Abstract
Most cellular processes are carried out by protein complexes, but it is still largely unknown how the subunits of lowly expressed complexes find each other in the crowded cellular environment. Here, we will describe a working model where RNA-binding proteins in cytoplasmic condensates act as matchmakers between their bound proteins (called protein targets) and newly translated proteins of their RNA targets to promote their assembly into complexes. Different RNA-binding proteins act as scaffolds for various cytoplasmic condensates with several of them supporting translation. mRNAs and proteins are recruited into the cytoplasmic condensates through binding to specific domains in the RNA-binding proteins. Scaffold RNA-binding proteins have a high valency. In our model, they use homotypic interactions to assemble condensates and they use heterotypic interactions to recruit protein targets into the condensates. We propose that unoccupied binding sites in the scaffold RNA-binding proteins transiently retain recruited and newly translated proteins in the condensates, thus promoting their assembly into complexes. Taken together, we propose that lowly expressed subunits of protein complexes combine information in their mRNAs and proteins to colocalize in the cytoplasm. The efficiency of protein complex assembly is increased by transient entrapment accomplished by multivalent RNA-binding proteins within cytoplasmic condensates.
Keywords: protein–protein interaction, function of biomolecular condensates, cooperativity between mRNA and protein motifs, cytoplasmic compartmentalization: subcellular organization, localized processes
KNOWN MODES OF LOCALIZED PROTEIN COMPLEX ASSEMBLY IN CELLS
Most cellular processes are carried out by protein complexes (Alberts 1998). These complexes are often composed of two or more distinct subunits. Although we have a good understanding whether two protein surfaces can interact with each other, it is still largely unknown how the subunits find each other in living cells. An important concept that reduces the number of possible interactions is compartment-specific complex assembly. For example, multicomponent channels and receptors that contain transmembrane domains are synthesized and assembled in the endoplasmic reticulum (ER) (Bonifacino et al. 1990; Zerangue et al. 1999). Also a group of non-membrane proteins is known to assemble locally. Among the most highly abundant cellular protein complexes are ribosomes which are partially assembled in the nucleolus (Klinge and Woolford 2019; Lafontaine et al. 2021). Moreover, synaptic proteins are often translated and assembled locally at synapses (Biederer et al. 2002; Hafner et al. 2019; Holt et al. 2019; Biever et al. 2020). However, it is currently not known how the majority of non-membrane protein complexes colocalize for efficient and specific assembly in the cytoplasm.
HOW ARE PROTEIN COMPLEXES FORMED IN CELLS WHEN THEY ARE GENERATED FROM LOWLY EXPRESSED SUBUNITS?
Protein complex assembly is often studied in vitro with purified subunits, assuming that individually translated subunits assemble through diffusion and random encounter post-translationally in cells. This model may work for protein complexes that consist of highly expressed subunits. However, among the millions of proteins in the crowded cellular mileu, it is unclear how lowly expressed subunits establish faithful and efficient protein complexes.
The problem of finding a lowly expressed partner is best illustrated by the following analogy: If one protein is the size of a person, then the cellular space corresponds to 400 football fields that are packed with people. How do you find your friend? If both of you start walking, it is unlikely that you will meet in due time. One way to meet is to enter the football fields together. For proteins in cells, this means that two mRNAs that encode the subunits of a complex would enter the cytoplasm together. This strategy is widely used by operon-encoded proteins in prokaryotes (Shieh et al. 2015). As eukaryotic cells do not have operons, this strategy would require that the mRNAs already colocalize in the nucleus. Although there is some evidence for this model (Morf et al. 2019; Nair et al. 2021), systematic studies that rigorously test this idea are still lacking.
Another way to increase the chance of interactor colocalization is to restrict the search space (Klemm et al. 1998; Kuriyan and Eisenberg 2007). Returning to our football field analogy, you could tell your friend to meet you near the south goal of football field 23 to restrict the search space. Smaller volumes that promote subunit encounter can be provided by membrane-enclosed organelles such as the ER and mitochondria. In order for proteins to localize to these specialized compartments, they need to bring the information on a “meeting point” with them. The information can be stored within the proteins in the form of signal peptides or transmembrane domains that guide them to their respective compartments. The meeting point information can also be stored in the mRNAs that encode the proteins through binding sites for RNA-binding proteins that determine subcellular mRNA localization (Lawrence and Singer 1986; Sylvestre et al. 2003; Saint-Georges et al. 2008; Gadir et al. 2011; Buxbaum et al. 2015; Glock et al. 2017). Alternatively, mRNA-bound RNA-binding proteins can recruit effector proteins such as the signal recognition particle to the translating ribosomes to guide them to their correct location for localized protein synthesis on the ER (Chartron et al. 2016). After the proteins are localized, the surrounding membrane prevents the immediate escape of the subunits. This increases their local dwell time, thus giving the subunits time to encounter their respective partners.
In addition to membrane-enclosed organelles, we propose that membraneless organelles can provide compartments for the assembly of protein complexes that lack transmembrane domains. The initial localization of mRNAs and proteins using mRNA and protein motifs is similar to the mechanism described for membrane-enclosed organelles. However, it is currently largely unclear how cytoplasmic condensates allow transient retention of proteins to promote their assembly into protein complexes.
Here, we will describe a working model by which cytoplasmic condensates promote protein complex assembly. We propose that RNA-binding proteins that are scaffolds of cytoplasmic condensates act as matchmakers between proteins to promote their assembly into complexes. Whereas the term “RNA matchmaking” has been used to describe RNA base-pairing that is promoted by RNA-binding proteins (Portman and Dreyfuss 1994; Wu et al. 2020), here, we propose that RNA-binding proteins bring together proteins for complex assembly.
WORKING MODEL FOR RNA-BINDING PROTEINS THAT SERVE AS MATCHMAKERS BETWEEN SUBUNITS OF PROTEIN COMPLEXES
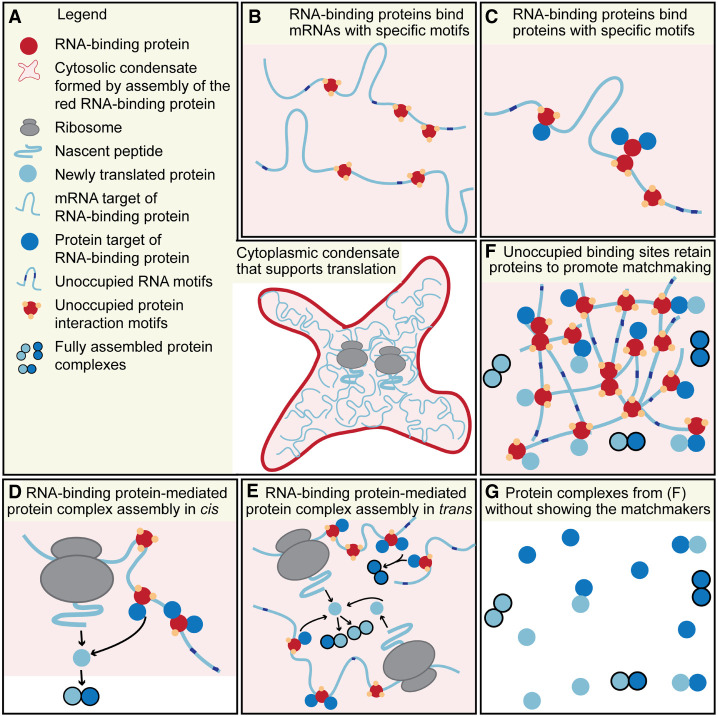
RNA-binding proteins assemble into translation-competent cytoplasmic condensates that enrich functionally related mRNAs and proteins. Therefore, cytoplasmic condensates might serve as organizational hubs that facilitate protein complex assembly between the enriched proteins and the proteins encoded by the enriched mRNAs (Fig. 1). The model consists of the following steps: (i) multivalent interaction of specific RNA-binding proteins with their bound mRNAs results in phase separation and the generation of cytoplasmic condensates that allow translation; (ii) Condensate RNA-binding proteins bind to specific mRNAs, thus recruiting and anchoring them to the condensates; (iii) RNA-binding proteins also bind specifically to proteins, thus bringing them into the condensates; (iv) Unoccupied binding sites within the RNA-binding proteins are used to transiently retain proteins in the condensates; (v) Colocalization of the recruited proteins with the newly translated proteins encoded by condensate-enriched mRNAs promotes protein–protein interactions of functionally related proteins. In this model, repeated motifs within mRNAs and proteins are used to increase their local dwell time to promote complex assembly.
FIGURE 1.
Model for condensate RNA-binding proteins to serve as protein matchmakers for complex assembly. (A) An RNA-binding protein (red) assembles into a cytoplasmic condensate that supports translation. (B) The RNA-binding protein (red) binds to specific mRNAs (mRNA target of the RNA-binding protein, light blue) which results in mRNA enrichment in the cytoplasmic condensate. (C) The RNA-binding protein (red) uses specific protein domains (yellow dots) to bind to specific proteins (protein target of the RNA-binding protein, blue dots) which results in their recruitment into the condensate. The protein interaction domains are used for homotypic interaction to assemble the condensate and for heterotypic interaction to recruit and retain protein targets in the condensate. The multivalent protein interaction domains provide specificity for the enriched proteins in the condensate which serve as potential subunits of complexes. (D) Translation of the mRNA target allows the newly translated protein to form protein–protein interactions with a protein target in cis. A fully formed protein complex is indicated by the black outline of the proteins and signifies that all potential retention sites are masked. This allows the fully formed complex to leave the condensate. (E) As in D, but the proximity of other mRNA and protein targets also allows interactions in trans between protein targets recruited by neighboring mRNAs and among newly translated mRNA targets. (F) Unoccupied multivalent interaction domains of the RNA-binding protein allow recruited proteins and newly translated proteins to be transiently retained within the condensate network. Repeated binding allows them to scan the condensate to increase the chance of encounter of specific interactors. After complex assembly, they leave the condensate through masking of the retention sites. (G) Without visualizing the protein matchmakers that consist of the condensate RNA-binding protein and the bound mRNAs, protein complex assembly appears to occur through diffusion and random encounter.
It seems that protein complexes formed by lowly expressed subunits that lack membrane domains would benefit the most from this assembly mode. Moreover, we hypothesize that the protein- and RNA-interaction domains of scaffold RNA-binding proteins each determine a unique condensate assembly mode. These interaction domains allow transient entrapment of proteins containing said domains, thus providing specificity for condensate-dependent protein complex assembly.
RNA-BINDING PROTEINS ASSEMBLE IN AN mRNA-DEPENDENT MANNER TO GENERATE CYTOPLASMIC CONDENSATES
Several cytoplasmic condensates have been found that are generated by mRNA-dependent phase separation (Fig. 1A). They include TIS granules, FXR1 condensates, and L-bodies, and they were detected in human cell lines, mouse myotubes, and in frog oocyte stages II–III, respectively (Ma and Mayr 2018; Smith et al. 2020; Neil et al. 2021). TIS granules are generated by assembly of the RNA-binding protein TIS11B. They have an irregularly shaped morphology and localize to the surface of the rough ER (Ma and Mayr 2018; Ma et al. 2021). FXR1 condensates are generated through RNA-dependent assembly of the RNA-binding protein FXR1. They were initially observed in muscle cells (Smith et al. 2020) but have also been found in human cell lines where they generate a large network in the cytoplasm, called FXR1 network (Chen X and Mayr C, unpubl.). In frog oocytes, irregularly shaped L-bodies are enriched for several RNA-binding proteins together with their bound mRNAs (Cabral and Mowry 2021; Neil et al. 2021).
RNA-BINDING PROTEINS BIND TO mRNAs WITH SPECIFIC MOTIFS THUS LEADING TO THEIR ENRICHMENT IN CYTOPLASMIC CONDENSATES
mRNAs often use specific sequence or structural motifs to localize to different cytoplasmic condensates (Zhang et al. 2015; Boke et al. 2016; Langdon et al. 2018; Ma and Mayr 2018; Neil et al. 2021). For example, mRNAs with binding sites for the RNA-binding protein Xvelo localize to the Balbiani body in frog oocytes. Injection of mRNAs that lacked these binding sites prevented their condensate localization, with localization restored when the Xvelo binding sites were added back (Boke et al. 2016). Similarly, the Vg1 mRNA localizes to L-bodies through binding sites for the RNA-binding proteins PTB and Vera. Mutation of the binding sites abrogates Vg1 mRNA localization to the condensates (Neil et al. 2021). The RNA-binding protein TIS11B binds to AU-rich elements (Hudson et al. 2004; Ma and Mayr 2018). mRNAs that contain several AU-rich elements localize to TIS granules, whereas the same mRNAs that lack these elements are excluded from TIS granules (Ma and Mayr 2018). These findings suggest that the mRNAs are locally entrapped in cytoplasmic condensates (Fig. 1B).
Local entrapment in condensates was first shown for the nanos mRNA which is produced at the anterior pole of fly oocytes and uses diffusion to translocate to the posterior pole. The nanos mRNA is then entrapped by the germ plasm at the posterior pole (Forrest and Gavis 2003). Although the entrapment is inefficient as only approximately 4% of nanos transcripts are found at the posterior pole, it is sufficient for body plan patterning (Forrest and Gavis 2003).
Determining the fraction of enriched transcripts in TIS granules revealed large differences between mRNAs that encode non-membrane and membrane proteins. Generally, mRNAs encoding non-membrane proteins only partially localize to TIS granules, whereas nearly all of the transcripts of mRNAs that encode membrane proteins localize to TIS granules (Ma and Mayr 2018). The data on TIS granules are consistent with a model where mRNAs translated by ribosomes are anchored to the ER surface by the signal recognition particle, thus stably entrapping AU-rich element-containing mRNAs that encode membrane proteins in TIS granules. AU-rich element-containing mRNAs that encode non-membrane proteins seem to be partially entrapped, likely by binding to RNA-binding proteins enriched in TIS granules, including the known AU-rich element binding proteins HuR and TIS11B (Ma and Mayr 2018). mRNAs that lack AU-rich elements are excluded from the condensate, presumably through lack of binding sites for TIS granule-enriched proteins, and by containing binding sites that localize them elsewhere.
SEVERAL CYTOPLASMIC CONDENSATES SUPPORT TRANSLATION
Protein synthesis was shown to occur in several cytoplasmic condensates, suggesting that mRNAs that are enriched in diverse cytoplasmic condensates are translated. For example, translation factor granules are sites of active translation in exponentially growing yeast cells and require translation for mRNA localization to the condensates (Pizzinga et al. 2019). Translation-dependent mRNA localization was also observed in mammalian cells (Voigt et al. 2017; Chouaib et al. 2020). Glycolytic mRNA granules, also called “core fermentation” or CoFe granules, were found in yeast and human cells and are translation factories for glycolytic proteins (Lui et al. 2014; Morales-Polanco et al. 2021).
Direct evidence for translation in cytoplasmic condensates can be obtained with the SunTag system which labels nascent peptides with GFP and their corresponding mRNAs with mCherry, thus visualizing active translation of single molecules (Yan et al. 2016). This system detected translation inside and outside of TIS granules and stress granules (Mateju et al. 2020; Ma W and Mayr C, unpubl.).
RNA-BINDING PROTEINS HAVE LARGE NUMBERS OF SPECIFIC PROTEIN INTERACTORS
RNA-binding proteins are defined as proteins that bind to RNA (Baltimore and Huang 1970; Baltz et al. 2012; Castello et al. 2012; Gerstberger et al. 2014; Caudron-Herger et al. 2019, 2021). Most RNA-binding proteins are characterized by a modular domain structure and contain repeated RNA-binding domains (Lunde et al. 2007). The repeated domains serve at least two purposes: (i) They allow the protein to contact a higher number of nucleotides, thus increasing the binding specificity to RNA and (ii) they are commonly used as protein interaction domains (Lunde et al. 2007). Structural analyses found that several RNA-binding domains bind equally well to RNA and protein (Chen et al. 1997; Kim et al. 2000; Ramos et al. 2002, 2006; Garcia-Mayoral et al. 2007; Clery et al. 2008; Chi et al. 2011; Tenzer et al. 2013; Pabis et al. 2019; Yan et al. 2019). RNA-binding domains that serve as protein binding sites include RRMs, KH domains, dsRNA-binding domains, zinc fingers, and intrinsically disordered regions. The capability for protein binding allows RNA-binding proteins to dimerize and to form extended RNA-binding surfaces (Lunde et al. 2007; Clery et al. 2008). It allows them to interact simultaneously with RNA and protein targets, thus enabling the recruitment of effector proteins to the mRNA. This was shown to determine the functional outcome of the bound mRNA or encoded protein (Dienstbier et al. 2009; Fabian et al. 2013; Berkovits and Mayr 2015; Lee and Mayr 2019; Fernandes and Buchan 2021). For example, when the RNA-binding protein binds a motor protein, it allows subcellular localization of the mRNA (Dienstbier et al. 2009). Or, when the RNA-binding protein recruits a trafficking factor while the mRNA is translated, it allows localization of the encoded protein (Berkovits and Mayr 2015).
In addition to structural analyses, genome-wide approaches have also revealed that RNA-binding proteins are among the protein classes with the largest number of protein–protein interactions and thus are considered as hubs for protein complex assembly (Baltz et al. 2012; Brannan et al. 2016). Through integration of more than 100 data sets that identified potential RNA-binding proteins, it was observed that the confidence to classify a protein as an RNA-binding protein increased with the number of protein interactors (Caudron-Herger et al. 2019, 2021). This finding emphasizes the fundamental role of RNA-binding proteins as protein interaction hubs. Importantly, the proteomic studies further revealed that typically up to half of the protein interactors bind in an RNA-independent manner, thus representing true protein interactors of RNA-binding proteins (Tenzer et al. 2013; Brannan et al. 2016). Taken together, these results indicate that for the functional characterization of RNA-binding proteins their capability for protein binding is as important as their capacity for RNA binding (Fig. 1C; Lunde et al. 2007). We call here the protein interactors of RNA-binding proteins their “protein targets.”
RNA-BINDING PROTEINS ACT AS MATCHMAKERS BETWEEN THEIR mRNA AND PROTEIN TARGETS, THUS COLOCALIZING RECRUITED AND NEWLY TRANSLATED PROTEINS
After having established that translation occurs in cytoplasmic condensates and that RNA-binding proteins pervasively, but specifically bind to mRNAs and proteins, it becomes evident that RNA-binding proteins that bind to mRNAs and proteins during translation act as recruiters of effector proteins to the site of translation (Fig. 1D; Berkovits and Mayr 2015; Ma and Mayr 2018; Lee and Mayr 2019; Fernandes and Buchan 2020). This facilitates protein–protein interactions between newly translated proteins and recruited proteins (Berkovits and Mayr 2015) and highlights the role of RNA-binding proteins as matchmakers between their protein and mRNA targets (Fig. 1D). During translation, the mRNA target is connected via the ribosome with its nascent peptide, therefore, RNA-binding proteins bring together their protein targets with the newly translated proteins encoded by their mRNA targets. The matchmaking principle of RNA-binding proteins has been experimentally demonstrated for a handful of proteins where one or two RNA-binding proteins bound to a scaffolding mRNA recruit effector proteins in cis to a newly translated protein (Fig. 1D; Berkovits and Mayr 2015; Chartron et al. 2016; Ma and Mayr 2018; Lee and Mayr 2019; Fernandes and Buchan 2020). Whereas these examples represent individual protein complexes, we wondered about the benefits of the matchmaking process if it occurred in cytoplasmic condensates.
PROXIMITY IN CYTOPLASMIC CONDENSATES ENABLES PROTEIN MATCHMAKING IN TRANS
Cytoplasmic condensates are enriched in RNA-binding proteins and in hundreds of bound mRNAs (Hubstenberger et al. 2017; Khong et al. 2017; Horste El, Zhen G, Mayr C, unpubl.). If we envision the matchmaking process to happen within a cytoplasmic condensate, the proximity between newly translated proteins and protein targets bound by neighboring mRNAs allows their potential interaction in trans (Fig. 1E). Because of the condensate-induced proximity, it is reasonable to assume that protein–protein interactions among recruited proteins or among two nascent proteins will also be promoted (Fig. 1E; Mayr 2018).
TRANSIENT RETENTION OF PROTEINS IN CYTOPLASMIC CONDENSATES BY BINDING TO UNOCCUPIED BINDING SITES IN RNA-BINDING PROTEINS
Protein complex assembly through matchmaking will work best if the scaffold RNA-binding protein of the condensate has a very high valency. As not all of the binding sites are used for condensate assembly, the RNA-binding protein will have a large number of unoccupied binding sites. The unoccupied binding sites will then be available for binding to protein targets and proteins encoded by mRNA targets. These proteins will become transiently retained in the condensate through repeated binding. This allows the proteins to scan the whole condensate for potential interaction partners that are also temporarily retained, thus resulting in preferential complex assembly between the retained proteins. If unoccupied binding sites are not present, the proteins will freely exchange between the condensate phase and the surrounding liquid phase (Banani et al. 2017; Shin and Brangwynne 2017).
Compartment-dependent protein retention was previously shown for the ER. The ER contains ER-resident proteins and at the same time is the site of protein synthesis for membrane proteins that are only temporarily present in the ER as they will traffic to their final destination. The ER has to distinguish between ER-resident proteins and proteins that are passing through. ER-resident proteins have sequences that bind to ER-resident chaperones and retain them in the ER (Nilsson et al. 1989; Bonifacino et al. 1990; Cosson and Letourneur 1994; Zerangue et al. 1999).
SPECIFICITY FOR TRANSIENT PROTEIN RETENTION IN CYTOPLASMIC CONDENSATES IS LIKELY PROVIDED BY THE MODE OF CONDENSATE ASSEMBLY
What protein domains could serve as protein retention signals for cytoplasmic condensates? Diverse domains were found to contribute to the high valency of RNA-binding proteins that act as scaffolds of cytoplasmic condensates: RNA-binding domains, intrinsically disordered regions, protein interaction domains, and binding motifs for post-translational modifications (PTMs) (Hofweber and Dormann 2019; Guillen-Boixet et al. 2020; Smith et al. 2020; Yang et al. 2020; Zhong et al. 2000) For G3BP1 it was shown that the amino-terminal NTF2 domain is used for homotypic binding that is necessary for stress granule assembly. Importantly, the same domain also engages in heterotypic interactions that recruit proteins into the condensates (Yang et al. 2020). Based on these results, we propose that the multivalent domains of RNA-binding proteins that are used to assemble specific condensates will serve as potential unoccupied binding sites for transient protein retention in cytoplasmic condensates.
Recent data suggest that each condensate uses a specific assembly mode. Whereas some RNA-binding proteins, including FUS and hnRNPA1, phase separate by using multivalent interactions between tyrosines and arginines, HP1α requires phosphorylation to form higher-order complexes (Larson et al. 2017; Wang et al. 2018). PML bodies assemble through multivalent interactions between SUMO-1 and SUMO-interaction motifs (Zhong et al. 2000; Lallemand-Breitenbach and de The 2010; Banani et al. 2016; Li et al. 2017), whereas the Balbiani body and Cajal bodies use multivalent interactions between dimethylated arginines and their corresponding binding sites in Tudor domains (Zhong et al. 2000; Tripsianes et al. 2011; Roovers et al. 2018; Courchaine et al. 2021). We propose that the unique combination of unoccupied binding sites that characterizes each cytoplasmic condensate provides specificity for protein complex assembly, as only certain groups of proteins can become retained in the condensates.
HOW CAN SPECIFICITY BE ACCOMPLISHED IF RNA-BINDING PROTEINS HAVE THOUSANDS OF mRNA TARGETS?
In the early 2000s, Jack Keene proposed the RNA regulon model which states that groups of mRNAs are coregulated by RNA-binding proteins (Keene and Tenenbaum 2002; Keene 2007). Subsequent CLIP analyses revealed that a single RNA-binding protein often has thousands of RNA targets that are involved in diverse biological processes (Lee and Ule 2018; Van Nostrand et al. 2020). For example, HuR has 4874 mRNA targets (Lebedeva et al. 2011; Mukherjee et al. 2011) and regulates mRNA modifications, splicing, 3′ end cleavage and polyadenylation, mRNA nuclear export, stability, translation, and protein localization (Fan and Steitz 1998; Gallouzi et al. 2001; Dai et al. 2012; Tiedje et al. 2012; Berkovits and Mayr 2015; Oktaba et al. 2015; Chen et al. 2019). These data suggest that the universe of RNA targets of a specific RNA-binding protein contains a large number of submodules with functionally related mRNAs.
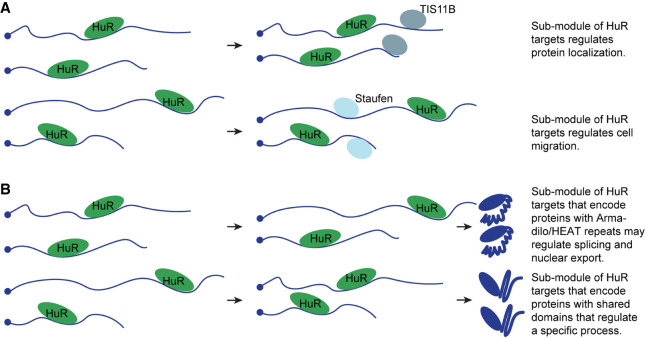
One way to identify functional submodules is to focus on mRNAs that are cobound by two or three RNA-binding proteins (Fig. 2A). RNA-binding proteins frequently bind to other RNA-binding proteins (Brannan et al. 2016) and it is well established that they often use cooperative interactions to regulate a specific process or function (Gregersen et al. 2014; Hennig et al. 2014; Hennig and Sattler 2015). For example, cobinding of HuR and TIS11B to mRNAs with AU-rich elements defines one of HuR's functions in the regulation of protein localization, whereas cobinding of HuR and Staufen to mRNAs was found to be important for the role of HuR in the regulation of cell migration (Ma and Mayr 2018; Lee and Mayr 2019).
FIGURE 2.
Functional submodules of RNA regulons. (A) We propose that cobinding of two or more RNA-binding proteins to groups of mRNAs define functionally related subgroups of mRNAs. A single RNA-binding protein, such as HuR, often has thousands of mRNA targets. The universe of mRNAs with HuR binding sites is shown on the left. Subgroups of these mRNAs are cobound by other RNA-binding proteins (for example TIS11B or Staufen; shown on the right), thus potentially representing groups of mRNA with shared functions, called functional submodules. (B) We propose that cooperative action between motifs in mRNAs and their encoded proteins defines functionally related groups of proteins. The universe of mRNAs with HuR binding sites is shown on the left. Groups of mRNAs with HuR binding sites may encode proteins containing shared domains, thus potentially reflecting functional submodules. Shown are HuR mRNA targets that encode proteins with Armadillo/HEAT repeats. These proteins may share functions such as the regulation of splicing and nuclear export. Another group of HuR mRNA targets that encode proteins with so far unknown shared motifs may regulate other functions known to be regulated by HuR.
In addition, we propose that functional submodules of RNA-binding proteins can be identified by co-occurrence of specific mRNA and protein motifs (Fig. 2B). To test this idea, we performed preliminary analyses and asked if certain protein motifs are enriched among the protein products of the mRNA targets of RNA-binding proteins. For example, we observed that HuR mRNA targets are significantly enriched in proteins with Armadillo/HEAT repeats (expected: 28, observed: 44, fold-enrichment: 1.6; χ-square, χ2 = 11.4; P = 0.0008). The enrichment was even stronger for the top 10% of targets with the highest number of HuR CLIP tags (expected: seven, observed: 19, fold-enrichment: 2.7; χ-square, χ2 = 22.2; P < 0.0001) (Mukherjee et al. 2011; Zhen G and Mayr C, unpubl.). Proteins with Armadillo/HEAT repeats are often involved in splicing, nuclear export/import, protein transport, and cell adherens junctions which represent functional categories that are over-represented among the HuR RNA targets.
Another example is the strong enrichment of zinc finger proteins among the mRNAs that contain CLIP tags for FUS (expected: 189, observed: 282, fold-enrichment: 1.5; χ-square, χ2 = 62.6; P < 0.0001). Importantly, the enrichment becomes stronger when focusing on the top 25% of targets with the highest number of FUS CLIP tags (expected: 45, observed: 83, fold-enrichment: 1.8; χ-square, χ2 = 36.6; P < 0.0001) (Hoell et al. 2011; Zhen G and Mayr C, unpubl.). These results make it possible that corresponding mRNA and protein motifs may have coevolved. Systematic analyses on such functional submodules that identify co-occurrence of RNA and protein motifs are currently lacking, but will likely increase our understanding of the regulatory logic of RNA-binding proteins.
TRANSIENT FORMATION VERSUS PERMANENT LOCALIZATION OF PROTEIN COMPLEXES IN CYTOPLASMIC CONDENSATES
Retention of proteins in cytoplasmic condensates may promote protein complex assembly by two mechanisms: (i) retention increases the local interactor concentration (Woodruff et al. 2017; King and Petry 2020) and (ii) repetitive binding to unoccupied binding sites increases the dwell time, which does not require a concentration increase of the interactor in the condensate to promote complex assembly. After having encountered the intended partner protein, their interaction should then mask the retention sites, thus allowing the protein complex to leave the condensates. In such cases, protein retention in cytoplasmic condensates would serve as a checkpoint for full assembly. This idea is again inspired by principles of protein complex assembly of multimeric surface receptors and channels in the ER (Bonifacino et al. 1990; Zerangue et al. 1999). The subunits of multimeric membrane complexes are translated at the ER. To ensure that only fully assembled channels or receptors leave the ER to be trafficked to the plasma membrane, the individual subunits contain ER retention sites (Nilsson et al. 1989; Cosson and Letourneur 1994). Upon full assembly, these sites are buried and are no longer accessible on the protein surface, thus allowing the multiprotein complex to leave the ER (Bonifacino et al. 1990; Zerangue et al. 1999). A similar mechanism was shown to take place in the nucleolus for ribosome assembly (Riback et al. 2020). Moreover, it could also take place in cytoplasmic condensates where protein motifs that interact with mRNA, RNA-binding proteins, or PTMs may serve as retention signals, thus only allowing fully assembled protein complexes to leave the condensates (Fig. 1F). RNA-binding proteins are usually not part of the assembled protein complexes (Berkovits and Mayr 2015; Lee and Mayr 2019), likely due to the fact that their high valency precludes their exit from the condensates.
A large fraction of protein complexes assemble in a cotranslational manner (Shiber et al. 2018; Kamenova et al. 2019; Panasenko et al. 2019). This means that a fully folded binding partner interacts with a polypeptide chain that is still being translated. Translation in cytoplasmic condensates where RNA-binding proteins act as matchmakers is an efficient way to bring the two subunits into proximity (Mayr 2018). The unfolded protein represents an mRNA target, whereas the fully folded protein is one of the protein targets. Alternatively, both subunits could be mRNA targets. In any case, transient retention of the fully folded subunit in the condensate is necessary for efficient cotranslational complex assembly.
Transient localization of proteins to cytoplasmic condensates for assembly and subsequent trafficking to their final destination has been observed for the chromatin factor Hmgn5 (Moretti et al. 2015). Hmgn5 mRNA is transported in a 3′UTR-dependent manner to the growth cones of dendrites. Neuronal activity changes the environment of the growth cones (Hornberg and Holt 2013; Sasaki 2020) and promotes the addition of specific PTMs to newly translated Hmgn5. Modified Hmgn5 protein traffics to the nucleus and regulates gene expression (Moretti et al. 2015). As only modified Hmgn5 is able to bind to chromatin, these results demonstrate a mechanism by which a change in the cellular environment leads to a change in gene expression. As a result, transient localization to cytoplasmic condensates allows proteins to take on new features such as protein interactors or PTMs to alter their function.
We hypothesize that some protein complexes may use cytoplasmic condensates as transient environments for complex assembly, whereas some signaling pathways may be set up within the cytoplasmic condensates (Peeples and Rosen 2021). For example, if all members of a signaling cascade contain condensate retention sites that are not being masked by complex assembly, then the cytoplasmic condensates may allow scaffolding or channeling of reactions (Yewdell et al. 2019). However, experimental evidence for this principle is currently lacking. So far, scaffolding of signaling pathways has only been observed in transiently generated, signaling-induced condensates that do not require mRNA or RNA-binding proteins to promote signaling reactions (Li et al. 2012; Su et al. 2016).
HOW TO TEST MATCHMAKING IN CYTOPLASMIC CONDENSATES?
RNA-binding proteins together with their bound mRNAs constitute the core components of the matchmaking process (Fig. 1A and F). When examining protein complex assembly in cis of individual complexes, it was shown that deletion of the matchmaking RNA-binding proteins prevented complex assembly, whereas tethering of the RNA-binding proteins to the 3′UTRs of the RNA targets restored it (Berkovits and Mayr 2015; Lee and Mayr 2019). In contrast, the requirement for RNA-binding proteins for complex assembly occurring in trans or as part of cytoplasmic condensates has not yet been shown. Therefore, it is currently often assumed that protein complex assembly in the cytoplasm occurs through diffusion and random encounter (Fig. 1G).
This viewpoint may change if we were able to visualize the proposed matchmakers in the future. Merfish or seqFISH+ could be used to directly detect groups of mRNAs within condensates (Chen et al. 2015; Eng et al. 2019). Moreover, depletion of condensate RNA-binding proteins followed by widespread disruption of protein complex assembly would provide indirect evidence for RNA-binding proteins serving as matchmakers for protein complex assembly. A more elegant experiment would be to identify the amino acids in RNA-binding proteins that are required for condensate assembly, followed by prime editing to mutate these residues in scaffold RNA-binding proteins to disrupt condensate assembly in cells (Anzalone et al. 2019). As transient retention of proteins in cytoplasmic condensates represents a collective property of the assembled RNA-binding protein, our model predicts that condensate disruption would impair protein complex assembly between the mRNA and protein targets of the RNA-binding protein. However, such experiments have not yet been reported.
It was previously reported that translation in TIS granules is required for establishing specific protein–protein interactions that cannot be formed when the proteins are translated outside of the condensate (Ma and Mayr 2018). In addition to TIS11B and FXR1, a large number of RNA-binding proteins localize predominantly to the cytoplasm (Thul et al. 2017; Caudron-Herger et al. 2021) and may assemble into additional cytoplasmic condensates. We envision the cytoplasm to be extensively compartmentalized by condensates and propose that condensate-dependent complex assembly is widespread.
ACKNOWLEDGMENTS
We thank all members of the Mayr laboratory for insightful discussions and Sibylle Mitschka for helpful comments on the manuscript. We thank Gang Zhen for the analysis of the CLIP data. This work was funded by the NIH Director′s Pioneer Award (DP1-GM123454), the Pershing Square Sohn Cancer Research Alliance, and the NCI Cancer Center Support Grant (P30 CA008748).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078995.121.
Freely available online through the RNA Open Access option.
REFERENCES
- Alberts B. 1998. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 92: 291–294. 10.1016/S0092-8674(00)80922-8 [DOI] [PubMed] [Google Scholar]
- Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, et al. 2019. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576: 149–157. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D, Huang AS. 1970. Interaction of HeLa cell proteins with RNA. J Mol Biol 47: 263–273. 10.1016/0022-2836(70)90301-3 [DOI] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. 2012. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell 46: 674–690. 10.1016/j.molcel.2012.05.021 [DOI] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK. 2016. Compositional control of phase-separated cellular bodies. Cell 166: 651–663. 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits BD, Mayr C. 2015. Alternative 3′UTRs act as scaffolds to regulate membrane protein localization. Nature 522: 363–367. 10.1038/nature14321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Sudhof TC. 2002. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science 297: 1525–1531. 10.1126/science.1072356 [DOI] [PubMed] [Google Scholar]
- Biever A, Glock C, Tushev G, Ciirdaeva E, Dalmay T, Langer JD, Schuman EM. 2020. Monosomes actively translate synaptic mRNAs in neuronal processes. Science 367: eaay4991. 10.1126/science.aay4991 [DOI] [PubMed] [Google Scholar]
- Boke E, Ruer M, Wuhr M, Coughlin M, Lemaitre R, Gygi SP, Alberti S, Drechsel D, Hyman AA, Mitchison TJ. 2016. Amyloid-like self-assembly of a cellular compartment. Cell 166: 637–650. 10.1016/j.cell.2016.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Cosson P, Klausner RD. 1990. Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell 63: 503–513. 10.1016/0092-8674(90)90447-M [DOI] [PubMed] [Google Scholar]
- Brannan KW, Jin W, Huelga SC, Banks CA, Gilmore JM, Florens L, Washburn MP, Van Nostrand EL, Pratt GA, Schwinn MK, et al. 2016. SONAR discovers RNA-binding proteins from analysis of large-scale protein–protein interactomes. Mol Cell 64: 282–293. 10.1016/j.molcel.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum AR, Haimovich G, Singer RH. 2015. In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol 16: 95–109. 10.1038/nrm3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral SE, Mowry KL. 2021. Multivalent interactions with RNA drive RNA binding protein recruitment and dynamics in biomolecular condensates in Xenopus oocytes. bioRxiv 10.1101/2021.06.21.449303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149: 1393–1406. 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- Caudron-Herger M, Rusin SF, Adamo ME, Seiler J, Schmid VK, Barreau E, Kettenbach AN, Diederichs S. 2019. R-DeeP: proteome-wide and quantitative identification of RNA-dependent proteins by density gradient ultracentrifugation. Mol Cell 75: 184–199.e10. 10.1016/j.molcel.2019.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron-Herger M, Jansen RE, Wassmer E, Diederichs S. 2021. RBP2GO: a comprehensive pan-species database on RNA-binding proteins, their interactions and functions. Nucleic Acids Res 49: D425–D436. 10.1093/nar/gkaa1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron JW, Hunt KC, Frydman J. 2016. Cotranslational signal-independent SRP preloading during membrane targeting. Nature 536: 224–228. 10.1038/nature19309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Damaj BB, Herrera C, Lasko P, Richard S. 1997. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: role of the KH domain. Mol Cell Biol 17: 5707–5718. 10.1128/MCB.17.10.5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. 2015. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348: aaa6090. 10.1126/science.aaa6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X, Chen RX, Wei WS, Liu Y, Gao CC, et al. 2019. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol 21: 978–990. 10.1038/s41556-019-0361-y [DOI] [PubMed] [Google Scholar]
- Chi YH, Semmes OJ, Jeang KT. 2011. A proteomic study of TAR-RNA binding protein (TRBP)-associated factors. Cell Biosci 1: 9. 10.1186/2045-3701-1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouaib R, Safieddine A, Pichon X, Imbert A, Kwon OS, Samacoits A, Traboulsi AM, Robert MC, Tsanov N, Coleno E, et al. 2020. A dual protein-mRNA localization screen reveals compartmentalized translation and widespread co-translational RNA targeting. Dev Cell 54: 773–791.e75. 10.1016/j.devcel.2020.07.010 [DOI] [PubMed] [Google Scholar]
- Clery A, Blatter M, Allain FH. 2008. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol 18: 290–298. 10.1016/j.sbi.2008.04.002 [DOI] [PubMed] [Google Scholar]
- Cosson P, Letourneur F. 1994. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science 263: 1629–1631. 10.1126/science.8128252 [DOI] [PubMed] [Google Scholar]
- Courchaine EM, Barentine AES, Straube K, Lee DR, Bewersdorf J, Neugebauer KM. 2021. DMA–tudor interaction modules control the specificity of in vivo condensates. Cell 184: 3612–3625.e17. 10.1016/j.cell.2021.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Zhang G, Makeyev EV. 2012. RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucleic Acids Res 40: 787–800. 10.1093/nar/gkr783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienstbier M, Boehl F, Li X, Bullock SL. 2009. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev 23: 1546–1558. 10.1101/gad.531009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng CL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun J, Cronin C, Karp C, Yuan GC, et al. 2019. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568: 235–239. 10.1038/s41586-019-1049-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, Blackshear PJ, Nagar B, Sonenberg N. 2013. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol 20: 735–739. 10.1038/nsmb.2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan XC, Steitz JA. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J 17: 3448–3460. 10.1093/emboj/17.12.3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes N, Buchan JR. 2020. RPS28B mRNA acts as a scaffold promoting cis-translational interaction of proteins driving P-body assembly. Nucleic Acids Res 48: 6265–6279. 10.1093/nar/gkaa352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes N, Buchan JR. 2021. RNAs as regulators of cellular matchmaking. Front Mol Biosci 8: 634146. 10.3389/fmolb.2021.634146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest KM, Gavis ER. 2003. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol 13: 1159–1168. 10.1016/S0960-9822(03)00451-2 [DOI] [PubMed] [Google Scholar]
- Gadir N, Haim-Vilmovsky L, Kraut-Cohen J, Gerst JE. 2011. Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA 17: 1551–1565. 10.1261/rna.2621111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Brennan CM, Steitz JA. 2001. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA 7: 1348–1361. 10.1017/S1355838201016089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mayoral MF, Hollingworth D, Masino L, Diaz-Moreno I, Kelly G, Gherzi R, Chou CF, Chen CY, Ramos A. 2007. The structure of the C-terminal KH domains of KSRP reveals a noncanonical motif important for mRNA degradation. Structure 15: 485–498. 10.1016/j.str.2007.03.006 [DOI] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15: 829–845. 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glock C, Heumuller M, Schuman EM. 2017. mRNA transport & local translation in neurons. Curr Opin Neurobiol 45: 169–177. 10.1016/j.conb.2017.05.005 [DOI] [PubMed] [Google Scholar]
- Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M. 2014. MOV10 is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol Cell 54: 573–585. 10.1016/j.molcel.2014.03.017 [DOI] [PubMed] [Google Scholar]
- Guillen-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlussler R, Kim K, Trussina I, Wang J, Mateju D, et al. 2020. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181: 346–361.e17. 10.1016/j.cell.2020.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner AS, Donlin-Asp PG, Leitch B, Herzog E, Schuman EM. 2019. Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments. Science 364: eaau3644. 10.1126/science.aau3644 [DOI] [PubMed] [Google Scholar]
- Hennig J, Sattler M. 2015. Deciphering the protein–RNA recognition code: combining large-scale quantitative methods with structural biology. Bioessays 37: 899–908. 10.1002/bies.201500033 [DOI] [PubMed] [Google Scholar]
- Hennig J, Militti C, Popowicz GM, Wang I, Sonntag M, Geerlof A, Gabel F, Gebauer F, Sattler M. 2014. Structural basis for the assembly of the Sxl–Unr translation regulatory complex. Nature 515: 287–290. 10.1038/nature13693 [DOI] [PubMed] [Google Scholar]
- Hoell JI, Larsson E, Runge S, Nusbaum JD, Duggimpudi S, Farazi TA, Hafner M, Borkhardt A, Sander C, Tuschl T. 2011. RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol 18: 1428–1431. 10.1038/nsmb.2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofweber M, Dormann D. 2019. Friend or foe—post-translational modifications as regulators of phase separation and RNP granule dynamics. J Biol Chem 294: 7137–7150. 10.1074/jbc.TM118.001189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Martin KC, Schuman EM. 2019. Local translation in neurons: visualization and function. Nat Struct Mol Biol 26: 557–566. 10.1038/s41594-019-0263-5 [DOI] [PubMed] [Google Scholar]
- Hornberg H, Holt C. 2013. RNA-binding proteins and translational regulation in axons and growth cones. Front Neurosci 7: 81. 10.3389/fnins.2013.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A, Courel M, Benard M, Souquere S, Ernoult-Lange M, Chouaib R, Yi Z, Morlot JB, Munier A, Fradet M, et al. 2017. P-body purification reveals the condensation of repressed mRNA regulons. Mol Cell 68: 144–157 e145. 10.1016/j.molcel.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. 2004. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 11: 257–264. 10.1038/nsmb738 [DOI] [PubMed] [Google Scholar]
- Kamenova I, Mukherjee P, Conic S, Mueller F, El-Saafin F, Bardot P, Garnier JM, Dembele D, Capponi S, Timmers HTM, et al. 2019. Co-translational assembly of mammalian nuclear multisubunit complexes. Nat Commun 10: 1740. 10.1038/s41467-019-09749-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. 2007. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8: 533–543. 10.1038/nrg2111 [DOI] [PubMed] [Google Scholar]
- Keene JD, Tenenbaum SA. 2002. Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell 9: 1161–1167. 10.1016/S1097-2765(02)00559-2 [DOI] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R. 2017. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell 68: 808–820.e5. 10.1016/j.molcel.2017.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Hahm B, Kim YK, Choi M, Jang SK. 2000. Protein–protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J Mol Biol 298: 395–405. 10.1006/jmbi.2000.3687 [DOI] [PubMed] [Google Scholar]
- King MR, Petry S. 2020. Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat Commun 11: 270. 10.1038/s41467-019-14087-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm JD, Schreiber SL, Crabtree GR. 1998. Dimerization as a regulatory mechanism in signal transduction. Annu Rev Immunol 16: 569–592. 10.1146/annurev.immunol.16.1.569 [DOI] [PubMed] [Google Scholar]
- Klinge S, Woolford JL Jr. 2019. Ribosome assembly coming into focus. Nat Rev Mol Cell Biol 20: 116–131. 10.1038/s41580-018-0078-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyan J, Eisenberg D. 2007. The origin of protein interactions and allostery in colocalization. Nature 450: 983–990. 10.1038/nature06524 [DOI] [PubMed] [Google Scholar]
- Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. 2021. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol 22: 165–182. 10.1038/s41580-020-0272-6 [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, de The H. 2010. PML nuclear bodies. Cold Spring Harb Perspect Biol 2: a000661. 10.1101/cshperspect.a000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann CA, Gerbich TM, Smith JA, Crutchley JM, Termini CM, Weeks KM, et al. 2018. mRNA structure determines specificity of a polyQ-driven phase separation. Science 360: 922–927. 10.1126/science.aar7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ. 2017. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547: 236–240. 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH. 1986. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45: 407–415. 10.1016/0092-8674(86)90326-0 [DOI] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. 2011. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell 43: 340–352. 10.1016/j.molcel.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Lee SH, Mayr C. 2019. Gain of additional BIRC3 protein functions through 3′-UTR-mediated protein complex formation. Mol Cell 74: 701–712.e9. 10.1016/j.molcel.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FCY, Ule J. 2018. Advances in CLIP technologies for studies of protein–RNA interactions. Mol Cell 69: 354–369. 10.1016/j.molcel.2018.01.005 [DOI] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. 2012. Phase transitions in the assembly of multivalent signalling proteins. Nature 483: 336–340. 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Peng Q, Wan X, Sun H, Tang J. 2017. C-terminal motifs in promyelocytic leukemia protein isoforms critically regulate PML nuclear body formation. J Cell Sci 130: 3496–3506. 10.1242/jcs.202879 [DOI] [PubMed] [Google Scholar]
- Lui J, Castelli LM, Pizzinga M, Simpson CE, Hoyle NP, Bailey KL, Campbell SG, Ashe MP. 2014. Granules harboring translationally active mRNAs provide a platform for P-body formation following stress. Cell Rep 9: 944–954. 10.1016/j.celrep.2014.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. 2007. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 8: 479–490. 10.1038/nrm2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Mayr C. 2018. A membraneless organelle associated with the endoplasmic reticulum enables 3′UTR-mediated protein–protein interactions. Cell 175: 1492–1506.e19. 10.1016/j.cell.2018.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Zheng G, Xie W, Mayr C. 2021. In vivo reconstitution finds multivalent RNA–RNA interactions as drivers of mesh-like condensates. Elife 10: e64252. 10.7554/eLife.64252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateju D, Eichenberger B, Voigt F, Eglinger J, Roth G, Chao JA. 2020. Single-molecule imaging reveals translation of mRNAs localized to stress granules. Cell 183: 1801–1812.e13. 10.1016/j.cell.2020.11.010 [DOI] [PubMed] [Google Scholar]
- Mayr C. 2018. Protein complexes assemble as they are being made. Nature 561: 186–187. 10.1038/d41586-018-05905-4 [DOI] [PubMed] [Google Scholar]
- Morales-Polanco F, Bates C, Lui J, Casson J, Solari CA, Pizzinga M, Forte G, Griffin C, Garner KEL, Burt HE, et al. 2021. Core fermentation (CoFe) granules focus coordinated glycolytic mRNA localization and translation to fuel glucose fermentation. iScience 24: 102069. 10.1016/j.isci.2021.102069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti F, Rolando C, Winker M, Ivanek R, Rodriguez J, Von Kriegsheim A, Taylor V, Bustin M, Pertz O. 2015. Growth cone localization of the mRNA encoding the chromatin regulator HMGN5 modulates neurite outgrowth. Mol Cell Biol 35: 2035–2050. 10.1128/MCB.00133-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morf J, Wingett SW, Farabella I, Cairns J, Furlan-Magaril M, Jimenez-Garcia LF, Liu X, Craig FF, Walker S, Segonds-Pichon A, et al. 2019. RNA proximity sequencing reveals the spatial organization of the transcriptome in the nucleus. Nat Biotechnol 37: 793–802. 10.1038/s41587-019-0166-3 [DOI] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M Jr, Tuschl T, Ohler U, Keene JD. 2011. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell 43: 327–339. 10.1016/j.molcel.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair RR, Zabezhinsky D, Gelin-Licht R, Haas BJ, Dyhr MC, Sperber HS, Nusbaum C, Gerst JE. 2021. Multiplexed mRNA assembly into ribonucleoprotein particles plays an operon-like role in the control of yeast cell physiology. Elife 10: e66050. 10.7554/eLife.66050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil CR, Jeschonek SP, Cabral SE, O'Connell LC, Powrie EA, Otis JP, Wood TR, Mowry KL. 2021. L-bodies are RNA-protein condensates driving RNA localization in Xenopus oocytes. Mol Biol Cell mbcE21030146T. 10.1091/mbc.E21-03-0146-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Jackson M, Peterson PA. 1989. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell 58: 707–718. 10.1016/0092-8674(89)90105-0 [DOI] [PubMed] [Google Scholar]
- Oktaba K, Zhang W, Lotz TS, Jun DJ, Lemke SB, Ng SP, Esposito E, Levine M, Hilgers V. 2015. ELAV links paused Pol II to alternative polyadenylation in the Drosophila nervous system. Mol Cell 57: 341–348. 10.1016/j.molcel.2014.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabis M, Popowicz GM, Stehle R, Fernandez-Ramos D, Asami S, Warner L, Garcia-Maurino SM, Schlundt A, Martinez-Chantar ML, Diaz-Moreno I, et al. 2019. HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs. Nucleic Acids Res 47: 1011–1029. 10.1093/nar/gky1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasenko OO, Somasekharan SP, Villanyi Z, Zagatti M, Bezrukov F, Rashpa R, Cornut J, Iqbal J, Longis M, Carl SH, et al. 2019. Co-translational assembly of proteasome subunits in NOT1-containing assemblysomes. Nat Struct Mol Biol 26: 110–120. 10.1038/s41594-018-0179-5 [DOI] [PubMed] [Google Scholar]
- Peeples W, Rosen MK. 2021. Mechanistic dissection of increased enzymatic rate in a phase-separated compartment. Nat Chem Biol 17: 693–702. 10.1038/s41589-021-00801-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzinga M, Bates C, Lui J, Forte G, Morales-Polanco F, Linney E, Knotkova B, Wilson B, Solari CA, Berchowitz LE, et al. 2019. Translation factor mRNA granules direct protein synthetic capacity to regions of polarized growth. J Cell Biol 218: 1564–1581. 10.1083/jcb.201704019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman DS, Dreyfuss G. 1994. RNA annealing activities in HeLa nuclei. EMBO J 13: 213–221. 10.1002/j.1460-2075.1994.tb06251.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Hollingworth D, Major SA, Adinolfi S, Kelly G, Muskett FW, Pastore A. 2002. Role of dimerization in KH/RNA complexes: the example of Nova KH3. Biochemistry 41: 4193–4201. 10.1021/bi011994o [DOI] [PubMed] [Google Scholar]
- Ramos A, Hollingworth D, Adinolfi S, Castets M, Kelly G, Frenkiel TA, Bardoni B, Pastore A. 2006. The structure of the N-terminal domain of the fragile X mental retardation protein: a platform for protein–protein interaction. Structure 14: 21–31. 10.1016/j.str.2005.09.018 [DOI] [PubMed] [Google Scholar]
- Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, Wei MT, Kriwacki RW, Brangwynne CP. 2020. Composition-dependent thermodynamics of intracellular phase separation. Nature 581: 209–214. 10.1038/s41586-020-2256-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers EF, Kaaij LJT, Redl S, Bronkhorst AW, Wiebrands K, de Jesus Domingues AM, Huang HY, Han CT, Riemer S, Dosch R, et al. 2018. Tdrd6a regulates the aggregation of Buc into functional subcellular compartments that drive germ cell specification. Dev Cell 46: 285–301.e89. 10.1016/j.devcel.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, Lemoine S, Tanty V, Devaux F, Jacq C. 2008. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS ONE 3: e2293. 10.1371/journal.pone.0002293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y. 2020. Local translation in growth cones and presynapses, two axonal compartments for local neuronal functions. Biomolecules 10: 668. 10.3390/biom10050668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiber A, Doring K, Friedrich U, Klann K, Merker D, Zedan M, Tippmann F, Kramer G, Bukau B. 2018. Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 561: 268–272. 10.1038/s41586-018-0462-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh YW, Minguez P, Bork P, Auburger JJ, Guilbride DL, Kramer G, Bukau B. 2015. Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science 350: 678–680. 10.1126/science.aac8171 [DOI] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP. 2017. Liquid phase condensation in cell physiology and disease. Science 357: eaaf4382. 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- Smith JA, Curry EG, Blue RE, Roden C, Dundon SER, Rodriguez-Vargas A, Jordan DC, Chen X, Lyons SM, Crutchley J, et al. 2020. FXR1 splicing is important for muscle development and biomolecular condensates in muscle cells. J Cell Biol 219: e201911129. 10.1083/jcb.201911129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD. 2016. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352: 595–599. 10.1126/science.aad9964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre J, Vialette S, Corral Debrinski M, Jacq C. 2003. Long mRNAs coding for yeast mitochondrial proteins of prokaryotic origin preferentially localize to the vicinity of mitochondria. Genome Biol 4: R44. 10.1186/gb-2003-4-7-r44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzer S, Moro A, Kuharev J, Francis AC, Vidalino L, Provenzani A, Macchi P. 2013. Proteome-wide characterization of the RNA-binding protein RALY-interactome using the in vivo-biotinylation-pulldown-quant (iBioPQ) approach. J Proteome Res 12: 2869–2884. 10.1021/pr400193j [DOI] [PubMed] [Google Scholar]
- Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Bjork L, Breckels LM, et al. 2017. A subcellular map of the human proteome. Science 356: eaal3321. 10.1126/science.aal3321 [DOI] [PubMed] [Google Scholar]
- Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, Kotlyarov A, Gaestel M. 2012. The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS Genet 8: e1002977. 10.1371/journal.pgen.1002977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripsianes K, Madl T, Machyna M, Fessas D, Englbrecht C, Fischer U, Neugebauer KM, Sattler M. 2011. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat Struct Mol Biol 18: 1414–1420. 10.1038/nsmb.2185 [DOI] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Yee BA, Wheeler EC, Blue SM, Mueller J, Park SS, Garcia KE, Gelboin-Burkhart C, Nguyen TB, et al. 2020. Principles of RNA processing from analysis of enhanced CLIP maps for 150 RNA binding proteins. Genome Biol 21: 90. 10.1186/s13059-020-01982-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt F, Zhang H, Cui XA, Triebold D, Liu AX, Eglinger J, Lee ES, Chao JA, Palazzo AF. 2017. Single-molecule quantification of translation-dependent association of mRNAs with the endoplasmic reticulum. Cell Rep 21: 3740–3753. 10.1016/j.celrep.2017.12.008 [DOI] [PubMed] [Google Scholar]
- Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. 2018. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174: 688–699.16. 10.1016/j.cell.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA. 2017. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169: 1066–1077.e10. 10.1016/j.cell.2017.05.028 [DOI] [PubMed] [Google Scholar]
- Wu SK, Roberts JT, Balas MM, Johnson AM. 2020. RNA matchmaking in chromatin regulation. Biochem Soc Trans 48: 2467–2481. 10.1042/BST20191225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Hoek TA, Vale RD, Tanenbaum ME. 2016. Dynamics of translation of single mRNA molecules in vivo. Cell 165: 976–989. 10.1016/j.cell.2016.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Sun L, Li J, Yu H, Lin H, Yu T, Zhao F, Zhu M, Liu L, Geng Q, et al. 2019. RNA-binding protein KHSRP promotes tumor growth and metastasis in non-small cell lung cancer. J Exp Clin Cancer Res 38: 478. 10.1186/s13046-019-1479-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Mathieu C, Kolaitis RM, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q, et al. 2020. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181: 325–345.e28. 10.1016/j.cell.2020.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell JW, Dersh D, Fahraeus R. 2019. Peptide channeling: the key to MHC class I immunosurveillance? Trends Cell Biol 29: 929–939. 10.1016/j.tcb.2019.09.004 [DOI] [PubMed] [Google Scholar]
- Zerangue N, Schwappach B, Jan YN, Jan LY. 1999. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron 22: 537–548. 10.1016/S0896-6273(00)80708-4 [DOI] [PubMed] [Google Scholar]
- Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. 2015. RNA controls polyQ protein phase transitions. Mol Cell 60: 220–230. 10.1016/j.molcel.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Muller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95: 2748–2752. 10.1182/blood.V95.9.2748.009k31a_2748_2752 [DOI] [PubMed] [Google Scholar]