Abstract
Background
HIV intervention activities directed towards both those most likely to transmit and their HIV-negative partners have the potential to substantially disrupt HIV transmission. Using HIV sequence data to construct molecular transmission clusters can reveal individuals whose viruses are connected. The utility of various cluster prioritization schemes measuring cluster growth have been demonstrated using surveillance data in New York City and across the United States (U.S.), by the Centers for Disease Control and Prevention (CDC).
Methods
We examined clustering and cluster growth prioritization schemes using Illinois HIV sequence data that includes cases from Chicago, a large urban center with high HIV prevalence, to compare their ability to predict future cluster growth.
Results
We found that past cluster growth was a far better predictor of future cluster growth than cluster membership alone but found no substantive difference between the schemes used by CDC and the relative cluster growth scheme previously utilized in New York City (NYC). Focusing on individuals selected simultaneously by both the CDC and the NYC schemes did not provide additional improvements.
Conclusion
Growth-based prioritization schemes can easily be automated in HIV surveillance tools, and can be used by health departments to identify and respond to clusters where HIV transmission may be actively occurring.
Keywords: HIV, phylogenetics, genetic, network, MSM, cluster
2. Introduction
HIV is transmitted through sexual and injection drug use contact networks. Interventions, such as linkage to care, initiation of antiretroviral treatment, and pre-exposure prophylaxis (PrEP) for uninfected partners, that prioritize individuals who are part of groups with active transmission can more effectively reduce HIV transmission than delivering services ad hoc 1-3. Genetic clustering approaches offer a route to rapidly identify individuals who are highly connected based on their viral sequences.
The viruses of individuals involved in recent transmission events, and viruses from known outbreaks, are closely related genetically 4,5. These transmission chains form clusters within genetic transmission networks. Interventions in Vancouver, Canada, illustrated the potential for public health departments to prioritize clusters for interventions to reduce onward transmission 6. A retrospective analysis of U.S. data determined that the transmission rate within clusters was eight times higher than the national average 7. However, clustering approaches have been criticized for their potential bias towards selecting densely sampled sub-populations 8,9. Another potential problem is that existing clusters could reflect past transmission without predicting future transmission. Thus, recent analyses have focused on identifying growing clusters within transmission networks 7,10-12. Growing clusters are those to which newly diagnosed cases are linked over time. We emphasize that this growth does not necessarily indicate onward transmission. New cases may reflect incident infections or the new diagnosis of previously infected prevalent infections. Nonetheless, growing clusters within national cohorts comprise individuals with high transmission rates 7 and better predict future growth 11 than clustering alone and may also point to groups where undiagnosed cases may be found.
Multiple measures of cluster growth have been developed, but the predictive abilities of these measures have not been systematically compared. In New York City, estimated monthly cluster growth over an 8-year period indicated that relative cluster growth was a better predictor of future growth than any other measure 11. The Centers for Disease Control and Prevention (CDC) classifies clusters as priority based on recent formation and the number of new diagnoses joining the cluster within the previous year 13. In this paper we compare these and other prioritization schemes to determine which best predicted future cluster growth using Illinois HIV genotypes collected as part of routine public health surveillance activities since 2012 14.
3. Methods
3.1. Data
HIV-1 protease and reverse transcriptase (pol) genetic sequences generated for antiretroviral resistance testing for individuals diagnosed with or receiving treatment for HIV in the state of Illinois have been routinely reported to Illinois Department of Public Health since 2012. Illinois is the sixth most populous state in the United States as of 2018 15, and has 7,000 people living with HIV.
For each case with a sequence reported to surveillance, we considered additional data, including race/ethnicity (American Indian/Alaska Native, Asian/Pacific Islander, Black/African-American, Hispanic/Latino, White, or mixed race), sex assigned at birth, transmission risk factor (men who have sex with men [MSM], people who inject drugs [PWID], MSM/PWID, heterosexual, perinatal, other, unknown), age at diagnosis, and date of diagnosis. Age at diagnosis was stratified into a categorical variable (0-12, 13-19, 20-29, 30-39, 40-49, 50-59, 60+).
At the time of analysis, genotypes were only available for individuals diagnosed up until June 2018, making 2017 our most recent complete year. Completeness of sequence reporting (the proportion of diagnoses with a genotype) has improved dramatically over time, and for the past 5 years has reached 48.0% for the state of Illinois. Previous to 2014, data available were insufficient to make inferences as older sequences were much less likely to cluster.
This study was approved by the Institutional Review Boards of the Northwestern University Feinberg School of Medicine and the University of California, San Diego with a human subjects exemption. The data analysed here were collected as part of routine HIV surveillance activities and are protected by local statute. The data cannot be submitted to public databases.
3.2. Transmission network construction
A molecular transmission network was constructed from genetic sequences using HIV-TRACE 16. HIV pol sequences were aligned to an HXB2 reference sequence and pairwise genetic distances were calculated under the Tamura Nei 93 model 17. Each individual is represented by a node in the network, and nodes were linked to each other if their pairwise distance was below a pre-specified threshold. Distances between ambiguous nucleotides were resolved (i.e., Y is 0 substitutions from both C and T) when the fraction of ambiguities across an entire sequence was ≤1.5%; when the fraction of ambiguities was >1.5%, distances from ambiguities were averaged (i.e., Y is 0.5 substitutions from both C and T). Drug resistance sites were not excluded from alignments as these have been demonstrated not to affect transmission reconstruction 18. Nodes linked to at least one other node are considered clustered in the transmission network. As described below, different genetic distance thresholds for reconstructing the network were utilized.
3.3. prioritization schemes for HIV interventions
We analyzed the molecular transmission network to find prioritization schemes capable of identifying clusters most likely to give rise to genetically-linked newly diagnosed cases in the subsequent 12 months.
Ten prioritization schemes were evaluated (Table 1) based on clustering in the network at various genetic distance thresholds (c1.5%, c1%, c0.5%), relative growth of clusters inferred at various genetic distance thresholds (g1.5%, g1%, g0.5%) 11, and clusters with recent and rapid growth as defined by the CDC 7. The original CDC definition for priority clusters with recent and rapid growth isbased on a network restricted to individuals diagnosed within the three most recent years, linked at 0.5%, and having at least five individuals in that cluster diagnosed in the past year (RR5 in Table 1). Here we extended this definition to include clusters with ≥4 (RR4), ≥3 (RR3), and ≥2 cases (RR2) diagnosed within the previous 12 months.
Table 1:
Cluster definitions for prioritization schemes
| Prioritization Scheme |
Scheme number |
Clustering method name |
Maximum genetic distance between nodes in cluster |
Nodes included in network |
Cluster definition |
|---|---|---|---|---|---|
| Clustering | 1 | c1.5%# | 1.5% | All | Linked at threshold |
| 2 | c1% | 1% | All | Linked at threshold | |
| 3 | c0.5% | 0.5% | All | Linked at threshold | |
| Relative Cluster Growth | 4 | g1.5%$ | 1.5% | All | Linked at threshold, and high growth* |
| 5 | g1% | 1% | All | Linked at threshold, and high growth* | |
| 6 | g0.5% | 0.5% | All | Linked at threshold, and high growth* | |
| Recent and Rapid Growth | 7 | RR5+ | 0.5% | Diagnosed in previous 3 years | Linked and ≥ 5 cases diagnosed in the previous year |
| 8 | RR4 | 0.5% | Diagnosed in previous 3 years | Linked and ≥ 4 cases diagnosed in the previous year | |
| 9 | RR3 | 0.5% | Diagnosed in previous 3 years | Linked and ≥3 cases diagnosed in the previous year | |
| 10 | RR2 | 0.5% | Diagnosed in previous 3 years | Linked and ≥2 cases diagnosed in the previous year |
c: clustered
g: growth
RR: recent and rapid.
We calculated growth based on the equation below, then ranked clusters based on their growth and selected those with the highest growth.
Relative cluster growth was calculated as the number of new individuals diagnosed with HIV that joined the cluster in the most recent year relative to the square root of cluster size at the time of prioritization 11. For example, for any given cluster in 2015, its relative growth would be calculated as:
Where N2015 is the number of individuals in the cluster in 2015 and ΔN2014/2015 is the number of new individuals who joined the cluster between 2014 and 2015.
We analyzed the years 2014-2016, each year making predictions for the following year. For each year, we selected clusters that met each prioritization scheme definition and reconstructed the network the following year to count the number of linked new diagnoses. For all prioritization schemes, linked new diagnoses were defined as those that linked to at least one sequence in the original cluster at a 0.5% genetic distance. The number of new diagnoses was then corrected for cluster size, so that a new case linked to a cluster of size 2 would count as 0.5 cases per prioritized case. For the prioritization schemes based on high cluster growth (4-6 in Table 1), the growth of each cluster was calculated as explained above, then clusters were sorted by growth. Individuals from the highest growth clusters were added to the priority group, one entire cluster at a time, until the total number of prioritized individuals met or exceeded 150. We chose 150 because this was the number of individuals selected under the RR3 cluster growth definition so as to facilitate downstream comparisons. As a sensitivity analysis, we repeated analyses selecting 250 individuals, the number selected under the RR2 scheme.
3.4. CORRELATES OF CLUSTER GROWTH
Multivariable logistic regression was used to examine demographic characteristics associated with cluster membership for each of the schemes: age at diagnosis, race/ethnicity and sex-specific transmission groups. The aim of this analysis was to explain, rather than predict, cluster membership, and all predictor variables are independently associated with our outcome variable (cluster membership).
4. Results
4.1. Data
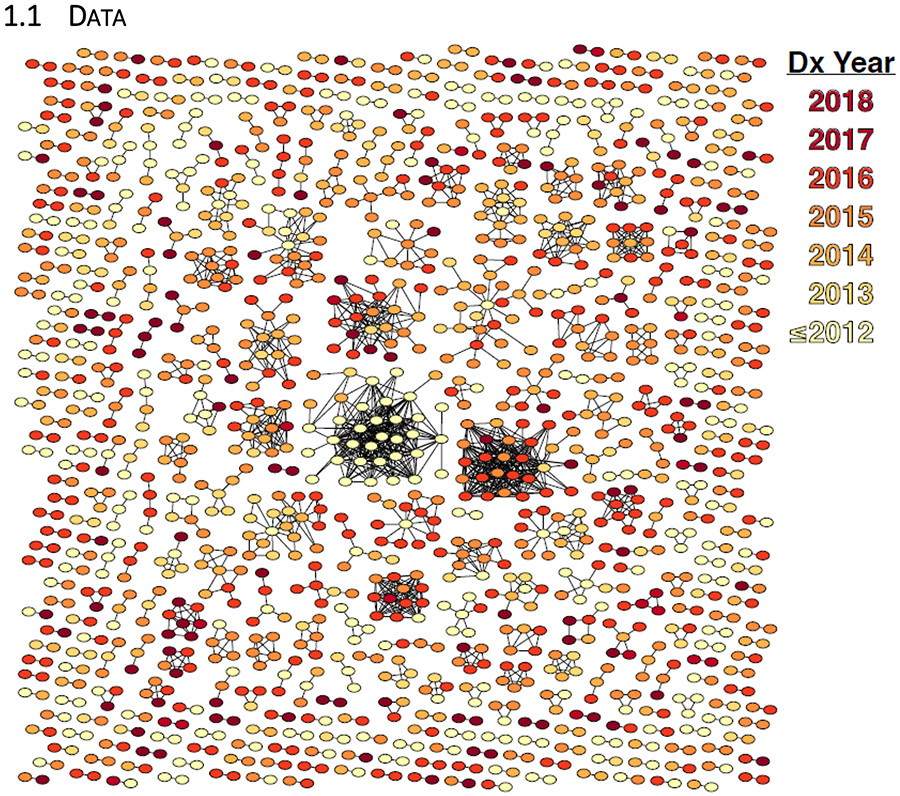
HIV sequences from 9,500 unique individuals diagnosed between 1980 through June 2018 were reported to Illinois Department of Public Health and were included in our analysis. Using a genetic distance of 0.5% to reconstruct the network, 1,405/9,500 (14.8%) individuals were linked to at least one other in the network (Figure 1). At the time of analysis, 7,846 individuals with a reported sequence were diagnosed prior to the end of 2015, 8,874 prior to the end of 2016, and 9,374 prior to the end of 2017.
Figure 1:
HIV-1 molecular transmission clusters in Illinois. Individuals are represented by a node in the network if they were linked to at least one other person at a genetic distance ≤0.5%.
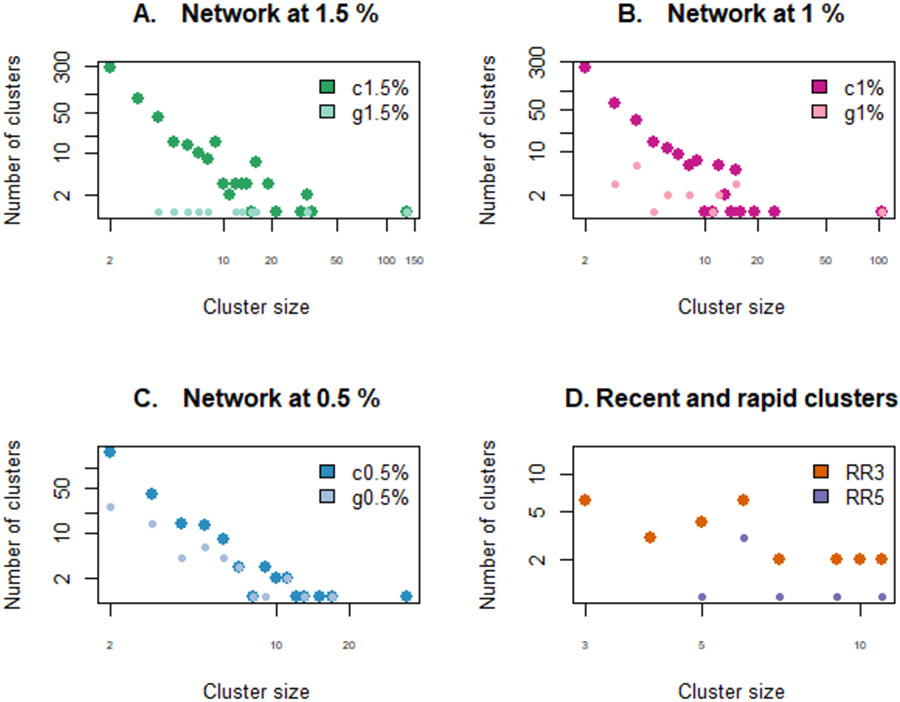
Our aim was to identify the prioritization scheme most likely to select clusters that would continue to grow the following year. For each of the prioritization schemes, we selected clusters in 2014, 2015 and 2016 that met that definition of clustering or cluster growth. Cluster size distributions under each scheme for year 2015 are shown in Figure 2. We then reconstructed the network the following year (2015, 2016, and 2017) and counted the number of newly diagnosed cases genetically linked to clusters selected under each prioritization scheme, allowing us to calculate the percent increase for each cluster (Figure 3; see Sup Fig 1 for 2016/2017).
Figure 2:
Cluster size distribution for each prioritization scheme in year 2015. All axes are log scales. (A.) Clusters with high growth at 1.5% (g1.5%) are a subset of clusters at 1.5% (c1.5%), and the same holds for (B.) 1% and (C.) 0.5%. RR5 clusters are a subset of RR4 clusters, themselves a subset of RR3 clusters, themselves a subset of RR2 clusters. Different distributions are shown with different sized characters for visibility of overlapping data points. Cluster prioritization schemes are defined in Table 1.
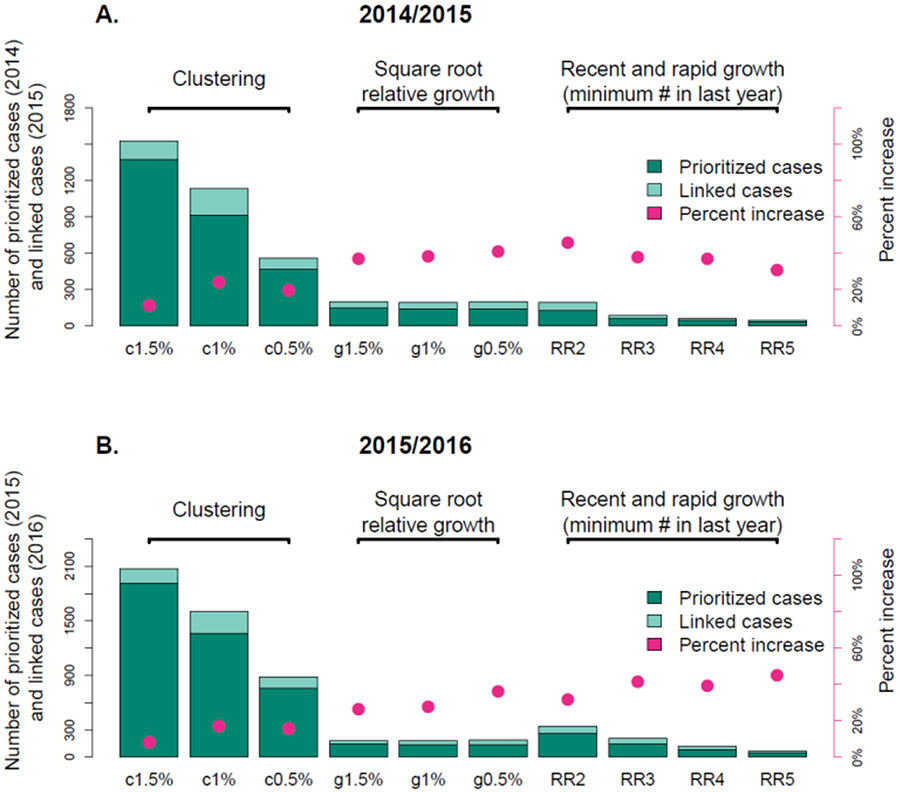
Figure 3:
Number of prioritized (dark green) and linked (pale green) cases and percent increase (pink) for each cluster prioritization scheme for years (A.) 2014/2015 and (B.) 2015/2016. Note that the number of individuals prioritized in the cluster growth schemes is determined by the number of individuals in category RR3 (150). Cluster prioritization schemes are defined in Table 1.
4.2. Percent increase for each prioritization scheme
The number of individuals selected under the clustering schemes (1-3) was substantially larger than for the recent and rapid schemes (7-10; Figure 3). The number of individuals selected by the relative growth schemes (4-6) was set according to the number of individuals selected by the recent and rapid schemes (specifically RR3, although as a sensitivity analysis we also used RR2). Cluster growth schemes all returned much higher percent increases than clustering schemes (e.g. in 2015/2016, 34.0% increase per prioritized individual on average, compared to 12.8%), and the number of individuals prioritized was dramatically smaller. This result was consistent across years (Figure 3; see Sup Fig 1 for 2016/2017), although the specific scheme with the highest percent increase varied across years. Our result was unaffected by the selected number of prioritized nodes (150 in Figure 3, 250 in Sup Fig 2). However, no single relative growth nor recent and rapid scheme consistently had a greater percent increase across years. Within the relative growth scheme, no genetic distance threshold performed consistently better than another. Among the recent and rapid definitions, the minimum number of recent diagnoses (2 – 5) did not consistently affect percent increase, indicating that percent increase was high even among clusters with only two new diagnoses in the previous year. Nonetheless, the number of individuals that would be prioritized varied widely across the different schemes. For example, the number of individuals to be prioritized under the RR5 definition was five times smaller than the number prioritized under the RR2 definition (Figure 3, Table 2).
Table 2:
Adjusted Odds Ratios from logistic regression models comparing individuals selected under each prioritization scheme in 2015 to those in clusters at 1.5% (2101 individuals, 517 clusters, 8.4 percent increase).
| Clustering | Relative Growth | Recent and Rapid Growth | |||||||
|---|---|---|---|---|---|---|---|---|---|
| c1% | c0.5% | g1.5% | g1% | g0.5% | RR2 | RR3 | RR4 | RR5 | |
| Number of priority clusters | 407 | 269 | 3 | 5 | 24 | 71 | 27 | 14 | 7 |
| Number of cases to be prioritized | 1499 | 838 | 158 | 154 | 152 | 283 | 161 | 94 | 51 |
| Percent increase | 17.2 | 15.8 | 26.6 | 27.9 | 36.2 | 31.8 | 41.6 | 39.4 | 45.1 |
| AGE AT DIAGNOSIS | |||||||||
| 14 - 19 | 4.1*** | 2.85*** | 2.22** | 3.02** | |||||
| 20 - 24 | 2.8*** | 2.04** | 1.64* | ||||||
| 25 - 29 | REF | REF | REF | REF | REF | REF | REF | REF | REF |
| 30 - 39 | |||||||||
| 40 - 49 | |||||||||
| 50 - 100 | 0.57* | ||||||||
| RACE/ETHNICITY | |||||||||
| African American | 0.62** | 0.55* | 0.6** | 0.6* | |||||
| Latino | 0.52* | 0.09** | |||||||
| White | REF | REF | REF | REF | REF | REF | REF | REF | REF |
| Unknown | |||||||||
| SEX/RISK GROUP | |||||||||
| MSM | 1.51* | 6.43** | 2.16* | 4.06* | 7.6* | ||||
| Female heterosexual | REF | REF | REF | REF | REF | REF | REF | REF | REF |
| Female PWID | 0.28* | ||||||||
| Female unknown risk | |||||||||
| Male heterosexual | |||||||||
| Male PWID | |||||||||
| MSM/PWID | 6.84* | ||||||||
| Male unknown risk | |||||||||
Cluster prioritization schemes are defined in Table 1. Results are shown only if they were significant (p<0.05).
p<0.05
p<0.01
p<0.001. MSM: men who have sex with men, PWID: people who inject drugs.
4.3. Demographic correlates of prioritized groups
We compared the demographic characteristics of individuals in every category of prioritized clusters (schemes 2 to 10 in Table 1; Sup Fig 2-12) to those of individuals clustering at 1.5% (c1.5%, n=2,101) who were not part of that group using logistic regression. Demographic characteristics varied across schemes, with a tendency for relative growth and recent and rapid clusters (schemes 4-10) to comprise more MSM, younger individuals and fewer African Americans than individuals clustering at 1.5% (Table 2).
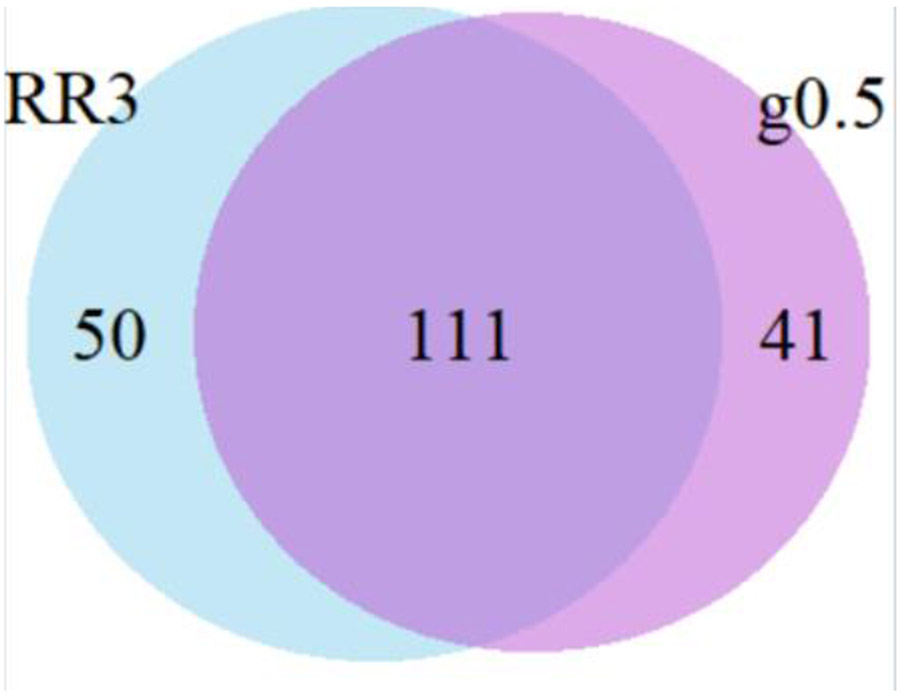
We noted that the demographics and transmission risk of individuals in each category of cluster differed and therefore examined to what extent individuals selected under each scheme overlapped. RR5 are a subset of RR4 individuals, themselves a subset of RR3, themselves a subset of RR2. Because the number of individuals selected under the growth models was based on the number of individuals in the RR3 category, and because the RR3 clustered individuals are linked at 0.5%, we examined the overlap between the RR3 and g0.5% groups (Figure 4). All members of a cluster are always picked together, so the number of individuals selected under each scheme varies: in RR3 there were 161 and in the g0.5% group, there were 152. In addition, the RR3 group includes only diagnoses within the previous three years.
Figure 4:
Overlap between individuals selected under two prioritization schemes: clusters with high growth at 0.5% (g0.5%) and recent and rapid clusters with at least 3 diagnoses in the previous year (RR3). Note that the number of individuals selected under the growth schemes was determined based on the number of individuals in the RR3 group (~150), but the RR3 group can only include individuals diagnosed within the previous three years.
We questioned whether the individuals selected by both the RR3 and the g0.5% schemes might be in the fastest growing clusters. We identified the 111 individuals prioritized under both the RR3 and the g0.5% schemes, isolated the clusters comprising those individuals and calculated percent increase for those clusters. We found no evidence that these individuals were associated with the most growth; clusters selected by both schemes grew by 38.5%, compared with 41.6% for RR3 and 36.2% for g0.5%.
5. Discussion
With limited public health resources, it is important to direct public health response towards individuals most associated with active HIV transmission. By investigating the growth of the HIV genetic transmission network in Illinois over a three-year period, we found that clusters exhibiting recent growth would identify nearly three times more cases the following year than other clusters. These results were consistent across the years considered in the analysis. The specific prioritization scheme measuring recent growth varied, and the characteristics of the individuals selected by each scheme varied, but all schemes that considered clusters with a high growth rate in the previous year predicted high growth the following year.
We hypothesized that the clusters comprising individuals selected under both the growth and the recent and rapid definitions might display the highest growth of all, but this turned out not to be the case: their growth was equivalent to that under each of the respective cluster growth definitions. We used square root relative cluster growth because it was previously demonstrated to be more predictive of future growth than cluster size, relative growth, or absolute growth11. The CDC recent and rapid definitions select sequences separated by genetic distances ≤0.5%, reflecting short transmission intervals (rapid transmission) and clusters with growth within the last year (recent cluster growth) 7. Other than the difference in the method for calculating growth, the schemes differ in that the CDC enforces a three-year cut-off on diagnosis dates to focus on recent transmissions. Our results indicate that prioritizing clusters with rapid growth is crucial, but the exclusion of diagnoses more than three years prior was inconsequential in our dataset.
As we did not find substantial differences in percent increase between the recent and rapid cluster definitions with varying minimum numbers of diagnoses in the previous year (2-5 diagnoses), the scheme chosen should depend on resource availability to respond to the number individuals in clusters resulting from the particular prioritization scheme. The number of individuals prioritized under the RR5 definition was five times smaller than the number prioritized under the RR2 definition. Thus, in places where resource limits preclude every single person receiving a timely partner elicitation interview, we suggest that public health departments might benefit from creating an ordered list of individuals for service prioritization based on recent cluster growth. Different thresholds may be more appropriate in rural versus urban thresholds, to take into account population density and local transmission dynamics.
We did not find meaningful differences in the percent increase between the cluster growth schemes at 0.5%, 1% and 1.5%, but there were stark differences in the number of clusters that would be prioritized under each of those schemes. Overall, our results support the hypothesis that the network at 0.5% is more informative in terms of recent and ongoing transmission19, and we would encourage public health responses to be directed towards clusters defined at this threshold – especially as it will be possible to target more of them. But even clusters defined at 1.5% displaying growth within the previous year are associated with ongoing diagnosis, in agreement with previous results10. Therefore, public health departments could consider conducting retrospective analyses comparing cluster growth schemes on their data, to establish the scheme best suited to their local epidemic. As such analyses become more common, it may be possible to develop schemes that are more widely generalizable. Although cluster growth in our analysis was calculated year-on year, for maximum impact, cluster-guided prioritization should be conducted in near real-time, and monthly reviews are a realistic objective6,11.
The demographic characteristics of high growth clusters were not as distinct as in a national analysis of molecular surveillance data7. In that analysis, recent and rapid clusters were more likely to include young Latino MSM. In our analysis, young MSM made up the majority of cases in high growth clusters, but this result was not consistently significant across prioritization schemes. The finding that high growth clusters could not be characterized demographically highlights the importance of the cluster growth schemes for prioritizing individuals that may not be identified from standard epidemiological/ demographic analyses of routine HIV surveillance data. Their independence from patient demographic traits may be a strength of cluster-guided approaches, meaning that they can capture at-risk populations who would not be prioritized based on demographic makeup alone. Concordantly, the New York study concluded that demographic characteristics were far less predictive of future growth than were past cluster growth dynamics 11.
The impact of cluster-guided prioritization will depend on the local epidemic. In places where individuals are diagnosed with HIV at a later stage of infection, the aim of such a strategy will be to decrease diagnosis delays. To account for the delays between infection and diagnosis, less conservative genetic thresholds may need to be applied. In places where diagnosis delays are already minimal, cluster-guided interventions may have the potential to prevent transmission by flagging recently infected cases before they go on to transmit3,20.
Our ability to distinguish between schemes may have been affected by small sample size and/or genotype reporting completeness. For example, a lack of completeness due to delays in reporting undermined our ability to predict cluster growth in 2017; and the best model selected varied slightly across the years examined. Independent analyses have shown that fewer clusters of concern are detected when sequence data completeness is lower, but that inferences made remain meaningful21. Our results are consistent with those from analyses of larger datasets7, thus it is possible that both sets of prioritization schemes perform equally well. Nonetheless, populations that are systematically less likely to be linked to care will not have a genotype and thus will not be identified by cluster-guided prioritization schemes. Improving sequence data completeness is a priority for the state of Illinois and analyses should be repeated.
Cluster-guided approaches are agnostic to the intricacies of disease transmission dynamics. As explained above, one benefit is that they consequently do not depend on pre-classification of subpopulations as being at-risk. However, they can only detect transmission that has occurred and cannot make predictions about future growth that might result from a shift in disease dynamics. Finally, we stress that it is essential that HIV programs continue to focus on inclusion and reducing inequities alongside cluster-guided approaches. Providing public health interventions to marginalized populations who are disproportionately affected is central to ending the HIV epidemic.
Rapid, simple, and automated tools to identify growing clusters are essential to assist health departments in developing a comprehensive, targeted response where there is active transmission. The growth-based prioritization schemes presented here are straightforward to communicate and easy to implement by public health departments, can be calculated on large surveillance datasets with tens of thousands of sequences, and can be automated within HIV-TRACE or similar frameworks16,22. Even more importantly, cluster-guided prioritization has the potential to better identify people in need of services to reduce new infections towards the goal of ending the HIV epidemic in the United States 23,24.
Supplementary Material
Supplementary Figure 1: Number of prioritized and linked cases and percent increase for each cluster prioritization scheme for years 2016/2017. Note that the number of individuals prioritized in the cluster growth schemes is determined by the number of individuals in category RR3 (n≈150). Cluster prioritization schemes are defined in Table 1.
Supplementary Figure 2: Sensitivity Analysis. Number of prioritized and linked cases and percent increase for each cluster prioritization scheme for years (A.) 2014/2015, (B.) 2015/2016 and (C.) 2015/2016. The number of individuals prioritized in the cluster growth schemes is determined by the number of individuals in category RR2 (n≈250), instead of in RR3 (n≈150). Cluster prioritization schemes are defined in Table 1.
Supplementary Figures 3-12: Genetic networks for the year 2015 under every cluster definition in Table 1, showing the breakdown by A. Race/ethnicity, B. Transmission risk group, C. Age group and D. Date of diagnosis. The year 2015 was chosen for consistency with Table 2.
Conflicts of Interest and Source of Funding
JOW received funding from Gilead Sciences, LLC as a grant paid to his institution. All other authors declare no competing interests.
This work was supported by an administrative supplement to an NIH-NIAID P30 AI117943 award; an NIH-NIAID K01 Career Development Award (K01AI110181), and an NIH-NIAID R01 (AI135992). MRC is jointly funded by the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreement and this award is part of the EDCTP2 programme supported by the European Union (MR/R015600/1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Meetings at which parts of the data were presented
Conference on Retroviruses and Opportunistic Infections; March 4-7, 2018; Boston, MA, USA.
6. Data availability
The datasets analysed during the current study are not available. The data analysed here were collected as part of routine HIV surveillance activities and are protected by local statute. The data cannot be submitted to public databases.
7 References
- 1.Jones JH, Handcock MS. An assessment of preferential attachment as a mechanism for human sexual network formation. Proceedings of the Royal Society B-Biological Sciences. 2003;270(1520):1123–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman ME. Spread of epidemic disease on networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2002;66(1 Pt 2):016128. [DOI] [PubMed] [Google Scholar]
- 3.Little SJ, Kosakovsky Pond SL, Anderson CM, et al. Using HIV networks to inform real time prevention interventions. PLoS One. 2014;9(6):e98443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peters PJ, Pontones P, Hoover KW, et al. HIV Infection Linked to Injection Use of Oxymorphone in Indiana, 2014-2015. N Engl J Med. 2016;375(3):229–239. [DOI] [PubMed] [Google Scholar]
- 5.Ragonnet-Cronin M, Jackson C, Bradley-Stewart A, et al. Recent and Rapid Transmission of HIV Among People Who Inject Drugs in Scotland Revealed Through Phylogenetic Analysis. J Infect Dis. 2018;217(12):1875–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poon AF, Gustafson R, Daly P, et al. Near real-time monitoring of HIV transmission hotspots from routine HIV genotyping: an implementation case study. Lancet HIV. 2016;3(5):e231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oster AM, France AM, Panneer N, et al. Identifying Clusters of Recent and Rapid HIV Transmission Through Analysis of Molecular Surveillance Data. J Acquir Immune Defic Syndr. 2018;79(5):543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poon AF. Impacts and shortcomings of genetic clustering methods for infectious disease outbreaks. Virus Evol. 2016;2(2):vew031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dearlove BL, Xiang F, Frost SDW. Biased phylodynamic inferences from analysing clusters of viral sequences. Virus Evol. 2017;3(2):vex020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Billock RM, Powers KA, Pasquale DK, et al. Prediction of HIV Transmission Cluster Growth With Statewide Surveillance Data. J Acquir Immune Defic Syndr. 2019;80(2):152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wertheim JO, Murrell B, Mehta SR, et al. Growth of HIV-1 Molecular Transmission Clusters in New York City. J Infect Dis. 2018;218(12):1943–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chato C, Kalish ML, Poon AFY. Public health in genetic spaces: a statistical framework to optimize cluster-based outbreak detection. Virus Evol. 2020;6(1):veaa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. Detecting and Responding to HIV Transmission Clusters: a guide for public health departments. 2018. [Google Scholar]
- 14.Illinois Department of Public Health. Illinois Integrated HIV Prevention and Care Plan 2017–2021: A Roadmap for Collective Action in Illinois. Office of Health Protection, HIV/AIDS Section;2016. [Google Scholar]
- 15.U.S. Census Bureau Population Division. Annual Estimates of the Resident Population: April 1, 2010 to July 1, 2018. 2018. [Google Scholar]
- 16.Kosakovsky Pond SL, Weaver S, Leigh Brown AJ, Wertheim JO. HIV-TRACE (TRAnsmission Cluster Engine): a Tool for Large Scale Molecular Epidemiology of HIV-1 and Other Rapidly Evolving Pathogens. Mol Biol Evol. 2018;35(7):1812–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–526. [DOI] [PubMed] [Google Scholar]
- 18.Hue S, Clewley JP, Cane PA, Pillay D. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. Aids. 2004;18(5):719–728. [DOI] [PubMed] [Google Scholar]
- 19.Le Vu S, Ratmann O, Delpech V, et al. Comparison of cluster-based and source-attribution methods for estimating transmission risk using large HIV sequence databases. Epidemics. 2018;23:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Little SJ, Chen T, Wang R, et al. Effective HIV Molecular Surveillance Requires Identification of Incident Cases of Infection. Clin Infect Dis. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dasgupta S, France AM, Brandt MG, et al. Estimating Effects of HIV Sequencing Data Completeness on Transmission Network Patterns and Detection of Growing HIV Transmission Clusters. AIDS Res Hum Retroviruses. 2019;35(4):368–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ragonnet-Cronin M, Hodcroft E, Hue S, et al. Automated analysis of phylogenetic clusters. BMC Bioinformatics. 2013;14:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.France AM, Oster AM. The Promise and Complexities of Detecting and Monitoring HIV Transmission Clusters. J Infect Dis. 2020;221(8):1223–1225. [DOI] [PubMed] [Google Scholar]
- 24.Fauci AS, Redfield RR, Sigounas G, Weahkee MD, Giroir BP. Ending the HIV Epidemic: A Plan for the United States. JAMA. 2019;321(9):844–845. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Number of prioritized and linked cases and percent increase for each cluster prioritization scheme for years 2016/2017. Note that the number of individuals prioritized in the cluster growth schemes is determined by the number of individuals in category RR3 (n≈150). Cluster prioritization schemes are defined in Table 1.
Supplementary Figure 2: Sensitivity Analysis. Number of prioritized and linked cases and percent increase for each cluster prioritization scheme for years (A.) 2014/2015, (B.) 2015/2016 and (C.) 2015/2016. The number of individuals prioritized in the cluster growth schemes is determined by the number of individuals in category RR2 (n≈250), instead of in RR3 (n≈150). Cluster prioritization schemes are defined in Table 1.
Supplementary Figures 3-12: Genetic networks for the year 2015 under every cluster definition in Table 1, showing the breakdown by A. Race/ethnicity, B. Transmission risk group, C. Age group and D. Date of diagnosis. The year 2015 was chosen for consistency with Table 2.
Data Availability Statement
The datasets analysed during the current study are not available. The data analysed here were collected as part of routine HIV surveillance activities and are protected by local statute. The data cannot be submitted to public databases.