Abstract
Objective: To investigate the specific roles of linc00662 and miR-199a-5p in bladder cancer (BC). Methods: A total of 104 cases of BC tissues and 52 cases of normal para-cancerous tissues were included to detect the expression of linc00662 and miR-199-5p by real-time quantitative PCR. The expression of linc00662 and miR-199a-5p in BC cells T24 was regulated to observe the changes in apoptosis, proliferation, adhesion, invasion, and migration. The nude mice bearing a BC cell transplanted xenograft was constructed, and the expression of linc00662 in rats was regulated. Tumor size and quality were observed within 24 days. The relationship between linc00662 and patients’ survival was observed. The targeting relationship between linc00662 and miR-199a-5p was verified by dual luciferase reporter gene assay. Results: Linc00662 was enhanced and miR-199a-5p was decreased in BC patients. Linc00662 targeted and negatively regulated the expression of miR-199a-5p. Down-regulation of linc00662 could reduce proliferation, migration, invasion, and adhesion activities of BC cells, but enhance the apoptosis. Down-regulation of miR-199a-5p counteracted the cell biological changes caused by linc00662. Down-regulating linc00662 cinduced the expression of miR-199a-5p in BC and suppressed tumor growth. Conclusion: Linc00662 plays an oncogenic role in BC by sponging miR-199a-5p.
Keywords: Bladder cancer, linc00662, miR-199a-5p, oncogene
Introduction
BC (bladder cancer) is the tenth most pervasive cancer in the world and the second most pervasive in the urinary tract [1]. According to statistics, approximately 550,000 patients were diagnosed with BC in 2018 [2]. Worldwide, the incidence of BC in men is four times higher than that in women [3]. BC is a heterogeneous disease, challenging in diagnosis, therapy, and prognosis [4]. Non-coding RNAs have attracted more and more attention in tumor research because of their regulatory role and evaluation value. MicroRNA (miRNA) and long-chain non-coding RNA (lncRNA) are the main types of non-coding RNA.
miRNA, a non-coding RNA, can participate in the operation of cell biologic functions by regulating target genes, while abnormally expressed miRNA may participate in the formation, growth, metastasis, and invasion of malignancy as oncogenes or tumor suppressor genes [5]. Abnormally expressed miRNAs are widely found in BC tissues, urine and blood samples, and they are closely related to the pathologic characteristics and chemical sensitivity of BC [6]. miR-199a-5p is a mature splice variant of miR-199a, that plays a role as gene epigenetic regulator in a variety of diseases. Chen et al. [7] believe that the imbalance of miR-199a-5p in TNBC (triple negative breast cancer) is involved in regulating epithelial-mesenchymal transition and cell stemness. Wang et al. [8] have revealed that miR-199a-5p suppresses the excessive proliferation of hemangioma cells by down-regulating the HIF1A gene. miR-199a-5p may be an inhibitor in the progression of BC. Zhou et al. [9] have proposed that the down-regulation of miR-199a-5p causes malignant metastasis of BC cells. In addition, miR-199a-5p may inhibit the formation and growth of malignant tumors through the MLK3/NF-κB pathway [10]. Although studies have revealed the role of miR-199a-5p in BC, its value in BC is still uncertain.
LncRNA can bind to the target gene through 3’-UTR, thus regulating the target gene. The downstream target gene of lncRNA can be either miRNA or mRNA. Generally, the targeted regulation between lncRNA and miRNA is referred as sponging. LncRNA is involved in chromatin remodeling, transcription process and post-transcriptional regulation. LncRNA is involved in chromatin remodeling, transcription process, and post-transcriptional regulation, and plays an indispensable role in carcinoma [11]. Linc00662 is an important member of the long-chain non-coding RNA family, that plays a vital role in the development and progression of liver carcinoma, gastric carcinoma, and prostate carcinoma [12-14]. Recent studies have shown that linc00662 is highly expressed in BC and has a ceRNA (competing endogenous RNAs) relationship with miR-199-5p [15]. It is unclear whether linc00662 can regulate miR-199-5p in BC.
In this study, 104 cases of BC tissues and 52 cases of normal para-cancerous tissues were included, and the expression of linc00662 was detected by real-time quantitative PCR. The results showed that linc00662 was abnormally up-regulated in BC tissues, and the 3-year survival rate of patients with high expression of linc00662 was obviously worse than that of patients with low expression of linc00662. This suggests that linc00662 may be related to the development and progression of BC. At the same time, it was found that miR-199a-5p might be a downstream target gene of linc00662 through the bioinformatics tool. Therefore, this study was designed to investigate the specific role of linc00662 and miR-199a-5p in the progression of BC.
Methods
Inclusion and exclusion criteria of BC
A total of 104 cases of BC (bladder cancer) tissues and 52 cases of normal para-cancerous tissues were collected.
Inclusion criteria: The patient was diagnosed with BC according to frozen section or pathological examination.
Exclusion criteria: The patient was accompanied by tumors other than BC. The patient had a history of radiotherapy and chemotherapy or surgical resection of BC. The patient also had other chronic bladder diseases.
In the study, all participants provided their informed consent, and this research was ratified by the hospital ethics committee. Tissue samples were sliced and stored at -80°C for testing. In this research, all patients were followed up by telephone and electronic pathology review in the outpatient department. They were followed up every 3 months in the first year and every 4 months in the second and third years.
Cell culture and transfection
Human BC cell strains 5637 and T24 were obtained from American Type Culture Collection (ATCC) and cultivated in Dulbecco’s modified eagle medium (DMEM) medium comprising 10% fetal bovine serum and 1% streptomycin-penicillin. This was cultured in an incubator at 37°C and 5% CO2 until it grew well. Cell transfection: The cells were inoculated into 6-hole plates at 1 × 105 cells/hole. Cell strains were transfected with Lipofectamine 3000 transfection kit (Invitrogen, USA). The procedure followed the kit specifications. After transfection for 8 h, the fresh medium was replaced. The NC Small interfering RNA (siRNA, 5’-CCTAGGCGTACAACGCTAA-3’), NC mimics (5’-CCCAGUGUUCAGACUACCUGUUC-3’), NC inhibitor (5’-CAGUACUUUUGUGUAGUACAA-3’), linc00662 siRNA (5’-CCTGCAGGCGTACAACTAA-3’), miR-199a-5p mimics (5’-CCCAGUGUUCAGACUACCUGUUC-3’) and miR-199a-5p inhibitor (5’-GAACAGGUAGUCUGAACACUGGG-3’) plasmids were purchased from Solarbio company.
Tumor formation of BC in nude mice
T24 cells (1 × 105) stably expressing sh-linc00662 were injected subcutaneously into the right side of 4-week-old male nude mice. T24 cells (1 × 105) were injected subcutaneously into the left side of nude mice as negative control. The tumor size (0.5 × length × width2) of mice was measured every 7 days for 21 days, and then every 3 days. After inoculation for 4 weeks, the mice were killed, and the tumors were removed and photographed. A 4% paraformaldehyde solution was used to immobilize for immunohistochemistry. All animal studies were ratified by the hospital ethics committee.
Adhesion experiment
Cell adhesion detection kit (BestBio, Nanjing, China) was used to detect the adhesion of BC cells. First, the 96-hole plates were taken out, and the coating solution was added into the holes with the specification of 100 μL per hole for 2-8°C overnight. Then, the cells were inoculated at 5 × 104 per hole, and cultured at 37°C in 5% CO2 for 1 hour. Finally, the dye solution was added to the plate at 10 μL per hole, and the cells were cultured at 37°C in 5% CO2 for 30 min. The absorbance value was detected at the peak of 450 nm, and the cell adhesion rate was calculated. Cell adhesion rate = (absorbance of the hole in the experimental group - blank hole absorbance)/(absorbance of the hole in the control group - blank hole absorbance) × 100%.
Cell invasion and migration experiment
The cells (2 × 104 cells/hole) were inoculated in the upper Transwell chamber (200 μL of mixed solution comprising 10% fetal bovine serum and 1% DMEM medium was added into the upper chamber in advance), and DMEM medium (comprising 10% fetal bovine serum, with a total volume of 500 μL) was added into the lower chamber. The migration pool was cultured in 5% CO2 at 37°C for 24 h, and then the liquid in the upper chamber was removed and the parietal cell was wiped off. Cells in the transwell chamber were fixed with 4% paraformaldehyde for 20 min. The cells were stained with crystal violet for 15 min, and the Transwell chamber was washed with PBS buffer solution. The photographs of cell migration were collected under a 200-fold microscope, and the number of cells was calculated by randomly selecting three fields of vision. Next, the average value was calculated as the number of transmembrane cells. The experiment was reduplicated three times. Invasion: on the basis of the above steps, 8% matrigel was placed, and the number of cells per well was increased to 5 × 104.
Cell proliferation experiment
4-hole plates were taken out. Then, BC cells were inoculated into 24-hole plates, and cultured in 5% CO2 at 37°C. Next, one-hole plate was taken out every 24 h, and the cells were dyed with 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reagent kit. Finally, the absorbance of the sample was detected at the peak of 570 nm by spectrophotometer, and the cell proliferation activity was assessed by the absorbance value.
Apoptosis experiment
The apoptosis of BC cells was detected by Annexin V-FITC/PI fluorescence double staining cell apoptosis detection kit (Procell, Wuhan, China) and flow cytometry (BD Biosciences, USA). First, the cell suspension was prepared and centrifuged for 5 min, and then the supernatant was discarded. Then, 1 × Annexin V Binding buffer (500 μL), Annexin V-FITC (5 μL) and propidium iodide (5 μL) were added in sequence and cultured for 20 min at ambient temperature in the dark. Finally, the apoptosis rate was analyzed by flow cytometry and CellQuest (BD Biosciences).
Real-time quantitative PCR
Tumor tissue samples were milled and prepared into homogenate before real-time quantification of linc00662 and miR-199a-5p expression. The cancer cells in each group were prepared into cell suspension. Total RNA in samples was extracted by Trizol reagent, and its RNA purity was detected by ultraviolet spectrophotometer. Then, the samples with proper purity were selected as subsequent quantitative objects. The fluorescence of linc00662 and miR-199a-5p was quantified by TaqMan One Step RT-qPCR kit (Solarbio, Beijing, China), and the primers were constructed by Solarbio company. The reaction system and program settings were referenced from the kit specifications. After obtaining Ct values, the expression level was standardized by 2-ΔΔCt method. GAPDH was used as lncRNA and mRNA control and U6 as miRNA control. Linc00662 upstream primer was 5’-TGGACATCTGTCTGGAGG-3’, and downstream primer was 5’-GGCTGAGGCATAAGAATCG-3’. miR-199a-5p upstream primer was 5’-GCCAAGCCCAGTGTTCAGAC-3’, and downstream primer was 5’-GTGCAGGGTCCGAGGTATTC-3’. GAPDH upstream primer was 5’-CGACTTATACATGGCCTTA-3’, and downstream primer was 5’-TTCCGATCACTGTTGGAAT-3’. U6 upstream primer was 5’-CTCGCTTCGGCAGCACA-3’, and downstream primer was 5’-AACGCTTCACGAATTTGCGT-3’.
Western blot
The cell sample to be detected was prepared into cell suspension, and RIPA lysis buffer was used to lyse the cells for 20 min. After centrifugation, the precipitate was discarded, and the supernatant was obtained. BCA protein kit was used to detect the protein concentration in the supernatant, and the sample volume was calculated according to the protein concentration. Separation gel and concentrated gel were prepared before electrophoresis. The total proteins were isolated by SDS-PAGE (SolarBio) electrophoresis at 90 V constant pressure for 45 min, followed by 110 V constant pressure for 60 min. The gel was carefully taken down and moved to the film transfer clip. The film was transferred to an ice bath at 100 V for 1 h. After the film transfer, the sealing solution was added for 2 h. After that, the antibodies of the protein to be detected and the internal reference protein were added and cultured overnight at 4°C. After washing with PBS solution, the second antibody was supplemented and cultured for 1 h at ambient temperature. Finally, PBS solution was used to clean the transfer film, and ECL luminescent agent was used to visualize the protein bands on the film. The strip was analyzed by Bio-Rad gel imaging system. The internal reference protein was GAPDH protein. E-cadherin (ab40772, 1:1000), N-cadherin (ab76011, 1:1000), β-catenin (ab32572, 1:5000), Bax (ab32503, 1:1000), Bcl-2 (ab32124, 1:1000), Caspase 3 (ab32150, 1:1000) and Cleaved Caspase 3 (ab32042, 1:1000) were the proteins to be detected. The above protein antibodies were all from Abcam. Caspase 9 (mAb 9508, 1:1000) and Cleaved Caspase 9 (mAb 9508, 1:1000) were from Cell Signaling Technology.
Statistics and analysis
The results of all experimental indicators were in the form of mean ± standard deviation (Mean ± SD). K-S test was used to verify the normal distribution of experimental results. The data between the two groups were compared by independent sample T-test. The data between multiple groups were compared by one-way ANOVA followed with Dunnett-t test. The confidence interval used in this paper was 95%. P<0.05 indicated a statistical difference.
Results
Expression of linc00662 and miR-199a-5p in BC
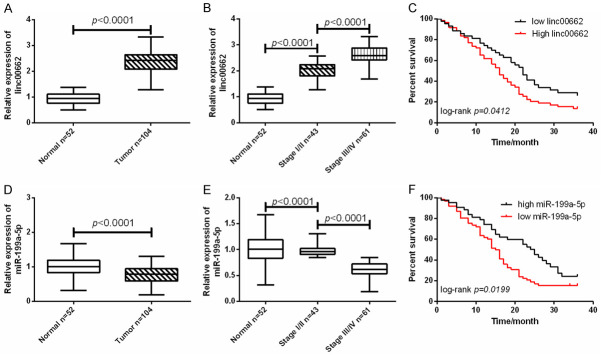
In this research, 104 cases of BC tissues were included, including 43 cases of stage I/II BC tissues and 61 cases of III/IV BC tissues. Another 52 cases of normal para-cancerous tissues were selected as control. Figure 1 shows that linc00662 was enhanced in BC tissues, and the expression of linc00662 in stage III/IV tumor was higher than that in stage I/II tumor. However, miR-199a-5p was decreased in BC, and its expression in stage III/IV tumor was lower than that in stage I/II tumor. The results showed poor 3-year survival outcomes in patients with low expression of miR-199a-5p and high expression of linc00662. In addition, by analyzing the relationship between linc00662, miR-199a-5p and clinical data, it was found that patients with high expression of linc00662 and low expression of mir-199a-5p had high TNM stage and increased probability of lymph node metastasis (Table 1). The above results indicated that up-regulated linc00662 and down-regulated miR-199a-5p might be involved in the progression of BC.
Figure 1.
Expression of linc00662 and miR-199a-5p in bladder cancer (BC). A. Linc00662 was up-regulated in BC tissues (Normal =52, Tumor =104). B. The expression of linc00662 was the highest in stage III/IV BC tissues (Normal =52, Stage I/II =43, Stage III/IV =61). C. The overall 3-year survival outcome of patients with high expression of linc00662 was worse than that of patients with low expression of linc00662 (Low =52, High =52). D. miR-199a-5p was down-regulated in BC tissues (Normal =52, Tumor =104). E. The expression of miR-199a-5p was the lowest in stage III/IV BC tissues (Normal =52, Stage I/II =43, Stage III/IV =61). F. The overall 3-year survival outcome of patients with low expression of miR-199a-5p was worse than that of patients with high expression of miR-199a-5p (Low =52, High =52).
Table 1.
Relationship between Linc00662 and miR-199a-5p and clinical data of patients with bladder cancer
| Factor | Linc00662 | P | miR-199a-5p | P | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| High expression | Low expression | High expression | Low expression | |||
| Age | 0.311 | 0.156 | ||||
| ≥60 years old (n=65) | 35 | 30 | 29 | 36 | ||
| <60 years old (n=39) | 17 | 22 | 23 | 16 | ||
| Gender | 0.152 | 0.539 | ||||
| Male (n=67) | 30 | 37 | 35 | 32 | ||
| Female (n=37) | 22 | 15 | 17 | 20 | ||
| Tumor stage | 0.001 | 0.001 | ||||
| I/II (n=43) | 15 | 28 | 30 | 13 | ||
| III/IV (n=61) | 37 | 24 | 22 | 39 | ||
| Lymph node metastasis | 0.039 | 0.010 | ||||
| Metastasis (n=79) | 44 | 35 | 34 | 46 | ||
| Non-metastasis (n=25) | 8 | 17 | 18 | 7 | ||
| Tumor size | 0.676 | 0.403 | ||||
| ≥3 cm (n=70) | 36 | 34 | 33 | 37 | ||
| <3 cm (n=34) | 16 | 18 | 19 | 15 | ||
miR-199a-5p was the downstream target gene of linc00662
In this research, Pearson analysis was used to study the internal relationship between linc00662 and miR-199a-5p in 104 patients with BC. The results revealed that there was a negative correlation between linc00662 and miR-199a-5p. The two base sequences were compared by Starbase3.0, and the results uncovered that there was a binding site between miR-199a-5p in the 3’-UTR of linc00662. Therefore, the linc00662 mutant was constructed according to this prediction site, and the targeting relationship between the two was verified by RNA co-immunoprecipitation method and dual luciferase reporter gene assay using the linc00662 wild type as the control. The results indicated that miR-199a-5p could be enriched on linc00662 by predicting sites, and linc00662 could negatively regulate miR-199a-5p. The above results revealed that miR-199a-5p was the downstream target gene of linc00662 and it was negatively regulated by it. Detailed results are shown in Figure 2.
Figure 2.
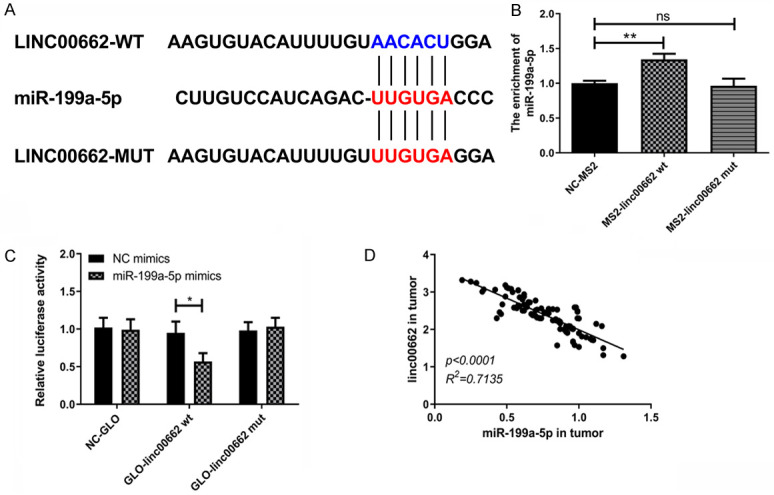
miR-199a-5p is a downstream target gene of linc00662. A. Starbase3.0 database predicted that miR-199a-5p was a target gene of linc00662. B. The relative luciferase activity decreased when linc00662 wt was co-transfected with miR-199a-5p mimics (n=3). C. miR-199a-5p was enriched on linc00662 (n=3). D. Linc00662 was negatively correlated with miR-199a-5p in BC (n=3). *P<0.05; ns means P>0.05.
Linc00662 promoted migration and invasion of BC cells through sponging miR-199a-5p
Cell migration and invasion are important basis for cancer cell metastasis. In this part, the regulatory effects of linc00662 and miR-199a-5p on cell migration and invasion were studied, and the results are shown in Figure 3. The down-regulation of linc00662 resulted in the up-regulation of miR-199a-5p, which was offset by miR-199a-5p in this research. The down-regulation of linc00662 resulted in the decrease of cell migration and invasiveness, and the down-regulation of miR-199a-5p could counteract this trend. Cell adhesion is an important condition for cell migration and invasiveness. Down-regulation of linc00662 decreased cell adhesion activity, while down-regulation of miR-199a-5p offset this change. E-cadherin, N-cadherin and β-catenin are important protein molecules in cell invasion and migration. Down-regulation of linc00662 could lead to the up-regulation of E-cadherin and the down-regulation of N-cadherin and β-catenin, while miR-199a-5p could counteract this process. The above results indicated that linc00662 promoted the migration and invasion of BC cells through miR-199a-5p.
Figure 3.
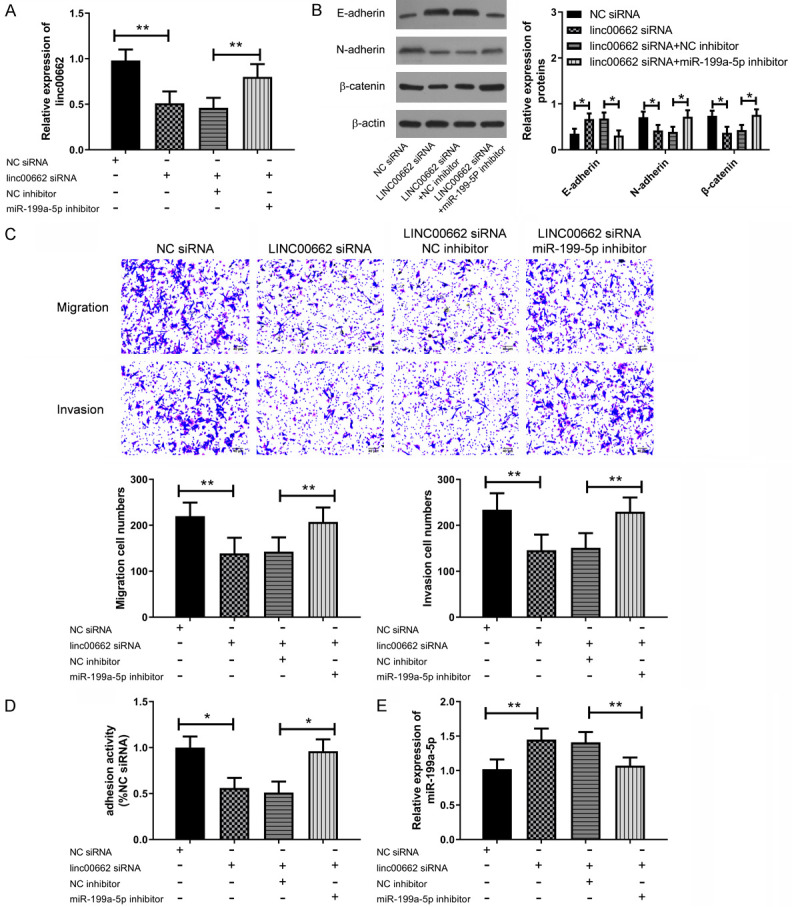
Linc00662 regulated the migration and invasion of BC cells through miR-199a-5p. A. Down-regulation of linc00662 led to the up-regulation of miR-199a-5p, which could be offset by a miR-199a-5p inhibitor (n=3). B. Down-regulation of linc00662 caused an up-regulation of E-cadherin protein, and the down-regulation of N-cadherin and β-catenin protein, while the down-regulation of miR-199a-5p could offset this change (n=3). C. Down-regulation of miR-199a-5p could offset the decrease of cell migration and invasion caused by down-regulation of linc00662, with scale =40 m (n=3). D. Down-regulation of miR-199a-5p could offset the decrease of cell adhesion activity caused by down-regulation of linc00662 (n=3). E. Down-regulation of linc00662 caused the up-regulation of E-cadherin protein, and the down-regulation of N-cadherin and β-catenin protein, while the down-regulation of miR-199a-5p offset this change. *P<0.05, **P<0.01; ns means P>0.05.
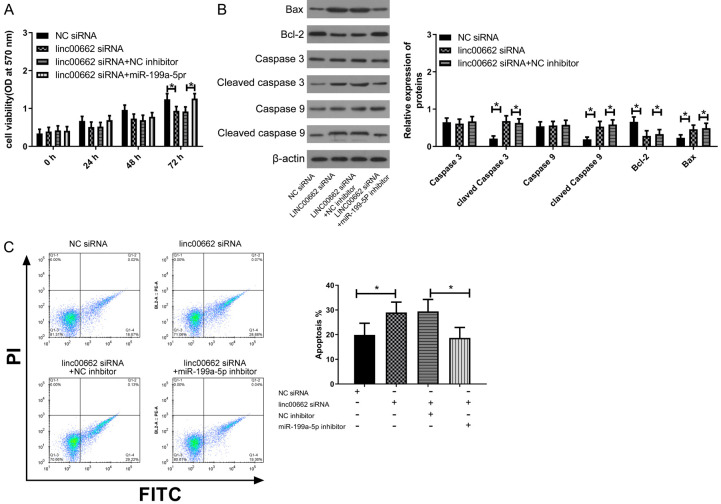
Linc00662 regulated apoptosis and proliferation of BC cells through miR-199a-5p
Apoptosis and proliferation are prerequisites for malignant expansion of BC cells. Thus, the regulatory effects of linc00662 and miR-199a-5p on cell proliferation and apoptosis were studied, and the results are shown in Figure 4. The down-regulation of Linc006662 could lead to the decrease of cell proliferation and the increase of apoptosis, while miR-199a-5p could counteract these changes. Caspase 3, Caspase 9, Bax and Bcl-2 proteins are important proteins for apoptosis and proliferation. Down-regulation of linc00662 could lead to increased cleavage of Caspase 3 and Caspase 9, up-regulation of Bax protein and down-regulation of Bcl2 protein, but down-regulation of miR-199a-5p could offset this change. The above results indicated that linc00662 suppressed apoptosis and promoted cell proliferation through sponging miR-199a-5p.
Figure 4.
Linc00662 regulated proliferation and apoptosis of BC cells through miR-199a-5p. A. After inoculation for 72 h, down-regulation of miR-199a-5p could offset the decrease of cell proliferation caused by down-regulation of linc00662 (n=3). B. Down-regulation of linc00662 caused increased cleavage of Caspase 3 and Caspase 9, up-regulation of Bax protein, and down-regulation of Bcl2 protein, while down-regulation of miR-199a-5p offset this change (n=3). C. Down-regulation of miR-199a-5p offset the increase of apoptosis caused by down-regulation of linc00662 (n=3). *P<0.05.
Linc00662/miR-199a-5p axis regulated the formation and growth of BC in vivo
In these experiments, the regulating effect of linc00662/miR-199a-5p axis on BC in vivo was studied, and the specific results are shown in Figure 5. Down-regulation of linc00662 could lead to up-regulation of miR-199a-5p in BC, while inhibiting the size and mass transplanted tumors. This indicated that linc00662/miR-199a-5p axis regulated the formation and growth of BC in vivo.
Figure 5.
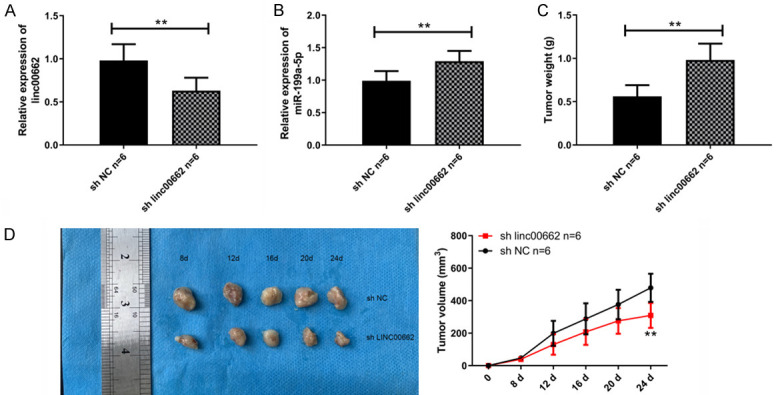
Linc00662/miR-199a-5p axis regulates the formation and growth of BC in vivo. A. Linc00662 siRNA down-regulated the expression of linc00662 in BC (n=6). B. Down-regulation of linc00662 caused the up-regulation of miR-199a-5p in BC tumor (n=6). C. Down-regulating linc00662 inhibited tumor quality (n=6). D. Down-regulating linc00662 inhibited an increase of tumor size (n=6). **P<0.01.
Discussion
BC (bladder cancer) is not only one of the most pervasive cancers, but also has a very high recurrence rate [16]. Therefore, there is an urgent need for a highly effective targeted therapy to inhibit the progression of BC. Treatment strategies based on lncRNA and miRNA focus on regulating the growth and survival of carcinoma cells from a molecular perspective.
BC cells have strong diffusion, and this malignant diffusion behavior is closely bound up with cell migration and invasion [17]. Many studies have revealed that non-coding RNA participates in the development and progression of carcinoma by regulating cell migration and invasiveness. For example, the novel lncRNA LINC00844 regulates migration and invasion of prostate cancer cells through AR signals, and microenvironment-regulated lncRNA-HAL is able to promote stemness in breast cancer cells [18,19]. The results of this research showed that linc00662 could act as the ceRNA of miR-199a-5p, and down-regulation of miR-199a-5p could suppress the reduction of cell migration and invasion induced by down-regulation of linc00662. Cell adhesion is responsible for supervising and regulating many cell processes [20], and the internal environment stability of tissues in vivo also depends on cell adhesion [21], so it controls the development and progression of many diseases. Cell adhesion provides an important prerequisite for the outward migration and invasion of carcinoma cells. Collective migration and invasion of carcinoma cells cannot be separated from the dynamic balance of cell-cell connection and cell-matrix adhesion [22,23]. The results of this research revealed that down-regulation of Linc00662 caused a decrease of cell adhesion activity, while down-regulation of miR1-199a-5p offset this decreasing tendency. E-cadherin, N-cadherin, and β-catenin are important protein molecules of cell adhesion, migration, and invasion [24-26], which can be used to evaluate the changes in cell adhesion, migration, and invasion at the molecular level through expression levels of these three proteins. The above results indicated that linc00662 regulates cell adhesion through sponging miR-199a-5p in BC cells, thus promoting malignant migration and invasion of BC cells.
Metastasis of cancer cells mainly depends on the changes of apoptosis and proliferation activity. Apoptosis defects and excessive proliferation may induce or promote cancer progression [27]. The results showed that down-regulation of linc00662 resulted in an increase in apoptosis activity and a decrease in proliferation capacity, while down-regulation of miR-199a-5p counteracted the above changes. Caspase 3, Caspase 9, Bax, and Bcl-2 proteins are important proteins for apoptosis and proliferation. In order to evaluate the process of apoptosis and proliferation at the molecular level, the expression levels of these proteins were detected. The results revealed that down-regulation of linc00662 resulted in increased cleavage of Caspase 3 and Caspase 9, up-regulation of Bax protein and down-regulation of Bcl-2 protein, while down-regulation of miR-199a-5p could offset this change. These results suggested that linc00662 regulated the apoptosis pathway mediated by Caspase 3 and Caspase 9 through sponging miR-199a-5p and promoted cell proliferation, ultimately leading to malignant diffusion of BC cells.
In this research, the relationship between lin00662, miR-199a-5p, and the 3-year survival of BC was investigated and showed that high expression of Linc00662 and low expression of miR-199a-5p may lead to poor 3-year survival. This indicates that the high expression of Linc00662 and the low expression of miR-199a-5p may be risk factors for the progression of BC, but the deficiency of this study is that the risk model cannot be used to evaluate the prognostic value of both. In cell processes, miRNA plays a role in gene regulation by regulating the degradation or stabilization of downstream mRNA [28], so miR-199a-5p may have many target genes in BC. Linc00662 may indirectly regulate a target gene through sponging miR-199a-5p, which will cause changes in related cell biologic functions. Moreover, we only used 5637 cells for in vitro experiments in this study, which is less than optimal. The following research will be designed to discuss the downstream target genes of linc00662/miR-199a-5p axis and explore the related genes and signaling pathways. In addition, we can also study whether targeted therapy focused on linc00662/miR-199a-5p axis can improve the chemotherapy effect of BC, and more cell lines will be used to verify our experimental results.
In sum, this research has revealed that linc00662 is an oncogene of BC, that can promote the progression of malignant tumor through sponging miR-199a-5p. Linc00662 and miR-199a-5p are potential targets for anti-cancer treatment of BC. It is feasible to use their abnormal expression in BC for prognosis or early diagnosis, which is worthy of further study.
Acknowledgements
Clinical study and application of transurethral non-muscular invasive bladder tumor dissection (project number: Zykjjsc20-yyjc-2019-04).
Disclosure of conflict of interest
None.
References
- 1.Kalan Farmanfarma K, Mahdavifar N, Salehiniya H. Bladder cancer in Iran: an epidemiological review. Res Rep Urol. 2020;12:91–103. doi: 10.2147/RRU.S232417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saginala K, Barsouk A, Aluru JS, Rawla P, Padala SA, Barsouk A. Epidemiology of bladder cancer. Med Sci (Basel) 2020;8:15. doi: 10.3390/medsci8010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richters A, Aben KKH, Kiemeney LALM. The global burden of urinary bladder cancer: an update. World J Urol. 2020;38:1895–1904. doi: 10.1007/s00345-019-02984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soria F, Krabbe LM, Todenhofer T, Dobruch J, Mitra AP, Inman BA, Gust KM, Lotan Y, Shariat SF. Molecular markers in bladder cancer. World J Urol. 2019;37:31–40. doi: 10.1007/s00345-018-2503-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enokida H, Yoshino H, Matsushita R, Nakagawa M. The role of microRNAs in bladder cancer. Investig Clin Urol. 2016;57(Suppl 1):S60–76. doi: 10.4111/icu.2016.57.S1.S60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong F, Xu T, Shen Y, Zhong S, Chen S, Ding Q, Shen Z. Dysregulation of miRNAs in bladder cancer: altered expression with aberrant biogenesis procedure. Oncotarget. 2017;8:27547–27568. doi: 10.18632/oncotarget.15173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Shin VY, Siu MT, Ho JC, Cheuk I, Kwong A. miR-199a-5p confers tumor-suppressive role in triple-negative breast cancer. BMC Cancer. 2016;16:887. doi: 10.1186/s12885-016-2916-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Dai YX, Wang SQ, Qiu MK, Quan ZW, Liu YB, Ou JM. miR-199a-5p inhibits proliferation and induces apoptosis in hemangioma cells through targeting HIF1A. Int J Immunopathol Pharmacol. 2018;31:394632017749357. doi: 10.1177/0394632017749357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou M, Wang S, Hu L, Liu F, Zhang Q, Zhang D. miR-199a-5p suppresses human bladder cancer cell metastasis by targeting CCR7. BMC Urol. 2016;16:64. doi: 10.1186/s12894-016-0181-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song T, Zhang X, Yang G, Song Y, Cai W. Decrement of miR-199a-5p contributes to the tumorigenesis of bladder urothelial carcinoma by regulating MLK3/NF-kappaB pathway. Am J Transl Res. 2015;7:2786–2794. [PMC free article] [PubMed] [Google Scholar]
- 11.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian X, Wu Y, Yang Y, Wang J, Niu M, Gao S, Qin T, Bao D. Long noncoding RNA LINC00662 promotes M2 macrophage polarization and hepatocellular carcinoma progression via activating Wnt/beta-catenin signaling. Mol Oncol. 2020;14:462–483. doi: 10.1002/1878-0261.12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Yao Y, Huang S, Li L, Jiang B, Guo H, Lei W, Xiong J, Deng J. LINC00662 promotes gastric cancer cell growth by modulating the Hippo-YAP1 pathway. Biochem Biophys Res Commun. 2018;505:843–849. doi: 10.1016/j.bbrc.2018.09.191. [DOI] [PubMed] [Google Scholar]
- 14.Li N, Zhang LY, Qiao YH, Song RJ. Long noncoding RNA LINC00662 functions as miRNA sponge to promote the prostate cancer tumorigenesis through targeting miR-34a. Eur Rev Med Pharmacol Sci. 2019;23:3688–3698. doi: 10.26355/eurrev_201905_17792. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Niu X, Jiang H, Mao F, Zhong B, Jiang X, Fu G. Long non-coding RNA DLX6-AS1 facilitates bladder cancer progression through modulating miR-195-5p/VEGFA signaling pathway. Aging (Albany NY) 2020;12:16021–16034. doi: 10.18632/aging.103374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian DW, Wu ZL, Jiang LM, Gao J, Wu CL, Hu HL. KIF5A promotes bladder cancer proliferation in vitro and in vivo. Dis Markers. 2019;2019:4824902. doi: 10.1155/2019/4824902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duff D, Long A. Roles for RACK1 in cancer cell migration and invasion. Cell Signal. 2017;35:250–255. doi: 10.1016/j.cellsig.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Lingadahalli S, Jadhao S, Sung YY, Chen M, Hu L, Chen X, Cheung E. Novel lncRNA LINC00844 regulates prostate cancer cell migration and invasion through AR signaling. Mol Cancer Res. 2018;16:1865–1878. doi: 10.1158/1541-7786.MCR-18-0087. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Venzor A, Mandujano-Tinoco EA, Lizarraga F, Zampedri C, Krotzsch E, Salgado RM, Davila-Borja VM, Encarnacion-Guevara S, Melendez-Zajgla J, Maldonado V. Microenvironment-regulated lncRNA-HAL is able to promote stemness in breast cancer cells. Biochim Biophys Acta Mol Cell Res. 2019;1866:118523. doi: 10.1016/j.bbamcr.2019.118523. [DOI] [PubMed] [Google Scholar]
- 20.Murai T, Kawashima H, Naor D. Editorial: cell-cell and cell-matrix adhesion in immunobiology and cancer. Front Immunol. 2019;10:3126. doi: 10.3389/fimmu.2019.03126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janiszewska M, Primi MC, Izard T. Cell adhesion in cancer: beyond the migration of single cells. J Biol Chem. 2020;295:2495–2505. doi: 10.1074/jbc.REV119.007759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laubli H, Borsig L. Altered cell adhesion and glycosylation promote cancer immune suppression and metastasis. Front Immunol. 2019;10:2120. doi: 10.3389/fimmu.2019.02120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendonsa AM, Na TY, Gumbiner BM. E-cadherin in contact inhibition and cancer. Oncogene. 2018;37:4769–4780. doi: 10.1038/s41388-018-0304-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derycke LD, Bracke ME. N-cadherin in the spotlight of cell-cell adhesion, differentiation, embryogenesis, invasion and signalling. Int J Dev Biol. 2004;48:463–476. doi: 10.1387/ijdb.041793ld. [DOI] [PubMed] [Google Scholar]
- 25.Shang S, Hua F, Hu ZW. The regulation of beta-catenin activity and function in cancer: therapeutic opportunities. Oncotarget. 2017;8:33972–33989. doi: 10.18632/oncotarget.15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldar S, Khaniani MS, Derakhshan SM, Baradaran B. Molecular mechanisms of apoptosis and roles in cancer development and treatment. Asian Pac J Cancer Prev. 2015;16:2129–2144. doi: 10.7314/apjcp.2015.16.6.2129. [DOI] [PubMed] [Google Scholar]
- 27.Beresford MJ, Wilson GD, Makris A. Measuring proliferation in breast cancer: practicalities and applications. Breast Cancer Res. 2006;8:216. doi: 10.1186/bcr1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]