Abstract
Bacteria have evolved sophisticated mechanisms to deliver potent toxins into bacterial competitors or into eukaryotic cells in order to destroy rivals and gain access to a specific niche or to hijack essential metabolic or signaling pathways in the host. Delivered effectors carry various activities such as nucleases, phospholipases, peptidoglycan hydrolases, enzymes that deplete the pools of NADH or ATP, compromise the cell division machinery, or the host cell cytoskeleton. Effectors categorized in the family of polymorphic toxins have a modular structure, in which the toxin domain is fused to additional elements acting as cargo to adapt the effector to a specific secretion machinery. Here we show that Photorhabdus laumondii, an entomopathogen species, delivers a polymorphic antibacterial toxin via a type VI secretion system. This toxin inhibits protein synthesis in a NAD+-dependent manner. Using a biotinylated derivative of NAD, we demonstrate that translation is inhibited through ADP-ribosylation of the ribosomal 23S RNA. Mapping of the modification further showed that the adduct locates on helix 44 of the thiostrepton loop located in the GTPase-associated center and decreases the GTPase activity of the EF-G elongation factor.
Graphical Abstract
Graphical Abstract.
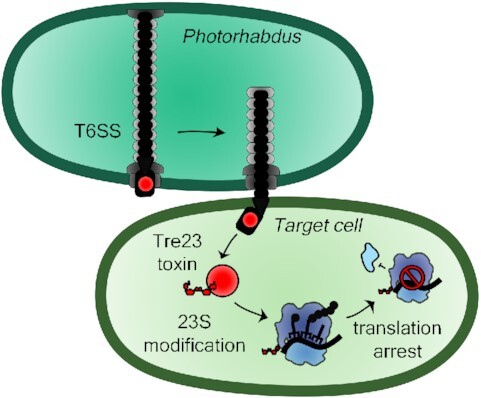
Photorhabdus laumondii uses its type VI secretion system (T6SS) to deliver a Rhs-linked toxin that ADP-ribosylates the 23S ribosomal RNA in the target bacterial cell to arrest translation.
INTRODUCTION
Bacteria compete with each other to have a privileged access to nutrients, to preserve their niche, or to colonize a new environment (1–3). They have evolved a broad repertoire of antibacterial weapons, such as antibiotics, bacteriocins and toxins delivered by dedicated secretion systems (2,4–6). The activities of antibacterial toxins are also very diverse, targeting essential cellular molecules, macromolecules or machines, such as nucleic acids, phospholipids, peptidoglycan, or the cell division machinery (7–9). These toxins and their delivery systems have been studied in many bacterial pathogens and gut symbionts (10–15), but little is known about environmental strains. Entomopathogenic bacteria of the genus Photorhabdus are Gram-negative and bioluminescent rods comprising the widely studied type strain Photorhabdus laumondii TT01 (16,17). P. laumondii has a complex cell cycle: it enters a symbiotic relationship with entomopathogenic nematodes, with which it forms a mutualistic consortium that will invade and destroy insect larvae (18,19). This property is of great importance for pest control in agriculture (20). As the result of the complex cell cycle P. laumondii colonizes various niches, such as the soil, the nematode intestine and the insect larvae. In addition to well-characterized insecticidal toxins such as Tc-toxins, P. laumondii produces a vast array of putative toxins of unknown functions (21).
One of these toxins, encoded by locus tag plu0353, is a polymorphic Rhs toxin carrying a predicted ADP-ribosyltransferase ART-HYD1 family (pfam: PF15633), that belongs to the H-Y-[EDQ] clade (22,23). Polymorphic toxins are defined as multidomain proteins with a modular organization, composed of a C-terminal toxin domain fused to a N-terminal cargo, which adapts the toxin to a specific delivery system. Polymorphic toxins comprise diverse families including bacteriocins, contact-dependent growth inhibition (CDI) CdiA, Multiple adhesin family (Maf), Rearrangement hot spots (Rhs), and specialized type VI secretion system (T6SS) Hcp, VgrG and PAAR proteins (22,24). The plu0353 gene is located within a gene cluster encoding the full set of subunits required to assemble a functional T6SS. The T6SS is a multiprotein machine that uses a contractile mechanism to inject an effector-loaded needle into bacterial or eukaryotic target cells (25–27). The T6SS comprises a membrane complex on which is anchored an assembly platform responsible for the polymerization of a tail structure composed of a needle engulfed into a contractile sheath (28,29). The tail is assembled under a metastable extended conformation, and once in contact with the target, its spring-like contraction propels the needle into the target (25,30). Effectors are either domains fused to the components of the needle, such as the Hcp tail tube protein, the VgrG spike, or PAAR, a small protein that sharpens the tip of the needle, or individual proteins that are recruited to the needle via direct interactions or through the action of specific adaptor proteins (8,31,32). Genes encoding effectors are usually clustered with genes encoding their cargo, and for those having antibacterial activity, they are usually co-transcribed with a gene encoding an immunity protein that protects the producing cell (8).
Here, we characterize the ART-HYD1 toxin fused to a Rearrangement hot spot (Rhs) domain in Photorhabdus. Rhs proteins are comprised of an N-terminal region responsible for the attachment to the secretion machinery, a central domain composed of Tyrosine/Aspartate-rich (YD) repeats that are predicted to form a β-cage encapsulating the C-terminal toxin domain (33,34). We demonstrate that the polymorphic Rhs-ART-HYD1 toxin interacts with the T6SS VgrG spike, suggesting that it is delivered into target cells by the T6SS. We further define that the T6SS and the effector are required for P. laumondii antibacterial activity. We then determine that the C-terminal ART-HYD1 domain blocks protein synthesis by ADP-ribosylation of helix 44 of the 23S ribosomal RNA, a region that is part of the GTPase-associated center, hence impairing activity of elongation factors.
MATERIALS AND METHODS
Strains, media and growth conditions
Bacterial strains, plasmids and oligonucleotides used in this study are listed in Table S1. Escherichia coli strains BW25113 and DH5α were used for cloning, BW25113 for toxicity tests, microscopy, and in vivo replication, transcription and translation assays, and BL21(DE3) for protein production. P. laumondii TT01 and its mutant derivatives were used for antibacterial competition assays. E. coli and P. laumondii cells were routinely grown in Lysogeny Broth (LB), with agitation at 37°C and 28°C, respectively. When required, media were supplemented with gentamycin (15 μg·mL–1), chloramphenicol (15–40 μg·ml–1 for E. coli, 8 μg·ml–1 for P. luminescens), ampicillin (25–100 μg·ml–1), streptomycin (100 μg·ml–1) or kanamycin (50 μg·ml–1). Gene expression was induced by the addition of l-arabinose (0.1–0.5%) or IPTG (50–500 μM).
P. laumondii strain construction
P. laumondii mutants in which the chloramphenicol cassette has replaced the T6SS-1 promoter region, the effector-immunity genes, or inserted on the rpmE/glmS unrelated locus, as a control (35), were constructed by allelic exchange using the suicide vector pJQ200 (36), as previously described (35,37). Briefly, regions downstream and upstream the fragment to be deleted were PCR-amplified using P. laumondii chromosomal DNA, digested, and ligated with the Cam_Plac_GFP cassette into pJQ200. Allelic exchange was performed as previously described (38). pJQ200 derivative plasmids were transformed into E. coli WM3064 and conjugated into P. laumondii TT01. Transconjugants were selected on LB agar plates supplemented with chloramphenicol and gentamycin. After streaking on LB plates supplemented with sucrose and chloramphenicol, sucrose-resistant, chloramphenicol-resistant, gentamycin-sensitive clones were selected, and verified by PCR and DNA sequencing (Eurofins).
Antibacterial competition assay
Overnight cultures were diluted 100-fold in LB at 28°C and grown until A600 = 0.6–0.8. One mL of each strain was centrifuged, washed in LB and resuspended to an A600 of 0.1. Cells were mixed with a 1:1 competitor:recipient ratio in a final volume of 100 μl, spotted on a nitrocellulose filter placed on GNO agar plates. After incubation at 28°C for 48 h, cells were resuspended in 1 ml of LB, washed with LB and serially diluted to 10–7. Dilutions from 10–4 to 10–7 were plated on plain GNO agar plates and GNO agar plates supplemented with chloramphenicol (8 μg·ml–1) and incubated at 28°C for 48 h. Colony-forming units (CFU) were counted, and competitive indexes (CI), i.e. chloramphenicol-resistant CFU (mutant) divided by the CFU on plain LB agar plates (all strains), were calculated. The experiment has been done in triplicate, with technical triplicates for each biological replicate. Statistical analyses of CIs were performed using Shapiro-Wilk (to evaluate normal distribution) and Levene (to evaluate variance homogeneity) tests. Statistical differences were determined by ANOVA and Tukey's range tests.
Plasmid construction
Cloning was performed by standard restriction-ligation procedures. Oligonucleotides were obtained from Sigma-Aldrich or Eurogentec. DNA fragments coding for Tre23, Tri23, VgrG, EagR and Rhs were amplified from P. laumondii TT01 using Q5 polymerase (NEB). PCR fragments were purified on NucleoSpin Gel and PCR Clean-up columns (Macherey-Nagel), digested as recommended by the manufacturer (NEB), and purified before ligation. After transformation into E. coli DH5α cells, constructions were first verified by colony-PCR. Plasmids were extracted using Wizard Plus SV Minipreps kit (Promega) and verified by DNA sequencing (Eurofins). Tre23 and Tri23 were amplified with a synthetic RBS, and cloned into the pBAD33 vector. Clones were selected on LB agar plates supplemented with chloramphenicol and 1% glucose. Tre23 clones gave small colonies due to its high toxicity. Tre23-encoding fragments were thus re-cloned into the low-copy number pNDM220 vector to yield pNDM-Tre23. The amplicon included the RBS site from the pBAD33 construction, and clones were selected on ampicillin and 1% glucose. Tre23 variants were engineered by site-directed mutagenesis using complementary oligonucleotides bearing the desired mutation. The gfp-lva gene was amplified from the pPROBE-gfp-lva plasmid (39) and cloned into the pBAD33 vector to yield pBAD33-gfp-lva. Then, pPJ23104-gfp-lva was constructed by amplification of the pBAD33-gfp-lva vector without AraC and the pAraBAD promoter but introducing the pJ23104 synthetic promoter (iGEM parts), and re-circularization by phosphorylation and ligation. Sequences coding for VgrG, EagR and Rhs were PCR-amplified and cloned into T7 promoter-driven pET-Duet™-1, pRSF-Duet™-1 and pCDF-Duet™-1, respectively, to yield pET-His-VgrG (encoding N-terminally His6-TEV-tagged VgrG), pRSF-EagR-ST (encoding C-terminally Strep-tagged EagR), and pCDF-Rhs-FL (encoding C-terminally FLAG-tagged Rhs). Toxicity of full-length Rhs was counteracted by introducing substitutions in the Y9 (Y9A) and Y33 (Y33A) conserved amino-acids of the ART-HYD1 domain (Y1351 and Y1375 in Rhs) and in the putative C-terminal Rhs autocleavage site (D1338N) by site-directed mutagenesis using complementary oligonucleotides bearing the desired mutation. For protein production, sequences coding for Tri23 and Tre23 were PCR-amplified and cloned into pCDF-Duet™-1 and pRSF-Duet™-1 respectively, to yield pCDF-Strep-TEV-Tri23 (encoding N-terminally Strep-TEV-tagged Tri23), pCDF-Tri23-FLAG (encoding C-terminally FLAG-tagged Tri23) and pRSF-Tre23-Strep (encoding C-terminally Strep-tagged Tre23). Toxicity of Tre23 was counteracted by introducing the Y9A and Y33A substitutions as described above. All plasmids were verified by colony-PCR and DNA sequencing.
Pull-down of the VgrG-Rhs-EagR complex
An overnight culture of E. coli BL21(DE3) freshly transformed with pET-His-VgrG, pRSF-EagR-FL and pCDF-Rhs-St was diluted 1/100 into 50 ml of LB supplemented with the required antibiotics, grown to A600 = 0.8 and induced with 0.5 mM IPTG for 18 h at 16°C. Cells were collected by centrifugation at 4000× g, resuspended in buffer A (50 mM Tris–HCl pH8.5, 250 mM NaCl, 1 mM TCEP) supplemented with cOmplete™ protease inhibitor cocktail (Sigma-Aldrich). After cell lysis by sonication, the cell extract was cleared by centrifugation for 30 min at 20 000 × g and incubated for 1 h at 4°C with gentle mixing with 100 μl of Strep-Tactin Superflow resin (IBA Technology) equilibrated in buffer A. The resin was washed 5 times with 300 μl of buffer A and proteins were eluted with 100 μl of buffer B (50 mM Tris–HCl pH8.5, 250 mM NaCl, 1 mM TCEP, 2.5 mM desthiobiotin). Ten microliter of the elution material was separated by 12.5%-acrylamide SDS-PAGE, stained using InstantBlue™ (Sigma-Aldrich) or transferred onto nitrocellulose membrane, immunodetected with His-Tag (clone 1B7G5, Proteintech catalogue #66005-1-Ig), Strep-Tag Classic (clone Strep-tag II, Bio-Rad catalogue #MCA2489) or anti-FLAG (clone M2, Sigma-Aldrich catalogue #F3165) monoclonal antibodies, and secondary antibodies coupled to the Alkaline Phosphate, and revealed using 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) in presence of 10 mM MgCl2 in alkaline buffer (pH 9).
Tre23-Tri23 co-purification
E. coli BL21(DE3) cells freshly transformed with pCDF-Tri23-FLAG and empty pRSF-Duet™-1 vector or pCDF-Tri23-FLAG and pRSF-Tre23-Strep vector were grown and proteins were induced as described above. Cells were collected and lysed, and proteins were purified on 100 μl of Strep-Tactin Superflow resin (IBA Technology) as described above. The Strep affinity eluted material was then loaded on 25 μl of anti-FLAG M2 Affinity Gel (Sigma-Aldrich), incubated for 1 h at 4°C with gentle mixing, washed five times with 300 μl of buffer A, and proteins were eluted in 40 μl of SDS-gel loading buffer. Elution material was analyzed by 12.5%-acrylamide SDS-PAGE, stained using InstantBlue™ or immunodetected with anti-Strep-Tag Classic and anti-FLAG M2 antibodies.
In vivo toxicity tests in heterologous host
E. coli BW25113 cells were transformed with pBAD33 and pNDM220 vector pairs carrying immunity or toxin genes respectively, or empty vectors as controls, and selected on LB agar plates supplemented with chloramphenicol (15 μg·ml–1), ampicillin (25 μg·ml–1) and 1% glucose. Overnight cultures of transformants grown in presence of antibiotics and glucose were diluted in 10-fold series and 10 μl drops were spotted on LB agar supplemented with antibiotics and 1% glucose, or 0.5% arabinose and 0.05 mM IPTG. Pictures were taken after incubation at 37°C for 16 h.
In vivo translation, transcription and replication assays
Overnight cultures of E. coli BW25113 carrying pNDM-Tre23 or its empty pNDM220 parental vector were diluted to M9 minimal media supplemented with amino acids. After growth with shaking at 37°C to an A600 = 0.3 (time 0), tre23 expression was induced with 0.05 mM IPTG. At time 0 and after 1 h of induction 1-ml aliquots were incubated with 3 μCi of [35S]-methionine, 1 μCi of [3H]‐uridine, or 1 μCi of [3H]‐thymidine (Perkin-Elmer) for 5 min. Samples were precipitated with 5 mL of cold 10% TCA for 30 min on ice, filtered on 0.45 μm nitrocellulose, washed twice with 10 ml of 10% TCA, air dried, immersed in 10 ml of scintillation liquid, and counted in Optisafe-3 (Fisher) scintillation cocktail using a liquid scintillation counter (Hidex 300SL super low-level TDCR).
Microscopy
Overnight cultures of E. coli BW25113 producing GFP-LVA under synthetic constitutive promoter from plasmid pPJ23104-GFP-lva were diluted 1/100 into fresh LB supplemented with glucose and antibiotics, and grown to an A600 = 0.3. Cells were concentrated to an A600 of 10, and spotted on LB agar pads supplemented with antibiotics and 0.05 mM IPTG prepared with Frame-Seal™ slide chambers (Bio-Rad) and covered with a cover slip. Fluorescence microscopy was performed on a Nikon Eclipse Ti microscope equipped with an Orcaflash 4.0 LT digital camera (Hamamatsu) and a perfect focus system. Fluorescence images were acquired with an exposure time of 30 ms for phase contrast, and 100 ms for GFP.
Purification of Tre23 and Tri23 for in vitro activity tests
C-terminally Strep-tagged Tre23 (Tre23ST) was synthesized in a coupled in vitro transcription-translation reaction (PURExpress Kit, NEB). The reaction was then diluted to 1 mL with buffer A and Tre23ST was purified on Strep-Tactin Superflow resin (IBA Technology) as described above. Strep-tagged Tri23 was purified from 500 mL of BL21(DE3) cells transformed with pCDF-Strep-TEV-Tri23 on 500 μl Strep-Tactin Superflow resin as described above. The Strep-TEV tag was cleaved off with TEV protease (1:100 molar ratio of protease:strep-tagged protein) overnight at 4°C. Tri23 was then purified by size exclusion chromatography using Superdex200increase column (GE Healthcare) pre-equilibrated with buffer (20 mM Tris–HCl pH 8, 250 mM NaCl, 1 mM TCEP).
In vitro translation inhibition
In vitro translation inhibition assays were performed for 2 h at 37°C in presence of a PCR fragment encoding for Strep-tagged GFP expressed from the T7 promoter and 100 nM of purified Tre23ST using the PURExpress® In Vitro Protein Synthesis Kit (NEB). When indicated, reactions included the addition of 0.1 mM NAD+ or NAD+ derivatives, and 2 mM of purified Tri23. When assaying ribosylation of stand-alone ribosomes or ribosomes in presence of factors and other reaction components, the PURExpress ΔRibosome Kit (NEB) was used instead. Translation reactions were then separated by SDS-PAGE, transferred onto nitrocellulose membranes and GFPST was immunodetected by Strep-Tag Classic antibodies (Bio-Rad).
70S ribosomes extracted from strains producing or not Tre23 were tested for translation activity using the coupled in vitro transcription-translation PURExpress® ΔRibosome Kit (NEB). Reactions were performed as described above, in presence of 150 AU of toxin-exposed or control ribosomes.
Effect on in vitro eukaryotic translation was determined using TnT® T7 Quick coupled transcription-translation system based on rabbit reticulocyte lysate (Promega). Ten μl-reactions were assembled according to the manufacturer recommendations in presence of 0.1 mM NAD+, with or without Tre23ST, and incubated for 2 h at 30°C. Luciferase activity was measured in black flat bottom 96-well plates (Microfluor 1, Nunc) using in microplate reader (Tecan M200 Infinite). 100 μl of substrate (25 mM HEPES pH 7.5, 10 mM MgCl2, 0.5 mM ATP, 200 μM D-luciferin) was injected into 2.5 μl of reaction, mixed for 3 s, and luminescence was recorded for 1 min (6 cycles of 10 s). Average of the 10 s-integrated signals was then plotted. The average and standard deviation of triplicates is also shown.
Native PAGE and mass spectrometry
Native polyacrylamide gels were prepared by layering 12.5%, 7.5% and 5% concentration separating polyacrylamide gel layers and topping them with 3% stacking polyacrylamide gel without SDS. The SDS was also omitted from standard Tris-glycine running buffer. Aliquots of 2.5 μl of in vitro translation reactions were mixed with native loading buffer (60 mM Tris–HCl pH 6.8, 10% glycerol) deposited in pre-washed wells and migrated in pre-chilled buffer in the cold room overnight at constant 30 V current. Gel was stained using InstantBlue™ (Sigma-Aldrich), or transferred onto nitrocellulose membrane for detection with streptavidin-alkaline phosphatase conjugate (catalog #S921, Invitrogen). Bands at the size corresponding to biotin signal were excised from stained gel using sterile scalpel, digested with Trypsin/LysC and analysed by mass spectrometry.
Preparation of rRNA fragment library
Fifteen microliters of in vitro translation reactions were prepared in presence of 0.1 mM of biotinylated NAD+ derivative, 6-biotin-17-NAD+. Control reaction was supplemented with dsDNA coding for GFPST, while toxin reaction was supplemented with dsDNA coding for Tre23ST. After incubation for 4 h, reactions were diluted to 500 μl, and RNAs were extracted using phenol and chloroform. The aqueous phase was precipitated with isopropanol, the pellet was washed with 70% ethanol and resuspended in 500 μl of binding buffer (5 mM Tris–HCl pH 7.5, 0.5 mM EDTA, 1 mM NaCl). After sonication for 3 min with a duty cycle of 50% (Branson Sonifier 450 equipped with a microtip), the RNA preparation was incubated for 30 min with 0.5 mg of Dynabeads™ M-270 streptavidin beads (Invitrogen) equilibrated in binding buffer. The beads were washed twice with binding buffer and once with T4 RNA Ligase Reaction Buffer (NEB). Ligation of enriched biotinylated RNA fragments to a linker was performed overnight at 25°C by incubating the RNA bound to the streptavidin beads with 1 μM of Universal miRNA Cloning Linker (NEB), 20 units of T4 RNA Ligase 1 (NEB, M0204S), 40 units of Murine RNase inhibitor (NEB), 10% PEG8000 in T4 RNA Ligase Reaction buffer in a 50-μl volume. After heat-inactivation at 65°C for 15 min, beads were washed twice with binding buffer and once with Reverse Transcription buffer (Thermofisher). Reverse transcription was assembled in 25 μl and performed as recommended by the manufacturer (Thermofisher). RNAs bound to beads were mixed with dNTPs and reverse transcription primer (RT-uni-NEB-Hind, NEB) and incubated at 65°C for 5 min for primer annealing. After incubation on ice for 5 min, the Reverse Transcriptase (SuperScript IV, Thermofisher), its reaction buffer and 5 mM of DTT were added. Reverse transcription was allowed at 55°C for 20 min and heat-inactivated for 10 min at 80°C. The beads were then washed 3 times with binding buffer, resuspended to a final volume of 25 μl. cDNA was extracted by resuspending the beads in RNase H buffer and RNA strands were digested with RNase H (NEB) for 2 h at 37°C. RNaseH was heat-inactivated at 65°C for 20 min, and the cDNA elution was used for ligation of the second Linker (phosphorylated ss-linker oligonucleotide) as described before, in presence of 0.5 mM of ATP. Five microliter of ligation reactions were mixed in 50-μl PCR reactions with oligonucleotides annealing to the linkers (F-sslinker-Eco and R-NEB-Hind) and introducing EcoRI and HindIII restrictions sites, and Q5 DNA polymerase (NEB). After verification on agarose gel, PCR products were digested with 10 U of EcoRI and HindIII (NEB) for 2 h at 37°C, heat-inactivated and ligated overnight at 16°C with T4 DNA polymerase (NEB) into EcoRI/HindIII-digested pKK223.3 vector. The ligation was transformed into DH5α competent cells. After colony-PCR to screen vectors containing inserts, plasmids were extracted, inserts were sequenced using the F-pKK-seq primer, and the sequences of the inserts were mapped on the 23S RNA.
Ribosylation position mapping by protection with DNA fragments
RNAs were extracted from the in vitro translation reaction supplemented with Tre23ST and 6-biotin-17-NAD+ as described above. The rRNA protection protocol was performed as previously described (40) with modifications. Briefly, 200 pmoles of purified RNAs were annealed with 2 nmoles of DNA oligonucleotides complementary to specific fragments of 23S rRNA, in hybridization buffer (250 mM HEPES pH 7, 500 mM KCl) by heating the mixture for 7 min at 90°C and slowly cooling down to 45°C for 3 h. Then, 30 U of Mung Bean Nuclease (NEB) and 0.5 μg of RNase A (PureLink™, Sigma Aldrich) were added in Mung Bean Nuclease buffer (30 mM NaCl, 50 mM sodium acetate, 1 mM ZnSO4, pH 5) and incubated for 1 h at 37°C, to digest single-stranded DNA and RNA. Protected RNA-DNA duplexes were purified by phenol–chloroform extraction, precipitated with cold ethanol, resuspended in water and biotinylated fragments were detected by Streptavidin blot as described in the following section.
Transfer of nucleic acids and detection of biotinyl-ribosylated RNA by streptavidin Blot
Purified RNAs or DNA-RNA duplex fragments were resuspended in water and run in 1.5–2% agarose gels in TAE buffer for 45–60 min at 50 V. The gel was photographed under UV light, and RNAs were transferred onto nylon membrane (Hybond™-N+, Amersham) overnight by capillarity in 10 × SCC buffer (150 mM citrate sodium pH 7, 1.5 M NaCl) and fixed by UV light. The nylon membrane was blocked with 3% BSA in PBST (PBS, 0.1% Tween) for 30 min and incubated with streptavidin-alkaline phosphatase conjugate (catalog #S921, Invitrogen) for 1 h. The membrane was washed three times with PBST and biotinyl-ribosylated RNA were revealed with BCIP/NBT in alkaline buffer (pH 9).
Polysome analysis by sucrose density gradient centrifugation
An overnight culture of E. coli BW25113 cells freshly transformed with the pNDM220 empty vector or pNDM-Tre23 were diluted to an A600 = 0.005 in 200 ml of LB supplemented with ampicillin and 0.2% glucose. tre23 expression was induced at A600 = 0.35 with 0.25 mM IPTG. At given time points, 50 ml of cells were harvested by centrifugation for 15 min at 10 000 × g. Cell pellets were resuspended in 0.5 ml of cell lysis buffer (95 mM KCl, 5 mM NH4Cl, 20 mM HEPES pH 7.5, 1 mM DTT, 5 mM Mg(OAc)2, 0.5 mM CaCl2, 8 mM putrescine, 1 mM spermidine, 1 tablet of cOmplete™ EDTA-free Protease Inhibitor Cocktail (Roche) per 10 mL of buffer), and lysed using a FastPrep homogeniser (MP Biomedicals) with 0.1 mm Zirconium beads (Techtum) in four 20-s cycles followed by 1 min on ice. Cell debris were removed after centrifugation for 15 min at 14 800 × g and lysates were stored at -80°C. After thawing the frozen lysates on ice, 3 A260 units of each sample was loaded onto 5–25% (w/v) sucrose density gradients in HEPES:Polymix buffer (41), 5 mM Mg(OAc)2. Gradients were resolved at 165 000 × g for 3 h at 4°C using a SW41 rotor (Beckman), and analyzed and fractionated using Biocomp Gradient Station (BioComp Instruments) with A280 as a readout.
Ribosome purification
E. coli 70S ribosomes were prepared from RNase I-deficient E. coli strain MRE600 (42) as previously described (43,44). This protocol was further optimized in order to purify small-scale 70S ribosome preparations from E. coli BW25113 expressing the toxin, or harboring the empty vector. In brief, 500 ml of exponentially growing E. coli BW25113 cells grown in LB supplemented with ampicillin and 0.2% glucose, were induced with 0.25 mM IPTG for 60 min. Cells were lysed via French press in 25 mL of lysis buffer (20 mM Tris–HCl pH 7.5, 100 mM NH4Cl, 15 mM Mg(OAc)2, 0.5 mM EDTA, 3 mM β-mercaptoethanol). 12.5 ml of cleared lysate were then applied to two 40-mLlsucrose cushions (37.6% sucrose equilibrated in lysis buffer) and pelleted via centrifugation in a Ti45 rotor for 19 h at 70 000 × g at 4°C. Ribosomes were then suspended in 1 ml of HEPES:Polymix buffer (1× Polymix, 20 mM HEPES pH 7.5, 5 mM Mg(OAc)2, 0.1 mM DTT) (41) and 25 A260 units were separated on six 12-ml 5–25% sucrose gradients, equilibrated in HEPES:Polymix buffer, at 4°C for 3 h at 220 000 × g using a SW41 rotor. Gradients were fractionated and fractions containing 70S were pooled and applied onto a second 25%-sucrose cushion equilibrated in HEPES:Polymix buffer. Purified ribosomes were resuspended in HEPES:Polymix, aliquoted and stored at –80°C.
GTPase assays with E. coli EF-G
Purification and GTPase activity measurements of EF-G were performed as previously described (45). In brief, reaction mixture containing 0.5 μM of E. coli 70S ribosomes, 0.1 μM of EF-G, in HEPES:Polymix buffer were pre-incubated for 2 min at 37°C prior to addition of 300 μM of [3H]-GTP substrate (Hartman). Subsequently, 5-μl time point samples were quenched with 4 μl of 70% formic acid and analyzed by Thin-Layer Chromatography (TLC) in 0.5 M KH2PO4 pH 3.5 buffer. TLC plates were air-dried, and [3H] radioactivity was quantified by scintillation counting in Optisafe-3 (Fisher) scintillation cocktail.
Replicates and statistical analyses
All experiments have been performed at least in triplicate. Statistical analyses of competitive indexes of antibacterial assays were performed using Shapiro-Wilk (to evaluate normal distribution) and Levene (to evaluate variance homogeneity) tests. Statistical differences were determined by ANOVA, and Tukey's range or Student's t tests.
RESULTS AND DISCUSSION
The C-terminal domain of the Rhs Plu0353 protein is a T6SS-dependent cytoplasmic-acting ADP-ribosyltransferase toxin
Examination of the genetic context of the ART-HYD1 domain shows that it constitutes the C-terminal extension of a Rhs (Rearrangement hot-spot) polymorphic toxin, a subunit encoded within a type VI secretion system (T6SS) gene cluster, T6SS-1 (Figure 1A and Supplementary Figure S1). The T6SS is a multiprotein machine that uses a contractile mechanism to deliver effectors into target cells, including bacterial and eukaryotic cells (8,25–27,46,47). The Plu0353 protein presents the canonical architectural organization of Rhs proteins: a N-terminal domain (RhsNT) that comprises a PAAR motif, the central YD-repeat shell (RhsShell) and the C-terminal toxin domain (RhsCT) (34,48). RhsCT is demarcated from RhsCore by a conserved motif that undergoes autoproteolysis (34). The RhsCT is predicted to have ADP-ribosyltransferase (ART) and shares the H-Y conserved catalytic residues (His8 and Tyr33) of the H-Y-[EDQ] ART clade. The rhs gene (plu0353) is located downstream of a gene encoding a T6SS chaperone of the EagR family (31,48) (plu0354) and upstream the plu0352 gene encoding a small protein that could correspond, based on its location, to a cognate immunity protein (Figure 1A). Because all T6SS Rhs effectors characterized so far have been shown to have antibacterial activity (31,33,34,48–55), we tested the ability of the P. laumondii T6SS-1 machine and its potential Rhs toxin to confer a competitive advantage in challenge experiments. Figure 1B shows that P. laumondii cells deleted of the promoter region of the T6SS-1 gene cluster or of the Rhs toxin and its putative immunity genes were outcompeted by the parental strain, demonstrating that the RhsCT ART-HYD1 toxin has antibacterial activity. Following the T6SS nomenclature, and because this ART-HYD1 toxin belongs to a different family of the previously characterized Tre1 ADP-ribosyltransferase (56), the P. laumondii RhsCT, was renamed Tre23 (for Type VI secretion ADP-ribosyltransferase effector 23). The sequence encoding the Tre23 RhsCT was cloned into a low-copy vector under the Para promoter. Figure 1C shows that Tre23 presents a cytoplasmic-acting antibacterial activity as its production in the cytoplasm of the heterologous strain E. coli caused growth defects. This growth defect is likely to be due to its ADP-ribosyl transferase activity as Tre23 carrying substitutions of the His8 and Tyr33 putative catalytic residues did not inhibit the growth of E. coli (Figure 1C). In addition, the Plu0352 protein conferred protection against Tre23, suggesting that Plu0352 acts as an immunity protein against the Tre23 toxin (Figure 1D), and was hence renamed Tri23 (for Type VI secretion ADP-ribosyltransferase immunity 23). In agreement with the antibacterial assays, and as previously shown in P. fluorescens, S. marcescens and A. dhakensis (28,34,48), the Tre23-domain Rhs toxin is likely to be mounted on and delivered by the T6SS-1 machine. To test this hypothesis, a Strep-tag pull-down experiment was performed from E. coli cells producing full-length FLAG-tagged Rhs, His-tagged VgrG and the Strep-tagged EagR chaperone. Coomassie blue staining showed the presence of four major bands, confirmed as Rhs, truncated Rhs, VgrG and EagR by immunodetection (Figure 1E).
Figure 1.
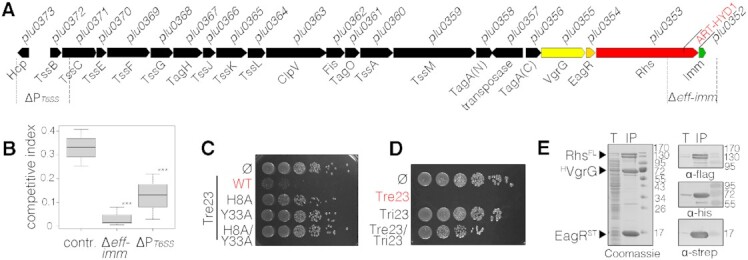
The ART-HYD1 toxin domain of Rhs is an antibacterial toxin delivered by the type VI secretion system. (A) Schematic representation of the Photorhabdus laumondii TT01 T6SS cluster. Genes encoding core and accessory T6SS components are depicted as black arrows. Genes encoding the VgrG spike protein, the putative EagR chaperone, the Rhs effector protein and its putative immunity protein are shown in yellow, orange, red, and green, respectively. The repertoire of T6SS gene clusters and T6SS islands encoded on the P. laumondii TT01 genome is shown in Supplementary Figure S1. (B) Intra-species competition assay between P. laumondii with mutants deleted for effector/immunity (Δeff-imm) or promoter region (ΔP-T6SS), or inserted in an unrelated control locus (contr.) (recipients) and the wild-type parental strain (donor). The deleted regions are depicted under the scheme in (A). Recipient and donor competitors were mixed in a 1:1 ratio and the competitive index (surviving recipient mutant cells relative to total cell number (recipient + donor)) is calculated after 48 hours of co-incubation on LB agar plates. *** indicates statistically significant differences between Control and Δeff-imm (Tukey's range test, P = 0.0012) or ΔPT6SS (P = 0.00182). Difference between Δeff-imm and ΔPT6SS is not statistically significant (P = 0.597). (C) Toxicity assay in the heterologous host E. coli. E. coli cells producing the Tre23 toxin (ART-HYD1 domain of Rhs encoded by plu0353 as shown in (A)), or its variants in the H8 and Y33 putative catalytic residues, from pBAD33 vectors were serially diluted and spotted on LB-agar plates supplemented with glucose. (D) Tri23 protects cells against the action of Tre23. E. coli cells producing the Tre23 toxin from the low-copy number vector pNDM220 and Tri23 from the pBAD33 vector were serially diluted and spotted on LB agar supplemented with 0.05 mM IPTG and 0.5% arabinose to induce expression from pNDM220 and pBAD33, respectively. (E) Pull-down assay. Total cell extracts from E. coli BL21(DE3) cells producing His6-tagged VgrG (HVgrG), strep-tagged EagR (STEagR) and FLAG-tagged Rhs (RhsFL) (T) were subjected to purification on streptactin-agarose beads. Strep-tagged and co-precipitated proteins were eluted with desthiobiotin (IP). The two fractions were analyzed by SDS-PAGE and the co-purified proteins were stained by Coomassie blue (left panel) or immunodetected using anti-His, anti-Strep and anti-FLAG antibodies (right panels).
Tre23 inhibits protein synthesis in a NAD+-dependent manner
A recent report described Tre1, a T6SS-associated ADP-ribosyltransferase of the R-S-E clade that inhibits cell division by modification of the septum protein FtsZ (56). While P. laumondii Tre23 did not cause cell elongation when produced in E. coli cells producing an unstable variant of GFP, we observed a rapid loss of the GFP signal (Figure 2A), suggesting that expression/production of GFP is altered. The impact of Tre23 on replication, transcription and translation was measured by in vivo incorporation of radioactive thymidine, uridine, and methionine, respectively. Figure 2B shows that Tre23 strongly inhibited protein synthesis without any significant impact on replication and transcription. The inhibitory effect of the Tre23 toxin on translation was further confirmed by an in vitro bacterial transcription-translation assay using a DNA fragment encoding the Strep-tagged GFP reporter protein (Figure 2C). Interestingly, Western blot analyses demonstrated that the addition of purified Tre23 inhibited protein synthesis only in presence of NAD+ (Figure 2C, lane 3), in agreement with the predicted ADP-ribosyltransferase activity of Tre23. Pre-incubation of Tre23 with Tri23, the putative immunity protein encoded by plu0352, inactivated the toxin (Figure 2C, lane 4), likely by protein-protein interaction as a Tre23/Tri23 complex can be readily purified from E. coli cells (Figure 2D). Contrarily to the Tri1 antitoxin, which also showed ADP-hydrolase activity (56), addition of purified Tri23 after toxin/NAD+ action did not resume protein synthesis (Figure 2C, lane 5), demonstrating that Tri23 does not repair the ADP-ribosylation adduct caused by Tre23. Taken together, these results demonstrate that Tre23 arrests translation in a NAD+-dependent manner, and that Tri23 inhibits its activity via direct contact with Tre23.
Figure 2.
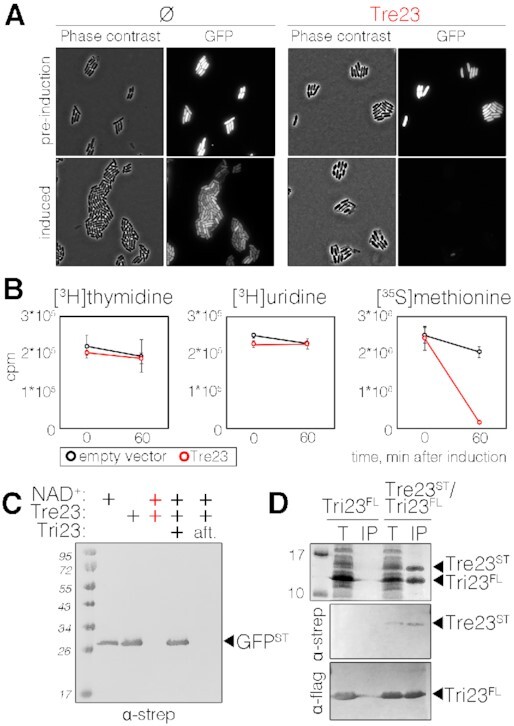
Tre23 inhibits protein synthesis. (A) Phase contrast and fluorescence microscopy recordings of E. coli cells constitutively producing short-lived green fluorescent protein (GFP-LVA) and the Tre23 toxin from the low-copy number pNDM220 vector, before and 100 min post toxin induction. Cells bearing the empty pNDM220 vector (Ø) were used as control. (B) Replication, transcription and translation rates before (time 0) and 60 min after the induction of Tre23 toxin from pNDM220 vector (red line) as compared to control carrying empty vector (black line) were measured by the incorporation (in count per minute, cpm) of [3H] thymidine, [3H] uridine and [35S] methionine, respectively. (C) Coupled in vitro transcription-translation reaction. In vitro transcription/translation of the Strep-tagged GFP reporter protein was estimated by immunodetection. Reactions were supplemented with NAD+, Tre23 and Tri23, as indicated. In the last reaction, Tri23 was added 1 h after incubating the reaction with Tre23 (aft) and reaction proceeded for 2 h. (D) Tre23-Tri23 co-purification. Total cell lysates of E. coli BL21(DE3) cells producing FLAG-tagged Tri23 (Tri23FL) alone or co-producing Strep-tagged Tre23 (Tre23ST) (T) were subjected to purification on streptactin agarose beads followed by immunoprecipitation on anti-FLAG affinity gel (IP). Proteins were separated by SDS-PAGE and stained by Coomassie (upper panel) or immunodetected using anti-Strep and anti-FLAG antibodies (middle and bottom panels, respectively).
Tre23 transfers ADP-ribose on the 23S rRNA on actively translating ribosomes
To identify the target of the Tre23 toxin, we used 6-biotin-17-NAD+, a biotinylated derivative of NAD+, in the in vitro transcription-translation assay. Native gel analyses revealed Tre23-dependent biotinylation of a high molecular weight complex (Figure 3A), corresponding to ribosomal proteins and translation factors, as identified by mass spectrometry. However, no biotinylated protein was detected on denaturing gels (Figure 3B); rather, probing RNAs extracted from the reaction with streptavidin-AP conjugate showed that ribosomal 23S RNA was ADP-ribosylated by Tre23 in a NAD+-dependent manner (Figure 3C). ADP-ribosylation of 23S rRNA also occurs in vivo: first, ribosomes purified from E. coli cells producing Tre23 were inactive in a transcription-translation assay (Figure 3D); second, while ribosomes purified from E. coli cells could be readily ADP-ribosylated in vitro, ribosomes purified from E. coli cells producing Tre23 could not be ADP-ribosylated in vitro by purified Tre23, suggesting that they already carried the modification (Figure 3E). Our analyses further demonstrated that Tre23-dependent 23S rRNA ADP-ribosylation requires actively-translating ribosomes as Tre23 did not ADP-ribosylate purified 23S rRNA or ribosome preparations missing tRNAs and/or elongation factors (Figure 3F).
Figure 3.
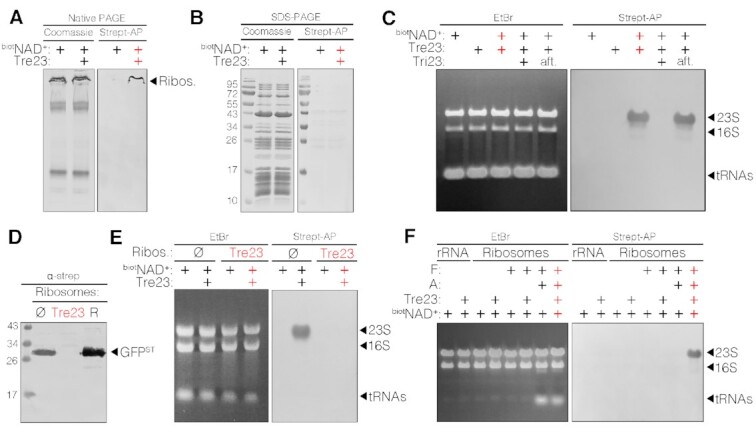
Tre23 ADP-ribosylates the 23S RNA of actively translating ribosomes. (A and B) Native PAGE (A) and SDS-PAGE (B) analyses of in vitro couple translation-transcription reaction supplemented with 6-biotin-17-NAD+ (biotNAD+) and with Tre23, as indicated. Samples were stained with Coomassie blue (left panels) or detected using streptavidin-AP (right panels). Mass spectrometry analysis of the indicated band identified subunits of 30S and 50S ribosome (Ribos.). (C) RNA extracted from of in vitro transcription-translation reactions supplemented with 6-biotin-17-NAD+ (biotNAD+), Tre23 and Tri23, as indicated were stained with ethidium bromide (EtBr, left panel) or detected using streptavidin-AP conjugate (right panel). The positions of tRNAs, 16S and 23S ribosomal RNAs are indicated. (D) Western-blot analyses with anti-Strep antibodies of GFP-strepII in vitro transcription-translation in presence of ribosomes purified from wild-type E. coli cells carrying empty pNDM220 vector (Ø), or cells producing Tre23. Ribosomes (R) included in the coupled transcription-translation kit were used as control. Ribosome profile changes over time after toxin induction are shown in Supplementary Figure S2. (E) RNA analyses of in vitro transcription-translation reactions supplemented with 6-biotin-17-NAD+ (biotNAD+), and Tre23, as indicated, using ribosomes purified from wild-type E. coli cells bearing empty vector (Ø), or cells producing Tre23. RNAs were stained with EtBr (left panel) or detected using streptavidin-AP (right panel). (F) EtBr staining (left panel) and Streptavidin-AP (right panel) RNA analyses of purified ribosomal RNAs (rRNA), or purified ribosomes alone or supplemented with translation factors (F) or translation reagents (tRNAs, amino acids, nucleotides; A), in presence of 6-biotin-17-NAD+ (biotNAD+) and Tre23, as indicated.
Tre23 modifies 23S rRNA GTPase-associated center helix 44 hence impairing the GTPase activity of elongation factor G
We hypothesized that Tre23 transfers ADP-ribose to amine group on a specific base of 23S rRNA, or to its 2′-OH group, similarly to poly-ADP-ribosyltransferases (PARPs), which also belong to the H-Y-E clade of ADP-ribosyltransferases (57). To further understand the molecular basis for Tre23-mediated ribosome inhibition, we first sought to identify the position of the modification on the 23S rRNA. 6-biotin-17-ADP-ribosylated 23S RNAs were fragmented by sonication, and biotinylated fragments were enriched on streptavidin beads and ligated to assemble a library (see Materials and Methods). Library sequencing showed that all fragments correspond to the flexible L11 stalk of the 23S rRNA, also known as thiostrepton loop (TL) (58), and cover the minimal region between bases C1043 and C1123 (Figure 4A, B). Together with the sarcin-ricin loop (SRL), this region of 23S forms the GTPase-associated center (GAC) that serves as binding sites for translation factors (59). To better define the modified site, biotinylated 23S RNAs were incubated with single-stranded complementary DNA fragments covering different sites in the GAC region. After digestion of the single-stranded extremities, RNA-DNA duplex regions were purified and blotted with streptavidin-AP. Figure 4C shows that the protected RNA fragments still bearing the biotin-ADP-ribosylation signal overlap with bases 1095 to 1098, identifying helix 44 of 23S rRNA (Figures 4B, C) as the target site of Tre23. The GAC region constitutes the scaffold for the EF-Tu and EF-G translational GTPases, the elongation factors that catalyze translocation of tRNA and mRNA down the ribosome after each round of polypeptide elongation (59). We thus tested the impact of helix 44 ADP-ribosylation on EF-G GTPase activity. Figure 4E shows that EF-G GTPase activity is severely impaired in presence of toxin-modified ribosomes as compared to control ribosomes. These results indicate that ADP-ribosylation of 23S helix 44 by Tre23 likely inhibits translation elongation by preventing proper EF-G binding, positioning or correct conformation for GTP-hydrolysis. In agreement with the fact that helix 44 is not conserved in the eukaryotic 28S rRNA (60), Tre23 did not have any effect in a rabbit reticulocyte lysate-based transcription-translation assay (Figure 4F), highlighting a remarkable specificity of Tre23 for the bacterial ribosome.
Figure 4.
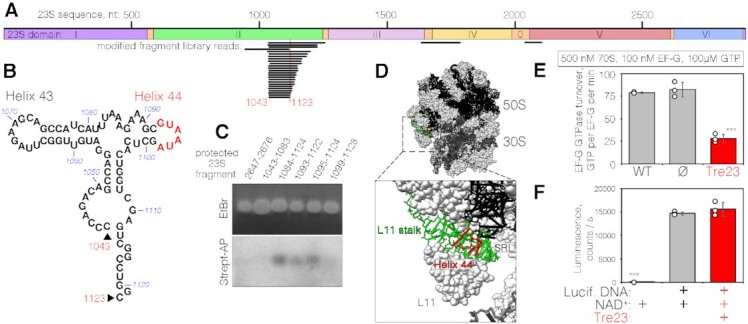
Tre23 ADP-ribosylates helix 44 of 23S rRNA of the GAC center. (A) Mapping of Tre23 ADP-ribosylation by biotinylated fragment enrichment. Sequenced library reads are shown below the diagram representing the architecture of 23S rRNA. The minimal overlapping sequence (nucleotides 1043–1123) is indicated below. (B) Secondary structure of the fragment corresponding to nucleotides 1043–1123. Positions of helices 43 and 44 are indicated, as well as the biotinylated region (red). (C) RNA Protection assay. rRNA-DNA duplexes corresponding to the protected region indicated on top were stained with EtBr (top panel) or detected using streptavidin-AP (bottom panel). (D) Location of the Tre23 ADP-ribosylated 23S region within the 70S ribosome (top, PDB: 6O9J (70)) and a zoom-in on the detected modification position (bottom). The region corresponding to library mapping is shown in green while the modified region, identified by the protection assays, is shown in red. (E) GTPase activity of 100 nM of EF-G was assayed in the presence of 500 nM 70S ribosomes purified from either E. coli MRE600 (WT), E. coli BW25113 cells harboring the empty pNDM220 vector (Ø) or E. coli BW25113 producing Tre23 toxin. Error bars represent standard deviations of the mean of three independent experiments (open circles). *** indicates statistically significant differences between measurements of WT and Tre23-affected ribosomes (unpaired t-test, P <0.0001) or ribosomes purified from cells carrying empty expression vectors and Tre23-affected ribosomes (P = 0.0006). Difference between wild-type and ribosomes from cells carrying empty vector is not statistically significant. (F) Effect of Tre23 on eukaryotic translation in rabbit reticulocyte lysate-based transcription-translation system in presence of DNA encoding the luciferase and purified Tre23 toxin, as indicated. All reactions were supplemented with 0.1 mM NAD+. Luciferase activity is reported in counts per second. Error bars represent standard deviations of the mean of three independent experiments (open circles). *** indicates statistically significant difference between negative control reaction omitting the DNA coding for luciferase and other reactions producing luciferase control in absence or in presence of Tre23 toxin (unpaired t-tests, P < 0.0001). Difference between reactions in absence or in presence of Tre23 toxin is not statistically significant.
CONCLUSIVE REMARKS
In this study, we have identified and characterized Tre23, an antibacterial toxin delivered by the Photorhabdus laumondii T6SS that inhibits translation through ADP-ribosylation of 23S ribosomal RNA. The modification occurs on helix 44 located in the GTPase associated center and affects GTP hydrolysis by the EF-G elongation factor, thus causing arrest of protein synthesis. So far, the only characterized ADP-ribosyl transferases that target protein synthesis are specific to eukaryotic translation; however, they all modify elongation factors, such as diphtheria toxin that ADP-ribosylates the eukaryotic elongation factor eEF2, a orthologue of bacterial EF-G (61). While Tre23 is, to our knowledge, the first secreted antibacterial toxin that targets the 23S, ribosomal RNA is a major target for antibiotics—such as chloramphenicol, lincomycin and clindamycin, and thiopeptides micrococcin, nosiheptide and thiostrepton—as well as toxins, such as ricin or VapC and MazF (62–64). MazF-mt6, VapC-mt20 and VapC-mt26—the toxins of a Mycobacterium tuberculosis MazFE and VapCB toxin-antitoxin pairs—all cleave the 23S (65–67), whereas ricin is a RNA N-glycosidase that depurinates the 23S at a specific position in the SRL (68). By binding to the 23S, thiopeptides prevent proper assembly of the ribosome or recruitment of elongation factors (64). Thiostrepton, nosiheptide and micrococcin are the only antimicrobials that target the GAC by binding to 23S helices 43 and 44 (64,69). While thiostrepton prevents EF-G binding, micrococcin stimulates EF-G GTPase activity by stabilizing the L11-L7 interaction (64). By targeting a highly conserved and functional region of the essential 23S RNA of the ribosome, Tre23 activity may constitute a novel antibacterial strategy. Finally, our data also showed that Tre23 is the C-terminal extension domain of a Rhs polymorphic toxin. In agreement with a putative PAAR motif located at the P. laumondii Rhs N-terminus, pull-down experiments have demonstrated that Rhs forms a complex with the T6SS VgrG spike and the EagR chaperone, suggesting that Tre23 is delivered into target cells by Rhs loading onto the T6SS needle spike. Further experiments will be needed to provide information on the architecture of this complex and on his mounting for delivery.
DATA AVAILABILITY
All data generated or analyzed in this study are included in the published article and its supplementary information.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the Cascales laboratory for discussions, Régine Lebrun and Rémy Puppo (Proteomic facility of the Mediterranean Institute of Microbiology, Marseille) for mass spectrometry analyses, Thierry Doan for microscopy analyses, Bérengère Ize and Olivier Genest for eukaryotic translation measurements, and Moly Ba, Isabelle Bringer, Annick Brun, Audrey Gozzi and Mathilde Valade for technical support.
Notes
Present address: Amaury Payelleville, Cellular and Molecular Microbiology, Faculté des Sciences, Université libre de Bruxelles, Gosselies, Belgium.
Present address: Mohammad Roghanian, Department of Clinical Microbiology, Rigshospitalet, 2200 Copenhagen, Denmark.
Present address: Kathryn J. Turnbull, Department of Clinical Microbiology, Rigshospitalet, 2200 Copenhagen, Denmark.
Contributor Information
Dukas Jurėnas, Laboratoire d’Ingénierie des Systèmes Macromoléculaires (LISM), Institut de Microbiologie, Bioénergies et Biotechnologie (IM2B), Aix-Marseille Université – CNRS, UMR 7255, Marseille, France.
Amaury Payelleville, Laboratoire d’Ingénierie des Systèmes Macromoléculaires (LISM), Institut de Microbiologie, Bioénergies et Biotechnologie (IM2B), Aix-Marseille Université – CNRS, UMR 7255, Marseille, France; DGIMI, Univ Montpellier, INRAE, Montpellier, France.
Mohammad Roghanian, Department of Molecular Biology, Umeå University, 901 87 Umeå, Sweden; Laboratory for Molecular Infection Medicine Sweden, Umeå University, 901 87 Umeå, Sweden.
Kathryn J Turnbull, Department of Molecular Biology, Umeå University, 901 87 Umeå, Sweden.
Alain Givaudan, DGIMI, Univ Montpellier, INRAE, Montpellier, France.
Julien Brillard, DGIMI, Univ Montpellier, INRAE, Montpellier, France.
Vasili Hauryliuk, Department of Molecular Biology, Umeå University, 901 87 Umeå, Sweden; Laboratory for Molecular Infection Medicine Sweden, Umeå University, 901 87 Umeå, Sweden; Department of Experimental Medical Science, Lund University, 221 00 Lund, Sweden; University of Tartu, Institute of Technology, 50411 Tartu, Estonia.
Eric Cascales, Laboratoire d’Ingénierie des Systèmes Macromoléculaires (LISM), Institut de Microbiologie, Bioénergies et Biotechnologie (IM2B), Aix-Marseille Université – CNRS, UMR 7255, Marseille, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique; Aix-Marseille Université; Agence Nationale de la Recherche [ANR-17-CE11-0039]; Fondation pour la Recherche Médicale [FRM, DEQ20180339165]; Fondation Bettencourt Schueller (to E.C.); D.J. was supported by FRM post-doctoral fellowship [SPF201809007142]; V.H. acknowledges financial support by the European Regional Development Fund through the Centre of Excellence for Molecular Cell Technology as well as by the Swedish Research council [2017-03783 to V.H.]; M.R. was supported by MIMS Excellence by Choice Postdoctoral Fellowship Programme grant 2018. Funding for open access charge: ANR [ANR-17-CE11-0039].
Conflict of interest statement. None declared.
REFERENCES
- 1. Stubbendieck R.M., Vargas-Bautista C., Straight P.D.. Bacterial communities: interactions to scale. Front. Microbiol. 2016; 7:1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chassaing B., Cascales E.. Antibacterial weapons: targeted destruction in the microbiota. Trends Microbiol. 2018; 26:329–338. [DOI] [PubMed] [Google Scholar]
- 3. Allsopp L.P., Bernal P., Nolan L.M., Filloux A.. Causalities of war: the connection between type VI secretion system and microbiota. Cell. Microbiol. 2020; 22:e13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stubbendieck R.M., Straight P.D.. Multifaceted interfaces of bacterial competition. J. Bacteriol. 2016; 198:2145–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coulthurst S. The Type VI secretion system: a versatile bacterial weapon. Microbiology (Reading). 2019; 165:503–515. [DOI] [PubMed] [Google Scholar]
- 6. Klein T.A., Ahmad S., Whitney J.C.. Contact-Dependent interbacterial antagonism mediated by protein secretion machines. Trends Microbiol. 2020; 28:387–400. [DOI] [PubMed] [Google Scholar]
- 7. Hernandez R.E., Gallegos-Monterrosa R., Coulthurst S.J.. Type VI secretion system effector proteins: Effective weapons for bacterial competitiveness. Cell. Microbiol. 2020; 22:e13241. [DOI] [PubMed] [Google Scholar]
- 8. Jurėnas D., Journet L.. Activity, delivery, and diversity of Type VI secretion effectors. Mol. Microbiol. 2020; 111:383–394. [DOI] [PubMed] [Google Scholar]
- 9. Ruhe Z.C., Low D.A., Hayes C.S.. Polymorphic toxins and their immunity proteins: diversity, evolution, and mechanisms of delivery. Annu. Rev. Microbiol. 2020; 74:497–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fu Y., Waldor M.K., Mekalanos J.J.. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe. 2013; 14:652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sana T.G., Flaugnatti N., Lugo K.A., Lam L.H., Jacobson A., Baylot V., Durand E., Journet L., Cascales E., Monack D.M.. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E5044–E5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chatzidaki-Livanis M., Geva-Zatorsky N., Comstock L.E.. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wexler A.G., Bao Y., Whitney J.C., Bobay L.-M., Xavier J.B., Schofield W.B., Barry N.A., Russell A.B., Tran B.Q., Goo Y.A.et al.. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:3639–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hecht A.L., Casterline B.W., Earley Z.M., Goo Y.A., Goodlett D.R., Bubeck Wardenburg J.. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep. 2016; 17:1281–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coyne M.J., Comstock L.E.. Type VI secretion systems and the gut microbiota. Microbiol. Spectr. 2019; 7: 10.1128/microbiolspec.PSIB-0009-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duchaud E., Rusniok C., Frangeul L., Buchrieser C., Givaudan A., Taourit S., Bocs S., Boursaux-Eude C., Chandler M., Charles J.-F.et al.. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 2003; 21:1307–1313. [DOI] [PubMed] [Google Scholar]
- 17. Machado R.A.R., Wüthrich D., Kuhnert P., Arce C.C.M., Thönen L., Ruiz C., Zhang X., Robert C.A.M., Karimi J., Kamali S.et al.. Whole-genome-based revisit of Photorhabdus phylogeny: proposal for the elevation of most Photorhabdus subspecies to the species level and description of one novel species Photorhabdus bodei sp. nov., and one novel subspecies Photorhabdus laumondii subsp. clarkei subsp. nov. Int. J. Syst. Evol. Microbiol. 2018; 68:2664–2681. [DOI] [PubMed] [Google Scholar]
- 18. Nielsen-LeRoux C., Gaudriault S., Ramarao N., Lereclus D., Givaudan A.. How the insect pathogen bacteria Bacillus thuringiensis and Xenorhabdus/Photorhabdus occupy their hosts. Curr. Opin. Microbiol. 2012; 15:220–231. [DOI] [PubMed] [Google Scholar]
- 19. Clarke D.J. Photorhabdus: a tale of contrasting interactions. Microbiology (Reading). 2020; 166:335–348. [DOI] [PubMed] [Google Scholar]
- 20. Kupferschmied P., Maurhofer M., Keel C.. Promise for plant pest control: root-associated pseudomonads with insecticidal activities. Front Plant Sci. 2013; 4:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McQuade R., Stock S.P.. Secretion systems and secreted proteins in Gram-negative entomopathogenic bacteria: their roles in insect virulence and beyond. Insects. 2018; 9:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang D., de Souza R.F., Anantharaman V., Iyer L.M., Aravind L.. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct. 2012; 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aravind L., Zhang D., de Souza R.F., Anand S., Iyer L.M.. The natural history of ADP-ribosyltransferases and the ADP-ribosylation system. Curr. Top. Microbiol. Immunol. 2015; 384:3–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jamet A., Nassif X.. New players in the toxin field: polymorphic toxin systems in bacteria. mBio. 2015; 6:e00285-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brackmann M., Nazarov S., Wang J., Basler M.. Using force to punch holes: mechanics of contractile nanomachines. Trends Cell Biol. 2017; 27:623–632. [DOI] [PubMed] [Google Scholar]
- 26. Cherrak Y., Flaugnatti N., Durand E., Journet L., Cascales E.. Structure and activity of the type VI secretion system. Microbiol. Spectr. 2019; 7: 10.1128/microbiolspec.PSIB-0031-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang J., Brodmann M., Basler M.. Assembly and subcellular localization of bacterial type VI secretion systems. Annu. Rev. Microbiol. 2019; 73:621–638. [DOI] [PubMed] [Google Scholar]
- 28. Cianfanelli F.R., Monlezun L., Coulthurst S.J.. Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 2016; 24:51–62. [DOI] [PubMed] [Google Scholar]
- 29. Zoued A., Brunet Y.R., Durand E., Aschtgen M.-S., Logger L., Douzi B., Journet L., Cambillau C., Cascales E.. Architecture and assembly of the Type VI secretion system. Biochim. Biophys. Acta. 2014; 1843:1664–1673. [DOI] [PubMed] [Google Scholar]
- 30. Basler M., Pilhofer M., Henderson G.P., Jensen G.J., Mekalanos J.J.. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012; 483:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alcoforado Diniz J., Coulthurst S.J.. Intraspecies competition in serratia marcescens is mediated by type VI-secreted Rhs effectors and a conserved effector-asociated accessory protein. J. Bacteriol. 2015; 197:2350–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Unterweger D., Kostiuk B., Pukatzki S.. Adaptor proteins of type VI secretion system effectors. Trends Microbiol. 2017; 25:8–10. [DOI] [PubMed] [Google Scholar]
- 33. Donato S.L., Beck C.M., Garza-Sánchez F., Jensen S.J., Ruhe Z.C., Cunningham D.A., Singleton I., Low D.A., Hayes C.S.. The β-encapsulation cage of rearrangement hotspot (Rhs) effectors is required for type VI secretion. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:33540–33548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pei T.-T., Li H., Liang X., Wang Z.-H., Liu G., Wu L.-L., Kim H., Xie Z., Yu M., Lin S.et al.. Intramolecular chaperone-mediated secretion of an Rhs effector toxin by a type VI secretion system. Nat. Commun. 2020; 11:1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Payelleville A., Blackburn D., Lanois A., Pagès S., Cambon M.C., Ginibre N., Clarke D.J., Givaudan A., Brillard J.. Role of the Photorhabdus Dam methyltransferase during interactions with its invertebrate hosts. PLoS One. 2019; 14:e0212655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Quandt J., Hynes M.F.. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993; 127:15–21. [DOI] [PubMed] [Google Scholar]
- 37. Glaeser A., Heermann R.. A novel tool for stable genomic reporter gene integration to analyze heterogeneity in Photorhabdus luminescens at the single-cell level. BioTechniques. 2015; 59:74–81. [DOI] [PubMed] [Google Scholar]
- 38. Brillard J., Duchaud E., Boemare N., Kunst F., Givaudan A.. The PhlA hemolysin from the entomopathogenic bacterium Photorhabdus luminescens belongs to the two-partner secretion family of hemolysins. J. Bacteriol. 2002; 184:3871–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller W.G., Leveau J.H., Lindow S.E.. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 2000; 13:1243–1250. [DOI] [PubMed] [Google Scholar]
- 40. Stojković V., Fujimori D.G.. Radical SAM-mediated methylation of ribosomal RNA. Methods Enzymol. 2015; 560:355–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Takada H., Roghanian M., Murina V., Dzhygyr I., Murayama R., Akanuma G., Atkinson G.C., Garcia-Pino A., Hauryliuk V.. The C-terminal RRM/ACT domain is crucial for Fine-Tuning the activation of ‘Long’ RelA-SpoT homolog enzymes by ribosomal complexes. Front Microbiol. 2020; 11:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kurylo C.M., Alexander N., Dass R.A., Parks M.M., Altman R.A., Vincent C.T., Mason C.E., Blanchard S.C.. Genome sequence and analysis of Escherichia coli MRE600, a colicinogenic, nonmotile strain that lacks RNase I and the Type I methyltransferase, EcoKI. Genome Biol Evol. 2016; 8:742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Turnbull K.J., Dzhygyr I., Lindemose S., Hauryliuk V., Roghanian M.. Intramolecular interactions dominate the autoregulation of Escherichia coli stringent factor RelA. Front. Microbiol. 2019; 10:1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Murina V., Kasari M., Hauryliuk V., Atkinson G.C.. Antibiotic resistance ABCF proteins reset the peptidyl transferase centre of the ribosome to counter translational arrest. Nucleic Acids Res. 2018; 46:3753–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kudrin P., Varik V., Oliveira S.R.A., Beljantseva J., Del Peso Santos T., Dzhygyr I., Rejman D., Cava F., Tenson T., Hauryliuk V.. Subinhibitory concentrations of bacteriostatic antibiotics induce relA-Dependent and relA-Independent tolerance to β-lactams. Antimicrob. Agents Chemother. 2017; 61:e02173-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jana B., Salomon D.. Type VI secretion system: a modular toolkit for bacterial dominance. Future Microbiol. 2019; 14:1451–1463. [DOI] [PubMed] [Google Scholar]
- 47. Monjarás Feria J., Valvano M.A.. An overview of Anti-Eukaryotic T6SS effectors. Front Cell Infect Microbiol. 2020; 10:584751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ahmad S., Tsang K.K., Sachar K., Quentin D., Tashin T.M., Bullen N.P., Raunser S., McArthur A.G., Prehna G., Whitney J.C.. Structural basis for effector transmembrane domain recognition by type VI secretion system chaperones. Elife. 2020; 9:e62816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Poole S.J., Diner E.J., Aoki S.K., Braaten B.A., Roodenbeke C.t’K, Low D.A., Hayes C.S.. Identification of functional Toxin/Immunity genes linked to Contact-Dependent growth inhibition (CDI) and Rearrangement Hotspot (Rhs) systems. PLoS Genet. 2011; 7:e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wenren L.M., Sullivan N.L., Cardarelli L., Septer A.N., Gibbs K.A.. Two independent pathways for self-recognition in Proteus mirabilis are linked by type VI-dependent export. mBio. 2013; 4:e00374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Whitney J.C., Beck C.M., Goo Y.A., Russell A.B., Harding B.N., De Leon J.A., Cunningham D.A., Tran B.Q., Low D.A., Goodlett D.R.et al.. Genetically distinct pathways guide effector export through the type VI secretion system. Mol. Microbiol. 2014; 92:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cianfanelli F.R., Diniz J.A., Guo M., Cesare V.D., Trost M., Coulthurst S.J.. VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog. 2016; 12:e1005735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Koskiniemi S., Lamoureux J.G., Nikolakakis K.C., Roodenbeke C.t’K, Kaplan M.D., Low D.A., Hayes C.S.. Rhs proteins from diverse bacteria mediate intercellular competition. Proc. Natl Acad. Sci. U.S.A. 2013; 110:7032–7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jones C., Hachani A., Manoli E., Filloux A.. An rhs gene linked to the second type VI secretion cluster is a feature of the Pseudomonas aeruginosa strain PA14. J. Bacteriol. 2014; 196:800–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ma J., Sun M., Dong W., Pan Z., Lu C., Yao H.. PAAR-Rhs proteins harbor various C-terminal toxins to diversify the antibacterial pathways of type VI secretion systems. Environ. Microbiol. 2017; 19:345–360. [DOI] [PubMed] [Google Scholar]
- 56. Ting S.-Y., Bosch D.E., Mangiameli S.M., Radey M.C., Huang S., Park Y.-J., Kelly K.A., Filip S.K., Goo Y.A., Eng J.K.et al.. Bifunctional immunity proteins protect bacteria against FtsZ-Targeting ADP-Ribosylating toxins. Cell. 2018; 175:1380–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pinto A.F., Schüler H.. Comparative structural analysis of the putative mono-ADP-ribosyltransferases of the ARTD/PARP family. Curr. Top. Microbiol. Immunol. 2015; 384:153–166. [DOI] [PubMed] [Google Scholar]
- 58. Gonzalo P., Reboud J.-P.. The puzzling lateral flexible stalk of the ribosome. Biol. Cell. 2003; 95:179–193. [DOI] [PubMed] [Google Scholar]
- 59. Allen G.S., Zavialov A., Gursky R., Ehrenberg M., Frank J.. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005; 121:703–712. [DOI] [PubMed] [Google Scholar]
- 60. Doris S.M., Smith D.R., Beamesderfer J.N., Raphael B.J., Nathanson J.A., Gerbi S.A.. Universal and domain-specific sequences in 23S-28S ribosomal RNA identified by computational phylogenetics. RNA. 2015; 21:1719–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Simon N.C., Aktories K., Barbieri J.T.. Novel bacterial ADP-ribosylating toxins: structure and function. Nat. Rev. Microbiol. 2014; 12:599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hong W., Zeng J., Xie J.. Antibiotic drugs targeting bacterial RNAs. Acta Pharm Sin B. 2014; 4:258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Douthwaite S. Interaction of the antibiotics clindamycin and lincomycin with Escherichia coli 23S ribosomal RNA. Nucleic Acids Res. 1992; 20:4717–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Harms J.M., Wilson D.N., Schluenzen F., Connell S.R., Stachelhaus T., Zaborowska Z., Spahn C.M.T., Fucini P.. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell. 2008; 30:26–38. [DOI] [PubMed] [Google Scholar]
- 65. Schifano J.M., Edifor R., Sharp J.D., Ouyang M., Konkimalla A., Husson R.N., Woychik N.A.. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8501–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Winther K.S., Brodersen D.E., Brown A.K., Gerdes K.. VapC20 of Mycobacterium tuberculosis cleaves the sarcin-ricin loop of 23S rRNA. Nat. Commun. 2013; 4:2796. [DOI] [PubMed] [Google Scholar]
- 67. Winther K., Tree J.J., Tollervey D., Gerdes K.. VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res. 2016; 44:9860–9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Grela P., Szajwaj M., Horbowicz-Drożdżal P., Tchórzewski M.. How ricin damages the ribosome. Toxins. 2019; 11:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Polikanov Y.S., Aleksashin N.A., Beckert B., Wilson D.N.. The mechanisms of action of ribosome-targeting peptide antibiotics. Front Mol Biosci. 2018; 5:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kaledhonkar S., Fu Z., Caban K., Li W., Chen B., Sun M., Gonzalez R.L., Frank J.. Late steps in bacterial translation initiation visualized using time-resolved cryo-EM. Nature. 2019; 570:400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed in this study are included in the published article and its supplementary information.


