Abstract
The pathogenic fungus Cryptococcus neoformans must overcome iron limitation to cause disease in mammalian hosts. Previously, we reported a screen for insertion mutants with poor growth on heme as the sole iron source. In this study, we characterized one such mutant and found that the defective gene encoded a Vam6/Vps39/TRAP1 domain-containing protein required for robust growth on heme, an important iron source in host tissue. We designated this protein Vps3 based on reciprocal best matches with the corresponding protein in Saccharomyces cerevisiae. C. neoformans encodes a second Vam6/Vps39/TRAP1 domain-containing protein designated Vam6/Vlp1, and we found that this protein is also required for robust growth on heme as well as on inorganic iron sources. This protein is predicted to be a component of the homotypic fusion and vacuole protein sorting (HOPS) complex involved in endocytosis. Further characterization of the vam6Δ and vps3Δ mutants revealed perturbed trafficking of iron acquisition functions (e.g., the high affinity iron permease Cft1) and impaired processing of the transcription factor Rim101, a regulator of heme and iron acquisition. The vps3Δ and vam6Δ mutants also had pleiotropic phenotypes including loss of virulence in a mouse model of cryptococcosis, reduced virulence factor elaboration, and increased susceptibility to stress, indicating pleiotropic roles for Vps3 and Vam6 beyond heme use in C. neoformans.
Graphical Abstract
The pathogenic fungus Cryptococcus neoformans must overcome iron limitation to cause disease in mammalian hosts. Two Vam6/Vps39/TRAP1 domain-containing proteins, Vps3 and Vam6, are required for robust growth on heme as well as proper regulation of iron homeostasis. Two independent deletion mutants for each gene, or double mutants lacking both genes, are unable to cause disease in a mouse inhalation model of cryptococcosis.
1. Introduction
The pathogenic fungus Cryptococcus neoformans causes life-threatening meningoencephalitis in immunocompromised individuals including those suffering from HIV/AIDS (May et al., 2016; Mayer and Kronstad, 2019; Park et al. 2009; Rajasingham et al., 2017). Normally, infection is asymptomatic in immunocompetent hosts, and C. neoformans remains dormant or is eliminated. However, in immunocompromised individuals, C. neoformans can disseminate to the central nervous system to cause death if untreated (May et al., 2016). In fact, C. neoformans causes ~180,000 cases of meningoencephalitis per year and is thought to be responsible for 15% of HIV-related deaths (Okurut et al., 2020; Park et al. 2009; Rajasingham et al., 2017).
In general, there is a pressing need for new antifungal drugs, drug targets, and therapeutic approaches because effective treatment options are limited for cryptococcosis and other fungal infections of humans. Pathogen sensing and acquisition of key nutrients such as iron in host tissue are potential therapeutic targets. Iron is an essential nutrient as it is involved in a variety of biochemical processes such as respiration, electron transport, and the biosynthesis of some amino acids, lipids, and nucleotides. Free iron is scarce in mammalian hosts because it is sequestered from microbial pathogens by binding to transferrin, lactoferrin, or other proteins (Kronstad et al., 2013; Legrand et al., 2008). Consequently, pathogens have evolved competitive systems for uptake of iron to overcome nutritional immunity (Cassat and Skaar, 2013; Schaible and Kaufmann, 2004). Additionally, iron availability influences virulence factor elaboration by pathogens, as demonstrated by the regulation of the polysaccharide capsule of C. neoformans (Jung et al. 2006; Vartivarian et al., 1993).
C. neoformans possesses several systems to acquire iron including cell surface reductases, exported reductants, and melanin, which have been shown to reduce ferric (Fe3+) to ferrous iron (Fe2+), the form that is bioavailable and that can be acquired by low-affinity iron permeases (Cadieux et al. 2013; Horianopoulos and Kronstad, 2019; Jung et al., 2008, 2009; Kronstad et al., 2013). A high-affinity uptake system also exists in which ferrous iron is oxidized to ferric iron by a ferroxidase, Cfo1, and then transported into the cytoplasm of the cell by an iron permease, Cft1 (Jung and Kronstad, 2008, Kronstad et al., 2013). Although C. neoformans cannot synthesize siderophores, which are high affinity iron binding molecules, the fungus does have a family of siderophore transporters for ferric iron uptake (Jung and Kronstad, 2008, Kronstad et al., 2013). C. neoformans can also acquire iron from heme although the mechanisms are not well understood. A number of components required for heme uptake have been identified including an extracellular Cig1 mannoprotein, clathrin mediated endocytosis, and ESCRT (endosomal sorting complex required for transport) proteins (Bairwa et al., 2019; Cadieux et al., 2013; Hu et al., 2013, 2015). In Candida albicans, heme is taken up in endosomes and sorted to the vacuole for storage and utilization (Navarathna and Roberts, 2010; Pendrak et al., 2004; Weissman et al., 2008). Accumulating evidence indicates that a similar mechanism involving endomembrane trafficking exists in C. neoformans, and our goal is to understand the roles of specific components in this process.
Iron acquisition by fungal pathogens may depend in part on endocytic functions (Bairwa et al., 2019; Weissman et al. 2008). The homotypic fusion and vacuole protein sorting (HOPS) tethering complex is a key component of the endosomal pathway, mediating the fusion events between the late endosome and the vacuole (Bröcker et al., 2012). This complex consists of six subunits: Vam6 (Vps39), Vps11, Vps18, Vps16, Vps33, and Vps41 (Bröcker et al., 2012). Vam6 and Vps41 are located at opposite ends of the HOPS complex and interact with Ypt7, a Rab7 GTPase present on late endosomes (Bröcker et al., 2012). Vps33 is suggested to mediate accessory interactions with SNARE proteins, which assist in vesicle fusion, and Vps11, Vps16, and Vps18 are structural components (Bröcker et al., 2012). A connection with iron acquisition is indicated by the finding that loss of the HOPS component Vps41 results in defective high affinity iron uptake in Saccharomyces cerevisiae due to impaired activity of the multicopper oxidase Fet3p (Radisky et al., 1997). A previous study showed that Vps41 is involved in virulence and intracellular survival in C. neoformans, although a connection to iron acquisition could not be established (Liu et al., 2006). A HOPS-related complex, the class C core vacuole/endosome tethering (CORVET) complex, shares Vps11, Vps16, Vps18, and Vps33 and is involved in endosome-endosome fusion, acting earlier in the endocytic pathway than the HOPS complex (Balderhaar and Ungermann 2013). The CORVET complex contains Vps3 and Vps8 instead of Vam6 and Vps41, and interacts with a Rab5 GTPase, characteristic of early endosomes, instead of the Rab7 GTPase (Balderhaar and Ungermann 2013). Individual proteins of the HOPS and CORVET complexes also participate in establishing membrane contact sites between organelles and possess complex-independent functions (Prinz et al., 2020). For example, Vam6 is a component of the vacuole and mitochondria patch (vCLAMP) that connects these organelles and allows the transport of phospholipids (Elbaz-Alon et al., 2014; Honscher et al., 2014; Gonzales-Montoro et al., 2018; Ladarola et al., 2020).
We previously employed Agrobacterium tumefaciens T-DNA insertion mutagenesis to identify C. neoformans genes involved in the use of heme as an iron source (Hu et al., 2013). Subsequent mutant characterization identified proteins with roles in heme use including components of ESCRT complexes and P4-ATPase subunit of the Cdc50 family (Hu et al., 2013, 2015, 2017). Here we report that this approach identified a protein, Vps3, in the Vam6/Vps39/TRAP1 family that was distinct from a known member of this protein family in C. neoformans, the predicted HOPS complex protein Vam6/Vlp1. The latter protein was recently demonstrated to play a role in pathogenesis and the response to stress (Liu et al., 2008; Fan and Liu, 2021; Lee et al. 2010; Tseng et al., 2012). As demonstrated here, mutants lacking VPS3 and/or VAM6 display defects in vacuolar morphology and endomembrane trafficking, and are impaired for growth on medium with low levels of inorganic iron or heme. These mutants are also unable to cause cryptococcosis in mice thus supporting the conclusion that endomembrane trafficking is important for iron acquisition and fungal pathogenesis.
2. RESULTS
2.1. Identification of Vam6/Vps39/TRAP1-domain proteins in C. neoformans
One of the mutants from our previous screen of Agrobacterium T-DNA insertion mutants (Hu et al., 2013) contained an insertion in the gene CNAG_07328 encoding a predicted protein with a citron homology (CNH) domain and a Vps39–2 domain (Supplemental Figure S1A). The latter domain, along with a Vps39–1 domain, is found in proteins of the Vam6/Vps39/TRAP1 family including the Vam6 protein of the HOPS complex in S. cerevisiae (Bröcker et al., 2012). However, the reciprocal best match for CNAG_07328 protein in the S. cerevisiae database was the CORVET complex component Vps3, although the E value was high (3e-07) and sequence similarity was limited to the CNH domain (Supplemental Figure S1B). A further analysis using PANTHER classified the CNAG_07328 protein as vacuolar protein sorting-associated protein 3 with HMM E value score of 4.2e-258 indicating likely correct family assignment (Mi et al., 2021). An examination of the C. neoformans genome database revealed one other protein with a Vam6/Vps39/TRAP1 domain (CNAG_05395); this protein was previously designated Vam6 or Vlp1 and was shown to contribute to C. neoformans pathogenesis (Liu et al., 2008; Fan and Liu, 2021; Lee et al., 2010; Tseng et al., 2012). The domain organization for the proteins is shown in Supplemental Figure S1. The connection between CNAG_07328 (designated Vps3) and heme use prompted a more detailed examination of the roles of the Vam6/Vps39/TRAP1 domain proteins in iron-related processes in C. neoformans.
2.2. Vam6 and Vps3 influence cell and vacuole morphology, and localize to punctate structures in the cytoplasm.
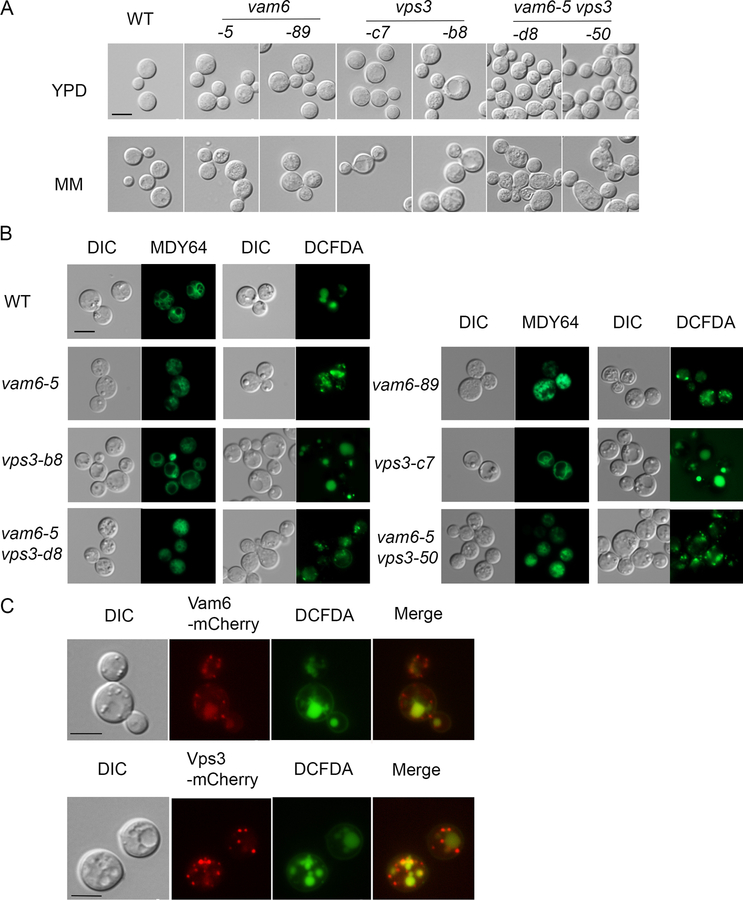
Initially, we constructed two independent deletion mutants each for VPS3 (designated vps3-c7 and vps3-b8) and VAM6 (designated vam6–5 and vam6–89) as well as mutants lacking both genes, and investigated the impact on cell and vacuolar morphology (Figure 1A, B). Loss of either Vps3 or Vam6 did not cause marked defects in cell morphology compared with the wild type (WT) strain in rich (YPD) or minimal (MM) media (Figure 1A). However, the vps3Δ vam6Δ double mutants exhibited enlarged, elliptical cells with defects in cell separation and this phenotype was more pronounced upon growth in MM. To determine whether the vps3Δ and vam6Δ mutants displayed vacuolar morphological changes consistent with roles in endocytic function, we stained the cells grown in MM with the lipophilic vacuolar membrane dye MDY64 and with the vacuole-sequestered dye c-DCFDA. We found that cells lacking Vam6 had multiple small and/or fragmented vacuoles similar to the class B or C vacuolar phenotypes in S. cerevisiae, and a mutant lacking Vps3 displayed enlarged vacuoles similar to the class D vacuolar phenotype in S. cerevisiae (Figure 1B) (Arlt et al., 2011; Raymond et al., 1992). The vacuolar morphology of the vps3Δ vam6Δ double mutant resembled that of the vam6Δ mutant. Overall, these results suggest that Vps3 and Vam6 function in vacuole biogenesis and/or stability, a finding consistent with participation endomembrane trafficking.
Figure 1. Vps3 and Vam6 are localized on endosomes and influence vacuolar and cell morphology.
(A) Differential interference microscopy (DIC) to examine the cell morphology of the WT, vps3Δ, vam6Δ and vps3Δ vam6Δ deletion strains. Cells were harvested from the overnight cultures in either yeast extract peptone dextrose (YPD) or minimum medium (MM). The absence of either VPS3 or VAM6 resulted in a similar morphology to the WT strain. Cells lacking both VPS3 and VAM6 displayed swollen or ecliptic shape and tended to be unseparated. (B) Vacuolar morphology in the WT strain and the indicated mutants. Vacuoles were stained with MDY64 or 5-(and-6)-carboxy-2’,7-dicholorofluorescein diacetate (carboxy-DCFDA). (C) Localization of Vam6-mCherry and Vps3-mCherry in WT cells upon growth of cells in minimum medium (MM). Vacuoles were stained with carboxy-DCFDA. Bar = 5 μm in all images.
We next examined the subcellular localization of Vps3 or Vam6 by fusing mCherry at the C-terminus of each protein with expression of the fusions from the native promoters. The resulting strains did not show any phenotypic differences compared with the parental WT strain indicating that the Vps3-mCherry and Vam6-mCherry fusion proteins were functional (G. Hu, data not shown). Fluorescent microscopy revealed that Vam6-mCherry was associated with punctated structures and with vacuoles, while Vps3-mCherry was localized on multiple punctated structures (Figure 1C). These patterns are reminiscent of those seen for Vam6 and Vps3 in S. cerevisiae (Cabrera et al., 2013; Nakamura et al., 1997). The vacuoles were identified by staining with the dye c-DCFDA and by examination of the DIC images (Figure 1C). Given that Vps3 (CORVET) and Vam6 (HOPS) in S. cerevisiae are involved in vesicle trafficking of early- and late- endosomes to vacuoles, respectively, we hypothesize that the observed punctate structures for Vps3-mCherry and Vam6-mCherry may represent endosomal vesicles in C. neoformans.
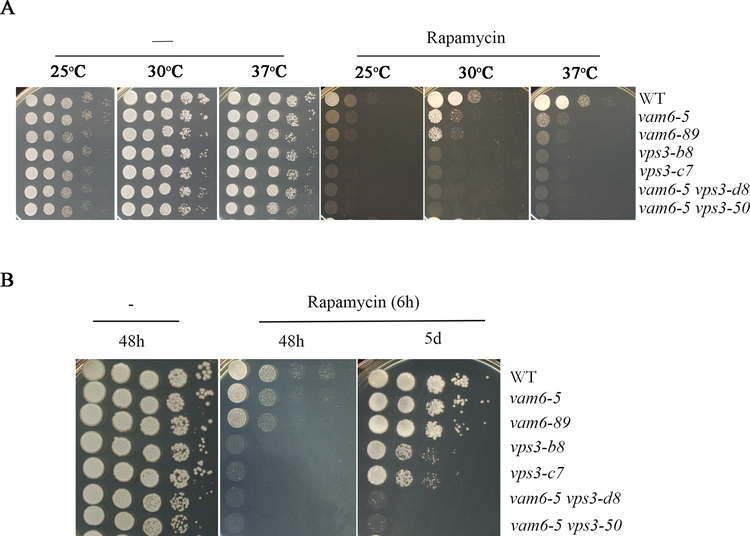
An inability to recover from rapamycin-induced growth arrest is one characteristic of class C vps mutants in S. cerevisiae (Zurita-Martinez et al., 2007). Rapamycin is a macrocyclic lactone antibiotic that targets the TOR pathway that regulates metabolism and proliferation (Heitman et al., 1991). We tested the sensitivity of the vps3Δ, vam6Δ and double mutants to rapamycin on solid medium and found that all of the mutants displayed increased sensitivity across a range of temperatures, compared with the WT strain (Figure 2A). We further examined the ability of the strains to resume the growth after a 6 h exposure to rapamycin. Interestingly, in contrast to S. cerevisiae, the mutant lacking VAM6 resumed the growth after rapamycin exposure, but the deletion mutant lacking VPS3 was unable to fully recover after drug treatment (Zurita-Martinez et al., 2007). The double vps3Δ vam6Δ mutant showed a more pronounced growth defect after rapamycin exposure compared to the single mutants (Figure 2B). Taken together, the characterization of the mutants revealed phenotypes consistent with endocytic trafficking and vacuole biogenesis.
Figure 2. Loss of Vps3 and/or Vam6 results in enhanced sensitivity to rapamycin and a double mutant fails to recover from rapamycin-induced growth arrest.
(A) Growth of the WT strain and the mutants on YPD medium supplemented with rapamycin (30 μg/ml). Tenfold serial dilutions of each strain (labeled on the right) were spotted on the agar and the plates were incubated at the indicated temperatures for two days before being photographed. (B) The cells of each strain were cultured with 30 μg/ml of rapamycin for 6 h at 30°C, washed twice, and serial dilutions of cells were then spotted on YPD plates without drug. The plates were incubated at 30°C for 2 and 5 days before being photographed.
2.3. Vam6 and Vps3 are required for robust growth on heme and inorganic iron sources
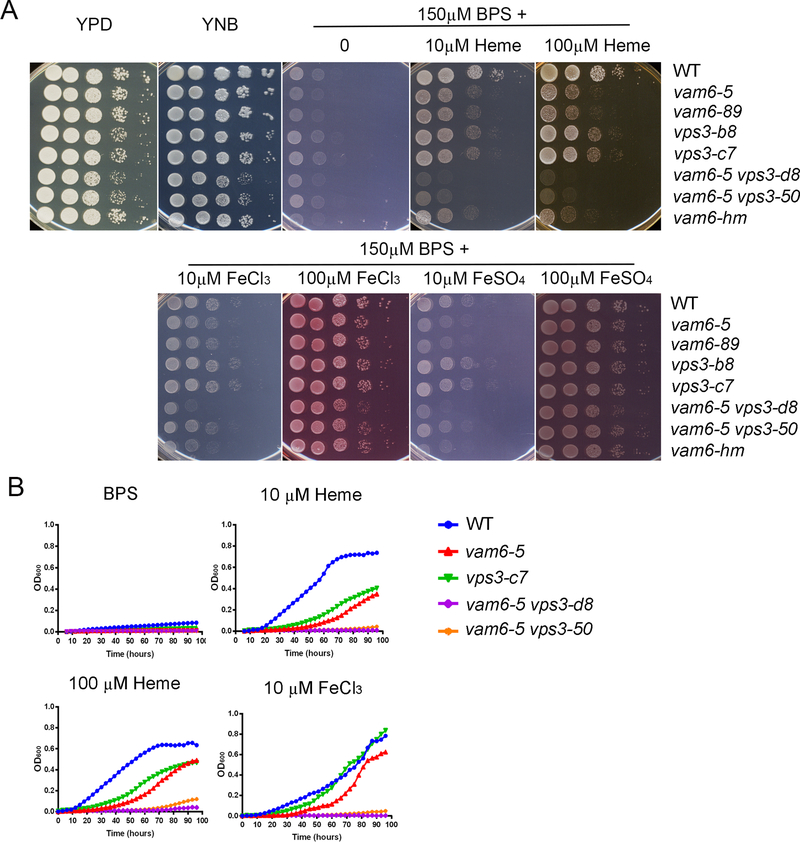
We next confirmed the iron and heme-related phenotypes of the T-DNA insertion mutant for VPS3 by examining the growth of the WT strain and targeted deletion mutants of VPS3 (vps3-c7 and vps3-b8) on medium with heme as the sole iron source (Figure 3A). Deletion mutants for VAM6 and double mutants were also tested. Each of the strains was pre-cultured in liquid low iron media (150 μM BPS) for 2 days to induce iron starvation. None of the strains (WT or mutant) were able to grow when cells were spotted on low iron medium after 2 days, and the WT strain grew on low iron medium with heme or inorganic iron sources (FeCl3 and FeSO4) at either 10 μM or 100 μM. Compared to WT, deletion of VPS3 or VAM6 caused reduced growth on medium with heme at either 10 μM or 100 μM, and we noted greater impairment for the vam6Δ mutants (Figure 3A). We also tested an independent deletion mutant, vam6-hm in the background of the WT strain KN99, from the deletion collection (Liu et al. 2008) and observed impaired growth on heme. Moreover, cells of two independent strains lacking both VPS3 and VAM6 exhibited even greater impaired growth on heme compared with the single mutants (Figure 3A), indicating an additive contribution to iron utilization from heme. Although the double mutants did show slight growth defects on YPD and YNB media, impaired growth was much more prominent on the media with heme compared to WT. We also found that the growth of the vam6Δ mutants on the inorganic iron sources FeCl3 or FeSO4 was impaired at 10 μM, but not at 100 μM, while the vps3Δ mutants grew much like the WT strain at 100 μM of FeCl3 or FeSO4, but somewhat better than WT at the 10 μM concentrations of these iron sources (Figure 3A). We also examined the growth of the WT strain and the vps3Δ and vam6Δ mutants in a liquid low iron medium supplemented with different iron source. The results were consistent with the spot assays, and revealed growth defects for the mutants on heme at either 10 μM or 100 μM, or with FeCl3 at 10 μM (Figure 3B). Again, the double mutants showed more marked growth defects than the single mutants.
Figure 3. Vam6 and Vps3, are required for growth on heme and inorganic iron sources.
(A) The growth of vps3Δ, vam6Δ and vps3Δ vam6Δ double mutants and the WT strain was tested on yeast extract peptone dextrose (YPD), yeast nitrogen base (YNB) and YNB plus bathophenanthrolinedisulfonic acid (BPS) supplemented with either heme or FeCl3 at 10 μM or 100 μM at pH 7.0. Tenfold serial dilutions of each strain (labeled on the right) were spotted on the indicated media after iron starvation and the plates were incubated at 30 °C for 2 days before being photographed. (B) Iron-starved cells of the indicated strains were inoculated in liquid YNB medium plus 150 μM BPS with and without supplementation with iron sources. A representative single mutant is shown for each gene and both double mutants are included. The cultures were incubated at 30°C, and OD600 was measured. WT = wild type
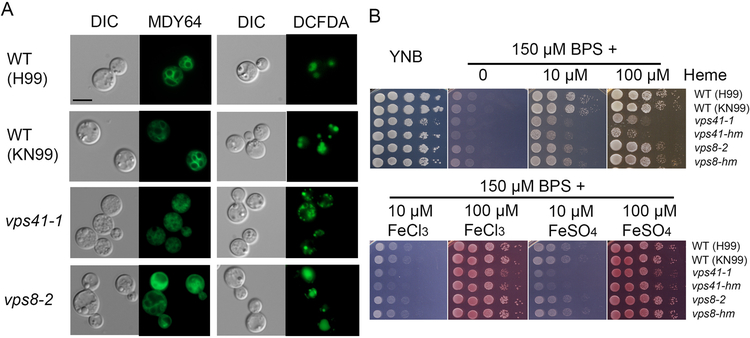
Although our focus was on Vps3 and Vam6, we also examined the vacuolar morphology and iron/heme-related phenotypes for mutants lacking the candidate HOPS component Vps41 and the candidate CORVET component Vps8 (Figure 4). The genes for these proteins were identified by comparisons with the corresponding S. cerevisiae genes (Supplemental Figure S1B). We tested two independent mutants for each component and representative mutant phenotypes are shown. The mutant lacking Vps41 had small or fragmented vacuoles (a class B or C phenotype), and the vps8 mutant displayed enlarged vacuoles similar to the class A or D phenotype in yeast (Figure 4A) (Arlt et al., 2011; Raymond et al., 1992). We found that loss of Vps41 but not Vps8 caused reduced growth on medium with heme (Figure 4B). Similar to the vam6Δ mutants, deletion of VPS41 led to growth defects on medium with FeCl3 or FeSO4 as the sole iron sources at 10 μM (and to a lesser extent with 100 μM) (Figure 4B). Taken together, the results revealed that Vam6 and Vps41 (as candidate components of the HOPS complex), but not Vps3 and Vps8, play important roles in iron utilization from heme. Additionally, Vam6 and Vps41 were found to be important for the use of inorganic iron sources.
Figure 4. Other candidate CORVET and HOPS complex components influence vacuolar morphology and growth on heme and inorganic iron sources.
(A) Analysis of vacuolar morphology of the WT (both H99 and KN99 background strains) and the deletion strains lacking VPS41 or VPS8 by differential interference microscopy and fluorescent microscopy. The vps41–1 and vps8–2 mutants were constructed in the H99 background for this study, and the vps41-mh vps8-mh were derived from the KN99 strain background (Liu et al., 2008). Cells were harvested from the overnight cultures in minimum medium (MM) and vacuoles were stained with MDY-64 or c-DCFDA. Representative mutants are shown. Bar = 5 μm. (B) The growth of vps41 and vps8 mutants and the WT strain was tested on yeast extract peptone dextrose (YPD), yeast nitrogen base (YNB) and YNB plus BPS supplemented with either heme at 10 μM or 100 μM at pH 7.0, or with FeCl3 or FeSO4 at 10 μM or 100 μM at pH 7.0. For all spot assays, tenfold serial dilutions of each strain (labeled on the right) were spotted on the indicated media after iron starvation and the plates were incubated at 30 °C for 2 days before being photographed.
To more broadly assess and compare the roles of Vps3 and Vam6, we tested the sensitivity of the WT, vps3Δ, vam6Δ and vps3Δ vam6Δ strains to agents related to iron, heme and vacuolar function (Supplemental Figure S2). We found that loss of both Vam6 and Vps3 caused minor growth defects on the hypoxia-mimicking agent CoCl2 and on the drug chloroquine which influences vacuolar acidification (a result consistent with the observed alteration in vacuolar morphology (Figure 1B)). Loss of Vam6 also caused a minor growth defect on chloroquine. We also noted increased sensitivity of the vam6Δ mutants to the antifungal drug fluconazole that targets ergosterol biosynthesis (a process that involves heme-dependent enzymes). The vps3Δ mutant had similar growth to WT, and the double mutant had similar phenotypes to the vam6Δ mutant on fluconazole and chloroquine but slightly increased sensitivity to CoCl2. The mutants also all showed increased sensitivity to the iron-dependent drug bleomycin suggesting impaired iron homeostasis, although the vps3Δ mutant was less sensitive compared to the other mutants. Taken together, the data are consistent with roles for Vps3 and Vam6 in iron- and vacuole-associated functions.
2.4. Vam6 or Vps3 are required for proper sorting of the iron permease Cft1
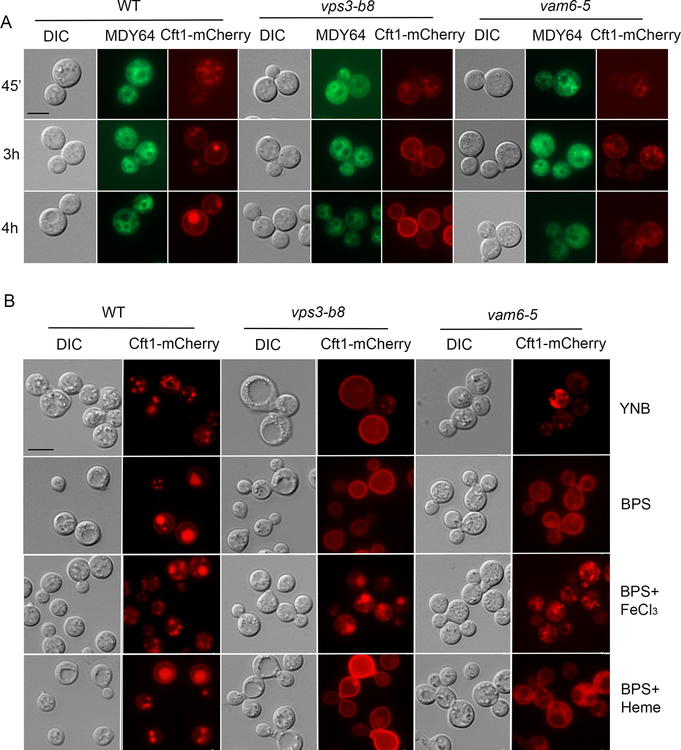
To investigate the underlying mechanisms of Vps3 and Vam6 contributions to iron and heme acquisition, we examined the subcellular localization of the iron permease Cft1 in the mutants. Cft1 functions with the ferroxidase Cfo1 as a high-affinity reductive iron uptake system in C. neoformans and is important for iron acquisition and pathogenesis (Caza et al., 2018; Jung et al., 2008, 2009). We hypothesized that the growth defects of the vps3Δ and vam6Δ mutants on iron sources might perturb iron homeostasis and lead to impaired trafficking of high affinity iron uptake functions. This idea is supported by the finding that iron availability influences the localization of the iron permease/ferroxidase proteins Ftr1 and Fet3 by recycling them to the plasma membrane (low iron) or targeting them to the vacuole for degradation (high iron) in S. cerevisiae (Felice et al., 2005). Additionally, loss of the HOPS component Vps41 results in defective high affinity transport due to impaired Fet3p activity (Radisky et al., 1997). We therefore constructed a gene for a Cft1-mCherry fusion and introduced it into the WT strain and the vps3Δ or vam6Δ mutants. After pre-growth in YPD medium and transfer of the cells to iron-chelated medium, Cft1-mCherry was initially and predominantly distributed in intracellular vesicles in the cytoplasm, and the fusion protein eventually accumulated in vacuoles (identified by staining with MDY64) and at the plasma membrane (by 3 – 4 hours) (Figure 5A). This result may indicate cycling of the protein between vacuoles and the plasma membrane (Figure 5A). For the vps3Δ mutant, the location of the fusion protein in punctate structures in the cytoplasm was initially similar to the WT strain, but with time the protein accumulated at the plasma membrane. In contrast to the WT strain, the Cft1-mCherry protein in the vps3Δ mutant failed to locate to the vacuoles even after 3 – 4 hours of incubation in the low iron medium, and the signal remained in the plasma membrane (Figure 5A). For the vam6Δ mutant, the Cft1-mCherry signal was generally weaker compared with the WT strain and the vps3Δ mutant. After transfer of the mutant to low iron medium, Cft1-mCherry accumulated in punctate structures and the signal was visible both on the plasma membrane and dispersed in the cytoplasm at 3 h. The Cft1-mCherry signal remained primarily dispersed by 4h (Figure 5A), perhaps indicating defective cycling between vacuoles and plasma membrane.
Figure 5. Localization of the high affinity iron permease Cft1 is influenced by iron availability and deletion of either VPS3 or VAM6.
(A) Localization of Cft1-mCherry in low iron medium. Strains (WT, and representative vps3Δ and vam6Δ mutants) containing a Cft1-mCherry construct were cultured overnight in YPD, washed 3 times and counted. Subsequently, 1X106 cells/mL were inoculated in YNB-BPS and incubated at 30°C for the indicated time intervals. A vacuole membrane stain MDY-64 was employed to detect the vacuole. Bar = 5 μm. (B) Localization of Cft1-GFP after 16h incubation in YNB, YNB + 150 μM BPS, and YNB + BPS + 100 μM FeCl3 YNB + BPS + 100 μM Heme at 30°C. DIC indicates differential interference contrast microscopy. Bar = 5 μm.
We also examined the subcellular localization of Cft1-mCherry in the mutants in response to different iron sources. In this experiment, cells expressing Cft1-mCherry were pre-cultured in YPD overnight and transferred into yeast nitrogen base (YNB) medium that has an intermediate level of iron, YNB with BPS, or YNB with BPS and heme or FeCl3, and incubated for 16h. Upon incubation in YNB, Cft1-mCherry was mainly found in punctate structures in the WT strain and the vam6Δ mutant, while the protein was mainly present at the plasma membrane in the vps3Δ mutant. In iron-chelated medium containing BPS, a strong Cft1-mCherry signal was observed both in vacuoles and on plasma membrane in the WT strain, while Cft1-mCherry was mainly on the plasma membrane in the vps3Δ and vam6Δ mutants, with some association with punctate structures in the latter mutant (Figure 5B). Addition of 100 μM FeCl3 to the YNB with BPS medium resulted in a predominantly vacuolar localization for Cft1-mCherry in the WT strain, vacuolar and plasma membrane signals in the vps3Δ mutant, and association with multiple small punctate structures in the vam6Δ mutant (Figure 5B). In contrast, addition of heme promoted Cft1-mCherry accumulation mainly on the plasma membrane and in vacuoles in the WT strain, predominantly on the plasma membrane in the vps3Δ mutant, and on the plasma membrane and punctate structures in the cytoplasm in the vam6Δ mutant (Figure 5B). It was notable that Cft1-mCherry was similarly internalized in the WT strain and both mutants upon iron repletion, but distinctions were clear upon addition of heme. That is, Cft1-mCherry was maintained at the plasma membrane in the vps3Δ mutant in the medium with heme but some internalization was observed in the WT strain and the vam6Δ mutant. The different responses to heme may parallel the more pronounced growth defects of the vam6Δ mutant on heme compared to the vps3Δ mutant (Figure 3A). Overall, the observations support the hypothesis that Vps3 and Vam6 play a role in iron homeostasis by influencing Cft1 localization between the plasma membrane, punctate structures which may be endocytic vesicles, and vacuoles.
2.5. Loss of Vam6 or Vps3 caused mis-localization of Rim101 and increased sensitivity to alkaline pH and ionic stress
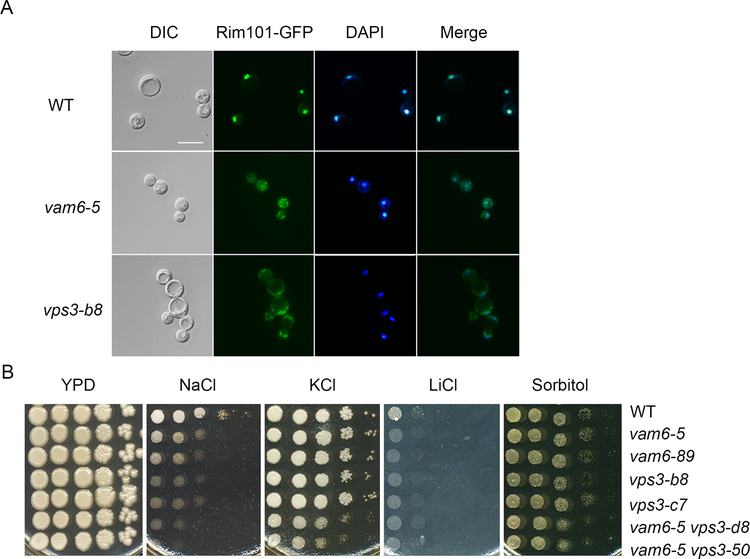
Iron utilization in fungi is influenced by environmental pH, and the pH-responsive transcription factor Rim101 (PacC) is known to regulate iron uptake functions and growth on heme in C. neoformans (Cadieux et al., 2013; Hu, et al., 2015; Kronstad et al., 2013; O’Meara et al., 2010). Given that the vps3Δ and vam6Δ deletion mutants exhibited growth defects on heme, we next tested the hypothesis that the mutants were impaired in the Rim101-regulated pH response pathway. The regulation of the pH response by Rim101 is influenced by the localization of the protein. For example, nuclear Rim101-GFP is mis-localized to the cytoplasm in response to alkaline pH, and in rim and ESCRT mutants in C. neoformans (Hu et al., 2015, 2017; Ost et al., 2015). We therefore assessed the localization of Rim101-GFP in the vps3Δ and vam6Δ mutants. While Rim101-GFP was localized in the nucleus in the WT strain, as verified by comparison with DAPI staining, we found that Rim101-GFP was mis-localized to the cytoplasm in the vps3Δ and vam6Δ mutants (Figure 6A). Taken together, these suggest that part of the defect in growth on iron and heme for the vps3Δ and vam6Δ mutants may be due to impaired function of the Rim101 pathway.
Figure 6. Vam6 and Vps3 influence Rim101 localization, growth at alkaline pH and sensitivity to salt stress.
(A) Localization of Rim101-GFP in the WT strain and in representative vps3Δ and vam6Δ mutants. Nuclear DNA was stained with DAPI. Bar = 10 μm. WT = wild type. (B) Tenfold serial dilutions of the strains were spotted onto solid YPD with or without 1.2 M NaCl, 1.5 M KCl, 1.5 M sorbitol, or 100 mM LiCl. The plates were incubated at 30 °C for 5 days before photographed.
To further test the link to the Rim101 pathway, we examined the vps3Δ, vam6Δ and double mutants for the known phenotypes of the rim101Δ mutant including the impaired growth at alkaline pH and under stress conditions (Hu et al., 2015; O’Meara et al., 2010). We found that the WT, vps3Δ and vam6Δ strains grew equally well on YPD from pH 5 to pH 7, and that the vam6Δ mutants did not show significant differences from WT at pH 8 and pH 9 (Supplemental Figure S3). However, the vps3Δ and the vps3Δ vam6Δ double mutants displayed reduced growth at both pH 8 and pH 9, indicating a requirement for robust growth at alkaline pH. The Rim101 pathway is also involved in the response to salt stress (Hu et al., 2015; O’Meara et al., 2010; Ost et al., 2015). We therefore tested the growth of the WT, vps3Δ, vam6Δ and vps3Δ vam6Δ mutant strains on medium supplemented with 1.2 M KCl, 1.2 M NaCl, or 0.2 M LiCl (or with 1.5 M sorbitol) (Figure 6B). As expected, the WT strain grew well on all of the media, but deletion of VPS3 or VAM6 caused increased sensitivity to NaCl and LiCl, similar to the phenotypes of the rim101Δ mutant (Hu et al., 2015; O’Meara et al., 2010; Ost et al., 2015). Loss of either VPS3 or VAM6 did not cause a growth defect on 1.2M KCl but loss of both VPS3 and VAM6 caused a defect (Figure 6B). Notably, none of the mutants had a growth defect on sorbitol indicating sensitivity to osmotic stress similar to WT (Figure 6B). Overall, we found that the vps3Δ and vam6Δ mutants shared some of the phenotypes reported for a rim101Δ mutant.
2.6. Vps3 influences the expression of iron-regulated genes
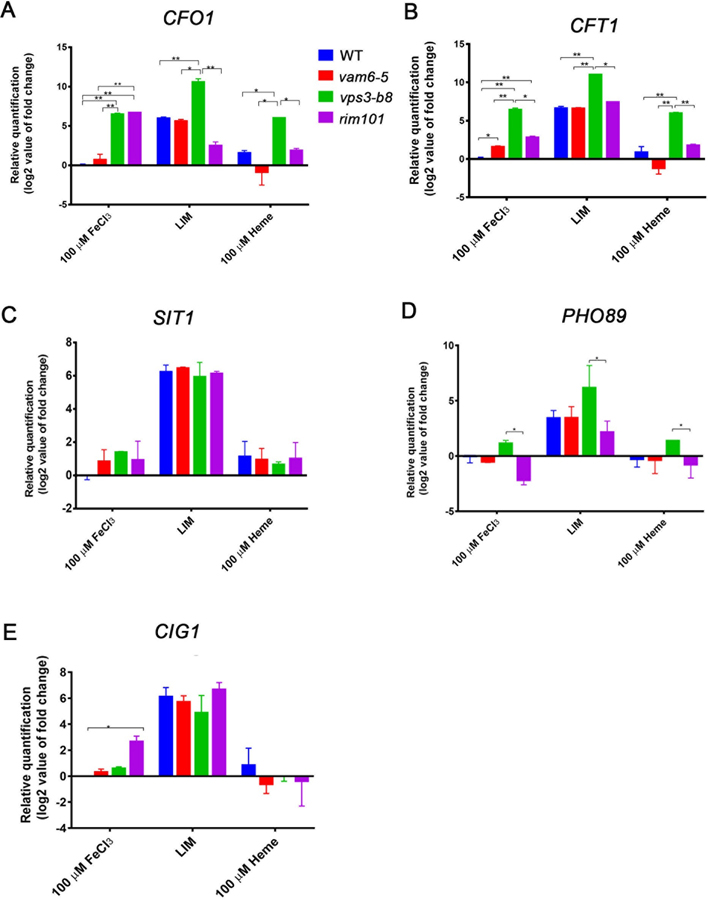
We further examined the influence of Vam6 or Vps3 on the transcript levels for iron-regulated genes, including CFT1 and CFO1 encoding the high affinity iron transport system, in response to iron availability (low iron, FeCl3 or heme). Our goal was to develop a more detailed understanding of the impact of Vam6 and Vps3 on iron homeostasis by evaluating the levels of transcripts that are known to be influence by iron limitation. We also included the rim101Δ mutant to further evaluate the potential impact of mis-localization of Rim101 in the vps3Δ and vam6Δ mutants, and we also evaluated additional iron/Rim101-regulated genes (CIG1, SIT1 and PHO89) (O’Meara et al., 2010). As expected, the levels of the CFO1, CFT1, CIG1, SIT1 and PHO89 transcripts were elevated in cells grown in iron-limiting condition for all strains, compared with growth with FeCl3 or heme (Figure 7A–E) (Jung et al, 2006; Lian et al, 2005; O’Meara et al., 2010; Tangen et al., 2007). The influence of the vam6Δ mutation was minimal in all conditions in comparison with the WT strain, with the exception that the CFT1 transcript was slightly higher in cells grown with FeCl3 (Fig. 7B). Notably, loss of Vps3 had a more pronounced influence in that the transcripts for CFO1 and CFT1 are up-regulated in the mutant in all conditions compared with all other strains (i.e., the WT strain, and the vam6Δ and rim101Δ mutants), indicating an involvement of Vps3 in regulation of the high affinity iron transport system (Fig. 7A, B). The vps3Δ mutant also had a higher level of transcripts for PHO89 in the low iron condition compared with the WT strain (Fig. 7D). Loss of Rim101 had an influence on the transcripts of the CFO1, CFT1 and CIG1 genes such that higher levels were observed in the FeCl3 condition compared with the WT strain (Fig. 7A, B, E). A somewhat lower level was found for CFO1 in the rim101Δ mutant in the low iron condition, and for PHO89 in the FeCl3 condition. Taken together, these results indicate a substantial impact for Vps3 but only a partial overlap with Rim101 in the regulation of iron-responsive genes. Additional regulatory mechanisms may influence the growth of the vam6Δ mutants on inorganic and heme iron sources given the minimal influence on transcript levels.
Figure 7. The relative expression of iron-regulated genes is altered in vps3Δ, vam6Δ and rim101Δ mutants.
(A-E) Analysis of transcript levels of target genes in Rim101 pathway (CIG1, PHO89 and SIT1) as well as the genes for high affinity transport (CFO1 and CFT1) in response to iron availability in the WT strain, and representative vps3Δ, vam6Δ and rim101Δ mutants by RT- qPCR. The data were from three biological replicates, the analyses were performed twice, and each gene was normalized to WT in 10 μM FeCl3. * indicates significance at P<0.05, and ** at P<0.01.
2.7. Vps3 and Vam6 each contribute to pathogenesis
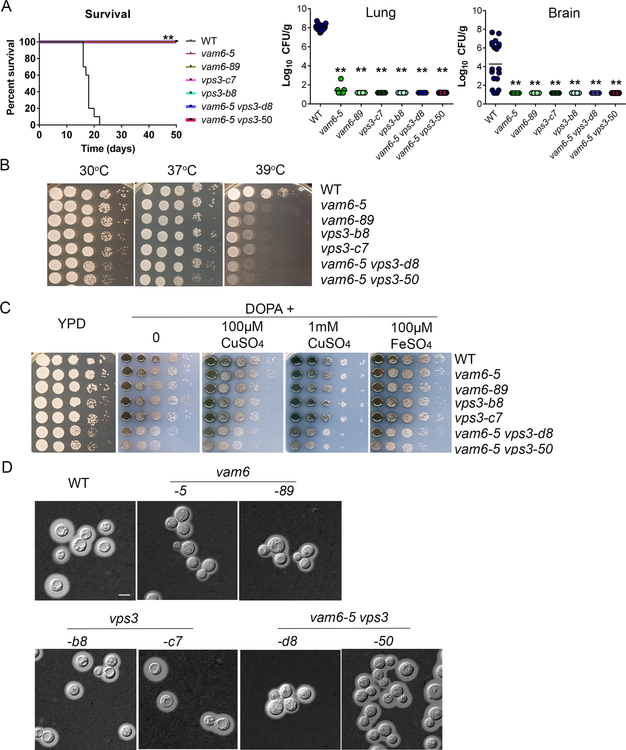
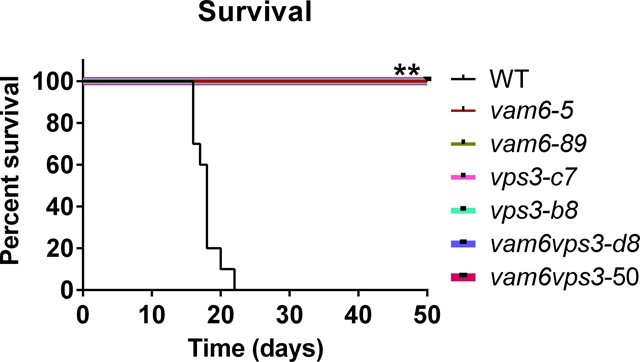
We hypothesize that Vps3 and Vam6 would be required for virulence in mice given the reduced growth of the mutants on heme and inorganic iron sources and previous findings that mutants with impaired iron acquisition have attenuated virulence (Attarian et al., 2018; Hu et al., 2015, 2017; Jung et al., 2008). We therefore challenged mice by intranasal inoculation with the WT strain or two independent mutants for vps3Δ, vam6Δ, and the vps3Δ vam6Δ mutant, and monitored disease progression. In contrast to the WT strain, which caused disease in all mice by day 22, each of the mutants was avirulent in this model; i.e., the infected mice did not show disease symptoms and survived to the end of the experiment at day 50 post-inoculation (Figure 8A). An examination of fungal burden in organs harvested from infected mice revealed much lower colony forming units in the lungs and brains of mice infected with the mutants (Figure 8A). In contrast, high numbers of fungal cells were retrieved from these organs for mice infected with the WT strain (Figure 8A). Overall, the mutants were unable proliferate in mice and to disseminate to other organs indicating that both Vps3 and Vam6 are required for virulence in mice.
Figure 8. Vps3 and Vam6 are required for virulence in mice and elaboration of virulence factors.
(A) BALB/c mice were inoculated intranasally with cells of the wild type (WT) strain, two independent vps3Δ mutants, two vam6Δ mutants, or two vps3Δ vam6Δ mutants, and the survival of the mice was monitored daily. Survival differences between groups of mice were evaluated by log-rank tests. The p values for the mice infected with the WT and mutant strains were statistically significantly different (p < 0.01). Fungal burden was determined in lung and brain for all mice infected with the strains at the end of the experiment. The Mann-Whitney U test was used for statistical analysis. Differences in the fungal loads between the WT and deletion mutants in each organ examined were statistically significant (p < 0.01). (B) The vps3Δ and vam6Δ mutants are sensitive to elevated temperatures (37°C and 39°C). Serial 10-fold dilutions of the indicated strains were spotted on yeast extract peptone dextrose (YPD). The plates were incubated at indicated temperature for 2 days before being photographed. WT = wild type strain H99. Two independent mutants of either VPS3 or VAM6, or both VPS3 and VAM6 were included in the assay. (C) Deletion of either Vps3 or Vam6 or both has an effect on melanin production. Melanin production was tested after growth at 30°C for 2 days by spotting serial 10-fold dilutions of the indicated strains onto L-3,4-dihydroxyphenylalanine plates. Addition of either 10 or 100 μM of copper remediates the defects of melanin production of the single mutants. (D) Cells were grown in defined low-iron medium at 30°C for 48 hr, and capsule formation was assessed by India ink staining for the indicated strains. Bar = 5 μm.
In light of the avirulence of the vps3Δ and vam6Δ mutants, we postulated that the proteins might also influence the elaboration of the major virulence factors of C. neoformans as well as iron acquisition. We examined the strains for the phenotypes related to virulence including growth at elevated temperature, and production of melanin and formation of the polysaccharide capsule. We found that the single and double mutants displayed poor growth at 39°C (Figure 8B) and that loss of VPS3 or VAM6 resulted in reduced production of melanin on medium containing L-DOPA as a substrate (Figure 8C). The double mutant appeared to have an additive defect for melanin formation. Addition of 1 mM CuSO4 or 100 μM FeSO4 partially remediated the melanin production of the vps3Δ mutants (Figure 8C). The capsule size of the vps3Δ mutant was similar to the WT strain with subtle reductions in size for the vam6Δ and vps3Δ vam6Δ mutants (Figure 8D). Overall, it appears that Vam6 and Vps3 make additional contributions such as thermotolerance that may contribute to virulence.
3. DISCUSSION
In this study, we demonstrate that two Vam6/Vps39/TRAP domain proteins, Vps3 and Vam6, contribute to robust growth on heme, and that Vam6 is important for growth in media with a low level of inorganic iron. The Vps3 protein displays quite weak sequence similarity to the corresponding best hit in S. cerevisiae, although Vam6 has greater similarity to the yeast ortholog and has been identified previously in C. neoformans (Liu et al., 2008; Fan and Liu, 2021; Lee et al. 2010; Tseng et al., 2012). We hypothesize that these proteins are components of the CORVET and HOPS complexes, respectively, because the vam6Δ mutant and the vps3Δ vam6Δ double mutant showed fragmented vacuoles as found with vam6Δ mutants of S. cerevisiae and C. albicans (Balderhaar and Ungermann, 2013; Mao et al., 2021; Nakamura et al., 1997; Raymond et al., 1992). Loss of another candidate HOPS subunit Vps41 from C. neoformans resulted in a similar phenotype. The vps3Δ deletion mutant displayed enlarged vacuoles similar to the class D phenotype of the vps3Δ mutant of S. cerevisiae and a defect in another candidate CORVET-specific subunit, Vps8, resulted in a similar vacuolar morphology (Arlt et al., 2011; Balderhaar and Ungermann, 2013; Raymond et al., 1992). Additionally, both Vps3 and Vam6 localized to punctate structures in the cytoplasm, a phenotype consistent with association with endosomes (Cabrera et al., 2013; Nakamura et al., 1997). As discussed below, the impact of Vps3 and Vam6 on the trafficking of Cft1 and Rim101 also supports proposed roles in endocytosis. However, additional work is needed to directly link Vps3 to CORVET complex-mediated fusion events at early endosomes, and Vam6 function to the HOPS complex role in fusion of late endosomes, and endosomes with vacuoles. In particular, it will be important to test interactions with the CORVET and HOPS core components Vps11, 16, 18, and 33.
It is also possible that Vps3 and Vam6 (as well as Vps41 and Vps8) have complex-independent trafficking activities in C. neoformans, as previously demonstrated in yeast and mammalian cells. For example, Vps41 has a HOPS-complex-independent function in transporting lysosomal proteins to endosomes (Asensio et al. 2013; Pols et al 2013). Similarly, Vam6 has roles in the regulation of TORC1, in vacuole-mitochondrial connections (vCLAMP), and in phospholipid transport independent of HOPS (Binda et al., 2009; Elbaz-Alon et al., 2014; Iadarola et al., 2020; Mao et al., 2021). In mammalian cells, Vps3 and Vps8 also have roles independent of the CORVET complex and participate in a specialized endosome recycling pathway (Jonker et al., 2018; Perini et al., 2014; van der Beek et al., 2019).
We hypothesized that impaired iron homeostasis resulting in the mis-localization of iron and heme uptake proteins represented a potential mechanism underlying the iron-related growth defects of the vps3Δ and vam6Δ mutants. We tested this idea by examining the localization of the iron permease Cft1 that partners with the ferroxidase Cfo1 in a high affinity iron transport system for iron from FeCl3 or transferrin (Jung et al., 2008, 2009). Previously, we found that iron availability and cAMP signaling influence the localization of Cfo1; the protein localizes to the plasma membrane after transfer into low iron condition and accumulates in vacuoles before recycling back to plasma membrane (Caza et al., 2018; Jung et al., 2009). Similarly, we demonstrated here that Cft1-mCherry accumulated both in vacuoles and on the plasma membrane of WT cells after incubation in low iron medium. Loss of either Vps3 or Vam6 altered the intracellular distribution of the protein such that the Cft1-mCherry signal generally remained in the plasma membrane in the low iron condition, but was not located in vacuoles. The presence of inorganic iron resulted in internalization of the signal to punctate structures in the vam6Δ mutant and partially to a vacuolar structure in the vps3Δ mutant. Addition of heme generally resulted in plasma membrane localization in the vps3Δ mutant, or at the plasma membrane and in dispersed punctate structures in the cytoplasm for the vam6Δ mutant. Overall, these results indicated that loss of Vps3 or Vam6 perturbed iron homeostasis. However, Cft1-mCherry generally appeared to be more prominently maintained at the plasma membrane in the vps3Δ mutant, compared to the vam6Δ mutant, and this may account for the better growth of vps3Δ mutants on concentrations of inorganic iron sources (10 μM) that would require high affinity transport (Figure 3). In this regard, the more pronounced growth defects on heme and inorganic iron sources for the vam6Δ mutant may reflect impaired trafficking of uptake proteins due to the fragmentation of the vacuole leading to general interference with iron use.
Importantly, our observations are consistent with the influence of endocytic sorting of the iron permease/ferroxidase proteins Ftr1 and Fet3 in S. cerevisiae (Radisky et al, 1997; Strochlic et al., 2007, 2008). In this yeast, iron deprivation results in sorting of the proteins back to the plasma membrane and iron addition causes targeting to the vacuole for degradation via the ESCRT protein mediated multivesicular body pathway. Additionally, the HOPS subunit Vps41 is required for growth in the low iron condition and proper transport and function of the ferroxidase Fet3 (Jo et al, 2009; Radisky et al., 1997). In general, impaired vacuolar function in the mutants may also impact iron homeostasis as demonstrated by the influence of mutants with defects in vacuole assembly in S. cerevisiae (Szczypka et al., 1997). The involvement of the HOPS subunit Vps41 in iron utilization was previously found in other fungi including C. albicans, and in the parasite protozoan Trypanosoma brucei (Lu et al., 2007; Nakamura et al.,1997; Radisky et al., 1997; Weissman et al., 2008). Notably, loss of Vps41 resulted in defective growth in iron utilization from haemoglobin in C. albicans (Weissman et al., 2008). Furthermore, cycling of the Arn1 transporter for the siderophore ferrichrome between endosomes and the plasma membrane is important for its function in S. cerevisiae (Kim et al., 2002). In this case, mutants with defects in endocytosis can maintain Arn1 at the plasma membrane but demonstrate reduced uptake of ferrichrome. Our previous studies on ESCRT complex functions, the Sec1/Munc18 (SM) protein Vps45, and the clathrin heavy chain Chc1 also indicate a role for endocytosis and endomembrane trafficking of heme (Bairwa et al., 2019; Caza et al., 2018; Kronstad et al., 2013, Hu et al., 2013, 2015). In light of these findings, we speculate that proper endocytosis and recycling of Cfo1 and Cft1 contributes to robust iron uptake, correct intracellular distribution, and overall iron homeostasis.
Deletion of VPS3, VAM6, or both genes revealed shared and distinct phenotypes in response to alkaline pH conditions, compared with the rim101Δ mutant, and with the ESCRT complex mutants that we previously described (Hu et al., 2013, 2015; O’Meara et al., 2010). Shared phenotypes included retarded growth at alkaline pH, and in the presence of salt stresses (e.g., NaCl and LiCl). Interestingly, the vps3Δ vam6Δ double mutant exhibited additive phenotypes under these conditions suggesting that they may influence parallel processes in conditions of stress sensitivity. Moreover, both Vps3 and Vam6 have a major influence on Rim101 localization similar to the impact of ESCRT complex mutations (Hu et al., 2015). Specifically, deletion of either VPS3 or VAM6 caused a Rim101-GFP fusion to remain in the cytoplasm, and prevented the localization of the protein to the nucleus as seen in WT strain. However, in contrast with the influence of ESCRT and Rim101, the vps3Δ, vam6Δ or vps3Δ vam6Δ mutants did not show marked reduced capsule formation in defined low iron medium, indicating a distinct role. In general, our findings for the role of Vam6 are similar to those of Fan and Liu (2021) with regard to sensitivity to salt stress and capsule formation. The impact of the vam6Δ mutation on Rim101 trafficking may also be due to impaired acidification in endocytic compartments (Raymond et al., 1992; Solinger, J.A., & Spang, 2013).
One potential impact of impaired translocation of Rim101 to the nucleus in vps3Δ and vam6Δ mutants would be altered expression of genes dependent on the transcription factor, including genes for iron uptake and heme use. It is known that Rim101 in C. neoformans regulates the transcript levels of iron-responsive genes in the cells grown in Dulbecco’s modified Eagle’s media with NaHCO3 or in low iron medium (Hu et al. 2015; O’Meara et al 2010). Furthermore, we previously showed that a rim101Δ mutant has impaired growth on heme and that Rim101 regulates expression of Cig1, a protein required for growth on heme (Cadieux et al., 2013; Hu et al., 2015). An analysis of the transcript levels of genes (CFO1, CFT1, CIG1, SIT1 and PHO89) known to be regulated by Rim101 in mutants lacking Vps3, Vam6 or Rim101 did not reveal similar patterns of regulation. Specifically, higher levels for the transcripts of the CFO1, CFT1 and CIG1 genes were observed in the FeCl3 condition for the rim101Δ mutant compared with the WT strain. In contrast, loss of Vam6 had little impact of transcript levels, and Vps3 had a greater influence such that the transcripts for CFO1 and CFT1 were up-regulated in the mutant in all conditions compared with all other strains (WT, vam6Δ and rim101Δ mutants), indicating an involvement of Vps3 in regulation of the high affinity iron transport system. These results suggest that the mis-localization of Rim101 in the vps3Δ and vam6Δ mutants does not entirely account for the growth defects on iron and heme. Rather, Vps3 appears to influence the repression of iron-responsive genes upon iron addition suggesting an iron starved state for the mutant even in the presence of iron. These findings are interesting in light of the influences of Vam6 and Vps3 on Cft1-mCherry localization such that impaired trafficking of uptake functions may play a greater role in the growth defects of the mutants.
We hypothesize that the inability of the vps3Δ, vam6Δ and vps3Δ vam6Δ mutants to cause disease results from the combination of defects in iron acquisition from heme and inorganic iron sources, a defect in virulence traits such as melanin formation, and increased susceptibility to the stress conditions. Additionally, proper vacuolar function and vesicular acidification are needed for C. neoformans to causes disease (Erickson et al., 2001). Previous studies showed that a vam6Δ mutant in C. neoformans has impaired infectivity and transmigration into the central nervous system in mice (Liu et al., 2008; Tseng et al., 2012), decreased survival in cerebral spinal fluid and in macrophages (Lee et al. 2010; Fan and Liu, 2021), and increased sensitivity to SDS and NaCl (Fan and Liu, 2021). Vam6 and Vps41 are also known to play a role in the virulence of several plant pathogenic fungi (Li et al., 2017, 2018; Ramanujam et al., 2013; Yang et al., 2016; Zhang et al., 2017). As revealed in our study, Vps3 is also important for the ability of C. neoformans to cause disease in a mouse model of cryptococcosis. The contributions to proliferation in infected mice may be similar given the shared phenotypes with the vam6Δ mutant although the greater impact of the vps3Δ mutation on the expression of iron-regulated genes may suggest additional contributions of Vps3.
In summary, Vam6 and Vps3 are each required for robust growth on heme in C. neoformans, but display distinct properties. Mutants lacking Vps3 have a less severe growth defect on heme but better growth on medium with a low level of iron, have a single large vacuole, display impaired cycling of the iron permease Cft1 away from the plasma membrane, show a lack of repression of iron-responsive gene transcription and are avirulent. In contrast, vam6Δ mutants are impaired for growth both on heme and low levels of inorganic iron, have fragmented vacuoles, show impaired cycling of Cft1 between the plasma membrane and the vacuole, have little change in the transcription of iron-regulated genes, but are also avirulent in mice. Given the observed differences, further work is needed to understand the roles of these two proteins, their potential participation in CORVET and HOPS complexes, and their possible functions independent of the complexes.
4. EXPERIMENTAL PROCEDURES
4.1. Strains, plasmids, and media.
Strains used for all experiments were derivatives of C. neoformans var. grubii (serotype A) strain H99. Strains were propagated and maintained on YPD rich medium (1% yeast extract, 2% peptone, 2% dextrose, 2% agar). Plasmid pCH233 was the source of a nourseothricin resistance cassette, pJAF15 was the source of a hygromycin resistance cassette, and pJAF1 was the source of a neomycin resistance cassette. YPD plates containing either neomycin G418 (200 μg/ml), or nourseothricin (ClonNAT) (100 μg/ml), or hygromycin (200 μg/ml) were used to select C. neoformans HOPS and CORVET complex deletion transformants. All chemicals used were from Sigma-Aldrich (St. Louis, MO) unless otherwise stated. Solid plate assays for iron-associated phenotype assessment were made with yeast nitrogen base (YNB) (Difco) supplemented with the iron chelator, bathophenanthroline disulphonate (BPS), adjusted to pH 7.0 with 1M 3-(N-Morpholino) propanesulfonic acid (MOPS). Iron-chelated dH2O was prepared through passage of dH2O through a column containing Chelex-100 resin (BIORAD Chelex-100) and used in the preparation of YNB-BPS (Jung et al., 2010). Defined low-iron media (LIM) (0.5% glucose, 38 mM L-asparagine, 2.3 mM K2HPO4, 1.7 mM CaCl2·2H2O, 0.3 mM MgSO4·7H2O, 20 mM HEPES, 22 mM NaHCO3, 1 ml of 1000X salt solution (0.005 g/L CuSO4·5H2O, 2 g/L ZnSO4·7H2O, 0.01 g/L MnCl3·4H2O, 0.46 g/L sodium molybdate, 0.057 g/L boric acid), in iron-chelated dH2O adjusted to pH 7.4 with 0.4 mg/L sterile thiamine added post-filtering) was prepared as described previously (Griffiths et al., 2012; Lian et al., 2005).
4.2. Construction of gene deletion constructs and deletion mutants.
The sequences of the VPS3, VAM6, VPS41 and VPS8 genes and corresponding proteins in C. neoformans were identified using the H99 genome database (https://fungidb.org/fungidb/app) (Stajich et al., 2012) and compared with the reciprocal best matches from S. cerevisiae, as listed in Supplemental Figure S1. All deletion mutants were constructed by homologous recombination using gene-specific knockout cassettes, which were amplified by three-step overlapping PCR (Hu et al., 2008) with the primers listed in Supplemental Table S1. The resistance markers for neomycin (NEO), nourseothricin (NAT) and hygromycin (HYG) were amplified by PCR using primers 2 and 5 and the plasmids pJAF1, pCH233 and pJAF15, respectively, as the templates. In general, the gene-specific knockout primers 1 and 3 and 4 and 6 were used to amplify the flanking sequences of their respective genes; and primers 1 and 6 were used to amplify the gene-specific deletion construct containing the resistance marker. All constructs for deletions were introduced into the H99 WT strain, or a vam6Δ mutant (for generation of vps3Δ vam6Δ double deletion mutants) of the H99 strain, by biolistic transformation, as described previously (Davidson et al., 2000). Colony PCR was performed to screen positive transformants using primers 7NE and 8NE, 9PO and NAT3L, and 10PO and NAT5R, respectively, and confirmed by genomic hybridization analysis (Supplemental Figure S4). Primers used in screening are also summarized in Supplemental Table S1. For all experiments, two independent mutants of each gene were used, with the exception of a previously characterized vps41–1 mutant (M. Scofield, unpublished results), in order to validate the results seen from phenotypic assays. The deletions were confirmed by colony PCR and DNA hybridization (Supplemental Figure S4). The vps41-mh and vps8-mh mutants were obtained from the whole genome deletion collection described by Liu et al. (2008).
4.3. Construction of mCherry and GFP fusion alleles.
The Rim101-GFP fusion was constructed and nuclei were stained with DAPI (4’,6-diamidino-2-phenylindole) as previously described (Hu et al., 2017). The C-terminal region of the Cft1, Vps3 and Vam6 proteins was tagged with mCherry to examine the subcellular localization of Cft1, Vps3 and Vam6, respectively. A modified overlapping PCR strategy was used to generate the constructs for the Cft1-mCherry, Vps3-mCherry and Vam6-mCherry strains. Briefly, to generate Cft1-mCherry tagging construct, the upstream sequence (598 bps) and downstream sequence (622 bps) for the fusion construct were amplified from WT gDNA using the primer set Cft1-mCherry-P1F and Cft1-mCherry-P1R and the primer set Cft1-mCherry-P3F and Cft1-mCherry-P3R, respectively. The gene encoding mCherry and the neomycin resistance gene (Neo) were amplified from the plasmid pGH025 using primers Cft1-mCherry-P2F and Cft1-mCherry-P2R. Overlap PCR was performed using primers Cft1-mCherry-P1F and Cft1-mCherry-P3R to yield the ~5 kb construct. The construct was then used to transform either the WT (H99), or vps3Δ, or vam6Δ mutant strains by biolistic transformation. Following biolistic transformation, mutants were screened for resistance to hygromycin and G418, and the proper location and orientation of mCherry were determined by PCR. Primer sequences for Cft1-mCherry tagging construct are listed in Supplemental Table S2.
To generate Vps3-mCherry construct, the left and right arms for the Vps3-mCherry fusion construct were amplified from WT genomic DNA using the primer set Vps3-mCherry-P1F and Vps3-mCherry-P1R and the primer set Vps3-mCherry-P3F and Vps3-mCherry-P3R, respectively. The mCherry gene and the neomycin (Neo) resistance gene were amplified from the plasmid pGH025 using primers Vps3-mCherry-P2F and Vps3-mCherry-P2R. Overlap PCR was performed using primers Vps3-mCherry-P1F and Vps3-mCherry-P3R to yield a 5-kb construct. The Vps3-mCherry constructs were then used to transform the WT strain by biolistic transformation. Transformants were screened for resistance to hygromycin and G418, and the proper location and orientation of the gene fusions at VPS3 loci were determined by PCR. Primer sequences for the Vps3-mCherry fusion construct are listed in Supplemental Table S2.
To generate Vam6-mCherry construct, the left and right arms for the Vam6-mCherry fusion construct were amplified from WT genomic DNA using the primer set Vam6-mCherry-P1F and Vam6-mCherry-P1R and the primer set Vam6-mCherry-P3F and Vam6-mCherry-P3R, respectively. The mCherry gene and the neomycin (Neo) resistance gene were amplified from the plasmid pGH025 using primers Vam6-mCherry-P2F and Vam6-mCherry-P2R. Overlap PCR was performed using primers Vam6-mCherry-P1F and Vam6-mCherry-P3R to yield a 5-kb construct. The Vam6-mCherry constructs were then used to transform the WT strain by biolistic transformation. Transformants were screened for resistance to hygromycin and G418, and the proper location and orientation of the gene fusions at the VAM6 loci were determined by PCR. Primer sequences for the Vam6-mCherry fusion construct are listed in Supplemental Table S2.
4.4. Capsule formation and melanin production, and visualization of vacuole morphology.
Formation of polysaccharide capsule was examined by differential interference microscopy (DIC) on an Axioplan 2 imaging microscope (Zeiss), after incubation for 24 h at 30°C in defined low-iron medium (LIM) and staining with India ink. Melanin production was examined on L-3,4-dihydroxyphenylalanine (L-DOPA) plates containing 0.1% glucose (Tangen et al., 2007). The influence of copper on melanin production was investigated on L-DOPA plates supplemented with 100 μM or 1 mM of CuSO4. To observe vacuole morphology, cells from overnight cultures were incubated with lipophilic vacuole dye MDY-64 (Invitrogen, USA, 2.5 μM final concentration) for 15 minutes on ice and washed with liquid YPD medium. Cells were incubated at 30°C in YPD for an additional 30 minutes before visualized under fluorescence and DIC microscopy on an Axioplan 2 imaging microscope (Zeiss) with magnification 1000X, and a Zen Lite software. An additional vacuole-sequestered dye, carboxy-DCFDA (5-(and-6)-carboxy-2’,7’-dichlorofluorescein diacetate, Invitrogen, USA, at 10 μM final concentration), was used to assess vacuole morphology (Harrison et al., 2002).
4.5. Analysis of growth upon iron limitation
Strains were grown overnight at 30°C in YPD. Cells were then washed with iron-chelated dH2O and inoculated in defined LIM at a 1/100 dilution and grown for 48 hours at 30°C. Cells were then washed in sterile iron-chelated dH2O and adjusted to 2×107 cells/ml using a hemocytometer. 5 μl 10-fold serial dilutions were spotted on solid agar-based media from 105 to 100 cells. Plates were incubated at 30°C or 37°C for 2–3 days before being photographed. Growth of the strains was also assessed in liquid media. Cells for growth assays in liquid media were pre-grown overnight at 30 °C with shaking in YPD. The cells were then washed twice with low-iron water, inoculated into YNB-LIM at 4 × 106 cells per milliliter, and grown at 30 °C for 2 days to starve the cells for iron. After starvation, the cells were harvested, washed, and inoculated in YNB-LIM with or without supplemented iron sources to a final concentration of 5 × 104 cells per milliliter. Cultures were incubated at 30°C, and growth was monitored by measuring the optical density at 600 nm using a microplate reader (Infinite M200, Tecan).
4.6. Stress and drug response assays.
To examine the response of the WT, vps3Δ, vam6Δ and vps3Δ vam6Δ strains to various stress conditions, exponentially growing cultures were washed, resuspended in H2O, and adjusted to a concentration of 2 × 104 cells per milliliter. The cell suspensions were diluted 10-fold serially, and 5 μl of each dilution was spotted onto YPD and/or YNB plates supplemented with different compounds. Plates were incubated for 2–10 days at 30 or 37 °C and photographed. The responses of strains to oxidative, salt, or osmotic stress were examined. The specific assays were performed on YPD and/or YNB plates supplemented with or without 1.2 M KCl, 1.2 M NaCl, or 100 mM LiCl. Sensitivity of strains to rapamycin was tested on YPD containing either 15 or 30 μg/ml of rapamycin dissolved in ethanol.
4.7. Real-time PCR.
Real-time PCR analysis was conducted as previously described (Hu et al., 2007, 2015) using primers designed with Primer Express (Applied Biosystems, http://www.appliedbiosystems.com). Briefly, total RNA from frozen cells was extracted the using RNeasy mini kit (Qiagen), DNA was removed by treatment with Turbo DNase (Ambion) for 30 min at 25°C, and cDNA was synthesized using a mixture of anchored oligo dT and random hexamers (3:1) and Superscript transcriptase II (Invitrogen, Canada). The resulting cDNA was used for real-time PCR with Green-2-Go qPCR Mastermix – low ROX (Bio Basic, Canada) according to manufacturer’s recommendations. An Applied Biosystems 7500 Fast Real-Time PCR System was used to detect and quantify the PCR products using the following conditions: incubation at 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The cDNA of the actin gene was used to normalize the data. Dissociation analysis on all PCRs confirmed the amplification of a single product for each primer pair and the lack of primer dimerization (Applied Biosystems). The primers for each gene are listed in Supplemental Table S3. Relative gene expression was quantified using SDS software 1.3.1 (Applied Biosystems) and the 2-ΔΔCt method (Livak and Schmittgen, 2001).
4.8. Assessment of virulence
The virulence of the WT, two independent vps3Δ mutants, two vam6Δ mutants, and two vps3Δ vam6Δ double deletion mutant strains was also tested with female BALB/c mice. Female BALB/c mice, 4 to 6 weeks old, were obtained from Charles Rivers Laboratories (Pointe-Claire, Quebec, Canada). Fungal cells were cultured in 5 ml YPD at 30°C overnight, washed twice with PBS, and resuspended in PBS (pH 7.4). A cell suspension of 2 X 105 cells in 50 μl was intranasally instilled. The status of the mice was monitored once per day post-inoculation. Mice reaching the humane end point were euthanized by CO2 inhalation. For determination of the fungal load in organs, the brains and lungs were excised, weighed, and homogenized in 2 volumes of phosphate-buffered saline using a MixerMill (Retsch, Cole-Parmer, Montreal, Canada). Serial dilutions of the homogenates were plated on YPD plates containing 50 μg/ml chloramphenicol, and colony-forming units were counted after an incubation for 48 hr at 30 °C. Differences in virulence were statistically assessed by log rank tests using the GraphPad Prism 7 software program (GraphPad Software, San Diego, CA). All experiments with mice were conducted in accordance with the guidelines of the Canadian Council on Animal Care and the protocols for the virulence assays (protocol A17–0117) were approved by the University of British Columbia Committee on Animal Care.
Supplementary Material
Figure S1. Domain organization for Vps3 and Vam6, and sequence comparisons with proteins from S. cerevisiae.
The amino acid sequences of the indicated proteins were retrieved from https://fungidb.org/fungidb/app (accessed 31 August 2021). Protein families and domains (Vam6/VPS39/TRAP1 for Vps3, and Vam6/VPS39/TRAP1-TGF beta receptor associated domains) were identified in Fungidb, InterProScan (https://www.ebi.ac.uk), and PANTHER (http://www.pantherdb.org). B) The S. cerevisiae sequences with the highest similarities with the proteins from C. neoformans were identified by BLASTp analysis on the Saccharomyces Genome Database (https://www.yeastgenome.org) (accessed 1 September 2021).
Figure S2. Vps3 and Vam6 influence sensitivity to CoCl2, fluconazole, chloroquine and bleomycin.
(A) Tenfold serial dilutions of each strain were spotted on media with the indicated drugs and the plates were incubated at 30 °C for 2 days before being photographed. (B) Susceptibility to bleomycin was tested by spotting cells onto YPD plus 2 μg/ml bleomycin in the absence or presence of 150 μM bathophenanthrolinedisulfonic acid (BPS), or 100 μM FeCl3 as indicated. YPD = yeast extract peptone dextrose; WT = wild type
Figure S3. Influence of the pH on the growth of the vps3Δ, vam6Δ, and vps3Δ vam6Δ mutants.
Cells from tenfold serial dilutions of each strain were spotted onto yeast extract peptone dextrose (YPD) at the indicated pH, and the plates were incubated at 30°C for 2 days before being photographed.
Figure S4. Structure of deletion constructs and confirmation of mutant genotypes by PCR and genomic hybridization.
(A) Diagram of the replacement of the open reading frame of each gene by transformation of a deletion cassette containing the selectable resistance marker for either hygromycin (HYGr in vam6Δ, vps3Δ, and vps8Δ) or neomycin (NEOr in vps41Δ). (B) Diagram of the approach to identify positive transformants by PCR screening using the target-specific gene primer P9 and the cassette-specific primer Nat3L for left arm, and P10 and Nat5R for right arm, respectively. (C) The two vam6Δ mutants were positive for deletion by PCR. (D) Diagram of the genomic organization for the WT and vam6–5 and vam6–89 alleles showing the expected fragments upon digestion with PstI, and the probe location for hybridzation. (E) Genomic hybridization of the digested genomic DNA hybridized with the probe as indicated in (D) to confirm VAM6 deletion in the mutants (Δvam6–5 and Δvam6–89) identified by PCR. (F) Two vps3Δ mutants were positive for deletion by PCR. (G) Diagram of the genomic organization for WT and vps3-b8 and vps3-c7 mutants showing the expected fragments upon digestion with EcoRI, and the probe location for hybridization. (H) Genomic hybridization of the digested genomic DNA hybridized with the probe as indicated in (G) confirmed the VPS3 deletion. (I) The vps8–2 mutant is positive for deletion by PCR. (J) Diagram of the genomic organization for the WT and vps8 mutant showing the expected fragments upon digestion with BamHI, and the probe location for hybridization. (K) Genomic hybridization of the digested genomic DNA hybridized with the probe as indicated in (J) to confirm the VPS8 deletion. (L) The vps41Δ mutant is positive for deletion by PCR. (M) Diagram of the genomic organization for the WT and vps41Δ upon digestion with NcoI and SpeI, and the location of the probe for hybridization. (N) Genomic hybridization of the digested genomic DNA hybridized with the probe as indicated in (M) confirmed the VPS41 deletion. Note that one additional (independent) mutant each for VPS8 and VPS41 was obtained from the genomic mutant collection (Liu et al., 2008).
Take Away.
Two Vam6/Vps39/TRAP1-domain proteins, Vps3 and Vam6, support the growth of C. neoformans on heme.
Loss of Vps3 and Vam6 influences the trafficking and expression of iron uptake proteins.
Loss of Vps3 or Vam6 eliminates the ability of C. neoformans to cause disease in a mouse model of cryptococcosis.
ACKNOWLEDGEMENTS
The authors thank Michael Murphy for discussions and advice. This work is supported by the National Institute of Allergy and Infectious Diseases (RO1 AI053721), the Canadian Institutes of Health Research (PJT-166043) (JWK), and a doctoral scholarship from the National Sciences and Engineering Research Council of Canada (to L.C.H.). JWK is a Burroughs Wellcome Fund Scholar in Molecular Pathogenic Mycology, and a Canadian Institute for Advanced Research (CIFAR) Fellow in the Fungal Kingdom: Threats & Opportunities Program.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- Arlt H, Perz A, & Ungermann C (2011). An overexpression screen in Saccharomyces cerevisiae identifies novel genes that affect endocytic protein trafficking. Traffic (Copenhagen, Denmark), 12(11), 1592–1603. 10.1111/j.1600-0854.2011.01252.x [DOI] [PubMed] [Google Scholar]
- Asensio CS, Sirkis DW, Maas JW Jr, Egami K, To TL, Brodsky FM, Shu X, Cheng Y, & Edwards RH (2013). Self-assembly of VPS41 promotes sorting required for biogenesis of the regulated secretory pathway. Developmental Cell, 27(4), 425–437. 10.1016/j.devcel.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attarian R, Hu G, Sánchez-León E, Caza M, Croll D, Do E, Bach H, Missall T, Lodge J, Jung WH, & Kronstad JW (2018). The monothiol glutaredoxin Grx4 regulates iron homeostasis and virulence in Cryptococcus neoformans. mBio, 9(6), e02377–18. 10.1128/mBio.02377-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairwa G, Caza M, Horianopoulos L, Hu G, & Kronstad J (2019). Role of clathrin-mediated endocytosis in the use of heme and hemoglobin by the fungal pathogen Cryptococcus neoformans. Cellular Microbiology, 21(3), e12961. 10.1111/cmi.12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairwa G, Sánchez-León E, Do E, Jung WH, & Kronstad JW (2020). A cytoplasmic heme sensor illuminates the impacts of mitochondrial and vacuolar functions and oxidative stress on heme-iron homeostasis in Cryptococcus neoformans. mBio, 11(4), e00986–20. 10.1128/mBio.00986-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar HJ, & Ungermann C (2013). CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. Journal of Cell Science, 126(Pt 6), 1307–1316. 10.1242/jcs.107805 [DOI] [PubMed] [Google Scholar]
- Binda M, Péli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, & De Virgilio C (2009). The Vam6 GEF controls TORC1 by activating the EGO complex. Molecular Cell, 35(5), 563–573. 10.1016/j.molcel.2009.06.033 [DOI] [PubMed] [Google Scholar]
- Bröcker C, Kuhlee A, Gatsogiannis C, Balderhaar HJ, Hönscher C, Engelbrecht-Vandré S, Ungermann C, & Raunser S (2012). Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proceedings of the National Academy of Sciences of the United States of America, 109(6), 1991–1996. 10.1073/pnas.1117797109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M, Arlt H, Epp N, Lachmann J, Griffith J, Perz A, Reggiori F, & Ungermann C (2013). Functional separation of endosomal fusion factors and the class C core vacuole/endosome tethering (CORVET) complex in endosome biogenesis. The Journal of Biological Chemistry, 288(7), 5166–5175. 10.1074/jbc.M112.431536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadieux B, Lian T, Hu G, Wang J, Biondo C, Teti G, Liu V, Murphy ME, Creagh AL, & Kronstad JW (2013). The Mannoprotein Cig1 supports iron acquisition from heme and virulence in the pathogenic fungus Cryptococcus neoformans. The Journal of Infectious Diseases, 207(8), 1339–1347. 10.1093/infdis/jit029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat JE, & Skaar EP (2013). Iron in infection and immunity. Cell Host & Microbe, 13(5), 509–519. 10.1016/j.chom.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caza M, Hu G, Nielson ED, Cho M, Jung WH, & Kronstad JW (2018). The Sec1/Munc18 (SM) protein Vps45 is involved in iron uptake, mitochondrial function and virulence in the pathogenic fungus Cryptococcus neoformans. PLoS Pathogens, 14(8), e1007220. 10.1371/journal.ppat.1007220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Cruz MC, Sia RA, Allen B, Alspaugh JA, & Heitman J (2000). Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genetics and Biology : FG & B, 29(1), 38–48. 10.1006/fgbi.1999.1180 [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, & Schuldiner M (2014). A dynamic interface between vacuoles and mitochondria in yeast. Developmental Cell, 30(1), 95–102. 10.1016/j.devcel.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Erickson T, Liu L, Gueyikian A, Zhu X, Gibbons J, & Williamson PR (2001). Multiple virulence factors of Cryptococcus neoformans are dependent on VPH1. Molecular Microbiology, 42(4), 1121–1131. 10.1046/j.1365-2958.2001.02712.x [DOI] [PubMed] [Google Scholar]
- Fan CL, & Liu TB (2021). The vacuolar morphogenesis protein Vam6-like protein Vlp1 Is required for pathogenicity of Cryptococcus neoformans. Journal of Fungi (Basel, Switzerland), 7(6), 418. 10.3390/jof7060418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felice MR, De Domenico I, Li L, Ward DM, Bartok B, Musci G, & Kaplan J (2005). Post-transcriptional regulation of the yeast high affinity iron transport system. The Journal of Biological Chemistry, 280(23), 22181–22190. 10.1074/jbc.M414663200 [DOI] [PubMed] [Google Scholar]
- González Montoro A, Auffarth K, Hönscher C, Bohnert M, Becker T, Warscheid B, Reggiori F, van der Laan M, Fröhlich F, & Ungermann C (2018). Vps39 Interacts with Tom40 to Establish One of Two Functionally Distinct Vacuole-Mitochondria Contact Sites. Dev Cell. 45(5):621–636.e7. doi: 10.1016/j.devcel.2018.05.011. [DOI] [PubMed] [Google Scholar]
- Griffiths EJ, Hu G, Fries B, Caza M, Wang J, Gsponer J, Gates-Hollingsworth MA, Kozel TR, De Repentigny L, & Kronstad JW (2012). A defect in ATP-citrate lyase links acetyl-CoA production, virulence factor elaboration and virulence in Cryptococcus neoformans. Molecular Microbiology, 86(6), 1404–1423. 10.1111/mmi.12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison TS, Chen J, Simons E, & Levitz SM (2002). Determination of the pH of the Cryptococcus neoformans vacuole. Medical Mycology, 40(3), 329–332. 10.1080/mmy.40.3.329.332 [DOI] [PubMed] [Google Scholar]
- Heitman J, Movva NR, & Hall MN (1991). Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science (New York, N.Y.), 253(5022), 905–909. 10.1126/science.1715094 [DOI] [PubMed] [Google Scholar]
- Hönscher C, Mari M, Auffarth K, Bohnert M, Griffith J, Geerts W, van der Laan M, Cabrera M, Reggiori F, & Ungermann C (2014). Cellular metabolism regulates contact sites between vacuoles and mitochondria. Developmental Cell, 30(1), 86–94. 10.1016/j.devcel.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Horianopoulos LC, & Kronstad JW (2019). Connecting iron regulation and mitochondrial function in Cryptococcus neoformans. Current Opinion in Microbiology, 52, 7–13. 10.1016/j.mib.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Steen BR, Lian T, Sham AP, Tam N, Tangen KL, & Kronstad JW (2007). Transcriptional regulation by protein kinase A in Cryptococcus neoformans. PLoS Pathogens, 3(3), e42. 10.1371/journal.ppat.0030042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Caza M, Cadieux B, Chan V, Liu V, & Kronstad J (2013). Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence. Infection and Immunity, 81(1), 292–302. 10.1128/IAI.01037-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Caza M, Cadieux B, Bakkeren E, Do E, Jung WH, & Kronstad JW (2015). The endosomal sorting complex required for transport machinery influences haem uptake and capsule elaboration in Cryptococcus neoformans. Molecular Microbiology, 96(5), 973–992. 10.1111/mmi.12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Cheng PY, Sham A, Perfect JR, & Kronstad JW (2008). Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Molecular Microbiology, 69(6), 1456–1475. 10.1111/j.1365-2958.2008.06374.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Caza M, Bakkeren E, Kretschmer M, Bairwa G, Reiner E, & Kronstad J (2017). A P4-ATPase subunit of the Cdc50 family plays a role in iron acquisition and virulence in Cryptococcus neoformans. Cellular Microbiology, 19(6), 10.1111/cmi.12718 . 10.1111/cmi.12718https://doi.org/10.1111/cmi.12718. https://doi.org/10.1111/cmi.12718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadarola DM, Basu Ball W, Trivedi PP, Fu G, Nan B, & Gohil VM (2020). Vps39 is required for ethanolamine-stimulated elevation in mitochondrial phosphatidylethanolamine. Biochimica et Biophysica Acta. Molecular and Cell Biology of Lipids, 1865(6), 158655. 10.1016/j.bbalip.2020.158655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo WJ, Kim JH, Oh E, Jaramillo D, Holman P, Loguinov AV, Arkin AP, Nislow C, Giaever G, & Vulpe CD (2009). Novel insights into iron metabolism by integrating deletome and transcriptome analysis in an iron deficiency model of the yeast Saccharomyces cerevisiae. BMC Genomics, 10, 130. 10.1186/1471-2164-10-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonker C, Galmes R, Veenendaal T, Ten Brink C, van der Welle R, Liv N, de Rooij J, Peden AA, van der Sluijs P, Margadant C, & Klumperman J (2018). Vps3 and Vps8 control integrin trafficking from early to recycling endosomes and regulate integrin-dependent functions. Nature Communications, 9(1), 792. 10.1038/s41467-018-03226-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Hu G, Kuo W, & Kronstad JW (2009). Role of ferroxidases in iron uptake and virulence of Cryptococcus neoformans. Eukaryotic Cell, 8(10), 1511–1520. 10.1128/EC.00166-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, & Kronstad JW (2008). Iron and fungal pathogenesis: a case study with Cryptococcus neoformans. Cellular Microbiology, 10(2), 277–284. 10.1111/j.1462-5822.2007.01077.x [DOI] [PubMed] [Google Scholar]
- Jung WH, Sham A, White R, & Kronstad JW (2006). Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biology, 4(12), e410. 10.1371/journal.pbio.0040410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, Lian T, Singh A, Kosman DJ, & Kronstad JW (2008). Iron source preference and regulation of iron uptake in Cryptococcus neoformans. PLoS Pathogens, 4(2), e45. 10.1371/journal.ppat.0040045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Saikia S, Hu G, Wang J, Fung CK, D’Souza C, White R, & Kronstad JW (2010). HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathogens, 6(11), e1001209. 10.1371/journal.ppat.1001209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Yun CW, & Philpott CC (2002). Ferrichrome induces endosome to plasma membrane cycling of the ferrichrome transporter, Arn1p, in Saccharomyces cerevisiae. The EMBO Journal, 21(14), 3632–3642. 10.1093/emboj/cdf382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad JW, Hu G, & Jung WH (2013). An encapsulation of iron homeostasis and virulence in Cryptococcus neoformans. Trends in Microbiology, 21(9), 457–465. 10.1016/j.tim.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Toffaletti DL, Tenor J, Soderblom EJ, Thompson JW, Moseley MA, Price M, & Perfect JR (2010). Survival defects of Cryptococcus neoformans mutants exposed to human cerebrospinal fluid result in attenuated virulence in an experimental model of meningitis. Infection and Immunity, 78(10), 4213–4225. 10.1128/IAI.00551-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand D, Pierce A, Elass E, Carpentier M, Mariller C, & Mazurier J (2008). Lactoferrin structure and functions. Advances in Experimental Medicine and Biology, 606, 163–194. 10.1007/978-0-387-74087-4_6 [DOI] [PubMed] [Google Scholar]
- Li B, Liu L, Li Y, Dong X, Zhang H, Chen H, Zheng X, & Zhang Z (2017). The FgVps39-FgVam7-FgSso1 Complex mediates vesicle trafficking and is important for the development and virulence of Fusarium graminearum. Molecular Plant-Microbe Interactions : MPMI, 30(5), 410–422. 10.1094/MPMI-11-16-0242-R [DOI] [PubMed] [Google Scholar]
- Li B, Dong X, Li X, Chen H, Zhang H, Zheng X, & Zhang Z (2018). A subunit of the HOPS endocytic tethering complex, FgVps41, is important for fungal development and plant infection in Fusarium graminearum. Environmental Microbiology, 20(4), 1436–1451. 10.1111/1462-2920.14050 [DOI] [PubMed] [Google Scholar]
- Lian T, Simmer MI, D’Souza CA, Steen BR, Zuyderduyn SD, Jones SJ, Marra MA, & Kronstad JW (2005). Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans. Molecular Microbiology, 55(5), 1452–1472. 10.1111/j.1365-2958.2004.04474.x [DOI] [PubMed] [Google Scholar]
- Liu X, Hu G, Panepinto J, & Williamson PR (2006). Role of a VPS41 homologue in starvation response, intracellular survival and virulence of Cryptococcus neoformans. Molecular Microbiology, 61(5), 1132–1146. 10.1111/j.1365-2958.2006.05299.x [DOI] [PubMed] [Google Scholar]
- Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, & Noble SM (2008). Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell, 135(1), 174–188. 10.1016/j.cell.2008.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, & Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif.), 25(4), 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- López-Berges MS, Arst HN, Pinar M, & Peñalva MA (2017). Genetic studies on the physiological role of CORVET in Aspergillus nidulans. FEMS Microbiology Letters, 364(7), fnx065. 10.1093/femsle/fnx065 [DOI] [PubMed] [Google Scholar]
- Lu S, Suzuki T, Iizuka N, Ohshima S, Yabu Y, Suzuki M, Wen L, & Ohta N (2007). Trypanosoma brucei vacuolar protein sorting 41 (VPS41) is required for intracellular iron utilization and maintenance of normal cellular morphology. Parasitology, 134(Pt 11), 1639–1647. 10.1017/S0031182007003046 [DOI] [PubMed] [Google Scholar]
- Mao X, Yang L, Liu Y, Ma C, Ma T, Yu Q, & Li M (2021). Vacuole and mitochondria patch (vCLAMP) protein Vam6 is involved in maintenance of mitochondrial and vacuolar functions under oxidative stress in Candida albicans. Antioxidants (Basel, Switzerland), 10(1), 136. 10.3390/antiox10010136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May RC, Stone NR, Wiesner DL, Bicanic T, & Nielsen K (2016). Cryptococcus: from environmental saprophyte to global pathogen. Nature Reviews. Microbiology, 14(2), 106–117. 10.1038/nrmicro.2015.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer FL, & Kronstad JW (2020). Cryptococcus neoformans. Trends in Microbiology, 28(2), 163–164. 10.1016/j.tim.2019.10.003 [DOI] [PubMed] [Google Scholar]
- Mi H, Ebert D, Muruganujan A, Mills C, Albou LP, Mushayamaha T, & Thomas PD (2021). PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 49(D1):D394–D403. doi: 10.1093/nar/gkaa1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Hirata A, Ohsumi Y, & Wada Y (1997). Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membranes and involved in the vacuolar assembly in the yeast Saccharomyces cerevisiae. The Journal of Biological Chemistry, 272(17), 11344–11349. 10.1074/jbc.272.17.11344 [DOI] [PubMed] [Google Scholar]
- Navarathna DH, & Roberts DD (2010). Candida albicans heme oxygenase and its product CO contribute to pathogenesis of candidemia and alter systemic chemokine and cytokine expression. Free Radical Biology & Medicine, 49(10), 1561–1573. 10.1016/j.freeradbiomed.2010.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okurut S, Boulware DR, Olobo J, & Meya DB (2020). Landmark clinical observations and immunopathogenesis pathways linked to HIV and Cryptococcus fatal central nervous system co-infection. Mycoses, 63(8), 840–853. 10.1111/myc.13122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, Norton D, Price MS, Hay C, Clements MF, Nichols CB, & Alspaugh JA (2010). Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathogens, 6(2), e1000776. 10.1371/journal.ppat.1000776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ost KS, O’Meara TR, Huda N, Esher SK, & Alspaugh JA (2015). The Cryptococcus neoformans alkaline response pathway: identification of a novel rim pathway activator. PLoS Genetics, 11(4), e1005159. 10.1371/journal.pgen.1005159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, & Chiller TM (2009). Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS (London, England), 23(4), 525–530. 10.1097/QAD.0b013e328322ffac [DOI] [PubMed] [Google Scholar]
- Perini ED, Schaefer R, Stöter M, Kalaidzidis Y, & Zerial M (2014). Mammalian CORVET is required for fusion and conversion of distinct early endosome subpopulations. Traffic (Copenhagen, Denmark), 15(12), 1366–1389. 10.1111/tra.12232 [DOI] [PubMed] [Google Scholar]
- Pendrak ML, Chao MP, Yan SS, & Roberts DD (2004). Heme oxygenase in Candida albicans is regulated by hemoglobin and is necessary for metabolism of exogenous heme and hemoglobin to alpha-biliverdin. The Journal of Biological Chemistry, 279(5), 3426–3433. 10.1074/jbc.M311550200 [DOI] [PubMed] [Google Scholar]
- Pols MS, van Meel E, Oorschot V, ten Brink C, Fukuda M, Swetha MG, Mayor S, & Klumperman J (2013). hVps41 and VAMP7 function in direct TGN to late endosome transport of lysosomal membrane proteins. Nature Communications, 4, 1361. 10.1038/ncomms2360 [DOI] [PubMed] [Google Scholar]
- Prinz WA, Toulmay A, & Balla T (2020). The functional universe of membrane contact sites. Nature Reviews. Molecular Cell Biology, 21(1), 7–24. 10.1038/s41580-019-0180-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, & Boulware DR (2017). Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. The Lancet. Infectious Diseases, 17(8), 873–881. 10.1016/S1473-3099(17)30243-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanujam R, Calvert ME, Selvaraj P, & Naqvi NI (2013). The late endosomal HOPS complex anchors active G-protein signaling essential for pathogenesis in Magnaporthe oryzae. PLoS Pathogens, 9(8), e1003527. 10.1371/journal.ppat.1003527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, & Stevens TH (1992). Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Molecular Biology of the Cell, 3(12), 1389–1402. 10.1091/mbc.3.12.1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisky DC, Snyder WB, Emr SD, & Kaplan J (1997). Characterization of VPS41, a gene required for vacuolar trafficking and high-affinity iron transport in yeast. Proceedings of the National Academy of Sciences of the United States of America, 94(11), 5662–5666. 10.1073/pnas.94.11.5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaible UE, & Kaufmann SH (2004). Iron and microbial infection. Nature Reviews. Microbiology, 2(12), 946–953. 10.1038/nrmicro1046 [DOI] [PubMed] [Google Scholar]
- Solinger JA, & Spang A (2013). Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J. 280(12):2743–57. doi: 10.1111/febs.12151. [DOI] [PubMed] [Google Scholar]
- Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS, Kissinger JC, Li W, Nayak V, Pinney DF, Stoeckert CJ, & Roos DS Jr. (2012). FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res 40: D675–D681. doi: 10.1093/nar/gkr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic TI, Setty TG, Sitaram A, & Burd CG (2007). Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. The Journal of Cell Biology, 177(1), 115–125. 10.1083/jcb.200609161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic TI, Schmiedekamp BC, Lee J, Katzmann DJ, & Burd CG (2008). Opposing activities of the Snx3-retromer complex and ESCRT proteins mediate regulated cargo sorting at a common endosome. Molecular Biology of the Cell, 19(11), 4694–4706. 10.1091/mbc.e08-03-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczypka MS, Zhu Z, Silar P, & Thiele DJ (1997). Saccharomyces cerevisiae mutants altered in vacuole function are defective in copper detoxification and iron-responsive gene transcription. Yeast (Chichester, England), 13(15), 1423–1435. [DOI] [PubMed] [Google Scholar]
- Tangen KL, Jung WH, Sham AP, Lian T, & Kronstad JW (2007). The iron- and cAMP-regulated gene SIT1 influences ferrioxamine B utilization, melanization and cell wall structure in Cryptococcus neoformans. Microbiology (Reading, England), 153(Pt 1), 29–41. 10.1099/mic.0.2006/000927-0 [DOI] [PubMed] [Google Scholar]
- Tseng HK, Liu CP, Price MS, Jong AY, Chang JC, Toffaletti DL, Betancourt-Quiroz M, Frazzitta AE, Cho WL, & Perfect JR (2012). Identification of genes from the fungal pathogen Cryptococcus neoformans related to transmigration into the central nervous system. PloS One, 7(9), e45083. 10.1371/journal.pone.0045083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Beek J, Jonker C, van der Welle R, Liv N, & Klumperman J (2019). CORVET, CHEVI and HOPS - multisubunit tethers of the endo-lysosomal system in health and disease. Journal of Cell Science, 132(10), jcs189134. 10.1242/jcs.189134 [DOI] [PubMed] [Google Scholar]
- Vartivarian SE, Anaissie EJ, Cowart RE, Sprigg HA, Tingler MJ, & Jacobson ES (1993). Regulation of cryptococcal capsular polysaccharide by iron. The Journal of Infectious Diseases, 167(1), 186–190. 10.1093/infdis/167.1.186 [DOI] [PubMed] [Google Scholar]
- Weissman Z, Shemer R, Conibear E, & Kornitzer D (2008). An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Molecular Microbiology, 69(1), 201–217. 10.1111/j.1365-2958.2008.06277.x [DOI] [PubMed] [Google Scholar]
- Yang X, Cui H, Cheng J, Xie J, Jiang D, Hsiang T, & Fu Y (2016). A HOPS protein, CmVps39, is required for vacuolar morphology, autophagy, growth, conidiogenesis and mycoparasitic functions of Coniothyrium minitans. Environmental Microbiology, 18(11), 3785–3797. 10.1111/1462-2920.13334 [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang G, Yang C, Huang J, Chen X, Zhou J, Li G, Norvienyeku J, & Wang Z (2017). A HOPS Protein, MoVps41, Is Crucially Important for Vacuolar Morphogenesis, Vegetative Growth, Reproduction and Virulence in Magnaporthe oryzae. Frontiers in Plant Science, 8, 1091. 10.3389/fpls.2017.01091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita-Martinez SA, Puria R, Pan X, Boeke JD, & Cardenas ME (2007). Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics, 176(4), 2139–2150. 10.1534/genetics.107.072835 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Domain organization for Vps3 and Vam6, and sequence comparisons with proteins from S. cerevisiae.
The amino acid sequences of the indicated proteins were retrieved from https://fungidb.org/fungidb/app (accessed 31 August 2021). Protein families and domains (Vam6/VPS39/TRAP1 for Vps3, and Vam6/VPS39/TRAP1-TGF beta receptor associated domains) were identified in Fungidb, InterProScan (https://www.ebi.ac.uk), and PANTHER (http://www.pantherdb.org). B) The S. cerevisiae sequences with the highest similarities with the proteins from C. neoformans were identified by BLASTp analysis on the Saccharomyces Genome Database (https://www.yeastgenome.org) (accessed 1 September 2021).
Figure S2. Vps3 and Vam6 influence sensitivity to CoCl2, fluconazole, chloroquine and bleomycin.
(A) Tenfold serial dilutions of each strain were spotted on media with the indicated drugs and the plates were incubated at 30 °C for 2 days before being photographed. (B) Susceptibility to bleomycin was tested by spotting cells onto YPD plus 2 μg/ml bleomycin in the absence or presence of 150 μM bathophenanthrolinedisulfonic acid (BPS), or 100 μM FeCl3 as indicated. YPD = yeast extract peptone dextrose; WT = wild type
Figure S3. Influence of the pH on the growth of the vps3Δ, vam6Δ, and vps3Δ vam6Δ mutants.
Cells from tenfold serial dilutions of each strain were spotted onto yeast extract peptone dextrose (YPD) at the indicated pH, and the plates were incubated at 30°C for 2 days before being photographed.
Figure S4. Structure of deletion constructs and confirmation of mutant genotypes by PCR and genomic hybridization.
(A) Diagram of the replacement of the open reading frame of each gene by transformation of a deletion cassette containing the selectable resistance marker for either hygromycin (HYGr in vam6Δ, vps3Δ, and vps8Δ) or neomycin (NEOr in vps41Δ). (B) Diagram of the approach to identify positive transformants by PCR screening using the target-specific gene primer P9 and the cassette-specific primer Nat3L for left arm, and P10 and Nat5R for right arm, respectively. (C) The two vam6Δ mutants were positive for deletion by PCR. (D) Diagram of the genomic organization for the WT and vam6–5 and vam6–89 alleles showing the expected fragments upon digestion with PstI, and the probe location for hybridzation. (E) Genomic hybridization of the digested genomic DNA hybridized with the probe as indicated in (D) to confirm VAM6 deletion in the mutants (Δvam6–5 and Δvam6–89) identified by PCR. (F) Two vps3Δ mutants were positive for deletion by PCR. (G) Diagram of the genomic organization for WT and vps3-b8 and vps3-c7 mutants showing the expected fragments upon digestion with EcoRI, and the probe location for hybridization. (H) Genomic hybridization of the digested genomic DNA hybridized with the probe as indicated in (G) confirmed the VPS3 deletion. (I) The vps8–2 mutant is positive for deletion by PCR. (J) Diagram of the genomic organization for the WT and vps8 mutant showing the expected fragments upon digestion with BamHI, and the probe location for hybridization. (K) Genomic hybridization of the digested genomic DNA hybridized with the probe as indicated in (J) to confirm the VPS8 deletion. (L) The vps41Δ mutant is positive for deletion by PCR. (M) Diagram of the genomic organization for the WT and vps41Δ upon digestion with NcoI and SpeI, and the location of the probe for hybridization. (N) Genomic hybridization of the digested genomic DNA hybridized with the probe as indicated in (M) confirmed the VPS41 deletion. Note that one additional (independent) mutant each for VPS8 and VPS41 was obtained from the genomic mutant collection (Liu et al., 2008).