Exercise-induced ROS mediates antioxidant defense to attenuate oxidative damage and insulin resistance in aging and obesity.
Abstract
Reactive oxygen species (ROS) generated during exercise are considered integral for the health-promoting effects of exercise. However, the precise mechanisms by which exercise and ROS promote metabolic health remain unclear. Here, we demonstrate that skeletal muscle NADPH oxidase 4 (NOX4), which is induced after exercise, facilitates ROS-mediated adaptive responses that promote muscle function, maintain redox balance, and prevent the development of insulin resistance. Conversely, reductions in skeletal muscle NOX4 in aging and obesity contribute to the development of insulin resistance. NOX4 deletion in skeletal muscle compromised exercise capacity and antioxidant defense and promoted oxidative stress and insulin resistance in aging and obesity. The abrogated adaptive mechanisms, oxidative stress, and insulin resistance could be corrected by deleting the H2O2-detoxifying enzyme GPX-1 or by treating mice with an agonist of NFE2L2, the master regulator of antioxidant defense. These findings causally link NOX4-derived ROS in skeletal muscle with adaptive responses that promote muscle function and insulin sensitivity.
INTRODUCTION
Throughout evolution, exercise in the form of food gathering, shelter seeking, and predator avoidance has been essential for human survival and necessitated the development of adaptive mechanisms to support muscle function and overall health. In the modern era, technological advances have markedly diminished the need for physical activity. Physical inactivity contributes to the development of a myriad of human diseases, including insulin resistance and type 2 diabetes, that have reached epidemic proportions throughout the world (1). Although the benefits of physical activity or exercise are irrefutable, the precise mechanisms by which physical activity promotes metabolic health are incompletely understood (1, 2).
Exercise results in skeletal muscle exposure to acute stresses that stimulate compensatory and adaptive physiological responses (1, 2). These adaptive responses increase tolerance and protect against subsequent stressors and are considered important for the health-promoting effects of exercise (1, 2). These favorable adaptation to stressors that would otherwise be harmful at higher doses is known as “hormesis.” A fundamental hormetic response to exercise is the induction of mitochondrial biogenesis, which serves to enhance respiratory capacity and endurance (2, 3). The induction of PGC1α (peroxisome proliferator–activated receptor gamma coactivator 1α) after exercise can promote mitochondrial biogenesis by driving the expression of nuclear- and mitochondrial-encoded genes. In particular, PGC1α promotes the expression of NRF (nuclear respiratory factor)–1 and NRF-2, which induce the expression of nuclear-encoded genes, as well as TFAM (mitochondrial transcription factor A), which facilitates mitochondrial DNA transcription and replication (2, 3). Another important hormetic response is the enhancement of insulin sensitivity. Exercise promotes glucose uptake and glycogen synthesis and increases insulin sensitivity, and this persists long after the cessation of exercise (1, 2). The enhanced skeletal muscle insulin sensitivity after exercise affords metabolic flexibility and a means for replenishing glycogen to deal with subsequent bouts of intense physical activity (1, 2). Precisely how exercise drives mitochondrial biogenesis remains incompletely understood (2, 3). Moreover, the mechanisms by which exercise elicits long-lasting effects on insulin sensitivity remain unknown. One possibility is that both may involve the generation of reactive oxygen species (ROS) and the induction of the KEAP1 (Kelch-like ECH-associated protein 1)/NFE2L2 (nuclear factor erythroid 2–related factor 2) pathway. The KEAP1/Cullin-3 E3 ligase complex normally binds and targets NFE2L2 for degradation (4). However, after exercise, ROS oxidize Cys residues in KEAP1 to facilitate the release and stabilization of NFE2L2, which translocates to the nucleus to drive the expression of >200 endogenous antioxidant and xenobiotic detoxifying enzymes (4). Previous studies have shown that antioxidant supplements or the deletion of NFE2L2 can block acute exercise–induced increases in PGC1α and mitochondrial biogenesis (5–7). Similarly, previous studies have shown that increasing H2O2 in skeletal muscle (8) or the deletion of KEAP1 (9) enhances insulin sensitivity or improves glucose tolerance. Moreover, although debated, antioxidant supplements can negate the beneficial effects of exercise training on glucose metabolism (6, 10). It remains unclear how the induction of NFE2L2 in skeletal muscle after exercise might affect insulin sensitivity or glucose homeostasis. Fundamentally, the NFE2L2 pathway serves to prevent the oxidative damage of macromolecules, including proteins and lipids (4). Because oxidative stress can contribute to the development of insulin resistance and type 2 diabetes (11–18), it is possible that the NFE2L2 response may serve to ameliorate insulin resistance, but this remains to be formally established.
ROS, such as superoxide (O2•−), are produced by all living organisms as natural by-products of metabolism and cellular respiration (18). O2•− can react with nitric oxide (NO) produced by NO synthase during muscle contraction to generate the toxic oxidant peroxynitrite (ONOO−) that damages protein, lipids, and DNA (19). O2•− can also promote the conversion of H2O2 into highly reactive and damaging hydroxyl radicals. However, O2•− is normally dismutated to the nonradical H2O2 by superoxide dismutase (SOD) and thereon eliminated by catalase, peroxiredoxins (PRDXs), and glutathione peroxidases (GPXs) (17, 18). Mitochondria generate most steady-state O2•− in muscle cells. However, mitochondria are not responsible for ROS generation during exercise (20–22). H2O2 production in the mitochondrial matrix during exercise may actually be decreased (23). Instead, NADPH (reduced nicotinamide adenine dinucleotide phosphate) oxidases (NOXs) have been suggested to be the primary source of ROS during exercise. NOXs transfer electrons across membranes, from NADPH to molecular oxygen, to generate O2•− (24). The human genome encodes seven NOXs that function in diverse biological processes (24). NOX2 and NOX4 are the primary NOXs expressed in skeletal muscle (21). NOX4 is distinct from NOX2 in its ability to generate directly both O2•− and H2O2 (25). Moreover, the two NOXs may be differentially localized in skeletal muscle, with the catalytic subunit of NOX2 localizing to the plasma membrane and transverse tubules and NOX4 to the sarcoplasmic reticulum, transverse tubules, and the inner mitochondrial membrane (21, 26). Recent studies have shown that skeletal muscle NOX2 expression is increased by acute exercise (27), whereas others have shown that NOX2 is activated and contributes to skeletal muscle cytosolic ROS generation and glucose uptake during moderate-intensity exercise (23). However, defective NOX2 activation, due to loss of function mutations in the regulatory p47phox subunit (23), or the inducible deletion of the NOX2 subunit Rac1 in adult mice (28), does not alter skeletal muscle mitochondrial content. Moreover, Rac1 is dispensable for the insulin-sensitizing effects of exercise (29). Thus, although NOX2 may be required for the promotion of glucose uptake during exercise, it is not important for exercise-induced hormetic responses that enhance muscle function, nor is it required for eliciting long-lasting insulin-sensitizing effects.
In this study, we report that NOX4 expression is increased after exercise and that this facilitates exercise-induced adaptive responses that attenuate the decline in insulin sensitivity associated with aging and obesity. We demonstrate that the generation of H2O2 by NOX4 in skeletal muscle and the induction of NFE2L2 are essential for promoting antioxidant defense and mitochondrial biogenesis, preventing mitochondrial oxidative stress, and maintaining insulin sensitivity. Moreover, we report that skeletal muscle NOX4 is decreased in aged or obese mice and that this contributes to the development of insulin resistance. Our findings demonstrate that skeletal muscle NOX4 is of fundamental importance in exercise-induced hormetic responses that promote muscle function and metabolic health.
RESULTS
Skeletal muscle NOX4 is required for exercise-induced ROS
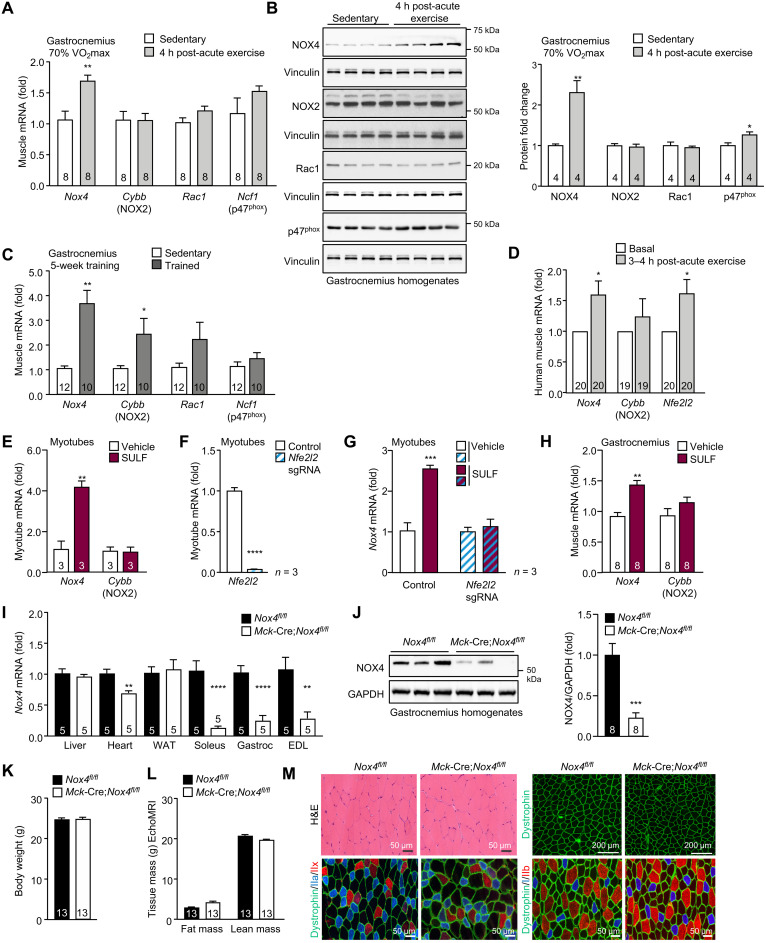
Although low levels of ROS are constantly produced by skeletal muscle, NOXs, rather than mitochondria, are thought to be primarily responsible for ROS production during exercise (20–22). Here, we assessed the contributions of skeletal muscle NOX4 to ROS generation following moderate or high-intensity acute exercise or exercise training. First, we examined the expression of NOX4, NOX2, and the NOX2 regulatory subunits p47phox (encoded by Ncf1) and RAC1 by quantitative real-time polymerase chain reaction (qPCR) in gastrocnemius muscle and soleus. For acute exercise studies, C57BL/6 mice were run for 50 min at moderate intensity [70% maximal rate of oxygen consumption (VO2max)] or high intensity (90 to 100% VO2max). For exercise training, mice were run for five consecutive weeks (5 days/week) at 50, 60, 70, 80, and 90% of their maximum pretraining exercise capacity in each successive week. We found that the expression of NOX4 increased by 1.5- to 2-fold in both gastrocnemius and soleus 4 hours after moderate or high-intensity exercise and by approximately 3- to 4-fold after exercise training (Fig. 1, A to C, and fig. S1, A to C). By contrast, the expression of Cybb (encoding NOX2) was only increased in gastrocnemius muscle after high-intensity exercise (fig. S1C) or after exercise training (Fig. 1C); Ncf1 (p47phox) and Rac1 were increased moderately, if at all, after exercise (Fig. 1, B and C). We found that NOX4, but not CYBB, expression was also increased in human muscle (vastus lateralis) 3 to 4 hours following acute high-intensity interval training (Fig. 1D). As expected, the increased NOX4 expression after exercise was accompanied by an increased expression of the gene encoding the transcription factor NFE2L2 (Fig. 1D), the master regulator of antioxidant defense (4). Therefore, skeletal muscle NOX4 expression is induced after exercise in mice and humans.
Fig. 1. Skeletal muscle NOX4 is induced after exercise.
Twelve-week-old C57BL/6 male mice were subjected to an acute bout of exercise on multilane treadmill for 50 min at moderate intensity (70% VO2max). After 4 hours, gastrocnemius muscles were collected from sedentary and exercised mice and processed for (A) quantitative real-time PCR (qPCR) or (B) immunoblotting. (C) Twelve-week-old C57BL/6 male mice underwent 5 weeks of treadmill exercise training; 4 hours after the last training session, gastrocnemius muscle was extracted and processed for qPCR. (D) Young middle-aged male humans (n = 20) were subjected to a bout of acute, high-intensity interval exercise. Thigh muscle (vastus lateralis) biopsies were taken serially before and 3 to 4 hours after exercise and processed for qPCR. (E) Myotubes were treated with vehicle or sulforaphane (2 μM) for 15 hours and processed for qPCR. (F and G) NFE2L2 was deleted in primary myoblasts using CRISPR-Cas9 gene editing. Control and NFE2L2-deficient myoblasts were differentiated into myotubes and (F) processed for qPCR or treated with vehicle or sulforaphane (2 μM) for 15 hours and processed for qPCR. (H) Twelve-week-old C57BL/6 male mice were administered vehicle or sulforaphane (0.5 mg/kg, intraperitoneally) for five consecutive days, and gastrocnemius muscle was extracted and processed for qPCR. (I to M) Twelve-week-old Nox4fl/fl and Mck-Cre;Nox4fl/fl male mice were fed a chow diet, and tissues were extracted and processed for (I) qPCR to assess Nox4 mRNA expression or (J) immunoblotting to assess NOX4 protein. Alternatively, (K) body weights and (L) body composition (EchoMRI) were determined. (M) Gastrocnemius muscle transverse sections (10 nm) were processed for H&E staining and dystrophin and fiber-type immunostaining. Representative and quantified results are shown (means ± SEM) for the indicated number of (A to C and H to M) mice, (D) humans, or (E to G) experiments; significance determined using a Student’s t test.
The increase in Nox4 expression after both acute exercise and exercise training in different muscles prompted us to examine whether the induction of NOX4 might be part of an adaptive physiological response to exercise. Previous studies have suggested that the potential deleterious effects of ROS after exercise can be ameliorated by the ROS-mediated induction of NFE2L2 (30) that induces the expression of antioxidant response element (ARE)–driven genes that encode endogenous antioxidant and xenobiotic detoxifying enzymes (4). Moreover, previous studies have shown that the NOX4 promoter consists of two AREs that bind NFE2L2 (31). NFE2L2 can be activated by the naturally occurring isothiocyanate sulforaphane, which has been reported to protect skeletal muscle and other tissues from oxidative stress and increase exercise capacity (32, 33). We found that the treatment of myotubes (differentiated from primary murine myoblasts) with sulforaphane increased Nox4, but not Cybb, expression by approximately four-fold (Fig. 1E). We found that the deletion of NFE2L2 in myoblasts, using CRISPR-Cas9 gene editing and a synthetic guide RNA (sgRNA) targeting Nfe2l2 (Fig. 1F), abrogated the sulforaphane-induced increased in Nox4 in myotubes (Fig. 1G). Moreover, treatment of 12-week-old C57BL/6 mice with sulforaphane for five consecutive days increased the expression of Nox4, but not Cybb, in skeletal muscle (Fig. 1H). Thus, NOX4 is induced in skeletal muscle after exercise, and this is mediated by NFE2L2 as part of an adaptive antioxidant defense response.
We reasoned that the induction of NOX4 after exercise may contribute to ROS generation as part of a feed-forward mechanism for accentuating the adaptive antioxidant defense response. To explore the extent to which NOX4 may contribute to exercise-induced ROS generation, we sought to delete NOX4 in muscle. To this end, we crossed Nox4fl/fl mice (34) with those expressing Cre under the control of muscle creatinine kinase (Mck) promoter (Mck-Cre); Cre expression occurs predominantly in skeletal muscle and, to a lesser extent, in cardiac muscle (35). We found that Nox4 expression, as assessed by qPCR in gastrocnemius, soleus, and extensor digitorum longus muscle homogenates (Fig. 1I) and by immunoblotting gastrocnemius muscle homogenates, was decreased by as much as 80% (Fig. 1J); any remaining Nox4 likely reflected its presence in other cell types, including endothelial and immune cells. Nox4 expression was not affected in other tissues, including liver and white adipose tissue (WAT), but was reduced by approximately 32% in cardiac muscle (Fig. 1I). The deletion of Nox4 did not affect body weight or body composition (lean or fat mass) in 12-week-old adult male mice (Fig. 1, K and L). Consistent with this, Nox4 deletion did not alter the weights of non-muscle tissues, such as liver or adipose tissue [inguinal WAT, epididymal WAT, brown adipose tissue (BAT)], cardiac tissue, or skeletal muscles composed of mixed [gastrocnemius and quadriceps] or predominantly glycolytic [tibialis anterior (TA) and triceps] myofibers (fig. S2A). However, Nox4 deletion moderately decreased the weight of soleus muscle in adult mice (fig. S2A). The decreased soleus weight was not accompanied by gross differences in muscle development or overt signs of disease pathology or atrophy (fig. S2, B to H). No alterations were evident in myofiber size (otherwise decreased in atrophied muscle or muscle undergoing degeneration/regeneration) or the abundance of oxidative [expressing type I myosin heavy chain (MHC)] and glycolytic (expressing type IIa and type IIb MHC) muscle fibers (fig. S2, B to H). In addition, subsarcolemmal nuclei were peripherally localized (fig. S2B; as opposed to centrally located nuclei evident in damaged and regenerating muscle) and the expression of dystrophin, which marks the perimeter of muscle fibers and is frequently decreased in muscular dystrophies, was uniform and uninterrupted (fig. S2C). As in soleus, we noted no differences in myofiber diameter/size and no change in fiber type or signs of nuclear centrality in gastrocnemius muscle (Fig. 1M and fig. S2, I to P). Thus, Nox4 deletion is not accompanied by gross differences in skeletal muscle development or muscle pathology/atrophy.
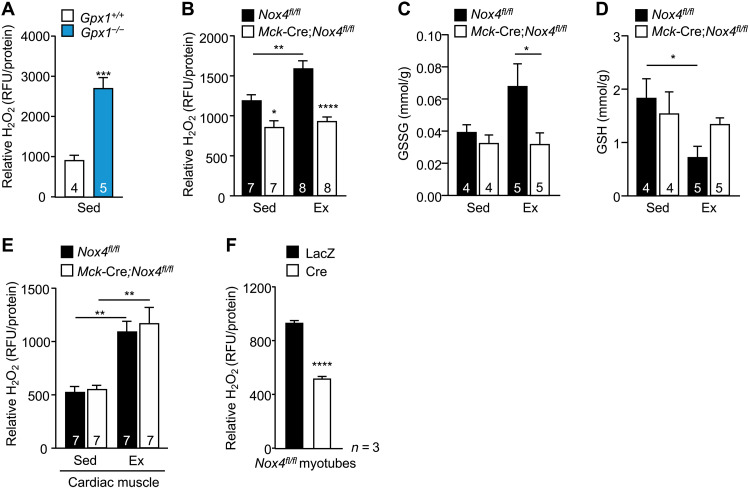
To assess the extent to which Nox4 deletion may affect exercise-induced ROS generation, we monitored for H2O2 levels in muscle that was rapidly excised, snap-frozen, and then homogenized in the presence of the H2O2-selective probe Amplex Red. To test whether this approach could be used to detect changes in H2O2 in muscle, we measured Amplex Red fluorescence in muscle homogenates from mice deficient for the H2O2-detoxifying enzyme GPX-1; Amplex Red fluorescence was readily detected in muscle homogenates from the sedentary C57BL/6 male mice and increased by the deletion of the H2O2-detoxifying enzyme GPX-1 in muscle, consistent with the detection of H2O2 (Fig. 2A). We thereon monitored for H2O2 in gastrocnemius muscle from 12-week-old sedentary Nox4fl/fl versus Mck-Cre;Nox4fl/fl mice and those exercised acutely at moderate intensity (70% VO2max) for 50 min. Nox4 deletion reduced skeletal muscle H2O2 in sedentary mice and abrogated the increase in skeletal muscle H2O2 that otherwise occurred after exercise (Fig. 2B). Consistent with this, the exercise-induced increase in skeletal muscle oxidized glutathione (GSSG) that follows ROS generation and the accompanying decrease in reduced glutathione (GSH) were abrogated by NOX4 deficiency (Fig. 2, C and D). By contrast, Nox4 deletion in Mck-Cre;Nox4fl/fl mice had no effect on cardiac H2O2 levels in sedentary mice and did not affect the approximately twofold increase in cardiac H2O2 detected using this approach after exercise (Fig. 2E). To determine whether the effects of NOX4 deficiency on skeletal muscle H2O2 generation may be cell intrinsic, we also assessed H2O2 production by live myotubes generated from fluorescence-activated cell sorting (FACS)–purified (CD11b−CD31−CD45−Sca1−α7-integrin+) Nox4fl/fl myoblasts (fig. S3A) transduced with adenoviruses expressing lacZ β-galactosidase (LacZ) as a control or Cre recombinase to delete Nox4. Nox4 was efficiently deleted in myoblasts (fig. S3B) and had no overt effect on the formation of myotubes in cell culture, as assessed by gross morphology and the expression of MHC and myogenin (fig. S3, C and D). However, Nox4 deletion decreased the emission of H2O2 by myotubes in culture (assessed by including Amplex Red in the culture medium; Fig. 2F). Together, our findings indicate that skeletal muscle NOX4 is required for ROS generation. Moreover, our findings indicate that the increased expression of Nox4 in skeletal muscle after exercise is required for the induction of ROS.
Fig. 2. NOX4 is required for exercise-induced H2O2 generation in skeletal muscle.
(A) Twelve-week-old Gpx1+/+ and Gpx1−/− male mice were fed a standard chow diet, and gastrocnemius muscle was excised, snap-frozen, and processed for the measurement of H2O2 using Amplex Red; relative fluorescence normalized to total protein. RFU, relative fluorescence units. (B to E) Twelve-week-old Nox4fl/fl and Mck-Cre;Nox4fl/fl male mice fed a chow diet were subjected to an acute bout of moderate-intensity exercise for 50 min (70% VO2max). Gastrocnemius muscle from sedentary (Sed) and exercised (Ex) mice was excised immediately after exercise, snap-frozen, and processed for (B) H2O2 measurements using Amplex Red or for analysis of (C) oxidized (GSSG) and (D) reduced glutathione (GSH) levels. (E) Cardiac muscle from sedentary and exercised mice was excised immediately after exercise, snap-frozen, and processed for H2O2 measurements using Amplex. (F) Skeletal muscle myoblasts from Nox4fl/fl mice were transduced with β-galactosidase (LacZ) control or Cre recombinase–expressing adenoviruses to delete Nox4; the resultant cells were differentiated into myotubes, and extracellular H2O2 levels were assessed in live myotubes using Amplex Red. Representative and quantified results are shown (means ± SEM) for the indicated number of (A to E) mice or (F) experiments; significance determined using (A and F) a Student’s t test or (B to E) a two-way ANOVA.
NOX4-derived H2O2 in skeletal muscle enhances exercise performance
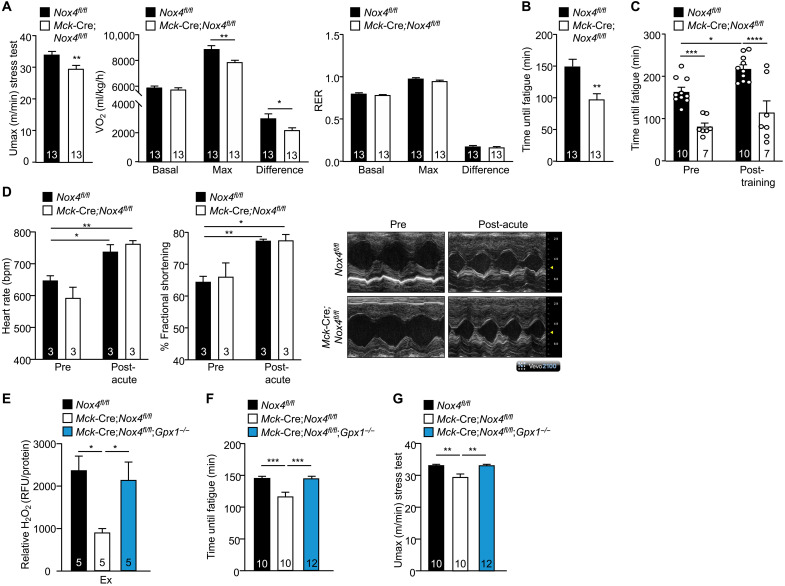
The production of ROS in response to acute bouts of exercise or chronic exercise training is considered essential for the adaptive responses necessary to meet the energy demands of exercise (36). Recent studies have shown that the global deletion of NOX4 compromises exercise capacity (37). The effects on exercise capacity were ascribed to diminished glucose and fatty acid oxidation in endothelial cells in the skeletal muscle vasculature (37). As Nox4 expression in skeletal muscle is increased after exercise and NOX4 is required for exercise-induced ROS generation, we reasoned that skeletal muscle NOX4 might be required for optimal muscle function. To explore the impact of Nox4 deletion on muscle function, we first performed exercise stress tests on 12-week-old chow-fed male Nox4fl/fl versus Mck-Cre;Nox4fl/fl mice, where mice were run at incrementally increasing speeds until exhaustion using a single-lane treadmill connected to an indirect calorimeter. The maximal speed upon which mice were exhausted (Umax) was decreased in Mck-Cre;Nox4fl/fl mice (Fig. 3A); this was accompanied by decreased oxygen consumption (VO2max) and heat production (Fig. 3A and fig. S4A). Carbohydrate and fatty acid oxidation were not different between genotypes at rest or during exercise, as reflected by the similar respiratory exchange ratios (RERs; Fig. 3A). The VO2max is predominantly a measure for the rate at which oxygen is delivered and used by muscle mitochondria to generate adenosine triphosphate (ATP). The decreased VO2max is consistent with Mck-Cre;Nox4fl/fl mice having decreased aerobic capacity. To explore further the impact of NOX4 deficiency on muscle function and exercise capacity, we performed an endurance test to exhaustion, which measures how long mice can run when exercised at the same relative intensity (70% VO2max). Endurance was assessed in both untrained mice and those that had been subjected to 5 weeks of exercise training, with progressive increases in training intensity. We found that exercise endurance (time until fatigue) was decreased by as much as ~50% in untrained Mck-Cre;Nox4fl/fl mice (Fig. 3, B and C) and that NOX4 deficiency prevented any significant increase in endurance with exercise training (Fig. 3C). Therefore, these findings are consistent with muscle NOX4 deficiency compromising muscle function and aerobic fitness and the adaptation to endurance exercise training in adult mice.
Fig. 3. NOX4-derived H2O2 in skeletal muscle enhances exercise performance.
(A) Twelve-week-old Nox4fl/fl and Mck-Cre;Nox4fl/fl male mice fed a standard chow diet were subjected to an exercise stress test in an enclosed treadmill connected to a Comprehensive Lab Animal Monitoring System for respiratory assessments during exercise. The maximal velocity (Umax), the maximal rate of oxygen consumption (VO2), and RERs during exercise were determined. (B) Twelve-week-old Nox4fl/fl and Mck-Cre;Nox4fl/fl chow-fed male mice were subjected to an endurance test on a multilane treadmill, and the time until fatigue was determined. (C) Nox4fl/fl and Mck-Cre;Nox4fl/fl chow-fed male mice were subjected to endurance tests before and after 5 weeks of treadmill exercise training (5 days/week, 60 min/day). (D) Echocardiographs from conscious 12-week-old Nox4fl/fl and Mck-Cre;Nox4fl/fl male mice before and immediately after an exercise stress test; heart rates and fractional shortening were assessed. Representative M-mode images acquired through a short-axis view at the papillary muscle level are shown. (E to G) Twelve-week-old Nox4fl/fl, Mck-Cre;Nox4fl/fl, and Mck-Cre;Nox4fl/fl;Gpx1−/− mice, fed a chow diet (4.8% fat), were subjected to an acute bout of moderate-intensity exercise for 50 min (70% VO2max), and gastrocnemius muscle was extracted and processed for (E) H2O2 measurements using Amplex Red. Twelve-week-old Nox4fl/fl, Mck-Cre;Nox4fl/fl, and Mck-Cre;Nox4fl/fl;Gpx1−/− male, fed a chow diet (4.8% fat), were subjected to endurance tests (F) and exercise stress tests (G). Representative and quantified results are shown (means ± SEM) for the indicated number of mice; significance determined using a Student’s t test (A and B) or a two-way ANOVA (A and C to G).
Previous studies have shown that in the heart, NOX4 promotes adaptive cardiac remodeling in a disease setting of chronic pressure overload (34, 38), whereas more recent studies have shown that the deletion of NOX4 in cardiomyocytes impairs heart function and exercise endurance (39). Although NOX4 deletion in Mck-Cre;Nox4fl/fl mice did not affect cardiac ROS production after an acute bout of exercise, the partial reduction in cardiac Nox4 expression prompted us to examine heart function more closely. To this end, we assessed cardiac contractile function by echocardiography in 10- to 12-week-old sedentary and anesthetized chow-fed male Nox4fl/fl versus Mck-Cre;Nox4fl/fl mice (fig. S4B), or in conscious mice before and after an exercise stress test where mice were run until exhaustion (Fig. 3D). We found no differences in left ventricular (LV) thickness, fractional shortening (FS), or heart rate in either case, consistent with cardiac function being unaltered by NOX4 deficiency in Mck-Cre;Nox4fl/fl mice (fig. S4B and Fig. 3D). The absence of a cardiac phenotype is consistent with the reduced VO2max and exercise capacity in Mck-Cre;Nox4fl/fl mice being attributed to compromised skeletal muscle function. Nonetheless, to substantiate our assertion that the decreased exercise capacity and endurance in Mck-Cre;Nox4fl/fl mice could be ascribed to the specific deletion of Nox4 in skeletal muscle, we also crossed Nox4fl/fl mice onto the human α-skeletal actin gene promoter (HSA)–MerCreMer (MCM) background that allows for the tamoxifen-inducible deletion of floxed alleles exclusively in skeletal muscle (40). Nine-week-old male Nox4fl/fl and HSA-MCM;Nox4fl/fl littermates were treated with tamoxifen for five consecutive days and analyzed at 12 weeks of age. We found that Nox4 was effectively deleted in the skeletal muscles of tamoxifen-treated HSA-MCM;Nox4fl/fl mice but was not affected in other tissues including heart (fig. S5A). Nox4 deletion in tamoxifen-treated HSA-MCM;Nox4fl/fl mice did not affect body weight, body composition (fat and lean mass), or tissue weights, including the weights of oxidative (soleus), glycolytic (TA and triceps), and mixed (gastrocnemius and) fiber skeletal muscles (fig. S5, B and D). However, as noted in Mck-Cre;Nox4fl/fl mice, the ablation of skeletal muscle Nox4 in adult HSA-MCM;Nox4fl/fl mice decreased exercise capacity and endurance (fig. S5, E and F). As the deletion of Nox4 in HSA-MCM;Nox4fl/fl mice occurred at adulthood, these findings reaffirm that the effects on exercise occur independently of any potential developmental differences. Moreover, these findings substantiate our assertion that the deletion of NOX4 in skeletal muscle compromises muscle function.
To determine whether NOX4 deletion may affect muscle function by abrogating exercise-induced ROS generation, we additionally deleted the H2O2-detoxifying enzyme GPX-1. Deletion of GPX-1 in the context of muscle NOX4 deletion (Mck-Cre;Nox4fl/fl;Gpx1−/−) corrected the otherwise reduced H2O2 levels, as assessed in gastrocnemius muscle homogenates from exercised mice using Amplex Red (Fig. 3E). The deletion of GPX-1 in Mck-Cre;Nox4fl/fl mice (Mck-Cre;Nox4fl/fl;Gpx1−/−) and the restoration of H2O2 levels in exercised muscle corrected the otherwise decreased exercise endurance and reduced exercise capacity (Umax) in Mck-Cre;Nox4fl/fl mice (Fig. 3, F and G). Therefore, these results causally link the decreased NOX4 and exercise-induced H2O2 generation in skeletal muscle with the compromised muscle function in Mck-Cre;Nox4fl/fl mice.
NOX4 in skeletal muscle is required for exercise-induced mitochondrial biogenesis
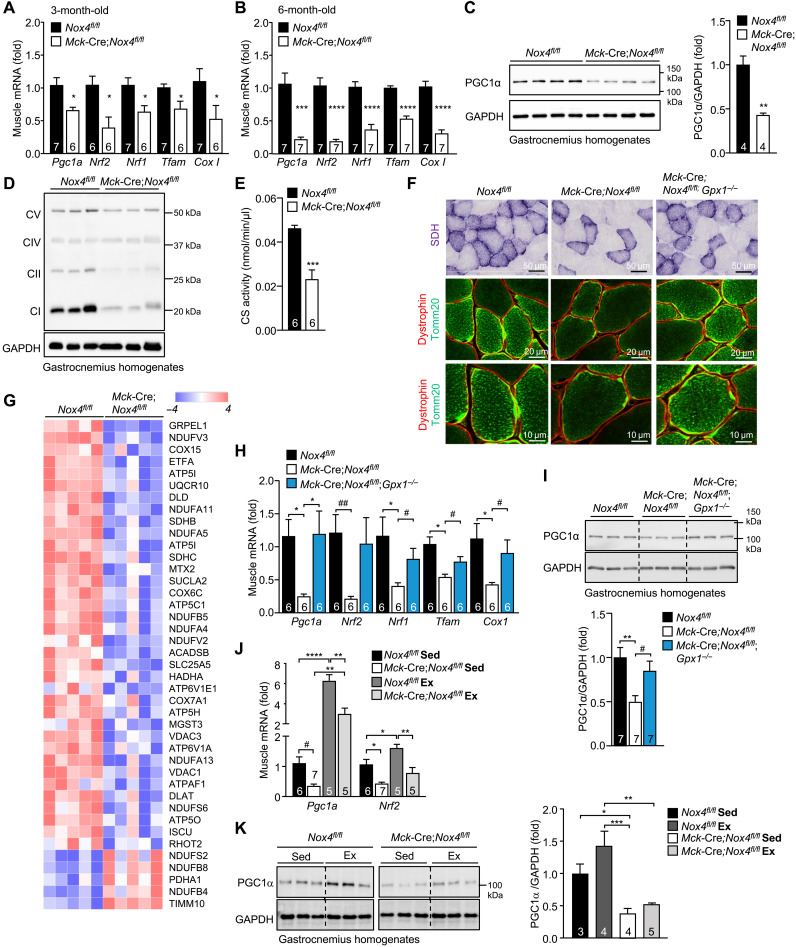
A fundamental response to exercise is the induction of mitochondrial biogenesis, which is considered essential for the promotion of respiratory capacity and endurance (2, 3). Mitochondrial biogenesis can be driven by PGC1α, which promotes the expression of NRF-1, NRF-2, and TFAM (2, 3). As exercise capacity and endurance were decreased in Mck-Cre;Nox4fl/fl mice, as well as in tamoxifen-treated HSA-MCM;Nox4fl/fl mice, we assessed the impact of muscle Nox4 deficiency on mitochondrial biogenesis. We found that mitochondrial biogenesis genes, including Pcg1a, Nrf1, Nrf2, and Tfam, were reduced in gastrocnemius muscle from 3-month-old adult Mck-Cre;Nox4fl/fl mice and reduced further by 6 months of age (Fig. 4, A and B). Similarly, expression of the Cox1 gene, encoding the respiratory complex protein cytochrome c oxidase subunit 1 (COX1), was decreased in gastrocnemius muscle from adult Mck-Cre;Nox4fl/fl mice (Fig. 4, A and B). In 6-month-old sedentary mice, the decreased gene expression was accompanied by more than 50% reduction in PGC1α protein (Fig. 4C) and a decreased abundance of mitochondrial complex proteins as assessed by immunoblotting (Fig. 4D), as well as decreased citrate synthase activity (Krebs cycle enzyme) (Fig. 4E) in gastrocnemius muscle homogenates. Moreover, although no differences were evident in oxidative and glycolytic fiber-type distribution (fig. S6A), the subsarcolemmal abundance of succinate dehydrogenase (SDH) that serves as complex II of the electron transport chain, pyruvate dehydrogenase (PDH) that carries out pyruvate decarboxylation in the mitochondrial matrix, and Tomm20, a mitochondrial import receptor protein, were reduced in muscles from 6-month-old Mck-Cre;Nox4fl/fl male mice (fig. S6B), consistent with decreased mitochondrial content. In addition, an unbiased proteomic analysis of gastrocnemius muscles from 6-month-old Nox4fl/fl versus Mck-Cre;Nox4fl/fl mice revealed that NOX4 deficiency was associated with an overt and significant decrease in mitochondrial proteins, including proteins involved in electron transfer such as Etfa, mitochondrial import proteins such as Grpel1 and Mtx2, and components of respiratory complexes I (Ndufv3, Ndufa11, Ndufa5, Ndufb5, Ndufa4, and Ndufv2), II (Sdhb and Sdhc,), III (Uqcr10), IV (Cox15, Cox6c, and Cox7a1), and V (Atp5i, Atp1b1, Atp5l, and Atp5c1) (Fig. 4G and fig. S6, C and D). The decreased SDH and Tomm20 in muscle from 6-month-old Mck-Cre;Nox4fl/fl mice (Fig. 4F) and the decreased expression of mitochondrial biogenesis genes and PGC1α protein were largely corrected in mice that were additionally deficient for GPX-1 (Mck-Cre;Nox4fl/fl;Gpx1−/−; Fig. 4, H and I), where the otherwise decreased H2O2 levels were corrected (Fig. 3E). Consistent with the importance of H2O2, we found that GPX-1 deficiency on its own was sufficient to promote the expression of some of the mitochondrial biogenesis genes, such as Pgc1a, and enhance exercise endurance when compared to Gpx1+/+ mice (fig. S7, A and C). Therefore, these findings demonstrate that NOX4 deficiency and decreased H2O2 result in decreased mitochondrial content in skeletal muscle. NOX4 deficiency not only decreased mitochondrial content in sedentary mice, but also attenuated the otherwise robust induction in mitochondrial biogenesis (as assessed by Pgc1a gene and PGC1α protein levels) that occurred after an acute bout of exercise (70% VO2max, 50 min) (Fig. 4, J and K). Therefore, NOX4-derived ROS are fundamentally important for the induction of mitochondrial biogenesis and the adaptive increases in respiratory capacity accompanying exercise.
Fig. 4. NOX4-derived H2O2 in skeletal muscle is required for mitochondrial biogenesis.
(A to B) Gastrocnemius muscles from 3- or 6-month-old Nox4fl/fl and Mck-Cre;Nox4fl/fl chow-fed male mice were processed for qPCR monitoring for the expression of mitochondrial biogenesis genes. (C to F) Gastrocnemius muscle from 6-month-old Nox4fl/fl and Mck-Cre;Nox4fl/fl chow-fed male mice was homogenized and immunoblotted for (C) PGC1α and (D) total OXPHOS proteins [complexes (C) I to V] or processed for the analysis of (E) citrate synthase (CS) activity. (F) Gastrocnemius muscles from 6-month-old male Nox4fl/fl, Mck-Cre;Nox4fl/fl, and Mck-Cre;Nox4fl/fl;Gpx1−/− chow-fed (4.8% fat) mice were frozen, and transverse sections were processed for SDH staining and Tomm20 along with dystrophin immunostaining to define mitochondria within individual muscle fibers. (G) Gastrocnemius muscle from 6-month-old male Nox4fl/fl and Mck-Cre;Nox4fl/fl chow-fed (4.8% fat) mice was homogenized. Proteins were digested with trypsin and analyzed on a QExactive HF mass spectrometer. KEGG and differentially regulated proteins were identified considering a P value and log2 fold-change cutoff of ≤0.05 and >|0.3|, respectively. A heatmap of selected differentially expressed proteins associated with OXPHOS is shown. (H and I) Gastrocnemius muscles from 6-month-old male Nox4fl/fl, Mck-Cre;Nox4fl/fl, and Mck-Cre;Nox4fl/fl;Gpx1−/− chow-fed (4.8% fat) mice were processed for (I) qPCR or (J) immunoblotting. (J and K) Twelve-week-old Nox4fl/fl and Mck-Cre;Nox4fl/fl chow-fed male mice were subjected to an acute bout of exercise on multilane treadmill for 50 min at moderate intensity (70% VO2max). After 4 hours, gastrocnemius muscles were excised from sedentary (Sed) and exercised (Ex) mice and processed for (J) qPCR or (K) immunoblotting. Representative and quantified results are shown (means ± SEM) for the indicated number of mice. Significance determined using (A to C and E) a Student’s t test, (H and I) a one-way ANOVA, or (J and K) a two-way ANOVA; # indicates significance using a Student’s t test.
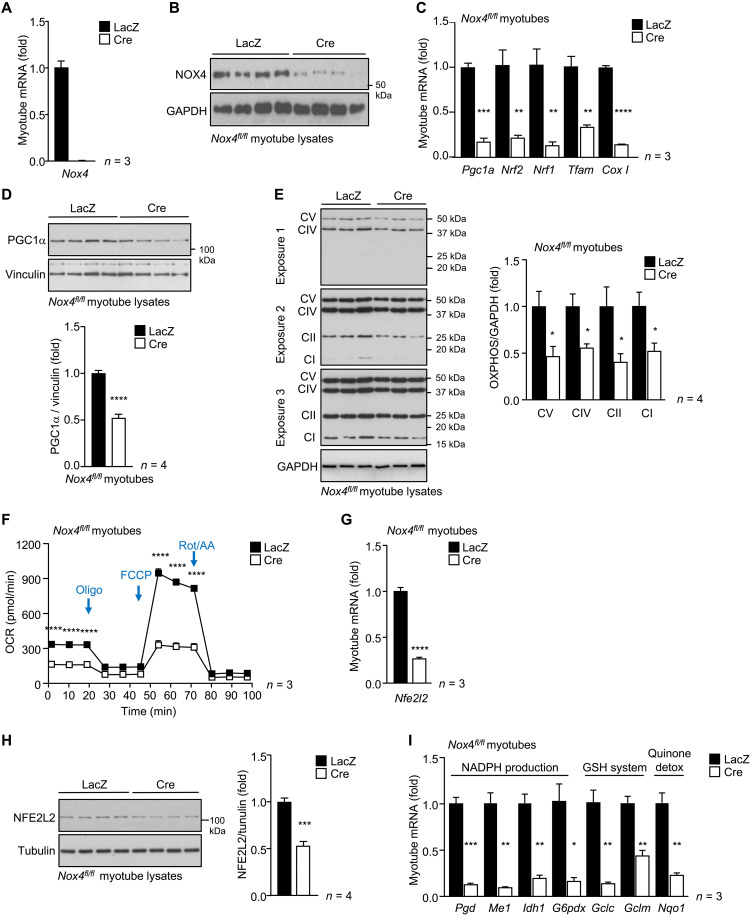
To determine whether the effects of NOX4 deficiency on mitochondrial biogenesis may be cell autonomous, we also assessed mitochondrial biogenesis in the myotubes generated from Nox4fl/fl myoblasts (fig. S3) transduced with control (Adeno-lacZ)– or Cre (Adeno-Cre)–expressing adenoviruses to delete Nox4 (Fig. 5, A and B). Nox4 deletion markedly repressed the expression of Cox1 and mitochondrial biogenesis genes (Pgc1a, Nrf1, Nrf2, and Tfam) (Fig. 5C and fig. S8A), decreased total PGC1α and oxidative phosphorylation (OXPHOS) protein complexes as assessed by immunoblotting (Fig. 5, D and E), and repressed mitochondrial function as assessed by measuring oxygen consumption rates (OCRs) (basal respiration and maximal respiratory capacity) using a Seahorse assay in myoblasts and myotubes (Fig. 5F and fig. S8B). Therefore, these results demonstrate that the effects of NOX4 deficiency on mitochondrial biogenesis are cell autonomous and independent of extrinsic factors such as neurological or endocrine inputs.
Fig. 5. NOX4 in myotubes is essential for mitochondrial biogenesis and NFE2L2-mediated antioxidant defense.
(A to I) Skeletal muscle myoblasts were FACS-purified from Nox4fl/fl mice and transduced with β-galactosidase (LacZ) control or Cre recombinase–expressing adenoviruses to delete Nox4. The resultant cells were differentiated into myotubes and processed for (A) qPCR or (B) immunoblotting to assess NOX4 levels, (C) qPCR to monitor mitochondrial biogenesis gene expression, (D) immunoblotting to monitor for PGC1α, or (E) OXPHOS protein levels [complexes (C) I to V; protein levels were quantified by densitometry]. (F) Mitochondrial respiration was assessed in live myotubes by performing the Seahorse XF Cell Mito Stress Test and measuring the oxygen consumption rate (OCR); basal respiration and maximal respiration, after inhibiting ATP synthase with oligomycin and uncoupling respiration with FCCP, were assessed. Myotubes were processed for (G) qPCR and (H) immunoblotting to assess NFE2L2 levels or (I) qPCR to assess antioxidant defense gene expression. Representative and quantified results are shown (means ± SEM) for the indicated number of experiments; significance determined using (A, C, D, and F to I) a Student’s t test or (E) two-way ANOVA.
Skeletal muscle NOX4 is essential for NFE2L2-mediated antioxidant defense and mitochondrial biogenesis
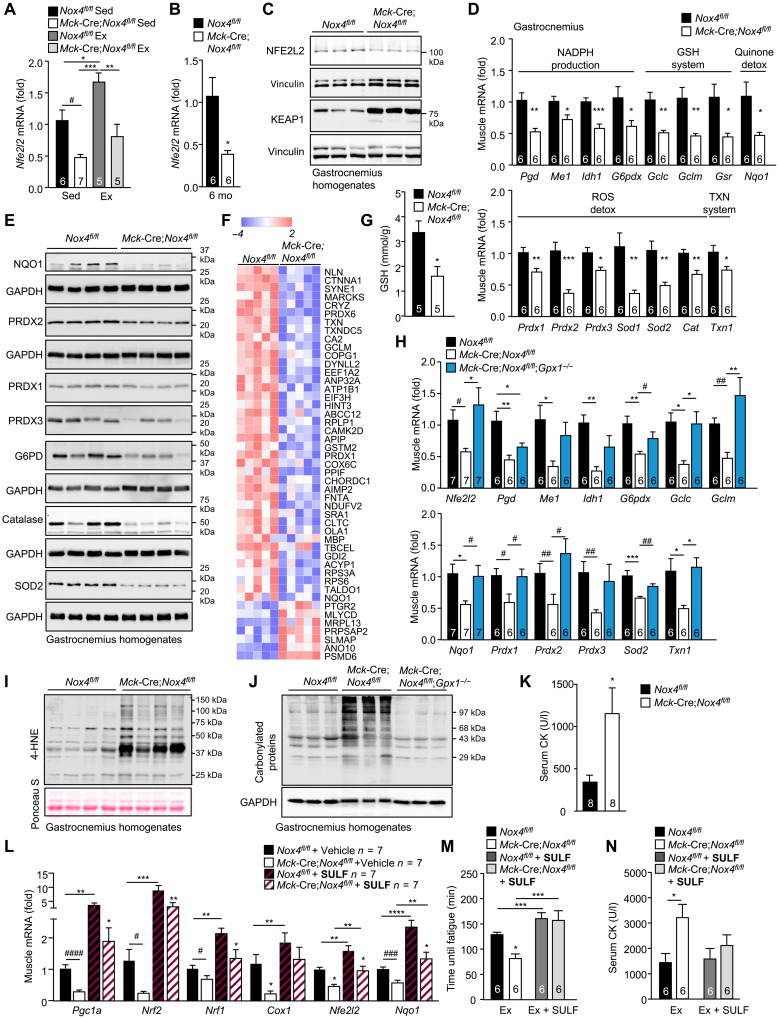
It is well established that contracting muscle generates ROS and that this is accompanied by increased antioxidant defense orchestrated by NFE2L2 (2, 7, 36). Given that NOX4 was required for basal and exercise-induced ROS, we assessed the impact of NOX4 deficiency on antioxidant defense. Exercise induces Nfe2l2 gene expression (7), whereas ROS generated during exercise would be expected to increase NFE2L2 stability (4, 30). NOX4 deficiency markedly reduced the skeletal muscle expression of Nfe2l2 in sedentary mice and abrogated the induction of Nfe2l2 after an acute bout of exercise (70% VO2max, 50 min; Fig. 6A). Moreover, we found that Nfe2l2 mRNA and NFE2L2 protein were reduced (Fig. 6, B and C, and fig. S9A) and KEAP1 protein was increased (Fig. 6C and fig. S9B) in gastrocnemius muscle homogenates from 6-month-old Mck-Cre;Nox4fl/fl mice. Consistent with this, we found a significant reduction in the expression of NFE2L2 target genes in muscle homogenates from 6-month-old Mck-Cre;Nox4fl/fl mice (Fig. 6D). Similarly, we noted that the deletion of Nox4 in myoblasts and/or myotubes was accompanied by marked reductions in Nfe2l2 gene and protein expression, as well as reduced expression of NFE2L2 target genes (Fig. 5, G to I, and fig. S8, C and D), consistent with the effects being cell intrinsic. The reduced NFE2L2 target genes included those encoding enzymes involved in (i) NADPH production, necessary for the reduction of GSSG to GSH, including phosphoglycerate dehydrogenase (Phgdh), malic enzyme 1 (Me1), isocitrate dehydrogenase 1 (Idh1), and glucose-6-phosphate dehydrogenase (G6PD; G6pd), a key enzyme in NADPH production; (ii) GSH production and regeneration, including glutamate-cysteine ligase (GCL) catalytic subunit (Gclc), GCL complex modifier subunit (Gclm), and glutathione reductase (Gsr); (iii) thioredoxin reduction, including thioredoxin reductase (Txnrd1); (iv) quinone detoxification, including NAD(P)H dehydrogenase (quinone 1) (NQO1; Nqo1); and (iv) ROS detoxification, including SOD1 (Sod1) and SOD2 (Sod2) that dismutate O2•− into H2O2, and PRDX1 (Prdx1) and catalase that eliminate H2O2 (Fig. 6D). In addition, the reduced NFE2L2 abundance in muscle from 6-month-old Mck-Cre;Nox4fl/fl mice was accompanied by decreased G6PD protein and decreased NQO1, which is exclusively regulated by NFE2L2 in muscle (41), and a decreased abundance in proteins involved in antioxidant defense, including mitochondrial SOD2, PRDX1 to PRDX3, and catalase (Fig. 6E and fig. S9C). Moreover, our unbiased proteomic analysis of gastrocnemius muscles from 6-month-old Nox4fl/fl versus Mck-Cre;Nox4fl/fl mice (fig. S6, C and D) further revealed that NOX4 deficiency was accompanied by an overt reduction in the abundance of NFE2L2 transcriptional targets, including those involved in antioxidant defense, such as GCLM, PRDX6, PRDX1, and NQO1 (Fig. 6F), and ROS metabolism (fig. S9D). The reductions in skeletal muscle NFE2L2 and antioxidant defense proteins in sedentary 6-month-old Mck-Cre;Nox4fl/fl mice were accompanied by reductions in reduced GSH (Fig. 6G). We found that the decreased Nfe2l2 and antioxidant gene expression in Mck-Cre;Nox4fl/fl mice could be largely rescued by superimposing GPX-1 deficiency onto the muscle NOX4 deficiency (Mck-Cre;Nox4fl/fl;Gpx1−/−), thereby causally linking the defective antioxidant defense response with the decreased ROS generation (Fig. 6H).
Fig. 6. NOX4-derived H2O2 in skeletal muscle is essential for NFE2L2-mediated antioxidant defense and mitochondrial biogenesis.
(A) Twelve-week-old Nox4fl/fl and Mck-Cre;Nox4fl/fl chow-fed male mice were subjected to an acute bout of exercise (50 min, 70% VO2max), and gastrocnemius muscles were processed for qPCR. (B to G) Gastrocnemius muscle from 6-month-old Nox4fl/fl and Mck-Cre;Nox4fl/fl chow-fed male mice processed for (B and D) qPCR, (C and E) immunoblotting, (F) proteomics, monitoring for differentially expressed transcriptional targets of NFE2L2, or (G) an analysis of GSH levels. (H to J) Gastrocnemius muscle from 6-month-old Nox4fl/fl, Mck-Cre;Nox4fl/fl, or Mck-Cre;Nox4fl/fl;Gpx1−/− male mice, fed a chow diet, was processed for (H) qPCR, or immunoblotting monitoring for (I) 4-HNE levels, or (J) protein carbonylation. (K) Serum CK levels in 6-month-old Nox4fl/fl and Mck-Cre;Nox4fl/fl chow-fed male mice. (L) Twelve-week-old Nox4fl/fl and Mck-Cre;Nox4fl/fl chow-fed male mice were administered vehicle [dimethyl sulfoxide (DMSO)] or sulforaphane (SULF; 0.5 mg/kg per day, intraperitoneally) for 5 days, and gastrocnemius muscle was extracted and processed for qPCR. (M and N) Twelve-week-old Nox4fl/fl and Mck-Cre;Nox4fl/fl chow-fed male mice were subjected to endurance tests before and after 5 days of administering sulforaphane (0.5 mg/kg per day, intraperitoneally), and (M) time until fatigue was determined. (N) Serum was collected 1 hour after exercise for analysis of CK levels. Representative and quantified results are shown (means ± SEM) for the indicated number of mice; significance determined using (A and L to N) two-way ANOVA, (B to E, G, and K) a Student’s t test, or (H) one-way ANOVA. # indicates significance for Nox4fl/fl versus Mck-Cre;Nox4fl/fl or Mck-Cre;Nox4fl/fl versus Mck-Cre;Nox4fl/fl;Gpx1−/− using a Student’s t test.
A potential outcome of defective skeletal muscle antioxidant defense in NOX4-deficient muscle might be an increased potential for oxidative damage (16, 19, 32, 42). Consistent with this, oxidative muscle damage, as assessed by immunoblotting gastrocnemius muscle homogenates for 4-hydroxynonenal (4-HNE), a marker of lipid peroxidation, and for protein carbonylation, a marker of protein oxidation and damage (18), was increased in 6-month-old Mck-Cre;Nox4fl/fl mice (Fig. 6, I and J); the increase in lipid peroxidation approximated that seen in high fat–fed obese mice (fig. S9E) that are characterized by systemic oxidative stress (13, 17). Moreover, although well below that seen in mice with overt muscle pathology, serum muscle protein creatine kinase (CK), a marker of muscle damage, was increased in 6-month-old Mck-Cre;Nox4fl/fl mice (Fig. 6K). The increased oxidative damage as assessed by protein carbonylation in NOX4-deficient mice was corrected when GPX-1 deficiency was superimposed onto muscle NOX4 deficiency (Mck-Cre;Nox4fl/fl;Gpx1−/−) (Fig. 6J). Therefore, these results indicate that the defective H2O2-induced antioxidant defense accompanying the deletion of NOX4 in muscle results in the oxidative damage of proteins and lipids.
NFE2L2 is thought to be required for the induction of mitochondrial biogenesis after exercise and for optimal exercise performance (3, 7). Therefore, we reasoned that the defective skeletal muscle NFE2L2-mediated antioxidant defense in Mck-Cre;Nox4fl/fl mice may also compromise muscle function by decreasing exercise-induced mitochondrial biogenesis and thereby respiratory capacity and exercise performance. To explore the extent to which defective NFE2L2 activity may be responsible for the defective mitochondrial biogenesis and exercise capacity, we treated mice with the NFE2L2 agonist sulforaphane, because previous studies have shown that treatment of rodents with sulforaphane can increase endurance capacity via the NFE2L2-ARE pathway (32, 33). To assess the impact of NFE2L2 on mitochondrial biogenesis, we treated 12-week-old Nox4fl/fl and Mck-Cre;Nox4fl/fl mice for five consecutive days with sulforaphane and then extracted tissues for an analysis of gene expression. Although sulforaphane treatment had greater effects in Nox4fl/fl mice, it nonetheless largely, if not completely, corrected the otherwise reduced skeletal muscle Nfe2l2 and Nqo1 gene expression in Mck-Cre;Nox4fl/fl mice so that they were indistinguishable from untreated controls and similarly rescued the reduced expression of mitochondrial biogenesis genes (Pgc1a, Nrf1, and Nrf2) (Fig. 6L). Thus, these results suggest that decreased mitochondrial biogenesis in Mck-Cre;Nox4fl/fl muscle might be ascribed to the defective NFE2L2 response. To determine whether the defective NFE2L2-induced mitochondrial biogenesis might be responsible for the compromised muscle function, we assessed exercise performance in 12-week-old Nox4fl/fl and Mck-Cre;Nox4fl/fl mice before and after 5 days of treatment with sulforaphane. To this end, we subjected mice to endurance tests before and after sulforaphane treatment. Notably, we found that treatment with sulforaphane rescued the otherwise decreased exercise endurance in Mck-Cre;Nox4fl/fl mice (Fig. 6M). Moreover, although muscle damage as assessed by measuring serum CK was significantly increased in Mck-Cre;Nox4fl/fl mice after the endurance tests, this was prevented by sulforaphane treatment (Fig. 6N). Together these results are consistent with NOX4-derived ROS being integral in instigating the NFE2L2 response to prevent oxidative damage, increase respiratory capacity, and ensure optimal muscle performance.
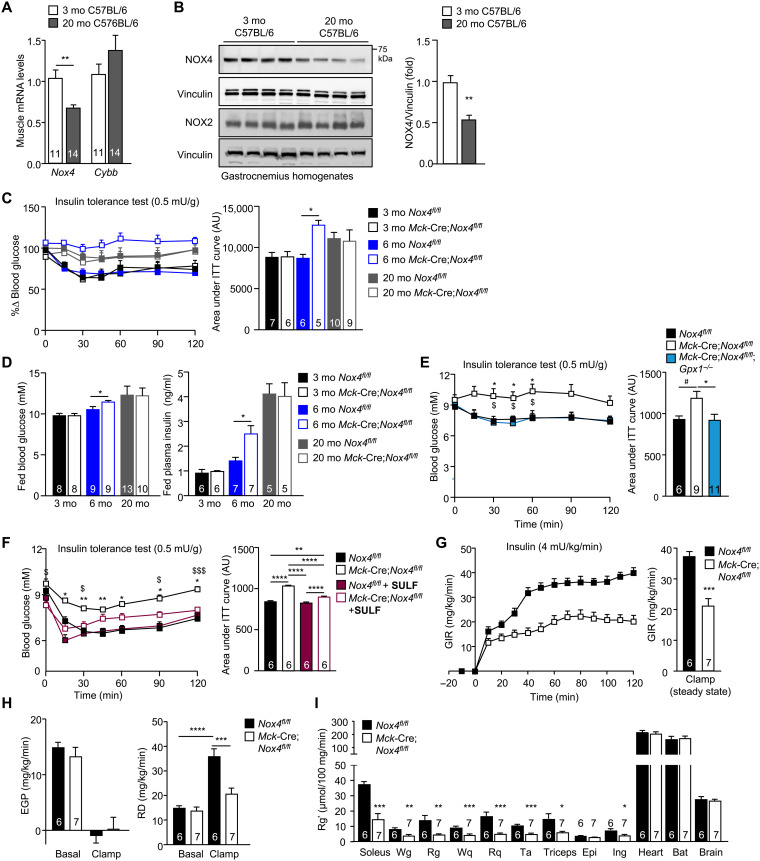
NOX4 deletion in muscle exacerbates the age-associated decline in insulin sensitivity
A sedentary lifestyle accompanying aging, and aging itself, can promote the development of insulin resistance and type 2 diabetes independent of any changes in body weight (1). Our studies show that NOX4 expression is induced after exercise and that NOX4 prevents oxidative damage. Given the well-established potential for oxidative stress and consequent oxidative damage to drive insulin resistance (11–18), we reasoned that the development of insulin resistance in the sedentary aged mice may be attributed to decreased NOX4 abundance or activity. To test this, we first assessed skeletal muscle NOX4 levels in young and old mice. We found that skeletal muscle (gastrocnemius) NOX4 expression, as assessed by real-time PCR (Fig. 7A) or immunoblotting (Fig. 7B), was reduced by as much as 46% in 20-month-old male mice when compared to 3-month-old male mice. By contrast, NOX2 expression was not significantly affected (Fig. 7, A and B). Next, we assessed whether the deletion of NOX4 and the accumulation of skeletal muscle oxidative damage in chow-fed mice might accelerate the onset of insulin resistance. To explore this, we assessed the impact of muscle NOX4 deficiency on insulin sensitivity in 3-, 6-, and 20-month-old Nox4fl/fl and Mck-Cre;Nox4fl/fl male mice fed a standard chow diet (Fig. 7, C and D). As noted in 3-month-old mice (Fig. 1, K and L, and fig. S2A), body weights, body composition/adiposity, and tissue weights were not altered by NOX4 deficiency in 6- or 20-month-old Mck-Cre;Nox4fl/fl male mice (although soleus weights were modestly decreased as in younger mice) (fig. S10, A to C). Although oxygen consumption, energy expenditure, and voluntary wheel running were decreased across all age groups (RERs were unaltered; fig. S11, A to D and F to I), this was accompanied by decreased food intake (fig. S11, E and J). Moreover, as in younger mice (fig. S2, B to P), NOX4 deficiency had no gross effect on muscle histomorphometry in older mice (fig. S6A).
Fig. 7. NOX4-derived H2O2 in skeletal muscle is essential for attenuating the age-associated development of insulin resistance.
(A) Gastrocnemius muscle Nox4 and Cybb (encoding NOX2) expression in 3- versus 20-month-old chow-fed C57BL/6 male mice assessed by qPCR. (B) Gastrocnemius muscle NOX4 and NOX2 proteins in 3- versus 20-month-old chow-fed C57BL/6 male mice were assessed by immunoblotting. (C) Insulin tolerance tests (ITTs; 0.5 mU/g) or (D) fed (satiated) blood glucose and plasma insulin levels in 3-, 6-, and 20-month-old Nox4fl/fl and Mck-Cre;Nox4fl/fl chow-fed male mice; areas under ITT curves were determined, and arbitrary units (AU) are shown. (E) ITTs in 6-month-old Nox4fl/fl, Mck-Cre;Nox4fl/fl, and Mck-Cre;Nox4fl/fl;Gpx1−/− chow-fed (4.8% fat) male mice. (F) Six-month-old male Nox4fl/fl and Mck-Cre;Nox4fl/fl chow-fed male mice were administered vehicle (DMSO) or sulforaphane (SULF; 0.5 mg/kg per day, intraperitoneally) for 5 days and subjected to ITTs. (G to I) Six-month-old male Nox4fl/fl and Mck-Cre;Nox4fl/fl chow-fed male mice were fasted for 6 hours, and mice were subjected to hyperinsulinemic-euglycemic clamps; [14C]2-deoxy-d-glucose was administered at the end of the clamp. (G) The GIR, (H) EGP and RD were assessed under basal and clamped conditions. (I) Tissues were extracted, and [14C]2-deoxy-d-glucose uptake (Rg′) was assessed. Representative and quantified results are shown (means ± SEM) for the indicated number of mice; significance determined using (E, F, and H) two-way ANOVA, (E) one-way ANOVA, or (A, C, D, G, and I) a Student’s t test. In (E), * indicates significance for Nox4fl/fl versus Mck-Cre;Nox4fl/fl mice, and $ indicates significance for Nox4fl/fl versus Mck-Cre;Nox4fl/fl;Gpx1−/− mice. In (F), * indicates significance for Nox4fl/fl versus Mck-Cre;Nox4fl/fl mice, and $ indicates significance for Mck-Cre;Nox4fl/fl versus Mck-Cre;Nox4fl/fl + SULF mice.
To monitor for effects on insulin sensitivity, we subjected mice to insulin tolerance tests (ITTs) and measured fed (satiated) blood glucose and plasma insulin levels. The deletion of NOX4 had no effect on insulin sensitivity, as assessed in ITTs (0.5 mU/g insulin) and by measuring blood glucose and plasma insulin levels, in 3-month-old mice when overt differences were otherwise evident in exercise performance (Fig. 7, C and D). Thus, the diminished respiratory capacity and exercise performance associated with muscle NOX4 deficiency per se are not sufficient to affect insulin sensitivity. By contrast, in 6-month-old mice, NOX4 deficiency promoted insulin resistance, as reflected by the diminished response in ITTs (0.5 mU/g insulin) and the heightened fed blood glucose and plasma insulin levels (Fig. 7, C and D). ITT responses in 6-month-old Mck-Cre;Nox4fl/fl mice resembled those in 20-month-old Nox4fl/fl mice that were largely unresponsive to 0.5 mU/g of insulin, consistent with NOX4 deficiency accelerating the age-associated decline in insulin sensitivity; NOX4 deficiency had no further effect on ITTs and blood glucose levels in 20-month-old mice that are already markedly insulin resistant (Fig. 7, C and D). The decreased insulin sensitivity in 6-month-old Mck-Cre;Nox4fl/fl mice was not accompanied by increases in body weight or adiposity (fig. S10, A to C), which are known to promote insulin resistance, or reductions in key insulin signaling intermediates, including the insulin receptor β subunit (IRβ), IR substrate-1 (IRS-1), p85 subunit of phosphatidylinositol 3-kinase (PI3K), and protein kinase AKT (fig. S10D). The development of insulin resistance in NOX4-deficient mice was also evident in 6-month-old HSA-MCM;Nox4fl/fl male mice, in which NOX4 was inducibly deleted at 10 weeks of age (fig. S5G). The effects of NOX4 deficiency on insulin sensitivity in 6-month-old mice could be reversed by superimposing GPX1 deficiency onto the Mck-Cre;Nox4fl/fl background (Mck-Cre;Nox4fl/fl;Gpx1−/−; Fig. 7E) to correct the otherwise decreased H2O2 in NOX4-deficient muscle (Fig. 3E). Moreover, the effects of NOX4 deficiency on insulin sensitivity in 6-month-old mice could be at least partially reversed by treating mice for 5 days with sulforaphane (Fig. 7F). Therefore, these results are consistent with the age/physical inactivity–associated decline in NOX4 abrogating the NFE2L2 antioxidant defense response to promote oxidative stress/damage and ensuing insulin resistance.
To explore further the impact of NOX4 deficiency on glucose homeostasis, we subjected 6-month-old Nox4fl/fl versus Mck-Cre;Nox4fl/fl chow-fed male mice that were conscious and free-moving to hyperinsulinemic-euglycemic clamps, a gold-standard measure of insulin sensitivity and glucose homeostasis. We found that the glucose infusion rate (GIR) necessary to maintain euglycemia during the insulin clamp was markedly reduced (Fig. 7G and fig. S10, E and F), consistent with the development of insulin resistance. Although endogenous glucose production (EGP) was similarly repressed in Nox4fl/fl and Mck-Cre;Nox4fl/fl mice, consistent with unaltered hepatic glucose production and hepatic insulin sensitivity (Fig. 7H), the rate of glucose disappearance (RD), a measure primarily of muscle and adipose tissue glucose uptake, was significantly repressed (Fig. 7H). To determine the extent to which the diminished RD may reflect decreased glucose uptake (Rg′) into muscle, we administered a bolus of [14C]2-deoxy-d-glucose at the end of hyperinsulinemic-euglycemic clamps and assessed uptake in varied tissues. NOX4 deficiency significantly repressed glucose uptake in both oxidative and glycolytic muscles [soleus, white gastrocnemius (Wg), red gastrocnemius (Rg), red quadriceps (Rq), white quadriceps (Wq), TA, and triceps], consistent with the development of marked skeletal muscle insulin resistance (Fig. 7I). NOX4 deficiency in 6-month-old chow-fed mice also repressed glucose uptake in subcutaneous WAT, consistent with signs of systemic insulin resistance, but had no effect on glucose uptake in epididymal WAT (a visceral fat depot) or interscapular BAT and similarly had no effect on uptake in brain and heart, where glucose uptake is insulin independent (Fig. 7I). Therefore, these results are consistent with decreased NOX4-derived ROS in Mck-Cre;Nox4fl/fl muscle exacerbating the age-associated decline in insulin sensitivity and promoting the development of insulin resistance as a result of diminished insulin-induced muscle glucose uptake.
NOX4 deletion in muscle exacerbates insulin resistance in obesity
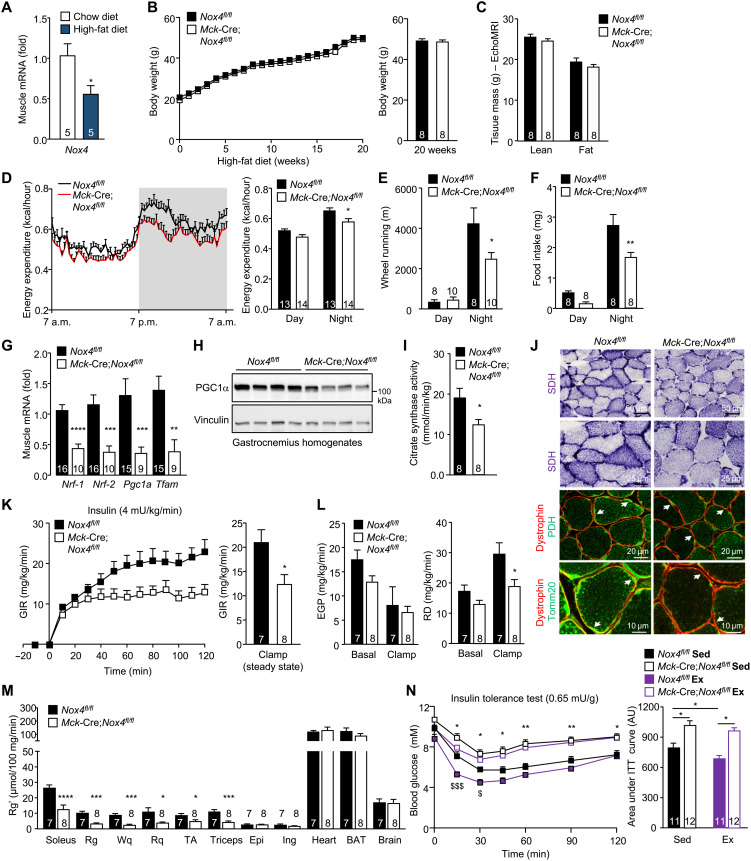
Obesity is associated with systemic oxidative stress (43, 44), which promotes insulin resistance (11–18). As in aging, we found that diet-induced obesity was accompanied by a 44% reduction in skeletal muscle Nox4 expression (Fig. 8A). Accordingly, we sought to determine whether NOX4 deficiency might similarly exacerbate the development of insulin resistance in diet-induced obesity. Eight-week-old Nox4fl/fl and Mck-Cre;Nox4fl/fl male mice were fed a high-fat diet (23% fat; 45% energy from fat) for 20 weeks, and effects on body weight, body composition, muscle development, and glucose homeostasis were assessed. NOX4 deficiency had no effect on body weight (Fig. 8B) or body composition (Fig. 8C and fig. S12A) and, apart from soleus, which was slightly smaller, had no effect on tissue weights (fig. S12B). As in chow-fed mice, NOX4 deficiency was associated with decreased energy expenditure and voluntary wheel running, as well as decreased food intake (Fig. 8, D to F); energy expenditure was also decreased when running wheels were omitted from the metabolic cages. NOX4 deficiency had no gross effect on muscle histomorphometry (fig. S12, C to I) but decreased mitochondrial biogenesis and content (Fig. 8, G to J, and fig. S13A), as also seen in chow-fed mice. In addition, NOX4 deficiency was accompanied by decreased NFE2L2 and the decreased expression of Nfe2l2 target genes (fig. S13, B and C), as seen in the skeletal muscle of chow-fed mice. NOX4 deficiency was accompanied by overt hyperglycemia and hyperinsulinemia in the fed state (fig. S13D). Moreover, NOX4 deficiency exacerbated the development of insulin resistance, as reflected by the diminished repression of fasted blood glucose in ITTs (0.5 mU/g), and promoted glucose intolerance, as reflected by the increased glucose excursions in glucose tolerance tests (GTTs; 2 mg/g) (fig. S13, E and F). Consistent with this, hyperinsulinemic-euglycemic clamps revealed that NOX4 deficiency was associated with a marked reduction in GIR and RD and decreased skeletal muscle glucose uptake (Fig. 8, K to M, and fig. S13, G and H). The diminished insulin-induced muscle glucose uptake in high fat–fed Mck-Cre;Nox4fl/fl mice was associated with a reduction in insulin-induced PI3K/AKT signaling as assessed by immunoblotting muscle homogenates from 12-week high fat–fed mice for the phosphorylation of AKT on Ser473 after administering a bolus of insulin (fig. S13I). Together, our findings demonstrate that the decreased NOX4-derived ROS in muscle not only accelerate the age-associated decline in insulin sensitivity, but also exacerbate the development of insulin resistance and the ensuing hyperinsulinemia and hyperglycemia in diet-induced obesity independent of body weight.
Fig. 8. Nox4 deletion in muscle exacerbates insulin resistance in obesity and abrogates the beneficial effects of exercise on insulin sensitivity.
(A) Gastrocnemius muscle Nox4 expression in 8-week-old C57BL/6 male mice fed a chow diet or a high-fat diet (23% fat) for 20 weeks was assessed by qPCR. (B to N) Eight-week-old Nox4fl/fl and Mck-Cre;Nox4fl/fl male mice were fed a high-fat diet (23% fat) for 20 weeks, and (B) body weight, (C) body composition (EchoMRI), (D) energy expenditure, (E) voluntary wheel running, and (F) diurnal food intake were assessed. (G to I) Gastrocnemius muscle was extracted and processed for (G) qPCR, (H) immunoblotting, or (I) assessment of citrate synthase activity. (J) Transverse sections of soleus muscle were processed for SDH staining and either PDH or Tomm20 immunostaining along with dystrophin immunostaining to define mitochondria within individual muscle fibers. (K to M) Alternatively, after a 6-hour fast, mice were subjected to hyperinsulinemic-euglycemic clamps with [14C]2-deoxy-d-glucose being administered at the end. (K) GIR, (L) EGP, RD, and (M) [14C]2-deoxy-d-glucose uptake (Rg′) were determined. (N) Eight-week-old Nox4fl/fl and Mck-Cre;Nox4fl/fl male mice were fed a high-fat diet (23% fat) for 12 weeks, fasted for 4 hours, and subjected to an acute exercise bout (60% VO2max) for 30 min, and after 1 hour, sedentary (Sed) and exercised (Ex) mice were subjected to ITTs (0.65 mU/g, intraperitoneally); areas under ITT curves were determined and AUs are shown. Representative and quantified results are shown (means ± SEM) for the indicated number of mice; significance determined using (D to F, L, and N) two-way ANOVA or (G to I, K, and M) a Student’s t test. In (N), * indicates significance for Nox4fl/fl Sed versus Mck-Cre;Nox4fl/fl Sed mice, and $ indicates significance for Nox4fl/fl Sed versus Mck-Cre;Nox4fl/fl Ex mice.
NOX4 deletion in muscle attenuates the beneficial effects of exercise on insulin sensitivity
Exercise not only promotes contraction-induced and insulin-independent glucose uptake into muscle, but also increases muscle insulin sensitivity that persists long after the cessation of exercise (1). Accordingly, we explored whether the deletion of NOX4 in skeletal muscle and the defective antioxidant defense response might abrogate the beneficial effects of exercise on insulin sensitivity. Specifically, we tested whether NOX4 was required for the ability of exercise to enhance insulin sensitivity and ameliorate insulin resistance in otherwise high fat–fed and insulin-resistant mice. Eight-week-old Nox4fl/fl and Mck-Cre;Nox4fl/fl male mice were fed a high-fat diet (23% fat) for 12 weeks to promote insulin resistance and either left untreated or subjected to an acute bout of exercise at the same relative intensity (60% VO2max for 30 min). We then assessed insulin sensitivity by performing ITTs. As noted already, NOX4-deficient mice exhibited marked insulin resistance, as reflected by the comparatively diminished ability of insulin to lower fasted blood glucose levels (Fig. 8N). Notably, we found that although a single bout of exercise was sufficient to enhance insulin sensitivity in high fat–fed Nox4fl/fl mice, exercise had no effect on insulin sensitivity in high fat–fed Mck-Cre;Nox4fl/fl mice (Fig. 8N). Therefore, the deletion of NOX4 in muscle not only exacerbates the development of insulin resistance with aging and obesity, but also diminishes the beneficial effects of acute exercise on insulin action.
NOX4 deletion in muscle promotes mitochondrial oxidative stress, attenuates insulin signaling, and promotes insulin resistance
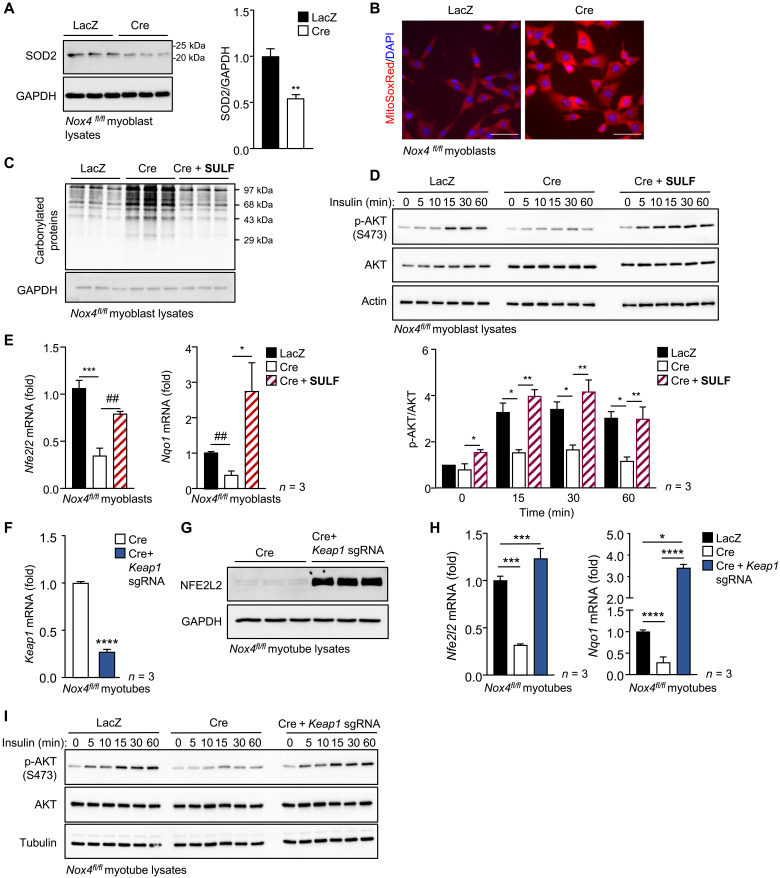
Our studies indicate that the deletion of NOX4 in skeletal muscle decreases antioxidant defense, results in the oxidative damage of proteins and lipids, and promotes the development of insulin resistance. The defective antioxidant defense included overt reductions in mitochondrial SOD2 protein and reductions in the abundance of several H2O2-detoxifying enzymes, including catalase, PRDX1, PRDX2, and mitochondrial PRDX3. It is well established that heightened mitochondrial ROS production attenuates insulin signaling and promotes insulin resistance (12–14, 45). The heterozygous deletion of mitochondrial SOD2 in mice on a chow diet is sufficient to promote insulin resistance, whereas its overexpression or the expression of a mitochondrial-targeted catalase can attenuate insulin resistance in high fat–fed mice (12–4). Therefore, we reasoned that the abrogated NOX4- and H2O2-mediated antioxidant defense, in particular, reductions in SOD2, would promote mitochondrial oxidative stress to diminish insulin signaling and promote insulin resistance. To explore this, we assessed mitochondrial ROS, protein oxidation, and insulin signaling in Nox4fl/fl muscle cells transduced with Adeno-lacZ as a control (LacZ), or Adeno-Cre (Cre) to delete Nox4. As indicated already, Nox4 was efficiently deleted in myoblasts, and this did not overtly influence their differentiation into myotubes (fig. S3) or the expression of key insulin molecules including IRβ, IR, IRS-1, p85 PI3K, and AKT (fig. S14A). However, as described earlier, NOX4 deletion was accompanied by reduced H2O2 production in myotubes (Fig. 2F) and decreased antioxidant defense, as reflected by the decreased expression of antioxidant defense genes, including Nfe2l2 and Nqo1 (Fig. 5, H and I, and fig. S8, C and D), and decreased SOD2 protein (Fig. 9A). The decreased antioxidant defense was accompanied by increased mitochondrial O2•− levels in NOX4-deficient myoblasts, as detected with the mitochondrial O2•− probe MitoSOX Red (Fig. 9B). This, in turn, was associated with increased protein carbonylation in NOX4-deficient myoblasts (Fig. 9C), consistent with increased oxidative damage. The deletion of NOX4 and the increased mitochondrial O2•− and oxidative damage were accompanied by a reduction in insulin signaling, as monitored by AKT Ser473 phosphorylation in myoblasts and myotubes (Fig. 9D and fig. S14B). Fifteen-hour treatment with the NFE2L2 agonist sulforaphane not only restored the decreased antioxidant defense gene expression (Nfe2l2 and Nqo1) (Fig. 9E) and reduced protein carbonylation (Fig. 9C) but also restored insulin signaling in NOX4-deficient myoblasts (Fig. 9D). Moreover, the deletion of KEAP1 in NOX4-deficient myoblasts using CRISPR-Cas9 gene editing and a sgRNA targeting Keap1 increased NFE2L2 protein (Fig. 9, F and G) and gene expression (Fig. 9H), increased antioxidant defense, as reflected by the increased Nqo1 expression (Fig. 9H), and restored insulin signaling (Fig. 9I). Therefore, our results indicate that NOX4-derived H2O2 is important for promoting NFE2L2-mediated antioxidant defense to prevent mitochondrial oxidative stress and maintain insulin sensitivity in a cell-autonomous manner.
Fig. 9. NOX4-dependent antioxidant defense prevents mitochondrial oxidative stress and oxidative protein damage and promotes insulin signaling in muscle cells.
FACS-purified Nox4fl/fl myoblasts were transduced with β-galactosidase or Cre-expressing adenoviruses, and the resultant LacZ or Cre cells used thereafter. (A and B) Laz or Cre myoblasts were processed for (A) immunoblotting or (B) stained with 1 μM MitoSOX Red, and mitochondrial superoxide levels were assessed by confocal microscopy (nuclei were counterstained with DAPI). (C to E) Laz or Cre myoblasts were treated with vehicle or 5 μM sulforaphane for 15 hours and then either (C) processed for the analysis of protein carbonylation, (D) serum-starved (6 hours), stimulated with 1 nM insulin for the indicated times, and processed for immunoblotting to monitor for AKT Ser473 phosphorylation (p-AKT), or (E) processed for qPCR. (F to I) Cre myoblasts or those in which Keap1 was deleted using the CRISPR RNP approach were differentiated into myotubes, and (F) Keap1 mRNA and (G) NFE2L2 protein levels were assessed by qPCR or immunoblotting, respectively. (H and I) Laz or Cre myoblasts or Cre myoblasts in which Keap1 was deleted using the CRISPR RNP approach were differentiated into myotubes and processed for (H) qPCR to assess antioxidant gene expression and (H) immunoblotting to assess protein carbonylation or (I) serum-starved (6 hours) and stimulated with 1 nM insulin and processed for immunoblotting to monitor for p-AKT. Representative results are shown for at least three independent experiments. Quantified results are shown (means ± SEM) for the indicated number of experiments; significance determined using (D, E, and H) one-way ANOVA or (A and F) a Student’s t test; # indicates significance using a Student’s t test.
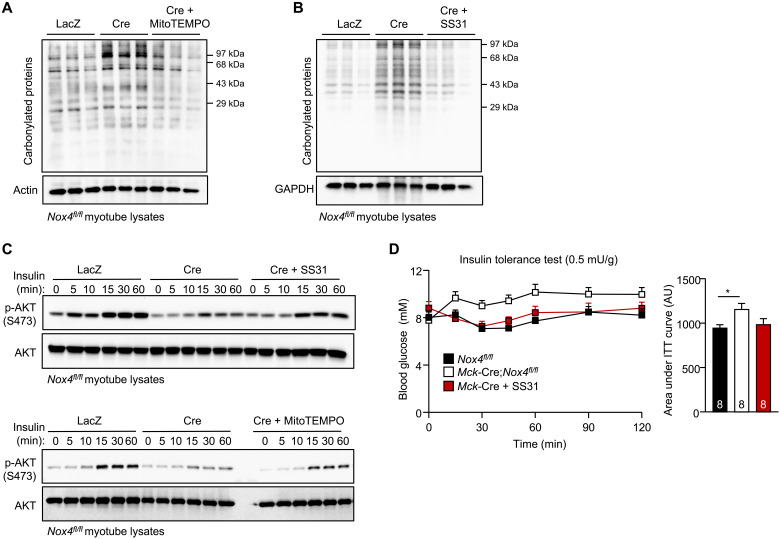
To determine whether the diminished insulin signaling in NOX4-deficient muscle cells could be attributed to the increased mitochondrial oxidative stress, we cultured NOX4-deficient muscle cells in the presence of either the mitochondrial-targeted antioxidant and SOD mimetic mitoTEMPO or the cell-permeable and mitochondrial-targeted tetrapeptide SS31 (elamipretide) that reduces mitochondrial O2•– generation/release (13, 46), and assessed effects on oxidative protein damage and insulin signaling. Culturing NOX4-deficient muscle cells in the presence of mitoTEMPO or SS31 reduced protein carbonylation (Fig. 10, A and B) and restored the defective insulin signaling, as monitored by AKT Ser473 phosphorylation (Fig. 10C). Last, to assess whether mitochondrial oxidative stress associated with skeletal muscle NOX4 deficiency may be responsible for the development of insulin resistance in vivo, we administered 7-month-old Mck-Cre;Nox4fl/fl mice SS31 for five consecutive days and assessed insulin sensitivity by performing ITTs. We found that the administration of SS31 reinstated insulin sensitivity in NOX4-deficient mice so that they were indistinguishable from controls (Fig. 10D). Together, our findings are consistent with NOX4 deficiency in muscle abrogating the antioxidant defense response to promote mitochondrial oxidative stress and the development of insulin resistance.
Fig. 10. Mitochondrial-targeted antioxidants ameliorate oxidative stress and insulin resistance associated with NOX4 deficiency.
FACS-purified Nox4fl/fl myoblasts were transduced with β-galactosidase or Cre-expressing adenoviruses, and the resultant LacZ or Cre cells were differentiated into myotubes. (A and B) Laz or Cre myotubes were treated with vehicle (LacZ, Cre) and either 20 μM mitoTEMPO (Cre) or 10 μM SS31 (Cre) for 72 hours and then either (A and B) processed for the analysis of protein carbonylation or (C) serum-starved (6 hours) and stimulated with 1 nM insulin for the indicated times and processed for immunoblotting to monitor for p-AKT. (D) Seven-month-old male Nox4fl/fl and Mck-Cre;Nox4fl/fl chow-fed male mice were administered vehicle (PBS) or SS31 (2 mg/kg per day, intraperitoneally) for five consecutive days and subjected to insulin tolerance tests (ITTs); areas under ITT curves were determined and AUs are shown. Representative results are shown for at least two independent experiments. Quantified results in (D) are means ± SEM for the indicated number of mice; significance determined using a two-way ANOVA or one-way ANOVA (for assessing significance for areas under ITT curves).
DISCUSSION
Our success as a species and the resulting diminished need for physical exertion coupled with the “on-demand” availability of energy-dense food has been a primary factor underpinning the obesity and type 2 diabetes epidemics. This is exacerbated further by aging, which markedly elevates the risk for type 2 diabetes and the metabolic syndrome independent of body weight (1). Precisely how physical activity/exercise promotes metabolic health and attenuates the development of insulin resistance and, conversely, how aging promotes insulin resistance and type 2 diabetes remain unclear (1). In this study, we demonstrate that the generation of ROS by skeletal muscle NOX4, the expression of which is increased by exercise and conversely is reduced with age or obesity, induces adaptive responses that prevent oxidative damage, maintain muscle function and exercise capacity, and attenuate the age- and obesity-associated development of insulin resistance.
The dysregulation of redox homeostasis is thought to contribute to the development of many human diseases (18). In particular, there is a strong body of evidence linking oxidative stress with the development of insulin resistance and type 2 diabetes (11–16). Both pharmacological (13) and genetic interventions (12–14) specifically implicate mitochondrial ROS in the development of insulin resistance that is induced by a high fat diet/obesity. The precise reason(s) why mitochondria produce more ROS in diet-induced obesity and type 2 diabetes remain(s) unclear. The increased availability of nutrients, especially fatty acids, can lead to increased mitochondrial ROS (13), but other studies suggest that there may be changes within mitochondria to increase ROS production or decrease scavenging (15). Mitochondrial dysfunction, especially in aged muscle, may also contribute to increased ROS generation (47–49). Notwithstanding, it is important to recognize that ROS such as H2O2 also have important physiological functions (18). A quantitative mapping of the mouse cysteine redox proteome recently identified some 34,000 unique sites across >9000 proteins that were reversibly oxidized in different tissues in young healthy mice (50). H2O2 can function as a second messenger to oxidize and inactivate protein tyrosine phosphatases and facilitate signaling in response to physiological stimuli such as insulin (8, 17). ROS generated in the context of exercise is also thought to promote mitochondrial biogenesis to meet the increased energy demands of contracting muscle and the NFE2L2 antioxidant defense response to prevent muscle damage (1, 5, 7, 19). However, the extent to which exercise-induced mitochondrial biogenesis and/or antioxidant defense in muscle affect insulin sensitivity or the development of insulin resistance has remained unclear. Previous studies have shown that NOX4 can promote the NFE2L2 response in cardiomyocytes (39, 51), but to the best of our knowledge, ours is the first to causally link NOX4-derived H2O2 in skeletal muscle with the control of NFE2L2 and thereby insulin sensitivity. Specifically, our studies indicate that NOX4-derived H2O2 and the stabilization of NFE2L2 in muscle are instrumental for preventing oxidative stress and attenuating insulin resistance otherwise induced by aging or diet-induced obesity. Skeletal muscle NOX4 expression declined in aged sedentary mice and in obese mice. The deletion of NOX4 and the reduced H2O2 and antioxidant defense in muscle increased mitochondrial ROS and oxidative protein and lipid damage, attenuated insulin signaling, and accelerated the age-associated decline in insulin sensitivity while also exacerbating the development of insulin resistance in diet-induced obesity. The deletion of NOX4 also abrogated the ability of exercise to enhance insulin sensitivity in otherwise high fat–fed and insulin-resistant mice. These effects were attributed to reductions in H2O2, defective NFE2L2-mediated antioxidant defense, and mitochondrial oxidative stress, as they were corrected by GPX1 deficiency or when mice were treated with either the NFE2L2 agonist sulforaphane or the mitochondrial-targeted SS31 that reduces mitochondrial O2•–. Consistent with this, we found that insulin signaling was diminished in NOX4-deficient muscle cells and that this was accompanied by defective antioxidant defense, mitochondrial oxidative stress, and oxidative protein damage, all of which were corrected when cells were cultured in the presence sulforaphane or mitochondrial-targeted antioxidants, or when KEAP1 was deleted to promote antioxidant defense. Precisely why NOX4-derived ROS promote antioxidant defense and insulin sensitivity in muscle and, conversely, why mitochondrial-derived ROS promote oxidative damage and insulin resistance remain unclear. Nonetheless, our studies argue against NOX4-derived ROS and the NFE2L2 antioxidant defense response directly influencing insulin sensitivity, because skeletal muscle NOX4 deficiency had no effect on insulin sensitivity in 3-month-old mice when reductions in skeletal muscle H2O2 levels, mitochondrial biogenesis, antioxidant defense, and exercise capacity/endurance were readily evident. Instead, we propose that the decreased antioxidant defense results in mitochondrial oxidative stress and the accumulation of oxidative damage, which over time diminishes the insulin response and contributes to the development of insulin resistance. In line with this, previous studies have shown that the global deletion of NFE2L2 and the decreased expression of antioxidant defense genes are accompanied by increased protein and lipid damage in skeletal muscle in aged mice (16), whereas heterozygosity in the NFE2L2 target gene Sod2 in mice promotes mitochondrial oxidative stress and the development of insulin resistance (12). Conversely, the transgenic overexpression of SOD2 attenuates the development of insulin resistance in high fat–fed mice (12). The potential for SOD2 deficiencies to contribute to insulin resistance and type 2 diabetes in humans is underscored by studies showing that SOD2 polymorphisms that reduce SOD2 activity are found in patients with type 2 diabetes (45). SOD2 localizes to the mitochondrial matrix and converts O2•− into O2 and H2O2 (18). We found that SOD2 protein was reduced by more than 50% in NOX4-deficient skeletal muscle and myoblasts. NOX4 deficiency was also accompanied by overt reductions in PRDX3 that localizes to mitochondria and catalase that is found in mitochondria, peroxisomes, and the cytosol and eliminate H2O2. The reduction in SOD2, in particular, would lead to increased O2•− in mitochondria, as we found in NOX4-deficient myoblasts, and ultimately facilitate the formation of toxic ONOO− during exercise and highly reactive hydroxyl radicals to damage proteins and lipids (19). Collectively, our findings are consistent with the concept that the development of insulin resistance may be driven largely by endogenous defects in antioxidant defense, or rather declining NOX4-dependent antioxidant defense, due to physical inactivity and aging rather than extrinsic insults such as hypernutrition, which inherently might be expected to elicit adaptive responses, such as those orchestrated by NFE2L2.
It remains unclear whether the reduced mitochondrial biogenesis in NOX4-deficient mice, which was also corrected by GPX1 deficiency and sulforaphane, might also contribute to the development of the insulin resistance in aging or obesity. Although previous studies have shown that the deletion of PGC1α and decreased mitochondrial biogenesis per se are not sufficient to alter insulin sensitivity in 3-month-old unstressed mice (52, 53), PGC1α has been reported to also promote the expression of SOD2 (54), as well as GPX-1 and catalase (54), whereas PGC1α deficiency in the heart is accompanied by myocardial oxidative stress in the context of pressure overload (55). Therefore, we propose that concomitant reductions in skeletal muscle NFE2L2 and PGC1α in NOX4-deficient mice might function in concert to decrease antioxidant defense and increase mitochondrial oxidative stress to promote oxidative damage and drive the development of insulin resistance during aging and in the context of obesity. Moreover, although defective mitochondrial biogenesis per se does not promote insulin resistance (52, 53), any reductions in muscle function/respiratory capacity associated with the decline in NOX4 might be expected to create a vicious cycle, further compromising exercise-induced antioxidant defense and contributing to the age-associated decline in insulin sensitivity.
The results of this study indicate that skeletal muscle NOX4 is instrumental in orchestrating hormetic responses that attenuate the age-associated development of insulin resistance, hyperinsulinemia, and hyperglycemia, hallmarks of type 2 diabetes. Our findings underscore the importance of redox balance in metabolic health and provide important insight into pathophysiological mechanisms contributing to the diabetes epidemic in our aging and increasingly sedentary populations.
MATERIALS AND METHODS
Mice
Age-matched male mice were used for all experimental conditions. Mice were maintained on a 12-hour light-dark cycle in a temperature-controlled high-barrier facility (Monash ARL) with ad libitum food and water. Mice were fed a standard chow diet (8.5% fat; Barastoc, Ridley AgriProducts, Australia) or, where indicated, a 4.8% fat chow diet (4.8% fat, 20% protein, and 4.8% crude fiber; Specialty Feeds), or otherwise fed a high-fat diet [18.4% protein, 23.5% fat (46% energy from fat), 4.7% fiber; Specialty Feeds, no. SF04-027]. Nox4fl/fl (C57BL/6J), Mck-Cre (C57BL/6J; IMSR catalog no. JAX:006475, RRID:IMSR_JAX:006475), and Gpx1−/− (C57BL/6J) mice have been described previously (8, 34, 56), and HSA-MCM (C57BL/6J; catalog no. JAX:025750, RRID:IMSR_JAX:025750) mice were from The Jackson Laboratory (Bar Harbor, ME). Mck-Cre mice or HSA-MCM mice were bred with Nox4fl/fl mice to generate Mck-Cre;Nox4fl/fl or HSA-MCM;Nox4 fl/fl mice, respectively. Gpx1−/− mice were bred with Nox4fl/fl mice to generate Nox4fl/fl;Gpx1−/− mice and then with Mck-Cre;Nox4fl/fl mice to generate Mck-Cre;Nox4fl/fl;Gpx1+/− mice, which were bred with Nox4fl/fl;Gpx1−/− to generate Mck-Cre;Nox4fl/fl;Gpx1−/− mice. All mice were on a C57BL/6/J background. C57BL/6J mice were purchased from Monash Animal Research Platform. Experiments were approved by the Monash University School of Biomedical Sciences Animal Ethics Committee (project IDs: 23077, 22138, and14368) and performed in accordance with the National Health and Medical Research Council Australian Code of Practice for the Care and Use of Animals.
Human participants and biopsies
Twenty healthy untrained males (age: 35 ± 3 years old, body mass index: 25 ± 0.7, VO2peak: 40.2 ± 1.6) were recruited in Auckland, New Zealand. Males met the criteria of nonsmokers and free of cardiovascular, metabolic disease and any injuries that could impair their ability to perform high-intensity aerobic training. All participants gave written informed consent before the commencement of the study, which was approved by the Health and Disability Ethics Committee, New Zealand (16/STH/116 and 17/STH/200). To assess VO2 peak (peak oxygen consumption), participants completed a ramped exercise protocol to exhaustion (initially at 60 W and subsequently increasing by 15 W every min) in an electromagnetically braked cycle ergometer (Velotron, RacerMate, WA). Oxygen consumption was measured using open-circuit spirometry (Parvo Medics True One 2400, Sandy, UT), at least 48 hours after VO2 peak measurements following overnight fasting. Participants completed an acute bout of short-term high-intensity interval exercise consisting of 10 × 60-s cycling intervals at VO2peak power output, interspersed with 75-s low-intensity (30 W) intervals on the cycle ergometer. Percutaneous muscle biopsies were obtained from the vastus lateralis muscle (thigh) before and at 3 to 4 hours after an acute bout of exercise, using a suction-modified Bergstrom biopsy needle under local anesthesia. Muscle biopsy samples were snap-frozen in liquid N2 and subsequently stored at −80°C until analysis.
Cultured cells
Primary skeletal muscle myoblasts were isolated from 3- to 4-week-old Nox4fl/fl mice. Hindlimb skeletal muscles were rinsed with phosphate-buffered saline (PBS; Sigma-Aldrich, St. Louis, MO), minced with scalpel blades under aseptic conditions, and digested enzymatically for 30 min at 37°C with collagenase D (1.5 U/ml; Roche, Germany) and dispase II (7.2 U/ml; Roche, Germany) in PBS plus 2.5 mM CaCl2. Samples were agitated every 10 min and then pelleted and resuspended in PBS and filtered through 100-μm and then 70-μm cell strainers (BD Biosciences, San Jose, CA). Cells were stained with fluorophore-conjugated biotinylated antibodies for CD45 [CD45–fluorescein isothiocyanate (FITC) (1:1000; 553079, RRID:AB_394609), BD Biosciences, San Jose, CA], CD31 [CD31-FITC (1:400; 558738, RRID:AB_397097), BD Biosciences, San Jose, CA], CD-11b [CD-11b–phycoerythrin (PE) (1:400; 557397, RRID:AB_396680), BD Biosciences, San Jose, CA], and Sca1 [Sca1-PE-Cy7/Ly-6A/E (1:600; 558162, RRID:AB_647253), BD Biosciences, San Jose, CA] to stain immune and endothelial cells and for α-7 integrin [α-7-integrin–allophycocyanin (FAB3518A, RRID:AB_1026275), R&D Bio-Techne, MN] to stain for myoblasts and propidium iodide–negative CD45−CD31−CD-11b−Sca1−α-7-integrin+ myoblasts purified by flow cytometry. FACS-purified myoblasts were then cultured in entactin-collagen-laminin cell attachment matrix (Merck, Millipore, CA)–coated tissue culture dishes in Dulbecco’s modified Eagle’s medium (DMEM)–low glucose (Sigma-Aldrich, St. Louis, MO) buffered with Hepes (pH 7.4) supplemented with 20% (v/v) fetal bovine serum (FBS; GIBCO Life Technologies, Carlsbad, CA), human basic fibroblast growth factor (hFGFb; 25 ng/ml; Sigma-Aldrich, St. Louis, MO, USA), penicillin (50 U/ml), streptomycin (50 μg/ml), and 2 mM l-glutamine at 37°C, 5% O2, and 5% CO2.
To delete Nox4, FACS-purified Nox4fl/fl myoblasts were transduced with either β-galactosidase (LacZ) control or Cre recombinase–expressing adenoviruses (Vector Biolabs, Malvern, PA) at a multiplicity of infection of 100 in the presence of polybrene (1 μg/ml). Where indicated, myoblasts that were 100% confluent were differentiated into myotubes over 3 days by culturing cells in DMEM–low glucose supplemented with 5% (v/v) horse serum (Sigma-Aldrich, St. Louis, MO), penicillin (50 U/ml), streptomycin (50 μg/ml), and 2 mM l-glutamine.
Nfe2l2 or Keap1 was deleted in LacZ or Cre-expressing myoblasts, respectively, using CRISPR-Cas9 ribonucleoprotein (RNP) gene editing. Cells were transfected with recombinant Cas9 (74 pmol Alt-R S.p. Cas9 Nuclease V3; Integrated DNA Technologies, Coralville, IA) precomplexed with short guide RNAs (sgRNAs; 600 pmol; Synthego, Menio Park, CA) targeting Keap1 (5′-GCUUGGGUUCGGGCUGCAUG-3′) or Nfe2l2 (5′-AUUUGAUUGACAUCCUUUGG-3′) using the P3 Primary Cell 4D-Nucleofector X Kit (Lonza, Basel, Switzerland) according to the manufacturer’s instructions.
Rodent exercise studies
Mice were acclimated to treadmill running for 3 days before the initiation of the experiments. Mice were placed on a multilane treadmill (Columbus Instruments, Columbus, OH) for 10 min and run for 5 min at 10 m/min and 1 min at 15 m/min at 0% slope. All animals were randomized before the initiation of exercise tests.
Exercise stress test
Mice were placed in an enclosed single-lane treadmill connected to Oxymax O2 and CO2 sensors (Columbus Instruments, Columbus, OH). On the day of the experiment, before the initiation of the exercise stress test, mice were acclimated in the chamber at rest for 20 min (basal O2 consumption and CO2 generation assessed for the last 5 min). The mice were then subjected to running at 10 m/min at 0% incline, and the running speed (U) was increased by 4 m/min every 3 min until the mice reached fatigue; fatigue was determined as the time point where the mice could not be prompted to continue running for at least 5 s. VO2max was assessed at the Umax. Differences between the exercised and basal states were calculated, and gas exchange data were used to determine energy expenditure, heat, and the RER (RER = VCO2/VO2).
Endurance test
Mice were exercised on a multilane treadmill (Columbus Instruments International) at 0% slope. The endurance test was performed with a warm-up period of 10 min at 10 m/min, followed by running at 60% of the Umax until fatigue was reached. The time until fatigue was considered the representative measure of endurance capacity. Where indicated, mice were administered vehicle or sulforaphane (Sigma-Aldrich, St. Louis, MO) at 0.5 mg/kg per day (intraperitoneally) for 5 days before endurance tests.
Exercise training
Mice were allocated into sedentary and training groups. The training group underwent exercise training for 5 weeks, 5 days per week, while the sedentary group remained sedentary on the treadmill for the same period of time. Mice were subjected to a gradual overload protocol, in which running speed was increased so that by the end of training weeks 1, 2, 3, 4, and 5 mice were running for up to 60 min at 50, 60, 70, 80, and 90% of their maximum pretraining speed.
Metabolic and blood measurements
Mouse body weights were monitored weekly, and body composition was assessed by EchoMRI (Echo Medical Systems, Houston, TX) or dual-energy x-ray absorptiometry (DEXA, Lunar PIXImus2, GE Healthcare) as described previously (8, 57). ITTs and GTTs were performed as described previously (8). Where indicated, mice were administered vehicle or SS31 (Selleckchem, Houston, TX) at 2 mg/kg per day (intraperitoneally) for five consecutive days before ITTs. Blood was collected from conscious fed [satiated, 11:00 p.m. (57, 58)] and 6-hour fasted mice by submandibular bleeding; plasma insulin levels were determined using the mouse insulin enzyme-linked immunosorbent assay (ELISA; ALPCO, Salem, NH) or an in-house ELISA (Monash Antibody Technologies Facility), and the corresponding blood glucose levels were determined with an Accu-Check glucometer. Food intake, wheel running, and energy expenditure were assessed over 72 hours after 24-hour acclimation using the Promethion Metabolic Screening System (Sable Systems International, North Las Vegas, NV) fitted with indirect open circuit calorimetry, running wheels, and food consumption and activity monitors.
For post-exercise ITTs, 12-week high fat–fed mice were acclimatized to the treadmill for 3 days before ITTs were carried out with a crossover design over 2 days with 3 days rest in between. Mice were fasted for 4 hours before exercise on the multilane treadmill (Columbus Instruments, Columbus, OH). Exercised mice were subjected to an acute bout of exercise for 30 min (55% Umax, 15% slope) on the multilane treadmill (Columbus Instruments, Columbus, OH). Sedentary mice were housed adjacent to the treadmill. Mice were allowed to rest without food for a further 1 hour and subjected to ITTs (0.65 mU/g insulin).
Echocardiography
Basal cardiac morphology and function were assessed by echocardiography in sedentary mice under 1.5% (v/v) isoflurane anesthesia with heart rates in the range of 400 to 450 beats per minute (bpm) using a Vevo2100 system equipped with a 40-MHz transducer (FUJIFILM/VisualSonics, Canada). Alternatively, heart rates and cardiac function in conscious mice were assessed before and immediately after an acute exercise stress test. For conscious scans, mice were acclimated to handling over 5 days. Mice were held firmly by the nape of the neck in the palm of a hand with the tail tightly secured between the last two fingers, and an ultrasound transducer was passed over the chest region. Images were acquired in M mode through a short-axis view at the papillary muscle level. Quantitative measurements were made using VevoLAB analytical software (FUJIFILM/VisualSonics). FS was calculated as FS% = [(LVIDd −LVIDs)/LVIDd] × 100, where LVIDs is the LV internal diameter in systole and LVIDd is the LV internal diameter in diastole.
Hyperinsulinemic-euglycemic clamps
Hyperinsulinemic-euglycemic clamps were performed as described previously (59). Briefly, mice were anesthetized under 2% (v/v) isoflurane (250 ml/min O2), and the left common carotid artery and the right jugular vein were catheterized for sampling and infusions. On the day of the clamp, food was removed, and after 3.5 hours of fasting, a primed (2 min, 0.5 μCi/min) continuous infusion (0.05 μCi/min) of [3-3H]glucose was administered to measure basal glucose turnover. After 5 hours of fasting, mice received a continuous insulin infusion (4 mU/kg per minute), and blood glucose was maintained at basal levels (euglycemia) by a variable infusion of a 50% (w/v) glucose solution. Arterial blood samples were collected at basal and during steady-state conditions and at 80, 90, 100, 110, and 120 min. At 120 min, a 13 μCi bolus of [14C]2-deoxy-d-glucose was injected into the jugular vein and arterial blood was sampled up to 145 min. Mice were then euthanized, and tissues were extracted and frozen for subsequent gene expression and glucose uptake determinations.
Cell stimulations
Myoblasts or myotubes were pretreated with vehicle control or sulforaphane (5 μM for myoblasts and 2 μM for myotubes; Sigma-Aldrich, St. Louis, MO) for 15 hours, and cells were used for either RNA isolation or insulin stimulation. Where indicated, myoblasts were treated with vehicle control, mitoTEMPO (20 μM) (Sigma-Aldrich, St. Louis, MO), or SS31 (10 μM) (Selleckchem, Houston, TX) for 72 hours and then processed for immunoblotting or insulin stimulation. For insulin stimulations, cells were serum-starved in DMEM–low glucose supplemented with 0.1% (v/v) FBS for 6 hours and stimulated with 1 to 2 nM insulin for the indicated times. Cells were lysed using radioimmunoprecipitation assay (RIPA) buffer [50 mM Hepes (pH 7.4), 1% (v/v) Triton X-100, 1% (v/v) sodium deoxycholate, 0.1% (w/v) SDS, 150 mM NaCl, 10% (v/v) glycerol, 1.5 mM MgCl2, 1 mM EGTA, 50 mM NaF, leupeptin (5 μg/ml), pepstatin A (1 μg/ml), 1 mM benzamidine, 2 mM phenylmethysulfonyl fluoride, 1 mM sodium vanadate, and 10 μM MG-132] and processed for immunoblotting.
Muscle cells were subjected to mitochondrial stress tests to measure OCRs using a Seahorse XFe24 extracellular flux analyzer (Seahorse, Agilent, Santa Clara, USA) according to the manufacturer’s recommendations. Cells (5 × 103 per well or 15 × 103 for analyses of differentiated myotubes) were seeded, differentiated (for myotube analyses), and subsequently processed for OCR determinations. On the day of the experiment, the medium was changed to assay medium [Seahorse XF DMEM (Seahorse, Agilent, Santa Clara, USA) with Hepes (pH 7.4) and supplemented with 10 mM glucose, 1 mM pyruvate, and 2 mM l-glutamine]. Continuous OCR measurements were recorded, starting at baseline and followed by measurements after the sequential addition of 1.5 μM oligomycin, 1 to 2 μM carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP; 1 μM for myotubes and 2 μM for myoblasts), and 0.5 μM rotenone/antimycin A (Seahorse XF Cell Mito Stress Test Kit, Agilent, Santa Clara, USA) according to the manufacturer’s instructions.
Mitochondrial superoxide (O2•−) production was monitored using MitoSOX Red (Invitrogen, Carlsbad, CA), a live-cell permeable dye that is selectively targeted to the mitochondria and fluoresces red after oxidation by superoxide. Primary myoblasts were serum-starved in DMEM–low glucose supplemented with 0.1% (v/v) FBS for 4 to 6 hours. Myoblasts were incubated with 1 μM MitoSOX Red plus 4′,6-diamidino-2-phenylindole (DAPI) to stain nuclei at 37°C for 5 min, washed twice with PBS, and fixed in 4% (w/v) paraformaldehyde (Thermo Fisher Scientific, Waltham, MA) for 20 min. MitoSOX Red fluorescence was visualized by confocal microscopy (Leica SP8 confocal microscope, Leica Microsystems, Wetzlar, Germany) at an excitation of 510 nm and an emission of 580 nm.
Real-time PCR
RNA was extracted using RNAzol (Sigma-Aldrich, St. Louis, MO), and total RNA quality and quantity were determined using NanoDrop 3300 (Thermo Fisher Scientific, Waltham MA). mRNA was reverse-transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) and processed for qPCR using either the TaqMan Universal PCR Master Mix and TaqMan Gene Expression probes (Applied Biosystems, Foster City, CA) or Quantinova SYBR Green Master Mix (Qiagen, Hilden, Germany) and Bio-Rad PrimePCR primers or validated oligonucleotides. Gapdh was used as an internal control for all murine studies, with the exception of aging studies where Rn18s was used instead. Rplp0 was used as an internal control for all cell studies. For analyzing human genes, RPLP0 and β2-microglobulin (B2M) were used as internal controls. The real-time PCR primers/probes that were used included (i) TaqMan Gene Expression Assays (Thermo Fisher Scientific, Waltham MA) for Nox4 (Mm00479246_m1), Catalase (Mm00437992_m1), Cybb (Mm01287743_m1), Gpx1 (Mm00656767_g1), Rac1 (Mm01201653_mH), Ncf1 (Mm00447921_m1), Sod1 (Mm01344233_g1), Sod2 (Mm01313000_m1), Gapdh (Mm99999915_g1), Keap1 (Mm00497268_m1) Rplp0 (Mm00725448_s1), Rn18s (Mm03928990_g1), and Txn1 (Mm00726847_s1); (ii) PrimePCR SYBR Green Gene Expression Assays (Bio-Rad, Hercules, CA) for Gapdh (qMmuCED0027497), Gclc (qMmuCID0019217), Gclm (qMmuCID0017192), G6pdx (qMmuCID0023829), Idh1 (qMmuCID0017698), Me1 (qMmuCED0047190), Gsr (qMmuCID0005414), Nfe2l2 (qMmuCID0021433), Nqo1 (qMmuCID0017192), Pgd (qMmuCID0019196), Prdx1 (qMmuCED0047858), Prdx2 (qMmuCID0024306), Prdx3 (qMmuCID0017576), and Rplp0 (qMmuCED0040751); or (iii) validated oligonucleotides (Sigma-Aldrich, St. Louis, MO) for Cox1 (TGCAACCCTACACGGAGGTAATA, ATGTATCGTGAAGCACGATGTCA), Nrf1 (ACATTGGCTGATGCTTCAGAA, TGCGTCGTCTGGATGGTCAT), Nrf2 (CCGCTACACCGACTACGATT, ACCTTCATCACCAACCCAAG), Pgc1α (AACGATGACCCTCCTCACAC, TCTGGGGTCAGAGGAAGAGA), Tfam (AGGCTTGGAAAAATCTGTCTC, TGCTCTTCCCAAGACTTCATT), NFE2L2 (GGTTGCCCACATTCCCAAATC, CGTAGCCGAAGAAACCTCA), NOX4 (CCGGCTGCATCAGTCTTAACC, TCGGCACAGTACAGGCACAA), CYBB (TTGCTGGAAACCCTCCTATGA, AAAACCGCACCAACCTCTCA), B2M (GGCTATCCAGCGTACTCCAA, GATGAAACCCAGACACATAGCA), and RPLP0 (GTGATGTGCAGCTGATCAAGACT, GATGACCAGCCCAAAGGAGA). Thermal cycling and fluorescent readouts were measured using the CFX384 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Relative quantification was achieved using the ΔΔCT method.
Skeletal muscle histology and immunostaining
Gastrocnemius and soleus muscles were dissected from mice, snap-frozen in liquid nitrogen–cooled isopentane, and stored at −80°C until required. Frozen transverse muscle sections (10 μm) were prepared and used for all histology and immunostaining experiments. Hematoxylin and eosin (H&E) staining was performed using a standard approach. For SDH staining to detect mitochondria, fresh transverse muscle sections were air-dried and then incubated in freshly prepared staining solution for 40 min at 37°C [for 10 ml; 5 mg of nitrotetrazolium blue chloride (NBT; N6876, Sigma-Aldrich), 2.5 ml of 0.2 M phosphate buffer (pH 7.6) (13 ml of 0.2 M NaH2PO4 and 87 ml of 0.2 M Na2HPO4), and 2.5 ml of 0.2 M sodium succinate]. Sections were washed thoroughly in distilled water, incubated in formal saline (4% formaldehyde, 145 mM NaCl in distilled water) for 1 min, washed again in water, and coverslip-mounted. For immunostaining to detect mitochondria, transverse muscle sections were fixed in ice-cold acetone for 5 min, washed thoroughly in PBS, and permeabilized by incubation in 0.1% (w/v) Triton X-100 in PBS for 5 min. Sections were blocked in blocking buffer [1% (w/v) bovine serum albumin, 10% (v/v) donkey serum in PBS] for 1 hour before the addition of primary antibodies to stain mitochondria [translocase of outer mitochondrial membrane 20 (TOMM20; 1:100; ab186735, RRID:AB_2889972) or PDH (1:200; ab110333, RRID:AB_10862029); Abcam, Cambridge, UK] and incubated overnight at 4°C. Coimmunostaining for dystrophin (1:200; ab15277, RRID:AB_301813, Abcam, Cambridge, UK) or syntrophin (1:200; ab11425, RRID:AB_298030, Abcam, Cambridge, UK) was used to stain the sarcolemma and define the boundaries of individual muscle fibers. Sections were washed thoroughly in PBS, incubated in Alexa Fluor 488– or Alexa Fluor 555–conjugated secondary antibodies [(A21206, RRID:AB_2535792), (A21202, RRID:AB_141607), (A31570, RRID:AB_2536180), and (A31572, RRID:AB_162543), 1:500; Thermo Fisher Scientific, Waltham, MA] for 1 hour at room temperature before washing again in PBS, and coverslip-mounted using Fluoromount-G (00-4958-02, Thermo Fisher Scientific, Waltham, MA). All antibodies were diluted in blocking buffer. For fiber typing, the immunostaining was performed as described above but with 10% (v/v) goat serum in lieu of donkey serum and with the addition of 0.05% (w/v) Triton X-100 to the blocking buffer and antibody diluent. The following fiber typing antibodies were used and were previously developed by the listed investigators and obtained from the Developmental Studies Hybridoma Bank, created by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health, and maintained at the University of Iowa, Department of Biology, Iowa City, IA 52242: type I fibers [BA-F8, MHC slow, mouse immunoglobulin G2b (IgG2b), RRID:AB_10572253, 1:100, S. Schiaffino, University of Padova, Italy], type IIa (SC-71, MHC 2A, mouse IgG1, RRID:AB_2147165, 1:100, S. Schiaffino, University of Padova, Italy), type IIb (BF-F3, MHC 2B, mouse IgM, RRID:AB_2266724, 1:100, S. Schiaffino, University of Padova, Italy), and type IIx (6H1, MHC fast, mouse IgM, 1:50, C. Lucas, University of Sydney, Australia). Isotype-specific, anti-mouse secondary antibodies were from Thermo Fisher Scientific (Waltham, MA) as follows: A21141 (RRID:AB_141626), A21044 (RRID:AB_141424), and A21121 (RRID:AB_2535764) diluted 1:500.
H&E, SDH, or fiber-type staining was viewed using an Olympus DotSlide microscope, whereby individual images were stitched together using the VS-ASW program (Olympus) to generate a composite image of the entire muscle section. Alternatively, SDH staining was imaged using a CX33 brightfield microscope, fitted with a DP23 camera (Olympus). TOMM20 and PDH immunostaining of mitochondria in muscle sections was imaged using a Leica SP5 5-channel confocal microscope. Microscopy was performed at Monash MicroImaging, Monash University, Australia. Measurement of the minimal Feret’s diameter for muscle fibers (n = 500 fibers per muscle) was undertaken using the “measure” function of ImageJ. The variability coefficient is defined as the SD of muscle fiber diameter measurements and is used to identify variability in muscle fiber size, which can relate to muscle atrophy, hypertrophy, or degeneration. For fiber-type measurements, the number of individual fiber types was quantified using the “cell counter” plug-in of ImageJ and expressed relative to the total number of fibers per muscle section.
Mass spectrometry
Mouse muscle tissue was homogenized in liquid nitrogen and lysed in 4% (w/v) SDS, 100 mM Hepes (pH 8.5), which was then heated at 95°C for 5 min and sonicated three times for 10 s each. Lysates were collected by centrifuging at 16,000g for 10 min, and protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA). Equal amounts of protein were denatured and alkylated using tris(2-carboxyethyl)-phosphine-hydrochloride) and 2-chloroacetamide at a final concentration of 10 mM and 40 mM, respectively, and incubated at 95°C for 5 min. SDS was removed by suspension trapping (60). Sequencing-grade trypsin was added at an enzyme to protein ratio of 1:100, and the reaction was incubated overnight at 37°C. The digested peptide from each sample was labeled with TMTpro (Thermo Fisher Scientific, Waltham MA), combined and fractionated with basic pH C18. The dried fractions were analyzed with a QExactive HF mass spectrometer (Thermo Fisher Scientific, Waltham, MA) operated in data-dependent acquisition mode, coupled by a Dionex UltiMate 3000 RSLCnano system equipped with a Dionex UltiMate 3000 RS autosampler. The samples were loaded via an Acclaim PepMap 100 trap column (100 μm × 2 cm, nanoViper, C18, 5 μm, 100 Å; Thermo Fisher Scientific, Waltham, MA) onto an Acclaim PepMap RSLC analytical column (75 μm × 50 cm, nanoViper, C18, 2 μm, 100 Å; Thermo Fisher Scientific, Waltham, MA). The instrument was operated in the data-dependent acquisition mode to automatically switch between full-scan mass spectrometry (MS) and MS/MS acquisition. Each survey full scan [mass/charge ration (m/z) 375 to 1575] was acquired in the Orbitrap with 120,000 resolution (at m/z 200) after accumulation of ions to a 3 × 106 target value with a maximum injection time of 54 ms. Dynamic exclusion was set to 90 s, and isolation window is set to 1.2 m/z. The 15 most intense multiply charged ions (z ≥ 2) were sequentially isolated and fragmented in the collision cell by higher-energy collisional dissociation with a fixed injection time of 54 ms, 30,000 resolution, and automatic gain control target of 2 × 105. The raw data files were analyzed in ProtemeDiscoverer 2.4 (Thermo Fisher Scientific, Waltham, MA), and statistical analysis was performed in Perseus 1.6.2.3.
To identify differentially expressed proteins associated with molecular pathways, gene set enrichment was performed using GSEA version 4.1.0 (https://gsea-msigdb.org/gsea/downloads.jsp). Kyoto Encyclopedia of Genes and Genomes (KEGG) and Hallmarks gene sets were used to identify significantly differentially represented pathways based on t test ranked order gene sets from our proteomic dataset. GSEA cutoff values were determined by published recommended cutoff values using a nominal P value ≤ 0.01 and false discovery rate ≤ 0.25. To identify potentially enriched transcription target genes/proteins of NFE2L2, we used the Enrichr chromatin immunoprecipitation sequencing dataset and matched overlapping proteins with corresponding gene symbols that were shown to have NFE2L2 binding sites in the gene promoter region (https://maayanlab.cloud/Enrichr/).
Biochemical assays
Mouse tissues were dissected and immediately snap-frozen in liquid N2. Tissues were homogenized with a bead homogenizer (Bead Ruptor 12, Omni International, GA) using 1-mm-diameter zirconia/silica beads (BioSpec Products, OK) for 30 to 60 s in 10 to 20 volumes of ice-cold RIPA lysis buffer [50 mM Hepes (pH 7.4), 1% (v/v) Triton X-100, 1% (v/v) sodium deoxycholate, 0.1% (w/v) SDS, 150 mM NaCl, 10% (v/v) glycerol, 1.5 mM MgCl2, 1 mM EGTA, 50 mM NaF, leupeptin (5 μg/ml), pepstatin A (1 μg/ml), 1 mM benzamidine, 2 mM phenylmethysulfonyl fluoride, and 1 mM sodium vanadate], incubated for 30 min on ice, and clarified by centrifugation (16,000g for 30 min at 4°C) as described previously (8). Clarified tissue homogenates were resolved by SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride and immunoblotted. The primary antibodies used for immunoblotting include rabbit monoclonal anti–β-actin (A5441, RRID:AB_476744), mouse monoclonal anti–α-tubulin (T5168, RRID:AB_477579), mouse monoclonal anti-vinculin (V9131, RRID:AB_477629), and rabbit monoclonal anti-NFE2L2 (SAB2701989) from Sigma-Aldrich (St. Louis, MO); rabbit monoclonal anti–phospho-AKT (p-AKT) (473) (4060, RRID:AB_2315049), mouse monoclonal anti-AKT (40D4, RRID:AB_1147620), and rabbit polyclonal anti-PRDX1 (8732) from Cell Signaling Technology (Danvers, MA); rabbit polyclonal anti-catalase (ab1877, RRID:AB_302649), rabbit polyclonal anti-G6PD (ab993, RRID:AB_296714), rabbit anti–4-HNE (ab46545, RRID:AB_722490), rabbit monoclonal anti-KEAP1 (ab139729, RRID:AB_2891077), mouse monoclonal anti-myogenin (ab1835, RRID:AB_302633), goat polyclonal anti-NCF1/p47phox (ab166930, RRID:AB_2832946), mouse monoclonal anti-PRDX2 [EPR5154] (ab109367, RRID:AB_10862524), rabbit polyclonal anti-PRDX3 (ab222807), rodent anti-OXPHOS cocktail (ab110413, RRID:AB_2629281), rabbit monoclonal anti-NFE2L2 (ab62352, RRID:AB_944418), and rabbit polyclonal anti-PGC1α antibody (ab54481, RRID:AB_881987) from Abcam (Cambridge, UK); rabbit polyclonal anti-NOX4 (sc-30141, RRID:AB_2151703), rabbit polyclonal anti-IRβ (sc-711, RRID:AB_631835), and mouse monoclonal anti-SOD2 (E-10, RRID:AB_2191808) from Santa Cruz Biotechnology (Santa Cruz, CA); rabbit monoclonal anti–IRS-1 (06-248, RRID:AB_2127890) and rabbit monoclonal anti-PI3K p85 (06-497, RRID:AB_310141) from Upstate Biotechnology (Lake Placid, NY); mouse monoclonal anti-gp91phox (611415, RRID:AB_398937) and mouse monoclonal anti-RAC1 (610651, RRID:AB_397978) from BD Biosciences (San Jose, CA); mouse monoclonal anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (AM4300, RRID:AB_2536381) from Thermo Fisher Scientific (Waltham, MA); mouse monoclonal anti-NQO1 (NB200-209, RRID:AB_10002706) from Novus Biologicals (Littleton, CO); and mouse monoclonal anti-MHC (MYH1E) (MF20) from J. M. Ziermann Laboratory (Howard University College of Medicine, Developmental Studies Hybridoma Bank, University of Iowa, USA).
Carbonylated proteins in gastrocnemius skeletal muscle and myoblasts were assessed by immunoblotting using the OxyBlot Protein Oxidation Detection Kit (Merck Millipore, Burlington, MA) according to the manufacturer’s instructions. Muscle citrate synthase activity was assessed using the Citrate Synthase Assay Kit (Abcam, Cambridge, UK).
H2O2 levels in gastrocnemius or cardiac muscles were measured using an Amplex Red hydrogen peroxide assay kit (Invitrogen, Carlsbad, CA) with minor modifications from the manufacturer’s instructions. Muscle was excised rapidly, snap frozen in liquid N2 and used immediately thereafter for the analysis of H2O2 levels. Muscle tissue was homogenized with a bead homogenizer in ice-cold lysis buffer [50 mM tris (pH 7.4), 50 mM sodium phosphate, 5 mM N-ethylmaleimide, 5 mM iodoacetamide, 12.5 mM amino-1,2,4-triazole, 0.035% sodium azide, pepstatin A (1 μg/ml), leupeptin (5 μg/ml), aprotinin (5 μg/ml), 1 mM benzamidine] containing 5 μM Amplex Red reagent (Thermo Fisher Scientific, Waltham MA) and horseradish peroxidase (HRP; 1 U/ml) = for 60 s (3 × 20 s pulses). Homogenates were incubated in the dark at room temperature for 10 min and then clarified by centrifugation at 16,000g for 10 min at room temperature. Amplex Red reacts with H2O2 at a stoichiometry of 1:1 to produce the fluorescent oxidation product resorufin. Fluorescence was measured on a BMG Labtech Clariostar plus plate reader (Hamilton, Reno, NV) at an excitation of 545 nm and an emission of 590 nm over 30 min, and fluorescence within the linear range was recorded and normalized to total protein.
For assessing cellular H2O2 production, myoblasts were differentiated into myotubes on the PerkinElmer 96-well View Plate (Waltham, MA) and serum-starved in DMEM–low glucose supplemented with 0.1 (v/v) FBS for 4 hours. Myotubes were incubated in PBS containing 200 μM Amplex Red reagent and HRP (0.5 U/ml; Amplex Red hydrogen peroxide assay kit; Invitrogen, Carlsbad, CA), and fluorescence was measured over 30 min on a BMG Labtech Clariostar plus plate reader (Hamilton, Reno, NV) at an excitation of 545 nm and an emission of 590 nm, and fluorescence within the linear range was recorded and normalized to total protein.
For assessing reduced GSH and GSSG, snap-frozen gastrocnemius muscle was homogenized with a bead homogenizer in ice-cold lysis buffer [50 mM Hepes (pH 7.4), 10 mM EDTA, 1 mM EGTA, 100 mM NaF, 50 mM sodium pyrophosphate, pepstatin A (1 μg/ml), leupeptin (5 μg/ml), aprotinin (5 μg/ml), 1 mM benzamidine, 1 mM phenylmethysulfonyl fluoride, and 5% (w/v) 3-metaphosphoric acid] containing 3 mM methyl-2-vinylpyridinium triflate for GSSG samples and clarified by centrifugation (16,000g, 30 min, 4°C). GSH and GSSG levels were determined using a GSH/GSSG assay kit (Biocytech GSH/GSSG-412, Oxis Research International Inc., Foster City, CA) according to the manufacturer’s instructions.
Quantification and statistical analysis
All data are represented as means ± SEM. Statistical significance was determined with a two-tailed Student’s t test for groups consisting of two experimental groups, or a one-way or two-way analysis of variance (ANOVA) with multiple comparisons (Tukey or Sidak, respectively) when comparing three or more experimental groups or a two-way ANOVA when comparing two experimental groups with multiple conditions. The level of significance was set at P < 0.05: * or #P < 0.05, ** or ##P < 0.01, ***P < 0.001, and ****P < 0.0001. Statistical details for individual experiments such as exact values of n can be found in figures and figure legends.
Supplementary Material
Acknowledgments
We thank R. S. Lee-Young and J. Eliades (Monash Metabolic Phenotyping Facility), M. J. DeVeer (Monash Biomedical Imaging), and the Monash Proteomics and Metabolomics Facility for technical support. This study used BPA-enabled (Bioplatforms Australia)/NCRIS-enabled (National Collaborative Research Infrastructure Strategy) infrastructure located at the Monash Proteomics and Metabolomics Facility.
Funding: This work was supported by the National Health and Medical Research Council (NHMRC) of Australia to T.T. (APP1162798) and C.A.M. (APP1082253) and Diabetes Australia to T.T. (Y18G-TIGT).
Author contributions: T.T. conceived and conceptualized the study, designed the experiments, wrote the manuscript, and interpreted the data with intellectual input and approval from all authors. C.E.X. conceptualized and designed experiments, conducted and analyzed most of the experiments, and contributed to the reviewing and editing of the manuscript. Other authors performed and/or analyzed experiments and/or contributed to the reviewing and editing of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Correction (11 March 2022): In an earlier version, the authors erroneously labeled the Y-axis of Figure 1G. The figure has been corrected in the updated online and PDF versions. The change does not affect any of the results or conclusions in the article.
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
REFERENCES AND NOTES
- 1.Cartee G. D., Hepple R. T., Bamman M. M., Zierath J. R., Exercise promotes healthy aging of skeletal muscle. Cell Metab. 23, 1034–1047 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egan B., Zierath J. R., Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 17, 162–184 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Hawley J. A., Hargreaves M., Joyner M. J., Zierath J. R., Integrative biology of exercise. Cell 159, 738–749 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Holmstrom K. M., Kostov R. V., Dinkova-Kostova A. T., The multifaceted role of Nrf2 in mitochondrial function. Curr. Opin. Toxicol 1, 80–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomez-Cabrera M. C., Domenech E., Romagnoli M., Arduini A., Borras C., Pallardo F. V., Sastre J., Vina J., Oral administration of vitamin C decreases muscle mitochondrial biogenesis and hampers training-induced adaptations in endurance performance. Am. J. Clin. Nutr. 87, 142–149 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Ristow M., Zarse K., Oberbach A., Kloting N., Birringer M., Kiehntopf M., Stumvoll M., Kahn C. R., Bluher M., Antioxidants prevent health-promoting effects of physical exercise in humans. Proc. Natl. Acad. Sci. U.S.A. 106, 8665–8670 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merry T. L., Ristow M., Nuclear factor erythroid-derived 2-like 2 (NFE2L2, Nrf2) mediates exercise-induced mitochondrial biogenesis and the anti-oxidant response in mice. J. Physiol. 594, 5195–5207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loh K., Deng H., Fukushima A., Cai X., Boivin B., Galic S., Bruce C., Shields B. J., Skiba B., Ooms L. M., Stepto N., Wu B., Mitchell C. A., Tonks N. K., Watt M. J., Febbraio M. A., Crack P. J., Andrikopoulos S., Tiganis T., Reactive oxygen species enhance insulin sensitivity. Cell Metab. 10, 260–272 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uruno A., Yagishita Y., Katsuoka F., Kitajima Y., Nunomiya A., Nagatomi R., Pi J., Biswal S. S., Yamamoto M., Nrf2-mediated regulation of skeletal muscle glycogen metabolism. Mol. Cell. Biol. 36, 1655–1672 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yfanti C., Nielsen A. R., Akerstrom T., Nielsen S., Rose A. J., Richter E. A., Lykkesfeldt J., Fischer C. P., Pedersen B. K., Effect of antioxidant supplementation on insulin sensitivity in response to endurance exercise training. Am. J. Physiol. Endocrinol. Metab. 300, E761–E770 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Houstis N., Rosen E. D., Lander E. S., Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature 440, 944–948 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Hoehn K. L., Salmon A. B., Hohnen-Behrens C., Turner N., Hoy A. J., Maghzal G. J., Stocker R., Van Remmen H., Kraegen E. W., Cooney G. J., Richardson A. R., James D. E., Insulin resistance is a cellular antioxidant defense mechanism. Proc. Natl. Acad. Sci. U.S.A. 106, 17787–17792 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson E. J., Lustig M. E., Boyle K. E., Woodlief T. L., Kane D. A., Lin C. T., Price J. W. III, Kang L., Rabinovitch P. S., Szeto H. H., Houmard J. A., Cortright R. N., Wasserman D. H., Neufer P. D., Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. J. Clin. Invest. 119, 573–581 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H. Y., Lee J. S., Alves T., Ladiges W., Rabinovitch P. S., Jurczak M. J., Choi C. S., Shulman G. I., Samuel V. T., Mitochondrial-targeted catalase protects against high-fat diet-induced muscle insulin resistance by decreasing intramuscular lipid accumulation. Diabetes 66, 2072–2081 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fazakerley D. J., Chaudhuri R., Yang P., Maghzal G. J., Thomas K. C., Krycer J. R., Humphrey S. J., Parker B. L., Fisher-Wellman K. H., Meoli C. C., Hoffman N. J., Diskin C., Burchfield J. G., Cowley M. J., Kaplan W., Modrusan Z., Kolumam G., Yang J. Y., Chen D. L., Samocha-Bonet D., Greenfield J. R., Hoehn K. L., Stocker R., James D. E., Mitochondrial CoQ deficiency is a common driver of mitochondrial oxidants and insulin resistance. eLife 7, e32111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitaoka Y., Tamura Y., Takahashi K., Takeda K., Takemasa T., Hatta H., Effects of Nrf2 deficiency on mitochondrial oxidative stress in aged skeletal muscle. Physiol. Rep. 7, e13998 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiganis T., Reactive oxygen species and insulin resistance: The good, the bad and the ugly. Trends Pharmacol. Sci. 32, 82–89 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Sies H., Jones D. P., Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 21, 363–383 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Powers S. K., Jackson M. J., Exercise-induced oxidative stress: Cellular mechanisms and impact on muscle force production. Physiol. Rev. 88, 1243–1276 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pye D., Palomero J., Kabayo T., Jackson M. J., Real-time measurement of nitric oxide in single mature mouse skeletal muscle fibers during contractions. J. Physiol. 581, 309–318 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakellariou G. K., Vasilaki A., Palomero J., Kayani A., Zibrik L., McArdle A., Jackson M. J., Studies of mitochondrial and nonmitochondrial sources implicate nicotinamide adenine dinucleotide phosphate oxidase(s) in the increased skeletal muscle superoxide generation that occurs during contractile activity. Antioxid. Redox Signal. 18, 603–621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson T., Kabayo T., Ng R., Chamberlain J., McArdle A., Jackson M. J., Skeletal muscle contractions induce acute changes in cytosolic superoxide, but slower responses in mitochondrial superoxide and cellular hydrogen peroxide. PLOS ONE 9, e96378 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henriquez-Olguin C., Knudsen J. R., Raun S. H., Li Z., Dalbram E., Treebak J. T., Sylow L., Holmdahl R., Richter E. A., Jaimovich E., Jensen T. E., Cytosolic ROS production by NADPH oxidase 2 regulates muscle glucose uptake during exercise. Nat. Commun. 10, 4623 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bedard K., Krause K. H., The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol. Rev. 87, 245–313 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Martyn K. D., Frederick L. M., von Loehneysen K., Dinauer M. C., Knaus U. G., Functional analysis of Nox4 reveals unique characteristics compared to other NADPH oxidases. Cell. Signal. 18, 69–82 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Sakellariou G. K., Jackson M. J., Vasilaki A., Redefining the major contributors to superoxide production in contracting skeletal muscle. The role of NAD(P)H oxidases. Free Radic. Res. 48, 12–29 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Osorio Alves J., Matta Pereira L., Cabral Coutinho do Rego Monteiro I., Pontes Dos Santos L. H., Soares Marreiros Ferraz A., Carneiro Loureiro A. C., Calado Lima C., Leal-Cardoso J. H., Pires Carvalho D., Soares Fortunato R., Marilande Ceccatto V., Strenuous acute exercise induces slow and fast twitch-dependent NADPH oxidase expression in rat skeletal muscle. Antioxidants 9, 57 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sylow L., Nielsen I. L., Kleinert M., Moller L. L., Ploug T., Schjerling P., Bilan P. J., Klip A., Jensen T. E., Richter E. A., Rac1 governs exercise-stimulated glucose uptake in skeletal muscle through regulation of GLUT4 translocation in mice. J. Physiol. 594, 4997–5008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sylow L., Moller L. L. V., D’Hulst G., Schjerling P., Jensen T. E., Richter E. A., Rac1 in muscle is dispensable for improved insulin action after exercise in mice. Endocrinology 157, 3009–3015 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Done A. J., Traustadottir T., Nrf2 mediates redox adaptations to exercise. Redox Biol. 10, 191–199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pendyala S., Moitra J., Kalari S., Kleeberger S. R., Zhao Y., Reddy S. P., Garcia J. G., Natarajan V., Nrf2 regulates hyperoxia-induced Nox4 expression in human lung endothelium: Identification of functional antioxidant response elements on the Nox4 promoter. Free Radic. Biol. Med. 50, 1749–1759 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oh S., Komine S., Warabi E., Akiyama K., Ishii A., Ishige K., Mizokami Y., Kuga K., Horie M., Miwa Y., Iwawaki T., Yamamoto M., Shoda J., Nuclear factor (erythroid derived 2)-like 2 activation increases exercise endurance capacity via redox modulation in skeletal muscles. Sci. Rep. 7, 12902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malaguti M., Angeloni C., Garatachea N., Baldini M., Leoncini E., Collado P. S., Teti G., Falconi M., Gonzalez-Gallego J., Hrelia S., Sulforaphane treatment protects skeletal muscle against damage induced by exhaustive exercise in rats. J. Appl. Physiol. 107, 1028–1036 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Kuroda J., Ago T., Matsushima S., Zhai P., Schneider M. D., Sadoshima J., NADPH oxidase 4 (Nox4) is a major source of oxidative stress in the failing heart. Proc. Natl. Acad. Sci. U.S.A. 107, 15565–15570 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruning J. C., Michael M. D., Winnay J. N., Hayashi T., Horsch D., Accili D., Goodyear L. J., Kahn C. R., A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol. Cell 2, 559–569 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Merry T. L., Ristow M., Mitohormesis in exercise training. Free Radic. Biol. Med. 98, 123–130 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Specht K. S., Kant S., Addington A. K., McMillan R. P., Hulver M. W., Learnard H., Campbell M., Donnelly S. R., Caliz A. D., Pei Y., Reif M. M., Bond J. M., DeMarco A., Craige B., Keaney J. F. Jr., Craige S. M., Nox4 mediates skeletal muscle metabolic responses to exercise. Mol. Metab. 45, 101160 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ago T., Kuroda J., Pain J., Fu C., Li H., Sadoshima J., Upregulation of Nox4 by hypertrophic stimuli promotes apoptosis and mitochondrial dysfunction in cardiac myocytes. Circ. Res. 106, 1253–1264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hancock M., Hafstad A. D., Nabeebaccus A. A., Catibog N., Logan A., Smyrnias I., Hansen S. S., Lanner J., Schroder K., Murphy M. P., Shah A. M., Zhang M., Myocardial NADPH oxidase-4 regulates the physiological response to acute exercise. eLife 7, e41044 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCarthy J. J., Srikuea R., Kirby T. J., Peterson C. A., Esser K. A., Inducible Cre transgenic mouse strain for skeletal muscle-specific gene targeting. Skelet. Muscle 2, 8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada M., Iwata M., Warabi E., Oishi H., Lira V. A., Okutsu M., p62/SQSTM1 and Nrf2 are essential for exercise-mediated enhancement of antioxidant protein expression in oxidative muscle. FASEB J. 33, 8022–8032 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Nobrega-Pereira S., Fernandez-Marcos P. J., Brioche T., Gomez-Cabrera M. C., Salvador-Pascual A., Flores J. M., Vina J., Serrano M., G6PD protects from oxidative damage and improves healthspan in mice. Nat. Commun. 7, 10894 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furukawa S., Fujita T., Shimabukuro M., Iwaki M., Yamada Y., Nakajima Y., Nakayama O., Makishima M., Matsuda M., Shimomura I., Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Invest. 114, 1752–1761 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gurzov E. N., Tran M., Fernandez-Rojo M. A., Merry T. L., Zhang X., Xu Y., Fukushima A., Waters M. J., Watt M. J., Andrikopoulos S., Neel B. G., Tiganis T., Hepatic oxidative stress promotes insulin-STAT-5 signaling and obesity by inactivating protein tyrosine phosphatase N2. Cell Metab. 20, 85–102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flekac M., Skrha J., Hilgertova J., Lacinova Z., Jarolimkova M., Gene polymorphisms of superoxide dismutases and catalase in diabetes mellitus. BMC Med. Genet. 9, 30 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao K., Zhao G. M., Wu D., Soong Y., Birk A. V., Schiller P. W., Szeto H. H., Cell-permeable peptide antioxidants targeted to inner mitochondrial membrane inhibit mitochondrial swelling, oxidative cell death, and reperfusion injury. J. Biol. Chem. 279, 34682–34690 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Kelley D. E., He J., Menshikova E. V., Ritov V. B., Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51, 2944–2950 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Hancock C. R., Han D. H., Chen M., Terada S., Yasuda T., Wright D. C., Holloszy J. O., High-fat diets cause insulin resistance despite an increase in muscle mitochondria. Proc. Natl. Acad. Sci. U.S.A. 105, 7815–7820 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montgomery M. K., Turner N., Mitochondrial dysfunction and insulin resistance: An update. Endocr. Connect. 4, R1–R15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao H., Jedrychowski M. P., Schweppe D. K., Huttlin E. L., Yu Q., Heppner D. E., Li J., Long J., Mills E. L., Szpyt J., He Z., Du G., Garrity R., Reddy A., Vaites L. P., Paulo J. A., Zhang T., Gray N. S., Gygi S. P., Chouchani E. T., A quantitative tissue-specific landscape of protein redox regulation during aging. Cell 180, 968–983.e24 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smyrnias I., Zhang X., Zhang M., Murray T. V., Brandes R. P., Schroder K., Brewer A. C., Shah A. M., Nicotinamide adenine dinucleotide phosphate oxidase-4-dependent upregulation of nuclear factor erythroid-derived 2-like 2 protects the heart during chronic pressure overload. Hypertension 65, 547–553 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Finley L. W., Lee J., Souza A., Desquiret-Dumas V., Bullock K., Rowe G. C., Procaccio V., Clish C. B., Arany Z., Haigis M. C., Skeletal muscle transcriptional coactivator PGC-1α mediates mitochondrial, but not metabolic, changes during calorie restriction. Proc. Natl. Acad. Sci. U.S.A. 109, 2931–2936 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sczelecki S., Besse-Patin A., Abboud A., Kleiner S., Laznik-Bogoslavski D., Wrann C. D., Ruas J. L., Haibe-Kains B., Estall J. L., Loss of Pgc-1α expression in aging mouse muscle potentiates glucose intolerance and systemic inflammation. Am. J. Physiol. Endocrinol. Metab. 306, E157–E167 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.St-Pierre J., Drori S., Uldry M., Silvaggi J. M., Rhee J., Jager S., Handschin C., Zheng K., Lin J., Yang W., Simon D. K., Bachoo R., Spiegelman B. M., Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 127, 397–408 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Lu Z., Xu X., Hu X., Fassett J., Zhu G., Tao Y., Li J., Huang Y., Zhang P., Zhao B., Chen Y., PGC-1α regulates expression of myocardial mitochondrial antioxidants and myocardial oxidative stress after chronic systolic overload. Antioxid. Redox Signal. 13, 1011–1022 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loh K., Merry T. L., Galic S., Wu B. J., Watt M. J., Zhang S., Zhang Z. Y., Neel B. G., Tiganis T., T cell protein tyrosine phosphatase (TCPTP) deficiency in muscle does not alter insulin signalling and glucose homeostasis in mice. Diabetologia 55, 468–478 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dodd G. T., Andrews Z. B., Simonds S. E., Michael N. J., DeVeer M., Bruning J. C., Spanswick D., Cowley M. A., Tiganis T., A hypothalamic phosphatase switch coordinates energy expenditure with feeding. Cell Metab. 26, 375–393.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Dodd G. T., Kim S. J., Méquinion M., Xirouchaki C. E., Brüning J. C., Andrews Z. B., Tiganis T., Insulin signaling in AgRP neurons regulates meal size to limit glucose excursions and insulin resistance. Sci. Adv. 7, eabf4100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dodd G. T., Lee-Young R. S., Bruning J. C., Tiganis T., TCPTP regulates insulin signaling in agrp neurons to coordinate glucose metabolism with feeding. Diabetes 67, 1246–1257 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Zougman A., Selby P. J., Banks R. E., Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. Proteomics 14, 1006–1000 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14