Abstract
The Mad family comprises four basic-helix-loop-helix/leucine zipper proteins, Mad1, Mxi1, Mad3, and Mad4, which heterodimerize with Max and function as transcriptional repressors. The balance between Myc-Max and Mad-Max complexes has been postulated to influence cell proliferation and differentiation. The expression patterns of Mad family genes are complex, but in general, the induction of most family members is linked to cell cycle exit and differentiation. The expression pattern of mad3 is unusual in that mad3 mRNA and protein were found to be restricted to proliferating cells prior to differentiation. We show here that during murine development mad3 is specifically expressed in the S phase of the cell cycle in neuronal progenitor cells that are committed to differentiation. To investigate mad3 function, we disrupted the mad3 gene by homologous recombination in mice. No defect in cell cycle exit and differentiation could be detected in mad3 homozygous mutant mice. However, upon gamma irradiation, increased cell death of thymocytes and neural progenitor cells was observed, implicating mad3 in the regulation of the cellular response to DNA damage.
The ability of proliferating cells to exit the cell cycle is crucial to the ordered growth and development of tissues and organisms, to tissue homeostasis, and to a coordinated response to stress (46). Failure to exit the cell cycle in response to differentiation signals and cellular stress, such as DNA damage, is likely to play a role in oncogenesis (13, 14, 51). The Mad protein family members, as well as the MNT/ROX proteins, are transcriptional repressors that are thought to antagonize the functions of the Myc family (c-, N-, L-, and s-Myc) and to be involved in the control of cell cycle exit upon differentiation (20, 25, 26, 36, 38). The Mad proteins are encoded by four paralogous genes, mad1, mxi1, mad3, and mad4 (3, 20, 27, 60). Like Myc and the more recently characterized MNT (25, 26, 38), the Mad proteins belong to a family of basic-helix-loop-helix/leucine zipper (bHLHZ) transcription factors which require heterodimerization with the stable and widely expressed adapter protein Max in order to bind the E-box (CACGTG) DNA recognition site and related sites (4, 7, 18, 27, 60). The presence of such sequences in synthetic and naturally occurring promoters permits transactivation by the Myc-Max heterocomplexes in transient-transfection assays (1, 2, 31). By contrast, in similar contransfection assays, Mad-Max heterocomplexes act as transcriptional repressors (4, 6, 27, 60).
Mad repression is mediated through the interaction between a conserved amino-terminal region (Sin3 interaction domain [SID]) of the Mad protein with the mSin3A and mSin3B corepressors. These corepressors exist as multiprotein complexes comprising histone deacetylases (HDACs) and other factors whose functions are still unclear (6, 19, 28, 32, 49). The function of the HDACs is crucial in mediating repression, most likely through deacetylation of the lysine residues within the amino-terminal tails of nucleosomal histones H3 and H4. Deacetylation leads to increased interaction of the histone tails with the DNA backbone, forming a repressive chromatin structure (28). Thus, Mad-Max complexes serve to recruit HDACs, through association with the mSin3 co repressors, to specific target genes. This is consistent with the fact that the mSin3 interaction domain, and hence the ability of Mad to repress transcription, is required for its biological activities. Indeed, disruption of the putative amphipathic helix in the SID of Mad1 blocks its ability to inhibit transformation of mouse embryo fibroblasts by Myc and Ras, to induce a G1 arrest in the macrophage cell line U937, or to promote erythroid differentiation of murine erythroleukemia cells (3–6, 8, 9, 11, 29, 33, 47, 58).
Targeted deletion of mad1 and mxi1 in mice has provided evidence of their roles in cell cycle exit (15, 16, 50). mad1 mutant mice display an increased proliferative capacity of late myeloid progenitors (16). Mice deficient in mxi1 show a more generalized phenotype, as progressive hyperplasia is detected in tissues such as the spleen and prostate and degenerative changes are detected in the kidney. Deletion of mxi1 also increases sensitivity to carcinogens and results in accelerated tumorigenesis in collaboration with Ink4a locus deletion. Mouse embryo fibroblasts derived from mxi1-null mice are also more prone to transformation by the Myc and Ras oncogenes (50). By contrast, mice engineered to express mad1 under the control of the beta-actin promoter display multiorgan hypoplasia and a reduced proliferative capacity of hematopoietic cells and mouse embryo fibroblasts in vitro (45). Taken together with the results of studies demonstrating induction of Mad proteins during differentiation, these results suggest that a balance between the Myc-Max and Mad-Max complexes regulates cell proliferation and cell cycle exit upon differentiation.
The expression pattern of mad3 remains the most perplexing of those of the Mad family genes. By Northern blotting mad3 RNA is undetectable in adult tissues with the exception of the testis and the thymus. In these tissues and in mouse embryos, mad3 transcripts were restricted to proliferating cells. In addition, within a population of proliferating cells, the expression of mad3 was not uniform, suggesting the possibility that mad3 expression is cell cycle regulated (27, 44). In this paper, we further investigate the expression and function of mad3 using in situ hybridization and targeted deletion of the mad3 gene.
MATERIALS AND METHODS
Generation of mad3 mutant mice.
The mad3 cDNA (32) was used to screen a murine 129/Sv genomic library (a kind gift from P. Soriano, Fred Hutchinson Cancer Research Center); the resulting clone was mapped by restriction analysis, and exon-intron boundaries were sequenced. A gene-targeting vector was prepared as follows: an 845-bp NheI fragment containing the 5′ end of the first coding exon in mad3 and a 5.8-kb BamH1-SacII fragment were cloned in the NheI and the SacII sites, respectively, of the vector pPGKneobpAPGKdtabpA (a kind gift of P. Soriano). The positive selection cassette PGKneobpA replaced a genomic region encompassing the translation initiation codon, the SID, and the basic helix-loop-helix region. The negative selection marker, PGKdtabpA, was placed at the 5′ end of the 845-bp short arm (see Fig. 2A). A mock PCR-positive control for the targeted mad3 locus was made by cloning the 1.6-kb HindIII-NheI fragment corresponding to the 5′ region of the mad3 locus into the HindIII-NheI sites of pPGKneobpAPGKdtabpA.
FIG. 2.
Targeted disruption of the mad3 gene. (A) Map of the mad3 locus, showing the exon-intron structure of the gene. From 5′ to 3′, the exons encode the SID (E1), the conserved HGYAS motif (E2), the basic region (E3), the HLH (E4), the LZ (E5), and the C-terminal segment (E6), respectively. The targeting construct comprised the pPGKneobpAPGKdtabpA cassette flanked by an 845-bp NheI fragment and a 5.8-kb BamHI-SacII fragment for recombination arms. Negative selection was provided by PGKdtabpA. Homologous recombination yielding to the targeted locus was selected by PCR using the primers M3133 and YZ29 (arrows) and later confirmed by Southern blotting using probes A and B. (B) Genomic Southern blot analysis confirming the correct 5′ (probe A) and 3′ (probe B) junctions. (C) PCR genotyping of mad3+/+, mad3+/−, and mad3−/− mice using the primers depicted as arrowheads in panel A. (D) Detection of mad3 by RT-PCR. In embryos obtained from the breeding of mad3+/− mice, RT-PCR confirmed the absence of mad3 expression in mad3−/− mice and demonstrated that a functionally null mutation was generated. Amplification of the S16 mRNA was used as a control for efficient cDNA synthesis. wt, wild type.
A Pvu1-linearized targeting vector was electroporated into the AK7 embryonic stem (ES) cells and placed under G418 selection (16; P. Soriano, Letter, Nat. Genet. 21:70–71, 1999). Surviving clones after 9 days were analyzed by PCR using the primers M3133 (5′-TTCATCGCCACACCTTGCCT-3′; sense primer, 1,608 bp 5′ of mad3 translation initiation codon) and YZ39 (5′-TCGCAGCGCATCGCCTTCTA-3′; antisense primer in the 3′ end of the neo gene) according to the method described in reference 16. Positive clones were expanded and frozen. Correct targeting was ascertained by Southern blotting with probes diagnostic for the 5′ (probe A) and the 3′ (probe B) ends of the expected homologous recombination event (see Fig. 2B) and for the neomycin-selectable marker (data not shown).
ES cell clones were injected into C57BL/6 blastocysts and then transplanted into pseudopregnant females according to standard procedures. Two clones gave rise to chimeric founders that transmitted the targeted allele through the germ line. Heterozygote progeny was interbred to produce homozygous mice. Genotyping was performed by Southern blotting (see above) or by PCR on DNA isolated from toes (clipped for numbering) using the protocol described in reference 16 and the primers M31186 (5′-ATCGCCGTCTCTCGTAGCTCG-3′) M31897 (5′-GACCCTTTCCCCAGTCACTCC-3′), and YZ39 (see Fig. 2C).
RNA preparation and analysis.
Total RNA was prepared from embryos at embryonic day 10.5 with TRIzol reagent (Gibco BRL) according to the manufacturer's instructions. Reverse transcription-PCR (RT-PCR) was carried out as described previously (30). The S16 control was amplified with the sense primer 5′-AGGAGCGATTTGCTGGTGTGGA-3′ and the antisense primer 5′-GCTACCAGGCCTTTGAGATGGA-3′ (35) (20 cycles; primers generated a 102-bp fragment). mad3 was amplified with a primer consisting of 5′-CAGCTGAAGCGGTGCTTAG-3′ (forward in the helix-loop-helix coding exon) and 5′-CAGGCCTGAAGAGTCCAAG-3′ (reverse in the leucine zipper coding exon; 30 cycles yielded a 263-bp fragment).
In situ hybridization and detection of BrdU.
5-Bromo-2"-deoxyuridine (BrdU) incorporation was done essentially as in described in reference 39. Briefly, 100 μg per kg of body weight was injected intraperitoneally into a pregnant mother. Embryos were harvested 1 h after injection, fixed in 4% paraformaldehyde, paraffin embedded, and sectioned. Sections were then treated for in situ hybridization using 35S-labeled cRNA probes as described previously (27, 44). BrdU was revealed using a monoclonal antibody to BrdU (Becton Dickinson) and detection with Vectastain avidin-biotin-peroxidase complex reagent and diaminobenzidine without nickel (Vector Laboratories) before the slides were dipped into NTB2 emulsion (Kodak). After a 7-day exposure at 4°C, the slides were developed, counterstained with bisbenzidine (Hoechst 33258), and examined under dark-field and epifluorescence illumination with a Zeiss Axioplan microscope.
TUNEL assay.
The terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) reaction to detect incorporation of biotinylated dUTP mediated by terminal transferase was carried out on sectioned embryos as described previously (17) except that the slides were microwaved for 2 min in 10 mM sodium citrate, pH 6.0. Biotinylated dUTP was revealed with the Vectastain avidin-biotin-peroxidase complex reagent (Vector) and the diaminobenzidine substrate (Vector) according to the manufacturer's recommendations. Sections were counterstained with Gill's hematoxylin.
Analysis of hematopoiesis and lymphocyte proliferation.
Hematopoietic cells from peripheral blood, bone marrow, spleen, and thymus were analyzed by examination of cellular morphology, flow cytometry, and culture in semisolid medium as described previously (16). To analyze cell proliferation of mature T and B lymphocytes, 5 × 104 splenocytes were stimulated in suspension with 20 μg of Escherichia coli lipopolysaccharide (Sigma) per ml or with 2 μg of concanavalin A (Sigma) per ml (in RPMI 1640 medium containing 10% fetal bovine serum [HyClone], 10 mM l-glutamine, and 1 mM melanocyte-stimulating hormone). BrdU was added after 16 h, and cells were harvested after 24, 48, or 72 h. Following BrdU labeling, lymphocytes were fixed with 86% ethanol in phosphate-buffered saline (and stored if necessary at 4°C) and stained with anti-BrdU antibody (Becton Dickinson) and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (Cappel) according to the manufacturer's protocols. Following staining with 10 μg of propidium iodide per ml, cells were analyzed on a FACScan cell sorter. The fraction of cells in S phase prior to stimulation with mitogens was determined by staining splenocytes with anti-CD45R or anti-CD8 and anti-CD4 directly conjugated to FITC followed by ethanol fixation, staining with propidium iodide, and analysis on a FACScan cell sorter.
Cell survival analysis.
Primary thymocytes were isolated from 8-week-old male mad3+/+ and mad3−/− mice in the 129/Sv inbred background and cultured in Dulbecco modified Eagle medium supplemented with 250 mM l-asparagine, 50 mM 2-mercaptoethanol, and 10% fetal bovine serum (HyClone) as described previously (55). Cell viability at 24 and 48 h after treatment was determined either by trypan blue (Gibco) exclusion and counting in a hemocytometer or by flow cytometric analysis of annexin V-FITC staining (Clontech) using a FACScan cell sorter (Becton Dickinson). Lymphotoxicity was induced by gamma irradiation at doses of 100, 200, 500, and 1,000 rads at a rate of 340 rads/min, by phorbol-12-myristate-13-acetate (PMA) at 2 ng/ml, by ionomycin at 1 μg/ml, and by dexamethasone at 1 μM.
RESULTS
mad3 is expressed in the S phase of the cell cycle.
In our initial description of mad3 expression (27, 44), we were surprised to find this gene predominantly expressed in cycling cells. In adult mice, mad3 was detected only in the proliferating areas of the thymus and the testis. In the developing embryo, mad3 RNA was detected from day 9.5 to day 12.5 postcoitus (p.c.) especially in the ventricular zone (VZ) of the neuroepithelia, in the progression zone of the limb buds, and in the aortic arches and liver. In all these tissues, mad3 was present in only a fraction of cells, producing a patchwork expression pattern similar to that of genes known to be expressed in specific cell cycle phases (see, for example, reference 54). To investigate this possibility further, we employed in situ hybridization to detect mad3 expression in parallel with immunohistochemistry to measure incorporation of the thymidine analogue BrdU into DNA. We concentrated our analysis on the spinal cord, where neuronal progenitors are generated in the VZ and migrate out to the intermediate zone (IZ) where they differentiate (41). The VZ consists of actively dividing neuronal progenitors. As they differentiate, these precursors exit from the cell cycle and migrate away from the VZ and into the IZ. Differentiation proceeds in a specific sequence with respect to both time and position along the dorsoventral and the rostrocaudal axis.
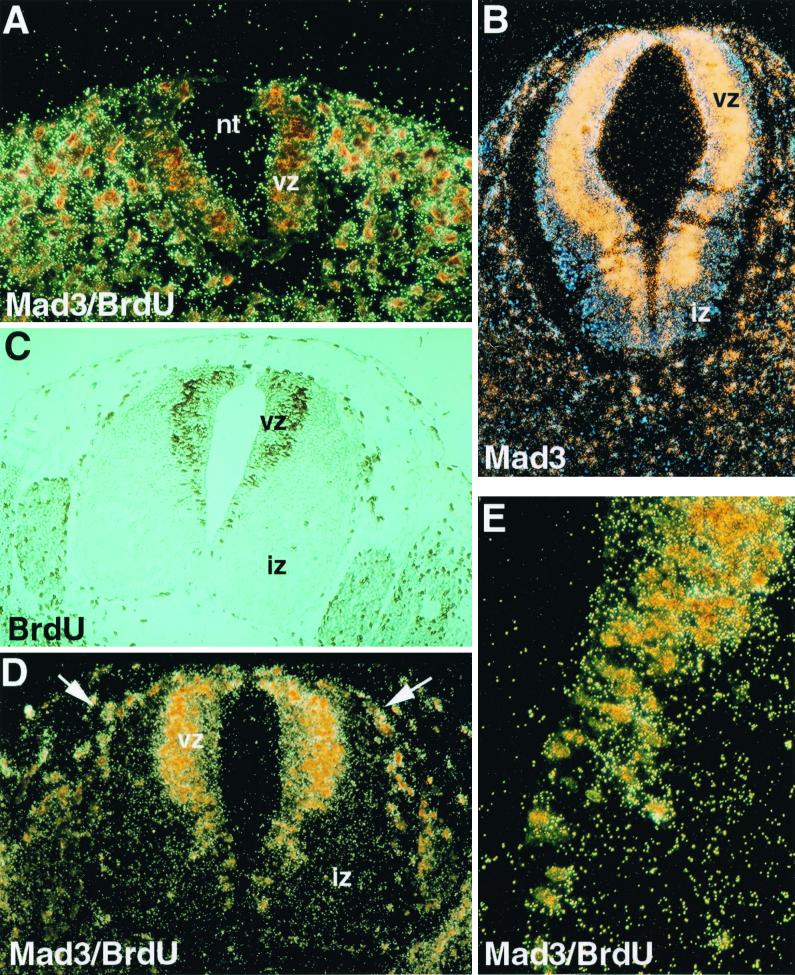
In the spinal cord before day 9.5 p.c. and in caudal regions between day 10.5 and 12.5 p.c., differentiation had not commenced, as evidenced by the absence of an IZ and the intense BrdU labeling detected all over the transverse section of the VZ (Fig. 1A). At that stage, mad3 expression was nearly undetectable in the neural tube, although it was apparent in the lateral mesenchyme (Fig. 1A and data not shown). By contrast, beginning in the anterior part of the spinal cord at day 10.5 p.c., mad3 expression was intense and restricted to the outermost periphery of the VZ (Fig. 1B). As we had shown previously (27), mad3 expression was absent from the IZ, where mad1 was highly expressed. To determine if cells at the periphery of the VZ were undergoing DNA synthesis, BrdU was injected into pregnant females 1 h before harvesting of embryos. This analysis revealed that BrdU-positive nuclei existed mainly in neural progenitors situated at the periphery of the VZ. These findings are consistent with previous studies demonstrating apical-basal nuclear migration of neuronal progenitors as a function of cell cycle phase (reviewed in references 37 and 56) (Fig. 1C). Double labeling for mad3 expression and BrdU incorporation revealed a striking superposition of the two signals (Fig. 1D and E). Colocalization was not limited to the neural progenitors; it was also observed in neural crest cells migrating from the roof of the neural tube, in the sclerotome, in the limb bud, and in the liver (Fig. 1D, arrows, and data not shown). Because the BrdU pulse was for only 1 h, the coincidence of mad3 signal with BrdU-positive nuclei strongly suggested that mad3 was expressed during the S phase of the cell cycle of neuronal progenitor cells (estimated S phase in neural progenitors is 4 h with a 10-h cell cycle time [see reference 7a]). Yet mad3 was not expressed in all S-phase progenitors, as its transcripts were absent from the embryo before day 9.5 p.c. and in the caudal neural tube, where differentiation had not yet commenced. Thus, mad3 expression would appear to be restricted to neuronal progenitors during the last S phase prior to terminal differentiation. To investigate the function of mad3, we generated a targeted mutation of the murine mad3 gene by homologous recombination in ES cells.
FIG. 1.
Expression of mad3 in neural progenitor cells. (A) Caudal section of a day 11.5 p.c. mouse embryo. The neural tube (nt) shows little sign of differentiation, as evidenced by the absence of the IZ, and consists primarily of proliferating neural progenitors. Brown-stained nuclei indicate cells that have incorporated BrdU for 1 h and are in the S phase of the cell cycle. The section was also hybridized with a mad3 antisense riboprobe. mad3 signal is realtively weakly detected in the proliferating neural progenitors but can be seen in tissue surrounding the neural tube (see panel D). (B) The expression of mad3 is restricted to the periphery of the VZ. No signal is detected in the IZ with the mad3 probe on this day 10.5 p.c. transverse section. (C) Transverse section though the neural tube in a truncal location at day 11.5 p.c. Cells in S phase of the cell cycle are labeled with BrdU (1 h) (brown stain). In the neural tube, the labeled nuclei are localized at the periphery of the VZ. The IZ contains the postmitotic neurons unlabeled with BrdU. Occasional endothelial cells are BrdU labeled. (D) Colocalization of BrdU-positive cells and mad3 in situ hybridization signal. Shown is a transverse section of the neural tube at day 11.5 p.c. processed for immunocytochemistry for the detection of BrdU-incorporating cells and for in situ hybridization to detect mad3. The nuclei at the periphery of the VZ are labeled by BrdU and are also highlighted by the mad3 hybridization signal. Doubly labeled cells can also be observed in the dorsolateral part of the neural tube and are likely to be neural crest cells (arrows). (E) Higher magnification of the VZ pictured in panel D, showing the juxtaposition of mad3 expression and the BrdU-labeled neural progenitors.
Targeted disruption of the murine mad3 gene.
To inactivate the mad3 gene in mice, we isolated the genomic fragment encoding MAD3 and determined its exon-intron structure. The mad3 gene comprises six exons (Fig. 2A), with each of the functional domains of mad3 (the SID, the basic region, the helix-loop-helix domain, and the leucine zipper) being encoded by separate exons (4–6). The vector used for targeted disruption of the mad3 gene contained 0.845 kb of genomic DNA encompassing a segment of exon 1 5′ to the translation initiation codon and 5.8 kb downstream of exon 4 (Fig. 2A). Homologous recombination between the targeting vector and the mad3 locus replaced the exons encoding the translation initiation codon, the SID, and the basic region by the positive selection marker PGKneobpA, inactivating the ability of MAD3 to initiate translation, repress transcription, and interact with MAX and DNA, thus creating a functionally null allele. Following standard procedures, the ES cells were electroporated with the linearized targeting vector and placed under G418 selection. The surviving colonies were screened by PCR and Southern blotting of their DNA (Fig. 2B). Correct targeting occurred in 5% of the surviving colonies. Eight correctly targeted ES cell clones were injected into mouse blastocysts, yielding founder chimeric mice, and two ES clones contributed to the germ line in the chimeras, giving rise to mad3 heterozygote mice. These lines were derived on the hybrid 129/Sv-C57BL/6J and on the inbred 129/Sv background, the latter being used in this study. The progeny of mating between heterozygotes were identified by PCR (Fig. 2C). mad3 homozygote mutants (mad3−/−) were found to be present at the expected Mendelian ratio (mad3+/+; 24%; mad3+/−; 48%; mad3−/−, 28% [n = 420] in hybrid 129/Sv × C57BL/6J mice; mad3+/+; 27%; mad3+/−; 51%; mad3−/−; 22% [n = 377] in inbred 129/Sv mice), indicating no significant embryonic lethality. mad3−/− mice were viable, appeared outwardly normal, and showed no obvious differences in size, behavior, reproductive ability, incidence of neoplasia, or life span compared with wild-type littermates.
The absence of mad3 mRNA was confirmed by RT-PCR on RNA prepared from day 10.5 p.c. embryos (Fig. 2D) and by mRNA in situ hybridization on different embryonic and adult tissues (data not shown).
mad3-null mice do not display obvious defects in cellular proliferation or differentiation.
Because the members of the Myc family of transcription factors are key regulators of cell proliferation, differentiation, and death, we investigated the mad3−/− mice for phenotypes related to these processes, both in vivo and in vitro. In vivo, we failed to find any significant differences in the number of cells incorporating BrdU in day 11.5 p.c. embryos or in cells that could be labeled by TUNEL, a hallmark of apoptosis (data not shown). Special scrutiny was applied to proliferation and differentiation in the central nervous system, where mad3 expression was well documented (references 27 and 44 and this study). Staining for the neural markers (Delta1, NeuroD1, NeuroD3, Mash1, Jagged2, and Pax3) or for other members of the Myc family (N-Myc, mad1, mxi1, and mad4) was identical in mad3+/+ and mad3−/− mice (data not shown). Because of abnormalities in the hematopoietic system in mad1-transgenic (45) and mad1−/− (16) mice, we carefully examined the hematopoietic systems of mad3−/− mice. No significant differences were observed between mad3+/+ and mad3−/− mice in the numbers or types of cells in peripheral blood, spleen, thymus, or bone marrow. Moreover, there were no differences in the frequency or number of hematopoietic precursor cells of the erythroid or myeloid lineages as determined by in vitro colony assays of bone marrow cells (data not shown). In vitro, the growth curves and the cell cycle distribution of mouse embryo fibroblasts prepared from wild-type and mutant mad3 129/Sv mice did not indicate deregulated proliferation due to the inactivation of mad3 (data not shown).
Increased sensitivity of mad3−/− mice to gamma irradiation.
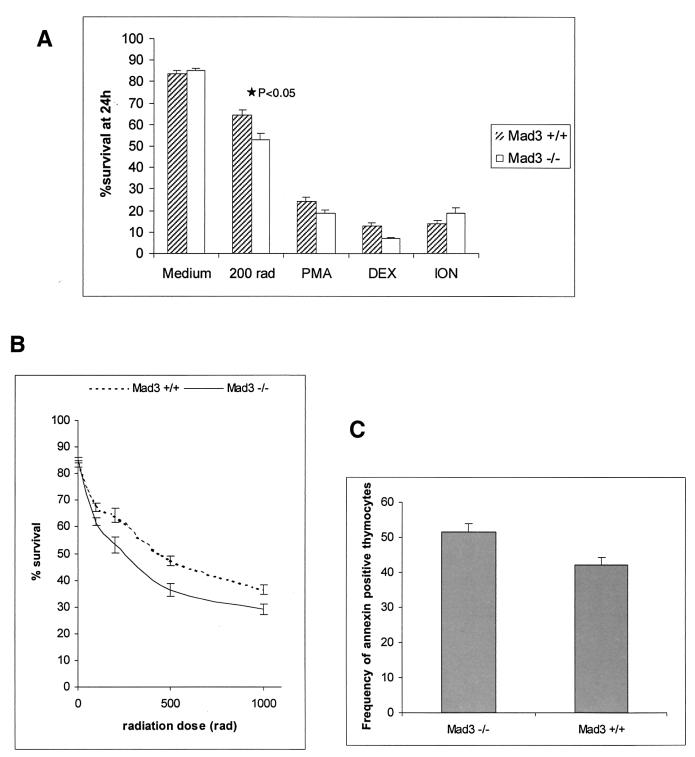
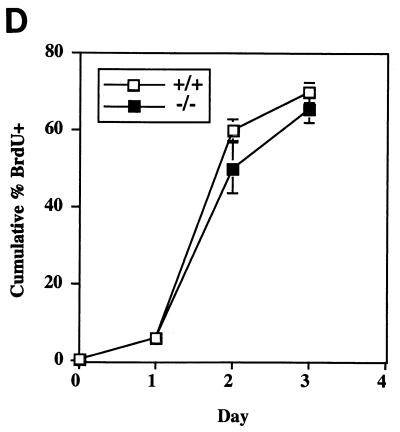
Because mad3 and c-Myc were coexpressed in the thymus (44), where Myc has been shown elsewhere to influence apoptosis in T lymphocytes (22, 42, 53, 57), we examined the response of mad3+/+ and mad3−/− T cells to apoptotic stimuli. After dissection of thymi (n = 9 for wild type; n = 9 for mad3−/−), the thymocytes were placed in culture and subjected to treatment with different apoptosis-inducing agents: PMA, ionomycin, dexamethasone, or gamma irradiation. As determined by trypan blue exclusion, no significant difference in the ratio between dead and living cells could be observed between wild-type and mutant cells in medium alone or supplemented with PMA, ionomycin, or dexamethasone (Fig. 3A). By contrast, in three separate experiments each employing three mad3+/+ and three mad3−/− mice, a 10% increase in the number of dead thymocytes following irradiation was observed (P < 0.05) (Fig. 3A). This increased cell death in response to gamma irradiation was observed with doses of 200, 500, and 1,000 rads (Fig. 3B) at 24 or 48 h after treatment (data not shown). The annexin V assay was used as an independent measure of apoptotic cell death and gave similar results, i.e., a 10% increase in annexin V-positive thymocytes following irradiation (Fig. 3C). Interestingly, this effect was specific for mad3 inactivation. No difference in the wild-type rate of death in response to gamma irradiation was observed in thymocytes isolated from mad1−/− mice (n = 2) or transgenic [BAP-mad1] mice (n = 3) (data not shown). Thymocytes isolated from mice mutant for both the mad1 and the mad3 genes (n = 3) behaved similarly to the one from the single mad3−/− mice (data not shown). The effect of mad3 loss of function on thymocyte death did not correlate with an increased rate of T-cell cycling. Stimulation of resting T cells from mad3−/− mice with concanavalin A did not induce these cells to enter S phase more rapidly than T cells from wild-type mice (Fig. 3D). Indeed, there was a trend toward slower entry of mad3−/− cells into S phase following stimulation with lectins for both T cells (Fig. 3D) and B cells (data not shown). Although, as expected, we could readily detect Mad3 protein in wild-type thymocytes, irradiation did not lead to a significant increase in Mad3 protein levels. Thus, Mad3 protein is not induced as a response to damage but may function at normal endogenous levels to inhibit apoptosis.
FIG. 3.
Increased sensitivity of mad3−/− thymocytes to gamma irradiation. (A) Thymocytes from mad3+/+ and mad3−/− mice were left untreated (medium), treated with gamma irradiation (200 rads), and then incubated in tissue culture medium containing no addition (medium or 200 rads), PMA (2 ng/ml) (PMA), dexamethasone (1 μM) (DEX), or ionomycin (1 μg/ml) (ION). Death was determined 24 h after culture by the trypan blue exclusion method. (B) Dose response of mad3+/+ and mad3−/− thymocytes to gamma irradiation. Decreased survival of mad3−/− thymocytes is observed at 100, 200, 500, and 1,000 rads at 24 h after culture. (C) Determination of the proportion of annexin-positive cells is an independent measurement of the increased apoptosis in mad3−/− mice in response to gamma irradiation. Thymocytes harvested from mad3+/+ and mad3−/− mice were subjected to 200-rad irradiation. The binding of annexin and the extent of apoptosis were determined by fluorescence-activated cell sorting 24 h after irradiation. (D) Loss of mad3 does not influence cell cycle entry in thymocytes. Thymocytes harvested from mad3+/+ and mad3−/− mice were stimulated by concanavalin A. Cell cycle entry was monitored by BrdU incorporation analyzed by fluorescence-activated cell sorting.
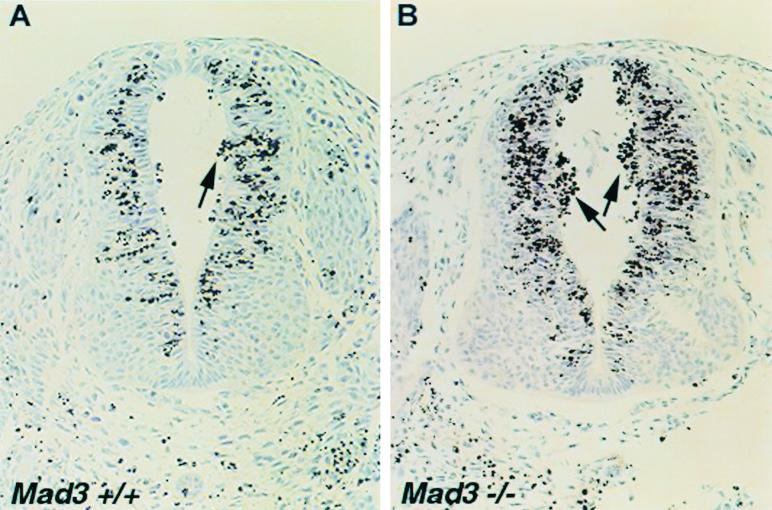
To investigate whether other cell types in mad3−/− mice displayed this increased sensitivity to apoptosis, we subjected 10.5-day pregnant mad3+/− mice that were bred to mad3+/− males to 200 rads of gamma irradiation. Embryos were harvested 3 h later, embedded, sectioned, and stained for TUNEL (17). Very few apoptotic cells could be observed in both wild-type and mad3 mutant mouse embryos without irradiation (data not shown). By contrast, as early as 3 h after irradiation, numerous cells were labeled with TUNEL and had fragmented nuclei characteristic of apoptosis (Fig. 4). In the neural tube, TUNEL-positive cells were mainly localized in the periphery of the VZ, in the cells that expressed mad3, and were in the S phase of the cell cycle when irradiated. TUNEL-positive cellular debris also accumulated in the lumen of the neural tube as previously reported (40). In these two regions, an increased number of cells or amount of debris was found in the mad3−/− embryos compared to that in wild-type embryos (Fig. 4). This result indicates that mad3−/− neural progenitor cells are also more sensitive to apoptosis induced by gamma irradiation.
FIG. 4.
mad3−/− embryos show increased sensitivity to gamma irradiation. Shown are results of TUNEL detection of apoptotic neural precursor cells in transverse sections of mad3+/+ (A) and mad3−/− (B) neural tube 3 h after irradiation at 200 rads in utero. The counterstain is hematoxylin. Brown apoptotic nuclei accumulated at the periphery of the VZ and in the lumen of the neural tube more readily in mad3-deficient embryos. Cell counting indicated a significant increase in dead cells in the mad3−/− neural tubes over those in the wild type (P < 0.05). Wild-type littermate control mice contained an average of 11.8 ± 2.7 apoptotic nuclei while mad3-null mice contained an average of 57.5 ± 5.8 apoptotic nuclei in the lumen of the neural tube.
DISCUSSION
We and others have shown previously that the expression of Mad family genes occurs in a sequential manner during cell cycle exit associated with differentiation (3, 10–12, 23, 24, 27, 34, 43, 44, 58). Whereas postmitotic cells in the developing mouse embryo clearly expressed mxi1, mad1, and mad4, mad3 expression could not be detected. In contrast, mad3 transcripts were found exclusively in proliferating cells overlapping with c-Myc or N-Myc expression (27, 43, 44). By combining in situ hybridization and immunological detection of BrdU, we show here that mad3 transcripts were detected exclusively in cells that have incorporated BrdU. Because the embryos used in this study were harvested 1 h after the injection of BrdU, the labeled cells were predominantly in the S phase of the cell cycle. This is consistent with the characteristically patchy expression pattern of mad3 previously observed in proliferating cells and agrees with the protein expression pattern observed for P19 cells (44). The results suggest that expression of mad3 is restricted to the S phase of the cell cycle in many cell types. Interestingly, mad3 was not expressed in all cells in S phase for a given cell lineage. This was best seen in the central nervous system. During the early phase of multiplication of the neural stem cell pool, mad3 was not detected. Instead, mad3 expression was concomitant with the appearance of postmitotic neuron precursors in the IZ. Interestingly, in the adipogenic 3T3L1 cells, mad3 expression correlated also with entry into the S phase of the cell cycle during the proliferative burst preceding terminal differentiation (43). Taken together, these data show that, even though mad3 expression is restricted to proliferating cells, it is closely linked to commitment to cell cycle exit and terminal differentiation.
The only significant phenotype detected in the mad3-deficient mice was an increased sensitivity to gamma irradiation. This could be documented in thymocytes ex vivo as well as in neural progenitor cells in embryo (Fig. 3 and 4). This altered sensitivity to irradiation was not observed in mad1-deficient thymocytes or in thymocytes prepared from mad1-overexpressing transgenic mice (data not shown). However, in mad1-knockout mice granulocytic progenitor cells exhibited significantly reduced survival when grown in limiting amounts of cytokines as well as after treatment with apoptosis-inducing drugs (15, 16). mad1 overexpression, by contrast, decreased the proliferative capacity of these cells and increased their survival when cytokine levels were reduced (45). Interestingly, the effect of mad3 loss on gamma irradiation-induced cell death was not associated with increased cell proliferation (Fig. 3D). Myc proteins were also shown previously to play a role in apoptosis in thymocytes and fibroblasts, apparently independent of their effects on cell cycle and progression into G1 (see references 42 and 57 for review). A number of studies have linked Myc-induced apoptosis to p53 function (21, 48, 52, 59, 61). Inactivation of p53 indeed prevented the death of thymocytes in response to a variety of signals including gamma irradiation (55). Furthermore, decreased apoptosis in neural progenitors in response to gamma irradiation has been also documented for p53−/− embryos (40). Since an increased sensitivity to irradiation was observed for mad3-deficient mice, it is tempting to speculate that deletion of mad3 altered the transcriptional balance on Myc-Mad target genes in favor of the activating Myc-Max complexes, leading in turn to a potentiation of p53-dependent cell death. Further evidence of genetic interaction between mad3 and p53 during the response to DNA insults awaits the generation and characterization of mouse double mutants for mad3 and p53.
Targeted mutation of mad3 in mice did not produce any evident phenotype associated with cell cycle exit and differentiation. This absence of phenotype is in contrast to the recent reports for mad1- and mxi1-deficient mice (15, 16, 50). As mentioned in the introduction, inactivation of mad1 deregulated cell cycle exit in granulocyte precursors while mxi1-deficient mice displayed multiorgan hyperplasia (16, 50). In mad3-knockout mice, however, no increase in cell proliferation could be detected in embryos, in T and B cells, in hematopoietic precursor cells, or in mouse embryo fibroblasts (Fig. 3D and data not shown). Differentiation of hematopoietic and neural precursor cells was especially scrutinized, but no precocious or delayed differentiation could be detected. Redundancy among Mad family members could be regarded as a potential explanation for the lack of detectable phenotype in cell cycle and differentiation in the mad3−/− mice. Indeed, in mad1-deficient mice we observed ectopic expression of mxi1 and mad3 in the spleen (16). However, in mad3 mutant mice, no change in the expression of any other Mad or Myc genes could be detected. Another possibility is that MAD3 function is redundant with the function of proteins other than those within the MAD family. One example is the recent finding of synthetic effects between mad1-null and p27KIP1-null mice, suggesting that the proteins encoded by these two genes may function in parallel to influence differentiation (G. A. McArthur et al., submitted for publication). In this regard, it is conceivable that MAD3 function is redundant with that of proteins involved in cell cycle regulation or in apoptosis. These possibilities can be further explored using multiple deletions of Mad family genes as well as of other genes involved in regulation of differentiation.
ACKNOWLEDGMENTS
We are indebted to Leni Sue Carlos and Pei Feng Cheng for technical assistance and to Keesook Lee for expert work in gene targeting. We are grateful to Philippe Soriano for advice and reagents. We are also thankful to to George Sale and the pathology service and to Barbara Johnston and the technicians of the animal facility.
This work was supported by grants CA57138 and HL54881 to R.N.E., a fellowship from the Damon Runyon-Walter Winchell Foundation (DRG-076) and a Special Fellowship of the Leukemia Society of America to G.A.M., and an NIH Mentored Clinical Investigator Award (K08 AJ01445) to B.M.I. R.N.E. is a Research Professor of the American Cancer Society.
REFERENCES
- 1.Amati B, Dalton S, Brooks M W, Littlewood T D, Evan G I, Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992;359:423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- 2.Amin C, Wagner A J, Hay N. Sequence-specific transcriptional activation by Myc and repression by Max. Mol Cell Biol. 1993;13:383–390. doi: 10.1128/mcb.13.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayer D E, Eisenman R N. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 1993;7:2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- 4.Ayer D E, Kretzner L, Eisenman R N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 5.Ayer D E, Laherty C D, Lawrence Q A, Armstrong A P, Eisenman R N. Mad proteins contain a dominant transcription repression domain. Mol Cell Biol. 1996;16:5772–5781. doi: 10.1128/mcb.16.10.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 7.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 7a.Caviness V S J, Takahashi T, Nowakowski R S. Numbers, time and neocortical neurogenesis: a general developmental and evolutionary model. Trends Neurosci. 1995;18:379–383. doi: 10.1016/0166-2236(95)93933-o. [DOI] [PubMed] [Google Scholar]
- 8.Cerni C, Bousset K, Seelos C, Burkhardt H, Henriksson M, Luscher B. Differential effects by Mad and Max on transformation by cellular and viral oncoproteins. Oncogene. 1995;11:587–596. [PubMed] [Google Scholar]
- 9.Chen J, Willingham T, Margraf L R, Schreiber-Agus N, DePinho R A, Nisen P D. Effects of the Myc oncogene antagonist, MAD, on proliferation, cell cycling and the malignant phenotype of human brain tumour cells. Nat Med. 1995;1:638–643. doi: 10.1038/nm0795-638. [DOI] [PubMed] [Google Scholar]
- 10.Chin L, Schreiber-Agus N, Pellicer I, Chen K, Lee H W, Dudast M, Cordon-Cardo C, DePinho R A. Contrasting roles for Myc and Mad proteins in cellular growth and differentiation. Proc Natl Acad Sci USA. 1995;92:8488–8492. doi: 10.1073/pnas.92.18.8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cultraro C M, Bino T, Segal S. Function of the c-Myc antagonist mad1 during a molecular switch from proliferation to differentiation. Mol Cell Biol. 1997;17:2353–2359. doi: 10.1128/mcb.17.5.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delgado M D, Lerga A, Canelles M, Gomez-Casares M T, Leon J. Differential regulation of Max and role of c-Myc during erythroid and myelomonocytic differentiation of K562 cells. Oncogene. 1995;10:1659–1665. [PubMed] [Google Scholar]
- 13.Fero M L, Randel E, Gurley K E, Roberts J M, Kemp C J. The murine gene P27(Kip1) is haplo-insufficient for tumour suppression. Nature. 1998;396:177–180. doi: 10.1038/24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fero M L, Rivkin M, Tasch M, Porter P, Carow C E, Firpo E, Polyak K, Tsai L H, Broudy V, Perlmutter R M, Kaushansky K, Roberts J M. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 15.Foley K P, Eisenman R N. Two MAD tails: what the recent knockouts of mad1 and mxi1 tell us about the Myc/MAX/MAD network. Biochim Biophys Acta. 1999;1423:M37–M47. doi: 10.1016/s0304-419x(99)00012-8. [DOI] [PubMed] [Google Scholar]
- 16.Foley K P, McArthur G A, Queva C, Hurlin P J, Soriano P, Eisenman R N. Targeted disruption of the Myc antagonist MAD1 inhibits cell cycle exit during granulocyte differentiation. EMBO J. 1998;17:774–785. doi: 10.1093/emboj/17.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gavrieli Y, Sherman Y, Ben-Sasson S A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grandori C, Eisenman R N. Myc target genes. Trends Biochem Sci. 1997;22:177–181. doi: 10.1016/s0968-0004(97)01025-6. [DOI] [PubMed] [Google Scholar]
- 19.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 20.Henriksson M, Luscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 21.Hermeking H, Eick D. Mediation of c-Myc-induced apoptosis by p53. Science. 1994;265:2091–2093. doi: 10.1126/science.8091232. [DOI] [PubMed] [Google Scholar]
- 22.Hueber A O, Zornig M, Lyon D, Suda T, Nagata S, Evan G I. Requirement for the CD95 receptor-ligand pathway in c-Myc-induced apoptosis. Science. 1997;278:1305–1309. doi: 10.1126/science.278.5341.1305. [DOI] [PubMed] [Google Scholar]
- 23.Hurlin P J, Ayer D E, Grandori C, Eisenman R N. The Max transcription factor network: involvement of Mad in differentiation and an approach to identification of target genes. Cold Spring Harbor Symp Quant Biol. 1994;59:109–116. doi: 10.1101/sqb.1994.059.01.014. [DOI] [PubMed] [Google Scholar]
- 24.Hurlin P J, Foley K P, Ayer D E, Eisenman R N, Hanahan D, Arbeit J M. Regulation of Myc and Mad during epidermal differentiation and HPV-associated tumorigenesis. Oncogene. 1995;11:2487–2501. [PubMed] [Google Scholar]
- 25.Hurlin P J, Queva C, Eisenman R N. Mnt, a novel Max-interacting protein, is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev. 1997;11:44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- 26.Hurlin P J, Queva C, Eisenman R N. Mnt: a novel Max-interacting protein and Myc antagonist. Curr Top Microbiol Immunol. 1997;224:115–121. doi: 10.1007/978-3-642-60801-8_11. [DOI] [PubMed] [Google Scholar]
- 27.Hurlin P J, Queva C, Koskinen P J, Steingrimsson E, Ayer D E, Copeland N G, Jenkins N A, Eisenman R N. mad3 and mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995;14:5646–5659. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knoepfler P S, Eisenman R N. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 29.Koskinen P J, Ayer D E, Eisenman R N. Repression of Myc-Ras cotransformation by Mad is mediated by multiple protein-protein interactions. Cell Growth Differ. 1995;6:623–629. [PubMed] [Google Scholar]
- 30.Kraut N, Snider L, Chen C M, Tapscott S J, Groudine M. Requirement of the mouse I-mfa gene for placental development and skeletal patterning. EMBO J. 1998;17:6276–6288. doi: 10.1093/emboj/17.21.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kretzner L, Blackwood E M, Eisenman R N. Myc and Max proteins possess distinct transcriptional activities. Nature. 1992;359:426–429. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- 32.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 33.Lahoz E G, Xu L, Schreiber-Agus N, DePinho R A. Suppression of myc, but not E1a, transformation activity by Max-associated proteins, Mad and mxi1. Proc Natl Acad Sci USA. 1994;91:5503–5507. doi: 10.1073/pnas.91.12.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsson L G, Pettersson M, Oberg F, Nilsson K, Luscher B. Expression of mad, mxi1, max and c-myc during induced differentiation of hematopoietic cells: opposite regulation of mad and c-myc. Oncogene. 1994;9:1247–1252. [PubMed] [Google Scholar]
- 35.Leonard M W, Lim K C, Engel J D. Expression of the chicken GATA factor family during early erythroid development and differentiation. Development. 1993;119:519–531. doi: 10.1242/dev.119.2.519. [DOI] [PubMed] [Google Scholar]
- 36.McArthur G A, Laherty C D, Queva C, Hurlin P J, Loo L, James L, Grandori C, Gallant P, Shiio Y, Hokanson W C, Bush A C, Cheng P F, Lawrence Q, Pulverer B, Koskinen P, Foley K P, Ayer D E, Eisenman R N. The Mad protein family links transcriptional repression to cell differentiation. Cold Spring Harbor Symp Quant Biol. 1998;63:423–433. doi: 10.1101/sqb.1998.63.423. [DOI] [PubMed] [Google Scholar]
- 37.McConnell S K. Constructing the cerebral cortex: neurogenesis and fate determination. Neuron. 1995;15:761–768. doi: 10.1016/0896-6273(95)90168-x. [DOI] [PubMed] [Google Scholar]
- 38.Meroni G, Reymond A, Alcalay M, Borsani G, Tanigami A, Tonlorenzi R, Nigro C L, Messali S, Zollo M, Ledbetter D H, Brent R, Ballabio A, Carrozzo R. Rox, a novel bHLHZip protein expressed in quiescent cells that heterodimerizes with Max, binds a non-canonical E box and acts as a transcriptional repressor. EMBO J. 1997;16:2892–2906. doi: 10.1093/emboj/16.10.2892. . (Erratum, 16:6055.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller M W, Nowakowski R S. Use of bromodeoxyuridine-immunohistochemistry to examine the proliferation, migration and time of origin of cells in the central nervous system. Brain Res. 1988;457:44–52. doi: 10.1016/0006-8993(88)90055-8. [DOI] [PubMed] [Google Scholar]
- 40.Norimura T, Nomoto S, Katsuki M, Gondo Y, Kondo S. p53-dependent apoptosis suppresses radiation-induced teratogenesis. Nat Med. 1996;2:577–580. doi: 10.1038/nm0596-577. [DOI] [PubMed] [Google Scholar]
- 41.Nornes H O, Carry M. Neurogenesis in spinal cord of mouse: an autoradiographic analysis. Brain Res. 1978;159:1–6. doi: 10.1016/0006-8993(78)90105-1. [DOI] [PubMed] [Google Scholar]
- 42.Prendergast G C. Mechanisms of apoptosis by c-Myc. Oncogene. 1999;18:2967–2987. doi: 10.1038/sj.onc.1202727. [DOI] [PubMed] [Google Scholar]
- 43.Pulverer B, Sommer A, McArthur G A, Eisenman R N E, Luscher B. Analysis of Myc/Max/Mad network members in adipogenesis: inhibition of the proliferative burst and differentiation by ectopically expressed mad1. J Cell Physiol. 2000;183:399–410. doi: 10.1002/(SICI)1097-4652(200006)183:3<399::AID-JCP13>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 44.Quéva C, Hurlin P J, Foley K P, Eisenman R N. Sequential expression of the MAD family of transcriptional repressors during differentiation and development. Oncogene. 1998;16:967–977. doi: 10.1038/sj.onc.1201611. [DOI] [PubMed] [Google Scholar]
- 45.Queva C, McArthur G A, Ramos L S, Eisenman R N. Dwarfism and dysregulated proliferation in mice overexpressing the Myc antagonist MAD1. Cell Growth Differ. 1999;10:785–796. [PubMed] [Google Scholar]
- 46.Raff M C. Size control: the regulation of cell numbers in animal development. Cell. 1996;86:173–175. doi: 10.1016/s0092-8674(00)80087-2. [DOI] [PubMed] [Google Scholar]
- 47.Roussel M F, Ashmun R A, Sherr C J, Eisenman R N, Ayer D E. Inhibition of cell proliferation by the mad1 transcriptional repressor. Mol Cell Biol. 1996;16:2796–2801. doi: 10.1128/mcb.16.6.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rupnow B A, Alarcon R M, Giaccia A J, Knox S J. p53 mediates apoptosis induced by c-Myc activation in hypoxic or gamma irradiated fibroblasts. Cell Death Differ. 1998;5:141–147. doi: 10.1038/sj.cdd.4400328. [DOI] [PubMed] [Google Scholar]
- 49.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi A I, DePinho R A. An amino-terminal domain of mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 50.Schreiber-Agus N, Meng Y, Hoang T, Hou H, Jr, Chen K, Greenberg R, Cordon-Cardo C, Lee H-W, DePinho R A. Role of mxi1 in ageing organ systems and the regulation of normal and neoplastic growth. Nature. 1998;393:483–487. doi: 10.1038/31008. [DOI] [PubMed] [Google Scholar]
- 51.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 52.Sherr C J. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 53.Shi Y, Glynn J M, Guilbert L J, Cotter T G, Bissonnette R P, Green D R. Role for c-myc in activation-induced apoptotic cell death in T cell hybridomas. Science. 1992;257:212–214. doi: 10.1126/science.1378649. [DOI] [PubMed] [Google Scholar]
- 54.Sitzmann J, Noben-Trauth K, Kamano H, Klempnauer K H. Expression of B-Myb during mouse embryogenesis. Oncogene. 1996;12:1889–1894. [PubMed] [Google Scholar]
- 55.Strasser A, Harris A W, Jacks T, Cory S. DNA damage can induce apoptosis in proliferating lymphoid cells via p53-independent mechanisms inhibitable by Bcl-2. Cell. 1994;79:329–339. doi: 10.1016/0092-8674(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 56.Takahashi T, Nowakowski R S, Caviness V S., Jr Interkinetic and migratory behavior of a cohort of neocortical neurons arising in the early embryonic murine cerebral wall. J Neurosci. 1996;16:5762–5776. doi: 10.1523/JNEUROSCI.16-18-05762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson E B. The many roles of c-Myc in apoptosis. Annu Rev Physiol. 1998;60:575–600. doi: 10.1146/annurev.physiol.60.1.575. [DOI] [PubMed] [Google Scholar]
- 58.Västrik I, Kaipainen A, Pentillä T-L, Lymboussakis A, Alitalo R, Parvinen M, Alitalo K. Expression of the mad gene during cell differentiation in vivo and its inhibition of cell growth in vitro. J Cell Biol. 1995;128:1197–1208. doi: 10.1083/jcb.128.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner A J, Kokontis J M, Hay N. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- 60.Zervos A S, Gyuris J, Brent R. mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. . (Erratum, 79:389, 1994.) [DOI] [PubMed] [Google Scholar]
- 61.Zindy F, Eischen C M, Randle D H, Kamijo T, Cleveland J L, Sherr C J, Roussel M F. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]