Abstract
Yersinia enterocolitica accounts for 50% of the clinical sepsis episodes caused by the transfusion of contaminated red blood cells. A 5′ nuclease TaqMan PCR assay was developed to detect Y. enterocolitica in blood. Primers and a probe based on the nucleotide sequence of the 16S rRNA gene from Y. enterocolitica were designed. Whole-blood samples were spiked with various numbers of Y. enterocolitica cells, and total chromosomal DNA was extracted. When the TaqMan PCR assay was performed, as few as six bacteria spiked in 200 μl of blood could be detected. The assay was specific and did not detect other Yersinia species. The TaqMan assay is easy to perform, takes 2 h, and has the potential for use in the rapid detection of Y. enterocolitica contamination in stored blood units.
Yersinia enterocolitica, a gram-negative bacterium, is responsible for 50% of all the clinical sepsis episodes that occur as a result of transfusion of contaminated red blood cells (RBCs) (19). It is the major bacterial contaminant found in RBC concentrates, and the result of such contamination has proven to be fatal in 61% of all the reported cases of Y. enterocolitica sepsis resulting from transfusion (19). Pseudomonas fluorescens is responsible for a further 26% of the transfusion-associated sepsis episodes. Serratia liquefaciens has become another major bacterial pathogen to contaminate blood product units, and since 1992, five cases have been reported to the Centers for Disease Control and Prevention (27). Other bacteria implicated in RBC contamination are Pseudomonas putida, Campylobacter jejuni, Enterobacter jejuni, Escherichia coli, and Flavobacterium (32). The ability of Y. enterocolitica to contaminate RBCs can be attributed to its being a psychrophilic bacterium that can survive well at refrigerator temperatures, using dextrose and iron from the blood. Thus, packed red cells, which are usually stored at 4°C for up to 42 days, can allow the growth of these bacteria (32). The bacteria go through an initial lag period of 7 to 32 days during storage at 4°C and then show exponential growth, with an 18- to 20-h doubling time (3, 8, 18, 29, 32). Y. enterocolitica secretes an endotoxin, which is probably responsible for much of the morbidity and mortality (9). While the bacterium may lose its virulence during storage, since the plasmid that encodes virulence factors that lead to cellular invasion and resistance to the complement-mediated lysis is often lost during storage (19, 24), the endotoxin released and the bacterium itself act as sources of toxicity. Conventional methods for detection of this bacterium in blood include detection of the endotoxin it produces by the Limulus amebocyte lysate assay (30) and staining of the bacterial cells with hematologic stains such as acridine orange or Giemsa or Wright-Giemsa (10, 23). However, the identity of the bacterium cannot be established by these methods. In addition, these methods depend on the growth of the bacteria to a density of 104 to 106 CFU per ml, a level that may take several days to achieve. A PCR-based assay for detection of Y. enterocolitica in blood was described by Feng et al. (11). They could detect 500 bacteria seeded in 100 μl of blood, or 5,000 bacteria/ml. This was a significant improvement in the detection sensitivity and specificity and suggested that PCR could be useful for detecting bacterial contamination in stored blood units. This assay, however, may not be able to detect early contamination, when only a few bacteria are present in blood. Therefore, newer nucleic acid-based methodology was sought, to achieve this level of detection sensitivity.
The 5′ nuclease fluorogenic TaqMan assay has been recently described (21). The method exploits the property of Taq polymerase to act as a 5′-3′ exonuclease (15). Briefly, an oligonucleotide probe that has a reporter fluorescent dye attached to its 5′ end and a quencher dye attached to its 3′ end is used in the assay. Initially, the unbound probe is not able to emit a fluorescent signal because of the proximity of the reporter and quencher dyes. When the probe hybridizes to its target template, the reporter dye is cleaved by the 5′ nuclease activity of Taq polymerase and becomes capable of emitting a fluorescent signal without the suppression activity of the quencher dye. With increasing cycles of amplification more fluorescent signal is generated by binding of the probe to more available target, which can be detected in real time by the ABI 7700 sequence detector (PE Applied Biosystems, Foster City, Calif.). The sequence detector contains a thermocycler, laser detection system, and analysis software system. Analysis of the signal takes only about a minute after the PCR is completed. Because the generation of the fluorescent signal depends on the hybridization of the probe to a specific template, which is being amplified, there is less scope for false signals from nonspecific amplification. It is necessary to detect a very few bacteria in blood, which may have contaminated a unit, in order to avoid a sepsis reaction, and this assay seemed to have the potential for such detection. In this report, a TaqMan PCR assay is described which is rapid and shows specific and sensitive detection of Y. enterocolitica in blood.
MATERIALS AND METHODS
Bacterial species and culture conditions.
Y. enterocolitica isolates of serotypes Y288, O:3, O:1,2,3, O:5,27, and O:20 were obtained from P. Feng (Food and Drug Administration, Washington, D.C.). Serotypes O:3, O:1,2,3, O:5,27, and O:20 were previously implicated in blood endotoxemia resulting from transfusion (30). The bacteria were grown in brain heart infusion (BHI) broth (Sigma, St. Louis, Mo.) at 30°C, with continuous shaking. O:3 was used in serial dilution and spiking of sodium citrate-preserved human whole blood. O:3 was grown to an optical density at 600 nm of 0.6 and then diluted in phosphate-buffered saline. Typically, 10-μl aliquots were spiked into 190 μl of blood. Another 10 μl was plated on BHI agar to determine the viable cell count. The number of bacteria spiked in the 10-μl volume ranged from 4 to 100,000. Other bacterial species used are listed in Table 1.
TABLE 1.
Specificity of the Y. enterocolitica TaqMan assaya
| Species | Serotype or strain | % Homology with primers and/or probe | CT value |
|---|---|---|---|
| Yersinia enterocolitica | O:3 | 100 with primers and probe | 18.94 |
| Yersinia enterocolitica | O:1,2,3 | 100 with primers and probe | 19.2 |
| Yersinia enterocolitica | O:5,27 | 100 with primers and probe | 18.18 |
| Yersinia enterocolitica | O:20 | 100 with primers and probe | 20.9 |
| Yersinia enterocolitica | Y288 | 100 with primers and probe | 18.69 |
| Yersinia pseudotuberculosis | ATCC 29833 | 100 with forward and reverse primer | 45 |
| Yersinia frederiksenii | ATCC 33641 | 100 with reverse primer and 84% with forward primer | 45 |
| Hafnia alvei | ATCC 13337 | 100 with reverse primer and probe | 45 |
| Serratia entomophila | ATCC 43705 | 100 with reverse primer and probe | 45 |
| Serratia ficaria | ATCC 33105 | 100 with reverse primer and 87 with probe | 45 |
| Serratia grimesii | ATCC 14460 | 100 with reverse primer and probe | 45 |
| Pseudomonas fluorescens | ATCC 13525 | 100 with reverse primer and 75 with probe | 45 |
| Serratia liquefaciens | ATCC 35551 | 100 with reverse primer and 84 with probe | 45 |
Total DNA was isolated from the bacteria by the Puregene kit. For the Y. enterocolitica strains, 20 ng of template DNA was used per 50 μl of PCR mixture. For the other bacterial strains, 200 ng of template DNA was used per 50 μl of PCR mixture.
Preparation of DNA.
Chromosomal DNA was prepared from spiked blood by the QIAamp blood kit (Qiagen Corp., Santa Clarita, Calif.) or the Dynabeads DNA Direct system II kit (Dynal, Oslo, Norway). For the QIAamp blood kit, 190 μl of blood was spiked with a 10-μl volume containing 5 to 100,000 bacteria. Proteinase K and the ALW buffer supplied in the kit were added, and the protocol was followed exactly as specified by the manufacturer. DNA was extracted in a final volume of 100 μl of Tris-Cl, pH 8. For the Dynal kit, 2 to 50,000 bacteria in a 5-μl volume were spiked into 95 μl of blood. The DNA was extracted in a final volume of 75 μl. When comparisons between the two kits were done, a 400-μl sample was spiked with bacteria; a 200-μl aliquot of this was used for the QIAamp blood kit and 100 μl was used for the Dynal kit. Total DNA was also extracted from overnight cultures of different Y. enterocolitica strains by the Puregene kit (Gentra, Minneapolis, Minn.). In addition, chromosomal DNA was prepared from other bacterial species by Midi Labs (Newark, Del.), using the Puregene kit. Plasmid DNA was purified with the Wizard Miniprep DNA purification kit (Promega, Madison, Wis.).
Design of primers and probes.
The 16S rRNA gene has been sequenced from several Y. enterocolitica strains. All the available partial and full-length 16S rRNA gene sequences from GenBank were aligned, and the conserved sequences were identified, by the PileUp program of the Genetics Computer Group (Madison, Wis.) software package. The approximate location of the specific probe was first determined. This was done by searching the GenBank databases for uniqueness of the probe to Y. enterocolitica 16S ribosomal DNA (rDNA), using the BLAST database search program (2). The final selection of the primers and the exact length of the probe were determined, using the ABI Primer Express program (PE Applied Biosystems). This program selects probe and primer sets with optimized melting temperatures, secondary structure, base composition, and amplicon lengths. The length of the amplicon is an important consideration in the sensitivity and the reproducibility of the assay. The forward primer, 16SF, had the sequence 5′CGGCAGCGGGAAGTAGTTT3′, and the reverse primer, 16SR, had the sequence 5′GCCATTACCCCACCTACTAGCTAA3′. Both of these primers recognized 16S rDNA sequences from all Y. enterocolitica strains. The primers were made by the Biotechnology Core Facility, Center for Biologics Evaluation and Research, Food and Drug Administration, and amplified a fragment of 201 bp spanning nucleotides 47 to 247 of the 16S rRNA gene (accession no. Z49830). The TaqMan fluorescent probe YE1 had the sequence 5′FAM-AAGGTCCCCCACTTTGGTCCGAAG-TAMRA3′ and was made by PE Applied Biosystems. It was located from nucleotides 166 to 190 (reverse complement) of the 16S rRNA gene of Y. enterocolitica. FAM (6-carboxyfluorescein) is the reporter dye, and TAMRA (6-carboxytetramethylrhodamine) is the quencher dye. The 3′ end was phosphorylated to prevent extension by Taq polymerase.
TaqMan assay.
Reactions were performed in 50-μl volumes in 0.5-ml optical-grade PCR tubes (PE Applied Biosystems). Each 50 μl of reaction mixture contained a 400 nM concentration of primers, an 80 nM concentration of the probe, 200 μM (each) dTTP, dUTP, dATP, and dGTP, 1 U of AmpliTaq Gold polymerase, and 1× PCR buffer (10× supplied with the enzyme). MgCl2 was added to a final concentration of 3 mM when the Puregene kit was used to extract the DNA from pure cultures and was added to a final concentration of 5 mM when either the QIAamp or the Dynal kit was used to isolate the DNA from blood. When the QIAamp blood kit was used to extract the DNA, 20 μl of the template was added, and 30 μl was used when DNA was isolated by the Dynal kit. Cycling conditions consisted of an initial single cycle at 95°C for 10 min to activate AmpliTaq Gold, followed by 40 to 50 cycles of two-temperature cycling consisting of 15 s at 95°C and 1 min at 60°C. PCR was performed with the ABI 7700 sequence detector, as per instructions in the instrument's manual.
Post-PCR analysis.
Data were analyzed by SDS software (PE Biosystems). The software calculates a value for ΔRn using the equation ΔRn = Rn+ − Rn−. Rn+ is the emission intensity of the reporter divided by the emission intensity of the quencher at any given time in a reaction tube; Rn− is the emission intensity of the reporter divided by the emission intensity of the quencher of the same reaction prior to PCR amplification. The ΔRn values were plotted on the y axis, and the time, represented by the cycle number, was plotted on the x axis. The threshold cycle (CT) value is the noninteger calculation of the number of cycles required for the reporter dye fluorescence to become significantly higher than the background, which can happen when a sufficient amount of the hybridization probe has been cleaved (12, 14). This will also indicate the increased formation of the PCR product. The CT values for each reaction were automatically calculated by the default parameters of the program. As expected, the amplification plots shift to the right, to higher CT values, with diminishing numbers of template copies.
The correct size of the PCR product from each assay was verified by running a sample from each reaction tube on agarose gels stained with ethidium bromide.
RESULTS
Theoretical detection limit by TaqMan PCR.
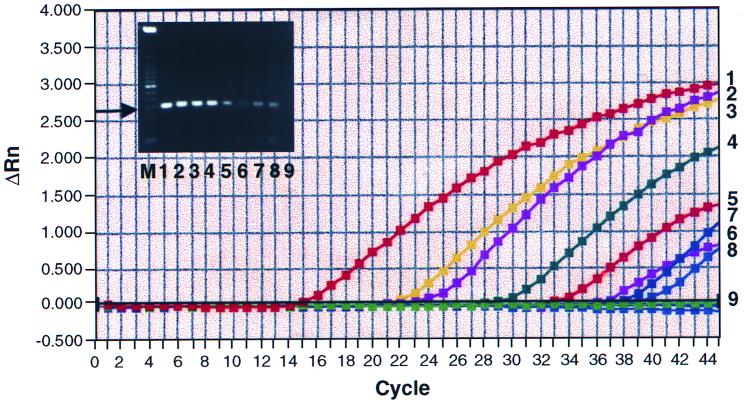
In order to determine the sensitivity of the assay, pure chromosomal DNA from serotype O:3 was isolated by the Puregene kit from overnight cultures of bacteria grown in BHI medium at 30°C. TaqMan PCR was performed by using 100 ng to 1 fg of chromosomal DNA as a template and the primers and probes described above. A DNA concentration of ≥5 fg could be detected (Fig. 1). Since the size of the Y. enterocolitica genome is not known, this number could not be used to calculate the exact number of bacteria that could be detected. However, based on the size of the Yersinia pestis genome, which is 4,398 kb (22), and that of the Yersinia ruckeri genome, which is 4,460 to 4,770 kb (26), if we assume the size of the Y. enterocolitica genome to be 4,400 kb, then 5 fg of DNA would amount to approximately one genome. If there were 1 copy of the 16S rRNA gene per cell, this would translate to 1 template, or 10 templates if there were 10 copies of the gene per cell, and so on. The 201-bp PCR fragment was also cloned into E. coli using the TopoTA cloning vector pCR 2.1-Topo, of a length of 3.9 kb, from the Topo TA cloning kit (Invitrogen Corp., Carlsbad, Calif.). When the plasmid pKSY, having a length of 4.101 kb (3.9 + 0.201 kb) and containing the cloned insert, was used as a template, the detection limit was 0.0061 fg. This number amounted to a copy number of 1.3. This was calculated as follows: 4.101 kb is equal to 2.7 × 106 g/mol, and 0.006 fg of a 4.101-kb plasmid would contain 2.2 × 10−24 mol. Multiplying this number by Avogadro's number, 6 × 1023, gives the number of molecules or the copy number value of 1.3. Thus, the primers and the probe combination as well as the length of the amplicon which was being amplified were optimal, since the greatest TaqMan PCR sensitivity could be achieved, which was 1 CFU.
FIG. 1.
Detection limit of the TaqMan assay for Y. enterocolitica. Chromosomal DNA from a pure culture of serotype O:3 was isolated as described in Materials and Methods. Quantities of 100 ng, 10 ng, 1 ng, 100 pg, 10 pg, 1 pg, 100 fg, 5 fg, and 1 fg, represented by amplification plots 1 to 9, respectively, were used as template in each 50 μl of PCR mixture. The gel inset shows 5 μl of the corresponding product from each reaction, which was examined on a 2.5% agarose gel. Lane M is the molecular size standard, consisting of a 50-kb DNA ladder. Lanes 1 to 9 correspond to plots 1 to 9. The arrow indicates the position of the 201-bp product obtained as a result of amplification of the region from nucleotides 47 to 247 of the 16S rRNA gene with primers 16SF and 16SR.
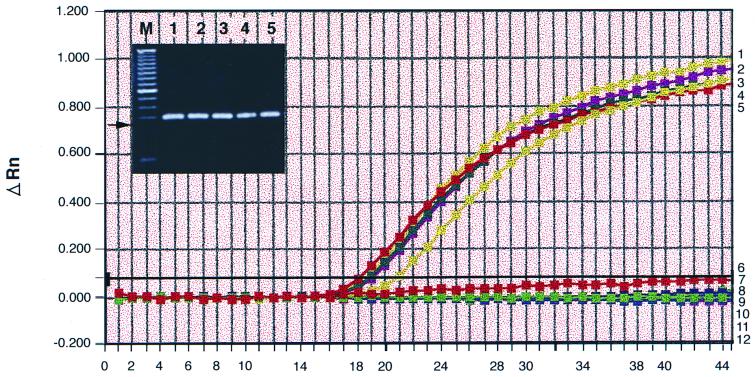
Specificity of TaqMan PCR.
The specificity of the TaqMan PCR assay for Y. enterocolitica strains was examined by isolating genomic DNA from five different Y. enterocolitica strains. In PCR assays, 20 ng of this pure DNA was used. A CT value in the range from 18.2 to 20.9 was obtained with each of the Y. enterocolitica strains (Fig. 2; Table 1). When 200-ng samples of chromosomal DNA from the related species Yersinia frederiksenii and Yersinia pseudotuberculosis were used, the CT values obtained were 45 (Fig. 2; Table 1). The CT values were also 45 when two other major bacteria that contaminate RBC units, P. fluorescens and S. liquefaciens, were examined by this set of primers and probe (Fig. 2; Table 1). The TaqMan probe YE1, in addition to recognizing all Y. enterocolitica strains, also showed 100% homology with 16S rDNA of Hafnia alvei, some Serratia spp., and Erwinia spp. TaqMan PCR was therefore done with chromosomal DNA isolated from H. alvei, Serratia ficaria, and Serratia grimesii. The CT values obtained were 45. The primers and probe could not be tested with Erwinia species because a sample chromosomal DNA could not be obtained. But it is unlikely that the assay would recognize Erwinia species because a set of three oligonucleotides is used in the TaqMan assay, and for a positive reaction, all three would have to show substantial homology. Thus, even though Y. frederiksenii showed 100% homology with the forward primer and 84% homology with the reverse primer, since the probe did not show any major sequence homology, the CT value was 45 (Fig. 2; Table 1).
FIG. 2.
Specificity of the TaqMan assay for Y. enterocolitica. Chromosomal DNAs (20 ng each) from five Y. enterocolitica serotypes (Y288; O:1,2,3; O:3; O:5,27; and O:20) were used as templates in TaqMan PCR, and their amplifications are represented in plots 1 to 5, respectively. The gel inset shows the 201-bp product from each PCR, and lanes 1 to 5 correspond to plots 1 to 5. The arrow indicates the position of the 201-bp product. Chromosomal DNAs (200 ng each) from seven other bacterial species listed in Table 1 were also tested with primers 16SF and 16SR and probe YE1, and their amplifications are represented in plots 6 to 12. The cycle numbers are on the x axis.
Blood samples.
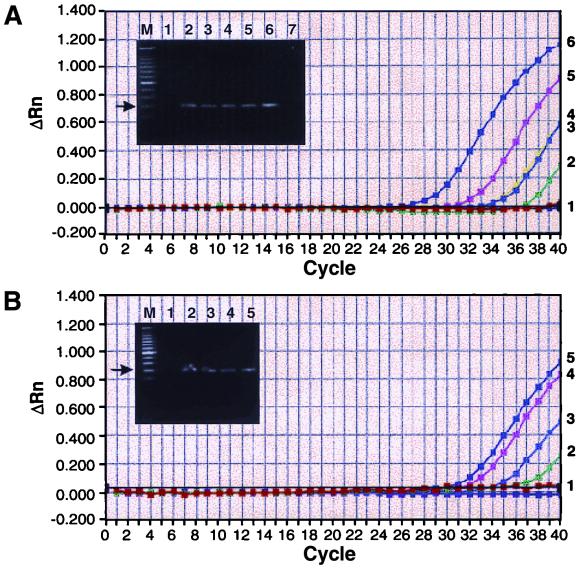
Logarithmic-phase Y. enterocolitica O:3 cells were spiked into blood at different concentrations. Six different kits were tried initially for extraction of the total DNA, containing DNA from blood and bacteria. The DNA extracted by the QIAamp kit and the Dynal kit proved to be equivalent for the TaqMan assay (Fig. 3). In addition, both methods were fast; they took only 20 min for extraction of the DNA. In four different spiking experiments, the minimum threshold of detection was six bacteria spiked into 200 μl of blood. Representative data, of a dilution series with 6 to 6,000 bacteria spiked into 200 μl of blood, are shown in Fig. 3. The CT values obtained typically were between 36 and 37. Optimization experiments were performed initially, to choose the right probe concentration and magnesium concentration, so that the highest ΔRn values were obtained without compromising the specificity of the signal. The cutoff CT value was taken to be 39, and any signal higher than this was not considered a positive signal because the unspiked blood sometimes gave a signal at a CT value of 40. However, in these samples the ΔRn value was less than 0.05.
FIG. 3.
Detection limit of the TaqMan assay for Y. enterocolitica in blood samples. Different amounts of Y. enterocolitica cells were spiked into blood, and the total DNA was isolated as described in Materials and Methods. (A) Bacteria were spiked into 200 μl of blood, and DNA was isolated by the QIAamp blood kit in a final volume of 100 μl. The corresponding numbers of cells spiked are as follows: plot 1, none (unseeded blood); plot 2, 30 cells/ml (1.2 cell equivalent/50 μl of PCR mixture); plot 3, 60 cells/ml (2.4 cell equivalent/50 μl of PCR mixture); plot 4, 300 cells/ml; plot 5, 3,000 cells/ml; and plot 6, 30,000 cells/ml. The gel inset shows the products obtained, and lanes 1 to 6 correspond to plots 1 to 6. Lane 7 is a no-template control. (B) Bacteria were spiked into 100 μl of blood, and DNA was extracted by the Dynal DNA Direct kit in a final volume of 75 μl. The corresponding cell numbers are as follows: plot 1, none (unseeded blood); plot 2, 30 cells/ml (1.2 cell equivalent/PCR mixture); plot 3, 300 CFU/ml (12 cell equivalents/PCR mixture); plot 4, 3,000 cells; and plot 5, 30,000 cells. The gel inset represents the products obtained, and lanes 1 to 5 correspond to plots 1 to 5. M is the 50-bp molecular size DNA ladder, and the arrow indicates the 201-bp product.
DISCUSSION
The need to identify a very small number of bacteria in blood that is to be transfused is critical. In this report the development of a PCR-based 5′ nuclease assay to detect a small number of Y. enterocolitica organisms in blood is described. The use of PCR to identify the presence of microbial DNA in a variety of clinical specimens has been reported by several laboratories (4, 17, 25). A factor that has limited the use of PCR-based diagnostic methods to detect microbial contamination in blood directly is the inhibitory effects of blood on Taq polymerase. This includes the hemoglobin in blood itself and the preservatives used to store blood (1, 33). In this study several DNA extraction methods have been evaluated. Furthermore, since the DNA extracted was to be used in an assay measuring fluorescence, some of the extraction methods needing the use of reagents that quenched fluorescence had to be eliminated. Both the QIAamp blood kit and the Dynal DNA kit proved to be superior in providing DNA suitable for the assay. The results with the QIAamp blood kit appeared to be more reproducible from day to day.
In PCR methods, if the template targeted is in several copies then the sensitivity of the assay should increase, which in this case would translate into detection of fewer CFU of bacteria. In bacteria there are several copies of the 16S rRNA gene (28), and therefore this gene was chosen over the invasion gene ail, the virF gene present in the virulence plasmid pYV, or the heat-stable enterotoxin (yst) gene, which have been used by other groups (13, 16, 20). The detection threshold in blood using the virF and ail genes was 5,000 bacteria/ml (11). The pYV plasmid is often lost during storage and growth and therefore is not suitable as a target (24). Although the ail and the yst genes are specific for virulence and would be present only in a pathogenic strain, it was argued that the presence of any species of Y. enterocolitica would be reason for removing a blood unit. The goal was to achieve the greatest sensitivity with respect to the number of bacteria detected, regardless of whether the strain was pathogenic or not. Targeting of the 16S rRNA gene by a seminested PCR approach has also been used by another group recently (31). Their method's detection level was 100 CFU/ml. However, their samples were from pure bacterial cell suspensions. The present assay was able to detect 5 fg of Y. enterocolitica bacterial DNA from pure cultures, which would be equivalent to 1 CFU and to 30 CFU of Y. enterocolitica per ml of blood. The assay could be developed further by increasing the efficiency of the purification of DNA from blood or by concentrating the extracted DNA. As of now, 20 μl of DNA out of a final volume of 100 μl, extracted by the QIAamp kit, was used per PCR. This would amount to 1.2 bacteria per 50 μl of PCR mixture, if originally 6 bacteria were spiked into 200 μl of blood. The question of whether the in vitro spiking experiments represent the true viability of bacteria in blood has not been addressed in this assay. The TaqMan PCR assay, using DNA as a template, cannot distinguish between live and dead bacteria. Use of the TaqMan assay in reverse transcription-PCR, using the 16S rRNA as a template, may be able to detect live bacteria. With this reverse transcription-PCR assay, one could then hope to study the viability of bacteria in blood.
Since the 16S rRNA gene has regions of conserved sequences in all bacterial species, targeting the ubiquitous sequences on this template by PCR has some inherent problems. Contamination of samples by bacteria from the laboratory environment or by translocation of bacterial DNA from the gut to the blood could lead to false positives. Precautions were taken to use sterile reagents and conditions wherever possible. Dedicated pre- and post-PCR pipettes and rooms were used. The PCR reagents were added in a UV-irradiated hood. It is unlikely other bacterial species would be detected by this assay, since the assay uses three oligonucleotides; all of them would have to have substantial homology with the template being amplified. Thus, even though the TaqMan probe and the reverse primer show 100% homology with the corresponding region of 16S rDNA of H. alvei, no signal was generated when the TaqMan PCR was performed with this set of oligonucleotides. The same was true with S. grimesii. However, the unspiked blood sometimes showed an unspecific amplification around cycle 40. Increasing the annealing temperature to 62°C or changing the forward and reverse primer set did not solve the problem. However, the ΔRn was very small in the unspiked blood.
Although a detection sensitivity of 6 bacteria/200 μl (30 bacteria/ml) of blood is better than that for any previously published methods, this assay still perhaps cannot be used to test donors or donated blood on day 0 or 1. However, it can be used for testing of blood shortly after it is processed or during the early days of storage. The earliest time at which this detection level would be useful remains to be established.
The initial cost for equipment may pose a problem for the widespread use of this method. However, technological advances are being made, and smaller, field-oriented thermocyclers and spectrofluorometers, which use silicon chips, are being developed (6). The advanced nucleic acid analyzer recently described by Belgrader et al. (7), besides cutting the assay time to minutes and being portable, would also help decrease the cost. It is also easily adaptable to automation. Furthermore, with increased use of nucleic acid testing for detection of viral markers in blood, the technical base for such molecular testing is already being developed and will soon be in place.
In conclusion, the TaqMan assay is rapid and eliminates the use of multiplex PCR, Southern blotting, or agarose gel electrophoresis. The entire test can be completed in 3 h, which includes the DNA extraction step. In addition, only 100 to 200 μl of blood is needed for analysis. Coupled with the current development of automated systems for DNA preparation that are capable of handling DNA from 96 blood samples, such as the QIAamp BioRobot 9604 and QIAamp 9600 Biorobot kits, this assay could be considered for incorporation into high-throughput testing.
ACKNOWLEDGMENTS
I thank Soren Kamstrup, Chiang Syin, Paul Mied, Ursula Utz, Mary Beth Jacobs, Stephen Wagner, and Gilliam Conley for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K. Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstains, a major inhibitor of polymerase chain reaction (PCR) amplification. J Forensic Sci. 1994;39:362–372. [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arduino M J, Bland L A, Tipple M A, Aguero S M, Favero M S, Jarvis W R. Growth and endotoxin production of Yersinia enterocolitica and Enterobacter agglomerans in packed erythrocytes. J Clin Microbiol. 1989;27:1483–1485. doi: 10.1128/jcm.27.7.1483-1485.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backman A, Lantz P, Radstrom P, Olcen P. Evaluation of an extended diagnostic PCR assay for detection and verification of the common causes of bacterial meningitis in CSF and other biological samples. Mol Cell Probes. 1999;13:49–60. doi: 10.1006/mcpr.1998.0218. [DOI] [PubMed] [Google Scholar]
- 5.Bassler H A, Flood S J A, Livak K J, Marmaro J, Knorr R, Batt C A. Use of a fluorogenic probe in a PCR-based assay for the detection of Listeria monocytogenes. Appl Environ Microbiol. 1995;61:3724–3728. doi: 10.1128/aem.61.10.3724-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belgrader P, Benett W, Hadley D, Richards J, Stratton P, Mariella R, Jr, Milanovich F. PCR detection of bacteria in seven minutes. Science. 1999;284:449–450. doi: 10.1126/science.284.5413.449. [DOI] [PubMed] [Google Scholar]
- 7.Belgrader P, Benett W, Hadley D, Long G, Mariella R, Jr, Milanovich F, Nasarabadi S, Nelson W, Richards J, Stratton P. Rapid pathogen detection using a microchip PCR array instrument. Clin Chem. 1998;44:2191–2194. [PubMed] [Google Scholar]
- 8.Bradley R M, Gander R M, Patel S K, Kaplan H S. Inhibitory effect of 0 degree C storage on the proliferation of Yersinia enterocolitica in donated blood. Transfusion. 1997;37:691–695. doi: 10.1046/j.1537-2995.1997.37797369443.x. [DOI] [PubMed] [Google Scholar]
- 9.Buchholz D H, AuBuchon J P, Snyder E L, Kandler R, Edberg S, Piscitelli V, Pickard C, Napychank P. Removal of Yersinia enterocolitica from AS-1 red cells. Transfusion. 1992;32:667–672. doi: 10.1046/j.1537-2995.1992.32792391043.x. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. Epidemiologic notes and reports update: Yersinia enterocolitica bacteremia and endotoxin shock associated with red blood cell transfusions—United States, 1991. Morb Mortal Wkly Rep. 1991;40:176–178. [PubMed] [Google Scholar]
- 11.Feng P, Keasler S P, Hill W E. Direct identification of Yersinia enterocolitica in blood by polymerase chain reaction amplification. Transfusion. 1992;32:850–854. doi: 10.1046/j.1537-2995.1992.32993110759.x. [DOI] [PubMed] [Google Scholar]
- 12.Gibson U E, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 13.Harnett N, Lin Y P, Krishnan C. Detection of pathogenic Yersinia enterocolitica using the multiplex polymerase chain reaction. Epidemiol Infect. 1996;117:59–67. doi: 10.1017/s0950268800001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 15.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′----3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibrahim A, Liesack W, Griffiths M W, Robins-Browne R M. Development of a highly specific assay for rapid identification of pathogenic strains of Yersinia enterocolitica based on PCR amplification of the Yersinia heat-stable enterotoxin gene (yst) J Clin Microbiol. 1997;35:1636–1638. doi: 10.1128/jcm.35.6.1636-1638.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kane T D, Alexander J W, Johannigman J A. The detection of microbial DNA in the blood: a sensitive method for diagnosing bacteremia and/or bacterial translocation in surgical patients. Ann Surg. 1998;227:1–9. doi: 10.1097/00000658-199801000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D M, Brecher M E, Bland L A, Estes T J, McAllister S K, Aguero S M, Carmen R A, Nelson E J. Prestorage removal of Yersinia enterocolitica from red cells with white cell-reduction filters. Transfusion. 1992;32:658–662. doi: 10.1046/j.1537-2995.1992.32792391041.x. [DOI] [PubMed] [Google Scholar]
- 19.Klein H G, Dodd R Y, Ness P M, Fratantoni J A, Nemo G J. Current status of microbial contamination of blood components: summary of a conference. Transfusion. 1997;37:95–101. doi: 10.1046/j.1537-2995.1997.37197176958.x. [DOI] [PubMed] [Google Scholar]
- 20.Kwaga J, Iversen J O, Misra V. Detection of pathogenic Yersinia enterocolitica by polymerase chain reaction and digoxigenin-labeled polynucleotide probes. J Clin Microbiol. 1992;30:2668–2673. doi: 10.1128/jcm.30.10.2668-2673.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak K J, Flood S J, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 22.Lucier T S, Brubaker R R. Determination of genome size, macrorestriction pattern polymorphism, and nonpigmentation-specific deletion in Yersinia pestis by pulsed-field gel electrophoresis. J Bacteriol. 1992;174:2078–2086. doi: 10.1128/jb.174.7.2078-2086.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarthy L R, Senne J E. Evaluation of acridine orange stain for detection of microorganisms in blood cultures. J Clin Microbiol. 1980;11:281–285. doi: 10.1128/jcm.11.3.281-285.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller V L, Farmer III J J, Hill W E, Falkow S. The ail locus is found uniquely in Yersinia enterocolitica serotypes commonly associated with disease. Infect Immun. 1989;57:121–131. doi: 10.1128/iai.57.1.121-131.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris T, Robertson B, Gallagher M. Rapid reverse transcription-PCR detection of hepatitis C virus RNA in serum by using the TaqMan fluorogenic detection system. J Clin Microbiol. 1996;34:2933–2936. doi: 10.1128/jcm.34.12.2933-2936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romalde J L, Iteman I, Carniel E. Use of pulsed field gel electrophoresis to size the chromosome of the bacterial fish pathogen Yersinia ruckeri. FEMS Microbiol Lett. 1991;68:217–225. doi: 10.1016/0378-1097(91)90130-3. [DOI] [PubMed] [Google Scholar]
- 27.Roth, V. R., M. Arduino, J. Nobiletti, S. Holt, L. Carson, C. E. F. Wolf, B. Lenes, P. Allison, and W. R. Jarvis. Transfusion-related sepsis due to Serratia liquefaciens in the United States. Transfusion, in press. [DOI] [PubMed]
- 28.Smith T J, Maher M, Gannon F, Dawson M T. Development of bacterial species-specific DNA probes based on ribosomal RNA genes using PCR. Methods Mol Biol. 1995;46:247–256. doi: 10.1385/0-89603-297-3:247. [DOI] [PubMed] [Google Scholar]
- 29.Stenhouse M A, Milner L V. Yersinia enterocolitica. A hazard in blood transfusion. Transfusion. 1982;5:396–398. doi: 10.1046/j.1537-2995.1982.22583017466.x. [DOI] [PubMed] [Google Scholar]
- 30.Tipple M A, Bland L A, Murphy J J, Arduino M J, Panlilio A L, Farmer III J J, Tourault M A, Macpherson C R, Menitove J E, Grindon A J. Sepsis associated with transfusion of red cells contaminated with Yersinia enterocolitica. Transfusion. 1990;30:207–213. doi: 10.1046/j.1537-2995.1990.30390194338.x. [DOI] [PubMed] [Google Scholar]
- 31.Trebesius K, Harmsen D, Rakin A, Schmelz J, Heesemann J. Development of rRNA-targeted PCR and in situ hybridization with fluorescently labelled oligonucleotides for detection of Yersinia species. J Clin Microbiol. 1998;36:2557–2564. doi: 10.1128/jcm.36.9.2557-2564.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner S J, Friedman L I, Dodd R Y. Transfusion-associated bacterial sepsis. Clin Microbiol Rev. 1994;7:290–302. doi: 10.1128/cmr.7.3.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokota M, Tatsumi N, Nathalang O, Yamada T, Tsuda I. Effects of heparin on polymerase chain reaction for blood white cells. J Clin Lab Anal. 1999;13:133–140. doi: 10.1002/(SICI)1098-2825(1999)13:3<133::AID-JCLA8>3.0.CO;2-0. [DOI] [PMC free article] [PubMed] [Google Scholar]