Abstract
Cell-free DNA(cfDNA) methylation profiling is considered promising and potentially reliable for liquid biopsy to study progress of diseases and develop reliable and consistent diagnostic and prognostic biomarkers. There are several different mechanisms responsible for the release of cfDNA in blood plasma, and henceforth it can provide information regarding dynamic changes in the human body. Due to the fragmented nature, low concentration of cfDNA, and high background noise, there are several challenges in its analysis for regular use in diagnosis of cancer. Such challenges in the analysis of the methylation profile of cfDNA are further aggravated due to heterogeneity, biomarker sensitivity, platform biases, and batch effects. This review delineates the origin of cfDNA methylation, its profiling, and associated computational problems in analysis for diagnosis. Here we also contemplate upon the multi-marker approach to handle the scenario of cancer heterogeneity and explore the utility of markers for 5hmC based cfDNA methylation pattern. Further, we provide a critical overview of deconvolution and machine learning methods for cfDNA methylation analysis. Our review of current methods reveals the potential for further improvement in analysis strategies for detecting early cancer using cfDNA methylation.
Keywords: Cell free DNA, Cancer heterogeneity, Diagnosis, Computation
Abbreviations: cfDNA, cell free DNA; ctDNA, circulating tumor DNA; MSRE, methylation sensitive restriction enzymes; HELP-seq, HpaII-tiny fragment Enrichment by Ligation-mediated PCR sequencing; MSCC, Methylation Sensitive Cut Counting; scCGI, methylated CGIs at single cell level; WGBS, Whole Genome Bisulfite Sequencing; RRBS, Reduced-Representation Bisulfite Sequencing; MCTA-seq, Methylated CpG tandems amplification and sequencing; DMR, Differentially methylated regions; DMP, Differentially methylated base position; MeDIP-seq, Methylated DNA Immunoprecipitation Sequencing; MBD-seq, Methyl-CpG Binding Domain Protein Capture Sequencing; dPCR, digital polymerase chain reaction; ddPCR, droplet digital polymerase chain reaction; ddMCP, droplet digital methylation-specific PCR
1. Introduction
Traditional clinical diagnostic methods such as bone marrow or tissue biopsies are invasive in nature and possess sampling bias; consequently, researchers are looking for alternative molecular biomarkers. In recent years liquid biopsy-based disease diagnosis techniques have gained importance due to their safer and faster approach in contrast to tissue-based studies [1]. One such liquid biopsy-derived method uses cancer traces obtained from cell-free DNA (cfDNA). These fragments are called circulating tumor DNA (ctDNA) and have shown the potential to help in the field of cancer diagnosis, and prognosis [2].
The hematopoietic system is the major origin of cfDNA in healthy subjects, while in clinical patients (e.g., cancer), the affected cells/tissues contribute more to it. The plasma of a healthy individual contains 0–100 ng/ml of cfDNA, while in the case of late-stage cancer patients, it can go up to 1000 ng/ml [3]. Following cfDNA discovery in 1948 in autoimmune diseases, applications of cfDNA have now been extended to the diagnosis of many types of abnormalities. Some of the applications include identification of fetal chromosomal abnormalities (NIPT), early graft rejection, and detection and monitoring of cancer [4]. Besides genetic alterations, epigenetic changes in cfDNA have also been found to be useful as diagnostic biomarkers in different types of cancers[5], [6]. One of the most robust epigenetic markers is DNA methylation which is obtained by the addition of a methyl group through DNA methyltransferases (DNMTs) to the fifth carbon of cytosine [6]. A high composition of unmethylated CpGs is found in promoter regions of genes (CpG islands), while 70–80% CpGs are found to be globally methylated in the case of somatic cells.
One application of cfDNA methylation patterns has been in the identification of tissue of origin [4]. Moreover, various research findings show that DNA methylation-based biomarkers are more consistent in comparison to those based on mutational profiles [7], [8]. Detection of lung cancer with the help of EGFR mutation test V2 (Roche Molecular Diagnostics) and Epi procolon (Epigenomics AG) for colorectal cancer are some examples of cfDNA based FDA-approved tests [9].
A few large-scale prospective clinical trials are underway for the early detection of multiple types of cancer. The names of some of such multi-center trial studies are CCGA (Circulating Cell-free Genome Atlas), STRIVE, SUMMIT, and PATHFINDER by GRAIL Inc. [10]. An early report from these large-scale studies indicates low sensitivity in the detection of stage-I (18%) and stage-II (43%) cancer at a specificity of 0.7 % [10]. Such low sensitivity for early cancer detection highlights the importance of reviewing various steps involved in cfDNA methylation analysis. There have been a few reviews on profiling and analysis of 5mC based DNA methylation patterns in cfDNA [6], [5], [11]. Each review has its own unique aspect in target disease, description of experimental protocols, and analysis procedures. In our review, besides exploring the cfDNA methylation origin and analysis techniques, we have highlighted the usability of markers and their sensitivity in light of heterogeneity found in tumors. We have also provided a new dimension of sensitivity of 5hmC based cfDNA methylation pattern for liquid biopsy. Finally, we highlight the benefits and limitations of deconvolution and machine learning methods to analyze cfDNA methylation profiles.
2. Understanding cfDNA sources and features
Despite the extensive available literature on cfDNA, the biological insight behind the actual molecular origin of cfDNA is still poorly understood. Recent research has shown that multiple mechanisms work behind the release of cfDNA in the blood such as apoptosis, necrosis, pyroptosis, autophagy, NETosis, erythroblast enucleation, and cf-mtDNA [12], [13]. Several lines of evidence also suggest the role of cellular secretions in the release of cfDNA.The length of such cfDNA fragments lies in a range of 1000–3000 bp, in contrast to snippets generated via apoptosis (90 bp to 166 bp) [14]. Moreover, cfDNA in the blood could be present in the naked form (unbound DNA) or streaming as complex bounded to nucleosomes, membrane fragments, or vitrosomes or encased inside extracellular vesicles (EVs) like exosomes, microvesicles, and apoptotic bodies [15]. Disease diagnosis can be made based on the signals derived from cfDNA fragmentation pattern, nucleosome positioning, binding of transcription factors, transcription start site regions, cfDNA ended positions, as well as peripheral cellular alterations. The inherent property of information derived from cfDNA like sensitivity and noise and DNA fragment length affect the pattern inference process in the downstream computational analysis [16].
Also, in the case of cancer, tumor cells alone are not only the producers of cfDNA, but other non-cancerous cells also play an essential role in its release. The release of cfDNA from non-cancerous cells creates aberration in the signal from cancerous cells, as a result the data becomes more noisy and heterogeneous [17]. Among other contributing factors to cfDNA, its clearance rate from plasma also plays a vital role in its detection [18].
3. Computational problems associated with different cfDNA methylation profiling techniques
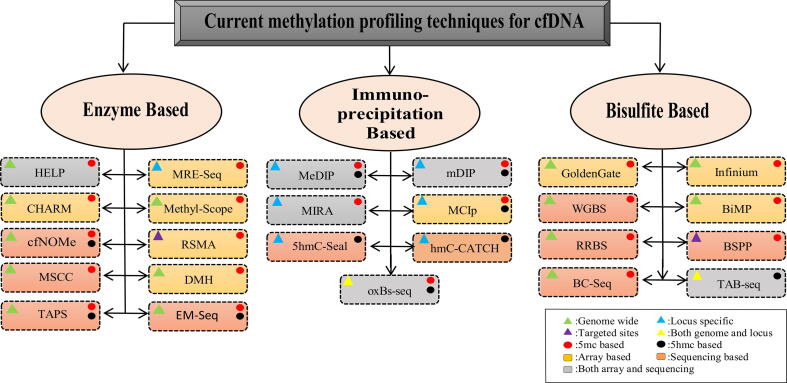
In order to tackle computational challenges associated with cancer detection using cfDNA methylation, it is crucial to understand different techniques used to profile it. Based on the mechanism to differentiate methylated cytosine from unmethylated one, the experimental assays for studying cfDNA methylation can be of three major types, i.e., restriction enzyme-based, bisulfite conversion-based, and enrichment/immuno-precipitation based [Fig. 1]. In addition there are many assay-specific pipelines for computational analysis of cfDNA methylation data as well [19].While currently, bisulfite-based conversion methods are more common, the selection of the method however, should be based on the proposed hypothesis, required resolution, cost, and nature of the experiment [20].
Fig. 1.
An overview of techniques for profiling DNA methylation which are also useful for detecting cfDNA. The triangular and circular symbols reveal further details of different methods. The expanded form of abbreviations for different methods are as such:- HELP: HpaII-tiny fragment enrichment by ligation-mediated PCR, CHARM: comprehensive high-throughput arrays for relative methylation, cfNOMe: cell-free DNA-based Nucleosome Occupancy and Methylation profiling, MSCC: methyl-sensitive cut counting, qPCR: Quantitative polymerase chain reaction, TAPS: TET-assisted pyridine borane sequencing, MRE-Seq: methylation restriction enzyme sequencing, RSMA: methylation-sensitive restriction enzyme-based assay, DMH: differential methylation hybridization, ddPCR: droplet digital PCR, EM-Seq: Enzymatic Methyl-seq, MeDip: methylation DNA immunoprecipitation sequencing, MIRA: methylated CpG island recovery assay, mDIP: methylated DNA immunoprecipitation, oxBs-seq: oxidative bisulphite sequencing, WGBS: whole-genome bisulphite sequencing, RRBS: reduced representation bisulphite sequencing, BC-Seq: bisulphite conversion followed by capture and sequencing, BiMP: bisulphite methylation profiling, BSPP: bisulphite padlock probe, TAB-seq: TET-assisted bisulphite sequencing).
3.1. Restriction enzyme based methods
The use of restriction enzymes has been a classical approach for profiling methylation patterns in cfDNA. Restriction enzymes are used to cleave DNA strands at the point bearing a particular nucleotide sequence; conversely, the presence of the methyl group might prevent digestion. Broadly, two categories of enzymes are used here: methylation-sensitive restriction enzymes (MSRE) such as HpaII, McrBC, AciI, and Hin6I, which can cleave only the unmethylated regions, while methylation-insensitive enzymes (e.g., MspI, ApeKI, and TaqI) cut DNA sequences without taking into consideration the methylation status of concerned sequences [14]. There are a few variations of basic MSRE techniques for genome-wide non-methylated region identification such as HELP-seq (HpaII-tiny fragment Enrichment by Ligation-mediated PCR sequencing), MSCC (Methylation Sensitive Cut Counting), Methyl-seq, scCGI (methylated CGIs at single-cell level), etc. [5].
However, the computational difficulty lies in distinguishing true and false negatives due to read loss caused by enzymatic digestion. Alternatively, analysis can be done using single-tube enzymatic methods such as DARE (DNA Analysis by Restriction Enzymes), where both can be quantified in the same sample [21]. Moreover, MSRE sequencing provides low methylome coverage due to limited CpG-containing cleavage sites, and it is also possible that some of the restriction enzymes might have been destroyed, leading to the non-trivial problem of identifying true negatives during computational analysis [22]. Besides since MRE-seq approach is relatively uncommon and most tools are inadequate to extract total read mapping to a given recognition site, there exist a gap in modern computational pipelines for studying MRE-seq generated DNA methylation data [20], [23].
3.2. Bisulfite based conversion methods
Since 1992, the application of bisulfite treatment has been a significant milestone in analyzing DNA methylation status. In this approach, all the unmethylated cytosines on reaction with bisulfite get converted to uracil, while methylated cytosines remain unchanged. Consequently, the comparison of methylation levels before and after bisulfite treatment gives an estimate of DNA methylation [24]. In addition, bisulfite-based conversion has been the foundation of many techniques such as WGBS, RRBS, MCTA-seq, targeted bisulfite sequencing, methylation array, MSP, etc. Whole Genome Bisulfite Sequencing (WGBS) is currently the most comprehensive technique for the identification of Genome-wide DNA methylation patterns [25]. Anyhow, since the whole of the genome is targeted in this approach, the cost of bisulfite conversion becomes extremely high [26], [27]. In contrast, RRBS (Reduced-Representation Bisulfite Sequencing) is a balanced combination of sequencing costs, genomic fold coverage, and CpG sites measured. However, the application of RRBS on highly fragmented DNA is yet to be determined [28]. MCTA-Seq (Methylated CpG tandems amplification and sequencing) is a very sensitive technology used to detect cfDNA hypermethylated sites in conditions such HCC and cirrhosis [29], [30]. However, one of the drawbacks is that it only recognizes CpG tandem regions, which means it may overlook certain non-CpG methylation sites. For routine diagnostic and target validations, TBS (Targeted Bisulfite Sequencing) has nowadays become a well-known approach in terms of epigenome-wide methylation profiling. It allows analysis of specific DNA locations while still retaining each single CpG resolution, which needs less DNA than the WGBS approach. The Bisulfite conversion step alters sequence complexity via non- complementarity and asymmetrical alignments, which makes the processing of bisulfite sequencing data difficult [20]. In order to reduce sequence complexity and allow adaption of conventional alignment algorithms, many bisulfite sequencing-based tools have been developed [Table 1]. Another non-trivial computational challenge with bisulfite-based DNA methylation profiling is finding DMR (Differentially methylated regions). The DNA fragments interrogated with bisulfite-based conversion methods are mostly small and have few cytosine positions; therefore, calling significant statistical DMR becomes more challenging than detecting DMP (Differentially methylated base position) [31]. A recent study by Erger et al., presented an assay named as cfNOMe that makes use of enzymatic cytosine conversion approach as a substituent to bisulfite based conversion to reduce the degradation loss and GC bias caused by later. The computational analysis of cfNOMe profile also helps in calculating nucleosome occupancy pattern at tissue-specific regulatory sites, making it a more efficient and comprehensive method for studying the epigenetic landscape of cfDNA [32].
Table 1.
Read alignment and Data visualization Tools.
| S.No | Tools | Advantages | Disadvantages | References |
|---|---|---|---|---|
| 1 | BatMeth2 | Indel-sensitive mapping | Removes some parts of reads (soft-clipping) | [129] |
| 2 | BSMAP | Good performance and flexibility due to seeding and hashing | Can detect indels with length less than 3 nucleotides only | [130] |
| 3 | Bismark | Flexible, easy to use and interpret | Increased run time | [131] |
| 4 | BS-Seeker2 | Supports both local and gapped alignments | Local alignment leads to longer CPU times | [132] |
| 5 | BWA-meth | Direct useable output, less storage requirements | doesn’t facilitate data visualization, only supports 3-letter alignment mode | [133] |
| 6 | BSmooth | Ability to handle low coverage experimental data | Assumes methylation profiles to be smooth, not able to detect single CpG sites | [134] |
| 7 | MethylCoder | Allows fast and sensitive mapping in both color and nucleotide space | Uses only short read aligners | [135] |
| 8 | Segemehl | Efficiently handles 3’ and 5’ contaminants along with mismatches and indels | Large memory requirements | [136] |
| 9 | GSNAP | SNP tolerant alignment, splicing and multiple mismatches can be detected | Might be slow for long positions | [137] |
| 10 | BRAT-BW | Runs faster on longer reads | Allows at most one mismatch in user defined reads | [138] |
| 11 | ERNE-BS5 | Analysis of methylation pattern at repeats, skillfully handles multiple mapping reads | Chances of false positives are higher | [139] |
| 12 | GEM3 | Exhaustive search model, fast, scalable, and gapped matches can also be found | some pruning methods are sensitive to mismatches | [140] |
| 13 | Last | High sensitivity and speed | Requires removal of poor quality bases | [141] |
| 14 | Msuite | supports bisulfite-free techniques,4-letter mode of alignment and computationally less expensive | analysis on irregular CpG sites needs additional validation | [142] |
| 15 | TAMeBS | Filters ambiguous read alignments and reduces bias in context of methylated cytosines | Memory requirements and running time are high | [143] |
3.3. Enrichment/immuno-precipitation based methods
The basic strategy behind enrichment-based methods is the use of anti methylcytosines antibodies for extraction of methylated regions from the cellular genome [33]. Methylated DNA Immunoprecipitation Sequencing (MeDIP-seq) and Methyl-CpG Binding Domain Protein Capture Sequencing (MBD-seq) are examples of techniques derived from affinity enrichment based array analysis. MeDIP uses antibodies directed against mC and mCG to extract methylated DNA fragments and has been used in several cases such as trisomy detection, cancer, and cardiology [34], [35]. High-quality methylomes can be obtained by combining MeDIP with NGS, which provides 1 to 300 bp resolution at costs comparable to other enrichment techniques [36]. MBD-seq, on the other hand, uses magnetic beads to pull out methylated-CpG binding domain (MBD) of DNA fragments. A study reports that MBD-seq can outperform MeDIP-seq in the identification of CGIs proportion [37]. Enrichment-based methods are cost-effective and have high discrimination power due to protein-binding specificity.
However, MBD-seq is sensitive for highly methylated regions with high CpG densities. Such properties of the enrichment-based method create a computational challenge of correctly identifying differential methylation at sites with high tissue specificity but low CpG densities. These methods also have a low resolution in comparison to bisulfite-based methods, and the estimated confidence score is highly influenced by the depth of sequencing [36]. Besides, some of the tools based on enrichment methods, such as Batman and MEDIPS [Table 2], require the user to perform prior quality control and reads mapping for data preparation which becomes time-consuming and computationally challenging [38], [39]. In addition, computational analysis of enrichment-based DNA methylation profiles with early-stage cancer becomes tough when the fraction of cfDNA non–hematopoietic cells is microscopic.
Table 2.
DNA Methylation Calling Software.
| Applicability | Tool | Advantages | Disadvantages | Statistical model | Reference |
|---|---|---|---|---|---|
| MeDIP-seq | Batman | High resolution and cost-effective whole genome methylome can be obtained | Time-consuming to run even with multiple processors | Bayesian model | [38] |
| MEDME | Provides both relative as well as absolute methylation levels, Can also be used for microarray designs of different platforms | Poor resolution in comparison to bisulfite based methods | Logistic model | [144] | |
| MEDIPS | More user friendly, cost and time effective | Difficult to detect methylation based on single end short reads | T-test, Wilcoxen test | [39] | |
| MeDUSA | Complete analysis of MeDIP-seq data from quality control to DMR calling | Approach employed is less efficient in terms of time and computation | Fisher’s exact test | [145], [146] | |
| MBD-seq | MethylAction | Applicable on larger study designs (four group comparisons), detects DMR’s through bootstrapping | Chances of type one error | Negative binomial and ANODEV (Analysis of Deviance) | [147] |
| Bisulfite-based | RnBeads | High computational efficiency and cross platform analysis | Limited genome annotation packages | Bayes framework and Bartlett test | [148] |
| DMRcate | Easy integration with other bioconductor tools, de novo based method | Make use of 450 k array only | F statistics | [149] | |
| DMRcaller | Detects DMRs in both CpG and non-CpG contexts | Sensitivity and specificity depends on window sizes, based on assumptions | Fisher’s exact test, Z test, Beta regression | [150] | |
| methylKit | Includes clustering functions along with DMRs visualisation | Limited by the memory of computer | Logistic regression and Fisher’s exact test | [151] | |
| MethylSig | Incorporates local information for estimating biological variation | Difficulty in handling heterogeneous data | Beta binomial model | [152] | |
| DSS | Capacity to handle multi factorial experimentation and data without biological replicates | Not suitable for paired design and longitudinal data type | Beta binomial distribution | [153] | |
| MRE-seq | msgbsR | Removes fallacious mapped reads, explores differential methylation | Requires pre-processed raw data | Negative binomial model | [154] |
| 5-hydroxymethylation | BiQ HiMod | user-friendly GUI, locus based methylation analysis and comprehensive analysis pipeline | pre-processed FASTA files are needed | Multiple statistical models | [155] |
3.4. 5-hydroxymethylation profiling
DNA demethylation by ten-eleven translocation (TET) enzymes can lead to oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), and further to 5-carboxylcytosine (5caC) and 5-formylcytosine (5fC) [40], [41]. Studies show the emerging role of 5hmC as a prominent epigenetic marker, and it has been found to be associated with tumor progression. It is also found to be enriched in enhancers, promoters and changes in 5hmC level are linked to changes in gene expression levels as well [42], [43], [44]. A variety of techniques have been developed such as 5hmC-Seal [45], hmC–CATCH [46], oxBS-seq [47], TAB-seq [48] and hMeDIP-seq [49] etc. which makes use of 5 hydroxymethylation profiling techniques.
The main weakness with 5hmc detection is its low frequency, making it more challenging in nature than 5mc. Also, 5hmc derived protocols possess low resolution (100–300 bp), are biased towards hypermethylated regions, and require relatively large DNA input. Hence for early-stage cancer detection where the contribution from non-blood sources of cfDNA is small, the output of 5hmC enrichment-based methylation profiles might suffer due to low sensitivity for relevant sites. Bergamaschi et al., suggests that to avoid model-based discrepancy, 5hmc based molecular classifiers for cancer should be interpreted in an integrative manner by combining demographic and disease comorbidity knowledge with tumor histology and pathology [50].
4. Computational issues related to cfDNA methylation detection techniques
After processing of samples according to different protocols for isolation or enrichment of methylated cfDNA, several detection techniques could be used to measure their quantity. However, each detection technique has its own analytical issues as discussed below:
4.1. Polymerase chain reaction based methods
Due to the low concentration of methylated DNA of non–hematopoietic origin in plasma cfDNA, digital polymerase chain reaction (dPCR) is preferred for cfDNA detection over traditional PCR. Digital PCR has shown to be 103–104 fold sensitive in having a lower limit of detection in comparison to the traditional version [51]. Digital PCR includes systems such as BEAMing (beads, emulsions, amplification, and magnetics) and droplet digital PCR (ddPCR). BEAMing was one of the first approaches for quantitatively detecting cfDNA and possess great sensitivity and specificity. However, its workflow is complex, necessitating oligonucleotides for each location, and is costly for typical clinical work [52], [53].
ddPCR is based on the technique of water–oil emulsion droplet and has got several applications like identification of the tissue origin [54], cancer detection [55], diagnosis of infectious diseases [56] among others. ddPCR is one of the most frequently used techniques these days with multiplex quantification. Various automated algorithms have been developed for ddPCR data analysis namely ‘definetherain’ [57], ‘ddpcRquant’ [58], ‘ddpcr’ [59], ‘twoddpcr’ [60], ‘ddPCRclust’ [61], ‘ddPCRmulti’ [62] etc. According to Dobnik et al., [61] the data analysis of such multiplex assays becomes difficult and noisy due to several possible target combinations along with probes cross hybridization in a single droplet. Brink et al., [61] reports that in the case of partially degraded DNA, multiplexing can also result in higher-order cluster disappearance and overlap.
Alternatively, methylation-specific PCR [MSP] can also be used to amplify DNA of interest by using methylation-specific PCR primer sets. MSP requires a small quantity of DNA and is sensitive to even 0.1% methylated regions of a given CpG island. The MSP technique has been used to identify hypermethylated promoter regions associated with tumor suppressor genes. With significant improvements in droplet digital PCR (ddPCR), droplet digital methylation-specific PCR (ddMCP) tools have also been established for early detection of cancer using cfDNA [63]. As methylation-specific PCR is qualitative, the sensitivity can only be tested via the ratio of methylated and unmethylated DNA. Such results show a lack of agreement between dilution ratio and band intensity, with many scenarios exhibiting quite similar bands despite differing levels of DNA methylation [64]. MethyLight, MethylQuant, and HeavyMethyl are some of the quantitative versions of the MSP with enhanced performance in quantifying DNA methylation. As these methods are able to investigate only one or two CpGs methylation levels, some of the sites remain unexplored, providing limited data for computational algorithm and downstream analysis [65].
Real-time PCR is one of the affordable rapid methods for nucleic acid amplification, and in the past, several different methods have been developed based on this technique. For instance, Allele-Specific amplification (AS-PCR), Peptide Nuclei Acid-Locked Nucleic Acid (PNA-LNA) PCR clamp, co-amplification at lower denaturation temperature (COLD-PCR), and Allele-Specific Non-Extendable Primer Blocker PCR (AS-NEPB-PCR) are some of the techniques that evolved from the RT-PCR approach. The main advantage of this method is that there is no need for post-PCR steps; hence chances of cross-contamination are reduced, which is beneficial for diagnostic purposes [66]. Besides, MethyLight can be used along with Real-time PCR as a quantitative assay where relative fluorescence units (RFUs) represent the methylation percentage. However, it is unable to correctly analyze a heterogeneous sample because the primers are designed in such a way to detect only specific fully methylated patterns [67]. Despite being among the most effective methods, the quality of the results of real-time PCR can hold variations due to insufficient quality control steps, inappropriate use of reference genes and data normalization methods, and batch effects [68], [69]. In addition, for data normalization, the choice of reference genes, their stability, and amplification efficiency also play a significant role during data analysis. Kuang et al., demonstrated that usage of unstable reference genes could create variations in the final output and proposed cDNA as an alternative for normalizing data [70]. Reference genes can be evaluated by applying some statistical tests on Cq or with the help of various analytical methods such as NormFinder [71], BestKeeper [72], GeNorm [73], RefFinder [74].
4.2. Next-generation sequencing
Although multiple studies have reported detection of ctDNA in different stages with high sensitivity by using ddPCR or BEAMing, yet limited clinical applications of PCR have led to the development of other assays based on Next-generation sequencing (NGS) [75]. NGS has emerged as an excellent technique for high throughput DNA sequencing and has revolutionized the concept of clinical samples analysis [3]. This technology has become a powerful tool for identifying biomarkers pertaining to its high sensitivity, specificity, and scalability. Since the resolution at the single-base level by NGS allows accurate mapping of disease-specific regions, consequently it has been applied for genome-wide profiling of plasma from various cancers [76], [77], [78]. The sensitivity and specificity of NGS analysis depend upon the type of platform used, such as deep sequencing, Tam-seq, Safe-SEQs, CAPP-Seq, MCTA-Seq, FASTSeqS, etc [79]. A study by Liang et al., demonstrated that a combination of deep methylation sequencing with machine learning can provide better efficiency concerning cancer identification in comparison to ultradeep sequencing[80].
However, despite its appreciable performance, a random error rate of 0.1% and 1% by NGS technology creates a challenge in reliable detection of methylation and mutation profile with non–hematopoietic origin in plasma cfDNA [81]. Moreover, the occurrence of repetitive sequences and indels (insertions and deletions) can also be one of the contributing factors for sequence misalignment, influencing variant analysis. Data processing also relies on several other parameters such as filtering variants, the NGS technology’s nature, VAFs (variant allelic frequency), quality of sequencing, and bioinformatics pipeline. Henceforth the routine clinical applicability of NGS workflows need special precautions to ensure its authenticity, especially in case of dispersed, fragmented ctDNA within the background of normal cfDNA [82]. The complex and large size NGS data obtained from repeated experimentation creates additional challenges for statisticians in terms of deciding lower limits of detection based on assay due to lack of standard pipeline. An additional challenge is building a classification model for a high feature and small sample size dataset without overfitting or bias [79].
4.3. Methylation array
Before the popularity of NGS, HM450k (Illumina Infinium HumanMethylation450 BeadChip) had been the most desirable choice for investigators when it came to studying cancer methylomes. HM450k contains pre-designed probes for methylation sites that cover 96% of CpG islands in 450k array and additional CpG sites of enhancer regions in 850K array. Currently plenty of HM450k datasets are available on The Cancer Genome Atlas (TCGA) [83] and Gene Expression Omnibus (GEO) [84] that are being used for discovery and validation of biomarkers along with the analysis of deconvolution based cfDNA tissue of origin [4].
The main limitation of array-based methods is the inadequate genome-wide coverage, causing dissipation of some other essential methylation regions [85]. In addition, the cost of the technique is highly dependent upon the input data amount along with genome coverage, besides the required assay expertise for the experiment and subsequent downstream computational analysis [86]. Occurrences of too many false positives, probes and samples quality control, bogus cross-hybridization of probes, rescaling of probes, platform specific background correction, data normalization to reduce technical, experimental, and systematic variations are some of the other concerning issues associated with the use of methylation array [87]. Methylation arrays are also susceptible to experimental conditions and laboratory environments, leading to batch effects in data from various studies. Many batch correction algorithms can reduce the effect of known confounding factors, but since the true source of confounding factors is often unknown, even this task become non-trivial during statistical modelling of array-based cfDNA methylation profiles. Moreover, several studies report that there exists a high correlation of methylation levels among the adjacent CpG loci; consequently, statistical analysis of array-based data with the notion of independence among each CpG methylation may be misleading [88].
5. Computational difficulties in cfDNA methylation data analysis
The basic workflow of computational analysis of cfDNA methylation data includes (i) reads pre-processing and quality assessment, (ii) alignment and visualization, (iii) statistical analysis and interpretation. Sample pre-processing makes sure that raw data is structured and there is no bias in it. Different programs have been developed based on various algorithms to perform quality analysis such as FastQC, NGS QC, QC–Chain, ClinQC [89], [90]. Once the raw data is analyzed, low-quality bases and adapters can be removed by programs such as Trim Galore. Wild card and three-letter are two types of algorithms used to align sequencing data to the reference genome. While wild card algorithm (e.g., GSNAP, BSMAP) allows mapping of both Cs and Ts of reads to Cs in the reference genome, the three-letter algorithm (e.g., BisMark, BS-Seeker2, BRAT-BW) changes all Cs of reference and reads into Ts so that standard alignment tools can be applied [Table 1]. In order to inspect the global distribution of methylation profiles, data visualization can be done through various approaches such as UCSC Genome Browser [91], DNMIVD [92], Methylation plotter [93], Integrative Genomics Viewer (IGV) [94] and Web Service for Bisulfite Sequencing Data Analysis (WBSA) [95]. For restriction enzyme and enrichment affinity-based methods (MRE-seq, MeDIP-seq), relative read-count is estimated. However, for bisulfite sequencing (WGBS and RRBS), methylation level at individual cytosine residues is estimated. Many recent DNA methylation calling software (e.g., RnBeads, MeDUSA, MEDME, Batman) have used different statistical models to quantify DNA methylation coverage [Table 2]. However, sequencing depth, which depends on the assay used, is a critical factor to consider before making any choices for the same.
5.1. Tumour heterogeneity and dependency on markers
Inter and intra-tumor heterogeneity has been in existence for decades due to the morphological, genetic, epigenetic, and phenotypic diversity in cell populations. Nowadays, cellular heterogeneity is among the primary causes of disease resistance and targeted therapy failure [96]. While the studies based on whole-cell populations may represent the dynamics of majority cells, they may mask the role of critical sub-populations and hence the fundamental biology behind it. Also, such cellular heterogeneity poses tough challenges in diagnostics and treatments of disease in studies based on population-averaged measurements [97]. While tissue biopsies may only capture a part of this heterogeneity, liquid biopsies are more useful in such a scenario [98]. Tumor heterogeneity is also one of the leading causes of therapeutic resistance, treatment failure, and poor survival rate of cancer patients. Often cancer diagnostics depend on the presence of specific biomarkers. However, due to the dynamic nature of tumor cells, the predicted biomarkers are found on a non-uniform scale causing an impediment to the treatment of disease [99]. Literature shows multiple instances when the non–homogeneous nature of the druggable targets is observed, namely gastric adenocarcinoma, lung adenocarcinoma, breast cancer, melanoma, etc. Consequently, applying the biomarker-based targeted therapies in heterogeneous neoplasms leads to recurrence in the long run [100]. Many different computational pipelines and algorithms are being developed for estimation of cellular heterogeneity as a pre-processing step so that more meaningful insights can be achieved [101], [102], [103].
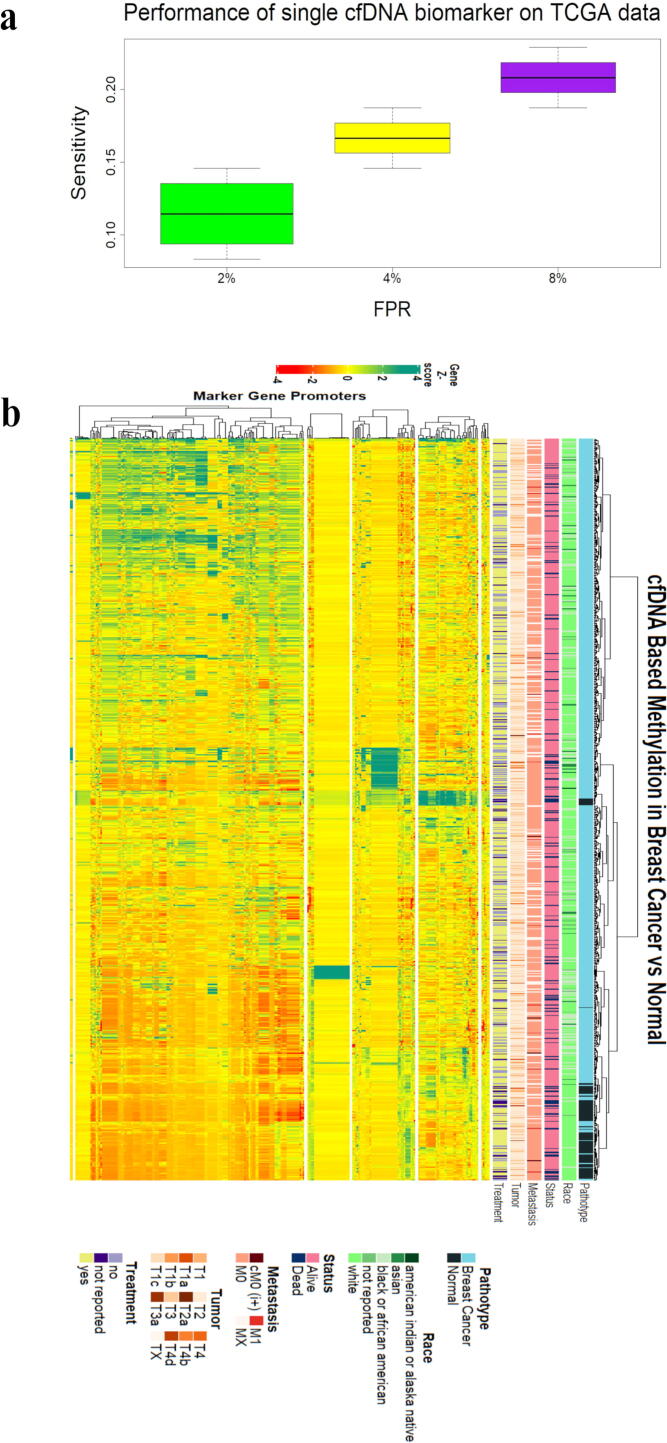
In order to analyze the consistency of some known cfDNA methylation literature-based biomarkers, we checked their expression in a set of 848 TCGA samples consisting of 96 normal and 752 breast cancer patients. It was found that the heterogeneity among the biomarkers was sufficiently large to hamper the process of diagnostics and therapeutics. Along with the heterogeneity arising from markers used for disease detection, other sources for the same could be some confounding factors. It can be also be seen from the box plot that the idea of using a single marker-based approach for disease detection does not seem to provide an acceptable level of sensitivity when applied to a classification model of 192 TCGA 450k methylation samples (96 normal, 96 breast cancer patients) [Fig. 2] (see supplementary material). Given the small amount of cfDNA produced, the power of a single marker may not be fully capable of distinguishing the cancerous state from non-cancerous. However, the sensitivity can be augmented by using a set of multiple markers.
Fig. 2.
cfDNA methylation based markers performance on TCGA data. Illumina 450k methylation data-set for bulk tissue (Breast Cancer) was retrieved from TCGA (The Cancer Genome Atlas) database and processed for manually curated literature based markers. (a) boxplot of FPR (False positive rate) vs sensitivity showing performance of single marker for sample class prediction (Breast Cancer vs Normal). Based on LDA (Linear Discriminant Analysis) fitting of TCGA samples for one marker, values for sensitivity and FPR were obtained and presented in the form of box-plot. It can be observed from the plot that a single marker-based approach for detection of disease delivers quite less sensitivity. (b) heatmap showing heterogeneity among biomarkers for the same cancer type. Markers based normalised beta scores for all the TCGA observations were visualised as a heatmap for differential analysis of cancer and non-cancerous observations. This figure demonstrates that such level of heterogeneity among biomarkers can be one of the influencing factors for disease diagnostics and therapeutics.
5.2. Multi-marker based detection: opportunities and obstacles
Although rogue cfDNA methylation level in cancer has been known for more than a decade, it has yet not fully established its importance as a diagnostic tool in clinical practice. A significant drawback with conventional biomarkers is that most of the time, the marker’s utility is limited to only metastatic and late-stage cancer [63]. Barault et al., showed that individual biomarkers have a relatively low prevalence in patients, which can be increased if they are used in combination [104]. Perhaps each of these markers may be informative alone; the multiparametric scenario could improve its discriminating power for cancer and healthy individuals. Mouliere et al., studied the use of multi markers (Intplex) in colorectal cancer for cfDNA, and it was found to be quite sensitive, specific, and easy to implement. Also, it was shown to be adaptable to repetitive examination, henceforth making the follow-up studies easy if one talks about in terms of personalized medicine [105]. However, there seem to be some weaknesses in using a multi-marker panel. Firstly, the performance of markers varies based on the population, test data, experimental assay, and analysis of the result. Due to these reasons, such biomarker panels hold less confidence of clinicians. Also, studies aimed to prove cfDNA marker’s robustness are often retrospective and possess inadequate sample size and statistical competency. In an effort to avoid such anomalies, comprehensive studies are required to abide by the standard guidelines for reporting the diagnostic accuracy [106].
5.3. 5hmc based detection: success and limitations
The human genome contains a large number of 5-hydroxymethylcytosines (5hmC) based epigenetic modifications as the oxidized form of 5-methyl-cytosines (5mc) and is proposed to act as ideal markers for reflecting the chromatin activation state. In a similar fashion to 5mc based studies, 5hmc modifications have also been reported as crucial factors for understanding different types of cancer pathology and tissue-specific origin [45]. However, in contrast to 5mc, 5hmc based profiles are shown to possess more stability and robustness, which provides better specificity in terms of cancerous vs. normal individuals. Besides, while 5mc is believed to have a repressive effect, 5hmc got permissive ramifications on the gene expression [107]. Also, since enhancers, promoters, and other regulatory elements are found to be enriched with 5hmc, it is also expected to be in more correlation with cellular gene expression [108]. 5hmc has recently been linked to many biological processes and disorders, including brain development, malignant melanoma, breast cancer, bladder cancer, and non-small cell lung cancer [108], [109], [110]. Although, in comparison to extensive cfDNA research on 5mc, 5hmc has yet to be thoroughly investigated in the realm of cancer diagnosis. Given the minute amount of cell-free DNA, obtaining noise-free signals and lack of highly sensitive DNA sequencer for 5hmc is one of the challenges faced by researchers while using 5hmc as an epigenetic biomarker (10-to 100-fold less than 5mC) [107].
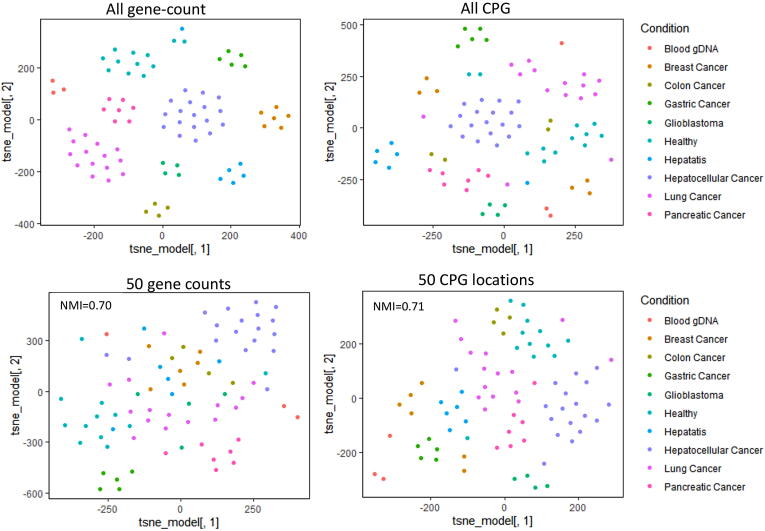
In order to evaluate the possibility of using markers for the 5hmC profile of cfDNA, we performed an analysis using data published by Song et al., for mostly advanced-stage cancer. For their study, Song et al., performed analysis using read-count on a large number of genes, and they did not report any classification based on fewer number of markers. Therefore, we evaluated the classification using the 5hMC profile of cfDNA with a reduced number of genomic loci as markers. Our result revealed that the classification accuracy reduces with a lower number of markers, but it was sufficient to group similar phenotype samples together. Our analysis used the top 50 marker locations using feature importance achieved by applying random forest-based classification on gene and CpG island read-counts (see supplementary material). Using top 50 markers, it was possible to achieve good separability among different phenotypes in the 2D embedding plot (see Fig. 3). Application of density-based clustering (see supplementary material) on the 2D embedding using top 50 markers resulted in clustering-purity above 0.70 NMI (Normalized Mutual Information) score (see Fig. 3). Thus the utilization of 5hmC profiles on selected markers for detection could be feasible to some extent for an advanced stage of cancer. As Song et al. generated 5hmC profile using cfDNA of patient with mid or late stage cancer, the challenge of sensitivity with 5hmC for detecting early cancer still remains as open problem.
Fig. 3.
The visualisation of low dimensional embedding of 5hmC profile of cell free DNA of samples from patients with different types of cancer. Here 2D embedding (using tSNE) of 5hmC profile is shown for samples either using read-count of genes or CpG islands. The results of embedding are shown for read-counts on all genes or only 50 selected genes. Similarly the results of embedding done using all CpG island or only 50 selected CpG islands are also shown. The 5hmC profiles used here published by Song et al. 2017. Using large number of genomic loci (all genes or all CpG island) can provide good separability among samples according to type of cancer. With 5hmc profile, top 50 chosen marker CpG island provide slightly better separability among different pathological condition in comparison to top 50 gene. The purity of clustering after embedding using top 50 markers (genes or CpG islands) is also shown terms of Normalized mutual information (NMI).
5.4. Deconvolution: pros and cons
Considering high levels of heterogeneity among tissues, reports suggest the use of tissue-specific biomarkers. For plasma DNA-based testing as well, tissue-specific markers are found to be more consistent in nature [111]. In order to map the origin of tumor tissue from cfDNA, one of the commonly used methods is the deconvolution algorithm, which recovers the original signal from a mixture of signals. Deconvolution algorithms are basically of two kinds: reference-based and reference-free. Reference-based deconvolution algorithms are based on supervised methods utilizing cell-type-specific differentially methylated regions (DMRs). On the other hand, reference-free algorithms do not need cell-type-specific DMRs as reference but estimate cellular proportion using unsupervised deconvolution approaches [112]. One of the earliest and most widely used algorithms, based on reference dataset, is constrained projection [CP] (also known as quadratic programming [QP]) which operates through least square minimization. For reference-free approaches, there are frameworks such as removing unwanted variation (RUV), non-negative matrix factorization (NMF) [113]. Recently many more reference-based [EpiDISH, CIBERSORT] and reference-free approaches [CellMix, CDSeq, TOAST, RefFreeEWAS, EWASher, SVA] for cfDNA deconvolution have emerged.[114], [115], [116], [117], [118], [119], [120]. Studies show that disease prediction accuracy increases by incorporating tissue proportion factors and more interpretative biological output is obtained. According to Moss et al., the use of only defined sets of significant CpG sites in deconvolution gives greater resolution and less noise in comparison to using the entire methylome, even with a low amount of DNA. [4].
Most of the reference-based deconvolution methods suffer from two main limitations. First, they often need a prior guess about the organ from which DNA could be found in plasma. Although with a correct estimation of organ, the calculation of the proportion of contribution from different cell types is reasonably satisfactory to some extent. The second limitation of reference-based deconvolution is the difference in technical batch-effect in reference cell methylome profile and cfDNA methylation profile. In actual practice, the prediction of cellular proportion can be more complicated due to some biological or technical artifacts. Hence there is a need for such computational methods which can accurately project the information in lower dimension space without being influenced by a reference methylation panel [1], [111].
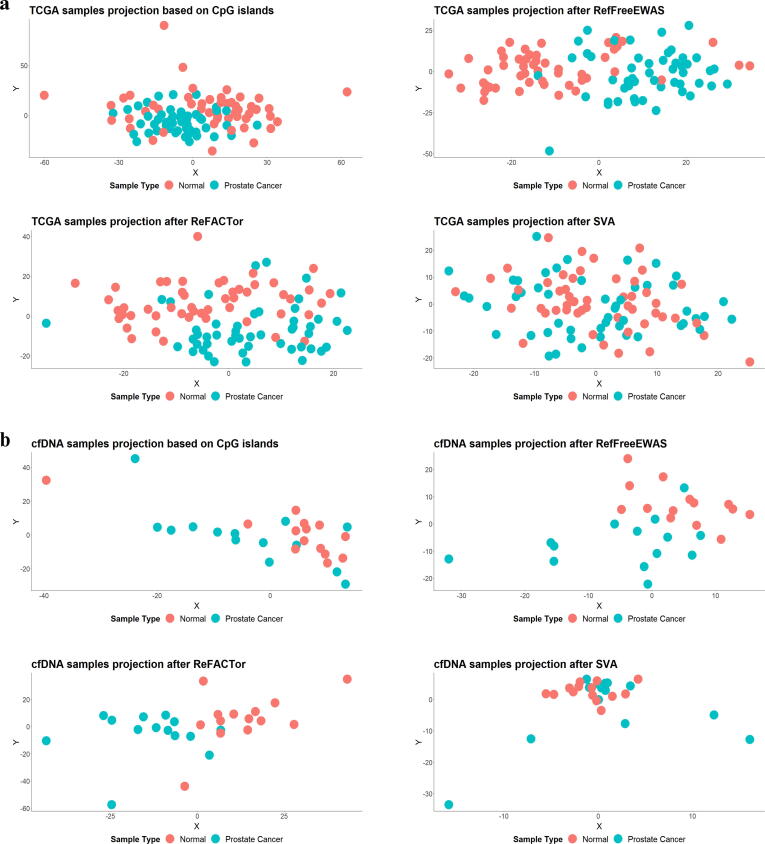
To analyse the data separability of reference-free deconvolution methods, we applied three most commonly used approaches such as RefFreeEWAS [119], ReFACTor [121], and SVA [122] on 450 k methylation profile from prostrate cancer and normal samples of TCGA (100 samples) and cfDNA (28 samples). In the current study, a comparison of the deconvolution techniques on randomly selected 100 CpG sites showed that the performance of a specific approach depends partially on the dataset itself; for example, in TCGA samples, RefFreeEWAS was able to do a better classification among others and in the case of cfDNA dataset RefFreeEWAS and ReFACTor showed similar separation [Fig. 4] (see supplementary material). Other limitations include batch effects, small datasets, unaccountable covariates related to CpG islands methylation etc.
Fig. 4.
Applying different deconvolution techniques on the DNA methylation profiles of cancer and normal samples. Reference-free deconvolution methods such as RefFreeEWAS, ReFACTor and SVA were applied to DNA methylation profiles and projected as tSNE coordinates to analyze sample separability. (a) Here DNA methylation profiles available in the TCGA portal for solid tissue from prostate cancer were used. (b) Deconvolution methods were applied to DNA-methylation profiles of cell-free DNA (cfDNA) extracted from the plasma of individuals with normal and prostate cancer pathotypes from CFEA. The comparative analysis is based on 100 randomly selected CpG sites of the samples.
5.5. Machine learning based approaches: strengths and weaknesses
With computational advancements in the field of liquid biopsy, the role of machine learning in diagnostics and therapeutics seems quite promising. Recently a few studies have applied machine learning approaches for cfDNA methylation analysis [123], [124], [1], [125], [126]. Machine learning techniques can be applied using whole-genome features or selected markers scores with or without deconvolution. Such as Shu et al., used meDIP-seq profile and first identified the top 300 DMRs among patients and non-patients before applying the binomial generalized linear model [123]. On the other hand, Feng et al., applied machine learning using three scenarios: 1) just using markers, 2) after NMF based reference-free deconvolution, and 3) after reference-based tissue proportion estimation using QP. With WGBS profile from cfDNA (liver cancer and normal), Feng et al., achieved higher accuracy using by training machine learning model after reference-based proportion estimation (accuracy = 0.79) in comparison to reference-free deconvolution (accuracy = 0.7) or using marker signal directly (accuracy = 0.75) [1]. It is not trivial to judge the usefulness of reports of high classification accuracy with smaller data sets from previous studies. Provided a large data size, machine learning algorithms may develop solutions to learn disease-related patterns directly from a patient’s whole genome or targeted sites (multi-marker) signal.
For cfDNA methylation-based predictions, machine learning techniques have their own limitations. Such as the requirement of a large number of samples to train, bias in classification due to imbalance in training data-set, batch effect [11]. Especially in the case of cfDNA methylation data-set, when the relevant signal is overwhelmed with the epigenetic signature of blood cells, suppressing batch effect for correct prediction in target sample is very challenging. It is reflected by the performance of classifier in detecting 50 types of cancer by CCGA consortium [127] using large training (1654 cancer + 1375 normal) and validation set (703 cancer + 605 normal). With such a large training set, the classifier used by CCGA consortium could achieve average sensitivity of 44.2 for cancer stages I, II and III [127]. Even for 12 predefined high signal cancer types, CCGA consortium could achieve a sensitivity of only 39% for stage I samples. Such results highlight the limitation caused by the low concentration of cfDNA from non–hematopoietic origin and heterogeneity among patients [127].
6. Discussion
Here we have described the strengths and weaknesses of several procedures involved in detecting cancer using cfDNA methylation. By analyzing existing DNA methylation profiles from tumor samples and cfDNA, we showed limitations in using individual markers due to cancer heterogeneity. However, there is yet another kind of bias, which adds to the computational challenge. The bias in different ways of detection of DNA methylation reduces the significance of detection of specific markers. Such as many markers detected using HM450k methylation array might be completely non-detectable by RRBS based cfDNA methylation profiling. Therefore despite the availability of a few data-sets of cfDNA methylation profiles from cancer patients, it is not trivial to finalize markers for any cancer type that could be used globally with multiple cfDNA methylation profiling techniques. In other fields of genomics, such as single-cell expression profile analysis, there have been a few attempts to perform integrative analysis irrespective of bias of platform and protocol used. However, rarely such attempts have been made to solve the computational problem of integrative analysis using cfDNA methylation profiles. The reason could be that single-cell expression profiles are not mixtures of unknown cell types, whereas cfDNA methylation profiles have mixed signals from several cell types.
The approach used by different clinical trials to learn machine-learning models on a data-set and to validate on another data-set is often called transfer learning. There has been substantial development in making transfer learning more adaptive [128] to new data-set to avoid the batch effect. However, adaptive transfer learning often needs small samples from target data to adjust itself. There could be day-to-day variation in the profiling of cfDNA methylation even from the same patient. Hence it remains to be seen how adaptive transfer learning can be used to identify the tissue of origin using cfDNA methylation, irrespective of batch effect and variation in signal-dilution by blood cells.
Even though a few clinical trials have reported good accuracy for detecting late-stage cancer, detection of early-stage is still a challenge [30], [125], [63]. The low accuracy on early cancer detection reduces the utility of liquid biopsy as advanced-stage tumors are often non-treatable. Hence there is still a demand for novel computational approaches to improve early-stage cancer detection using cfDNA methylation profiles.
Funding information
This work was supported by Department of Biotechnology and Indian Council of Medical Research (ICMR).
Availability of data and materials
The datasets used for analysis in the current study can be found at The Cancer Genome Atlas (TCGA) https://portal.gdc.cancer.gov/ and Cell Free Epigenome Atlas (CFEA) http://www.bio-data.cn/CFEA/ repositories.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank our institute (Indraprastha Institute of Information Technology (IIIT) Delhi) for providing the computing support.
Footnotes
Supplementary data associated with this article can be found, in the online version, athttps://doi.org/10.1016/j.csbj.2021.12.001.
Supplementary data
The following are the Supplementary data to this article:
References
- 1.Feng H., Jin P., Wu H. Disease prediction by cell-free DNA methylation. Briefings in Bioinformatics. 2019;20(2):585–597. doi: 10.1093/bib/bby029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu S., Wu J., Xia Q., Liu H., Li W., et al. Finding new cancer epigenetic and genetic biomarkers from cell-free DNA by combining SALP-seq and machine learning: Esophageal cancer as an example. Cancer Biology. 2020 doi: 10.1101/2020.01.18.911172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elazezy M., Joosse S.A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management, Computational and Structural. Biotechnology Journal. 2018;16:370–378. doi: 10.1016/j.csbj.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moss J., Magenheim J., Neiman D., Zemmour H., Loyfer N., et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nature Communications. 2018;9(1):5068. doi: 10.1038/s41467-018-07466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Z., Wang Z., Jia E., Ouyang T., Pan M., et al. Analysis of genome-wide in cell free DNA methylation: Progress and prospect. The Analyst. 2019;144(20):5912–5922. doi: 10.1039/C9AN00935C. [DOI] [PubMed] [Google Scholar]
- 6.Huang C.-C., Du M., Wang L. Bioinformatics Analysis for Circulating Cell-Free DNA in Cancer. Cancers. 2019;11(6):805. doi: 10.3390/cancers11060805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan S., Chi W. Methods for genome-wide DNA methylation analysis in human cancer. Briefings in Functional Genomics. 2016:elw010. doi: 10.1093/bfgp/elw010. [DOI] [PubMed] [Google Scholar]
- 8.Warton K., Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Frontiers in Molecular Biosciences. Apr. 2015;2 doi: 10.3389/fmolb.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan Y.-Y., Guo Q.-R., Wang F.-H., Adhikari R., Zhu Z.-Y., et al. Cell-Free DNA: Hope and Potential Application in Cancer. Frontiers in Cell and Developmental Biology. 2021 doi: 10.3389/fcell.2021.639233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ofman J.J., Hall M., Aravanis A., Park M. Grail and the quest for earlier multi-cancer detection. Nature. 2018 [Google Scholar]
- 11.Huang Wang. Cell-Free DNA Methylation Profiling Analysis—Technologies and Bioinformatics. Cancers. 2019;11(11):1741. doi: 10.3390/cancers11111741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aucamp J., Bronkhorst A.J., Badenhorst C.P.S., Pretorius P.J. The diverse origins of circulating cell-free DNA in the human body: A critical re-evaluation of the literature. Biological Reviews. 2018;93(3):1649–1683. doi: 10.1111/brv.12413. [DOI] [PubMed] [Google Scholar]
- 13.Grabuschnig S., Bronkhorst A.J., Holdenrieder S., Rosales Rodriguez I., Schliep K.P., et al. Putative Origins of Cell-Free DNA in Humans: A Review of Active and Passive Nucleic Acid Release Mechanisms. International Journal of Molecular Sciences. 2020;21(21):8062. doi: 10.3390/ijms21218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L., Feng J., Polimeni J., Zhang M., Nguyen H., et al. Characterization of Cell Free Plasma Methyl-DNA From Xenografted Tumors to Guide the Selection of Diagnostic Markers for Early-Stage Cancers. Frontiers in Oncology. 2021;11:503. doi: 10.3389/fonc.2021.615821. [DOI] [Google Scholar]
- 15.Panagopoulou M., Esteller M., Chatzaki E. Circulating Cell-Free DNA in Breast Cancer: Searching for Hidden Information towards Precision Medicine. Cancers. 2021;13(4):728. doi: 10.3390/cancers13040728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng H., Zhu M.S., Liu Y. FinaleDB: A browser and database of cell-free DNA fragmentation patterns. Bioinformatics. 2021;37(16):2502–2503. doi: 10.1093/bioinformatics/btaa999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bronkhorst A.J., Ungerer V., Holdenrieder S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomolecular Detection and Quantification. 2019;17 doi: 10.1016/j.bdq.2019.100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khier S., Lohan L. Kinetics of circulating cell-free DNA for biomedical applications: Critical appraisal of the literature. Future Science OA. 2018;4(4):FSO295. doi: 10.4155/fsoa-2017-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galardi F., De Luca F., Romagnoli D., Biagioni C., Moretti E., et al. Cell-Free DNA-Methylation-Based Methods and Applications in Oncology. Biomolecules. 2020;10(12):1677. doi: 10.3390/biom10121677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rauluseviciute I., Drabløs F., Rye M.B. DNA methylation data by sequencing: Experimental approaches and recommendations for tools and pipelines for data analysis. Clinical Epigenetics. 2019;11(1):193. doi: 10.1186/s13148-019-0795-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viswanathan R., Cheruba E., Cheow L.F. DNA Analysis by Restriction Enzyme (DARE) enables concurrent genomic and epigenomic characterization of single cells. Nucleic Acids Research. 2019;47(19) doi: 10.1093/nar/gkz717. e122–e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Z., Bai Y., Cheng Z., Liu F., Wang P., et al. Absolute quantification of DNA methylation using microfluidic chip-based digital PCR. Biosensors and Bioelectronics. 2017;96:339–344. doi: 10.1016/j.bios.2017.05.021. [DOI] [PubMed] [Google Scholar]
- 23.B.T. Mayne, S.Y. Leemaqz, S. Buckberry, C.M. Rodriguez Lopez, C.T. Roberts, T. others, J. Breen, msgbsR: An R package for analysing methylation-sensitive restriction enzyme sequencing data, Scientific Reports 8 (1) (2018) 2190. doi:10.1038/s41598-018-19655-w. [DOI] [PMC free article] [PubMed]
- 24.Werner B., Yuwono N.L., Henry C., Gunther K., Rapkins R.W., et al. Circulating cell-free DNA from plasma undergoes less fragmentation during bisulfite treatment than genomic DNA due to low molecular weight. PLOS ONE. 2019;14(10) doi: 10.1371/journal.pone.0224338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuart T., Buckberry S., Lister R. In: Jeltsch A., Rots M.G., editors. Vol. 1767. Springer New York; New York, NY: 2018. Approaches for the Analysis and Interpretation of Whole Genome Bisulfite Sequencing Data; pp. 299–310. (Epigenome Editing). [DOI] [Google Scholar]
- 26.E.-J. Lee, J. Luo, J.M. Wilson, H. Shi, Analyzing the cancer methylome through targeted bisulfite sequencing, Cancer letters 340 (2) (2013) 10.1016/j.canlet.2012.10.040. doi:10.1016/j.canlet.2012.10.040. [DOI] [PMC free article] [PubMed]
- 27.Suzuki M., Liao W., Wos F., Johnston A.D., DeGrazia J., et al. Whole-genome bisulfite sequencing with improved accuracy and cost. Genome Research. 2018;28(9):1364–1371. doi: 10.1101/gr.232587.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanić M., Beck S. Epigenome-wide association studies for cancer biomarker discovery in circulating cell-free DNA: Technical advances and challenges. Current Opinion in Genetics & Development. 2017;42:48–55. doi: 10.1016/j.gde.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 29.Wen L., Li J., Guo H., Liu X., Zheng S., et al. Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell Research. 2015;25(11):1250–1264. doi: 10.1038/cr.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J., Zhou X., Liu X., Ren J., Wang J., et al. Detection of Colorectal Cancer in Circulating Cell-Free DNA by Methylated CpG Tandem Amplification and Sequencing. Clinical Chemistry. 2019;65(7):916–926. doi: 10.1373/clinchem.2019.301804. [DOI] [PubMed] [Google Scholar]
- 31.Paun O., Verhoeven K.J., Richards C.L. Opportunities and limitations of reduced representation bisulfite sequencing in plant ecological epigenomics. New Phytologist. 2019;221(2):738–742. doi: 10.1111/nph.15388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erger F., Nörling D., Borchert D., Leenen E., Habbig S., et al. cfNOMe —A single assay for comprehensive epigenetic analyses of cell-free DNA. Genome Medicine. 2020;12(1):54. doi: 10.1186/s13073-020-00750-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan R.F., Shabalin A.A., Xie L.Y., Adkins D.E., Zhao M., et al. Enrichment methods provide a feasible approach to comprehensive and adequately powered investigations of the brain methylome. Nucleic Acids Research. 2017;45(11) doi: 10.1093/nar/gkx143. e97–e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papageorgiou E.A., Karagrigoriou A., Tsaliki E., Velissariou V., Carter N.P., et al. Fetal-specific DNA methylation ratio permits noninvasive prenatal diagnosis of trisomy 21. Nature Medicine. 2011;17(4):510–513. doi: 10.1038/nm.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen S.Y., Burgener J.M., Bratman S.V., De Carvalho D.D. Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA. Nature Protocols. 2019;14(10):2749–2780. doi: 10.1038/s41596-019-0202-2. [DOI] [PubMed] [Google Scholar]
- 36.Taiwo O., Wilson G.A., Morris T., Seisenberger S., Reik W., et al. Methylome analysis using MeDIP-seq with low DNA concentrations. Nature Protocols. 2012;7(4):617–636. doi: 10.1038/nprot.2012.012. [DOI] [PubMed] [Google Scholar]
- 37.Nair S.S., Coolen M.W., Stirzaker C., Song J.Z., Statham A.L., et al. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics. 2011;6(1):34–44. doi: 10.4161/epi.6.1.13313. [DOI] [PubMed] [Google Scholar]
- 38.Down T.A., Rakyan V.K., Turner D.J., Flicek P., Li H., et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nature Biotechnology. 2008;26(7):779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chavez L., Jozefczuk J., Grimm C., Dietrich J., Timmermann B., et al. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Research. 2010;20(10):1441–1450. doi: 10.1101/gr.110114.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M. Tahiliani, K.P. Koh, Y. Shen, W.A. Pastor, H. Bandukwala, others., Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1, Science 324 (5929) (2009) 930–935. doi:10.1126/science.1170116. [DOI] [PMC free article] [PubMed]
- 41.He Y.-F., Li B.-Z., Li Z., Liu P., Wang Y., et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bachman M., Uribe-Lewis S., Yang X., Williams M., Murrell A., et al. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nature Chemistry. 2014;6(12):1049–1055. doi: 10.1038/nchem.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasanthakumar A., Godley L.A. 5-hydroxymethylcytosine in cancer: Significance in diagnosis and therapy. Cancer Genetics. 2015;208(5):167–177. doi: 10.1016/j.cancergen.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 44.Li W., Zhang X., Lu X., You L., Song Y., et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Research. 2017;27(10):1243–1257. doi: 10.1038/cr.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai J., Chen L., Zhang Z., Zhang X., Lu X., et al. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut. 2019;68(12):2195–2205. doi: 10.1136/gutjnl-2019-318882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian X., Sun B., Chen C., Gao C., Zhang J., et al. Circulating tumor DNA 5-hydroxymethylcytosine as a novel diagnostic biomarker for esophageal cancer. Cell Research. 2018;28(5):597–600. doi: 10.1038/s41422-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Booth M.J., Branco M.R., Ficz G., Oxley D., Krueger F., et al. Quantitative Sequencing of 5-Methylcytosine and 5-Hydroxymethylcytosine at Single-Base Resolution. Science. 2012;336(6083):934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 48.Yu M., Hon G.C., Szulwach K.E., Song C.-X., Zhang L., et al. Base-Resolution Analysis of 5-Hydroxymethylcytosine in the Mammalian Genome. Cell. 2012;149(6):1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gabrieli T., Sharim H., Nifker G., Jeffet J., Shahal T., et al. Epigenetic Optical Mapping of 5-Hydroxymethylcytosine in Nanochannel Arrays. ACS Nano. 2018;12(7):7148–7158. doi: 10.1021/acsnano.8b03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bergamaschi A., Ning Y., Ku C.-J., Ellison C., Collin F., et al. Pilot study demonstrating changes in DNA hydroxymethylation enable detection of multiple cancers in plasma cell-free DNA, Preprint. Genetic and Genomic Medicine. Jan. 2020 doi: 10.1101/2020.01.22.20018382. [DOI] [Google Scholar]
- 51.Cui X., Cao L., Huang Y., Bai D., Huang S., et al. In Vitro diagnosis of DNA methylation biomarkers with digital PCR in breast tumors. The Analyst. 2018;143(13):3011–3020. doi: 10.1039/C8AN00205C. [DOI] [PubMed] [Google Scholar]
- 52.Volik S., Alcaide M., Morin R.D., Collins C. Cell-free DNA (cfDNA): Clinical Significance and Utility in Cancer Shaped By Emerging Technologies. Molecular Cancer Research. 2016;14(10):898–908. doi: 10.1158/1541-7786.MCR-16-0044. [DOI] [PubMed] [Google Scholar]
- 53.Richardson A.L., Iglehart J.D. BEAMing Up Personalized Medicine: Mutation Detection in Blood. Clinical Cancer Research. 2012;18(12):3209–3211. doi: 10.1158/1078-0432.CCR-12-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shemer R., Magenheim J., Dor Y. Digital Droplet PCR for Monitoring Tissue-Specific Cell Death Using DNA Methylation Patterns of Circulating Cell-Free DNA. Current Protocols in Molecular Biology. 2019;127(1):(Jun. doi: 10.1002/cpmb.90. [DOI] [PubMed] [Google Scholar]
- 55.Udesen P.B., Sørensen A.E., Joglekar M.V., Hardikar A.A., Wissing M.L.M., et al. Levels of circulating insulin cell-free DNA in women with polycystic ovary syndrome – a longitudinal cohort study. Reproductive Biology and Endocrinology. 2019;17(1):34. doi: 10.1186/s12958-019-0478-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.H. Li, R. Bai, Z. Zhao, L. Tao, M. Ma, et al., Application of droplet digital PCR to detect the pathogens of infectious diseases, Bioscience Reports 38 (6) (2018) BSR20181170. doi:10.1042/BSR20181170. [DOI] [PMC free article] [PubMed]
- 57.Jones M., Williams J., Gärtner K., Phillips R., Hurst J., et al. Low copy target detection by Droplet Digital PCR through application of a novel open access bioinformatic pipeline, ‘definetherain’. Journal of Virological Methods. 2014;202:46–53. doi: 10.1016/j.jviromet.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trypsteen W., Vynck M., De Neve J., Bonczkowski P., Kiselinova M., et al. ddpcRquant: Threshold determination for single channel droplet digital PCR experiments. Analytical and Bioanalytical Chemistry. 2015;407(19):5827–5834. doi: 10.1007/s00216-015-8773-4. [DOI] [PubMed] [Google Scholar]
- 59.D. Attali, R. Bidshahri, C. Haynes, J. Bryan, Ddpcr: An R package and web application for analysis of droplet digital PCR data, F1000Research 5 (2016) 1411. doi:10.12688/f1000research.9022.1. [DOI] [PMC free article] [PubMed]
- 60.Chiu A., Ayub M., Dive C., Brady G., Miller C.J. Twoddpcr: An R/Bioconductor package and Shiny app for Droplet Digital PCR analysis. Bioinformatics. 2017;33(17):2743–2745. doi: 10.1093/bioinformatics/btx308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brink B.G., Meskas J., Brinkman R.R. ddPCRclust: An R package and Shiny app for automated analysis of multiplexed ddPCR data. Bioinformatics. 2018;34(15):2687–2689. doi: 10.1093/bioinformatics/bty136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dobnik D., Štebih D., Blejec A., Morisset D., Žel J. Multiplex quantification of four DNA targets in one reaction with Bio-Rad droplet digital PCR system for GMO detection. Scientific Reports. 2016;6(1):35451. doi: 10.1038/srep35451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uehiro N., Sato F., Pu F., Tanaka S., Kawashima M., Kawaguchi K., Sugimoto M., Saji S., Toi M. Circulating cell-free DNA-based epigenetic assay can detect early breast cancer. Breast Cancer Research. 2016;18(1):129. doi: 10.1186/s13058-016-0788-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Häfner N., Diebolder H., Jansen L., Hoppe I., Dürst M., et al. Hypermethylated DAPK in serum DNA of women with uterine leiomyoma is a biomarker not restricted to cancer. Gynecologic Oncology. 2011;121(1):224–229. doi: 10.1016/j.ygyno.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 65.Jiang M., Zhang Y., Fei J., Chang X., Fan W., et al. Rapid quantification of DNA methylation by measuring relative peak heights in direct bisulfite-PCR sequencing traces. Laboratory Investigation. 2010;90(2):282–290. doi: 10.1038/labinvest.2009.132. [DOI] [PubMed] [Google Scholar]
- 66.Klein D. Quantification using real-time PCR technology: Applications and limitations. Trends in Molecular Medicine. 2002;8(6):257–260. doi: 10.1016/S1471-4914(02)02355-9. [DOI] [PubMed] [Google Scholar]
- 67.Hernández H.G., Tse M.Y., Pang S.C., Arboleda H., Forero D.A. Optimizing methodologies for PCR-based DNA methylation analysis. BioTechniques. 2013;55(4):(Oct. doi: 10.2144/000114087. [DOI] [PubMed] [Google Scholar]
- 68.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clinical Chemistry. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 69.Bustin S.A., Nolan T. Pitfalls of Quantitative Real-Time Reverse-Transcription Polymerase Chain Reaction. Journal of Biomolecular Techniques: JBT. 2004;15(3):155–166. [PMC free article] [PubMed] [Google Scholar]
- 70.Kuang J., Yan X., Genders A.J., Granata C., Bishop D.J. An overview of technical considerations when using quantitative real-time PCR analysis of gene expression in human exercise research. PLOS ONE. 2018;13(5) doi: 10.1371/journal.pone.0196438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andersen C.L., Jensen J.L., Ørntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 72.Pfaffl M.W., Tichopad A., Prgomet C., Neuvians T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnology Letters. 2004;26(6):509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 73.J. Vandesompele, K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, et al., Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes, Genome Biology 3 (7) (2002) research0034.1. doi:10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed]
- 74.Xie F., Xiao P., Chen D., Xu L., Zhang B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Molecular Biology. 2012;80(1):75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- 75.Gauri S., Ahmad M.R. ctDNA Detection in Microfluidic Platform: A Promising Biomarker for Personalized Cancer Chemotherapy. Journal of Sensors. 2020;2020 doi: 10.1155/2020/8353674. [DOI] [Google Scholar]
- 76.Chan K.A., Jiang P., Zheng Y.W., Liao G.J., Sun H., et al. Cancer Genome Scanning in Plasma: Detection of Tumor-Associated Copy Number Aberrations, Single-Nucleotide Variants, and Tumoral Heterogeneity by Massively Parallel Sequencing. Clinical Chemistry. 2013;59(1):211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 77.Couraud S., Vaca-Paniagua F., Villar S., Oliver J., Schuster T., et al. Noninvasive Diagnosis of Actionable Mutations by Deep Sequencing of Circulating Free DNA in Lung Cancer from Never-Smokers: A Proof-of-Concept Study from BioCAST/IFCT-1002. Clinical Cancer Research. 2014;20(17):4613–4624. doi: 10.1158/1078-0432.CCR-13-3063. [DOI] [PubMed] [Google Scholar]
- 78.Madic J., Kiialainen A., Bidard F.-C., Birzele F., Ramey G., et al. Circulating tumor DNA and circulating tumor cells in metastatic triple negative breast cancer patients. International Journal of Cancer. 2015;136(9):2158–2165. doi: 10.1002/ijc.29265. [DOI] [PubMed] [Google Scholar]
- 79.Chen M., Zhao H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Human Genomics. 2019;13(1):34. doi: 10.1186/s40246-019-0220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liang N., Li B., Jia Z., Wang C., Wu P., et al. Ultrasensitive detection of circulating tumour DNA via deep methylation sequencing aided by machine learning. Nature Biomedical Engineering. 2021;5(6):586–599. doi: 10.1038/s41551-021-00746-5. [DOI] [PubMed] [Google Scholar]
- 81.Glenn T.C. Field guide to next-generation DNA sequencers: FIELD GUIDE TO NEXT-GEN SEQUENCERS. Molecular Ecology Resources. 2011;11(5):759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 82.Singh R.R. Next-Generation Sequencing in High-Sensitive Detection of Mutations in Tumors: Challenges. Advances, and Applications, The Journal of Molecular Diagnostics. 2020;22(8):994–1007. doi: 10.1016/j.jmoldx.2020.04.213. [DOI] [PubMed] [Google Scholar]
- 83.The Cancer Genome Atlas Program - National Cancer Institute, https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga (06/13/2018 - 08:00).
- 84.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., et al. NCBI GEO: Archive for functional genomics data sets—update. Nucleic Acids Research. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moran S., Arribas C., Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8(3):389–399. doi: 10.2217/epi.15.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stirzaker C., Taberlay P.C., Statham A.L., Clark S.J. Mining cancer methylomes: Prospects and challenges. Trends in Genetics. 2014;30(2):75–84. doi: 10.1016/j.tig.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 87.Wilhelm-Benartzi C.S., Koestler D.C., Karagas M.R., Flanagan J.M., Christensen B.C., et al. Review of processing and analysis methods for DNA methylation array data. British Journal of Cancer. 2013;109(6):1394–1402. doi: 10.1038/bjc.2013.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.J.T. Leek, R.B. Scharpf, H.C. Bravo, D. Simcha, B. Langmead, et al., Tackling the widespread and critical impact of batch effects in high-throughput data, Nature reviews. Genetics 11 (10) (2010) 10.1038/nrg2825. doi:10.1038/nrg2825. [DOI] [PMC free article] [PubMed]
- 89.Patel R.K., Jain M., Toolkit N.G.S.Q.C. A Toolkit for Quality Control of Next Generation Sequencing Data. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pandey R.V., Pabinger S., Kriegner A., Weinhäusel A. ClinQC: A tool for quality control and cleaning of Sanger and NGS data in clinical research. BMC Bioinformatics. 2016;17(1):56. doi: 10.1186/s12859-016-0915-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Haeussler M., Zweig A.S., Tyner C., Speir M.L., Rosenbloom K.R., et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Research. 2019;47(D1):D853–D858. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ding W., Chen J., Feng G., Chen G., Wu J., et al. DNMIVD: DNA methylation interactive visualization database. Nucleic Acids Research. 2020;48(D1):D856–D862. doi: 10.1093/nar/gkz830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mallona I., Díez-Villanueva A., Peinado M.A. Methylation plotter: A web tool for dynamic visualization of DNA methylation data. Source Code for Biology and Medicine. 2014;9(1):11. doi: 10.1186/1751-0473-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Briefings in Bioinformatics. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liang F., Tang B., Wang Y., Wang J., Yu C., et al. WBSA: Web Service for Bisulfite Sequencing Data Analysis. PLOS ONE. 2014;9(1) doi: 10.1371/journal.pone.0086707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.L. Gay, A.-M. Baker, T.A. Graham, Tumour Cell Heterogeneity, F1000Research 5 (2016) 238. doi:10.12688/f1000research.7210.1. [DOI] [PMC free article] [PubMed]
- 97.Altschuler S.J., Wu L.F. Cellular Heterogeneity: Do Differences Make a Difference? Cell. 2010;141(4):559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Castro-Giner F., Gkountela S., Donato C., Alborelli I., Quagliata L., et al. Cancer Diagnosis Using a Liquid Biopsy: Challenges and Expectations. Diagnostics. 2018;8(2):31. doi: 10.3390/diagnostics8020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Russano M., Napolitano A., Ribelli G., Iuliani M., Simonetti S., et al. Liquid biopsy and tumor heterogeneity in metastatic solid tumors: The potentiality of blood samples. Journal of Experimental & Clinical Cancer Research. 2020;39(1):95. doi: 10.1186/s13046-020-01601-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.S. Ramón y Cajal, M. Sesé, C. Capdevila, T. Aasen, L. De Mattos-Arruda, et al., Clinical implications of intratumor heterogeneity: Challenges and opportunities, Journal of Molecular Medicine 98 (2) (2020) 161–177. doi:10.1007/s00109-020-01874-2. [DOI] [PMC free article] [PubMed]
- 101.Huan Q., Zhang Y., Wu S., Qian W. HeteroMeth: A Database of Cell-to-cell Heterogeneity in DNA Methylation. Genomics, Proteomics & Bioinformatics. 2018;16(4):234–243. doi: 10.1016/j.gpb.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scherer M., Nebel A., Franke A., Walter J., Lengauer T., et al. Quantitative comparison of within-sample heterogeneity scores for DNA methylation data. Nucleic Acids Research. 2020;48(8) doi: 10.1093/nar/gkaa120. e46–e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim M.-C., Kim N.-Y., Seo Y.-R., Kim Y. An Integrated Analysis of the Genome-Wide Profiles of DNA Methylation and mRNA Expression Defining the Side Population of a Human Malignant Mesothelioma Cell Line. Journal of Cancer. 2016;7(12):1668–1679. doi: 10.7150/jca.15423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Barault L., Amatu A., Siravegna G., Ponzetti A., Moran S., et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut. 2018;67(11):1995–2005. doi: 10.1136/gutjnl-2016-313372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mouliere F., El Messaoudi S., Pang D., Dritschilo A., Thierry A.R. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Molecular Oncology. 2014;8(5):927–941. doi: 10.1016/j.molonc.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Salvi S., Gurioli G., De Giorgi U., Conteduca V., Tedaldi G., et al. Cell-free DNA as a diagnostic marker for cancer: Current insights. OncoTargets and Therapy. 2016;9:6549–6559. doi: 10.2147/OTT.S100901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Song C.-X., Yin S., Ma L., Wheeler A., Chen Y., et al. 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Research. 2017;27(10):1231–1242. doi: 10.1038/cr.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu T., Gao H. Hydroxymethylation and tumors: Can 5-hydroxymethylation be used as a marker for tumor diagnosis and treatment? Human Genomics. 2020;14(1):15. doi: 10.1186/s40246-020-00265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang J., Han X., Gao C., Xing Y., Qi Z., et al. 5-Hydroxymethylome in Circulating Cell-free DNA as A Potential Biomarker for Non-small-cell Lung Cancer. Genomics, Proteomics & Bioinformatics. 2018;16(3):187–199. doi: 10.1016/j.gpb.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dong C., Chen J., Zheng J., Liang Y., Yu T., et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic and predictive biomarkers for coronary artery disease. Clinical Epigenetics. 2020;12(1):17. doi: 10.1186/s13148-020-0810-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gai W., Ji L., Lam W.K.J., Sun K., Jiang P., et al. Liver- and Colon-Specific DNA Methylation Markers in Plasma for Investigation of Colorectal Cancers with or without Liver Metastases. Clinical Chemistry. 2018;64(8):1239–1249. doi: 10.1373/clinchem.2018.290304. [DOI] [PubMed] [Google Scholar]
- 112.Titus A.J., Gallimore R.M., Salas L.A., Christensen B.C. Cell-type deconvolution from DNA methylation: A review of recent applications. Human Molecular Genetics. 2017;26(R2):R216–R224. doi: 10.1093/hmg/ddx275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Teschendorff A.E., Zheng S.C. Cell-type deconvolution in epigenome-wide association studies: A review and recommendations. Epigenomics. 2017;9(5):757–768. doi: 10.2217/epi-2016-0153. [DOI] [PubMed] [Google Scholar]
- 114.Teschendorff A.E., Breeze C.E., Zheng S.C., Beck S. A comparison of reference-based algorithms for correcting cell-type heterogeneity in Epigenome-Wide Association Studies. BMC Bioinformatics. 2017;18(1):105. doi: 10.1186/s12859-017-1511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., et al. Robust enumeration of cell subsets from tissue expression profiles. Nature Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gaujoux R., Seoighe C. Cell Mix: A comprehensive toolbox for gene expression deconvolution. Bioinformatics. 2013;29(17):2211–2212. doi: 10.1093/bioinformatics/btt351. [DOI] [PubMed] [Google Scholar]
- 117.Kang K., Meng Q., Shats I., Umbach D.M., Li M., et al. CDSeq: A novel complete deconvolution method for dissecting heterogeneous samples using gene expression data. PLOS Computational Biology. 2019;15(12) doi: 10.1371/journal.pcbi.1007510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li Z., Wu H. TOAST: Improving reference-free cell composition estimation by cross-cell type differential analysis. Genome Biology. 2019;20(1):190. doi: 10.1186/s13059-019-1778-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Houseman E.A., Molitor J., Marsit C.J. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics. 2014;30(10):1431–1439. doi: 10.1093/bioinformatics/btu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zou J., Lippert C., Heckerman D., Aryee M., Listgarten J. Epigenome-wide association studies without the need for cell-type composition. Nature Methods. 2014;11(3):309–311. doi: 10.1038/nmeth.2815. [DOI] [PubMed] [Google Scholar]
- 121.Rahmani E., Zaitlen N., Baran Y., Eng C., Hu D., et al. Sparse PCA corrects for cell type heterogeneity in epigenome-wide association studies. Nature Methods. 2016;13(5):443–445. doi: 10.1038/nmeth.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.J.T. Leek, J.D. Storey, Capturing Heterogeneity in Gene Expression Studies by Surrogate Variable Analysis, PLoS Genetics 3 (9) (Sep. 2007). doi:10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed]
- 123.Shen S.Y., Singhania R., Fehringer G., Chakravarthy A., Roehrl M.H.A., et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018;563(7732):579–583. doi: 10.1038/s41586-018-0703-0. [DOI] [PubMed] [Google Scholar]
- 124.Wan N., Weinberg D., Liu T.-Y., Niehaus K., Ariazi E.A., et al. Machine learning enables detection of early-stage colorectal cancer by whole-genome sequencing of plasma cell-free DNA. BMC Cancer. 2019;19(1):832. doi: 10.1186/s12885-019-6003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Luo H., Zhao Q., Wei W., Zheng L., Yi S., et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Science Translational Medicine. 2020;12(524):eaax7533. doi: 10.1126/scitranslmed.aax7533. [DOI] [PubMed] [Google Scholar]
- 126.Panagopoulou M., Karaglani M., Balgkouranidou I., Biziota E., Koukaki T., et al. Circulating cell-free DNA in breast cancer: Size profiling, levels, and methylation patterns lead to prognostic and predictive classifiers. Oncogene. 2019;38(18):3387–3401. doi: 10.1038/s41388-018-0660-y. [DOI] [PubMed] [Google Scholar]
- 127.Liu M.C., Oxnard G.R., Klein E.A., Swanton C., Seiden M.V., et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Annals of Oncology. 2020;31(6):745–759. doi: 10.1016/j.annonc.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.B. Cao, S.J. Pan, Y. Zhang, D.-Y. Yeung, Q. Yang, Adaptive transfer learning, in: proceedings of the AAAI Conference on Artificial Intelligence, Vol. 24, 2010.
- 129.Zhou Q., Lim J.-Q., Sung W.-K., Li G. An integrated package for bisulfite DNA methylation data analysis with Indel-sensitive mapping. BMC Bioinformatics. 2019;20(1):47. doi: 10.1186/s12859-018-2593-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xi Y., Li W. BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10(1):232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Krueger F., Andrews S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Guo W., Fiziev P., Yan W., Cokus S., Sun X., et al. BS-Seeker2: A versatile aligning pipeline for bisulfite sequencing data. BMC Genomics. 2013;14(1):774. doi: 10.1186/1471-2164-14-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.B.S. Pedersen, K. Eyring, S. De, I.V. Yang, D.A. Schwartz, Fast and accurate alignment of long bisulfite-seq reads 8.
- 134.Hansen K.D., Langmead B., Irizarry R.A. BSmooth: From whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biology. 2012;13(10):R83. doi: 10.1186/gb-2012-13-10-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pedersen B., Hsieh T.-F., Ibarra C., Fischer R.L. MethylCoder: Software pipeline for bisulfite-treated sequences. Bioinformatics. 2011;27(17):2435–2436. doi: 10.1093/bioinformatics/btr394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hoffmann S., Otto C., Kurtz S., Sharma C.M., Khaitovich P., et al. Fast Mapping of Short Sequences with Mismatches, Insertions and Deletions Using Index Structures. PLoS Computational Biology. 2009;5(9) doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wu T.D., Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26(7):873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Harris E.Y., Ponts N., Le Roch K.G., Lonardi S. BRAT-BW: Efficient and accurate mapping of bisulfite-treated reads. Bioinformatics. 2012;28(13):1795–1796. doi: 10.1093/bioinformatics/bts264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.N. Prezza, C. Del Fabbro, F. Vezzi, E. De Paoli, A. Policriti, ERNE-BS5: Aligning BS-Treated Sequences by Multiple Hits on a 5-Letters Alphabet, 2012. doi:10.1145/2382936.2382938.
- 140.Marco-Sola S., Sammeth M., Guigó R., Ribeca P. The GEM mapper: Fast, accurate and versatile alignment by filtration. Nature Methods. 2012;9(12):1185–1188. doi: 10.1038/nmeth.2221. [DOI] [PubMed] [Google Scholar]
- 141.Frith M.C., Mori R., Asai K. A mostly traditional approach improves alignment of bisulfite-converted DNA. Nucleic Acids Research. 2012;40(13) doi: 10.1093/nar/gks275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sun K., Li L., Ma L., Zhao Y., Deng L., et al. Msuite: A High-Performance and Versatile DNA Methylation Data-Analysis Toolkit. Patterns. 2020;1(8) doi: 10.1016/j.patter.2020.100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sun R., Tian Y., Chen X. TAMeBS: A sensitive bisulfite-sequencing read mapping tool for DNA methylation analysis, in. IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2014;2014:176–181. doi: 10.1109/BIBM.2014.6999148. [DOI] [Google Scholar]
- 144.Pelizzola M., Koga Y., Urban A.E., Krauthammer M., Weissman S., et al. MEDME: An experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Research. 2008;18(10):1652–1659. doi: 10.1101/gr.080721.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wilson G.A., Dhami P., Feber A., Cortázar D., Suzuki Y., et al. Resources for methylome analysis suitable for gene knockout studies of potential epigenome modifiers. GigaScience. 2012;1(1):3. doi: 10.1186/2047-217X-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.G.A. Wilson, S. Beck, Computational Analysis and Integration of MeDIP-seq Methylome Data, in: J.K. Kulski (Ed.), Next Generation Sequencing - Advances, Applications and Challenges, InTech, 2016. doi:10.5772/61207. [PubMed]
- 147.Bhasin J.M., Hu B., Ting A.H. MethylAction: Detecting differentially methylated regions that distinguish biological subtypes. Nucleic Acids Research. 2016;44(1):106–116. doi: 10.1093/nar/gkv1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Müller F., Scherer M., Assenov Y., Lutsik P., Walter J., et al. RnBeads 2.0: Comprehensive analysis of DNA methylation data. Genome Biology. 2019;20(1):55. doi: 10.1186/s13059-019-1664-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Peters T.J., Buckley M.J., Statham A.L., Pidsley R., Samaras K., et al. De novo identification of differentially methylated regions in the human genome. Epigenetics & Chromatin. 2015;8(1):6. doi: 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Catoni M., Tsang J.M., Greco A.P., Zabet N.R. DMRcaller: A versatile R/Bioconductor package for detection and visualization of differentially methylated regions in CpG and non-CpG contexts. Nucleic Acids Research. Jul. 2018 doi: 10.1093/nar/gky602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., et al. methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biology. 2012;13(10):R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Park Y., Figueroa M.E., Rozek L.S., Sartor M.A. MethylSig: A whole genome DNA methylation analysis pipeline. Bioinformatics. 2014;30(17):2414–2422. doi: 10.1093/bioinformatics/btu339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Feng H., Wu H. Differential methylation analysis for bisulfite sequencing using DSS. Quantitative Biology. 2019;7(4):327–334. doi: 10.1007/s40484-019-0183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mayne B.T., Leemaqz S.Y., Buckberry S., Lopez C.M.R., Roberts C.T., et al. msgbsr: An r package for analysing methylation-sensitive restriction enzyme sequencing data. Scientific reports. 2018;8(1):1–8. doi: 10.1038/s41598-018-19655-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.D. Becker, P. Lutsik, P. Ebert, C. Bock, T. Lengauer, et al., BiQ Analyzer HiMod: An interactive software tool for high-throughput locus-specific analysis of 5-methylcytosine and its oxidized derivatives, Nucleic Acids Research 42 (Web Server issue) (2014) W501–W507. doi:10.1093/nar/gku457. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used for analysis in the current study can be found at The Cancer Genome Atlas (TCGA) https://portal.gdc.cancer.gov/ and Cell Free Epigenome Atlas (CFEA) http://www.bio-data.cn/CFEA/ repositories.