Abstract
Ionizing radiation (IR) and chemotherapy are mainstays of treatment for patients with rhabdomyosarcoma (RMS), yet the molecular mechanisms that underlie the success or failure of radiotherapy remain unclear. The transcriptional repressor SNAI2 was previously identified as a key regulator of IR sensitivity in normal and malignant stem cells through its repression of the proapoptotic BH3-only gene PUMA. Here, we demonstrate a clear correlation between SNAI2 expression levels and radiosensitivity across multiple RMS cell lines. Modulating SNAI2 levels in RMS cells through its overexpression or knockdown altered radiosensitivity in vitro and in vivo. SNAI2 expression reliably promoted overall cell growth and inhibited mitochondrial apoptosis following exposure to IR, with either variable or minimal effects on differentiation and senescence, respectively. Importantly, SNAI2 knockdown increased expression of the proapoptotic BH3-only gene BIM, and ChIP-seq experiments established that SNAI2 is a direct repressor of BIM. Since the p53 pathway is nonfunctional in the RMS cells used in this study, we have identified a new, p53-independent SNAI2/BIM signaling axis that could potentially predict clinical responses to IR treatment and be exploited to improve RMS therapy.
Introduction
Ionizing radiation (IR), chemotherapy and surgery comprise the current standard of care for patients with rhabdomyosarcoma (RMS), a pediatric malignancy of the muscle, and lead to greater than 70% tumor-free survival in children with this disease (1–3). However, the survival rate for patients with disease relapse remains dismal at less than 30% (1–4). IR is used to treat both primary tumors and metastatic lesions in relapsed RMS patients (5). Remarkably, these patients often receive a cumulative dose of 36 to 50.4 Gy (5); yet, an understanding of the pathways that regulate the IR-induced DNA damage response in RMS tumors remains incomplete. In this study, we identify SNAI2 as a critical radioprotector of RMS tumor cells and define the pathways downstream of SNAI2 signaling that regulate the response to IR in RMS.
SNAIL genes comprise a family of transcriptional repressors important for epithelial morphogenesis during development (e.g., the epithelial-mesenchymal transition) and for cell survival (6–8). The role of SNAI2 in protecting normal hematopoietic stem cells (HSCs) from IR-induced apoptosis is well established (9,10). In radiosensitive cells (e.g., the lymphoid lineage), exposure to IR causes DNA double-strand breaks that trigger the activation of P53. P53-mediated induction of the BH3-only gene PUMA then leads to mitochondrial apoptosis (11). However, HSCs are uniquely protected from IR-induced apoptosis due to a concomitant P53-mediated induction of SNAI2 in these cells, which directly represses the expression of PUMA (10). Recent studies also implicate SNAI2 as a regulator of the DNA damage response in normal mammary stem cells (12), and SNAIL family members SNAI1 and TWIST have been shown to regulate the IR-induced DNA damage response in breast cancer cells through regulation of ZEB1 (13). These studies suggest that the SNAIL family may have widespread importance in regulating the response to IR.
Not surprisingly, adult cancers and relapse disease often present with mutations in or loss of TP53 (14,15). In a subset of these tumors that are still radiosensitive, IR has been shown to induce cell death through P53-independent mechanisms involving cell cycle checkpoint proteins and alternative DNA damage response pathways (Reviewed in (16,17)). Interestingly, childhood cancers including RMS often retain wild-type TP53 (18–21). Mutations in TP53 account for <6% of RMS primary tumors, yet they can be acquired during relapse and are associated with a poor outcome (19–21). Since RMS tumors harboring either wild-type or mutant P53 are sensitive to IR (19–21), both P53-dependent and -independent mechanisms of IR-induced cell death appear to be active in this disease. Our analysis show that SNAI2 protects RMS tumors from IR both in vitro and in vivo. Using RMS cell lines that express varying levels of SNAI2 and a dysfunctional P53 pathway (22,23), we show that levels of SNAI2 establish the degree of protection of RMS cell lines from IR, regardless of TP53 mutation status, and that the proapoptotic BH3-only gene BIM is directly repressed by SNAI2 to confer protection from radiation. Our results suggest that SNAI2 is a major player in the response to IR in RMS and represents a promising target for the radiosensitization of RMS tumors during IR therapy.
Materials and Methods
Animals
Animal studies were approved by the University of Texas – Health San Antonio Committee on Research Animal Care under protocol #20150015AR. C.B-Igh-1b/IcrTac-Prkdcscid (SCID) female mice, aged 6–8 weeks, were used for in vivo xenograft experiments.
Mouse xenograft and in vivo IR experiments
Rh30, Rh18, and RD cells with scrambled (shScr), SNAI2 knockdown (shSNAI2) and RD cells with scrambled (shScr), SNAI2 knockdown (shSNAI2), BIM knockdown (shBIM) and double BIM/SNAI2 knockdown (shBIM/SNAI2) treatment conditions were collected, counted, and analyzed by flow cytometry to determine viability using DAPI. Equal numbers of viable cells were then embedded into Matrigel at a final concentration of 1×106 (Rh30) or 5×106 (Rh18) cells per 100 μl and injected subcutaneously into anesthetized mice. Tumor growth was monitored and measured weekly using a caliper scale to measure the greatest diameter and length, which were then used to calculate tumor volume. While a subset of tumors was monitored without any treatment, another subset was subjected to low-dose IR therapy for 3 weeks (2 Gy/day; 5 days a week), receiving a total of 30-Gy IR (PXi Precision X-Ray X-RAD 320). Tumor volume was monitored throughout the treatment and during the weeks following treatment. Comparisons between groups was performed using a Student’s t test. Rh18 pBabe and SNAI2-Flag xenografts were performed as above, with the treatment arm receiving 2 weeks of IR therapy (2 Gy/day for 5 days/week) for a total of 20 Gy. Relative tumor volume (RTV) was assessed in the RD Scr, BIM sh, BIM/SNAI2 sh1, and SNAI2 sh1 tumors treated with 30 Gy of radiation and tumor responses were analyzed using the guidelines outlined by Houghton et al. (24). Progressive disease (PD): RTV>0.50 during the study period and RTV>1.25 at end of study; Stable disease (SD): RTV>0.50 during the study period and RTV≤1.25 at end of study; Partial response (PR): 0<RTV<0.50 for at least one time point; Complete response (CR): RTV=0 for at least one time point.
Human rhabdomyosarcoma (RMS) cell lines
The human RMS cell lines RD and SMS-CTR were a gift from Dr. Corinne Linardic, Duke University, Durham, NC. The JR-1 cell line was a gift from Dr. Marielle Yohe, NCI, Frederick, MD. The Rh28, RH30, RH36, Rh41, and Rh18 lines were obtained from Dr. Peter Houghton, GCCRI, San Antonio, TX. All lines except RD and SMS-CTR were maintained in RPMI supplemented with 10% fetal bovine serum (VWR) at 37°C with 5% CO2. RD and SMS-CTR cells were maintained in DMEM supplemented with 10% FBS at 37°C with 5% CO2. Cell lines utilized were between passage 10 and 25. For knockdown assays, cells were not passaged more than 5 passages after stable lines were generated. Cell lines were authenticated by genotyping and STR analyses. All RMS cell lines were tested and confirmed to be negative for mycoplasma.
Lentiviral/siRNA/retroviral knockdown assays
Scrambled-control and SNAI2 -specific shRNAs were delivered via the pLKO.1-background vector and packaged using 293T cells. siSlug2 (SNAI2 sh1) was a gift from Bob Weinberg (Addgene plasmid # 10904 ; http://n2t.net/addgene:10904 ; RRID:Addgene_10904) (25). siSlug3 (SNAI2 sh2) was a gift from Bob Weinberg (Addgene plasmid # 10905 ; http://n2t.net/addgene:10905 ; RRID:Addgene_10905) (25). pMKO shRNA Bim was a gift from Joan Brugge (Addgene plasmid # 17235 ; http://n2t.net/addgene:17235 ; RRID:Addgene_17235) (26). Retroviral particles were made in Plat-A packaging cells using TranstIT-LT1 (Mirus). RMS cells were infected with viral particles for 24 hr at 37oC using 8 mg/mL of polybrene (EMD Millipore).
Cell confluence and colony formation assays
Rh18, JR-1, Rh36, RD, SMS-CTR, Rh28, Rh30, and Rh41 parental cells were seeded into 24-well plates at 20–50% confluency and stored at 37oC in the Incucyte ZOOM (Essen Bioscience). After 24h, cells were subjected to varying degrees of IR (0 Gy – 20 Gy) and placed back in the Incucyte. Total confluency over time was monitored every 4 hours over a period of 5 days. Incucyte confluency assays were performed similarly for scrambled, SNAI2 shRNA, TP53 shRNA, BIM shRNA, and SNAI2/BIM double knockdown shRNA RMS cells with and without exposure to IR. For colony formation assays, RMS cells were seeded in 12 well plates (~1,250 – 10,000 cells/well for cells receiving radiation and 300 – 600 cells/well for cell receiving no treatment). After 24h, cells were subjected to varying degrees of radiation (2 – 8 Gy). Incubation time for colony formation assays between cell lines varied from 3 to 6 weeks. When colonies were sufficiently large, media was gently removed from each plate by aspiration, and colonies were fixed with 50% methanol for 10 – 15 minutes RT. Colonies were then stained with 3% (w/v) crystal violet in 25% methanol for 10 – 15 minutes RT, and excess crystal violet was washed with dH2O with plates being allowed to dry. Colony formation was analyzed using ImageJ (Fiji). Significance was calculated by one-way ANOVA with Dunnett’s multiple comparisons tests. All assays were performed in triplicates.
Western blot analysis
Total cell lysates from human RMS cell lines and human myoblasts were obtained following lysis in RIPA lysis buffer supplemented with protease inhibitors (Roche). Western blot analysis was performed similar to Ignatius et. al., 2017 (27). Membranes were developed using an ECL reagent (Western Lightning Plus ECL, PerkinElmer; or sensitive SuperSignal West Femto Maximum Sensitivity Substrate, Thermo Scientific). Membranes were stripped, rinsed, and re-probed with the respective internal control antibodies. List of primary and secondary antibodies is included in supplementary data (Supplemental Table 1). Western blots were quantified using ImageJ software, normalizing proteins of interest to their respective β-tubulin expression and the control (either hSKMC or Scr of the same time point). If control expression was zero, quantifications shown are shown as a ratio of the protein of interest expression to its respective β-tubulin expression.
Immunohistochemistry
Once tumors reached 4X the initial volume, mice were euthanized and tumors were fixed with 4% paraformaldehyde/PBS, sectioned, blocked, and stained with H&E, Ki67, Myogenin, and MF20 antibodies (see Supplemental Table 1).
Caspase-Glo 3/7 assay
Rh18, Rh30, and RD Scr- and SNAI2-knockdown cells were seeded at 20% confluency in 24-well plates and placed in the Incucyte ZOOM instrument. After reaching ~40% confluency, media was supplemented with Caspase-Glo 3/7 reagent (1:1,000, Essen Bioscience) and Nuclight reagent (1:500, Essen Bioscience). Cells were then subjected to a range of doses of IR (0 Gy – 15 Gy) and placed back in the Incucyte. Images taken at 48h and 72h were processed using Adobe Photoshop and analyzed using ImageJ Cell Counter to determine percent caspase 3/7 events. Significance was determined using a two-way ANOVA with Sidak’s multiple comparisons test or one-way ANOVA with a post-hoc Tukey test accordingly.
Flow Cytometry
Rh18, Rh30, and RD Scr- and SNAI2-knockdown cells were seeded in 6-well plates and irradiated (PXi Precision X-Ray X-RAD 320). Cells were collected at varying time points (48, 72, 96, and 120 hours). A negative control of cells that did not receive IR were collected as well. Cells were centrifuged and resuspended in annexin-binding buffer. After determining cell density and diluting to 1 × 106 cells/mL with annexin-binding buffer, annexin V conjugate, and propidium iodide were added to sample aliquots and left to incubate at room temperature in the dark for 15 minutes. After incubation, aliquots were mixed gently while adding annexin-binding buffer on ice and analyzed by flow cytometry (LSRFortessa X-20; BD Biosciences). Cell cycle was assessed using the same cells and conditions described above with Click-iT EdU Alexa Flour 647 Flow Cytometry Assay (ThermoFisher) according to the provided protocol. Significance was determined using a Two-Proportion Z-test.
RNA-seq
RNA was extracted using the RNeasy mini kit (Qiagen). Poly-A-selected RNA libraries were prepared and sequenced on an Illumina HiSeq2000. QC was performed using FastQC version 0.11.2 and Picard’s version 1.127 RNASeqMetrics function with the default parameters. PCR duplicates were marked using Picard’s MarkDuplicates function. RNA-seq reads were aligned to the UCSC hg19 reference genome using TopHat version 2.0.13. Significance was defined as having FDR q-value <0.01 and FWER p-value of <0.05. Gene set enrichment analysis (http://www.broadinstitute.org/gsea/index.jsp) was performed using default parameter settings.
ChIP-seq and ChIP-qPCR
ChIP-seq data used, was published previously (28) and performed as follows. 1% Formaldehyde-fixed chromatin from RD and SMS-CTR cells were sheared to 200–700 bp with Active Motif EpiShear Sonicator. Chromatin-IP with SNAI2 Ab (CST, Catalogue # 9585) was performed O/N, using ChIP-IT High Sensitivity kit (Active Motif). Drosophila chromatin (Active Motif, Catalogue #53083) and H2Av ab (Active Motif, Catalogue #61686) was used for spike-in normalization across samples. ChIP-seq libraries were prepared using Illumina TruSeq ChIP Library Prep Kit and sequenced on NextSeq500. Reads were mapped to reference genome (version hg19) using BWA. High-confidence ChIP-seq peaks were called by MACS2.1. Raw sequencing data and processed files are available through GEO (GSE137168). For real-time qPCR immunoprecipitated DNA was amplified with BCL2L11 ChIP primers (SNAI2 binding peak 1 & 2) along with negative controls. All signals were normalized against input by the percentage input method. Significance was calculated by Unpaired t test.
Gene expression analysis
Real-time qPCR was completed using the QuantStudio 7 Flex system (Applied Biosciences). PCR primers are provided in Supplemental Table 2 (28). RNA isolation and cDNA preparation were performed as previously described(27,28). Significance was calculated by a two-way ANOVA with a Sidak’s multiple comparisons test. RNAseq data which includes alternative splice variants from Shern et al. (21) was used for corelation analyses. Non-coding and non-sense mediated decay isoforms were excluded from the analyses. Pairwise Pearson correlation of gene expression was calculated using R (version 4.0.5).
Results:
SNAI2 expression directly correlates with protection from radiation in RMS cells
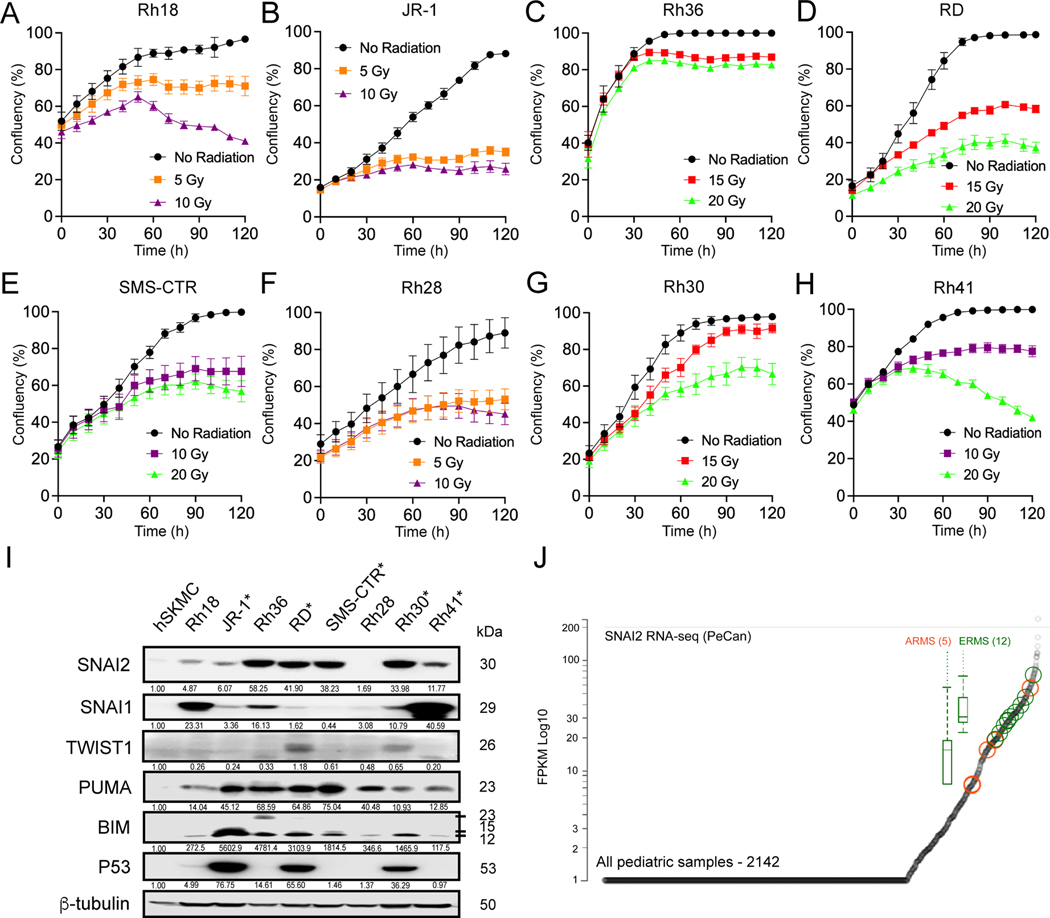
To better understand the factors regulating sensitivity to IR in RMS tumors, we used a panel of 8 representative human RMS cell lines namely JR1, RD, Rh18, Rh28, Rh30, Rh36, Rh41 and SMS-CTR. These cell lines include both Embryonal RMS (ERMS) and Alveolar RMS (ARMS) subtypes in which TP53 is both mutant and wild-type (Supplemental Table 3). Using an imaging-based platform to test different doses of IR on cell number/confluence, we found that Rh18, JR1, Rh28 and Rh41 cells are relatively more sensitive to IR while RD, SMS-CTR, Rh36, and Rh30 the more radioresistant (Figure 1A–H, Supplemental Figure 1A–B). Since SNAI2 is known to protect cells from IR (9,10), we analyzed SNAI2 expression levels across RMS cell lines. Interestingly, the expression of SNAI2 was correlated with the degree of protection from radiation, with Rh18, JR1, Rh28 and Rh41 showing low SNAI2 expression levels while RD, SMS-CTR, Rh36, and Rh30 show relatively high levels of SNAI2 protein. In contrast, the expression of proteins known to be involved post IR including SNAI1, TWIST1, ZEB1, CHEK1 and CHEK2 were not correlated with radiosensitivity (Figure 1I, Supplemental Figure 2A). Analysis of SNAI2 expression across RMS tumors and cell lines indicates high SNAI2 expression with a trend toward higher SNAI2 expression in ERMS tumors compared to ARMS, but that expression is variable across tumors (Figure 1J) (28). Finally, analysis of the St. Jude PeCan Data Portal and The Cancer Genome Atlas (TCGA) for SNAI2 expression showed that sarcomas, including RMS, are among the cancers that express the highest levels of SNAI2 and have higher expression levels compared to control tissue (Figure 1J and Supplemental Figure 2B–C). These results suggest that SNAI2 may have a pro-tumorigenic function in RMS.
Figure 1. SNAI2 expression directly correlates with radiosensitivity in RMS cells.
A-H. Rh18, JR-1, Rh36, RD, SMS-CTR, Rh28, Rh30, and Rh41 cells were radiated at 24h post imaging with varying levels of radiation and cell confluency (%) was assessed using Incucyte Zoom software based on phase-contrast images acquired from 0 h to 120 h. Error bars represent ±1 SD.
I. Western blot showing protein levels of SNAI2, SNAI1, TWIST1, PUMA, BIM, and P53 in parental RMS cell lines with skeletal human myoblast cells (hSKMCs) as a control. Asterisks (*) note RMS cell lines with known P53 mutations.
J. PeCan SNAI2 RNA-seq data for ARMS (orange) and ERMS (green) tumors.
SNAI2 protects RMS cells from IR in vitro
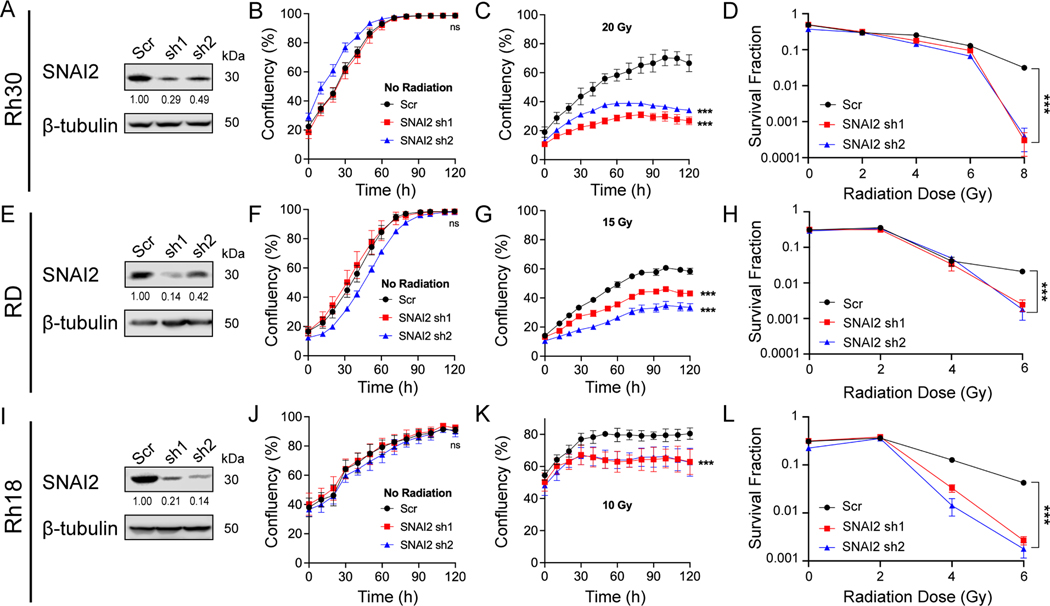
To assess whether SNAI2 protects RMS cells from IR, we performed SNAI2-knockdown experiments using validated shRNAs in Rh30, RD and Rh18 cells (25), which were chosen as representative lines with low, moderate, and high levels of radiosensitivity (Figure 1A–H, Figure 2A–L). Compared to the scrambled control (Scr) shRNA, two SNAI2 shRNAs (sh1 and sh2) reduced SNAI2 protein expression by 51–86% in Rh30, RD and Rh18 cells (Figures 2A, 2E, and 2I). Interestingly, while transient SNAI2 knockdown initially slowed cell proliferation (28), this effect was no longer observed once stable lines were generated (Figure 2B, 2F, 2J). Each cell line was then exposed to an appropriate dose of IR (see Figure 1A–H) and analyzed for confluency every 4 hours for 5 days (Figure 2B, 2F, 2J). Compared to control-knockdown cells at 120 hours, SNAI2-knockdown cells became sensitized to IR across all three cell lines (Figure 2C, 2G, 2K). Similar results were observed in clonogenic colony-forming assays, where the surviving fraction of colonies was assessed in RMS cells exposed to a range of IR doses between 2 and 8 Gy. At lower doses of IR exposure, differences in survival and colony formation were minimal compared to higher doses of IR where there was a clear separation between the shScr and shSNAI2 treated cells (Figure 2D, 2H, 2L and Supplemental Figure 3A). Next, to test whether overexpression of SNAI2 could promote radioresistance, we transfected control pBabe vector and SNAI2-Flag constructs into the highly radiosensitive Rh18 cell line and tested the response to IR (Supplemental Figure 3B–D). While SNAI2 overexpression in Rh18 cells had no effect on proliferation in the absence of IR (Supplemental Figure 3C), the cells became less sensitive to 10 Gy IR at 120 hours (Supplemental Figure 3D). Since in SNAI2 knockdown RMS cells SNAI1 is induced and some RMS cells express high SNAI1, we assessed the effect of loss of SNAI1 on sensitivity to radiation in Rh18, RD and Rh30 cells and show that SNAI1 ablated cells have similar survival to control cells post IR (Supplemental Figure 3E–G). Thus, SNAI2 protects RMS cells from IR in vitro.
Figure 2. SNAI2 protects RMS cells from IR in vitro.
A. Western blot showing protein expression for SNAI2 of control (Scr shRNA) or SNAI2 knockdown (sh1 or sh2) in Rh30 cells.
B, C. Cell Confluency measured as a % of the total of Rh30 cells with no IR or IR at 20 Grays (Gy) with either control or SNAI2 knockdown was assessed using phase-contrast images acquired from 0 h to 120 h. Error bars represent ±1 SD. Ns = not significant, ***p<0.001 by two-way ANOVA with Sidak’s multiple comparisons test.
D. Survival fractions of Rh30 Scr and SNAI2 knockdown colony formation assays were assessed at increasing IR dose exposures. Error bars represent ±1 SD. Statistical differences were observed at 8 Gy. ***p<0.0001 by one-way ANOVA with Dunnett’s multiple comparisons test.
E. Western blots showing protein expression for SNAI2 after control (Scr) or SNAI2 knockdown (sh1 or sh2) in RD cells.
F, G. Cell Confluency measured as a % of the total of RD cells with no IR or IR at 15 Gy with either control or SNAI2 knockdown was assessed using phase-contrast images acquired from 0 to 120 h. Error bars represent ±1 SD. Ns = not significant, ***p<0.001by two-way ANOVA with Sidak’s multiple comparisons test.
H. Survival fractions of RD Scr and SNAI2 knockdown colony formation assays were assessed at increasing IR dose exposures. Error bars represent ±1 SD. Statistical differences were observed at 6 Gy. ***p<0.0001 by one-way ANOVA with Dunnett’s multiple comparisons test.
I. Western blot showing protein expression of SNAI2 in control (Scr) or SNAI2 knockdown (sh1 or sh2) Rh18 cells.
J, K. Cell Confluency measured as a % of the total of Rh18 cells with no IR or IR at 10 Gy in either control or SNAI2 knockdown cells was assessed using phase-contrast images acquired from 0 to 120 h. Error bars represent ±1 SD. Ns = not significant, ***p<0.001 by two-way ANOVA with Sidak’s multiple comparisons test.
L. Survival fractions of Rh18 Scr and SNAI2 knockdown colony formation assays were assessed at increasing IR dose exposures. Error bars represent ±1 SD. Statistical differences were observed at 6 Gy. ***p<0.0001 by one-way ANOVA with Dunnett’s multiple comparisons test.
SNAI2 protects RMS tumors from IR in vivo
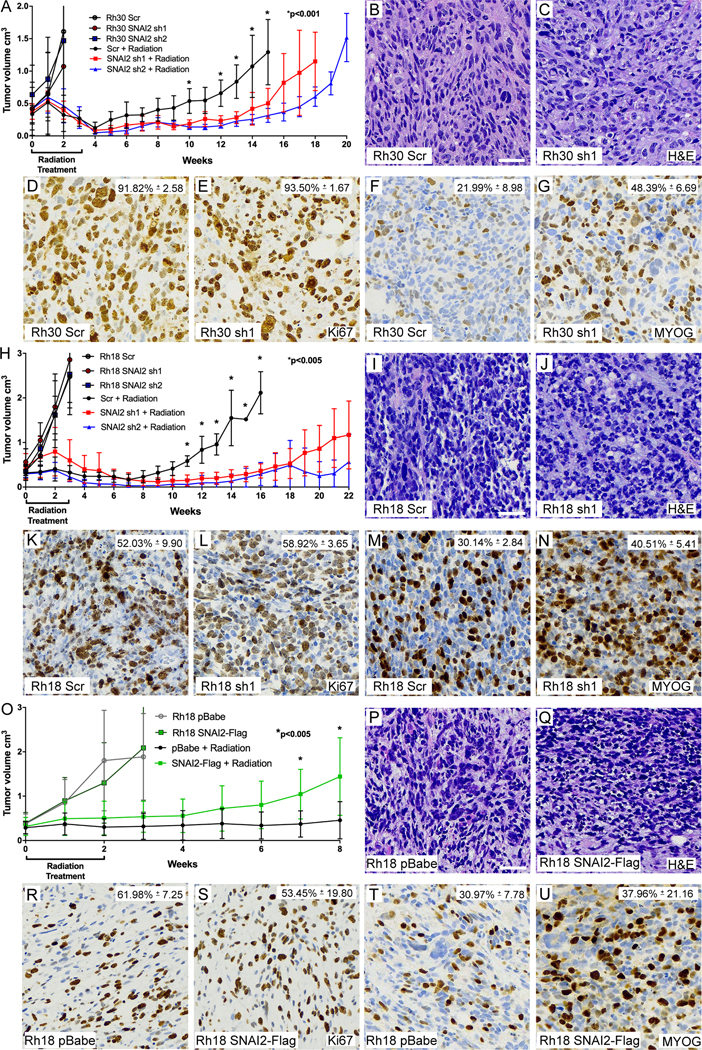
We next questioned whether SNAI2 could protect RMS tumor cells from IR in vivo. We created murine xenografts of Rh30 and Rh18 cells with SNAI2 knockdown and Rh18 cells with SNAI2 overexpression, as well as appropriate controls, and performed irradiation experiments after each group of mice developed palpable tumors (200–400 mm3). Rh30 and Rh18 tumors with SNAI2 (or control) knockdown were subjected to a cumulative 30-Gy dose of IR (2 Gy/day, 5 days per week, for 3 weeks). Following the completion of the IR regimen, tumors were analyzed weekly for relapse (Figure 3 A–N). Relapse was defined by re-growth of tumors to four times their size prior to IR treatment (29). In the absence of IR, there were no significant differences in the growth rates between xenografts derived from Rh30 control- and SNAI2-knockdown cells (1×106 cells injected/mouse, Figure 3A, Supplemental Figure 4A, n = 5). However, control Rh30 xenografts that were exposed to IR gave rise to relapse tumors significantly earlier (5 weeks post-IR) than SNAI2-knockdown Rh30 xenografts (11–14 weeks post-IR: Figure 3A, Supplemental Figure 4B, n = 8–10). Similar to Rh30 xenografts, there were no differences in the growth rates between xenograft tumors derived from Rh18 control- and SNAI2-knockdown cells (5×106 cells injected/mouse) in the absence of IR (Figure 3H, Supplemental Figure 4C), and post IR control Rh18 xenografts gave rise to relapsed tumors significantly earlier (7 weeks post-IR) than SNAI2-knockdown Rh18 xenografts (13–14 weeks post-IR: Figure 3H, Supplemental Figure 4D, n = 8–10). Importantly, a subset of Rh18 SNAI2 shRNA knockdown tumors did not relapse until approximately 21 weeks post IR. Finally, Rh18 xenografts (5×106 cells injected/mouse) with control or SNAI2 overexpression showed no differences in growth rate in the absence of IR (Figure 3O–U, Supplemental Figure 4E, n = 4), but following a cumulative 20-Gy dose of IR (2 Gy/day, 5 days per week, for 2 weeks), SNAI2-overexpressing Rh18 xenografts relapsed more rapidly than control Rh18 xenografts (Figure 3O, Supplemental Figure 4F, n = 8–9).
Figure 3. SNAI2 protects RMS tumors from IR in vivo.
A. Growth curves of Rh30 xenografts including Scr shRNA and SNAI2 shRNA 1 and 2 (sh1, sh2) engrafted in mice. Xenograft growth was assessed under no IR and 30 Gy IR treatments. IR was given for 3 weeks at 2 Gy/day, 5 days a week. Error bars represent ±1 SD. *p<0.001 by Student’s t test.
B, C. H&E staining showing histology of Rh30 Scr and SNAI2 knockdown tumor sections. Scale bar = 100 μm.
D-G. Immunohistochemistry analysis of Ki67 (D, E) staining to assess proliferation and Myogenin (MYOG) staining (F, G) in Rh30 xenografts with either Scr shRNA or SNAI2 sh1. Ki67 –Scr vs. sh1 p. MYOG – Rh30 Scr vs. Rh30 sh1 p<0.05 by Welch’s t test. Magnification same as B, C.
H. Growth curves of Rh18 xenografts including Rh18 Scr shRNA and SNAI2 shRNA 1 and 2 (sh1, sh2) engrafted in mice. Xenograft growth was assessed under no IR and 30 Gy IR treatments. IR was given for 3 weeks at 2 Gy/day for 5 days a week. Error bars represent ±1 SD. *p<0.005 by Student’s t test.
I, J. H&E staining showing histology of Rh18 Scr and SNAI2 knockdown tumor sections. Scale bar = 100 μm.
K-N. Immunohistochemistry analysis of Ki67 (K, L) staining to assess proliferation and Myogenin (MYOG) staining (M, N) in Rh18 xenografts with either Scr shRNA or SNAI2 sh1. MYOG – Scr vs. sh1 p=0.0519 by Welch’s t test. Magnification same as I, J.
O. Growth curves of Rh18 xenografts expressing control vector (pBabe) and SNAI2-Flag engrafted in mice. Xenograft growth was assessed under no IR and 20 Gy IR treatments. IR was given for 2 weeks at 2 Gy/day for 5 days a week. Error bars represent ±1 SD. *p<0.005 by Student’s t test.
P, Q. H&E staining showing histology of Rh18 pBabe and SNAI2-Flag tumor sections. Scale bar = 100 μm.
R-U. Immunohistochemistry analysis of Ki67 (R, S) staining to assess proliferation and Myogenin (MYOG) staining (T, U) in Rh18 xenografts with either pBabe or SNAI2-Flag expression. Ki67 – pBabe vs. SNAI2-Flag not significantly different. MYOG – pBabe vs. SNAI2-Flag p=0.0519 by Welch’s t test. Magnification same as P, Q.
To investigate the effect of SNAI2 knockdown on tumor histology, proliferation, and differentiation in Rh30 and Rh18 relapsed tumors, xenografts with Scrambled or SNAI2 knockdown were harvested from mice once they reached 4X their initial volume and tumors were processed, sectioned, and assessed for histology (H&E), proliferation (Ki67), and differentiation (MYOG). H&E analysis did not show significant differences between Scrambled vs. SNAI2-knockdown tumors (Figure 3B–C, 3I–J). Next, Ki67 staining showed Control (shScr) and SNAI2 knockdown xenografts proliferate at similar rates (Figure 3D–E, 3K–L). In Rh30 and Rh18 tumors with SNAI2 shRNA knockdown, there was a trend toward areas showing increased MYOG expression (Figure 3F–G, 3M–N). However, this increase in MYOG did not result in terminal differentiation as assessed by MyHC (MF20) staining (Supplemental Figure 4G–J). Similarly, there was not a significant effect on tumor histology, proliferation, or differentiation observed when comparing Rh18 controls to Rh18 SNAI2 overexpressing xenografts (Figure 3P–U). Thus, while the H&E and MYOG staining showed that SNAI2 may also inhibit myogenic differentiation in some RMS tumors; however, this effect is not as prominent compared to our previously described results in RAS mutant ERMS (28). Altogether, our data suggest that the major conserved effect of SNAI2 post IR is to protect RMS tumors from IR in vivo.
Loss of SNAI2 promotes IR-mediated apoptosis and blocks irradiated RMS cells from exiting the cell cycle
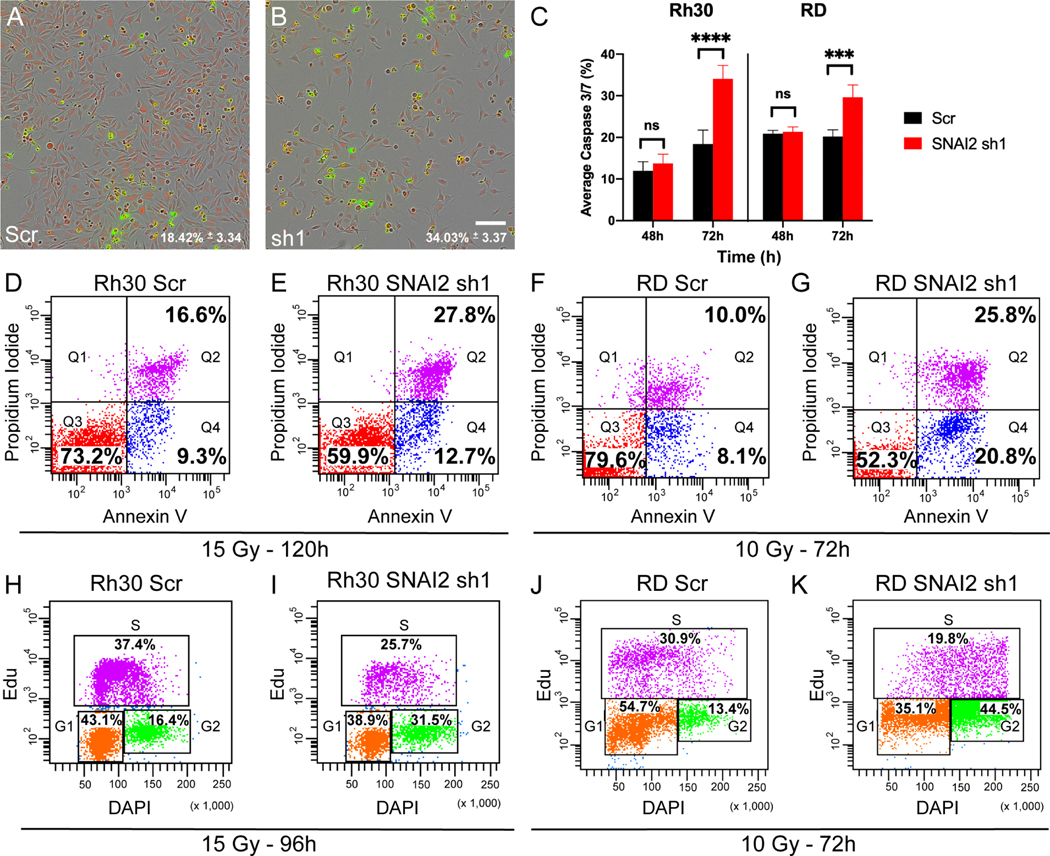
To better understand how SNAI2 protects RMS cells from IR, we assessed the effects of SNAI2 knockdown on apoptosis and the cell cycle in RMS cell lines. We first analyzed live cells for signs of apoptosis using a Caspase-Glo assay in Rh30, RD and Rh18 cells. In the absence of IR, control- and SNAI2-knockdown cells grew at similar rates (see Figure 2B, 2F, 2J) and had significantly lower Caspase3/7 staining until they reached confluence in all three RMS cell lines (Supplemental Figure 5A). However, at 72 hours post-IR (hpIR), SNAI2-knockdown cells exhibited a significant increase in apoptosis compared to controls (Figure 4A–C, Supplemental Figure 5B). We then performed independent apoptosis assays using annexin V/propidium iodide staining to confirm these findings by analyzing live cells post IR by flow cytometry. Consistent with the Caspase-Glo assay, there was a significant increase in early and late apoptotic cells in the SNAI2-knockdown Rh30, RD and Rh18 populations between 72 to 120 hpIR compared to control-knockdown cells (Figure 4D–G, Supplemental Figure 5C–D). Importantly, in the absence of IR, there were no significant differences in apoptosis between control- and SNAI2-knockdown cells for each of the cell lines (Supplemental Figure 5E–J). These experiments indicate that SNAI2 protects RMS cells from IR-induced apoptosis.
Figure 4. Loss of SNAI2 promotes IR-mediated apoptosis and blocks irradiated RMS cells from exiting the cell cycle.
A, B. Representative images of Caspase-Glo assay in Rh30 cells (either control or SNAI2 knockdown) at 72h post IR exposure (15 Gy) with red labeling cells/nuclei and green labeling caspase 3/7; average caspase 3/7 levels (%) were quantified in C. Scale bar = 150 μm.
C. Average Caspase 3/7 percentage (mean ± 1 SD) in Rh30 and RD Scr and SNAI2 sh1 cells 48h and 72h after IR exposure of 15 or 10 Gy respectively. ***p<0.001, ****p<0.0001 by two-way ANOVA with Sidak’s multiple comparison.
D-G. Flowcytometry plots showing Propidium iodide vs. Annexin V staining of Rh30 and RD Scr or SNAI2 sh1 cells and treated with indicated IR doses. Q4 represents cells undergoing early apoptosis, whereas Q2 represents cells undergoing late apoptosis. Q3 represents live cells not undergoing apoptosis. Rh30 early apoptosis (Q4): Scr 9.3% vs. sh1 12.7%, p<0.0001, late apoptosis (Q2): Scr 16.6% vs. sh1 27.8%, p<0.0001; RD early apoptosis (Q4): Scr 8.1% vs. sh1 20.8%, p<0.0001, late apoptosis (Q2): Scr 10.0% vs. sh1 25.8%, p<0.0001 by Two Proportions Z-test.
H-K. Flowcytometry plots of EdU vs. DAPI staining in Rh30 and RD cells with either Scr shRNA or SNAI2 sh1 after exposure to indicated IR doses. Rh30 Scr vs. sh1 G2 phase p<0.0001; RD Scr vs. sh1 G2 phase p<0.0001 by Two Proportions Z-test.
We next analyzed whether SNAI2 regulates the cell cycle after exposure to IR using EdU labeling followed by flow cytometry. Following IR treatment, SNAI2-knockdown cells showed a significant reduction in cells in the G1 and S phases and an accumulation of cells in the G2-M phase of the cell cycle, indicative of a G2/M block (Figure 4H–K, Supplemental Figure 5K–L). In the absence of IR, there were no differences in proliferation between control- and SNAI2-knockdown cells (Supplemental Figure 5M–R). These experiments suggest that loss of SNAI2 may prevent mitosis or alter progression through the M-phase of cell cycle following exposure to IR.
SNAI2 represses the expression of the BH3-only BIM in RMS cells
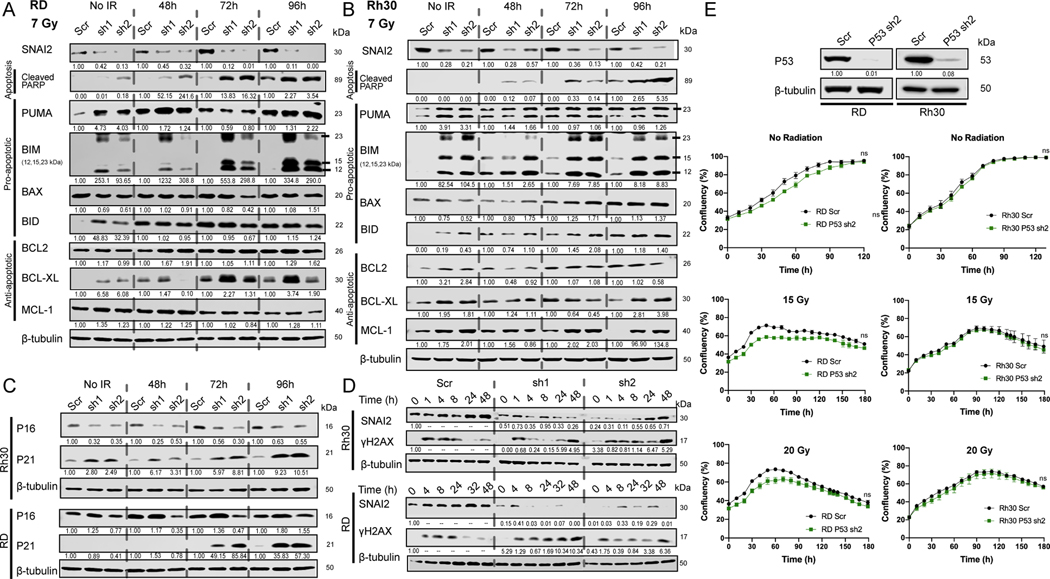
To investigate the mechanisms by which SNAI2 protects RMS cells from IR-induced apoptosis, we analyzed the expression of both pro- and anti-apoptotic regulators of mitochondrial apoptosis across RMS cells (Supplemental Figure 2C) and more specifically in Rh30, RD, and Rh18 cells expressing shScr vs. shSNAI2 with and without IR (Figure 5A–B, Supplemental Figure 6A). In response to IR, SNAI2-knockdown cells showed increased expression of the apoptosis marker cleaved-PARP, which is prominent especially between 72 to 96 hpIR and consistent with the annexin-V and Caspase-Glo analyses (Figure 4D–G, Supplemental Figure 5C–D). In the absence of IR, expression of the proapoptotic BH3-only regulator PUMA/BBC3 appears to be upregulated upon SNAI2 knockdown in all cell lines. Interestingly, SNAI2 knockdown also elicited a prominent increase in the proapoptotic BH3-only BIM across all three cell lines as well as in BID and BAX in Rh18 and BID in Rh30 lines. With respect to anti-apoptotic regulators, BCL-2 showed a modest increase in both Rh30 and Rh18 in response to SNAI2 knockdown in the absence of IR, while BCL-XL expression was elevated in RD and Rh30. Moreover, in response to IR treatment, BCL-2, MCL-1, and BCL-XL varied across cell lines and failed to correlate with SNAI2 knockdown. Among the regulators of apoptosis, only BIM expression was found to be consistently elevated across all cell lines in response to SNAI2 knockdown in IR-exposed cells. These experiments indicate that SNAI2, in addition to its known function as a transcriptional repressor of PUMA (10,30), appears to repress the expression of BIM in RMS cells.
Figure 5. SNAI2 represses the expression of BH3-only BIM in RMS cells.
A, B. Western blot analyses to determine protein expression of SNAI2, Cleaved PARP, PUMA, BIM, BAX, BID, BCL2, BCL-XL, and MCL-1 in RD and Rh30 cells under no IR or at 48-, 72-, or 96-hpIR after 7 Gy treatment.
C. Western blot analysis to determine protein expression of P16 and P21 in Rh30 and RD cells under no IR or at 48-, 72-, or 96-hpIR with 7 Gy. Protein used for RD cell line western blots was from the same analyses for experiments in Figure 5A (see above), therefore the β-tubulin blot is the same as in Figure 5A.
D. Western blot analysis of γH2AX over time after exposure to 7 Gy IR in Rh30 and RD control (Scr) and SNAI2 knockdown (sh1 and sh2) cells.
E. Western blot analysis to determine protein levels of P53 in RD and Rh30 cells (either Scr control or P53 sh2 knockdown). Confluency (%) of non-IR or IR-treated (15 or 20 Gy) RD and Rh30 cells (with either Scr control or P53 shRNA knockdown) was assessed on phase-contrast images acquired from 0 to 180 h. No statistical differences were observed. Error bars represent ±1 SD. ns = not significant by Unpaired t test.
To determine the mechanism by which SNAI2 influences the cell cycle in RMS cells, we analyzed the expression of the P21/CDKN1A cell cycle checkpoint inhibitor and found that SNAI2 knockdown leads to upregulation of P21 in both Rh30 and RD cells (Figure 5C). Since P21 is also a marker of cells undergoing senescence, we analyzed P16 expression and performed β-gal staining in Rh30 and RD cells. There were no differences in P16 expression or β-gal staining between control and SNAI2 knockdown in either cell type, suggesting that senescence is not regulated by SNAI2 in these cells (Figure 5C, Supplemental Figure 6B–E). In RMS tumors, CDKN1A/P21 expression is often repressed, and re-expression of P21 promotes differentiation (31,32). Moreover, SNAI2 has been shown by ChIP-seq experiments to indirectly block CDKN1A expression in ERMS RD and SMS-CTR cells, and co-knockdown of CDKN1A and SNAI2 in RD and JR1 RMS cells results in the loss of differentiation-positive, myosin-heavy-chain-expressing cells (28). We also assayed the effect of SNAI2 knockdown on differentiation post IR in Rh30 and RD cells and in another ERMS cell line with wild-type P53, Rh36. Both RD and Rh36 are RAS mutant ERMS cell lines. Following exposure to IR, RD and Rh36 cells with SNAI2 knock down exhibited a significant increase in differentiation as determined by the expression of differentiated myosin MF-20 staining (Supplemental Figure 6F–K). In contrast, in Rh30 (ARMS) cells this effect was not as prominent (Supplemental Figure 6L), suggesting that SNAI2 may be more important for the suppression of muscle differentiation in RAS mutant ERMS tumors consistent with our previous findings (28).
We next questioned whether SNAI2 regulates IR-induced DNA repair. Since IR primarily causes DNA double-stranded breaks, we analyzed the expression of a well-established marker for these DNA lesions, γH2AX(33), during a time course following IR in Rh30 and RD cells via western blot analysis (Figure 5D). As expected, γH2AX levels increased rapidly following exposure to IR in both cell lines with SNAI2-knockdown cells showing similar increases compared to control-knockdown cells; however, γH2AX expression was retained as late as 48h in SNAI2 knockdown cells, suggesting a delay in the ability of SNAI2 knockdown cells to repair damaged DNA (Figure 5D). However, SNAI2-knockdown RD and Rh30 cells have little if any differences in expression of DNA damage checkpoint regulators pCHEK1 and pCHEK2 following IR (Supplemental Figure 6M–N). These findings suggest that SNAI2 might have additional roles on influencing the timing of repair of DNA double-strand breaks in RMS cells.
Both Rh30 and RD cells have TP53 mutations that aberrantly stabilize the P53 protein (Figure 1E)(34,35). To determine if the expression of mutant P53 is important for IR-mediated inhibition of cell growth in RD and Rh30 cells, we performed an shRNA-mediated knockdown of P53 and tested its effects on the RD and Rh30 lines (Figure 5E). In both lines, knockdown of mutant P53 failed to affect sensitivity to IR (Figure 5E). Additionally, in both Rh30 and RD cells, SNAI2 is slightly induced or maintained following IR suggesting alternate mechanisms by which SNAI2 expression is modulated in RMS (Figure 5D).
Direct repression of BIM by SNAI2 blocks apoptosis in irradiated RMS cells
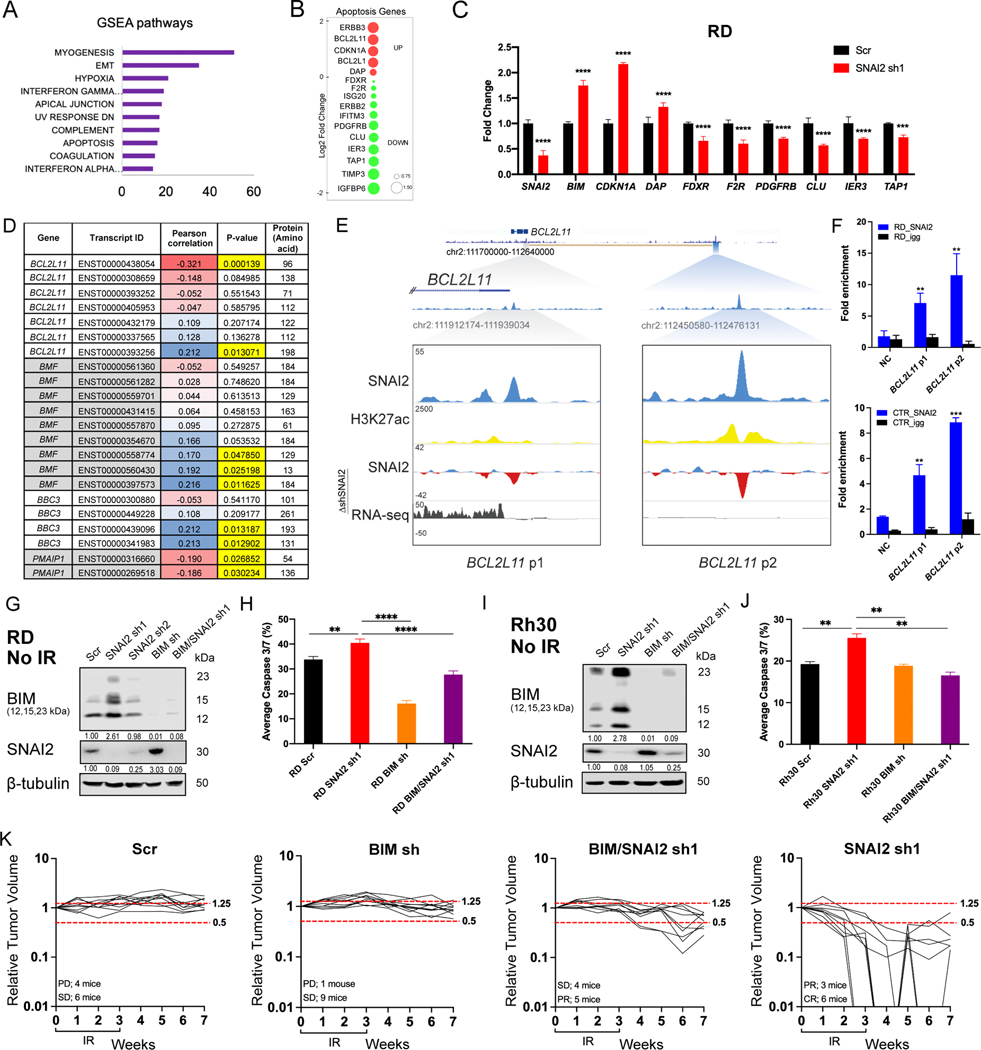
Since BIM was consistently and robustly induced by loss of SNAI2 in RMS cells and apoptosis post IR is the major effect of SNAI2 ablation across all RMS cells assessed, (Figure 5A–B, Supplemental Figure 6A) we questioned whether SNAI2 could directly repress expression of BH3 pro-apoptotic regulator BIM. We first performed RNA-seq analysis comparing control- and SNAI2-knockdown in RD cells at 24 hpIR (5 Gy). GSEA pathway analysis revealed that in addition to differences in myogenic differentiation, several stress response pathways were modulated in response to IR. These pathways included “hypoxia”, “UV response down”, and “apoptosis” (Figure 6A). Several apoptotic pathway genes were increased or decreased in SNAI2 knockdown cells post IR (Figure 6B). We performed qRT-PCR analysis using RD and Rh30 cells for a subset of the apoptotic regulators and validated the upregulation of CDKN1A (only RD), BCL2L11(BIM), and DAP and downregulation of FDXR, F2R, PDGFRB, CLU, IER3, and TAP1 (Figure 6C, Supplemental Figure 7A). We also analyzed the correlation between SNAI2 expression and BIM/BCL2L11 and other pro and antiapoptotic regulators in human RMS tumors and find that SNAI2 expression is significantly negatively correlated with at least one alternatively spliced transcript coding for BIM/BCL2L11 but not for PUMA/BBC3 or BMF transcripts (Figure 6D; Supplemental Tables 4–5). Finally, ChIP-seq analysis of SNAI2 binding in SMS-CTR and RD cells with shScr and shSNAI2-knockdown treatments was used to assess SNAI2-chromatin binding peaks in BH3 containing proapoptotic regulators. Our analyses show that SNAI2 binds to enhancers associated with BIM/BCL2L11 (Figure 6E)(28) and BMF (Supplemental Figure 7B). In contrast, no SNAI2 shared binding peaks in SMS-CTR and RD cells were found associated with the promoter enhancer regions of CDKN1A (Supplemental Figure 7B)(28), PUMA/BBC3 or other pro and anti-apoptotic regulators (Supplemental Figure 7B). To validate the ChIP-seq findings, we performed ChIP-qPCR analyses at the SNAI2 binding sites downstream of BIM/BCL2L11 and confirmed that SNAI2 enrichment is relatively increased at the distal enhancer contained within the BIM/BCL2L11 Topologically Associated Domain (TAD) (Figure 6F, Supplemental Figure 8) consistent with our ChIP-seq results. Together, these experiments establish that SNAI2 is a direct repressor of pro-apoptotic BIM/BCL2L11.
Figure 6. Direct repression of BIM by SNAI2 blocks apoptosis in irradiated RMS cells.
A. GSEA pathway analysis comparing mRNA expression (RNA-seq) in control/Scr and SNAI2 shRNA treated RD cells 24 h post IR (5 Gy). Enriched pathways in shSNAI2 cells (GSEA Hallmark pathways) with number of up-regulated genes in each pathway class (x-axis).
B. RNA-seq data showing genes highly upregulated or downregulated when comparing control/ Scr- and SNAI2-shRNA knockdown RD cells 24 h post IR (5 Gy). Red circles denote up-regulated genes and Green denotes down-regulated genes; size of circles represent Log2 fold change compared to shScr.
C. Real time qPCR analysis (mean ± 1 SD) of various cell cycle and apoptosis genes in RD cells 48h after irradiation with 5 Gy. ***p<0.001, ****p<0.0001 by two-way ANOVA with a Sidak’s multiple comparisons test.
D. Pearson correlation between the expression of SNAI2 and pro-apoptotic regulators BIM/BCL2L11, BMF, BBC3, and PMAIP1 in primary human RMS assessed by RNA-sequencing analysis. Blue cells indicate positive correlation, while red cells indicate negative correlation.
E. ChIP-seq tracks of SNAI2 (Blue), H3K27ac (Yellow) binding at the BIM/BCL2L11 locus in SMS-CTR cells in shScr cells and delta (Δ) enrichment value (shSNAI2 sh1 minus shScr, Blue and Red) for SNAI2 and gene expression (RNA-seq, Black). Boxed area corresponds to SNAI2 binding region and shaded areas represent relative position of the peaks to the TAD containing BCL2L11. Values on Y-axis represent fold enrichment.
F. ChIP-qPCR of SNAI2 enriched regions down stream of BIM/BCL2L11 gene in RD and SMS-CTR cells. Fold enrichment value of SNAI2 at the BCL2L11-peak1 & peak2, is plotted along with Negative Control region (NC) and IgG controls, **p<0.01, ***p<0.0001 by Unpaired t test compared to NC.
G. Western blot analyses of BIM and SNAI2 expression in non-IR treated RD cells under either Scr control, SNAI2 shRNA, BIM shRNA, or double BIM/SNAI2 shRNA conditions.
H. Average Caspase 3/7 (%) (mean ± 1 SD) in RD Scr, SNAI2 sh1, BIM sh, and BIM/SNAI2 sh1 cells 72h after IR exposure (15 Gy). **p<0.01, ****p<0.0001 by one-way ANOVA with Tukey’s multiple comparisons test.
I. Western blot analyses of BIM and SNAI2 expression in non-IR treated Rh30 cells under either Scr control, SNAI2 shRNA, BIM shRNA, or double BIM/SNAI2 shRNA conditions.
J. Average Caspase 3/7 (%) (mean ± 1 SD) in Rh30 Scr, SNAI2 sh1, BIM sh, and BIM/SNAI2 sh1 cells 72h after IR exposure (15 Gy). **p<0.01 by one-way ANOVA with Tukey’s multiple comparisons test.
K. Individual relative tumor volumes of RD Scr, SNAI2 sh1, BIM sh, and BIM/SNAI2 sh1 xenografts after 30 Gy (2 Gy/day, 5x a week, for 3 weeks). Change in volume 2 weeks post-IR: shScr = 0.0295 cm3/week, shBIM = 0.01125 cm3/week; no significant difference by one-way ANOVA with Tukey’s multiple comparisons test. Change in volume 2 weeks post-IR: shSNAI2 = −0.067 cm3/week, shSNAI2/BIM = −0.0105 cm3/week; p<0.0001 by one-way ANOVA with Tukey’s multiple comparisons test.
We next questioned whether reducing BIM expression would abrogate the SNAI2-knockdown-induced radiosensitivity in RMS cells. We therefore generated RD and Rh30 cells with stable lentiviral infections for 1) control shRNA, 2) SNAI2 shRNA, 3) BIM shRNA and 4) both SNAI2 and BIM shRNAs (Figure 6G–K) (26). Consistent with our earlier findings, knockdown of SNAI2 increased the levels of all three forms of BIM, which was reversed by concomitant knockdown of BIM (Figure 6G, 6I). As expected, IR treatment of SNAI2-knockdown RD and Rh30 cells with a single dose of 15 Gy resulted in selective loss of confluency (Supplemental Figure 9A–B) and a significant increase in Caspase 3/7 positive cells compared to irradiated control-knockdown cells. However, this effect on apoptosis and confluency was significantly reversed in SNAI2/BIM- double knockdown cells post-IR (Figure 6H, 6J, Supplemental Figure 9A–B). Of note, in the absence of IR, no differential effects on confluency or apoptosis were observed across all four groups (Supplemental Figure 9C–D). Since the SNAI2/BIM-knockdown-mediated rescue of radiosensitivity in Rh30 cells was only partial, it is possible that other SNAI2-regulated genes may also contribute to IR-induced effects (Supplemental Figure 9B). Finally, we tested in RD xenografts the effect of combined knockdown of SNAI2 and BIM on response to IR. We created murine xenografts of RD with shScr, shBIM, shSNAI2, and shSNAI2/BIM double knockdown conditions. After each group of mice developed palpable tumors (200–400 mm3), they were subjected to a cumulative 30-Gy dose of IR (2 Gy/day, 5 days per week, for 3 weeks). Following the completion of the IR regimen, tumors were analyzed weekly for changes in tumor volume. While shScr and shBIM tumors showed no effect on relative change in tumor volume post-IR compared to their initial volumes, with animals demonstrating either stable or progressive disease, in the shSNAI2 group, 6 of 9 tumors showed a complete response, while the other 3 of 9 show a partial response 4 weeks post-IR (Figure 6K). In contrast, in the double SNAI2/BIM shRNA group none of the tumors resulted in a complete response. Instead, only 5 of 9 tumors resulted in a partial response, whereas the remaining 4 mice demonstrated stable disease. Indeed, the relative change in tumor volume post-IR compared to their initial volumes in shSNAI2 xenografts were significantly decreased compared to double SNAI2/BIM knockdown xenografts (Figure 6K). Together, these experiments show that SNAI2-mediated repression of BIM protects RMS cells from the effects of IR.
DISCUSSION
Radiation therapy is an important component of RMS treatment, especially during the management of metastases in high-risk patients (2,3,5). Identifying pathways that regulate the response to IR therapy could potentially provide biomarkers of resistance or sensitivity to IR as well as targets for therapeutic intervention. Here, we show that SNAI2 directly represses the proapoptotic BH3-only gene BIM to protect RMS tumors from IR.
SNAI2 protects RMS tumor cells from IR despite the fact that the P53 pathway is nonfunctional or is mutant and consequently PUMA induction is not robust in the TP53 mutant RMS cell lines tested in this study. In lymphocytes and other highly radiosensitive cell types, P53 becomes rapidly activated in response to IR, and triggers mitochondrial apoptosis through induction of the expression of PUMA (10). Our finding that SNAI2 expression levels appear to dictate radiosensitivity of RMS cells in a manner that is independent of TP53 mutant status and possibly PUMA expression, but rather by direct repression of BIM that is not directly regulated by P53, suggests an important role for SNAI2 in the radiation response in cancer cells that can be dependent or independent of P53. Experiments in vivo in mice mutant for Puma and Bim demonstrate that both Puma and Bim can potently trigger the mitochondrial apoptosis response post radiation. Further, the loss of Bim protects lymphocytes from radiation and decreases the time to tumor initiation in thymocytes compared to wild-type controls (11,36–38). This effect of BIM protecting cells from radiation is seen by correlation in renal cell carcinomas and also in KRAS mutant lung cancer cell lines. Correlation studies in renal cell carcinomas that often have mutations in VHL express low levels of BIM (EL) and are more resistant to several apoptotic stimuli, including UV-radiation (39). Also, in a subset of KRAS mutant lung cancer cell lines, low BIM expression was associated with relative resistance to radiation (40). Thus, BIM is a bona fide BH3 pro-apoptotic regulator that can be induced post IR to mediate mitochondrial apoptosis. Our study shows that in RMS tumors SNAI2 is a potent repressor of BIM expression, yet its expression can be independent of TP53. In support of this assertion, we have recently found in the ERMS sub-type that both MEK signaling and muscle specific regulator MYOD maintains SNAI2 expression; while in the ARMS subtype the PAX3-FOXO fusion oncogene is also known to be required for SNAI2 expression (28,41,42).
Despite the presence of different genetic drivers namely, the PAX3-FOXO fusion oncogene in ARMS lines and mutant RAS and amplified MDM2 in the ERMS lines among the cell lines used in this study, the radiosensitivity of each cell line correlated with expression of SNAI2 rather than oncogene/tumor suppressor status. This suggests that SNAI2 expression levels could be used to predict the degree of radiosensitivity across different RMS tumors, and perhaps multiple tumor types. For example, most solid tumors have intermediate levels of SNAI2 and intermediate sensitivities to IR (43,44) (Supplemental Figure 1C). In contrast, melanomas and osteosarcomas, which are generally not treated with IR in the clinic due to their inherent radioresistance and radiation when administered is at high doses for local control of disease and in palliative care (45–47), have the highest levels of SNAI2 expression. This makes sense from a developmental perspective since sarcomas originate from mesoderm(48,49), a tissue that expresses high levels of SNAI2 during development and has roles in muscle, bone and cartilage tissues (Reviewed in (8)). The analysis of SNAI2 expression levels could therefore be potentially informative for the treatment of multiple different cancers.
RMS cells with stable SNAI2 knockdown while showing relatively little differences in proliferation, apoptosis or cell viability compared to controls, nevertheless have consistent increased expression of pro-apoptotic modulators BIM, PUMA, and BID at baseline even in cells not exposed to IR. Additionally, our studies show that SNAI2 directly represses BIM expression. This suggests that SNAI2 knockdown cells are primed for mitochondrial apoptosis and an important oncogenic role for SNAI2 is to prevent IR induced mitochondrial apoptosis. In contrast to the consistent expression of BIM and PUMA, in untreated cells different anti- apoptotic regulators, BCL2, BCLXL and MCL-1, are expressed at varying levels in the Rh30, RD, and Rh18 RMS lines to balance pro apoptotic factor expression in SNAI2 knockdown cells. Additionally, we find relatively different protein levels of several pro and anti-apoptotic regulators across commonly studied RMS cell lines (Supplemental Figure 2C. Based on these observations, one would predict that treating RMS cells with inhibitors of BCL2, BCLXL, and MCL1 in the context of SNAI2 knockdown would have variable effects on mitochondrial apoptosis and might be an important factor when considering combination treatments, whereas increasing BIM activity would further prime RMS tumors to undergo apoptosis.
In summary, our study implicates SNAI2 as a potential biomarker for IR sensitivity in RMS, with an inverse correlation between SNAI2 expression levels and radiosensitivity of the tumor cells. Post IR in conditions where SNAI2 expression is reduced, both ERMS and ARMS cell lines exhibit significantly reduced cell growth in addition to increased levels of apoptosis. ERMS cell lines may also undergo differentiation following exposure to IR. Differences in other known pathways that could explain reduced cell proliferation, such as senescence and dysfunctional DNA repair, were either variable or not observed in the SNAI2-knockdown cells compared to control RMS cells. Indeed, the finding that SNAI2 directly represses BIM, a potent inducer of mitochondrial apoptosis, supports the existence of an exploitable SNAI2/BIM signaling axis in RMS or potentially other tumors with high SNAI2 expression, which could ultimately improve the efficacy of IR therapy in the clinic.
Supplementary Material
Significance.
SNAI2 is identified as a major regulator of radiation-induced apoptosis in rhabdomyosarcoma through previously unknown mechanisms independent of p53.
Acknowledgements:
This project has been funded with federal funds from NIH grants M. Ignatius and P. Houghton (R00CA175184, NCI P01 CA165995) and CPRIT Scholar grant to M. Ignatius (RR160062). M. Ignatius is a recipient of the Max and Minnie Tomerlin Voelcker Fund Young Investigator Award. K. Baxi is a T32 and TL1 fellow (T32CA148724, TL1TR002647). N. Hensch is a Greehey CCRI Graduate Student Fellow and a CPRIT Predoctoral Fellow (RP 170345).
Footnotes
Conflicts of Interest: The authors declare no potential conflicts of interest.
References
- 1.Arndt CA, Crist WM. Common musculoskeletal tumors of childhood and adolescence. N Engl J Med 1999;341:342–52 [DOI] [PubMed] [Google Scholar]
- 2.Hawkins DS, Gupta AA, Rudzinski ER. What is new in the biology and treatment of pediatric rhabdomyosarcoma? Curr Opin Pediatr 2014;26:50–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hawkins DS, Spunt SL, Skapek SX, Committee COGSTS. Children’s Oncology Group’s 2013 blueprint for research: Soft tissue sarcomas. Pediatr Blood Cancer 2013;60:1001–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weigel BJ, Lyden E, Anderson JR, Meyer WH, Parham DM, Rodeberg DA, et al. Intensive Multiagent Therapy, Including Dose-Compressed Cycles of Ifosfamide/Etoposide and Vincristine/Doxorubicin/Cyclophosphamide, Irinotecan, and Radiation, in Patients With High-Risk Rhabdomyosarcoma: A Report From the Children’s Oncology Group. J Clin Oncol 2016;34:117–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borinstein SC, Steppan D, Hayashi M, Loeb DM, Isakoff MS, Binitie O, et al. Consensus and controversies regarding the treatment of rhabdomyosarcoma. Pediatr Blood Cancer 2018;65 [DOI] [PubMed] [Google Scholar]
- 6.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell 2016;166:21–45 [DOI] [PubMed] [Google Scholar]
- 7.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 2017;14:611–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou W, Gross KM, Kuperwasser C. Molecular regulation of Snai2 in development and disease. J Cell Sci 2019;132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue A, Seidel MG, Wu W, Kamizono S, Ferrando AA, Bronson RT, et al. Slug, a highly conserved zinc finger transcriptional repressor, protects hematopoietic progenitor cells from radiation-induced apoptosis in vivo. Cancer Cell 2002;2:279–88 [DOI] [PubMed] [Google Scholar]
- 10.Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, et al. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell 2005;123:641–53 [DOI] [PubMed] [Google Scholar]
- 11.Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L, et al. BH3-only proteins Puma and Bim are rate-limiting for gamma-radiation- and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood 2005;106:4131–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross KM, Zhou W, Breindel JL, Ouyang J, Jin DX, Sokol ES, et al. Loss of Slug Compromises DNA Damage Repair and Accelerates Stem Cell Aging in Mammary Epithelium. Cell Rep 2019;28:394–407 e6 [DOI] [PubMed] [Google Scholar]
- 13.Zhang P, Wei Y, Wang L, Debeb BG, Yuan Y, Zhang J, et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat Cell Biol 2014;16:864–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas M, et al. Integrated Analysis of TP53 Gene and Pathway Alterations in The Cancer Genome Atlas. Cell Rep 2019;28:1370–84 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2010;2:a001008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Begg AC, Stewart FA, Vens C. Strategies to improve radiotherapy with targeted drugs. Nat Rev Cancer 2011;11:239–53 [DOI] [PubMed] [Google Scholar]
- 17.Kirsch DG, Diehn M, Kesarwala AH, Maity A, Morgan MA, Schwarz JK, et al. The Future of Radiobiology. J Natl Cancer Inst 2018;110:329–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grobner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, et al. The landscape of genomic alterations across childhood cancers. Nature 2018;555:321–7 [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Stewart E, Shelat AA, Qu C, Bahrami A, Hatley M, et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell 2013;24:710–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seki M, Nishimura R, Yoshida K, Shimamura T, Shiraishi Y, Sato Y, et al. Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nat Commun 2015;6:7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 2014;4:216–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinson AR, Jones R, Crose LE, Belyea BC, Barr FG, Linardic CM. Human rhabdomyosarcoma cell lines for rhabdomyosarcoma research: utility and pitfalls. Front Oncol 2013;3:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokolowski E, Turina CB, Kikuchi K, Langenau DM, Keller C. Proof-of-concept rare cancers in drug development: the case for rhabdomyosarcoma. Oncogene 2014;33:1877–89 [DOI] [PubMed] [Google Scholar]
- 24.Houghton PJ, Morton CL, Tucker C, Payne D, Favours E, Cole C, et al. The pediatric preclinical testing program: description of models and early testing results. Pediatr Blood Cancer 2007;49:928–40 [DOI] [PubMed] [Google Scholar]
- 25.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet 2005;37:1047–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmelzle T, Mailleux AA, Overholtzer M, Carroll JS, Solimini NL, Lightcap ES, et al. Functional role and oncogene-regulated expression of the BH3-only factor Bmf in mammary epithelial anoikis and morphogenesis. Proc Natl Acad Sci U S A 2007;104:3787–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ignatius MS, Hayes MN, Lobbardi R, Chen EY, McCarthy KM, Sreenivas P, et al. The NOTCH1/SNAIL1/MEF2C Pathway Regulates Growth and Self-Renewal in Embryonal Rhabdomyosarcoma. Cell Rep 2017;19:2304–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pomella S, Sreenivas P, Gryder BE, Wang L, Milewski D, Cassandri M, et al. Interaction between SNAI2 and MYOD enhances oncogenesis and suppresses differentiation in Fusion Negative Rhabdomyosarcoma. Nat Commun 2021;12:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woods GM, Bondra K, Chronowski C, Leasure J, Singh M, Hensley L, et al. Radiation therapy may increase metastatic potential in alveolar rhabdomyosarcoma. Pediatr Blood Cancer 2015;62:1550–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao L, Sun Y, Zhang Z, Feng W, Gao Y, Cai Z, et al. Deletion of proapoptotic Puma selectively protects hematopoietic stem and progenitor cells against high-dose radiation. Blood 2010;115:4707–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacQuarrie KL, Yao Z, Fong AP, Diede SJ, Rudzinski ER, Hawkins DS, et al. Comparison of genome-wide binding of MyoD in normal human myogenic cells and rhabdomyosarcomas identifies regional and local suppression of promyogenic transcription factors. Mol Cell Biol 2013;33:773–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otten AD, Firpo EJ, Gerber AN, Brody LL, Roberts JM, Tapscott SJ. Inactivation of MyoD-mediated expression of p21 in tumor cell lines. Cell Growth Differ 1997;8:1151–60 [PubMed] [Google Scholar]
- 33.Sharma A, Singh K, Almasan A. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol Biol 2012;920:613–26 [DOI] [PubMed] [Google Scholar]
- 34.Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev 2004;18:2614–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Felix CA, Kappel CC, Mitsudomi T, Nau MM, Tsokos M, Crouch GD, et al. Frequency and Diversity of p53 Mutations in Childhood Rhabdomyosarcoma. Cancer Research 1992;52:2243. [PubMed] [Google Scholar]
- 36.Erlacher M, Labi V, Manzl C, Bock G, Tzankov A, Hacker G, et al. Puma cooperates with Bim, the rate-limiting BH3-only protein in cell death during lymphocyte development, in apoptosis induction. J Exp Med 2006;203:2939–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly PN, White MJ, Goschnick MW, Fairfax KA, Tarlinton DM, Kinkel SA, et al. Individual and overlapping roles of BH3-only proteins Bim and Bad in apoptosis of lymphocytes and platelets and in suppression of thymic lymphoma development. Cell Death Differ 2010;17:1655–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Labi V, Bertele D, Woess C, Tischner D, Bock FJ, Schwemmers S, et al. Haematopoietic stem cell survival and transplantation efficacy is limited by the BH3-only proteins Bim and Bmf. EMBO Mol Med 2013;5:122–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo Y, Schoell MC, Freeman RS. The von Hippel-Lindau protein sensitizes renal carcinoma cells to apoptotic stimuli through stabilization of BIM(EL). Oncogene 2009;28:1864–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang M, Han J, Marcar L, Black J, Liu Q, Li X, et al. Radiation Resistance in KRAS-Mutated Lung Cancer Is Enabled by Stem-like Properties Mediated by an Osteopontin-EGFR Pathway. Cancer Res 2017;77:2018–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan J, Bittner ML, Saal LH, Teichmann U, Azorsa DO, Gooden GC, et al. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci U S A 1999;96:13264–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gryder BE, Yohe ME, Chou HC, Zhang X, Marques J, Wachtel M, et al. PAX3-FOXO1 Establishes Myogenic Super Enhancers and Confers BET Bromodomain Vulnerability. Cancer Discov 2017;7:884–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen EL, Yoo CH, Gutkin PM, Merriott DJ, Avedian RS, Steffner RJ, et al. Outcomes for pediatric patients with osteosarcoma treated with palliative radiotherapy. Pediatr Blood Cancer 2020;67:e27967 [DOI] [PubMed] [Google Scholar]
- 44.Zuch D, Giang AH, Shapovalov Y, Schwarz E, Rosier R, O’Keefe R, et al. Targeting radioresistant osteosarcoma cells with parthenolide. J Cell Biochem 2012;113:1282–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown LC, Lester RA, Grams MP, Haddock MG, Olivier KR, Arndt CA, et al. Stereotactic body radiotherapy for metastatic and recurrent ewing sarcoma and osteosarcoma. Sarcoma 2014;2014:418270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeLaney TF, Park L, Goldberg SI, Hug EB, Liebsch NJ, Munzenrider JE, et al. Radiotherapy for local control of osteosarcoma. Int J Radiat Oncol Biol Phys 2005;61:492–8 [DOI] [PubMed] [Google Scholar]
- 47.Matsunobu A, Imai R, Kamada T, Imaizumi T, Tsuji H, Tsujii H, et al. Impact of carbon ion radiotherapy for unresectable osteosarcoma of the trunk. Cancer 2012;118:4555–63 [DOI] [PubMed] [Google Scholar]
- 48.Nieto MA, Sargent MG, Wilkinson DG, Cooke J. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science 1994;264:835–9 [DOI] [PubMed] [Google Scholar]
- 49.Ros MA, Sefton M, Nieto MA. Slug, a zinc finger gene previously implicated in the early patterning of the mesoderm and the neural crest, is also involved in chick limb development. Development 1997;124:1821–9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.