Abstract
The mitochondrion-associated RNase P activity (mtRNase P) was extensively purified from HeLa cells and shown to reside in particles with a sedimentation constant (∼17S) very similar to that of the nuclear enzyme (nuRNase P). Furthermore, mtRNase P, like nuRNase P, was found to process a mitochondrial tRNASer(UCN) precursor [ptRNASer(UCN)] at the correct site. Treatment with micrococcal nuclease of highly purified mtRNase P confirmed earlier observations indicating the presence of an essential RNA component. Furthermore, electrophoretic analysis of 3′-end-labeled nucleic acids extracted from the peak of glycerol gradient-fractionated mtRNase P revealed the presence of a 340-nucleotide RNA component, and the full-length cDNA of this RNA was found to be identical in sequence to the H1 RNA of nuRNase P. The proportions of the cellular H1 RNA recovered in the mitochondrial fractions from HeLa cells purified by different treatments were quantified by Northern blots, corrected on the basis of the yield in the same fractions of four mitochondrial nucleic acid markers, and shown to be 2 orders of magnitude higher than the proportions of contaminating nuclear U2 and U3 RNAs. In particular, these experiments revealed that a small fraction of the cell H1 RNA (of the order of 0.1 to 0.5%), calculated to correspond to ∼33 to ∼175 intact molecules per cell, is intrinsically associated with mitochondria and can be removed only by treatments which destroy the integrity of the organelles. In the same experiments, the use of a probe specific for the RNA component of RNase MRP showed the presence in mitochondria of 6 to 15 molecules of this RNA per cell. The available evidence indicates that the levels of mtRNase P detected in HeLa cells should be fully adequate to satisfy the mitochondrial tRNA synthesis requirements of these cells.
The unique mode of transcription of the mammalian mitochondrial DNA in the form of giant polycistronic molecules, containing tRNA sequences regularly interspersed between the individual rRNA and mRNA sequences and in most cases butt-joined to them (39, 42, 43), demands the existence of a complex RNA-processing apparatus. The tRNA sequences presumably function as signals for RNA-processing enzymes, which carry out the endonucleolytic cleavages that eventually lead to the formation of the mature rRNA, mRNA, and tRNA species (43). One of these enzymatic activities would be expected to cut precisely the polycistronic transcripts on the 5′ side of each tRNA sequence and therefore to be analogous to the RNase P, an RNA-containing enzyme first identified in Escherichia coli (3), and subsequently found ubiquitously in prokaryotic and eukaryotic organisms (4). In previous work from this laboratory, an endoribonuclease which cleaves the precursor of the E. coli suppressor tRNATyr at the same site as E. coli RNase P, producing the mature 5′ end of this tRNA, has been identified. The enzyme was partially purified from HeLa cell mitochondria (and is henceforth referred to as mtRNase P) (15). An analogous enzyme activity occurs in HeLa cell nuclei, and some of it leaks to the cytosol during cell fractionation (15, 21). This activity will be referred to as nuRNase P. Enzymatic activities involved in HeLa cell mitochondrial tRNA processing, in particular an RNase P-like activity, a 3′-tRNA precursor-processing endonuclease, and an ATP (CTP)-tRNA-specific nucleotidyltransferase, have been described and partially characterized by another group (51). A 5′-tRNA precursor-processing endonuclease with similar properties to those of the HeLa cell enzyme, as well as a 3′-tRNA precursor-processing endonuclease, had been previously reported to occur also in rat liver mitochondria (37).
The HeLa cell mtRNase P identified in this laboratory exhibited sensitivity to micrococcal nuclease (MN) and pronase, indicating that this enzyme, like the prokaryotic and eukaryotic counterparts (4), has essential RNA and protein components (15). These must be encoded in the nucleus, since the functions of all RNA species and proteins encoded in human mtDNA have been identified (6). Surprisingly, the second report of a HeLa cell mitochondrion-associated RNase P activity claimed that this is MN resistant but contains degraded RNA molecules (51, 52). In the present work, the analysis of the RNA species associated with an mtRNase P preparation extensively purified from HeLa cell mitochondria has allowed the identification of a 340-nucleotide (nt) RNA species identical in sequence to the H1 RNA component of the HeLa cell nuRNase P (9). After normalization for the recovery of four different mitochondrial RNA and DNA markers in HeLa cell mitochondria purified by different treatments, a fraction of the cell H1 RNA corresponding to ∼33 to ∼175 intact molecules per cell in variously treated mitochondria has been estimated to be intrinsically associated with these organelles.
MATERIALS AND METHODS
Isolation and treatment of mitochondria.
A 5,000 × gav mitochondrial fraction was isolated, as previously described (33), from late-exponential-phase HeLa cells (5 × 109 to 1 × 1010 cells) and extensively washed. The final pellet was resuspended at 25°C in one-quarter of the volume of packed cells used for isolation of mitochondria in 0.25 M sucrose–20 mM Tris-HCl (pH 8.0)–0.1 mM EDTA–1 mM dithiothreitol (DTT)–0.2 mM phenylmethylsulfonyl fluoride (PMSF) (buffer A) containing 2 mM EGTA and washed twice in the above buffer. Further treatments were carried out as described below.
(i) D treatment.
Digitonin (D), purified as previously described (32), was added, at 90 μg per mg of mitochondrial protein, to the mitochondrial suspension, which was then gently stirred on ice for 15 min. The mitoplasts were sedimented at 12,600 × g for 15 min, resuspended in buffer A, and repelleted twice under the same conditions.
(ii) MN treatment.
To the mitochondrial suspension in buffer A, CaCl2 was added to 2 mM and MN was added to 300 U/ml, unless otherwise specified. After 30 min on ice, the MN was inactivated by the addition of 10 mM EGTA to chelate the available Ca2+. The mitochondrial suspension was then diluted with 3 volumes of buffer A containing 2 mM EGTA and centrifuged at 12,600 × gav for 15 min. The resulting mitochondrial pellet was washed twice in buffer A plus 2 mM EGTA.
(iii) D-plus-MN treatment.
The D-treated mitochondria were resuspended in buffer A and treated with MN as described above.
The final mitochondrial pellet from each of the treatments described above was resuspended in a suitable volume of 20 mM Tris-HCl (pH 8.0)–10 mM MgCl2–1 mM EDTA–1 mM DTT–0.2 mM PMSF–10% glycerol (buffer B), supplemented with 0.25 mM KCl, and stored at −20°C.
Isolation of the postmitochondrial fraction.
The supernatant after the initial pelleting of the crude mitochondrial fraction from ∼30 ml of packed cells was centrifuged for 30 min at 20,000 × gav. The supernatant, designated the postmitochondrial S20 fraction (pmS20), was carefully decanted; adjusted to 20 mM Tris-HCl (pH 8.0), 20 mM KCl, 1 mM EDTA, and 20% glycerol; and stored at −70°C.
Purification of mtRNase P and nuRNase P.
Each frozen mitochondrial suspension was quickly thawed, placed on ice, and homogenized in an Elvejem-Potter homogenizer. The nonionic detergent NP-40 was then added to 2% (wt/vol), and the suspension was rehomogenized and centrifuged for 1 h at 100,000 × gav. The pellet was resuspended in approximately one-half volume of the first lysate in buffer B plus 0.25 M KCl, NP-40 was added to 2%, and the suspension was homogenized and centrifuged as described above. The two supernatants were combined, diluted with 10 volumes of buffer B, and applied to a 50-ml DEAE-cellulose column (DE52; Whatman) equilibrated with buffer B containing 0.2% NP-40 and 20 mM KCl. Following sample application, the column was sequentially washed with 2 column volumes of buffer C (buffer B plus 0.05% NP-40) containing 20 mM KCl and 3 column volumes of buffer C containing 0.07 M KCl. The bound material was further eluted with a 600-ml linear gradient of 0.07 to 0.6 M KCl in buffer C. Fractions (6 ml) were collected and assayed for RNase P activity as described below, with the KCl concentration in the reaction mixtures varying between ∼80 and 125 mM. The active fractions were pooled, diluted with buffer C to 100 mM KCl, and then loaded onto a 10-ml DEAE-Sepharose column equilibrated with buffer C plus 100 mM KCl. The RNase P activity was eluted, using 60 ml of buffer C plus 0.6 M KCl, in 1-ml fractions.
The active fractions from the DEAE-Sepharose column were combined, dialyzed against buffer C plus 100 mM KCl, and loaded onto a fast performance liquid chromatography (FPLC) Mono-S column, and RNase P activity was eluted in 1-ml fractions with a 0.1 to 0.6 M KCl gradient in buffer C. No activity was detected in a higher-salt (1.0 M KCl) wash. The RNase P activity recovered from the Mono-S column appeared in both the unbound (US) fractions and bound (BS) fractions. The active US and BS fractions were pooled separately, adjusted to 0.1 M KCl, and loaded onto columns equilibrated with buffer C containing 0.1 M KCl, and the bound material on each column was eluted with a 0.1 to 0.6 M KCl gradient in buffer C. The active fractions derived from the US and BS pools which were eluted from the Mono-Q columns were pooled (UQ and BQ fractions), and 1-ml portions were overlaid on glycerol gradients (15 to 35%). These were made in 20 mM Tris-HCl (pH 8.0)–75 mM KCl–5 mM MgCl2–0.1 mM EDTA–0.25 mM DTT–0.05 M PMSF–0.05% NP-40. The glycerol gradients were centrifuged on an SW41 rotor at 36,000 rpm (∼150,000 × gav) for 16 h at 5°C and decelerated without brake. Fractions of 350 to 700 μl were collected from the bottom and assayed for RNase P activity.
For the purification of the nuRNase P, the frozen pmS20 was thawed and distributed in 20-ml aliquots, which were then adjusted to 10 mM MgCl2 and 0.5% NP-40 and centrifuged for 1 h at 100,000 × gav. The supernatant (pmS20 S100) was applied directly onto a 50-ml column of DEAE-cellulose equilibrated with buffer B plus 0.5% NP-40 and 20 mM KCl. Sequential chromatography through DEAE-cellulose, DEAE-Sepharose, FPLC Mono-S, and FPLC Mono-Q columns and centrifugation through a glycerol gradient were carried out as described above for mtRNase P.
RNA-processing assays.
RNase P activity during purification of mtRNase P and nuRNase P was measured as described previously (15), using a precursor of E. coli suppressor tRNATyr (ptRNATyr) transcribed in vitro, in the presence of [α−32P]CTP, with SP6 RNA polymerase from an artificial gene cloned in the pGEM-1 vector (Promega).
To test the capacity of mitochondrial precursors to function as substrates for the mtRNase P and nuRNase P, an artificial mitochondrial tRNASer(UCN) precursor [ptRNASer(UCN)] was cloned in the vector pGEM.4Z (Promega). For this purpose, a segment of HeLa cell mtDNA encompassing the whole tRNASer(UCN) coding sequence assigned in the Cambridge sequence (between positions 7445 and 7516) (5) and a 69-bp stretch upstream of it in the direction of L-strand transcription (between positions 7517 and 7585) was amplified by PCR. In particular, for the purpose of cloning this fragment and at the same time creating a 3′-terminal -CCA (57), a 3′-end 34-nt oligodeoxyribonucleotide, encompassing the 3′-end-proximal 22 nt assigned in the Cambridge sequence to the tRNASer(UCN) with an additional 13 nt at its 3′ end containing overlapping BamHI and BstNI sites (5′GCGC GGAT CCTGG3′), and a 5′-end 33-nt oligodeoxyribonucleotide, encompassing the 25-nt mtDNA segment between positions 7561 and 7585 with an additional 10 nt at its 5′ end containing an EcoRI site (5′ ATCG GAATTC 3′), were used as primers. The amplified PCR product was digested with EcoRI and BamHI and cloned in similarly digested pGEM.4Z. The sequence of the cloned product was verified by DNA sequencing. The plasmid was cut with the enzyme BstNI and transcribed with SP6 RNA polymerase in the presence of [α-32P]CTP or [35S]CTP. The 32P-labeled product, expected to contain the encoded tRNASer(UCN) (72 nt in length on the basis of the Cambridge sequence [5]), with an added 3′-terminal -CCA (57) and an 81-nt extension at its 5′ end, was purified by electrophoresis in a 5% polyacrylamide–7 M urea gel and processed with mtRNase P, using the samples of UQ-fractionated mtRNase P that exhibited activity with ptRNATyr as substrate or glycerol gradient-fractionated nuRNase P. The reaction products were proteinase K digested, phenol extracted, ethanol precipitated, and analyzed on a 5% polyacrylamide–7 M urea gel. The UQ fraction which exhibited peak activity and the nuRNase P were also used for processing a [35S]CTP-labeled ptRNASer(UCN). A portion of the products of this reaction was analyzed on a gel to verify the processing reaction, and the remaining portion was utilized for primer extension (63). In particular, to map the processing site, primer extension was carried out on the SP6 transcript processed by mtRNase P or nuRNase P, using reverse transcriptase (Stratagene) and Ser-oligo 1 (L-strand 5′ [7445] → 3′ [7468]) and Ser-oligo 2 (L-strand 5′ [7465] → 3′ [7488]) as primers.
Nucleic acid extraction.
For total nucleic acid extraction, whole cells or the mitochondrial fractions were treated at 37°C for 1 h with proteinase K (350 μg/ml) in a buffer containing 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5 mM EDTA, and 0.5% sodium dodecyl sulfate (SDS). The samples were then extracted three times with phenol-chloroform-isoamyl alcohol (25:24:1), and the nucleic acids were precipitated twice with ethanol. Total RNA was isolated by the acid guanidinium thiocyanate-phenol-chloroform extraction method (12).
RNA analysis.
The electrophoretic properties of the RNA components associated with glycerol gradient-purified mtRNase P and nuRNase P were analyzed by labeling the RNA with [32P]pCp and RNA ligase (17) and fractionating it on a 5% polyacrylamide–7 M urea gel. For RNA-sequencing analysis, individual components were eluted from the gel, phenol-chloroform extracted, and subjected to digestion with different RNases (RNase A [C+U specific], RNase T1 [G specific], RNase PhyM [A+U specific], and RNase CL3 [C specific]) under the conditions recommended by the supplier (Bethesda Research Laboratories), and the products of digestion were resolved by electrophoresis on a 5% polyacrylamide–7 M urea sequencing gel.
RNA transfer hybridization analysis to detect mtRNase P, MRP RNase, and U2 and U3 RNAs was carried out using complementary oligodeoxyribonucleotides labeled at their 5′ ends with [γ-32P]ATP and polynucleotide kinase (Promega). The RNAs were resolved on 5% polyacrylamide–7 M urea gels, electrotransferred to nylon membranes (Immobilon-N; Millipore Corp., Bedford, Mass.), and then fixed to the membrane by baking the filters at 80°C under vacuum for 90 min. Prior to prehybridization, the filters were washed for 1 h at 50°C with 0.1× SSPE (1× SSPE is 180 mM NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) containing 0.2% SDS. Prehybridization was carried out at 50°C for 4 h in 5× SSPE–5× Denhardt's solution (1× Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% bovine serum albumin) containing 150 μg of denatured salmon sperm DNA per ml and 0.2% SDS. Hybridization was initiated by adding the 32P-labeled specific oligodeoxyribonucleotide at ∼5 × 106 cpm/ml. After 16 to 18 h at 50°C, the filters were sequentially washed at 50°C for 30 min each with 5×, 2×, and 0.4× SSPE containing 0.2% SDS. The following oligodeoxyribonucleotides were used: 5′-CCTTCCCAAGGGACATGGGAGTGGAGTG-3′ (H1 RNA-specific probe)(9), 5′-GTAACTAGAGGGAGCTGACGGATGACGCCCCCG-3′ (MRP RNA-specific probe) (22), 5′-GAGTGGACGGAGCAAGCTCCTATTCCATCTCC-3′ (U2 RNA-specific probe) (49), and CGCTACCTCTCTTCCTCGTGGTTTTCGGTGCTCTACA (U3 RNA-specific probe) (49).
Hybridization with the probes listed above was carried out either simultaneously with more than one probe or sequentially, after stripping the blot, as specified in the text and legend to Fig. 6. Stripping of the blot was performed by incubating it at 70°C for 2 h in 50 ml of 0.1× SSPE containing 0.2% SDS, with a change of buffer after the first hour.
FIG. 6.
Quantification, by transfer hybridization analysis, of H1 RNA, MRP RNA, U2 RNA, U3 RNA, mitochondrial DNA, 12S rRNA, ND1 mRNA, and COII mRNA in whole HeLa cells and variously treated mitochondrial fractions. A mitochondrial fraction was prepared from 44 ml of packed HeLa cells and subjected to various treatments, as described in Materials and Methods. (a and b) The indicated amounts of nucleic acids from total HeLa cells and from washed (C) or MN-, D-, or D-plus-MN-treated mitochondrial (Mit.) fractions and 1 or 5 ng of H1 cDNA (NuH1) digested with EcoRI and BamHI were run on a 5% polyacrylamide–7 M urea gel, electrotransferred to a nylon membrane, and hybridized simultaneously with γ-32P-labeled oligodeoxynucleotide probes specific for H1 RNA and MRP RNA (a). After appropriate exposure and quantification of the bands by PhosphorImager analysis, the blot was stripped and rehybridized sequentially with γ-32P-labeled oligodeoxynucleotides specific for U3 and U2 RNA (b). In other experiments, the hybridization with the U3 and U2 RNA probes was carried out on independent blots, with similar results. (c) The indicated amounts of nucleic acids from total-cell (T) or from washed (C) or MN-, D-, or D-plus-MN-treated mitochondrial fractions (Mit. fr.) were digested with RNase A and with EcoRV, fractionated on a 1% agarose gel, transferred to a nylon membrane, and hybridized with HeLa cell mitochondrial DNA 32P labeled by random priming. (d to f) The indicated amounts of total nucleic acids from total cells or variously treated mitochondrial fractions (Mit. fr.) were fractionated on formaldehyde–1.2% agarose gels, transferred to nylon membranes, and hybridized with 32P-labeled probes specific for COII (HCOII-pGEM1-α) (d), 12S rRNA (p12ssf) (e), or ND1 mRNA (pKS2ND1) (f). M, HinfI-digested and 3′-end-labeled pBR322 marker; RNA 19, processing intermediate of heavy-strand transcripts containing sequences of 16S rRNA, tRNALeu(UUR), and ND1 mRNA (58).
To provide a marker for quantification of the total cell- or mitochondrion-associated H1 RNA, plasmid NuH1, containing the complete nuH1 cDNA sequence in the vector pGEM.4Z (9), was digested with EcoRI and BamHI, and the insert was purified using Geneclean II (Bio 101, La Jolla, Calif.) and quantified spectrophotometrically. To provide an RNA marker for the same purpose, NuH1 was cut with EcoRI and transcribed with the T7 RNA polymerase, and the product was then DNase treated, phenol-chloroform extracted, purified by electrophoresis in a 5% urea–polyacrylamide gel, phenol-chloroform extracted, and quantified spectrophotometrically.
For RNA transfer hybridization analysis of mitochondrial ND1, 12S, and COII RNAs, nucleic acids from the various samples were fractionated by electrophoresis through formaldehyde–1.2% agarose gels (13) and transferred to nylon membranes under standard conditions, as described previously (55). Prehybridization of the membranes, previously washed with 0.1× SSPE–0.2% SDS at 50°C for 1 h, was carried out for 4 h at 42°C in a solution containing 50% deionized formamide, 5× SSPE, 0.5% SDS, and 150 μg of salmon sperm DNA per ml. Specific probes for 12S rRNA, ND1 mRNA, and COII mRNA were labeled by random priming (19), using [α-32P]CTP, of plasmids containing a 12S rRNA gene fragment (between positions 764 and 1466) (5) (p12SSf), an ND1 gene fragment (between positions 3312 and 4121) (pKS2ND1), or a COII gene fragment (between positions 7441 and 8287) (HCOII-pGEM1-α). These probes were added at 2 × 106 to 3 × 106 cpm/ml to the prehybridization solution, the solution was supplemented with dextran sulfate to a final concentration of 10%, and hybridization was carried out at 42°C for 18 to 20 h. The filters were washed at room temperature three times with 5× SSPE–0.2% SDS and thereafter sequentially at 65°C for 30 min each with 1× (twice), 0.5×, and 0.2× SSPE containing 0.2% SDS. In all hybridization experiments described in this section, the intensities of the bands were quantified using a PhosphorImager (Molecular Dynamics) and the ImageQuant program.
Southern blot analysis of mitochondrial DNA.
Nucleic acids from the various samples were digested with EcoRV and then with RNase A and resolved by electrophoresis through a 1% agarose gel in TBE (89 mM Tris-borate [pH 8.3] [25°C], 2 mM disodium EDTA). Southern transfer onto a nylon membrane was performed as described previously (55). The filters were UV cross-linked in a Stratagene UV cross-linker and washed as described above. Prehybridization was carried out at 65°C for 4 h using a medium containing 5× SSPE, 2× Denhardt's solution, 10% dextran sulfate, 150 μg of salmon sperm DNA per ml, and 0.5% SDS. Hybridization was performed in the same solution containing 1.5 × 106 to 2 × 106 cpm of purified mtDNA from HeLa cells per ml, labeled by random priming with [α-32P]CTP. After an 18-h incubation at 65°C, the filters were washed, and the intensities of the bands were quantified as detailed above for the ND1, 12S rRNA, and COII blots.
Cloning and sequencing of the cDNA of mtRNase P H1 RNA.
The H1 RNA contained in total RNA extracted from D-plus-MN-treated mitochondria was reverse transcribed and PCR amplified using two appropriate primers carrying an EcoRI site or a BamHI site, digested with EcoRI and BamHI, and cloned directionally in the pGEM.4Z vector (Promega) cut with EcoRI and BamHI. The DNA was sequenced completely on both strands from three independent clones using the Sequenase enzyme system (U.S. Biochemical).
Other assays.
The distribution of bovine liver catalase (Sigma) after sedimentation in a glycerol gradient was determined by spectrophotometric analysis at 405 nm (56). Protein was measured with the Coomassie Plus protein assay reagent, as recommended by the manufacturer (Pierce, Rockford, Ill.).
RESULTS
Purification of the mtRNase P from a D-treated or D-plus- MN-treated mitochondrial fraction.
In the present work, a HeLa cell mitochondrial fraction isolated by centrifugation at 5,000 × gav for 10 min (33), which is enriched in “heavy mitochondria,” transcriptionally the most active organelles (61), was used throughout this study. In our previous work (15), a crucial step in the purification of mtRNase P was the treatment of the mitochondrial fraction with a high concentration of MN to destroy any contaminating nuRNase P activity. In the present experiments, the purification procedure was modified by replacing, or by preceding or following, the MN treatment of the mitochondrial fraction with D treatment. The latter treatment was aimed at disrupting and partially solubilizing the outer mitochondrial membrane, as well as at lysing other membranaceous structures contaminating the mitochondrial fraction, in particular cytoplasmic vesicles trapping cytosolic components and lysosomes, which carry potentially harmful nucleases. The extensive washing of the D-resistant structures was expected to largely eliminate the soluble nucleases.
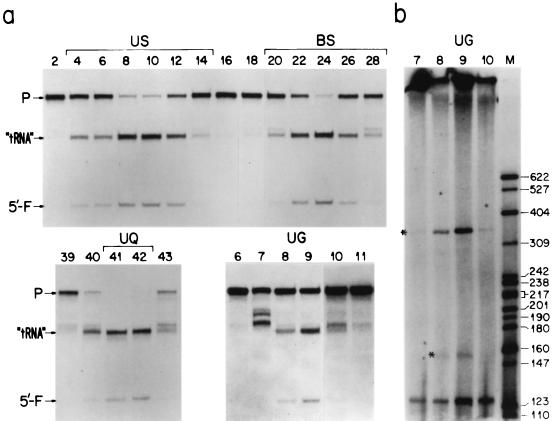
In the subsequent purification of the enzyme, after the DEAE-cellulose chromatography, the previously used octyl-Sepharose chromatography step, which gave only a low recovery, was replaced by sequential chromatography through a DEAE-Sepharose column, a Mono-S FPLC column, and a Mono-Q FPLC column, followed by sedimentation through a 15 to 35% glycerol gradient. The purification scheme and the detection of the RNase P activity after each step, in a representative experiment involving a simple D treatment of the mitochondrial fraction, are shown in Fig. 1. In these experiments, as a substrate for the RNase P assay, an in vitro-transcribed E. coli ptRNATyr was used (see Materials and Methods). The RNase P activity eluted from a DEAE-cellulose column (between 140 and 190 mM KCl) was concentrated by binding to a DEAE-Sepharose column and elution in a single step with 0.6 M KCl and then passed through a Mono-S FPLC column (Fig. 1a). As shown in Fig. 1b, a major portion (∼60%) of the RNase P activity did not bind to the Mono-S column and was recovered in the flowthrough. The retained portion of the activity was eluted between 140 and 190 mM KCl. Both the US and the BS activities were completely retained on a Mono-Q FPLC column and were eluted as fairly sharp peaks at between 0.35 and 0.4 M KCl. In the fractions flanking the peak of activity in the BQ fraction and, much less pronounced, in the UQ fraction, there was evidence of a weak cleavage of the substrate upstream of the canonical cleavage (Fig. 1b). This was presumably due to a contaminant activity. When the fractions UQ and BQ were run through 15 to 35% glycerol gradients, the RNase P activity in both fractions sedimented as a fairly sharp peak to the same fraction volume (Fig. 1b). In the purification scheme illustrated above, no RNase P activity on ptRNATyr was detected in any fractions other than those described, and the activities of the U and B fractions were identical.
FIG. 1.
Purification of mtRNase P from HeLa cell mitoplasts. (a) Fractionation scheme. (b) 5′-End processing of in vitro-synthesized tRNATyr uniformly labeled with [α-32P]CTP by 50-μl samples of fractions from Mono-S and Mono-Q chromatography and glycerol gradient centrifugations. The D-treated mitochondrial fraction from 70 ml of packed HeLa cells was lysed with 2% NP-40, and the S100 supernatant of the lysate was run through a DEAE-cellulose column. The eluted RNase P activity was concentrated on a DEAE-Sepharose column, and the active fractions from this fraction were loaded on a Mono-S FPLC column. After collection of the flowthrough and the buffer wash, a 0.1 to 0.6 M KCl gradient was applied. Fractions 2 to 14, containing unbound activity (US), and fractions 20 to 24, containing bound activity eluted between 0.14 and 0.19 M KCl (BS), were pooled separately and each loaded onto a Mono-Q FPLC column at 0.1 M KCl. In both cases, activity was eluted as a fairly sharp peak between 0.35 and 0.4 M Cl (UQ and BQ) and fractionated through a 15 to 35% linear glycerol gradient (UG and BG). See Materials and Methods for details. S, substrate; P, precursor; “tRNA,” 5′-end-cleaved tRNATyr precursor; 5′-F, 5′ fragment of precursor cleaved off by RNase P.
Results similar to those shown above were obtained in six other experiments in which the purification scheme of mtRNase P involved either D-prepared or D-plus-MN-prepared mitoplasts. The distribution of RNase P activity in a glycerol gradient, after purification of the enzyme from D-plus- MN-treated mitochondria by the scheme illustrated in Fig. 1a (fraction BQ), is shown in Fig. 2a. The results of an experiment in which mtRNase P samples from the UQ fractions of D-treated and D-plus-MN-treated mitochondria were run in a glycerol gradient, in parallel with bovine liver catalase, are shown in Fig. 3. Using a value of 11.2S for the sedimentation constant of bovine liver catalase (56), a value of ∼17S was calculated for mtRNase P. In a parallel experiment, the same sedimentation constant was determined for the mtRNase P derived from the BQ fraction of D-plus-MN-treated mitochondria.
FIG. 2.
RNA associated with purified mtRNase P. The mitochondrial fraction was isolated from 64 ml of packed HeLa cells, D treated, and then MN treated, and the mtRNase P was purified from this preparation following the scheme of Fig 1a. (a) Enzyme activity distribution after glycerol gradient centrifugation of the BQ fraction. (b) RNA species isolated by proteinase K digestion, phenol extraction, and ethanol precipitation from the individual glycerol gradient fractions corresponding to the peak of enzyme activity and side fractions, 3′-end labeled with [32P]pCp and RNA ligase, and run on a 5% polyacrylamide–7 M urea gel. M, MspI-digested and 3′-end-labeled pBR322 DNA marker; other symbols as in Fig. 1. The asterisks indicate the 340-and ∼155-nt RNA species.
FIG. 3.
Sedimentation properties of mtRNase P. Two equal samples (1 ml) of the UQ fraction from D-treated mitochondria (■) and two equal samples (1 ml) of the UQ fraction from D-plus-MN-treated mitochondria (□) were run in parallel with two samples of bovine liver catalase (1 ml of a 1-mg/ml solution) through 15 to 35% glycerol gradients. After the enzyme activity in the individual fractions of the mtRNase P gradients and the absorbance at 405 nm (Abs405) of the fractions of the catalase gradients were determined, the combined values of the activities of the two mtRNase P samples derived from D-treated mitochondria and of the two samples derived from D-plus-MN-treated mitochondria and the values of absorbance of the two catalase samples were plotted against migration.
An RNA component is associated with highly purified mtRNase P.
Previous work had shown that the HeLa cell mtRNase P activity, partially purified by sequential DEAE-cellulose and octyl-Sepharose chromatography, was sensitive to pretreatment with MN, suggesting the presence of an essential RNA component. In the present work, the enzyme retained on DEAE-cellulose and the UQ and BQ fractions were also tested and found to be completely MN sensitive. It has, however, been reported that in a crude in vitro system from spinach chloroplasts, the sensitivity of a pre-tRNA 5′-end-processing activity to MN does not reflect the existence of an RNA component in the enzyme but rather the capacity of EGTA-inactivated MN to form a complex with the pre-tRNA substrate, which thus becomes inaccessible to the enzyme (64). To test this possibility, a series of assays were carried out on highly purified mtRNase P as shown in Fig. 1 (fraction UG in Fig. 1b). In agreement with the previous observations (15), pretreatment of the enzyme with MN in the presence of Ca2+, but not pretreatment in the absence of Ca2+ or pretreatment with Ca2+ alone, nearly completely inhibited pre-tRNA processing activity, as tested in the presence of EGTA. That the inhibition of RNase P activity was not due to a complex formation between EGTA-inactivated MN and the pre-tRNA substrate was strongly suggested by an experiment in which preincubation of the mtRNase P with MN in the presence of EGTA reduced only slightly (by∼20%) tRNA-processing activity during the subsequent exposure of the RNase P to the substrate (data not shown). This small decrease in RNase P activity in the last experiment may be due to the presence in the mtRNase P preparation of a contaminating protease or nuclease. Similar results were obtained when the mtRNase P from the BG fraction was analyzed.
To investigate the nature of the RNA species associated with the mtRNase P, the two glycerol gradient fractions corresponding to the peak of RNase P activity of the BQ fraction (Fig. 2a) and one fraction on each side of the peak were individually dialyzed. Their components were ethanol precipitated in the presence of 20 μg of mussel glycogen, proteinase K treated, and phenol extracted, and the RNA was then 3′-end -labeled with [32P]pCp and T4 RNA ligase (17) and fractionated by electrophoresis in a 5% polyacrylamide–7 M urea gel. As shown in Fig. 2b, the labeled RNA extracted from the two fractions corresponding to the peak of RNase P activity exhibited a band with the mobility expected for a species of ∼340 nt and a band of similar abundance corresponding to a size of ∼155 nt. Some higher-molecular-weight components in the same fractions presumably resulted from ligation of the smaller components, as strongly suggested by their comigration with the ∼340-nt and ∼155-nt species. In addition, there was labeling of RNA species of 120 to 125 nt, which were, however, also present in the side fractions that did not exhibit any RNase P activity. The 340-nt RNA species corresponded in size to the H1 RNA species, which is associated with the nuRNase P (9). Furthermore, it seemed possible that the ∼155-nt RNA species corresponded to the 170-nt RNA species H2a, previously found to copurify with the H1 RNA and to represent the 3′-end half of this RNA resulting from its breakdown (9).
After glycerol gradient centrifugation of the UQ fraction, the RNA from the active fractions, labeled with [32P]pCp and T4 RNA ligase, exhibited a very similar electrophoretic pattern to that of the BQ fraction RNA (data not shown).
Purification of nuRNase P.
To investigate how the nuRNase P behaved during the various steps of the purification procedure used for mtRNase P, the same procedure was used for its isolation. As shown in Fig. 4a, the nuRNase P activity present in the postmitochondrial supernatant (pmS20), and separated by DEAE-cellulose and DEAE-Sepharose chromatography, was eluted from a Mono-S FPLC column similarly to the mtRNase P activity and was largely (∼55%) recovered in the flowthrough of the column (fraction US); a minor part was retained and eluted between 0.13 M and 0.19 M KCl (fraction BS). Both the US and BS activities were retained on a Mono-Q FPLC column and were eluted between 0.35 and 0.4 M KCl, as was the mtRNase P activity. When the UQ fraction of nuRNase P was run in a 15 to 35% glycerol gradient, it sedimented similarly to its mitochondrial counterpart (Fig. 4a). A sedimentation constant of ∼16S was estimated in a glycerol gradient centrifugation run using catalase as a sedimentation marker (data not shown). Figure 4b shows the analysis in a 5% polyacrylamide–7 M urea gel of the RNA components extracted from the two individual glycerol gradient fractions corresponding to the peak of RNase P activity and from one fraction on each side and labeled with [32P]pCp and T4 RNA ligase. The two fractions of the peak of RNase P activity showed a strong band corresponding to a 340-nt RNA species and a weak band corresponding to a ∼160-nt RNA species, representing, respectively, the H1 RNA and its 3′-end half H2a (Fig. 4b). Also present was an RNA component of ∼125 nt, whose distribution in the gradient, however, did not correlate with the nuRNase P activity.
FIG. 4.
Purification of the nuRNase P and analysis of the RNA associated with it. (a) The postmitochondrial S20 fraction (pmS20) from 30 ml of packed HeLa cells was processed for RNase P purification as previously described (15) and chromatographed sequentially through DEAE-cellulose, DEAE-Sepharose, and Mono-S FPLC columns, as described in Materials and Methods. The Mono-S fractions containing unbound RNase P activity were pooled and run through a Mono-Q FPLC column, and the activity eluted between 0.35 and 0.41 M KCl (UQ) was fractionated through a 15 to 30% glycerol gradient (UG). (b) RNA was extracted from individual glycerol gradient fractions exhibiting RNase P activity, 3′-end labeled with [32P]pCp and RNA ligase, and run through a 5% polyacrylamide–7 M urea gel. M, MspI-digested and 3′-end-labeled pBR322 DNA marker; other symbols as in Fig. 1. The asterisks indicate the 340- and ∼ 160-nt species.
The RNA component of mtRNase P is identical in sequence to that of nuRNase P.
The size correspondence between the 340- and 155-nt RNA species identified in the mtRNase P and the equivalent species detected in the nuRNase P strongly pointed to their being sequence related. To obtain direct sequencing information on these RNA components, an RNA-sequencing analysis was carried out on a 3′ end-proximal 20-nt segment of these RNA species, using RNases A, T1, PhyM, and CL3. This analysis revealed that the two RNA species isolated from mtRNase P were closely related or identical in sequence to the RNA species (340 and 160 nt) isolated from the nuRNase P (data not shown), confirming their correspondence to the H1 and H2a RNAs of the nuclear enzyme, as suggested above. Definitive evidence for the identity of the RNA components of the mtRNase P and nuRNase P was provided by the cloning and sequencing of the cDNA of the 340-nt RNA from the mitochondrial enzyme. The two sequences turned out to be identical (data not shown).
Correct processing activity of the mtRNase P and the nuRNase P on a mitochondrial ptRNASer(UCN) with an added 3′-terminal -CCA.
In the work described so far, the activities of mtRNase P and nuRNase P were tested on E. coli ptRNATyr. To investigate the capacity of the two enzymes to act on a mitochondrial substrate, an artificial ptRNASer(UCN), consisting of the complete mtDNA-encoded tRNA sequence (72 nt) with an added -CCA at its 3′ end and an 81-nt stretch at its 5′ end (see Materials and Methods), was used as a substrate. The results are shown in Fig. 5.
FIG. 5.
Processing of ptRNASer(UCN) with an added 3′-terminal -CCA by mtRNase P and nuRNase P. (a) Processing activity on [32P]CTP-labeled E. coli ptRNATyr and HeLa mt-ptRNASer(UCN) of samples of the UQ fraction of mtRNase P from D-plus-MN-treated mitochondria and of a sample of glycerol gradient-fractionated nuRNase P. (b) The products of similar processing reactions carried out on [35S]CTP-labeled ptRNASer(UCN) were subjected to primer extension using reverse transcriptase and Ser-oligo 1 and Ser-oligo 2 as primers and [α-32P]dCTP as the labeled nucleotide. Therefore, the primer extension products are 32P labeled and the 5′ fragments (5′-F) are 35S labeled. Sequencing reactions to generate the tRNASer(UCN) sequence were carried out with the Sequenase kit, using the ptRNASer(UCN)-carrying pGEM.4Z plasmid as a template and the two oligodeoxynucleotides mentioned above as primers. S, substrate; P, products.
The Mono-Q-fractionated US fraction from D-plus-MN-treated mitochondria had a clear processing activity on E. coli ptRNATyr labeled with [α-32P]CTP and also had a processing activity on the mitochondrial ptRNASer(UCN) (Fig. 5a). In the latter case, as expected, two fragments of slightly different sizes were produced, with the faster-moving one being significantly more strongly labeled than the slower-moving one. A similar processing activity was detected on ptRNASer(UCN) when the glycerol gradient-purified nuRNase P was tested (Fig. 5a). The slower-and faster-moving bands presumably represented the 5′-end stretch of the precursor and the mature tRNASer(UCN), with the difference in labeling between the two fragments being accounted for by the difference in the number of C residues between the two fragments (9 and 24 residues in the 5′ and 3′ fragments, respectively). This interpretation was fully confirmed by the reverse transcriptase primer extension of the processing products obtained from in vitro-transcribed [35S]CTP-labeled ptRNASer(UCN) and by the parallel sequencing of the ptRNASer(UCN) gene, carried out using, in both reactions, two different oligodeoxynucleotides as primers (Fig. 5b). Surprisingly, however, this analysis revealed that the cleavage of the precursor, rather than occurring 5′ to the U residue at position 7516, as expected from the Cambridge sequence, occurred 2 nt downstream, i.e., 5′ to the G residue at position 7514 [Fig. 5b; notice that the sequences shown in the figure are complementary to the tRNASer(UCN) sequence]. However, this result is in full agreement with recent sequencing data on mitochondrial tRNASer(UCN) from bovine (65) and human (23) sources, which have shown that the original interpretation of the Cambridge sequence was incorrect. Therefore, these experiments demonstrated clearly that both the mtRNase P and the nuRNase P recognized correctly the expected 5′-end-processing site of the mitochondrial tRNASer(UCN) precursor.
Yield of H1 RNA and MRP RNA from variously treated mitochondrial fractions.
The evidence presented in the previous sections, which indicated the identities of the RNA components of the mtRNase P and nuRNase P and the close similarity in sedimentation and enzymatic properties of the two types of ribonucleoprotein particles, raised the question of whether and to what extent the RNase P detected in the mitochondrial fraction reflected contamination of this fraction by nuRNase P. As a preliminary approach to this question, the effect of the various treatments used in the present work to purify mitochondria on the yield of H1 RNA from these organelles was investigated by RNA transfer hybridization experiments using specific oligodeoxynucleotide probes. To reduce the tendency of the 340-nt H1 RNA to be degraded to two halves during the lengthy procedure involving sequential DEAE-cellulose, DEAE-Sepharose, Mono-S, and Mono-Q chromatography and glycerol gradient fractionation, the RNA extracted from variously treated mitochondria was used directly for the transfer experiments. Indeed, under these conditions, the previously observed amount of the 155- to 160-nt component (Fig. 2b and 4) was drastically decreased (data not shown). In the same experiments, the recovery from the variously treated mitochondrial fractions of the 7.2/MRP RNA, a 260-nt RNA evolutionarily related to H1 RNA (20), was also analyzed using the corresponding specific probe. MRP RNA is a component of ribonucleoprotein particles mainly associated with the nucleolus (11, 30), but it has also been reported to occur in mitochondria (11, 34, 62).
Figure 6a shows the results of a blot hybridization analysis of total nucleic acids extracted from the washed mitochondrial fraction or from the same fraction treated with D, MN, or D followed by MN; transferred to nylon membranes; and hybridized simultaneously with an H1 RNA- and an MRP RNA-specific oligodeoxynucleotide, as described in Materials and Methods. The chosen oligodeoxynucleotide probes corresponded to sequences outside the regions of sequence similarity between H1 RNA and MRP RNA (22). Furthermore, control experiments failed to reveal any cross-hybridization between either of the two probes and the nonhomologous RNA species (data not shown). In the present work, total nucleic acids extracted from the various fractions were used for RNA transfer experiments; however, in the polyacrylamide gel electrophoresis system utilized for analysis, the DNA would not penetrate into the gel and only small RNA species would be separated.
It appears from Fig. 6a that equivalent amounts of nucleic acids from the untreated and MN-treated mitochondrial fractions gave hybridization signals of the 340-nt band of similar intensities when the H1 RNA-specific probe was used. In contrast, the nucleic acids from the D-treated and the D-plus- MN-treated fractions showed somewhat reduced and more markedly decreased hybridization, respectively, of the 340-nt band with the same probe. In striking contrast, the MRP RNA-specific probe produced a much stronger signal of the expected 260-nt band with the nucleic acids from the untreated or D-treated mitochondrial fraction than the signal obtained with the nucleic acids from the MN-treated fraction or, even more so, than the signal obtained with the nucleic acids from the D-plus-MN-treated mitochondrial fraction. In other experiments, D treatment of the mitochondrial fraction following the MN treatment or treatment of the same fraction with 15 mM EDTA prior to the MN treatment (300 or 600 U/ml) did not reduce to any significant extent the signal obtained with the H1 RNA or the MRP RNA probe, compared to that obtained with the simple MN treatment (300 U/ml) (data not shown).
In the experiment shown in Fig. 6a, the signal obtained by the hybridization of the specific probe with total-cell H1 RNA was compared with that produced by the same probe with known amounts of the H1 cDNA. This allowed a quantification of the total H1 RNA content per cell. In several experiments, after a small correction for the somewhat higher efficiency (by ∼6%) of formation of RNA-DNA hybrids than of DNA-DNA hybrids (see Materials and Methods), an average value of 54,400 ± 2,100 molecules per cell was obtained (Table 1).
TABLE 1.
Recovery of H1 RNA, MRP RNA, U2 RNA, and U3 RNA in variously treated mitochondrial fractions
| Sample | Amt (molecules/per cell) of RNA species (%)a
|
|||
|---|---|---|---|---|
| U2 | U3 | H1 | MRP | |
| Whole cells | 500,000b | 200,000b | 54,400 ± 2,100c | 30,000b |
| Mitochondrial fraction | ||||
| Washed | 12,960 ± 890 (100%) | 5,740 ± 590 (100%) | 288 ± 32 (100%) | 176 ± 26 (100%) |
| MN treated | 257 ± 67 (1.98%) | 67 ± 22 (1.16%) | 96 ± 10 (33.3%), 174 ± 19 (60.2%) | 8 ± 2 (4.5%), 14.5 ± 4 (8.2%) |
| D treated | 4,640 ± 210 (35.8%) | 4,050 ± 88 (70.6%) | 82 ± 12 (28.3%), 107 ± 17 (37.0%) | 90 ± 18 (51.1%) |
| D plus MN treated | 11.7 ± 0.6 (0.09%) | 6.7 ± 0.6 (0.12%) | 12 ± 2 (4.0%), 33 ± 6 (11.0%) | 2.3 ± 0.8 (1.3%), (6.1 ± 2.3 (3.5%) |
The values represent the means (±2 standard errors of the mean) of nine, six, two, and three independent gel determinations (from two, two, two, and one preparation, respectively) for H1, MRP, U2, and U3 RNAs. The data for whole cells and the variously treated mitochondrial fractions were normalized to the total nucleic acids recovered from each fraction per milliliter of packed cells. The data for the average number of molecules per cell of H1 RNA, MRP RNA, U2 RNA, and U3 RNA for the variously treated mitochondrial fractions were determined from the yield of the RNA in each fraction relative to the whole-cell content and corrected for the average recovery of the mitochondrial nucleic acid markers in the washed mitochondrial fraction (17.9% [see Table 2]). The data for the average number of H1 RNA molecules per cell in the variously treated washed mitochondrial fractions and those for the average number of MRP RNA molecules per cell in the MN-treated and the D-plus-MN-treated mitochondrial fractions were also corrected for the average recovery of the mitochondrial nucleic acid markers in each fraction relative to the washed mitochondrial fraction (bold numbers). The percentages of the content of each RNA species in the washed mitochondrial fraction which are found to be associated, after the normalization and correction mentioned above, with the MN-, D-, or D-plus-MN-treated mitochondrial fractions are shown in parentheses.
From reference 31.
Mean (±2 standard errors of the mean) of determinations made by comparison of the H1 RNA signal obtained for the whole-cell sample, normalized to total nucleic acids recovered per milliliter of packed cells (=1.5 × 108 cells), with the signal obtained for known amounts of H1 cDNA using the same H1 RNA probe, after a small correction (∼6%) for the difference in efficiency of the formation of RNA-DNA hybrids and DNA-DNA hybrids. See the text for details.
Recovery of mitochondrial markers in the variously treated mitochondrial fractions.
To determine the fraction of the original mitochondria recovered in the washed mitochondrial fraction analyzed, as well as to establish the effects on the organelles of the various treatments subsequently used for their purification, the yield of known mitochondrial markers in the variously treated mitochondrial fractions was determined by carrying out RNA or DNA transfer hybridization experiments with appropriate probes, measuring the signals produced by these probes by PhosphorImager analysis, and normalizing the results per milliliter of packed cells. Mitochondrial DNA, 12S rRNA, ND1 mRNA, and COII mRNA were chosen as nucleic acid markers.
As shown in Table 2, the recovery of total-cell mitochondrial DNA, ND1 mRNA, and COII mRNA in the washed mitochondrial fraction ranged between 19 and ∼24%, reflecting mostly the moderate degree of cell breakage chosen for cell homogenization and the selective centrifugation conditions. The recovery of 12S rRNA, in a single experiment, was considerably lower (7.5%), for unknown reasons. MN treatment of the washed mitochondrial fraction decreased the yield of 12S rRNA, ND1 mRNA, and COII mRNA by 46 to 61%, while that of mitochondrial DNA was decreased by only 26%. Digitonin treatment of the washed mitochondrial fraction decreased the recovery of all four mitochondrial nucleic acid markers by a smaller factor (16 to 30%). In contrast, a greater decrease in the yields of the four nucleic acid markers (58 to 67%) was observed after D-plus-MN treatment of the washed mitochondria, presumably reflecting a more extensive attack of the markers by MN in organelles damaged by the D treatment.
TABLE 2.
Recovery of mitochondrial nucleic acid markers
| Sample | Relative amt of RNA speciesa
|
Mean recovery | |||
|---|---|---|---|---|---|
| mtDNA | 12S rRNA | ND1 mRNA | COII mRNA | ||
| Whole cells | 100 | 100 | 100 | 100 | 100 |
| Mitochondrial fraction | |||||
| Washed | 21.5 | 7.5 | 23.6 ± 2.1 | 19.0 ± 1.2 | 17.9 |
| MN treated | 16 | 4 | 9.2 ± 1.8 | 10.2 ± 0.9 | 9.9 |
| D treated | 18 | 6 | 16.5 ± 2.6 | 14.5 ± 1.0 | 13.7 |
| D plus MN treated | 9 | 3 | 7.7 ± 1.5 | 6.7 ± 1.1 | 6.6 |
The data for total cells and the variously treated mitochondrial fractions were normalized to the total nucleic acids recovered in total cells and in each fraction per milliliter of packed cells. The values reported represent the results of single gel determinations for mitochondrial DNA (mtDNA) and 12S rRNA, and the averages (±2 standard errors of the mean) of five independent determinations (from two preparations) for ND1 mRNA and COII mRNA.
Contamination of the variously treated mitochondrial fractions by U2 and U3 small nuclear RNAs.
To determine to what extent residual nuclear and cytosolic contamination contributed to the RNase P activity found in the variously treated mitochondrial fractions from HeLa cells, the quantitative behavior in the same fractions and in whole cells of the nuclear U2 RNA- or U3 RNA-containing particles was investigated by RNA transfer hybridization experiments using specific probes. From Fig. 6b it appears that there was an appreciable contamination of both the extensively washed and the D-treated mitochondrial fractions by U2 RNA and U3 RNA, whereas treatment of the mitochondrial fraction with MN and, especially, with D plus MN almost completely eliminated this contamination. The data obtained by PhosphorImager analysis of the signals in the experiments for Fig. 6b were normalized to the total nucleic acid content in the cells or in each mitochondrial fraction, expressed per milliliter of packed cells (≃1.5 × 108 cells). The data for the mitochondrial samples were further corrected for the average yield of the four different mitochondrial nucleic acid markers in the washed mitochondrial fraction (17.9%; Table 2), taken as an indicator of the recovery of mitochondria, which reflected mainly the extent of cell breakage.
It appears from Table 1 that ∼98 and 99% of the U2 RNA- and U3 RNA-containing particles, respectively, were sensitive to MN treatment of the mitochondrial fraction, indicating that very few of these particles were trapped inside cytoplasmic vesicles during cell homogenization. Both types of particles were ∼99.9% sensitive to D-plus-MN treatment. On the other hand, a large fraction of the U2 RNA- or U3 RNA-containing particles (∼36 and ∼71%, respectively) was not removed by D treatment of the mitochondrial fraction, suggesting their association with contaminating nucleus-derived structures.
In vivo content of H1 RNA and MRP RNA in HeLa cell mitochondria.
To estimate the amounts per cell of H1 and MRP RNAs associated with the variously treated mitochondrial fractions, the yields of the two RNAs in the above fractions were determined by PhosphorImager analysis of the signals in the experiments shown in Fig. 6a and by normalization of the results to the amount of total nucleic acids recovered in those fractions per milliliter of packed cells. Furthermore, the normalized H1 and MRP RNA data from the experiments shown in Fig. 6a were also corrected for the average yield of the four chosen mitochondrial markers in the washed mitochondrial fraction (17.9%; Table 2). From these corrected values, the numbers of H1 and MRP RNA molecules in each of the variously treated mitochondrial fractions were calculated from the total H1 RNA or MRP RNA hybridization signal in this fraction, relative to that obtained in whole cells, on the basis of a total content of 54,400 H1 RNA molecules per cell (this work) and 30,000 7-2/MRP RNA molecules per cell (31).
It appears from Table 1 that ∼33% of the H1 RNA associated with the washed mitochondrial fraction was resistant to MN treatment of this fraction and that a similar proportion (∼28%) was resistant to D treatment of the fraction. Treatment of the mitochondrial fraction with D followed by MN caused a much greater decrease in the content of H1 RNA in this fraction. This decrease most probably reflected an action of MN on H1 RNA associated with organelles damaged by D, as strongly suggested by the marked reduction (63%) in the amounts of mitochondrial nucleic acids. In view of the substantial effects of the various treatments of the washed mitochondrial fraction on the yields of the four chosen mitochondrial markers (Table 2), it was deemed appropriate to correct further the values for H1 RNA in the differently treated fractions for the incomplete recovery of the mitochondrial markers. The corrected values, shown in Table 1, indicated that the number of molecules between a minimum of ∼33 molecules (D-plus-MN resistant) and a maximum of ∼175 molecules (MN resistant) of H1 RNA per cell was intrinsically associated with HeLa cell mitochondria.
In comparison with the results obtained for H1 RNA, a much lower proportion (∼5%) of the MRP RNA associated with the washed mitochondrial fraction was found to be resistant to MN treatment and a higher proportion (∼51%) was resistant to D treatment (Table 1). These results pointed to the occurrence in the D-treated fraction of a major portion of extramitochondrial MRP RNA, presumably associated with nucleolus-derived structures: this would be consistent with the main nucleolar localization of the MRP RNA-containing particles (34, 35). The yields of MRP RNA in the MN-treated and the D-plus-MN-treated mitochondrial fractions, after correction for the incomplete recovery of the mitochondrial nucleic acid markers, indicated that an amount of MRP RNA between a minimum of 6 molecules and a maximum of 15 molecules per cell was associated with HeLa cell mitochondria (Table 1).
DISCUSSION
In the present work, a variety of cell fractionation, biochemical, and molecular approaches have led to the extensive purification and partial characterization of mtRNase P from HeLa cells and to the identification and cloning of an RNA component which is essential for its activity and its quantification.
Characterization of the mtRNase P.
The ribonucleoprotein particles with mtRNase P activity purified in the present work have been found to sediment in a glycerol gradient with a sedimentation constant of ∼17S. A similar value (∼16S) was found for nuRNase P, in reasonable agreement with a previous estimate (9). Furthermore, both the mtRNase P and the nuRNase P were found to process correctly the E. coli ptRNATyr and the mitochondrial ptRNASer(UCN).
At least seven proteins have been found to be associated with highly purified nuRNase P from human cells (16, 26, 35). For the mtRNase P, in yeast mitochondria a 105-kDa protein is a subunit of the enzyme and is required for its activity (14, 40), and its gene has been cloned and sequenced (14). No information is available about the protein composition of mammalian mtRNase P. From the close similarity of the sedimentation properties determined for the human mtRNase P and nuRNase P, one can predict that the mitochondrial enzyme is, like the nuclear enzyme, richer in protein than are the homologous bacterial enzymes.
HeLa cell mtRNase P has an essential RNA component identical in sequence to the nuRNase P H1 RNA.
Several lines of evidence obtained in the present work strongly support the conclusion that HeLa cell mtRNase P has an essential RNA component with a sequence identical to that of nuRNase P RNA, as discussed below.
(i) Treatment with MN of highly purified mtRNase P, under conditions excluding substrate masking by EGTA-inactivated nuclease, caused an almost complete loss of RNase P activity.
(ii) A comparison of the quantitative behavior of the mitochondrion-associated H1 RNA, after different purification treatments of the organelles, with that of bona fide extramitochondrial ribonucleoprotein particles or of intramitochondrial nucleic acid markers revealed a much closer similarity to the behavior of the latter. Thus, an extensive treatment of the washed mitochondrial fraction with MN, which eliminated all but 1 to 2% of the small nuclear RNAs U2 and U3 originally contaminating that fraction, caused a loss of H1 RNA from the mitochondrial fraction (∼67%) which was only moderately higher than the average loss of four mitochondrial nucleic acid markers (∼45%). After correction for the average loss of the mitochondrial markers from the MN-treated mitochondrial fraction, it could be calculated that about 60% of the H1 RNA, corresponding to ∼175 molecules per cell, remained associated with that fraction. Similarly, the much harsher treatment of the washed mitochondrial fraction with D and MN, which eliminated all but ∼0.1% of the contaminating U2 and U3 RNAs, left in that fraction, after correction for the ∼63% loss of the mitochondrial nucleic acid markers, about 11% of the H1 RNA originally present: this corresponded to ∼33 molecules per cell. It should be noted that the somewhat lower sensitivity to D-plus-MN treatment of the mitochondrial markers than that of the H1 RNA may reflect different accessibilities of the different substrates to MN. In fact, the very high sensitivity to degradation of the H1 RNA has been observed in previous work (9) and was confirmed here. Conversely, the resistance to RNase attack of the mRNAs associated with mitochondrial polysomes had been previously reported (44). The dramatic effects of the D-plus-MN treatment on the recovery of the mitochondrial nucleic acid markers strongly suggest that the in vivo content of H1 RNA in HeLa cell mitochondria may be much closer to the upper estimate (∼175 molecules per cell) than to the minimum estimate (∼33 molecules per cell) obtained in the present work.
(iii) A comparison of the number of mtRNase P-associated H1 RNA molecules per cell estimated in the present work and of the known amount and rate of synthesis of mitochondrial tRNA molecules per cell in HeLa cells with the amount of RNase P RNA molecules and number and rate of synthesis of tRNA molecules per cell in yeast mitochondria and in E. coli revealed that even the minimum estimated number of mtRNase P enzyme molecules per cell should be fully adequate to satisfy the tRNA synthesis requirements of HeLa cell mitochondria (Table 3). It should be mentioned here that the average number of mitochondria per HeLa cell has been estimated to vary between ∼400 in the early G1 phase of the cell cycle and 500 to 600 in the late S and G2 phases (47). Considering that the major part of mitochondrial DNA transcription in HeLa cells occurs in the G2 phase of the cell cycle (45) and that ∼15% of the cells are in G2 phase in exponentially growing HeLa cells (48), the upper, more reliable estimate of the number of mtRNase P RNA molecules per cell obtained in the present work (∼175) should exceed the number of mitochondria in the transcriptionally active cells. It may indeed be adequate to secure in these cells an average steady-state number of 2 or 3 mtRNase P RNPs per 10 to 20 mtDNA molecules, which are present on average in each HeLa cell mitochondrion (29).
TABLE 3.
Levels of RNase P RNA and amounts and rates of synthesis of tRNAs in the HeLa cell nucleocytoplasmic compartment and mitochondria, S. cerevisiae mitochondria, and E. coli
| Cell | No. of tRNA molecules/cell | Rate of tRNA synthesis (molecules/min/cell)c | No. of RNase P molecules/cell | Ratio of RNase P molecules/cell to rate of tRNA synthesis |
|---|---|---|---|---|
| HeLa cells | 7.7 × 107a | 8.0 × 104 | 54.4 × 103d | 0.678 |
| Nucleocytoplasmic compartment | ||||
| Mitochondria | 6 × 105a | 630 | 33–175d | 0.052–0.28 |
| S. cerevisiae mitochondria | 5 × 104b | ≈4 × 102 | 40b | 0.11 |
| E. coli | 3 × 105a | 104 | 500e | 0.05 |
In recent studies, an RNase P activity different in some properties from nuRNase P and, in particular, residing in particles lacking intact H1 RNA but containing degraded RNA molecules, has been claimed to be associated with HeLa cell mitochondria (51, 52). However, in that work, the procedure used for isolation of mitochondria from frozen cells (which would be expected to yield damaged mitochondria, as well as damaged lysosomes, which could release a variety of nucleases and proteases) would not have guaranteed a good recovery of intact mitochondrial enzyme and contaminating nuRNase P. Indeed, in the cited work, the finding that the extract of the original untreated mitochondrial fraction contained enormous amounts of degradation products of H1 RNA (51) indicated a heavy contamination by damaged nuRNase P. Therefore, it is a distinct possibility that the decrease in the sedimentation constant of the “mitochondrial” enzyme of Rossmanith and Karwan (52), compared to the nuclear enzyme, and the modification of some substrate specificities of the enzyme (51) pertained in reality to the contaminating nuclear enzyme that was damaged in its RNA and possibly in its proteins during the isolation procedure but still retained at least part of its activity, as previously observed for Saccharomyces cerevisiae mtRNase P (52). Furthermore, concerning the reported MN resistance of the enzyme, there is abundant evidence that this property is not a reliable indicator of the absence of an essential RNA component (52). Finally, the lack of any characterization of the subcellular fractions analyzed in the above studies for contamination by other cellular components and the absence of any quantification of the H1 RNA and degraded RNA and of the residual MN-resistant enzymatic activities in the fractions tested prevent any evaluation of the comparative data reported.
In the present work, all the fractions obtained during the long procedure used for the purification of mtRNase P were analyzed. In all of those which exhibited RNase P activity, such activity was characterized, revealing sedimentation properties, MN sensitivity, and RNA components identical to those of the highly purified mtRNase P. This evidence argues strongly against the possibility of a putative RNA-free mtRNase P (51) being lost during the fractionation procedure. Furthermore, the failure of the enzyme described by Rossmanith et al. to process correctly the E. coli ptRNATyr (51) is in striking contrast with the finding in the present work that the mtRNase P activity, assayed at the earliest stage of purification (i.e., after binding to DEAE-cellulose) and at subsequent stages, cleaved this tRNA correctly and in a manner identical to that of nuRNase P. These results support the interpretation that the enzyme used in the earlier studies was damaged. Furthermore, no trace of RNase P activity was ever found to sediment with a sedimentation constant of 9S, as reported in reference 52.
Occurrence of MRP RNA in HeLa cell mitochondria.
The present work has also demonstrated the presence of MRP RNA molecules in extensively purified HeLa cell mitochondria. The amount of MRP RNA in the D-plus-MN- and MN-treated mitochondrial fractions, as estimated after correction for the average recovery of the four mitochondrial nucleic acid markers, corresponds to ∼6 and 15 molecules per cell, respectively. These amounts represent a proportion of the RNA found in the washed mitochondrial fraction which exceeds by more than an order of magnitude and by a factor of 4 to 6, respectively, the recovery of U2 RNA and U3 RNA in the D-plus-MN- and MN-treated mitochondrial fractions.
Although originally recognized as an endonuclease involved in cleaving in vitro an RNA primer for heavy-strand mtDNA synthesis (11), the majority of the MRP ribonucleoprotein particles (RNPs) are generally agreed to occur in the nucleolus (30, 34, 62), where they play an important role in rRNA processing, as demonstrated in S. cerevisiae (35). In particular, the MRP RNA has been shown (22) to be identical to the previously identified nucleolar 7-2 RNA (25, 50). In a previous quantitative study of the level of MRP RNA associated with mitochondria in HeLa cells, it has been reported that the amounts of detectable full-length MRP RNA (4 molecules per cell in MN-treated and 0.4 molecule per cell in D-plus-MN-treated mitochondria) were too small to attribute a function in mitochondria to RNase MRP (31). However, in this work, no correction was made for the recovery of mitochondria in the original subcellular fractionation and for the losses of intramitochondrial markers due to the drastic D and/or MN treatment of the mitochondrial fraction and the attendant centrifugation steps. In the present study, the detected levels of MRP RNA, without correction for the yield of mitochondrial markers in the subcellular fractionation and purification procedure, were similar to those reported by Kiss and Filipowicz (31). However, when the correction for the losses mentioned above was applied, the values increased by more than an order of magnitude (i.e., to 15 and 6 molecules per cell, respectively).
While there are no data about the rate of import of the RNase MRP into the organelles, its half-life, and the turnover number of the enzyme in the primer cleavage step, it is reasonable to assume that the small number of heavy-strand synthesis initiation events required in HeLa cells (∼8 per min) could well be produced by 6 to 15 RNase MRP RNPs per cell. Furthermore, a nonuniform rate of mtDNA replication throughout the cell cycle would increase the concentration of MRP RNPs in the cells undergoing mtDNA replication. It has indeed been shown that the rate of mtDNA replication in HeLa cells is greatly accelerated in the late S and G2 phases of the cell cycle (46). In addition, if RNase MRP plays a rate-limiting role in mtDNA synthesis, as has been suggested (62), it can be anticipated that the partitioning of RNase MRP to mitochondria may increase in cells with a large amount of mtDNA and high rate of mtDNA synthesis. Indeed, in a study of the subcellular partitioning of MRP RNA, carried out by transfection and in situ hybridization experiments on mouse cardiomyocytes and mouse C2C12 myogenic cells, which are actively respiring cells very rich in mitochondria and mtDNA, evidence was obtained for the preferential location of MRP RNA in both nucleoli and mitochondria (34).
Nuclear-mitochondrial partitioning of H1 RNA and its possible regulatory role.
The occurrence in mitochondria of nucleus-encoded RNA species has been previously reported, for tRNAs, in plants (59), Tetrahymena thermophila (53, 54), Leishmania (1), Trypanosoma brucei (24), and S. cerevisiae (38), for MRP RNA, in mouse L cells (11) and mouse cardiomyocytes and myogenic cells (34), and, more recently, for 5S rRNA, in mammalian cells (36). In addition human immunodeficiency virus RNA has been detected in mitochondria of infected cells (60). Furthermore, a partitioning between the nuclear-cytosolic compartment and mitochondria of nucleus-encoded tRNAs has been previously observed. Thus, it has been shown that in S. cerevisiae, the nucleus-encoded tRNALys(CUU) is unequally distributed between the cytosol (95%) and mitochondria (5%) (18). Furthermore, in T. thermophila, ∼10% of the nucleus-encoded tRNAGln(UUG) is imported into mitochondria while the rest functions in the cytosol (53, 54).
The evidence presented here that the very small amount of mitochondrion-associated H1 RNA in HeLa cells should be adequate to satisfy the requirements for mitochondrial tRNA synthesis, without being in excess, suggests the possibility that the imported RNase P may play a regulatory role, being rate limiting for mitochondrial tRNA synthesis. Therefore, it is a reasonable assumption that the import into mitochondria of mtRNase P is itself a highly regulated process. It should be noted that the mtDNA light-strand transcripts, which contain the sequences of eight tRNAs, are synthesized at a rate at least 10 to 15 times higher than required to account for the steady-state levels of these tRNAs (7). Furthermore, it is known that these transcripts have a much shorter half-life than the heavy-strand transcripts and do not accumulate to any significant extent (2, 10). It is therefore a plausible hypothesis that the vast majority of these transcripts decay before the occurrence of any processing, which would lead to a maturation and stabilization of the tRNAs (7). The limiting amount of mtRNase P may prevent any excess processing of the L-strand transcripts, avoiding the accumulation of a 10- to 20-fold excess of light-strand-encoded tRNAs over most of the heavy-strand-encoded tRNAs. This extreme imbalance of tRNAs, from the evidence available in bacterial systems, would negatively affect mitochondrial translation (28).
It is known that processing by RNase P of the polycistronic transcripts on the 5′ side of each tRNA sequence makes the 3′ ends of the upstream mRNA or noncoding sequence available for polyadenylation (43), a step that would tend to stabilize them. Therefore, the limiting amount of RNase P would also prevent an undue stabilization of light-strand transcripts, which may function as antisense RNAs and thus affect heavy-strand gene expression. The idea that mtRNase P may be limiting for tRNA synthesis in mitochondria would also be consistent with other evidence pointing to a tight control of respiration by mitochondrial gene expression in mammalian cells. In fact, it has recently been shown that mitochondrial protein synthesis in cultured mouse cells is rate limiting for respiration (8).
The mechanism of RNA import into mitochondria is largely unknown, although some progress is being made in identifying the RNA sequences and proteins involved in this process (1, 18, 24, 34, 54). In light of the structural and functional similarities between RNase P and MRP RNAs (20) and of their sharing some protein subunits (27), it is tempting to speculate that a similar mechanism could be operative for the import of both RNAs into human mitochondria. This import could be promoted by a factor(s) binding to the RNA or to a protein(s) of the complex associated with the RNA and transferring it into the mitochondrial matrix. It is clear that elucidation of the mechanism of RNase P and MRP RNase import into mitochondria would help in understanding the factors operating in the partitioning of the two enzymes between the extramitochondrial and the mitochondrial compartments and in the control of these processes.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM11726 to G.A.
We thank S. Altman for providing the NuH1 cDNA clone and S. Altman, C. Takada, A. Chomyn, K. Puranam, and P. Sethna for critically reading the manuscript. The excellent technical help of A. Drew, B. Keeley, and R. Kinzel is gratefully acknowledged. R.S.P. especially thanks W. Kibbe for the suggestion to use FPLC for enzyme purification and for his constant support and friendship. R.S.P. also owes thanks to members of G. Attardi's and J.-P. Revel's laboratory for their constant support and help.
REFERENCES
- 1.Adhya S, Ghosh T, Das A, Bera S K, Mahaptra S. Role of an RNA-binding protein in import of tRNA into Leishmania mitochondria. J Biol Chem. 1997;272:21396–21402. doi: 10.1074/jbc.272.34.21396. [DOI] [PubMed] [Google Scholar]
- 2.Aloni Y, Attardi G. Symmetrical in vivo transcription of mitochondrial DNA in HeLa cells. Proc Natl Acad Sci USA. 1971;68:1757–1761. doi: 10.1073/pnas.68.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altman S, Smith J D. Tyrosine tRNA precursor molecule polynucleotide sequence. Nat New Biol. 1971;233:35–39. doi: 10.1038/newbio233035a0. [DOI] [PubMed] [Google Scholar]
- 4.Altman S, Baer M, Guerrier-Takada C, Vioque A. Enzymatic cleavage of RNA by RNA. Trends Biochem Sci. 1986;11:515–518. [Google Scholar]
- 5.Anderson S, Bankier A T, Barrell B G, de Bruijn M H L, Coulson A R, Drouin J, Eperon E I, Nierlich D P, Roe B A, Sanger F, Schreier P H, Smith A J H, Staden R, Young I G. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 6.Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 7.Attardi G, Chomyn A, King M P, Kruse B, Loguercio-Polosa P, Narasimhan Murdter N. Regulation of mitochondrial gene expression in mammalian cells. Biochem Soc Trans. 1990;18:509–513. doi: 10.1042/bst0180509. [DOI] [PubMed] [Google Scholar]
- 8.Bai Y, Shakely R M, Attardi G. Tight control of respiration by NADH dehydrogenase ND5 subunit gene expression in mouse mitochondria. Mol Cell Biol. 2000;20:805–815. doi: 10.1128/mcb.20.3.805-815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartkiewicz M, Gold H A, Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989;3:489–499. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- 10.Cantatore P, Attardi G. Mapping of nascent light and heavy strand transcripts on the physical map of HeLa cell mitochondrial DNA. Nucleic Acids Res. 1980;8:2605–2625. doi: 10.1093/nar/8.12.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang D D, Clayton D A. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987;235:1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- 12.Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 13.Chomyn A, Lai S S-A T. cDNA of the 24 kDa subunit of the bovine respiratory chain NADH dehydrogenase: high sequence conservation in mammals and tissue-specific and growth-dependent expression. Curr Genet. 1989;16:117–126. doi: 10.1007/BF00393404. [DOI] [PubMed] [Google Scholar]
- 14.Dang Y L, Martin N C. Yeast mitochondrial RNase P. Sequence of the RPM2 gene and demonstration that its product is a protein subunit of the enzyme. J Biol Chem. 1993;268:19791–19796. [PubMed] [Google Scholar]
- 15.Doersen C J, Takada C G, Altman S, Attardi G. Characterization of an RNAse P activity from HeLa cell mitochondria. Comparison with the cytosol RNase P activity. J Biol Chem. 1985;260:5942–5949. [PubMed] [Google Scholar]
- 16.Eder P S, Kekuda R, Stolc V, Altman S. Characterization of two scleroderma autoimmune antigens that copurify with human ribonuclease P. Proc Natl Acad Sci USA. 1997;94:1101–1106. doi: 10.1073/pnas.94.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.England T E, Bruce A G, Uhlenbeck O C. Specific labeling of 3′-termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65:65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- 18.Entelis N S, Kieffer S, Kolesnikova O A, Martin R P, Tarassov I A. Structural requirements of tRNALys for its import into yeast mitochondria. Proc Natl Acad Sci USA. 1998;95:2838–2843. doi: 10.1073/pnas.95.6.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 20.Forster A C, Altman S. Similar cage-shaped structure for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990;62:407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- 21.Gold H A. Studies of RNase P from HeLa cells. Ph. D. thesis. New Haven, Conn: Yale University; 1988. [Google Scholar]
- 22.Gold H A, Topper J N, Clayton D A, Craft J. The RNA processing enzyme RNase MRP is identical to the Th RNP and related to RNase P. Science. 1989;245:1377–1380. doi: 10.1126/science.2476849. [DOI] [PubMed] [Google Scholar]
- 23.Guan M X, Enriquez A, Fischel-Ghodsian N, Puranam R S, Lin C P, Maw M A, Attardi G. The deafness-associated mitochondrial DNA mutation at position 7445, which affects tRNASer (UCN) precursor processing, has long-range effects on NADH dehydrogenase subunit ND6 gene expression. Mol Cell Biol. 1998;18:5868–5879. doi: 10.1128/mcb.18.10.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hancock K, LeBlanc A J, Donze D, Hajduk S L. Identification of nuclear encoded precursor tRNAs within the mitochondrion of Trypanosoma brucei. J Biol Chem. 1992;267:23963–23971. [PubMed] [Google Scholar]
- 25.Hashimoto C, Steitz J A. Sequential association of nucleolar 7–2 RNA with-two different autoantigens. J Biol Chem. 1983;258:1379–1382. [PubMed] [Google Scholar]
- 26.Jarrous N, Eder P S, Guerrier-Takada C, Hoog C, Altman S. Autoantigenic properties of some protein subunits of catalytically active complexes of human ribonuclease P. RNA. 1998;4:407–417. [PMC free article] [PubMed] [Google Scholar]
- 27.Jarrous N, Wolenski J S, Wesolowski D, Lee C, Altman S. Localization in the nucleolus and coiled bodies of protein subunits of the ribonucleoprotein ribonuclease P. J Cell Biol. 1999;146:559–572. doi: 10.1083/jcb.146.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King M P, Attardi G. Post-transcriptional regulation of the steady-state levels of mitochondrial tRNAs in HeLa cells. J Biol Chem. 1993;268:10228–10237. [PubMed] [Google Scholar]
- 29.King M P, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 30.Kiss T, Marshallsay C, Filipowicz W. 7-2/MRP RNAs in plant and mammalian cells: association with higher order structures in the nucleolus. EMBO J. 1992;11:3737–3746. doi: 10.1002/j.1460-2075.1992.tb05459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiss T, Filipowicz W. Evidence against a mitochondrial location of the 7-2/MRP RNA in mammalian cells. Cell. 1992;70:11–16. doi: 10.1016/0092-8674(92)90528-k. [DOI] [PubMed] [Google Scholar]
- 32.Kun E, Kirsten E, Piper W N. Stabilization of mitochondrial functions with digitonin. Methods Enzymol. 1979;55:115–118. doi: 10.1016/0076-6879(79)55016-2. [DOI] [PubMed] [Google Scholar]
- 33.Lederman M, Attardi G. In vitro protein synthesis in a mitochondrial fraction from HeLa cells, sensitivity to antibiotics and ethidium bromide. Biochem Biophys Res Commun. 1970;40:1492–1500. doi: 10.1016/0006-291x(70)90037-9. [DOI] [PubMed] [Google Scholar]
- 34.Li K, Smagula C S, Parsons W J, Richardson J A, Gonzalez M, Hagler H K, Williams R S. Subcellular partitioning of MRP RNA assessed by ultrastructural and biochemical analysis. J Cell Biol. 1994;124:871–882. doi: 10.1083/jcb.124.6.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lygerou Z, Pluk H, van Venrooij W J, Seraphin B. hPop1: an autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO J. 1996;15:5936–5948. [PMC free article] [PubMed] [Google Scholar]
- 36.Magalhaes P J, Andreu A L, Schon E A. Evidence for the presence of 5S rRNA in mammalian mitochondria. Mol Biol Cell. 1998;9:2375–2382. doi: 10.1091/mbc.9.9.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manam S, van Tuyle G C. Separation and characterization of 5′-and 3′-tRNA processing nucleases from rat liver mitochondria. J Biol Chem. 1987;262:10272–10279. [PubMed] [Google Scholar]
- 38.Martin R P, Schneller J M, Stahl A J C, Dirheimer G. Import of nuclear deoxyribonucleic acid coded lysine-accepting transfer ribonucleic acid (anticodon C-U-U) into yeast mitochondria. Biochemistry. 1979;18:4600–4605. doi: 10.1021/bi00588a021. [DOI] [PubMed] [Google Scholar]
- 39.Montoya J, Ojala D, Attardi G. Distinctive features of the 5′ terminal sequences of human mitochondrial mRNAs. Nature. 1981;290:465–470. doi: 10.1038/290465a0. [DOI] [PubMed] [Google Scholar]
- 40.Morales M J, Dang Y L, Lou Y C, Sulo P, Martin N C. A 105-kDa protein is required for yeast mitochondrial RNase P activity. Proc Natl Acad Sci USA. 1992;89:9875–9879. doi: 10.1073/pnas.89.20.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller D M, Getz G S. Steady-state analysis of mitochondrial RNA after growth of yeast Saccharomyces cerevisiae under catabolite repression and derepression. J Biol Chem. 1986;261:11816–11822. [PubMed] [Google Scholar]
- 42.Ojala D, Merkel C, Gelfand R, Attardi G. The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell. 1980;22:393–403. doi: 10.1016/0092-8674(80)90350-5. [DOI] [PubMed] [Google Scholar]
- 43.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 44.Ojala D, Attardi G. Expression of the mitochondrial genome in HeLa cells. X. Properties of mitochondrial polysomes. J Mol Biol. 1972;65:273–289. doi: 10.1016/0022-2836(72)90282-3. [DOI] [PubMed] [Google Scholar]
- 45.Pica-Mattoccia L, Attardi G. Expression of the mitochondrial genome in HeLa cells. V. Transcription of mitochondrial DNA in relationship to the cell cycle. J Mol Biol. 1971;57:615–621. doi: 10.1016/0022-2836(71)90113-6. [DOI] [PubMed] [Google Scholar]
- 46.Pica-Mattoccia L, Attardi G. Expression of the mitochondrial genome in HeLa cells. IX. Replication of mitochondrial DNA in relationship to the cell cycle. J Mol Biol. 1972;64:465–484. doi: 10.1016/0022-2836(72)90511-6. [DOI] [PubMed] [Google Scholar]
- 47.Posakony J W, England J M, Attardi G. Mitochondrial growth and cell division during the cell cycle in HeLa cells. J Cell Biol. 1977;74:468–491. doi: 10.1083/jcb.74.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao P N, Engelberg J. Effects of temperature on the mitotic cycle of normal and synchronized mammalian cells. In: Cammeron I L, Padilla G M, editors. Cell synchrony. New York, N.Y: Academic Press, Inc.; 1996. pp. 332–352. [Google Scholar]
- 49.Reddy R, Busch H. Small nuclear RNAs. RNA sequences, structure and modifications. In: Birnstiel M L, editor. Small nuclear ribonucleoprotein particles. New York, N.Y: Springer-Verlag; 1988. pp. 1–37. [Google Scholar]
- 50.Reddy R, Tan E M, Henning D, Nohga K, Busch H. Detection of nucleolar 7-2 ribonucleoprotein and cytoplasmic 8-2 ribonucleoprotein with autoantibodies from patients with scleroderma. J Biol Chem. 1983;258:1383–1386. [PubMed] [Google Scholar]
- 51.Rossmanith W, Tullo A, Potuschak T, Karwan R, Sbisà E. Human mitochondrial t-RNA processing. J Biol Chem. 1995;270:12885–12891. doi: 10.1074/jbc.270.21.12885. [DOI] [PubMed] [Google Scholar]
- 52.Rossmanith W, Karwan R M. Characterization of human mitochondrial RNase P: novel aspects in tRNA processing. Biochem Biophys Res Commun. 1998;247:234–241. doi: 10.1006/bbrc.1998.8766. [DOI] [PubMed] [Google Scholar]
- 53.Rusconi C P, Cech T R. Mitochondrial import of only one of three nuclear-encoded glutamine tRNAs in Tetrahymena thermophila. EMBO J. 1996;15:3286–3295. [PMC free article] [PubMed] [Google Scholar]
- 54.Rusconi C P, Cech T R. The anticodon is the signal sequence for mitochondrial import of glutamine tRNA in Tetrahymena. Genes Dev. 1996;10:2870–2880. doi: 10.1101/gad.10.22.2870. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook J E, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 56.Samejima T, Shibata K. Denaturation of catalase by formamide and urea related to the subunit make up of the molecule. Arch Biochem Biophys. 1961;93:407–412. doi: 10.1016/0003-9861(61)90286-7. [DOI] [PubMed] [Google Scholar]
- 57.Sampson J R, Uhlenbeck O C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schon E A, Koga Y, Davidson M, Moraes C T, King M P. The mitochondrial tRNA (Leu) (UUR) mutation in MELAS: a model for pathogenesis. Biochim Biophys Acta. 1992;1101:206–209. [PubMed] [Google Scholar]
- 59.Small I, Marechal-Drouard L, Masson J, Pelletier G, Cosset A, Weil J H, Dietrich A. In vivo import of a normal or mutagenized heterologous transfer RNA into the mitochondria of transgenic plants: towards novel ways of influencing mitochondrial gene expression? EMBO J. 1992;11:1291–1296. doi: 10.1002/j.1460-2075.1992.tb05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Somasundaran M, Zapp M L, Beattie L K, Pang L, Byron K S, Bassell G J, Sullivan J L, Singer R H. Localization of HIV RNA in mitochondria of infected cells: potential role in cytopathogenicity. J Cell Biol. 1994;126:1353–1360. doi: 10.1083/jcb.126.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Storrie B, Attardi G. Expression of the mitochondrial genome in HeLa cells. XIII. Effect of selective inhibition of cytoplasmic or mitochondrial protein synthesis on mitochondrial nucleic acid synthesis. J Mol Biol. 1972;71:177–199. doi: 10.1016/0022-2836(72)90345-2. [DOI] [PubMed] [Google Scholar]
- 62.Topper J N, Bennett J L, Clayton D A. A role for RNase MRP in mitochondrial RNA processing. Cell. 1992;70:16–20. doi: 10.1016/0092-8674(92)90529-l. [DOI] [PubMed] [Google Scholar]
- 63.Triezenburg S J. Primer extension. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1994. pp. 4.8.1–4.8.5. [Google Scholar]
- 64.Wang M J, Davis W N, Gegenheimer P. Novel mechanisms for maturation of chloroplast transfer RNA precursors. EMBO J. 1988;7:1567–1574. doi: 10.1002/j.1460-2075.1988.tb02981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yokogawa T, Watanabe Y, Kumazawa Y, Udea T, Hirao I, Miura K, Watanabe K. A novel cloverleaf structure found in mammalian mitochondrial tRNA (Ser) (UCN) Nucleic Acids Res. 1991;19:6101–6105. doi: 10.1093/nar/19.22.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]