Time-restricted feeding improves glucose homeostasis through epigenetic control of pancreatic β cell function.
Abstract
Circadian rhythm disruption (CD) is associated with impaired glucose homeostasis and type 2 diabetes mellitus (T2DM). While the link between CD and T2DM remains unclear, there is accumulating evidence that disruption of fasting/feeding cycles mediates metabolic dysfunction. Here, we used an approach encompassing analysis of behavioral, physiological, transcriptomic, and epigenomic effects of CD and consequences of restoring fasting/feeding cycles through time-restricted feeding (tRF) in mice. Results show that CD perturbs glucose homeostasis through disruption of pancreatic β cell function and loss of circadian transcriptional and epigenetic identity. In contrast, restoration of fasting/feeding cycle prevented CD-mediated dysfunction by reestablishing circadian regulation of glucose tolerance, β cell function, transcriptional profile, and reestablishment of proline and acidic amino acid–rich basic leucine zipper (PAR bZIP) transcription factor DBP expression/activity. This study provides mechanistic insights into circadian regulation of β cell function and corresponding beneficial effects of tRF in prevention of β T2DM.
INTRODUCTION
The circadian system coordinates ~24-hour periodicity in essential cellular, physiological, and behavioral functions and thus represents a fundamental component of animal homeostasis (1). In mammals, the circadian system is organized as a hierarchical oscillator network with the master pacemaker being localized in the suprachiasmatic nucleus (SCN) of the hypothalamus, which coordinates the network of peripheral tissue oscillators (2). A key property of the circadian system is the ability to integrate responses to environmental cues (e.g., zeitgebers) such as light and food, which tune circadian physiology to daily changes in the environment (2). Whereas light entrains the oscillator network within the SCN, food availability and corresponding fasting/feeding cycles are dominant zeitgebers of peripheral tissue oscillators (3–5).
At the cellular level, circadian rhythms are driven by cell-autonomous transcriptional-translational feedback loops. The core components of the primary feedback loop include transcriptional activators: brain muscle aryl nuclear translocase 1 (BMAL1) [encoded by aryl hydrocarbon receptor nuclear translocator like (Arntl)] and its heterodimeric partner, circadian locomotor output cycles kaput (CLOCK), which bind to E-box elements in the promoters of their own repressor genes Period 1-3 (Per1, Per2, and Per3) and Cryptochrome 1 and 2 (Cry1 and Cry2) (6). Once translated, PERIOD and CRYPTOCHROME proteins bind to the CLOCK:BMAL1 complex to repress their own transcription, thus completing the feedback loop (6). This primary feedback loop is further modulated by secondary regulatory loops involving the nuclear hormone transcription factors REV-ERBα/β and RAR-related orphan receptor α and β (RORα/β) as well as the proline and acidic amino acid–rich basic leucine zipper (PAR bZIP) transcription factors: D site of albumin promoter binding protein (DBP), thyrotroph embryonic factor (TEF), and hepatic leukemia factor (HLF) (7). Circadian transcription factors coordinate expression of thousands of genes that orchestrate circadian rhythmicity and optimize regulation of essential cellular functions such as substrate metabolism, cell cycle, and mitochondrial function (8–11).
Although the circadian system undoubtedly provides evolutionary advantage, modern lifestyle and work conditions impose temporal constraints that produce disruptions in daily light/dark, rest/activity, and fasting/feeding cycles, as seen in a large proportion of the population worldwide (1, 12, 13). Most notably, chronic circadian rhythm disruptions (CDs) are associated with increased incidence of metabolic diseases such as type 2 diabetes mellitus (T2DM) (14–17). Consistently, genetic disruption of the core elements of the molecular clock (e.g., Arntl and Clock) in mice leads to characteristic features of T2DM such as hyperglycemia and loss of circadian regulation of glucose tolerance and insulin secretion (18–20). While the link between circadian disruption and T2DM remains unclear, there is accumulating research suggesting that disrupted fasting/feeding circadian cycles are a key mediator of metabolic dysfunction (21). Consequently, reinforcement of circadian fasting/feeding patterns through time-restricted feeding (tRF) augments the magnitude of circadian gene expression and improves metabolic function as demonstrated in animal models (22–25) and, more recently, in human clinical trials (26).
Despite accumulating evidence of the broad health benefits of tRF on human health, the physiological and molecular mechanisms underlying these effects are not fully understood. Subsequently, in the current study, we performed an integrative study to analyze the systemic behavioral, physiological, transcriptomic, and single-cell epigenomic effects of isocaloric tRF under conditions of global circadian disruption in mice. Our study demonstrates that tRF prevents metabolic derangements associated with global CD induced by exposure to light at night, a condition that leads to desynchronization of circadian oscillators in the SCN, resulting in the loss of behavioral and endocrine circadian rhythms (27, 28). We found that the beneficial metabolic effects of tRF were mediated through enhancement of pancreatic β cell secretory function, restoration of normal β cell circadian transcriptional and epigenomic profiles, and, in particular, reestablishment of the circadian PAR bZip transcription factor DBP expression and activity in β cells.
RESULTS
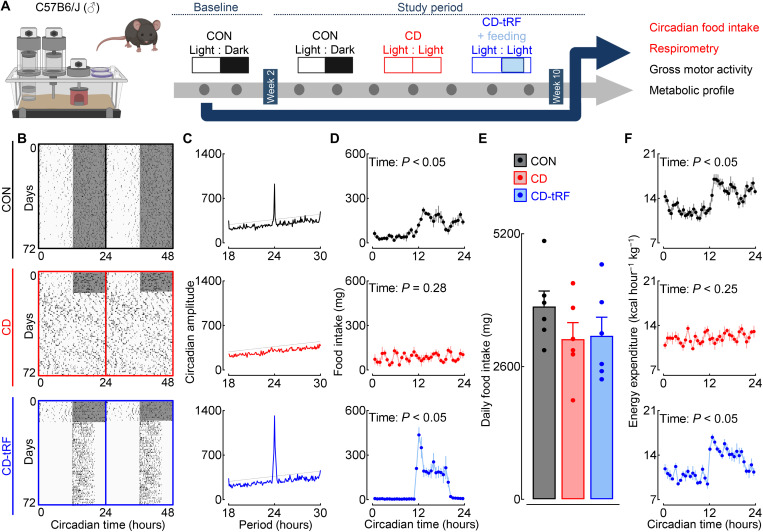
tRF restores circadian fasting/feeding cycles under conditions of global circadian disruption without altering cumulative food intake and activity
Circadian disruption associated with continuous exposure to light at night produces disruptions in daily light/dark, rest/activity, and fasting/feeding circadian cycles that lead to the dysregulation of glucose homeostasis and increased risk for development of T2DM (17). To gain insights into the biological mechanisms underlying this phenomenon, C57B6/J male mice were exposed for 8 weeks to either control conditions (CON; 12-hour light/12-hour dark circadian cycles), circadian disruption (CD; 24-hour continuous exposure to light), or circadian disruption with concomitant selective restoration of circadian fasting/feeding cycles through tRF (CD-tRF) (Fig. 1A). To validate our model, we used high-precision (3-mg resolution), continuous noninvasive food intake monitoring (5-min intervals for 10 weeks) to assess circadian feeding behavior in mice (Fig. 1, B and C). As expected, CON mice consumed ~80% of their food during the dark/active phase of the circadian cycle (P < 0.05), displaying a robust ~24-hour circadian period of feeding activity (Fig. 1, B to D). Consistent with previous observations, exposure to CD phenocopied the effects of SCN lesioning by initially increasing the period of circadian activity, followed by induction of near-complete circadian arrhythmicity in gross motor activity and feeding behavior (Fig. 1, B to D, and fig. S1).
Fig. 1. tRF restores circadian fasting/feeding cycles under conditions of global circadian disruption without altering cumulative food intake.
(A) Overview of study design. C57B6/J male mice were exposed for 8 weeks to either (i) control conditions (CON; 12-hour light/12-hour dark circadian cycles), (ii) circadian disruption (CD; 24-hour exposure to constant light), or (iii) circadian disruption with concomitant selective restoration of fasting/feeding cycles through tRF during CT12 to CT20 (CD-tRF). Experimental parameters assessed and displayed in the figure are highlighted in red. (B) Representative double-plotted foodograms and corresponding χ2 periodograms (C) in CON (highlighted in black), CD (red), and CD-tRF (blue) mice monitored for 2 weeks at baseline followed by 8 weeks of experimental conditions. Shaded areas represent periods of dark. (D) Average 24-hour food intake binned into 30-min intervals in CON (black), CD (red), and CD-tRF (blue) mice (repeated-measures one-way ANOVA, effect of time; n = 6 mice per condition). (E) Average cumulative 24-hour food intake in CON (black), CD (red), and CD-tRF (blue) mice (n = 6 mice per condition). (F) Average 24-hour energy expenditure binned into 30-min intervals in CON (black), CD (red), and CD-tRF (blue) mice (repeated-measures one-way ANOVA, effect of time; n = 6 per group). Respirometry recordings were made using a comprehensive laboratory animal monitoring system. All values represent means ± SEM.
By design, tRF led to restoration of rhythmic feeding cycles (P < 0.05) and enhancement of circadian amplitude of feeding activity in CD-tRF mice (Fig. 1, B to D). Total daily (24 hours) food intake was not significantly different between the groups (P > 0.05; Fig. 1E). In concert with normalized circadian feeding patterns, CD-tRF animals also demonstrated restoration of the circadian regulation of energy expenditure assessed by indirect calorimetry (effect of time, P < 0.05, CON and CD-tRF; Fig. 1F), which was disrupted under CD conditions (effect of time, P > 0.05; Fig. 1F). Despite restoration of circadian fasting/feeding cycles in CD-tRF, circadian activity patterns were significantly disrupted under both CD and CD-tRF conditions compared to CON (effect of time, P > 0.05 for CD and CD-tRF and P < 0.05 for CON; fig. S1, A and B). However, cumulative daily activity was not different between CON, CD, and CD-tRF conditions (P > 0.05; fig. S1C). These results indicate that exposure to CD results in disruption of light/dark, rest/activity, and fasting/feeding circadian cycles, whereas CD-tRF selectively restores daily cycles of fasting/feeding behavior without altering cumulative daily food intake and/or overall activity in mice.
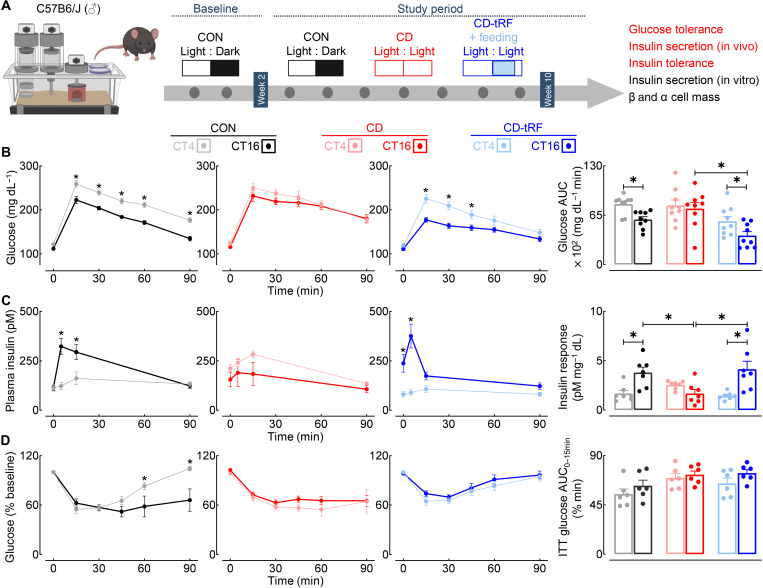
tRF restores diurnal glucose homeostasis through modulation of pancreatic β cell function under conditions of global circadian disruption in mice
Exposure to CD resulted in expected metabolic dysfunction characterized by a modest increase in total body mass (~10% versus CON, P < 0.05; fig. S2, A to C), fat mass, and abrogation of circadian rhythmicity in key metabolic intermediaries such as plasma glucose, insulin, and insulin-to-glucose ratio (effect of time, P > 0.05 for CD; fig. S2, D to F). Notably, restoration of fasting/feeding cycle in CD-tRF not only normalized circadian rhythmicity in plasma glucose and insulin concentrations (effect of time, P < 0.05 for CD-tRF; fig. S2, D to F) but also prevented the increase in body fat mass accumulation in response to CD (P < 0.05, CD-tRF versus CD; fig. S2C).
In light of this information, we next aimed to further delineate the physiological effects of CD and CD-tRF on the circadian control of glucose homeostasis by assessing the diurnal regulation of in vivo glucose tolerance, insulin secretion, and insulin sensitivity (Fig. 2A). All three parameters were examined at two distinct time points corresponding to the middle of the inactive/light [circadian time 4 (CT4)] and/or active/dark (CT16) circadian cycle in the CON group. The CON group demonstrated robust diurnal rhythmicity characterized by markedly enhanced glucose tolerance and β cell secretory function (insulin response) at the CT16 time point (P < 0.05, CT4 versus CT16; Fig. 2, B and C). Notably, diurnal regulation of glucose tolerance and β cell secretory function was lost under CD conditions (P > 0.05, CT4 versus CT16; Fig. 2, B and C) but was fully restored under CD-tRF (P < 0.05, CT4 versus CT16; Fig. 2, B and C). Furthermore, glucose tolerance and insulin secretory response were significantly enhanced (~2-fold) in CD-tRF versus CD group at the CT16 time point (P < 0.05 CD-tRF versus CD; Fig. 2, B and C). In our study CD-tRF failed to significantly affect insulin tolerance, suggesting that tRF-dependent restoration of fasting/feeding cycles primarily enhanced β cell secretory functionality (Fig. 2D). To further confirm this observation, we isolated pancreatic islets from CON, CD, and CD-tRF mice at CT4 and CT16 time points and subsequently assessed the diurnal capacity for glucose-stimulated insulin secretion (GSIS) in vitro (fig. S3, A and B). Consistent with in vivo observations, isolated CON islets exhibited an enhanced capacity (~2-fold) for GSIS at CT16 compared to the CT4 time point (P < 0.05, CT4 versus CT16; fig. S3B). In concert with previous findings, exposure to CD abrogated (P > 0.05 for CT4 versus CT16; fig. S3B), whereas CD-tRF restored, the diurnal regulation of GSIS in isolated islets (P < 0.05 for CT4 versus CT16; fig. S3B). Last, note that we did not observe differences in the overall pancreatic β or α cell mass or changes in islet cell composition between the three groups (P > 0.05 for all groups; fig. S3C), providing further evidence for tRF-dependent circadian control of β cell secretory function.
Fig. 2. tRF preserves diurnal glucose tolerance and β cell function under conditions of global circadian disruption in mice.
(A) Overview of study design. Diurnal glucose tolerance, in vivo and in vitro glucose-stimulated insulin secretion (GSIS), insulin tolerance, and β/α cell mass were assessed in CON, CD, and CD-tRF mice at the end of the 8-week study period. Experimental parameters assessed and displayed in the figure are highlighted in red. (B) Blood glucose concentrations sampled during intraperitoneal glucose tolerance tests (GTT) and corresponding glucose area under the curve (AUC) during GTT performed at CT4 and CT16 in CON (gray/black), CD (pink/red), and CD-tRF (light blue/dark blue) mice (n = 9 mice per condition). *P < 0.05 denotes statistical significance for CT4 versus CT16 (paired, two-tailed Student’s t test with Holm-Sidak correction for intragroup comparison and one-way ANOVA with Tukey correction for intergroup comparison). (C) Plasma insulin concentrations collected at 0, 5, 15, and 90 min after glucose administration during GTT and corresponding insulin response (expressed as insulin AUC/glucose AUC during GTT) performed at CT4 and CT16 in CON, CD, and CD-tRF mice (n = 7 mice per condition). *P < 0.05 denotes statistical significance for CT4 versus CT16 (paired, two-tailed Student’s t test with Holm-Sidak correction for intragroup comparison and one-way ANOVA with Tukey correction for intergroup comparison). (D) Blood glucose concentrations sampled during intraperitoneal insulin tolerance tests (ITT) and corresponding glucose AUC from 0 to 15 min performed at CT4 and CT16 in CON, CD, and CD-tRF mice (n = 6 mice per condition). *P < 0.05 denotes statistical significance for CT4 versus CT16 (paired, two-tailed Student’s t test with Holm-Sidak correction). All values represent means ± SEM.
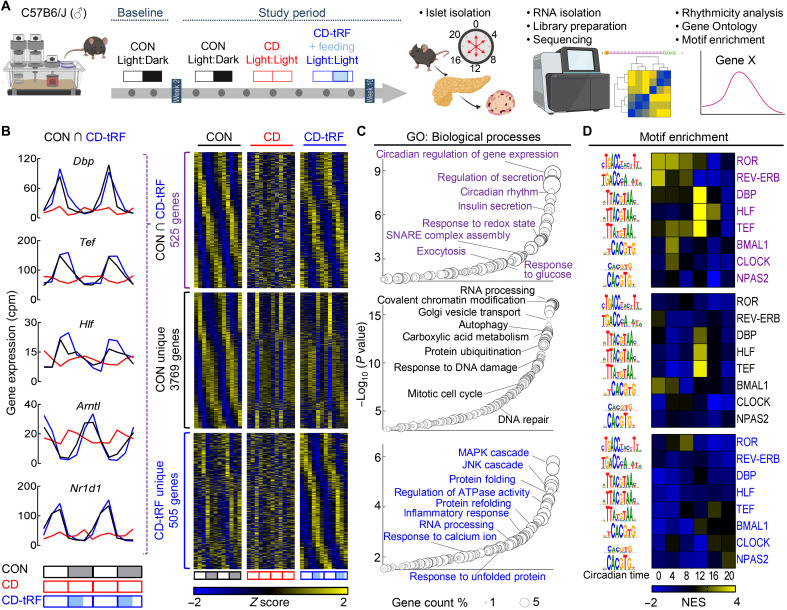
tRF restores circadian islet transcriptional profile under conditions of global circadian disruption in mice
To begin investigating the mechanistic link between the restoration of fasting/feeding cycles and the corresponding circadian control of islet secretory function, we next assessed the islet’s circadian transcriptional profile by performing RNA sequencing (RNA-seq) of CON, CD, and CD-tRF islets isolated at 4-hour intervals for two independent replicates in vivo (Fig. 3A and fig. S4A). Subsequently, using the harmonic regression algorithm ARSER [false discovery rate (FDR) ≤ 0.10] (29–31), we identified 4294 transcripts with significant circadian rhythmicity in mRNA expression under CON conditions, which is consistent with previous observations reporting ~4000 to 6000 circadian cycling transcripts in mouse islets/β cells (32, 33). Exposure to CD resulted in a complete loss of circadian rhythmicity in islet mRNA expression, whereas exposure to CD-tRF was able to restore the circadian rhythmicity for 525 of these transcripts (Fig. 3B, data S1, and fig. S4B). Gene ontology (GO) of these 525 transcripts revealed the top enriched pathways to be associated with regulation of circadian rhythms (GO: ~circadian regulation of gene expression, GO: ~circadian rhythm; Fig. 3C) and β cell insulin secretory function (GO: ~regulation of secretion, GO: ~insulin secretion; Fig. 3C). Most notably, tRF resulted in complete restoration in the rhythmic expression of key circadian transcription factors such as the PAR bZip family (Dbp, Tef, and Hlf), Arntl encoding for BMAL1, and nuclear receptor subfamily 1 group D member 1 (Nr1d1) encoding for REV-ERBα (Fig. 3B). Moreover, tRF preserved rhythmic expression of transcripts essential for insulin secretion such as glucose-6-phosphatase catalytic subunit 2 (G6pc2), synaptosomal-associated protein 25 (Snap25), vesicle-associated membrane protein 2 (Vamp2), and NK6 homeobox 1 (Nkx6.1).
Fig. 3. tRF preserves circadian regulation of gene expression in islets under conditions of global circadian disruption in mice.
(A) Overview of study design. Islets were isolated for RNA-seq from CON, CD, and CD-tRF mice at 4-hour intervals for two independent replicates in vivo. Rhythmic circadian gene expression was determined using MetaCycle (30) with GO and motif enrichment assessed using WebGestalt and i-Cis Target, respectively. (B) Heatmaps representing genes commonly cycling (FDR ≤ 0.1) in CON and CD-tRF islets (top), uniquely cycling in CON islets (middle), and uniquely cycling in CD-tRF islets (bottom). Gene expression was normalized by row Z score. Each column depicts one time point, sampled every 4 hours, starting at CT0 for two independent replicates (n = 12 independent samples per group from n = 2 mice per time point/per group). Inset shows circadian gene expression profiles for Dbp, Tef, Hlf, and Arntl encoding for BMAL1 and Nr1d1 encoding for REV-ERBα commonly cycling in CON and CD-tRF but arrhythmic under CD conditions. (C) Enriched GO: Biological Processes annotated from genes commonly cycling in CON and CD-tRF islets (top), uniquely cycling in CON islets (middle), and uniquely cycling in CD-tRF islets (bottom). Key pathways associated with β cell function are highlighted. (D) Heatmaps representing predicted i-Cis Target enrichment of circadian regulatory motifs (colored by Z-normalized enrichment score) in cis-regulatory regions of rhythmic genes with a peak phase centering around 4-hour intervals starting from CT0. Motif enrichment analysis was assessed in genes commonly cycling in CON and CD-tRF islets (top), uniquely cycling in CON islets (middle), and uniquely cycling in CD-tRF islets (bottom). Highly enriched motifs within cis-regulatory regions of cycling genes are colored yellow.
We also found that there were 3769 islet transcripts that demonstrated circadian mRNA cycling under CON conditions but remained arrhythmic under both CD and CD-tRF (Fig. 3, B and C, and fig. S4B). These tRF-independent transcripts annotated to biological pathways regulating cell survival and proliferation such as ~RNA processing, ~autophagy, ~response to DNA damage, and ~mitotic cell cycle (Fig. 3, B and C). Meanwhile, tRF appeared to induce de novo rhythmicity in a subset of 505 genes that were arrhythmic under both CON and CD conditions (Fig. 3, B and C). These transcripts were enriched for genes annotating to pathways associated with cellular stress such as the ~mitogen-activated protein and c-Jun N-terminal kinase cascades (MAPK and JNK, respectively), ~protein folding, and ~rRNA (ribosomal RNA) processing (Fig. 3, B and C).
Next, we set out to extend these findings by identifying potential regulatory elements that mediated temporal control over circadian islet transcription in response to CD and CD-tRF. In the 525 transcripts cycling in CON and restored in CD-tRF islets, we found that genes with a peak phase at the onset of feeding (e.g., CT12) were highly enriched for DNA binding motifs associated with PAR bZip transcription factor family activity (Fig. 3D and fig. S5), highlighting the potential role for these transcription factors in the β cell response to tRF. Conversely, we observed enrichment of DNA binding motifs for upstream clock elements such as REV-ERB, ROR, and BMAL1 at genes with a peak phase at CT0 and CT4 (Fig. 3D and fig. S5). Consistent with GO, genes uniquely cycling in CD-tRF islets exhibited enrichment for motifs associated with the erythroblast transformation specific (ETS) transcription factor family [ETS-like protein (ELK) and ETS variant transcription factor (ETV)] at CT12 and CT16, which have been demonstrated to be activated in response to cellular stress downstream of MAPK and JNK pathways (fig. S5) (34, 35). Overall, these data suggest that tRF-dependent reestablishment of circadian glucose tolerance and β cell function is mediated, in part, through restoring the islet’s circadian transcriptional identity and, particularly, the temporal expression and activity of the PAR bZip transcription factor family (e.g., Dbp, Tef, and Hlf).
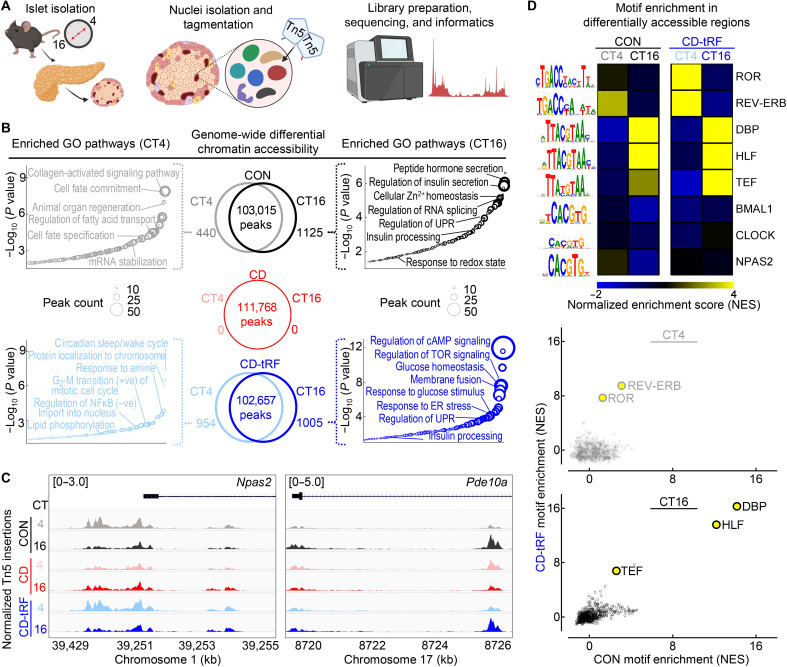
tRF maintains diurnal islet epigenomic identity under conditions of global circadian disruption in mice
We next set out to determine whether the observed transcriptional changes were regulated at the level of the epigenome by investigating which transcription factors could mediate the islet response to tRF. To accomplish this at a genome-wide level in CON, CD, and CD-tRF islets, we diurnally performed the assay for transposase-accessible chromatin using sequencing (ATAC-seq) at CT4 and CT16 time points (Fig. 4A and fig. S6A). Globally, we observed ~100,000 peaks per condition and time point (Fig. 4B) that each annotated to a similar proportion of genomic regions (fig. S6B) and were equally enriched for transcription factor motifs associated with islet cell identity such as NKX6.1, forkhead box A1 (FOXA1), and neuronal differentiation 1 (NEUROD1) (fig. S6C). Upon differential analysis of diurnally accessible chromatin regions [fold change (FC) > 1.5, FDR < 0.1], we found that exposure to CD completely abrogated diurnal regulation of islet chromatin accessibility (Fig. 4B), providing further evidence supporting the loss of circadian rhythmicity of the CD islet transcriptome. In contrast, in CON and CD-tRF islets, ~1000 chromatin regions were differentially accessible at CT16 with 440 and 954 differentially accessible at the CT4 time point, respectively (Fig. 4B). Upon annotating the differentially accessible regions at CT16 in CON and CD-tRF, we found that key pathways regulating β cell insulin secretory function were enriched within these regions such as ~regulation of insulin secretion, ~insulin processing, ~regulation of cyclic adenosine monophosphate (cAMP) signaling, and ~response to glucose stimulus (Fig. 4, B and C). On the other hand, differentially accessible chromatin regions enriched at the CT4 time point in CON and CD-tRF islets annotated to biological pathways associated with cell survival, maintenance, and proliferation including ~regulation of cell fate commitment, ~regulation of fatty acid transport, ~circadian sleep/wake cycle, and ~positive G2-M transition of mitotic cell cycle (Fig. 4, B and C). To extend these findings, we next assessed which cis-regulatory elements are enriched within these differentially accessible chromatin regions to define potential transcription factors regulating the diurnal response to tRF. Consistent with motif enrichment observed in transcriptomic analysis, we identified that motifs for PAR bZip (DBP, HLF, and TEF) transcription factors were among the topmost enriched motifs within differentially accessible regions at CT16 in both CON and CD-tRF islets (Fig. 4D). At CT4, we observed enrichment for REV-ERB and ROR binding motifs in the differentially accessible chromatin regions, also mirroring data obtained through our transcriptomic analysis (Fig. 4D).
Fig. 4. tRF maintains diurnal regulation of chromatin accessibility in islets under conditions of global circadian disruption in mice.
(A) Overview of study design. Islet cell nuclei were isolated from CON, CD, and CD-tRF mice at CT4 and CT16 time points and subjected to assay for transposase-accessible chromatin using sequencing (ATAC-seq). (B) Venn diagrams depict diurnal (CT4 versus CT16) differentially accessible chromatin regions (FC > 1.5; FDR < 0.1 using Benjamini-Hochberg correction) from ATAC-seq performed in CON (black), CD (red), and CD-tRF (blue) isolated islets (n = 2 independent samples per time point/group from n = 2 mice per time point/group) (center). Enriched GO: Biological Process pathways annotated from differentially accessible chromatin regions at CT4 and CT16 in CON (left) and CD-tRF (right). Key pathways associated with β cell function are highlighted. (C) Integrative Genomics Viewer browser tracks of representative open chromatin region at the Npas2 (left) and Pde10a (right) transcriptional start site CON (gray/black), CD (pink/red), and CD-tRF (light blue/dark blue) samples for CT4 and CT16. Signal is normalized to counts per million reads (cpm) from 0 to 5 cpm (Pde10a) and 0 to 3 cpm (Npas2). Shaded regions represent areas of accessible chromatin. (D) Heatmap representing predicted i-Cis Target enrichment of circadian regulatory motifs (colored by Z-normalized enrichment score) in differentially accessible chromatin regions at CT4 and CT16 time points from CON and CD-tRF islets. Highly enriched motifs in differentially accessible chromatin regions are colored yellow (top). Correlation comparing normalized motif enrichment score from CON and CD-tRF islets isolated at CT4 (middle) and CT16 (bottom). Highly correlated motifs common to CON and CD-tRF are denoted in yellow.
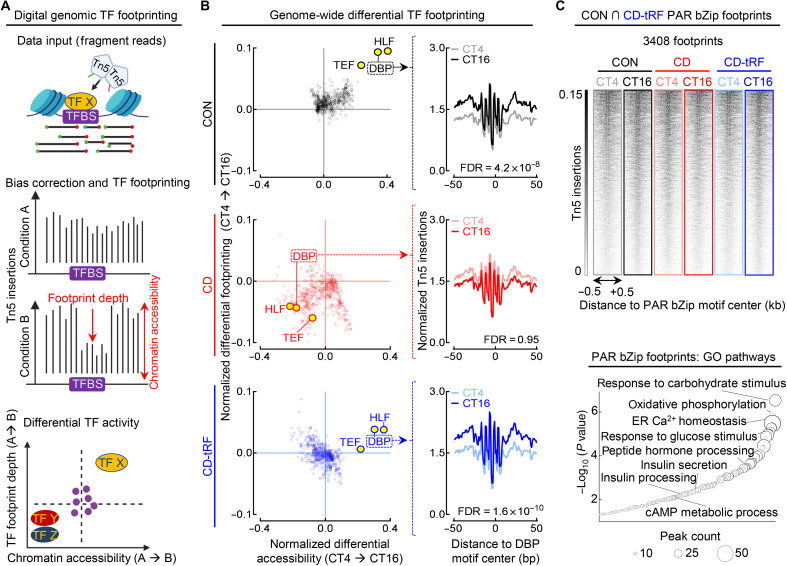
To further validate these findings, we next used an unbiased genome-wide bioinformatic approach, digital genomic transcription factor footprinting, to quantify diurnal changes in global transcription factor activity in islets isolated from CON, CD, and CD-tRF mice (Fig. 5). Since bound transcription factors impede DNA cleavage, this creates areas of lower Tn5 insertion in larger accessible chromatin regions (e.g., “footprints”) (Fig. 5A). In turn, computational approaches have been developed/validated to identify and differentially analyze transcription factor binding and activity between biological samples at a genome-wide level (36, 37). Using this method, we observed significant differences in PAR bZip transcription factor activity at CT16 versus CT4 in CON islets (FDR ≤ 0.05 for DBP, HLF, and TEF; Fig. 5B), which was completely disrupted with CD (FDR > 0.05) and fully restored with tRF (FDR ≤ 0.05 for DBP, HLF, and TEF). Notably, PAR bZip footprints for DBP, TEF, and HLF were all found to be in the top 10 most significantly diurnal transcription factors under both CON and CD-tRF conditions out of the 681 transcription factors tested [BagFootR default; Catalog of Inferred Sequence Binding Preferences (cis-BP) motifs]. To investigate the potential biological role of PAR bZip transcription factor activity in islets, we annotated the 3408 footprints where DBP, TEF, and HLF were predicted to bind in both CON and CD-tRF islets (Fig. 5C). We found that these regions represented loci associated with key pathways instrumental for β cell function such as ~response to carbohydrate stimulus, ~oxidative phosphorylation, ~insulin secretion, and ~insulin processing (Fig. 5C). We subsequently investigated potential unique roles for each PAR bZip transcription factor. By annotating unique DBP, TEF, and HLF footprints commonly enriched in CON and CD-tRF islets, we found that while DBP and TEF footprints annotated to regions associated with regulation of insulin secretion and synthesis (fig. S7, A to D), HLF produced footprints in genomic regions associated with cell survival and proliferation (fig. S7, E and F). Together, this suggests that DBP and TEF may play a larger role in regulating circadian islet secretory function, especially given their ~5-fold higher peak-phase mRNA expression relative to Hlf under CON conditions.
Fig. 5. Digital genomic footprinting reveals that tRF protects diurnal PAR bZip transcription factor activity in islets under conditions of circadian disruption.
(A) Overview of transcription factor (TF) footprinting. Regulatory TFs protect against tagmentation, resulting in a signal decrease (footprint), and allow for identification of bound TFs. Differential changes in footprint depth or accessibility are associated with increased/decreased TF activity between conditions. (B) Global changes in TF footprint depth from CT4 to CT16 representing diurnal differential TF activity (y axis) and flanking accessibility from CT4 to CT16 representing diurnal differential global chromatin accessibility around each TF motif tested (x axis) in CON (black), CD (red), and CD-tRF (blue) islets. PAR bZip (DBP, HLF, and TEF) TFs are highlighted in yellow (left). Mean diurnal DBP footprints in CON (gray/black), CD (pink/red), and CD-tRF (light blue/dark blue) islets at CT4 and CT16. Signal is normalized to the total read depth of each sample and is plotted ±50 bp from DBP motif center (FDR = Hotelling’s T squared test with Benjamini-Hochberg correction) (right). (C) ATAC-seq signal at PAR bZip footprints (DBP, HLF, and TEF) detected in both CON and CD-tRF samples and subsequently visualized in CON (gray/black), CD (pink/red), and CD-tRF (light blue/dark blue) samples at CT4 and CT16. Signal is normalized to reads per genomic content and is plotted ±0.5 kb from PAR bZip motif center. Shaded dark regions represent areas of accessible chromatin (top). Enriched GO: Biological Process pathways annotated from common PAR bZip footprints (DBP, HLF, and TEF) detected in both CON and CD-tRF. Key pathways associated with β cell secretory function are highlighted (bottom). TFBS, Transcription factor binding sites.
Circadian PAR bZip transcription factor activation defines a subset of β cells in a state of increased insulin production and secretion
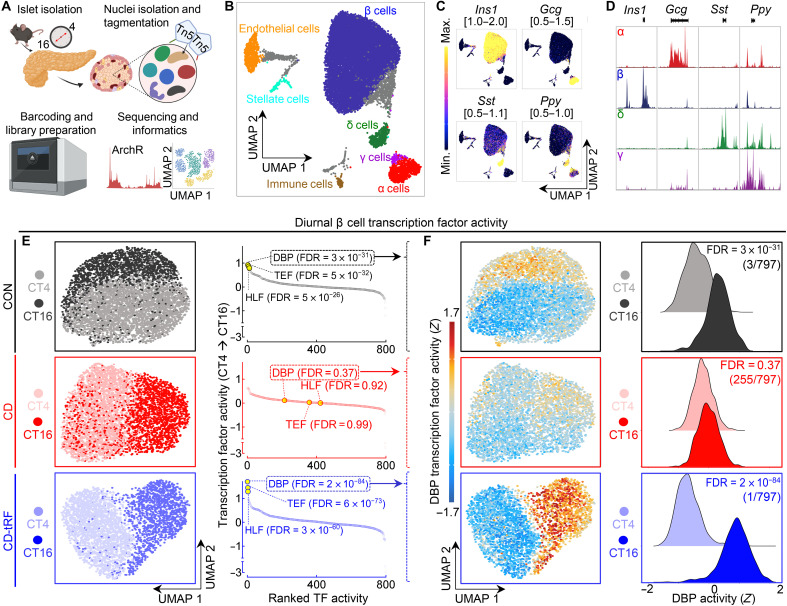
Given the evidence that the islet response to tRF is associated with the activation of PAR bZip transcription factors, we next set out to (i) confirm these findings at the level of individual β cells and (ii) determine whether there is heterogeneity in the circadian regulation of PAR bZip activity among populations of β cells. To accomplish this, we performed single-cell ATAC-seq (scATAC-seq) in CON, CD, and CD-tRF dispersed islet cells isolated at CT4 and CT16 time points (Fig. 6A and fig. S8A). Following stringent quality control, we recovered a total of 16,470 cells with a median transcription start site (TSS) enrichment score of 19.9 and a median unique fragment count of 12,009 (fig. S8B). The cells clustered into 17 groups using iterative latent semantic indexing and were subsequently assigned to pancreatic cell types based on gene activity scores (chromatin accessibility) of canonical cell-specific markers (fig. S8, C to E) (38–40). We were able to successfully identify the main islet cell types: β cells (78%), α cells (5.6%), δ cells (3.0%), and γ cells (0.5%), along with immune (0.7%), endothelial (3.2%), and stellate cells (1.0%) (Fig. 6, B to D). Four clusters were enriched for multiple cell-specific markers and were considered likely unclassified doublets for downstream analysis (41). Following clustering, cell identification, and peak calling, we proceeded to use ChromVar (42), an R package that calculates transcription factor motif activity at a single-cell resolution with the goal of understanding the heterogeneous enrichment of diurnal transcription factor activity in CON, CD, and CD-tRF β cells. Consistent with bulk ATAC-seq data, we found that the PAR bZip transcription factor family was among the top diurnally active transcription factors (out of 797 tested; ArchR default cis-BP transcription factor motifs) during feeding at CT16 relative to CT4 in CON β cells, with DBP as the 3rd most active (FDR = 3 × 10−31 CT16 versus CT4), TEF as the 8th most active (FDR = 5 × 10−32), and HLF as the 12th most active transcription factor at CT16 (FDR = 5 × 10−26; Fig. 6, E and F). Conversely, diurnal PAR bZip activity in single β cells was abrogated with CD (all FDR > 0.05; CT16 versus CT4), with DBP, TEF, and HLF ranking 215th, 358th, and 422nd most active, respectively, at CT16. tRF completely counteracted these effects and enhanced the activity of DBP (1st; FDR = 2 × 10−84 CT16 versus CT4), TEF (3rd; FDR = 6 × 10−73), and HLF (4th; FDR = 3 × 10−60) at CT16 in CD-tRF β cells.
Fig. 6. tRF promotes diurnal PAR bZip transcription factor activity in single β cells under conditions of circadian disruption.
(A) Overview of study design. Islet cell nuclei were isolated from CON, CD, and CD-tRF mice at CT4 and CT16 and subjected to single-cell ATAC-sequencing (scATAC-seq). (B) Uniform Manifold Approximation and Projection (UMAP) of 16,470 islet cells with each dot representing one cell. Identified islet cell types are labeled (n = 2 mice per condition), and gray shaded cells are unidentified because of activity of multiple canonical islet cell markers. (C) UMAP of gene activity scores for canonical endocrine islet cell markers Insulin 1 (Ins1), Glucagon (Gcg), Somatostatin (Sst), and Pancreatic Polypeptide (Ppy). Gene activity score ranges are noted on top of each UMAP. Yellow represents high activity (open chromatin) at noted gene loci, while blue/purple represents low activity (closed chromatin). (D) Corresponding track plotting of pseudo-bulk chromatin accessibility at Ins1, Gcg, Sst, and Ppy gene loci in identified endocrine cells (β, α, δ, and γ cells). Gene loci are depicted below each genome-wide browser track. Signal is normalized to maximal accessibility of each loci across the displayed cell types. (E) UMAP of diurnal β cells under CON (gray/black), CD (pink/red), and CD-tRF (light blue/dark blue) conditions at CT4 and CT16 (left). Global diurnal changes in Z-normalized TF activity (out of 797 cis-BP TFs tested) in CON, CD, and CD-tRF β cells (FDR = Wilcoxon rank sum test with Benjamini-Hochberg correction) (right). Note that PAR bZip (DBP, HLF, and TEF) TFs are highlighted in yellow with corresponding FDR values presented in parentheses. (F) UMAP of Z-normalized diurnal β cell DBP activity in CON, CD, and CD-tRF β cells assessed using ChromVar at CT4 and CT16 (left). Histograms depicting diurnal differences in Z-normalized DBP activity in CON, CD, and CD-tRF β cells at CT4 and CT16 (right).
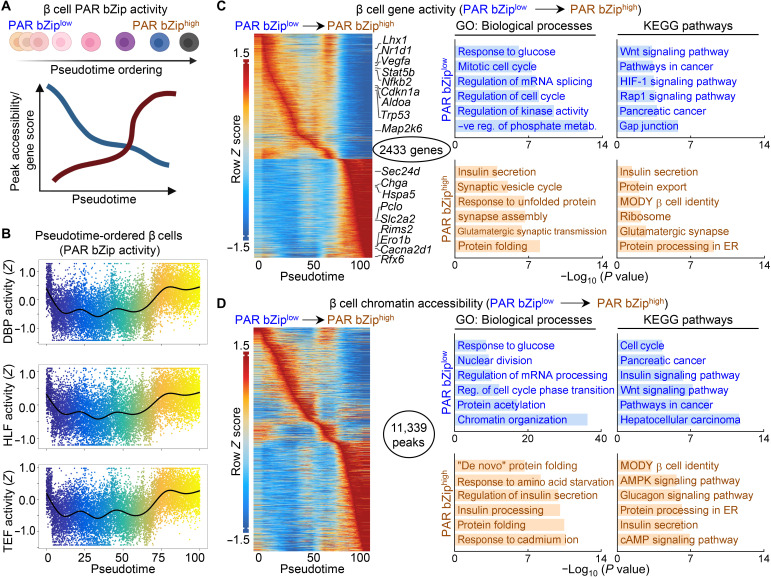
Notably, during our analysis, we observed both intersample and intrasample heterogeneity of PAR bZip activity in β cells and thus speculated that the magnitude of PAR bZip activity could be related to states of β cell secretory function. Using pseudotime trajectory analysis, we grouped β cells based on quartiles of PAR bZip activity from low (bottom quartile) to intermediate (second and third quartiles) and high activity (top quartile) and subsequently ordered them along this trajectory based on their Euclidean distance between the states (Fig. 7A) (40). Following trajectory analysis, we were successfully able to identify a clear bimodal pattern in PAR bZip activity (Fig. 7B). Correspondingly, we identified a subset of chromatin regions and highly active genes enriched in β cells with low (PAR bZiplow) and high (PAR bZiphigh) Par bZip activity (Fig. 7, C and D). Genes highly active in PAR bZiphigh β cells included those essential for insulin processing and secretion such as solute carrier family 2, member 2 (Slc2a2) encoding key glucose transporter 2 (GLUT2), heat shock protein A5 (Hspa5) encoding immunoglobulin (Ig) heavy-chain binding protein (Bip), chromogranin A (Chga), regulating synaptic membrane exocytosis 2 (Rims2), and endoplasmic oxidoreductin-1–like protein B (Ero1lb), while genes associated with β cell stress including nuclear factor kappa B subunit 2 (Nfkb2), signal transducer and activator of transcription 5B (Stat5b), and vascular endothelial growth factor A (Vegfa) were highly active in PAR bZiplow β cells. Annotation of highly active genes identified in PAR bZiphigh β cells against GO: Biological Process and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways revealed enrichment for pathways such as ~insulin secretion, ~protein processing/folding in the endoplasmic reticulum (ER), ~β cell identity [maturity onset diabetes of the young (MODY)], and ~synaptic vesicle cycle (Fig. 7C). Conversely, pathways enriched in genes active in PAR bZiplow cells included ~regulation of cell cycle, ~mRNA splicing, ~pancreatic cancer, and ~hypoxia-inducible factor 1 (HIF-1) signaling (Fig. 7C). Similar pathways were differentially enriched in chromatin regions more accessible in PAR bZiplow versus PAR bZiphigh β cells and vice versa (Fig. 7D). Overall, these data suggest that PAR bZip transcription factor activation plays an important role in the β cell’s response to tRF and is likely part of a transcriptional program activated in a subset of β cells to maintain adequate circadian control of insulin production and secretion in response to daily fasting/feeding cycles.
Fig. 7. Circadian PAR bZip transcription factor activation defines a subset of β cells in a state of increased insulin production and secretion.
(A) Overview of computational approach. All identified β cells by scATAC-seq were subjected to semisupervised pseudotime ordering by PAR bZip TF activity. β cells were grouped into quartiles of PAR bZip activity from low (bottom quartile) to intermediate (second and third quartile) and high (top quartile). Accessible chromatin regions and gene activation scores that varied across pseudotime were identified and annotated. (B) Trajectory analysis of PAR bZip TF (DBP, HLF, and TEF) activity along pseudotime in β cells from CON, CD, and CD-tRF mice isolated at CT4 and CT16 and subjected to scATAC-seq. (C and D) Heatmaps depicting the top 10% of gene activity scores (C) (left) and the top 5% of accessible chromatin regions (D) (left), which varied along pseudotime. Key genes controlling β cell function, identity, and stress are highlighted in (C) (left). Ontology of enriched GO: Biological Process and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways from genes (C) (right) and annotated chromatin regions (D) (right) with maximal activity/accessibility in PAR bZiplow β cells (top in blue; pseudotime, 5 to 50) or in PAR bZiphigh cells (bottom in orange; pseudotime, 70 to 100).
Circadian PAR bZip transcription factor DBP regulates GSIS and β cell functional identity
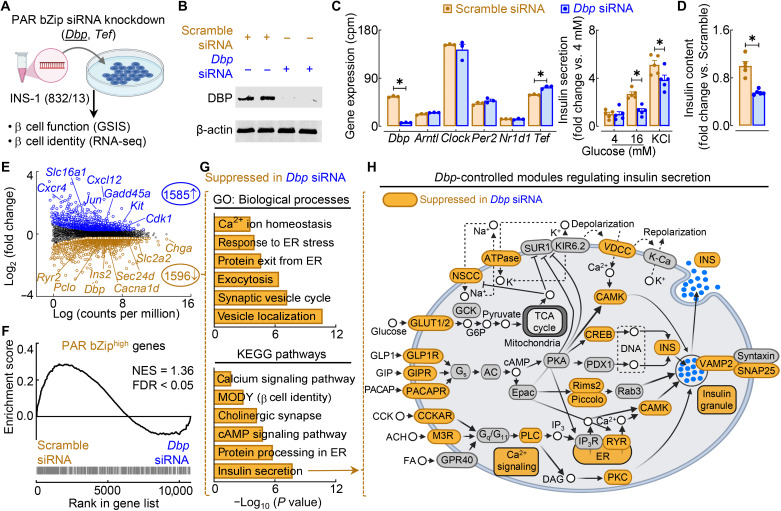
Next, we aimed to determine whether circadian PAR bZip transcription factors play a direct role in the regulation of β cell insulin secretory function and identity. To accomplish this, we used a common glucose-responsive β cell line (INS-1 832/13) in which we individually knocked down the PAR bZip transcription factors Dbp and Tef using a small interfering RNA (siRNA) approach (Fig. 8, A to C, and fig. S9, A to C) and subsequently assessed β cell function and transcriptional profiles. Knockdown of Dbp did not significantly affect the mRNA expression of core circadian clock elements Arntl, Clock, Per2, and Nr1d1 (Fig. 8C). Knockdown of Dbp in β cells resulted in marked blunting of GSIS, maximal insulin secretory capacity, and total insulin content relative to control (scrambled siRNA–treated) cells (P < 0.05 versus scramble; Fig. 8D). In contrast, knockdown of Tef alone did not affect the normal regulation of β cell function or insulin content (fig. S9D).
Fig. 8. PAR bZip transcription factor DBP is required for GSIS through control of key gene regulatory modules underlying the insulin secretory pathway.
(A) Overview of study design. PAR bZip TFs (Dbp and Tef) were knocked down using siRNA in INS-1 832/13 rat β cells. GSIS was assessed in each knockdown, and RNA-seq was performed in Dbp siRNA–treated β cells. (B) Representative Western blot of DBP and β-actin expression in control (20 nM scrambled siRNA) and Dbp knockdown (20 nM Dbp siRNA) β cells (n = 2 independent experiments). (C) Circadian clock gene expression in scramble siRNA– and Dbp siRNA–treated cells normalized to cpm. *FDR < 0.05 denotes statistical significance (n = 3 independent samples per group). Values represent means ± SEM. (D) GSIS (16 mM glucose) and maximal insulin secretion (30 mM KCl) of scramble siRNA– and Dbp siRNA–treated β cells normalized to basal insulin secretion (4 mM glucose; left). Total insulin content normalized to scramble control (right). *P < 0.05 denotes statistical significance (unpaired, two-tailed Student’s t test; n = 5 independent experiments per group). Values represent means ± SEM. (E) Volcano plot identifying differentially expressed genes (FC > 1.2; FDR < 0.05 by Benjamini-Hochberg method) from RNA-seq of scramble siRNA– versus Dbp siRNA–treated β cells. Key genes regulating β cell function and stress are highlighted (n = 3 independent samples per group). (F) Gene set enrichment analysis comparing gene expression in scramble siRNA– versus Dbp siRNA–treated β cells with genes active in PAR bZiphigh β cells (FDR < 0.05 by Benjamini-Hochberg method). (G) Ontology of enriched GO: Biological Process (top) and KEGG pathways (bottom) in differentially expressed genes suppressed in Dbp siRNA–treated β cells. (H) Schematic of KEGG insulin secretion pathway with gene modules significantly suppressed (FDR < 0.05, FC > 1.2) in control versus Dbp siRNA–treated cells highlighted in orange.
Guided by these results, we subsequently performed RNA-seq of Dbp siRNA–treated versus scramble siRNA–treated INS-1 cells to gain insights into the role of Dbp in modulating β cell’s transcriptional profile. Differential analysis of Dbp knockdown β cells (relative to scramble siRNA) revealed down-regulation of key genes associated with β cell insulin secretory function such as Slc2a2, Chga, Sec24-related gene family, member D (Sec24d) encoding for a component of the coat protein complex II (COPII), and Insulin 2 (Ins2), while genes associated with β cell stress and dedifferentiation including Proto-oncogene c-KIT (Kit), Jun proto-oncogene (Jun), solute carrier family 16 member 1 (Slc16a1) encoding for the monocarboxylate transporter 1 (MCT1), and C-X-C motif chemokine 12 (Cxcl12) were robustly enriched in Dbp knockdown cells (FDR < 0.05, FC > 1.2; Fig. 8E). Moreover, gene set enrichment analysis of genes previously identified by scATAC-seq as active in PAR bZiphigh β cells demonstrated significant overlap with genes repressed in Dbp knockdown β cells (FDR < 0.05; Fig. 8F). Annotation of genes suppressed in Dbp knockdown to both GO:BP and KEGG pathways suggested the importance of Dbp in regulating the transcription for genes fundamental for β cell function including ~protein processing in the ER, ~exocytosis, ~cAMP signaling, and ~insulin secretion (Fig. 8G). This is further illustrated upon visualizing the extent to which insulin secretory modules are directly affected by Dbp knockdown in β cells (Fig. 8H) and implicates Dbp as a transcriptional regulator of tRF-mediated circadian control of insulin secretory response.
Circadian PAR bZip transcription factor DBP binds to active enhancers critical for the regulation of β cell function
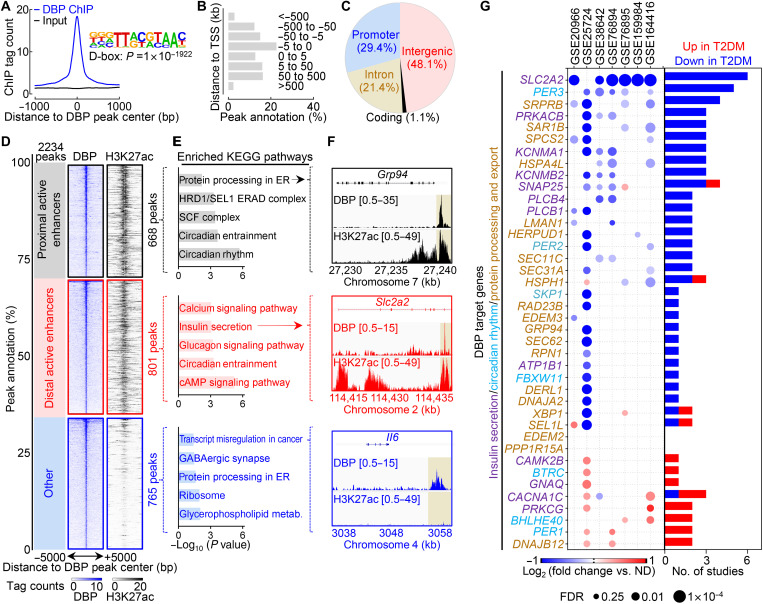
To establish the direct role of DBP in regulating expression of genes critical for β cell function, we performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) in INS-1 832/13 cells using a previously validated DBP antibody (Fig. 9) (43). Using the model-based analysis of ChIP-seq algorithm version 2 (MACS2) to categorize DBP-bound regions (FDR < 0.01), we identified 2234 statistically significant DBP peaks that were highly enriched for the canonical D-box binding motif (P = 1 × 10−1922; Fig. 9A). Among the identified peaks, nearly one-third were located <5 kb from transcriptional start sites in promoter regions, with the remainder annotating to primarily intergenic (48.1%) and intronic regions (21.1%) (Fig. 9, B and C). To elucidate the functional connection between DBP binding and gene activation/regulation, we next integrated our ChIP-seq data with previously identified active proximal (<3 kb from TSS) and distal enhancer regions in INS-1 832/13 β cells (44, 45). We found that more than two-thirds of DBP binding sites occurred in active enhancer regions, consistent with its role as a transcriptional activator (Fig. 9D). Upon annotating DBP-bound active enhancers to the nearest transcriptional start site of corresponding target genes, we found enrichment for KEGG pathways regulating circadian rhythmicity and protein/insulin processing [e.g., glucose-regulated protein 94 (Grp94)] within proximally active enhancers (Fig. 9, E and F), whereas distal active enhancers were enriched for β cell functional pathways including cAMP signaling, calcium signaling, and insulin secretion (e.g., Slc2a2/Glut2) (Fig. 9, E and F). Last, we aimed to determine whether the expression of identified DBP target genes was attenuated in T2DM human islets, which are characterized by β cell functional failure. We analyzed whole-genome array (46–50) and RNA-seq (51, 52) data from seven independent studies of nondiabetic (ND) versus T2DM human islets and assessed the differential expression of potential DBP target genes annotated to key β cell functional pathways (Fig. 9G). Notably, we found that the expression of DBP target genes was significantly (P = 2 × 10−4) down-regulated in T2DM islets relative to ND controls, highlighting the potential importance of DBP for the proper regulation of β cell function in humans (Fig. 9G).
Fig. 9. DBP binds to active enhancers critical for the regulation of insulin processing and secretion.
(A) Normalized ChIP-seq signal for DBP and input centered ±1 kb around DBP binding sites. HOMER motif enrichment analysis of the D-box motif in DBP binding sites (inset; binomial test, P = 1 × 10−1922). (B) Histogram illustrating the genomic distribution of identified DBP binding sites (relative to TSS). (C) Pie chart illustrating the genomic distribution of identified DBP binding sites against promoter (≤3 kb from TSS), intronic, coding, and intergenic regions. (D) Distribution illustrating the proportion of DBP peaks occurring in H3K27ac active enhancer regions from INS-1 832/13 β cells (GSE126556) (44). DBP peaks found ≤3 kb from TSS were considered proximal. Corresponding visualization of DBP and H3K27ac ChIP-seq signal at DBP peaks (±5 kb) occurring at proximal active enhancers (top), distal active enhancers (center), and non-H3K27ac regions (bottom). (E) Enriched KEGG pathways of genes annotated to DBP-bound proximal active enhancers (top), distal active enhancers (middle), and non-H3K27ac regions (bottom). (F) Genome browser tracks of DBP and H3K27ac within the Grp94 (top), Slc2a2 (middle), and Il6 (bottom) gene loci. (G) Dot plot visualization of DBP target gene expression for genes regulating insulin secretion, circadian rhythm, and protein processing and export KEGG pathways from seven ND/T2DM human islet studies (left). Bar graph summarizing the number of studies where each gene is differentially expressed (FC ≥ 1.2; FDR ≤ 0.25) in ND or T2DM islets (right; P = 2 × 10−4; paired, two-tailed Student’s t test). Note the attenuated expression of DBP target genes in T2DM versus ND human islets.
Glucose regulates β cell Dbp expression through modulation of FOXO1 signaling
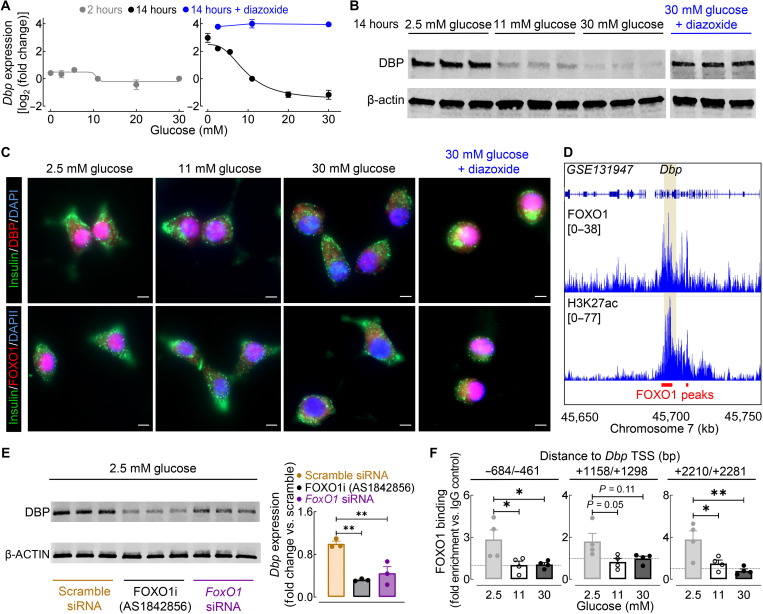
Last, we set out to elucidate potential cellular mechanisms that account for the impairment of β cell DBP activity in response to CD and corresponding rescue by tRF. Consistent with previous studies, we noted that CD results in the abrogation of circadian rhythmicity in key metabolic intermediaries such as plasma glucose and insulin, which was restored upon tRF. Given that glucose is a critical mediator of β cell gene transcription, we first evaluated the regulation of Dbp gene (and protein) expression by glucose in β cells (Fig. 10). We noted robust time (2 versus 14 hours) and dose-dependent (0 to 30 mM) regulation of Dbp by glucose (Fig. 10, A to C). Most notably, Dbp showed the highest expression in response to chronic (14 hours) exposure to low physiological glucose levels (e.g., <5 mM glucose), a condition designed to mimic fasting circadian phase under tRF. Notably, Dbp expression demonstrated near-exponential decline with increasing glucose concentrations (e.g., >10-fold change in Dbp mRNA between 2.5 and 20 mM glucose, P < 0.01; Fig. 10, A to C). Since the β cell’s transcriptional response to glucose is attributed in part to insulin secretion and corresponding autocrine insulin signaling (53, 54), we next evaluated Dbp expression in the presence of diazoxide (a potent inhibitor of insulin release) (Fig. 10, A to C). Diazoxide completely abrogated glucose-mediated changes in β cell Dbp expression, implicating insulin signaling as the primary regulator (and notably repressor) of Dbp in β cells (e.g., ~ 30-fold change in Dbp mRNA between 30 mM glucose and 30 mM glucose + diazoxide; Fig. 10, A to C). To further delineate the mechanisms of glucose/insulin-dependent regulation of Dbp, we focused on the role of the insulin-dependent FOXO1 transcriptional factor, a key regulator of β cell function in health and T2DM (55). In particular, FOXO1 levels and nuclear localization in β cells mirrored that of DBP in response to varying glucose concentrations and the addition of diazoxide (Fig. 10C). FOXO1 nuclear localization and transcriptional activity in β cells (and other tissues) are maximal under fasting (insulin-deficient) conditions, whereas insulin signaling leads to nuclear to cytoplasmic translocation and subsequent inactivation of FOXO1’s transcriptional activity (56). Direct FOXO1 binding to an active enhancer region on the Dbp gene locus was recently reported [Fig. 10D and (55)]. Consistent with these findings, inhibition of FOXO1 activity using either chemical inhibitor AS1842856 (57) or FoxO1 siRNA resulted in a robust (~65%) reduction in β cell DBP protein expression (P < 0.05 versus scramble siRNA; Fig. 10E). Last, ChIP quantitative polymerase chain reaction (qPCR) analysis confirmed that FOXO1 binds to the Dbp gene locus in β cells cultured in low-glucose concentrations, but the interactions significantly decrease under hyperglycemic conditions (P < 0.05 for 2.5 mM versus 11 mM and 30 mM glucose; Fig. 10F). Together, these data strongly implicate FOXO1 as a molecular mediator of fasting-induced Dbp expression/activity in β cells.
Fig. 10. Glucose regulates β cell Dbp expression through modulation of FoxO1.
(A) Dbp mRNA expression of INS-1 832/13 β cells cultured in varying glucose concentrations (0 to 30 mM glucose) for either 2 hours (left, gray) or 14 hours (right, black). A subset of β cells was treated with 2.5, 11, or 30 mM glucose + 250 μM diazoxide for 14 hours (blue; n = 3 to 4 experiments per condition/time). (B) DBP and β-actin protein expression in INS-1 832/13 β cells treated with 2.5, 11, 30, or 30 mM glucose + 250 μM diazoxide for 14 hours (n = 3 experiments per condition). (C) Representative examples of INS-1 832/13 β cells treated for 14 hours with 2.5, 11, 30, or 30 mM glucose + 250 μM diazoxide (out of n = 3 experiments per condition) and immunostained for DBP (red, top) or FOXO1 (red, bottom), insulin (green), and DAPI nuclear stain (blue). Scale bars, 5 μm (×40 magnification). (D) Genome browser tracks of mouse islet FOXO1 and H3K27ac ChIP-seq from GSE131947 (55). FOXO1 binding sites are highlighted in red with the Dbp locus highlighted in yellow. (E) DBP and β-actin protein expression in INS-1 832/13 β cells transfected with FoxO1 or scramble siRNA and exposed to 2.5 mM glucose + dimethyl sulfoxide vehicle or 100 nM AS1842856 for 14 hours (left). Quantification of DBP expression is normalized to scramble siRNA control (right; n = 3 experiments per condition). (F) ChIP qPCR analysis of FOXO1 binding to three regions adjacent to the Dbp TSS in INS-1 832/13 β cells cultured for 14 hours in 2.5, 11, or 30 mM glucose (n = 4 experiments per condition). *P < 0.05 and **P < 0.01 denote statistical significance (one-way ANOVA with Tukey post hoc test). Values represent means ± SEM.
DISCUSSION
The current digital era is accompanied by nearly continuous exposure to light and corresponding daily disruptions in light/dark, rest/activity, and fasting/feeding circadian cycles. Disruption of normal circadian physiology adversely affects human health and, in particular, leads to the dysregulation of glucose homeostasis and increased risk of T2DM (16). Our study reports that restoration of normal fasting/feeding circadian cycles (through isocaloric tRF) in mice prevents the induction of metabolic derangements associated with circadian disruption. This outcome was shown to be mediated through effects of tRF on circadian control of glucose tolerance, pancreatic β cell function, and the β cell’s circadian transcriptional and epigenetic profiles. Most notably, our study identified that the PAR bZip circadian transcription factor DBP plays an important role in mediating the β cell’s circadian response to tRF and is required for maintenance of GSIS. These data provide a critical link between circadian disruption and T2DM and offer a mechanistic underpinning to observations that disrupted fasting/feeding cycles are important mediators of metabolic dysfunction (4, 5, 21, 58).
Clinical and preclinical studies have started to unravel the physiological and molecular dysfunction associated with global circadian disruption (14, 59). Consistent with our observations, exposure to constant light and/or shift work has been demonstrated to disrupt circadian energy balance, glucose metabolism, and GSIS (21, 60–63). Deleterious metabolic effects of environmental circadian disruption are phenocopied in rodent models of β cell–specific deletion of core circadian clock components (e.g., Arntl), highlighting the importance of maintaining circadian gene expression in pancreatic β cells for proper regulation of insulin secretion and the overall glucose homeostasis (19, 20). In line with these observations, our study identified 4294 islet transcripts with significant circadian rhythmicity in mRNA expression in CON mice. These transcripts were annotated to genes regulating key pathways associated with circadian rhythms, insulin secretion, exocytosis, and insulin processing, among others. Moreover, CON conditions were also associated with robust diurnal changes in islet (and single β cell) chromatin accessibility, with differentially accessible DNA regions annotated to key pathways regulating insulin secretion and biosynthesis.
In particular, our study demonstrated that CD was associated with complete abrogation of rhythmic islet gene expression and diurnal chromatin accessibility, consistent with recent circadian transcriptome analysis of the liver in response to arrhythmic feeding (64). In contrast to our findings, the expression pattern of core components of the circadian clock was not perturbed in the liver in response to arrhythmic feeding, which indicates that the islet clock is potentially more sensitive to disruptions in daily fasting/feeding cycles. Although it was previously postulated that the core circadian clock would be required for the circadian response to feeding (and other zeitgebers), evidence is beginning to demonstrate that expression of circadian transcription factors alone, such as BMAL1, may not be required for the rhythmic control of gene expression (65, 66). Rather, it has been suggested that rhythmic gene expression is mediated by time-dependent control of the cistrome and requires phased recruitment of cis-regulatory elements (67, 68). Consistent with this notion, our study found that circadian disruption resulted in complete loss of all 1565 diurnally regulated chromatin regions and blunted diurnal circadian transcription factor activity in β cells.
There is growing evidence that tRF is a viable therapeutic approach to prevent and/or treat a variety of chronic diseases including cancer, cardiovascular disease, and T2DM (25, 69–71). In the context of T2DM, initial studies in animal models demonstrated that tRF could reverse metabolic dysfunction associated with obesity and clock deficiency, consistent with our results (23, 25, 69). Although it has long been known that food availability can entrain peripheral tissue oscillators outside of the SCN, the mechanism underlying this response, especially in the β cell, remains unclear (4, 5, 22). Circadian transcriptomic analysis of CD-tRF islets highlighted that reestablishment of normal fasting/feeding cycles was associated with circadian reentrainment of 525 islet transcripts preferentially involved in the regulation of circadian clock function and insulin secretion. In particular, our analysis of cis-regulatory elements of genes with a peak phase expression at the onset of feeding (e.g., CT12) revealed enrichment for motifs associated with circadian PAR bZip transcription factor family binding (e.g., DBP, TEF, and HLF), highlighting a previously underappreciated role of these transcription factors in the β cell response to tRF and the regulation of insulin secretion. Diurnal analysis of chromatin accessibility in both whole islets and single β cells confirmed this finding, consistent with recent reports demonstrating activation of D-box elements during feeding cycles in the liver (66, 68).
Pseudotime trajectory analysis in β cells combined with RNA-seq revealed that PAR bZip transcription factor activity, and specifically activity of DBP, is highly enriched in β cells in a state of increased insulin production and secretion. Consistently, siRNA-mediated knockdown of Dbp in cultured β cells results in the loss of GSIS, diminished insulin content, and down-regulation of genes regulating insulin secretion. Furthermore, ChIP-seq analysis in cultured β cells reveals 2234 DBP peaks with more than two-thirds of DBP binding sites localized to active enhancer regions, consistent with its role as a transcriptional activator. Annotation of DBP-bound active enhancers shows enrichment for pathways regulating circadian rhythmicity, insulin secretion, and protein/insulin processing.
Cumulatively, these observations strongly implicate DBP as a transcriptional regulator of β cell function and raise the question related to which molecular mechanisms account for the impairment of β cell DBP activity in response to CD (and corresponding rescue by tRF). To this end, our results implicate FOXO1 signaling as a molecular regulator of fasting/feeding-induced Dbp expression in β cells. Specifically, integration of our in vivo and in vitro data suggests that CD-mediated abrogation of circadian feeding cycles results in a continuous state of β cell insulin hypersecretion associated with inactivation of β cell’s FOXO1 transcriptional activity (56), leading to loss of Dbp/DBP expression/activity. In contrast, restoration of the normal fasting circadian period in CD-tRF leads to a state of “β cell rest” (e.g., insulin hyposecretion) and corresponding induction of FOXO1-mediated Dbp/DBP expression/activity. Therefore, our findings suggest that a prolonged fasting period is a prerequisite for the activation of β cell Dbp expression and prepares the β cell for the subsequent feeding period by promoting expression of genes regulating insulin processing, synthesis, and secretion. It is intriguing to postulate that our observations provide the molecular underpinnings behind the previously described therapeutic concept of β cell rest, long known to enhance insulin secretory function in obesity and T2DM (72).
It is also important to acknowledge the potential limitations of our study. Diurnal sampling of bulk and single-cell chromatin accessibility provides a snapshot into the rhythmic control of the cistrome and corresponding transcription factor activity. Although currently technically challenging, future studies are warranted to characterize the complete circadian nature of the islet and single β cell epigenomic landscape in vivo. Moreover, although digital genomic transcription factor footprinting provides clear advantages for identifying genome-wide differential transcription factor activity from small cell numbers in physiological relevant systems, it remains limited by the sparsity of both bulk and scATAC-seq and the limited specificity within transcription factor motif families (36–38, 42). To validate these findings, we integrated a siRNA knockdown system with a physiological relevant output (insulin secretion) together with RNA-seq and ChIP-seq. This allowed us to overcome this limitation and identify DBP as the key PAR bZip transcription factor regulating β cell function in response to tRF. Last, although the focus of our study was on the effects of CD/CD-tRF on the pancreatic β cell, CD also negatively affects the expression of circadian clocks and corresponding physiological functions in other tissues/organs contributing to the regulation of glucose homeostasis (e.g., liver, skeletal muscle, and adipose tissue) (73). There is strong experimental evidence that skeletal muscle circadian clocks control insulin-mediated glucose uptake and oxidation (74), whereas hepatic clocks regulate insulin-mediated glucose production and uptake (75). Since the beneficial metabolic effects of tRF have been shown to affect multiple organ systems (76), it is important to acknowledge the potential of tRF to restore circadian function in other metabolic tissues of CD-tRF mice in our study.
In conclusion, our results provide an integrative picture of the physiological, transcriptional, and epigenetic mechanisms underlying circadian disruption–induced impairments in glucose homeostasis and its apparent reversal upon restoration of fasting/feeding cycles. Specifically, we provide mechanistic insights into how tRF can reverse metabolic dysfunction highlighted by β cell activation and synchronization of DBP expression/activity during fasting/feeding cycles. Overall, these studies support the hypothesis that circadian disruption drives β cell dysfunction through mistimed and continuous feeding cycles; however, further studies are warranted to evaluate the direct molecular link between feeding, circadian disruption, and development of T2DM in humans. To this end, our observation of reduced expression of DBP target genes in T2DM human islets highlights the potential importance of DBP for the proper regulation of insulin secretion in health and T2DM.
MATERIALS AND METHODS
Animals and experimental study design
All animal procedures were approved by the Mayo Clinic Institutional Animal Care and Use Committee. In total, this study used 87 3-month-old male C57BL/6J mice (the Jackson Laboratory, Bar Harbor, ME). All mice were individually housed in the Promethion metabolic cage system (Sable Systems International, Las Vegas, NV) within external light-proof cabinets (Phenome Technologies, Lincolnshire, IL) and fed standard chow diet (14% fat, 32% protein, and 54% carbohydrates; Harlan, Indianapolis, IN). By convention, CT0 was denoted as the onset of light cycle under control (CON) conditions, and CT12 was denoted as the onset of dark cycles, before experimental circadian disruption. As an initial 2-week acclimatization period, all animals were entrained under control conditions: 12-hour light, 12-hour dark with ad libitum access to water and food. Following the 2-week period, the animals were randomly assigned to one of three experimental groups for 8 weeks: (i) control 12-hour light, 12-hour dark cycle with ad libitum access to water and chow diet (CON); (ii) exposure to 24-hour constant light conditions (>100-lux light intensity) with ad libitum access to water and chow diet to induce CD; and (iii) exposure to 24-hour constant light with 8 hours of restricted feeding window (CT12 to CT20) of chow diet (CD-tRF) to maintain circadian food intake rhythms despite disruption of global circadian rhythms. Temporal access to feeding hopper was controlled by MetaScreen v2.3.14 (Sable Systems International), which allowed remote/noninvasive opening and closing of the feeding trough in individual mouse cages.
Assessment of circadian food intake, motor activity, and indirect calorimetry
Circadian rhythms in feeding were assessed in mice individually housed in Promethion metabolic cages outfitted with a Promethion MM-1 food intake load cell (at 3-mg resolution). Food intake events were exported by ExpeData v1.9.27 (Sable Systems International) and subsequently binned into 10-s or 5-min intervals using MATLAB 2015b (MathWorks, Natick, MA). Foodograms were constructed using ClockLab software (Actimetrics, Wilmette, IL). Corresponding circadian rhythmicity analysis was calculated using χ2 periodogram analysis in ClockLab. Circadian rhythms in activity were assessed in mice individually housed in Promethion metabolic cages outfitted with BXY beambreak activity monitor system to track gross motor activity and analyzed by ExpeData and MATLAB 2015b as noted. Assessment of energy expenditure was performed using the comprehensive laboratory animal monitoring system (equipped with an Oxymax open-circuit calorimeter system, Columbus Instruments, Columbus, OH). Volume of carbon dioxide production (V.CO2), Volume of oxygen uptake (V.O2), and food intake were monitored and energy expenditure (kilocalories per hour per kilogram) was calculated using Oxymax Software V5.40.14 (Columbus Instruments).
Assessment of diurnal in vivo glucose tolerance, insulin secretion, and insulin tolerance
Diurnal in vivo glucose tolerance, insulin secretion, and insulin tolerance were assessed in each mouse at CT4 and CT16 time points. For glucose tolerance testing, mice were injected in the intraperitoneal space with a bolus of 50% dextrose (1 g/kg of body mass). Blood glucose was sampled at baseline and subsequently at 15, 30, 45, 60, and 90 min after dextrose injection with parallel collection of plasma for insulin measurement at baseline and 5, 15, and 90 min after injection. For insulin tolerance, mice received a bolus of insulin (0.75 mU/g of body mass) via intraperitoneal injection, and blood glucose was measured at baseline and subsequently at 15, 30, 45, 60, and 90 min after injection.
Mouse islet isolation, culture, and measurements of in vitro insulin secretion
For diurnal assessment of in vitro insulin secretion in mouse islets, mice were euthanized at CT4 and CT16 time points, and islets were isolated using standard collagenase method. Islets were allowed to recover for 2 hours in standard RPMI 1640 medium at 37°C and 5% CO2. Groups of 5 to 10 islets were washed with basal (4 mM glucose) Krebs-Ringer-bicarbonate buffer (KRB; pH 7.4) for 1 hour at 37°C and 5% CO2. GSIS was then assessed by static incubation at basal KRB buffer for 1 hour followed by hyperglycemic (16 mM glucose) KRB buffer for 1 hour at 37°C and 5% CO2. An aliquot of KRB supernatant was obtained after each incubation step for subsequent measure of secreted insulin. Islets were lysed overnight at 4°C with 0.18 N HCl for assessment of total insulin content.
Immunofluorescence analysis
Mice were euthanized, and pancreata were harvested for immediate fixation in 4% paraformaldehyde overnight at 4°C. The pancreata were embedded in paraffin, sectioned into 4-μm slices, and immunostained for insulin (ab7842, Abcam, Cambridge, UK) and glucagon (ab92517, Abcam) with VECTASHIELD-DAPI (4′,6-diamidino-2-phenylindole) mounting medium (Vector Laboratories, Burlingame, CA). Complete pancreatic sections were imaged at 5× using a Zeiss Axio Observer Z1 microscope (Carl Zeiss Microscopy, Oberkochen, Germany) and processed using ZenPro software (Carl Zeiss Microscopy). β cell and α cell mass were calculated as the product of pancreatic mass and the insulin- or glucagon-positive pancreatic area, respectively. For the assessment of DBP/FOXO1 expression and localization in cultured β cells, ~1 × 105 INS-1 832/13 cells were plated in Nunc Lab-Tek II four-well chamber slides (Thermo Fisher Scientific). Cells were incubated with 2.5, 11, and 30 mM glucose-containing media or 30 mM glucose and 250 μM diazoxide for 14 hours. Cells were fixed with 4% formaldehyde for 10 min at room temperature. The cells were then immunostained for insulin (MAB1417, R&D Systems, Minneapolis, MN), FOXO1 (C29H4, Cell Signaling Technologies), and/or DBP (12662-1-AP, Proteintech, Rosemont, IL). The slides were mounted with VECTASHIELD-DAPI medium and subsequently imaged at 40× using the Zeiss Axio Observer Z1 microscope.
Cell culture, siRNA knockdown, and measurements of in vitro insulin secretion
INS-1 832/13 immortalized rat β cells, provided by C. Newgard (Duke University), were cultured in standard RPMI 1640 media supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, 10 mM Hepes, 1 mM sodium pyruvate, and 50 μM β-mercaptoethanol. For the assessment of varying glucose concentration on Dbp expression, cells were treated with 0, 2.5, 5.5, 11, 20, and 30 mM glucose-containing RPMI 1640 media for 2 or 14 hours and subjected to RT-PCR or Western blot analysis as described. A subset of cells was treated with glucose media supplemented with 250 μM diazoxide (J66010, Alfa Aesar, Ward Hill, MA). For siRNA knockdown, SMARTpool ON-TARGETplus siRNA was obtained specifically targeting rat FoxO1, Dbp, and Tef mRNA or scrambled nontargeting siRNA as a control (Dharmacon, Lafayette, CO). Gene knockdown of INS-1 832/13 β cells was performed using 20 nM experimental or control siRNA via Lipofectamine RNAiMAX–mediated transfection (Invitrogen, Carlsbad, CA) for 24 to 36 hours, according to the manufacturer’s instructions. For the assessment of FoxO1’s role in regulating Dbp expression, ~1 × 106 cells transfected with 20 nM FoxO1 or scramble siRNA for 36 hours were washed with 1× phosphate-buffered saline (PBS), exposed to 2.5 mM glucose media in the presence of vehicle (dimethyl sulfoxide) or 100 nM FOXO1 inhibitor AS1842856 (344355, EMD Millipore) for 14 hours, and subjected to Western blot analysis as described. For the assessment of in vitro insulin secretion, 100,000 cells per well were seeded into a 24-well plate and allowed to attach overnight. Following Dbp or Tef knockdown as noted above, cells were washed for 2 hours with basal KRB buffer at 37°C and 5% CO2. Insulin secretion was measured sequentially following 1-hour basal (4 mM glucose) KRB, 1-hour hyperglycemic (16 mM glucose) KRB, and 1-hour basal KRB + 30 mM KCl to assess maximal insulin secretion. Cells were lysed overnight at 4°C with 0.18 N HCl for assessment of total insulin content.
Western blot analysis
INS-1 832/13 cells (~1 × 106) were lysed using NP-40 supplemented with protease inhibitor cocktail (Cell Signaling Technologies, Danvers, MA). Assessment of protein expression was performed by Western blot protein analysis with antibodies against DBP (12662-1-AP), TEF (PA5-106495, Invitrogen), and β-actin (MAB1501, EMD Millipore, Burlington, MA) as previously described (77).
RNA isolation, quantitative RT-PCR, and RNA-seq
Total RNA was isolated from freshly picked mouse islets or transfected INS-1 832/13 cells using the RNeasy mini kit (Qiagen, Hilden, Germany). For quantitative real-time PCR, mRNA was converted into complementary DNA (cDNA) using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA), and qRT-PCR was performed on the StepOnePlus machine (Applied Biosystems, Carlsbad, CA) under default thermal cycling conditions. Data were analyzed using the 2−ΔΔCT method with mRNA levels normalized to genes β-actin for every sample. For RNA-seq, RNA size and quality were assessed using the Tape Station Bionalyzer (Agilent, Santa Clara, CA), and concentration was determined by Qubit fluorometry. All samples had DV200 scores (percentage of RNA fragments >200 nucleotides) ≥70% and were subsequently approved for library preparation and sequencing by GENEWIZ (South Plainfield, NJ). RNA libraries were prepared using the polyA selection method (Illumina, San Diego, CA) from 100 ng of total RNA. Paired-end, 150–base pair (bp) sequencing of the libraries (>25 million reads per sample) was performed using an Illumina HiSeq instrument according to the manufacturer’s instructions. All sequenced samples had a Q30 score (percentage of bases with a quality score > 30) ≥93% with a mean quality score ≥35. RNA-seq bioinformatic analysis was performed as described in the Supplementary Materials.
Bulk ATAC-seq
Mouse islets were freshly isolated at CT4 and CT16 (n = 2 independent samples per CT and group). Fifty islets (~50,000 cells) per reaction were then immediately counted, washed with ice-cold 1× PBS, and lysed for 20 to 25 min on ice [10 mM tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, and 0.1% Igepal CA-630] (78). Nuclei were subsequently obtained following centrifugation for 15 min at 500g and 4°C and subjected to the Omni ATAC-seq protocol (79). DNA library size following PCR amplification was determined by Fragment Analysis (Agilent), and enrichment of accessible regions was verified by RT-PCR using the FC between a positive control locus (AT-Dusp6; forward, GGCTTATCCGGAGCGGAAAT; reverse, GGCTGGAACAGGTTGTGTTG) and a negative control locus (AT-Vmn2r17; forward, TCCCCTTTACTGTTTTCCTCTAC; reverse, GGATTGATGAGGAAACAGCCTC) before sequencing. Paired-end, 50-bp sequencing of the libraries was performed using an Illumina HiSeq instrument according to the manufacturer’s settings (Illumina). Bulk ATAC-seq bioinformatic analysis was performed as described in the Supplementary Materials.
Single-cell ATAC-seq
Mouse islets were freshly isolated at CT4 and CT16 (n = 2 mice per CT and group), >100 islets were counted, and nuclei isolation was performed as noted above using ice-cold 1× lysis buffer from 10X Genomics (Pleasanton, CA). A portion of isolated nuclei were stained with Trypan Blue for manual counting and to ensure a single-nucleus suspension before library preparation. Approximately 5000 nuclei per sample were subsequently aliquoted for scATAC-seq library preparation using the Chromium Next GEM Single-Cell ATAC Kit (10X Genomics). Ten microliters of 1× Tn5 Transposition enzyme was mixed with 5 μl of the diluted nuclei suspension and was incubated at 50°C for 1 hour. The tagmented nuclei were partitioned into Gel Bead-In Emulsions (GEMs) and barcoded by unique molecular identifiers using the Chromium Next GEM Single-Cell ATAC library and Gel Bead kit and the C1 Chromium Instrument (10X Genomics). scATAC-seq libraries were then prepared and indexed using the Chromium i7 Sample Index kit (10X Genomics) with library size determined by Fragment Analysis (Agilent) before sequencing. Paired-end, dual-indexed sequencing of the libraries (50-bp read #1, 8-bp i7 index, 16-bp i5 index, and 50-bp read#2) was performed using an Illumina HiSeq instrument. scATAC-seq bioinformatic analysis was performed as described in the Supplementary Materials.
Chromatin immunoprecipitation sequencing
INS-1 832/13 cells (~40 × 106) were cross-linked with 4% formaldehyde for 10 min, followed by quenching with 125 mM glycine for 5 min at room temperature. Fixed cells were washed with 1× tris-buffered saline and kept at −80°C. ChIP-seq was performed as previously described (80). Briefly, fixed cells were resuspended in 2 ml of cell lysis buffer [10 mM tris-HCl (pH 7.5), 10 mM NaCl, and 0.5% NP-40] and incubated on ice for 10 min. The lysates were washed with MNase digestion buffer [20 mM tris-HCl (pH 7.5), 15 mM NaCl, 60 mM KCl, and 1 mM CaCl2] and incubated with 3000 or 5000 gel units of MNase (M0247S, New England Biolabs, Ipswich, MA) for 20 min at 37°C. After adding the same volume of sonication buffer [100 mM tris-HCl (pH 8.1), 20 mM EDTA, 200 mM NaCl, 2% Triton X-100, and 0.2% sodium deoxycholate], the lysate was sonicated for 3.5 min (30 s on/30 s off) in a Diagenode bioruptor and centrifuged at 15,000 rpm for 10 min. The cleared supernatant equivalent to about 40 million cells was incubated with 2.5 μg of anti-DBP antibody (PM079, MBL Life Science, Woburn, MA) previously validated for ChIP-seq (43). Subsequent immunoprecipitation and library preparation were performed as previously described (80). The libraries were sequenced to 51 bp from both ends on an Illumina NextSeq instrument (Illumina) according to the manufacturer’s instructions. ChIP-seq bioinformatic analysis was performed as described in the Supplementary Materials.
Chromatin immunoprecipitation qPCR
A total of ~10 × 106 INS-1 832/13 cells per replicate were treated for 14 hours with 2.5, 11, or 30 mM glucose-containing media and were cross-linked as described. ChIP was subsequently performed using the iDeal ChIP-qPCR (Diagenode, Denville, NJ) kit according to the manufacturer’s instructions using either 1 μg of FOXO1 (C29H4, Cell Signaling Technologies) or IgG antibody (C15410206, Diagenode). Immunoprecipitated DNA (2.5 μl) was mixed with SYBR Green Master Mix and primers targeting regulatory regions of the Dbp gene (−684/−461; forward, CGCCTGTTCTGAGTTTCCTC; reverse, CGCGTGTTCACAGACTCATT′; +1158/+1298; forward, GACCGTCTCTATTGCTGGGG; reverse, GAGACGGGAGACGTAGGGTA; and +2210/+2281; forward, GGAGCTCCCCATATTCCTGC; reverse, CAAGCTCTTGGCCATCCAGA) for a total reaction volume of 20 μl. qPCR analysis was performed using the ABI StepOnePlus Real-Time PCR System, and the results were normalized to a background intergenic region (forward, GACAACCCTGGGGTAAGCTC; reverse, TCTGAGCGTCCACATCAGTG) with enrichment calculated as FC versus IgG control.
Analytical methods
Blood glucose was measured using a Freestyle Lite glucometer (Abbott, Abbott Park, IL). Plasma insulin, islet insulin content, and secreted mouse islet insulin were measured using Ultrasensitive Mouse Insulin enzyme-linked immunosorbent assay (ELISA) (Alpco Diagnostics, Salem, NH) as per the manufacturer’s instructions. INS-1 832/13 secreted insulin and cellular insulin content were measured using High Range Rat Insulin ELISA (Alpco Diagnostics).
Statistical analysis
Results presented are expressed as the means ± SE. Statistical analysis was completed using GraphPad Prism v8.4.2 (San Diego, CA), RStudio v3.6.2, and OriginPro2020 (OriginLab Corporation, Northampton, MA). Statistical analysis was performed by either two-tailed Student’s t test for comparison of two independent groups or one-way analysis of variance (ANOVA) and/or two-way ANOVA for comparison of three or more groups with Tukey method for multiple comparisons correction. A value of P < 0.05 was considered statistically significant, unless otherwise noted. Statistical analyses for RNA-seq, bulk ATAC-seq, scATAC-seq, and ChIP-seq are described in the respective informatics analyses sections.
Acknowledgments
We thank T. A. White and P. Zhang for technical assistance with respirometry experiments. We thank D. King for technical assistance with processing transcriptomic samples and G. Xian (Grant) Shi for technical assistance with ChIP-seq experiments. Illustrations of study design and analysis were created with Biorender.com premium service.
Funding: We acknowledge funding support from the NIH (R01DK098468 to A.V.M., F99DK123834 to M.R.B., and R01AG53832 to N.K.L.), the Mayo Clinic Graduate School of Biomedical Sciences (Biomedical Engineering and Physiology Graduate Program), and the Center for Regenerative Medicine (Mayo Clinic, Rochester, MN).
Author contributions: M.R.B. contributed to study design, conducted experiments, and assisted with the data analysis, interpretation, and preparation of the manuscript. S.K.S., A.M., T.K.H., and Y.X. conducted experiments and assisted with the data analysis, interpretation, and preparation of the manuscript. K.R., J.-H.L., N.J., C.S.C., N.K.L., A.G.-M., and T.O. contributed to study design, data analysis, interpretation, and preparation of the manuscript. A.V.M. designed, interpreted the studies, and wrote the manuscript with M.R.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Genomic data were deposited to the GEO under accession number GSE182938.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S9
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Bass J., Lazar M. A., Circadian time signatures of fitness and disease. Science 354, 994–999 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Schibler U., Gotic I., Saini C., Gos P., Curie T., Emmenegger Y., Sinturel F., Gosselin P., Gerber A., Fleury-Olela F., Rando G., Demarque M., Franken P., Clock-Talk: Interactions between central and peripheral circadian oscillators in mammals. Cold Spring Harb. Symp. Quant. Biol. 80, 223–232 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Zeitzer J. M., Dijk D. J., Kronauer R., Brown E., Czeisler C., Sensitivity of the human circadian pacemaker to nocturnal light: Melatonin phase resetting and suppression. J. Physiol. 526 (Pt. 3), 695–702 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stokkan K. A., Yamazaki S., Tei H., Sakaki Y., Menaker M., Entrainment of the circadian clock in the liver by feeding. Science 291, 490–493 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Damiola F., Le Minh N., Preitner N., Kornmann B., Fleury-Olela F., Schibler U., Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14, 2950–2961 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi J. S., Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim Y. H., Lazar M. A., Transcriptional control of circadian rhythms and metabolism: A matter of time and space. Endocr. Rev. 41, 707–732 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaix A., Zarrinpar A., Panda S., The circadian coordination of cell biology. J. Cell Biol. 215, 15–25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G. Z., Hickey S. L., Shi L., Huang H. C., Nakashe P., Koike N., Tu B. P., Takahashi J. S., Konopka G., Cycling transcriptional networks optimize energy utilization on a genome scale. Cell Rep. 13, 1868–1880 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z., Odstrcil E. A., Tu B. P., McKnight S. L., Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science 316, 1916–1919 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Eckel-Mahan K. L., Patel V. R., De Mateo S., Orozco-Solis R., Ceglia N. J., Sahar S., Dilag-Penilla S. A., Dyar K. A., Baldi P., Sassone-Corsi P., Reprogramming of the circadian clock by nutritional challenge. Cell 155, 1464–1478 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerhart-Hines Z., Lazar M. A., Circadian metabolism in the light of evolution. Endocr. Rev. 36, 289–304 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lunn R. M., Blask D. E., Coogan A. N., Figueiro M. G., Gorman M. R., Hall J. E., Hansen J., Nelson R. J., Panda S., Smolensky M. H., Stevens R. G., Turek F. W., Vermeulen R., Carreon T., Caruso C. C., Lawson C. C., Thayer K. A., Twery M. J., Ewens A. D., Garner S. C., Schwingl P. J., Boyd W. A., Health consequences of electric lighting practices in the modern world: A report on the National Toxicology Program’s workshop on shift work at night, artificial light at night, and circadian disruption. Sci. Total Environ. 607-608, 1073–1084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arble D. M., Bass J., Behn C. D., Butler M. P., Challet E., Czeisler C., Depner C. M., Elmquist J., Franken P., Grandner M. A., Hanlon E. C., Keene A. C., Joyner M. J., Karatsoreos I., Kern P. A., Klein S., Morris C. J., Pack A. I., Panda S., Ptacek L. J., Punjabi N. M., Sassone-Corsi P., Scheer F. A., Saxena R., Seaquest E. R., Thimgan M. S., Van Cauter E., Wright K. P., Impact of sleep and circadian disruption on energy balance and diabetes: A summary of workshop discussions. Sleep 38, 1849–1860 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian J., Scheer F. A. J. L., Circadian system and glucose metabolism: Implications for physiology and disease. Trends Endocrinol. Metab. 27, 282–293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kivimaki M., Batty G. D., Hublin C., Shift work as a risk factor for future type 2 diabetes: Evidence, mechanisms, implications, and future research directions. PLoS Med. 8, e1001138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan A., Schernhammer E. S., Sun Q., Hu F. B., Rotating night shift work and risk of type 2 diabetes: Two prospective cohort studies in women. PLoS Med. 8, e1001141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., Eckel R. H., Takahashi J. S., Bass J., Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308, 1043–1045 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcheva B., Ramsey K. M., Buhr E. D., Kobayashi Y., Su H., Ko C. H., Ivanova G., Omura C., Mo S., Vitaterna M. H., Lopez J. P., Philipson L. H., Bradfield C. A., Crosby S. D., JeBailey L., Wang X., Takahashi J. S., Bass J., Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466, 627–631 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rakshit K., Hsu T. W., Matveyenko A. V., Bmal1 is required for beta cell compensatory expansion, survival and metabolic adaptation to diet-induced obesity in mice. Diabetologia 59, 734–743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fonken L. K., Workman J. L., Walton J. C., Weil Z. M., Morris J. S., Haim A., Nelson R. J., Light at night increases body mass by shifting the time of food intake. Proc. Natl. Acad. Sci. 107, 18664–18669 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vollmers C., Gill S., DiTacchio L., Pulivarthy S. R., Le H. D., Panda S., Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl. Acad. Sci. 106, 21453–21458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaix A., Lin T., Le H. D., Chang M. W., Panda S., Time-restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metab. 29, 303–319.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tognini P., Murakami M., Liu Y., Eckel-Mahan K. L., Newman J. C., Verdin E., Baldi P., Sassone-Corsi P., Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metab. 26, 523–538.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Hatori M., Vollmers C., Zarrinpar A., DiTacchio L., Bushong E. A., Gill S., Leblanc M., Chaix A., Joens M., Fitzpatrick J. A., Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15, 848–860 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gill S., Panda S., A smartphone app reveals erratic diurnal eating patterns in humans that can be modulated for health benefits. Cell Metab. 22, 789–798 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohta H., Yamazaki S., McMahon D. G., Constant light desynchronizes mammalian clock neurons. Nat. Neurosci. 8, 267–269 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Granados-Fuentes D., Prolo L. M., Abraham U., Herzog E. D., The suprachiasmatic nucleus entrains, but does not sustain, circadian rhythmicity in the olfactory bulb. J. Neurosci. 24, 615–619 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang R., Su Z., Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. Bioinformatics 26, i168–i174 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu G., Anafi R. C., Hughes M. E., Kornacker K., Hogenesch J. B., MetaCycle: An integrated R package to evaluate periodicity in large scale data. Bioinformatics 32, 3351–3353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laloum D., Robinson-Rechavi M., Methods detecting rhythmic gene expression are biologically relevant only for strong signal. PLoS Comput. Biol. 16, e1007666 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perelis M., Marcheva B., Ramsey K. M., Schipma M. J., Hutchison A. L., Taguchi A., Peek C. B., Hong H., Huang W., Omura C., Allred A. L., Bradfield C. A., Dinner A. R., Barish G. D., Bass J., Pancreatic β cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 350, aac4250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrenko V., Saini C., Giovannoni L., Gobet C., Sage D., Unser M., Masson M. H., Gu G., Bosco D., Gachon F., Philippe J., Dibner C., Pancreatic α-and β-cellular clocks have distinct molecular properties and impact on islet hormone secretion and gene expression. Genes Dev. 31, 383–398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camunas-Soler J., Dai X.-Q., Hang Y., Bautista A., Lyon J., Suzuki K., Kim S. K., Quake S. R., MacDonald P. E., Patch-seq links single-cell transcriptomes to human islet dysfunction in diabetes. Cell Metab. 31, 1017–1031.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suriben R., Kaihara K. A., Paolino M., Reichelt M., Kummerfeld S. K., Modrusan Z., Dugger D. L., Newton K., Sagolla M., Webster J. D., Liu J., Hebrok M., Dixit V. M., β-cell insulin secretion requires the ubiquitin ligase COP1. Cell 163, 1457–1467 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Baek S., Goldstein I., Hager G. L., Bivariate genomic footprinting detects changes in transcription factor activity. Cell Rep. 19, 1710–1722 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bentsen M., Goymann P., Schultheis H., Klee K., Petrova A., Wiegandt R., Fust A., Preussner J., Kuenne C., Braun T., Kim J., Looso M., ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat. Commun. 11, 4267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rai V., Quang D. X., Erdos M. R., Cusanovich D. A., Daza R. M., Narisu N., Zou L. S., Didion J. P., Guan Y., Shendure J., Parker S. C. J., Collins F. S., Single-cell ATAC-seq in human pancreatic islets and deep learning upscaling of rare cells reveals cell-specific type 2 diabetes regulatory signatures. Mol. Metab. 32, 109–121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Dijk D., Sharma R., Nainys J., Yim K., Kathail P., Carr A. J., Burdziak C., Moon K. R., Chaffer C. L., Pattabiraman D., Bierie B., Mazutis L., Wolf G., Krishnaswamy S., Pe’er D., Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729.e27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Granja J. M., Corces M. R., Pierce S. E., Bagdatli S. T., Choudhry H., Chang H. Y., Greenleaf W. J., ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corces M. R., Shcherbina A., Kundu S., Gloudemans M. J., Frésard L., Granja J. M., Louie B. H., Eulalio T., Shams S., Bagdatli S. T., Mumbach M. R., Liu B., Montine K. S., Greenleaf W. J., Kundaje A., Montgomery S. B., Chang H. Y., Montine T. J., Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat. Genet. 52, 1158–1168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schep A. N., Wu B., Buenrostro J. D., Greenleaf W. J., chromVAR: Inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshitane H., Asano Y., Sagami A., Sakai S., Suzuki Y., Okamura H., Iwasaki W., Ozaki H., Fukada Y., Functional D-box sequences reset the circadian clock and drive mRNA rhythms. Commun. Biol. 2, 300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van de Velde S., Wiater E., Tran M., Hwang Y., Cole P. A., Montminy M., CREB promotes beta cell gene expression by targeting its coactivators to tissue-specific enhancers. Mol. Cell. Biol. 39, e00200–e00219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramírez F., Dündar F., Diehl S., Grüning B. A., Manke T., deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown M. R., Holmes H., Rakshit K., Javeed N., Her T., Stiller A., Sen S. K., Shull G. E., Prakash Y., Romero M. F., Matveyenko A. V., Electrogenic sodium bicarbonate cotransporter NBCe1 regulates pancreatic β cell function in type 2 diabetes. J. Clin. Invest. 131, e142365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taneera J., Lang S., Sharma A., Fadista J., Zhou Y., Ahlqvist E., Jonsson A., Lyssenko V., Vikman P., Hansson O., Parikh H., Korsgren O., Soni A., Krus U., Zhang E., Jing X.-J., Esguerra J. L. S., Wollheim C. B., Salehi A., Rosengren A., Renström E., Groop L., A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metab. 16, 122–134 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Dominguez V., Raimondi C., Somanath S., Bugliani M., Loder M. K., Edling C. E., Divecha N., da Silva-Xavier G., Marselli L., Persaud S. J., Turner M. D., Rutter G. A., Marchetti P., Falasca M., Maffucci T., Class II phosphoinositide 3-kinase regulates exocytosis of insulin granules in pancreatic β cells. J. Biol. Chem. 286, 4216–4225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marselli L., Thorne J., Dahiya S., Sgroi D. C., Sharma A., Bonner-Weir S., Marchetti P., Weir G. C., Gene expression profiles of β-cell enriched tissue obtained by laser capture microdissection from subjects with type 2 diabetes. PLOS ONE 5, e11499 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Solimena M., Schulte A. M., Marselli L., Ehehalt F., Richter D., Kleeberg M., Mziaut H., Knoch K.-P., Parnis J., Bugliani M., Siddiq A., Jörns A., Burdet F., Liechti R., Suleiman M., Margerie D., Syed F., Distler M., Grützmann R., Petretto E., Moreno-Moral A., Wegbrod C., Sönmez A., Pfriem K., Friedrich A., Meinel J., Wollheim C. B., Baretton G. B., Scharfmann R., Nogoceke E., Bonifacio E., Sturm D., Meyer-Puttlitz B., Boggi U., Saeger H.-D., Filipponi F., Lesche M., Meda P., Dahl A., Wigger L., Xenarios I., Falchi M., Thorens B., Weitz J., Bokvist K., Lenzen S., Rutter G. A., Froguel P., von Bülow M., Ibberson M., Marchetti P., Systems biology of the IMIDIA biobank from organ donors and pancreatectomised patients defines a novel transcriptomic signature of islets from individuals with type 2 diabetes. Diabetologia 61, 641–657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wigger L., Barovic M., Brunner A.-D., Marzetta F., Schöniger E., Mehl F., Kipke N., Friedland D., Burdet F., Kessler C., Lesche M., Thorens B., Bonifacio E., Legido-Quigley C., Hilaire P. B. S., Delerive P., Dahl A., Klose C., Gerl M. J., Simons K., Aust D., Weitz J., Distler M., Schulte A. M., Mann M., Ibberson M., Solimena M., Multi-omics profiling of living human pancreatic islet donors reveals heterogeneous beta cell trajectories towards type 2 diabetes. Nat. Metab. 3, 1017–1031 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Marselli L., Piron A., Suleiman M., Colli M. L., Yi X., Khamis A., Carrat G. R., Rutter G. A., Bugliani M., Giusti L., Ronci M., Ibberson M., Turatsinze J.-V., Boggi U., De Simone P., De Tata V., Lopes M., Nasteska D., De Luca C., Tesi M., Bosi E., Singh P., Campani D., Schulte A. M., Solimena M., Hecht P., Rady B., Bakaj I., Pocai A., Norquay L., Thorens B., Canouil M., Froguel P., Eizirik D. L., Cnop M., Marchetti P., Persistent or transient human β cell dysfunction induced by metabolic stress: Specific signatures and shared gene expression with type 2 diabetes. Cell Rep. 33, 108466 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Ohsugi M., Cras-Méneur C., Zhou Y., Warren W., Bernal-Mizrachi E., Permutt M. A., Glucose and insulin treatment of insulinoma cells results in transcriptional regulation of a common set of genes. Diabetes 53, 1496–1508 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Leibiger I. B., Leibiger B., Berggren P.-O., Insulin signaling in the pancreatic β-cell. Annu. Rev. Nutr. 28, 233–251 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Kuo T., Kraakman M. J., Damle M., Gill R., Lazar M. A., Accili D., Identification of C2CD4A as a human diabetes susceptibility gene with a role in β cell insulin secretion. Proc. Natl. Acad. Sci. 116, 20033–20042 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kitamura T., Nakae J., Kitamura Y., Kido Y., Biggs W. H. III, Wright C. V., White M. F., Arden K. C., Accili D., The forkhead transcription factor Foxo1 links insulin signaling to Pdx1 regulation of pancreatic β cell growth. J. Clin. Invest. 110, 1839–1847 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagashima T., Shigematsu N., Maruki R., Urano Y., Tanaka H., Shimaya A., Shimokawa T., Shibasaki M., Discovery of novel forkhead box O1 inhibitors for treating type 2 diabetes: Improvement of fasting glycemia in diabetic db/db mice. Mol. Pharmacol. 78, 961–970 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Fonken L. K., Nelson R. J., The effects of light at night on circadian clocks and metabolism. Endocr. Rev. 35, 648–670 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Rüger M., Scheer F. A. J. L., Effects of circadian disruption on the cardiometabolic system. Rev. Endocr. Metab. Disord. 10, 245–260 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coomans C. P., van den Berg S. A. A., Houben T., van Klinken J.-B., van den Berg R., Pronk A. C. M., Havekes L. M., Romijn J. A., van Dijk K. W., Biermasz N. R., Meijer J. H., Detrimental effects of constant light exposure and high-fat diet on circadian energy metabolism and insulin sensitivity. FASEB J. 27, 1721–1732 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Qian J., Dalla Man C., Morris C. J., Cobelli C., Scheer F. A. J. L., Differential effects of the circadian system and circadian misalignment on insulin sensitivity and insulin secretion in humans. Diabetes Obes. Metab. 20, 2481–2485 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morris C. J., Yang J. N., Garcia J. I., Myers S., Bozzi I., Wang W., Buxton O. M., Shea S. A., Scheer F. A. J. L., Endogenous circadian system and circadian misalignment impact glucose tolerance via separate mechanisms in humans. Proc. Natl. Acad. Sci. 112, E2225–E2234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qian J., Block G. D., Colwell C. S., Matveyenko A. V., Consequences of exposure to light at night on the pancreatic islet circadian clock and function in rats. Diabetes 62, 3469–3478 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greenwell B. J., Trott A. J., Beytebiere J. R., Pao S., Bosley A., Beach E., Finegan P., Hernandez C., Menet J. S., Rhythmic food intake drives rhythmic gene expression more potently than the hepatic circadian clock in mice. Cell Rep. 27, 649–657.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Ray S., Valekunja U. K., Stangherlin A., Howell S. A., Snijders A. P., Damodaran G., Reddy A. B., Circadian rhythms in the absence of the clock gene Bmal1. Science 367, 800–806 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Weger B. D., Gobet C., David F. P. A., Atger F., Martin E., Phillips N. E., Charpagne A., Weger M., Naef F., Gachon F., Systematic analysis of differential rhythmic liver gene expression mediated by the circadian clock and feeding rhythms. Proc. Natl. Acad. Sci. 118, e2015803118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koike N., Yoo S.-H., Huang H.-C., Kumar V., Lee C., Kim T.-K., Takahashi J. S., Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fang B., Everett L. J., Jager J., Briggs E., Armour S. M., Feng D., Roy A., Gerhart-Hines Z., Sun Z., Lazar M. A., Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 159, 1140–1152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chaix A., Zarrinpar A., Miu P., Panda S., Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 20, 991–1005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilkinson M. J., Manoogian E. N., Zadourian A., Lo H., Fakhouri S., Shoghi A., Wang X., Fleischer J. G., Navlakha S., Panda S., Taub P. R., Ten-hour time-restricted eating reduces weight, blood pressure, and atherogenic lipids in patients with metabolic syndrome. Cell Metab. 31, 92–104.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sancar A., Van Gelder R. N., Clocks, cancer, and chronochemotherapy. Science 371, eabb0738 (2021). [DOI] [PubMed] [Google Scholar]
- 72.Laedtke T., Kjems L., Pørksen N., Schmitz O., Veldhuis J., Kao P. C., Butler P. C., Overnight inhibition of insulin secretion restores pulsatility and proinsulin/insulin ratio in type 2 diabetes. Am. J. Physiol.-Endocrinol. Metab. 279, E520–E528 (2000). [DOI] [PubMed] [Google Scholar]
- 73.Javeed N., Matveyenko A. V., Circadian etiology of type 2 diabetes mellitus. Phys. Ther. 33, 138–150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dyar K. A., Ciciliot S., Wright L. E., Biensø R. S., Tagliazucchi G. M., Patel V. R., Forcato M., Paz M. I. P., Gudiksen A., Solagna F., Albiero M., Moretti I., Eckel-Mahan K. L., Baldi P., Sassone-Corsi P., Rizzuto R., Bicciato S., Pilegaard H., Blaauw B., Schiaffino S., Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol. Metab. 3, 29–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jacobi D., Liu S., Burkewitz K., Kory N., Knudsen N. H., Alexander R. K., Unluturk U., Li X., Kong X., Hyde A. L., Gangl M. R., Mair W. B., Lee C.-H., Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metab. 22, 709–720 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Longo V. D., Panda S., Fasting, circadian rhythms, and time-restricted feeding in healthy lifespan. Cell Metab. 23, 1048–1059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Javeed N., Brown M. R., Rakshit K., Her T., Sen S. K., Matveyenko A. V., Pro-inflammatory cytokine interleukin 1β disrupts β cell circadian clock function and regulation of insulin secretion. Endocrinology 162, bqaa084 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.H. Raurell-Vila, M. Ramos-Rodríguez, L. Pasquali, Assay for transposase accessible chromatin (ATAC-Seq) to chart the open chromatin landscape of human pancreatic islets, in CpG Islands (Springer, 2018), vol. 1766, pp. 197–208. [DOI] [PubMed] [Google Scholar]
- 79.Corces M. R., Trevino A. E., Hamilton E. G., Greenside P. G., Sinnott-Armstrong N. A., Vesuna S., Satpathy A. T., Rubin A. J., Montine K. S., Wu B., Kathiria A., Cho S. W., Mumbach M. R., Carter A. C., Kasowski M., Orloff L. A., Risca V. I., Kundaje A., Khavari P. A., Montine T. J., Greenleaf W. J., Chang H. Y., An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhong J., Ye Z., Lenz S. W., Clark C. R., Bharucha A., Farrugia G., Robertson K. D., Zhang Z., Ordog T., Lee J.-H., Purification of nanogram-range immunoprecipitated DNA in ChIP-seq application. BMC Genomics 18, 985 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S9
Legend for data S1
Data S1