Abstract
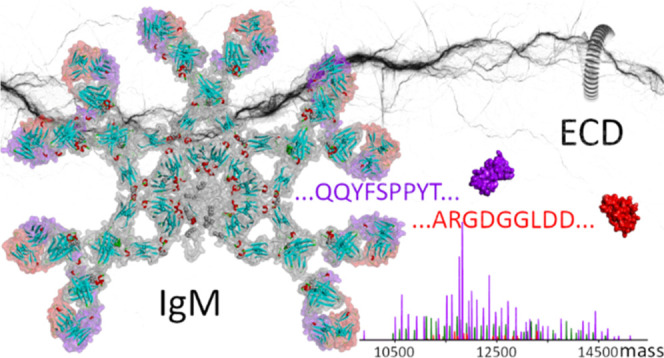
Native top-down mass spectrometry (MS) is gaining traction for the analysis and sequencing of intact proteins and protein assemblies, giving access to their mass and composition, as well as sequence information useful for identification. Herein, we extend and apply native top-down MS, using electron capture dissociation, to two submillion Da IgM- and IgG-based oligomeric immunoglobulins. Despite structural similarities, these two systems are quite different. The ∼895 kDa noncovalent IgG hexamer consists of six IgG subunits hexamerizing in solution due to three specifically engineered mutations in the Fc region, whereas the ∼935 kDa IgM oligomer results from the covalent assembly of one joining (J) chain and 5 IgM subunits into an asymmetric “pentamer” stabilized by interchain disulfide bridges. Notwithstanding their size, structural differences, and complexity, we observe that their top-down electron capture dissociation spectra are quite similar and straightforward to interpret, specifically providing informative sequence tags covering the highly variable CDR3s and FR4s of the Ig subunits they contain. Moreover, we show that the electron capture dissociation fragmentation spectra of immunoglobulin oligomers are essentially identical to those obtained for their respective monomers. Demonstrated for recombinantly produced systems, the approach described here opens up new prospects for the characterization and identification of IgMs circulating in plasma, which is important since IgMs play a critical role in the early immune response to pathogens such as viruses and bacteria.
Introduction
Early control of viral and bacterial infections is dependent on innate natural antibodies. Among those, immunoglobulins M (IgM) are critical to the initial humoral immune response.1−3 Via their ability to recognize modified self-components and altered cells displaying specific patterns such as carbohydrates, glycolipids, and repetitive structures, IgMs also contribute to immunosurveillance mechanisms against precancerous and cancerous cells.4,5 Consequently, IgMs are promising agents for immunotherapy.4,6
IgM’s high molecular weight, high number of isoforms, and rather complex structure poses, however, a challenge to both therapeutic and diagnostic applications. The vast majority of IgMs consists of five identical subunits and one joining (J) chain assembled into an asymmetric pentamer.7−9 Besides being present in the gastrointestinal tract, lymphatic vessels, mucosal tissues, bone marrow, and so forth, pentameric IgMs (with J-chain) also represent about 30% of the blood-circulating immunoglobulins.10 Similarly to other isotypes, IgM’s subunits are formed by two heavy chains (HCs), each paired to one light chain (LC) and stabilized by intra- and interchain disulfide bonds.11 Stabilization of the pentameric assembly relies on interactions of the C-terminal of the HCs involving inter-subunit disulfide bonds, the formation by the C-terminal 18 amino-acid-long secretory tailpieces of a central β-sandwich structure, and bonding to the J-chain.8,9,12 The mature J-chain contains eight cysteine residues with two involved in disulfide bonds with an IgM HC and the other six forming intrachain disulfide bridges.13
IgM naturally forms oligomers, whereas IgGs are thought to predominantly exist as monomers. The introduction of specific mutations in recombinant IgG1s can, however, induce the formation of stable IgG hexamers in solution.14−16 Compared to disulfide-stabilized IgMs, these hexameric IgG1s are characterized by more mobile Fab regions—reduced steric hindrance related to the long IgG hinge—which may facilitate binding to low-accessibility or close-to-the-membrane epitopes.14 The therapeutic potential of these induced IgG hexamers is currently being investigated, especially for their role in complement activation.17−20
Here, we aim at analyzing these very large and important, albeit structurally complicated, oligomeric immunoglobulins, through the application of native top-down mass spectrometry (nTDMS).21−24 Relying on the fact that native MS transfers the molecules to the gas phase under conditions retaining their structural features, native top-down proteomics should provide access to the sequence of the proteins’ subunits and an insight into its post-translational modifications and higher-order structure. It has become apparent that inducing fragmentation in native top-down proteomics requires moving beyond the well-established activation method of collision-induced dissociation (CID) as this rather slow heating process often induces too little fragmentation when applied to these very high mass systems.25−27 Several groups have explored alternative fragmentation methods for nTDMS, such as surface-induced dissociation,28 photon-induced dissociation (UVPD and IRMPD),29−36 electron transfer dissociation,36,37 and electron capture dissociation (ECD).22,34,35,38−46
Here, we discuss the performance and characteristics of ECD (comparing it with CID) in the native top-down characterization of two ∼1 MDa immunoglobulin oligomers, making use of (1) a recombinant IgM pentamer with J-chain targeting the wall teichoic acids (WTA) antigen (molecular weight of approximately 937,500 Da) and (2) an engineered recombinant IgG1-RGY hexamer (molecular weight of approximately 895,300 Da) targeting CD52.
We a priori hypothesized that the native top-down analysis of these two systems would be analytically very challenging or even impossible, for various reasons. First, their size and high mass (close to 1 MDa) make them challenging to ionize and analyze by MS. Second, their vast structural complexity—especially for the (IgM)5J pentamer with J-chain— resulting from disulfide bonds and glycans, leads to heterogeneous mass distributions and complicates fragment ion formation. Third, being built of roughly 7000 amino acids, extensive backbone cleavage by nTDMS may theoretically result in overwhelmingly complex fragmentation spectra.
We showed in previous work that ECD without supplemental activation of monomeric IgG (∼150 kDa) and IgA (∼165 kDa) variants can result in very clean and interpretable fragmentation spectra dominated by (c/z·) fragment pairs without side-chain cleavages.45,46 Targeting a single type of bond for fragmentation reduces spectral congestion and the dilution of signal intensity in the (n)TDMS spectra of native intact antibodies.47,48 The conservation of inter- and intrachain disulfide bridges under ECD conditions also results in straightforward-to-read c-ion sequence ladders from sequence segments outside regions bridged by disulfide-bridged loops. For the system studied, the fragments cover the LC’s and HC’s CDR3s, as well as the sequence segments, immediately downstream without interference of fragments from other parts of the protein. Testing the ECD nTDMS approach on all IgG subtypes (IgG1, IgG2, IgG3, and IgG4) and an IgA1 proved the method to be generally applicable to monomeric immunoglobulins even when they are heavily glycosylated.47,49
As an ambitious follow-up, we here explore the application of ECD without supplemental activation to the 5–6 times larger covalently-linked J-chain-coupled IgM pentamers and the noncovalently associated IgG1 hexamers. ECD without supplemental activation of these ∼900–950 kDa assemblies primarily leads to extensive electron capture without dissociation (ECnoD) with sometimes up to 40 electrons captured by the precursor ions. However, in the low m/z range of both the J-chain-coupled IgM pentamers and IgG1-RGY hexamers, informative fragment ions are observed, which yield sequence information for the complementarity-determining regions (CDRs), similarly to the monomers. When comparing ECD and CID fragmentation behaviors for these systems, we observe that informative sequence tags from the variable regions of oligomeric IgG and IgM are favorably generated by native top-down ECD. The different nature of these two immunoglobulin oligomers, one, a highly glycosylated covalently linked heterohexamer and the other a noncovalent homohexamer, does not seem to significantly impact the formation of the CDR sequence-informative low m/z fragments. As in the case of refs (45) and (46), ECD appears here directed by charge localization in the vicinity of the N-terminal regions. Consequently, the present approach suggests that serum-purified IgM molecules could also, in principle, be partly de novo sequenced by this native top-down approach, thereby facilitating the analysis of this important class of antibodies.
Materials and Methods
Samples
The monoclonal aWTA (IgM)5J was provided by S. Rooijakers (Medical Microbiology, UMCU, The Netherlands). The aCD52 hexamer-forming IgG1-RGY mutant14,16 was provided by Genmab (Utrecht, The Netherlands).
Preparation of Intact (IgM)5J and RGY-IgG1 Hexamer Samples for Native Top-Down MS Analysis
Prior to the native top-down analysis, the storage buffer was exchanged to aqueous 150 mM ammonium acetate (pH ∼ 7.0) through six consecutive dilutions and concentration steps at 4 °C using Amicon Ultra centrifugal filters with a 10 kDa molecular weight cutoff (Merck KGaA, Darmstadt, Germany). (IgM)5J and hexameric IgG-RGY concentrations used for native electrospray ionization MS were typically around 2 μM (concentration of the monomer).
Native Top-Down ECD and CID MS
Top-down MS of native immunoglobulin oligomers (and monomers) was performed on an ultrahigh mass-range (UHMR) Q-Exactive Orbitrap (Thermo Fisher Scientific, Bremen, Germany) using a built-in collision cell and an ECD cell developed by e-MSion.50 Electrospray involved a nanospray source using emitters produced from borosilicate capillaries by a P-97 Sutter puller (Novato, CA, USA). ESI voltages were in the 1.0–1.4 kV range, the source temperature was set at 275 °C, and the S-lens RF was set at 200 (service mode). The in-source trapping desolvation voltage was set to −100 V as it corresponds to an optimum in terms of fragmentation.51 Using ion transfer parameters optimized for high m/z ions, the (IgM)5J or (IgG)6 intact precursor ions were guided to the quadrupole, where they underwent mass selection using windows narrowed to the intact precursor charge distribution (see Table S1). The ions were then transferred into the ECD cell, where they were transmitted in the absence of electrons to the HCD cell for CID or subjected to electron capture dissociation for ECD. The small permanent magnets responsible for the radial confinement of electrons emitted by a cathode and the additional electrodes responsible for the electron cloud longitudinal confinement ensured efficient ion transfer to the HCD cell in both cases. Upon transfer from the ECD cell to the HCD cell following electron capture, potential post-ECD collisional activation was kept to a minimum (HCD Direct eV setting = 1 or 0) to avoid (b/y) ion formation and to concentrate the fragment signal intensity into the c- and z-ions of interest, while for CID, an HCD direct eV setting = 200 was used (Figure S1 and Table S1). The analyzer injection parameter, CE-Inject (V) UHMR, was initially kept to its standard value of 3200 for high m/z ions. This ensured optimal detection of the high m/z precursor ions but proved detrimental to the recording of fragments in the 1000–3000 m/z range as the third harmonic fraction of the precursor ion signal then overlapped with the fragment ions. To reduce interference via the optimization of trajectories in the analyzer for low m/z ions, we set the CE-Inject (V) UHMR parameter to 3700, which considerably increased the low m/z ion signal while suppressing the signal (third harmonic included) corresponding to the intact precursor. The 3700 value reduces the recording time and facilitates processing (e.g., it removes the need for baseline subtraction), but it does not affect the number of detected fragments for high-quality spectra. It is worth noting that operating at a low pressure—the UHV readout is under the range, indicative of a pressure below 5 × 10–11 mbar close to the analyzer region of the instrument—is also beneficial to the recording of high-resolution spectra as it enables the detection of long transients with limited dephasing by collisions. Overall, the HCD cell trapping and extraction parameters were optimized for operation under low nitrogen collision gas pressure conditions. All spectra were acquired with the noise threshold parameter set to 3.64 at a set resolution of 280,000 @ m/z 200. Between 5000 and 10,000 scans were averaged for each spectrum.
Data Analysis
Processing of the ECD fragmentation spectra involved the conversion of raw files to the mzML format by Proteowizard.52 We used the MSDeisotope Python library (Joshua Klein, Boston University CBMS)53,54 with a minimum_score = 10.0 and mass_error_tolerance = 0.02 to generate a charge-deconvoluted spectrum with all the isotopic peaks retained.53,54 Fragments were assigned by applying LcMsSpectator (Pacific Northwest National Laboratory)55,56 to the charge-deconvoluted spectra generated by MSDeisotope. The accuracy threshold was set to 3 ppm for all assignments following recalibration of the fragment’s m/z via shifting by the average error on assignable c-ion fragments. The results were exported as tsv files for further analysis. Sequence assignment accommodated the major ECD (a·, c, x, z, z·) or CID (b,y) ion types without considering H2O and NH3 neutral losses, except when explicitly mentioned.
Results and Discussion
Structures and Native Mass Spectra of the Studied IgM- and IgG-Based Oligomeric Immunoglobulin Molecules
The here-studied immunoglobulin oligomers, the (IgM)5J pentamer (with J-chain) and (IgG1)6 hexamer, are quite different in structural organization, with the first one being a highly glycosylated and covalently linked heterohexamer (pentamer with J-chain) and the second one being a noncovalent homohexamer, as schematically depicted in Figure 1. The recombinant production of the hexamerizing IgG1-RGY is reasonably straightforward and similar to the production of normal IgG1 antibodies, whereby the inclusion of the RGY mutations promotes extensive hexamerization in solution, as previously described.14,20,57 The production of recombinant IgM, especially in its (IgM)5J “pentamer” format, is less straightforward as it requires the appropriate coexpression of the joining J chain and IgM subunits and the correct formation of all inter- and intrachain disulfide bridges (Figure 1A,B). Here, a pure aWTA (IgM)5J pentamer (with J-chain) was recombinantly expressed and purified by SEC.
Figure 1.
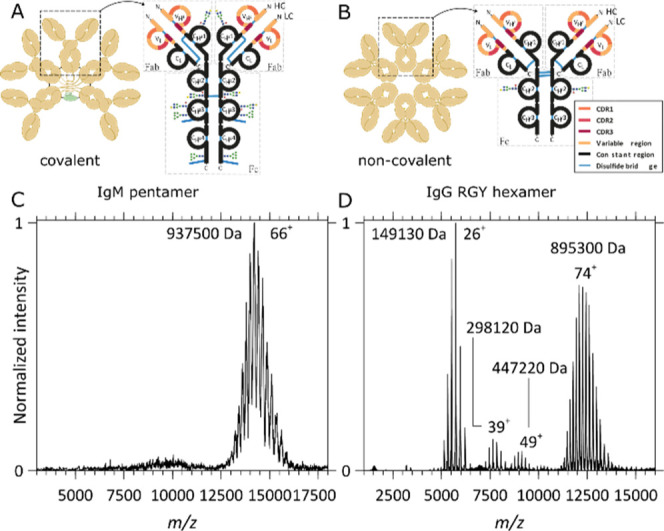
Schematic structures and native MS1 spectra of oligomeric immunoglobulins. (A,C) Structure and native MS1 spectrum of the aWTA IgM pentamer with the J-chain, respectively, and (B,D) structure and native MS1 spectrum of the aCD52 IgG1-RGY hexamer, respectively. While, in (IgM)5J, the IgM’s monomers and J-chain are connected by interchain disulfide bonds, the IgG1 monomers forming the IgG1 hexamers assemble noncovalently. (C) The MS1 spectrum of the aWTA IgM displays a single charge distribution with charge state broadening resulting from the presence of multiple heterogeneous glycans on each monomer. Although heterogeneity and glycan lability hamper an accurate mass analysis, a mean average mass of 937,500 Da could be extracted from this high-resolution data set. (D) In the MS1 spectrum of the aCD52 IgG1-RGY, monomers, dimers, and trimers co-occur with the hexamers, a direct consequence of the noncovalent interactions stabilizing the assembly and of the dynamical equilibria taking place in solution. An average mass of 895,300 Da could be extracted from these data for the hexamer, indeed being within the experimental error 6 times that of the monomer mass.
In Figure 1, we depict native MS1 spectra of the aWTA IgM pentamer coupled to the J-chain and spectra of the aCD52 IgG1-RGY hexamer. For the aWTA (IgM)5J, a single charge-state distribution is observed in the 12,500 < m/z < 16,000 range. Glycosylation of the aWTA IgM is quite heterogeneous, resulting in a broadening of the peaks. Although the presence of all these glycans hampers an accurate mass analysis, a mean average mass of 937,500 Da could be extracted from these data, which is in line with the theoretically expected mass of the (IgM)5J. The native MS spectra of the aCD52 IgG1-RGY mutant reveal charge states corresponding to IgG monomers and lower abundant dimers and trimers that co-occur with the IgG1 hexamers, a direct consequence of the noncovalent interactions stabilizing the assembly of the dynamical equilibria taking place in solution. Compared to the aWTA IgM, the aCD52 hexameric IgG1 contains a much more homogeneous glycosylation profile, as can be expected from the smaller number of glycosylation sites: IgG1’s HCs harbor one N-linked glycosylation site in the fragment-crystallizable (Fc) region. Therefore, the ion signals in Figure 1D are much sharper than those in Figure 1C. The spectra shown in Figure 1D and the extracted masses are in excellent agreement with previously reported data for the IgG1 RGY constructs.20
Native Top-Down MS of Oligomeric Immunoglobulins by CID
As displayed in Figure 2, we first performed CID on mass-selected aWTA (IgM)5J and aCD52 hexameric IgG1 assemblies. In peptide and protein centric MS, CID is still the most ubiquitous activation technique. Applied to large noncovalent assemblies, it typically results in the release of one or more subunits carrying most of the complex charge: a process called asymmetric dissociation.58,59 Here, we could only achieve CID by using the highest available collision energies, that is, CID of such large systems is demanding. Applied to precursor ions of aWTA (IgM)5J (Figures 2A and S2A) carrying 66 charges, CID primarily results in the release of the intact IgM LC—as a result of disulfide bond cleavage. Additionally, limited backbone cleavage yields small sequence tags formed by b-ions covering part of the LC’s constant regions (in the m/z window below 5000). Contrastingly, for the precursor ions of the aCD52 hexameric IgG1 assembly which carry ∼74 charges (Figures 2B and S3A), the primary observed CID channel is the ejection of IgG monomer ions, as expected from this noncovalent assembly. These monomer ions take, upon release, on average, one-third of all charges of the precursor ions. Secondary lower abundant dissociation channels correspond, for the hexamer, to dimer ejection and the formation of a small sequence tag made of b-ions covering part of the LC’s constant region (in the m/z window below 5000). Overall, obtaining sequence information from large (∼900 kDa) Ig complexes using CID is challenging because of the many observed competitive dissociation channels, overlapping ion signals—the result of extensive water and ammonia losses—as well as signal broadening, when recording ion signals (transients) at a resolution of 280,000 @m/z 200 and a pressure setting of 1.0. Short CID sequence tags determined at a 3 ppm accuracy (Figures S2B and S3B) can nevertheless contribute valuable information by confirming ECD de novo assignments.
Figure 2.
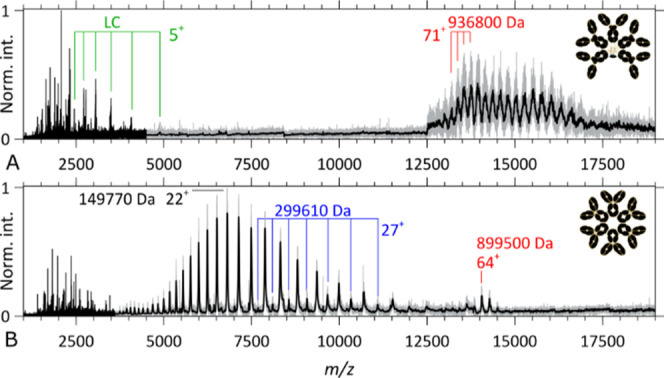
Native top-down CID MS2 spectra of IgM- and IgG-based oligomeric immunoglobulins. (A) CID MS2 spectrum of the aWTA IgM precursor ion signal (z ∼ 74) overlapping with high m/z product ions in the upper m/z range, while isotopically resolved intact LCs and backbone fragments are detected below m/z 5000. (B) CID MS2 spectrum of the aCD52 hexameric IgG1 precursor ion (z ∼ 66) at high m/z, intact IgG1 monomers, and lower abundant IgG dimers in the 3500 ≤ m/z ≤ 12,500 range, and backbone fragments below m/z 3500. A Gaussian filter has been applied to the high m/z range with the resulting spectrum overlapping the raw data (in gray).
Native Top-Down MS of Oligomeric Immunoglobulins by ECD
While CID of large protein assemblies mostly results in subunit ejection, ECD is known to preferentially generate backbone fragments.60 Practically, the addition of an electron to a cationic protein increases its internal energy by 6–7 eV of which about 3 eV is used to cleave the backbone and form (c, z·)-ion pairs, while the residual energy contributes to fragment separation (the intermolecular hydrogen bond energies range from −6 to −3 kJ/mol or −0.06 to −0.03 eV).61,62 The efficiency of fragment generation by ECD can however be low, especially for large protein assemblies, as precursor ions are known to capture up to tens of electrons without fragmenting, a process sometimes referred to as ECnoD.23 Applied to the precursor ions of the aWTA IgM and aCD52 hexameric IgG1, ECD yields precursor ions having captured tens of electrons, as shown in Figures 3A,B and S4, as well as Figures 3C and S10 (m/z regions above the original ion signals). These ECnoD ions are unfortunately not very useful for sequencing and structural elucidation. Gratifyingly, in the 500 < m/z < 3000 region, a variety of fragment ions are observed, which are the result of ECD cleavages in the backbone of the immunoglobulin oligomers, as discussed below.
Figure 3.
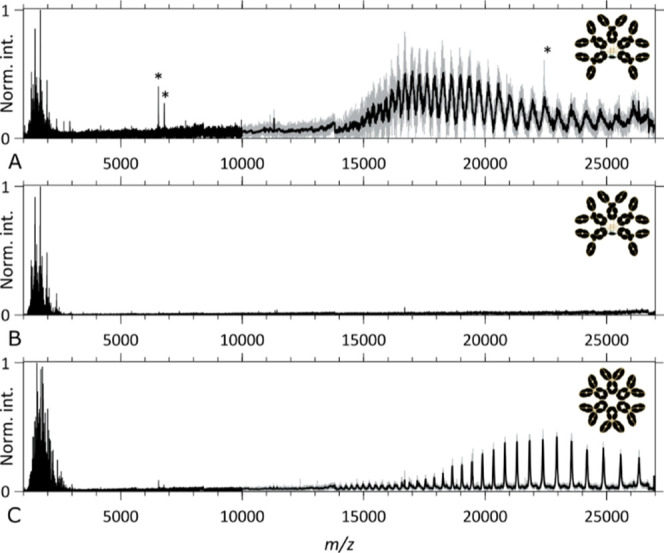
Native top-down ECD MS2 spectra of IgM- and IgG-based oligomeric immunoglobulins. (A,B) ECD MS2 spectra of the aWTA IgM assembly and (C) ECD MS2 spectra of the aCD52 hexameric IgG1 assembly. (A,C) ECD MS2 spectra taken with the “Analyzer CE-Inject (V) UHMR” at its default value of 3200, displaying the precursor ion signals and the ECnoD product ions at high m/z and backbone fragments below m/z 3000, respectively, for the aWTA IgM and the aCD52 hexameric IgG1 assemblies. (B) ECD MS2 spectrum of the aWTA IgM assembly taken with the “Analyzer CE-Inject (V) UHMR” at a value of 3700, optimized for “low m/z” fragment ion detection. * denotes noise peaks.
Optimizing ECD on a UHMR Q-exactive requires reducing the kinetic energy of the ions in order to maximize residence time in the ECD cell to facilitate efficient electron capture. A significant fraction of the kinetic energy transferred to the ions by the electrospray process and the drag force exerted by the gas entering the instrument can be removed by performing extensive in-source trapping. On our setup, in-source trapping also serves to increase the internal energy of the precursor ions. We determined that by using a desolvation potential of −100 V, ECD fragment abundances were maximized for compounds ranging from Fab, F(ab’)2, and intact Igs to the here-studied close-to-MDa oligomeric Igs. There are likely two reasons contributing to this observation. First, as described by Loo and co-workers, ion-pair/salt-bridge rearrangements can occur upon collisional activation,59 which in turn impacts the hydrogen bonding interactions that contribute some of the radical hydrogens ultimately responsible for backbone cleavages and the more efficient formation of c and z· fragments.63 Second, by increasing the ions’ internal energy, we bring them closer to the dissociation threshold of the noncovalent interactions stabilizing the c and z· ion pairs and thereby facilitate their separation.21,44
Upon release from the source trap, minimized potential differences transfer the precursor ions first to the quadrupole—where they are mass-selected—then to the ECD cell where they undergo electron capture. Following their transmission through the ECD cell and C-trap, the ions—at this stage, a mixture of precursors and fragments—are trapped in the HCD cell. Reduced kinetic energies and pressure allow efficient trapping of fragments with the m/z up to a few thousands, while the precursor ions and the high m/z product ions responsible for the third harmonic interference, interfering with the low m/z fragment signal, are suppressed. Orbitrap measurements of large protein assemblies in the hundreds-of-kDa range are known to yield spurious frequencies, of which the third harmonic is the most problematic one as it overlaps, for precursors with m/z ranging from 10,000 to 30,000, with fragments about the 1–3 kDa range (Figure S13). High m/z signal suppression at low pressure is a consequence of the limited stopping power of the low-pressure buffer gas used to trap ions in the HCD cell.
In Figure 3A, we demonstrate that high-quality ECD fragment signals can be obtained for an aWTA IgM while retaining the detection of the charge-reduced precursor. Upon decreasing the analyzer’s “CE-Inject (V) UHMR” setting from 3700 to 3200, we obtain the spectra as shown in Figure 3B with significantly improved signal-to-noise ratio and complete suppression of the precursor signal and its third harmonics. Charge deconvolution of the spectra of Figure 3A,B yield the m/[z = 1] spectra displayed in Figures (S5, S6) and (4, S8, and S9), respectively.
Figure 4.
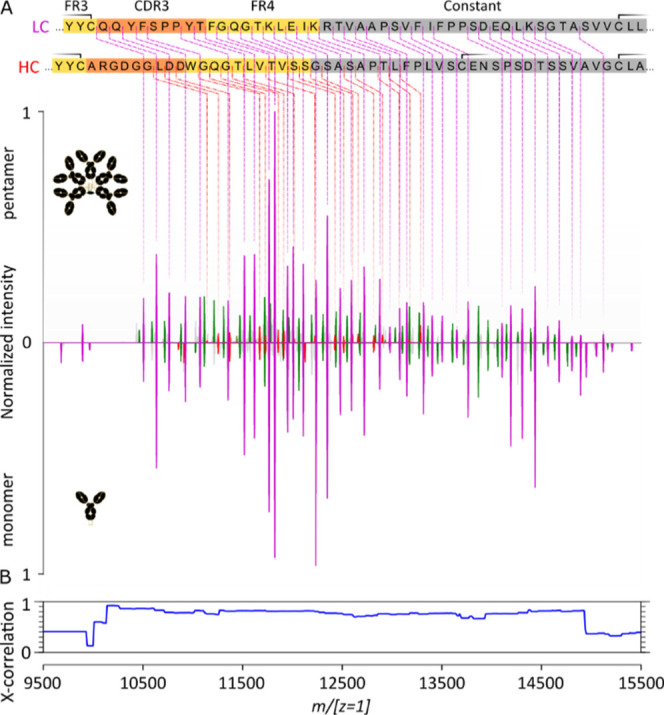
Comparison of native top-down ECD fragment ion spectra of (IgM)5J and the corresponding monomeric IgM. (A) Charge-deconvoluted (m/[z = 1]) native top-down ECD spectra of the aWTA IgM pentamer with the J-chain and the aWTA IgM monomer (“Analyzer CE-Inject (V) UHMR” at 3700 in both cases). Note that the sequence coverage of the HC stops before the cysteine bridging to the LC (the second of the three cysteine parts of the displayed HC sequence segment). HC c-ions are in red, LC c-ions are in purple, and the green peaks correspond to LC a-ions. (B) Cross-correlation analysis of the two native top-down ECD fragment ion spectra.
Remarkably, similar to other monomeric Ig types and their subclasses, ECD of these very large ∼MDa oligomeric Igs, also results in backbone cleavages primarily in the LC’s and HC’s sequence segment outside the disulfide loops bridging the variable and the CHμ1 regions. For both the HC and LC, the sequence tags cover close to the whole range outside the disulfide loops with the gaps mostly filled, in the LC case, by a-ions. This remarkable feature illustrates that even for a ∼930 kDa “heterohexamer” consisting of five intact IgMs and one J-chain, all extensively and covalently linked with each other in the (IgM)5J assembly, the native top-down ECD can produce informative sequence tags from the variable regions of IgMs. The (IgM)5J assembly studied here was recombinantly produced by coexpressing the IgM and J chains together, whereafter the full assembly was purified. By expressing IgM without coexpression of the J-chain (data not shown), IgM is formed in a variety of oligomeric states, with the tetramer, pentamer, and hexamer being the most abundant57 and also with quite some IgM monomers expressed. Using this sample, we were able to produce and study, by native top-down ECD, the fragmentation behavior of the ∼184 kDa aWTA IgM monomer, that is, having exactly the same sequence as the aWTA IgM present in the here-studied ∼935 kDa pentamer (+J chain). Comparison of the native top-down ECD m/[z = 1] spectra for the intact aWTA IgM with the J-chain and the aWTA IgM monomer (Figure 4, top and bottom spectra) reveals the high similarity between these ECD spectra. Notably, here, the precursor ions were quite different, that is, the 72–60 charge envelope of the ∼935 kDa and the 30–25 charge envelope of the ∼184 kDa IgM monomer. Correlation values between these spectra are in excess of 70%. It is to be noted that the monomer was sprayed without making use of in-source trapping but relying on a “source fragmentation (eV)” setting of 120 instead.
In Figure 5, the results of a similar approach are shown comparing the ECD spectra of the aCD52 IgG1-RGY hexamer and monomer. As in the case of the aWTA IgM with the J-chain, high-quality ECD fragment signals can be obtained for the aCD52 IgG1-RGY hexamer. Charge deconvolution of the ECD fragment spectra yields ECD m/[z = 1] spectra characterized by nearly complete sequence coverage of both the LC’s and HC’s sequence segments outside the disulfide loops bridging the variable and the constant regions (Figures 5, S11, and S12). For both the HC and LC, a-ions fill most of the gaps in the sequence tag, as observed previously.45,46
Figure 5.
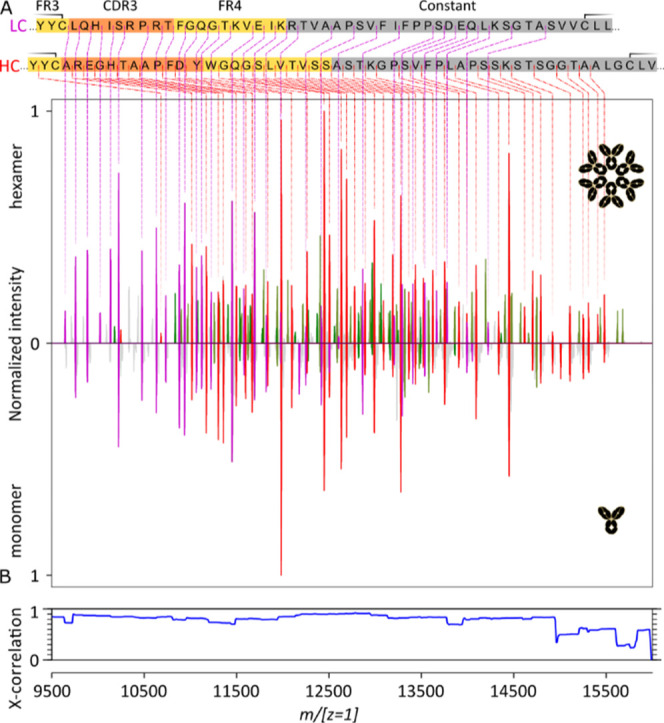
Comparison of the native top-down ECD fragment ion spectra of (IgG)6 and the corresponding monomeric IgG. (A) Charge-deconvoluted (m/[z = 1]) ECD spectra of the aCD52 IgG1 hexamer and its monomer (“Analyzer CE-Inject (V) UHMR” at 3200 in both cases). HC c-ions are in red, LC c-ions are in purple, and the green peaks correspond to LC a-ions. (B) Cross-correlation of the two native top-down ECD fragment ion spectra.
Comparison of the ECD m/[z = 1] spectra for the intact aCD52 IgG1-RGY hexamer and a monomer (Figure 5, top and bottom spectra) establishes again the high similarity between these ECD spectra and the absence of an effect of the IgG oligomerization on the obtained sequence coverage. Here also, the precursor ions were quite different, that is, the 78–67 charge envelope of the ∼890 kDa and the 29–23 charge envelope of the ∼149 kDa IgG1-RGY monomer. Correlation values between the two spectra are in excess of 80%. It is to be noted that the monomer was readily obtained from an aCD52 IgG1-RGY hexamer sample (Figure 1), and acquisition was performed under identical conditions.
Conclusions
Over the last decades, proteomics has expanded from the traditional peptide-centric approaches (i.e., bottom-up or shotgun proteomics with precursor masses typically being 500 < Mw < 5000 Da) to the direct analysis of intact proteins by the so-called top-down proteomics (with precursor masses typically in the range 5 kDa <Mw < 50 kDa). In the past years, this trend has been expanding even further to cover much larger protein assemblies by the native top-down proteomics (with precursor masses typically in the range 50 kDa < Mw < 0.5 MDa). These larger and more complex precursors require dedicated approaches to separate, fragment, and characterize analytes, tackled by native top-down proteomics. Here, we successfully demonstrate that native top-down proteomics can be extended to the important class of oligomeric immunoglobulins, with MWs reaching close to 1 MDa, and that ECD implemented on an Orbitrap UHMR can yield valuable sequence information to characterize and partially identify these important immune molecules. Specifically, the present approach allowed us to get straightforward-to-read isotopically resolved amino acid sequence ladders of c-ions (9000–16,000 Da) spanning the CDR3s and FR4s. The analysis of the intact oligomers was validated by performing ECD of their monomers. Only small differences (<20%) were observed between the native top-down ECD spectra of the intact oligomers and their monomers, suggesting a low impact of the assembly on fragment formation.
Overall, we have demonstrated that the complexity and size of the pentameric IgM’s (with J chain) and hexameric IgG1’s structures do not adversely impact their ECD compared to smaller-monomeric Igs. As in the case of IgGs and IgAs,45,46 we showed that ECD without supplemental activation could be deliberately restricted to the highly informative hypervariable regions outside the disulfide loops of both the LC and HC. ECD without supplemental activation thereby proved to be uniquely suited to the screening of all isotypes and variants of immunoglobulins. This, in turn, may impact regulatory requirements for the characterization of therapeutic antibodies ranging from IgGs, IgAs, to IgMs, or, in a diagnostic context, facilitate the screening of complex mixtures in combination with orthogonal separation methods.
Acknowledgments
We thank the members of the Heck laboratory for general support, especially Arjan Barendregt. We acknowledge the group of Janine Schuurman at Genmab for providing the recombinant RGY-mutant IgG samples and the Suzan Rooijakkers group at the Medical Microbiology Department, University Medical Center Utrecht (The Netherlands), in particular, P. Aerts and C. Gosselar-de Haas, for providing the recombinant IgM samples. This research received funding through the Netherlands Organization for Scientific Research (NWO) TTW project 15575 (Structural analysis and position-resolved imaging of macromolecular structures using novel mass spectrometry-based approaches), the ENPPS.LIFT.019.001 project (A.J.R.H., S.-H.L., and J.-F.G.), the NACTAR project 16442 (A.J.R.H. and M.A.d.B.), and the Spinoza Award SPI.2017.028 to A.J.R.H. This project received additional funding from the European Union’s Horizon 2020 research and innovation program under the grant agreement 686547 (EPIC-XS) for A.J.R.H.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.1c03740.
ECD parameter used, the native top-down CID and ECD MS2 spectra of the aWTA IgM and hexameric aCD52 IgG1, full-range charge-deconvoluted spectra of the aWTA IgM and hexameric aCD52 IgG, and a spectrum highlighting the third harmonic interferences occurring when the signal is optimized for high m/z detection, and link to raw data files (PDF)
Author Present Address
∥ Bruker Daltonics GmbH & Co. KG Fahrenheitstraße 4 28359 Bremen
Author Contributions
All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): J.C. is an employee of MSVision, a company that offers instrument modifications as a service, and installs ECD modules in a variety of mass analyzers. All other authors have no conflict of interest to report.
Supplementary Material
References
- Klingler J.; Weiss S.; Itri V.; Liu X.; Oguntuyo K. Y.; Stevens C.; Ikegame S.; Hung C.-T.; Enyindah-Asonye G.; Amanat F.; Baine I.; Arinsburg S.; Bandres J. C.; Kojic E. M.; Stoever J.; Jurczyszak D.; Bermudez-Gonzalez M.; Nádas A.; Liu S.; Lee B.; Zolla-Pazner S.; Hioe C. E. Role of Immunoglobulin M and A Antibodies in the Neutralization of Severe Acute Respiratory Syndrome Coronavirus 2. J. Infect. Dis. 2020, 223, 957. 10.1093/infdis/jiaa784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud E.; Mastrandrea C.; Rochereau N.; Paul S. Human Secretory IgM: An Elusive Player in Mucosal Immunity. Trends Immunol. 2020, 41, 141–156. 10.1016/j.it.2019.12.005. [DOI] [PubMed] [Google Scholar]
- Ehrenstein M. R.; Notley C. A. The Importance of Natural IgM: Scavenger, Protector and Regulator. Nat. Rev. Immunol. 2010, 10, 778–786. 10.1038/nri2849. [DOI] [PubMed] [Google Scholar]
- Brandlein S.; Pohle T.; Ruoff N.; Wozniak E. Natural IgM Antibodies and Immunosurveillance Mechanisms against Epithelial Cancer Cells in Humans. Cancer Res. 2003, 63, 7995. [PubMed] [Google Scholar]
- Vollmers H. P.; Brändlein S. Natural IgM Antibodies: From Parias to Parvenus. Histol. Histopathol. 2006, 21, 1355–1366. 10.14670/HH-21.1355. [DOI] [PubMed] [Google Scholar]
- Kretschmer A.; Schwanbeck R.; Valerius T.; Rösner T. Antibody Isotypes for Tumor Immunotherapy. Transfus. Med. Hemotherapy 2017, 44, 320–326. 10.1159/000479240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramoto E.; Tsutsumi A.; Suzuki R.; Matsuoka S.; Arai S.; Kikkawa M.; Miyazaki T. The IgM Pentamer Is an Asymmetric Pentagon with an Open Groove That Binds the AIM Protein. Sci. Adv. 2018, 4, eaau1199 10.1126/sciadv.aau1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N.; Arthur C. P.; Ciferri C.; Matsumoto M. L. Structure of the Human Secretory Immunoglobulin M Core. Structure 2021, 29, 594. 10.1016/j.str.2021.01.002. [DOI] [PubMed] [Google Scholar]
- Li Y.; Wang G.; Li N.; Wang Y.; Zhu Q.; Chu H.; Wu W.; Tan Y.; Yu F.; Su X.-D.; Gao N.; Xiao J. Structural Insights into Immunoglobulin M. Science 2020, 367, 1014–1017. 10.1126/science.aaz5425. [DOI] [PubMed] [Google Scholar]
- Vollmers H. P.; Brändlein S. Natural IgM Antibodies: The Orphaned Molecules in Immune Surveillance. Adv. Drug Delivery Rev. 2006, 58, 755–765. 10.1016/j.addr.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Perkins S. J.; Nealis A. S.; Sutton B. J.; Feinstein A. Solution Structure of Human and Mouse Immunoglobulin M by Synchrotron X-Ray Scattering and Molecular Graphics Modelling: A Possible Mechanism for Complement Activation. J. Mol. Biol. 1991, 221, 1345–1366. 10.1016/0022-2836(91)90937-2. [DOI] [PubMed] [Google Scholar]
- Reddy P. S.; Corley R. B. The Contribution of ER Quality Control to the Biologic Functions of Secretory IgM. Immunol. Today 1999, 20, 582–588. 10.1016/S0167-5699(99)01542-X. [DOI] [PubMed] [Google Scholar]
- Johansen; Braathen; Brandtzaeg Role of J Chain in Secretory Immunoglobulin Formation. Scand. J. Immunol. 2000, 52, 240–248. 10.1046/j.1365-3083.2000.00790.x. [DOI] [PubMed] [Google Scholar]
- de Jong R. N.; Beurskens F. J.; Verploegen S.; Strumane K.; van Kampen M. D.; Voorhorst M.; Horstman W.; Engelberts P. J.; Oostindie S. C.; Wang G.; Heck A. J. R.; Schuurman J.; Parren P. W. H. I. A Novel Platform for the Potentiation of Therapeutic Antibodies Based on Antigen-Dependent Formation of IgG Hexamers at the Cell Surface. PLoS Biol. 2016, 14, e1002344 10.1371/journal.pbio.1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. I.; Coloma M. J.; Morrison S. L. Addition of a Mu-Tailpiece to IgG Results in Polymeric Antibodies with Enhanced Effector Functions Including Complement-Mediated Cytolysis by IgG4. J. Immunol. 1995, 154, 2226–2236. [PubMed] [Google Scholar]
- Diebolder C. A.; Beurskens F. J.; de Jong R. N.; Koning R. I.; Strumane K.; Lindorfer M. A.; Voorhorst M.; Ugurlar D.; Rosati S.; Heck A. J. R.; van de Winkel J. G. J.; Wilson I. A.; Koster A. J.; Taylor R. P.; Ollmann Saphire E.; Burton D. R.; Schuurman J.; Gros P.; Parren P. W. H. I. Complement Is Activated by IgG Hexamers Assembled at the Cell Surface. Science 2014, 343, 1260–1263. 10.1126/science.1248943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; de Jong R. N.; Beurskens F. J.; Labrijn A. F.; Ugurlar D.; Gros P.; Schuurman J.; Parren P. W. H. I.; Heck A. J. R.; Heck A. J. R. Molecular Basis of Assembly and Activation of Complement Component C1 in Complex with Immunoglobulin G1 and Antigen. Mol. Cell 2016, 63, 135–145. 10.1016/j.molcel.2016.05.016. [DOI] [PubMed] [Google Scholar]
- Lubbers R.; Oostindie S. C.; Dijkstra D. J.; Parren P. W. H. I.; Verheul M. K.; Abendstein L.; Sharp T. H.; Ru A.; Janssen G. M. C.; Veelen P. A.; den Bremer E. T. J.; Bleijlevens B.; Kreuk B. J.; Beurskens F. J.; Trouw L. A. Carbamylation Reduces the Capacity of IgG for Hexamerization and Complement Activation. Clin. Exp. Immunol. 2020, 200, 1–11. 10.1111/cei.13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B.; Gao X.; Cadang L.; Izadi S.; Liu P.; Zhang H.-M.; Hecht E.; Shim J.; Magill G.; Pabon J. R.; Dai L.; Phung W.; Lin E.; Wang C.; Whang K.; Sanchez S. Jr.; Oropeza J. Jr.; Camperi J.; Zhang J.; Sandoval W.; Zhang Y. T.; Jiang G. Fc Galactosylation Follows Consecutive Reaction Kinetics and Enhances Immunoglobulin G Hexamerization for Complement Activation. mAbs 2021, 13, 1893427. 10.1080/19420862.2021.1893427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz A. R.; Boer M. A. d.; Strasser J.; Zwarthoff S. A.; Beurskens F. J.; de Haas C. J. C.; Aerts P. C.; Wang G.; de Jong R. N.; Bagnoli F.; van Strijp J. A. G.; van Kessel K. P. M.; Schuurman J.; Preiner J.; Heck A. J. R.; Rooijakkers S. H. M. Staphylococcal Protein A Inhibits Complement Activation by Interfering with IgG Hexamer Formation. Proc. Natl. Acad. Sci. U.S.A. 2021, 118, e2016772118 10.1073/pnas.2016772118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Cui W.; Gross M. L. Mass Spectrometry for the Biophysical Characterization of Therapeutic Monoclonal Antibodies. FEBS Lett. 2014, 588, 308–317. 10.1016/j.febslet.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Wolff J. J.; Van Orden S. L.; Loo J. A. Native Top-Down Electrospray Ionization-Mass Spectrometry of 158 KDa Protein Complex by High-Resolution Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Anal. Chem. 2014, 86, 317–320. 10.1021/ac4033214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M.; Lantz C.; Brown K. A.; Ge Y.; Paša-Tolić L.; Loo J. A.; Lermyte F. Higher-Order Structural Characterisation of Native Proteins and Complexes by Top-down Mass Spectrometry. Chem. Sci. 2020, 11, 12918–12936. 10.1039/D0SC04392C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives A. N.; Su T.; Durbin K. R.; Early B. P.; dos Santos Seckler H.; Fellers R. T.; LeDuc R. D.; Schachner L. F.; Patrie S. M.; Kelleher N. L. Using 10,000 Fragment Ions to Inform Scoring in Native Top-down Proteomics. J. Am. Soc. Mass Spectrom. 2020, 31, 1398–1409. 10.1021/jasms.0c00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konijnenberg A.; Bannwarth L.; Yilmaz D.; Koçer A.; Venien-Bryan C.; Sobott F. Top-down Mass Spectrometry of Intact Membrane Protein Complexes Reveals Oligomeric State and Sequence Information in a Single Experiment: Top-down MS of Membrane Proteins. Protein Sci. 2015, 24, 1292–1300. 10.1002/pro.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale O. J.; Cooper H. J. Native Mass Spectrometry Imaging and In Situ Top-Down Identification of Intact Proteins Directly from Tissue. J. Am. Soc. Mass Spectrom. 2020, 31, 2531–2537. 10.1021/jasms.0c00226. [DOI] [PubMed] [Google Scholar]
- Skinner O. S.; Haverland N. A.; Fornelli L.; Melani R. D.; Do Vale L. H. F.; Seckler H. S.; Doubleday P. F.; Schachner L. F.; Srzentić K.; Kelleher N. L.; Compton P. D. Top-down Characterization of Endogenous Protein Complexes with Native Proteomics. Nat. Chem. Biol. 2018, 14, 36–41. 10.1038/nchembio.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder D. T.; Panczyk E. M.; Somogyi A.; Kaplan D. A.; Wysocki V. Simple and Minimally Invasive SID Devices for Native Mass Spectrometry. Anal. Chem. 2020, 92, 11195–11203. 10.1021/acs.analchem.0c01657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J. P.; Li W.; Zhang Y.; Brodbelt J. S. Characterization of Native Protein Complexes Using Ultraviolet Photodissociation Mass Spectrometry. J. Am. Chem. Soc. 2014, 136, 12920–12928. 10.1021/ja505217w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashyal A.; Sanders J. D.; Holden D. D.; Brodbelt J. S. Top-Down Analysis of Proteins in Low Charge States. J. Am. Soc. Mass Spectrom. 2019, 30, 704–717. 10.1007/s13361-019-02146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greisch J.-F.; Tamara S.; Scheltema R. A.; Maxwell H. W. R.; Fagerlund R. D.; Fineran P. C.; Tetter S.; Hilvert D.; Heck A. J. R. Expanding the Mass Range for UVPD-Based Native Top-down Mass Spectrometry. Chem. Sci. 2019, 10, 7163–7171. 10.1039/C9SC01857C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greisch J.-F.; van der Laarse S. A. M.; Heck A. J. R. Enhancing Top-Down Analysis Using Chromophore-Assisted Infrared Multiphoton Dissociation from (Phospho)Peptides to Protein Assemblies. Anal. Chem. 2020, 92, 15506–15516. 10.1021/acs.analchem.0c03412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehaffey M. R.; Xia Q.; Brodbelt J. S. Uniting Native Capillary Electrophoresis and Multistage Ultraviolet Photodissociation Mass Spectrometry for Online Separation and Characterization of Escherichia Coli Ribosomal Proteins and Protein Complexes. Anal. Chem. 2020, 92, 15202–15211. 10.1021/acs.analchem.0c03784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M.; Liu W.; Shaw J. B. Charge Movement and Structural Changes in the Gas-Phase Unfolding of Multimeric Protein Complexes Captured by Native Top-Down Mass Spectrometry. Anal. Chem. 2020, 92, 1788–1795. 10.1021/acs.analchem.9b03469. [DOI] [PubMed] [Google Scholar]
- Li H.; Nguyen H. H.; Ogorzalek Loo R. R.; Campuzano I. D. G.; Loo J. A. An Integrated Native Mass Spectrometry and Top-down Proteomics Method That Connects Sequence to Structure and Function of Macromolecular Complexes. Nat. Chem. 2018, 10, 139–148. 10.1038/NCHEM.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srzentić K.; Fornelli L.; Tsybin Y. O.; Loo J. A.; Seckler H.; Agar J. N.; Anderson L. C.; Bai D. L.; Beck A.; Brodbelt J. S.; van der Burgt Y. E. M.; Chamot-Rooke J.; Chatterjee S.; Chen Y.; Clarke D. J.; Danis P. O.; Diedrich J. K.; D’Ippolito R. A.; Dupré M.; Gasilova N.; Ge Y.; Goo Y. A.; Goodlett D. R.; Greer S.; Haselmann K. F.; He L.; Hendrickson C. L.; Hinkle J. D.; Holt M. V.; Hughes S.; Hunt D. F.; Kelleher N. L.; Kozhinov A. N.; Lin Z.; Malosse C.; Marshall A. G.; Menin L.; Millikin R. J.; Nagornov K. O.; Nicolardi S.; Paša-Tolić L.; Pengelley S.; Quebbemann N. R.; Resemann A.; Sandoval W.; Sarin R.; Schmitt N. D.; Shabanowitz J.; Shaw J. B.; Shortreed M. R.; Smith L. M.; Sobott F.; Suckau D.; Toby T.; Weisbrod C. R.; Wildburger N. C.; Yates J. R.; Yoon S. H.; Young N. L.; Zhou M. Interlaboratory Study for Characterizing Monoclonal Antibodies by Top-Down and Middle-Down Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2020, 31, 1783–1802. 10.1021/jasms.0c00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley N. M.; Coon J. J. The Role of Electron Transfer Dissociation in Modern Proteomics. Anal. Chem. 2018, 90, 40–64. 10.1021/acs.analchem.7b04810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuker K.; McLafferty F. W. Native Electron Capture Dissociation for the Structural Characterization of Noncovalent Interactions in Native Cytochrome c. Angew. Chem., Int. Ed. 2003, 42, 4900–4904. 10.1002/anie.200351705. [DOI] [PubMed] [Google Scholar]
- Zubarev R. A. Electron-Capture Dissociation Tandem Mass Spectrometry. Curr. Opin. Biotechnol. 2004, 15, 12–16. 10.1016/j.copbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Cui W.; Wen J.; Blankenship R. E.; Gross M. L. Native Electrospray and Electron-Capture Dissociation in FTICR Mass Spectrometry Provide Top-Down Sequencing of a Protein Component in an Intact Protein Assembly. J. Am. Soc. Mass Spectrom. 2010, 21, 1966–1968. 10.1016/j.jasms.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadkari V. V.; Ramírez C. R.; Vallejo D. D.; Kurulugama R. T.; Fjeldsted J. C.; Ruotolo B. T. Enhanced Collision Induced Unfolding and Electron Capture Dissociation of Native-like Protein Ions. Anal. Chem. 2020, 92, 15489–15496. 10.1021/acs.analchem.0c03372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. P.; Morrison L. J.; Brown J. M.; Beckman J. S.; Voinov V. G.; Lermyte F. Top-Down Characterization of Denatured Proteins and Native Protein Complexes Using Electron Capture Dissociation Implemented within a Modified Ion Mobility-Mass Spectrometer. Anal. Chem. 2020, 92, 3674–3681. 10.1021/acs.analchem.9b04763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Loo R. R. O.; Loo J. A. Structural Characterization of a Thrombin-Aptamer Complex by High Resolution Native Top-Down Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2017, 28, 1815–1822. 10.1007/s13361-017-1751-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermyte F.; Valkenborg D.; Loo J. A.; Sobott F. Radical Solutions: Principles and Application of Electron-Based Dissociation in Mass Spectrometry-Based Analysis of Protein Structure. Mass Spectrom. Rev. 2018, 37, 750–771. 10.1002/mas.21560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boer M. A.; Tamara S.; Bondt A.; Heck A. J. R.; Heck A. J. R. Selectivity over Coverage in de Novo Sequencing of IgGs. Chem. Sci. 2020, 11, 11886–11896. 10.1039/d0sc03438j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greisch J.-F.; den Boer M. A.; Beurskens F.; Schuurman J.; Tamara S.; Bondt A.; Heck A. J. R. Generating Informative Sequence Tags from Antigen-Binding Regions of Heavily Glycosylated IgA1 Antibodies by Native Top-Down Electron Capture Dissociation. J. Am. Soc. Mass Spectrom. 2021, 32, 1326. 10.1021/jasms.0c00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y.; Valeja S. G.; Rouse J. C.; Hendrickson C. L.; Marshall A. G. Top-Down Structural Analysis of an Intact Monoclonal Antibody by Electron Capture Dissociation-Fourier Transform Ion Cyclotron Resonance-Mass Spectrometry. Anal. Chem. 2013, 85, 4239–4246. 10.1021/ac303525n. [DOI] [PubMed] [Google Scholar]
- Shaw J. B.; Malhan N.; Vasil’ev Y. V.; Lopez N. I.; Makarov A.; Beckman J. S.; Voinov V. G. Sequencing Grade Tandem Mass Spectrometry for Top–Down Proteomics Using Hybrid Electron Capture Dissociation Methods in a Benchtop Orbitrap Mass Spectrometer. Anal. Chem. 2018, 90, 10819–10827. 10.1021/acs.analchem.8b01901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. B.; Liu W.; Vasil′ev Y. V.; Bracken C. C.; Malhan N.; Guthals A.; Beckman J. S.; Voinov V. G. Direct Determination of Antibody Chain Pairing by Top-down and Middle-down Mass Spectrometry Using Electron Capture Dissociation and Ultraviolet Photodissociation. Anal. Chem. 2020, 92, 766–773. 10.1021/acs.analchem.9b03129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort K. L.; Cramer C. N.; Voinov V. G.; Vasil’ev Y. V.; Lopez N. I.; Beckman J. S.; Heck A. J. R. Exploring ECD on a Benchtop Q Exactive Orbitrap Mass Spectrometer. J. Proteome Res. 2018, 17, 926–933. 10.1021/acs.jproteome.7b00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacholarz K. J.; Peters S. J.; Garlish R. A.; Henry A. J.; Taylor R. J.; Humphreys D. P.; Barran P. E. Molecular Insights into the Thermal Stability of MAbs with Variable-Temperature Ion-Mobility Mass Spectrometry. ChemBioChem 2016, 17, 46–51. 10.1002/cbic.201500574. [DOI] [PubMed] [Google Scholar]
- Chambers M. C.; Maclean B.; Burke R.; Amodei D.; Ruderman D. L.; Neumann S.; Gatto L.; Fischer B.; Pratt B.; Egertson J.; Hoff K.; Kessner D.; Tasman N.; Shulman N.; Frewen B.; Baker T. A.; Brusniak M.-Y.; Paulse C.; Creasy D.; Flashner L.; Kani K.; Moulding C.; Seymour S. L.; Nuwaysir L. M.; Lefebvre B.; Kuhlmann F.; Roark J.; Rainer P.; Detlev S.; Hemenway T.; Huhmer A.; Langridge J.; Connolly B.; Chadick T.; Holly K.; Eckels J.; Deutsch E. W.; Moritz R. L.; Katz J. E.; Agus D. B.; MacCoss M.; Tabb D. L.; Mallick P. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J.; Carvalho L.; Zaia J. Application of Network Smoothing to Glycan LC-MS Profiling. Bioinformatics 2018, 34, 3511–3518. 10.1093/bioinformatics/bty397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J.; Heckendorf C.; Lukauskas S.. Mobiusklein/Ms_deisotope: Release v0.0.10; Zenodo, 2019.
- Wilkins C.; Gibbons B.. PNNL-Comp-Mass-Spec/LCMS-Spectator; Computational Mass Spectrometry; Pacific Northwest National Laboratory, 2020.
- Park J.; Piehowski P. D.; Wilkins C.; Zhou M.; Mendoza J.; Fujimoto G. M.; Gibbons B. C.; Shaw J. B.; Shen Y.; Shukla A. K.; Moore R. J.; Liu T.; Petyuk V. A.; Tolić N.; Paša-Tolić L.; Smith R. D.; Payne S. H.; Kim S. Informed-Proteomics: Open-Source Software Package for Top-down Proteomics. Nat. Methods 2017, 14, 909–914. 10.1038/nmeth.4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wörner T. P.; Snijder J.; Bennett A.; Agbandje-McKenna M.; Makarov A. A.; Heck A. J. R. Resolving Heterogeneous Macromolecular Assemblies by Orbitrap-Based Single-Particle Charge Detection Mass Spectrometry. Nat. Methods 2020, 17, 395–398. 10.1038/s41592-020-0770-7. [DOI] [PubMed] [Google Scholar]
- Metwally H.; Duez Q.; Konermann L. Chain Ejection Model for Electrospray Ionization of Unfolded Proteins: Evidence from Atomistic Simulations and Ion Mobility Spectrometry. Anal. Chem. 2018, 90, 10069–10077. 10.1021/acs.analchem.8b02926. [DOI] [PubMed] [Google Scholar]
- Loo R. R. O.; Loo J. A. Salt Bridge Rearrangement (SaBRe) Explains the Dissociation Behavior of Noncovalent Complexes. J. Am. Soc. Mass Spectrom. 2016, 27, 975–990. 10.1007/s13361-016-1375-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurov K. O.; Fornelli L.; Wodrich M. D.; Laskay Ü. A.; Tsybin Y. O. Principles of Electron Capture and Transfer Dissociation Mass Spectrometry Applied to Peptide and Protein Structure Analysis. Chem. Soc. Rev. 2013, 42, 5014–5030. 10.1039/C3CS35477F. [DOI] [PubMed] [Google Scholar]
- Breuker K.; Oh H.; Lin C.; Carpenter B. K.; McLafferty F. W. Nonergodic and Conformational Control of the Electron Capture Dissociation of Protein Cations. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 14011–14016. 10.1073/pnas.0406095101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler K.; Thar J.; Zahn S.; Kirchner B. Estimating the Hydrogen Bond Energy. J. Phys. Chem. A 2010, 114, 9529–9536. 10.1021/jp103470e. [DOI] [PubMed] [Google Scholar]
- Ganisl B.; Breuker K. Does Electron Capture Dissociation Cleave Protein Disulfide Bonds?. Chemistryopen 2012, 1, 260–268. 10.1002/open.201200038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


