Abstract
Macroscopic membraneless organelles containing RNA such as the nucleoli, germ granules, and the Cajal body have been known for decades. These biomolecular condensates are liquid-like bodies that can be formed by a phase transition. Recent evidence has revealed the presence of similar microscopic condensates associated with the transcription of genes. This brief article summarizes thoughts about the importance of condensates in the regulation of transcription and how RNA molecules, as components of such condensates, control the synthesis of RNA. Models and experimental data suggest that RNAs from enhancers facilitate the formation of a condensate that stabilizes the binding of transcription factors and accounts for a burst of transcription at the promoter. Termination of this burst is pictured as a nonequilibrium feedback loop where additional RNA destabilizes the condensate.
Keywords: RNA, transcription, transcription factors, condensate, phase separation, intrinsically disordered domains
RNA functions as the messenger of the genetic code, can catalyze numerous transesterification reactions, and binds specifically to diverse small and large molecules. Added to this list now is the role of RNA in the formation and regulation of biomolecular condensates. This brief article summarizes recent thoughts about the importance of condensates in the regulation of transcription and how RNA molecules, as components of such condensates, may control the synthesis of RNA.
Membraneless subcellular organelles, termed condensates or biomolecular condensates, are formed by phase transitions (Banani et al. 2017). Two examples of condensate prototypes, germ cell granules in Caenorhabditis elegans and nucleoli, consist of both proteins and RNA. If a condensate has liquid-like properties, its protein components can dynamically exchange with those in the surrounding environment in seconds. The RNA components concentrated in the condensate can exchange with those in the external environment as well, but typically do so more slowly (Aumiller et al. 2016). At equilibrium, phase transitions leading to the formation of a condensate occur when the energetic advantage of concentrating components that interact favorably with each other outweighs the entropic cost of confining these components in a condensate. Many proteins found in condensates in vivo will associate and form droplets when concentrated in vitro. This reflects favorable weak, cooperative, multivalent interactions between amino acids in intrinsically disordered domains (IDRs), and in small modular interacting domains (MIDs) (Banani et al. 2017; Hnisz et al. 2017). Long RNAs can also generate condensates when concentrated in vitro, but these are less dynamic than IDR droplets. It is likely that in condensates containing both RNA and proteins, RNA provides a negatively charged scaffold bound by positive charges in the proteins. In many cases, these proteins with IDRs and MIDs also have domains that associate, with high affinity, with specific sequences, or folded domains of RNA (Roden and Gladfelter 2021). Thus, proteins and RNAs could be concentrated in condensates containing RNA and proteins through various types of favorable RNA–protein interactions as well as IDR or MID-mediated, protein–protein interactions.
Regulation of transcription is central to both normal and abnormal physiology. In mammalian systems, DNA sequence-specific transcription factors (TFs) are the primary source of specificity for gene activation or suppression (Lambert et al. 2018). These factors function within the context of the continuous life cycle of cells and organisms and their specificity and activity are dependent upon the cell state. Thus, the binding of a transcription factor to a high-affinity site is influenced by the chromatin status of the site. In any specific cell, only a very small fraction of a TF's cognate high-affinity genome sites are bound by a TF. A combination of properties is thought to determine the subset of sites occupied by a TF: its affinity, its ability to bind to sequences in contact with the nucleosome, and the nature of other factors or processes mediated by flanking sequences (Spitz and Furlong 2012). TFs possess two domains—a structured sequence-specific binding domain and an IDR commonly named the activation domain (AD). The AD facilitates downstream processes resulting in the initiation of transcription through association with IDRs of complexes such as Mediator (bound to Pol II), the Pol II holocomplex, BRD-type complexes, and components of the preinitiation complex (PIC) including basal transcription factors such as TFIID. As we shall describe, the AD also plays a key role in forming condensates involved in transcription.
TFs regulate the transcription of genes by binding to multiple short sequences (4–12 bp) in two types of elements—promoters and enhancers. Promoters encompass the transcriptional start sites for genes, and TFs bound to promoter-proximal regions can promote a basal level of transcription. Enhancer elements are generally bound by multiple cell-type-specific TFs, can be found at various distances from a gene, and can function in orientations independent of the promoter. Enhancer-bound TFs and their bound coactivator complexes contact promoter-bound complexes through DNA looping. Multiple enhancers can stimulate a promoter simultaneously. In fact, genes expressed at high levels in specific cell types commonly have clusters of enhancers ranging over 5–20 kbs called super enhancers (SEs), which can cooperatively stimulate transcription from a promoter (Whyte et al. 2013).
A property shared by enhancers and promoters is that both are sites of divergent transcription (Core et al. 2008; Seila et al. 2008; Sigova et al. 2013; Field and Adelman 2020). Enhancers produce short RNAs (eRNAs) radiating from their center that are generally degraded over minutes, although a small fraction is more stable, falling into the class of long noncoding RNAs (lncRNAs). Promoters are also sites of divergent transcription, producing unstable upstream antisense-transcripts (uaRNAs), and pre-mRNAs. Hence, the RNA environment of an active gene may consist of eRNAs, uaRNAs, lncRNAs, and both un-spliced and partially spliced RNA from gene sequences. Some of these RNA species have been shown to contribute to gene regulation (Kaikkonen and Adelman 2018), but the functions of most of these have yet to be deciphered.
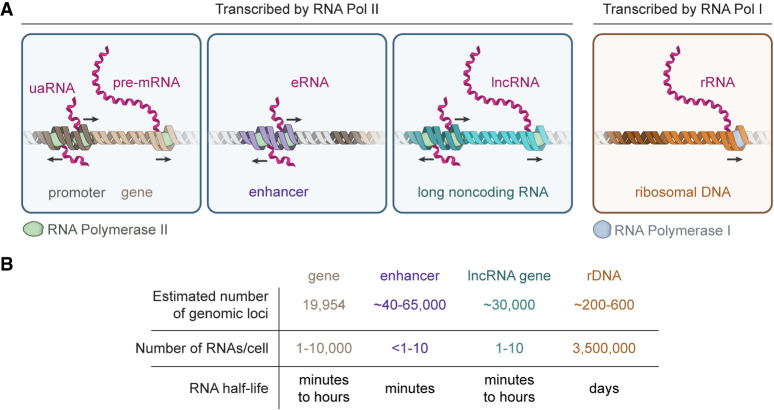
RNA molecules are key components of well-studied biomolecular condensates, including the nucleolus, nuclear speckles, paraspeckles, and stress granules (Fay and Anderson 2018; Roden and Gladfelter 2021). RNA molecules produced by RNA polymerase I and II are summarized in Figure 1. RNA polymerase I transcribes the rRNA precursor, which becomes a key component of the nucleolus, a well-studied multiphase biomolecular condensate. Similarly, RNA polymerase II transcribes both genes and their regulatory elements, producing eRNAs, uaRNAs, lncRNAs, and pre-mRNAs, all of which can become components of transcriptional and splicing condensates.
FIGURE 1.
RNA molecules produced by RNA polymerase I and RNA polymerase II. (A) Various types of RNA molecules synthesized by either RNA polymerase I or RNA polymerase II ([uaRNA] upstream antisense RNA, [eRNA] enhancer RNA, [lncRNA] long noncoding RNA, [rRNA] ribosomal RNA). (B) Properties of various RNA species—numbers and properties were derived from the following sources (McStay and Grummt 2008; Li et al. 2016; Schwalb et al. 2016; Hon et al. 2017; Frankish et al. 2019).
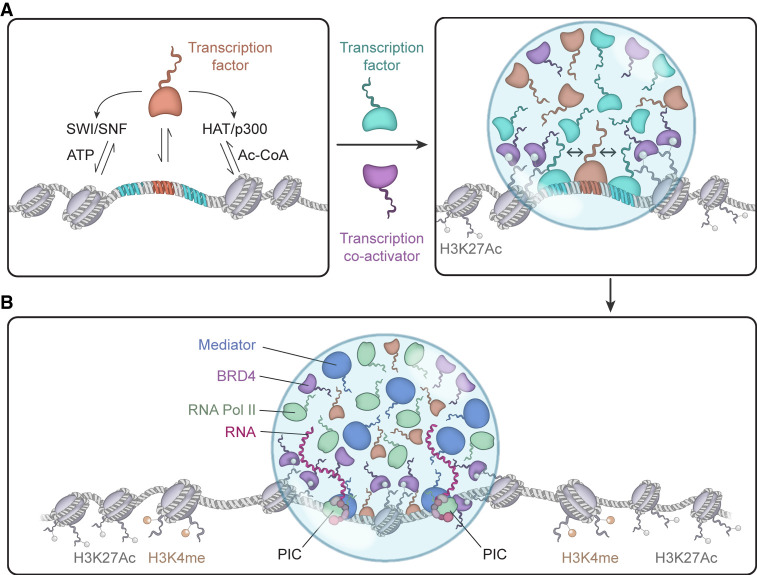
Figure 2 illustrates a simplified model of activation of an enhancer by a TF. In this model, a TF interacts with a high-affinity site, recruiting factors that mobilize nucleosomes such as the SWI/SNF complex, and factors that modify histones such as HAT/p300. This event likely occurs in concert with other TFs, which through their AD IDRs interact with coactivators such as Mediator and BRD4, and can be present in numbers sufficient to produce an assembly of molecules that, through weak multivalent interactions, can form a condensate (Boija et al. 2018; Cho et al. 2018; Sabari et al. 2018; Shrinivas et al. 2019). A subset of TFs is cell-type-specific master transcription factors that occupy SEs with their coactivators and are especially prone to the formation of large assemblies capable of undergoing phase separation. The genes encoding master transcription factors are themselves also controlled by SE-bound master transcription factors, creating auto-regulatory networks for feed-forward and backward regulation, thus regulating the gene expression programs that define cell state (Boyer et al. 2005). Although the study of SEs has provided much of the evidence for transcriptional condensate formation, the same principles should apply to any regulatory DNA element that effectively crowds sufficient numbers of transcription factors.
FIGURE 2.
A model for enhancer-driven condensate formation and gene activation. (A) Initial stages of transcription factor binding, which recruits chromatin remodelers, chromatin modifiers, and transcriptional coactivators. (B) Formation of transcriptional condensates by the concerted interaction of transcription factors, coactivators (BRD4, Mediator), RNA polymerase II, and modified chromatin. Compartmentalization and concentration of these factors promotes the assembly of the PIC.
The model in Figure 2 posits that a condensate consisting of enhancer DNA bound by TFs, together with cofactors, forms a dynamic condensate prior to transcription of eRNA, and that subsequent eRNA transcription then contributes to the composition and behavior of the condensate. The dynamic stability of this complex is pictured as being dependent upon the stability of the eRNA that typically has a half-life of minutes. The total activity of the enhancer might then reflect the composition and lifetime of the condensate, which provides the reservoir of RNA polymerase II molecules that engage in a burst of transcription (Rodriguez and Larson 2020).
With this model in mind, we can delve more deeply into the contribution of DNA sequences and DNA-associated proteins to the formation of condensate assemblies and then return to the subject of RNA. Modeling of TF binding to the IFN beta core enhancer suggests that a 50-bp portion of an enhancer element can be bound cooperatively by half a dozen TF molecules (Kim and Maniatis 1997). Several mechanisms have been proposed to contribute to this cooperativity. A canonical model is that the binding of a single TF molecule can alter the structure of adjacent DNA and create a higher affinity binding site for another TF molecule (Lambert et al. 2018). Furthermore, cooperativity can be obtained when the ADs of multiple TFs bind to coactivator complexes. Recent studies argue that the binding of multiple TFs to a cluster of cognate binding sites on DNA and interactions of their ADs with each other and IDRs of Mediator can result in favorable interactions that are sufficient to outweigh the entropic cost of concentrating these molecules relative to their surroundings, and thus push the assembly across a threshold for condensate formation (Boija et al. 2018; Shrinivas et al. 2019). In essence, the role of the enhancer is to crowd transcription factors to enable the favorable interactions required to exceed the threshold required for condensation. Such condensates, in turn, can facilitate further recruitment of TFs and other transcriptional machinery into the condensate compartment, even when these components are not bound directly to DNA. In this manner, condensate formation provides a general mechanism to account for the binding and clustering of transcription factors.
How might the enhancer condensate cause the burst of transcription from the promoter of a target gene? Interaction between an enhancer-associated and a promoter-associated condensate might occur through transient condensate interactions or fusion of the two condensates. We do not quite understand how an enhancer condensate “finds” its correct target promoter. Nonetheless, such interactions could provide the levels of active transcriptional apparatus needed to assemble multiple RNA polymerase II molecules on promoter DNA in rapid succession (Rodriguez and Larson 2020).
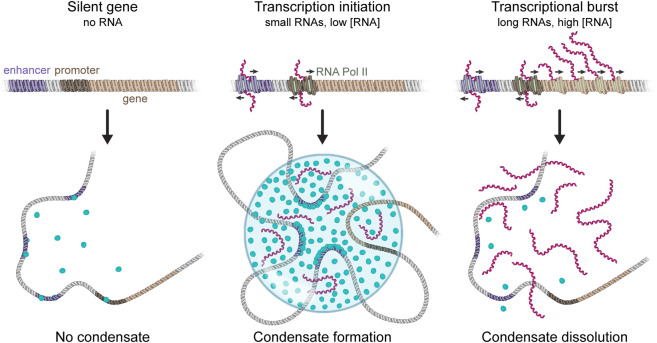
RNA can be a powerful regulator of condensates that are formed by electrostatic forces because of its high negative charge density (Maharana et al. 2018; Boeynaems et al. 2019). We have described evidence that transcriptional regulation involves a nonequilibrium feedback mechanism mediated by electrostatic forces whereby low levels of RNAs transcribed from enhancers initially stimulate transcription by promoting transcriptional condensate formation, whereas bursts of transcription of the enhancer-associated gene ultimately arrest the process through RNA-mediated condensate dissolution (Fig. 3; Henninger et al. 2021). The mechanism underlying this phenomenon is nonequilibrium regulation of complex coacervation, a type of liquid–liquid phase separation mediated by electrostatic interactions between oppositely charged polyelectrolytes (Overbeek and Voorn 1957; Srivastava and Tirrell 2016; Sing 2017; Lin et al. 2019). In the context of transcription, one of the polyelectrolytes is negatively charged RNA, and the other is comprised of transcriptional molecules with IDRs containing a net positive charge. Low levels of RNA can enhance condensate formation because of favorable interactions between oppositely charged species. Addition of RNA beyond the point where the opposite charges compensate each other, results in an excess of negatively charged RNA molecules that repel each other. Thus, high RNA levels can cause condensate dissolution (Banerjee et al. 2017; Milin and Deniz 2018). Condensate formation and subsequent dissolution with increasing RNA concentration is an example of reentrant phase behavior. The condensed fraction of protein peaks at the RNA concentration at which the charges between protein and RNA are balanced, while alteration of this charge balance in either direction decreases the condensed fraction. The low levels of short-lived RNA molecules transcribed from regulatory regions by RNA polymerase II at early times can enhance the formation of condensates controlling initiation. In contrast, the much higher local density of RNA produced by a transcriptional burst can later dissolve condensates. In this model, transcription is a nonequilibrium process that provides dynamic feedback through its RNA product, doing so by altering the charge balance in transcriptional condensates (Henninger et al. 2021).
FIGURE 3.
A nonequilibrium feedback mechanism for transcription mediated by RNA. Early stages of transcription produce low levels of RNA that help stimulate transcriptional condensate formation. Upon a burst of RNA synthesis during transcription elongation, the high levels of local RNA promote condensate dissolution. The stimulation and dissolution of transcriptional condensates by RNA define an auto-regulatory nonequilibrium feedback loop for transcription initiation and arrest.
RNA binding proteins are found associated with enhancers and promoters and are likely to further influence condensate behaviors. For example, the methyl-adenosine reader protein YTHDC1 promotes condensate formation at enhancers by binding enhancer RNAs, which harbor high levels of m6A methylation (Lee et al. 2021). Some transcription factors can bind RNA as well as DNA, and the presence of nascent RNA molecules elevates their occupancy of gene regulatory elements (Weintraub et al. 2017).
In this article, we have made the case that DNA elements crowd transcription factors to a threshold for condensate formation, creating a compartment that concentrates these and additional TFs and transcriptional molecules, and this is a general mechanism that is superimposed on other specific factors that have been described before. The RNA molecules transcribed from regulatory regions can facilitate the formation of condensates controlling initiation. Enhancer condensates can fuse or pair with promoter sites and produce the burst in transcription. RNA-mediated feedback can account for the termination of the burst in transcription.
Although the RNA-mediated feedback model warrants further investigation, it has implications for possible roles of lncRNAs near enhancers or promoters (Guttman et al. 2009; Ulitsky and Bartel 2013; Bhat et al. 2021). Interactions of such RNAs with enhancers could stimulate the rate of formation of a condensate resulting in an increase in the frequency and possible size of the associated transcriptional burst. Alternatively, accumulation of lncRNA in the vicinity of a gene could dissolve or limit the size of such a condensate by the feedback process outlined above. Finally, not discussed here, lncRNA could also influence transcription through interaction with CTCF bound to insulators, thus helping to stabilize boundaries, and limiting the genomic territory over which an enhancer or super-enhancer can operate (Hansen et al. 2019; Saldaña-Meyer et al. 2019).
ACKNOWLEDGMENTS
This work was supported by United States National Science Foundation grants PHY-1743900 and MCB-2044895 (to P.A.S., A.K.C., and R.A.Y.) and 1F32CA254216-01 (to J.E.H.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078997.121.
Freely available online through the RNA Open Access option.
REFERENCES
- Aumiller WM, Pir Cakmak F, Davis BW, Keating CD. 2016. RNA-based coacervates as a model for membraneless organelles: formation, properties, and interfacial liposome assembly. Langmuir 32: 10042–10053. 10.1021/acs.langmuir.6b02499 [DOI] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee PR, Milin AN, Moosa MM, Onuchic P, Deniz AA. 2017. Reentrant phase transition drives dynamic substructure formation in ribonucleoprotein droplets. Angew Chem Int Ed Engl 56: 11354–11359. 10.1002/anie.201703191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat P, Honson D, Guttman M. 2021. Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat Rev Mol Cell Biol 22: 653–670. 10.1038/s41580-021-00387-1 [DOI] [PubMed] [Google Scholar]
- Boeynaems S, Holehouse AS, Weinhardt V, Kovacs D, Lindt JV, Larabell C, Bosch LVD, Das R, Tompa PS, Pappu RV, et al. 2019. Spontaneous driving forces give rise to protein−RNA condensates with coexisting phases and complex material properties. Proc Natl Acad Sci 116: 7889–7898. 10.1073/pnas.1821038116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall'Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. 2018. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175: 1842–1855. 10.1016/j.cell.2018.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956. 10.1016/j.cell.2005.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, Cisse II. 2018. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361: 412–415. 10.1126/science.aar4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848. 10.1126/science.1162228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay MM, Anderson PJ. 2018. The role of RNA in biological phase separations. J Mol Biol 430: 4685–4701. 10.1016/j.jmb.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field A, Adelman K. 2020. Evaluating enhancer function and transcription. Annu Rev Biochem 89: 213–234. 10.1146/annurev-biochem-011420-095916 [DOI] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. 2019. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47: D766–D773. 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin M, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227. 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Hsieh TS, Cattoglio C, Pustova I, Saldaña-Meyer R, Reinberg D, Darzacq X, Tjian R. 2019. Distinct classes of chromatin loops revealed by deletion of an RNA-binding region in CTCF. Mol Cell 76: 395–411. 10.1016/j.molcel.2019.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henninger JE, Oksuz O, Shrinivas K, Sagi I, LeRoy G, Zheng MM, Andrews JO, Zamudio AV, Lazaris C, Hannett NM, et al. 2021. RNA-mediated feedback control of transcriptional condensates. Cell 184: 207–225. 10.1016/j.cell.2020.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. 2017. A phase separation model for transcriptional control. Cell 169: 13–23. 10.1016/j.cell.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJL, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, et al. 2017. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543: 199–204. 10.1038/nature21374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Adelman K. 2018. Emerging roles of non-coding RNA transcription. Trends Biochem Sci 43: 654–667. 10.1016/j.tibs.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Kim TK, Maniatis T. 1997. The mechanism of transcriptional synergy of an in vitro assembled interferon-β enhanceosome. Mol Cell 1: 119–129. 10.1016/s1097-2765(00)80013-1 [DOI] [PubMed] [Google Scholar]
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT. 2018. The human transcription factors. Cell 172: 650–665. 10.1016/j.cell.2018.01.029 [DOI] [PubMed] [Google Scholar]
- Lee JH, Wang R, Xiong F, Krakowiak J, Liao Z, Nguyen PT, Moroz-Omori EV, Shao J, Zhu X, Bolt MJ, et al. 2021. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol Cell 81: 3368–3385.e9. 10.1016/j.molcel.2021.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Rosenfeld MG. 2016. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet 17: 207–223. 10.1038/nrg.2016.4 [DOI] [PubMed] [Google Scholar]
- Lin Y, McCarty J, Rauch JN, Delaney KT, Kosik KS, Fredrickson GH, Shea JE, Han S. 2019. Narrow equilibrium window for complex coacervation of tau and RNA under cellular conditions. Elife 8: e42571. 10.7554/eLife.42571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillén-Boixet J, Franzmann TM, et al. 2018. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360: 918–921. 10.1126/science.aar7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B, Grummt I. 2008. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol 24: 131–157. 10.1146/annurev.cellbio.24.110707.175259 [DOI] [PubMed] [Google Scholar]
- Milin AN, Deniz AA. 2018. Reentrant phase transitions and non-equilibrium dynamics in membraneless organelles. Biochemistry 57: 2470–2477. 10.1021/acs.biochem.8b00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek JTG, Voorn MJ. 1957. Phase separation in polyelectrolyte solutions. Theory of complex coacervation. J Cell Physiol Suppl 49: 7–26. 10.1002/jcp.1030490404 [DOI] [PubMed] [Google Scholar]
- Roden C, Gladfelter AS. 2021. RNA contributions to the form and function of biomolecular condensates. Nat Rev Mol Cell Biol 22: 183–195. 10.1038/s41580-020-0264-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Larson DR. 2020. Transcription in living cells: molecular mechanisms of bursting. Annu Rev Biochem 89: 189–212. 10.1146/annurev-biochem-011520-105250 [DOI] [PubMed] [Google Scholar]
- Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. 2018. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361: eaar3958. 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldaña-Meyer R, Rodriguez-Hernaez J, Escobar T, Nishana M, Jácome-López K, Nora EP, Bruneau BG, Tsirigos A, Furlan-Magaril M, Skok J, et al. 2019. RNA interactions are essential for CTCF-mediated genome organization. Mol Cell 76: 412–422.e5. 10.1016/j.molcel.2019.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalb B, Michel M, Zacher B, Frühauf K, Demel C, Tresch A, Gagneur J, Cramer P. 2016. TT-seq maps the human transient transcriptome. Science 352: 1225–1228. 10.1126/science.aad9841 [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. 2008. Divergent transcription from active promoters. Science 322: 1849–1851. 10.1126/science.1162253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrinivas K, Sabari BR, Coffey EL, Klein IA, Boija A, Zamudio AV, Schuijers J, Hannett NM, Sharp PA, Young RA, et al. 2019. Enhancer features that drive formation of transcriptional condensates. Mol Cell 75: 549–561.e7. 10.1016/j.molcel.2019.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, et al. 2013. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci 110: 2876–2881. 10.1073/pnas.1221904110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing CE. 2017. Development of the modern theory of polymeric complex coacervation. Adv Colloid Interface Sci 239: 2–16. 10.1016/j.cis.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Spitz F, Furlong EEM. 2012. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13: 613–626. 10.1038/nrg3207 [DOI] [PubMed] [Google Scholar]
- Srivastava S, Tirrell MV. 2016. Polyelectrolyte complexation. In Advances in chemical physics (Rice SA, Dinner AR), pp. 499–544. Wiley, Hoboken, NJ. [Google Scholar]
- Ulitsky I, Bartel DP. 2013. lincRNAs: genomics, evolution, and mechanisms. Cell 154: 26–46. 10.1016/j.cell.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL, et al. 2017. YY1 is a structural regulator of enhancer-promoter loops. Cell 171: 1573–1588.e28. 10.1016/j.cell.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]