Abstract
Two-component systems (TCSs) are a ubiquitous family of signal transduction pathways that enable bacteria to sense and respond to diverse physical, chemical, and biological stimuli outside and inside the cell. Synthetic biologists have begun to repurpose TCSs for applications in optogenetics, materials science, gut microbiome engineering, and soil nutrient biosensing, among others. New engineering methods including genetic refactoring, DNA-binding domain swapping, detection threshold tuning, and phosphorylation cross-talk insulation are being used to increase the reliability of TCS sensor performance and tailor TCS signaling properties to the requirements of specific applications. There is now potential to combine these methods with large-scale gene synthesis and laboratory screening to discover the inputs sensed by many uncharacterized TCSs and develop a large new family of genetically-encoded sensors that respond to an unrivaled breadth of stimuli.
Introduction
Synthetic biologists program cells to sense and respond to extra- and intracellular stimuli for applications in medicine[1], agriculture[2], chemicals synthesis[3] and many other areas. For example, a probiotic strain of E. coli that naturally colonizes tumors was recently engineered to lyse and release immune checkpoint-inhibiting nanobodies upon reaching high intra-tumoral density[4]. By enabling local delivery of these potent biologic drugs, this approach could improve the efficacy of cancer immunotherapy while decreasing side effects. In other work, researchers are engineering bacteria to sense stresses imposed by heterologous metabolic and genetic pathways and activate the expression of enzymes or stress-response systems that ameliorate these effects [5]. Such ‘host-aware’ design strategies could increase the stabilities of engineered genetic systems and improve the yields of industrial fermentations, among other benefits.
To program cells to sense and respond to stimuli, synthetic biologists utilize genetically-encoded sensors. The canonical synthetic biological sensor is an RNA or protein that binds to a ligand or perceives a biochemical or physical property of the environment and responds by altering gene expression. Riboswitches are a well-studied family of RNA-based sensors. The prototypical riboswitch is encoded on the 5’ end of a messenger RNA and comprises an aptamer domain linked to a gene regulatory domain. In the presence of a cognate stimulus (a.k.a. input), the aptamer domain undergoes a conformational rearrangement that is transmitted to the gene regulatory domain, resulting in a change in the level of expression of the gene encoded on the mRNA in cis [6]. Riboswitch inputs are often involved in fundamental biological processes and include metal ions, molecules involved in RNA metabolism, and amino acids[7]. The gene regulatory domains of riboswitches most often modulate transcription of the downstream mRNA by exposing or obscuring transcriptional terminators, translation of the downstream open reading frame by exposing or obscuring a ribosome binding site, or mRNA splicing[8]. Synthetic riboswitches that sense inputs not known to be detected by their natural counterparts have been engineered by rational design[9-11], directed evolution[12], and machine learning[13]. One notable recent study utilized a physics-based model of riboswitch structure and function to engineer translation-regulating sensors of theophylline, tetramethylrosamine, fluoride, dopamine, thyroxine, and 2,4 dinitrotoluene with activation ratios up to several hundred-fold [14]. Though good progress is being made on riboswitch design, the repertoire of inputs that these RNA-based sensors can detect remains small. On the other hand, nature has provided a wealth of protein-based sensors that sense diverse inputs relevant to synthetic biology applications.
One-component systems (OCSs) are the largest family of bacterial signal transduction pathways [15,16] and the most frequently-used family of genetically-encoded sensors. The typical OCS comprises an allosteric transcription factor (aTF) and a target (a.k.a. output) promoter. The aTF usually consists of an N-terminal sensor domain linked to a C-terminal DNA-binding domain (DBD). In the presence of the input, the aTF sensor domain allosterically modulates the activity of the DBD, and thus the rate of transcription from the output promoter. A handful of well-characterized OCSs (e.g. LacI, TetR, AraC, LuxR) are often used as model sensors in synthetic biology studies. Recent efforts based on genome mining, directed evolution, protein engineering, and computational protein design have generated several dozen new OCS sensors, including those that detect inputs linked to bacterial or host physiology [17-21]. It is likely that many new OCSs will be characterized and engineered for synthetic biology applications going forward.
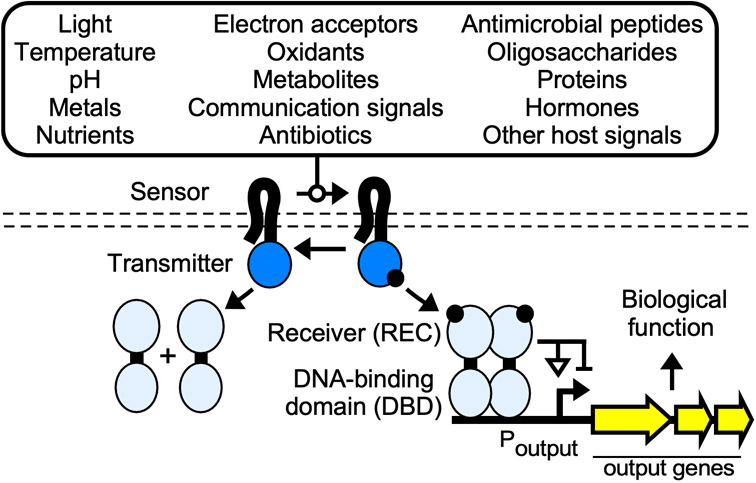
Two-component systems (TCSs) are the largest class of multi-step signal transduction pathways in nature[22] and an important family of sensors for synthetic biology. The classical TCS sensor comprises a sensor histidine kinase (SHK), a response regulator (RR), and an output promoter (Fig. 1). A given SHK contains an N-terminal sensor domain linked to C-terminal transmitter domain, often by a transmembrane region[23]. The presence of the input causes the SHK sensor domain to undergo a conformational rearrangement that is relayed to the transmitter domain, typically increasing kinase activity. In the activated state, the transmitter domain phosphorylates the partner RR on a conserved aspartate residue within an N-terminal receiver (REC) domain. Similar to aTFs, most RRs contain C-terminal DBDs that regulate transcription[24]. Phosphorylation induces a conformational switch that activates the DBD, often via REC-mediated homodimerization[25-27]. Most SHK transmitter domains also dephosphorylate their cognate RRs in the absence of input, thus de-activating the TCS response. A spectrum of signal transduction pathways that elaborate upon the core TCS architecture are present in bacteria, archaea, and non-animal eukaryotes[28-30]. However, the prototypical TCS is predominantly found in bacteria.
Figure 1.
Bacterial two-component systems and their function. Inputs are shown inside of the rounded box. Sensor histidine kinase (SHK): Black/dark blue. Phosphoryl group: black circle. Response regulator (RR): light blue. Poutput: output promoter.
TCS performance features
Though OCSs are simpler and more abundant, TCSs offer several advantages for synthetic biology. First, SHKs can be membrane-bound or cytoplasmic while OCSs are almost exclusively cytoplasmic[15]. Accordingly, TCSs can sense extracellular, intra-membrane, or intracellular inputs while OCSs typically sense intracellular inputs. From a sensor design perspective, a transporter must be co-expressed alongside an OCS if the input does not naturally diffuse or is not naturally transported across the membrane [31]. As transporters are not readily available for many compounds (e.g. large molecular weight species), OCSs tend to sense a more restricted range of inputs than TCSs. Second, the bi-functional kinase/phosphatase activity of SHKs makes TCS output signals (phosphorylated RR and thus transcription rate) relatively insensitive to changes in the expression levels of SHKs and RRs[32,33]. This built-in robustness can buffer TCS responses against gene expression noise or fluctuations in SHK and RR expression that arise from changing growth conditions. Third, we recently demonstrated that SHK phosphatase activity acts as a built-in knob for tuning TCS detection threshold[34]. In particular, by introducing transmitter domain mutations that specifically reduce SHK phosphatase activity, TCSs can be made to respond to their inputs at up to two orders of magnitude lower concentrations. Furthermore, we demonstrated that the first variable residue in the transmitter domain GXGXG motif, which is present in 64% of SHKs, can be mutated to different hydrophobic residues to tune the detection thresholds of TCSs even in the absence of well-characterized phosphatase-altering mutations. This phosphatase tuning method is simpler to implement than computational design and directed evolution, which are frequently used to tune OCS sensitivity.
TCSs have evolved to sense a remarkable assortment of inputs. Classes of known TCS inputs include light, temperature, pH, metals, nutrient availability, respiratory electron acceptors, oxidizing agents, small molecule metabolites, inter-bacterial communication signals, antibiotics, antimicrobial peptides, oligosaccharides, proteins, hormones, and other host-derived signals (Table 1). Some SHKs are highly specific for a single input. For example, E. coli NarX and Shewanella halifaxensis ThsS are activated by the terminal electron acceptors nitrate (NO3−) and thiosulfate (S2O32−), but discriminate against closely-related compounds such as nitrite (NO2−) and tetrathionate (S4O62−), respectively[35,36]. Other SHKs sense multiple inputs characteristic of a specific environment. For example, Salmonella Typhimurium PhoQ is activated by low divalent cation concentrations, acidic pH, and antimicrobial peptides – three distinct stimuli that are likely to be encountered by the bacterium during the infection of a host[37]. SHKs can detect multiple inputs via multiple binding sites in a single sensor domain[37], or via multiple sensor domains[38]. Finally, many SHKs sense general phenomena such as membrane disruption that occur in the presence of a wide range of inputs[39].
Table 1.
Available two-component system (TCS) sensors. Often, numerous TCSs sense a given input and a given response regulator regulates numerous output promoters. In these cases, one well-characterized TCS or output promoter is listed for simplicity.
| Input | Native organism(s) | SHK | RR | Output promoter | Performance notes | Reference |
|---|---|---|---|---|---|---|
| Light | ||||||
| UV-violet light | Synechocystis PCC6803 | UirS | UirR | PcsiR1 | 5-fold activationAlso reported to sense ethylene | [54] |
| Blue light | B. subtilis, B. japonicum | YF1 | FixJ | PfixK2 | Chimeric YtvA-FixL SHK. 460-fold activation when coupled to transcriptional inverter (pDushk system) | [80] |
| Green light | Synechocystis PCC6803 | CcaSmini#10 | CcaR | PcpcG2-172 | 600-fold activation | [81] |
| Red light | Synechocystis PCC6803, E. coli | Cph8* | OmpR | PompF112 | 80-fold de-activation | [60] |
| Near infrared light | Rps. palustris, B. japonicum BTAi1 | BphP1 | PpsR2 | PBr_crtE | Signaling based on RR sequestration. 2-fold activation | [62] |
| Temperature | ||||||
| 25°C | B. subtilis | DesK | DesR | Pdes | Fully activated and repressed at 25°C and 37°C, respectively | [82] |
| pH | ||||||
| Acidic pH (<6.2) | S. oneidensis, B. subtilis | SO_4387 | SO_4388REC-PsdRDBD 137 | PpsdA110 | Used to sense small intestinal inflammation in mice | [52,61] |
| Acidic pH (extracellular) | H. pylori | ArsS | ArsR | PamiE, PamiF | Activated in the human stomach | [83,84] |
| Metals | ||||||
| As3+ (extracellular) | A. tumefaciens | AioS | AioR | PaioB | Requires AioX inner membrane accessory protein | [85] |
| Ca2+ (extracellular) | P. aeruginosa PAO1 | CarS | CarR | PcarO | CarS is related to PhoQ but senses the presence not absence of the divalent cation | [86] |
| Cu+ (extracellular) | E. coli | CusS | CusR | PcusC | Also activated by Ag (I) | [87-89] |
| Cu2+ (extra- and intracellular) | Synechocystis PCC 6803 | CopS | CopR | PcopM | CopS partially localizes to thylakoid membranes | [90] |
| Cu2+ (extracellular) | M. xanthus | CorS | CorR | PcuoA | [91] | |
| Fe2+, Fe3+ (extracellular) | S. marcescens | RssA | RssB | PpvcA | Activity can be tuned with the natural product 2-isocyano-6,7-dihydroxycoumarin | [92] |
| K+ (extracellular, intracellular) | E. coli | KdpD | KdpE | PkdpF | Inactivated by K+ | [93,94] |
| Ni2+ | Synechocystis PCC6803 | NrsS | NrsR | PnrsB | 10-fold activation. Also weakly activated by Co2+ | [95] |
| U | E. coli | UzcS | UzcR | PurcA | Sensitivity and specificity improved by coupling to UrpRS via AND gate | [96] |
| Zn2+ | E. coli | ZraS | ZraR | PzraP | PzraP is σ54-dependent | [97] |
| Nutrient availability | ||||||
| Nitrogen limitation | E. coli | NtrB | NtrC | Pddp | Requires the accessory protein GlnBog1. | [98] |
| Respiratory electron acceptors | ||||||
| O2 | S. meliloti | FixL | FixJ | PnifA | O2 inhibits pathway. O2 binds FixL via covalently attached heme. | [99] |
| Thiosulfate | S. halifaxensis | ThsS | ThsR | PphsA342 | Activated proportional to DSS-induced inflammation in mouse colon | [36] |
| Tetrathionate | S. baltica | TtrS | TtrR | PttrB185-269 | 100-fold dynamic range | [36] |
| Nitrate | E. coli | NarX | NarLREC-YdfIDBD 131 | PydfJ115 | 1300-fold activation in B. subtilis. | [61] |
| Nitrate OR Nitrite | E. coli | NarQ | NarP | PnrfA | NarQ also responds to nitrate. NarX cross-phosphorylates NarP. NarL binds NarP output promoters. | [100] |
| Trimethyl amine N-oxide (TMAO) | E. coli | TorS | TorRREC-PsdRDBD 137 | PpsdA110 | DBD swapping eliminates O2 cross repression at native output promoter. Requires periplasmic TorT accessory protein | [61] |
| Oxidizing agents | ||||||
| O2, H2O2, NO | S. aureus | AirS | AirR | PcrtO | AirS requires a [2Fe–2S]2+ cluster | [101,102] |
| Small molecule metabolites | ||||||
| α-ketoglutarate (extracellular) | P. aeruginosa PAO1 | MifS | MifR | PPA5530 | 10-fold activation. Weak response to glutarate | [103] |
| Butanol | C. acetobutylicum | BtrK | BtrR | PbtrT | Involved in butanol tolerance | [104] |
| C4-dicarboxylates (extracellular) | E. coli | DcuS | DcuR | PfrdA | 22-fold activation | [105] |
| Citrate | K. pneumoniae | CitA | CitB | PcitC | Requires anaerobic conditions | [106] |
| Fucose | E. coli | Fushk | FusR | Pz0461 | Fucose represses transcriptional output | [107] |
| Fumarate | E. coli | DcuSZ | OmpR | PompC | Chimeric DcuS-EnvZ SHK. < 5-fold activation | [66] |
| Glucose-6-phosphate | E. coli | UhpB | UhpA | PuhpT99 | Requires UhpC inner-membrane accessory protein | [61,108] |
| l-Glutamate | P. aeruginosa PAO1 | AauS | AauR | PaatJ | Very weakly activated by aspartate, glutamine, and asparagine | [109] |
| Heme (extracellular) | S. aureus | HssS | HssR | PhrtA | >100-fold activation | [110] |
| Indole | E. coli | BaeS | BaeR | PacrD | Presence of CpxAR TCS amplified the indole response | [111] |
| Malate | B. subtilis | YufL | YufM | PmaeN381 | 100-fold activation | [112] |
| Methanol | P. denitrificans/E. coli | FlhS-EnvZ | OmpR | PompC | 2-fold activation | [65] |
| Pyruvate (extracellular) | E. coli | BtsS | BtsR | PyjiY | Active in uropathogenic E. coli during urinary-tract infections | [113] |
| Ribose | E. coli | Trg-EnvZ | OmpR | PompC | Chimeric SHK. 20-fold activation | [71] |
| d-xylose (extracellular) | C. beijerinckii | LytS | YesN | PxylF | d-xylose recognized by outer membrane transporter-like protein XylFII | [114] |
| Styrene | Pseudomonas sp.strain Y2 | StyS | StyR | PstyA | StyS has non-canonical HK-REC-HK structure | [115] |
| Inter-bacterial communication signals | ||||||
| AIP-I (autoinducer peptide) | S. aureus | AgrC | AgrA | PagrB | AIP-II inhibits AgrC kinase activity | [116] |
| CAI-1 ((S)-3-hydroxytridecan-4-one) | V. cholerae | CqsS | LuxO | Ptpqrr4 | Requires intermediate phosphotransfer protein LuxU | [117] |
| CSP (Competence Stimulating Peptide) | S. gordonii | ComD | ComE | PcomC | [118] | |
| ComX (extracellular pheromone) | B. subtilis | ComP | ComA | PsrfA | [119] | |
| Antibiotics | ||||||
| β-lactams | V. cholerae | VxrA | VxrB | PmurJ | Generally activated by cell envelope damage | [120] |
| Linearmycins (intra-membrane) | B. subtilis | LnrJ | LnrK | PlnrL | Can also detect the antifungal polyene amphotericin B. | [121,122] |
| Vancomycin (extracellular) | S. coelicolor | VanS | VanR | PvanJ | Phosphorylated VanR additionally activates the VanSR operon | [123] |
| Antimicrobial peptides | ||||||
| Antimicrobial peptides produced by the oral pathogen S. mutans (extracellular) | Streptococcus sp.A12 | PcfK | PcfR | PpcfF | Nearly 100-fold activation | [124] |
| Bacitracin (extracellular) | B. subtilis | LiaS | LiaR | PliaI(opt) | 1000-fold activation. Activation occurs in a 1–2 h pulse. Requires accessory membrane protein LiaF. | [125] |
| Nisin (extracellular) | L. lactis | Nishk | NisR | PnisA | 1000-fold activation. | [126] |
| Subtilin (extracellular) | B. subtilis | SpaK | SpaR | PspaS | 110-fold induction | [126,127] |
| Oligosaccharides | ||||||
| Arabinogalactan | B. thetaiotaomicron | BT0267 | BT0267 | PBT0268 | BT0267 is a hybrid TCS wherein the SHK and RR are fused | [128] |
| Chondroitin sulfate | B. thetaiotaomicron | BT3334 | BT3334 | PBT3324 | BT3334 is a hybrid TCS | [128] |
| Mucin glycans | P. aeruginosa | GacS | GacA | PrsmY | Mucin glycans are sensed via the accessory histidine kinase RetS | [129] |
| Oligo-Arabinose | B. thetaiotaomicron | BT0366 | BT0366 | PBT0365 | BT0366 is a hybrid TCS | [130] |
| Proteins | ||||||
| PilA | P. aeruginosa | PilS | PilR | PpilA | PilA is the major Type IV pilin protein | [131] |
| Host signals | ||||||
| Antimicrobial peptides, divalent cation limitation, and acidic pH produced by mammalian hosts during infection | S. Typhimurium | PhoQ | PhoP | PvirK | 70-fold activation in E. coli | [132] |
| Epinephrine, norepinephrine | E. coli O157:H7 | QseC | QseB | PflhD | 2-fold activation by epinephrine. 1.5-fold repression by norepinephrine. Also activated by bacterial autoinducer 3. | [133] |
| Indole-3-acetic acid (auxin) | P. phytofirmans PsJN | IacS | IacR1 | PiacA | Dioxindole-3-acetic acid amplifies the signal | [134] |
| 2-isopentenyladenine (cytokinin) | X. campestris | PcrK | PcrR | PctrA | 3-fold induction | [135] |
| Phenolics, monosaccharides, and acidic pH present at plant wounds | A. tumefaciens | VirA | VirG | Pvir | Monosaccharide sensing requires the periplasmic accessory protein ChvE. | [136] |
| Trans-zeatin (cytokinin) | A. thaliana, E. coli | AQ4* | PhoP4* | PmgrB | AQ4* is a fusion of the sensing domain of A. thaliana AHK4 and E. coli PhoQ. This TCS is insulated against phosphorylation cross-talk with all E. coli TCSs | [70] |
Repurposing TCSs as sensors for synthetic biology
Researchers have begun to utilize TCSs as sensors for synthetic biology applications. For example, light-responsive TCSs (Table 1) have been used to spatially manipulate gene expression across two-dimensional bacterial lawns[40-42], program bacteria to perform the image processing function of edge detection[43], pattern biofilm deposition on ceramics, polystyrene, and cotton[44], introduce single-base pair edits in genomic DNA[45], characterize the input/output dynamics of transcriptional regulatory circuits[46], dynamically control metabolic pathway flux[47-49], and characterize how gut bacterial metabolite secretion can impact host health and longevity[50]. In another series of studies, TCSs activated by thiosulfate, tetrathionate, and acidic pH (Table 1) have been used to program bacteria to sense and report intestinal inflammation in mouse models of colitis and Crohn’s Disease [36,51,52]. Such diagnostic gut bacteria could be advanced to enable long-term monitoring and treatment of inflammatory bowel diseases with less invasiveness and fewer side-effects compared to current standards of care.
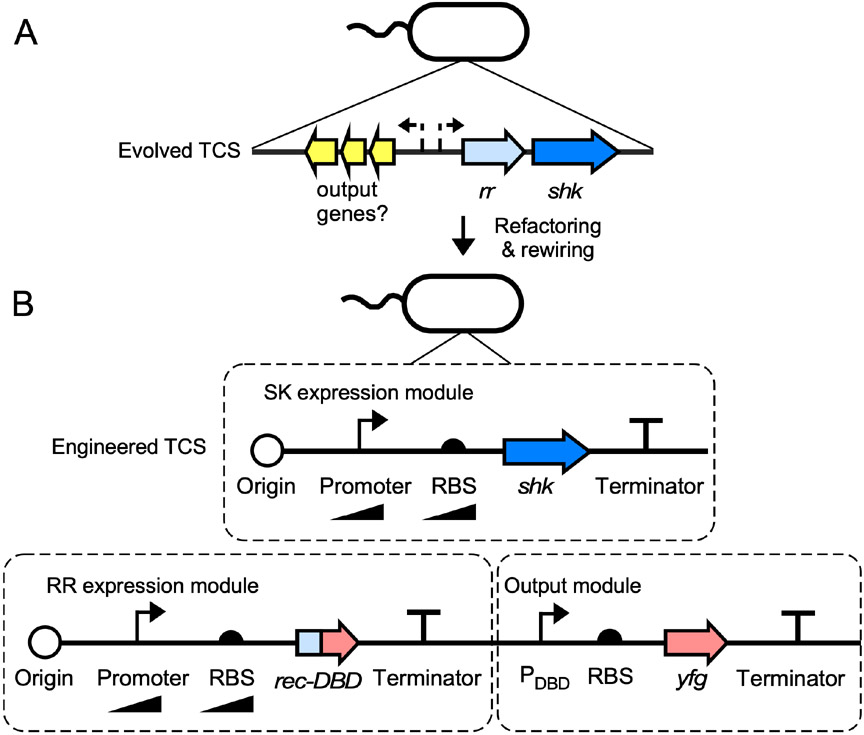
It is often necessary to replace the evolved gene regulatory systems that govern TCS function prior to using them as sensors (Fig. 2a). First, shk and rr genes are generally regulated by multiple interacting pathways[53]. These evolved TCS regulatory networks can cause problems such as silencing[54] or transient input responses[55] that can compromise TCS sensor function. These challenges can often be overcome using genetic refactoring (Fig. 2b). Here, evolved promoters, ribosome binding sites (RBSs), and terminators are replaced with well-characterized alternatives that function reliably in a bacterium of interest. The open reading frames (ORFs) of shk, rr, and any required auxiliary genes can also be computationally redesigned to eliminate known and unknown regulation while increasing translational efficiency in a heterologous host[56]. Though TCSs are relatively insensitive to the levels of their component proteins, it is still important to optimize shk and rr expression levels during refactoring. TCSs are generally more sensitive to RR than SHK levels. If total RR abundance is too low, there will be too few phosphorylated RRs to bind to output promoters and activate transcription in response to the input. Conversely, if total RR abundance is too high, output promoters will often exhibit strong activity in the absence of any input. This problem can arise due to promoter binding by non-phosphorylated RRs, which exist in an equilibrium between inactive and active conformations[24,57]. It can also arise due to residual SHK kinase activity in the absence of input, or alternative sources of RR phosphorylation such as small molecule donors[58] or non-cognate SHKs[59], the effects of which become pronounced at high total RR expression. Very low and very high SHK expression often degrade TCS responses[34,36,52,60,61] and should be avoided. SHK and RR abundances can be co-optimized using orthogonal gene expression inducers (e.g. IPTG, aTc) or libraries of constitutive promoters or RBSs of different strengths[34,36,52,54,60-62].
Figure 2.
Refactoring and rewiring TCSs. (A) Evolved TCSs can be computationally identified in bacterial genomes due to the conservation of SHK and RR domain architectures and the fact that the genes encoding interacting SHK-RR pairs often reside adjacent to one another. However, the promoters (dashed bent lines) and RBSs driving the expression of shk and rr genes can be difficult to predict and characterize from sequence information. This problem is exacerbated for output promoters, which may or may not reside adjacent to shk and rr genes. (B) TCSs can be engineered to function more reliably by replacing native promoters and RBSs with well-characterized synthetic versions. Promoters and RBSs of different strengths should be screened to achieve optimal SHK and RR expression levels. If an evolved output promoter is unknown or has undesirable features, it can be replaced by DBD swapping, wherein the native rr gene is replaced by a chimeric rec-DBD gene and the native output promoter is replaced by a well-characterized promoter that responds to the Rec-DBD protein (PDBD). yfg: your favorite gene.
Unfortunately, many evolved TCS output promoters can only be activated to a small extent. Furthermore, TCS output promoters are often cross-regulated by alternative pathways or silent in heterologous hosts[36,54,61,62]. These challenges can be addressed by introducing mutations or truncations into output promoters to remove unwanted regulatory sites or nested constitutive promoters that generate leaky transcription[36,54,60,61]. TCSs can also be rewired to non-cognate output promoters with superior performance features. The traditional approach to TCS rewiring is to make a chimera between an SHK sensor domain of interest and a non-native transmitter domain. Here, the chimeric SHK controls the phosphorylation of a non-cognate RR and thereby transcription from a non-cognate output promoter. Though sensor domain swapping has been used to engineer numerous TCS sensors[40,63-71], the allosteric mechanisms that enable communication between sensor and transmitter domains are intricate and incompletely understood. As a result, general strategies for TCS sensor domain swapping remain elusive. Approaches based SHK-RR interface swapping[72] and protein scaffolds that redirect SHK phosphorylation to non-cognate RRs[73] have also shown promise for TCS rewiring. However, these approaches tend to yield TCSs whose activity is not dependent on the presence of the input.
Recently, we developed a general method for rewiring TCSs to well-characterized output promoters by modularly swapping RR DBDs[61]. In particular, we identified standard amino acids at which the DBDs of RRs from the OmpR/PhoB or NarL/FixJ sub-families can be removed and replaced with those from structurally-related but functionally unrelated RRs. In addition, we developed standard output modules (e.g. the CcaR DBD and its PcpcG2-172 output promoter for OmpR/PhoB family RRs, and the YdfI DBD and its PydfJ115 output promoter for NarL/FixJ RRs) to which TCSs from those families can be rewired with high rates of success. OmpR/PhoB and NarL/FixJ constitute over 70% of all transcription-regulating RRs[24], suggesting that thousands of TCSs could be characterized and potentially deployed as sensors using this approach.
Due to similarities in the sequences and structures of their interaction interfaces, SHKs and RRs from different TCSs may cross-talk with one another in the same cell. Such phospho-signaling cross-talk could compromise the fidelity of a given TCS sensor in the complex cellular environment. However, work by Laub and colleagues has demonstrated that SHKs generally exhibit a high degree of specificity toward their cognate RRs in the cell due to a large and relatively unoccupied interaction sequence space[70]. This result suggests that synthetic biologists can utilize multiple TCS sensors in a single cell without a high risk of phospho-signaling cross-talk. In the event that phoshpo-signaling cross-talk is a problem, this group also demonstrated that the evolved interaction interfaces can be replaced with insulated versions that have been shown not to cross-talk with other TCSs. The authors used this approach to engineer a system based on a sensor domain-swapped version of PhoQ that responds to the plant cytokinin trans-zeatin and does not cross-talk with any native TCS in E. coli[70] (Table 1).
Porting TCSs into eukaryotes
There is substantial interest in using TCSs to endow eukaryotic cells with novel sensing capabilities. In early work, an engineered E. coli Trg-PhoR system was used to sense 2,4,6-trinitrotoluene via an interaction with the computationally-designed extracellular accessory protein TNT.R3 in A. thaliana[74,75]. To achieve transcription regulation in plants, the RR PhoB was fused to a VP64 transactivation domain and used to activate a minimal plant promoter engineered to contain multiple Pho operator sites. Porting TCSs into plants more broadly remains a challenge. In other work, Benenson and colleagues expressed the E. coli TCSs EnvZ-OmpR, NarX-NarL, and DcuS-DcuR in mammalian cells[76]. Similar RR:transactivator domain fusions and synthetic eukaryotic output promoter design strategies were used to enable control of transcription. While these three TCSs were capable of phospho-signaling and transcriptional activation, they did not respond to their cognate inputs. The reasons for sensing failure are unclear and warrant further investigation. One likely source of the problem is an incompatibility between bacterial SHK transmembrane regions and eukaryotic membranes. In an interesting recent follow-up study, this group fused two NarX transmitter domain mutants that must be brought into close physical proximity to phosphorylate NarL to a G-protein coupled receptor-β arrestin pair that heterodimerize in the presence of ligands including procaterol[77]. This engineered pathway generates large transcriptional responses to these ligands in mammalian cells. This approach may be useful for engineering synthetic mammalian signaling pathways that do not cross-talk with endogenous systems.
Harnessing nature’s treasure trove of TCS sensors
Despite exciting progress, TCSs remain a largely untapped treasure trove of sensors for synthetic biology. The number of TCSs with well-characterized inputs is on the order of one hundred (Table 1). However, many thousands of TCSs are present in bacterial genomes[34,78] and the inputs of most of these systems cannot currently be predicted. A major impediment to identifying the inputs of these orphan TCSs is that most bacteria are intractable; they cannot be cultured nor genetically-manipulated in the laboratory. Synthetic biology methods are helping to overcome this challenge. If an output promoter of an orphan TCS is known or can be inferred from genomic context, the system can be introduced into a tractable organism such as E. coli or B. subtilis[36]. If an output promoter is not known or does not function well in laboratory conditions, sensor domain swapping or DBD-swapping can be used to replace it with an alternative that functions reliably. In either case, the orphan TCS can then be screened against targeted input panels designed by analyzing the function of genes residing adjacent to the TCS or the environment in which the native organism lives[36], complex samples representative of those environments, or even large panels of untargeted chemicals. If inputs of interest are known, commercial gene synthesis can be used to apply this process to hundreds or thousands of orphan TCS pathways in order to identify novel sensors. For example, libraries of orphan TCSs from the human gut microbiome could be synthesized and screened against disease biomarkers found in the gut. If a TCS responsive to the biomarker is found, it can be converted into a high-performance sensor using genetic refactoring and phospho-signaling insulation. Finally, the detection threshold of the new sensor can be matched to the needs of applications such as engineering bacteria that diagnose and treat disease[79]. Similar approaches are being taken to engineer bacteria to detect soil nitrate levels [34], which could eventually be coupled with engineered nitrogen fixation pathways[2] to maintain nitrogen homeostasis in soil without fertilizer.
Conclusions
Though they are more complex than alternatives such as riboswitches and OCSs, TCSs offer a number of benefits as sensors for synthetic biology. First, due to the transmembrane architecture of most SHKs, TCSs can sense inputs that are both accessible and inaccessible to cytoplasmic sensors. Additionally, TCS phospho-signaling increases the reliability of sensor function in the face of variable protein expression levels while also providing a built-in knob for tuning TCSs to respond to different input concentrations. It is likely that bacteria have exploited these and other features of TCSs to better adapt to diverse environmental conditions. Synthetic biologists are increasingly taking advantage of these same properties to endow bacteria with artificial sense and respond capabilities for new engineering applications. However, we have only reached the tip of the iceberg. Bacterial genomes host a huge number of uncharacterized TCSs that likely sense inputs of agricultural, biotechnological, environmental, medical, physiological, and scientific relevance for which no biosensors are currently available. Recent methods for porting TCSs into laboratory bacteria are accelerating the pace at which their inputs can be discovered. Recapitulating TCS function in eukaryotes remains an important challenge that will likely require both new biological insights and new engineering approaches to solve. Overall, the breadth of inputs that they sense combined with the robustness and programmability of their performance will make TCSs an important family of sensors for synthetic biology in the future.
Highlights:
Two-component systems (TCSs) are a large class of bacterial signaling pathways that can detect an incredibly wide variety of stimuli
Recent synthetic biology methods can reduce cross-talk, increase dynamic range, and adjust TCS detection thresholds
Many TCSs can now be reliably rewired to synthetic output promoters, facilitating elucidation of their inputs
TCSs are being applied to the gut microbiome, metabolic engineering, and biomaterial production among other applications.
Acknowledgements
This work was supported by the Welch Foundation (C-1856), the National Science Foundation (CAREER 1553317), the NIH/National Institutes of Allergy and Infectious Diseases (R01AI155586), the Office of Naval Research (N00014-17-1-2642, N00014-18-1-2611) and the Defense Advanced Research Projects Agency (HR00111920019). JTL is supported by a US National Defense Science and Engineering Graduate Fellowship. The content is solely the responsibility of the authors and does not necessarily represent the views of the funding agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McNerney MP, Doiron KE, Ng TL, Chang TZ, Silver PA: Theranostic cells: emerging clinical applications of synthetic biology. Nat Rev Genet 2021, doi: 10.1038/s41576-021-00383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryu M-H, Zhang J, Toth T, Khokhani D, Geddes BA, Mus F, Garcia-Costas A, Peters JW, Poole PS, Ané J-M, et al. : Control of nitrogen fixation in bacteria that associate with cereals. Nat Microbiol 2020, 5:314–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ni C, Dinh CV, Prather KLJ: Dynamic Control of Metabolism. Annu Rev Chem Biomol 2021, 12:1–23. [DOI] [PubMed] [Google Scholar]
- 4.Gurbatri CR, Lia I, Vincent R, Coker C, Castro S, Treuting PM, Hinchliffe TE, Arpaia N, Danino T: Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci Transl Med 2020, 12:eaax0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boo A, Ellis T, Stan G-B: Host-Aware Synthetic Biology. Curr Opin Syst Biology 2019, 14:66–72. [Google Scholar]
- 6.Roth A, Breaker RR: The Structural and Functional Diversity of Metabolite-Binding Riboswitches. Annu Rev Biochem 2009, 78:305–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCown PJ, Corbino KA, Stav S, Sherlock ME, Breaker RR: Riboswitch diversity and distribution. Rna 2017, 23:995–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breaker RR: Riboswitches and Translation Control. Csh Perspect Biol 2018, 10:a032797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berens C, Suess B: Riboswitch engineering — making the all-important second and third steps. Curr Opin Biotech 2015, 31:10–15. [DOI] [PubMed] [Google Scholar]
- 10.Wu MJ, Andreasson JOL, Kladwang W, Greenleaf W, Das R: Automated Design of Diverse Stand-Alone Riboswitches. Acs Synth Biol 2019, 8:1838–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werstuck G, Green MR: Controlling Gene Expression in Living Cells Through Small Molecule-RNA Interactions. Science 1998, 282:296–298. [DOI] [PubMed] [Google Scholar]
- 12.Boussebayle A, Torka D, Ollivaud S, Braun J, Bofill-Bosch C, Dombrowski M, Groher F, Hamacher K, Suess B: Next-level riboswitch development—implementation of Capture-SELEX facilitates identification of a new synthetic riboswitch. Nucleic Acids Res 2019, 47:4883–4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt CM, Smolke CD: A convolutional neural network for the prediction and forward design of ribozyme-based gene-control elements. Elife 2021, 10:e59697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espah Borujeni A, Mishler DM, Wang J, Huso W, Salis HM: Automated physics-based design of synthetic riboswitches from diverse RNA aptamers. Nucleic Acids Res 2016, 44:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulrich LE, Koonin EV, Zhulin IB: One-component systems dominate signal transduction in prokaryotes. Trends Microbiol 2005, 13:52–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckweiler D, Dudek C-A, Hartlich J, Brötje D, Jahn D: PRODORIC2: the bacterial gene regulation database in 2018. Nucleic Acids Res 2017, 46:gkx1091-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer AJ, Segall-Shapiro TH, Glassey E, Zhang J, Voigt CA: Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nat Chem Biol 2019, 15:196–204. [DOI] [PubMed] [Google Scholar]
- 18.Hanko EKR, Paiva AC, Jonczyk M, Abbott M, Minton NP, Malys N: A genome-wide approach for identification and characterisation of metabolite-inducible systems. Nat Commun 2020, 11:1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng J, Jester BW, Tinberg CE, Mandell DJ, Antunes MS, Chari R, Morey KJ, Rios X, Medford JI, Church GM, et al. : A general strategy to construct small molecule biosensors in eukaryotes. Elife 2015, 4:e10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor ND, Garruss AS, Moretti R, Chan S, Arbing MA, Cascio D, Rogers JK, Isaacs FJ, Kosuri S, Baker D, et al. : Engineering an allosteric transcription factor to respond to new ligands. Nat Methods 2016, 13:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juárez JF, Lecube-Azpeitia B, Brown SL, Johnston CD, Church GM: Biosensor libraries harness large classes of binding domains for construction of allosteric transcriptional regulators. Nat Commun 2018, 9:3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao R, Stock AM: Biological insights from structures of two-component proteins. Annu Rev Microbiol 2009, 63:133–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacob-Dubuisson F, Mechaly A, Betton J-M, Antoine R: Structural insights into the signalling mechanisms of two-component systems. Nat Rev Microbiol 2018, 16:585–593. [DOI] [PubMed] [Google Scholar]
- 24.Gao R, Bouillet S, Stock AM: Structural Basis of Response Regulator Function. Annu Rev Microbiol 2019, 73:1–23. [DOI] [PubMed] [Google Scholar]
- 25.Menon S, Wang S: Structure of the Response Regulator PhoP from Mycobacterium tuberculosis Reveals a Dimer through the Receiver Domain. Biochemistry-us 2011, 50:5948–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbieri CM, Wu T, Stock AM: Comprehensive Analysis of OmpR Phosphorylation, Dimerization, and DNA Binding Supports a Canonical Model for Activation. J Mol Biol 2013, 425:1612–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leonard PG, Golemi-Kotra D, Stock AM: Phosphorylation-dependent conformational changes and domain rearrangements in Staphylococcus aureus VraR activation. P Natl Acad Sci Usa 2013, 110:8525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papon N, Stock AM: What do archaeal and eukaryotic histidine kinases sense? F1000research 2019, 8:2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bourret RB, Kennedy EN, Foster CA, Sepúlveda VE, Goldman WE: A Radical Reimagining of Fungal Two-Component Regulatory Systems. Trends Microbiol 2021, doi: 10.1016/j.tim.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grefen C, Harter K: Plant two-component systems: principles, functions, complexity and cross talk. Planta 2004, 219:733–742. [DOI] [PubMed] [Google Scholar]
- 31.Mimee M, Nadeau P, Hayward A, Carim S, Flanagan S, Jerger L, Collins J, McDonnell S, Swartwout R, Citorik RJ, et al. : An ingestible bacterial-electronic system to monitor gastrointestinal health. Science 2018, 360:915–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batchelor E, Goulian M: Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc National Acad Sci 2003, 100:691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinar G, Milo R, Martínez MR, Alon U: Input–output robustness in simple bacterial signaling systems. Proc National Acad Sci 2007, 104:19931–19935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **34. Landry BP, Palanki R, Dyulgyarov N, Hartsough LA, Tabor JJ: Phosphatase activity tunes two-component system sensor detection threshold. Nat Commun 2018, 9:1433. Here, we show that the phosphatase activity of an SHK affects the detection threshold of a TCS sensor. We identified the first X in the GXGXG motif as a general mutation target for tuning TCS detection threshold. Finally, we express unmodified and sensitivity-enhanced engineered versions of NarX-NarL in B. subtilis to sense 3 orders of magnitude nitrate levels in soil.
- 35.Rabin RS, Stewart V: Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J Bacteriol 1993, 175:3259–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daeffler KN, Galley JD, Sheth RU, Ortiz-Velez LC, Bibb CO, Shroyer NF, Britton RA, Tabor JJ: Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol Syst Biol 2017, 13:923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prost LR, Miller SI: The Salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cell Microbiol 2008, 10:576–582. [DOI] [PubMed] [Google Scholar]
- 38.Song J-Y, Cho HS, Cho J-I, Jeon J-S, Lagarias JC, Park Y-I: Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Proc National Acad Sci 2011, 108:10780–10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mascher T, Helmann JD, Unden G: Stimulus Perception in Bacterial Signal-Transducing Histidine Kinases. Microbiol Mol Biol R 2006, 70:910–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, et al. : Engineering Escherichia coli to see light. Nature 2005, 438:441–442. [DOI] [PubMed] [Google Scholar]
- 41.Tabor JJ, Levskaya A, Voigt CA: Multichromatic control of gene expression in Escherichia coli. J Mol Biol 2010, 405:315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandez-Rodriguez J, Moser F, Song M, Voigt CA: Engineering RGB color vision into Escherichia coli. Nat Chem Biol 2017, 13:706–708. [DOI] [PubMed] [Google Scholar]
- 43.Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, Voigt CA, Ellington AD: A synthetic genetic edge detection program. Cell 2009, 137:1272–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *44. Moser F, Tham E, González LM, Lu TK, Voigt CA: Light-Controlled, High-Resolution Patterning of Living Engineered Bacteria Onto Textiles, Ceramics, and Plastic. Adv Funct Mater 2019, 29:1901788. This group utilized 3 light-responsive TCSs (CcaS-CcaR, Cph8-OmpR, Yf1-FixJ) to control three separate genes involved in curli biofilm formation in order to pattern bacteria onto commercially-relevant materials.
- 45.Tang W, Liu DR: Rewritable multi-event analog recording in bacterial and mammalian cells. Science 2018, 360:eaap8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olson EJ, Hartsough LA, Landry BP, Shroff R, Tabor JJ: Characterizing bacterial gene circuit dynamics with optically programmed gene expression signals. Nat Methods 2014, 11:449–55. [DOI] [PubMed] [Google Scholar]
- 47.Davidson EA, Basu AS, Bayer TS: Programming Microbes Using Pulse Width Modulation of Optical Signals. J Mol Biol 2013, 425:4161–4166. [DOI] [PubMed] [Google Scholar]
- *48. Tandar ST, Senoo S, Toya Y, Shimizu H: Optogenetic switch for controlling the central metabolic flux of Escherichia coli. Metab Eng 2019, 55:68–75. This group utilized the green/red photoreversible sensor CcaS-CcaR to control the direction of metabolic flux at the pgi glycolytic branchpoint with green and red light.
- *49. Senoo S, Tandar ST, Kitamura S, Toya Y, Shimizu H: Light-inducible flux control of triosephosphate isomerase on glycolysis in Escherichia coli. Biotechnol Bioeng 2019, 116:3292–3300. These authors placed the tpiA gene under control of CcaS-CcaR to control flux distribution between the glycolysis and the methylglyoxal pathway.
- **50. Hartsough LA, Park M, Kotlajich MV, Lazar JT, Han B, Lin C-CJ, Musteata E, Gambill L, Wang MC, Tabor JJ: Optogenetic control of gut bacterial metabolism to promote longevity. Elife 2020, 9:e56849. This paper is the first report of using optogenetics to manipulate the gut microbiome. CcaS-CcaR was used to control Colanic Acid production in the gut of C. elegans with an external green light stimuli in order to study its impact on mitochondrial health and longevity.
- 51.Riglar DT, Giessen TW, Baym M, Kerns SJ, Niederhuber MJ, Bronson RT, Kotula JW, Gerber GK, Way JC, Silver PA: Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat Biotechnol 2017, 35:653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *52. Cartwright IM, Dowdell AS, Lanis JM, Brink KR, Mu A, Kostelecky RE, Schaefer REM, Welch N, Onyiah JC, Hall CHT, et al. : Mucosal acidosis elicits a unique molecular signature in epithelia and intestinal tissue mediated by GPR31-induced CREB phosphorylation. Proc National Acad Sci 2021, 118:e2023871118. In this work, the acidic pH-activated DBD-swapped TCS SO_4387-SO_4388REC-PsdRDBD 137 was used to detect mucosal acidification in inflamed intestinal tissue in mice.
- 53.Bijlsma JJE, Groisman EA: Making informed decisions: regulatory interactions between two-component systems. Trends Microbiol, 2003, 11:359–366. [DOI] [PubMed] [Google Scholar]
- 54.Ramakrishnan P, Tabor JJ: Repurposing Synechocystis PCC6803 UirS-UirR as a UV-Violet/Green Photoreversible Transcriptional Regulatory Tool in E. coli. Acs Synth Biol 2016, 5:733–40. [DOI] [PubMed] [Google Scholar]
- 55.Salazar ME, Laub MT: Temporal and evolutionary dynamics of two-component signaling pathways. Carr Opin Microbiol 2015, 24:7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song M, Sukovich DJ, Ciccarelli L, Mayr J, Fernandez-Rodriguez J, Mirsky EA, Tucker AC, Gordon DB, Marlovits TC, Voigt CA: Control of type III protein secretion using a minimal genetic system. Nat Commun 2017, 8:14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Volkman BF, Lipson D, Wemmer DE, Kern D: Two-State Allosteric Behavior in a Single-Domain Signaling Protein. Science 2001, 291:2429–2433. [DOI] [PubMed] [Google Scholar]
- 58.Lukat GS, McCleary WR, Stock AM, Stock JB: Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc National Acad Sci 1992, 89:718–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Podgornaia AI, Laub MT: Determinants of specificity in two-component signal transduction. Curr Opin Microbiol 2013, 16:156–162. [DOI] [PubMed] [Google Scholar]
- 60.Schmidl SR, Sheth RU, Wu A, Tabor JJ: Refactoring and Optimization of Light-Switchable Escherichia coli Two-Component Systems. Acs Synth Biol 2014, 3:820–831. [DOI] [PubMed] [Google Scholar]
- **61. Schmidl SR, Ekness F, Sofjan K, Daeffler KN-M, Brink KR, Landry BP, Gerhardt KP, Dyulgyarov N, Sheth RU, Tabor JJ: Rewiring bacterial two-component systems by modular DNA-binding domain swapping. Nat Chem Biol 2019, 15:690–698. Here we demonstrate that RR DBDs can be modularly interchanged and develop standard output modules for rewiring TCSs with OmpR/PhoB and NarL/FixJ family DBDs to well0-characterized output promoters. This method can increase TCS dynamic range, eliminated cross-regulation at output promoters, enabling TCS porting between distant species, and facilitate TCS input discovery.
- 62.Ong NTX, Olson EJ, Tabor JJ: Engineering an E. coli near-infrared light sensor. Acs Synth Biol 2017, 7:240–248. [DOI] [PubMed] [Google Scholar]
- 63.Utsumi R, Brissette R, Rampersaud A, Forst S, Oosawa K, Inouye M: Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science 1989, 245:1246–1249. [DOI] [PubMed] [Google Scholar]
- 64.Hori M, Oka S, Sugie Y, Ohtsuka H, Aiba H: Construction of a photo-responsive chimeric histidine kinase in Escherichia coli. J Gen Appl Microbiol 2017, 63:2016.07.005. [DOI] [PubMed] [Google Scholar]
- 65.Selvamani V, Ganesh I, Maruthamuthu M kannan, Eom GT, Hong SH: Engineering chimeric two-component system into Escherichia coli from Paracoccus denitrificans to sense methanol. Biotechnol Bioproc E 2017, 22:225–230. [Google Scholar]
- 66.Ganesh I, Ravikumar S, Lee SH, Park SJ, Hong SH: Engineered fumarate sensing Escherichia coli based on novel chimeric two-component system. J Biotechnol 2013, 168:560–566. [DOI] [PubMed] [Google Scholar]
- 67.Ganesh I, Ravikumar S, Yoo I, Hong SH: Construction of malate-sensing Escherichia coli by introduction of a novel chimeric two-component system. Bioproc Biosyst Eng 2015, 38:797–804. [DOI] [PubMed] [Google Scholar]
- 68.Möglich A, Ayers RA, Moffat K: Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol 2008, 385:1433–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lehning CE, Heidelberger JB, Reinhard J, Nørholm MHH, Draheim RR: A Modular High-Throughput In Vivo Screening Platform Based on Chimeric Bacterial Receptors. Acs Synth Biol 2017, 6:1315–1326. [DOI] [PubMed] [Google Scholar]
- **70. McClune CJ, Alvarez-Buylla A, Voigt CA, Laub MT: Engineering orthogonal signalling pathways reveals the sparse occupancy of sequence space. Nature 2019, 574:702–706. This study demonstrates that the sequence space of SHK-RR interaction domains is sparsely occupied. This result implies that TCS sensors will generally not cross-talk with one another or native TCSs in a cell. A new trans-zeatin sensing TCS is engineered and given a synthetic SHK-RR interaction interface shown to be insulated from cross-talk with all E. coli TCSs.
- 71.Baumgartner JW, Kim C, Brissette RE, Inouye M, Park C, Hazelbauer GL: Transmembrane signalling by a hybrid protein: communication from the domain of chemoreceptor Trg that recognizes sugar-binding proteins to the kinase/phosphatase domain of osmosensor EnvZ. J Bacteriol 1994, 176:1157–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT: Rewiring the Specificity of Two-Component Signal Transduction Systems. Cell 2008, 133:1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whitaker WR, Davis SA, Arkin AP, Dueber JE: Engineering robust control of two-component system phosphotransfer using modular scaffolds. Proc National Acad Sci 2012, 109:18090–18095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Antunes MS, Morey KJ, Tewari-Singh N, Bowen TA, Smith JJ, Webb CT, Hellinga HW, Medford JI: Engineering key components in a synthetic eukaryotic signal transduction pathway. Mol Syst Biol 2009, 5:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Antunes MS, Morey KJ, Smith JJ, Albrecht KD, Bowen TA, Zdunek JK, Troupe JF, Cuneo MJ, Webb CT, Hellinga HW, et al. : Programmable Ligand Detection System in Plants through a Synthetic Signal Transduction Pathway. Plos One 2011, 6:e16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hansen J, Mailand E, Swaminathan KK, Schreiber J, Angelici B, Benenson Y: Transplantation of prokaryotic two-component signaling pathways into mammalian cells. Proc National Acad Sci 2014, 111:15705–15710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *77. Mazé A, Benenson Y: Artificial signaling in mammalian cells enabled by prokaryotic two-component system. Nat Chem Biol 2020, 16:179–187. This study demonstrates artificial NarXL TCS signaling in HEK293 cells by fusing the GPCR beta-arrestin to the sensor kinase NarX, resulting in a transcriptional output in the presence of epinephrine/procaterol.
- 78.Salvado B, Vilaprinyo E, Sorribas A, Alves R: A survey of HK, HPt, and RR domains and their organization in two-component systems and phosphorelay proteins of organisms with fully sequenced genomes. Peerj 2015, 3:e1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Landry BP, Tabor JJ: Engineering Diagnostic and Therapeutic Gut Bacteria. Microbiol Spectr 2017, 5. [DOI] [PubMed] [Google Scholar]
- 80.Ohlendorf R, Vidavski RR, Eldar A, Moffat K, Möglich A: From Dusk till Dawn: One-Plasmid Systems for Light-Regulated Gene Expression. J Mol Biol 2012, 416:534–542. [DOI] [PubMed] [Google Scholar]
- 81.Ong NT, Tabor JJ: A Miniaturized Escherichia coli Green Light Sensor with High Dynamic Range. Chembiochem 2018, 19:1255–1258. [DOI] [PubMed] [Google Scholar]
- 82.Aguilar PS, Hernandez-Arriaga AM, Cybulski LE, Erazo AC, Mendoza D de: Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. Embo J 2001, 20:1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wen Y, Marcus EA, Matrubutham U, Gleeson MA, Scott DR, Sachs G: Acid-Adaptive Genes of Helicobacter pylori. Infect Immun 2003, 71:5921–5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wen Y, Feng J, Scott DR, Marcus EA, Sachs G: The HP0165-HP0166 Two-Component System (ArsRS) Regulates Acid-Induced Expression of HP1186 α-Carbonic Anhydrase in Helicobacter pylori by Activating the pH-Dependent Promoter ▿. J Bacteriol 2007, 189:2426–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu G, Liu M, Kim E, Maaty WS, Bothner B, Lei B, Rensing C, Wang G, McDermott TR: A periplasmic arsenite-binding protein involved in regulating arsenite oxidation. Environ Microbiol 2012, 14:1624–1634. [DOI] [PubMed] [Google Scholar]
- 86.Guragain M, King MM, Williamson KS, Pérez-Osorio AC, Akiyama T, Khanam S, Patrauchan MA, Franklin MJ: The Pseudomonas aeruginosa PAO1 Two-Component Regulator CarSR Regulates Calcium Homeostasis and Calcium-Induced Virulence Factor Production through Its Regulatory Targets CarO and CarP. J Bacteriol 2016, 198:951–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ravikumar S, Pham VD, Lee SH, Yoo I, Hong SH: Modification of CusSR bacterial two-component systems by the introduction of an inducible positive feedback loop. J Ind Microbiol Biot 2012, 39:861–868. [DOI] [PubMed] [Google Scholar]
- 88.Gudipaty SA, Larsen AS, Rensing C, McEvoy MM: Regulation of Cu(I)/Ag(I) efflux genes in Escherichia coli by the sensor kinase CusS. Fems Microbiol Lett 2012, 330:30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yamamoto K, Ishihama A: Transcriptional response of Escherichia coli to external copper. Mol Microbiol 2005, 56:215–227. [DOI] [PubMed] [Google Scholar]
- 90.Giner-Lamia J, López-Maury L, Reyes JC, Florencio FJ: The CopRS Two-Component System Is Responsible for Resistance to Copper in the Cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 2012, 159:1806–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sánchez-Sutil MC, Marcos-Torres FJ, Pérez J, Ruiz-González M, García-Bravo E, Martínez-Cayuela M, Gómez-Santos N, Moraleda-Muñoz A, Muñoz-Dorado J: Dissection of the sensor domain of the copper-responsive histidine kinase CorS from Myxococcus xanthus. Env Microbiol Rep 2016, 8:363–370. [DOI] [PubMed] [Google Scholar]
- 92.Lin C-S, Tsai Y-H, Chang C-J, Tseng S-F, Wu T-R, Lu C-C, Wu T-S, Lu J-J, Horng J-T, Martel J, et al. : An iron detection system determines bacterial swarming initiation and biofilm formation. Sci Rep-uk 2016, 6:36747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Heermann R, Jung K: The complexity of the ‘simple’ two-component system KdpD/KdpE in Escherichia coli. Fems Microbiol Lett 2010, 304:97–106. [DOI] [PubMed] [Google Scholar]
- 94.Laermann V, Ćudić E, Kipschull K, Zimmann P, Altendorf K: The sensor kinase KdpD of Escherichia coli senses external K+. Mol Microbiol 2013, 88:1194–1204. [DOI] [PubMed] [Google Scholar]
- 95.López-Maury L, García-Domínguez M, Florencio FJ, Reyes JC: A two-component signal transduction system involved in nickel sensing in the cyanobacterium Synechocystis sp. PCC 6803. Mol Microbiol 2002, 43:247–256. [DOI] [PubMed] [Google Scholar]
- *96. Park DM, Taffet MJ: Combinatorial Sensor Design in Caulobacter crescentus for Selective Environmental Uranium Detection. Acs Synth Biol 2019, 8:807–817. Here, UzcR-UzcS and UrpR-UrpS were used to detect uranium in the presence of other elemental metals.
- 97.Petit-Härtlein I, Rome K, Rosny E de, Molton F, Duboc C, Gueguen E, Rodrigue A, Covès J: Biophysical and physiological characterization of ZraP from Escherichia coli, the periplasmic accessory protein of the atypical ZraSR two-component system. Biochem J 2015, 472:205–216. [DOI] [PubMed] [Google Scholar]
- 98.Zimmer DP, Soupene E, Lee HL, Wendisch VF, Khodursky AB, Peter BJ, Bender RA, Kustu S: Nitrogen regulatory protein C-controlled genes of Escherichia coli: Scavenging as a defense against nitrogen limitation. Proc National Acad Sci 2000, 97:14674–14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.David M, Daveran M-L, Batut J, Dedieu A, Domergue O, Ghai J, Hertig C, Boistard P, Kahn D: Cascade regulation of nif gene expression in Rhizobium meliloti. Cell 1988, 54:671–683. [DOI] [PubMed] [Google Scholar]
- 100.Darwin AJ, Tyson KL, Busby SJW, Stewart V: Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K-12 depends on DNA binding site arrangement. Mol Microbiol 1997, 25:583–595. [DOI] [PubMed] [Google Scholar]
- 101.Sun F, Ji Q, Jones MB, Deng X, Liang H, Frank B, Telser J, Peterson SN, Bae T, He C: AirSR, a [2Fe-2S] Cluster-Containing Two-Component System, Mediates Global Oxygen Sensing and Redox Signaling in Staphylococcus aureus. J Am Chem Soc 2012, 134:305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hall JW, Yang J, Guo H, Ji Y: The Staphylococcus aureus AirSR Two-Component System Mediates Reactive Oxygen Species Resistance via Transcriptional Regulation of Staphyloxanthin Production. Infect Immun 2017, 85:e00838–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sarwar Z, Wang MX, Lundgren BR, Nomura CT: MifS, a DctB family histidine kinase, is a specific regulator of α-ketoglutarate response in Pseudomonas aeruginosa PAO1. Microbiology+ 2020, 166:867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *104. Yang Y, Lang N, Zhang L, Wu H, Jiang W, Gu Y: A novel regulatory pathway consisting of a two-component system and an ABC-type transporter contributes to butanol tolerance in Clostridium acetobutylicum. Appl Microbiol Biot 2020, 104:5011–5023. This study reports the characterization of the novel butanol-sensing TCS BtrK-BtrR that can be used for in vivo biosensing in the process of butanol bioproduction.
- 105.Golby P, Davies S, Kelly DJ, Guest JR, Andrews SC: Identification and Characterization of a Two-Component Sensor-Kinase and Response-Regulator System (DcuS-DcuR) Controlling Gene Expression in Response to C4-Dicarboxylates in Escherichia coli. J Bacteriol 1999, 181:1238–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.YAMAMOTO K, MATSUMOTO F, OSHIMA T, FUJITA N, OGASAWARA N, ISHIHAMA A: Anaerobic Regulation of Citrate Fermentation by CitAB in Escherichia coli. Biosci Biotechnology Biochem 2008, 72:3011–3014. [DOI] [PubMed] [Google Scholar]
- 107.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V: Fucose sensing regulates bacterial intestinal colonization. Nature 2012, 492:113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Olekhnovich IN, Kadner RJ: Mutational Scanning and Affinity Cleavage Analysis of UhpA-Binding Sites in the Escherichia coli uhpT Promoter. J Bacteriol 2002, 184:2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lundgren BR, Shoytush JM, Scheel RA, Sain S, Sarwar Z, Nomura CT: Utilization of L-glutamate as a preferred or sole nutrient in Pseudomonas aeruginosa PAO1 depends on genes encoding for the enhancer-binding protein AauR, the sigma factor RpoN and the transporter complex AatJQMP. Bmc Microbiol 2021, 21:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stauff DL, Torres VJ, Skaar EP: Signaling and DNA-binding Activities of the Staphylococcus aureus HssR-HssS Two-component System Required for Heme Sensing*. J Biol Chem 2007, 282:26111–26121. [DOI] [PubMed] [Google Scholar]
- 111.Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A: Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol Microbiol 2005, 55:1113–1126. [DOI] [PubMed] [Google Scholar]
- 112.Tanaka K, Kobayashi K, Ogasawara N: The Bacillus subtilis YufLM two-component system regulates the expression of the malate transporters MaeN (YufR) and YflS, and is essential for utilization of malate in minimal medium. Microbiology+ 2003, 149:2317–2329. [DOI] [PubMed] [Google Scholar]
- 113.Behr S, Kristoficova I, Witting M, Breland EJ, Eberly AR, Sachs C, Schmitt-Kopplin P, Hadjifrangiskou M, Jung K: Identification of a High-Affinity Pyruvate Receptor in Escherichia coli. Sci Rep-uk 2017, 7:1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sun Z, Chen Y, Yang C, Yang S, Gu Y, Jiang W: A novel three-component system-based regulatory model for d-xylose sensing and transport in Clostridium beijerinckii. Mol Microbiol 2015, 95:576–589. [DOI] [PubMed] [Google Scholar]
- 115.Velasco A, Alonso S, García JL, Perera J, Díaz E: Genetic and Functional Analysis of the Styrene Catabolic Cluster of Pseudomonas sp. Strain Y2. J Bacteriol 1998, 180:1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang B, Zhao A, Novick RP, Muir TW: Activation and Inhibition of the Receptor Histidine Kinase AgrC Occurs through Opposite Helical Transduction Motions. Mol Cell 2014, 53:929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jayaraman P, Holowko MB, Yeoh JW, Lim S, Poh CL: Repurposing a Two-Component System-Based Biosensor for the Killing of Vibrio cholerae. Acs Synth Biol 2017, 6:1403–1415. [DOI] [PubMed] [Google Scholar]
- 118.Davey L, Halperin SA, Lee SF: Mutation of the Thiol-Disulfide Oxidoreductase SdbA Activates the CiaRH Two-Component System, Leading to Bacteriocin Expression Shutdown in Streptococcus gordonii. J Bacteriol 2016, 198:321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Guan C, Cui W, Cheng J, Zhou L, Guo J, Hu X, Xiao G, Zhou Z: Construction and development of an auto-regulatory gene expression system in Bacillus subtilis. Microb Cell Fact 2015, 14:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shin J, Choe D, Ransegnola B, Hong H, Onyekwere I, Cross T, Shi Q, Cho B, Westblade LF, Brito IL, et al. : A multifaceted cellular damage repair and prevention pathway promotes high-level tolerance to β-lactam antibiotics. Embo Rep 2021, 22:e51790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stubbendieck RM, Straight PD: Linearmycins Activate a Two-Component Signaling System Involved in Bacterial Competition and Biofilm Morphology. J Bacteriol 2017, 199:e00186–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *122. Revilla-Guarinos A, Dürr F, Popp PF, Döring M, Mascher T: Amphotericin B Specifically Induces the Two-Component System LnrJK: Development of a Novel Whole-Cell Biosensor for the Detection of Amphotericin-Like Polyenes. Front Microbiol 2020, 11:2022. Here, Lnr J-LnrK is refactored and engineered to enable amphotericin B detection in B. subtilis.
- 123.Lockey C, Edwards RJ, Roper DI, Dixon AM: The Extracellular Domain of Two-component System Sensor Kinase VanS from Streptomyces coelicolor Binds Vancomycin at a Newly Identified Binding Site. Sci Rep-uk 2020, 10:5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *124. Lee K, Kaspar JR, Rojas-Carreño G, Walker AR, Burne RA: A single system detects and protects the beneficial oral bacterium Streptococcus sp. A12 from a spectrum of antimicrobial peptides. Mol Microbiol 2021, doi: 10.1111/mmi.14703. This group demonstrates that the TCS PcfK-PcfR is activated by oral antimicrobial peptides.
- 125.Toymentseva AA, Schrecke K, Sharipova MR, Mascher T: The LIKE system, a novel protein expression toolbox for Bacillus subtilis based on the liaI promoter. Microb Cell Fact 2012, 11:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wolf D, Mascher T: The applied side of antimicrobial peptide-inducible promoters from Firmicutes bacteria: expression systems and whole-cell biosensors. Appl Microbiol Biot 2016, 100:4817–4829. [DOI] [PubMed] [Google Scholar]
- 127.Bongers RS, Veening J-W, Wieringen MV, Kuipers OP, Kleerebezem M: Development and Characterization of a Subtilin-Regulated Expression System in Bacillus subtilis: Strict Control of Gene Expression by Addition of Subtilin. Appl Environ Microb 2005, 71:8818–8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mimee M, Tucker AC, Voigt CA, Lu TK: Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota. Cell Syst 2015, 1:62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang BX, Wheeler KM, Cady KC, Lehoux S, Cummings RD, Laub MT, Ribbeck K: Mucin Glycans Signal through the Sensor Kinase RetS to Inhibit Virulence-Associated Traits in Pseudomonas aeruginosa. Carr Biol 2021, 31:90–102.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schwalm ND, Townsend GE, Groisman EA: Multiple Signals Govern Utilization of a Polysaccharide in the Gut Bacterium Bacteroides thetaiotaomicron. Mbio 2016, 7:e01342–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kilmury SLN, Burrows LL: Type IV pilins regulate their own expression via direct intramembrane interactions with the sensor kinase PilS. Proc National Acad Sci 2016, 113:6017–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Richards SM, Strandberg KL, Conroy M, Gunn JS: Cationic antimicrobial peptides serve as activation signals for the Salmonella Typhimurium PhoPQ and PmrAB regulons in vitro and in vivo. Front Cell Infect Mi 2012, 2:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V: The QseC sensor kinase: A bacterial adrenergic receptor. Proc National Acad Sci 2006, 103:10420–10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Donoso R, Leiva-Novoa P, Zúñiga A, Timmermann T, Recabarren-Gajardo G, González B: Biochemical and Genetic Bases of Indole-3-Acetic Acid (Auxin Phytohormone) Degradation by the Plant-Growth-Promoting Rhizobacterium Paraburkholderia phytofirmans PsJN. Appl Environ Microb 2017, 83:e01991–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang F-F, Cheng S-T, Wu Y, Ren B-Z, Qian W: A Bacterial Receptor PcrK Senses the Plant Hormone Cytokinin to Promote Adaptation to Oxidative Stress. Cell Reports 2017, 21:2940–2951. [DOI] [PubMed] [Google Scholar]
- 136.Lin Y-H, Pierce BD, Fang F, Wise A, Binns AN, Lynn DG: Role of the VirA histidine autokinase of Agrobacterium tumefaciens in the initial steps of pathogenesis. Front Plant Sci 2014, 5:195. [DOI] [PMC free article] [PubMed] [Google Scholar]