Summary
Electrical synapses are established between specific neurons and within distinct subcellular compartments, but the mechanisms that direct gap junction assembly in the nervous system are largely unknown. Here we show that a developmental program tunes cAMP signaling to direct the neuron-specific assembly and placement of electrical synapses in the C. elegans motor circuit. We used live cell imaging to visualize electrical synapses in vivo and an optogenetic assay to confirm that they are functional. In VA motor neurons, the UNC-4 transcription factor blocks expression of cAMP antagonists that promote gap junction miswiring. In unc-4 mutants, VA electrical synapses are established with an alternative synaptic partner and are repositioned from the VA axon to soma. cAMP counters these effects by driving gap junction trafficking into the VA axon for electrical synapse assembly. Thus, our experiments establish that cAMP regulates gap junction trafficking for the biogenesis of functional electrical synapses.
Keywords: Electrical synapse, gap junction, synaptic specificity, cAMP, subcellular targeting, synaptic choice
eTOC Blurb
The molecular players that control the specificity of electrical synapses are largely unknown. Studying a C. elegans motor circuit, Palumbos et al. determine that cAMP promotes the neuron-specific targeting and subcellular location of electrical synapses by controlling the trafficking of the gap junction component UNC-9.
Graphical Abstract
INTRODUCTION
Neuronal function depends on the neuron-specific assembly of both Chemical and electrical synapses. In comparison to chemical synapses (Margeta and Shen, 2010; Sanes and Yamagata, 2009), strikingly little is known of the pathways that direct the formation of electrical synapses between specific neurons (Hendi et al., 2019; Hestrin and Galarreta, 2005). This disparity is significant as electrical synapses account for nearly 20% of connections in mature nervous systems (Connors and Long, 2004; Cook et al., 2019; White et al., 1986) and are required for diverse neural processes (Allen et al., 2011; Kawano et al., 2011; Phelan et al., 1998; Song et al., 2016; Walker and Schafer, 2020). In addition, the differential placement of electrical synapses in specific subcellular compartments (e.g., axon vs. soma) strongly influences neuronal output (Tamás et al., 2000; Wang et al., 2017). Thus, a deeper understanding of mechanisms that direct assembly of electrical synapses within a given circuit is of fundamental importance to developmental neuroscience.
Electrical synapses, or gap junctions, mediate the rapid transfer of ions and small molecules, thus coupling connected neurons (Pereda et al., 2013). In vertebrates, gap junctions are assembled from connexins which oligomerize to form hexagonal hemichannels in each neuron (Maeda et al., 2009; Sosinsky and Nicholson, 2005). Hemichannels in adjacent neurons appose one another to form a gap junction array. In an intriguing example of convergent evolution, the invertebrate gap junction subunits, innexins, adopt a topology and function that is strikingly similar to that of connexins despite lacking clear sequence homology (Skerrett and Williams, 2016). The likelihood of conserved mechanisms for gap junction assembly is supported by the finding that innexin-containing gap junctions can function in vertebrate cells (Dykes et al., 2004; Phelan et al., 1998; Starich et al., 2009), and conversely, that connexins can form gap junctions when expressed in invertebrate cells (Meng and Yan, 2020; Rabinowitch et al., 2014). It follows that invertebrate model organisms, with their powerful genetic tools and accessibility to live-cell imaging, can be exploited to identify regulators of gap junction biogenesis that may also specify electrical synapses in mammalian neurons (Meng and Yan, 2020; Schneider et al., 2012; Von Stetina et al., 2007).
Early experiments in cultured cells suggested a straightforward model in which neuron-specific electrical synapses arise solely from differential expression of gap junction subunits. This idea derived from the observation that overexpression of either connexins or innexins is sufficient to drive assembly of gap junctions at the interface between random pairs of adjacent cells (Elfgang et al., 1995; Rabinowitch et al., 2014; Teubner et al., 2000). Investigations of this question in vivo, however, have revealed that assembly of neuron-specific electrical synapses requires additional regulatory mechanisms. For example, in many instances, gap junctions do not assemble between adjacent neurons that express compatible gap junction subunits (Bhattacharya et al., 2019; Fukuda, 2017; Greb et al., 2017; White et al., 1992; Yao et al., 2016). Thus, developmental programs that direct trafficking and assembly of gap junction components are likely necessary for the placement of electrical synapses between specific neurons.
A wide range of molecular regulators of gap junction homeostasis have been defined by experiments in cultured cells. For example, specific protein kinases modulate gap junction function and trafficking (Lampe and Lau, 2004; Solan and Lampe, 2005, 2009, 2016). Notably, Protein Kinase A (PKA), a 3’,5’-cyclic adenosine monophosphate (cAMP) dependent kinase, can promote gap junction coupling, potentially by regulating trafficking of Cx43 connexons to the plasma membrane (Atkinson et al., 1995; Burghardt et al., 1995; Holm et al., 1999; Ouyang et al., 2005; Paulson et al., 2000; TenBroek et al., 2001). Few studies, however, have asked whether similar mechanisms regulate gap junction placement in an intact nervous system.
Here, we exploit the simplicity and accessibility of the C. elegans nervous system to identify components that direct the formation of neuron-specific gap junctions. We used a genetic screen to identify two negative-regulators of cAMP signaling, a phosphodiesterase (PDE-1) and a GPCR (FRPR-17), that direct the neuron-specificity and subcellular placement of gap junctions in the motor circuit. Both pde-1 and frpr-17 are normally turned off in Ventral A Class (VA) motor neurons by the UNC-4 transcription factor. Ectopic expression of PDE-1 and FRPR-17 in an unc-4 mutant, however, results in the creation of electrical synapses between VAs and a new interneuron partner and also repositions gap junctions from the VA axon to the cell soma. We performed genetic and pharmacologic experiments to confirm that cAMP acts within a critical developmental period to regulate assembly of functional electrical synapses. Our findings show that elevated cAMP promotes trafficking of gap junction components from the VA cell soma for assembly of electrical synapses in the VA axon. In addition, we find that this trafficking mechanism facilitates the creation of additional neuron-specific electrical synapses as VA neurons expand in size during larval development. Thus, cAMP regulates a trafficking mechanism that controls the neuron specificity, subcellular placement and developmental scaling of electrical synapses in the nervous system.
RESULTS
UNC-4-regulated genes direct gap junction specificity
In the C. elegans motor circuit, VA motor neurons adopt both Chemical and electrical synapses with the interneuron AVA (VA→AVA), whereas VBs, the sister cells of VAs, exclusively establish electrical synapses with the interneuron AVB (VB→AVB) (White et al., 1986). These connections are required to drive either backward (VA→AVA) or forward (VB→AVB) locomotion (Chalfie et al., 1985; Kawano et al., 2011; Wen et al., 2012). Gap junctions in this circuit are heterotypic, with the innexin UNC-7 contributed by AVA and AVB and the innexin UNC-9 derived from VA and VB motor neurons (Figure 1A) (Starich et al., 2009). Importantly, both AVA and AVB are in physical contact with VA and VB neurons (White et al., 1986, 1992), suggesting that a developmental program is required to direct the neuron-specific assembly of electrical synapses in this circuit. The transcription factor UNC-4 is selectively expressed in VA motor neurons where it controls synaptic specificity (Miller and Niemeyer, 1995; Miller et al., 1992). unc-4 mutants are unable to move backward because chemical and electrical synapses with AVA are replaced with ectopic electrical input from AVB (VA→AVB) (Figure 1A–C) (White et al., 1992). The specificity of these electrical synapses is readily detectable utilizing the GFP-tagged innexin UNC-7 (UNC-7S::GFP) expressed in the interneuron AVB (Starich et al., 2009; Von Stetina et al., 2007) (Figure 1B). In addition to the misassembly of VA electrical synapses with AVB, VA gap junctions are also repositioned from the VA axon to the cell soma in unc-4 mutants (Figure 1A) (Von Stetina et al., 2007; White et al., 1992). To identify the molecular players that influence these effects, we used a combination of cell-specific RNA-Seq profiling and genetic tests to detect transcripts regulated by UNC-4 that control the neuron-specific assembly of electrical synapses.
Figure 1: UNC-4 functions as a transcriptional repressor to direct gap junction specificity.
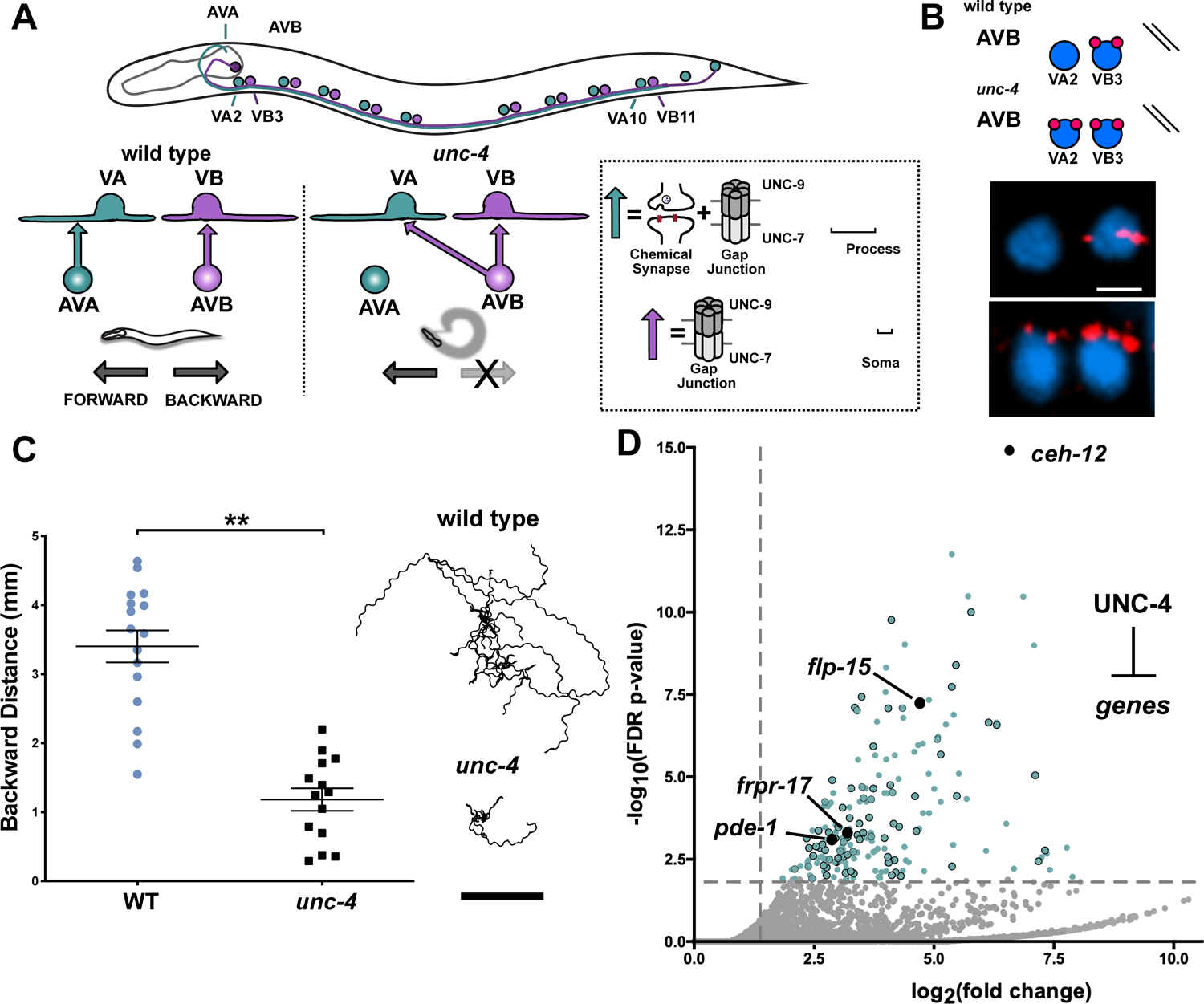
(A) Motor circuit regulated by UNC-4. In the wild type, the AVA interneuron establishes Chemical and electrical synapses with the VA motor neuron axon (VA→AVA), whereas AVB forms gap junctions on the VB cell soma. unc-4 mutants are miswired with AVB electrical synapses on VA cell soma (VA→AVB). Gap junctions in this circuit are heterotypic with UNC-7 provided by AVA and AVB and UNC-9 contributed by VA and VB. (B) (Top) UNC-7s::GFP expressed in AVB labels AVB gap junctions. (Bottom) AVB gap junctions are labeled with UNC-7s::GFP (red) and VA2 and VB3 soma are stained with DAPI (blue). In unc-4 mutants UNC-7s::GFP labels ectopic VA→AVB gap junctions. Scale bar = 2.5μm. (C) (Left) Backward distance traveled by wild type (n = 15) and unc-4 (n = 13) in a 3-minute period. Student’s t-test, ** = p< .01. Data are mean +/− SE. (Right) Representative tracks of 10 wild type and 10 unc-4 L4 worms in a 3-minute period. Scale bar = 2 mm. (D) Volcano plot of upregulated transcripts (> 2X, FDR p-value < .01) detected in unc-4(e120) VAs. RNAi or genetic mutants of 80/214 upregulated targets (blue dots) identified four suppressors of the Unc-4 movement defect (black). unc-4(e2323) allele used unless otherwise noted.
We used an intersectional labeling strategy for FACS-isolation of VA neurons from L2 larval stage animals, the developmental period in which unc-4 function is required (Miller et al., 1992; Spencer et al., 2014). RNA-seq analysis identified >2000 transcripts that are significantly enriched in wild-type VAs. Robust expression of known VA cell marker genes (e.g., unc-4, unc-3, del-1, cfi-1) in this data set validates FACS enrichment of VA motor neurons (GEO: GSE173287) (Kerk et al., 2017; Miller and Niemeyer, 1995; Von Stetina et al., 2007; Taylor et al., 2021). Comparison of wild-type vs. unc-4 mutant VAs datasets detected over 300 transcripts (98 down-regulated, 214 up-regulated) as differentially expressed in unc-4 mutant VAs (Figure S1). We focused on the 214 transcripts that are up-regulated in unc-4 mutant VAs because UNC-4 normally functions as a transcriptional repressor (Pflugrad et al., 1997; von Stetina et al., 2007; Winnier et al., 1999) (Figure 1D). Consistent with previous findings, the homeodomain transcription factor gene, ceh-12, was the most significantly upregulated transcript in unc-4 mutant VAs. Importantly, ectopic expression of ceh-12 in unc-4 mutants is restricted to a subset of VA neurons in the posterior ventral nerve cord (VNC) where it regionally drives gap junction miswiring (Schneider et al., 2012; Von Stetina et al., 2007). The local effect of ceh-12 on gap junction specificity predicts that UNC-4 regulates other targets that act in anterior VAs to sustain the wild-type pattern of electrical connectivity.
To identify additional regulators of VA gap junction specificity, we used RNAi and available loss-of-function mutants in a “suppressor” screen of upregulated UNC-4 target genes. Knockdown of an UNC-4 target that drives gap junction miswiring is predicted to partially restore backward movement in an unc-4 mutant, an outcome we describe as Unc-4 suppression (Figure S1). We tested 80 (Table S1) upregulated genes with corresponding mammalian homologs and detected three “suppressor” loci (pde-1, frpr-17, flp-15) that significantly improved backward locomotion in an unc-4 mutant (Figure 2B–C, Figure S1). Notably, pde-1 and frpr-17 are both predicted to antagonize cAMP signaling (Figure 2A). PDE-1 is functionally homologous to the mammalian phosphodiesterase PDE1A (77.4%, BLAST e-value 3e-173) with calcium-dependent enzymatic activity that degrades both cAMP and cGMP (Lugnier, 2006). FRPR-17 corresponds to an orphan GPCR that is predicted to bind FMRF-type neuropeptides and couple to Gαi/o (99% coupling score, PredCouple2) (Figure S1). C. elegans expresses a single Gαi/o, GOA-1/Gαo, which is homologous to the mammalian GNAI2 (100%, BLAST e-value 0) and GNAO1 (82.2%, BLAST e-value 0) that antagonize (AC) Adenylyl Cyclase-dependent synthesis of cAMP (Muntean et al., 2021). Therefore, ectopic expression of FRPR-17 in unc-4 mutant VAs could activate GOA-1 with a consequent reduction in cAMP. Taken together, these results suggest that UNC-4 may act to preserve cAMP signaling in VA neurons by blocking expression of both PDE-1, which degrades cAMP, and FRPR-17, which limits cAMP synthesis.
Figure 2: UNC-4 maintains cAMP with distinct mechanisms in different VA motor neurons.
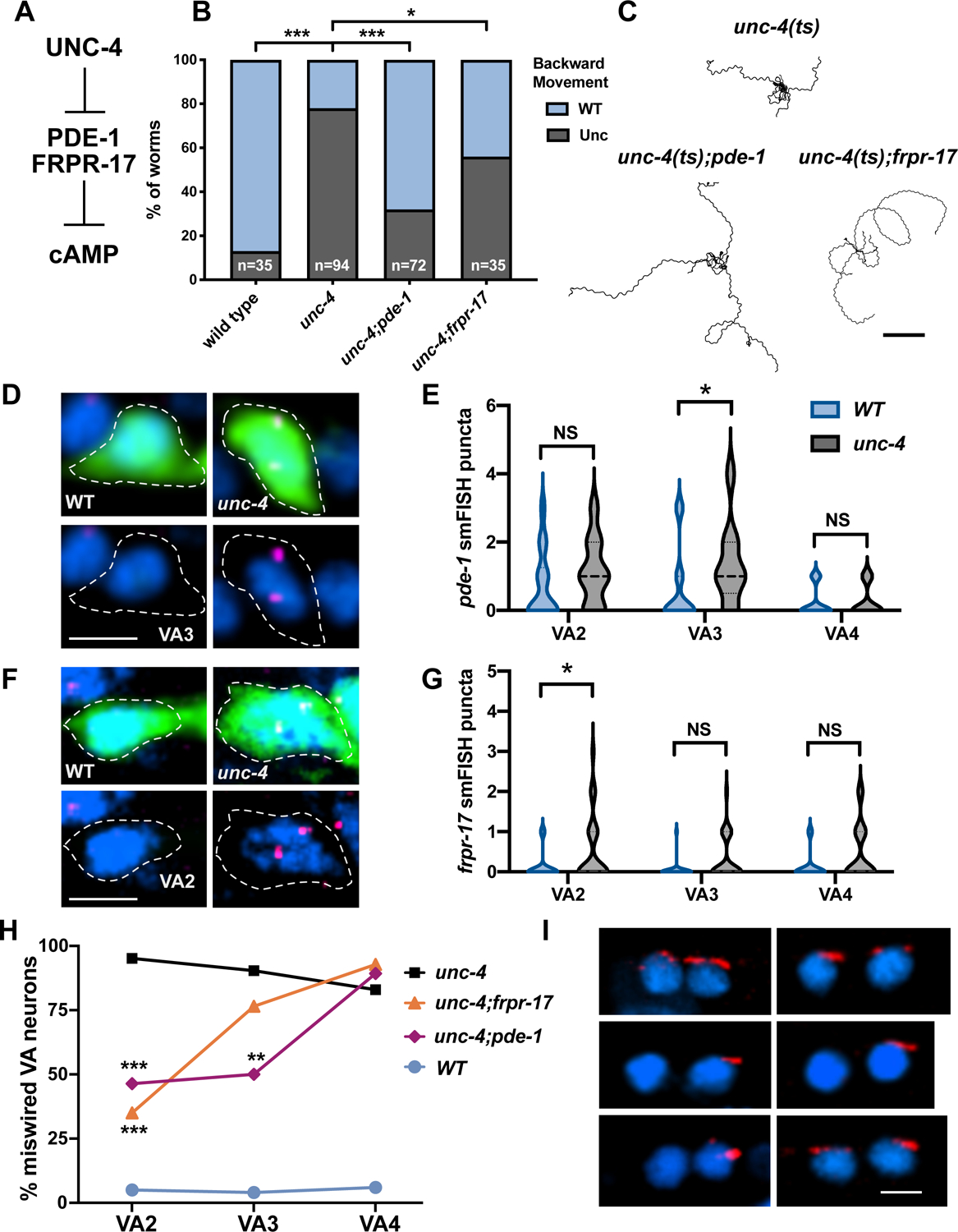
(A) UNC-4 blocks expression of PDE-1/Phosphodiesterase and FRPR-17/GPCR, both predicted negative-regulators of cAMP. (B) Backward movement scored (23°C) as either wild type (WT) or uncoordinated (Unc) for unc-4(ts), unc-4(ts);pde-1 and unc-4(ts);frpr-17. Fisher’s Exact test versus unc-4(ts), * = p< 0.05, *** = p<0.001. (C) Representative tracks of L4 larvae for each genotype at 23°C in a 3-minute period. Scale bar = 2 mm. unc-4(ts) = unc-4(e2322). (D) Representative images of VA3 in wild type (left) and unc-4(e120) (right). Dashed lines denote cell soma marked with GFP (top), labeled with DAPI (blue) and pde-1 smFISH probe (magenta). (E) Violin plots of pde-1 smFISH puncta in WT and unc-4(e120) in L2 stage VA neurons. Dashed line denotes median. Mann-Whitney test, * p = 0.037, n>20 per VA. (F) Representative images of VA2 in wild type (left) and unc-4(e120) (right). Dashed lines denote cell soma marked with GFP (top), labeled with DAPI (blue) and frpr-17 smFISH probe (magenta). (G) Quantification of frpr-17 smFISH puncta in VA motor neurons. Dashed line denotes median. Mann-Whitney test, * p= 0.039. n>25 per VA. (H) Ectopic VA→AVB gap junctions plotted as percent of each anterior VA neuron (VA2, VA3, VA4) in unc-4 (black), unc-4;frpr-17 (orange), unc-4;pde-1 (magenta) and wild type (WT) (light blue). Lines connect genotype. Fisher’s exact versus unc-4. N>15 for each VA, ** = p<0.01, *** = p<0.001. (I) Representative images of UNC-7S::GFP marking AVB gap junctions (red) with VA and VB motor neurons in unc-4 (top) unc-4;pde-1 (middle) and unc-4;frpr-17 (top). DAPI (blue) labels VA and VB nuclei. Scale bar = 2.5 μm. unc-4(e2323) was used for H and I.
UNC-4 regulates cAMP signaling through neuron-specific gene regulation
Our RNA-sequencing analysis indicates that pde-1 (4.5X, FDR p<.01) and frpr-17 (8.2X, FDR p<.01) are upregulated in unc-4 mutant VAs (Figure 1D). As an independent confirmation of this finding, we used single molecule fluorescent in situ hybridization (smFISH) to visualize pde-1 and frpr-17 transcripts in VA neurons. We focused on VAs in the anterior VNC (VA2, VA3, VA4) because we have previously shown that ceh-12 is ectopically expressed in posterior VA neurons (VA8, VA9, VA10) to drive miswiring in unc-4 mutants (von Stetina et al., 2007). In the wild type, we detected pde-1 smFISH puncta in VA2 and VA3. In unc-4 mutants, we observed a significant increase in pde-1 transcripts in VA3 (Figure 2D–E). pde-1 is also ectopically expressed in a subset of posterior VA neurons in unc-4 mutants (Figure S1). In contrast, frpr-17 was selectively elevated in unc-4 mutant VA2 with little frpr-17 transcript detected in wild-type VA2–4 (Figure 2F–G). These findings confirm that UNC-4 negatively regulates frpr-17 and pde-1 expression in VA neurons.
UNC-4 blocks the formation of ectopic electrical synapses through differential neuron-specific gene regulation
As noted above, ectopic expression of pde-1 and frpr-17 contributes to the impaired backward movement of unc-4 mutants (Figure 2B, C). We next utilized the UNC-7S::GFP marker to ask if pde-1 and frpr-17 are necessary for the VA→AVB wiring defect in unc-4 mutants. If ectopic pde-1 expression is required for the Unc-4 miswiring defect, then the occurrence of VA→AVB gap junctions should be reduced in unc-4; pde-1 double mutants compared to unc-4 alone. As predicted, we observed significantly fewer VA→AVB gap junctions in unc-4; pde-1 mutants for VA2 and VA3 but not for VA4 (Figure 2 H–I). Our smFISH assay showed that pde-1 transcripts are detectable in VA2 and elevated in VA3 in unc-4 mutants (Figure 2D–E), suggesting that pde-1 expression in both VA2 and VA3 favors miswiring.
We also used the UNC-7S::GFP marker to show that VA→AVB gap junctions are reduced in VA2 in unc-4; frpr-17 double mutants, but not in VA3 or in VA4 (Figure 2H–I). This selective effect of the frpr-17 mutation on VA2 wiring is consistent with our smFISH results showing that the frpr-17 transcript is exclusively detected in unc-4 mutant VA2 neurons in the anterior nerve cord (Figure 2F–G). Together, our findings point to a model in which UNC-4 antagonizes the formation of ectopic VA→AVB gap junctions by blocking expression of different target genes in specific VA neurons (e.g., frpr-17 in VA2 and pde-1 in VA3) that antagonize cAMP signaling.
UNC-4 preserves cAMP to direct the neuron specificity of electrical synapses
Having shown that UNC-4 prevents expression of two negative-regulators of cAMP signaling (PDE-1 and FRPR-17) to regulate the neuron specificity of gap junction assembly, we hypothesized that elevated cAMP signaling prevents the formation of ectopic VA→AVB electrical synapses. In that case, other pharmacological or genetic manipulations that elevate cAMP should similarly restore backward locomotion to an unc-4 mutant. As an initial test of this prediction, we utilized the cell-permeable, non-hydrolyzable cAMP analog, 8-Bromo-cAMP (8-Br-cAMP), to phenocopy global elevation of cAMP (Hussey et al., 2017). unc-4 mutant worms fed 8-Br-cAMP exhibited significant restoration of backward locomotion (Figure S2), suggesting that elevated cAMP is sufficient to prevent miswiring of the backward movement circuit.
To further delineate the role of cAMP in electrical synaptic specificity, we used a series of genetic approaches to elevate cAMP levels in vivo. In C. elegans, the biosynthetic enzyme adenylyl cyclase (ACY-1/AC) is regulated by antagonistic G-protein pathways that either stimulate (GSA-1/GαS) or reduce (GOA-1/GαO) cAMP production (Figure 3A) (Govindan et al., 2006). Therefore, activation of either ACY-1/AC or GSA-1/GαS should increase cAMP. cAMP levels should also be elevated by genetic ablation of GOA-1/GαO. First, we asked if the loss of GOA-1/GαO function could suppress the Unc-4 movement defect. We determined that backward movement in unc-4; goa-1(lof) mutants was significantly improved in comparison to unc-4 (Figure 3B–C, Video S1). We next used the UNC-7S::GFP marker to score ectopic VA→AVB gap junctions in unc-4; goa-1(lof) double mutants and confirmed a significant reduction of VA→AVB miswiring in all three anterior VAs (VA2, VA3, VA4) in comparison to unc-4 mutant VAs (Figure 3D–E). We also observed that VA→AVB miswiring is similarly suppressed in multiple posteriorly located VAs in unc-4; goa-1(lof) (Skelton, 2012). These observations suggest that the specificity of VA gap junction formation is globally sensitive to cAMP signaling.
Figure 3: cAMP is required in VA neurons to maintain electrical synapses for backward locomotion.
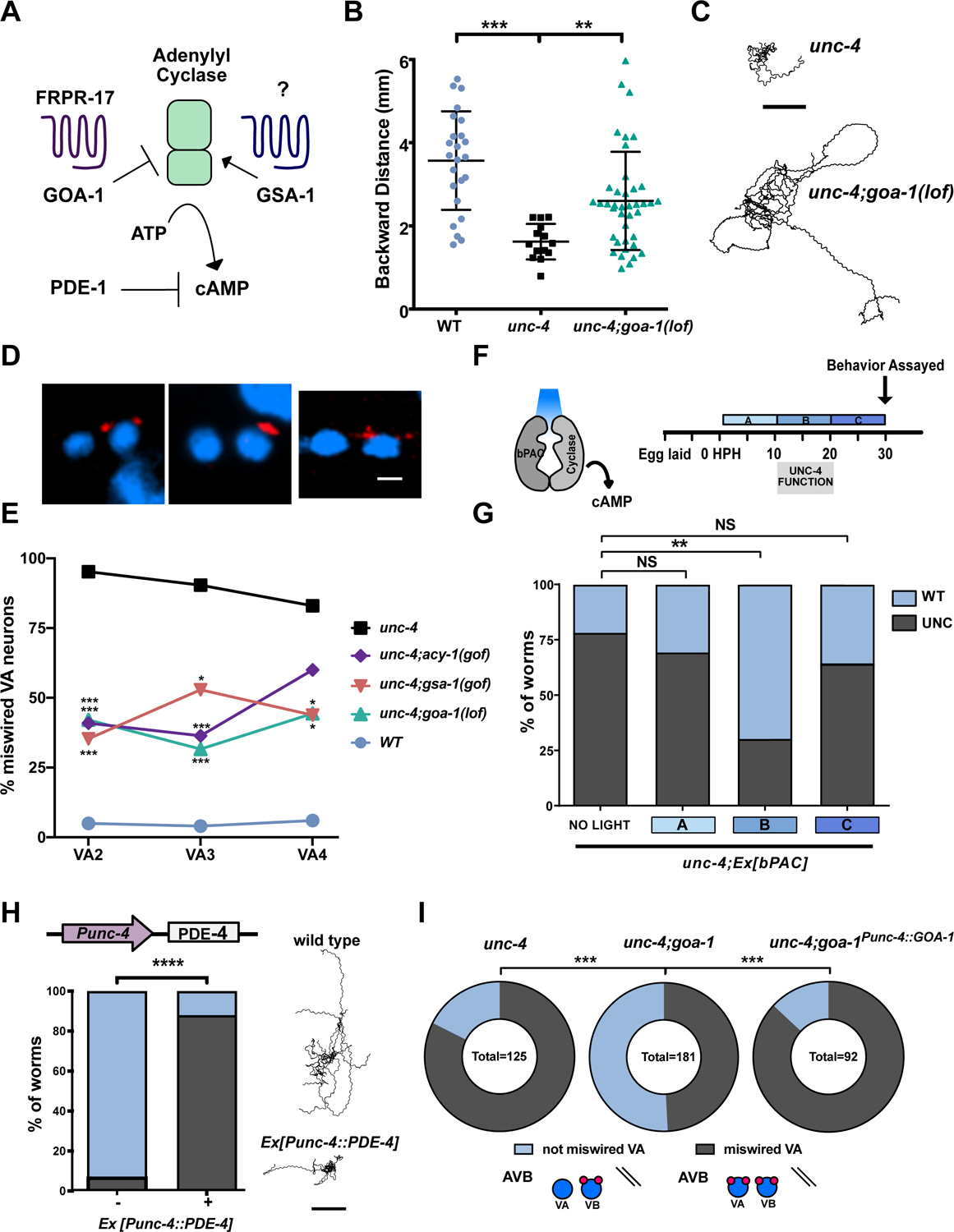
(A) Schematic of cAMP regulation. The G-protein GSA-1/GaS promotes Adenylyl Cyclase-dependent synthesis of cAMP whereas GOA-1/GaO antagonizes cAMP production. The GPCR, FRPR-17, is predicted to couple to GOA-1. PDE-1/phosphodiesterase degrades cAMP. (B) Backward distance traveled by wild type, unc-4 and unc-4;goa-1(lof). One-way ANOVA, N > 15 for each genotype, ** p=.0089, **** = p<.0001. (C) Representative tracks of ten unc-4 (left) and ten unc-4;goa-1(lof) (right) L4 larvae in a 3-minute period. Scale bar = 2mm. (D) UNC-7s::GFP (red) marks AVB gap junctions in unc-4;goa-1(lof), unc-4;gsa-1(gof) and unc-4;acy-1(gof). DAPI (blue) labels VA and VB nuclei. Scale bar = 2.5 μm. (E) Quantification of ectopic VA→AVB gap junctions marked with UNC-7s::GFP. Data are shown as percent of miswired VA motor neurons (VA2, VA3, VA4) in wild type, unc-4 and in double mutants unc-4;goa-1(lof), unc-4;gsa-1(gof) and unc-4;acy-1(gof). Fisher’s exact test vs. unc-4, n > 15 for each VA, * = p<.05, ***= p<.001. (F) Strategy for optogenetic activation of cAMP synthesis. unc-4; Ex[bPAC] worms were exposed to blue light during one of three developmental Windows (A) 0–10 HPH, (B) 10–20 HPH, or (C) 20–30 HPH. HPH = hours post hatching. (G) Backward locomotion of unc-4;Ex[bPAC] worms exposed to blue light during one developmental window, (A), (B) or (C). Fisher’s Exact test vs unc-4, ** = p< 0.01, NS = not significant. N > 40 for each group. (H) (left) The Punc-4 promoter was used to drive expression of PDE-4/phosphodiesterase in VAs. Backward movement was scored as either wild type (WT) or Uncoordinated (Unc). Fisher’s Exact test vs wild type, **** = p< 0.0001, N>50. (Right) Representative tracks of ten wild type and ten Punc-4::PDE-4 worms in a 3-minute period. Scale bar = 2 mm. (I) GOA-1/GαO activation in VA motor neurons drives miswiring. Quantification of ectopic VA→AVB electrical synapses marked with UNC-7s::GFP. Data are percent of total VAs (VA2–4, VA8–10) miswired with AVB electrical synapses vs. VAs that are not miswired in unc-4(e120), unc-4(e120); goa-1(lof), and unc-4(e120); goa-1(lof); Ex[Punc-4::GOA-1]. Fisher’s exact test vs unc-4(e120);goa-1(lof), *** = p<0.001. unc-4(e2323) was used unless otherwise noted.
Next, we utilized a hyperactive allele of GSA-1/GαS to elevate cAMP and again observed that ectopic VA→AVB gap junctions are reduced in VA2-VA4 (Figure 3D–E). As both GSA-1 and GOA-1 also affect cholinergic signaling, we considered the possibility that excess ACh release from VA motor neurons could be sufficient to account for suppression of the VA→AVB gap junction miswiring defect in unc-4; goa-1(lof) and unc-4; gsa-1(gof) mutants. However, genetic experiments with downstream components involved in cholinergic signaling did not alter VA gap junction specificity (Figure S3). Thus, our findings favor a model in which GOA-1/Gαo and GSA-1/Gαs regulation of cAMP signaling, and not ACh release, mediates the specificity of electrical synapses in this circuit. This idea is also supported by the finding that a gain-of-function adenylate cyclase allele, acy-1(gof), which increases production of cAMP, suppresses the Unc-4 gap junction miswiring defect in VA2-VA3 (Figure 3D–E). Together, these results suggest that elevated cAMP antagonizes the formation of ectopic VA→AVB electrical synapses and that UNC-4 turns off specific target genes (e.g., pde-1, frpr-17) to preserve cAMP signaling in VA neurons.
cAMP acts within a temporal developmental window to promote gap junction specificity
Previous studies established that UNC-4 function is required in VAs during a defined period of larval development (L2-L3), or 10–20 hours post hatching (HPH) (Miller et al., 1992). To ask whether cAMP is also required during this developmental window to establish VA inputs, we used an optogenetic tool for temporal and tissue-specific elevation of cAMP. The Beggiatoa-photoactivated adenylyl cyclase (bPAC) produces cAMP in response to blue light (Steuer Costa et al., 2017) (Figure 3F). We used the unc-17 promoter to drive bPAC (Punc-17::bPAC) expression in cholinergic neurons, including VAs, in unc-4 mutant animals. We exposed different groups of unc-4;pUnc-17::bPAC animals to blue light for separate 10-hour periods during three developmental stages: (A) 0–10 HPH (B) 10–20 HPH (C) 20–30 HPH (Figure 3F). These experiments revealed that photostimulation of bPAC during 10–20 HPH (Period B) resulted in improved backward locomotion in unc-4;pUnc-17::bPAC worms, whereas transient exposure to blue light at either an earlier (Period A) or later (Period C) developmental stage did not enhance backward locomotion (Figure 3G). These findings suggest that UNC-4 acts within a critical developmental window to preserve cAMP levels and thus maintain VA wiring in the motor circuit.
Reduced cAMP in VA neurons disrupts the backward movement circuit and induces miswiring with VA→AVB electrical synapses.
Having established that forced elevation of cAMP is sufficient to rescue the movement and wiring defects of an unc-4 mutant, we next asked if the reciprocal effect of reduced cAMP in VAs could induce the Unc-4 movement and miswiring phenotypes. We used the unc-4 promoter to drive ectopic expression of the cAMP-specific phosphodiesterase, PDE-4, in VA motor neurons to lower cAMP levels. Worms expressing Punc-4::PDE-4 showed a strong Unc-4 like backward movement defect (Figure 3H). We also observed that expression of the constitutively active allele GOA-1(Q205L), which should antagonize cAMP synthesis, in VA neurons enhanced the movement defect of the temperature-sensitive allele unc-4(ts) (Miller et al., 1992) (Figure S2). These findings argue that lowered cAMP in VA neurons is sufficient to disrupt the backward movement circuit.
We next asked whether reduced cAMP in VA neurons is sufficient to induce VA→AVB miswiring. We used the UNC-7S::GFP marker to confirm that the majority (83%) of VA neurons are miswired with VA→AVB gap junctions in the null allele, unc-4(e120), and that VA→AVB miswiring is substantially reduced (49%) in unc-4(e120); goa-1(lof) double mutants (Figure 3I). Moreover, forced expression of GOA-1 in VA motor neurons (Punc-4::GOA-1) in unc-4(e120); goa-1(lof) double mutants resulted in a robust VA→AVB miswiring defect (86%) comparable to that of unc-4(120) (Figure 3I). These observations indicate that cAMP acts cell autonomously within VA motor neurons to prevent miswiring with VA→AVB electrical synapses.
cAMP drives neuron-specific gap junction assembly
Our results show that cAMP antagonizes the formation of ectopic VA→AVB gap junctions. Because manipulations that elevate cAMP also restore backward locomotion to unc-4 mutants (Figure 3B, Figure S2), we predicted that cAMP should also promote the formation of wild-type VA→AVA gap junctions that function in the backward movement circuit (Figure 1). VA→AVA gap junctions are composed of the innexins UNC-7 (expressed in AVA) and UNC-9 (expressed in VAs) (Starich et al., 2009). The resultant heterotypic gap junctions are composed of homomeric arrays of UNC-7 in AVA vs UNC-9 in VA neurons. We utilized a two-color approach for live-cell imaging of UNC-7 in AVA and UNC-9 in VAs (Figure 4A). First, we expressed an N-terminal GFP fusion with UNC-9 in VA neurons (Punc-4::GFP::UNC-9) (Meng et al., 2016). We observed that GFP::UNC-9 puncta largely reside in VA neuronal processes in the wild type but are preferentially displaced to VA soma in unc-4, consistent with previous EM reconstructions (Figure S4) (White et al., 1986, 1992). Next, we used a flp/frt strategy for specific labeling of endogenous UNC-7 with TagRFP in AVA neurons (Schwartz and Jorgensen, 2016). With this two-color approach we could visualize both innexins, GFP::UNC-9 and UNC-7::TagRFP, that contribute to heterotypic VA→AVA gap junctions (Figure 4A–B).
Figure 4: cAMP promotes neuron-specific assembly of electrical synapses.
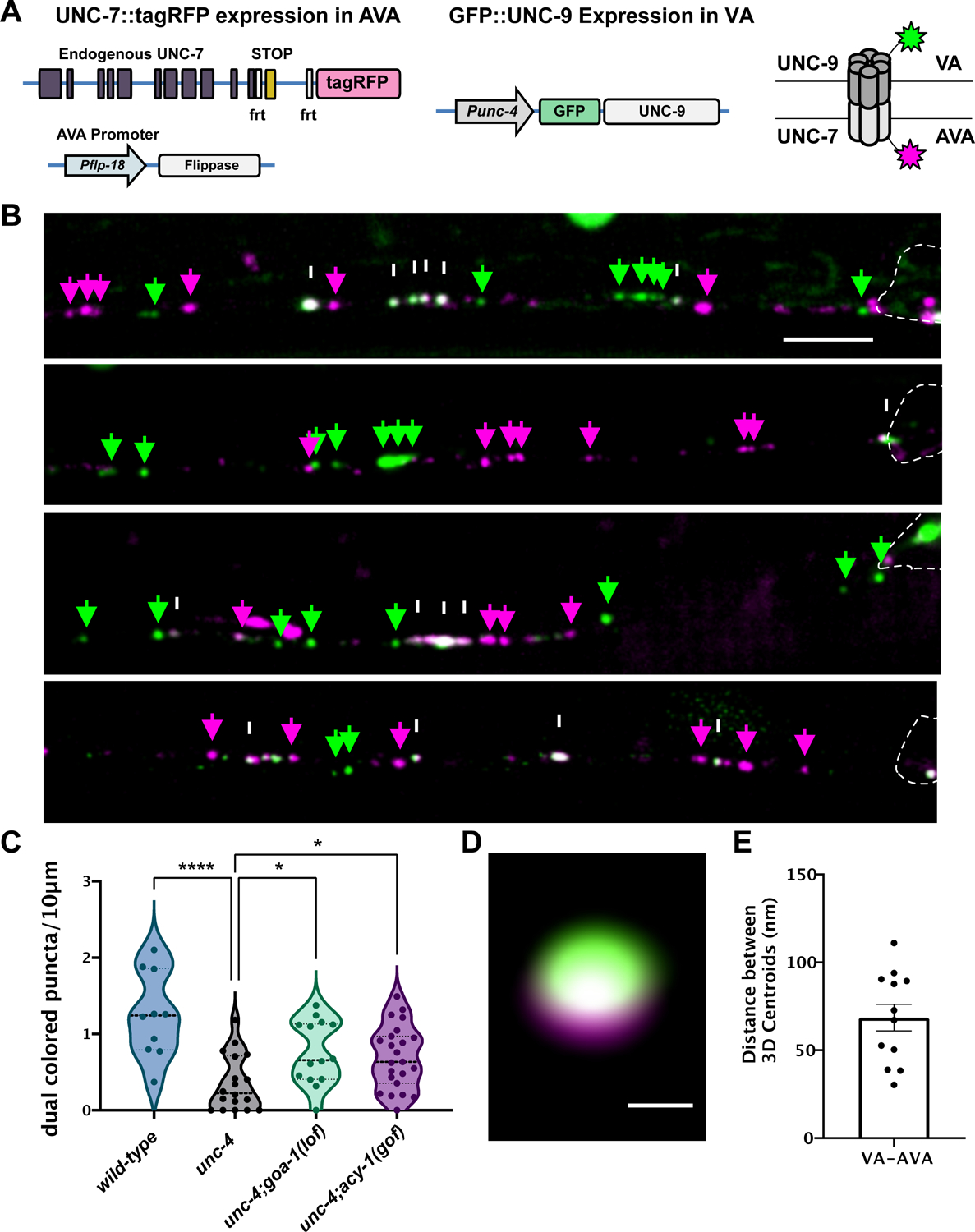
(A) Two-color labeling of UNC-7 and UNC-9 innexins in heterotypic VA→AVA gap junctions. VA→AVA gap junctions contain UNC-7 (AVA) and UNC-9 (VA). AVA promoter (Pflp-18) drives flippase resulting in expression of endogenous UNC-7::tagRFP (magenta) in AVA. VA promoter (Punc-4) drives expression of GFP::UNC-9 (green) in VAs. Schematic of dual-colored heterotypic gap junction. (B) Representative images of AVA::UNC-7::tagRFP (magenta arrows), GFP::UNC-9 (green arrows) and co-localized UNC-7 and UNC-9 (white arrows) puncta in wild-type, unc-4 and unc-4;goa-1(lof) in the ventral nerve cord of L4 larvae. VA cell soma denoted by dashed outline. Scale bar = 2.5 μm. (C) Violin plots for density of co-localized UNC-7::tagRFP and GFP::UNC-9 puncta in wild type, unc-4 and unc-4;goa-1(lof) VAs. Dashed line represents median. 2-way ANOVA. * = p < .05. **** = p <.0001. (D) Super resolution image of dual-colored gap junction. Scale bar = 200 nm. (E) 3D distance between centroids of UNC-7::tagRFP (magenta) and GFP::UNC-9 (green) images at dual-colored puncta. unc-4(e2323) was used in all experiments.
We first used this strategy to monitor VA→AVA gap junctions in wild-type VAs. At the L4 stage, we observed multiple dual-color puncta in each VA (Figure 4B–C). We performed live-cell imaging to determine that dual-color puncta are largely immobile and thus likely correspond to intact electrical synapses (Figure S4). Super resolution microscopy confirmed that GFP::UNC-9 in VA neurons and UNC-7::tagRFP in AVA are closely apposed (~70nm) as expected for bona fide VA→AVA gap junctions (Figure 4D–E, Video S2) (Marsh et al., 2017; Oshima et al., 2013). Next, we tracked GFP::UNC-9/UNC-7::TagRFP puncta in unc-4 mutant VAs and detected a significant decrease in the number of dual-color puncta in the VA axon vs. wild type (Figure 4B–C). This observation confirmed previous EM results showing that VA→AVA gap junctions are largely absent in unc-4 mutants (White et al., 1992). Finally, we utilized our two-color labeling strategy to ask if elevated cAMP could restore VA→AVA gap junction assembly. We counted GFP::UNC-9/UNC-7::TagRFP puncta in both unc-4; goa-1(lof) and unc-4;acy-1(gof) mutants and detected significant restoration of dual-color puncta in the VA axon in both cases (Figure 4B–C). Because both the goa-1(lof) and acy-1(gof) alleles are predicted to elevate cAMP, these observations support the hypothesis that cAMP normally promotes VA→AVA gap junction biogenesis.
cAMP promotes the formation of functional neuron-specific VA→AVA electrical synapses
Functional gap junctions facilitate the flow of ions between communicating neurons. Having determined that cAMP promotes the assembly of heterotypic VA→AVA electrical synapses composed of UNC-9 and UNC-7, we next asked if these VA→AVA gap junctions are functional. Our approach relies on a previous finding that VA→AVA gap junctions are antidromic (Liu et al., 2017); ions flow unidirectionally from VA→AVA in opposition to the direction of cholinergic signaling from AVA to VA via chemical synapses. Thus, excitation of VAs is predicted to activate AVAs via functional VA→AVA gap junctions and result in visible Ca2+ transients in AVA mediated by voltage-gated calcium channels (VGCCs). To test this idea, we built a transgenic line expressing red-shifted channelrhodopsin Chrimson (Klapoetke et al., 2014)in VA neurons (VA::Chrimson) and the Ca2+ sensor GCaMP6s (Tian et al., 2009) in AVA (AVA::GCaMP6s) (Figure 5A–B). We determined that optogenetic activation of VA::Chrimson in anteriorly placed VA neurons (VA2–4) triggers a robust GCaMP response in the AVA axon in the wild-type as predicted from previous electrophysiological recordings (Figure 5C, Video S3) (Liu et al., 2017). This response was abrogated in unc-7 mutants in which VA→AVA gap junctions are disabled (Starich et al., 2009), and in the absence of ATR, a necessary co-factor for optogenetic activation of Chrimson (Figure S5). Finally, an evoked AVA GCaMP response was still detectable in unc-13 mutants in which neurotransmitter release at chemical synapses is disabled (Figure S5) (Richmond et al., 1999). We therefore concluded that our optogenetic assay can reliably detect functional VA→AVA gap junctions.
Figure 5: cAMP promotes assembly of functional VA→AVA electrical synapses.
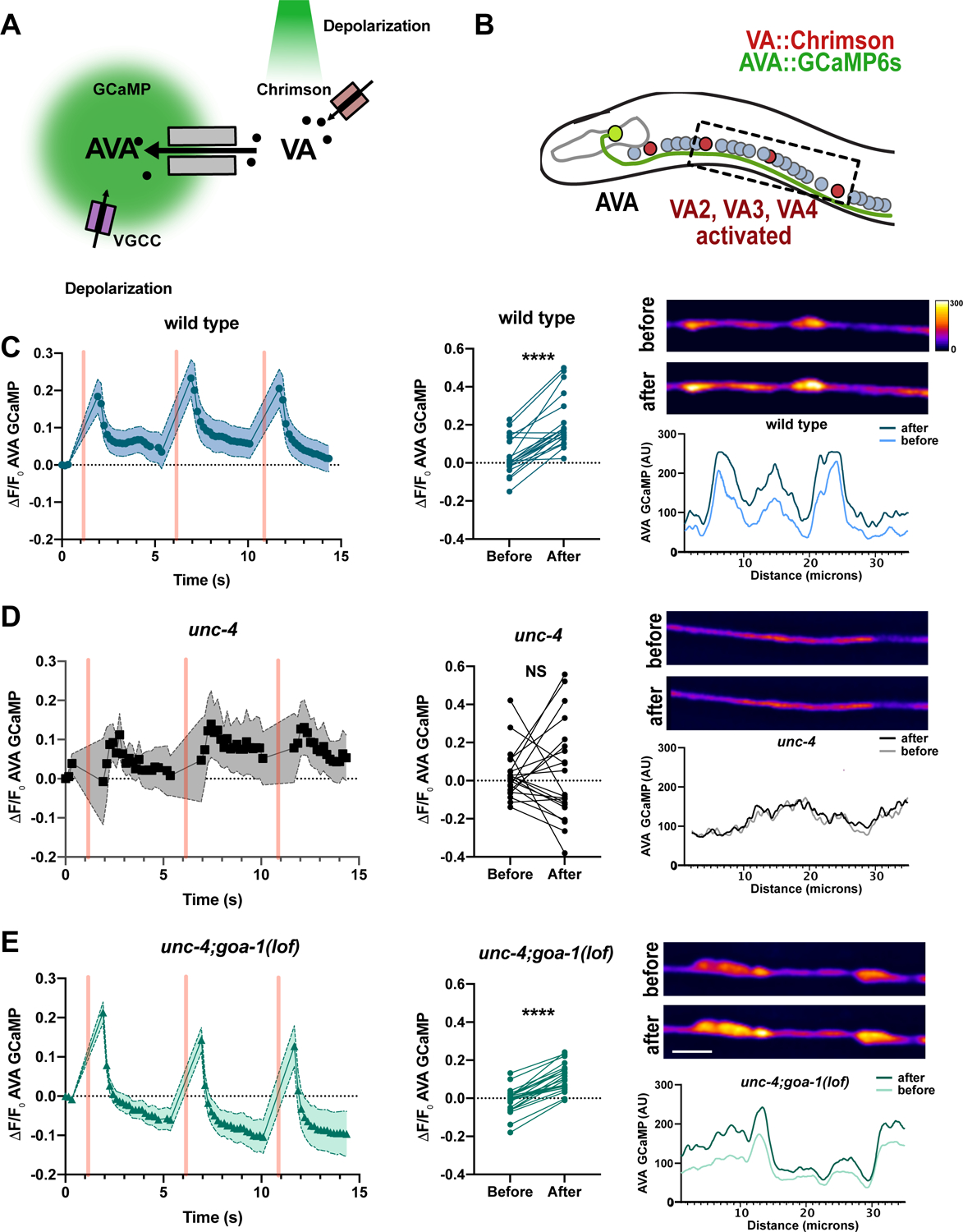
(A) Monitoring functional VA→AVA gap junctions. Channelrhodopsin, Chrimson (red rectangles) is expressed in VA motor neurons and the calcium sensor GCaMP6s is expressed in AVA. Green light (561 nm) activates Chrimson and depolarizes VAs leading to cation (black circles) flux through antidromic gap junctions with AVA (grey rectangles). AVA depolarization activates voltage gated calcium channels (VGCC) (purple rectangles) and calcium influx (open circles) for detection by AVA-GCaMP. (B) AVA neuron (green) establishes antidromic (VA→AVA) electrical synapses in the ventral nerve cord. VA::Chrimson labels VAs (red) and AVA::GCaMP6s (green) marks AVA neurons. Box denotes activated VA neurons (VA2-VA4) and adjacent AVA axon for recording Ca++ transients. Detection of AVA::GCaMPs response in the AVA axon in (C) wild type (D) unc-4 and (E) unc-4;goa-1. (left) Quantification of ΔF/F0 of AVA::GCaMP6s fluorescence over time. Three successive VA activations (500 ms) are denoted by pink vertical bars. Shapes (e.g. circles, squares, triangles) mark 488 timepoints connected by lines. Shaded area = SEM. N = 10 worms. (middle) Quantification of GCaMP6s ΔF/F0 before versus after 561 stimulation. N = 10 worms, 20 activations. Paired t-test. **** = p< 0.0001. NS = Not Significant (Right) Representative images and line scans of AVA::GCaMP6s signal before and after VA activation. Scale bar = 5μm. Heatmap for arbitrary units (AU) of fluorescence. unc-4(e2323) was used in all experiments.
We next used the optogenetic assay to confirm that functional VA→AVA gap junctions are eliminated in unc-4 mutants (White et al., 1992). As expected, activation of VA::Chrimson in an unc-4 mutant failed to trigger a detectable increase in AVA::GCaMP6s fluorescence (Figure 5D). Finally, we used this assay to ask: Is cAMP sufficient to promote the formation of competent VA→AVA gap junctions? We assayed unc-4; goa-1(lof) double mutants and confirmed that activation of VA::Chrimson triggered a robust GCaMP response in AVA (Figure 5E). Taken together, these results argue that cAMP is sufficient to promote the assembly of functional VA→AVA electrical synapses.
cAMP promotes gap junction trafficking in VA axons
In addition to the misassembly of VA electrical synapses with AVB, VA gap junctions are also relocated to a different cellular compartment in unc-4 mutants (Figure 1A) (Von Stetina et al., 2007; White et al., 1992). This change in the intracellular position of VA gap junctions led us to hypothesize that the translocation of gap junction subunits from the VA soma to the VA axon could be perturbed in unc-4 mutants. To test this idea, we utilized GFP::UNC-9 to monitor gap junction trafficking in VAs. Time-lapse imaging revealed rapid bidirectional movement of GFP::UNC-9-labeled puncta in wild-type VA axons (Figure 6A–D). In striking contrast, GFP::UNC-9 puncta were largely stationary in unc-4 mutants and predominantly localized to the VA cell soma (Figure 6A–B, Figure S4, Video S4). To ask whether cAMP drives gap junction trafficking, we monitored GFP::UNC-9 puncta in unc-4; goa-1(lof) and unc-4; acy-1(gof) double mutants in which cAMP levels should be elevated (Figure 6A). This experiment resulted in the partial restoration of GFP::UNC-9 mobility in both unc-4; goa-1(lof) and unc-4; acy-1(gof) mutants, findings consistent with the model that cAMP promotes UNC-9 trafficking in VAs (Figure 6A–D). Finally, having previously shown that cAMP is selectively required during a specific developmental stage (i.e., L2-L3 larvae) for the formation of a functional VA circuit (Figure 3G), we treated unc-4 mutant worms with 8-Br-cAMP for 10-hours during the L2-L3 transition and detected partial restoration of GFP::UNC-9 directed movement, suggesting that developmental elevation of cAMP facilitates UNC-9 trafficking (Figure S6).
Figure 6: cAMP promotes axonal trafficking of UNC-9 gap junction components.
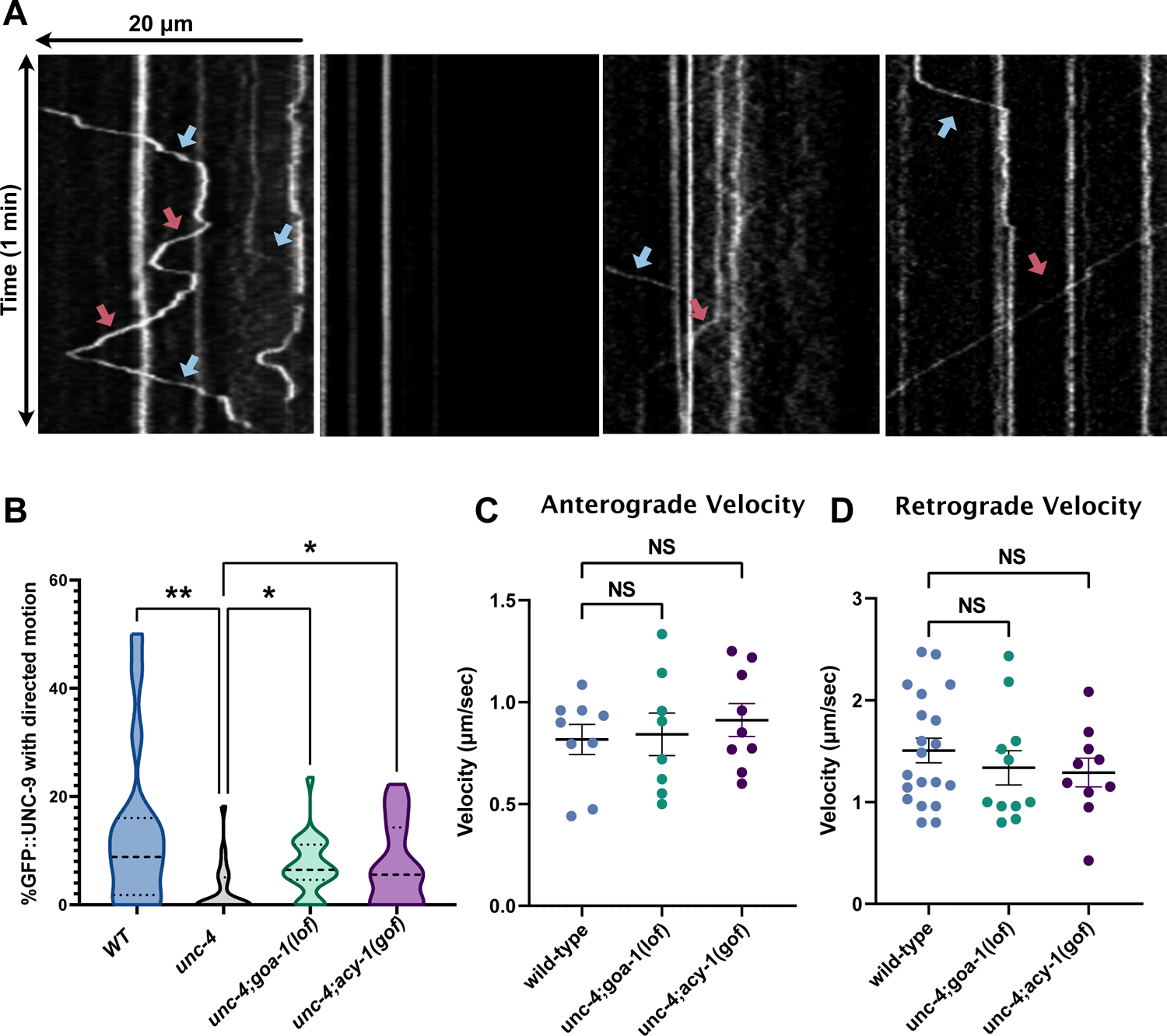
(A) Punc-4::GFP::UNC-9 imaged for 3-minutes. Kymographs show retrograde (blue arrow) and anterograde (red arrow) trafficking of GFP::UNC-9 in wild-type, unc-4; goa-1(lof) and unc-4;acy-1(gof) L4 worms. (B) Quantification of GFP::UNC-9 movement in wild type, unc-4, unc-4;goa-1(lof) and unc-4;acy-1(gof) for VA2–4. Data are percent of motile puncta in 3-minutes. N > 13. Kruskal-Wallis test, *** p= 0.0005, * p< 0.05, NS= Not Significant. Quantification of the (C) anterograde or (D) retrograde velocity of individual GFP::UNC-9 puncta in wild-type, unc-4;goa-1(lof) and unc-4;acy-1(gof) VA motor neurons. N>9 for each genotype. Student’s t-test, NS= not significant. unc-4(e2323) was used in all experiments.
UNC-4 promotes gap junction assembly in VA axons during larval development
VA motor neurons are generated during the first Larval stage (L1) and wired into the motor circuit by the L1-L2 larval molt (Sulston and Horvitz, 1977). Interestingly, UNC-4 function is required at a later stage, in L2-L3 larvae, to prevent VA miswiring (Miller et al., 1992). These observations suggest that UNC-4 might not be necessary for the initial assembly of VA→AVA gap junctions but rather could be required for their maintenance. We used our dual-color labeling strategy to confirm that VA→AVA gap junctions are detectable in L2-stage wild-type larvae. Interestingly, we observed a comparable number of VA→AVA electrical synapses at the L2-stage in unc-4 mutants (Figure 7A–B). This finding argues that UNC-4 is not required for the initial formation of VA→AVA gap junctions. In the wild type, a substantial number of additional VA→AVA gap junctions are detectable later in development by the L4 stage (Figure 7B). However, we observed no comparable increase in VA→AVA gap junctions in unc-4 mutants (Figure 7B). Together, these findings suggest that UNC-4 is critical for developmental scaling, i.e., the formation of additional VA→AVA gap junctions during larval development as VA neurons expand in size.
Figure 7: UNC-4 promotes gap junction assembly in VA axon during larval development.
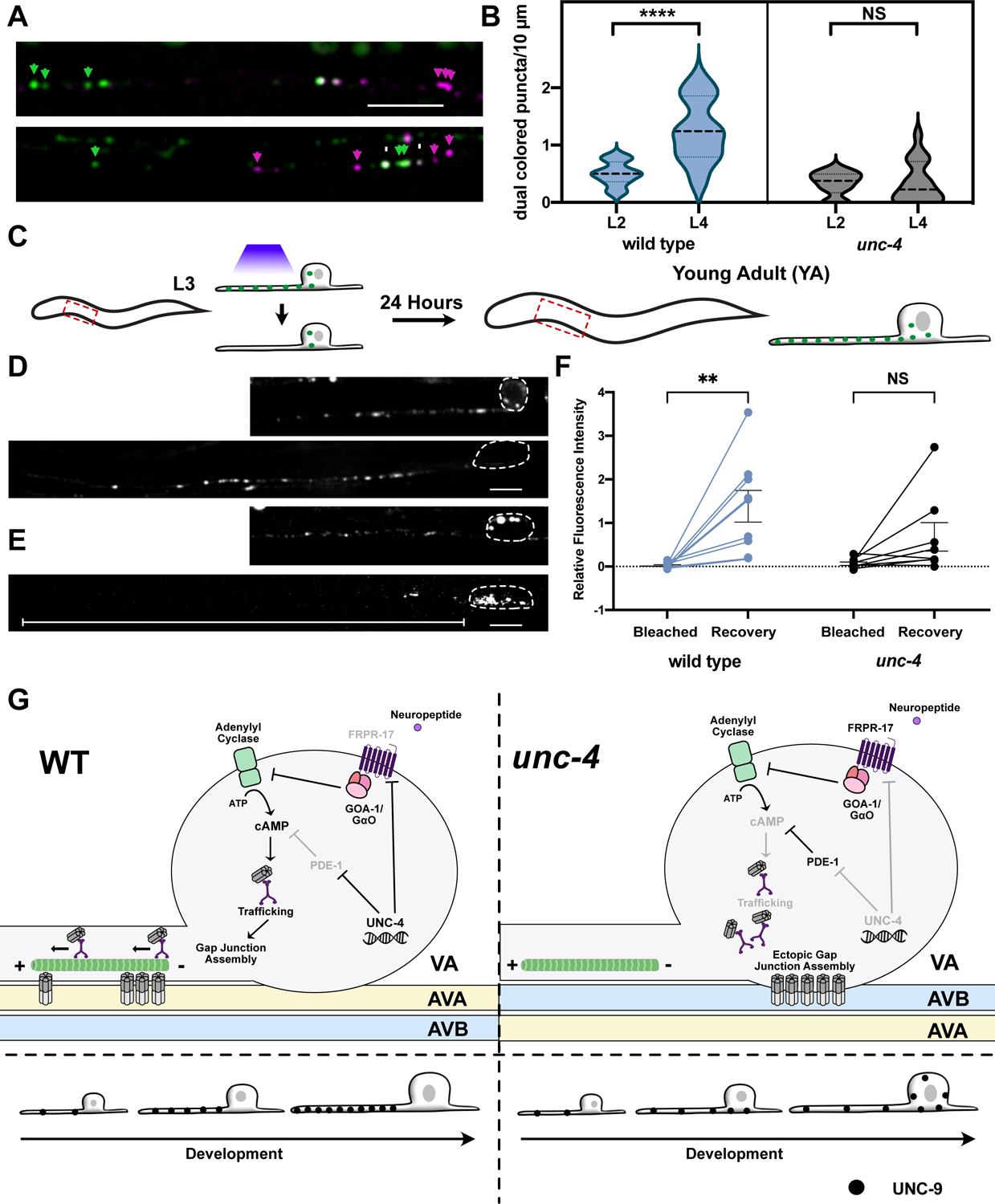
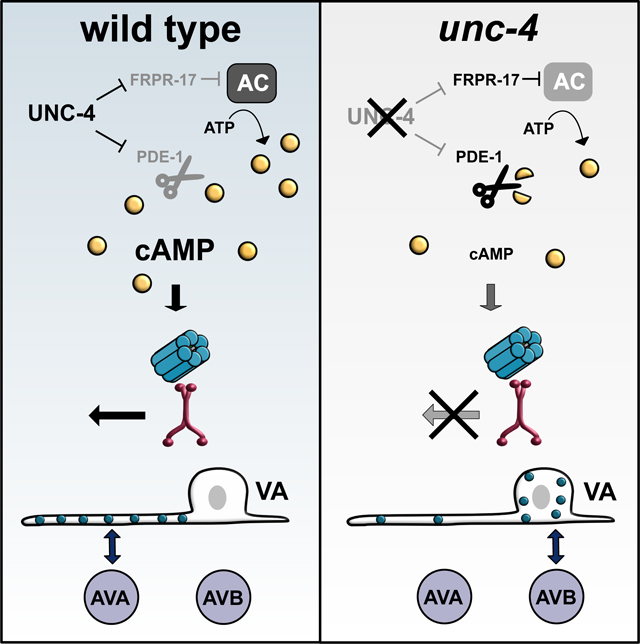
(A) AVA::UNC-7::tagRFP (magenta) and VA::GFP::UNC-9 (green) and co-localized UNC-7 and UNC-9 puncta (white) in VA axons of wild-type and unc-4 mutant L2 larvae. Scale bar = 2.5 μm.(B) Violin plots for density of co-localized UNC-7::tagRFP and GFP::UNC-9 puncta in wild-type and unc-4 VAs in L2 and L4 larvae. L4 data for wild type and unc-4 from Figure 4C. Dashed line represents median, 2-way ANOVA, **** = p < 0.0001. NS = Not Significant. (C) Schematic of FRAP strategy. A single VA axon (VA2, VA3, or VA4) is bleached in individual L3 larvae. Each treated animal is re-imaged 24-hours later (young adult) to record fluorescence intensity from photobleached VA axon. VA::GFP::UNC-9 puncta in (D) wild type and (E) unc-4 in larvae (L3) before photobleaching and at the young adult (YA) stage. Dashed outline denotes VA soma. Scale bars = 2.5 μm. GFP::UNC-9 in soma of unc-4 mutant VA (arrowheads). White line denotes unc-4 axon showing little fluorescence recovery. (F) Relative fluorescence intensity of individual VAs immediately after photobleaching (Bleach) and after 24 hours (Recovery) in wild type and unc-4. Intensity values are normalized to VA axon length and to fluorescence prior to bleaching. 2-way ANOVA, ** p = 0.0023. NS = Not Significant. (G) UNC-4 maintains cAMP to direct trafficking, neuron-specific assembly, subcellular placement and developmental scaling of electrical synapses. (Left) In wild-type VAs, UNC-4 blocks expression of cAMP antagonists, PDE-1 (phosphodiesterase) and FRPR-17(GOA-1-coupled GPCR), to maintain cAMP. cAMP promotes trafficking of UNC-9 in the VA axon for assembly with UNC-7 at heterotypic VA→AVA electrical synapses. Additional UNC-9 puncta accumulate in the wild-type VA axon during larval development. (Right) Ectopic expression of PDE-1 and FRPR-17 in unc-4 mutant VAs reduces cAMP levels and limits UNC-9 trafficking. UNC-9 accumulates in VA soma for assembly with UNC-7 at heterotypic VA→AVB gap junctions.
We hypothesized that the failure to assemble additional VA→AVA gap junctions in unc-4 mutants could be due to defective gap junction trafficking during development. To test this idea, we performed a Fluorescence Recovery After Photobleaching (FRAP) experiment. If gap junction components are actively trafficked into wild-type VA motor axons during development, then GFP::UNC-9 fluorescence should recover after an early photobleaching event. We tracked GFP::UNC-9 at three timepoints: 1) in L3 larvae prior to photobleaching; 2) in L3 larvae immediately following photobleaching; and 3) in young adults, 24-hour after photobleaching (Figure 7C). This experiment revealed that the GFP::UNC-9 signal was fully restored in wild-type VAs in young adults. In fact, average GFP::UNC-9 fluorescence actually exceeded initial values before bleaching in the L3 which we attribute to enhanced trafficking of GFP::UNC-9 during development (Figure 7D, F). In unc-4 mutants, however, we observed no significant recovery of fluorescence following photobleaching (Figure 7E–F), thus indicating that little GFP::UNC-9 is transported into unc-4 mutant VA axons during this period. Together, these results suggest that UNC-4 preserves the specificity of VA electrical synapses by promoting transport of UNC-9 into VA axons for the formation of additional VA→AVA gap junctions during larval development (Figure 7G).
Discussion
Here, we demonstrate that cAMP directs both the specificity and placement of electrical synapses in the C. elegans motor circuit. Because experiments in cultured mammalian cells have also identified cAMP as a regulator of gap junction biogenesis (Atkinson et al., 1986; Holm et al., 1999; Paulson et al., 2000; TenBroek et al., 2001), we suggest that cAMP directs evolutionarily conserved pathways that control the assembly of electrical synapses in the brain (Figure 7G).
The neuron-specificity of electrical synapses is tightly regulated.
The formation of functional electrical synapses depends on expression of compatible gap junction subunits (e.g., connexins/innexins) in each of the coupled neurons. Although necessary for gap junction assembly, connexin/innexin expression is not sufficient to explain the striking neuron specificity of electrical synapses (Martin et al., 2020). For example, in the mouse retina, photoreceptor and bipolar neurons are closely apposed but are not electrically coupled despite the joint expression of connexin Cx36 for gap junction assembly with other adjacent neurons (Asteriti et al., 2017; Deans et al., 2002; Trenholm and Awatramani, 1995). Similar examples of selective gap junction assembly among neurons that express compatible connexins have been observed in the carp retina and mouse neocortex (Greb et al., 2017; Yao et al., 2016). To investigate this question in C. elegans, we relied on uniquely available comprehensive catalogs of neuron-specific synapses and innexin expression for the entire nervous system (Bhattacharya et al., 2019; Cook et al., 2019; Taylor et al., 2021; White et al., 1986). Utilizing these datasets, we identified over 600 pairs of adjacent neurons that create chemical synapses but fail to form gap junctions despite their close proximity and co-expression of UNC-7 and UNC-9 (Table S2). Thus, the neuron specificity of electrical synapses across species likely depends on regulatory pathways that control gap junction assembly.
Recent studies in vivo have identified molecular determinants of gap junction biogenesis. In the zebrafish, the connexin-associated protein, Zona Occludens 1 (ZO1), localizes to sites of gap junction formation in the Mauthner neuron where it is proposed to function as a scaffolding protein for gap junction assembly (Lasseigne et al., 2021; Marsh et al., 2017). In C. elegans, the membrane protein, NLR-1/CASPR, is similarly proposed to recruit innexins to sites of gap junction assembly in multiple tissues including neurons (Meng and Yan, 2020). It is unclear, however, if either ZO-1 or NLR-1/CASPR are integral components of all gap junctions or are uniquely required for directing the assembly of gap junctions between specific cell types.
cAMP directs the neuron specificity of electrical synapses
Both cAMP synthesis and degradation are actively modulated to mediate the myriad of roles of cAMP in neural development (Lee, 2015). G-protein-coupled receptors (GPCRs) influence cAMP production by either promoting (GαS) or inhibiting (Gαi/o) adenylyl cyclase-dependent conversion of ATP to cAMP (Figure 3A). Phosphodiesterases (PDEs) down-regulate cAMP signaling by cleaving cyclic phosphodiester bonds to convert cAMP to AMP. Our results show that the UNC-4 transcription factor represses negative-regulators of cAMP signaling, a phosphodiesterase (PDE-1) and a GPCR (FRPR-17), in VA motor neurons to maintain neuron-specific electrical synapses required for wild-type movement. Further, we determined that experimental manipulations that elevate cAMP signaling, e.g.., treatment with the cAMP analog, 8-Br-cAMP (Figure S2), optogenetic activation of adenylyl cyclase (Figure 3G), loss-of-function mutation in GαO and gain-of-function mutations in either adenylyl cyclase or GαS (Figure 3E) were sufficient to prevent the formation of ectopic VA→AVB gap junctions that disrupt locomotion. Overall, our findings demonstrate that cAMP signaling normally favors the wild-type pattern (VA→AVA) of electrical synapses in VA motor neurons.
Our results suggest that cAMP in VA motor neurons is modulated by antagonistic G-protein signaling pathways to control electrical synaptic specificity. GSA-1/GαS promotes wild-type VA→AVA electrical synapse formation, whereas GOA-1/Gαi/o favors ectopic VA→AVB gap junction assembly. These antagonistic pathways are reminiscent of a previously described program that regulates meiotic diapause in C. elegans. During ovulation, gap junctions between sheath cells and oocytes are destabilized to enable oocyte maturation. This effect is regulated by opposing G-proteins which are proposed to either promote (GSA-1/GαS) or antagonize (GOA-1/Gαi/o) gap junction destabilization (Govindan et al., 2006, 2009). Thus, the antagonistic action of G-protein signaling mechanisms could be a consistent motif for modulating cAMP control of gap junction assembly and function.
Although cAMP signaling appears to be broadly required in VA neurons to block the formation of dysfunctional VA→AVB gap junctions (Figure 3E), our results suggest that UNC-4 preserves cAMP signaling by repressing distinct cAMP negative-regulators in different VA neurons. For example, in unc-4 mutants, VA3 adopts VA→AVB gap junctions due to ectopic expression of Phosphodiesterase/PDE-1 whereas de-repression of the UNC-4 target and GPCR, FRPR-17 selectively promotes VA2 miswiring (Figure 2). Our finding that genetic ablation of either pde-1 or frpr-17 failed to fully restore functional VA connectivity (Figure 2H) suggests that VA wiring also depends on additional unc-4-regulated genes. This idea is consistent with our previous finding that the homeodomain transcription factor, CEH-12, is down-regulated by UNC-4 in a subset of posterior VAs to prevent gap junction miswiring (Schneider et al., 2012; von Stetina et al., 2007). Interestingly, ectopic ceh-12 expression in unc-4 mutant VA neurons depends on the diffusible cue, EGL-20/Wnt. Although FRPR-17 is selectively regulated by UNC-4 in VA2 (Figure 2H), the miswiring role of GOA-1/Gαi/o throughout the ventral nerve cord (Figure 3E) suggests that the specificity of VA electrical synapses is likely to depend on UNC-4 regulation of additional GPCRs in other VAs. We thus suggest the intriguing possibility that UNC-4 could effectively tune the sensitivity of VA motor neurons to external cues (e.g., neuropeptides, Wnts) to regulate the neuron specificity of electrical synapses in the motor circuit.
Subcellular placement and maintenance of electrical synapses
The placement of electrical synapses in neural circuits must depend on the shipment of gap junction components to the site of assembly, but the mechanisms that regulate gap junction trafficking in the nervous system are largely unknown. Studies in cultured cells have established that connexins are exported from the Golgi apparatus in vesicles which are then transported along microtubules for assembly at gap junction plaques. Kinesin motors likely drive microtubule transport of connexin-containing vesicles although evidence for this role is indirect (Flores et al., 2012; Fort et al., 2011; Shaw et al., 2007). Additional studies in vitro have reported cAMP-dependent expansion of gap junction plaques which could result from enhanced trafficking of gap junction components (Paulson et al., 2000; TenBroek et al., 2001). Our findings parallel these observations and suggest that cAMP signaling promotes assembly of electrical synapses in the nervous system by regulating gap junction trafficking.
The specific subcellular location of an electrical synapse is linked to its role in neuronal function. For example, the axon initiation segment is densely populated with voltage gated ion channels and gap junctions in this region can amplify local action potentials, a property important for fast network oscillations (Schmitz et al., 2001; Simon et al., 2014; Thomas et al., 2020; Traub et al., 2002). Dendro-dendritic gap junctions can be similarly positioned near voltage-gated ion channels for synergistic interactions that amplify currents (Connors, 2017; Zsiros et al., 2007). In some cases, specific connexins are targeted to different subcellular compartments for gap junction assembly (Miller et al., 2015, 2017). Our results show that cAMP is required for active transport of the innexin UNC-9 from the VA neuron cell soma into the axon where it is assembled into VA→AVA electrical synapses. Important unanswered questions include the cell biological mechanism of cAMP-dependent trafficking and whether additional factors are required for the neuron specificity of UNC-9 gap junction assembly in the VA axon (AVA) vs cell soma (AVB).
Studies of developing neural circuits have determined that initial patterns of connectivity are maintained with the addition of new synapses as neurons expand in size. For example, in the Drosophila nociceptive circuit, the number of chemical synapses among established synaptic partners increases with larval growth (Gerhard et al., 2017). Similarly, existing connections in the C. elegans nervous system are strengthened during larval development with the addition of new synapses that scale in proportion to increasing neurite length (Witvliet et al., 2021). Our findings point to a similar developmental scaling mechanism for electrical synapses. In the wild type, the number and density of VA→AVA gap junctions increase during larval development. Interestingly, a few VA→AVA gap junctions are initially established in unc-4 mutants in early L2 larvae, but these fail to expand with larval growth, likely due to defective trafficking. This finding is consistent with the previous observation that UNC-4 function is required after the initial establishment of connectivity (Miller et al., 1992). Together, these results suggest that UNC-4 controls a genetic program that promotes insertion of additional neuron-specific gap junctions in the VA axon to sustain functional electrical connectivity as the motor circuit expands in size.
In conclusion, we have used a multifaceted approach including live-cell imaging, an in vivo assay of electrical synaptic function, behavioral assays and genetic analysis to establish that cAMP controls a trafficking mechanism for the subcellular placement and neuron specificity of electrical synapses in the developing C. elegans motor circuit. The widely reported role of cAMP signaling in gap junction assembly and function in vertebrate cells (Thévenin et al., 2013) suggests that cAMP-dependent pathways are also likely to govern the formation and maintenance of electrical synapses in mammalian neural circuits.
Limitations of Study:
Although our results suggest that a cAMP-dependent mechanism directs the subcellular placement and neuron specificity of electrical synapses, this model would be strengthened by answers to additional questions:
What are the downstream effectors of cAMP that promote trafficking of gap junction components?
Electron microscopy and immunostaining suggest that ectopic VA→AVB neuron electrical synapses assemble when cAMP signaling is impaired. Are these aberrant VA→AVB gap junctions functional?
Elevation of cAMP partially rescues the unc-4 miswiring defect. What are the additional UNC-4-regulated genes that function in tandem with cAMP signaling to specify gap junction assembly?
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents should be directed to and will be fulfilled by the lead contact, David M. Miller III (david.miller@vanderbilt.edu)
Materials availability
C. elegans strains used in this study are available on request.
Data and code availability
The RNA-seq datasets generated during this study are publicly available at GEO (GSE173287). Microscopy data can be shared on request.
No original code was developed in this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Strains and Genetics
C. elegans strains were grown at 23° C unless otherwise noted on OP50–1 Escherichia coli-seeded nematode growth medium plates (Brenner, 1974). For RNAseq experiments, C. elegans strains were grown on 8P nutrient agar seeded with E. coli strain NA22. The N2 strain was used as the wild-type reference. Mutant alleles and strains used in this study are described in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-GFP | Roche | 11814460001 |
| goat-anti-mouse-Cy3 | Jackson ImmunoLaboratories | AB_2338680 |
| Bacterial and virus strains | ||
| E. coli OP50-1 | Caenorhabditis Genetics Center (CGC) | Wormbase ID: OP50-1 |
| E. coli NA22 | Caenorhabditis Genetics Center (CGC) | Wormbase ID: OP50-1 |
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| All-trans retinal (ATR) | Sigma-Aldrich | R2500 |
| 8-Bromo-cAMP | Sigma-Aldrich | B7880 |
| Critical commercial assays | ||
| SMART-Seq V3 Ultra Low Input RNA Kit | Clontech-Takara | 634888 |
| Deposited data | ||
| RNA-Seq data | This Paper | GEO: GSE173287 |
| Experimental models: cell lines | ||
| Experimental models: C. elegans strains | ||
| Listed in Table S3 | ||
| Oligonucleotides | ||
| Recombinant DNA | ||
| pUnc-4::mCherry | This work | N/A |
| pUnc-4C::GFP | This work | N/A |
| pUnc-4::GOA-1(GOF) | This work | N/A |
| pUnc-4::GOA-1 | This work | N/A |
| pUnc-4::GFP::UNC-9 | This work | N/A |
| pUflp-18::flppase | This work | N/A |
| pUnc-4::PDE-4 | This work | N/A |
| pUnc-4::Chrimson | This work | N/A |
| Software and algorithms | ||
| CLC Genomics Workbench v.11 | QIAGEN Bioinformatics | https://www.qiagenbioinformatics.com/product-downloads/ |
| NIS Elements | Nikon | https://www.microscope.healthcare.nikon.com/products/software/nis-elements |
| ImageJ 1.8.0 | NIH | https://imagej.nih.gov/ij/download.html |
| KymoButler | Wolfram Cloud | https://www.wolframcloud.com/objects/deepmirror/Projects/KymoButler/KymoButlerForm |
| Other | ||
METHOD DETAILS
Molecular Biology
We used the In-Fusion cloning kit (Takara) to build all transgenes in this study (Key Resources Table). Plasmids were injected into N2 lines before crossing into a given genotype. The chromosomal integrants, wdIs117(pUnc-4::Chrimson), ufis26[pUnc-4::mCherry] and wdIs90[pUnc-4C::GFP] were obtained by x-ray irradiation (Miller and Niemeyer, 1995) and outcrossed for three generations.
Bulk RNA sequencing of FACS-isolated cells
We used Fluorescence Activated Cell Sorting (FACS) to isolate VA neurons labeled by an intersectional strategy. An integrated strain expressing ufis26[pUnc-4::mCherry] (VAs, DAs, SAB, I5, AVF) and wdIs90[pUnc-4C::GFP] (DAs, SAB, I5) labeled VA and AVF exclusively with mCherry in wild-type and unc-4 L2 larvae (NC2957, NC2958). With 12 VA neurons (VA1-VA12) vs 2 AVFs (AVFL, AVFR) in each animal, we reasoned that VAs would be enriched relative to AVF with this strategy. Cell dissociation and FACS were performed as previously described (Spencer et al., 2014; Taylor et al., 2021). Briefly, synchronized L1 larvae were plated on 8P plates seeded with NA22 and grown overnight at 23 C. L2 larvae were then dissociated by successive treatments with 0.25% SDS, 0.2M DTT followed by 15mg/mL pronase and passed through a 5μm filter to remove debris before FACS. Dead cells were excluded by DAPI staining. mCherry (+), GFP(−) VA and AVF neurons were collected in Trizol for RNA extraction. Three biological replicates for each genotype were performed with >50,000 cells/sample. For the wild-type VA expression profile, a reference sample of all cells was obtained from quick frozen aliquots of synchronized L2 larvae. The Clontech-Takara SMART-Seq V3 Ultra Low Input RNA Kit was used for cDNA synthesis and amplification. Paired-end-100 (PE-100) reads were collected in an Illumina NovaSeq 6000. ≥ 50 million reads/sample were obtained for three independently-isolated VA samples from both wild-type and unc-4 larvae and for three L2 whole animal reference samples. Reads were analyzed using CLC Genomics Workbench Version 11. Differentially-expressed transcripts were obtained using the RNA-Seq Differential Expression analysis pipeline in CLC, which utilizes a negative binomal GLM model. Genes were scored as differentially expressed >2-fold difference and FDR-p-value <.01 compared to control.
Feeding RNA Interference Experiments
Bacteria producing double-stranded RNA for each target gene (Table S1) were seeded on NGM plates (Kamath and Ahringer, 2003). unc-4(e2323);eri-1;lin-15 adults were plated on the seeded plates, allowed to lay eggs for two hours and progeny were grown at 23°C. L4 progeny were picked to a new plate for backward movement assays (see below). Experimenters were blinded to genotype. RNAi of either goa-1 or ceh-12 were used in each experiment as positive controls for RNAi-dependent suppression of the Unc-4 backward movement defect. The hypomorphic allele, unc-4(e2323) (Winnier et al., 1999) was used to sensitize the RNAi screen for Unc-4 suppression.
Pharmacological elevation of cAMP signaling
NGM plates were seeded with OP-50 containing 0.5mM 8-Bromoadenosine 3’,5’-cyclic monophosphate (8-Br-cAMP) (Sigma Aldrich) (Hussey et al., 2017). Plates were kept in the dark to prevent degradation of 8-Br-cAMP and used within one week. unc-4(e2323) adults were allowed to lay eggs on 8-Br-cAMP-containing plates for 2-hours and then removed. Progeny were grown to the L4 stage on 8-Br-cAMP-containing plates and transferred to NGM-plates seeded with OP-50 for behavioral analysis. Controls were unc-4(e2323) L4 larvae grown on NGM-plates seeded with OP-50. The experimentalist was blinded to growth condition.
Optogenetic activation of cAMP
NC3815 worms (unc-4(e2323);lite-1(ce314); zxIs53[punc-17::bPAC::YFP, pmyo-2::mCherry]) were grown on ATR-containing NGM-plates (Steuer Costa et al., 2017) were allowed to lay eggs for 2 hours to produce tightly synchronized larvae. Larvae were subjected to blue light activation (A) 0–10 HPH (Hours Post Hatch), (B) 10–20 HPH, and (C) 20–30 HPH. Larvae were maintained in a darkened room for a series of pulses of blue light (70 μW/mm2) of 10s on followed by 10s off for the 10 hr duration of each treatment period (Steuer Costa et al., 2017). Following stimulation, L4 worms were transferred to a new NGM plate for a tapping assay performed at the young adult stage (see below) for backward movement. For each treatment, backward locomotion was examined later, at the adult stage, to rule out potential acute effects of cAMP elevation on motor circuit function (Steuer Costa et al., 2017). NC3815 kept in the dark were used as negative controls. The experimentalist was blinded to genotype. Fisher’s exact test was used to determine significance.
Behavioral Assays
Backward movement was assessed by either 1) a “tapping assay” or 2) video tracking software (WormLab MBF Bioscience).
Tapping Assay to assay backward locomotion
L4 larvae were tapped on the head with a platinum wire to evoke backward locomotion (Von Stetina et al., 2007). unc-4 mutants typically coil dorsally with head tap (White et al., 1992) and are unable to sustain backward movement. Movement was scored as “wild-type” for sustained sinusoidal backward locomotion (two full bends) or “Unc” for failed backward locomotion. Experimenters were blinded to genotype. N> 50 for each genotype. Fisher’s exact test was used to determine significance. (Figures 2B, 4B–D).
Video Tracking
L4 larvae were placed on a NGM plate lightly-seeded with OP-50. WormLab was used to track the worms over a three-minute period with a 1s blue light pulse at 5s intervals throughout to promote movement. unc-4 mutants exhibit a readily detectable reduction in backward distance traveled in a three-minute period compared to wild-type (Figure 1). We therefore used this assay to identify suppression of the Unc-4 backward movement defect in RNAi feeding and genetic experiments that tested UNC-4 targets identified by RNA seq analysis. Worms were included in analysis if they were captured and tracked for the entire 3-minute period of the video. A 2-way-ANOVA was used to determine significance. (Figures 1C, 3B, 3E).
Microscopy and Image Analysis
Immunostaining to detect AVB gap junctions with ventral cord motor neurons
AVB gap junctions in the ventral nerve cord were marked with wdIs54[Punc7::UNC-7S::GFP and immunostained to detect the dim GFP signal (Starich et al., 2009; Von Stetina et al., 2007) after fixation (Finney and Ruvkun, 1990). Briefly, L4 larvae were successively immersed in sealed microfuge tubes in liquid nitrogen for three freeze-thaw cycles before incubating in 1% paraformaldehyde for 40 minutes. Following fixation, a series of treatments (1% BME, 10 mM DTT, 0.3% H2O2, 0.1% Triton-X) were performed to permeabilize the cuticle. Treated larvae were incubated with 1:500 anti-GFP primary antibody (Roche, 11814460001) overnight at 4°C. Following washes with Antibody Buffer B (AbB), larvae were incubated with 1:500 goat-anti-mouse-Cy3 (Jackson ImmunoLaboratories, AB_2338680) for 2-hours at room temperature, washed with AbB, stained with DAPI (1:1000) for 30 minutes and mounted with VectaShield (Vector Labs). Z-stack (0.2 μm steps) images of DAPI (405 nm excitation) and Cy3 (561 nm excitation) were obtained in a Nikon A1R confocal microscope with a 60X objective (Plan Apo Lambda Oil, 1.40=NA). Images were 3D-deconvolved using Nikon NIS elements. Because AVB gap junctions are characteristically positioned on motor neuron cell soma (White et al., 1986, 1992), ectopic gap junctions with VA neurons (VA→AVB) can be determined by scoring the co-localization of UNC-7S::GFP puncta with the nuclei of ventral cord motor neurons marked with the DNA-specific dye, DAPI (4’,6-diamidino-2-phenylindole). VA neurons were identified by position in the DAPI-stained queue of ventral cord nuclei (Miller and Niemeyer, 1995). Cy3-stained puncta adjacent to VA nuclei were scored as gap junctions with AVB. To distinguish UNC-7s::GFP puncta from background, we imposed a size and fluorescence intensity cutoff using NIS Elements. N> 20 VA neurons were scored for each group. Fisher’s exact test was used to determine significance.
Single molecule mRNA Fluorescence In Situ Hybridization (smFISH)
smFISH was performed with custom pde-1 and frpr-17 probes linked to Quasar® 670 or 561 respectively (Biosearch Technologies). Synchronized L2 larvae were collected by washing plates with M9, fixed in 4% paraformaldehyde in 1X PBS for 45 min and permeabilized in 70% ethanol for 24–48 h. Hybridization followed manufacturer’s instructions (http://www.biosearchtech.com/stellarisprotocols) and was performed at 37°C for 16h in Stellaris RNA FISH hybridization buffer (Biosearch Technologies Cat# SMF-HB1–10) containing pde-1 or frpr-17 probe at 1:100. VA neurons were marked with Pbnc-1::GFP (OH15624). Cell nuclei were stained with DAPI. Z-stacks were collected in a Nikon spinning disk confocal microscope equipped with optical filters for DAPI, Quasar® 670 or 651 and GFP using a 100X objective (NA=1.49) in 0.2 μm steps spanning the cell body and merged for quantification following 3D-deconvolution in NIS elements. smFISH puncta were counted if they corresponded to circular fluorescent spots, exceeded the Quasar® 670/561 background signal and were located within a GFP-labeled VA cell body. At least 20 worms were scored for each group and the Mann-Whitney test was used to determine significance. The wild type N2 strain and null allele, unc-4(e120), were used in all smFISH experiments.
Dual-colored heterotypic VA→AVA gap junction
Worms expressing endogenous AVA::UNC-7::tagRFP and transgenic VA::GFP::UNC-9 were imaged to identify VA→AVA gap junctions. UNC-7 was endogenously tagged at the N-terminus with frt-STOP-UTR-frt-tagRFP (syb2341) (Schwartz and Jorgensen, 2016). The Pflp-18 promoter was used to drive filppase in AVA resulting in tagged UNC-7::tagRFP selectively in AVA. C-terminal-GFP-fused UNC-9 (Meng et al., 2016) was expressed in VAs using the UNC-4 promoter (Punc-4::GFP::UNC-9). L4 worms were placed on 10% agarose pads and immobilized with 50 mM muscimol. Z-stacks were captured (0.2 μm/step) spanning the VA process (VA2, VA3, VA4) for GFP (488 nm excitation) and tagRFP (561 nm excitation) Stacks were 3D-deconvolved in NIS elements. Images were thresholded by fluorescence intensity, circularity, and size to create masks of puncta in each fluorophore in NIS elements. Puncta were counted as “dual-colored” if masks overlapped. The number of dual-colored puncta was normalized the length of each VA axonal region to yield a density value (# puncta/10 μm) (density). A 2-way ANOVA was used to determine significance.
Structured Illumination Microscopy
Animals fixed with 4% paraformaldehyde for thirty minutes (NC3775 [unc-7(syb2341);Ex[Pflp-19::flppase, Punc-4::GFP::UNC-9]) were mounted with Vectashield and # 1.5 coverslips. Z-stacks (0.07 μm/step) of GFP (488 nm excitation) and tagRFP (561 nm excitation) were captured with Nikon N-SIM microscope 100X SR Apo TIRF (1.49 NA) objective in 3D-SIM mode. To ensure that channels were accurately aligned, FocalCheck™ Microspheres (Invitrogen F14806) were included in each sample. Slice reconstruction was performed using NIS Elements. Following reconstruction, a 3D mask for each fluorophore was created based on intensity. The centroid of each 3D mask was determined and the distance between the centroids was measured to estimate the proximity of UNC-7 vs UNC-9 gap junction arrays.
Monitoring functional VA→AVA gap junctions
We constructed a transgenic line (NC3666 [wdIs117(Punc-4::Chrimson);Ex1148[pFlp-18::GCaMP6S]) in which Punc-4 drives expression of the red-shifted opsin Chrimson in VAs and the Pflp-18 promoter drives expression of the Ca2+ sensor GCaMP6s in AVA. Worms were grown on plates containing All-trans-Retinol (ATR), a necessary cofactor for Chrimson. L4 worms were mounted on 10% agarose pads and anesthetized with 50 mM muscimol in a slurry of 0.5 μm Polybead® Carboxylate Microspheres (Polysciences). We captured AVA::GCaMPs fluorescence from the AVA process in a region adjacent to VA2-VA4 at 10 frames/sec (100ms) with a 100X SR Apo TIRF (1.49 NA) objective on a Nikon Spinning Disk. VA::Chrimson was activated with a 500 ms burst of 561 nm light every 5 seconds (Basu et al., 2015; Klapoetke et al., 2014). Following Chrimson activation, the next AVA::GCaMPs frame was captured within 500 ms. Importantly, t1/2 = 600ms for GCaMP6s (Chen et al., 2013), therefore ensuring that our sampling interval would capture a transient GCaMPs signal. A single Z-plane was captured for 1-minute. Movies were 2D deconvolved and aligned using NIS elements. AVA fluorescence intensity was corrected by subtracting the value of a background ROI adjacent to the AVA process of the same size for each time point. Change in fluorescence intensity was calculated as ΔF/F0 = (Ft-F0)/F0 (F0 = baseline fluorescence intensity of timepoint three frames before first Chrimson activation. Ft = Fluorescence intensity of a given timepoint). To detect evoked changes in AVA::GCaMPs, we compared the ΔF/F0 of AVA::GCaMPs immediately before Chrimson activation versus the ΔF/F0 of AVA::GCaMPs immediately following Chrimson activation. A paired t-test was performed for AVA GCaMPs fluorescence time points before vs after 561 activation of Chrimson in VA neurons.
Trafficking of GFP::UNC-9 particles
L4 larvae were immobilized using 10% agarose pads, 50 mM muscimol and 0.5 μm Polybead® Carboxylate Microspheres (Polysciences). VA::GFP::UNC-9 was captured using 488 nm excitation 5 frames/sec for three-minutes on a Nikon Spinning Disk microscope with a 100X SR Apo TIRF (1.49 NA) objective. Movies were obtained from a single plane and were deconvolved with Nikon 2D deconvolution software. A 5-pixel line was drawn through the VA process to create a kymograph in Nikon elements. The kymograph was then analyzed using KymoButler Premium (Jakobs et al., 2019). The number of GFP::UNC-9 puncta was determined based on fluorescence intensity and size. Puncta were considered motile if they moved at a speed > 0.4 μm/sec for three consecutive frames. The percentage of puncta that moved in a given VA was calculated and velocity, direction, and displacement were calculated for each punctum with directed locomotion. A Mann-Whitney was used to determine significance between groups.
Fluorescence Recovery After Photobleaching (FRAP) of GFP::UNC-9 particles
Individual L3 larvae were placed on 10% agarose pads and immobilized with 50 mM muscimol. A single anterior VA neuron (VA2, VA3, or VA4) in each animal was imaged with 488 nm excitation to collect a Z-stack (0.2 μm/step) on a Nikon Spinning Disk with a 100X SR Apo TIRF (1.49 NA) objective. A 405 nm laser (15% power, 15 ms dwell time) was used to bleach an ROI encompassing the anteriorly directed VA axon but excluding the VA cell soma which was not bleached. An additional Z-stack with 488 nm excitation was captured immediately after photobleaching. The treated worm was recovered by washing the slide with M9 and allowed to recover on a bacterially seeded (OP50–1) NGM plate until reaching the adult stage (24 hours at 20°C). Each animal was then placed on a 10% agarose pad and immobilized with 50 mM muscimol. GFP::UNC-9 signal was imaged from the previously photobleached region of each treated VA neuron. GFP::UNC-9 signal was summed from a 5-pixel-wide line drawn along the VA process. The total amount of corrected fluorescence (background subtracted) was measured and divided by the length of the process to account for growth of the VA process during development. The resultant GFP::UNC-9 puncta density (Total fluorescence/10 μm) was normalized to the GFP::UNC-9 puncta density at the initial timepoint at the L3 larval stage before photobleaching. A paired t-test was used to determine significance between groups.
QUANTIFICATION AND STATISTICAL ANALYSIS
For all categorical data, we used a Fisher’s exact test. For all quantitative data, we used Prism9 to determine if a sample was normally distributed. For normally distributed samples, a Student’s t-test (2 groups) or one-way ANOVA with multiple comparisons correction (3 or more groups) was used. If a sample in a given analysis was not normally distributed, a Mann-Whitney (2 groups) or Kruskal-Wallis test (3 or more groups) was used. Figure legends specify the statistical test and N used in each experiment.
Supplementary Material
Video S1: unc-4 mutants exhibit a backward locomotion defect in the tapping assay, related to Figures 1 and 3. Representative examples of tapping assay for wild type, unc-4(e2323), and unc-4(e2323);goa- 1(lof). Individual L4 worms are tapped on the head and backward locomotion is scored as wild type or Unc. unc-4(e2323) mutants are unable to move backward. Backward locomotion is partially restored in unc-4(e2323);goa-1(lof).
Table S2: Adjacent neurons that fail to form electrical synapses in C. elegans, related to Figure 1.
Video S2: Super resolution microscopy of dual-color gap junction, related to Figure 4. 3D reconstruction of SIM Z-stack of UNC-7::tagRFP in AVA and GFP::UNC-9 in VA. UNC-7 and UNC-9 are closely localized, but slightly displaced as expected for UNC-7 vs UNC-9 hemichannels at a heterotypic gap junction.
Video S3: Evoked AVA GCaMP response, related to Figure 5. Individual AVA axon expressing GCaMP in wild type, unc-4(e2323), and unc-4(e2323);goa-1(lof) double mutant. VAs expressing Chrimson are optogenetically activated with a pulse (500 ms) of 561 nm light. Two subsequent frames are noted with a red circle in the left-hand corner. In wild type and unc-4(e2323);goa-1(lof) worms, a detectable AVA GCaMP response is detected following VA activation
Video S4: GFP::UNC-9 trafficking in VA axon, related to Figure 6. Representative videos of GFP::UNC-9 trafficking in wild-type and unc-4(e2323) mutant VA axon. Wild-type worms exhibit rapid, bidirectional trafficking of GFP::UNC-9 puncta which is largely disabled in unc-4(e2323) mutants. Scale bar = 5 μm.
Highlights.
UNC-4 controls the specificity and location of electrical synapses in VA motor neurons.
UNC-4 maintains cAMP signaling by blocking expression of cAMP antagonists.
cAMP promotes trafficking of innexins into the VA axon for gap junction assembly.
cAMP drives assembly of new electrical synapses as VA neurons grow during development.
Acknowledgments
We thank O Hobert, A Gottschalk and D Yan for reagents, B Millis, L Sundararajan, B Kesler, E Tross, K Peterson, M Carter, S He and E Tataru for their help and John White for encouragement. Funded by NIH (R01NS113559) to DMM and AHA (19PRE34380582) and NSF (DGE:1445197) to SP. Experiments were performed in the Vanderbilt Flow Cytometry Shared Resource which is supported by the VICC (P30 CA68485) and VDDKC (DK058404) and in the Cell Imaging Shared Resource which is supported by NIH grant DK020593. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Allen K, Fuchs EC, Jaschonek H, Bannerman DM, and Monyer H (2011). Gap junctions between interneurons are required for normal spatial coding in the hippocampus and short-term spatial memory. Journal of Neuroscience 31, 6542–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asteriti S, Gargini C, and Cangiano L (2017). Connexin 36 expression is required for electrical coupling between mouse rods and cones. Visual Neuroscience 34. [DOI] [PubMed] [Google Scholar]
- Atkinson MM, Anderson SK, and Sheridan JD (1986). Modification of gap junctions in cells transformed by a temperature-sensitive mutant of Rous sarcoma virus. The Journal of Membrane Biology 91, 53–64. [DOI] [PubMed] [Google Scholar]
- Atkinson MM, Lampe PD, Lin HH, Kollander R, Li XR, and Kiang DT (1995). Cyclic AMP modifies the cellular distribution of connexin43 and induces a persistent increase in the junctional permeability of mouse mammary tumor cells. [DOI] [PubMed]
- Basu R, Taylor MR, and Williams ME (2015). The classic cadherins in synaptic specificity. Cell Adhesion and Migration 9, 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Aghayeva U, Berghoff EG, and Hobert O (2019). Plasticity of the Electrical Connectome of C. elegans. Cell 176, 1174–1189.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghardt RC, Barhoumi R, Sewall TC, and Bowen JA (1995). Cyclic AMP induces rapid increases in gap junction permeability and changes in the cellular distribution of connexin43. The Journal of Membrane Biology 148, 243–253. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, and Brenner S (1985). The neural circuit for touch sensitivity in Caenorhabditis elegans. Journal of Neuroscience 5, 956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, and Jayaraman V (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors BW (2017). Synchrony and so much more: Diverse roles for electrical synapses in neural circuits. Developmental Neurobiology 77, 610–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors BW, and Long MA (2004). Electrical synapses in the mammalian brain. Annual Review of Neuroscience 27, 393–418. [DOI] [PubMed] [Google Scholar]
- Cook SJ, Jarrell TA, Brittin CA, Wang Y, Bloniarz AE, Yakovlev MA, Nguyen KCQ, Tang LTH, Bayer EA, Duerr JS, et al. (2019). Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 571, 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans MR, Volgyi B, Goodenough DA, Bloomfield SA, and Paul DL (2002). Connexin36 is essential for transmission of rod-mediated visual signals in the mammalian retina. Neuron 36, 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykes IM, Freeman FM, Bacon JP, and Davies JA (2004). Molecular Basis of Gap Junctional Communication in the CNS of the Leech Hirudo medicinalis. Journal of Neuroscience 24, 886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfgang C, Eckert R, Lichtenberg-Fraté H, Butterweck A, Traub O, Klein RA, Hülser DF, and Willecke K (1995). Specific permeability and selective formation of gap junction channels in connexin-transfected HeLa cells. Journal of Cell Biology 129, 805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M, and Ruvkun G (1990). The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63, 895–905. [DOI] [PubMed] [Google Scholar]
- Flores CE, Nannapaneni S, Davidson KGV, Yasumura T, Bennett MVL, Rash JE, and Pereda AE (2012). Trafficking of gap junction channels at a vertebrate electrical synapse in vivo. Proceedings of the National Academy of Sciences of the United States of America 109, E573–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort AG, Murray JW, Dandachi N, Davidson MW, Dermietzel R, Wolkoff AW, and Spray DC (2011). In vitro motility of liver connexin vesicles along microtubules utilizes kinesin motors. Journal of Biological Chemistry 286, 22875–22885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T (2017). Structural organization of the dendritic reticulum linked by gap junctions in layer 4 of the visual cortex. Neuroscience 340, 76–90. [DOI] [PubMed] [Google Scholar]
- Gerhard S, Andrade I, Fetter RD, Cardona A, and Schneider-Mizell CM (2017). Conserved neural circuit structure across Drosophila larval development revealed by comparative connectomics. Elife 6, e29089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan JA, Cheng H, Harris JE, and Greenstein D (2006). Gαo/i and Gαs Signaling Function in Parallel with the MSP/Eph Receptor to Control Meiotic Diapause in C. elegans. Current Biology 16, 1257–1268. [DOI] [PubMed] [Google Scholar]
- Govindan JA, Nadarajan S, Kim S, Starich TA, and Greenstein D (2009). Somatic cAMP signaling regulates MSP-dependent oocyte growth and meiotic maturation in C. elegans. Development 136, 2211–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb H, Hermann S, Dirks P, Ommen G, Kretschmer V, Schultz K, Zoidl G, Weiler R, and Janssen-Bienhold U (2017). Complexity of gap junctions between horizontal cells of the carp retina. Neuroscience 340, 8–22. [DOI] [PubMed] [Google Scholar]
- Hendi A, Kurashina M, and Mizumoto K (2019). Intrinsic and extrinsic mechanisms of synapse formation and specificity in C. elegans. Cellular and Molecular Life Sciences 76, 2719–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hestrin S, and Galarreta M (2005). Electrical synapses define networks of neocortical GABAergic neurons. Trends in Neurosciences 28, 304–309. [DOI] [PubMed] [Google Scholar]
- Holm I, Mikhailov A, Jillson T, and Rose B (1999). Dynamics of gap junctions observed in living cells with connexin43-GFP chimeric protein. European Journal of Cell Biology 78, 856–866. [DOI] [PubMed] [Google Scholar]
- Hussey R, Stieglitz J, Mesgarzadeh J, Locke TT, Zhang YK, Schroeder FC, and Srinivasan S (2017). Pheromone-sensing neurons regulate peripheral lipid metabolism in Caenorhabditis elegans. PLoS Genetics 13, e1006806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs MA, Dimitracopoulos A, and Franze K (2019). Kymobutler, a deep learning software for automated kymograph analysis. ELife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, and Ahringer J (2003). Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321. [DOI] [PubMed] [Google Scholar]
- Kawano T, Po MD, Gao S, Leung G, Ryu WS, and Zhen M (2011). An imbalancing act: Gap junctions reduce the backward motor circuit activity to bias C. elegans for forward locomotion. Neuron 72, 572–586. [DOI] [PubMed] [Google Scholar]
- Kerk SY, Kratsios P, Hart M, Mourao R, and Hobert O (2017). Diversification of C. elegans Motor Neuron Identity via Selective Effector Gene Repression. Neuron 93, 80–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, et al. (2014). Independent optical excitation of distinct neural populations. Nature Methods 11, 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe PD, and Lau AF (2004). The effects of connexin phosphorylation on gap junctional communication. International Journal of Biochemistry and Cell Biology 36, 1171–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasseigne AM, Echeverry FA, Ijaz S, Michel JC, Martin EA, Marsh AJ, Trujillo E, Marsden KC, Pereda AE, and Miller AC (2021). Electrical synaptic transmission requires a postsynaptic scaffolding protein. ELife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D (2015). Global and local missions of cAMP signaling in neural plasticity, learning, and memory. Frontiers in Pharmacology 6, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Chen B, Mailler R, and Wang ZW (2017). Antidromic-rectifying gap junctions amplify chemical transmission at functionally mixed electrical-chemical synapses. Nature Communications 8, 14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugnier C (2006). Cyclic nucleotide phosphodiesterase (PDE) superfamily: A new target for the development of specific therapeutic agents. Pharmacology and Therapeutics 109, 366–398. [DOI] [PubMed] [Google Scholar]
- Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, and Tsukihara T (2009). Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature 458, 597–602. [DOI] [PubMed] [Google Scholar]
- Margeta MA, and Shen K (2010). Molecular mechanisms of synaptic specificity. Molecular and Cellular Neuroscience 43, 261–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh AJ, Michel JC, Adke AP, Heckman EL, and Miller AC (2017). Asymmetry of an Intracellular Scaffold at Vertebrate Electrical Synapses. Current Biology 27, 3561–3567.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EA, Lasseigne AM, and Miller AC (2020). Understanding the Molecular and Cell Biological Mechanisms of Electrical Synapse Formation. Frontiers in Neuroanatomy 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, and Yan D (2020). NLR-1/CASPR Anchors F-Actin to Promote Gap Junction Formation. Developmental Cell 55, 574–587.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Chen CH, and Yan D (2016). Regulation of Gap Junction Dynamics by UNC-44/ankyrin and UNC-33/CRMP through VAB-8 in C. elegans Neurons. PLoS Genetics 12, e1005948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, and Niemeyer CJ (1995). Expression of the unc-4 homeoprotein in Caenorhabditis elegans motor neurons specifies presynaptic input. Development 121, 2877–2886. [DOI] [PubMed] [Google Scholar]
- Miller AC, Voelker LH, Shah AN, and Moens CB (2015). Neurobeachin is required postsynaptically for electrical and chemical synapse formation. Current Biology 25, 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AC, Whitebirch AC, Shah AN, Marsden KC, Granato M, O’Brien J, and Moens CB (2017). A genetic basis for molecular asymmetry at vertebrate electrical synapses. ELife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Shen MM, Shamu CE, Bürglin TR, Ruvkun G, Dubois ML, Ghee M, and Wilson L (1992). C. elegans unc-4 gene encodes a homeodomain protein that determines the pattern of synaptic input to specific motor neurons. Nature 355, 841–845. [DOI] [PubMed] [Google Scholar]
- Muntean BS, Masuho I, Dao M, Sutton LP, Zucca S, Iwamoto H, Patil DN, Wang D, Birnbaumer L, Blakely RD, et al. (2021). Gαo is a major determinant of cAMP signaling in the pathophysiology of movement disorders. Cell Reports 34, 108718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima A, Matsuzawa T, Nishikawa K, and Fujiyoshi Y (2013). Oligomeric structure and functional characterization of caenorhabditis elegans innexin-6 gap junction protein. Journal of Biological Chemistry 288, 10513–10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X, Winbow VM, Patel LS, Burr GS, Mitchell CK, and O’Brien J (2005). Protein kinase A mediates regulation of gap junctions containing connexin35 through a complex pathway. Molecular Brain Research 135, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson AF, Lampe PD, Meyer RA, TenBroek E, Atkinson MM, Walseth TF, and Johnson RG (2000). Cyclic AMP and LDL trigger a rapid enchancement in gap junction assembly through a stimulation of connexin trafficking. [DOI] [PubMed]
- Pereda AE, Curti S, Hoge G, Cachope R, Flores CE, and Rash JE (2013). Gap junction-mediated electrical transmission: Regulatory mechanisms and plasticity. Biochimica et Biophysica Acta - Biomembranes 1828, 134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugrad A, Meir JYJ, Barnes TM, and Miller DM (1997). The Groucho-like transcription factor UNC-37 functions with the neural specificity gene unc-4 to govern motor neuron identity in C. elegans. Development 124, 1699–1709. [DOI] [PubMed] [Google Scholar]
- Phelan P, Stebbings LA, Baines RA, Bacon JP, Davies JA, and Ford C (1998). Drosophila shaking-B protein forms gap junctions in paired Xenopus oocytes. Nature 391, 181–184. [DOI] [PubMed] [Google Scholar]
- Rabinowitch I, Chatzigeorgiou M, Zhao B, Treinin M, and Schafer WR (2014). Rewiring neural circuits by the insertion of ectopic electrical synapses in transgenic C. elegans. Nature Communications 5, 4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond JE, Davis WS, and Jorgensen EM (1999). UNC-13 is required for synaptic vesicle fusion in C. elegans. Nature Neuroscience 1999 2:11 2, 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, and Yamagata M (2009). Many Paths to Synaptic Specificity. [DOI] [PubMed]
- Schmitz D, Schuchmann S, Fisahn A, Draguhn A, Buhl EH, Petrasch-Parwez E, Dermietzel R, Heinemann U, and Traub RD (2001). Axo-axonal coupling: a novel mechanism for ultrafast neuronal communication. Neuron 31, 831–840. [DOI] [PubMed] [Google Scholar]
- Schneider J, Skelton RL, von Stetina SE, Middelkoop TC, van Oudenaarden A, Korswagen HC, and Miller DM (2012). UNC-4 antagonizes Wnt signaling to regulate synaptic choice in the C. elegans motor circuit. Development 139, 2234–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz ML, and Jorgensen EM (2016). SapTrap, a toolkit for high-throughput CRISPR/Cas9 gene modification in Caenorhabditis elegans. Genetics 202, 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RM, Fay AJ, Puthenveedu MA, von Zastrow M, Jan Y-N, and Jan LY (2007). Microtubule plus-end-tracking proteins target gap junctions directly from the cell interior to adherens junctions. Cell 128, 547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A, Traub RD, Vladimirov N, Jenkins A, Nicholson C, Whittaker RG, Schofield I, Clowry GJ, Cunningham MO, and Whittington MA (2014). Gap junction networks can generate both ripple-like and fast ripple-like oscillations. European Journal of Neuroscience 39, 46–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelton RL (2012). Molecular analysis of UNC-4 pathway genes that regulate synaptic choice.
- Solan JL, and Lampe PD (2005). Connexin phosphorylation as a regulatory event linked to gap junction channel assembly. Biochimica et Biophysica Acta - Biomembranes 1711, 154–163. [DOI] [PubMed] [Google Scholar]
- Solan JL, and Lampe PD (2009). Connexin43 phosphorylation: structural changes and biological effects. Biochem. J 419, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solan JL, and Lampe PD (2016). Kinase programs spatiotemporally regulate gap junction assembly and disassembly: Effects on wound repair. Seminars in Cell and Developmental Biology 50, 40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Ampatzis K, Bjornfors ER, and El Manira A (2016). Motor neurons control locomotor circuit function retrogradely via gap junctions. Nature 529, 1–5. [DOI] [PubMed] [Google Scholar]
- Sosinsky GE, and Nicholson BJ (2005). Structural organization of gap junction channels. Biochimica et Biophysica Acta - Biomembranes 1711, 99–125. [DOI] [PubMed] [Google Scholar]
- Spencer WC, McWhirter R, Miller T, Strasbourger P, Thompson O, Hillier LW, Waterston RH, and Miller DM (2014). Isolation of specific neurons from C. Elegans larvae for gene expression profiling. PLoS ONE 9, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich TA, Xu J, Skerrett IM, Nicholson BJ, and Shaw JE (2009). Interactions between innexins UNC-7 and UNC-9 mediate electrical synapse specificity in the Caenorhabditis elegans locomotory nervous system. Neural Development 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Stetina SE, Fox RM, Watkins KL, Starich TA, Shaw JE, and Miller DM (2007). UNC-4 represses CEH-12/HB9 to specify synaptic inputs to VA motor neurons in C. elegans. Genes and Development 21, 332–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuer Costa W, Yu S, Liewald JF, and Gottschalk A (2017). Fast cAMP Modulation of Neurotransmission via Neuropeptide Signals and Vesicle Loading. Current Biology 27, 495–507. [DOI] [PubMed] [Google Scholar]
- Tamás G, Buhl EH, Lörincz A, and Somogyi P (2000). Proximally targeted GABAergic synapses and gap junctions synchronize cortical interneurons. Nature Neuroscience 3, 366–371. [DOI] [PubMed] [Google Scholar]
- Taylor SR, Santpere G, Weinreb A, Barrett A, Reilly MB, Xu C, Varol E, Oikonomou P, Glenwinkel L, McWhirter R, et al. (2021). Molecular topography of an entire nervous system. Cell 184, 4329–4347.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TenBroek EM, Lampe PD, Solan JL, Reynhout JK, and Johnson RG (2001). Ser364 of connexin43 and the upregulation of gap junction assembly by cAMP. The Journal of Cell Biology 155, 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teubner B, Degen J, Söhl G, Güldenagel M, Bukauskas FF, Trexler EB, Verselis VK, De Zeeuw CI, Lee CG, Kozak CA, et al. (2000). Functional Expression of the Murine Connexin 36 Gene Coding for a Neuron-Specific Gap Junctional Protein. The Journal of Membrane Biology 176, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenin AF, Kowal TJ, Fong JT, Kells RM, Fisher CG, and Falk MM (2013). Proteins and Mechanisms Regulating Gap-Junction Assembly, Internalization, and Degradation. Physiology 28, 93–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Senecal JMM, Lynn BD, Traub RD, and Nagy JI (2020). Connexin36 localization along axon initial segments in the mammalian CNS. International Journal of Physiology, Pathophysiology and Pharmacology 12, 153. [PMC free article] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, et al. (2009). Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nature Methods 6, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub RD, Draguhn A, Whittington MA, Baldeweg T, Bibbig A, Buhl EH, and Schmitz D (2002). Axonal gap junctions between principal neurons: a novel source of network oscillations, and perhaps epileptogenesis. Reviews in the Neurosciences 13, 1–30. [DOI] [PubMed] [Google Scholar]
- Trenholm S, and Awatramani GB (1995). Myriad roles for gap junctions in retinal circuits (University of Utah Health Sciences Center; ). [PubMed] [Google Scholar]
- Walker DS, and Schafer WR (2020). Distinct roles for innexin gap junctions and hemichannels in mechanosensation. ELife 9, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Borst A, Zaslavsky N, Tishby N, and Segev I (2017). Efficient encoding of motion is mediated by gap junctions in the fly visual system. PLoS Computational Biology 13, e1005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Q, Po MD, Hulme E, Chen S, Liu X, Kwok SW, Gershow M, Leifer AM, Butler V, and Fang-Yen C (2012). Proprioceptive coupling within motor neurons drives C. elegans forward locomotion. Neuron 76, 750–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, and Brenner S (1986). The Structore of the Nervous System of the Nematode Caenorhabditis elegans. Phil. Trans. Royal Soc. London. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, and Thomson JN (1992). Mutations in the Caenorhabditis elegans unc-4 gene alter the synaptic input to ventral cord motor neurons. Nature 355, 838–841. [DOI] [PubMed] [Google Scholar]
- Winnier AR, Meir JYJ, Ross JM, Tavernarakis N, Driscoll M, Ishihara T, Katsura I, and Miller DM (1999). UNC-4/UNC-37-dependent repression of motor neuron-specific genes controls synaptic choice in Caenorhabditis elegans. Genes and Development 13, 2774–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witvliet D, Mulcahy B, Mitchell JK, Meirovitch Y, Berger DR, Wu Y, Liu Y, Koh WX, Parvathala R, Holmyard D, et al. (2021). Connectomes across development reveal principles of brain maturation. Nature 2021 596:7871 596, 257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X-H, Wang M, He X-N, He F, Zhang S-Q, Lu W, Qiu Z-L, and Yu Y-C (2016). Electrical coupling regulates layer 1 interneuron microcircuit formation in the neocortex. Nature Communications 7, 12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsiros V, Aradi I, and Maccaferri G (2007). Propagation of postsynaptic currents and potentials via gap junctions in GABAergic networks of the rat hippocampus. Journal of Physiology 578, 527–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1: unc-4 mutants exhibit a backward locomotion defect in the tapping assay, related to Figures 1 and 3. Representative examples of tapping assay for wild type, unc-4(e2323), and unc-4(e2323);goa- 1(lof). Individual L4 worms are tapped on the head and backward locomotion is scored as wild type or Unc. unc-4(e2323) mutants are unable to move backward. Backward locomotion is partially restored in unc-4(e2323);goa-1(lof).
Table S2: Adjacent neurons that fail to form electrical synapses in C. elegans, related to Figure 1.
Video S2: Super resolution microscopy of dual-color gap junction, related to Figure 4. 3D reconstruction of SIM Z-stack of UNC-7::tagRFP in AVA and GFP::UNC-9 in VA. UNC-7 and UNC-9 are closely localized, but slightly displaced as expected for UNC-7 vs UNC-9 hemichannels at a heterotypic gap junction.
Video S3: Evoked AVA GCaMP response, related to Figure 5. Individual AVA axon expressing GCaMP in wild type, unc-4(e2323), and unc-4(e2323);goa-1(lof) double mutant. VAs expressing Chrimson are optogenetically activated with a pulse (500 ms) of 561 nm light. Two subsequent frames are noted with a red circle in the left-hand corner. In wild type and unc-4(e2323);goa-1(lof) worms, a detectable AVA GCaMP response is detected following VA activation
Video S4: GFP::UNC-9 trafficking in VA axon, related to Figure 6. Representative videos of GFP::UNC-9 trafficking in wild-type and unc-4(e2323) mutant VA axon. Wild-type worms exhibit rapid, bidirectional trafficking of GFP::UNC-9 puncta which is largely disabled in unc-4(e2323) mutants. Scale bar = 5 μm.
Data Availability Statement
The RNA-seq datasets generated during this study are publicly available at GEO (GSE173287). Microscopy data can be shared on request.
No original code was developed in this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


