Abstract
The effects of brassinosteroid signaling on shoot and root development have been characterized in great detail but a simple consistent positive or negative impact on a basic cellular parameter was not identified. In this study, we combined digital 3D single-cell shape analysis and single-cell mRNA sequencing to characterize root meristems and mature root segments of brassinosteroid-blind mutants and wild type. The resultant datasets demonstrate that brassinosteroid signaling affects neither cell volume nor cell proliferation capacity. Instead, brassinosteroid signaling is essential for the precise orientation of cell division planes and the extent and timing of anisotropic cell expansion. Moreover, we found that the cell-aligning effects of brassinosteroid signaling can propagate to normalize the anatomy of both adjacent and distant brassinosteroid-blind cells through non-cell-autonomous functions, which are sufficient to restore growth vigor. Finally, single-cell transcriptome data discern directly brassinosteroid-responsive genes from genes that can react non-cell-autonomously and highlight arabinogalactans as sentinels of brassinosteroid-dependent anisotropic cell expansion.
Key words: Arabidopsis, root, meristem, brassinosteroid, single-cell mRNA sequencing, segmentation, morphology, BRI1, digital single-cell analysis
This study combines digital 3D single-cell shape analysis and single-cell mRNA sequencing to characterize roots of brassinosteroid-blind mutants. The data demonstrate that brassinosteroid signaling is essential for precise cell division plane orientation and the extent and timing of anisotropic cell expansion. These cell-aligning brassinosteroid effects can propagate non-cell-autonomously, manifesting in arabinogalactan expression as sentinels of brassinosteroid-dependent anisotropic cell expansion.
Introduction
Steroids are found throughout the tree of life and have acquired hormone function in animals as well as in plants (Tarkowska, 2019; Ferreira-Guerra et al., 2020). The brassinosteroids comprise a few bioactive molecules, among which brassinolide is typically the natural, endogenous ligand in dicotyledons (Nomura et al., 2005; Roh et al., 2020). In the model plant Arabidopsis thaliana (Arabidopsis), brassinolide is sensed by the extra-cellular domains of the receptor kinase BRASSINOSTEROID-INSENSITIVE 1 (BRI1) and its close homologs, BRI1-LIKE 1 (BRL1) and BRL3 (Li and Chory, 1997; Cano-Delgado et al., 2004). Their interaction with brassinolide triggers a phospho-transfer cascade, which eventually increases the nuclear abundance of downstream transcriptional effectors and thereby modulates the expression of their target genes (Wang et al., 2002; Yin et al., 2002; Tang et al., 2016; Kim and Russinova, 2020). Among them, brassinosteroid biosynthesis pathway genes are prominent, which establishes a negative feedback that maintains brassinosteroid signaling homeostasis (Wang et al., 2002). Loss-of-function mutations in genes encoding brassinosteroid receptors, biosynthetic enzymes, or downstream effectors lead to severe growth retardation, which can vary in its extent depending on allele strength and redundancies (Szekeres et al., 1996; Li and Chory, 1997; Choe et al., 1998; Cano-Delgado et al., 2004; Chen et al., 2019). The dwarf phenotype of brassinosteroid-related mutants has been interpreted from various angles (Oh et al., 2020). Although overall reduced cell size appears to be a consistent phenotype, effects of brassinosteroids on cell proliferation might be context dependent. For example, although a brassinosteroid signaling gain of function slightly promotes cell proliferation in Arabidopsis leaves (Gonzalez et al., 2010; Vanhaeren et al., 2014), it represses cell proliferation in rice leaf lamina (Sun et al., 2015). However, detailed analysis of a biosynthesis mutant suggests that brassinosteroids promote the exit from cell division, and thereby also the transition to cell expansion and differentiation (Zhiponova et al., 2013). Moreover, it has been proposed that brassinosteroid signaling is required for the differentiation of vascular tissues (Yamamoto et al., 2001; Cano-Delgado et al., 2004; Kang et al., 2017; Holzwart et al., 2018; Tamaki et al., 2020).
Leaves are initiated from founder cells at the flanks of shoot apical meristems. The size of the founder cell pool largely determines final leaf size, which appears to be remarkably well buffered against fluctuation in cell proliferation or expansion by compensatory mechanisms (Hisanaga et al., 2015; Vercruysse et al., 2020). By contrast, root growth is essentially continuous and driven by an apical stem cell niche (SCN) in the root meristem tip, which is maintained by the so-called quiescent center (QC). Proximal to the SCN, daughter cells undergo repeated meristematic as well as a few highly controlled formative divisions before they eventually expand and differentiate (supplemental Figure 1A). These divisions thus give rise to precisely aligned cell files that differentiate into the individual root tissues in a stereotypic pattern of overall radial symmetry, and bilateral symmetry in the vascular cylinder (supplemental Figure 1B). Finally, the timing of differentiation is not uniform across cell files and tissues. Thus, although the overall shape and organization of the root meristem is more recognizable and stereotypic than in the shoot meristem, cell proliferation, elongation, and differentiation are by comparison more intimately intertwined.
The role of brassinosteroid signaling in root development appears to be complex (Singh and Savaldi-Goldstein, 2015). Opposing, action-site-dependent effects on cellular differentiation have been reported (Vragovic et al., 2015), and, with regard to cell proliferation, interpretations of phenotypes vary. For example, root meristem size in bri1 mutants could be reduced as indicated by the number of meristematic cells in cortex or epidermal cell files (Gonzalez-Garcia et al., 2011; Hacham et al., 2011), but such reduction was not observed in a brassinosteroid biosynthesis mutant (Chaiwanon and Wang, 2015). Moreover, brassinosteroid signaling appears to restrict formative divisions (Kang et al., 2017). Cell proliferation defects together with generally decreased cell elongation could explain the strongly reduced root growth vigor in brassinosteroid mutants, and it has been proposed that brassinosteroid signaling promotes cell cycle progression (Gonzalez-Garcia et al., 2011; Hacham et al., 2011) and sets the absolute cell size in a “sizer” mechanism of cell elongation (Pavelescu et al., 2018). Indeed, the maximum elongation rate in bri1 mutants seems to be reduced (Hacham et al., 2011), whereas the average time cells spend elongating remains similar to wild type (Cole et al., 2014). Moreover, brassinosteroid signaling can at least in part act non-cell-autonomously, as initially proposed from the observation that BRI1 expression restricted to the epidermis could largely rescue the bri1 shoot growth defect (Savaldi-Goldstein et al., 2007). Restricted epidermal BRI1 also appears to promote both cell proliferation and elongation in roots (Hacham et al., 2011), and this effect was exaggerated in bri1 brl1 brl3 triple receptor null mutant background (briTRIPLE mutants) (Vragovic et al., 2015). The observation that overall root growth is only partially rescued in such lines (Kang et al., 2017) might reflect a trade-off in the balance between cell proliferation and cell growth, which generally determines overall root meristem size and growth vigor (Svolacchia et al., 2020). However, restoration of brassinosteroid signaling exclusively in the two developing protophloem sieve element cell files of briTRIPLE mutant root meristems, the first proximal tissue to differentiate (Furuta et al., 2014; Rodriguez-Villalon et al., 2014), can rescue root growth vigor to nearly wild-type levels (Kang et al., 2017), likely via non-canonical signaling outputs (Graeff et al., 2020). Thus, brassinosteroid can act non-cell-autonomously in various ways, including through a phloem-derived, non-cell-autonomous organizing signal.
In this study, we set out to map the effects of brassinosteroid signaling in the root by the characterization of briTRIPLE mutants at single-cell resolution. Our data suggest that brassinosteroid signaling affects neither cell proliferation nor cellular growth capacity per se, but rather enforces accurate cell division plane orientation and cellular anisotropy (i.e., an increased ratio between longitudinal and radial cell expansion).
Results
Because brassinosteroid biosynthesis is a branched pathway (Ohnishi et al., 2006), and because brassinosteroid signaling is subject to negative feedbacks (Wang et al., 2002; Ruan et al., 2018) and can additionally act through non-canonical outputs (Caesar et al., 2011; Cho et al., 2014; Kondo et al., 2014; Wolf et al., 2014; Holzwart et al., 2018; Tamaki et al., 2020), the brassinosteroid receptor triple null mutant represents the consequences of absent brassinosteroid action most stringently. We therefore concentrated our efforts on the briTRIPLE mutant and contrasted it against wild type and briTRIPLE mutants in which brassinosteroid signaling was exclusive restored in the developing protophloem sieve elements (briT-RESCUE) (Kang et al., 2017; Graeff et al., 2020).
Brassinosteroid signaling promotes anisotropy of mature root cells
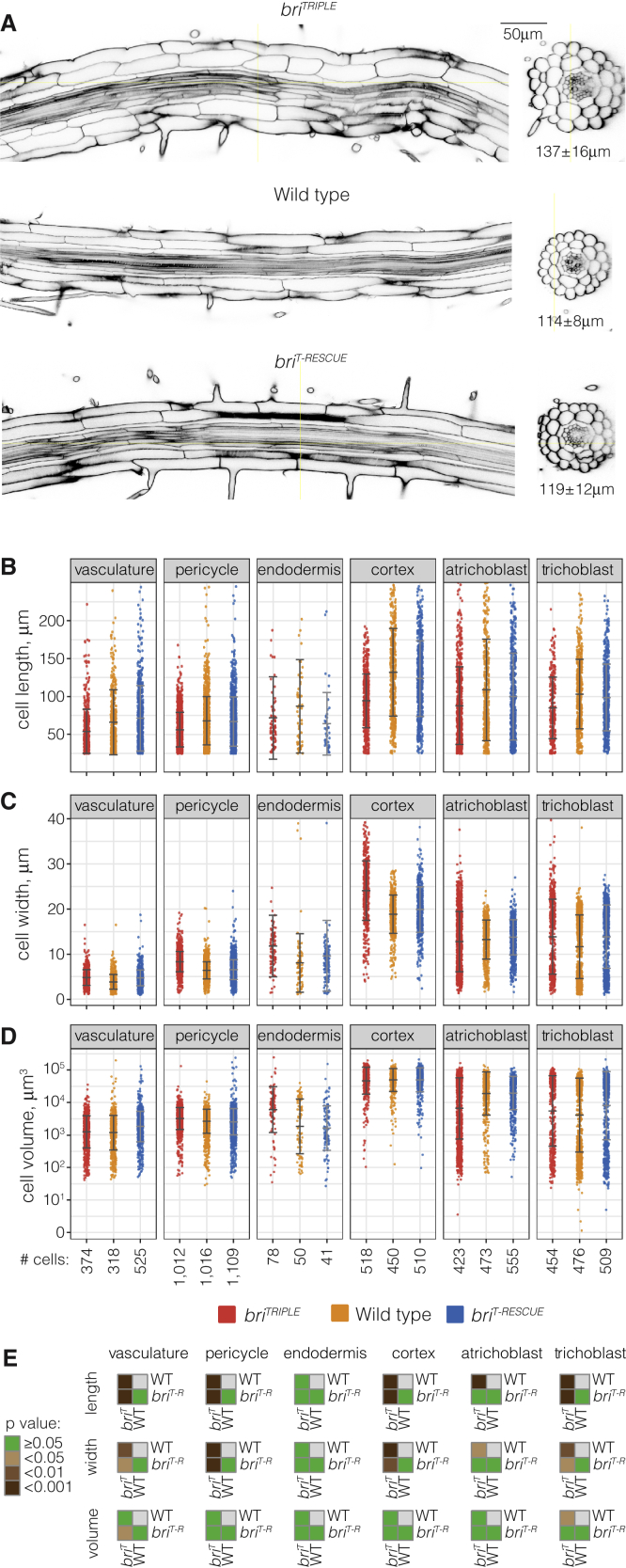
Typically, effects of brassinosteroid mutants on cell size have been described in terms of cell elongation in the root meristem, with a focus on cortex cells because of their considerable size and uniformity compared with other root tissues (Chaiwanon and Wang, 2015; Hacham et al., 2011; Kang et al., 2017; Pavelescu et al., 2018). Here we sought to determine the impact of brassinosteroid signaling on root cell growth comprehensively, in the first instance by measuring cellular parameters of mature root cells across all tissues. To preserve root anatomy, we prepared roots according to the ClearSee protocol (Kurihara et al., 2015). They were then mounted on microscopy slides in medium supplemented with 0.3% agar and with distancers to prevent their physical deformation during imaging by confocal microscopy. High-resolution 3D stacks were acquired of the mature parts of 10 to 11 roots per genotype (Figure 1A; supplemental Videos 1, 2, and 3). Each image stack was then processed with the PlantSeg segmentation software (Wolny et al., 2020) to identify and label individual cells, followed by analysis in MorphoGraphX (Barbier de Reuille et al., 2015) to extract cellular parameters. For subsequent single-cell 3D geometric analyses, 3DCellAtlas (Montenegro-Johnson et al., 2015) was used, which both identified cell types and measured cellular parameters in defined cellular positions along the length of the root axis. With this approach, cells could be grouped into six categories that reflected distinct cell types, with the exception of the vascular tissues that were lumped together in one category. The quantitative analysis of hundreds of cells showed that they were overall significantly shorter in briTRIPLE than in wild type or in briT-RESCUE in all tissues (Figure 1B and 1E). At the same time, they were also significantly wider with respect to the radial or circumferential dimensions (Figure 1C and 1E), which likely contributed to the overall increased thickness of briTRIPLE roots (Figure 1A). The only outliers were mature endodermal cells, whose lignin- and suberin-rich secondary walls sometimes caused uneven, oversaturated calcofluor cell wall staining that interfered with cell boundary prediction. This might have thwarted some measurements, although they displayed the same tendencies as in the other tissues. However, with very few, borderline significant exceptions, no significant differences were observed for cell volume between the three genotypes (Figure 1D and 1E). In summary, our high-throughput data thus indicate that loss of brassinosteroid signaling results in reduced cellular anisotropy but does not affect cell volume.
Figure 1.
Comparison of mature wild-type, briTRIPLE and briT-RESCUE root tissues.
(A) Transverse and horizontal (optical) sections of representative 8-day-old primary roots, ~1 cm above the root meristem, illustrating reduced anisotropy of mature cells in briTRIPLE roots. Average root diameters (n = 33 each) and their SDs are indicated.
(B–D) Comparative quantitative analyses for different cell morphology parameters obtained by processing of 3D stacks with the PlantSeg-MorphoGraphX-3DCellAtlas pipeline, 10 roots per genotype. Overall cell length (B) is reduced in the briTRIPLE roots, whereas cell width (C) is increased. Average cell volume (D) is similar between the three genotypes, indicating that it is not cell expansion but rather cellular anisotropy that is reduced in briTRIPLE mutants. Whiskers indicate mean and SD.
(E) Statistical comparison between cell types of different genotypes (ANOVA, averages per root). Note that the lignin- and suberin-rich secondary walls of mature endodermal cells caused uneven calcofluor cell wall staining, which oversaturated some images and interfered with cell boundary prediction by the segmentation software. Obviously fused endodermis cells were discarded from the analysis. The attributes of the remaining endodermis cells might still be thwarted by these difficulties, which could explain why differences in (A) and (B) were not statistically significant although the measurements display the same tendencies as in the other tissues.
Brassinosteroid signaling does not affect cell division capacity of the root meristem
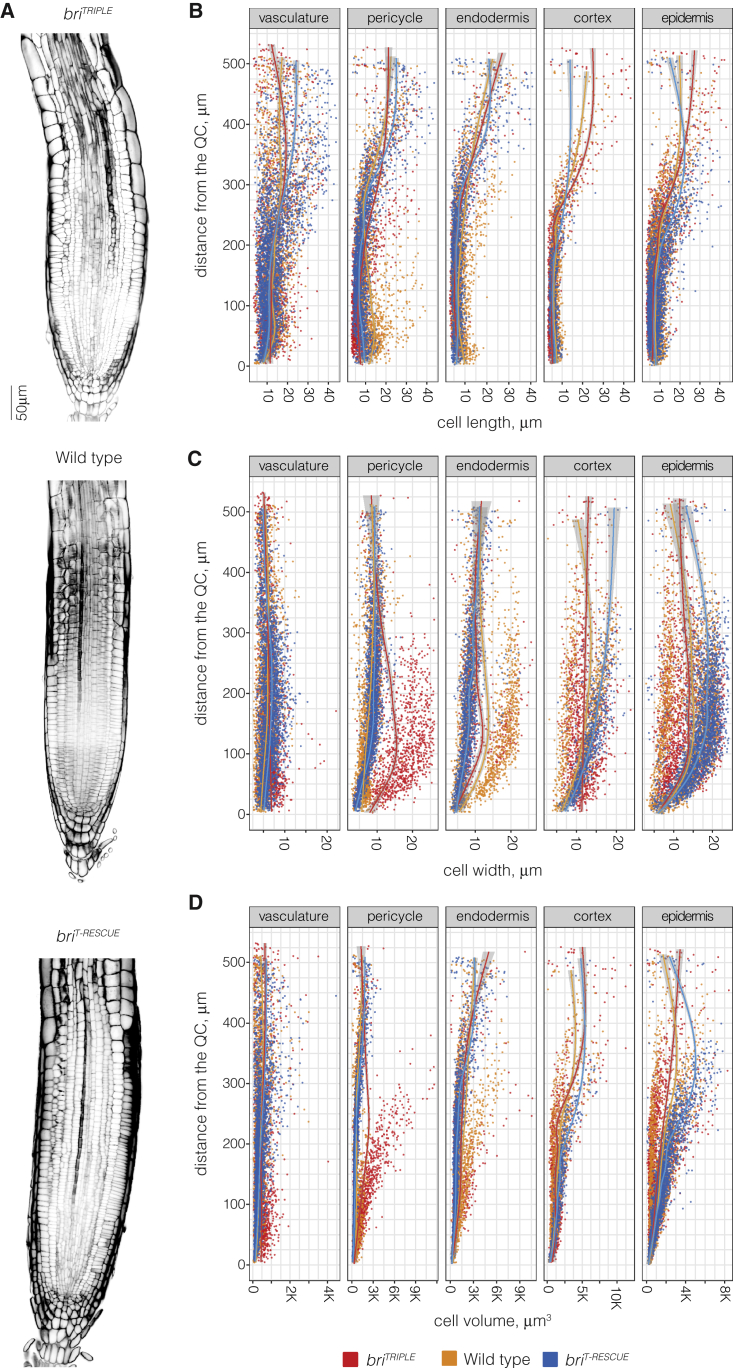
The fact that the reduction in briTRIPLE root growth vigor cannot be explained by reduced cell elongation alone has been established previously, and additive effects from reduced cell proliferation as well as delays due to supernumerary formative divisions were deemed responsible for the extent of the macroscopic root phenotype (Gonzalez-Garcia et al., 2011; Hacham et al., 2011; Kang et al., 2017). Indeed, the estimated number of cells in 7-day-old briTRIPLE roots appeared to be reduced (supplemental Figure 1C and 1D). We therefore extended our 3D cellular morphometric analysis to the root meristems (Figure 2A). Again, we obtained high-resolution 3D stacks (supplemental Videos 4, 5, and 6) and processed them through the PlantSeg-MorphoGraphX-3DCellAtlas pipeline, with automated annotation of cell identities that were close to the SCN to assign tissue identity to individual cell files. As we were interested in the question how the growth vigor differences between the investigated genotypes arise, we concentrated our analyses on the proximal, permanent root tissues and disregarded both the columella and lateral root cap cells, which eventually undergo cell death and are shed (Fendrych et al., 2014; Xuan et al., 2016). The analysis indicated that cellular anisotropy is already reduced in meristematic briTRIPLE cells, because close to the QC they were in general already shorter (Figure 2B) but also wider than in wild type (Figure 2C), and therefore less anisotropic. Moreover, briTRIPLE cells again did not appear to be consistently smaller in terms of volume (Figure 2D). Finally, we sought to determine the number of actively dividing cells per root meristem, which can be visualized by antibody immuno-staining of cell plates by the cytokinesis-specific syntaxin KNOLLE (KN) (Lauber et al., 1997). Based on these immunostainings (supplemental Figure 1E–1G), the number of actively dividing cells was indeed not significantly different between briTRIPLE and wild type (supplemental Figure 1H). Moreover, cell plates were in all cases (n = 21–25 meristems per genotype) observed within ∼200 μm from the QC, suggesting that the spatial window for cell divisions is neither substantially shortened nor prolonged in briTRIPLE meristems as compared with wild type. In summary, these analyses indicate that cells in briTRIPLE meristems do not display a generic reduction in their proliferation capacity.
Figure 2.
Comparison of wild-type, briTRIPLE and briT-RESCUE root meristems.
(A) Transverse sections of representative 8-day-old primary root meristems of the three genotypes. Note the comparatively disorganized morphology of briTRIPLE meristems.
(B–D) Scatterplots of cell length (B), width (C), and volume (D) for the root meristem cell types analyzed through the PlantSeg-MorphoGraphX-3DCellAtlas pipeline. Models were fitted to the measurements for each genotype using a general additive model with a shrinking cubic regression spline interpolation; gray areas indicate the SE. Variation within roots of the same genotype is generally high, making comparisons of the cellular properties difficult.
Brassinosteroid signaling guides the transition to volumetric cell growth
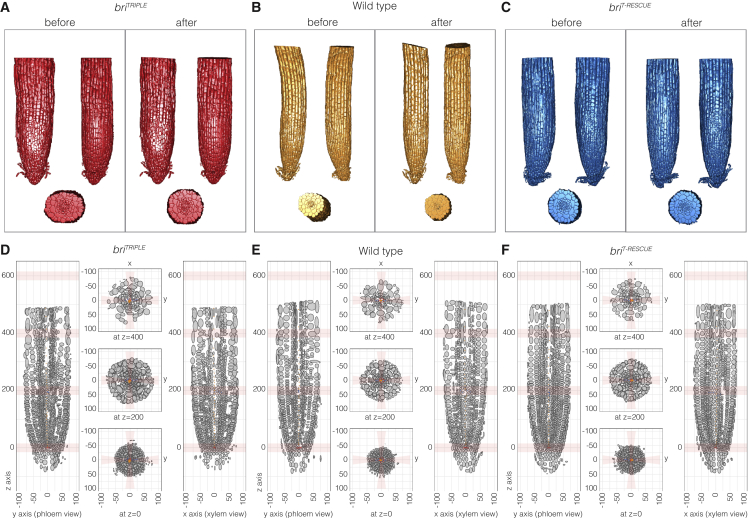
Our initial cellular morphometric analyses produced statistically robust outputs per root for the mature parts (Figure 1B–1E). However, for the root meristems, we sometimes observed substantial variation across and within genotypes between individual samples (supplemental Figure 1J–1L). Such effects were compounded with increasing distance from the QC, where morphology became more variable because root meristems are dynamic, growing structures (Thompson and Holbrook, 2004; Taylor et al., 2021). Their developmental plasticity was, for instance, evident in the fluctuation of growth orientation (Thompson and Holbrook, 2004), which led to both within-root and between-root variation in cellular parameters of the same tissue. Moreover, because of frequently disorganized cell files in briTRIPLE meristems (Figure 2A), positional attributes were often distorted and thus difficult to compare between roots. Therefore, we devised a method to permit more fine-grained, standardized comparisons across several roots and genotypes. In brief, we first manually marked the QC, the central xylem pole, and the orientation of the xylem axis in each segmentation to define its biological main axes. These 3D meshes were then realigned by straightening of the main axis, by restoring the overall radial symmetry, and by aligning the main axis with the z axis, and the xylem axis with the x axis (Figure 3A–3C). The y axis, perpendicular to the x axis, consequently passed approximately through the two phloem poles. Importantly, the original cellular parameters obtained from MorphoGraphX were not altered by these realignments. Altogether, these transformations gave rise to idealized representations of the samples (Figure 3D–3F), which could be directly compared due to their straight, cylindrical layout around a central z axis, their alignment with respect to the bilateral symmetry of the stele, and the reference point set to the center of the QC.
Figure 3.
Standardization of root meristem segmentations for quantitative comparisons.
(A–C) Illustration of the straightening of root meristem 3D meshes for examples of the different genotypes, before and after realignment.
(D–F) Transverse and horizontal virtual sections of simplified root meristem models obtained by re-orientation (straightening) of 3D meshes along the defined central and radial axes. Cells are shown as ellipsoids with semi-axis proportional to the cell length in the corresponding direction. Cells of the central xylem pole are marked by orange dots, of the QC by red dots, and of the xylem axis by blue dots. For each root, the central xylem pole was realigned with the vertical z axis, the xylem axis with the radial y axis, and the phloem poles with the radial x axis. The original cellular parameters were maintained as cells were re-oriented along the respective axis.
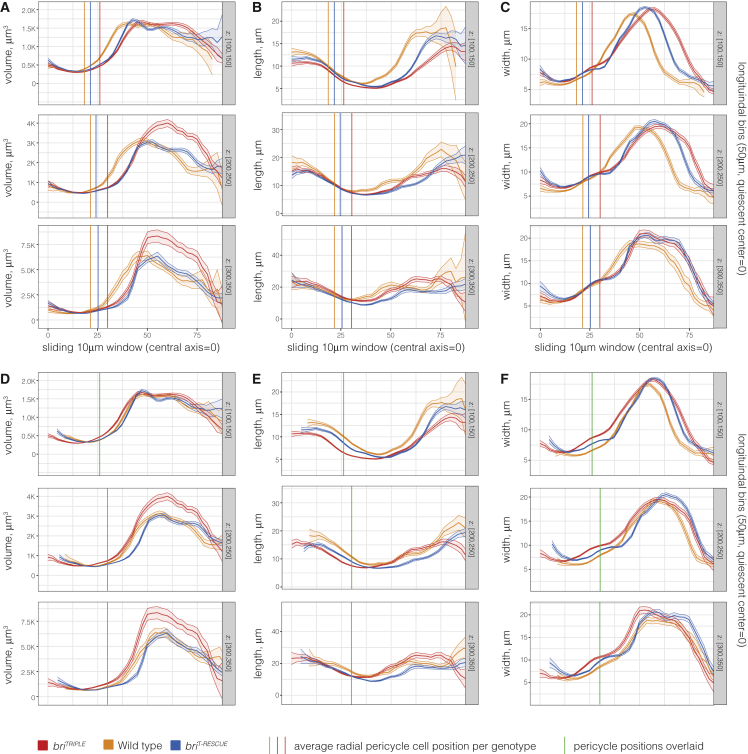
Our method allowed us to acquire comparable per-position parameters, and a pool of 11 to 12 roots per genotype was used for comparison of the genotypes. Unbiased quantifications were performed by radial and longitudinal sliding window analyses using concentric cylindrical shells (centered on the z axis) of 10 μm thickness in the radial direction and 50 μm height. Analyses in the radial direction were performed by varying the radius of the shell while keeping its z position fixed (supplemental Figure 2 and 3). These data gave a general impression of cellular features along the x and y axes and across tissues. They corroborated that meristematic briTRIPLE cells were already shorter in the longitudinal dimension and wider in both the radial and circumferential dimensions close to the SCN (supplemental Figure 2A–2C), whereas cell volume was comparable with the wild type (supplemental Figure 3A). For analyses in the longitudinal direction, the radius of the shell was kept fixed while sliding its z position from the QC to the top of the meristem (supplemental Figures 4 and 5). These data gave a general impression of the progressive changes of cellular features in individual or neighboring tissues (depending on bin position) along the z axis. Again, they confirmed the reduced anisotropy of briTRIPLE cells from early on (supplemental Figure 4A–4C), but also suggested an earlier transition to volumetric growth than in the wild type (supplemental Figure 5A). Because of the increased width of briTRIPLE roots, especially the wider stele (supplemental Figure 6), this transition was offset in the strictly per-position comparisons (Figure 4A–4C). However, when sliding window curves were aligned with the average position of pericycle cells as the reference point for the border between the smaller vascular and the considerably bigger ground tissue cells (Figure 4D–4F), it became evident that cells in briTRIPLE mutants gained in volume earlier than in wild type (Figure 4D). This coincided with cells approaching wild-type length (Figure 4E) while maintaining an increased width (Figure 4F). In summary, our analyses confirmed that briTRIPLE cells across tissues already displayed reduced anisotropy at the outset, and also suggested that their volumetric expansion preceded wild type.
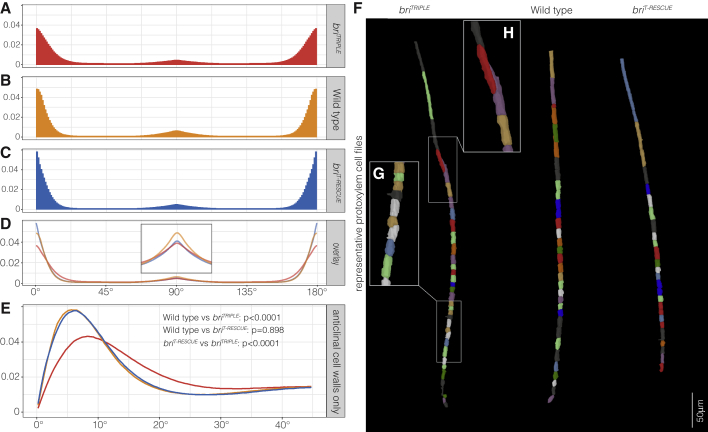
Figure 5.
Analysis of global cell wall orientation.
(A–C) Distribution of angles θ between z axis and vectors normal to the 3D meshes triangles per unit of solid angle (weighted by triangle area) of 11–12 briTRIPLE(A), wild-type (B), and briT-RESCUE(C) roots. 90° signifies normal vector perpendicular to the z axis, 0° signifies normal vector parallel to the z axis. See supplemental methods 1 (Figure 10 therein) for details on angle orientation.
(D) Superimposition of the three plots highlights the wider distribution of angles in briTRIPLE cells, indicating more surfaces that are poorly aligned with the main axes.
(E) Distribution of angles θ (weighted by triangle area) limited to triangles with θ < 45°. The spread of the distribution away from θ = 0 was estimated as the median of the θ distribution (weighted by triangle area and considering only triangles with θ < 45°), which was evaluated for each root separately. Indicated p values were obtained by Welch two-sided t-tests of median θ grouped by genotype.
(F–H) Representative protoxylem cell file segmentations (F), illustrating oblique cell division planes in briTRIPLE meristems (G) and the consequent ballooning effect of cells sliding next to each other, out of file, upon elongation (H).
Figure 4.
Comparative radial sliding window analysis of standardized root models.
(A–C) Extract of quantitative features of each simplified root meristem model (supplemental Figures 4 and 5). Models were analyzed using concentric 10 μm thick and 50 μm high cylindrical shells with increasing radius (x axis) fixed z position (indicated on the right). Average parameters were calculated by taking into account cells whose centers (after realignment) fell into the corresponding shell. The graphs indicate average cell volume (μm3) (A), length (μm) (B) and width (μm) (C) for 11–12 roots per genotype combined. Shaded regions indicate ± SE of the mean. Note the displacement of the briTRIPLE measurements as compared with wild type, due to the increased diameter of briTRIPLE meristems. Vertical lines indicate the average position of pericycle cells with respect to the central xylem axis in the different genotypes 100 μm, 200 μm, or 300 μm above the QC.
(D–F) Graphs from (A)–(C) with curves shifted in order to center them on the average pericycle position (green vertical lines). Note the comparatively premature transition of briTRIPLE cells to volumetric growth with growing distance from the QC.
Brassinosteroid signaling is required for precise cell division orientation
One conspicuous feature of briTRIPLE meristems is their disorganized appearance (Figure 2A), which can be mainly attributed to the occurrence of oblique cell division planes that break up the stereotypic, precise alignment of cell files observed in wild type (supplemental Figure 7A–7D). For an organ-wide quantification of this phenomenon, we determined the orientation of the triangles that defined the cell surfaces in the 3D meshes of the segmented roots by evaluating the angle between their normal vector and the z axis. Plotting the distribution of angles (weighted by triangle area) demonstrated that, in wild type, cell walls are overwhelmingly oriented in anticlinal or periclinal orientation as expected (Figure 5A). By contrast, the distribution of cell wall orientation angles was substantially wider in briTRIPLE meristems (Figure 5B and 5D), matching their visual appearance. A focused analysis on anticlinal cells walls (angles smaller than 45°) corroborated the notion that this deviation was frequent and statistically significant (Figure 5E). Such imperfect alignments also sometimes appeared to cause a “ballooning” phenomenon, where neighboring cells in a file that were separated by an oblique division plane slid next to each other as they elongated (Figure 5F–5H). In briTRIPLE meristems, such imperfectly aligned cell files could be observed routinely and sometimes created the deceptive impression of the presence of additional cell files. However, we also observed late formative divisions in briTRIPLE meristems (supplemental Figure 1I). These findings suggest that extra cell files in cross sections (supplemental Figure 8A) (Kang et al., 2017) could reflect a combination of ballooning effects and late or isolated periclinal divisions. However, the consistently wider stele in briTRIPLE meristems (supplemental Figure 6), the steady increase of cell number per z-axis position from the start (supplemental Figure 8B), and the increased number of cell files in the mature region (supplemental Figure 8C) reiterate the major contribution of supernumerary formative divisions. In summary, the analyses suggest that although cell proliferation capacity is not impaired in briTRIPLE meristems, the precision of cell divisions is.
Phloem-specific BRI1 imparts an intermediate compensatory state on briTRIPLE meristems
Interestingly, despite the macroscopically nearly wild-type growth vigor of briT-RESCUE roots (supplemental Figure 1C) and the essentially normal morphology of their mature cells (Figure 1A–1E), their meristems displayed distinct abnormalities (Figure 2A) that became more apparent in the 3D characterization (Figure 3F and supplemental Video 6). Throughout the analyses, briT-RESCUE cells were overall quantitatively situated between briTRIPLE and wild type (Figure 2B–2D and supplemental Figures 2–6). However, they generally appeared more similar to briTRIPLE in the meristematic region, and transitioned to a more wild type-like state as cells elongated and differentiated. That is, although cells in briT-RESCUE meristems appeared initially less anisotropic than in wild type, they eventually caught up and did not undergo the comparatively earlier transition to volumetric growth observed in briTRIPLE meristems, but rather approached the growth trajectory observed in wild type (Figure 4D). In line with the overall visual impression of normalized tissue organization as compared with briTRIPLE meristems (Figure 2A), cell wall orientations were largely restored with respect to the main axes (Figure 5C and 5D) and anticlinal cell wall orientations were indistinguishable from wild type (Figure 5E). Finally, although briT-RESCUE meristems also displayed supernumerary periclinal or formative divisions (supplemental Figures 6, 8A, and 8B), this only translated into a modest increase of cell files in the mature roots (supplemental Figure 8C). This could be mainly attributed to extra vascular cell files, matching the in-between diameter of the stele (supplemental Figure 6), whereas the phenomenon was less pronounced in the bigger ground tissue cells (supplemental Figure 8A and 8C). In combination with the restored anisotropy (Figure 1B–1D), this resulted in a mature root diameter that was close to wild type (Figure 1A). In summary, the data suggest that restriction of brassinosteroid signaling to the developing phloem cell files predominantly affects the trajectory of cell growth and differentiation and normalizes the cellular anatomy even in distant brassinosteroid-blind cell files through non-cell-autonomous functions.
The intermediate compensatory state is associated with a unique transcriptomic signature
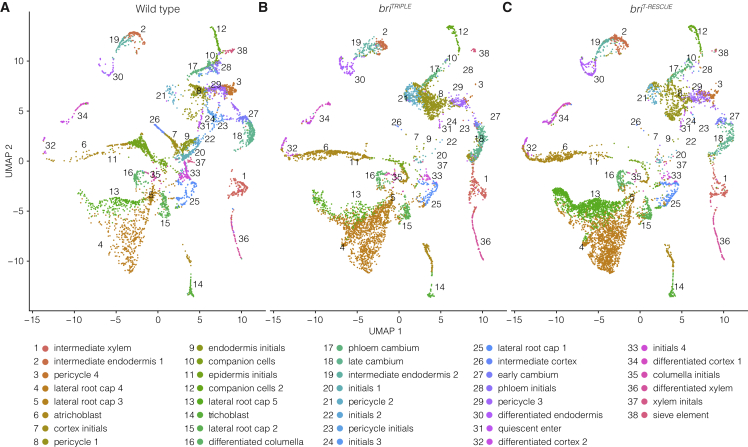
To determine whether the observed morphological differences could be correlated to distinct gene activities, we performed single-cell mRNA sequencing (scRNA-seq) of briTRIPLE and briT-RESCUE meristems. After quality control and filtering, 5767 and 6724 high-quality cells, respectively, were obtained and next compared with a wild-type root meristem dataset of 5061 cells produced on the same platform (Wendrich et al., 2020). Cell types were assigned through established marker gene expression (supplemental Figure 9). An additional comparison with another wild-type dataset, independently produced on a different platform (Denyer et al., 2019), showed that on average 91% and 94% of briTRIPLE and briT-RESCUE cells, respectively, received the same cell type label, indicating the robustness of the annotation method (supplemental Figure 10). Unsupervised clustering identified 28 distinct groups of cells, which could be further sorted into 38 subclusters (Figure 6A–6C, supplemental Tables 1, 2, and 3) based on known tissue- as well as stage-specific wild-type marker genes (Wendrich et al., 2020). The relative abundance of cells within the clusters was overall comparable with wild type, although several subclusters contained very few cells (supplemental Figure 11A). The transcriptomics data suggest that individual tissues differentiate correctly in the absence of brassinosteroid signaling, and, despite their (relatively mild) differentiation defects (Cano-Delgado et al., 2004; Kang et al., 2017), vascular tissues of briTRIPLE roots could be readily identified. Globally, the differences between briT-RESCUE and briTRIPLE were less pronounced than between either of those two genotypes and wild type (supplemental Figure 12A), corroborating earlier observations from bulk mRNA sequencing (Graeff et al., 2020). Thus, the phenotype of briT-RESCUE meristems was associated with a transcriptomic signature that was intermediate between briTRIPLE and wild type.
Figure 6.
scRNA-seq overview.
(A–C) UMAP of wild-type (A), briTRIPLE(B), and briT-RESCUE(C) single-cell transcriptomes (see supplemental Tables 1, 2, and 3). Colors indicate the assigned cell identities based on established cell type- and stage-specific marker genes (Wendrich et al., 2020).
To specifically monitor known brassinosteroid-responsive genes, we compared our data with two reference sets. On the one hand, we specifically investigated the overlap with a high-confidence list of generically brassinosteroid-regulated genes (Liu et al., 2020); on the other hand, we looked at a set of particularly pertinent genes that were reported to respond to brassinosteroid specifically in root tips (Chaiwanon and Wang, 2015). For a first analysis, the stage-specific scRNA-seq subclusters were merged to represent the 13 cell types (Wendrich et al., 2020) (supplemental Figures 11B and 12B–12D and supplemental Table 4). The overlap was substantial; for example, of the 1385 brassinolide-downregulated and 2312 brassinolide-upregulated genes reported for the root tip of a brassinosteroid biosynthesis mutant (Chaiwanon and Wang, 2015), 576 and 1218 transcripts, respectively, were detected among the differentially expressed genes (DEG) in at least one cluster comparison (adjusted p < 0.05) (supplemental Figure 13A and 13B, supplemental Tables 5 and 6). Among them, 525 and 1040, respectively, showed differential expression between briTRIPLE and wild type (supplemental Figure 14A and 14B). These findings matched the expected behavior of known brassinosteroid-responsive genes in briTRIPLE roots; i.e., brassinosteroid-downregulated genes were typically over-expressed in the mutant, whereas brassinosteroid-upregulated genes typically showed reduced expression levels. Behavior was also consistent across clusters; for example, among 572 brassinosteroid-upregulated genes that each were differentially expressed in at least three different clusters, changes were entirely coherent across clusters for 402 genes (supplemental Table 6). A substantial portion (∼39%–43%) of the affected genes were also differentially expressed in briT-RESCUE meristems compared with briTRIPLE meristems (supplemental Figures 13A, 13B, 14A, and 14B), and consistently resembled the expression in wild type. However, an equally large portion (∼43%–48%) of genes was still differentially expressed between briT-RESCUE and wild-type meristems (supplemental Figure 13A and 13B). Because brassinosteroid-responsive genes in our set were particularly prominent in the cortex, possibly because of its comparatively strong cellular growth, we tested whether this skewed our analyses. However, our observations also held up when cortex cells were excluded from the comparison (supplemental Figure 13C and 13D). Finally, the trends described above were confirmed by similar comparisons with the generic set of brassinosteroid-responsive genes (Liu et al., 2020) (supplemental Figure 13E, 13F, and 15). In summary, the briT-RESCUE genotype again represented an intermediate state between briTRIPLE and wild type.
Brassinosteroid-responsive genes can be controlled via non-cell-autonomous signals
Including the subset of 732 brassinosteroid-responsive reference genes (Chaiwanon and Wang, 2015), the briTRIPLE and briT-RESCUE scRNA-seq transcriptomes displayed 2157 genes that were differentially expressed in at least one cell type (adjusted p < 0.05). Interestingly, a set of 61 core cell cycle genes (Vandepoele et al., 2002) was nearly absent from this sample (supplemental Figure 16). In-depth comparison of tissue-specific differential gene expression yielded some insight into the nature of the rescue. For instance, we found that 310 genes showed robust differential expression in phloem tissues; however, only a minority (n = 32) were specific for the phloem and, of those, even fewer (n = 9) were also known brassinosteroid-regulated genes. This pattern was also observed in the subcluster analysis (p < 0.01), where 63 of 278 genes that were differentially expressed in sieve elements were also known as brassinosteroid responsive. Even less were sieve-element-specific (10 and 109, respectively), but included a few known protophloem sieve element markers such as ALTERED PHLOEM DEVELOPMENT (Bonke et al., 2003), BREVIS RADIX (Rodriguez-Villalon et al., 2014), or CLAVATA3/EMBRYO SURROUNDING REGION-RELATED 26 (CLE26) (Anne et al., 2018). The differentially expressed phloem and sieve element transcripts spanned a variety of encoded proteins, which might include factors that are involved in the generation or transmission of the non-cell-autonomous signal and could thereby aid in its discovery.
Importantly, in briT-RESCUE meristems, the expression of many brassinosteroid-responsive genes was normalized not only in the phloem but also in directly neighboring as well as distant tissue layers (supplemental Figures 14 and 15). This was also evident in the more fine-grained analysis across the stage-specific clusters (supplemental Figure 17). That is, the expression of brassinosteroid-responsive genes was fully normalized not only in sieve elements and phloem initials as expected but also in other clusters like xylem initials or intermediate cortex. Moreover, in clusters in which a higher number of brassinosteroid-responsive genes could be detected, the expression of a sizable portion was normalized to the same degree as in the overall analyses (supplemental Figure 17A and 17B). These results suggest that brassinosteroid can control many genes indirectly, through non-cell-autonomous signals.
Arabinogalactan-encoding genes are sentinels of brassinosteroid-dependent cellular anisotropy
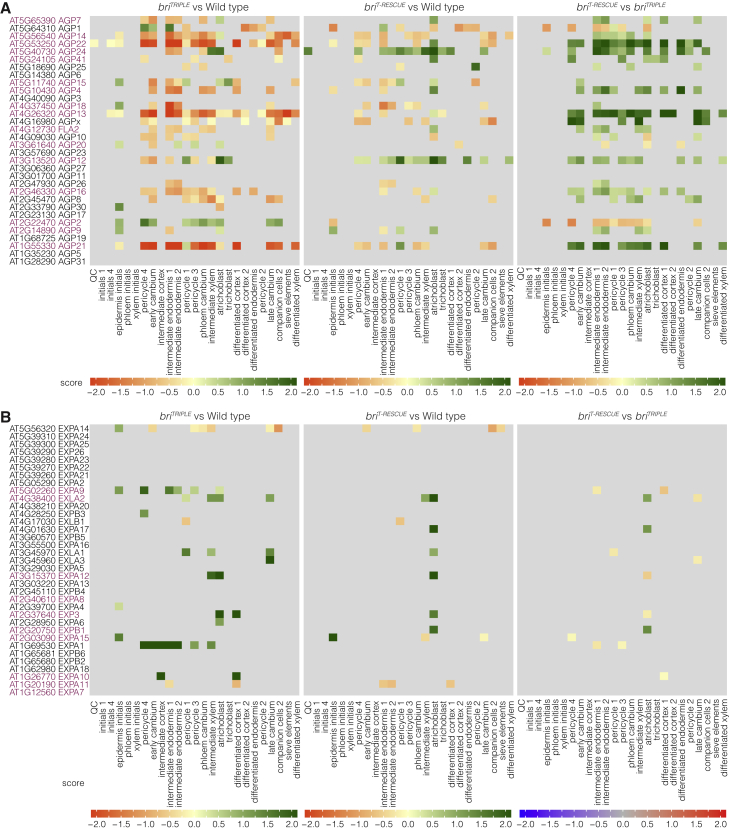
Apart from known brassinosteroid-responsive genes, transcripts whose expression was fully or partially normalized in briT-RESCUE meristems can be considered a systemic readout of such non-cell-autonomous signals. We sought to determine whether they comprise particular gene classes that are differentially expressed throughout tissues in a coherent manner. However, gene ontology analyses did not reveal any consistent pattern, but genes encoding arabinogalactan proteins and peptides (AGPs) clearly stood out because of their robust, comparatively high expression in briT-RESCUE tissues compared with the briTRIPLE mutant (supplemental Figure 18A and supplemental Table 7). This observation is in line with the proposed role of AGPs in cell growth in various systems (Seifert and Roberts, 2007; van Hengel and Roberts, 2002; Willats and Knox, 1996), and the positive correlation between their expression level and cell elongation (Pacheco-Villalobos et al., 2016). For example, interference with AGP activity results in shorter, swollen, and therefore likely less anisotropic root cells (van Hengel and Roberts, 2002; Willats and Knox, 1996). We corroborated and quantified this notion experimentally (supplemental Figure 19A), using the PlantSeg-MorphoGraphX-3DCellAtlas pipeline (supplemental Figure 19B). By contrast, differential expression of the classic cell expansion genes, the expansins, which facilitate cell growth by promoting cell wall loosening (Cosgrove, 2005), was only observed sporadically (supplemental Figure 18B and supplemental Table 8). Moreover, those expansins that displayed differential expression were nearly exclusively over-expressed in the briTRIPLE meristems, which might reflect the premature transition to volumetric growth described above. These observations were corroborated in the subcluster analysis, where most AGPs (23 out of 32) showed differential expression between briTRIPLE mutant and wild-type or briT-RESCUE tissues, and generally also displayed stronger expression at later stages of tissue ontogeny (Figure 7A). This included many AGPs (15 out of 23) that were part of the brassinosteroid-responsive set (Chaiwanon and Wang, 2015), like AGP21, which was recently described as a mediator of brassinosteroid effects in root hair patterning (Borassi et al., 2020). Again, such strong and spatially graded expression differences were not observed for expansins (Figure 7B), although several (10 out of 35) have also been described as brassinosteroid target genes. In summary, our single-cell transcriptomics identified a strong correlation between brassinosteroid-dependent root cell morphology and the expression of AGPs, matching their biological role in cellular growth (Seifert and Roberts, 2007; Yang et al., 2007). In light of our finding that volumetric growth is not affected in briTRIPLE mutants, but cellular growth orientation is, AGPs could therefore be considered sentinels of brassinosteroid-dependent cellular anisotropy and represent the most prominent readout of non-cell-autonomous brassinosteroid functions.
Figure 7.
AGP gene expression is a sentinel of brassinosteroid-dependent cellular anisotropy.
(A and B) Heatmap indicating the differential gene expression score of AGPs (A) or expansins (B) in scRNA-seq subclusters and ordered with respect to stage-specific markers in increasing distance from the QC. Known brassinosteroid-responsive gene family members (Chaiwanon and Wang, 2015) are highlighted in purple.
Discussion
Brassinosteroid signaling controls cellular growth orientation
In summary, our combined analyses reveal that brassinosteroid signaling in root development is essential for the precise orientation of cell division planes and the extent and timing of anisotropic cell expansion. Our conclusions highlight the strength of quantitative 3D digital single-cell analyses and show that they are necessary to arrive at non-obvious and quantitatively confirmed conclusions.
The observed absence of effects on cell volume and proliferation capacity versus the statistically significant effects on cellular anisotropy and division plane orientation open new perspectives on previous analyses. For instance, the notion that cell division frequency is reduced in roots of brassinosteroid signaling mutants was largely deduced from the abundance of cell division markers relative to 1D meristem size expressed as the number of meristematic cells in individual cell files. However, this method is prone to error, especially when absolute differences are small (Perilli and Sabatini, 2010), which might explain why effects on meristem size are not consistent across studies (Gonzalez-Garcia et al., 2011; Hacham et al., 2011; Chaiwanon and Wang, 2015; Kang et al., 2017). Despite the normal frequency of dividing cells, our estimations suggest that the number of cells produced per time unit is indeed reduced in briTRIPLE roots. This might reflect slowed down root growth due to the production of extra cell files (De Rybel et al., 2013) as previously proposed (Kang et al., 2017) or slower progression of the cell cycle (Gonzalez-Garcia et al., 2011; Hacham et al., 2011), but it could also be a consequence of sub-optimal cell-cell communication due to the disorganized meristem. High-resolution live imaging of briTRIPLE roots that express pertinent dynamic cell cycle markers might be required to gain ultimate insight. Furthermore, based on 1D cortex cell length measurements and modeling, it has been proposed that brassinosteroid signaling is required to set absolute cell size (Pavelescu et al., 2018). Our 3D data are consistent with this proposition with regard to cell elongation, but also show that the sizer mechanism applies to cellular anisotropy rather than a reduction of cell size in terms of volume. The same study suggested that brassinosteroid signaling suppresses a timer mechanism of cell elongation (Pavelescu et al., 2018), which resonates with the premature onset of volumetric growth in briTRIPLE meristems observed here. Interestingly, this result matches the graded activity of canonical brassinosteroid signaling in the root meristem, which increases as cells start to elongate and differentiate (Chaiwanon and Wang, 2015). Thus, timing the transition to anisotropic growth might be a primary effect of brassinosteroid signaling in the root, and also an effect that can be conveyed by brassinosteroid-dependent non-cell-autonomous signals.
Brassinosteroid-controlled genes can respond to non-cell-autonomous brassinosteroid action
The latter conclusion is derived from our finding that the systemic rescue of briTRIPLE roots through phloem-specific brassinosteroid signaling manifests in a normalized anatomy of mature cells. However, it is also associated with an intermediate meristem phenotype. Although briT-RESCUE meristems thus do not entirely resemble wild-type meristems, the cell-aligning effects of brassinosteroid signaling specifically in the developing phloem can obviously normalize the anatomy of both adjacent and distant brassinosteroid-blind cells to a degree that is sufficient to impart nearly wild-type root growth vigor. In line with this key finding, the single-cell mRNA sequencing data demonstrate that many brassinosteroid-controlled genes can respond to the postulated non-cell-autonomous signals. In this context, the expression of genes encoding AGPs was a particularly robust systemic readout, and our data therefore qualify AGPs as sentinels of brassinosteroid-dependent cellular anisotropy rather than of cell growth per se. AGPs can therefore be of use to trace the activity of non-cell-autonomous brassinosteroid-triggered signals. Given the strong effect of the rescue, one could assume that these signals are self-reinforcing but need to be activated by brassinosteroid perception in the first place. Such propagation could explain the far reach of the signal, including its apparently growing impact as cells transition to elongation and differentiation across cell layers, although the developing phloem sieve elements differentiate very early and no longer express the brassinosteroid receptor once they differentiate (Graeff et al., 2020). The present comprehensive analysis of brassinosteroid effects in the root might aid in the discovery of the non-cell-autonomous signals and can also serve as a blueprint for comparative analyses to determine whether brassinosteroid action in the shoot is based on similar principles.
Methods
Plant materials and growth conditions
Seeds were surface sterilized using 3% sodium hypochlorite, sown onto half-strength Murashige and Skoog agar medium (0.9% agarose) supplemented with 0.3% sucrose, and stratified for 3 days at 4°C. For Yariv reagent treatments, the α-galactosyl or β-glucosyl Yariv reagents (Biosupplies, Australia) were added to the media. Plants were grown under continuous white light (intensity ∼120 μE) at 22°C. All mutants and marker lines were in the A. thaliana Columbia-0 (Col-0) wild-type background. The briTRIPLE mutant and briT-RESCUE lines have been described previously (Graeff et al., 2020).
Tissue fixation and clearing
For confocal microscopy, roots of 7-day-old seedlings were fixed in 4% PFA in PBS and transferred into a vacuum of 25–30 mm Hg/Torr for 15–30 min, then washed three times in PBS for 5 min. For clearing of the samples, seedlings were transferred into ClearSee solution (Kurihara et al., 2015; Ursache et al., 2018) for 7 days at 4°C with regular changes of the clearing solution. Cleared roots were stained with 0.25 mg/ml Calcofluor White (CCFW; Sigma, product no. F3543), washed with ClearSee solution and mounted on microscope slides with distancers in ClearSee solution that contained 0.3% agarose to prevent moving of the samples.
Confocal microscopy
Samples were imaged on a Zeiss LSM880 confocal microscope with a 40× objective, using a 405-nm laser for CCFW excitation and recording of the cell wall signal in the 450–480-nm range. Tile scans and z stacks were set to image 500 μm of root meristem or 1000 μm of mature root (located ∼1 cm shootward from the root tip), respectively. The distance between the z scans was set to 0.42 μm for the root meristems and 0.5 μm for the mature roots, with a pinhole diameter of 33 μm, and 512 × 512-pixel 16-bit images were recorded with a scanning speed of 2.06 μs per pixel.
PlantSeg-MorphoGraphX-3DCellAtlas workflow
Segmentation of cells in the images
For segmentation of root micrographs, the channel of the stained cell walls was extracted as a 16-bit.tiff image. The PlantSeg workflow (Wolny et al., 2020) was used to predict cell boundaries and label the cells in the image stacks. The re-scaling factor for the images was calculated based on their resolution and the model used for cell boundary prediction ([1.70, 2.8, 2.8] to fit root meristems to the ≪ confocal_unet_bce_dice_ds2x≫ model; and [1.74, 2.04, 2.04] to fit the mature root images to the ≪confocal_unet_bce_dice_ds3x≫ model). Graphics processing unit (GPU)-based prediction was used for cell boundary prediction, followed by segmentation using the generalized algorithm for signed graph partitioning (GASP) segmentation algorithm with beta: 0.5, 2D watershed, and a watershed threshold of 0.5. Segments smaller than 1000 were discarded. After segmentation, images were re-scaled with the appropriate factors and saved as .tiff image files.
Creation of 3D meshes from the segmented images and extraction of cell properties
Segmentations were imported into MorphoGraphX software (Barbier de Reuille et al., 2015). 3D meshes of each root were created using the marching cubes 3D workflow with a cube size of two and a minimal cell size of 500 voxels. Cell properties were extracted using the 3DCellAtlas plugin as described (Montenegro-Johnson et al., 2015). Cell types were assigned based on the radial and circumferential coordinate of the cells and manually corrected. Attributes and properties of all cells were exported for comparative analysis. Images of the labeled root meshes using horizontal or transverse cutting planes and of the original image were generated with MorphoGraphX. Videos of 3D image stacks and renderings were generated using Imaris software.
Digital single-cell analysis using 3DCellAtlas
Cell types and 3D cell geometries in roots were determined using 3DCellAtlas (Montenegro-Johnson et al., 2015) implemented within MorphoGraphX (Barbier de Reuille et al., 2015) using the methodology described previously (Stamm et al., 2017). Cell type annotation was manually verified and corrected in instances where errors were present. Data describing cell size and shape were exported as CSV files for further statistical analyses.
Root comparison based on MorphoGraphX-3DCellAtas output
The comparison of cell properties for different genotypes was performed in R (4.0.4). Attribute data of the mature root segments were imported and cells with length, width, or circumference ≤0 μm or a volume <1000 μm3 were removed, leaving a dataset with 9571 cells. Cell properties were analyzed by the assigned cell type. For statistical comparison, mean values for each cell type were calculated per root and one-sided ANOVA was performed to analyze the significance of the observed variance between roots. Tuckey-HSD (honestly significant difference) tests were performed to compare the variances for each tissue between the genotypes. For comparative analyses of the root meristems, cell types of four roots per genotype were assigned using the 3DCellAtlas plugin and manually corrected. To compare dimensional properties, each root was re-oriented so that the y axis ran along the longitudinal axis of the root and x and z axes defined the horizontal dimensions. The distance of each cell centroid to the center of the SCN was calculated. For cell length, width, circumference, volume, and wall area, the data distribution was analyzed and cells below the 2.5% quantile and above the 97.5% quantile were discarded to remove segmentation artifacts. Roots were sliced into 5-μm bins along the longitudinal axis, and each cell was assigned to the bin containing its centroid. The dataset contained 46 559 cells in total (13 778 from wild type, 15 508 from the briTRIPLE mutant, and 17 273 from the briT-RESCUE line). Cell properties were plotted as scatterplots, and a regression curve was fitted to the data for each genotype using a general additive model with a shrinking cubic regression spline interpolation.
Straightening of 3D meshes, creation of simplified root models, and per-position comparisons
The procedure for the straightening and alignment of 3D meshes to produce simplified root meristem models for per-position cell parameter comparisons is described in detail in the separate supplemental methods 1 file.
10X Genomics sample preparation, library construction, and sequencing
Single-cell sequencing was performed on protoplasts from briTRIPLE and briT-RESCUE primary seedling roots (processed in parallel) as described (Wendrich et al., 2020). Briefly, 6-day-old briTRIPLE and briT-RESCUE roots were cut and protoplasts were isolated, stained for live/dead using 4′,6-diamidino-2-phenylindole (DAPI) at 14 μM final concentration, and sorted on a BD Aria II instrument. For each genotype, DAPI-negative protoplasts were selected for further analysis. Sorted cells were centrifuged at 400 g at 4°C and resuspended in protoplast solution A to yield an estimated concentration of 1000 cells/μl. Cellular suspensions were loaded on a Chromium Single Cell 3′ GEM, Library & Gel Bead Kit (V3 chemistry, 10X Genomics) according to the manufacturer's instructions. Libraries were sequenced on an Illumina NovaSeq600 instrument following recommendations of 10X Genomics at the VIB Nucleomics Core facility (VIB, Leuven). The sequencing data are available at the NCBI Gene Expression Omnibus repository under accession number GSE181178.
Raw data processing and generation of the gene expression matrix
Demultiplexing of the raw sequencing data was performed with the 10x CellRanger (version 3.1.0) software cellranger mkfastq. The fastq files obtained after demultiplexing were used as the input for cellranger count, which aligns the reads to the A. thaliana reference genome (Ensemble TAIR10.40) using STAR and collapses them to unique molecular identifier (UMI) counts. The result is a large digital expression matrix with cell barcodes as rows and gene identities as columns. Initial filtering in CellRanger recovered 10 574 cells for the briTRIPLE sample, and 12 335 cells for the briT-RESCUE sample. This corresponded to 54 593 mean reads and 4214 median genes per cell in the briTRIPLE sample, and 40 520 mean reads and 3525 median genes per cell in the briT-RESCUE sample. To ensure only high-quality cells were further analyzed, the filtered data provided by CellRanger were used as input for further filtering steps.
Data analysis (clustering, identity assignment, DEG, and quality control)
All analyses were performed in R (version 3.6.0). Data pre-processing was performed by the scater (version 1.10.1) package following a recommended workflow (Lun et al., 2016). Outlier cells were identified based on two metrics (library size and number of expressed genes) and tagged as outliers if they were five median absolute deviations (MADs) away from the median value of these metrics across all cells. After removing the outliers, this resulted in a final number of 5767 cells for the briTRIPLE sample and 6724 cells for the briT-RESCUE sample. Normalizing the raw counts, detecting highly variable genes, finding clusters, and creating Uniform Manifold Approximation and Projection (UMAP) plots was done using the Seurat pipeline (version 3.2.3). Differential expression analysis for marker gene identification per subpopulation was based on the non-parametric Wilcoxon rank sum test implemented within the Seurat pipeline. Clusters with the same cell annotation based on gene expression analysis were combined to generate a more comprehensible dataset. Samples were merged with the published wild-type root dataset (Wendrich et al., 2020) and annotations were transferred using the LabelTransfer method from the Seurat package. To validate the reliability of our single-cell analysis, the datasets were compared with another independent wild-type sample (Denyer et al., 2019) that was processed through our analysis pipeline. Cell types based on this reference were then annotated in the briTRIPLE and briT-RESCUE cells using the LabelTransfer method from the Seurat package. On average, 91% and 94% of briTRIPLE and briT-RESCUE cells, respectively, received the same cell type label from the Denyer et al. and the Wendrich et al. reference datasets, indicating the robustness of the annotation method. Feature plots of the mutated brassinosteroid receptor genes are shown in supplemental Figure 20.
Visualization of scRNA-seq expression data in heatmaps
Lists of Arabidopsis gene identifier (AGI) for brassinosteroid-responsive genes in the root tip or the whole seedling were obtained from the literature (Chaiwanon and Wang, 2015; Liu et al., 2020). Each gene was referenced to the cluster datafiles and LogFC, adjusted p value, and score were extracted to a new list. If no entry could be found for the respective cluster, N/A values were added. Summarized data were imported into R and tile plots for each comparison were created with tile color indicating the score value using the ggplot2 and scales libraries. Gene entries with an adjusted p value > 0.05 or N/A were displayed in gray. The scales were fixed to a range of −2 to 2 for better comparability. Bigger datasets were restricted to genes with a significant logFC in at least one cluster. Venn diagrams comparing the overlap between the different datasets were created using the ggVennDiagram library.
Funding
This work was funded by core funding from the University of Lausanne, the Swiss National Science Foundation (grant 310030B_185379, awarded to C.S.H.), The Research Foundation - Flanders (FWO; post-doc fellowship 1215820N, awarded to T.E.), the European Research Council (ERC Starting Grant TORPEDO; 714055, awarded to B.D.R.), the BBSRC (grant BB/S002804/1 to G.W.B.), and the Deutsche Forschungsgemeinschaft (DFG; post-doctoral fellowship GR 5009/1-1, awarded to M.G.).
Author contributions
Conceptualization, M.G., S.R., G.W.B., B.d.R., and C.S.H.; methodology, M.G., S.R., J.R.W., J.D., T.E., and N.G.; investigation, M.G., S.R., A.C.A.F., J.R.W., and T.E.; writing – original draft, M.G. and C.S.H.; writing – review & editing, all authors; supervision, G.W.B., B.d.R., and C.S.H.
Acknowledgments
We would like to thank Prof. Thomas Berleth for helpful comments on the manuscript, Prof. Gerd Juergens and Dr. Ulrike Mayer for a gift of anti-KN antibody, and Emma Picot for technical support in image analysis. No conflict of interest declared.
Published: August 3, 2021
Footnotes
Published by the Molecular Plant Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Molecular Plant Online.
Supplemental information
1
References
- Anne P., Amiguet-Vercher A., Brandt B., Kalmbach L., Geldner N., Hothorn M., Hardtke C.S. CLERK is a novel receptor kinase required for sensing of root-active CLE peptides in Arabidopsis. Development. 2018;145:dev162354. doi: 10.1242/dev.162354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier de Reuille P., Routier-Kierzkowska A.L., Kierzkowski D., Bassel G.W., Schupbach T., Tauriello G., Bajpai N., Strauss S., Weber A., Kiss A., et al. MorphoGraphX: a platform for quantifying morphogenesis in 4D. eLife. 2015;4:05864. doi: 10.7554/eLife.05864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonke M., Thitamadee S., Mahonen A.P., Hauser M.T., Helariutta Y. APL regulates vascular tissue identity in Arabidopsis. Nature. 2003;426:181–186. doi: 10.1038/nature02100. [DOI] [PubMed] [Google Scholar]
- Borassi C., Gloazzo Dorosz J., Ricardi M.M., Carignani Sardoy M., Pol Fachin L., Marzol E., Mangano S., Rodriguez Garcia D.R., Martinez Pacheco J., Rondon Guerrero Y.D.C., et al. A cell surface arabinogalactan-peptide influences root hair cell fate. New Phytol. 2020;227:732–743. doi: 10.1111/nph.16487. [DOI] [PubMed] [Google Scholar]
- Caesar K., Elgass K., Chen Z., Huppenberger P., Witthoft J., Schleifenbaum F., Blatt M.R., Oecking C., Harter K. A fast brassinolide-regulated response pathway in the plasma membrane of Arabidopsis thaliana. Plant J. 2011;66:528–540. doi: 10.1111/j.1365-313X.2011.04510.x. [DOI] [PubMed] [Google Scholar]
- Cano-Delgado A., Yin Y., Yu C., Vafeados D., Mora-Garcia S., Cheng J.C., Nam K.H., Li J., Chory J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development. 2004;131:5341–5351. doi: 10.1242/dev.01403. [DOI] [PubMed] [Google Scholar]
- Chaiwanon J., Wang Z.Y. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr. Biol. 2015;25:1031–1042. doi: 10.1016/j.cub.2015.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.G., Gao Z., Zhao Z., Liu X., Li Y., Zhang Y., Liu X., Sun Y., Tang W. BZR1 family transcription factors function redundantly and indispensably in BR signaling but exhibit BRI1-independent function in regulating anther development in Arabidopsis. Mol. Plant. 2019;12:1408–1415. doi: 10.1016/j.molp.2019.06.006. [DOI] [PubMed] [Google Scholar]
- Cho H., Ryu H., Rho S., Hill K., Smith S., Audenaert D., Park J., Han S., Beeckman T., Bennett M.J., et al. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat. Cell Biol. 2014;16:66–76. doi: 10.1038/ncb2893. [DOI] [PubMed] [Google Scholar]
- Choe S., Dilkes B.P., Fujioka S., Takatsuto S., Sakurai A., Feldmann K.A. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole R.A., McInally S.A., Fowler J.E. Developmentally distinct activities of the exocyst enable rapid cell elongation and determine meristem size during primary root growth in Arabidopsis. BMC Plant Biol. 2014;14:386. doi: 10.1186/s12870-014-0386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005;6:850–861. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- De Rybel B., Moller B., Yoshida S., Grabowicz I., Barbier de Reuille P., Boeren S., Smith R.S., Borst J.W., Weijers D. A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev. Cell. 2013;24:426–437. doi: 10.1016/j.devcel.2012.12.013. [DOI] [PubMed] [Google Scholar]
- Denyer T., Ma X., Klesen S., Scacchi E., Nieselt K., Timmermans M.C.P. Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev. Cell. 2019;48:840–852 e845. doi: 10.1016/j.devcel.2019.02.022. [DOI] [PubMed] [Google Scholar]
- Fendrych M., Van Hautegem T., Van Durme M., Olvera-Carrillo Y., Huysmans M., Karimi M., Lippens S., Guerin C.J., Krebs M., Schumacher K., et al. Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr. Biol. 2014;24:931–940. doi: 10.1016/j.cub.2014.03.025. [DOI] [PubMed] [Google Scholar]
- Ferreira-Guerra M., Marques-Bueno M., Mora-Garcia S., Cano-Delgado A.I. Delving into the evolutionary origin of steroid sensing in plants. Curr. Opin. Plant Biol. 2020;57:87–95. doi: 10.1016/j.pbi.2020.06.005. [DOI] [PubMed] [Google Scholar]
- Furuta K.M., Yadav S.R., Lehesranta S., Belevich I., Miyashima S., Heo J.O., Vaten A., Lindgren O., De Rybel B., Van Isterdael G., et al. Plant development. Arabidopsis NAC45/86 direct sieve element morphogenesis culminating in enucleation. Science. 2014;345:933–937. doi: 10.1126/science.1253736. [DOI] [PubMed] [Google Scholar]
- Gonzalez N., De Bodt S., Sulpice R., Jikumaru Y., Chae E., Dhondt S., Van Daele T., De Milde L., Weigel D., Kamiya Y., et al. Increased leaf size: different means to an end. Plant Physiol. 2010;153:1261–1279. doi: 10.1104/pp.110.156018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Garcia M.P., Vilarrasa-Blasi J., Zhiponova M., Divol F., Mora-Garcia S., Russinova E., Cano-Delgado A.I. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development. 2011;138:849–859. doi: 10.1242/dev.057331. [DOI] [PubMed] [Google Scholar]
- Graeff M., Rana S., Marhava P., Moret B., Hardtke C.S. Local and systemic effects of brassinosteroid perception in developing phloem. Curr. Biol. 2020;30:1626–1638.e23. doi: 10.1016/j.cub.2020.02.029. [DOI] [PubMed] [Google Scholar]
- Hacham Y., Holland N., Butterfield C., Ubeda-Tomas S., Bennett M.J., Chory J., Savaldi-Goldstein S. Brassinosteroid perception in the epidermis controls root meristem size. Development. 2011;138:839–848. doi: 10.1242/dev.061804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisanaga T., Kawade K., Tsukaya H. Compensation: a key to clarifying the organ-level regulation of lateral organ size in plants. J. Exp. Bot. 2015;66:1055–1063. doi: 10.1093/jxb/erv028. [DOI] [PubMed] [Google Scholar]
- Holzwart E., Huerta A.I., Glockner N., Garnelo Gomez B., Wanke F., Augustin S., Askani J.C., Schurholz A.K., Harter K., Wolf S. BRI1 controls vascular cell fate in the Arabidopsis root through RLP44 and phytosulfokine signaling. Proc. Natl. Acad. Sci. U S A. 2018;115:11838–11843. doi: 10.1073/pnas.1814434115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.H., Breda A., Hardtke C.S. Brassinosteroid signaling directs formative cell divisions and protophloem differentiation in Arabidopsis root meristems. Development. 2017;144:272–280. doi: 10.1242/dev.145623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.J., Russinova E. Brassinosteroid signalling. Curr. Biol. 2020;30:R294–R298. doi: 10.1016/j.cub.2020.02.011. [DOI] [PubMed] [Google Scholar]
- Kondo Y., Ito T., Nakagami H., Hirakawa Y., Saito M., Tamaki T., Shirasu K., Fukuda H. Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF-TDR signalling. Nat. Commun. 2014;5:3504. doi: 10.1038/ncomms4504. [DOI] [PubMed] [Google Scholar]
- Kurihara D., Mizuta Y., Sato Y., Higashiyama T. ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development. 2015;142:4168–4179. doi: 10.1242/dev.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber M.H., Waizenegger I., Steinmann T., Schwarz H., Mayer U., Hwang I., Lukowitz W., Jurgens G. The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 1997;139:1485–1493. doi: 10.1083/jcb.139.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Liu X., Yang H., Wang Y., Zhu Z., Zhang W., Li J. Comparative transcriptomic analysis to identify brassinosteroid response genes. Plant Physiol. 2020;184:1072–1082. doi: 10.1104/pp.20.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A.T., Bach K., Marioni J.C. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro-Johnson T.D., Stamm P., Strauss S., Topham A.T., Tsagris M., Wood A.T., Smith R.S., Bassel G.W. Digital single-cell analysis of plant organ development using 3DCellAtlas. Plant Cell. 2015;27:1018–1033. doi: 10.1105/tpc.15.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Kushiro T., Yokota T., Kamiya Y., Bishop G.J., Yamaguchi S. The last reaction producing brassinolide is catalyzed by cytochrome P-450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. J. Biol. Chem. 2005;280:17873–17879. doi: 10.1074/jbc.M414592200. [DOI] [PubMed] [Google Scholar]
- Oh M.H., Honey S.H., Tax F.E. The control of cell expansion, cell division, and vascular development by brassinosteroids: a historical perspective. Int. J. Mol. Sci. 2020;21:1743. doi: 10.3390/ijms21051743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T., Szatmari A.M., Watanabe B., Fujita S., Bancos S., Koncz C., Lafos M., Shibata K., Yokota T., Sakata K., et al. C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell. 2006;18:3275–3288. doi: 10.1105/tpc.106.045443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco-Villalobos D., Diaz-Moreno S.M., van der Schuren A., Tamaki T., Kang Y.H., Gujas B., Novak O., Jaspert N., Li Z., Wolf S., et al. The effects of high steady state auxin levels on root cell elongation in Brachypodium. Plant Cell. 2016;28:1009–1024. doi: 10.1105/tpc.15.01057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelescu I., Vilarrasa-Blasi J., Planas-Riverola A., Gonzalez-Garcia M.P., Cano-Delgado A.I., Ibanes M. A Sizer model for cell differentiation in Arabidopsis thaliana root growth. Mol. Syst. Biol. 2018;14:e7687. doi: 10.15252/msb.20177687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S., Sabatini S. Analysis of root meristem size development. Methods Mol. Biol. 2010;655:177–187. doi: 10.1007/978-1-60761-765-5_12. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Villalon A., Gujas B., Kang Y.H., Breda A.S., Cattaneo P., Depuydt S., Hardtke C.S. Molecular genetic framework for protophloem formation. Proc. Natl. Acad. Sci. U S A. 2014;111:11551–11556. doi: 10.1073/pnas.1407337111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh J., Moon J., Youn J.H., Seo C., Park Y.J., Kim S.K. Establishment of biosynthetic pathways to generate castasterone as the biologically active brassinosteroid in Brachypodium distachyon. J. Agric. Food Chem. 2020;68:3912–3923. doi: 10.1021/acs.jafc.9b07963. [DOI] [PubMed] [Google Scholar]
- Ruan Y., Halat L.S., Khan D., Jancowski S., Ambrose C., Belmonte M.F., Wasteneys G.O. The microtubule-associated protein CLASP sustains cell proliferation through a brassinosteroid signaling negative feedback loop. Curr. Biol. 2018;28:2718–2729.e5. doi: 10.1016/j.cub.2018.06.048. [DOI] [PubMed] [Google Scholar]
- Savaldi-Goldstein S., Peto C., Chory J. The epidermis both drives and restricts plant shoot growth. Nature. 2007;446:199–202. doi: 10.1038/nature05618. [DOI] [PubMed] [Google Scholar]
- Seifert G.J., Roberts K. The biology of arabinogalactan proteins. Annu. Rev. Plant Biol. 2007;58:137–161. doi: 10.1146/annurev.arplant.58.032806.103801. [DOI] [PubMed] [Google Scholar]
- Singh A.P., Savaldi-Goldstein S. Growth control: brassinosteroid activity gets context. J. Exp. Bot. 2015;66:1123–1132. doi: 10.1093/jxb/erv026. [DOI] [PubMed] [Google Scholar]
- Stamm P., Strauss S., Montenegro-Johnson T.D., Smith R., Bassel G.W. In: Kleine-Vehn J., Sauer M., editors. Vol. 1497. Humana Press; New York, NY: 2017. In silico methods for cell annotation, quantification of gene expression, and cell geometry at single-cell resolution using 3DCellAtlas; pp. 99–123. (Plant Hormones. Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- Sun S., Chen D., Li X., Qiao S., Shi C., Li C., Shen H., Wang X. Brassinosteroid signaling regulates leaf erectness in Oryza sativa via the control of a specific U-type cyclin and cell proliferation. Dev. Cell. 2015;34:220–228. doi: 10.1016/j.devcel.2015.05.019. [DOI] [PubMed] [Google Scholar]
- Svolacchia N., Salvi E., Sabatini S. Arabidopsis primary root growth: let it grow, can't hold it back anymore! Curr. Opin. Plant Biol. 2020;57:133–141. doi: 10.1016/j.pbi.2020.08.005. [DOI] [PubMed] [Google Scholar]
- Szekeres M., Nemeth K., Koncz-Kalman Z., Mathur J., Kauschmann A., Altmann T., Redei G.P., Nagy F., Schell J., Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Tamaki T., Oya S., Naito M., Ozawa Y., Furuya T., Saito M., Sato M., Wakazaki M., Toyooka K., Fukuda H., et al. VISUAL-CC system uncovers the role of GSK3 as an orchestrator of vascular cell type ratio in plants. Commun. Biol. 2020;3:184. doi: 10.1038/s42003-020-0907-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Han Z., Chai J. Q&A: what are brassinosteroids and how do they act in plants? BMC Biol. 2016;14:113. doi: 10.1186/s12915-016-0340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkowska D. Plants are capable of synthesizing animal steroid hormones. Molecules. 2019;24:2585. doi: 10.3390/molecules24142585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I., Lehner K., McCaskey E., Nirmal N., Ozkan-Aydin Y., Murray-Cooper M., Jain R., Hawkes E.W., Ronald P.C., Goldman D.I., et al. Mechanism and function of root circumnutation. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2018940118. e2018940118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M.V., Holbrook N.M. Root-gel interactions and the root waving behavior of Arabidopsis. Plant Physiol. 2004;135:1822–1837. doi: 10.1104/pp.104.040881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursache R., Andersen T.G., Marhavy P., Geldner N. A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J. 2018;93:399–412. doi: 10.1111/tpj.13784. [DOI] [PubMed] [Google Scholar]
- van Hengel A.J., Roberts K. Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. Plant J. 2002;32:105–113. doi: 10.1046/j.1365-313x.2002.01406.x. [DOI] [PubMed] [Google Scholar]
- Vandepoele K., Raes J., De Veylder L., Rouze P., Rombauts S., Inze D. Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell. 2002;14:903–916. doi: 10.1105/tpc.010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaeren H., Gonzalez N., Coppens F., De Milde L., Van Daele T., Vermeersch M., Eloy N.B., Storme V., Inze D. Combining growth-promoting genes leads to positive epistasis in Arabidopsis thaliana. eLife. 2014;3:e02252. doi: 10.7554/eLife.02252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruysse J., Baekelandt A., Gonzalez N., Inze D. Molecular networks regulating cell division during Arabidopsis leaf growth. J. Exp. Bot. 2020;71:2365–2378. doi: 10.1093/jxb/erz522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vragovic K., Sela A., Friedlander-Shani L., Fridman Y., Hacham Y., Holland N., Bartom E., Mockler T.C., Savaldi-Goldstein S. Translatome analyses capture of opposing tissue-specific brassinosteroid signals orchestrating root meristem differentiation. Proc. Natl. Acad. Sci. U S A. 2015;112:923–928. doi: 10.1073/pnas.1417947112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- Wendrich J.R., Yang B., Vandamme N., Verstaen K., Smet W., Van de Velde C., Minne M., Wybouw B., Mor E., Arents H.E., et al. Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science. 2020;370:eaay4970. doi: 10.1126/science.aay4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats W.G., Knox J.P. A role for arabinogalactan-proteins in plant cell expansion: evidence from studies on the interaction of beta-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. Plant J. 1996;9:919–925. doi: 10.1046/j.1365-313x.1996.9060919.x. [DOI] [PubMed] [Google Scholar]
- Wolf S., van der Does D., Ladwig F., Sticht C., Kolbeck A., Schurholz A.K., Augustin S., Keinath N., Rausch T., Greiner S., et al. A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc. Natl. Acad. Sci. U S A. 2014;111:15261–15266. doi: 10.1073/pnas.1322979111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolny A., Cerrone L., Vijayan A., Tofanelli R., Barro A.V., Louveaux M., Wenzl C., Strauss S., Wilson-Sanchez D., Lymbouridou R., et al. Accurate and versatile 3D segmentation of plant tissues at cellular resolution. eLife. 2020;9:e57613. doi: 10.7554/eLife.57613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W., Band L.R., Kumpf R.P., Van Damme D., Parizot B., De Rop G., Opdenacker D., Moller B.K., Skorzinski N., Njo M.F., et al. Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis. Science. 2016;351:384–387. doi: 10.1126/science.aad2776. [DOI] [PubMed] [Google Scholar]
- Yamamoto R., Fujioka S., Demura T., Takatsuto S., Yoshida S., Fukuda H. Brassinosteroid levels increase drastically prior to morphogenesis of tracheary elements. Plant Physiol. 2001;125:556–563. doi: 10.1104/pp.125.2.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Sardar H.S., McGovern K.R., Zhang Y., Showalter A.M. A lysine-rich arabinogalactan protein in Arabidopsis is essential for plant growth and development, including cell division and expansion. Plant J. 2007;49:629–640. doi: 10.1111/j.1365-313X.2006.02985.x. [DOI] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Zhiponova M.K., Vanhoutte I., Boudolf V., Betti C., Dhondt S., Coppens F., Mylle E., Maes S., Gonzalez-Garcia M.P., Cano-Delgado A.I., et al. Brassinosteroid production and signaling differentially control cell division and expansion in the leaf. New Phytol. 2013;197:490–502. doi: 10.1111/nph.12036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1