Abstract
Background & Aims
We aim to describe the liver immune microenvironment by analyzing liver biopsies from patients with chronic HBV infection (CHB). Host immune cell signatures and their corresponding localization were characterized by analyzing the intrahepatic transcriptome in combination with a custom multiplex immunofluorescence panel.
Method
Matching FFPE and fresh frozen liver biopsies were collected from immune active patients within the open-label phase IV study GS-US-174-0149. RNA-Seq was conducted on 53 CHB liver biopsies from 46 patients. Twenty-eight of the 53 samples had matched FFPE biopsies and were stained with a 12-plex panel including cell segmentation, immune and viral biomarkers. Corresponding serum samples were screened using the MSD Human V-plex Screen Service to identify peripheral correlates for the immune microenvironment.
Results
Using unsupervised clustering of the transcriptome, we reveal two unique liver immune signatures classified as immune high and immune low based on the quantification of the liver infiltrate gene signatures. Multiplex immunofluorescence analysis demonstrated large periportal lymphoid aggregates in immune high samples consisting of CD4 and CD8 T cells, B cells and macrophages. Differentiation of the high and low immune microenvironments was independent of HBeAg status and peripheral viral antigen levels. In addition, longitudinal analysis indicates that treatment and normalization of ALT correlates with a decrease in liver immune infiltrate and inflammation. Finally, we screened a panel of peripheral biomarkers and identified ICAM-1 and CXCL10 as biomarkers that strongly correlate with these unique immune microenvironments.
Conclusion
These data provide a description of immune phenotypes in patients with CHB and show that immune responses are downregulated in the liver following nucleotide analogue treatment. This may have important implications for both the safety and efficacy of immune modulator programs aimed at HBV cure.
Lay summary
Liver biopsies from patients with chronic hepatitis B were submitted to RNA-Seq and multiplex immunofluorescence and identified two different liver immune microenvironments: immune high and immune low. Immune high patients showed elevated immune pathways, including interferon signaling pathways, and increase presence of immune cells. Longitudinal analysis of biopsies from treatment experienced patients showed that treatment correlates with a marked decrease in inflammation and these findings may have important implications for both safety and efficacy of immune modulator programs for HBV cure.
Keywords: Immune Microenvironment, Hepatitis B, Chronic HBV, Intrahepatic transcriptome, multiplex immunofluorescence
Abbreviations: ALT, alanine aminotransferase; BCR, B-cell receptor; CHB, chronic HBV infection; DEG, differentially expressed gene; FFPE, formalin-fixed paraffin-embedded; IHC, immunohistochemistry; mIF, multiplex immunofluorescence; PEG-IFNα, pegylated-interferon-α; ssGSEA, single sample gene set enrichment analysis; TCR, T-cell receptor; TDF, tenofovir disoproxil fumarate; TLS, tertiary lymphoid structures
Graphical abstract
Highlights
-
•
Two different liver immune microenvironments were identified in patients with chronic hepatitis B: immune high and immune low.
-
•
Immune high patients had elevated immune pathway activity and immune cell signatures corresponding to B cells, T cells and macrophages.
-
•
Antiviral treatment and normalization of ALT correlates with a marked decrease in liver immune infiltrate and inflammation.
-
•
CXCL10 and ICAM-1 were identified as peripheral biomarkers that correlated with these differentiated immune microenvironments.
Introduction
HBV chronically infects over 257 million people worldwide and is a leading cause of liver-related mortality.1 HBV can cause lifelong chronic infection that results in progression through various stages of liver disease including fibrosis, cirrhosis and ultimately hepatocellular carcinoma.2,3 Progression to end-stage liver disease is mediated largely through chronic liver inflammation, which is thought to be highly variable throughout the natural history of disease.4 Long-term chronic inflammation of the liver results in hepatocyte death and regeneration, slowly leading to liver scarring and fibrosis, and eventually leading to advanced liver disease. Hepatocyte death is measured in the periphery using a serum biomarker, alanine aminotransferase (ALT), which is released from ruptured hepatocytes into the circulation.5
CHB infection typically progresses through several stages of natural history, ultimately leading to hepatocellular carcinoma.6 The immune tolerant phase is characterized by patients that are positive for HBeAg, have very high viral DNA levels, but have ALT levels that remain normal.6 Patients that contract HBV at birth often stay in the immune tolerant phase of disease for several decades and fail to mount an anti-HBV immune response despite high viral burden.7 Patients eventually experience elevated ALT and transition to the immune active phase of disease, however, the immune triggers that mediate this switch are unknown. The presence of viral antigens alone is insufficient to trigger this immune switch, as they are present at high levels throughout the immune tolerant phase of disease. Patients with elevated ALT are indicated for treatment based on the current treatment guidelines.8 Treatment with pegylated-interferon-α (PEG-IFNα) leads to higher cure rates than other regimens, but still cures less than 10% of patients.9 Several studies characterizing the response signature in patients treated with PEG-IFNα revealed interferon signaling in the liver and elevated CXCL10 and CXCL9 in serum of patients that experience cure.10,11
We examined liver biopsies from the GS-US-174-0149 clinical trial.9 Liver biopsies were collected from volunteers at baseline and week 96 (tenofovir disoproxil fumarate [TDF] and/or PEG-IFNα). RNA-Seq was performed on baseline biopsies and revealed two immune signatures, immune high and immune low. The immune high transcriptome resembles the known PEG-IFNα response signature and includes elevated interferon signaling pathways and increased intrahepatic lymphoid signatures. RNA-Seq from the week 96 biopsies demonstrated even lower intrahepatic immune cell levels and transcriptional signatures. Multiplex immunofluorescence (mIF) demonstrated that immune high samples had significantly higher levels of T cells and B cells that accumulated into periportal lymphoid aggregates. Finally, we demonstrate that peripheral ICAM-1 and CXCL10 correlate with the immune high and immune low signatures, indicating that these are potential biomarkers and may help stratify patients based on immune microenvironment.
Materials and methods
Liver biopsy collection
Liver biopsies were obtained from treatment-naïve HBeAg-positive and -negative patients enrolled in a phase IV clinical trial of TDF ± PEG-IFNα (GS-US-174-0149).9 63 patients participated in voluntary liver biopsy donation including 56 formalin-fixed paraffin-embedded (FFPE) and 67 fresh frozen liver biopsies. Thirty-one samples had matched FFPE and fresh frozen biopsies. Ten patients were biopsied longitudinally at baseline and week 96. Patient samples analyzed were collected from 8 countries (US, Korea, Turkey, Hong Kong, Poland, Netherlands, Greece and Germany). All patients signed an informed consent form prior to screening and in accordance with local regulatory and ethics committee requirements. The experimental protocol in these trials was approved by Gilead Sciences and all local regulatory agencies (see ClinicalTrials.gov: NCT01940471).
Transcriptomic analyses
Total RNA was isolated from all fresh frozen samples using the Allprep kit (Qiagen) and RNA-Seq analysis was performed at Expression Analysis – Q2 Solutions (Morrisville, NC). 54 of 67 samples met the quantity and quality standards for RNA-Seq. Sequencing libraries were prepared using TruSeq Stranded Total Gold with RiboZero and sequenced on an Illumina HiSeq with 2∗150 bp reads. Reads were aligned to reference genome hg38 using STAR and quantified using protein coding gene annotations from Gencode. Gene count data were converted to an RPKM matrix of all samples combined with flow-sorted gene expression data from Blueprint Epigenome Project to calculate cell type enrichment scores using xCell for 43 cell types that may reside in the liver.12 Baseline samples were grouped using unsupervised hierarchical clustering of lymphoid cell xCell scores including the following cell types: activated dendritic cells, CD4+ memory T cells, CD8+ naïve T cells, CD8+ central memory T cells, T helper 1 cells and T helper 2 cells. Subsequent cell deconvolution analyses were performed using EPIC.13,14 Single sample gene set enrichment analysis (ssGSEA) scores were calculated using gene set variation analysis for hallmark, canonical and immune pathways from MSigDB v7.0.[15], [16], [17] Statistical analyses for ssGSEA scores, xCell scores, and log transformed gene expression were performed using a moderated t-test.18
Immunohistochemistry
FFPE blocks were sectioned at 5 μm and mounted on charged microscope slides for staining. Slides were stained with anti-HBcAg (25-7, Gilead), anti-HBsAg (polyclonal, Novus Biologics cat# NB100-62652), anti-PD-1 (NAT105, Abcam cat# ab52587), or anti-PD-L1 (28-8, Abcam cat# ab205921) on the Ventana Discovery Ultra using routine immunoperoxidase methods. Stained slides were scanned on the Leica Aperio AT2 at 40x. Quantitative morphometric analysis of whole slide images was performed using Definiens Developer and Definiens Tissue Studio.
Multiplex immunofluorescence
Liver biopsy slides were shipped to Neogenomics (Fort Myers, FL). The MultiOmyx™ platform involves serial staining, imaging and dye-inactivation of two antigens at a time, one on Cy3 and another on Cy5 (Fig. S2).19 The least robust targets are visualized first while the more robust targets are visualized last to limit the effects of dye-inactivation. Slides were stained in the following order: Round 1: anti-PD-L1 (73-10, Cell Signaling Technologies) and anti-CD299 (EPR11211, Abcam), Round 2: anti-PanCK (AE-1/AE-3, Biolegend) and anti-HBsAg (XTL17, Gilead), Round 3: anti-CD8 (EP1150Y, Abcam) and anti-HBcAg (366-2, Gilead), Round 4: anti-CD4 (EPR6855, Abcam) and anti-PD-1 (EH33, Cell Signaling Technologies), Round 5: anti-CD3 (97707, Abcam) and anti-FoxP3 (326A/E7, Abcam), Round 6: anti-CD19 (LE-CD19, Dako) and anti-Ki67 (MIB-1, Dako), Round 7: Anti-CD20 (L26, Dako) and anti-CD68 (KP-1, Diagnostic Biosystems). Images were collected on the GE InCell scanner at 20x. Quantitative morphometric image analysis was performed both at Neogenomics using a proprietary software platform, and at Gilead using Visiopharm.
Peripheral cytokine analysis
HBsAg, HBeAg, HBV DNA and ALT were collected during the GS-US-174-0149 clinical trial.9 Patient plasma matched to each liver biopsy was shipped to DDL Diagnostic Labs (Rijswijk, The Netherlands) for quantitation of HBcrAg (Fujiribio, Lumipulse) and HBV RNA (DDL Diagnostics).20
Human peripheral biomarkers were analyzed from serum at Meso Scale Diagnostics (Gaithersburg, MD). sPD-1 and sPD-L1 were analyzed using the MSD R-plex platform. Human V-plex Screening Service 1 panel is commercially available and consists of 39 analytes.
Results
Identification of unique liver immune microenvironments in treatment-naïve liver biopsies
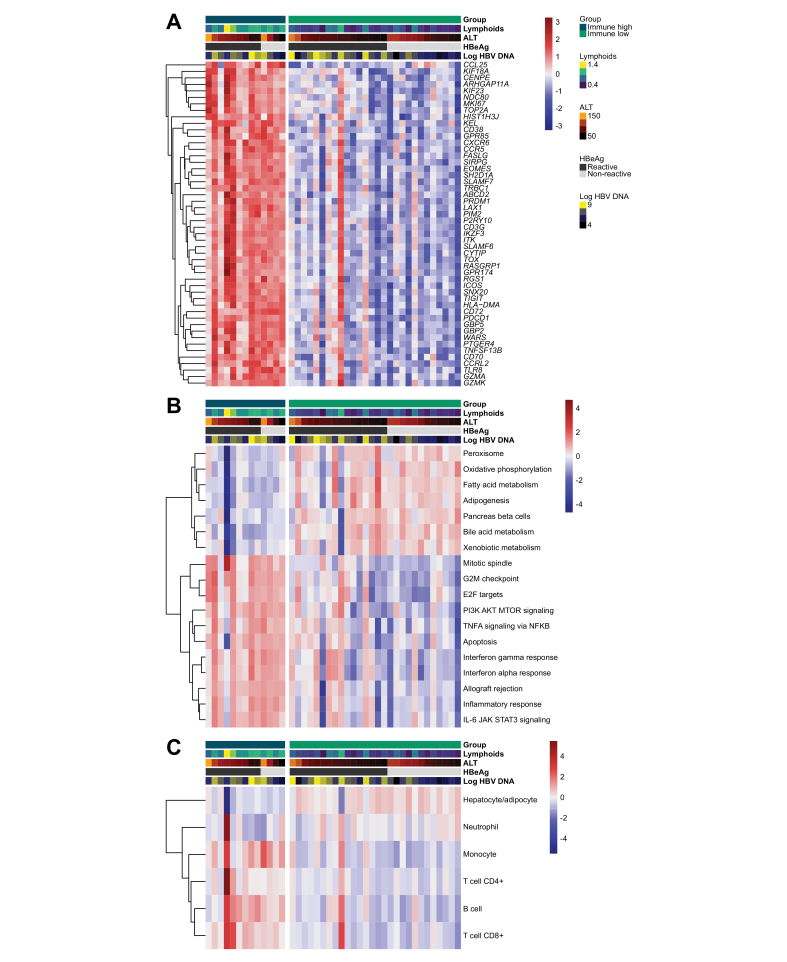
We analyzed fresh frozen and FFPE liver biopsies from a sub-study of the Gilead GS-US-174-0149 clinical trial. Patients were enrolled based on elevated HBV DNA and ALT independent of HBeAg status. Fifty-three liver biopsies were analyzed by RNA-Seq: 41 samples from baseline and 12 samples from week 96, including 7 longitudinal pairs. To characterize the liver immune microenvironment, we used the deconvolution algorithm, xCell, to estimate immune cell types infiltrating the liver of each sample.21 Unsupervised hierarchical clustering of lymphoid xCell scores resolved two unique liver immune microenvironments within baseline samples that we classified as immune high and immune low. No baseline clinical characteristics differentiated these two clusters including age, race, HBV genotype, BMI or sex (Table S1). To characterize the transcriptional changes between immune high and immune low samples we highlight individual genes (Fig. 1A), signaling pathways (Fig. 1B) and intrahepatic immune cell signatures (Fig. 1C) that are the most differentiated between these two groups. Differentially expressed gene (DEG) analysis demonstrated that the top DEGs are mostly immune genes that are upregulated in immune high vs. immune low samples (Fig. 1A). Next, we analyzed pathways that are differentially regulated between immune high and immune low biopsies (Fig. 1B). A variety of immune pathways were upregulated in immune high samples, including those related to interferon and other inflammatory signaling pathways. In contrast, specific metabolic pathways were downregulated in immune high samples compared to immune low samples. This likely represents a lower relative abundance of hepatocyte gene signatures as a proportion of the total sample transcriptome. We quantified cell type estimates using an alternative algorithm, EPIC, to determine the relative composition of immune cell types in each sample (Fig. 1C).13,14 Consistent with the pathway analysis, we observed an increase in cell signatures representing T cells, B cells and monocytes and a decrease in hepatocyte signatures in the immune high samples. Surprisingly, these immune signatures did not correlate with serum HBV DNA or HBsAg levels, HBeAg status or ALT levels (Fig. S1).
Fig. 1.
Comparison of intrahepatic immune cell and pathway signatures across CHB liver biopsies.
Gene expression patterns were analyzed by xCell (lymphoids on x-axis) and unsupervised hierarchical clustering to define two groups called immune high and immune low. (A) List of the top differentially expressed genes between immune high and immune low clusters. (B) Differentiated Hallmark pathways between immune high and immune low. (C) EPIC cell deconvolution shows an increase of T-cell, B-cell, and monocyte signatures in immune high samples.13 Patient order is consistent in heatmaps A-C. ALT, alanine aminotransferase; CHB, chronic HBV infection.
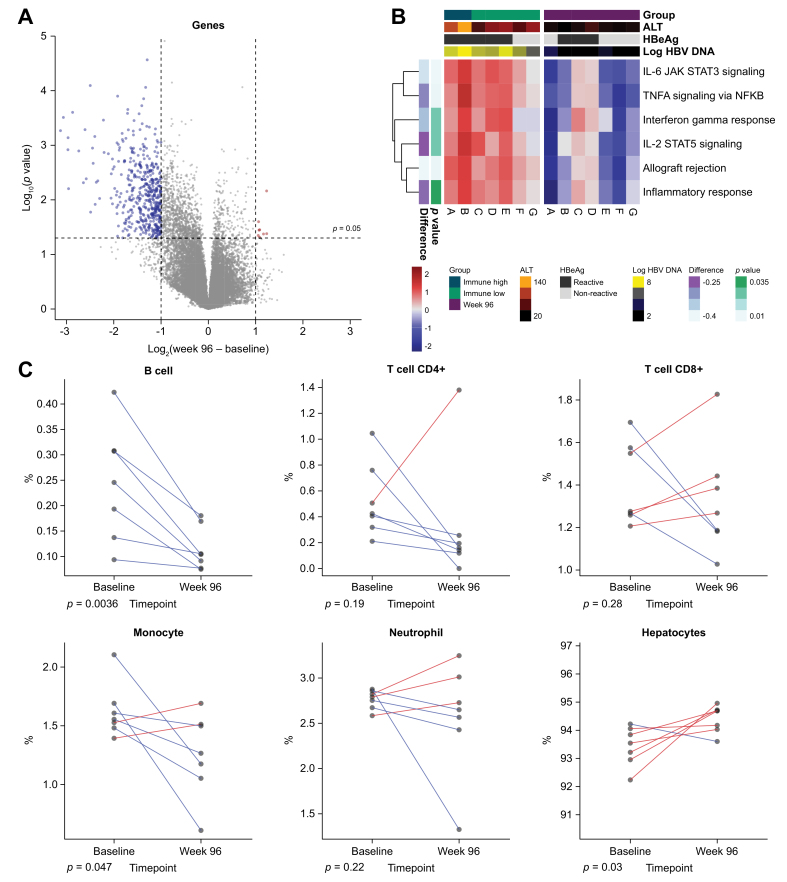
Our biopsy collection also included 12 samples post-treatment (week 96). Patients were treated for the first 48 weeks with a combination of TDF ± PEG-IFNα. During treatment-free follow-up, if patients experienced HBV DNA rebound to an established threshold or had an ALT flare, then patients were re-treated with TDF alone for the remainder of the trial. While the week 96 samples do not represent the end-of-treatment per se, all samples in our collection came from patients with normalized ALT and 8 of 12 achieved full viral suppression. Of these 12 patients, 7 of them had matching baseline biopsies such that we could assay the liver immune microenvironment longitudinally (Fig. 2). Changes in gene expression were calculated between baseline and week 96 and plotted vs. their adjusted p values (Fig. 2A). Most DEGs were significantly downregulated at week 96 and consisted of immune genes. Pathway analysis confirmed that many of the immune pathways were downregulated at week 96 (Fig. 2B). We used EPIC cell deconvolution to monitor the reduction of a variety of immune cell types following treatment. Intrahepatic CD4 T-cell and B-cell signatures were significantly reduced in treated samples, and likewise, there was an increase in hepatocytes (Fig. 2C).
Fig. 2.
Tenofovir disoproxil fumarate treatment suppresses liver inflammation.
(A) Seven patients donated longitudinal liver biopsies from baseline and week 96. Differential gene expresssion analysis between baseline and matched week 96 samples. P values are from limma's moderated t test. (B) Immune pathways were scored for each patient at baseline and week 96. Each row is a unique immune-related pathway from MSigDB which is normalized to the mean of the 7 week 96 samples. (C) Longitudinal analysis using EPIC cell deconvolution.13P values represent pairwise t tests. ALT, alanine aminotransferase.
Multiplex immunofluorescence reveals the architecture of the liver immune microenvironment
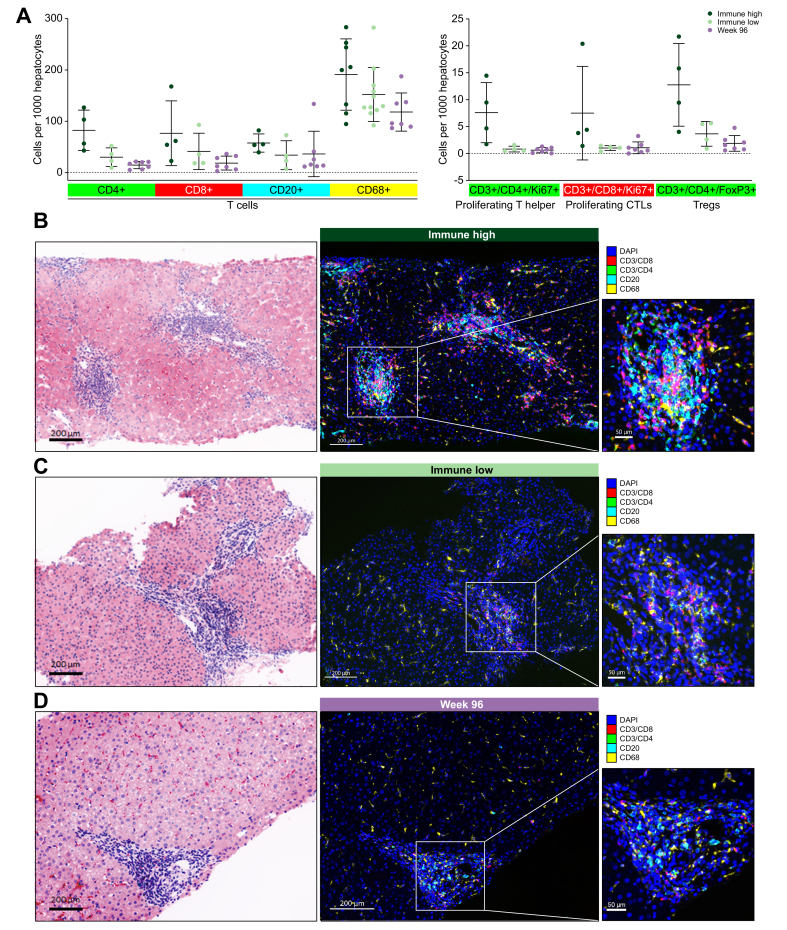
The transcriptome indicated that the immune high samples had significantly increased numbers of T cells, B cells and macrophages. To determine the localization of these immune cells, matched FFPE biopsies were imaged using mIF and subjected to multiple rounds of staining using the MultiOmyx™ platform (Fig. S2).19 In line with the transcriptional analysis, image quantitation demonstrated that immune high biopsies had greater numbers of intrahepatic T cells, B cells and macrophages (Fig. 3A). Immune high liver biopsies had clusters of immune cells in and near portal regions (Fig. 3B). These periportal lymphoid aggregates consisted of CD4 T cells, CD8 T cells, B cells and CD68-positive macrophages. In addition, there was an increase in CD68-positive Kupffer cells and macrophages in the liver sinusoids. These aggregates of lymphocytes in the liver portal tracts appear similar in structure to tertiary lymphoid structures (TLS) observed in many other tissues and tumors. Indeed, immune high patients do appear to have elevated expression of TLS-associated gene signatures including T follicular helper cell signatures and TLS cytokines (Fig. S3).22 In addition, we quantified T-cell receptor (TCR) and B-cell receptor (BCR) clonality using RNA-Seq (Fig. S3C). Although there are more intrahepatic T and B cells in immune high samples, there does not appear to be enrichment of specific T and B cell clones, and instead this likely represents general recruitment of T and B cells from the periphery. Week 96 samples were also imaged using mIF. These images confirmed the transcriptome findings, demonstrating that treated liver biopsies have lower numbers of intrahepatic T cells, B cells and macrophages than baseline patient livers (Fig. 3).
Fig. 3.
Immune high liver biopsies contain increased B-cell, T-cell and Kupffer cell numbers in portal regions.
(A) mIF was performed on FFPE slides from 28 samples with matched RNA-Seq. Immune cell types and markers were quantified and normalized per 1,000 hepatocytes. (B) Representative images from an immune high, immune low and week 96 sample are shown. CD3/8 (red), CD3/4 (green), CD20 (cyan), CD68 (yellow) and DAPI (blue). Virtual H&E stains were generated following all rounds of fluorescent staining. FFPE, formalin-fixed paraffin-embedded; mIF, multiplex immunofluorescence.
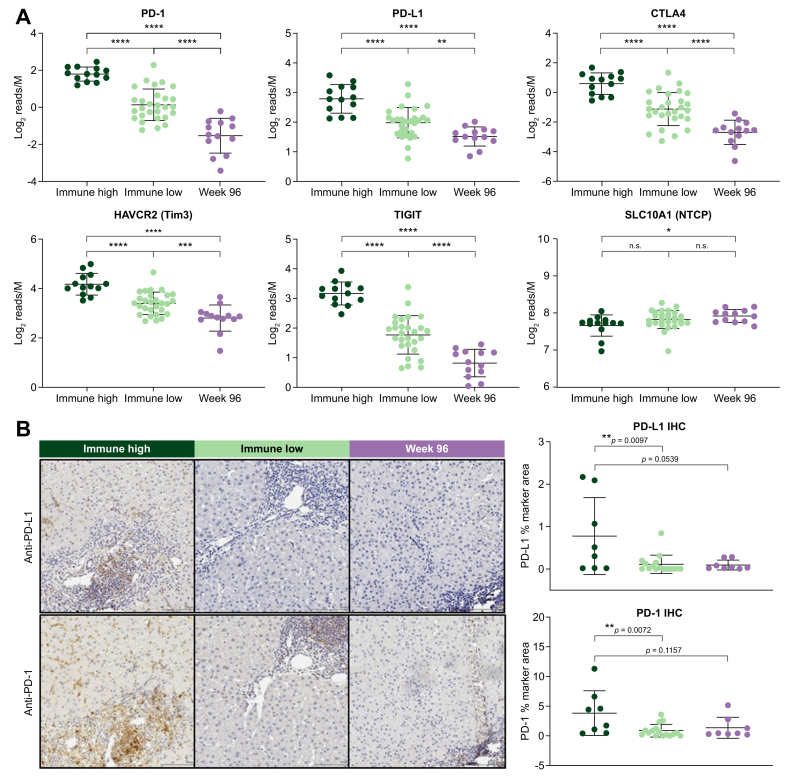
Immune high samples demonstrate elevated expression of immune checkpoints
Immune checkpoint blockade improves T-cell function and leads to positive outcomes for cancer patients and is now being explored for HBV cure.23,24 We examined the expression of a variety of checkpoint genes in our liver biopsy collection and found that immune high patients had significantly increased expression of PDCD1 (PD-1), CD274 (PD-L1), CTLA4, TIGIT and HAVCR2 (TIM3) (Fig. 4A). Immune high liver biopsies showed significant expression of both PD-1 and PD-L1 as detected by immunohistochemistry (IHC), largely driven by half of the samples with larger than 1% marker area expression (Fig. 4B). In contrast, immune low and week 96 biopsies demonstrated low or undetectable PD-1 and PD-L1 protein expression by IHC (Fig. 4B). In the 8 longitudinal biopsy pairs, there was a significant decrease in PD-1 staining at week 96 compared to baseline biopsies (Fig. S4). In addition, two patients with elevated PD-L1 staining at baseline displayed a sharp decrease in PD-L1 staining at week 96 (0.7% and 0.93% marker area decrease at week 96) (Fig. S4).
Fig. 4.
Immune high cluster demonstrates significantly higher expression of immune checkpoints.
(A) Gene expression patterns for a variety of checkpoint receptors (and ligands) plotted to compare expression in immune high (blue) vs. immune low (green) vs. week 96 (purple) samples. Statistical significance was calculated by unpaired t test. (B) FFPE slides were analyzed by single-plex IHC with anti-PD-1 and anti-PD-L1. Images from a representative immune high, immune low and week 96 sample are displayed. PD-1 and PD-L1 staining was quantified by % marker area staining. Staining patterns between sample groups were analyzed using unpaired t tests. FFPE, formalin-fixed paraffin-embedded; IHC, immunohistochemistry.
The PD-L1 staining pattern observed by IHC appeared to localize mostly to liver sinusoids and Kupffer cells, and not directly on hepatocytes. To confirm which cell types expressed PD-L1 in the liver of patients with CHB, we developed a 4-plex fluorescence assay. Our panel consisted of pan cytokeratin (PanCK) as a marker for hepatocytes, CD299 as a marker for liver sinusoidal endothelial cells, CD68 as a marker for Kupffer cells and PD-L1. Colocalization analysis on images acquired by confocal microscopy determined a strong correlation between PD-L1 with both Kupffer cells and liver sinusoidal endothelial cells and an inverse correlation with hepatocytes (Fig. S4).
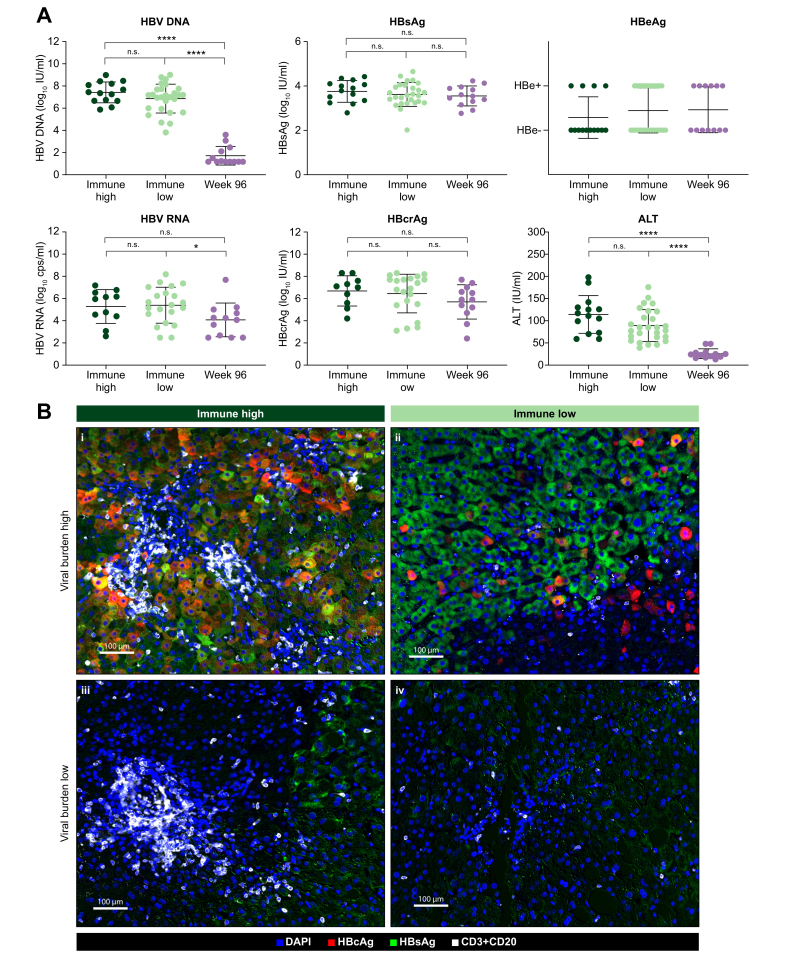
Viral antigen burden is heterogeneous and does not correlate with the liver immune microenvironment
Next, we determined if there was a correlation between viral load and immune activity in the liver. Surprisingly, none of the viral biomarkers (HBV DNA, HBV RNA, HBcrAg, HBsAg or HBeAg) correlated with liver immune microenvironment (Fig. 5A). We have also observed a lack of correlation between HBsAg and the immune microenvironment in previous studies.25 However, ALT was lower in the immune low patients and the p value (p = 0.054) neared statistical significance (Fig. 5A). Analysis of the HBcAg and HBsAg burden in the liver demonstrated significant heterogeneity in the number of positive hepatocytes (Fig. S5). HBcAg ranged from undetectable in some samples to a high of 87% HBcAg-positive hepatocytes (mean is 14.8% for HBeAg+ and 0.52% for HBeAg-). HBsAg ranged from 2% to 56% HBsAg-positive hepatocytes (mean is 25.44% for HBeAg+ and 20.24% for HBeAg-)(Fig. S5). Correlation analyses confirmed that HBcAg staining correlated with peripheral viral antigens more strongly than HBsAg staining, likely due to the presence of HBsAg+/HBcAg- hepatocytes with HBV DNA integrations.26 Image overlays consisting of both lymphocytes (CD3 and CD20 merged into one channel, white) and viral antigens (HBcAg in red and HBsAg in green) demonstrate that periportal lymphoid aggregates appear in patients irrespective of viral burden (Fig. 5B, i vs. iii). Additionally, two liver biopsies with similarly high antigen burden had markedly different levels of periportal lymphoid infiltrate (Fig. 5B, i vs. ii and iii vs. iv). In addition, single-plex IHC was performed on a larger cohort of samples and demonstrated a high heterogeneity in viral antigen burden in the liver and no correlation to the liver immune microenvironment (Fig. S5).
Fig. 5.
Immune group clusters are not differentiated by viral antigen burden.
(A) Patient sera matched to each liver biopsy was analyzed for viral biomarkers and ALT levels. The viral biomarkers HBV DNA, HBV RNA, HBcrAg, HBsAg and HBeAg were analyzed from each patient and timepoint and compared between immune groups. P values were calculated using unpaired t tests. (B) mIF channels for HBcAg, HBsAg, CD20 and CD3 were overlaid to visualize intrahepatic immune signatures in relation to viral antigens. Representative images from 4 individual samples representing immune high and immune low signatures at baseline with corresponding high or low viral antigen burdens are shown. HBcAg (red), HBsAg (green), CD3/CD20 merged (white) and DAPI (blue). IHC, immunohistochemistry; mIF, multiplex immunofluorescence.
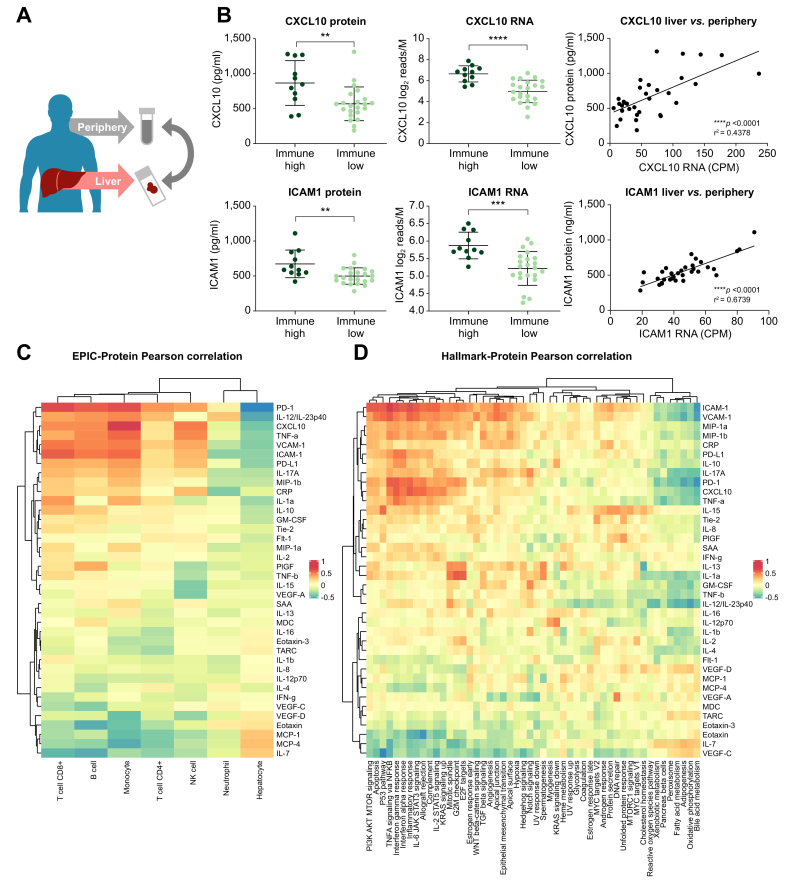
ICAM-1 and CXCL10 levels in the periphery correlate with liver immune gene signatures
Since the heterogeneity of the liver immune microenvironment did not correlate with established viral biomarkers, we performed a targeted screen for peripheral biomarkers of liver inflammation. Our screen consisted of 41 host cytokines and chemokines including 39 analytes in the MSD Human V-plex Screening Panel 1 biomarker set as well as soluble PD-1 (sPD-1) and soluble PD-L1 (sPD-L1). The level of each serum biomarker was correlated to its corresponding liver gene expression (Fig. 6A). We then determined if biomarkers with strong correlation to their intrahepatic expression levels could identify the immune high and immune low populations. This analysis yielded two chemokines: ICAM-1 and CXCL10 (Fig. 6B). Intrahepatic gene expression for CXCL10 (p <0.0001) and ICAM-1 (p = 0.0004) was significantly higher in immune high vs. immune low patients. In addition, both CXCL10 (p = 0.0001) and ICAM-1 (p <0.0001) liver gene expression correlated to the peripheral chemokine values. Next, we correlated all the peripheral biomarkers with the EPIC immune cell types and hallmark immune pathways and found that CXCL10 correlates with intrahepatic interferon signaling pathways (Fig. 6C and D). In addition, sPD-1 levels correlated with intrahepatic T-cell signatures (Fig. 6C). sPD-1 also correlated with intrahepatic PDCD1 gene expression but did not reach statistical significance with immune high and immune low sample sets.
Fig. 6.
Targeted biomarker screen for peripheral correlates of the liver immune clusters.
(A) Patient sera were analyzed using Meso-Scale Discovery platform. (B) Serum protein levels of each analyte were compared to the corresponding liver expression levels using Pearson’s correlation analysis. CXCL10 and ICAM-1 protein significantly correlate with their liver gene expression and differentiate the immune groups. (C) Peripheral biomarkers were correlated to intrahepatic immune pathway signatures using Hallmark analysis. (D) Peripheral biomarkers were correlated to intrahepatic cell signatures using EPIC analysis.
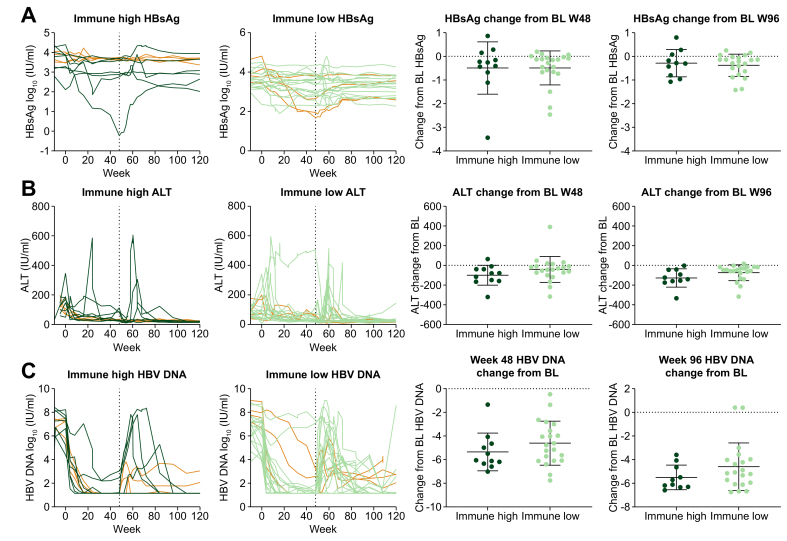
As this liver biopsy collection was associated with a clinical trial, we integrated clinical outcomes to the liver immune microenvironment analysis. HBV DNA, HBsAg and ALT were plotted from 33 patients who completed treatment from baseline to week 96 and divided by immune high and immune clusters (Fig. 7). Eight patients from our collection experienced HBeAg loss (Fig. 7, orange lines) including 4 immune high and 4 immune low patients. Unfortunately, no patients from which we have biopsies experienced HBsAg loss. Although we did not observe differentiated outcomes between immune high and immune low clusters, this analysis is confounded by the fact that patients were evenly randomized across 4 different treatment arms and most receive TDF. We would not anticipate that TDF treatment response would be influenced by the liver immune microenvironment.
Fig. 7.
Clinical outcomes analysis for immune high vs. immune low patients.
Clinical parameters (A) HBsAg, (B) ALT, (C) HBV DNA were plotted over time between baseline and week 96 for patients from immune high (n = 11) and immune low (n = 21) groups. Changes from baseline to week 48 or week 96 were also calculated. Eight patients that experienced HBeAg loss are highlighted in orange. ALT, alanine aminotransferase.
Discussion
We examined the intrahepatic immune microenvironment in biopsies collected through voluntary donation within the GS-US-174-0149 clinical trial.9 Biopsies were predominantly from baseline treatment-naïve patients, although a small number of biopsies were obtained post-treatment. Using hierarchical unsupervised clustering of the transcriptome, two different liver immune microenvironments were identified. Immune high patients had elevated immune pathways including interferon signaling pathways, and elevated immune cell signatures corresponding to B cells, T cells and macrophages (Fig. 1) as opposed to immune low patients. Additionally, these patients had periportal lymphoid aggregates, as shown by mIF. Post-treatment samples demonstrated even further suppressed immune pathways and cell type signatures compared to immune low samples (Fig. 2). Surprisingly, neither liver nor peripheral viral burden correlated with the immune microenvironment at baseline (Fig. 5). However, suppression of HBV DNA did correlate with decreased liver inflammation.
Immune high and immune low samples were assessed by mIF to determine the location of the intrahepatic immune cells. We observed CD3+, CD4+, CD8+, CD20+ and CD68+ cells aggregating in periportal regions of immune high samples (Fig. 3). Although we did not quantify T follicular helper cells or follicular dendritic cells, the organization of these immune cells in tissue, outside of lymph nodes, resembles TLS. In support of this observation, we noticed an increase in gene signatures associated with TLS in immune high samples (Fig. S3).22,[27], [28], [29], [30] We have previously observed similar follicular structures assembling in CHB chimpanzees treated with the TLR7 agonist, GS-9620, and formation of these structures was temporally correlated with antiviral response to treatment.31 The immune high samples trended towards elevated ALT, suggesting that the intrahepatic immune aggregates were leading to hepatocyte killing. Whether this is non-specific or HBV-specific hepatocyte death is unknown. However, TCR and BCR clonality was assessed using RNA-Seq and did not suggest that the infiltrating T and B cells were enriched for any specific clonotypes (Fig. S3C). Studies have shown that non-specific intrahepatic T cells are a driver of liver inflammation, and that patients without liver inflammation have a higher proportion of HBV-specific T cells and a lower intrahepatic viral burden.32,33 This further suggests that the intrahepatic T cells observed here are unlikely to represent a largely HBV-specific population. In addition to traditional T-cell subsets, we were also able to use Ki67 and FOXP3 staining to determine that immune high samples had increased numbers of Tregs and proliferating CD4 and CD8 T-cell subsets (Fig. 3A). We were unable to differentiate whether this reflects a specific change in intrahepatic immune cell subsets or if this is reflective of the overall increase in total immune cells.
The immune high samples demonstrated a significantly higher expression level for several key immune checkpoints, as well as elevated PD-1 and PD-L1 staining by IHC (Fig. 4). Elevated expression of checkpoint receptors and ligands during chronic infection is typically associated with T-cell exhaustion and dysfunctional immune responses. Suppression of checkpoint inhibition in cancer has demonstrated significant clinical outcomes, and this is more apparent in “hot” tumors with elevated immune activity.23,24 One study, investigating nivolumab (anti-PD-1), demonstrated safety and modest efficacy in patients with CHB, as indicated by HBsAg declines from baseline.23 However, this study only enrolled nucleos(t)ide analogue-suppressed individuals. Our data demonstrate that PD-1 expression is suppressed in this patient cohort and may limit target engagement and T-cell activation. Nucleos(t)ide analogue-treated patients with CHB may mimic “cold” tumors which correlate with decreased efficacy of checkpoint inhibition. Although treatment of viremic patients with elevated immune checkpoint expression may lead to the highest target engagement, balancing efficacy with safety in these studies will be imperative as that patient population will likely have elevated ALT at baseline (Fig. 5 and Table S1).
HBV liver burden was determined using both mIF and single-plex IHC analyses and was found to be highly heterogeneous among patients. HBcAg was expressed at higher levels in HBeAg-positive samples compared to HBeAg-negative samples, however no such relationship was observed for HBsAg staining. HBeAg-negative samples contained many cells that were HBsAg-positive, but HBcAg-negative, suggesting that these cells may be expressing HBsAg from integrated HBV. We observed no correlation between the viral load, as measured in the liver or the periphery, to the immune microenvironment (Fig. 4 and Fig. S5). As HBsAg has been demonstrated to regulate the immune response we anticipated that we may observe an inverse correlation between HBsAg burden and intrahepatic inflammation; however, this was not observed.34 We have also observed this lack of correlation between HBsAg and intrahepatic immune signatures in other sample collections.25 Immune tolerant patients are characterized as having high levels of HBV DNA and HBsAg in serum and a high burden of infected hepatocytes, yet have limited intrahepatic immune activity, suggesting that the presence of virus alone is not sufficient to activate an immune response. However, treatment and the corresponding decrease in HBV DNA levels did correlate with ALT reduction and a reduction in liver immune cells suggesting an underlying relationship. The fact that elevated levels of HBV DNA can be recognized by macrophages may represent part of the link between nucleos(t)ide analogue suppression and ALT normalization.35
We performed a targeted biomarker screen to correlate peripheral chemokines and cytokines to the liver immune microenvironment. We identified serum CXCL10 and ICAM-1 as peripheral biomarkers that correlated with their liver gene expression and differentiated immune high and immune low microenvironments (Fig. 6). CXCL10 also correlated with intrahepatic immune cell signatures for T cells, B cells and monocytes as well as the interferon-α and interferon-γ signaling pathways. Elevated baseline CXCL10 has been demonstrated to correlate with response to PEG-IFNα.11 These data further characterize this relationship and imply a PEG-IFNα response mechanism that relies on preexisting intrahepatic immune cells – stimulation of these immune cells may drive HBV clearance. The exact immunological mechanism underlying response still requires further clarification. The periportal lymphoid aggregate structures that we observed may function similar to TLS found in other tissues.22 We are unsure if T-cell and B-cell priming is occurring within these structures. Future mIF studies that incorporate markers for T follicular helper cells and follicular dendritic cells may help uncover the function of these lymphoid aggregates. At baseline, we do not see evidence for TCR and BCR clone enrichment that would indicate intrahepatic expansion (Fig. S3). However, one hypothesis is that treatment with PEG-IFNα, or other immune modulators, may drive expansion of HBV-specific T and B cells from within these structures and drive HBV cure.
As HBV cure strategies continue to explore immune modulator therapy, further characterization of the liver immune microenvironment, its heterogeneity, how it is affected by nucleos(t)ide analogue treatment, and the continued identification of peripheral biomarkers will be critical to ensure that the most suitable patients are enrolled in these early studies. These data indicate that nucleotide analogue treatment decreases the level of intrahepatic immune activity, including downregulation of immune checkpoints. The intrahepatic immune microenvironment is likely to have implications for both the safety and efficacy of many of these future investigational medicines.
Financial support
This study was funded by Gilead Sciences Inc.
Authors’ contributions
Experimental design and analysis: NvB, RR, ST, DC, VS, BF. Bioinformatics: RR, SK. Pathology support and image analysis: ST, AA, CM, DK. Sample Management: NB, NB, VS. Sample collection: HC, PM, MB. Writing and Revisions: NvB, RR, ST, DC, JW, AG, SF, LD, LL, HM and BF.
Data availability statement
Sequencing data is available upon request.
Conflict of interest
All authors are either employees of Gilead Sciences or funded by Gilead Sciences for clinical trial sample collection.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
We thank the patients for voluntary liver biopsy donation within GS-US-174-0149.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2021.100388.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Organization WH . 2017. Hepatitis B.https://www.who.int/news-room/fact-sheets/detail/hepatitis-b [cited; Available from: [Google Scholar]
- 2.Iloeje U.H., Yang H.I., Jen C.L., Su J., Wang L.Y., You S.L., et al. Risk and predictors of mortality associated with chronic hepatitis B infection. Clin Gastroenterol Hepatol. 2007;5:921–931. doi: 10.1016/j.cgh.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 3.Fattovich G., Stroffolini T., Zagni I., Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35–S50. doi: 10.1053/j.gastro.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 4.Hou J., Brouwer W.P., Kreefft K., Gama L., Price S.L., Janssen H.L.A., et al. Unique intrahepatic transcriptomics profiles discriminate the clinical phases of a chronic HBV infection. PLoS One. 2017;12 doi: 10.1371/journal.pone.0179920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karmen A., Wroblewski F., Ladue J.S. Transaminase activity in human blood. J Clin Invest. 1955;34:126–131. doi: 10.1172/JCI103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yim H.J., Lok A.S. Natural history of chronic hepatitis B virus infection: what we knew in 1981 and what we know in 2005. Hepatology. 2006;43:S173–S181. doi: 10.1002/hep.20956. [DOI] [PubMed] [Google Scholar]
- 7.Liaw Y.F., Chu C.M. Hepatitis B virus infection. Lancet. 2009;373:582–592. doi: 10.1016/S0140-6736(09)60207-5. [DOI] [PubMed] [Google Scholar]
- 8.European Association for the Study of the Liver EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67:370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 9.Marcellin P., Ahn S.H., Ma X., Caruntu F.A., Tak W.Y., Elkashab M., et al. Combination of tenofovir disoproxil fumarate and peginterferon alpha-2a increases loss of hepatitis B surface antigen in patients with chronic hepatitis B. Gastroenterology. 2016;150:134–144 e110. doi: 10.1053/j.gastro.2015.09.043. [DOI] [PubMed] [Google Scholar]
- 10.Wu H.L., Hsiao T.H., Chen P.J., Wong S.H., Kao J.H., Chen D.S., et al. Liver gene expression profiles correlate with virus infection and response to interferon therapy in chronic hepatitis B patients. Sci Rep. 2016;6:31349. doi: 10.1038/srep31349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willemse S.B., Jansen L., de Niet A., Sinnige M.J., Takkenberg R.B., Verheij J., et al. Intrahepatic IP-10 mRNA and plasma IP-10 levels as response marker for HBeAg-positive chronic hepatitis B patients treated with peginterferon and adefovir. Antivir Res. 2016;131:148–155. doi: 10.1016/j.antiviral.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Blueprint Epigenome Project. 2015. http://www.blueprint-epigenome.eu/ [cited; Available from: [Google Scholar]
- 13.Racle J., Gfeller D. EPIC: a tool to estimate the proportions of different cell types from bulk gene expression data. Methods Mol Biol. 2020;2120:233–248. doi: 10.1007/978-1-0716-0327-7_17. [DOI] [PubMed] [Google Scholar]
- 14.Racle J., de Jonge K., Baumgaertner P., Speiser D.E., Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife. 2017;6 doi: 10.7554/eLife.26476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerdes M.J., Sevinsky C.J., Sood A., Adak S., Bello M.O., Bordwell A., et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A. 2013;110:11982–11987. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Bommel F., Bartens A., Mysickova A., Hofmann J., Kruger D.H., Berg T., et al. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology. 2015;61:66–76. doi: 10.1002/hep.27381. [DOI] [PubMed] [Google Scholar]
- 21.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sautes-Fridman C., Petitprez F., Calderaro J., Fridman W.H. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat Rev Cancer. 2019;19:307–325. doi: 10.1038/s41568-019-0144-6. [DOI] [PubMed] [Google Scholar]
- 23.Gane E., Verdon D.J., Brooks A.E., Gaggar A., Nguyen A.H., Subramanian G.M., et al. Anti-PD-1 blockade with nivolumab with and without therapeutic vaccination for virally suppressed chronic hepatitis B: a pilot study. J Hepatol. 2019;71:900–907. doi: 10.1016/j.jhep.2019.06.028. [DOI] [PubMed] [Google Scholar]
- 24.Topalian S.L., Drake C.G., Pardoll D.M. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rico Montanari N., Ramirez R., Van Buuren N., van den Bosch T.P.P., Doukas M., Debes J.D., et al. Transcriptomic analysis of livers of Inactive Carrier HBV patients with differential HBsAg. J Infect Dis. 2021 doi: 10.1093/infdis/jiab381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez R., van Buuren N., Gamelin L., Soulette C., May L., Han D., et al. Targeted long-read sequencing reveals comprehensive architecture, burden and transcriptional signatures from HBV-associated integrations and translocations in HCC cell lines. J Virol. 2021:JVI0029921. doi: 10.1128/JVI.00299-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hennequin A., Derangere V., Boidot R., Apetoh L., Vincent J., Orry D., et al. Tumor infiltration by Tbet+ effector T cells and CD20+ B cells is associated with survival in gastric cancer patients. Oncoimmunology. 2016;5 doi: 10.1080/2162402X.2015.1054598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becht E., de Reynies A., Giraldo N.A., Pilati C., Buttard B., Lacroix L., et al. Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin Cancer Res. 2016;22:4057–4066. doi: 10.1158/1078-0432.CCR-15-2879. [DOI] [PubMed] [Google Scholar]
- 29.Messina J.L., Fenstermacher D.A., Eschrich S., Qu X., Berglund A.E., Lloyd M.C., et al. 12-Chemokine gene signature identifies lymph node-like structures in melanoma: potential for patient selection for immunotherapy? Sci Rep. 2012;2:765. doi: 10.1038/srep00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coppola D., Nebozhyn M., Khalil F., Dai H., Yeatman T., Loboda A., et al. Unique ectopic lymph node-like structures present in human primary colorectal carcinoma are identified by immune gene array profiling. Am J Pathol. 2011;179:37–45. doi: 10.1016/j.ajpath.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L., Barry V., Daffis S., Niu C., Huntzicker E., French D.M., et al. Anti-HBV response to toll-like receptor 7 agonist GS-9620 is associated with intrahepatic aggregates of T cells and B cells. J Hepatol. 2018;68:912–921. doi: 10.1016/j.jhep.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maini M.K., Boni C., Lee C.K., Larrubia J.R., Reignat S., Ogg G.S., et al. The role of virus-specific CD8(+) cells in liver damage and viral control during persistent hepatitis B virus infection. J Exp Med. 2000;191:1269–1280. doi: 10.1084/jem.191.8.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertoletti A., Maini M.K. Protection or damage: a dual role for the virus-specific cytotoxic T lymphocyte response in hepatitis B and C infection? Curr Opin Immunol. 2000;12:403–408. doi: 10.1016/s0952-7915(00)00108-4. [DOI] [PubMed] [Google Scholar]
- 34.Jiang M., Broering R., Trippler M., Poggenpohl L., Fiedler M., Gerken G., et al. Toll-like receptor-mediated immune responses are attenuated in the presence of high levels of hepatitis B virus surface antigen. J Viral Hepat. 2014;21:860–872. doi: 10.1111/jvh.12216. [DOI] [PubMed] [Google Scholar]
- 35.Cheng X., Xia Y., Serti E., Block P.D., Chung M., Chayama K., et al. Hepatitis B virus evades innate immunity of hepatocytes but activates cytokine production by macrophages. Hepatology. 2017;66:1779–1793. doi: 10.1002/hep.29348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data is available upon request.