Abstract
The transcriptional coactivator CREB binding protein (CBP) possesses intrinsic histone acetyltransferase (HAT) activity that is important for gene regulation. CBP binds to and cooperates with numerous nuclear factors to stimulate transcription, but it is unclear if these factors modulate CBP HAT activity. Our previous work showed that CBP interacts with the Epstein-Barr virus-encoded basic region zipper (b-zip) protein, Zta, and augments its transcriptional activity. Here we report that Zta strongly enhances CBP-mediated acetylation of nucleosomal histones. Zta stimulated the HAT activity of CBP that had been partially purified or immunoprecipitated from mammalian cells as well as from affinity-purified, baculovirus expressed CBP. Stimulation of nucleosome acetylation required the CBP HAT domain, the Zta DNA binding and transcription activation domain, and nucleosomal DNA. In addition to Zta, we found that two other b-zip proteins, NF-E2 and C/EBPα, strongly stimulated nucleosomal HAT activity. In contrast, several CBP-binding proteins, including phospho-CREB, JUN/FOS, GATA-1, Pit-1, and EKLF, failed to stimulate HAT activity. These results demonstrate that a subset of transcriptional activators enhance the nucleosome-directed HAT activity of CBP and suggest that nuclear factors may regulate transcription by altering substrate recognition and/or the enzymatic activity of chromatin modifying coactivators.
Eukaryotic gene expression is inhibited by higher order chromatin structures that limit the access of transcription factors to regulatory DNA sequences (reviewed in references 43, 45, and 67). These higher order structures are thought to be regulated by posttranslational modification of chromatin components (66). The acetylation of lysine residues in the core histone amino terminal tails is one chromatin modification that correlates with increased transcription activity (reviewed in references 36, 40, 56, 66, and 70). Several transcriptional coactivators possess intrinsic histone acetyltransferase (HAT) activity, including the CREB binding protein (CBP), its close relative p300, the SAGA-associated GCN5, the GCN5-related P/CAF, the MYST family protein Tip60, and the TATA-binding protein-associated factor TAFII250 (7, 15, 35, 39, 59, 61). Numerous transcriptional activators bind HAT-containing coactivators through interaction domains that are essential for transcription function (68). One important question is whether the coactivator HAT activity is constitutive or whether it can be modulated by interaction with transcriptional activators.
CBP was isolated by virtue of its ability to bind the transcriptionally active phosphorylated form of CREB (34). Subsequently, CBP has been shown to function as a transcriptional coactivator for numerous cellular transcription factors, including the tumor suppressor p53, proto-oncoproteins c-JUN and c-MYB, nuclear hormone receptors, the hematopoietic transcription factors EKLF, GATA-1, and NF-E2, and the viral proteins, such as the adenovirus E1A, the simian virus 40 T antigen, the human papilloma virus E6, and the Epstein-Barr virus Zta (1, 8, 13, 19, 24, 26, 28, 54, 63, 72, 73). CBP and p300 are essential for early embryogenesis, and aberrations in these genes have been implicated in developmental abnormalities and human cancers (reviewed in references 33, 34, and 64). Both CBP and p300 have intrinsic HAT activity for all four histones, and the HAT domain of CBP can stimulate transcription when tethered upstream of some core promoters (7, 57, 61). In addition to acetylating histone tails, CBP is also capable of acetylating nonhistone nuclear factors, including the tumor suppressor p53 (37), the erythroid differentiation factor GATA-1 (14, 41), and the architectural protein HMGI/Y (60). Acetylation of p53 increases its ability to bind DNA and consequently to stimulate transcription, while the acetylation of HMGI/Y may inhibit DNA binding and limit the duration of an activation signal. Moreover, CBP associates with other HAT-containing coactivators, including P/CAF (71), ACTR (18), and SRC-1 (65), suggesting that acetylation may be a signaling mechanism coupled to transcription activation.
Epstein-Barr virus is a human herpesvirus that establishes a latent infection in B lymphocytes (reviewed in references 46 and 62). During latency the viral genome is maintained as an extrachromosomal episome that is repressed for transcription by chromatin packaging (27). Latency can be disrupted by treatment with sodium butyrate, which is known to affect histone acetylation, as well as by expression of Zta, the viral immediate-early protein (46). Zta is a member of the basic region zipper (b-zip) family of DNA binding proteins that stimulates transcription of several viral and cellular genes essential for viral lytic replication (16, 23, 29, 30, 52). Zta activates transcription on naked DNA templates in vitro by recruiting general transcription factors TFIIA and TFIID (20, 50). Transcription activation of the viral chromosome is stimulated by CBP coexpression, and Zta can bind to two domains of CBP, referred to as the cysteine-histidine (C/H)-rich regions 1 and 3 (72). Both the activation domain and the DNA binding domain of Zta have been implicated in the binding to CBP.
Promoter regulatory regions consist of a collection of binding sites for transcription factors with nonredundant functions. The locus control region (LCR) of the β-globin gene cluster, for example, contains multiple sites for the hematopoietic transcription factors GATA-1, NF-E2, and EKLF and is thought to contribute to the formation of an open chromatin domain at the globin gene locus (reviewed in reference 48). GATA-1, NF-E2, and EKLF can all bind DNA and recruit CBP independently, but it is the concerted action of all three that triggers LCR activity (reviewed in reference 12). The interferon enhancer is another example of a transcriptional control region that requires the concerted activity of several factors that individually can all bind CBP but only in concert can trigger a transcription response (58). While the stereospecific and cooperative binding of these factors are required for the stable association of CBP with the promoter control region, it is not clear whether these factors are simply redundant recruiting modules or whether they contribute mechanistically distinct interactions that alter CBP activity.
A second major question in gene regulation is how chromatin-modifying activities are targeted to specific sites in the genome. Chromatin-modifying activities, like CBP, do not bind DNA directly and are thought to be recruited to specific sites in the genome by DNA bound nuclear factors. However, it remains unclear how sequence-specific factors can recruit chromatin-modifying activities to sites obstructed by higher order chromatin structures. While some transcription factors can bind to their cognate sites in phased mononucleosomes (10), it seems more problematic for these factors to access their sites on the highly compact 30-nm chromatin fiber more characteristic of repressed heterochromatin in vivo. One possibility is that some transcriptional activators initiate the unfolding of chromatin by stimulating chromatin modification in a sequence-independent manner. Several CBP interacting proteins that lack DNA binding activity, like E1A, have been shown to alter the activity of CBP in a sequence-independent manner (17, 38). In this work we explore the possibility that some CBP-interacting nuclear proteins can promote histone acetylation. We found that a class of b-zip proteins, represented by Zta, NF-E2, and C/EBPα, stimulate the acetylation of nucleosomal histones by CBP in a reaction dependent upon the CBP HAT domain and the presence of nucleosomal DNA.
MATERIALS AND METHODS
Plasmid and recombinant proteins.
Zta, ZtaΔ(2-141) and ZtaΔ(141-245) were prepared as described previously (50). Zta m.1 has alanine substitution at F22, F26, W74, and F75. Zta m.2 has alanine substitution at L48 and W49 and was prepared as described previously (53). Zta-dbd has alanine substitution at R187, K188, and C189. CBP-C/H1, containing CBP amino acids (aa) 301 to 585, was prepared as already described (72). BHLF1CAT was described previously (51). The expression plasmids for GST-GATA-1 were also described previously (13). GST-NF-E2 is a tethered heterodimer of p45 and MafG (a gift from V. Blank [11]). GST-EKLF contains the zinc finger DNA binding domain of EKLF (aa 272 to 376) in the pGEX2T vector. Rat C/EBPα was cloned as an NcoI fragment into pRSETB (Invitrogen) and purified by Ni-nitrilotriacetic acid (NTA) agarose chromatography. GST-GCN4 protein was a gift of Shelley Berger. CREB was a gift from Ramin Shiekhattar. Full-length c-JUN and c-FOS proteins were gifts of Tom Kerppola. Pit-1 and CREM protein were purchased from Santa Cruz Biotechnology. Flag-tagged full-length CBP and Flag-tagged CBPΔHAT have been described previously (41). CBPΔN(700-2441) and CBPΔNΔH were generated by PCR amplification with Vent DNA polymerase using CBP wild type (wt) or CBPΔHAT as templates and were cloned into pCMV-FLAG2 (Sigma) as BamHI-HindIII fragments. The BZLF1 promoter-luciferase construct was generated by amplification of BZLF1 −220 to +12 as a NheI-HindIII fragment in pGL3BASIC (Promega).
HeLa nuclear extract.
HeLa nuclear extracts were prepared as described in Dignam et al. (25). Extracts were fractionated on a P11 phosphocellulose (PC) column and 0.1, 0.3, 0.5, and 1 M KCl step elutions were collected.
Baculovirus CBP.
Hexa-histidine-tagged CBP baculovirus was obtained from Dimitrios Thanos. SF-9 cells were seeded on 100-mm plates with 107 cells and were infected with His-tagged CBP virus at a multiplicity of infection of 5. Cells were harvested 72 h postinfection. Cell lysates were prepared using a Dounce homogenizer (12 strokes) in buffer H (10 mM Tris, 10% glycerol, 0.5 M NaCl, 15 mM imidazole, 0.1% NP-40, 2 mM β-mercaptoethanol, 2 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg of leupeptin/ml, 1 μg of pepstatin/ml). Cell lysates were incubated with Ni-NTA agarose at 4°C for 2 h and washed with washing buffer (buffer H containing 0.3 M NaCl and 5 mM imidazole). His-tagged CBP was eluted with elution buffer (buffer H containing 250 mM imidazole and 0.2 M NaCl) and analyzed by Western analysis using CBP antibody (A-22; Santa Cruz Biotechnology).
SON and core histone preparation.
Small oligonucleosomes (SONs) from HeLa cell nuclear pellets were prepared as described previously (22). Free histones were purchased from Sigma (type III-S). Core histones were generated by hydroxyapatite chromatography of HeLa nuclear pellets as described before (22).
HAT assay.
SONs (100 to 200 ng) were incubated with 0.25 μCi [3H]acetyl coenzyme A ([3H]acetyl CoA) (Amersham) and approximately 250 μg of HeLa nuclear extract or PC fractions in the presence or absence of Zta (300 ng unless otherwise indicated) in a 30-μl HAT buffer (50 mM Tris [pH 8.0], 5% glycerol, 0.1 mM EDTA, 50 mM KCl, 1 mM dithiothreitol (DTT), 1 mM PMSF, and 10 mM sodium butyrate) at 30°C for 1 h. Zta mutants m.1, m.2, and Zta-dbd and other transcription activators were typically used at 300 ng for HAT assays. The reactions were resolved by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis (SDS–15% PAGE). The gel was enhanced using Entensify (NEN) and was analyzed by autoradiography. The 30-bp double-stranded oligonucleotides ZRE-wt and ZRE-m were included as competitors in some reactions, as indicated. Oligonucleotide sequences of the top strand are as follows: ZRE-wt oligonucleotide (5′-GATCTTCTAGACCAAATGTGCAAAGGTGAG); and ZRE-m oligonucleotide (5′-GATCTTCTAGACCAAATCTCCGGGGGTGAG). To determine if DNA of SONs is essential for Zta stimulation, 200 ng of SONs was digested with 0.2, 2, and 20 U of micrococcal nuclease (MNase)/ml in HAT buffer at 25°C for 1 min prior to HAT assays.
Immunoprecipitation and Western analysis.
Approximately 2.5 mg of HeLa nuclear extract was used for each immunoprecipitation to incubate with 10 μl of anti-CBP-NT antibody (A-22; Santa Cruz Biotechnology) or control antibody at 4°C for 1 h. Protein A-Sepharose CL4B was added to the reaction for 1 h at 4°C. The beads containing immunoprecipitates were washed three times with 0.5 ml of washing buffer (20 mM HEPES, 20% glycerol, 400 mM KCl, 0.2 mM EDTA, 1 mM DTT, 0.05% NP-40, and protease inhibitors). Approximately 150 μg of crude HeLa nuclear extract or PC fractions was used for Western analysis. Anti-CBP-NT antibody (A-22) or anti-Flag M2 (Sigma) was used to detect CBP or transfected Flag-tagged CBP. For immunoprecipitation HAT assays, NIH 3T3 cells were transfected with Flag-tagged CBP constructs using Lipofectamine (Gibco-BRL). Nuclear extracts were prepared according to Andrews and Faller (5). High-salt nuclear extracts were diluted with water containing 0.5 mM DTT, 10 mM sodium butyrate, and protease inhibitors to reduce NaCl concentrations to 150 mM. Anti-Flag antibodies (5 μg) were used for immunoprecipitation. Immunoprecipitation reactions contained 5 μg of anti-Flag antibody for 2 h, followed by protein G-Sepharose beads for another 2 h of incubation at 4°C on a wheel. Beads were washed five times in 150 mM NaCl–50 mM Tris [pH 7.5]–0.1% Igepal–0.5 mM DTT–10 mM butyrate–protease inhibitors. Prior to the HAT assay, Western analysis was performed to ensure that equal amounts of CBP protein were present in each reaction.
EMSAs.
Magnesium-agarose and acrylamide electrophoretic mobility shift assays (EMSAs) were performed as described previously (50). Acrylamide EMSA was performed with 7% polyacrylamide for Zta binding or 4% polyacrylamide for binding to SONs. Agarose (1.4%) was used for activator binding to SONs. Approximately 50 to 150 ng of various activators was incubated with 32P-labeled SONs in the presence of 80 μg of poly(dIdC)/ml at 30°C for 30 min.
Chromatin assembly on biotinylated DNA templates.
Biotinylated PCR products were generated using a 5′ chemically biotinylated oligonucleotide. Two micrograms of 400-bp PCR products containing five ZREs or five GREs were assembled by salt titration using HeLa cell oligonucleosomes. HAT assays were performed in the presence of donor oligonucleosomes and 80 μg of poly dldC/ml. After the HAT reaction (30 min, 30°C), the biotinylated nucleosomes were purified using 10 μl of Dynabeads (Dynal) and were washed three times in HAT reaction buffer. Acetylated histones were eluted by boiling the beads in sample loading buffer and were visualized by fluorography of SDS-PAGE gels.
RESULTS
CBP HAT domain contributes to Zta coactivation.
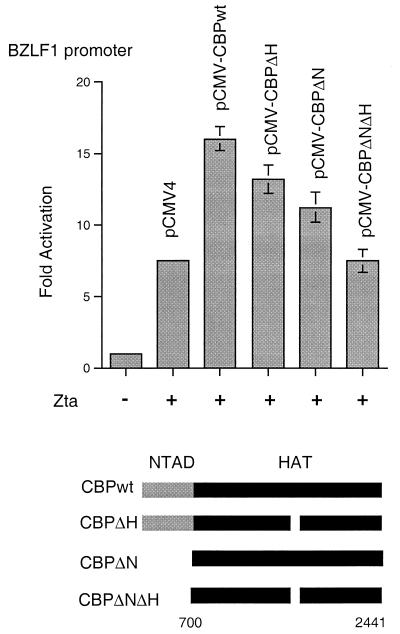
Previous work has shown that CBP cooperates with Zta to stimulate reactivation of latent EBV (1, 72). We had found that the CBP amino terminal activation domain (NTAD [aa 1 to 700]) was sufficient for transcription coactivation of the viral EA-D promoter and a synthetic promoter in transient transfections. In contrast to our findings, Adamson and Kenney found that the CBP HAT domain was essential for coactivation of the EBV BRLF1 promoter (1). Additionally, Jenkins et al. showed that the BZLF1 promoter regulating Zta expression undergoes changes in histone H4 acetylation during viral reactivation (44). To determine the relative contribution of the CBP NTAD and HAT domains to CBP coactivation, we assayed the BZLF1 promoter in transient transfection assays with Zta and various CBP deletion mutants (Fig. 1). We found that wild-type CBP potentiated Zta activation of BZLF1 by two- to threefold. A deletion in the CBP HAT domain (aa 1458 to 1475) that eliminates HAT activity reduced but did not eliminate CBP coactivation (Fig. 1, CBPΔH). Deletion of the CBP NTAD (aa 1 to 699) similarly reduced but did not eliminate coactivation of BZLF1 (Fig. 1, CBPΔN). In contrast, deletion of the NTAD and the HAT domain completely eliminated CBP coactivation of BZLF1 (Fig. 1, CBPΔNΔH). Zta and CBP deletion mutants were expressed in these assays to similar levels, as determined by Western blot analysis (data not shown). These results suggest that CBP potentiates Zta transcription by more than one mechanism, and CBP HAT activity contributes to BZLF1 coactivation in transient transfection assays.
FIG. 1.
CBP HAT domain contributes to Zta coactivation. HeLa cells were cotransfected with 1 μg of BZLF1-luciferase reporter plasmid and expression plasmids for Zta, CBPwt, CBPΔH, CBPΔN, CBPΔNΔH, or the pCMV4 control vector as indicated in the figure. Luciferase units were expressed as fold activation. Values are the averages of at least three independent transfections.
HAT activity in nuclear extracts.
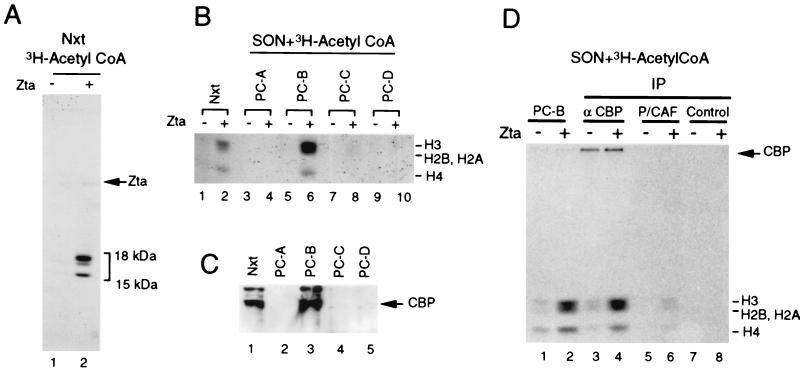
To determine if Zta altered the acetylation of cellular substrates or was itself a target of acetylation, HeLa nuclear extracts were incubated with [3H]acetyl CoA in the absence or presence of Zta and then assayed by SDS-PAGE and fluorography (Fig. 2A). We found that the addition of Zta to HeLa nuclear extracts stimulated the acetylation of polypeptides with mobilities similar to those of histones H3, H2A, H2B, and H4 (Fig. 2A, lane 2). To further characterize this acetylation reaction, HeLa nuclear extracts were fractionated over PC and then assayed for Zta-dependent acetylation (Fig. 2B). When purified SON substrate was supplemented to each fraction, Zta-dependent acetylase activity was detected in the PC-B fraction (0.3 M KCl step elution) (Fig. 2B). In the absence of exogenous SONs, Zta-dependent acetylation was undetectable, indicating that the acetylation substrate is indeed core histones H3, H2A, H2B, and H4 (data not shown). The PC fractions were then assayed by Western blot to determine if CBP cofractionated with the Zta-regulated HAT activity. The majority of CBP coeluted with the HAT activity found in the PC-B fraction (Fig. 2C). The PC-B fraction was then subjected to immunoprecipitation with anti-CBP specific antisera (A22; Santa Cruz Biotechnology), with anti-P/CAF antibody (D20; Santa Cruz Biotechnology), or as the control, with protein A-Sepharose beads (Fig. 2D). Zta stimulated HAT activity in the PC-B fraction, as above (Fig. 2D, lanes 1 and 2). Beads from the CBP-specific immunoprecipitates also retained Zta-stimulated HAT activity (Fig. 2D, lanes 3 and 4), whereas immunoprecipitates from P/CAF-specific antibodies or control reactions did not reveal significant acetylase activity in the absence or presence of Zta (Fig. 2D, lanes 5 to 8), indicating that CBP is the predominant HAT activity in this fraction. An acetylated polypeptide with a mobility similar to that of CBP was found in the anti-CBP immunoprecipitate, indicating that CBP autoacetylates in vitro. However, the addition of Zta did not further stimulate CBP autoacetylation (Fig. 2D, lanes 3 and 4).
FIG. 2.
Zta stimulates CBP-associated HAT activity. (A) HeLa nuclear extract (Nxt) (250 μg) was incubated with [3H]acetyl CoA in the absence (lane 1) or presence (lane 2) of Zta (300 ng) and then assayed by SDS-PAGE and fluorography. (B) HeLa nuclear extract was incubated with [3H]acetyl CoA without (lane 1) or with Zta (lane 2). PC fractions of HeLa nuclear extracts were incubated with [3H]acetyl CoA and purified SONs with or without Zta as indicated. Acetylation of nucleosomes was assayed by SDS-PAGE and fluorography. The positions of core histones H3, H2B, H2A, and H4 are indicated at the right. (C) Western blot of HeLa nuclear extract and PC fractions probed with CBP-specific antisera. (D) HAT activity of CBP-specific immunoprecipitates can be stimulated by Zta. HAT assays containing SONs, [3H]acetyl CoA and either the PC-B fraction (lanes 1 and 2), CBP-specific immunoprecipitates (lanes 3 and 4), P/CAF-specific immunoprecipitates (lanes 5 and 6), or protein A only immunoprecipitates (lanes 7 and 8) were incubated in the absence (−) or presence (+) of Zta. Acetylated CBP and histones are indicated.
Zta stimulates CBP HAT activity.
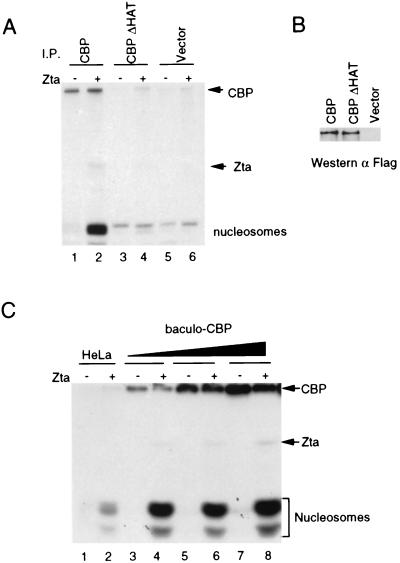
CBP binds several proteins with intrinsic HAT activity, including P/CAF, SRC1, and ACTR1, all of which might account for the Zta-mediated stimulation of HAT activity. To determine if the HAT activity of CBP was essential for the responsiveness to Zta, we transfected NIH 3T3 cells with Flag-tagged full-length CBP or with Flag-tagged CBP containing an 18-amino-acid deletion (aa 1458 to 1475) in the HAT domain (ΔHAT) (57) (Fig. 3A). Immunoprecipitates derived from transfected cell extracts were then assayed for HAT activity directed against small oligonucleosomes in the presence or absence of Zta. We found that the HAT activity of immunoprecipitated wild-type CBP was stimulated by Zta (lane 1 and 2), whereas CBP ΔHAT (lanes 3 and 4), or control (lanes 5 and 6) showed no response to Zta. The presence of equal amounts of wild-type and HAT-deficient CBP proteins was confirmed by Western blotting analysis (Fig. 3B). These results indicate that the CBP HAT domain is essential for Zta mediated stimulation of nucleosomal histone acetylation.
FIG. 3.
CBP HAT activity is necessary and sufficient for activation by Zta. (A) Immunoprecipitates derived from NIH 3T3 cells transfected with Flag-tagged full-length CBP (lanes 1 and 2), Flag-tagged CBP ΔHAT (lanes 3 and 4) or vector (lanes 5 and 6) were assayed for HAT activity with purified SONs in the absence (−) or presence (+) of Zta. (B) Western blot of immunoprecipitates derived from NIH 3T3 cells transfected with Flag CBP, Flag CBPΔHAT, or vector that were used for the HAT assay in panel A. (C) Ni-NTA-purified His-tagged CBP expressed and purified from baculovirus was assayed for acetylation of SONs in the absence (−) or presence (+) of Zta. CBP was increased by threefold increments up to 200 ng. Acetylated products were visualized by fluorography of SDS-PAGE gels.
To determine if CBP was sufficient for Zta mediated stimulation, we expressed hexa-His-tagged CBP to high levels in baculovirus and purified it to near homogeneity by affinity chromatography on Ni-NTA agarose. Affinity purified CBP was tested for its ability to acetylate small oligonucleosomes in the absence or presence of Zta (Fig. 3C). At all concentrations of CBP tested, we found that Zta stimulated acetylation of nucleosomes by CBP. Zta similarly stimulated CBP protein that was expressed in baculovirus as a hemagglutinin-tagged protein and isolated by 12CA5 monoclonal antibody affinity purification (data not shown). Based on these results, stimulation of HAT activity by Zta is very likely due to CBP and not to other HATs which may copurify with CBP at low concentrations.
Stimulation of HAT activity depends on transcriptional activation and DNA binding domain of Zta.
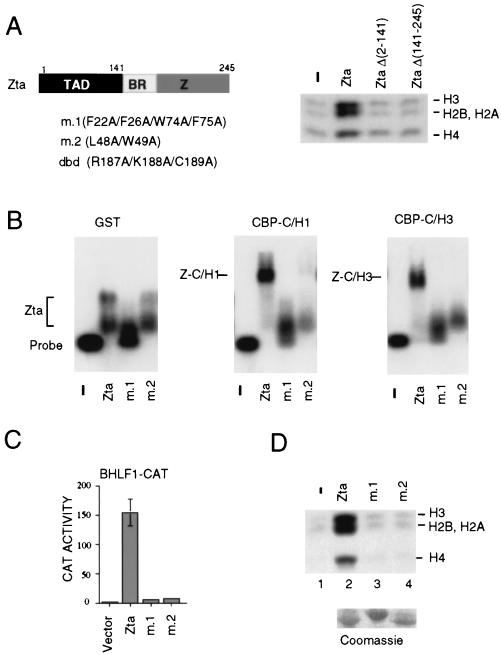
Zta is a member of the b-zip family of DNA binding proteins that forms a stable homodimer through the C-terminal basic region and zipper domain. It was previously found that the amino-terminal activation domain of Zta was required for binding to CBP (72). To determine which domains of Zta were important for the stimulation of HAT activity, we tested Zta deletion mutants lacking the amino-terminal activation domain (Δ2-141) or the DNA binding-dimerization domain (Δ141-245). While full-length Zta stimulated HAT activity, the amino-terminal activation domain and the carboxy-terminal DNA binding-dimerization domain by themselves failed to stimulate HAT activity (Fig. 4A). Previously published work identified several aromatic residues in the Zta amino-terminal activation domain that were essential for transcription stimulation of the EBV-BHLF1 promoter (53). Alanine substitutions at Zta aa F22, F26, W74, and F75 (m.1) or at L48 and W49 (m.2) were previously shown to reduce transcription activation but had no effect on Zta dimerization or DNA binding function (53). Mutants m.1 and m.2 were tested for their ability to form a complex with the C/H1 and C/H3 domains of CBP in an EMSA. Using magnesium agarose EMSA with a probe containing 5 Zta response elements (ZREs), Zta bound to CBP C/H1 and CBP C/H3, consistent with our previous results, but neither m.1 nor m.2 formed a stable complex with CBP-C/H1 or CBP-C/H3 (Fig. 4B). The transcriptional activities of m.1 and m.2 were compared to wild-type Zta in transient transfection assays with the Zta-responsive viral BHLF1 promoter-CAT reporter construct. As expected, m.1 and m.2 were defective for transcription activation relative to Zta (Fig. 4C). These experiments establish that m.1 and m.2 do not bind the CBP-C/H1 or C/H3 domain and suggest that interaction with CBP is essential for transcription activation. The m.1 and m.2 mutants of Zta were then assayed along with wild-type Zta for their ability to stimulate HAT activity (Fig. 4D). We found that m.1 and m.2 were unable to stimulate CBP HAT activity (Fig. 4D, lanes 3 and 4), suggesting that physical interaction of Zta with CBP is required for stimulation of HAT activity. These results further suggest that stimulation of HAT activity might constitute one mechanism by which Zta activates transcription.
FIG. 4.
Transcriptional activation domain of Zta is required for stimulation of HAT activity. (A) Schematic indicating Zta functional domains. The transcriptional activation domain (TAD) maps to amino acid residues 1 to 141. The DNA binding basic region (BR) and zipper-like dimerization domain (Z) map to residues 141 to 245. Alanine substitution mutations in the activation domains m.1 and m.2 and the DNA binding (dbd) domain are indicated. HeLa nuclear extracts were incubated alone, with full-length Zta, with Zta (Δ2-141), or with Zta (Δ141-245) and were assayed for HAT activation. (B) Zta mutants m.1 and m.2 abrogate CBP recruitment in magnesium agarose EMSA. The Z5E4T promoter probe alone (lane 1, at left) and with wild-type Zta (lane 2), m.1 (lane 3), or m.2 (lane 4) are shown with GST (left panel), GST-CBP-C/H1 (middle panel), or GST-CBP-C/H3 (right panel). (C) Transiently transfected HeLa cells were assayed for CAT activity from the BHLF1 promoter after cotransfection of vector, wild-type Zta, m.1, or m.2. (D) Nucleosome acetylation was assayed with HeLa nuclear extract and [3H]acetyl CoA alone (lane 1) or with wild-type Zta (lane 2), m.1 (lane 3), or m.2 (lane 4). Wild-type Zta, m.1, and m.2 were compared by Coomassie brilliant blue staining of SDS-polyacrylamide gels (lower panel).
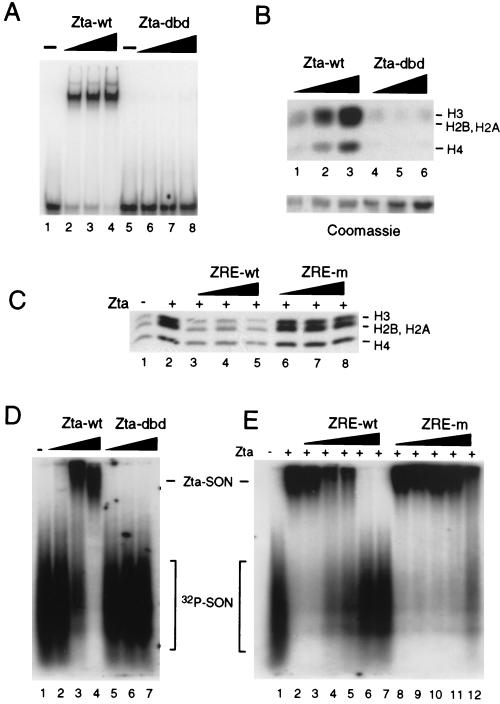
The DNA binding domain of Zta was also found to be important for Zta-dependent HAT activity. To determine whether DNA binding of Zta is required for stimulation of CBP HAT activity, we designed a Zta construct that lacks the ability to bind DNA but contains an otherwise largely intact b-zip domain capable of assuming other possible functions associated with the b-zip domain, such as protein-protein interactions. To this end, three highly conserved residues, R187, K188, and C189, in the basic region of Zta were mutated to alanine. This mutant (Zta-dbd) was purified from Escherichia coli and shown to lack DNA binding activity (Fig. 5A, lanes 6 to 8). Zta-dbd was then compared to wild-type Zta for its ability to stimulate HAT activity. Zta, but not Zta-dbd, stimulated HAT activity in a dose-dependent manner (Fig. 5B). This indicates that residues essential for DNA binding function were essential for the stimulation of HAT activity. To demonstrate that Zta must bind DNA to stimulate HAT activity, we performed HAT assays with increasing concentrations of oligonucleotides containing a single Zta response element (ZRE-wt) or a mutated ZRE element with an ∼20-fold lower affinity for Zta (ZRE-m) (data not shown). We found that the ZRE-wt oligonucleotide was a potent inhibitor of Zta stimulation of HAT activity (Fig. 5C, lanes 3 to 5). In contrast, ZRE-m did not inhibit Zta stimulation of HAT activity (Fig. 5C, lanes 6 to 8), showing that only competition with high-affinity Zta binding sites abrogates nucleosome-directed acetylation. Consistent with these results, control reactions containing poly(dGdC) competitor DNA did not inhibit Zta-mediated acetylation (data not shown). These results indicate that the DNA binding domain of Zta is essential for stimulation of HAT activity and further indicate that DNA binding per se is required for stimulation of SON acetylation by Zta.
FIG. 5.
DNA binding activity of Zta is required for stimulation of HAT activity. (A) Increasing concentrations (33, 100, and 300 ng) of wild-type Zta (lanes 1 to 4) and Zta-dbd (lanes 5 to 8) were compared for their ability to bind radiolabeled ZRE in EMSA. (B) Zta-wt (lanes 1 to 3) and Zta-dbd (lanes 4 to 6) were compared at 33, 100, and 300 ng for stimulation of nucleosomal HAT activity in HeLa nuclear extracts with [3H]acetyl CoA. Zta and Zta-dbd visualized by Coomassie brilliant blue staining of SDS-polyacrylamide gels (lower panel). (C) HeLa nuclear extracts with [3H]acetyl-CoA were incubated with Zta (lanes 2 to 8) and increasing concentrations of ZRE-wt oligonucleotide (lanes 3 to 5) or ZRE-m oligonucleotide (lanes 6 to 8). Acetylated histones were visualized by SDS-PAGE and fluorography. (D) 32P-labeled SONs (20 ng) were incubated with Zta (10, 30, or 90 ng) or Zta-dbd (10, 30, or 90 ng) and assayed by EMSA. (E) Zta (30 ng) and 32P-labeled SONs (20 ng) were incubated with ZRE or ZRE-m oligonucleotide (fivefold dilutions up to 1.0 μg) and assayed by EMSA.
Zta binds small oligonucleosomes.
The requirement for the Zta DNA binding function suggests that Zta stimulates CBP HAT by interacting with nucleosomal DNA. To directly test this possibility, SONs were radiolabeled with T4 polynucleotide kinase and assayed for interaction with Zta in EMSA (Fig. 5D and E). Since SONs are a heterogeneous population of sonicated HeLa cell DNA associated with histones, the radiolabeled SONs appear after electrophoresis as a heterogeneous smear. Zta bound SONs under conditions similar to that required for specific DNA binding (33 nM Zta, and 40 μg of poly(dldC)/ml (Fig. 5D, lanes 3 and 4). Zta-dbd did not bind SONs (Fig. 5D, lanes 5 to 7). To determine the relative specificity of Zta for SONs, we compared the ability of specific (ZRE) and nonspecific (ZRE-m) oligonucleotides to compete for Zta binding. We found that a molar excess of high-affinity ZRE (6, 30, and 150 μM) could compete for Zta (33 nM) binding to 50 nM SONs, whereas the same concentration of ZRE-m oligonucleotide failed to compete (Fig. 5E). Complete competition for SON binding did not occur until an ∼1,000-fold excess of specific ZRE competitor was included in the reaction, suggesting that Zta is binding to nucleosomes with relatively high specificity. Nonspecific competitor DNA [poly(dGdC) and poly(dldC)] was also a poor competitor for Zta binding to SONs (data not shown). These results indicate that Zta can bind SONs with an affinity greater than that for random double-stranded DNA.
Nucleosomal DNA is required for Zta-dependent stimulation of HAT activity.
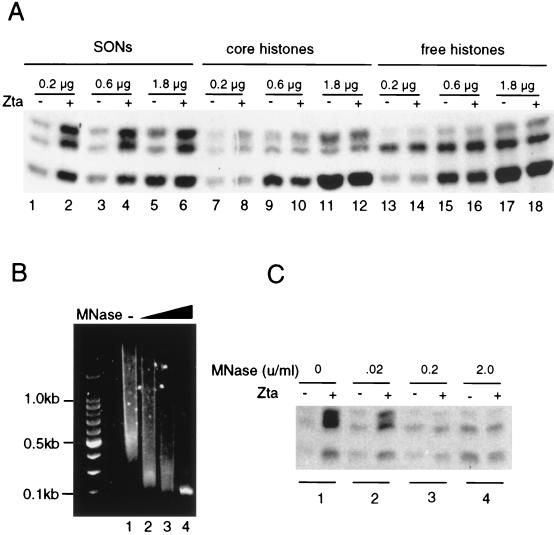
Previous experiments indicate that Zta can bind nucleosomes in vitro. To determine whether oligonucleosomes might contain architectural determinants important for mediating the effects of Zta, which might be lacking in free histones, we compared the acetylation of SONs with core histones depleted of DNA by hydroxyapatite chromatography, or with commercial preparations of acid-extracted histones (free histones) (Fig. 6A). As before, Zta stimulated acetylation of SONs (Fig. 6A). In contrast to SONs, Zta did not stimulate acetylation of the core histones (Fig. 6A) or acid-extracted free histones (Fig. 6A). To further determine if the DNA in the SON substrate was required for Zta-stimulated HAT, we treated the SON fraction with micrococcal nuclease (MNase) (Fig. 6B and C). In the absence of MNase treatment, the average size of nucleosomal DNA was ∼500 bp (Fig. 6B, lane 1). Treatment of SONs with 0.02, 0.2, and 2.0 U of MNase/ml reduced the average DNA size to a range between 200 and 100 bp (Fig. 6B, lanes 2 to 4). The same digested samples were assayed in parallel for acetylation in the absence or presence of Zta (Fig. 6C). Zta stimulated nucleosome acetylation in the absence of MNase treatment (Fig. 6C, set 1) and weakly stimulated acetylation of nucleosomes treated with 0.02 U of MNase/ml (set 2). Treatment of SONs with 0.2 and 2.0 U of MNase/ml completely eliminated the stimulation of acetylation by Zta (sets 3 and 4). These results suggest that nucleosomes with DNA less than 200 bp fail to support Zta-mediated stimulation of acetylation.
FIG. 6.
Activation of oligonucleosome-specific acetylation by Zta. (A) Increasing amounts of SONs (lanes 1 to 6), hydroxyapatite purified core histones (lanes 7 to 12), or acid-extracted free histones (lanes 13 to 18) were assayed for acetylation by phosphocellulose B fraction in the absence (−) or presence (+) of Zta. The amount of histones in each reaction is indicated above each set of reactions. (B) Characterization of DNA in oligonucleosomes. The DNA purified from SONs (lane 1) or SONs treated with 0.2 (lane 2), 2 (lane 3), or 20 (lane 4) U of MNase/ml were extracted, electrophoresed on a 1.5% agarose gel, and visualized by ethidium bromide staining. (C) MNase digestion inhibits HAT activation by Zta. SONs alone (lane 1) or SONs digested with 0.2 (lanes 2), 2.0 (lanes 3), or 20 (lanes 4) U of MNase/ml (as shown in panel B) were assayed as substrates for acetylation in the absence (−) or presence (+) of 300 ng of Zta.
Sequence-specific targeting of CBP HAT activity by Zta.
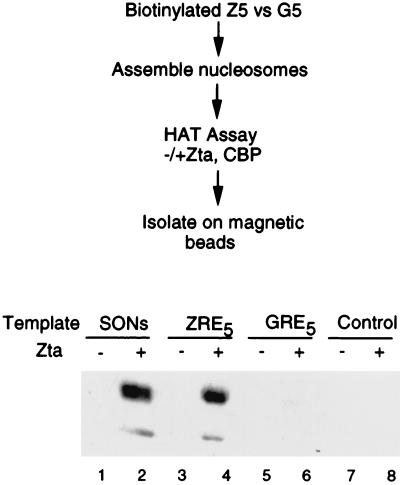
The above results suggest that Zta stimulates acetylation of oligonucleosomes in a sequence-independent manner. However, it is also likely in vivo that Zta directs HAT activity to specific promoter sequences containing high-affinity ZREs. To determine whether Zta can target CBP HAT activity to oligonucleosomes containing high-affinity ZREs, we assembled synthetic dinucleosomes on biotinylated DNA templates containing either five ZREs or, as a control, five GAL4 recognition elements (GREs) (Fig. 7). Biotinylated templates containing five ZREs or five GREs were assembled by salt titration with HeLa-derived oligonucleosomes. Assembled ZRE- and GRE-containing dinucleosomes were acetylated in the presence of excess competitor donor HeLa oligonucleosomes. Zta stimulated acetylation of unpurified SONs as expected (Fig. 7, lanes 1 and 2). Nucleosomes assembled on ZRE-containing templates were stimulated by Zta after DNA affinity purification with strepavidin-conjugated magnetic beads (lanes 3 and 4). In contrast, nucleosomes assembled on GRE-containing templates were not stimulated by Zta after DNA affinity purification (lanes 5 and 6). Nonbiotinylated ZRE templates were not recovered after DNA affinity purification demonstrating the specificity of the binding reaction (control lanes 7 and 8). Equivalent amounts of assembled nucleosomes were precipitated by the magnetic beads in lanes 3 to 6 as determined by silver staining of reaction products (data not shown). These results indicate that Zta can direct CBP acetylation to ZRE-containing templates in a complex mixture of competitor oligonucleosomes and DNA.
FIG. 7.
Template-specific targeting of CBP acetylation. Biotinylated ZRE and GRE templates (400 bp) were assembled into dinucleosomes and then incubated with CBP and [3H]acetyl CoA in the presence (+) or absence (−) of Zta. Biotinylated templates were isolated on magnetic beads after the HAT reaction and were assayed by autoradiography of SDS-polyacrylamide gels. The control lane contains a nonbiotinylated ZRE template, and a reaction not purified on magnetic beads is shown in the lanes marked SONs.
Subset of cellular transcription factors stimulates histone acetylation in vitro.
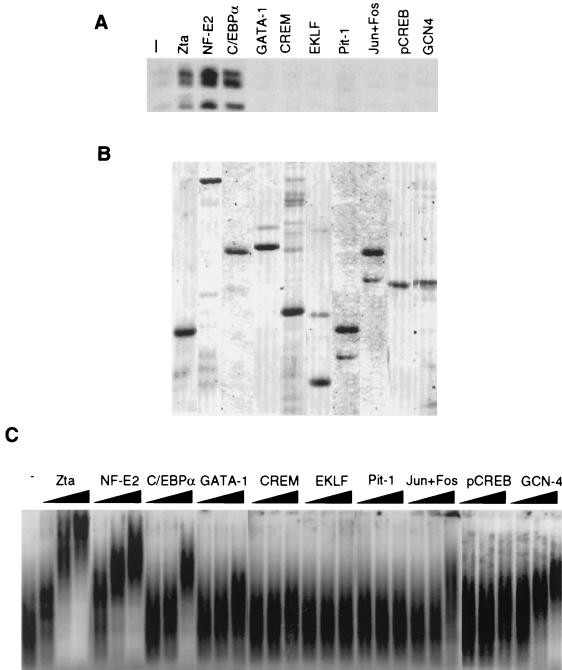
To determine if the stimulation of HAT activity was a feature shared by other transcriptional activators, we compared Zta to several other mammalian and yeast-derived transcription factors in the in vitro HAT assay (Fig. 8). The hematopoietic transcription factors EKLF, GATA-1, and NF-E2 bind multiple sites in the locus control region of the β-globin gene cluster and directly interact with CBP. EKLF and GATA-1 are zinc finger DNA binding proteins (12). NF-E2 is a heterodimer of two b-zip proteins that consists of the hematopoietic restricted subunit p45 and the more widely expressed small subunit mafG. In these experiments, we used a tethered heterodimer of NF-E2 in which the two subunits are connected by a flexible linker polypeptide. This form of NF-E2 binds DNA, activates transcription, and is completely functional in assays that measure NF-E2 function in its physiological context (11). C/EBPα is a b-zip protein involved in cellular differentiation of various cell types (47). CREB, the cyclic AMP response element binding protein, is a member of the b-zip family that binds CBP with high affinity when phosphorylated by protein kinase A (21). Jun and Fos are b-zip proteins that form the heterodimeric transcription factor AP1. CREM 1 is a germ-cell variant of CREB (3). Pit-1 is a member of the POU domain family (3). GCN4 is a yeast b-zip protein with structural motifs similar to the Zta DNA binding domain and activation domain (42). All proteins were expressed in E. coli and purified to near homogeneity (Fig. 8B). Purified activators were incubated with HeLa nuclear extracts supplemented with partially purified SONs and [3H]acetyl CoA (Fig. 8A). As expected, Zta stimulated nucleosome acetylation. In addition to Zta, we found that NF-E2 and C/EBPα stimulated CBP HAT. In contrast, GATA-1, CREM, EKLF, Pit-1, Jun+Fos, and PKA phosphorylated CREB did not stimulate nucleosome acetylation even though all of these proteins can bind CBP and DNA with high affinity. The yeast GCN4 had no effect on histone acetylation, despite some sequence similarities to Zta. NF-E2 and C/EBPα also stimulated the activity of affinity-purified CBP expressed in baculovirus, indicating that CBP was the active acetylase in these reactions (data not shown).
FIG. 8.
A class of transcription activators stimulate nucleosomal HAT activity. (A) For each protein, 0.5 μg of purified recombinant Zta, NF-E2, C/EBPα, GATA-1, CREM, EKLF, Pit-1, Jun+Fos, phospho-CREB, and GCN4 was tested for its ability to stimulate acetylation of SONs using PC-B as the source of acetylase. (B) Coomassie brilliant blue staining of SDS-polyacrylamide gels containing 1.0 μg of the various recombinant activator proteins used in panel A. (C) Oligonucleosome binding of transcription factors was assayed by agarose gel EMSA with 32P-labeled HeLa-derived SONs. Proteins were assayed as threefold dilutions from 20 to 180 ng for each reaction.
To determine if the ability to stimulate CBP HAT activity correlated with the binding to oligonucleosome substrates, we assayed the panel of DNA binding proteins for their ability to alter the mobility of labeled HeLa oligonucleosomes in agarose gel EMSA. SONs were radiolabeled by T4 polynucleotide kinase and incubated with the equivalent protein concentrations of the transcription factors assayed above in the presence of excess poly(dldC) competitor. We found that Zta, NF-E2, and C/EBPα bound oligonucleosomes with similar efficiency (Fig. 8C). Jun+Fos, GATA-1, and GCN4 bound weakly to these oligonucleosomes, but almost no detectable binding was observed for CREM, EKLF, Pit-1, and pCREB (Fig. 8C). The random DNA binding activity of these proteins did not correlate well with their ability to stimulate nucleosomal HAT activity (data not shown). Together, these results indicate that a subset of b-zip proteins can stimulate the HAT activity of CBP and that this activity correlates with the ability of these transcription factors to bind oligonucleosomes in vitro.
DISCUSSION
Histone acetylation correlates well with transcription activation and may be essential for transcription factor access to chromatin repressed genes (36, 67, 70). The coactivator HAT proteins CBP and p300 bind numerous transcriptional activators and are thought to mediate their transcription activation function. While activators recruit HAT-containing coactivators to specific sequences to promote local histone acetylation (69), it is not clear if nuclear factors modulate the intrinsic enzymatic HAT activity or the substrate specificity of these coactivator HATs. We have shown here that three members of the b-zip family of transcription activators can stimulate the nucleosome-specific HAT activity of CBP in vitro. We showed that Zta stimulated the HAT activity of CBP immunoprecipitated from transfected cells on small oligonucleosomes isolated from HeLa nuclear pellets (Fig. 2). Acetylation was dependent on an intact CBP HAT domain (Fig. 3A) and could be reconstituted with affinity-purified baculovirus-expressed CBP (Fig. 3B). The transcriptional activation and DNA binding functions of Zta were required for the stimulation of HAT activity (Fig. 4 and 5). Interestingly, Zta could not stimulate acetylation of free histones or core histones stripped of nucleosomal DNA, and Zta was capable of binding directly to oligonucleosomes in EMSA (Fig. 6). The importance of the nucleosomal DNA was further demonstrated by MNase digestion of small oligonucleosomes, which resulted in the inhibition of Zta-stimulated HAT activity. Nucleosomes assembled on templates containing ZRE binding sites were preferentially acetylated relative to templates lacking ZRE sites, indicating that specific targeting is likely to occur in vivo (Fig. 7). Finally, we show that a subset of b-zip proteins can stimulate CBP nucleosome acetylation (Fig. 8A) and that the ability to stimulate nucleosome acetylation correlates well with the ability of these activators to bind nucleosomes in EMSA (Fig. 8C). Together, these data suggest that a class of activators represented by Zta, C/EBPα, and NF-E2 can stimulate the nucleosomal HAT activity of CBP by directing CBP to nucleosomes.
Several earlier studies have demonstrated that nuclear factors can modulate HAT activity. Adenovirus E1A has been shown to stimulate CBP HAT activity in a cell cycle- and phosphorylation-dependent manner, but the biochemical basis for this activation is unclear (2). At high concentrations, E1A has also been shown to inhibit CBP-HAT activity by binding directly to the HAT domain of CBP (17, 38). The discrepancy between these reports might be the result of differences in the amounts of E1A (49). The cellular differentiation factor Twist can also bind the CBP HAT domain and inhibit histone acetylation (38). DNA-dependent protein-kinase can inhibit GCN5 HAT activity by a phosphorylation-associated mechanism (9). In contrast to these inhibitory associations through the CBP HAT domain, Utley et al. demonstrated that the synthetic activator GAL4-VP16 can increase the yeast SAGA and NuA4-associated acetylation of nucleosomes bound to chromatinized templates containing GAL4 binding sites (69). Our results are similar to those found by Utley et al., since Zta stimulates HAT activity by interacting with both CBP and nucleosomes directly. However, our observations are importantly distinct from this earlier study in several respects. Most intriguingly, we have used natural oligonucleosome substrates rather than mononucleosomes reconstituted on synthetic templates containing an accessible activator recognition site. Thus, Zta, C/EBPα, and NF-E2 must possess a capacity to bind oligonucleosomal substrates with relatively high affinity. This was demonstrated for Zta (Fig. 5D and E) and for NF-E2 and C/EBPα (Fig. 8C). We have also shown that HAT activity was targeted to templates containing high affinity Zta binding sites under conditions of nucleosome excess (Fig. 7). But it remains intriguing that these activators could strongly stimulate nucleosomal HAT without a homogeneous population of substrates containing an accessible high-affinity DNA binding site, suggesting that this is an important feature of their biological activities in vivo.
The stimulation of nucleosome acetylation by Zta and NF-E2 is likely to represent a novel mechanism by which transcription factors modulate HAT activity. In our experiments with Zta, DNA binding and CBP interactions were essential for stimulation of acetylation, suggesting that recruitment of CBP to the nucleosome is necessary. The primary feature shared by Zta, C/EBPα, and NF-E2 is the ability to bind oligonucleosomes with high affinity. The ability to interact with oligonucleosomes did not correspond well with the random DNA binding affinity of these proteins (data not shown). Based on these observations, we suggest that these members of the b-zip family share a structural motif that allows them to bind with relatively high affinity to nucleosomal DNA. A phylogenetic tree of b-zip proteins shows that Zta, C/EBPα, and the two components of NF-E2 (MafG and p45) are closer phylogenetically than are the other members of the b-zip family, including Jun, Fos, CREB, CREM and ATF2. Moreover, we have found that a chimeric Zta protein containing the Fos b-zip domain fails to stimulate CBP HAT activity (data not shown). This suggests that distinct structural features of the b-zip domain confer the ability to stimulate CBP HAT activity and presumably, the ability to bind oligonucleosomes.
In addition to the recruitment of CBP to nucleosomes, it is also possible that Zta increases the intrinsic enzymatic rate of CBP by altering the enzyme or substrate conformation. Zta can bind two domains of CBP (C/H1 and C/H3) that straddle the HAT domain of CBP (Fig. 4). It is possible that simultaneous binding to both the C/H1 and C/H3 domains of CBP is important for the stimulation of CBP HAT activity and that this binding induces an open conformation of the HAT domain. CBP mutants containing the HAT and C/H3 domains were not stimulated by Zta (data not shown), supporting the possibility that two interactions sites are important for stimulation of CBP HAT. We have also found that Zta stimulated acetylation of oligonucleosomes with an average DNA size of 500 bp, suggesting that the dinucleosome was the minimal substrate for HAT activation (Fig. 6). It is likely that the histone tails are positioned differently in oligonucleosomes than in mononucleosomes or on free histones (55, 56). One potential mechanism to explain this substrate specificity is that Zta alters the conformation of the histone tails in dinucleosomes to increase their access for CBP acetylation. Future experiments will help distinguish between these potential mechanisms of HAT activation.
Several other lines of evidence suggest that Zta possesses a chromatin-specific activation function. We have shown that Zta can bind to oligonucleosomes in the presence of substantial amounts of nonspecific, double-stranded DNA competitor, suggesting that Zta binds oligonucleosomes with physiologically significant affinity (Fig. 5D and E). Others have shown that Zta can activate numerous viral and some cellular genes that lack obvious Zta binding sites in their promoters (16). Francis and colleagues have identified a single amino acid substitution mutation in Zta (S186A) that abrogates the transcription activation of latent viral chromosomes but has no detectable effect on the transcription activation of transiently transfected reporter plasmids (32). Additionally, a DNA binding defective mutant of Zta could stimulate transcription from a subset of viral promoters, suggesting Zta activates transcription at some promoters by a DNA binding independent mechanism (31). Together, these observations indicate that Zta can regulate transcription by diverse mechanisms, and modulation of CBP HAT activity, as we have described here, may be an important component of Zta-mediated transcription activation.
Like Zta, NF-E2 is thought to play an important role in relieving chromatin repression (4). NF-E2 binding sites are essential components of the human β-globin locus control region (LCR), which may contribute to the formation of an open chromatin structure at the globin gene locus extending over 100 kb (6, 12). The LCR consists of multiple binding sites for NF-E2, GATA-1, and EKLF. All three of these proteins bind CBP, and it is thought that the cooperative binding of these proteins to CBP results in a highly stable association of CBP with the LCR (13, 19, 73). Both subunits of NF-E2 can bind directly to CBP (H. L. Hung and G. A. Blobel, unpublished data). It is not clear, however, whether the interactions of these proteins with CBP provide nonredundant activation functions. NF-E2 is capable of binding to its specific recognition site in the LCR reconstituted in vitro with chromatin (6). NF-E2 binding leads to nucleosome disruption, thereby facilitating the binding of GATA-1 to a chromatin assembled template (6). In our experiments shown in Fig. 8, we found that NF-E2, but not GATA-1 and EKLF, could stimulate CBP HAT activity. Together, these results suggest that transcriptional control regions, like the LCR, assemble multiple factors that provide mechanistically distinct activation functions. We propose that stimulation of the nucleosome-directed HAT activity of CBP is one such nonredundant activation function provided by NF-E2.
Stimulation of nucleosome-directed histone acetylation is likely to be important for initiating gene activity in highly repressed chromatin structures, like the 30-nm fiber, where sequence-specific DNA binding is obstructed. It is possible that nuclear factors, like Zta, C/EBPα, and NF-E2, initiate their search for sequence-specific binding by promoting a transient wave of histone acetylation. Since histone acetylation is in dynamic equilibrium with active histone deacetylation, it is likely that stable acetylation is not established until activators dock at high-affinity DNA binding sites. Nucleosome-directed acetylation might be important for the initial search by sequence-specific activators for their cognate DNA binding sites in highly repressed chromatin environments. These mechanisms are likely to be important for the NF-E2-dependent alteration of chromatin structure in the β-globin locus and for the Zta-mediated reactivation of latent EBV.
ACKNOWLEDGMENTS
We thank Dimitrios Thanos for generously providing baculovirus H6-CBP and Shelley Berger and members of her laboratory for instruction in acetylation assays. We are grateful to Volker Blank for providing the tethered NF-E2 construct, Ramin Shiekhattar for CREB protein, Xiangyuan Wang for purification of core histones, T. Kerppola for Jun and Fos proteins, and Hsiao-Ling Hung for GST fusion proteins.
This work was supported by grants from NIH (GM 54687), the Edward Mallinckrodt, Jr. Foundation, and the Leukemia & Lymphoma Society (to P.M.L.) and an NCI Core Grant to the Wistar Institute. G.A.B. was supported by a grant from NIH (RO1 DK54937-01) and by the American Society of Hematology Scholar Award.
REFERENCES
- 1.Adamson A L, Kenney S. The Epstein-Barr virus BZLF1 protein interacts physically and functionally with the histone acetylase CREB-binding protein. J Virol. 1999;73:6551–6558. doi: 10.1128/jvi.73.8.6551-6558.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ait-Si-Ali S, Ramirez S, Barre F-X, Dkhissi F, Magnaghi-Jaulin L, Girault J A, Robin P, Knibiehler M, Pritchard L L, Ducommun B, Trouche D, Harel-Bellan A. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- 3.Andersen B, Rosenfeld M G. Pit-1 determines cell types during development of the anterior pituitary gland. A model for transcriptional regulation of cell phenotypes in mammalian organogenesis. J Biol Chem. 1994;269:29335–29338. [PubMed] [Google Scholar]
- 4.Andrews N C. The NF-E2 transcription factor. Int J Biochem Cell Biol. 1998;30:429–432. doi: 10.1016/s1357-2725(97)00135-0. [DOI] [PubMed] [Google Scholar]
- 5.Andrews N C, Faller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armstrong J A, Emerson B M. NF-E2 disrupts chromatin structure at human β-globin locus control region hypersensitive site 2 in vivo. Mol Cell Biol. 1996;16:5634–5644. doi: 10.1128/mcb.16.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 8.Bannister A J, Oehler T, Wilhelm D, Angel P, Kouzarides T. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- 9.Barlev N A, Poltoratsky V, Owen-Hughes T, Ying C, Liu L, Workman J L, Berger S L. Repression of GCN5 histone acetyltransferase activity via bromodomain-mediated binding and phosphorylation by the Ku-DNA-dependent protein kinase complex. Mol Cell Biol. 1998;18:1349–1358. doi: 10.1128/mcb.18.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beato M, Eisfeld K. Transcription factor access to chromatin. Nucleic Acids Res. 1997;25:3559–3563. doi: 10.1093/nar/25.18.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blank V, Kim M J, Andrews N C. Human MafG is a functional partner for p45 NF-E2 in activating globin gene expression. Blood. 1997;89:3925–3935. [PubMed] [Google Scholar]
- 12.Blobel G. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood. 2000;95:745–755. [PubMed] [Google Scholar]
- 13.Blobel G A, Nakajima T, Eckner R, Montminy M, Orkin S H. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 15.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 16.Cayrol C, Flemington E K. Identification of cellular target genes of the Epstein-Barr virus transactivator Zta: activation of transforming growth factor βigh3 (TGF-βigh3) and TGF-β1. J Virol. 1995;69:4206–4212. doi: 10.1128/jvi.69.7.4206-4212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 18.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 19.Cheng X, Reginato M J, Andrews N C, Lazar M A. The transcriptional integrator CREB-binding protein mediates positive cross talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol Cell Biol. 1997;17:1407–1416. doi: 10.1128/mcb.17.3.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chi T, Carey M. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 21.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 22.Cote J, Utley R T, Workman J L. Analysis of transcription factor binding to nucleosomes. Methods Mol Genet. 1995;6:108–128. doi: 10.1016/s0076-6879(96)74024-7. [DOI] [PubMed] [Google Scholar]
- 23.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai P, Akimaru H, Tanaka Y, Hou D X, Yasukawa T, Kanei I C, Takahashi T, Ishii S. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 25.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorsman J C, Teunisse A F, Zantema A, van der Eb A J. The adenovirus 12 E1A proteins can bind directly to proteins of the p300 transcription coactivator family, including the CREB-binding protein CBP and p300. J Gen Virol. 1997;78:423–426. doi: 10.1099/0022-1317-78-2-423. [DOI] [PubMed] [Google Scholar]
- 27.Dyson P J, Farrell P J. Chromatin structure of Esptein-Barr virus. J Gen Virol. 1985;66:1931–1940. doi: 10.1099/0022-1317-66-9-1931. [DOI] [PubMed] [Google Scholar]
- 28.Eckner R, Ludlow J W, Lill N L, Oldread E, Arany Z, Modjtahedi N, DeCaprio J A, Livingston D M, Morgan J A. Association of p300 and CBP with simian virus 40 large T antigen. Mol Cell Biol. 1996;16:3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farrell P J, Rowe D T, Rooney C M, Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fixman E D, Hayward G S, Hayward S D. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol. 1995;69:2998–3006. doi: 10.1128/jvi.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flemington E K, Lytle J P, Cayrol C, Borras A M, Speck S H. DNA-binding-defective mutants of the Epstein-Barr virus lytic switch activator Zta transactivate with altered specificities. Mol Cell Biol. 1994;14:3041–3052. doi: 10.1128/mcb.14.5.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis A L, Gradoville L, Miller G. Alteration of a single serine in the basic domain of Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J Virol. 1997;71:3054–3061. doi: 10.1128/jvi.71.4.3054-3061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giles R H, Peters D J M, Bruening M H. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 34.Goldman P S, Tran V K, Goodman R H. The multifunctional role of the co-activator CBP in transcriptional regulation. Recent Prog Horm Res. 1997;52:103–119. [PubMed] [Google Scholar]
- 35.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen H T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 36.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 37.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 38.Hamamori Y, Sartorelli V, Ogryzko V, Puri P L, Wu H-Y, Wang J Y J, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein Twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 39.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi J C. mof, a putative acetyltransferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howe L, Brown C E, Lechner T, Workman J L. Histone acetyltransferase complexes and their link to transcription. Crit Rev Eukaryot Gene Expr. 1999;9:231–243. doi: 10.1615/critreveukargeneexpr.v9.i3-4.80. [DOI] [PubMed] [Google Scholar]
- 41.Hung H L, Lau J, Kim A Y, Weiss M J, Blobel G A. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol Cell Biol. 1999;19:3496–3505. doi: 10.1128/mcb.19.5.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson B M, Drysdale C M, Nataran K, Hinnebusch A G. Identification of seven hydrophobic clusters in GCN4 making redundant contributions to transcriptional activation. Mol Cell Biol. 1996;16:5557–5571. doi: 10.1128/mcb.16.10.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacobson S, Pillus L. Modifying chromatin and concepts of cancer. Curr Opin Genet Dev. 1999;9:175–184. doi: 10.1016/S0959-437X(99)80027-6. [DOI] [PubMed] [Google Scholar]
- 44.Jenkins P J, Binne U K, Farrell P J. Histone acetylation and reactivation of Epstein-Barr virus from latency. J Virol. 2000;74:710–720. doi: 10.1128/jvi.74.2.710-720.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 46.Kieff E. Epstein-Barr virus and its replication. In: Knipe D, Howley P M, editors. Field's virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 47.Lekstrom J, Xanthoupoulos K G. Biological role of the CCAAT/Enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273:28545–28546. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 48.Li Q, Harju S, Peterson K R. Locus control regions: coming of age at a decade plus. Trends Genet. 1999;15:403–408. doi: 10.1016/s0168-9525(99)01780-1. [DOI] [PubMed] [Google Scholar]
- 49.Li Q, Imhof A, Collingwood T N, Urnov F D, Wolffe A P. p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step subsequent to chromatin disruption. EMBO J. 1999;18:5634–5652. doi: 10.1093/emboj/18.20.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lieberman P M, Berk A J. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 51.Lieberman P M, Hardwick J M, Hayward S D. Responsiveness of the Epstein-Barr virus NotI repeat promoter to the Z transactivator is mediated in a cell-type-specific manner by two independent signal regions. J Virol. 1989;63:3040–3050. doi: 10.1128/jvi.63.7.3040-3050.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lieberman P M, Hardwick J M, Sample J, Hayward G S, Hayward S D. The zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J Virol. 1990;64:1143–1155. doi: 10.1128/jvi.64.3.1143-1155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lieberman P M, Ozer J, Gursel D B. Requirement for TFIIA-TFIID recruitment by an activator depends on promoter structure and template competition. Mol Cell Biol. 1997;17:6624–6632. doi: 10.1128/mcb.17.11.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 55.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 56.Luger K, Richmond T J. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 57.Martinez-Balbas M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merika M, Williams A J, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFNb enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 59.Mizzen C A, Yang X-J, Kokubo T, Brownell J E, Bannister A J, Wang L, Berger S, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 60.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 61.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 62.Rickinson A B, Kieff E. Epstein-Barr virus. In: Knipe D, Howley P M, editors. Field's virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 63.Scolnick D M, Chehab N H, Stavridi E S, Lien M C, Caruso L, Moran E, Berger S L, Halazonetis T D. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 1997;57:3693–3696. [PubMed] [Google Scholar]
- 64.Shikama N, Lyon J, La Thangue N B. The p300/CBP family: integration signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 65.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 66.Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 67.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 68.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 69.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcription activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 70.Wade P, Wolffe A. Histone acetyltransferases in control. Curr Biol. 1997;7:R82–R84. doi: 10.1016/s0960-9822(06)00042-x. [DOI] [PubMed] [Google Scholar]
- 71.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 72.Zerby D, Chen C-J, Poon E, Lee D, Shiekhattar R, Lieberman P M. The amino-terminal C/H1 domain of CREB binding protein mediates Zta transcription activation of latent Epstein-Barr virus. Mol Cell Biol. 1999;19:1617–1626. doi: 10.1128/mcb.19.3.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang W, Bieker J J. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]