Abstract
TFIID recognizes multiple sequence elements in the hsp70 promoter of Drosophila. Here, we investigate the function of sequences downstream from the TATA element. A mutation in the initiator was identified that caused an eightfold reduction in binding of TFIID and a fourfold reduction in transcription in vitro. Another mutation in the +24 to +29 region was somewhat less inhibitory, but a mutation in the +14 to +19 region had essentially no effect. The normal promoter and the mutants in the initiator and the +24 to +29 region were transformed into flies by P element-mediated transformation. The initiator mutation reduced expression an average of twofold in adult flies, whereas the mutation in the +24 to +29 region had essentially no effect. In contrast, a promoter combining the two mutations was expressed an average of sixfold less than the wild type. The results suggest that the initiator and the +24 to +29 region could serve overlapping functions in vivo. Protein-DNA cross-linking was used to identify which subunits of TFIID contact the +24 to +29 region and the initiator. No specific subunits were found to cross-link to the +24 to +29 region. In contrast, the initiator cross-linked exclusively to dTAF230. Remarkably, dTAF230 cross-links approximately 10 times more efficiently to the nontranscribed strand than to the transcribed strand at the initiator.
DNA sequences contributing to transcription of protein-encoding genes can be separated into two categories. Gene-specific regulatory sequences compose one category, and they are typically situated at various distances upstream from the transcription start site. These sequences are recognition elements for a variety of transcriptional activators and repressors. These regulatory elements play major roles in governing gene-specific patterns of expression. The other category is located in the region contacted by the basal transcription machinery. These sequences are often referred to as the core promoter elements (38). The best-characterized core promoter elements consist of a TATA box located approximately 30 nucleotides upstream from the start site (3), an initiator element located at the transcription start site (37), and a downstream promoter element (DPE) located approximately 30 nucleotides downstream from the transcription start (17). All three regions are recognized by TFIID (4, 10, 16, 29, 38). The TATA-binding protein (TBP) subunit of TFIID recognizes the TATA box, whereas TBP-associated factors (TAFs) appear to be responsible for the recognition of sequences downstream from the TATA box (5, 41).
Numerous studies provide evidence that the core promoter elements could be important in establishing specific patterns of gene expression. It was observed that a Gal4-fusion protein containing the glutamine-rich activation domains of Sp1 stimulated transcription from an Inr-only core promoter, but not from a TATA-only core promoter (9). The TdT gene has a promoter that lacks a consensus TATA box. Insertion of a consensus TATA box upstream from the Inr increased the strength of transcription when the DNA was transiently expressed in a T-cell line. Removing the initiator, but leaving the TATA box, results in no expression. This result suggests that the initiator is required for at least one of the activators of this promoter to function (12). The alcohol dehydrogenase gene in Drosophila has two promoters. The distal promoter is used primarily during early embryogenesis and in adults, whereas the proximal promoter is used late in embryogenesis and during larval development. In vitro transcription analyses suggest that the preferential use of the distal promoter in early embryos depends in part on TAF150 and the sequence at the Inr (15). More recently, however, a repressor protein has been identified that binds the initiator region of the proximal promoter, and this repressor is thought to cause transcription to shift from the proximal to the distal promoter in adults (31). Promoter competition experiments in which the ability of one enhancer to act on promoters placed on each side of the enhancer was determined also demonstrate that various core promoters can respond differently to an enhancer (27). Chalkley and Verrijzer recently found that certain sequences placed in the initiator region inhibit basal expression, but not activation by Sp1 (6). If this situation occurs naturally, it could significantly contribute to the degree of induction caused by an activator. Finally, sequence and biochemical analyses indicate that sequences in the region 30 nucleotides downstream from the transcription start may be important for expression of a wide spectrum of genes (17).
A missing-nucleoside analysis provided evidence that TFIID makes specific contact with at least four regions of the hsp70 promoter of Drosophila: the TATA box, the initiator, position +18, and position +28 (29). Here, we set out to determine what contribution the regions downstream from the TATA box make towards expression from the hsp70 promoter in Drosophila. The hsp70 promoter is rapidly induced in response to heat shock (20). We have mutated specific sites contacted by TFIID and used P element-mediated transformation to analyze the effects of the mutations on expression in a normal chromosomal context. UV cross-linking was used to identify which subunits of TFIID are likely to recognize the downstream elements, and DNase I footprinting was used to further explore the nature of the TFIID interaction downstream from the transcription start.
MATERIALS AND METHODS
Plasmids and construction of the mutants.
The hsp70 promoter spanning −194 to +84 was used for this study. The mutant clones were constructed with the Clontech site-directed mutagenesis kit according to the manufacturer's instructions. The different mutations (underlined) were produced with the primers described below:
For the Inr mutation, the primer was 5′ CGGAGCGCACCCTCAATTCAA 3′. For the +14/+19 mutation, the primer was 5′ CAATTCAAACTTACTTAGTGAACACGTCGC 3′. For the+24/+29 mutation, the primer was 5′ AGTGACCTAGGCGCTAAGCGAAAG 3′.
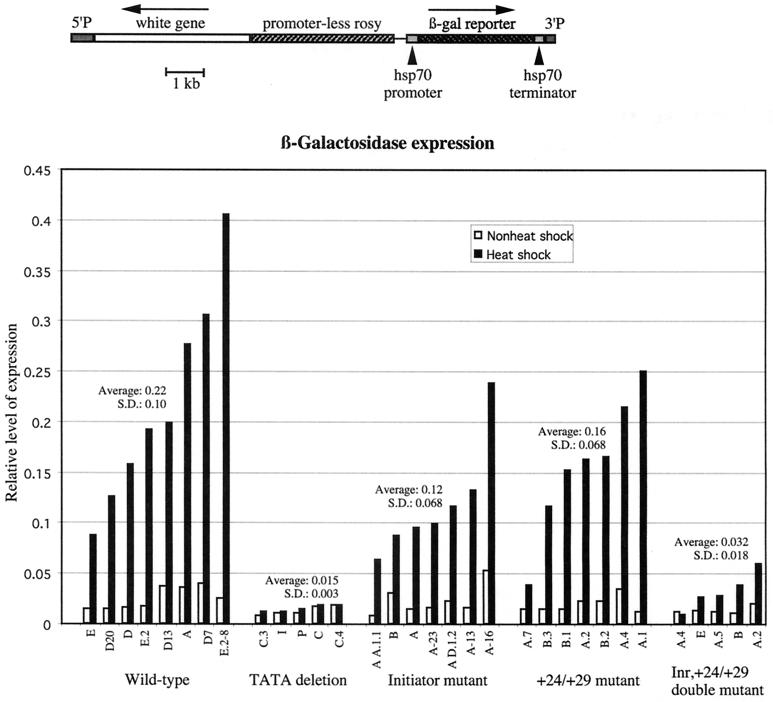
For P element transposition, the promoters were subcloned into the transformation vector CaSpeR 4 (40). This vector contains the white gene as a selectable marker for transposition. The hsp70 promoters were situated upstream from sequences encoding Escherichia coli β galactosidase. A schematic of the region that is transposed into the genome is provided in Fig. 2.
FIG. 2.
β-Galactosidase expression from transgenes in adult flies. The top drawing illustrates the DNA that was inserted into the fly genome. Transformants were identified by expression of the white gene. The large piece of the rosy gene is leftover from subcloning manipulations; it lacks a promoter and so is not expected to be transcribed. Each promoter is inserted upstream from sequences encoding E. coli β galactosidase. The graph summarizes the relative amounts of β-galactosidase expressed in the various fly lines. Adult flies from the indicated lines were heat shocked for 2 h at 37°C, and whole-fly lysates were assayed for β-galactosidase activity. In addition, lysates from non-heat-shocked flies were also assayed for β-galactosidase activity. β-Galactosidase assays were performed as previously described by Simon and Lis (35). The results were normalized for small differences in total protein concentrations for the various lysates. S.D., standard deviation.
In vitro transcription.
Nuclear extracts were prepared from Drosophila embryos as described by Biggin and Tjian (2). In vitro transcription reactions were carried out with 40-μl reaction volumes containing 50 mM KCl, 50 mM HEPES (pH 7.6), 6.25 mM MgCl2, 5% glycerol, 0.5 mM dithiothreitol (DTT), 10 μl of nuclear extract, 10 ng of supercoiled hsp70 DNA template, 1 μg of HaeIII-cut Escherichia coli DNA, and 0.3 mM nucleoside triphosphates (NTPs). The reaction mixture was incubated at 21°C for 25 min, and transcription was stopped with 80 μl of stop buffer (20 mM EDTA [pH 8.0], 0.2 M NaCl, 1% sodium dodecyl sulfate [SDS], 0.25 mg of yeast tRNA per ml, 0.1 mg of proteinase K per ml). After proteinase K digestion at 42°C for 30 min, the reaction mixture was extracted once with 100 μl of a phenol-chloroform-isoamyl alcohol mixture (25:24:1 ratio), and the RNA was precipitated with ethanol.
The RNA was dissolved in 10 μl of annealing mixture consisting of 2 mM Tris-Cl (pH 7.8), 0.2 mM EDTA, 250 mM KCl, and 0.03 pmol of radioactive primer. The mixture was heated to 75°C and allowed to cool to 37°C. Forty microliters of reverse transcriptase mixture containing 62.5 mM Tris-Cl (pH 8.3), 30 mM MgCl2, 12.5 mM DTT, 62.5 μM dNTPs, and 15 U of Moloney murine leukemia virus reverse transcriptase (RNase H−) (Promega) was added, and the mixture was incubated at 37°C for 1 h. The nucleic acid was ethanol precipitated and analyzed on an 8% denaturing polyacrylamide gel. The gel was then quantified on a Molecular Dynamics PhosphorImager.
DNA binding analysis with immunoprecipitated TFIID.
Nuclear extracts were prepared from Drosophila embryos and fractionated over a DEAE column as previously described (30). For the experiments shown in Fig. 1B, 4, and 5, the DEAE fraction was fractionated further on phosphocellulose. TFIIDs immunoprecipitated from the DEAE fraction or the phosphocellulose fractions have the same polypeptide composition, as detected by SDS-polyacrylamide gel electrophoresis (PAGE), and produce the same DNase I footprints and patterns of cross-linking to dTAF150 and dTAF230.
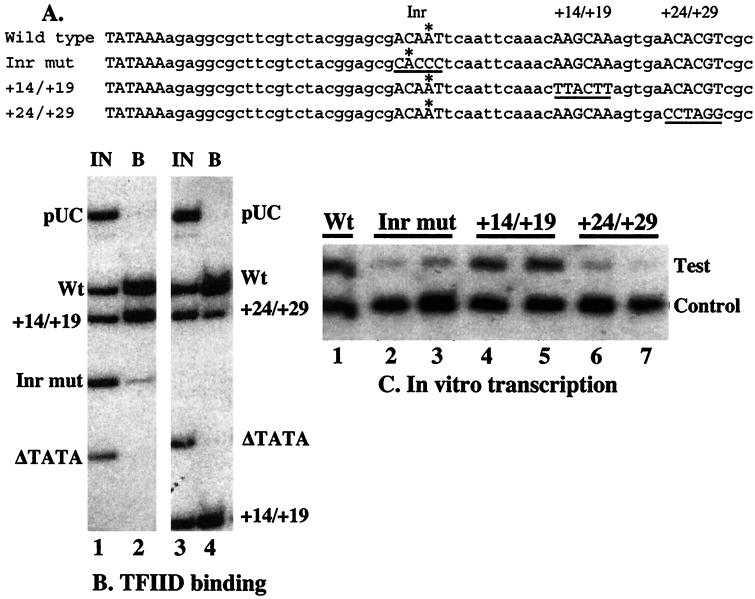
FIG. 1.
In vitro analyses of the initiator mutation. (A) The sequences represent the core promoter regions of the normal and mutant promoters examined in this work. An A marked with an asterisk represents the major transcription start site detected in vitro. The underlined sequences designate the locations where mutations have been made. The entire promoter region in the plasmids used for transcription studies in vitro and in vivo extended from −194 to +84. (B) A mixture of end-labeled DNA fragments was incubated with TFIID immobilized on beads with a monoclonal antibody against TBP. The promoter fragments were end labeled at an NruI site located at −50 relative to the transcription start site of the wild-type (Wt) promoter. To generate labeled fragments of various sizes, fragments were subsequently cut 65 to 100 nucleotides downstream from the transcription start with PvuII, PstI, or SalI. Negative controls were provided by an hsp70 fragment that lacked the TATA element and a HindIII-NarI fragment from pUC18. Lanes 1 and 3 show the mixture of fragments in the input sample (IN) for two separate binding reactions. Lanes 2 and 4 show the DNA that bound (B) to TFIID. The amount of radioactive material loaded in each lane was equal. The average ratios and standard deviations (SDs) of results from the TFIID binding analysis as measured with a PhosphorImager were as follows: Inr/wild-type, 0.13 (SD = 0.03, three experiments); +14 to +19/wild type, 0.8 (SD = 0.03, two experiments); +24/+29/wild type, 0.34 (SD = 0.03, three experiments); TATA deletion/wild type, 0.055 (SD = 0.02, four experiments). (C) Mixtures of two plasmids were transcribed in nuclear extracts from Drosophila embryos. The resulting transcripts were detected by primer extension with reverse transcriptase. Test plasmids contained the normal promoter (Wt), initiator mutant (Inr mut), the +14/+19 mutant (+14/+19 mut), or the +24/+29 mutant (+24/+29 mut). Each reaction mixture contained an internal control that produced a shorter transcript. Duplicate transcription reactions were performed with each of the mutants. The average ratios and SDs of results from the in vitro transcription reactions after correction for differences in the control signal were as follows: Inr/wild type, 0.23 (SD = 0.06, four experiments); +14/+19/wild type, 0.74 (SD = 0.06, two experiments); +24/+29/wild type, 0.25 (SD = 0.035, two experiments).
For the experiment shown in Fig. 1B, the 0.5 M KCl fraction from the phosphocellulose column was used for immunoprecipitation. Forty microliters of the 0.5 M phosphocellulose fraction and 1.0 μl of anti-TBP monoclonal antibody (14C-F4) in mouse ascites fluid were incubated together on ice for 2 h. To this mixture, 25 μl of protein G-Sepharose beads, which had been washed in 1 ml of 200 mM KCl-HGEDPN (HGEDPN is 25 mM HEPES [pH 7.6], 0.1 mM EDTA, 10% glycerol, 0.5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.1% NP-40), was added. The protein G-antibody mixture was rocked at 4°C for 2 h. The beads were then washed four times with 1-ml portions of 200 mM KCl-HGEDPN and transferred to a new tube. The slurry was washed once more with 1 ml of CGSBB (CGSBB is composed of 10 mM HEPES [pH 7.6], 5 mM MgCl2, 0.8 mM DTT, 0.1 mM PMSF, 10% glycerol, and 90 mM KCl). A 40-μl mixture consisting of radioactively labeled DNA fragments from the different promoters and 1 μg of HaeIII-cut E. coli DNA was added to the immunoprecipitated TFIID, and this mixture was then incubated at room temperature for 2 h on a rocker. Some of the radioactive DNA mixture was retained as the input sample. The beads were collected by brief centrifugation. The supernatant was removed. The beads were then washed three times with 1 ml of cold CGSBB. Two hundred microliters of TFIID stop mixture containing 50 mM Tris-Cl (pH 7.9), 10 mM EDTA, 0.5% SDS, and 10 ng of sonicated salmon sperm DNA per μl was added to the beads. The beads were treated with proteinase K for 1 h and extracted once with phenol-chloroform-isoamyl alcohol at a 25:24:1 ratio. The DNA was recovered by ethanol precipitation and analyzed along with DNA from the input sample on an 8% denaturing polyacrylamide gel.
The DNase I footprinting analyses shown in Fig. 4 and 5 were performed with TFIID that had been immunoprecipitated from the phosphocellulose fraction with monoclonal antibodies against dTAF230 or dTBP. The monoclonal antibodies were in hybridoma supernatants and were designated 14C-F4 for dTBP and 30H9 for dTAF230. The DNase I analysis was performed as previously described (10, 29).
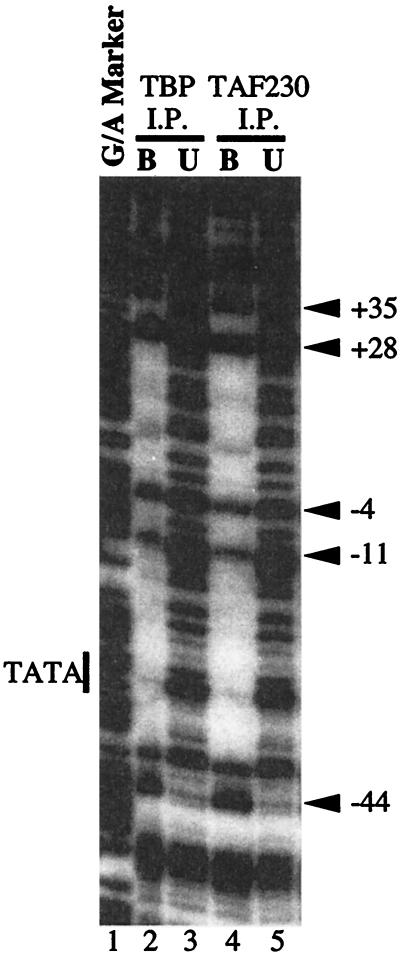
FIG. 4.
DNase I footprinting analysis of DNA bound to immobilized TFIID. End-labeled DNA was incubated with TFIID that had been immunoprecipitated with monoclonal antibody against dTBP (lane 2) or dTAF230 (lane 4). The complexes were subjected to DNase I cutting. The patterns of cutting for DNA bound to TFIID (B, lanes 2 and 4) were compared to the patterns of cutting for DNA that remained unbound (U, lanes 3 and 5). Lane 1 provides molecular mass markers generated by cutting DNA at purines. The DNA fragment spanned −194 to +84 of the hsp70 promoter and was radiolabeled near −194 on the nontranscribed strand.
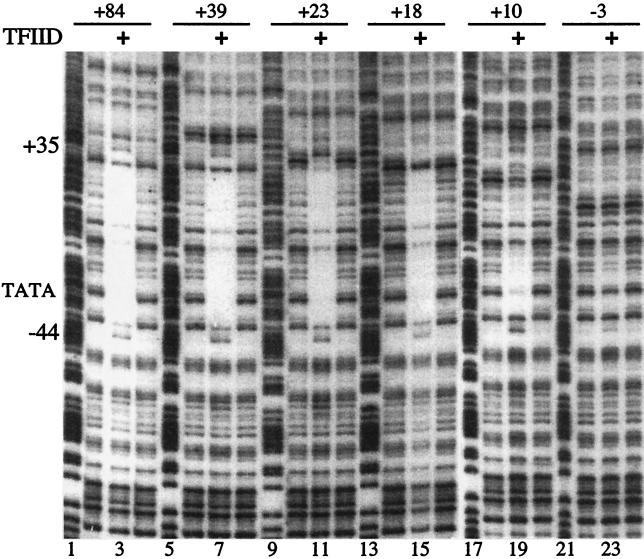
FIG. 5.
DNase I footprinting analysis of 3′ deletions bound to immobilized TFIID. DNA fragments corresponding to the wild-type promoter and a series of 3′ deletions were end labeled on the nontranscribed strand near −194. The other end of each fragment was established by a restriction enzyme cut in the vector located well beyond the region contacted by TFIID. Samples in lanes 3, 7, 11, 15, 19, and 23 are DNA isolated from TFIID after DNase I treatment. The lanes flanking each side of the TFIID-bound samples were derived by DNase I treatment of unbound DNA. Lanes 1, 5, 9, 13, 17, and 21 correspond to purine-cutting patterns for each of the respective promoter fragments.
UV cross-linking analysis.
Radiolabeled DNA was prepared by pulse-chase primer elongation on a single-stranded M13 phage DNA template (14). The following primers were used to generate DNA fragments selectively labeled in small regions (the NruI and PvuII sites are underlined): −59 to −2 of the nontranscribed strand, 5′GAATGTTCGCGAAAAGAGCGCCGGAGTATAAATAGAGGCGCTTCGTCTACGGAGCGACA3′; +74 to +4 of the transcribed strand, 5′CTTGTTCAGCTGCGCTTGTTTATTTGCTTAGCTTTCGCTTAGCGACGTGTTCACTTTGCTTGTTTGAATTG3′; +72 to +30 of the transcribed strand, 5′TGTTCAGCTGCGCTTGTTTATTTGCTTAGCTTTCGCTTAGCGA3′; and +71 to +43 of the transcribed strand, 5′GTTCAGCTGCGCTTGTTTATTTGCTTAGC3′. Three picomoles of oligonucleotide primer was annealed to 1 pmol of single-stranded hsp70-M13 DNA for 2 h at 37°C in 7.5 μl of a mixture containing 10 mM Tris-HCl (pH 8), 1 mM EDTA, and 40 mM NaCl. Limited extensions were carried out by adding 1.5 μl of 0.1 M Tris-HCl (pH 8), 0.1 M MgCl2, 1.5 μl of 100 mM DTT, 1 μl of 0.1 mM bromodeoxyuridine (BrdU), and 1 μl of 1 mM dGTP or dCTP or by adding 10 mM Tris-HCl (pH 8), 1 mM EDTA; 2 μl of [α-32P]dCTP, [α-32P]dGTP, or [α-32P]dATP (3,000 Ci/mmol, 25 mCi/ml); and 0.5 μl of Klenow fragment (2.5 U). Incubation was carried out for 10 min at 37°C. After the pulse, all four nucleotide triphosphates were added to allow unlimited extension for another 15 min. The reaction was stopped by heating at 65oC for 15 min. The resulting double-stranded hsp70 promoters were excised as PvuII-NruI restriction fragments and isolated on an 8% polyacrylamide gel. Purified DNA fragments were bound to immunoprecipitated TFIID.
Immunoprecipitation of TFIID was carried out by mixing 40 μl of the 0.25 M DEAE fraction with 20 μl of hybridoma supernatant containing monoclonal antibody against dTAF230. After incubation for 2 h on ice, 20 μl of 50% protein G-Sepharose was added to the mixture, which was then rotated in the cold room for 1 h. The beads were washed three times with 1 ml of 100 mM KCl-HGEMNDP (25 mM HEPES [pH 7.6], 10% glycerol, 0.1 mM EDTA, 12.5 mM MgCl2, 0.1% NP-40, 1 mM DTT, 0.1 mM PMSF) and then washed once with 1 ml of CGSBB. The material was briefly centrifuged and brought to 20 μl.
Binding DNA to immunoprecipitated TFIID was performed by adding 500,000 cpm of labeled DNA with 1 μg of HaeIII-cut E. coli DNA in a total volume of 30 μl of CGSBB to 20 μl of immunoprecipitated TFIID. The mixture was rotated at room temperature for 2 h. The beads were washed three times with 1 ml of cold CGSBB and briefly centrifuged, and all but 25 μl of the mixture was removed. The remaining mixture was UV irradiated from a distance of 4 cm for 2 min with a long-wavelength UV transilluminator with the filter removed (La Jolla Scientific Co., Inc., La Jolla, Calif.). One microliter of 10 mM CaCl2 and 1 μl of DNase I (10 U/μl) were added to each sample, and the samples were incubated at 37°C for 1 h. Samples were analyzed by 4 to 15% polyacrylamide gradient SDS-PAGE. The radiolabeled polypeptides were detected with the PhosphorImager.
RESULTS
Mutation of the initiator or the +24 to +29 region debilitates promoter functions in vitro.
Previously, we had identified a high-affinity consensus element for TFIID in the region of the transcription start site by using the selected and amplified binding (SAAB) procedure (29). Sequencing of another 10 clones from this earlier study refined our consensus to the sequence (−3)G/A/T, T/C, A, G/T, T, G/A/T/C(+3). Visual inspection of the selected sequences suggested that certain nucleotides at particular positions might inhibit TFIID binding. For example, a C at position −3 and an A at position −2 only appeared once in the 20 clones that were sequenced. After careful consideration of the sequences, we mutated the initiator region to CACCC as a possible candidate for inhibiting TFIID binding (Fig. 1A). Such a mutation does not fit our consensus in any of the positions from −3 to +3. To test if this mutation inhibited TFIID binding, a relative measure of TFIID affinities for various DNA fragments was determined by incubating a mixture of DNA fragments with TFIID that had been immunoprecipitated with TBP antibody. The relative amounts of various fragments bound to the immobilized TFIID were compared to the amount in the initial mixture of fragments (Fig. 1B, lanes 1 and 2). The fragment corresponding to the initiator mutant was depleted eightfold relative to the fragment corresponding to the normal promoter. Fragments corresponding to a part of the pUC cloning vector or a TATA deletion were depleted even more.
The design of the +24/+29 and +14/+19 mutations was aided by use of a Drosophila promoter database (1). In the +24 to +29 region of hsp70, there are several nucleotide quadruplets that are overrepresented in the database; these are ACAC, ACGT, and TCGC. All of these were disrupted by replacing the sequence ACACGT with CCTAGG, which also generated an AvrII restriction site. None of the quadruplets or triplets generated by this mutation displayed any degrees of overrepresentation in the promoter database. In the case of the +14 to +19 region, the hsp70 promoter does not exhibit any overrepresented arrays of nucleotides. Hence, this region was mutated to change the original hsp70 sequence and to avoid generating an overrepresented array of triplets or quadruplets of nucleotides. The +24/+29 mutation inhibited binding by approximately threefold (Fig. 1B, lanes 3 and 4), whereas the +14/+19 mutation had only a slight effect (Fig. 1B).
Note that the +14/+19 promoter fragment was used in both binding experiments, but its size varied (Fig. 1B). This variation in size did not affect binding, indicating that the differences in binding observed for other fragments are not simply due to differences in fragment size. This is in accord with our previous observations (10). The relative amounts of each fragment binding TFIID are a measure of the affinity of each DNA relative to each other for TFIID. The associations of the DNA fragments are in competition with excess E. coli DNA—<10% of the input radioactive DNA associates with the immobilized TFIID. Changing the amount of E. coli DNA (10) or the amount of TFIID (22) does not change the relative amount of each fragment that binds to the TFIID.
We next determined if the mutations affected transcription in vitro. The various hsp70 promoters were transcribed in nuclear extracts from Drosophila embryos, and the level of transcription in vitro from the different hsp70 mutants was measured by primer extension analysis. The primer anneals to the region located 110 nucleotides downstream from the transcription start of the test promoters (Fig. 1C, test). A truncated version of the normal promoter was used as an internal control (Fig. 1C, control). The transcription levels of the Inr and +24/+29 mutants were found to be decreased by fourfold. In contrast, the +14/+19 mutation decreased transcription by 25%.
Mutation in the initiator alone inhibits expression in vivo, but a double mutation is required to observe a contribution by the +24/+29 region.
Having identified mutations in the initiator and the +24/+29 region that debilitated the promoter in vitro, we wanted to investigate what effect these mutations would have on expression in vivo. P element-mediated transformation was used to generate transgenic flies containing the normal or mutant promoters (33). This experimental approach would allow us to analyze the effects of the mutations in a chromosomal context. The promoters were inserted upstream from a β-galactosidase reporter gene (Fig. 2). We also generated transgenic flies that contained an hsp70 promoter lacking the TATA element to serve as a negative control. This deletion removes 20 nucleotides encompassing the TATA element. Previous work had indicated that this deletion rendered the promoter inactive both in vitro and in vivo (10, 13, 42). Eight wild-type, seven initiator mutant, and seven +24/+29 mutant lines were obtained. Southern blotting analysis indicated that the lines contained single-copy inserts located at different positions in the genome. Following heat shock treatment, all of the wild-type, initiator mutant, and +24/+29 mutant lines expressed β galactosidase at levels greater than those of the TATA deletions, indicating that most of the β-galactosidase detected was due to transcription from the hsp70 promoter. On average, the Inr mutant promoter was expressed at approximately one-half the level of the normal promoter, and the majority of the Inr mutant lines expressed β-galactosidase at levels lower than the majority of wild-type lines (Fig. 2). The +24/+29 mutation had little effect, because the majority of the +24/+29 mutant lines expressed β-galactosidase at levels comparable to those of the majority of wild-type lines.
Given the modest effect of the mutations, we combined the initiator mutation and the +24/+29 mutation and tested this double mutant in the transgenic assay. The double-mutant lines clearly reduced expression by more than the majority of single-mutant lines.
Cross-linking reveals that TAF230 is in close proximity to the initiator.
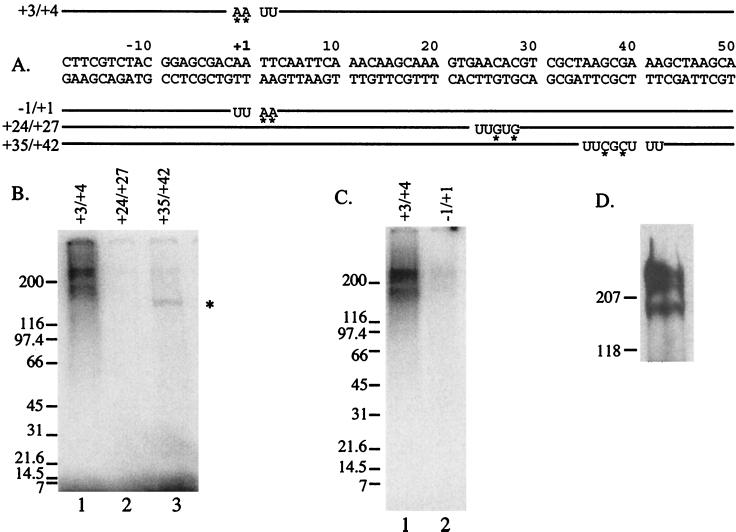
We wanted to identify which subunits of TFIID might be responsible for recognizing the initiator and the region from +24 to +29. DNA fragments were synthesized that contained radionucleotides and BrdU in selected regions (Fig. 3A). Each fragment was incubated with TFIID immobilized by a monoclonal antibody against dTAF230. The immobilized complexes were irradiated with long-wavelength UV light to induce cross-links, and then the DNA was degraded with DNase I. The resulting polypeptides were separated by SDS-PAGE, and radioactively tagged polypeptides were identified. When radioactivity was incorporated into the nontranscribed strand in the initiator, there was strong labeling of several large polypeptides (Fig. 3B, lane 1). Western blot analysis showed that the large polypeptides comigrated with dTAF230 (Fig. 3D). Presumably the largest fragment is intact dTAF230, whereas the smaller fragments are proteolytic breakdown products. Interestingly, dTAF230 was only weakly labeled when the radioactivity was incorporated into the transcribed strand of the initiator (Fig. 3C, compare lanes 1 and 2). When the radioactivity was incorporated elsewhere in the fragment, a different pattern of polypeptides was observed. Incorporation into the +24 to +27 region failed to consistently label any polypeptides (Fig. 3B, lane 2). We also analyzed contacts in the region from +35 to +42, since previous analyses of a crude preparation of TFIID had shown that a 150-kDa polypeptide cross-linked to this region (39). In accord with this earlier work, dTAF150 was strongly labeled, whereas dTAF230 was weakly labeled (Fig. 3B, lane 3). The identity of dTAF150 was determined by doing a Western blot analysis with antibody against dTAF150 (data not shown). The labeling of each of the polypeptides in Fig. 3 is due to specific binding, since deletion of the TATA element from the fragments results in no labeling of any of these polypeptides (data not shown). Specificity is also indicated by the variation in the pattern of labeled polypeptides that resulted when the radiolabel was placed in different locations in the DNA.
FIG. 3.
UV cross-linking analysis of TFIID. TFIID was immunoprecipitated from a DEAE fraction with a monoclonal antibody against dTAF230. DNA fragments containing radiolabel and BrdU incorporated in small patches were bound to the immunoprecipitated TFIID, and unbound DNA was washed away. Each fragment spans an NruI site cut at −50 to a PvuII site cut at +65. The immunoprecipitated complexes were UV irradiated, DNA was degraded with DNase I, and the polypeptides were fractionated on 4 to 15% polyacrylamide gradient SDS-polyacrylamide gels. The radiolabeled polypeptides were detected with a PhosphorImager. (A) Labeled fragments. BrdU is designated by U, and the asterisks mark radionucleotides. The numbers to the left designate nucleotides encompassed by the photoactive deoxyuridines. (B) Results for cross-linking to the nontranscribed strand in the region from +3 to +4 (lane 1), the transcribed strand in the region from +24 to +27 (lane 2), or the transcribed strand in the region from +35 to +42 (lane 3). The strong bands seen above and below the gap delineated by the 200-kDa molecular mass marker are intact and proteolyzed dTAF230 (D). The asterisk next to lane 3 delineates dTAF150. (C) Cross-linking to the initiator on the nontranscribed strand at +2 and +3 (lane 1) and transcribed strand at −1 and +1 (lane 2). (D) Western blot analysis performed by binding the dTAF230 monoclonal antibody to TFIID from the DEAE fraction. The bound antibody was detected by chemiluminescence. The slight differences in molecular masses seen in panel D (207 versus 200 kDa and 118 versus 116 kDa) reflect differences in the molecular mass markers used in the analyses.
For the cross-linking analysis, TFIID was immobilized with a monoclonal antibody against dTAF230. To be certain that immobilization of TFIID with this antibody did not alter the interaction between TFIID and DNA, complexes were subjected to DNase I footprinting. TFIID immobilized with the dTAF230 antibody produced a DNase I footprint that spanned from −44 to +35 (Fig. 4, lane 4). This footprint was indistinguishable from the footprint produced by a less pure, soluble form of TFIID (10, 14) or by TFIID that had been immobilized with antibody against TBP (Fig. 4, lane 2). The good agreement between footprinting and cross-linking data for crude and immobilized TFIID indicates that the immobilization strategy does not disturb the interaction between TFIID and the hsp70 promoter DNA.
DNase I footprinting analysis of complexes formed between TFIID and various 3′ deletions of the hsp70 promoter.
As shown in Fig. 4, TFIID makes extensive contact with DNA downstream from the transcription start site. In some situations, this downstream contact seems to be dependent on the presence of upstream activators, and the extended contact correlates with activated transcription (7, 8). We wondered if the extended contact of TFIID with the hsp70 promoter required specific sequences located downstream from the transcription start, since the results could bear on the failure of our +24/+29 mutation to affect expression in vivo.
We examined the interaction between TFIID and a series of 3′ deletions, since these deletions had previously been shown to inhibit TFIID binding, and the effect of some of these deletions exceeded the impact that our +24/+29 mutation had on binding (10). These deletions placed sequences from the cloning vector downstream from the 3′ deletion endpoint. The end-labeled promoter fragments were allowed to bind immobilized TFIID. Unbound DNA was washed away, and the remaining material was subjected to DNase I. Equal amounts of radioactive material from each bound fraction were analyzed on the gel. Note that the deletions extending to +23, +18, +10, or −3 reduce the affinity of the fragments for TFIID (10), but the unbound material has been washed away. Hence, the DNase I footprinting is being performed on DNA adhering to the beads via specific interaction with TFIID or via other nonspecific interactions, as appears to be the case for the −3 deletion.
Figure 5 shows the results of the analysis. A large footprint spanning −44 to +35 is clearly evident on fragments containing hsp70 sequences from −194 to +84 or +39. TFIID also clearly footprints over the vector sequences associated with the +23 and +18 deletions. The +10 deletion also shows evidence of downstream contact, although the degree of protection is less complete. There is very little evidence of protection on the −3 deletion, suggesting that much of this DNA is adhering nonspecifically to the immobilized TFIID or to the beads. We conclude that TFIID is able to make extensive contact with the downstream region of the hsp70 promoter, even after the sequence in the downstream region has been significantly altered. This interaction is not dependent on the activator and could help explain why a small mutation such as the +24/+29 mutation had no impact on expression in flies.
It is interesting that the DNA is hypersensitive to cutting around +45 for all of the promoter fragments except the −3 mutant. This hypersensitivity suggests that TFIID makes contact with the DNA beyond +35. This contact might be due to TAF150, which was found to cross-link to the region encompassing +35 to +42 (Fig. 3B, lane 3).
DISCUSSION
We have investigated the functions of two regions situated downstream from the TATA element in the hsp70 promoter that are recognized by TFIID. Although the functions of sequences corresponding to this location in several genes have been investigated, the majority of these studies have been directed at TATA-less promoters. Most studies of both TATA-containing and TATA-less promoters have relied on either in vitro transcription or transient expression assays to assess the impact of mutation of sequence elements. Neither the in vitro transcription assay nor the transient expression assay ensures that the promoter function is being evaluated in a normal chromosomal context. In addition, many of the studies have not provided data directly assessing the relationship between these sequences and TFIID.
For the hsp70 promoter, it had already been shown that deletions in the region downstream from the TATA box reduced expression (18). We were interested in assessing the function of individual sequences that were recognized by TFIID. Mutations were directed at three regions based on chemical interference studies (29, 30). Two mutations were identified that reduced binding: one in the initiator and another located +24 to +29 nucleotides downstream from the transcription start site. A third mutation was made in the region from +14 to +19, but this had essentially no impact on TFIID binding. Unfortunately, there is little sequence information to guide design of the mutation in the +14 to +19 region, so we chose to focus on the other two mutations.
The use of P element-mediated transformation allowed us to assess the contribution of the initiator and +24 to +29 region when the promoter resides in the chromosome. One shortcoming of this approach is that the transposed DNA inserts randomly into the chromosome, and sequences flanking the insertion can affect the level of expression. We attempted to reduce the magnitude of this problem by placing the hsp70 promoter at least 3 kb from the DNA flanking the insertion. At present, techniques for targeted insertion of DNA into the Drosophila genome are not well established. Nevertheless, examination of numerous lines carrying the insertions in different locations provides a way to assess the function of specific elements in vivo (21).
Our results show that the individual mutations have only a modest impact on expression. The initiator mutation, which reduced binding of TFIID by eightfold, only reduced expression in vivo by an average of twofold. The +24 to +29 mutation had virtually no effect, despite the observation that this mutation did reduce TFIID binding and inhibited transcription in vitro. The double mutant reduces expression by an average of sixfold. We suggest that both of these elements contribute to expression and that one compensates for the absence of the other in vivo. It is clear from the DNase I footprinting analysis that TFIID can still contact the downstream region even when the sequences in the downstream region are drastically changed.
The modest impact of our individual mutations is in accord with two other recent transgenic studies that have restricted their mutations to small regions in the core promoter region. It was observed that mutating the initiator in the β2-tubulin promoter caused a 25% reduction in the tissue-specific expression of this promoter (34). The mutation of a downstream region reduced expression by 75% (threefold), and the combination of the two mutations did not inhibit transcription to any greater extent. In another study, mutation of the initiator element of the yellow gene was found to cause a slight decrease in body pigment (25). This indicates that the level of yellow gene expression has been diminished, but the amount was not quantified.
An important point to emerge from the small number of transgenic studies (including this work) is that individual sequences located at the initiation site and further downstream can make rather modest contributions to the level of expression when the genes are evaluated in a chromosomal context. The results with the β2-tubulin gene are particularly noteworthy, because this promoter lacks a TATA box. Thus, one might anticipate that this promoter would be particularly sensitive to mutations in the downstream region, and yet it is not.
The results of the transgenic studies with Drosophila are a warning that the contributions of individual core promoter elements need to be evaluated in a chromosomal context to fully assess the contribution of these elements to the function of normal promoters. In this regard, it is worth noting that an early transgenic analysis of the vermilion promoter in Drosophila showed that mutation of sequences from +19 to +36 reduced expression of this promoter by 100-fold (11). This promoter lacks a TATA element. Hence there are likely to exist cases in which the contribution of an individual element to expression could be quite large. Of course, even if the contributions of individual core promoter sequences turn out to be small, these contributions could have significant physiological impact. For example, mutations in the promoter proximal region of the adult β-globin gene might be the basis for two kinds of human β-thalassemia (19).
We sought to identify the subunits of TFIID that might be responsible for recognizing different regions of the promoter. Our cross-linking procedure failed to detect any candidates contacting the +24 to +27 region. Previous work by Burke and Kadonaga showed that dTAF60 and dTAF40 cross-linked to the downstream region of a TATA-less promoter (5). The cross-linking agent used in their study had a length of 10 Å, whereas our cross-linking agent (BrdU) has a length of 1 or 2 Å. It is possible that TAF40 and TAF60 are not in intimate contact with the bases in DNA. Another possibility is that the interaction between TFIID and the hsp70 promoter is different from the interaction that occurs with a TATA-less promoter. Our chemical interference data support this possibility (29, 30). Our data showed that TFIID makes intimate contact at nucleotide +28, yet this region does not contain a sequence that matches the consensus sequence for the downstream promoter element described by Kutach and Kadonaga (17).
The results of our cross-linking analysis indicate that the initiator is recognized by dTAF230. It was previously observed that human TAFs (hTAFs) 250 and 135 cross-linked to the initiator region when TFIID was bound to the adenovirus major late promoter in the presence of TFIIA (26). TFIIA copurifies with Drosophila TFIID (43), so we anticipate that our interaction should be similar. hTAF250 is the homologue of dTAF230. Some of the hTAF135 detected in the previous cross-linking experiments could correspond to dTAF150 (24). There are two distinct TAFs in human TFIID that comigrate at a position of 135 kDa, and only one of these appears to be functionally and structurally related to dTAF150. The previously reported cross-linking experiments were performed with a cross-linker with a length of 10 Å, so the question of which TAF is most likely responsible for directly contacting the initiator was left unanswered.
Recently a complex containing only dTAF150 and dTAF230, but neither TAF alone, was found to recognize the initiator sequence (6). Binding site selection analysis with this dimeric complex identifies an initiator consensus that matches well with the consensus we found for TFIID (29). This observation, in combination with our cross-linking data, suggests that dTAF230 is responsible for recognizing the initiator, and dTAF150 might stabilize DNA binding by making DNA contacts outside of the initiator. Alternatively, dTAF150 could induce a conformational change in dTAF230 that is necessary for dTAF230 to recognize the initiator. We favor the former possibility, because we observed dTAF150 making contact in the region 35 to 42 nucleotides downstream from the initiator. Mutations downstream from +33 do not affect the affinity of TFIID, suggesting that the contact of dTAF150 with DNA downstream from +35 is not dependent on the sequence (10).
An intriguing result from our cross-linking analysis is that dTAF230 preferentially cross-links to the nontranscribed strand of the initiator. This is not simply due to differences in the labeling of the nontranscribed strand and transcribed strand, since the modified region in the DNA is exactly the same: AATT. Moreover, we did not detect any difference in the amount of DNA binding to the immobilized TFIID, suggesting the incorporation of BrdU into either strand of the initiator does not interfere with the association of TFIID. The biased cross-linking of TAF230 with the nontranscribed strand might simply be a consequence of the spatial distribution of groups that are capable of participating in the photochemistry. However, the more interesting possibility is that this cross-linking is indicative of a strand-specific association of dTAF230. Thirty years ago, sigma 70 was found to cross-link preferentially to the nontranscribed strand in an RNA polymerase-promoter complex (28, 36). Subsequently, sigma 70 was found to recognize the nontranscribed strand of the Pribnow box (23, 32). It will be interesting to see if dTAF230 has a similar property.
ACKNOWLEDGMENTS
We thank Kate Cilli and Chris Bell for assistance in generating plasmids and characterizing transformants. We thank Peter Verrijzer, Robert Weinzierl, and Robert Tjian for antibodies against TFIID.
This work was supported by Public Health Service grant GM47477 from the NIH.
REFERENCES
- 1.Arkhipova I R. Promoter elements in Drosophila melanogaster revealed by sequence analysis. Genetics. 1995;139:1359–1369. doi: 10.1093/genetics/139.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biggin M D, Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53:699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- 3.Breathnach R, Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- 4.Burke T W, Kadonaga J T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 5.Burke T W, Kadonaga J T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalkley G E, Verrijzer C P. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the Initiator. EMBO J. 1999;18:4835–4845. doi: 10.1093/emboj/18.17.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi T, Carey M. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 8.Chi T, Lieberman P, Ellwood K, Carey M. A general mechanism for transcriptional synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 9.Emami K H, Navarre W W, Smale S T. Core promoter specificities of the Sp1 and VP16 transcriptional activation domains. Mol Cell Biol. 1995;15:5906–5916. doi: 10.1128/mcb.15.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emanuel P A, Gilmour D S. TFIID recognizes DNA sequences downstream of the TATA element in the hsp 70 heat shock gene. Proc Natl Acad Sci USA. 1993;90:8449–8453. doi: 10.1073/pnas.90.18.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fridell Y-W C, Searles L L. In vivo transcriptional analysis of the TATA-less promoter of the Drosophila melanogaster vermillion gene. Mol Cell Biol. 1992;12:4571–4577. doi: 10.1128/mcb.12.10.4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garraway I P, Semple K, Smale S T. Transcription of the lymphocyte-specific terminal deoxynucleotidyltransferase gene requires a specific core promoter structure. Proc Natl Acad Sci USA. 1996;93:4336–4341. doi: 10.1073/pnas.93.9.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilmour D S, Dietz T J, Elgin S C R. TATA box-dependent protein-DNA interactions are detected on heat shock and histone gene promoters in nuclear extracts derived from Drosophila melanogaster embryos. Mol Cell Biol. 1988;8:3204–3214. doi: 10.1128/mcb.8.8.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilmour D S, Dietz T J, Elgin S C R. UV-cross-linking identifies four polypeptides that require the TATA box to bind to the Drosophila hsp70 promoter. Mol Cell Biol. 1990;10:4233–4238. doi: 10.1128/mcb.10.8.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen S K, Tjian R. TAFs and TFIIA mediate differential utilization of the tandem Adh promoters. Cell. 1995;82:565–575. doi: 10.1016/0092-8674(95)90029-2. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 17.Kutach A K, Kadonaga J T. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol Cell Biol. 2000;20:4754–4764. doi: 10.1128/mcb.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H, Kraus K W, Wolfner M F, Lis J T. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes Dev. 1992;60:284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- 19.Lewis B A, Kim T K, Orkin S H. A downstream element in the human beta-globin promoter: evidence of extended sequence-specific transcription factor IID contacts. Proc Natl Acad Sci USA. 2000;97:7172–7177. doi: 10.1073/pnas.120181197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harbor Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- 21.Lu Q, Wallrath L L, Elgin S C R. The use of Drosophila P-element mediated germline transformation to examine the chromatin structure and expression of in vitro modified genes. Methods Mol Genet. 1993;1:333–357. [Google Scholar]
- 22.Lu Q, Wallrath L L, Emanuel P A, Elgin S C R, Gilmour D S. Insensitivity of the preset hsp26 chromatin structure to a TATA box mutation in Drosophila. J Biol Chem. 1994;269:15906–15911. [PubMed] [Google Scholar]
- 23.Marr M T, Roberts J W. Promoter recognition as measured by binding of polymerase to nontemplate strand oligonucleotide. Science. 1997;276:1258–1260. doi: 10.1126/science.276.5316.1258. [DOI] [PubMed] [Google Scholar]
- 24.Martinez E, Ge H, Tao Y, Yuan C-X, Palhan V, Roeder R G. Novel cofactors and TFIIA mediate functional core promoter selectivity by the human TAFII150-containing TFIID complex. Mol Cell Biol. 1998;18:6571–6583. doi: 10.1128/mcb.18.11.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris J R, Geyer P K, Wu C T. Core promoter elements can regulate transcription on a separate chromosome in trans. Genes Dev. 1999;13:253–258. doi: 10.1101/gad.13.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oelgeschlager T, Chiang C M, Roeder R G. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 27.Ohtsuki S, Levine M, Cai H N. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park C S, Hillel Z, Wu C W. DNA strand specificity in promoter recognition by RNA polymerase. Nucleic Acids Res. 1980;8:5895–5912. doi: 10.1093/nar/8.23.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purnell B A, Emanuel P A, Gilmour D S. TFIID sequence-recognition of the initiator and sequences further downstream in Drosophila class II genes. Genes Dev. 1994;8:830–842. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- 30.Purnell B A, Gilmour D S. Contribution of sequences downstream of the TATA element to a protein/DNA complex containing the TATA-binding protein. Mol Cell Biol. 1993;13:2593–2603. doi: 10.1128/mcb.13.4.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren B, Maniatis T. Regulation of Drosophila Adh promoter switching by an initiator-targeted repression mechanism. EMBO J. 1998;17:1076–1086. doi: 10.1093/emboj/17.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts C W, Roberts J W. Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 33.Rubin G, Spradling A C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 34.Santel A, Kaufmann J, Hyland R, Renkawitz-Pohl R. The initiator element of the Drosophila beta2 tubulin gene core promoter contributes to gene expression in vivo but is not required for male germ-cell specific expression. Nucleic Acids Res. 2000;28:1439–1446. doi: 10.1093/nar/28.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon J A, Lis J T. A germline transformation analysis reveals flexibility in the organization of heat shock consensus elements. Nucleic Acids Res. 1987;15:2971–2988. doi: 10.1093/nar/15.7.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simpson R B. Contacts between Escherichia coli RNA polymerase and thymines in the lac UV5 promoter. Proc Natl Acad Sci USA. 1979;76:3233–3237. doi: 10.1073/pnas.76.7.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smale S T. Core promoter architecture for eucaryotic protein-coding genes. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press, Ltd.; 1994. pp. 63–81. [Google Scholar]
- 38.Smale S T. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 39.Sypes M A, Gilmour D S. Protein/DNA cross-linking of a TFIID complex reveals novel interactions downstream of the transcription start. Nucleic Acids Res. 1994;22:807–814. doi: 10.1093/nar/22.5.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thummel C S, Boulet A M, Lipshitz H D. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]
- 41.Verrijzer C P, Chen J L, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 42.Weber J A, Taxman D J, Lu Q, Gilmour D S. Molecular architecture of the hsp70 promoter after deleting the TATA box or the upstream regulatory region. Mol Cell Biol. 1997;17:3799–3808. doi: 10.1128/mcb.17.7.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yokomori K, Admon A, Goodrich J A, Chen J-L, Tjian R. Drosophila TFIIA-L is processed into two subunits that are associated with the TBP/TAF complex. Genes Dev. 1993;7:2235–2245. doi: 10.1101/gad.7.11.2235. [DOI] [PubMed] [Google Scholar]