Abstract
Nuclei are central hubs for information processing in eukaryotic cells. The need to fit large genomes into small nuclei imposes severe restrictions on genome organization and the mechanisms that drive genome-wide regulatory processes. How a disordered polymer such as chromatin, which has vast heterogeneity in its DNA and histone modification profiles, folds into discernibly consistent patterns is a fundamental question in biology. Outstanding questions include how genomes are spatially and temporally organized to regulate cellular processes with high precision and whether genome organization is causally linked to transcription regulation. The advent of next-generation sequencing, super-resolution imaging, multiplexed fluorescent in situ hybridization, and single-molecule imaging in individual living cells has caused a resurgence in efforts to understand the spatiotemporal organization of the genome. In this review, we discuss structural and mechanistic properties of genome organization at different length scales and examine changes in higher-order chromatin organization during important developmental transitions.
Keywords: chromosome topology, cohesin and condensin complexes, loop extrusion, phase separation, gametogenesis, zygotic genome activation, neural development
MOTIVATION FOR UNDERSTANDING THE SPATIOTEMPORAL DYNAMICS OF GENOME ORGANIZATION
Regulatory complexes that control RNA transcription require dynamic restructuring of chromatin to gain access to genes in a densely packed genome (Woodcock & Ghosh 2010). A well-choreographed, dynamic organization of the genome is critical for the accurate implementation of such genome-wide regulatory processes. The topological manipulation of chromatin by ATP-dependent molecular machines and local compartmentalization of the genome into structural domains based on DNA and histone modifications play key roles in genome organization. Although a general structural framework of genome folding has emerged from averaging ensembles of chromosomal conformations, genomes examined at single-cell resolution exhibit vast heterogeneity in their organization. As a result, considerable skepticism remains about the role of genome organization in controlling gene expression (Ghavi-Helm et al. 2019).
The minimal packing unit of a standard eukaryotic genome, the nucleosome (Kornberg 1974), is composed of ∼147 bp of double-helical DNA wrapped around an octamer of core histones. Within living cells, these nucleosomes are packed into chromatin fibers. While the core nucleosome structure was solved at atomic resolution almost two decades ago (Davey et al. 2002), the nature of the next higher level of chromatin organization has remained marred in controversy. For decades, an elusive 30-nm chromatin fiber was proposed to represent the second tier of packing (Woodcock & Ghosh 2010), but its presence in native chromatin has never been established beyond doubt. Instead, recent electron microscopy (Ou et al. 2017) and super-resolution imaging (Ricci et al. 2015) in vivo have shown that chromatin fibers are polymorphic. By combining electron microscopy tomography with a novel labeling method (ChromEM), Ou et al. (2017) showed that chromatin fibers lack regular nucleosomal arrangements in vivo and vary in diameter between 5 and 24 nm. This variability would result in a wide range of chromatin-packing densities.
The arrangement of chromatin fibers in three-dimensional (3D) space can be explained adequately using basic polymer physical models that were initially developed for homopolymers constituting a system at equilibrium (Rosa & Zimmer 2014). More complex and realistic models of chromatin structure and dynamics such as ATP-dependent chromatin remodeling can account for chromatin processes that deviate from equilibrium. These models have their foundations in contact-frequency matrices generated using techniques that capture genome-wide chromosomal conformations (Kempfer & Pombo 2020).
In the last decade, huge strides in computational modeling, genome editing, and precision mapping of chromosomal loci at high genomic and optical resolutions have helped address longstanding questions regarding the role of chromatin organization in gene regulation, including the following:
Is chromatin folding deterministic or stochastic?
What rules govern chromatin folding?
What are the spatial scales of chromatin folding?
To what extent do these spatial scales vary across developmental stages, cell types, and organisms?
What are the major cis and trans determinants of chromosomal association?
What orchestrates precision and accuracy in genomic interactions at different length scales?
What prevents spurious interactions between genes and noncognate regulatory elements while promoting proper interactions with cognate regulatory sites?
GENOME ORGANIZATION: AN OVERVIEW
Answering these questions about genome organization requires a thorough understanding of mechanisms regulating chromatin topology and the relationship between topology and gene regulation. Energetics of DNA–protein and protein–protein interactions impose a strict limit on the distances over which two distinct sets of protein factors bound to two distinct genomic loci can stably interact with one another to initiate loop-capture. The range of such an affinity-based loop-capture process is usually a few nanometers. Therefore, while transcription-driven looping may be deterministic at a local scale, long-range enhancer-promoter (E-P) interactions are likely to be stochastic. However, the probability of stochastic interaction between two genomic loci separated by even a moderate distance of, for example, 100 Kb is estimated to be incredibly low during a mammalian cell cycle (Dekker & Mirny 2016). Thus, for two distant genomic loci to interact within a functionally relevant timescale, the chromatin needs to be compacted over intermediate genomic length scales (10 Kb–1 Mb) and needs to explore its surroundings dynamically (Di Pierro et al. 2018, Ghosh et al. 2019).
While a century of cytogenetic, light microscopic, and ultrastructural studies has provided invaluable insights into chromosome structure at two extremes of genomic length scales, single nucleosomes (∼10 nm) and chromosomal territories (CTs) (∼1–2 μm in mammalian nuclei), only the recently invented Hi-C technology (Lieberman-Aiden et al. 2009) provides a quantifiable metric to measure contact probability among all genomic loci at intermediate length scales. Long before the advent of super-resolution imaging and deep sequencing, Carl Rabl (1885) and Theodor Boveri (1909) proposed that each interphase chromosome occupied a discrete territory. Rabl later proposed that all cell types in an organism have a constant number of chromosomes, and each chromosome occupies a distinct volume in an interphase nucleus (see Cremer & Cremer 2006). More recently, 3D chromosome painting using fluorescent in situ hybridization (3D FISH) proved the presence of CTs (Cremer et al. 2012).
In agreement with the CT organization of large chromosomal domains, Hi-C data revealed two physically and functionally distinct multimegabase-scale subchromosomal compartments in mammalian nuclei: the transcriptionally active A compartment and the transcriptionally repressive B compartment (Lieberman-Aiden et al. 2009, Zhang et al. 2012); see the sidebar titled Chromosome Compartments and Domains. Biochemically, the A compartment differs from the B compartment in having more open chromatin, a set of histone modifications associated with active transcription, and higher gene density. Initially, A and B compartments were defined as structural units formed by homotypic associations between biochemically similar chromosomal regions, but later work (Mirny et al. 2019) showed A and B compartment organization to be less homogeneous and instead composed of a broad continuum of transcriptional states.
Recent high-resolution Hi-C data revealed fine-scale organizational domains within the A/B compartments termed compartmental domains that are as small as a few kilobases and are defined by uniform chromatin marks and/or transcriptional state (Rowley et al. 2017). These domains generally encompass one or two genes and frequently result from interactions between initiation and termination sites of transcriptionally active genes, resulting in gene loops. Compartmental domains can be embedded inside a single CCCTC-binding factor (CTCF)-mediated chromatin loop, can reside in the linker region connecting two such consecutive chromatin loops, or can be spread over two adjacent CTCF-chromatin loops (see the sidebar titled Chromosome Compartments and Domains). Interactions between compartmental domains with similar chromatin marks and/or transcriptional states contribute to the formation of larger A and B compartments. These interactions are proposed to be driven and stabilized by liquid–liquid unmixing of distinct sets of polyvalent and intrinsically disordered proteins (IDPs) that bind to distinct sets of posttranslational marks on nucleosomal core histones (Erdel & Rippe 2018, Singh & Newman 2020).
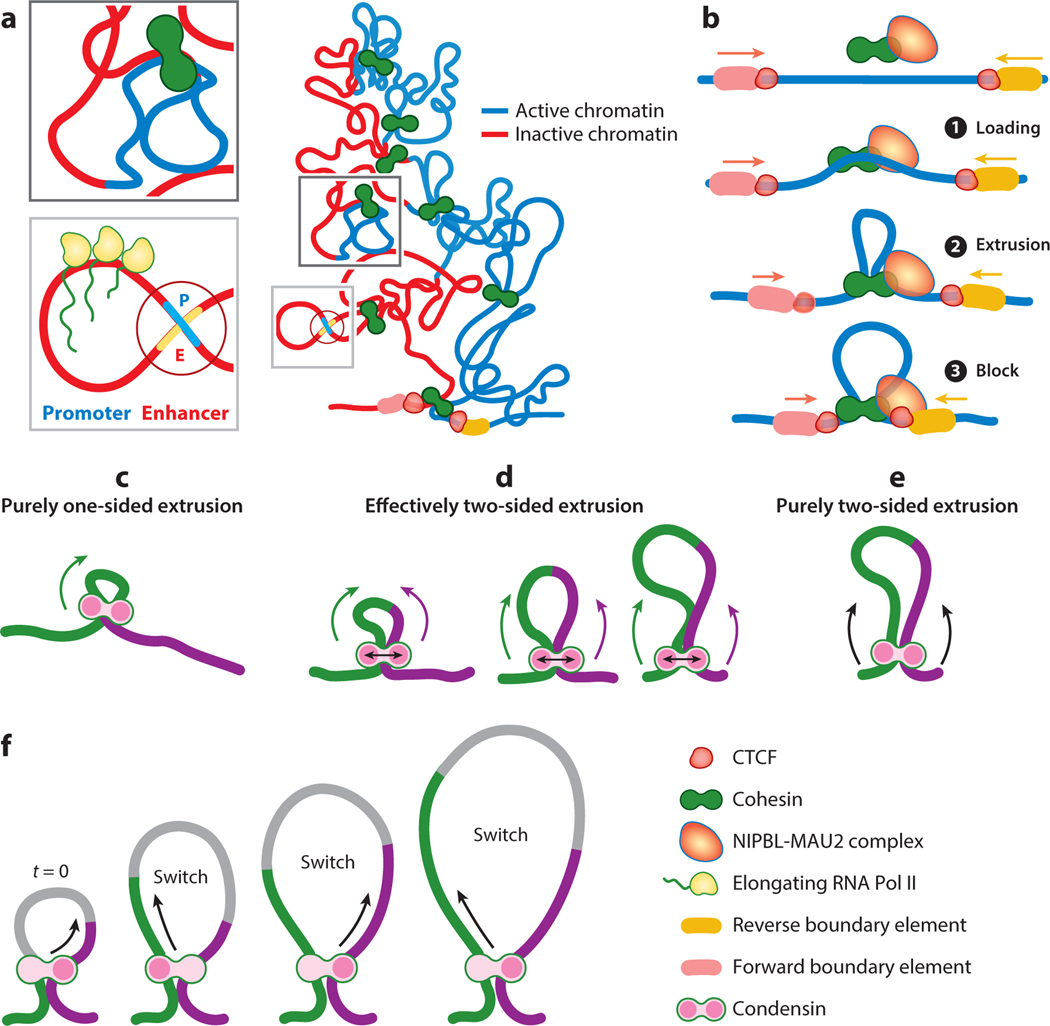
Compartmental domains present a sharp contrast to topologically associated domains (TADs) (Figure 1a). A TAD is a submegabase-scale block [median size of 880 Kb in mouse embryonic stem cells (ESCs)] (Dixon et al. 2012) of globular chromatin domains abutted by linear boundary elements that is characterized by higher interaction frequency between DNA within the block than between sequences outside the block. Unlike compartmental domains, TADs typically lack a uniform transcriptional state or histone modification profile and are instead demarcated by insulator protein enrichment at their boundaries. The most common insulator protein at TAD boundaries is CTCF, a highly conserved transcription factor (TF) implicated in genome organization at kilobase-to-megabase-length scales, even though the exact mechanism by which CTCF enforces boundaries remains unclear.
Figure 1.
Formation of topologically associated domains (TADs). (a, right) Shown is a TAD that includes both active (blue) and inactive (red) chromatin. TADs are flanked by forward- and reverse-oriented boundary elements facing each other in a convergent orientation. CCCTC-binding factor (CTCF) binds to the flanking boundaries. (Top left) This inset shows an enlargement of the dark gray boxed region that highlights a chromatin loop extruded by cohesin. The loop contains stretches of active and inactive chromatin. (Bottom left) This inset shows an enlargement of the light gray boxed region that highlights a chromatin loop extruded by cohesin. The loop joins an enhancer and a promoter to initiate transcription by RNA polymerase II (Pol II). (b) Loop extrusion of chromatin by cohesin occurs in a stepwise fashion. ❶ Cohesin is loaded onto chromatin by the nipped B-like protein (NIPBL)-MAU2 complex. The cohesin loader remains engaged with cohesin throughout the loop-extrusion process and plays a role in loop stability. ❷ After loading, cohesin extrudes chromatin loops in a two-sided manner until ❸ it is blocked by CTCF or any other barrier protein. Cohesin is then released with the aid of the cohesin-release factor WAPL (not shown). (c–e) Condensin-mediated loop extrusion is predominantly one sided. As shown in panel c, purely one-sided loop extrusion by condensin generates much smaller loops than purely two-sided loop extrusion (see panel e) and is unable to achieve the levels of compaction required for mitotic condensation. As shown in panel d, effectively two-sided loop extrusion is proposed as an alternate model for efficient loop extrusion. The effectively two-sided model matches the values of the extrusion parameters. In this model, condensin subunits stochastically switch between extruding from one side and then extruding from the other. In any one instant, condensin extrudes in a one-sided manner, but on average the switching causes the asymmetric one-sided extrusion to be effectively two sided. Panel d shows three different levels of loop extrusion, in which the size of the extruded loop scales with the switching frequency. (f) A detailed schematic representation of stepwise asymmetric strand extrusion by condensin. The chromatin is color coded (green and purple) to help visualize the chromatin segments that are being alternately extruded.
Ultra-high-resolution contact matrices revealed nested sub-TAD structures that generally have weaker insulation scores and more homogeneous chromatin marks than TADs and exhibit greater heterogeneity across cells. For an in-depth discussion on TAD and sub-TAD structure, see Beagan & Phillips-Cremins (2020).
Spontaneous and stochastic collisions between distant genomic sites and subsequent capture by CTCF and other boundary-located proteins provide an appealing rationale for TAD formation and stabilization. However, the model does not explain several features: (a) why inter-TAD interactions are underrepresented in the genome, (b) why CTCF-binding sites require convergent orientations at TAD flanks for stable TAD formation (Tang et al. 2015), (c) how specific interactions between distant loci are temporally controlled, and (d) how two genomic loci that flank the edges of a TAD boundary maintain robust insulation despite being in close one-dimensional (1D) space.
THE MOLECULAR FRAMEWORK FOR CHROMATIN LOOPING
CTCF and Cohesin Collaborate to Generate Interphase Chromatin Loops
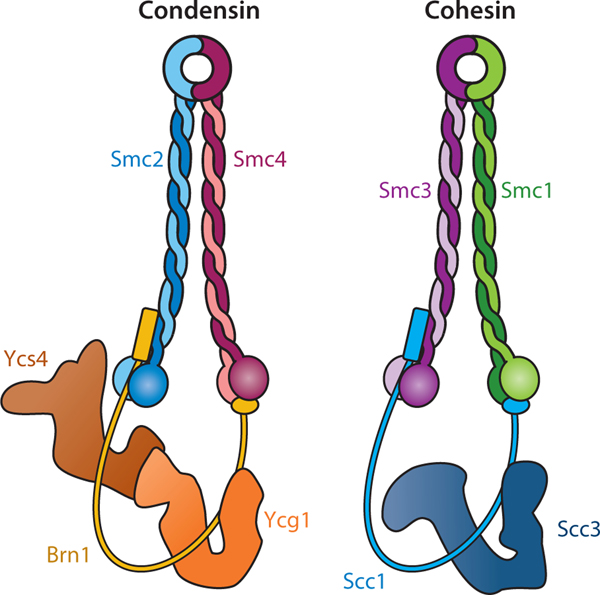
While studying the role of CTCF in establishing human genome architecture, Tang et al. (2015) showed that ∼99% of chromatin loops were anchored by CTCF and ∼80% of loop-anchor sites were co-occupied with the SMC cohesin complex (see the sidebar titled Architectural Proteins That Create Chromosome Structure Utilize Chromatin Loop Extrusion). Cohesin was initially identified as the core molecular complex required for sister chromatid cohesion but was subsequently implicated in the interphase genome organization of higher eukaryotes (Sanborn et al. 2015; Fudenberg et al. 2016, 2017). Cohesin (Figure 2) is composed of an SMC (structural maintenance of chromosomes) heterodimer (Smc1 and Smc3), an α-kleisin subunit Scc1 (Gligoris et al. 2014) that bridges the ATP-binding domain of each SMC subunit, and Scc3, an α-kleisin interacting subunit that was proposed to topologically entrap chromatin fibers and also translocate along chromatin (Kim et al. 2019, Yatskevich et al. 2019). In agreement with earlier in situ Hi-C data (Rao et al. 2014), Tang et al. (2015) showed that CTCF/cohesin dual-occupancy loops have a predominantly inward orientation of CTCF-binding motifs, with the 5 and 3 motifs facing each other. Furthermore, in these loops, the 3 side of the CTCF anchor is preferentially bound by cohesin, suggesting that the molecular mechanism for chromatin looping is coded into the genome. The asymmetric enrichment of cohesin at the 3 anchor of CTCF/cohesin loops further suggests that (a) 3D interactions between genomic sites could be driven by 1D linear tracking along the length of a chromatin fiber, and (b) CTCF constitutes an insurmountable barrier to translocating cohesin complexes only when approached from the 3 end. A compelling model of local genome organization that emerged over the last decade is that TADs are formed through chromatin loop extrusion, in which protein complexes such as cohesin extrude a chromatin fiber until the loop extruder dissociates from chromatin or encounters a barrier protein such as CTCF (Figure 1a, b). An efficient barrier would therefore promote interloop interactions inside a TAD. Thus, cell-specific variations in genome topology might be achieved through the differential expression of regulators that set loop-extrusion rate, extruder loading efficiency, loop extruder concentration, and binding strength and polarity of barrier proteins.
Figure 2.
Subunit composition of Saccharomyces cerevisiae condensin and cohesin complexes. Figure adapted with permission from Yatskevich et al. (2019).
Condensin I and II Collaborate to Generate Nested Chromatin Loops During Prometaphase
The multisubunit SMC condensin complexes (condensin I and II) (Figure 2) are key mediators of mitotic and meiotic chromosome assembly and segregation. They compact chromosomes by anchoring radial loops of DNA to a central scaffold (Hirano 1995).
In the late 1970s and early 1980s, Ulrich Laemmli (Paulson & Laemmli 1977, Marsden & Laemmli 1979, Earnshaw & Laemmli 1983) showed that histone-depleted metaphase chromosomes were composed of a central proteinaceous scaffold surrounded by a halo of DNA loops in which the exit and entry points of each loop were tethered to the scaffold. John Marko (2009) then proposed a novel mechanism of chromosome condensation called lengthwise condensation or catenation condensation. By combining imaging, Hi-C, and polymer simulations, Gibcus et al. (2018) showed that condensins I and II play crucial roles in mitotic chromosome compaction. The authors showed that mitotic compaction is achieved through the formation of loop arrays in which consecutive loops are angularly oriented along a helical scaffold. The most striking feature of this compaction path is the transition from a flexible ∼60 Kb loop array in prophase to a nested-loop array arranged on a helical scaffold in prometaphase. Using an auxin-degron depletion system, the authors showed that (a) condensin I generated prophase loop arrays and ∼80-Kb inner loops of the prometaphase nested-loop array, and (b) condensin II generated the ∼400-Kb outer loops and the central helical scaffold. The difference in size between the inner and outer loops of the nested-loop arrays has been attributed to differences in the exchange kinetics of condensins I and II. Condensin II remains longer on chromatin than does condensin I, thus extruding larger loops.
Beyond their canonical role in mitotic chromosome compaction, condensins have been implicated in a wide variety of interphase chromosomal regulations, including but not limited to X-chromosome dosage compensation in Caenorhabditis elegans (Chuang et al. 1994, Csankovszki et al. 2009, Meyer 2018), interphase CT intermixing (Rosin et al. 2018) and disassembly of polytene chromosomes during Drosophila (Hartl et al. 2008) mid-oogenesis, ribosomal DNA maintenance in Saccharomyces cerevisiae (Johzuka et al. 2006), and gene regulation and DNA damage repair (Wallace & Bosco 2013).
A Recent Deluge of Single-Molecule Data Provides Resounding Evidence for Loop Extrusion
Proof of the direct involvement of SMC protein complexes in loop extrusion and TAD organization has come from protein depletion studies. The partial depletion of cohesin (Zuin et al. 2014) caused the loss of short-range intra-TAD interactions, whereas acute depletion caused the genome-wide disruption of TADs followed by rapid recovery upon cohesin restoration (Rao et al. 2017). Similar results were obtained with the depletion of the cohesin-loading protein nipped B-like protein (NIPBL) (Schwarzer et al. 2017). CTCF depletion in mouse ESCs (mESCs) revealed an obligatory role of CTCF in creating TAD boundaries (Nora et al. 2017).
Despite resounding support for the role of loop extrusion in forming TADs, the mechanistic details of loop extrusion have remained speculative from its conception almost a decade ago (Alipour & Marko 2012). Only recently, several independent studies (Ganji et al. 2018, Davidson et al. 2019, Banigan & Mirny 2020, Golfier et al. 2020, Kim et al. 2020) using single-molecule detection and tracking of cohesin- and condensin-mediated loop dynamics have provided unequivocal evidence for the role of SMC proteins in loop extrusion.
Mechanochemical translocation of condensin was first demonstrated in a DNA curtain setup (Terakawa et al. 2017), but the first unequivocal demonstration of loop extrusion and its potential for large-scale compaction of mitotic chromosomes came from Ganji et al. (2018) showing that yeast condensin extrudes tens of kilobases of DNA loops asymmetrically in an ATP-dependent manner. In yeast, condensin reeled in the DNA only from one end of a growing loop, contrary to the symmetric extrusion scheme used in polymer simulations (Figure 1c, d). Subsequent work by Golfier et al. (2020) using Xenopus metaphase-egg extract showed condensin-driven loop extrusion to be predominantly asymmetric. However, cohesin in the presence of Xenopus interphase-egg extract extruded DNA loops almost exclusively in a two-sided symmetric manner. Further work by Davidson et al. (2019) using human cohesin showed that the NIPBL-MAU2 heterodimeric cohesin-loading complex was required for loop initiation as well as loop maintenance.
Still perplexing is how one-sided asymmetric DNA extrusion by condensin can lead to the thousandfold compaction of human chromosomes in metaphase. For a detailed discussion on effectively two-sided loop extrusion by condensins (Kim et al. 2020), either through dimerization or strand switching (Hassler et al. 2018), see Banigan & Mirny (2020). Effective two-sided loop extrusion through stochastic segment/loop-capture, in which only one subunit of a loop extruder can actively reel in DNA at any one time, is a particularly suitable model for chromosome compaction through one-sided loop extrusion. Polymer simulations showed that stochastic strand switching can efficiently recapitulate chromosome compaction levels achieved through a two-sided extrusion model (Banigan et al. 2020) (Figure 1c–f). While several studies provided exquisite molecular details of loop extrusion, future work must focus on creating strategies for visualizing loop extrusion in vivo to determine whether condensin and cohesin complexes can bypass steric obstacles such as nucleosomes, RNA polymerases, and other chromatin-bound complexes. Also important will be to expand existing single-molecule tracking strategies to test if strand switching is a viable alternative for two-sided extrusion.
How Important Are Topologically Associated Domains for Gene Regulation? The Local Versus Global Regulation Conundrum
While a mechanistic understanding of TAD formation is emerging through the directvisualization of loop extrusion, the extent to which TADs play a role in gene regulation remains controversial. Here we discuss the current state of understanding of the role of TADs in regulating both local and global gene regulation.
Local disruption of topologically associated domains can lead to spurious enhancer-promoter contacts and aberrant gene expression.
Support for a role of TADs in gene regulation comes from pathogenic structural variations that perturb the integrity of TADs. Lupiáñez et al. (2015) showed that duplications, deletions, and inversions that alter the boundaries of TADs spanning the extended WNT6/IHH/EPHA4/PAX3 region caused limb malformations in humans. Using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 genome editing to replicate corresponding DNA rearrangements in mice, the authors showed that disruption of TAD boundaries led to inappropriate E-P contacts and pathogenic phenotypes due to aberrant expression of genes.
In addition, several studies showed that structural disruption of TADs can activate oncogenes. For example, depletion of CTCF can drive oncogene activation through nonspecific association between a constitutive enhancer and a promoter for a glioma oncogene normally located in two separate TADs (Flavahan et al. 2016). Similar activation of oncogenes in T cell acute lymphoblastic leukemia (T-ALL) occurs through disruption of TAD boundaries surrounding a topological domain containing several important T-ALL proto-oncogenes in nonmalignant cells (Hnisz et al. 2016). For a more detailed account of how disruption of TADs causes cancer, see Valton & Dekker (2016).
Another example showing the importance of topological domains in gene regulation comes from the analysis of Hox genes, in which CTCF is essential to maintain active and inactive domain boundaries within the Hox cluster. CTCF loss resulted in the spreading of active chromatin into repressed regions and the aberrant expression of Hox genes (Narendra et al. 2015). In most cases, fusion of topological domains upon deletion of TAD boundaries generates novel E-P contacts through inter-TAD interactions (enhancer hijacking) while having minimal effects on native E-P contacts (Hanssen et al. 2017, Cavalheiro et al. 2021). Because E-P interactions are highly dynamic, a mechanism must constrain the spatial reach of enhancers and promoters, thereby preventing ectopic gene activation. Indeed, chromosomal variants such as deletions and duplications that can mediate spurious contacts between distant genomic elements are specifically depleted from TAD boundaries and CTCF-binding sites, which is indicative of a purifying selection against enhancer hijacking (Fudenberg & Pollard 2019). Similarly, a strong association was found between disruption of topological domains and changes in gene expression across broad evolutionary time (Krefting et al. 2018).
The connection between TADs and local gene regulation, however, remains tenuous due to studies showing local decoupling of chromatin topology and gene expression. For example, pairs of genes residing inside a TAD do not exhibit transcriptional coordination any more than predicted by sequence proximity alone (Long et al. 2020). In the context of the Shh locus, which contains an array of tissue-specific enhancers embedded within one evolutionarily conserved TAD (Dixon et al. 2012, Jin et al. 2013), the local 3D genomic configuration was proposed to be essential for achieving contact specificity and a much higher contact probability than expected on the basis of sheer genomic distance. However, work by Williamson et al. (2019) contradicted this view by showing that wide-scale changes in chromatin conformation in the Shh locus induced by internal deletions, CTCF-binding-site deletions, and deletions or inversions of sequences at TAD boundaries had no discernible impact on Shh gene expression during development. Additionally, Kragesteen et al. (2018) showed that the permissive forelimb-specific E-P configuration of the Pen enhancer and its target gene Pitx1 occurred without well-defined TADs or CTCF-dependent chromatin looping.
Genome-wide disruption of topologically associated domains has a relatively meager impact on global gene expression.
Unlike targeted deletions or inversions at TAD boundaries that result in aberrant gene expression through spurious E-P contacts, global disruption of TADs does not correlate well with changes in gene expression. Capitalizing on the abundant DNA rearrangements inherent in Drosophila melanogaster balancer chromosomes, Ghavi-Helm et al. (2019) showed that while large nested inversions and small inversions, deletions, and duplications caused major changes in chromatin topology, they barely affected gene expression. This finding suggested that topological control of gene expression is highly locus specific.
A further example of decoupling between gene expression and chromosome-wide TAD structure came from X-chromosome dosage compensation in C. elegans (Anderson et al. 2019). A dosage compensation condensin (DCC) complex compensates for the difference in X-chromosome number between males (XO) and hermaphrodites (XX) by binding to both hermaphrodite X chromosomes and reducing gene expression by half (Meyer 2018). The DCC complex establishes megabase-sized TADs by binding to its highest-occupancy recruitment elements on X (rex) sites located at TAD boundaries (Crane et al. 2015). The DCC complex also enriches histone modification H4K20me1 on dosage-compensated X chromosomes. The loss of H4K20me1 reduces X compaction, disrupts TADs, and causes overexpression of X-linked genes (Brejc et al. 2017). However, disruption of TADs by the loss of H4K20me1 is not the cause of elevated X expression. Anderson et al. (2019) showed that disrupting TAD boundaries on hermaphrodite X chromosomes by deleting high-occupancy rex sites had virtually no effect on gene expression during dosage compensation in embryos. Mutating rex sites did, however, cause a reduction in the life span of adult hermaphrodites but not of males. Loss of TADs caused accelerated aging of hermaphrodites. These results indicate that dosage compensation and TAD formation are two independent aspects of DCC function.
Using a single-cell spatial genomics approach, Espinola et al. (2020) showed that promoter interactions with cis-regulatory modules (CRMs) such as enhancers and silencers are established prior to the formation of TADs during Drosophila development and that spatial juxtaposition of promoters and CRMs is not a universal requirement for transcriptional activity. Similarly, studies in Drosophila by Ing-Simmons et al. (2020) of dorsoventral patterning and its relationship to developmentally regulated gene expression showed that tissue-specific differences in gene expression are not reflected in the spatial organization of chromatin.
Using Micro-C, Hsieh et al. (2020) showed that transcription elongation drives the formation of local sub-TAD chromatin domains through E-P and promoter-promoter interactions. Inhibition of promoter melting or transcription elongation resulted in loss of these finer gene-level structures, but higher-order structures remained intact. Furthermore, independent studies (Rao et al. 2017, Schwarzer et al. 2017) showed that acute depletion of cohesin and CTCF had a minimal effect on gene expression while causing genome-wide changes in TAD structure. These findings provide further support for the idea that TADs constitute a structural framework for the genome with only a minor role in regulating gene expression.
Populational heterogeneity of topologically associated domains and allelic variability in chromatin structure.
The dynamic nature of TADs revealed by single-cell Hi-C, FISH, and super-resolution imaging studies has further called into question the idea of invariant TAD boundaries and their role in gene regulation. The large cell-to-cell variation in TAD boundaries indicates that TADs are an ensemble average of the entropic mix of configurations (Bintu et al. 2018). For a detailed discussion of this point, see Luppino & Joyce (2020).
The transcriptional state of two alleles of the same gene can vary in the same cell, demonstrating the generally stochastic nature of allelic transcription. Even looping events appear to be highly dynamic and variable and rarely stand out upon population averaging (Symmons & Raj 2016, Cattoni et al. 2017). These observations suggest that functionally relevant interactions between genomic elements are transient and may form and dissolve multiple times within one cell cycle. Indeed, Finn et al. (2019) showed that wide variation occurs in genome organization patterns in individual cells within a cell population. At the single-cell level, considerable variability occurs in spatial organization of alleles. Measurement of the spatial distance between two homologous loci for 125 different pairs of loci showed no obvious coordination between alleles within a cell for the minimum distance separating one locus from its partner locus.
MECHANISTIC BASIS OF CHROMOSOME COMPARTMENTALIZATION
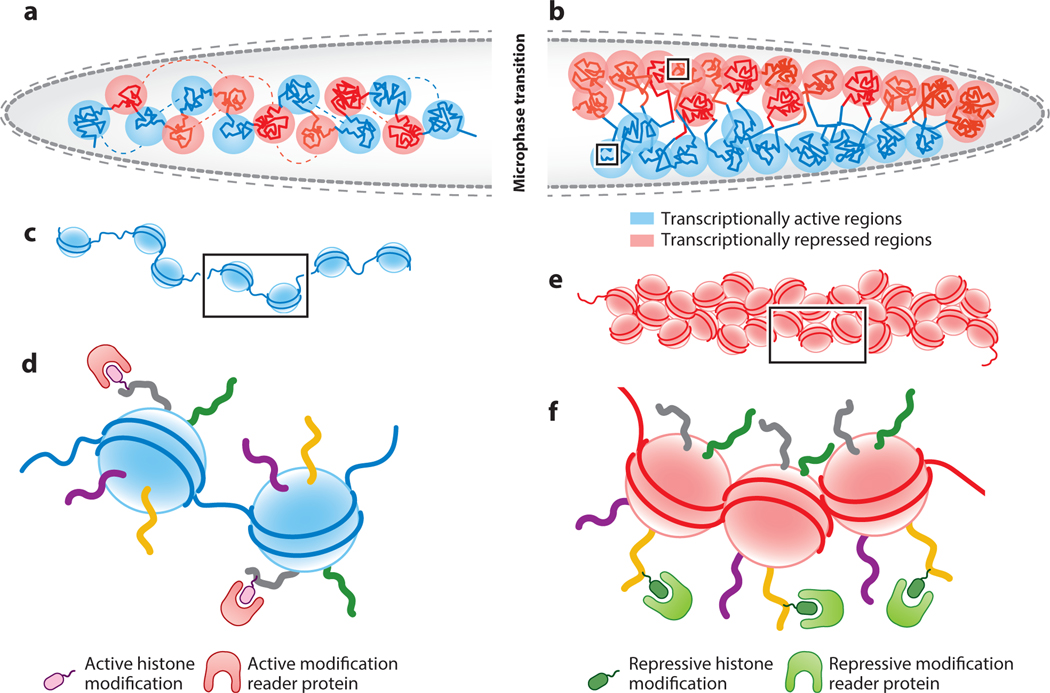
While loop extrusion by cohesin complexes is a strong model for the formation of TADs and nested sub-TADs, the biophysical mechanisms driving the formation of transcriptionally active and repressive compartments remain unclear. TADs and compartments are proposed to have distinct mechanistic origins. Degradation of CTCF, which causes genome-wide disruption of chromatin loops and TADs, has minimal effects on A/B compartment stability (Nora et al. 2017), whereas cohesin depletion strengthens compartmentalization (Schwarzer et al. 2017, Wutz et al. 2017). Depletion of the cohesin loader NIPBL, which abolishes TADs, enhances compartment strength by ∼1. 8-fold, revealing a finer set of subcompartments that are obscured in wild-type cells (Schwarzer et al. 2017). These smaller subcompartments are composed primarily of small B-like compartments embedded inside A compartments. These small B-like compartments interact preferentially with other small B-like compartments. These observations suggest that TADs can span compartments having different chromatin marks and that subcompartments are intrinsic genomic features dependent solely on local chromatin marks and transcriptional activity (Rowley et al. 2017). Spatial segregation of subcompartments with distinct chromatin marks into immiscible phases is akin to microphase separation of block copolymers (Figure 3). Phase separation may mediate genome compartmentalization by utilizing and reinforcing chromatin modifications (Hildebrand & Dekker 2020) (see the sidebar titled Chromatin Organization Through Phase Separation). In fact, methylation of histone H3K9 promotes the formation of genome compartments in C. elegans by enhancing chromosome compaction and by anchoring chromosome arms to the nuclear envelope. The loss of H3K9 methylation weakens compartments, but compartment attenuation fails to cause significant changes in gene expression (Bian et al. 2020).
Figure 3.
Microphase separation leads to the formation of active and inactive chromatin domains. (a) Chromatin can be envisioned as a block copolymer of alternative repeating units of transcriptionally active (blue) and repressed (red) regions. (b) Units of the same activity state interact with one another (blue with blue, red with red) to undergo microphase separation to form small domains of active and inactive chromatin. (c, d) Panel c shows an enlargement of the boxed area in the active block in panel b and represents an open conformation of nucleosomes (blue nucleosomes). These nucleosomes are marked with active histone modifications that are bound by proteins harboring recognition domains for active modifications shown in panel d. (e, f) Panel e shows an enlargement of the boxed area in the repressed block of panel b and represents compactly arranged nucleosomes (red nucleosomes). These nucleosomes are marked with repressive histone modifications that are bound by proteins harboring recognition domains for repressive modifications shown in panel f.
Phase Separation: The Other Major Player in Genome Organization?
The role of phase separation in genome organization has recently garnered significant attention regarding the formation of heterochromatin and super-enhancer (SE) assemblies. Phase separation is a process by which a homogeneous mixture of molecules spontaneously sorts into distinct phases with unique material properties. While the idea of colloidal phase separation as an operational principle for the formation of membrane-less cellular compartments dates back to William Hardy (1899) and Edmund Wilson (1899) in the late nineteenth century, a resurgence of this idea has occurred recently regarding the role of liquid–liquid phase separation (LLPS) in driving macromolecular assembly (Hyman et al. 2014). LLPS is a specific type of phase transition in which a homogeneous mixture of molecules spontaneously sorts into two immiscible liquid phases, one denser than the other due to preferential partitioning of the molecules. For a detailed discussion of the molecular underpinnings of phase separation in biological assembly, see Alberti (2017), Shin & Brangwynne (2017), McSwiggen et al. (2019b), and Chen et al. (2020).
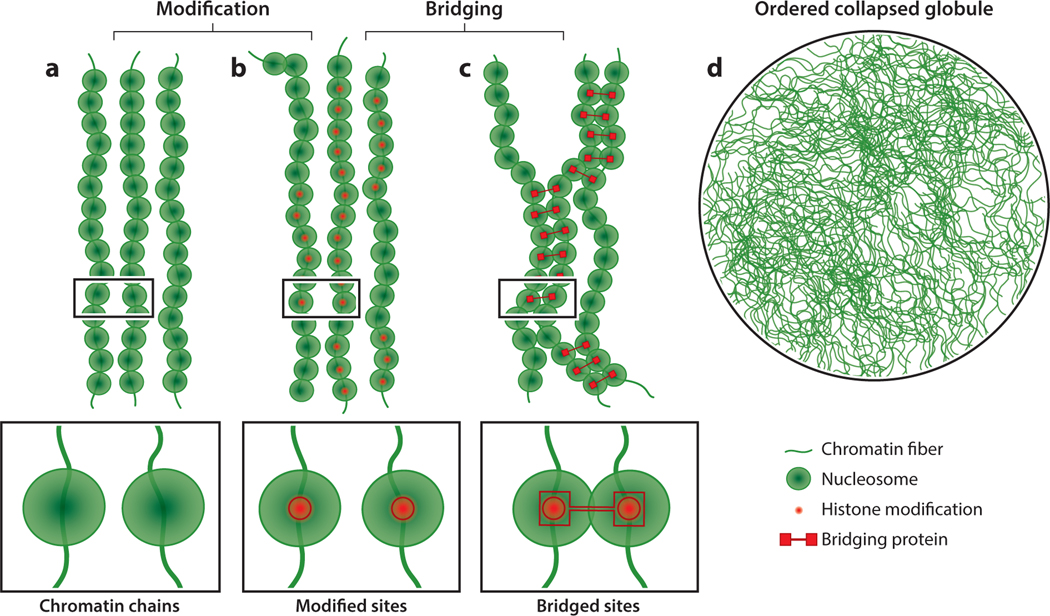
Genomic phase separation can occur through two mechanisms. The first is polymer–polymer phase separation (PPPS), in which a network of chromatin fibers harboring similar chromatin marks can be noncovalently bridged by proteins with multiple binding domains or through adaptor molecules that simultaneously engage multiple chromatin-bound proteins, resulting in the formation of a collapsed polymer globule (Barbieri et al. 2012, Brackley et al. 2016, Michieletto et al. 2016) (Figure 4); see the sidebar titled Chromatin Organization Through Phase Separation. The second is through LLPS (Figure 5a–e) in which proteins self-assemble through (a) well-defined interaction surfaces with a propensity for oligomerization, (b) the arrangement of multiple binding domains in series, or (c) intrinsically disordered regions (IDRs). The extent to which each of these mechanisms contributes to phase separation is difficult to gauge experimentally and remains a matter of intense debate.
Figure 4.
Polymer–polymer phase separation (PPPS) of chromatin. (a, b) Chromatin fibers are shown with nucleosomes and histone modifications. Enlargements of the black boxed areas are shown at the bottom. (c) Histone modifications can act as recruitment platforms for proteins that bridge chromatin fibers using multiple modular chromatin-binding domains (bridging proteins). An enlargement of the black boxed area is shown at the bottom. (d) Cross-linking of chromatin fibers by bridging proteins leads to PPPS and the formation of an ordered collapsed globule.
Figure 5.
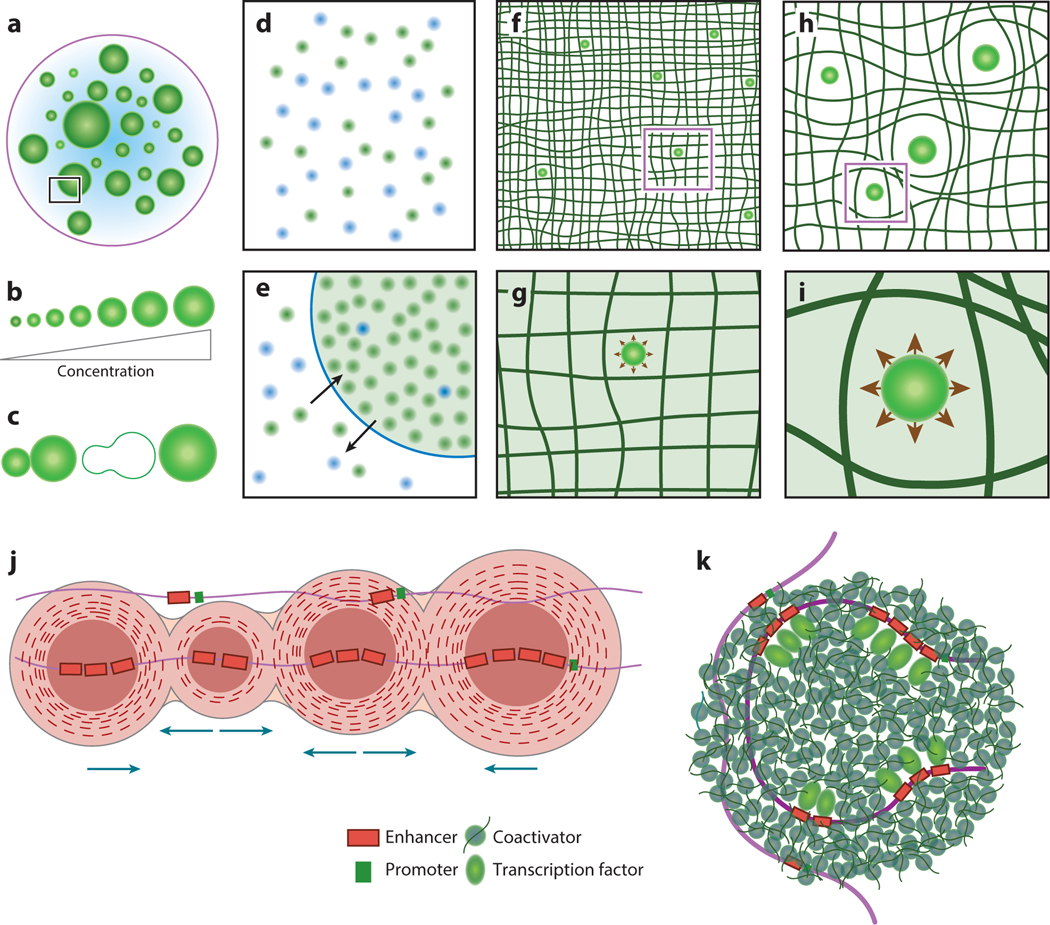
Liquid–liquid phase separation (LLPS) in chromatin organization. (a–c) Characteristics of droplets formed by LLPS. As shown in panel a, cells contain membrane-less bodies of varying sizes that form through LLPS. These liquid droplets have spherical shapes due to surface tension. As shown in panel b, the size of these liquid droplets scales with the concentration of the component molecules. As shown in panel c, droplets can undergo spontaneous fusion or fission. (d, e) During LLPS, a homogeneous mixture of molecules shown in panel d sorts into two liquid phases, a dense phase with a higher concentration of molecules and a dilute phase surrounding the dense phase shown in panel e. Panel e is an enlargement of the boxed area in panel a. Molecules in liquid phases can rearrange dynamically. (f–i) Chromatin mechanically regulates droplet growth. Droplet growth dynamics can be regulated by the stiffness of surrounding chromatin. Heterochromatin, shown in panel f, is stiffer than euchromatin, shown in panel h, and is less permissive of droplet growth. Growing droplets can, in turn, mechanically deform the surrounding chromatin. Panel g shows an enlargement of the purple boxed area in panel f, while panel i shows an enlargement of the purple boxed area in panel h. (j, k) Super-enhancer (SE) clusters can drive coactivator condensation. As shown in panel j, adjacent enhancers in an SE cluster can nucleate multiple coactivator droplets, where coactivators recruited by transcription factors can phase separate through homotypic polyvalent binding with other coactivators. As shown in panel k, these droplets can coalesce to generate a regulatory hub for transcriptional control of genes without the need for physical juxtaposition of distal enhancers and promoters.
Nuclear bodies formed by LLPS and PPPS have fundamental differences. In chromatin condensates formed by PPPS, soluble bridging proteins inside a collapsed chromatin globule exist in a dynamic equilibrium with those present outside the condensate, as the nucleoplasm surrounding these condensates can freely percolate the interior of the chromatin bodies. In contrast, for chromatin condensates formed by LLPS, the surrounding nucleoplasm forms a second less dense liquid phase. Nuclear bodies born out of LLPS can nucleate and grow without binding to chromatin fibers and can diffuse through the nucleus.
While LLPS can lead to nucleation independently of binding to chromatin, binding sites on chromatin with high recruitment potential facilitate stable droplet formation (Shin et al. 2018). At concentrations below a critical threshold, the energy gained through multivalent interactions cannot outweigh the energetic cost of liquid–liquid demixing. However, once stable droplets are formed through LLPS, they are more resistant to fluctuations in the number of chromatin binding sites and concentrations of constituent binders than are condensates formed by PPPS. For example, fluctuations in the density of local chromatin marks can lead to sharp changes in the concentration of available binding sites. Sizes of condensates formed by LLPS scale with the change in concentration of multivalent binders (Figure 5b). Such concentration-dependent size scaling means that to maintain a constant chromatin-mass-to-droplet-size ratio, growing droplets would have to incorporate adjacent chromatin domains of similar compaction state (Wachsmuth et al. 2008, Erdel & Rippe 2018). While droplet coalescence is a common feature of both LLPS and PPPS (Figure 5c), the speed of coalescence and geometry of fusion products are likely to differ for these two processes (Wachsmuth et al. 2008, Lappala & Terentjev 2013, Nozaki et al. 2017). Dynamic imaging of chromatin droplets at high spatiotemporal resolution may help discriminate between LLPS and PPPS in vivo. Because condensates formed via LLPS and PPPS differ markedly in their origin, composition, and dynamics, fluctuations in the composition of the nucleoplasm and in chromatin marks may lead to considerable differences in local genome compartmentalization depending on the precise type of phase separation at play.
Phase separation in heterochromatinization.
A central theme of nuclear structure and dynamics is the self-organization of nuclear proteins and RNAs into membrane-less bodies. Over the years, many nuclear condensates have been studied, including nucleoli, Cajal bodies, promyelocytic leukemia bodies, nuclear speckles, paraspeckles, transcription factories, and others (Mao et al. 2011). Heterochromatin protein 1 (HP1) is the condensate-inducing factor that has received the most attention, owing primarily to its capacity to translate specific marks on chromatin (H3K9me3) to a constitutive heterochromatic state. Both HP1a in Drosophila (Strom et al. 2017) and HP1α in humans (Larson et al. 2017) undergo LLPS, albeit with different molecular reactivities. While HP1a can autonomously partition into a liquid phase, HP1α cannot phase separate on its own and demixes only upon N-terminal phosphorylation or upon binding to DNA. Larson et al. (2017) showed that an HP1α-dominated phase preferentially solvated its interaction partners. A DNA curtain experiment revealed that HP1α can bridge DNA strands separated by micrometers (Larson et al. 2017). This result suggested the existence of mesoscopic nucleoprotein networks composed of ∼100 HP1α dimers capable of coalescing distant DNA strands without forming large droplets.
Even though HP1a can form droplets spontaneously, genomic sites with high concentrations of H3K9me2/3 most likely promote condensation by facilitating HP1a nucleation. Localized binding-induced nucleation and a slower supersaturation rate would preferentially promote droplet growth at specific genomic loci over spontaneous generation of multiple seeds (Onuki 2002). While HP1α can undergo LLPS, its role in directly bridging DNA strands suggests it may also mediate genomic compartmentalization through PPPS. Indeed, Erdel et al. (2020) suggested that facultative mouse pericentromeric heterochromatin has characteristics of collapsed chromatin globules but not of LLPS. Furthermore, the heterochromatic state of chromocenters is insensitive to HP1 concentration but instead undergoes a switchlike transition to a less compact state upon exposure to a strong activator. The collapsed chromatin globule state of chromocenters is potentially mediated through bridging proteins specific to pericentromeric heterochromatin and through the propensity of pericentromeric repeats to self-associate.
Heterochromatinization through phase separation is not limited to HP1 condensates. Reconstituted Polycomb group (PcG)-repressive complex 1 (PRC1) undergoes LLPS in vitro. PRCs are essential regulators of development and include a number of variants (PRC2 and PRC1) with fundamentally different activities. PRC2 catalyzes H3K27me3, which acts as a recruitment platform for canonical PRC1 (cPRC1) complexes, which in turn compact chromatin and silence gene transcription. cPRC1 constitutes phase-separated Polycomb bodies in mESCs (Saurin et al. 1998). Using an in vitro reconstitution system, Plys et al. (2019) showed that in vitro phase separation of reconstituted PRC1 is driven by a heterodimer of two core components: CBX2, a chromodomain protein with IDRs, and RING1b, a ubiquitin ligase. This phase-separation process was independent of CBX2’s binding to H3K27me3 and efficiently incorporated DNA, RNA, and nucleosomal arrays into the condensates. Mutation of a lysine/arginine (K/R) cluster in a CBX IDR essential for condensate formation abrogates LLPS. Furthermore, K/R mutations that induce different degrees of defects in PRC1-LLPS correlate strongly with nucleosome-compaction defects in vitro and the severity of axial patterning defects in mice (Lau et al. 2017). While PRC1 condensates are dynamic and are formed de novo during each cell cycle, both H3K27me3 deposited by PRC2 and H2AK119ub1 deposited by the RING1b subunit of PRC1 can be inherited through replication and cell division.
The key discrepancy of how condensates, which are transient in nature, can lead to long-lasting impacts on chromatin in the form of epigenetic memory has been addressed by Eeftens et al. (2020) using an optogenetically induced oligomerization platform to initiate the assembly and disassembly of multicomponent condensates involving the exposure of chromatin to PRC1 condensates. That chromatin can retain its compaction even after the dissolution of PRC1 condensates argues against a direct mechanical role of condensates in chromatin compaction and in favor of an indirect role in chromatin self-organization (L. Wang et al. 2019) through repressive chromatin modification. Higher concentrations of reactants in a condensate have been proposed to boost biochemical reactions. Recently, CBX-PRC1 condensates were found to promote stable CBX2-genome interactions by constraining the search space and mitigating nonspecific interactions, resulting in markedly higher CBX2 genome occupancy inside condensates (Kent et al. 2020).
Transcription hubs: an LLPS-based model for mediating distant enhancer-promoter contacts without actual physical bridging.
Phase separation of chromatin-bound activators into large transcriptional hubs provides an interesting mechanism for the coordinated regulation of genes without physically bridging promoters and enhancers. Phase separation has been proposed to mediate transcriptional activation through the assembly of nanocondensates of transcriptional activators at SE clusters (Cho et al. 2018, Chong et al. 2018, Sabari et al. 2018) (Figure 5j, k). SEs are clusters of putative enhancers that exhibit both a significantly higher level of activity than regular enhancers and much higher occupancy of RNA polymerase II (Pol II), transcriptional coactivators, components of the Mediator complex, and master regulators of transcription. Many of these proteins, including subunits of Mediator, BRD4 (which is an epigenetic reader molecule containing two tandem bromodomains), and RNA Pol II, undergo LLPS due to polyvalent interactions mediated by their IDRs. Many TFs such as the homeodomain TF Oct4, yeast GCN4, and the estrogen receptor also generate cocondensates with Mediator proteins through weak multivalent interactions driven by IDRs. Dynamic assembly and disassembly of these nanocondensates provide a probable regulatory platform for coordinated control of multiple E-P interactions upon addition of an external cue. For example, the estrogen receptor–MED1 phase equilibrium can be controlled through estrogen stimulation (Boija et al. 2018).
SE clusters, which have disproportionately high levels of BRD4 compared to standard enhancers, are believed to act as nucleation sites for Mediator condensates. The large size of Mediator clusters at select enhancers may recruit a large number of promoters to a common regulatory hub (Cho et al. 2018, Sabari et al. 2018). The large size of such condensates (>300 nm) could recruit distant promoters to the SE hub without the need for direct contact between promoters and distal enhancers. As these supramolecular condensates are highly dynamic, adaptive genome reorganizations are likely to be highly dynamic as well. Mediator and RNA Pol II colocalization in shared condensates (Cho et al. 2018) may further explain how multiple genes in a gene-expression program could be regulated through interactions between enhancer-located TF-Mediator cocondensates and RNA Pol II condensates at promoters without the need for E-P juxtaposition.
Despite these recent developments, considerable skepticism remains regarding the role of LLPS in transcriptional regulation, primarily due to the extremely short half-lives of TF clusters and considerable heterogeneity in sizes of TF hubs, which seem uncorrelated with TF concentration in the nucleus (McSwiggen et al. 2019b). Furthermore, McSwiggen et al. (2019a) showed that RNA polymerase clusters formed through large-scale nonspecific DNA–protein interactions in viral replication compartments could be easily mistaken for LLPS condensates. Resolving TF cluster dynamics at a molecular scale is therefore of paramount importance to distinguish between mechanisms driving localized TF assembly. Finally, emergent properties of liquid condensates formed at SE clusters are difficult to reconcile with the additive rather than collective impact of enhancers in SE clusters, as seen in the case of globin genes (Hay et al. 2016).
CHROMATIN ARCHITECTURE IN DEVELOPMENT
Multiphasic Reorganization of Higher-Order Chromatin Structure During Gametogenesis
The 3D conformation of the genome is never more dynamic than during gametogenesis, zygote formation, and the early stages of embryogenesis. Formation of haploid gametes from primordial germ cells, creation of zygotes through the fusion of male and female pronuclei, and zygotic genome activation (ZGA) during maternal-to-zygotic transition (MZT) are marked by rapid transformations in the epigenome and in global gene expression, providing unique opportunities to probe causality between transcription and higher-order chromatin structure (Vallot & Tachibana 2020). However, only recently have the structural conformations of paternal and maternal genomes been determined and their contributions to zygotic genome organization analyzed. In 2017, three independent studies revealed novel details regarding chromatin reconfiguration during developmental transitionsusing Hi-C analysis of libraries made from a single nucleus (Flyamer et al. 2017) or from hundreds of cells via optimized low-input in situ Hi-C (Ke et al. 2017) and small-scale in situ Hi-C (Du et al. 2017).
Spermatogenesis: reprogramming chromatin structure through the stages of meiosis.
Precise knowledge of chromatin structure in gametes is essential to determine whether embryos configure their genomes de novo or inherit features of maternal and paternal chromatin. In Meiosis I, during the conversion of primary spermatocytes (4n) to secondary spermatocytes (2n), homologous chromosomes undergo dramatic changes in structure. In prophase, replicated homologous chromosomes begin to align and form a proteinaceous axial element between them (leptotene, stage 1). In zygotene (stage 2), a synaptonemal complex (SC) starts to form between homologs and is completed in pachytene (stage 3), during which DNA recombination occurs between homologous chromosomes. In diplotene (stage 4), the SC dissolves but the recombined homologs remain attached. In diakinesis (stage 5), paired recombined chromosomes condense. The nuclear membrane then breaks down in preparation for homolog alignment at the metaphase plate and subsequent segregation (Handel & Schimenti 2010). Segregation of homologous chromosomes in Meiosis I is followed by segregation of sister chromatids in Meiosis II, thereby producing haploid spermatids. The transition to round and then to elongated spermatids is marked by the replacement of core nuclear histones by protamine (Rathke et al. 2014).
During spermatogenesis, extensive transcriptome and epigenome reprogramming occurs (Jung et al. 2017) as well as stepwise restructuring of TADs and chromatin compartments. TADs dissolve progressively from leptotene to pachytene, followed by a gradual reestablishment phase during which TADs increase in strength and consolidate into an interphase-like organization in round spermatids and mature sperm (Y. Wang et al. 2019).
While large A/B compartments become depleted in pachytene chromosomes of Rhesus monkeys, fine-scale transcriptionally coupled A/B-like compartments (refined A/B) remain. This pattern is specific to autosomes. In male germlines, X and Y chromosomes undergo meiotic sexchromosome inactivation (MSCI) in which chromosome-wide condensation and transcriptional silencing occur (Turner 2007). X chromosomes lack refined A/B compartments, suggesting that these compartments are formed by the transcription-coupled partitioning of active and silent chromatin. However, two lines of evidence undermine a role for transcription in establishing refined A/B compartments in pachytene chromosomes: (a) complete absence of refined A/B compartments near X-chromosome genes that escape MSCI, and (b) overall retention of higher-order chromatin structure in mouse pachytene chromosomes after global inhibition of transcription with α-amanitin (Y. Wang et al. 2019).
The SC participates in establishing the unique architecture of pachytene chromosomes in part by preventing loop extrusion by cohesin and CTCF. Pachytene chromosomes of mice lacking the core SC component SYCP2 exhibit increased TAD strength, attenuation of refined A/B compartments, and concomitant restoration of conventional A/B compartments (Y. Wang et al. 2019). Pachytene chromatin structure of wild-type mice differs from the structure of mature sperm chromatin in its strong depletion of long-range interactions. Pachytene chromatin has an “interaction insulation boundary” (Y. Wang et al. 2019, p. 548) in which intrachromosomal contact frequency increases sharply beyond 1 Mb and is enriched over the 1–10 Mb range, indicating a metaphaselike loop-array organization. Thus, in pachytene chromosomes, as in metaphase chromosomes, the tethering of loop arrays along a proteinaceous scaffold may restrict chromatin motion, resulting in a loss of compartments and TADs. In the pachytene stage, highly transcribed loci exhibit long-range Hi-C contacts, indicating a propensity of transcriptionally active loci to congregate (Patel et al. 2019). However, how pachytene chromosomes balance the need for compaction with the need to maintain a relaxed and active transcriptional landscape is not yet known.
Mature sperm chromatin shares many features of interphase chromatin.
Contrary to the general belief that sperm are transcriptionally inert, ∼60% of sperm promoters are maintained in an active epigenetic state (Jung et al. 2017), raising the question of whether sperm that are poised in a transcriptionally permissive epigenetic state can instruct embryonic gene-expression programs postfertilization. A mature sperm has substantially different nuclear morphology than that of a somatic cell. Furthermore, sperm DNA is typically wrapped around protamines instead of histones, unlike somatic cells. Despite these differences, mature sperm chromatin bears a tremendous resemblance to interphase chromatin, including (a) well-positioned nucleosomes bearing active histone modifications at promoters, (b) a histone-modification profile emblematic of enhancers and SEs in embryonic and adult tissues, (c) CTCF and cohesin enrichment at genomic sitesthat are shared with ESCs, and (d) robust organization of the genome into TADs and A/B compartments (Jung et al. 2017). Whether interphase-like chromatin features of mature sperm chromosomes harbor instructions to rapidly shape the embryonic genome remains to be determined.
Multiphasic chromatin reconfiguration in developing oocytes.
Much like spermatogenesis, oogenesis involves multiphasic genome organization dynamics. Oocytes, termed germinal vesicles (GVs) when stalled at diplotene of Meiosis I, have typical interphase-like higher-order chromatin organization. Using a novel single-nucleus Hi-C protocol, Flyamer et al. (2017) studied the topological transformation of the mouse genome during the transition of GVs with immature nonsurrounded nucleoli (NSN) to mature surrounded nucleoli (SN). The SN stage differs from the NSN stage by the presence of a heterochromatin mass called the karyosphere, which includes a rim of condensed chromatin surrounding a central nucleolus-like body (Bogolyubova & Bogolyubov 2020). Transcriptional silencing, which marks the transition of NSN to SN oocytes, is accompanied by substantial attenuation of TADs, loops, and compartments. Following a surge in luteinizing hormone levels (Arroyo et al. 2020), mature GVs reenter the meiotic cycle. GV breakdown occurs, followed by completion of Meiosis I, entry into Meiosis II, and subsequent metaphase II arrest. Unlike mature sperm, for which TADs and compartments are reestablished after Meiosis I prophase, oocytes paused at metaphase II lack TADs and compartments (Du et al. 2017). Metaphase II oocytes instead have a metaphase-like, uniform contact pattern across entire chromosomes. Moreover, genome organization in metaphase II oocytes, much like genome organization in pachytene sperm, displays an “interaction insulation boundary” (Du et al. 2017, p. 232) in which intrachromosomal contact frequencies decrease sharply beyond 4 MB. Metaphase II oocyte chromatin is arranged into a linear array of loops, albeit with a shorter interaction unit (4 Mb) than that of human mitotic chromatin (10 Mb). Whether loop extrusion by condensins I and II and axial scaffolding help organize the metaphase II oocyte genome is currently unknown.
Higher-Order Chromatin Reorganization During Zygotic Genome Activation and Early Embryogenesis
The transition from a totipotent zygote to a lineage-committed embryo begins with ZGA, which contributes to the complex developmental shift known as the maternal-to-zygotic transition (MZT). Questions at the core of understanding MZT and early embryonic development include the following:
To what extent and for how long are the topological signatures and chromatin marks of male and female gametes retained in the developing embryo?
What is the temporal sequence of epigenetic reprogramming, chromatin topological remodeling, and transcriptional reprogramming in early embryos?
What are the functional interdependencies of these early remodeling steps?
Are topologically associated domains part of the zygotic genome architecture?
To track chromatin reorganization during the oocyte-to-zygote transition and to assess whether chromatin structure of maternal and paternal genomes in mouse zygotes is inherently different, Flyamer et al. (2017) examined chromatin organization in paternal and maternal nuclei from zygotes using single-nucleus Hi-C. The zygotic genome contains twice the number of contacts as do somatic cells, and TADs have similar strengths in maternal and paternal nuclei. These findings were in stark contrast to the results of Ke et al. (2017) and Du et al. (2017), who showed through low-input bulk Hi-C analysis that TAD structures were considerably weaker in zygotes and two-cell-stage embryos and that the paucity of TAD peaks was not restricted to any particular zygotic cell cycle stage. However, upon reanalysis of bulk Hi-C data from Ke et al. (2017) and Du et al. (2017), Gassler et al. (2017) detected TADs in zygotes and two-cell-stage embryos and attributed their blurring of aggregate signals to the lack of rescaling and differences in data normalization strategies.
Maternal and paternal genomes differ in the extent of cohesin-driven looping in zygotes.
Detection of TADs and loops in zygotes and two-cell-stage embryos begets the question of whether cohesin-mediated loop extrusion is an essential mechanism for genome organization in early-stage embryos. Gassler et al. (2017) showed that Scc1-cohesin depletion caused nearcomplete loss of TADs and loops in both maternal and paternal nuclei. Similar results were obtained in zebrafish, in which cohesin depletion resulted in severely compromised ZGA and higher-order chromatin structure (Kaaij et al. 2018). Depletion of Wapl, the cohesin-release factor in mice, produced larger loops, albeit to different extents in paternal and maternal genomes, suggesting differences in the density and processivity of cohesin in the two genomes. However, these differences diminished as zygotes progressed toward ZGA. These results suggest that cohesin-mediated loop extrusion is important for genome organization in early-stage embryos.
A/B compartmentalization in early embryos.
While Ke et al. (2017) and Du et al. (2017) differ from Flyamer et al. (2017) and Gassler et al. (2017) in their conclusions regarding the presence of TADs and chromatin loops in zygotes, they all agree on a monotonic stepwise increase in TAD strength during early embryonic development and on parent-specific differences in A/B compartment organization. While paternal chromosomes show clear segregation of A/B compartments by pronuclear stage 3, formation of maternal A/B compartments occurs later (Du et al. 2017). Unlike in mice, human A/B compartmentalization does not become apparent until the morula stage and is fully established only at the blastocyst stage (Chen et al. 2019). A substantial fraction of the human genome that is enriched in genes controlling developmental processes undergoes A/B compartment switching during transition from the morula to the 6-week-stage embryo (Chen et al. 2019).
Does zygotic genome activation play a direct role in establishing topologically associated domains?
ZGA is the major turning point in embryonic development, when the maternal regulatory machinery is replaced by de novo embryonic regulators. To test if zygotic transcription during ZGA plays a role in chromatin reorganization, several studies (Du et al. 2017, Flyamer et al. 2017, Ke et al. 2017) analyzed genome structure in early mouse embryos treated with transcriptional inhibitors. No studies found a substantive role for ZGA in early murine embryonic chromatin reconfiguration. In zebrafish, genome organization follows a unique timeline in which prominent compartments and TAD structures that are present before the onset of zygotic transcription are lost upon ZGA, followed by their reestablishment through early development (Kaaij et al. 2018). Unlike in mice, Drosophila, and zebrafish, for which transcription is dispensable for early embryonic chromatin organization, in two-cell-stage human embryos, transcriptional inhibition blocks TAD establishment, suggesting a role for ZGA, and more specifically CTCF expression during ZGA, in promoting TADs (Chen et al. 2019).
Replication Domains: Developmentally Regulated Coupling of Replication-Timing and Three-Dimensional Chromatin Interactions
Unlike transcription inhibition, replication inhibition by treatment of two-cell-stage mouse embryos with the replication inhibitor aphidicolin completely abolished TAD structures, suggesting a definitive role of replication in establishing higher-order structure during early embryogenesis (Ke et al. 2017). In the last decade, a link between topological organization of the genome and replication-timing (RT) has emerged. Linear stretches of the genome spanning 400 to 800 Kb, dubbed replication domains (RDs), undergo synchronous replication due to simultaneous initiation at multiple intradomain origins called replicon clusters (Jackson & Pombo 1998); see the sidebar titled Replication and Chromatin: An Effector-Responder Interaction Module.
While RDs in specific cell types have highly stable boundaries, replication-timing of RDs in half of the genome switches between early and late stages during cell-fate transitions. This result is consistent with early- and late-replicating chromatin being highly correlated with A and B compartments, respectively (Ryba et al. 2010), which themselves switch during ESC differentiation. Pope et al. (2014) showed that TADs and RDs bear an almost one-to-one correspondence. Constant-timing regions (CTRs), which span a 1–5 Mb genomic stretch and contain multiple adjacent RDs with similar RT, can be interrupted by timing transition regions (TTRs). TTRs represent genomic regions that divide early and late CTRs. In mouse ESCs, TAD boundaries delineate junctions between early CTRs and TTRs, whereas TTRs and adjacent late CTRs share the same TAD (Pope et al. 2014). ESC differentiation is accompanied by an increase in the synchronous replication of adjacent RDs, thereby consolidating the genome into larger units of early and late CTRs (Hiratani et al. 2008). Moreover, the timing of TAD formation upon mitotic exit matches closely with the establishment of the replication program in early G1.
While RT and genome organization are highly correlated in time, depletion of CTCF or cohesin and perturbation of TAD structure by deleting TAD boundary sequences had no impact on the replication program. The link between RT and genome architecture was clarified by the identification within TADS of cis-acting elements termed early-replication cis-elements (ERCEs) that were critical for preservation of RT (Sima et al. 2019).
While disruption of TAD boundaries or depletion of CTCF had no effect on RT, disruption of ERCEs perturbed TAD organization, early-replication-coupled chromatin compartmentalization, and transcription. These results suggest a replication-based genome organization model in which ERCE-driven loops can organize TADs independently of canonical boundary elements and CTCF. While ∼20% of ERCE sites overlap with SEs in mESCs, depletion of YY1, which mediates E-P loops, did not alter RT, suggesting that the role of ERCEs in RT is independent of their role in transcription. Clarifying the link between ERCEs, RT, and chromatin organization will require identification of trans-acting factors that bind ERCEs and mediate looping.
Megadomains: Unique Chromosomal Architecture Established During X-Chromosome Inactivation
Mammalian sex chromosomes determine sexual fate and also control neurodevelopment and cognition (Skuse 2005). To compensate for the difference in X-chromosome dose between males (XY) and females (XX), one female X chromosome is randomly inactivated. This random Xchromosome inactivation (XCI) is mediated by the long noncoding RNA (lncRNA) Xist (Brown et al. 1991) and is one of the most intensively studied models of epigenetic reprogramming (Lee 2011; Galupa & Heard 2015, 2018; Jégu et al. 2017; Loda & Heard 2019). Unlike MSCI, XCI is zygotically driven and is established early in development, at the epiblast stage in mouse embryos (Lee 2011). Here we focus on the connection between XCI and inactive X-chromosome topology and discuss evidence suggesting a potential role for phase separation in XCI.
XCI is driven by a master cis control element called the X-inactivation center (XIC) and a set of trans-acting proteins and lncRNAs. The functional unit of the XIC is a 450-Kb region of the X chromosome that is organized into two neighboring TADs and includes genes encoding lncRNAs. The XIC has been implicated in the stochastic selection of X chromosomes for inactivation (Monkhorst et al. 2008), in X-chromosome counting (Avner & Heard 2001), and in the control of Xist expression.
The bipartite organization of the XIC into two adjacent TADs is critical for the regulation of Xist expression. The Xist promoter and most of its positive regulators are embedded in one TAD, while most of its negative cis regulators, including the antisense regulator Tsix, are located in the adjacent TAD (Galupa & Heard 2018). When promoters of Xist and Tsix were transferred to their noncognate TADs, the promoters lost their native interactions within the XIC and caused the ectopic expression of Xist (van Bemmel et al. 2019). The promoter of the Linx lncRNA resides inside the Tsix TAD, yet it represses Xist expression in a Tsix-independent manner, suggesting a role for inter-TAD interactions in controlling Xist expression (Galupa et al. 2020), contrary to the common perception of TADs being strong inhibitors of cross-boundary interactions.
Super-resolution imaging has showed that differences in shape between active X chromosomes (Xas) and inactive X chromosomes (Xis) are due to differences in higher-order chromatin structure rather than to differences in nucleosome compaction (Smeets et al. 2014). Xis differ topologically from Xas in three critical ways. (a) The Xa maintains an autosome-like TAD organization, but Xi shows global erasure of TADs and is instead organized into two megadomains that exhibit frequent intrachromosomal interactions (Giorgetti et al. 2016). (b) The boundary between the megadomains in an Xi is defined by the macrosatellite repeat element DXZ4, which impairs megadomain organization when deleted. (c) Xi chromatin exhibits long-distance looping over megabase-length scales, termed superloops (Rao et al. 2014). Superloops occur between Xist, DXZ4, and FIRRE loci, all of which encode lncRNAs. Loops between DXZ4 and FIRRE can be 25 Mb in length, far longer than other loops in the mammalian genome.
Topological asymmetry between Xis and Xas may arise from the preferential binding of CTCF and cohesin to the Xas (Minajigi et al. 2015). The allele-specific binding is prominent for cohesin, which binds to ∼600 unique sites on an Xa and only ∼20 unique sites on an Xi. This allele specificity is lost upon conditional deletion of Xist. Both DXZ4, which is critical for megadomain formation, and FIRRE, which mediates intra-megadomain interactions, are dispensable for XCI (Froberg et al. 2018), suggesting that megadomain formation and XCI are decoupled. Indeed, mice lacking FIRRE and DXZ4 have functional XCI (Andergassen et al. 2019). Xist collaborates with the SMC variant called SMCHD1 (structural-maintenance-of-chromosomes hinge domain–containing 1) to attenuate TADs through the exclusion of CTCF and cohesin. Without SMCHD1, CTCF and cohesin bind similarly to Xis and Xas (C. -Y. Wang et al. 2018).
While the mechanisms shaping Xi topology during the early stages of XCI (Zylicz et al. 2019)˙ are not fully understood, the timeline of epigenetic reprogramming following early Xist accumulation provides some insights. The preloaded histone deacetylase HDAC3 is responsible for loss of H3K27ac immediately upon loading of Xist. Rapid accumulation of H2AK119Ub occurs at X-chromosome locations proximal to Xist RNA entry sites (XESs) in a PRC1-dependent manner. These early sites of accumulation coincide with H3K27me3 deposition by PcG. The preferential enrichment of H3K27me3 in spatial proximity to XESs suggests that PcG distribution along the X chromosome may instruct the early Xi topology required for Xist spreading and subsequent transcriptional silencing (Zylicz et al. 2019).
Phase separation has been proposed as a mechanism for XCI (Cerase et al. 2019). While this hypothesis needs rigorous testing, at least three strong parallels exist between XCI assembly and membrane-less bodies, such as paraspeckles and stress granules. (a) The size and morphology of these assemblies are similar; (b) Xist contains structured repeats like other scaffold RNAs that drive formation of RNA–protein condensates; (c) and both Xist and the lncRNA Neat1, the primary structural component of paraspeckles, interact with many IDPs (Cerase et al. 2019).
Role of Genome Architecture in Neural Development and Cognition
In recent years, a common theme has emerged from studies on genome organization in neurons. The 3D chromatin-contact network presents a tunable parameter in neurodevelopment and cognition. Neuronal activity-based restructuring of the 3D genome has also been implicated in learning and plasticity. In the next few sections, we discuss our current understanding of how dynamic changes in the 3D genome structure translate to specific changes in neural circuitry.
Neuronal enhancers: a model for activity-driven genome reconfiguration.
E-P interactions are at the core of cell-fate determination. Dynamics of E-P interactions are particularly complicated during neuronal-fate determination because neurons have much longer life spans than do other cell types and must have adaptable responses to different signals over time. Maintaining a stable fate while being synaptically plastic and adaptable to the changing nature of stimuli over a long lifetime imposes a unique set of constraints on gene expression and, by extension, on genome organization in neurons (Nord & West 2020). Unique subsets of E-P contacts, activated exclusively in different types of neurons in response to a universal stimulus, can initiate different gene-expression programs in different regions of the brain (Mardinly et al. 2016, Hrvatin et al. 2018). This form of permissive genome configuration, in which unique subsets of E-P pairs are poised for interaction in different types of neurons, sets the stage for rapid shifts in the transcriptional landscape upon the reception of a stimulus. Indeed, Lu et al. (2020) showed that chromatin loops rather than higher-order genomic compartments form the structural unit of neuronal differentiation. Neuronal differentiation involved strengthening of chromatin loops at 174 enhancer aggregates, which showed a progressive increase in strength from induced pluripotent stem cells to neurons and then to primary brain tissues, suggesting that enhancers aggregate and consolidate through the stages of neuronal maturation.
Enhancer activity can also be modulated during neuronal signaling through epigenetic means to alter E-P contact profiles. In mouse cortical neurons, activity-dependent recruitment of RNA PoI II to thousands of enhancers can cause bidirectional transcription of enhancer RNAs (eRNAs) (Kim et al. 2010). eRNAs promote transcription in neurons by several means, including release of transcriptional pauses (Schaukowitch et al. 2014) and acetylation (Ac) of H3K27 by histone acetyltransferases p300 and CBP (Kim et al. 2010, Bose et al. 2017). H3K27Ac then recruits BRD4 (Zippo et al. 2009), a primary mediator of chromatin organization through phase separation. Evidence of a direct role for eRNAs in chromosomal enhancer assembly through phase separation of enhancer-ribonucleoprotein complexes with condensins comes from studies of human breast cancer cells (Nair et al. 2019). Whether eRNAs play a direct role in mediating neuronal E-P contacts through genome reconfiguration by phase separation remains to be determined.
Three-dimensional genome reconfiguration during neural development.
Using ultra-deep Hi-C analysis of genome-wide chromosome contacts in models of neural differentiation and cortical development, Bonev et al. (2017) showed that neural differentiation involves changes in genome folding over multiple scales that occur through a variety of mechanisms. For example, loops demarcated by convergent CTCF-binding motifs grew in strength through differentiation, even though the chromatin-bound fraction of CTCF decreased during this time. The antithetical nature of these observations may be reconciled by the fact that the expression of cohesin-unloading factors also decreased during neural differentiation, thereby increasing loop size through enhancement of loop-extrusion processivity. Furthermore, bodies of genes with high levels of transcription interacted in cis as well as in trans to generate loop clusters, with the strength of interactions between gene bodies scaling with the frequency of splicing events per gene. The mechanistic basis of the correlation between splicing frequency and loop-interaction strength remains unclear. Whether a direct link exists between loop-interaction strength and cotranscriptional splicing is unknown. Splicing factors can cause phase separation at highly expressed genes associated with SEs (Guo et al. 2019).
Neuronal differentiation is accompanied by massive pruning of loops.
Loss and gain of contacts during 3D gene reconfiguration are common mechanisms for differential regulation of genes involved in cell-fate transitions. Alternate loop clusters within a TAD can provide a framework for differential gene regulation during cell-fate transitions. Rajarajan et al. (2018) showed that genomes of neural progenitor cells (NPCs) undergo extensive reconfiguration during the transition of these cells to neurons or glia. A prominent aspect of the NPC-to-neuron transition is the pruning of loops that harbor genes involved in proliferation, morphogenesis, and neurogenesis. This pruning reflects elimination of spatial contacts specific to the precursor stage. Similarly, the transition of NPCs to the nonneuronal glial lineage is accompanied by pruning of loops related to neuronal functions. Loss of short-range contacts was accompanied by gain of long-range (>100–200 Kb) contacts. Although the transition of NPCs to neurons or glia generally conforms to the positive correlation between gene activity and chromatin looping, Rajarajan et al. (2018) found that postmitotic neurons and adult cerebral cortex had from 25% to 50% fewer genome-wide contacts than did NPCs and fetal cortex. The reason for such unexpectedly large-scale pruning of genome-wide spatial contacts during neural development remains unclear.
Rajarajan et al. (2018, p. 7) also found that the “schizophrenia-related chromosomal connectome” varied considerably between NPCs, glia, and neurons. For example, in NPCs, a schizophrenia locus located upstream of the clustered protocadherin gene locus, which resides in a 1. 2-Mb TAD with >70 genes, showed two sets of distinct loop clusters formed through interactions with sequences upstream or downstream of the risk locus. In neurons, the upstream loop bundle was pruned, but glia showed the inverse pattern (Rajarajan et al. 2018).
OUTLOOK
In the last decade, advances in chromatin-conformation-capture technologies, super-resolution imaging, single-cell genomics, and multiplexed FISH have greatly facilitated our understanding of spatial genome organization. Moreover, recent developments in high-throughput, multiscale, and multimodal imaging for high-resolution chromatin tracing and transcript analysis in situ now provide tremendous promise for resolving complex E-P interactions and for deciphering the link between cell-specific chromatin conformations and gene expression (Su et al. 2020, Zhuang 2021). However, these technologies only generate static snapshots of the genome. To generate associative maps of genome configuration and transcriptional states, as well as to track the dynamics of these associations through developmental stages, future work must focus on simultaneous spatiotemporal analysis of genome organization and transcription at endogenous loci. Simultaneous multicolor detection and prolonged tracking of genomic loci, coupled with live detection of nascent transcription of genes, will provide invaluable information about the relationship between chromatin-folding dynamics and transcriptional output. Targeting of nuclease-deficient Cas9 fused to numerous bright, sustainable fluorescent probes, such as the ArrayG tag (Ghosh et al. 2019) or the Sun tag (Tanenbaum et al. 2014), to specific genomic loci holds promise for prolonged and simultaneous tracking of multiple genomic sites at high spatiotemporal resolution. A recent strategy that combines computer vision and diffraction-limited optical microscopy to build nanoscale correlative maps of chromatin motion (Shaban et al. 2018, 2020; Agbleke et al. 2020) provides a method to track global dynamics of chromosomes at subpixel resolution.
These approaches are destined to reveal both new organizing principles for conventional chromosomes and new chromosome structures, such as in dinoflagellates (Marinov et al. 2020, Nand et al. 2020). Their chromosomes exist in a liquid crystalline state, lack conventional histone-based nucleosome structures, and remain highly condensed throughout the cell cycle.
Future directions for developing spatiotemporal maps of heritable changes in genome architecture will entail error-robust reconstruction of cell lineages in intact tissue. Recently, a surge has occurred in synthetic cellular-recording systems (Kretzschmar & Watt 2012, Woodworth et al. 2017, Sheth & Wang 2018, Baron & van Oudenaarden 2019, Chan et al. 2019, Salvador-Martínez et al. 2019), but their reliance on sequencing-based readout of heritable stochastic mutations precludes their use in deciphering the spatial context of lineages. The key to building fine-scale maps of genome architecture and transcription on a cell-by-cell basis in multicellular organisms without losing essential lineage and spatial information is to combine imaging-based clonal analysis (Frieda et al. 2017, Chow et al. 2020) in intact tissues or whole organisms with both imagingbased multiplexed analysis of transcriptional states (X. Wang et al. 2018, Eng et al. 2019, Rodriques et al. 2019) and chromatin-interaction maps (Mateo et al. 2019, Nguyen et al. 2020, Su et al. 2020).
CHROMOSOME COMPARTMENTS AND DOMAINS.
Chromosome organization arises from many factors, including the segregation of genomic DNA into active and inactive compartments and the compaction of chromatin into topological domains.
A/B Compartments
A/B compartments are multimegabase-scale compartments detected in Hi-C interaction matrices that are categorized into two classes based on whether they contain transcriptionally active open chromatin (A compartment) or transcriptionally repressed closed chromatin (B compartment). The A compartment is usually gene rich and is enriched with active histone marks such as H3K36me3, whereas the B compartment is gene poor and is enriched in repressive histone marks such as H3K9me3.
Topologically Associated Domains
Topologically associated domains (TADs) are megabase-scale blocks of DNA sequences detected in Hi-C interaction matrices that exhibit considerably higher frequencies of interactions between sequences within the block than outside the block. A TAD is demarcated by boundaries that are frequently enriched with CCCTC-binding factor (CTCF) and the SMC complex called cohesin.
CCCTC-Binding Factor
CTCF binds to DNA via its zinc-finger domains and plays diverse roles in gene regulation, including, but not limited to, transcriptional activation, transcriptional repression, and chromatin insulation. In its role as a chromatin architectural protein, CTCF collaborates with cohesin to create TADs through loop extrusion.
Compartmental Domains
Unlike A/B compartments, compartmental domains are genomic folding units smaller than TADs revealed through principal component analysis of high-resolution Hi-C data. Compartmental domains arise predominantly out of interactions between initiation and termination sites of transcriptionally active genes, resulting in gene loops. These domains can be embedded inside a CTCF-dependent loop, reside between loops, or partially overlap with adjacent loops.
ARCHITECTURAL PROTEINS THAT CREATE CHROMOSOME STRUCTURE UTILIZE CHROMATIN LOOP EXTRUSION.
Chromosome structure is organized through the action of specific structural maintenance of chromosomes (SMC) complexes.
The SMC cohesin complex is a multisubunit chromatin architectural protein complex that mediates sister chromatid cohesion and is required for chromosome segregation and DNA damage repair through homologous recombination. Cohesin is also one of the primary players in the generation of topologically associated domains (TADs).
SMC condensin complexes are large multisubunit chromatin architectural protein complexes that are critical for mitotic and meiotic chromosome assembly, compaction, and segregation. Condensins also play significant roles in regulating interphase chromosome structure and function. In their roles as chromatin architectural protein complexes, condensin I and II generate nested loop arrays in mitotic chromosomes and extrude DNA loops in vitro.
One of the most widely acclaimed models for the formation of topological domains is the loop-extrusion model. This model proposes that TADs are formed by the activity of loop-extruding factors such as cohesins, which progressively extrude large loops of chromatin through an ATP-dependent reeling of DNA until they dissociate from chromatin or encounter a barrier such as CCCTC-binding factor. Single-molecule experiments have provided direct visual evidence for the loop-extrusion process.
CHROMATIN ORGANIZATION THROUGH PHASE SEPARATION.
Genome compartmentalization into A/B compartments depends on the local chromatin state. Recently, phase separation of chromosomal regions with similar histone and DNA modification profiles has emerged as a leading mechanism that drives genome compartmentalization. Two primary modes have been invoked for phase separation, liquid–liquid phase separation (LLPS) and polymer–polymer phase separation (PPPS).
LLPS is a process that spontaneously sorts a homogeneous mixture of molecules into two separate liquid phases, which differ in the densities of their constituent molecules. For LLPS of biomolecules to occur, multivalent weak interactions between molecules are critical.
PPPS is a type of phase transition in which an ordered collapsed globule is formed via the cross-linking of chromatin fibers by chromatin-binding proteins.
Phase separation has also been implicated in the more targeted control of gene expression through its role in assembling super-enhancer (SE) clusters. SEs are arrays of enhancers in close genomic proximity spanning a large genomic sequence block. SEs show considerably higher levels of Med1, RNA polymerase II, and histone acetyltransferases p300 and CBP than do canonical enhancers and also bear substantially higher levels of H3K27ac, H3K4me2, and H3K4me1 histone marks. While certain SEs form liquid-like condensates, whether enhancers in SEs act synergistically remains contentious.
REPLICATION AND CHROMATIN: AN EFFECTOR-RESPONDER INTERACTION MODULE.
Recent experiments have revealed that the three-dimensional genome organization plays a critical role in the selection of replication origins and in the specification of temporal order of replication at these origins. Eukaryotic DNA replication follows a temporal pattern known as replication-timing (RT) in which different regions of the genome are replicated in a specific spatiotemporal order. The unit of the RT program is called a replication domain, which is composed of several replication origins firing in near synchrony and corresponds closely to a topologically associated domain. Constant-timing regions (CTRs) are multimegabase-scale regions of the genome that contain several adjacent replicons having synchronized activity at a particular stage of S phase (e. g., early CTRs and late CTRs). CTRs are punctuated by timing transition regions, which are genomic regions that bridge early-replicating regions to late-replicating regions and exhibit gradients of progressive delay in replication-timing, ranging from their borders with the early-replicating regions to their borders with the late-replication regions.
Insulation score: a metric for determining TAD boundaries; local minima on the insulation profile represent a sharp increase in insulation, which denotes a TAD boundary
Zygotic genome activation (ZGA): a multiphasic switch in the transcriptional program of an embryo during the maternal-to-zygotic transition in which different sets of embryonic genes are turned on in waves in close coordination with the degradation of maternal RNAs
ACKNOWLEDGMENTS
This work was supported by funding to B. J. M. from the Howard Hughes Medical Institute and a grant to B. J. M. from the National Institutes of Health (R35GM131845).
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Glossary
- Insulation score
a metric for determining TAD boundaries; local minima on the insulation profile represent a sharp increase in insulation, which denotes a TAD boundary
- Zygotic genome activation (ZGA)
a multiphasic switch in the transcriptional program of an embryo during the maternal-to-zygotic transition in which different sets of embryonic genes are turned on in waves in close coordination with the degradation of maternal RNAs
LITERATURE CITED
- Agbleke AA, Amitai A, Buenrostro JD, Chakrabarti A, Chu L, et al. 2020. Advances in chromatin and chromosome research: perspectives from multiple fields. Mol. Cell 79(6):881–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S. 2017. Phase separation in biology. Curr. Biol 27(20):R1097–102 [DOI] [PubMed] [Google Scholar]
- Alipour E, Marko JF. 2012. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 40(22):11202–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andergassen D, Smith ZD, Lewandowski JP, Gerhardinger C, Meissner A, Rinn JL. 2019. In vivo firre and Dxz4 deletion elucidates roles for autosomal gene regulation. eLife 8:e47214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EC, Frankino PA, Higuchi-Sanabria R, Yang Q, Bian Q, et al. 2019. X chromosome domain architecture regulates Caenorhabditis elegans lifespan but not dosage compensation. Dev. Cell 51(2):192–207. e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo A, Kim B, Yeh J. 2020. Luteinizing hormone action in human oocyte maturation and quality: signaling pathways, regulation, and clinical impact. Reprod. Sci 27(6):1223–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avner P, Heard E. 2001. X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet 2(1):59–67 [DOI] [PubMed] [Google Scholar]
- Banigan EJ, Mirny LA. 2020. Loop extrusion: theory meets single-molecule experiments. Curr. Opin. Cell Biol 64:124–38 [DOI] [PubMed] [Google Scholar]
- Banigan EJ, van den Berg AA, Brandão HB, Marko JF, Mirny LA. 2020. Chromosome organization by one-sided and two-sided loop extrusion. eLife 9:e53558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri M, Chotalia M, Fraser J, Lavitas LM, Dostie J, et al. 2012. Complexity of chromatin folding is captured by the strings and binders switch model. PNAS 109(40):16173–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron CS, van Oudenaarden A. 2019. Unravelling cellular relationships during development and regeneration using genetic lineage tracing. Nat. Rev. Mol. Cell Biol 20(12):753–65 [DOI] [PubMed] [Google Scholar]
- Beagan JA, Phillips-Cremins JE. 2020. On the existence and functionality of topologically associating domains. Nat. Genet 52(1):8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Q, Anderson EC, Yang Q, Meyer BJ. 2020. Histone H3K9 methylation promotes formation of genome compartments in Caenorhabditis elegans via chromosome compaction and perinuclear anchoring. PNAS 117(21):11459–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu B, Mateo LJ, Su JH, Sinnott-Armstrong NA, Parker M, et al. 2018. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362(6413):eaau1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogolyubova I, Bogolyubov D. 2020. Heterochromatin morphodynamics in late oogenesis and early embryogenesis of mammals. Cells 9(6):1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, et al. 2018. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175(7):1842–55. e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, et al. 2017. Multiscale 3D genome rewiring during mouse neural development. Cell 171(3):557–72. e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose DA, Donahue G, Reinberg D, Shiekhattar R, Bonasio R, Berger SL. 2017. RNA binding to CBP stimulates histone acetylation and transcription. Cell 168(1–2):135–49. e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T. 1909. Die Blastomerenkerne von Ascaris megalocephala und die Theorie der Chromosomenindivid-ualität [The blastomeric nuclei of Ascaris megalocephala and the theory of chromosome individuality]. Arch. Zellforsch 3:181–268 [Google Scholar]
- Brackley CA, Johnson J, Kelly S, Cook PR, Marenduzzo D. 2016. Simulated binding of transcription factors to active and inactive regions folds human chromosomes into loops, rosettes and topological domains. Nucleic Acids Res. 44(8):3503–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brejc K, Bian Q, Uzawa S, Wheeler BS, Anderson EC, et al. 2017. Dynamic control of X chromosome conformation and repression by a histone H4K20 demethylase. Cell 171(1):85–102. e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, et al. 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349(6304):38–44 [DOI] [PubMed] [Google Scholar]
- Cattoni DI, Gizzi AMC, Georgieva M, Di Stefano M, Valeri A, et al. 2017. Single-cell absolute contact probability detection reveals chromosomes are organized by multiple low-frequency yet specific interactions. Nat. Commun 8(1):1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalheiro GR, Pollex T, Furlong EE. 2021. To loop or not to loop: what is the role of TADs in enhancer function and gene regulation? Curr. Opin. Genet. Dev 67:119–29 [DOI] [PubMed] [Google Scholar]
- Cerase A, Armaos A, Neumayer C, Avner P, Guttman M, Tartaglia GG. 2019. Phase separation drives X-chromosome inactivation: a hypothesis. Nat. Struct. Mol. Biol 26(5):331–34 [DOI] [PubMed] [Google Scholar]
- Chan MM, Smith ZD, Grosswendt S, Kretzmer H, Norman TM, et al. 2019. Molecular recording of mammalian embryogenesis. Nature 570(7759):77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ke Y, Wu K, Zhao H, Sun Y, et al. 2019. Key role for CTCF in establishing chromatin structure in human embryos. Nature 576(7786):306–10 [DOI] [PubMed] [Google Scholar]
- Chen X, Wu X, Wu H, Zhang M. 2020. Phase separation at the synapse. Nat. Neurosci 33(3):301–10 [DOI] [PubMed] [Google Scholar]
- Cho WK, Spille JH, Hecht M, Lee C, Li C, et al. 2018. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361(6400):412–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, et al. 2018. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361(6400):eaar2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow K-H, Budde M, Granados A, Cabrera M, Yoon S, et al. 2020. Imaging cell lineage with a synthetic digital recording system. bioRxiv 958678. 10.1101/2020.02.21.958678 [DOI] [PubMed] [Google Scholar]
- Chuang PT, Albertson DG, Meyer BJ. 1994. DPY-27: a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell 79(3):459–74 [DOI] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, et al. 2015. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523(7559):240–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer M, Grasser F, Lanctôt C, Müller S, Neusser M, et al. 2012. Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. In The Nucleus, Volume 1: Nuclei and Subnuclear Components, ed. Hancock R, pp. 205–39. Totowa, NJ: Humana; [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. 2006. Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. The concept of chromosome territories falls in disgrace. Eur. J. Histochem 50(4):223–72 [PubMed] [Google Scholar]
- Csankovszki G, Collette K, Spahl K, Carey J, Snyder M, et al. 2009. Three distinct condensin complexes control C. elegans chromosome dynamics. Curr. Biol 19(1):9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. 2002. Solvent mediated interactions in the structure of the nucleosome core particle at 1. 9 A resolution. J. Mol. Biol 319(5):1097–113 [DOI] [PubMed] [Google Scholar]
- Davidson IF, Bauer B, Goetz D, Tang W, Wutz G, Peters JM. 2019. DNA loop extrusion by human cohesin. Science 366(6471):1338–45 [DOI] [PubMed] [Google Scholar]
- Dekker J, Mirny L. 2016. The 3D genome as moderator of chromosomal communication. Cell 164(6):1110–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pierro M, Potoyan DA, Wolynes PG, Onuchic JN. 2018. Anomalous diffusion, spatial coherence, and viscoelasticity from the energy landscape of human chromosomes. PNAS 115(30):7753–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, et al. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485(7398):376–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zheng H, Huang B, Ma R, Wu J, et al. 2017. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547(7662):232–35 [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Laemmli UK. 1983. Architecture of metaphase chromosomes and chromosome scaffolds. J. Cell Biol 96(1):84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeftens JM, Kapoor M, Brangwynne CP. 2020. Epigenetic memory as a time integral over prior history of Polycomb phase separation. bioRxiv 254706. 10.1101/2020.08.19.254706 [DOI] [Google Scholar]
- Eng C-HL, Lawson M, Zhu Q, Dries R, Koulena N, et al. 2019. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 568(7751):235–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F, Rademacher A, Vlijm R, Tünnermann J, Frank L, et al. 2020. Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol. Cell 78(2):236–49. e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F, Rippe K. 2018. Formation of chromatin subcompartments by phase separation. Biophysical J. 114(10):2262–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinola SM, Götz M, Fiche J-B, Bellec M, Houbron C, et al. 2020. Cis-regulatory chromatin loops arise before TADs and gene activation, and are independent of cell fate during development. bioRxiv 191015. 10.1101/2020.07.07.191015 [DOI] [PubMed] [Google Scholar]
- Finn EH, Pegoraro G, Brandão HB, Valton AL, Oomen ME, et al. 2019. Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 176(6):1502–15. e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, et al. 2016. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529(7584):110–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov SV, et al. 2017. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544(7648):110–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieda KL, Linton JM, Hormoz S, Choi J, Chow K-HK, et al. 2017. Synthetic recording and in situ readout of lineage information in single cells. Nature 541(7635):107–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froberg JE, Pinter SF, Kriz AJ, Jégu T, Lee JT. 2018. Megadomains and superloops form dynamically but are dispensable for X-chromosome inactivation and gene escape. Nat. Commun 9(1):5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Abdennur N, Imakaev M, Goloborodko A, Mirny LA. 2017. Emerging evidence of chromosome folding by loop extrusion. Cold Spring Harb. Symp. Quant. Biol 82:45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. 2016. Formation of chromosomal domains by loop extrusion. Cell Rep. 15(9):2038–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Pollard KS. 2019. Chromatin features constrain structural variation across evolutionary timescales. PNAS 116(6):2175–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galupa R, Heard E. 2015. X-chromosome inactivation: new insights into cis and trans regulation. Curr. Opin. Genet. Dev 31:57–66 [DOI] [PubMed] [Google Scholar]
- Galupa R, Heard E. 2018. X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu. Rev. Genet 52:535–66 [DOI] [PubMed] [Google Scholar]
- Galupa R, Nora EP, Worsley-Hunt R, Picard C, Gard C, et al. 2020. A conserved noncoding locus regulates random monoallelic Xist expression across a topological boundary. Mol. Cell 77(2):352–67. e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, et al. 2018. Real-time imaging of DNA loop extrusion by condensin. Science 360(6384):102–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassler J, Brandão HB, Imakaev M, Flyamer IM, Ladstätter S, et al. 2017. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J. 36(24):3600–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Jankowski A, Meiers S, Viales RR, Korbel JO, Furlong EEM. 2019. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat. Genet 51(8):1272–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh RP, Franklin JM, Draper WE, Shi Q, Beltran B, et al. 2019. A fluorogenic array for temporally unlimited single-molecule tracking. Nat. Chem. Biol 15(4):401–9 [DOI] [PubMed] [Google Scholar]
- Gibcus JH, Samejima K, Goloborodko A, Samejima I, Naumova N, et al. 2018. A pathway for mitotic chromosome formation. Science 359(6376):eaao6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, et al. 2016. Structural organization of the inactive X chromosome in the mouse. Nature 535(7613):575–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gligoris TG, Scheinost JC, Bürmann F, Petela N, Chan KL, et al. 2014. Closing the cohesin ring: structure and function of its Smc3-kleisin interface. Science 346(6212):963–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golfier S, Quail T, Kimura H, Brugués J. 2020. Cohesin and condensin extrude DNA loops in a cell-cycle dependent manner. eLife 9:e53885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall’Agnese A, et al. 2019. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572(7770):543–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel MA, Schimenti JC. 2010. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet 11(2):124–36 [DOI] [PubMed] [Google Scholar]
- Hanssen LLP, Kassouf MT, Oudelaar AM, Biggs D, Preece C, et al. 2017. Tissue-specific CTCF-cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat. Cell Biol 19(8):952–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy WB. 1899. On the structure of cell protoplasm: Part I. The structure produced in a cell by fixative and post-mortem change. The structure of colloidal matter and the mechanism of setting and of coagulation. J. Physiol 24(2):158–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl TA, Smith HF, Bosco G. 2008. Chromosome alignment and transvection are antagonized by condensin II. Science 322(5906):1384–87 [DOI] [PubMed] [Google Scholar]
- Hassler M, Shaltiel IA, Haering CH. 2018. Towards a unified model of SMC complex function. Curr. Biol 28(21):R1266–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay D, Hughes JR, Babbs C, Davies JOJ, Graham JB, et al. 2016. Genetic dissection of the α-globin super-enhancer in vivo. Nat. Genet 48(8):895–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand EM, Dekker J. 2020. Mechanisms and functions of chromosome compartmentalization. Trends Biochem. Sci 45(5):385–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. 1995. Biochemical and genetic dissection of mitotic chromosome condensation. Trends Biochem. Sci 20(9):357–61 [DOI] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, et al. 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLOS Biol. 6(10):2220–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintrau AS, Day DS, Valton AL, Bak RO, et al. 2016. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351(6280):1454–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S, Hochbaum DR, Nagy MA, Cicconet M, Robertson K, et al. 2018. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat. Neurosci 21(1):120–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh THS, Cattoglio C, Slobodyanyuk E, Hansen AS, Rando OJ, et al. 2020. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol. Cell 78(3):539–53. e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Jülicher F. 2014. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol 30:39–58 [DOI] [PubMed] [Google Scholar]
- Ing-Simmons E, Vaid R, Mannervik M, Vaquerizas JM. 2020. Independence of 3D chromatin conformation and gene regulation during Drosophila dorsoventral patterning. bioRxiv 186791. 10.1101/2020.07.07.186791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. 1998. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol 140(6):1285–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jégu T, Aeby E, Lee JT. 2017. The X chromosome in space. Nat. Rev. Genet 18(6):377–89 [DOI] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, et al. 2013. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503(7475):290–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka K, Terasawa M, Ogawa H, Ogawa T, Horiuchi T. 2006. Condensin loaded onto the replication fork barrier site in the rRNA gene repeats during S phase in a FOB1-dependent fashion to prevent contraction of a long repetitive array in Saccharomyces cerevisiae. Mol. Cell. Biol 26(6):2226–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YH, Sauria MEG, Lyu X, Cheema MS, Ausio J, et al. 2017. Chromatin states in mouse sperm correlate with embryonic and adult regulatory landscapes. Cell Rep. 18(6):1366–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaaij LJT, van der Weide RH, Ketting RF, de Wit E. 2018. Systemic loss and gain of chromatin architecture throughout zebrafish development. Cell Rep. 24(1):1–10. e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Xu Y, Chen X, Feng S, Liu Z, et al. 2017. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell 170(2):367–81. e20 [DOI] [PubMed] [Google Scholar]
- Kempfer R, Pombo A. 2020. Methods for mapping 3D chromosome architecture. Nat. Rev. Genet 21(4):207–26 [DOI] [PubMed] [Google Scholar]
- Kent S, Brown K, Yang C, Alsaihati N, Tian C, et al. 2020. Phase-separated transcriptional condensates accelerate target-search process revealed by live-cell single-molecule imaging. Cell Rep. 33(2):108248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Kerssemakers J, Shaltiel IA, Haering CH, Dekker C. 2020. DNA-loop extruding condensin complexes can traverse one another. Nature 579(7799):438–42 [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465(7295):182–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Shi Z, Zhang H, Finkelstein IJ, Yu H. 2019. Human cohesin compacts DNA by loop extrusion. Science 366(6471):1345–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. 1974. Chromatin structure: a repeating unit of histones and DNA. Science 184(4139):868–71 [DOI] [PubMed] [Google Scholar]
- Kragesteen BK, Spielmann M, Paliou C, Heinrich V, Schöpflin R, et al. 2018. Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat. Genet 50(10):1463–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krefting J, Andrade-Navarro MA, Ibn-Salem J. 2018. Evolutionary stability of topologically associating domains is associated with conserved gene regulation. BMC Biol. 16:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar K, Watt FM. 2012. Lineage tracing. Cell 148(1–2):33–45 [DOI] [PubMed] [Google Scholar]
- Lappala A, Terentjev EM. 2013. “Raindrop” coalescence of polymer chains during coil-globule transition. Macromolecules 46(3):1239–47 [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, et al. 2017. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547(7662):236–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau MS, Schwartz MG, Kundu S, Savol AJ, Wang PI, et al. 2017. Mutation of a nucleosome compaction region disrupts Polycomb-mediated axial patterning. Science 355(6329):1081–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. 2011. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat. Rev. Mol. Cell Biol 12(12):815–26 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326(5950):289–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loda A, Heard E. 2019. Xist RNA in action: past, present, and future. PLOS Genet. 15(9):e1008333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HS, Powell G, Greenaway S, Mallon AM, Lindgren CM, Simon MM. 2020. Making sense of the linear genome, gene function and TADs. bioRxiv 316786. 10.1101/2020.09.28.316786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Liu X, Huang W-K, Giusti-Rodríguez P, Cui J, et al. 2020. Robust Hi-C maps of enhancer-promoter interactions reveal the function of non-coding genome in neural development and diseases. Mol. Cell 79(3):521–34. e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161(5):1012–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luppino JM, Joyce EF. 2020. Single cell analysis pushes the boundaries of TAD formation and function. Curr. Opin. Genet. Dev 61:25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL. 2011. Biogenesis and function of nuclear bodies. Trends Genet. 27(8):295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinly AR, Spiegel I, Patrizi A, Centofante E, Bazinet JE, et al. 2016. Sensory experience regulates cortical inhibition by inducing IGF1 in VIP neurons. Nature 531(7594):371–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinov G, Trevino A, Xiang T, Kundaje A, Grossman A, Greenleaf W. 2020. Transcription-dependent domain-scale 3D genome organization in dinoflagellates. bioRxiv 181685. 10.1101/2020.07.01.181685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko JF. 2009. Linking topology of tethered polymer rings with applications to chromosome segregation and estimation of the knotting length. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 79(5 Part 1):051905. [DOI] [PubMed] [Google Scholar]
- Marsden MPF, Laemmli UK. 1979. Metaphase chromosome structure: evidence for a radial loop model. Cell 17(4):849–58 [DOI] [PubMed] [Google Scholar]
- Mateo LJ, Murphy SE, Hafner A, Cinquini IS, Walker CA, Boettiger AN. 2019. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568(7750):49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggen DT, Hansen AS, Teves SS, Marie-Nelly H, Hao Y, et al. 2019a. Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. eLife 8:e47098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggen DT, Mir M, Darzacq X, Tjian R. 2019b. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev. 33(23–24):1619–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BJ. 2018. Sex and death: from cell fate specification to dynamic control of X-chromosome structure and gene expression. Mol. Biol. Cell 29(22):2616–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michieletto D, Orlandini E, Marenduzzo D. 2016. Polymer model with epigenetic recoloring reveals a pathway for the de novo establishment and 3D organization of chromatin domains. Phys. Rev. X 6(4):041047 [Google Scholar]
- Minajigi A, Froberg JE, Wei C, Sunwoo H, Kesner B, et al. 2015. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349(6245):aab2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA, Imakaev M, Abdennur N. 2019. Two major mechanisms of chromosome organization. Curr. Opin. Cell Biol 58:142–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monkhorst K, Jonkers I, Rentmeester E, Grosveld F, Gribnau J. 2008. X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell 132(3):410–21 [DOI] [PubMed] [Google Scholar]
- Nair SJ, Yang L, Meluzzi D, Oh S, Yang F, et al. 2019. Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol 26(3):193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nand A, Zhan Y, Salazar OR, Aranda M, Voolstra CR, Dekker J. 2020. Chromosome-scale assembly of the coral endosymbiont Symbiodinium microadriaticum genome provides insight into the unique biology of dinoflagellate chromosomes. bioRxiv 182477. 10.1101/2020.07.01.182477 [DOI] [Google Scholar]
- Narendra V, Rocha PP, An D, Raviram R, Skok JA, et al. 2015. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347(6225):1017–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HQ, Chattoraj S, Castillo D, Nguyen SC, Nir G, et al. 2020. 3D mapping and accelerated super-resolution imaging of the human genome using in situ sequencing. Nat. Methods 17(8):822–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, et al. 2017. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169(5):930–44. e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, West AE. 2020. Neurobiological functions of transcriptional enhancers. Nat. Neurosci 23(1):5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki T, Imai R, Tanbo M, Nagashima R, Tamura S, et al. 2017. Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging. Mol. Cell 67(2):282–93. e7 [DOI] [PubMed] [Google Scholar]
- Onuki A. 2002. Phase Transition Dynamics. Cambridge, UK: Cambridge Univ. Press [Google Scholar]
- Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O’Shea CC. 2017. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357(6349):eaag0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel L, Kang R, Rosenberg SC, Qiu Y, Raviram R, et al. 2019. Dynamic reorganization of the genome shapes the recombination landscape in meiotic prophase. Nat. Struct. Mol. Biol 26(3):164–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson JR, Laemmli UK. 1977. The structure of histone-depleted metaphase chromosomes. Cell 12(3):817–28 [DOI] [PubMed] [Google Scholar]
- Plys AJ, Davis CP, Kim J, Rizki G, Keenen MM, et al. 2019. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 33(13–14):799–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, et al. 2014. Topologically associating domains are stable units of replication-timing regulation. Nature 515(7527):402–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl C. 1885. Über Zelltheilung [About cell healing]. Morph. Jahrb 10:214–330 [Google Scholar]
- Rajarajan P, Borrman T, Liao W, Schrode N, Flaherty E, et al. 2018. Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science 362(6420):eaat4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, St Hilaire BG, Engreitz JM, Perez EM, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171(2):305–20. e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159(7):1665–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R. 2014. Chromatin dynamics during spermiogenesis. Gene Regul. Mech 1839(3):155–68 [DOI] [PubMed] [Google Scholar]
- Ricci MA, Manzo C, García-Parajo MF, Lakadamyali M, Cosma MP. 2015. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 160(6):1145–58 [DOI] [PubMed] [Google Scholar]
- Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, et al. 2019. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363(6434):1463–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Zimmer C. 2014. Computational models of large-scale genome architecture. Int. Rev. Cell Mol. Biol 307:275–349 [DOI] [PubMed] [Google Scholar]
- Rosin LF, Nguyen SC, Joyce EF. 2018. Condensin II drives large-scale folding and spatial partitioning of interphase chromosomes in Drosophila nuclei. PLOS Genet. 14(7):e1007393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Nichols MH, Lyu X, Ando-Kuri M, Rivera ISM, et al. 2017. Evolutionarily conserved principles predict 3D chromatin organization. Mol. Cell 67(5):837–52. e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, et al. 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 20(6):761–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, et al. 2018. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361(6400):eaar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador-Martínez I, Grillo M, Averof M, Telford MJ. 2019. Is it possible to reconstruct an accurate cell lineage using CRISPR recorders? eLife 8:e40292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SSP, Huang SC, Durand NC, Huntley MH, et al. 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. PNAS 112(47):E6456–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ, Shiels C, Williamson J, Satijn DPE, Otte AP, et al. 1998. The human Polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol 142(4):887–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. 2014. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell 56(1):29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, et al. 2017. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551(7678):51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaban HA, Barth R, Bystricky K. 2018. Formation of correlated chromatin domains at nanoscale dynamic resolution during transcription. Nucleic Acids Res. 46(13):e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaban HA, Barth R, Recoules L, Bystricky K. 2020. Hi-D: nanoscale mapping of nuclear dynamics in single living cells. Genome Biol. 21(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth RU, Wang HH. 2018. DNA-based memory devices for recording cellular events. Nat. Rev. Genet 19(11):718–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP. 2017. Liquid phase condensation in cell physiology and disease. Science 357(6357):eaaf4382 [DOI] [PubMed] [Google Scholar]
- Shin Y, Chang Y-C, Lee DSW, Berry J, Sanders DW, et al. 2018. Liquid nuclear condensates mechanically sense and restructure the genome. Cell 175(6):1481–91. e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sima J, Chakraborty A, Dileep V, Michalski M, Klein KN, et al. 2019. Identifying cis elements for spatiotemporal control of mammalian DNA replication. Cell 176(4):816–30. e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PB, Newman AG. 2020. On the relations of phase separation and Hi-C maps to epigenetics. R. Soc. Open Sci 7(2):191976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse DH. 2005. X-linked genes and mental functioning. Hum. Mol. Genet 14(1):R27–32 [DOI] [PubMed] [Google Scholar]
- Smeets D, Markaki Y, Schmid VJ, Kraus F, Tattermusch A, et al. 2014. Three-dimensional super-resolution microscopy of the inactive X chromosome territory reveals a collapse of its active nuclear compartment harboring distinct Xist RNA foci. Epigenetics Chromatin 7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. 2017. Phase separation drives heterochromatin domain formation. Nature 547(7662):241–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su JH, Zheng P, Kinrot SS, Bintu B, Zhuang X. 2020. Genome-scale imaging of the 3D organization and transcriptional activity of chromatin. Cell 182(6):1641–59. e26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons O, Raj A. 2016. What’s luck got to do with it: single cells, multiple fates, and biological nondeterminism. Mol. Cell 62(5):788–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. 2014. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159(3):635–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, et al. 2015. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163(7):1611–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakawa T, Bisht S, Eeftens JM, Dekker C, Haering CH, Greene EC. 2017. The condensin complex is a mechanochemical motor that translocates along DNA. Science 358(6363):672–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JMA. 2007. Meiotic sex chromosome inactivation. Development 134(10):1823–31 [DOI] [PubMed] [Google Scholar]
- Vallot A, Tachibana K. 2020. The emergence of genome architecture and zygotic genome activation. Curr. Opin. Cell Biol 64:50–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton AL, Dekker J. 2016. TAD disruption as oncogenic driver. Curr. Opin. Genet. Dev 36:34–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bemmel JG, Galupa R, Gard C, Servant N, Picard C, et al. 2019. The bipartite TAD organization of the X-inactivation center ensures opposing developmental regulation of Tsixand Xist. Nat. Genet 51(6):1024–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsmuth M, Caudron-Herger M, Rippe K. 2008. Genome organization: balancing stability and plasticity. Mol. Cell Res 1783(11):2061–79 [DOI] [PubMed] [Google Scholar]
- Wallace HA, Bosco G. 2013. Condensins and 3D organization of the interphase nucleus. Curr. Genet. Med. Rep 1(4):219–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-Y, Jégu T, Chu H-P, Oh HJ, Lee JT. 2018. SMCHD1 merges chromosome compartments and assists formation of super-structures on the inactive X. Cell 174(2):406–21. e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Gao Y, Zheng X, Liu C, Dong S, et al. 2019. Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol. Cell 76(4):646–59. e6 [DOI] [PubMed] [Google Scholar]
- Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, et al. 2018. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361(6400):eaat5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang H, Zhang Y, Du Z, Si W, et al. 2019. Reprogramming of meiotic chromatin architecture during spermatogenesis. Mol. Cell 73(3):547–61. e6 [DOI] [PubMed] [Google Scholar]
- Williamson I, Kane L, Devenney PS, Flyamer IM, Anderson E, et al. 2019. Developmentally regulated Shh expression is robust to TAD perturbations. Development 146(19):dev179523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EB. 1899. The structure of protoplasm. Science 10(237):33–45 [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Ghosh RP. 2010. Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect. Biol 2(5):a000596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth MB, Girskis KM, Walsh CA. 2017. Building a lineage from single cells: genetic techniques for cell lineage tracking. Nat. Rev. Genet 18(4):230–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz G, Várnai C, Nagasaka K, Cisneros DA, Stocsits RR, et al. 2017. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36(24):3573–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatskevich S, Rhodes J, Nasmyth K. 2019. Organization of chromosomal DNA by SMC complexes. Annu. Rev. Genet 53:445–82 [DOI] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, et al. 2012. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148(5):908–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X. 2021. Spatially resolved single-cell genomics and transcriptomics by imaging. Nat. Methods 18:18–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. 2009. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138(6):1122–36 [DOI] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MIJA, Ye Z, Kolovos P, et al. 2014. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. PNAS 111(3):996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylicz JJ, Bousard A, Žumer K, Dossin F, Mohammad E, et al. 2019. The implication of early chromatin˙ changes in X chromosome inactivation. Cell 176(1–2):182–97. e23 [DOI] [PMC free article] [PubMed] [Google Scholar]