Significance
Microbes secrete a diversity of molecules into their environment to mediate niche colonization. During host ingress, plant pathogenic microbes secrete effector proteins that facilitate disease development, many of which deregulate host immune responses. We recently demonstrated that plant pathogens additionally exploit effectors with antibacterial activities to manipulate beneficial plant microbiota to promote host colonization. Here, we show that the fungal pathogen Verticillium dahliae has co-opted an ancient antimicrobial protein, which likely served in microbial competition in terrestrial environments before land plants existed, as effector for the manipulation of fungal competitors during host colonization. Thus, we demonstrate that pathogen effector repertoires comprise antifungal proteins and speculate that such effectors could be exploited for the development of antimycotics.
Keywords: plant pathogenic fungus, microbiome, effector, antimicrobial, mycobiome
Abstract
Microbes typically secrete a plethora of molecules to promote niche colonization. Soil-dwelling microbes are well-known producers of antimicrobials that are exploited to outcompete microbial coinhabitants. Also, plant pathogenic microbes secrete a diversity of molecules into their environment for niche establishment. Upon plant colonization, microbial pathogens secrete so-called effector proteins that promote disease development. While such effectors are typically considered to exclusively act through direct host manipulation, we recently reported that the soil-borne, fungal, xylem-colonizing vascular wilt pathogen Verticillium dahliae exploits effector proteins with antibacterial properties to promote host colonization through the manipulation of beneficial host microbiota. Since fungal evolution preceded land plant evolution, we now speculate that a subset of the pathogen effectors involved in host microbiota manipulation evolved from ancient antimicrobial proteins of terrestrial fungal ancestors that served in microbial competition prior to the evolution of plant pathogenicity. Here, we show that V. dahliae has co-opted an ancient antimicrobial protein as effector, named VdAMP3, for mycobiome manipulation in planta. We show that VdAMP3 is specifically expressed to ward off fungal niche competitors during resting structure formation in senescing mesophyll tissues. Our findings indicate that effector-mediated microbiome manipulation by plant pathogenic microbes extends beyond bacteria and also concerns eukaryotic members of the plant microbiome. Finally, we demonstrate that fungal pathogens can exploit plant microbiome-manipulating effectors in a life stage–specific manner and that a subset of these effectors has evolved from ancient antimicrobial proteins of fungal ancestors that likely originally functioned in manipulation of terrestrial biota.
Microbes are found in a wide diversity of niches on our planet. To facilitate establishment within microbial communities, microbes secrete a multitude of molecules to manipulate each other. Many of these molecules exert antimicrobial activities and are exploited to directly suppress microbial coinhabitants in order to outcompete them for the limitedly available nutrients and space of a niche. Microbially secreted antimicrobials encompass diverse molecules including peptides and lytic enzymes but also nonproteinaceous molecules such as secondary metabolites. Soils are among the most biologically diverse and microbially competitive environments on earth. Microbial proliferation in the soil environment is generally limited by the availability of organic carbon (1), for which soil microbes continuously compete. Consequently, numerous saprophytic soil-dwelling microbes secrete potent antimicrobials that promote niche protection or colonization. Notably, these microbes are the primary source of our clinically used antibiotics (2, 3).
Like free-living microbes, microbial plant pathogens also secrete a multitude of molecules into their environment to mediate niche colonization (4, 5). The study of molecules secreted by microbial plant pathogens has been largely confined to the context of binary interactions between pathogens and hosts. To establish disease, plant pathogenic microbes secrete a plethora of so-called effectors, molecules of various kinds that promote host colonization and that are typically thought to mainly deregulate host immune responses (4, 6, 7). Upon host colonization, plant pathogens encounter a plethora of plant-associated microbes that collectively form the plant microbiota, which represent a key factor for plant health. Beneficial plant-associated microbes are found in and on all organs of the plant and help to mitigate (a)biotic stresses (8–13). Plants shape their microbiota and specifically attract beneficial microbes to suppress pathogens (14–16). Hence, the plant microbiome can be considered an inherent, exogenous layer that complements the plant’s endogenous innate immune system. We previously hypothesized that plant pathogens not only utilize effectors to target components of host immunity as well as other aspects of host physiology to support host colonization but also to target the host microbiota in order to establish niche colonization (4, 5). We recently provided experimental evidence for this hypothesis by showing that the ubiquitously expressed effector VdAve1 that is secreted by the soil-borne fungal plant pathogen Verticillium dahliae acts as a bactericidal protein that promotes host colonization through the selective manipulation of host microbiomes by suppressing microbial antagonists (17, 18). Additionally, we demonstrated that VdAve1 and a further antibacterial effector named VdAMP2 are exploited by V. dahliae for microbial competition in soil and promote virulence of V. dahliae in an indirect manner (18). Collectively, these observations demonstrate that V. dahliae dedicates part of its effector catalog toward microbiota manipulation. Likely, the V. dahliae genome encodes further effectors that act in microbiome manipulation.
Evidently, bacterial and fungal evolution on land preceded land plant evolution. As a consequence, fungal pathogen effectors involved in the manipulation of (host-associated) microbial communities may have evolved from ancestors that served in microbial competition in terrestrial niches hundreds of millions of years ago prior to land plant evolution. However, evidence for this hypothesis is presently lacking.
V. dahliae is an asexual xylem-dwelling fungus that causes vascular wilt disease on hundreds of plant species (19). The fungus survives in the soil in the form of multicellular melanized resting structures, called microsclerotia, that offer protection against (a)biotic stresses and can persist in the soil for many years (20). Microsclerotia represent the major inoculum source of V. dahliae in nature, and their germination is triggered by carbon- and nitrogen-rich exudates from plant roots (21). Following microsclerotia germination, fungal hyphae grow through the soil and rhizosphere toward the roots of host plants. Next, V. dahliae colonizes the root cortex and crosses the endodermis, from which it invades xylem vessels. Once the fungus enters those vessels, it forms conidiospores that are transported with the water flow until they get trapped, for instance, by vessel end walls. This triggers germination of the conidiospores followed by penetration of cell walls, hyphal growth, and renewed sporulation, leading to systematic colonization of the plant (22). Once tissue necrosis commences and plant senescence occurs, host immune responses fade and V. dahliae enters a saprophytic phase in which it emerges from the xylem vessels to invade adjacent host tissues, which is accompanied by the production of microsclerotia. Upon littering and decomposition of plant tissues, these microsclerotia are released into the soil (23).
Results
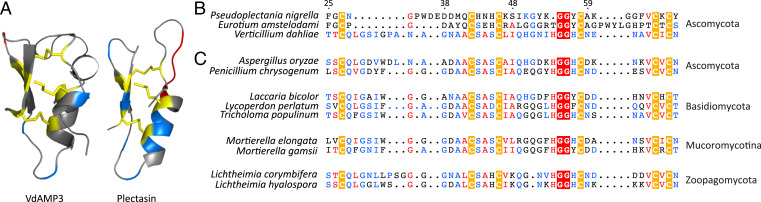
To identify effectors potentially acting in microbiome manipulation, we recently queried the V. dahliae secretome for structural homologs of known antimicrobial proteins (AMPs), which led to the identification of 10 candidates including the functionally characterized VdAMP2 (18). Among the remaining nine candidates, we now identified a small cysteine-rich protein of ∼4.9 kDa, which we name VdAMP3 (Ensembl: VDAG_JR2_Chr3g05620a). As a first step in the characterization of VdAMP3, we assessed its predicted structure. Interestingly, VdAMP3 is predicted to adopt a cysteine-stabilized αβ (CSαβ) fold that is also found in defensin-like proteins (Fig. 1A) (24–26). CSαβ defensins represent a widespread and well-characterized family of antimicrobial proteins that are presumed to share a single ancient origin in the last common ancestor of animals, plants, and fungi that produce these proteins today (24–27). It is important to note, however, that many typical small cysteine-rich pathogen effectors adopt AMP-like confirmations and that tertiary structures of several AMP families strongly resemble each other (27, 28). Hence, structure prediction can easily lead to false-positive classifications as AMP or allocation to the wrong AMP family.
Fig. 1.
The V. dahliae effector VdAMP3 evolved from an ancient fungal protein. (A) VdAMP3 (Left) is predicted to adopt a CSαβ defensin-like fold. The structure of the CSαβ defensin plectasin (Right) of the fungus P. nigrella is included as reference. The disulfide bonds stabilizing the antiparallel β-sheets and the α-helix are highlighted in yellow. Positively and negatively charged amino acid residues are highlighted in blue and red, respectively. (B) Protein sequence alignment with CSαβ defensins plectasin and eurocin (E. amstelodami) supports the structure prediction of VdAMP3. (C) VdAMP3 homologs are widespread in the fungal kingdom. Protein sequence alignment of VdAMP3 with a subset of its homologs identified in higher (Ascomycota and Basidiomycota) and lower fungi (Mucoromycotina and Zoopagomycota). The alignment as shown in B and C displays the most conserved region of the CSαβ defensin protein family and was performed using HMMER and visualized with Espript3. The highly conserved cysteine and glycine residues that contribute to the CSαβ defensin structure are highlighted by yellow and red backgrounds, respectively. The numbers on top of the alignment indicate the corresponding residue numbers of VdAMP3. The homologs displayed in C were identified using blastP in the predicted proteomes of the respective fungi included in the JGI 1000 Fungal Genomes Project (32).
CSαβ defensins, or so-called cis-defensins, owe their structure to highly conserved cis-orientated disulfide bonds that establish an interaction between a double- or triple-stranded antiparallel β-sheet with an α-helix (25, 27). To validate the prediction of VdAMP3 as a member of this ancient antimicrobial protein family, we aligned its amino acid sequence with the antibacterial CSαβ defensins plectasin and eurocin, from the saprophytic Ascomycete species Pseudoplectania nigrella and Eurotium amstelodami (formerly Aspergillus amstelodami), respectively (29–31). Although the biological relevance of these defensins for the respective fungi remains unclear, their antibacterial activity and protein structure have been well characterized, which led to their recognition as genuine CSαβ defensins (29–31). Although the overall identity between the three proteins was rather low (25 to 40%), protein sequence alignment revealed that VdAMP3 contains the six highly conserved cysteine residues that are considered crucial for the structure of CSαβ defensins (Fig. 1B) (27). To further substantiate the emerging picture that VdAMP3 belongs to this particular protein family and that the detected similarities with plectasin and eurocin are not the result of convergent protein evolution, we queried the predicted proteomes of the fungi from the Joint Genome Institute (JGI) 1000 Fungal Genomes Project (32) for homologs of VdAMP3 with higher sequence identity and included a subset of those in the protein alignment (Fig. 1C). Interestingly, besides homologs in Ascomycota and Basidiomycota, our sequence similarity search also revealed homologs in early-diverging fungi from the subphyla Mucoromycotina and Zoopagomycota [both formerly classified as Zygomycota (33)] (Fig. 1C). Importantly, this divergence is estimated to have taken place ∼900 million years ago (34), indicating it preceded the evolution of the first land plants ∼450 million years later (34–37). Consequently, this analysis indicates that VdAMP3 evolved from an ancestral fungal gene hundreds of millions of years ago before land plants existed.
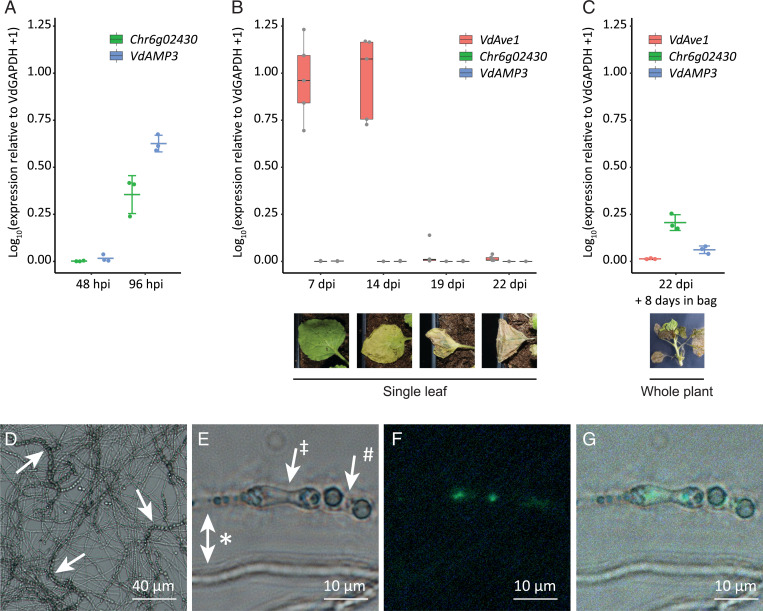
As a first step to determine a potential role of VdAMP3 in V. dahliae infection biology, we assessed whether we could find evidence for VdAMP3 expression during host colonization. Analysis of previously generated transcriptome datasets of diverse V. dahliae strains during colonization of a diversity of hosts did not reveal in planta expression of VdAMP3 (17, 38–40). However, strong induction of this effector gene was reported during microsclerotia formation in a transcriptome analysis of V. dahliae strain XS11 grown in vitro (24). To validate this finding, we analyzed in vitro expression of VdAMP3 in V. dahliae strain JR2. To this end, V. dahliae conidiospores were spread on nitrocellulose membranes placed on top of solid minimal medium and fungal material was harvested prior to microsclerotia formation after 48 h of incubation and after the onset of microsclerotia formation after 96 h of incubation. Expression of VdAMP3 was determined at both time points with real-time PCR alongside expression of the Chr6g02430 gene that encodes a putative cytochrome P450 enzyme that acts as a marker for microsclerotia formation (24, 41). Consistent with the observations for V. dahliae strain XS11 (24), no VdAMP3 expression was detected at 48 h, when Chr6g02430 was also not expressed and no visual microsclerotia formation could be observed on the growth medium (Fig. 2A). However, induction of VdAMP3, as well as Chr6g02430, was observed after 96 h of incubation, at which time point the formation of microsclerotia on the growth medium also became apparent (Fig. 2A). Collectively, these data demonstrate that expression of VdAMP3 coincides with microsclerotia formation in vitro also for V. dahliae strain JR2.
Fig. 2.
VdAMP3 is specifically expressed in hyphal cells that develop into microsclerotia. (A) Expression of VdAMP3 and the marker gene for microsclerotia development Chr6g02430, relative to the household gene VdGAPDH at 48 and 96 h of in vitro cultivation (n = 3). (B) Expression of VdAve1, VdAMP3, and Chr6g02430 in N. benthamiana leaves from 7 to 22 dpi (n = 5). (C) Expression of VdAve1, VdAMP3, and Chr6g02430 in tissue of N. benthamiana plants harvested at 22 dpi after 8 d of incubation in sealed plastic bags (n = 3). (D) Microsclerotia formation of a pVdAMP3::eGFP reporter mutant as detected after 7 d of cultivation in Czapek Dox medium. Typical chains of microsclerotia (42, 43) are indicated by arrows. (E) Bright-field image of various V. dahliae cell types after 7 d of cultivation in Czapek Dox, including hyphae (*), swollen hyphal cells developing into microsclerotia (‡), and mature microsclerotia cells (#). (F) GFP signal for the image as shown in E, indicative for activity of the VdAMP3 promoter, is exclusively detected in the swollen hyphal cells developing into microsclerotia. (G) Overlay of E and F.
Although previous transcriptome analyses failed to detect in planta expression of VdAMP3, we realized that these analyses were predominantly performed for infection stages when the fungus was still confined to the xylem vessels and microsclerotia formation had not yet been initiated. Accordingly, in planta expression of VdAMP3 may have been missed. Thus, we inoculated Nicotiana benthamiana with V. dahliae and determined expression of VdAMP3 in leaves and petioles sampled at different time points and displaying different disease phenotypes, ranging from asymptomatic at 7 d postinoculation (dpi) to complete necrosis at 22 dpi. As expected, a strong induction of the previously characterized VdAve1 effector gene was detected at 7 and 14 dpi (Fig. 2B) (17, 18). In contrast, however, no expression of VdAMP3 was recorded, even at the latest time point, when the leaf tissue had become completely necrotic (Fig. 2B). Importantly, no Chr6g02430 expression was detected at any of these time points either (Fig. 2B), suggesting that microsclerotia formation had not yet started in these tissues. Indeed, visual inspection of the necrotic plant tissue collected at 22 dpi did not reveal microsclerotia presence. To induce microsclerotia formation, V. dahliae–inoculated N. benthamiana plants harvested at 22 dpi were sealed in plastic bags and incubated in the dark to increase the relative humidity and mimic conditions that occur during tissue decomposition in the soil. Interestingly, after 8 d of incubation, the first microsclerotia could be observed and induction of VdAMP3, as well as Chr6g02430, was detected (Fig. 2C). Notably, the induction of both genes in planta is markedly weaker when compared with their expression in vitro (Fig. 2A). However, this is likely explained by a much smaller proportion of the total population of V. dahliae cells undergoing synchronized development into microsclerotia, also because the time window from conidial germination through hyphal growth to microsclerotia formation is much smaller in vitro than in planta. Collectively, our findings suggest that in planta expression of VdAMP3 coincides with microsclerotia formation, similar to our observations in vitro. Moreover, our data suggest that VdAMP3 expression primarily depends on a developmental stage of V. dahliae rather than on host factors such as tissue necrosis.
To determine more precisely where VdAMP3 is expressed and to improve our understanding of how V. dahliae may benefit from effector expression during microsclerotia formation, we generated a V. dahliae reporter strain expressing eGFP under control of the VdAMP3 promoter. Intriguingly, microscopic analysis of the reporter strain during microsclerotia formation stages in vitro (Fig. 2D) revealed that VdAMP3 is expressed by swollen hyphal cells that act as primordia that subsequently develop into microsclerotia but not by the adjacent hyphal cells or recently developed microsclerotia cells (Fig. 2 E–G). This highly specific expression of VdAMP3 suggests that the effector protein may facilitate the formation of microsclerotia in decaying host tissue. Given its presumed antimicrobial activity, VdAMP3 may be involved in antagonistic activity against opportunistic decay organisms in this microbially competitive niche.
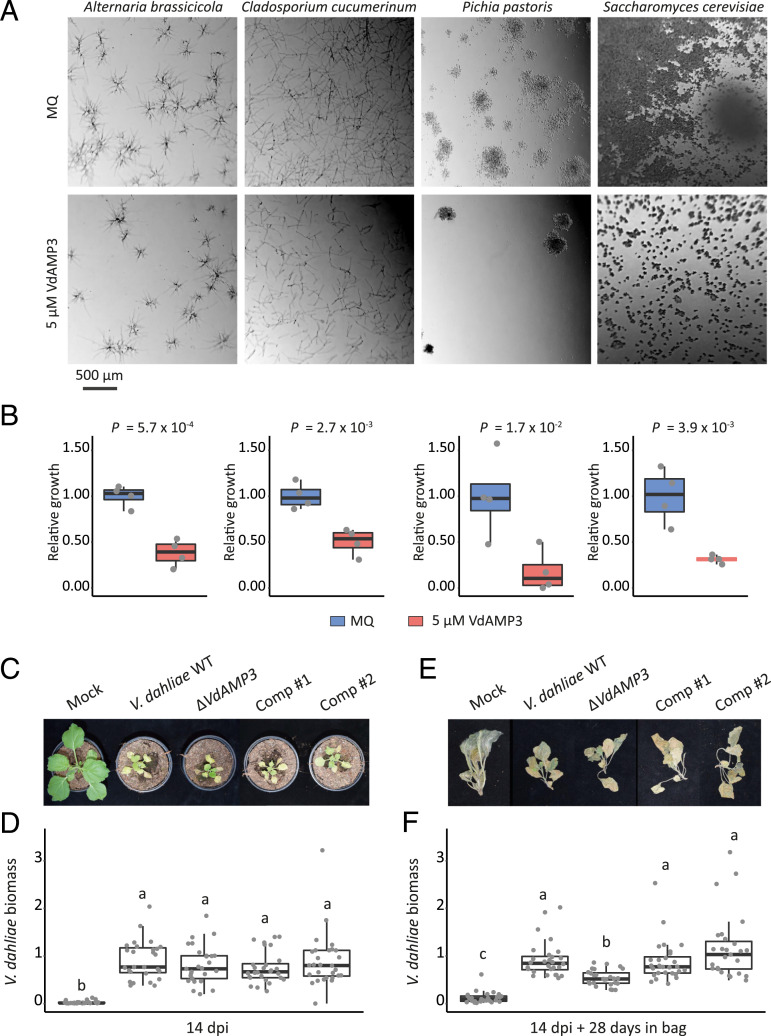
To determine if VdAMP3 indeed exerts antimicrobial activity, we tried to produce VdAMP3 heterologously in the yeast Pichia pastoris and in the bacterium Escherichia coli, but these attempts failed, indicative of potential antimicrobial activity of the effector protein. Therefore, chemical synthesis of VdAMP3 was pursued. Next, we incubated a randomly selected panel of bacterial isolates with the effector protein and monitored their growth in vitro. VdAMP3 concentrations as high as 20 µM resulted in no or only marginal bacterial growth inhibition (SI Appendix, Fig. 1). A similar assay with fungal isolates showed that incubation with 5 µM VdAMP3 already markedly affected growth of the filamentous fungi Alternaria brassicicola and Cladosporium cucumerinum and the yeasts P. pastoris and Saccharomyces cerevisiae (Fig. 3 A and B). This finding suggests that VdAMP3 displays more potent activity against fungi than against bacteria. Importantly, a thorough heat treatment involving boiling of VdAMP3 abolished its antifungal activity (SI Appendix, Fig. 2), indicating that the specificity of this activity depends on its correct three-dimensional confirmation.
Fig. 3.
VdAMP3 is an antifungal protein that contributes to V. dahliae biomass accumulation in the decaying host phyllosphere. (A) Microscopic pictures of fungal isolates grown in 5% PDB supplemented with 5 µM VdAMP3 or ultrapure water (Milli-Q). VdAMP3 impairs growth of A. brassicicola, C. cucumerinum, P. pastoris, and S. cerevisiae. Pictures were taken after 24 (A. brassicicola, C. cucumerinum, and S. cerevisiae) or 64 (P. pastoris) h of incubation. (B) Fungal growth as displayed in A was quantified using ImageJ (unpaired two-sided Student’s t test; n = 4). (C) VdAMP3 does not contribute to establishment of Verticillium wilt disease in N. benthamiana. Photos display representative phenotypes of N. benthamiana plants infected by wild-type V. dahliae (WT), the VdAMP3 deletion (ΔVdAMP3), and two complementation (Comp) mutants 14 dpi. (D) Relative V. dahliae biomass in aboveground N. benthamiana tissues determined with real-time PCR. Different letter labels represent significant differences (one-way ANOVA and Tukey’s post hoc test; P < 0.05; n ≥ 27. (E) Representative phenotypes of N. benthamiana plants as shown in C after 28 d of incubation in plastic bags. (F) Relative V. dahliae biomass in N. benthamiana tissues as displayed in E. Letters represent significant differences (one-way ANOVA and Tukey’s post hoc test; P < 0.05; n ≥ 27).
Considering its antifungal activity, but also the highly controlled timely and topical expression of VdAMP3, we tested if exogenous VdAMP3 application negatively impacts hyphal growth of V. dahliae. Interestingly, incubation of V. dahliae with 5 µM VdAMP3 markedly affected its growth (SI Appendix, Fig. 3 A and B). However, it needs to be realized that this effector protein is produced by the time when most hyphae of the fungus have lost their function, as the host tissue has become senescent and will soon decompose, and the fungus produces microsclerotia for long-term survival. Next, to verify if growth or development of V. dahliae is affected by VdAMP3, we generated a VdAMP3 deletion mutant (SI Appendix, Fig. 4), which we cultivated in vitro alongside wild-type (WT) V. dahliae. As anticipated, deletion of VdAMP3 did not accelerate growth of the fungus (SI Appendix, Fig. 3C), confirming that the effector gene does not compromise the development of the fungus during the life stages prior to microsclerotia formation. Moreover, deletion of VdAMP3 also did not impair the ability of V. dahliae to form resting structures, nor their ability to infect new plants and cause disease (SI Appendix, Fig. 3 C–E). Next, we aimed to determine if the antifungal activity of VdAMP3 contributes to Verticillium wilt disease development. To this end, N. benthamiana plants were inoculated with V. dahliae WT as well as with VdAMP3 complementation and deletion mutants (SI Appendix, Fig. 4). In line with our inability to detect expression during early infection stages, disease phenotypes and V. dahliae biomass quantification using real-time PCR did not reveal a contribution of VdAMP3 to host colonization up to 2 wk after inoculation (Fig. 3 C and D). Considering the cell type–specific expression of VdAMP3 in developing microsclerotia, we speculated that the effector protein contributes to V. dahliae niche establishment during host plant senescence when the fungus has emerged from the xylem and has colonized the mesophyll. To test this hypothesis, we performed additional disease assays using V. dahliae WT and the VdAMP3 deletion mutant and sealed the N. benthamiana plants in plastic bags after harvesting to stimulate the onset of tissue decomposition and microsclerotia formation. Intriguingly, when we visually inspected the plants after 4 wk of incubation, we detected dispersed patches of dark mycelium, typical for V. dahliae microsclerotia, on the surface of plants colonized by V. dahliae WT (SI Appendix, Fig. 5). Strikingly, we did not identify such patches on plants colonized by the VdAMP3 deletion mutant, suggesting that V. dahliae depends on the antifungal activity of VdAMP3 to form microsclerotia in decaying host phyllospheres. It needs to be noted that an experimental setup that depends on a largely unpredictable occurrence of visibly detectable patches of microsclerotia on the surface of decaying plant parts that are colonized by diverse assemblages of opportunistic microbes that seize their opportunity to prosper while plant defenses fade is hardly feasible for standardized, robust quantification of microsclerotia formation. Also, this setup does not permit assessment of microsclerotia formation deeper in the decaying tissues. Instead, we quantified V. dahliae biomass using real-time PCR. As anticipated, we detected a significant reduction in biomass of the VdAMP3 deletion mutant when compared with V. dahliae WT and the complementation mutants (Fig. 3 E and F), confirming that VdAMP3 indeed is essential during microsclerotia formation in planta presumably by acting in self-protection against other microbes.
To investigate if the effects of VdAMP3 are limited to N. benthamiana or whether those also extend to other hosts, we inoculated Arabidopsis thaliana plants with V. dahliae WT and the VdAMP3 deletion mutant. Consistent with our observations for N. benthamiana, deletion of VdAMP3 did not affect establishment of Verticillium wilt in A. thaliana (SI Appendix, Fig. 6 A and B). However, V. dahliae biomass quantification in aboveground A. thaliana tissues at 3 wk postinoculation revealed reduced accumulation of V. dahliae in the absence of VdAMP3 (SI Appendix, Fig. 6C). Thus, the effects of VdAMP3 are not restricted to a single host.
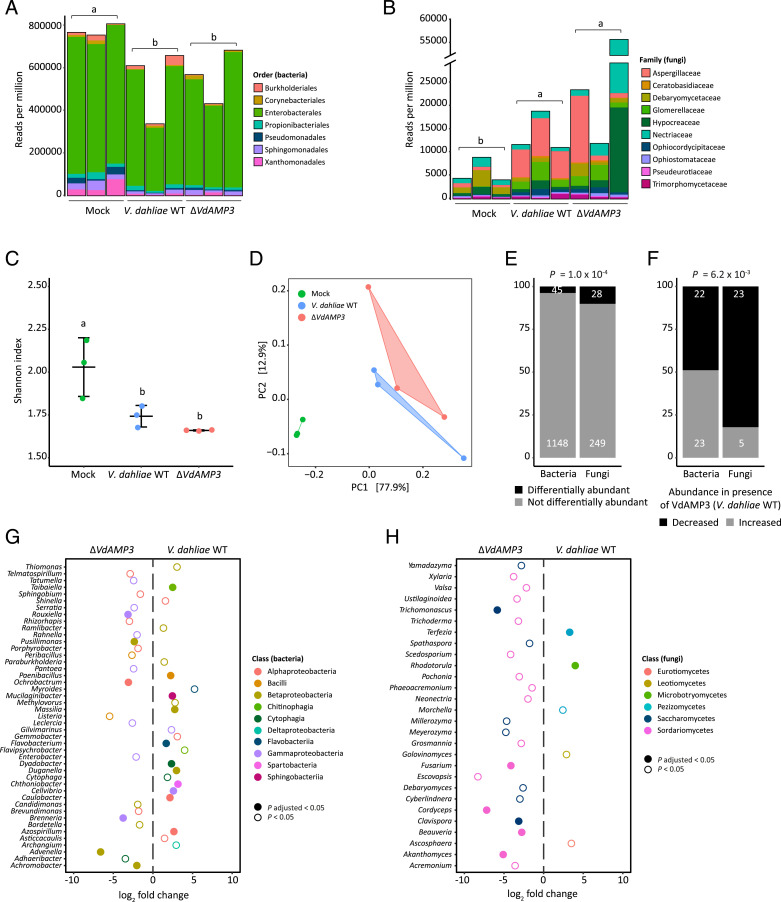
As in vitro antimicrobial activity assays pointed toward fungi as the primary targets of VdAMP3, we speculated that V. dahliae exploits VdAMP3 to suppress fungal competitors in decomposing host tissues to safeguard the formation of its resisting structures. To characterize the microbiota associated with N. benthamiana decomposition and to determine the impact of VdAMP3 on these microbial communities, we characterized the phyllosphere microbiota of fresh mock-inoculated N. benthamiana plants, and decaying plants diseased by V. dahliae WT or the VdAMP3 deletion mutant incubated in plastic bags, through shotgun metagenomic sequencing. Consistent with a primary role for fungi in the decomposition of dead plant material (44–48), we detected a significant increase of fungi and decrease of bacteria in the phyllosphere of the N. benthamiana plants diseased by the V. dahliae strains when compared with healthy mock-treated plants (Fig. 4 A and B). These changes are accompanied by a reduced alpha diversity in the decaying phyllospheres (Fig. 4C). Additionally, principal coordinate analysis (PCoA) based on Bray–Curtis dissimilarities (beta diversity) uncovered clear separation of the microbiota of the healthy plants from those in decay (Fig. 4D). The PCoA also revealed a weaker, yet potentially relevant, separation of the microbiota colonized by V. dahliae WT and the VdAMP3 deletion mutant, which suggests that secretion of VdAMP3 manipulates microbiome compositions (Fig. 4D). Intriguingly, when we compared the abundances of the identified microbial genera between the microbiomes colonized by V. dahliae WT and the VdAMP3 deletion mutant, we detected significantly more differentially abundant fungi (10.1%) than bacteria (3.8%) (Fig. 4E) (SI Appendix, Tables 1 and 2). Interestingly, whereas the number of bacterial genera that display an increased or a decreased abundance in the presence of VdAMP3 is more or less equal, the vast majority of the differentially abundant fungal genera (82.1%) are repressed in the presence of VdAMP3 (Fig. 4F). Moreover, while no consistent suppression of bacterial genera from the same class could be detected, we exclusively identified suppression of the differentially abundant fungal genera from the Saccharomycetes or Sordariomycetes in the presence of VdAMP3 (Fig. 4 G and H). Thus, these observations indicate that V. dahliae VdAMP3 mainly acts as an antifungal effector protein that displays selective activity that predominantly impacts the mycobiome in the decaying host phyllosphere.
Fig. 4.
VdAMP3 manipulates the mycobiome of the decaying N. benthamiana phyllosphere. (A and B) V. dahliae–induced decay of the N. benthamiana phyllosphere is associated with a decreased bacterial and increased fungal abundance. Relative abundance of bacteria (A) and fungi (B), excluding V. dahliae, in the phyllosphere of decaying N. benthamiana plants colonized by WT V. dahliae (WT) or the VdAMP3 deletion mutant (14 dpi and after 28 d of incubation in plastic bags) and in the phyllosphere of fresh N. benthamiana plants (mock). Letters represent significant differences in total bacterial/fungal abundance between the three treatments (one-way ANOVA and Tukey’s post hoc test; P < 0.05; n = 3). (C) V. dahliae–induced decay of N. benthamiana plants impacts alpha diversity of the phyllosphere. The plot displays the average Shannon index ± SD; letters represent significant differences (one-way ANOVA and Tukey’s post hoc test; P < 0.05; n = 3). (D) PCoA based on Bray–Curtis dissimilarities (beta diversity) reveals separation of the microbiomes based on the three different treatments. (E) Differential abundance analysis of microbial genera between the microbiomes colonized by V. dahliae WT and the VdAMP3 deletion mutant indicates that secretion of VdAMP3 significantly impacts a larger proportion of the fungi than of the bacteria (two-tailed Fisher’s exact test). (F) Of the differentially abundant microbial genera, significantly more fungi display a decreased abundance in the presence of VdAMP3 when compared with the bacteria (two-tailed Fisher’s exact test). (G and H) Overview of the differentially abundant bacterial (G) and fungal (H) genera. The plots display increased (positive log2 fold change) or decreased (negative log2 fold change) abundance in the presence of V. dahliae WT when compared with the VdAMP3 deletion mutant (Wald test, P adjusted < 0.05 and P < 0.05, n = 3). Differentially abundant fungal genera from the Saccharomycetes or Sordariomycetes are consistently suppressed in the presence of VdAMP3 (i.e., by V. dahliae WT).
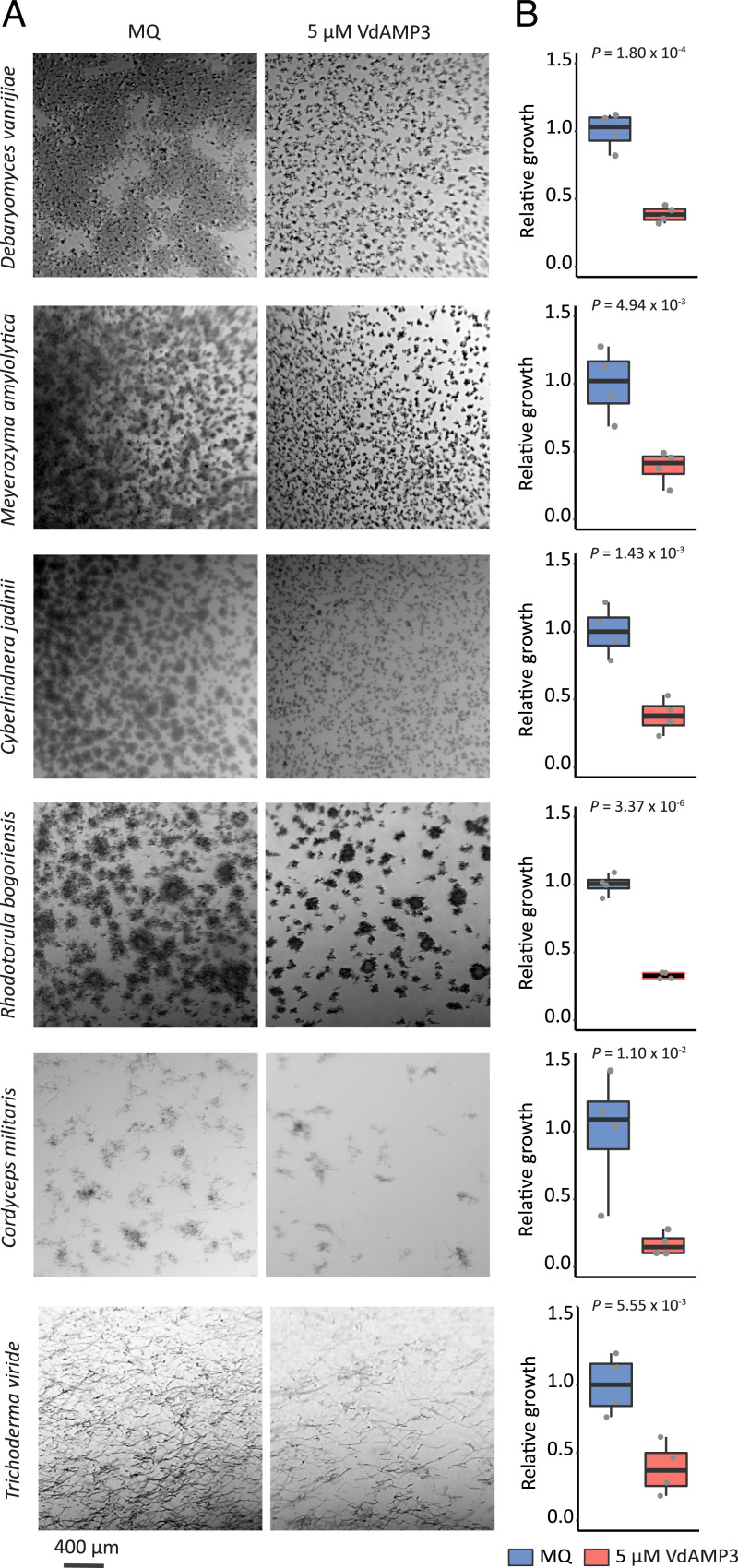
To further substantiate that the suppression of the Saccharomycetes and Sordariomycetes is a direct consequence of the VdAMP3 activity, we incubated fungal species belonging to the suppressed genera with the effector to determine their sensitivity. In line with the previously observed sensitivity of the Saccharomycetes P. pastoris and S. cerevisiae, the Saccharomycete species Cyberlindnera jadinii, Debaryomyces vanrijiae, Rhodotorula bogoriensis, and Meyerozyma amylolytica also displayed markedly reduced growth in the presence of VdAMP3 (Fig. 5 A and B). Similarly, growth of the Sordariomycetes Cordyceps militaris and Trichoderma viride was inhibited by the effector (Fig. 5 A and B). Hence, these findings support the observed suppression of the Saccharomycetes and Sordariomycetes in the N. benthamiana phyllosphere mycobiome as a direct consequence of VdAMP3 activity.
Fig. 5.
VdAMP3 negatively affects Saccharomycetes and Sordariomycetes. (A) Microscopic pictures of fungal isolates grown in 5% PDB supplemented with 5 µM VdAMP3 or ultrapure water (Milli-Q). VdAMP3 impairs growth of D. vanrijae, M. amylolytica, C. jadinii, R. bogoriensis, C. militaris, and T. viride. Pictures were taken after 10 (D. vanrijae and C. jadinii), 24 (M. amylolytica and R. bogoriensis), or 30 (C. militaris and T. viride) h of cultivation. (B) Fungal growth as displayed in A was quantified using ImageJ (unpaired two-sided Student’s t test; n = 4).
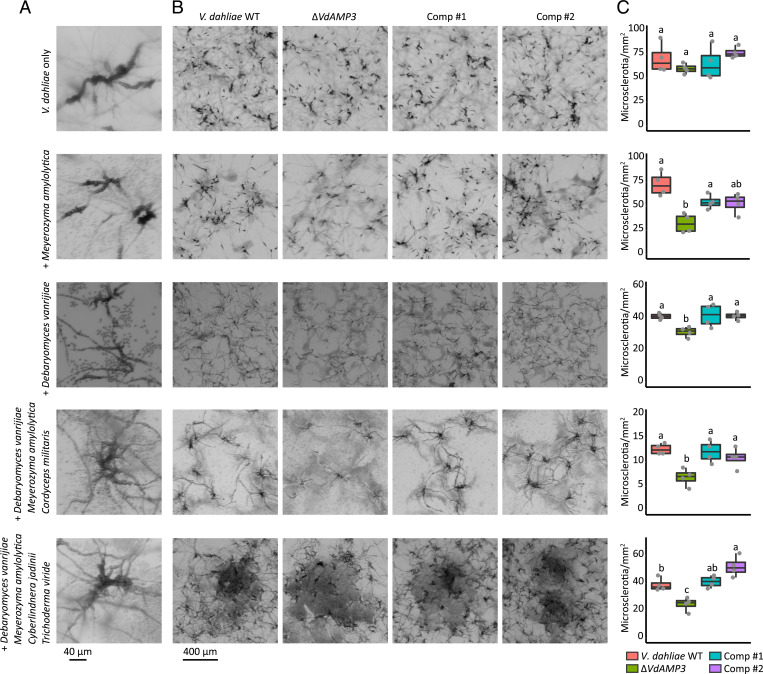
The cell type–specific expression of VdAMP3, combined with its role in mycobiome manipulation, strongly suggests that VdAMP3 is exploited to ward off fungal niche competitors in planta to safeguard the formation of V. dahliae microsclerotia. To test if VdAMP3 indeed is essential for V. dahliae microsclerotia formation in the presence of other fungi, we cocultivated V. dahliae WT and the VdAMP3 deletion and complementation mutants with D. vanrijiae and M. amylolytica. Once microsclerotia formation by V. dahliae WT became apparent (Fig. 6A), we quantified the number of resting structures that were formed by the different V. dahliae genotypes. As anticipated, we detected a significant reduction of microsclerotia formed by the VdAMP3 deletion mutant when compared with V. dahliae WT and the complementation mutants in the presence of both fungal species, confirming that V. dahliae relies on the antifungal activity of VdAMP3 to form microsclerotia in the presence of particular fungal niche competitors (Fig. 6 B and C). Additionally, to confirm that this activity is not only relevant in the presence of a single microbial interactor but also facilitates microsclerotia formation in the presence of fungal communities, we performed similar experiments using two synthetic communities that, besides D. vanrijiae and M. amylolytica, also comprised the filamentous fungus C. militaris or the yeast C. jadinii plus the filamentous mycoparasite T. viride. Also in these experiments, we detected a significant reduction of microsclerotia formed by the VdAMP3 deletion mutant when compared with V. dahliae WT and the complementation mutants (Fig. 6 B and C). Collectively, these findings underpin the idea that V. dahliae exploits the antifungal activity of VdAMP3 to safeguard the formation of its resting structures by warding off fungal niche competitors in senescing host mesophyll tissues.
Fig. 6.
VdAMP3 contributes to V. dahliae microsclerotia formation in the presence of fungal niche competitors. (A) Close-up of V. dahliae microsclerotia formed during cultivation in the presence of D. vanrijae (6 dpi), M. amylolytica (6 dpi), a syncom comprising D. vanrijae, M. amylolytica, and C. militaris (6 dpi), or a syncom comprising D. vanrijae, M. amylolytica, C. jadinii, and T. viride (9 dpi). (B) VdAMP3 contributes to V. dahliae microsclerotia formation in the presence of other fungal species. Representative microscopic pictures displaying V. dahliae WT, the VdAMP3 deletion mutant (ΔVdAMP3), and two complementation mutants (Comp) cultivated in the presence of the fungal species/communities as detailed in A. (C) Number of microsclerotia formed by V. dahliae in the presence of the fungal species or communities (one-way ANOVA and Tukey’s post hoc test; P < 0.05; n = 4).
Discussion
Microbes secrete a plethora of molecules to promote niche colonization (4). Free-living microbes are well-known producers of antimicrobials that are secreted to outcompete microbial coinhabitants to establish themselves in a microbial community. Microbial plant pathogens secrete a diversity of so-called effector molecules during host ingress, many of which are small cysteine-rich proteins that deregulate host immune responses to promote colonization (4, 6, 7). While investigating the vascular wilt fungus V. dahliae, we recently demonstrated that plant pathogens not only exploit effector proteins to promote disease establishment through direct host manipulation but also through the manipulation of plant microbiota by means of antibacterial activities (18). Considering that the advent of fungi on earth preceded land plant evolution, we speculated that a subset of the pathogen effectors involved in host microbiota manipulation may have evolved from antimicrobial proteins that originally functioned in microbial competition in terrestrial niches before the first land plants appeared and plant pathogenicity evolved. Here, we demonstrated that the soil-borne fungal plant pathogen V. dahliae has co-opted an ancient antimicrobial protein as effector for mycobiome manipulation in planta to safeguard the formation of its resting structures. Thus, our findings indicate that plant pathogenicity in fungi is not exclusively associated with the evolution of novel effectors that manipulate plants or their associated microbial communities but also with the co-option of previously evolved secreted proteins that initially served alternative lifestyles, such as saprotrophism, as effectors to promote host colonization. Moreover, our findings indicate that effector-mediated manipulation of plant microbiota by microbial plant pathogens is not confined to bacterial targets but extends to eukaryotic microbes.
Functional characterization of VdAMP3 unveiled that the effector evolved to play a life stage–specific role in microbiome manipulation during microsclerotia formation by V. dahliae. Recently, we described the characterization of the first microbiome-manipulating effectors secreted by V. dahliae, VdAve1 and VdAMP2 (18). VdAve1 is a ubiquitously expressed bactericidal effector that promotes V. dahliae host colonization through the selective manipulation of host microbiota in the roots as well as in the xylem by suppressing microbial antagonists. Moreover, VdAve1 is also expressed in the soil biome, where it similarly contributes to niche colonization. Intriguingly, VdAMP2 is exclusively expressed in soil and, like VdAve1, exerts antibacterial activity that contributes to niche establishment. Interestingly, VdAMP2 and VdAve1 display divergent activity spectra and, therefore, likely complement each other for optimal soil colonization. In decaying host tissue, neither VdAve1 nor VdAMP2 are expressed, yet VdAMP3 expression occurs. Collectively, our findings for VdAve1, VdAMP2, and VdAMP3 demonstrate that V. dahliae dedicates a substantial part of its catalog of effector proteins toward microbiome manipulation and that each of these effectors act in a life stage–specific manner.
The life stage–specific exploitation of the in planta secreted antimicrobial effectors VdAve1 and VdAMP3 is well reflected by their antimicrobial activities and by the microbiota of the niches in which they act. Contrary to previous V. dahliae transcriptome analyses that repeatedly identified VdAve1 as one of the most highly expressed effector genes in planta (17, 38–40), we detected a repression of the effector gene in decomposing N. benthamiana tissues (Fig. 1 B and C). Characterization of the antimicrobial activity exerted by VdAve1 previously uncovered that the protein exclusively affects bacteria and does not impact fungi (18). Thanks to their ability to produce a wide diversity of hydrolytic enzymes, fungi are the primary decomposers of plant debris on earth (44). The phyllosphere of plants comprises a diversity of fungi (49–51). Importantly, upon plant senescence, these fungi are provided the first access to decaying material on which they can act opportunistically once host immune responses have faded. Accordingly, we detected an increased abundance of fungi in the phyllosphere of the decomposing N. benthamiana plants diseased by V. dahliae when compared with healthy plants (Fig. 4B). The observed repression of VdAve1 and the subsequent induction of VdAMP3 in a niche in which V. dahliae encounters more fungal competition underscores the notion that V. dahliae tailors the expression of its microbiome-manipulating effectors according to the various microbiota that it encounters during the different life stages. Along these lines, it is tempting to speculate that during saprotrophism in soil, V. dahliae exploits antimicrobial effector proteins to ward off other eukaryotic competitors including soil-dwelling parasites such as fungivorous nematodes or protists. However, evidence for this hypothesis is presently lacking.
Antimicrobial resistance in bacteria and fungi is posing an increasing threat to human health. Possibly, microbiome-manipulating effectors represent a valuable source for the identification and development of novel antimicrobials that can be deployed to treat microbial infections. Arguably, our findings that microbiome-manipulating effectors secreted by plant pathogens also comprise antifungal proteins open up opportunities for the identification and development of antimycotics. Most fungal pathogens of mammals are saprophytes that generally thrive in soil or decaying organic matter but can opportunistically cause disease in immunocompromised patients (52–54). Azoles are an important class of antifungal agents that are used to treat fungal infections in humans. Unfortunately, agricultural practices involving massive spraying of azoles to control fungal plant pathogens, but also the extensive use of azoles in personal care products, ultraviolet stabilizers, and anticorrosives in aircrafts, for instance, gives rise to an enhanced evolution of azole resistance in opportunistic pathogens of mammals in the environment (52, 55). For instance, azole resistant Aspergillus fumigatus strains are ubiquitous in agricultural soils and in decomposing crop waste material, where they thrive as saprophytes (56, 57). Thus, fungal pathogens of mammals, like A. fumigatus, comprise niche competitors of fungal plant pathogens. Hence, we speculate that, like V. dahliae, other plant pathogenic fungi may also carry potent antifungal proteins in their effector catalogs that aid in niche competition with these fungi. Possibly, the identification of such effectors could contribute to the development of novel antimycotics.
Materials and Methods
Gene Expression Analyses.
In vitro cultivation of V. dahliae strain JR2 for analysis of VdAMP3 and Chr6g02430 expression was performed as described previously (24). Additionally, for in planta expression analyses, total RNA was isolated from individual leaves or complete N. benthamiana plants harvested at different time points after V. dahliae root dip inoculation. To induce microsclerotia formation, N. benthamiana plants were harvested at 22 dpi and incubated in sealed plastic bags (volume = 500 mL) for 8 d prior to RNA isolation. RNA isolations were performed using the the Maxwell 16 LEV Plant RNA Kit (Promega). Real-time PCR was performed as described previously using the primers listed in SI Appendix, Table 3 (17).
Generation of V. dahliae Mutants.
The VdAMP3 deletion and complementation mutants, as well as the eGFP expression mutant, were generated as described previously using the primers listed in SI Appendix, Table 3 (18). To generate the VdAMP3 complementation construct, the VdAMP3 coding sequence was amplified with flanking sequences (∼0.9 kb upstream and ∼0.8 kb downstream) and cloned into pCG (58). Finally, the construct was used for Agrobacterium tumefaciens–mediated transformation of V. dahliae as described previously (59). In vitro growth and microsclerotia production of the VdAMP3 deletion mutant were tested and quantified as described previously (18).
In Vitro Microbial Growth Assays.
Bacterial isolates were grown on lysogeny broth agar at 28 °C. Single colonies were selected and grown overnight in low-salt lysogeny broth (LB) (10 g/L tryptone, 5 g/L yeast extract, and 0.5 g/L sodium chloride) at 28 °C while shaking at 200 rpm. Overnight cultures were resuspended to optical density (OD)600 = 0.025 in fresh low-salt LB supplemented with 20 μM VdAMP3 or ultrapure water (Milli-Q). In vitro growth was quantified using a CLARIOstar plate reader (BMG Labtech) as described previously (18).
Fungal isolates were grown on potato dextrose agar (PDA) at 22 °C. For yeasts, single colonies were selected and grown overnight in 0.05× potato dextrose broth (PDB) at 28 °C while shaking at 200 rpm. Overnight cultures were resuspended to OD600 = 0.01 in fresh 5% PDB supplemented with ultrapure water (Milli-Q), 5 μM VdAMP3, or 5 μM VdAMP3 that was incubated in a PCR thermocycler at 95 °C for 16 h. Alternatively, for filamentous fungi, spores were harvested from PDA and suspended in 5% PDB supplemented with 5 μM VdAMP3 or ultrapure water (Milli-Q) to a final concentration of 104 spores/mL. Next, 200 μL of the fungal suspensions was aliquoted in clear 96-well flat-bottom polystyrene tissue-culture plates. Plates were incubated at 28 °C, and fungal growth was imaged using an SZX10 stereo microscope (Olympus) with EP50 camera (Olympus).
Microbiome Analysis.
Inoculation and incubation of N. benthamiana plants were performed as described above. Subsequent genomic DNA isolation and V. dahliae biomass quantification were performed as previously described using the primers listed in SI Appendix, Table 3 (60). After 4 wk of incubation in plastic bags at room temperature in the dark, the decaying N. benthamiana phyllosphere samples colonized by V. dahliae WT and the VdAMP3 deletion mutant were collected. The phyllospheres of fresh 3-wk-old N. benthamiana plants were included as controls. All samples were flash-frozen in liquid nitrogen and ground using mortar and pestle, and genomic DNA was isolated using the DNeasy PowerSoil Kit (Qiagen). Sequencing libraries were prepared using the TruSeq DNA Nano kit (Illumina), and paired-end 150-bp sequencing was performed on the Illumina NextSeq500 platform at the Utrecht Sequencing Facility.
The sequencing data were processed as follows. Quality control of the reads, adapter trimming, and removal of N. benthamiana reads were performed with the ATLAS metagenomic workflow using the default parameters of the configuration file (61). Reads of the different samples were combined and assembled using metaSPAdes (used k-mer sizes: 21, 33, and 55) to obtain a single metagenome cross-assembly (62). Subsequently, the cross-assembled contigs were taxonomically classified using Contig Annotation Tool and binned per genus (63). The reads of the individual samples were mapped to the binned contigs using Burrows-Wheeler Aligner Maximal Exact Match (64). Next, the mapping files were converted to bam format using SAMtools (65) version 1.10, and the number of reads mapped to the contigs of a single genus were converted to “reads per million” for the individual samples. The generated taxonomy table and abundance table were subsequently transformed into a phyloseq (66) object (version 1.30.0) in R (version 3.6.1) to facilitate analysis of the microbiomes. The alpha diversity (Shannon index) and beta diversity (Bray–Curtis dissimilarity) were determined as described previously (66, 67). The DESeq2 extension of phyloseq was used to identify differentially abundant microbial genera (68). To this end, a parametric model was applied to the data and a negative binomial Wald test was used to test for significant differential abundance.
Fungal Cocultivation Assays.
Fungal isolates were grown on PDA at room temperature. For D. vanrijiae, M. amylolytica, and C. jadinii, single colonies were selected and grown overnight in 5% PDB at 28 °C while shaking at 200 rpm. The overnight cultures of D. vanrijiae and M. amylolytica were resuspended to OD600= 0.0001 in fresh 5% PDB. For the synthetic communities, D. vanrijiae, M. amylolytica, and C. jadinii were resuspended to OD600 = 0.001 and spores of C. militaris and T. viride were harvested from PDA and resuspended to 104 spores/mL. Next, equal volumes of the various fungal suspensions were mixed to obtain two syncoms: (A) D. vanrijae, M. amylolytica, and C. militaris and (B) D. vanrijae, M. amylolytica, C. jadinii, and T. viride, which were stored at −20 °C in 5% PDB supplemented with 10% glycerol (wt/vol). Upon use, the syncoms were thawed at room temperature and diluted 10× (A) or 25× (B) in fresh 5% PDB, after which they were mixed with V. dahliae. To this end, conidiospores of V. dahliae strain JR2 and the VdAMP3 deletion and complementation mutants were harvested from PDA plates and diluted in ultrapure water (Milli-Q) to a final concentration of 104 or 105 conidiospores/mL. Next, 150 μL of the fungal suspensions was mixed with 150 μL of the V. dahliae condiospore suspensions (104 conidiospores/mL for cultivation with syncom A or M. amylolytica and 105 conidiospores/mL for cultivation syncom B or D. vanrijae) in clear 24-well flat-bottom polystyrene tissue-culture plates. Finally, after six to nine d of incubation at 22 °C, fungal growth was imaged using an SZX10 stereo microscope (Olympus) with EP50 camera (Olympus). The number of microsclerotia formed by the different V. dahliae strains was quantified through counting.
Supplementary Material
Acknowledgments
B.P.H.J.T. is supported by the Research Council Earth and Life Sciences of the Netherlands Organization of Scientific Research (NWO). B.P.H.J.T. acknowledges funding by the Alexander von Humboldt Foundation in the framework of an Alexander von Humboldt Professorship endowed by the German Federal Ministry of Education, and research is furthermore supported by the Deutsche Forschungsgemeinschaft (German Research Foundation) under Germany’s Excellence Strategy—EXC 2048/1—Project identification: 390686111. We thank the Utrecht Sequencing Facility, subsidized by the University Medical Center Utrecht, Hubrecht Institute, Utrecht University, and The Netherlands X-omics Initiative (NWO Project 184.034.019) for providing sequencing service.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110968118/-/DCSupplemental.
Data Availability
The metagenomics data have been deposited in the National Center for Biotechnology Information GenBank database under BioProject accession no. PRJNA728211 (69) (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA728211).
References
- 1.Demoling F., Figueroa D., Bååth E., Comparison of factors limiting bacterial growth in different soils. Soil Biol. Biochem. 39, 2485–2495 (2007). [Google Scholar]
- 2.Katz L., Baltz R. H., Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 43, 155–176 (2016). [DOI] [PubMed] [Google Scholar]
- 3.van der Meij A., Worsley S. F., Hutchings M. I., van Wezel G. P., Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 41, 392–416 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Rovenich H., Boshoven J. C., Thomma B. P. H. J., Filamentous pathogen effector functions: Of pathogens, hosts and microbiomes. Curr. Opin. Plant Biol. 20, 96–103 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Snelders N. C., Kettles G. J., Rudd J. J., Thomma B. P. H. J., Plant pathogen effector proteins as manipulators of host microbiomes? Mol. Plant Pathol. 19, 257–259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giraldo M. C., Valent B., Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11, 800–814 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Lo Presti L., et al. , Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66, 513–545 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Berendsen R. L., Pieterse C. M. J., Bakker P. A. H. M., The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Dimkpa C., Weinand T., Asch F., Plant-rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 32, 1682–1694 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Castrillo G., et al. , Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzpatrick C. R., et al. , Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. U.S.A. 115, E1157–E1165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai Y., et al. , Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Durán P., et al. , Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175, 973–983.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lombardi N., et al. , Root exudates of stressed plants stimulate and attract Trichoderma soil fungi. Mol. Plant Microbe Interact. 31, 982–994 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Berendsen R. L., et al. , Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 12, 1496–1507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrión V. J., et al. , Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 366, 606–612 (2019). [DOI] [PubMed] [Google Scholar]
- 17.de Jonge R., et al. , Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 109, 5110–5115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snelders N. C., et al. , Microbiome manipulation by a soil-borne fungal plant pathogen using effector proteins. Nat. Plants 6, 1365–1374 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Inderbitzin P., et al. , Phylogenetics and taxonomy of the fungal vascular wilt pathogen Verticillium, with the descriptions of five new species. PLoS One 6, e28341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klosterman S. J., et al. , Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 7, e1002137 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mol L., Van Riessen H. W., Effect of plant roots on the germination of microsclerotia of Verticillum dahliae. Eur. J. Plant Pathol. 101, 673–678 (1995). [Google Scholar]
- 22.Klosterman S. J., Atallah Z. K., Vallad G. E., Subbarao K. V., Diversity, pathogenicity, and management of verticillium species. Annu. Rev. Phytopathol. 47, 39–62 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Schnathorst W. C., “Life cycle and epidemiology of Verticillium” in Fungal Wilt Diseases of Plants, M. E. Mace, A. A. Bell, C. H. Beckman, Eds. (Elsevier, 1981), vol. 82. [Google Scholar]
- 24.Xiong D., et al. , Deep mRNA sequencing reveals stage-specific transcriptome alterations during microsclerotia development in the smoke tree vascular wilt pathogen, Verticillium dahliae. BMC Genomics 15, 324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dias Rde. O., Franco O. L., Cysteine-stabilized αβ defensins: From a common fold to antibacterial activity. Peptides 72, 64–72 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shafee T. M. A., Lay F. T., Hulett M. D., Anderson M. A., The defensins consist of two independent, convergent protein superfamilies. Mol. Biol. Evol. 33, 2345–2356 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Dal Peraro M., van der Goot F. G., Pore-forming toxins: Ancient, but never really out of fashion. Nat. Rev. Microbiol. 14, 77–92 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Mygind P. H., et al. , Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature 437, 975–980 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Schneider T., et al. , Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science 328, 1168–1172 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Oeemig J. S., et al. , Eurocin, a new fungal defensin: Structure, lipid binding, and its mode of action. J. Biol. Chem. 287, 42361–42372 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grigoriev I. V., et al. , MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 42, D699–D704 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spatafora J. W., et al. , A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108, 1028–1046 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar S., Stecher G., Suleski M., Hedges S. B., TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Heckman D. S., et al. , Molecular evidence for the early colonization of land by fungi and plants. Science 293, 1129–1133 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Berbee M. L., Taylor J. W., “Fungal molecular evolution: Gene trees and geologic time” in Systematics and Evolution, D. J. McLaughlin, E. G. McLaughlin, P. A. Lemke, Eds. (Springer, 2001), pp. 229–245. [Google Scholar]
- 37.Morris J. L., et al. , The timescale of early land plant evolution. Proc. Natl. Acad. Sci. U.S.A. 115, E2274–E2283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faino L., de Jonge R., Thomma B. P. H. J., The transcriptome of Verticillium dahliae-infected Nicotiana benthamiana determined by deep RNA sequencing. Plant Signal. Behav. 7, 1065–1069 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Depotter J., et al. , The interspecific fungal hybrid Verticillium longisporum displays subgenome-specific gene expression. mBio 12: e01496-01421 (2021). [DOI] [PMC free article] [PubMed]
- 40.Gibriel H., Li J., Zhu L., Seidl M. F., Thomma B. P. H. J., Verticillium dahliae strains that infect the same host plant display highly divergent effector catalogs. bioRxiv [Preprint] (2019). https://www.biorxiv.org/content/10.1101/528729v1. Accessed 10 June 2021.
- 41.Duressa D., et al. , RNA-seq analyses of gene expression in the microsclerotia of Verticillium dahliae. BMC Genomics 14, 607 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klimes A., Dobinson K. F., A hydrophobin gene, VDH1, is involved in microsclerotial development and spore viability in the plant pathogen Verticillium dahliae. Fungal Genet. Biol. 43, 283–294 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Smith H. C., The morphology of Verticillium albo-atrum, V. dahliae, and V. tricorpus. N. Z. J. Agric. Res. 8, 450–478 (1965). [Google Scholar]
- 44.Mäkelä M. R., Donofrio N., de Vries R. P., Plant biomass degradation by fungi. Fungal Genet. Biol. 72, 2–9 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Voříšková J., Baldrian P., Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J. 7, 477–486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korkama-Rajala T., Müller M. M., Pennanen T., Decomposition and fungi of needle litter from slow- and fast-growing Norway spruce (Picea abies) clones. Microb. Ecol. 56, 76–89 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Osono T., Role of phyllosphere fungi of forest trees in the development of decomposer fungal communities and decomposition processes of leaf litter. Can. J. Microbiol. 52, 701–716 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Osono T., Phyllosphere fungi on leaf litter of Fagus crenata: Occurrence, colonization, and succession. Can. J. Bot. 80, 460–469 (2002). [Google Scholar]
- 49.Sapkota R., Knorr K., Jørgensen L. N., O’Hanlon K. A., Nicolaisen M., Host genotype is an important determinant of the cereal phyllosphere mycobiome. New Phytol. 207, 1134–1144 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Gomes T., Pereira J. A., Benhadi J., Lino-Neto T., Baptista P., Endophytic and epiphytic phyllosphere fungal communities are shaped by different environmental factors in a Mediterranean ecosystem. Microb. Ecol. 76, 668–679 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Arnold A. E., Understanding the diversity of foliar endophytic fungi: Progress, challenges, and frontiers. Fungal Biol. Rev. 21, 51–66 (2007). [Google Scholar]
- 52.Verweij P. E., Snelders E., Kema G. H. J., Mellado E., Melchers W. J. G., Azole resistance in Aspergillus fumigatus: A side-effect of environmental fungicide use? Lancet Infect. Dis. 9, 789–795 (2009). [DOI] [PubMed] [Google Scholar]
- 53.May R. C., Stone N. R. H., Wiesner D. L., Bicanic T., Nielsen K., Cryptococcus: From environmental saprophyte to global pathogen. Nat. Rev. Microbiol. 14, 106–117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein E., Ofek M., Katan J., Minz D., Gamliel A., Soil suppressiveness to fusarium disease: Shifts in root microbiome associated with reduction of pathogen root colonization. Phytopathology 103, 23–33 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Buil J. B., et al. , The fading boundaries between patient and environmental routes of triazole resistance selection in Aspergillus fumigatus. PLoS Pathog. 15, e1007858 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang J., et al. , Dynamics of Aspergillus fumigatus in azole fungicide-containing plant waste in the Netherlands (2016–2017). Appl. Environ. Microbiol. 87, e02295-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schoustra S. E., et al. , Environmental hotspots for azole resistance selection of Aspergillus fumigatus, the Netherlands. Emerg. Infect. Dis. 25, 1347–1353 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou L., Zhao J., Guo W., Zhang T., Functional analysis of autophagy genes via Agrobacterium-mediated transformation in the vascular Wilt fungus Verticillium dahliae. J. Genet. Genomics 40, 421–431 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Santhanam P., “Random insertional mutagenesis in fungal genomes to identify virulence factors” in Plant Fungal Pathogens, Thomma B. P. H. J., Bolton M. D., Eds. (Springer, 2012), pp. 509–517. [DOI] [PubMed] [Google Scholar]
- 60.Song Y., et al. , Transfer of tomato immune receptor Ve1 confers Ave1-dependent Verticillium resistance in tobacco and cotton. Plant Biotechnol. J. 16, 638–648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kieser S., Brown J., Zdobnov E. M., Trajkovski M., McCue L. A., ATLAS: A Snakemake workflow for assembly, annotation, and genomic binning of metagenome sequence data. BMC Bioinformatics 21, 257 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nurk S., Meleshko D., Korobeynikov A., Pevzner P. A., metaSPAdes: A new versatile metagenomic assembler. Genome Res. 27, 824–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.von Meijenfeldt F. A. B., Arkhipova K., Cambuy D. D., Coutinho F. H., Dutilh B. E., Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol. 20, 217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint] (2013). https://arxiv.org/abs/1303.3997. Accessed 10 June 2021.
- 65.Li H., et al. , 1000 Genome Project Data Processing Subgroup, The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McMurdie P. J., Holmes S., phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Callahan B. J., Sankaran K., Fukuyama J. A., McMurdie P. J., Holmes S. P., Bioconductor Workflow for Microbiome Data Analysis: From raw reads to community analyses. F1000 Res. 5, 1492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.N. C. Snelders et al. , Phyllosphere microbiome of Nicotiana benthamiana plants infected by Verticillium dahliae. National Center for Biotechnology Information GenBank database. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA728211. Deposited 8 May 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metagenomics data have been deposited in the National Center for Biotechnology Information GenBank database under BioProject accession no. PRJNA728211 (69) (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA728211).