Significance
Current acute myeloid leukemia (AML) risk assessment relies on cytogenetics and gene-sequencing studies but is imperfect, especially for patients with normal karyotypes and intermediate risk. To understand factors associated with excellent responses in these patients, we compared genetic and transcriptional data from patients with first remissions lasting more than 5 y after chemotherapy, to matched controls with first remissions lasting fewer than 2 y. AML cells from patients with early relapse displayed an immunosuppressive phenotype that blocked CD4 T cell activation via the T cell receptor; the long first-remission AML cells did not display this phenotype. Inhibiting LAG3 reversed this immunosuppression in most tested cases. This immunosuppressive phenotype may help to stratify risk of relapse at presentation, and outcomes.
Keywords: acute myeloid leukemia, immunosuppression, checkpoints, chemotherapy, cancer genomics
Abstract
Acute myeloid leukemia (AML) patients rarely have long first remissions (LFRs; >5 y) after standard-of-care chemotherapy, unless classified as favorable risk at presentation. Identification of the mechanisms responsible for long vs. more typical, standard remissions may help to define prognostic determinants for chemotherapy responses. Using exome sequencing, RNA-sequencing, and functional immunologic studies, we characterized 28 normal karyotype (NK)-AML patients with >5 y first remissions after chemotherapy (LFRs) and compared them to a well-matched group of 31 NK-AML patients who relapsed within 2 y (standard first remissions [SFRs]). Our combined analyses indicated that genetic-risk profiling at presentation (as defined by European LeukemiaNet [ELN] 2017 criteria) was not sufficient to explain the outcomes of many SFR cases. Single-cell RNA-sequencing studies of 15 AML samples showed that SFR AML cells differentially expressed many genes associated with immune suppression. The bone marrow of SFR cases had significantly fewer CD4+ Th1 cells; these T cells expressed an exhaustion signature and were resistant to activation by T cell receptor stimulation in the presence of autologous AML cells. T cell activation could be restored by removing the AML cells or blocking the inhibitory major histocompatibility complex class II receptor, LAG3. Most LFR cases did not display these features, suggesting that their AML cells were not as immunosuppressive. These findings were confirmed and extended in an independent set of 50 AML cases representing all ELN 2017 risk groups. AML cell–mediated suppression of CD4+ T cell activation at presentation is strongly associated with unfavorable outcomes in AML patients treated with standard chemotherapy.
A normal karyotype (NK) is the most common cytogenetic finding in acute myeloid leukemia (AML) cells at presentation (NK-AML), and has long been associated with an intermediate risk for relapse (1). Although most NK-AML patients achieve a complete remission following standard induction chemotherapy (2, 3), the majority of patients experience relapse within 2 y, and fewer than 10% remain in remission beyond 5 y without an allogeneic transplant (4). Refinements introduced with the European LeukemiaNet (ELN) 2017 criteria (presence or absence of mutations in NPM1, FLT3-ITD, ASXL1, RUNX1, TP53, or biallelic CEBPA mutations) have helped to reclassify some NK-AML patients into the favorable- or adverse-risk group categories. However, the mutational heterogeneity of NK-AML, combined with our limited understanding of the biological consequences deriving from the interplay among mutations, still poses a major challenge for risk stratification at presentation, and for postremission treatment decisions (5, 6).
The genetic and epigenetic characteristics of NK-AML patients with very long first remissions (LFRs) have not been defined, and it is not clear whether these patients are living free from disease, or whether they harbor “dormant” AML cells that may be held in check by immune surveillance or cell-intrinsic mechanisms. Predictive algorithms for identifying these very long responders at presentation do not yet exist, but clearance of all AML-associated mutations assessed after induction chemotherapy is strongly associated with prolonged relapse-free survival (RFS) and overall survival (OS) (7–12). Persistent ancestral clones have been detected in patients in complete morphologic remission within the first 2 to 3 y after treatment (13, 14), but similar studies of patients with very LFRs have not yet been described.
Most published AML studies have justifiably focused on the mechanisms responsible for treatment failure. However, the reasons for chemotherapy successes are underexplored, even though they hold the potential to uncover determinants that are especially important for excellent outcomes that might be identifiable at presentation. To address this, we have studied two well-matched groups of NK-AML patients that were selected retrospectively for dramatically different outcomes after chemotherapy (long vs. standard first remissions, [SFRs]). In this study, we show that the outcomes of AML patients are not only related to their mutational status, but also to an immunosuppressive phenotype of AML cells at presentation (or lack thereof), a finding that was predicted 50 y ago by Freireich and colleagues (15) and extended in this report.
Methods
Patient Selection.
All clinical data are available in the SI Appendix, sections A and B. NK-AML patients were selected for this study because their outcomes are highly variable and still difficult to predict at presentation. We defined LFRs as lasting more than 5 y to ensure the durability of treatment responses, which increases the chance of identifying robust biomarkers associated with excellent responses. The comparator set of SFRs were defined as having an initial relapse within 2 y of presentation, since ∼70% of NK-AML cases relapse within 2 y. We did not use the ELN 2017 criteria to select the initial sample sets for the study, since many of the patients were treated before gene testing for common AML mutations was routinely performed, and it did not guide therapeutic decisions. We required the following inclusion criteria for all patients: 1) morphologically documented de novo AML with adequate bone marrow (BM) and matched control (skin) DNA for sequencing studies, 2) at least 18 y of age, 3) NK of AML cells at presentation, 4) received “7 + 3” (or similar variants) for induction chemotherapy, and 5) received high or intermediate-dose AraC as the primary consolidation therapy (i.e., no allogeneic transplant in first remission). For the LFR cases, the initial remission had to last at least 5 y. For the SFR cases, patients either had a morphologically documented relapse occurring within 2 y of starting therapy (28 patients) or primary refractory disease with 7 + 3 (3 patients), prior to any form of transplant (none of these patients were transplanted in first remission).
To identify the LFR samples for study, 1,579 cases were screened from our AML collection at Washington University in St. Louis, and 19 cases meeting all criteria were identified (1.2%). Nine additional LFR cases matching the above criteria were obtained after screening 846 NK-AML samples (1.06%) at the Alliance collection housed at The Ohio State University (but matched normal DNA and RNA was not available for these samples). Thirty-one well-matched NK-AML cases with SFRs were selected as comparators. Post hoc analyses confirmed that there were no significant differences in age, sex, BM blast percentage at diagnosis, karyotype, or induction/consolidation treatments between LFR and SFR patients. Samples were acquired as part of studies that were approved by the Human Research Protection Office at both institutions. All the patients provided written informed consent that included explicit permission for genetic studies, under Institutional Review Board–approved protocols (#201011766 for Washington University and CALGB/Alliance 9665 [NCT0089922] and 20202 [NCT00900224] for samples obtained from The Ohio State University).
Survival Analysis.
OS was defined as the time from diagnosis to death. Patients who died in remission were censored at the time of death. RFS was defined as the time from diagnosis to the first AML relapse or death from leukemia, whichever occurred first. Patients who were alive and disease-free were censored at last follow-up. Thirteen of 50 cases in the extension cohort who were transplanted in first remission were censored at the time of transplant. The distributions of OS or RFS between different groups were described using Kaplan–Meier product limit methods and compared by a log-rank test. Survival curves were visualized with GraphPad Prism (v9.0.2).
Molecular, Immunologic, and Statistical Analyses.
DNA, RNA, and viably cryopreserved cells were obtained from the BM aspirates of normal donors or AML patients at presentation or at various time points during remission. The LFR and SFR samples that were used for sequencing and functional studies are listed in the SI Appendix. For details regarding T cell activation studies, flow cytometry, cell culture, sequencing, bioinformatic, and statistical analyses, refer to the SI Appendix.
Results
Mutational Spectrum of NK-AML Cases with LFRs vs. SFRs.
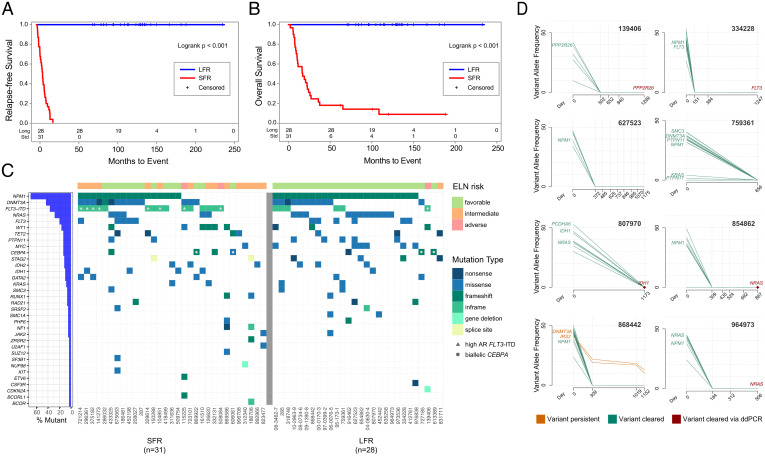
The clinical characteristics of the 28 LFR and 31 SFR cases are summarized in Table 1 and detailed in Dataset S1. All patients had an NK defined by standard cytogenetics, and received standard-of-care induction chemotherapy and consolidation, as detailed in Patient Selection. At the time of this writing, none of the LFR patients from Washington University in St. Louis had relapsed (range 5.8 to 19+ y, median follow-up 9.5 y). The two groups were comparable in terms of age, sex, percentage of BM blasts, and white blood cell (WBC) counts at diagnosis (Table 1). The Kaplan–Meier distribution of RFS and OS of the LFR and SFR cases is shown in Fig. 1 A and B). The mutational landscape of these cases was defined by sequencing the exomes from the presentation BM samples from all cases. Matched normal (skin) samples were available for the 19 LFR and 31 SFR cases banked at Washington University in St. Louis. Somatic mutation status was inferred for the nine Alliance samples by limiting mutation calling to known, recurrently mutated AML genes (16) (Fig. 1C and Dataset S2).
Table 1.
Clinical characteristics of SFR vs. LFR patients
| SFR (n = 31) | LFR (n = 28) | P value | |
| Age at diagnosis (y) | 0.93 | ||
| Median | 51 | 55 | |
| Range | 21–76 | 19–71 | |
| Female sex (%) | 48.40 | 42.90 | 0.79 |
| WBC count | 0.09 | ||
| Median | 52.9 | 26.8 | |
| Range | 1.5–222.8 | 1.6–298.4 | |
| Blasts in the BM (%) | 0.28 | ||
| Median | 72 | 72 | |
| Range | 30–97 | 23–91 | |
| RFS (mo) | <0.001 | ||
| Median | 7.7 | 126 | |
| Range | 1.1–20.6 | 70.3–231.9 | |
| OS (mo) | <0.001 | ||
| Median | 17 | 126 | |
| Range | 2.2–188.1 | 70.3–231.9 | |
| ELN category, n (%) | |||
| Favorable | 14 (45) | 26 (93) | <0.001 |
| Intermediate | 15 (48) | 1 (3.5) | <0.001 |
| Adverse | 2 (8) | 1 (3.5) | 1 |
| Occurrence of mutation, n (%) | |||
| NPM1 | 17 (54.8) | 24 (85.7) | 0.01 |
| DNMT3A | 14 (45.2) | 11 (39.3) | 0.79 |
| FLT3-ITD | 14 (45.2) | 5 (17.9) | 0.03 |
| Other FLT3 | 5 (16.1) | 6 (21.4) | 0.74 |
| MYC | 1 (3.2) | 6 (21.4) | 0.05 |
| NRAS | 5 (16.1) | 11 (39.3) | 0.08 |
| WT1 | 6 (19.4) | 3 (10.7) | 0.48 |
| TET2 | 4 (12.9) | 3 (10.7) | 1 |
| PTPN11 | 2 (6.5) | 5 (17.9) | 0.24 |
| IDH1 | 4 (12.9) | 1 (3.6) | 0.36 |
| IDH2 | 3 (9.7) | 3 (10.7) | 1 |
| CEBPA | 4 (12.9) | 3 (10.7) | 1 |
Fig. 1.
Clinical and genomic features of AML patients at presentation and in remission. (A) RFS and (B) OS curves for NK-AML patients who were treated with chemotherapy only for induction and consolidation. The blue line represents the LFR cases (n = 28) and the red line the SFR cases (n = 31). (C) The mutational landscape and the ELN classification for each case. Each column represents a patient, and each row represents a gene that is mutated in at least one of these cases. Every case had one or more recognized AML driver mutations, with a median of 11 (range 1 to 37) protein-altering somatic mutations per case in the Washington University in St. Louis samples. Color indicates the type of mutation, as specified in the legend. Cases with a high FLT3-ITD allelic ratio are indicated by the gray triangle in the figure. Blue bars at left indicate the mutation frequency in the sample set. (D) Clearance plots displaying the variant allele frequencies (VAFs) of recurrently mutated AML genes, plotted at presentation (day 0) and at available time points during clinically defined remissions (days), assessed with error-corrected sequencing. The average coverage of all variants for the remission samples was 3,042×, with a range of 161 to 13,266×. Every sample had at least one AML-specific mutation assayed with a coverage depth greater than 5,000×, yielding a sensitivity of 1 AML cell in ∼2,500 (0.04%). In the remission samples, seven of eight patients demonstrated clearance of all mutations in all samples tested. In one case (868442), a persistent ancestral clone was detected in all remission samples, harboring a DNMT3AR882H (VAF 10.19% at day 1,152) and an IRS2D106Y mutation (VAF 12.82% at day 1,152). Mutation clearance of the genes highlighted in red was confirmed with digital droplet PCR, at a sensitivity of 1 AML per 100,000 cells tested (SI Appendix, Fig. S2).
Using these data, we classified patients according to the ELN 2017 genetic risk categories (Fig. 1C, Table 1, and Dataset S1). The ELN criteria classified 26 of the 28 (93%) LFR cases as favorable risk, 1 of 28 (3.5%) as intermediate risk, and 1 of 28 (3.5%) as adverse risk. For the 31 SFR cases, 14 of 31 cases (45%) were classified as favorable risk (median RFS 301 d), 15 (48%) were classified as intermediate risk (median RFS 195 d), and 2 cases (8%) were classified as adverse risk by ELN criteria (median RFS 330 d). Kaplan–Meier plots for the ELN-classified SFR cases are shown in SI Appendix, Fig. S1. The fact that nearly half of the SFR patients were classified as favorable risk by ELN criteria suggested that other factors must play a role in the fates of these patients.
Evaluation of Persistent Molecular Disease in LFR Cases.
For eight of the LFR cases, we had access to remission samples obtained 506 to 1,299 d after presentation. We performed error-corrected sequencing (sensitivity of 4 AML cells in 10,000 total cells) of the presentation and remission samples, targeting 5 to 18 known somatic variants per case (17) (Dataset S3). For seven of the eight cases, no AML-associated mutations could be detected in any remission sample (Fig. 1D). In one case (868442), a DNMT3AR882H and an IRS2D106Y mutation persisted in remission, suggesting that the patient retained a preleukemic, ancestral clone. To increase the detection sensitivity of persistent AML cells to ∼1 in 100,000 (0.001%), we performed digital-droplet PCR for a single founding clone mutation (for which commercial reagents were available) for five patients in remission; no persistent AML cells bearing these mutations were detected in any of these samples (SI Appendix, Fig. S2).
RNA-Sequencing Analyses of SFR vs. LFR Cases.
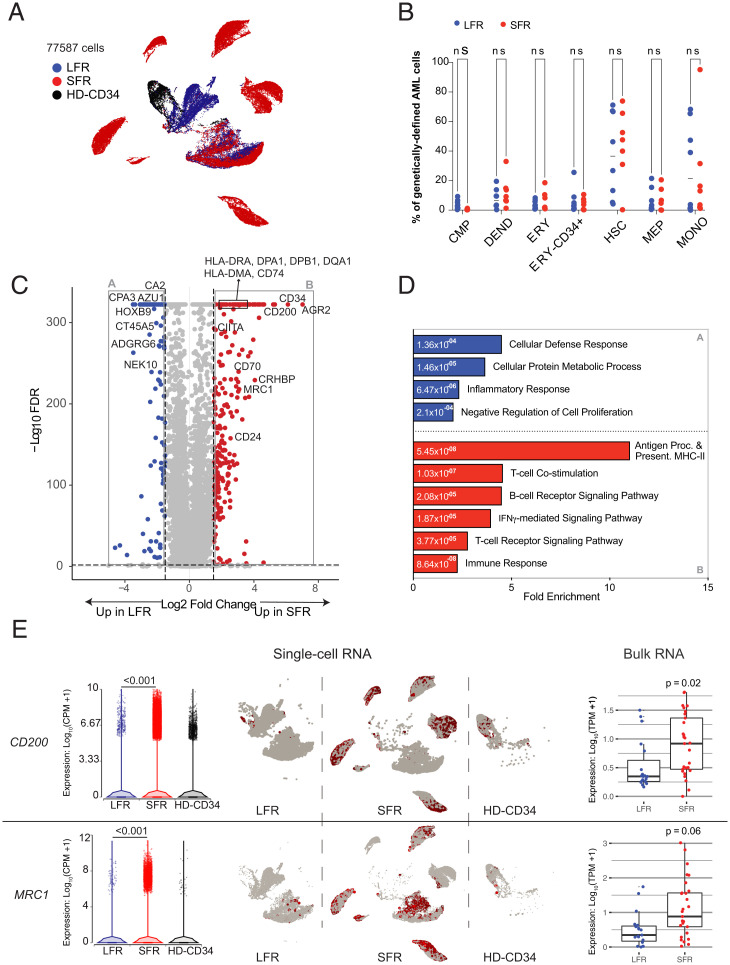
Because bulk RNA-sequencing cannot accurately define the expression patterns of cellular subsets in AML samples, we performed single-cell RNA-sequencing (scRNA-seq) of unfractionated BM from the presentation samples of eight LFR and seven SFR cases that were chosen based on high cellular viability (>50%) in available cryovials; the two sets had comparable fractions of myeloblasts based on BM aspirates flow cytometry at diagnosis (mean for LFR 71.1% vs. SFR 66.3%, P = 0.51). In addition, whole BM and CD34+ flow-sorted cells were evaluated from two healthy donors (ages 33 and 36 y). SI Appendix, Fig. S3 A and B display Uniform Manifold Approximation and Projection (UMAP) projections of normalized expression data from all cells, labeled by cell type (18) and sample, respectively. Nonmalignant cells, such as immune cells and erythroid cells, formed common clusters containing cells from all 17 samples. For each AML sample, we identified cells expressing known AML-associated somatic mutations (defined by prior exome sequencing), allowing us to restrict our analysis to genetically defined AML cells (Fig. 2A) (19). Determination of cell type proportions in these AML cells (Fig. 2B), and pathways expected to be relevant for chemotherapy sensitivity (i.e., cell cycle, DNA repair, or AraC responsiveness), revealed no significant differences between the genetically defined LFR and SFR AML cells (SI Appendix, Fig. S3 C–F and Dataset S4).
Fig. 2.
RNA-seq studies of LFR and SFR AML samples. (A) UMAP projection of 77,587 genetically defined AML cells from 8 LFR and 7 SFR samples at presentation, and purified CD34+ cells from the BM samples of 2 healthy donors (HD-CD34). Colors reflect the sample groups, as indicated in the legend. Cells were identified as AML by requiring expression of a AML-specific somatic mutation in that cell; relevant mutations were defined for each sample by exome sequencing. (B) Proportion of the genetically defined AML cells composed of different cell types across each AML sample for LFR cases (blue) vs. SFR cases (red). Horizontal lines indicate median values. (C) DEGs in genetically defined AML cells from the presentation BM samples of SFR vs. LFR patients. Dashed lines indicate log2 FC of ±2 and FDR < 0.001. Red points represent genes with significantly higher expression in the SFR cases, and blue points represent genes that are more highly expressed in the LFR cases. (D) Enrichment for gene ontology terms in the DEGs. Blue and red bars are pathways enriched in the LFR or SFR cases, respectively; numeric values indicate FDR. (E, Left) Expression of CD200, and MRC1 in single-cell data, with each point representing a cell. (Center) UMAP plots (split by category) showing single-cell expression of the corresponding gene. (Right) Expression from bulk RNA-seq datasets with additional AML samples (LFR, blue dots, n = 19; SFR, red dots, n = 31).
By comparing genetically defined AML cells from LFR vs. SFR cases using ANOVA, we identified 11,657 differentially expressed genes (DEGs) (false-discovery rate [FDR] = 0.001, log fold-change [FC] > ±1) (Fig. 2C and Dataset S5). Functional enrichment analysis showed that morphogenesis, chemotaxis, and inflammation pathways were enriched in the genes that were more highly expressed in the LFR AML cells, while genes involved in major histocompatibility complex (MHC) class II-mediated antigen presentation, T cell receptor (TCR) signaling, and immune responses were expressed at significantly higher levels in the AML cells from SFR samples (SI Appendix, Fig. S4 A and B). Because NPM1 mutations are known to influence MHC class II (20, 21) and HOX gene expression (22, 23), we repeated the ANOVA comparison after restriction to the NPM1 mutant cases (n = 8 LFR cases, and n = 3 SFR cases) (Dataset S6). We identified 8,191 DEGs that were shared with the previous analysis (FDR = 0.001, log FC ± 1) (SI Appendix, Fig. S4C and Dataset S6). Repeat gene ontology analysis of the overlapping DEGs eliminated the significance of HOX gene pathways but increased the significance of the MHC class II signature found in the SFR AML cells (Fig. 2D and Dataset S7).
To extend these findings, we evaluated MHC class II gene expression using bulk RNA-seq data from the 31 SFR vs. 19 LFR cases. We found that CIITA (the master regulator of MHC class II expression) and several class II genes were expressed at significantly higher levels in the SFR cases (SI Appendix, Fig. S4D), even after restricting the comparison to the cases that did not have NPM1 mutations (SFR n = 13, LFR n = 4) (SI Appendix, Fig. S4E). The combined data from single-cell and bulk RNA-seq corroborated the MHC class II phenotype, and suggested that NPM1 mutations do not define this phenotype per se. Two other immunomodulatory genes were found to be expressed at significantly higher levels in SFR AML cells: CD200 and MRC1 (Fig. 2E). CD200 has been reported to be associated with immunosuppression and worse outcomes in AML (24–26). MRC1 encodes CD206, a marker of tumor-promoting macrophages in chronic inflammatory diseases (27, 28) and solid tumors (29, 30); it was recently identified as an independent negative prognostic indicator in AML (31). The expression differences for CD200 and MRC1 were similar in the bulk RNA-sequencing data from the extended sample set (Fig. 2E). The lack of an apparent explanation for chemotherapy susceptibility in the LFR cases, and the detection of dysregulated immune-related genes (and genes involved in resistance to NK/T cell–mediated killing) in SFR cases, suggested that immunomodulatory activities of the tumor microenvironment could influence chemotherapy responses in these cases.
Expression Signatures in Marrow-Infiltrating Immune Cells.
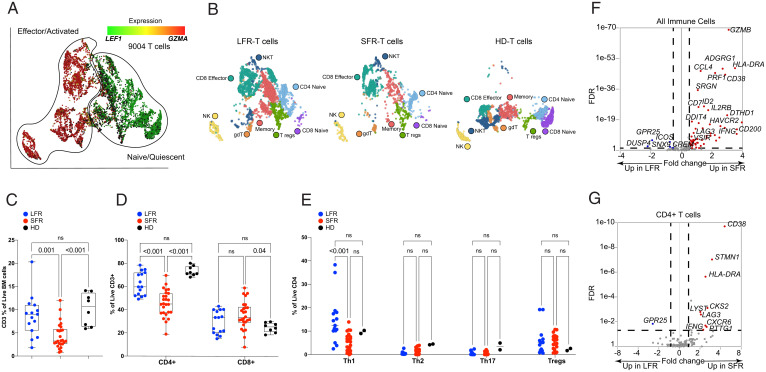
We therefore sought to compare the expression signatures of the infiltrating immune cells in the leukemic marrow samples. Populations of B cells, natural killer cells, regulatory T cells (Tregs), and γ/δ T cells were too small to allow for meaningful studies of differential gene expression. However, we identified 9,004 T cells after pooling all samples, shown in the UMAP projection of Fig. 3A. To identify and label specific T cell subpopulations, we applied a graph-based clustering approach (Fig. 3B, SI Appendix, Fig. S5A, and Dataset S8). SFR marrows at presentation had a smaller proportion of total T cells (SI Appendix, Fig. S5B). Flow cytometric evaluation of 15 LFR, 26 SFR, and 8 healthy adult donor marrow samples (age range 25 to 40 y) confirmed that the fraction of CD3+ T cells in SFR AML BM was significantly lower than that of LFR and healthy donor marrows (Fig. 3C); specifically, SFR BM samples contained a significantly smaller fraction of CD4+ T cells (Fig. 3D). Among the CD4 T cell subsets, the SFR marrow samples had significantly fewer Th1 cells, but no statistical differences were detected in the proportions of Th2, Th17, or Treg cells, which were all rare populations in these AML samples (Fig. 3E).
Fig. 3.
scRNA-seq and flow cytometric studies of T cells from LFR vs. SFR cases. (A and B) UMAP projections of 9,004 T cells from the total BM samples of 2 healthy donors and the 15 AML patients described in Fig. 2. (A) T cells are colored by the relative expression of LEF1, representing naïve or quiescent cells (green), vs. GZMA, representing activated or effector cells (red). (B) T cell subsets are labeled by subtypes, identified by applying graph-based clustering, then identifying enriched biomarkers for each cluster. (C–E) Boxplots displaying flow cytometric data from BM samples from healthy donors (HD) or LFR vs. SFR cases, for (C) percentages of CD3+ cells, (D) percentages of CD4+ and CD8+ cells, and (E) percentages of Th1, Th2, Th17, and Treg subsets. Means were analyzed for significance with two-way ANOVA and Tukey correction for multiple comparisons. (F and G) The results of an ANOVA comparison of 142 previously described activation/exhaustion markers (32, 33) in all 9,004 T cells (F) and in CD4+ T cells (G), comparing SFR to LFR cases. The x axis shows the log FC for each gene, with no change (N/C) as the midpoint. The y axis shows FDR in descending values. DEGs (defined by log FC ± 1.3, FDR < 0.05) are highlighted in red for SFR cases, or blue for LFRs.
We next tested the differential expression of the genes encoding 143 previously described T cell–specific activation/exhaustion markers (32, 33) in the total immune cells of SFR vs. LFR samples. Notably, 57 of the 62 DEGs were expressed at significantly higher levels in the SFR T cells (Fig. 3F and Dataset S9). These included genes encoding several putative immune checkpoint proteins, including LAG3, a T cell inhibitory receptor engaged by MHC class II (log FC = 1.59, FDR < 0.001), CD38 (log FC = 3.28, FDR < 0.001), and HAVCR2 (which encodes TIM3; log FC = 1.51, FDR < 0.01), although the latter was expressed by only a very small number of T cells (SI Appendix, Fig. S5C). The degree of differential expression for these genes varied across T cell subpopulations; for both LAG3 and CD38 it was more prominent in the CD4+ T cell compartment (Fig. 3G) (LAG3, log FC = +2.06, FDR < 0.001; CD38, log FC = +4.58, FDR < 0.001). CD4+ T cells did not express significant levels of HAVCR2 (SI Appendix, Fig. S5D and Dataset S10). Other genes encoding potentially relevant immune checkpoint proteins—such as PDCD1, TIGIT, CTLA4, and CD200R1—were not differentially expressed by the immune cells of LFR vs. SFR samples (SI Appendix, Fig. S5D and Dataset S9).
To determine whether the proteins encoded by these immune checkpoint genes may be relevant for the suppression of T cell function in AML marrows, we examined the expression of genes encoding known ligands for these receptors. MHC class II proteins serve as ligands for LAG3 (34), and as noted above, many MHC class II genes were more highly expressed in the SFR AML cells (Fig. 2D and SI Appendix, Fig. S4 D and E). FGL1, a recently described LAG3 ligand (35), was not expressed by AML cells (SI Appendix, Fig. S6A). The gene encoding PECAM1, the ligand for CD38, was expressed similarly in both LFR and SFR AML cells (SI Appendix, Fig. S6B). Of the known TIM3 ligands, the gene encoding CEACAM 1 was expressed in only a small fraction of AML cells (SI Appendix, Fig. S6C), while LGALS9 was more highly expressed in SFR AML cells (SI Appendix, Fig. S6D) (log FC = +1.3, FDR < 0.001). Thus, when considering the potential relevance of immune checkpoint inhibition for relieving T cell suppression in NK-AML, LAG3 emerged as a leading candidate because of its high expression in SFR T cells, and because the genes encoding its major ligands (MHC class II) are highly expressed in the AML cells of most SFR cases.
T Cell Function in AML BM Samples.
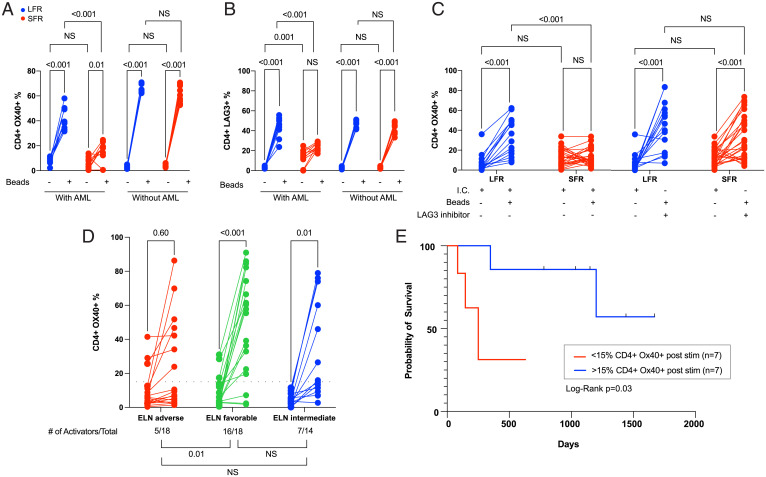
We next assessed the ability of AML cells from cryopreserved LFR and SFR BM samples to influence the activation of their own infiltrating T cells over 5 d in vitro, using CD3/CD28 stimulatory beads to activate the TCR. First, we evaluated unfractionated BM samples from LFR vs. SFR cases (i.e., containing both AML cells and T cells) obtained at presentation. In the presence of AML cells, bead-activated CD4+ T cells from the LFR cases significantly (P < 0.001) up-regulated the activation markers OX40 and inducible T cell costimulator, while activation was blunted in the SFR cases (Fig. 4A and SI Appendix, Fig. S7A, graphs “with AML”). LFR CD4 T cells activated the LAG3 marker in the presence of AML cells (Fig. 4B) (P < 0.001), while SFR cases had higher baseline percentages of CD4+LAG3+ cells (which corroborated the scRNA-seq data) that activated less robustly in the presence of AML cells (Fig. 4B). PD-1 levels increased with T cell stimulation, but there was no significant difference between LFR and SFR samples (SI Appendix, Fig. S7B). We then tested the activation potential of CD3+ T cells (purified by bead separation) from the same BM samples; more than 95% of the AML cells were removed by this procedure. In the absence of AML cells, the activation of SFR CD4+ T cells was restored, and LAG3 baseline levels decreased to levels similar to that of LFR samples (Fig. 4 A and B, “without AML”). CD8+ cells displayed similar trends (SI Appendix, Fig. S7 C–F), although endogenous CD8+ T cells suffered disproportionate losses in numbers after removal of AML cells, making measurements more difficult.
Fig. 4.
CD4+ T cell activation studies from the BM samples of AML cases at presentation. For the data shown in A–C, cryovials from presentation BM AML samples from LFR vs. SFR cases were thawed, and the fraction of CD3+ T cells was immediately defined by flow cytometry. These unfractionated samples were placed in media containing human SCF, IL-3, FLT3L, TPO, and IL-2 (10 ng/mL), and also CD3/CD28 T cell receptor agonist beads in a 1:1 ratio with the previously defined number of T cells (“with AML”). An identical experiment was performed using BM-derived CD3+ T cells enriched from the same samples (“without AML”); CD3/CD28 beads were added at a 1:1 ratio after a 24 h “washout” period. T cell activation and inhibition markers were then quantified by flow cytometry 5 d later in both sets of experiments. The lines show the change in percentage of CD4+ cells expressing the activation marker OX40 (A) vs. the inhibitory marker LAG3 (B) in samples treated with or without CD3/CD28 beads, which activate via the T cell receptor (as indicated by the legend below each graph). Red lines represent the SFR samples and blue lines represent LFR samples. Two-way ANOVA and Tukey multiple comparison tests were used to test for significance differences between groups. Results represent the summary of three independent experiments (n = 10 unique samples from both the LFR and SFR sets). (C) Similar levels of activation (as defined by OX40 expression) can be achieved in CD4+ T cells from SFR samples in the presence of a LAG3 blocking antibody at day 5 poststimulation. The negative controls (I.C. = isotype control) were treated with an isotype matched antibody. Two-way ANOVA and Tukey multiple comparison tests were used to test for significance differences between groups (n = 15 for LFR and n = 26 for SFR cases). (D) Changes in T cell activation, measured by the percentage of CD4+ cells expressing OX40, in 50 novel AML cases from an extension set. Unfractionated BM samples from the presentation samples were evaluated 5 d after activation with CD3/CD28 T cell receptor agonist beads (in a 1:1 ratio with the number of measured T cells in each sample). The AML samples are grouped according to ELN category. Two-tailed t tests were used to calculate significance between pre- and postactivation samples. The threshold for CD4+ cell activation was defined as the median difference in activation (poststimulation over baseline) for the 50 samples tested with this assay (indicated by the black dotted line, at y = 15%). The number of AML samples that exhibited T cell activation in each ELN risk category, and a statistical comparison of the three groups, are shown below the graphs. Pearson’s χ2 test with Yates’ continuity correction was used to calculate differences in T cell activators among the groups. (E) The RFS for the intermediate ELN risk cases from the extension set (D) stratified by CD4+ cell activation status. The blue and red lines represent samples with CD4+ cell activation above and below the activation threshold (defined as >15% CD4+ OX40+ cells, and an FC from baseline CD4+ OX40 expression ≥2.0). Vertical black lines indicate subjects at the time of censoring, as further defined in Methods. Log-rank (Mantel–Cox) test was used to estimate the difference in RFS (P = 0.03).
We repeated the 5-d CD3/CD28 bead stimulation protocol using unfractionated BM samples from presentation (i.e., in the presence of AML cells), with or without a well-characterized LAG3 inhibitor known to selectively block the LAG3:MHCII interaction (clone 17B4) (36–38), or an isotype control antibody. With LAG3 inhibition, endogenous CD4+ T cell activation was restored in most SFR samples to levels that were similar to that of the LFR samples (Fig. 4C and SI Appendix, Fig. S7G).
To determine whether these findings were more broadly applicable, we performed the 5-d T cell stimulation assay using presentation BM samples from 50 additional de novo AML patients that were selected based on ELN 2017 classifications from DNA sequencing and cytogenetics, and on the availability of high-quality cryovials from presentation BM (see SI Appendix, Fig. S8 and Dataset S11 for patient characteristics, mutational profiles, and ELN classifications). The fraction of activated ( i.e., OX40+) CD4+ T cells before and after stimulation for each sample are shown in Fig. 4D. Activation was defined as >15% CD4+ OX40+ cells with stimulation, and an FC from baseline CD4+ OX40 expression ≥2.0, which represented the median difference in activation (comparing poststimulation over baseline) for the 50 samples tested. Five of 18 (28%) ELN adverse-risk patients exhibited activation; RFS in these 5 cases was 1.2, 1.4, 4.5, 9.3, and 33.6 mo. None of these patients was transplanted in first remission. In contrast, 14 of 18 (78%) favorable-risk patients exhibited CD4 activation. The strong correlation of CD4 activation with these ELN risk groups suggests that the mutational profile must influence the immunosuppressive properties of AML cells, and can in fact supersede them in some adverse-risk cases. For intermediate-risk cases, 7 of 14 (50%) displayed CD4 activation; importantly, the “activators” in this risk group had significantly longer RFS (median survival not yet reached) compared with the nonactivators (median survival of 250 d, Mantel–Cox log-rank = 0.03) (Fig. 4E). These data suggest that T cell activation induced by CD3/28 beads in vitro may provide novel prognostic information for the intermediate-risk group that is not detected by the current, genetically based classifications at presentation. We performed a multivariate analysis (using Firth logistic regression) to determine whether any covariate at presentation (including sex, age at diagnosis, WBC count, BM blast percentage, ELN classification, or somatic mutations in AML-associated genes) was associated with CD4 activation status. Although ELN favorable-risk patients were 8.2 times more likely to be CD4 activators, this difference did not reach significance due to wide confidence intervals. None of the other covariates were predictive of CD4+ T cell activation status at presentation (Dataset S12).
Discussion
By evaluating a group of NK-AML patients with LFRs after chemotherapy alone, we have been able to identify mechanisms that may be relevant for AML cell eradication after chemotherapy. Current prognostic tools, such as the ELN 2017 classification algorithm, have been especially helpful for guiding therapeutic decisions for favorable- and adverse-risk patients, but lose some resolution in the intermediate-risk group, perhaps because these patients are more genetically heterogenous. The analysis of remission blood and BM samples from LFR patients revealed that many do not appear to be living with persistent disease; one had a persistent ancestral clone, which is a risk factor for the redevelopment of AML. Since recent studies have shown that AML relapses often arise from ancestral clones that were present at diagnosis (10, 12), these combined observations suggest that eradication of virtually all AML cells after initial therapy may be essential for long-term remissions.
Data from this study suggest that an immunosuppressive phenotype at presentation may negatively influence the effectiveness of standard chemotherapy. Specifically, we showed that BM samples from LFR patients have well-preserved CD4+ and CD8+ T cell populations that can react appropriately to TCR-mediated activation signals, even in the presence of their own AML cells. In contrast, SFR AML cells express high levels of MHC class II, and genes associated with immune suppression. Their marrows have reduced numbers of immune cells (especially CD4+ Th1 cells) and their remaining T cells often express an exhaustion signature (39). Importantly, T cell activation was blocked in SFR samples by the presence of the AML cells themselves, and was reversible in most cases by removing the AML cells, or blocking the MHC class II inhibitory receptor, LAG3.
Several mechanisms could potentially explain these findings. For example, the AML cells of SFR cases may express neoantigen-creating mutations that cause prolonged antigen-specific T cell activation, and eventual exhaustion (perhaps because these AML cells have developed mechanisms to resist T cell–mediated killing). However, using predictive algorithms, we did not detect substantial differences in putative neoantigen burdens or human leukocyte antigen (HLA) alleles between LFR vs. SFR cases (SI Appendix, Fig. S9 and Dataset S13). However, peptides derived from mutant NPM1 proteins can be presented by HLA molecules (40) and effectively recognized by T cells (41, 42). Since the vast majority of LFR cases had NPM1 mutations (and since CD4+ T cell numbers and activation are often normal in LFR cases), it is possible that NPM1-specific T cell responses after chemotherapy may also contribute to the favorable outcomes of the LFR cases.
Our data also suggest a T cell suppression phenotype in the SFR cases that may be mediated by the engagement of inhibitory T cell receptors (i.e., checkpoints) by ligands that are expressed by AML cells. Although inhibition of PD1 and CTLA4 checkpoint inhibitors have not yet yielded clinical benefits for AML patients (43–45), this study suggests that alternative checkpoints may be relevant. We and others have now identified LAG3, CD200 (24, 25, 46), and MRC1 (31) as potentially relevant targets for inhibition in AML. CD200, which is known to suppress T cell function (25) and directly inhibit NK-cell (47) and Treg function (24), was significantly overexpressed in SFR cases. While the Treg and Natural Killer–cell populations in the LFR and SFR marrow aspirates were small and highly variable, studies have shown that these cells play a role in AML immune surveillance and responses (48, 49) and they may also contribute to the immunosuppressive phenotype of SFR cases. The means to inhibit these pathways currently exist, and are being explored in preclinical models and early-phase clinical trials (50–52). Additional mechanisms (e.g., inhibitory chemokines, metabolic inhibitors of T cell function, and others) could also be relevant for this phenotype, and will need to be further explored.
Previous studies have shown that a dysfunctional CD4+ T cell population can dampen immune responses and lead to CD8 T cell exhaustion (53–57); CD4-depleted tumors have decreased responsiveness to PD1 inhibition (58, 59) that may in part be dependent on MHC class II–LAG3 interactions, especially for tumors with high expression of MHC class II (like SFR AML cells) (60). Additionally, several groups have reported variable degrees of T cell dysfunction in the BM samples of AML patients (61–65), possibly caused by a suppressive effect of AML cells themselves [shown with AML cell lines (66), and also with primary AML samples (67)]; here, we demonstrate that the absence of this immunosuppressive phenotype is strongly correlated with favorable clinical outcomes in both the LFR cases, and also in many ELN favorable- and intermediate-risk patients. Among patients with intermediate risk, RFS was significantly longer in samples with preserved T cell activation. Importantly, the immune activation phenotype seems to be independent of other known prognostic factors in AML, such as age and ELN status.
Remarkably, the concept that well-preserved cell-mediated immunity can predict responses to chemotherapy in AML patients (both at diagnosis and during treatment) was proposed 50 y ago by Freireich and colleagues (15). In that study, AML patients with impaired delayed type IV hypersensitivity skin testing, and impaired in vitro T cell responses to mitogens, had higher rates of chemotherapy induction failure and relapse; interestingly, type IV reactions are now known to require the function of effector Th1 cells and macrophages (68), both of which appear to be altered in the SFR cases.
In summary, the immunological phenotype of LFR patients strongly suggests that overcoming the immunosuppressive environment of “typical” NK-AML cases at presentation (perhaps by targeting relevant immune checkpoints) could increase the chances for chemotherapy successes and long remissions. The ability to integrate immunological phenotype assessment with current “up-front” risk stratification approaches [for example, by measuring CD3/CD28-mediated T cell activation in BM aspirates at presentation or, as Freireich and colleagues (15) proposed, skin testing for type IV hypersensitivity] may also prevent unnecessary allotransplantation, with its attendant complications. Future trials of immunologic modulation for the therapy of AML should therefore be guided both by the genotypes and phenotypes of AML cells, and of their immune microenvironments.
Supplementary Material
Acknowledgments
We thank Dr. Allegra Petti for insightful discussions about the single-cell data analysis; the Siteman Flow Cytometry Core and the Tissue Procurement Core, supported by the Siteman Cancer Center support Grant P30 CA091842; and our patients for their participation. This work was supported by National Cancer Institute (NCI) K12 Program Grant CA167540 and K08 Grant CA252632 (to F.F.); NCI Research Specialist Awards R50 CA211466 (to M.P.R.) and R50 CA211782 (to C.A.M.); Genomics of Acute Myeloid Leukemia Program Project Grant P01 CA101937 (to T.J.L.); the Edward P. Evans Foundation (to M.J.W.); NCI Outstanding Investigator Awards R35 CA197561 (to T.J.L.) and R35 CA210084 (to J.F.D.); Barnes-Jewish Hospital Foundation Grant 00335-0505-02 (to T.J.L.); Specialized Program of Research Excellence in Acute Myeloid Leukemia Grant P50 CA171963 (to D.C.L.); and NCI grants to the Alliance for banked samples from that entity, including Grants U10 CA180821 (to Dr. Bertagnoli), U10 CA180882 (to Dr. Mandrekar), and U24 CA196171 (to Dr. Watson).
Footnotes
Reviewers: B.F., Universita degli Studi di Perugia; and T.R.G., University of Cambridge.
The authors declare no competing interest.
2Deceased March 1, 2020.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116427118/-/DCSupplemental.
Data Availability
Sequencing data for all patients have been deposited in dbGaP, https://www.ncbi.nlm.nih.gov/gap/ (phs000159 v.11) (69).
References
- 1.Döhner H., Weisdorf D. J., Bloomfield C. D., Acute myeloid leukemia. N. Engl. J. Med. 373, 1136–1152 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Lichtman M. A., A historical perspective on the development of the cytarabine (7 days) and daunorubicin (3 days) treatment regimen for acute myelogenous leukemia: 2013 the 40th anniversary of 7 + 3. Blood Cells Mol. Dis. 50, 119–130 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Mrózek K., et al. , Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J. Clin. Oncol. 30, 4515–4523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasu S., et al. , Ten-year outcome of patients with acute myeloid leukemia not treated with allogeneic transplantation in first complete remission. Blood Adv. 2, 1645–1650 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larkin K. T. M., Byrd J. C., Whole-genome sequencing for myeloid disease: One assay to stratify them all? Nat. Rev. Clin. Oncol. 18, 543–544 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Westermann J., Bullinger L., Precision medicine in myeloid malignancies. Semin. Cancer Biol., 10.1016/j.semcancer.2021.03.034 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Klco J. M., et al. , Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA 314, 811–822 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morita K., et al. , Clearance of somatic mutations at remission and the risk of relapse in acute myeloid leukemia. J. Clin. Oncol. 36, 1788–1797 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothenberg-Thurley M., et al. , Persistence of pre-leukemic clones during first remission and risk of relapse in acute myeloid leukemia. Leukemia 32, 1598–1608 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yilmaz M., et al. , Late relapse in acute myeloid leukemia (AML): Clonal evolution or therapy-related leukemia? Blood Cancer J. 9, 7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivey A., et al. , UK National Cancer Research Institute AML Working Group, Assessment of minimal residual disease in standard-risk AML. N. Engl. J. Med. 374, 422–433 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Bertoli S., et al. , More than ten percent of relapses occur after five years in AML patients with NPM1 mutation. Leuk. Lymphoma 61, 1226–1229 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Bhatnagar B., et al. , Persistence of DNMT3A R882 mutations during remission does not adversely affect outcomes of patients with acute myeloid leukaemia. Br. J. Haematol. 175, 226–236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jongen-Lavrencic M., et al. , Molecular minimal residual disease in acute myeloid leukemia. N. Engl. J. Med. 378, 1189–1199 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Hersh E. M., Whitecar J. P. Jr., McCredie K. B., Bodey G. P. Sr., Freireich E. J., Chemotherapy, immunocompetence, immunosuppression and prognosis in acute leukemia. N. Engl. J. Med. 285, 1211–1216 (1971). [DOI] [PubMed] [Google Scholar]
- 16.Ley T. J., et al. , Cancer Genome Atlas Research Network, Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duncavage E. J., et al. , Mutation clearance after transplantation for myelodysplastic syndrome. N. Engl. J. Med. 379, 1028–1041 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novershtern N., et al. , Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144, 296–309 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petti A. A., et al. , A general approach for detecting expressed mutations in AML cells using single cell RNA-sequencing. Nat. Commun. 10, 3660 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dufva O., et al. , Immunogenomic landscape of hematological malignancies. Cancer Cell 38, 380–399.e13 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Chauhan P. S., et al. , Mutation of NPM1 and FLT3 genes in acute myeloid leukemia and their association with clinical and immunophenotypic features. Dis. Markers 35, 581–588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunetti L., et al. , Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell 34, 499–512.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spencer D. H., et al. , Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells. Leukemia 29, 1279–1289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coles S. J., et al. , Increased CD200 expression in acute myeloid leukemia is linked with an increased frequency of FoxP3+ regulatory T cells. Leukemia 26, 2146–2148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coles S. J., et al. , Expression of CD200 on AML blasts directly suppresses memory T-cell function. Leukemia 26, 2148–2151 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damiani D., et al. , Clinical impact of CD200 expression in patients with acute myeloid leukemia and correlation with other molecular prognostic factors. Oncotarget 6, 30212–30221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki Y., et al. , Macrophage mannose receptor, CD206, predict prognosis in patients with pulmonary tuberculosis. Sci. Rep. 8, 13129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaku Y., et al. , Overexpression of CD163, CD204 and CD206 on alveolar macrophages in the lungs of patients with severe chronic obstructive pulmonary disease. PLoS One 9, e87400 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komohara Y., Jinushi M., Takeya M., Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 105, 1–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haque A. S. M. R., et al. , CD206+ tumor-associated macrophages promote proliferation and invasion in oral squamous cell carcinoma via EGF production. Sci. Rep. 9, 14611 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z. J., et al. , The M2 macrophage marker CD206: A novel prognostic indicator for acute myeloid leukemia. OncoImmunology 9, 1683347 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo X., et al. , Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 24, 978–985 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Zheng C., et al. , Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 169, 1342–1356.e16 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Maruhashi T., et al. , LAG-3 inhibits the activation of CD4+ T cells that recognize stable pMHCII through its conformation-dependent recognition of pMHCII. Nat. Immunol. 19, 1415–1426 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Wang J., et al. , Fibrinogen-like protein 1 is a major immune inhibitory ligand of LAG-3. Cell 176, 334–347.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baixeras E., et al. , Characterization of the lymphocyte activation gene 3-encoded protein. A new ligand for human leukocyte antigen class II antigens. J. Exp. Med. 176, 327–337 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maçon-Lemaître L., Triebel F., The negative regulatory function of the lymphocyte-activation gene-3 co-receptor (CD223) on human T cells. Immunology 115, 170–178 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huard B., et al. , Characterization of the major histocompatibility complex class II binding site on LAG-3 protein. Proc. Natl. Acad. Sci. U.S.A. 94, 5744–5749 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crawford A., et al. , Molecular and transcriptional basis of CD4+ T cell dysfunction during chronic infection. Immunity 40, 289–302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liso A., et al. , Nucleophosmin leukaemic mutants contain C-terminus peptides that bind HLA class I molecules. Leukemia 22, 424–426 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Greiner J., et al. , Mutated regions of nucleophosmin 1 elicit both CD4(+) and CD8(+) T-cell responses in patients with acute myeloid leukemia. Blood 120, 1282–1289 (2012). [DOI] [PubMed] [Google Scholar]
- 42.van der Lee D. I., et al. , Mutated nucleophosmin 1 as immunotherapy target in acute myeloid leukemia. J. Clin. Invest. 129, 774–785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stahl M., Goldberg A. D., Immune checkpoint inhibitors in acute myeloid leukemia: Novel combinations and therapeutic targets. Curr. Oncol. Rep. 21, 37 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Bewersdorf J. P., Zeidan A. M., Randomized trials with checkpoint inhibitors in acute myeloid leukaemia and myelodysplastic syndromes: What have we learned so far and where are we heading? Best Pract. Res. Clin. Haematol. 33, 101222 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Bewersdorf J. P., Shallis R. M., Zeidan A. M., Immune checkpoint inhibition in myeloid malignancies: Moving beyond the PD-1/PD-L1 and CTLA-4 pathways. Blood Rev. 45, 100709 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Ho J. M., et al. , CD200 expression marks leukemia stem cells in human AML. Blood Adv. 4, 5402–5413 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coles S. J., et al. , CD200 expression suppresses natural killer cell function and directly inhibits patient anti-tumor response in acute myeloid leukemia. Leukemia 25, 792–799 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carlsten M., Järås M., Natural killer cells in myeloid malignancies: Immune surveillance, NK cell dysfunction, and pharmacological opportunities to bolster the endogenous NK cells. Front. Immunol. 10, 2357 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lion E., Willemen Y., Berneman Z. N., Van Tendeloo V. F., Smits E. L., Natural killer cell immune escape in acute myeloid leukemia. Leukemia 26, 2019–2026 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Jaynes J. M., et al. , Mannose receptor (CD206) activation in tumor-associated macrophages enhances adaptive and innate antitumor immune responses. Sci. Transl. Med. 12, eaax6337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scodeller P., et al. , Precision targeting of tumor macrophages with a CD206 binding peptide. Sci. Rep. 7, 14655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahadevan D., et al. , Phase I study of samalizumab in chronic lymphocytic leukemia and multiple myeloma: Blockade of the immune checkpoint CD200. J. Immunother. Cancer 7, 227 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matloubian M., Concepcion R. J., Ahmed R., CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J. Virol. 68, 8056–8063 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wherry E. J., Kurachi M., Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ozkazanc D., Yoyen-Ermis D., Tavukcuoglu E., Buyukasik Y., Esendagli G., Functional exhaustion of CD4+ T cells induced by co-stimulatory signals from myeloid leukaemia cells. Immunology 149, 460–471 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oh D. Y., et al. , Intratumoral CD4(+) T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell 181, 1612–1625.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sato Y., et al. , CD4+ T cells induce rejection of urothelial tumors after immune checkpoint blockade. JCI Insight 3, e121062 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kagamu H., et al. , CD4+ T-cell immunity in the peripheral blood correlates with response to anti-PD-1 therapy. Cancer Immunol. Res. 8, 334–344 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Homet Moreno B., et al. , Response to programmed cell death-1 blockade in a murine melanoma syngeneic model requires costimulation, CD4, and CD8 T cells. Cancer Immunol. Res. 4, 845–857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagasaki J., et al. , The critical role of CD4+ T cells in PD-1 blockade against MHC-II-expressing tumors such as classic Hodgkin lymphoma. Blood Adv. 4, 4069–4082 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knaus H. A., et al. , Signatures of CD8+ T cell dysfunction in AML patients and their reversibility with response to chemotherapy. JCI Insight 3, e120974 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian T., et al. , The profile of T helper subsets in bone marrow microenvironment is distinct for different stages of acute myeloid leukemia patients and chemotherapy partly ameliorates these variations. PLoS One 10, e0131761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams P., et al. , The distribution of T-cell subsets and the expression of immune checkpoint receptors and ligands in patients with newly diagnosed and relapsed acute myeloid leukemia. Cancer 125, 1470–1481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deng M., et al. , LILRB4 signalling in leukaemia cells mediates T cell suppression and tumour infiltration. Nature 562, 605–609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z., Philip M., Ferrell P. B., Alterations of T-cell-mediated immunity in acute myeloid leukemia. Oncogene 39, 3611–3619 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Galen P., et al. , Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell 176, 1265–1281.e24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lamble A. J., et al. , Reversible suppression of T cell function in the bone marrow microenvironment of acute myeloid leukemia. Proc. Natl. Acad. Sci. U.S.A. 117, 14331–14341 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mak T. W., Saunder M. E., Jett B. D., “Immune hypersensitivity” in Primer to the Immune Response (Academic Cell, ed. 2, 2014) pp. 487–516. [Google Scholar]
- 69.T. Ley, Genomics of acute myeloid leukemia. The Database of Genotypes and Phenotypes. https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000159.v11.p5. Deposited 22 July 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data for all patients have been deposited in dbGaP, https://www.ncbi.nlm.nih.gov/gap/ (phs000159 v.11) (69).