Abstract
Introduction
Immunotherapy has improved outcomes for patients with non-small cell lung cancer (NSCLC), yet durable clinical benefit (DCB) is experienced in only a fraction of patients. Here, we test the hypothesis that radiomic features from baseline pre-treatment 18F-FDG-PET/CT scans can predict clinical outcomes of NSCLC patients treated with checkpoint blockade immunotherapy.
Methods
This study included 194 patients with histologically confirmed stage IIIB-IV NSCLC with pre-treatment PET/CT images. Radiomics features were extracted from PET, CT, and PET+CT fusion images based on minimum Kullback–Leibler Divergence (KLD) criteria. The radiomic features from 99 retrospective patients were used to train a multiparametric radiomics signature (mpRS) to predict DCB using an improved least absolute shrinkage and selection operator (LASSO) method, which was subsequently validated in both retrospective (N=47) and prospective test cohorts (N=48). Using these cohorts, the mpRS was also used to predict progression-free survival (PFS) and overall survival (OS) by training nomogram models using multivariable Cox regression analyses with additional clinical characteristics incorporated.
Results
The mpRS could predict patients who will receive DCB, with areas under receiver operating characteristics curves (AUCs) of 0.86 (95%CI:0.79–0.94), 0.83 (95%CI:0.71–0.94), and 0.81 (95%CI:0.68–0.92) in the training, retrospective test, and prospective test cohorts, respectively. In the same three cohorts, respectively, nomogram models achieved C-indices of 0.74 (95%CI:0.68–0.80), 0.74 (95%CI:0.66–0.82), and 0.77 (95%CI:0.69–0.84) to predict PFS, and C-indices of 0.83 (95%CI:0.77–0.88), 0.83 (95%CI:0.71–0.94) and 0.80 (95%CI:0.69–0.91) to predict OS.
Conclusion
PET/CT-based signature can be used prior to initiation of immunotherapy to identify NSCLC patients most likely to benefit from immunotherapy. As such, these data may be leveraged to improve more precise and individualized decision support in the treatment of patients with advanced NSCLC.
Keywords: Non-small cell lung cancer (NSCLC), Immunotherapy, PET/CT, Radiomics, machine learning
INTRODUCTION
Lung cancer is the most commonly diagnosed cancer and the leading cause of cancer-related death in the world[1], with non-small cell lung cancer (NSCLC) being the most commonly diagnosed histologic subtype[2]. Over the past 30 years, the 5-year overall survival (OS) of patients with metastatic disease has remained at 5%[3]. More recently, checkpoint blockade antibodies targeting PD-(L)1 have revolutionized cancer treatment and improved long-term survival among some patients with advanced NSCLC[4–6]. However, current published evidence showed that PD-1/PD-L1 antibody monotherapies yield durable (>6mo.) clinical benefit (DCB) for only a subgroup of patients (15%~19.4% in phase I/II clinical trials[4, 7]). As such, robust biomarkers that are predictive of response immune-checkpoint blockades at baseline are needed to avoid immune-related-toxicities in patients unlikely to achieve durable clinical benefit.
Currently, PD-L1 status is the only approved diagnostic biomarker for immunotherapy; patients with positive PD-L1 status generally have higher objective response rates (ORR) [8, 9]. However, patients with PD-L1-negative tumors can still benefit from anti-PD-(L)1 therapies[6, 10]. A recent study showed that a combination of pembrolizumab and chemotherapy achieved objective radiographic response rates, ORRs, of 22.9% to 61.4% regardless of PD-L1 status[11]. Additionally, intra-tumor heterogeneity of PD-L1 staining across biopsies is prevalent, leading to sampling bias[12]. There have been concerted efforts to utilize molecular characteristic biomarkers for prediction and, along these lines, total mutational burden (TMB) exhibited a sensitivity of 86% and a specificity of 75% in predicting DCB in response to PD-1 checkpoint blockade[13]. However, TMB by whole exome sequencing is not clinically approved in the immunotherapy setting and is similarly subject to sampling bias. Hence, we seek to identify alternative predictive biomarkers that do not suffer from these weaknesses.
Radiological images are routinely available in clinic. In particular 18F-FDG PET/CT imaging is widely used for the staging of patients with advanced NSCLC. These medical images can be analyzed quantitatively with machine learning or “radiomics”, which is the process of converting medical images into high-dimensional mineable data. Prior studies have shown a strong correlation of radiomic signatures with genetics in NSCLC[14]. Recent advances in PET/CT radiomics have provided insights into precision medicine related to therapeutic response assessment[15, 16]. A Radiomics approach has multiple advantages, including: 1) being non-invasive and thus available for longitudinal sampling, 2) based on standard-of-care images and thus widely available, and 3) sampling the entire tumor and thus not subject to sampling bias[17].
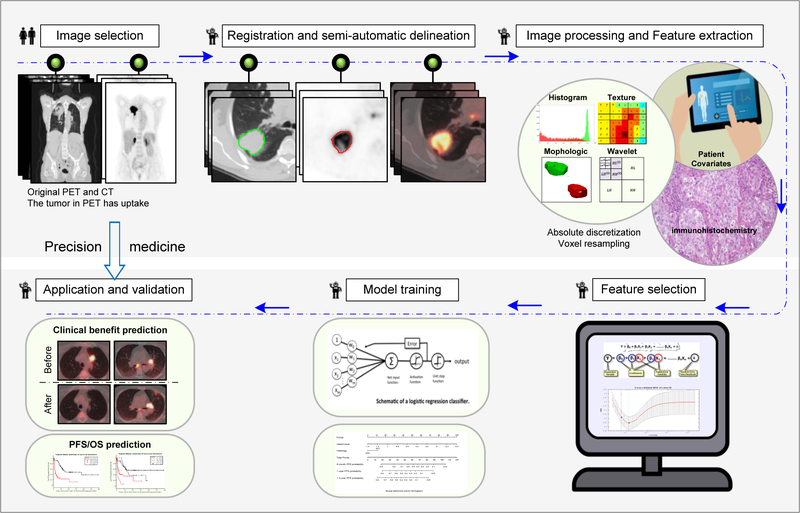
The goal of this study was to determine if PET/CT radiomic features at baseline, alone or in combination with clinical factors, can predict subsequent immunotherapy response leading to DCB, as well as to predict PFS and OS in patients with advanced NSCLC and secondarily, whether the PET/CT radiomic feature could be as a prediction biomarker in real time. The workflow is presented in Fig.1.
Fig 1. Radiomics Workflow.
The workflow includes image selection (only images with slice thickness≤5 mm no artifacts, and the tumor in PET images has FDG uptake were included), registration and automatic delineation, imaging preprocessing and feature extraction, feature selection, model training and model validation.
PATIENTS AND METHODS
Patients
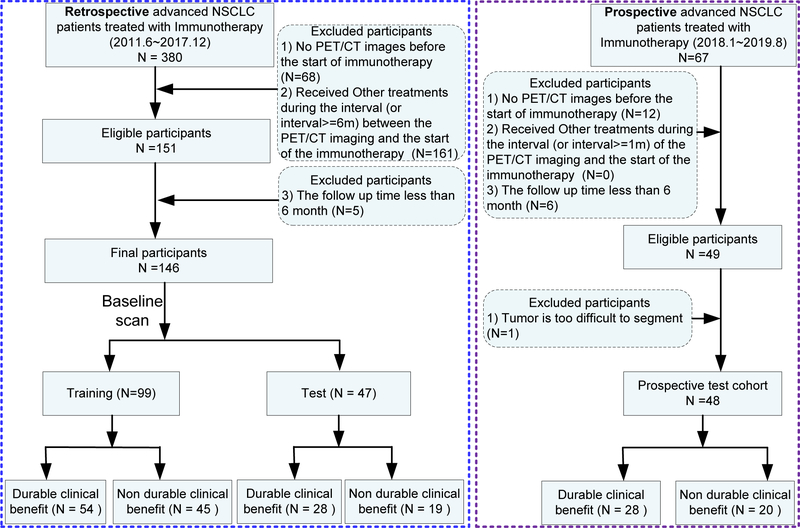
Inclusion criteria for this study included 1) patients with histologically-confirmed advanced stage (stage IIIB and IV) NSCLC who were treated with anti-PD-(L)1 immunotherapy, 2) PET/CT images were acquired during the interval (less than 6 months) of the last treatment (or diagnosis) and the start of immunotherapy, 3) no other treatment were provided during the interval and 4) follow-up time from initiation of immunotherapy treatment was greater than 6 months (Fig.2). Initially, 146 retrospective patients were enrolled who initiated therapy between June 2011 and December 2017. These patients were randomized into training (N=99) and test (N=47) cohorts, with the conditions that these two cohorts were not significantly different in terms of demographics, as well as FDG-PET avidity, as measured by the maximum standard uptake value (SUVmax). Using the same inclusion criteria, a prospective cohort was subsequently accrued, consisting of 48 patients who initiated immunotherapy between January 2018 and June 2019. This was used as an additional independent (prospective) test cohort.
Fig 2. Inclusion and exclusion diagram.
The training cohort comprised clinical data and the corresponding imaging data of the retrospective patients were used to train the radiomics signature, the DCB, PFS and OS nomogram models, which were further validated using the test cohort of the retrospective patients and the prospective cohort enrolled according to the same inclusion and exclusion criterion.
The baseline clinical characteristics (age, sex, histology, smoke, chronic obstructive pulmonary disease (COPD) status, family history, ECOG scale, distant metastasis (M stage), brain metastasis and prior treatments), gene (EGFR, ALK and ROS1) mutation status were obtained from the medical records. The main endpoints of this study were: DCB (durable benefit), PFS, and OS. Clinically, immunotherapy response is frequently measured as DCB or NDB (no durable benefit) using binary cutoff of PFS at 6-months[18, 19]. PFS and OS, were defined as the time from the start date of immunotherapy to progression (or death), and patients free of progression (or alive) or lost to follow-up which were censored at the time of last confirmed contact. Response Evaluation Criteria in Solid Tumors (RECIST1.1)[20] was used to define progression.
This study was approved by the Institutional Review Board at the University of South Florida (USF), and was conducted in accordance with ethical standards of the 1964 Helsinki Declaration and its later amendments. The requirement for informed consent was waived, as PHI was not revealed.
PET/CT imaging
18F-FDG PET/CT imaging was performed as standard diagnostic work-up before treatment with immunotherapy. Details of the retrospective PET/CT images obtained from 9 different scanner and the prospective PET/CT images obtained from 2 different scanners are shown in Supplemental Methods S1. Heterogeneity in scanner parameters was deliberately chosen to ensure generalizability of the derived predictive models. All PET images were converted into SUV units by normalizing the activity concentration to the dosage of 18F-FDG injected and the patient body weight after decay correction. Further, all the PET and CT images were resampled to 1×1×1mm3 voxels using 3-dimensional Lagrange Interpolating Polynomials.
Radiomic feature extraction
The primary lung tumors of PET and CT images were semi-automatically segmented with an improved level-set method based on gradient fields [19] were further reviewed and corrected by a radiologist with 16 years of experience (JQ) who was blinded to the outcome label. After spatial registration using a rigid transformation by maximizing the Dice Similarity Coefficients on the condition that the maximal axial cross sections of the nodules were aligned, KLD images were generated from the fused PET and CT images on a voxel-wise basis using Kullback–Leibler Divergence (KLD) criteria[21]. 790 features including PET features, CT features, and KLD features were then extracted from these segmented tumors and scaled into the range [0 1] with unity-based normalization, as shown in Supplemental Methods S2–S4. After dimensionality reduction according to the internal stability (Supplemental Methods S5), we investigated the radiomics expression patterns with unsupervised clustering to observe correlations among different features and different patients.
Statistical analyses
The Wilcoxon signed-rank test and Fisher’s exact test were used to test of differences for continuous variables and categorical variables, respectively. For PFS and OS comparisons, a log-rank test was used. To determine the association of the radiomics expression patterns with clinical characteristics, a χ2 test was used. P values less than 0.05 were regarded as significant. Statistical analyses were performed with R 3.5.2 and MATLAB R2019a (Natick, MA)
Feature selection and radiomics signature building
We improved the least absolute shrinkage and selection operator LASSO method[22] by performing analyses separately on Squamous Carcinomas or Adenocarcinoma (Supplemental Methods S6) to develop minimal feature sets for each that retained predictive information, which were then combined into a multiparametric radiomic signature (mpRS) weighted by their respective coefficients. To avoid overfitting, 100 times 5-fold cross validation was performed in the training cohort to generate a prioritized list of the most parsimonious sets of predictive features.
Improved diagnostic validation of mpRS
To investigate the importance of the KLD features, digital phantom simulation experiments were performed. Two different digital phantoms were generated to have the same heterogeneity distribution (measured by Entropy and Inverse Difference calculated from 3D co-occurrence matrix) and used to detect the different heterogeneity distribution of the KLD fusion images.
Additionally, three different radiomics signatures generated from PET features (PETRS), CT features (CTRS), and the linear combination of PET and CT features (PETCTRS) using the above method were used for the comparison with mpRS according to their discrimination performances measured by the areas under receiver operating characteristics curves (AUC), classification accuracy (ACC), sensitivity (SEN), and specificity (SPEC). Furthermore, the total net reclassification improvement (NRI) was used to investigate the added value of the KLD features to PET and CT features.
ANOVA analysis was performed to compare the distribution of the radiomics signatures among the different scanner types.
DCB prediction nomogram model building
Univariable logistical regression analysis was initially conducted to identify radiomics signatures, clinical factors, and common metrics (including the image-derived features of volume, SUVmax, and metabolic tumor volume*23+) that are associated with a DCB. Covariates that yielded a significant Wald’s Statistic from multivariable logistical regression analysis were used for developing the DCB prediction nomogram model.
The goodness-of-fit for the models were evaluated with Akaike Information Criteria (AIC) and the Hosmer-Lemeshow (HL) tests[24]. The AUC, ACC, SEN, and SPEC were also used to evaluate the discrimination performances of different models. To compare the clinical usefulness of the different models, a decision curve analysis was performed by quantifying the added benefits at different threshold probabilities[25].
PFS and OS estimation nomogram model building
The potential of the radiomics signature to predict PFS and OS was assessed and optimized in the training cohort and then validated in the two independent test cohorts by using Kaplan-Meier survival analysis. The patients were classified into high-risk or low-risk groups according to the radiomics score cutoff that maximized Youden’s index based on the training cohort.
Univariable Cox regression analyses were conducted using the radiomics signature, clinical factors, SUVmax, MTV (metabolic tumor volume), and volume. Statistically significant hazard ratios (HRs) were included in PFS and OS nomogram models using multivariable Cox regression analysis and a backward step-wise selection with AIC as the stopping rule. The C-index and AIC were used to evaluate the prediction ability of the models, and Z test was applied to determine whether the differences between different models were significant.
Pilot study for longitudinal assessment of the radiomic signature
In order to validate the dynamic characteristic and investigate whether the radiomics signature could identify the patients receiving benefit over the ensuing six months, patients with follow-up PET/CT scans after the initiation of the immunotherapy were used to investigate the longitudinal trend of the radiomics signature. The patients who had DCB since the start of the immunotherapy but progressed within 6 month since the follow-up scan time were regarded as follow-up NDB, and the patients didn’t progress within 6 months since the follow-up scan time were regarded as follow-up DCB. The AUC was calculated to evaluate the follow-up DCB and NDB discrimination performances of the radiomic signature, and the distribution of the radiomic signature across the baseline time and follow-up time were used to reveal the trend of the radiomic signature.
Finally, to assess the quality of this radiomics study, the radiomic quality score (RQS) was calculated according to Lambin et al.[26]
RESULTS
Clinical characteristics
The demographic and clinical characteristics of the three patient cohorts are presented in Table 1. Among the 146 retrospective patients, there were 88 men and 58 women and the overall mean age was 65.72 (±12.88), and the median PFS and OS were 7.52 and 10.38 months. The retrospective training and test cohorts had identical distributions of SUVmax, and statistically insignificant differences in their clinical characteristics, PFS (p=0.62) and OS (p=0.42) (Supplemental Fig. S1). The 48 prospective patients curated from standard of care regimens had median PFS and OS of 6.78 and 9.95 months, respectively. Although slightly shorter compared to the retrospective cohorts, the PFS and OS were not significantly different in the prospective cohort. There were also no significant differences in histology, smoking history, sex, age, and mutation status. 22 patients had follow up PET/CT scans between 1 and 53 months of therapy, and these were used to investigate the longitudinal trend of the radiomics signature in a pilot study. Five of these patients had NDB since the start of the immunotherapy. For the remaining 17 patients who had DCB since the start of the immunotherapy, 6 of them progressed within 6 month since the follow-up scan time, while 11 patients did not progress and continued to experience clinical benefit within 6 month since the follow-up scan time.
Table 1.
Demographic and clinical characteristics of patients
| Characteristic | Training Cohort | Test Cohort | Pǂ | Prospective Cohort | P§ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| DCB(N=54) | NDB(N=45) | P# | DCB(N=28) | NDB(N=19) | P# | DCB(N=28) | NDB(N=20) | P# | |||
| Age(y) | 0.12 | .015* | 0.38 | 0.43 | 0.72 | ||||||
| Mean ± SD | 66.76±13.64 | 63.04±14.24 | 70.43±6.08 | 62.05±12.95 | 65.32±8.92 | 68.00±10.90 | |||||
| Sex, NO. (%) | 0.54 | 1.00 | 0.59 | 0.24 | 0.19 | ||||||
| Male | 30 (55.56) | 28 (62.22) | 18 (64.29) | 12 (63.16) | 11 (39.29) | 12 (60.00) | |||||
| Female | 24 (44.44) | 17 (37.78) | 10 (35.71) | 7 (36.84) | 17 (60.71) | 8 (40.00) | |||||
| BMI | .008* | 0.20 | 0.44 | 0.12 | 0.83 | ||||||
| Mean ± SD | 26.84±4.86 | 24.3±5.06 | 27.03±5.71 | 25.26±3.53 | 24.91±3.77 | 27.69±6.19 | |||||
| ECOG scale | <.001* | .019* | 0.89 | 0.57 | 0.78 | ||||||
| 0 | 20 (37.04) | 2 (4.44) | 9 (32.14) | 0(0) | 6 (21.43) | 3 (15.00) | |||||
| 1 | 34 (62.96) | 36 (80) | 18 (64.29) | 17 (89.47) | 21 (75.00) | 17 (85.00) | |||||
| >=2 | 0 (0) | 7 (15.56) | 1 (3.57) | 2 (10.53) | 1 (3.57) | 0 (0) | |||||
| Distant Metastasis | 0.12 | .003* | 0.49 | 0.025 | <.001* | ||||||
| M0 | 7 (12.96) | 2 (4.44) | 1 (3.57) | 1 (5.26) | 6 (21.43) | 0 (0) | |||||
| M1a | 28 (51.85) | 19 (42.22) | 18 (64.29) | 3 (15.79) | 13 (46.43) | 7 (35.00) | |||||
| M1b | 19 (35.19) | 24 (53.33) | 9 (32.14) | 15 (78.95) | 9 (32.14) | 13 (65.00) | |||||
| Brain Metastasis | 0.66 | 0.40 | 0.66 | 0.49 | <.009* | ||||||
| NO. (%) | 2(3.70) | 3(6.67) | 0 | 1(5.26) | 7(25.00) | 3(15.00) | |||||
| EGFR, NO. (%) | 0.12 | 0.61 | 0.28 | 0.53 | 0.60 | ||||||
| Mutation | 4(7.41) | 0(0) | 2(7.14) | 2(10.53) | 2(7.14) | 0(0) | |||||
| Wild | 35(64.81) | 32(46.67) | 19(67.86) | 10(52.63) | 23(82.14) | 14(70.00) | |||||
| ALK, NO. (%) | 0.44 | 1.00 | 0.29 | 1.00 | 0.86 | ||||||
| Mutation | 0(0) | 1(2.22) | 1(3.57) | 0 | 1(3.57) | 0(0) | |||||
| Wild | 40(74.07) | 30(66.67) | 20(71.43) | 12(63.16) | 23(82.14) | 15(75.00) | |||||
| ROS1, NO. (%) | NaN | 0.25 | 0.66 | NaN | 1.00 | ||||||
| Mutation | 0(0) | 0(0) | 1(3.57) | 0(0) | 0(0) | 0(0) | |||||
| Wild | 15(27.78) | 8(17.78) | 1(3.57) | 6(31.58) | 20(71.43) | 13(65.00) | |||||
| Histology (baseline), NO. (%) | 0.53 | .011* | 1.00 | <.001 | 0.11 | ||||||
| ADC | 37 (68.52) | 28 (62.22) | 23 (82.14) | 8 (42.11) | 23 (82.14) | 4 (20.00) | |||||
| SCC | 17 (31.48) | 17 (37.78) | 5 (17.86) | 11 (57.89) | 5 (17.86) | 16 (80.00) | |||||
| Historical treatment, NO. (%) | |||||||||||
| SURG | 9 (16.67) | 8 (17.78) | 1.00 | 4 (14.29) | 2 (10.53) | 1.00 | 0.63 | 19 (67.86) | 8 (40.00) | 0.079 | <.001 |
| RT | 22 (40.74) | 17 (37.78) | 0.84 | 8 (28.57) | 10 (52.63) | 0.13 | 1.00 | 14 (50.00) | 7 (35.00) | 0.38 | 0.57 |
| Chemo | 34 (62.96) | 28 (62.22) | 1.00 | 17 (60.71) | 11 (57.89) | 1.00 | 0.72 | 21 (75.00) | 18 (90.00) | 0.27 | 0.012 |
| Smoke, NO. (%) | 0.44 | 0.28 | 0.17 | 0.34 | 0.59 | ||||||
| Non | 24 (44.44) | 16 (35.56) | 11 (39.29) | 5 (26.32) | 6 (21.43) | 7 (35.00) | |||||
| Former | 28 (51.85) | 25 (55.56) | 16 (57.14) | 11 (57.89) | 22 (78.57) | 13 (65.00) | |||||
| Current | 2 (3.7) | 4 (8.89) | 1 (3.58) | 3 (15.79) | 0 | ||||||
| COPD | 0.78 | 0.17 | 0.36 | 0.25 | 0.94 | ||||||
| NO. (%) | 9 (16.67) | 6 (13.33) | 8 (28.57) | 2 (10.53) | 3 (10.71) | 5 (25.00) | |||||
| Family History, NO. (%) | 0.48 | 0.57 | 0.17 | 0.54 | 0.11 | ||||||
| negative | 15 (27.78) | 9 (20) | 9 (32.14) | 8 (42.11) | 10 (35.71) | 5 (25.00) | |||||
| positive | 39 (72.22) | 36 (80) | 19 (67.86) | 11 (57.89) | 18 (64.29) | 15 (75.00) | |||||
| Progression-free Survival | <.001* | <.001* | 0.61 | <.001* | 0.56 | ||||||
| Rate (%) | 62.96 | 8.89 | 60.71 | 0 | 71.43 | 5.00 | |||||
| Time to progression (month) | |||||||||||
| Median(Mean) | 12.55(17.45) | 1.83 (2.40) | 12.85(19.46) | 3.13 (3.31) | 10.30(11.32) | 3.38 (3.87) | |||||
| Overall Survival | <.001* | <.001* | 0.42 | <.001* | 0.85 | ||||||
| Rate (%) | 83.33 | 33.33 | 89.29 | 42.11 | 89.29 | 50.00 | |||||
| Overall survival time (month) | |||||||||||
| Median(Mean) | 19.37(20.34) | 5.47 (7.06) | 15.27(20.11) | 6.57 (7.37) | 12.02(12.05) | 5.25 (6.88) | |||||
| mpRS | <.001* | <.001* | <.001* | ||||||||
| Mean ± SD | 0.68±0.24 | 0.32±0.24 | 0.56±0.25 | 0.27±0.20 | 0.57±0.21 | 0.28±0.23 | |||||
Note.- :
means P value <.05. RT is short for radiotherapy; Chemo is short for chemotherapy; SD: standard deviation. The interval between PET/CT and treatment initiation of ranged from 0 to 6 months (median: 1.65 month) for training and test cohorts, and was less than 1month for prospective cohort.
The comparasion of age and BMI between two groups was performed with Wikcoxon sign rank test, and the rest variables were compared with Fisher’s test. The P value for PFS and OS are derived from the log-rank test; P# value is derived between DCB and NDB; Pǂ is derived between training and test cohorts. P§ is derived between the retrospective and prospective cohorts. Data are patient numbers, with percentages in parentheses. The groups of the variables are mutually exclusive except historical treament. The historical treatment of a patients could include any of the three treatments. NaN means no statistics are computed because the variable is a constant. The missing data in the groups of EGFR, ALK and ROS1 was not listed.
Feature extraction
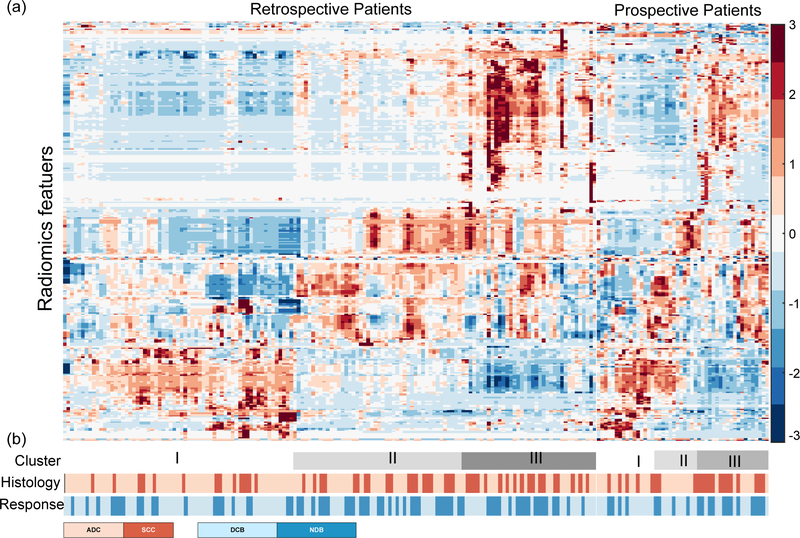
Of the original 790 extracted features, 324 remained after filtering for internal stability. Unsupervised clustering revealed 3 clusters of patients with similar radiomic expression patterns (Fig.3a), which were significantly associated with histology (p=0.008, χ2 test) and response (p=0.028, χ2 test). The prospective patients further showed similar radiomic expression patterns and validated this association of these radiomic feature patterns with histology (p=0.041, χ2 test) and response (p=0.085, χ2 test) (Fig. 3b).
Fig 3. Expression patterns of Radiomic features.
(a) Unsupervised clustering of all the retrospective and prospective patients on the row direction and radiomics feature expression on the column direction, revealed clusters of patients with similar radiomics expression patterns. (b) Clinical patient parameters for showing significant association of the radiomics expression patterns with Histology (retrospective: p = 0.008, prospective: p = 0.041, χ2 test) and response (retrospective: p = 0.029, prospective: p = 0.085, χ2 test)
Feature Selection and Radiomics Signature building
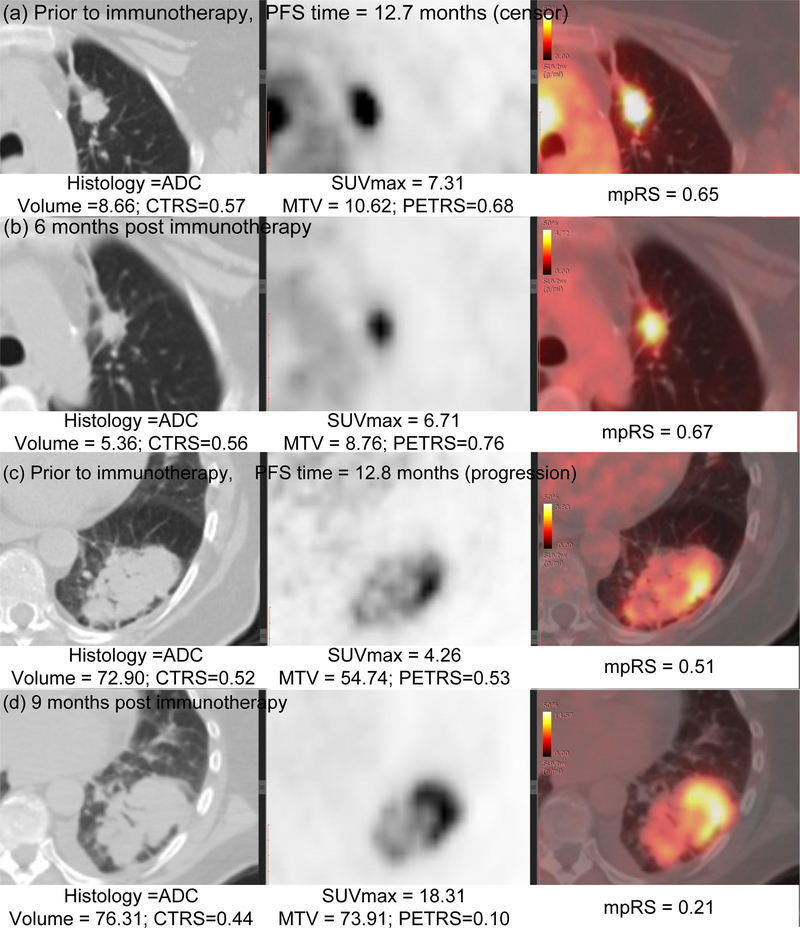
Pearson grouping was used to eliminate redundant features, resulting in 21 uncorrelated features (10 PET features, 4 CT features, and 7 KLD features). These were then used as inputs into the LASSO method. Through 5-fold cross validation on the training cohort (Supplemental Fig.S2), 8 features emerged as the best features to construct radiomics signature, and these are shown in Supplemental data S7. Representative radiomics signatures of two patients from baseline PET/CT scan and follow-up PET/CT scan are shown Fig.4.
Fig 4. Radiomics signatures of NSCLC patients.
(a~b) The CT, PET, and fusion images for a patient with ADC NSCLC obtained 1 month before and 6 month after immunotherapy, which means the patient would have DCB since the start of immunotherapy and 6 month post immunotherapy; (c)~(d) The CT, PET, and fusion images for a patient with ADC NSCLC obtained 1 month before and 9 month after immunotherapy, which means the patient would have DCB since the start of immunotherapy, but would have NDB after 9 month immunotherapy. The corresponding clinical feature and radiomics scores are shown in the bottom of each image.
Validation of the radiomics signature
Improved diagnostic validation of mpRS
From the simulated PET images (the first column) and simulated CT images (the second column) shown in Fig. S3, the two phantoms are classified as having the same heterogeneity and homogeneity distribution. However, from the fusion images (the third column), the two phantoms were classified as having different heterogeneity and homogeneity, which means the KLD features could identify the relative different positional relationship of the heterogeneity.
There were significant differences in the four radiomics signature scores between DCB and NDB patients in the training cohort (PETRS: p<0.001; CTRS: p=0.020; PETCTRS: p<0.001; mpRS: p<0.001). Except for the CTRS, the other three radiomics signatures had significant differences between DCB and NDB patients in the retrospective (PETRS: p=0.006; PETCTRS: p=0.003; mpRS: p<0.001) and prospective (PETRS: p=0.019; PETCTRS: p=0.009; mpRS: p<0.001) test cohorts. The mpRS achieved the highest AUCs of 0.86 (95%CI:0.79–0.93), 0.83 (95%CI:0.71–0.94), and 0.81 (95%CI:0.68–0.92) in the training, retrospective test, and prospective test cohorts, respectively (Supplemental Table S1 and Fig S4). Compared to PET+CT features, the inclusion of the KLD features yielded a total net reclassification improvement (NRI) of 0.50 (95%CI:0.11–0.88,p=0.011), 0.55 (95%CI:−0.012–1.11,p=0.055), and 0.94 (95%CI:0.44–1.45,p<0.001) in the training, test, and prospective test cohorts, respectively, which showed significantly improved classification accuracy for response prediction. Therefore, only mpRS was used for the subsequent analyses.
Box plots and ANOVA analyses of each of the radiomic signatures are shown in Supplemental Fig.S5, which illustrates that these signatures are stable across 9 different equipment manufacturers (p>0.05), with mpRS being the most stable signature with the fewest outliers.
DCB prediction nomogram
Univariable logistical regression analysis on the training cohort identified mpRS, distant metastasis, and ECOG status as strong predictors for response, and these were also validated in the test cohorts (p<0.05, Supplemental Table S2). Through multivariable logistical regression analysis (Supplemental Table S3), ECOG scale and Distant Metastasis were predictive of a DCB response combined with the mpRS (Fig.5a), and adding these clinical variables generated a higher AUC of 0.89, 0.86 and 0.86 in the training, retrospective test and prospective test cohorts, respectively (Supplemental Table S1 and Fig. S4). This model was well calibrated in all three cohorts (Fig.5c). The inclusion of ECOG scale and Distant Metastasis yielded a significant total NRI of 0.79 (95%CI: 0.47–1.01, p<0.001), 1.05 (95%CI: 0.55–1.54, p<.001), and 1.20 (95%CI: 0.75–1.65, p<.001) in the training, retrospective test and prospective test cohorts, respectively.
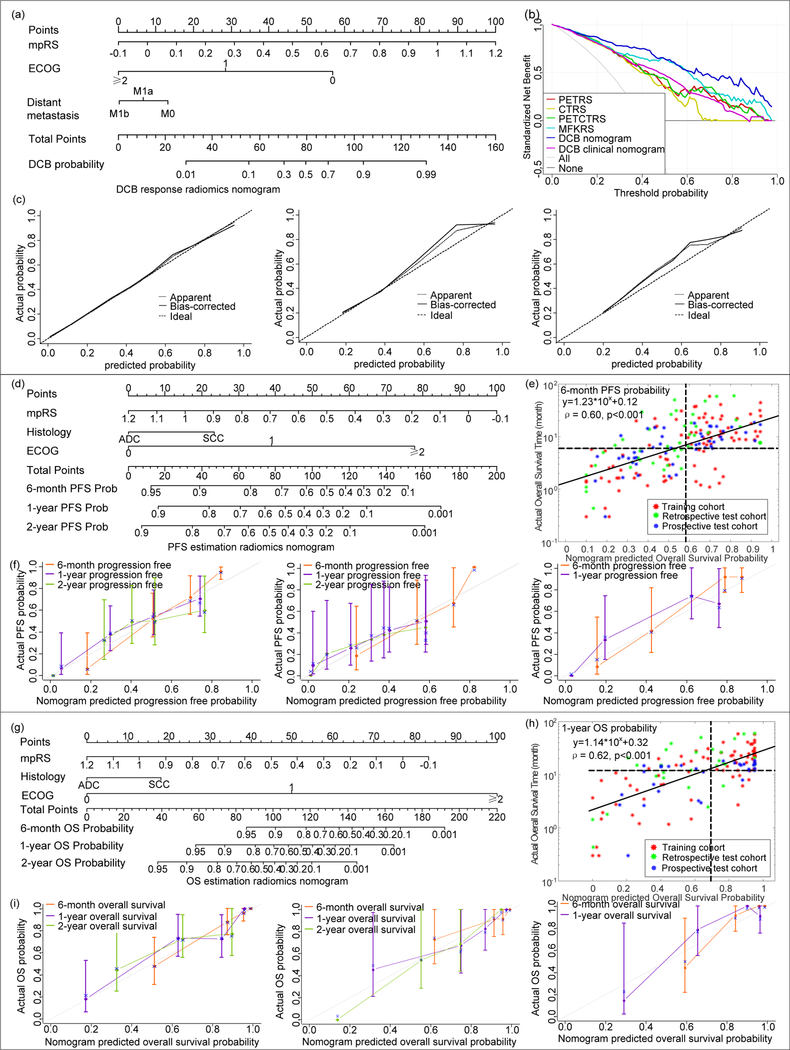
Fig 5. Nomograms and the corresponding calibration curves.
(a) and (c) are the nomograms constructed with mpRS and clinical characteristics to estimate the probability of DCB, along with the assessment of the model calibration in the training cohort, retrospective test cohort and prospective cohort, respectively. (e.g. For a patient with MRS of 0.6, ECOG 1 at stage M0, his total point is 95 (MRS 0.6 corresponding to point 54, ECOG 1 corresponding to point 28, no distant metastasis corresponding to point 13, 54+28+13=95), which corresponds to a DCB probability of 0.79). (b) is the decision curves for different radiomics signatures and DCB response prediction models on all the patients. (d) and (f) are the nomograms constructed with mpRS and clinical characteristics to estimate the risk of progression, along with the assessment of the model calibration in the training cohort, retrospective test cohort and prospective cohort, respectively. (e.g. For a ADC patient with MRS of 0.6 and ECOG 1, his total point is 85 (MRS 0.6 corresponding to point 46, ECOG 1 corresponding to point 39, ADC corresponding to point 0, 46+39+0=85), which corresponds to a 6-month PFS probability of 0.71, 1-year PFS probability of 0.5, and 2-year PFS probability of 0.42). (e) is the scatter plots of nomograms predicted 6-month PFS probability of individual patients versus PFS time of the corresponding patients. (g) and (i) are the nomograms constructed with mpRS and clinical characteristics to estimate the risk of death, along with the assessment of the model calibration in the training cohort, retrospective test cohort and prospective cohort, respectively. (e.g. For a ADC patient with MRS of 0.6 and ECOG 1, his total point is 89 (MRS 0.6 corresponding to point 39, ECOG 1 corresponding to point 50, ADC corresponding to point 0, 39+50+0=89), which corresponds to a 6-month OS probability of 0.94, 1-year OS probability of 0.81, and 2-year OS probability of 0.59). (h) is the scatter plots of nomograms predicted 1-year OS probability of individual patients versus OS time of the corresponding patients. For (e) and (h), the red points represent the training cohort, the green points represent the test cohort, and the blue points represent the prospective cohort. The vertical dotted line and the horizontal dotted line mean the actual PFS and OS probability obtained with Kaplan-Meier estimate at the certain time point, and the horizontal dotted line means the 6 month and 1 year time point. The pair of the vertical and horizontal dotted lines divided the graph into 4 quadrants, the points in the lower left and top right quadrants correspond to the accurate prediction.
The decision curves shown in Fig.5b indicate that the combined (clinical + radiomics) DCB nomogram model had the highest overall net benefit across the majority of the range of reasonable threshold probabilities in all the patients compared to radiomics or clinical signatures alone.
PFS and OS prediction nomogram
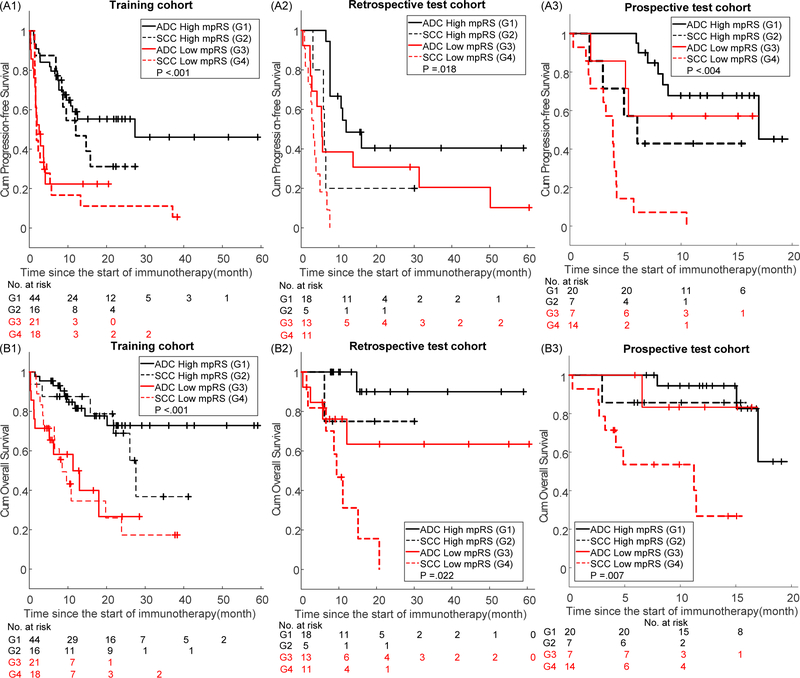
Through Kaplan-Meier survival analysis (cutoff=0.41), mpRS was able to significantly predict PFS and OS in training (p<0.001, p<0.001), retrospective test (p=0.001, p=0.002) and prospective test cohorts (p<0.001, p=0.002), respectively. Patients with higher radiomics scores had longer PFS and OS (Supplemental Fig.S6). According to univariable Cox regression analysis (Supplemental Tables S4 and S6), mpRS, histology, and ECOG scale were significantly associated with PFS and OS in the training and test cohorts. To investigate the prognostic value of the mpRS within the different subgroups of histology, stratified Kaplan-Meier survival analyses were also performed. As shown in Fig. 6, even though patients with adenocarcinoma (ADC) had better PFS than those with squamous cell carcinoma (SCC), the mpRS was predictive of PFS and OS for both histologies. Further multivariate Cox proportional hazards models (Supplemental Tables S5 and S7) including mpRS, histology and ECOG scale were built for PFS and OS estimation, which are expressed as nomograms shown in Fig. 5d and g. The calibration curves of the mpRS nomogram on training, retrospective test and prospective test cohorts are shown in Fig.5f and i for PFS and OS, respectively, which show excellent agreements between the nomogram predictions and actual observation. Additionally, the scatter plots of nomogram-predicted 6-month PFS and 1-year OS probability for individual patients versus the corresponding PFS and OS times are provided in Fig 5e and h. The Spearman’s rank correlation coefficients for individualized predictions were > 0.6 with p<0.001, suggesting a strong positive correlation.
Fig 6. Stratified Kaplan-Meier survival curves.
(a-c) are stratified Kaplan-Meier survival curves of PFS according to mpRS on training, retrospective test and prospective test cohorts within the different subgroups of histology. (d-f) are stratified Kaplan-Meier survival curves of OS according to mpRS on training, retrospective test and prospective test cohorts within the different subgroups of histology.
The quantitative results of different models are shown in Table 2, which shows the combined (clinical+radiomic) nomograms yielded significantly strong prediction results (p<0.05) with C-indexes of 0.74 (95% CI: 0.68–0.80), 0.74 (95% CI: 0.66–0.82) and 0.77(95%CI: 0.69–0.84) for PFS estimation, and C-indexes of 0.83 (95%CI:0.77–0.88), 0.83 (95%CI: 0.71–0.94), and 0.80 (95%CI: 0.69–0.91) for OS estimation in the training, test and prospective test cohorts, respectively.
Table 2.
Performance of different models in PFS and OS estimation
| Category | Training cohort | Test cohort | Prospective cohort | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| C-Index (95%CI) | AIC | p | C-index (95%CI) | AIC | p | C-index (95%CI) | AIC | p | |
| PFS estimation | |||||||||
| mpRS | 0.70 (0.64–0.76) | 475.42 | 0.37§ | 0.67 (0.58–0.75) | 231.03 | 0.81§ | 0.68 (0.59–0.78) | 177.68 | 0.69§ |
| PFS nomogram | 0.74 (0.68–0.80) | 464.21 | 0.12# | 0.74 (0.66–0.82) | 224.74 | 0.03# | 0.77 (0.69–0.84) | 175.02 | .051# |
| clinical nomogram | 0.66 (0.60–0.78) | 479.93 | .006ǂ | 0.68 (0.61–0.76) | 226.61 | 0.01ǂ | 0.66 (0.57–0.76) | 186.46 | .027ǂ |
| OS estimation | |||||||||
| mpRS | 0.74 (0.67–0.81) | 291.99 | 0.92§ | 0.71 (0.56–0.85) | 90.82 | 0.36§ | 0.76 (0.64–0.89) | 78.46 | 0.38§ |
| OS nomogram | 0.83 (0.77–0.88) | 266.82 | <.001# | 0.83 (0.71–0.94) | 79.38 | 0.07# | 0.80 (0.69–0.91) | 78.62 | 0.55# |
| clinical nomogram | 0.74 (0.67–0.82) | 286.77 | <.001ǂ | 0.79 (0.67–0.90) | 79.40 | 0.16ǂ | 0.67 (0.53–0.81) | 87.00 | .020ǂ |
The C-index was compared between the mpRS and the mpRS nomogram
The C-index was compared between the mpRS nomogram and clinical nomogram
The C-index was compared between the mpRS and clinical nomogram
Pilot study for longitudinal assessment of the radiomic signature
A further analysis using subsequent follow-up scans, when available, showed the mpRS generated from the follow-up PET/CT images during treatment could also predict follow-up DCB with an AUC of 0.82 (95%CI:0.63–1.00). Further, it had a decreasing trend with time (Supplemental Fig S7), suggesting that the risk of progression increased with time.
Radiomic quality score
Radiomics is a rapidly maturing field and qualities of study designs and their results can bechallenging to evaluate. To address this, Lambin et al. developed a 36 point “Radiomics Quality Score” (RQS) metric[26]. The criteria are described in Supplemental Table S8, which shows that the current study had a RQS of 24. To put this in perspective, a recent meta-analysis[27] analyzed 77 radiomics publications and documented that the mean RQS across all studies was 9.4 + 5.6, indicating that the current study is in the upper 5 percentile of radiomics study designs.
A TRIPOD Checklist following reporting guidelines for prediction model development and validation (https://www.equator-network.org/reporting-guidelines/tripod-statement/) has also been provided in Supplemental Table S9, which further validated the integrity of the work.
DISCUSSION
In this work, we developed a radiomics signature to predict DCB in immunotherapy-treated NSCLC patients, which was successfully validated in independent retrospective and prospective test cohorts. In addition, combining this signature with ECOG status and histology into a nomogram further facilitated the individualized prediction of PFS and OS prior to initiation of checkpoint blockade immunotherapy.
PET/CT radiomics studies typically extract image-derived quantitative features independently from PET or corresponding CT images *28–32+ and mutual information between PET and CT images have only assessed by qualitative metrics rather than quantitative descriptors [33–36]. By contrast, in this work, we constructed KLD features as quantitative descriptors of mutual information between co-registered PET and CT, and these significantly improved the prediction results. To illustrate the importance the KLD features, digital phantom simulation experiments were performed. Supplemental Fig. S3 shows that found that different phantoms (a and b) could not be distinguished by PET or CT heterogeneity features. However, the KLD fusion images, which reflect the relative different positional relationship of the heterogeneity, could distinguish these phantoms.
When investigating the informative components of mpRS formula, we found multiple texture features (PET_SRLGE, KLD_SZE) were positively correlated, suggesting that the more heterogeneous tumors had a larger probability to have a DCB. This was a bit surprising, as prior studies have shown that more heterogeneous tumors with CT textures had worse response to radiation or chemotherapy[14]. A recent study showing the patients who had faster growing tumors before initiation of immunotherapy had better responses[37] may suggest that the more aggressive tumors that respond worse to conventional therapies are more susceptible to immune modulation. In terms of shape, those tumors with more convexity (smaller CHDensity) had a higher probability of a DCB. This could be explained by Saeed-vafa’s study[38] that PD-L1 cells are able to form a more rounded mass due to the ability of evading immune attack and coexistence with the vasculature, and high PD-L1 expression is associated with significantly longer PFS and longer OS[8, 9]. In terms of metabolic activity, tumors with a lower mean SUV and HU (KLDiv_mean) had a larger probability of having a DCB. This is expected, as more metabolically active tumors will produce more lactic acid [39] and lactic acid is a potent inhibitor of effector T cell function[40].
Notably, only 4 CT features were remained after Pearson grouping and internal stability comparison, and the CT radiomics score constructed with these 4 features did not perform as well compared to mpRS, with an AUC of 0.69 and 0.64 in test and prospective test cohorts, respectively. These AUCs are smaller than those observed in a recent Annals of Oncology paper, which generated an AUC=0.79 in the NSCLC primary tumors with contrast enhanced computed tomography (CE-CT) scans[41]. This was also surprising, given our prior radiomic analyses of CT images to predict immunotherapy response[3]. A possible explanation for the low predictive power of CT in the current study is that the resolution of CT images in PET/CT is lower and non-contrast enhanced, compared to diagnostic CTs, and this also may lead to lower discrimination ability. Secondly, given no inter-scanner-correction was used for CT features, only a few CT features remained after filtering for internal stability. Additionally, other possible reasons that the 4 CT features were not selected in the mpRS formula were the KLD features had included the morphological information, and some studies had shown that the metabolic modifications on PET are more predictive than morphological modifications on CT especially in early response prediction[42, 11].
Moreover, as Moffitt Cancer Center is a referral hospital, many patients’ PET/CT images were acquired from different institutions, leading to large variability in acquisition and reconstruction methods. As we first filtered for internal stability, we selected for features that would be stable across different scanners. We contend that this is a strength of the current approach, as it allows for a more generalizable and transportable model (Supplemental Fig. S5).
SUVmax and MTV were shown effective in predicting efficacy and survival at 1 month after immunotherapy in some studies[42]. However, according to Supplemental Table S2, these factors were not significant factors in predicting DCB in our current study. On one hand, this may be because the time points were different (1 month vs 6 month), and SUVmax and MTV may play different roles at different time points. On the other hand, this may be due to the different cohorts. Those prior studies were single institution with rigorous acquisition protocols, whereas our study utilized data from multiple scanners, multiple institutions, acquired under standard of care. We have previously shown that reproducibility of SUVmax and SUVmean under standard of care multi-institutional PET+CT imaging is lower than that which can be achieved at a single institution under a research protocol[43].
The present study also possesses some limitations. First, PD-L1 status of most patients was unavailable in this dataset, and hence couldn’t be used for comparing with the mpRS. Second, given PET/CT is not commonly used in clinical follow up, and only 22 patients had follow up PET/CT scans that could be further analyzed. Their radiomics scores had a decreasing trend with time (Supplemental Fig. S7), suggesting that the risk of progression increases with time, which may be caused by the emergence of resistance. And the significant prediction results (AUC=0.82) showed the radiomics signature might be used as a prediction biomarker in real time to guide the following immunotherapy pending further prospective validation with larger cohorts. Third, in order to capture enough data for training, an interval time of 6 months in inclusion criterion was fairly broad. However, the median (average) interval time of these data was 1.73 (2.13) months for the retrospective cohort, and most of the PET/CT images (76%) were acquired within 3 months. Through ANOVA analysis (Fig.S8), there was no significant difference of mpRS among different subgroups divided by interval time for DCB and NDB patients on both training and test cohorts. Additionally, the AUCs were decreased with the interval time larger than 3months and 2 months in the training and test cohorts, respectively, but the differences were not significant. One possible reason is the small number of patients with the interval time larger than 3 months. And another possible reason could be the mpRS was not significantly correlated with the volume of the tumor (Spearman’s rho = −0.20, p=0.06 for training cohort, Spearman’s rho=−0.17, p=0.27 for test cohort), which means the increased volume within the interval time may not lead to the significant variations of mpRS in predicting DCB. Given the subgroup patients with 0–1 month interval time has the largest AUC in both training and test cohorts, the interval time of all the patients in the prospective cohort was less than 1 month. Lastly, there were significant differences in Distant Metastasis between the retrospective and prospective cohorts, but this did not affect the final prediction of DCB due to the incorporation of Distant Metastasis into the DCB nomogram model. Given Distant Metastasis is not a significant factor for PFS and OS prediction, this difference also would not affect the PFS and OS prediction.
CONCLUSIONS
In conclusion, an effective and stable radiomics signature combing PET and KLD features were identified and may serve as a predictive biomarker for immunotherapy response. Furthermore, radiomics nomograms well demonstrated the incremental value of the radiomics signature for individualized DCB response, PFS and OS estimation, and have a potential to be used to guide individual pre and post immunotherapy pending further external validation with larger cohorts.
Supplementary Material
Acknowledgments
Funding: This study was funded by U.S. Public Health Service research grant U01 CA143062 and R01 CA190105 (awarded to Dr. Gillies).
Disclosure:
This paper is supported by U.S. Public Health Service research grant U01 CA143062 and R01 CA190105 (principal investigator Dr. Gillies). Robert James Gillies declared a potential conflict with HealthMyne, Inc [Investor, Board of Advisors]. The remaining authors declare no conflict of interest.
Footnotes
Conflict of Interest: Robert James Gillies declared a potential conflict with HealthMyne, Inc [Investor, Board of Advisors]. The remaining authors declare no conflict of interest.
COMPLIANCE WITH ETHICAL STANDARDS
Ethical approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the Institutional Review Board at the University of South Florida (USF) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent: The requirement for informed consent was waived.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES
- 1.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Lung Cancer and Personalized Medicine. Springer; 2016. p. 1–19. [DOI] [PubMed] [Google Scholar]
- 2.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S et al. Tracking the Evolution of Non–Small-Cell Lung Cancer. New England Journal of Medicine 2017. p. 2109–21. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Rizvi NA, Mazières J, Planchard D, Stinchcombe TE, Dy GK, Antonia SJ et al. Activity and safety of nivolumab, an anti-PD-1 immune checkpoint inhibitor, for patients with advanced, refractory squamous non-small-cell lung cancer (CheckMate 063): a phase 2, single-arm trial. Lancet Oncol. 2015;16(3):257–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A et al. Pembrolizumab versus chemotherapy for PD-L1–positive non–small-cell lung cancer. New Engl J Med. 2016;375(19):1823–33. [DOI] [PubMed] [Google Scholar]
- 6.Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WEE, Poddubskaya E et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non–Small-Cell Lung Cancer. New Engl J Med. 2015;373(2):123–35. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP et al. Pembrolizumab for the treatment of non–small-cell lung cancer. New Engl J Med. 2015;372(21):2018–28. [DOI] [PubMed] [Google Scholar]
- 8.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF et al. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. New Engl J Med. 2012;366(26):2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, Von Pawel J et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. The Lancet. 2017;389(10066):255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herbst RS, Soria J-C, Kowanetz M, Fine GD, Hamid O, Gordon MS et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F et al. Pembrolizumab plus Chemotherapy in Metastatic Non–Small-Cell Lung Cancer. New Engl J Med. 2018;378(22):2078–92. doi: 10.1056/NEJMoa1801005. [DOI] [PubMed] [Google Scholar]
- 12.Meng X, Huang Z, Teng F, Xing L, Yu J. Predictive biomarkers in PD-1/PD-L1 checkpoint blockade immunotherapy. Cancer Treat Rev. 2015;41(10):868–76. [DOI] [PubMed] [Google Scholar]
- 13.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. 2015;348(6230):124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beukinga RJ, Hulshoff JB, Mul VE, Noordzij W, Kats-Ugurlu G, Slart RH et al. Prediction of Response to Neoadjuvant Chemotherapy and Radiation Therapy with Baseline and Restaging 18F-FDG PET Imaging Biomarkers in Patients with Esophageal Cancer. Radiology. 2018:172229. [DOI] [PubMed] [Google Scholar]
- 16.Schwarzenberg J, Czernin J, Cloughesy TF, Ellingson BM, Pope WB, Grogan T et al. Treatment Response Evaluation Using 18F-FDOPA PET in Patients with Recurrent Malignant Glioma on Bevacizumab Therapy. Clin Cancer Res. 2014;20(13):3550–9. doi: 10.1158/1078-0432.ccr-13-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Napel S, Mu W, Jardim-Perassi BV, Aerts H, Gillies RJ. Quantitative imaging of cancer in the postgenomic era: Radio(geno)mics, deep learning, and habitats. Cancer. 2018;124(24):4633–49. doi: 10.1002/cncr.31630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. 2015;348(6230):124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campesato LF, Barroso-Sousa R, Jimenez L, Correa BR, Sabbaga J, Hoff PM et al. Comprehensive cancer-gene panels can be used to estimate mutational load and predict clinical benefit to PD-1 blockade in clinical practice. Oncotarget. 2015;6(33):34221–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–47. [DOI] [PubMed] [Google Scholar]
- 21.Kullback S, Leibler RA. On information and sufficiency. Ann Math Stat. 1951;22(1):79–86. [Google Scholar]
- 22.Tibshirani R Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society Series B (Methodological). 1996:267–88. [Google Scholar]
- 23.Crivellaro C, Signorelli M, Guerra L, De Ponti E, Buda A, Dolci C et al. 18F-FDG PET/CT can predict nodal metastases but not recurrence in early stage uterine cervical cancer. Gynecologic oncology. 2012;127(1):131–5. [DOI] [PubMed] [Google Scholar]
- 24.Taniguchi K, Okami J, Kodama K, Higashiyama M, Kato K. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci. 2008;99(5):929–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitzgerald M, Saville BR, Lewis RJ. Decision curve analysis. Jama. 2015;313(4):409–10. [DOI] [PubMed] [Google Scholar]
- 26.Gambhir SS. Molecular imaging of cancer with positron emission tomography. Nature Reviews Cancer. 2002;2(9):683. [DOI] [PubMed] [Google Scholar]
- 27.Park JE, Kim D, Kim HS, Park SY, Kim JY, Cho SJ et al. Quality of science and reporting of radiomics in oncologic studies: room for improvement according to radiomics quality score and TRIPOD statement. Eur Radiol. 2019. doi: 10.1007/s00330-019-06360-z. [DOI] [PubMed] [Google Scholar]
- 28.Wu J, Aguilera T, Shultz D, Gudur M, Rubin DL, Loo BW Jr et al. Early-Stage Non–Small Cell Lung Cancer: Quantitative Imaging Characteristics of 18F Fluorodeoxyglucose PET/CT Allow Prediction of Distant Metastasis. Radiology. 2016;281(1):270–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vallières M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol. 2015;60(14):5471. [DOI] [PubMed] [Google Scholar]
- 30.Carvalho S, Leijenaar R, Troost E, van Elmpt W, Muratet J-P, Denis F et al. Early variation of FDG-PET radiomics features in NSCLC is related to overall survival-the “delta radiomics” concept. Radiother Oncol. 2016;118:S20–S1. [Google Scholar]
- 31.Oikonomou A, Khalvati F, Tyrrell PN, Haider MA, Tarique U, Jimenez-Juan L et al. Radiomics analysis at PET/CT contributes to prognosis of recurrence and survival in lung cancer treated with stereotactic body radiotherapy. Sci Rep. 2018;8(1):4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirienko M, Cozzi L, Antunovic L, Lozza L, Fogliata A, Voulaz E et al. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imag. 2018;45(2):207–17. [DOI] [PubMed] [Google Scholar]
- 33.Fukunaga H, Sekimoto M, Ikeda M, Higuchi I, Yasui M, Seshimo I et al. Fusion image of positron emission tomography and computed tomography for the diagnosis of local recurrence of rectal cancer. Ann Surg Oncol. 2005;12(7):561–9. doi: 10.1245/Aso.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Nakamoto Y, Senda M, Okada T, Sakamoto S, Saga T, Higashi T et al. Software-based fusion of PET and CT images for suspected recurrent lung cancer. Mol Imaging Biol. 2008;10(3):147–53. doi: 10.1007/s11307-008-0131-x. [DOI] [PubMed] [Google Scholar]
- 35.Schaarschmidt BM, Heusch P, Buchbender C, Ruhlmann M, Bergmann C, Ruhlmann V et al. Locoregional tumour evaluation of squamous cell carcinoma in the head and neck area: a comparison between MRI, PET/CT and integrated PET/MRI. European Journal of Nuclear Medicine and Molecular Imaging. 2016;43(1):92–102. doi: 10.1007/s00259-015-3145-z. [DOI] [PubMed] [Google Scholar]
- 36.Bar-Shalom R, Yefremov N, Guralnik L, Gaitini D, Frenkel A, Kuten A et al. Clinical performance of PET/CT in evaluation of cancer: additional value for diagnostic imaging and patient management. Journal of nuclear medicine. 2003;44(8):1200–9. [PubMed] [Google Scholar]
- 37.Champiat S, Dercle L, Ammari S, Massard C, Hollebecque A, Postel-Vinay S et al. Hyperprogressive disease is a new pattern of progression in cancer patients treated by anti-PD-1/PD-L1. Clinical Cancer Research. 2017;23(8):1920–8. [DOI] [PubMed] [Google Scholar]
- 38.Saeed-Vafa D, Bravo R, Dean JA, El-Kenawi A, Père NM, Strobl M et al. Combining radiomics and mathematical modeling to elucidate mechanisms of resistance to immune checkpoint blockade in non-small cell lung cancer. bioRxiv. 2017:190561. [Google Scholar]
- 39.Longo DL, Bartoli A, Consolino L, Bardini P, Arena F, Schwaiger M et al. In Vivo Imaging of Tumor Metabolism and Acidosis by Combining PET and MRI-CEST pH Imaging. Cancer Res. 2016;76(22):6463–70. doi: 10.1158/0008-5472.CAN-16-0825. [DOI] [PubMed] [Google Scholar]
- 40.Pilon-Thomas S, Kodumudi KN, El-Kenawi AE, Russell S, Weber AM, Luddy K et al. Neutralization of Tumor Acidity Improves Antitumor Responses to Immunotherapy. Cancer Res. 2016;76(6):1381–90. doi: 10.1158/0008-5472.CAN-15-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yip SS, Kim J, Coroller TP, Parmar C, Velazquez ER, Huynh E et al. Associations between somatic mutations and metabolic imaging phenotypes in non–small cell lung cancer. J Nucl Med. 2017;58(4):569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaira K, Higuchi T, Naruse I, Arisaka Y, Tokue A, Altan B et al. Metabolic activity by 18F–FDG-PET/CT is predictive of early response after nivolumab in previously treated NSCLC. European journal of nuclear medicine and molecular imaging. 2018;45(1):56–66. [DOI] [PubMed] [Google Scholar]
- 43.Kumar V, Nath K, Berman CG, Kim J, Tanvetyanon T, Chiappori AA et al. Variance of SUVs for FDG-PET/CT is greater in clinical practice than under ideal study settings. Clinical nuclear medicine. 2013;38(3):175–82. doi: 10.1097/RLU.0b013e318279ffdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.