Abstract
Exciting recent work has highlighted that numerous cellular compartments lack encapsulating lipid bilayers (often called “membraneless organelles”), and that their structure and function are central to the regulation of key biological processes, including transcription, RNA splicing, translation, and more. These structures have been described as “biomolecular condensates” to underscore that biomolecules can be significantly concentrated in them. Many condensates, including RNA granules and processing bodies, are enriched in proteins and nucleic acids. Biomolecular condensates exhibit a range of material states from liquid- to gel-like, with the physical process of liquid–liquid phase separation implicated in driving or contributing to their formation. To date, in vitro studies of phase separation have provided mechanistic insights into the formation and function of condensates. However, the link between the often micron-sized in vitro condensates with nanometer-sized cellular correlates has not been well established. Consequently, questions have arisen as to whether cellular structures below the optical resolution limit can be considered biomolecular condensates. Similarly, the distinction between condensates and discrete dynamic hub complexes is debated. Here we discuss the key features that define biomolecular condensates to help understand behaviors of structures containing and generating RNA.
Keywords: phase separation, dynamic hub, ribonucleoprotein particle, simulation
INTRODUCTION: KEY DISTINGUISHING FEATURES OF BIOMOLECULAR CONDENSATES
The ability of polymers to self-organize into discrete liquid-like droplets (de-mix) from a solvent solution has been readily observed and extensively studied in polymer chemistry for decades (Flory 1941). This phenomenon, called liquid–liquid phase separation (LLPS), occurs when dynamic multivalent interactions between components create an exchanging network in which the sum of interactions overcomes the entropic free energy that favors a fully mixed solution. As a result, a phase that is rich in these interacting components (here termed a condensate) segregates from its surroundings with the formation of a physical boundary defined by surface tension (Brangwynne et al. 2009; Li et al. 2012; Banani et al. 2017). Cells contain significant concentrations of biopolymers (e.g., proteins, nucleic acids, lipids, carbohydrates), and accordingly numerous condensates have been shown to form, including intracellular membraneless organelles, membranes, lipid rafts, extracellular biomaterials, and bacterial biofilms, to name a few (Heberle and Feigenson 2011; Muiznieks et al. 2018; Bratek-Skicki et al. 2020; Seviour et al. 2020). The physics of in vitro LLPS has been found to reasonably mimic the manner in which a number of biomolecular condensates assemble and disassemble in cells (Riback et al. 2020; Shimobayashi et al. 2021). There is increasing evidence that biomolecular condensates play significant roles in regulating central enzymatic functions, such as transcription, RNA processing, and translation (Cho et al. 2018; Lu et al. 2018; Sabari et al. 2018; Guo et al. 2019). Because of their importance in RNA biology, here we focus on biomolecular condensates containing RNA and protein, such as “transcriptional condensates” including RNA polymerase II (RNA Pol II) and enhancer proteins that activate transcription, nuclear speckles and Cajal bodies involved in splicing, and germ granules, processing bodies (P-bodies), stress granules, and mRNA transport granules that regulate cytoplasmic RNA processing and translation.
Biomolecular condensates are non-membrane-delimited biological structures that have a physically defined boundary generated by surface tension (via LLPS or otherwise) creating distinct inside versus outside environments and that are dynamic and can be reversibly assembled and disassembled. The insides of condensates are chemically distinct microenvironments that are densely concentrated with certain protein and RNA molecules that together solvate and enrich for specific sets of biomolecules, while excluding others (Jain et al. 2016; Su et al. 2016; Woodruff et al. 2017; Langdon et al. 2018). The inside condensate environments often contain lower water concentrations than the outside, as measured within simple in vitro systems. While this may also be the case for biomolecular condensates in cells, it is not necessarily the case; selective concentration of certain biomolecules inside could leave similar overall biomolecular concentrations outside, consistent with the overall 70% water by mass found in cells (i.e., the cell is highly concentrated in biomolecules). Condensates exhibit fluid dynamic material properties like liquids or gels, and are not solid or irreversible structures. They have been described as tunable viscoelastic fluids, varying from low to high viscosity depending on the protein and RNA composition, sequence and structure, as well as conditions (Alshareedah et al. 2021; Zhou 2021). Biomolecules inside and outside the condensate are usually able to exchange across the boundary, with exceptions involving interfacial physical forces or other factors that inhibit this, as observed in some gel-like condensates (Brady et al. 2017) and those with a nondynamic shell (Schmidt et al. 2021). Liquid- and gel-like (or complex fluid) material properties of these condensates are supported by dynamically exchanging multivalent interactions, including between folded protein domains, intrinsically disordered protein regions (IDRs) and nucleic acids. On the other hand, stable interactions that do not exchange with similar partners in the context of multivalency do not give rise to a fluid network of interactions, but form either defined complexes with unique partners for each interface, such as a ribosome (also see next section), or static large-scale associated states or aggregates, such as tau aggregates. Increasing evidence suggests that tuning of interaction strengths can shift the properties of initially fluid-like condensates to more stable structures, including solid aggregates or fibrous states. This transition has been observed with many proteins that form insoluble protein aggregates in neurodegenerative diseases (Lin et al. 2015; Patel et al. 2015; Wegmann et al. 2018; Hardenberg et al. 2021).
Condensates can exhibit various sizes, simple spherical shapes or complex multiphase or multilayer architectures with nonspherical topologies (Feric et al. 2016; Jain et al. 2016; Kim et al. 2019; Marmor-Kollet et al. 2020; Sanders et al. 2020; Schmidt et al. 2021). In addition, they can be rapidly assembled and disassembled in seconds to minutes, or persist over many hours to days (nuclear pore, extracellular matrix components). Despite the many possible variations, there are key features that distinguish condensates from other cellular assemblies, such as hubs of interacting proteins: the presence of a unique fluid “solvent environment” inside created from a mixture of biomolecules and water, which concentrates or excludes certain molecules inside, with these specific components relatively depleted or enriched, respectively, outside, and with the inside/outside boundary defined by surface tension. Here we use “fluid inside” as a short reference to these distinguishing features.
WHAT CAN WE EASILY RULE OUT AS A CONDENSATE AND WHAT IS DEBATABLE?
Based on these condensate-defining features, it is crucial to consider the roles of internal dynamics (whether it is fluid) and of component mass and size (whether it is large enough to support an inside–outside regime) in determining whether something can be called a condensate, particularly in relation to how many molecules of protein or RNA are required. Many types of complexes arising from protein interactions have been described, including (i) molecular machines and supramolecular complexes, which are stably folded multisubunit structures (primarily enzymes) composed of protein and RNA or only protein (e.g., ribosome, proteasome, mitochondrial complex 1) (Yusupov et al. 2001; Leggett et al. 2002; Zickermann et al. 2015), (ii) stable homo- and hetero-oligomeric complexes (e.g., for homo-oligomers, insulin, hemerythrin) (Schlichtkrull 1956; Sieker et al. 1981), and (iii) discrete dynamic complexes involving intrinsically disordered protein regions (e.g., Fbw7/Jun, prothymosin α/histone H1α) (Csizmok et al. 2018; Feng et al. 2018). While stable complexes can often have aqueous internal cavities and have significant functional dynamic modes, the proteins (or nucleic acids) themselves are not part of the fluid solvent, making a clear distinction between these assemblies and condensates. Furthermore, discrete dynamic complexes that have multivalent interaction sites on only one component of a heteromeric complex cannot lead to large-scale networks. On the other hand, complexes that have multivalent sites on two or more components can, in principle, support the formation of large-scale networks depending on the balance of solvation/association energetics. In the cases where the energetics for condensate formation are not favorable, however, multivalent components can form discretely defined oligomers (i.e., dimers, trimers, n-mers with n being a defined integer) (e.g., the prothymosin α/histone H1α dimer [Borgia et al. 2018]). Thus, stable, folded complexes can be ruled out, but dynamic complexes are more challenging.
Proteins containing multiple interaction sites are often critical for forming either stable scaffolds (with unique interfaces that do not exchange), or “dynamic hubs” (defined below). The stable scaffold alone can be ruled out as a condensate, although scaffolds and other stable complexes may be incorporated into condensates through bridging interactions. However, it is less clear what discriminates between a dynamic hub and a condensate. The phrase “dynamic hub” has been used for complexes with exchanging multivalent interactions that are considered to be smaller than what is required for a condensate, including both discrete and larger-scale associated dynamic states (Chong et al. 2018; Mir et al. 2018). If the dynamic hub is a larger-scale (i.e., a structure with an undetermined but large number, n, of components and no defined upper size limit), this may merge with the definition of a condensate and raises questions relating to how small a “true” condensate can be. If the hub is discrete, with one or a defined number of molecules of a particular component, this raises questions as to whether any discrete defined oligomer state of a dynamic complex can be considered a condensate, or whether a condensate must be composed of n units, with n large and not specifically definable. One approach to answering these questions from a theoretical perspective is in terms of density fluctuations. Thus, the distinctions could be stated as the inside of a condensate should be liquid/fluid-like with a significant component of the biomolecules “solvating” themselves or other biomolecules, while a dynamic hub would be more gas-like, referring to the spatially sparse distribution of biomolecules and the large fluctuations of their densities at a given spatial position, with greater water solvation. On the other hand, the nondynamic, stable complexes would be solid.
A biological perspective suggests that the distinction between a condensate and dynamic hub lies in whether its function requires the generation of a unique internal environment (condensate) or whether it functions optimally in the surrounding aqueous solvent without requiring the inside vs. outside distinction found in condensates (dynamic hub). To reiterate, any state having a physically defined boundary leading to an inside and outside, with the inside being a fluid solvent that is rich in certain proteins and/or RNAs and the outside depleted in these specific components, can fulfill the definition of a condensate, while states lacking these do not. We propose that “dynamic hubs” may be a valuable term for complexes that function without forming a compartment having a fluid inside.
RNA AND RIBONUCLEOPROTEIN PARTICLES IN CONDENSATES
RNA-containing structures pose a valuable focus for asking these questions about the nature of condensates, due to the large sizes of many pre- and mRNA molecules and their frequent association with self and with proteins (both folded domains and IDRs). From the moment it is transcribed, an RNA is decorated with various RNA-binding proteins (RBPs) to form a ribonucleoprotein particle (RNP). An RNP travels through different cellular locations during its lifetime: from transcription sites, often to nuclear speckles for splicing, through the nuclear pore, and in certain cells into cytoplasmic granules, including transport granules, P-bodies for degradation, and stress granules under stress conditions (McKnight 2019). All of these cellular locations have been described as biomolecular condensates (Patel et al. 2015; Schmidt and Gorlich 2016; Cho et al. 2018; Lu et al. 2018; Sabari et al. 2018; Guo et al. 2019; Kim et al. 2019; Sachdev et al. 2019; Tsang et al. 2019; Liao and Regev 2021).
Some of the cellular bodies or structures with which RNA have been associated, including transcription, splicing, nuclear transport, and mRNA transport condensates, range in size from 50 to 500 nm in diameter. One example is mRNA transport granules, a type of RNP prevalent in neurons. These “granules,” which are ∼100 to 250 nm in diameter, may contain a single or very few mRNA molecules with some associated proteins, and transport the mRNAs from the soma along axons or dendrites to the synapse to regulate activity-dependent local translation in neurons (Batish et al. 2012). These compartments enrich certain RNA-binding proteins, which are dynamically exchanging by live-cell imaging, but not others (Gopal et al. 2017). These constituents, including FMRP, CAPRIN1, and TDP-43, dynamically exchange in and out of the granules and have all been shown to phase separate in vitro (Conicella et al. 2016; Kim et al. 2019; Tsang et al. 2019). These proteins are also components of other micron-sized biomolecular condensates, such as stress granules and P-bodies. Another example is paraspeckles, nuclear bodies with a lower size estimates of ∼50 nm that appear to regulate gene expression by sequestrating certain proteins or mRNAs (Fox et al. 2002; Prasanth et al. 2005; Clemson et al. 2009). Specifically, paraspeckles are composed of the long noncoding RNA NEAT1 and certain proteins such as FUS, NONO, and SFPQ that dynamically exchange with the nucleoplasm. Advanced microscopic imaging of RNA Pol II-enhancer clusters at transcriptional activation sites suggests that these also possess characteristic features of condensates, such as fusion and internal dynamics, while being as small as ∼100 nm in diameter (Cho et al. 2018). These distinct condensate-like characteristics of submicron-sized cellular structures raise a number of questions regarding the lower size limit of condensates. Does an RNP have to reach a “critical” size to achieve an inside–outside condensate distinction or can a single RNA molecule in the nucleus or cytoplasm that interacts with a limited number of protein molecules be considered a condensate?
WHAT ARE THE SIZE REQUIREMENTS OF A CONDENSATE AND HOW MANY MOLECULES ARE NEEDED?
Work from the Pomès group (Rauscher and Pomès 2017) demonstrated, via molecular dynamics simulations, that 27 copies of a 35-residue disordered elastin-like peptide (ELP) could self-associate in an aqueous solvent to create liquid condensates with clear differences between their inside and outside. The condensate had 40% water content by mass, in agreement with experimental in vitro measurements for elastin of 40%–60% (Chalmers et al. 1999). A plateau in water density representing a homogeneous “inside” indicates that the resulting structure is large enough to exhibit bulk-like properties of a separate liquid phase. Indeed, they observed quantifiable differences in the peptides inside and outside of the condensates. The number of water molecules contacting each peptide residue in the condensate was fewer than half those contacting a monomer freely diffusing in the dilute solution. Peptides had a significantly larger average radius of gyration (Rg) in condensates, consistent with less hydrogen-bonded turn formation and reduced contact between amino- and carboxyl termini. Together, this ELP model system clearly describes a state with a distinct inside and outside “solvent” environment that can impact conformational equilibria. We use these results to suggest a lower bound on what size is needed to create such distinct environments in the rest of this perspective, recognizing the potential limitations of protein molecular dynamics to model cellular phenomena as well as their value in providing a concrete molecular description and estimate of the size that can generate such condensate properties. So how big/small is it? While not exactly spherical, the distance from the center of mass of the ELP condensates to the inflection point of peptide and water density is ∼30 Å, leading to a diameter of ∼60 Å, or 6 nm (Fig. 1).
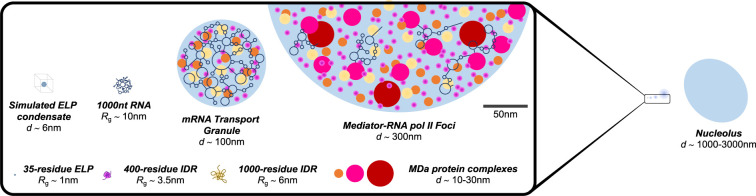
FIGURE 1.
Scales of biomolecular condensates and component biomolecules. Left (expanded view). (Top row from left to right) Elastin-like peptide (ELP) condensates as characterized by molecular dynamics simulations to be ∼6 nm in diameter (Rauscher and Pomès 2017); single mRNA molecule (1 kb or 1000 nt), ∼10 nm radius of gyration (Rg) (Yoffe et al. 2008; Gopal et al. 2012; Borodavka et al. 2016); RNP condensate, such as of a mRNA transport granule, composed of a single mRNA with some RBPs, ∼100 nm diameter (Batish et al. 2012); mediator-RNA Pol II foci, containing growing RNA chains and multiple components of the transcriptional machinery, ∼300 nm in diameter (Cho et al. 2018). (Bottom row from left to right) 35-residue elastin-like peptide (ELP), Rg ∼ 1 nm; 400-residue intrinsically disordered protein region (IDR), Rg ∼ 3.5 nm; 1000-residue IDR, Rg ∼ 6 nm; megadalton (MDa) protein complexes, ∼10 to 30 nm diameters (depending on geometry). (Right) Comparison of the scales of the condensates on the top row to the nucleolus, ∼1000 to 3000 nm in diameter.
More recent coarse-grained simulations of biomolecular condensates (Dignon et al. 2018; Wessén et al. 2021) also demonstrate a distinction between inside and outside for droplet sizes that are comparable to the ELP condensates simulated using all-atom and explicit-water molecular dynamics (Rauscher and Pomès 2017). In one of these simulations, 100 chains of 50-residue IDRs with mixed-charged sequence created a droplet of about 15 nm in diameter (Wessén et al. 2021). The system was used to explore the impact of the condensed environment on the dielectric constant, a measure of how a fluid or another material screens electrostatic interactions, which has significant impact on biomolecular interactions and catalysis in biology. When contributions from the charged sequences are included, the dielectric constant experienced by a molecule partitioned inside the droplet was found to be significantly higher than that of the bulk solvent (Wessén et al. 2021). The presence of the protein inside the simulated condensate thus creates a different solvent environment within the physical boundary defined by the surface of the droplet. This result provides additional support for the idea that even a nanoscale condensate environment can have significant biochemical impact.
Given that this nanometer size bound is small, a question arises about whether a condensate could be formed from a single large polymer. The term “condensate” is usually considered to imply a phase transition involving multiple molecules rather than a conformational state of a single chain, and the concentration of molecules needed to drive the phase transition under certain conditions is a key parameter. However, from the point of view of polymer theory, there is a relatively trivial difference (relating to the configurational entropy) between generating a condensed state from multiple shorter homopolymeric chains or a single long chain that is the sum of these lengths (consider the thought experiment of cutting up a long chain into a number of shorter chains); in addition, the interaction term that drives phase separation in the highly used Flory polymer theory does not depend on number of chains (Flory 1953). It is also possible to draw a parallel between condensates and single chain polymers or disordered proteins in “collapsed globule” states in which there are significant dynamic chain–chain interactions, distinct from highly extended or random coil states with fewer chain–chain and more chain–solvent interactions (Matsuyama and Tanaka 1981; Kolinski and Madziar 1998; Holehouse and Pappu 2018). The patterning of charged and aromatic residues has been shown to correlate with both individual chain collapse (in terms of Rg) and phase behavior; this suggests that the Rg of a long chain could be thought of as a measure of condensation of a single chain (Harmon et al. 2017; Lin and Chan 2017; Dignon et al. 2018; Martin et al. 2020; Zeng et al. 2020). These arguments support the idea that a single, long biomolecule can have condensate properties (e.g., a distinct fluid inside of a physically defined boundary, as defined above).
CAN AN RNA OR AN RNP CONSTITUTE A CONDENSATE?
How does the simulated ELP condensate size compare to proteins and RNA? Eukaryotic proteins come in a wide variety of sizes, from under 50 amino acid residues to Titin at ∼34,000 residues (canonical splice isoform) with an average of ∼400 residues. An mRNA is approximately nine times larger in mass than the protein for which it codes, with the average mRNA being ∼1 to 2 kilobases (kb). Introns in pre-mRNAs can be five times larger than the processed mRNA (Novikova et al. 2012; Palazzo and Lee 2015). The average size of long noncoding RNAs (lncRNAs) is around 1 kb and small nuclear and nucleolar RNAs (sn- and snoRNAs, respectively) range from 60 to 300 bases (Liang et al. 2019). On the other hand, microRNAs (miRNAs) and Piwi-interacting RNAs (piRNAs) are smaller, <35 nt (Starega-Roslan et al. 2011). Beyond the number of residues or bases of proteins and RNA, the physical dimensions for these molecules are highly dependent on their structural conformations, with stable globular (and packed) proteins or tRNAs having smaller Rg values per residue or base than dynamic and disordered states, which sample mixtures of compact (lower Rg) and extended (higher Rg) conformations. Taking this into consideration, average Rg values of a 400-residue protein range from 2 to 4 nm (Erickson 2009) with shapes ranging from spherical to rod-like to long disordered chains (Sommer and Cohen 1980; Tiessen et al. 2012; Borodavka et al. 2016). The Rg values of a 1 kb mRNA range from 9 to 11 nm depending on secondary and tertiary RNA structure and solvent conditions (Yoffe et al. 2008; Gopal et al. 2012; Borodavka et al. 2016). All in all, pre-mRNA, mRNA, and lncRNA are much larger than proteins and they can easily form structures that are larger than 6 nm, the size of the simulated ELP condensate.
Single-stranded RNAs, such as pre-mRNA, mRNA, and lncRNA, form intramolecular hydrogen-bonds and base-pairing interactions to generate secondary and tertiary structures. These single-stranded RNAs, like disordered proteins, have immense multivalent interaction potential via pi, hydrogen-bonding and electrostatic interactions involving bases, the phosphate backbone, and metal or other counter ions. Such interactions support condensate formation, most likely requiring cations to neutralize the negative charge (or the presence of RNA-binding proteins, see the following). Indeed, single-stranded RNAs (including polyU/G/A/Cs and tandem repeats of CAGs and CUGs) undergo phase separation in vitro without proteins (Jain and Vale 2017; Van Treeck et al. 2018). In addition, RNA in the cell, from the time of transcription, is almost always coated with proteins (Duss et al. 2018), which further enhance multivalency via their folded RNA-binding domains and IDRs. More than 60% of proteins have significant intrinsically disordered protein regions (Tsang et al. 2020) and there are estimates that 40% of proteins have sequence features facilitating phase separation (Hardenberg et al. 2020). A significantly higher percentage of RBPs contain IDRs than the general proteome (Varadi et al. 2015) with over 20% of them containing 80% or more disorder (Jarvelin et al. 2016). Many RBPs undergo phase separation in vitro via multivalent interactions involving both IDR–IDR and promiscuous IDR–RNA interactions, as demonstrated by the phase separation of RGG-rich and other low complexity IDRs alone or with simple oligoribonucleotides and longer RNAs (Kato et al. 2012; Elbaum-Garfinkle et al. 2015; Molliex et al. 2015; Zhang et al. 2015; Saha et al. 2016; Banerjee et al. 2017; Langdon et al. 2018; Maharana et al. 2018; Tsang et al. 2019; Sanders et al. 2020). In the context of full-length RBPs, RNAs can interact more specifically with folded RNA-binding domains, including via secondary and tertiary structures or with posttranscriptionally modified bases, such as with m6A methylation (Ries et al. 2019). The combination of interactions involving long RNA molecules, with IDRs and folded domains of RBPs, including protein oligomerization domains, facilitates dynamic multivalent interactions leading to phase separation.
Given these considerations, the size and potential multivalency of a single pre-mRNA, mRNA, or lncRNA (here grouped as “long” RNA) molecule appears to be enough to support a condensate with distinct inside and outside regions, compared to the ELP model. But can a single RNA generate an internal fluid chemical environment that can solvate, enrich or exclude other molecules? The answer likely lies in the length and stability of interactions and folded nature within the RNAs.
RNAs that form significant base-pairing interactions, such as tRNAs, are similar to folded protein domains in that they do not present the dynamic fluctuating interactions that are required for liquid or fluid behavior by themselves. tRNAs are also relatively short at 70 to 100 bases and have similar sizes as globular proteins, further limiting their ability to facilitate dynamic interactions. Single-stranded long RNAs, however, are highly dynamic, with multiple energetically accessible secondary and tertiary structures that rapidly fluctuate (Šponer et al. 2018). This presents significant multivalent interaction potential that could exchange and be fluid. Drawing a parallel with folded protein domains that do not offer as many interacting sites, or as much flexibility, per residue to support fluid states compared to disordered regions, the same is likely true for RNA. Regions exhibiting stable structures are less likely to support dynamic multivalent interactions required for condensate formation. Thus, the propensity of a single long RNA molecule to form a condensate will depend on the amount of stable secondary and tertiary structure in the RNA, with a wide range of changing structural propensities observed in RNA sequences, with some single long RNAs having highly dynamic structures potentially able to generate a “fluid inside” region, as defined above (Fig. 1).
Adding to the possibility that some long RNAs could form condensates on their own, RNPs—with estimates of a dozen or more RBPs per mRNA (Hogan et al. 2008)—certainly meet the size criterion set by the ELP condensate simulation that provides a fluid inside zone (Fig. 1). As noted before, most RBPs have significant IDRs that are known to or highly predicted to phase separate to form fluid condensates in vitro, including with RNA, that can enrich or exclude other biomolecules (Zhang et al. 2015; Langdon et al. 2018; Maharana et al. 2018; Wang et al. 2018; Guillen-Boixet et al. 2020; Yang et al. 2020). Thus, these are strongly suggestive of the potential condensate properties of a single RNA with associated RBPs. One possibility is that an mRNA acts as a long scaffold that nucleates RNP condensates, which in turn concentrate additional proteins to build enzymatic compartments. For example, an RBP, Argonaute, recruits GW182 and Pumilio proteins, deadenylases, and cap binding complexes (4EHP/GiGYF2), all of which have IDRs (Braun et al. 2011; Chapat et al. 2017; Sheu-Gruttadauria and MacRae 2018; Sternburg et al. 2018). On the other hand, if the RBPs form stable interactions that hinder the internal dynamics of the RNA, this would not support a condensate. Based on the size of long single-stranded RNAs and RNPs and their dynamic properties, we argue that these could meet the characteristic features of condensates, even if they are smaller than observable using light microscopes.
WHAT NEW INSIGHTS CAN WE GLEAN FROM CONSIDERING RNAs AND RNPs AS CONDENSATES?
While experiments have not conclusively shown that some single RNA molecules or RNPs are condensates, it is worth considering how this view may be valuable for providing insights into RNA metabolism. One expectation provided by the lens of RNA or RNP condensation is that the sharp transition from condensed to mixed states dictated by the physics of phase separation provides the means for their rapid assembly and disassembly. While the formation of all macromolecular complexes depends on the concentration of components and conditions, the concentration- and condition-dependence of phase transitions is much steeper than the transition between monomer and discrete complex. This has the potential to lead to powerfully fine-tuned biological regulation, generating rapid switch-like responses in the assembly, disassembly, composition, and function of RNPs. One such example may be the regulation of mRNA translation by neuronal RNA transport granules containing FMRP. The sharp dependence of in vitro translational inhibition on the concentration of an FMRP IDR (Hill coefficient ∼5) is completely unlike that of small molecule translational inhibitors, such as puromycin, that have a noncooperative dose dependence (Hill coefficient ∼1), and can be explained by formation of protein–mRNA condensates, not by binding of the protein to mRNA (Tsang et al. 2019). For condensates formed by one or a small number of molecules, the steep dependence on conditions would be more relevant than on concentration.
Understanding RNPs as condensates can also help to clarify or unify apparently contradictory literature about the role of RBPs in RNA regulation. Continuing to use the example of translational regulation, there are conflicting views presented and numerous questions in the literature about the mechanism of FMRP's translational inhibition and the role of phosphorylation (Santoro et al. 2012; Davis and Broadie 2017). FMRP may have multiple mechanisms of translational repression, including by inhibition of translational initiation, stalling of translocating ribosomes (Chen et al. 2014), and interacting with miRNA and the RNA-induced silencing complex (Edbauer et al. 2010). The solvated, mixed-phase FMRP IDR in the context of the in vitro assay used for that study (Tsang et al. 2019) is not a translational inhibitor, and neither is phosphorylated FMRP. But the condensed state of the FMRP IDR, which phosphorylation facilitates, inhibits translation ∼10- to 20-fold relative to the solvated, mixed-phase state and 100-fold relative to buffer (Kim et al. 2019; Tsang et al. 2019). This condensed state also concentrates 4E-BP2, a known inhibitor of translation initiation, and a miRNA involved in translational inhibition, even without a canonical miRNA-binding domain (Tsang et al. 2019). A subcompartmentalized condensate of a phosphorylated FMRP IDR with a CAPRIN1 IDR inhibits translation to an even greater extent, by four orders of magnitude relative to buffer, likely by partitioning RNA away from certain components of the translational regulatory machinery (Kim et al. 2019). Thus, depending on the conditions of a particular cellular experiment, FMRP may or may not appear to repress translation via various mechanisms and phosphorylation may or may not impact translation; consideration of whether the protein is in a condensed state, however, can provide clarity and potential insights into the multiple mechanisms through which FMRP can act (Davis and Broadie 2017).
Single RNP “condensates” have the potential to form compartments that modulate enzymatic activities relating to RNA metabolism, as this example for translation suggests. Indeed, condensates can enhance/reduce activities by concentrating/excluding reaction substrates, products and/or enzymes, and by creating conditions that can modulate conformational equilibria of proteins (O'Flynn and Mittag 2021; Peeples and Rosen 2021). The regulation of mRNA decapping in P-bodies, which are storage and decay sites, provides another illustration of some of these enzymatic regulatory mechanisms, and another instance where the condensate perspective clarifies data. Recent work demonstrates that both storage and decay outcomes are possible in the context of Dcp1/Dcp2-containing condensates that can significantly stabilize an inactive Dcp2 conformation relative to dilute solution (inhibiting decapping enables RNA storage); Dcp2 can be activated by the addition of Edc3 (stimulating decapping promotes RNA decay) (Tibble et al. 2021). The conformational switch between inactive and active states of Dcp2 is impacted by the composition of the condensate environment, coupling regulation of enzymatic function to reorganization of interactions spanning the condensate. These results clarify complex data and demonstrate how an “activator” can change the interactions within a condensate to control conformational equilibria to give opposing outcomes.
Another idea that emerges from the view of RNA or RNPs as condensates is the notion that most of the RNA “life-cycle” may exist in the context of various condensates, that we briefly alluded to above. In this model, which requires additional experimental validation, RNA is initially generated within RNA Pol II-enhancer condensates that function as transcription initiation reaction vessels; from there it moves into transcription elongation condensates as the post-translational modification state of RNA Pol II changes, then into splicing condensates such as nuclear speckles, then into the nuclear pore condensate and then cytoplasmic mRNA transport granules and P-bodies (Kwon et al. 2013; Lu et al. 2018; Guo et al. 2019; McKnight 2019). One way to view this is that the composition of the RNP (i.e., which RBPs interact with the RNA) represents partitioning of specific RBPs to an RNA or RNP condensate. Accordingly, one could view this as droplet fusion and fission events, with specific RNPs merging with or emerging from different larger condensates that mediate RNA metabolism (such as nuclear speckles and P-bodies). In this manner, the physics of condensate partitioning and solvation may provide a means for the right RBPs to concentrate with RNAs at specific stages to facilitate RNA processing. In addition, these transitions can be sharply controlled through post-translational or post-transcriptional modifications of protein and RNA, respectively, and by the available concentrations of various components. Indeed, these modifications affect condensate composition in various contexts (Ditlev et al. 2018; Langdon et al. 2018; Ries et al. 2019; Sanders et al. 2020). Some of the RNP condensate regulators could also be other RNAs, including the lncRNAs NEAT1 and MALAT1 that are important for paraspeckle and nuclear speckle formation, respectively (Statello et al. 2021), and smaller RNAs that could possibly modulate the phase behavior of these RNPs (Sheu-Gruttadauria and MacRae 2018; Manage et al. 2020). In this context, RNA can modulate the size, morphology and composition of RNP condensates as demonstrated by artificial granules with RBPs (Garcia-Jove Navarro et al. 2019). Conversely, certain RBPs may solvate mRNAs outside of condensates. For example, ribosome-bound mRNAs are readily solvated in the cytoplasm and generally also do not incorporate into condensates (Delarue et al. 2018). Thus, specific RNAs and RBPs determine whether or not condensates are generated; and the composition, material properties, unique solvent environment and function of condensates that are formed will be controlled by the RNAs and RNPs that partition into or fuse with them.
CAN CONDENSATE-LIKE PROPERTIES OF RNAs AND RNPs BE OBSERVED IN CELLS?
Regardless of the conceptual value of this condensate lens for RNAs and RNPs, it is critical to examine how experimental data supports or argues against this view and consider alternative explanations (Alberti et al. 2019; McSwiggen et al. 2019; Peng and Weber 2019). For instance, if small cellular RNAs and RNPs are fluid-like condensates, it may be expected that they fuse to grow to be microscopically observable through the process of “coarsening” that is seen for in vitro and micron-sized cellular condensates (Brangwynne et al. 2009, 2011; Molliex et al. 2015; Nott et al. 2015; Patel et al. 2015; Smith et al. 2016; Wheeler et al. 2016; Kistler et al. 2018). However, this behavior is not frequently observed in cells. This raises the question, if RNPs are indeed condensates, as is predicted by their size, multivalency, and in vitro counterparts, why is coarsening not more widely observed in cells? Several potential explanations for this observation exist. Because cells are not in a state of equilibrium, various factors that control the formation and growth of condensates are in constant flux which limits growth (Zwicker et al. 2016; Kirschbaum and Zwicker 2021). Examples of such factors are potentially limiting concentrations of condensing protein and RNA, as well as the concentration of other molecules that may solubilize condensate components. In addition, some cellular components or types of protein sequences act as surfactants to decrease the energy of the interface between condensates and the solvent, thereby limiting growth (Cuylen et al. 2016). Forces generated by motors and/or the cytoskeleton can also act on condensates to prevent their fusion or promote fission (Brangwynne et al. 2011; Ditlev et al. 2019). Chemical reactions can theoretically control droplet sizes (Kirschbaum and Zwicker 2021). Finally, the presence of component clusters at the interface can regulate the size of condensates by tuning the exchange of materials between the cytoplasm and condensates or fusion of condensates (Folkmann et al. 2021). Any one or combination of these reasons can explain why RNPs, like other types of condensates (Ranganathan and Shakhnovich 2020), do not form larger structures in cells. Interestingly, RNP condensate growth might be regulated in a cell-type and condition specific manner, as certain RNPs travel in the form of microscopically observable, larger granules in neurons and germ cells or form micron-sized stress granules in response to stress (Knowles et al. 1996; Carson and Barbarese 2005; Brangwynne et al. 2009; Lee et al. 2020).
The inside/outside distinction and fluid nature of various RNA-containing condensates have been observed for micron-sized condensates in cells. For example, the enrichment of specific components has been observed for numerous condensates using imaging and proximity labeling approaches. These techniques report on the spatial distribution of and relationships between components that may not have specific high-affinity binding partners, thereby identifying which factors “partition” into condensates. Not only RNAs and proteins can partition; there is increasing work demonstrating that small molecules can be enriched in or excluded from biomolecular condensates, including drugs and nucleotides (Klein et al. 2020; Folkmann et al. 2021; Kim et al. 2021). Techniques that assess diffusion, including photobleaching/photoactivation-based approaches and fluorescence correlation spectroscopy, have also been used to probe certain aspects of fluidity in cells. However, in-depth studies of these features in condensates smaller than 100 nm in diameter is quite limiting for these techniques; many condensates in mammalian cells, and most in bacteria, are simply too small to easily probe their biophysical properties with these tools (e.g., fluorescent microscopy). In fact, the nature of imaging methods being limited by the resolution of light has almost led to a de facto definition of the size of biomolecular condensates based on the ability to resolve them by microscopy. The 6 nm diameter ELP condensates would elude microscopic observation, yet they fulfill the definitions we propose of a fluid inside a physical boundary. Cryo-electron tomography may enable insights with resolution bridging optical microscopy and molecular imaging techniques, but there have not been significant studies to date (Franzmann et al. 2018). So, what are some of the latest advances that are enabling us to extract information about condensates below the standard optical resolution?
Two recent studies have used innovative techniques to break through the resolution barrier that traditionally defines biomolecular condensates. The Cissé laboratory has modified super-resolution photoactivated localization microscopy techniques to study the dynamics of condensate-dependent gene regulation in living cells. Using this technique, they demonstrated that RNA Pol II clusters in the nucleus display the behaviors, such as coalescence and rapid fluorescence recovery after photobleaching, expected of liquid-like condensates (Cho et al. 2018). The Brangwynne group found that in vitro condensate nucleation is well modeled by classical nucleation theory (CNT), which states that the degree of condensate formation depends on component saturation while the location of condensate formation depends on the heterogenous interactions between surfaces of condensate seeds. They found that, in cells, CNT can be used to describe both model condensate formation and endogenous condensate formation. Thus, CNT can be used to describe cellular events that are not resolvable using traditional light microscopy. These results also support the notion that in vitro condensate formation can be representative of cellular condensate formation (Shimobayashi et al. 2021). In principle, these techniques with theoretical modeling could be adapted to study groups of RNPs throughout living cells to better understand their properties regardless of their size.
CONCLUSION
In summary, we propose that RNPs and possibly some single long RNA molecules could be understood as condensates, forming fluid states with unique inside environments dependent on the nucleotide sequence and the composition of RBPs that decorate it. The life-cycle of RNAs can be correlated with changes in RNP condensate compositions and material properties, impacting their localization and, ultimately, their function. The RNA life-cycle could include transcription initiation and then transcription elongation condensates where RNA is generated; splicing condensates; the nuclear pore condensate; and various cytoplasmic RNA condensates that control RNA processing/modifications, mRNA translation and finally degradation. New experimental and theoretical methods will hopefully provide more clear evidence for the presence of cellular RNA and RNP condensates. As these tools are being developed and critical data emerge, we argue that this condensate view can provide a valuable context for interpreting experimental results and a conceptual foundation for understanding RNA and RNP regulation and the mechanisms of their biological functions.
ACKNOWLEDGMENTS
The authors appreciate stimulating discussions and comments on the manuscript from Hue Sun Chan, Alex Palazzo, and Andrew Chong. J.D.F.-K. and H.O.L. acknowledge funding from the Canadian Institutes for Health Research (CIHR), the Natural Sciences and Engineering Research Council of Canada (NSERC), and the Canada Research Chair Program. H.O.L. also receives support from the Medicine by Design initiative, which receives funding from the Canada First Research Excellence Fund. M.L.N. is funded by an Alexander Graham Bell Canada Graduate Scholarship—Doctoral from NSERC. J.A.D. is supported by the Hospital for Sick Children Research Institute.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079026.121.
Freely available online through the RNA Open Access option.
REFERENCES
- Alberti S, Gladfelter A, Mittag T. 2019. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176: 419–434. 10.1016/j.cell.2018.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alshareedah I, Thurston GM, Banerjee PR. 2021. Quantifying viscosity and surface tension of multicomponent protein-nucleic acid condensates. Biophys J 120: 1161–1169. 10.1016/j.bpj.2021.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee PR, Milin AN, Moosa MM, Onuchic PL, Deniz AA. 2017. Reentrant phase transition drives dynamic substructure formation in ribonucleoprotein droplets. Angew Chem Int Ed Engl 56: 11354–11359. 10.1002/anie.201703191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batish M, van den Bogaard P, Kramer FR, Tyagi S. 2012. Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci 109: 4645–4650. 10.1073/pnas.1111226109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgia A, Borgia MB, Bugge K, Kissling VM, Heidarsson PO, Fernandes CB, Sottini A, Soranno A, Buholzer KJ, Nettels D, et al. 2018. Extreme disorder in an ultrahigh-affinity protein complex. Nature 555: 61–66. 10.1038/nature25762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodavka A, Singaram SW, Stockley PG, Gelbart WM, Ben-Shaul A, Tuma R. 2016. Sizes of long RNA molecules are determined by the branching patterns of their secondary structures. Biophys J 111: 2077–2085. 10.1016/j.bpj.2016.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady JP, Farber PJ, Sekhar A, Lin YH, Huang R, Bah A, Nott TJ, Chan HS, Baldwin AJ, Forman-Kay JD, et al. 2017. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc Natl Acad Sci 114: E8194–E8203. 10.1073/pnas.1706197114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324: 1729–1734. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA. 2011. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci 108: 4334–4339. 10.1073/pnas.1017150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratek-Skicki A, Pancsa R, Meszaros B, Van Lindt J, Tompa P. 2020. A guide to regulation of the formation of biomolecular condensates. FEBS J 287: 1924–1935. 10.1111/febs.15254 [DOI] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Fauser M, Izaurralde E. 2011. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell 44: 120–133. 10.1016/j.molcel.2011.09.007 [DOI] [PubMed] [Google Scholar]
- Carson JH, Barbarese E. 2005. Systems analysis of RNA trafficking in neural cells. Biol Cell 97: 51–62. 10.1042/BC20040083 [DOI] [PubMed] [Google Scholar]
- Chalmers GWG, Gosline JM, Lillie MA. 1999. The hydrophobicity of vertebral elastins. J Exp Biol 202: 301–314. 10.1242/jeb.202.3.301 [DOI] [PubMed] [Google Scholar]
- Chapat C, Jafarnejad SM, Matta-Camacho E, Hesketh GG, Gelbart IA, Attig J, Gkogkas CG, Alain T, Stern-Ginossar N, Fabian MR, et al. 2017. Cap-binding protein 4EHP effects translation silencing by microRNAs. Proc Natl Acad Sci 114: 5425–5430. 10.1073/pnas.1701488114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Sharma MR, Shi X, Agrawal RK, Joseph S. 2014. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol Cell 54: 407–417. 10.1016/j.molcel.2014.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube H, Cisse II. 2018. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361: 412–415. 10.1126/science.aar4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, Heckert A, Banala S, Lavis L, Darzacq X, et al. 2018. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361: eaar2555. 10.1126/science.aar2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–726. 10.1016/j.molcel.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conicella AE, Zerze GH, Mittal J, Fawzi NL. 2016. ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure 24: 1537–1549. 10.1016/j.str.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csizmok V, Montecchio M, Lin H, Tyers M, Sunnerhagen M, Forman-Kay JD. 2018. Multivalent interactions with Fbw7 and Pin1 facilitate recognition of c-Jun by the SCFFbw7 ubiquitin ligase. Structure 26: 28–39.e2. 10.1016/j.str.2017.11.003 [DOI] [PubMed] [Google Scholar]
- Cuylen S, Blaukopf C, Politi AZ, Muller-Reichert T, Neumann B, Poser I, Ellenberg J, Hyman AA, Gerlich DW. 2016. Ki-67 acts as a biological surfactamt to disperse mitotic chromosomes. Nature 535: 308–312. 10.1038/nature18610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JK, Broadie K. 2017. Multifarious functions of the Fragile X mental retardation protein. Trends Genet 33: 703–714. 10.1016/j.tig.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M, Brittingham GP, Pfeffer S, Surovtsev IV, Pinglay S, Kennedy KJ, Schaffer M, Gutierrez JI, Sang D, Poterewicz G, et al. 2018. mTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell 174: 338–349.e20. 10.1016/j.cell.2018.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignon GL, Zheng W, Best RB, Kim YC, Mittal J. 2018. Relation between single-molecule properties and phase behavior of intrinsically disordered proteins. Proc Natl Acad Sci 115: 9929–9934. 10.1073/pnas.1804177115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditlev JA, Case LB, Rosen MK. 2018. Who's in and who's out—compositional control of biomolecular condensates. J Mol Biol 430: 4666–4684. 10.1016/j.jmb.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditlev JA, Vega AR, Koster DV, Su X, Tani T, Lakoduk AM, Vale RD, Mayor S, Jaqaman K, Rosen MK. 2019. A composition-dependent molecular clutch between T cell signaling condensates and actin. Elife 8: e42695. 10.7554/eLife.42695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duss O, Stepanyuk GA, Grot A, O'Leary SE, Puglisi JD, Williamson JR. 2018. Real-time assembly of ribonucleoprotein complexes on nascent RNA transcripts. Nat Commun 9: 5087. 10.1038/s41467-018-07423-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. 2010. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65: 373–384. 10.1016/j.neuron.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP. 2015. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci 112: 7189–7194. 10.1073/pnas.1504822112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP. 2009. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol Proced Online 11: 32–51. 10.1007/s12575-009-9008-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Zhou BR, Bai Y. 2018. Binding affinity and function of the extremely disordered protein complex containing human linker histone H1.0 and its chaperone ProTα. Biochemistry 57: 6645–6648. 10.1021/acs.biochem.8b01075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. 2016. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165: 1686–1697. 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory PJ. 1941. Thermodynamics of high polymer solutions. J Chem Phys 9: 660. 10.1063/1.1750971 [DOI] [Google Scholar]
- Flory PJ. 1953. Principles of polymer chemistry. Cornell University Press, Ithaca, NY. [Google Scholar]
- Folkmann AW, Putnam A, Lee CF, Seydoux G. 2021. Regulation of biomolecular condensates by interfacial protein clusters. Science 373: 1218–1224. 10.1126/science.abg7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AKL, Lyon CE, Andersen J, Mann M, Lamond AI. 2002. Paraspeckles: a novel nuclear domain. Curr Biol 12: 13–25. 10.1016/S0960-9822(01)00632-7 [DOI] [PubMed] [Google Scholar]
- Franzmann TM, Jahnel M, Pozniakovsky A, Mahamid J, Holehouse AS, Nuske E, Richter D, Baumeister W, Grill SW, Pappu RV, et al. 2018. Phase separation of a yeast prion protein promotes cellular fitness. Science 359: eaao5654. 10.1126/science.aao5654 [DOI] [PubMed] [Google Scholar]
- Garcia-Jove Navarro M, Kashida S, Chouaib R, Souquere S, Pierron G, Weil D, Gueroui Z. 2019. RNA is a critical element for the sizing and the composition of phase-separated RNA-protein condensates. Nat Commun 10: 3230. 10.1038/s41467-019-11241-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal A, Zhou ZH, Knobler CM, Gelbart WM. 2012. Visualizing large RNA molecules in solution. RNA 18: 284–299. 10.1261/rna.027557.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal PP, Nirschl JJ, Klinman E, Holzbaur EL. 2017. Amyotrophic lateral sclerosis-linked mutations increase the viscosity of liquid-like TDP-43 RNP granules in neurons. Proc Natl Acad Sci 114: E2466–E2475. 10.1073/pnas.1614462114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillen-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlussler R, Kim K, Trussina I, Wang J, Mateju D, et al. 2020. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181: 346–361.e17. 10.1016/j.cell.2020.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall'Agnese A, Hannett NM, Spille JH, Afeyan LK, Zamudio AV, Shrinivas K, et al. 2019. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572: 543–548. 10.1038/s41586-019-1464-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardenberg M, Horvath A, Ambrus V, Fuxreiter M, Vendruscolo M. 2020. Widespread occurrence of the droplet state of proteins in the human proteome. Proc Natl Acad Sci 117: 33254–33262. 10.1073/pnas.2007670117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardenberg MC, Sinnige T, Casford S, Dada ST, Poudel C, Robinson EA, Fuxreiter M, Kaminksi CF, Kaminski Schierle GS, Nollen EAA, et al. 2021. Observation of an α-synuclein liquid droplet state and its maturation into Lewy body-like assemblies. J Mol Cell Biol 13: 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon TS, Holehouse AS, Rosen MK, Pappu RV. 2017. Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife 6: e30294. 10.7554/eLife.30294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle FA, Feigenson GW. 2011. Phase separation in lipid membranes. Cold Spring Harb Perspect Biol 3: a004630. 10.1101/cshperspect.a004630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. 2008. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol 6: e255. 10.1371/journal.pbio.0060255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holehouse AS, Pappu R. 2018. Collapse transitions of proteins and the interplay among backbone, sidechain, and solvent interactions. Annu Rev Biophys 47: 19–39. 10.1146/annurev-biophys-070317-032838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Vale RD. 2017. RNA phase transitions in repeat expansion disorders. Nature 546: 243–247. 10.1038/nature22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. 2016. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164: 487–498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvelin AI, Noerenberg M, Davis I, Castello A. 2016. The new (dis)order in RNA regulation. Cell Commun Signal 14: 9. 10.1186/s12964-016-0132-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. 2012. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149: 753–767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Tsang B, Vernon RM, Sonenberg N, Kay LE, Forman-Kay JD. 2019. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 365: 825–829. 10.1126/science.aax4240 [DOI] [PubMed] [Google Scholar]
- Kim TH, Payliss BJ, Nosella ML, Lee ITW, Toyama Y, Forman-Kay JD, Kay LE. 2021. Interaction hot spots for phase separation revealed by NMR studies of a CAPRIN1 condensed phase. Proc Natl Acad Sci 118: e2104897118. 10.1073/pnas.2104897118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschbaum J, Zwicker D. 2021. Controlling biomolecular condensates via chemical reactions. J R Soc Interface 18: 20210255. 10.1098/rsif.2021.0255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler KE, Trcek T, Hurd TR, Chen R, Liang FX, Sall J, Kato M, Lehmann R. 2018. Phase transitioned nuclear Oskar promotes cell division of Drosophila primordial germ cells. Elife 7: e37949. 10.7554/eLife.37949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein IA, Boija A, Afeyan LK, Hawken SW, Fan M, Dall'Agnese A, Oksuz O, Henninger JE, Shrinivas K, Sabari BR, et al. 2020. Partitioning of cancer therapeutics in nuclear condensates. Science 368: 1386–1392. 10.1126/science.aaz4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, Kosik KS. 1996. Translocation of RNA granules in living neurons. J Neurosci 16: 7812–7820. 10.1523/JNEUROSCI.16-24-07812.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolinski A, Madziar P. 1998. Collapse transitions in protein-like lattice polymers: the effect of sequence patterns. Biopolymers 42: 537–548. [DOI] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. 2013. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155: 1049–1060. 10.1016/j.cell.2013.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon EM, Qiu Y, Niaki AG, McLaughlin GA, Weidmann CA, Gerbich TM, Smith JA, Crutchley JM, Termini CM, Weeks KM, et al. 2018. mRNA structure determines specificity of a polyQ-driven phase separation. Science 360: 922–927. 10.1126/science.aar7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Putnam A, Lu T, He S, Ouyang JPT, Seydoux G. 2020. Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. Elife 9: e52896. 10.7554/eLife.52896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. 2002. Multiple associated proteins regulate proteasome structure and function. Mol Cell 10: 495–507. 10.1016/S1097-2765(02)00638-X [DOI] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. 2012. Phase transitions in the assembly of multivalent signalling proteins. Nature 483: 336–340. 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wen J, Huang Z, Chen XP, Zhang BX, Chu L. 2019. Small nucleolar RNAs: insight into their function in cancer. Front Oncol 9: 587. 10.3389/fonc.2019.00587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao SE, Regev O. 2021. Splicing at the phase-separated nuclear speckle interface: a model. Nucleic Acids Res 49: 636–645. 10.1093/nar/gkaa1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YH, Chan HS. 2017. Phase separation and single-chain compactness of charged disordered proteins are strongly correlated. Biophys J 112: 2043–2046. 10.1016/j.bpj.2017.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R. 2015. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell 60: 208–219. 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Yu D, Hansen AS, Ganguly S, Liu R, Heckert A, Darzacq X, Zhou Q. 2018. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558: 318–323. 10.1038/s41586-018-0174-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillen-Boixet J, Franzmann TM, et al. 2018. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360: 918–920. 10.1126/science.aar7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manage KI, Rogers AK, Wallis DC, Uebel CJ, Anderson DC, Nguyen DAH, Arca K, Brown KC, Cordeiro Rodrigues RJ, de Albuquerque BF, et al. 2020. A tudor domain protein, SIMR-1, promotes siRNA production at piRNA-targeted mRNAs in C. elegans. Elife 9: e56731. 10.7554/eLife.56731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmor-Kollet H, Siany A, Kedersha N, Knafo N, Rivkin N, Danino YM, Moens TG, Olender T, Sheban D, Cohen N, et al. 2020. Spatiotemporal proteomic analysis of stress granule disassembly using APEX reveals regulation by SUMOylation and links to ALS pathogenesis. Mol Cell 80: 876–891.e6. 10.1016/j.molcel.2020.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, Grace CR, Soranno A, Pappu RV, Mittag T. 2020. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367: 694–699. 10.1126/science.aaw8653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, Tanaka F. 1981. Theory of solvation-induced reentrant coil-globule transition of an isolated polymer chain. J Chem Phys 94: 781–786. 10.1063/1.460296 [DOI] [Google Scholar]
- McKnight SL. 2019. Passing through. Trends Biochem Sci 44: 899–901. 10.1016/j.tibs.2019.09.004 [DOI] [PubMed] [Google Scholar]
- McSwiggen DT, Mir M, Darzacq X, Tjian R. 2019. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev 33: 1619–1634. 10.1101/gad.331520.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M, Stadler MR, Ortiz SA, Hannon CE, Harrison MM, Darzacq X, Eisen MB. 2018. Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. Elife 7: e40497. 10.7554/eLife.40497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. 2015. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163: 123–133. 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muiznieks LD, Sharpe S, Pomes R, Keeley FW. 2018. Role of liquid-liquid phase separation in assembly of elastin and other extracellular matrix proteins. J Mol Biol 430: 4741–4753. 10.1016/j.jmb.2018.06.010 [DOI] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. 2015. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 57: 936–947. 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova IV, Hennelly SP, Sanbonmatsu KY. 2012. Sizing up long non-coding RNAs: do lncRNAs have secondary and tertiary structure? Bioarchitecture 2: 189–199. 10.4161/bioa.22592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Flynn BG, Mittag T. 2021. The role of liquid-liquid phase separation in regulating enzyme activity. Curr Opin Cell Biol 69: 70–79. 10.1016/j.ceb.2020.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, Lee ES. 2015. Non-coding RNA: what is functional and what is junk? Front Genet 6: 2. 10.3389/fgene.2015.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. 2015. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162: 1066–1077. 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- Peeples W, Rosen MK. 2021. Mechanistic dissection of increased enzymatic rate in a phase-separated compartment. Nat Chem Biol 17: 693–702. 10.1038/s41589-021-00801-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng A, Weber SC. 2019. Evidence for and against liquid–liquid phase separation in the nucleus. Non-coding RNA 5: 50. 10.3390/ncrna5040050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. 2005. Regulating gene expression through RNA nuclear retention. Cell 123: 249–263. 10.1016/j.cell.2005.08.033 [DOI] [PubMed] [Google Scholar]
- Ranganathan S, Shakhnovich EI. 2020. Dynamic metastable long-living droplets formed by sticker-spacer proteins. Elife 9: e56159. 10.7554/eLife.56159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauscher S, Pomès R. 2017. The liquid structure of elastin. Elife 6: e26526. 10.7554/eLife.26526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, Wei MT, Kriwacki RW, Brangwynne CP. 2020. Composition-dependent thermodynamics of intracellular phase separation. Nature 581: 209–214. 10.1038/s41586-020-2256-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee JH, Jaffrey SR. 2019. m6A enhances the phase separation potential of mRNA. Nature 571: 424–428. 10.1038/s41586-019-1374-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. 2018. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361: eaar3958. 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev R, Hondele M, Linsenmeier M, Vallotton P, Mugler CF, Arosio P, Weis K. 2019. Pat1 promotes processing body assembly by enhancing the phase separation of the DEAD-box ATPase Dhh1 and RNA. Elife 8: e41415. 10.7554/eLife.41415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Weber CA, Nousch M, Adame-Arana O, Hoege C, Hein MY, Osborne-Nishimura E, Mahamid J, Jahnel M, Jawerth L, et al. 2016. Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell 166: 1572–1584 e1516. 10.1016/j.cell.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A, et al. 2020. Competing protein-RNA interaction networks control multiphase intracellular organization. Cell 181: 306–324.e28. 10.1016/j.cell.2020.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro MR, Bray SM, Warren ST. 2012. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu Rev Pathol 7: 219–245. 10.1146/annurev-pathol-011811-132457 [DOI] [PubMed] [Google Scholar]
- Schlichtkrull J. 1956. Insulin crystals. II. Shape of rhombohedral zinc-insulin crystals in relation to species and crystallization media. Acta Chem Scand 10: 1459–1464. 10.3891/acta.chem.scand.10-1459 [DOI] [Google Scholar]
- Schmidt HB, Gorlich D. 2016. Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem Sci 41: 46–61. 10.1016/j.tibs.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Schmidt H, Putnam A, Rasoloson D, Seydoux G. 2021. Protein-based condensation mechanisms drive the assembly of RNA-rich P granules. Elife 10: e63698. 10.7554/eLife.63698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seviour T, Wong LL, Lu Y, Mugunthan S, Yang Q, Shankari U, Segaran C, Bessarab I, Liebl D, Williams RBH, et al. 2020. Phase transitions by an abundant protein in the Anammox extracellular matrix mediate cell-to-cell aggregation and biolfilm formation. MBio 11: e02052. 10.1128/mBio.02052-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu-Gruttadauria J, MacRae IJ. 2018. Phase transitions in the assembly and function of human miRISC. Cell 173: 946–957.e16. 10.1016/j.cell.2018.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimobayashi SF, Ronceray P, Sanders DW, Haataja MP, Brangwynne CP. 2021. Nucleation landscape of biomolecular condensates. Nature 10.1038/s41586-021-03905-5 [DOI] [PubMed] [Google Scholar]
- Sieker LC, Bolles L, Stenkamp RE, Jensen LH, Appleby CA. 1981. Preliminary X-ray study of a dimeric form of hemerythrin from Phascolosoma arcuatum. J Mol Biol 148: 493–494. 10.1016/0022-2836(81)90189-3 [DOI] [PubMed] [Google Scholar]
- Smith J, Calidas D, Schmidt H, Lu T, Rasoloson D, Seydoux G. 2016. Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. Elife 5: e21337. 10.7554/eLife.21337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer SS, Cohen JE. 1980. The size distribution of proteins, mRNA, and nuclear RNA. J Mol Evol 15: 37–57. 10.1007/BF01732582 [DOI] [PubMed] [Google Scholar]
- Šponer J, Bussi G, Krepl M, Banáš P, Bottaro S, Cunha RA, Gil-Ley A, Pinamonti G, Poblete S, Jurečka P, et al. 2018. RNA structural dynamics as captured by molecular simulations: a comprehensive overview. Chem Rev 118: 4177–4338. 10.1021/acs.chemrev.7b00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starega-Roslan J, Krol J, Koscianska E, Kozlowski P, Szlachcic WJ, Sobczak K, Krzyzosiak WJ. 2011. Structural basis of microRNA length variety. Nucleic Acids Res 39: 257–268. 10.1093/nar/gkq727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statello L, Guo CJ, Chen LL, Huarte M. 2021. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22: 96–118. 10.1038/s41580-020-00315-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternburg EL, Estep JA, Nguyen DK, Li Y, Karginov FV. 2018. Antagonistic and cooperative AGO2-PUM interactions in regulating mRNAs. Sci Rep 8: 15316. 10.1038/s41598-018-33596-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD. 2016. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352: 595–599. 10.1126/science.aad9964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibble RW, Depaix A, Kowalska J, Jemielity J, Gross JD. 2021. Biomolecular condensates amplify mRNA decapping by biasing enzyme conformation. Nat Chem Biol 17: 615–623. 10.1038/s41589-021-00774-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiessen A, Perez-Rodrigues P, Delaye-Aredondo LJ. 2012. Mathematical modeling and comparison of protein size distribution in different plant, animal, fungal and microbial species reveals a negative correlation between protein size and protein number, thus providing insight into the evolution of proteomes. BMC Res Notes 5: 85. 10.1186/1756-0500-5-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang B, Arsenault J, Vernon RM, Lin H, Sonenberg N, Wang LY, Bah A, Forman-Kay JD. 2019. Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proc Natl Acad Sci 116: 4218–4227. 10.1073/pnas.1814385116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang B, Pritisanac I, Scherer SW, Moses AM, Forman-Kay JD. 2020. Phase separation as a missing mechanism for interpretation of disease mutations. Cell 183: 1742–1756. 10.1016/j.cell.2020.11.050 [DOI] [PubMed] [Google Scholar]
- Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R. 2018. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci 115: 2734–2739. 10.1073/pnas.1800038115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi M, Zsolyomi F, Guharoy M, Tompa P. 2015. Functional advantages of conserved intrinsic disorder in RNA-binding proteins. PLoS One 10: e0139731. 10.1371/journal.pone.0139731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. 2018. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174: 688–699.e16. 10.1016/j.cell.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann S, Eftekharzadeh B, Tepper K, Zoltowska KM, Bennett RE, Dujardin S, Laskowski PR, MacKenzie D, Kamath T, Commins C, et al. 2018. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J 37: e98049. 10.15252/embj.201798049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessén J, Pal T, Das S, Lin YH, Chan HS. 2021. A simple explicit-solvent model of polyampholyte phase behaviors and its ramifications for dielectric effects in biomolecular condensates. J Phys Chem B 125: 4337–4358. 10.1021/acs.jpcb.1c00954 [DOI] [PubMed] [Google Scholar]
- Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. 2016. Distinct stages in stress granule assembly and disassembly. Elife 5: e18413. 10.7554/eLife.18413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA. 2017. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169: 1066–1077 e1010. 10.1016/j.cell.2017.05.028 [DOI] [PubMed] [Google Scholar]
- Yang P, Mathieu C, Kolaitis RM, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q, et al. 2020. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181: 325–345.e28. 10.1016/j.cell.2020.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoffe AM, Prinsen P, Gopal A, Knobler CM, Gelbart WM, Ben-Shaul A. 2008. Predicting the sizes of large RNA molecules. Proc Natl Acad Sci 105: 16153–16158. 10.1073/pnas.0808089105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. 10.1126/science.1060089 [DOI] [PubMed] [Google Scholar]
- Zeng X, Holehouse AS, Chilkoti A, Mittag T, Pappu RV. 2020. Connecting coil-to-globule transitions to full phase diagrams for intrinsically disordered proteins. Biophys J 119: 402–418. 10.1016/j.bpj.2020.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. 2015. RNA controls polyQ protein phase transitions. Mol Cell 60: 220–230. 10.1016/j.molcel.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX. 2021. Viscoelasticity of biomolecular condensates conforms to the Jeffreys model. J Chem Phys 154: 041103. 10.1063/5.0038916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickermann V, Wirth C, Nasiri H, Siegmund K, Schwalber H, Hunte C, Brandt U. 2015. Mechanistic insight from the crystal structure of mitochondrial complex I. Science 347: 44–49. 10.1126/science.1259859 [DOI] [PubMed] [Google Scholar]
- Zwicker D, Seyboldt R, Weber CA, Hyman AA, Jülicher F. 2016. Growth and division of active droplets provides a model for protocells. Nat Phys 13: 408–413. 10.1038/nphys3984 [DOI] [Google Scholar]