Abstract
Human insulin (INS) gene diverged from the ancestral genes of invertebrate and mammalian species millions of years ago. We previously found that mouse insulin gene (Ins2) isoforms are expressed in brain choroid plexus (ChP) epithelium cells, where insulin secretion is regulated by serotonin and not by glucose. We further compared human INS isoform expression in postmortem ChP and islets of Langerhans. We uncovered novel INS upstream open reading frame isoforms and their protein products. In addition, we found a novel alternatively spliced isoform that translates to a 74–amino acid (AA) proinsulin containing a shorter 19-AA C-peptide sequence, herein designated Cα-peptide. The middle portion of the conventional C-peptide contains β-sheet (GQVEL) and hairpin (GGGPG) motifs that are not present in Cα-peptide. Islet amyloid polypeptide (IAPP) is not expressed in ChP, and its amyloid formation was inhibited in vitro more efficiently by Cα-peptide than by C-peptide. Of clinical relevance, the ratio of the 74-AA proinsulin to proconvertase-processed Cα-peptide was significantly increased in islets from type 2 diabetes mellitus autopsy donors. Intriguingly, 100 years after the discovery of insulin, we found that INS isoforms are present in ChP from insulin-deficient autopsy donors.
Introduction
The human insulin (INS) gene underwent significant variations during evolution as its tissue expression migrated from neurons of invertebrates to islets of Langerhans in pancreas of vertebrates, and the functional spectrum of its peptide products diversified over time from growth and development, learning and memory, and reproduction to primarily metabolism and regulation of anabolism, including its well-known function of regulating cellular uptake of glucose (1). The primary source for brain insulin is presumed to be insulin secreted from β-cells in islets (2). However, this is not the sole source, because it is now established that mouse choroid plexus (ChP) epithelium cells (EChP) also express insulin. Mouse ChP contains several Ins2 isoforms (Ins2-V1, -V2, -V3, and -V4) that are processed into mature insulin that is then secreted into cerebrospinal fluid (CSF) proportional to CSF serotonin concentrations; glucose does not regulate insulin secretion from isolated EChP (3). Because of ongoing interest in actions of insulin in brain and the fact that insulin is produced in ChP, we undertook further investigation of the human INS isoforms and their protein products.
Human INS is typically described as having three exons that encode a preproinsulin of 110 amino acids (AA) that undergoes extensive posttranslational modifications. Proinsulin of 86 AA, a 24-AA signal peptide having been removed, exits the endoplasmic reticulum and is delivered to the Golgi apparatus for packaging in secretory granules. As granules become mature and acidify, proinsulin is Remain A-chain and B-chain typically then cleaved, freeing a 30-AA B-chain, a 31-AA C-peptide, and a 21-AA A-chain. In the process, four AA (RR and RK) are lost from the ends of C-peptide. Secretory granules in β-cells therefore contain various proportions of proinsulin, partially processed insulin, C-peptide, and mature insulin. Biologically active human and mouse insulin in circulation is composed of single A- and B-chains connected by disulfide bonds and small amounts of cleaved and noncleaved proinsulin with varying degrees of biological activity (4).
Humans possess one INS gene, while mice have two: Ins1 and Ins2. Ins1 is a retrogene derived from partially spliced Ins2 (5). Mouse islets express both genes in a three-to-two ratio (6), but only Ins2, the evolutionarily older gene, is present in ChP, while its expression is barely if at all detectable in other brain areas; Ins1 is not detected in any brain area (3). Invertebrate genomes encode multiple insulin-related peptides in neurons, including pro–molluscan insulin-related peptides, and posttranslational processing produces A- and B-chains and long and short C-peptides (7). Multiple human INS mRNA isoforms, all of which encode the known 110-AA preproinsulin, are reported to be regulated by glucose in pancreatic islets (8), but interestingly, insulin secretion from ChP is not controlled by glucose (3). However, in an extensive literature search, we found no reports of human-specific INS upstream open reading frames (uORF) or spliced isoforms encoding novel polypeptides (8,9). At least 50% of human genes contain uORF that add layers of gene regulation and protein products for functional diversity (10). As INS evolved, expansion of actions of its isoform products in islets, ChP, and brain areas may have resulted in divergence of posttranscription and translational products and divergence of functions. We uncovered novel human-specific uORF isoforms and a novel intraexon 3 splicing variant (here designated INS3B) that encodes a short 19-AA Cα-peptide at both mRNA and protein levels. We found that protease cleavage of INS3B was reduced in islets from individuals with type 2 diabetes mellitus (T2DM). We also found that INS isoforms and Cα-peptide are present in human ChP, including in those whose β-cells were presumed absent because of β-cell destruction (11).
Research Design and Methods
Source of Tissue Samples
Islets from human nondiabetic (n = 13) and T2DM (n = 9) postmortem pancreata (human islet checklist provided in Supplementary Table 1) were provided by the National Institute of Diabetes and Digestive and Kidney Diseases–funded Integrated Islet Distribution Program at City of Hope (National Institutes of Health grant 2UC4DK098085) (https://iidp.coh.org/). A total of 1,000–2,000 islets were ordered for each sample, and ∼200 islets were handpicked for proteomics and the rest for gene expression experiments. Frozen human ChP sections and blood samples (nondiabetic n = 5; T1DM n = 6; T2DM n = 4) were from the Human Brain Collection Core, National Institute of Mental Health (12) (Supplementary Table 2). Mouse pancreata, islets, and ChP were dissected as we previously described (3).
We measured insulin levels in plasma of adults without diabetes after overnight fast (fasting glucose) (n = 7) and 2 h after continuous intravenous glucose (2h-IVG) (n = 15) administration (glucose clamp methodology as previously described (13), whereby circulating glucose was clamped at fasting levels plus 98 mg/dL glucose during glucose infusion). The protocol has institutional review board approval (National Institutes of Health Institutional Review Board 15-AG-0063). This was undertaken to confirm that our new mass spectrometry (MS)-based assay was capable of quantifying endogenous INS products in controlled settings and for comparison with measurements in the same samples by ELISA.
Bioinformatics and Sequencing of Insulinoma cDNA Clones
We used the Sequencher 5.4.6 software (Gene Codes Corporation, Ann Arbor, MI) to assemble pancreatic islet and insulinoma expressed sequence tag sequences (https://www.ncbi.nlm.nih.gov/dbEST/) that are homologous to human INS gene. Human Splicing Finder (HSF) (https://www.umd.be/HSF/HSF.shtml) was used to predict potential INS exon 3 intraexonal splicing events (14). We used GlobPlot 2 (https://globplot.embl.de/) (15) to predict intrinsic protein disorder domain of INS isoforms. IMAGE cDNA clones were ordered from Source Bioscience (https://www.sourcebioscience.com/), and the inserts were sequenced by the Sanger sequencing service of Eurofins Genomics (Louisville, KY).
RNA Isolation, cDNA Synthesis, Quantitative RT-PCR, and RNAscope In Situ Hybridization
RNA isolation, cDNA synthesis, quantitative RT-PCR, and RNAscope in situ hybridization (ISH) procedures were described previously (3). We designed splicing junction–specific TaqMan probes (Supplementary Table 3) for primary ORF (pORF) isoforms (INS1A, -1B, -1C, -I1, and -3B). FAM-labeled TaqMan probes of INS exons 2 and 3A junction (INS3A; Hs00355773_m1) and β-cell autoantigen and transcription factor probes were ordered from Thermo Fisher Scientific (Waltham, MA). We designed the uORF-specific forward primer overlapping the INSU isoform translation initiation methionine and the TaqMan probes at the splicing junctions of exon 1UA/exon 2 (INSUA), exon 1UB/exon 2 (INSUB), exon 1UC/exon 2 (INSUC), and exon 1UC/intron 1 (INSU1) (Fig. 1) to measure expression of uORF in human tissues. Because the exon-intron junction size of INSU2 (intron 2 retention) is larger than the amplicon limit for the TaqMan assay, INSU2 transcription levels were measured by averaged values of INSU1 and EX2-I2 probes (Fig. 1A and Supplementary Table 3). The duplex fluorescent TaqMan assay was performed in replicates (StepOnePlus Real-Time PCR System), and the relative fold change was calculated using the formula: 2−ΔΔCt normalized with GAPDH (VIC labeled; catalog number 4326317E) (16). Droplet digital PCR (ddPCR) absolute values were derived from Poisson distribution of positive and negative droplets (QX200 ddPCR System; Bio-Rad Laboratories, Philadelphia, PA) that were normalized with endogenous control β2 microglobulin (VIC labeled; catalog number 4326319E).
Figure 1.
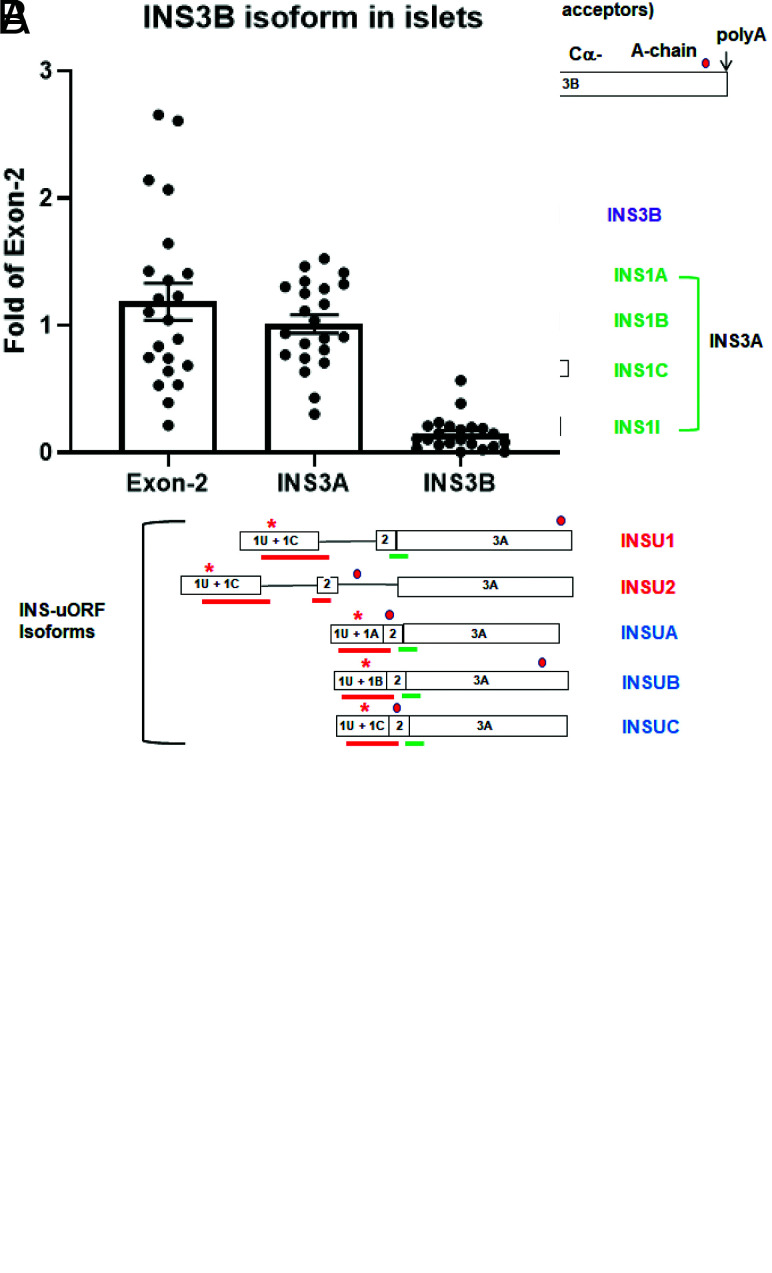
A: Human INS gene structures and their alternatively spliced isoforms. Open boxes represent exons, and solid lines represent introns. Downward arrows and capital letters are at the intraexonal splicing donor sites of exon 1 and acceptor sites of exon 3 for human and polyadenylation sites. Red and green asterisks are the translation initiation sites for uORF and pORF, respectively, while the red dot is at the translational stop codon. The blue bar is the Exon-2 TaqMan Probe that hybridizes to all the isoforms, the red bars are the isoform-specific probes, the green bar is the INS3A probe that hybridizes to all the INS isoforms except INS3B and INSU2, and the purple bar is the probe unique to the INS3B isoform. Red and blue names are the respective novel nonspliced and spliced uORF isoforms. The purple name is the intraexon 3 spliced INS3B isoform, while the green names are the respective spliced and intron 1 retention pORF isoforms. B: TaqMan quantitative RT-PCR of INS3A and INS3B in human islets using total exon 2 as a reference. y-axis is fold change, and each individual dot represents a single islet sample.
RNAscope ISH probes were custom designed for INSU isoform (11 ZZ pairs targeting 2–272 of upstream transcription start site [uTSS] and 273–732 of primary TSS nucleotide sequences of MT335691 in C1 or C2 channel). Transthyretin (TTR) (18 ZZ pairs targeting 2–917 nucleotides of NM_000371.3 in C1 channel), islet amyloid polypeptide (IAPP) (16 ZZ pairs targeting 424–1,947 nucleotides of NM_000415.2 in C3 channel), and RBFOX3 (NEUN) (20 ZZ pairs targeting region 720–2,217 nucleotides of NM_001082575.2 in C3 channel) were from cataloged probes from Advanced Cell Diagnostics, Inc. (Hayward, CA). Human islets were individually handpicked and embedded with Shandon M-1 Embedding Matrix (Thermo Fisher Scientific), frozen in ethanol/dry ice bath, and stored at −80°C. Pretreatment of ChP sections, probe reactions, and labeling were performed according to the RNAscope Multiplex Fluorescent Detection Kit v2 protocol (17).
Thioflavin T In Vitro Assay for IAPP Amyloid Formation
Thioflavin T (ThT) was from MilliporeSigma (catalog number T3516; Rockville, MD). IAPP (37 AA) (catalog number LT110006) (amidated C-terminal and disulfide bridge between Cys2 and Cys7) was from LifeTein (Hillsborough, NJ), and C-peptide (catalog number AS-61127) was from AnaSpec (Fremont, CA). Cα-peptide was custom made (Genemed Synthesis, Inc., San Antonio, TX). Black 96-well microplates (chimney well) were from Greiner Bio-One (Frickenhausen, Germany). Soluble and monomeric IAPP was made according to published protocols (18,19), and the AnaSpec Manual of SensoLyte Thioflavin T Aggregation Kit (catalog number AS-72214) was used for the IAPP aggregation assay, with final concentration of 1% DMSO and 25 μmol/L ThT in component A buffer. The buffer was used as blank and IAPP without inhibitors, and the final concentration of IAPP and C- and Cα-peptides was 50 μmol/L each in the inhibition assays. Amyloid kinetics were measured by increasing ThT fluorescent amyloid binding intensity (λex 440 nm; λem 485 nm; height 3 mm; flashes 12) for 36 cycles, 5 min per cycle, at 37°C with 15 s of shaking (100 rpm) between the reads in the EnSpire Multimode Plate Reader (PerkinElmer, Inc., Waltham, MA).
ELISA and Selected Reaction Monitoring Assays
For a full overview, see Supplementary Material for ELISA and selected reaction monitoring (SRM)–MS, Supplementary Tables 4–6, and Supplementary Fig. 1A–K.
Statistical Data Analysis
GraphPad Prism v9 software was used for statistical analysis, and data are presented as means ± SEM. For the ThT amyloid dye assay, area under the curve was calculated using the trapezoidal rule, and the data were analyzed using linear regression and one-way ANOVA with Tukey multiple comparison correction (20). The normalized expression values of INS isoforms in TaqMan fold changes and ddPCR percentages of target/β2 microglobulin droplets, ELISA, and SRM quantitative signals were analyzed with one-way ANOVA, two-way ANOVA (repeated measures; subject matching with Sidak correction), and unpaired Student t test, and P < 0.05 was considered significant.
Data and Resource Availability
The data sets and resources (TaqMan probes and SRM-MS peptides) generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request. National Center for Biotechnology Information GenBank accession numbers: INSU1 (MT335687), INSU2 (MT335688), INSUA (MT335689), INSUB (MT335690), INSUC (MT335691), and INS3B (MT335692)
Results
Identification of Novel Human Insulin Isoform INS3B and a 19-AA Cα-Peptide
Using the HSF bioinformatic tool (14), we found that the third exon contains a potential intraexonal splicing acceptor (>85% HSF matrix) site 36 bp downstream of the conventional exon 3A splicing acceptor site. We named the intraexon 3 spliced isoform INS3B (Fig. 1A, purple lettering), which translates to a 74-AA proinsulin (i.e., CαK-peptide), within which resides a 19-AA Cα-peptide instead of the classical 86-AA proinsulin with its 31-AA C-peptide encoded by INS3A (Fig. 1A and Fig. 2A). The Cα-peptide (EAEDLQGSLQPLALEGSLQ) does not contain the β-sheet (GQVEL) and hairpin (GGGPG) motifs that are present in the middle potion of the 31-AA C-peptide (21); however, they have identical estimated pI of 3.34 (22). Additionally, the 74-AA proinsulin of INS3B is predicted to have a soluble globular structure (15) without an intrinsically disordered protein region (VELGGGPGAGSLQP) (Fig. 2C). The classical insulin is transcribed from exons 1A (INS1A), 1B (INS1B), and 1C (INS1C), including one with intron 1 retention (INS1I) (Fig. 1A, green lettering). The combination is named INS3A and translates to a 110-AA preproinsulin and can be detected by TaqMan probe INS3A (Fig. 1A, green bar and bracket, and Supplementary Table 3). INS3A (31-AA C-peptide) and INS3B (19-AA Cα-peptide) expression in islets made up 85.2 ± 7.2% (n = 22) and 14.6 ± 2.7% (n = 22), respectively, of the total INS expression (Fig. 1B) measured by using the Exon-2 TaqMan Probe (Fig. 1A, blue bar under exon 2 that exists in all INS isoforms).
Figure 2.
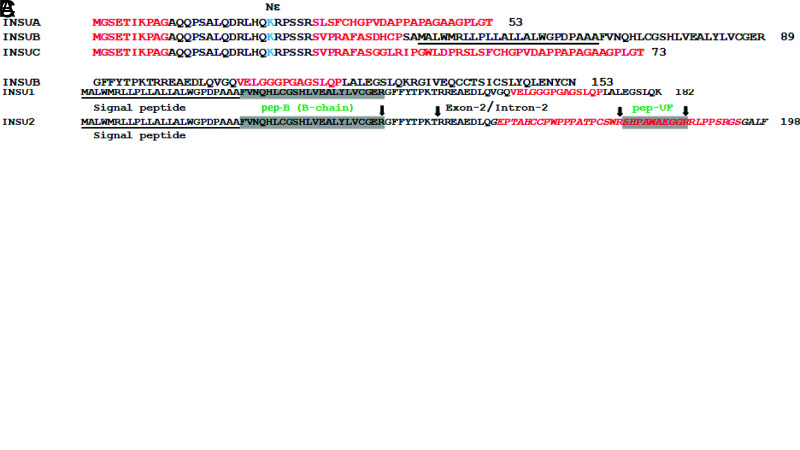
A–C: AA alignment of INS and INS3B (A); INSU1 and INSU2 (B); and INSUA, INSUB, and INSUC (C). Arrows indicate the tryptic cutting sites of lysine or arginine; the shaded regions are the SRM-MS–quantified peptides and are labeled with green lettering. The AA sequence numbers are on the right, and the signal peptide sequences are underlined. C-peptide β-sheet (GQVEL) and hairpin (GGGPG) motifs are marked in yellow and blue, respectively. The dashed lines represent 12 absent AA in Cα-peptide, and the trypsin cutting site K of CαK-peptide is in purple. Red AA are the intrinsically disordered protein regions; lysine (K) with potential Nε-glycated Nε-carboxymethyllysine are in light blue; the red AA sequence in italics is the 42-AA polypeptide of the frameshifted INSU2 encoded by intron 2; and exon-intron and exon-exon junctions are respectively marked with / and |, respectively.
uORF of INS (INSU) Isoforms
Exons 1 and 3 of INS are alternatively spliced, while exon 2 is a constitutive exon, within which a translation initiation site resides (9) (Fig. 1A, green asterisk). We assembled 3,544 human insulinoma and islet expressed sequence tags and found that many of them retain introns 1 and 2. In addition, several cDNA clones contain alternative uTSS upstream of the conventional INS 5′-cap site (21). Furthermore, we found that the uTSS nucleotide sequence contains a consensus human Kozak ribosomal binding site (23) (TGGGAGATGGGC) for an alternative translation initiation 45 bp upstream of the 5′-cap site (Fig. 1, red asterisk). The uORF of INSU1 is in frame with the pORF with retention of intron 1 and could be potentially translated to a 204-AA polypeptide, while the uORF of INSU2 (insulinoma BioSample: SAMN00164222) retains introns 1 and 2, potentially producing a frameshifted 198-AA polypeptide (Fig. 1A, red lettering, and Fig. 2B). The presence of INSU1 clones was validated by Sanger sequencing of IMAGE clones (https://www.imageconsortium.org/) of IMAGp998A2012483Q (BM85746) and IMAGp998O0113413Q (BU782803) in both directions. The uORF containing exons 1UA, 1UB, and 1UC (Fig. 1A, blue lettering) could be potentially spliced to exon 2, generating INSUA, INSUB, and INSUC, encoding polypeptides of 53, 153, and 73 AA, respectively (Fig. 2C).
Tissue Expression of INS Isoforms
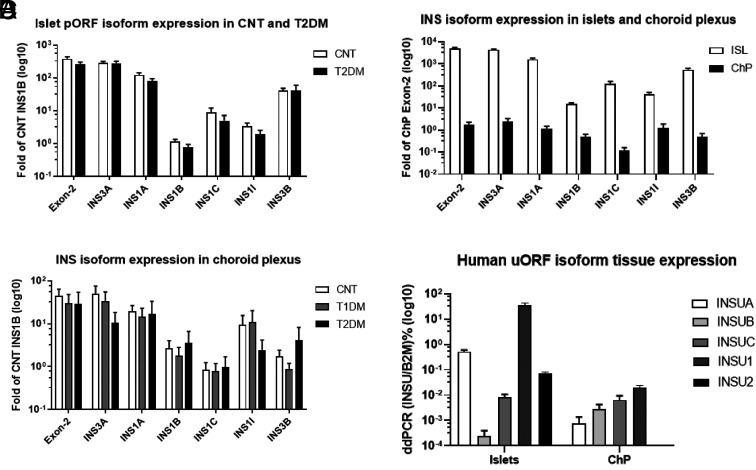
We detected pORF isoforms that had similar expression levels in both control and T2DM islets (Fig. 3A) by quantitative RT-PCR. All the INS pORF were expressed in both islets and ChP, indicating that ChP contains INS precursor mRNA splicing machinery similar to that of β-cells (Fig. 3B). The total INS (Exon-2 TaqMan Probe) mRNA level was >2,000-fold higher in islets than in ChP. The abundant INS1A, INS1C, and INS3B mRNA levels were ∼1,000-fold greater in islets than in ChP, while the lower-expressed INS1B and INS1I mRNA levels were ∼30-fold higher in islets than in ChP (Fig. 3B). We also found that postmortem ChP from insulin-deficient T1DM and T2DM contained INS isoforms at similar levels (Fig. 3C); we verified by ELISA in the stored blood samples of those who were receiving insulin therapy that C-peptide was absent. Additionally, by ddPCR, we detected uORF spliced isoforms (INSUA, -UB, -UC, and -U1) in both islets and ChP. On the other hand, the nonspliced isoform INSU2 was not detected in ChP (Fig. 3D).
Figure 3.
A–D: TaqMan quantitative RT-PCR of the isoforms in islets (ISL) and ChP. A: INS pORF isoforms in control (CNT) (n = 13) and T2DM islets (n = 9) using INS1B as a reference. B: INS pORF isoforms in human islets (n = 13) and ChP (n = 15) using exon 2 as a reference. C: Comparison of INS isoforms in ChP samples from CNT, T1DM, and T2DM. D: ddPCR quantification of INS uORF isoforms in ISL and ChP. B2M, β2 microglobulin.
Expression of Known Autoantigens, IAPP, and β-Cell Transcription Factors in ChP
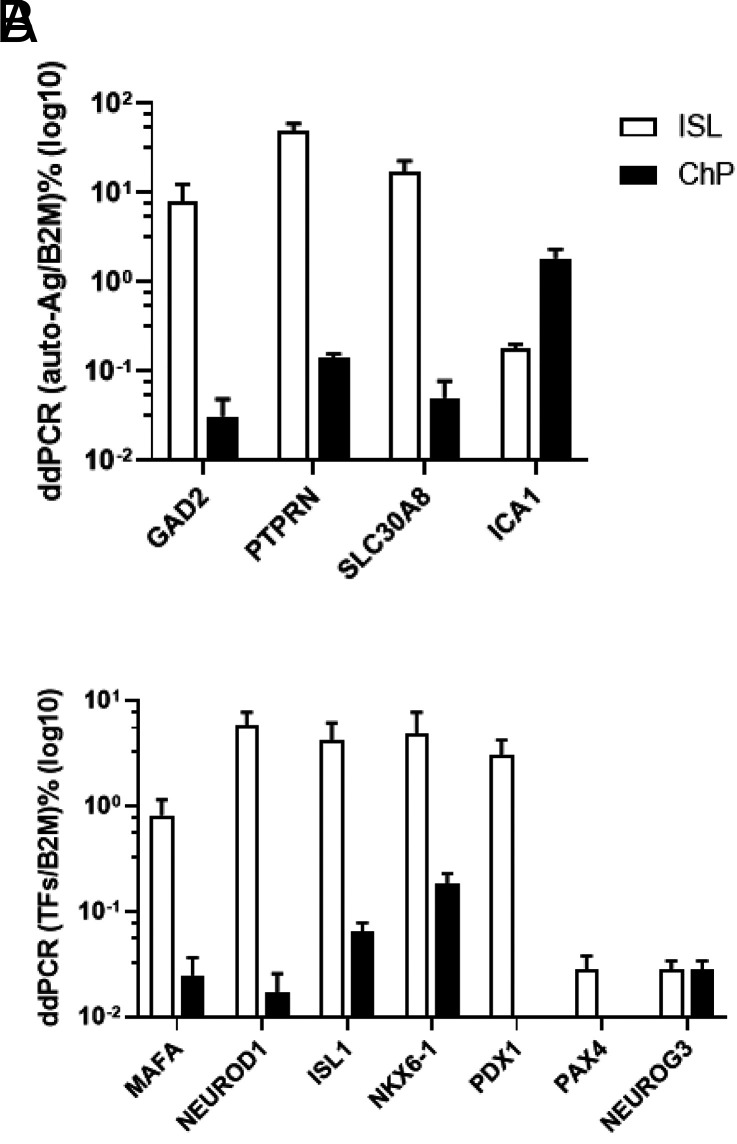
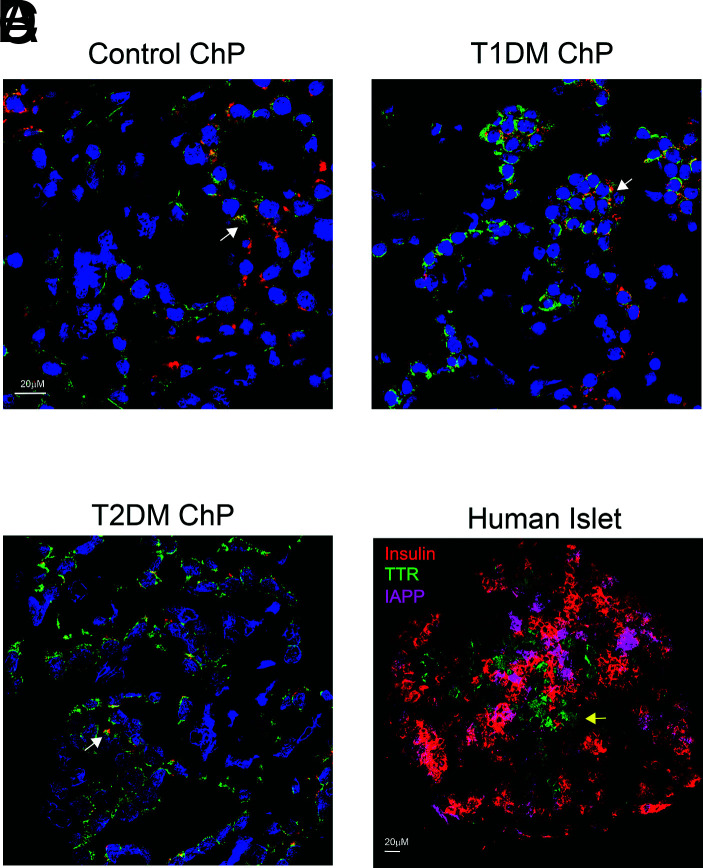
By ddPCR, we compared expression of β-cell autoantigens and transcription factors between islets and ChP. Except for islet cell autoantigen 1 (ICA1), expression of which was 10-fold higher in ChP than in control islets, expression of other known autoantigens (GAD2, PTPRN, and SLC30A8) was 253-, 346-, and 339-fold higher in islets (Fig. 4A), respectively. Endocrine differentiation factor neurogenin-3 (NEUROG3) mRNA levels were similar, while expression levels of several other critical β-cell transcription factors (MAFA, NEUROD1, ISL1, and NKX6–1) were 33-, 344-, 66-, and 27-fold higher in islets (Fig. 4B), respectively. Pancreatic and duodenal homeobox 1 transcription factor (PDX1) and paired box gene 4 transcription factor (PAX4) were absent from ChP (Fig. 4B). Using a triplex RNAscope ISH assay, we found that INS mRNA colocalized with TTR (a thyroid hormone transporter and present in all EChP) in control, T1DM, and T2DM ChP samples (Fig. 5, white arrows), as in mice (3); the IAPP gene was not expressed in ChP. This observation contrasts with pancreatic islets where INS colocalized with IAPP, as expected in β-cells, but did not colocalize with TTR (Fig. 5, yellow arrow); in islets, TTR was present in α-cells, as previously reported (24).
Figure 4.
A–B: ddPCR quantification of autoantigens and transcription factors. A: Comparison of autoantigen expression in islets (ISL) and ChP. B: Comparison of INS gene transcription and differentiation factors in ISL and ChP. y-axis is logarithm values of the target gene as percentages of β2 microglobulin (B2M) (normalized droplets).
Figure 5.
A–D: Confocal microscopy images of triplex RNAscope ISH in control ChP (A), T1DM ChP (B), and T2DM ChP (C) (×40 with DAPI staining) and human islets (D) (×20 omitting DAPI staining). Red represents INSU; green, TTR; and magenta, IAPP probes. White arrows indicate colocalization of INSU with TTR in ChP, and yellow arrow shows that TTR does not colocalize with either insulin or IAPP in islet.
SRM-MS Quantification of A- and B-chains, Cα-Peptide, and 74-AA Proinsulin (CαK-peptide) in Human Plasma, Islets, and ChP
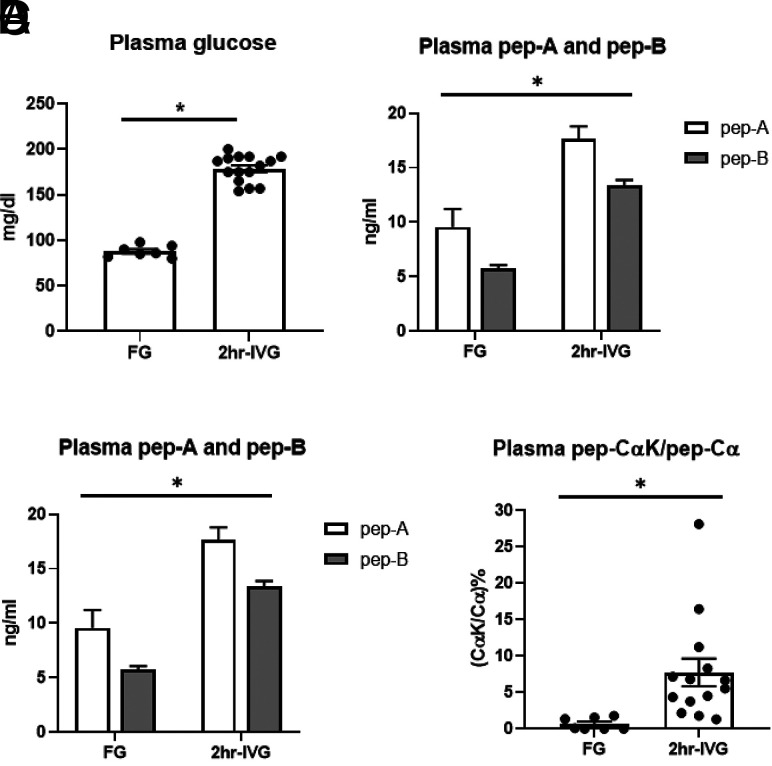
To evaluate the robustness of our SRM-MS assay, we measured A-chain, B-chain, and Cα-peptide in plasma under controlled conditions (i.e., after an intravenous hyperglycemic glucose clamp), during which circulating glucose levels were held constant at each participant’s fasting level plus 98 mg/dL; none had diabetes. Fasting plasma glucose and glucose after 2h-IVG were 88 ± 2 and 179 ± 4 mg/dL, respectively (Fig. 6A). There were significant increases after 2h-IVG, when measured by SRM, in both A- and B-chains (P < 0.0001) (Fig. 6B). ELISA measures total insulin, and as expected, it was also significantly increased after 2h-IVG, in concordance with the SRM results (P < 0.0001) (Supplementary Fig. 2A). The changes in Cα-peptide and CαK-peptide after 2h-IVG were much smaller (P = 0.0579) (Fig. 6C). However, there was a significant difference with ELISA measurement of C-peptide (P = 0.002) (Supplementary Fig. 2B), again as expected, demonstrating that Cα-peptide and 74-AA proinsulin were not as responsive to glucose as the classical C-peptide. We then calculated the sample-matched ratio of CαK-peptide to Cα-peptide and found it to be significantly increased after 2h-IVG (P = 0.019) (Fig. 6D), demonstrating that 2 h after continuous hyperglycemia, a significant amount of 74-AA proinsulin is released, likely from immature granules.
Figure 6.
A–D: Plasma glucose concentration (A), SRM A-peptide (A-chain) and B-peptide measurement (B-chain) (B), SRM quantification of processed Cα-peptide and nonprocessed CαK-peptide (C), and ratio of CαK-peptide to Cα-peptide (ratio of 74-AA proinsulin surrogate to mature insulin surrogate) (D) after an overnight fast (FG) (62.9 ± 1.1 years) (male n = 3; female n = 4) and 2h-IVG (57.9 ± 5.3 years) (male n = 9; female n = 6) infusion (continuous glucose infusion to maintain steady-state FG level + 98 mg/dL for 2 h). *P < 0.05.
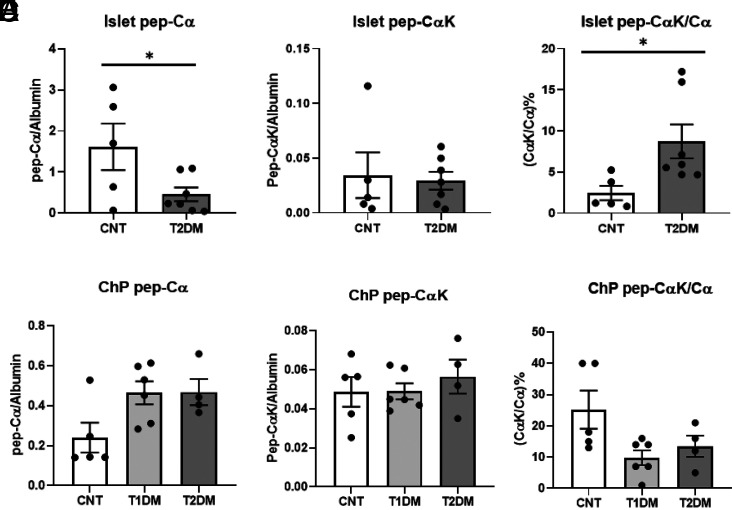
We did not find significant changes in A- or B-chain or uORF U-peptide levels extracted from islets between control and T2DM islets (Supplementary Fig. 3A–C). However, we did find that the amount of Cα-peptide was significantly lower in T2DM islets compared with control islets (P = 0.047) (Fig. 7A). The amounts of the 74-AA proinsulin represented by CαK-peptide were similar in control and T2DM islets (Fig. 7B), although the sample-matched ratio of CαK-peptide to Cα-peptide was significantly increased (P = 0.035) (Fig. 7C) in T2DM islets, implying the proinsulin protease process is compromised in T2DM β-cells. There were no significant changes in Cα-peptide, CαK-peptide, or ratio of CαK-peptide to Cα-peptide in nondiabetic, T1DM, or T2DM ChP samples (Fig. 7D–F). SRM detection of Cα-peptide again demonstrated insulin presence in postmortem ChP from individuals treated with lifelong exogenous insulin and whose peripheral blood lacked C-peptide.
Figure 7.
A–C: SRM relative quantification (normalized with islet endogenous albumin) of Cα-peptide (A), CαK-peptide (B), and ratio of CαK-peptide to Cα-peptide (C) in human control (CNT) and T2DM islets. D–F: SRM relative quantification (normalized with ChP endogenous albumin) of Cα-peptide (D), CαK-peptide (E), and ratio of CαK-peptide to Cα-peptide (F) in CNT, T1DM, and T2DM ChP samples. *P < 0.05.
Inhibition of IAPP Amyloid Fibrillation by Cα- and C-Peptides
Figure 8A is a representative experiment of three replicates showing IAPP fibrillation dynamics with lag time of 90 min. We observed that Cα-peptide inhibited IAPP fibrillation more efficiently than C-peptide by one-way ANOVA (P = 0.032) (Fig. 8B) and by regression analysis (P = 0.002) (Fig. 8C) in the range of 90–130 min. The ThT reporter was not affected by C- and Cα-peptides themselves (data not shown). Cα-peptide, but not C-peptide, also inhibits β-amyloid fibrillation (Supplementary Fig. 5).
Figure 8.
A–C: Inhibition of IAPP fibrillation by C- and Cα-peptides. A: Representative IAPP fibrillation dynamics in three replicates. B: One-way ANOVA (box-whisker plot) showed significant different inhibitory effects of C- and Cα-peptides (P = 0.032) on IAPP fibrillation. C: Linear regression analysis showed significant different inhibitory effects of C- and Cα-peptides (P = 0.002) on IAPP fibrillation. Area under the curve (AUC) calculated by trapezoid rule. Black represents 50 μmol/L IAPP plus buffer; blue, 50 μmol/L IAPP plus 50 μmol/L C-peptide; and red, 50 μmol/L IAPP plus 50 μmol/L Cα-peptide. *P < 0.05. F.I., fluorescence intensity.
Discussion
We uncovered novel uORF in specific INS isoforms that are transcribed from an alternative INS upstream promoter and uncovered a shorter 19-AA connecting peptide, Cα-peptide, in islets and ChP. Furthermore, this is the first report of the presence of insulin, either its mRNA or protein, in human ChP of insulin-deficient patients. Using our newly developed SRM-MS method, we validated translation of the novel human INS isoforms, and we quantified the distinctive uORF- and pORF-derived A- and B-chains and processed Cα- and nonprocessed CαK-peptides at low nanogram to picogram per milliliter biofluids. Commercial ELISA cannot distinguish between A- and B-chains, nor between Cα- and C-peptides. The ratio of CαK-peptide to Cα-peptide reflects islet proinsulin processing status and confirms insulin presence in ChP. The limit of quantitation of subnanogram per milliliter of the uORF- and pORF-derived peptides allows for detection of low quantities of insulin that cannot be accurately quantified via ELISA in CSF (25).
The conventional 86-AA proinsulin is prefolded into its correct conformation in the Golgi apparatus, together with C-peptide (26). The N-terminus of the 31-AA C-peptide (EAEDLQVGQ) acts as a chaperone (27) to promote correct insulin folding, while its C-terminus (EGSLQ), in circulation, is known to bind the orphan G-protein–coupled receptor GPR146 (28). Furthermore, and intriguingly, its middle region contains β-sheet (GQVEL) and hairpin (GGGPG) motifs that prevent proinsulin fibrillation (29,30). The novel human Cα-peptide has the same estimated isoelectric point as that of C-peptide (pI 3.34), but it is devoid of those motifs and therefore acts more efficiently than C-peptide as a trans-chaperone against IAPP amyloid formation. The shortened isoform might also even function as a cis-chaperone to mitigate any amyloidogenicity of insulin itself (30). The increased ratio of nonprocessed to processed Cα-peptide in hyperglycemic plasma and T2DM islets implies more proinsulin secretion, a phenomenon already described (31). The expression of proinsulin-processing enzymes (convertase PC2, carboxypeptidase E, and especially convertase PC1/3) is reduced in T2DM islets (32).
Of no less interest, IAPP is not present in ChP, nor is PDX1. PDX1 is a necessary transcription factor for glucose-mediated INS regulation and for IAPP gene expression (33), and its absence explains why glucose does not regulate INS (3) and why IAPP is lacking in ChP. IAPP packaging in granules with insulin within β-cells points to its later evolution in islets, most likely at approximately the same time as INS expression in the periphery became confined to β-cells. Teleologically, it is indeed a good thing that IAPP is absent in ChP, because IAPP-based amyloidosis, which was described as far back as 1901 by Opie (34) as being pathological to islets of patients with T2DM, would in all likelihood be detrimental to neuronal function. It is a highly fibrillogenic peptide, and it is not known exactly how its fibril formation is inhibited within granules. Within granules, as seen on electron micrographic images, insulin is stored in the core and IAPP in the halo region. C-peptide within proinsulin is also in the halo and, like insulin itself, inhibits IAPP fibril formation (35). The alignment of Cα-peptide would place it in the halo, and therefore, it likely has a role in preventing not only insulin but also IAPP-based fibril formation. Decreasing amounts of Cα-peptide in T2DM might therefore make IAPP deposition more likely in β-cells. Cα-peptide therapeutics could possibly prevent IAPP-based islet amyloidosis.
Over the span of evolution, and with the enlargement and increased sophistication of neural networks, neurotrophic factors such as BDNF and GDNF that are not present in invertebrates (36,37) made their appearance and likely superseded and/or added to the many original insulin growth functions in the brain. Insulin gene duplications and deletions also occurred many times (38) and were permissive to the appearance of the gene in vertebrate endodermal cells such as β-cells (39), with residual remains in the ectoderm (40) such as EChP. We have already reported three isoforms in mice, Ins2-V1, -V2, and -V3, and we recently uncovered an additional mouse Ins2 isoform, Ins2-V4 (Supplementary Fig. 4A), in which uTSS (without uORF) is encoded by exon 1A, which is located almost 20 kb upstream of the classical exon 1B. The Ins2-V4 isoform mRNA was not detected in mouse islets but was seen in mouse ChP and total pancreas (Supplementary Fig. 4B), providing us with another example of selectivity in isoform expression. It is now accepted that during evolution, the endocrine and nervous systems overlapped. Phylogenetically, neurons preceded typical endocrine glands; neurons first emerged in cnidarians, while endocrine glands were first seen in primitive vertebrates, such as the hagfish, the insulin of which is surprisingly similar to that of humans (41). Vertebrates benefitted greatly in evolution by separation of metabolic properties of islet-derived insulin in peripheral organs, thereby allowing increases in body size and storage of energy supplies, such as glycogen and fat, for later use by the evolving brain. However, remnant brain INS is still provided by ChP, but its regulation, unlike islet-derived insulin, is not by glucose, because ChP is devoid of PDX1 and PAX4 transcription factors; ChP insulin synthesis and secretion are regulated by a neurotransmitter, serotonin, as we stated earlier, through the 5-HT2C receptor (3).
If ChP-derived insulin is critical for certain postnatal brain functions (which have not been adequately determined) or has any biological relevance independent of peripherally derived insulin, it seems reasonable to assume that loss of this source of insulin, should insulin-containing EChP be subject to autoimmune destruction or other critical insults, would have detrimental effects. Additionally, EChP, which are terminally differentiated cells (42), have many essential functions, not least of which are transportation of thyroid hormone into brain via TTR and formation of CSF. Yet, distinct central nervous system effects other than what can be ascribed to classical disordered metabolism are not hallmarks of T1DM, not even prior to initiation of insulin therapy or during the insulitis phase of the disease. Moreover, genetic susceptibility to T1DM does not confer certainty of its development, and it is presumed that environmental factors are also required; EChP might be protected, even though they express low levels of triggering antigens. Regardless, it seems clear that EChP still maintain insulin expression when there is peripheral insulin deficiency.
In summary, we now report that insulin is produced in human ChP. We uncovered a novel cis-chaperone and nonamyloidogenic Cα-peptide, and we conclude that proinsulin is produced and processed in EChP in a similar manner to β-cells. However, IAPP and PDX1 are not expressed by EChP. We also introduce a novel MS-based assay for quantifying low amounts of proinsulin products.
Article Information
Acknowledgments. The authors thank Drs. Ruin Moaddel, Zhixing Yuan, and Maire E. Doyle, Laboratory of Clinical Investigation, National Institute on Aging, National Institutes of Health, for critical review and input for the liquid chromatography with tandem MS–SRM methods, results, and images. The authors thank Professor Daniel Raleigh, Stony Brook University, for invaluable advice on the IAPP ThT fibrillation assay, without which we could not have carried out the experiments in Fig. 8 and Supplementary Fig. 5.
Funding. Q.-R.L., M.Z., P.Z., C.H.M., N.S.H., D.L.L., Q.C., J.F.O., L.F., and J.M.E. are supported by the Intramural Research Program (IRP) of the National Institute on Aging, and P.A. and S.M. are supported by the IRP of the National Institute of Mental Health (ZIC MH002903). Postmortem pancreata were provided by the National Institute of Diabetes and Digestive and Kidney Diseases–funded Integrated Islet Distribution Program at City of Hope (National Institutes of Health grant 2UC4DK098085) (https://iidp.coh.org/). The tissue/data used in this research were obtained from the Human Brain Collection Core, IRP, National Institute of Mental Health (https://www.nimh.nih.gov/hbcc).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. Q.-R.L. and J.M.E. conceptualized the study and wrote the manuscript. Q.-R.L. performed bioinformatic search and molecular biological studies. M.Z., P.Z., and L.F. performed proteomic SRM experiments and data analysis. C.H.M. dissected mouse ChP and performed mouse gene expression assay. N.S.H. and D.L.L. performed RNAscope ISH of human islets and ChP. Q.C. performed ELISA experiments. P.A. and S.M. dissected human postmortem ChP samples, extracted clinical information, and organized Supplementary Table 1. J.F.O. arranged human islet collection and dissected mouse pancreatic islets. C.W.C. performed the hyperglycemic clamps and plasma collection. All authors reviewed and approved the publication of this manuscript. J.M.E. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.16640458.
References
- 1. Smit AB, Vreugdenhil E, Ebberink RH, Geraerts WP, Klootwijk J, Joosse J. Growth-controlling molluscan neurons produce the precursor of an insulin-related peptide. Nature 1988;331:535–538 [DOI] [PubMed] [Google Scholar]
- 2. Griffith CM, Eid T, Rose GM, Patrylo PR. Evidence for altered insulin receptor signaling in Alzheimer’s disease. Neuropharmacology 2018;136(Pt B):202–215 [DOI] [PubMed] [Google Scholar]
- 3. Mazucanti CH, Liu QR, Lang D, et al. Release of insulin produced by the choroid plexis is regulated by serotonergic signaling. JCI Insight 2019;4:e131682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peavy DE, Abram JD, Frank BH, Duckworth WC. In vitro activity of biosynthetic human proinsulin. Receptor binding and biologic potency of proinsulin and insulin in isolated rat adipocytes. Diabetes 1984;33:1062–1067 [DOI] [PubMed] [Google Scholar]
- 5. Shiao MS, Liao BY, Long M, Yu HT. Adaptive evolution of the insulin two-gene system in mouse. Genetics 2008;178:1683–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leroux L, Desbois P, Lamotte L, et al. Compensatory responses in mice carrying a null mutation for Ins1 or Ins2. Diabetes 2001;50(Suppl. 1):S150–S153 [DOI] [PubMed] [Google Scholar]
- 7. Smit AB, van Kesteren RE, Li KW, et al. Towards understanding the role of insulin in the brain: lessons from insulin-related signaling systems in the invertebrate brain. Prog Neurobiol 1998;54:35–54 [DOI] [PubMed] [Google Scholar]
- 8. Evans-Molina C, Garmey JC, Ketchum R, Brayman KL, Deng S, Mirmira RG. Glucose regulation of insulin gene transcription and pre-mRNA processing in human islets. Diabetes 2007;56:827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bell GI, Pictet RL, Rutter WJ, Cordell B, Tischer E, Goodman HM. Sequence of the human insulin gene. Nature 1980;284:26–32 [DOI] [PubMed] [Google Scholar]
- 10. de Klerk E, ’t Hoen PA. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet 2015;31:128–139 [DOI] [PubMed] [Google Scholar]
- 11. Roep BO, Peakman M. Antigen targets of type 1 diabetes autoimmunity. Cold Spring Harb Perspect Med 2012;2:a007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoffman GE, Bendl J, Voloudakis G, et al. CommonMind Consortium provides transcriptomic and epigenomic data for schizophrenia and bipolar disorder. Sci Data 2019;6:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mager DE, Abernethy DR, Egan JM, Elahi D. Exendin-4 pharmacodynamics: insights from the hyperglycemic clamp technique. J Pharmacol Exp Ther 2004;311: 830–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 2009;37:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res 2003;31:3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu QR, Rubio FJ, Bossert JM, et al. Detection of molecular alterations in methamphetamine-activated Fos-expressing neurons from a single rat dorsal striatum using fluorescence-activated cell sorting (FACS). J Neurochem 2014;128:173–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu QR, Huang NS, Qu H, et al. Identification of novel mouse and rat CB1R isoforms and in silico modeling of human CB1R for peripheral cannabinoid therapeutics. Acta Pharmacol Sin 2019;40:387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stine WB, Jungbauer L, Yu C, LaDu MJ. Preparing synthetic Aβ in different aggregation states. Methods Mol Biol 2011;670:13–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tu LH, Young LM, Wong AG, Ashcroft AE, Radford SE, Raleigh DP. Mutational analysis of the ability of resveratrol to inhibit amyloid formation by islet amyloid polypeptide: critical evaluation of the importance of aromatic-inhibitor and histidine-inhibitor interactions. Biochemistry 2015;54:666–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chia CW, Odetunde JO, Kim W, Carlson OD, Ferrucci L, Egan JM. GIP contributes to islet trihormonal abnormalities in type 2 diabetes. J Clin Endocrinol Metab 2014;99:2477–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bell GI, Pictet R, Rutter WJ. Analysis of the regions flanking the human insulin gene and sequence of an Alu family member. Nucleic Acids Res 1980;8:4091–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wilkins MR, Lindskog I, Gasteiger E, et al. Detailed peptide characterization using PEPTIDEMASS—a World-Wide-Web-accessible tool. Electrophoresis 1997;18:403–408 [DOI] [PubMed] [Google Scholar]
- 23. Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 1987;15:8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Su Y, Jono H, Misumi Y, et al. Novel function of transthyretin in pancreatic alpha cells. FEBS Lett 2012;586:4215–4222 [DOI] [PubMed] [Google Scholar]
- 25. Liu QR, Zhu M, Chia CW, et al. Novel mass spectrometry based selected reaction monitoring proteomics for analysis of low abundant insulin levels in cerebrospinal fluid (Abstract). Diabetes 2020;69(Suppl. 1):1719-P [Google Scholar]
- 26. Tsiolaki PL, Louros NN, Zompra AA, Hamodrakas SJ, Iconomidou VA. Unraveling the aggregation propensity of human insulin C-peptide. Biopolymers 2017;108:108. [DOI] [PubMed] [Google Scholar]
- 27. Steiner DF. The proinsulin C-peptide—a multirole model. Exp Diabesity Res 2004;5:7–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kolar GR, Grote SM, Yosten GL. Targeting orphan G protein-coupled receptors for the treatment of diabetes and its complications: C-peptide and GPR146. J Intern Med 2017;281:25–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang K, Dong J, Phillips NB, Carey PR, Weiss MA. Proinsulin is refractory to protein fibrillation: topological protection of a precursor protein from cross-beta assembly. J Biol Chem 2005;280:42345–42355 [DOI] [PubMed] [Google Scholar]
- 30. Yang Y, Hua QX, Liu J, et al. Solution structure of proinsulin: connecting domain flexibility and prohormone processing. J Biol Chem 2010;285:7847–7851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen YC, Taylor AJ, Verchere CB. Islet prohormone processing in health and disease. Diabetes Obes Metab 2018;20(Suppl. 2):64–76 [DOI] [PubMed] [Google Scholar]
- 32. Teitelman G. Heterogeneous expression of proinsulin processing enzymes in beta cells of non-diabetic and type 2 diabetic humans. J Histochem Cytochem 2019;67:385–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang H, Maechler P, Ritz-Laser B, et al. Pdx1 level defines pancreatic gene expression pattern and cell lineage differentiation. J Biol Chem 2001;276:25279–25286 [DOI] [PubMed] [Google Scholar]
- 34. Opie EL. On the relation of chronic interstitial pancreatitis to the islands of Langerhans and to diabetes melutus. J Exp Med 1901;5:397–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Westermark P. Amyloid in the islets of Langerhans: thoughts and some historical aspects. Ups J Med Sci 2011;116:81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chao MV. Trophic factors: an evolutionary cul-de-sac or door into higher neuronal function? J Neurosci Res 2000;59:353–355 [PubMed] [Google Scholar]
- 37. Liu QR, Lu L, Zhu XG, Gong JP, Shaham Y, Uhl GR. Rodent BDNF genes, novel promoters, novel splice variants, and regulation by cocaine. Brain Res 2006;1067:1–12 [DOI] [PubMed] [Google Scholar]
- 38. Shuldiner AR, Phillips S, Roberts CT Jr, LeRoith D, Roth J. Xenopus laevis contains two nonallelic preproinsulin genes. cDNA cloning and evolutionary perspective. J Biol Chem 1989;264:9428–9432 [PubMed] [Google Scholar]
- 39. Slack JM. Developmental biology of the pancreas. Development 1995;121:1569–1580 [DOI] [PubMed] [Google Scholar]
- 40. Johansson PA. The choroid plexuses and their impact on developmental neurogenesis. Front Neurosci 2014;8:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peterson JD, Coulter CL, Steiner DF, Emdin SO, Falkmer S. Structural and crystallographic observations on hagfish insulin. Nature 1974;251:239–240 [DOI] [PubMed] [Google Scholar]
- 42. Redzic ZB, Preston JE, Duncan JA, Chodobski A, Szmydynger-Chodobska J. The choroid plexus-cerebrospinal fluid system: from development to aging. Curr Top Dev Biol 2005;71:1–52 [DOI] [PubMed] [Google Scholar]
- 44. Chia CW, Carlson OD, Liu DD, González-Mariscal I, Santa-Cruz Calvo S, Egan JM. Incretin secretion in humans is under the influence of cannabinoid receptors. Am J Physiol Endocrinol Metab 2017;313:E359–E366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Palazzoli F, Citti C, Licata M, et al. Development of a simple and sensitive liquid chromatography triple quadrupole mass spectrometry (LC-MS/MS) method for the determination of cannabidiol (CBD), Δ9-tetrahydrocannabinol (THC) and its metabolites in rat whole blood after oral administration of a single high dose of CBD. J Pharm Biomed Anal 2018;150:25–32 [DOI] [PubMed] [Google Scholar]
- 46. Zhang P, Zhu M, Geng-Spyropoulos M, et al. A novel, multiplexed targeted mass spectrometry assay for quantification of complement factor H (CFH) variants and CFH-related proteins 1-5 in human plasma. Proteomics 2017;17:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhu M, Zhang P, Geng-Spyropoulos M, Moaddel R, Semba RD, Ferrucci L. A robotic protocol for high-throughput processing of samples for selected reaction monitoring assays. Proteomics 2017;17:XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]