Abstract
Pluripotent stem cells are known to shift their mitochondrial metabolism upon differentiation, but the mechanisms underlying such metabolic rewiring are not fully understood. We hypothesized that during differentiation of human induced pluripotent stem cells (hiPSCs), mitochondria undergo mitophagy and are then replenished by the biogenesis of new mitochondria adapted to the metabolic needs of the differentiated cell. To evaluate mitophagy during iPSC differentiation, we performed live cell imaging of mitochondria and lysosomes in hiPSCs differentiating into vascular endothelial cells using confocal microscopy. We observed a burst of mitophagy during the initial phases of hiPSC differentiation into the endothelial lineage, followed by subsequent mitochondrial biogenesis as assessed by the mitochondrial biogenesis biosensor MitoTimer. Furthermore, hiPSCs undergoing differentiation showed greater mitochondrial oxidation of fatty acids and an increase in ATP levels as assessed by an ATP biosensor. We also found that during mitophagy, the mitochondrial phosphatase PGAM5 is cleaved in hiPSC-derived endothelial progenitor cells and in turn activates β-catenin-mediated transcription of the transcriptional coactivator PGC-1α, which upregulates mitochondrial biogenesis. These data suggest that mitophagy itself initiates the increase in mitochondrial biogenesis and oxidative metabolism through transcriptional changes during endothelial cell differentiation. In summary, these findings reveal a mitophagy-mediated mechanism for metabolic rewiring and maturation of differentiating cells via the β-catenin signaling pathway. We propose that such mitochondrial-nuclear cross talk during hiPSC differentiation could be leveraged to enhance the metabolic maturation of differentiated cells.
Keywords: mitophagy, mitochondrial metabolism, induced pluripotent stem cells, cell differentiation, β-catenin
Abbreviations: CM, cardiomyocyte; EC, endothelial cell; hiPSCs, human induced pluripotent stem cells; OCR, oxygen consumption rate; PGAM5, phosphoglycerate mutase family member 5
Pluripotent stem cells such as human induced pluripotent stem cells (hiPSCs) rely mainly on glycolysis for cellular ATP (1, 2, 3) and use glutamine metabolism to sustain pluripotency (4, 5). However, when stem cells differentiate, their metabolism also shifts toward enhanced oxidative phosphorylation and mitochondrial ATP generation (6). Importantly, the metabolic shift can reinforce cell differentiation (7), thus suggesting there is a positive feedback loop between cell differentiation and metabolic rewiring. One such example of the cross talk between cell metabolism and the pluripotency state occurs when pluripotent cells downregulate glutamine metabolism, which in turn increases ROS generation and reduces activity of the pluripotency transcription factor OCT4, thus further stabilizing differentiation and the shift away from pluripotency (4). The recognition that metabolic reprogramming is an essential feature of stem cell differentiation (8, 9) raises the important question of how mitochondria in pluripotent stem cells are reprogrammed during differentiation.
Mitophagy (the autophagy of mitochondria) is the selective elimination of depolarized or damaged mitochondria. Generally, mitophagy occurs as a method of quality control, eliminating damaged mitochondria with mutant mtDNA or misfolded proteins in order to maintain mitochondrial health (10, 11). The canonical pathway for mitophagy involves the serine/threonine kinase PINK1 (PTEN-induced kinase 1) and the E3 ubiquitin ligase Parkin (12). PINK1 continuously cycles from the cytosol to the mitochondrial outer membrane. If the mitochondria are healthy, PINK1 is cleaved and is released back into the cytosol (13). When mitochondria are damaged and the mitochondrial membrane is depolarized, then PINK1 is stabilized and accumulates at the membrane. PINK1 phosphorylates the mitochondrial outer membrane protein Mitofusin 2, which in turn serves as a receptor for Parkin (14). Parkin then translocates to the mitochondria to ubiquitinate multiple proteins and trigger autophagy mechanisms to remove the mitochondria (13, 15).
While mitophagy has primarily been studied in the setting of culling damaged mitochondria in the setting of injury or disease, we posited that mitophagy may serve as a physiologic mechanism for the metabolic rewiring of differentiating stem cells by removing mitochondria adapted to the pluripotent state and replacing them with new mitochondria configured for the differentiated cell state. The vast majority of mitochondrial proteins are encoded by the nuclear genome; therefore, any initiation of compensatory mitochondrial biogenesis in response to mitophagy would require communication between mitochondria and the nucleus. One such potential mediator is the mitochondrial protein PGAM5 (Phosphoglycerate Mutase Family Member 5), a serine/threonine phosphatase that is embedded in the mitochondrial outer membrane (16) and is cleaved and released into the cytosol during mitophagy (17, 18). It enhances signaling via the β-catenin pathway by dephosphorylating β-catenin, thus stabilizing it and allowing it to translocate to the nucleus to transcribe Wnt/β-catenin pathway genes (17).
In this study, we show that mitophagy and subsequent mitochondrial biogenesis are required for the differentiation of hiPSCs to endothelial cells through the interaction of cleaved PGAM5 and β-catenin. Furthermore, we show that PGAM5 cleavage leads to differentiation of progenitor cells into more mature endothelial cells and the biogenesis of new mitochondria that exhibit a shift from mitochondrial glutamine metabolism to fatty acid metabolism. This novel feedback mechanism of mitochondrial-nuclear cross talk in differentiating pluripotent stem cells provides new insights into the intersection of metabolic and developmental signaling.
Results
There is an initial burst of mitophagy during differentiation
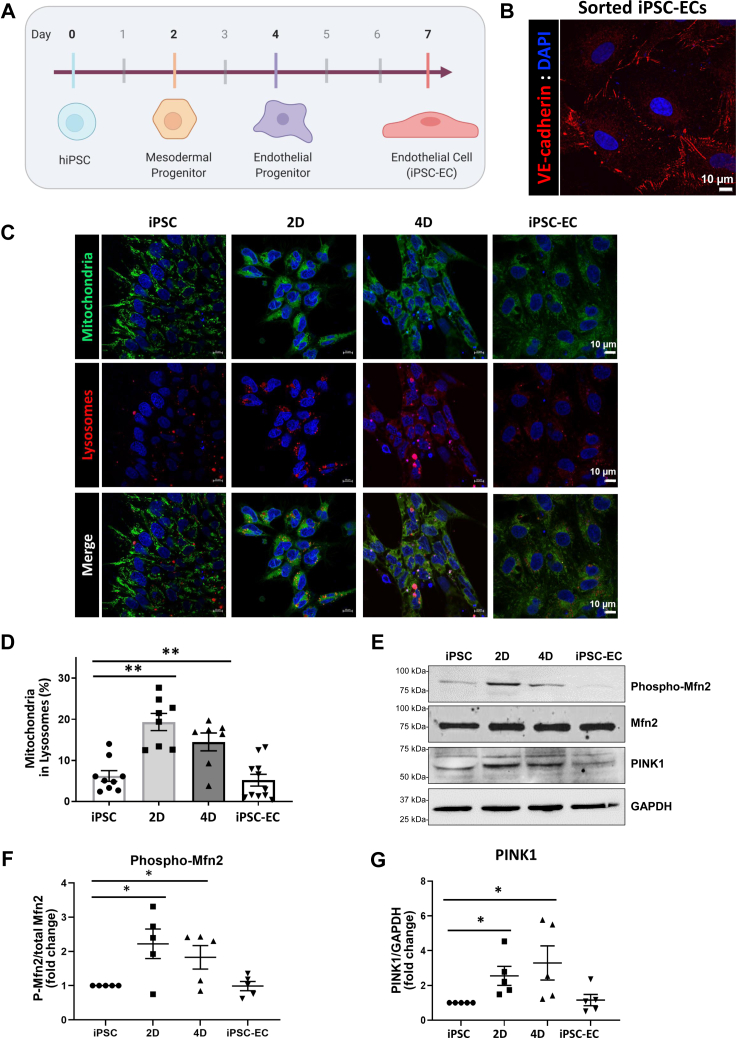
To investigate whether mitophagy is activated during differentiation, we first differentiated hiPSCs to ECs (endothelial cells) using a three-step protocol (Fig. 1A). Cells were initially differentiated toward a mesodermal lineage, then an endothelial progenitor stage, and lastly to a mature differentiated endothelial state in which the cells express the endothelial adherens junction molecule VE-Cadherin (Fig. 1B). iPSC-ECs represent only VE-Cadherin positive cells. To further characterize iPSC-ECs, two iPSC lines (273 and C2) were differentiated to ECs, and the iPSC-EC identity was seen in the form of cobblestone morphology (Fig. S1A), as well as flow cytometry (Fig. S1B), which showed that more than 90% of cells express the cell surface proteins VE-Cadherin and CD31. A capillary-like tube formation assay was performed (Fig. S1C), which demonstrated that iPSC-ECs but not iPSCs are able to form a vascular-like network. Moreover, mRNA levels of VE-Cadherin, CD31, and VEGFR2 (Fig. S1D) showed a significant increase in these markers. Together these results show that iPSC-ECs in this study exhibit endothelial cell traits. We used this endothelial differentiation approach because human pluripotent stem cells demonstrate a clear metabolic shift characterized by downregulation of mitochondrial glutamine metabolism when pluripotent stem cells differentiate into mature endothelial cells (4).
Figure 1.
There is an initial burst of mitophagy during differentiation. A, schematic of differentiation protocol. hiPSCs were differentiated for 7 days in serum-free media toward an endothelial lineage. Cells were collected at days 0 (iPSC), 2, 4, and 7 after sorting for VE-Cadherin positive cells (iPSC-EC). B, representative confocal image of VE-Cadherin after VE-Cadherin (CD144) sorting of day 7 cells. iPSC-ECs express VE-Cadherin at the junctions between cells. DAPI depicts nuclei staining. Scale bar = 10 μm. C, live cells were stained with MitoTracker (mitochondria, green) and Lysotracker (lysosomes, red) and imaged at specified timepoints with confocal microscopy. iPSC indicates cells at day 0, 2D = day 2, 4D = day 4, and iPSC-EC indicates 7 days differentiated cells, sorted for VE-Cadherin. Scale bar = 10 μm. D, quantification of (C) indicating the percent of overall mitochondria within lysosomes. Mitophagy is increased on days 2 and 4. Mean ± SEM; n = 8 fields of view, image shown is representative of three independent experiments, with ∗∗p < 0.01. E, cells were lysed at different stages of differentiation. Western blots show Ser443 Mitofusin 2 phosphorylation, overall Mfn2 levels, PINK1 levels, and GAPDH as loading control. F, quantification of (E), which shows a higher level of phosphorylated Mfn2 on days 2 and 4 of differentiation. Mean ± SEM; n = 5 with ∗p < 0.05. G, quantification of (E), which shows a higher level of PINK1 on days 2 and 4 of differentiation. Mean ± SEM; n = 5 with ∗p < 0.05.
To determine whether mitophagy occurs during differentiation, the cells at the defined differentiation stages were stained with MitoTracker (green) and Lysotracker (red), followed by confocal microscopy. Mitophagy was evaluated by quantifying colocalization of lysosomes and mitochondria (Fig. 1, C and D). Mitophagy increased early in differentiation (at days 2 and 4) and then returned to baseline after cells reached the differentiated EC stage at day 7. We also examined the change in Ser443 Phospho-Mfn2 and PINK1, key mediators of the canonical PINK1/Parkin/Mitofusin 2 pathway. As shown in Figure 1, E–G, Phospho-Mfn2 and PINK1 protein levels increased on days 2 and 4, concomitantly with the observed increase in mitophagy, suggesting that the canonical mitophagy pathway was activated during differentiation of hiPSCs.
To understand whether the increase in mitophagy during differentiation was also seen during differentiation into other cell lineages, we differentiated two iPSC cell lines (273 and C2) to cardiomyocytes using a 15-day cardiomyocyte (CM) differentiation protocol (19). We used Mitotracker (green) and Lysotracker (red) colabeling to assess mitophagy on day 2, day 5, and day 9. While the observed increase in mitophagy was not as prominent as during endothelial differentiation, there was a significant increase in mitophagy early on at day 2 (Fig. S2, A and B), corresponding to increases in the mitophagy mediators PINK1 and Phospho-Mfn2 (Fig. S2, C and D).
Mitophagy is followed by a compensatory rise in mitochondrial biogenesis
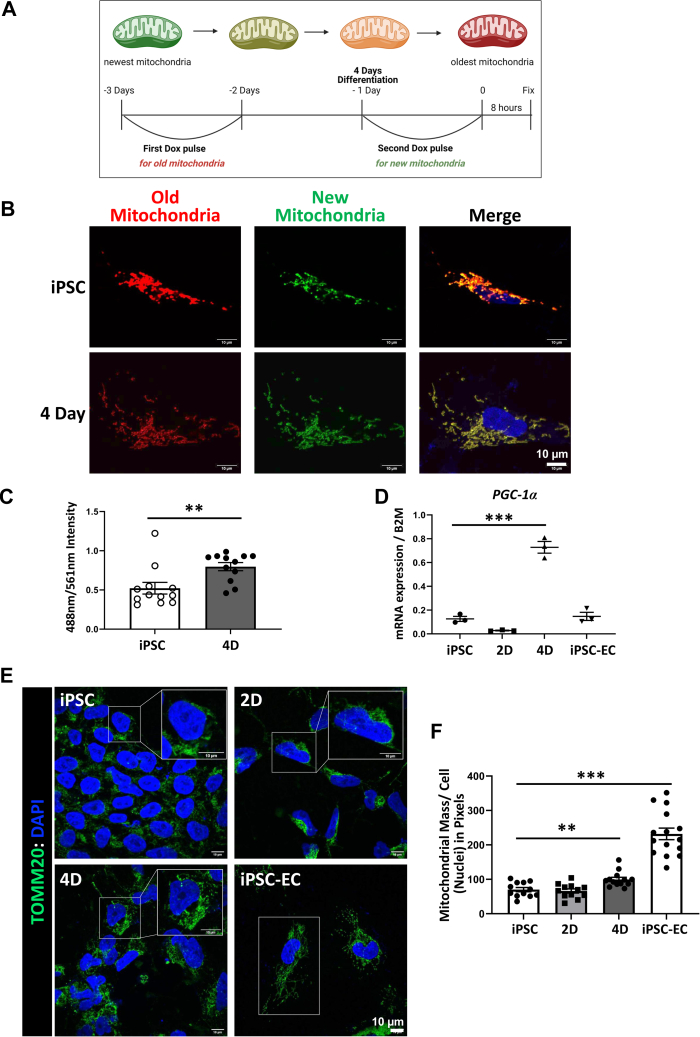
To determine whether mitophagy was followed by compensatory mitochondrial biogenesis to replenish the culled mitochondria, we utilized the doxycycline inducible MitoTimer construct (20). In cells expressing the doxycycline-inducible fluorescent protein construct MitoTimer, mitochondrial biogenesis can be quantified as a ratio of fluorescence emission spectra. MitoTimer in newly synthesized mitochondria exhibit green fluorescence but over time, aging mitochondria have increased red fluorescence (Fig. 2A). MitoTimer is used to track mitochondrial aging and biogenesis. We examined whether mitophagy in differentiating hiPSCs was followed by mitochondrial biogenesis subsequent to the early mitophagy during differentiation. Using this MitoTimer construct, we compared control hiPSCs with cells differentiated for 4 days. We first stimulated the cells with a pulse of 2 μg/ml doxycycline for 24 h. After 48 h, all the mitochondria were red fluorescent indicative of mitochondrial aging. The cells were given a second pulse of doxycycline for the 24 h of day 4 differentiation, and then newly synthesized mitochondria were evaluated by monitoring green mitochondria using confocal microscopy in fixed cells. De novo mitochondrial biogenesis was analyzed as the ratio of green over red fluorescence intensity. We found a doubling of newly synthesized mitochondria in day four differentiated endothelial progenitor cells as compared with undifferentiated hiPSCs indicating more de novo mitochondrial biogenesis after the initial mitophagy (Fig. 2, B and C).
Figure 2.
There is a subsequent burst of mitochondrial biogenesis after mitophagy.A, schematic of MitoTimer doxycycline-inducible construct. Mitochondria fluoresce green as they are newly synthesized and then gradually fluoresce red as they age over 72 h. Doxycycline pulses depicted for experiment in (B). Three days before endpoint, cells were pulsed for 24 h with 2 μg/ml of doxycycline. After doxycycline is removed, mitochondria age and the network emit a red fluorescence. On day 4 of differentiation, a second 2 μg/ml doxycycline pulse was added for 24 h and then removed. Eight hours is sufficient time for these mitochondria to fluoresce green. Afterward, the cells were fixed with 4% PFA. B, MitoTimer confocal images of fixed iPSCs and 4-day differentiated cells. 2 μg/ml doxycycline pulses are outlined in (A). Red mitochondria represent old mitochondria and green mitochondria represent new mitochondria. Merged image includes both channels as well as DAPI nuclear staining. Scale bar= 10 μm. C, quantification of (B) indicating the intensity of green fluorescence (488 nm) over the intensity of red fluorescence (561 nm). Mean ± SEM, with n = 12 fields of view from three independent experiments, ∗p < 0.05. D, mRNA was extracted from cells at different stages of differentiation and mRNA levels of PGC-1α, which is increased on day 4 as compared with iPSCs. Mean ± SEM, n = 3 with ∗∗∗p < 0.001. E, immunohistochemistry of cells fixed at different days of differentiation on coverslips and imaged with confocal microscopy. TOMM20 is shown in green and the nuclei are in blue. Top right inset is a zoom image of one cell. Scale bar = 10 μm. F, quantification of (E) as the amount of total pixels of TOMM20 over the amount of nuclei per frame. There are more mitochondria on day 4 of differentiation followed by a larger increase of mitochondria in iPSC-ECs. Mean ± SEM with n = 12 fields of view from three independent experiments, ∗∗p < 0.01, ∗∗∗p < 0.001.
We next assessed PGC-1α, a key regulator of mitochondrial biogenesis, in differentiating hiPSCs. On day 4 of endothelial differentiation, there is a significant increase in PGC1α expression (Fig. 2D). To visualize this increase in mitochondrial mass, we fixed cells at defined stages of differentiation and stained for TOMM20 (Fig. 2, E and F). Interestingly, we found that mitochondrial mass steadily increases after the initial mitophagy phase, suggesting that the de novo mitochondrial biogenesis outpaces mitophagy.
We also examined PGC-1α mRNA levels during cardiomyocyte differentiation and found that the levels also increase following mitophagy (Fig. S2E), concurrent with an increase in the cardiomyocyte markers TNNT2 and RYR2 (Fig. S2F), which suggests that cardiomyocytes increase mitochondrial mass while differentiating into cardiomyocytes.
Mitophagy is required for metabolic reprogramming during differentiation
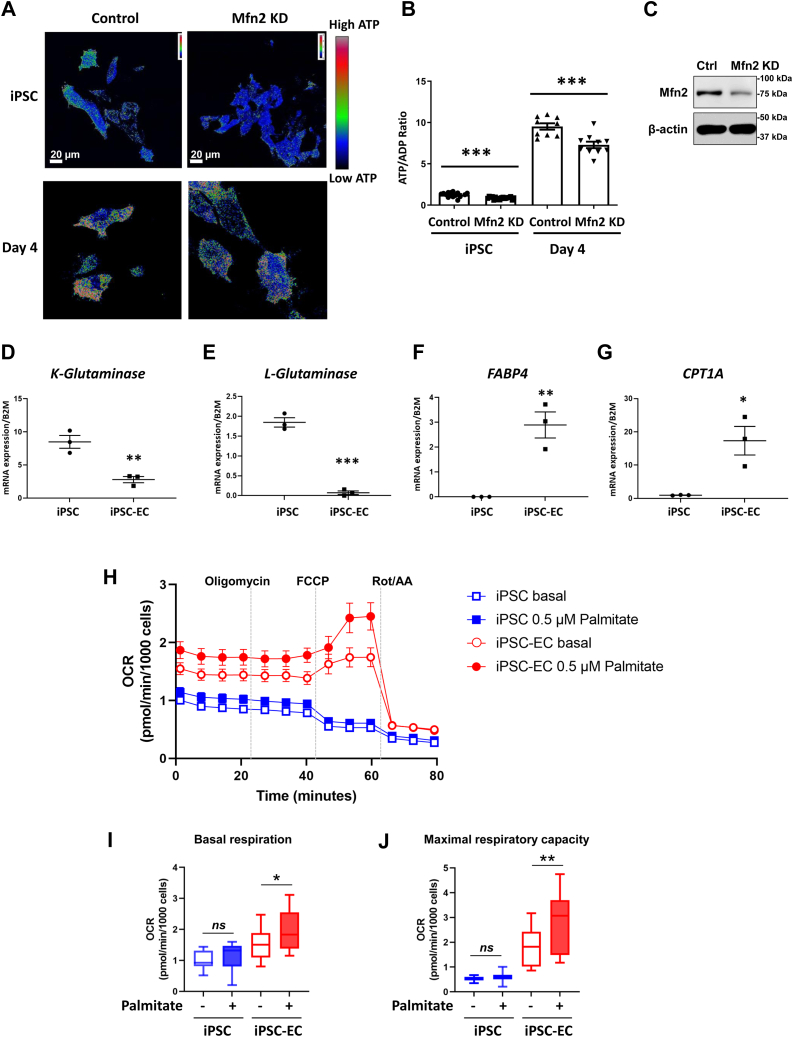
We next examined whether the axis of mitophagy- mitochondrial biogenesis directs a metabolic shift in the differentiation of hiPSCs to iPSC-ECs. We used the fluorescence ATP sensor PercevalHR to quantify the ratio of ATP to ADP (21). Interestingly, we found that ATP production was significantly increased in hiPSCs differentiating to endothelial progenitor (day 4) cells (Fig 3A, left panel and Fig. 3B) concomitant with the increase in mitochondrial biogenesis. This suggests that upon differentiation of hiPSCs to endothelial progenitor cells (day 4), the cells are able to generate greater amounts of ATP as compared with undifferentiated iPSCs. We next tested whether suppressing mitophagy by depletion of Mfn2 (Fig. 3C) would impact the metabolic function of differentiating hiPSCs. hiPSCs were infected with lentiviral doxycycline inducible Mfn2 shRNA construct to prevent mitophagy. The efficacy of this approach to decrease Mfn2-mediated mitophagy was confirmed using mitophagy assessment by Lysotracker and Mitotracker. iPSCs with and without Mitofusin 2 knockdown were treated with the mitophagy inducers oligomycin and antimycin (O/A). We observed a marked attenuation of mitophagy induction in Mfn2 depleted cells (Fig. S3A) indicating that Mfn2 is critical for mitophagy to occur in iPSCs. As shown in Figure 3, A and B, Mfn2-depleted endothelial progenitor cells at day 4 have significantly decreased ATP production compared with control cells. These data suggest that mitophagy regulates metabolic activity by activating the production of ATP.
Figure 3.
Mitophagy is required for metabolic reprogramming in differentiation.A, cells were transduced with the lentivirus PercevalHR, which has an excitation wavelength at 405 nm for ADP, an excitation wavelength at 488 nm for ATP, and one emission wavelength of 529 nm. Cells were also transduced with a doxycycline-inducible Mfn2 shRNA lentivirus. Cells were first treated with 200 ng/ml doxycycline (or DMSO) for 3 days to knockdown Mfn2. They were differentiated for 4 days and then imaged live with confocal microscopy. Ratiometric images created with ImageJ are shown. The calibration bar shows that red has the highest ATP-to-ADP ratio followed by green and then blue. Scale bar = 20 μm. B, quantification of (A) where control is DMSO vehicle treated and Mfn2 KD is doxycycline treated. Day 4 cells have a significantly higher ATP-to-ADP ratio, whereas Mfn2 KD cells have a blunted increase in ATP generation. Mean ± SEM with n = 36 fields of view, representative of three independent experiments with ∗∗∗p < 0.001. C, Western blot image of knockdown of Mitofusin 2 on Day 3 after doxycycline induction in iPSCs. D–G, mRNA levels of (D) K-Glutaminase, (E) L-Glutaminase, (F) Fatty Acid Binding Protein 4 (FABP4), and (G) CPT1A were evaluated in iPSCs and VE-Cadherin sorted cells (iPSC-ECs) by RT-qPCR. B2M (β-2 microglobulin) was used as housekeeping control. Mean ± SEM, n = 3 with ∗∗p < 0.01. H, Seahorse Analyzer assessment depicting mitochondrial OCR (oxygen consumption rate) of iPSC and iPSC-ECs with previous addition of either vehicle (0.1% BSA, basal) or 0.5 μM palmitate. I and J, quantification of the basal respiration and maximal respiration in (H) showing a significant increase of OCR in iPSCs as compared with iPSC-ECs at basal levels and after the addition of palmitate. n = 22 ∗∗∗∗p < 0.0001.
Next, we determined whether the iPSC-ECs changed metabolism as compared with the parental iPSCs. Human stem cells rely on glutamine metabolism for their metabolic needs (4, 22). We therefore measured the mRNA levels of 2 K and L isoforms of glutaminase, which converts glutamine to glutamate using RT-qPCR. The levels of both glutaminases markedly decreased in differentiated ECs (Fig. 3, D and E) indicating a shift away from glutamine metabolism. We also investigated fatty acid oxidation which mature ECs use to provide acetyl-CoA to the TCA cycle (23, 24). In contrast to the glutaminases, Fatty Acid Binding Protein 4 (FABP4), a cytosolic protein that binds fatty acids, is significantly increased in iPSC-ECs but not in hiPSCs (Fig. 3F) We also observed a similar upregulation for CPT1A, an essential mitochondrial enzyme involved in fatty acid beta-oxidation (Fig. 3G). We also tested this metabolic shift in another iPSC line (C2 cells) (Fig. S3, B–D) and found a similar decrease in glutamine metabolism gene expression by showing mRNA levels of K-Glutaminase and L-Glutaminase and corresponding upregulation of the mRNA levels of the fatty acid metabolism regulator FABP4.
Moreover, to assess the metabolic shift in iPSC-ECs, we measured the mitochondrial oxygen consumption rate (OCR) using the Seahorse Analyzer (Fig. 3H). At basal levels, iPSC-ECs had a significantly greater basal respiration and maximal respiratory capacity than iPSCs (Fig. 3, I and J) indicating increased utilization of mitochondrial respiration. There was also an increase in OCR in iPSC-ECs after the addition of the fatty acid palmitate and carnitine (for mitochondrial transport of the palmitate). However, this increase is not present in iPSCs suggesting that mitochondria in iPSC-ECs were able to increasingly utilize fatty acids as a fuel when compared with undifferentiated iPSCs.
The mitochondrial phosphatase PGAM5 activates the Wnt/β-catenin pathway during differentiation to increase mitochondrial mass
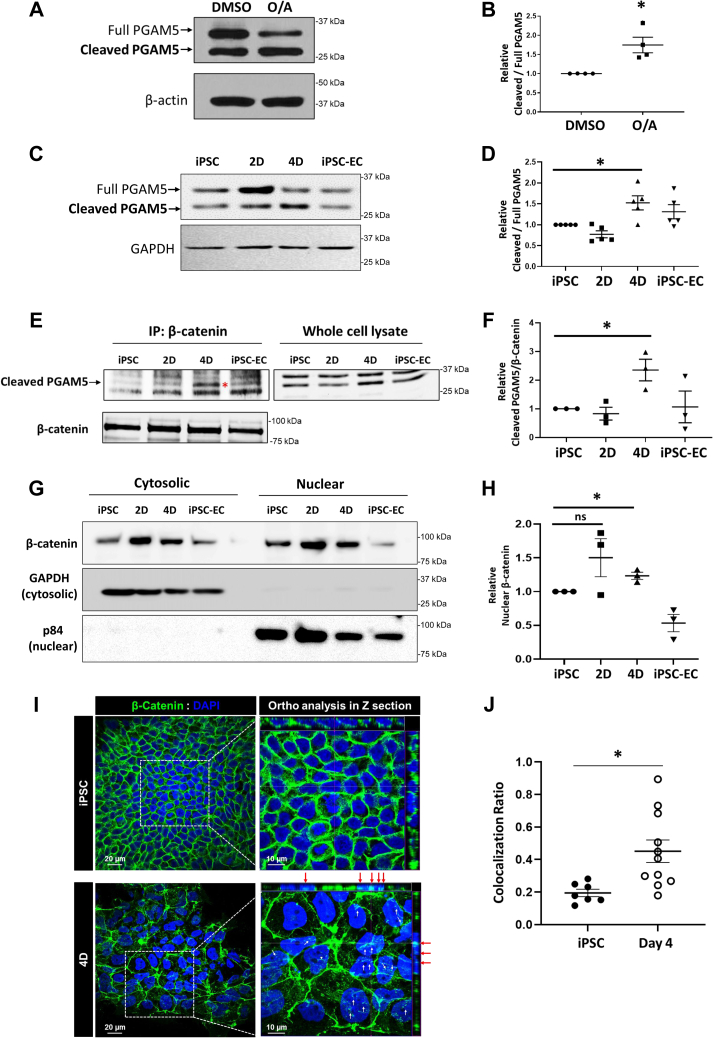
We next investigated the underlying mechanism of how mitophagy could induce the transcriptional programs involved in mitochondrial biogenesis. One way that the mitochondria can communicate with transcriptional programs is through the mitochondrial phosphatase PGAM5. This protein can be cleaved upon mitophagy and dephosphorylates β-catenin, which triggers its translocation to the nucleus to activate Wnt target genes (17). We first examined whether this PGAM5 cleavage occurs in mitophagy induced-iPSC differentiation. iPSCs were treated with the mitophagy inducers oligomycin/antimycin for 3 h. We found that PGAM5 is indeed cleaved as shown through an increase of the bottom (cleaved) band over the top (full) band of PGAM5 (Fig. 4, A and B). We next investigated if PGAM5 is cleaved during differentiation. While mitophagy is increased on days 2 and 4, we found an increase in cleaved PGAM5 only on day 4 (Fig. 4, C and D), which is also the day that mRNA levels of PGC-1α are increased. This suggests that cleaved PGAM5 might be involved with mitochondrial biogenesis by regulating transcription factors.
Figure 4.
The mitochondrial phosphatase PGAM5 activates the Wnt/β-catenin pathway during differentiation.A, iPSCs were treated with 10 μM oligomycin and 5 μM antimycin (or DMSO vehicle) for 3 h to induce mitophagy. Cells were then lysed and run on a SDS-PAGE gel. Membranes were immunoblotted for PGAM5. Full PGAM5 is the upper band and cleaved PGAM5 is the lower band. β-actin was used for loading control. B, quantification of (A), which shows a higher level of cleaved PGAM5 after mitophagy induction. Mean ± SEM, n = 4 with ∗p < 0.05. C, cells were lysed at different stages of differentiation. Western blots are shown of full and cleaved PGAM5. D, quantification of (C), which shows a higher level of cleaved PGAM5 on day 4 of differentiation. Mean ± SEM, n = 5 with ∗p < 0.05. E, cells were lysed at different stages of differentiation and PGAM5 was immunoprecipitated with 1 μg β-catenin specific antibody followed by Western blotting for PGAM5 and β-catenin. F, quantification of (E), which shows a higher level of interaction between cleaved PGAM5 from IP and β-catenin on Day 4 of differentiation. Mean ± SEM, n = 3 with ∗p < 0.05. G, cells were collected at different stages of differentiation and then subjected to a subcellular fractionation. An equal amount of nuclear and cytosolic extracts was loaded onto an SDS-PAGE gel and then probed for β-catenin. p84 and GAPDH were used as nuclear and cytosolic loading controls, respectively. H, quantification of (G), which shows an increased amount of nuclear β-catenin on day 4. Mean ± SEM, n = 3 with ∗p < 0.05. I, immunohistochemistry of cells fixed at days 0 and 4 of differentiation on coverslips and imaged by Z-sectioning with confocal microscopy. β-catenin is shown in green and the nuclei are in blue. Scale bar = 20 μm. A zoomed image of the white box is shown on the right. Scale bar = 10 μm. J, quantification of (I) as the colocalization of nuclei to β-catenin. There is more nuclear β-catenin on day 4 of differentiation. Mean ± SEM, n = 7 fields of view, three independent experiments with ∗p < 0.05.
It is known that PGAM5 interacts with β-catenin under mitochondrial stress in HEK293T and U2OS cells (17). Therefore, we evaluated whether the cleaved PGAM5 binds with β-catenin during differentiation to activate β-catenin transcriptional activity using an immunoprecipitation assay. We found that β-catenin interacted with cleaved PGAM5 on day 4 (Fig. 4, E and F). Next, to evaluate if the stabilized β-catenin through interaction with PGAM5 translocates to the nucleus on day 4, we performed a subcellular fractionation assay of cells at different days of differentiation. Consistent with the increased levels of cleaved PGAM5, the nuclear β-catenin increased on days 2 and 4 (Fig. 4, G and H). To confirm these findings, we also performed immunohistochemistry staining for β-catenin and nuclei to visualize nuclear β-catenin (Fig. 4, I and J). We found an increase in nuclear β-catenin on day 4 as compared with control iPSCs. This further demonstrates increased β-catenin activity on day 4.
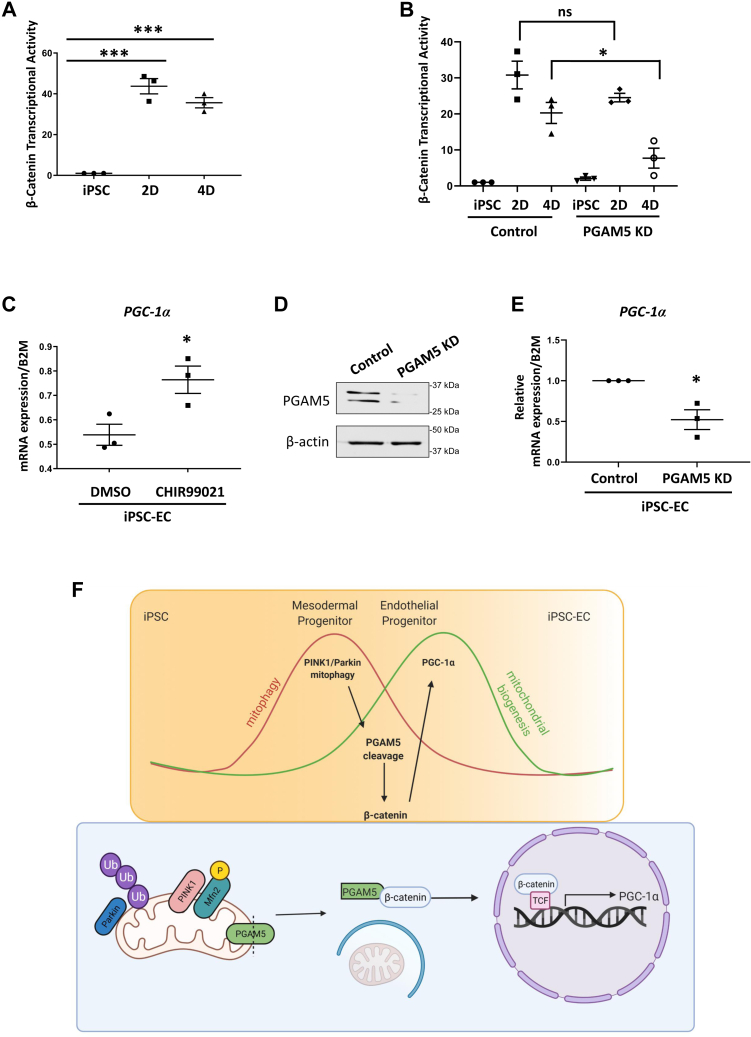
Furthermore, we examined the β-catenin transcriptional activity during differentiation. hiPSCs were transfected with 8xTopFlash (TCF/LEF promoter) reporter luciferase and Renilla (control for transfection) and then differentiated for 2 or 4 days followed by luciferase assay. We found that β-catenin transcriptional activity on days 2 and 4 corresponded with an increase in nuclear β-catenin (Fig. 5A). These data suggest that mitophagy induces PGAM5 cleavage during differentiation and that cleaved PGAM5 dephosphorylates β-catenin, which promotes its translocation into the nucleus to express target genes. As we observed increased β-catenin activity on days 2 and 4, we performed a luciferase assay on cells with PGAM5 knockdown. We found that while transcriptional activity levels on day two did not change, there was a significant decrease in β-catenin activity on day 4 with PGAM5 knockdown (Fig. 5B), which coincided with PGAM5 cleavage.
Figure 5.
PGAM5 induces PGC-1α expression to increase mitochondrial mass.A, cells were transfected with 1 μg Topflash (β-catenin reporter containing the TCF promoter) and 35 ng of pRL/TK (Renilla) as transfection control. Cells were then differentiated for 0, 2, or 4 days. Luciferase activity was determined by the dual luciferase reporter assay system. Transcriptional activity of β-catenin is presented for fold change by normalizing with the value of control iPSCs. Days 2 and 4 show a significant increase of β-catenin activity. Mean ± SEM, n = 3 with ∗∗∗p < 0.001. B, cells were treated with 200 ng/ml doxycycline or DMSO to induce PGAM5 knockdown for 3 days and then transfected with Topflash and Renilla. Cells were then differentiated for 2 or 4 days with or without doxycycline. Luciferase activity was determined with the dual luciferase reagent assay. While there is no significant change on day 2 with PGAM5 knockdown, there is a decrease in β-catenin activity on day 4 indicating that cleaved PGAM5 is regulating β-catenin transcriptional activity. Mean ± SEM with ∗p < 0.05. C, iPSC-ECs were treated with the GSK3 inhibitor and Wnt activator CHIR99021 (or DMSO vehicle) for 24 h. mRNA was extracted and levels of PGC-1α were quantified with qPCR. There is an increase in PGC-1α mRNA levels after Wnt activation. B2M (β2-Microglobulin) was used as housekeeping control. Mean ± SEM, n = 3 with ∗p < 0.05. D, Western blot of PGAM5 in hiPSCs transduced with doxycycline-inducible PGAM5 shRNA lentivirus and treated with 200 ng/ml doxycycline for 3 days. β-actin is used as a loading control. E, cells transduced with PGAM5 shRNA lentivirus were differentiated for 7 days. Control cells were treated with DMSO vehicle while KD cells were given doxycycline. mRNA was extracted and levels of PGC-1α were quantified with RT-qPCR. PGAM5 knockdown cells show a decrease in PGC-1α mRNA levels. B2M was used as housekeeping control. Mean ± SEM, n = 3 with ∗p < 0.05. F, as iPSCs differentiate into iPSC-ECs, there is a burst of mitophagy in mesodermal progenitor cells (day 2) followed by a burst of mitochondrial biogenesis in endothelial progenitor cells (day 4). PINK1/Parkin mitophagy in mesodermal progenitors leads to a cleavage of the phosphatase PGAM5 in endothelial progenitors. PGAM5 dephosphorylates β-catenin, which stabilizes the protein and allows it to translocate to the nucleus, interact with TCF, and transcribe for PGC-1α. This results in an increase in mitochondrial mass that contributes to an alteration of metabolism from glutamine metabolism in iPSCs to fatty acid oxidation in iPSC-ECs.
We next investigated whether β-catenin transcriptional activity promotes mitochondrial biogenesis. iPSC-ECs were treated with CHIR99021 (which activates Wnt/β-catenin signaling by inhibiting GSK3β) for 24 h and then evaluated expressional levels of the key regulator of mitochondrial biogenesis, the transcriptional coactivator PGC-1α. Upon Wnt/β-catenin activation, PGC-1α levels were significantly increased (Fig. 5C). To determine whether PGC-1α expression was dependent on PGAM5-induced β-catenin transcriptional activity, we examined the effect of PGAM5 depletion. hiPSCs were transduced with lentivirus encoding doxycycline inducible PGAM5 shRNA and differentiated for 7 days into iPSC-ECs. Following PGAM5 depletion, differentiated ECs had significantly lower PGC-1α when compared with control cells (Fig. 5, D and E). Taken together, our results suggest that PGAM5 is required for β-catenin transcriptional activity to induce PGC-1α expression, a known inducer for mitochondrial biogenesis during differentiation.
Discussion
The main objective of this study was to understand the role of mitophagy and mitochondrial biogenesis in mediating metabolic reprogramming during iPSC differentiation. We show that (1) mitophagy is increased early on during iPSC differentiation toward the endothelial lineage; (2) mitochondrial biogenesis increases after the initial phase of increased mitophagy during differentiation; (3) during iPSC differentiation, the phosphatase PGAM5 is cleaved during mitophagy and stabilizes β-catenin, thus signaling the initiation of compensatory mitochondrial biogenesis (Fig. 5F).
Traditionally, mitophagy is considered to serve as a quality control mechanism, which eliminates damaged or dysfunctional mitochondria and is the setting of aging or stress (11, 12, 25). Inadequate mitophagy due to genetic mutations in mediators such as Mfn2 or Parkin (26, 27, 28, 29) or due to downregulation of mitophagy mediators can lead to disease because there is inadequate quality control of mitochondria, thus leading to the accumulation of dysfunctional mitochondria, which release deleterious reactive oxygen species and are unable to meet the biosynthetic and bioenergetic needs to the cells (30). However, our findings suggest that mitophagy has an important additional role where it serves as an adaptive mechanism in differentiating cells, thus allowing them to remove mitochondria as part of the differentiation process. With other cell types such as cardiac cells or myoblasts, mitophagy has been found to occur during differentiation (31, 32, 33). In these studies, immature cells were found to upregulate mitochondrial quality control mechanisms and, in some cases, new mitochondria replaced those that were degraded. Importantly, we found that the mitophagy elimination works in tandem with mitochondrial replenishment via de novo mitochondrial biogenesis in endothelial differentiation.
We were able to determine the kinetics of mitochondrial biogenesis using the MitoTimer fluorescent biosensor which labels mitochondria based on age (20). We found that as differentiation progresses to the endothelial progenitor cell stage, mitochondrial biogenesis increases and thus compensates for the preceding loss of mitochondrial mass as a consequence of mitophagy. We also observed an increase in the expression of the transcriptional coactivator PGC-1α, which is known to initiate mitochondrial biogenesis in several cell types (34), and the increase in mitochondrial biogenesis was further confirmed by direct visualization of mitochondrial mass via confocal microscopy.
To interrogate the function of these newly synthesized mitochondria, we also assessed whether mitochondria generated during advanced stages of differentiation were distinct from those during the pluripotent state because differentiation is known to shift metabolic pathways (35). To achieve this, we used the biosensor PercevalHR, which quantifies the ATP:ADP ratio by imaging live cells (21) and found that a depletion of Mitofusin 2 blunted ATP generation in differentiation, thus suggesting that mitophagy is essential in the tuning of metabolic function. Importantly, upon differentiation, cells downregulated glutaminase, which is essential for mitochondrial glutamine metabolism and instead upregulated fatty acid oxidation mediators FABP4 and CPT1A. Our functional assessment of mitochondrial fatty acid oxidation showed increased mitochondrial oxidation of fatty acids in differentiated iPSC-ECs when compared with undifferentiated iPSCs, which rely on glutamine as a mitochondrial carbon source (4, 5). The increase in mitochondrial fatty acid metabolism is especially relevant for endothelial cells, which upregulate fatty acid metabolism mediators such as FABP4 and CPT1 during angiogenesis (36). Our findings raise the intriguing possibility that the metabolic shift during stem cell differentiation is due to the coupling of mitophagy and mitochondrial biogenesis, a form of wholesale recycling of mitochondria from the pluripotent state into de novo synthesized mitochondria, which are better adapted to the needs of the maturing cell.
Mitochondria communicate with the nucleus through mediators such as reactive oxygen species and metabolites (37), but it has been recently reported that mitophagy can signal to the nucleus by releasing the phosphatase PGAM5, which dephosphorylates the transcriptional activator β-catenin in HEK293T and U2OS cells (17). We therefore examined whether PGAM5- β-catenin interaction plays a similarly critical role in mitophagy-induced mitochondrial biogenesis in differentiating iPSCs. We showed that PGAM5 is cleaved during mitophagy in the endothelial progenitor cell stage and that depletion of PGAM5 reduced β-catenin transcriptional activity. PGAM5 depletion also reduced PGC-1α levels, consistent with the notion that PGAM5 serves as a mediator driving mitochondrial biogenesis. β-catenin activity increases prior to the peak mitophagy stage, which could be due to the fact that endothelial differentiation requires addition of the Wnt activator/β-catenin activator CHIR99021 during the first 24 h to induce transition to the mesodermal progenitor stage. Thus, mitophagy-induced PGAM5 release may occur as an endogenous mechanism to sustain β-catenin activity even when the exogenous Wnt/β-catenin activator CHIR99021 is removed.
The recent report that differentiation of skeletal myoblasts also involves mitophagy and mitochondrial biogenesis (32) suggests that this phenomenon may indeed be present in a wider range of differentiation lineages. Therefore, we also examined whether a similar process of mitophagy occurs during iPSC differentiation into cardiomyocytes. We found that there cardiomyocyte differentiation of iPSCs also demonstrates an increase in mitophagy followed by an increase in PGC-1α, a similar tandem coupling of mitophagy and mitochondrial biogenesis that we had seen in iPSC-EC differentiation. However, the increase in mitophagy during cardiomyocyte differentiation was not as great as that seen in iPSC-ECs, suggesting that the kinetics and degree of differentiation-induced mitophagy may vary depending on the cell type and may also depend on the external cues that could differ for distinct cell types. Cardiomyocytes generally have more mitochondria than ECs, which we validated through their large augmentation of mitochondrial biogenesis.
Our study focused on the downstream effects of mitophagy during iPSC differentiation such as the initiation of compensatory mitochondrial biogenesis via the PGAM5 pathway, but one limitation of our work is that the upstream initiators of mitophagy during cell differentiation were not identified. Potential initiators of mitophagy could include growth factors that induce differentiation, but they could also involve additional environmental cues such as matrix signaling, cell–cell cross talk, or mechano-signaling. These cues could be cell type specific and may also in part explain differences in the kinetics and extent of differentiation-induced mitophagy across cell types. Future studies will be needed to address how such differentiation cues initiate mitophagy signaling.
In conclusion, our findings suggest that during endothelial differentiation of iPSCs, mitophagy coupled with compensatory mitochondrial biogenesis serves as a mechanism for metabolic reprogramming that is mediated via β-catenin signaling to the nucleus. Leveraging this signaling from the mitochondria undergoing mitophagy to the nucleus could help generate mature, differentiated cells ideally suited for tissue regeneration.
Experimental procedures
Reagents
Oligomycin A (75351), FCCP (C2920) Antimycin A (A8674), Rotenone (R8875) Sodium Palmitate (P9767), and L-carnitine hydrochloride (C0283) were obtained from Sigma.
Cell culture
Human iPSCs were reprogrammed from 273 human fibroblasts by Dr Glenn Marsboom at the University of Illinois at Chicago (38). Cells were cultured on hESC-qualified Matrigel (Corning #354277) in mTeSR1 media (Stem Cell Technologies 85850) and kept in a 37 °C incubator with 5% CO2. C2 iPSC cells (19) were cultured in E8 Medium (Fisher A1517001). Colonies were routinely split using 1 U/ml Dispase (Stem Cell Technologies 7923) and 5 μM Y-27632 (Stem Cell Technologies 72304).
iPSC endothelial differentiation
To induce differentiation, iPSCs were split using Accumax (Stem Cell Technologies 7921) and plated at 10% confluency with 5 μM Y-27632 for 24 h on Matrigel. iPSCs were then differentiated for 7 days on Matrigel using serum-free media: 75% IMDM (Gibco 12440053), 24% F12 media (Gibco 11765054), 0.5% N2 (Gibco 12587), 1% B27 (Gibco 17502), and 0.05% BSA-lipid rich (Gibco 11021029). On day 1, 5 μM CHIR99021 (Sigma SML1046) and 20 ng/ml BMP4 (PeproTech 120-05ET) were added. On day 2, 50 ng/ml bFGF (PeproTech AF-100-18B), 50 ng/ml Activin A (PeproTech 120-14P), and 20 ng/ml BMP4 were added. On day 3, cells were split 1:2 on Matrigel to reduce confluency. On day 4, 50 ng/ml VEGF (PeproTech 100-20), 10 μM SB431542 (Sigma 616464), and 20 ng/ml BMP4 were added. Media was changed every day. On day 7, cells were dissociated with 0.05% trypsin and sorted using CD144 (VE-Cadherin) MicroBeads (Miltenyi Biotec 130-097-857) using MACS magnetic sorting. CD144+ cells were plated on 0.2% gelatin culture dishes in EGM2-MV media (Lonza CC-3202) + 15% FBS (Hyclone).
Cardiomyocyte differentiation
iPSC cardiomyocytes are differentiated according to a previous reported method (19). Briefly, iPSCs cultured in E8 Medium (Fisher A1517001) were split onto Matrigel coated dishes using ReLeSR (Stem Cell Technologies 100-0484). When the cells reach 80% confluency, media is switched to RPMI 1640 (Fisher 11875093) with B27 minus insulin supplement (Fisher A18956-01). On the day media is switched (day 0), 8 μM CHIR 99021 is added for 2 days. On day 2, media is replaced with RPMI 1640 with B27 minus insulin supplement. On day 3, 5 μM IWR-1 (Sigma I0161) is added for 2 days. On day 5, media is replaced with B27 minus insulin. On day 7, media is replaced with RPMI 1640 with B27 + insulin (Fisher A18956-01). By day 8, we see rhythmic beating of differentiated cardiomyocytes. On day 10, glucose starvation begins when media is replaced with RPMI 1640 No Glucose Medium (Fisher 11879020) with B27 + insulin. Media is changed every other day until day 15 when fully differentiated cardiomyocytes are cultured in RPMI 1640 + B27 + insulin.
Generation of shRNA lines
Lentivirus shRNA constructs were generated in our laboratory. shRNA oligos were inserted into a lentiviral Tet-pLKO-puro backbone vector (Addgene #21915). An shRNA construct was generated for Mfn2 (5′-CCG GGC TCA GTG CTT CAT CCC ATT TCT CGA GAA ATG GGA TGA AGC ACT GAG CTT TTT -3′, 3′- AATTAA AAA GCT CAG TGC TTC ATC CCA TTT CTC GAG AAA TGG GAT GAA GCA CTG AGC -5′) and an shRNA construct was generated for PGAM5 (5′-CCG GGG CGA GAA CCA CTG TCT CTG ACT CGA GTC AGA GAC AGT GGT TCT CGC CTT TTT-3′, 3′ AAT TAA AAA GGC GAG AAC CAC TGT CTC TGA CTC GAG TCA GAG ACA GTG GTT CTC GCC-5′). hiPSCs were transduced with these shRNA viruses and then selected with 0.5 μg/ml puromycin for at least 4 days. For optimal knockdown of protein, cells were treated with 200 ng/ml doxycycline for 3 days before and throughout differentiation.
Lentivirus generation
HEK293T cells in DMEM +Pen/Strep +10% FBS were cultured to generate lentiviruses at 37 °C with 5% CO2. Each lentiviral plasmid + psPAX2 (Addgene #12260) and VSV-G (Addgene # 8454) were transfected using jetPRIME (Polypus) and media was changed the next day. After 2 days, the supernatant was concentrated with Lenti-X Concentrator (Takara 631231).
Immunofluorescence
Cells were fixed on glass coverslips (EMS 72229-01) with 4% paraformaldehyde for 10 min and permeabilized with 1:400 (0.25%) Triton-X for 10 min. The coverslips were washed with PBS with 0.05% Tween-20 and incubated with primary antibody 1:300 TOMM20 (Santa Cruz sc-11415), 1:300 VE-Cadherin (Cayman 160840), or 1:500 β-catenin (Abcam 32572) in 2% BSA overnight at 4 °C. The next day, the coverslips were incubated with Alexa Flour- 488 or -568 (Thermo Fisher A11034) and 1:2000 DAPI was added. Cells were imaged on a Zeiss LSM 880 microscope. For TOMM20 staining, the area of pixels was quantified with ImageJ FIJI 1.52d (Java 1.8.0_172, 64 bit). For β-catenin and DAPI colocalization, the ImageJ plugin JACoP was used with M1 and M2 coefficients. The amount of DAPI that colocalized with β-catenin was measured to account for increases in overall β-catenin levels and to analyze the amount of nuclear β-catenin. Three independent experiments were performed with 12 fields of view (multiple cells per field of view) quantified per experiment.
Mitophagy using MitoTracker and Lysotracker
hiPSCs, day 2 and 4 differentiated cells, and iPSC-ECs were plated onto glass bottom dishes (MatTek P35GC-1.5-14-C) using Accumax or Trypsin. Cells were stained with 75 nM Lysotracker Red DND-99 (Thermo Fisher L7528) for 1 h and 0.5 mM MitoTracker Deep Red FM (Thermo Fisher M22426) for 30 min in their media. Cells were washed with Ca+ Mg+ PBS and then imaged on a confocal Zeiss LSM 710 at 37 °C with 5% CO2 in Hank's Balanced Salt Solution (Gibco 14025092). Quantification of colocalization of mitochondria and lysosomes was generated using ImageJ FIJI with the JACoP plugin using M1 and M2 coefficients after thresholding images. The same method was used with iPSC-CMs during differentiation. Three independent experiments were performed with five fields of view per experiment quantified.
Capillary-like tube formation assay
Growth factor reduced Matrigel (Corning 354230) was coated onto 48 well plates for 1 h at 37 °C. The iPSCs and iPSC-ECs (50,000 cells) were plated onto the Matrigel in EGM2-MV media at 37 °C with 5% CO2. Four hours later, brightfield images were obtained using an ECHO Revolve microscope (10× objective) to visualize the network formation of capillary-like tubes.
Flow cytometry
iPSC-ECs were cultured in EGM2-MV media for three passages. Cells were dissociated with Trypsin and were resuspended in Antibody Binding Buffer (PBS + 2% BSA). For 30 min on ice, cells were incubated with 1:100 VE-Cadherin FITC (BD Biosciences 560874), 1:100 CD31-PE (R&D FAB3567P), and 1:100 DAPI. FITC-mouse IgG1 (BD Biosciences 555748) and PE-mouse IgG1 (R&D IC002P) isotype controls were also used. Data presented are VE-Cadherin+, CD31+, and DAPI− to exclude dead cells. Samples were run on a Gallios flow cytometer (Beckman Coulter) and analyzed by Kaluza software (Beckman Coulter).
Mitochondrial biogenesis using MitoTimer
hiPSCs were transduced with MitoTimer (Addgene #50547) and rtTA (VectorBuilder Tet3G/Hygromycin) plasmids together. Cells were either kept in hiPSC media or differentiated for 4 days. On day 2 of differentiation, cells were treated with 2 μg/ml doxycycline for 24 h to turn all mitochondria red. On day 4 of differentiation, cells were treated with 2 μg/ml doxycycline for 24 h. Eight hours later, cells were fixed with 4% PFA, stained with DAPI, and mounted onto glass slides. Fixed cells were imaged on a LSM 880 Zeiss microscope. Three independent experiments were performed with 12 fields of view quantified.
ATP imaging
hiPSCs were transduced with FUGW-PercevalHR (Addgene #49083). hiPSCs and 4-day differentiated cells with (control) or without shMfn2 (Mfn2 KD) were plated on glass bottom dishes using Accumax or trypsin. Cell media was changed to Hank's Balanced Salt Solution (Gibco 14025092) 1 h before imaging. Cells were imaged on a Zeiss LSM 710 at 37 °C with 5% CO2. The wavelengths 405 and 488 nm were used to image ADP and ATP, respectively. Ratiometric images and quantitative analysis were generated with ImageJ 1.52d (Java 1.8.0_172, 64 bit). Three independent experiments were performed with 3–5 fields of view (multiple cells) per condition per experiment quantified.
Mitochondria oxygen consumption rate by Seahorse XFe96 analyzer
iPSCs and iPSC-ECs were plated on a Seahorse XFe96 96 well plate on 1 % Matrigel or 0.2% gelatin, respectively, 1 day before measurement. Directly before analysis, either 0.1% BSA or 0.5 μM palmitate was added to the media. OCR was measured using the Seahorse XF Cell Mito Stress Kit (103015-100). The OCR values were normalized by cell number.
Western blotting
Cells were lysed in RIPA buffer with 1% Triton-X and protease/phosphatase inhibitors (Millipore 539134/524625). Protein was quantified with Bradford Dye (Bio-Rad 500-0006), and 20 μg was loaded onto a SDS-polyacrylamide gel (SDS-PAGE) with SDS/Tris/Glycine running buffer. The antibodies used for blotting were: 1:1000 PGAM5 (Abcam ab126534), 1:2000 β-actin-HRP (Santa Cruz sc-47778), 1:500 β-catenin (Santa Cruz sc-7963), 1:1000 Mitofusin 2 (Abcam ab56889), 1:1000 Phospho-Mfn2 (Millipore ABC963), 1:1000 PINK1 (Abcam ab23707), 1:1000 THOC1/p84 (Santa Cruz sc-514123), and 1:1000 GAPDH (Proteintech 60004-I-AP). Membranes were incubated in primary antibody in 0.1% TBST with 5% milk at 4 °C overnight. Membranes were incubated in 1:2000 secondary antibody in 0.1% TBST for 1 h and then imaged on an iBright CL1500 machine (Thermo Fisher). Chemiluminescence intensity was quantified in ImageJ 1.52d (Java 1.8.0_172, 64 bit) with 3–5 blots assessed for each experiment.
Immunoprecipitation
Cells were lysed in lysis buffer (50 mM HEPES pH 7.5, 120 mM NaCl, 5 mM EDTA, 10 mM Na pyrophosphate, 50 mM NaF, 1 mM Na3VO4, 1% Triton X-100) and protease/phosphatase inhibitors. The supernatant was quantified with Bradford Dye and 500 μg of protein was incubated with 1 μg β-catenin antibody (Santa Cruz sc-7963) and kept on a rotator at 4 °C overnight. The sample was incubated with Protein A/G PLUS-Agarose beads (Santa Cruz sc-2003) for 2 h at 4 °C. Samples were then washed and loaded onto a 12% SDS–polyacrylamide gel. Three blots were quantified using ImageJ 1.52d (Java 1.8.0_172, 64 bit).
Subcellular fractionation
Cells were collected at days 0, 2, 4, and 7 (sorted) of differentiation. The subcellular fractionation was performed with a previously reported method (39). Briefly, the cells were lysed with lysis buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, and EDTA free protease inhibitor cocktail). The lysate was added to 10% NP-40 and then vortexed and centrifuged for 13,000 rpm for 1 min at 4 °C. The pellet was washed with lysis buffer and then nuclear extract buffer (25 mM HEPES, pH 7.9, 0.4 M NaCl, 0.5 mM EDTA, 0.5 mM EGTA, and EDTA-free protease inhibitor cocktail) was added. The nuclear fraction was obtained by centrifugation for 5 min at 13,000 rpm. The same amount of cytosol and nuclear fraction was loaded in an SDS-PAGE gel followed by Western blotting. GAPDH and p84 were used as controls for cytosolic or nuclear fraction, respectively. Three blots were quantified using ImageJ 1.52d (Java 1.8.0_172, 64 bit).
Luciferase assay
hiPSCs were transfected with a β-catenin reporter (M50 super 8× TOPflash containing TCF/LEF sites upstream of a luciferase reporter) and pRL/TK (Renilla) reporter, with Lipofectamine 3000 (Thermo Fisher L3000015). Firefly and Renilla luciferase activity was determined by the dual luciferase reporter assay system Promega. The relative luciferase activity represents as the mean value of the Firefly/Renilla luciferase. Three independent experiments were performed.
Quantitative real-time PCR
Total RNA was isolated using Trizol (Invitrogen). Reverse transcription was done with high-capacity cDNA reverse transcription kit (Applied Biosystems 4368814). qPCR was run with fast start universal SYBR green master (ROX) PCR Kit (Roche 04913914001) using QuantStudio7 (Thermo Fisher). Samples were run in triplicate for 3–4 independent experiments.
RT-qPCR human primer sequences
| Gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| CD31 | AGGGGCCACGATGTGGCTTG | TTGCACCGTCCAGTCCGGCA |
| CPT1A | TCCAGTTGGCTTATCGTGGTG | CTAACGAGGGGTCGATCTTGG |
| FABP4 | ACTGGGCCAGGAATTTGACG | CTCGTGGAAGTGACGCCTT |
| K-Glutaminase | TGTGATTCCTGACTTTATGTCTTTTACCT | GACACACCCCACAAATCGG |
| L-Glutaminase | TTCAATTTCCACAACTATGACAACCT | ACAGTCTTGTTCCGAATTTCTGC |
| PGC-1α | ACTTACAAGCCAAACCAACAACTTTAT | CTTTATGAGGAGGAGTGGTGGGT |
| RYR2 | CTGAGGAAGACCACCTGAAAG | AAGAGAGGGTAGAAGGCATAGA |
| TNNT2 | TTCGACCTGCAGGAGAAGTT | GCGGGTCTTGGAGACTTTCT |
| VE-Cadherin | AAGTGTGTGAGAACGCTGTCCAT | ACGGGTAGGAAGTGGACCTTG |
| VEGFR2 | ATAGAAGGTGCCCAGGAAAAG | GTCTTCAGTTCCCCTCCATTG |
Statistical analysis
Colocalization microscopy analysis was performed using JACoP plugin in ImageJ 1.52d (Java 1.8.0_172, 64 bit) with Manders' coefficient. Band intensity analysis of Western blots and immunoprecipitations was performed with ImageJ 1.52d (Java 1.8.0_172, 64 bit). Data is presented as mean ± SEM with significance levels expressed as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. The Student's t test or ANOVA was used for analysis of statistical significance using GraphPad Prism 9 software.
Data availability
Data described in the article is provided with Source data.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Schematics were created with BioRender.com.
Author contributions
S. K., G. M., and J. R. conceptualization; S. K. data curation; S. K., Y.-M. K., and P. T. T. formal analysis; S. K. and J. R. funding acquisition; S. K., Y.-M. K., S. S., J. W. L., and J. R. investigation; S. K., Y.-M. K., S. S., J. W. L., P. T. T., and G. M. methodology; Y.-M. K. and J. R. project administration; Y.-M. K., G. M., and J. R. supervision; P. T. T. visualization; S. K. and J. R. writing–original draft; S. K., Y.-M. K., S. S., J. W. L., and J. R. writing–review and editing.
Funding and additional information
The studies were supported in part by NIH grants P01-HL60678 (J. R.), T32-HL007829 (to S. K. and J. W. L.), T32-HL139439 (S. K.), R01-HL126516 (to J. R.), and AHA CDA grant 19CDA34680000 (to Y.-M. K.). We were assisted by the Fluorescence Imaging Core at the Research Resources Center of the University of Illinois at Chicago. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by John Denu
Contributor Information
Young-Mee Kim, Email: youngmee@uic.edu.
Jalees Rehman, Email: jalees@uic.edu.
Supporting information
References
- 1.Varum S., Momcilovic O., Castro C., Ben-Yehudah A., Ramalho-Santos J., Navara C.S. Enhancement of human embryonic stem cell pluripotency through inhibition of the mitochondrial respiratory chain. Stem Cell Res. 2009;3:142–156. doi: 10.1016/j.scr.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Folmes C.D., Nelson T.J., Martinez-Fernandez A., Arrell D.K., Lindor J.Z., Dzeja P.P., Ikeda Y., Perez-Terzic C., Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panopoulos A.D., Yanes O., Ruiz S., Kida Y.S., Diep D., Tautenhahn R., Herrerias A., Batchelder E.M., Plongthongkum N., Lutz M., Berggren W.T., Zhang K., Evans R.M., Siuzdak G., Izpisua Belmonte J.C. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsboom G., Zhang G.F., Pohl-Avila N., Zhang Y., Yuan Y., Kang H., Hao B., Brunengraber H., Malik A.B., Rehman J. Glutamine metabolism regulates the pluripotency transcription factor OCT4. Cell Rep. 2016;16:323–332. doi: 10.1016/j.celrep.2016.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palm W., Thompson C.B. Nutrient acquisition strategies of mammalian cells. Nature. 2017;546:234–242. doi: 10.1038/nature22379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathieu J., Ruohola-Baker H. Metabolic remodeling during the loss and acquisition of pluripotency. Development. 2017;144:541–551. doi: 10.1242/dev.128389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung S., Arrell D.K., Faustino R.S., Terzic A., Dzeja P.P. Glycolytic network restructuring integral to the energetics of embryonic stem cell cardiac differentiation. J. Mol. Cell Cardiol. 2010;48:725–734. doi: 10.1016/j.yjmcc.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteban-Martinez L., Boya P. BNIP3L/NIX-dependent mitophagy regulates cell differentiation via metabolic reprogramming. Autophagy. 2018;14:915–917. doi: 10.1080/15548627.2017.1332567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng X., Boyer L., Jin M., Mertens J., Kim Y., Ma L., Ma L., Hamm M., Gage F.H., Hunter T. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife. 2016;5 doi: 10.7554/eLife.13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esteban-Martinez L., Sierra-Filardi E., McGreal R.S., Salazar-Roa M., Marino G., Seco E., Durand S., Enot D., Grana O., Malumbres M., Cvekl A., Cuervo A.M., Kroemer G., Boya P. Programmed mitophagy is essential for the glycolytic switch during cell differentiation. EMBO J. 2017;36:1688–1706. doi: 10.15252/embj.201695916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng M.Y.W., Wai T., Simonsen A. Quality control of the mitochondrion. Dev. Cell. 2021;56:881–905. doi: 10.1016/j.devcel.2021.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Palikaras K., Lionaki E., Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 2018;20:1013–1022. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 13.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., Magrane J., Moore D.J., Dawson V.L., Grailhe R., Dawson T.M., et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U. S. A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y., Dorn G.W., 2nd. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., Tanaka K. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jedrzejas M.J. Structure, function, and evolution of phosphoglycerate mutases: Comparison with fructose-2,6-bisphosphatase, acid phosphatase, and alkaline phosphatase. Prog. Biophys. Mol. Biol. 2000;73:263–287. doi: 10.1016/s0079-6107(00)00007-9. [DOI] [PubMed] [Google Scholar]
- 17.Bernkopf D.B., Jalal K., Bruckner M., Knaup K.X., Gentzel M., Schambony A., Behrens J. Pgam5 released from damaged mitochondria induces mitochondrial biogenesis via Wnt signaling. J. Cell Biol. 2018;217:1383–1394. doi: 10.1083/jcb.201708191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sekine S., Kanamaru Y., Koike M., Nishihara A., Okada M., Kinoshita H., Kamiyama M., Maruyama J., Uchiyama Y., Ishihara N., Takeda K., Ichijo H. Rhomboid protease PARL Mediates the mitochondrial membrane potential loss-induced cleavage of PGAM5. J. Biol. Chem. 2012;287:34635–34645. doi: 10.1074/jbc.M112.357509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon Y., Nukala S.B., Srivastava S., Miyamoto H., Ismail N.I., Jousma J., Rehman J., Ong S.B., Lee W.H., Ong S.G. Detection of viral RNA fragments in human iPSC cardiomyocytes following treatment with extracellular vesicles from SARS-CoV-2 coding sequence overexpressing lung epithelial cells. Stem Cell Res. Ther. 2020;11:514. doi: 10.1186/s13287-020-02033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernandez G., Thornton C., Stotland A., Lui D., Sin J., Ramil J., Magee N., Andres A., Quarato G., Carreira R.S., Sayen M.R., Wolkowicz R., Gottlieb R.A. MitoTimer: A novel tool for monitoring mitochondrial turnover. Autophagy. 2013;9:1852–1861. doi: 10.4161/auto.26501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tantama M., Martinez-Francois J.R., Mongeon R., Yellen G. Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat. Commun. 2013;4:2550. doi: 10.1038/ncomms3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carey B.W., Finley L.W., Cross J.R., Allis C.D., Thompson C.B. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoors S., Bruning U., Missiaen R., Queiroz K.C., Borgers G., Elia I., Zecchin A., Cantelmo A.R., Christen S., Goveia J., Heggermont W., Godde L., Vinckier S., Van Veldhoven P.P., Eelen G., et al. Fatty acid carbon is essential for dNTP synthesis in endothelial cells. Nature. 2015;520:192–197. doi: 10.1038/nature14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falkenberg K.D., Rohlenova K., Luo Y., Carmeliet P. The metabolic engine of endothelial cells. Nat. Metab. 2019;1:937–946. doi: 10.1038/s42255-019-0117-9. [DOI] [PubMed] [Google Scholar]
- 25.Lahiri V., Hawkins W.D., Klionsky D.J. Watch what you (self-) eat: Autophagic mechanisms that modulate metabolism. Cell Metab. 2019;29:803–826. doi: 10.1016/j.cmet.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y.J., Cao Y.L., Feng J.X., Qi Y., Meng S., Yang J.F., Zhong Y.T., Kang S., Chen X., Lan L., Luo L., Yu B., Chen S., Chan D.C., Hu J., et al. Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset. Nat. Commun. 2019;10:4914. doi: 10.1038/s41467-019-12912-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gladkova C., Maslen S.L., Skehel J.M., Komander D. Mechanism of parkin activation by PINK1. Nature. 2018;559:410–414. doi: 10.1038/s41586-018-0224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schrepfer E., Scorrano L. Mitofusins, from mitochondria to metabolism. Mol. Cell. 2016;61:683–694. doi: 10.1016/j.molcel.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 29.Rocha A.G., Franco A., Krezel A.M., Rumsey J.M., Alberti J.M., Knight W.C., Biris N., Zacharioudakis E., Janetka J.W., Baloh R.H., Kitsis R.N., Mochly-Rosen D., Townsend R.R., Gavathiotis E., Dorn G.W., 2nd. MFN2 agonists reverse mitochondrial defects in preclinical models of Charcot-Marie-Tooth disease type 2A. Science. 2018;360:336–341. doi: 10.1126/science.aao1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schofield J.H., Schafer Z.T. Mitochondrial reactive oxygen species and mitophagy: A complex and nuanced relationship. Antioxid. Redox Signal. 2021;34:517–530. doi: 10.1089/ars.2020.8058. [DOI] [PubMed] [Google Scholar]
- 31.Gong G., Song M., Csordas G., Kelly D.P., Matkovich S.J., Dorn G.W., 2nd. Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science. 2015;350:aad2459. doi: 10.1126/science.aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sin J., Andres A.M., Taylor D.J., Weston T., Hiraumi Y., Stotland A., Kim B.J., Huang C., Doran K.S., Gottlieb R.A. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy. 2016;12:369–380. doi: 10.1080/15548627.2015.1115172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lampert M.A., Orogo A.M., Najor R.H., Hammerling B.C., Leon L.J., Wang B.J., Kim T., Sussman M.A., Gustafsson A.B. BNIP3L/NIX and FUNDC1-mediated mitophagy is required for mitochondrial network remodeling during cardiac progenitor cell differentiation. Autophagy. 2019;15:1182–1198. doi: 10.1080/15548627.2019.1580095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gureev A.P., Shaforostova E.A., Popov V.N. Regulation of mitochondrial biogenesis as a way for active longevity: Interaction between the Nrf2 and PGC-1alpha signaling pathways. Front. Genet. 2019;10:435. doi: 10.3389/fgene.2019.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakrabarty R.P., Chandel N.S. Mitochondria as signaling organelles control mammalian stem cell fate. Cell Stem Cell. 2021;28:394–408. doi: 10.1016/j.stem.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X., Kumar A., Carmeliet P. Metabolic pathways fueling the endothelial cell drive. Annu. Rev. Physiol. 2019;81:483–503. doi: 10.1146/annurev-physiol-020518-114731. [DOI] [PubMed] [Google Scholar]
- 37.Mottis A., Herzig S., Auwerx J. Mitocellular communication: Shaping health and disease. Science. 2019;366:827–832. doi: 10.1126/science.aax3768. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L., Jambusaria A., Hong Z., Marsboom G., Toth P.T., Herbert B.S., Malik A.B., Rehman J. SOX17 regulates conversion of human fibroblasts into endothelial cells and erythroblasts by dedifferentiation into CD34(+) progenitor cells. Circulation. 2017;135:2505–2523. doi: 10.1161/CIRCULATIONAHA.116.025722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim Y.M., Krantz S., Jambusaria A., Toth P.T., Moon H.G., Gunarathna I., Park G.Y., Rehman J. Mitofusin-2 stabilizes adherens junctions and suppresses endothelial inflammation via modulation of beta-catenin signaling. Nat. Commun. 2021;12:2736. doi: 10.1038/s41467-021-23047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data described in the article is provided with Source data.