Summary
In order to combat molecular damage, most cellular proteins undergo rapid turnover. We have previously identified large nuclear protein assemblies that can persist for years in post-mitotic tissues and are subject to age-related decline. Here we report that mitochondria can be long-lived in the mouse brain and reveal that specific mitochondrial proteins have half-lives longer than the average proteome. These mitochondrial long-lived proteins (mitoLLPs) are core components of the electron transport chain (ETC) and display increased longevity in respiratory supercomplexes. We find that COX7C, a mitoLLP that forms a stable contact site between Complexes I and IV, is required for Complex IV and supercomplex assembly. Remarkably, even upon depletion of COX7C transcripts, ETC function is maintained for days, effectively uncoupling mitochondrial function from ongoing transcription of its mitoLLPs. Our results suggest that modulating protein longevity within the ETC is critical for mitochondrial proteome maintenance and the robustness of mitochondrial function.
Graphical Abstract
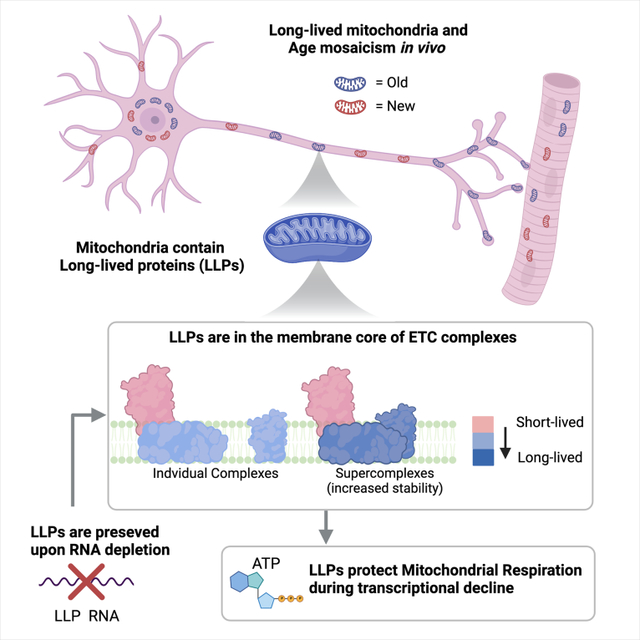
eTOC Blurb:
Krishna et al., discover that mitochondria in neurons and muscle fibers exhibit slow turnover and contain many long-lived proteins that can be part of large protein complexes. The longevity of these proteins in the electron transport chain can maintain oxphos complex assembly and mitochondrial function despite their transcriptional decline.
Introduction
Protein homeostasis is critical for maintaining cell function and organismal health. As proteins age, they accumulate molecular damage that causes them to dysfunction and aggregate (Hipp et al., 2019). For example, mitochondrial protein aggregation has been observed during aging in C. elegans (David et al., 2010). Also, the mitochondrial matrix proteases Lon and ClpXP that degrade misfolded proteins show reduced activity with age in humans (Bezawork-Geleta et al., 2015). This loss of proteostasis could contribute to the age-related decline in mitochondrial function and respiratory capacity (Lopez-Otin et al., 2013), particularly since the mitochondrial proteome is vulnerable to damage due to the high levels of reactive oxygen species (ROS) generated from the ETC (Moehle et al., 2019). Damaged mitochondrial proteins can lead to ETC dysfunction and further ROS production that would promote mtDNA mutations and compromise organelle function (Moehle et al., 2019). To counteract this, mitochondria have evolved several protein quality control mechanisms. These include resident mitochondrial chaperones and AAA-proteases, cytosolic ubiquitin/proteasomal machinery, mitochondrial unfolded protein response and lysosomal degradation of mitochondrial-derived vesicles (Picca et al., 2020; Shpilka and Haynes, 2018; Song et al., 2021). Finally, clearance of whole mitochondria or specific regions can occur through mitophagy that is in part regulated by PINK1 and Parkin, both of which are highly implicated in Parkinson’s Disease (Palikaras et al., 2018).
Another critical protein quality control mechanism is protein turnover or recycling. To combat the accumulation of damaged proteins, the majority of the cellular proteome turns over rapidly, a process that is crucial for post-mitotic cells like neurons and muscle fibers as they are unable to dilute the deleterious proteins via cell division (Toyama and Hetzer, 2013). Interestingly, we have recently identified proteins in the rat brain that are extremely long-lived and can persist for months or even years (Savas et al., 2012; Toyama et al., 2013). These long-lived proteins (LLPs) are found primarily in the nucleus or the plasma membrane/extracellular matrix. Many of the nuclear LLPs are components of large protein complexes, like histones H3.1/H4 in the nucleosome, Lamin-B1/B2 in the nuclear lamina and nucleoporins (nups) of the nuclear pore complex (NPC), which is required for nuclear membrane transport (Toyama et al., 2013). By imaging cortical neurons using correlated multi-isotope imaging mass spectroscopy and scanning electron microscopy (MIMS-EM), we have also established that NPCs are long-lived structures in vivo (Toyama et al., 2019). Interestingly, we have found that not all NPCs are long-lived, and that protein turnover within the NPC itself is also heterogeneous, with peripheral nups being short-lived and scaffold nups being long-lived (Toyama et al., 2013). This molecular age mosaicism raises key questions about the role of protein longevity in macromolecular complexes. LLPs could be critical for maintaining a scaffold for the structural and functional integrity of complexes. They may also be prone to damage due to their long-lived nature, leading to impaired function with age. Indeed, aged neurons show more damage in long-lived nups and have impaired nuclear transport that results in accumulation of cytosolic proteins in the nucleus (D’Angelo et al., 2009). Such intranuclear aggregates have also been seen in neurons from patients with neurodegenerative disease, providing an unexpected link between LLPs and age-related disorders (D’Angelo et al., 2009).
While extreme protein longevity has been seen in the nucleus – an organelle that is not replaced in post-mitotic cells, the turnover of mitochondria is poorly understood. Although our original studies primarily detected nuclear LLPs (Toyama et al., 2013), there have been reports that certain mitochondrial proteins can have half-lives of one month in the mouse brain (Fornasiero et al., 2018; Price et al., 2010). Some of these proteins were also components of the ETC. Interestingly, it was recently shown that the ETC exists as individual complexes (ICs) as well as higher-order quaternary structures called “supercomplexes” (SCs) (Gu et al., 2016; Letts et al., 2016). However, there are no reports of protein half-lives in the SCs compared to the ICs, as the previous studies used tissue homogenates without separating the different complexes (Fornasiero et al., 2018; Price et al., 2010). Also, the precise role of SCs in the ETC is unclear, with some reports suggesting that they could increase the stability of Complex I (CI) (Letts and Sazanov, 2017). Hence the identification of LLPs and characterization of their role in in the ETC is critical to understanding mitochondrial protein homeostasis.
In this study we find that neurons and muscle fibers have mitochondria that are long-lived and can persist up to 6-months in vivo. Moreover, we find considerable age mosaicism between different mitochondria, a phenomenon that we also demonstrate in neurons in vitro. We further determine in vitro that mitochondria turn over slower compared to the cellular proteome, with many proteins exhibiting longer than average lifespans. These mitoLLPs are core components of the ETC, specifically the membrane arm of CI and Complex III (CIII). Remarkably, we find that these LLPs display increased stability in the SCs compared to ICs. One of the critical interactions between CI and Complex IV (CIV) is stabilized by COX7C, a mitoLLP that shows an age-dependent decline in transcription (Su et al., 2015). We recapitulate this decline and find that despite depletion of COX7C transcripts, the protein still persists and is able to protect myotubes from loss-of-function defects in CIV and SC assembly. These data show that mitoLLPs could potentially be required for maintaining stability and function of the ETC in times of transcriptional decline during cellular stress or aging.
Results
Discovery of long-lived mitochondria in post-mitotic tissues of 6-month old mice
While there are thousands of mitochondria in each cell, and even more in highly metabolic tissues, how long individual mitochondria can persist is unknown. To study the lifespan of mitochondria in post-mitotic tissues, we performed 15N-SILAM labeling of mice coupled with MIMS-EM, which can help visualize protein, organelle and cell turnover in vivo (Arrojo et al., 2019). Briefly, female mice were fed an 15N diet, followed by a chase with 14N until 6 months of age, when mitochondria-dense tissues like brain and skeletal muscle were harvested and imaged (see methods). MIMS images represent the 15N-to-14N ratio, and since 15N is lost during cell and/or protein turnover, the 15N/14N is a reporter of “old” vs “new” for cells and protein complexes (Toyama et al., 2019). The MIMS images were colorized based on the fold increase over the natural ratio (NR) of 15N/14N at 37×10−4 (Figure S1A). As seen before with post-mitotic cells (Arrojo et al., 2019), the nuclei of cerebellar granule cell neurons show a 15N/14N >50x above the NR (Figure 1A–B, black arrow). In contrast, structures like blood vessel lumens in the cerebellum or intercostal muscle (ICM) show minimal 15N/14N signal (Figure 1C,1F–G). As this was the lowest signal seen, we used a 15N/14N of 55 ×10−4 (1.5xNR) as our background. These data support our previous observations that most 15N signal is retained in the nucleus of non-dividing cells, which includes DNA and nuclear LLPs (Toyama et al., 2013).
Figure 1. Discovery of long-lived mitochondria in post-mitotic tissues of 6-month old mice.
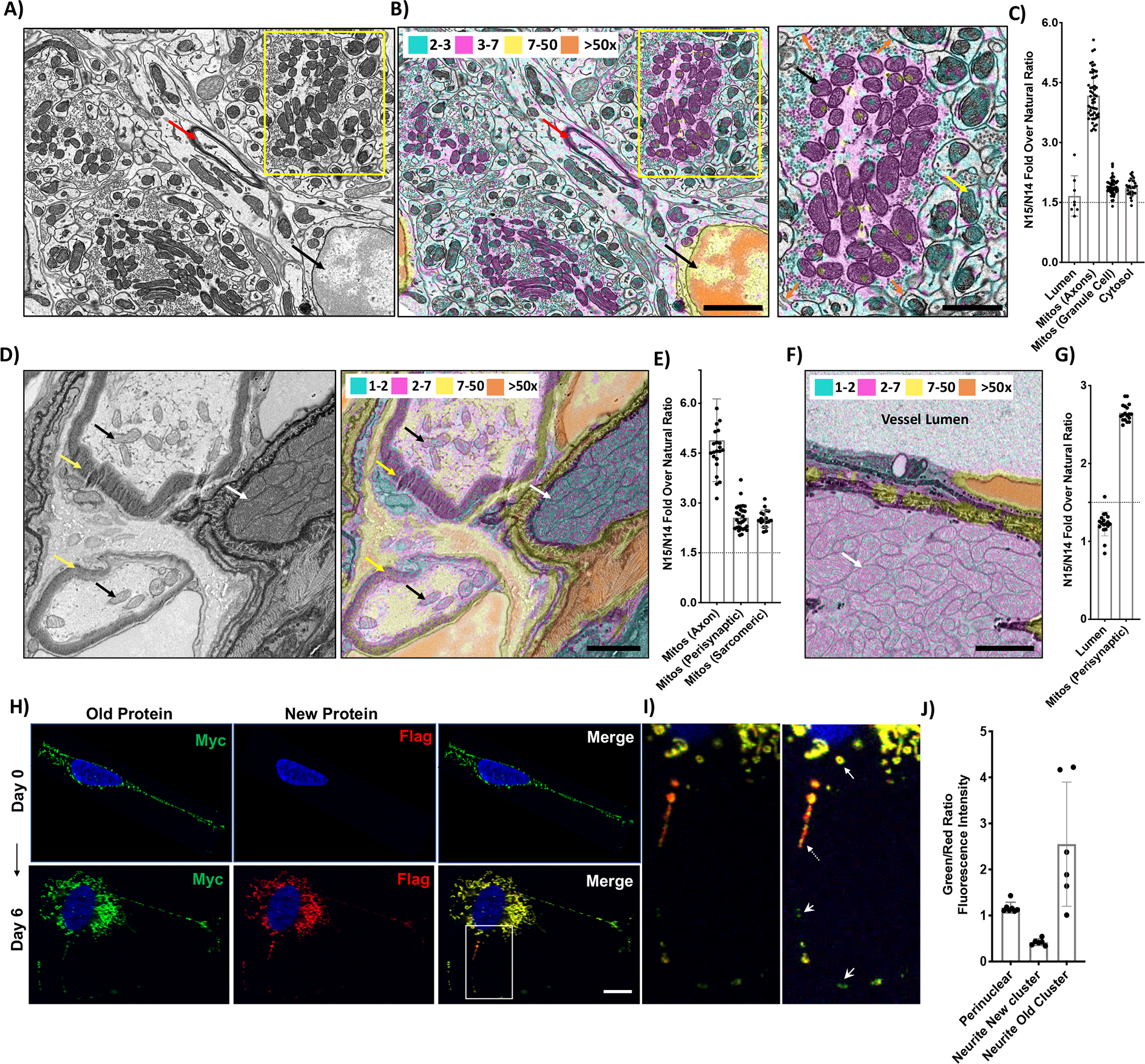
A) SEM image and B) MIMS overlay of cerebellar neurons from a 6-month chase mouse. Nucleus of a granule cell neuron (black arrow), myelin sheath (red arrows) and mitochondria (inset, B-right panel) are indicated. Inset shows mitochondria in mossy fibers axons containing “higher” (black arrow) and granule cell dendrites containing “lower” (yellow arrow) signal. Orange arrows show the boundary between mossy fiber axons and granule cell dendrites.15N/14N signal thresholds over the natural ratio (NR) of 37×10−4: 2–3x (cyan), 3–7x (magenta), 7–50x (yellow), >50x (vermillion). C) Dot plot overlaid with a bar graph shows the 15N/14N fold over the NR for blood vessel lumen (Lumen, n=8), mitochondria from mossy fiber axons (Mitos-Axons, n=53), mitochondria from granule cell dendrites (Mitos-Granule cells, n=50) and the cytosol (Cytosol, n=29). For the mitos plots, each dot represents the 15N/14N fold ratio for individual mitochondria whereas for the lumen and cytosol, each dot represents different regions of those structures. D) SEM image (left) and MIMS overlay (right) of myelinated axons near a neuromuscular junction in intercostal muscle from a 6-month chase mouse. Individual mitochondria in two myelinated axons (mitochondria – black arrows, myelin - yellow arrows) and perisynaptic mitochondria aggregates in a muscle fiber (white arrow) are indicated. 15N/14N thresholds: 1–2x (cyan), 2–7x (magenta), 7–50x (yellow), >50x (vermillion). E) Dot plot overlaid with a bar graph shows the 15N/14N fold over NR for mitochondria from axons (n=23) and perisynaptic aggregates (n=28), and sarcomeric mitochondria in the muscle fiber (n=18). F) MIMS overlay of intercostal muscle fiber containing mitochondria (white arrow) next to a vessel lumen (indicated). G) Graph shows the 15N/14N fold over NR for regions of the blood vessel lumen (Lumen, n=18) and mitochondria (Mitos, n=18). H) Turnover of ATP5C1 in SHSY-5Y neurons as measured by RITE. Images show green Myc tag (old protein, left panel) and red Flag tag (new protein, middle panel) at Day 0 (top panel) and Day 6 in (bottom panel). Merge is shown in right panel. I) An inset from the merge at lower (left image) and higher fluorescent intensity (right image) shows green and red signal in mitochondria that are peri-nuclear (white arrow) or in neurites with old (white arrowhead) and new (white dashed arrow) clusters indicated. J) Graph shows the Green/Red Fluorescence Intensity over background for the mitochondria shown in the inset (Perinuclear n=7, neurites new cluster n=6, neurites old cluster n=6).
For all MIMS-EM graphs, a dotted line is drawn at the background 15N/14N of 1.5x NR and error bars indicate SD. Scale bars: 5μm (B), 2.5μm (B, inset, D, F), 10μm (H).
Remarkably, we found that the majority of mitochondria in cerebellar neurons show an increased 15N/14N signal in the range of 2-to-3x NR, with many clusters that are as high as 3-to-7x NR (Figure 1A–B, yellow rectangle). Indeed, these clusters show signals similar to the myelin sheath itself, which is known to contain LLPs and is consistently labeled in MIMS-EM studies (Figure 1A–B, red arrows) (Arrojo et al., 2019; Toyama et al., 2013). Interestingly, these clusters of old mitochondria specifically reside in the axonal terminals and synaptic boutons of mossy fiber neurons (Figure 1B, inset, black arrow). These axonal processes form large presynaptic terminals with the postsynaptic processes of granule cell dendrites, which themselves primarily contain mitochondria with lower signal (Figure 1B, inset, yellow arrow). These data demonstrate considerable heterogeneity in mitochondrial age in the same tissue. Quantification of the 15N/14N of individual mitochondria shows that granule cell dendritic mitochondria have “lower” signal at ~1.9x NR while the axonal mitochondria have “higher” signal at 4xNR (Figure 1C). These results reveal 15N retention in neuronal mitochondria, signifying the presence of mitochondrial components that are long-lived and have limited turnover in 6-month old mice. Given that mitochondria have always been considered a dynamic organelle undergoing constant fission/fusion, the presence of these clusters of long-lived mitochondria is an important finding. Moreover, the “higher” signal axonal population shows a significant level of variability in the 15N/14N signal (Figure 1C), suggesting that mitochondria in the same cell can also have different lifespans. This data indicates age mosaicism within mitochondria, similar to what has been observed at the cellular and protein level in the brain, liver and pancreas (Toyama et al., 2013). We next looked at myelinated axons near a neuromuscular junction (NMJ) in the ICM as they have specialized mitochondria in different regions like the neuronal axoplasm, the terminal boutons of the motor synapse, the non-sarcomeric and the sarcomeric regions of the muscle fiber (Figure S1B). The mitochondria in myelinated axons have high levels of 15N/14N, comparable to the signal from the myelin sheath (Figure 1D–E) as well as the cerebellar mitochondria. We also detect increased 15N/14N in perisynaptic mitochondria aggregates at the periphery and sarcomeric mitochondria interspersed within the muscle fibers (Figure 1D–E, S1B–C). The axonal mitochondria contain more 15N, indicating that they are older than mitochondria in the muscle fibers. We also observe increased 15N/14N in mitochondria specifically in ICM muscle fibers that are not near NMJ or neurons (Figure 1F–G), and in perisynaptic as well as the sarcomeric mitochondria in the fibers (Figure S1D–E). These data show that there are mitochondrial components that are long-lived and have limited turnover in neurons and muscle fibers in 6-month old mice, with the former having older mitochondria, as well as more heterogeneity.
The 15N signal in the long-lived mitochondria in vivo could be from mitochondrial proteins or mtDNA. To determine specifically if mitochondrial proteins display longevity and mosaicism, we looked at protein turnover of ATP synthase subunit ATP5C1 in cultured SHSY-5Y neurons. We used a previously published technique called Recombinant Induced Tag Exchange (RITE), where a protein expresses a Myc tag, which upon Cre-recombination gets replaced by a Flag tag, facilitating the imaging of old (Myc) and new (Flag) protein simultaneously (Toyama et al., 2019). This previous study showed that while nuclear LLPs turn over rapidly within 2 days in dividing cells, they persist for 1–2 weeks in post-mitotic cells in vitro (Toyama et al., 2019). We find that at Day 0 all the ATP5C1 protein is Myc-tagged (Figure 1H) and the old protein persists by Day 6, despite production of new protein (Figure 1H,S1F). Importantly, we find in the merge image that the mitochondria have differing green and red signals (Figure 1H, Inset,1I). The perinuclear mitochondria have similar green to red signal (~1), whereas the neurites have separate clusters of red and green signal indicating new and old mitochondria respectively (Figure 1I–J). As seen in vivo, the old cluster shows considerable variability in the old/new signal. These data suggest that mitochondrial age mosaicism and heterogeneity can occur within a cell, among different cell types and tissues within the same animal, as well as in human neurons in vitro.
Identification of LLPs in neuronal and myotube mitochondria
Given the longevity of mitochondria and ETC proteins in post-mitotic tissues, we wanted to conduct an unbiased and comprehensive analysis of mitochondrial proteome turnover in vitro to determine if mitochondria contain LLPs. We use stable isotope amino acid labeling (SILAC) in human ES (H9) cells differentiated to neurons and C2C12 myoblasts differentiated to myotubes (Figure 2A). We find that the heavy label in the whole cell proteome went up to 60% in neurons and 80% in myotubes by Day 8 (Figure S2A–B), indicating that while majority proteins incorporate the heavy label during the timecourse, neurons on average turn over slower than myotubes. Upon calculating %old protein and half-lives (see methods, Table S1), we find that the whole-cell, ER, lysosomal and cytosolic proteins in the neurons turn over rapidly with a median half-life of ~4–5 days (Figure S2C,2B). Notably, the nuclear proteins showed a slower turnover rate due to long-lived histones, the removal of which brings nuclear turnover down to the whole cell level (Figure S2C). Consistent with this the median half-life of the nucleus is similar to the whole cell (Figure 2B). Remarkably, mitochondrial proteins show slower turnover than all the other organelles with a half-life of 7 days in neurons (Figure S2C,2B). We see similar results in myotubes, with the whole cell, nucleus, ER, cytosol and lysosomal half-lives at ~3.5 days, whereas mitochondrial proteins are at 5.5 days (Figure S2D,2C). These data suggest that neuronal and myotube mitochondrial proteins are indeed significantly more long-lived than those residing in other organelles. Intriguingly, within the mitochondria, the outer mitochondrial membrane (OMM) proteins show similar turnover to the whole-cell, whereas matrix, intermembrane space (IMS) and inner mitochondrial membrane (IMM), particularly ETC proteins, are longer-lived in both neurons and myotubes (Figure S2E–H). This difference could be due to the OMM being recycled by cytosolic degradative pathways, while the inner compartments are dependent on mitochondrial degradative mechanisms (Vazquez-Calvo et al., 2020). Surprisingly, nucleoid proteins in neurons exhibit extremely slow turnover with half-lives up to 16 days, which was not seen in myotubes (Figure S2E–F,S2H), suggesting perhaps a specialized role for these proteins in neurons.
Figure 2. Identification of LLPs in neuronal and myotube mitochondria using heavy isotope labeling.
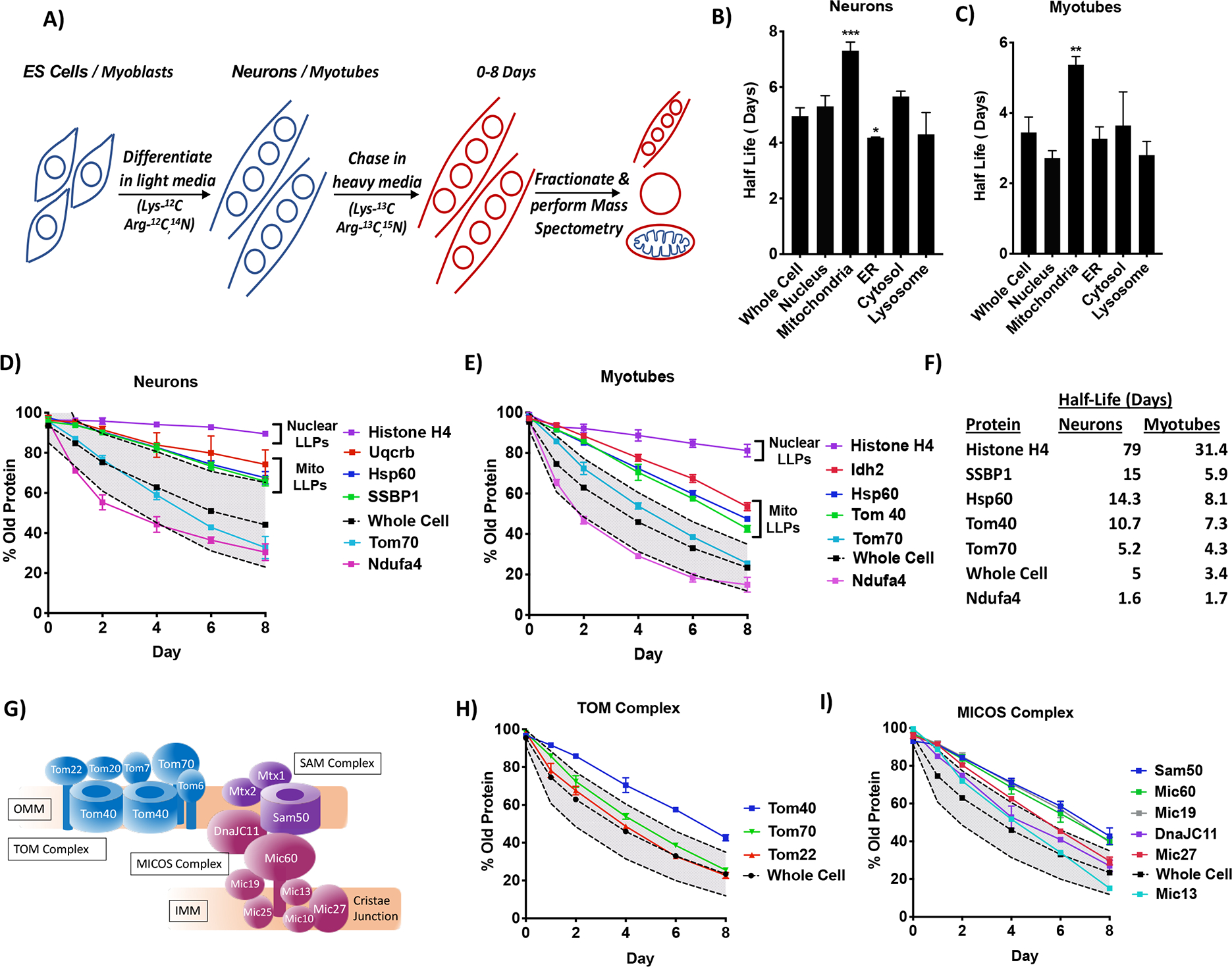
A) Schematic of the SILAC: Human ES cells or C2C12 mouse myoblasts were differentiated into neurons or myotubes in light media and then switched to heavy media for 0–8 days. Whole-cell, nuclear and mitochondrial fractions were subject to mass spectrometry. Diagram specifically shows myotubes. B) Graph shows median half-lives for Whole Cell, Nucleus, Mitochondria, ER, Cytosol and Lysosome in neurons. C) Graph shows median half-lives for Whole Cell, ER, Cytosol and Lysosome (from whole cell fraction), Nucleus (from the nuclear fraction) and Mitochondria (from the mitochondrial fraction) in myotubes. D) Graph shows %old protein for Day 0–8 for Histone H4 and mitochondrial proteins Uqcrb, Hsp60, SSBP1, Tom70 and Ndufa4 in neurons. Nuclear and MitoLLPs are indicated. E) Graph shows %old protein for Day 0–8 for the Histone H4 (nuclear fraction) and mitochondrial proteins Idh2, Hsp60, Tom40, Tom70 and Ndufa4 (mitochondrial fraction) in myotubes. Nuclear and MitoLLPs are indicated. F) Table showing the half-lives (in days) of key nuclear and mitochondrial proteins in neurons and myotubes. G) Model showing the TOM and SAM Complexes in the OMM, and the MICOS Complex in the IMM near the cristae junction. Tom40, Sam50 and Mic60 are in the core of the complexes, with Sam50 and Mic60 interacting with each other. H) Graph shows %old protein for Day 0–8 for Tom40, Tom70 and Tom22 in myotubes. I) Graph shows %old protein for Day 0–8 for Sam50, Mic60, Mic19, DnaJC11, Mic27 and Mic13 in myotubes.
For all graphs, the average whole cell proteome is the dashed black line and the shaded grey area within dotted lines is the SD for the fraction. Error bars indicate SD. Data are an n=3. *p <0.05, **p<0.001, ***p<0.0001
To validate that we could indeed identify LLPs, we analyzed turnover of a known nuclear LLP Histone H4. In neurons, Histone H4 had 90% old protein at Day 8, with a half-life of 79 days, suggesting minimal turnover (Figure 2D,2F). The average whole cell fraction in neurons has ~50% old protein remaining at Day 8, with a standard deviation (SD) spread of ~20% (Figure 2D). While many mitochondrial proteins lie within this spread, we identified a class of mitochondrial proteins that has about 75% old protein at Day 8, with half-lives ranging from 10–15 days (Figure 2D,2F). We term this class as “mitochondrial LLPs” or “mitoLLPs” (see methods), and it contains many integral mitochondrial proteins like CIII subunit Uqcrb, chaperone Hsp60 and nucleoid protein SSBP1 (Figure 2D,2F). We observe similar results in myotubes, albeit with faster turnover and shorter half-lives. Histone H4 has a half-life of 31 days and the average whole cell fraction has 25% old protein at Day 8, with an SD spread of ~15%, with a class of mitochondrial proteins lie above this spread (Figure 2E–F). Remarkably, 40% of neuronal and myotube mitochondrial proteins detected in our study fall into the category of mitoLLPs. These mitoLLPs while longer-lived than the average proteome, still turn over faster than nuclear LLPs that seem to be extremely stable. Moreover, the whole cell and short-lived proteins (SLPs) in the neurons and myotubes have similar half-lives, suggesting that the primary difference between the two cell types is due to LLPs. Since we use human neurons and mouse myotubes, some of the slower turnover in neurons could be due to species-specific differences. However, comparison of our dataset to a previous study in cultured mouse neurons (Mathieson et al., 2018) demonstrates good correlation of half-lives for whole cell (r=0.65) and ETC proteins (r=0.71), suggesting that our dataset is indeed comparable to mouse neurons.
To determine if mitoLLPs, similar to nuclear LLPs, can be part of large protein complexes, we looked at the TOM (Translocase of the Outer Membrane), SAM (Sorting and Assembly Machinery) and MICOS (Mitochondrial contact site and Cristae Organizing System) complexes, which are required for protein import, insertion and cristae formation respectively (Figure 2G). Due to better coverage of OMM proteins in myotubes compared to neurons, we used those datasets. We find that Tom40, a beta-barrel protein forming the central scaffold of the TOM complex is longer-lived (T1/2: 7.4d) compared to peripheral subunits Tom70 and Tom22 (T1/2: 3.5d) (Figure 2H). Similarly, Sam50 and Mic60, which are found in the central core of the SAM and MICOS complexes respectively, are longer-lived (T1/2~8d) compared to the peripheral DnaJC11 and Mic13 (T1/2~4d) (Figure 2I). Thus, similar to the NPC, these critical mitochondrial complexes also show heterogeneity in the turnover rate of their components with cores that are relatively stable (Figure S2I) (Toyama et al., 2013). We also find that the mito-ribosomal complex contains LLPs, with the 28S and 39S subunits clustering around a half-life of 6 days (Figure S2I). Interestingly, we find that ETC complexes CI and CIV show heterogenous turnover with multiple LLPs and SLPs (Figure S2J,S2L,S2N). In contrast, CIII and CV contain only LLPs, suggesting homogeneous turnover of the entire complex (Figure S2K,S2M–N). Importantly, we see the same trend in neurons (Figure S2O), with a strong correlation between the half-lives of neuronal vs myotube ETC proteins (Figure S2P). Our data show that neuronal and myotube mitochondria exhibit higher longevity compared to other organelles and contain many LLPs that can be part of protein complexes. While some complexes have heterogeneous turnover that would require unique mechanisms to turn over the peripheral SLPs, others turn over homogeneously as a single unit.
Mitochondrial LLPs are core components of ETC complexes
Since the ETC contains multiple LLPs, we wanted to perform a detailed proteomic study of the respiratory chain that exists as ICs and higher-order SCs (Gu et al., 2016; Letts et al., 2016). While there is some previous evidence that SCs can increase the stability of the CI (Letts and Sazanov, 2017), the actual turnover rate of the ETC proteins in SCs has never been studied. Since our SILAC of mitochondrial fractions contained data from both ICs and SCs together, we used a blue-native page to isolate and identify SCs (Jha et al., 2016). Given the technical challenges in scaling up neuronal for SC studies, we used myotubes for all further experiments. Consistent with previous studies, we observe a cluster of 4–5 bands at 1000kDa of higher-order SCs with varying stoichiometries of CI, CIII and CIV (SCI,IIIn,IVn), along with individual CI, intermediate supercomplex (SCIII2,IV), dimers of CIII & CIV (CIII2, CIV2), individual CIV and CII (Figure 3A, left gel, MYO) (Jha et al., 2016). We also confirmed the identity of these bands with an anti-Oxphos cocktail, as well as CI, CIII and CIV antibodies (Figure 3A, right blots). Interestingly, while dividing myoblasts also contain SCs, myotubes show an enrichment of both ICs and SCs (Figure 3A, DIV vs MYO). Differentiating myoblasts are known to undergo mitophagy and mitochondrial remodeling to increase oxphos (Sin et al., 2016), and this SC enrichment could be another adaptation to increase mitochondrial respiration. This idea is supported by recent studies that show an increase in SCs during adipocyte differentiation and with exercise in skeletal muscles in human adults (Greggio et al., 2017; Hofmann et al., 2012).
Figure 3. Mitochondrial LLPs are core components of the ETC complexes.
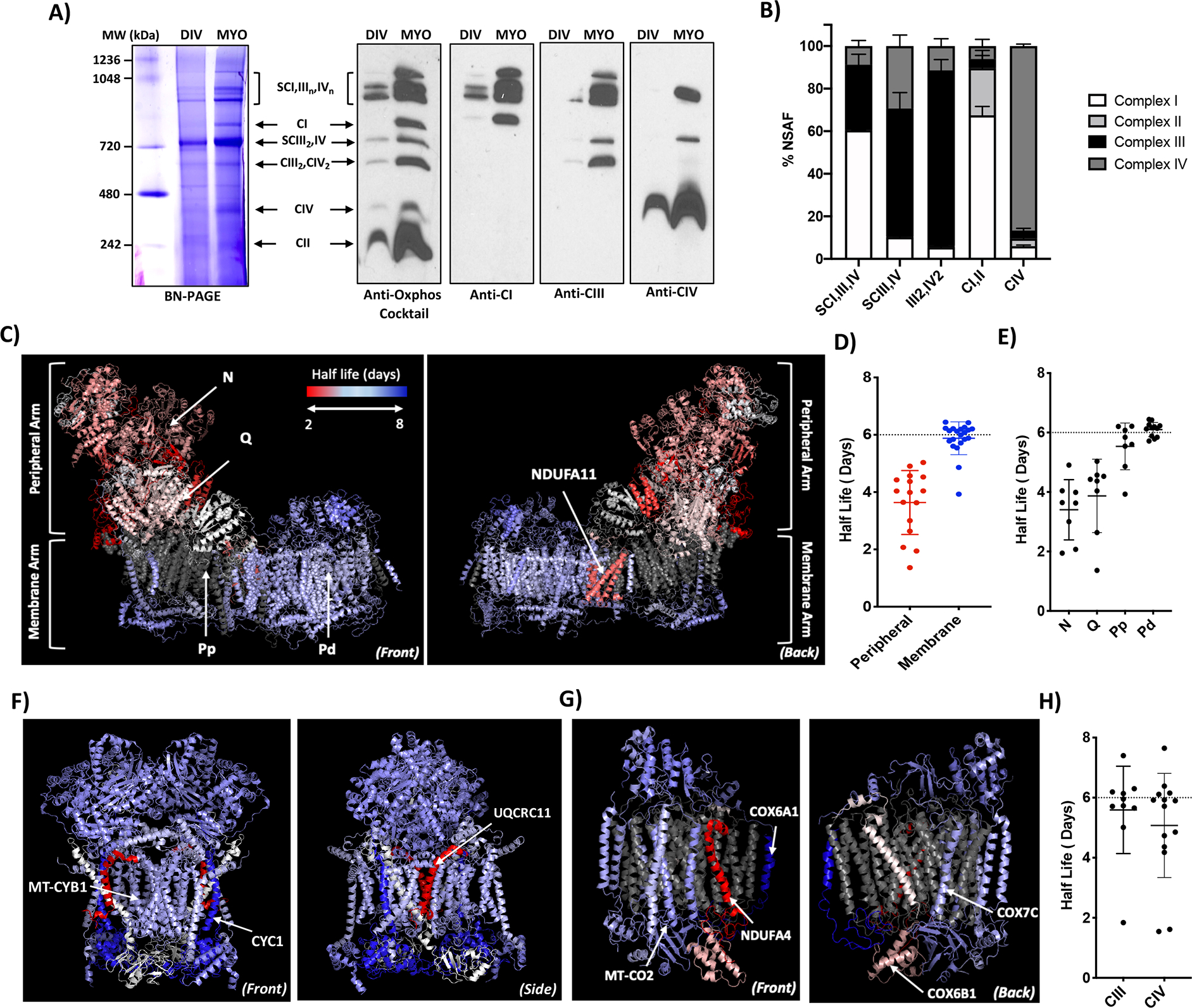
A) ETC complexes and supercomplexes in dividing myoblasts vs myotubes. BN-PAGE gel (left) and western blots (right) with Anti-Oxphos cocktail, CI, CIII & CIV antibodies show the bands corresponding to higher-order SCs (SCI,IIIn,IVn), individual CI (CI), Intermediate SC (SCIII2,IV), dimers of CIII & CIV (CIII2, CIV2), individual CIV (CIV) and individual CII (CII). B) Graph shows %NSAF abundance values of individual ETC complexes in the five gel band samples (SCI,III,IV), (SCIII,IV), (III2,IV)2, (CI,II),(CIV). C) Turnover of CI subunits in the individual complex. Images show CI structure color-coded with protein half-lives from the front (left image) and back (right image). The peripheral and membrane arm, the N, Q, Pp, Pd modules, and subunit NDUFA11 are indicated. D) Dot plot shows the half-lives of the individual protein subunits in the peripheral and membrane arms of CI respectively. E) Dot plot shows half-lives of individual protein subunits in the N, Q, Pp and Pd modules of CI. F) Turnover of CIII2 subunits in the individual dimeric complex. Images show CIII2 structure color-coded with protein half-lives from the front (left image) and side (right image). Subunits MT-CYB1, CYC1 and UQCRC11 are indicated. G) Turnover of CIV subunits in the individual complex. Images show CIV structure color-coded with protein half-lives from the front (left image) and back (right image). Subunits MT-CO2, COX6A1, NDUFA4, COX7C and COX6B1 are indicated. H) Dot plot shows half-lives of the individual subunits of CIII and CIV.
Scale of half-lives for all images is 2–8 days color-coded from dark red to white to dark blue. Proteins with no coverage are colored grey. For all graphs, a dotted line is drawn at the half-life of 6 days. Error bars indicate SD. Data are an n=3.
To measure turnover, we ran the SILAC samples on blue-native gels, followed by mass spectrometry on higher-order SCs (SCI,III,IV), intermediate SC (SCIII,IV), dimers (III2,IV2), individual CI & CII pooled (CI,II) and individual CIV (CIV). We confirmed their identity by quantifying protein abundance in each complex in the samples using normalized spectral abundance factor (NSAF) (Figure 3B) and determined their half-lives (Table S2). To effectively visualize the turnover rate, we color-coded the crystal structures of the complexes based on their half-lives ranging from 2 to 8 days (red to blue). We observed that subunits in the peripheral and membrane arms of the individual CI turnover at distinct rates, with the former turning over in a heterogeneous albeit rapid fashion with half-lives at ~3.7 days, while the latter turns over slower and homogenously with half-lives clustering around 6 days (Figure 3C–D). CI has 4 functional modules: N (NADH dehydrogenase), Q (electron transfer), Pp and Pd (proton pumping) (Wirth et al., 2016). Our data shows that the Pp & Pd modules in the membrane arm are long-lived, with the Pd module half-lives clustering very tightly at 6 days (Figure 3E). Interestingly, the only protein in the membrane arm that shows rapid turnover is NDUFA11 (Figure 3C, back). It is worth noting that NDUFA11 interacts with CIII within the SCs, therefore it is conceivable that its rapid turnover could play a role in regulating this interaction and consequently the assembly/disassembly of SCs (Letts and Sazanov, 2017). CIII, which exists as a dimer (CIII2) shows homogenously blue subunits with core proteins MT-CYB1 and CYC1 being particularly long-lived and UQCRC11 being the only short-lived red subunit (Figure 3F,3H). Interestingly, CIV shows heterogenous turnover with some very short-lived proteins (NDUFA4, COX6B1), but also many LLPs like COX6A1, MT-CO2 and COX7C (Figure 3G–H). Our data demonstrate that the ICs in myotubes have many LLPs and show unique turnover patterns - while CI has a stable core membrane arm and a short-lived peripheral arm, CIII is homogeneously long-lived, and CIV shows generally heterogeneous turnover.
Differential stability of mitochondrial LLPs in ETC supercomplexes
We next generated half-lives and visualized higher-order and intermediate SC turnover by color-coding the structure of the porcine respirasome (SCI,III2,IV) since the mouse or human respirasome is unavailable (Figure 4A,S3F). We find that similar to the ICs, the peripheral arm of CI in the respirasome SC shows heterogeneous and fast turnover (Figure 4A). The membrane arm of CI, as well as CIII2 and CIV, which are all membrane-embedded, exhibit slower turnover (Figure 4A,S3A–D). Similar to the individual CI, NDUFA11 is the only membrane arm subunit that shows fast turnover (Figure S3A–B). NDUFA11 is shown to interact with UQCRB, UQCRQ and UQCRH from CIII2 (Gu et al., 2016; Letts et al., 2016), all of which are all long-lived stable proteins (Figure S3E, middle image). Indeed, many other proteins forming interactions between the different complexes within the respirasome are long-lived and form stable contacts sites (Figure S3E, left, right image). In particular, COX7C, one of the most stable proteins in CIV, binds three LLPs in CI - MTND5, NDUFB9, and NDUFB7, suggesting a stable contact site for CI and CIV (Figure 5A).
Figure 4. Differential stability of mitochondrial LLPs in ETC supercomplexes.
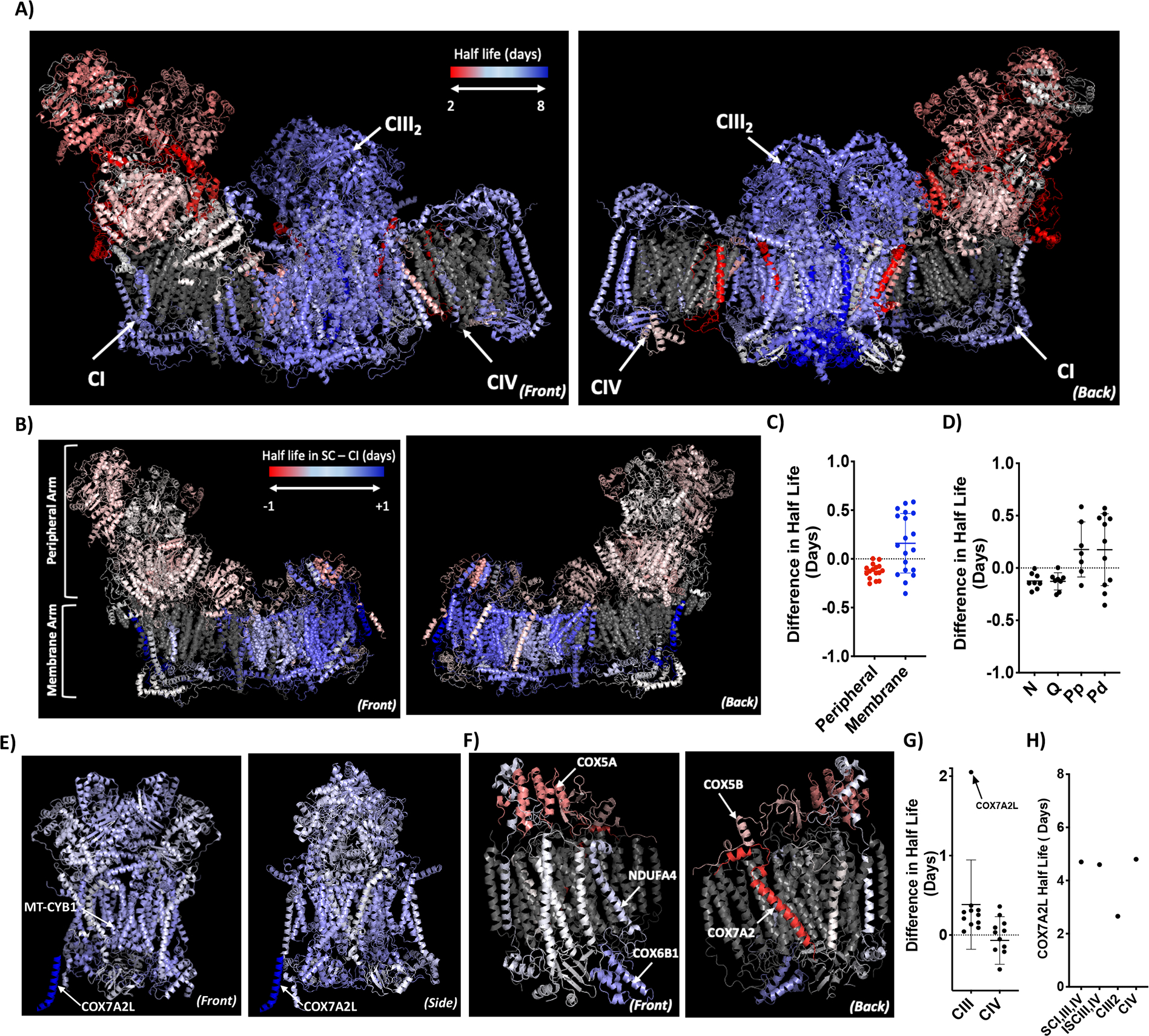
A) Turnover of subunits in the respirasome SC. Images show the porcine respirasome structure with protein half-lives color-coded in the range of 2–8 days from dark red to dark blue from the front (left image) and back (right image). CI, CIII2 and CIV are indicated. B) Differential turnover between SCs and individual CI. Images show CI structure color-coded using a half-life subtraction of SC-CI. The peripheral and membrane arms are indicated. C) Dot plot shows the difference in half-lives of the individual protein subunits in the peripheral and membrane arms between SC-CI. D) Dot plot shows difference in half-lives of the individual protein subunits in the N, Q, Pp and Pd modules between SC-CI. E) Differential turnover between SCs and CIII2. Images show CIII2 structure color-coded using a half-life subtraction of SC-CIII2. COX7A2L is not present in the PDB structure and is added manually at the side of CIII2. MT-CYB1 and COX7A2L are indicated. F) Differential turnover between SCs and CIV. Images show CIV structure color-coded using a half-life subtraction of SC-CIV. Subunits COX5A, NDUFA4, COX6B1, COX5B, COX7A2 are indicated G) Dot plot shows the difference in half-lives of the protein subunits in the CIII2 and CIV between SC-CIII2 and SC-CIV respectively. COX7A2L is indicated on the graph. H) Half-life of COX7A2L in supercomplexes (SCI,IIIn,IVn), intermediate supercomplex (ISCIII,IV), CIII2 and CIV.
Scale for differential turnover structures is −1 to +1 days from dark red to white to dark blue representing faster turnover in SCs to slower turnover in SCs. Proteins with no coverage are colored grey. For all graphs, a dotted line is drawn at the half-life difference of 0 days. Error bars indicate SD. Data are an n=3.
Figure 5. Longevity of COX7C protects myotubes from loss of function defects in Complex IV and Supercomplex assembly.
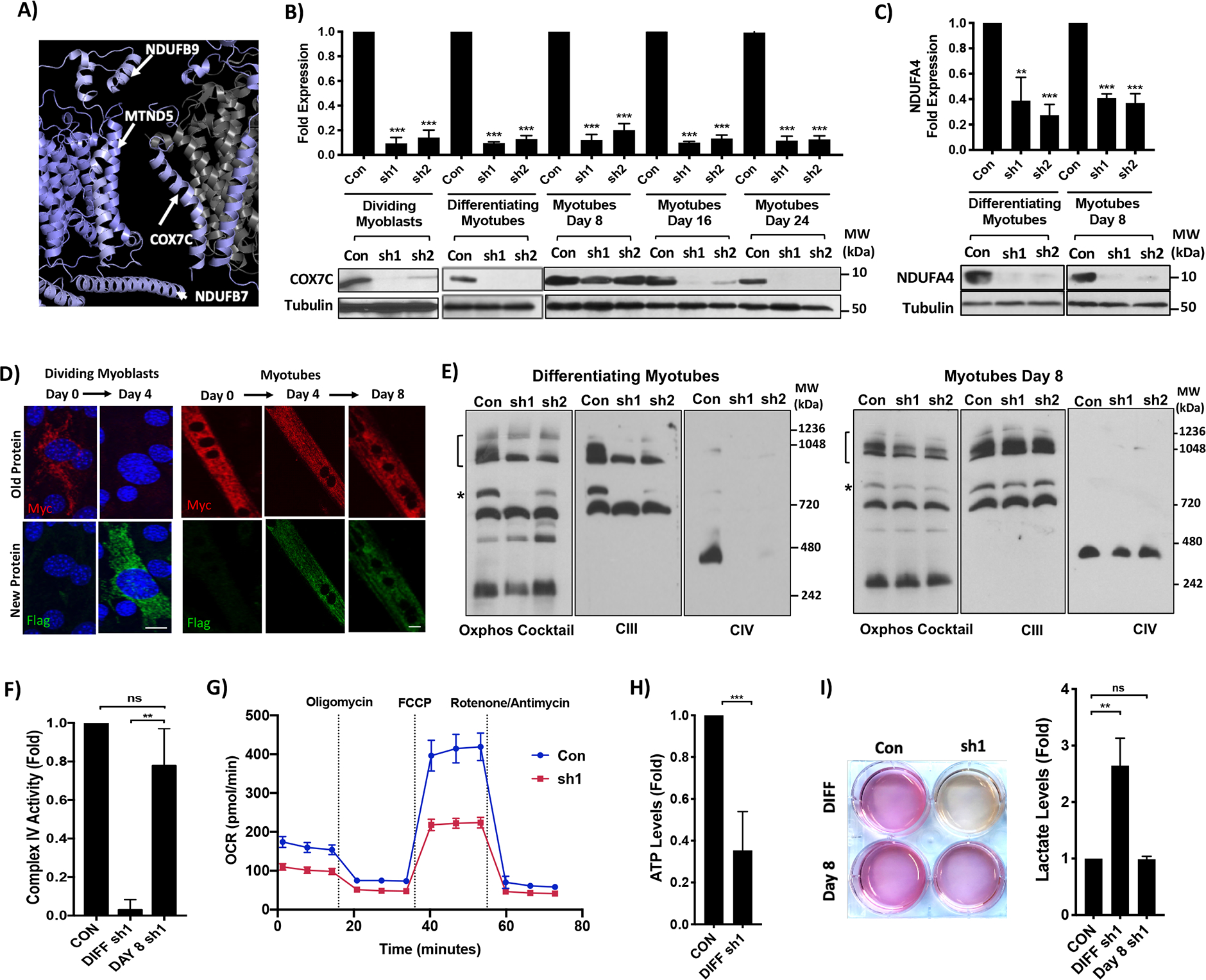
A) LLP COX7C from CIV forms a stable contact site with CI by interacting with long-lived subunits NDUFB9, MTND5 and NDUFB7 in the respirasome SC. Image shows subunits in the contact site with half-lives color-coded from 2–8 days from dark red to white to dark blue. Proteins with no coverage are colored grey. B) COX7C mRNA levels (qPCR, top panel) and protein levels (western blot, bottom panels) with control and COX7C-hairpins (sh1,sh2) in dividing myoblasts, differentiating myotubes, fully differentiated myotubes at Day 8, Day 16 and Day 24. C) NDUFA4 mRNA levels (top panel) and protein levels (bottom panel) with control and NDUFA4-hairpins (sh1,sh2) in differentiating myotubes and fully differentiated myotubes at Day 8. Western blots show Tubulin as loading control. D) COX7C turnover as measured by RITE. Images show Myc tag (old protein, top panel) and Flag tag (new protein, bottom panel) at Day 0 and 4 in dividing myoblasts (left) and Day 0, 4 and 8 in myotubes (right). E) ETC SC and IC levels in control and COX7C hairpins. Western blots show levels of all the complexes (Oxphos cocktail), CIII and CIV containing complexes in differentiating myotubes (left blots) and Day 8 myotubes (right blots). Bracket indicates higher-order SCs and asterisk indicates the intermediate SC. F) CIV activity in control and sh1-mediated COX7C knockdown in differentiating myotubes or Day 8 myotubes. G) Oxygen Consumption Rates (OCR) in pmol/min over a seahorse assay of 80 minutes and in control and sh1-COX7C knockdowns in differentiating myotubes. Injection timing of Oligomycin, FCCP and Rotenone/Antimycin is indicated with dotted lines. H) ATP levels in control and sh1-COX7C knockdowns in differentiating myotubes. I) Left, Image of cell culture media in a 6-well plate and Right, graph of fold change in lactate levels in control and sh1-COX7C knockdowns in differentiating myotubes or Day 8 myotubes.
For all graphs, error bars represent SD from an n=3. **p<0.01, ***p<0.001. Scale bars: 10μm. Western blot markers are indicated.
To compare the turnover of SCs vs ICs, we subtracted the half-lives of proteins in the SCs from their respective half-lives in the ICs, and then color-coded them on the scale of −1 to +1 days (red to blue). Remarkably, we find that while turnover in the peripheral arm of CI is relatively similar between the SCs and ICs, the majority of the membrane arm of CI exhibits more longevity in the SCs (Figure 4B). Indeed, most subunits in the membrane arm (consisting of Pp and Pd modules) show a 0.5d or 12h delay in turnover (Figure 4C–D). Interestingly, only the LLPs in CI display this increased stability, while the shorter-lived proteins do not. Similarly, we find that CIII2 is also more stable in SCs as well as in the intermediate SC, with many subunits exhibiting the 0.5d delay (Figure 4E,4G,S4A). This is the first direct evidence based on protein turnover data establishing that CI and CIII2 are more stable in SCs. Moreover, we find that the subunit COX7A2L is more stable in the SCs by 2 days, the largest difference observed for any ETC subunit (Figure 4E,4G). Interestingly, COX7A2L has been reported to bind to CIII2 to promote the assembly of intermediate SCIII2,IV and the respirasome SCI,III2,IV, and its loss-of-function leads to reduced SC formation (Cogliati et al., 2016; Lobo-Jarne et al., 2018). We see that COX7A2L has a half-life of 2.5 days in CIII2, which increases to 4.8 days in SCI,III,IV, ISCIII,IV and CIV (Figure 4H). This increased stability could be due to its structural location as it binds directly with CIII2 to facilitates its interaction with CIV and CI, making it peripheral in CIII2 but buried deep inside the SCs and less accessible for degradation. Unlike CI and CIII2, CIV shows a different stability pattern in the SCs vs IC. We find that SLPs like COX6A2, COX6B1, and NDUFA4 are more stable in the SCs, while LLPs like COX5A and COX5B are more stable in the IC (Figure 4F–G). We also find that CIV subunits are more stable in the SCs compared to the intermediate SC, while the CIV2 is more stable than monomeric CIV (Figure S4B–E). These results show that LLPs are core components of SCs, also forming stable contact sites between the different complexes that could be essential for maintaining its quaternary structure. Our study also shows that ETC proteins exhibit differential stability in SCs vs ICs, particularly CI and CIII2, where the LLPs show increased longevity in the SCs.
Longevity of COX7C protects myotubes from loss of function defects in Complex IV and Supercomplex assembly
Since LLPs are core components of protein complexes, we hypothesize that under conditions that lead to a decline in their transcription, they could function as a scaffold for the structural and functional integrity of the complex. We decided to uncouple protein levels and function of a mitoLLP in the ETC from its transcription by depleting its mRNA. We picked COX7C as it is one of the longest-lived proteins in CIV (Figure 3G, T1/2: 6 days), and also forms critical interactions with three LLPs from CI in the respirasome, suggesting that this is one of the most stable contact points within the SC (Figure 5A). Also, the function of COX7C is currently unknown and it has been reported to undergo an age-dependent decline in transcription in human skeletal muscles (Su et al., 2015).
To deplete COX7C, we used miRE-inducible knockdowns in three cell systems - dividing myoblasts, differentiating myotubes, and fully differentiated myotubes (knockdowns were induced post-differentiation for 8,16,24 days) and determined COX7C RNA and protein levels. Dividing and differentiating cells show a dramatic reduction in COX7C RNA as well as protein levels (Figure 5B). Interestingly, while myotubes at Day 8 clearly show a reduction of COX7C at the transcript level, they show minimal change in the protein level (Figure 5B). Importantly, by Day 16 we start seeing a reduction of COX7C protein, with complete depletion by Day 24, to the same extent as seen with differentiating myotubes (Figure 5B). To validate that this effect was indeed due to the long-lived nature of the protein and not due to any defects in COX7C degradation or the RNAi machinery, we knocked down another LLP as well as an SLP from CIV. Similar to COX7C, COX5A knockdowns (T1/2:6d) show a clear reduction of RNA and protein levels in differentiating myotubes, and knockdowns in Day 8 myotubes show a reduction of RNA levels while having minimal effect on the protein levels (Figure S5A). Importantly, knockdowns of short-lived CIV protein NDUFA4 (T1/2:1.8d) show a clear reduction of RNA as well as protein levels in both differentiating and Day 8 myotubes (Figure 5C). These data demonstrate that COX7C and COX5A are indeed long-lived in post-mitotic myotubes. To further validate COX7C’s longevity we imaged old COX7C protein in the cell using RITE. We find that in dividing myoblasts by Day 4 old COX7C protein (Myc) is down to less than 20% and mostly new protein is present (Flag) (Figure 5D,S5B–C), indicating a short half-life. However, in myotubes, there is a persistence of old protein at Day 4 and even Day 8 (55% and 40% respectively) despite production of new protein (Figure 5D,S5B–C), validating that COX7C has a longer half-life in myotubes.
We next examined the function of COX7C, and the role of its longevity in maintaining that function in myotubes. Intriguingly, we find that COX7C knockdowns in differentiating myotubes show a substantial depletion of CIV, intermediate as well as higher-order SC levels (Figure 5E,S5D). This indicates that COX7C might be involved in the assembly or stability of CIV, which subsequently leads to loss of SCs as well. We also find a dramatic reduction in CIV enzymatic activity, oxygen consumption and ATP levels with the knockdowns in differentiating myotubes (Figure 5F–H). Additionally, we also noted that the color of the culture media changed from pink to yellow with the knockdowns due to increased lactate production (Figure 5I) and extracellular acidification rate (Figure S5H). This indicates that COX7C-depleted cells shunt to glycolysis because of impaired ETC function. Importantly, these cells also show a reduction in myotube thickness (Figure S5I–J) indicating that COX7C-dependent respiration is critical for myotube health. Interestingly, knockdowns in Day 8 myotubes do not show any reduction in CIV or SC levels (Figure 5E,S5E). Also, the Day 8 knockdown has the same the CIV enzymatic activity as control cells (Figure 5F) and the culture media stays pink, with lactate levels similar to control cells (Figure 5I). Thus, despite depletion of COX7C transcripts, the longevity of this protein is able to protect myotubes from its loss-of-function defects in CIV/SC assembly, thereby maintaining CIV activity and ETC function. Indeed, only at Day 16 and 24 when the levels of the protein start to get depleted do we see a reduction in the levels of CIV and SCs (Figure S5F–G). Therefore, we have demonstrated a critical role for COX7C in CIV and SC assembly and have established that its longevity can maintain ETC integrity despite a decline in its mRNA. Given that the majority of the ETC is long-lived and is known to undergo age-dependent transcriptional decline in post-mitotic cells (Su et al., 2015), our study provides an exciting link between mitoLLPs in protein complexes and aging.
Discussion
A critical finding of our study is that LLPs show differential stability in SCs. Previous studies have shown that mutations in CIII and CIV can affect CI activity, suggesting that SCs can increase CI stability (Acin-Perez et al., 2004; Diaz et al., 2006). However, we provide the first half-life measurements that demonstrate increased stability of CI as well as CIII2 in the SCs. These data establish a clear function of respiratory SCs in myotubes. Although it is only an increase of 0.5d, given that the ETC functions in the scale of milliseconds (electron transfer takes 5–20ms), a 12h difference could easily lead to increased respiratory output. While SCs don’t affect the actual rate of electron transfer between the complexes (Fedor and Hirst, 2018), their increased stability could cause electron transfer to occur for longer. Interestingly, only the LLPs of CI and CIII2 show this increased stability, lending further support to our hypothesis that LLPs are critical for protein complex stability and function. Also, in CIV the shorter-lived subunits are more stable in the SCs, and vice versa, suggesting that there could be specialized functions of these proteins in the SCs and ICs respectively.
We also provide the first observation of age mosaicism in mitochondria, similar to what we have previously observed with NPCs (Toyama et al., 2019), primary cilia and pancreatic beta cells (Arrojo et al., 2019). This raises a key question – do the distinct mitochondrial populations have differences in structure, composition and function? Interestingly, mitochondria in the same cell can exhibit extensive heterogeneity in membrane potential, respiratory capacity and ROS production (Kuznetsov and Margreiter, 2009). Indeed, perinuclear clustering of aged mitochondria has been observed in stem cells before cell division (Katajisto et al., 2015). Moreover, perinuclear mitochondria in cardiomyocytes can drive ATP synthesis for nuclear import (Dzeja et al., 2002). It is conceivable that the clusters of old mitochondria we observe in neurons could have specialized functions, particularly for the axonal mitochondria at synapses. While we observed that axonal mitochondria tend to be older in vivo, we did not observe a consistent similar polarization in the cultured neurons, most likely because they require further maturation or glial cell co-culture to establish this polarity. Our MIMS-EM data also shows that neurons have older mitochondria than muscle fibers, which could be because muscle fibers have quiescent satellite cells that when activated during injury and development can divide and fuse into the fiber (Yin et al., 2013). Since quiescent myoblasts in vitro display faster turnover of most nuclear LLPs (Toyama et al., 2019), the satellite cells in vivo could have newer mitochondria that undergo fusion/fission cycles with the muscle mitochondria to reduce the 15N/14N.
An important finding from our study is the distinction between nuclear and mitoLLPs, as the former turnover extremely slowly with half-lives of a month, while the latter turn over in a week. This is corroborated in vivo where nuclear proteins persist for a year, and mitochondrial proteins can persist for a month (Fornasiero et al., 2018; Karunadharma et al., 2015; Toyama et al., 2013). Our half-lives are in contrast with the in vivo studies because while our cells are post-mitotic, they are not fully mature neurons or muscle fibers and can only be cultured for a few weeks. Indeed, a previous study that compared in vivo and in vitro data showed that half-lives are lower in the latter (Fornasiero et al., 2018). While the half-lives generated in our system may be scaled down, the relative turnover among LLPs is similar. Indeed, an in vivo study categorized proteins that are above 98th percentile in terms of stability as “Exceptional LLPs (histones and nups), whereas the 95–98th percentile were “Normal LLPs” (containing signaling, SNARE and mitochondrial proteins) (Fornasiero et al., 2018). We also find a strong correlation in the half-lives of ETC proteins between our in vitro and this in vivo dataset of brain and heart tissues (r=0.75, p<0.001) (Fornasiero et al., 2018). We speculate that some of the differences in the data could be due to the cell systems used or by the use of tissue homogenates in previous studies instead of separating the mitochondrial fraction and ETC complexes. Since proteins show differential stability in SCs, it is indeed critical to separate the complexes for ETC turnover studies.
A key finding is that 40% of mitochondrial proteins are LLPs. This suggests that mitochondria have evolved to be longer-lived than the cellular proteome, an adaptation that has vital implications for the role of LLPs. While previous work has described a link between nuclear LLPs and their role in aging (D’Angelo et al., 2009), an open question is – Why would a cell have mitoLLPs, given that their longevity makes them more susceptible to oxidative damage from the ETC? Our COX7C suggests that mitoLLPs are insulated against changes in their gene expression as the complexes will still contain the old protein and therefore function effectively. Thus, we speculate that a critical role for LLPs could be maintaining protein complex function in conditions of cellular stress where global transcription/translation is diminished. Indeed, transient stressors like heat-shock and starvation can lead to a decline in transcriptional and translational rates (Mahat et al., 2016; Pain, 1994). More importantly, during aging there is a global decline in gene expression, particularly of mitochondrial and ETC proteins (Su et al., 2015). It is conceivable that during these conditions the peripheral SLPs in a complex would fall off, but the core LLPs would be present and could maintain a functional subcomplex to slow down cellular deterioration. Moreover, such heterogeneity in the complex could also function to modulate the activity of protein complexes for adapting to different conditions without having to dismantle the long-lived scaffold.
Limitations of Study
While we identify mitoLLPs in vitro, a limitation is that the study lacks correlative proteomics data in vivo. Also, it is unclear if longevity of LLPs compared to SLPs can protect mitochondrial function during specific cellular stress conditions and aging. Some of these questions could not be addressed due to technical and experimental delays caused by the COVID-19 pandemic.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
All requests for further information, reagents and resources should be directed to the Lead Contact, Martin W. Hetzer (hetzer@salk.edu).
Materials availability
All materials generated in this study are available upon request to the Lead Contact.
Data and code availability
All mass spectrometry proteomic datasets have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository and are publicly available as of the date of publication. DOIs are listed in the key resources table.
All original code has been deposited at Mendeley and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| OxPhos Antibody Cocktail Rodent | Invitrogen | Cat#458099, RRID:AB_2533835 |
| NDUFA9 Monoclonal Antibody | Invitrogen | Cat#459100, RRID:AB_2532223 |
| UQCRC1 Monoclonal Antibody | Invitrogen | Cat#459140, RRID:AB_2532227 |
| COX IV (3E11) Rabbit mAb | Cell Signaling | Cat#4850S, RRID:AB_2085424 |
| COX7C Polyclonal Antibody | Invitrogen | Cat#PA551284 RRID:AB_2636731 |
| NDUFA4 Polyclonal Antibody | Invitrogen | Cat#PA599439, RRID:AB_2818372 |
| COX5A Polyclonal Antibody | Proteintech | Cat# 11448–1-AP, RRID:AB_2085429 |
| α/β-Tubulin Antibody | Cell Signaling | Cat#2148S, RRID:AB_2288042 |
| Myc-Tag (9B11) Mouse mAb | Cell Signaling | Cat# 2276S, RRID:AB_331783 |
| Monoclonal ANTI-FLAG® M2 antibody | Sigma | Cat#F3165, RRID:AB_259529 |
| Goat anti-Mouse IgG2a Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat# A21131, RRID:AB_141618 |
| Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Invitrogen | Cat# A21124, RRID:AB_141611 |
| Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat# A21121, RRID:AB_2535764 |
| Goat anti-Mouse IgG2a Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Invitrogen | Cat# A21134, RRID:AB_1500825 |
|
Chemicals, peptides, and recombinant proteins | ||
| 15N-spirulina algae | Cambridge isotope Laboratories | Cat#MF-SPIRULINA-N-S |
| 2.5% Glutaraldehyde | Ted Pella Inc., Redding, CA | Cat#18426 |
| 2% Formaldehyde | Ted Pella Inc., Redding, CA | Cat#18200 |
| Sodium Cacodylate | Ted Pella Inc., Redding, CA | Cat#18851 |
| Osmium Tetroxide | Electron Microscopy Sciences | Cat#19190 |
| Potassium Ferrocyanide | Electron Microscopy Sciences | Cat#3114 |
| Thiocarbohydrazide | Electron Microscopy Sciences | Cat#21900 |
| Durcupan ACM Resin | Sigma-Aldrich | Cat#44611 (Comp A), Cat# 44612 (Comp B), Cat#44613 (Comp C), Cat#44614 (Comp D). |
| Matrigel (Cultrex) | R&D Systems | Cat#3433–005-01 |
| DMEM:F12 for SILAC | Fisher Scientific | Cat#88370 |
| Dialyzed FBS | Gibco | Cat#26400044 |
| DMEM for SILAC | Fisher Scientific | Cat#A33822 |
| L-Lysine-13C6 hydrochloride | Sigma | Cat#643459 |
| L-Arginine-13C6,15N4 hydrochloride | Sigma | Cat#608033 |
| TCEP-HCL | Sigma-Aldrich | Cat#C4706 |
| Chloroacetamide | Sigma-Aldrich | Cat#C0267 |
| Trypsin | Promega | Cat# V5111 |
| NativePAGE™ Sample Prep Kit | Invitrogen | Cat#BN2008 |
| Native PAGE 3–12% gradient gel | Invitrogen | Cat# BN1001BOX |
| Dark Blue Cathode Buffer | Invitrogen | Cat# BN2002 |
| Running Anode Buffer | Invitrogen | Cat#BN2001 |
| Trizol | Invitrogen | Cat#15596026 |
| SuperSignal West Pico | Fisher Scientific | Cat#PI34078 |
| SYBR Green PCR Master Mix | Applied Biosystems | Cat#4309155 |
| Ibidi u-Slide chambers | Ibidi | Cat#80826 |
| 4-hydroxytamoxifen (4OHT) | Sigma-Aldrich | Cat#H6278 |
| Critical commercial assays | ||
| RNeasy Mini Kit | Qiagen | Cat#74106 |
| QuantiTect Reverse Transcriptase Kit | Qiagen | Cat#205311 |
| Complex IV Rodent Enzyme Activity Microplate Assay Kit | Abcam | Cat# ab109911 |
| Seahorse XF Cell Mito Stress Test | Agilent | Cat#103015–100 |
| Seahorse XF Glycolysis Stress Test Kit | Agilent | Cat#103020–100 |
| ATP Chemiluminescence Detection Assay Kit | Cayman Chemicals | Cat# NC1357058 |
| L-Lactate Assay Kit | Abcam | Cat# ab65330 |
| Deposited Data | ||
| Mass spectrometry Proteomic Datasets | ProteomeXchange Consortium via PRIDE (Perez-Riverol et al., 2019) | DOI: 10.6019/PXD028963 |
| Experimental models: Cell lines | ||
| SH-SY5Y | ATCC | Cat#CRL-2266 |
| C2C12 Myoblasts | ATCC | Cat#CRL-1772 |
| Human ES Cells (H9) | WiCell | Cat#WA09 |
| Experimental models: Organisms/strains | ||
| Mouse:FVB (Male) | The Jackson Laboratory, Bar Harbor,ME | Stock No: 001800 |
| Oligonucleotides | ||
| COX7C sh1 targeting sequence - TGCTGTTGACAGTGAGCGACCGCACCTTTCTTTATAGTAATAGTGAAGCCACAGATGTATTACTATAAAGAAAGGTGCGGCTGCCTACTGCCTCGGA | This paper | Eton Biosciences; Sequences generated using shERWOOD algorithm (Knott et al., 2014) |
| COX7C sh2 targeting sequence - TGCTGTTGACAGTGAGCGCCACCAGCTACTTAAAAAATAATAGTGAAGCCACAGATGTATTATTTTTTAAGTAGCTGGTGTTGCCTACTGCCTCGGA | This paper | Eton Biosciences; Sequences generated using shERWOOD algorithm (Knott et al., 2014) |
| COX7C qPCR Primer: Forward - AGCATGTTGGGCCAGAGT | This paper | Eton Biosciences |
| COX7C qPCR Primer: Reverse-ACTGAAAACGGCAAATTCTT | This paper | Eton Biosciences |
| NDUFA4 qPCR Primer: Forward - CGCTTGGCACTGTTTAATCCA | This paper | Eton Biosciences |
| NDUFA4 qPCR Primer: Reverse - TCCATGGCTCTGGGTTGTTC | This paper | Eton Biosciences |
| COX5A qPCR Primer: Forward - CTGCCGCTGTCTGTTCCATTCG | This paper | Eton Biosciences |
| COX5A qPCR Primer: Reverse - TGTCACCCAGCGAGCATCAAACT | This paper | Eton Biosciences |
| Beta-actin qPCR Primer: Forward - CTGTCCCTGTATGCCTCTG | This paper | Eton Biosciences |
| Beta-actin qPCR Primer: Reverse - ATGTCACGCACGATTTCC | This paper | Eton Biosciences |
| Recombinant DNA | ||
| UNG Construct | Addgene | Cat#127288 |
| Plasmid: pRITE Myc to Flag (pRITE-MF) | (Toyama et al., 2019) | N/A |
| Plasmid: ATP5C1 RITE-MF pLentiCMVBlast | This paper | N/A |
| Plasmid: COX7C RITE-MF pLentiCMVBlast | This paper | N/A |
| shERWOOD UltramiR Lentiviral Inducible shRNA for NDUFA4 | Transomic technologies | Cat#TLMSU2300–17992 |
| Set of 3 SMARTvector Inducible Mouse COX5A (mCMV-TurboGFP shRNA) SMARTvector Inducible Lentiviral Control |
Dharmacon | Cat#V3SM11256–01EG12858 Cat# VSC11651 |
| Software and algorithms | ||
| OpenMIMS | (Steinhauser et al, 2012) | https://github.com/BWHCNI/OpenMIMS/ |
| Unwarp Plugin for ImageJ | (Schneider et al., 2012; Sorzano et al., 2005) | N/A |
| MesoFusion Plugin and Code for ImageJ | This paper; Mendeley Data | https://data.mendeley.com/datasets/b3hww8ng7w/1; DOI:10.17632/b3hww8ng7w.1 |
| Integrated Proteomics Pipeline Version 6.0.5 IP2 | N/A | http://www.integratedproteomics.com/ |
| Census2 | (Park et al., 2014) | N/A |
| nl2sol algorithm | R package | http://www.netlib.org/port/ |
| modelr algorithm | R package | https://CRAN.R-project.org/package=modelr |
| Proteomics Data Analysis Code (Mass spectrometry and half-life calculation) | This paper; Mendeley Data | https://data.mendeley.com/datasets/b3hww8ng7w/1; DOI:10.17632/b3hww8ng7w.1 |
| RCSB Protein Data Bank | https://www.rcsb.org/ | mouse Complex I: 6G2J, human Complex III: 5XTE, human Complex IV: 5Z62, porcine respirasome: 5XTH |
| PyMOL | PyMOL by Schrödinger | https://pymol.org/2/ |
| Half-life Color Code for PDB crystal structures in PyMOL | This paper; Mendeley Data | https://data.mendeley.com/datasets/b3hww8ng7w/1; DOI:10.17632/b3hww8ng7w.1 |
| Graphpad Prism 8.0 | Graphpad | https://www.graphpad.com/scientific-software/prism/; RRID: SCR_002798 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse
The mouse used in this study was of the FVB/NJ strain (Stock No: 001800), male and was acquired from the Jackson Laboratory (Maine, USA). The housing conditions used were 12/12h light-dark cycles, water and food provided ad-libitum. For the SILAM breeding, as described in the methods details section, 6-week old FVB/NJ females (Jackson Laboratories) were fed ad-libitum with chow containing 15N-spirulina algae (Cambridge isotope Laboratories, Inc.) for 70 days prior to mating with a male. Mating period was of 1 week. While female mice were used for the SILAM, the final MIMS-EM imaging study was conducted on 1 male mouse. All animal experimentation was approved by the University of California San Diego IACUC (protocol number S03172M).
Cell Culture
SH-SY5Y and C2C12 cells were all obtained from American Type Culture Collection (ATCC). SH-SY5Y cells were cultured in DMEM:F-12 with 10%FBS with Pen-Strep (growth media). For neuronal differentiation, SH-SY5Y cells were plated in growth media on ibidi chamber slides that had been previously coated with 10μg/ml polyornithine solution for 1 day, followed by 10μg/ml laminin for another day. The cells were grown until 80–90% confluent, then washed twice with PBS and switched to DMEM:F12 with 0.5%FBS, 3μM Retinoic Acid and Pen-Strep for 5 days to generate neurons as described previously (Cheung et al., 2009). Human ES H9 Cells (WiCell) cells with UNG construct (Addgene, 127288) were a kind gift from the Fred Gage lab. H9 cells were grown on Matrigel coated plates (R&D Systems, 3433–005-01) with iPS-Brew XF (Miltenyi Biotec, 130–104-368) for 24hours before induction with 2μg/ml Doxycycline (Sigma) for 3 days. For further maturation, induced neurons were switched to Neuronal Maturation Medium (NMM) containing 1:1 DMEM/F12 and Neurobasal Media, supplemented with GlutaMAX, B27, N2, 0.5mM dbcAMP, 20ng/ml GDNF and 20ng/ml BDNF, 10μg/ml laminin as well as 2μg/ml Doxycyline. At Day 7 cells were treated with 2μM AraC for 2 days to induce cell death of non-neuronal cells, following which cells were kept in the NMM till Day 11 to generate mature neurons as described previously (Schafer et al., 2019). C2C12 cells were cultured in DMEM with 20%FBS with Pen-Strep (growth media). For myotube differentiation, C2C12 cells were grown in growth media until fully confluent, then washed twice with PBS and switched to DMEM with 2% Horse Serum with Pen-Strep (differentiation media) for 5 days to generate mature myotubes as described previously (D’Angelo et al., 2009). All cells were cultured at 37°C with 5% CO2, and media was changed every other day.
METHOD DETAILS
Stable isotope metabolic labelling of mammals (SILAM)
SILAM was performed as described previously (Arrojo et al., 2019). Briefly, 6-week old FVB/NJ females (Jackson Laboratories) were fed ad-libitum with chow containing 15N-spirulina algae (Cambridge isotope Laboratories, Inc.) for 70 days prior to mating with a male. Mating period was of 1 week. 15N-chow was provided throughout the mating period as well as gestation and lactation period until the 15N-labelled pups were 21 days (P21) in post-natal age. Pups were weaned at P21 and fed 15N-chow until they were P45, after which their food source was replaced with 14N-chow. The pups were then euthanized, perfused with fixative as previously described (Sosinsky et al., 2005) at 6-months after the start of feeding with 14N-chow. The perfusion process was as follows: mice were anesthetized with a ketamine:xylazine mix and transcardially perfused with a solution of 2.5% glutaraldehyde, 2% PFA in 0.15M sodium cacodylate buffer containing 2mM calcium chloride warmed to 37°C. Perfusion time was of 10 minutes and the brain and skeletal muscle depots were dissected, the brain was further cut into 150-micron thick slices with a vibratome (Leica VT100S), and both tissue types prepared for electron microscopy.
Sample processing for Scanning Electron Microscopy imaging
After dissection and sectioning, the samples were post-fixed in the same fixative at 4°C overnight. The brain slices were washed with cacodylate buffer and then fixed in a solution containing 2% osmium tetroxide, 1.5% potassium ferrocyanide in cacodylate buffer for 1 hour at room temp (25°C). Samples were washed with water and placed in a solution of 0.5% thiocarbohydrazide for 30 minutes and washed with water. Next, samples were placed in 2% aq. osmium tetroxide for 1 hour, washed with water and then placed in 2% aq. uranyl acetate at 4°C overnight. Samples were again washed with water and placed into Walton’s lead aspartate solution for 30 min at 60°C, followed by washes in water and sequential dehydration with 70% EtOH, 90% EtOH, 100% EtOH, 100% EtOH, and dry acetone (10 minutes each step) on ice. Next, the samples were placed into 1:3, 1:1, and 3:1 solutions of Durcupan ACM:acetone for 12 hours each. Lastly, the samples were placed into 3 changes of 100% Durcupan ACM for 1 day each and baked at 60°C for 48 hours and stored in aluminum foil dishes until sectioning in 80nm-thick sections using an ultra-microtome (Leica).
Multi-isotope mass spectroscopy and scanning electron microscopy
Correlated Multiple isotope mass spectrometry imaging (MIMS) and scanning electron microscopy imaging (called MIMS-EM) was used to quantify the concentration of 15N and 14N in brain tissue sections from 15N-labelled mice in this study. First, 80nm-thick sections of 15N-labelled brain and intercostal muscle (ICM) from animals chased for 6-months were placed on a 5×7mm Si wafer (TedPella). Next, we used the Atlas 5 engine (Fibics, Ottawa, Canada) to create large field of view maps with 5nm pixels in X-Y of sections of the mouse cerebellum and ICM using scanning electron microscopy (GeminiSEM, MerlinSEM (Zeiss, Germany). We targeted the granular cell layer in the cerebellum and the neuromuscular junction region in the ICM. MIMS image acquisition of mapped regions of interest was done as previously described (Arrojo et al., 2019; Steinhauser et al., 2012; Zhang et al., 2012). Here, sections of cerebellum and ICM tissue sections containing mapped cells and organelles were imaged with a NanoSIMS 50L (Cameca, France) using a cesium (Cs−) beam, 1024×1024 image raster and 15N and 14N levels were detected simultaneously.
Stable isotope labeling by amino acids in cell culture (SILAC)
For the H9 ES cell SILAC, light media was regular DMEM:F12 supplemented with GlutaMAX, B27, N2, 0.5mM dbcAMP, 20ng/ml GDNF and 20ng/ml BDNF, 10μg/ml laminin and 2μg/ml Doxycyline. Heavy media was DMEM:F12 for SILAC (Fisher Scientific, 88370) with heavy amino acids L-Lysine-13C6 hydrochloride (Sigma, 643459) and L-Arginine-13C6,15N4 hydrochloride (Sigma, 608033), also supplemented with above mentioned factors. For the SILAC timecourse, H9 cells were differentiated into neurons for 11 days as mentioned before, and this day was treated as the beginning of the timecourse (Day 0). Neurons were switched to heavy media for the Day 8 timepoint and light media for the other timepoints on this day. The media was switched to heavy for the other timepoints on respective days and the neurons for all timepoints were harvested 8 days after the beginning of the timecourse. Media was refreshed every other day. For the C2C12 SILAC, light media was regular DMEM with 2% Dialyzed FBS (Gibco, 26400044) with Pen-Strep, and heavy media was DMEM for SILAC (Fisher Scientific, A33822) with 2% Dialyzed FBS with Pen-Strep and heavy amino acids L-Lysine-13C6 hydrochloride and L-Arginine-13C6,15N4 hydrochloride. For the SILAC timecourse, C2C12 cells were differentiated for 5 days in differentiation media, and this day was treated as the beginning of the timecourse (Day 0). The rest of the timecourse was conducted exactly like the neuronal SILAC mentioned before.
Fractionation and LC/MS Analysis
Neurons were harvested at the end of the SILAC timecourse by washing the plate with 1xPBS and spinning down the pellet at 1200rpm for 5min. They were then fractionated into crude nuclear, mitochondrial and cytosolic fractions as described previously (Clayton and Shadel, 2014). Briefly, neuronal cell pellets were resuspended in Mannitol-Sucrose (MS) Buffer (210mM Mannitol, 70mM Sucrose, 5mM Tris HCL pH 7.5, 5mM EDTA pH 8 and protease inhibitors), followed by mechanical lysis through a 271/2-gauge needle and syringe. The homogenate was centrifuged at 1000g for 5min for crude nuclear pellet, and the supernatant was centrifuged at 10,000g for 20min to get the crude mitochondrial pellet. The remaining supernatant was kept as the cytosolic fraction. Myotubes were harvested at the end of the SILAC timecourse through selective trypsinization for 5mins to separate them from the mononuclear reserve cell population. The harvested myotubes were either kept as whole cell pellets or fractionated into crude nuclear and mitochondrial pellets only. The different fractions for neurons (nuclear, mitochondria and cytosolic) and myotubes (whole cell, nuclear and mitochondrial) were precipitated by methanol/chloroform and redissolved in 8M urea/100mM TEAB, pH 8.5. Proteins were reduced with 5 mM tris(2-carboxyethyl) phosphine hydrochloride (TCEP-HCL, Sigma-Aldrich, C4706) and alkylated with 10 mM chloroacetamide (Sigma-Aldrich, C0267). Proteins were digested overnight at 37°C in 2M urea/100mM TEAB, pH 8.5, with trypsin (Promega, V5111) and digestion was quenched with formic acid at 5% final concentration. The samples were then subject to electrospray ionization (ESI) and tandem mass spectrometry (MS/MS), to identify peptides and determine their heavy to light (13C6/12C6 and 15N4/14N4) isotope ratios as described previously (Buchwalter et al., 2019). Briefly, digested samples were run with CID fragmentation on a Thermo Orbitrap Fusion Lumos Tribrid MS/MS by direct injection onto a column (30cm, 75uM ID column with BEH 1.7um C18 resin). A flow rate of 300nl/min was used on a nLC 1200 for separation of the injected samples. 0.1% formic acid (water) was used as Buffer A and 0.1% formic acid (90% acetonitrile) was used as Buffer B. A total run time of 240 minutes was used, with the first 180 minutes run on a gradient of 1–25% B, the next 40 minutes was an increase to 50% B, the next 10 minutes was an increase to 100% B and the final 10 minutes was held at 100%B. Before sample injection, re-equilibration of the column was performed with Buffer A and peptide elution was conducted directly from the column tip. By using 2.8 kV voltage at the back of the column, the eluted peptides were then nanosprayed into the mass spectrometer. The Lumos MS/MS was operated in data dependent mode and the collection of full MS1 scans was performed in the Orbitrap. The data collection was conducted with a resolution of 120K, a mass range of 400–1500 m/z and a target of 4e5 for the AGC. For each scan, the most abundant ions were selected for CID MS/MS within a cycle time of 3s. An AGC target of 4e5 and minimum intensity of 5000 was used in the ion trap. A maximum fill time of 50ms was used for the MS scans and 100ms for the MS/MS scans. Monoisotropic precursor selection was allowed and a quadruple isolation of 1.6 m/z was used. 2–7 charge states were chosen and a 5s exclusion duration was used with dynamic exclusion.
BN-PAGE and western blots
Blue-native PAGE gels were conducted as described previously (Jha et al., 2016). Briefly, crude mitochondria were isolated as mentioned in the previous section from dividing myoblasts and differentiated myotubes, or for control and COX7C-knockdowns for differentiating myotubes, Day 8,16 and 24 myotubes. The mitochondrial pellets were subject to digitonin solubilization at 8g/g digitonin/protein ratio using NativePAGE™ Sample Prep Kit (Invitrogen, BN2008). For BN-PAGE gels subject to Coomassie staining (for dividing myoblasts, myotubes, and the SILAC samples) 100μg mitochondrial pellet protein was used for each sample to clearly visualize the supercomplex and individual complex bands. For the BN-PAGE that was transferred onto a membrane for western blots 50ug mitochondrial pellet protein was used for each sample. Upon addition of digitonin, the mitochondria were put on ice for 20min. The solubilized mitochondria were then centrifuged at 20,000g for 10 mins at 4°C, and the supernatant collected. Coomassie G-250 was added to the supernatant at a 1:7.5 ratio (2μl G-250/15μl supernatant) before loading the samples in a Native PAGE 3–12% gradient gel (Invitrogen, BN1001BOX) with dark blue cathode buffer (Invitrogen, BN2002) in the inner chamber and running anode buffer (Invitrogen, BN2001) in the outer chamber. The gel was run for 30 minutes at 150V, after which the dark blue cathode buffer in the inner chamber was replaced with running anode buffer and run for another 60 minutes at 250V. The gel was either stained with Coomassie and destained for BN-PAGE or transferred onto PVDF membrane. Post-transfer the PVDF membrane was destained with 100% methanol before blocking and adding primary antibodies against Oxphos Cocktail (Invitrogen, 458099), Complex I subunit NDUFA9 (Invitrogen, 459100), Complex III subunit UQCRC1 (Invitrogen, 459140) and Complex IV subunit COXIV (Cell Signaling, 4850S) at 1:1000 dilution in 5% BSA/TBST overnight at 4 degrees. Blots were washed 3x in TBST and incubated with HRP-conjugated secondary antibodies for 1 hour, and developed using SuperSignal West Pico (Thermo Scientific, PI34078).
Gel Band Extraction and LC/MS Analysis
The SILAC samples from Day 0–8 were all subject to mitochondrial fractionation and digitonin solubilization and run on BN-PAGE gels as described above. The gels were stained in Coomassie and destained, and the bands on the gels were cut as labelled in Figure 3A and put in tubes as follows: Sample 1 – SCI,IIIn,IVn, Sample 2 – SCIII2,IV, Sample 3 – CIII2,CIV2, Sample 4 – CI and CII, Sample 5 – CIV. Each tube containing the gels was destained with 50% methanol/5% acetic acid overnight and dehydrated with 100% acetonitrile (ACN) twice. The gels were air dried, then 5mM TCEP-HCl was added for 30mins and removed, followed by treatment with 50mM chloroacetamide for 30 mins in the dark. The chloroacetamide was removed, and the gels washed twice in ACN, and dried. 20ng/ul trypsin (Promega, V5111) in 100mM ammonium bicarbonate was then added to barely cover the gels and incubated overnight. The gels were then incubated with 5% formic acid (without removing the liquid from the tube) for 10 mins, and the supernatant stored in a new tube. The gels were incubated with 50% ACN/5% formic acid twice and the supernatant stored in the same tube. The eluate was then dried down, and peptides were resuspended in Buffer A (0.1% formic acid in water). The digested samples were then run on a Thermo Orbitrap Fusion Tribrid MS/MS with a flow rate of 200nl/min on a nLC 1000. The rest of the LC/MS was conducted following the same protocol as described above for the fractionated and whole cell samples.
shRNA construction, lentiviral generation and knockdowns
shRNA’s targeting mouse COX7C were designed using the shERWOOD algorithm (Knott et al., 2014), with sequences listed in the Key Resources Table. The shRNA’s were cloned into the miR-E lentiviral plasmid LT3GEPIR (Fellmann et al., 2013) using EcoRI and XhoI restriction sites for doxycycline-inducible knockdowns. The shRNA’s targeting mouse NDUFA4 and COX5A were purchased from Transomic technologies (shERWOOD UltramiR Lentiviral Inducible shRNA, Cat#TLMSU2300–17992) and Dharmacon (SMARTvector Inducible Lentiviral shRNA, Cat#V3SM11256–01EG12858) respectively. 293T cells were transfected with 3rd generation packaging plasmids and the control or knockdown lentiviral plasmids, and virus was collected at 48 and 72 hours. C2C12 cells were transduced with virus diluted 1:1 with growth media containing 6ug/mL polybrene for 24 hours and selected with 5ug/ml puromycin for 48–72 hours to generated stable cell lines. For knockdowns in dividing myoblasts C2C12 control, sh1 and sh2 lines were exposed to doxycycline for 4 days. For differentiating myotubes, dividing cells were exposed to dox for 4 days and then switched to differentiation media with dox for 5 days. For differentiated myotubes, cells were differentiated into myotubes first (as described before) and then exposed to dox for 8,16 and 24 days. In all conditions, media was changed every other day. At the end of the timepoints, myoblasts or myotubes were harvested using trypsinization and subject to qPCR and western blots.
qPCR
Control and COX7C-knockdowns in myoblasts and myotubes were homogenized in Trizol (Invitrogen, 15596026) and total RNA extracted using RNeasy Mini Kit (Qiagen, 74106). 1ug of RNA was then reverse transcribed into cDNA using the QuantiTect Reverse Transcriptase Kit (Qiagen, 205311). The cDNA samples were diluted 1:10 and qPCR performed with SYBR Green PCR Master Mix (Applied Biosystems, 4309155) and primers against mouse COX7C, NDUFA4, COX5A and beta-Actin using the Bio-Rad iCycler real-time system. qPCR primers used are listed in the Key Resources Table.
Western Blotting
Control and COX7C-knockdowns in myoblasts and myotubes were lysed by boiling the samples in SDS lysis buffer (4% SDS, 100mM Tris pH 6.8) for 10 minutes. Lysates were quantified using BCA protein assay, and loaded on 10% gels, transferred on nitrocellulose membrane (Protran NC, GE Healthcare Biosciences), blocked with 5% BSA/TBST and incubated with anti-COX7C (Invitrogen, PA551284), NDUFA4 (Invitrogen, PA599439), COX5A (Proteintech, 11448–1-AP) and Tubulin (Cell Signaling, 2148S) primary antibodies at 1:1000 dilution in 5% BSA/TBST overnight at 4 degrees. Blots were washed 3x in TBST and incubated with HRP-conjugated secondary antibodies for 1 hour, and developed using SuperSignal West Pico (Thermo Scientific, PI34078).
RITE plasmid generation, tag switching and immunofluorescence
pRITE Myc to Flag (pRITE-MF) construct was generated previously (Toyama et al., 2019). Human ATP5C1 ORF sequence or Mouse COX7C ORF sequence was cloned into the pRITE-MF constructs using NheI and XhoI restriction sites. The ATP5C1 and COX7C RITE-MF constructs were then amplified using degenerate gateway primers, which were then cloned into pDONR207 constructs using Gateway cloning BP reaction (ThermoFisher). The ATP5C1 and COX7C RITE-MF pDONR207 gateway constructs were then recombined into lentiviral vector pLentiCMVBlast (http://www.ericcampeau.com) using the LR reaction (Gateway Cloning, ThermoFisher). 293T cells were transfected with 3rd generation packaging plasmids and the ATP5C1 and COX7C RITE-MF pLentiCMVBlast plasmid and virus was collected at 48 and 72 hours. SHSY-5Y and C2C12’s were transduced with the ATP5C1 RITE-MF and COX7C RITE-MF viruses respectively at a dilution of 1:1 with growth media containing 6ug/mL polybrene for 24 hours, and selected with 10ug/ml blasticidin for 48–72 hours to generated stable cell lines. Full sequences are available upon request.
ATP5C1 RITE-MF SHSY-5Y cells were plated on Polyornithine and Laminin coated ibidi u-Slide chambers and differentiated into neurons as described before. Tag exchange was initiated by adding concentrated adenovirus expressing Cre (Ad-Cre) to the media. Media was changed for cells every other day, and cells were fixed 6 days after Ad-Cre exposure. COX7C RITE-MF C2C12 dividing myoblasts were plated on ibidi u-Slide chambers (80826, Ibidi), and tag exchange was initiated by adding concentrated adenovirus expressing Cre (Ad-Cre) to the media. Media was changed for cells every other day, and cells were fixed 4 days after Ad-Cre exposure. For RITE exchange in myotubes, COX7C RITE-MF myoblasts were plated with Cre-expressing CreER2 C2C12 cell line in a 5:1 ratio on ibidi slides as described previously (Toyama et al., 2019). When the cells were fully confluent, differentiation media was added for 5 days to generate myotubes. Tag exchange was initiated by addition of 1uM of 4-hydroxytamoxifen (4OHT) (H6278, Sigma) 4 or 8 days before fixation for Day 4 and Day 8 timepoints respectively. Media was changed every other day. At the final timepoints for SHSY-5Y neurons, COX7C dividing cells and myotubes, the entire slide (all timepoints) was fixed with 4% paraformaldehyde (Electron Microscopy Sciences) for 5mins, and blocked in IF buffer (0.1% Triton X-100, 0.02% SDS, 20mg/mL BSA, in PBS) for 20 mins at room temperature (RT). The slide was then stained for anti-Myc (9B11, Cell Signaling) and anti-Flag (M2, Sigma) antibodies for 2 hours at 1:1000 dilution in IF buffer at RT. The slides were washed 3x in PBS, and secondary antibodies anti-mouse IgG2a-488 (A21131, Invitrogen) and anti-mouse IgG1–568 (A21124, Invitrogen) for SHSY-5Y cells and anti-mouse IgG1–488 (A21121, Invitrogen) and anti-mouse IgG2a-568 (A21134, Invitrogen) for C2C12 cells were added at 1:1000 dilution in IF buffer at RT in the dark. The slides were washed 3x in PBS and Hoechst dye was added to stain the nuclei. The slides were imaged using confocal microscopy on a Leica SP8.
Mitochondrial Functional Assays
Complex IV Assay was performed using the Complex IV Rodent Enzyme Activity Microplate Assay Kit (Abcam, ab109911). Briefly, control and COX7C-knockdowns in myotubes were fractionated for mitochondria as described before. The mitochondrial pellets were digitonin-permeabilized as described before, and the supernatant loaded on the 96-well microplate from the kit for 3h at RT. The wells were washed, and the substrate reduced cytochrome c was added to the wells before measuring the absorbance at 550nm at 30°C for 120 minutes. The data was quantified by plotting the absorbance over time, fitting a line through the curve and measuring the slope, which was normalized to the controls. Mitochondrial respiration was measured using the Seahorse XFe96 Assay and Analyzer (Agilent). Briefly, control and COX7-knockdowns in differentiating myotubes grown in 96-well Seahorse XF Cell Culture Microplate and the Seahorse XF Cell Mito Stress Test for OCR, (Agilent, 103015–100) and the Seahorse XF Glycolysis Stress Test Kit for ECAR, (Agilent, 103020–100) were conducted using the manufacturer’s protocol. OCR was conducted using the following injections: 1) Oligomycin 2μM, 2) FCCP 2μM 3) Rotenone 2μM + Antimycin-A 2μM, while ECAR was conducted with 1) Glucose 10μM 2) Oligomycin 2μM 3) 2-DG 50μM. ATP Levels were measured using a luminescence ATP Detection Assay Kit (Cayman Chemicals, NC1357058). Briefly, control and COX7C-knockdowns in differentiating myotubes were homogenized in ATP sample buffer and loaded on a 96-well microplate along with the standards. The reaction mix containing D-luciferin and Luciferase in the ATP assay buffer was added to samples and standards, and the plate was incubated for 15mins at RT in the dark. The luminescence was then measured using a plate reader, the ATP concentration in samples determined using the standard curve, and the values were normalized to protein concentration for each sample. Lactate Assays was performed using the L-Lactate Assay Kit (Abcam, ab65330). Briefly, cell culture medium from control and COX7C-knockdowns was diluted 1:500 with Lactate Assay buffer and loaded on a 96-well microplate along with L-Lactate standards. The reaction mix containing the probe and enzyme was then added to the plate and incubated for 30mins at RT in the dark. The lactate levels were then measured using a plate reader at absorbance of 570nm, the lactate concentration in samples determined using the standard curve, and the values were normalized to protein concentration for each sample.
QUANTIFICATION AND STATISTICAL ANALYSIS
MIMS Image Analysis, Alignment and Quantification
With MIMS-EM, the correlation and overlay of 15N/14N ratio images atop SEM micrographs has the advantage of displaying the chemical information of the sample while retaining the resolution and ultrastructural detail afforded by the SEM. An SEM image on UCSD’s Zeiss platform can be acquired for a raster size of up to 32k × 32k pixels. In comparison, the raster size on Caltech’s Cameca NanoSIMS can go up to a maximum of only 1024 × 1024 pixels. Beyond differences in scale, the signal collection mechanisms for the two instruments are completely different. When imaging sections in the SEM (to be later interrogated by nano-SIMS), signal collection is non-destructive, while in the NanoSIMS the signals are collected destructively by the sputtering atoms from the sample (Benninghoven, 1987). The sputter rates are vastly different for the different atomic species, resulting in a non-linear distortion of the NanoSIMS image with respect to the SEM ultrastructural reference image, complicating the fusion of these datasets. Manual alignment of the two images can insert operator bias into the correlation process. Automated alignment using simple affine transformations is also insufficient given the non-linear nature of the distortions. For proper alignment, the 15N/14N ratio image needs to be automatically adjusted for warping with respect to the SEM image. There are freely available warp adjustment softwares, such as the ‘UnwarpJ’ plugin of ImageJ (Schneider et al., 2012; Sorzano et al., 2005) that adjusts for distortions in one image with respect to the undistorted image. However, the plugin expects that the images be somewhat similar i.e. they are from the same imaging modalities.
To facilitate the seamless alignment of the MIMS image to the SEM image, we developed a plugin called ‘MesoFusion’, which stands for Mesoscale Image Fusion. The plugin has 4 subroutines: 1. Rescale and Resize 2. Create Layout 3. Categorize Ratio map 4. Colorize Ratio map, the utility of each of the sub-routine will be shown with the aid of the Figure S1A. The Figure S1Aa, shows the SEM image (6500 × 7000 pixels) of cerebellar granule neurons from a 6-month chase mouse, acquired at a pixel size of 400nm. Figure S1Ab, shows the N15/N14 ratio map (512 × 512 pixels) of the same region, acquired at a pixel size of 400nm. It is clearly evident that at a macro level, these two images differ markedly in detail, scale, orientation and field of view (FOV). The Rescale and Resize subroutine, up-scales the MIMS image to match the pixel size of the SEM image, and roughly aligns both the images for any linear shifts, orientation and image flip. It also matches the two images such that they have roughly the same FOV, see Figures S1Ac and S1Ad. The subroutine Create Layout applies appropriate threshold and skeletonizes these two images so as to convert them to somewhat similar looking images, as shown in Figures S1Ae and S1Af. Now these two images can be fed into already available warping programs like UnwarpJ (Sorzano et al., 2005), and compensated for any shrinkage and distortion. Figures S1Ag and S1Ah, show the macro and micro distortion map, when the skeletonized MIMS image (Figure S1Af) was adjusted to the skeletonized SEM image (Figure S1Ae) using UnwarpJ. The transformations can be saved and applied to the originally aligned MIMS image prior to skeletonization (Figure S1Ad). The subroutine Categorize Ratio map divides the completely aligned 15N/14N ratio map into different categories based on the signal intensity i.e. concentration of the N15 signal with respect to the natural ratio. The Figures S1Ai, j, k and l, show the 15N/14N ratio map categorized into 2–3, 3–7, 7–50 and > 50 times the natural ratio. Finally, these four images are colorized and overlaid over the conventional SEM image, by the Colorize Ratio map subroutine as shown in Figure S1Am. The algorithm used in Colorize Ratio map has been described in detail elsewhere (Adams et al., 2016), but new features have been added to accommodate color schemes for people with color vision deficiencies (Keene, 2015). The MesoFusion plugin will be made available to the scientific community at no cost and can be requested from the authors. For quantification of the 15N/14N signal, the raw ratio was calculated by manually drawing around individual mitochondria, and similarly sized circles in the lumen and cytosol in the final aligned 15N/14N image. The raw values were then divided by 37×10−4 to determine the 15N/14N fold over natural ratio. This plugin along with an instruction manual is available online (Mendeley Data DOI:10.17632/b3hww8ng7w.1).
Mass spectrometry data analysis and peptide filtration
Post-mass spectrometry, the identification, quantification and preliminary analysis of the peptides and proteins was performed on the Integrated Proteomics Pipeline Version 6.0.5 IP2 (http://www.integratedproteomics.com/). The tandem mass spectra were extracted using RawConverter from raw files (He et al., 2015) and searched against a nonredundant mouse UniProt database using ProLUCID (Xu et al., 2015). The search space included all fully-tryptic and half-tryptic peptide candidates, and cysteine-specific static modification of 57 was allowed for carbamidomethylation. SILAC labeling at K (6.0201) and R (10.0083) were searched as heavy metabolic labeling. A high-resolution precursor/peptide mass tolerance of 50ppm was used with the precursor mass range from 600 to 6000ppm. DTASelect software (Tabb et al., 2002) was used to filter the identified proteins which were searched against a target-decoy database to keep the false discovery rate to 1% at the protein level (Peng et al., 2003). A minimum of 1 peptide per protein and 1 tryptic end per peptide was required. A false positive rate of 0.01 and precursor delta mass cutoff of 10ppm was used to filter the data.
Quantification of SILAC-labeled peptides was performed using the software Census2 (Park et al., 2014). The peptides were further filtered based on rigorous quality control criteria of a minimum profile score of 0.8, and peptides with area ratios from 0.111–9 were required to have a minimum regression score of 0.8. Peptides with extreme area ratios <0.111 or >9 were only retained if the regression score was >0. The percent heavy label (percent new protein) was then calculated by transforming the area ratio values using the following equation:
The mean, median, standard deviation and interquartile range was calculated for each peptide and outlier peptides were removed if they fell outside 20 times the interquartile range. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD028963 and 10.6019/PXD028963.
Half-life calculation
The individual peptide data for each protein was then pooled together and we restricted our analysis to proteins that had at least 2 peptides detected at 3 different timepoints during the timecourse. To assess protein turnover, we used the following exponential plateau decay equation, (t) = yf + (y0 − yf) e−αt, where: y = % light at time t, α = rate of turnover, yf = minimum value of % light, y0 = starting value of % light. To estimate the parameters of this nonlinear model, we used nonlinear least-squares (NLS) regression via the nls function in R with the nl2sol algorithm from the Port library (http://www.netlib.org/port/). For very long-lived proteins, like histones, that barely show any turnover during the SILAC timecourse, the available data does not encompass enough timepoints to allow for an accurate fit of an exponential decay curve within this timeframe. Hence, in these cases, an alternate linear model was used to estimate the turnover rate α, as follows: log(y) = β + αt, where β = intercept and other parameters as described above. To assess model quality, the value of R-square was calculated for all models using the rsquare function from the modelr package in R (https://CRAN.R-project.org/package=modelr) and estimates were filtered for R-square values above 0.9. The estimated rate of turnover (α), was then used to calculate protein half-life with the following formula, Thalf = ln(2)/−α. For neurons, the linear and port model showed very similar results, hence the port model was chosen to maintain consistency for comparisons of half-lives of mitochondrial proteins with myotubes.
For neurons, the data from nuclear, mitochondrial and cytosolic fractions was pooled together in order to improve the coverage of proteins. The pooled data was then separated based on human databases of different organelles (Nucleus, Mitochondria, ER, Lysosome, Cytosol) and submitochondrial compartments (Outer Mitochondrial Membrane, Inner Mitochondrial Membrane, Intermitochondrial Membrane Space, Nucleoid, Matrix) downloaded from Uniprot. For myotubes, the whole cell pellet, nuclear and mitochondrial fractions the replicates were analyzed separately, and the whole cell data was separated into different organelles using mouse Uniprot databases. Nuclear and mitochondrial proteins were taken from their respective fractions for most of the analysis as they had sufficient coverage (They showed similar data to nuclear and mitochondrial proteins from whole cell data). ETC proteins were collated by hand for both human and mouse. For both neurons and myotubes, 3 independent biological replicates were performed, and the average %old protein and median half-life values across the 3 replicates shown for the graphs. However, for gel bands, in order to increase coverage of the ETC proteins, the 3 individual replicate experiments were pooled at the peptide level after calculating the % heavy label. Outlier peptide removal was then performed using the interquartile range and the remaining analysis of half-life values was done similarly. We used half-lives that were ~2-fold higher than the respective average whole cell fractions as the cut-off value for a mitoLLP (>8.5 days in neurons and > 6 days in myotubes). These mass spectrometry data sets and the Uniprot databases used are available in Table S1 and Table S2. The computer code developed in this paper for analyzing SILAC mass spectrometry datasets generated after Census2 and calculating half-lives is available online (Mendeley Data DOI:10.17632/b3hww8ng7w.1).
NSAF Analysis
NSAF values for all the proteins in the Day 0 timepoint of 5 samples of higher-order SCs (SCI,III,IV), intermediate SC (SCIII,IV), dimers (III2,IV2), individual CI & CII pooled (CI,II) and individual CIV (CIV) were calculated using the ID Compare function on IP2. NSAF values of proteins in CI, CII, CIII and CIV were then subset and summed up to calculate the total NSAF value for each complex. The NSAF values of each complex was then divided by the total NSAF for all the four complexes CI-CIV and multiplied by 100 to generate %NSAF values for each complex in each gel band sample.
Half-life visualization of crystal structures
In order to color code the crystal structures of proteins in complexes with their respective half-lives, the structure of the complex of interest was downloaded from RCSB Protein Data Bank (https://www.rcsb.org/). The accession code for the complexes were as follows - mouse Complex I: 6G2J, human Complex III: 5XTE, human Complex IV: 5Z62, porcine respirasome: 5XTH. (Human CIII and CIV, and porcine respirasome structures were used as mouse structures are not available). Using a script, the downloaded structures were color-coded with half-lives ranging from 2 days to 8 days, from dark red to white to dark blue for regular turnover rates. For the differential turnover between supercomplexes and individual complexes, the subtracted half-lives ranged from −1 to +1 days, from dark red to white to dark blue. The subunits without any coverage in the mass spectrometry and half-life data were labelled grey. The computer code developed in this paper for downloading the PDB crystal structure file into Pymol and color coding the crystal structures with half-lives is available online (Mendeley Data DOI:10.17632/b3hww8ng7w.1).
RITE & Myotube Image Analysis
The RITE images for SHSY-5Y and COX7C were quantified using ImageJ (with FIJI). Each image was set to maximum projection and average intensity, and an outline was drawn manually around the cells, followed by measurement of the area, mean intensity and integrated density of the green and red channels using the measure function of ImageJ. Outlines were also drawn to take a background signal reading for each image. For quantification the background signal was subtracted from the cell signal to generate the total fluorescent intensity for the red and green channels. For ratio measurements the green to red intensities were divided, whereas for %old protein the individual channel intensity was divided by the total intensity (green+red) for each cell. Myotube thickness was measured by drawing lines across each myotube every 1 inch and then measuring the pixel length. The values for all myotubes in images were averaged and compared between control and knockdowns.
Statistical Methods and Representative Figures
Error bars show mean ± SD from n=3 replicates for all graphs except when indicated. For MIMS-EM images, data are from 1 mouse and error bars show mean ± SD of N15/N14 ratio for multiple mitochondria and other regions in the images shown. p-Values were obtained using an unpaired two-tailed Student’s t test. All correlation coefficients represent Spearman’s coefficient. Graphpad prism was used to conduct statistical analysis and make graphs. Images and western blots are representatives of at least 3 independent experiments unless otherwise stated.
Supplementary Material
Table S1. Mass spectrometry data from subcellular fractions in neurons and myotubes, Related to Figure 2: Tables contain filtered protein level data for each replicate of neurons all fractions combined (Neu_AllFraction_Replicates), myotube whole cell pellet (Myo_WholeCell_Replicates), nuclear (Myo_NucFraction_Replicates) and mitochondrial fractions (Myo_MitoFraction_Replicates) for Days 0–8. The tables contain the UniprotID, protein name (Description), mean (Day 0–8_mean), standard deviation (Day 0–8_SD), number of peptides (Day 0–8_N), α (alpha), R2 (Rsq) and half-lives (Thalf) values. The half-life values for neurons are from the port model. For the myotubes, the half-lives of the whole cell pellet, nuclear and mitochondrial fractions are from the NLS model and the nuclear LLPs in the whole cell and nuclear fractions are from the linear model (see Methods). Each table contains the data from 3 replicates in different columns. The list of proteins in mitochondria, nucleus, ER, cytosol, lysosome (all from Uniprot databases) and ETC proteins (curated manually) for humans (Human uniprot protein lists) and mouse (Mouse uniprot protein lists) are also provided.
Table S2. Mass spectrometry data for ETC complexes and supercomplexes from gel bands in myotubes, Related to Figures 3 and 4. Tables contain filtered protein level data for ETC complex proteins for Complexes I-IV for Gel Band 1, 2, 3, 4 and 5, for Days 0–8. The tables contain the Complex, UniprotID, Entry name, protein name (Description), Gene name, mean (Day 0–8_mean), standard deviation (Day 0–8_SD), number of peptides (Day 0–8_N), α (alpha), R2 (Rsq) and half-lives (Thalf) values. The half-life values are from the NLS model (see Methods). Data are pooled from 3 independent replicates.
Highlights:
Mitochondria in the mouse brain are long-lived and show considerable age mosaicism
Mitochondria contain many long-lived proteins (LLPs) as part of protein complexes
LLPs in the electron transport chain show increased stability within supercomplexes
Longevity of LLP COX7C preserves oxphos function despite transcriptional decline
Acknowledgements
This work is supported by NIH Transformative Research Award grant R01 NS096786, Keck Foundation, NOMIS Foundation, NGS Core Facility, Razavi Newman Integrative Genomics, Bioinformatics Core Facility of the Salk Institute (NIH-NCI CCSG: P30 014195), Chapman Foundation, Helmsley Charitable Trust, Flow Cytometry Core Facility of the Salk Institute (NIH-NCI CCSG: P30 014195), Mass Spectrometry Core of the Salk Institute (NIH-NCI CCSG: P30 014195), Helmsley Center for Genomic Medicine and Salk Women and Science. This work is also supported by NIH R24GM137200, U24NS120055 and NSF - NSF2014862-UTA20-000890. The authors thank J. Moresco, J. Diedrich and A. Pinto for technical support with Mass Spectometry (Salk Institute), J. Letts (University of California, Davis) and D. Butler (Salk Institute) for codes for Pymol half-life structures, S. Schafer and F. Gage (Salk Institute) for hES cells and members of the Shadel Lab (Salk Institute) for help with seahorse assays. Graphical abstract created with Biorender.com.
Footnotes
Declaration of Interests
Authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Acin-Perez R, Bayona-Bafaluy MP, Fernandez-Silva P, Moreno-Loshuertos R, Perez-Martos A, Bruno C, Moraes CT, and Enriquez JA (2004). Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol Cell 13, 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams SR, Mackey MR, Ramachandra R, Palida Lemieux SF, Steinbach P, Bushong EA, Butko MT, Giepmans BNG, Ellisman MH, and Tsien RY (2016). Multicolor Electron Microscopy for Simultaneous Visualization of Multiple Molecular Species. Cell Chem Biol 23, 1417–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrojo EDR, Lev-Ram V, Tyagi S, Ramachandra R, Deerinck T, Bushong E, Phan S, Orphan V, Lechene C, Ellisman MH, et al. (2019). Age Mosaicism across Multiple Scales in Adult Tissues. Cell Metab 30, 343–351 e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benninghoven A, Rudenauer FG & Werner HW (1987). Secondary ion mass spectrometry : basic concepts, instrumental aspects, applications, and trends. Wiley. [Google Scholar]
- Bezawork-Geleta A, Brodie EJ, Dougan DA, and Truscott KN (2015). LON is the master protease that protects against protein aggregation in human mitochondria through direct degradation of misfolded proteins. Sci Rep 5, 17397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter A, Schulte R, Tsai H, Capitanio J, and Hetzer M (2019). Selective clearance of the inner nuclear membrane protein emerin by vesicular transport during ER stress. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung YT, Lau WK, Yu MS, Lai CS, Yeung SC, So KF, and Chang RC (2009). Effects of all-trans-retinoic acid on human SH-SY5Y neuroblastoma as in vitro model in neurotoxicity research. Neurotoxicology 30, 127–135. [DOI] [PubMed] [Google Scholar]
- Clayton DA, and Shadel GS (2014). Isolation of mitochondria from cells and tissues. Cold Spring Harb Protoc 2014, pdb top074542. [DOI] [PubMed] [Google Scholar]
- Cogliati S, Calvo E, Loureiro M, Guaras AM, Nieto-Arellano R, Garcia-Poyatos C, Ezkurdia I, Mercader N, Vazquez J, and Enriquez JA (2016). Mechanism of super-assembly of respiratory complexes III and IV. Nature 539, 579–582. [DOI] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, and Hetzer MW (2009). Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136, 284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, and Kenyon C (2010). Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol 8, e1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F, Fukui H, Garcia S, and Moraes CT (2006). Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol Cell Biol 26, 4872–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzeja PP, Bortolon R, Perez-Terzic C, Holmuhamedov EL, and Terzic A (2002). Energetic communication between mitochondria and nucleus directed by catalyzed phosphotransfer. Proc Natl Acad Sci U S A 99, 10156–10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor JG, and Hirst J (2018). Mitochondrial Supercomplexes Do Not Enhance Catalysis by Quinone Channeling. Cell Metab 28, 525–531 e524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IA, Kwon JS, Guan Y, et al. (2013). An optimized microRNA backbone for effective single-copy RNAi. Cell Rep 5, 1704–1713. [DOI] [PubMed] [Google Scholar]
- Fornasiero EF, Mandad S, Wildhagen H, Alevra M, Rammner B, Keihani S, Opazo F, Urban I, Ischebeck T, Sakib MS, et al. (2018). Precisely measured protein lifetimes in the mouse brain reveal differences across tissues and subcellular fractions. Nat Commun 9, 4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio C, Jha P, Kulkarni SS, Lagarrigue S, Broskey NT, Boutant M, Wang X, Conde Alonso S, Ofori E, Auwerx J, et al. (2017). Enhanced Respiratory Chain Supercomplex Formation in Response to Exercise in Human Skeletal Muscle. Cell Metab 25, 301–311. [DOI] [PubMed] [Google Scholar]
- Gu J, Wu M, Guo R, Yan K, Lei J, Gao N, and Yang M (2016). The architecture of the mammalian respirasome. Nature 537, 639–643. [DOI] [PubMed] [Google Scholar]
- He L, Diedrich J, Chu YY, and Yates JR 3rd (2015). Extracting Accurate Precursor Information for Tandem Mass Spectra by RawConverter. Anal Chem 87, 11361–11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp MS, Kasturi P, and Hartl FU (2019). The proteostasis network and its decline in ageing. Nat Rev Mol Cell Biol 20, 421–435. [DOI] [PubMed] [Google Scholar]
- Hofmann AD, Beyer M, Krause-Buchholz U, Wobus M, Bornhauser M, and Rodel G (2012). OXPHOS supercomplexes as a hallmark of the mitochondrial phenotype of adipogenic differentiated human MSCs. PLoS One 7, e35160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha P, Wang X, and Auwerx J (2016). Analysis of Mitochondrial Respiratory Chain Supercomplexes Using Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE). Curr Protoc Mouse Biol 6, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunadharma PP, Basisty N, Chiao YA, Dai DF, Drake R, Levy N, Koh WJ, Emond MJ, Kruse S, Marcinek D, et al. (2015). Respiratory chain protein turnover rates in mice are highly heterogeneous but strikingly conserved across tissues, ages, and treatments. FASEB J 29, 3582–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katajisto P, Dohla J, Chaffer CL, Pentinmikko N, Marjanovic N, Iqbal S, Zoncu R, Chen W, Weinberg RA, and Sabatini DM (2015). Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science 348, 340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene DR (2015). A review of color blindness for microscopists: guidelines and tools for accommodating and coping with color vision deficiency. Microsc Microanal 21, 279–289. [DOI] [PubMed] [Google Scholar]
- Knott SRV, Maceli A, Erard N, Chang K, Marran K, Zhou X, Gordon A, Demerdash OE, Wagenblast E, Kim S, et al. (2014). A computational algorithm to predict shRNA potency. Mol Cell 56, 796–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov AV, and Margreiter R (2009). Heterogeneity of mitochondria and mitochondrial function within cells as another level of mitochondrial complexity. Int J Mol Sci 10, 1911–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts JA, Fiedorczuk K, and Sazanov LA (2016). The architecture of respiratory supercomplexes. Nature 537, 644–648. [DOI] [PubMed] [Google Scholar]
- Letts JA, and Sazanov LA (2017). Clarifying the supercomplex: the higher-order organization of the mitochondrial electron transport chain. Nat Struct Mol Biol 24, 800–808. [DOI] [PubMed] [Google Scholar]
- Lobo-Jarne T, Nyvltova E, Perez-Perez R, Timon-Gomez A, Molinie T, Choi A, Mourier A, Fontanesi F, Ugalde C, and Barrientos A (2018). Human COX7A2L Regulates Complex III Biogenesis and Promotes Supercomplex Organization Remodeling without Affecting Mitochondrial Bioenergetics. Cell Rep 25, 1786–1799 e1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, and Kroemer G (2013). The hallmarks of aging. Cell 153, 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Salamanca HH, Duarte FM, Danko CG, and Lis JT (2016). Mammalian Heat Shock Response and Mechanisms Underlying Its Genome-wide Transcriptional Regulation. Mol Cell 62, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson T, Franken H, Kosinski J, Kurzawa N, Zinn N, Sweetman G, Poeckel D, Ratnu VS, Schramm M, Becher I, et al. (2018). Systematic analysis of protein turnover in primary cells. Nat Commun 9, 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle EA, Shen K, and Dillin A (2019). Mitochondrial proteostasis in the context of cellular and organismal health and aging. J Biol Chem 294, 5396–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain VM (1994). Translational control during amino acid starvation. Biochimie 76, 718–728. [DOI] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, and Tavernarakis N (2018). Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol 20, 1013–1022. [DOI] [PubMed] [Google Scholar]
- Park SK, Aslanian A, McClatchy DB, Han X, Shah H, Singh M, Rauniyar N, Moresco JJ, Pinto AF, Diedrich JK, et al. (2014). Census 2: isobaric labeling data analysis. Bioinformatics 30, 2208–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, and Gygi SP (2003). Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2, 43–50. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picca A, Guerra F, Calvani R, Coelho-Junior HJ, Bossola M, Landi F, Bernabei R, Bucci C, and Marzetti E (2020). Generation and Release of Mitochondrial-Derived Vesicles in Health, Aging and Disease. J Clin Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JC, Guan S, Burlingame A, Prusiner SB, and Ghaemmaghami S (2010). Analysis of proteome dynamics in the mouse brain. Proc Natl Acad Sci U S A 107, 14508–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas JN, Toyama BH, Xu T, Yates JR 3rd, and Hetzer MW (2012). Extremely long-lived nuclear pore proteins in the rat brain. Science 335, 942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer ST, Paquola ACM, Stern S, Gosselin D, Ku M, Pena M, Kuret TJM, Liyanage M, Mansour AA, Jaeger BN, et al. (2019). Pathological priming causes developmental gene network heterochronicity in autistic subject-derived neurons. Nat Neurosci 22, 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka T, and Haynes CM (2018). The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat Rev Mol Cell Biol 19, 109–120. [DOI] [PubMed] [Google Scholar]
- Sin J, Andres AM, Taylor DJ, Weston T, Hiraumi Y, Stotland A, Kim BJ, Huang C, Doran KS, and Gottlieb RA (2016). Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy 12, 369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Herrmann JM, and Becker T (2021). Quality control of the mitochondrial proteome. Nat Rev Mol Cell Biol 22, 54–70. [DOI] [PubMed] [Google Scholar]
- Sorzano CO, Thevenaz P, and Unser M (2005). Elastic registration of biological images using vector-spline regularization. IEEE Trans Biomed Eng 52, 652–663. [DOI] [PubMed] [Google Scholar]
- Sosinsky GE, Deerinck TJ, Greco R, Buitenhuys CH, Bartol TM, and Ellisman MH (2005). Development of a model for microphysiological simulations: small nodes of ranvier from peripheral nerves of mice reconstructed by electron tomography. Neuroinformatics 3, 133–162. [DOI] [PubMed] [Google Scholar]
- Steinhauser ML, Bailey AP, Senyo SE, Guillermier C, Perlstein TS, Gould AP, Lee RT, and Lechene CP (2012). Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature 481, 516–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Ekman C, Oskolkov N, Lahti L, Strom K, Brazma A, Groop L, Rung J, and Hansson O (2015). A novel atlas of gene expression in human skeletal muscle reveals molecular changes associated with aging. Skelet Muscle 5, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, and Yates JR 3rd (2002). DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Arrojo EDR, Lev-Ram V, Ramachandra R, Deerinck TJ, Lechene C, Ellisman MH, and Hetzer MW (2019). Visualization of long-lived proteins reveals age mosaicism within nuclei of postmitotic cells. J Cell Biol 218, 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, and Hetzer MW (2013). Protein homeostasis: live long, won’t prosper. Nat Rev Mol Cell Biol 14, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR 3rd, and Hetzer MW (2013). Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 154, 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Calvo C, Suhm T, Buttner S, and Ott M (2020). The basic machineries for mitochondrial protein quality control. Mitochondrion 50, 121–131. [DOI] [PubMed] [Google Scholar]
- Wirth C, Brandt U, Hunte C, and Zickermann V (2016). Structure and function of mitochondrial complex I. Biochim Biophys Acta 1857, 902–914. [DOI] [PubMed] [Google Scholar]
- Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D, Lu B, Liao L, Hewel J, Han X, et al. (2015). ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteomics 129, 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Price F, and Rudnicki MA (2013). Satellite cells and the muscle stem cell niche. Physiol Rev 93, 23–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DS, Piazza V, Perrin BJ, Rzadzinska AK, Poczatek JC, Wang M, Prosser HM, Ervasti JM, Corey DP, and Lechene CP (2012). Multi-isotope imaging mass spectrometry reveals slow protein turnover in hair-cell stereocilia. Nature 481, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Mass spectrometry data from subcellular fractions in neurons and myotubes, Related to Figure 2: Tables contain filtered protein level data for each replicate of neurons all fractions combined (Neu_AllFraction_Replicates), myotube whole cell pellet (Myo_WholeCell_Replicates), nuclear (Myo_NucFraction_Replicates) and mitochondrial fractions (Myo_MitoFraction_Replicates) for Days 0–8. The tables contain the UniprotID, protein name (Description), mean (Day 0–8_mean), standard deviation (Day 0–8_SD), number of peptides (Day 0–8_N), α (alpha), R2 (Rsq) and half-lives (Thalf) values. The half-life values for neurons are from the port model. For the myotubes, the half-lives of the whole cell pellet, nuclear and mitochondrial fractions are from the NLS model and the nuclear LLPs in the whole cell and nuclear fractions are from the linear model (see Methods). Each table contains the data from 3 replicates in different columns. The list of proteins in mitochondria, nucleus, ER, cytosol, lysosome (all from Uniprot databases) and ETC proteins (curated manually) for humans (Human uniprot protein lists) and mouse (Mouse uniprot protein lists) are also provided.
Table S2. Mass spectrometry data for ETC complexes and supercomplexes from gel bands in myotubes, Related to Figures 3 and 4. Tables contain filtered protein level data for ETC complex proteins for Complexes I-IV for Gel Band 1, 2, 3, 4 and 5, for Days 0–8. The tables contain the Complex, UniprotID, Entry name, protein name (Description), Gene name, mean (Day 0–8_mean), standard deviation (Day 0–8_SD), number of peptides (Day 0–8_N), α (alpha), R2 (Rsq) and half-lives (Thalf) values. The half-life values are from the NLS model (see Methods). Data are pooled from 3 independent replicates.
Data Availability Statement
All mass spectrometry proteomic datasets have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository and are publicly available as of the date of publication. DOIs are listed in the key resources table.
All original code has been deposited at Mendeley and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| OxPhos Antibody Cocktail Rodent | Invitrogen | Cat#458099, RRID:AB_2533835 |
| NDUFA9 Monoclonal Antibody | Invitrogen | Cat#459100, RRID:AB_2532223 |
| UQCRC1 Monoclonal Antibody | Invitrogen | Cat#459140, RRID:AB_2532227 |
| COX IV (3E11) Rabbit mAb | Cell Signaling | Cat#4850S, RRID:AB_2085424 |
| COX7C Polyclonal Antibody | Invitrogen | Cat#PA551284 RRID:AB_2636731 |
| NDUFA4 Polyclonal Antibody | Invitrogen | Cat#PA599439, RRID:AB_2818372 |
| COX5A Polyclonal Antibody | Proteintech | Cat# 11448–1-AP, RRID:AB_2085429 |
| α/β-Tubulin Antibody | Cell Signaling | Cat#2148S, RRID:AB_2288042 |
| Myc-Tag (9B11) Mouse mAb | Cell Signaling | Cat# 2276S, RRID:AB_331783 |
| Monoclonal ANTI-FLAG® M2 antibody | Sigma | Cat#F3165, RRID:AB_259529 |
| Goat anti-Mouse IgG2a Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat# A21131, RRID:AB_141618 |
| Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Invitrogen | Cat# A21124, RRID:AB_141611 |
| Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat# A21121, RRID:AB_2535764 |
| Goat anti-Mouse IgG2a Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Invitrogen | Cat# A21134, RRID:AB_1500825 |
|
Chemicals, peptides, and recombinant proteins | ||
| 15N-spirulina algae | Cambridge isotope Laboratories | Cat#MF-SPIRULINA-N-S |
| 2.5% Glutaraldehyde | Ted Pella Inc., Redding, CA | Cat#18426 |
| 2% Formaldehyde | Ted Pella Inc., Redding, CA | Cat#18200 |
| Sodium Cacodylate | Ted Pella Inc., Redding, CA | Cat#18851 |
| Osmium Tetroxide | Electron Microscopy Sciences | Cat#19190 |
| Potassium Ferrocyanide | Electron Microscopy Sciences | Cat#3114 |
| Thiocarbohydrazide | Electron Microscopy Sciences | Cat#21900 |
| Durcupan ACM Resin | Sigma-Aldrich | Cat#44611 (Comp A), Cat# 44612 (Comp B), Cat#44613 (Comp C), Cat#44614 (Comp D). |
| Matrigel (Cultrex) | R&D Systems | Cat#3433–005-01 |
| DMEM:F12 for SILAC | Fisher Scientific | Cat#88370 |
| Dialyzed FBS | Gibco | Cat#26400044 |
| DMEM for SILAC | Fisher Scientific | Cat#A33822 |
| L-Lysine-13C6 hydrochloride | Sigma | Cat#643459 |
| L-Arginine-13C6,15N4 hydrochloride | Sigma | Cat#608033 |
| TCEP-HCL | Sigma-Aldrich | Cat#C4706 |
| Chloroacetamide | Sigma-Aldrich | Cat#C0267 |
| Trypsin | Promega | Cat# V5111 |
| NativePAGE™ Sample Prep Kit | Invitrogen | Cat#BN2008 |
| Native PAGE 3–12% gradient gel | Invitrogen | Cat# BN1001BOX |
| Dark Blue Cathode Buffer | Invitrogen | Cat# BN2002 |
| Running Anode Buffer | Invitrogen | Cat#BN2001 |
| Trizol | Invitrogen | Cat#15596026 |
| SuperSignal West Pico | Fisher Scientific | Cat#PI34078 |
| SYBR Green PCR Master Mix | Applied Biosystems | Cat#4309155 |
| Ibidi u-Slide chambers | Ibidi | Cat#80826 |
| 4-hydroxytamoxifen (4OHT) | Sigma-Aldrich | Cat#H6278 |
| Critical commercial assays | ||
| RNeasy Mini Kit | Qiagen | Cat#74106 |
| QuantiTect Reverse Transcriptase Kit | Qiagen | Cat#205311 |
| Complex IV Rodent Enzyme Activity Microplate Assay Kit | Abcam | Cat# ab109911 |
| Seahorse XF Cell Mito Stress Test | Agilent | Cat#103015–100 |
| Seahorse XF Glycolysis Stress Test Kit | Agilent | Cat#103020–100 |
| ATP Chemiluminescence Detection Assay Kit | Cayman Chemicals | Cat# NC1357058 |
| L-Lactate Assay Kit | Abcam | Cat# ab65330 |
| Deposited Data | ||
| Mass spectrometry Proteomic Datasets | ProteomeXchange Consortium via PRIDE (Perez-Riverol et al., 2019) | DOI: 10.6019/PXD028963 |
| Experimental models: Cell lines | ||
| SH-SY5Y | ATCC | Cat#CRL-2266 |
| C2C12 Myoblasts | ATCC | Cat#CRL-1772 |
| Human ES Cells (H9) | WiCell | Cat#WA09 |
| Experimental models: Organisms/strains | ||
| Mouse:FVB (Male) | The Jackson Laboratory, Bar Harbor,ME | Stock No: 001800 |
| Oligonucleotides | ||
| COX7C sh1 targeting sequence - TGCTGTTGACAGTGAGCGACCGCACCTTTCTTTATAGTAATAGTGAAGCCACAGATGTATTACTATAAAGAAAGGTGCGGCTGCCTACTGCCTCGGA | This paper | Eton Biosciences; Sequences generated using shERWOOD algorithm (Knott et al., 2014) |
| COX7C sh2 targeting sequence - TGCTGTTGACAGTGAGCGCCACCAGCTACTTAAAAAATAATAGTGAAGCCACAGATGTATTATTTTTTAAGTAGCTGGTGTTGCCTACTGCCTCGGA | This paper | Eton Biosciences; Sequences generated using shERWOOD algorithm (Knott et al., 2014) |
| COX7C qPCR Primer: Forward - AGCATGTTGGGCCAGAGT | This paper | Eton Biosciences |
| COX7C qPCR Primer: Reverse-ACTGAAAACGGCAAATTCTT | This paper | Eton Biosciences |
| NDUFA4 qPCR Primer: Forward - CGCTTGGCACTGTTTAATCCA | This paper | Eton Biosciences |
| NDUFA4 qPCR Primer: Reverse - TCCATGGCTCTGGGTTGTTC | This paper | Eton Biosciences |
| COX5A qPCR Primer: Forward - CTGCCGCTGTCTGTTCCATTCG | This paper | Eton Biosciences |
| COX5A qPCR Primer: Reverse - TGTCACCCAGCGAGCATCAAACT | This paper | Eton Biosciences |
| Beta-actin qPCR Primer: Forward - CTGTCCCTGTATGCCTCTG | This paper | Eton Biosciences |
| Beta-actin qPCR Primer: Reverse - ATGTCACGCACGATTTCC | This paper | Eton Biosciences |
| Recombinant DNA | ||
| UNG Construct | Addgene | Cat#127288 |
| Plasmid: pRITE Myc to Flag (pRITE-MF) | (Toyama et al., 2019) | N/A |
| Plasmid: ATP5C1 RITE-MF pLentiCMVBlast | This paper | N/A |
| Plasmid: COX7C RITE-MF pLentiCMVBlast | This paper | N/A |
| shERWOOD UltramiR Lentiviral Inducible shRNA for NDUFA4 | Transomic technologies | Cat#TLMSU2300–17992 |
| Set of 3 SMARTvector Inducible Mouse COX5A (mCMV-TurboGFP shRNA) SMARTvector Inducible Lentiviral Control |
Dharmacon | Cat#V3SM11256–01EG12858 Cat# VSC11651 |
| Software and algorithms | ||
| OpenMIMS | (Steinhauser et al, 2012) | https://github.com/BWHCNI/OpenMIMS/ |
| Unwarp Plugin for ImageJ | (Schneider et al., 2012; Sorzano et al., 2005) | N/A |
| MesoFusion Plugin and Code for ImageJ | This paper; Mendeley Data | https://data.mendeley.com/datasets/b3hww8ng7w/1; DOI:10.17632/b3hww8ng7w.1 |
| Integrated Proteomics Pipeline Version 6.0.5 IP2 | N/A | http://www.integratedproteomics.com/ |
| Census2 | (Park et al., 2014) | N/A |
| nl2sol algorithm | R package | http://www.netlib.org/port/ |
| modelr algorithm | R package | https://CRAN.R-project.org/package=modelr |
| Proteomics Data Analysis Code (Mass spectrometry and half-life calculation) | This paper; Mendeley Data | https://data.mendeley.com/datasets/b3hww8ng7w/1; DOI:10.17632/b3hww8ng7w.1 |
| RCSB Protein Data Bank | https://www.rcsb.org/ | mouse Complex I: 6G2J, human Complex III: 5XTE, human Complex IV: 5Z62, porcine respirasome: 5XTH |
| PyMOL | PyMOL by Schrödinger | https://pymol.org/2/ |
| Half-life Color Code for PDB crystal structures in PyMOL | This paper; Mendeley Data | https://data.mendeley.com/datasets/b3hww8ng7w/1; DOI:10.17632/b3hww8ng7w.1 |
| Graphpad Prism 8.0 | Graphpad | https://www.graphpad.com/scientific-software/prism/; RRID: SCR_002798 |


