Abstract
In sorghum (Sorghum bicolor) and other C4 grasses, brown midrib (bmr) mutants have long been associated with plants impaired in their ability to synthesize lignin. The brown midrib 30 (Bmr30) gene, identified using a bulk segregant analysis and next-generation sequencing, was determined to encode a chalcone isomerase (CHI). Two independent mutations within this gene confirmed that loss of its function was responsible for the brown leaf midrib phenotype and reduced lignin concentration. Loss of the Bmr30 gene function, as shown by histochemical staining of leaf midrib and stalk sections, resulted in altered cell wall composition. In the bmr30 mutants, CHI activity was drastically reduced, and the accumulation of total flavonoids and total anthocyanins was impaired, which is consistent with its function in flavonoid biosynthesis. The level of the flavone lignin monomer tricin was reduced 20-fold in the stem relative to wild type, and to undetectable levels in the leaf tissue of the mutants. The bmr30 mutant, therefore, harbors a mutation in a phenylpropanoid biosynthetic gene that is key to the interconnection between flavonoids and monolignols, both of which are utilized for lignin synthesis in the grasses.
Keywords: lignin biosynthesis, flavonoid biosynthesis, tricin lignin, mutant, NMR, monolignol
Introduction
Sorghum bicolor is an economically important C4 grass, the fifth most important cereal crop in the world, which is grown as a grain, forage, sugar, and lignocellulosic bioenergy crop. Although sorghum can grow in a wide range of environments, its production is usually associated with hot and dry regions because of its high water-use efficiency and drought tolerance. Sorghum is, therefore, being developed as a potential bioenergy crop due to its ability to grow under sub-optimal conditions and climate resiliency (van der Weijde et al., 2013). Utilizing plant cell wall biomass as a renewable resource for the production of energy and fuels has become a major research focus (Vermerris et al., 2007). Plant cell walls predominantly consist of the polysaccharides cellulose and hemicelluloses along with the phenolic polymer lignin. Lignin is polymerized through radical coupling processes in which hydroxycinnamyl alcohols, mainly p-coumaryl, coniferyl, and sinapyl alcohols derived from monolignol biosynthesis, form radicals catalyzed by cell wall laccases and peroxidases. These radicals undergo coupling reactions during the process of lignification to form p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) lignin units, respectively. In addition to monolignol biosynthesis, the flavone tricin [5,7-dihydroxy-2-(4-hydroxy-3,5-dimethoxyphenyl)-4H-chromen-4-one] was recently established as a lignin monomer in grass cell walls in which it potentially functions as a nucleation site for lignin polymerization (del Río et al., 2012; Lan et al., 2015, 2016a). Thus, this discovery linked together two branches of phenylpropanoid metabolism, monolignol and flavonoid biosynthesis, in grass lignification.
Lignocellulosic biomass from brown midrib mutants has consistently contained less lignin than their wild-type (WT) counterparts, resulting in forage with increased digestibility for livestock (Porter et al., 1978; Cherney et al., 1991; Vogel and Jung, 2001; Sattler et al., 2010). The brown midrib mutants, derived spontaneously or through chemical mutagenesis, have played an important role in identifying a non-redundant set of genes whose products are required in cell wall lignification. In maize and sorghum collectively, five Brown midrib (Bm in maize; Bmr in sorghum) loci have been cloned and characterized, which encode enzymes in either monolignol biosynthesis or S-adenosyl methionine (SAM) metabolism. Bm1 and Bmr6 encode a cinnamyl alcohol dehydrogenase (CAD) that catalyzes the reduction of cinnamaldehydes to alcohols in the last step of monolignol biosynthesis (Halpin et al., 1998; Saballos et al., 2009; Sattler et al., 2009; Chen W. et al., 2012). Bm3 and Bmr12 encode a caffeic acid O-methyltransferase (COMT) that catalyzes the methylation of 5-hydroxyguaiacyl groups to form syringyl groups in monolignol biosynthesis (Vignols et al., 1995; Humphreys et al., 1999; Osakabe et al., 1999; Bout and Vermerris, 2003). Bm5 and Bmr2 encode a 4-coumarate: coenzyme A ligase that catalyzes the formation of p-coumaroyl-CoA, an intermediate in both monolignol and flavonoid biosynthesis (Saballos et al., 2012; Xiong et al., 2019). Bm2 encodes methylenetetrahydrofolate reductase (MTHFR), and Bm4 and Bmr19 encode folylpolyglutamate synthase (FPGS); both enzymes catalyze reactions in SAM synthesis (Tang et al., 2014; Li et al., 2015; Adeyanju et al., 2021). The cofactor SAM is a methyl donor for the two methyltransferases, caffeoyl-CoA O-methyltransferase (CCoAOMT) and COMT in monolignol biosynthesis. The genes encoded at four brown midrib loci in maize and sorghum, bm6, bmr29, bmr30, and bmr31 remain to be identified (Ali et al., 2010; Chen Y. et al., 2012; Sattler et al., 2014).
Classical map-based cloning approaches for delineating the genes underlying a given phenotype are potentially informative, but these approaches are low-throughput and time-consuming. Bulked segregant analysis (BSA) provides a simple approach for rapidly identifying molecular markers tightly linked to the causal gene (Michelmore et al., 1991). This approach requires the creation of a segregating population from the progenies; two bulked DNA samples are generated with contrasting phenotypes and further genotyped with molecular markers. The BSA technique has been used to map many important genes in various crop species (Quarrie et al., 1999; Asad et al., 2012; Wang et al., 2012). The development of next-generation sequencing (NGS) technology, with its ease and cost efficiency, has dramatically accelerated the process of identifying causal genes (Hartwig et al., 2012; James et al., 2013; Krothapalli et al., 2013; Nordström et al., 2013). This approach has been successfully used to identify candidate genes for important traits or phenotypes in rice (Tang et al., 2016; Zheng et al., 2016; Wambugu et al., 2018), sorghum (Krothapalli et al., 2013; Jiao et al., 2018), maize (Liu et al., 2012; Haase et al., 2015), and soybean (Kawashima et al., 2016; Song et al., 2017).
In the current study, the gene encoded at the sorghum Brown midrib 30 (Bmr30) locus was identified and characterized using a BSA–NGS approach with the bmr30 mutant previously isolated by Sattler et al. (2014). Bmr30 encodes a chalcone isomerase (CHI) and, unlike other brown midrib mutants of maize and sorghum, bmr30 mutants appear to not directly impair monolignol synthesis. Rather, the loss of CHI activity affects the synthesis of the flavonoid tricin, which ultimately results in reduced lignin deposition.
Materials and Methods
Germplasm and Genetic Stocks
The bmr30-1 mutant was isolated and characterized from an ethyl methane sulfonate (EMS)-mutagenized TILLING population of, BTx623, sorghum (S. bicolor) (Xin et al., 2008; Sattler et al., 2014). The mutant referred to as bmr30-2 (PI 678335) was identified using Sorghum Genomics, Gene Discovery Platform at Purdue University1 from a sequenced EMS-mutagenized TILLING population of BTx623, and the seed was obtained through GRIN-Global2. Near-isogenic lines of bmr30-1 in two genetic backgrounds (Wheatland and RTx430; Brown et al., 1936; Miller, 1984) were developed through three cycles of backcrossing bmr30-1 in BTx623 with these two parental lines as recurrent parent and the leaf midrib phenotype as the genetic marker.
Plants Growth and Care
Seeds were planted in a soil mixture with a 1:2:1:1 ratio of soil, peat moss, vermiculite, and sand and arranged in a randomized complete block design at the University of Nebraska–Lincoln greenhouse facility. Plants were grown under a 12:12 h light:dark cycle and supplemented with high-pressure sodium lighting; greenhouse temperatures were maintained at 29–30°C during the day and 26–27°C at night, respectively. Watering was conducted daily or as needed and Dyna Green All Purpose 12-12-12 fertilizer was applied weekly. Plants were harvested 5–6 weeks after planting for microscopy, RT-qPCR, and protein analyses. Plants grown for stover analyses were grown to maturity.
Generation of the Mapping Population
The bmr30-1 mutant was backcrossed to BTx623 for three generations creating a BC3F2 mapping population. BC3F2 bmr30 plants were scored for the midrib phenotype when plants were approximately 0.5 m in height. Digital images were collected to document the leaf midrib phenotype that was continuously monitored throughout the growth of the plants. Individuals whose leaf midribs were not clearly brown (bmr) or green (WT) were not included in the study.
Fully expanded leaves from 25 individuals from each phenotypic class were collected for genomic DNA (gDNA) extraction and whole-genome sequencing (WGS). gDNA was extracted from each individual leaf using a cetyl-trimethyl-ammonium bromide-based (CTAB) DNA extraction buffer (Rogers and Bendich, 1985). A total of 300 ng gDNA per individual was pooled to create each bmr and WT pool for sequencing. The individual and pooled DNA samples were analyzed by 0.8% agarose gel-electrophoresis to visualize the integrity of the DNA. DNA pools were sonicated on the Covaris LE200 (Covaris, MA, United States) using a protocol designed to achieve a target size of 350 bp. One microgram of total gDNA per pooled sample was used for Nextera Mate Pair library preparation (Illumina, San Diego, CA, United States) and WGS on an Illumina HiSeq X platform (Illumina, San Diego, CA, United States), generating 150-bp paired-end reads. The barcoded libraries were multiplexed and sequenced at Hudson-Alpha Institute for Biotechnology (Huntsville, AL, United States)3. Approximately 59.5 and 74.2 Gb of high-quality 150-bp paired-end sequence data was obtained for the bmr and WT pools, respectively. The gDNA datasets analyzed for this study are available at NCBI’s Sequence Read Archive under PRJNA736969.
Variant Calling
Paired-end Illumina reads from each WT and bmr pooled sample were aligned to the BTx623 S. bicolor reference genome (version 3.14) using Bowtie 2 (v2.3.4.1) mem algorithm with default parameters (Langmead and Salzberg, 2012). Alignment files were converted from sam to bam files and subsequently sorted using SAMtools (v1.8) view and sort commands, respectively. SAMtools mpileup (v1.8) (Li, 2011) was used to output variants using the following parameters: “-B -Q 20 -P Illumina -C50 -uf,” results were piped to BCFtools (v1.7) and variants were called with the view command (Li, 2011). Single nucleotide polymorphisms (SNPs) were filtered by the following criteria: (1) coverage ≥5 and ≤100, (2) EMS generated mutations result in G:C to A:T transition mutations, therefore only G to A and C to T single nucleotide changes were retained, and (3) SNPs homozygous and heterozygous for the mutant and WT pools, respectively. The effect of each SNP was annotated using SnpEff (v4.3) based on gene models from the S. bicolor reference genome (version 3.15). SNPs with large effects on genes (missense, nonsense, splice site acceptor, and splice site donor) were predicted using SnpEff (Cingolani et al., 2012) and retained as candidate causal mutations.
Confirmation of Mutation in Mapping Population
Derived cleaved amplified polymorphic sequence (dCAPS) markers were designed to interrogate the causal mutations for both the bmr30-1 and bmr30-2, a G to A transition at position 1252 bp in bmr30-1 and an insertion of ATGA at position 1029 bp in bmr30-2 of Sobic.001G035600 based on the genomic sequence from BTx623 S. bicolor reference genome v3.1 in Phytozome. PCR primers 5′-GCTGGAGTCCATCATCAGGGAGCACG-3′ (forward) and 5′-CGTGCTCCCTGATGATGGACTCCAGC-3′ (reverse) were used to amplify the bmr30-1 region and introduce an AvaI restriction site in the 209 bp product. PCR primers 5′-GAGAATTGCGTGGCGTTCTG-3′ (forward) and 5′-ACA GGCAGGTAGGGTATAGTACCCA-3′ (reverse) were used to amplify the bmr30-2 region and introduce an NcoI restriction site in the 176 bp product. The bmr30-1 and bmr30-2 amplified products were restriction enzyme digested with AvaI and NcoI, respectively (New England Biolabs) for 2 h at 37°C following the manufacturer’s conditions. The digested samples were analyzed using 4.0% agarose gel-electrophoresis. The AvaI restriction enzyme cleaves only the WT PCR product resulting in the 183-bp product, whereas the 209-bp product containing the mutation remained uncleaved. The NcoI restriction enzyme cleaves only the WT PCR product resulting in the 149-bp product, whereas the 176-bp product containing the mutation remained uncleaved. Controls included a negative control without DNA template and a heterozygous mix (equal parts homozygous mutant and WT gDNA).
Allelism Test
Cross-pollinations were made between bmr30-2 as female parent and bmr30-1 pollen parent. F1 seeds from the complementation test, their parents (bmr30-1 and bmr30-2) and WT (BTx623) were planted in the greenhouse in summer 2021. The plants were visually classified as being brown midrib (bmr) or WT when the plants were 6-weeks old. Digital images were collected to document the leaf midrib phenotype from the complementation test. DNA analyses were performed to verify the progeny were the result of cross-pollination.
RT-qPCR
At 6–7 weeks after germination, the fifth leaf from the base and 10 cm of stalk tissue were harvested, immediately flash-frozen in liquid nitrogen, ground using a freezer mill (SPEX SamplePrep) and stored at −80°C. Total RNA was extracted from tissue from four individual plants per genotypic class. Approximately 100 mg of homogenized plant material was added to 1 mL of TriPure Isolation Reagent (Sigma-Aldrich) followed by RNA extraction and purification using the RNA Clean and Concentrator Kit (Zymo Research). RNA was treated with an on-column DNase treatment (Zymo Research). RNA integrity was confirmed using a 1.8% denaturing agarose gel stained with ethidium bromide (EtBr). RNA quantity was determined using a Synergy Microplate (BioTek Instruments). Total RNA (900 ng) was used for cDNA synthesis with the Transcriptor First Strand cDNA Synthesis Kit (Roche Life Science) and RT-qPCR was conducted using SsoAdvanced SYBR Green Supermix (Bio-Rad) using the Bio-Rad CFX Connect Real Time System (Bio-Rad, Inc.). Primers used for CHI (Sobic.001G035600) were 5′-TCAGATCGTTAGTTGGGCGG-3′ (forward) and 5′-CAAACACGACGCACAGACAG-3′ (reverse). The Bio-Rad data were analyzed using the housekeeping gene α-tubulin (Sobic.001G1070200.1) for normalization and ΔCt values, which were subsequently used for statistical evaluation as described below. No-template and no-reverse transcription controls were included to verify the absence of DNA contamination. Four biological replicates were analyzed for each genotypic class in duplicate.
Western Blot and Immunodetection
Proteins from bmr30 and WT plants were isolated from ground leaf and stalk tissue collected from the first set of greenhouse-grown plants. Proteins were extracted using an extraction buffer containing protease inhibitor (Sigma-Aldrich Co., P9599) (Sattler et al., 2009). Protein concentrations were measured using the Pierce 660 nm Protein Assay (Thermo Fisher Scientific). Western blot analysis was conducted as previously described in Sattler et al. (2009). Briefly, the membrane was probed with primary antibody raised against the tomato CHI (polyclonal rabbit) at a 1:5,000 dilution (Kang et al., 2014). Actin content was used as a loading control, and determined using a mouse anti-Actin monoclonal antibody (Sigma-Aldrich Co., A0480) at a 1:20,000 dilution. The secondary antibodies goat anti-rabbit (Sigma-Aldrich Co., A0545) and goat anti-mouse (Actin) IgG + horseradish peroxidase (Sigma-Aldrich Co., A4416) were used at dilutions of 1:8,000 and 1:20,000, respectively. The secondary antibody was detected using chemiluminescence with Amersham ECL western blotting reagent (GE Healthcare). Imaging of chemiluminescence was performed on a Bio-RAD ChemiDoc XRS+ instrument (Bio-RAD).
Chalcone Isomerase Activity
The Bmr30 coding region (Sobic.001G035600) was synthesized (GenScript) in expression vector pET-30a (EMD Biosciences) into KpnI and XhoI restriction sites. The plasmid was introduced into Escherichia coli Rosetta R2 cells for protein expression. Cultures inoculated from a single colony were grown to log phase at 37°C, transferred to 18°C, and induced to produce protein for approximately 18 h following addition of 0.1 mM isopropyl β-D-1-thiogalactopyranoside. Soluble protein was extracted by sonication. The expressed protein contained an N-terminal 6×-his tag and was captured on a nickel resin column and eluted using imidazole. Induction of the expressed protein and protein purification were monitored by SDS-PAGE. The mutant version (Gly191Arg; bmr30-1) was introduced through site-directed mutagenesis and purified as described above.
The bottom 10 cm of stalk tissue from 6-week-old greenhouse-grown plants were ground to a fine powder under liquid nitrogen. Cold extraction buffer containing 50 mM potassium phosphate pH 8.0, 1.4 mM 2-mercaptoethanol was added to the ground tissue, and pulse-sonicated (Branson Digital Sonifier) on ice (Robbins and Dixon, 1984). The samples were centrifuged at 18,620× g for 15 min and the supernatant was collected for the activity assay.
Chalcone isomerase activity was measured using both purified recombinant protein and plant extracts at 22°C in a 0.2 mL reaction volume containing 50 mM HEPES (pH 7.5) and 50 μM naringenin-chalcone dissolved in ethanol (Jez et al., 2000). The assays with plant extracts included 40 mM sodium cyanide to inhibit chalcone peroxidase activity (Bednar and Hadcock, 1988). Disappearance of the substrate naringenin-chalcone was monitored at 390 nm with a (BioTek Synergy H1) spectrophotometer. The protein content in reactions was determined using the Pierce 660 nm protein assay with bovine serum albumin as a protein standard. Velocity was calculated as a pmol s–1 mg–1 of protein.
Anthocyanins and Total Flavonoids
Anthocyanins and total flavonoids were extracted from nutrient-deprived seedlings using methods described in Li et al. (2006) with minor modifications. In brief, seedlings were ground in liquid nitrogen and anthocyanins were extracted with HCl/methanol (1:99 v/v) at three times volume to sample weight for 24 h at 4°C. Samples were centrifuged at 19,000× g and supernatants measured at 530 nm. Total flavonoids were determined from ground tissue by extracting in 80% methanol for 24 h at 4°C at the same sample to liquid ratio as above. After centrifugation, 10% AlCl3 was added to the supernatant to a final concentration of 1% and total flavonoids measured at 420 nm.
Histochemical Staining
When plants were 7 weeks old, midribs from the fifth leaf and stalk from the top internode under the peduncle were collected and fixed in 25:75 acetic acid:ethanol overnight and stored in 25:75 dH2O:ethanol then embedded in 7% agarose. Leica VT1200s vibratome (Leica Microsystems) was used to make 100 μm transverse sections. Sections were stained for 1–2 m in phloroglucinol-20% HCl. For vanillin-HCl staining, sections were treated for 1–2 min in ethanolic vanillin [10% (w/v)], followed with 1 volume of concentrated HCl. Sections were imaged using an Olympus BX-51 light microscope (Olympus Co.).
Chemical Analyses of Stover
Harvested stover (stalk and leaf tissue) was dried in forced-air ovens at 50°C and subsequently ground in a Wiley mill fitted with a 2-mm mesh screen (Arthur H. Thomas Co.), followed by grinding on a cyclone mill fitted with a 1-mm mesh screen (UDY Co.). Fiber analysis was performed on ground stover to determine cell wall components using a detergent digestion protocol as described by Vogel et al. (1999). Neutral detergent fiber (NDF), acid detergent fiber (ADF), and acid detergent lignin (ADL) concentrations were determined using the ANKOM 200 fiber analyzer (ANKOM Tech Co.). Relative percentage of cell wall components were calculated using component concentrations extracted on a dry weight basis (Sarath et al., 2007). Stover from four biological replicates was analyzed in duplicate.
Stover from bmr30-1, bmr30-2, and WT plants was treated for thioacidolysis followed by gas chromatography-mass spectrometry (GC–MS) to determine relative lignin subunit composition [p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) lignin units]. Samples were prepared and analyzed as described in Palmer et al. (2008). Analysis was performed in duplicate on four biological replicates per line.
Bomb Calorimetry
The energy concentration of ground stover samples (see above) were determined using a Parr 6400 bomb calorimeter (Parr Instrument Co.). Approximately 200 mg of dried, ground stover combined with 600 mg of mineral oil was combusted to estimate calories per gram of dry weight. Energy values were calculated by subtracting the energy released from combustion of the mineral oil alone from the combined mineral oil and stover, which was standardized to the sample weight.
Nutrient Deprivation of Seedlings
Seeds from WT, bmr30-1, and bmr30-2 were germinated on filter paper moistened with autoclaved purified water (NanoPure Technology) at 26°C for 5 days in the dark. After germination, the seeds were transplanted into trays containing sand moistened with reverse-osmosis (RO)-purified water. The seedlings were grown at 26°C with a 12-h day/night cycle in a growth chamber and watered with RO water as needed. After 3 weeks, the seedlings at three-leaf stage were photographed to document the accumulation of red pigments.
Nuclear Magnetic Resonance Sample Preparation and Enzyme Lignin Isolation
Samples were prepared for nuclear magnetic resonance (NMR) analysis by essentially following methods previously published (Kim et al., 2008, 2017; Kim and Ralph, 2010; Mansfield et al., 2012; Lu et al., 2013; Landucci et al., 2020).
Coarse-Milling and Removal of Extractives
Four biological replicates were obtained for each line, the stems and leaves of WT, bmr30-1, and bmr30-2, a total of 24 samples. Each vacuum-dried leaf or stem sample was ground using a shaker mill (Retch MM400, 50 mL hardened steel jar, 1 mm × 15 mm hardened steel grinding ball, 30 Hz for 2 min). The fine powder was solvent-extracted with RO water (4 × 40 mL), 80% ethanol (4 × 40 mL), and acetone (3 × 40 mL), cycling through the solvent extractions by suspending the solids in the solvent, sonicating the suspension for 20 min, centrifuging to pellet the solids (4500× g, 20 min, 4°C), and decanting the solvent. Following solvent extraction, each sample was dried under high-vacuum (Freezemobile 35EL SP Scientific, 15 mT, 48 h) to provide an extract-free cell-wall powder.
Ball-Milling
Each sample (500–600 mg) was then planetary ball-milled (Fritsch Pulverisette 7, 20 mL agate jars, 10 × 10 mL agate grinding balls, 22 grinding cycles at 600 rpm for 10 min, with 10 min rest time between cycles, and reversing direction each cycle). An aliquot (40–50 mg) of this material was put aside for whole-cell-wall NMR analysis (not reported here).
Enzyme Lignin Preparation
The remainder of the ball-milled powder was transferred to a 50 mL falcon tube, suspended in 40 mL of sodium acetate buffer (25.5 mM, pH 5.0), and treated with 40 mg of crude cellulases (Cellulysin®, CALBIOCHEM®). Each sample was incubated at 35°C on a shaker at 250 rpm for 48 h. After incubation, the solids were pelleted by centrifugation (4500× g, 20 min, 4°C) and the buffer was decanted. The acetate buffer (40 mL) was replenished, the sample vortexed to resuspend the solids, fresh cellulase (40 mg) was then added, and the solids were incubated on a shaker for 48 h. The sample was again pelleted, spent buffer decanted, and then the pelleted solids were washed with RO water (3 × 40 mL) by suspending the solids, pelleting the solids, and decanting the water. The washed solids were frozen (–20°C) and then freeze-dried (Freezemobile 35EL SP Scientific, 15 mT, 24 h) to yield the enzyme lignin (EL) that comprises essentially all of the (polymeric) lignin in the sample, without fractionation, but also containing some polysaccharide and some protein.
Nuclear Magnetic Resonance Analysis
Nuclear magnetic resonance experiments were performed on both whole-cell-wall gel and EL samples as previously described (Kim et al., 2008, 2017; Kim and Ralph, 2010; Mansfield et al., 2012; Lu et al., 2013; Landucci et al., 2020). The whole-cell-wall gel samples were prepared by suspending 40–50 mg of sample in 0.6 mL DMSO-d6:pyridine-d5 (4:1, v/v) and sonicating the samples, with occasional mixing by vortexing, until a uniform gel was formed. The EL samples were prepared by dissolving 10–20 mg of sample in 0.6 mL DMSO-d6:pyridine-d5 and sonicating them, with occasional vortexing, until the solids dissolved. NMR experiments were performed on a Bruker Biospin (Billerica, MA, United States) AVANCE NEO 700 MHz spectrometer equipped with a 5-mm QCI 1H/31P/13C/15N cryoprobe with inverse geometry (proton coils closest to the sample). The central DMSO solvent peak was used as the internal reference (δC 39.5, δH 2.49 ppm). The 1H–13C correlation experiment was an adiabatic HSQC experiment (Bruker standard pulse sequence “hsqcetgpsisp2.2”; phase-sensitive gradient-edited-2D HSQC using adiabatic pulses for inversion and refocusing) (Kupce and Freeman, 2007). HSQC experiments for the EL and whole-cell-wall samples were carried out using the following parameters: acquired from 11.66 to −0.66 ppm in F2 (1H) with 3448 data points (acquisition time, 200 ms) and 215 to −5 ppm in F1 (13C) with 618 increments (F1 acquisition time, 8 ms) of 24 scans with a 1 s interscan delay; the d24 delay was set to 0.89 ms (1/8J, J = 140 Hz). The total acquisition time for a sample was 5 h. After zero-filling to 2k × 1k datapoints, processing used typical matched Gaussian apodization (GB = 0.001, LB = −0.5) in F2 and squared cosine-bell in F1 (without using linear prediction). Volume integration of contours in HSQC plots used TopSpin 4.1.1 Mac software, and no correction factors were used; that is, the data represent volume-integrals only. The aromatic signals composition on a ½ S2/6 + G2 = 100% basis. The sidechains are reported on an Aα + Bα = 100% basis, in which A = β-ether, B = phenylcoumaran; C = resinol/tetrahydrofuran peaks were too small to integrate and are not reported.
Statistical Analysis
Data were analyzed using JMP 12.2.0 (SAS Institute Inc.). Data were tested for normality using the Wilkes–Shapiro test and were log-transformed if the data failed to meet normality. Pairwise comparisons among lines were performed using Tukey’s Honest Significant Differences test at α ≤ 0.05.
Results
Bmr30 Encodes a Chalcone Isomerase
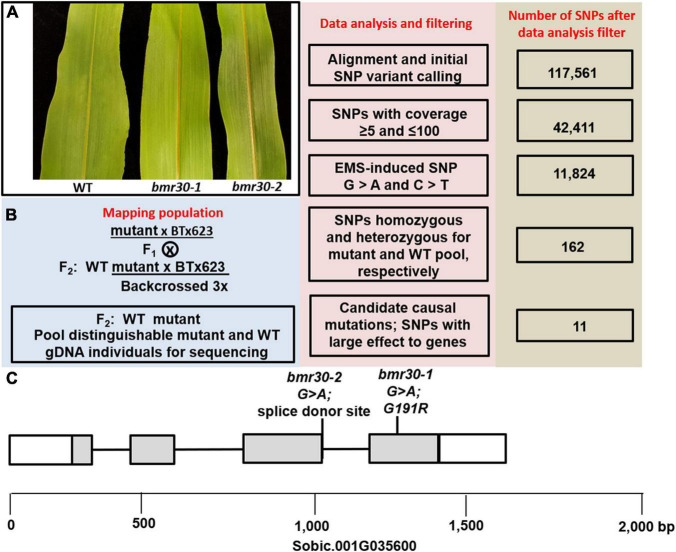
A BC3F2 population segregating for bmr30-1 was used for a bulked segregate analysis (BSA) and NGS (Figures 1A,B). Pooled gDNA was generated by bulking 25 mutant or 25 WT individuals and was subsequently subjected to high-throughput whole genome resequencing (Illumina HiSeq X platform), which yielded 74 and 59 Gb of 150-bp paired-end data for the mutant and WT pools, respectively. Over 86.8 and 86.0% of the total reads were properly and uniquely mapped to the S. bicolor v3.1 reference genome (see text footnote 5), corresponding to an average genome coverage of 80-fold for the mutant and WT pools. Based on alignment to the sorghum reference genome 117,561 SNPs were identified in the mutant and WT pools (Figure 1B). After background mutations were filtered using our data analysis pipeline, 11 homozygous mutations remained for the bulked F2 of the bmr30-1 mutant (Supplementary Table 1). Investigation of the short list with potential candidates contained Sobic.001G035600, which was annotated as a CHI. In bmr30-1, a G-to-A transition mutation at position 2,676,019 on chromosome 1 introduced a missense mutation at position 1252 bp in the CHI gene, resulting in a substitution of arginine for glycine at amino acid 191 (G191R). To confirm the genetic linkage between the leaf midrib phenotype of bmr30 and this mutation, a dCAPS marker designed to detect this mutation was used to analyze the individual DNA samples, which were pooled for high-throughput whole-genome resequencing. All the bmr30-1 individuals were confirmed to be homozygous for the G-to-A mutation (Supplementary Figure 1A). The individuals with the WT midrib phenotype were expected to segregate in a 2:1 ratio (heterozygous:homozygous WT), and 18 heterozygous and 7 homozygous WT for the polymorphism out of 25 individuals (Chi-squared 0.184; P > 0.50) were obtained. The bmr30-1 mutation was introduced into two other sorghum backgrounds, Wheatland and Tx430, through phenotype-based backcross breeding, and DNA marker anlysis confirmed the genetic linkage between this mutation and the leaf brown-midrib phenotype in different lineages (Supplementary Figure 1B).
FIGURE 1.
(A) The leaf midrib phenotype of the wild-type BTx623 (WT), bmr30-1, and bmr30-2. (B) Overview for gene identification of the mapping population, flowchart of the workflow and number of SNPs from the WT and bmr30-1 pooled and sequenced gDNA. (C) Location of the bmr30-1 and bmr30-2 mutations in the chalcone isomerase gene, Sobic.001G035600. Boxes and gray shading denote exons and coding sequences, respectively.
To corroborate these findings, a second independent mutation was identified in Sobic.001G035600 through an electronic search of the Purdue sorghum TILLING population. A G-to-A mutation at position 1029 bp of Sobic.001G035600, which is the splice donor site (GU) of the third intron (Figure 1C). Seeds from this TILLING line were planted in the greenhouse, and plants with the brown midrib phenotype were observed (Figure 1A), which confirms that loss of the Sobic.001G035600 function results in the brown midrib leaf phenotype. To determine the impact of the loss of the splice donor site, cDNA was synthesized, and a portion of the cDNA containing the junction between the third and fourth exons was amplified and sequenced from bmr30-2 plants, which showed loss of the endogenous splice donor site and resulted in an insertion of 4 bp (ATGA) at this junction. This insertion altered the reading frame of the entire fourth exon and changed amino acids 160–231. To confirm the genetic linkage between the leaf midrib phenotype of bmr30-2 and this mutation, a dCAPS marker designed to detect this mutation was used to analyze an F2 population. All the bmr30-2 individuals were confirmed to be homozygous for the G-to-A mutation at 1091 (Supplementary Figure 2A). To confirm allelism, bmr30-1 and bmr30-2 were cross-pollinated, and the leaf midrib phenotype of the F1 progeny (bmr30-2 × bmr30-1) were visually assessed when the plants were 6-weeks old. All six plants were determined to have the brown midrib phenotype (Supplementary Figure 3).
In the sorghum genome, five genes were previously identified as encoding CHI based on predicted amino acid sequence similarity to the characterized enzyme that converts naringenin chalcone to naringenin in the flavonoid pathway (Nielsen et al., 2016). Phylogenetic analysis of these genes with homologous genes from other flowering plants indicated that Bmr30 (Sobic.001G035600) resides in a clade with CHIs from other grasses, and the other four sorghum genes were in separate clades (Nielsen et al., 2016). Likewise, the pigments associated with the flavonoid pathway were not completely absent in the bmr30 mutant tissues, but visible decreases in purple pigmentation were observed in glumes surrounding the seeds (Supplementary Figure 4). To induce flavonoid biosynthesis, the seeds were germinated under nutrient deprivation, which resulted in seedling with red pigmented hypocotyls in WT seedling, but this coloration was visibly reduced in bmr30-1 and bmr30-2 seedling (Supplementary Figure 5). Further analysis showed that total flavonoids and total anthocyanin were significantly reduced both by approximately two-fold in bmr30-2 seedlings relative to WT ones, and a similar trend was observed in bmr30-1-seedlings (Supplementary Figures 5A,B). In addition, bmr30-2 plants had a lesion mimic phenotype, which only became apparent in late vegetative stages (Supplementary Figures 4A,B).
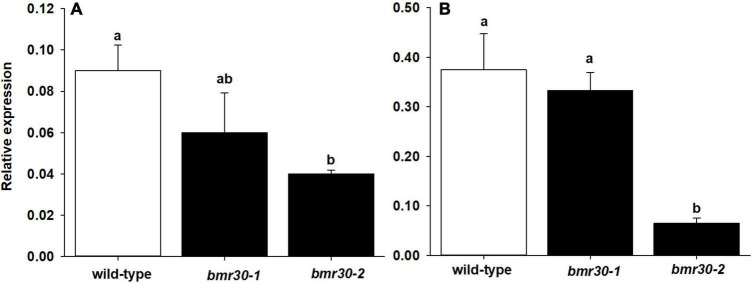
The expression levels of Bmr30 in WT, bmr30-1, and bmr30-2 plants were analyzed by quantitative RT-PCR (Figure 2) from leaf and stalk tissue. Bmr30 was expressed at low basal levels in WT leaf tissue relative to the control gene α-tubulin, and its expression was not significantly different in bmr30-1 (Figure 2A). Bmr30 expression in leaf was decreased by 60% in bmr30-2 compared to WT (Figure 2A). Bmr30 expression in stalk tissue was not significantly different between WT and bmr30-1, whereas expression in bmr30-2 stalk was significantly decreased, by 83%, compared to WT (Figure 2B). Primers used for RT-PCR were located at the fourth exon just beyond the splice site mutation in bmr30-2. Thus, the splice site mutation in bmr30-2, which is predicted to cause a loss in function for the gene, appeared to reduce expression of this transcript. The JGI Plant Gene Atlas Project available on Phytozome v136 establishes that expression of CHI (Sobic.001G035600) exists across various tissue types and developmental stages of S. bicolor (Supplementary Figure 6). CHI levels in leaves and stalk protein extracts were detected using a polyclonal antibody against a tomato CHI. The levels of CHI were barely detectable and differences among WT and the two bmr30 alleles were not observed (Supplementary Figure 7), which is consistent with the low expression levels observed by RT-qPCR.
FIGURE 2.
Relative expression levels of CHI in (A) leaf and (B) stalk tissue of wild-type, bmr30-1, and bmr30-2 plants determined using RT-qPCR. Relative expression was determined using the ΔCt method with the α-tubulin transcript (Sobic.001G1070200.1) for normalization. Error bars represent standard error. Samples with different letters are statistically different at α ≤ 0.05 using Tukey’s HSD test.
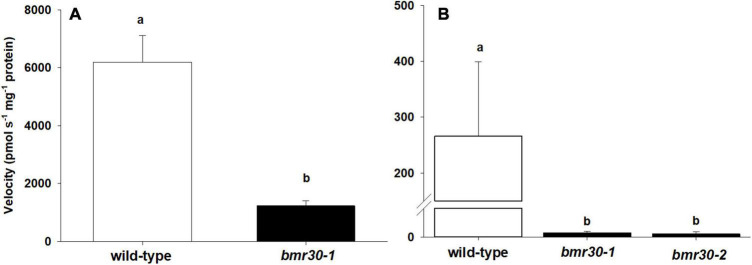
The Bmr30 coding region was cloned, heterologously expressed in E. coli, and the protein assayed for CHI activity. In addition, a version containing a G191R amino acid change to replicate the bmr30-1 allele was also expressed and purified from E. coli for a CHI activity assay. Bmr30 catalyzed the conversion of the substrate naringenin chalcone to naringenin. The CHI activity of the G191R version was reduced relative to the WT version of Bmr30 by five-fold (Figure 3A; p > 0.0017). Thus, the product of bmr30-1 may retain some residual CHI activity. Activity was assayed from crude protein extract from both stalks and leaves. CHI activity were detected in stalk extracts from WT, bmr30-1, and bmr30-2 (Figure 3B), but no activity was detectable in leaf extracts. CHI activity in bmr30 stalk extracts was reduced approximately 100-fold relative to WT levels (p < 0.0001). The undetectable levels of enzyme activity in leaf tissue were consistent with the basal level of gene expression observed with RT-PCR for Bmr30.
FIGURE 3.
Chalcone isomerase activity from (A) purified wild-type recombinant protein and a version containing the bmr30-1 mutation, and (B) plant stalk extracts of wild type, bmr30-1, and bmr30-2. CHI activity levels were assayed using naringenin chalcone substrate. The enzyme velocity was normalized to the amount of protein (mg) added to the reaction. Error bars represent standard error. Samples with different letters are statistically different at α ≤ 0.05 using Tukey’s HSD test.
Microscopy and Cell Wall Composition
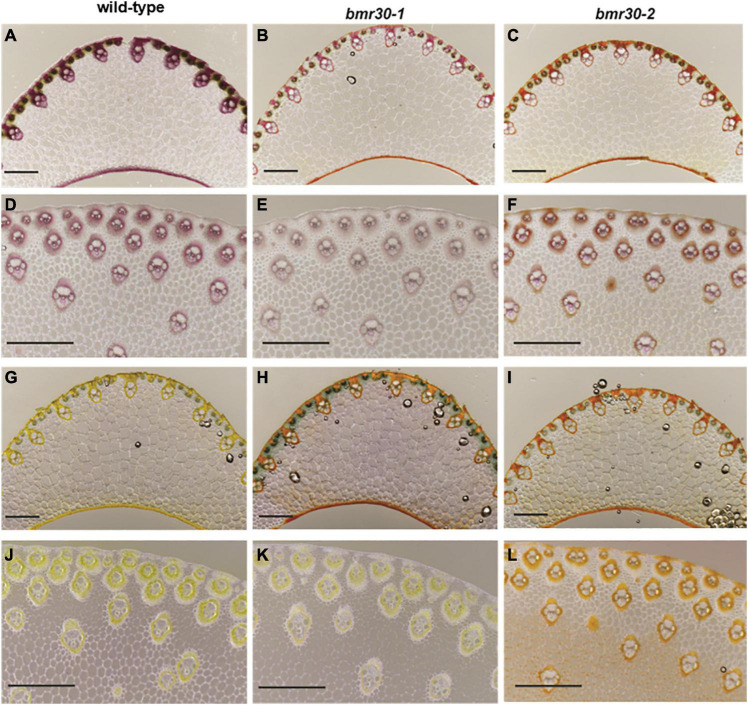
Microscopy following histochemical staining was used to examine how the loss of Bmr30 activity affected the cell walls of leaf midribs and stalks. The cell morphology of both bmr30 mutants, including fiber cells and xylem of the vascular bundles, closely resembled those cells in WT plants (Figure 4). Secondary cell walls of vascular bundles in both leaf midrib and stalk sections of WT were stained with phloroglucinol-HCl, which reacts primarily with p-hydroxycinnamaldehyde end-groups of lignin polymers (Pomar et al., 2002; Figures 4A,D). However, the staining intensity in the vascular bundles of both bmr30 mutants was reduced compared to WT vascular bundles, which indicated a potential decreased lignin deposition or an alteration in its composition (Figures 4B,C,E,F). Transverse sections were also stained with vanillin-HCl, a staining reagent that reacts with flavonoids (Gardner, 1975). The cell walls of WT leaf midrib and stalk stained yellow in color, which indicated the presence of flavonoids, presumably tricin, within them (Figures 4G,J). In contrast, the cell walls of midribs and stalks from bmr30 mutants displayed vanillin staining that was considerably different in color and intensity from that in WT, and there were also distinct differences in color and intensity in the staining between the two bmr30 alleles.
FIGURE 4.
Visualization of lignification using phloroglucinol (A–F) and vanillin-HCl (G–L) staining of leaf midrib (A–C,G–I) and stalk (D–F,J–L) tissues from wild-type (BTx623), bmr30-1, and bmr30-2 plants. Scale bar = 500 μm for both leaf midrib and stalk tissue. Leaf midrib tissue was observed at 40× magnification, stalk tissue was observed at 80× magnification using an Olympus BX-51 light microscope (Olympus Co.).
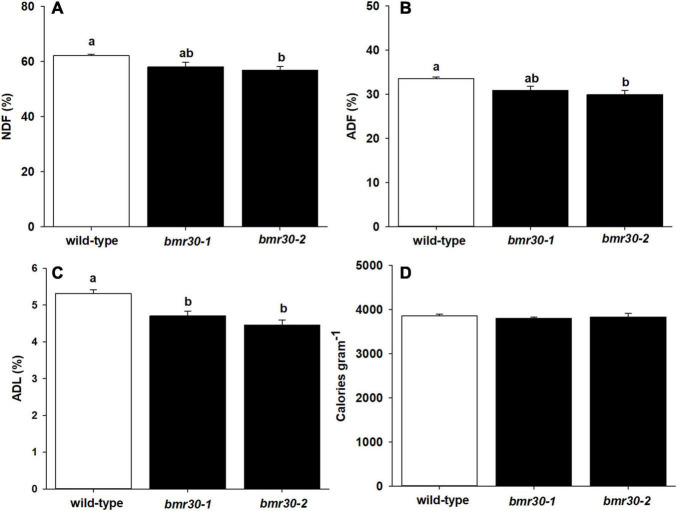
To evaluate how the loss of Bmr30 activity affects biomass composition, fiber analysis was performed on mature stover from WT, bmr30-1 and bmr30-2 plants to measure the levels of NDF, ADF, and ADL. Overall NDF, ADF and ADL were all significantly decreased in the bmr mutants compared to WT. Specifically, NDF levels were lower in both bmr30 mutants than in the WT (p = 0.0363; Figure 5A). Decreases of 6 and 10% relative to WT were observed in bmr30-1 and bmr30-2, respectively. Similarly, ADF levels were also decreased in bmr30-1 and bmr30-2 compared to WT, by 10 and 13%, respectively (p = 0.0264; Figure 5B). ADL levels were 11 and 15% lower than WT in bmr30-1 and bmr30-2, respectively (p = 0.0024; Figure 5C). The energy density of the biomass was measured using bomb calorimetry. Energy concentrations of stover were not statistically different for either mutant compared to WT (p = 0.7909; Figure 5D). The observed reduction in lignin concentration in these bmr30 mutants is consistent with the previously described effects of bmr30 on cell wall lignification (Sattler et al., 2014).
FIGURE 5.
Stover from mature wild-type (BTx623), bmr30-1, and bmr30-2 plants was subjected to fiber analysis to determine percent of (A) neutral detergent fiber (NDF), (B) acid detergent fiber (ADF), and (C) acid detergent lignin (ADL). The NDF, ADF, and ADL were determined using ANKOM fiber analyzer. Stover was also subjected to (D) bomb calorimetry to determine total energy using a Parr 6400 bomb calorimeter. Values presented are least squares means (±1 SE). Samples with different letters are statistically different from one another at α ≤ 0.05 using Tukey’s HSD test.
Thioacidolysis was performed to determine the composition of β-O-4-linked p-hydroxyphenyl (H), syringyl (S), and guaiacyl (G) subunits within the lignin polymer (Figure 6). The levels of H-lignin were significantly decreased in the bmr30-1 and bmr30-2 compared to WT (p < 0.0001; Figure 6A), with a 31 and 54% decrease, respectively. Levels of G-lignin were significantly different (p = 0.0011), with a 25 and 30% decrease in bmr30-1 and bmr30-2 compared to WT, respectively (Figure 6B). Similarly, S-lignin was significantly different (p = 0.0343), with 18 and 26% decrease in bmr30-1 and bmr30-2 relative to WT plants, respectively (Figure 6C). Overall, these two mutations in this CHI gene caused a significant decrease in lignin observed through fiber analysis, which was also detected as decreased levels in the three major β-O-4-linked lignin subunits (Figure 5C).
FIGURE 6.
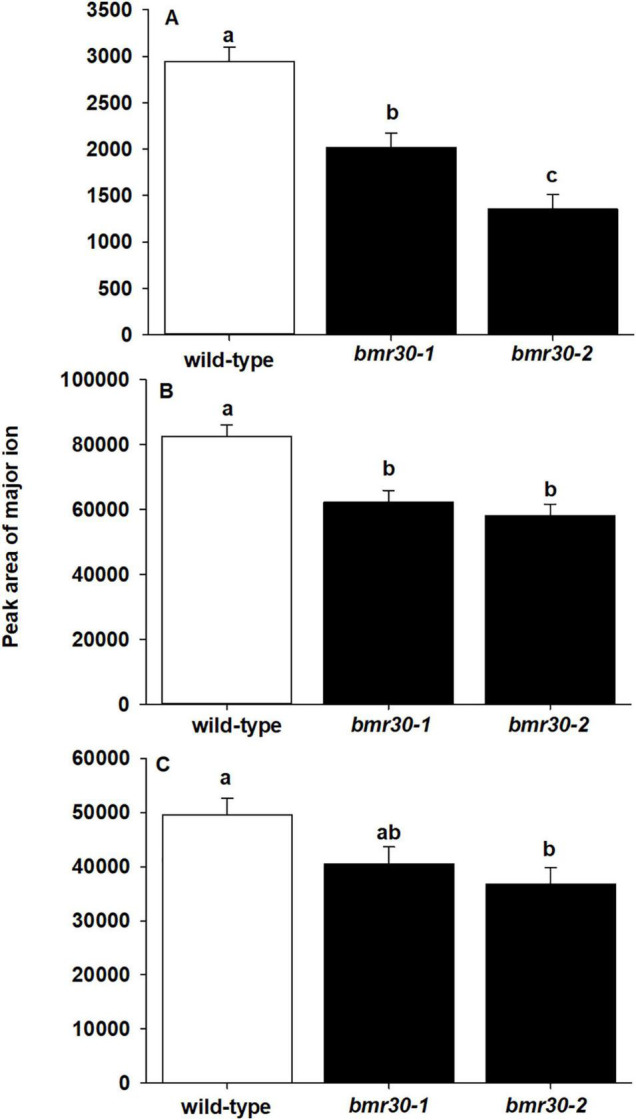
Mutations in CHI induced changes in lignin subunit composition. The abundance of (A) p-hydroxyphenyl (H), (B) syringyl (S), and (C) guaiacyl (G) lignin subunits from stover of mature wild-type (BTx623), bmr30-1, and bmr30-2 plants was measured using thioacidolysis and gas chromatography–mass spectrometry (GC–MS). Values presented are least square means (±1 SE). Samples with different letters are statistically different from one another at α ≤ 0.05 using Tukey’s HSD test.
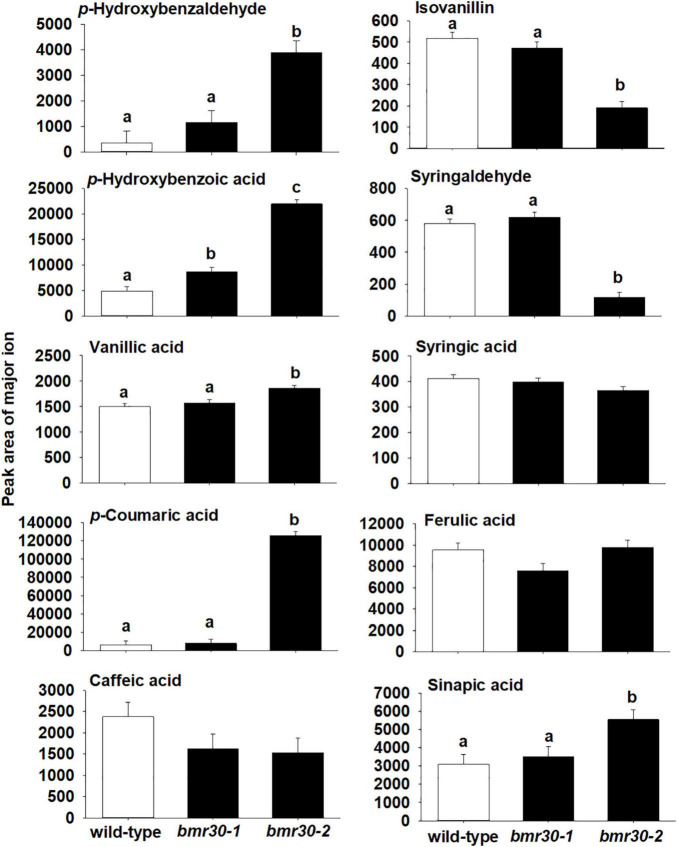
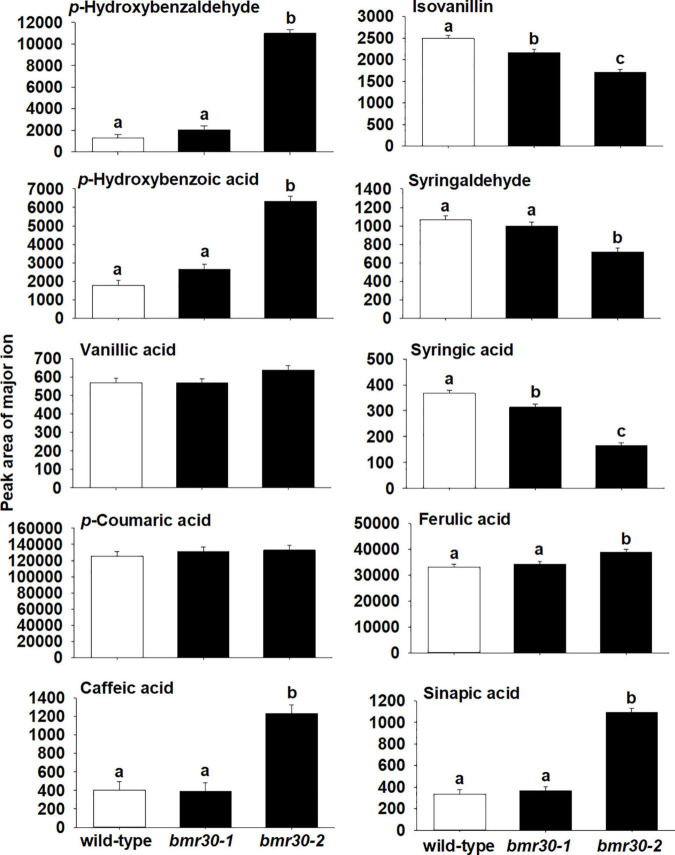
To further determine how bmr30 affects monolignol biosynthesis, soluble and cell-wall-bound phenolic compounds were extracted from stover collected from mature plants. Relative abundances of several phenolic compounds derived from the monolignol pathway were measured by GC–MS (Figures 7, 8). Several soluble phenolic compounds were different among WT and the bmr30 mutants. For example, bmr30-2 had significantly higher amounts of p-hydroxybenzaldehyde (11-fold greater), p-hydroxybenzoic acid (4.5-fold greater), vanillic acid (1.2-fold greater), p-coumaric acid (19-fold greater), and sinapic acid (1.7-fold greater) when compared to WT (Figure 8), whereas isovanillin (2.7-fold less) and syringaldehyde (4.9-fold less) were significantly reduced in bmr30-2 compared to WT (Figure 7). The only soluble phenolic compound that differed between WT and bmr30-1 was p-hydroxybenzoic acid, for which there was 3.3-fold greater amount in bmr30-1 than in WT. Cell-wall-bound phenolics were also significantly different between WT and bmr30 mutants (Figure 8). Specifically, p-hydroxybenzaldehyde (8.5-fold greater), p-hydroxybenzoic (3.6-fold greater), caffeic (3-fold greater), ferulic (2-fold greater), and sinapic (3.3-fold greater) acids were increased in bmr30-2 compared to WT. Isovanillin (1.3- to 1.5-fold less), syringaldehyde (1.5-fold less), and syringic acid (1.8- to 2.2-fold less) were lower in bmr30 mutants compared to WT. Taken together, loss of Bmr30 activity affected accumulation of the compounds derived from monolignol biosynthesis.
FIGURE 7.
Relative abundance of soluble phenolic compounds in stover from wild-type, bmr30-1, and bmr30-2 plants. Phenolic compounds were analyzed via GC–MS. Values presented are least square means; error bars represent standard error. Samples with different letters for soluble fractions are statistically different from one another at α ≤ 0.05 using Tukey’s HSD test.
FIGURE 8.
Relative abundance of wall-bound phenolic compounds in stover from wild-type, bmr30-1, and bmr30-2 plants. Phenolic compounds were analyzed via GC–MS. Values presented are least square means; error bars represent standard error. Samples with different letters for wall-bound fractions are statistically different from one another at α ≤ 0.05 using Tukey’s HSD test.
Lignin Polymer Analysis
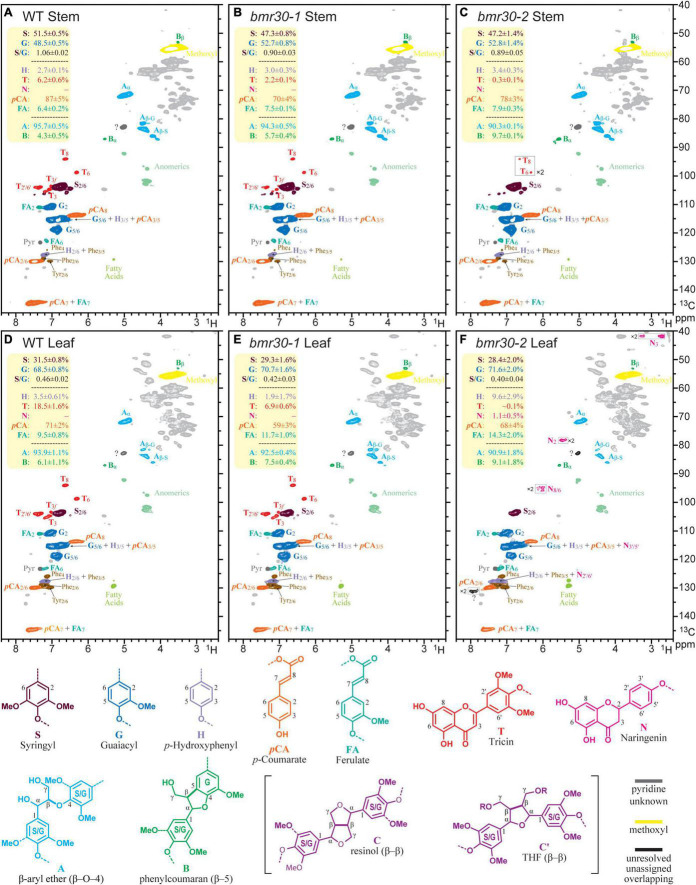
Changes to the lignin polymeric composition and structure were discerned from NMR profiling (Ralph and Landucci, 2010; Tobimatsu et al., 2019). Extractions for ELs, representing essentially the entire lignin component, were performed by digesting away the majority of the polysaccharides using polysaccharidases (crude cellulase) (Chang et al., 1975). NMR spectra from such lignins are cleaner and sharper than those spectra obtained from whole-cell-wall samples, and were useful here to glean diagnostic details. As shown in Figure 9, the 2D HSQC spectra disperse and resolve various aromatic components in the polymer, including the H, G, and S units in the core lignin, the p-coumarates (pCA) acylating the lignin sidechain in grasses, the ferulates (FA) that may be on residual arabinoxylans or analogously acylating lignin sidechains, and the flavone tricin (T) that has been found on all Poaceae lignins studied prior to the publication date (Lan et al., 2016b). In the leaf tissue EL of the bmr30-2 mutant, the intermediary flavanone naringenin (N) was also identified.
FIGURE 9.
Partial 2D HSQC NMR spectra (in 4:1 DMSO-d6/pyridine-d5) of the enzyme lignin (EL) preparations isolated from the wild-type and bmr30 lines. (A–C) Spectra of ELs from the stems of wild type (WT), and the bmr30-1, and bmr30-2 mutants. (D–F) Analogous spectra from the leaf ELs. Each plotted spectrum is a representative from a set of 4 biological replicates for each; the integral data in the top-left yellow-backgrounded panel of each presents the average and standard deviation from the analysis of all 4 replicates of each line, for a total of 24 samples. The main structural features are, where resolved, colored to match the structures below; no attempt is made to individually color the more minor components in overlapping peaks. The T denotes the tricin peaks, and N the naringenin peaks. Volume integration was used to determine the relative abundances of the G and S lignin units (and the S/G ratio); the other aromatic units including the H, p-coumarate (pCA), ferulate (FA), tricin (T), and naringenin (N) are reported on an S + G = 100% basis. The H-levels are not reliable because of the overlap with significant phenylalanine (Phe) peaks from proteins, particularly in the leaves, but are corrected by subtracting the volume of the Phe2/6 from the volume of the H + Phe3/5 peak. Only two of the lignin sidechain structures, characterized by their inter-unit linkages (A, B), were readily determined here, and are expressed as fractions of the sum A + B = 100%. The β-ether content in these samples are not considered to be over 90% of all total units. Resinols C (see structures below) were found at low contour levels (not shown), and the correlation peak labeled? is where the major tetrahydrofuran peak occurs. However, the β and γ correlations were not evident (even from TOCSY-HSQC spectra, not shown), so the large peak here is overlapped by another unknown peak; therefore, measurement was not attempted. As previously noted (Mansfield et al., 2012; Abu-Omar et al., 2021), endgroup units such as T and pCA, are over-represented in these spectra, but the relative levels remain useful for comparisons.
HSQC NMR data is not strictly quantitative and the following should be noted: (1) Despite being strictly non-quantitative, the S:G data are considered to be reliable because of the similar environments of the G2 and S2/6 proton/carbon pairs (Mansfield et al., 2012). (2) The H-level data are distinctly unreliable because of overlap with a significant phenylalanine component (Kim et al., 2017). (3) End-units such as pCA and tricin, and perhaps FA, are significantly overestimated due to the longer relaxation of more mobile units than the backbone units in the polymer, but the integrals are useful for comparative analysis (Ralph and Landucci, 2010; Mansfield et al., 2012).
Bearing the above in mind, the following observations are particularly relevant results on the composition and structure of the different lignins. First, the S/G ratio was significantly lower in both the stems and the leaves of the bmr30 mutants than in the WT, and was essentially equivalent in both mutants. Second, pCA levels were similarly lower, which reflected the lower S contents of the mutants and pCA units were predominantly (∼90%) on S-units in these samples. Third, ferulate (FA) levels appeared to be higher in the mutants. Fourth, consistent with the loss of CHI activity, the tricin (T) contents were strikingly lower in the mutants, and particularly so in bmr30-2 (0.1%), than in the WT (18.5%). Naringenin (N) units were detected in the leaf EL from bmr30-2 (Figure 9E), and perhaps were also detectable in the bmr30-2 stem EL. This finding is unexpected and appears to be inconsistent with loss of CHI (Sobic.001G035600) function in bmr30-2, because naringenin is the product of the CHI enzyme. The incorporation of naringenin, a flavonoid pathway intermediate has been previously documented in a rice flavone synthase (FNSII) mutant (Lam et al., 2017) and most recently in a poplar CHS transgenic (Mahon et al., 2021) and in papyrus (Rencoret et al., 2021).
Discussion
This study demonstrated that the sorghum Bmr30 locus encodes a CHI, with loss of function affecting monolignol and flavonoid biosynthesis and lignin deposition. Many genes encoding CHI from angiosperm plant species have been isolated and characterized. The brown midrib phenotype, which led to the isolation of the bmr30-1 mutant (Sattler et al., 2014), has long been linked to C4 grasses impaired in cell wall lignification (Jorgenson, 1931; Kuc and Nelson, 1964; Gee et al., 1968; Kuc et al., 1968). Previously identified bmr/bm loci of monolignol biosynthesis have all encoded enzymes of the monolignol pathway or enzymes involved in the synthesis/recycling of S-adenosylmethionine (SAM), a required cofactor for the two methyltransferases of this pathway. However, Bmr30, a CHI, breaks this precedent and links two branches of phenylpropanoid metabolism, monolignol and flavonoid biosynthetic pathways, as being required for lignin deposition.
Chalcone synthase (CHS) catalyzes the first committed step in flavonoid biosynthesis that combines p-coumaroyl-CoA derived from monolignol biosynthesis and malonyl-CoA from the acetate/malonate-derived polyketide pathway into naringenin-chalcone, which is then isomerized by CHI (CHI) within the cytosol. Bmr30 has CHI activity as demonstrated in this study using versions of the recombinant protein expressed in E. coli, and the version G191R to recapitulate the missense mutation bmr30-1 also retained residual activity, albeit with a 49-fold reduction from WT levels in vitro (Figure 3A). The amino acid glycine at position 191 of CHI is highly conserved among Phaseolus vulgaris, Medicago sativa, Pisum sativum, Zea maize, Vitis vinifera, Ipomoea purpurea, Petunia hybrida, and Arabidopsis thaliana and is two amino acids downstream from residues of the (2S)-naringenin binding cleft (Jez et al., 2000). The substitution of arginine, a charged amino acid with a large side chain for glycine without a sidechain proximal to the substrate binding cleft would most likely impair substrate binding in bmr30-1. Likewise, bmr30-2 contains a 4-bp insertion resulting in a frameshift, which eliminates several secondary structural motifs found in the last 72 amino acids and that includes strands e and f of the third β-sheet and α-helices 6 and 7. α-Helix 6 is highly conserved across all CHIs, and it is part of the active site cleft. Amino acids 188 and 189 of this helix are proposed to confer substrate specificity (Jez et al., 2000). Thus, this allele encodes a protein which is most likely misfolded and rapidly degraded; hence bmr30-2 is most likely a null (amorphic) allele. The Bmr30 gene (Sobic.001G035600; previously designated Sb01g003330) transgenically complemented the CHI mutant transparent testa 5 (tt5) restoring anthocyanin pigmentation in Arabidopsis (Liu et al., 2010). Together these results demonstrate that the bmr30 mutant contains a defective CHI. However, the expression of the gene is relatively low in leaves not experiencing stress, which is consistent with our CHI assays, the RT-qPCR analysis and previously published RT-qPCR analysis (Liu et al., 2010). There is some genetic redundancy that is responsible for the residual flavonoids and anthocyanins present especially bmr30-2 under stress (Supplementary Figures 4, 5), and one or more of four other CHI-like sequences in the sorghum genome may be responsible. These results suggest that the function of Bmr30 requires its presence at relatively low levels in plant tissue not under stress conditions, which is consistent with the sorghum gene expression atlas for this gene (Supplementary Figure 6). Alternatively, Bmr30 may only be present in a limited subset of cell types within plants, which could also explain the results observed. Our data indicate that a loss or reduction of Bmr30 activity results in reduced lignin deposition and flavonoid biosynthesis. However, accumulation of pigment in bmr30-1 (designated line 100) in response to peduncle inoculation of the fungal pathogen Fusarium thapsinum was not different from WT (Funnell-Harris et al., 2014). The awns surrounding the kernels were one tissue where the intensity of pigmentation was visibly reduced for both bmr30 mutants compared to WT (Supplementary Figure 4). Similarly, nutrient-deficiency also showed synthesis of total flavonoids and total anthocyanins were decreased in bmr30 seedlings compared to WT seedlings (Supplementary Figure 5).
Loss of Bmr30 reduced lignin deposition in both mutants and across four lines of experimental evidence. Cytologically, the phloroglucinol staining showed reduced amounts of lignin in bmr30 cell walls relative to WT. Likewise, chemical analyses showed reduced ADL and decreased levels of major lignin monomers released by thioacidolysis in bmr30 relative to WT, which together corroborate this reduction. Recent studies of monocotyledonous plant cell walls using NMR have identified the flavone tricin as an endogenous subunit of the lignin polymer in wheat (del Río et al., 2012), maize (Lan et al., 2015), and sorghum (Eudes et al., 2017), and is predicted to be in all species in the Poaceae (Lan et al., 2016b). Tricin links flavonoid metabolism to lignin deposition, and has been shown to be an authentic monomer of lignin, which acts as an initiation site for the lignin chain (del Río et al., 2012; Lan et al., 2015, 2016a). In addition, tricin may have potential health benefits as an antioxidant, anti-aging, anticancer, and cardioprotective compound (Oyama et al., 2009; Zhou and Ibrahim, 2010; Chambers and Valentova, 2015). In the maize CHS Colorless2 (C2-Idf) mutant, tricin levels were significantly reduced in the lignin polymer (Eloy et al., 2017). Indeed, in the current study, NMR profiling also showed alteration to the composition and structure of the different lignins in both bmr30 mutants relative to WT. The S contents were lower in the mutants, which led to reduced levels of p-coumarate and increased levels of ferulate. The tricin (T) contents were strikingly lower in the mutants as anticipated (Figure 9).
Vanillin-HCl stains flavonoid compounds yellow (Gardner, 1975), as was observed in WT leaf midrib and stalk sections. In contrast to WT, staining was greatly reduced in bmr30-1 tissue sections, which is consistent with the substantial reduction of tricin in lignins determined by NMR profiling. However, the more intense orange-brown vanillin-HCl staining in bmr30-2 cell walls may suggest increased accumulation of other flavonoids or phenylpropanoids in this mutant. Likewise, there were substantial increases in several soluble and wall-bound phenylpropanoids in bmr30-2 stover, particularly in mature tissues. Paradoxically, the presence of naringenin, the product of CHI, in lignin from bmr30-2 mature leaves (Figure 9E) is not consistent with results from the bmr30-1 mutant and requires further examination. The most plausible explanation is that an additional, unidentified mutation, present in the bmr30-2 line causes the accumulation in phenylpropanoids described above. The gene discovery platform did not identify any mutations in other flavonoid-related genes for this EMS-generated line, which included the FNSII gene (see text footnote 1). Potentially, the lesion-mimic phenotype observed in the bmr30-2 may be associated with the accumulation of phenylpropanoid compounds as a response to lesion development. The lesion-mimic phenotype is most likely due to a mutation in a gene other than CHI (Sobic.001G035600) because the bmr30-1 mutant does not exhibit this phenotype and neither do CHI mutants from other plants (Shirley et al., 1995; Hong et al., 2012; Gurdon et al., 2019). A backcrossing strategy has been initiated to attempt to separate the brown midrib phenotype from the lesion-mimic phenotype, which should resolve whether loss of CHI function results in the accumulation of naringenin and hydroxycinnamates in bmr30-2 leaves and stover.
In summary, the identification of Bmr30 provides new avenues for the investigation of phenylpropanoid metabolism in sorghum and other C4 grasses. The bmr30 mutants represent a new class of tools to alter lignin deposition to improve forage for livestock, biofuels, and green chemistry utilization. A future goal will be to combine bmr30 with other characterized bmr mutants that directly impact monolignol biosynthesis to explore the effect on lignin and cell walls on sorghum biomass.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA736969.
Author Contributions
HT and SS designed the research and wrote the first draft of the manuscript. HT, TG, JT, SL, and JR performed the experiments. HT, TG, JT, DF-H, WV, JR, SL, ZX, and SS analyzed and interpreted the data. All authors reviewed and revised the manuscript prior to publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Alison Coomer for technical assistance, Nathan Palmer for expertise with GC/MS, Gregg Howe (Michigan State University) for the gift of the polyclonal antibody against the tomato chalcone isomerase and Heather Van Buskirk for critically reviewing the manuscript. The United States Department of Agriculture, Agricultural Research Service, is an equal opportunity/affirmative action employer and all agency services are available without discrimination. Mention of commercial products and organizations in this manuscript is solely to provide specific information. It does not constitute endorsement by USDA-ARS over other products and organizations not mentioned.
Footnotes
Funding
This research was supported by the United States Department of Agriculture, Agriculture Research Service Project 3042-21220-033-00-D (SS and DF-H), and the DOE Great Lakes Bioenergy Research Center, DOE Office of Science BER DE-SC0018409 (JR and SL). The bmr30 mutant was first identified as part of United States Department of Energy Office of Science (BER) grant DE-FG02-07ER64458 (SS and WV).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.732307/full#supplementary-material
The derived cleaved amplified polymorphic sequence (dCAPS) marker for the bmr30-1 allele was used to verify genetic linkage between brown midrib phenotype and the bmr30-1 mutation from F2 individuals that were pooled for next generation DNA sequencing. (A) Genomic DNA from 25 individuals with the brown midrib phenotype and 25 individuals with the wild-type (green; wt) midribs were screened with dCAPS marker for bmr30-1, which was designed to create an AvaI restriction site in only the wild-type allele that resulted in a 183 bp product upon restriction digestion. The bmr30-1 allele lacked this restriction site due to the transition and the 209 bp product was not cleaved by AvaI. All individuals with the brown midrib phenotype were homozygous for the bmr30-1 allele, whereas seven individuals were homozygous for the wild-type allele and 18 individuals heterozygous. The latter two groups had normal midribs. (B) bmr30-1 was backcrossed into three different parental lines (Tx623, Wheatland, Tx430) and progeny were phenotyped based on the leaf midrib. These individuals were further tested and confirmed using the dCAPS marker and compared to the original mutant line (bmr30-1). PCR amplification controls were performed with DNA from bmr30-1 (bmr), wild-type (wt), 1:1 mixture of wild-type and bmr30-1 DNA to simulate a heterozygous plant (het) and no DNA template (-).
The derived cleaved amplified polymorphic sequence (dCAPS) marker for the bmr30-2 allele was used to verify genetic linkage between brown midrib phenotype and the bmr30-2 mutation from F2 individuals. Genomic DNA from (A) 25 individuals with the brown midrib phenotype and (B) 25 individuals with the wild-type (green; wt) midribs were screened with dCAPS marker for bmr30-2, which was designed to create an NcoI restriction site in only the wild-type allele that resulted in a 149 bp product upon restriction digestion. The bmr30-2 allele lacked this restriction site due to the insertion and the 176 bp product was not cleaved by NcoI. All individuals with the brown midrib phenotype were homozygous for the bmr30-2 allele, whereas eight individuals were homozygous for the wild-type allele and 15 individuals heterozygous. The latter two groups had normal midribs. PCR amplification controls were performed with DNA from bmr30-2, 1:1 mixture of wild-type and bmr30-2 DNA to simulate a heterozygous plant (het), wild-type (wt), and no DNA template (-).
The leaf midrib phenotype of the wild-type (BTx623), bmr30-1, bmr30-2, and F1 plants form the complementation test bmr30-2 × bmr30-1. The F1 progeny were confirmed to result from cross-pollination using DNA marker analysis. The eighth leaf was photographed from 6-week-old plants.
(A) Ten week old plants grown in the greenhouse, (B) seventh leaf from the bottom of 10 week old bmr30-2 plant, displaying the disease lesion mimic phenotype, and (C) seeds and glumes from wild-type (WT), bmr30-1, and bmr30-2. The top row is the abaxial side and bottom row is the adaxial side. The grain was harvested from the bottom of the panicles at approximately 40 days after anthesis.
Sorghum seedlings were germinated and grown under nutrient-depleted conditions to induce flavonoid accumulation. The absorbance of (A) total flavonoids and (B) anthocyanins were measured from the (C) seedlings. The seedlings from left to right are WT, bmr30-1, and bmr30-2, photographed 21 days after germination.
Expression abundance of chalcone isomerase (CHI; Sobic.001G035600), across diverse tissues from Sorghum bicolor. Gene expression data was obtained from The JGI Plant Gene Atlas Project available on Phytozome v13 (https://phytozome-next.jgi.doe.gov/phytomine/aspect.do?name=Expression).
Immunoblot detection of CHI from leaves (top) and stalks (bottom). Protein extracts from wild-type (WT), bmr30-1, and bmr30-2 were separated by SDS-PAGE, transferred to membrane, and probed with polyclonal antibodies raised against the recombinant tomato CHI protein. The recombinant SbCHI protein (SbCHI) was included as a positive control. Polyclonal antibodies raised against actin protein were used as a loading control.
References
- Abu-Omar M. M., Barta K., Beckham G. T., Luterbacher J. S., Ralph J., Rinaldi R., et al. (2021). Guidelines for performing lignin-first biorefining. Energy Environ. Sci. 14 262–292. [Google Scholar]
- Adeyanju A. O., Sattler S. E., Rich P. J., Rivera-Burgos L. A., Xu X., Ejeta G. (2021). Sorghum brown midrib19 (Bmr19) gene links lignin biosynthesis to folate metabolism. Genes (Basel) 12:660. 10.3390/genes12050660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali F., Scott P., Bakht J., Chen Y., Lübberstedt T. (2010). Identification of novel brown midrib genes in maize by tests of allelism. Plant Breed. 129 724–726. 10.1111/j.1439-0523.2010.01791.x [DOI] [Google Scholar]
- Asad M. A., Xia X., Wang C., He Z. (2012). Molecular mapping of stripe rust resistance gene YrSN104 in Chinese wheat line Shaannong 104. Hereditas 149 146–152. 10.1111/j.1601-5223.2012.02261.x [DOI] [PubMed] [Google Scholar]
- Bednar R. A., Hadcock J. R. (1988). Purification and characterization of chalcone isomerase from soybeans. J. Biol. Chem. 263 9582–9588. [PubMed] [Google Scholar]
- Bout S., Vermerris W. (2003). A candidate-gene approach to clone the sorghum Brown midrib gene encoding caffeic acid O-methyltransferase. Mol. Genet. Genomics 269 205–214. 10.1007/s00438-003-0824-4 [DOI] [PubMed] [Google Scholar]
- Brown H., Clark J., Gaines E., Hayes H., Morse W., Parker J., et al. (1936). Varietal standardization and registration. J. Am. Soc. Agron. 28:1027. 10.18699/VJ20.652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers C., Valentova K. (2015). “Non-taxifolin” derived flavonolignans: phytochemistry and biology. Curr. Pharm. Design 21 5489–5500. 10.2174/1381612821666151002112720 [DOI] [PubMed] [Google Scholar]
- Chang H. M., Cowling E. B., Brown W., Adler E., Miksche G. (1975). Comparative studies on cellulolytic enzyme lignin and milled wood lignin of sweetgum and spruce. Holzforschung 29 153–159. [Google Scholar]
- Chen W., Vanopdorp N., Fitzl D., Tewari J., Friedemann P., Greene T., et al. (2012). Transposon insertion in a cinnamyl alcohol dehydrogenase gene is responsible for a brown midrib1 mutation in maize. Plant Mol. Biol. 80 289–297. 10.1007/s11103-012-9948-4 [DOI] [PubMed] [Google Scholar]
- Chen Y., Liu H., Ali F., Scott M. P., Ji Q., Frei U. K., et al. (2012). Genetic and physical fine mapping of the novel brown midrib gene bm6 in maize (Zea mays L.) to a 180 kb region on chromosome 2. Theor. Appl. Genet. 125 1223–1235. 10.1007/s00122-012-1908-5 [DOI] [PubMed] [Google Scholar]
- Cherney J., Cherney D., Akin D., Axtell J. (1991). Potential of brown-midrib, low-lignin mutants for improving forage quality. Adv. Agron. 46 157–198. [Google Scholar]
- Cingolani P., Platts A., Wang Le L., Coon M., Nguyen T., Wang L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río J. C., Rencoret J., Prinsen P., Martínez ÁT., Ralph J., Gutiérrez A. (2012). Structural characterization of wheat straw lignin as revealed by analytical yyrolysis, 2D-NMR, and reductive cleavage methods. J. Agric. Food Chem. 60 5922–5935. 10.1021/jf301002n [DOI] [PubMed] [Google Scholar]
- Eloy N. B., Voorend W., Lan W., Saleme M. D. L. S., Cesarino I., Vanholme R., et al. (2017). Silencing CHALCONE SYNTHASE in maize impedes the incorporation of tricin into lignin and increases lignin content. Plant Physiol. 173 998–1016. 10.1104/pp.16.01108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudes A., Dutta T., Deng K., Jacquet N., Sinha A., Benites V. T., et al. (2017). SbCOMT (Bmr12) is involved in the biosynthesis of tricin-lignin in sorghum. PLoS One 12:e0178160. 10.1371/journal.pone.0178160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell-Harris D. L., Sattler S. E., Pedersen J. F. (2014). Response of Fusarium thapsinum to sorghum brown midrib lines and to phenolic metabolites. Plant Dis. 98 1300–1308. 10.1094/PDIS-09-13-0980-RE [DOI] [PubMed] [Google Scholar]
- Gardner R. (1975). Vanillin-hydrochloric acid as a histochemical test for tannin. Stain Technol. 50 315–317. 10.3109/10520297509117081 [DOI] [PubMed] [Google Scholar]
- Gee M. S., Nelson O. E., Kuæ J. (1968). Abnormal lignins produced by the brown-midrib mutants of maize: II. Comparative studies on normal and brown-midrib-1 dimethylformamide lignins. Arch. Biochem. Biophys. 123 403–408. 10.1016/0003-9861(68)90151-3 [DOI] [PubMed] [Google Scholar]
- Gurdon C., Poulev A., Armas I., Satorov S., Tsai M., Raskin I. (2019). Genetic and phytochemical characterization of lettuce flavonoid biosynthesis mutants. Sci. Rep. 9:3305. 10.1038/s41598-019-39287-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase N. J., Beissinger T., Hirsch C. N., Vaillancourt B., Deshpande S., Barry K., et al. (2015). Shared genomic regions between derivatives of a large segregating population of maize identified using bulked segregant analysis sequencing and traditional linkage analysis. G3 Genes Genomes Genet. 5 1593–1602. 10.1534/g3.115.017665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin C., Holt K., Chojecki J., Oliver D., Chabbert B., Monties B., et al. (1998). Brown-midrib maize (bm1)–a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J. 14 545–553. 10.1046/j.1365-313x.1998.00153.x [DOI] [PubMed] [Google Scholar]
- Hartwig B., James G. V., Konrad K., Schneeberger K., Turck F. (2012). Fast isogenic mapping-by-sequencing of ethyl methanesulfonate-induced mutant bulks. Plant Physiol. 160 591–600. 10.1104/pp.112.200311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L., Qian Q., Tang D., Wang K., Li M., Cheng Z. (2012). A mutation in the rice chalcone isomerase gene causes the golden hull and internode 1 phenotype. Planta 236 141–151. 10.1007/s00425-012-1598-x [DOI] [PubMed] [Google Scholar]
- Humphreys J. M., Hemm M. R., Chapple C. (1999). New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. U.S.A. 96 10045–10050. 10.1073/pnas.96.18.10045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James G. V., Patel V., Nordstrom K. J., Klasen J. R., Salome P. A., Weigel D., et al. (2013). User guide for mapping-by-sequencing in Arabidopsis. Genome Biol. 14:R61. 10.1186/gb-2013-14-6-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jez J. M., Bowman M. E., Dixon R. A., Noel J. P. (2000). Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat. Struct. Biol. 7 786–791. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Burow G., Gladman N., Acosta-Martinez V., Chen J., Burke J., et al. (2018). Efficient identification of causal mutations through sequencing of bulked F2 from two allelic bloomless mutants of Sorghum bicolor. Front. Plant Sci. 8:2267. 10.3389/fpls.2017.02267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson L. R. (1931). Brown midrib in maize and its linkage relations. J. Am. Soc. Agron. 23 549–557. 10.1186/1471-2164-8-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.-H., McRoberts J., Shi F., Moreno J. E., Jones A. D., Howe G. A. (2014). The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol. 164 1161–1174. 10.1104/pp.113.233395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima C. G., Guimarães G. A., Nogueira S. R., Maclean D., Cook D. R., Steuernagel B., et al. (2016). A pigeonpea gene confers resistance to Asian soybean rust in soybean. Nat. Biotechnol. 34:661. 10.1038/nbt.3554 [DOI] [PubMed] [Google Scholar]
- Kim H., Ralph J. (2010). Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6/pyridine-d5. Org. Biomol. Chem. 8 576–591. 10.1039/b916070a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Padmakshan D., Li Y., Rencoret J., Hatfield R. D., Ralph J. (2017). Characterization and elimination of undesirable protein residues in plant cell wall materials for enhancing lignin analysis by solution-state nuclear magnetic resonance spectroscopy. Biomacromolecules 18 4184–4195. 10.1021/acs.biomac.7b01223 [DOI] [PubMed] [Google Scholar]
- Kim H., Ralph J., Akiyama T. (2008). Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6. BioEnergy Res. 1 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krothapalli K., Buescher E. M., Li X., Brown E., Chapple C., Dilkes B. P., et al. (2013). Forward genetics by genome sequencing reveals that rapid cyanide release deters insect herbivory of Sorghum bicolor. Genetics 195 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuc J., Nelson O. E. (1964). The abnormal lignins produced by the brown-midrib mutants of maize: I. The brown-midrib-1 mutant. Arch. Biochem. Biophys. 105 103–113. 10.1016/0003-9861(64)90240-1 [DOI] [PubMed] [Google Scholar]
- Kuc J., Nelson O. E., Flanagan P. (1968). Degradation of abnormal lignins in the brown-midrib mutants and double mutants of maize. Phytochemistry 7 1435–1436. [Google Scholar]
- Kupce E., Freeman R. (2007). Compensated adiabatic inversion pulses: broadband INEPT and HSQC. J. Magn. Reson. 187 258–265. 10.1016/j.jmr.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Lam P. Y., Tobimatsu Y., Takeda Y., Suzuki S., Yamamura M., Umezawa T., et al. (2017). Disrupting flavone synthase II alters lignin and improves biomass digestibility. Plant Physiol. 174 972–985. 10.1104/pp.16.01973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Lu F., Regner M., Zhu Y., Rencoret J., Ralph S. A., et al. (2015). Tricin, a flavonoid monomer in monocot lignification. Plant Physiol. 167 1284–1295. 10.1104/pp.114.253757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Morreel K., Lu F., Rencoret J., Carlos Del Río J., Voorend W., et al. (2016a). Maize tricin-oligolignol metabolites and their implications for monocot lignification. Plant Physiol. 171 810–820. 10.1104/pp.16.02012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Rencoret J., Lu F., Karlen S. D., Smith B. G., Harris P. J., et al. (2016b). Tricin-lignins: occurrence and quantitation of tricin in relation to phylogeny. Plant J. 88 1046–1057. 10.1111/tpj.13315 [DOI] [PubMed] [Google Scholar]
- Landucci L., Smith R. A., Liu S., Karlen S. D., Ralph J. (2020). Eudicot nutshells: cell-wall composition and biofuel feedstock potential. Energy Fuels 34 16274–16283. [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Jin Z., Qu W., Zhao D., Ma F. (2006). Cloning of a cDNA encoding the Saussurea medusa chalcone isomerase and its expression in transgenic tobacco. Plant Physiol. Biochem. 44 455–461. 10.1016/j.plaphy.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Li H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Hill-Skinner S., Liu S., Beuchle D., Tang H. M., Yeh C.-T., et al. (2015). The maize brown midrib4 (bm4) gene encodes a functional folylpolyglutamate synthase. Plant J. 81 493–504. 10.1111/tpj.12745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Du Y., Chu H., Shih C. H., Wong Y. W., Wang M., et al. (2010). Molecular dissection of the pathogen-inducible 3-deoxyanthocyanidin biosynthesis pathway in sorghum. Plant Cell Physiol. 51 1173–1185. 10.1093/pcp/pcq080 [DOI] [PubMed] [Google Scholar]
- Liu S., Yeh C.-T., Tang H. M., Nettleton D., Schnable P. S. (2012). Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS One 7:e36406. 10.1371/journal.pone.0036406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Li Q., Wei H., Chang M. J., Tunlaya-Anukit S., Kim H., et al. (2013). Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. U.S.A. 110 10848–10853. 10.1073/pnas.1308936110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield S. D., Kim H., Lu F., Ralph J. (2012). Whole plant cell wall characterization using solution-state 2D NMR. Nat. Protoc. 7 1579–1589. 10.1038/nprot.2012.064 [DOI] [PubMed] [Google Scholar]
- Michelmore R. W., Paran I., Kesseli R. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88 9828–9832. 10.1073/pnas.88.21.9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller F. R. (1984). Registration of RTx430 sorghum parental line. Crop Sci. 24:1224. [Google Scholar]
- Mahon E. L., De Vries L., Jang S.-K., Middar S., Kim H., Unda F., et al. (2021). Expression of chalcone synthase in developing xylem results in the incorporation of naringenin in lignins. Plant Physiol. 10.1093/plphys/kiab499 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen L. J., Stuart P., Pièmanová M., Rasmussen S., Olsen C. E., Harholt J., et al. (2016). Dhurrin metabolism in the developing grain of Sorghum bicolor (L.) Moench investigated by metabolite profiling and novel clustering analyses of time-resolved transcriptomic data. BMC Genomics 17:1021. 10.1186/s12864-016-3360-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K. J., Albani M. C., James G. V., Gutjahr C., Hartwig B., Turck F., et al. (2013). Mutation identification by direct comparison of whole-genome sequencing data from mutant and wild-type individuals using k-mers. Nat. Biotechnol. 31 325–330. 10.1038/nbt.2515 [DOI] [PubMed] [Google Scholar]
- Osakabe K., Tsao C. C., Li L., Popko J. L., Umezawa T., Carraway D. T., et al. (1999). Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc. Natl. Acad. Sci. U.S.A. 96 8955–8960. 10.1073/pnas.96.16.8955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T., Yasui Y., Sugie S., Koketsu M., Watanabe K., Tanaka T. (2009). Dietary tricin suppresses inflammation-related colon carcinogenesis in male Crj: CD-1 mice. Cancer Prev. Res. 2 1031–1038. 10.1158/1940-6207.capr-09-0061 [DOI] [PubMed] [Google Scholar]
- Palmer N. A., Sattler S. E., Saathoff A. J., Funnell D., Pedersen J. F., Sarath G. (2008). Genetic background impacts soluble and cell wall-bound aromatics in brown midrib mutants of sorghum. Planta 229:115. 10.1007/s00425-008-0814-1 [DOI] [PubMed] [Google Scholar]
- Pomar F., Merino F., Barceló A. R. (2002). O-4-Linked coniferyl and sinapyl aldehydes in lignifying cell walls are the main targets of the Wiesner (phloroglucinol-HCl) reaction. Protoplasma 220 0017–0028. 10.1007/s00709-002-0030-y [DOI] [PubMed] [Google Scholar]
- Porter K. S., Anxtell J. D., Lechtenberg V. L., Colenbrander V. F. (1978). Phenotype, fiber composition, and in vitro dry matter disappearance of chemically induced brown midrib (bmr) mutants of sorghum. Crop Sci. 18 205–208. 10.2135/cropsci1978.0011183x001800020002x 34798789 [DOI] [Google Scholar]
- Quarrie S. A., Laziæ-Janèiæ V., Kovaèeviæ D., Steed A., Pekiæ S. (1999). Bulk segregant analysis with molecular markers and its use for improving drought resistance in maize. J. Exp. Bot. 50 1299–1306. 10.1093/jxb/50.337.1299 12432039 [DOI] [Google Scholar]
- Ralph J., Landucci L. L. (2010). “NMR of lignins,” in Lignin and Lignans; Advances in Chemistry, eds Heitner C., Dimmel D. R., Schmidt J. A. (Boca Raton, FL: CRC Press; ), 137–234. 10.1201/ebk1574444865-c5 [DOI] [Google Scholar]
- Rencoret J., Rosado M. J., Kim H., Timokhin V. I., Gutiérrez A., Bausch F., et al. (2021). Flavonoids naringenin chalcone, naringenin, dihydrotricin, and tricin are lignin monomers in papyrus. Plant Physiol. 10.1093/plphys/kiab469 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins M. P., Dixon R. A. (1984). Induction of chalcone isomerase in elicitor-treated bean cells. Comparison of rates of synthesis and appearance of immunodetectable enzyme. Eur. J. Biochem. 145 195–202. 10.1111/j.1432-1033.1984.tb08540.x [DOI] [PubMed] [Google Scholar]
- Rogers S. O., Bendich A. J. (1985). Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 5 69–76. 10.1007/BF00020088 [DOI] [PubMed] [Google Scholar]
- Saballos A., Ejeta G., Sanchez E., Kang C. H., Vermerris W. (2009). A genomewide analysis of the cinnamyl alcohol dehydrogenase family in Sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics 181 783–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saballos A., Sattler S. E., Sanchez E., Foster T. P., Xin Z., Kang C., et al. (2012). Brown midrib2 (Bmr2) encodes the major 4-coumarate:coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor (L.) Moench). Plant J. 70 818–830. 10.1111/j.1365-313X.2012.04933.x [DOI] [PubMed] [Google Scholar]
- Sarath G., Baird L. M., Vogel K. P., Mitchell R. B. (2007). Internode structure and cell wall composition in maturing tillers of switchgrass (Panicum virgatum. L). Bioresour. Technol. 98 2985–2992. 10.1016/j.biortech.2006.10.020 [DOI] [PubMed] [Google Scholar]
- Sattler S. E., Funnell-Harris D. L., Pedersen J. F. (2010). Brown midrib mutations and their importance to the utilization of maize, sorghum, and pearl millet lignocellulosic tissues. Plant Sci. 178 229–238. 10.1016/j.plantsci.2010.01.001 [DOI] [Google Scholar]
- Sattler S. E., Saathoff A. J., Haas E. J., Palmer N. A., Funnell-Harris D. L., Sarath G., et al. (2009). A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the sorghum brown midrib6 phenotype. Plant Physiol. 150 584–595. 10.1104/pp.109.136408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler S. E., Saballos A., Xin Z., Funnell-Harris D. L., Vermerris W., Pedersen J. F. (2014). Characterization of novel Sorghum brown midrib mutants from an EMS-Mutagenized population. G3 4 2115–2124. 10.1534/g3.114.014001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley B. W., Kubasek W. L., Storz G., Bruggemann E., Koornneef M., Ausubel F. M., et al. (1995). Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8 659–671. [DOI] [PubMed] [Google Scholar]
- Song J., Li Z., Liu Z., Guo Y., Qiu L.-J. (2017). Next-generation sequencing from bulked-segregant analysis accelerates the simultaneous identification of two qualitative genes in soybean. Front. Plant Sci. 8:919. 10.3389/fpls.2017.00919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. M., Liu S., Hill-Skinner S., Wu W., Reed D., Yeh C.-T., et al. (2014). The maize brown midrib2 (bm2) gene encodes a methylenetetrahydrofolate reductase that contributes to lignin accumulation. Plant J. Cell Mol. Biol. 77 380–392. 10.1111/tpj.12394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Wu T., Ye J., Sun J., Jiang Y., Yu J., et al. (2016). SNP-based analysis of genetic diversity reveals important alleles associated with seed size in rice. BMC Plant Biol. 16:93. 10.1186/s12870-016-0779-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobimatsu Y., Takano T., Umezawa T., Ralph J. (2019). “Solution-state multidimensional NMR of lignins: approaches and applications,” in Lignin: Biosynthesis, Functions, and Economic Significance, eds Lu F., Yue F. (Hauppauge, NY: Nova Science Publisher; ), 79–110. [Google Scholar]
- van der Weijde T., Alvim Kamei C., Torres A., Vermerris W., Dolstra O., Visser R., et al. (2013). The potential of C4 grasses for cellulosic biofuel production. Front. Plant Sci. 4:107. 10.3389/fpls.2013.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermerris W., Saballos A., Ejeta G., Mosier N. S., Ladisch M. R., Carpita N. C. (2007). Molecular breeding to enhance ethanol production from corn and sorghum stover. Crop Sci. 47 S-142–S-153. [Google Scholar]
- Vignols F., Rigau J., Torres M. A., Capellades M., Puigdomenech P. (1995). The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7 407–416. 10.1105/tpc.7.4.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel K. P., Jung H.-J. G. (2001). Genetic modification of herbaceous plants for feed and fuel. Crit. Rev. Plant Sci. 20 15–49. 10.1016/s0735-2689(01)80011-3 [DOI] [Google Scholar]
- Vogel K. P., Pedersen J. F., Masterson S. D., Toy J. J. (1999). Evaluation of a filter bag system for NDF, ADF, and IVDMD forage analysis. Crop Sci. 39 276–279. 10.2135/cropsci1999.0011183x003900010042x 34798789 [DOI] [Google Scholar]
- Wambugu P., Ndjiondjop M.-N., Furtado A., Henry R. (2018). Sequencing of bulks of Segregants allows dissection of genetic control of amylose content in rice. Plant Biotechnol. J. 16 100–110. 10.1111/pbi.12752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-H., Bible P., Loganantharaj R., Upadhyaya H. D. (2012). Identification of SSR markers associated with height using pool-based genome-wide association mapping in sorghum. Mol. Breed. 30 281–292. 10.1139/g11-055 [DOI] [PubMed] [Google Scholar]
- Xin Z., Li Wang M., Barkley N. A., Burow G., Franks C., Pederson G., et al. (2008). Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol. 8:103. 10.1186/1471-2229-8-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W., Wu Z., Liu Y., Li Y., Su K., Bai Z., et al. (2019). Mutation of 4-coumarate: coenzyme A ligase 1 gene affects lignin biosynthesis and increases the cell wall digestibility in maize brown midrib5 mutants. Biotechnol. Biofuels 12:82. 10.1186/s13068-019-1421-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Wang Y., Wang L., Ma Z., Zhao J., Wang P., et al. (2016). Genetic mapping and molecular marker development for Pi65 (t), a novel broad-spectrum resistance gene to rice blast using next-generation sequencing. Theor. Appl. Genet. 129 1035–1044. 10.1007/s00122-016-2681-7 [DOI] [PubMed] [Google Scholar]
- Zhou J.-M., Ibrahim R. K. (2010). Tricin—a potential multifunctional nutraceutical. Phytochem. Rev. 9 413–424. 10.1007/s11101-009-9161-5 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The derived cleaved amplified polymorphic sequence (dCAPS) marker for the bmr30-1 allele was used to verify genetic linkage between brown midrib phenotype and the bmr30-1 mutation from F2 individuals that were pooled for next generation DNA sequencing. (A) Genomic DNA from 25 individuals with the brown midrib phenotype and 25 individuals with the wild-type (green; wt) midribs were screened with dCAPS marker for bmr30-1, which was designed to create an AvaI restriction site in only the wild-type allele that resulted in a 183 bp product upon restriction digestion. The bmr30-1 allele lacked this restriction site due to the transition and the 209 bp product was not cleaved by AvaI. All individuals with the brown midrib phenotype were homozygous for the bmr30-1 allele, whereas seven individuals were homozygous for the wild-type allele and 18 individuals heterozygous. The latter two groups had normal midribs. (B) bmr30-1 was backcrossed into three different parental lines (Tx623, Wheatland, Tx430) and progeny were phenotyped based on the leaf midrib. These individuals were further tested and confirmed using the dCAPS marker and compared to the original mutant line (bmr30-1). PCR amplification controls were performed with DNA from bmr30-1 (bmr), wild-type (wt), 1:1 mixture of wild-type and bmr30-1 DNA to simulate a heterozygous plant (het) and no DNA template (-).
The derived cleaved amplified polymorphic sequence (dCAPS) marker for the bmr30-2 allele was used to verify genetic linkage between brown midrib phenotype and the bmr30-2 mutation from F2 individuals. Genomic DNA from (A) 25 individuals with the brown midrib phenotype and (B) 25 individuals with the wild-type (green; wt) midribs were screened with dCAPS marker for bmr30-2, which was designed to create an NcoI restriction site in only the wild-type allele that resulted in a 149 bp product upon restriction digestion. The bmr30-2 allele lacked this restriction site due to the insertion and the 176 bp product was not cleaved by NcoI. All individuals with the brown midrib phenotype were homozygous for the bmr30-2 allele, whereas eight individuals were homozygous for the wild-type allele and 15 individuals heterozygous. The latter two groups had normal midribs. PCR amplification controls were performed with DNA from bmr30-2, 1:1 mixture of wild-type and bmr30-2 DNA to simulate a heterozygous plant (het), wild-type (wt), and no DNA template (-).
The leaf midrib phenotype of the wild-type (BTx623), bmr30-1, bmr30-2, and F1 plants form the complementation test bmr30-2 × bmr30-1. The F1 progeny were confirmed to result from cross-pollination using DNA marker analysis. The eighth leaf was photographed from 6-week-old plants.
(A) Ten week old plants grown in the greenhouse, (B) seventh leaf from the bottom of 10 week old bmr30-2 plant, displaying the disease lesion mimic phenotype, and (C) seeds and glumes from wild-type (WT), bmr30-1, and bmr30-2. The top row is the abaxial side and bottom row is the adaxial side. The grain was harvested from the bottom of the panicles at approximately 40 days after anthesis.
Sorghum seedlings were germinated and grown under nutrient-depleted conditions to induce flavonoid accumulation. The absorbance of (A) total flavonoids and (B) anthocyanins were measured from the (C) seedlings. The seedlings from left to right are WT, bmr30-1, and bmr30-2, photographed 21 days after germination.
Expression abundance of chalcone isomerase (CHI; Sobic.001G035600), across diverse tissues from Sorghum bicolor. Gene expression data was obtained from The JGI Plant Gene Atlas Project available on Phytozome v13 (https://phytozome-next.jgi.doe.gov/phytomine/aspect.do?name=Expression).
Immunoblot detection of CHI from leaves (top) and stalks (bottom). Protein extracts from wild-type (WT), bmr30-1, and bmr30-2 were separated by SDS-PAGE, transferred to membrane, and probed with polyclonal antibodies raised against the recombinant tomato CHI protein. The recombinant SbCHI protein (SbCHI) was included as a positive control. Polyclonal antibodies raised against actin protein were used as a loading control.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA736969.