Abstract
The cotranslational incorporation of the unusual amino acid selenocysteine (Sec) into both prokaryotic and eukaryotic proteins requires the recoding of a UGA stop codon as one specific for Sec. The recognition of UGA as Sec in mammalian selenoproteins requires a Sec insertion sequence (SECIS) element in the 3′ untranslated region as well as the SECIS binding protein SBP2. Here we report a detailed analysis of SBP2 structure and function using truncation and site-directed mutagenesis. We have localized the RNA binding domain to a conserved region shared with several ribosomal proteins and eukaryotic translation termination release factor 1. We also identified a separate and novel functional domain N-terminal to the RNA binding domain which was required for Sec insertion but not for SECIS binding. Conversely, we showed that the RNA binding domain was necessary but not sufficient for Sec insertion and that the conserved glycine residue within this domain was required for SECIS binding. Using glycerol gradient sedimentation, we found that SBP2 was stably associated with the ribosomal fraction of cell lysates and that this interaction was not dependent on its SECIS binding activity. This interaction also occurred with purified components in vitro, and we present data which suggest that the SBP2-ribosome interaction occurs via 28S rRNA. SBP2 may, therefore, have a distinct function in selecting the ribosomes to be used for Sec insertion.
There are many examples of translational regulation that involve RNA binding proteins interacting with specific sequences in the 3′ untranslated regions (UTRs) of various mRNAs (10, 18). A specialized case of this is found in the incorporation of selenocysteine (Sec) into a select group of eukaryotic proteins where a sequence specific 3′ UTR binding protein is required for Sec insertion at its cognate UGA codon (6). The Sec-containing proteins that have been characterized perform myriad biological functions including oxidant defense and hormone maturation (reviewed in references 9 and 20). While mice deficient in the selenoprotein glutathione peroxidase are viable (11), one or more selenoproteins are apparently required for early development, as elimination of the tRNASec gene in mice causes early embryo lethality (3).
While the incorporation of Sec into the Escherichia coli formate dehydrogenase isozymes is fairly well characterized (2), the system which governs mammalian Sec insertion has only been partially elucidated. Several essential components of what we here term the Sec insertion complex (SIC) have been identified. These include the UGA codon that encodes Sec (13), the Sec insertion sequence (SECIS) element found in all selenoprotein 3′ UTRs (1), and the recently identified SECIS binding protein (SBP2), which we demonstrated to be required for Sec insertion in vitro (6). A fourth component of the SIC that is also likely to be required for Sec incorporation is the Sec-specific elongation factor (eEFsec) that has been recently shown to be functional in transfected cells (7, 21). Because Sec is encoded by what is ordinarily a stop codon, the mechanism of Sec insertion is likely to involve direct competition with translation termination. The SIC may therefore interact with or regulate the interactions of the release factors required for translation termination.
This work focuses on the RNA binding protein SBP2, a novel 94-kDa protein that shares a 32-amino acid motif with several ribosomal proteins and eukaryotic translation termination release factor 1 (eRF-1). eRF-1 functionally and structurally mimics a tRNA molecule that is specific for stop codons and in this fashion binds the ribosomal A site and terminates chain elongation (19). The fact that SBP2 and eRF-1 share this sequence suggests that they also share a common class of targets, which may shed light on the molecular basis for the competition between Sec insertion and termination. This conserved sequence has been proposed to be a novel RNA binding motif (12) and has recently been demonstrated to be involved in the binding of ribosomal protein L30 to its own mRNA (15). In this report, we establish that this motif, which we will refer to as the L30 RNA binding domain, is required for SBP2 RNA binding activity and function as measured by the ability to incorporate Sec in vitro. In addition, we have identified a nonoverlapping functional domain that is required for Sec insertion but not RNA binding. Further analysis indicates that SBP2 is stably associated with the ribosomal fraction in transfected cells and in vitro, and that this interaction may be mediated by 28S rRNA. We hypothesize that SBP2 may be involved in selecting a subset of ribosomes which are competent for Sec insertion.
MATERIALS AND METHODS
Mutagenesis.
All truncated SBP2 constructs were derived from internal PCR primers (Table 1) which added a methionine to the N terminus. Annealing sites were chosen so that the optimal initiation sequence ATGG would be present for all truncation mutants except TM535–846, which used a naturally occurring ATG. PCR fragments were TA cloned into pCR3.1 (Invitrogen). Point mutations were made with an Altered Sites II Ex-1 mutagenesis kit (Promega). Mutant constructs were sequenced in their entirety by automated DNA sequencing. The human SBP2-like protein (hSLP) was derived from human hypothetical protein KIAA0256 (GenBank accession no. 6634006) obtained from the Kazusa DNA Research Institute (Kisarazu, Japan). This study made use of the C-terminal half of hSLP, which is similar to SBP2. It was obtained by PCR using primers hSLP-5′ and hSLP-3′ (Table 1).
TABLE 1.
Oligonucleotides used in this study
| Primer name | Sequencea |
|---|---|
| TM399–846-5′ | CCATGGAATTCCCCAACCTGTCAGTTGCA |
| TM459–846-5′ | CCATGGCCAAGCCATCCTCCAGACCCGTC |
| TM517-846-5′ | CCATGGCCAAGAAGCCCACCTCACTGAAG |
| TM535-846-5′ | ATGCAGCAGCGACTCCAAG |
| TM585-846-5′ | CCATGGTGGTTGAGGGTGAGTCAGAAGAG |
| TM399-777-3′ | TTACTGCTCCTGCCGCATCGTCTC |
| TM399-728-3′ | TTACCCCAGTGCCTTGCGGTTGAG |
| G669R | CGGCTTGTGCTGAGGCTGAGGGAG |
| G669A | CGGCTTGTGCTGGCGCTGAGGGAG |
| C684L | AAGCTGAAGTTGATCATCATCTC |
| C684W | AAGCTGAAGTGGATCATCATCTC |
| hSLP-5′ | ATGGAGCAAAAAAAATTACAGGAAGC |
| hSLP-3′ | TTACGTAGTTTGCGTTGTGTAATAG |
Residues that vary from the wild-type target sequence are in italics.
RNA probes and binding assays.
32P-labeled wild-type and mutant phospholipid hydroperoxide glutathione peroxidase (PHGPx) 3′ UTR probes were synthesized exactly as described elsewhere (5). Electrophoretic mobility shift assays (EMSA) included 4.4 fmol of in vitro-translated SBP2 as indicated for the figures and 20 fmol of wild-type or mutant PHGPx 3′ UTR in 1× phosphate-buffered saline (PBS) supplemented with 250 μg of E. coli tRNA per ml. 10 mM dithiothreitol (DTT), and 5 μg of soybean trypsin inhibitor (Sigma) per ml. Complexes were formed at 37°C for 30 min and then resolved on 4% nondenaturing polyacrylamide gels, which were dried and exposed to a PhosphorImager cassette (Molecular Dynamics).
In vitro translation.
Plasmid DNAs containing wild-type and mutant SBP2 constructs were linearized with XbaI and used as templates for in vitro transcription with T7 RNA polymerase (Ribomax T7; Promega) in the presence of m7G(5′)ppp(5′)G (AP Biotech). A 50-ng aliquot of each SBP2 RNA was used in 12.5 or 25-μl rabbit reticulocyte lysate in vitro translation reactions in the presence of [35S]Met as described by the manufacturer (Promega). Translation products were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% gel and quantitated by PhosphorImager analysis. The amount of each protein was determined by quantitation of known amounts of [35S]Met spotted on 3-mm filter paper and calculated based on an endogenous concentration of 5 μM cold Met in the lysate (as specified by the manufacturer).
Expression of recombinant SBP2.
For the construction of (Strep-tagged C-terminal SBP2, the original ATCC clone corresponding to expressed sequence tag H31811 was digested with EcoRI and XhoI and subcloned into the EcoRI and XhoI sites of pASK-IBA7 (Strep-tag II; Genosys). This vector added to the N terminus of the SBP2 clone the amino acid sequence MASWSHPQFEKIEGRRDRGP (the Strep tag sequence is underlined), which begins with amino acid 399.
Glycerol gradients.
McArdle 7777 cells, a rat hepatoma cell line, were transiently transfected with wild-type and mutant SBP2 clones using LipofectAMINE (Life Technologies); 40 h posttransfection, the cells were washed and then harvested by scraping into PBSD (1× PBS, 2 mM DTT). The cells were disrupted by 15 strokes with a high-clearance Dounce homogenizer. The extracts were spun at 14,000 × g for 10 min at 4°C. The supernatants were diluted to 1 mg/ml, and 500 μl of each was loaded onto 10 to 30% linear glycerol gradients made in PBSD plus or minus NaCl or EDTA as indicated. For in vitro complex assembly, 5 μg of purified Strep-tagged SBP2 was incubated with or without 2.0 A260 units of salt-washed ribosomes purified from rabbit reticulocyte lysate (generously provided by W. Merrick, Case Western Reserve University) in a total volume of 0.5 ml diluted with PBSD. The mixture was incubated at 37°C for 5 min, placed on ice for 10 to 15 min, and then loaded onto a 10 to 30% linear glycerol gradient made in PBSD. All gradients were spun at 210,000 × g in a SW41 rotor for 3.5 h (McArdle cell gradients) or 4.5 h (in vitro complexes) at 4°C. Fractions (0.6 ml) were pulled from the top of each gradient, and 200 μl from each fraction was subjected to trichloroacetic acid (TCA) precipitation (10% TCA, 0.05% Tween 20). Fractions from gradients that contained Strep-tagged recombinant SBP2 alone were supplemented with 5 μg of pure soybean trypsin inhibitor (Sigma) as a carrier. Precipitated proteins were resolved by SDS-PAGE (12% gel), blotted to nitrocellulose, blocked with 3% BSA in PBST (1× PBS plus 0.2% Tween 20), and, for Strep-tagged proteins, probed with a 1:4,000 dilution of alkaline phosphatase-conjugated streptavidin (AP Biotech). For untagged proteins, the blots were probed with a 1:2,000 dilution of anti-SBP2 polyclonal antibody followed by a 1:5,000 dilution of alkaline phosphatase-conjugated anti-rabbit immunoglobulin G secondary antibody. Blots were developed in nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (Promega).
PHGPx translation assay.
PHGPx translation was monitored essentially as described previously (6), except that the protein was labeled with [35S]Met instead of [75Se]Sec. Our previous results showed that the incorporation of 75Se-labeled Sec was both codon and SECIS element dependent (6). PHGPx translation can also be assayed by [35S]Met labeling, which is also codon and SECIS element dependent and which yields exactly the same results as 75Se labeling. Briefly, 4.4-fmol aliquots of in vitro-translated SBP2 proteins were added in a total volume of 2 μl (brought to volume with fresh rabbit reticulocyte lysate) to a standard 12.5-μl assay including 5 μCi of [35S]Met. PHGPx translation products were purified from the entire reaction using bromosulfophthalein S-glutathione (BSP)–agarose, and the bound proteins were resolved by SDS-PAGE (15% gel) followed by autoradiography or PhosphorImager analysis.
RESULTS
Analysis of SBP2 truncation mutants.
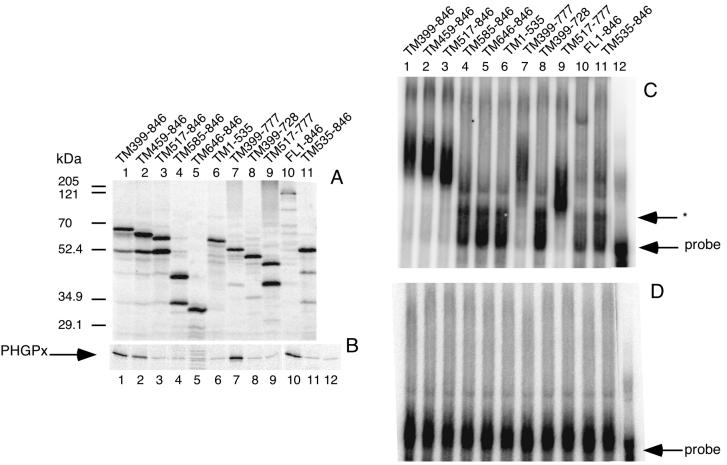
In earlier work, we showed that the N-terminal half of SBP2 is dispensable for both SECIS binding and selenoprotein synthesis (6). To gain more insight into the mechanism of SBP2 action, we constructed a series of truncation mutants in the C-terminal half of SBP2 in an effort to define domains necessary for RNA binding and functional activity. The truncation mutants (diagrammed in Fig. 2) are termed TMx-y, where x-y refers to the positions of the remaining amino acids derived from full-length SBP2. All constructs were efficiently translated in rabbit reticulocyte lysate (Fig. 1A) except for full-length SBP2 (lane 10). This is consistent with our difficulty in expressing full-length SBP2 in bacteria, but the basis for reduced translation is unknown. Excess SBP2 (4.4 fmol) protein was used in the following assays; the exception was the full-length protein, of which only 0.9 fmol was used. This amount of full-length SBP2 is still in excess for the functional assay but is within the lower portion of the linear range of the RNA binding assay (data not shown). As we previously reported, the apparent and predicted molecular masses of SBP2 are not in agreement and differ by approximately 26 kDa. The truncation mutants described above help to delineate the origin of this anomaly, as the difference in molecular mass between TM517–846 and TM585–846 is predicted to be only 7.4 kDa but is observed to be approximately 20 kDa by SDS-PAGE (Fig. 1). It should be noted that the major contaminating lower-molecular-mass band at 52 kDa is likely the result of internal initiation at Met 535. To test the contribution of this contaminant to SBP2 RNA binding and functional activity, TM535–846 (Fig. 1A, lane 11) was engineered to begin with Met 535 and runs at exactly the same molecular weight as the lower-molecular-weight band in lanes 1 to 3.
FIG. 2.
Summary of SBP2 truncation mutant data. RNA binding and functional activity (Fig. 1) and ribosome binding activity (Fig. 6) are summarized to the right of the diagram of each SBP2 truncation mutant. The values for functional activity represent the percentage of the wild-type activity normalized to that found with the addition of C-terminal SBP2 (TM399-846); those for binding activity represent the percentage of wild-type probe shifted to slower-migrating species normalized to the shift observed with TM399–846. All quantitation was performed by PhosphorImager analysis. The putative functional domain as determined from the mutant data is indicated at the bottom. The hatched box on the full-length diagram represents the conserved L30 RNA binding domain. Constructs that were not tested in the ribosome binding assay are marked “ND.”
FIG. 1.
Expression and activity of SBP2 truncation mutants. (A) [35S]Met-labeled in vitro-translated SBP2 resolved by SDS-PAGE. Mutants are identified by the amino acid positions indicated. (B) In vitro PHGPx translation assay in which 4.4 fmol of each SBP2 protein was added in the presence of [35S]Met. PHGPx was purified from the reaction using BSP-agarose. Lane 12 contains no SBP2. (C) EMSA for the mutants listed in panel A. 35S-labeled SBP2 (4.4 fmol) was incubated with 20 fmol of wild-type 32P-labeled PHGPx 3′ UTR, and the complexes were resolved on a 4% nondenaturing gel. The asterisk indicates the position of a SECIS-specific complex in nonsupplemented reticulocyte lysate. (D) Same as panel C except that the probe was an AUGA deletion mutant of the PHGPx 3′ UTR (14).
To assay SBP2 function, equimolar amounts of in vitro-translated SBP2 protein were added to a PHGPx translation assay. The results are shown in Fig. 1B and summarized in Fig. 2, where the data are expressed as a percentage of the wild-type activity derived from the addition of C-terminal SBP2 (TM399–846). As mentioned above, the elimination of amino acids 1 to 398 has no effect on activity. Elimination of amino acids 1 to 458 (mutant TM459–846; lane 2) results in a 50% reduction in activity, and the elimination of amino acids 1 to 516 (mutant TM517–846; lane 3) results in a nearly complete loss of activity. Interestingly, neither of these mutations has an effect on SECIS binding activity (see below). Elimination of the C-terminal amino acids 778 to 846 (mutant TM399–777; lane 7) has no effect on activity, but removal of 49 additional amino acids (TM399–728; lane 8) eliminates both function and RNA binding activity. From these data we conclude that amino acids 399 to 517 are specifically required for Sec insertion and as such can be considered a distinct functional domain.
To assay RNA binding, in vitro-translated SBP2 was incubated with radiolabeled PHGPx 3′ UTR RNA and analyzed by EMSA. RNA binding activities of wild-type and mutant SBP2 proteins are shown in Fig. 1C. All of the expressed proteins that bind the PHGPx 3′ UTR do so specifically, as shown by the lack of binding to a mutant 3′ UTR from which the conserved AUGA element has been deleted (Fig. 1D). The elimination of amino acids 1 to 534 (mutant TM535–846; lane 11) as well as 778–846 (mutant TM399–728; lane 8) severely reduces binding efficiency, indicating that the RNA binding domain encompasses the region from 517 to 777. The SECIS binding data are presented numerically in Fig. 2 as the percentage of wild-type probe shifted to slower-migrating species normalized to the binding activity of TM399–846. It is noteworthy that any mutation which eliminates RNA binding also eliminates Sec insertion activity, suggesting that RNA binding is absolutely required for Sec insertion. The mutant which corresponds to an internal initiation at M535 does not contribute significantly to the RNA binding activity (Fig. 1C, lane 11) and lacks any detectable functional activity (Fig. 1B, lane 11). Interestingly, the region that appears to be responsible for the aberrant migration during SDS-PAGE overlaps with part of the RNA binding domain, and the modification or unique structure which causes the aberration may be necessary for proper SBP2 function. Overall, these results indicate that the RNA binding domain resides between amino acids 517 and 777, a region which includes the putative RNA binding motif known to be required for ribosomal protein L30 RNA binding activity (15). In addition, it appears that the SBP2 functional domain (amino acids 399 to 517) does not overlap with the RNA binding domain and may represent the site of interaction with another component of the Sec insertion machinery.
Analysis of SBP2 point mutants.
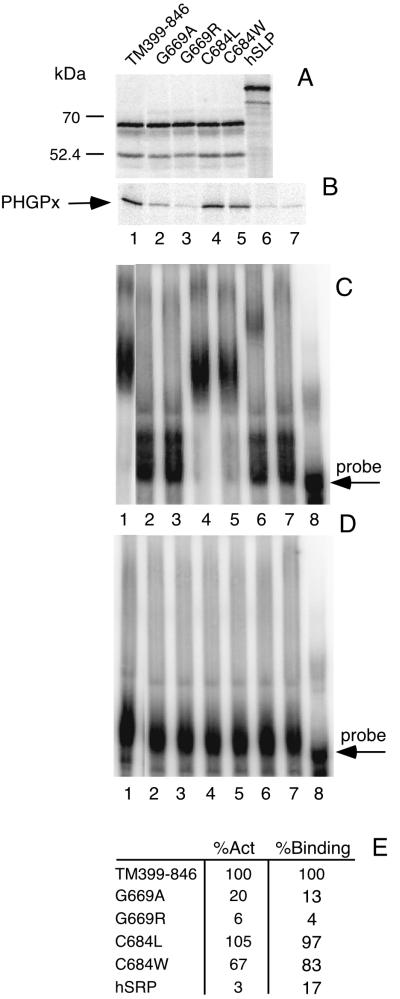
To further characterize the RNA binding domain, we introduced point mutations in the stretch of amino acids that is conserved among the sequences shown in the alignment of what we here term the L30 RNA binding motif (Fig. 3). No other significant similarity among these proteins exists. As shown in Fig. 3, SBP2 contains a Cys residue at position 25 (SBP2 position 684) that is unique among the group. To test if this unique Cys residue is required for binding, C684 was changed to the more commonly observed Leu or to a potentially disruptive Trp residue. As shown in Fig. 4B and C, neither mutant was significantly diminished in its ability to bind the PHGPx 3′ UTR or enhance PHGPx translation. Quantitation of the effects indicates a slightly lower amount of both activities for the C684W mutant (Fig. 4). We suspect that the introduction of a bulky aromatic side group into the conserved RNA binding domain may cause some disruption of the local structure. Binding specificity is also not affected by these mutations, as shown in Fig. 4D, in which a PHGPx 3′ UTR with a mutant SECIS element was used. These data indicate that the unique Cys residue found in SBP2 is not required for RNA binding activity.
FIG. 3.
Alignment of amino acids contained within the L30 RNA binding motif. Sequences are grouped according to similarity. The top group consists of the omnipotent suppressors of translation termination (SUP1) which have since been identified as eRF-1. The second group consists of the ribosomal protein L30 sequences. The third group consists of SBP2 and hSLP. The remaining groups consist of ribosomal proteins and RimK. Conserved amino acids are in boldface. Species noted include Trypanosoma cruzi, Methanococcus vannielii, Suefolobus acidocaldarius, Haloarcula marismortui, Bacillus subtilis, and Trypanosoma brucei.
FIG. 4.
Analysis of SBP2 point mutants and hSLP. (A) [35S]Met-labeled SBP2 proteins were resolved by SDS-PAGE. (B) Assay of PHGPx expression as describe for Fig. 1B. Lane 8 contains no SBP2. (C) EMSA for SBP2 mutant proteins as described for Fig. 1C. (D) Same as panel C except that the probe was an AUGA deletion mutant of the PHGPx 3′ UTR. Lane 7 contains 2 μl of rabbit reticulocyte lysate without SBP2; lane 8 contains probe in the absence of rabbit reticulocyte lysate. (E) Quantitation of the results in panels B and C expressed as percentage of activity (%Act) or percentage of binding (%Binding) as described for Fig. 2.
We also wished to determine if the invariant Gly residue at position 10 (SBP2 position 669) is required for RNA binding activity. Alteration of this Gly residue to Arg (G669R) completely abrogated both binding activity and function, while a G669A mutation eliminated all detectable binding activity but not all of the detectable enhancement of PHGPx translation (Fig. 4B and C). To test the idea that any of the conserved domains shown in Fig. 3 might suffice for PHGPx 3′ UTR binding, the SBP2 analog known as human hypothetical protein KIAA0256 (here termed human SBP2-like protein, or hSLP) was subcloned and expressed in vitro. This does not appear to be the human SBP2 homologue, as sequences which are greater than 90% identical to rat SBP2 are present in the human expressed sequence tag database. The hSLP construct used here consists of the C-terminal 428 amino acids which are 46% identical to the C-terminal 447 amino acids of SBP2, with 75% identity in the L30 RNA binding domain (6). As with the SBP2 mutants, hSLP was expressed in rabbit reticulocyte lysate and tested for its ability to bind the PHGPx 3′ UTR and enhance PHGPx translation. Expression of hSLP in rabbit reticulocyte lysate yields a protein with an apparent molecular mass exceeding 85 kDa (Fig. 4A), which is almost double the predicted molecular mass. As mentioned above, the C-terminal portion of SBP2 is involved in its anomalous migration during SDS-PAGE, and this appears to be the case for hSLP as well. As shown in Fig. 4C, lane 6, hSLP binds the PHGPx 3′ UTR very weakly, albeit specifically (compare to Fig. 4D, lane 6), and does not support PHGPx translation (Fig. 4B). Thus, the possession of a conserved motif is not sufficient for full RNA binding activity. These results are consistent with the truncation data, which indicated that a significant amount of sequence surrounding the conserved motif is required for RNA binding activity.
Interaction of SBP2 with ribosomes.
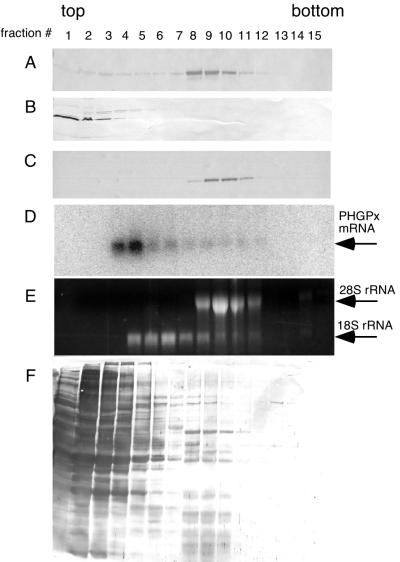
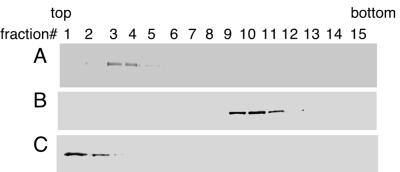
Our current model of Sec incorporation employs SBP2 in preventing termination while perhaps simultaneously delivering a functional SBP2–eEFsec–Sec-tRNASec complex to the ribosomal A site. To test whether SBP2 is stably associated with ribosomes, we used glycerol gradients to study SBP2 sedimentation after transient transfection of wild-type and mutant SBP2 constructs. Our initial attempts to use standard sucrose-based polysome gradients failed due to the sensitivity of the SBP2 complex to the artificially high magnesium concentration necessary to stabilize polysomes (data not shown). Therefore, the conditions used for this experiment do not show an analysis of mRNAs stably associated with the ribosomal fraction. Strep-tagged C-terminal SBP2 (TM399–846) was transiently transfected into the rat hepatoma cell line McArdle 7777. After 40 h, extracts from these cells were fractionated on 10 to 30% glycerol gradients. Fractions were collected and precipitated with TCA followed by Western blot analysis using an anti-Strep tag probe. Figure 5 shows the results of three gradients run under various conditions. Figure 5A indicates that the vast majority of Strep-tagged SBP2 sediments with the heavier fractions (numbers 8 to 10) under low-stringency conditions (PBS–2 mM DTT). The addition of 0.5 M NaCl to the extraction and gradient buffers resulted in complete disruption of the complex, as shown in Fig. 5B. Complex formation was not sensitive to 5 mM EDTA (Fig. 5C), suggesting that divalent metal ions are not necessary for this interaction. Reprobing of these blots with anti-SBP2 antibody indicates that all detectable endogenous SBP2 is found to cofractionate with fractions 8 to 10 (data not shown). It is unlikely that transfected SBP2 is “pulling” endogenous SBP2 onto the ribosomes because our analysis of cells transfected with the N-terminal half of SBP2, which is not ribosome associated, also shows that the endogenous protein sediments with ribosomes (data not shown). Staining of the blot in Fig. 5A for total protein demonstrated that most protein sedimented in the first five fractions of the gradient (Fig. 5F). To ascertain whether or not a SECIS element was involved in this interaction, we extracted RNA from the EDTA-containing gradient fractions and analyzed the distribution of PHGPx mRNA by Northern blot analysis. PHGPx mRNA was chosen because it is expressed to relatively high levels in McArdle cells (8) and because GPx mRNA was not detectable in this experiment, which contains limited amounts of mRNA. As shown in Fig. 5D, the bulk of PHGPx mRNA does not cosediment with SBP2, suggesting that SBP2 is not stably associated with SECIS elements and the ribosome simultaneously. This was a surprising result and may reflect an effect of the methods used, but it may suggest that SBP2 preferentially binds to ribosomes and interacts with SECIS elements only after translation initiation. PHGPx mRNA sedimentation was the same as that of nonselenoprotein (glyceraldehyde-3-phosphate dehydrogenase) mRNA (data not shown). To analyze the positions of the ribosomal subunits, the Northern gel described above was stained with ethidium bromide to allow visualization of the positions of the 18S and 28S rRNAs in the gradient, which indicates the positions of the 40 and 60/80S ribosomal subunits (Fig. 5E). These results suggest that SBP2 is stably associated with either 60S or 80S, but not isolated 40S, ribosomal subunits.
FIG. 5.
Sedimentation of SBP2 in glycerol gradients. Strep-tagged TM399-846 was transiently transfected into McArdle 7777 cells, and 500-μg aliquots of extracted proteins were loaded onto 10 to 30% linear glycerol gradients. Fractions (0.6 ml) were collected from the top of each gradient. Proteins were TCA precipitated, resolved by SDS-PAGE, blotted to nitrocellulose, and probed with alkaline phosphatase-conjugated streptavidin, which recognizes the Strep tag. (A to C) Gradients run in PBSD, PBSD plus 0.5 M NaCl, and PBSD plus 5 mM EDTA respectively. (D) RNA was extracted from the fractions run in 5 mM EDTA, resolved on a 1% denaturing agarose gel, blotted to nylon, and probed for PHGPx mRNA. (E) Gel in panel D stained with ethidium bromide. (F) Blot in panel A stained with India ink.
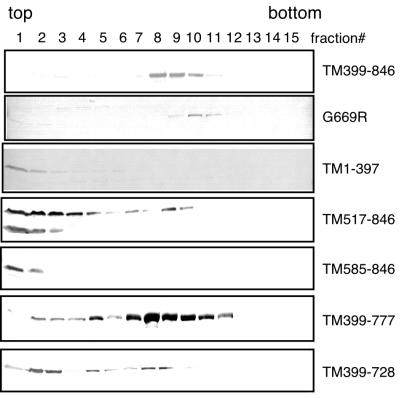
To determine which domain of SBP2 is required for ribosome interaction, we expressed several SBP2 mutant proteins in McArdle 7777 cells, and analyzed complex formation on glycerol gradients as described above. The results from these experiments are shown in Fig. 6 and are summarized in Fig. 2. As best indicated by the G669R mutant, SECIS binding activity is not required for complex formation. Mutations which eliminate activity but not RNA binding (e.g., TM517-846) are impaired for complex formation, as the majority of the protein is not associated with fractions 8 to 10. In addition, the mutation which eliminates part of the RNA binding domain (TM399-728) is also reduced in its ability to form complexes. These results suggest that both the putative functional domain and the RNA binding domain are crucial for the SBP2-ribosome interaction. Because the G669R mutant still binds ribosomes, however, we conclude that the RNA binding domain and the ribosome binding domain are not identical.
FIG. 6.
Sedimentation of SBP2 mutants in glycerol gradients. McArdle 7777 cell extracts containing the mutant SBP2 proteins indicated to the right were loaded onto glycerol gradients as described for Fig. 5. TM399–846, G669R, and TM1-397 are Strep-tagged proteins detected with alkaline phosphatase-conjugated streptavidin. The remainder of the proteins were untagged and detected with anti-SBP2 antibodies.
To verify that the complex we have identified in glycerol gradients is due to ribosomes and not self-association or other interactions, purified recombinant Strep-tagged TM399–846 (6) was analyzed in the presence or absence of purified salt-washed ribosomes derived from rabbit reticulocytes (16). Interestingly, purified TM399-846 alone sediments in fractions 3 and 4 (Fig. 7A), while the addition of 2 A260 units of ribosomes moves the SBP2 to fractions 10 to 12 (Fig. 7B; SBP2 is found further down the gradient than described above due to a longer spinning time used to increase resolution). The addition of 0.5 M NaCl to the gradient results in a complete disruption of the complex and moves SBP2 to the first two fractions of the gradient. These results suggest that SBP2 is, indeed, interacting with ribosomes and is also self-associated into a salt-sensitive complex in the absence of ribosomes.
FIG. 7.
Sedimentation of recombinant SBP2 and purified ribosomes. (A) Purified Strep-tagged C-terminal SBP2 (TM399-846; 5 μg) was layered onto a 10 to 30% glycerol gradient. Fractions processed as described for Fig. 5. (B) Purified SBP2 (TM399-846; 5 μg) was incubated with 2.0 A260 units of purified salt-washed ribosomes from rabbit reticulocyte lysate and then layered onto a glycerol gradient and processed as for panel A. (C) Same as panel B except that 0.5 M NaCl was added to the reaction mix.
SBP2 interacts with rRNA.
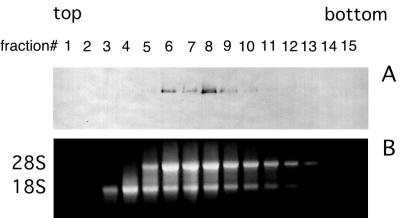
Because the RNA binding domain of SBP2 is required for ribosome binding and because ribosomal protein L30 uses a similar domain to interact with its own mRNA as well as 28S rRNA (15, 22), we decided to test for the ability of SBP2 to bind rRNA in vitro. Toward this end, Strep-tagged TM399-846 was incubated with rRNA extracted from the purified salt-washed ribosomes used for Fig. 7 and subjected to glycerol gradient fractionation. As shown in Fig. 8A, SBP2 is not found in the early fractions (numbers 3 and 4) where the 18S rRNA alone is present. However, SBP2 does form a complex with 28S rRNA or the combination of 28S and 18S rRNAs which, as shown in Fig. 8B, is present in fractions 6 to 9. While we cannot rule out that this interaction requires the presence of both rRNA species, it is likely that the SBP2-ribosome interaction depends on SBP2 binding to 28S rRNA.
FIG. 8.
Cosedimentation of SBP2 and purified rRNA. Purified Strep-tagged C-terminal SBP2 (TM399-846; 5 μg) was incubated with purified total rRNA and fractionated on a 10 to 30% glycerol gradient. Gradient fractions were analyzed for SBP2 content by Western blotting (A) and rRNA content by agarose gel electrophoresis and ethidium bromide staining (B).
DISCUSSION
The recent discovery of two novel factors involved in mammalian selenoprotein synthesis, SBP2 and eEFSec, has shed significant light on the mechanism of Sec incorporation. The latter of these factors has been described as the Sec-specific elongation factor which is required for delivering the Sec-tRNASec to the ribosomal A site. The role of SBP2 is much less clear, and we have previously hypothesized that it is involved in the other major dynamic presumed to be required for Sec insertion, competition with translation termination. Here we describe the beginning of a detailed investigation of the structure and function of SBP2 in order to elucidate its role in Sec insertion. From this and previous work, we know that SBP2 is a multifunctional protein that binds to SECIS elements, interacts with eEFSec, and also binds stably to ribosomes.
As shown in Fig. 3, SBP2 shares a sequence motif with a variety of eukaryotic and prokaryotic proteins. The best-studied member of this family is ribosomal protein L30 which is involved in its own splicing and translational regulation by means of binding to its own pre-mRNA and mRNA, respectively. The fact that the L30 RNA binding domain is involved in RNA binding has been established by extensive nuclear magnetic resonance analysis on the L30 protein and mRNA target (15). While there is no sequence similarity between the L30 mRNA and a SECIS element, the overall structures of both SBP2 and L30 binding sites can be described as a bulge flanked on both sides by paired sequences (5, 15), and both sequences may form the GA quartet that is known to be essential for Sec insertion. Interestingly, the L30 target site on 25S rRNA from Saccharomyces cerevisiae has also recently been determined and found to be very similar to the L30 mRNA binding site in both primary and secondary structures (22). The truncation and point mutants described in this report establish that the L30 RNA binding domain is required for SBP2 RNA binding and functional activity. However, mutations which eliminate SECIS binding do not eliminate ribosome association, suggesting that this motif is not strictly relegated to ribosome-rRNA interactions. In addition, the conserved motif is not sufficient for sequence specific binding, as demonstrated by the truncation mutants which still retain the motif but are unable to sustain binding activity. Furthermore, the SBP2-related protein that was tested here was barely able to bind the PHGPx 3′ UTR, suggesting that even a related sequence is not sufficient for specific binding. This point is further supported by the fact that the L30 nuclear magnetic resonance data indicate that residues C terminal to the conserved region are involved in protein-RNA contacts (15). These residues are not conserved in SBP2.
In terms of SBP2 function, it is clear from the data presented here that RNA binding activity is required for function and that a functional domain can be identified as a discrete stretch of amino acids. This region, however, is not a member of any known sequence families and thus does not shed light on its mechanism of action. It is likely that this region is involved in the interactions necessary for Sec insertion, and so our current efforts are involved in identifying the critical residues in this region and testing mutants for their ability to interact with eEFSec. It is possible, for example, that this region is involved directly with preventing termination either by binding to the release factor(s) directly or by blocking access of eRF-1 to the ribosomal A site. We have thus far been unable to demonstrate any interaction between SBP2 and either of the eukaryotic release factors (eRF-1 or eRF-3), but work is still in progress on that front.
Perhaps the most significant finding from this work is that SBP2 is stably associated with the ribosomal fraction of glycerol gradients both in vivo and in vitro. This interaction appears to be direct, as purified SBP2 is able to interact with salt-washed purified ribosomes. It is striking that the point mutant that fails to bind the SECIS element still associates with the ribosome, but that deletions which encroach on the RNA binding domain greatly reduce ribosome association. In light of these results, we propose that the ribosome binding domain and SECIS binding domain overlap but are not identical. Considering that SBP2 appears to form a homomeric complex based on its sedimentation in glycerol gradients in the absence of any other factors, it is tempting to suggest that an SBP2 dimer in a head-to-head orientation uses one RNA binding domain to interact with the ribosome and the other to interact with the SECIS element.
These results lead us to speculate that SBP2 may be involved in selecting ribosomes for Sec insertion. That is, by binding to a subset of ribosomes without discrimination, SBP2 is making that pool of ribosomes competent for Sec insertion. This model would predict, therefore, that selenoproteins translated on ribosomes lacking SBP2 will terminate at the Sec codon, while those being translated on SBP2-containing ribosomes will bypass termination and insert Sec. While the data presented here do not directly support the idea that SBP2 is involved in preventing termination, we anticipate that future work which will define the site of the SBP2-ribosome interaction should shed light on potential interactions with the termination mechanism. The binding of SBP2 to the ribosome being the first step, a second step will likely consist of quaternary complex formation (SBP2, SECIS element, eEFSec, and Sec-tRNASec), and that this step will also be regulated by SBP2 by means of differential affinity for the various SECIS elements. Thus, it is conceivable that SBP2 is the major determinant of the efficiency of selenoprotein synthesis, and that it may be involved in the differential stability of selenoprotein mRNA that is known to be highly regulated during selenium deficiency (4, 17). In reticulocyte lysates SBP2 is the clearly limiting factor, and this may also be true in tissues that have limited selenoprotein synthesis capacity.
ACKNOWLEDGMENTS
We thank Bill Merrick for providing purified reticulocyte ribosomes as well as Julia Fletcher and Carri Gerber for a critical review of the manuscript.
This work was supported by Public Health Service grants HL29582 (D. M. D.) and F32 DK09878-01 (P.R.C.) from the National Institutes of Health.
REFERENCES
- 1.Berry M J, Banu L, Chen Y Y, Mandel S J, Kieffer J D, Harney J W, Larsen P R. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 2.Bock A. Biosynthesis of selenoproteins—an overview. Biofactors. 2000;11:77–78. doi: 10.1002/biof.5520110122. [DOI] [PubMed] [Google Scholar]
- 3.Bosl M R, Takaku K, Oshima M, Nishimura S, Taketo M M. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp) Proc Natl Acad Sci USA. 1997;94:5531–5534. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brigelius-Flohe R. Tissue-specific functions of individual glutathione peroxidases. Free Radic Biol Med. 1999;27:951–965. doi: 10.1016/s0891-5849(99)00173-2. [DOI] [PubMed] [Google Scholar]
- 5.Copeland P R, Driscoll D M. Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J Biol Chem. 1999;274:25447–25454. doi: 10.1074/jbc.274.36.25447. [DOI] [PubMed] [Google Scholar]
- 6.Copeland P R, Fletcher J E, Carlson B A, Hatfield D L, Driscoll D M. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher J E, Copeland P R, Driscoll D M. Polysome distribution of phospholipid hydroperoxide glutatione peroxidase mRNA: evidence for a block in elongation at the UGA/selenocysteine codon. RNA. 2000;6:1573–1584. doi: 10.1017/s1355838200000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gladyshev V N, Hatfield D L. Selenocysteine-containing proteins in mammals. J Biomed Sci. 1999;6:151–160. doi: 10.1007/BF02255899. [DOI] [PubMed] [Google Scholar]
- 10.Gray N K, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 11.Ho Y S, Magnenat J L, Bronson R T, Cao J, Gargano M, Sugawara M, Funk C D. Mice deficient in cellular glutathione peroxidase develop normally and show no increased sensitivity to hyperoxia. J Biol Chem. 1997;272:16644–16651. doi: 10.1074/jbc.272.26.16644. [DOI] [PubMed] [Google Scholar]
- 12.Koonin E V, Bork P, Sander C. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 1994;22:2166–2167. doi: 10.1093/nar/22.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee B J, Worland P J, Davis J N, Stadtman T C, Hatfield D L. Identification of a selenocysteyl-tRNA (Ser) in mammalian cells that recognizes the nonsense codon, UGA. J Biol Chem. 1989;264:9724–9727. [PubMed] [Google Scholar]
- 14.Lesoon A, Mehta A, Singh R, Chisolm G M, Driscoll D M. An RNA-binding protein recognizes a mammalian selenocysteine insertion sequence element required for cotranslational incorporation of selenocysteine. Mol Cell Biol. 1997;17:1977–1985. doi: 10.1128/mcb.17.4.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao H, White S A, Williamson J R. A novel loop-loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nat Struct Biol. 1999;6:1139–1147. doi: 10.1038/70081. [DOI] [PubMed] [Google Scholar]
- 16.Merrick W C. Assays for eukaryotic protein synthesis. Methods Enzymol. 1979;60:108–123. doi: 10.1016/s0076-6879(79)60011-3. [DOI] [PubMed] [Google Scholar]
- 17.Moriarty P M, Reddy C C, Maquat L E. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol. 1998;18:2932–2939. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 19.Song H, Mugnier P, Das A K, Webb H M, Evans D R, Tuite M F, Hemmings B A, Barford D. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 20.Stadtman T C. Selenium biochemistry. Mammalian selenoenzymes. Ann NY Acad Sci. 2000;899:399–402. doi: 10.1111/j.1749-6632.2000.tb06203.x. [DOI] [PubMed] [Google Scholar]
- 21.Tujebajeva R M, Copeland P R, Xu X-M, Carlson B A, Harney J W, Driscoll D M, Hatfield D L, Berry M J. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 2000;1:1–6. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vilardell J, Yu S J, Warner J R. Multiple functions of an evolutionarily conserved RNA binding domain. Mol Cell. 2000;5:761–766. doi: 10.1016/s1097-2765(00)80255-5. [DOI] [PubMed] [Google Scholar]