Abstract
Several MYB transcription factors are known to play important roles in plant resistance to environmental stressors. However, the mechanism governing the involvement of MYBs in regulating tobacco mosaic virus (TMV) resistance in plants is still unclear. In this study, we found that not only is Nicotiana benthamiana MYB4‐like involved in defence against TMV, but also that the ethylene pathway participates in MYB4L‐mediated resistance. Transcription of NbMYB4L was up‐regulated in N. benthamiana infected with TMV. Silencing of NbMYB4L led to intensified TMV replication, whereas overexpression of NbMYB4L induced significant resistance to TMV. Transcription of NbMYB4L was greater in 1‐aminocyclopropanecarboxylic acid (ACC, ethylene precursor)‐pretreated plants but lower when the ethylene signalling pathway was blocked during TMV infection. Gene expression analysis showed that the transcription of NbMYB4L was largely suppressed in ETHYLENE INSENSITIVE 3‐like 1(EIL1)‐silenced plants. The results of electrophoretic mobility shift assay and chromatin immunoprecipitation‐quantitative PCR (ChIP‐qPCR) experiments indicated that NbEIL1 could directly bind to two specific regions of the NbMYB4L promoter. Furthermore, a luciferase assay revealed that NbEIL1 significantly induced the reporter activity of the MYB4L promoter in N. benthamiana. These results point to NbEIL1 functioning as a positive regulator of NbMYB4L transcription in N. benthamiana against TMV. Collectively, our work reveals that EIL1 and MYB4L constitute a coherent feed‐forward loop involved in the robust regulation of resistance to TMV in N. benthamiana.
Keywords: Ethylene insensitive 3‐like 1, ethylene, Nicotiana benthamiana, tobacco mosaic virus, transgenic, V‐myb avian myeloblastosis viral oncogene homolog 4‐like
Tobacco mosaic virus infection triggers ethylene accumulation in the host cells, which may activate the ethylene signal transduction pathway in systemically infected leaves, leading to activation of NbMYB4L‐dependent defence responses via NbEIL1.
1. INTRODUCTION
During growth and development, plants have evolved multiple defence mechanisms to withstand the attack of pathogens and pests. Plant hormones, which control all aspects of growth and development, also induce important plant defence pathways against pathogens (Zhao & Li, 2021), leading to the production of various pathogenesis‐related (PR) proteins (Clarke et al., 1998; Hibi et al., 2007; Murray et al., 2005). Also implicated in the activation of defence pathways are transcription factors.
Plant v‐myb avian myeloblastosis viral oncogene homolog (MYB) is one of the largest transcription factor (TF) families. In Arabidopsis thaliana, more than 198 genes have been identified in the MYB superfamily, which includes 126 R2R3‐MYB, five R1R2R3‐MYB, 64 MYB‐related, and three atypical MYB genes (Chen et al., 2006). This gene family has been implicated in controlling cell development, cell cycling, hormone responses, secondary metabolism, and environmental stress responses (Azuma et al., 2008; Lea et al., 2007; Yang & Klessig, 1996; Yang et al., 2007) and several MYBs have also been implicated in various forms of plant defence. For example, AtMYB72 was shown to be required in the early signalling steps in rhizobacteria‐induced systemic resistance in Arabidopsis (Ent et al., 2008). AtMYB96 was found to serve as a molecular link that integrates abscisic acid (ABA) and salicylic acid (SA) signals during pathogen induction of PR genes in Arabidopsis (Seo & Park, 2010) and AtMYB15 is required for the induction of lignin as a component of the basal defence mechanism in the plant innate immune response (Chezem et al., 2017; Kim et al., 2020). Other MYB TFs, including AtMYB30 and poplar MYB115, are also involved in plant pathogen responses (Vailleau et al., 2002; Wang et al., 2017).
In Nicotiana species, MYB1 plays an important role in N gene‐mediated resistance to tobacco mosaic virus (TMV) (Liu et al., 2004). Nicotiana tabacum MYB1 is a typical R2R3 MYB transcription factor. Early studies showed that tobacco MYB1 participated in transcriptional activation of PR genes and TMV resistance (Yang & Klessig, 1996). Although it is clear that MYBs participate in plant disease resistance, the mechanism of action of MYBs and the relationship between MYBs and phytohormones in plant defence against TMV are still unclear.
Plant hormones, including SA and jasmonic acid (JA), have been implicated in plant responses to viruses for many years (Jameson & Clarke, 2002; Zhao & Li, 2021). Ethylene production in infected plants has been associated with defence responses and exogenous application of ethylene or ethylene‐releasing compounds have generally, but not always, been shown to enhance resistance to viruses (Alazem & Lin, 2015; Jameson & Clarke, 2002; Zhao & Li, 2021). Plant resistance responses to TMV infection of Nicotiana species are associated with accumulation of ethylene. For example, in N. tabacum, the precursor of ethylene, 1‐aminocyclopropane‐1‐carboxylic acid (ACC), accumulates locally around TMV‐induced necroses, along with an early increase in the activity of the rate‐limiting enzyme of ethylene biosynthesis, 1‐aminocyclopropane‐1‐carboxylic acid‐synthase (ACS), leading to a burst of ethylene production concomitant with necrotic lesion formation (Knoester et al., 2001; Ohtsubo et al., 1999).
While it is clear that supplementation of the endogenous pool of ACC had a suppressive effect on the replication of white clover mosaic virus (WClMV) (Clarke et al., 1998), in their review Alazem and Lin (2015) suggest that ethylene does not appear to be essential for plant resistance against viruses. This may indeed be the case in Arabidopsis. Recent studies indicated ACC treatment dramatically increases the accumulation of TMV‐cg in systemically infected leaves in Arabidopsis. In the upper systemically infected leaves of infected plants, viral RNAs were detected at 4 days postinoculation (dpi) in ACC‐treated plants but at 5 dpi in control plants. The accumulation of viral RNA was also significantly greater in plants pretreated with ACC. Thus, ethylene contributes to virus susceptibility in Arabidopsis. Furthermore, mutation in ethylene‐related genes greatly inhibits the accumulation of TMV‐cg in systemic leaves (Chen et al., 2013). It appears that ethylene is involved in promoting both symptom development and systemic movement in the case of TMV‐cg and also turnip mosaic virus infection in Arabidopsis and N. benthamiana, respectively (Chen et al., 2013; Wang et al., 2019), in contrast to repressing TMV infection of Nicotiana (Zhao & Li, 2021).
Signalling of systemic acquired resistance (SAR) in tobacco depends on the perception of ethylene (Knoester et al., 2001; Verberne et al., 2003). In contrast, in Arabidopsis, the ethylene transcription factor EIN3/EIL1 was shown to target and repress SID2, which encodes isochorismate synthase during pathogen‐induced biosynthesis of SA, to negatively regulate plant innate immunity (Chen et al., 2009). These findings demonstrate that ethylene may work antagonistically on SA in plant immunity in Arabidopsis. In addition, the timing of ethylene treatment affects plant defence against viral infection (Alazem & Lin, 2015). These results demonstrate that ethylene plays a complicated role in defence against pathogens.
The ethylene signal transduction pathway is initiated by ethylene negatively regulating its receptors, which are a small family of five receptors (ETR1, ETR2, ERS1, ERS2, and EIN4). This then removes the repressing effect of CTR1 on downstream elements including EIN2 and EIN3/EILs. Ethylene stabilizes EIN3/EIL1 by inducing proteasomal degradation of EBF1/2 (An et al., 2010). The nuclear protein EIN3/EILs regulates the transcription of other transcription factors such as Ethylene Response Factor1, ERF1, a transcription factor that belongs to the large APETALA2 gene family. ERF1 binds to a GCC or dehydration‐responsive element (DRE) box present in the promoters of many ethylene inducible/repressed defence‐related genes (Guo & Ecker, 2004). Notably, Fischer and Dröge‐Laser (2004) showed that tobacco plants overexpressing ERF5 show enhanced resistance to TMV with a reduction in the size of local lesions and impaired systemic spread of the virus, as well as reduced viral RNA replication. They concluded that NtERF5‐regulated gene expression controlled resistance to viral propagation (Fischer & Dröge‐Laser, 2004).
Previous studies have shown that N. benthamiana MYB1 could directly regulate the SA‐dependent defence pathway (Liu et al., 2004), which means NbMYB1 acts in a central role in plant defence responses against viruses. Therefore, we hypothesized that MYBs play a potential role in the crosstalk between phytohormones and defence responses in TMV resistance in N. benthamiana. Using genetic and molecular biology approaches, we found that the MYB4L‐dependent signalling pathway is involved in TMV defence responses in N. benthamiana. Furthermore, the relationship between the ethylene signalling pathway and MYB4L in plant defence against TMV was also investigated. Our results cast light on the possibility that the ethylene‐mediated NbMYB4L signalling pathway affects the susceptibility of N. benthamiana to TMV, and might function as an SA‐independent resistance mechanism.
2. RESULTS
2.1. Role of NbMYB4L in TMV infection of N. benthamiana
NbMYB4L (Sequence ID: Niben101Scf03570Ctg037; https://solgenomics.net) has 837 nucleotides in an open reading frame encoding 278 amino acids. Sequence alignment showed that NbMYB4L protein had 96% identity to NbMYB1, 95.2% identity to NtMYB1, and at least 49% identity to AtMYB15 (data not shown). The expression of NbMYB4L was significantly greater in leaves than in stems, roots, or flowers (Figure 1a). At 3 dpi with TMV, the transcription of NbMYB4L in leaves of N. benthamiana was markedly up‐regulated (Figure 1b).
FIGURE 1.
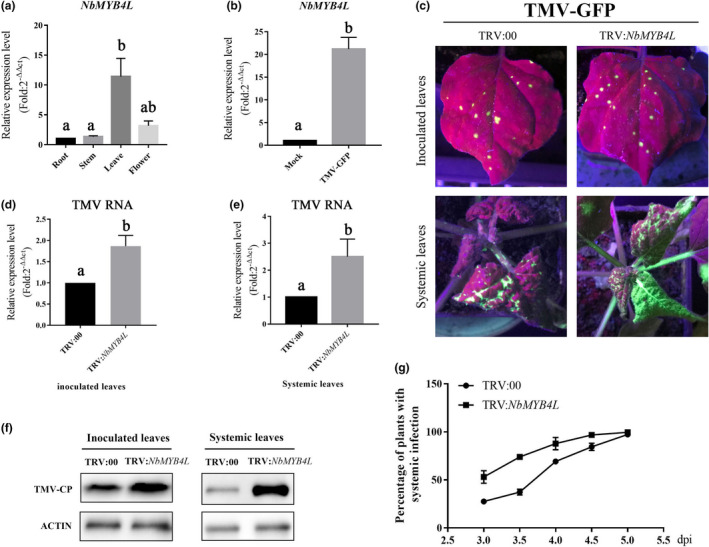
NbMYB4L expression was induced by tobacco mosaic virus (TMV) in Nicotiana benthamiana and its silencing facilitated infection by TMV. (a) Reverse transcription quantitative PCR (RT‐qPCR) showing that NbMYB4L was expressed at higher levels in leaves than in flowers, stems, and roots. (b) RT‐qPCR showing that the transcription of NbMYB4L was significantly induced by TMV at 3 days postinoculation (dpi). Results from three independent biological replicates are shown; Actin was used as the internal reference gene. (c) Leaves of plants inoculated with TMV‐GFP and examined under ultraviolet light at 3 dpi. On NbMYB4L‐silenced plants (TRV:NbMYB4L) there were more fluorescent spots (depicting infection foci) on the inoculated leaves and a larger fluorescent area on systemically infected leaves than on the controls (TRV:00). (d, e) RT‐qPCR analysis of TMV RNA accumulation in the inoculated leaves and the systemic leaves collected at 3 and 7 dpi, respectively. Actin was used as the internal reference. Error bars show the mean ± SD of three replicates (at least 20 plants per replicate). Different letters on histograms indicate significant differences (p < 0.05). (f) Western blotting analysis of coat protein accumulation of TMV at 3 dpi in inoculated leaves and 7 dpi in systemic leaves of different N. benthamiana plants infected with TMV‐GFP. The samples were detected with primary antibody to GFP‐tag and secondary anti‐rabbit antibody. The antigen‐antibody (for secondary anti‐rabbit) was visualized using ECL horseradish peroxidase chemiluminescent substrate under a chemiluminescence analyser. Actin proteins were used as loading controls and were labelled with β‐actin antibody. (g) Percentage of plants (more than 20 per replicate, n > 20) with GFP fluorescence on newly emerged leaves (i.e., systemically infected) at different times after inoculation. Error bars show the mean ± SD of three replicates with at least 20 plants per replicate
To investigate the potential role of NbMYB4L in TMV infection, we used tobacco rattle virus (TRV)‐induced gene silencing (VIGS) to silence NbMYB4L. Following inoculation with TMV, the transcription of NbMYB4L was reduced in the silenced plants to less than 25% of normal levels (Figure S1). There were no obvious phenotypic differences between control (TRV:00) and NbMYB4L‐silenced plants. The TRV:00 and TRV:NbMYB4L plants were inoculated with TMV linked to the green fluorescent protein (GFP) gene (TMV‐GFP) and subsequent viral infection was monitored by detecting GFP. There were more fluorescent spots on the inoculated leaves of NbMYB4L‐silenced plants (Figure 1c) compared to the control. From 4 dpi, GFP began to appear on newly emerged leaves, indicating systemic infection. Initially, the NbMYB4L‐silenced plants had a higher incidence of systemic infection and a larger area of GFP than the controls, indicating a faster spread of systemic infection (Figure 1c,g). Additionally, both viral RNA and coat protein (CP) accumulated to much higher levels in the inoculated leaves of NbMYB4L‐silenced plants than in the TRV:00‐treated controls (Figure 1d–f). We concluded that silencing of NbMYB4L facilitated infection by TMV.
2.2. NbMYB4L‐mediated TMV‐GFP resistance in N. benthamiana
To understand the role of NbMYB4L in defence against TMV, transgenic N. benthamiana overexpressing NbMYB4L (OE) and suppressing NbMYB4L (RNAi) were generated. The production of the NbMYB4L‐hemagglutinin (HA) fusion protein in the transgenic OE lines was checked by western blot using an anti‐HA antibody. Four OE transgenic lines exhibited specific bands, located around 30 kDa, the expected size of the NbMYB4L‐HA fusion protein (Figure S2). To rule out whether silencing of NbMYB4L led to co‐silencing of genes closely related to NbMYB4L, we also checked other NbMYBs in NbMYB4L‐RNAi plants using reverse transcription quantitative PCR (RT‐qPCR) and showed that there was little influence of NbMYB4L‐RNAi on the transcription of other NbMYB genes (Figure S3).
To investigate whether NbMYB4L mediates TMV resistance, four OE transgenic lines and two NbMYB4L‐RNAi lines of N. benthamiana plants were inoculated with TMV‐GFP on the fifth and sixth leaves. Virus accumulation was determined by direct observation of GFP (Figure 2a), which showed that systemic TMV‐GFP accumulation was clearly delayed in the OE lines, whereas TMV‐GFP clearly accumulated to much greater levels in both local and systemic leaves of NbMYB4L‐RNAi plants relative to the control. The extent of viral replication was monitored by RT‐qPCR and western blot analysis. In both local and systemic leaves of two of the four OE lines, the level of TMV‐coat protein (CP) RNA was significantly reduced compared to the wild‐type (WT) plants, which suggests that the OE lines may have enhanced resistance to TMV‐GFP (Figure 2b,c). In contrast, the TMV‐CP transcript level in NbMYB4L‐RNAi leaves was significantly greater than that of WT plants in both local and systemic leaves (Figure 2b,c). The transcript data were confirmed by western blot analysis, which also showed a reduction in CP in the OE lines, and an increase in CP in NbMYB4L‐RNAi plants in both local and systemic leaves of N. benthamiana inoculated with TMV‐GFP (Figure 2d). As the OE‐69 line showed particularly strong viral resistance, OE‐69 was used in subsequent experiments.
FIGURE 2.
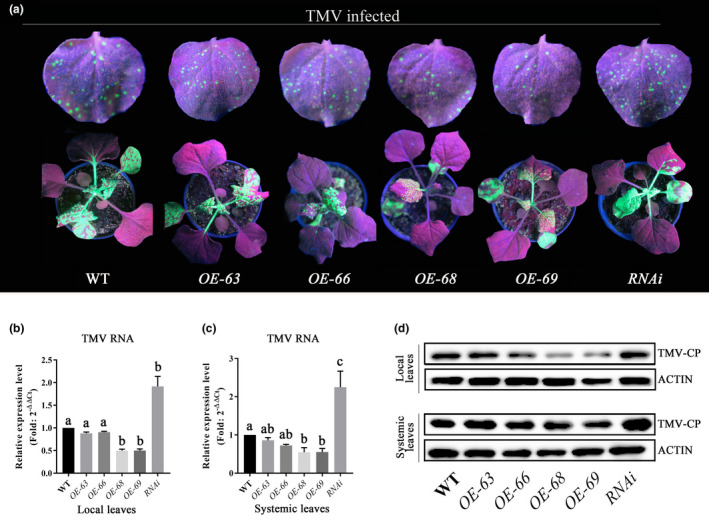
The role of NbMYB4L in Nicotiana benthamiana resistance to tobacco mosaic virus (TMV). (a) Leaves of plants inoculated with TMV‐GFP and examined under ultraviolet light at 3 days postinoculation (dpi) in inoculated leaves and 7 dpi in systemic leaves of transgenic N. benthamiana MYB4L overexpression and RNAi transgenic lines. N. benthamiana plants were inoculated with TMV‐GFP on local leaves. TMV‐GFP spread in the inoculated leaves and systemic leaves was photographed at 7 dpi. Experiments were repeated three times, with similar results. WT, wild type. (b, c) Reverse transcription quantitative PCR (RT‐qPCR) analysis of TMV accumulation in the inoculated leaves and the systemic leaves collected at 3 and 7 dpi, respectively. Actin was used as the internal reference gene. Error bars show the mean ± SD of three replicates (at least 20 plants per replicate). Different letters on histograms indicate significant differences (p < 0.05). (d) Western blot showing accumulation of TMV coat protein at 3 dpi in inoculated leaves and 7 dpi in systemic leaves of different N. benthamiana plants infected with TMV‐GFP. The samples were detected with primary antibody to the GFP‐tag and secondary anti‐rabbit antibody. The antigen‐antibody (for secondary anti‐rabbit) was visualized using ECL horseradish peroxidase chemiluminescent substrate under a chemiluminescence analyser. Actin protein was used as the internal control and was detected with an anti‐β‐actin antibody. Experiments were repeated three times with similar results
To further analyse the defence responses, antioxidant activity was examined in different transgenic N. benthamiana lines. The activities of both superoxide dismutase (SOD) and catalase (CAT) were greater in the OE‐69 line but lower in the RNAi line compared with WT (Figure S4). These results demonstrate that NbMYB4L increased resistance to TMV‐GFP in N. benthamiana.
2.3. The transcription of NbMYB4L is activated by ACC during TMV infection
To assess the response of defence‐related genes associated with NbMYB4L‐dependent responses, we chose NbPR1, NbPR2, and NbPDF1 as indicator genes for plant immunity (Wang et al., 2009). The transcription of NbPR1, NbPR2, and NbPDF1 was highly up‐regulated in the OE‐69 plants but repressed in the NbMYB4L RNAi plants compared with WT in TMV‐GFP inoculated leaves at 3 dpi (Figure 3a–c). In TMV‐GFP infected systemic leaves at 7 dpi, transcription of the three genes was significantly up‐regulated in OE‐69 but blocked in RNAi compared with WT plants (Figure 3d–f).
FIGURE 3.
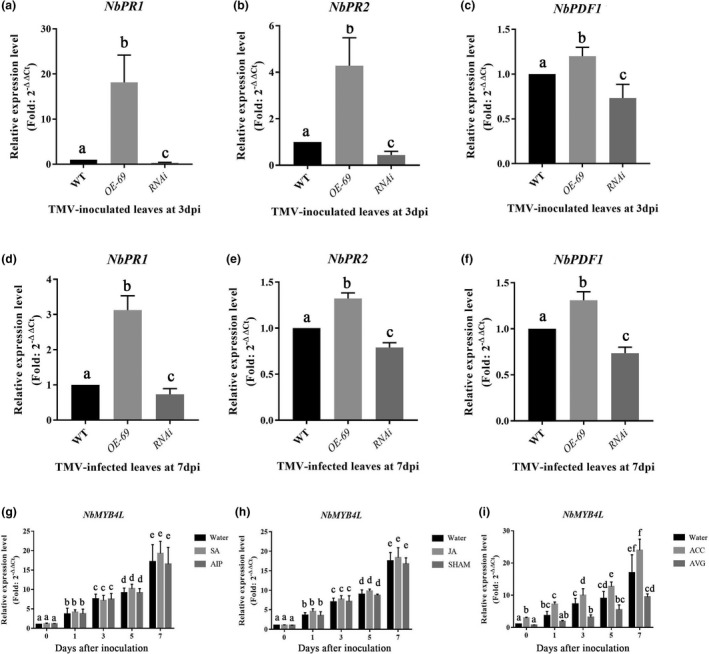
The transcription of NbMYB4L is influenced by ethylene. (a–f) Transcription of NbPR1 (a), NbPR2 (b), and NbPDF1 (c) in the inoculated leaves at 3 days postinoculation (dpi) and transcription of NbPR1 (d), NbPR2 (e), and NbPDF1 (f) in the systemic leaves at 7 dpi in different transgenic Nicotiana benthamiana lines during TMV‐GFP infection. OE‐69: overexpression of NbMYB4L; RNAi: reduced expression of NbMYB4L. (g–i) N. benthamiana plants were first pretreated with salicylic acid (SA), jasmonic acid (JA) or 1‐aminocyclopropanecarboxylic acid (ACC), or their inhibitors, AIP, SHAM, and AVG, respectively. After 12 h the plants were inoculated with TMV‐GFP, and NbMYB4L transcription recorded at 0, 1, 3, 5, and 7 dpi. (g) NbMYB4L transcription in SA‐ and AIP‐pretreated plants during TMV‐GFP infection. (h) NbMYB4L transcription in JA‐ and SHAM‐pretreated plants during TMV‐GFP infection. (i) NbMYB4L transcription in ACC‐ and AVG‐pretreated plants during TMV‐GFP infection. Bars represent mean and SD of values obtained from three biological replicates per genotype and time point. Different lower case letters indicate significant differences (p < 0.05)
Previous studies have shown that SA, ethylene, and JA are involved in defence against TMV (Jameson & Clarke, 2002; Verberne et al., 2003). To examine whether SA, JA, or ethylene influenced the transcription of NbMYB4L in response to TMV infection, we monitored the transcript level of NbMYB4L during TMV‐GFP infection following foliar application of the hormone or their inhibitor, or water. The plants were inoculated with TMV‐GFP 12 h after foliar application. The transcription of NbMYB4L in the inoculated leaves was measured by RT‐qPCR from the time of inoculation up to 7 dpi. N. benthamiana plants pretreated with SA or 2‐amidoindane‐2‐phosphonic acid (AIP, an SA biosynthesis inhibitor) appeared to have little influence on NbMYB4L transcription during TMV‐GFP infection (Figure 3g). The transcription of NbMYB4L was similar in JA‐ or salicylhydroxamic acid (SHAM, a JA biosynthesis inhibitor)‐treated N. benthamiana plants (Figure 3h). However, N. benthamiana plants pretreated with ACC resulted in significantly increased transcription of NbMYB4L during TMV‐GFP infection. In contrast, pretreatment with aminoethoxyvinylglycine (AVG, an ethylene biosynthesis inhibitor) led to suppression of transcription of NbMYB4L (Figure 3i). While SA and JA seemed to have little effect, these results demonstrated that ethylene may play an important role in mediating the expression of NbMYB4L during TMV infection. Furthermore, the levels of NbMYB4L transcript positively correlated with the levels of expression of defence genes.
2.4. Transcription of ethylene biosynthetic and signalling genes is up‐regulated during TMV‐GFP infection in transgenic N. benthamiana plants overexpressing NbMYB4L
To determine if NbMYB4L transcription moderated plant hormone accumulation and signalling pathways, we examined the content and the transcription of biosynthesis and signalling genes of SA, ethylene, and JA during NbMYB4L‐mediated viral resistance. We chose SA‐associated NbICS1, NbNPR1, ethylene‐associated NbACS6, NbEIN2, and JA‐associated NbOPR3, NbCOI1, as each pair represented a hormone biosynthesis gene and a signalling pathway gene, respectively. Each of these six genes has been associated with plant defence (Wang et al., 2009; Zhu et al., 2014). There was no significant difference in the content of SA or in the expression of NbICS1 or NbNPR1 relative to control plants in either local or systemic leaves. However, relative to the NbMYB4L‐RNAi plants, the OE‐69 plants showed significantly elevated SA and elevated expression of NbICS1 and NbNPR1 in local leaves (Figure 4a,d,g). The content of JA did not differ between control or transgenics, nor did the biosynthetic gene (NbOPR3) or signalling gene (NbCOI1) at 3 dpi. However, at 7 dpi there was a significant reduction in the expression of both NbOPR3 and NbCOI1 in the systemic leaves of the OE‐69 plants relative to the control (Figure 4c,f,i), but this was not reflected in the JA content.
FIGURE 4.
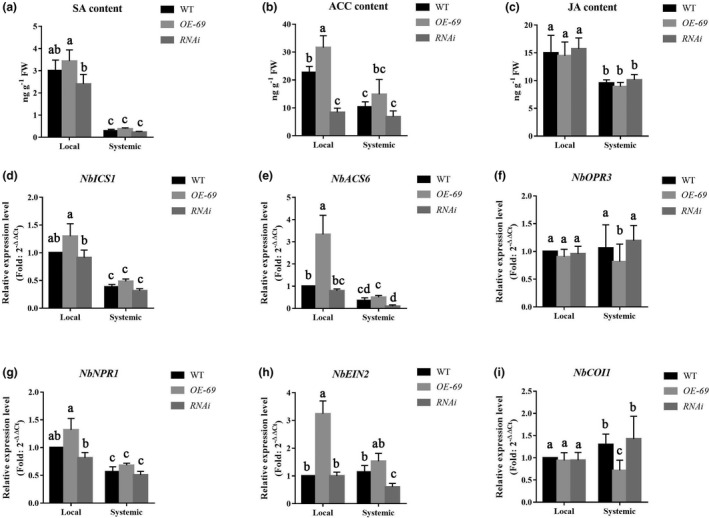
Hormone content and gene expression changes in transgenic Nicotiana benthamiana either over‐ (OE‐69) or underexpressing (RNAi) NbMYB4L during TMV‐GFP infection, measured in inoculated (local) and systemic leaves at 3 and 7 days postinoculation (dpi), respectively. (a) Salicylic acid (SA) content; (b) 1‐aminocyclopropanecarboxylic acid (ACC) content; (c) jasmonic acid (JA) content. (d–f) Transcription level of NbICS1, NbACS6, and NbOPR3; (g–i) transcription level of NbNPR1, NbEIN2, and NbCOI1 in inoculated and systemic leaves of NbMYB4L transgenic N. benthamiana plants during TMV‐GFP infection. Significant differences (p < 0.05) are denoted by different lower case letters
Notably, the ACC content was significantly increased in the OE‐69 plants and was suppressed in the NbMYB4L‐RNAi plants compared with WT plants in TMV‐GFP inoculated leaves at 3 dpi (Figure 4b). The transcription of both NbACS6 and NbEIN2 was significantly induced in OE‐69 plants, whereas the expression was significantly reduced in the NbMYB4L‐RNAi plants compared with WT plants (Figure 4e,h). Combined with the effect of ACC and AVG pretreatment on the transcription of NbMYB4L and the impact of NbMYB4L transcript levels on ACC synthesis and content, and ethylene signalling in transgenic N. benthamiana during TMV‐GFP infection, these results indicate that an ethylene‐associated signalling pathway plays a pivotal role in NbMYB4L‐induced TMV resistance.
2.5. Ethylene works upstream of NbMYB4L‐dependent TMV resistance
To determine the position of ethylene in NbMYB4L‐dependent TMV‐GFP resistance, OE‐69 plants were pretreated with water or AVG, and NbMYB4L‐RNAi plants were pretreated with water or ACC, prior to TMV‐GFP inoculation. Virus accumulation was determined by direct observation of GFP (Figure 5a), as well as by RT‐qPCR and western blot analysis of viral replication (Figure 5b–d). There was a decrease in resistance to TMV‐GFP in AVG‐pretreated OE‐69 plants relative to water‐pretreated OE‐69 plants (Figure 5a–d), confirming the requirement for ethylene. However, there was no difference in resistance between the water‐pretreated and ACC‐pretreated RNAi plants, that is, ACC did not rescue resistance in the NbMYB4L‐RNAi plants (Figure 5a–d). Moreover, neither was the reduced expression of SOD or CAT rescued by ACC pretreatment of the RNAi plants (Figure 5e,f). Taken together, we suggest that ethylene probably acts upstream of NbMYB4L and is required as a signalling molecule to activate the NbMYB4L‐dependent TMV defence response.
FIGURE 5.
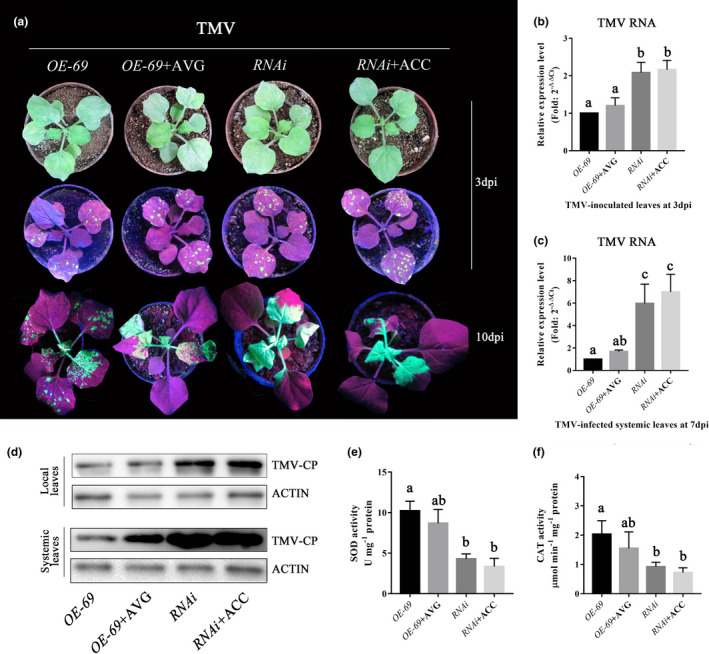
Ethylene works upstream of the NbMYB4L‐mediated defence response. (a) Plants were inoculated with TMV‐GFP and examined under ultraviolet light at 3 days postinoculation (dpi) for inoculated leaves and 7 dpi for systemic leaves of NbMYB4L overexpressing line OE‐69, OE‐69+ AVG, and NbMYB4L‐RNAi and RNAi + 1‐aminocyclopropanecarboxylic acid (ACC) plants. Experiments were repeated three times, with similar results. (b, c) Reverse transcription quantitative PCR (RT‐qPCR) analysis of tobacco mosaic virus (TMV) accumulation levels in the inoculated leaves and the systemic leaves collected at 3 and 7 dpi, respectively. Actin was used as the internal reference gene. Error bars show the mean ± SD of three replicates (at least 20 plants per replicate). Different letters on histograms indicate significant differences (p < 0.05). (d) Western blot analysis of coat protein (CP) accumulation of TMV at 3 dpi in inoculated leaves and 7 dpi in systemic leaves of different Nicotiana benthamiana plants infected with TMV‐GFP. The samples were detected with primary antibody to the GFP‐tag and secondary anti‐rabbit antibody. The antigen‐antibody (for secondary anti‐rabbit) was visualized using ECL horseradish peroxidase chemiluminescent substrate under a chemiluminescence analyser. Actin proteins were used as loading controls and were detected with anti‐β‐actin antibody. (e, f) Superoxide dismutase (SOD) and catalase (CAT) activity in different N. benthamiana plants. Significant differences (p < 0.05) are denoted by different lowercase letters
2.6. NbEIL1 activates the transcription of NbMYB4L
To uncover whether a downstream component of the ethylene signalling pathway directly regulates the transcription of NbMYB4L, we analysed the NbMYB4L promoter region to determine if any potential binding sequences for ethylene‐related genes were present. The most well‐documented ethylene‐responsive elements are the DRE and the GCC box, to which the ethylene response factors (ERF) bind, and the ATGTA motif, which is bound by ethylene insensitive 3 (EIN3)/EIN3‐like proteins (EILs) (Fujimoto et al., 2000; Liu et al., 2006; Qiu et al., 2015). No DRE elements or GCC box were found, but three ATGTA motifs were located at −917 to −913, −718 to −714, and −626 to −622 sites in the promoter region of NbMYB4L and named P1, P2, and P3, respectively. Consequently, we speculated that EIN3/EILs might be the key factors directly regulating the transcription of NbMYB4L (Figure 6b).
FIGURE 6.
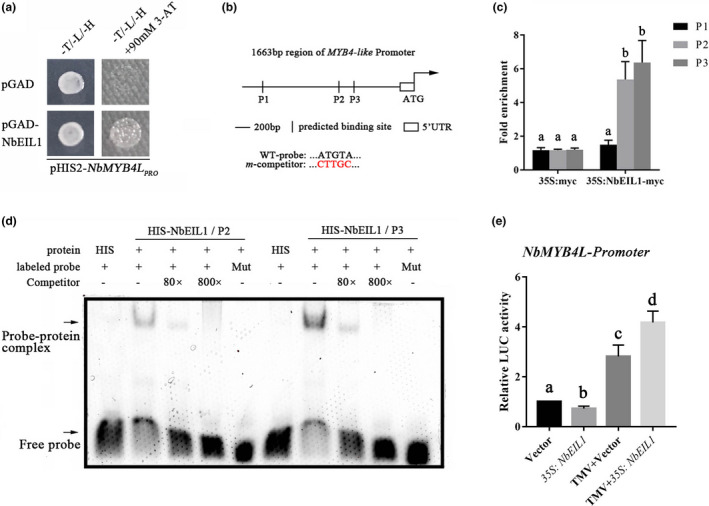
Binding of NbEIL1 to the NbMYB4L promoter. (a) Yeast one‐hybrid assay showing NbEIL1 interaction with the promoter of NbMYB4L. The promoter fragment of NbMYB4L was fused to the pHIS2 vector, and the NbEIL1 gene was fused to the pGAD vector. The columns represent the addition of the pHIS2‐NbMYB4L pro vector. The rows represent the addition of the pGAD and pGAD‐NbEIL1 vectors. (b) Schematic diagram of the NbMYB4L promoter showing the potential NbEIL1‐binding sites (P1 to P3). (c) Chromatin immunoprecipitation (ChIP)‐qPCR assay of NbEIL1 binding to the promoter of the NbMYB4L gene in Nicotiana benthamiana during tobacco mosaic virus (TMV) infection. Chromatin from the empty vector control (N‐myc) and 35S:NbEIL1‐myc transient N. benthamiana (NbEIL1‐myc) were immunoprecipitated with or without anti‐myc antibodies. Three regions (P1 to P3) were examined by reverse transcription quantitative PCR (RT‐qPCR). The enrichment of the wild type (WT) was set to 1. RT‐qPCR was performed with three technical replicates and three biological replicates to examine the enrichment of NbMYB4L fragments. (d) Electrophoretic mobility shift assay (EMSA) showing that the NbEIL1‐HIS fusion protein bound directly to the P2 and P3 site of the NbMYB4L promoter. Unlabelled probes were used as competitors. Mut represents a mutated probe in which the ATGTA motif was replaced by CTTGC. (e) Relative luciferase (LUC) activity of NbMYB4L promoters was detected under control or TMV stress condition. N. benthamiana plants were inoculated with TMV‐GFP for 3 days and then the leaves were transiently transformed with different constructs. The CaMV 35S promoter was fused to LUC as a control for variation in transformation rate. Bars represent mean and SD of values obtained from three biological repeats. Significant differences (p < 0.05) are denoted by different lower case letters
To verify our speculation, we searched for all related EIN3/EILs within Nicotiana tabacum (its genome is highly similar to N. benthamiana) in the NCBI database and found at least five EIN3/EIL proteins, NtEIL1–5. Subsequently, we used BLAST tools to search for these proteins in the Sol Genomics Network (https://solgenomics.net) using an N. benthamiana genome database to compare the five NtEILs sequences. Interestingly, the results showed more than 90% nucleotide similarity between NtEILs and NbEILs. We therefore designed primers and cloned the NbEILs genes according to the BLAST results. To further check which member(s) of the NbEILs could directly bind to the NbMYB4L promoter, a yeast‐one‐hybrid (Y1H) assay was performed. Each coding sequence of NbEIL1‐5 was cloned and inserted into a pGADT7 vector, and the promoter region of NbMYB4L was fused to the pHIS2 vector. The results showed that only NbEIL1 could directly interact with the NbMYB4L promoter region (Figures 6a and S5). These results demonstrate that NbEIL1 might be the upstream factor that directly mediates the transcription of NbMYB4L.
To verify the binding of NbEIL1 to the NbMYB4L promoter in vivo, a chromatin immunoprecipitation (ChIP)‐qPCR assay was conducted using a 35S:NbEIL1‐HA transient expression assay with the empty vector as control. The fragment containing P2 and P3 had higher enrichment in the NbEIL1 transient N. benthamiana leaves, suggesting that NbEIL1 bound to the promoter of NbMYB4L in vivo (Figure 6c).
To further confirm whether NbEIL1 bound to the two specific regions of the NbMYB4L promoter, electrophoretic mobility shift assays (EMSAs) were performed using an NbEIL1‐HIS fusion protein. NbEIL1 specifically bound to the P2 and P3 sites (Figure 6d). When unlabelled probes were added as competitors, the binding was reduced, whereas the band disappeared when the mutated probes were added (Figure 6d). These results indicate that NbEIL1 bound to the ATGTA motif of the NbMYB4L promoter in vitro.
In addition, a transient transfection assay was conducted to explore the transcription activity of the NbMYB4L promoter that was regulated by NbEIL1. NbEIL1 enhanced the activation of NbMYB4Lpro:LUC in N. benthamiana as compared to the control during TMV‐GFP infection (Figure 6e). Taken together, these results suggest that NbEIL1 could bind directly to the specific region of NbMYB4L and activate its transcription.
2.7. The NbEIL1‐dependent pathway is responsible for the defence response to TMV
Finally, to evaluate whether NbEIL1 is involved in NbMYB4L‐associated TMV‐GFP resistance, we constructed TRV:NbEIL1 and TRV:NbEIL1/OE‐69 VIGS vectors and agroinfiltrated these into N. benthamiana. The results showed that the transcription of the silenced targeted genes was reduced to less than 30% of the TRV:00 control plants (Figure S6). As shown in Figure 7a, TMV‐GFP resistance was impaired in the TRV:NbEIL1 plants compared with TRV: 00 control plants, and TMV‐GFP resistance was not fully recovered in the TRV:NbEIL1/OE‐69 plants. Partial recovery was shown in the RT‐qPCR assay but viral transcription was still elevated in the TRV:NbEIL1/OE‐69 plants relative to OE‐69 and the control, particularly in systemic leaves 7 dpi (Figure 7b,c). In addition, we checked the antioxidant enzyme activities in TRV:NbEIL1 and TRV:NbEIL1/OE‐69 plants during TMV‐GFP infection. The SOD and CAT activities are somewhat consistent with the virus resistance in the different transgenic N. benthamiana lines (Figure 7d,e). As the VIGS suppression of NbEIL1 was incomplete, some activity of NbMYB4L could be expected, leading to elevation of SOD and CAT activity in the TRV:NbEIL1/OE‐69 plants, and the nonsignificant reduction in activity in the TRV:NbEIL1 plants. Overall, these data indicate that NbEIL1 is upstream of the NbMYB4L‐dependent defence pathway and that NbEIL1 is essential for NbMYB4L‐mediated TMV‐GFP resistance.
FIGURE 7.
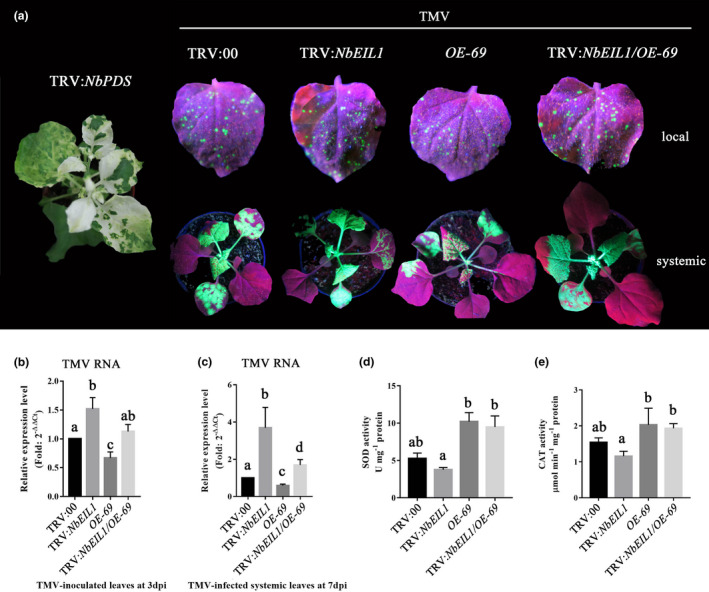
NbEIL1 is involved in NbMYB4L‐associated TMV‐GFP resistance. (a) Nicotiana benthamiana plants silenced/not silenced for NbEIL1 exhibited various levels of resistance to tobacco mosaic virus (TMV). TMV‐GFP spread in the local and systemic leaves photographed at 7 days postinoculation (dpi). Experiments were repeated three times with similar results. (b, c) Reverse transcription quantitative PCR (RT‐qPCR) analysis of TMV RNA replication in the inoculated leaves and the systemic leaves collected at 3 and 7 dpi, respectively. Actin was used as the internal reference gene. (d, e) Superoxide dismutase (SOD) and catalase (CAT) activity in different N. benthamiana plants, respectively. Error bars show the mean ± SD of three replicates (at least 20 plants per replicate). Different letters on histograms indicate significant differences (p < 0.05)
3. DISCUSSION
Plant MYB TFs have been shown to be involved in plant defence. Early studies showed that NtMYB1 and NbMYB1 were TMV‐inducible and participated in the N‐mediated defence pathway in N. tabacum (Yang & Klessig, 1996) and N. benthamiana (Liu et al., 2004), respectively. A recent study showed that AtMYB15 acts as a positive regulator of defence‐induced lignification and basal immunity. The myb15 mutant showed increased susceptibility to the bacterial pathogen Pseudomonas syringae (Chezem et al., 2017; Kim et al., 2020). Interestingly, through a BLAST search of NCBI, we found that AtMYB15 is homologous to N. benthamiana MYB4L (data not shown) and, in our study, we found that NbMYB4L plays a positive role in TMV resistance in N. benthamiana. Infection by TMV‐GFP elevated the transcription of NbMYB4L (Figure 1) and N. benthamiana overexpressing NbMYB4L was less susceptible to TMV (Figure 2), whereas when NbMYB4L was knocked down by VIGS or RNAi, N. benthamiana was much more susceptible to TMV (Figures 1 and 2). Additionally, key pathogenesis‐related genes (PR‐1, PR‐2, and PDF1) were up‐regulated when NbMYB4L was overexpressed and showed reduced expression in NbMYB4L‐RNAi plants (Figure 3a–f), indicating that the defence signal transduction pathway had been affected when the transcription levels of NbMYB4L were manipulated.
In plants, it is well known that the SA‐associated signalling pathway plays a pivotal role in SAR (Fu & Dong, 2013; Seyfferth & Tsuda, 2014). However, there is also a subset of defence‐related genes whose induction in response to TMV is independent of SA (Guo et al., 2000; Knoester et al., 2001). We found that the transcription of NbMYB4L was unaffected by pretreatment of N. benthamiana with SA or its inhibitor, but was significantly induced by pretreatment with the ethylene precursor, ACC, whereas pretreatment with the ethylene biosynthetic inhibitor, AVG, reduced the transcription of NbMYB4L (Figure 3). Accumulation of ACC synthase (ACS6), ACC, and ethylene signal transduction gene, EIN2, were all induced in plants overexpressing NbMYB4L (Figure 4b,e,h), which suggested that ethylene‐mediated TMV resistance is partially dependent on NbMYB4L. This becomes more apparent when plants overexpressing NbMYB4L were pretreated with AVG. These plants were particularly susceptible to TMV. However, the TMV‐susceptible NbMYB4L‐RNAi plants were not rescued by ACC, which suggests NbMYB4L acts downstream of the ethylene transduction pathway and works positively to confer resistance against TMV (Figures 4 and 5). In other words, ethylene participates in the induction of NbMYB4L in N. benthamiana during TMV‐GFP infection. However, as ACC synthase and ACC accumulation are themselves induced by NbMYB4L, we speculate that TMV infection has probably triggered ethylene emission from the local leaves and, potentially, also from necrotic lesions of the host cells due to wound‐induced ethylene (Ohtsubo et al., 1999). As gaseous ethylene can diffuse externally around the whole plant, it may activate the ethylene signalling pathway, which might further enhance the NbMYB4L‐dependent responses in antagonizing the replication of the virus through a feed‐forward loop.
Ethylene is clearly an important hormone in regulating plant biotic stress responses but the study by Chen et al. (2013) revealed that it is the mutants of acs6 (1‐aminocyclopropane‐1‐carboxylate synthase 6), erf106 (ethylene responsive transcription factor 106), and ein2 (ethylene insensitive 2) that are resistant to TMV‐cg in Arabidopsis. In addition, ACC application enhanced TMV‐cg accumulation in treated plants (Chen et al., 2013). That study also showed that ACS6 and ERF104 were significantly up‐regulated in wrky8 mutants, and that WRKY8 negatively regulates these genes by binding to W‐box clusters within their promoter (Chen et al., 2013). It seems likely that a SA‐dependent signalling pathway dominates the virus resistance process in Arabidopsis (Alazem & Lin, 2015; Kim et al., 2003). Indeed, Knoester et al. (2001) had concluded much earlier that, while in tobacco SAR is stimulated by ethylene perception, SAR is differently regulated in tobacco and Arabidopsis.
Published evidence showed ethylene might act as an important signalling agent in N. benthamiana TMV resistance (Chen et al., 2009; Hibi et al., 2007; Knoester et al., 2001; Ohtsubo et al., 1999). In this study, we did not examine TMV resistance following ACC or AVG pretreatment of WT N. benthamiana. However, our results demonstrate that ACC‐pretreatment increased the transcription of NbMYB4L during TMV‐GFP infection, and plants overexpressing NbMYB4L showed enhanced TMV resistance, accompanied with the raised antioxidant enzyme activities (Figure S4) and defence‐associated genes (Figure 3). The latter genes are generally associated with SAR (Durrant & Dong, 2004) and SA can induce antioxidants (Fodor et al., 1997; Jameson & Clarke, 2002; Király et al., 2002), but whether these were elevated by the ethylene‐induced NbMYB4L pathway independently of SA‐induced SAR remains to be determined. There appeared to be no antagonism between the ethylene‐mediated NbMYB4L pathway and the SA‐dependent defence pathway: SA biosynthesis, SA content, and signal transduction were not negatively impacted in NbMYB4L‐OE plants. Although our data reveal a new pathway and mechanism of plant virus resistance in N. benthamiana, we still need additional genetic evidence to determine whether NbMYB4L‐mediated and SA‐mediated virus resistance responses are indeed independent of each other. Using SA‐deficient (NahG) transgenic N. benthamiana plants would assist in this determination (Künstler et al., 2019).
Our investigation of the link between ethylene and downstream activation of NbMYB4L showed that EIL1 bound to the NbMYB4L promoter. An earlier study had shown that the tobacco EIN3 homologue, TEIL, could induce basic PR gene expression but suppress acidic PR gene expression (Hibi et al., 2007). In Arabidopsis, EIN3 and EIL1 have been shown to repress SID2, which encodes isochorismate synthase required for pathogen‐induced biosynthesis of SA, by directly target SID2 and down‐regulating its transcription (Chen et al., 2009). Therefore, EIN3/EILs were regarded as negatively regulating the SA‐associated signalling pathway, but this is in Arabidopsis, where ethylene enhances virus susceptibility (Chen et al., 2009).
Ethylene regulates the protein levels of EIN3/EILs by stabilizing them through inducing the degradation of the upstream EBF1/2 factor by ubiquitin/26S proteasome (An et al., 2010). To sum up, it is highly plausible that TMV contributed to the increased ethylene biosynthesis and emission in host cells, after which the activated ethylene transcription factors, such as EIN3/EILs, directly induced the transcription of several defence response genes and other metabolic changes during TMV infection. By analysing the possible binding sites on the NbMYB4L promoter, the ERFs were excluded but an ATGTA motif, to which EIN3/EILS can bind (Qiu et al., 2015), was detected. Of the five NbEILs tested in the Y1H assay, only NbEIL1 interacted with the NbMYB4L promoter.
In addition, through analysis of promoter activity and EMSA assay, NbEIL1 was further confirmed to bind directly to the specific region of the NbMYB4L promoter and to mediate the transcription of NbMYB4L (Figure 6). Binding of EILs to MYB promoters has been shown previously. For example, AtEIL3 was shown to interact with AtMYB72 during the early signalling steps of rhizobacteria‐induced systemic resistance in Arabidopsis (Ent et al., 2008). An EIL1 was isolated and its involvement in stress‐inducible DcMYB1 expression in suspension‐cultured carrot cells identified. DcEIL bound to a region of the DcMYB1 promoter containing a putative cis‐element, which suppressed DcMYB1 promoter transcription (Miyahara et al., 2010). A recent study reported that MdEIL1 could promote anthocyanin accumulation by directly inducing MdMYB1 transcription in apple fruit (An et al., 2018). In our study, we found NbMYB4L to be directly induced by NbEIL1. Furthermore, genetic evidence showed that TMV resistance was decreased in both NbEIL1‐VIGS knockdown plants and NbEIL1‐VIGS/NbMYB4L‐OE plants relative to NbMYB4L‐OE plants and controls. The different levels of TMV resistance were also consistent with antioxidant enzyme activity (Figure 7). Based on our evidence, we suggest that, during TMV infection, the ethylene‐NbEIL1 signal induces NbMYB4L‐mediated resistance responses and antioxidant enzymes systems. This signal cascade explains, at least partially, the regulatory network for lesion formation and virus resistance mediated by ethylene.
In summary, we propose a model showing the underlying mechanism that regulates the ethylene signalling pathway and the MYB4L‐mediated resistance during TMV infection of N. benthamiana (Figure 8). In this model, at the onset of TMV attack, the ethylene signal is activated, which enhances the stability of the NbEIL1 protein. NbEIL1 then interacts with the promoter of NbMYB4L and induces its transcription, which contributes to necrotic lesion formation and senescence of infected tissues in local leaves. We suggest NbEIL1 and NbMYB4L constitute a coherent feed‐forward loop, as NbMYB4L also stimulates ethylene production. Simultaneously, in uninfected and systemic leaf cells, the presence of ethylene again stabilizes EIL1 and potentially other TFs that could directly trigger the transcription of NbMYB4L. The activation of NbMYB4L leads to changing transcription of several defence‐related genes. SA also has a pivotal role in the SAR, and we speculate that NbMYB4L‐dependent responses do not conflict with SA. Other upstream molecules or transcription factors targeting NbMYB4L and the secondary metabolites controlled by NbMYB4L need further exploration. Overall, our findings provide insight into the transcriptional regulatory mechanism of the interaction of the ethylene signal with NbMYB4L to regulate plant disease resistance. These results provide important information and clues to further understand the mechanism of plant defence against viral pathogens.
FIGURE 8.
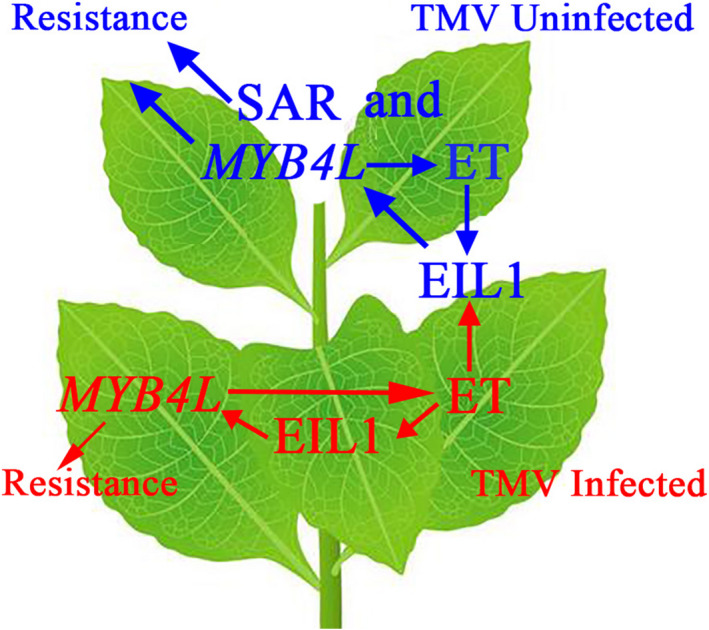
Proposed model for NbMYB4L‐mediated defence signalling. In local leaves, TMV infection triggers ethylene accumulation in the host cells, which also stabilizes EIL1. Spatially, ethylene released from infected leaves may activate the ethylene signal transduction pathway in systemic leaves, leading to NbMYB4L‐dependent defence responses being activated via NbEIL1. In TMV uninfected cells, the activated transcription of defence‐related genes means that there is no conflict between NbMYB4L‐dependent defence responses and salicylic acid (SA)‐dependent systemic acquired resistance (SAR). Additionally, the changing activity of antioxidant enzymes demonstrated that NbMYB4L probably triggers the modification of some secondary metabolic pathways. These series of changes result in enhancement of virus resistance
4. EXPERIMENTAL PROCEDURES
4.1. Plant growth and virus inoculation
The N. benthamiana plants were grown in a controlled growth chamber at 25°C under 16 h light/8 h darkness. When seedlings were 5 weeks old they were infiltrated with Agrobacterium tumefaciens carrying the binary vector with the TMV‐GFP replicon. At 7 dpi, the systemically infected leaves were collected, and 100 mg fresh tissue was ground in 10 ml of phosphate‐buffered saline (PBS; 8 g/L NaCl, 0.2 g/L KCl, 1.44 g/L Na2HPO4, 0.24 g/L KH2PO4, pH 7.4). The tissue suspension was used for mechanical virus inoculation, which was carried out on plants that were 3 weeks old. The fifth and sixth fully expanded leaves were inoculated. Plants were examined daily and the numbers of plants with symptoms recorded. After the symptoms appeared on the upper leaves, the leaves were photographed and harvested. GFP was photographed under ultraviolet (UV) light using a Canon digital camera and a B‐100AP longwave‐UV lamp (Ultra‐Violet Products) (Deng et al., 2016).
4.2. Total RNA extraction and RT‐qPCR
N. benthamiana leaves were frozen in liquid nitrogen and ground to a fine powder in a pestle and mortar. RNA was extracted using the Spectrum Plant Total RNA kit (Sigma‐Aldrich) following the manufacturer's recommendations. RNA was quantified by measuring absorbance at wavelengths of 260 and 280 nm using a NanoDrop 1000 spectrophotometer (Thermo Scientific). RNA was treated with DNase I (Promega) and cDNA synthesis conducted using SuperScript II reverse transcriptase (Invitrogen) following the manufacturer's guidelines. One microgram of total RNA was used for cDNA synthesis and RT‐qPCR analysis was performed. The qPCR assay was conducted as described previously (Zhu et al., 2018), using SYBR Green master mix (Applied Biosystems), and used for qPCR with a Rotor‐Gene‐Q (Qiagen). Amplification was followed by a melt curve analysis. The 2−ΔΔ C t method was used for relative quantification (Livak & Schmittgen, 2001). To detect transcript levels, oligonucleotides for specific genes were used (Table S1). Oligonucleotides amplifying Actin were used for normalization.
4.3. Generation and selection of transgenic plants
For overexpressing NbMYB4L, the coding region of NbMYB4L (Niben101Scf03570Ctg037) was inserted into pCAMBIA1307 vector, which contains the CaMV 35S promoter and an HA‐tag. The inserted fragment was obtained by PCR using a pair of primers linked to SalI and KpnI sequences on either side of the fused pMD19‐T vector, which carried the coding region of NbMYB4L. The pCM1307 vector was digested with SalI and KpnI, then the digested vector and PCR product were treated with T4 DNA ligase. Transgenic plants were generated by Agrobacterium tumefaciens EHA105‐mediated transformation according to the method described previously (Xu et al., 2012), and transformed lines were first selected for kanamycin (70 mg/L) resistance and then analysed by western blot to determine the presence of the HA‐tag.
To suppress NbMYB4L, a cDNA fragment was used for the double‐stranded RNA interference (RNAi) experiments. A segment of NbMYB4L was obtained by PCR and the fragment and its inverted repeat fragment were inserted downstream of the CaMV 35S promoter at the BamHI and SacI restriction sites of the modified pBI121 vector (Xu et al., 2012). The construct NbMYB4L‐RNAi was thus generated. The resulting constructs were verified by sequencing and the NbMYB4L‐RNAi‐containing vector was transformed into A. tumefaciens. Transformed lines were first selected for kanamycin (70 mg/L) resistance, and then the silencing efficiency was analysed by RT‐qPCR. All primers used are listed in Table S1.
4.4. TRV‐mediated virus‐induced gene silencing assay
For construction of VIGS vectors, partial cDNA of NbMYB4L (248 bp) and NbEIL1 (241 bp) were amplified by reverse transcription PCR from an N. benthamiana cDNA library using specific primers (Table S1). These PCR products were then inserted into the tobacco rattle virus‐derived (TRV)‐RNA2 vector. The VIGS assay was performed as described previously (Zhu et al., 2016, 2018).
4.5. Protein extraction and western blot analysis
Total protein was extracted using extraction buffer (50 mM Tris.HCl, pH 6.8, 5% mercaptoethanol, 10% glycerol, 4% SDS, 4 M urea). For western blot analysis, each sample was subjected to SDS‐PAGE and transferred to a nitrocellulose membrane. Reactive proteins were then labelled with antibodies against the GFP protein (Genscript; cat. no. A01704; 1:5000), and against HA (Genscript, cat. no. A01244, 1:10,000). Actin was used as the internal control and labelled with anti‐β‐actin antibody (Genscript, cat. no. A00702, 1:10,000).
4.6. Quantification of SA, JA, and ACC
The content of SA and JA was determined from samples of fresh leaves of WT, OE‐69, and RNAi plants. For each sample of different plants 0.5 g was ground in liquid nitrogen using a mortar and pestle. After the addition of 5 ml of extraction buffer (isopropanol, 5 M HCl), the suspension was stirred for 30 min at 4°C. Dichloromethane (10 ml) was then added and the mixture stirred again for 30 min at 4°C. After centrifugation at 13,000 × g for 5 min at 4°C, the supernatant was collected and the pellet re‐extracted. Subsequently, the supernatants were pooled and dried under a stream of nitrogen. The dried supernatant was then dissolved in 400 μl of methanol containing 0.1% carboxylic acid. Samples were analysed by HPLC‐electrospray ionization/MS‐MS using a Waters ACQUITY UPLC coupled to a Xevo TQ. Chromatographic separation was carried out on a Waters ACQUITY UPLC R BEH C18 100 mm × 2.1 mm × 1.7 μm column at 40°C. The solvent gradient used was 100% A (98/2 = H2O/CH3OH (vol/vol) + 0.05% CHOOH + 5 mM CH3COONH4) to B (CH3CN) over 10 min. Solvent B was held at 100% for 5 min then the solvent returned to 100% A for 10 min equilibration prior to the next injection. The solvent flow rate was 0.3 ml/min. The MS was operated in the negative mode using electrospray ionization as the ion source.
ACC was extracted from 0.5g of the same leaf tissues of WT, OE‐69, and RNAi plants that were ground in liquid nitrogen, then transferred into 50 ml tubes with 5 ml of deionized water and treated in the ultrasonic cleaning instrument for 20 min. After centrifugation at 10,000 × g for 5 min at 4°C, the supernatants were collected and the pH adjusted to 4.0 with HCl. Twenty millilitres of trichloromethane was added, the mixture was centrifuged at 10,000 × g for 5 min, the supernatant was collected, and the pellet re‐extracted. The supernatants were then collected and filtrated through a MCX polybase that had been pre‐activated with 3 ml of CH3OH followed by 3 ml of deionized water. The residue was eluted with 4 ml of 1 M NH3.H2O. The ACC concentration in the supernatant was determined by HPLC‐electrospray ionization/MS‐MS (Wang et al., 2019).
4.7. Yeast one‐hybrid assays
The coding region of NbEIL1 was inserted into the pGADT7 vector to generate the recombined constructs pGAD‐NbEIL1, while the promoter fragment of NbMYB4L was cloned into the pHIS2 vector. The co‐transformed Y187 yeast strains were plated on SD/−Trp/−Leu/−His medium, supplemented by appropriate concentration of 3‐amino‐1,2,4‐triazole (3‐AT) (Hickman et al., 2013).
4.8. Characterization of promoter activity
To determine the promoter activity, fragments of the NbMYB4L promoter region were amplified using specific primers and fused independently to the luciferase (LUC) reporter gene in the pGreenII 0800‐LUC vectors to generate the reporter constructs. The transient expression assays were performed using N. benthamiana leaves (An et al., 2018). The effectors were generated by recombining the NbEIL1 genes into the pGreenII 62‐SK vector. The recombinant plasmids were transformed into A. tumefaciens GV3101::pSOUP‐p19 (Weidi Co.). The bacteria were mixed and co‐injected into the N. benthamiana leaves. Transient expression was analysed in N. benthamiana leaves as described previously (Zhu et al., 2018).
4.9. Protein purification and EMSA assay
The NbEIL1 recombinant protein was prepared as previously described (Zhu et al., 2018). Briefly, the coding region of NbEIL1 was digested with BamHI and SalI and ligated into the same sites of pET‐32a vector. The recombinant vector was transformed into Escherichia coli BL21 (DE3). Expression of the recombinant proteins was induced by isopropyl β‐d‐1‐thiogalactopyranoside (IPTG) and purified according to the instructions of the Novagen pET purification system. The probes are shown in Table S1. EMSA of the protein–DNA complexes was performed according to the instructions of electrophoretic mobility shift assay (EMSA) Kit, with SYBR Green & SYPRO Ruby EMSA stains (Invitrogen).
4.10. ChIP‐qPCR assays
ChIP‐qPCR assays were performed as described by An et al. (2018). The 35S: NbEIL1 transiently transformed N. benthamiana leaves containing the HA targets were applied to the ChIP‐qPCR assays using the EpiTect ChIP OneDay Kit (Qiagen). The empty vector pCM1307‐HA was used as a control. An HA‐specific antibody was used in this study. The enriched DNA fragments were examined by RT‐qPCR using the primers shown in Table S1.
4.11. Measurement of the enzyme activities
For the enzyme assays, 500 mg of fresh leaves were homogenized in 5 ml of 25 mM PBS (pH 7.8) containing 0.2 mM EDTA, 2 mM ascorbic acid, and 2% polyvinylpyrrolidone (PVP). The homogenate was centrifuged at 12,000 × g for 20 min at 4°C, and the supernatant was immediately used for the determination of enzymatic activity. SOD activity was assayed by measuring the ability to inhibit the photochemical reduction of nitroblue tetrazolium (NBT) following the method of Zhu et al. (2016). CAT activity was measured as the decline in the absorbance at 240 nm due to the decrease of extinction of H2O2 using the method of Zhu et al. (2016).
4.12. Application of plant hormones and inhibitors
N. benthamiana leaves were treated on both sides of the fifth and sixth leaves with plant hormones or inhibitors then, 12 hours later, inoculated with TMV‐GFP on the pretreated leaves. The expression of NbMYB4L in the inoculated leaves was measured by RT‐qPCR from the time of inoculation up to 7 dpi. SA, JA, salicylhydroxamic acid (SHAM), ACC, and AVG were purchased from Sigma‐Aldrich. 2‐amidoindane‐2‐phosphonic acid (AIP) was purchased from Matrix‐Scientific. All hormone and inhibitor solutions were prepared in water containing 0.02% (vol/vol) Tween 20. The concentrations used were as follows: SA 300 μM, AIP 50 μM, JA 100 μM, SHAM 500 μM, ACC 10 μM, AVG 50 μM (Clarke et al., 1998; Lemarié et al., 2015; Zhu et al., 2014). Distilled water containing 0.02% vol/vol Tween 20 was used as a control treatment.
4.13. Quantification and statistical analysis
Three replicates, each of 20 plants per treatment and genotype, were used to assess the rate of systemic infection and three independent plants per treatment and genotype were sampled for RT‐qPCR analyses in all experiments. All results were obtained from three independent experiments. Analyses of variance with type II sum of squares were performed on log10‐transformed values to assess statistical differences of data sets. Unpairwise comparisons of treatment versus control values were performed with a two‐tailed Student's t test in Prism 7 and Holm–Sidak's multiple comparisons test was used for multiple comparison.
Supporting information
ACKNOWLEDGEMENTS
This work was financially supported by the Shandong Nature Science Foundation (ZR2020QC032), the “Bohai Sea Granary” Science and Technology Demonstration Project of Shandong province (2019 BHLC001), the National Natural Science Foundation of China (31772288, 31370296), and the Major Science and Technology Project of Yunnan province (2019ZG002). P.J. was financially supported by the “Double Hundred” Plan for Foreign Experts in Shandong Province, China. TMV‐GFP and useful suggestions were given by Professor Lili Shen of the Tobacco Research Institute, Chinese Academy of Agricultural Sciences. Writing skill suggestions were given by Dr John Clemens of the School of Biological Sciences, University of Canterbury.
Zhu, T. , Zhou, X. , Zhang, J.‐L. , Zhang, W.‐H. , Zhang, L.‐P. , You, C.‐X. , et al (2022) Ethylene‐induced NbMYB4L is involved in resistance against tobacco mosaic virus in Nicotiana benthamiana . Molecular Plant Pathology, 23, 16–31. 10.1111/mpp.13139
Contributor Information
Tong Zhu, Email: tzhu@ytu.edu.cn.
Peng‐Tao Ma, Email: ptma@ytu.edu.cn.
Shan‐Li Guo, Email: gsl@ytu.edu.cn.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Alazem, M. & Lin, N.S. (2015) Roles of plant hormones in the regulation of host–virus interactions. Molecular Plant Pathology, 16, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, F. , Zhao, Q. , Ji, Y. , Li, W. , Jiang, Z. , Yu, X. et al. (2010) Ethylene‐induced stabilization of ETHYLENE INSENSITIVE3 and EIN3‐LIKE1 is mediated by proteasomal degradation of EIN3 Binding F‐box 1 and 2 that requires EIN2 in Arabidopsis . The Plant Cell, 22, 2384–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, J.‐P. , Wang, X.‐F. , Li, Y.‐Y. , Song, L.‐Q. , Zhao, L.‐L. , You, C.‐X. et al. (2018) EIN3‐LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiology, 178, 808–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma, A. , Kobayashi, S. , Mitani, N. , Shiraishi, M. , Yamada, M. , Ueno, T. et al. (2008) Genomic and genetic analysis of Myb‐related genes that regulate anthocyanin biosynthesis in grape berry skin. Theoretical and Applied Genetics, 117, 1009–1019. [DOI] [PubMed] [Google Scholar]
- Chen, H. , Xue, L. , Chintamanani, S. , Germain, H. , Lin, H. , Cui, H. et al. (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3‐LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis . The Plant Cell, 21, 2527–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Yang, X. , He, K. , Liu, M. , Li, J. , Gao, Z. et al. (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology, 60, 107–124. [DOI] [PubMed] [Google Scholar]
- Chen, L. , Zhang, L. , Li, D. , Wang, F. & Yu, D. (2013) WRKY8 transcription factor functions in the TMV‐cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis . Proceedings of the National Academy of Sciences USA, 110, E1963–E1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chezem, W.R. , Memon, A. , Li, F.S. , Weng, J.K. & Clay, N.K. (2017) SG2‐type R2R3‐MYB transcription factor MYB15 controls defense‐induced lignification and basal immunity in Arabidopsis . The Plant Cell, 29, 1907–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, S.F. , Burritt, D.J. , Jameson, P.E. & Guy, P.L. (1998) Influence of plant hormones on virus replication and pathogenesis‐related proteins in Phaseolus vulgaris L. infected with white clover mosaic potexvirus. Physiological and Molecular Plant Pathology, 53, 195–207. [Google Scholar]
- Deng, X.G. , Tong, Z. , Zou, L.J. , Han, X.Y. , Xue, Z. , Xi, D.H. et al. (2016) Orchestration of hydrogen peroxide and nitric oxide in brassinosteroid‐mediated systemic virus resistance in Nicotiana benthamiana . The Plant Journal, 85, 478–493. [DOI] [PubMed] [Google Scholar]
- Durrant, W.E. & Dong, X. (2004) Systemic acquired resistance. Annual Review of Phytopathology, 42, 185–209. [DOI] [PubMed] [Google Scholar]
- Ent, S.V.D. , Verhagen, B.W.M. , Doorn, R.V. , Bakker, D. , Verlaan, M.G. , Pel, M.J.C. et al. (2008) MYB72 Is required in early signaling steps of Rhizobacteria‐induced systemic resistance in Arabidopsis . Plant Physiology, 146, 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, U. & Dröge‐Laser, W. (2004) Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to Tobacco mosaic virus . Molecular Plant‐Microbe Interactions, 17, 1162–1171. [DOI] [PubMed] [Google Scholar]
- Fodor, J. , Gullner, G. , Adam, A.L. , Barna, B. , Komives, T. & Kiraly, Z. (1997) Local and systemic responses of antioxidants to Tobacco mosaic virus infection and to salicylic acid in tobacco. Role in systemic acquired resistance. Plant Physiology, 114, 1443–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Z.Q. & Dong, X. (2013) Systemic acquired resistance: turning local infection into global defense. Annual Review of Plant Biology, 64, 839–863. [DOI] [PubMed] [Google Scholar]
- Fujimoto, S.Y. , Ohta, M. , Usui, A. , Shinshi, H. & Ohme‐Takagi, M. (2000) Arabidopsis ethylene‐responsive element binding factors act as transcriptional activators or repressors of GCC box‐mediated gene expression. The Plant Cell, 12, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, A. , Salih, G. & Klessig, D.F. (2000) Activation of a diverse set of genes during the tobacco resistance response to TMV is independent of salicylic acid; induction of a subset is also ethylene independent. The Plant Journal, 21, 409–418. [DOI] [PubMed] [Google Scholar]
- Guo, H. & Ecker, J.R. (2004) The ethylene signaling pathway: new insights. Current Opinion in Plant Biology, 7, 40–49. [DOI] [PubMed] [Google Scholar]
- Hibi, T. , Kosugi, S. , Iwai, T. , Kawata, M. , Seo, S. , Mitsuhara, I. et al. (2007) Involvement of EIN3 homologues in basic PR gene expression and flower development in tobacco plants. Journal of Experimental Botany, 58, 3671–3678. [DOI] [PubMed] [Google Scholar]
- Hickman, R. , Hill, C. , Penfold, C.A. , Breeze, E. , Bowden, L. , Moore, J.D. et al. (2013) A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. The Plant Journal, 75, 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson, P.E. & Clarke, S.F. (2002) Hormone–virus interactions in plants. Critical Reviews in Plant Sciences, 21, 205–228. [Google Scholar]
- Kim, C.Y. , Liu, Y. , Thorne, E.T. , Yang, H. , Fukushige, H. , Gassmann, W. et al. (2003) Activation of a stress‐responsive mitogen‐activated protein kinase cascade induces the biosynthesis of ethylene in plants. The Plant Cell, 15, 2707–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H. , Lam, P.Y. , Lee, M.‐H. , Jeon, H.S. , Tobimatsu, Y. & Park, O.K. (2020) The Arabidopsis R2R3 MYB transcription factor MYB15 is a key regulator of lignin biosynthesis in effector‐triggered immunity. Frontiers in Plant Science, 11, 583153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Király, Z. , Barna, B. , Kecskés, A. & Fodor, J. (2002) Down‐regulation of antioxidative capacity in a transgenic tobacco which fails to develop acquired resistance to necrotization caused by TMV. Free Radical Research, 36, 981–991. [DOI] [PubMed] [Google Scholar]
- Knoester, M. , Linthorst, H.J.M. , Bol, J.F. & Van Loon, L.C. (2001) Involvement of ethylene in lesion development and systemic acquired resistance in tobacco during the hypersensitive reaction to Tobacco mosaic virus . Physiological and Molecular Plant Pathology, 59, 45–57. [Google Scholar]
- Künstler, A. , Király, L. , Kátay, G. , Enyedi, A.J. & Gullner, G. (2019) Glutathione can compensate for salicylic acid deficiency in tobacco to maintain resistance to Tobacco mosaic virus . Frontiers in Plant Science, 10, 1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea, U.S. , Slimestad, R. , Smedvig, P. & Lillo, C. (2007) Nitrogen deficiency enhances expression of specific MYB and bHLH transcription factors and accumulation of end products in the flavonoid pathway. Planta, 225, 1245–1253. [DOI] [PubMed] [Google Scholar]
- Lemarié, S. , Robert‐Seilaniantz, A. , Lariagon, C. , Lemoine, J. , Marnet, N. , Jubault, M. et al. (2015) Both the jasmonic acid and the salicylic acid pathways contribute to resistance to the biotrophic clubroot agent Plasmodiophora brassicae in Arabidopsis . Plant and Cell Physiology, 56, 2158–2168. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. & Dinesh‐Kumar, S.P. (2004) Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N‐mediated resistance to Tobacco mosaic virus . The Plant Journal, 38, 800–809. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Zhao, T.‐J. , Liu, J.‐M. , Liu, W.‐Q. , Liu, Q. , Yan, Y.‐B. et al. (2006) The conserved Ala37 in the ERF/AP2 domain is essential for binding with the DRE element and the GCC box. FEBS Letters, 580, 1303–1308. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. & Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Miyahara, T. , Satoh, S. , Maeda, K. , Kimura, S. , Sasaki, N. & Ozeki, Y. (2010) Isolation of Daucus carota ethylene insensitive3‐like (DcEIL) involved in stress‐inducible DcMYB1 expression in suspension‐cultured carrot cells. Plant Biotechnology, 27, 91–97. [Google Scholar]
- Murray, S.L. , Adams, N. , Kliebenstein, D.J. , Loake, G.J. & Denby, K.J. (2005) A constitutive PR‐1:luciferase expression screen identifies Arabidopsis mutants with differential disease resistance to both biotrophic and necrotrophic pathogens. Molecular Plant Pathology, 6, 31–41. [DOI] [PubMed] [Google Scholar]
- Ohtsubo, N. , Mitsuhara, I. , Koga, M. , Seo, S. & Ohashi, Y. (1999) Ethylene promotes the necrotic lesion formation and basic PR gene expression in TMV‐infected tobacco. Plant and Cell Physiology, 40, 808–817. [Google Scholar]
- Qiu, K. , Li, Z. , Yang, Z. , Chen, J. , Wu, S. , Zhu, X. et al. (2015) EIN3 and ORE1 accelerate degreening during ethylene‐mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis . PLoS Genetics, 11, e1005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, P.J. & Park, C.M. (2010) MYB96‐mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis . New Phytologist, 186, 471–483. [DOI] [PubMed] [Google Scholar]
- Seyfferth, C. & Tsuda, K. (2014) Salicylic acid signal transduction: the initiation of biosynthesis, perception and transcriptional reprogramming. Frontiers in Plant Science, 5, 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vailleau, F. , Daniel, X. , Tronchet, M. , Montillet, J.L. , Triantaphylidès, C. & Roby, D. (2002) A R2R3‐MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proceedings of the National Academy of Sciences USA, 99, 10179–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberne, M.C. , Hoekstra, J. , Bol, J.F. & Linthorst, H.J.M. (2003) Signaling of systemic acquired resistance in tobacco depends on ethylene perception. The Plant Journal, 35, 27–32. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Ran, L. , Hou, Y. , Tian, Q. , Li, C. , Liu, R. et al. (2017) The transcription factor MYB115 contributes to the regulation of proanthocyanidin biosynthesis and enhances fungal resistance in poplar. New Phytologist, 215, 351–367. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Han, K. , Peng, J. , Zhao, J. , Jiang, L. , Lu, Y. et al. (2019) NbALD1 mediates resistance to Turnip mosaic virus by regulating the accumulation of salicylic acid and the ethylene pathway in Nicotiana benthamiana . Molecular Plant Pathology, 20, 990–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Goregaoker, S.P. & Culver, J.N. (2009) Interaction of the Tobacco mosaic virus replicase protein with a NAC domain transcription factor is associated with the suppression of systemic host defenses. Journal of Virology, 83, 9720–9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, F. , Yuan, S. , Zhang, D.W. , Lv, X. & Lin, H.H. (2012) The role of alternative oxidase in tomato fruit ripening and its regulatory interaction with ethylene. Journal of Experimental Botany, 63, 5705–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.Y. , Li, J.G. , Pei, M. , Gu, H. , Chen, Z.L. & Qu, L.J. (2007) Over‐expression of a flower‐specific transcription factor gene AtMYB24 causes aberrant anther development. Plant Cell Reports, 26, 219–228. [DOI] [PubMed] [Google Scholar]
- Yang, Y. & Klessig, D.F. (1996) Isolation and characterization of a Tobacco mosaic virus‐inducible myb oncogene homolog from tobacco. Proceedings of the National Academy of Sciences USA, 93, 14972–14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, S. & Li, Y. (2021) Current understanding of the interplays between host hormones and plant viral infections. PLoS Pathogens, 17, e1009242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, F. , Xi, D.H. , Yuan, S. , Xu, F. & Lin, H.H. (2014) Salicylic acid and jasmonic acid are essential for systemic resistance against Tobacco mosaic virus in Nicotiana benthamiana . Molecular Plant‐Microbe Interactions, 27, 567–577. [DOI] [PubMed] [Google Scholar]
- Zhu, T. , Deng, X.G. , Tan, W.R. , Zhou, X. , Luo, S.S. , Han, X.Y. et al. (2016) Nitric oxide is involved in brassinosteroid‐induced alternative respiratory pathway in Nicotiana benthamiana seedlings’ response to salt stress. Physiologia Plantarum, 156, 150–163. [DOI] [PubMed] [Google Scholar]
- Zhu, T. , Zou, L. , Li, Y. , Yao, X. , Xu, F. , Deng, X. et al. (2018) Mitochondrial alternative oxidase‐dependent autophagy involved in ethylene‐mediated drought tolerance in Solanum lycopersicum . Plant Biotechnology Journal, 16, 2063–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


