Abstract
Aging is associated with many deleterious changes at the cellular level, including the accumulation of potentially toxic components that can have devastating effects on health. A key protective mechanism to this end is the cellular recycling process called autophagy. During autophagy, damaged or surplus cellular components are delivered to acidic vesicles called lysosomes, that secure degradation and recycling of the components. Numerous links between autophagy and aging exist. Autophagy declines with age, and increasing evidence suggests that this reduction plays important roles in both physiological aging and the development of age-associated disorders. Studies in pharmacologically and genetically manipulated model organisms indicate that defects in autophagy promote age-related diseases, and conversely, that enhancement of autophagy has beneficial effects on both healthspan and lifespan. Here, we review our current understanding of the role of autophagy in different physiological processes and their molecular links with aging and age-related diseases. We also highlight some recent advances in the field that could accelerate the development of autophagy-based therapeutic interventions.
Keywords: Aging, autophagy, neurodegeneration, lifespan, healthspan, mTOR, AMPK, C. elegans
1. INTRODUCTION TO AGING
Aging can be defined as the time-dependent decline in function of a living organism that ultimately leads to death (López-Otín et al., 2013). Aging is a universal phenomenon throughout the animal kingdom, but the lifespans of diverse species can differ dramatically. For example, mayflies generally survive for only hours or days, while bowhead whales can live for >200 years (Cohen, 2018). Indeed, even closely related animal species may have quite different lifespans, as illustrated by mice and mole rats, which are similarly sized rodents but have markedly different lifespans of 1–2 years and >30 years, respectively (Ruby et al., 2018). Interestingly, and undoubtedly related, mole rats are highly resistant to disease, including cancer (Buffenstein and Ruby, 2021). Individuals within the same species can also display diverse lifespans and healthspans, a phenomenon known as heterogeneity of organismal aging. Inhabitants of some regions of the globe have longer than average lifespans, which may be at least partially linked to diet and lifestyle, but isolated cases of individuals with exceptional longevity have also been documented, such as the French woman Jeanne Calment, who lived for ~122 years (Robin-Champigneul, 2020). Conversely, people suffering from progeroid syndromes experience profoundly accelerated aging and severe health issues, culminating in death at ~20 years of age, or even earlier (Gordon et al., 2014). Understanding the intrinsic and extrinsic factors that lead to such diversity of lifespan and healthspan in the animal kingdom is of fundamental importance in comprehending the biology of aging in humans.
The average lifespan of the global human population is steadily increasing, generating significant concern about the rapidly expanding public health burden. Aging is the main risk factor for numerous devastating diseases that require intensive and/or long-term treatment, such as neurodegenerative disorders, cancer, and cardiovascular diseases (https://www.nia.nih.gov/research/dbsr/global-aging). It is therefore imperative to reduce the socioeconomic pressure brought about by the aging society, requiring the involvement of a broad spectrum of stakeholders including politicians, economists, gerontologists, and researchers (Fang et al., 2020). Moreover, understanding the molecular mechanisms underlying the aging process will be of crucial importance to support the development of novel therapies for age-related disorders and to prolong human healthspan.
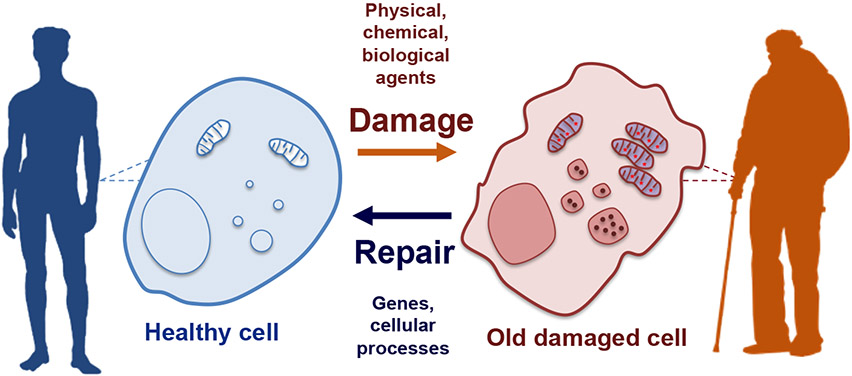
There are several theories behind the molecular basis of aging, of which the most widely accepted is the progressive accumulation of damaged cellular components (Figure 1) (Johnson et al., 1999). Throughout our lives, we are exposed to stressful or damaging insults that affect the structure and function of the complex molecular machinery required for cell and organismal health. These insults may be extrinsic, such as radiation, chemicals, and pathogens, or intrinsic, such as inherited mutations in the case of some progeroid syndromes. Regardless of the source, insults that modify the properties of cellular organelles or macromolecules can start a chain of events that perpetuate the damage, thereby endangering cellular integrity, function, and survival. While the accumulation of cellular damage may be initiated as a localized event, it can also be communicated distally throughout the organism in a cell non-autonomous manner, and thus contribute to the overall decline in organismal health. This is usually mediated via inflammatory responses, that can become chronic and uncontrolled eventually affecting distal tissues/organs within a given organism, accelerating the age-related organismal changes (López-Otín et al., 2013). Living organisms have evolved several mechanisms to ensure the timely and efficient removal of potentially deleterious cellular components, including DNA repair pathways, the ubiquitin-proteasome system (UPS) of protein degradation, and various forms of the major cellular recycling process, autophagy. These protective molecular pathways and repair mechanisms play essential roles in maintaining homeostasis, but they too are susceptible to extrinsic and intrinsic insults that affect their activity (López-Otín and Kroemer, 2021). Thus, the rate at which cells age can be viewed as a function of the cumulative effects of damaging insults balanced by the efficiency of the mechanisms dedicated to damage repair/removal.
Figure 1. Damage accumulation and the molecular basis of aging.
In everyday life, humans are exposed to many stressors that can damage cellular components. Under normal circumstances, homeostatic mechanisms clear damaged material from the cell, but they can accumulate with time and eventually render the cells non-functional and/or induce a pathogenic state. To counteract these challenging insults, cells have evolved diverse cytoprotective mechanisms that include various types of autophagy. Cellular aging is thus dictated by the balance between the rate of damage infliction/accumulation and repair/clearance.
Among the protective pathways that maintain cellular and organismal homeostasis (López-Otín and Kroemer, 2021), autophagy has emerged as a central mechanism (Hansen et al., 2018). Accordingly, the autophagy machinery and autophagy-regulating pathways have been identified as modulators of longevity, and not surprisingly, dysregulation of autophagy is recognized as a key factor in the development of many age-related diseases (Hansen et al., 2018). This review aims to discuss our current understanding of the molecular basis of the cytoprotective and longevity-promoting roles of autophagy, the events that underlie the age-associated decline in the pathway, and interventions that may extend both lifespan and healthspan in an autophagy-dependent manner.
2. INTRODUCTION TO AUTOPHAGY
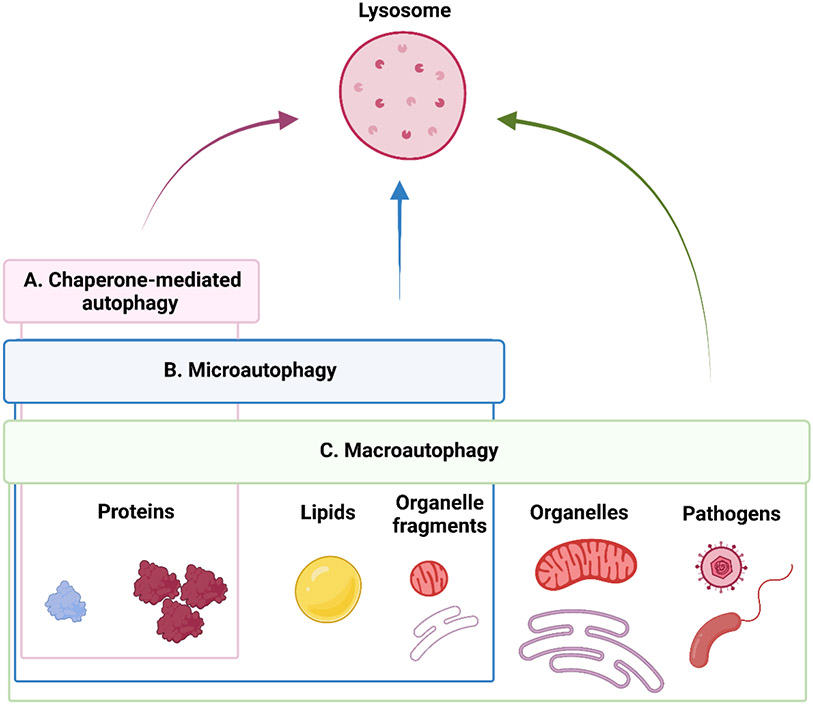
Autophagy, from the Greek words for “self” and “eating”, is a catabolic process that enables the degradation and recycling of diverse cellular components (Dikic and Elazar, 2018), including surplus constituents, aberrantly aggregated or damaged macromolecules and organelles, and invading pathogens (Figure 2). Consequently, autophagy operates continually to maintain cellular homeostasis and is also responsive to triggers that increase pathway activity under stressful conditions, such as nutrient deprivation, growth factor limitation, genotoxic stress, and organelle damage (Kroemer et al., 2010).
Figure 2. Pathways of autophagy-mediated lysosomal degradation.
Autophagy allows the degradation of diverse intracellular components via delivery to the lysosomes. Three types of autophagy exist that are defined by the molecular mechanisms that facilitate cargo delivery: chaperone-mediated autophagy, microautophagy, and macroautophagy. Each autophagy type is specialized to enable the degradation of different cargos, with macroautophagy mediating the disposal of the most diverse array of cytoplasmic components, from proteins to organelles. This review focus on macroautophagy in the context of aging.
2.1. Types of autophagy
Autophagic degradation involves the targeting of cellular components for recycling and their subsequent delivery via alternative mechanisms to lysosomes (Figure 2), which are cellular organelles containing hydrolases that operate under low pH conditions to degrade biomolecules and transform them into their building blocks (Yim and Mizushima, 2020). There are different molecular routes that allow the delivery of cellular components to lysosomes for autophagic degradation. Accordingly, autophagy can be classified as chaperone-mediated autophagy (CMA), microautophagy, and macroautophagy (Figure 2). CMA has been described in mammals and birds (Kaushik and Cuervo, 2018), while microautophagy and macroautophagy are conserved in nearly all eukaryotes (Schuck, 2020). CMA is the process by which select protein cargos are internalized into the lysosome lumen through a transmembrane protein translocation complex, guided by specialized proteins known as chaperones (Figure 2A) (Kaushik and Cuervo, 2018). CMA-degraded proteins are characterized by the presence of a common “KFERQ-like” amino acid motif, which is widely represented in the human proteome and is recognized by the chaperone proteins (Kaushik and Cuervo, 2018). Microautophagy implies the invagination of the lysosomal membrane to engulf an array of different cargos, from proteins to lipids and organelle fragments in a bulk or selective manner (Figure 2B) (Schuck, 2020). In mammals, a related process named endosomal microautophagy (eMI) involves delivery of cargo via inward budding of endosomes, instead of lysosomes, to ensure cargo degradation (Sahu et al., 2011). Notably, the chaperone Hsc70, involved in CMA, can also participate in cargo delivery during eMI (Tekirdag et al., 2018; Mukjerhee et al., 2016). Finally, macroautophagy mediates the degradation of diverse, often large, cargo, including proteins, lipids, organelles, and pathogens (Figure 2C). For this, cargo is sequestered into a specialized de-novo double-membrane vesicle called autophagosome, which originates from a membranous precursor called a phagophore. Mature autophagosomes loaded with cargo are transported toward the nuclear periphery of the cell to encounter and fuse with lysosomes which concentrate there, to ultimately allow cargo degradation, which happens in a hybrid organelle called the autolysosome (reviewed in [Dikic and Elazar, 2018]). CMA plays a crucial role in aging and age-related processes and has been comprehensively reviewed (Kaushik and Cuervo, 2018). Little is known about the relevance of microautophagy in organismal aging, particularly in mammals, and this pathway remains understudied. This review will focus predominantly on macroautophagy (hereafter referred to as autophagy), which has been widely studied in the context of aging.
2.2. Stages of autophagy and the autophagy machinery
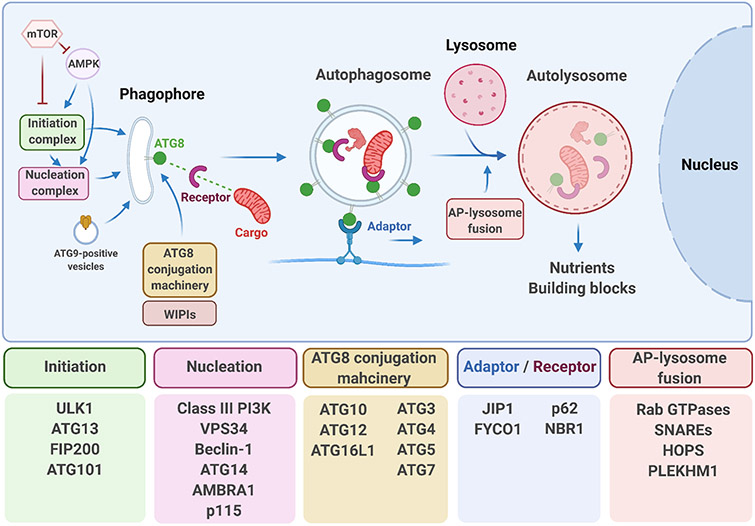
Autophagosomes are formed through a complex cascade of events supported by multiple groups of proteins, collectively referred to as the autophagy machinery. The overall multistep process and the main proteins involved are shown in Figure 3. Autophagosome biogenesis starts with the activity of the initiation complex composed of the ULK1 serine/threonine kinase together with additional regulatory proteins (reviewed in [Dikic and Elazar, 2018]). The ULK1 complex triggers various phosphorylation events that lead to the activation of the phospho-inositol tri-phosphate PI3KC3 complex I that operates during the nucleation stage of the process (Figure 3), which culminates in the formation of a membranous autophagosome precursor called the phagophore (Figure 3). PI3KC3 complex I catalyzes the production of the membrane lipid phosphatidylinositol 3-phosphate (PI3P), which promotes recruitment of PI3P-binding proteins such as members of the WIPI protein family (WD-repeat protein interacting with phosphoinositides) to the site of autophagosome formation (reviewed in [Dikic and Elazar, 2018]). This event, in conjunction with the delivery of lipids via vesicles containing the only transmembrane protein of the core autophagy machinery, ATG9, contributes to the growth and expansion of the phagophore (reviewed in [Dikic and Elazar, 2018]) (Figure 3). The binding of WIPIs to the growing phagophore facilitates conjugation of the ATG8/LC3 family of proteins to the autophagosome membranes, which is a hallmark event in the autophagy process. In humans, the ATG8 family of proteins comprises seven members LC3A (2 splice variants), LC3B, LC3C, GABARAP, GABARAPL1, and GABARAPL2 (reviewed in [Schaaf et al., 2016]). Microscopic detection of foci containing ATG8/LC3 or its orthologs is a common experimental readout of autophagic function. A diverse subset of proteins, around one-third of the core autophagy machinery, participates in the processing and lipid conjugation of ATG8 to the phagophore membrane via a cascade of ubiquitin-like conjugation reactions (Figure 3). Once conjugated to the growing phagophore membrane, ATG8 proteins allow the recruitment of a set of key proteins that will differentially decorate the inner and the outer membrane of the closed autophagosome via diverse key protein-protein interactions. In the inner part of the vesicle, ATG8 binds receptor proteins, such as the protein SQSTM-1/p62, that simultaneously recruits the cargo targeted for degradation into the autophagosome lumen. In the outer membrane, ATG8s recruit adaptor proteins involved in the transport, such as FYCO1 (Pankiv et al., 2010), and also the fusion, such as PLEKHM1 (McEwan et al., 2015) of the autophagosomes with the lysosomes (reviewed in [Nieto-Torres et al., 2021a]). Once the autophagosome fuses with the lysosome, a complex process that involves small Rab GTPases, soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) including the protein syntaxin 17 (STX17), and the HOPS-tethering complex (Jiang et al., 2014), a hybrid organelle called the autolysosome is formed (Figure 3). The lysosomal hydrolases will then degrade the autophagosomal content to recycle it and produce building blocks that will be exported to the cell cytoplasm and can be used in new biosynthetic reactions (Yim and Mizushima, 2020).
Figure 3. Overview of the molecular events underlying macroautophagy.
Autophagy is a multistep process broadly defined by the formation of a double-membraned vesicle called autophagosome in which cargo is recruited. Autophagosomes originate from the progression and growth of a membranous structure called phagophore. The fusion of autophagosomes with lysosomes, finally allows degradation of the cargo. Prominent examples of the autophagy molecular machinery members implicated in the different steps are shown in the boxes.
2.3. Autophagy regulatory networks
The autophagy process is tightly controlled by signaling networks that play major roles in sensing and integrating metabolic cues such as altered nutrient, energy, and growth factor levels, among others. The two main regulatory pathways involve the mechanistic target of rapamycin (mTOR) and AMP-activated protein kinase (AMPK) (Figure 3), kinases that sense nutrient availability and intracellular energy status, respectively. When nutrient levels are high, mTOR inhibits autophagy by phosphorylating and inactivating the initiation kinase ULK1 (reviewed in [Kim and Guan, 2015]). Conversely, in low nutrient conditions, mTOR activity is inhibited, and autophagy activity increases to restore nutrient availability. AMPK is activated by low energy levels. Specifically, a high intracellular AMP/ATP ratio triggers AMPK, which increases autophagy by activating ULK1, the PI3KC3 complex I components Beclin-1, and VPS34, and by inactivating mTOR (reviewed in [Kim and Guan, 2015]). Acute nutrient depletion also reduces cellular levels of acetyl coenzyme A, a key metabolite formed during glycolysis and fatty acid oxidation, which plays crucial roles as an acetylation cofactor in many pathways. This triggers the deacetylation of multiple autophagy proteins and their concomitant activation, in which the deacetylase Sirtuin-1 (also a downstream member of the AMPK pathway) and the acetyltransferase EP300 participate (Madeo et al., 2014; Mariño et al., 2014; Su et al., 2017). Because post-translational modifications occur rapidly, regulation of autophagy machinery proteins via phosphorylation and acetylation allows the cell to respond promptly to autophagy-inducing insults. However, in the long term, autophagy machinery availability needs to be sustained by increased transcriptional/translation of key autophagy components.
The transcription factor EB (TFEB), a member of the microphthalmia transcription factor E (MiT/TFE) family, is a master regulator of the expression of key genes involved in autophagy and lysosomal biogenesis. TFEB activity is controlled by its nuclear/cytoplasmic localization, which is largely regulated by mTOR (Napolitano et al., 2018). Another major family of autophagy-regulating proteins is the forkhead box O (FOXO) transcription factors (Webb and Brunet, 2014). FOXO activity, like TFEB, is regulated predominantly by its subcellular localization, which is controlled mainly by the growth factor- and insulin-sensing pathway kinase AKT. Notably, FOXO activity can be further boosted via Sirtuin-1-mediated deacetylation (Brunet et al., 2004). Studies using Caenorhabditis elegans, a short-lived and genetically tractable nematode that is extensively used for longevity studies (Apfeld and Alper, 2018), have revealed that HLH-30 (TFEB ortholog) regulates transcription of autophagy genes (Lapierre et al., 2013; O’Rourke et al., 2013), at least in part in cooperation with DAF-16 (FOXO ortholog) (Lin et al., 2018). Besides this, epigenetic modifications may also contribute to the maintenance of autophagic activity over the longer term (Lapierre et al., 2015). Nutrient-sensing pathways that regulate histone methylation and acetylation have also been reported to control the transcription of autophagy-related and lysosomal genes (reviewed in [di Malta et al., 2019]). Importantly, these regulatory pathways as well as the autophagy machinery described above, have been linked to aging and lifespan regulation which will be described next.
3. MOLECULAR LINKS BETWEEN LONGEVITY AND AUTOPHAGY
An extensive body of evidence suggests that autophagy, aging, and the onset of age-related diseases are closely interconnected. Model organisms, especially unicellular budding yeast (Saccharomyces cerevisiae), invertebrates such as nematodes (C. elegans) and fruit flies (Drosophila melanogaster) and vertebrates such as mice, have been instrumental in aging research, due in large part to their relatively short lifespans and genetic tractability (Hansen et al., 2018). Different reports indicate that increased autophagic function correlates with better health and lifespan, and boosting autophagy can have longevity-promoting effects, as determined via genetic experiments done in multiple model organisms. In turn, the age-dependent decline of autophagic activity has been observed in different organisms.
Longevity paradigms in S. cerevisiae, C. elegans, and Drosophila show increased expression of autophagy and lysosomal genes, as well as the potentially higher autophagic capacity. Conversely, impairment of the expression or function of autophagy and lysosomal genes abrogates the lifespan extensions observed in these paradigms (Hansen et al., 2018). These findings strongly suggest that the beneficial effects on lifespan in these models are autophagy-dependent. Whole-body genetic deletion of autophagy genes results in mice that fail to survive neonatal starvation (Komatsu et al., 2005; Kuma et al., 2004), and deletion of autophagy genes during adulthood leads to rapid lethality of mice, which succumb to infections and suffer from neurodegeneration, consistent with the need for autophagy as a pathogen- and neuroprotective pathway (Karsli-Uzunbas et al., 2014). On a related note, overexpression of autophagy-related genes increases lifespan in model organisms. For example, overexpression of HLH-30/TFEB in C. elegans extends lifespan in an autophagy-dependent manner (Lapierre et al., 2013), and overexpression of ATG8 or ATG1 (ULK1 ortholog) in Drosophila or of Atg5 in mice similarly promotes longevity, although it is not yet formally known whether autophagy induction is required to observe lifespan extension in the latter models (Pyo et al., 2013; Simonsen et al., 2008; Ulgherait et al., 2014). Interestingly, the presence of a point mutation in murine Beclin-1 that renders it more resistant to BCL2-mediated inhibition was shown to confer systemic health benefits in wild-type mice and to significantly prolong lifespan in mouse models of accelerated aging (Fernández et al., 2018). Overall, longevity paradigms in multiple model organisms rely on autophagy for their lifespan benefits, and overexpression of autophagy components can promote lifespan. This suggests a correlation between autophagy levels and longevity, although this has to be formally tested. Some reports suggest that this may be conserved in human individuals. For example, autophagic activity in T cells from individuals belonging to families with reported high longevity is increased as compared to T cells derived from age-matched people (Raz et al., 2017). Whether the autophagy–longevity link represents causation or correlation in humans and its potential tissue/organ specificity remains to be further investigated. Overall, these data suggest that sustaining autophagic activity and preventing its age-related decline, at levels probably no higher than a physiologically tolerated level, may be beneficial to health.
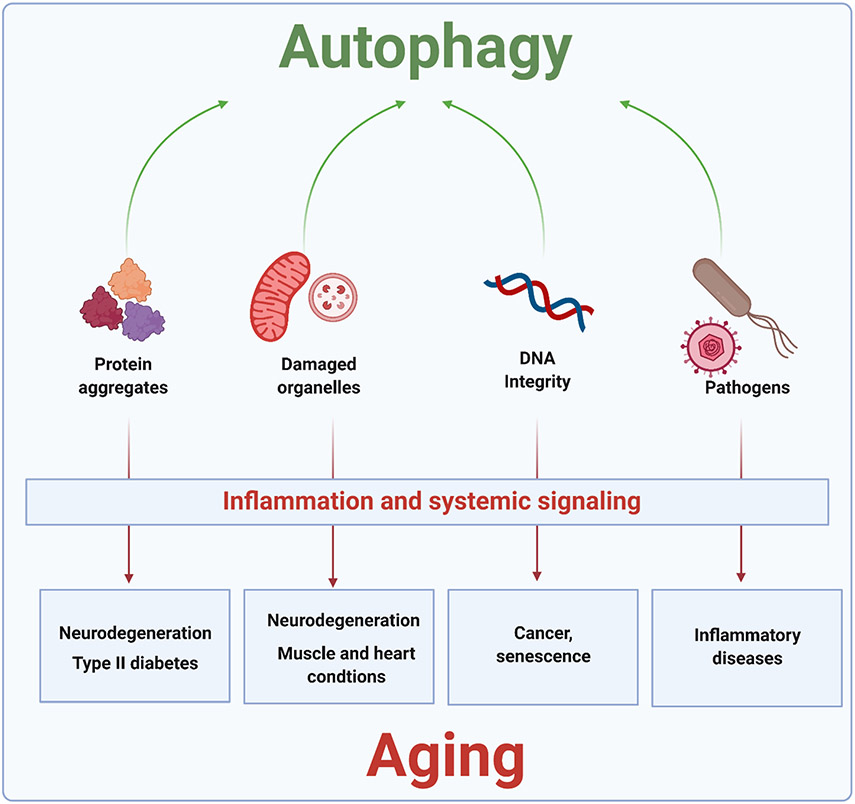
Autophagy plays a pivotal role in the maintenance of physiological homeostasis and in protection from pathogenic states, both of which are crucial to optimal healthspan and lifespan. In the following sections, we review some of the main homeostatic/protective functions of autophagy (summarized in Figure 4).
Figure 4. Protective roles of autophagy in promoting cellular and organismal health.
By orchestrating the degradation of potentially harmful cellular components, including protein aggregates, dysfunctional organelles (including mitochondria and lysosomes), cytoplasmic DNA, and pathogens, autophagy plays a major role in maintaining cellular homeostasis. Aging and the development of many age-related diseases correlate with a decline in the rate and/or efficiency of autophagy and recycling of specific types of debris, leading to their accumulation. This can trigger cellular dysfunction, followed by tissue/organismal damage which is frequently amplified by inflammatory signaling leading to different diseases. See text for details.
3.1. Proteostasis
The human proteome comprises >10,000 different proteins that drive most of the physiological functions of our cells (Hipp et al., 2019). Proteins are susceptible to damage that may render them nonfunctional and even pathogenic, as is the case for protein misfolding and aggregation in some neurodegenerative disorders (Hipp et al., 2019). Toxic and dysfunctional proteins are targeted for degradation, and together with the UPS, autophagy constitutes a major proteolytic pathway (Vilchez et al., 2014). Accordingly, perturbations that affect the core autophagy machinery in model organisms lead to an increase in the age-associated protein aggregation, shorten lifespan, and exacerbates pathological features, while increasing autophagy has the opposite effects on proteostasis and health (Boland et al., 2018; Rubinsztein et al., 2011). Besides the core autophagy proteins, autophagy receptors may suffer age-related alterations in their function leading to compromised degradation of select cargos. The expression levels of the autophagy receptor SQSTM-1/p62 decrease with age in mice (Kwon et al., 2012), and the protein levels and subcellular localization are dysregulated in mouse models of Huntington’s disease (Rue et al., 2013). Furthermore, dysregulation of p62 promoter and decreased p62 levels correlate with neurodegenerative diseases in humans (Du et al., 2009a; Du et al., 2009b). p62 recruits proteins and aggregates that have been targeted by ubiquitination for destruction, including advanced glycation end products, which accumulate with age and in several diseases (Aragonès et al., 2020), and has been implicated in longevity regulation in several species. Specifically, overexpression of the p62 ortholog SQST-1 in C. elegans is sufficient to promote proteostasis, induce autophagy, and extend lifespan (Kumsta et al., 2019), and overexpression of the p62 homolog Ref(2)p in Drosophila has similar effects on lifespan and autophagy (Aparicio et al., 2019). These findings suggest that autophagy of select forms of cargo (e.g., ubiquitinated proteins) mediated by specialized receptors may play an important role in aging and longevity, an open question that needs to be studied in more detail.
The deposition of protein aggregates is a hallmark of many age-related diseases (Iadanza et al., 2018) (Figure 4). Neuronal tissue is especially susceptible to defects in protein homeostasis, as the majority of human neurons are postmitotic cells, and therefore they are not renewed via stem cell progenitors. Not surprisingly, the accumulation of protein aggregates of different origins is a hallmark of neurodegenerative processes, such as Alzheimer’s disease (amyloid β, tau), Parkinson’s disease (α-synuclein), Huntington’s disease (Huntingtin), and amyotrophic lateral sclerosis (SOD1, TDP-43) (reviewed in [Monaco and Fraldi, 2020; Lautrup et al., 2019]). Accordingly, inhibition of autophagy in mouse neurons via Atg5 or Atg7 depletion leads to the appearance of cytoplasmic aggregates in the brain as well as accelerated neurodegeneration in mice (Hara et al., 2006; Komatsu et al., 2006). In the long run, the accumulation of protein aggregates can affect autophagic functions, creating a vicious cycle that amplifies the detrimental effects of malfunctioning protein homeostasis. For example, α-synuclein aggregates affect lysosomal function and signaling, compromising the autophagy process, and polyglutamine expansions in the protein huntingtin that make it prone to aggregation, affect autophagosome biogenesis and transport (reviewed in [Monaco and Fraldi, 2020]).
3.2. Lipid metabolism
Lipid droplets are subcellular lipid storage structures that play key roles in energy mobilization in many eukaryotic cells. Upon energy depletion, part of the cellular lipid content stored in lipid droplets can be directly metabolized in an autophagy-dependent manner, also known as lipophagy (Singh et al., 2009). Lipid droplets are coated with proteins that play essential roles in lipophagy. Perilipins (PLN) are one of the most abundant lipid droplet-associated proteins, with PLN5 promoting lipophagy (Zhang et al., 2020), while PLN2 coating protects lipid droplets from autophagic degradation (Griffin et al., 2021). Interestingly, both PLN2 and PLN3 are substrates of CMA, and therefore CMA-based degradation of these proteins, in turn, promotes lipophagy (Kaushik et al., 2015). Finally, direct transfer of lipids from lipid droplets to lysosomes, bypassing the need of autophagosome formation, has also been discovered (Schulze et al., 2020). Lipophagy may be especially important in the liver, which is a major organ for lipid storage in vertebrates. Consistent with this possibility, autophagy depletion in mouse liver cells increases the number and size of lipid droplets, whereas activation of autophagy by drugs such as the mTOR inhibitor rapamycin promotes co-localization of lipids with endolysosomal and autophagy markers, indicative of lipophagy (Singh et al., 2009). Deficient lipid homeostasis can lead to devasting conditions such as liver steatosis and cardiovascular diseases, for which autophagy-related function may play an important role.
Genetic inhibition of autophagy via Atg7 depletion in mouse liver leads to histological changes resembling non-alcoholic fatty liver disease in humans (Singh et al., 2009). Other studies in model organisms and humans have identified a tight link between autophagy and lipid stores in aging (Liu and Czaja, 2013). For example, age-dependent increases in lipid stores, as well as ectopic fat deposition, have been associated with autophagy dysfunction in C. elegans (Palikaras et al., 2017), and the age-associated failure in autophagy in the liver of mice leads to fatty acid accumulation (Singh et al., 2009). Finally, impaired autophagy has also been connected to the accumulation of toxic peroxidized lipids, which can be severely damaging and potentially lethal to cells (Ding et al., 2011).
3.3. Organelle homeostasis
3.3.1. Mitochondria
Cellular organelles are also susceptible to damage that may accumulate with time, rendering them nonfunctional and even pathogenic. While damaged proteins can be cleared by either the UPS or autophagy, malfunctioning organelles are mostly degraded by autophagy. Particularly important from a health perspective is the role of autophagy in the turnover of dysfunctional mitochondria, which entirely relies on the autophagy machinery. Mitochondria are highly metabolically and synthetically active organelles and are the central source of energy production through oxidative phosphorylation (reviewed in [Chen et al., 2020]). Mitochondria are also a source of endogenous reactive oxygen species, which play important physiological roles in signal transduction but can also induce oxidative damage to macromolecules when present at abnormally high levels. In addition, damage of mitochondrial membranes can result in the cytoplasmic release of mitochondrial components, including cytochrome c and mitochondrial DNA, which are sensed as danger signals and trigger proinflammatory and cell death pathways (reviewed in [López-Otín and Kroemer, 2021]).
Mitophagy, the selective autophagic turnover of mitochondria, is deceased in normal aging, many age-related diseases, and in progeroid disorders (Bakula and Scheibye-Knudsen, 2020; Palikaras et al., 2018; Fang et al., 2019a). For example, impaired mitophagy is observed in human patients with Werner syndrome, as well as in C. elegans and Drosophila models of this progeroid syndrome (Fang et al., 2019a). Defective mitophagy has been linked with neurodegeneration in ataxia telangiectasia, a rare recessive human disease (Fang et al., 2016), as well as in xeroderma pigmentosum group A disorder (Fang et al., 2014). Similarly, the accumulation of dysfunctional mitochondria in dopaminergic neurons is a hallmark of Parkinson's disease (reviewed in [Pickrell and Youle, 2015]), and impaired removal of damaged mitochondria leading to neuroinflammation is a key event in Alzheimer’s disease pathogenesis (Fang et al., 2019b). Mitophagy is facilitated by PTEN-induced kinase 1 (PINK) and Parkin, a ubiquitin ligase, and mutations in these genes in humans correlate with early-onset Parkinson's disease (reviewed in [Pickrell and Youle, 2015]). In addition, there are PINK/Parkin-independent mitophagy pathways that rely on the recognition of dysfunctional mitochondria by autophagy receptors such as NIX, BNIP3, p62, and FUNDC1 (Bakula and Scheibye-Knudsen, 2020). Furthermore, other pathways that rely on the autophagy machinery but are different from mitophagy, operate to maintain mitochondrial homeostasis, such as the “piecemeal” mitophagy pathway that allows basal degradation of mitochondrial proteins (le Guerroué et al., 2017), and the mitochondrial-derived vesicles that allow the delivery of mitochondrial fractions to the lysosome and may play important roles in cancer cells (Towers et al., 2020).
Besides its important role in neuronal health, regulation of mitochondrial turnover is also relevant to tissues that have high energy demands, such as skeletal muscle cells and cardiomyocytes. A decline in mitophagy has been detected in the hearts of aging mice, and deficiencies in mitophagy correlate with cardiovascular diseases such as cardiomyopathy and atherosclerosis (Bravo-San Pedro et al., 2017; Hoshino et al., 2013). Similarly, skeletal muscle stem cell progenitors from elderly humans show accumulation of mitochondria and p62 (Garcϭa-Prat et al., 2016), suggesting that mitophagy may be compromised.
3.3.2. Lysosomes
Lysosomal homeostasis is crucial to the maintenance of cellular function and organismal health, not only because of its central role in autophagy and other metabolic pathways but also because damage to lysosomes can result in leakage of the potentially harmful contents into the cytoplasm (Papadopoulos et al., 2020). Leaky lysosomes can lead to the activation of apoptotic pathways (Gómez-Sintes et al., 2016) and the presence of lysosomal enzymes in the cell cytoplasm correlates with aging and age-related diseases such as neurodegeneration (reviewed in [Papadopoulos et al., 2020]). Endogenous repair mechanisms exist to ensure normal lysosomal function and membrane integrity. However, if these mechanisms fail, defective lysosomes are degraded by a selective form of autophagy known as lysophagy, which implies lysosome encapsulation in autophagosomes and their subsequent degradation via lysosomal hydrolases provided by healthy lysosomes (reviewed in [Papadopoulos et al., 2020]).
3.3.3. Endoplasmic reticulum
The endoplasmic reticulum (ER) is a vast cellular organelle that supports protein translation, modification, and folding, lipid metabolism, and other critical cellular functions (Wilkinson, 2019). Although the ER has the ability to upregulate its activity in response to stressors, degradation of the ER is required to maintain normal homeostasis under both basal and stressful conditions. Autophagy-mediated degradation of the ER, or ERphagy, may have implications in aging, as suggested by ERphagy deficiencies identified in mouse models of progeroid syndromes (Peng et al., 2018).
3.4. DNA integrity and senescence
Lysosomes can actively degrade both RNA and DNA, which can be delivered to the lysosomal lumen via the interaction of nucleic acid-associated proteins with proteins in the lysosomal membrane through a process resembling CMA (Fujiwara et al., 2013). Furthermore, autophagosome-mediated transfer of DNA to lysosomes has been observed in certain pathological conditions (Luo et al., 2016; Park et al., 2009). Normally the genetic material of eukaryotic cells is contained within the nuclear membrane, and similarly mitochondrial DNA is well protected inside of mitochondria. However, aberrant events or pathogenic states may lead to the release of DNA fragments to the cell cytoplasm, which can activate inflammatory and pro-aging pathways, as happens in senescent cells (Lan et al., 2019; Yang et al., 2021). Upon different types of severe stress, including DNA damage, cells can activate the senescence program that results in an irreversible proliferation arrest. This has important roles in physiology, including prevention of cancer, but the accumulation of senescent cells within tissues, has been associated with aging and detrimental tissue/organ function (Song et al., 2020). Accordingly, selective elimination of senescent cells holds promising therapeutic potential (van Deursen, 2019). Senescent cells extrude cytoplasmic-chromatin fragments (CCFs) that contribute to the activation of a complex secretory program of proinflammatory mediators termed SASP (secretory-associated senescent phenotype) with pro-aging effects (Kumari and Jat, 2021). The autophagy machinery has been tightly linked to CCFs. Specifically, autophagy participates in the degradation of these DNA fragments, which prevents activation of the GMP-SMP synthase (cGAS) and stimulator of interferon genes (STING) inflammatory pathway (Dou et al., 2017). Thus, autophagy may possess an anti-senescent role. Accordingly, accumulation of senescent cells with age in the joints leads to osteoarthritis, and genetic autophagy depletion in chondrocytes predisposes mice to age-dependent osteoarthritis (Bouderlique et al., 2016; Zhang et al., 2015). Despite the potential protective anti-senescence role of autophagy, as happens with cancer, the senescence-autophagy connection may be more complex, as once formed, senescent cells seem to rely on enhanced autophagic activity, and the autophagy machinery itself may participate in the formation of CCFs, as well as the promotion of the SASP (Dou et al., 2017).
3.5. Pathogens and inflammation
Pathogens can cause extensive cellular and organismal damage. Bacteria and viruses that access the cytoplasm of eukaryotic cells can be targeted for degradation via autophagy (Levine et al., 2011). Not surprisingly, multiple pathogen recognition and innate immunity pathways lead to the activation of autophagy (Deretic, 2021). Recurrent infections are more frequent in older individuals, often showing less proficient immune systems that lead not only to defects in pathogen killing but also to deregulated inflammatory responses (Cuervo and Macian, 2014) events that may have a connection with the autophagy machinery, as explained below.
Inflammatory responses are integral to the physiology of organisms and participate in diverse processes ranging from wound healing to defense against pathogens. However, when inflammatory responses are uncontrolled or become chronic, they can lead to a profound systemic amplification of damaging responses throughout the whole organism leading to diseases such as type 2 diabetes, cardiovascular disease, cancers, and dementia. Interestingly, there is a clear correlation between a progressive inflammatory increase with age and the detrimental effects of this in the organisms, promoting the term “inflammaging” as an active area of research (Franceschi et al., 2018).
Impaired autophagy is tightly linked with chronic inflammatory responses (Deretic, 2021; Rubinsztein et al., 2011). As described above, inefficient autophagy-mediated degradation of defective organelles, cytoplasmic or mitochondrial DNA, or pathogens can lead to the activation of diverse innate immunity pathways via the activation of receptors that recognize damage-associated molecular patterns (DAMPs) or pathogen associated-molecular pathogens (PAMPs). Therefore autophagy provides the cells with a natural anti-inflammatory mechanism. While these pathways are highly relevant for every single cell type within an organism, they become even more important in immune cells, which control and amplify systemic inflammatory responses. Striking examples of this include how periodontal disease can lead to cardiovascular disease amplification via the action of specific subsets of T cells and the effects of gut microbiota-related inflammatory cell-mediated responses in the brain (reviewed in [Franceschi et al., 2018]). A subset of immune cells, such as macrophages, dendritic cells, and neutrophils, contain specific DAMPs/PAMPS sensors such as the NLRP3 inflammasome. NLRP3 constitutes a tightly regulated system that operates to control the release of IL-1β, one of the major inflammatory markers. Selective knockdown of key autophagy components leads to unrestricted activation of the inflammasome, and macrophages with defective autophagy are linked with increased IL-1β secretion and damage of endothelial tissue (Razani et al., 2012). Overall, decreased autophagy in phagocytic cells has been suggested to cause deregulated activation of the inflammasome promoting age-dependent increased inflammation (Cuervo and Macian, 2014).
3.6. Non-canonical degradation pathways that utilize the autophagy machinery
Interestingly, components of the autophagy machinery are also implicated in the lysosomal degradation of pathogens and toxic proteins, uptaken from the extracellular media via endocytic pathways (reviewed in [Nieto-Torres et al., 2021a]). To facilitate these processes, ATG8 proteins incorporate into single-membrane vesicles of endosomal origin (as opposed to newly formed double-membrane autophagosomes during canonical autophagy) also via the ATG8 conjugation machinery, including the proteins ATG3, ATG4, ATG5, ATG7, ATG10, ATG12, and ATG16L1. Through its WD40 domain, which is dispensable for canonical autophagy, the protein ATG16L1 is able to recognize and target ATG8 conjugation into these vesicles of endosomal origin (Fletcher et al., 2018). Examples of these processes involving non-canonical functions of the autophagy machinery include LC3-associated phagocytosis (LAP), a crucial degradative process that allows engulfment of extracellular pathogens via phagocytosis and their subsequent clearance via lysosomal function. LAP is used by macrophages, among other cell types, to counteract infections and mount appropriate defensive inflammatory responses and therefore is a key immunity-related pathway (Heckmann and Green, 2019). Accordingly, LAP is important to control Streptococcus pneumoniae infections, an invasive pneumonia-causing disease that is particularly serious in the elderly. Interestingly, experiments done in mice revealed that macrophages from old individuals showed deficient LAP, bactericidal capacity, and a concomitant increased pro-inflammatory response, as opposed to their young counterparts (Inomata et al., 2021). LAP deficiency in professional phagocytic cells of the immune system may also have important health implications for other life-threatening pathogens such as Legionella neumophila and Mycobacterium tuberculosis, which cause severe diseases in the aged (Rajagopalan and Yoshikawa, 2000; Sopena et al., 2007). Likewise, LAP is extremely important in blood-separated tissues like the retina, testis, and ovarian follicles to eliminate apoptotic bodies and maintain homeostasis (Yefimova et al., 2021). In the ocular tissue, LAP is critical for the removal of photoreceptor outer segments by the retina pigment epithelium, and defects in this process have been observed during age-related macular degeneration (Saito et al., 2020; Kim et al., 2013). While connections between LAP and age-related diseases exist, the mechanisms leading to LAP deregulation with age involving the autophagy machinery are not fully understood.
A second non-canonical cellular degradation and clearance pathway involving the autophagy machinery is LC3-associated endocytosis (LANDO). This pathway is particularly important in microglia, the main phagocytic cells of the central nervous system. LANDO plays a crucial role in the prevention of neurodegenerative diseases not only via uptake and clearance of toxic extracellular molecules such as β-amyloid but also via recycling of cell surface β-amyloid receptors. Consistent with this role, defects in LANDO in microglia leads to β-amyloid deposition, proinflammatory signaling, and accelerated neurodegeneration in mouse models (Heckmann et al., 2019). Of note, aged mice lacking the WD40 domain of ATG16L1, which is required for non-canonical autophagy, show enhanced age-related accumulation of β-amyloid and neuroinflammation as well as accelerated neurodegeneration (Heckmann et al., 2020). Mechanistically, the ATG16L1 WD40 domain is required for LANDO-mediated recycling of β-amyloid receptors in microglia and prevention of detrimental proinflammatory signaling in neural tissue impacting neurodegeneration.
4. MOLECULAR BASIS OF THE AGE-ASSOCIATED DECLINE IN AUTOPHAGY
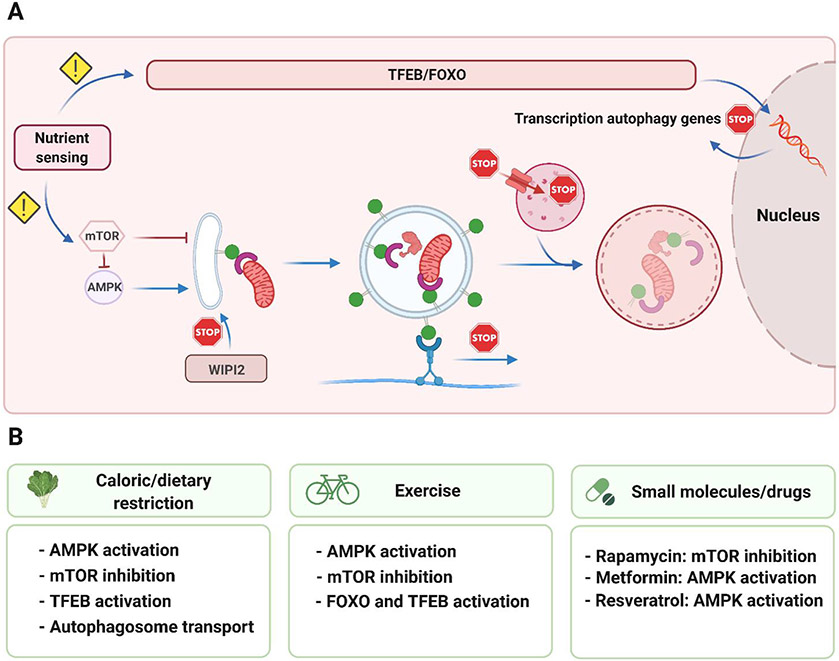
Considerable evidence suggests that reduced autophagy contributes to aging. Studies in many organisms have shown that autophagic activity naturally declines with age, and cells from long-lived animals, including humans, tend to have elevated basal levels of autophagy. Moreover, defects resulting in reduced autophagic activity have been linked to accelerated aging and the development of age-related disorders (reviewed in [Hansen et al., 2018]). Consistently, aging has also been associated with the appearance of aberrant autophagic structures and altered dynamics between the pools of autophagic vesicles, indicative of pathway dysregulation. A systematic study of autophagy in C. elegans of various ages identified an age-dependent accumulation of stalled autophagic vesicles, including autophagosomes and autolysosomes, in many tissues, suggesting a systemic progressive decline in the autophagy degradative capacity, also known as autophagic flux (Chang et al., 2017). Likewise, ultrastructural analysis of rodent tissues revealed a similar age-related accumulation of autophagic vesicles, in some cases showing aberrant or incomplete morphologies (del Roso et al., 2003; Donati et al., 2001; Stavoe et al., 2019). In the following sections, we discuss some possible molecular mechanisms to explain the observed appearance of aberrant autophagic vesicle structure, dynamics, and function with age (Figure 5).
Figure 5. Molecular events linked to the age-associated decline in autophagy.
(A) The main steps of macroautophagy are depicted together with the events that may fail during aging and in age-related diseases (indicated with a ‘stop’ sign). These include deregulated nutrient signaling, defects in autophagosome biogenesis and transport, as well as failure to maintain low lysosomal pH and lysosomal proteolytic activity. Shown below are several interventions (B), known to mitigate the age-associated loss of autophagy, such as caloric restriction, exercise, and small molecules, and the pathways by which they are thought to act. See text for details.
4.1. Decline in the availability of autophagy machinery components
The sustainability of autophagy, especially under stressful conditions, relies on the inducible expression of core autophagy components. However, aging is accompanied by decreased expression of autophagy-related genes in Drosophila, mice, and humans (Demontis and Perrimon, 2010; Kaushik et al., 2012; Lipinski et al., 2010; Simonsen et al., 2008), suggesting that defects in the transcriptional program regulating autophagy-related gene expression may be a cause of or contribute to the failure of autophagy with age. In support of this idea, and as noted earlier, overexpression of the autophagy transcriptional regulator HLH-30/TFEB increases autophagic flux and extends the lifespan of C. elegans (Lapierre et al., 2013), and nuclear translocation of TFEB, indicative of enhanced activity, is increased in hepatic cells from dietary-restricted long-lived mice (Lapierre et al., 2013). Mechanistically, enhanced TFEB activity may be important to maintain autophagic flux during aging and to protect cells from age-related disease-promoting stressors such as infection and protein aggregation, as has been demonstrated in cellular and mouse models of neurodegenerative and lysosomal storage diseases (Visvikis et al., 2014).
As noted earlier, FOXO transcription factors also regulate the expression of autophagy and lysosomal genes. Dysregulation of FOXO activity has been implicated in human aging and the development of age-related diseases affecting muscle and bone tissues (Rached et al., 2010; Sandri et al., 2004). For example, FOXO activity is reduced and correlates with decreased expression of autophagy genes in the cartilage from elderly individuals and patients with osteoarthritis, indicating that the loss of autophagy in this tissue may play a role in cartilage degeneration (Lotz and Caramés, 2011). Studies in Drosophila identified a parallel age-dependent decline in FOXO activity and muscle function, and conversely, overexpression of FOXO protected against the decline, in part via autophagy-mediated regulation of proteostasis (Demontis and Perrimon, 2010). Interestingly, genetic variants of FOXO and insulin-like growth factor-I (IGF-I) receptor, which controls FOXO activity, have been identified in some human populations with exceptional longevity (Suh et al., 2008; Willcox et al., 2008). However, it remains to be determined whether enhanced autophagy plays a role in the extended lifespans of these individuals, bearing in mind that FOXO regulates multiple other homeostatic pathways that may contribute to longevity, including proteostasis, oxidative stress resistance, and stem cell renewal (Martins et al., 2016).
4.2. Alterations in upstream autophagy-regulating pathways
Efficient sensing of nutrient and growth factor availability plays a key role in maintaining and regulating autophagic activity via transcriptional and post-translational regulation of the autophagy machinery. One hallmark of aging is the progressive failure of nutrient-sensing pathways (López-Otín et al., 2013), which, in turn, leads to the age-associated deregulation of the autophagy machinery.
The insulin/IGF-1 signaling pathway is a conserved glucose-sensing mechanism that integrates nutrient availability with autophagy, as well as other pathways, via FOXO and mTOR. Constitutively decrease of insulin/IGF-1 signaling is a known and conserved intervention known to extend lifespan in several model organisms that confers metabolic and protective beneficial effects preventing damage accumulation, in part via autophagy. Although it may look contradictory, decreased insulin/IGF-1 signaling has also been shown to accompany both natural and accelerated aging in mouse models (reviewed in [López-Otín et al., 2013]). This phenomenon may reflect an attempt done by already damaged tissues/organisms to decrease metabolism and upregulate FOXO-regulated protective pathways, including autophagy.
Decreased and increased mTOR activity upregulates and downregulates, respectively, autophagic flux. Aberrant activation of mTOR has been reported in senescent human cells (Carroll et al., 2017; Leontieva et al., 2014). Increased signaling downstream of mTOR has been observed in muscle tissues of elderly mice and humans (Sandri et al., 2013), tissues from mice with progeroid syndromes, and neuronal tissue from patients with severe Alzheimer’s disease (Sun et al., 2014). Moreover, genetic or pharmacological inactivation of mTOR results in lifespan extension in several model organisms, including yeast, C. elegans, and Drosophila (Nacarelli et al., 2015). Interestingly, rapamycin-induced inhibition of mTOR in cells from patients with Hutchinson–Gilford progeria syndrome delays the onset of cellular senescence and increases the degradation of progeria-related proteins (Cao et al., 2011). In C. elegans, genetic inactivation of mTOR fails to extend the lifespan of autophagy-deficient animals, indicating that lifespan extension induced by reduced mTOR activity is mediated, at least in part, via autophagy (Tóth et al., 2008). Similarly, this has been observed in S. cerevisiae and Drosophila (reviewed in [Hansen et al., 2018]).
4.3. Aberrant regulation of autophagosome dynamics
An imbalance between the biogenesis and degradation of autophagy-related structures may contribute to the decline in autophagy observed in age-associated diseases and aging. The age-associated dysregulation of post-translational regulatory pathways that operate on autophagy proteins to control autophagosome dynamics, and/or the aberrant interplay of these autophagy proteins with other cellular factors, can lead to defects in the autophagy process. For example, studies performed in mouse dorsal root ganglion neurons have revealed a decrease in autophagosome biogenesis, which was not due to changes in the kinetics of autophagy initiation or nucleation. Instead, these defects were connected to the PI3P-binding protein WIPI2 and its phosphorylation status (Stavoe et al., 2019). In aged mice neurons, deficient WIPI signaling has been connected with the appearance of multilamellar aberrant autophagic structures, often also seen in Alzheimer’s patients (Nixon, 2005). Strikingly, overexpression of WIPI2 is sufficient to restore proficient autophagosome biogenesis rates (Stavoe et al., 2019). Whether age-associated autophagy impairment associated with defects in WIPI2 post-translational modification is specific for mouse neurons or also occurs in other species and cell types remains to be elucidated. Given the particular importance of efficient autophagic activity in neurons, modulation of WIPI2 levels may hold promise as a target for therapeutic interventions for age-related neuronal defects.
Movement of mature cargo-containing autophagosomes towards the perinuclear region of the cell for fusion with lysosomes is a crucial step in autophagy. Not surprisingly, the failure of the molecular pathways that regulate this directed transport can compromise the entire autophagy process. For example, alanine mutation of autophagosome protein ATG8/LC3B in its Thr50 phosphorylation site or knockdown of the ATG8-phosphorylating Hippo kinase STK4 leads to autophagy block in mouse embryonic fibroblasts (Wilkinson et al., 2015). This phenotype is likely caused by aberrant recruitment of transport-related proteins and impaired retrograde transport, resulting in reduced autophagosome-lysosome association, as observed in human HeLa cells and primary hippocampal mouse neurons (Nieto-Torres et al., 2021b). The potential implications of this pathway during aging still remain to be elucidated, however, recent experimental evidence indicates that age-dependent autophagy deficiencies may be connected with inefficient or dysregulated recruitment of the cell transport machinery to autophagosomes and lysosomes. Specifically, in isolated fibroblasts and hepatocytes from aged mice, the transport-related proteins dynein and KIF3C associate less with autophagosomes and lysosomes, respectively, leading to deficient retrograde transport of autophagosomes and compromised autophagosome-lysosome fusion (Bejarano et al., 2018). Age-associated autophagosome transport defects may be especially relevant in neurons, not only because, in general, they are postmitotic cells that rely extensively on autophagy but also because neurons are highly polarized cells in which a very important fraction of autophagosome is synthesized at the axon tip, whereas the majority of lysosomes resides in the soma (Hill and Colón-Ramos, 2020). In fact, deficiencies in retrograde transport have been reported in mouse models of Alzheimer's disease, where β-amyloid oligomers prevent proper recruitment of the transport-related protein dynein in neurons (Tammineni et al., 2017).
4.4. Alterations in lysosomal function
Lysosomal activity is essential for the completion of the autophagy process (Yim and Mizushima, 2020). Therefore, lysosomal function may be one of the major steps of the autophagy process that upon alteration can have negative consequences on cellular homeostasis and organismal health, noting that not only macroautophagy but also CMA and microautophagy converge at this endpoint stage. Age-dependent malfunctioning of lysosomes has been reported in many different organisms, which correlates with the accumulation of damaged/unfolded proteins within cells (reviewed in [Martinez-Vicente et al., 2005]). Many times, this also parallels with the accumulation of non-degradable pigmented components within lysosomes, known as lipofuscin. Accumulation of lipofuscin seems to be slower in long-lived animals (Nakano et al., 1995). The abundance of lysosomal lipofuscin is therefore considered to be an indicator of cell and tissue aging.
The lysosomal function is directly related to its proteolytic capacity. Cathepsins constitute the main family of lysosomal proteases (Stoka et al., 2016). Pharmacological inhibition of cathepsins in the brains of young rats induces phenotypic features reminiscent of brains from aged animals, including lipofuscin accumulation (Nunomura and Miyagishi, 1993). Brain samples from aged rats exhibit reduced activity, but not protein levels, of cathepsin L compared with brain samples from young rats (Nakanishi et al., 1994). Interestingly, other studies have shown that the activity of other lysosomal cathepsins is either unaltered (Nakanishi et al., 1994) or increases (reviewed in [Stoka et al., 2016]) with age in the brains of rats. Although it is not known which cell types were responsible for the age-related alterations in cathepsins in those brain samples, an age-associated increase in cathepsin activity has been reported in microglia, which, in turn, was associated with detrimental inflammatory responses in the brain (reviewed in [Stoka et al., 2016]). The presence of lysosomal cathepsins in the cytosol of neuronal cells in rats has been suggested to be an age-associated phenotype reflective of a breakdown in homeostasis (Sato et al., 2006).
Control of lysosomal pH is critical for the optimal activity of proteases as well as other key processes such as nutrient transport (Yim and Mizushima, 2020). Studies done in yeast showed that lysosomal pH increases with age, compromising mitochondrial degradation. Interestingly, pH is restored in ‘young’ daughters of aged yeast cells (Hughes and Gottschling, 2012). Moreover, caloric restriction, which is a well-characterized promoter of longevity in many species (see section below), prevents an increase in vacuolar pH in aging yeast cells via effects on V-ATPase, the main regulator of lysosomal acidification (Hughes and Gottschling, 2012). Accordingly, overexpression of Vma1p, a V-ATPase component, or Vph2p, which facilitates V-ATPase assembly, extends the lifespan of yeast (Hughes and Gottschling, 2012; Ruckenstuhl et al., 2014), firmly establishing the importance of proper lysosomal function in longevity regulation. Similarly, studies in C. elegans have identified a number of age-associated defects in lysosomal biology in the hypodermis and intestine, including increased pH and aberrant morphology, volume, motility, and degradative capacity. Interestingly, these defects were suppressed in various long-lived C. elegans mutants that require an increased expression of lysosomal genes for lifespan extension (Sun et al., 2020). Neurodegeneration has been associated with loss-of-function mutations in V-ATPase components in Drosophila, and with reduced activity of V-ATPase secondary to depletion of the regulator ATp6ap2 in mouse and fly models (Dubos et al., 2015). While the expression of many V-ATPase subunits and regulators is controlled by TFEB (Sardiello et al., 2009), it remains to be tested whether age-dependent malfunction of lysosomal pH homeostasis mostly relies on this regulatory axis. Regulation of this signaling pathway as well as V-ATPase activity may hold promising potential toward eventual therapeutic interventions for age-related disorders.
5. PHARMACOLOGICAL AND BEHAVIORAL INTERVENTIONS TO MODULATE AUTOPHAGY AND PROMOTE HEALTHY AGING
Given the many beneficial effects of autophagic function on longevity and the delay of age-related diseases, there is intense interest in identifying interventions that can boost the protective and homeostatic roles of autophagy, such as behavioral modification and administration of small molecules or drugs.
5.1. Dietary regimens
Caloric restriction, defined as limiting nutrient intake without malnutrition, is known to increase both healthspan and lifespan in most, perhaps all, living organisms (Madeo et al., 2019). In higher mammals, caloric restriction usually represents a 20–40% reduction in total caloric intake (Bergamini et al., 2007; Mirzaei et al., 2014). Dietary restriction, defined as limiting the intake of nutrients without changing total calorie intake, is another diet-based regimen that benefits health and longevity. Dietary restriction encompasses a variety of strategies, including short-term starvation, intermittent fasting, and diets lacking specific macronutrients such as proteins or carbohydrates (Lee and Longo, 2016). The limitation of nutrients is a strong autophagy inducer, and autophagy plays a key role in the health and lifespan benefits conferred by decreased caloric intake. Nutrient deprivation activates the energy sensor and autophagy trigger AMPK (reviewed in [Cantó and Auwerx, 2011]), which in conjunction with the activity of the histone deacetylases sirtuins (reviewed in [Chen and Guarente, 2007]) activate autophagy. Via AMPK, and through direct sensing of nutrients, the activity of the nutrient sensor mTOR is also inhibited upon fasting, which in turn leads to direct activation of the autophagy machinery via post-translational mechanisms and the nuclear translocation of TFEB and the concomitant activation of a transcriptional program to support autophagic function.
5.2. Physical activity
Exercise has been proven beneficial for the maintenance of health. Various reports suggest that exercise is a potent autophagy inducer and that the duration and the intensity of exercise are important contributing factors (Andreotti et al., 2020). Mechanistically, exercise leads to activation of AMPK secondary to energy depletion, which is then followed by inactivation of mTOR, activation of ULK1, and initiation of autophagy. In addition, AMPK upregulates the activity of FOXO and thus the transcription of autophagy-related genes (reviewed in [Andreotti et al., 2020]). Prolonged exercise has also been connected with increased nuclear translocation of TFEB (reviewed in [Andreotti et al., 2020]). Importantly, autophagy induction by exercise in mice has been detected beyond skeletal muscle tissue, including in the nervous system, where it may confer beneficial effects (He et al., 2012). Moderate exercise in rats protects against early neurodegeneration, which correlates with enhanced levels of autophagy and mitophagy (Almeida et al., 2018). Swimming also promotes better cognitive function in rats, and this is associated with enhanced lysosomal degradation in the hippocampus (Luo et al., 2017). Thus, the beneficial effects of exercise on autophagic activity, including in the brain, suggest that it plays a positive role in delaying/preventing the onset of age-related disorders.
5.3. Small molecule, drugs, and supplements
Identification of drugs and small compounds that can improve autophagic function is an active field of research, and some of these candidates are currently being tested in clinical trials for diseases of the elderly such as neurodegeneration (Towers and Thorburn, 2016). Also, drugs with potential lifespan/heathspan promoting effects, including autophagy modulators, are actively being tested in mice (https://www.nia.nih.gov/research/dab/interventions-testing-program-itp). These small-molecule autophagy modulators are often found to interfere or signal via nutrient-sensing pathways. A very extensive review on this topic is provided by (Madeo et al., 2019, Galluzzi et al., 2017, Morel et al., 2017). The US Food and Drug Administration-approved drug rapamycin, originally used as an immunomodulator to prevent organ transplant rejection in humans, is a potent inhibitor of mTOR activity and thus an autophagy inducer, besides its modulatory role in many other key pathways such as protein translation (reviewed in [Kaeberlein, 2017]). Several studies in model organisms have demonstrated that rapamycin treatment extends lifespan and improves healthspan in an autophagy-associated manner (reviewed in [Hansen et al., 2018]). Moreover, a recent research initiative is studying the effects of mTOR inhibition on the health of dogs (https://dogagingproject.org/). Given that rapamycin is a potent immunosuppressive agent with potentially severe side effects, testing for age-related pathologies has thus far been limited to elderly human subjects (Kaeberlein, 2017). However, it is possible that anti-aging benefits may be obtained with doses of rapamycin much lower than those that induce immunosuppression. In addition, efforts are being devoted to developing rapamycin-like compounds, “rapalogs” that can affect more specifically mTOR signaling and the autophagy pathway (Kaeberlein, 2017).
Several additional small molecules are known to activate autophagy through alternative mechanisms. Metformin is a glycemia-lowering drug currently used for the treatment of type 2 diabetes. Interestingly, a recent study of diabetic patients treated with metformin revealed a substantial reduction of different age-related pathologies suggesting that this drug may have utility in extending healthspan (Campbell et al., 2017). Metformin is currently being evaluated in clinical trials for its potential effects against aging (NCT02432287). Metformin is known to activate AMPK (Duca et al., 2015), and like rapamycin, it has been found to extend lifespan in several model organisms (Martin-Montalvo et al., 2013) in a manner at least partially involves autophagy induction (Li et al., 2017; Xie et al., 2011). Resveratrol, a polyphenol enriched in grapes, also has lifespan and health benefits in several organisms (reviewed in [Madeo et al., 2019]). Resveratrol increases autophagy by targeting several key upstream regulators, including AMPK and sirtuin 1 (Madeo et al., 2019). Spermidine (a polyamine) is another compound known to induce autophagy in diverse model organisms and to confer autophagy-dependent health and lifespan benefits (reviewed in [Madeo et al., 2019]). Spermidine can be synthesized by human cells and microbiota or obtained through the diet in foods such as nuts. Interestingly, higher than normal levels of spermidine have been detected in blood samples from human centenarians (Pucciarelli et al., 2012). Nicotinamide-adenine dinucleotide (NAD)+ is a central metabolite conserved in all living organisms that participate in many reduction-oxidation reactions (Verdin, 2015). NAD+ is relevant to autophagy through its function as an important cofactor for sirtuins. Notably, systemic NAD+ levels decline in elderly mice and humans, suggesting a possible correlation with the development of age-related diseases (Das et al., 2018). Several studies have reported healthspan benefits after administration of diverse NAD+ derivatives or compounds that promote NAD+ synthesis (reviewed in [Madeo et al., 2019]). Importantly, these beneficial effects may require, at least partially, the autophagy machinery as increased mitophagy upon administration of NAD+ precursors is required for cognitive improvement in Alzheimer’s disease models in C. elegans (Fang, 2019; Fang et al., 2019b). Similarly, NAD+ precursors ameliorate the mitochondrial defects in xeroderma pigmentosum group A human and mouse cells via promoting mitophagy and extends the lifespan of C. elegans models of this disease (Fang et al., 2014).
6. CONCLUSIONS AND FUTURE PERSPECTIVES
A large body of evidence, mostly from model organisms, supports that autophagy plays largely cytoprotective roles, promoting longevity and preventing/mitigating various age-related pathologies (Hansen et al., 2018). As detailed in this review, autophagy serves as a quality-control system that maintains the integrity of the cell by removing defective macromolecules and organelles and ensuring an ample supply of recycled building blocks for their replenishment. The importance of the optimal autophagic activity to long-term health is effectively illustrated by the contributions of altered autophagy to systemic and organ-specific diseases such as metabolic dysfunction, and cancer, non-alcoholic fatty liver disease, myopathies, and neurodegenerative disease, all of which have age as an additional risk factor. Thus, autophagy is vital not only to delay natural cellular aging but also to suppress the emergence of age-related pathologies.
Research over the past ~20 years has begun to elucidate the molecular events underlying the decline in autophagy observed during aging and in certain age-related diseases. Malfunctioning of upstream regulatory pathways, especially nutrient- and growth factor-sensing pathways, compromise autophagy through two routes: via modulation of autophagy initiation, and via regulation of the transcription factors that control autophagy and lysosomal gene expression. The malfunctioning of the upstream nutrient- and growth factor-sensing signaling pathways involving the insulin/IGF-1 as well as mTOR, in turn, compromise the activity of the transcription factors TFEB and FOXO leading to a decrease in the transcription of autophagy and lysosomal genes. The deregulation of these regulatory cascades may play a key role in the age-dependent autophagy decline, which may be especially relevant when autophagy needs to be upregulated in response to nutrient/growth factor deficiencies. There are also examples of the age-dependent decline in the post-translational regulation of different autophagy proteins, with mTOR-based inactivation of the initiation complex possibly being the best characterized one. Other post-translational regulatory pathways with roles in later stages of autophagosome biogenesis might also be susceptible to age-dependent deregulation. For example, Bcl-2 phosphorylation by JNK1 prevents the inhibitory association of Bcl-2 with Beclin-1, thus promoting autophagy nucleation (Wei et al., 2008). While aging has not yet been shown to directly affect this interaction, genetic manipulations that disrupt Bcl-2–Beclin-1 associations have beneficial effects on both healthspan and lifespan in mice (Fernández et al., 2018). Given that phosphorylation and other post-translational modifications play regulatory roles at multiple stages of autophagy such as cargo recruitment, autophagosome transport, and autophagosome-lysosome fusion, for which phosphorylation of ATG8 proteins play key roles (Herhaus et al., 2020; Nieto-Torres et al., 2021b; Shrestha et al., 2020), elucidating the potential impact of aging on autophagy regulation via post-translational modifications will be an exciting topic of future research. Although lysosomal function is known to decline with age, there is still much to learn about the underlying mechanisms. Understanding how age alters the expression of V-ATPase subunits and other regulators of pH, as well as the availability and recruitment of lysosomal proteases, are of particular interest for unraveling the precise contribution of these organelles to the overall decline in autophagy with age.
An emerging concept in the autophagy field, whose impact on aging and age-related diseases is still understudied, is the non-canonical functions of the autophagy machinery (reviewed in [Nieto-Torres et al., 2021a]). As explained above, non-canonical degradative functions displayed by certain autophagy proteins, play key roles in physiology and health, such as LAP and LANDO, and recent evidence suggests that they can be compromised with age (Inomata et al., 2021; Heckmann et al., 2020). This may be a reflection of deficiencies in lysosomal function or ATG8 conjugation/regulation, both of which are involved in canonical and non-canonical autophagic degradation, but further investigation will be needed to clarify this. Finally, and intriguingly, part of the autophagy machinery can also participate in the secretion of a variety of cargos via alternative mechanisms involving autophagosome-like intermediates (reviewed in [Nieto-Torres et al., 2021a]), small extracellular vesicles (Leidal et al., 2020), and large vesicles called exophers (Melentijevic et al., 2017; Nicolás-Ávila et al., 2020). The diverse cargos, which range from inflammatory mediators to RNA-binding proteins and defective mitochondria, affect cell non-autonomous signaling in a manner that may have both physiological and pathological implications (Leidal et al., 2020; Melentijevic et al., 2017; Nicolás-Ávila et al., 2020; Nieto-Torres et al., 2021a). Overall, the mechanisms that regulate this non-canonical processes, the autophagy machinery involved along with ATG8 conjugation, the connection to degradative functions, and the impact of age all remain to be further elucidated.
Whether the age-associated decline in autophagy affects all cells/tissues equally is another area of research that deserves attention. It is not unreasonable to assume that the dynamics of autophagy in various cell types will change not only during development and aging but also under different physiological and pathological conditions. Cells with a limited capacity for self-regeneration, such as neurons, are especially reliant on autophagy and thus become more vulnerable to age-associated perturbations in their homeostasis (Perrotta et al., 2020). Similarly, the ocular tissue in mammals, which contains quiescent and postmitotic cells, highly relies on autophagy for its homeostasis and is especially sensitive to autophagy deficiencies (Morishita et al., 2013; Zhang et al., 2017; Fernandez-Albarral et al., 2021). Furthermore, tissues with high metabolic and energy demands, such as the liver and muscle, are likely to be highly dependent on autophagy (Ueno and Komatsu, 2017; Xu et al., 2020). Along these lines, autophagy is known to be crucial to the dramatic remodeling and transformation of muscle tissue upon exercise (Perrotta et al., 2020). Model organisms will undoubtedly play a large role in understanding tissue-specific effects and the potential involvement of inter-tissue regulatory networks in the age-related decline in autophagy in this. Future work should also include the development of minimally invasive and reliable methods for measuring autophagic activity in humans, ideally in a tissue- and cell-specific manner. Such methods could have major clinical implications for the identification of individuals at risk for developing age-related diseases with known autophagy contributions, such as type 2 diabetes and neurodegenerative diseases.
Lastly, we described some of the behavioral interventions that promote efficient autophagy function and extend healthspan. They include certain dietary regimens, exercise, and small molecule drugs that modulate autophagy at various steps along the pathway. Unraveling the molecular basis of the deleterious effects of age on autophagy will undoubtedly facilitate the discovery of novel interventions that can delay or prevent age-related pathologies by promoting autophagy, preferably in a cell/tissue-specific manner. The optimal timing for autophagy-promoting interventions will be an important factor in their design. Recently, reports in mice revealed that systemic inhibition of autophagy via inducible Atg5 gene silencing leads to accelerated age-associated pathologies and subsequent autophagy restoration then provides significant improvement of health and lifespan. However, these mice are more prone to succumb due to spontaneous tumors, for which autophagy may be conferring stress-resistance properties (Cassidy et al., 2020). On a related note, a report from C. elegans showed that autophagy can be detrimental in cases of increased mitochondrial permeability, a deterioration of the mitochondrial network that can be age-dependent (Zhou et al., 2019). Similarly, in aged C. elegans, inactivation of autophagy genes with early-stage functions appears to provide beneficial effects on longevity (Wilhelm et al., 2017). Thus, therapeutic enhancement of autophagy may not always be beneficial and it is clear that several factors will need to be taken into account, including whether a cell is already irreversibly damaged and/or whether the autophagy process is irretrievably blocked such that further activation of the pathway may cause additional unwanted detrimental effects. An ideal autophagy-modulating intervention should thus consider timing, cell/tissue specificity, and the overall health status of the cell.
Overall, autophagy plays a fundamental role in the aging process and age-related conditions. This research field holds significant potential to help improve the quality of life of individuals via the modulation of deregulated autophagic activity. To reach this goal, the continuation of basic and translational autophagy research will be fundamental.
HIGHLIGHTS.
The recycling pathway autophagy prevents accumulation of harmful cellular components
Autophagy activity declines with age and is dysregulated in many age-related diseases
Autophagy is essential for lifespan extension observed in conserved longevity models
Interventions that promote autophagy have beneficial effects on health and lifespan
Acknowledgments
We thank Dr. Caroline Kumsta (SBP Medical Discovery Institute) and Drs. Ee Phie Tan and Anupama Singh from the Hansen lab for critical reading of the manuscript. Figures were created with BioRender.com. JNT was supported by a Fundacion Ramon Areces Postdoctoral Fellowship and an NIH K99/R00 pathway to independence grant (K99AG062774); MH was supported by NIH grants GM117466 and AG028664.
VITAE
Dr. Malene Hansen received her Ph.D. in Molecular Biology from Copenhagen University, Denmark, and continued her postdoctoral work in the laboratory of Professor Cynthia Kenyon at University of California, San Francisco, where she studied the molecular genetics of aging using the nematode Caenorhabditis elegans as a model organism. In 2007, Dr. Hansen started an independent research laboratory at Sanford Burnham Prebys Medical Discovery Institute, a non-profit research institute in La Jolla, California, USA. Current research in the Hansen lab focuses on the role and regulation of autophagy in aging, using both C. elegans and mammalian model systems.
Dr. Jose L. Nieto-Torres received his Ph.D. in Molecular Biology from the Autonomous University of Madrid, Spain, where he studied lethal human coronaviruses under the supervision of Professor Luis Enjuanes. Motivated by an interest in the involvement of aging, inflammation, and autophagy in coronavirus-related and other diseases, he joined Dr. Hansen’s laboratory as a postdoctoral fellow. Dr. Nieto-Torres studies the molecular mechanisms that regulate autophagy and plans to further explore their role in aging as an independent investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
The authors declare no competing interests.
References
- Almeida MF, Silva CM, Chaves RS, Lima NCR, Almeida RS, Melo KP, Demasi M, Fernandes T, Oliveira EM, Netto LES, Cardoso SM, Ferrari MFR, 2018. Effects of mild running on substantia nigra during early neurodegeneration. Journal of Sports Sciences 36, 1363–1370. 10.1080/02640414.2017.1378494 [DOI] [PubMed] [Google Scholar]
- Andreotti DZ, Silva J do N, Matumoto AM, Orellana AM, de Mello PS, Kawamoto EM, 2020. Effects of Physical Exercise on Autophagy and Apoptosis in Aged Brain: Human and Animal Studies. Frontiers in Nutrition. 10.3389/fnut.2020.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio R, Rana A, Walker DW, 2019. Upregulation of the Autophagy Adaptor p62/SQSTM1 Prolongs Health and Lifespan in Middle-Aged Drosophila. Cell Reports 28, 1029–1040.e5. 10.1016/j.celrep.2019.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, Alper S, 2018. What can we learn about human disease from the nematode C. Elegans?, in: Methods in Molecular Biology. Humana Press Inc., pp. 53–75. 10.1007/978-1-4939-7471-9_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragonès G, Dasuri K, Olukorede O, Francisco SG, Renneburg C, Kumsta C, Hansen M, Kageyama S, Komatsu M, Rowan S, Volkin J, Workman M, Yang W, Daza P, Ruano D, Dominguez-Martín H, Rodríguez-Navarro JA, Du XL, Brownlee MA, Bejarano E, Taylor A, 2020. Autophagic receptor p62 protects against glycation-derived toxicity and enhances viability. Aging Cell 19. 10.1111/acel.13257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakula D, Scheibye-Knudsen M, 2020. MitophAging: Mitophagy in Aging and Disease. Frontiers in Cell and Developmental Biology. 10.3389/fcell.2020.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano E, Murray JW, Wang X, Pampliega O, Yin D, Patel B, Yuste A, Wolkoff AW, Cuervo AM, 2018. Defective recruitment of motor proteins to autophagic compartments contributes to autophagic failure in aging. Aging Cell 17, 1–18. 10.1111/acel.12777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamini E, Cavallini G, Donati A, Gori Z, 2007. The role of autophagy in aging: Its essential part in the anti-aging mechanism of caloric restriction, in: Annals of the New York Academy of Sciences. Blackwell Publishing Inc., pp. 69–78. 10.1196/annals.1396.020 [DOI] [PubMed] [Google Scholar]
- Boland B, Yu WH, Corti O, Mollereau B, Henriques A, Bezard E, Pastores GM, Rubinsztein DC, Nixon RA, Duchen MR, Mallucci GR, Kroemer G, Levine B, Eskelinen EL, Mochel F, Spedding M, Louis C, Martin OR, Millan MJ, 2018. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nature Reviews Drug Discovery. 10.1038/nrd.2018.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouderlique T, Vuppalapati KK, Newton PT, Li L, Barenius B, Chagin AS, 2016. Targeted deletion of Atg5 in chondrocytes promotes age-related osteoarthritis. Annals of the Rheumatic Diseases 75, 627–631. 10.1136/annrheumdis-2015-207742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-San Pedro JM, Kroemer G, Galluzzi L, 2017. Autophagy and Mitophagy in Cardiovascular Disease. Circulation Research. 10.1161/CIRCRESAHA.117.311082 [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsy R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME, 2004. Stress-Dependent Regulation of FOXO Transcription Factors by the SIRT1 Deacetylase. Science 303, 2011–2015. 10.1126/science.1094637 [DOI] [PubMed] [Google Scholar]
- Buffenstein R, Ruby JG, 2021. Opportunities for new insight into aging from the naked mole-rat and other non-traditional models. Nature Aging 1, 3–4. 10.1038/s43587-020-00012-4 [DOI] [PubMed] [Google Scholar]
- Campbell JM, Bellman SM, Stephenson MD, Lisy K, 2017. Metformin reduces all-cause mortality and diseases of ageing independent of its effect on diabetes control: A systematic review and meta-analysis. Ageing Research Reviews. 10.1016/j.arr.2017.08.003 [DOI] [PubMed] [Google Scholar]
- Cantó C, Auwerx J, 2011. Calorie restriction: Is AMPK a key sensor and effector? Physiology. 10.1152/physiol.00010.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K, Graziotto JJ, Blair CD, Mazzulli JR, Erdos MR, Krainc D, Collins FS, 2011. Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson-Gilford progeria syndrome cells. Science Translational Medicine 3. 10.1126/scitranslmed.3002346 [DOI] [PubMed] [Google Scholar]
- Carroll B, Nelson G, Rabanal-Ruiz Y, Kucheryavenko O, Dunhill-Turner NA, Chesterman CC, Zahari Q, Zhang T, Conduit SE, Mitchell CA, Maddocks ODK, Lovat P, von Zglinicki T, Korolchuk VI, 2017. Persistent mTORC1 signaling in cell senescence results from defects in amino acid and growth factor sensing. Journal of Cell Biology 216, 1949–1957. 10.1083/jcb.201610113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy LD, Young ARJ, Young CNJ, Soilleux EJ, Fielder E, Weigand BM, Lagnado A, Brais R, Ktistakis NT, Wiggins KA, Pyrillou K, Clarke MCH, Jurk D, Passos JF, Narita M, 2020. Temporal inhibition of autophagy reveals segmental reversal of ageing with increased cancer risk. Nature Communications 11, 1–12. 10.1038/s41467-019-14187-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Kumsta C, Hellman AB, Adams LM, Hansen M, 2017. Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. eLife 6. 10.7554/eLife.18459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Guarente L, 2007. SIR2: a potential target for calorie restriction mimetics. Trends in Molecular Medicine. 10.1016/j.molmed.2006.12.004 [DOI] [PubMed] [Google Scholar]
- Chen G, Kroemer G, Kepp O, 2020. Mitophagy: An Emerging Role in Aging and Age-Associated Diseases. Frontiers in Cell and Developmental Biology. 10.3389/fcell.2020.00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AA, 2018. Aging across the tree of life: The importance of a comparative perspective for the use of animal models in aging. Biochimica et Biophysica Acta - Molecular Basis of Disease. 10.1016/j.bbadis.2017.05.028 [DOI] [PubMed] [Google Scholar]
- Cohen E, Dillin A, 2008. The insulin paradox: Aging, proteotoxicity and neurodegeneration. Nature Reviews Neuroscience. 10.1038/nrn2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Macian F, 2014. Autophagy and the immune function in aging. Current Opinion in Immunology. 10.1016/j.coi.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Huang GX, Bonkowski MS, Longchamp A, Li C, Schultz MB, Kim LJ, Osborne B, Joshi S, Lu Y, Treviño-Villarreal JH, Kang MJ, Hung T. tyng, Lee B, Williams EO, Igarashi M, Mitchell JR, Wu LE, Turner N, Arany Z, Guarente L, Sinclair DA, 2018. Impairment of an Endothelial NAD+-H2S Signaling Network Is a Reversible Cause of Vascular Aging. Cell 173, 74–89.e20. 10.1016/j.cell.2018.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Roso A, Vittorini S, Cavallini G, Donati A, Gori Z, Masini M, Pollera M, Bergamini E, 2003. Ageing-related changes in the in vivo function of rat liver macroautophagy and proteolysis. Experimental Gerontology 38, 519–527. 10.1016/S0531-5565(03)00002-0 [DOI] [PubMed] [Google Scholar]
- Demontis F, Perrimon N, 2010. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143, 813–825. 10.1016/j.cell.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, 2021. Autophagy in inflammation, infection, and immunometabolism. Immunity. 10.1016/j.immuni.2021.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Malta C, Cinque L, Settembre C, 2019. Transcriptional regulation of autophagy: Mechanisms and diseases. Frontiers in Cell and Developmental Biology. 10.3389/fcell.2019.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, Elazar Z, 2018. Mechanism and medical implications of mammalian autophagy. Nature Reviews Molecular Cell Biology. 10.1038/s41580-018-0003-4 [DOI] [PubMed] [Google Scholar]
- Ding WX, Li M, Yin XM, 2011. Selective taste of ethanol-induced autophagy for mitochondria and lipid droplets. Autophagy. 10.4161/auto.7.2.14347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati A, Cavallini G, Paradiso C, Vittorini S, Pollera M, Gori Z, Bergamini E, 2001. Age-related changes in the autophagic proteolysis of rat isolated liver cells: Effects of antiaging dietary restrictions. Journals of Gerontology - Series A Biological Sciences and Medical Sciences 56. 10.1093/gerona/56.9.B375 [DOI] [PubMed] [Google Scholar]
- Dou Z, Ghosh K, Vizioli MG, Zhu J, Sen P, Wangensteen KJ, Simithy J, Lan Y, Lin Y, Zhou Z, Capell BC, Xu C, Xu M, Kieckhaefer JE, Jiang T, Shoshkes-Carmel M, al Tanim KMA, Barber GN, Seykora JT, Millar SE, Kaestner KH, Garcia BA, Adams PD, Berger SL, 2017. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406. 10.1038/nature24050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Wooten MC, Gearing M, Wooten MW, 2009a. Age-Associated Oxidative Damage to the p62 Promoter: Implications for Alzheimer’s Disease. Free radical biology & medicine 46, 492. 10.1016/J.FREERADBIOMED.2008.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Wooten MC, Wooten MW, 2009b. Oxidative damage to the promoter region of SQSTM1/p62 is common to neurodegenerative disease. Neurobiology of disease 35, 302. 10.1016/J.NBD.2009.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos A, Castells-Nobau A, Meziane H, Oortveld MAW, Houbaert X, Iacono G, Martin C, Mittelhaeuser C, Lalanne V, Kramer JM, Bhukel A, Quentin C, Slabbert J, Verstreken P, Sigrist SJ, Messaddeq N, Birling MC, Selloum M, Stunnenberg HG, Humeau Y, Schenck A, Herault Y, 2015. Conditional depletion of intellectual disability and Parkinsonism candidate gene ATP6AP2 in fly and mouse induces cognitive impairment and neurodegeneration. Human molecular genetics 24, 6736–6755. 10.1093/hmg/ddv380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca FA, Côté CD, Rasmussen BA, Zadeh-Tahmasebi M, Rutter GA, Filippi BM, Lam TKT, 2015. Metformin activates a duodenal Ampk-dependent pathway to lower hepatic glucose production in rats. Nature Medicine 21, 506–511. 10.1038/nm.3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgan J, Lystad AH, Sloan K, Carlsson SR, Wilson MI, Marcassa E, Ulferts R, Webster J, Lopez-Clavijo AF, Wakelam MJ, Beale R, Simonsen A, Oxley D, Florey O, 2021. Non-canonical autophagy drives alternative ATG8 conjugation to phosphatidylserine. Molecular Cell 81, 2031–2040.e8. 10.1016/j.molcel.2021.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, 2019. Mitophagy and NAD+ inhibit Alzheimer disease. Autophagy 15, 1112. 10.1080/15548627.2019.1596497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Hou Y, Lautrup S, Jensen MB, Yang B, SenGupta T, Caponio D, Khezri R, Demarest TG, Aman Y, Figueroa D, Morevati M, Lee H-J, Kato H, Kassahun H, Lee J-H, Filippelli D, Okur MN, Mangerich A, Croteau DL, Maezawa Y, Lyssiotis CA, Tao J, Yokote K, Rusten TE, Mattson MP, Jasper H, Nilsen H, Bohr VA, 2019a. NAD+ augmentation restores mitophagy and limits accelerated aging in Werner syndrome. Nature Communications 10. 10.1038/S41467-019-13172-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Hou Y, Palikaras K, Adriaanse BA, Kerr JS, Yang B, Lautrup S, Hasan-Olive MM, Caponio D, Dan X, Rocktäschel P, Croteau DL, Akbari M, Greig NH, Fladby T, Nilsen H, Cader MZ, Mattson MP, Tavernarakis N, Bohr VA, 2019b. Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nature neuroscience 22, 401. 10.1038/S41593-018-0332-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Kassahun H, Croteau DL, Scheibye-Knudsen M, Marosi K, Lu H, Shamanna RA, Kalyanasundaram S, Bollineni RC, Wilson MA, Iser WB, Wollman BN, Morevati M, Li J, Kerr JS, Lu Q, Waltz TB, Tian J, Sinclair DA, Mattson MP, Nilsen H, Bohr VA, 2016. NAD+ replenishment improves lifespan and healthspan in Ataxia telangiectasia models via mitophagy and DNA repair. Cell metabolism 24, 566. 10.1016/J.CMET.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H, Mitchell JR, Croteau DL, Bohr VA, 2014. Defective Mitophagy in XPA via PARP1 Hyperactivation and NAD+/SIRT1 Reduction. Cell 157, 882. 10.1016/J.CELL.2014.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Xie C, Schenkel JA, Wu C, Long Q, Cui H, Aman Y, Frank J, Liao J, Zou H, Wang NY, Wu J, Liu X, Li T, Fang Y, Niu Z, Yang G, Hong J, Wang Q, Chen G, Li J, Chen H-Z, Kang L, Su H, Gilmour BC, Zhu X, Jiang H, He N, Tao J, Leng SX, Tong T, Woo J, 2020. A research agenda for ageing in China in the 21st century (2nd edition): Focusing on basic and translational research, long-term care, policy and social networks. Ageing Research Reviews 64, 101174. 10.1016/J.ARR.2020.101174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández ÁF, Sebti S, Wei Y, Zou Z, Shi M, McMillan KL, He C, Ting T, Liu Y, Chiang WC, Marciano DK, Schiattarella GG, Bhagat G, Moe OW, Hu MC, Levine B, 2018. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 558, 136–140. 10.1038/S41586-018-0162-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Albarral JA, Julián-López E. de, Soler-Domínguez C, Hoz R. de, López-Cuenca I, Salobrar-García E, Ramírez JM, Pinazo-Durán MD, Salazar JJ, Ramírez AI, 2021. The Role of Autophagy in Eye Diseases. Life 11. 10.3390/LIFE11030189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher K, Ulferts R, Jacquin E, Veith T, Gammoh N, Arasteh JM, Mayer U, Carding SR, Wileman T, Beale R, Florey O, 2018. The WD 40 domain of ATG 16L1 is required for its non-canonical role in lipidation of LC 3 at single membranes. The EMBO Journal 37, e97840. 10.15252/embj.201797840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Garagnani P, Parini P, Giuliani C, Santoro A, 2018. Inflammaging: a new immune–metabolic viewpoint for age-related diseases. Nature Reviews Endocrinology. 10.1038/S41574-018-0059-4 [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Kikuchi H, Aizawa S, Furuta A, Hatanaka Y, Konya C, Uchida K, Wada K, Kabuta T, 2013. Direct uptake and degradation of DNA by lysosomes. Autophagy 9, 1167. 10.4161/AUTO.24880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G, 2017. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nature Reviews Drug Discovery 2017 16:7 16, 487–511. 10.1038/nrd.2017.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Prat L, Martínez-Vicente M, Perdiguero E, Ortet L, Rodríguez-Ubreva J, Rebollo E, Ruiz-Bonilla V, Gutarra S, Ballestar E, Serrano AL, Sandri M, Muñoz-Cánoves P, 2016. Autophagy maintains stemness by preventing senescence. Nature 529, 37–42. 10.1038/nature16187 [DOI] [PubMed] [Google Scholar]
- Gómez-Sintes R, Ledesma MD, Boya P, 2016. Lysosomal cell death mechanisms in aging. Ageing Research Reviews. 10.1016/j.arr.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Gordon LB, Rothman FG, López-Otín C, Misteli T, 2014. Progeria: A paradigm for translational medicine. Cell. 10.1016/j.cell.2013.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JD, Bejarano E, Wang X-D, Greenberg AS, 2021. Integrated Action of Autophagy and Adipose Tissue Triglyceride Lipase Ameliorates Diet-Induced Hepatic Steatosis in Liver-Specific PLIN2 Knockout Mice. Cells 10. 10.3390/CELLS10051016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Rubinsztein DC, Walker DW, 2018. Autophagy as a promoter of longevity: insights from model organisms. Nature Reviews Molecular Cell Biology 19, 579–593. 10.1038/s41580-018-0033-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N, 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889. 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- He C, Sumpter R, Levine B, 2012. Exercise induces autophagy in peripheral tissues and in the brain. Autophagy 8, 1548–1551. 10.4161/auto.21327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann BL, Green DR, 2019. LC3-associated phagocytosis at a glance. Journal of cell science. 10.1242/jcs.222984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann BL, Teubner BJW, Boada-Romero E, Tummers B, Guy C, Fitzgerald P, Mayer U, Carding S, Zakharenko SS, Wileman T, Green DR, 2020. Noncanonical function of an autophagy protein prevents spontaneous Alzheimer’s disease, Sci. Adv [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann BL, Teubner BJW, Tummers B, Boada-Romero E, Harris L, Yang M, Guy CS, Zakharenko SS, Green DR, 2019. LC3-Associated Endocytosis Facilitates β-Amyloid Clearance and Mitigates Neurodegeneration in Murine Alzheimer’s Disease. Cell 178, 536–551.e14. 10.1016/j.cell.2019.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herhaus L, Bhaskara RM, Lystad AH, Gestal-Mato U, Covarrubias-Pinto A, Bonn F, Simonsen A, Hummer G, Dikic I, 2020. TBK1-mediated phosphorylation of LC3C and GABARAP-L2 controls autophagosome shedding by ATG4 protease. EMBO reports 21. 10.15252/embr.201948317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SE, Colón-Ramos DA, 2020. The Journey of the Synaptic Autophagosome: A Cell Biological Perspective. Neuron. 10.1016/j.neuron.2020.01.018 [DOI] [PubMed] [Google Scholar]
- Hipp MS, Kasturi P, Hartl FU, 2019. The proteostasis network and its decline in ageing. Nature Reviews Molecular Cell Biology. 10.1038/s41580-019-0101-y [DOI] [PubMed] [Google Scholar]
- Hoshino A, Mita Y, Okawa Y, Ariyoshi M, Iwai-Kanai E, Ueyama T, Ikeda K, Ogata T, Matoba S, 2013. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nature Communications 4, 1–12. 10.1038/ncomms3308 [DOI] [PubMed] [Google Scholar]
- Hughes AL, Gottschling DE, 2012. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492, 261–265. 10.1038/nature11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadanza MG, Jackson MP, Hewitt EW, Ranson NA, Radford SE, 2018. A new era for understanding amyloid structures and disease. Nature Reviews Molecular Cell Biology. 10.1038/S41580-018-0060-8 [DOI] [PubMed] [Google Scholar]
- Inomata M, Xu S, Chandra P, Meydani SN, Takemura G, Philips JA, Leong JM, 2021. Macrophage LC3-associated phagocytosis is an immune defense against Streptococcus pneumoniae that diminishes with host aging. Proceedings of the National Academy of Sciences of the United States of America 117, 33561–33569. 10.1073/PNAS.2015368117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N, 2014. The HOPS complex mediates autophagosome–lysosome fusion through interaction with syntaxin 17. Molecular Biology of the Cell 25, 1327. 10.1091/MBC.E13-08-0447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson FB, Sinclair DA, Guarente L,1999. Molecular biology of aging. Cell. 10.1016/S0092-8674(00)80567-X [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, 2017. Translational geroscience: A new paradigm for 21st century medicine. Translational Medicine of Aging. 10.1016/j.tma.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha S. v., Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD, White E, 2014. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discovery 4, 915–927. 10.1158/2159-8290.CD-14-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Arias E, Kwon H, Lopez NM, Athonvarangkul D, S., Schwartz GJ, Pessin JE, Singh R, 2012. Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Reports 13, 258–265. 10.1038/embor.2011.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM, 2015. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nature cell biology 17, 759. 10.1038/NCB3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM, 2018. The coming of age of chaperone-mediated autophagy. Nature Reviews Molecular Cell Biology. 10.1038/s41580-018-0001-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Guan KL, 2015. MTOR: A pharmacologic target for autophagy regulation. Journal of Clinical Investigation. 10.1172/JCI73939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-Y, Zhao H, Martinez J, Doggett TA, Kolesnikov A. v., Tang PH, Ablonczy Z, Chan CC, Zhou Z, Green DR, Ferguson TA, 2013. Non-canonical Autophagy Promotes the Visual Cycle. Cell 154, 365. 10.1016/J.CELL.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata JI, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K, 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884. 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T, 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. Journal of Cell Biology 169, 425–434. 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N, 2004. The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036. 10.1038/nature03029 [DOI] [PubMed] [Google Scholar]
- Kumari R, Jat P, 2021. Mechanisms of Cellular Senescence: Cell Cycle Arrest and Senescence Associated Secretory Phenotype. Frontiers in Cell and Developmental Biology 0, 485. 10.3389/FCELL.2021.645593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta C, Chang JT, Lee R, Tan EP, Yang Y, Loureiro R, Choy EH, Lim SHY, Saez I, Springhorn A, Hoppe T, Vilchez D, Hansen M, 2019. The autophagy receptor p62/SQST-1 promotes proteostasis and longevity in C. elegans by inducing autophagy. Nature Communications 10, 1–12. 10.1038/s41467-019-13540-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J, Han E, Bui C-B, Shin W, Lee J, Lee S, Choi Y-B, Lee A-H, Lee K-H, Park C, Obin MS, Park SK, Seo YJ, Oh GT, Lee H-W, Shin J, 2012. Assurance of mitochondrial integrity and mammalian longevity by the p62–Keap1–Nrf2–Nqo1 cascade. EMBO reports 13, 150–156. 10.1038/EMBOR.2011.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan YY, Heather JM, Eisenhaure T, Garris CS, Lieb D, Raychowdhury R, Hacohen N, 2019. Extranuclear DNA accumulates in aged cells and contributes to senescence and inflammation. Aging Cell 18, e12901. 10.1111/acel.12901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, de Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, Dillin A, Hansen M, 2013. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nature Communications 4, 1–8. 10.1038/ncomms3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, Kumsta C, Sandri M, Ballabio A, Hansen M, 2015. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy 11, 867–880. 10.1080/15548627.2015.1034410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautrup S, Sinclair DA, Mattson MP, Fang EF, 2019. Cell Metabolism Review NAD + in Brain Aging and Neurodegenerative Disorders. Cell Metabolism 30, 630–655. 10.1016/j.cmet.2019.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Guerroué F, Eck F, Jung J, Starzetz T, Mittelbronn M, Kaulich M, Behrends C, 2017. Autophagosomal Content Profiling Reveals an LC3C-Dependent Piecemeal Mitophagy Pathway. Molecular Cell 68, 786–796.e6. 10.1016/j.molcel.2017.10.029 [DOI] [PubMed] [Google Scholar]
- Lee C, Longo V, 2016. Dietary restriction with and without caloric restriction for healthy aging. F1000Research. 10.12688/f1000research.7136.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidal AM, Huang HH, Marsh T, Solvik T, Zhang D, Ye J, Kai FB, Goldsmith J, Liu JY, Huang YH, Monkkonen T, Vlahakis A, Huang EJ, Goodarzi H, Yu L, Wiita AP, Debnath J, 2020. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nature Cell Biology 22, 187–199. 10.1038/s41556-019-0450-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontieva O. v., Demidenko ZN, Blagosklonny M. v., 2014. Contact inhibition and high cell density deactivate the mammalian target of rapamycin pathway, thus suppressing the senescence program. Proceedings of the National Academy of Sciences of the United States of America 111, 8832–8837. 10.1073/pnas.1405723111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW, 2011. Autophagy in immunity and inflammation. Nature. 10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Sharma A, Yin C, Tan X, Xiao Y, 2017. Metformin ameliorates hepatic steatosis and improves the induction of autophagy in HFD-induced obese mice. Molecular Medicine Reports 16, 680–686. 10.3892/mmr.2017.6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XX, Sen I, Janssens GE, Zhou X, Fonslow BR, Edgar D, Stroustrup N, Swoboda P, Yates JR, Ruvkun G, Riedel CG, 2018. DAF-16/FOXO and HLH-30/TFEB function as combinatorial transcription factors to promote stress resistance and longevity. Nature Communications 9, 1–15. 10.1038/s41467-018-06624-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, Yuan J, 2010. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America 107, 14164–14169. 10.1073/pnas.1009485107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Czaja MJ, 2013. Regulation of lipid stores and metabolism by lipophagy. Cell Death and Differentiation. 10.1038/cdd.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G, 2013. The hallmarks of aging. Cell. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Kroemer G, 2021. Hallmarks of Health. Cell. 10.1016/j.cell.2020.11.034 [DOI] [Google Scholar]
- Lotz MK, Caramés B, 2011. Autophagy and cartilage homeostasis mechanisms in joint health, aging and OA. Nature Reviews Rheumatology. 10.1038/nrrheum.2011.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Dai JR, Guo SS, Lu AM, Gao XF, Gu YR, Zhang XF, Xu HD, Wang Y, Zhu Z, Wood LJ, Qin ZH, 2017. Lysosomal Proteolysis Is Associated with Exercise-Induced Improvement of Mitochondrial Quality Control in Aged Hippocampus. Journals of Gerontology - Series A Biological Sciences and Medical Sciences 72, 1342–1351. 10.1093/gerona/glw242 [DOI] [PubMed] [Google Scholar]
- Luo M, Zhao X, Song Y, Cheng H, Zhou R, 2016. Nuclear autophagy: An evolutionarily conserved mechanism of nuclear degradation in the cytoplasm. Autophagy 12, 1973. 10.1080/15548627.2016.1217381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Carmona-Gutierrez D, Hofer SJ, Kroemer G, 2019. Caloric Restriction Mimetics against Age-Associated Disease: Targets, Mechanisms, and Therapeutic Potential. Cell Metabolism. 10.1016/j.cmet.2019.01.018 [DOI] [PubMed] [Google Scholar]
- Madeo F, Pietrocola F, Eisenberg T, Kroemer G, 2014. Caloric restriction mimetics: Towards a molecular definition. Nature Reviews Drug Discovery. 10.1038/nrd4391 [DOI] [PubMed] [Google Scholar]
- Mariño G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, Zamzami N, Scoazec M, Durand S, Enot DP, Fernández ÁF, Martins I, Kepp O, Senovilla L, Bauvy C, Morselli E, Vacchelli E, Bennetzen M, Magnes C, Sinner F, Pieber T, López-Otín C, Maiuri MC, Codogno P, Andersen JS, Hill JA, Madeo F, Kroemer G, 2014. Regulation of Autophagy by Cytosolic Acetyl-Coenzyme A. Molecular Cell 53, 710–725. 10.1016/j.molcel.2014.01.016 [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Sovak G, Cuervo AM, 2005. Protein degradation and aging. Experimental Gerontology. 10.1016/j.exger.2005.07.005 [DOI] [PubMed] [Google Scholar]
- Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, Gomes AP, Ward TM, Minor RK, Blouin MJ, Schwab M, Pollak M, Zhang Y, Yu Y, Becker KG, Bohr VA, Ingram DK, Sinclair DA, Wolf NS, Spindler SR, Bernier M, de Cabo R, 2013. Metformin improves healthspan and lifespan in mice. Nature Communications 4, 1–9. 10.1038/ncomms3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins R, Lithgow GJ, Link W, 2016. Long live FOXO: Unraveling the role of FOXO proteins in aging and longevity. Aging Cell. 10.1111/acel.12427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda de Stegmann D, Bhogaraju S, Maddi K, Kirchof A, Gatti E, Helfrich MH, Wakatsuki S, Behrends C, Pierre P, Dikic I, 2015. PLEKHM1 Regulates Autophagosome-Lysosome Fusion through HOPS Complex and LC3/GABARAP Proteins. Molecular Cell 57, 39–54. 10.1016/J.MOLCEL.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Melentijevic I, Toth ML, Arnold ML, Guasp RJ, Harinath G, Nguyen KC, Taub D, Parker JA, Neri C, Gabel C. v., Hall DH, Driscoll M, 2017. C. elegans neurons jettison protein aggregates and mitochondria under neurotoxic stress. Nature 542, 367–371. 10.1038/nature21362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei H, Suarez JA, Longo VD, 2014. Protein and amino acid restriction, aging and disease: From yeast to humans. Trends in Endocrinology and Metabolism. 10.1016/j.tem.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco A, Fraldi A, 2020. Protein Aggregation and Dysfunction of Autophagy-Lysosomal Pathway: A Vicious Cycle in Lysosomal Storage Diseases. Frontiers in Molecular Neuroscience 13, 37. 10.3389/fnmol.2020.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel E, Mehrpour M, Botti J, Dupont N, Hamaï A, Nascimbeni AC, Codogno P, 2017. Autophagy: A Druggable Process. 10.1146/annurev-pharmtox-010716-104936 57, 375–398. [DOI] [PubMed] [Google Scholar]
- Morishita H, Eguchi S, Kimura H, Sasaki J, Sakamaki Y, Robinson ML, Sasaki T, Mizushima N, 2013. Deletion of Autophagy-related 5 (Atg5) and Pik3c3 Genes in the Lens Causes Cataract Independent of Programmed Organelle Degradation. The Journal of Biological Chemistry 288, 11436. 10.1074/JBC.M112.437103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Patel B, Koga H, Cuervo AM, Jenny A, 2016. Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy 12, 1984. 10.1080/15548627.2016.1208887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacarelli T, Azar A, Sell C, 2015. Aberrant mTOR activation in senescence and aging: A mitochondrial stress response? Experimental Gerontology 68, 66–70. 10.1016/j.exger.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Tominaga K, Amano T, Hirotsu I, Inoue T, Yamamoto K, 1994. Age-related changes in activities and localizations of cathepsins D, E, B, and L in the rat brain tissues. Experimental Neurology 126, 119–128. 10.1006/exnr.1994.1048 [DOI] [PubMed] [Google Scholar]
- Nakano M, Oenzil F, Mizuno T, Gotoh S, 1995. Age-related changes in the lipofuscin accumulation of brain and heart. Gerontology 41, 69–79. 10.1159/000213726 [DOI] [PubMed] [Google Scholar]
- Napolitano G, Esposito A, Choi H, Matarese M, Benedetti V, di Malta C, Monfregola J, Medina DL, Lippincott-Schwartz J, Ballabio A, 2018. mTOR-dependent phosphorylation controls TFEB nuclear export. Nature Communications 9, 1–10. 10.1038/s41467-018-05862-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolás-Ávila JA, Lechuga-Vieco A. v., Esteban-Martínez L, Sánchez-Díaz M, Díaz-García E, Santiago DJ, Rubio-Ponce A, Li JLY, Balachander A, Quintana JA, Martínez-de-Mena R, Castejón-Vega B, Pun-García A, Través PG, Bonzón-Kulichenko E, García-Marqués F, Cussó L, A-González N, González-Guerra A, Roche-Molina M, Martin-Salamanca S, Crainiciuc G, Guzmán G, Larrazabal J, Herrero-Galán E, Alegre-Cebollada J, Lemke G, Rothlin C. v., Jimenez-Borreguero LJ, Reyes G, Castrillo A, Desco M, Muñoz-Cánoves P, Ibáñez B, Torres M, Ng LG, Priori SG, Bueno H, Vázquez J, Cordero MD, Bernal JA, Enríquez JA, Hidalgo A, 2020. A Network of Macrophages Supports Mitochondrial Homeostasis in the Heart. Cell 183, 94–109.e23. 10.1016/j.cell.2020.08.031 [DOI] [PubMed] [Google Scholar]
- Nieto-Torres JL, Leidal AM, Debnath J, Hansen M, 2021a. Beyond Autophagy: The Expanding Roles of ATG8 Proteins. Trends in Biochemical Sciences. 10.1016/j.tibs.2021.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres JL, Shanahan S-L, Chassefeyre R, Chaiamarit T, Zaretski S, Landeras-Bueno S, Verhelle A, Encalada SE, Hansen M, 2021b. LC3B phosphorylation regulates FYCO1 binding and directional transport of autophagosomes. Current Biology. 10.1016/j.cub.2021.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA, 2005. Endosome function and dysfunction in Alzheimer’s disease and other neurodegenerative diseases, in: Neurobiology of Aging. Neurobiol Aging, pp. 373–382. 10.1016/j.neurobiolaging.2004.09.018 [DOI] [PubMed] [Google Scholar]
- Nunomura A, Miyagishi T, 1993. Ultrastructural observations on neuronal lipofuscin (age pigment) and dense bodies induced by a proteinase inhibitor, leupeptin, in rat hippocampus. Acta Neuropathologica 86, 319–328. 10.1007/BF00369443 [DOI] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, Tavernarakis N, 2018. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nature Cell Biology 2018 20:9 20, 1013–1022. 10.1038/s41556-018-0176-2 [DOI] [PubMed] [Google Scholar]
- Palikaras K, Mari M, Petanidou B, Pasparaki A, Filippidis G, Tavernarakis AN, 2017. Ectopic fat deposition contributes to age-associated pathology in caenorhabditis elegans. Journal of Lipid Research 58, 72–80. 10.1194/jlr.M069385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjorkoy G, Johansen T, 2010. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol 188, 253–269. https://doi.org/jcb.200907015 [pii] 10.1083/jcb.200907015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos C, Kravic B, Meyer H, 2020. Repair or Lysophagy: Dealing with Damaged Lysosomes. Journal of Molecular Biology. 10.1016/j.jmb.2019.08.010 [DOI] [PubMed] [Google Scholar]
- Park Y-E, Hayashi YK, Bonne G, Arimura T, Noguchi S, Nonaka I, Nishino I, 2009. Autophagic degradation of nuclear components in mammalian cells. 10.4161/auto.8901 5, 795–804. [DOI] [PubMed] [Google Scholar]
- Peng Y, Shapiro SL, Banduseela VC, Dieterich IA, Hewitt KJ, Bresnick EH, Kong G, Zhang J, Schueler KL, Keller MP, Attie AD, Hacker TA, Sullivan R, Kielar-Grevstad E, Arriola Apelo SI, Lamming DW, Anderson RM, Puglielli L, 2018. Increased transport of acetyl-CoA into the endoplasmic reticulum causes a progeria-like phenotype. Aging Cell 17. 10.1111/acel.12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotta C, Cattaneo MG, Molteni R, de Palma C, 2020. Autophagy in the Regulation of Tissue Differentiation and Homeostasis. Frontiers in Cell and Developmental Biology. 10.3389/fcell.2020.602901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell AM, Youle RJ, 2015. The roles of PINK1, Parkin, and mitochondrial fidelity in parkinson’s disease. Neuron. 10.1016/j.neuron.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucciarelli S, Moreschini B, Micozzi D, de Fronzo GS, Carpi FM, Polzonetti V, Vincenzetti S, Mignini F, Napolioni V, 2012. Spermidine and spermine are enriched in whole blood of nona/centenarians. Rejuvenation Research 15, 590–595. 10.1089/rej.2012.1349 [DOI] [PubMed] [Google Scholar]
- Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, Jung S, Jung YK, 2013. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nature Communications 4, 1–9. 10.1038/ncomms3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rached MT, Kode A, Silva BC, Jung DY, Gray S, Ong H, Paik JH, DePinho RA, Kim JK, Karsenty G, Kousteni S, 2010. FoxO1 expression in osteoblasts regulates glucose homeostasis through regulation of osteocalcin in mice. Journal of Clinical Investigation 120, 357–368. 10.1172/JCI39901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Yoshikawa TT, 2000. Tuberculosis in the elderly. Zeitschrift für Gerontologie und Geriatrie 2000 33:5 33, 374–380. 10.1007/S003910070034 [DOI] [PubMed] [Google Scholar]
- Raz Y, Guerrero-Ros I, Maier A, Slagboom PE, Atzmon G, Barzilai N, Macian F, 2017. Activation-Induced Autophagy Is Preserved in CD4+ T-Cells in Familial Longevity. The journals of gerontology. Series A, Biological sciences and medical sciences 72, 1201–1206. 10.1093/gerona/glx020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B, Feng C, Coleman T, Emanuel R, Wen H, Hwang S, Ting JP, Virgin HW, Kastan MB, Semenkovich CF, 2012. Autophagy links inflammasomes to atherosclerotic progression. Cell Metabolism 15, 534–544. 10.1016/j.cmet.2012.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin-Champigneul F, 2020. Jeanne Calment’s Unique 122-Year Life Span: Facts and Factors; Longevity History in Her Genealogical Tree. Rejuvenation Research 23, 19–47. 10.1089/rej.2019.2298 [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Mariño G, Kroemer G, 2011. Autophagy and aging. Cell. 10.1016/j.cell.2011.07.030 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Smith M, Buffenstein R, 2018. Naked mole-rat mortality rates defy gompertzian laws by not increasing with age. eLife 7. 10.7554/eLife.31157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckenstuhl C, Netzberger C, Entfellner I, Carmona-Gutierrez D, Kickenweiz T, Stekovic S, Gleixner C, Schmid C, Klug L, Sorgo AG, Eisenberg T, Büttner S, Mariño G, Koziel R, Jansen-Dürr P, Fröhlich KU, Kroemer G, Madeo F, 2014. Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification. PLoS Genetics 10, 1004347. 10.1371/journal.pgen.1004347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rué L, López-Soop G, Gelpi E, Martínez-Vicente M, Alberch J, Pérez-Navarro E, 2013. Brain region- and age-dependent dysregulation of p62 and NBR1 in a mouse model of Huntington’s disease. Neurobiology of Disease 52, 219–228. 10.1016/J.NBD.2012.12.008 [DOI] [PubMed] [Google Scholar]
- Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L, 2011. MICROAUTOPHAGY OF CYTOSOLIC PROTEINS BY LATE ENDOSOMES. Developmental cell 20, 131. 10.1016/J.DEVCEL.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Yako T, Otsu W, Nakamura S, Inoue Y, Muramatsu A, Nakagami Y, Shimazawa M, Hara H, 2020. A triterpenoid Nrf2 activator, RS9, promotes LC3-associated phagocytosis of photoreceptor outer segments in a p62-independent manner. Free Radical Biology and Medicine 152, 235–247. 10.1016/J.FREERADBIOMED.2020.03.012 [DOI] [PubMed] [Google Scholar]
- Sandri M, Barberi L, Bijlsma AY, Blaauw B, Dyar KA, Milan G, Mammucari C, Meskers CGM, Pallafacchina G, Paoli A, Pion D, Roceri M, Romanello V, Serrano AL, Toniolo L, Larsson L, Maier AB, Muñoz-Cánoves P, Musarò A, Pende M, Reggiani C, Rizzuto R, Schiaffino S, 2013. Signalling pathways regulating muscle mass in ageing skeletal muscle. the role of the IGF1-Akt-mTOR-FoxO pathway. Biogerontology 14, 303–323. 10.1007/s10522-013-9432-9 [DOI] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL, 2004. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412. 10.1016/S0092-8674(04)00400-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A, 2009. A gene network regulating lysosomal biogenesis and function. Science 325, 473–477. 10.1126/science.1174447 [DOI] [PubMed] [Google Scholar]
- Sato Y, Suzuki Y, Ito E, Shimazaki S, Ishida M, Yamamoto T, Yamamoto H, Toda T, Suzuki M, Suzuki A, Endo T, 2006. Identification and characterization of an increased glycoprotein in aging: Age-associated translocation of cathepsin D. Mechanisms of Ageing and Development 127, 771–778. 10.1016/j.mad.2006.07.001 [DOI] [PubMed] [Google Scholar]
- Schaaf MBE, Keulers TG, Vooijs MA, Rouschop KMA, 2016. LC3/GABARAP family proteins: autophagy-(un)related functions. The FASEB Journal 30, 3961–3978. 10.1096/FJ.201600698R [DOI] [PubMed] [Google Scholar]
- Schuck S, 2020. Microautophagy – distinct molecular mechanisms handle cargoes of many sizes. Journal of Cell Science. 10.1242/jcs.246322 [DOI] [PubMed] [Google Scholar]
- Schulze RJ, Krueger EW, Weller SG, Johnson KM, Casey CA, Schott MB, McNiven MA, 2020. Direct lysosome-based autophagy of lipid droplets in hepatocytes. Proceedings of the National Academy of Sciences of the United States of America 117, 32443. 10.1073/PNAS.2011442117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha BK, Rasmussen MS, Abudu YP, Bruun JA, Larsen KB, Alemu EA, Sjøttem E, Lamark T, Johansen T, 2020. NIMA-related kinase 9 –mediated phosphorylation of the microtubule-associated LC3B protein at Thr-50 suppresses selective autophagy of p62/sequestosome 1. Journal of Biological Chemistry 295, 1240–1260. 10.1074/jbc.RA119.010068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD, 2008. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4, 176–184. 10.4161/auto.5269 [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ, 2009. Autophagy regulates lipid metabolism. Nature 458, 1131–1135. 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Tchkonia T, Jiang J, Kirkland JL, Sun Y, 2020. Targeting Senescent Cells for a Healthier Aging: Challenges and Opportunities. Advanced Science. 10.1002/advs.202002611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopena N, Pedro-Botet L, Mateu L, Tolschinsky G, Rey-Joly C, Sabrià M, 2007. Community-Acquired Legionella Pneumonia in Elderly Patients: Characteristics and Outcome. Journal of the American Geriatrics Society 55, 114–119. 10.1111/J.1532-5415.2006.01021.X [DOI] [PubMed] [Google Scholar]
- Stavoe AKH, Gopal PP, Gubas A, Tooze SA, Holzbaur ELF, 2019. Expression of WIPI2B counteracts age-related decline in autophagosome biogenesis in neurons. eLife 8. 10.7554/eLife.44219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoka V, Turk V, Turk B, 2016. Lysosomal cathepsins and their regulation in aging and neurodegeneration. Ageing Research Reviews. 10.1016/j.arr.2016.04.010 [DOI] [PubMed] [Google Scholar]
- Su H, Yang F, Wang Q, Shen Q, Huang J, Peng C, Zhang Y, Wan W, Wong CCL, Sun Q, Wang F, Zhou T, Liu W, 2017. VPS34 Acetylation Controls Its Lipid Kinase Activity and the Initiation of Canonical and Non-canonical Autophagy. Molecular Cell 67, 907–921.e7. 10.1016/j.molcel.2017.07.024 [DOI] [PubMed] [Google Scholar]
- Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P, 2008. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proceedings of the National Academy of Sciences of the United States of America 105, 3438–3442. 10.1073/pnas.0705467105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li M, Zhao D, Li X, Yang C, Wang X, 2020. Lysosome activity is modulated by multiple longevity pathways and is important for lifespan extension in C. Elegans. eLife 9, 1–28. 10.7554/eLife.55745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YX, Ji X, Mao X, Xie L, Jia J, Galvan V, Greenberg DA, Jin K, 2014. Differential activation of mTOR Complex 1 Signaling in human brain with mild to severe Alzheimer’s disease. Journal of Alzheimer’s Disease 38, 437–444. 10.3233/JAD-131124 [DOI] [PubMed] [Google Scholar]
- Tammineni P, Ye X, Feng T, Aikal D, Cai Q, 2017. Impaired retrograde transport of axonal autophagosomes contributes to autophagic stress in Alzheimer’s disease neurons. eLife 6. 10.7554/eLife.21776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekirdag K, Cuervo AM, 2018. Chaperone-mediated autophagy and endosomal microautophagy: Jointed by a chaperone. The Journal of Biological Chemistry 293, 5414. 10.1074/JBC.R117.818237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth ML, Sigmond T, Borsos É, Barna J, Erdélyi P, Takács-Vellai K, Orosz L, Kovács AL, Csikós G, Sass M, Vellai T, 2008. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy 4, 330–338. 10.4161/auto.5618 [DOI] [PubMed] [Google Scholar]
- Towers CG, Thorburn A, 2016. Therapeutic Targeting of Autophagy. EBioMedicine. 10.1016/j.ebiom.2016.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers CG, Wodetzki D, Thorburn J, Smith KR, Caino MC, Thorburn A, 2020. Alternate Mitochondrial Pathways Compensate for Loss of LC3-Mediated Mitophagy. SSRN Electronic Journal. 10.2139/ssrn.3728139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai T-H, Chen E, Li L, Saha P, Lee H-J, Huang L-S, Shelness GS, Chan L, Chang BH-J, 2017. The constitutive lipid droplet protein PLIN2 regulates autophagy in liver. Autophagy 13, 1130. 10.1080/15548627.2017.1319544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Komatsu M, 2017. Autophagy in the liver: Functions in health and disease. Nature Reviews Gastroenterology and Hepatology. 10.1038/nrgastro.2016.185 [DOI] [PubMed] [Google Scholar]
- Ulgherait M, Rana A, Rera M, Graniel J, Walker DW, 2014. AMPK modulates tissue and organismal aging in a non-cell-autonomous manner. Cell Reports 8, 1767–1780. 10.1016/j.celrep.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deursen JM, 2019. Senolytic therapies for healthy longevity. Science. 10.1126/science.aaw1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, 2015. NAD+ in aging, metabolism, and neurodegeneration. Science. 10.1126/science.aac4854 [DOI] [PubMed] [Google Scholar]
- Vilchez D, Saez I, Dillin A, 2014. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nature Communications. 10.1038/ncomms6659 [DOI] [PubMed] [Google Scholar]
- Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves AMF, Wollenberg AC, Stuart LM, Stormo GD, Irazoqui JE, 2014. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity 40, 896–909. 10.1016/j.immuni.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AE, Brunet A, 2014. FOXO transcription factors: Key regulators of cellular quality control. Trends in Biochemical Sciences. 10.1016/j.tibs.2014.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B, 2008. JNK1-Mediated Phosphorylation of Bcl-2 Regulates Starvation-Induced Autophagy. Molecular Cell 30, 678–688. 10.1016/j.molcel.2008.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm T, Byrne J, Medina R, Kolundžic E, Geisinger J, Hajduskova M, Tursun B, Richly H, 2017. Neuronal inhibition of the autophagy nucleation complex extends life span in post-reproductive C. Elegans. Genes and Development 31, 1561–1572. 10.1101/gad.301648.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DS, Jariwala JS, Anderson E, Mitra K, Meisenhelder J, Chang JT, Ideker T, Hunter T, Nizet V, Dillin A, Hansen M, 2015. Phosphorylation of LC3 by the Hippo Kinases STK3/STK4 Is Essential for Autophagy. Mol Cell 57, 55–68. https://doi.org/S1097-2765(14)00914-9 [pii] 10.1016/j.molcel.2014.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S, 2019. ER-phagy: shaping up and destressing the endoplasmic reticulum. FEBS Journal. 10.1111/febs.14932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD, 2008. FOXO3A genotype is strongly associated with human longevity. Proceedings of the National Academy of Sciences of the United States of America 105, 13987–13992. 10.1073/pnas.0801030105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Lau K, Eby B, Lozano P, He C, Pennington B, Li H, Rathi S, Dong Y, Tian R, Kem D, Zou MH, 2011. Improvement of cardiac functions by chronic metformin treatment is associated with enhanced cardiac autophagy in diabetic OVE26 mice. Diabetes 60, 1770–1778. 10.2337/db10-0351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Hua C, Tautenhahn HM, Dirsch O, Dahmen U, 2020. The role of autophagy for the regeneration of the aging liver. International Journal of Molecular Sciences. 10.3390/ijms21103606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Dan X, Hou Y, Lee J, Wechter N, Krishnamurthy S, Kimura R, Babbar M, Demarest T, McDevitt R, Zhang S, Zhang Y, Mattson MP, Croteau DL, Bohr VA, 2021. NAD+ supplementation prevents STING-induced senescence in ataxia telangiectasia by improving mitophagy. Aging Cell 20. 10.1111/ACEL.13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yefimova MG, Ravel C, Rolland AD, Bourmeyster N, Jégou B, 2021. MERTK-Mediated LC3-Associated Phagocytosis (LAP) of Apoptotic Substrates in Blood-Separated Tissues: Retina, Testis, Ovarian Follicles. Cells 10. 10.3390/CELLS10061443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim WWY, Mizushima N, 2020. Lysosome biology in autophagy. Cell Discovery. 10.1038/s41421-020-0141-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang E, Cui W, Lopresti M, Mashek MT, Najt CP, Hu H, Mashek DG, 2020. Hepatic PLIN5 signals via SIRT1 to promote autophagy and prevent inflammation during fasting. Journal of Lipid Research 61, 338. 10.1194/JLR.RA119000336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cross SD, Stanton JB, Marmorstein AD, Le YZ, Marmorstein LY, 2017. Early AMD-like defects in the RPE and retinal degeneration in aged mice with RPE-specific deletion of Atg5 or Atg7. Molecular Vision 23, 228. [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Vasheghani F, Li YH, Blati M, Simeone K, Fahmi H, Lussier B, Roughley P, Lagares D, Pelletier JP, Martel-Pelletier J, Kapoor M, 2015. Cartilage-specific deletion of mTOR upregulates autophagy and protects mice from osteoarthritis. Annals of the Rheumatic Diseases 74, 1432–1440. 10.1136/annrheumdis-2013-204599 [DOI] [PubMed] [Google Scholar]
- Zhou B, Kreuzer J, Kumsta C, Wu L, Kamer KJ, Cedillo L, Zhang Y, Li S, Kacergis MC, Webster CM, Fejes-Toth G, Naray-Fejes-Toth A, Das S, Hansen M, Haas W, Soukas AA, 2019. Mitochondrial Permeability Uncouples Elevated Autophagy and Lifespan Extension. Cell 177, 299–314.e16. 10.1016/j.cell.2019.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]