Abstract
Objective.
Neural activity represents a functional readout of neurons that is increasingly important to monitor in a wide range of experiments. Extracellular recordings have emerged as a powerful technique for measuring neural activity because these methods do not lead to the destruction or degradation of the cells being measured. Current approaches to electrophysiology have a low throughput of experiments due to manual supervision and expensive equipment. This bottleneck limits broader inferences that can be achieved with numerous long-term recorded samples.
Approach.
We developed Piphys, an inexpensive open source neurophysiological recording platform that consists of both hardware and software. It is easily accessed and controlled via a standard web interface through Internet of Things (IoT) protocols.
Main Results.
We used a Raspberry Pi as the primary processing device along with an Intan bioamplifier. We designed a hardware expansion circuit board and software to enable voltage sampling and user interaction. This standalone system was validated with primary human neurons, showing reliability in collecting neural activity in near real-time.
Significance.
The hardware modules and cloud software allow for remote control of neural recording experiments as well as horizontal scalability, enabling long-term observations of development, organization, and neural activity at scale.
1. Introduction
Extracellular voltage recordings from in vitro cell cultures support the investigation of neural activity and dynamics. These recordings allow us to assess information processing in complex neuronal networks and enable discovery on a scale from single neuron firing patterns to local and long-range functional connectivity, network synchrony, and oscillatory activity [1–6].
Longitudinal recordings are essential to capture features of neurodevelopment and dynamics: basic physiological properties of neuron development, how 2D and 3D cultures grow and change activity patterns, and what rhythms the activity may follow [7–10]. Recordings across time are essential to study response to electrical or drug stimulus over weeks and months. Thus, longitudinal electrophysiology recordings can inform in vitro drug discovery and genetic screens [11, 12].
The further combination of longitudinal recordings and large numbers of parallel experimental replicates allow investigations to progress significantly faster and makes new experiments feasible [13]. Also, scaling up experiments generates the large volume of data necessary for taking advantage of Machine Learning algorithms and creates a faster turnaround between hypothesis, experiment, and re-testing [14]. In vitro culture models serve as a flexible system that is much easier to scale up than animal models, especially when paired with developments in robotic automation, microfluidics, and probes [15–18].
Longitudinal recordings from multi-channel experiments demand vast amounts of data and memory. The data is challenging to manage, especially since out-of-the-box hardware and software are often offline. Storage on physical disks usually requires manual monitoring to prevent running out of disk space and laborious transfer of data for backup or processing. Furthermore, many recording systems require a designated workspace for experiments with a physical computer nearby with cables or wireless transmission to stream data. Several open-source efforts have been created to provide more affordable and modifiable recording equipment [19–23]. However, no software solutions exist to easily manage and control a large amount of electrophysiology equipment and data at once.
Recent advances in commodity hardware allow for more affordable computing devices. The Internet of Things allows many devices to come online when needed and be relinquished when not needed, and protocols have been developed to effectively and securely manage and communicate with these devices. Affordable, internet-connected devices have already been developed for ECG, EEG, EMG, and heart rate variability monitoring [24–29]. Furthermore, commodity cloud compute from major companies as well as academic coalitions [30] has become widely available and many tools for downstream analysis to process voltage recordings are already offered online [16, 31–34]. However, data acquisition for in vitro cultures remains relatively isolated, as no platform exists to stream data online to link with these analysis infrastructures.
One solution is to write software add-ons for existing data acquisition systems. However, not all existing data acquisition systems are flexible or open in terms of data formats, programmability, and remote control. Additionally, channel count and price range are not always suitable for the desired application.
To address these issues we created Piphys, an all-in-one electrophysiology and processing system, that can simultaneously record data from multiple channels in the μV scale and stream it to the cloud. The user interacts with the device through a dashboard website to view data and control experiment parameters. Both hardware and software are made available as open source.
Piphys is based on a Raspberry Pi computer and eliminates the need for a desktop or laptop computer to manage an electrophysiology experiment or for an operator to be present in the lab to start a recording. The Raspberry Pi comes with a Unix-based operating system that can be easily programmed with many existing software libraries and tools. Overall, the low price and extreme flexibility of the Raspberry Pi significantly lowers the cost of the entire electrophysiology system, providing an opportunity for broader education and research opportunities.
Piphys can be used with a wide range of electrode probes including, but not limited to, rigid 2D and flexible 3D microelectrode arrays (MEAs) [35], silicon probes [36], and tetrodes. The system is built for long-term experiments with the goal of full automation using programs that can optimize experimental variables. Here we detail the Piphys system’s functionality and validate its accuracy and reliability for measuring neural activity.
2. Design
The Piphys hardware records from neural tissue remotely using our versatile circuit board connecting to Intan RHD series recording chips to perform highly sensitive analog to digital conversion. Data from the Intan can be optionally preprocessed on-site using a Raspberry Pi computer and streamed to a cloud service where deeper sorting and analysis of detected spikes can be performed. Spike sorting analysis measures neural activity changes over time in individual neurons and networks of neurons, using features like spike waveform, frequency of activity, and correlation to the activity of nearby neurons.
Hardware design
Design elements (choice of platform)
The Raspberry Pi Model 3 B+ is a low-cost, small-scale, single-board computer. It has a quad-core ARM Cortex-A53 processor with an Input/Output system. It can be programmed to interface with customized hardware with a standard data communication protocol. It also has an expandable memory space configured by a removable SD card.
The Intan RHD2132 bioamplifier chip is the key driver of the shield biopotential-sensing functionality. The chip amplifies voltage signals sensed by the electrodes and converts the analog signals to digital values for storage inside the Raspberry Pi computer.
Circuit design
The key physical innovation in Piphys is a hardware expansion board (“shield”) that enables a Raspberry Pi computer to interface with an Intan RHD2132 bioamplifier chip to perform electrophysiology.
Firstly, the expansion shield provides different levels of power derivative from the +5V source input. The system requires a +5V 2.5A DC power source which is provided via a micro-USB or barrel jack. In this set of experiments we used a power supply plugged into the wall outlet. The +5V input powers both the Pi and shield, and can be supplied either through the power barrel on the shield or through the micro-USB on the Pi for flexibility. On the shield, the power source is filtered through ferrite beads to remove high-frequency power line noise. The +5V source is converted to a +3.5V source for the Intan RHD2132 bioamplifier chip and a +3.3V for the SN65LVDT41 chip. Conversion is performed by low-noise linear voltage regulators to smooth and isolate any fluctuations from the power supply.
Secondly, the expansion shield provides translation between signal types. The SN65LVDT41 chip is configured to use low-voltage differential signaling (LVDS) to reduce the effects of noise and electromagnetic interference (EMI) and allow increased cable length. However, the Raspberry Pi communicates using complementary metal-oxide-semiconductor (CMOS) level logic. To translate between the two signal types, the expansion shield uses the SN65LVDT41 chip from Texas Instruments. The SN65LVDT41 chip has four LVDS line drivers and one LVDS line receiver to control data lines required to communicate with the Intan chip over its Serial Peripheral Interface (SPI).
Connection to electrodes
Electrodes are connected to the Intan RHD 32-channel recording headstage. For experiments reported here, we created a connection to a commercially available 6-well multi-electrode array (MEA) plate from Axion Biosystems. However, any other electrode system fitting an Omnetics 32-pin connector is compatible. The design can be adapted to custom and commercial MEAs of different form factors using adapter boards.
The Axion electrode plate mates its bottom contacts to spring finger pins on our designed adapter board. The parts are aligned using a custom holder consisting of a plastic interior surrounded by aluminum plates and compressed together by screws on four corners. The plastic holder has a slot to hold the adapter board and a groove to align the plate in the correct position. The aluminum plate casing prevents warping of the plastic and ensures even pressure compressing the plate and connector on both sides. The compressing holder provides consistent mating of spring finger pins to electrode contacts on the plate.
Software design
The Piphys system runs custom software to perform: (1) communication with the Intan RHD2132 bioamplifier chip, (2) buffering and file storage of recorded voltage data locally, (3) near real-time data streaming and plotting on the online dashboard, and (4) experiment control from the dashboard. In order to stream data, interact with data being recorded, and control the device, we deployed Redis, Amazon IoT, and S3, as described in Methods.
To perform an electrophysiology recording, the user can configure the sampling rate and start the experiment from the dashboard. Once started, the neural cell activity is firstly digitized and sampled by the Intan RHD2132 bioamplifier chip in 32 channels. Raspberry Pi stores the data on local memory and also streams it to Redis for near real-time visualization on the online dashboard. Since the Raspberry Pi computer has a 10 second streaming buffer, the data visualized on the dashboard is offset by 10 seconds from the data recorded. Therefore the data streaming is “near real-time”. During a recording, raw data is saved in chunks of 5 minutes to local memory and streamed in chunks of 10 seconds to Redis. Once the recording ends, all local data files are uploaded to S3 for permanent storage, and data is further backed up to Amazon Glacier for long-term archiving. Local data files on the Pi auto-erase every 14 days to release memory. To view a dated recording, the user can select and pull the data files from S3 to the dashboard for display (Figure 3). The Raspberry Pi has 4 CPU cores, and allows multicore and multithreading. According to resource monitoring with “htop”, the Piphys program runs on two cores. The software uses four threads to record and stream to the cloud simultaneously. One thread is used for hardware interfacing with the Intan chip; a second thread is for cloud streaming, the third one is for local saving and another is for experiment control that gets MQTT messages.
Figure 3. Software overview.
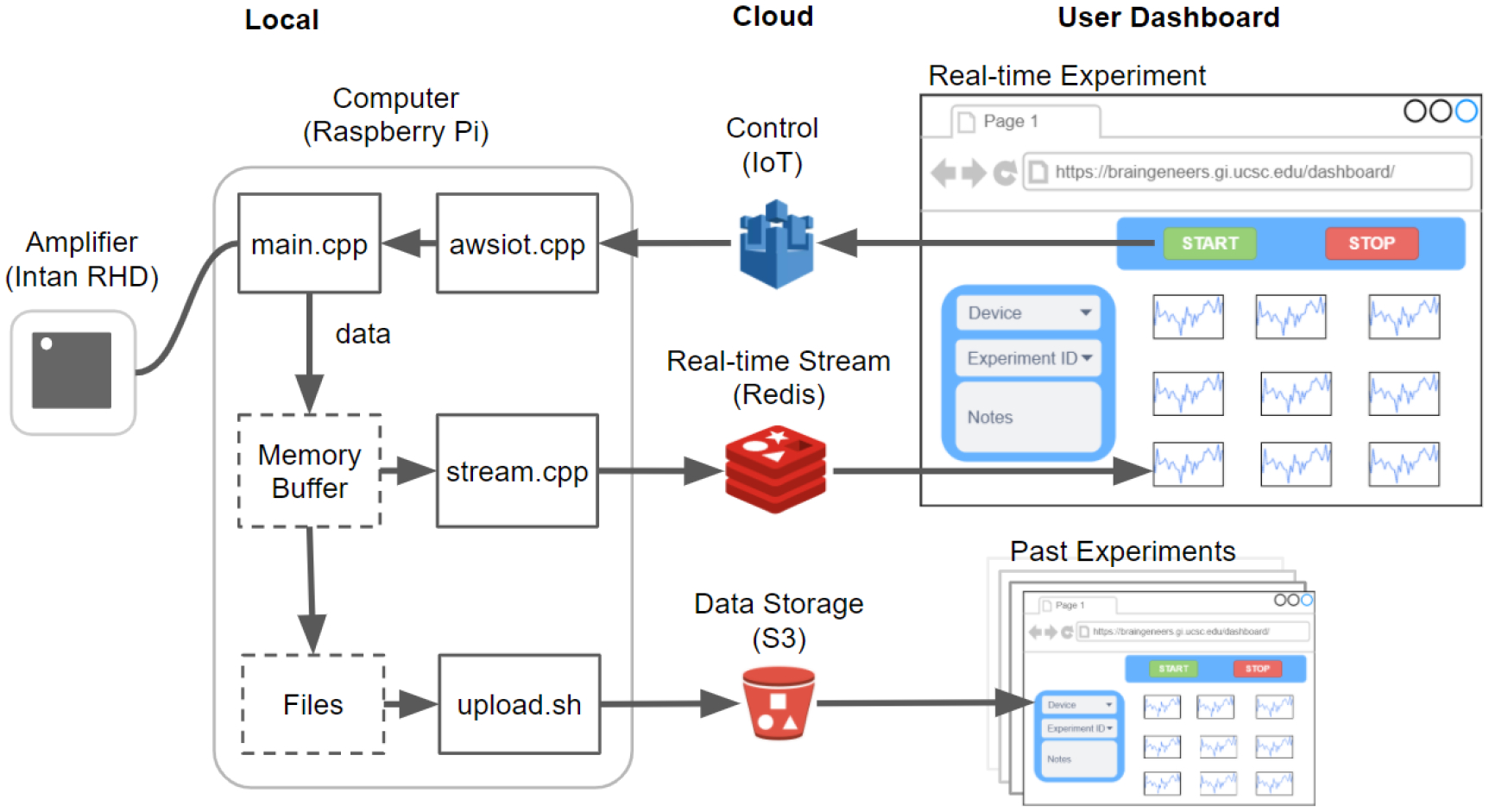
The software that runs on the local Raspberry Pi device communicates with the Intan RHD2132 bioamplifier chip to stream and store the digitized neural signal. Concurrently, it pushes the signal to Redis for near real-time visualization on the online dashboard. Datasets are also uploaded to S3 after each recording for permanent storage and access. Experimental control such as ‘start’, ‘stop’, and variable configuration is sent from the dashboard through Amazon IoT to the local device. Past experiment data can also be browsed using records from S3.
Communication with hardware
Communication between Raspberry Pi and Intan RHD2132 bioamplifier chip uses Serial Peripheral Interface (SPI). SPI is a fast and synchronous interface that is widely used in embedded systems for short-distance data streaming. It is a full-duplex master-slave-based interface where both master and slave can transmit data at the same time. The protocol for both Raspberry Pi and Intan RHD2132 bioamplifier chip is a four-wire interface: Clock (SCLK), Chip select , Master-Out-Slave-In (MOSI), and Master-In-Slave-Out (MISO). In Piphys, the Raspberry Pi acts as the master device and generates a clock signal and recording commands to configure the Intan RHD2132 bioamplifier chip through MOSI. The Intan chip responds as a slave and sends the digitized data back by MISO. The chip allows the configuration of sampling rate and bandwidth of the low-noise amplifiers. The 32 channels on the chip are sampled sequentially with available sampling rate options from 2 kHz to 15 kHz per channel. The SPI clock is divided from the core clock on Raspberry Pi. Performance and error of the SPI clock are discussed in the Supplementary Material. The amplifiers give 46 dB midband gain with lower bandwidth from 0.1 Hz to 500 Hz, and upper bandwidth from 100 Hz to 20 kHz.
Online dashboard
Users interact with Piphys devices through a web browser application, referred to as the Graphical User Interface (GUI). The GUI allows a user to initiate a recorded experiment and monitor electrical activity on each channel. Programatically, the GUI mimics an IoT device that sends messages to other devices (i.e., Piphys units) and listens to their corresponding data streams in a high-performance Redis database service. The Piphys device produces a single data stream to Redis, and many users can view the stream from the Redis server. Therefore, many users can monitor and interact with a particular Piphys device without additional overhead placed on that device.
Users can be located anywhere on the Internet without concern for where the physical Piphys device is or which network it is on. We routinely perform electrophysiology experiments from Santa Cruz on a Piphys-connected device that is located 90 miles away in San Francisco.
When a new user opens the browser GUI, the web application queries the AWS IoT service for online Piphys devices to populate a device dropdown list. When the user selects a device from the dropdown, an MQTT ‘ping’ message is sent to the relevant device every 30 seconds, indicating that a user is actively monitoring data from that device. As long as the Piphys device receives these pings, the Piphys device will continue to send its raw data stream to the central Redis service. When the Piphys device has not received any user messages for at least a minute, it will cease sending its raw data stream. This protocol ensures the proper decoupling of users from devices. The Piphys device is not dependent on a user gracefully shutting down.
While the Piphys device feeds raw data to the Redis service, data transformations are applied downstream by other IoT-connected processes. For example, the Piphys Control Panel displays a threshold spike sorted transformation of the raw data. This data transformation is an independent process that listens for MQTT requests for the raw data stream and transforms the raw stream into a stream containing the past ten spike events detected per channel. For channels with no detected spikes, a random sample of the channel is saved to the stream every 30 seconds to provide a sampling of the channel’s activity.
3. Validation Results
We tested the Piphys system for long-term recordings of human primary neurons. The goal of this work is to compare the neural signal recorded by the proposed apparatus to commercially available systems. Therefore, as a reference we choose two neural recording devices: (1) Axion Maestro Edge by Axion Biosystems, as one of the leading commercial instruments for neural recording and (2) Intan RHD interface board as one of the leading commercial open source neural recording instruments. These neurons were cultured in an Axion CytoView MEA 6-well plate ‡ (see Methods). We designed a set of adapters (see Supplementary Material), which allowed the same culture plate to be used by Piphys and Intan RHD interface board. As mentioned in the Discussion section, the proposed system can interface with any type of neural recording electrodes using the Omnetics connector. After recording, the raw data was ingested to SpyKING CIRCUS software [33] on a personal computer for analysis. SpyKING CIRCUS is a semi-automatic spike sorting software that uses thresholding, clustering, and greedy template match approaches to detect single cell action potentials. Here, we show two types of results, first for single neuron recordings and second for a bursting neural network.
Recording from primary neurons
After 14 days in culture, primary neurons were recorded with the Piphys system and two commercially available systems: the Intan RHD USB interface board and the Axion Maestro Edge. After recording, all three datasets were filtered with bandpass filtering from 300 Hz to 6000 Hz and spike sorted with a threshold of ± 6 μV. Figure 5 shows a ten-second spike train from Piphys with dots highlighting detected spikes in the raw data.
Figure 5. Detection of neuronal spike activity using Piphys.
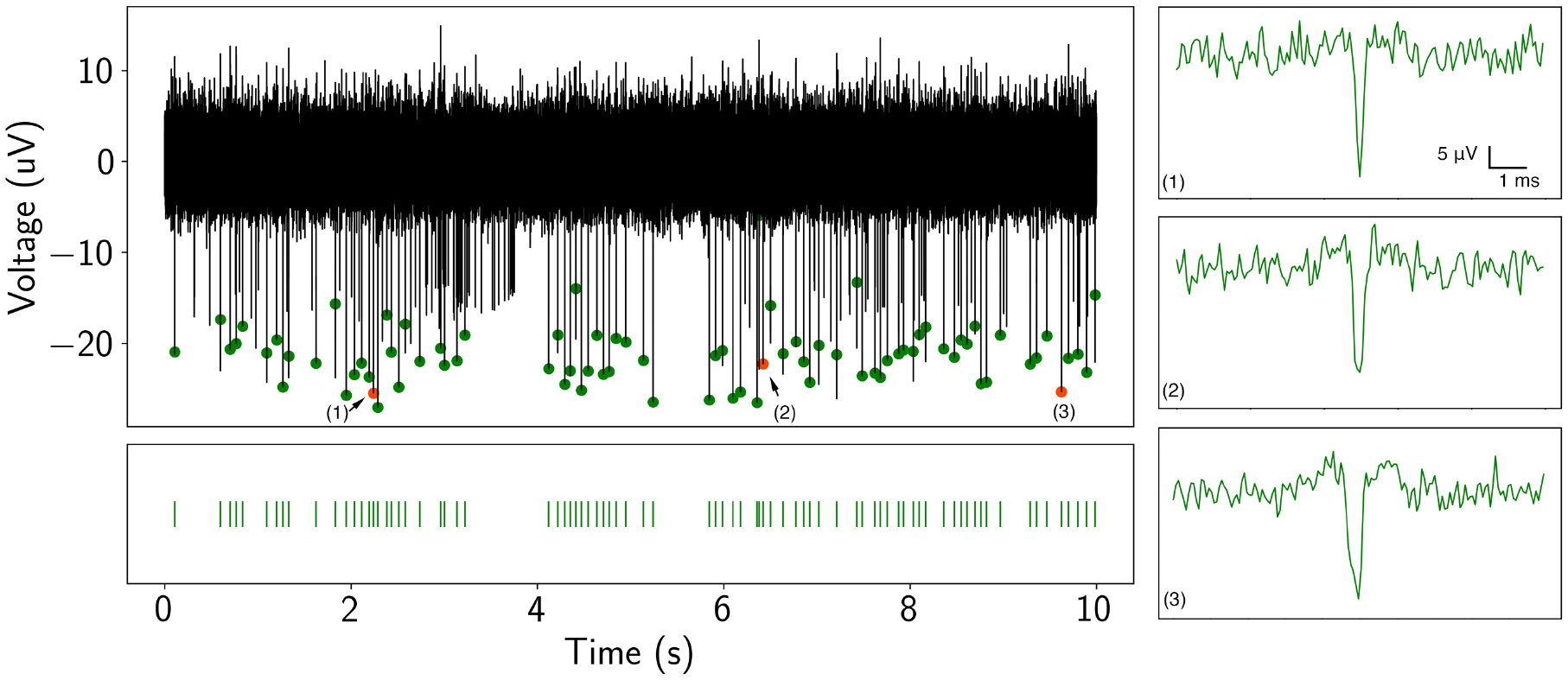
Spike train (black trace) from a recorded neuron in the time domain from Piphys. Spikes shown here are sorted from SpyKING CIRCUS software and labeled on the raw data with green and orange dots. Bottom: spike raster is aligned with the detected spikes showing firing activities at specific positions. (1)(2)(3) Individual spike examples randomly picked from the spike train. The analysis and quantification of the spike train and waveform are shown in Figure 6.
To further demonstrate the applicability of Piphys to primary neuron recording, we compare the shape of the detected action potential and quality metrics such as amplitude distribution, interspike interval distribution, and firing rate to commercially available systems (Figure 6). The data was recorded from the same channel in the same well of neurons by Piphys, Intan, and Axion systems in sequential order on the same day. The data recorded on Piphys corresponds to the data obtained from both commercial systems, with high similarity to Intan and overall consistency with Axion across metrics in Figure 6.
Figure 6. Piphys performance is similar to commercial systems.
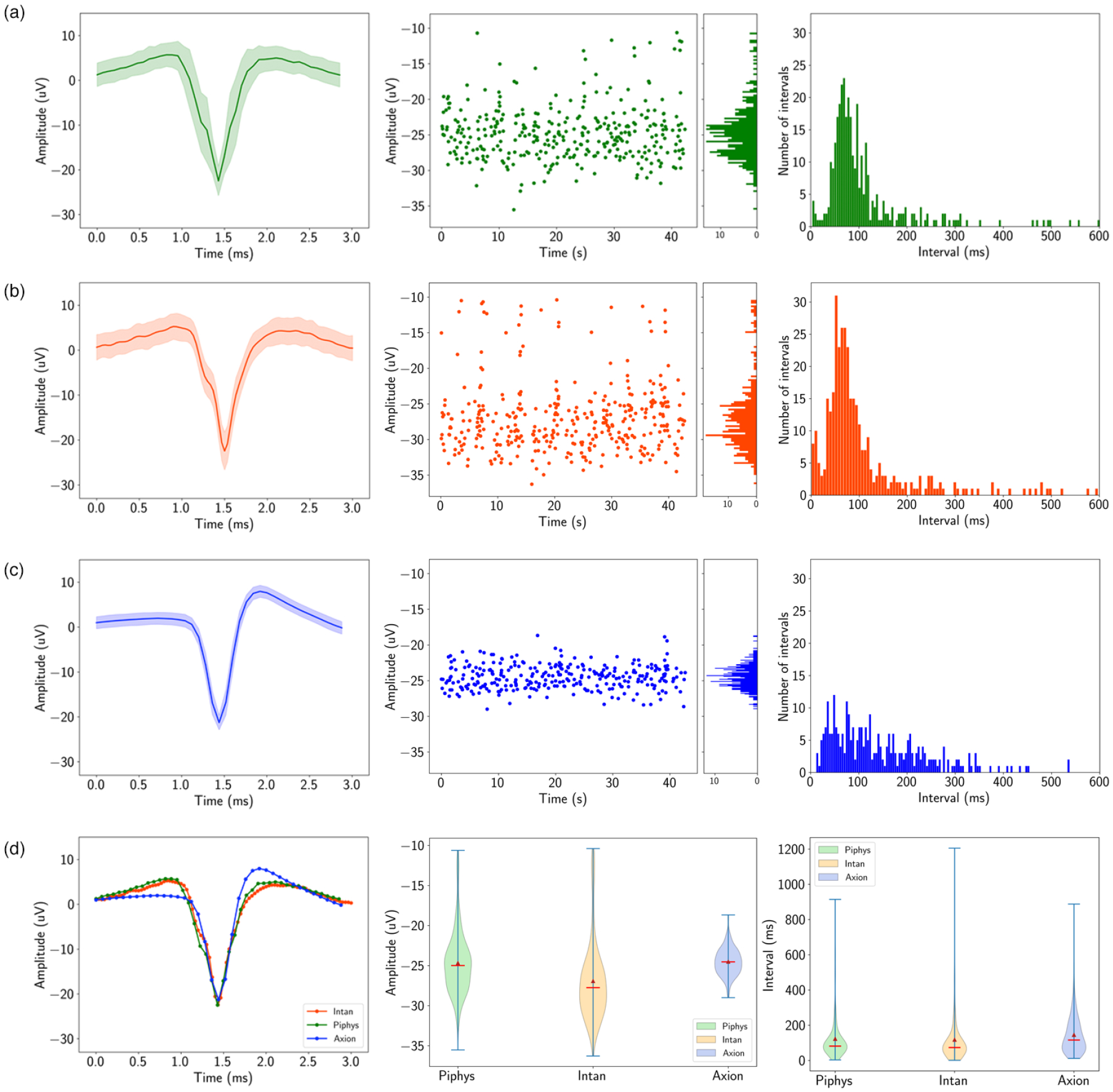
Spike sorting result for the same recording channel from Piphys, Intan RHD USB interface board, and Axion Maestro Edge. Shown from left to right are mean waveform with standard deviation (shaded area), amplitudes of the detected spikes over time, and interspike interval distribution. (Figure 5 shows a small sample of one of the channels measured using the Piphys system) (a) Piphys has −24.67 ±3.92μV for the mean waveform, firing rate of 8.05 spikes/s, and mean interspike interval of 122.79 ms. (b) Intan RHD USB interface board has −26.92 ±4.96μV for the mean waveform, firing rate of 8.44 spikes/s, and mean interspike interval of 118.15 ms. (c) Axion Maestro Edge has −24.50 ±1.69μV for the mean waveform, firing rate of 6.86 spikes/s, and mean interspike interval of 145.57 ms. (d) Comparison of the mean waveform, amplitude, and interspike interval distribution from three systems.
The mean spike waveform, shown in the first column of Figure 6, was determined by averaging the voltage in a 3 ms window centered around the point where the voltage crossed the spike threshold. Differences in Axion’s waveform shape are a flatter starting point and a higher upstroke before settling to resting state. The amplitudes for the mean waveform are −24.67 ± 3.92 μV for Piphys, −26.92 ± 4.96 μV for Intan, and −24.50 ± 1.69 μV for Axion. Axion has a smaller deviation than Piphys and Intan, showing lower noise in the recording system.
The amplitudes of the detected spikes over time, shown in the middle column of Figure 6 are more sparse for Axion than for Intan and Piphys. Firing rates in events per second over the recording period shown are 8.05 for Piphys, 8.44 for Intan, and 6.86 for Axion.
The interspike interval histograms, shown in the middle column of Figure 6, have similar longer-tail distributions for Piphys and Intan centered at 122.79 ms and 118.15 ms, and a tighter distribution for Axion centered at 145.57 ms. However, the interspike interval means for all three systems are significantly close together.
Detecting burst activity from primary neural network
On day 42 of culture, we recorded from the neurons with Piphys and found the primary neurons displayed synchronized network bursts, consistent with previous observations [37, 38]. Figure 7 shows the synchronous activity captured across four channels. After spike sorting, most detected spikes were arranged in short intervals with periods of silence in between. The spikes inside the bursts align among the channels, indicating that synchronized activity was present through the network. Quantitatively, the bursting has a general population rate of 0.13 bursts each second, with each burst lasting around 1 second. Within one burst, the number of spikes is 55 ± 17.58.
Figure 7. Bursting activity across four channels with channel mapping.
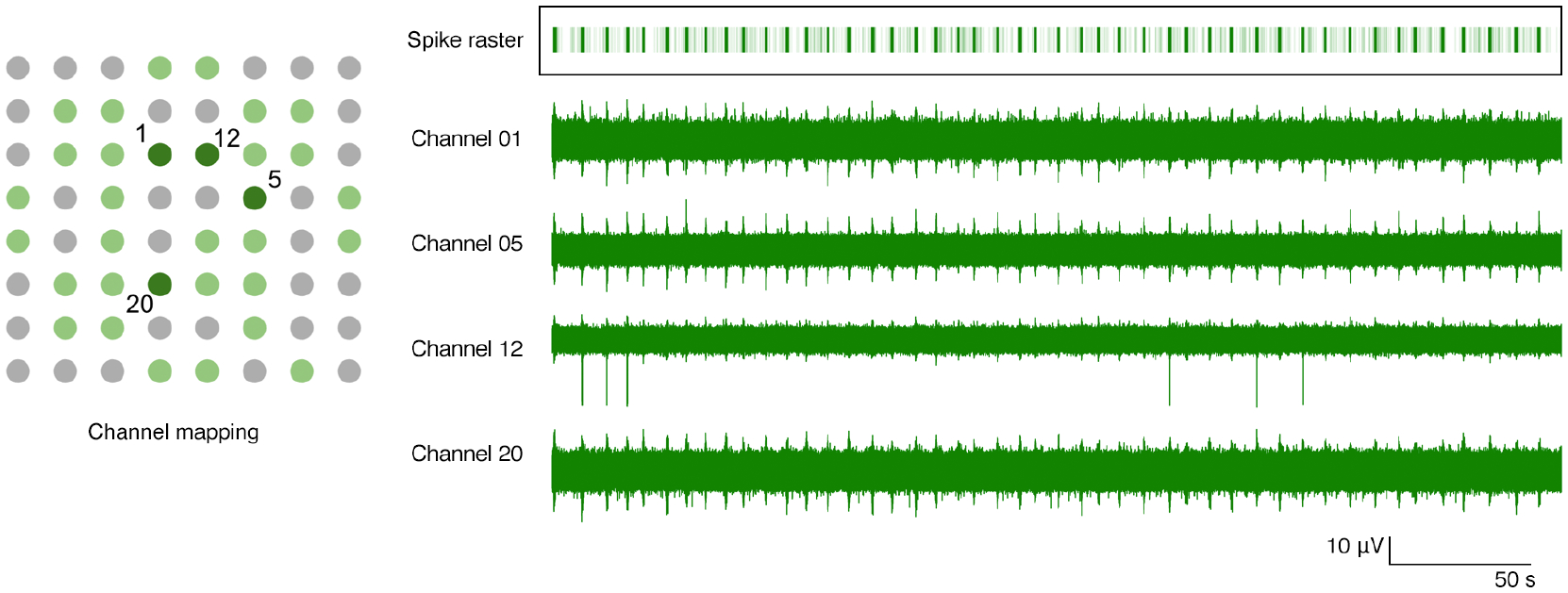
Channel mapping shows 64 electrodes in well B2 of the Axion plate. Light green dots are the 32 electrodes recorded by Piphys. Dark green dots mark channels 1, 5, 12 and 20 whose raw recording plots are on the right. The spike raster superimposes all the detected spikes in the shown channels. Each light green vertical line in the raster indicates a spike, and the dark green bar is the result of superimposing multiple spikes in the burst. The bars in the raster plot align with the bursts throughout these four channels. Burst population rate is 0.13 per second. The number of spikes in each burst is 55 ± 17.58.
To further characterize Piphys system’s performance, we compute the SNR of bursting activity by the following equation applied to the smoothed signal:
| (1) |
where μb and μn are the mean for the burst and baseline noise, respectively, σn is the standard deviation of the noise. In Figure 8, background signal in green represents the original recording. The signal in blue is the smoothed product by boxcar averaging with a window size of three times the standard deviation of the original. The median SNR across active channels is measured at 4.35 dB. The mean for baseline noise in the burst recording is around 2.13 μV RMS, consistent with the noise measurement for the experiments described in the Setup of validation and neural recording experiment section. Comparing the baseline noise to other systems, Intan and Axion, these experiments demonstrated that the Piphys system is sensitive and reliable in the relatively low amplitude neural signal recording range from tens of μV. In addition, with its open-source, light-weight, and remote monitoring capability through the IoT, Piphys adds unique value in extracellular electrophysiology.
Figure 8. Signal-to-noise ratio of the burst.
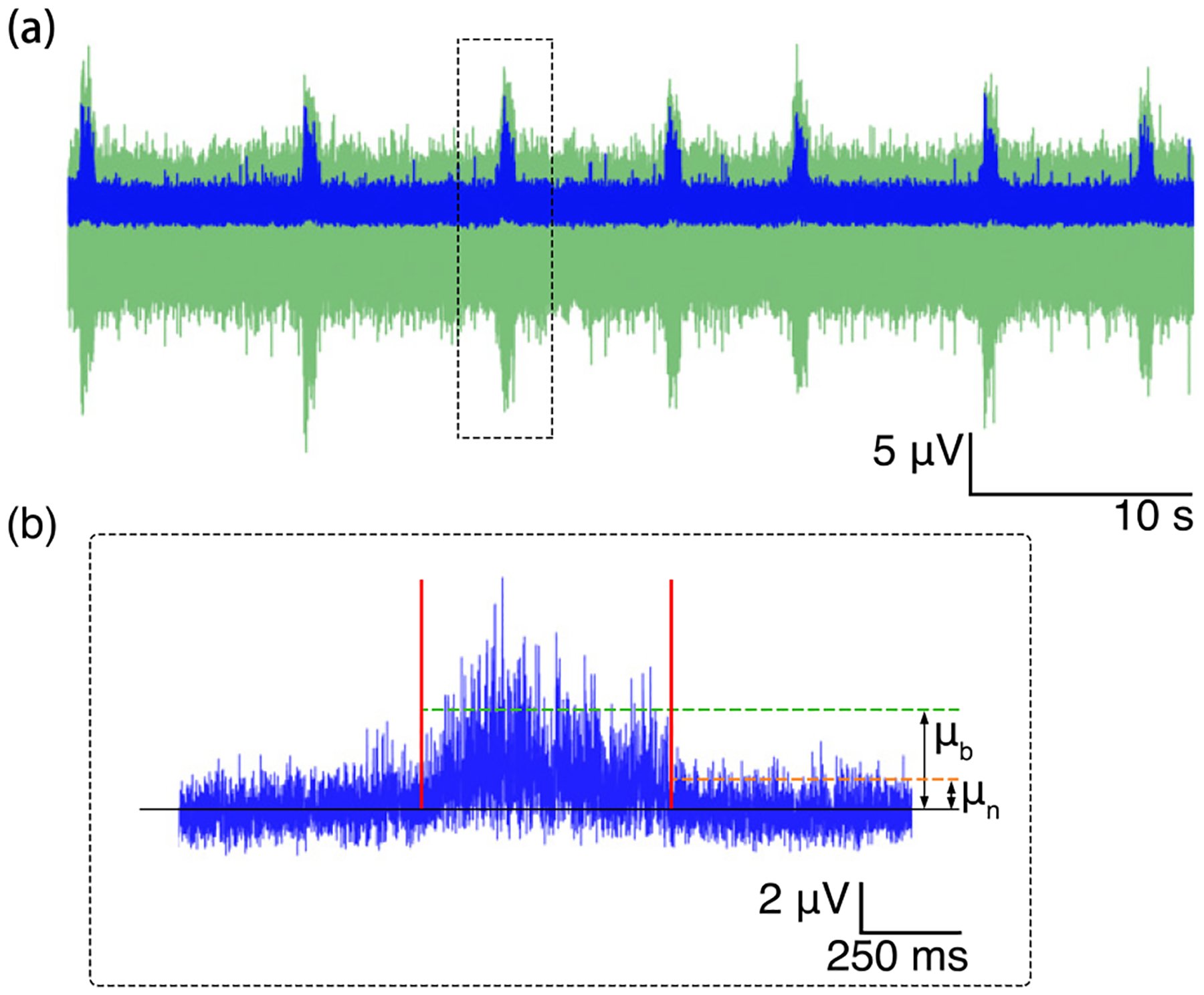
(a) Burst train (green) and the smoothed signal (blue). (b) Zoom in to the third smoothed burst showing means of the signal and the baseline noise for SNR calculation.
Discussion of Validation
The variation between Piphys and Axion shown in Figure 6 could be attributed to physical differences in the circuity and possible advanced filtering performed by Axion’s proprietary BioCore v4 chip §. The filtering could account for the smoothness and low variability of the signal (measured 1.12 ± 0.18 μV RMS noise baseline), resulting in a smaller number of identified firing events with a tighter distribution. Piphys and Intan systems both use the same amplifier chips (Intan RHD2000 series), where the optional on-chip filtering was disabled during recording ∥. The raw signal, therefore, has a larger noise margin (measured 3.21 ± 0.66 μV RMS noise baseline for Intan, 2.36 ± 0.4 μV RMS for Piphys), which may create more false-positive firing events. The tail of the amplitude distributions in Intan and Piphys is skewed towards lower-amplitude events, closer to the noise floor. The interspike intervals for Intan and Piphys register several events with near-zero intervals, likely suggesting false-positive spikes from noise contamination. Contamination from noise, which is likely symmetrical, could affect the shape of the mean waveform calculated by overlaying and averaging all registered spikes.
Overall, these results demonstrate that Piphys can record neural activity in a manner comparable to commercially available hardware and software.
Comparison to other platforms
Comparing electrophysiology platforms side by side is challenging because each system fits a specific niche and requirements for a particular workflow. Different platforms are targeted to particular problems and, therefore, have specific challenges and user needs. Piphys is intended to integration with other IoT sensors, and flexible recording equipment that can be used in a fleet for longitudinal study of many in vitro replicates. It should be noted that that the system proposed in this paper has an average signal to noise ratio (SNR) of 4.35 dB above the baseline noise in recording neuron burst, which is comparable with other similar systems [39, 40].
Table 1 summarizes electrophysiology systems comparable to Piphys. The Axion Maestro Edge is designed as an out-of-the-box benchtop electrophysiology system with maximum comfort and usability. Although it has the highest price per channel, it is also an incubator. The Intan RHD USB interface board and headstages require more effort to calibrate, ground, and shield. Unlike Axion, Intan designs and code are open source. Intan bioamplifier chips have been used in many open source systems, including Intsy, Willow, Open Ephys, and now Piphys. Both Intan and Axion systems provide valuable perspectives for comparison to Piphys. Axion produces the lowest noise baseline but has a different bioamplifier circuit. Piphys and Intan have the same bioamplifier chip; therefore Intan is a good reference for ensuring Piphys has the same noise floor and low EMI. Piphys and Intan RHD interface board differ in the way they sample the bioamplifier. Specifically, Intan has more stable sampling with FPGA, while Piphys samples the chip with a CPU, which has more clock jitter (discussed in Table S1, supplementary material). Overall the neural waveforms recorded on both systems are statistically comparable in shape for neural spikes for the detected neuron.
Table 1.
Comparison of Piphys features to several commercial and open source electrophysiology systems. Sampling Rate and Channels columns show the maximum numbers for all systems.
| Platform | System Noise (μV RMS) | Sample Rate (kHz) | Channels | Cost (USD) | Cost per Channel | Open Source | IoT |
|---|---|---|---|---|---|---|---|
| Piphys | 2.36 ± 0.4 † | 15 | 32 | $819 | $48 | Yes | Yes |
| Intsy [19] | 6–8 | 2 | 64 | $2,500 | $39 | Yes | No |
| Intan RHD ¶ | 3.21 ± 0.66 † | 30 | 256 | $10,295 | $40 | Yes | No |
| Open Ehpys [20, 41] | 2.4 * | 30 | 512 | $15,545 | $30 | Yes | No |
| Willow [22] | 3.9 | 30 | 1024 | $20,480 | $20 | Yes | No |
| Axion Maestro Edge + | 1.12 ± 0.18 † | 12.5 | 384 | $70,000 | $182 | No | No |
Noise shown on Open Ephys website is the amplifier input noise for Intan RHD2132 bioamplifier chip, not the whole system noise.
RMS noise recorded experimentally.
Other comparable platforms in the literature include Intsy, Willow, and Open Ephys. Intsy was designed for measuring gastrointestinal (EGG), cardiac (ECG), neural (EEG), and neuromuscular (EMG) signals [19]. Willow was designed for high channel count neural probes and resolved the need for many computers by writing data directly to hard drives [22]. Open Ephys is an alternative system to Intan integrating more features into their GUI for closed-loop experiments and plugin-based workflows [20] *. Noise measurements for Piphys, Intan, and Axion were experimentally recorded, while noise measurements for Intsy, Willow, and Open Ephys were cited. Intan claims 2.4 μV RMS as typical in the datasheet for their chips # which was likely inherited into Open Ephys documentation. The whole system noise for Open Ephys is not explicitly mentioned in the documentation.
5. Discussion
Remote longitudinal recording of neural circuits on an accessible platform will open up many exciting avenues for research into the physiology, organization, development, and adaptation of neural tissue. Integration with cloud software will allow in-depth experimentation and automation of analysis.
The proof of principle for Piphys has been shown on 2D cultures. As experiments with other devices have shown, it should be applicable to measurements of 3D brain organoids, which are becoming an increasingly popular model for studying human brain tissue development and function [5, 42–47]. One example application of Piphys would be monitoring how genotypes (such as NOTCH2NL variants) affect neural activity over the course of organoid development [48, 49]. More generally, IoT devices would allow less invasive and less laborious collection of longitudinal datasets of organoid development, to benchmark what wild-type organoid activity looks like throughout the first few months of growth. It would be interesting to compare whether different protocols and cell lines affect organoid activity over the course of development. IoT devices could be distributed and shared to compare whether organoid datasets are replicable and comparable between different labs, using the same low-cost hardware.
Many electrode probes have been designed to interface with tissues to provide measurement points for voltage recordings [17, 35, 36, 50, 51]. Future work on Piphys would involve expanding the number of different electrodes types for long-term culture of the biological sample through collaborations with other research groups.
Future work on Piphys also includes increasing sampling rate and precision of timing in between samples. Currently, the Raspberry Pi CPU samples the Intan RHD2132 bioamplifier chip, and the sampling rates are limited by the CPU’s ability to multitask. Future solutions may involve adding another CPU or FPGA to the hardware shield. The platform will continue to be improved, and its modularity allows adapting hardware and software components as different needs arise.
The current proof of concept design is based on a Raspberry Pi chip and uses one 32 channel chip attached to one of the SPI ports. The system can be easily extended to sample 64 channels (one 32 channel Intan chip for each of the two SPI ports available on the Raspberry Pi). The channel number can be doubled (128 channels) if the design would include an FPGA and alternative Intan chips that have 64 channels/chip (Intan RHD2164). However, the true scalability advantage of the proposed system lies in its open source and open hardware architecture. If the number of channels is insufficient, the shield board could be modified to accept multiple Raspberry Pi’s, therefore, increase the number of channels.
Piphys is the only electrophysiology device that supports Internet of Things (IoT) software integration out of the box. The IoT hardware modules and cloud software allow for horizontal scalability, enabling long-term observations of development, organization, and neural activity at scale, and integration with other IoT sensors. Piphys has a low entry cost, and the cost per channel can also be significantly lowered by increasing the number of channels supported per device. This would be accomplished by engineering an inexpensive FPGA into the controller shield to sample multiple bioamplifier chips and buffer those readings for the Pi. Piphys can have a large cost reduction if extra specialty connectors and adapters are removed (cutting roughly $300) and it is fitted with a USB cable which is less expensive.
The signal-to-noise ratio could be improved by enabling and tuning on-chip filtering, and improving Faraday cage shielding. In vitro cultures typically fire with amplitudes between 10 – 40 μV [5, 7, 52]. They demand sensitive recording equipment, as an increase of just a few μV in noise for spikes on the lower end of the spectrum can be considered a non-trivial variable.
Overall, the open source Piphys design, programmability, and extreme flexibility of the Raspberry Pi significantly lowers the entry barrier of the electrophysiology system, providing an opportunity for broader applications in education and research.
6. Methods
Tissue source
De-identified tissue samples were collected with previous patient consent in strict observance of the legal and institutional ethical regulations. Protocols were approved by the Human Gamete, Embryo, and Stem Cell Research Committee (institutional review board) at the University of California, San Francisco.
Primary neuron culture
Prior to cell culture, the electrode surfaces of 6-well Axion plates (Axion Biosystems, CytoView MEA 6) were coated with 10 mg/mL poly-D-lysine (Sigma, P7280) at room temperature overnight. The following day, plates were rinsed 4 times with water and dried at room temperature. Primary cells were obtained from human brain tissue at gestational week 21. Briefly, cortical tissue was cut into small pieces, incubated in 0.25% trypsin (Gibco, 25200056) for 30 minutes, then triturated in the presence of 10 mg/mL DNAse (Sigma Aldrich, DN25) and passed through a 40 μm cell strainer. Cells were spun down and resuspended in BrainPhys (StemCell Technologies, 05790) supplemented with B27 (Thermo Fisher, 17504001), N2 (Thermo Fisher, 17502001), and penicillin-streptomycin (Thermo Fisher, 15070063), then diluted to a concentration of 8,000,000 cells/mL. Laminin (Thermo Fisher, 23017015, final concentration 50 μg/mL) was added to the final aliquot of cells, and a 10 μL drop of cells was carefully pipetted directly onto the dried, PDL-coated electrodes, forming an intact drop. The plate was transferred to a 37 °C, 5% CO2 incubator for 1 hour to allow the cells to settle, then 200 μL of supplemented BrainPhys media was gently added to the drops. The following day, another 800 μL of media was added, and each well was kept at 1 mL media for the duration of the cultures, with half the volume exchanged with fresh media every other day. Activity was first observed at 14 days in culture, and the second recordings were performed on day 42 of culture.
Setup of validation and neural recording experiment
The power supplied to the Raspberry Pi is through a mains adapter plugged into the wall outlet. To reduce environmental noise and maximize the signal-to-noise ratio (SNR), we use a Faraday cage during recording. The Faraday cage is made of 1 mm thick steel and connected to the wall outlet ground.
For noise measurement benchmarks on Piphys, an empty Axion plate was filled with the same media used in cell culture and placed in the Faraday cage. The noise baseline of this media-only system was an average of 2.36 ± 0.4 μV RMS for all the channels with software filters. Comparison of the baseline noise we measured for Piphys, Intan, and Axion is in Table 1.
During the experiment, the systems were compared by measuring the same neural culture on the same plate within a similar time frame. Recordings occurred within 1 to 3 hours of each other. A 300–6000 Hz 3rd order Butterworth bandpass filter was used to attenuate frequency components outside the neural activity range after the recording. Data was analyzed by a spike sorting algorithm and shown side by side in Figure 6 over an identical time length.
Instructions and source files for construction of Piphys hardware (Figure 2) and software (Figure 3) are available open source on GitHub ††. All files are provided ‘as is’ and end-users are encouraged to freely use and adapt the system for their own application-specific protocols.
Figure 2. Piphys hardware components.
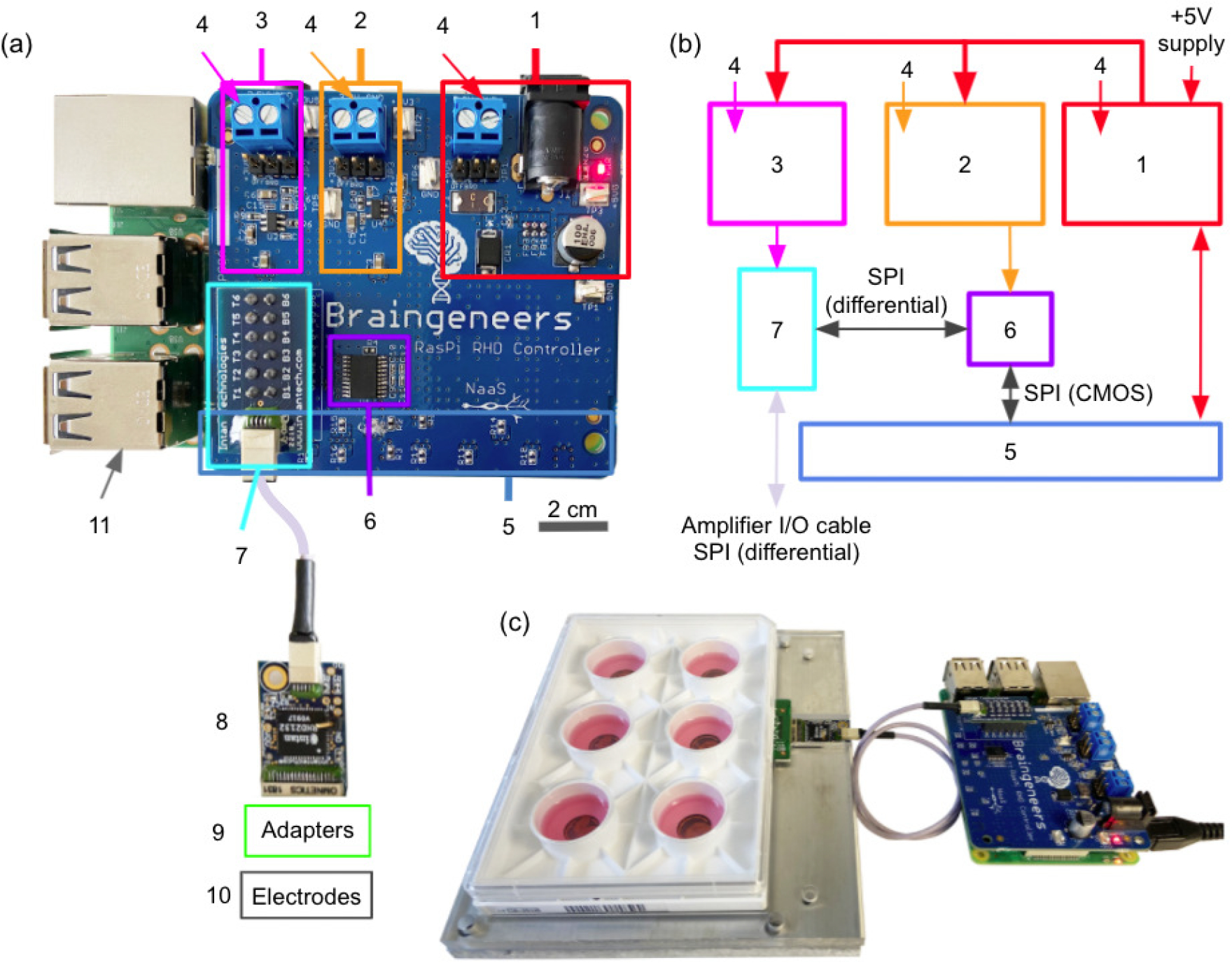
(a) Expansion shield (blue board) attached on top of Raspberry Pi (green board). (b) Logic level connection. (c) Example interface with standard 6-well electrode plate. (1) +5V logic, (2) +3.3V logic, (3) +3.5V logic, (4) External supply inputs, (5) Raspberry Pi input/output pins (bottom), (6) LVDS converter, (7) Intan RHD adapter, (8) Intan RHD 32-channel recording headstage containing Intan RHD2132 bioamplifier chip, (9) Optional adapter board to electrodes, (10) Multiple electrode types possible, (11) Raspberry Pi computer (bottom).
Circuit board design, reduction of noise and EMI
The printed circuit board was designed in Autodesk Eagle. The board has four layers of copper. The top and bottom layers of the board are GND, while the two layers inside are signal and power. Every signal via has a ground via next to it to sink EMI as signals switch layers. The layout of the circuit board is done in modules. Via stitching was done around the perimeter and throughout the board area to separate modules (highlighted by squares in Figure 2) and fill in areas with no components. The amplifier chip and Raspberry Pi computer are separated by a cable such that noise from the computer would not interfere with the sensitive neural signal recording. During data acquisition, all of the electronics and biology were shielded by a 1 mm thick steel faraday cage.
Cloud services integration
We deployed servers and cloud computing platforms to achieve permanent data storage and messaging between the local device and the dashboard. We used Remote Dictionary Server (Redis), Amazon Web Services Internet of Things (AWS IoT), and Simple Storage Service (S3). All services (except AWS IoT) are platform agnostic and can be hosted anywhere.
For our particular experimental setup, Redis and S3 were hosted on the Pacific Research Platform (PRP) [30]. The Internet of Things (IoT) service with MQTT messaging and device management was coordinated through Amazon Web Services (AWS). The dashboard was hosted on a server at UC Santa Cruz.
The thresholding spike detection shown in the dashboard (Figure 4) runs inside a Docker container, which reads from the Piphys data Redis stream and writes to another Redis stream shown in the dashboard. In our case the Docker, the Redis service and the dashboard run on the Pacific Research Platform (PRP). However, this could be transferred to any cloud storage provider (e.g., AWS). The IoT architecture of these cloud services is explicitly described in [53].
Figure 4. Dashboard.
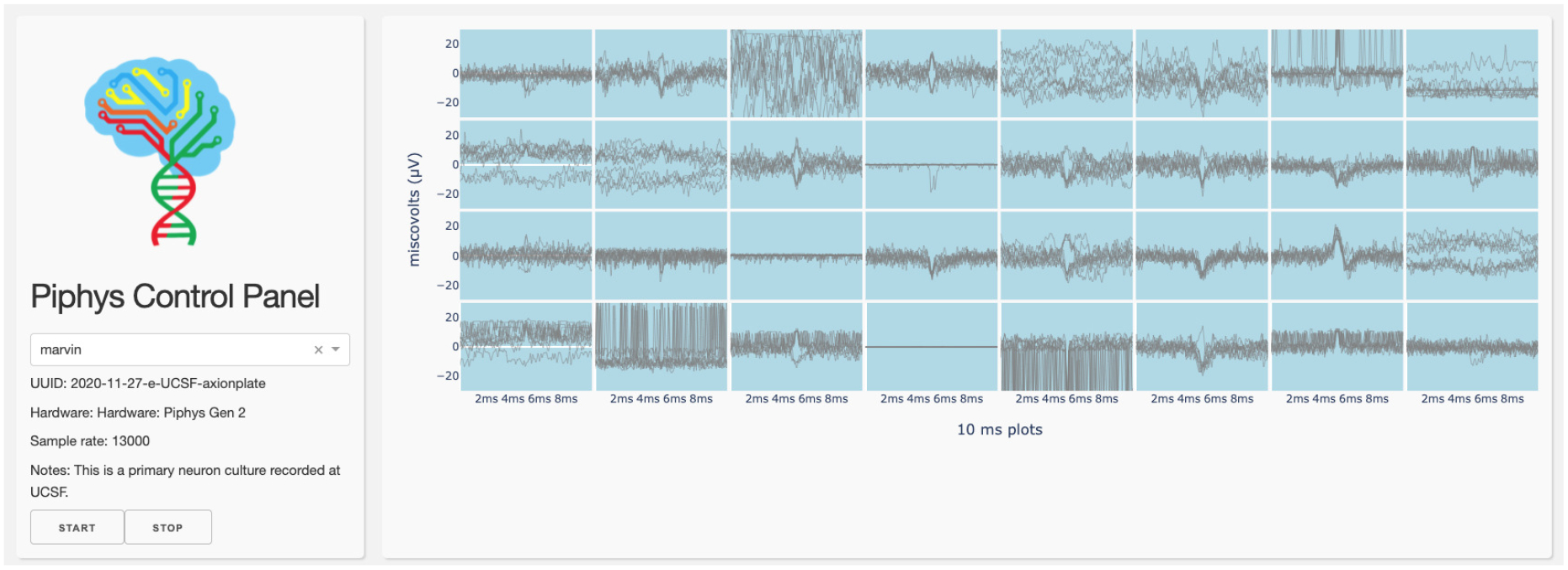
A control panel interface is displayed through the browser running spike detection by thresholding.
Redis, near real-time data stream
Neuronal action potential recording with a high sample rate and multiple channels requires a high throughput pipeline to make near real-time streaming possible. Remote Dictionary Server (Redis) is a good choice for the implementation of this objective. It is a high-speed cloud-based data structure store that can be used as a cache, message broker, and database. Based on benchmarking results, Redis can handle hundreds of thousands of requests per second. The highest data rate for every push from Piphys system to Redis is 7.68 Mb (0.96 MB) for each second (32 channels × 15 kHz sampling rate × 16 bits/sample).Pi data stream to Redis requires the network bandwidth to be at least 7.68 Mbps so that uploading to the Dashboard through Redis can be uninterrupted.
Internet of Things (IoT) communication
The dashboard is programmed to be an IoT device that sends Message Queuing Telemetry Transport (MQTT) messages to control and check the Piphys system. In response, the Piphys subscribes to a particular MQTT topic to wait for instructions. The AWS IoT supports the communication of hundreds of devices, making the Piphys system’s extension to a large scale possible in the future.
Simple Storage Service (S3)
The Simple Storage Service (S3) is the final data storage location. S3 is accessible from anywhere at any time on the internet. It supports both management from a terminal session and integration to a custom web browser application. After each experiment, a new identifier will be updated on the dashboard. When a user asks for a specific experiment result, the dashboard can pull the corresponding data file directly from S3 for visualization.
Supplementary Material
Figure 1.
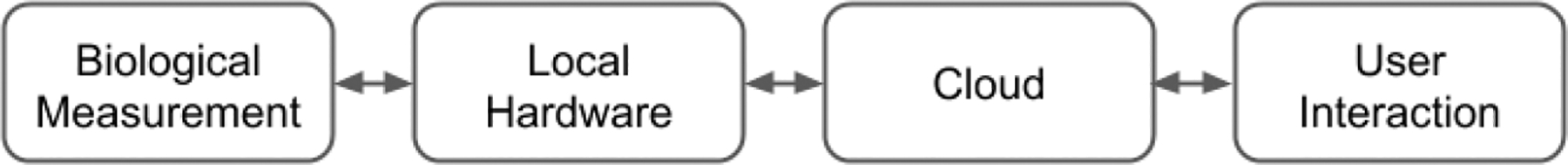
Cloud-based experiment paradigm: biological measurement and local hardware are presented to the user through the cloud, such that experiment management and control can be administrated remotely and may be automated by a computer program.
Acknowledgements
This work is supported by the Schmidt Futures Foundation SF 857 (D.H.). Research reported in this publication was also supported by the National Institute Of Mental Health of the National Institutes of Health (NIH) under award number R01MH120295 (S.R.S.), the National Science Foundation under award number NSF 2034037 (M.T.), the National Defense Science and Engineering Graduate Fellowship (00002116, M.G.K.), and gifts from Schmidt Futures and the William K. Bowes Jr Foundation (T.J.N). K.V. was supported by grant T32HG008345 from the National Human Genome Research Institute (NHGRI), part of the National Institutes of Health, USA. D.H. is a Howard Hughes Medical Institute Investigator.
We are thankful to the Pacific Research Platform, supported by the National Science Foundation under award numbers CNS-1730158, ACI-1540112, ACI-1541349, OAC-1826967, the University of California Office of the President, and the University of California San Diego’s California Institute for Telecommunications and Information Technology/Qualcomm Institute. Thanks to CENIC for the 100 Gpbs networks.
We would like to thank Erik Jung for encouragement and organizational support, Sergio A. Cordero and Pattawong Pansodtee for help with early electronics prototypes and CNC fabrication of the plate holder.
Footnotes
Competing interests
The authors declare no conflict of interest.
References
- [1].Hansel D, Mato G, and Meunier C. Synchrony in Excitatory Neural Networks. Neural Computation, 7(2):307–337, March 1995. Publisher: MIT Press. [DOI] [PubMed] [Google Scholar]
- [2].Beggs John M. and Plenz Dietmar. Neuronal Avalanches in Neocortical Circuits. Journal of Neuroscience, 23(35):11167–11177, December 2003. Publisher: Society for Neuroscience Section: Behavioral/Systems/Cognitive. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Poli Daniele, Pastore Vito P., and Massobrio Paolo. Functional connectivity in in vitro neuronal assemblies. Frontiers in Neural Circuits, 9, 2015. Publisher: Frontiers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Huang Yu-Ting, Chang Yu-Lin, Chen Chun-Chung, Lai Pik-Yin, and Chan CK. Positive feedback and synchronized bursts in neuronal cultures. PLOS ONE, 12(11):e0187276, November 2017. Publisher: Public Library of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Trujillo Cleber A., Gao Richard, Negraes Priscilla D., Gu Jing, Buchanan Justin, Preissl Sebastian, Wang Allen, Wu Wei, Haddad Gabriel G., Chaim Isaac A., Domissy Alain, Vandenberghe Matthieu, Devor Anna, Yeo Gene W., Voytek Bradley, and Muotri Alysson R.. Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development. Cell Stem Cell, 25(4):558–569.e7, October 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sharf Tal, van der Molen Tjitse, Guzman Elmer, Glasauer Stella M. K., Luna Gabriel, Cheng Zhouwei, Audouard Morgane, Ranasinghe Kamalini G., Kudo Kiwamu, Nagarajan Srikantan S., Tovar Kenneth R., Petzold Linda R., Hansma Paul K., and Kosik Kenneth S.. Intrinsic network activity in human brain organoids. bioRxiv, page 2021.01.28.428643, January 2021. Publisher: Cold Spring Harbor Laboratory Section: New Results. [Google Scholar]
- [7].Negri Joseph, Menon Vilas, and Young-Pearse Tracy L.. Assessment of Spontaneous Neuronal Activity In Vitro Using Multi-Well Multi-Electrode Arrays: Implications for Assay Development. eNeuro, 7(1), January 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yada Yuichiro, Mita Takeshi, Sanada Akihiro, Yano Ryuichi, Kanzaki Ryohei, Bakkum Douglas J., Hierlemann Andreas, and Takahashi Hirokazu. Development of neural population activity toward self-organized criticality. Neuroscience, 343:55–65, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zafeiriou Maria-Patapia, Bao Guobin, Hudson James, Halder Rashi, Blenkle Alica, Schreiber Marie-Kristin, Fischer Andre, Schild Detlev, and Zimmermann Wolfram-Hubertus. Developmental GABA polarity switch and neuronal plasticity in Bioengineered Neuronal Organoids. Nature Communications, 11(1):3791, July 2020. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zenke Friedemann and Gerstner Wulfram. Hebbian plasticity requires compensatory processes on multiple timescales. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 372(1715), 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Killian Nathaniel J., Vernekar Varadraj N., Potter Steve M., and Vukasinovic Jelena. A Device for Long-Term Perfusion, Imaging, and Electrical Interfacing of Brain Tissue In vitro. Frontiers in Neuroscience, 10, 2016. Publisher: Frontiers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang Yu Shrike, Aleman Julio, Shin Su Ryon, Kilic Tugba, Kim Duckjin, Mousavi Shaegh Seyed Ali, Massa Solange, Riahi Reza, Chae Sukyoung, Hu Ning, Avci Huseyin, Zhang Weijia, Silvestri Antonia, Nezhad Amir Sanati, Manbohi Ahmad, Ferrari Fabio De, Polini Alessandro, Calzone Giovanni, Shaikh Noor, Alerasool Parissa, Budina Erica, Kang Jian, Bhise Nupura, Ribas João, Pourmand Adel, Skardal Aleksander, Shupe Thomas, Bishop Colin E., Dokmeci Mehmet Remzi, Atala Anthony, and Khademhosseini Ali. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proceedings of the National Academy of Sciences, 114(12):E2293–E2302, March 2017. Publisher: National Academy of Sciences Section: PNAS Plus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dhawale Ashesh K, Poddar Rajesh, Wolff Steffen BE, Normand Valentin A, Kopelowitz Evi, and Ölveczky Bence P. Automated long-term recording and analysis of neural activity in behaving animals. eLife, 6:e27702, September 2017. Publisher: eLife Sciences Publications, Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Raghu Maithra and Schmidt Eric. A Survey of Deep Learning for Scientific Discovery. arXiv:2003.11755 [cs, stat], March 2020. arXiv: 2003.11755. [Google Scholar]
- [15].Nasiotis Konstantinos, Cousineau Martin, Tadel François, Peyrache Adrien, Leahy Richard M., Pack Christopher C., and Baillet Sylvain. Integrated open-source software for multiscale electrophysiology. Scientific Data, 6(1):231, October 2019. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Siegle Joshua H, Hale Gregory J, Newman Jonathan P, and Voigts Jakob. Neural ensemble communities: open-source approaches to hardware for large-scale electrophysiology. Current Opinion in Neurobiology, 32:53–59, June 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Putzeys Jan, Raducanu Bogdan C., Carton Alain, Ceulaer Jef De, Karsh Bill, Siegle Joshua H., Van Helleputte Nick, Harris Timothy D., Dutta Barundeb, Musa Silke, and Lopez Carolina Mora. Neuropixels Data-Acquisition System: A Scalable Platform for Parallel Recording of 10 000+ Electrophysiological Signals. IEEE Transactions on Biomedical Circuits and Systems, 13(6):1635–1644, December 2019. [DOI] [PubMed] [Google Scholar]
- [18].Abe Taiga, Kinsella Ian, Saxena Shreya, Paninski Liam, and Cunningham John P. Neuroscience Cloud Analysis As a Service. page 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Erickson Jonathan C., Hayes James A., Bustamante Mauricio, Joshi Rajwol, Rwagaju Alfred, Paskaranandavadivel Niranchan, and Angeli Timothy R.. Intsy: a low-cost, open-source, wireless multi-channel bioamplifier system. Physiological Measurement, 39(3):035008, March 2018. Publisher: IOP Publishing. [DOI] [PubMed] [Google Scholar]
- [20].Siegle Joshua H, López Aarón Cuevas, Patel Yogi A, Abramov Kirill, Ohayon Shay, and Voigts Jakob. Open Ephys: an open-source, plugin-based platform for multichannel electrophysiology. Journal of Neural Engineering, 14(4):045003, August 2017. [DOI] [PubMed] [Google Scholar]
- [21].Newman Jonathan Paul, Zeller-Townson Riley, Fong Ming-fai, Desai Sharanya Arcot, Gross Robert E., and Potter Steve M.. Closed-Loop, Multichannel Experimentation Using the Open-Source NeuroRighter Electrophysiology Platform. Frontiers in Neural Circuits, 6, 2013. Publisher: Frontiers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kinney Justin P., Bernstein Jacob G., Meyer Andrew J., Barber Jessica B., Bolivar Marti, Newbold Bryan, Scholvin Jorg, Moore-Kochlacs Caroline, Wentz Christian T., Kopell Nancy J., and Boyden Edward S.. A direct-to-drive neural data acquisition system. Frontiers in Neural Circuits, 9, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Garma Leonardo D., Matino Laura, Melle Giovanni, Moia Fabio, De Angelis Francesco, Santoro Francesca, and Dipalo Michele. Cost-effective and multifunctional acquisition system for in vitro electrophysiological investigations with multi-electrode arrays. PLOS ONE, 14(3):e0214017, March 2019. Publisher: Public Library of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hussein AF, kumar NA, Burbano-Fernandez M, Ramírez-González G, Abdulhay E, and De Albuquerque VHC. An Automated Remote Cloud-Based Heart Rate Variability Monitoring System. IEEE Access, 6:77055–77064, 2018. Conference Name: IEEE Access. [Google Scholar]
- [25].Yang Zhe, Zhou Qihao, Lei Lei, Zheng Kan, and Xiang Wei. An IoT-cloud Based Wearable ECG Monitoring System for Smart Healthcare. Journal of Medical Systems, 40(12):286, October 2016. [DOI] [PubMed] [Google Scholar]
- [26].Hassanalieragh M, Page A, Soyata T, Sharma G, Aktas M, Mateos G, Kantarci B, and Andreescu S. Health Monitoring and Management Using Internet-of-Things (IoT) Sensing with Cloud-Based Processing: Opportunities and Challenges. In 2015 IEEE International Conference on Services Computing, pages 285–292, June 2015. [Google Scholar]
- [27].Wai AAP, Dajiang H, and Huat NS. IoT-enabled multimodal sensing headwear system. In 2018 IEEE 4th World Forum on Internet of Things (WF-IoT), pages 286–290, February 2018. [Google Scholar]
- [28].Hermiz J, Rogers N, Kaestner E, Ganji M, Cleary D, Snider J, Barba D, Dayeh S, Halgren E, and Gilja V. A clinic compatible, open source electrophysiology system. In 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pages 4511–4514, August 2016. ISSN: 1558-4615. [DOI] [PubMed] [Google Scholar]
- [29].Yong PK and Wei Ho ET. Streaming brain and physiological signal acquisition system for IoT neuroscience application. In 2016 IEEE EMBS Conference on Biomedical Engineering and Sciences (IECBES), pages 752–757, December 2016. [Google Scholar]
- [30].Smarr Larry, Crittenden Camille, DeFanti Thomas, Graham John, Mishin Dmitry, Moore Richard, Papadopoulos Philip, and Würthwein Frank. The Pacific Research Platform: Making High-Speed Networking a Reality for the Scientist. In Proceedings of the Practice and Experience on Advanced Research Computing, PEARC ‘18, pages 1–8, Pittsburgh, PA, USA, July 2018. Association for Computing Machinery. [Google Scholar]
- [31].Wagenaar D, DeMarse TB, and Potter SM. MeaBench: A toolset for multi-electrode data acquisition and on-line analysis. In Conference Proceedings. 2nd International IEEE EMBS Conference on Neural Engineering, 2005., pages 518–521, March 2005. ISSN: 1948-3554. [Google Scholar]
- [32].Chung Jason E., Magland Jeremy F., Barnett Alex H., Tolosa Vanessa M., Tooker Angela C., Lee Kye Y., Shah Kedar G., Felix Sarah H., Frank Loren M., and Greengard Leslie F.. A Fully Automated Approach to Spike Sorting. Neuron, 95(6):1381–1394.e6, September 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yger Pierre, Spampinato Giulia LB, Esposito Elric, Lefebvre Baptiste, Deny Stéphane, Gardella Christophe, Stimberg Marcel, Jetter Florian, Zeck Guenther, Picaud Serge, Duebel Jens, and Marre Olivier. A spike sorting toolbox for up to thousands of electrodes validated with ground truth recordings in vitro and in vivo. eLife, 7:e34518, March 2018. Publisher: eLife Sciences Publications, Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lee JinHyung, Mitelut Catalin, Shokri Hooshmand, Kinsella Ian, Dethe Nishchal, Wu Shenghao, Li Kevin, Reyes Eduardo B., Turcu Denis, Batty Eleanor, Kim Young J., Brackbill Nora, Kling Alexandra, Goetz Georges, Chichilnisky EJ, Carlson David, and Paninski Liam. YASS: Yet Another Spike Sorter applied to large-scale multi-electrode array recordings in primate retina. bioRxiv, page 2020.03.18.997924, March 2020. Publisher: Cold Spring Harbor Laboratory Section: New Results. [Google Scholar]
- [35].Park Yoonseok, Franz Colin K., Ryu Hanjun, Luan Haiwen, Cotton Kristen Y., Kim Jong Uk, Chung Ted S., Zhao Shiwei, Vazquez-Guardado Abraham, Yang Da Som, Li Kan, Avila Raudel, Phillips Jack K., Quezada Maria J., Jang Hokyung, Kwak Sung Soo, Won Sang Min, Kwon Kyeongha, Jeong Hyoyoung, Bandodkar Amay J., Han Mengdi, Zhao Hangbo, Osher Gabrielle R., Wang Heling, Lee KunHyuck, Zhang Yihui, Huang Yonggang, Finan John D., and Rogers John A.. Three-dimensional, multifunctional neural interfaces for cortical spheroids and engineered assembloids. Science Advances, 7(12):eabf9153, March 2021. Publisher: American Association for the Advancement of Science Section: Research Article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yang Long, Lee Kwang, Villagracia Jomar, and Masmanidis Sotiris C.. Open source silicon microprobes for high throughput neural recording. Journal of Neural Engineering, 17(1):016036, January 2020. Publisher: IOP Publishing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wagenaar Daniel A., Madhavan Radhika, Pine Jerome, and Potter Steve M.. Controlling Bursting in Cortical Cultures with Closed-Loop Multi-Electrode Stimulation. The Journal of Neuroscience, 25(3):680–688, January 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bakkum Douglas J., Radivojevic Milos, Frey Urs, Franke Felix, Hierlemann Andreas, and Takahashi Hirokazu. Parameters for burst detection. Frontiers in Computational Neuroscience, 7, 2014. Publisher: Frontiers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Regalia Giulia, Biffi Emilia, Ferrigno Giancarlo, and Pedrocchi Alessandra. A Low-Noise, Modular, and Versatile Analog Front-End Intended for Processing In Vitro Neuronal Signals Detected by Microelectrode Arrays. Computational Intelligence and Neuroscience, 2015:1–15, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tambaro Mattia, Bisio Marta, Maschietto Marta, Leparulo Alessandro, and Vassanelli Stefano. FPGA Design Integration of a 32-Microelectrodes Low-Latency Spike Detector in a Commercial System for Intracortical Recordings. Digital, 1(1):34–53, March 2021. Number: 1 Publisher: Multidisciplinary Digital Publishing Institute. [Google Scholar]
- [41].Dunham James P., Sales Anna C., and Pickering Anthony E.. Ultrasound-guided, open-source microneurography: Approaches to improve recordings from peripheral nerves in man. Clinical Neurophysiology, 129(11):2475–2481, November 2018. [DOI] [PubMed] [Google Scholar]
- [42].Eiraku Mototsugu, Watanabe Kiichi, Matsuo-Takasaki Mami, Kawada Masako, Yonemura Shigenobu, Matsumura Michiru, Wataya Takafumi, Nishiyama Ayaka, Muguruma Keiko, and Sasai Yoshiki. Self-Organized Formation of Polarized Cortical Tissues from ESCs and Its Active Manipulation by Extrinsic Signals. Cell Stem Cell, 3(5):519–532, November 2008. Publisher: Elsevier. [DOI] [PubMed] [Google Scholar]
- [43].Lancaster Madeline A., Renner Magdalena, Martin Carol-Anne, Wenzel Daniel, Bicknell Louise S., Hurles Matthew E., Homfray Tessa, Penninger Josef M., Jackson Andrew P., and Knoblich Juergen A.. Cerebral organoids model human brain development and microcephaly. Nature, 501(7467):373–379, September 2013. Number: 7467 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Quadrato Giorgia, Nguyen Tuan, Macosko Evan Z., Sherwood John L., Yang Sung Min, Berger Daniel, Maria Natalie, Scholvin Jorg, Goldman Melissa, Kinney Justin, Boyden Edward S., Lichtman Jeff, Williams Ziv M., McCarroll Steven A., and Arlotta Paola. Cell diversity and network dynamics in photosensitive human brain organoids. Nature, 545(7652):48–53, May 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shcheglovitov Alex, Wang Yueqi, Bell Laura, Russell Chad, Armstrong Celeste, and Spampanato Jay. Human cortical organoids from single iPSC-derived neural rosettes for studying human cortical development and disorders. The FASEB Journal, 33(1_supplement):205.3–205.3, April 2019. Publisher: Federation of American Societies for Experimental Biology. [Google Scholar]
- [46].Sakaguchi Hideya, Ozaki Yuki, Ashida Tomoka, Matsubara Takayoshi, Oishi Naotaka, Kihara Shunsuke, and Takahashi Jun. Self-Organized Synchronous Calcium Transients in a Cultured Human Neural Network Derived from Cerebral Organoids. Stem Cell Reports, 13(3):458–473, June 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yakoub Abraam M.. Cerebral organoids exhibit mature neurons and astrocytes and recapitulate electrophysiological activity of the human brain. Neural Regeneration Research, 14(5):757, May 2019. Publisher: Wolters Kluwer – Medknow Publications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pollen Alex A., Bhaduri Aparna, Andrews Madeline G., Nowakowski Tomasz J., Meyerson Olivia S., Mostajo-Radji Mohammed A., Di Lullo Elizabeth, Alvarado Beatriz, Bedolli Melanie, Dougherty Max L., Fiddes Ian T., Kronenberg Zev N., Shuga Joe, Leyrat Anne A., West Jay A., Bershteyn Marina, Lowe Craig B., Pavlovic Bryan J., Salama Sofie R., Haussler David, Eichler Evan E., and Kriegstein Arnold R.. Establishing Cerebral Organoids as Models of Human-Specific Brain Evolution. Cell, 176(4):743–756.e17, February 2019. Publisher: Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Fiddes Ian T., Lodewijk Gerrald A., Mooring Meghan, Bosworth Colleen M., Ewing Adam D., Mantalas Gary L., Novak Adam M., van den Bout Anouk, Bishara Alex, Rosenkrantz Jimi L., Lorig-Roach Ryan, Field Andrew R., Haeussler Maximilian, Russo Lotte, Bhaduri Aparna, Nowakowski Tomasz J., Pollen Alex A., Dougherty Max L., Nuttle Xander, Addor Marie-Claude, Zwolinski Simon, Katzman Sol, Kriegstein Arnold, Eichler Evan E., Salama Sofie R., Jacobs Frank M. J., and Haussler David. Human-Specific NOTCH2NL Genes Affect Notch Signaling and Cortical Neurogenesis. Cell, 173(6):1356–1369.e22, May 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Li Qiang, Nan Kewang, Floch Paul Le, Lin Zuwan, Sheng Hao, and Liu Jia. Cyborg Organoids: Implantation of Nanoelectronics via Organogenesis for Tissue-Wide Electrophysiology. bioRxiv, page 697664, July 2019. Publisher: Cold Spring Harbor Laboratory Section: New Results. [DOI] [PubMed] [Google Scholar]
- [51].Lee Jung Min, Hong Guosong, Lin Dingchang, Schuhmann Thomas G., Sullivan Andrew T., Viveros Robert D., Park Hong-Gyu, and Lieber Charles M.. Nanoenabled Direct Contact Interfacing of Syringe-Injectable Mesh Electronics. Nano Letters, 19(8):5818–5826, August 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Schmunk Galina, Kim Chang N., Soliman Sarah S., Keefe Matthew G., Bogdanoff Derek, Tejera Dario, Ziffra Ryan S., Shin David, Allen Denise E., Chhun Bryant B., McGinnis Christopher S., Winkler Ethan A., Abla Adib A., Chang Edward F., Gartner Zev J., Mehta Shalin B., Piao Xianhua, Hengen Keith B., and Nowakowski Tomasz J.. Human microglia upregulate cytokine signatures and accelerate maturation of neural networks. bioRxiv, page 2020.03.24.006874, March 2020. Publisher: Cold Spring Harbor Laboratory Section: New Results. [Google Scholar]
- [53].Parks David F., Voitiuk Kateryna, Geng Jinghui, Elliott Matthew A. T., Keefe Matthew G., Jung Erik A., Robbins Ash, Baudin Pierre V., Ly Victoria T., Hawthorne Nico, Yong Dylan, Sanso Sebastian E., Rezaee Nick, Sevetson Jess, Seiler Spencer T., Currie Rob, Hengen Keith B., Nowakowski Tomasz J., Salama Sofie R., Teodorescu Mircea, and Haussler David. Internet of Things Architecture for High Throughput Biology. page 2021.07.29.453595, August 2021. Company: Cold Spring Harbor Laboratory Distributor: Cold Spring Harbor Laboratory Label: Cold Spring Harbor Laboratory Section: New Results Type: article. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


