Abstract
Acute B-lymphocytic leukemia (B-ALL) is associated with a high mortality rate, with no effective treatment strategies available. The identification of diagnostic and prognostic biomarkers of B-ALL can contribute to the development of novel therapeutic methods and drugs, which can improve the survival outcomes of patients with B-ALL. The present study aimed to identify downregulated circular RNAs (circRNAs) in patients with B-ALL. RNA sequencing was performed to construct the circRNA expression profiles in B-ALL cells and normal human lymphoblasts. The Database for Annotation, Visualization and Integrated Discovery was used to perform Gene Ontology enrichment and Kyoto Encyclopedia of Genes and Genomes pathway analyses. In addition, reverse transcription-quantitative (RT-q)PCR analysis was performed to detect the expression levels of the downregulated circRNAs. A total of 263 differentially expressed circRNAs were identified, including 76 upregulated and 187 downregulated circRNAs, respectively. The upregulated circRNAs were mainly enriched in ‘macromolecule modification’, ‘protein modification’ and ‘cellular protein modification processes’, while the downregulated circRNAs were mainly enriched in the ‘negative regulation of RNA biosynthetic processes’, ‘natural killer cell-mediated cytotoxicity’ and ‘viral carcinogenesis’. RT-qPCR analysis demonstrated that two of the downregulated circRNAs (hsa_circ_0000745 and chr15:87949594-87966067-), identified during microarray analysis were also significantly downregulated in Ball-1 cells and B-ALL bone marrow samples. Thus, these circRNAs may serve as biomarkers for patients with B-ALL.
Keywords: acute B-lymphocytic leukemia, circular RNAs, biomarkers, gene ontology, Kyoto Encyclopedia of Genes and Genomes, RNA sequencing
Introduction
Leukemia is a malignant cancer of the hemopoietic system (1). The morbidity rate of patients with leukemia remains high due to factors, such as the living environment and diet (2). Chemotherapy and bone marrow transplantation are the main treatment strategies for leukemia (3); however, patients with different types of leukemia may receive different treatments (4). Acute B-lymphocytic leukemia (B-ALL) has a high mortality rate (5) and no effective treatment strategies (6,7). Thus, identification of diagnostic and prognostic biomarkers of B-ALL can contribute to the development of novel therapeutic methods and drugs, which can improve the survival outcomes of patients with B-ALL.
Circular RNAs (circRNAs) are non-coding RNA molecules, with a closed ring structure that lacks the 5′ and 3′ ends of traditional linear RNA (8). circRNAs are conserved, tissue-specific, insensitive to RNase and more stable compared with linear RNA (9). These characteristics suggest that circRNAs may have great potential for cancer diagnosis, treatment and identification of novel methods to overcome human cancer (10,11). Previous studies have demonstrated that the expression levels of some circRNAs are higher in cancer tissues compared with normal tissues, and some circRNAs can serve as prognostic biomarkers (12–14). Thus, it is important to investigate differentially expressed circRNAs in B-ALL cells and normal human B lymphoblasts.
The present study aimed to investigate the expression profiles of circRNAs in B-ALL and HMy2.CIR cells via high throughput RNA sequencing, identify downregulated circRNAs in B-ALL cells compared with HMy2.CIR cells, and perform Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of differentially expressed circRNAs to determine their function. In addition, the present study aimed to validate these differentially expressed circRNAs using bone marrow samples from patients with and without B-ALL.
Materials and methods
Bone marrow samples
Bone marrow samples were collected from six patients treated at the Department of Hematology, Shengjing Hospital of China Medical University, (Shenyang, China), between September 2014 and December 2016. All patients were diagnosed with B-ALL and received the CCLG-ALL-2008 treatment plan (15). These patients included three boys and three girls (mean age, 5.8 years; age range, 4–8 years). Bone marrow samples were also collected from six individuals without B-ALL (control). These patients included three boys and three girls (mean age, 5.5 years; age range, 4–8 years). The inclusion criteria were as follows: Initial diagnosis was according to the 2008 World Health Organization classification standard of lymphoid and hematopoietic tissue tumors and bone marrow cytology, immunotyping, chromosome karyotype analysis and fusion gene analysis were performed (16). The exclusion criteria were as follows: i) Patients received anti-leukemia treatment prior to admission; ii) patients with mature B-ALL and mixed cell leukemia and iii) patients with secondary ALL or other tumors. The present study was approved by the China Medical University Ethics Committee (institution review board no. 2020028) and written informed consent was provided by all participants prior to the study start.
Cells and cell culture
The human B-ALL cell line, Ball-1, and the human B lymphoblast cell line, HMy2.CIR, were purchased from the Type Culture Collection of The Chinese Academy of Sciences. Cells were maintained in RPMI-1640 medium (Hyclone; Cytiva) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), at 37°C with 5% CO2.
The HMy2.CIR cell line was used as the control according to previous studies (17–20), which demonstrated that HMy2.CIR cells are comparable with human B lymphoblasts and have a lower malignancy potential than B-ALL cells.
circRNA expression profile analysis
Total RNA was extracted from Ball-1 and HMy2.CIR cells with TRIzol® reagent (Beijing Solarbio Science & Technology Co., Ltd.). A total of three samples were taken from each cell line. circRNAs were enriched from 5 µg total RNA using the circRNA Enrichment kit (CloudSeq, www.cloud-seq.com.cn). Subsequently, strand-specific RNA-seq libraries were prepared using the TruSeq Stranded Total RNA Library Prep kit (Illumina, Inc.), which were subjected to deep sequencing using BioAnalyzer 2100 (Agilent Technologies, Inc.).
Identification of circRNAs
The RNA-seq FASTQ reads were mapped to a human reference genome (GRCh37/hg19) using TopHat2 (21). The number of spliced reads were used as the expression level of each circRNA. The total number of reads was used to standardize the samples, and log2 transformation was performed to obtain the standardized number of reads. Using the standardized number of reads and the R software package DEGseq (22), differential expression of circRNAs between Ball-1 and HMy2.CIR cells were calculated. Log2 (fold change) >2.0 (or <-2.0) and P<0.05 were considered to indicate significantly differentially expressed circRNAs. Hierarchical clustering analysis was also performed to generate an overview of circRNA expression profiles between the two cell lines.
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from Ball-1 cells, HMy2.CIR cells, B-ALL bone marrow and non-B-ALL bone marrow using TRIzol® reagent, and reverse transcribed into cDNA using PrimeScript™ RT reagent Kit with gDNA Erase (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. qPCR was subsequently performed using the SYBR-Green PCR Master Mix (Takara Biotechnology Co., Ltd.) within the Fast-Real-Time PCR System (CFX96™ Optics Module; Bio-Rad Co., Ltd.). The following primer sequences were used for qPCR: GAPDH forward, 5′-GGCCTCCAAGGAGTAAGACC-3′ and reverse, 5′-AGGGGAGATTCAGTGTGGTG-3′. Primer sequences for the circRNAs are listed in Table SI. The following thermocycling conditions were used for qPCR: 95°C for 30 sec, followed by 40 cycles of 95°C for 3 sec and 60°C for 30 sec and a final extension step at 60°C for 7 min. Relative expression levels were calculated using the 2−ΔΔCq method (23) and normalized to the internal reference gene GAPDH.
Bioinformatic analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.ncifcrf.gov) was used to perform GO enrichment and KEGG pathway analyses of the differentially expressed circRNAs to determine the processes and pathways involved.
Cell transfection
Under normal culture conditions, cells were starved in serum-free RPMI-1640 medium (Hyclone, Cytiva) for 2 h after reaching 60–70% confluence. OE-CIRC and OE-negative control (NC) were respectively transfected into HMy2.CIR cells using Lipofectamine® 3000, according to the manufacturer's protocol. Cells were harvested for subsequent experimentation after 24–72 h. The concentration of plasmid OE-CIRC and OE-NC of hsa_circ_0000745 purchased from jtsbio company was 100 nM/l. The temperature and duration of transfection were 37°C and 24–72 h. Fluorescence intensity was detected via fluorescence microscopy (magnification, ×200; Nikon Corporation), according to the manufacturer's instructions.
Cell proliferation
After 24 h of transfection, cell suspensions of the experimental and control groups were inoculated into 96-well plates at a density of 3,000 cells/well/100 µl. The plates were precultured in the incubator at 37°C with 5% CO2. Cell Counting Kit-8 (10 µl) reagent (Cofitt Life Science Company; www.cofitt.com) was subsequently added to each well and incubated for an additional 4 h. The absorbance was measured at a wavelength of 450 nm, using a microplate reader (BioTek Instruments, lnc.). The OD value of each well was measured for 3 consecutive days.
Statistical analysis
Statistical analysis was performed using SPSS 23.0 software (IBM Corp.). Edge R 4.1.0 software (https://www.r-project.org) was used to establish a linear model of negative binomial distribution according to various factors in the experimental design, and the dispersion coefficient of each factor was calculated (Fig. 1). The Quasi-Likelihood F test (P<0.05 and fold change ≥2.0) was used to screen the differentially expressed circRNAs. The volcano map was constructed to compare differences between the two groups (P<0.05 and fold change ≥2.0). The Heatmap. 2 function of R 4.1.0 software (https://www.r-project.org) was used to cluster the differentially expressed circRNAs. Unpaired independent sample t-test was used to compare the expression of circRNAs in the different cell lines and samples. P<0.05 was considered to indicate a statistically significant difference.
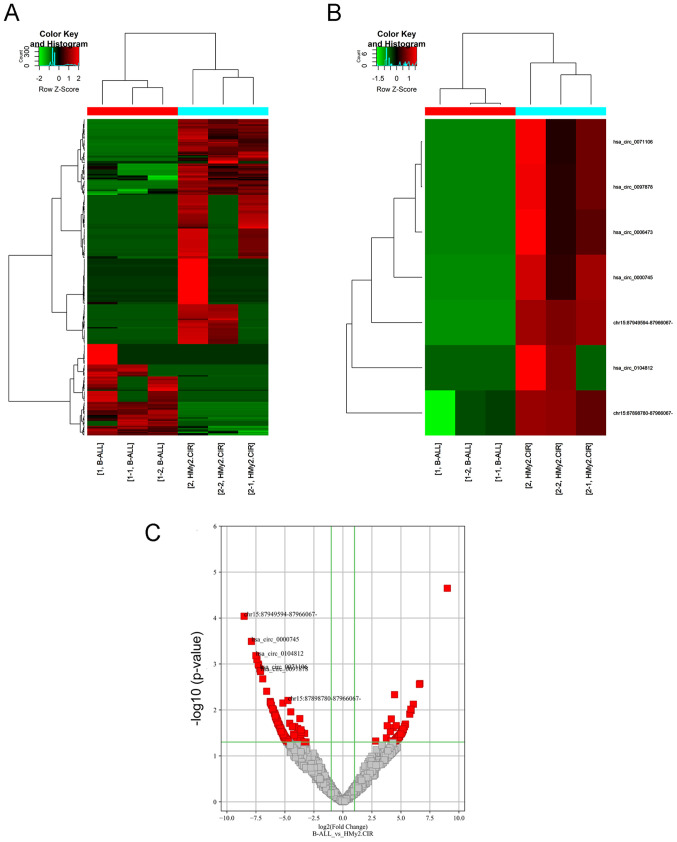
Figure 1.
circRNA expression profiles in Ball-1 and HMy2.CIR cells. (A) Clustered heatmap for differentially expressed circRNAs. (B) Clustered heatmap for the seven selected differentially expressed circRNAs (labeled). (C) Volcano plot. Vertical green lines correspond to 2-fold upregulation and downregulation, while the horizontal green line represents P<0.05. Red dots represent significantly differentially expressed circRNAs. These gray dots represent differentially expressed circRNAs that were not significant. circRNA, circular RNA; B-ALL, acute B-lymphocytic leukemia.
Results
Identification of differentially expressed circRNAs in Ball-1 and HMy2.CIR cells
To identify the differentially expressed circRNAs in Ball-1 and HMy2.CIR cells, the circRNA expression profiles were analyzed via high-throughput circRNA microarray analysis. A total of 263 differentially expressed circRNAs were identified [log2 (fold change) >2.0 (or ≥2.0) and P<0.05], of which 76 were upregulated and 187 were downregulated in Ball-1 cells compared with HMy2.CIR cells (Fig. 1A-C).
GO enrichment and KEGG pathway analyses of differentially expressed circRNAs
GO enrichment and KEGG pathway analyses of the differentially expressed circRNAs were performed using DAVID. GO enrichment analysis identified three terms, biological process (BP), cellular component (CC) and molecular function (MF).
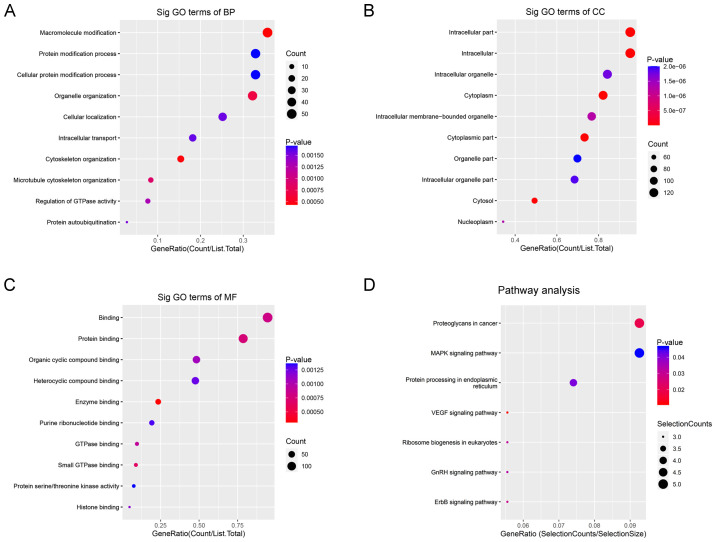
For BP, upregulated circRNAs were mainly enriched in ‘macromolecule modification’ (Fig. 2A). For CC, upregulated circRNAs were mainly enriched in ‘intracellular parts’ (Fig. 2B). For MF, upregulated circRNAs were mainly enriched in ‘protein binding’ (Fig. 2C). KEGG pathway analysis demonstrated that upregulated circRNAs were mainly enriched in ‘Proteoglycans in cancer’, ‘MAPK signaling pathway’ and ‘protein processing in endoplasmic reticulum’ (Fig. 2D).
Figure 2.
GO enrichment and KEGG pathway analyses of upregulated circular RNAs. GO (A) BP, (B) CC and (C) MF analyses. (D) KEGG pathway enrichment analysis. GO, Gene Ontology, KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological process; CC, cellular component; MF, molecular function.
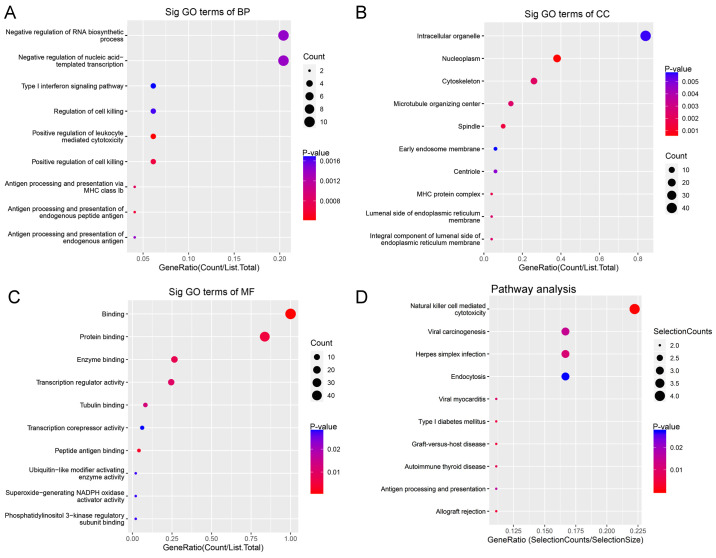
For BP, downregulated circRNAs were mainly enriched in ‘negative regulation of RNA biosynthetic processes’ and ‘negative regulation of nucleic acid templated transcription’ (Fig. 3A). For CC, downregulated circRNAs were mainly enriched in ‘intracellular organelles’ (Fig. 3B). For MF, downregulated circRNAs were mainly enriched in ‘transcription regulator activity and binding’ (Fig. 3C). KEGG pathway analysis demonstrated that downregulated circRNAs were mainly enriched in ‘natural killer cell-mediated cytotoxicity’ and ‘viral carcinogenesis’ (Fig. 3D).
Figure 3.
GO enrichment and KEGG pathway analyses of downregulated circular RNAs. GO (A) BP, (B) CC and (C) MF analyses. (D) KEGG pathway enrichment analysis. GO, Gene Ontology, KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological process; CC, cellular component; MF, molecular function.
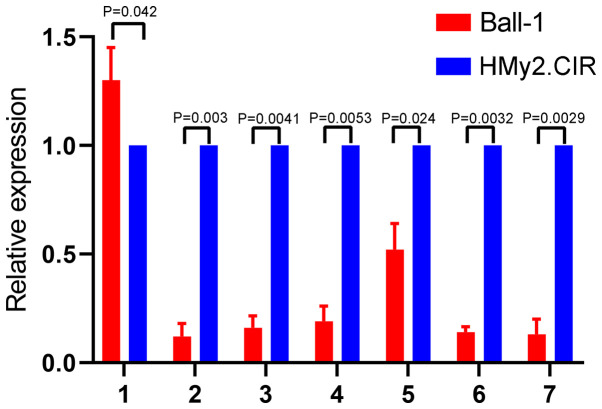
Validation of seven downregulated circRNAs in Ball-1 cells
To validate the microarray results, seven circRNAs downregulated in Ball-1 cells were selected and their expression was validated via RT-qPCR analysis. The seven differentially expressed circRNAs were selected according to the following criteria: i) High fold-change value, ii) significant P-value and iii) not reported in previous studies. The results are presented in Fig. 4. A total of six circRNAs (hsa_circ_0000745, chr15:87949594-87966067-, chr15:87898780-87966067-, hsa_circ_0006473, hsa_circ_0071106 and hsa_circ_0097878) were significantly downregulated in Ball-1 cells compared with HMy2.CIR cells (P=0.003; P=0.0041; P=0.0053; P=0.024; P=0.0032; P=0.0029), while has_circ_0104812 expression was slightly but significantly upregulated in Ball-1 cells compared with HMy2.CIR cells (P=0.042).
Figure 4.
Validation of seven downregulated circRNAs in Ball-1 cells. Reverse transcription-quantitative PCR analysis was performed to detect the expression levels of the seven downregulated circRNAs in Ball-1 and HMy2.CIR cells. The following circRNAs were selected for analysis: 1, hsa_circ_0104812; 2, hsa_circ_0000745; 3, chr15:87949594-87966067-; 4, chr15:87898780-87966067-; 5, hsa_circ_0006473; 6, hsa_circ_0071106 and 7, hsa_circ_0097878. circRNA, circular RNA.
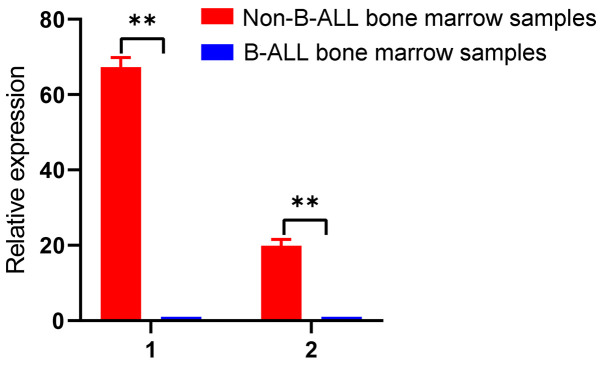
Validation of two downregulated circRNAs in B-ALL bone marrow samples
To validate the results from the cell lines, hsa_circ_0000745 and chr15:87949594-87966067- were selected to validate bone marrow samples from patients with B-ALL. The results demonstrated that these two circRNAs were significantly downregulated in B-ALL bone marrow samples compared with non-B-ALL bone marrow samples (P<0.01; Fig. 5). Therefore, these two circRNAs may serve as biomarkers for B-ALL. Detailed characteristics of the patients and controls are presented in Table SII.
Figure 5.
Validation of two downregulated circRNAs in bone marrow samples. circRNA expression in bone marrow samples of individuals with and without B-ALL. 1, hsa_circ_0000745 and 2, chr15:87949594-87966067-. **P<0.01. circRNA, circular RNA; B-ALL, acute B-lymphocytic leukemia.
Validation of the function of hsa_circ_0000745 in HMy2.CIR cells
hsa_circ_0000745 expression was overexpressed in HMy2.CIR cells, and the results demonstrated that high hsa_circ_0000745 expression significantly inhibited the proliferation of HMy2.CIR cells (P=0.034; Fig. S1). Transfection efficiency is presented in Fig. S2.
Discussion
The development of next-generation sequencing methods, such as RNA sequencing, has led to the discovery of non-coding RNAs, including circRNAs, that may contribute to the progression of cancer (24). circRNAs can regulate proliferation, clonal selection and somatic hypermutation, posing a risk of malignant transformation of normal B cells (25). In addition, deregulation of circRNAs, such as circESYT2, circFBXW4 and circCAMSAP1, promotes the development of B-cell malignancies, such as B-ALL (25). Further, circRNAs can function as microRNA sponges, thereby acting as ceRNAs to regulate the development of B-ALL (26,27).
The present study identified differentially expressed circRNAs in Ball-1 and HMy2.CIR cells using microarray. Downregulated circRNAs in Ball-1 cells were enriched in the regulation of leukocyte-mediated cytotoxicity, suggesting that these circRNAs may be involved in the progression of B-ALL. A total of seven downregulated circRNAs in Ball-1 cells were validated. The results demonstrated that six circRNAs (hsa_circ_0000745, chr15:87949594-87966067-, chr15:87898780-87966067-, hsa_circ_0006473, hsa_circ_0071106 and hsa_circ_0097878) were significantly downregulated in Ball-1 cells compared with HMy2.CIR cells. Similar results were obtained using B-ALL bone marrow samples, whereby hsa_circ_0000745 and chr15:87949594-87966067- were significantly downregulated in patients with B-ALL. These results suggest that these circRNAs can be used as biomarkers for patients with B-ALL.
B-ALL is the most common cancer diagnosed in children (28,29). Although multi-drugs chemotherapy has been widely applied in clinical practice, delayed diagnosis and the relapse of B-ALL are major causes of mortality (30,31). The pathogenesis and molecular mechanism of B-ALL occurrence and progression remain largely unknown. Thus, identifying diagnostic and prognostic biomarkers of B-ALL is important to improve the survival rate and quality of life of those affected. The results of the present study suggest that hsa_circ_0000745 and chr15:87949594-87966067- may serve as biomarkers for the diagnosis, and even treatment of B-ALL.
The present study is not without limitations. First, the validated circRNAs require further investigation to determine their roles in the development of B-ALL. Secondly, the prognostic effects of these circRNAs require further investigation using clinical samples.
In conclusion, the present study identified and validated six differentially expressed circRNAs in Ball-1 cells and further confirmed the downregulation of two circRNAs in bone marrow samples from patients with B-ALL. Thus, these circRNAs may serve as potential biomarkers in patients with B-ALL.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BZ and LZ performed most of the experiments and drafted the initial manuscript. LT, YZ and RW performed the experiments, and BZ performed statistical analysis. QH and YJZ conceived the present study, and participated in its design and coordination. BZ analyzed and interpreted the results. BZ and YJZ confirmed the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the China Medical University Ethics Committee (institution review board no. 2020028) and written informed consent was provided by all participants prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kantarjian HM, Keating MJ, Freireich EJ. Toward the potential cure of leukemias in the next decade. Cancer. 2018;124:4301–4313. doi: 10.1002/cncr.31669. [DOI] [PubMed] [Google Scholar]
- 2.Miranda-Filho A, Piñeros M, Ferlay J, Soerjomataram I, Monnereau A, Bray F. Epidemiological patterns of leukaemia in 184 countries: A population-based study. Lancet Haematol. 2018;5:e14–e24. doi: 10.1016/S2352-3026(17)30232-6. [DOI] [PubMed] [Google Scholar]
- 3.Atsuta Y, Kato S, Morishima Y, Ohashi K, Fukuda T, Ozawa Y, Eto T, Iwato K, Uchida N, Ota S, et al. HLA Working Group of the Japan Society for Hematopoietic Cell Transplantation Comparison of HLA allele mismatch and antigen mismatch in unrelated bone marrow transplantation in patients with leukemia. Biol Blood Marrow Transplant. 2019;25:436–442. doi: 10.1016/j.bbmt.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Chung C, Ma H. Driving toward precision medicine for acute leukemias: Are we there yet? Pharmacotherapy. 2017;37:1052–1072. doi: 10.1002/phar.1977. [DOI] [PubMed] [Google Scholar]
- 5.Fousek K, Watanabe J, Joseph SK, George A, An X, Byrd TT, Morris JS, Luong A, Martínez-Paniagua MA, Sanber K, et al. CAR T-cells that target acute B-lineage leukemia irrespective of CD19 expression. Leukemia. 2021;35:75–89. doi: 10.1038/s41375-020-0792-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davila ML, Riviere I, Wang X, Bartido S, Park J, Curran K, Chung SS, Stefanski J, Borquez-Ojeda O, Olszewska M, et al. Efficacy and toxicity management of 19–28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med. 2014;6:224ra25. doi: 10.1126/scitranslmed.3008226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tambaro FP, Garcia-Manero G, O'Brien SM, Faderl SH, Ferrajoli A, Burger JA, Pierce S, Wang X, Do KA, Kantarjian HM, et al. Outcomes for patients with chronic lymphocytic leukemia and acute leukemia or myelodysplastic syndrome. Leukemia. 2016;30:325–330. doi: 10.1038/leu.2015.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836. doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. circRNA: A novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 11.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Shuai M, Hong J, Huang D, Zhang X, Tian Y. Upregulation of circRNA_0000285 serves as a prognostic biomarker for nasopharyngeal carcinoma and is involved in radiosensitivity. Oncol Lett. 2018;16:6495–6501. doi: 10.3892/ol.2018.9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin G, Sheng H, Xie H, Zheng Q, Shen Y, Shi G, Ye D. circLPAR1 is a novel biomarker of prognosis for muscle-invasive bladder cancer with invasion and metastasis by miR-762. Oncol Lett. 2019;17:3537–3547. doi: 10.3892/ol.2019.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaichian S, Shafabakhsh R, Mirhashemi SM, Moazzami B, Asemi Z. Circular RNAs: A novel biomarker for cervical cancer. J Cell Physiol. 2020;235:718–724. doi: 10.1002/jcp.29009. [DOI] [PubMed] [Google Scholar]
- 15.Cui L, Li ZG, Chai YH, Yu J, Gao J, Zhu XF, Jin RM, Shi XD, Zhang LP, Gao YJ, et al. Chinese Children Leukemia Group (CCLG), corp-author Outcome of children with newly diagnosed acute lymphoblastic leukemia treated with CCLG-ALL 2008: The first nation-wide prospective multicenter study in China. Am J Hematol. 2018;93:913–920. doi: 10.1002/ajh.25124. [DOI] [PubMed] [Google Scholar]
- 16.Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM, Hellström-Lindberg E, Tefferi A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 17.He M, Dong C, Konishi T, Tu W, Liu W, Shiomi N, Kobayashi A, Uchihori Y, Furusawa Y, Hei TK, et al. Differential effects of p53 on bystander phenotypes induced by gamma ray and high LET heavy ion radiation. Life Sci Space Res (Amst) 2014;1:53–59. doi: 10.1016/j.lssr.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Ye S, Yuan D, Xie Y, Pan Y, Shao C. Role of DNA methylation in long-term low-dose γ-rays induced adaptive response in human B lymphoblast cells. Int J Radiat Biol. 2013;89:898–906. doi: 10.3109/09553002.2013.806832. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, Tan Q, Hu B, Wu H, Wang C, Liu R, Tang C. Somatostatin improved B cells mature in macaques during intestinal ischemia-reperfusion. PLoS One. 2015;10:e0133692. doi: 10.1371/journal.pone.0133692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye S, Yuan D, Xie Y, Pan Y, Shao C. Role of DNA methylation in the adaptive responses induced in a human B lymphoblast cell line by long-term low-dose exposures to γ-rays and cadmium. Mutat Res Genet Toxicol Environ Mutagen. 2014;773:34–38. doi: 10.1016/j.mrgentox.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 21.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 23.Adnan M, Morton G, Hadi S. Analysis of rpoS and bolA gene expression under various stress-induced environments in planktonic and biofilm phase using 2(−ΔΔCT) method. Mol Cell Biochem. 2011;357:275–282. doi: 10.1007/s11010-011-0898-y. [DOI] [PubMed] [Google Scholar]
- 24.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al. The landscape of circular RNA in cancer. Cell. 2019;176:869–881.e13. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dahl M, Kristensen LS, Grønbæk K. Long non-coding RNAs guide the fine-tuning of gene regulation in B-cell development and malignancy. Int J Mol Sci. 2018;19:2457. doi: 10.3390/ijms19092475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 27.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 28.Locatelli F, Schrappe M, Bernardo ME, Rutella S. How I treat relapsed childhood acute lymphoblastic leukemia. Blood. 2012;120:2807–2816. doi: 10.1182/blood-2012-02-265884. [DOI] [PubMed] [Google Scholar]
- 29.Bhojwani D, Pui CH. Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol. 2013;14:e205–e217. doi: 10.1016/S1470-2045(12)70580-6. [DOI] [PubMed] [Google Scholar]
- 30.Hunger SP, Lu X, Devidas M, Camitta BM, Gaynon PS, Winick NJ, Reaman GH, Carroll WL. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: A report from the children's oncology group. J Clin Oncol. 2012;30:1663–1669. doi: 10.1200/JCO.2011.37.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jing W, Li J. Identification of biomarkers for the prediction of relapse free survival in pediatric B precursor acute lymphoblastic leukemia. Oncol Rep. 2019;41:659–667. doi: 10.3892/or.2018.6846. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used analyzed during the current study are available from the corresponding author on reasonable request.