Abstract
In view of the potential of traditional plant-based remedies (or phytomedicines) in the management of COVID-19, the present investigation was aimed at finding novel anti-SARS-CoV-2 molecules by in silico screening of bioactive phytochemicals (database) using computational methods and drug repurposing approach. A total of 160 compounds belonging to various phytochemical classes (flavonoids, limonoids, saponins, triterpenoids, steroids etc.) were selected (as initial hits) and screened against three specific therapeutic targets (Mpro/3CLpro, PLpro and RdRp) of SARS-CoV-2 by docking, molecular dynamics simulation and drug-likeness/ADMET studies. From our studies, six phytochemicals were identified as notable ant-SARS-CoV-2 agents (best hit molecules) with promising inhibitory effects effective against protease (Mpro and PLpro) and polymerase (RdRp) enzymes. These compounds are namely, ginsenoside Rg2, saikosaponin A, somniferine, betulinic acid, soyasapogenol C and azadirachtin A. On the basis of binding modes and dynamics studies of protein–ligand intercations, ginsenoside Rg2, saikosaponin A, somniferine were found to be the most potent (in silico) inhibitors potentially active against Mpro, PLpro and RdRp, respectively. The present investigation can be directed towards further experimental studies in order to confirm the anti-SARS-CoV-2 efficacy along with toxicities of identified phytomolecules.
Keywords: SARS-CoV-2 infection, Phytomedicine, Molecular docking, Molecular dynamics, Phytochemicals, Drug repurposing
1. Introduction
In December 2019 at the city of Wuhan, China, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has begun the outbreak of coronavirus disease-2019 (COVID-19) (Ciotti et al., 2019, Rudrapal et al., 2020a, Rudrapal et al., 2020b, Bhat, 2021). The disease has led to a catastrophic situation with unprecedented health and economic burden of current times around the world. Infection with SARS-CoV-2 causes mild illness (fever, cough, sore throat) to severe respiratory disease (Tijjani et al., 2021, Kumar et al., 2021b, Rasmi et al., 2021, Rudrapal et al., 2021). This devastating infectious disease may even cause fatal systemic complications and eventually deaths. SARS-CoV-2 has affected lives of millions of people till date. Apart from certain prophylactic measures (antiviral therapy, use of antibacterial antibiotics, corticosteroids or immunosuppressants, and convalescent plasma therapy etc.) (Guo et al., 2020, Singhal, 2020), there are no currently available FDA approved drugs and/or drug therapy that can cure the disease. The discovery of novel antiviral agents against COVID-19 is, therefore, an urgent need of the hour.
There is an extensive research going on across the globe to discover and develop therapeutic candidates that can effectively treat SARS-CoV-2 infection. Many existing drugs/drug therapy/clinical candidates (for example, hydroxychloroquine/chloroquine, hydroxychloroquine/azithromycin, favipiravir, oseltamivir (tamiflu), lopinavir/ritonavir, remdesivir/baricitinib/tocilizumab, etc. (Hassan et al., 2020, Singhal, 2020) have been repurposed, but the clinical development of these drugs/drug regimens in confirmation of their efficacy and toxicity are still under trial. Drug repositioning (DR) utilizes computational and experimental approaches to explore new clinical indications of existing drugs on a rational basis. DR increases the efficiency of drug discovery and it reduces the time as well as cost of discovery as well (Adeoye et al., 2020, Singhal, 2020). Repurposing has investigated the clinical usefulness of many existing drugs as depicted above including some of the natural products such as ivermectin, colchicine etc. (Elmezayen et al., 2020, Harapan et al., 2020) as prophylactic agents. Researchers have also been investigation the repurposing of available antiviral molecules as possible therapeutic alternatives against coronavirus infection (Caly et al., 2020, Harapan et al., 2020, Rudrapal et al., 2020a, Rudrapal et al., 2020b). Apart from repurposing FDA approved and clinical candidates, phytomedicine-derived bioactive compounds (or simply called phytochemicals such as curcumin, quercetin, epigallocatechin gallate (EGCG) and many others) (Khan and Al-Balushi, 2020, Shah et al., 2020) have also been extensively investigated in search for potential lead molecules/drug candidates against COVID-19.
Phytomedicine have been an immense repository of bioactive molecules of medicinal importance with biological/pharmacological potential in diverse therapeutic areas. For instance, natural products like quinine, taxol, artemisinin have been derived from traditionally useful plants or phytomedicine. Phytomedicine is still, therefore, believed to be a potential resource for new drug molecules/drug candidates (Cherian et al., 2020, Kouznetsova et al., 2020). Many medicinal plants and their isolated compounds have also been claimed to exhibit antiviral activities (Swargiary et al.; 2020). Plant-derived compounds (phytochemicals) belonging to several classes of phytochemicals (such as polyphenols, flavonoids, limonoids, terpenoids, saponins, steroids etc.) obtained from several phytochemical database (PubChem, Zinc, Drug Bank, IMPPAT, COCONUT, NPBS etc.) have been investigated by computational methods using in silico approaches to identify possible leads that can inhibit biological (protein/enzymatic) molecules as targets in SARS-CoV-2 (Wang et al., 2015, Swargiary et al., 2020). In recent days, in sillico screening of phytochemical database has gained tremendous interest in drug discovery research for the identification of new drugs leads. In DR approach, the combined DR strategy of in silico approach i.e., high-throughput virtual screening/structure-based drug design (SBDD) and experimental assays is successfully implemented in the discovery of drug molecules (Tiwari et al., 2010, Rajasekaran et al., 2013). The combined DR strategy of in silico approach and experimental assays offers an effective alternative platform for the development of bioactive phytochemicals as possible drug candidates. Virtual screening approaches including molecular dynamics simulation studies are usually being adopted to screen out potential drug candidates from natural products/phytochemicals libraries using various phytochemical databases (mentioned above) available in the public domain. Recent studies also report virtual screening/docking/molecular dynamics of medicinal plants/phytochemicals having traditional medicinal importance with SARS-CoV-2 protease/polymerase inhibitory potential (Gupta et al., 2020, Vardhan and Sahoo, 2020).
Inhibition of protease (chymotrypsin-like protease, 3CLpro or main protease, Mpro) (Joshi et al., 2020, Rudrapal et al., 2020a, Rudrapal et al., 2020b, Umadevi et al., 2020, Sepay et al., 2020, Chowdhury, 2020, Gowrishankar et al., 2021), and RNA-dependent RNA polymerase (RdRp) polymerase (Motiwale et al., 2020, Bhat, 2021, Naik et al., 2021, Ishola et al., 2021, Shree et al., 2020, Schoeman and Fielding, 2019) enzymes has been attributed to be crucial for blocking maturation of viral particles, viral replication and further infection. Several research related to the development of natural products as inhibitors of protease and polymerase enzymes have already been reported using computational modeling and approaches (Prajapat et al., 2020a, Prajapat et al., 2020b, Motiwale et al., 2020, Schoeman and Fielding, 2019). In view of above considerations, our investigation was carried out to evaluate the anti-SARS-CoV-2 potential of biologically active phytochemicals particularly found in traditional medicines (Ayurvedic medicines, Chinese medicines etc.) with possible inhibitory activities against Mpro, PLpro and RdRp enzymes. In this paper, a total of 160 phytochemicals belonging to various phytochemical classes were selected and virtually screened against three aforementioned therapeutic targets of SARS-CoV-2 by DR approach with an aim to find out anti-SARS-CoV-2 molecules effective against SARS-CoV-2 infection with protease and polymerase inhibitory activities.
2. Materials and methods
2.1. Phytocompounds preparation
A total of 160 biologically active plant-derived compounds (phytochemicals) with a wide range of structural diversity belonging to different phytochemical classes (polyphenols, flavonoids, limonoids, triterpenoids, saponins and steroids) were selected based on their potential medicinal/biological interests (such as antiviral, antimicrobial, antibacterial activities etc.) as reported in traditional (Ayurvedic or Chinese medicines) as well as modern phytomedicines (Hussain et al., 2021, Isyaku et al., 2020, Rolta et al., 2020). The 3D structures of compounds were downloaded from the PubChem database and saved in sdf files. Ligands were energetically minimized (2000 steps, RMSD value = 0.01 kcal/mol) using the CHARMm-based minimizer on Biovia Discovery Studio (DS 2020).
2.2. Proteins preparation
The structures of SARS-CoV-2 proteins viz., Mpro (3CLpro) with resolution 2.16 Å (6LU7) (Prajapat et al., 2020a, Prajapat et al., 2020b), PLpro with resolution 1.66 Å (6WX4) (Shree et al., 2020), and RdRp with resolution 2.50 Å (7BV2) (Ibrahim et al., 2020) were retrieved from the RCSB-PDB database. DS 2020 program was used for the preparation of proteins. The co-crystallized inhibitor, water molecules and hetero atoms were removed. The protein molecules were energetically minimized (200 steps, RMSD value = 0.1 kcal/mol) using the CHARMm minimizer.
2.3. Molecular docking
Docking was performed using Biovia DS 2020 (Biovia, USA) software. The binding site residues were selected and thereby binding spheres were generated and parameters for the receptor grid were set (Kumar et al., 2022, Ibrahim et al., 2020; (Kalita et al., 2020). The radius for the sphere of binding site was set at 12.738 Å, 10.323 Å and 8.500 Å, with x, y, z dimensions of −10.897304, 13.066857, 68.557888; 8.904486, −27.443594, −37.926085; and 95.588588, 92.112297, 105.258816 for 6LU7, 6WX4 and 7BV2, respectively.
The docking was performed using CDocker of the DS 2020 (Larini et al., 2007, Holzinger et al., 2014). The calculations of CDocker energy and CDocker energy were done. Based upon docking score, top 25 phytochemicals were selected for further study. The binding modes of docked complexes were studied using the 3D receptor-ligand poses. 2D diagrams of receptor-ligand complexes were used to study various interactions like hydrogen bonding, hydrophobic etc.
2.4. Molecular dynamics (MD) simulation
MD simulation of docked complexes was executed on DS software 2020. Compounds which exhibited best binding affinity were further investigated for MD simulation. This study validates results of docking and evaluates the conformation of protein–ligand complexes. The original structures of the target proteins with co-crystal inhibitors (6LU7, 6WX4 and 7BV2) were treated as native compounds for the MD simulation. The best pose obtained in docking for each selected ligand having the highest dock score against each target protein was used for the MD simulation (Rolta et al., 2020, Singh et al., 2020, Larini et al., 2007). Basic parameters of trajectory analyses such as root mean square deviations (RMSD), root mean square fluctuation (RMSF), radius of gyration (ROG), number of hydrogen bonds (H-bonds) were analyzed for each protein–ligand complex (Noha et al., 2017).
After cleaning, the solvation of complexes was done in a cubic water box with 10 Å × 10 Å × 10 Å in size. During solvation 0.15 M NaCl was used for the neutralization of the system. The energy minimization was then carried out for 5000 steps with a RMSD gradient of 0.01 kcal/mol (Peele et al., 2020). Once the simulation is complete, RMSD, RMSF and ROG were computed. During simulation period, analysis of hydrogen bonding was also carried out (Wang et al., 2006, Zakaryan et al., 2017).
2.5. Molecular mechanics Poisson-Boltzmann surface area (MM-PBSA)-based binding free energies calculation
To calculate the binding free energy (ΔG), the MM-PBSA is applied (Rubenstein et al., 2018, Lai et al., 2011). The binding free energies were calculated using DS software. All generated conformations were analyzed and the individual binding free energy was calculated. The average binding free energy (ΔG) was finally calculated (Genheden and Ryde, 2015, Zhu et al., 2020).
2.6. Drug-likeness and ADMET calculations
Top 15 phytochemicals which showed higher docking score or better binding affinities were selected for in silico drug-likeness and ADME-Toxicity (ADMET) calculations using DS 2020. Parameters such as log of n-octanol/water partition coefficient (LogP), molecular weight (MW), hydrogen bond acceptors (nHBAs), hydrogen bond donors (nHBDs), molecular polar surface area (PSA) and number of rotable bonds (nRotBs) incorporated in Lipinski’s rule of five (Rastelli et al., 2010) and Veber rule (Lipinski et al., 1997, Pillaiyar et al., 2020) were calculated. In ADMET prediction, parameters such as aqueous solubility, blood–brain barrier penetration, cytochrome P450 (CYP) 2D6 inhibition, hepatotoxicity, intestinal absorption and plasma protein binding) were determined (Jorgensen and Duffy, 2002, Veber et al., 2002).
3. Results
3.1. Molecular docking
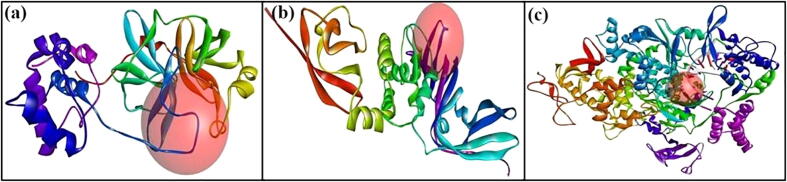
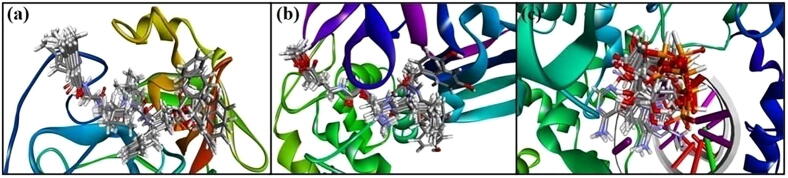
Prior to protein–ligand docking, the receptor grid models [Fig. 1(a)-(c)] of Mpro, PLpro and RdRp were generated/optimized in terms of predicting binding site spheres in order to achieve optimal molecular interactions between receptor molecules and test compounds. The validation of the protein models (6LU7/N3, 6WX4/VIR251 and 7BV2/Remdesivir) used for the docking study was done by re-docking procedure (Kousar et al., 2020). After docking, top 10 docked posed generated were superimposed within the binding site containing the original co-crystal ligand and RMSD values of docked poses were calculated using the co-crystal ligand as the reference ligand (Naik et al., 2021). Results showed superimposition of the complexes to the original structures of the protein molecules. Fig. 2 shows superimposition of co-crystal ligands (N3, VIR251 and Remdesivir) in the predicted binding site of the original structure of Mpro (6LU7), PLpro (6WX4) and RdRp (7BV2), respectively. The figure also demonstrates that the docked pose possesses binding mode similar to the co-crystal ligand.
Fig. 1.
Receptor grid models: (a) 6LU7, (b) 6WX4 and (c): 7BV2.
Fig. 2.
Redocked conformers of receptors: (a) 6LU7, (b) 6WX4, and (c) 7BV2.
Results of docking clearly reveal that almost all of the 160 phytochemicals were successfully docked into the protein molecules. Depending on the scores of CDocker and CDocker interaction energies, top 25 docked molecules were screened out and selected for the protein–ligand interaction analyses. Results of top 25 dock scored phytochemicals against the three target proteins are summarized in Table 1. A majority of phytochemicals exhibited binding affinities against all the three target proteins. These 25 phytochemicals were identified as active inhibitors (best hit molecules) of both protease (Mpro and PLpro) and polymerase (RdRp) enzymes. Out of 25 compounds, top three dock scored phytochemicals were selected as the best inhibitors against their respective protein molecule. Detailed explanation about the selected candidates protease or polymerase inhibitors are as follows:
Table 1.
Binding energies of top 25 dock scored compounds.
| Sl. No. | Name of compound (PubChem ID) | Binding energies (kcalmol−1) |
|||||
|---|---|---|---|---|---|---|---|
| 6LU7 |
6WX4 |
7BV2 |
|||||
| CDocker energy (–) | CDocker interaction energy (–) | CDocker energy (–) | CDocker interaction energy (–) | CDocker energy (–) | CDocker interaction energy (–) | ||
| 1 | Azadirachtin A (5281303) | 104.956 | 38.6349 | – | – | 100.087 | 68.8895 |
| 2 | Arachidic acid (10467) | 43.7174 | 42.0866 | 44.106 | 46.6142 | 46.7756 | 53.6177 |
| 3 | β-Amyrin (73145) | 60.3082 | 31.7661 | 65.1977 | 26.8483 | 57.1626 | 36.6992 |
| 4 | Andrographolide (5318517) | 43.3188 | 35.7343 | 42.1422 | 35.4204 | 32.0387 | 47.0894 |
| 5 | β-Sitosterol (222284) | 37.2309 | 34.7529 | 46.0316 | 25.772 | 27.6764 | 51.4468 |
| 6 | Betulinic acid (64971) | 79.5416 | 35.2337 | 87.0782 | 25.1481 | 72.1015 | 44.9878 |
| 7 | Betulinaldehyde (99615) | 81.7462 | 32.733 | −86.1886 | 25.5429 | 65.1237 | 51.4551 |
| 8 | Chebulagic acid (442674) | 54.1232 | 47.4416 | – | – | 48.8295 | 70.7162 |
| 9 | Curcumin (969516) | 14.7075 | 18.4052 | 28.1422 | 41.9514 | 38.3145 | 48.2425 |
| 10 | Diosgenin (99474) | 51.9098 | 36.9906 | −53.657 | 34.012 | 38.6811 | 48.9045 |
| 11 | EGCG (65064) | 43.7174 | 42.0866 | 47.5688 | 47.6269 | 59.0082 | 59.3958 |
| 12 | Glycyrrhetic acid (10114) | 45.2717 | 35.4892 | −53.2921 | 26.9553 | 35.8437 | 48.3936 |
| 13 | Ginsenoside Rg2 (75412551) | 136.969 | 38.2382 | – | – | – | – |
| 14 | Gingerol (442793) | 36.2828 | 38.7509 | 36.68 | 44.4235 | 40.7771 | 43.7796 |
| 15 | Nimbolide (100017) | 53.4119 | 44.6792 | 65.028 | 31.4749 | −45.2656 | 52.269 |
| 16 | Oleanolic acid (10494) | 63.1905 | 30.5139 | 64.5524 | 29.168 | −52.5498 | 41.367 |
| 17 | Ricinoleic acid (643684) | 31.6222 | 45.425 | 27.4482 | 52.2438 | 32.9493 | 43.1878 |
| 18 | Saikosaponin A (167928) | 115.685 | 48.6531 | 252.934 | 14.6437 | – | – |
| 19 | Salannin (6437066) | 77.1071 | 41.024 | – | – | 68.0723 | 59.5693 |
| 20 | β-Sitosterol (222284) | 34.8653 | 38.5059 | 43.3334 | 28.4243 | 28.3424 | 48.2521 |
| 21 | Soyasapogenol C (3083637) | 70.2491 | 34.214 | 77.4819 | 26.2634 | 63.1893 | 45.4958 |
| 22 | Sergeolide (134025) | 58.5221 | 41.0786 | 59.5372 | 42.1074 | 52.1866 | 50.3068 |
| 23 | Somniferine (14106343) | 189.968 | 45.0046 | – | – | 187.218 | 58.4079 |
| 24 | Ursolic acid (64945) | 57.0124 | 33.1967 | 62.7633 | 26.6306 | 46.1564 | 45.1468 |
| 25 | Withaferin A (265237) | 20.209 | 18.1592 | 17.3026 | 37.8881 | 33.248 | 57.2572 |
| 26 | Co-crystal inhibitor | 95.5007 | 81.2693 | 75.0382 | 62.4873 | 29.2231 | 63.6939 |
‘–‘: Not docked. 6LU7: 3CLpro or Mpro/N3; 6WX4: PLpro/VIR251; 7BV2: RdRp/Remdesivir.
3.1.1. Inhibitors for Mpro (3CLpro)
The Mpro (3CLpro) is a cysteine protease viral enzyme. The catalytic dyad i.e., His41-Cys148 is responsible for the proteolytic activity of Mpro (Chowdhury, 2020). From the docking study, the three phytochemicals which exhibited very good binding affinity against Mpro are namely, ginsenoside Rg2, saikosaponin A and somniferine. They exhibited better binding affinities against Mpro over PLpro than rest of the compounds. These three compounds interacted strongly with the Mpro through the formation of stable protein–ligand complexes. Analysis of docking interactions reveals that compounds interacted the Mpro predominantly by hydrogen bonding interactions. Ginsenoside Rg2 interacted strongly with the Mpro with a CDocker energy of −136.969 kcalmol−1 and a CDocker interaction energy of −38.2382 kcalmol−1. Saikosaponin A also interacted prominently with the Mpro with a CDocker energy of −115.685 kcalmol−1 and a CDocker interaction energy of −48.6531 kcalmol−1. Somniferine also interacted strongly with the Mpro with a CDocker energy of −189.968 kcalmol−1 and a CDocker interaction energy of −45.0046 kcalmol−1. Ginsenoside binds with the active site residues (Thr25, His41, Gly143, Gly143, Ser144, Ser144, Gly146, Glu166, Glu166, Thr 190 and Gln192) of Mpro through the formation of 11 conventional H-bonds. The interaction between saikosaponin A and Mpro was mediated to some extent through conventional H-bonds with Thr26 and Gln189. Somniferine formed four H-bonds with His163, His164, Glu166 and Gln189 of Mpro. The co-crystal ligand (N3) showed better binding affinity with a CDocker energy of −95.5007 kcalmol−1 and a CDocker interaction energy of −81.2693 kcalmol−1. It also interacted strongly with the Mpro molecule with the formation of five H bonds. Hydrogen bonds were formed with amino acid residues like Thr26, Asn142, Glu166, Gln189 and Ala191. The extent of interaction of co-crystal ligand with the Mpro enzyme was comparatively less than that of phytochemical inhibitors. Docking interaction diagrams 2D of ginsenoside Rg2 along with their co-crystal inhibitor, N9 showing the participation of active site amino acids of Mpro are presented in Fig. 7 (a, b). Details of hydrogen bonding interactions are given in Table 2 (a).
Fig. 7.
Different non-bond interactions formed for various docked complexes after the simulation period: (a) Mpro-ginsenoside Rg2, (b) Mpro-N3, (c) PLpro-saikosaponin A, (d) PLpro-VIR251, (e) RdRp-somniferine, and (f) RdRp-Remdesivir.
Table 2.
Hydrogen bonding details for top three dock scored compounds.
| Compound | H-bond(s) | Ligand |
Receptor |
H-bond distance (Å) | |||
|---|---|---|---|---|---|---|---|
| Element | Type | Residue | Element | Type | |||
| (a) Interactions with Mpro | |||||||
| Ginsenoside Rg2 | 11 | O | A | Thr25 | H | D | 2.09931 |
| H | D | His41 | O | A | 2.96313 | ||
| O | A | Gly143 | H | D | 2.45716 | ||
| O | A | Gly143 | H | D | 2.56430 | ||
| H | D | Ser144 | O | A | 2.23076 | ||
| O | A | Ser144 | H | D | 2.49087 | ||
| O | A | Gly146 | H | D | 2.12785 | ||
| H | D | Glu166 | O | A | 2.56784 | ||
| H | D | Glu166 | O | A | 2.90532 | ||
| H | D | Thr190 | O | A | 2.45679 | ||
| H | D | Gln192 | O | A | 2.5623 | ||
| Saikosaponin A | 2 | H | D | Thr26 | O | A | 2.30434 |
| H | D | Gln189 | O | A | 2.68592 | ||
| Somniferine | 4 | O | A | His163 | H | D | 2.30379 |
| O | A | His164 | H | D | 2.87922 | ||
| H | D | Glu166 | O | A | 1.92774 | ||
| H | D | Gln189 | O | A | 2.12622 | ||
| N3 | 5 | O | A | Thr26 | H | D | 2.35774 |
| H | D | Asn142 | O | A | 2.92344 | ||
| H | D | Glu166 | O | A | 2.01679 | ||
| H | D | Gln189 | O | A | 2.01203 | ||
| O | A | Ala191 | H | D | 2.14907 | ||
| Compound | H-bond(s) | Ligand |
Receptor |
H-bond distance (Å) | |||
|---|---|---|---|---|---|---|---|
| Element | Type | Residue | Element | Type | |||
| (b) Interactions with PLpro | |||||||
| Betulinic acid | 0 | – | – | – | – | – | – |
| Saikosaponin A | 8 | O | A | Asn109 | H | D | 2.33903 |
| H | D | Asn109 | O | A | 2.56439 | ||
| O | A | Tyr112 | H | D | 2.05147 | ||
| H | D | Gln269 | O | A | 2.00272 | ||
| H | D | Gln269 | O | A | 1.88363 | ||
| O | A | Gln269 | H | D | 1.56782 | ||
| O | A | Gln269 | H | D | 1.23452 | ||
| H | D | Tyr273 | O | A | 2.34567 | ||
| Soyasapogenol C | 0 | – | – | – | – | – | – |
| VIR251 | 13 | O | A | Asn110 | H | D | 2.13183 |
| O | A | Cys111 | H | D | 2.58805 | ||
| O | A | Gly163 | H | D | 2.74516 | ||
| H | D | Gly163 | O | A | 2.12056 | ||
| H | D | Asp164 | O | A | 2.46426 | ||
| O | A | Val165 | H | D | 2.17347 | ||
| H | D | Ala246 | O | A | 1.92975 | ||
| O | A | Tyr264 | H | D | 1.23456 | ||
| H | D | Tyr264 | O | A | 2.67833 | ||
| O | A | Asn267 | H | D | 2.56788 | ||
| O | A | Tyr268 | H | D | 1.37890 | ||
| O | A | Gln269 | H | D | 1.34567 | ||
| O | A | Gln269 | H | D | 2.45675 | ||
| Compound | H-bond(s) | Ligand | Receptor | H-bond distance (Å) | |||
|---|---|---|---|---|---|---|---|
| Element | Type | Residue | Element | Type | |||
| (c) Interactions with RdRp | |||||||
| Azadirachtin A | 5 | O | A | Lys551 | H | D | 2.85043 |
| O | A | Lys551 | H | D | 1.95257 | ||
| O | A | Lys551 | H | D | 2.1227 | ||
| O | A | Arg553 | H | D | 1.72225 | ||
| O | A | Ser814 | H | D | 2.63803 | ||
| H | D | Thr556 | O | A | 1.91562 | ||
| Betulinic acid | 2 | O | A | Arg553 | H | D | 2.04290 |
| O | A | Arg624 | H | D | 2.76787 | ||
| Somniferine | 3 | O | A | Lys551 | H | D | 2.01788 |
| O | A | Arg553 | H | D | 1.82204 | ||
| H | D | Asp623 | O | A | 1.95053 | ||
| Remdesivir | 5 | O | A | Ile548 | A | O | 2.35364 |
| O | A | Arg555 | D | H | 1.87313 | ||
| H | D | Thr680 | O | A | 2.72405 | ||
| H | D | Thr680 | O | A | 2.67892 | ||
| H | D | Ser681 | O | A | 2.56783 | ||
A: acceptor, D: donor.
3.1.2. Inhibitors for PLpro
The PLpro is another important cysteine protease enzyme responsible for the maturation of viral particles (Shree et al., 2020). From the docking study, it was revealed that betulinic acid, saikosaponin A and soyasapogenol C exhibited better binding affinities as compared to rest of the compounds. Betulinic acid interacted strongly with the PLpro with a CDocker energy of −87.0782 kcalmol−1 and a CDocker interaction energy of −25.1481 kcalmol−1. Saikosaponin A also interacted prominently with PLpro with a CDocker energy of −252.934 kcalmol−1 and a CDocker interaction energy of −14.6437 kcalmol−1. Soyasapogenol C interacted strongly with PLpro with a CDocker energy of −77.4819 kcalmol−1 and a CDocker interaction energy of −26.2634 kcalmol−1. Betulinic acid formed no conventional H-bonds. The interaction between saikosaponin A and PLpro was mediated to some extent through conventional H-bonds with eight amino acid residues (Asn109, Tyr112, Gln269 and Tyr273). Soyasapogenol C formed no H-bonds. Docking study indicates that these three compounds possess notable inhibitory effects against PLpro. The co-crystal ligand (VIR251) showed good binding affinity with a CDocker energy of −75.0382 kcalmol−1 and a CDocker interaction energy of −62.4873 kcalmol−1. It also interacted strongly with the PLpro molecule with the formation of 13 H bonds. Hydrogen bonds were formed with binting site residues such as Asn110, Cys111, Gly163, Asp164, Val165, Ala246, Tyr264, Asn267, Tyr268 and Gln269. The extent of interaction of co-crystal ligand, VIR251 with the PLpro enzyme was comparatively more than that of phytochemical inhibitors. Docking interaction diagrams 2D of saikosaponin A along with its co-crystal inhibitor, VIR251 are displayed in Fig. 7 (c, d). Details of hydrogen bonding interactions are given in Table 2.
3.1.3. Inhibitors for RdRp
The RdRp is believd to be an indispensable enzyme for the replication of viral genomes (Chowdhury, 2020). Docking study against PLpro revealed that azadirachtin, betulinic acid and somniferine exhibited betters binding affinity compared to rest of the compounds. Azadirachtin interacted strongly with the RdRp with a CDocker energy of −100.087 kcalmol−1 and a CDocker interaction energy of −68.8895 kcalmol−1. Betulinic acid also interacted intensely with the RdRp with a CDocker energy of −72.1015 kcalmol−1 and a CDocker interaction energy of −44.9878 kcalmol−1. Somniferine also interacted firmly with the RdRP with a CDocker energy of −187.218 kcalmol−1 and a CDocker interaction energy of −58.4079 kcalmol−1. Azadirachtin binds with the active site residues (Lys551, Arg553, Ser814 and Thr556) of RdRp through the formation of 6 conventional H-bonds. The interaction between betulinic acid and RdRp was mediated to some extent through conventional H-bonds with Srg553 and Arg624. Somniferine formed three H-bonds with Lys551, Arg553 and Asp623 of RdRp. The co-crystal ligand (Remdesivir) showed better binding profile with a CDocker energy of −29.2231 kcalmol−1 and a CDocker interaction energy of −63.6939 kcalmol−1. It also interacted strongly with the RdRp molecule with the formation of five H bonds. Hydrogen bonds were formed with amino acid residues like Ile548, Arg555, Thr680 and Ser681. The extent of interaction of co-crystal ligand with the RdRp enzyme was more than that of the phytochemical inhibitors. Docking interaction diagrams 2D of somniferine along with their co-crystal inhibitor, Remdesivir showing the involvement of active site residues of RdRp are presented in Fig. 7 (e, f). Details of hydrogen bonding interactions are given in Table 2.
3.2. MD simulation
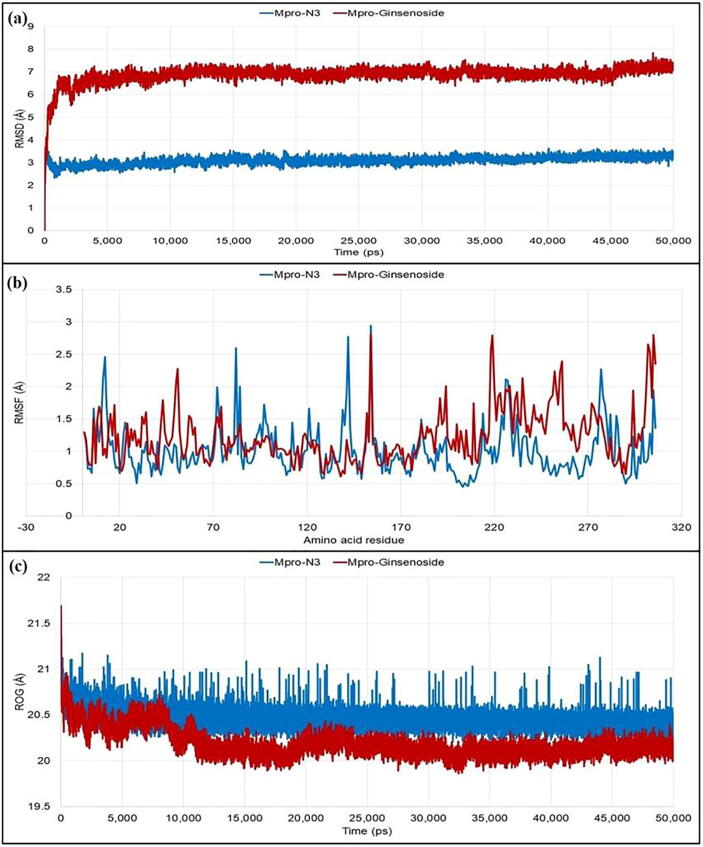
The RMSD value of Mpro-N3 complex was found stable without significant deviations during the 50 ns simulation period which was maintained below 4 Å. In the case of Mpro-ginsenoside Rg2, the deviation was more and maintained below 8 Å. In both the cases, the plateu state of protein–ligand complexes reached within 3 ns and thereafter no significant deviations were observed. Fluctuations for each of the individual amino acid of the target protein in the case of the co-crystal inhibitor (N3) and the phytocompound (ginsenoside Rg2) were then observed from the RMSF values. The average fluctuation of the amino acid residues for Mpro-N3 complex was 1.009 Å, whereas it was 1.230 Å for the Mpro-ginsenoside Rg2 complex. The catalytic residue His41 showed slightly higher difference in the fluctuation (0.381 Å) in comparison to the Cys145 (0.164 Å). Thereafter, the compactness of the protein–ligand complexes analyzed from recording the ROG values was also obtained. There was no significant difference between the average ROG values of Mpro-N3 (20.462 Å) and Mpro-ginsenoside Rg2 (20.192 Å) observed which indicated that the complexes were formed with equal compactness (Fig. 3).
Fig. 3.
Different stability related parameters of docked complexes using Mpro obtained from MD analysis: (a) RMSD, (b) RMSF, and (c) ROG.
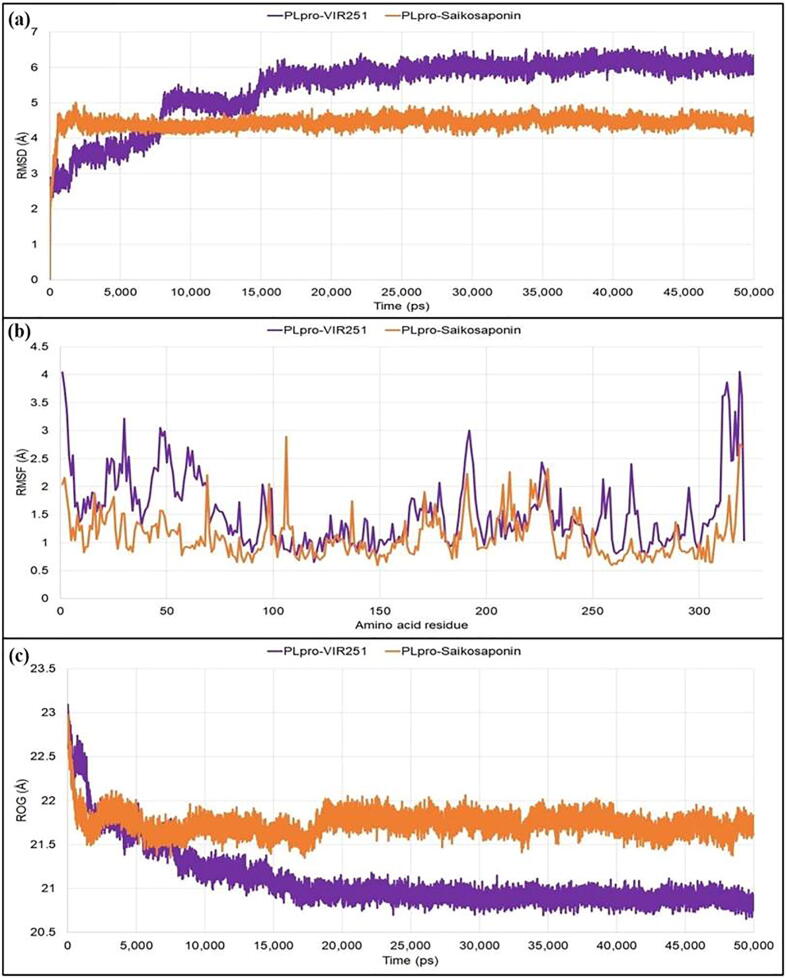
For PLpro, the PLpro-VIR251 reached the plateau state after 15 ns and then maintained the deviation below 7 Å, whereas the PLpro-saikosaponin complex reached the plateau state within 4 ns during the simulation period and maintained the deviations within 5 Å. The fluctuations of the individual amino acids were then determined from RMSF analysis. With PLpro-VIR251, the average fluctuations were 1.549 Å, while the average fluctuation for the PLpro-saikosaponin A was recorded to be 1.105 Å during the 50 ns simulation period. In the analysis of the compactness, initially both the complexes had similar kind of property, but later on the PLpro-VIR251 complex became more compact with average ROG value 21.101 Å in comparison to the PLpro-saikosaponin A complex (average ROG value of 21.737 Å) during the simulation period of 50 ns (Fig. 4).
Fig. 4.
Different stability related parameters of docked complexes using PLpro obtained from MD analysis: (a) RMSD, (b) RMSF, and (c) RO.
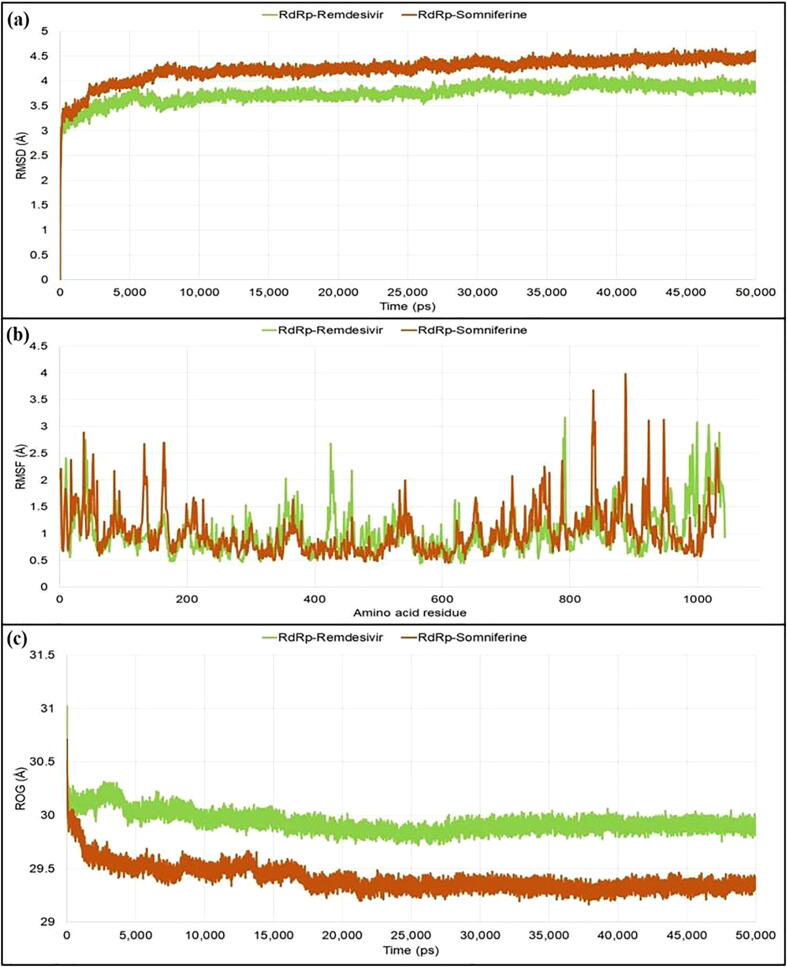
For RdRp protein, the plateau state was reached after 8 ns for both the protein–ligand complexes and thereafter the RdRP-Remdesivir maintained the deviations within 4 Å, whereas the RdRp-somniferine complex maintained deviations within 4.5 Å. From RMSF analysis, the amino acid residues of RdRp-Remdesivir complex fluctuated within an average range of 1.005 Å, whereas the amino acid residues of the RdRp-somniferine complex fluctuated within an average range of 1.025 Å. From ROG values, the RdRp-somniferine (29.406 Å) formed slightly more compact and tighter complex as compared to the RdRp-Remdesivir system (29.937 Å) (Fig. 5).
Fig. 5.
Different stability related parameters of docked complexes using RdRp obtained from MD analysis: (a) RMSD, (b) RMSF, and (c) ROG.
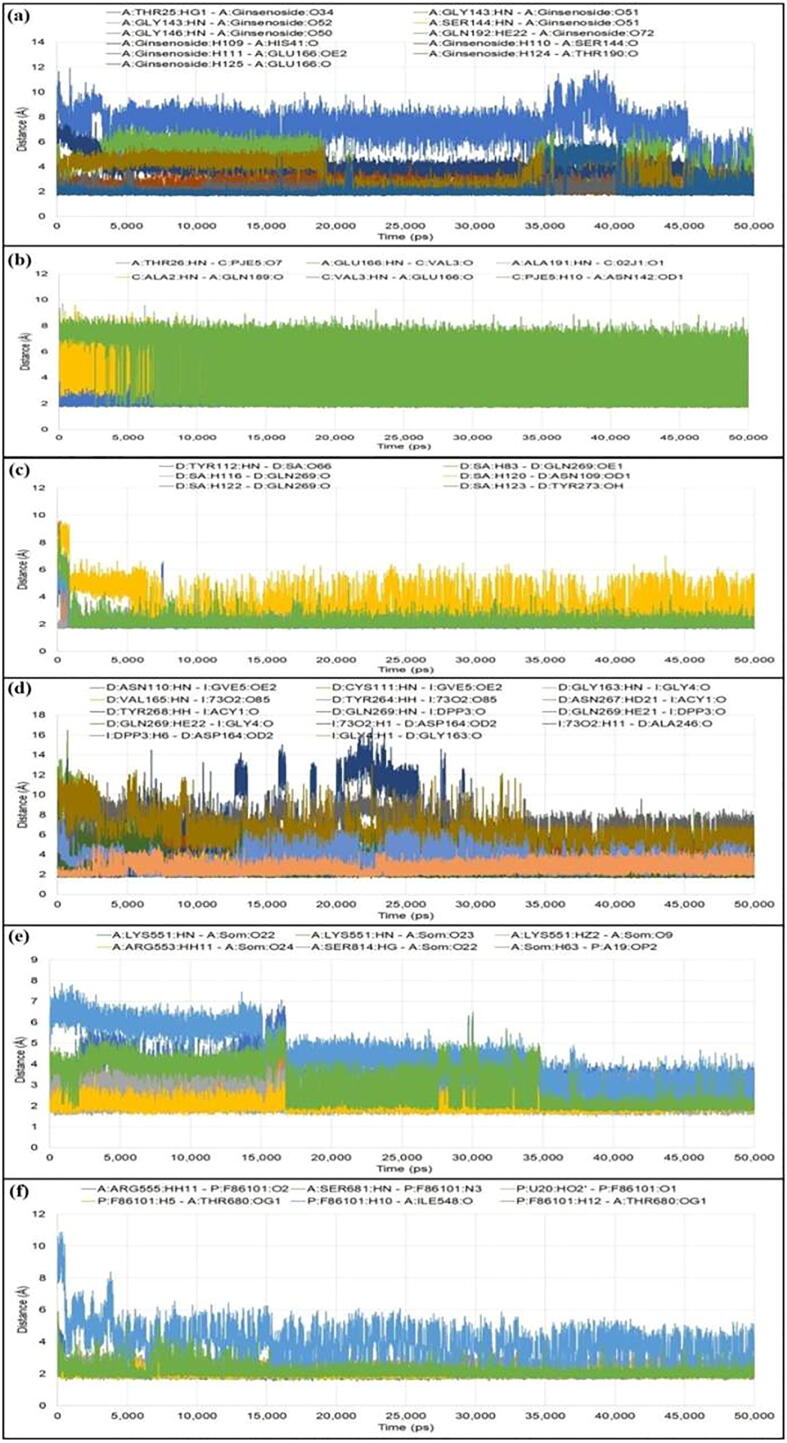
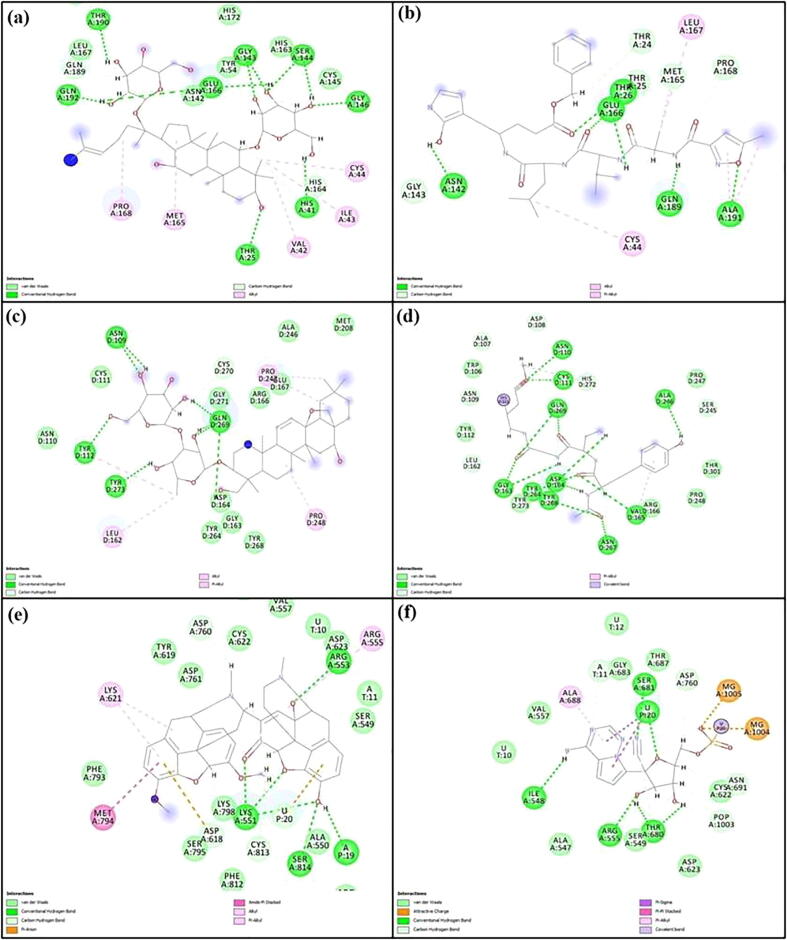
The number of H-bonds formed and the deviations of their distances during the simulation period were also evaluated (Fig. 6). Along with H-bonds various other non-bonding interactions were also investigated. In Mpro, N3 formed five conventional H-bonds with Thr26, Asn142, Glu166, Gln189 and Ala191; seven carbon H-bonds with Thr24, Thr25, Gly143, Met165 and Pro168; and five hydrophobic bonds with Cys44, Met165, Leu167, Gln189 and Ala191. On the other side, ginsenoside Rg2 formed eleven conventional H-bonds with Thr25, His41, Ser144, Gly143, Gly146, Glu166, Thr190 and Gln192; six C-H bonds with His41, His164, Glu166, Leu167 and Gln189; and five hydrophobic bonds with Val42, Ile43, Cys44, Met165 and Pro168. For PLpro protein, VIR251 formed thirteen H-bonds with Asn110, Cys111, Gly163, Asp164, Val165, Ala246, Tyr264, Asn267 and Tyr268; seven C-H bonds with Ala107, Asp108, Asn109, Leu162, Ser245 and Asn267; and two interactions of hydrophobic type with Val165 and His272. With the same protein, saikosaponin formed eight H-bonds with Asn109, Tyr112, Gln269 and Tyr273; one C-H bond with Cys270; and five hydrophobic bonds with Tyr112, Leu162, Pro247 and Pro248. In the case of RdRp protein, Remdesivir formed five H-bonds with Ile548, Arg555, Thr680 and Ser681; three carbon hydrogen bonds with Cys622, Asn691 and Asp760; and one interaction of hydrophobic type with Ala688. It also formed one H-bond with U20, two C-H bonds with A11 and POP1003 and two charged interaction with MG1004 and MG1005. Somniferine formed five conventional hydrogen bonds with Lys551, Arg553 and Ser814; three C-H bonds with Asp760, Ser795 and Cys815; and four hydrophobic interactions with Arg555, Lys621 and Met794; and one charged interaction with Asp618. It also formed one H-bond with A19; one carbon hydrogen bond with U20 and one charged interaction with U20. Details of protein–ligand interactions are given in Fig. 7.
Fig. 6.
Fluctuations of distances of hydrogen bonds for various docked complexes from MD analysis: (a) Mpro-N3, (b) Mpro-ginsenoside Rg2, (c) PLpro-VIR251, (d) PLpro-saikosaponin A, (e) RdRp-Remdesivir, and (f) RdRp-somniferine.
3.3. MM-PBSA-based binding energies
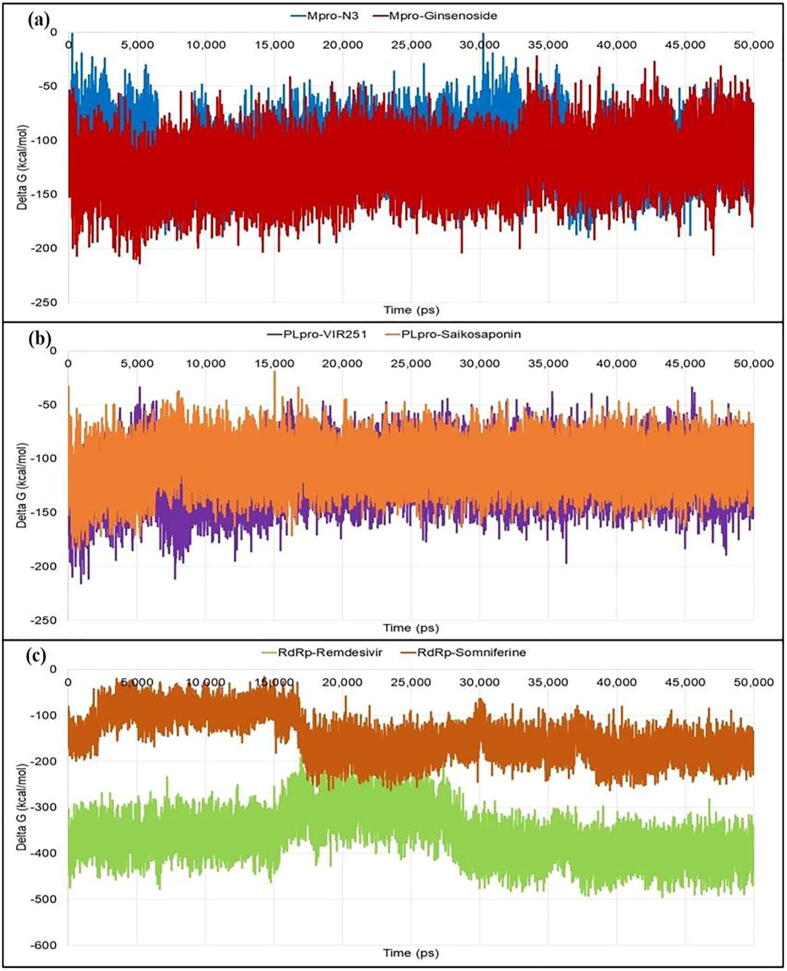
The binding energies reveal the thermodynamic stability of docked protein–ligand complexes. It is an important property which assesses the bioactivity of ligands. From MM-PBSA analyses (Fig. 8), the average energy of the Mpro-N3 complex (−111.077 kcal/mol) was observed to be higher than the Mpro-ginsenoside Rg2 (−123.319 kcal/mol). The binding energy of PLpro-VIR251 (−116.234 kcal/mol) was less than the PLpro-saikosaponin A (−105.912 kcal/mol). The binding free energy of RdRp-Remdesivir (−361.626 kcal/mol) was even less than the RdRp-somniferine (−146.026 kca/mol). MM-PBSA analyses (Table 3) indicate that the Mpro-ginsenoside Rg2 was thermodynamically more stable as compared to the co-crystal inhibitor (Mpro-N3). In other two cases, phtochemicals (saikosaponin A and somniferine) formed comparatively less stable complexes with the target proteins in comparison to their corresponding co-crystal inhibitors (VIR251 and Remdesivir, respectively).
Fig. 8.
Fluctuations of the binding free energy (Delta G) during the simulation period for various complexes: (a) Mpro-ligands, (b) PLpro-ligands, and (c) RdRp-ligands.
Table 3.
MM-PBSA binding energies of docked complexes.
| Complex | MM-PBSA binding energy (kcal/mol) |
|---|---|
| Mpro-ginsenoside Rg2 | −123.319 |
| Mpro-N3 | −111.077 |
| PLpro-saikosaponin A | −105.912 |
| PLpro-VIR251 | −116.234 |
| RdRp-somniferine | −146.026 |
| RdRp-Remdesivir | −361.626 |
3.4. Drug-likeness/ADMET
Results of drug-likeness parameters for top 15 dock scored compounds are displayed in Table 4. All drug-likeness data were found to be within the considerable range indicating the good drug-likeness behaviour of the screened phytochemicals. LogP, MW, and molecular PSA indicate good membrane permeability, intestinal absorption and oral bioavailability, whereas, other parameters such as nHBAs, nHBDs, and nRotb bonds facilitate drug metabolism and pharmacokinetics (DMPK) (Rudrapal and Sowmya, 2019). Some compounds showed relatively higher lipophilicity, which could be accounted for the better biological activities due to the increased permeation/absorption of biological membranes (Rudrapal et al., 2017).
Table 4.
Molecular and drug-likeness properties of top 15 dock scored compounds.
| Compound | Parameters |
|||||
|---|---|---|---|---|---|---|
| LogP | Mol. wt. | nHBA | nHBD | nRotB | Mol. PSA | |
| Azadirachtin A | −1.451 | 720.714 | 16 | 3 | 10 | 215.33 |
| β-Amyrin | 7.303 | 426.717 | 1 | 1 | 0 | 20.23 |
| Betulinic caid | 6.546 | 456.70 | 3 | 2 | 2 | 57.53 |
| Betulonaldehyde | 6.568 | 440.701 | 2 | 1 | 2 | 37.29 |
| Chebulagic acid | 1.714 | 954.661 | 27 | 13 | 5 | 447.09 |
| Diosgenin | 4.633 | 414.621 | 3 | 1 | 0 | 38.69 |
| EGCG | 3.097 | 458.372 | 11 | 8 | 4 | 197.36 |
| Ginsenoside Rg2 | 1.126 | 801.013 | 14 | 10 | 10 | 239.21 |
| Glycyrrhetic acid | 5.656 | 470.684 | 4 | 2 | 1 | 74.59 |
| Nimbolide | 2.758 | 466.523 | 6 | 0 | 4 | 92.04 |
| Oleanolic acid | 6.447 | 456.70 | 3 | 2 | 1 | 57.53 |
| Saikosaponin A | 1.107 | 780.982 | 13 | 8 | 6 | 207.98 |
| Salannin | 3.708 | 596.708 | 8 | 0 | 9 | 110.50 |
| Sargeolide | −0.751 | 504.483 | 11 | 2 | 4 | 154.88 |
| Soyasapogenol C | 5.768 | 440.701 | 2 | 2 | 1 | 40.46 |
| Somniferine | 2.801 | 608.68 | 9 | 2 | 3 | 100.93 |
| Ursolic acid | 6.492 | 456.70 | 3 | 2 | 1 | 57.53 |
LogP: log of n-octanol/water partition coefficient; Mol. wt: molecular weight; nHBA: number of hydrogen bond acceptor(s); nHBD: number of hydrogen bond donor(s); nRotB: number of rotable bond(s); Mol. PSA- molecular polar surface area.
The predicted ADMET data of top 15 compounds are presented in Table 5. All the compounds exhibited good aqueous solubility and gastrointestinal absorption, which could facilitate compounds to attain increased concentration in the blood for optimal biological action. These compounds also exhibited poor blood–brain barrier (BBB) penetration indicating less probability of producing CNS toxicity. Most of the compounds were found to be not inhibitors of cytochrome P450 2D6 (CYP 2D6). Compounds showed no predictive hepatotoxicity, which means that compounds are not expected to possess significant liver toxicity. Plasma protein binding (PPB) data revealed that some of the compounds were strongly bound, while some were less strongly bound. However, overall predicted ADMET data proved satisfactory ADMET parameters of phytochemicals confirming their drug-like or lead-like properties.
Table 5.
Predicted ADMET properties of top 15 dock scored compounds.
| Compound | AS level | BBB level | CYP 2D6 inhibition | Hepatox prediction | IA level | PPB level |
|---|---|---|---|---|---|---|
| Azadirachtin A | 3 | 4 | F | T | 3 | F |
| β-Amyrin | 0 | 4 | F | F | 3 | T |
| Betulinic caid | 1 | 4 | F | F | 2 | T |
| Betulonaldehyde | 0 | 0 | F | F | 1 | T |
| Chebulagic acid | 0 | 4 | F | T | 3 | F |
| Diosgenin | 1 | 1 | F | F | 0 | T |
| EGCG | 1 | 4 | F | T | 3 | F |
| Ginsenoside Rg2 | 2 | 4 | F | F | 3 | F |
| Glycyrrhetic acid | 1 | 4 | F | F | 1 | T |
| Nimbolide | 2 | 3 | F | F | 0 | T |
| Oleanolic acid | 1 | 4 | F | F | 1 | T |
| Saikosaponin A | 2 | 4 | F | F | 3 | F |
| Salannin | 2 | 4 | F | F | 0 | T |
| Sargeolide | 3 | 4 | F | F | 3 | F |
| Soyasapogenol C | 1 | 0 | F | F | 0 | T |
| Somniferine | 2 | 3 | F | F | 0 | F |
| Ursolic acid | 1 | 4 | F | F | 1 | T |
AS (aqueous solubility level): 3- good, 2- low, 1- very low, 0- extremely low; BBB (blood brain barrier) penetration level: 4- very low, 3- low, 2- medium, 1- high, 0- very high; CYP2D6 (cytochrome P450 2D6) inhibition level: F (false)- non-inhibitor, T (true)- inhibitor; Hepatox (hepatotoxicity): T (true)- toxic, F (false)- non-toxic; IA (intestinal absorption level): 0- good, 1- moderate, 2- poor, 3-very poor; PPB (plasma protein binding): T (true)-highly bound, F (false)-poorly bound.
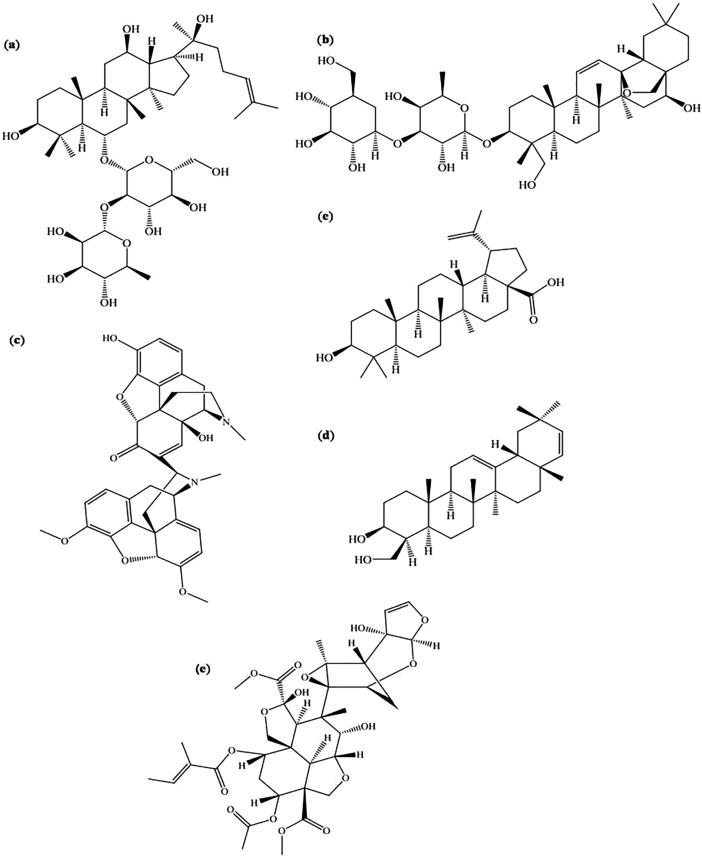
A more detailed explanation about the phytochemicals including their phytochemical type/nature of structural skeleton, plant source and inhibitory activity against specific enzymes are summarized in Table 6. Fig. 9 depicts chemical structures of individual compounds identified as protease/polymerase inhibitors.
Table 6.
Promising Mpro, PLpro and RdRp inhibitors.
| Compound name (PubChem ID) | Phytochemical type | Plant source | Activity against |
|---|---|---|---|
| Ginsenoside Rg2 (75412551) | Steroid glycoside/Triterpenoid saponin | Panax ginseng (Asian ginseng)# | Mpro |
| Saikosaponin A (167928) | Triterpenoid saponin | Bupleuri radix (Chai-hu)# | Mpro, PLpro |
| Somniferine (14106343) | Steroidal lactone triterpenoid | Withania somnifera (Ashwagandha)* | Mpro, RdRp |
| Betulinic acid (64971) | Pentacyclic triterpenoid | Syzygium aromaticum | PLpro, RdRp |
| Soyasapogenol C (3083637) | Soyasaponin | Glycine max (Soybean, Food plant)$ | PLpro |
| Azadirachtin A (5281303) | Tetranortriterpenoid Limonoid | Azadirachta indica (Neem)* | RdRp |
Indian system of medicine (Ayurvedic medicine).
Traditional Chinese medicine.
Grown in China, Japan.
Fig. 9.
Structure of some phytochemicals identified as promising protease (Mpro and PLpro) and polymerase (RdRp) inhibitors: (a) Ginsenoside Rg2, (b) Saikosaponin A, (c) Somniferine, (d) Betulinic acid, (e) Soyasapogenol C, (f) Azadirachtin A.
4. Discussion
Medicinal plants (turmeric, neem, ashwagandha, tea, holy basil, licorice, dioscorea, sesame etc.) that are reported in traditional remedies and herbal medicine (Ayurvedic or Chinese medicine) having antiviral, anti-HIV, antibacterial, antifungal, antiprotozoal, antimutagenic and immunomodulatory activities were extensively reviewed for the selection of phytochemicals (flavonoids, limonoids, saponins, triterpenoids, steroids etc.) with plausible protease and/or polymerase inhibitory effects. A total of 16 phytochemicals (initial hits) were selected and virtually screened by molecular docking, MD and drug-likeness/ADMET studies in order to find out potential protease and polymerase inhibitors, particularly against 3CLpro, PLpro and RdRp enzymes.
In validation study, the RMSD values were obtained as 1.57 Å, 1.47 Å, 2.45 Å for Mpro, PLpro and RdRp, respectively, which indicated the robustness of the docking procedure. Superimposed docked complexes showed lower RMSD values as compared to the original co-crystal structures. It further demonstrated the stability of the complexes. The validation study further substantiates the strong binding affinity of ligands for their corresponding protein molecules. Results of validation study also confirmed the experimental binding modes of co-crystal inhibitors (N3, VIR251 and Remdesivir) in the active site residues of respective protein molecule (6LU7, 6WX4 and 7BV2) with sufficient reproducibility of the predicted protein–ligand interactions.
Molecular docking is a structure-based virtual screening approach which identifies active inhibitors based upon the prediction of binding affinity as well as molecular interactions of ligand molecules (or inhibitors) with their corresponding target proteins or enzymes (Cheng et al., 2007, Ford et al., 2020). Flexible docking simulation protocol is usually performed to assess the binding affinity of active inhibitors. The most favourable binding mode of docked poses is investigated for the protein–ligand complex formed with low energy conformation. Docking finds out the best binding orientation of ligands for their corresponding target molecules. With the prediction of binding affinity against the target protein, docking helps to assess the biological efficacy of small molecules (Zhang et al., 2020). In docking study, CDocker program docked all the phytocompounds into the predicted active site of three different protein molecules. CDocker, a simulation based algorithm which uses a CHARMm-based molecular dynamics (Swargiary et al., 2020, Ferrin, 2004). Compounds showed predictable binding affinity against target protein molecules (Mpro, PLpro and RdRp) with well defined molecular interactions with several active site residues. Higher binding affinities were observed for docked compounds as compared to the co-crystal inhibitor. CDcoker energy represents the minimum binding energy of a protein–ligand complex, whereas CDocker interaction energy is the minimum energy of interactions taken place between the protein and ligand molecule. The more negative the interaction energy, the stronger this interaction will be. Affinity therefore depends on the energy of interaction. Binding energies thus depicts the strength of interactions as well as the affinity of a ligand molecule for its receptor molecule. Formation of stable complexes with well-defined interaction details predicts the significance of molecular docking and further molecular modeling studies.
From MD simulation, the stability and flexibility of complexes can be assessed (Salmaso and Moro, 2018). From results of MD simulation and MM-PBSA analyses, it is claimed that ginsenoside Rg2 possesses superior binding affinity against Mpro than that of of saikosaponin A and somniferine against PLpro and RdRp, respectively. Medicinal plants containing polyphenolic substances, saponins and steroids have been reported to possess antiviral activities (Buonaguro et al., 2020).
Results of drug-likeness revealed that all these fifteen compounds exhibited acceptable drug-like properties. Lipinski’s rule of five (Rudrapal et al., 2021) and Veber rule (Zhang and Lazim, 2017) were obeyed. Compounds with LogPo/w ≤ 5, Mol. wt. ≤ 500, nHBAs ≤ 10 and nHBDs ≤ 5 (Zhang and Lazim, 2017) are satisfactory. Further, compounds having ≤ 10 RotB and molecular PSA of ≤140 A2 behave to be drug-like to exhibit optimal permeability across membrane with acceptable oral bioavailability. In ADMET prediction, good intestinal absorption is probably owing to their satisfactory LogP property. Unsatisfactory ADMET profile accounts for the failure of most drug candidates in the late phase of drug discovery (Ibrahim et al., 2020). Poor solubility and inadequate intestinal absorption are attributed to lower the extent of drug absorption and oral bioavailability (Kousar et al., 2020, Othman et al., 2021, Ghosh et al., 2021, Khan et al., 2021). CYP 2D6 is the key metabolic enzyme in drug metabolism (Ibrahim et al., 2020). Non-inhibition of CYP 2D6 suggests their ease of metabolism by this enzyme. The extent of PPB signifies the therapeutic action of drugs. Fraction of drug in a protein bound state does not confer any action, whereas the unbound fraction exerts the therapeutic action (Isyaku et al., 2020, Junejo et al., 2021).
From docking and MD simulation studies, six phytochemicals were found to exhibit remarkable SARS-CoV-2 inhibitory activities (best hit compounds), particularly against Mpro, PLpro and RdRp. These compounds are namely, ginsenoside Rg2, saikosaponin A, somniferine, betulinic acid, soyasapogenol C and azadirachtin A. These phytochemicals are found in traditional Indian i.e., Ayurvedic and Chinese medicines such as neem, ashwagandha, ginseng including food plants like soybean. All the identified compounds are basically tri-/tetra-terpenoids, saponins or steroids with their wide natural abundance in traditional Ayurvedic and/or Chinese medicines. However, upon critical analysis of MD results, it is apparent that the interaction occurred between ginsenoside Rg2 and the Mpro protein is comparatively more significant over the interactions involved between saikosaponin and PLpro and somniferine and RdRp. Conventional hydrogen bonding predominates over other non-bonding interactions like carbon hydrogen bonds and hydrophobic interaction. Amino acid residues involved in various interactions come from predicted catalytic/active sites of protein molecules. It is evident that the interaction of ginsenoside Rg2 with Mpro protein is much more promising than the interaction between N3 and Mpro. In case of saikosaponin and somniferine, the interactions with their corresponding protein targets, PLpro and RdRp, respectively occurs to a lesser extent comapred to VIR251 and Remdesivir, respectively.
5. Conclusion
Our study identifies six bioactive phytomolecules with promising anti-SARS-CoV-2 potential, particularly effective against SARS-CoV-2 Mpro, PLpro (protease) and RdRp (polymerase) enzymes. The identified phytocompounds include ginsenoside Rg2, saikosaponin A, somniferine, betulinic acid, soyasapogenol C and azadirachtin A. They possess triterpenoid-/limonoid-/saponin-/steroid-like structural framework (considered as main pharmacophoric moiety) with unique biologically relevant physicochemical and structural/stereochemical properties. These phytochemicals are found in various traditional medicines such as Ayurvedic medicine (neem, Azadirachta sp.; ashwagandha, Withania sp.) and Chinese medicine (ginseng, Panax sp.; Chai-hu, Bupleuri sp.), medicines including Syzygium sp. and food plants like soybean. Amongst six compounds, ginsenoside Rg2, saikosaponin A, somniferine were found to be the most potent anti-SARS-CoV-2 molecules with inhibitory activity against Mpro, PLpro and RdRp, respectively. The present investigation can be directed towards further experimental studies in order to confirm the antiviral efficacy along with toxicities of identified phytomolecules.
6. Author’s contributions
MR contributed in the conceptualization, methodology, investigation, and writing of original draft. NG worked on validation, writing and review of the manuscript. MR and JK edited the final version of the manuscript. DC supervised the project. JK, SB, BA, MAA, UDL and SJK reviewed the manuscript and managed acquisition of funds. All authors read and approved the final version of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank the Deanship of Scientific Research at Majmaah University, Saudi Arabia for supporting this work under Project number (R-2021-282). Authors would like to thank Dr. Sagarika Chandra for her needful help towards editing the images incorporated in the manuscript.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Mithun Rudrapal, Email: rsmrpal@gmail.com.
Johra Khan, Email: j.khan@mu.edu.sa.
References
- Adeoye A.O., Oso B.J., Olaoye I.F., Tijjani H., Adebayo A.I. Repurposing of chloroquine and some clinically approved antiviral drugs aseffective therapeutics to prevent cellular entry and replication of coronavirus. J. Biomol. Struct. Dyn. 2020;1–14 doi: 10.1080/07391102.2020.1765876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat E.A. SARS-CoV-2: Insight in genome structure, pathogenesis and viral receptor binding analysis – an updated review. Int. Immunopharmacol. 2021;95:107493. doi: 10.1016/j.intimp.2021.107493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonaguro L., Tagliamonte M., Tornesello M.L., Buonaguro F.M. SARS-CoV-2 RNA polymerase as target for antiviral therapy. J. Transl. Medi. 2020;18:1–8. doi: 10.1186/s12967-020-02355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral. Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbi. Rev. 2007;20(4):660. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian S.S., Agrawal M., Basu A., Abraham P., Gangakhedkar, Bhargava B. Perspectives for repurposing drugs for the coronavirus disease 2019. Indian J. Med. Res. 2020;151(2–3):160. doi: 10.4103/ijmr.IJMR_585_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury P. In silico investigation of phytoconstituents from Indian medicinal herb Tinospora cordifolia (giloy)’against SARS-CoV-2 (COVID-19) by molecular dynamics approach. J. Biomol. Struct. Dyn. 2020;39(17):6792–6809. doi: 10.1080/07391102.2020.1803968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciotti M., Angeletti S., Minieri M., Giovannetti M., Benvenuto D., Pascarella S., Sagnelli C., Bianchi M., Bernardini S., Ciccozzi M. COVID-19 outbreak: an overview. Chemotherapy. 2019;64(5–6):215–223. doi: 10.1159/000507423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A.D., Al-Obaidi A., Şahin A.T., Yelekçi K. Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J. Biomol. Struct. Dyn. 2020;39(8):2980–2992. doi: 10.1080/07391102.2020.1758791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrin U. Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Ford N., Vitoria M., Rangaraj A., Norris S.L., Calmy A., Doherty M. Systematic review of the efficacy and safety of antiretroviral drugs against SARS, MERS or COVID-19: initial assessment. J. Int. AIDS Soc. 2020;23(4):e25489. doi: 10.1002/jia2.25489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Exp. Opi. Drug Dis. 2015;10(5):449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Chetia D., Gogoi N., Rudrapal Design, molecular docking, drug-likeness, and molecular dynamics studies of 1, 2, 4-trioxane derivatives as novel Plasmodium falciparum falcipain-2 (FP-2) inhibitors. BioTechnologia. 2021;102(3):257–275. doi: 10.5114/bta.2021.108722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar S., Muthumanickam S., Kamaladevi A., Karthika C., Jothi K.S., Boomi R., Maniazhagu P., Pandian D. Promising phytochemicals of traditional Indian herbal steam inhalation therapy to combat COVID-19–An in silico study. F. Chem. Tox. 2021;148:111966. doi: 10.1016/j.fct.2020.111966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Tan, Wang D.Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil Med. Res. 2020;7(1):1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Singh A.K., Kushwaha P.P., Prajapati K.S., Shuaib M., Senapati S., Kumar S. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J. Biomol. Struct. Dyn. 2020;1–12 doi: 10.1080/07391102.2020.1776157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harapan H., Itoh N., Yufika A., Winardi W., Keam S., Te H., Megawati D., Hayati Z., Wagner A.L., Mudatsir M. Coronavirus disease 2019 (COVID-19): a literature review. J. Infect. Publ. Health. 2020;13(5):667–673. doi: 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S.A., Sheikh F.N., Jamal S., Ezeh J.K., Akhtar A. Coronavirus (COVID-19): a review of clinical features, diagnosis, and treatment. Cureus. 2020;12(3):e7355. doi: 10.7759/cureus.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger A., Dehmer M., Jurisica I. Knowledge discovery and interactive data mining in bioinformatics-state-of-the-art, future challenges and research directions. BMC Biol. 2014;15(6):1–9. doi: 10.1186/1471-2105-15-S6-I1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain N., Kakoti B.B., Rudrapal M., Sarwa K.K., Celik I., Attah E.I. Bioactive antidiabetic flavonoids from the stem bark of Cordia dichotoma forst.: identification, docking and ADMET studies. Molbank. 2021;2021:M1234. [Google Scholar]
- Ibrahim M.A., Abdelrahman A.H., Hussien T.A., Badr E.A., Mohamed T.A., El-Seedi H.R., Pare P.W., Efferth T., Hegazy M.E.F. In silico drug discovery of major metabolites from spices as SARS-CoV-2 main protease inhibitors. Comput. Biol. Med. 2020;126:104046. doi: 10.1016/j.compbiomed.2020.104046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishola A.A., Adewole K.E., Tijjani H., Abdulai S.I., Asogwa N.T. Phylogenic analysis of coronavirus genome and molecular studies on potentialanti-COVID-19 agents from selected FDA-approved drugs. J. Biomol. Struct. Dyn. 2021;1–18 doi: 10.1080/07391102.2021.1902392. [DOI] [PubMed] [Google Scholar]
- Isyaku Y., Uzairu A., Uba S. Computational studies of a series of 2-substituted phenyl-2-oxo-, 2-hydroxyl-and 2-acylloxyethylsulfonamides as potent anti-fungal agents. Heliyon. 2020;6(4):e03724. doi: 10.1016/j.heliyon.2020.e03724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W.L., Duffy E.M. Prediction of drug solubility from structure. Adv. Drug Deliv. Rev. 2002;54(3):355–366. doi: 10.1016/s0169-409x(02)00008-x. [DOI] [PubMed] [Google Scholar]
- Joshi T., Joshi T., Sharma P., Mathpal S., Pundir V., Bhatt, Chandra S. In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking. Eur. Rev. Med. Pharmacol. Sci. 2020;24(8):4529–4536. doi: 10.26355/eurrev_202004_21036. [DOI] [PubMed] [Google Scholar]
- Junejo J.A., Zaman K., Rudrapal M., Celik I., Attah E.I. Antidiabetic bioactive compounds from Tetrastigma angustifolia (Roxb.) Deb and Oxalis debilis Kunth.: Validation of ethnomedicinal claim by in vitro and in silico studies. S. Afr. J. Bot. 2021;143:164–175. [Google Scholar]
- Kalita J., Chetia D., Rudrapal M. Design, synthesis, antimalarial activity and docking study of 7-chloro-4-(2-(substituted benzylidene) hydrazineyl) quinolines. Med. Chem. 2020;16(7):928–937. doi: 10.2174/1573406415666190806154722. [DOI] [PubMed] [Google Scholar]
- Khan J., Asoom L.I.A., Khan M., Chakrabartty I., Dandoti S., Rudrapal M., Zothantluanga J.H. Evolution of RNA viruses from SARS to SARS-CoV-2 and diagnostic techniques for COVID-19: a review. Beni-Suef Univ. J. Basic Appl. Sci. 2021;10(1):1–14. doi: 10.1186/s43088-021-00150-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S.A., Al-Balushi K. Combating COVID-19: The role of drug repurposing and medicinal plants. J. Infect. Public Health. 2020;14(4):495–503. doi: 10.1016/j.jiph.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousar K., Majeed A., Yasmin F., Hussain W., Rasool N. Phytochemicals from selective plants have promising potential against SARS-CoV-2: investigation and corroboration through molecular docking, MD simulations, and quantum computations. BioMed Res. Int. 2020 doi: 10.1155/2020/6237160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P.P., Shaik R.A., Khan J., Alaidarous M.A., Rudrapal M., Khairnar S.J., Sahoo R., Zothantluanga J.H., Walode S.G. Cerebroprotective effect of Aloe Emodin: in silico and in vivo studies. Saudi J. Biol. Sci. 2022;29(2):998–1005. doi: 10.1016/j.sjbs.2021.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouznetsova V.L., Zhang A., Tatineni M., Miller M.A., Tsigelny I.F. Potential COVID-19 papain-like protease PLpro inhibitors: repurposing FDA-approved drugs. PeerJ. 2020;8:e9965. doi: 10.7717/peerj.9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Jahan S., Khan A., Siddiqui A.J., Redhu N.S., Khan J., Banwas S., Alshehri B., Alaidarous M. Neurological manifestation of SARS-CoV-2 induced inflammation and possible therapeutic strategies against COVID-19. Mol. Neurobiol. 2021:1–18. doi: 10.1007/s12035-021-02318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C., Chang Y.T.T., Sun M.F., Chen H.Y., Tsai F.J., Lin J.G., Chen C.Y.C. Molecular dynamics analysis of potent inhibitors of M2 proton channel against H1N1 swine influenza virus. Mol. Simul. 2011;37(03):250–256. [Google Scholar]
- Larini L., Mannella R., Leporini D. Langevin stabilization of molecular-dynamics simulations of polymers by means of quasisymplectic algorithms. J. Chem. Phys. 2007;126(10):104101. doi: 10.1063/1.2464095. [DOI] [PubMed] [Google Scholar]
- Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23(1–3):3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Motiwale M., Yadav N.S., Kumar S., Kushwaha T., Choudhir G., Sharma S., Singour P.K. Finding potent inhibitors for COVID-19 main protease (Mpro): an in silico approach using SARS-CoV-3CL protease inhibitors for combating CORONA. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1829501. [DOI] [PubMed] [Google Scholar]
- Naik S.R., Bharadwaj P., Dingelstad N., Kalyaanamoorthy S., Mandal S.C., Ganesan A., Chattopadhyay D., Palit P. Structure-based virtual screening, molecular dynamics and binding affinity calculations of some potential phytocompounds against SARS-CoV-2. J. Biomol. Struct. Dyn. 2021:1–18. doi: 10.1080/07391102.2021.1891969. [DOI] [PubMed] [Google Scholar]
- Noha S.M., Schmidhammer H., Spetea M. Molecular docking, molecular dynamics, and structure-activity relationship explorations of 14-oxygenated N-methylmorphinan-6-ones as potent μ-opioid receptor agonists. ACS Chem. Neurosci. 2017;8(6):1327–1337. doi: 10.1021/acschemneuro.6b00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othman I.M., Mahross M.H., Gad-Elkareem M.A., Rudrapal M., Gogoi N., Chetia D., Aouadi K., Snoussi M., Kadri A. Toward a treatment of antibacterial and antifungal infections: design, synthesis and in vitro activity of novel arylhydrazothiazolylsulfonamides analogues and their insight of DFT, docking and molecular dynamic simulations. J. Mol. Struct. 2021;130862 [Google Scholar]
- Peele K.A., Durthi C.P., Srihansa T., Krupanidhi S., Ayyagari V.S., Babu D.J., Indira M., Reddy A.R., Venkateswarulu T. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: A computational study. Inf. Med. Unlocked. 2020;19:100345. doi: 10.1016/j.imu.2020.100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T., Meenakshisundaram S., Manickam M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov. Today. 2020;25(4):668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapat M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H., Kumar S., Bhattacharyya A., Kumar H., Bansal S. Drug targets for corona virus: a systematic review. Indian J. Pharmacol. 2020;52(1):56. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapat M., Sarma P., Shekhar N., Prakash A., Avti P., Bhattacharyya A., Kaur H., Kumar S., Bansal S., Sharma A.R. Update on the target structures of SARS-CoV-2: a systematic review. Indian J. Pharmacol. 2020;52(2):142. doi: 10.4103/ijp.IJP_338_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekaran D., Palombo E.A., Yeo T.C., Ley D.L.S., Tu C.L., Malherbe F., Grollo L. Identification of traditional medicinal plant extracts with novel anti-influenza activity. Plos One. 2013;8(11):e79293. doi: 10.1371/journal.pone.0079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmi Y., Li X., Khan J., Ozer T., Choi J.R. Emerging point-of-care biosensors for rapid diagnosis of COVID-19: current progress, challenges, and future prospects. Anal. Bioanal. Chem. 2021:1–23. doi: 10.1007/s00216-021-03377-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastelli G., Rio A.D., Degliesposti G., Sgobba M. Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA. J. Comput. Chem. 2010;31(4):797–810. doi: 10.1002/jcc.21372. [DOI] [PubMed] [Google Scholar]
- Rolta R., Yadav R., Salaria D., Trivedi S., Imran M., Sourirajan A., Baumler D.J., Dev K. In silico screening of hundred phytocompounds of ten medicinal plants as potential inhibitors of nucleocapsid phosphoprotein of COVID-19: an approach to prevent virus assembly. J. Biomol. Struct. Dyn. 2020:1–18. doi: 10.1080/07391102.2020.1804457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein A.B., Blacklock K., Nguyen H., Case D.A., Khare S.D. Systematic comparison of Amber and Rosetta energy functions for protein structure evaluation. J. Chem. Theory Comput. 2018;14(11):6015–6025. doi: 10.1021/acs.jctc.8b00303. [DOI] [PubMed] [Google Scholar]
- Rudrapal M., Chetia D., Singh V. Novel series of 1, 2, 4-trioxane derivatives as antimalarial agents. J. Enzyme Inhib. Med. Chem. 2017;32(1):1159–1173. doi: 10.1080/14756366.2017.1363742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrapal M., Khairnar S.J., Borse L.B., Jadhav A.G. Coronavirus disease-2019 (COVID-19): an updated review. Drug Res. 2020;70(9):389–400. doi: 10.1055/a-1217-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrapal M., Khairnar S.J., Jadhav A.G. In: Drug Repurposing-Hypothesis, Molecular Aspects and Therapeutic Applications. Badria F.A., editor. IntechOpen; London: 2020. Drug repurposing (DR): an emerging approach in drug discovery. [Google Scholar]
- Rudrapal M., Rashid I.A., Agoni C., Bendale A.R., Nagar A., Soliman M.E.S., Lokwani D. In Silico screening of phytopolyphenolics for the identification of bioactive compounds as novel protease inhibitors effective against SARS-CoV-2. J. Biomol. Struct. Dyn. 2021 doi: 10.1080/07391102.2021.1944909. [DOI] [PubMed] [Google Scholar]
- Rudrapal M., Sowmya M.P. Design, synthesis, drug-likeness studies and bio-evaluation of some new chalconeimines. Pharm. Chem. J. 2019;53(9):814–821. [Google Scholar]
- Salmaso V., Moro S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: an overview. Front. Pharmacol. 2018;9:923. doi: 10.3389/fphar.2018.00923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16(1):1–22. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepay N., Sepay N., Al Hoque A., Mondal R., Halder U.C., Muddassir M. In silico fight against novel coronavirus by finding chromone derivatives as inhibitor of coronavirus main proteases enzyme. Struct. Chem. 2020;31:1831–1840. doi: 10.1007/s11224-020-01537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah B., Modi P., Sagar S.R. In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sci. 2020;252:117652. doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shree P., Mishra P., Selvaraj C., Singh S.K., Chaube R., Garg N., Tripathi Y.B. Targeting COVID-19 (SARS-CoV-2) main protease through active phytochemicals of ayurvedic medicinal plants–Withania somnifera (Ashwagandha), Tinospora cordifolia (Giloy) and Ocimum sanctum (Tulsi)–a molecular docking study. J. Biomol. Struct. Dyn. 2020;1–14 doi: 10.1080/07391102.2020.1810778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Gautam A., Chandel S., Ghosh A., Dey D., Roy S., Ravichandiran V., Ghosh D. Protease inhibitory effect of natural polyphenolic compounds on SARS-CoV-2: an in silico study. Molecules. 2020;25(20):4604. doi: 10.3390/molecules25204604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020;87(4):281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swargiary A., Mahmud S., Saleh M.A. Screening of phytochemicals as potent inhibitor of 3-chymotrypsin and papain-like proteases of SARS-CoV2: an in silico approach to combat COVID-19. J. Biomol. Struct. Dyn. 2020;1–15 doi: 10.1080/07391102.2020.1835729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijjani H., Matinja A.I., Olatunde A., Zangoma M.H., Mohammed A., Akram M., Adeoye A.O., Lawal H. In silico insight into the inhibitory effects of active antidiabetic compounds frommedicinal plants against SARS-CoV-2 replication and post translational modification. Coronaviruses. 2021 doi: 10.2174/2666796702666210118154948. [DOI] [Google Scholar]
- Tiwari V., Darmani N.A., Yue B.Y., Shukla D. In vitro antiviral activity of neem (Azardirachta indica L.) bark extract against herpes simplex virus type-1 infection. Phytother. Res. 2010;24(8):1132–1140. doi: 10.1002/ptr.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umadevi P., Manivannan S., Fayad A.M., Shelvy S. In silico analysis of phytochemicals as potential inhibitors of proteases involved in SARS-CoV-2 infection. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1866669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardhan S., Sahoo S.K. In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Comut. Biol. Med. 2020;124:103936. doi: 10.1016/j.compbiomed.2020.103936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veber D.F., Johnson S.R., Cheng H.Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang W., Kollman P.A., Case D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Mod. 2006;25(2):247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Wang L., Yang R., Yuan B., Liu Y., Liu C. The antiviral and antimicrobial activities of licorice, a widely-used Chinese herb. Acta Pharma. Sin., B. 2015;5(4):310–315. doi: 10.1016/j.apsb.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakaryan H., Arabyan E., Oo A., Zandi K. Flavonoids: promising natural compounds against viral infections. Arch. Virol. 2017;162(9):2539–2551. doi: 10.1007/s00705-017-3417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Lazim R. Application of conventional molecular dynamics simulation in evaluating the stability of apomyoglobin in urea solution. Sci. Rep. 2017;7(1):1–12. doi: 10.1038/srep44651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld K. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Li J., Pang Z. Recent insights for the emerging COVID-19: drug discovery, therapeutic options and vaccine development. Asian J. Pharm. Sci. 2020;16(1):4–23. doi: 10.1016/j.ajps.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]