (4645-8CG) gave only [ure-o] cytoductants, showing again that [URE3] was lost and not merely unexpressed (data not shown).
TABLE 3.
Complementation of null mutants (hsp104::G418) by HSP104-carrying plasmidsa
| Plasmid | Growth on USA of cytoductants | Uracil feeding of cytoductantsb |
|---|---|---|
| None | − | |
| pRS313 (CEN vector) | − | 0/24 |
| pRS423 (2μm vector) | − | 0/24 |
| pH218 (CEN HSP104) | + | 24/24 |
| pH220 (2μm HSP104) | + | 24/24 |
The hsp104::G418 strain 4751-11A was transformed with the indicated plasmids, and [URE3-1] was introduced into the transformant by cytoduction from [URE3-1] donor strain 4645-8CU.
Numbers of colonies showing feeding/total number of colonies tested.
Crossing hsp104::G418 strain 4751-11A with ure2 strain 4131 showed that meiotic segregants with the hsp104 deletion were USA+ when carrying the ure2 deletion, and thus deficiency of Hsp104 did not interfere with the USA+ phenotype produced by deficiency of Ure2p.
Overproduction of Hsp104 does not cure [URE3].
To test the effects of overproduction of Hsp104 on [URE3] stability, we introduced single-copy (CEN-HSP104) or high-copy-number (2μm HSP104) plasmids, in which HSP104 was controlled by its own promoter, into [URE3] strain YHE835. Similar proportions of transformants with each plasmid were USA+ (Table 4). USA+ transformants were further grown on medium selective for the plasmid, and single colonies were tested for [URE3]. There was no increased loss of [URE3] compared to vector controls. Furthermore, the high-copy-number HSP104 plasmids did not affect the strength of the USA+ or uracil feeding phenotypes.
TABLE 4.
High-copy-number HSP104 plasmids do not cure [URE3]a
| Plasmid | No. of colonies positive for [URE3]/total no. tested
|
|
|---|---|---|
| Vector | HSP104 | |
| CEN TRP1 | 50/52 | 41/49 |
| CEN HIS3 | 49/55 | 40/44 |
| 2μm TRP1 | 40/42 | 43/45 |
| 2μm HIS3 | 41/44 | 41/41 |
Transformants of YHE835 with the CEN and 2μm plasmids with and without HSP104 driven by its own promoter were tested for [URE3] by the feeding test.
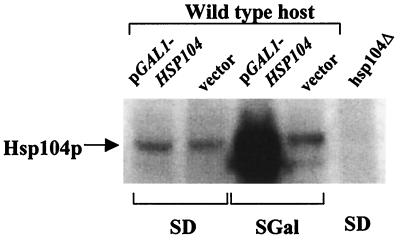
A plasmid overproducing Hsp104 from the strong GAL1 promoter was introduced into [URE3] strain YHE835, selecting transformants on dextrose media where the promoter is repressed. Confirmed USA+ transformants were then grown on media containing galactose or galactose plus the nonrepressing sugar raffinose. There was no detectable loss of [URE3] induced by these conditions (Table 5). Confirmation that Hsp104 was indeed dramatically overproduced was obtained by Western blot analysis using an antibody to Hsp104 (Fig. 2).
TABLE 5.
Overexpression of Hsp104 from the GAL1 promoter does not cure [URE3]a
| Mediumb | Plasmid | No. of positive colonies/total no. tested in:
|
|
|---|---|---|---|
| Growth test | Feeding test | ||
| SDextrose | Vector | 150/150 | 24/24 |
| GAL1-HSP104 | 150/150 | 24/24 | |
| SGalactose | Vector | 160/160 | 24/24 |
| GAL1-HSP104 | 160/160 | 24/24 | |
| SGalactose + raffinose | Vector | 95/95 | 24/24 |
| GAL1-HSP104 | 112/112 | 24/24 | |
Strain YHE835 was transformed on dextrose media with either the vector (pH316) or the same plasmid containing HSP104 under the control of the GAL1 promoter (pA). Confirmed USA+ colonies of each were pooled and grown to single colonies on the indicated media. These single colonies were then tested for the ability to use USA for growth and for uracil feeding to a lawn of a ura2/ura2 strain.
SDextrose, minimal medium with dextrose; SGalactose, minimal medium with galactose.
FIG. 2.
Overexpression of HSP104 from the GAL1 promoter. Strain YHE835 was transformed with control vector pH316 (CEN LEU2 pGAL1) or the plasmid carrying HSP104 under the control of GAL1 promoter pA (CEN LEU2 GAL1-HSP104). The transformants were grown on minimal medium (SD) or the same medium with galactose in place of glucose (SGal). After galactose induction, proteins were extracted and separated in SDS–10% polyacrylamide gels, transferred to the membranes, and probed with a polyclonal antibody to Hsp104. Twenty micrograms of protein of each extract was used. Deletion strain 4751-11A (hsp104Δ), which was cultured in minimal medium (SD), does not contain any Hsp104 protein.
As another control, the same plasmids were introduced into the [PSI+] strain DM628-3Aa+. [PSI] was lost from all transformants with the 2μm HSP104 plasmid (pH221) but not with the parent vector. [PSI] remained in glucose-grown transformants with the GAL1-promoted HSP104 plasmid but was uniformly lost after 1 day of growth on galactose-raffinose medium. Thus, the same conditions of Hsp104 overproduction that did not affect [URE3] stability uniformly cured [PSI+].
Different [URE3] prion strains are cured by hsp104Δ.
Most of the experiments reported here used [URE3-1], the strain of [URE3] that was isolated by F. Lacroute (31) and passed to various strains by cytoplasmic mixing (cytoduction). To determine whether Hsp104 is necessary for propagation of other independently isolated strains of [URE3], we used [URE3-2], isolated in an earlier study by overproduction of Ure2p (56) and transferred by cytoduction to strain 3686. When [URE3-2] was transferred by cytoduction from 3686 to the hsp104::G418 [rho0] strain 4751-11A, all cytoductants were found to be USA− (Table 6). When these cytoductants were then used as donors to strain 4645-8Cρ0 [ure-o], all 20 of these second-generation cytoductants were also USA− (Table 6). These results indicate that Hsp104 is necessary for propagation of this strain of [URE3] as well. That [URE3-1] and [URE3-2] are indeed distinct strains of [URE3] is suggested by the fact that the latter slows cell growth more in the same host than does the former (data not shown). Introduction into the [URE3-2] strain YHE64 of the high-copy-number HSP104 plasmid p221, which in control experiments cured [PSI+], did not result in the loss of [URE3-2].
TABLE 6.
Two independent [URE3] prion strains both require Hsp104a
| Donor strain | Recipient 1 | Results of growth test of cytoductantsb | Recipient 2 | Results of feeding test of cytoductantsb |
|---|---|---|---|---|
| 4645-8CU [URE3-1] | 4751-11AρOhsp104::G418 | 0/100 | 4645-8CρO [ure-o] | 0/20 |
| YHE64 [URE3-2] | 4751-11AρOhsp104::G418 | 0/100 | 4645-8CρO [ure-o] | 0/20 |
A two-stage cytoduction was carried out, like that in Table 2, with cytoplasm transferred from the donor to recipient 1 and then from those cytoductants to recipient 2.
Number of cytoductants positive for USA/total number tested.
[URE3] is cured by overexpression of Ydj1p.
The ability of Ydj1p, with Ssa1p, to renature proteins in vitro (34) and the interaction of Hsp104 with Hsp70s and with Ydj1p (25) suggested that Ydj1, like Hsp104, might be involved in [URE3] propagation. We introduced p901, in which YDJ1 was transcribed from the GAL1 promoter, or the parent vector, pH316, into [URE3-2] strain 3687 and [URE3-1] strain YHE835, selecting transformants on dextrose media. USA+ transformants were picked and grown for about 20 or 40 generations by subcloning on Leu− plates containing dextrose or galactose and raffinose. After 40 generations on glucose, each of 16 clones of each transformant retained [URE3], as indicated by the uracil feeding test, as did the vector transformants on either glucose or galactose media. After 20 generations on galactose-raffinose, most colonies with p901 were negative or weakly positive on the uracil feeding test. After 40 generations, each of 16 clones with p901 grown on galactose-raffinose had lost [URE3], as assessed by the uracil feeding test. In each case, the feeding test was done on dextrose-containing media on which overexpression of Ydj1p is repressed, so that Ydj1p interference with expression of [URE3] is not an issue. The extremely slow growth of ydj1Δ strains and their very inefficient mating made it difficult to test their ability to maintain [URE3].
DISCUSSION
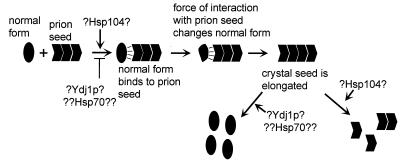
The biology of scrapie argues for the “crystal” view of prions (reviewed in reference 32). One characteristic of crystals is that closely related molecules interfere with each other's crystal formation. Indeed, overproduction of hamster PrP in transgenic mice slows development of mouse scrapie and vice versa (46). Likewise, certain PrP peptides interfere with propagation of scrapie in tissue culture cells (8). The curing of [URE3] by overexpression of fragments of Ure2p may be similarly explained (20). Curing of [URE3] by even low-level expression of fusion proteins consisting of part or all of Ure2p fused to another protein can likewise be explained as interference with growth of the amyloid crystal (20). The crystal model also offers an explanation of the propagation of the alteration of protein conformation which is believed to be central to prion propagation. According to this model, normal Ure2p is driven to change its conformation by the energy of interaction with other Ure2p molecules in the amyloid filaments (Fig. 3). Finally, the in vitro formation of amyloid by Ure2p driven by the prion domain suggests that this linear crystal is the basis of the [URE3] phenomenon (53).
FIG. 3.
The crystal seed model of prion generation and possible roles of chaperones Hsp104 and Ydj1. The energy of interaction between the free-normal-form and the abnormal-form molecules in the crystal drives the change of the normal form to the abnormal form. Hsp104 may promote conversion of normal Ure2p to an intermediate or break up aggregates to provide seeds for daughter cells. Ydj1 may be involved in dissolving aggregates or may block a step in aggregate formation.
The first demonstration of a role for chaperones in prion propagation was the finding that [PSI+] is lost from strains that either overproduce or are deficient in Hsp104 (9, 10). Because [URE3] was reported to be independent of Hsp104 (33), we directly screened for chromosomal mutants defective in its propagation. The first mutant identified had an insertion in the middle of the HSP104 gene. We showed that this mutant was actually unable to propagate the [URE3] genetic element. Moreover, the hsp104::G418 mutant with the coding sequence completely deleted showed the same behavior. The hsp104::G418 deletion is compatible with the USA+ phenotype resulting from a ure2 mutation, but the [URE3] prion cannot propagate in such a strain. We showed that a second independent [URE3] prion strain is also lost in hsp104 strains, indicating that this dependence is not unique to [URE3-1].
In contrast to what was found for [PSI+], we find that overproduction of Hsp104p does not eliminate [URE3]. Kushnirov et al. likewise found that a “synthetic” hybrid prion formed by the amino-terminal part of Pichia methanolica Sup35p and the C-terminal part of Saccharomyces Sup35p also requires Hsp104 for its propagation but is not cured by overexpression (30). [PIN+] is a nonchromosomal genetic element that is necessary for the induction of [PSI+] appearance by the overproduction of Sup35p (17). [PIN+] may be a prion and, like [URE3], is cured by deletion of HSP104 but is not cured by overexpression of Hsp104 (16).
The mechanisms by which Hsp104 is involved in prion propagation are not entirely understood. Since Hsp104 is capable of renaturing heat-denatured proteins both in vitro and in vivo (40), it is likely that overproduced Hsp104 can depolymerize the aggregated Sup35p and thereby reverse the [PSI+] state (9). While this is certainly the end result and while Sup35p interacts with Hsp104p in vitro (49), disassembly of Sup35p amyloid by Hsp104 has not yet been demonstrated in vitro. Hsp104 has been shown to cooperate with Hsp70s and Hsp40s (25), and multiple components may thus be required to reproduce this reaction in vitro. Moreover, it is not yet certain that amyloid is the prion form of either Ure2p or Sup35p.
The explanation for the requirement for Hsp104 for the propagation of [PSI+] and now for [URE3] is less clear. One model is that Hsp104 is necessary to convert the native protein into an intermediate form that is then susceptible to conversion to the prion (amyloid) form (9) (Fig. 3). An alternate model suggests that Hsp104 is necessary to ensure segregation of the aggregates to both daughter cells, thus ensuring the stability of the prion (42, 43). This model holds that without Hsp104's conversion of large aggregates to many small aggregates, daughter cells may fail to receive a “seed” to ensure propagation of the prion. The formation of amyloid in vitro in the absence of Hsp104 by both the recombinant prion domain of Sup35p (24, 28) and either the prion domain of or full-length native Ure2p (53) argues against a direct role for Hsp104 in the amyloid formation process. However, under physiological conditions or time constraints, there may be such a requirement.
Why does overproduction of Hsp104 efficiently cure [PSI] but does not detectably affect [URE3]? It is unlikely that Hsp104 specifically recognizes aggregates of Sup35p and not those of Ure2p. It is more likely that a difference in the form of the aggregates or in their stability or amount explains this difference. Possibly different chaperones or combinations of chaperones are responsible for surveillance of [URE3] prions.
The curing of [URE3] by Ydj1p (an Hsp40) may involve the interactions of Ydj1p with Hsp70s or with Hsp104 or both. Lu and Cyr showed that guanidine-denatured luciferase could be completely renatured by the in vitro action of Ydj1p and Ssa1p (34). Hsp104 can also promote the partial reactivation of denatured luciferase in a yeast crude extract in a reaction requiring Ydj1p and Ssa1p as well (25). Overproduced Ydj1p may directly block the formation of amyloid by Ure2p without cooperation with other chaperones and thereby prevent propagation of [URE3] (Fig. 3). Alternatively, it is possible that Ydj1p interferes with [URE3] propagation by occupying Hsp104 or Hsp70 in interactions not productive for [URE3] propagation.
The involvement of the chaperones Hsp104 and Ydj1 in [URE3] propagation indicates that the mechanism of [URE3] involves altered conformational states of Ure2p rather than a covalent modification. It will be of interest to determine the effects of these chaperones and others on in vitro amyloid formation by Ure2p and its fragments.
ACKNOWLEDGMENTS
We thank Daniel Masison for help with the [PSI] experiments, Giman Jung for help with Western blots, and B. Tibor Roberts for valuable advice. We are grateful to Magdalena Boguta (Warsaw, Poland) for pFL44L-HSP104.
REFERENCES
- 1.Alper T, Cramp W A, Haig D A, Clarke M C. Does the agent of scrapie replicate without nucleic acid? Nature. 1967;214:764–766. doi: 10.1038/214764a0. [DOI] [PubMed] [Google Scholar]
- 2.Atencio D, Yaffe M. MAS5, a yeast homolog of DnaJ involved in mitochondrial import. Mol Cell Biol. 1992;12:283–291. doi: 10.1128/mcb.12.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bechet J, Grenson M, Wiame J M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970;12:31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 4.Becker J, Walter W, Yan W, Craig E A. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 6.Burns N, Grimwade B, Ross-Macdonald P B, Choi E Y, Finberg K, Roeder G S, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 7.Caplan A, Douglas M G. Characterization of YDJ1: a yeast homolog of the bacterial dnaJ protein. J Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chabry J, Caughey B, Chesebro B. Specific inhibition of in vitro formation of protease-resistant prion protein by synthetic peptides. J Biol Chem. 1998;273:13203–13207. doi: 10.1074/jbc.273.21.13203. [DOI] [PubMed] [Google Scholar]
- 9.Chernoff Y O, Lindquist S L, Ono B-I, Inge-Vechtomov S G, Liebman S W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 10.Chernoff Y O, Ono B-I. Dosage-dependent modifiers of PSI-dependent omnipotent suppression in yeast. In: Brown A J P, Tuite M F, McCarthy J E G, editors. Protein synthesis and targeting in yeast. Berlin, Germany: Springer-Verlag; 1992. pp. 101–107. [Google Scholar]
- 11.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 12.Conde J, Fink G R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci USA. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper T G. Nitrogen metabolism in Saccharomyces cerevisiae. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces: metabolism and gene expression. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 39–99. [Google Scholar]
- 14.Coschigano P W, Magasanik B. The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione S-transferases. Mol Cell Biol. 1991;11:822–832. doi: 10.1128/mcb.11.2.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox B S. PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- 16.Derkatch I L, Bradley M E, Masse S V, Zadorsky S P, Polozkov G V, Inge-Vechtomov S G, Liebman S W. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? EMBO J. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derkatch I L, Bradley M E, Zhou P, Chernoff Y O, Liebman S W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derkatch I L, Chernoff Y O, Kushnirov V V, Inge-Vechtomov S G, Liebman S W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drillien R, Aigle M, Lacroute F. Yeast mutants pleiotropically impaired in the regulation of the two glutamate dehydrogenases. Biochem Biophys Res Commun. 1973;53:367–372. doi: 10.1016/0006-291x(73)90671-2. [DOI] [PubMed] [Google Scholar]
- 20.Edskes H K, Gray V T, Wickner R B. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci USA. 1999;96:1498–1503. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edskes H K, Hanover J A, Wickner R B. Mks1p is a regulator of nitrogen catabolism upstream of Ure2p in Saccharomyces cerevisiae. Genetics. 1999;153:585–594. doi: 10.1093/genetics/153.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edskes H K, Wickner R B. A protein required for prion generation: [URE3] induction requires the Ras-regulated Mks1 protein. Proc Natl Acad Sci USA. 2000;97:6625–6629. doi: 10.1073/pnas.120168697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 24.Glover J R, Kowal A S, Shirmer E C, Patino M M, Liu J-J, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 25.Glover J R, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 26.Griffith J S. Self-replication and scrapie. Nature. 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 27.Jung G, Jones G, Wegrzyn R D, Masison D C. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics, 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King C-Y, Tittmann P, Gross H, Gebert R, Aebi M, Wuthrich K. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci USA. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirschner D A, Teplow D B, Damas A M. Twist and sheet: variations on the theme of amyloid. J Struct Biol. 2000;130:87–130. doi: 10.1006/jsbi.2000.4293. [DOI] [PubMed] [Google Scholar]
- 30.Kushnirov V V, Kochneva-Pervukhova N V, Cechenova M B, Frolova N S, Ter-Avanesyan M D. Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J. 2000;19:324–331. doi: 10.1093/emboj/19.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacroute F. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J Bacteriol. 1971;106:519–522. doi: 10.1128/jb.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lansbury P T, Caughey B. The chemistry of scrapie infection: implications of the ‘ice 9’ metaphor. Curr Biol. 1995;2:1–5. doi: 10.1016/1074-5521(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 33.Liebman S W, Derkatch I L. The yeast [PSI+] prion: making sense out of nonsense. J Biol Chem. 1999;274:1181–1184. doi: 10.1074/jbc.274.3.1181. [DOI] [PubMed] [Google Scholar]
- 34.Lu Z, Cyr D M. Protein folding activity of Hsp70 is modified differentially by the Hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- 35.Ma H, Kunes S, Schatz P J, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 36.Magasanik B. Regulation of nitrogen utilization. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. 2nd ed. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 283–317. [Google Scholar]
- 37.Masison D C, Maddelein M-L, Wickner R B. The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc Natl Acad Sci USA. 1997;94:12503–12508. doi: 10.1073/pnas.94.23.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masison D C, Wickner R B. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 39.Meacham G C, Browne B L, Zhang W, Kellermayer R, Bedwell D M, Cyr D M. Mutations in the yeast Hsp40 chaperone protein Ydj1 cause defects in Axl1 biogenesis and pro-a-factor processing. J Biol Chem. 1999;274:34396–34402. doi: 10.1074/jbc.274.48.34396. [DOI] [PubMed] [Google Scholar]
- 40.Parsell D A, Kowal A S, Singer M A, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 41.Patino M M, Liu J-J, Glover J R, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 42.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. In vitro propagation of the prion-like state of yeast Sup35 protein. Science. 1997;277:381–383. doi: 10.1126/science.277.5324.381. [DOI] [PubMed] [Google Scholar]
- 43.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 44.Prusiner S B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 45.Prusiner S B. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prusiner S B, Scott M, Foster D, Pan K-M, Groth D, Mirenda C, Torchia M, Yang S-L, Serban D, Carlson G A, Hoppe P C, Westaway D, DeArmond S J. Transgenic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 47.Rai R, Genbauffe F, Lea H Z, Cooper T G. Transcriptional regulation of the DAL5 gene in Saccharomyces cerevisiae. J Bacteriol. 1987;169:3521–3524. doi: 10.1128/jb.169.8.3521-3524.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riles L, Dutchik J E, Baktha A, McCauley B K, Thayer E C, Leckie M P, Braden V V, Depke J E, Olson M V. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics. 1993;134:81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schirmer E C, Lindquist S. Interactions of the chaperone Hsp104 with yeast Sup35 and mammalian PrP. Proc Natl Acad Sci USA. 1997;94:13932–13937. doi: 10.1073/pnas.94.25.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 51.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stansfield I, Jones K M, Kushnirov V V, Dagkesamanskaya A R, Poznyakovski A I, Paushkin S V, Nierras C R, Cox B S, Ter-Avanesyan M D, Tuite M F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor K L, Cheng N, Williams R W, Steven A C, Wickner R B. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- 54.Turoscy V, Cooper T G. Ureidosuccinate is transported by the allantoate transport system in Saccharomyces cerevisiae. J Bacteriol. 1987;169:2598–2600. doi: 10.1128/jb.169.6.2598-2600.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weissmann C. Molecular genetics of transmissible spongiform encephalopathies. J Biol Chem. 1999;274:3–6. doi: 10.1074/jbc.274.1.3. [DOI] [PubMed] [Google Scholar]
- 56.Wickner R B. Evidence for a prion analog in S. cerevisiae: the [URE3] non-Mendelian genetic element as an altered URE2 protein. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 57.Wickner R B. Prion diseases of mammals and yeast: molecular mechanisms and genetic features. R. G. Austin, Tex: Landes Company; 1997. [Google Scholar]
- 58.Wickner R B, Chernoff Y. Prions of yeast and fungi: [URE3], [PSI] and [Het-s] discovered as heritable traits. In: Prusiner S B, editor. Prions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 229–272. [Google Scholar]
- 59.Wickner R B, Edskes H K, Maddelein M-L, Taylor K L, Moriyama H. Prions of yeast and fungi: proteins as genetic material. J Biol Chem. 1999;274:555–558. doi: 10.1074/jbc.274.2.555. [DOI] [PubMed] [Google Scholar]
- 60.Zhouravleva G, Frolova L, LeGoff X, LeGuellec R, Inge-Vectomov S, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]