Abstract
Simple Summary
Intra-cranial ependymoma (EPN) accounts for approximately 10% of pediatric brain tumors. The current therapeutic strategies have not significantly improved prognosis, which is still dismal in nearly 40% of patients. Major challenges for treatment are chemorefractoriness of EPN, tendency to recur, and high intra-tumoral heterogeneity (ITH). It is increasingly emerging that stalled neurodevelopmental programs driven by cancer stem cells (CSCs)/progenitor cells are at the root of oncogenesis and ITH of pediatric brain tumors, including EPN. This is the first review that examines how genetic and heritable epigenetic alterations and environmental selection forces drive ITH of pediatric intra-cranial EPN in the perspective of the CSC model. This review also summarizes how improvement in the single-cell technology has deepened the comprehension of the complexity, cell-of-origin, and developmental trajectories of EPN, paving the way for novel therapeutic options.
Abstract
Intra-tumoral heterogeneity (ITH) is a complex multifaceted phenomenon that posits major challenges for the clinical management of cancer patients. Genetic, epigenetic, and microenvironmental factors are concurrent drivers of diversity among the distinct populations of cancer cells. ITH may also be installed by cancer stem cells (CSCs), that foster unidirectional hierarchy of cellular phenotypes or, alternatively, shift dynamically between distinct cellular states. Ependymoma (EPN), a molecularly heterogeneous group of tumors, shows a specific spatiotemporal distribution that suggests a link between ependymomagenesis and alterations of the biological processes involved in embryonic brain development. In children, EPN most often arises intra-cranially and is associated with an adverse outcome. Emerging evidence shows that EPN displays large intra-patient heterogeneity. In this review, after touching on EPN inter-tumoral heterogeneity, we focus on the sources of ITH in pediatric intra-cranial EPN in the framework of the CSC paradigm. We also examine how single-cell technology has shed new light on the complexity and developmental origins of EPN and the potential impact that this understanding may have on the therapeutic strategies against this deadly pediatric malignancy.
Keywords: intra-tumoral heterogeneity, ependymoma, genetics, epigenetics, tumor microenvironment, cancer stem cells, single cell RNA seq, H3K27me3, H3K27M
1. Introduction
Tumors are complex ecosystems composed of non-malignant and malignant cell populations [1]. The malignant populations themselves are genetically and phenotypically heterogeneous and define the so-called intra-tumoral heterogeneity (ITH) that governs tumor evolution [2] and drug resistance [3]. Although ITH is a “contemporary concept” [4], its complex nature was highlighted back in the 1970s [5]. However, the mechanistic explanations and full understanding of the origins of ITH still have a long way to go.
The first model to explain ITH was the clonal evolution model, whereby stochastic DNA mutations confer a growth advantage to single cancer cells, which are selected for and clonally outcompete the other cells [6,7], overcoming spatial and temporal microenvironmental constraints in a Darwinian-like process [8]. Cancer cells may also adapt epigenetically to ever-changing tumor niches by virtue of high intrinsic cellular plasticity [9,10].
Besides the gene-centric view, another framework of ITH is the cancer stem cell (CSC) model, whereby a small subpopulation of cells become hierarchically organized, phenotypically diverse tumor cells or, alternatively, shift reversibly between stem-like and more committed cell states (CSC plasticity model) [11,12]. It is likely that the clonal evolution model and the CSC model coexist and act together in a cooperative manner to determine ITH.
In this light, tumors might be considered as an “organ system”, where the cellular subclones act as “tissue types” with distinct functions [13] and reciprocal signaling between tumor subpopulations [14,15,16] and between tumor and the surrounding microenvironment [17], with complex interactions to enhance tumor fitness and facilitate immune evasion [18], drug resistance [19], and metastasis [20].
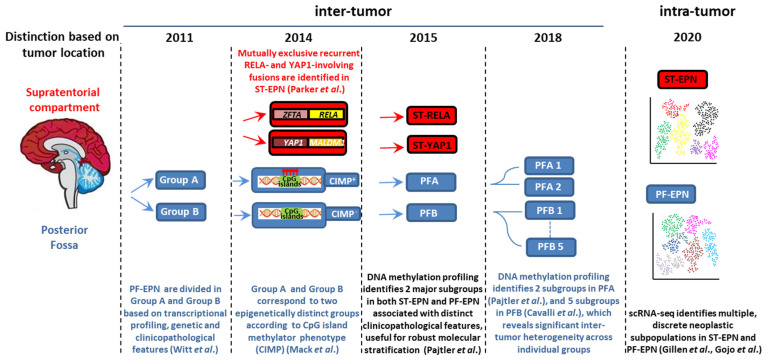
Pediatric brain tumors (PBTs) represent the leading cause of cancer-related morbidity and mortality in children [21]. The second most common PBT is ependymoma (EPN), a group of molecularly and clinically heterogeneous entities that in children arise almost exclusively intra-cranially. Despite advances in the understanding of EPN biology, the prognosis is still grim in approximately 40% of patients because of a high degree of ITH and intrinsic chemoresistance [22]. Over the last few years, whole-genome sequencing, gene-expression profiling, and genome-wide methylation at a whole-population level have stratified EPN into nine molecular groups, four of which represent the major types of intracranial pediatric EPN and differ in demographic, clinicopathological, and (epi)genetic profiles (Figure 1) [23].
Figure 1.
A timeline of the most important molecular findings which have contributed to uncovering inter-tumor and intra-tumor heterogeneity in intracranial pediatric ependymoma.
ST-EPN-RELA (ST-RELA) and ST-EPN-YAP1 (ST-YAP1) tumors arise in the supratentorial (ST) compartment, and are distinguished by mutually exclusive recurrent zinc finger translocation associated (ZFTA)-RELA proto-oncogene, NF-kB subunit (RELA) or Yes1 associated transcriptional regulator (YAP1)-involving fusions [24], whereas PF-EPN-A (PFA) and PF-EPN-B (PFB) tumors occur in the posterior fossa (PF) [25].
To date, the majority of research and treatment decisions in EPN have been based on analyses of bulk tumors that, however, return an averaged picture of all cell populations. The advances of sequencing technologies and single-cell omics over the last decade have deepened the knowledge of the bewildering heterogeneity of the tumor genome, transcriptome, epigenome, and proteome at an unprecedented scale [26,27].
After briefly summarizing EPN inter-tumoral heterogeneity, the goal of this review is to synthesize how the main genetic, epigenetic, and environmental factors drive ITH in pediatric intracranial EPN in the light of the CSC model. We also examine how single-cell RNA sequencing (scRNA-seq) has contributed to the understanding of the complexity and developmental origins of EPN, unraveling some common transcriptional programs across different EPN subtypes, and even across different pediatric brain tumors, which might help define potential druggable vulnerabilities.
2. Inter-Tumoral Heterogeneity and Clinicopathological Characteristics of Pediatric Intracranial EPN: A Brief Overview
EPNs are neuroepithelial tumors that account for approximately 10% of pediatric intra-cranial neoplasms. Surgery and radiotherapy are well-established treatments, whereas chemotherapy has not clearly demonstrated a survival benefit. According to the World Health Organization (WHO) classification for tumors of the central nervous system (CNS), the four major pediatric EPN entities are WHO grade II/III [28], although high ITH makes accurate grading challenging. The predominant groups in children are highly aggressive PFA and ST-RELA EPNs, whereas PFB and ST-YAP1 show an indolent behavior with a more favorable outcome [23]. Although molecular subgrouping shows better correlation with prognosis than histological grade alone, to date it has not informed treatment strategies [22], which have remained substantially unchanged over recent years and unsuccessful in approximately 40% of patients [21]. Very recently, these four EPN molecular variants have been incorporated into the fifth edition of the WHO Classification of Tumors of the CNS published in 2021 (WHO CNS5) [29], which integrates molecular diagnostics with more traditional histopathological approaches. According to WHO CNS5As, ST EPNs are now categorized into two types containing either ZFTA (formerly, C11orf95) fusions (because ZFTA may be fused with partners others than RELA), or YAP1 fusions.
Approximately 70% of childhood EPNs occur in the PF, whereas 20% occur in the supratentorium. PFAs arise in younger children (<5 years) and are characterized by dysregulation of numerous cancer-related networks, such as angiogenesis, receptor tyrosine kinase (RTK) signaling, and cell cycle (Table 1) [25]. Despite an aggressive behavior, PFAs display a balanced genetic profile and absence of highly recurrent oncogenic events. The most common copy number alterations are 1q gain [23] and 6q loss [23,30], both associated with unfavorable outcome. Instead, epigenetic modifications are hallmarks of PFAs, including CpG island methylator phenotype (CIMP) [31], DNA hypomethylation [32] and global reduction of the repressive histone H3 lysine 27 trimethylation (H3K27me3), with focal retention of H3K27me3 at CpG islands [32,33,34]. Recently, other alterations of the epigenetic landscape of PFA have been discovered, including infrequent histone H3K27M mutation (with a lysine 27-to-methionine exchange) [35], enhancer of zeste homologs inhibitory protein (EZHIP, formerly CXorf67) mutations, and overexpression of EZHIP in nearly all the cases [36] (see Section 4.2.1).
Table 1.
Summary of the main genetic/epigenetic alterations and clinicopathological characteristics of the major groups of pediatric intra-cranial EPN.
| Molecular Group | ST-RELA | ST-YAP1 | PFA | PFB | References |
|---|---|---|---|---|---|
| Location | ST, cerebral | ST, intra-periventricular | PF | PF | [25,40] |
| Age | children/adolescents median age 8 years |
young children median age 1.4 years |
young children median age 3 years |
all age groups median age 30 years |
[23] |
| Gender | [23] | ||||
| Male | 65% | 25% | 65% | 41% | |
| Female | 35% | 75% | 35% | 59% | |
| Molecular events | |||||
| Genetic | chromothripsis ZFTA-RELA fusions CDKN2a deletion loss of chromosome 9 |
YAP1-fusions | balanced genome 1q gain 6q loss infrequent H3K27M substitution infrequent EZHIP mutations |
chromosomal instability | [23] [23,24] [23,30] [35] [36] |
| Epigenetic | CIMP positive DNA hypomethylation H3K27me3 loss EZHIP overexpression |
CIMP negative H3K27me3 retention |
[31] [32] [32,34] [36] |
||
| Pathogenic impact | NF-κB pathway cell cycle cell migration MAPK pathway |
Hippo pathway | angiogenesis RTK pathways cell cycle cell migration derepression of PRC2 target genes |
ciliogenesis oxidative metabolism |
[24,39] [23,25] [31] |
| Outcome | poor | favorable | poor | favorable | [23] |
PFBs frequently occur in older children (5–18 years) and display many gains and losses of entire chromosomes [23], but dysregulation of a very restricted number of pathways controlling microtubule assembly and oxidative metabolism [25] (Table 1). DNA methylation profiling has evidenced significant inter-tumor heterogeneity in both PFAs and PFBs, distinguishing further subgroups with distinct patterns of genetic alterations and expression profiling, which suggests distinct histogenesis involving a cell of origin at different anatomic locations in the hindbrain [36,37].
More than two thirds of ST-EPNs harbor alternative ZFTA–RELA fusions [24] (Table 1), that lead to constitutively active NF-κB signaling, an established driver of solid tumors [38]. In addition, ST-RELA EPNs display other subgroup-specific genomic alterations, including frequent loss of chromosome 9 and homozygous INK4a-ARF (CDKN2a) deletions [23]. YAP1 fusions, the most common being YAP1-mastermind like domain containing 1 (MAMLD1), define the other clinically relevant subgroup of ST-EPN, rare tumors with relatively stable genomes, besides recurrent rearrangements involving YAP1 gene locus on chromosome 11 [23,39]. Compared to ST-RELA, YAP1-MAMLD1 tumors differ in demographic distribution (occurring mainly in children with a median age of 1.4 years vs. 8 years of RELA EPNs, and mostly restricted to female patients), anatomical location (intra-/periventricular in YAP1 vs. cerebral in RELA) and prognosis (favorable vs. unfavorable) [40].
There is increasing evidence of ST-EPNs with alternative gene fusions and ambiguous DNA methylation-based classification [41,42,43]. Pediatric supratentorial RELA fusion-negative EPNs show other fusion events, the majority involving ZFTA as a partner gene, such as ZFTA-mastermind like transcriptional coactivator 2 (MAML2) or ZFTA-nuclear receptor coactivator 1 (NCOA1). These tumors exhibit histopathological heterogeneity, no nuclear NF-κB expression, and epigenetic proximity to the RELA methylation class in some cases [44,45]. Recently, recurrent fusions involving the pleomorphic adenoma gene-like 1 (PLAGL1) gene have been discovered in a group of histopathologically diagnosed ST EPNs exhibiting a distinct DNA methylation profile [46], that occur mostly in children. To add complexity, rearrangements of ZFTA associated with RELA DNA methylation profiling have also been identified in EPN with an infratentorial location [47,48].
3. CSCs as a Source of ITH
3.1. The CSC Model
The CSC model was revived about two decades ago with the isolation of a subset of functionally distinct cells from hematologic and solid malignancies that uniquely drive tumor growth [49], and has rapidly emerged to explain the versatile features of tumor populations [50]. The isolation of clonogenic neural stem cells (NSCs) from human fetal brain tissue [51] corroborated the hypothesis that brain tumors may develop from transformed NSCs or progenitor cells [52].
CSCs are rare, relatively quiescent cells endowed with indefinite self-renewal, multilineage differentiation properties, and tumorigenicity, whereas transiently proliferating, more differentiated, non-tumorigenic non-CSCs form the bulk tumor [49,53]. Brain tumor SCs (BTSCs) are also functionally identified based on their ability to propagate serially in an undifferentiated state and to form floating clonally derived heterogeneous colonies called neurospheres (NSs) [54]. Central to the CSC paradigm is the identification of cancer-specific markers that allow unequivocally distinguishing of CSCs from non-CSCs. However, some controversies over universal stemness markers exist. For example, the cell surface glycoprotein CD133 has been proposed as a robust marker for BTSCs, although CD133-negative populations exhibit stemness functional features [55], and other proteins, such as nestin and SOX2, define the BTSC immunophenotypic profile [9], consistent with remarkable context-dependent inter-cellular heterogeneity among the BTSC population itself.
According to the classical paradigm, CSCs divide asymmetrically and give rise to daughter stem cells and non-stem progeny to drive unidirectional hierarchy-organized phenotypic differentiation that installs ITH (Figure 2a). More recently, accumulating evidence has revisited the traditional model into the plasticity model [12], whereby cancer cells possess the ability to bidirectionally transition between an SC and non-SC state. According to this model: (1) CSCs are not necessarily a small, slow-proliferating fraction of the bulk tumor, and (2) the functional features of both CSCs and non-CSCs are dynamically generated by combinatorial genetic and non-genetic factors. The observation that purified breast cancer [56] and glioblastoma (GBM) [9,57] stem and non-stem cells re-establish an equilibrium of mixed populations of all cellular states in cultures and in vivo is a proof of principle of the plasticity model.
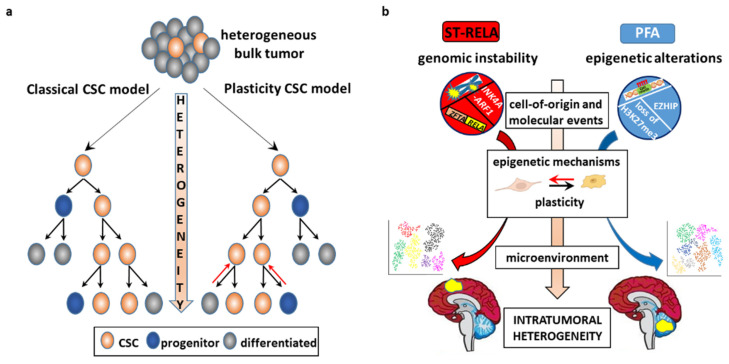
Figure 2.
Intra-tumoral heterogeneity (ITH) of EPN in the cancer stem cell (CSC) perspective. (a) Schematics of the classical (on the left) and plasticity (on the right) CSC model. The classical CSC model asserts that CSCs are a rare fraction of the bulk tumor, able to self-renew and to differentiate along multiple lineages, driving unidirectional hierarchy of heterogeneous cells. CSC plasticity model proposes that cancer cells can dynamically transition from a CSC state to a non-CSC state and vice versa. Combinatorial dynamics of genetic and epigenetic alterations and distinct microenvironmental contexts engender cell stemness and plasticity, installing ITH. (b) CSC model(s) in intra-cranial ependymomagenesis. Molecular events occurring in distinct cells of origin drive ependymomagenesis in the two compartments: genomic instability, leading to chromothripsis, gene fusions, and INK4a-ARF deletions, predominate in ST-RELA, whereas epigenetic factors (e.g., CIMP phenotype, global loss of H3K27me3, EZHIP overexpression) predominate in PFA.
Although cancers result from genetic and epigenetic events in interaction with the microenvironment, the cell(s) where these events occur are equally important determinants of tumorigenesis [58,59]. It is likely that CSCs derive from neoplastic transformation of healthy SCs, because pathways involved in the normal SC homeostasis are often hijacked and/or epigenetically altered in CSCs to bring about a “malignant reprogramming” that locks the cells into a state of self-renewal that persists beyond the timeframe of normal development (Figure 2b) [60]. Aberrant DNA hypermethylation in promoter regions displays a clonal pattern, being present in CSCs and their offspring [61], which suggests that epigenetic alterations play a causative role from very early tumorigenesis steps on and are then maintained. Alternatively, oncogenic lesions may be acquired by committed non-CSCs that undergo dedifferentiation to generate CSCs and initiate tumorigenesis [50,62].
Several lines of evidence have demonstrated that normal stem and progenitor cells are particularly susceptible to oncogenic transformation. Concurrent Ras overexpression and Tp53 inhibition transform NSCs and oligodendrocyte precursor cells (OPCs) that develop GBM-like tumors in the mouse brain, but fail to transform differentiated astrocytes [63]. Transgene expression of H3K27M is oncogenic in vivo when expressed in NSCs, but not always in OPCs [64,65,66]. By contrast, medulloblastomas with similar molecular features can be initiated by activation of Hedgehog signaling via genetic deletion of its receptor coding gene Ptch1 in NSCs or neuronal progenitors [67]. Hence, the transcriptional context in the cell(s) of origin governs susceptibility to transformation and acquisition of tumor-initiating capability in some cancers, while in some others, driver mutations, rather than the cell of origin, appear to dictate the tumor features [58] (see Section 3.2 and Section 5.2 for EPN cell of origin).
3.2. CSC-Driven Preclinical Models of EPN
The relevance of CSCs in ependymomagenesis was first highlighted by the isolation from EPN of rare populations of cells with features of radial glia cells (RGCs) [68,69,70], the progenitors that give rise to neurons, astrocytes, oligodendrocytes, and ependymal cells [71]. EPN SCs express the RGC markers CD133, nestin, RC2, and brain lipid binding protein (BLBP) and fulfill all benchmark functional assays for the characterization of CSCs. In differentiation media, EPN SCs show dramatic morphological and immunophenotypic changes towards glial, neuronal, and oligodendroglial lineages as well as reduction in tumor-propagating potential [70,72], thus recapitulating the ITH of the bulk tumor and positing RGCs at the root of EPN (see Section 5.2).
Cell lines have been established from EPN surgical samples by selection of the CSC component in NS-promoting conditions [72,73,74,75]. NS models mimic a 3D structure, which resembles the tumor microenvironment (TME) more faithfully than 2D cultures, preserving cell variability [76]. Compared to cell lines grown as monolayers, EPN 3D cultures transplanted in the mouse brain show better fidelity to the original tumor in terms of genetic, transcriptomic, and histopathological characteristics [74,75].
Drug treatment of patient-derived EPN cell lines results in preferential depletion of a stem-like cell population with a tumor-initiating property, as shown by a decrease in NSC markers, increase in differentiation-associated markers, and reduction in tumorigenicity in ex vivo transplantation assays [72,74]. Specific targeting of BLBP by PPAR antagonists lessens cell migration and invasion and promotes chemoresistance in vitro [77]. In comparison with EPN stem-like cells, differentiated cells are less sensitive to temozolomide because of differentiation-induced upregulation of MGMT [78], although others have also reported temozolomide resistance in undifferentiated EPN SCs [74].
High cellular variability within individual EPN cell lines has been reported. For instance, serial transplantation of EPN patient-derived lines in mice [74] or cultures in medium devoid of mitogens [79] select more tumorigenic cells. Moreover, mitogen-independent EPN cell lines display constitutive activation of EGFR, AKT, and STAT3 and sensitization to EGFR inhibitors in vitro and in vivo.
Clonal expansion of patient-derived cells by differential selective pressure exerted by culture conditions can help uncover genetic ITH in EPN. One mitogen-independent highly tumorigenic EPN line has been found to harbor protein-coding SEC61G–EGFR fusion genes, also found in one PFA out of 16 pediatric EPN cases by RT-PCR sequencing [79] and in glioblastoma (GBM) [80]. Similar to findings in EPN, ITH of GBM with a heterogeneous pattern of expression/amplification of RTKs is revealed by genotype selection under receptor-targeted ligand stimulation [81]. GBM SCs (GSCs) in EGF-free media retain EGFR amplification and EGFRvIII expression, which are usually lost in cells cultured in mitogen-enriched media [82]. Together, these data suggest that differential selection in vitro and in vivo may represent a complementary strategy to address ITH and its functional relevance in EPN.
4. Determinants of ITH
4.1. Genetic ITH
As “cancer is, in essence, a genetic disease” [83], the first recognized source of ITH is the inherent genomic instability of cancer cells that generates the progressive emergence of distinct genotypes upon which selection can act in a given microenvironmental context [84]. Based on functional activity, gene mutations are mainly distinguished as driver mutations and passenger mutations [85]. Driver mutations confer a selective advantage on cancer cells by the activation of oncogenic pathways and/or the inactivation of tumor suppressors, whereas passenger mutations are neutral. However, even usually silent mutations may become advantageous in the adaptive responses to certain selection pressures, such as resource deprivation, ligand stimulation, natural defenses, or chemo/radiotherapy [7,86].
Besides small-scale genetic changes, genomic instability includes large-scale genomic events [87], such as chromosomal instability [88], aneuploidy, chromothripsis [89], and extrachromosomal DNA (ecDNA) [90], that involve an ample number of genes and provide massive copy-number amplification. Large-scale DNA alterations are quite unstable through mitoses and can undergo strong selective pressures, accelerating tumor evolution [19].
ecDNAs exist in almost 50% of tumors [91], and are circular-shaped elements of DNA with high chromatin accessibility [92] that contain amplified oncogenes and drug resistance genes [93]. As they lack centromeres, ecDNAs segregate randomly among daughter cells and confer massive intercellular genetic heterogeneity and improved fitness, that might result in tumor aggressiveness and chemoresistance [94,95]. Loss of ecDNA carrying EGFRvIII induces resistance to EGFRvIII inhibitors in GBM models and patients [96]. Different evolution of ecDNA at diagnosis and relapse has been reported across multiple cancers, including pediatric high-grade gliomas (pHGGs) [97]. However, the contribution of ecDNA to ITH of EPN has not been addressed yet.
Chromothripsis is a single cellular catastrophic event in which hundreds of genomic rearrangements take place at once in one or a few chromosomes [89]. Generally considered as an early mutational phenomenon occurring in a minority of neoplasms, sequencing-based analyses at high coverage depth have demonstrated that chromothripsis is pervasive in cancers, reaching a frequency of more than 50% in some entities [98]. Moreover, longitudinal analysis in paired primary and relapsed tumors has shown that chromothripsis may occur only in the primary tumor, only at relapse, or, conversely, in both events in the same patient, which suggests subclonal heterogeneity and evolution occurring through all steps of tumor progression [99]. A causative role for chromothripsis has been inferred in ST-RELA EPNs, where ZFTA–RELA fusions result from a shattering event on chromosome 11, that juxtaposes ZFTA to the NF-κB master transcription factor RELA [24] (see Section 4.1.1). Remarkably, among the nine EPN molecular subgroups, chromothripsis is detected exclusively in ST-RELA, where it more frequently involves chromosome 11 [23].
4.1.1. CSCs and Genomic Instability: ST-Ependymomagenesis
Compelling evidence for a causal role of genetic alterations in CSC-driven EPN oncogenesis comes from studies in embryonic NSCs transduced with relevant EPN driver mutations, that demonstrate that regional distinct NSCs uniquely susceptible to specific mutations foster ependymomagenesis in the different anatomical compartments [100,101]. A cross-species study of human EPN and mouse NSCs isolated from different regions of embryonic and postnatal CNS with a wild-type or Ink4a/Arf-null genetic background has shown that the transcriptomes of human ST-EPNs with amplified EPHB2 and deleted INK4a/ARF match only that of embryonic cerebral Ink4a/Arf-null NSCs [100], and neither the hindbrain nor spine. Corroboratively, EPHB2 drives ependymal-like tumors only if expressed in embryonic cerebral Ink4a/Arf-null NSCs, but in neither adult NSCs nor embryonic NSCs from other CNS regions, or in the absence of Ink4a/Arf−/− deletions.
ZFTA–RELA fusions have been identified as the first pathogenic genes in ST-EPN, because they are sufficient to induce EPN-like tumors in allograft models of forebrain-derived murine Ink4a/Arf-null NSCs, whereas neither wild-type translocation partner alone does [24]. Corroboratively, de novo ependymomagenesis has been reported in neonatal mouse brain after RELA fusion transfer to NES-expressing cells by the RCAS/tv-a system [102] or lentivirus injection [103]. Tumors develop in all models with morphological, immunophenotypic, and transcriptomic features that echo those of human ST-RELA EPNs.
Unlike wild-type RELA, RELA fusion oncoproteins are constitutively localized in the nucleus, where they drive aberrant activation of NF-κB target gene transcription in vitro and in vivo [24]. Recent key studies integrating epigenomic and transcriptomic mapping have demonstrated that a number of non-canonical NF-κB transcriptional programs, such as Notch, MAPK, and focal adhesion networks, are critical actors of ST-RELA formation, in addition to the canonical NF-κB signaling [104,105,106]. Contrary to the initial hypothesis positing that the RELA partner drives the transcriptional activity of RELA fusion proteins, recent chromatin interaction-based analyses support the idea that it is the ZFTA moiety that shuttles the RELA component to the nucleus and dictates the RELA fusion binding affinity across the genome, so as to orchestrate the transcription of ependymoma-associated genes in collaboration with RELA targets [105,106,107,108].
YAP1 fusions involve the first exons of the YAP1 gene fused in frame with the 3′ coding portion of other translocation partner genes, most frequently the mastermind like domain containing 1 gene (MAMLD1), and seldom the family with sequence similarity 118 member B gene (FAM118B) [23,24]. YAP1 functions as a transcriptional cofactor of the Hippo signaling pathway, a tumor suppressor pathway that controls organ size and tumorigenesis by sequestering YAP1 in the cytosol [109], whereas YAP1-MAMLD1 accumulates predominantly in the nucleus in ST-YAP1 tumors [39]. The oncogenicity of YAP1-MAMLD1 and YAP1-FAM118B has been demonstrated via gene transfer of the full-length fusions or wild-type fusion partners to target RGCs/NSCs in the embryonal cerebral ventricular zone (VZ) using in utero electroporation [39], as well as the RCAS/tv-a system [110] or lentivirus injections [103] in mouse neonatal brains. The exogenous expression of the fusions alone drives the formation of murine tumors similar to human EPNs, identifying cells positive for the RGC marker PAX6 as the cell of origin of YAP1-MAMLD1 EPN [39]. Interestingly, ST-YAP1 tumors display the highest PAX6 expression of all EPN subgroups [111]. In the postnatal mouse brain, constitutive expression of YAP1-MAMLD1 impairs neural differentiation and migration of ventricular neural precursor cells that are forced instead into active proliferation. The MAMLD1 domain is necessary for translocation of the fusion in the nucleus and for the interaction with NFI transcription factors (TFs), that in turn recruit the fusion protein to enhancer regions enriched in TEAD and NFI-binding motifs to drive the transforming gene expression of YAP1-MAMLD1 EPN [39,110].
Overall, these studies demonstrate that mutually exclusive transforming fusions are likely the key event in ependymomagenesis of the ST compartment via a similar mechanism, whereby the fusion products accumulate constitutively in the nucleus of topographically restricted NSCs to disrupt developmental gene expression programs, ultimately leading to oncogenesis.
4.2. Epigenetic ITH
Variable phenotypes of cancer cells can also be mediated by epigenetic, transcriptional, and microenvironmental changes without concomitant genetic mutations. Non-genetic ITH is far more dynamic than genetic heterogeneity and is therefore increasingly recognized as a driving force of tumor evolution [112,113].
The term “epigenetic” describes the covalent modifications of DNA and histones that affect gene expression without intrinsic changes in the DNA sequence through modulation of the chromatin structure [112]. Epigenetic changes are inherited by offspring cells just like genetic alterations and provide an additional pool of selectable traits. An interplay between genetic and epigenetic alterations occurs in virtually all tumor types, where epigenetic lesions may precede or arise simultaneously with genetic mutations, or conversely be a consequential event [10,114]. PBTs display an overall low mutational burden, but there are a number of epigenetic dysregulations [115,116,117] that can drive tumorigenesis even in the absence of highly recurrent driver mutations, CIMP-positive PFA being a prominent example. Most of the few recurrent mutations of PBTs target epigenetic regulatory genes, such as H3.3A, ATRX, and enhancer of zeste homolog 2 (EZH2) [118]. For example, a hallmark of pHGG is H3K27M [119], that is associated with neuroanatomical specificity, DNA methylation pattern, and age distribution [120].
Epigenome regulation of intercellular heterogeneous gene expression is a dynamic condition between transcriptionally active and repressive chromatin states by virtue of cell-to-cell variation in DNA methylation at enhancers and promoters, covalent histone modifications [33], nucleosome positioning [121], and chromatin accessibility [122]. Prominent alterations of DNA methylation in cancers, including high-risk PFAs, are focal gains at normally unmethylated CpG islands and promoter regions, that heritably silence hundreds of genes that counteract tumor development, outnumbering gene mutations [123,124]. Posttranslational covalent histone modifications include methylation or acetylation at histone tails, such as H3K27me3 and H3K27ac, markers of repressed and active transcription, respectively [114]. H3K27 trimethylation is mediated by the Polycomb repressive complex 2 (PRC2) via the methyltransferase activity of the PRC2 catalytic subunit EZH2 [125].
Epigenetic ITH has primarily been assessed focusing on DNA methylation, because of its stability and mitotic heritability, and is found in regulatory regions that control the transcription of associated genes, contributing to gene expression heterogeneity relevant to cell identity and disease processes [123,126,127,128]. High epigenetic heterogeneity at enhancers has been reported in ESCs [129] and during progression from normal tissues to primary tumors and to metastases with a cancer-specific pattern [130], which indicates that enhancer DNA methylation may be primed to respond to microenvironmental cues and to increase cancer cell plasticity. In temporally distinct tumor specimens, DNA methylation levels are reported to be increased, equal, or decreased in primary vs. relapsed tumors [131], maybe because of variable epigenetic clonal dynamics in different cancers. Compared to primary EPNs, relapsed EPNs display neither significant differences in DNA methylation profiles nor in H3K27me3 levels, whereas major changes occur at CpG islands that show higher methylations in relapsed ST-RELA and PFA EPNs [132].
Spatiotemporal epigenetic heterogeneity in distinct areas of the same tumor has been described in a wide range of cancers and allows for building the evolutionary history of the tumor alongside genetic heterogeneity. Comparison between phylogenetic and epigenetic trees has usually shown similar and integrated patterns, which suggests a codependency of genetic and epigenetic mechanisms in tumor progression [131,133]. In primary low-grade gliomas and matched recurrent HGG, cell cycle genes are epigenetically upregulated through promoter hypomethylation during tumor progression, in parallel with genetic mutations that affect cell cycle checkpoints [134]. Multiplatform molecular profiling of spatially distinct meningioma shows regional alterations in chromosome structure that underpin clonal transcriptomic, epigenomic, and histopathologic signatures [135]. DNA methylation and RNA sequencing of six topographically distinct samples from one ST-RELA tumor reveal significant transcriptional and epigenetic heterogeneity [136]. Remarkably, the expression of the subgroup-specific markers L1CAM, CCND1, ZFTA, and RELA is similar across the sections, whereas DNA methylation-based and gene expression variability define three geographically distinct clusters enriched in stem-like, neuronal differentiation, and mature microglia signatures that recapitulate brain development.
4.2.1. CSCs and Epigenetic Alterations: PFA Ependymomagenesis
A link between aberrant epigenome and pediatric hindbrain tumorigenesis has increasingly been recognized [32,137]. Although the underlying mechanisms are different, they all converge on PRC2 function and targets during narrow developmental windows [138]. ESCs rely on PRC2 to reversibly silence genes required for differentiation and there is evidence that PRC2 targets similar sets of CpG-containing genes in ESCs and in cancer cells [139]. The PRC2 component EZH2 is essential for GSC maintenance and its pharmacological or molecular inhibition impairs GSC-driven tumor growth [140]. In pHGG, H3K27M competitively binds to and dominantly suppresses EZH2 function, that results in reduced H3K27me3 and spurious activation of earlier developmental programs in NSCs that are crucial for oncogenesis [141]. Some of these genes, such as Pbx3, Eya1, and Plag1, are regulated by bivalent promoters with both permissive and repressive histone marks that are poised for activation in stem and progenitor cell types [142]. Transgene expression of H3K27M in human ESC-derived neural progenitor cells [64] or human induced pluripotent SCs [66] synergizes with TP53 knockout and constitutive PDGFR activation to enhance self-renewal and drives in vivo and de novo gliomagenesis. Concordantly, removal of H3K27M in pHGG-derived cell lines using CRISPR-Cas9 restores full differentiation capabilities along the glial lineage [143].
In PFA EPN, CpG hypermethylation is enriched at the promoter regions of genes important for neurodevelopment that are silenced by PRC2-mediated trimethylation of H3K27 in ESCs [144], such as differentiation-associated genes, thereby locking cells in a perpetual proliferative state. Corroboratively, treatment of PFA CIMP-positive cultures with demethylating agents results in derepression of the PRC2 target genes in ESCs, and impaired proliferation of PFA cells in vitro and in vivo [31].
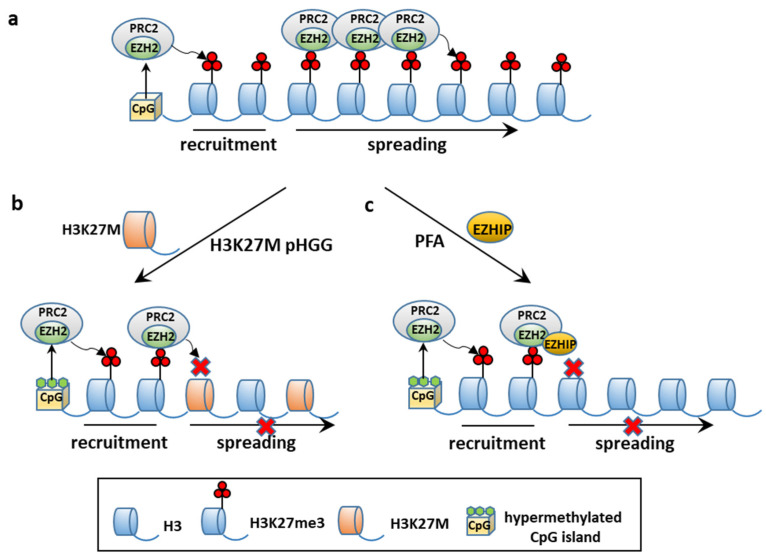
Like H3K27M mutant pHGG, a hallmark of the PFA epigenome is global reduction in H3K27me3. However, the mechanism(s) underlying H3K27 hypomethylation in PFAs that harbor infrequent H3K27M mutations have been elusive. Recently, several publications have demonstrated that EZHIP contains a short “K27M-like” sequence that inhibits EZH2, causing reduction in H3K27me3 and an overly permissive chromatin state [145,146,147]. Transgenic cell lines with expression of either H3K27M or EZHIP exhibit comparable genome-wide loss of H3K27me3 with focal gains at CpG islands and gene expression profiles that reflect dysregulation of PRC2-mediated gene repression. Consistently, ablation of EZHIP in cell lines by a CRISPR-Cas9 strategy results in increased H3K27me3 levels [36]. However, in vivo models of EZHIP-driven EPN tumorigenesis are still lacking, suggesting that other hit(s) are required [145].
Consistently with the similarities between EZHIP- and H3K27M-mediated mechanism, EZHIP overexpression shows a non-overlapping pattern with H3K27M in PFAs [36,148]. Remarkably, EZHIP missense mutations found in a small proportion of PFAs do not impair EZHIP-mediated inhibition of PRC2 activity [146]. No loss-of-function mutations of PRC2 have been found in PFA EPN and H3K27M mutant pHGG, which suggests that residual PRC2 activity is required for the development of these tumors. Cell-based and molecular assays in HGG models have shown that H3K27M and EZHIP impair the production and spread of H3K27me3 from PRC2 high-affinity sites, while sparing residual H3K27me3 at CpG sites (Figure 3) [146,149,150]. Corroboratively, removal of H3K27M restores H3K27me3 propagation associated with inhibition of cell proliferation and tumorigenicity. Analog mechanisms may occur in PFA, as hinted by the sensitiveness of PFA lines to EZH2 inhibitors [31,151].
Figure 3.
Schematics depicting mechanisms which mediate global loss of H3K27me3 in H3K27M pediatric high-grade glioma (pHGG) and PFA. (a) In normal cells, PRC2 is recruited to CpG islands and catalyzes H3K27 trimethylation and spreading of H3K27me3; (b) in H3K27M mutant pHGG, PRC2 is recruited at hypermethylated CpG islands, but H3K27M prevents spreading of H3K27Me3; (c) in PFA with EZHIP overexpression, PRC2 is recruited to hypermethylated CpG islands, but EZHIP competitively binds to and inhibits EZH2-mediated trimethylation of H3K27 and spreading. Both mechanisms in (b,c) determine loss of PRC2-mediated gene repression and transcription of normally silenced genes (Reprinted with permission from Siddhant U. Jain, (2020), Elsevier and Copyright Clearance Center, Licence Number 5200711451432, 2 December 2021).
The chromatin profile and gene signature of PFAs converge on genes involved in neurodevelopmental pathways and RGC functions [32,146]. Interestingly, during human PF neurogenesis, H3K27Me3 is reduced in RGCs in prenatal phases, while increasing postnatally [32], which indicates that dynamic gains and losses of H3K27me3 are necessary for normal neural differentiation and development. Reduced H3K27me3 in PFA tumors and in PF RGCs in early neurogenesis is consistent with RGCs as PFA presumptive cells of origin.
4.3. TME, CSCs, and EPN
The epigenome stands at the intersection of the genome and TME. Unlike genetic alterations, epigenetic modifications are reversible and less consistently transmitted through mitosis, and therefore play a major role in opportunistic adaptation to spatiotemporal fluctuations of the TME [3,152]. Whereas in healthy tissues the environment acts as the main barrier to counteract cancer initiation, in tumor tissues neoplastic cells subvert this organized architecture into a deranged tumor-sustaining milieu. These changes include matrix remodeling, development of tumor vasculature networks, recruitment of stromal and immune cells, and interactions between tumor and normal cells as well as between functionally different tumor subpopulations [17]. The complex tumor architecture creates topographical constraints, changeable blood flow [153], and heterogeneous microenvironmental conditions with a combinatorial dynamic of contextual cues that trigger a variety of signaling pathways and regulatory networks [13].
This paradigmatically occurs at the tumor core and tumor/host interface. Although region-specific driver mutations have been documented [154], contextual factors are equally important in shaping the zonal pattern, with high proliferation, signaling activities and invasion-promoting properties almost exclusively restricted to the leading edge of the tumor as opposed to a quiescent, apoptotic, and therapy-resistant phenotype predominating in the center. These distinct intrinsic signatures and phenotypes are driven by hypoxic [155] and/or acidic microenvironmental gradients [156,157] and paracrine cross-talk [158] between the distinct tumor populations.
Microenvironmental variability promotes commonly observed phenotypic cellular properties, such as stemness and epithelial-to-mesenchymal transition (EMT). There is an intricate interaction between CSCs and their microenvironment. CSCs are actively engaged in shaping their own supportive niche, but are in turn regulated by exogenous signals that affect their epigenome and cellular state [9,124] shaping tumor heterogeneity and evolution. Examples of the interconnections between EPN and TME are given below (Figure 4).
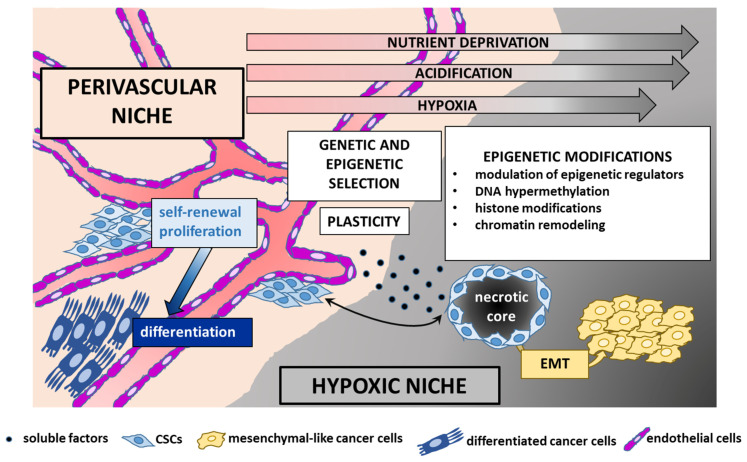
Figure 4.
Schematic of the main factors that contribute to intra-tumoral heterogeneity in ependymoma. Genetic alterations and epigenetic modifications are selected in distinct tumor microenvironments, such as the perivascular and the hypoxic niche, that create gradients of oxygen, nutrients, and acidification. Changing contextual cues affect epigenetic regulators and remodel the chromatin landscape that mediates dynamic cellular plasticity. CSCs are able to adapt bidirectionally to both normoxic and hypoxic niches, fostering tumor growth and progression. Cross-talk among the distinct tumor and normal cell populations (such as endothelial cells) mediated by soluble factors or extracellular vesicles contributes to intra-tumoral cell-to-cell diversity. Cancer cells of the same phenotype tend to cluster together in the same topographical location. Differentiated cell concentrate in perivascular niches, whereas CSCs and mesenchymal-like cells coexist in hypoxic microenvironments, suggesting cooperative interactions and interconversion between these cell states (epithelial-to-mesenchymal transition, EMT).
4.3.1. The Perivascular TME
One of the key structural changes of the host tissue that cancer cells bring about to create growth promoting niches is the organization of the vascular network [3]. Variable blood flow selects for highly plastic phenotypes, whereby cells can shift from dormancy to rapid proliferation and vice versa, migrate to escape harsh contextual conditions, adapt to low oxygen concentrations, and use a wide arrays of nutrients [153,159].
In perivascular environments rich in nutrients and oxygen, cancer cells rapidly divide and foster tumor growth, whereas hypoxic contexts sustain slow-cycling stem-like cells able to drive tumor progression and recurrence due to their intrinsic plasticity and epigenetic adaptation to harsher conditions. The molecular response to hypoxia is mainly mediated by the hypoxia-inducible factor (HIF) family of TFs, especially HIF1α, that is also involved in the maintenance of stemness features and induction of angiogenesis [160]. In low oxygen concentrations, CSCs secrete a number of angiogenetic factors, such as VEGF and PDGF [161] that in turn recruit endothelial cells to shape new blood vessels, followed by CSC epigenetic adaptation to the newly formed, normoxic niche.
EPNs display remarkable spatial heterogeneity, that reflects functional adaptation to different contextual conditions. Preoperative imaging of one ST-RELA tumor showed that regions in the tumor core are associated with low blood flow as well as enrichment in hypoxia-related genes and immune-related genes characteristic of microglia, which promote an inflammatory microenvironment [136], similar to that observed in PFAs [162]. In contrast, stem-like regions are associated with high blood flow and enrichment in the histone deacetylase gene HDAC9, hinting towards chromatin modification in the cells of the perivascular areas. This is in agreement with the critical role that the perivascular niche plays for EPN CSCs, because soluble factors released from endothelial cells maintain self-renewal and proliferation of EPN SCs, while counteracting their differentiation [163].
Immunophenotyping analyses with molecular markers specific for neurodevelopmental cell types have provided the histological spatial distribution of distinct EPN subpopulations and their possible reciprocal interactions. In general, differentiated ependymal-like cells and immature stem-like cells exhibit mutually exclusive localization with a patterning in clusters of cells of the same phenotype [164,165]. Immature cells tend to concentrate in perivascular or perinecrotic zones, often colocalized with mesenchymal-like cells, suggesting a potential supportive cross-talk between both cell types. In vitro, hypoxic conditions trigger the switch of EPN cells to a mesenchymal phenotype, with upregulation of stress-related programs, including angiogenesis [164]. Hence, it is tempting to speculate that mesenchymal-mediated vasculogenesis likely occurring in vivo may sustain a perivascular niche that supports EPN neoplastic populations.
4.3.2. The Hypoxic TME
Besides perivascular niches, the brain TME includes hypoxic areas [166], which result from inadequate vasculature and/or rapidly dividing cells that outstrip the local supply of oxygen and nutrients living behind many dying cells. Hypoxic areas are often acidic [167], although acidosis can also occur independently from hypoxia [168].
Studies in NSCs and embryonic neurodevelopment have supported the notion that the neural niche is relatively hypoxic, because the partial pressure of oxygen near the ependymal surface—where NSCs reside—is low [169], and hypoxic conditions in vitro maintain self-renewal and an undifferentiated state of NSCs [170]. However, perivascular [163] and hypoxic [160] areas have been reported to promote expansion and tumor-initiating properties of CSCs. These contrasting data may be reconciled by the intrinsic plasticity of CSCs, as shown in GSCs able to reversibly adapt to hypoxic and normoxic conditions [9]. Similarly, nutrient restriction enriches for GSCs able to adapt to low glucose supply by upregulating the high-affinity neuronal glucose transporter Glut3, used by cells with both a high glucose demand and a glucose-poor microenvironment, such that occurring in perinecrotic areas [171].
Hypoxia has recently been recognized as an oncogenic driver of PFA by reshaping its metabolic and epigenetic landscape [172]. Low oxygen concentration promotes optimal growth of PFA cells and induces glucose dependency and upregulation of glycolytic and hypoxia-related programs, that are prominent in PFA tumors with respect to normal brain and other EPN groups [23,151]. Hypoxia-induced epigenetic reprogramming is initiated by restrictions of metabolic intermediary products, that impact on modifications at H3K27, i.e., diminished methylation, and increased demethylation and acetylation. Mechanistically, hypoxia increases EZHIP expression, which blocks PRC2-mediated H3K27 trimethylation. Concurrently, hypoxia maintains H3K27 hyperacetylation and hypomethylation via an epigenetic mechanism that involves high levels of acetyl-CoA and metabolite-mediated activation of H3K27 histone demethylases KDM6A and KDM6B. Interestingly, a non-overlapping histological staining between the hypoxia-related marker carbonic anhydrase 9 [CA9] and H3K27me3 is observed in PFA, which underlines a link between hypoxia, metabolism, and epigenetics. Comparison with single-cell transcriptomics and metabolomics of the developing murine brain has shown that the hypoxic and glycolytic programs of PFAs mirror the signatures of gliogenic progenitors in the hindbrain that reside in low oxygen concentrations, hinting at gliogenic progenitors as the founder population of PFA [173].
Similar to findings in EPN, stress-related alterations of the homeostatic balance of chromatin via direct modulation of epigenetic regulators are observed in other cancers. In GSCs, RTK inhibition induces upregulation of histone demethylases KDM6A/B, that results in widespread redistribution of repressive H3K27me3 and chromatin remodeling and promotes drug tolerance and adaptive transition of cells to a slow-cycling state [174]. Permissive histone acetylation is under the control of heterogeneous contextual cues, such as intracellular acidification [175,176] and hypoxia [177]. In breast and lung adenocarcinoma, inhibition of the activity of oxygen-dependent TET demethylases results in DNA hypermethylation at promoters of tumor suppressor genes, associated with maladaptive oncogenic processes involved in the cell cycle, apoptosis, metastasis, and angiogenesis [178]. High TET1 levels correlating with Tet-dependent activity are observed in medulloblastoma and EPN cell lines, suggesting an involvement of TET1 in the pathogenesis of these hindbrain tumors [179].
4.3.3. EMT
EMT is a highly dynamic multistep program, whereby non-motile neoplastic epithelial cells, in response to pleiotropic extrinsic signaling factors, acquire mesenchymal characteristics accompanied by loss of epithelial cell–cell junctions and acquisition of migration and invasion properties [180]. The reverse of the process—mesenchymal–epithelial transition (MET)—is associated with the reacquisition of junctional complexes and loss of migratory capacity. EMT is executed by specific TFs and epigenetic regulators [181], some of which are involved in embryogenesis, suggesting a link between cellular plasticity in embryonic development, stemness, and cancer progression [182]. The bidirectional transition between CSC and non-CSC states is functionally related to MET and EMT programs, respectively, whereby CSCs may differentiate into non-CSCs by the activation of the MET programs, and non-CSCs may undergo dedifferentiation and acquire CSC-like features and tumor-initiating potential through EMT changes [183,184].
Recent findings describe an oncogenic role for EMT in ependymomagenesis [185]. A hallmark of EMT in epithelial cells is the concurrent transcriptional repression of E-cadherin and upregulation of N-cadherin, the so-called cadherin switch [186]. Cadherin switching and upregulation of the EMT-TFs SNAI1/Snail, SNAI2/Slug, and ZEB1 is observed in PFA, PFB, and ST-RELA tumors, correlating with a shorter progression-free survival [187]. Remarkably, Wani et al. first described a mesenchymal phenotype with expression of angiogenesis, migration, and adhesion gene ontologies in a subset of infratentorial EPNs similar to transcriptomal PFA and associated with a short recurrence-free survival [188], as observed in other cancers, where EMT promotes tumor progression and metastasis and is associated with a poor outcome [182].
5. ITH of EPN: A Single-Cell Perspective
To date, the molecular characteristics of PBTs have mostly been informed by large-scale “omic” analyses of bulk tumors. Recently, scRNA-seq strategies have overcome these technical barriers, providing resolution of the variation of bulk samples at the individual cell scale [189]. Analysis of scRNA-seq datasets from tumor samples is performed through the following main steps: (1) distinction of neoplastic from non-neoplastic cells; (2) clustering of transcriptional profiles to identify distinct tumor subpopulations; (3) comparison of the identified signatures (a) across patients, to discover common transcriptional programs relevant to the disease, and (b) with external datasets, including bulk tumor datasets and human and mouse reference atlas datasets of developing and adult brain cell types, to clarify their biological meaning [26].
Seminal studies leveraging scRNA-seq have begun to elucidate how distinct transcriptional signatures promote malignant transformation and also delineate the developmental trajectories across diverse types of PBTs, including pHGG [65,143,173,190] and EPN [164,165,173].
5.1. EPN Is Composed of Multiple Discrete Neoplastic Subpopulations
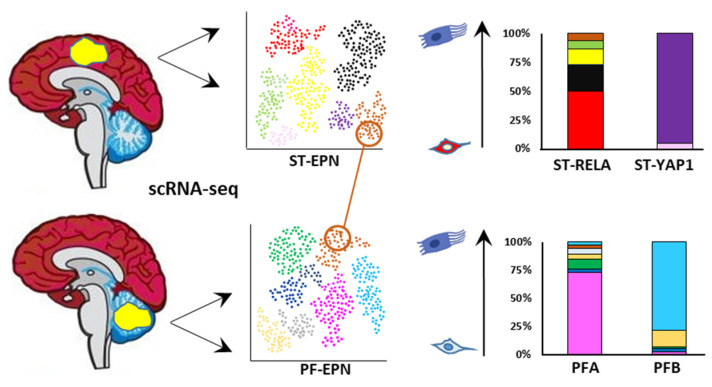
The four major childhood EPN subgroups dissected by scRNA-seq appear to be a composite mixture of multiple phenotypically discrete neoplastic subpopulations with divergent transcriptomic profiles. Although transcriptional signatures and their number differ across EPN subgroups, the common patterns of ITH that have been observed are mostly associated with cell cycle and neurodevelopmental programs. Contrasting the current classification paradigms, these studies have demonstrated that the relative proportions of the individual cell types dictate the molecular subgroup assignment, aggressiveness, and potential biomarkers of individual tumors, as reported in other brain tumors [191]. A high degree of ITH and enrichment for undifferentiated cell populations are associated with lower age and an unfavorable clinical outcome, as observed in ST-RELA and PFA, which might explain the profound difference in prognosis between these subtypes and their respective anatomical counterparts ST-YAP1 and PFB (Figure 5). Another commonality between ST-RELA and PFA is that cycling cells are specifically enriched in undifferentiated subpopulations, implying that progenitor subpopulations are more proliferative than more differentiated ones. Although, overall, cells separate according to the bulk tumor subgrouping, partially shared transcriptional programs are observed across all EPN molecular variants [164,165]. For instance, programs related to cell cycle, stress response, and ependymal differentiation are similar in ST-EPN and PF-EPN.
Figure 5.
Intra-tumoral heterogeneity in intracranial ependymoma by scRNA-seq. scRNA-seq of tumor cells is colored based on distinct gene signatures that define cell subpopulations. The relative frequency of each subpopulation is different in PFA tumors that are enriched for undifferentiated cells vs. PFB tumors enriched for ependymal-like cells. ST-RELA tumors also harbor high fractions of progenitor cell subpopulations, whereas a distinct ST-YAP1 gene signature is overrepresented in ST-YAP1. Some overlap between transcriptional signatures is observed across EPN groups.
5.1.1. PF-EPN and scRNA-seq
The more mature cell types of PF-EPN recapitulate the functional states of normal ependyma and express markers of ciliogenesis and cellular transport. Based on their function, these clusters have been named ciliated EPN cells (CECs) and transportive EPN cells (TECs) [164], and they share transcriptional commonalities with the clusters termed PF-Ependymal-like and PF-Astroependymal, respectively, in Gojo et al.’s study based on their neurodevelopmental phenotypes [165]. Both CECs and PF-Ependymal-like cells express cilia-related genes (e.g., DNAAF1 and RSPH1) and TF networks, including RFX2 [192] and FOXJ1 [193], that play a critical role in normal ependymal development. Likewise, TEC and PF-Astroependymal display expression of the AQP4 gene and of markers of the astrocytic differentiation lineage. AQP4 is a membrane water channel found in astrocytes and ependymal cells [194]. Increased AQP4 expression at mRNA and protein levels is observed in PF-EPN, but not ST-EPN [195,196], possibly identifying AQP4 as a compartment-specific marker of intra-cranial EPN. Other transcriptomes, referred to as mesenchymal EPN cells (MECs) [164] or PF metabolic [165], are defined by mesenchymal markers and contain stress response genes related to angiogenesis, hypoxia, and glycolysis, indicating that EMT may occur in these cells.
Undifferentiated PF-EPN cell subpopulations are distinguished by stemness transcriptional programs and markers of RGCs or early neural lineage commitment [70,100]. These clusters have been named undifferentiated ependymoma cells-1 (UEC-1) and cells-2 (UEC-2) in Gillen et al.’s study [164], and PF-Neural-Stem-Cell-like cells (PF-NSC-like), PF-Glial-Progenitor-like cells, and PF-Neuronal-Precursor-like cells [165] in Gojo et al.’s study. UEC-1 and PF-NSC-like show a broadly immature cell type and are driven by TF regulatory circuits that include JUN and FOS, which to date has never been associated with EPN tumorigenesis. Immature subpopulations in PF-EPN have also been described by Vladoiu et al. [173], who, however, failed to identify differentiated cell types, maybe due to the absence of differentiated cells in the mouse cerebellar developmental lineage comparators used.
5.1.2. ST-EPN and scRNA-seq
ST-EPN is also composed of multiple neoplastic subpopulations, such as cycling cells, differentiated ST-Ependymal-like cells with a ciliogenesis-related phenotype, and less differentiated clusters that map to RGCs (ST-Radial-Glia-like) or neuronal precursors (ST-Neuronal-Precursor-like) [164,165]. Parallel to MEC and PF metabolic signatures is a cluster of cells expressing genes involved in hypoxia and glycolysis, and is named, accordingly, ST-metabolic, whereas an ST-YAP transcriptomic signature associated with more quiescent, differentiated phenotypes is highly predominant in ST-EPN harboring this fusion.
Overall, the transcriptional programs in ST-RELA subpopulations are under the control of unfused RELA activity more than ZFTA-RELA, indicating that the transcriptional programs underlying the phenotypic diversification of the ST-RELA subpopulations are independent of ZFTA-RELA activity. Of interest, whereas Neuronal-Precursor-like population in PF-EPN and ST-EPN share the NEUROG1 regulatory circuitry, which is involved in neurogenesis [197], ependymal-like populations in the two compartments show selective TF networks. FOXJ1 target genes that mediate ciliogenesis [198] are enriched in the ST-YAP cluster, consistent with a cilia-associated program hallmark of this tumor. Corroboratively, the FOXJ1 TF network also characterizes differentiated CEC and ependymal-like subpopulations in the PF compartment, specifically in PFBs, that all express a strong ciliogenesis signature [25,199].
5.2. The Cell of Origin and Developmental Trajectories of EPN from an scRNA-seq Perspective
All cells of the nervous system originate from a common ancestor, the NSCs lining the neural tube, the primitive developmental tissue that gives rise to the CNS [71]. In the earlier stages of normal brain development, RGCs originate from and replace NSCs at the onset of neurogenesis [200]. Unlike NSCs, the majority of RGCs are regionally and fate restricted, giving rise to a single cell type—astrocytes, oligodendrocytes, neurons, or ependymal cells—based on both extrinsic signals and cell-intrinsic factors [201]. RGCs reside in two distinct niches of the developing cortex, the ventricular zone (VZ) and the outer subventricular zone (oSVZ), and are distinguished in ventricular RGCs (vRGCs) and outer RGCs (oRGCs) by a combination of position, gene expression, cell morphology, and function [202,203].
EPNs arising in the different compartments exhibit a gene signature that recapitulates that of RGCs in the corresponding CNS region [100,188]. For instance, the human ST-EPN signature matches that of RGCs in the murine SVZ of the lateral ventricles during neurogenesis when RGCs in this region of the brain give rise to ependymal cells. Likewise, the molecular profile of spinal EPN overlaps that of RGCs in the SVZ of the spinal canal [70].
scRNA-seq and trajectory inference (TI) analyses [204,205] have shed new light on the identity of the candidate SCs and the developmental trajectories of EPN variants at different anatomical sites. Comparison of scRNA-seq datasets of EPN subpopulations to those of defined cellular lineages in the developing human and murine brain has indicated a potential cell of origin residing in the VZ for PF-EPN [164,165,173], whereas an RG-like cell residing in the SVZ is at the root of ST-RELA [70,165]. The PF-EPN and ST-EPN progenitor cells share minimal transcriptomic overlap [164], supporting that regionally and developmentally restricted populations of RGCs are the candidate cells of origin of EPNs. Across scRNA-seq studies, the putative PF-EPN cell of origin is identified in progenitor-like cells at distinct stages of neurodevelopment, and indicated as NSC-like cells [165], vRG-like cells (the predominant signature in undifferentiated UEC-1) [164], or gliogenic progenitors [173]. However, these presumptive PF founder cells share some commonalities because both NSCs and vRGCs are located in the VZ of the embryonic brain, and UEC-1 show enrichment in both gliogenic progenitor and vRGC genes.
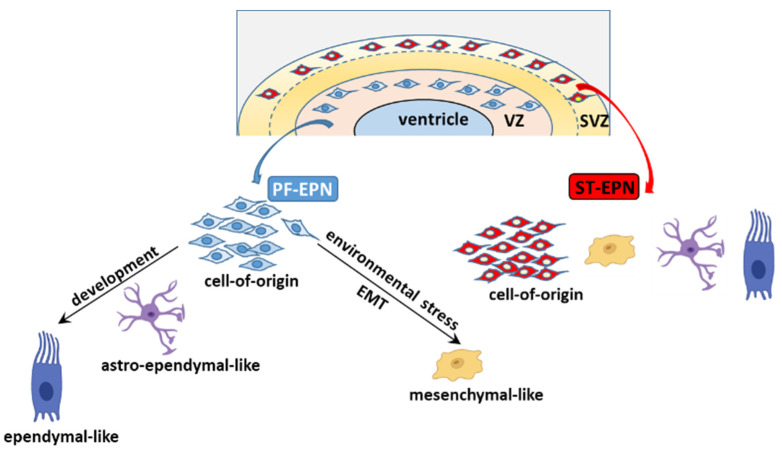
Leveraging scRNA-seq and TI analyses, it emerges that the distinct PF-EPN subpopulations are arranged in a neural tri-lineage cancer hierarchy driven by immature progenitor cells at the apex, that undergo impaired differentiation along neuronal, astrocytic, and ependymal-like trajectories [165]. Of the three branches, the predominant one sees the NSC-like population differentiate into less aggressive progenies, the astro-ependymal cells, and successively to ependymal-like cells, presumably in response to developmental or differentiation stimuli (Figure 6). This axis potentially overlaps with the differentiating trajectory described in Gillen et al.’s study, whereby the stem cell population of UEC-1 develops into TECs (which express markers of further differentiation, such as oRGC genes, as well as gliogenic progenitor and astrocytic progenitor genes), and then CECs characterized by an ependymal-like signature. In response to unfavorable microenvironmental cues, such as oxygen and/or nutrient deprivation in hypoxic areas, undifferentiated UEC-1 develop along a stress-associated trajectory and undergo EMT to give rise to mesenchymal MECs.
Figure 6.
The candidate cell of origin of PF-EPNs resides in the ventricular zone (VZ) of the developing brain, whereas that of ST-EPNs is located in the subventricular zone (SVZ). In PF-EPNs, tumor subpopulations are arranged in two major distinct lineage trajectories driven by undifferentiated progenitors, that either undergo impaired differentiation towards ependymal-like cells, or transition to mesenchymal-like cells in response to cellular stresses, e.g., hypoxia. In ST-EPNs, clear developmental trajectories have not been identified yet (Reprinted with permission from Austin E. Gillen et al., (2020), Elsevier and Copyright Clearance Center, Licence Number 5200810483356, 2 December 2021).
The cellular make-up of ST-EPN mostly includes immature cells stalled at the earlier phases of the neurodevelopment with a minor component of more differentiated cells. Therefore, ST-EPNs appear to be characterized by the coexistence of distinct progenitor populations rather than a hierarchical developmentally organized structure.
6. Therapeutic Applications
Cancer cell heterogeneity has long been recognized as a major cause of treatment failure. More effective chemotherapeutic strategies should consider targeting not only driver genes, but also different cell types and cell states [206]. This is urgently needed for the aggressive forms of EPN for which effective drugs are still lacking. The distinct inter-group and intra-group transcriptomic signatures identified in EPN may help define molecular dependencies and treatment vulnerabilities.
A potentially druggable pathway in PFA EPN is EZHIP, although direct targeting of EZHIP might prove difficult to achieve, because no enzymatic activity has hitherto been identified [207]. High EZHIP expression sensitizes to PARP inhibitors by inhibiting homologous recombination-mediated DNA repair, especially in combination with radiotherapy, indicating this treatment approach as potentially beneficial in PFA [208]. A recent publication has shown that PFA tumors and patient-derived cell lines with EZHIP overexpression exhibit enhanced glycolysis and tricarboxylic acid cycle (TCA) metabolism, associated with enrichment of H3K27ac at hexokinase-2, pyruvate dehydrogenase, and AMPKα-2 [209]. The antidiabetic AMPK activator metformin increases H3K27me3, while reducing EZHIP expression, TCA cycle metabolism, and tumor growth. As ST-EPNs also show cell subpopulations with increased glycolysis (e.g., ST-metabolic), it is conceivable to hypothesize that repurposing metformin as an antitumoral agent might have therapeutic efficacy in pediatric EPN. The opposite role of EZH2 in PFAs—whereby it is globally repressed, although its residual activity is essential for tumor development—calls for preclinical and clinical testing of EZH2 inhibitors as potential therapeutic interventions in PFA EPN. Numerous EZH2 inhibitors are currently undergoing phase 1 and phase 2 clinical testing in different tumors [210], and might have important implications for novel treatment protocols. Specifically, an advanced trial is evaluating the effectiveness of the EZH2 inhibitor tazemetostat in pediatric patients with recurrent EPN (NCT03213665).
Compounds aimed at blocking the oncogenic NF-κB pathway [211] are potential therapeutic agents against ST-EPNs harboring ZFTA–RELA fusion that contains the NF-κB subunit encoding gene RELA. The NF-κB subunit is activated via proteosomal degradation of its inhibitor IκB, thus suggesting proteasome inhibitors as candidate drugs in ST-RELA [212]. Marizomib, a pan-proteasome inhibitor with good brain-penetrating capacity [213], is currently undergoing a phase 2 clinical trial in adult patients with anaplastic EPN (NCT03727841), while in the pediatric setting it is being tested against diffuse intrinsic pontine glioma (DIPG) (NCT04341311).
scRNA-seq is expected to make significant breakthroughs in EPN and inform future therapeutic approaches. Since an increased proportion of differentiated cells is associated with a favorable clinical behavior in ST-RELA and PFA, differentiation-promoting agents might prove effective in these high-risk groups. Corroboratively, retinoids have demonstrated selective efficacy against EPN lines compared to other brain tumor-derived models in an in vitro drug screen [214].
In the context of PF-EPN, a druggable driver in the PF-NSC-like program is the Wnt pathway gene LGR5, a key mediator of cell proliferation and stemness features, as shown by small interfering RNA (siRNA)-mediated LGR5 knockdown that results in reduction of self-renewal [165]. In PF-Neuronal-Precursor-like cells, targetable pathways might be the epigenetic regulators HDAC2, DNMT3A, and BRD3. Indeed, the pan-HDAC inhibitor CN133 [215] and the HDAC2 inhibitor panobinostat [165], as well as the pan-BRD inhibitors JQ1 [199] and OTX012 [216], have been reported to decrease cell viability and tumor growth in patient-derived PFA cell lines. Emerging epigenetic therapies under evaluation in clinical trials for brain tumors, including EPN, have been reviewed recently [217].
As for ST-EPN, actionable vulnerabilities in ST-Radial-Glia-like cells are FGFR3 and IGF2, whereas in the ST-Neuronal-Precursor-like subpopulation they are CCND2 and HDAC2. FGFR3 mRNA levels are enriched in ST-RELA EPNs, and specifically in cycling and progenitor-like cell populations, mirroring FGFR3 expression in RGCs of the embryonic and adult brain [218]. Indeed, blockade of FGFR by dominant-negative and pharmacological inhibitors impairs cell survival and stemness features in ST-RELA cells [165,218] Simultaneous inhibition of CDK4/6-CCND2 (with palbociclib) and IGF2/IGF1R (with ceritinib) pathways results in combinatorial drug efficacy, highlighting that targeting distinct subpopulations may be a successful therapeutic option [165]. CDK4/6 has also been proposed as an actionable driver in PFA, because the tumor suppressor gene CDKN2A, which codes for the CDK4/6 inhibitor p16, is epigenetically silenced by H3K27 trimethylation in PFA [146,207]. A phase 1 trial is addressing the safety and tolerability of the CDK4/6 inhibitor ribociclib in children and young adults with recurrent brain tumors, including EPN (NCT03434262).
A link between the overexpression of strong growth-promoting IGF2 and members of the PLAG1/PLAG1L TF family is emerging in EPN. The PLAGL1 gene is developmentally regulated and is expressed in NSCs and developing neuroepithelial cells, with low expression in the adult brain [46,219]. Although the function of PLAGL1 in tumorigenesis is controversial, acting as either a tumor suppressor or an oncogene in a context-dependent manner, PLAG1L has been shown to foster progression of GBM [207]. In ST-RELA tumors, ZETA-RELA protein binds to PLAGL family TF motifs, indicating a possible corecruitment to drive ependymoma-related transcriptional programs [106]. PLAG1 is silenced during development by PRC2-mediated H3K27 trimethylation; however, in tumors with impaired PRC2 function, such as PFA and H3K27M mutant pHGG, PLAG1 is derepressed, leading to overexpression of its downstream targets, including IGF2 [207]. Therefore, it is conceivable to hypothesize that the PLAG1/PLAG1L-IGF2 axis might be therapeutically targeted in EPN.
7. Concluding Remarks
Despite the enormously increased understanding of the molecular drivers and biology of EPN, the treatment standards have essentially remained static over recent years. Gross total resection is still the strongest predictor of outcome [220]. The role for chemotherapy in young children to protect them from the side effects of radiation therapy is still debated [221,222], whereas no survival advantage with the use of chemotherapy in recurrent EPN has been found [223] despite intensive investigation.
At present, 108 clinical trials are ongoing in pediatric EPN (ClinicalTrials.gov [224], accessed on 22 November 2021), however, they do not take into account EPN variants. Moving forward, the next challenge is to go beyond tumor control and to include EPN molecular classification in treatment decisions so as to adapt therapeutic strategies based on risk stratification, reducing therapy-induced morbidity in low-risk patients, while intensifying treatment for high-risk patients [225]. Development of new treatments for patients with EPN, especially in the pediatric cohort, meets several challenges, including low investment by pharmaceutical companies and low incidence of patients with rare cancers, that hamper testing of new compounds in prospective clinical trials [212]. The establishment of appropriate preclinical models, which mirror the distinct EPN subgroups and even the distinct EPN subpopulations, is critical for drug testing and identification of drug response biomarkers [226]. Although there are many preclinical studies in EPN models [74,199,227,228], few studies have hitherto compared drug sensitivities in heterogeneous subpopulations of EPN cell lines [78,79,216].
scRNA-seq has just begun to be applied to translational research in EPN. Future studies are warranted to increase the number of EPN specimens dissected at the single-cell level, to sample anatomically and temporally distinct regions in order to address tumor heterogeneity and evolution at recurrence, with the ultimate goal to discover EPN sub-type specific drivers and druggable pathways. In addition, the diverse and complex extrinsic interactions of EPN cells with the tumor microenvironment should also be prospectively evaluated.
Although single-cell genomics have shown the complex intercellular variability that governs EPN biology and challenges the response to treatment, they have also evidenced coalescing commonalities shared across subgroups of tumors and even across tumors of disparate histologies. For instance, Neuronal-Precursor-like programs are strictly correlated in PF-EPN and ST-EPN, and share transcriptional overlaps with the Neuronal-Precursor-like cell programs described in GBM [165,229]. Likewise, the mesenchymal signatures in PF-EPN and ST-EPN are similar to that reported in GBM. In addition, the astro-ependymal program in PF-EPN resembles the astrocyte-like programs in both DIPG and GBM [165,190]. In addition to scRNA-seq, integrative proteogenomics analyses have identified common biological processes between and among PBTs of seven histological types, including HGG, and EPN, which suggests that tumors of disparate histologies may share common therapeutic vulnerabilities [230].
In conclusion, not only has scRNA-seq highlighted the bewildering heterogeneity of EPN, but it may also contribute to defining subtype- and subgroup-specific molecular vulnerabilities and new options for therapeutic interventions. Moreover, the observation that some transcriptomic signatures cross histological boundaries among EPN groups and even amongst disparate pediatric tumors suggests that some treatment opportunities may be effective in a larger group of diseases than that might have been expected.
Abbreviations
| BTSCs | brain tumor stem cells |
| CEC | ciliated ependymoma cells |
| CIMP | CpG island methylator phenotype |
| CSCs | cancer stem cells |
| DIPG | diffuse intrinsic pontine glioma |
| ecDNA | extrachromosomal DNA |
| EMT | epithelial-to-mesenchymal transition |
| EPN | ependymoma |
| ESCs | embryonic stem cells |
| GBM | glioblastoma |
| GSCs | glioblastoma stem cells |
| ITH | intra-tumoral heterogeneity |
| MEC | mesenchymal ependymoma cells |
| MET | mesenchymal epithelial transition |
| NSCs | neural stem cells |
| NSs | neurospheres |
| OPCs | oligodendrocyte precursor cells |
| oRGCs | outer radial glia cells |
| oSVZ | outer subventricular zone |
| PBTs | pediatric brain tumors |
| PF | posterior fossa |
| PF-NSC-like | posterior fossa-Neural-Stem-Cell-like |
| pHGG | pediatric high-grade gliomas |
| RGCs | radial glia cells |
| RTK | receptor tyrosine kinase |
| RT-PCR | reverse transcriptase polymerase chain reaction |
| scRNA-seq | single-cell RNA sequencing |
| ST | supratentorial |
| ST_EPN | supratentorial ependymoma |
| TCA | tricarboxylic acid cycle |
| TECs | transportive ependymoma cells |
| TF | transcription factor |
| TI | trajectory inference |
| TME | tumor microenvironment |
| UEC-1 | undifferentiated ependymoma cells-1 |
| vRGCs | ventricular radial glia cells |
| VZ | ventricular zone |
Author Contributions
Conceptualization, T.S. and A.R.; writing—original draft preparation, T.S.; writing—review and editing, T.S., D.L., P.N., A.S. and A.R.; visualization, T.S.; funding acquisition, R.R. and A.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fondazione per l’Oncologia Pediatrica.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.McGranahan N., Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell. 2017;168:613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 2.McGranahan N., Swanton C. Biological and Therapeutic Impact of Intratumor Heterogeneity in Cancer Evolution. Cancer Cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Junttila M.R., de Sauvage F.J. Influence of Tumour Micro-Environment Heterogeneity on Therapeutic Response. Nature. 2013;501:346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 4.Welch D.R. Tumor Heterogeneity—A “Contemporary Concept” Founded on Historical Insights and Predictions. Cancer Res. 2016;76:4–6. doi: 10.1158/0008-5472.CAN-15-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heppner G.H. Tumor Heterogeneity. Cancer Res. 1984;44:2259–2265. [PubMed] [Google Scholar]
- 6.Merlo L.M.F., Pepper J.W., Reid B.J., Maley C.C. Cancer as an Evolutionary and Ecological Process. Nat. Rev. Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 7.Marusyk A., Almendro V., Polyak K. Intra-Tumour Heterogeneity: A Looking Glass for Cancer? Nat. Rev. Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 8.Lloyd M.C., Cunningham J.J., Bui M.M., Gillies R.J., Brown J.S., Gatenby R.A. Darwinian Dynamics of Intratumoral Heterogeneity: Not Solely Random Mutations but Also Variable Environmental Selection Forces. Cancer Res. 2016;76:3136–3144. doi: 10.1158/0008-5472.CAN-15-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dirkse A., Golebiewska A., Buder T., Nazarov P.V., Muller A., Poovathingal S., Brons N.H.C., Leite S., Sauvageot N., Sarkisjan D., et al. Stem Cell-Associated Heterogeneity in Glioblastoma Results from Intrinsic Tumor Plasticity Shaped by the Microenvironment. Nat. Commun. 2019;10:1787. doi: 10.1038/s41467-019-09853-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flavahan W.A. Epigenetic Plasticity, Selection, and Tumorigenesis. Biochem. Soc. Trans. 2020;48:1609–1621. doi: 10.1042/BST20191215. [DOI] [PubMed] [Google Scholar]
- 11.Easwaran H., Tsai H.-C., Baylin S.B. Cancer Epigenetics: Tumor Heterogeneity, Plasticity of Stem-like States, and Drug Resistance. Mol. Cell. 2014;54:716–727. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batlle E., Clevers H. Cancer Stem Cells Revisited. Nat. Med. 2017;23:1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 13.Hinohara K., Polyak K. Intratumoral Heterogeneity: More Than Just Mutations. Trends Cell Biol. 2019;29:569–579. doi: 10.1016/j.tcb.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inda M.-M., Bonavia R., Mukasa A., Narita Y., Sah D.W.Y., Vandenberg S., Brennan C., Johns T.G., Bachoo R., Hadwiger P., et al. Tumor Heterogeneity Is an Active Process Maintained by a Mutant EGFR-Induced Cytokine Circuit in Glioblastoma. Genes Dev. 2010;24:1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polyak K., Marusyk A. Cancer: Clonal Cooperation. Nature. 2014;508:52–53. doi: 10.1038/508052a. [DOI] [PubMed] [Google Scholar]
- 16.Ricklefs F., Mineo M., Rooj A.K., Nakano I., Charest A., Weissleder R., Breakefield X.O., Chiocca E.A., Godlewski J., Bronisz A. Extracellular Vesicles from High-Grade Glioma Exchange Diverse Pro-Oncogenic Signals That Maintain Intratumoral Heterogeneity. Cancer Res. 2016;76:2876–2881. doi: 10.1158/0008-5472.CAN-15-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broekman M.L., Maas S.L.N., Abels E.R., Mempel T.R., Krichevsky A.M., Breakefield X.O. Multidimensional Communication in the Microenvirons of Glioblastoma. Nat. Rev. Neurol. 2018;14:482–495. doi: 10.1038/s41582-018-0025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janiszewska M., Tabassum D.P., Castaño Z., Cristea S., Yamamoto K.N., Kingston N.L., Murphy K.C., Shu S., Harper N.W., Del Alcazar C.G., et al. Subclonal Cooperation Drives Metastasis by Modulating Local and Systemic Immune Microenvironments. Nat. Cell Biol. 2019;21:879–888. doi: 10.1038/s41556-019-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marusyk A., Janiszewska M., Polyak K. Intratumor Heterogeneity: The Rosetta Stone of Therapy Resistance. Cancer Cell. 2020;37:471–484. doi: 10.1016/j.ccell.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naffar-Abu Amara S., Kuiken H.J., Selfors L.M., Butler T., Leung M.L., Leung C.T., Kuhn E.P., Kolarova T., Hage C., Ganesh K., et al. Transient Commensal Clonal Interactions Can Drive Tumor Metastasis. Nat. Commun. 2020;11:5799. doi: 10.1038/s41467-020-19584-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostrom Q.T., Gittleman H., Truitt G., Boscia A., Kruchko C., Barnholtz-Sloan J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro Oncol. 2018;20:iv1–iv86. doi: 10.1093/neuonc/noy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pajtler K.W., Mack S.C., Ramaswamy V., Smith C.A., Witt H., Smith A., Hansford J.R., von Hoff K., Wright K.D., Hwang E., et al. The Current Consensus on the Clinical Management of Intracranial Ependymoma and Its Distinct Molecular Variants. Acta Neuropathol. 2017;133:5–12. doi: 10.1007/s00401-016-1643-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pajtler K.W., Witt H., Sill M., Jones D.T.W., Hovestadt V., Kratochwil F., Wani K., Tatevossian R., Punchihewa C., Johann P., et al. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell. 2015;27:728–743. doi: 10.1016/j.ccell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker M., Mohankumar K.M., Punchihewa C., Weinlich R., Dalton J.D., Li Y., Lee R., Tatevossian R.G., Phoenix T.N., Thiruvenkatam R., et al. C11orf95-RELA Fusions Drive Oncogenic NF-ΚB Signalling in Ependymoma. Nature. 2014;506:451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witt H., Mack S.C., Ryzhova M., Bender S., Sill M., Isserlin R., Benner A., Hielscher T., Milde T., Remke M., et al. Delineation of Two Clinically and Molecularly Distinct Subgroups of Posterior Fossa Ependymoma. Cancer Cell. 2011;20:143–157. doi: 10.1016/j.ccr.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suvà M.L., Tirosh I. Single-Cell RNA Sequencing in Cancer: Lessons Learned and Emerging Challenges. Mol. Cell. 2019;75:7–12. doi: 10.1016/j.molcel.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Stuart T., Satija R. Integrative Single-Cell Analysis. Nat. Rev. Genet. 2019;20:257–272. doi: 10.1038/s41576-019-0093-7. [DOI] [PubMed] [Google Scholar]
- 28.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 29.Louis D., Perry A., Wesseling P., Brat D., Cree I., Figarella-Branger D., Hawkins C., Ng H., Pfister S., Reifenberger G., et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro Oncol. 2021;23:1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baroni L.V., Sundaresan L., Heled A., Coltin H., Pajtler K.W., Lin T., Merchant T.E., McLendon R., Faria C., Buntine M., et al. Ultra High-Risk PFA Ependymoma Is Characterized by Loss of Chromosome 6q. Neuro Oncol. 2021;23:1360–1370. doi: 10.1093/neuonc/noab034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mack S.C., Witt H., Piro R.M., Gu L., Zuyderduyn S., Stütz A.M., Wang X., Gallo M., Garzia L., Zayne K., et al. Epigenomic Alterations Define Lethal CIMP-Positive Ependymomas of Infancy. Nature. 2014;506:445–450. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bayliss J., Mukherjee P., Lu C., Jain S.U., Chung C., Martinez D., Sabari B., Margol A.S., Panwalkar P., Parolia A., et al. Lowered H3K27me3 and DNA Hypomethylation Define Poorly Prognostic Pediatric Posterior Fossa Ependymomas. Sci. Transl. Med. 2016;8:366ra161. doi: 10.1126/scitranslmed.aah6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baylin S.B., Jones P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016;8:a019505. doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panwalkar P., Clark J., Ramaswamy V., Hawes D., Yang F., Dunham C., Yip S., Hukin J., Sun Y., Schipper M.J., et al. Immunohistochemical Analysis of H3K27me3 Demonstrates Global Reduction in Group-A Childhood Posterior Fossa Ependymoma and Is a Powerful Predictor of Outcome. Acta Neuropathol. 2017;134:705–714. doi: 10.1007/s00401-017-1752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gessi M., Capper D., Sahm F., Huang K., von Deimling A., Tippelt S., Fleischhack G., Scherbaum D., Alfer J., Juhnke B.-O., et al. Evidence of H3 K27M Mutations in Posterior Fossa Ependymomas. Acta Neuropathol. 2016;132:635–637. doi: 10.1007/s00401-016-1608-3. [DOI] [PubMed] [Google Scholar]
- 36.Pajtler K.W., Wen J., Sill M., Lin T., Orisme W., Tang B., Hübner J.-M., Ramaswamy V., Jia S., Dalton J.D., et al. Molecular Heterogeneity and CXorf67 Alterations in Posterior Fossa Group A (PFA) Ependymomas. Acta Neuropathol. 2018;136:211–226. doi: 10.1007/s00401-018-1877-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavalli F.M.G., Hübner J.-M., Sharma T., Luu B., Sill M., Zapotocky M., Mack S.C., Witt H., Lin T., Shih D.J.H., et al. Heterogeneity within the PF-EPN-B Ependymoma Subgroup. Acta Neuropathol. 2018;136:227–237. doi: 10.1007/s00401-018-1888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taniguchi K., Karin M. NF-ΚB, Inflammation, Immunity and Cancer: Coming of Age. Nat. Rev. Immunol. 2018;18:309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 39.Pajtler K.W., Wei Y., Okonechnikov K., Silva P.B.G., Vouri M., Zhang L., Brabetz S., Sieber L., Gulley M., Mauermann M., et al. YAP1 Subgroup Supratentorial Ependymoma Requires TEAD and Nuclear Factor I-Mediated Transcriptional Programmes for Tumorigenesis. Nat. Commun. 2019;10:3914. doi: 10.1038/s41467-019-11884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andreiuolo F., Varlet P., Tauziède-Espariat A., Jünger S.T., Dörner E., Dreschmann V., Kuchelmeister K., Waha A., Haberler C., Slavc I., et al. Childhood Supratentorial Ependymomas with YAP1-MAMLD1 Fusion: An Entity with Characteristic Clinical, Radiological, Cytogenetic and Histopathological Features. Brain Pathol. Zurich Switz. 2019;29:205–216. doi: 10.1111/bpa.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukuoka K., Kanemura Y., Shofuda T., Fukushima S., Yamashita S., Narushima D., Kato M., Honda-Kitahara M., Ichikawa H., Kohno T., et al. Significance of Molecular Classification of Ependymomas: C11orf95-RELA Fusion-Negative Supratentorial Ependymomas Are a Heterogeneous Group of Tumors. Acta Neuropathol. Commun. 2018;6:134. doi: 10.1186/s40478-018-0630-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamai S., Nakano Y., Kinoshita M., Sabit H., Nobusawa S., Arai Y., Hama N., Totoki Y., Shibata T., Ichimura K., et al. Ependymoma with C11orf95-MAML2 Fusion: Presenting with Granular Cell and Ganglion Cell Features. Brain Tumor Pathol. 2021;38:64–70. doi: 10.1007/s10014-020-00388-6. [DOI] [PubMed] [Google Scholar]
- 43.Zheng T., Ghasemi D.R., Okonechnikov K., Korshunov A., Sill M., Maass K.K., Benites Goncalves da Silva P., Ryzhova M., Gojo J., Stichel D., et al. Cross-Species Genomics Reveals Oncogenic Dependencies in ZFTA/C11orf95 Fusion-Positive Supratentorial Ependymomas. Cancer Discov. 2021;11:2230–2247. doi: 10.1158/2159-8290.CD-20-0963. [DOI] [PubMed] [Google Scholar]
- 44.Tauziède-Espariat A., Siegfried A., Nicaise Y., Kergrohen T., Sievers P., Vasiljevic A., Roux A., Dezamis E., Benevello C., Machet M.-C., et al. Supratentorial Non-RELA, ZFTA-Fused Ependymomas: A Comprehensive Phenotype Genotype Correlation Highlighting the Number of Zinc Fingers in ZFTA-NCOA1/2 Fusions. Acta Neuropathol. Commun. 2021;9:135. doi: 10.1186/s40478-021-01238-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zschernack V., Jünger S.T., Mynarek M., Rutkowski S., Garre M.L., Ebinger M., Neu M., Faber J., Erdlenbruch B., Claviez A., et al. Supratentorial Ependymoma in Childhood: More than Just RELA or YAP. Acta Neuropathol. 2021;141:455–466. doi: 10.1007/s00401-020-02260-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sievers P., Henneken S.C., Blume C., Sill M., Schrimpf D., Stichel D., Okonechnikov K., Reuss D.E., Benzel J., Maaß K.K., et al. Recurrent Fusions in PLAGL1 Define a Distinct Subset of Pediatric-Type Supratentorial Neuroepithelial Tumors. Acta Neuropathol. 2021;142:827–839. doi: 10.1007/s00401-021-02356-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ellison D.W., Aldape K.D., Capper D., Fouladi M., Gilbert M.R., Gilbertson R.J., Hawkins C., Merchant T.E., Pajtler K., Venneti S., et al. CIMPACT-NOW Update 7: Advancing the Molecular Classification of Ependymal Tumors. Brain Pathol. Zurich Switz. 2020;30:863–866. doi: 10.1111/bpa.12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keenan C., Graham R.T., Harreld J.H., Lucas J.T., Finkelstein D., Wheeler D., Li X., Dalton J., Upadhyaya S.A., Raimondi S.C., et al. Infratentorial C11orf95-Fused Gliomas Share Histologic, Immunophenotypic, and Molecular Characteristics of Supratentorial RELA-Fused Ependymoma. Acta Neuropathol. 2020;140:963–965. doi: 10.1007/s00401-020-02238-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem Cells, Cancer, and Cancer Stem Cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 50.Prasetyanti P.R., Medema J.P. Intra-Tumor Heterogeneity from a Cancer Stem Cell Perspective. Mol. Cancer. 2017;16:41. doi: 10.1186/s12943-017-0600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uchida N., Buck D.W., He D., Reitsma M.J., Masek M., Phan T.V., Tsukamoto A.S., Gage F.H., Weissman I.L. Direct Isolation of Human Central Nervous System Stem Cells. Proc. Natl. Acad. Sci. USA. 2000;97:14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B. Identification of Human Brain Tumour Initiating Cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 53.Pardal R., Clarke M.F., Morrison S.J. Applying the Principles of Stem-Cell Biology to Cancer. Nat. Rev. Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 54.Vescovi A.L., Galli R., Reynolds B.A. Brain Tumour Stem Cells. Nat. Rev. Cancer. 2006;6:425–436. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- 55.Prestegarden L., Svendsen A., Wang J., Sleire L., Skaftnesmo K.O., Bjerkvig R., Yan T., Askland L., Persson A., Sakariassen P.Ø., et al. Glioma Cell Populations Grouped by Different Cell Type Markers Drive Brain Tumor Growth. Cancer Res. 2010;70:4274–4279. doi: 10.1158/0008-5472.CAN-09-3904. [DOI] [PubMed] [Google Scholar]
- 56.Gupta P.B., Fillmore C.M., Jiang G., Shapira S.D., Tao K., Kuperwasser C., Lander E.S. Stochastic State Transitions Give Rise to Phenotypic Equilibrium in Populations of Cancer Cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 57.Wang X., Prager B.C., Wu Q., Kim L.J.Y., Gimple R.C., Shi Y., Yang K., Morton A.R., Zhou W., Zhu Z., et al. Reciprocal Signaling between Glioblastoma Stem Cells and Differentiated Tumor Cells Promotes Malignant Progression. Cell Stem Cell. 2018;22:514–528.e5. doi: 10.1016/j.stem.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lytle N.K., Barber A.G., Reya T. Stem Cell Fate in Cancer Growth, Progression and Therapy Resistance. Nat. Rev. Cancer. 2018;18:669–680. doi: 10.1038/s41568-018-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kondo T. Glioblastoma-Initiating Cell Heterogeneity Generated by the Cell-of-Origin, Genetic/Epigenetic Mutation and Microenvironment. Semin. Cancer Biol. 2021 doi: 10.1016/j.semcancer.2020.12.003. [DOI] [PubMed] [Google Scholar]
- 60.Muñoz P., Iliou M.S., Esteller M. Epigenetic Alterations Involved in Cancer Stem Cell Reprogramming. Mol. Oncol. 2012;6:620–636. doi: 10.1016/j.molonc.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aryee M.J., Liu W., Engelmann J.C., Nuhn P., Gurel M., Haffner M.C., Esopi D., Irizarry R.A., Getzenberg R.H., Nelson W.G., et al. DNA Methylation Alterations Exhibit Intraindividual Stability and Interindividual Heterogeneity in Prostate Cancer Metastases. Sci. Transl. Med. 2013;5:169ra10. doi: 10.1126/scitranslmed.3005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Friedmann-Morvinski D., Bushong E.A., Ke E., Soda Y., Marumoto T., Singer O., Ellisman M.H., Verma I.M. Dedifferentiation of Neurons and Astrocytes by Oncogenes Can Induce Gliomas in Mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hide T., Takezaki T., Nakatani Y., Nakamura H., Kuratsu J., Kondo T. Combination of a Ptgs2 Inhibitor and an Epidermal Growth Factor Receptor-Signaling Inhibitor Prevents Tumorigenesis of Oligodendrocyte Lineage-Derived Glioma-Initiating Cells. Stem Cells. 2011;29:590–599. doi: 10.1002/stem.618. [DOI] [PubMed] [Google Scholar]
- 64.Funato K., Major T., Lewis P.W., Allis C.D., Tabar V. Use of Human Embryonic Stem Cells to Model Pediatric Gliomas with H3.3K27M Histone Mutation. Science. 2014;346:1529–1533. doi: 10.1126/science.1253799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagaraja S., Quezada M.A., Gillespie S.M., Arzt M., Lennon J.J., Woo P.J., Hovestadt V., Kambhampati M., Filbin M.G., Suva M.L., et al. Histone Variant and Cell Context Determine H3K27M Reprogramming of the Enhancer Landscape and Oncogenic State. Mol. Cell. 2019;76:965–980.e12. doi: 10.1016/j.molcel.2019.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haag D., Mack N., Benites Goncalves da Silva P., Statz B., Clark J., Tanabe K., Sharma T., Jäger N., Jones D.T.W., Kawauchi D., et al. H3.3-K27M Drives Neural Stem Cell-Specific Gliomagenesis in a Human IPSC-Derived Model. Cancer Cell. 2021;39:407–422.e13. doi: 10.1016/j.ccell.2021.01.005. [DOI] [PubMed] [Google Scholar]
- 67.Yang Z.-J., Ellis T., Markant S.L., Read T.-A., Kessler J.D., Bourboulas M., Schüller U., Machold R., Fishell G., Rowitch D.H., et al. Medulloblastoma Can Be Initiated by Deletion of Patched in Lineage-Restricted Progenitors or Stem Cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hemmati H.D., Nakano I., Lazareff J.A., Masterman-Smith M., Geschwind D.H., Bronner-Fraser M., Kornblum H.I. Cancerous Stem Cells Can Arise from Pediatric Brain Tumors. Proc. Natl. Acad. Sci. USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh S.K., Clarke I.D., Terasaki M., Bonn V.E., Hawkins C., Squire J., Dirks P.B. Identification of a Cancer Stem Cell in Human Brain Tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 70.Taylor M.D., Poppleton H., Fuller C., Su X., Liu Y., Jensen P., Magdaleno S., Dalton J., Calabrese C., Board J., et al. Radial Glia Cells Are Candidate Stem Cells of Ependymoma. Cancer Cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 71.Götz M., Huttner W.B. The Cell Biology of Neurogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 72.Servidei T., Meco D., Trivieri N., Patriarca V., Vellone V.G., Zannoni G.F., Lamorte G., Pallini R., Riccardi R. Effects of Epidermal Growth Factor Receptor Blockade on Ependymoma Stem Cells in Vitro and in Orthotopic Mouse Models. Int. J. Cancer. 2012;131:E791–E803. doi: 10.1002/ijc.27377. [DOI] [PubMed] [Google Scholar]
- 73.Yu L., Baxter P.A., Voicu H., Gurusiddappa S., Zhao Y., Adesina A., Man T.-K., Shu Q., Zhang Y.-J., Zhao X.-M., et al. A Clinically Relevant Orthotopic Xenograft Model of Ependymoma That Maintains the Genomic Signature of the Primary Tumor and Preserves Cancer Stem Cells in Vivo. Neuro Oncol. 2010;12:580–594. doi: 10.1093/neuonc/nop056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Milde T., Kleber S., Korshunov A., Witt H., Hielscher T., Koch P., Kopp H.-G., Jugold M., Deubzer H.E., Oehme I., et al. A Novel Human High-Risk Ependymoma Stem Cell Model Reveals the Differentiation-Inducing Potential of the Histone Deacetylase Inhibitor Vorinostat. Acta Neuropathol. 2011;122:637–650. doi: 10.1007/s00401-011-0866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amani V., Donson A.M., Lummus S.C., Prince E.W., Griesinger A.M., Witt D.A., Hankinson T.C., Handler M.H., Dorris K., Vibhakar R., et al. Characterization of 2 Novel Ependymoma Cell Lines with Chromosome 1q Gain Derived From Posterior Fossa Tumors of Childhood. J. Neuropathol. Exp. Neurol. 2017;76:595–604. doi: 10.1093/jnen/nlx040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bian S., Repic M., Guo Z., Kavirayani A., Burkard T., Bagley J.A., Krauditsch C., Knoblich J.A. Genetically Engineered Cerebral Organoids Model Brain Tumor Formation. Nat. Methods. 2018;15:631–639. doi: 10.1038/s41592-018-0070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sabnis D.H., Liu J.-F., Simmonds L., Blackburn S., Grundy R.G., Kerr I.D., Coyle B. BLBP Is Both a Marker for Poor Prognosis and a Potential Therapeutic Target in Paediatric Ependymoma. Cancers. 2021;13:2100. doi: 10.3390/cancers13092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meco D., Servidei T., Lamorte G., Binda E., Arena V., Riccardi R. Ependymoma Stem Cells Are Highly Sensitive to Temozolomide in Vitro and in Orthotopic Models. Neuro Oncol. 2014;16:1067–1077. doi: 10.1093/neuonc/nou008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Servidei T., Meco D., Muto V., Bruselles A., Ciolfi A., Trivieri N., Lucchini M., Morosetti R., Mirabella M., Martini M., et al. Novel SEC61G-EGFR Fusion Gene in Pediatric Ependymomas Discovered by Clonal Expansion of Stem Cells in Absence of Exogenous Mitogens. Cancer Res. 2017;77:5860–5872. doi: 10.1158/0008-5472.CAN-17-0790. [DOI] [PubMed] [Google Scholar]
- 80.Brennan C.W., Verhaak R.G.W., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., et al. The Somatic Genomic Landscape of Glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Szerlip N.J., Pedraza A., Chakravarty D., Azim M., McGuire J., Fang Y., Ozawa T., Holland E.C., Huse J.T., Jhanwar S., et al. Intratumoral Heterogeneity of Receptor Tyrosine Kinases EGFR and PDGFRA Amplification in Glioblastoma Defines Subpopulations with Distinct Growth Factor Response. Proc. Natl. Acad. Sci. USA. 2012;109:3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schulte A., Günther H.S., Martens T., Zapf S., Riethdorf S., Wülfing C., Stoupiec M., Westphal M., Lamszus K. Glioblastoma Stem-like Cell Lines with Either Maintenance or Loss of High-Level EGFR Amplification, Generated via Modulation of Ligand Concentration. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012;18:1901–1913. doi: 10.1158/1078-0432.CCR-11-3084. [DOI] [PubMed] [Google Scholar]
- 83.Vogelstein B., Kinzler K.W. Cancer Genes and the Pathways They Control. Nat. Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 84.Burrell R.A., McGranahan N., Bartek J., Swanton C. The Causes and Consequences of Genetic Heterogeneity in Cancer Evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 85.Watson I.R., Takahashi K., Futreal P.A., Chin L. Emerging Patterns of Somatic Mutations in Cancer. Nat. Rev. Genet. 2013;14:703–718. doi: 10.1038/nrg3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dagogo-Jack I., Shaw A.T. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 87.Negrini S., Gorgoulis V.G., Halazonetis T.D. Genomic Instability--an Evolving Hallmark of Cancer. Nat. Rev. Mol. Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 88.Siri S.O., Martino J., Gottifredi V. Structural Chromosome Instability: Types, Origins, Consequences, and Therapeutic Opportunities. Cancers. 2021;13:3056. doi: 10.3390/cancers13123056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A., et al. Massive Genomic Rearrangement Acquired in a Single Catastrophic Event during Cancer Development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Paulsen T., Kumar P., Koseoglu M.M., Dutta A. Discoveries of Extrachromosomal Circles of DNA in Normal and Tumor Cells. Trends Genet. 2018;34:270–278. doi: 10.1016/j.tig.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Verhaak R.G.W., Bafna V., Mischel P.S. Extrachromosomal Oncogene Amplification in Tumour Pathogenesis and Evolution. Nat. Rev. Cancer. 2019;19:283–288. doi: 10.1038/s41568-019-0128-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu S., Turner K.M., Nguyen N., Raviram R., Erb M., Santini J., Luebeck J., Rajkumar U., Diao Y., Li B., et al. Circular EcDNA Promotes Accessible Chromatin and High Oncogene Expression. Nature. 2019;575:699–703. doi: 10.1038/s41586-019-1763-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Turner K.M., Deshpande V., Beyter D., Koga T., Rusert J., Lee C., Li B., Arden K., Ren B., Nathanson D.A., et al. Extrachromosomal Oncogene Amplification Drives Tumour Evolution and Genetic Heterogeneity. Nature. 2017;543:122–125. doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.deCarvalho A.C., Kim H., Poisson L.M., Winn M.E., Mueller C., Cherba D., Koeman J., Seth S., Protopopov A., Felicella M., et al. Discordant Inheritance of Chromosomal and Extrachromosomal DNA Elements Contributes to Dynamic Disease Evolution in Glioblastoma. Nat. Genet. 2018;50:708–717. doi: 10.1038/s41588-018-0105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim H., Nguyen N.-P., Turner K., Wu S., Gujar A.D., Luebeck J., Liu J., Deshpande V., Rajkumar U., Namburi S., et al. Extrachromosomal DNA Is Associated with Oncogene Amplification and Poor Outcome across Multiple Cancers. Nat. Genet. 2020;52:891–897. doi: 10.1038/s41588-020-0678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nathanson D.A., Gini B., Mottahedeh J., Visnyei K., Koga T., Gomez G., Eskin A., Hwang K., Wang J., Masui K., et al. Targeted Therapy Resistance Mediated by Dynamic Regulation of Extrachromosomal Mutant EGFR DNA. Science. 2014;343:72–76. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu K., Ding L., Chang T.-C., Shao Y., Chiang J., Mulder H., Wang S., Shaw T.I., Wen J., Hover L., et al. Structure and Evolution of Double Minutes in Diagnosis and Relapse Brain Tumors. Acta Neuropathol. 2019;137:123–137. doi: 10.1007/s00401-018-1912-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cortés-Ciriano I., Lee J.J.-K., Xi R., Jain D., Jung Y.L., Yang L., Gordenin D., Klimczak L.J., Zhang C.-Z., Pellman D.S., et al. Comprehensive Analysis of Chromothripsis in 2,658 Human Cancers Using Whole-Genome Sequencing. Nat. Genet. 2020;52:331–341. doi: 10.1038/s41588-019-0576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Voronina N., Wong J.K.L., Hübschmann D., Hlevnjak M., Uhrig S., Heilig C.E., Horak P., Kreutzfeldt S., Mock A., Stenzinger A., et al. The Landscape of Chromothripsis across Adult Cancer Types. Nat. Commun. 2020;11:2320. doi: 10.1038/s41467-020-16134-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Johnson R.A., Wright K.D., Poppleton H., Mohankumar K.M., Finkelstein D., Pounds S.B., Rand V., Leary S.E.S., White E., Eden C., et al. Cross-Species Genomics Matches Driver Mutations and Cell Compartments to Model Ependymoma. Nature. 2010;466:632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mohankumar K.M., Currle D.S., White E., Boulos N., Dapper J., Eden C., Nimmervoll B., Thiruvenkatam R., Connelly M., Kranenburg T.A., et al. An in Vivo Screen Identifies Ependymoma Oncogenes and Tumor-Suppressor Genes. Nat. Genet. 2015;47:878–887. doi: 10.1038/ng.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ozawa T., Arora S., Szulzewsky F., Juric-Sekhar G., Miyajima Y., Bolouri H., Yasui Y., Barber J., Kupp R., Dalton J., et al. A De Novo Mouse Model of C11orf95-RELA Fusion-Driven Ependymoma Identifies Driver Functions in Addition to NF-ΚB. Cell Rep. 2018;23:3787–3797. doi: 10.1016/j.celrep.2018.04.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Takadera M., Satomi K., Szulzewsky F., Cimino P.J., Holland E.C., Yamamoto T., Ichimura K., Ozawa T. Phenotypic Characterization with Somatic Genome Editing and Gene Transfer Reveals the Diverse Oncogenicity of Ependymoma Fusion Genes. Acta Neuropathol. Commun. 2020;8:203. doi: 10.1186/s40478-020-01080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Almeida Magalhães T., Cruzeiro G.A.V., de Sousa G.R., da Silva K.R., Lira R.C.P., Scrideli C.A., Tone L.G., Valera E.T., Borges K.S. Notch Pathway in Ependymoma RELA-Fused Subgroup: Upregulation and Association with Cancer Stem Cells Markers Expression. Cancer Gene Ther. 2020;27:509–512. doi: 10.1038/s41417-019-0122-x. [DOI] [PubMed] [Google Scholar]
- 105.Zhu J.J., Jillette N., Li X.-N., Cheng A.W., Lau C.C. C11orf95-RELA Reprograms 3D Epigenome in Supratentorial Ependymoma. Acta Neuropathol. 2020;140:951–960. doi: 10.1007/s00401-020-02225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Arabzade A., Zhao Y., Varadharajan S., Chen H.-C., Jessa S., Rivas B., Stuckert A.J., Solis M., Kardian A., Tlais D., et al. ZFTA-RELA Dictates Oncogenic Transcriptional Programs to Drive Aggressive Supratentorial Ependymoma. Cancer Discov. 2021;11:2200–2215. doi: 10.1158/2159-8290.CD-20-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kupp R., Ruff L., Terranova S., Nathan E., Ballereau S., Stark R., Sekhar Reddy Chilamakuri C., Hoffmann N., Wickham-Rahrmann K., Widdess M., et al. ZFTA-Translocations Constitute Ependymoma Chromatin Remodeling and Transcription Factors. Cancer Discov. 2021;11:2216–2229. doi: 10.1158/2159-8290.CD-20-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ozawa T., Kaneko S., Szulzewsky F., Qiao Z., Takadera M., Narita Y., Kondo T., Holland E.C., Hamamoto R., Ichimura K. C11orf95-RELA Fusion Drives Aberrant Gene Expression through the Unique Epigenetic Regulation for Ependymoma Formation. Acta Neuropathol. Commun. 2021;9:36. doi: 10.1186/s40478-021-01135-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zeng Q., Hong W. The Emerging Role of the Hippo Pathway in Cell Contact Inhibition, Organ Size Control, and Cancer Development in Mammals. Cancer Cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 110.Szulzewsky F., Arora S., Hoellerbauer P., King C., Nathan E., Chan M., Cimino P.J., Ozawa T., Kawauchi D., Pajtler K.W., et al. Comparison of Tumor-Associated YAP1 Fusions Identifies a Recurrent Set of Functions Critical for Oncogenesis. Genes Dev. 2020;34:1051–1064. doi: 10.1101/gad.338681.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tabasaran J., Schuhmann M., Ebinger M., Honegger J., Renovanz M., Schittenhelm J. PAX6 Is Frequently Expressed in Ependymal Tumours and Associated with Prognostic Relevant Subgroups. J. Clin. Pathol. 2021 doi: 10.1136/jclinpath-2021-207526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brock A., Chang H., Huang S. Non-Genetic Heterogeneity—A Mutation-Independent Driving Force for the Somatic Evolution of Tumours. Nat. Rev. Genet. 2009;10:336–342. doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- 113.Carja O., Plotkin J.B. The Evolutionary Advantage of Heritable Phenotypic Heterogeneity. Sci. Rep. 2017;7:5090. doi: 10.1038/s41598-017-05214-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shen H., Laird P.W. Interplay between the Cancer Genome and Epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chalmers Z.R., Connelly C.F., Fabrizio D., Gay L., Ali S.M., Ennis R., Schrock A., Campbell B., Shlien A., Chmielecki J., et al. Analysis of 100,000 Human Cancer Genomes Reveals the Landscape of Tumor Mutational Burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Capper D., Jones D.T.W., Sill M., Hovestadt V., Schrimpf D., Sturm D., Koelsche C., Sahm F., Chavez L., Reuss D.E., et al. DNA Methylation-Based Classification of Central Nervous System Tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gröbner S.N., Worst B.C., Weischenfeldt J., Buchhalter I., Kleinheinz K., Rudneva V.A., Johann P.D., Balasubramanian G.P., Segura-Wang M., Brabetz S., et al. The Landscape of Genomic Alterations across Childhood Cancers. Nature. 2018;555:321–327. doi: 10.1038/nature25480. [DOI] [PubMed] [Google Scholar]
- 118.Huether R., Dong L., Chen X., Wu G., Parker M., Wei L., Ma J., Edmonson M.N., Hedlund E.K., Rusch M.C., et al. The Landscape of Somatic Mutations in Epigenetic Regulators across 1,000 Paediatric Cancer Genomes. Nat. Commun. 2014;5:3630. doi: 10.1038/ncomms4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu G., Broniscer A., McEachron T.A., Lu C., Paugh B.S., Becksfort J., Qu C., Ding L., Huether R., Parker M., et al. Somatic Histone H3 Alterations in Pediatric Diffuse Intrinsic Pontine Gliomas and Non-Brainstem Glioblastomas. Nat. Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mackay A., Burford A., Carvalho D., Izquierdo E., Fazal-Salom J., Taylor K.R., Bjerke L., Clarke M., Vinci M., Nandhabalan M., et al. Integrated Molecular Meta-Analysis of 1000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell. 2017;32:520–537.e5. doi: 10.1016/j.ccell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Carter B., Zhao K. The Epigenetic Basis of Cellular Heterogeneity. Nat. Rev. Genet. 2021;22:235–250. doi: 10.1038/s41576-020-00300-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Klemm S.L., Shipony Z., Greenleaf W.J. Chromatin Accessibility and the Regulatory Epigenome. Nat. Rev. Genet. 2019;20:207–220. doi: 10.1038/s41576-018-0089-8. [DOI] [PubMed] [Google Scholar]
- 123.Flavahan W.A., Gaskell E., Bernstein B.E. Epigenetic Plasticity and the Hallmarks of Cancer. Science. 2017;357:eaal2380. doi: 10.1126/science.aal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mack S.C., Hubert C.G., Miller T.E., Taylor M.D., Rich J.N. An Epigenetic Gateway to Brain Tumor Cell Identity. Nat. Neurosci. 2016;19:10–19. doi: 10.1038/nn.4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Comet I., Riising E.M., Leblanc B., Helin K. Maintaining Cell Identity: PRC2-Mediated Regulation of Transcription and Cancer. Nat. Rev. Cancer. 2016;16:803–810. doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 126.Shen L., Kondo Y., Rosner G.L., Xiao L., Hernandez N.S., Vilaythong J., Houlihan P.S., Krouse R.S., Prasad A.R., Einspahr J.G., et al. MGMT Promoter Methylation and Field Defect in Sporadic Colorectal Cancer. J. Natl. Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 127.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-Enhancers in the Control of Cell Identity and Disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Brocks D., Assenov Y., Minner S., Bogatyrova O., Simon R., Koop C., Oakes C., Zucknick M., Lipka D.B., Weischenfeldt J., et al. Intratumor DNA Methylation Heterogeneity Reflects Clonal Evolution in Aggressive Prostate Cancer. Cell Rep. 2014;8:798–806. doi: 10.1016/j.celrep.2014.06.053. [DOI] [PubMed] [Google Scholar]
- 129.Song Y., van den Berg P.R., Markoulaki S., Soldner F., Dall’Agnese A., Henninger J.E., Drotar J., Rosenau N., Cohen M.A., Young R.A., et al. Dynamic Enhancer DNA Methylation as Basis for Transcriptional and Cellular Heterogeneity of ESCs. Mol. Cell. 2019;75:905–920.e6. doi: 10.1016/j.molcel.2019.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bell R.E., Golan T., Sheinboim D., Malcov H., Amar D., Salamon A., Liron T., Gelfman S., Gabet Y., Shamir R., et al. Enhancer Methylation Dynamics Contribute to Cancer Plasticity and Patient Mortality. Genome Res. 2016;26:601–611. doi: 10.1101/gr.197194.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mazor T., Pankov A., Song J.S., Costello J.F. Intratumoral Heterogeneity of the Epigenome. Cancer Cell. 2016;29:440–451. doi: 10.1016/j.ccell.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang D., Holsten T., Börnigen D., Frank S., Mawrin C., Glatzel M., Schüller U. Ependymoma Relapse Goes along with a Relatively Stable Epigenome, but a Severely Altered Tumor Morphology. Brain Pathol. 2021;31:33–44. doi: 10.1111/bpa.12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Guo M., Peng Y., Gao A., Du C., Herman J.G. Epigenetic Heterogeneity in Cancer. Biomark. Res. 2019;7:23. doi: 10.1186/s40364-019-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mazor T., Pankov A., Johnson B.E., Hong C., Hamilton E.G., Bell R.J.A., Smirnov I.V., Reis G.F., Phillips J.J., Barnes M.J., et al. DNA Methylation and Somatic Mutations Converge on the Cell Cycle and Define Similar Evolutionary Histories in Brain Tumors. Cancer Cell. 2015;28:307–317. doi: 10.1016/j.ccell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Magill S.T., Vasudevan H.N., Seo K., Villanueva-Meyer J.E., Choudhury A., John Liu S., Pekmezci M., Findakly S., Hilz S., Lastella S., et al. Multiplatform Genomic Profiling and Magnetic Resonance Imaging Identify Mechanisms Underlying Intratumor Heterogeneity in Meningioma. Nat. Commun. 2020;11:4803. doi: 10.1038/s41467-020-18582-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu S.J., Magill S.T., Vasudevan H.N., Hilz S., Villanueva-Meyer J.E., Lastella S., Daggubati V., Spatz J., Choudhury A., Orr B.A., et al. Multiplatform Molecular Profiling Reveals Epigenomic Intratumor Heterogeneity in Ependymoma. Cell Rep. 2020;30:1300–1309.e5. doi: 10.1016/j.celrep.2020.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bender S., Tang Y., Lindroth A.M., Hovestadt V., Jones D.T.W., Kool M., Zapatka M., Northcott P.A., Sturm D., Wang W., et al. Reduced H3K27me3 and DNA Hypomethylation Are Major Drivers of Gene Expression in K27M Mutant Pediatric High-Grade Gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 138.Krug B., Harutyunyan A.S., Deshmukh S., Jabado N. Polycomb Repressive Complex 2 in the Driver’s Seat of Childhood and Young Adult Brain Tumours. Trends Cell Biol. 2021;31:814–828. doi: 10.1016/j.tcb.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schlesinger Y., Straussman R., Keshet I., Farkash S., Hecht M., Zimmerman J., Eden E., Yakhini Z., Ben-Shushan E., Reubinoff B.E., et al. Polycomb-Mediated Methylation on Lys27 of Histone H3 Pre-Marks Genes for de Novo Methylation in Cancer. Nat. Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 140.Suvà M.-L., Riggi N., Janiszewska M., Radovanovic I., Provero P., Stehle J.-C., Baumer K., Le Bitoux M.-A., Marino D., Cironi L., et al. EZH2 Is Essential for Glioblastoma Cancer Stem Cell Maintenance. Cancer Res. 2009;69:9211–9218. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- 141.Lewis P.W., Müller M.M., Koletsky M.S., Cordero F., Lin S., Banaszynski L.A., Garcia B.A., Muir T.W., Becher O.J., Allis C.D. Inhibition of PRC2 Activity by a Gain-of-Function H3 Mutation Found in Pediatric Glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Larson J.D., Kasper L.H., Paugh B.S., Jin H., Wu G., Kwon C.-H., Fan Y., Shaw T.I., Silveira A.B., Qu C., et al. Histone H3.3 K27M Accelerates Spontaneous Brainstem Glioma and Drives Restricted Changes in Bivalent Gene Expression. Cancer Cell. 2019;35:140–155.e7. doi: 10.1016/j.ccell.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jessa S., Blanchet-Cohen A., Krug B., Vladoiu M., Coutelier M., Faury D., Poreau B., De Jay N., Hébert S., Monlong J., et al. Stalled Developmental Programs at the Root of Pediatric Brain Tumors. Nat. Genet. 2019;51:1702–1713. doi: 10.1038/s41588-019-0531-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mills A.A. Throwing the Cancer Switch: Reciprocal Roles of Polycomb and Trithorax Proteins. Nat. Rev. Cancer. 2010;10:669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hübner J.-M., Müller T., Papageorgiou D.N., Mauermann M., Krijgsveld J., Russell R.B., Ellison D.W., Pfister S.M., Pajtler K.W., Kool M. EZHIP/CXorf67 Mimics K27M Mutated Oncohistones and Functions as an Intrinsic Inhibitor of PRC2 Function in Aggressive Posterior Fossa Ependymoma. Neuro Oncol. 2019;21:878–889. doi: 10.1093/neuonc/noz058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jain S.U., Do T.J., Lund P.J., Rashoff A.Q., Diehl K.L., Cieslik M., Bajic A., Juretic N., Deshmukh S., Venneti S., et al. PFA Ependymoma-Associated Protein EZHIP Inhibits PRC2 Activity through a H3 K27M-like Mechanism. Nat. Commun. 2019;10:2146. doi: 10.1038/s41467-019-09981-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Piunti A., Smith E.R., Morgan M.A.J., Ugarenko M., Khaltyan N., Helmin K.A., Ryan C.A., Murray D.C., Rickels R.A., Yilmaz B.D., et al. CATACOMB: An Endogenous Inducible Gene That Antagonizes H3K27 Methylation Activity of Polycomb Repressive Complex 2 via an H3K27M-like Mechanism. Sci. Adv. 2019;5:eaax2887. doi: 10.1126/sciadv.aax2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nambirajan A., Sharma A., Rajeshwari M., Boorgula M.T., Doddamani R., Garg A., Suri V., Sarkar C., Sharma M.C. EZH2 Inhibitory Protein (EZHIP/Cxorf67) Expression Correlates Strongly with H3K27me3 Loss in Posterior Fossa Ependymomas and Is Mutually Exclusive with H3K27M Mutations. Brain Tumor Pathol. 2021;38:30–40. doi: 10.1007/s10014-020-00385-9. [DOI] [PubMed] [Google Scholar]
- 149.Harutyunyan A.S., Krug B., Chen H., Papillon-Cavanagh S., Zeinieh M., De Jay N., Deshmukh S., Chen C.C.L., Belle J., Mikael L.G., et al. H3K27M Induces Defective Chromatin Spread of PRC2-Mediated Repressive H3K27me2/Me3 and Is Essential for Glioma Tumorigenesis. Nat. Commun. 2019;10:1262. doi: 10.1038/s41467-019-09140-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jain S.U., Rashoff A.Q., Krabbenhoft S.D., Hoelper D., Do T.J., Gibson T.J., Lundgren S.M., Bondra E.R., Deshmukh S., Harutyunyan A.S., et al. H3 K27M and EZHIP Impede H3K27-Methylation Spreading by Inhibiting Allosterically Stimulated PRC2. Mol. Cell. 2020;80:726–735.e7. doi: 10.1016/j.molcel.2020.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Michealraj K.A., Kumar S.A., Kim L.J.Y., Cavalli F.M.G., Przelicki D., Wojcik J.B., Delaidelli A., Bajic A., Saulnier O., MacLeod G., et al. Metabolic Regulation of the Epigenome Drives Lethal Infantile Ependymoma. Cell. 2020;181:1329–1345.e24. doi: 10.1016/j.cell.2020.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yuan Y. Spatial Heterogeneity in the Tumor Microenvironment. Cold Spring Harb. Perspect. Med. 2016;6:a026583. doi: 10.1101/cshperspect.a026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gillies R.J., Brown J.S., Anderson A.R.A., Gatenby R.A. Eco-Evolutionary Causes and Consequences of Temporal Changes in Intratumoural Blood Flow. Nat. Rev. Cancer. 2018;18:576–585. doi: 10.1038/s41568-018-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hoefflin R., Lahrmann B., Warsow G., Hübschmann D., Spath C., Walter B., Chen X., Hofer L., Macher-Goeppinger S., Tolstov Y., et al. Spatial Niche Formation but Not Malignant Progression Is a Driving Force for Intratumoural Heterogeneity. Nat. Commun. 2016;7:1–12. doi: 10.1038/ncomms11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hubert C.G., Rivera M., Spangler L.C., Wu Q., Mack S.C., Prager B.C., Couce M., McLendon R.E., Sloan A.E., Rich J.N. A Three-Dimensional Organoid Culture System Derived from Human Glioblastomas Recapitulates the Hypoxic Gradients and Cancer Stem Cell Heterogeneity of Tumors Found In Vivo. Cancer Res. 2016;76:2465–2477. doi: 10.1158/0008-5472.CAN-15-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Estrella V., Chen T., Lloyd M., Wojtkowiak J., Cornnell H.H., Ibrahim-Hashim A., Bailey K., Balagurunathan Y., Rothberg J.M., Sloane B.F., et al. Acidity Generated by the Tumor Microenvironment Drives Local Invasion. Cancer Res. 2013;73:1524–1535. doi: 10.1158/0008-5472.CAN-12-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Korenchan D.E., Flavell R.R. Spatiotemporal PH Heterogeneity as a Promoter of Cancer Progression and Therapeutic Resistance. Cancers. 2019;11:1026. doi: 10.3390/cancers11071026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bastola S., Pavlyukov M.S., Yamashita D., Ghosh S., Cho H., Kagaya N., Zhang Z., Minata M., Lee Y., Sadahiro H., et al. Glioma-Initiating Cells at Tumor Edge Gain Signals from Tumor Core Cells to Promote Their Malignancy. Nat. Commun. 2020;11:4660. doi: 10.1038/s41467-020-18189-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vander Heiden M.G., DeBerardinis R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Soeda A., Park M., Lee D., Mintz A., Androutsellis-Theotokis A., McKay R.D., Engh J., Iwama T., Kunisada T., Kassam A.B., et al. Hypoxia Promotes Expansion of the CD133-Positive Glioma Stem Cells through Activation of HIF-1alpha. Oncogene. 2009;28:3949–3959. doi: 10.1038/onc.2009.252. [DOI] [PubMed] [Google Scholar]
- 161.Peleli M., Moustakas A., Papapetropoulos A. Endothelial-Tumor Cell Interaction in Brain and CNS Malignancies. Int. J. Mol. Sci. 2020;21:7371. doi: 10.3390/ijms21197371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Griesinger A.M., Josephson R.J., Donson A.M., Mulcahy Levy J.M., Amani V., Birks D.K., Hoffman L.M., Furtek S.L., Reigan P., Handler M.H., et al. Interleukin-6/STAT3 Pathway Signaling Drives an Inflammatory Phenotype in Group A Ependymoma. Cancer Immunol. Res. 2015;3:1165–1174. doi: 10.1158/2326-6066.CIR-15-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Calabrese C., Poppleton H., Kocak M., Hogg T.L., Fuller C., Hamner B., Oh E.Y., Gaber M.W., Finklestein D., Allen M., et al. A Perivascular Niche for Brain Tumor Stem Cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 164.Gillen A.E., Riemondy K.A., Amani V., Griesinger A.M., Gilani A., Venkataraman S., Madhavan K., Prince E., Sanford B., Hankinson T.C., et al. Single-Cell RNA Sequencing of Childhood Ependymoma Reveals Neoplastic Cell Subpopulations That Impact Molecular Classification and Etiology. Cell Rep. 2020;32:108023. doi: 10.1016/j.celrep.2020.108023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gojo J., Englinger B., Jiang L., Hübner J.M., Shaw M.L., Hack O.A., Madlener S., Kirchhofer D., Liu I., Pyrdol J., et al. Single-Cell RNA-Seq Reveals Cellular Hierarchies and Impaired Developmental Trajectories in Pediatric Ependymoma. Cancer Cell. 2020;38:44–59.e9. doi: 10.1016/j.ccell.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Boyd N.H., Tran A.N., Bernstock J.D., Etminan T., Jones A.B., Gillespie G.Y., Friedman G.K., Hjelmeland A.B. Glioma Stem Cells and Their Roles within the Hypoxic Tumor Microenvironment. Theranostics. 2021;11:665–683. doi: 10.7150/thno.41692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chiche J., Brahimi-Horn M.C., Pouysségur J. Tumour Hypoxia Induces a Metabolic Shift Causing Acidosis: A Common Feature in Cancer. J. Cell. Mol. Med. 2010;14:771–794. doi: 10.1111/j.1582-4934.2009.00994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Fukumura D., Xu L., Chen Y., Gohongi T., Seed B., Jain R.K. Hypoxia and Acidosis Independently Up-Regulate Vascular Endothelial Growth Factor Transcription in Brain Tumors in Vivo. Cancer Res. 2001;61:6020–6024. [PubMed] [Google Scholar]
- 169.Mohyeldin A., Garzón-Muvdi T., Quiñones-Hinojosa A. Oxygen in Stem Cell Biology: A Critical Component of the Stem Cell Niche. Cell Stem Cell. 2010;7:150–161. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 170.Pistollato F., Chen H.-L., Schwartz P.H., Basso G., Panchision D.M. Oxygen Tension Controls the Expansion of Human CNS Precursors and the Generation of Astrocytes and Oligodendrocytes. Mol. Cell. Neurosci. 2007;35:424–435. doi: 10.1016/j.mcn.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 171.Flavahan W.A., Wu Q., Hitomi M., Rahim N., Kim Y., Sloan A.E., Weil R.J., Nakano I., Sarkaria J.N., Stringer B.W., et al. Brain Tumor Initiating Cells Adapt to Restricted Nutrition through Preferential Glucose Uptake. Nat. Neurosci. 2013;16:1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Englinger B., Hack O.A., Filbin M.G. Into Thin Air: Hypoxia Drives Metabolic and Epigenomic Deregulation of Lethal Pediatric Ependymoma. Dev. Cell. 2020;54:134–136. doi: 10.1016/j.devcel.2020.06.020. [DOI] [PubMed] [Google Scholar]
- 173.Vladoiu M.C., El-Hamamy I., Donovan L.K., Farooq H., Holgado B.L., Sundaravadanam Y., Ramaswamy V., Hendrikse L.D., Kumar S., Mack S.C., et al. Childhood Cerebellar Tumours Mirror Conserved Fetal Transcriptional Programs. Nature. 2019;572:67–73. doi: 10.1038/s41586-019-1158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liau B.B., Sievers C., Donohue L.K., Gillespie S.M., Flavahan W.A., Miller T.E., Venteicher A.S., Hebert C.H., Carey C.D., Rodig S.J., et al. Adaptive Chromatin Remodeling Drives Glioblastoma Stem Cell Plasticity and Drug Tolerance. Cell Stem Cell. 2017;20:233–246.e7. doi: 10.1016/j.stem.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.McBrian M.A., Behbahan I.S., Ferrari R., Su T., Huang T.-W., Li K., Hong C.S., Christofk H.R., Vogelauer M., Seligson D.B., et al. Histone Acetylation Regulates Intracellular PH. Mol. Cell. 2013;49:310–321. doi: 10.1016/j.molcel.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bulusu V., Tumanov S., Michalopoulou E., van den Broek N.J., MacKay G., Nixon C., Dhayade S., Schug Z.T., Vande Voorde J., Blyth K., et al. Acetate Recapturing by Nuclear Acetyl-CoA Synthetase 2 Prevents Loss of Histone Acetylation during Oxygen and Serum Limitation. Cell Rep. 2017;18:647–658. doi: 10.1016/j.celrep.2016.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kindrick J.D., Mole D.R. Hypoxic Regulation of Gene Transcription and Chromatin: Cause and Effect. Int. J. Mol. Sci. 2020;21:8320. doi: 10.3390/ijms21218320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Thienpont B., Steinbacher J., Zhao H., D’Anna F., Kuchnio A., Ploumakis A., Ghesquière B., Van Dyck L., Boeckx B., Schoonjans L., et al. Tumour Hypoxia Causes DNA Hypermethylation by Reducing TET Activity. Nature. 2016;537:63–68. doi: 10.1038/nature19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ramsawhook A., Lewis L., Coyle B., Ruzov A. Medulloblastoma and Ependymoma Cells Display Increased Levels of 5-Carboxylcytosine and Elevated TET1 Expression. Clin. Epigenet. 2017;9:18. doi: 10.1186/s13148-016-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Brabletz S., Schuhwerk H., Brabletz T., Stemmler M.P. Dynamic EMT: A Multi-Tool for Tumor Progression. EMBO J. 2021;40:e108647. doi: 10.15252/embj.2021108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Puisieux A., Brabletz T., Caramel J. Oncogenic Roles of EMT-Inducing Transcription Factors. Nat. Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 182.Nieto M.A., Huang R.Y.-J., Jackson R.A., Thiery J.P. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 183.Mani S.A., Guo W., Liao M.-J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shibue T., Weinberg R.A. EMT, CSCs, and Drug Resistance: The Mechanistic Link and Clinical Implications. Nat. Rev. Clin. Oncol. 2017;14:611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ma Q., Shu C., Wang J. Bioinformatics Analysis of Microarray Data Reveals Epithelial-Mesenchymal-Transition in Pediatric Ependymoma. Anticancer. Drugs. 2021;32:437–447. doi: 10.1097/CAD.0000000000001046. [DOI] [PubMed] [Google Scholar]
- 186.Kalluri R., Weinberg R.A. The Basics of Epithelial-Mesenchymal Transition. J. Clin. Investig. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Malgulwar P.B., Nambirajan A., Pathak P., Rajeshwari M., Suri V., Sarkar C., Singh M., Sharma M.C. Epithelial-to-Mesenchymal Transition-Related Transcription Factors Are up-Regulated in Ependymomas and Correlate with a Poor Prognosis. Hum. Pathol. 2018;82:149–157. doi: 10.1016/j.humpath.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 188.Wani K., Armstrong T.S., Vera-Bolanos E., Raghunathan A., Ellison D., Gilbertson R., Vaillant B., Goldman S., Packer R.J., Fouladi M., et al. A Prognostic Gene Expression Signature in Infratentorial Ependymoma. Acta Neuropathol. 2012;123:727–738. doi: 10.1007/s00401-012-0941-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lawson D.A., Kessenbrock K., Davis R.T., Pervolarakis N., Werb Z. Tumour Heterogeneity and Metastasis at Single-Cell Resolution. Nat. Cell Biol. 2018;20:1349–1360. doi: 10.1038/s41556-018-0236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Filbin M.G., Tirosh I., Hovestadt V., Shaw M.L., Escalante L.E., Mathewson N.D., Neftel C., Frank N., Pelton K., Hebert C.M., et al. Developmental and Oncogenic Programs in H3K27M Gliomas Dissected by Single-Cell RNA-Seq. Science. 2018;360:331–335. doi: 10.1126/science.aao4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L., et al. Single-Cell RNA-Seq Highlights Intratumoral Heterogeneity in Primary Glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chung M.-I., Peyrot S.M., LeBoeuf S., Park T.J., McGary K.L., Marcotte E.M., Wallingford J.B. RFX2 Is Broadly Required for Ciliogenesis during Vertebrate Development. Dev. Biol. 2012;363:155–165. doi: 10.1016/j.ydbio.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yu X., Ng C.P., Habacher H., Roy S. Foxj1 Transcription Factors Are Master Regulators of the Motile Ciliogenic Program. Nat. Genet. 2008;40:1445–1453. doi: 10.1038/ng.263. [DOI] [PubMed] [Google Scholar]
- 194.Nielsen S., Nagelhus E.A., Amiry-Moghaddam M., Bourque C., Agre P., Ottersen O.P. Specialized Membrane Domains for Water Transport in Glial Cells: High-Resolution Immunogold Cytochemistry of Aquaporin-4 in Rat Brain. J. Neurosci. Off. J. Soc. Neurosci. 1997;17:171–180. doi: 10.1523/JNEUROSCI.17-01-00171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wang D., Owler B.K. Expression of AQP1 and AQP4 in Paediatric Brain Tumours. J. Clin. Neurosci. Off. J. Neurosurg. Soc. Australas. 2011;18:122–127. doi: 10.1016/j.jocn.2010.07.115. [DOI] [PubMed] [Google Scholar]
- 196.Noell S., Fallier-Becker P., Mack A.F., Hoffmeister M., Beschorner R., Ritz R. Water Channels Aquaporin 4 and -1 Expression in Subependymoma Depends on the Localization of the Tumors. PLoS ONE. 2015;10:e0131367. doi: 10.1371/journal.pone.0131367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Han S., Dennis D.J., Balakrishnan A., Dixit R., Britz O., Zinyk D., Touahri Y., Olender T., Brand M., Guillemot F., et al. A Non-Canonical Role for the Proneural Gene Neurog1 as a Negative Regulator of Neocortical Neurogenesis. Dev. Camb. Engl. 2018;145:dev157719. doi: 10.1242/dev.157719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Stauber M., Weidemann M., Dittrich-Breiholz O., Lobschat K., Alten L., Mai M., Beckers A., Kracht M., Gossler A. Identification of FOXJ1 Effectors during Ciliogenesis in the Foetal Respiratory Epithelium and Embryonic Left-Right Organiser of the Mouse. Dev. Biol. 2017;423:170–188. doi: 10.1016/j.ydbio.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 199.Mack S.C., Pajtler K.W., Chavez L., Okonechnikov K., Bertrand K.C., Wang X., Erkek S., Federation A., Song A., Lee C., et al. Therapeutic Targeting of Ependymoma as Informed by Oncogenic Enhancer Profiling. Nature. 2018;553:101–105. doi: 10.1038/nature25169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hartfuss E., Galli R., Heins N., Götz M. Characterization of CNS Precursor Subtypes and Radial Glia. Dev. Biol. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- 201.Rowitch D.H., Kriegstein A.R. Developmental Genetics of Vertebrate Glial-Cell Specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 202.Pollen A.A., Nowakowski T.J., Chen J., Retallack H., Sandoval-Espinosa C., Nicholas C.R., Shuga J., Liu S.J., Oldham M.C., Diaz A., et al. Molecular Identity of Human Outer Radial Glia during Cortical Development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Thomsen E.R., Mich J.K., Yao Z., Hodge R.D., Doyle A.M., Jang S., Shehata S.I., Nelson A.M., Shapovalova N.V., Levi B.P., et al. Fixed Single-Cell Transcriptomic Characterization of Human Radial Glial Diversity. Nat. Methods. 2016;13:87–93. doi: 10.1038/nmeth.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Liu Z., Lou H., Xie K., Wang H., Chen N., Aparicio O.M., Zhang M.Q., Jiang R., Chen T. Reconstructing Cell Cycle Pseudo Time-Series via Single-Cell Transcriptome Data. Nat. Commun. 2017;8:22. doi: 10.1038/s41467-017-00039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Saelens W., Cannoodt R., Todorov H., Saeys Y. A Comparison of Single-Cell Trajectory Inference Methods. Nat. Biotechnol. 2019;37:547–554. doi: 10.1038/s41587-019-0071-9. [DOI] [PubMed] [Google Scholar]
- 206.Mack S.C., Bertrand K.C. Sub-Group, Sub-Type, and Cell-Type Heterogeneity of Ependymoma. Cancer Cell. 2020;38:15–17. doi: 10.1016/j.ccell.2020.06.010. [DOI] [PubMed] [Google Scholar]
- 207.Jenseit A., Camgöz A., Pfister S.M., Kool M. EZHIP: A New Piece of the Puzzle towards Understanding Pediatric Posterior Fossa Ependymoma. Acta Neuropathol. 2021:1–13. doi: 10.1007/s00401-021-02382-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Han J., Yu M., Bai Y., Yu J., Jin F., Li C., Zeng R., Peng J., Li A., Song X., et al. Elevated CXorf67 Expression in PFA Ependymomas Suppresses DNA Repair and Sensitizes to PARP Inhibitors. Cancer Cell. 2020;38:844–856.e7. doi: 10.1016/j.ccell.2020.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Panwalkar P., Tamrazi B., Dang D., Chung C., Sweha S., Natarajan S.K., Pun M., Bayliss J., Ogrodzinski M.P., Pratt D., et al. Targeting Integrated Epigenetic and Metabolic Pathways in Lethal Childhood PFA Ependymomas. Sci. Transl. Med. 2021;13:eabc0497. doi: 10.1126/scitranslmed.abc0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Duan R., Du W., Guo W. EZH2: A Novel Target for Cancer Treatment. J. Hematol. Oncol. J. Hematol. Oncol. 2020;13:104. doi: 10.1186/s13045-020-00937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Medeiros M., Candido M., Valera E., Brassesco M. The Multifaceted NF-KB: Are There Still Prospects of Its Inhibition for Clinical Intervention in Pediatric Central Nervous System Tumors? Cell Mol. Life Sci. 2021;78:6161–6200. doi: 10.1007/s00018-021-03906-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Armstrong T.S., Gilbert M.R. Clinical Trial Challenges, Design Considerations, and Outcome Measures in Rare CNS Tumors. Neuro Oncol. 2021;23:S30–S38. doi: 10.1093/neuonc/noab209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Di K., Lloyd G.K., Abraham V., MacLaren A., Burrows F.J., Desjardins A., Trikha M., Bota D.A. Marizomib Activity as a Single Agent in Malignant Gliomas: Ability to Cross the Blood-Brain Barrier. Neuro Oncol. 2016;18:840–848. doi: 10.1093/neuonc/nov299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Donson A.M., Amani V., Warner E.A., Griesinger A.M., Witt D.A., Levy J.M.M., Hoffman L.M., Hankinson T.C., Handler M.H., Vibhakar R., et al. Identification of FDA-Approved Oncology Drugs with Selective Potency in High-Risk Childhood Ependymoma. Mol. Cancer Ther. 2018;17:1984–1994. doi: 10.1158/1535-7163.MCT-17-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Antonelli R., Jiménez C., Riley M., Servidei T., Riccardi R., Soriano A., Roma J., Martínez-Saez E., Martini M., Ruggiero A., et al. CN133, a Novel Brain-Penetrating Histone Deacetylase Inhibitor, Hampers Tumor Growth in Patient-Derived Pediatric Posterior Fossa Ependymoma Models. Cancers. 2020;12:1922. doi: 10.3390/cancers12071922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Servidei T., Meco D., Martini M., Battaglia A., Granitto A., Buzzonetti A., Babini G., Massimi L., Tamburrini G., Scambia G., et al. The BET Inhibitor OTX015 Exhibits In Vitro and In Vivo Antitumor Activity in Pediatric Ependymoma Stem Cell Models. Int. J. Mol. Sci. 2021;22:1877. doi: 10.3390/ijms22041877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kukreja L., Li C.J., Ezhilan S., Iyer V.R., Kuo J.S. Emerging Epigenetic Therapies for Brain Tumors. Neuromol. Med. 2021:1–9. doi: 10.1007/s12017-021-08691-x. [DOI] [PubMed] [Google Scholar]
- 218.Lötsch D., Kirchhofer D., Englinger B., Jiang L., Okonechnikov K., Senfter D., Laemmerer A., Gabler L., Pirker C., Donson A.M., et al. Targeting Fibroblast Growth Factor Receptors to Combat Aggressive Ependymoma. Acta Neuropathol. 2021;142:339–360. doi: 10.1007/s00401-021-02327-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Vega-Benedetti A.F., Saucedo C., Zavattari P., Vanni R., Zugaza J.L., Parada L.A. PLAGL1: An Important Player in Diverse Pathological Processes. J. Appl. Genet. 2017;58:71–78. doi: 10.1007/s13353-016-0355-4. [DOI] [PubMed] [Google Scholar]
- 220.Jünger S.T., Mynarek M., Wohlers I., Dörner E., Mühlen A.Z., Velez-Char N., von Hoff K., Rutkowski S., Warmuth-Metz M., Kortmann R.-D., et al. Improved Risk-Stratification for Posterior Fossa Ependymoma of Childhood Considering Clinical, Histological and Genetic Features—A Retrospective Analysis of the HIT Ependymoma Trial Cohort. Acta Neuropathol. Commun. 2019;7:181. doi: 10.1186/s40478-019-0820-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Rudà R., Reifenberger G., Frappaz D., Pfister S.M., Laprie A., Santarius T., Roth P., Tonn J.C., Soffietti R., Weller M., et al. EANO Guidelines for the Diagnosis and Treatment of Ependymal Tumors. Neuro Oncol. 2018;20:445–456. doi: 10.1093/neuonc/nox166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Khatua S., Mangum R., Bertrand K.C., Zaky W., McCall D., Mack S.C. Pediatric Ependymoma: Current Treatment and Newer Therapeutic Insights. Future Oncol. 2018;14:3175–3186. doi: 10.2217/fon-2018-0502. [DOI] [PubMed] [Google Scholar]
- 223.Adolph J.E., Fleischhack G., Gaab C., Mikasch R., Mynarek M., Rutkowski S., Schüller U., Pfister S.M., Pajtler K.W., Milde T., et al. Systemic Chemotherapy of Pediatric Recurrent Ependymomas: Results from the German HIT-REZ Studies. J. Neurooncol. 2021:1–10. doi: 10.1007/s11060-021-03867-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Home-ClinicalTrials.Gov. [(accessed on 17 November 2021)]; Available online: https://clinicaltrials.gov/
- 225.Jünger S.T., Timmermann B., Pietsch T. Pediatric Ependymoma: An Overview of a Complex Disease. Childs Nerv. Syst. 2021;37:2451–2463. doi: 10.1007/s00381-021-05207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Arakaki A.K.S., Szulzewsky F., Gilbert M.R., Gujral T.S., Holland E.C. Utilizing Preclinical Models to Develop Targeted Therapies for Rare Central Nervous System Cancers. Neuro Oncol. 2021;23:S4–S15. doi: 10.1093/neuonc/noab183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Atkinson J.M., Shelat A.A., Carcaboso A.M., Kranenburg T.A., Arnold L.A., Boulos N., Wright K., Johnson R.A., Poppleton H., Mohankumar K.M., et al. An Integrated in Vitro and in Vivo High-Throughput Screen Identifies Treatment Leads for Ependymoma. Cancer Cell. 2011;20:384–399. doi: 10.1016/j.ccr.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Rogers H.A., Chapman R., Kings H., Allard J., Barron-Hastings J., Pajtler K.W., Sill M., Pfister S., Grundy R.G. Limitations of Current in Vitro Models for Testing the Clinical Potential of Epigenetic Inhibitors for Treatment of Pediatric Ependymoma. Oncotarget. 2018;9:36530–36541. doi: 10.18632/oncotarget.26370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Neftel C., Laffy J., Filbin M.G., Hara T., Shore M.E., Rahme G.J., Richman A.R., Silverbush D., Shaw M.L., Hebert C.M., et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell. 2019;178:835–849.e21. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Petralia F., Tignor N., Reva B., Koptyra M., Chowdhury S., Rykunov D., Krek A., Ma W., Zhu Y., Ji J., et al. Integrated Proteogenomic Characterization across Major Histological Types of Pediatric Brain Cancer. Cell. 2020;183:1962–1985.e31. doi: 10.1016/j.cell.2020.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]