Abstract
The Enhancing NeuroImaging Genetics through Meta‐Analysis copy number variant (ENIGMA‐CNV) and 22q11.2 Deletion Syndrome Working Groups (22q‐ENIGMA WGs) were created to gain insight into the involvement of genetic factors in human brain development and related cognitive, psychiatric and behavioral manifestations. To that end, the ENIGMA‐CNV WG has collated CNV and magnetic resonance imaging (MRI) data from ~49,000 individuals across 38 global research sites, yielding one of the largest studies to date on the effects of CNVs on brain structures in the general population. The 22q‐ENIGMA WG includes 12 international research centers that assessed over 533 individuals with a confirmed 22q11.2 deletion syndrome, 40 with 22q11.2 duplications, and 333 typically developing controls, creating the largest‐ever 22q11.2 CNV neuroimaging data set. In this review, we outline the ENIGMA infrastructure and procedures for multi‐site analysis of CNVs and MRI data. So far, ENIGMA has identified effects of the 22q11.2, 16p11.2 distal, 15q11.2, and 1q21.1 distal CNVs on subcortical and cortical brain structures. Each CNV is associated with differences in cognitive, neurodevelopmental and neuropsychiatric traits, with characteristic patterns of brain structural abnormalities. Evidence of gene‐dosage effects on distinct brain regions also emerged, providing further insight into genotype–phenotype relationships. Taken together, these results offer a more comprehensive picture of molecular mechanisms involved in typical and atypical brain development. This “genotype‐first” approach also contributes to our understanding of the etiopathogenesis of brain disorders. Finally, we outline future directions to better understand effects of CNVs on brain structure and behavior.
Keywords: brain structural imaging, copy number variant, diffusion tensor imaging, evolution, genetics‐first approach, neurodevelopmental disorders, psychiatric disorders
The enhancing neuroimaging genetics through meta‐analysis (ENIGMA) copy number variant (CNV) and 22q11.2 Working Groups focus on gaining insight into how rare genetic variants affect human brain development, cognition, and behavior. The two ENIGMA working groups have collated CNV and brain‐imaging data from numerous individuals, gathered by numerous international research centers, and analyzed this data with standardized processing and analysis pipelines. Future directions for the ENIGMA CNV and 22q11.2 working groups are to analyze CNVs with larger sample sizes and more imaging modalities to better understand how rare genetic variants affect the brain, and their clinical and behavioral consequences.
1. INTRODUCTION
Classical twin and family studies show that most complex human traits are moderately to highly heritable, including brain structure and function (Hilker et al., 2018; Jansen, Mous, White, Posthuma, & Polderman, 2015; Teeuw et al., 2019). Since 2009, the Enhancing NeuroImaging Genetics through Meta‐Analysis (ENIGMA) Consortium (Thompson et al., 2014; Thompson et al., 2020) and other large‐scale consortia such as Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) (Psaty & Sitlani, 2013) have made significant progress in identifying common genetic variants associated with variability in brain structure (Adams et al., 2016; Grasby et al., 2020; Hibar et al., 2015; Hibar et al., 2017; Knol et al., 2020; Satizabal et al., 2019; Stein et al., 2012) and function (Smit et al., 2018) through so‐called genome‐wide association studies (GWAS). The relatively common variants (genotyped in large numbers on single nucleotide polymorphism [SNP]) arrays in these studies are typically associated with minor variations in magnetic resonance imaging (MRI)‐derived brain measures, thus highlighting the polygenic nature of structural neuroanatomy. So far, our understanding of the biology including the impact of individual, single variants is limited. Therefore, identifying genetic variants with larger effects on MRI‐derived measures of brain structure or function may provide a path to help deduce molecular mechanisms contributing to brain development and diseases.
Copy number variants (CNVs) (Figure 1) result from the deletion or duplication of a segment of the genome (Feuk, Marshall, Wintle, & Scherer, 2006) (a glossary of genetic terms is found in Table 1). CNVs represent a promising approach to study neurogenetic mechanisms shaping human behavior, cognition, and development. There are several rationales for this: certain rare, recurrent CNVs are associated with high risk (odds ratio up to 67.7) for a wide range of medical and behavioral consequences including brain disorders (Hastings et al., 2009) and some display large macroscopic effects on brain structure. The same CNV may confer elevated risk for several different (brain) disorders while reciprocal CNVs (Figure 1) at each end of the gene dosage response may be associated with the same disorder. Such clues gleaned from CNV research suggest that brain disorders are highly interlinked. Consequently, the study of rare CNV carriers may help us to understand the mechanisms behind not only rare isolated syndromes, but also of interrelated disorders, including the interaction between rare and common variants in shaping brain and disease as well as the intersection between somatic and brain disorders. This may allow us to identify both resilience and risk factors in common variants with potential to improve individual disease management.
FIGURE 1.
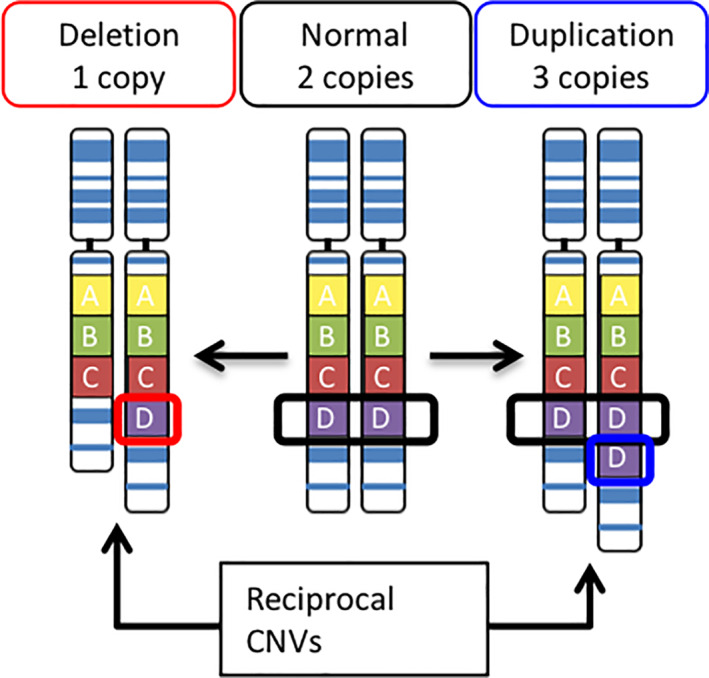
Copy number variants. CNV carriers may have a deletion (one copy of region D, red) or duplication (three copies of region D, blue) compared with the normal copy number (two copies of region D, black). Reciprocal CNVs are a deletion and duplication occurring at the same locus
TABLE 1.
Glossary table
| Term | Definition |
|---|---|
| Aneuploidy | The presence of an abnormal number of chromosomes in a cell. Examples are Down syndrome and monosomy X (Turner syndrome). |
| Anthropometric trait | A trait that describes body dimensions, such as head circumference, height, weight, girth, or body fat composition. |
| Array comparative genomic hybridization (aCGH) | A molecular cytogenetic method to detect copy number variants (CNVs) by comparing large fragments of DNA from a test individual to those from a reference sample. |
| Breakpoints (BP), chromosomal | A specific site of breakage, usually associated with a recurrent chromosomal abnormality. As in 16p11.2 distal CNV BP2‐BP3, where BP2‐BP3 refers to BP 2 to BP 3. For some CNVs, several low copy repeats (LCRs) in the region allow for multiple such BPs. |
| Copy number variant (CNV) | A type of structural genomic variation (Figure 1) that includes insertions, inversions, and translocations (Sharp, Cheng, & Eichler, 2006) in which segments of the genome are either deleted or duplicated. “Pathogenic” recurrent CNVs are of vastly different sizes and can span many genes (up to 90 for 22q11.2; McDonald‐McGinn et al., 2015) or just one (as in the case of NRXN1 CNVs; Lowther et al., 2017). Differences in BPs within the same locus add to the complexity of CNVs (e.g., in the 16p11.2 or 1q21.1 regions). In addition to recurrent CNVs, numerous ultra‐rare nonrecurrent, “one‐hit,” or single CNVs may also disrupt normal function. |
| CNV ‐ naming | A CNV is named based on its locus, that is, its specific position on the chromosome. The shorter arm of a chromosome is termed the p‐arm (petite = French for small), while the longer arm is the q‐arm. For example, the 16p11.2:16 = chromosome 16; p = p‐arm; 11 = region 1, band 1; 2 = sub‐band 2. Distal and proximal are used when two CNVs are present at the same locus (e.g., the 16p11.2 distal and proximal CNVs)—Distal is situated farther away from the centre of the chromosome (called the centromere) than the proximal which is closer to the centromere. |
| de novo | A genomic variation that occurs spontaneously in the offspring and thus is not inherited from the parents. |
| Fluorescence in situ hybridization (FISH) | A targeted molecular cytogenetic method used to detect and localize a chromosomal deletion or duplication using fluorescent probes corresponding to the DNA sequence targeted. |
| Gene dosage effect | The relationship between the number of copies of a gene and, for example, gene expression or brain volume. |
| Gene dose response | The effect of altering the amount of genetic material in a region/the magnitude of the response of an organism to changes in gene presence. |
| Genome assembly/build |
A reference genome assembly is a string of digital ATCG nucleotides representing the complete set of genes from an organism. It is assembled through a consensus of the genomes of different donors. The most recent human genome assembly, termed GRCh38 (also called “build 38”), was released in 2013 and is derived from 13 anonymous donors. Earlier human reference genome versions include: GRCh37 or hg19 (2009), NCBI36 or hg18 (2006), NCBI35 or hg17 (2004), and NCBI34 or hg16 (2003) |
| Genetics‐first approach | A strategy used in epidemiological studies to associate specific genotypes (such as a specific CNV) with apparent clinical phenotypes of a complex disease or trait. Also called “genotype‐first.” |
| Genotyping | The process of determining differences in the genetic make‐up (genotype) of an individual by examining the individual's DNA sequence using biological assays. The term is often used to refer to the identification of SNPs through (SNP) genotyping arrays. |
| Genetic heterogeneity | The same or similar phenotypes caused by different genetic mechanisms. |
| Idiopathic | Any disease or condition for which the cause is unknown. |
| Insertion | A structural variant that involves a mutation through the addition of genetic material to a chromosome. |
| Inversion | A structural variant in which a segment of a chromosome is reversed end to end. |
| Low copy repeats (LCRs) | Highly homologous sequence elements within the eukaryotic genome arising from segmental duplication and predisposing the genome to nonallelic homologous recombination (NAHR). LCRs mediate many of the chromosomal rearrangements that underlie genomic disorders by predisposition to recombination errors. |
| Multiplex ligation‐dependent probe amplification (MLPA) | A molecular cytogenetic method used to identify copy number variants. It is a variation of the multiplex polymerase chain reaction that permits amplification of multiple targets with only a single primer pair. |
| Nonallelic homologous recombination (NAHR) | A form of homologous recombination that occurs in two pieces of DNA that have similar sequences, often as a result of the presence of low copy repeats (LCRs). NAHR can occur within the same LCR or in an alternative LCR, and can result in a variety of chromosomal rearrangements, including deletion, duplication, translocation, and inversion. The presence of LCRs and resultant NAHR is believed to play a key role in molecular evolution in primates, as a mechanism involved in rapidly changing gene dosage (which may be advantageous) and even in the creation of new genes. |
| Noncarrier | In the context of CNVs, this is usually defined as an individual who does not carry the particular CNV being studied. |
| Penetrance | The proportion of people with a particular genotype/CNV who have any signs or symptoms of the disease. |
| Pleiotropy | The phenomenon whereby one allele (or a pair of alleles) influences multiple, independent phenotypes. |
| Polygenic trait | A phenotype that is influenced by multiple genetic variants at different genomic sites. |
| Rare CNV | Typically defined as a CNV with <1% frequency in the population. |
| Reciprocal CNVs | Deletions and duplications that occur at the same locus, usually flanked by LCRs. |
| Recurrent CNVs | CNVs that occur as spontaneous de novo events at the same sites in the genome repeatedly in unrelated individuals due to the presence of flanking low copy repeats, or LCRs) (Hastings, Lupski, Rosenberg, & Ira, 2009). In other words, they occur de novo in the first individual, and hence are not observed in the CNV carrier's parents but are potentially inherited in subsequent generations. |
| Single nucleotide polymorphism (SNP) | The substitution of a single base (A, T, C, or G) for another base at a specific genetic location that occurs in at least 1% of the population. A SNP may or may not have functional consequences on gene expression. |
| SNP genotyping array | DNA microarray used to detect SNPs within a population. |
| Somatic disease | A disease relating to the body, especially as distinct from the mind. |
| Translocation | A structural variant in which a portion of a chromosome breaks from its original location and reattaches to a different location in the genome. |
Despite their clinical relevance and evolutionary importance (Lauer & Gresham, 2019), effects of rare CNVs on human brain structure are poorly understood partially due to the rarity of these CNVs, which pose challenges in data collection. Several consortia including the 16p11.2 European consortium (Maillard et al., 2015) and Simons VIP/Searchlight (Qureshi et al., 2014) as well as individual projects (Meda, Pryweller, & Thornton‐Wells, 2012; Reiss et al., 2004; Stefansson et al., 2014; Ulfarsson et al., 2017) have addressed this. In addition, under the umbrella of ENIGMA, two groups—the ENIGMA 22q11.2 Deletion Syndrome Working Group (22q‐ENIGMA WG) and the ENIGMA‐CNV Working Group (ENIGMA‐CNV WG)—are devoted to increasing knowledge of the effect of CNVs on the brain. The 22q‐ENIGMA WG was founded in 2014 based on an extensive sample of 22q11.2 deletion carriers with brain MRI data (Figure 2). The ENIGMA‐CNV WG was formed in 2015 to study rare CNVs beyond the 22q11.2 locus and collated previously collected neuroimaging samples with genome‐wide individual genotyping (Figure 2). Both WGs aim to address some of the core limitations, especially those relating to low power and replicability, of prior brain imaging CNV studies and to foster collaborative discovery.
FIGURE 2.
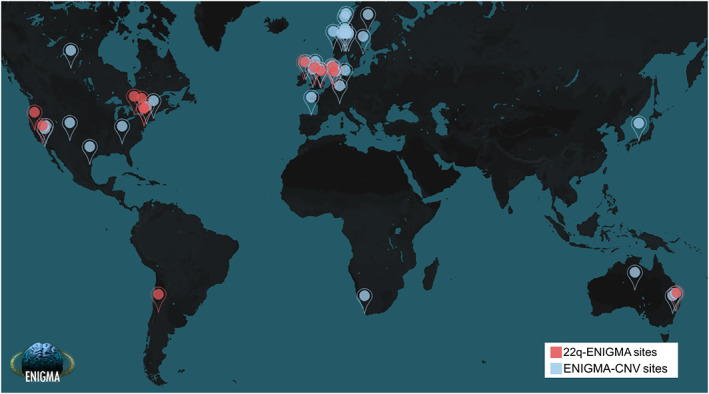
World map of the ENIGMA‐CNV and 22q‐ENIGMA WG study sites. A full list of participating cohorts and members for ENIGMA‐CNV and 22q‐ENIGMA may be found at the respective webpages: http://enigma.ini.usc.edu/ongoing/enigma‐cnv/enigma‐cnv‐co‐authors/ and http://enigma.ini.usc.edu/ongoing/enigma‐22q‐working‐group/22qwg/. Both working groups consist of international teams of clinicians, neuroscientists, engineers, bioinformaticians, statisticians, computer scientists, and geneticists who pool their resources to conduct large‐scale neuroimaging studies of CNVs
In this review, we focus on the work done by the ENIGMA WGs on CNVs. We first outline the significance of CNVs for elucidating genetic mechanisms underlying brain development and disease. We then describe the data collection, study design, and analytical methods used by the two WGs. Next, we review key findings of the 22q‐ENIGMA and ENIGMA‐CNV WGs on the 22q11.2, 16p11.2 distal, 15q11.2, and 1q21.1 CNVs and include results from other relevant work that has helped us to understand effects of CNVs on brain structure. We then discuss emerging principles that may govern how rare CNVs affect the brain. Finally, we summarize future plans to understand the neurobiology of CNVs for a broader range of brain phenotypes.
2. CNVS: HIGHLY RELEVANT FOR RISK FOR NEURODEVELOPMENTAL DISORDERS AND DRIVERS OF HUMAN BRAIN EVOLUTION
2.1. The role of CNVs in neurodevelopmental disorders
CNVs may account for up to 13% of the genome (Stankiewicz & Lupski, 2010), with the vast proportion being common across individuals and without any known negative effects (Iafrate et al., 2004). However, some CNVs can disrupt normal function in humans, causing psychiatric and neurodevelopmental disorders (NDs), somatic and neurological diseases, as well as cancer (Hastings et al., 2009). For instance, individuals with rare, recurrent CNVs are at much higher risk of NDs, including autism spectrum disorders (ASD), epilepsy, schizophrenia (SCZ), and intellectual disability (ID) (Kirov, Rees, & Walters, 2015) as well as Alzheimer's disease and other neurodegenerative diseases (Cervera‐Carles et al., 2016; Cuccaro, De Marco, Cittadella, & Cavallaro, 2017). De novo and inherited CNVs combined have been estimated to explain ~15% of neurodevelopmental disorder cases (Wilfert, Sulovari, Turner, Coe, & Eichler, 2017). Likewise, at least 9% of all ASD (Munnich et al., 2019) and 2.5% of SCZ cases carry a known pathogenic CNV (Rees et al., 2014). CNV carriers also have high rates of additional comorbid medical conditions (Crawford et al., 2018) and some display altered anthropometric traits (Mace et al., 2017; Owen et al., 2018). This high disease rate is often mirrored by reduced fecundity (Stefansson et al., 2014). Thus, altogether, a high impact CNV may represent a lifelong burden for the affected individuals and their caregivers, leading to substantial personal and societal costs.
The high odds ratio (>10) (Marshall et al., 2017) for neurodevelopmental disorders associated with specific CNVs is in contrast to the highest effect sizes identified for individual common genetic variants in SCZ (OR = 1.09; SCZ WG of the Psychiatric Genomics Consortium, 2014), bipolar disorder (OR = 1.13; Stahl et al., 2019), ASD (OR = 1.25; Grove et al., 2019), major depressive disorder (OR = 1.05; Wray et al., 2018) and attention deficit hyperactivity disorder (ADHD; OR = 1.12; Demontis et al., 2019). This has spurred considerable interest in studying CNVs as a genetics‐first approach to understand mechanisms of abnormal brain development as well as risk for disorders such as SCZ (Kirov et al., 2015), ASD (Stessman, Bernier, & Eichler, 2014) in addition to other medical comorbidities (Pierpont et al., 2018).
Such interest has been further encouraged by the diversity of CNVs: There are at least 93 known clinically relevant recurrent rare CNVs (Kendall et al., 2016), each with its own clinical profile/consequences (Girirajan et al., 2012; Rosenfeld & Patel, 2017). Some recurrent CNVs have moderate to small effects, for example, the more common 15q11.2 deletion, while others have large effects with near‐complete penetrance, such as the very rare Williams syndrome/7q11.23 deletion. Such high penetrance is positively correlated with the proportion of de novo occurrence in the population (Rosenfeld, Coe, Eichler, Cuckle, & Shaffer, 2013). In contrast, CNVs with small effects tend to be inherited more often, and may be identified in seemingly asymptomatic parents. Thus, different CNVs allow insight into different clinical risk profiles and their potential mechanisms.
Likewise, a specific CNV lacks diagnostic specificity and offers hugely diverse pleiotropic outcome. For instance, the same CNV may be associated with congenital defects, SCZ, ASD, ID, epilepsy, or early‐onset Parkinson's disease (Bijlsma et al., 2009; Shen et al., 2011; Stefansson et al., 2014; Tabet et al., 2012) as exemplified by the 22q11.2 deletion syndrome (22q11DS), which has been associated with all the above conditions (Boot et al., 2018; Butcher et al., 2013; Gudmundsson et al., 2019; Marshall et al., 2017). Furthermore, CNVs may lead to multiple disorders in the same individual (known as multimorbidity). For example, individuals with 22q11DS who have a psychiatric disorder are at increased risk for other psychiatric disorders, as well as motor coordination problems (Cunningham et al., 2018) and sleep problems (Moulding et al., 2020). Thus, few traits show evidence of genotypic specificity (Chawner et al., 2019; Cunningham, Hall, Einfeld, Owen, & van den Bree, 2020; Girirajan et al., 2012; Rosenfeld & Patel, 2017).
Harmful effects of CNVs may be partially explained by altered expression of genes in the affected region due to the difference in gene copy number, leading to higher or lower transcription levels (Hastings et al., 2009). This phenomenon is sometimes referred to as the “gene dosage effect” or “dose response per copy number.” CNVs can also modulate expression of genes outside of the region deleted or duplicated, either by addition or removal of regulatory elements, or by modifications of the 3D structure of the genome (Spielmann, Lupianez, & Mundlos, 2018). Thus, CNVs is a means for studying the effects of gene dosage alterations for many genes at a time and how these shape neurodevelopmental disease and brain structure.
The 22q11.2 region is an interesting region in this regard as it displays dose response with regard to SCZ risk: the deletion is associated with increased risk (Schneider et al., 2014) but the duplication appears to be associated with decreased risk (Marshall et al., 2017; Rees et al., 2014). In contrast, reciprocal CNVs may also carry risk for related disorders. For instance, the 22q11.2 deletion and duplication both confer high risk of ADHD (Gudmundsson et al., 2019), Likewise, the reciprocal 16p11.2 distal and proximal (Loviglio et al., 2016; Niarchou et al., 2019), 1q21.1 distal (Bernier et al., 2016; Mefford et al., 2008) and 22q11.2 loci (Lin et al., 2020) all confer risk of ASD. In this context, it is noteworthy, that population‐based studies overall suggest milder effects of duplication (vs. deletion) CNVs on cognition (Männik et al., 2015), which could suggest differences in the severity of, for example, ID in the reciprocal CNVs. Thus, CNVs allow investigations into how reciprocal CNVs at each end of the gene dosage response can cause both a “gene dose response” for disease risk but also similar disease risk.
The ultimate phenotype of a CNV likely depends on both environmental impacts and genetic background (Cleynen et al., 2020; Huguet et al., 2018; Kirov et al., 2014). Such influencing genetic factors likely include protective or disease‐enhancing genes located within the CNV region, or elsewhere in the genome. Educational attainment as a proxy for parental intelligence, for example, seems to modulate intellectual impairment related to a 22q11.2 deletion (Klaassen et al., 2016), indicating interplay of the CNV with common variants. The interactions between genetic factors as well as the environment will be key to a better understanding of CNV‐mediated disease risk. Investigations of interactions between CNVs and polygenic risk score as a proxy for common variants have already been initiated in disorders such as SCZ (Bergen et al., 2018; Davies et al., 2020; Tansey et al., 2016) and ADHD (Martin, O'Donovan, Thapar, Langley, & Williams, 2015). Thus, studies of CNV carriers may help disentangle the effects of the combination of rare and common variants as well as environment in shaping neurodevelopmental disease risk.
2.2. The role of CNVs in brain evolution
Changes in DNA—including CNVs—occur naturally and are a part of the evolutionary process and adaptation (Hastings et al., 2009) in all living organisms including animals and plants (Lauer & Gresham, 2019). Gene duplications provide a driving force in evolution (Bailey & Eichler, 2006) by allowing for the adaptation of new gene copies while maintaining the function of the old gene copy (Innan & Kondrashov, 2010). Even so, they also put the next generation at risk for re‐arrangements due to the presence of low copy repeats (LCRs), long clusters of related gene sequences with high sequence identity, that arise via duplication (Harel & Lupski, 2018). Interestingly, in the human and great ape lineage, there are proportionately more deletions and duplications observed in comparison to other mammals (Hahn, Demuth, & Han, 2007).
Some of these duplications have been hypothesized to be major driving forces in the rapid evolution of the human and great ape lineages (Dennis & Eichler, 2016) including brain enlargement and have given rise to entirely new human‐specific genes with novel characteristics. Examples include SRGAP2 (three copies in humans, one in nonhuman primates) (Dennis et al., 2012), NOTCH2NL (three–four copies in humans, one in primates) (Fiddes et al., 2018; Suzuki et al., 2018) and BOLA2 (Giannuzzi et al., 2019). The NOTCH2NL and SRGAP2 genes are particularly interesting in the context of brain development: The NOTCH2NL genes confer delayed neuronal differentiation and increased progenitor self‐renewal (Fiddes et al., 2018; Suzuki et al., 2018), their occurrence coincides with a time just before or during the early stages of the expansion of the human cortex and they have thus been hypothesized to have contributed to the rapid evolution of the human neocortex. Likewise, transient overexpression of SRGAP2C in culture and in vivo leads to human‐specific features, including neoteny of dendritic spine maturation, promotion of longer spines at a greater density, and sustained radial migration in the developing mouse neocortex. Thus, duplications in human evolution appear to have shaped the formation of the human brain.
To date, discoveries on CNV‐related phenotypes have been hindered by the low frequency of each single pathogenic CNV in the general population (from 1 in ~400 to 1 in ~50,000 for recurrent CNVs; Kendall et al., 2016; Stefansson et al., 2014), making it challenging to collect sufficiently large, well‐powered samples. Even so, new technologies have moved the field forward considerably during the last 10 years.
3. NEW TECHNOLOGY—BIG DATA ANALYTICS IN GENETICS AND IMAGING
3.1. Genotyping and CNV calling
Among the earliest genetic syndromes to be detected were those caused by aneuploidies, such as trisomy 21 (Down's syndrome) and monosomy X (Turner syndrome). Testing for such genetic syndromes was incorporated into clinical practice in the 1950s and involved counting the number of chromosomes per cell, a technique known as karyotyping (Durmaz et al., 2015). Since then, a number of techniques including targeted fluorescence in situ hybridization (FISH), genome‐wide array comparative genomic hybridization (aCGH) and SNP arrays have allowed detection of smaller aberrations including CNVs down to ~10 kb (Nowakowska, 2017).
In 2004, two landmark studies (Iafrate et al., 2004; Sebat et al., 2004) showed that submicroscopic variations (<500 kb in size) in DNA copy number are widespread across the human genome. In the last 10–15 years, it has become possible to obtain genome‐wide CNV “calls” for many individuals through massive population‐scale SNP genotyping followed by demanding computational analyses. Likewise, clinical investigations and detection have become standard for some disorders. These new developments in technology have been vital for the increased knowledge of CNV carriers obtained in recent years.
3.2. Neuroimaging as a tool to study CNV effects on the brain
For some time, clinical observations have indicated characteristic macroscopic brain alterations in specific CNV carriers: reciprocal carriers of 16p11.2 and 1q21.1 distal CNVs (Brunetti‐Pierri et al., 2008) display macro‐ and microcephaly, respectively, and 17p13.3 deletion carriers (causing Miller‐Dieker syndrome) present with lissencephaly (smooth cortex because of lack of development of gyri and sulci) (Blazejewski, Bennison, Smith, & Toyo‐Oka, 2018). Thus, clinical data indicate that rare CNV carriers can teach us valuable lessons about brain development.
More detailed mapping through MRI—a reliable, noninvasive technique for mapping macro brain structure and functional consequences—have dived deeper and shown wide‐reaching phenotypic impacts of CNVs with substantial structural and functional alterations in the brain; for example, 22q11.2 deletions and duplications (Lin et al., 2017; Sun et al., 2018), 7q11.23 deletion (Fan et al., 2017; Meda et al., 2012), 15q11.2 (Silva et al., 2018; Stefansson et al., 2014; Ulfarsson et al., 2017; van der Meer et al., 2020) and 16p11.2 proximal (Maillard et al., 2015; Martin‐Brevet et al., 2018; Qureshi et al., 2014) and 16p11.2 distal CNVs (Sonderby et al., 2018). The 22q‐ENIGMA and ENIGMA‐CNV WGs have contributed significantly to this effort, by combining already collected cohorts of clinically ascertained samples on 22q11.2DS (Ching et al., 2020; Sun et al., 2018), as well as primarily non‐clinically ascertained samples for brain CNV research, so far publishing on 16p11.2 distal, 15q11.2 BP1‐BP2 and 1q21.1 distal (Sønderby et al., 2021; Sonderby et al., 2018; van der Meer et al., 2020).
3.3. ENIGMA‐standardized image processing
A prerequisite for large imaging studies is the standardization of approaches. The publicly available ENIGMA imaging processing and analysis protocols make it possible to consistently extract brain measures, and perform quality assessment and statistical modeling across many international research centers (http://enigma.ini.usc.edu/protocols).
ENIGMA processing pipelines applied in the ENIGMA‐CNV (point 1) and 22q‐ENIGMA WG (points 1–3) include:
-
ROI brain measures: Subcortical and cortical regions of interest (ROI) measures are extracted with FreeSurfer software (Fischl et al., 2002).
FreeSurfer subcortical volumes (eight gross volumetric features for both hemispheres) including thalamus, hippocampus, amygdala, caudate, putamen, pallidum, nucleus accumbens, lateral ventricles, and estimated intracranial volume (ICV) (as measured in the ENIGMA2 GWAS; Hibar et al., 2017).
Global and regional cortical thickness and cortical surface area measures (34 features for each hemisphere for both) based on the Desikan–Killiany atlas (Desikan et al., 2006; as measured in the ENIGMA3 cortical GWAS; Grasby et al., 2020).
-
Vertex‐wise brain shape measures: ENIGMA Subcortical Shape and FreeSurfer protocols are used to derive local thickness and surface area measures across cortical and subcortical structures.
Subcortical vertex‐wise shape modeling uses the ENIGMA Shape Analysis Pipeline to more finely map the spatial distribution of volumetric alterations across subcortical structures. The method derives local thickness and surface area expansion/contraction metrics for up to 2,502 vertices along the aforementioned subcortical ROIs, mapping potentially complex morphometric alterations (Ching et al., 2020).
Cortical thickness and surface area metrics extracted with FreeSurfer across tens of thousands of cortical vertices provides fine mapping of CNV‐related subregional cortical alterations (Sun et al., 2018).
Diffusion‐weighted imaging and white matter microstructure: The ENIGMA DTI protocol uses the tensor model and standardized DTI template to calculate fractional anisotropy (FA), mean diffusivity (MD), radial diffusivity (RD), and axial diffusivity (AD) in the Tract‐Based Spatial Statistics (TBSS) framework (Jahanshad et al., 2013; Smith et al., 2006); values for each measure are averaged along the skeleton of each ROI from the Johns Hopkins University White Matter Atlas (JHU‐ICBM‐DTI‐81) and analyzed in brain space (Mori et al., 2008). 18–25 ROIs are typically included (Kochunov et al., 2020).
These standardized feature extraction pipelines lead to more unbiased investigations of brain metrics, in that they are consistently applied across many data sets and cohorts. This approach improves upon traditional meta‐analyses, which often attempt to combine published effect sizes derived from different processing and analysis protocols. By pooling data derived using standard image processing pipelines in a coordinated effort, the ENIGMA‐CNV and 22q‐ENIGMA WG studies boost statistical power by incorporating data sets that may have been underpowered to detect brain effects on their own. The standardization of protocols, now being applied in large prospective studies such as UK Biobank (Alfaro‐Almagro et al., 2018), allows large‐scale comparison of brain measures and profiles of disease effects across studies to better characterize common and distinct brain signatures across CNVs and major brain disorders from independently collected study samples.
4. THE 22Q‐ENIGMA WG: DEEP DIVE INTO A HIGHLY PENETRANT GENETIC RISK FACTOR FOR PSYCHOSIS
22q11DS is a prominent example of a highly penetrant, recurrent CNV for which detailed phenotypic data has been collected in multiple cohorts worldwide (Gur et al., 2017). The main goals of the 22q‐ENIGMA WG are threefold: (a) map robust and reproducible multimodal brain markers of 22q11DS in large cohorts; (b) investigate how genetic and neuroanatomic variability relate to variability in phenotypic expression; and (c) determine convergence (and/or divergence) of neuroanatomical effects of high‐penetrance CNV versus behaviorally defined neuropsychiatric disorders.
The 22q‐ENIGMA WG has built an international network of research programs and has centralized data from the largest available cohorts of 22q11.2 deletion carriers with brain imaging. The 22q‐ENIGMA WG consists of 12 international sites (Figure 2), and has analyzed data from over 533 individuals with molecularly confirmed 22q11.2 deletions and over 350 healthy controls. In addition, the UCLA lead site has collected 40 individuals with 22q11.2 duplications through a novel initiative. Age, sex, deletion size, IQ, history of psychosis and medication usage, along with structural and diffusion‐weighted imaging measures are collected and shared centrally for standardized processing and analysis (Figure 3).
FIGURE 3.
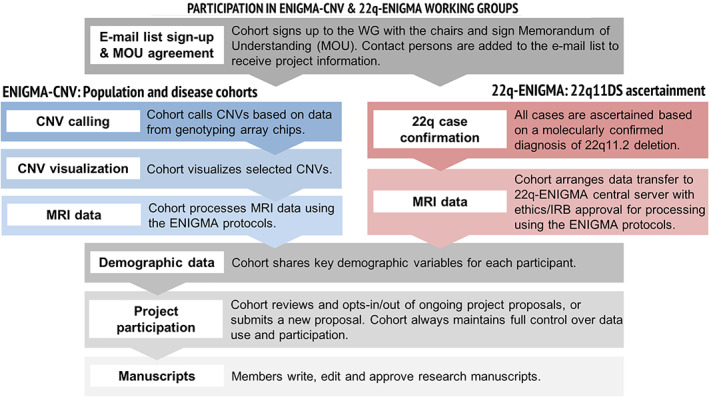
The overall procedure for participation in ENIGMA‐CNV and 22q‐ENIGMA
4.1. Collection of CNV information
A molecularly confirmed diagnosis of 22q11.2 deletion is necessary for study inclusion. The most common deletion subtype, known as the LCR22A‐LCR22D or A‐D deletion, is found in ~85% of cases and involves the loss of ~2.6 megabases (Mb) of DNA. A smaller 1.5 Mb deletion—called the LCR22A‐LCR22B or A‐B deletion—is the next most common subtype, found in ~10% of cases (McDonald‐McGinn et al., 2015).
4.2. Demographic data harmonization
History of psychotic disorder is established by a trained mental health professional at each 22q‐ENIGMA site via a structured diagnostic interview, collateral information, and medical records. A cross‐site reliability procedure is conducted by two investigators to independently review representative cases from each site and to ensure diagnostic reliability across sites (Gur et al., 2017).
5. THE ENIGMA‐CNV WORKING GROUP: STANDARDIZED DATA COLLATION, PROCESSING AND ANALYSIS TO EMPOWER LARGE‐SCALE STUDIES
The primary goal of the ENIGMA‐CNV WG is to identify CNVs that significantly influence the brain globally and regionally to gain insight into the neurobiology of CNVs. The WG follows the main philosophy of the wider ENIGMA Consortium, which is to leverage existing legacy data sets to their full potential by combining samples using standardized processing. Notably, few of the research groups in ENIGMA‐CNV could have performed well‐powered CNV‐brain imaging studies on their own due to the low prevalence of individual CNVs.
5.1. Data collection and coordination
The large‐scale international nature of ENIGMA requires coordination of data originally collected with vastly different study designs, so initial analyses tend to be simple, followed by more complex analyses. From the beginning, ENIGMA‐CNV, rather than focusing on predefined selection of CNVs, chose a pragmatic approach driven by data availability. One key to success is a unified approach across studies for CNV calling, imaging analysis and quality control (Figure 3), given the differences in original cohort data collection and study design.
5.2. Standardized CNV calling and visualization across cohorts
The low frequency of recurrent CNVs makes a mega‐analysis approach preferable to the original ENIGMA meta‐analysis approach (Boedhoe et al., 2018). Given the lack of experience in genetic analysis, in particular CNV calling, for many participating cohorts, ENIGMA‐CNV first developed an easy‐to‐follow protocol for CNV calling. Many SNP genotyping arrays exist that vary in the number of SNPs included and their coverage of the genome. The often nonuniform distribution of tagged SNPs across the genome means that there may be limited coverage in regions with segmental duplications or complex CNVs (Carter, 2007). Consequently, larger CNVs (> 500 kb) can be reliably detected by microarrays from most platforms, whereas variability between platforms is greater for smaller CNVs (10–100 kb). A number of different CNV calling methods exist (Pinto et al., 2011). PennCNV (Wang et al., 2007), a widely used CNV calling software platform (Macé et al., 2016), was chosen since it accommodates a wide selection of SNP‐based arrays (e.g., Affymetrix and Illumina) and is user friendly and fast (Macé et al., 2016)—a key advantage at a time when the number of available samples increases at an unprecedented rate.
Most participating cohorts call CNVs themselves. Alternatively, the ENIGMA‐CNV WG does the calling on their behalf based on raw genotype information provided by the respective participating cohort. To address regulatory issues, the CNV calling protocol includes a de‐identification step. Following CNV calling, individual cohorts follow a CNV visualization protocol based on the iPsychCNV R package (https://github.com/mbertalan/iPsychCNV/). Finally, the WG analysts do the final, manual QC of the visualized CNVs to ensure harmonized CNV calls across cohorts.
Cohorts with smaller sample sizes should feel encouraged to join the ENIGMA‐CNV WGs as the number and nature of CNV carriers is unknown prior to CNV calling. As the project has developed, samples verified by alternative genotyping such as aCGH, Multiplex Ligation‐dependent Probe Amplification (MLPA) or FISH have also been included in the study. These typically constitute clinical samples, so the corresponding noncarriers (used as controls) have typically only been checked for presence or absence of the CNV of interest.
5.3. Demographic data
A minimal number of demographic metrics are collected, including age at brain scan, sex, diagnosis (if applicable), scanner site, and multidimensional scaling (MDS) factors (when available) from the analysis of population structure in the genome‐wide data.
5.4. Study and analysis design
In disease studies, controls are typically defined at the outset of the individual studies. This contrasts to ENIGMA‐CNV where controls, dubbed noncarriers, are individuals who do not carry the particular CNV being studied nor any other potentially pathogenic CNV (as defined by a precompiled list; Kendall et al., 2016). The latter allows a truly blinded sampling as neither the recruiters, nor the participants, knew CNV status at the time of the analysis except for the few clinically ascertained carriers.
For primary data analyses, ENIGMA‐CNV applies both a linear regression, to test the effect of the CNV per copy number of the region in question, that is, the dose response, and a t test to compare the pairs of groups (deletion or duplication vs. noncarriers or deletion vs. duplication carriers). Imaging data are adjusted for age at brain scan, sex, and scanner site—both with and without adjusting for ICV. The number of noncarriers in ENIGMA‐CNV is an order of magnitude larger than carriers. This provides the opportunity to perform an estimate of the effect of the CNVs in comparison to the overall population. Separate “sensitivity” analyses are performed including a matched analysis (matching each carrier with a noncarrier based on, e.g., age, sex, affection status, and ICV) as well as separate analyses that take into account ancestry information (MDS factors) and diagnoses (if known). These sensitivity analyses allow testing of the robustness of the results in selected subsets of the sample.
5.5. Overview of the ENIGMA‐CNV working groups
The ENIGMA‐CNV sample currently comprises a total of 38 cohorts (Figure 2) with genotyping and MRI data comprised of core ENIGMA‐CNV based on clinical (mostly case–control) and population studies as well as publicly available data sets (currently the UK Biobank) and represent a broad spectrum of CNVs (Table 2). Part of the strength of the ENIGMA‐CNV sample, compared to clinical CNV studies, is a higher proportion of high‐functioning CNV carriers given the high proportion of nonclinical “volunteer”/population samples (~70% of core ENIGMA‐CNV). This advantage comes with the downside of an under‐representation or absence of CNVs with high penetrance, such as individuals with Prader‐Willi/Angelman syndrome (15q11.2–13.2 deletion carriers), Sotos syndrome (5q35 deletion), the 22q11.2 deletion (Table 2) as well as severely functionally affected individuals carrying CNVs with a broad phenotypic spectrum, such as 1q21.1 distal, 16p11.2, and 15q11.2 CNVs. The under‐representation is partially compensated by fruitful collaboration with more clinically focused studies such as the 16p11.2 European consortium (Martin‐Brevet et al., 2018) and the Cardiff ECHO‐DEFINE/IMAGINE‐study (Chawner et al., 2019) and case–control studies, for example, epilepsy, SCZ, bipolar disorders, and ADHD. Consequently, ~10% of individuals in the core ENIGMA‐CNV sample has a known clinical diagnosis. Thus, the current ENIGMA‐CNV sample represents extensive parts of the phenotypic spectrum of CNV carriers. We continue to add samples to broaden the scope of the studies.
TABLE 2.
Numbers of selected deletion (del) and duplication (dup) carriers and noncarriers (nc) in the current ENIGMA‐CNV sample including UK Biobank
| CNVs of interest | Deletion carriers | Noncarriers | Duplication carriers |
|---|---|---|---|
| 1q21 proximal (TAR) | 22 | 69 | |
| 1q21.1 distal | 34 | 25 | |
| 2p16.3 (NRXN1) | 1 | ||
| 3q29 | 1 | ||
| 10q11.22–23 | 5 | 1 | |
| 15q11.2 | 167 | 225 | |
| 15q11.2‐q13 | 1 | ||
| 16p11.2 proximal | 7 | 15 | |
| 16p11.2 distal | 10 | 12 | |
| 16p11.2 distal proximal a | 3 | 3 | |
| 16p12.1 | 25 | 25 | |
| 16p13.11 | 14 | 99 | |
| 17p12 | 33 | 18 | |
| 17q12 | 2 | 14 | |
| 17q21.31 | 1 | ||
| 22q11.2, 2.6MB | 2 | 22 | |
| Noncarriers | 53,879 |
The 16p11.2 distal proximal CNV spans both the distal and proximal region.
6. FINDINGS FROM THE 22Q‐ENIGMA AND ENIGMA‐CNV WGS
To date, the 22q‐ENIGMA WG has published three peer‐reviewed studies on alterations of cortical, subcortical, and white matter structure, respectively (Ching et al., 2020; Sun et al., 2018; Villalon‐Reina et al., 2019). The ENIGMA‐CNV WG has also published three peer‐reviewed studies on the 16p11.2 distal CNV (Sonderby et al., 2018), the 15q11.2 CNV (van der Meer et al., 2020) and the 1q21.1 distal CNV (Sønderby et al., 2021) while two secondary projects are underway. Main results and comparisons with idiopathic disease can be seen in Figures 4 and 5. Analysis is ongoing for several more CNV regions.
FIGURE 4.
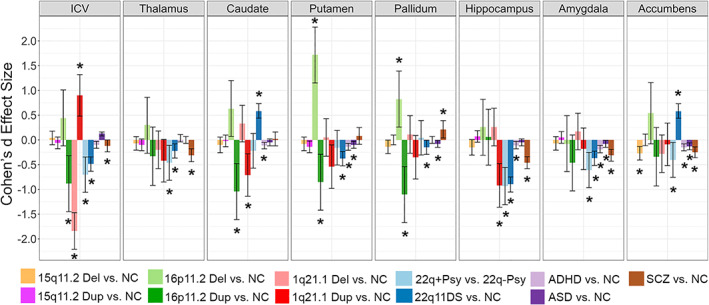
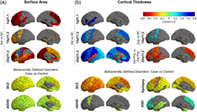
The subcortical findings from ENIGMA‐CNV, 22q‐ENIGMA and selected ENIGMA psychiatric working groups. Averaged left and right subcortical volume case versus non‐carriers (NC) Cohen's d effect size estimates for the ENIGMA SCZ (van Erp et al., 2016), ADHD (Hoogman et al., 2017), ASD (van Rooij et al., 2018), 22q11DS (Ching et al., 2020), 15q11.2 CNV (van der Meer, 2019), 16p11.2 distal CNV (Sønderby et al., 2018), and the 1q21.1 distal CNV (in review) studies. 22q+Psy vs. 22q‐Psy indicates a comparison from Ching et al. (2020) where a subset of individuals with 22q11.2 deletion syndrome with a history of psychosis were compared to a matched group of individuals with 22q11.2 deletion without a history of psychosis. Significant group differences are indicated by an asterisk (*); the plot includes vertical 95% confidence intervals
FIGURE 5.
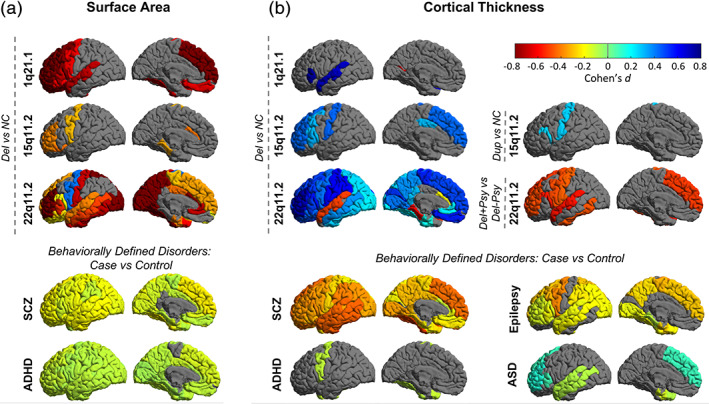
Cortical findings from the ENIGMA‐CNV, 22q‐ENIGMA, and selected ENIGMA psychiatric working groups. Copy number variant (CNV) analyses: for deletion or duplication carriers vs non‐carriers for the 15q11.2 CNVs (ICV‐corrected; van der Meer et al., 2019), 1q21.1 distal CNVs (ICV‐corrected; in review) and 22q11DS (Sun et al., 2018). 22q11DS results include 22q11DS psychosis deletion (Del+Psy) vs non psychosis deletion (Del‐Psy; left hemisphere shown). Behaviorally defined disorders analyses: Results are shown from case‐control studies from ASD's mega‐analysis (left hemisphere shown; van Rooij et al., 2018), all ages in ADHD combined (children, adolescents and adults; Hoogman et al., 2017), all types of epilepsies combined (left hemisphere shown; Whelan et al., 2018), and schizophrenia (SCZ; left hemisphere shown; van Erp et al., 2018). Only significant results are shown
6.1. 22q11.2 deletion syndrome
6.1.1. Cortical structure
The 22q‐ENIGMA WG analyzed the largest sample to date of brain images from individuals with 22q11DS, from 10 cohorts, including 474 individuals with 22q11DS (age = 18.2 ± 8.6 years; 46.9% female) and 315 matched, typically developing controls (age = 18.0 ± 9.2; 45.9% female) (Sun et al., 2018). Compared to controls, the 22q11DS group showed overall thicker cortex (left/right hemispheres: Cohen's d = 0.61/0.65) and widespread lower cortical surface area (left/right hemispheres: d = −1.01/−1.02), which was most prominent in parieto‐occipital and medial brain regions. Surface area decreases were less pronounced in deletion carriers with the smaller 1.5 Mb deletion (LCRA‐LCRB) compared to those with the larger, more typical ~2.6 Mb deletion (LCRA‐LCRD). This provided the first evidence of differential brain morphometry associated with 22q11DS deletion size. When applied to the cortical thickness and surface area measures, a machine learning method provided a high degree of accuracy (sensitivity 94.2%; specificity 93.3%) in classifying 22q11DS cases from healthy controls. Individuals with 22q11DS and a history of psychosis had a pattern of thinner cortex, particularly in frontotemporal regions (vs deletion carriers without a history of psychosis) that significantly overlapped with alterations reported in the largest study of cortical structure in idiopathic SCZ (van Erp et al., 2018). Importantly, the ENIGMA SCZ study (van Erp et al., 2018) and the 22q‐ENIGMA WG studies used the same image processing, quality control, and analysis protocols. These results lend further evidence that 22q11DS offers a biologically tractable framework to better understand the underlying mechanisms driving complex phenotypes such as psychosis.
6.1.2. Subcortical structure
The 22q‐ENIGMA WG performed a mega‐analysis of subcortical volume and shape analysis (Ching et al., 2020) that included 533 individuals with 22q11DS and 330 matched healthy controls (HC; age: 6–56 years) from 11 study sites. Compared to HC, 22q11DS individuals had, on average, smaller bilateral hippocampal, putamen, amygdala and left thalamus volumes, and larger bilateral ventricle, caudate and accumbens volumes. However, a novel shape analysis technique revealed complex local morphometric differences between groups. Shape analysis also revealed regions of the hippocampus, caudate, accumbens, thalamus, and putamen that were less affected in individuals with the smaller (LCR22A‐LCR22B) deletion—the first time that subcortical morphometric variations have been tied to deletion size. Deletion carriers with a history of psychosis had smaller thalamic, hippocampal and amygdala volumes compared with matched carriers without psychosis. These alterations overlapped with the subcortical effects observed in the largest neuroimaging study of SCZ (van Erp et al., 2016), including smaller overall ICV, amygdala, hippocampal, and thalamic volumes. Furthermore, when compared to other ENIGMA subcortical psychiatric studies using the same image processing pipelines, subcortical volume alterations in 22q11DS‐associated psychosis were strongly correlated with case–control effects found in studies of major depressive disorder (Schmaal et al., 2016), bipolar disorder (Hibar et al., 2016) and obsessive compulsive disorder (OCD; Boedhoe et al., 2017), but not with subcortical patterns reported in studies of ASD (van Rooij et al., 2018) or ADHD (Hoogman et al., 2017). Overall, 22q11DS and 22q11DS‐associated psychosis effect sizes were larger than those found in all other ENIGMA studies of idiopathic psychiatric disorders (Figure 4). This lends credence to the idea that a genetics‐first approach may provide greater power to detect biomarkers by providing larger effect sizes than those associated with more common genetic variation (Medland et al., 2020, this issue).
6.1.3. White matter structure
The first study of white matter microstructure from the 22q‐ENIGMA WG was a mega‐analysis of 334 deletion carriers and 260 healthy controls (age: 6–52 years) from 10 international sites (Villalon‐Reina et al., 2019). In the largest study of its kind, a widespread pattern of lower mean diffusivity, axial diffusivity and radial diffusivity and higher fractional anisotropy was detected in 22q11.2 deletion carriers compared to controls, with moderate to large effect sizes. Individuals with both 22q11DS and a history of psychosis displayed more pronounced abnormalities in diffusivity, which pointed to a pattern of white matter abnormalities that may reflect disrupted neurogenesis, particularly in outer layer cortical neurons. However, white matter alterations for individuals with 22q11DS and psychosis diverged from results reported in the largest study of idiopathic SCZ (1,963 SCZ and 2,359 healthy controls from Kelly et al., 2018), which used the same ENIGMA‐DTI processing and quality control pipelines. Whereas individuals with 22q11DS and a history of psychosis showed a general pattern of higher fractional anisotropy and lower diffusivity, people with idiopathic SCZ showed, on average, a pattern of lower fractional anisotropy and higher diffusivity, especially mean and radial diffusivity (Kuchonov et al., this issue). These opposing patterns in white matter variation (Bakker et al., 2016) stand in contrast to findings in cortical and subcortical gray matter, where brain alterations were largely convergent between 22q11DS psychosis and idiopathic SCZ. These findings suggest that different connectivity patterns in white matter may be associated with similar behavioral/clinical outcomes. Ongoing work using more advanced imaging protocols such as “multishell” diffusion MRI—combined with reliable biophysical models that estimate tissue microstructural properties—are being used to investigate the fiber tracts and cellular attributes leading to white matter vulnerabilities in 22q11DS (Villalon Reina et al., 2019; Villalon Reina, Nir, Kushan, Bearden, & Thompson, 2019).
6.2. Other CNVs
6.2.1. 16p11.2 distal CNV
This ENIGMA‐CNV study examined the impact of the 16p11.2 distal CNV on brain structure and function (Sønderby et al., 2018). The 16p11.2 distal CNV (BP2‐BP3, 28.7 to 28.9 Mb; hg18 genome assembly) predisposes carriers to psychiatric conditions including ASD and SCZ and had been associated with macro‐ and microcephaly in deletion and duplication carriers, respectively (Loviglio et al., 2016). It has a frequency of 0.02 and 0.04% for the deletion and duplication, respectively (Kendall et al., 2016; Smajlagić et al., 2020; Stefansson et al., 2014). The 16p11.2 distal CNV lies within a region with many LCRs that also give rise to the 16p11.2 proximal CNV (29.5–30.1, hg18, BP4–BP5) whose brain structural underpinnings have been studied in several studies (Cardenas‐de‐la‐Parra et al., 2019; Maillard et al., 2015; Martin‐Brevet et al., 2018; Qureshi et al., 2014). Both the 16p11.2 distal and proximal CNVs display a negative dose response for body mass index (BMI) (Mace et al., 2017; Owen et al., 2018) and head circumference (Jacquemont et al., 2011; Loviglio et al., 2016).
Based on 12 16p11.2 distal deletion, 12 duplication carriers and 6,882 noncarriers, we identified a negative dose response association of copy number, that is, greater volumes for deletions and lower volumes for the duplication, with ICV, caudate, pallidum and putamen volumes. The pallidum finding was replicated in a smaller sample from deCODE Genetics, Iceland. Further, the combined meta‐analysis of 15 16p11.2 distal deletion and 18 duplication carriers and 7,714 noncarriers identified a negative dose response on nucleus accumbens volume.
The minimal core segment of the 16p11.2 distal CNV is 200 kb in length and contains nine genes. A study in zebrafish found that only over‐expression of the LAT gene from the 16p11.2 distal region induced a decrease in cell proliferation in the brain with a concomitant microcephalic phenotype (Loviglio et al., 2017). LAT knockout mice also showed anatomical brain abnormalities (Loviglio et al., 2017) and brain regions expressing the highest levels of the LAT gene include basal ganglia (Hawrylycz et al., 2012), providing overlap with the brain structural changes identified in the ENIGMA‐CNV study. These findings provide converging evidence that LAT, an immune signaling adaptor, is a possible dosage‐dependent driver of the CNV‐associated brain phenotypes, including the alterations in the basal ganglia. These findings also fit well with a proposed role of the immune system in the development of psychiatric disorders such as SCZ (Khandaker et al., 2015). Notably, a recent GWAS on subcortical volumes identified a GWAS hit, rs1987471, for the caudate nucleus in the 16p11.2 distal region upstream of the ATXNL2 gene (Satizabal et al., 2019), indicating that several genes in the interval may be involved in the brain structural changes.
6.2.2. 15q11.2 CNV
In this study, ENIGMA‐CNV targeted a more frequent CNV, the 15q11.2 CNV (BP1‐BP2, 20.3–20.8 Mb, hg18 genome assembly) with a population prevalence around 0.3% (Crawford et al., 2018; Stefansson et al., 2014). The deletion has unequivocally been associated with an increased risk for SCZ, (OR = 1.6; Marshall et al., 2017). Overall, the effect sizes on disease, cognitive and behavioral phenotypes on the duplication are absent or small: In fact, the duplication has not been clearly associated with psychiatric or neurodevelopmental disorders and its carriers perform on par with controls on cognitive tests (Abdellaoui et al., 2015; Kendall et al., 2019; Stefansson et al., 2014). In contrast, the deletion is associated with a reduction in IQ of ~4 points (Huguet et al., 2018; Jønch et al., 2019) while deletion carriers unaffected by psychiatric or neurodevelopmental disorders have an increased risk of dyslexia and dyscalculia (Stefansson et al., 2014). Reflecting these small effects, the vast majority of carriers are not clinically affected, and the deletion is inherited in >90% of the cases (Cox & Butler, 2015; Jønch et al., 2019; Mohan et al., 2019). Finally, 15q11.2 dosage has been reported to be associated with white matter alterations (Silva et al., 2019; Stefansson et al., 2014; Ulfarsson et al., 2017).
We assessed the association of the 15q11.2 CNV with cognition, and cortical and subcortical morphology in over 45,000 individuals from 38 cohorts, including the UK Biobank (203 individuals with a 15q11.2 deletion, 45,247 noncarriers, and 306 duplication carriers; van der Meer et al., 2020). We identified a clear pattern of widespread poorer cognitive performance, smaller surface area, and thicker cortices for deletion carriers compared to noncarriers and duplication carriers, particularly across the frontal lobe, anterior cingulate, and pre‐ and postcentral gyri.
The 15q11.2 region contains four evolutionarily highly conserved genes: NIPA1, NIPA2, CYFIP1, and TUBGCP5 (Chai et al., 2003). The first three of these genes have known roles in neurodevelopment and contain polymorphisms associated with several brain disorders (Goytain, Hines, El‐Husseini, & Quamme, 2007; Goytain, Hines, & Quamme, 2008; Napoli et al., 2008; van der Zwaag et al., 2010). CYFIP1 and NIPA1 are highly expressed in the developing brain (van der Zwaag et al., 2010) and are key players in a number of processes contributing to brain plasticity, including axon outgrowth and dendritic spine formation (De Rubeis et al., 2013; Schenck et al., 2003; Wang, Shaw, Tsang, Reid, & O'Kane, 2007). Likewise, common CYFIP1 polymorphisms, that influence its expression levels, have been linked to variation in cortical surface area (Woo et al., 2016). Thus, the pattern of results fits well with known molecular functions of the genes in the 15q11.2 region, in particular CYFIP1, and suggests involvement of these genes in neuronal plasticity and cortical development.
6.2.3. 1q21.1 distal CNV
Carriers of the 1q21.1 distal CNVs (BP3‐BP4, 145–145.8 Mb, hg18 genome assembly) are at higher risk for several disorders including SCZ, ID, developmental delay, speech problems, ASD, motor impairment and epilepsy (Bernier et al., 2016; Chawner et al., 2019; Gourari, Schubert, & Prasad, 2018; Haldeman‐Englert & Jewett, 1993; Mefford et al., 2008) and separate risk for the duplication carriers for ADHD (Gudmundsson et al., 2019), bipolar disorder and major depressive disorder (Green et al., 2016; Kendall et al., 2019). The CNV has a frequency of 0.03% and 0.05% for the deletion and duplication, respectively, (Kendall et al., 2016; Stefansson et al., 2014). The 1q21.1 distal CNV has an effect on head circumference, as evident from a high prevalence of micro‐ and macrocephaly in deletion and duplication carriers, respectively (Bernier et al., 2016; Brunetti‐Pierri et al., 2008; Rosenfeld et al., 2012).
ENIGMA‐CNV systematically assessed brain structural MRI variation in 28 1q21.1 distal deletion and 22 duplication carriers and 37,088 noncarriers. We identified positive dosage effects of copy number on ICV and total cortical surface area, with the largest effects in frontal and cingulate cortices, and negative dosage effects on caudate and hippocampal volumes. The effects on subcortical volumes were also observed in a UK biobank exploratory study on six individuals with a 1q21.1 distal duplication (Warland, Kendall, Rees, Kirov, & Caseras, 2019). Carriers displayed distinctive deficits in cognitive tasks with intermediate decreases in duplication, and somewhat larger decreases in deletion carriers—the latter apparently mediated by the decrease in ICV and cortical surface area.
Despite the high effect sizes identified, at the time of writing, GWAS based on the hg19 genome assembly have not identified hits in the 1q21.1 genomic region for ICV (Adams et al., 2016; Knol et al., 2020), total cortical or regional surface area (Grasby et al., 2020; Hofer et al., 2020). Because of the many LCRs in the region (Brunetti‐Pierri et al., 2008; Sharp et al., 2006), assembly of the 1q21.1 region was faulty until version GRCh38 (Steinberg et al., 2014), likely inhibiting gene discovery and this may explain the current lack of GWAS hits in the region.
Given the different breakpoints of the 1q21.1 distal CNVs, estimates of the number of affected genes vary, but the core interval encompasses at least 12 protein‐coding genes including several human‐specific genes, such as HYDIN2 (Dolcetti et al., 2013; Rosenfeld et al., 2012), NOTCH2NLs (Fiddes et al., 2018; Suzuki et al., 2018) and the DUF1220/Olduvai domain‐containing NBPF‐encoding genes. The recently characterized NOTCH2NL genes are particularly interesting in the context of brain development, and as candidates for a dosage‐dependent amplifier of the CNV‐associated brain phenotypes. They are absent in humans' closest living relatives—nonhuman primates—and confer delayed neuronal differentiation and increased progenitor self‐renewal (Fiddes et al., 2018; Suzuki et al., 2018)—in line with the radial unit hypothesis of cortical development (Rakic, 1995). A neurodevelopmental effect on cell proliferation fits well with the overall directional effect of this CNV on cortical surface area and ICV.
6.3. Summary and implications of the findings
A common finding across all four CNVs studied by the 22q11‐ENIGMA and ENIGMA‐CNV WGs is the presence of a gene dosage response on several brain structures (Sønderby et al., 2021; Lin et al., 2017; Sønderby et al., 2018; van der Meer et al., 2020) whose direction may differ between CNVs: carriers of 16p11.2 CNVs display a negative dose response on ICV, while individuals with 1q21.1 CNVs show a positive dose response. Likewise, a dose response on subcortical volumes may be in the same direction (e.g., caudate in 16p11.2 distal) or opposite direction (e.g., caudate in 1q21.1 distal) as that of a macroscopic effect on ICV. Likewise, a dose response may not span both reciprocal CNVs—for example, nucleus accumbens is only altered in 15q11.2 deletion carriers, not in the duplication.
Many CNV carriers overlap in terms of symptoms and susceptibility to disease. Even reciprocal CNVs at each end of the gene dose response can cause both a “gene dose response” for disease risk (22q11.2 and SCZ, Marshall et al., 2017) but also similar disease risk (16p11.2 proximal and distal, 22q11.2, and 1q21.1 distal) for ID and ASD (Chawner et al., 2019). The general rule seems to be that the effect on brain structure fits an additive model for gene dosage formed by, for example, gene expression, whereas an inverted U‐shaped effect curve is observed for the phenotype (Deshpande & Weiss, 2018).
Importantly, brain structural findings in CNV carriers appear to overlap, to some extent, with patterns of brain alterations found in several major brain disorders including ADHD (Hoogman et al., 2017), ASD (van Rooij et al., 2018), SCZ (van Erp et al., 2016), bipolar disorder (Hibar et al., 2016), major depressive disorder (Schmaal et al., 2016), and epilepsy (Whelan et al., 2018). However, several CNVs clearly have effect sizes far greater than those of the idiopathic diseases (Figures 4 and 5). Likewise, so far, it is difficult to find a direct pattern in the overlap between known disease susceptibility for CNV carriers and brain structural effects—both in terms of specificity (actual overlap) and effect sizes. Thus, this makes it increasingly evident that vastly different brain alterations—for example, large macroscopic effects (e.g., in 16p11.2 and 1q21.1 CNVs) as well as small subtle effects (e.g.,15q11.2 CNVs)—can lead to similar phenotypes, underlining the heterogeneity in brain structure within diseases such as SCZ and a putative potential to stratify based on brain structure within specific diseases. Improved understanding of these types of relationships may prove important for understanding disease susceptibility and outcome.
7. ONGOING PROJECTS AND FUTURE DIRECTIONS
Results from the 22q‐ENIGMA and ENIGMA‐CNV WGs confirm that multiple CNVs are associated with differences in brain morphology. This effort is providing information on genetically determined variation in brain development and its relation to neurodevelopmental, psychiatric, and neurological disorders. Current and future CNV studies will benefit both from even larger samples with more ethnicities represented, based on broad collaborations and standardized data collection across samples and in smaller, more flexible studies with deeper phenotyping.
7.1. Increasing sample sizes
There is great potential to include additional samples in ENIGMA WGs on CNVs. First, additional cohorts can easily be incorporated and additional measures such as the corpus callosum, cerebellum, brain stem, and ventricles—not yet targeted in ENIGMA‐CNV—can be added to the protocol. Second, independent research projects performing targeted recruitment and MRI brain scans on CNV carriers can easily join. Third, clinical scans of CNV carriers may be leveraged, provided the MRI quality is sufficient for accurate morphometry, and a number of appropriate genotyped controls from the same scanner site is provided: part of the standard evaluation for children with developmental delay or ID may include brain MRI, in particular for cases where additional clinical indications such as epilepsy, head circumference abnormalities or focal neurological signs are present (Mithyantha, Kneen, McCann, & Gladstone, 2017). Finally, such independent samples can be supplemented with an increasing amount of data from open data sets such as the ABCD study (Casey et al., 2018) and UK Biobank (Littlejohns et al., 2020).
There are notably analytical challenges and interpretational risks involved in large‐scale neuroimaging studies (Smith & Nichols, 2018). The initial ENIGMA studies on CNVs were able to capture effects across diverse, heterogenous study samples, but they might lack sensitivity to subtype‐specific effects that may be better captured by smaller well‐controlled studies focusing on targeted phenotyping. Likewise, the latter studies might be more suited to test new technology or methods. Nevertheless, a considerable strength lies in the ability of big studies to discover if effects generalize across samples collected all over the world. In other words, large‐scale studies and well‐designed smaller scale neuroimaging studies on CNVs offer useful complementary approaches.
7.2. Expanding the generalizability of data
7.2.1. Clinical versus nonclinical effects and ancestry considerations
An increase in cross‐diagnostic data will allow a deeper insight into what distinguishes a CNV carrier with a severe phenotype (e.g., clinical diagnosis) from a well‐functioning carrier (typically with no clinical diagnosis). So far, effects of CNVs on the brain seem to be found in clinical and nonclinical carriers alike. With larger, more diverse samples that cover the entire phenotypic heterogeneity of all carriers, we may be able to deduce if a less severe clinical outcome is associated with a more moderate effect on the brain structural fingerprint.
Likewise, distinctions may be found for groups of different ancestry. So far, there is no evidence that CNVs have different effects across different ancestries but data are sparse, as most studies to date are based on white European samples; there is an urgent need to include individuals from diverse ancestries, to provide a broader and more inclusive view on this topic. Such cohorts are already being collected through several efforts, including initiatives on SCZ (Gulsuner et al., 2020) and neurodevelopment (de Menil et al., 2019).
7.2.2. Lifespan trajectories
Current investigations have focused on the overall impact of CNVs on the brain, mostly disregarding a potential dynamic or interactive effect of age on brain maturation. Gross structural brain alterations are likely present from an early stage of development—as exemplified by the macrocephaly observed in utero in a 1q21.1 distal duplication carrier (Verhagen et al., 2015). However, detailed knowledge on the development of brain structure in CNV carriers over the lifespan is lacking. Recently, the first study to address this—targeting the 16p11.2 proximal CNV—suggested that differences in brain structure in deletion and duplication carriers in comparison to noncarriers remain stable throughout childhood, adolescence and at least until around 23 years of age (Cardenas‐de‐la‐Parra et al., 2019). Other evidence suggests that there may be differences in the profiles of neurodegeneration at older ages when carrying a CNV (Gentile, La Cognata, & Cavallaro, 2020) highlighting the need to study individuals across the lifespan.
7.3. Expanding phenotypic information
7.3.1. Expanding to brain connectivity: Inclusion of DTI and resting state MRI
Accumulating evidence converges on brain dysconnectivity as a trans‐diagnostic phenotype in mental illness, based on aberrations in the “wiring” of the brain in individuals suffering from mental illness. To date, the 22q‐ENIGMA WG has targeted brain measures more broadly—including diffusion‐weighted MRI and surface‐based shape analysis—whereas the ENIGMA‐CNV WG has focused on ROI‐based measures of brain structure. Workflows for diffusion MRI analyses have already been developed and tested in various ENIGMA WGs (Jahanshad et al., 2013; Kochunov et al., 2016) including the 22q‐ENIGMA WG (Villalon‐Reina et al., 2019). Likewise, a resting‐state functional MRI processing pipeline was proposed recently for use in ENIGMA meta‐analyses (Adhikari et al., 2018). Future ENIGMA‐studies aimed at large scale investigations of structural and functional brain metrics aim to directly test the proposal of a “common symptom, common circuit” model of psychopathology in which genetic, epigenetic, and environmental risk factors affect connectivity in one or several neural circuits, producing cognitive, and emotional disturbances (Buckholtz & Meyer‐Lindenberg, 2012). So far, only a few studies have tested this concept in CNVs—either by doing analysis across pathogenic CNV carriers (Drakesmith et al., 2019), through analysis of severely affected individuals (22q11DS; Villalon‐Reina et al., 2019; Moreau et al., 2020), the 16p11.2 proximal (Chang et al., 2016; Moreau et al., 2020), or the most abundant low impact recurrent CNV, 15q11.2 (Silva et al., 2018). A combined effort within this arena would be likely to move the field forward.
7.3.2. Structural covariation and spatial gene expression
The primary ENIGMA‐CNV and 22q‐ENIGMA imaging studies have provided a better understanding of the spatial distribution of CNV‐related brain alterations across the brain (localized to lobes, gyri, sulci, etc.). Methods such as structural covariance may help to better understand CNV‐related disruptions in the developmental coordination between maturing brain regions (Alexander‐Bloch, Raznahan, Bullmore, & Giedd, 2013). Techniques such as “virtual histology,” which relate group differences in MRI‐derived cortical measures (e.g., 22q11.2 carrier vs. healthy control) to gradients in cell‐specific gene expression from the Allen Human Brain Atlas may provide a step toward bridging the gap between MRI‐brain alterations and underlying cell‐specific pathophysiology (Patel et al., 2018; Shin et al., 2017).
7.3.3. Adding clinical, cognitive, and behavioral data
The strength of the 22q‐ENIGMA and ENIGMA‐CNV WGs is combining large‐scale data on both CNV calls and imaging data. Adding deeper phenotyping information such as cognitive, mental, and behavioral data would be highly beneficial. The challenge with incorporating such data from independent studies is that the standardization of phenotypic information across the independently collected cohorts is lacking; other ENIGMA WGs have begun to deal with this challenge by harmonizing cognitive endpoints (Tate et al., 2021). Through organization and standardization of samples, we have the potential to deepen our knowledge regarding the relationships between CNV carriers, imaging measures and other phenotypes.
Publicly available data sets, such as the UK Biobank, already allow for large‐scale analysis of cross‐phenotypic traits—such as brain‐cognition mediation by combining cognition and brain imaging data. Given the continued recruitment of individuals for MRI studies, and the future availability of other large‐scale harmonized brain imaging data sets such as in the ABCD study (Casey et al., 2018), the potential of this type of analysis is continuously expanding. Future studies aim to move beyond case–control comparisons to better understand the underlying brain structure and functional mechanisms related to behavioral phenotype heterogeneity in CNV carriers (Marquand, Rezek, Buitelaar, & Beckmann, 2016). These types of analysis, taking advantage of “big data” samples, call for robust data‐driven approaches that do not simply maximize prediction accuracy but improve our interpretation and understanding of underlying mechanisms (Bzdok, Nichols, & Smith, 2019).
7.3.4. Identifying causal mechanisms—Mechanistic follow‐up studies
Candidate genes within the CNV regions can be linked to neuronal differentiation, axon outgrowth and dendritic spine formation, neurotransmitter and immune signaling, and other processes important for brain development. Moving forward, interest will no doubt focus on identifying and characterizing these driver genes causing the phenotypes or factors modifying the phenotypes. Many approaches can aid in this effort.
Studies of variable CNV breakpoints can narrow down potential driver genes. This is exemplified by the recent dismissal of HYDIN2 as a driver gene for the head circumference phenotype in 1q21.1 distal CNV: Several atypical 1q21.1 distal deletion and duplication carriers with normal copy number variation of HYDIN2, but still exhibiting the microcephaly or macrocephaly phenotype, were identified (Dougherty et al., 2017). A recent exome sequencing study of people with autism identified BCL11A as a potential driver gene for autism in the 2p15‐p16.1 CNV (Satterstrom et al., 2020). Likewise, gene expression studies have helped to narrow down the gene behind the SNP association for brain structure. Another option is expression of individual genes in model organisms such as mouse (Dominguez‐Iturza et al., 2019; Nielsen et al., 2017) or zebrafish (Loviglio et al., 2017). Such approaches may identify driver genes for brain phenotypes associated with CNVs, pinpointing the biological mechanisms involved.
Finally, future studies should try to disentangle the role of common genetic variants in moderating the phenotype caused by CNVs, through common and rare variant interplay analysis as done for instance in SCZ (Bergen et al., 2018; Tansey et al., 2016) and ADHD (Martin et al., 2015).
7.4. Secondary proposals
Several secondary projects are ongoing in the 22q‐ENIGMA and ENIGMA‐CNV WGs. In the 22q‐ENIGMA WG, a secondary project led by Fidel Vila‐Rodriguez (The University of British Columbia) is now investigating the structural covariance of gray matter volume using source‐based morphometry, and another study led by Jennifer Forsyth at UCLA is investigating specific 22q11.2 genes driving cortical surface area and thickness alterations.. In addition to recurrent CNVs, numerous single, nonrecurrent CNVs disrupting one or more genes may have a large impact on the brain and behavior but determining the impact and clinical interpretation of these single CNVs is even more challenging (Nowakowska, 2017). In ENIGMA‐CNV, “The effect of very rare CNVs on brain structure and function,” headed by the group of Sébastien Jacquemont at University of Montreal, investigates very rare CNVs by analyzing all CNVs in the genome—even single hit CNVs—and their effect on brain structure. Another secondary project in the ENIGMA‐CNV WG, headed by David Linden (Cardiff University/Maastricht University), addresses how brain changes across different pathogenic CNVs correlate with penetrance scores for SCZ and developmental delay, aiming to find common brain phenotypes that are most related to risk for disorders.
8. CONCLUSION
CNV analysis offers a genetics‐first approach to studying neurodevelopmental, neurological and neuropsychiatric disorders as well as related traits. Using convergent evidence from neuropsychological testing, structural and functional neuroimaging, the 22q‐ENIGMA and ENIGMA‐CNV WGs have mapped the effects of CNVs in some of the largest neuroimaging data sets ever analyzed, revealing consistent brain signatures of CNVs, overlap with idiopathic disorders and, in the case of 22q11DS, its relation to psychotic conditions. Although these studies have revealed consensus associations between varying CNVs and brain structure, much remains to be explored. Data sharing and international collaborations are essential for CNV studies, and the large‐scale efforts conducted by the 22q‐ENIGMA WG, the ENIGMA‐CNV WG and Psychiatric Genomics Consortium serve as an example of how team science can tackle some of these core challenges facing CNV research.
8.1. Join the CNV efforts of ENIGMA
The ENIGMA‐CNV and 22q‐ENIGMA WGs continue to recruit new participating members, and the infrastructure allows for new groups to be efficiently incorporated into ongoing and future analyses. Groups interested in joining or contributing data are encouraged to contact the WG Chairs. This needs to be a community effort—so we encourage everyone to join the effort. Because every CNV carrier counts.
9. CONTACT DETAILS
Those interested are invited to contact the coordinators:
ENIGMA‐CNV WG: Ida Elken Sønderby & Ole A. Andreassen. Webpage: http://enigma.ini.usc.edu/ongoing/enigma-cnv/.
22q‐ENIGMA WG: Carrie E. Bearden and Christopher R. K. Ching. Webpage: http://enigma.ini.usc.edu/ongoing/enigma-22q-working-group/
DISCLOSURE OF INTERESTS
O. A. A. is a Speaker's honorarium at Lundbeck, Consultant at HealthLytix. K. M. A. has received investigator‐initiated research funds from Shire and has participated in an advisory panel for Arbor Pharmaceuticals. C. A. has been a consultant to or has received honoraria or grants from Acadia, Angelini, Gedeon Richter, Janssen Cilag, Lundbeck, Minerva, Otsuka, Roche, Sage, Servier, Shire, Schering Plough, Sumitomo Dainippon Pharma, Sunovion, and Takeda. P. M. T. and C. R. K. C. have received partial research support from Biogen, Inc. (Boston) for work unrelated to the topic of this manuscript. D. P. H. is a full‐time employee of Genentech, Inc. A. M. D. is a Founder of and holds equity in CorTechs Labs, Inc, and serves on its Scientific Advisory Board. H. S., M. O. U., G. B. W., K. S. are employees of deCODE genetics/Amgen. H. J. G. has received travel grants and speakers honoraria from Fresenius Medical Care, Neuraxpharm, Servier, and Janssen Cilag as well as research funding from Fresenius Medical Care. JH has received speakers honoraria from Lilly, Biocodex, HB‐Pharma, Medice, Takeda, and Shire. D. J. S. has received research grants and/or consultancy honoraria from Lundbeck and Servier. F. V.‐R. receives research support from CIHR, Brain Canada, Michael Smith Foundation for Health Research, Vancouver Coastal Health Research Institute, and in‐kind equipment support for this investigator‐initiated trial from MagVenture. F. V.‐R. has also participated in an advisory board for Janssen.
ACKNOWLEDGMENTS
The ENIGMA Consortium is supported by the NIH Big Data to Knowledge (BD2K) program under consortium grant number U54 EB020403 (PI: Thompson). OAA is supported by the Research Council of Norway, South East Norway Health Authority, KG Jebsen Stiftelsen, EU H2020. C. A. has been funded by the Spanish Ministry of Science and Innovation; Instituto de Salud Carlos III (SAM16PE07CP1, PI16/02012, PI19/024), co‐financed by ERDF Funds from the European Commission, “A way of making Europe”, CIBERSAM; Madrid Regional Government (B2017/BMD‐3740 AGES‐CM‐2), European Union Structural Funds; European Union Seventh Framework Program under grant agreements FP7‐4‐HEALTH‐2009‐2.2.1‐2‐241,909 (Project EU‐GEI), FP7‐ HEALTH‐2013‐2.2.1‐2‐603,196 (Project PSYSCAN) and FP7‐ HEALTH‐2013‐2.2.1‐2‐602,478 (Project METSY); and European Union H2020 Program under the Innovative Medicines Initiative two Joint Undertaking (grant agreement No 115916, Project PRISM, and grant agreement No 777394, Project AIMS‐2‐TRIALS), Fundación Familia Alonso and Fundación Alicia Koplowitz. R. A‐A is funded by a Miguel Servet contract from the Carlos III Health Institute (CP18/00003). G. B. is supported by the Dutch Organization for Health Research and Development ZonMw (grants 91112002 & 91712394). A. S. B. is supported by the Dalglish Family Chair in 22q11.2 Deletion Syndrome, Canadian Institutes of Health Research (CIHR) grants MOP‐79518, MOP‐89066, MOP‐97800 and MOP‐111238, and NIMH grant number U01 MH101723–01(3/5). C. E. B. is also supported by the National Institute of Mental Health: RO1 MH085953, R01 MH100900 and 1U01MH119736. N. E. B. is granted the KNAW Academy Professor Award (PAH/6635). V. D. C. is supported by NIH R01 MH094524. S. C. is supported by the European Union's Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreement No. 945539 (Human Brain Project SGA3); Helmholtz Initiative and Networking Fund. C. R. K. C. is supported by NIA T32AG058507. E. W. C. C. is supported by the Canadian Institutes of Health Research, Ontario Mental Health Foundation grant MOP‐74631 and NIMH grant U01MH101723–01(3/5). S. Ci. has received funding from the European Union's Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreement No. 945539 (Human Brain Project SGA3). M. C. C. is supported by the Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King's College London. N. A. C. is supported by Agencia Nacional de Investigación y Desarrollo (ANID Chile) PIA ACT192064. GId. Z. is supported by the NHMRC. J. L. D. and D. E. J. L. are supported by the Wellcome Trust. T. B. C. is supported by NICHD grant PO1‐HD070454, NIH grant UO1‐MH191719, and NIMH grant R01 MH087636‐01A1. AMD is supported by U24DA041147. B. D. is supported by the Swiss National Science Foundation (NCCR Synapsy, project grant numbers 32003B_135679, 32003B_159780, 324730_192755 and CRSK‐3_190185), the Leenaards Foundation and the Roger De Spoelberch Foundation. SE is supported by the NARSAD‐Young Investigator Grant “Epigenetic Regulation of Intermediate Phenotypes in Schizophrenia”. B. E. S. is supported by the NIH (NIMH). D. C. G. is supported by NIH grant numbers MH078143, MH083824, AG058464. W. R. K. is supported by NIH/MH R0106824. R. E. G. is supported by NIH/NIMH grant numbers MH087626, MH119737. DMMcD‐McG is supported by National Institutes of Mental Health (NIMH), grant numbers MH119737‐02; MH191719; and MH087636‐01A1. S. E. M. is supported by NHMRC grants APP1103623; APP1158127; APP1172917. TM is supported by Research Council of Norway ‐ grant number 273345. D. G. M. is supported by the National Institute for Health Research Mental Health Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King's College London and EU‐AIMS (European Autism Interventions)/EU AIMS‐2‐TRIALS, a European Innovative Medicines Initiative Joint Undertaking under grant agreements 115300 and 777394. T. N. was supported by Stiftelsen KG Jebsen under grant number SKGJ‐MED‐021. R. A. O. is supported by NIMH R01 MH090553. S. Y. S. has been funded by the Canadain Institutes of Health Research. M. J. O. is supported by MRC Centre grant MR/L010305/1 and Wellcome Trust grant 100,202/Z/12/Z; Dr. Owen has received research support from Takeda. Z. P. is supported by CIHR, CFI, HSFC. B. G. P. is supported by CIHR FDN 143290 and CAIP Chair. G. M. R. is supported by Fondecyt‐Chile #1171014 and ANID‐Chile ACT192064. A. Re. was supported by a grant from the Swiss National Science Foundation (31003A_182632). DRR is supported by R01 MH120174 (PI: Roalf). This report represents independent research funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King's College London (to J. J. R). PSS is supported by NHMRC (Australia) program grant 1093083. J. E. S. is supported by NIH K01‐ES026840. S. M. S. is supported by the Epilepsy Society. T. J. S. is supported by NIH grants R01MH107108, R01HD042794, and HDU54079125. I. E. S. is supported by South‐Eastern Norway Regional Health Authority (#2020060), European Union's Horizon2020 Research and Innovation Programme (CoMorMent project; grant #847776) and the KG Jebsen Foundation (SKGJ‐MED‐021). V. M. S. is supported by Research Council of Norway (CoE funding scheme, grant number 223273). D. J. S. is supported by the SA MRC. C. K. T. is supported by Research Council of Norway (#230345, #288083, #223273) and South‐Eastern Norway Regional Health Authority (#2019069, #2021070, #500189). D. T.‐G. was supported by the Instituto de Salud Carlos III (PI14/00639 and PI14/00918) and Fundación Instituto de Investigación Marqués de Valdecilla (NCT0235832 and NCT02534363). Dvd. M. is supported by Research Council of Norway #276082. F. V. R. is supported by the Michael Smith Foundation for Health Research Scholar Award. deCODE genetics has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreements' no. 115008 (NEWMEDS) and no. 115300 (EUAIMS), of which resources are composed of EFPIA in‐kind contribution and financial contribution from the European Union's Seventh Framework Programme (EU‐FP7/2007–2013). L. T. W. is supported by Research Council of Norway, European Research Council. The IDIVAL neuroimage unit is supported by Instituto de Salud Carlos III PI020499, research funding SCIII‐INT13/0014, MICINN research funding SAF2010‐20840‐C02‐02, SAF2013‐46292‐R. The TOP/NORMENT study are supported by the Research Council of Norway (#223273). The GOBS study data collection was supported in part by the National Institutes of Health (NIH) grants: R01 MH078143, R01 MH078111, and R01 MH083824 with work conducted in part in facilities constructed under the support of NIH grant number C06 RR020547. The Sydney Memory and Ageing Study has been funded by three National Health & Medical Research Council (NHMRC) Program Grants (ID No. ID350833, ID568969, and APP1093083). We thank the participants and their informants for their time and generosity in contributing to this research. We also acknowledge the MAS research team: https://cheba.unsw.edu.au/research‐projects/sydney‐memory‐and‐ageing‐study. We acknowledge the contribution of the OATS research team (https://cheba.unsw.edu.au/project/older-australian-twins-study) to this study. The OATS study has been funded by a National Health & Medical Research Council (NHMRC) and Australian Research Council (ARC) Strategic Award Grant of the Aging Well, Aging Productively Program (ID No. 401162); NHMRC Project (seed) Grants (ID No. 1024224 and 1025243); NHMRC Project Grants (ID No. 1045325 and 1085606); and NHMRC Program Grants (ID No. 568969 and 1093083). We thank the participants for their time and generosity in contributing to this research. This research was facilitated through access to Twins Research Australia, a national resource supported by a Centre of Research Excellence Grant (ID No. 1079102) from the National Health and Medical Research Council. The NCNG sample collection was supported by grants from the Bergen Research Foundation and the University of Bergen, the Dr Einar Martens Fund, the KG Jebsen Foundation, the Research Council of Norway, to S. L. H., V. M. S., A. J. L., and T. E. The authors thank Dr. Eike Wehling for recruiting participants in Bergen, and Professor Jonn‐Terje Geitung and Haraldplass Deaconess Hospital for access to the MRI facility. Additional support by RCN grants 177458/V50 and 231286/F20. The Betula study was supported by a Wallenberg Scholar Grant (KAW). The HUNT Study is a collaboration between HUNT Research Centre (Faculty of Medicine and Health Sciences, NTNU—Norwegian University of Science and Technology), Nord‐Trøndelag County Council, Central Norway Health Authority, and the Norwegian Institute of Public Health. HUNT‐MRI was funded by the Liaison Committee between the Central Norway Regional Health Authority and the Norwegian University of Science and Technology, and the Norwegian National Advisory Unit for functional MRI. Research for the GAP cohort was supported by the Department of Health via the National Institute for Health Research (NIHR) Specialist Biomedical Research Center for Mental Health award to South London and Maudsley NHS Foundation Trust (SLaM) and the Institute of Psychiatry at King's College London, London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. S.J. is supported by Calcul Quebec (http://www.calculquebec.ca), Compute Canada (http://www.computecanada.ca), the Brain Canada Multi investigator research initiative (MIRI), the Institute of Data Valorization (Canada First Research Excellence Fund), CHIR, Canada Research Chairs and the Jeanne et Jean Louis Levesque Foundation.
The NTR cohort was supported by the Netherlands Organization for Scientific Research (NWO), MW904‐61‐193 (de Geus & Boomsma), MaGWnr: 400‐07‐080 (van 't Ent), MagW 480‐04‐004 (Boomsma), NWO/SPI 56‐464‐14,192 (Boomsma), the European Research Council, ERC‐230374 (Boomsma), and Amsterdam Neuroscience. Funding for genotyping was obtained from the National Institutes of Health (NIMH U24 MH068457‐06; Grand Opportunity grants 1RC2 MH089951, and 1RC2 MH089995); the Avera Institute for Human Genetics, Sioux Falls, South Dakota (USA). Part of the genotyping and analyses were funded by the Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health. The Brainscale study was supported by the Netherlands Organization for Scientific Research MagW 480‐04‐004 (Boomsma), 51.02.060 (Hilleke Hulshoff Pol), 668.772 (Boomsma & Hulshoff Pol); NWO/SPI 56‐464‐14192 (Boomsma), the European Research Council (ERC230374) (Boomsma), High Potential Grant Utrecht University (Hulshoff Pol), NWO Brain and Cognition 433‐09‐220 (Hulshoff Pol). SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs and the Social Ministry of the Federal State of Mecklenburg‐West Pomerania. Genome‐wide SNP typing in SHIP and MRI scans in SHIP and SHIP‐TREND have been supported by a joint grant from Siemens Healthcare, Erlangen, Germany and the Federal State of Mecklenburg‐West Pomerania. The ENIGMA‐22q11.2 Deletion Syndrome Working Group wishes to acknowledge our dear colleague Dr. Clodagh Murphy, who sadly passed away in April 2020. Open access funding enabled and organized by Projekt DEAL.
ENIGMA‐CNV Additional Members
Manon Bernard, Bloorview Research Institute, Holland Bloorview Kids Rehabilitation Hospital, Toronto, Ontario, Canada.
Nicholas B. Blackburn, Department of Human Genetics, School of Medicine, University of Texas Rio Grande Valley, Brownsville, Texas, USA; South Texas Diabetes and Obesity Institute, School of Medicine, University of Texas Rio Grande Valley, Brownsville, Texas, USA.
Rune Bøen, Department of Medical Genetics, Oslo University Hospital, Oslo, Norway; Norwegian Centre for Mental Disorders Research (NORMENT), Division of Mental Health and Addiction, Oslo University Hospital and University of Oslo, Oslo, Norway.
Eco de Geus, Department of Biological Psychology, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands; Amsterdam Public Health (APH) Research Institute, Amsterdam UMC, Amsterdam, The Netherlands.
Sonja M. C. de Zwarte, Department of Psychiatry, UMC Utrecht Brain Center, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
Marta Di Forti, Institute of Psychiatry, Psychology & Neuroscience, King's College London, London, UK.
Oleksandr Frei, Norwegian Centre for Mental Disorders Research (NORMENT), Division of Mental Health; Addiction, Oslo University Hospital and University of Oslo, Oslo, Norway and Center for Bioinformatics, Department of Informatics, University of Oslo, Oslo, Norway, Vincent Frouin, Université Paris‐Saclay, CEA, Neurospin, Gif‐sur‐Yvette, France.
Masaki Fukunaga, Division of Cerebral Integration, National Institute for Physiological Sciences, Okazaki, Japan.
Jayne Y. Hehir‐Kwa, Princess Máxima Center for Pediatric Oncology, Utrecht, The Netherlands.
Manon H. J. Hillegers, Department of Child and Adolescent Psychiatry/Psychology, Erasmus Medical Center, Rotterdam, The Netherlands.
Per Hoffmann, Institute of Human Genetics, University of Bonn, Bonn, Germany; Human Genomics Research Group, Department of Biomedicine, University of Basel, Basel, Switzerland.
Georg Homuth, Interfaculty Institute for Genetics and Functional Genomics, University Medicine Greifswald, Greifswald, Germany.
Neda Jahanshad, Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine, University of Southern California, Marina del Rey, CA, USA.
Sanne Koops, Department of Biomedical Sciences of Cells & Systems, Cognitive Neurosciences, University of Groningen, University Medical Center Groningen (UMCG), Groningen, The Netherlands.
Kuldeep Kumar, Sainte Justine Research Center, University of Montreal, Montreal, QC, Canada.
Masataka Kikuchi, Graduate School of Medicine, Osaka University, Osaka, Japan.
Stephanie Le Hellard, NORMENT, Department of Clinical Science, University of Bergen, Bergen, Norway; Dr. Einar Martens Research Group for Biological Psychiatry, Department of Medical Genetics, Haukeland University Hospital, Bergen, Norway.
Costin Leu, Genomic Medicine Institute, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA; Department of Clinical and Experimental Epilepsy, UCL Queen Square Institute of Neurology, London, UK; Stanley Center for Psychiatric Research, Broad Institute of Harvard and M.I.T, Cambridge, MA, USA.
Robin M Murray, Institute of Psychiatry, Psychology & Neuroscience, King's College London, London, UK.
Terje Nærland, KG Jebsen Centre for Neurodevelopmental Disorders, University of Oslo, Oslo, Norway; NevSom, Department of Rare Disorders, Oslo University Hospital, Oslo, Norway.
Lars Nyberg, Integrative Medical Biology (IMB) Umeå University, Umeå, Sweden; Department of Psychiatry, Erasmus University Medical Center, Rotterdam, The Netherlands.
Roel A. Ophoff, Center for Neurobehavioral Genetics, University of California Los Angeles, Los Angeles, CA, USA; Department of Psychiatry, Erasmus University Medical Center, Rotterdam, The Netherlands.
G Bruce Pike, Radiology and Clinical Neurosciences, University of Calgary, Calgary, AB, Canada.
Sigrid B. Sando, Department of Neuromedicine and Movement Science, Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology, Trondheim, Norway; Department of Neurology and Clinical Neurophysiology, University Hospital of Trondheim, Trondheim, Norway.
Jean Shin, The Hospital for Sick Children, Toronto, ON, Canada; Departments of Physiology and Nutritional Sciences, University of Toronto, Toronto, Ontario, Canada.
Elena Shumskaya, Donders Institute for Brain, Cognition, and Behavior, Nijmegen, The Netherlands.
Sanjay M. Sisodiya, Department of Clinical and Experimental Epilepsy, UCL Queen Square Institute of Neurology, London, UK; Chalfont Centre for Epilepsy, Chalfont‐St‐Peter, Bucks, UK.
Vidar M. Steen, NORMENT, Department of Clinical Science, University of Bergen, Bergen, Norway; Dr. Einar Martens Research Group for Biological Psychiatry, Department of Medical Genetics, Haukeland University Hospital, Bergen, Norway.
Alexander Teumer, Institute for Community Medicine, University Medicine Greifswald, Greifswald, Germany,
Anne Uhlmann, Department of Psychiatry and Mental Health, University of Cape Town, Cape Town, South Africa.
Margaret J. Wright, Queensland Brain Institute, The University of Queensland, Brisbane, QLD, Australia
ENIGMA 22q11.2 Deletion Syndrome Additional Members
Kevin M. Antshel, Department of Psychology, Syracuse University, Syracuse, NY, USA.
Linda E. Campbell, University of Newcastle, Newcastle, NSW, Australia.
Nicolas A. Crossley, Department of Psychiatry, Pontificia Universidad Católica de Chile, Santiago, Chile.
T. Blaine Crowley, Division of Human Genetics and 22q and You Center, Children's Hospital of Philadelphia, Philadelphia, PA, USA.
Eileen Daly, Department of Forensic and Neurodevelopmental Sciences, and the Sackler Institute for Translational Neurodevelopmental Sciences, Institute of Psychiatry, Psychology and Neuroscience, King's College, London, UK.
Ania M. Fiksinski, Clinical Genetics Research Program, Centre for Addiction and Mental Health, Toronto, ON, Canada; Dalglish Family 22q Clinic for Adults with 22q11.2 Deletion Syndrome, Toronto General Hospital, University Health Network, Toronto, ON, Canada; Department of Psychiatry, UMC Utrecht Brain Center, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
Jennifer K. Forsyth, Department of Psychiatry and Biobehavioral Sciences, University of California Los Angeles, Los Angeles, CA, USA.
Wanda Fremont, Department of Psychiatry and Behavioral Sciences, SUNY Upstate Medical University, Syracuse, NY, USA.
Naomi J. Goodrich‐Hunsaker, MIND Institute and Department of Psychiatry and Behavioral Sciences, University of California Davis, Davis, CA, USA; Department of Neurology, University of Utah, Salt Lake City, UT, USA.
Maria Gudbrandsen, Department of Forensic and Neurodevelopmental Sciences, and the Sackler Institute for Translational Neurodevelopmental Sciences, Institute of Psychiatry, Psychology and Neuroscience, King's College, London, UK.
Rachel K. Jonas, Semel Institute for Neuroscience and Human Behavior, Departments of Psychiatry and Biobehavioral Sciences and Psychology, University of California Los Angeles, Los Angeles, CA, USA.
Wendy R. Kates, Department of Psychiatry and Behavioral Sciences, State University of New York at Upstate Medical University, Syracuse, NY, USA.
Amy Lin, Semel Institute for Neuroscience and Human Behavior, University of California Los Angeles, Los Angeles, CA, USA.
Kathryn L. McCabe, Department of Psychiatry and Behavioral Sciences, State University of New York at Upstate Medical University, Syracuse, NY, USA; Department of Psychology, University of Newcastle, Newcastle, NSW, Australia.
Hayley Moss, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK.
Declan G. Murphy, Department of Forensic and Neurodevelopmental Sciences, and the Sackler Institute for Translational Neurodevelopmental Sciences, Institute of Psychiatry, Psychology and Neuroscience, King's College, London, UK; Behavioural Genetics Clinic, Adult Autism Service, Behavioural and Developmental Psychiatry Clinical Academic Group, South London and Maudsley NHS Foundation Trust, London, UK.
Kieran C. Murphy, Department of Psychiatry, Royal College of Surgeons in Ireland, Dublin, Ireland.
Michael J. Owen, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK.
Kosha Ruparel, Department of Psychiatry, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.
Tony. J. Simon, MIND Institute and Department of Psychiatry and Behavioral Sciences, University of California Davis, Davis, CA, USA.
Therese van Amelsvoort, Department of Psychiatry and Neuropsychology, Maastricht University, Maastricht, The Netherlands.
Jacob A. S. Vorstman, Department of Psychiatry, The Hospital for Sick Children, Toronto, ON, Canada.
Sønderby IE, Ching CRK, Thomopoulos SI, et al. Effects of copy number variations on brain structure and risk for psychiatric illness: Large‐scale studies from the ENIGMA working groups on CNVs . Hum Brain Mapp. 2022;43:300–328. 10.1002/hbm.25354
Members of ENIGMA‐CNV Working Group and ENIGMA 22q11.2 Deletion Syndrome Working Group are given in Appendix.
Funding information European Union's Horizon2020 Research and Innovation Programme, Grant/Award Number: CoMorMent project; Grant #847776; KG Jebsen Stiftelsen; National Institutes of Health, Grant/Award Number: U54 EB020403; Norges Forskningsråd, Grant/Award Number: #223273; South‐Eastern Norway Regional Health Authority, Grant/Award Number: #2020060
Contributor Information
Ida E. Sønderby, Email: i.e.sonderby@medisin.uio.no.
for the ENIGMA‐CNV Working Group:
Manon Bernard, Nicholas B. Blackburn, Rune Bøen, Eco de Geus, Sonja M. C. de Zwarte, Marta Di Forti, Oleksandr Frei, Masaki Fukunaga, Jayne Y. Hehir‐Kwa, Manon H. J. Hillegers, Per Hoffmann, Georg Homuth, Neda Jahanshad, Sanne Koops, Kuldeep Kumar, Masataka Kikuchi, Stephanie Le Hellard, Costin Leu, Robin M Murray, Terje Nærland, Lars Nyberg, Roel A. Ophoff, G Bruce Pike, Sigrid B. Sando, Jean Shin, Elena Shumskaya, Sanjay M. Sisodiya, Vidar M. Steen, Alexander Teumer, Anne Uhlmann, and Margaret J. Wright
for the ENIGMA 22q11.2 Deletion Syndrome Working Group:
Kevin M. Antshel, Linda E. Campbell, Nicolas A. Crossley, T. Blaine Crowley, Eileen Daly, Ania M. Fiksinski, Jennifer K. Forsyth, Wanda Fremont, Naomi J. Goodrich‐Hunsaker, Maria Gudbrandsen, Rachel K. Jonas, Wendy R. Kates, Amy Lin, Kathryn L. McCabe, Hayley Moss, Declan G. Murphy, Kieran C. Murphy, Michael J. Owen, Kosha Ruparel, Tony. J. Simon, Therese van Amelsvoort, and Jacob A. S. Vorstman
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Abdellaoui, A. , Ehli, E. A. , Hottenga, J. J. , Weber, Z. , Mbarek, H. , Willemsen, G. , … Boomsma, D. I. (2015). CNV concordance in 1,097 MZ twin pairs. Twin Research and Human Genetics, 18(1), 1–12. 10.1017/thg.2014.86 [DOI] [PubMed] [Google Scholar]
- Adams, H. H. , Hibar, D. P. , Chouraki, V. , Stein, J. L. , Nyquist, P. A. , Renteria, M. E. , … Thompson, P. M. (2016). Novel genetic loci underlying human intracranial volume identified through genome‐wide association. Nature Neuroscience, 19(12), 1569–1582. 10.1038/nn.4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari, B. M. , Jahanshad, N. , Shukla, D. , Glahn, D. C. , Blangero, J. , Fox, P. T. , … Kochunov, P. (2018). Comparison of heritability estimates on resting state fMRI connectivity phenotypes using the ENIGMA analysis pipeline. Human Brain Mapping, 39(12), 4893–4902. 10.1002/hbm.24331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander‐Bloch, A. , Raznahan, A. , Bullmore, E. , & Giedd, J. (2013). The convergence of maturational change and structural covariance in human cortical networks. The Journal of Neuroscience, 33(7), 2889–2899. 10.1523/JNEUROSCI.3554-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro‐Almagro, F. , Jenkinson, M. , Bangerter, N. K. , Andersson, J. L. , Griffanti, L. , Douaud, G. , … Vallee, E. (2018). Image processing and quality control for the first 10,000 brain imaging datasets from UKbiobank. NeuroImage, 166, 400–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J. A. , & Eichler, E. E. (2006). Primate segmental duplications: Crucibles of evolution, diversity and disease. Nature Reviews. Genetics, 7(7), 552–564. 10.1038/nrg1895 [DOI] [PubMed] [Google Scholar]
- Bakker, G. , Caan, M. W. , Schluter, R. S. , Bloemen, O. J. , da Silva‐Alves, F. , de Koning, M. B. , … van Amelsvoort, T. A. (2016). Distinct white‐matter aberrations in 22q11.2 deletion syndrome and patients at ultra‐high risk for psychosis. Psychological Medicine, 46(11), 2299–2311. 10.1017/s0033291716000970 [DOI] [PubMed] [Google Scholar]
- Bergen, S. E. , Ploner, A. , Howrigan, D. , O'Donovan, M. C. , Smoller, J. W. , Sullivan, P. F. , … Kendler, K. S. (2018). Joint contributions of rare copy number variants and common SNPs to risk for schizophrenia. The American Journal of Psychiatry, 176, 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier, R. , Steinman, K. J. , Reilly, B. , Wallace, A. S. , Sherr, E. H. , Pojman, N. , … Chung, W. K. (2016). Clinical phenotype of the recurrent 1q21.1 copy‐number variant. Genetics in Medicine, 18(4), 341–349. 10.1038/gim.2015.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma, E. K. , Gijsbers, A. C. , Schuurs‐Hoeijmakers, J. H. , van Haeringen, A. , Fransen van de Putte, D. E. , Anderlid, B. M. , … Ruivenkamp, C. A. (2009). Extending the phenotype of recurrent rearrangements of 16p11.2: Deletions in mentally retarded patients without autism and in normal individuals. European Journal of Medical Genetics, 52(2–3), 77–87. 10.1016/j.ejmg.2009.03.006 [DOI] [PubMed] [Google Scholar]
- Blazejewski, S. M. , Bennison, S. A. , Smith, T. H. , & Toyo‐Oka, K. (2018). Neurodevelopmental genetic diseases associated with microdeletions and microduplications of chromosome 17p13.3. Frontiers in Genetics, 9, 80. 10.3389/fgene.2018.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedhoe, P. S. , Schmaal, L. , Abe, Y. , Ameis, S. H. , Arnold, P. D. , Batistuzzo, M. C. , … van den Heuvel, O. A. (2017). Distinct subcortical volume alterations in pediatric and adult OCD: A worldwide meta‐ and mega‐analysis. The American Journal of Psychiatry, 174(1), 60–69. 10.1176/appi.ajp.2016.16020201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedhoe, P. S. W. , Heymans, M. W. , Schmaal, L. , Abe, Y. , Alonso, P. , Ameis, S. H. , … Twisk, J. W. R. (2018). An empirical comparison of meta‐ and mega‐analysis with data from the ENIGMA obsessive‐compulsive disorder working group. Frontiers in Neuroinformatics, 12, 102. 10.3389/fninf.2018.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boot, E. , Butcher, N. J. , Udow, S. , Marras, C. , Mok, K. Y. , Kaneko, S. , … Bassett, A. S. (2018). Typical features of Parkinson disease and diagnostic challenges with microdeletion 22q11.2. Neurology, 90(23), e2059–e2067. 10.1212/wnl.0000000000005660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti‐Pierri, N. , Berg, J. S. , Scaglia, F. , Belmont, J. , Bacino, C. A. , Sahoo, T. , … Patel, A. (2008). Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nature Genetics, 40(12), 1466–1471. 10.1038/ng.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckholtz, J. W. , & Meyer‐Lindenberg, A. (2012). Psychopathology and the human connectome: Toward a transdiagnostic model of risk for mental illness. Neuron, 74(6), 990–1004. 10.1016/j.neuron.2012.06.002 [DOI] [PubMed] [Google Scholar]
- Butcher, N. J. , Kiehl, T. R. , Hazrati, L. N. , Chow, E. W. , Rogaeva, E. , Lang, A. E. , & Bassett, A. S. (2013). Association between early‐onset Parkinson disease and 22q11.2 deletion syndrome: Identification of a novel genetic form of Parkinson disease and its clinical implications. JAMA Neurology, 70(11), 1359–1366. 10.1001/jamaneurol.2013.3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzdok, D. , Nichols, T. E. , & Smith, S. M. (2019). Towards algorithmic analytics for large‐scale datasets. Nature Machine Intelligence, 1, 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas‐de‐la‐Parra, A. , Martin‐Brevet, S. , Moreau, C. , Rodriguez‐Herreros, B. , Fonov, V. S. , Maillard, A. M. , … Louis Collins, D. (2019). Developmental trajectories of neuroanatomical alterations associated with the 16p11.2 copy number variations. NeuroImage, 203, 116155. 10.1016/j.neuroimage.2019.116155 [DOI] [PubMed] [Google Scholar]
- Carter, N. P. (2007). Methods and strategies for analyzing copy number variation using DNA microarrays. Nature Genetics, 39(7 Suppl), S16–S21. 10.1038/ng2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey, B. J. , Cannonier, T. , Conley, M. I. , Cohen, A. O. , Barch, D. M. , Heitzeg, M. M. , … Dale, A. M. (2018). The adolescent brain cognitive development (ABCD) study: Imaging acquisition across 21 sites. Developmental Cognitive Neuroscience, 32, 43–54. 10.1016/j.dcn.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera‐Carles, L. , Pagonabarraga, J. , Pascual‐Sedano, B. , Pastor, P. , Campolongo, A. , Fortea, J. , … Clarimón, J. (2016). Copy number variation analysis of the 17q21.31 region and its role in neurodegenerative diseases. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 171b(2), 175–180. 10.1002/ajmg.b.32390 [DOI] [PubMed] [Google Scholar]
- Chai, J. H. , Locke, D. P. , Greally, J. M. , Knoll, J. H. , Ohta, T. , Dunai, J. , … Nicholls, R. D. (2003). Identification of four highly conserved genes between breakpoint hotspots BP1 and BP2 of the Prader‐Willi/Angelman syndromes deletion region that have undergone evolutionary transposition mediated by flanking duplicons. American Journal of Human Genetics, 73(4), 898–925. 10.1086/378816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y. S. , Owen, J. P. , Pojman, N. J. , Thieu, T. , Bukshpun, P. , Wakahiro, M. L. , … Mukherjee, P. (2016). Reciprocal white matter alterations due to 16p11.2 chromosomal deletions versus duplications. Human Brain Mapping, 37(8), 2833–2848. 10.1002/hbm.23211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawner, S. , Owen, M. J. , Holmans, P. , Raymond, F. L. , Skuse, D. , Hall, J. , & van den Bree, M. B. M. (2019). Genotype‐phenotype associations in children with copy number variants associated with high neuropsychiatric risk in the UK(IMAGINE‐ID): A case‐control cohort study. Lancet Psychiatry, 6(6), 493–505. 10.1016/s2215-0366(19)30123-3 [DOI] [PubMed] [Google Scholar]
- Ching, C. R. K. , Gutman, B. A. , Sun, D. , Villalon Reina, J. , Ragothaman, A. , Isaev, D. , … Bearden, C. E. (2020). Mapping subcortical brain alterations in 22q11.2 deletion syndrome: Effects of deletion size and convergence with idiopathic neuropsychiatric illness. The American Journal of Psychiatry, 177, 589–600. 10.1176/appi.ajp.2019.19060583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleynen, I. , Engchuan, W. , Hestand, M. S. , Heung, T. , Holleman, A. M. , Johnston, H. R. , … Bassett, A. S. (2020). Genetic contributors to risk of schizophrenia in the presence of a 22q11.2 deletion. Molecular Psychiatry. 10.1038/s41380-020-0654-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, D. M. , & Butler, M. G. (2015). The 15q11.2 BP1‐BP2 microdeletion syndrome: A review. International Journal of Molecular Sciences, 16(2), 4068–4082. 10.3390/ijms16024068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, K. , Bracher‐Smith, M. , Owen, D. , Kendall, K. M. , Rees, E. , Pardinas, A. F. , … Kirov, G. (2018). Medical consequences of pathogenic CNVs in adults: Analysis of the UKbiobank. Journal of Medical Genetics, 56, 131–138. 10.1136/jmedgenet-2018-105477 [DOI] [PubMed] [Google Scholar]
- Cuccaro, D. , De Marco, E. V. , Cittadella, R. , & Cavallaro, S. (2017). Copy number variants in Alzheimer's disease. Journal of Alzheimer's Disease, 55(1), 37–52. 10.3233/jad-160469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, A. C. , Delport, S. , Cumines, W. , Busse, M. , Linden, D. E. J. , Hall, J. , … van den Bree, M. B. M. (2018). Developmental coordination disorder, psychopathology and IQ in 22q11.2 deletion syndrome. The British Journal of Psychiatry, 212(1), 27–33. 10.1192/bjp.2017.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, A. C. , Hall, J. , Einfeld, S. , Owen, M. J. , & van den Bree, M. B. M. (2020). Assessment of emotions and behaviour by the developmental behaviour checklist in young people with neurodevelopmental CNVs. Psychological Medicine. 10.1017/s0033291720002330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, R. W. , Fiksinski, A. M. , Breetvelt, E. J. , Williams, N. M. , Hooper, S. R. , Monfeuga, T. , … Vorstman, J. A. S. (2020). Using common genetic variation to examine phenotypic expression and risk prediction in 22q11.2 deletion syndrome. Nature Medicine, 26(12), 1912–1918. 10.1038/s41591-020-1103-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Menil, V. , Hoogenhout, M. , Kipkemoi, P. , Kamuya, D. , Eastman, E. , Galvin, A. , … Robinson, E. (2019). The NeuroDev study: Phenotypic and genetic characterization of neurodevelopmental disorders in Kenya and South Africa. Neuron, 101(1), 15–19. 10.1016/j.neuron.2018.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis, S. , Pasciuto, E. , Li, K. W. , Fernández, E. , Di Marino, D. , Buzzi, A. , … Bagni, C. (2013). CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron, 79(6), 1169–1182. 10.1016/j.neuron.2013.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan, R. S. , Ségonne, F., Fischl, B., Quinn, B. T., Dickerson, B. C., Blacker, D., … Killiany, R. J. (2006). An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage, 31(3), 968–980. 10.1016/j.neuroimage.2006.01.02 [DOI] [PubMed] [Google Scholar]
- Demontis, D. , Walters, R. K. , Martin, J. , Mattheisen, M. , Als, T. D. , Agerbo, E. , … Neale, B. M. (2019). Discovery of the first genome‐wide significant risk loci for attention deficit/hyperactivity disorder. Nature Genetics, 51(1), 63–75. 10.1038/s41588-018-0269-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, M. Y. , & Eichler, E. E. (2016). Human adaptation and evolution by segmental duplication. Current Opinion in Genetics & Development, 41, 44–52. 10.1016/j.gde.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, M. Y. , Nuttle, X. , Sudmant, P. H. , Antonacci, F. , Graves, T. A. , Nefedov, M. , … Eichler, E. E. (2012). Evolution of human‐specific neural SRGAP2 genes by incomplete segmental duplication. Cell, 149(4), 912–922. 10.1016/j.cell.2012.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande, A. , & Weiss, L. A. (2018). Recurrent reciprocal copy number variants: Roles and rules in neurodevelopmental disorders. Developmental Neurobiology, 78(5), 519–530. 10.1002/dneu.22587 [DOI] [PubMed] [Google Scholar]
- Dolcetti, A. , Silversides, C. K. , Marshall, C. R. , Lionel, A. C. , Stavropoulos, D. J. , Scherer, S. W. , & Bassett, A. S. (2013). 1q21.1 microduplication expression in adults. Genetics in Medicine, 15(4), 282–289. 10.1038/gim.2012.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez‐Iturza, N. , Lo, A. C. , Shah, D. , Armendariz, M. , Vannelli, A. , Mercaldo, V. , … Bagni, C. (2019). The autism‐ and schizophrenia‐associated protein CYFIP1 regulates bilateral brain connectivity and behaviour. Nature Communications, 10(1), 3454. 10.1038/s41467-019-11203-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty, M. L. , Nuttle, X. , Penn, O. , Nelson, B. J. , Huddleston, J. , Baker, C. , … Eichler, E. E. (2017). The birth of a human‐specific neural gene by incomplete duplication and gene fusion. Genome Biology, 18(1), 49. 10.1186/s13059-017-1163-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakesmith, M. , Parker, G. D. , Smith, J. , Linden, S. C. , Rees, E. , Williams, N. , … Linden, D. E. J. (2019). Genetic risk for schizophrenia and developmental delay is associated with shape and microstructure of midline white‐matter structures. Translational Psychiatry, 9(1), 102. 10.1038/s41398-019-0440-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmaz, A. A. , Karaca, E. , Demkow, U. , Toruner, G. , Schoumans, J. , & Cogulu, O. (2015). Evolution of genetic techniques: Past, present, and beyond. BioMed Research International, 2015, 461524–461527. 10.1155/2015/461524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, C. C. , Brown, T. T. , Bartsch, H. , Kuperman, J. M. , Hagler, D. J., Jr. , Schork, A. , … Dale, A. M. (2017). Williams syndrome‐specific neuroanatomical profile and its associations with behavioral features. NeuroImage: Clinical, 15, 343–347. 10.1016/j.nicl.2017.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk, L. , Marshall, C. R. , Wintle, R. F. , & Scherer, S. W. (2006). Structural variants: Changing the landscape of chromosomes and design of disease studies. Human Molecular Genetics, 15, R57–R66. 10.1093/hmg/ddl057 [DOI] [PubMed] [Google Scholar]
- Fiddes, I. T. , Lodewijk, G. A. , Mooring, M. , Bosworth, C. M. , Ewing, A. D. , Mantalas, G. L. , … Haussler, D. (2018). Human‐specific NOTCH2NL genes affect NOTCH signaling and cortical neurogenesis. Cell, 173(6), 1356–1369.e1322. 10.1016/j.cell.2018.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl, B. , Salat, D. H. , Busa, E. , Albert, M. , Dieterich, M. , Haselgrove, C. , … Dale, A. M. (2002). Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron, 33(3), 341–355. 10.1016/s0896-6273(02)00569-x [DOI] [PubMed] [Google Scholar]
- Gentile, G. , La Cognata, V. , & Cavallaro, S. (2020). The contribution of CNVs to the most common aging‐related neurodegenerative diseases. Aging Clinical and Experimental Research. 10.1007/s40520-020-01485-4 [DOI] [PubMed] [Google Scholar]
- Giannuzzi, G. , Schmidt, P. J. , Porcu, E. , Willemin, G. , Munson, K. M. , Nuttle, X. , … Reymond, A. (2019). The human‐specific BOLA2 duplication modifies iron homeostasis and anemia predisposition in chromosome 16p11.2 autism individuals. American Journal of Human Genetics, 105(5), 947–958. 10.1016/j.ajhg.2019.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan, S. , Rosenfeld, J. A. , Coe, B. P. , Parikh, S. , Friedman, N. , Goldstein, A. , … Eichler, E. E. (2012). Phenotypic heterogeneity of genomic disorders and rare copy‐number variants. The New England Journal of Medicine, 367(14), 1321–1331. 10.1056/NEJMoa1200395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourari, I. , Schubert, R. , & Prasad, A. (2018). 1q21.1 duplication syndrome and epilepsy: Case report and review. Neurology: Genetics, 4(1), e219. 10.1212/nxg.0000000000000219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goytain, A. , Hines, R. M. , El‐Husseini, A. , & Quamme, G. A. (2007). NIPA1(SPG6), the basis for autosomal dominant form of hereditary spastic paraplegia, encodes a functional Mg2+ transporter. The Journal of Biological Chemistry, 282(11), 8060–8068. 10.1074/jbc.M610314200 [DOI] [PubMed] [Google Scholar]
- Goytain, A. , Hines, R. M. , & Quamme, G. A. (2008). Functional characterization of NIPA2, a selective Mg2+ transporter. American Journal of Physiology. Cell Physiology, 295(4), C944–C953. 10.1152/ajpcell.00091.2008 [DOI] [PubMed] [Google Scholar]
- Grasby, K. L. , Jahanshad, N. , Painter, J. N. , Colodro‐Conde, L. , Bralten, J. , Hibar, D. P. , … Medland, S. E. (2020). The genetic architecture of the human cerebral cortex. Science, 367(6484), eaay6690. 10.1126/science.aay6690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, E. K. , Rees, E. , Walters, J. T. , Smith, K. G. , Forty, L. , Grozeva, D. , … Kirov, G. (2016). Copy number variation in bipolar disorder. Molecular Psychiatry, 21(1), 89–93. 10.1038/mp.2014.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove, J. , Ripke, S. , Als, T. D. , Mattheisen, M. , Walters, R. K. , Won, H. , … Børglum, A. D. (2019). Identification of common genetic risk variants for autism spectrum disorder. Nature Genetics, 51(3), 431–444. 10.1038/s41588-019-0344-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson, O. O. , Walters, G. B. , Ingason, A. , Johansson, S. , Zayats, T. , Athanasiu, L. , … Stefansson, K. (2019). Attention‐deficit hyperactivity disorder shares copy number variant risk with schizophrenia and autism spectrum disorder. Translational Psychiatry, 9(1), 258. 10.1038/s41398-019-0599-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsuner, S. , Stein, D. J. , Susser, E. S. , Sibeko, G. , Pretorius, A. , Walsh, T. , … McClellan, J. M. (2020). Genetics of schizophrenia in the South African Xhosa. Science, 367(6477), 569–573. 10.1126/science.aay8833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur, R. E. , Bassett, A. S. , McDonald‐McGinn, D. M. , Bearden, C. E. , Chow, E. , Emanuel, B. S. , … Morrow, B. (2017). A neurogenetic model for the study of schizophrenia spectrum disorders: The international 22q11.2 deletion syndrome brain behavior consortium. Molecular Psychiatry, 22(12), 1664–1672. 10.1038/mp.2017.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, M. W. , Demuth, J. P. , & Han, S. G. (2007). Accelerated rate of gene gain and loss in primates. Genetics, 177(3), 1941–1949. 10.1534/genetics.107.080077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldeman‐Englert, C. R. , & Jewett, T. (1993). 1q21.1 Recurrent Microdeletion. In Adam M. P., Ardinger H. H., Pagon R. A., Wallace S. E., Bean L. J. H., Stephens K., & Amemiya A. (Eds.), GeneReviews([R]). Seattle (WA): University of Washington, Seattle University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. [Google Scholar]
- Harel, T. , & Lupski, J. R. (2018). Genomic disorders 20 years on‐mechanisms for clinical manifestations. Clinical Genetics, 93(3), 439–449. 10.1111/cge.13146 [DOI] [PubMed] [Google Scholar]
- Hastings, P. J. , Lupski, J. R. , Rosenberg, S. M. , & Ira, G. (2009). Mechanisms of change in gene copy number. Nature Reviews. Genetics, 10(8), 551–564. 10.1038/nrg2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz, M. J. , Lein, E. S. , Guillozet‐Bongaarts, A. L. , Shen, E. H. , Ng, L. , Miller, J. A. , … Jones, A. R. (2012). An anatomically comprehensive atlas of the adult human brain transcriptome. Nature, 489(7416), 391–399. 10.1038/nature11405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar, D. P. , Adams, H. H. , Jahanshad, N. , Chauhan, G. , Stein, J. L. , Hofer, E. , … Ikram, M. A. (2017). Novel genetic loci associated with hippocampal volume. Nature Communications, 8, 13624. 10.1038/ncomms13624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar, D. P. , Stein, J. L. , Renteria, M. E. , Arias‐Vasquez, A. , Desrivieres, S. , Jahanshad, N. , … Medland, S. E. (2015). Common genetic variants influence human subcortical brain structures. Nature, 520(7546), 224–229. 10.1038/nature14101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar, D. P. , Westlye, L. T. , van Erp, T. G. , Rasmussen, J. , Leonardo, C. D. , Faskowitz, J. , … Andreassen, O. A. (2016). Subcortical volumetric abnormalities in bipolar disorder. Molecular Psychiatry, 21(12), 1710–1716. 10.1038/mp.2015.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilker, R. , Helenius, D. , Fagerlund, B. , Skytthe, A. , Christensen, K. , Werge, T. M. , … Glenthoj, B. (2018). Heritability of schizophrenia and schizophrenia spectrum based on the nationwide Danish twin register. Biological Psychiatry, 83(6), 492–498. 10.1016/j.biopsych.2017.08.017 [DOI] [PubMed] [Google Scholar]
- Hofer, E. , Roshchupkin, G. V. , Adams, H. H. H. , Knol, M. J. , Lin, H. , Li, S. , … Seshadri, S. (2020). Genetic correlations and genome‐wide associations of cortical structure in general population samples of 22,824 adults. Nature Communications, 11(1), 4796. 10.1038/s41467-020-18367-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogman, M. , Bralten, J. , Hibar, D. P. , Mennes, M. , Zwiers, M. P. , Schweren, L. S. J. , … Franke, B. (2017). Subcortical brain volume differences in participants with attention deficit hyperactivity disorder in children and adults: A cross‐sectional mega‐analysis. Lancet Psychiatry, 4(4), 310–319. 10.1016/s2215-0366(17)30049-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguet, G. , Schramm, C. , Douard, E. , Jiang, L. , Labbe, A. , Tihy, F. , … Jacquemont, S. (2018). Measuring and estimating the effect sizes of copy number variants on general intelligence in community‐based samples. JAMA Psychiatry, 75(5), 447–457. 10.1001/jamapsychiatry.2018.0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate, A. J. , Feuk, L. , Rivera, M. N. , Listewnik, M. L. , Donahoe, P. K. , Qi, Y. , … Lee, C. (2004). Detection of large‐scale variation in the human genome. Nature Genetics, 36(9), 949–951. 10.1038/ng1416 [DOI] [PubMed] [Google Scholar]
- Innan, H. , & Kondrashov, F. (2010). The evolution of gene duplications: Classifying and distinguishing between models. Nature Reviews. Genetics, 11(2), 97–108. 10.1038/nrg2689 [DOI] [PubMed] [Google Scholar]
- Jacquemont, S. , Reymond, A. , Zufferey, F. , Harewood, L. , Walters, R. G. , Kutalik, Z. , … Froguel, P. (2011). Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature, 478(7367), 97–102. 10.1038/nature10406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad, N. , Kochunov, P. V. , Sprooten, E. , Mandl, R. C. , Nichols, T. E. , Almasy, L. , … Glahn, D. C. (2013). Multi‐site genetic analysis of diffusion images and voxelwise heritability analysis: A pilot project of the ENIGMA‐DTI working group. NeuroImage, 81, 455–469. 10.1016/j.neuroimage.2013.04.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, A. G. , Mous, S. E. , White, T. , Posthuma, D. , & Polderman, T. J. (2015). What twin studies tell us about the heritability of brain development, morphology, and function: A review. Neuropsychology Review, 25(1), 27–46. 10.1007/s11065-015-9,278-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jønch, A. E. , Douard, E. , Moreau, C. , Van Dijck, A. , Passeggeri, M. , Kooy, F. , … Jacquemont, S. (2019). Estimating the effect size of the 15Q11.2 BP1‐BP2 deletion and its contribution to neurodevelopmental symptoms: Recommendations for practice. Journal of Medical Genetics, 56(10), 701–710. 10.1136/jmedgenet-2018-105,879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, S. , Jahanshad, N. , Zalesky, A. , Kochunov, P. , Agartz, I. , Alloza, C. , … Donohoe, G. (2018). Widespread white matter microstructural differences in schizophrenia across 4,322 individuals: Results from the ENIGMA schizophrenia DTI working group. Molecular Psychiatry, 23(5), 1261–1269. 10.1038/mp.2017.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall, K. M. , Bracher‐Smith, M. , Fitzpatrick, H. , Lynham, A. , Rees, E. , Escott‐Price, V. , … Kirov, G. (2019). Cognitive performance and functional outcomes of carriers of pathogenic copy number variants: Analysis of the UKbiobank. The British Journal of Psychiatry, 214(5), 297–304. 10.1192/bjp.2018.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall, K. M. , Rees, E. , Bracher‐Smith, M. , Legge, S. , Riglin, L. , Zammit, S. , … Walters, J. T. R. (2019). Association of Rare Copy Number Variants with Risk of depression. JAMA Psychiatry, 76(8), 818–825. 10.1001/jamapsychiatry.2019.0566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall, K. M. , Rees, E. , Escott‐Price, V. , Einon, M. , Thomas, R. , Hewitt, J. , … Kirov, G. (2016). Cognitive performance among carriers of pathogenic copy number variants: Analysis of 152,000 UKbiobank subjects. Biological Psychiatry, 82, 103–110. 10.1016/j.biopsych.2016.08.014 [DOI] [PubMed] [Google Scholar]
- Khandaker, G. M. , Cousins, L. , Deakin, J. , Lennox, B. R. , Yolken, R. , & Jones, P. B. (2015). Inflammation and immunity in schizophrenia: Implications for pathophysiology and treatment. Lancet Psychiatry, 2(3), 258–270. 10.1016/s2215-0366(14)00122-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov, G. , Rees, E. , & Walters, J. (2015). What a psychiatrist needs to know about copy number variants. BJPsych Advances, 21(3), 157–163. 10.1192/apt.bp.113.012039 [DOI] [Google Scholar]
- Kirov, G. , Rees, E. , Walters, J. T. R. , Escott‐Price, V. , Georgieva, L. , Richards, A. L. , … Owen, M. J. (2014). The penetrance of copy number variations for schizophrenia and developmental delay. Biological Psychiatry, 75(5), 378–385. 10.1016/j.biopsych.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen, P. , Duijff, S. , Swanenburg de Veye, H. , Beemer, F. , Sinnema, G. , Breetvelt, E. , … Vorstman, J. (2016). Explaining the variable penetrance of CNVs: Parental intelligence modulates expression of intellectual impairment caused by the 22q11.2 deletion. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 171(6), 790–796. 10.1002/ajmg.b.32441 [DOI] [PubMed] [Google Scholar]
- Knol, M. J. , Poot, R. A. , Evans, T. E. , Satizabal, C. L. , Mishra, A. , van der Auwera, S. , … Adams, H. H. H. (2020). Genetic variants for head size share genes and pathways with cancer. bioRxiv , 2020.2007.2015.191114. doi: 10.1101/2020.07.15.191114 [DOI] [PMC free article] [PubMed]
- Kochunov, P. , Fu, M. , Nugent, K. , Wright, S. N. , Du, X. , Muellerklein, F. , … Hong, L. E. (2016). Heritability of complex white matter diffusion traits assessed in a population isolate. Human Brain Mapping, 37(2), 525–535. 10.1002/hbm.23047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov, P. , Hong, L. E. , Dennis, E. L. , Morey, R. A. , Tate, D. F. , Wilde, E. A. , … Jahanshad, N. (2020). ENIGMA‐DTI: Translating reproducible white matter deficits into personalized vulnerability metrics in cross‐diagnostic psychiatric research. Human Brain Mapping. 10.1002/hbm.24998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer, S. , & Gresham, D. (2019). An evolving view of copy number variants. Current Genetics, 65(6), 1287–1295. 10.1007/s00294-019-00980-0 [DOI] [PubMed] [Google Scholar]
- Lin, A. , Ching, C. R. K. , Vajdi, A. , Sun, D. , Jonas, R. K. , Jalbrzikowski, M. , … Bearden, C. E. (2017). Mapping 22q11.2 gene dosage effects on brain Morphometry. The Journal of Neuroscience, 37(26), 6183–6199. 10.1523/jneurosci.3759-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, A. , Vajdi, A. , Kushan‐Wells, L. , Helleman, G. , Hansen, L. P. , Jonas, R. K. , … Bearden, C. E. (2020). Reciprocal copy number variations at 22q11.2 produce distinct and convergent neurobehavioral impairments relevant for schizophrenia and autism Spectrum disorder. Biological Psychiatry, 88(3), 260–272. 10.1016/j.biopsych.2019.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlejohns, T. J. , Holliday, J. , Gibson, L. M. , Garratt, S. , Oesingmann, N. , Alfaro‐Almagro, F. , … Allen, N. E. (2020). The UKbiobank imaging enhancement of 100,000 participants: Rationale, data collection, management and future directions. Nature Communications, 11(1), 2624. 10.1038/s41467-020-15,948-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loviglio, M. N. , Arbogast, T. , Jonch, A. E. , Collins, S. C. , Popadin, K. , Bonnet, C. S. , … Reymond, A. (2017). The immune signaling adaptor LAT contributes to the neuroanatomical phenotype of 16p11.2 BP2‐BP3 CNVs. American Journal of Human Genetics, 101(4), 564–577. 10.1016/j.ajhg.2017.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loviglio, M. N. , Leleu, M. , Mannik, K. , Passeggeri, M. , Giannuzzi, G. , van der Werf, I. , … Reymond, A. (2016). Chromosomal contacts connect loci associated with autism, BMI and head circumference phenotypes. Molecular Psychiatry, 22, 836–849. 10.1038/mp.2016.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowther, C. , Speevak, M. , Armour, C. M. , Goh, E. S. , Graham, G. E. , Li, C. , … Bassett, A. S. (2017). Molecular characterization of NRXN1 deletions from 19,263 clinical microarray cases identifies exons important for neurodevelopmental disease expression. Genetics in Medicine, 19(1), 53–61. 10.1038/gim.2016.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macé, A. , Tuke, M. A. , Beckmann, J. S. , Lin, L. , Jacquemont, S. , Weedon, M. N. , … Kutalik, Z. (2016). New quality measure for SNP array based CNV detection. Bioinformatics, 32(21), 3,298–3,305. 10.1093/bioinformatics/btw477 [DOI] [PubMed] [Google Scholar]
- Mace, A. , Tuke, M. A. , Deelen, P. , Kristiansson, K. , Mattsson, H. , Noukas, M. , … Kutalik, Z. (2017). CNV‐association meta‐analysis in 191,161 European adults reveals new loci associated with anthropometric traits. Nature Communications, 8(1), 744. 10.1038/s41467-017-00556-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard, A. M. , Ruef, A. , Pizzagalli, F. , Migliavacca, E. , Hippolyte, L. , Adaszewski, S. , … Jacquemont, S. (2015). The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Molecular Psychiatry, 20(1), 140–147. 10.1038/mp.2014.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Männik, K. , Mägi, R. , Macé, A. , Cole, B. , Guyatt, A. L. , Shihab, H. A. , … Reymond, A. (2015). Copy number variations and cognitive phenotypes in unselected populations. JAMA, 313(20), 2044–2054. 10.1001/jama.2015.4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquand, A. F. , Rezek, I. , Buitelaar, J. , & Beckmann, C. F. (2016). Understanding heterogeneity in clinical cohorts using normative models: Beyond case–control studies. Biological Psychiatry, 80, 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, C. R. , Howrigan, D. P. , Merico, D. , Thiruvahindrapuram, B. , Wu, W. , Greer, D. S. , … Sebat, J. (2017). Contribution of copy number variants to schizophrenia from a genome‐wide study of 41,321 subjects. Nature Genetics, 49(1), 27–35. 10.1038/ng.3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, J. , O'Donovan, M. C. , Thapar, A. , Langley, K. , & Williams, N. (2015). The relative contribution of common and rare genetic variants to ADHD. Translational Psychiatry, 5, e506. 10.1038/tp.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Brevet, S. , Rodriguez‐Herreros, B. , Nielsen, J. A. , Moreau, C. , Modenato, C. , Maillard, A. M. , … Jacquemont, S. (2018). Quantifying the effects of 16p11.2 copy number variants on brain structure: A multi‐site “genetic‐first” study. Biological Psychiatry, 84, 253–264. 10.1016/j.biopsych.2018.02.1176 [DOI] [PubMed] [Google Scholar]
- McDonald‐McGinn, D. M. , Sullivan, K. E. , Marino, B. , Philip, N. , Swillen, A. , Vorstman, J. A. , … Bassett, A. S. (2015). 22q11.2 deletion syndrome. Nature Reviews. Disease Primers, 1, 15071. 10.1038/nrdp.2015.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meda, S. A. , Pryweller, J. R. , & Thornton‐Wells, T. A. (2012). Regional brain differences in cortical thickness, surface area and subcortical volume in individuals with Williams syndrome. PLoS One, 7(2), e31913. 10.1371/journal.pone.0031913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medland, S. E. , Grasby, K. L. , Jahanshad, N. , Painter, J. N. , Colodro‐Conde, L. , Bralten, J. , … P.M., on behalf of the ENIGMA genetics working group . (2020). Ten years of enhancing neuro‐imaging genetics through meta‐analysis: An overview from the ENIGMA genetics working group. Human Brain Mapping: ENIGMA Consortium Special Issue, 43 (1), 292–299. 10.1002/hbm.25311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford, H. C. , Sharp, A. J. , Baker, C. , Itsara, A. , Jiang, Z. , Buysse, K. , … Eichler, E. E. (2008). Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. The New England Journal of Medicine, 359(16), 1685–1699. 10.1056/NEJMoa0805384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithyantha, R. , Kneen, R. , McCann, E. , & Gladstone, M. (2017). Current evidence‐based recommendations on investigating children with global developmental delay. Archives of Disease in Childhood, 102(11), 1071–1076. 10.1136/archdischild-2016-311271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan, K. N. , Cao, Y. , Pham, J. , Cheung, S. W. , Hoffner, L. , Ou, Z. Z. , … Beaudet, A. L. (2019). Phenotypic association of 15q11.2 CNVs of the region of breakpoints 1–2 (BP1‐BP2) in a large cohort of samples referred for genetic diagnosis. Journal of Human Genetics, 64(3), 253–255. 10.1038/s10038-018-0543-7 [DOI] [PubMed] [Google Scholar]
- Moreau, C. A. , Urchs, S. G. W. , Kuldeep, K. , Orban, P. , Schramm, C. , Dumas, G. , … Jacquemont, S. (2020). Mutations associated with neuropsychiatric conditions delineate functional brain connectivity dimensions contributing to autism and schizophrenia. Nature Communications, 11(1), 5,272. 10.1038/s41467-020-18,997-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, S. , Oishi, K. , Jiang, H. , Jiang, L. , Li, X. , Akhter, K. , … Mazziotta, J. (2008). Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. NeuroImage, 40(2), 570–582. 10.1016/j.neuroimage.2007.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulding, H. A. , Bartsch, U. , Hall, J. , Jones, M. W. , Linden, D. E. , Owen, M. J. , & van den Bree, M. B. M. (2020). Sleep problems and associations with psychopathology and cognition in young people with 22q11.2 deletion syndrome (22q11.2DS). Psychological Medicine, 50(7), 1191–1202. 10.1017/s0033291719001119 [DOI] [PubMed] [Google Scholar]
- Munnich, A. , Demily, C. , Frugere, L. , Duwime, C. , Malan, V. , Barcia, G. , … Assouline, M. (2019). Impact of on‐site clinical genetics consultations on diagnostic rate in children and young adults with autism spectrum disorder. Molecular Autism, 10, 33. 10.1186/s13229-019-0284-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli, I. , Mercaldo, V. , Boyl, P. P. , Eleuteri, B. , Zalfa, F. , De Rubeis, S. , … Bagni, C. (2008). The fragile X syndrome protein represses activity‐dependent translation through CYFIP1, a new 4E‐BP. Cell, 134(6), 1042–1054. 10.1016/j.cell.2008.07.031 [DOI] [PubMed] [Google Scholar]
- Niarchou, M. , Chawner, S. , Doherty, J. L. , Maillard, A. M. , Jacquemont, S. , Chung, W. K. , … Bree, M. (2019). Psychiatric disorders in children with 16p11.2 deletion and duplication. Translational Psychiatry, 9(1), 8. 10.1038/s41398-018-0339-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, J. , Fejgin, K. , Sotty, F. , Nielsen, V. , Mork, A. , Christoffersen, C. T. , … Didriksen, M. (2017). A mouse model of the schizophrenia‐associated 1q21.1 microdeletion syndrome exhibits altered mesolimbic dopamine transmission. Translational Psychiatry, 7(11), 1261. 10.1038/s41398-017-0011-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowska, B. (2017). Clinical interpretation of copy number variants in the human genome. Journal of Applied Genetics, 58(4), 449–457. 10.1007/s13353-017-0407-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen, D. , Bracher‐Smith, M. , Kendall, K. M. , Rees, E. , Einon, M. , Escott‐Price, V. , … Kirov, G. (2018). Effects of pathogenic CNVs on physical traits in participants of the UKbiobank. BMC Genomics, 19(1), 867. 10.1186/s12864-018-5,292-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, Y. , Shin, J. , Gowland, P. A. , Pausova, Z. , Paus, T. , & IMAGEN consortium . (2018). Maturation of the human cerebral cortex during adolescence: Myelin or dendritic arbor? Cerebral Cortex, 29(8), 3,351–3,362 10.1093/cercor/bhy204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpont, M. E. , Brueckner, M. , Chung, W. K. , Garg, V. , Lacro, R. V. , McGuire, A. L. , … Russell, M. W. (2018). Genetic basis for congenital heart disease: Revisited: A scientific statement from the American Heart Association. Circulation, 138(21), e653–e711. 10.1161/cir.0000000000000606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, D. , Darvishi, K. , Shi, X. , Rajan, D. , Rigler, D. , Fitzgerald, T. , … Feuk, L. (2011). Comprehensive assessment of array‐based platforms and calling algorithms for detection of copy number variants. Nature Biotechnology, 29(6), 512–520. 10.1038/nbt.1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psaty, B. M. , & Sitlani, C. (2013). The cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium as a model of collaborative science. Epidemiology, 24(3), 346–348. 10.1097/EDE.0b013e31828b2cbb [DOI] [PubMed] [Google Scholar]
- Qureshi, A. Y. , Mueller, S. , Snyder, A. Z. , Mukherjee, P. , Berman, J. I. , Roberts, T. P. , … Buckner, R. L. (2014). Opposing brain differences in 16p11.2 deletion and duplication carriers. The Journal of Neuroscience, 34(34), 11,199–11,11211. 10.1523/jneurosci.1366-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic, P. (1995). A small step for the cell, a giant leap for mankind: A hypothesis of neocortical expansion during evolution. Trends in Neurosciences, 18(9), 383–388. [DOI] [PubMed] [Google Scholar]
- Rees, E. , Walters, J. T. , Georgieva, L. , Isles, A. R. , Chambert, K. D. , Richards, A. L. , … Kirov, G. (2014). Analysis of copy number variations at 15 schizophrenia‐associated loci. The British Journal of Psychiatry, 204(2), 108–114. 10.1192/bjp.bp.113.131052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss, A. L. , Eckert, M. A. , Rose, F. E. , Karchemskiy, A. , Kesler, S. , Chang, M. , … Galaburda, A. (2004). An experiment of nature: Brain anatomy parallels cognition and behavior in Williams syndrome. The Journal of Neuroscience, 24(21), 5009–5015. 10.1523/jneurosci.5272-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld, J. A. , Coe, B. P. , Eichler, E. E. , Cuckle, H. , & Shaffer, L. G. (2013). Estimates of penetrance for recurrent pathogenic copy‐number variations. Genetics in Medicine, 15(6), 478–481. 10.1038/gim.2012.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld, J. A. , & Patel, A. (2017). Chromosomal microarrays: Understanding genetics of neurodevelopmental disorders and congenital anomalies. Journal of Pediatric Genetics, 6(1), 42–50. 10.1055/s-0036-1584306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld, J. A. , Traylor, R. N. , Schaefer, G. B. , McPherson, E. W. , Ballif, B. C. , Klopocki, E. , … Aylsworth, A. S. (2012). Proximal microdeletions and microduplications of 1q21.1 contribute to variable abnormal phenotypes. European Journal of Human Genetics, 20(7), 754–761. 10.1038/ejhg.2012.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satizabal, C. L. , Adams, H. H. H. , Hibar, D. P. , White, C. C. , Knol, M. J. , Stein, J. L. , … Ikram, M. A. (2019). Genetic architecture of subcortical brain structures in 38,851 individuals. Nature Genetics, 51(11), 1624–1636. 10.1038/s41588-019-0511-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterstrom, F. K. , Kosmicki, J. A. , Wang, J. , Breen, M. S. , De Rubeis, S. , An, J. Y. , … Buxbaum, J. D. (2020). Large‐scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell, 180(3), 568–584.e523. 10.1016/j.cell.2019.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck, A. , Bardoni, B. , Langmann, C. , Harden, N. , Mandel, J. L. , & Giangrande, A. (2003). CYFIP/Sra‐1 controls neuronal connectivity in drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron, 38(6), 887–898. 10.1016/s0896-6,273(03)00354-4 [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium . (2014). Biological insights from 108 schizophrenia‐associated genetic loci. Nature, 511(7510), 421–427. 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaal, L. , Veltman, D. J. , van Erp, T. G. , Samann, P. G. , Frodl, T. , Jahanshad, N. , … Hibar, D. P. (2016). Subcortical brain alterations in major depressive disorder: Findings from the ENIGMA major depressive disorder working group. Molecular Psychiatry, 21(6), 806–812. 10.1038/mp.2015.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, M. , Debbane, M. , Bassett, A. S. , Chow, E. W. , Fung, W. L. , van den Bree, M. , … Eliez, S. (2014). Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: Results from the international consortium on brain and behavior in 22q11.2 deletion syndrome. The American Journal of Psychiatry, 171(6), 627–639. 10.1176/appi.ajp.2013.13070864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat, J. , Lakshmi, B. , Troge, J. , Alexander, J. , Young, J. , Lundin, P. , … Wigler, M. (2004). Large‐scale copy number polymorphism in the human genome. Science, 305(5683), 525–528. 10.1126/science.1098918 [DOI] [PubMed] [Google Scholar]
- Sharp, A. J. , Cheng, Z. , & Eichler, E. E. (2006). Structural variation of the human genome. Annual Review of Genomics and Human Genetics, 7, 407–442. 10.1146/annurev.genom.7.080505.115618 [DOI] [PubMed] [Google Scholar]
- Sharp, A. J. , Hansen, S. , Selzer, R. R. , Cheng, Z. , Regan, R. , Hurst, J. A. , … Eichler, E. E. (2006). Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nature Genetics, 38(9), 1038–1,042. 10.1038/ng1862 [DOI] [PubMed] [Google Scholar]
- Shen, Y. , Chen, X. , Wang, L. , Guo, J. , Shen, J. , An, Y. , … Wu, B. L. (2011). Intra‐family phenotypic heterogeneity of 16p11.2 deletion carriers in a three‐generation Chinese family. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 156(2), 225–232. 10.1002/ajmg.b.31147 [DOI] [PubMed] [Google Scholar]
- Shin, J. , French, L. , Xu, T. , Leonard, G. , Perron, M. , Pike, G. B. , … Paus, T. (2017). Cell‐specific gene‐expression profiles and cortical thickness in the human brain. Cerebral Cortex, 28(9), 3267–3277. 10.1093/cercor/bhx197 [DOI] [PubMed] [Google Scholar]
- Silva, A. I. , Ulfarsson, M. O. , Stefansson, H. , Gustafsson, O. , Walters, G. B. , Linden, D. E. J. , … Stefansson, K. (2018). Reciprocal White matter changes associated with copy number variation at 15q11.2 BP1‐BP2: A diffusion tensor imaging study. Biological Psychiatry, 85, 563–572. 10.1016/j.biopsych.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smajlagić, D. , Lavrichenko, K. , Berland, S. , Helgeland, Ø. , Knudsen, G. P. , Vaudel, M. , … Johansson, S. (2020). Population prevalence and inheritance pattern of recurrent CNVs associated with neurodevelopmental disorders in 12,252 newborns and their parents. European Journal of Human Genetics, 29, 1–11. 10.1038/s41431-020-00707-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, D. J. A. , Wright, M. J. , Meyers, J. L. , Martin, N. G. , Ho, Y. Y. W. , Malone, S. M. , … Boomsma, D. I. (2018). Genome‐wide association analysis links multiple psychiatric liability genes to oscillatory brain activity. Human Brain Mapping, 39(11), 4183–4195. 10.1002/hbm.24238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. M. , Jenkinson, M. , Johansen‐Berg, H. , Rueckert, D. , Nichols, T. E. , Mackay, C. E. , … Behrens, T. E. (2006). Tract‐based spatial statistics: Voxelwise analysis of multi‐subject diffusion data. NeuroImage, 31(4), 1487–1505. 10.1016/j.neuroimage.2006.02.024 [DOI] [PubMed] [Google Scholar]
- Smith, S. M. , & Nichols, T. E. (2018). Statistical challenges in “big data” human neuroimaging. Neuron, 97, 263–268. [DOI] [PubMed] [Google Scholar]
- Sønderby, I. E. , Gustafsson, O. , Doan, N. T. , Hibar, D. P. , Martin‐Brevet, S. , Abdellaoui, A. , … Andreassen, O. A. (2018). Dose response of the 16p11.2 distal copy number variant on intracranial volume and basal ganglia. Molecular Psychiatry, 25(3), 584–602. 10.1038/s41380-018-0118-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby, I. E. , van der Meer, D. , Moreau, C. , Kaufmann, T. , Walters, G. B. , Ellegaard, M. , … Andreassen, O. A. (2021). (accepted). 1q21.1 distal copy number variants are associated with cerebral and cognitive alterations in humans. Translational Psychiatry. 10.1038/s41398-021-01213-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann, M. , Lupianez, D. G. , & Mundlos, S. (2018). Structural variation in the 3D genome. Nature Reviews. Genetics, 19(7), 453–467. 10.1038/s41576-018-0007-0 [DOI] [PubMed] [Google Scholar]
- Stahl, E. A. , Breen, G. , Forstner, A. J. , McQuillin, A. , Ripke, S. , Trubetskoy, V. , … Sklar, P. (2019). Genome‐wide association study identifies 30 loci associated with bipolar disorder. Nature Genetics, 51(5), 793–803. 10.1038/s41588-019-0397-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz, P. , & Lupski, J. R. (2010). Structural variation in the human genome and its role in disease. Annual Review of Medicine, 61, 437–455. 10.1146/annurev-med-100708-204735 [DOI] [PubMed] [Google Scholar]
- Stefansson, H. , Meyer‐Lindenberg, A. , Steinberg, S. , Magnusdottir, B. , Morgen, K. , Arnarsdottir, S. , … Stefansson, K. (2014). CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature, 505(7483), 361–366. 10.1038/nature12818 [DOI] [PubMed] [Google Scholar]
- Stein, J. L. , Medland, S. E. , Vasquez, A. A. , Hibar, D. P. , Senstad, R. E. , Winkler, A. M. , … Thompson, P. M. (2012). Identification of common variants associated with human hippocampal and intracranial volumes. Nature Genetics, 44(5), 552–561. 10.1038/ng.2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, K. M. , Schneider, V. A. , Graves‐Lindsay, T. A. , Fulton, R. S. , Agarwala, R. , Huddleston, J. , … Wilson, R. K. (2014). Single haplotype assembly of the human genome from a hydatidiform mole. Genome Research, 24(12), 2066–2076. 10.1101/g.180893.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stessman, H. A. , Bernier, R. , & Eichler, E. E. (2014). A genotype‐first approach to defining the subtypes of a complex disease. Cell, 156(5), 872–877. 10.1016/j.cell.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, D. , Ching, C. R. K. , Lin, A. , Forsyth, J. K. , Kushan, L. , Vajdi, A. , … Bearden, C. E. (2018). Large‐scale mapping of cortical alterations in 22q11.2 deletion syndrome: Convergence with idiopathic psychosis and effects of deletion size. Molecular Psychiatry, 25, 1822–1834. 10.1038/s41380-018-0078-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, I. K. , Gacquer, D. , Van Heurck, R. , Kumar, D. , Wojno, M. , Bilheu, A. , … Vanderhaeghen, P. (2018). Human‐specific NOTCH2NL genes expand cortical neurogenesis through Delta/NOTCH regulation. Cell, 173(6), 1370–1384.e1316. 10.1016/j.cell.2018.03.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabet, A. C. , Pilorge, M. , Delorme, R. , Amsellem, F. , Pinard, J. M. , Leboyer, M. , … Betancur, C. (2012). Autism multiplex family with 16p11.2p12.2 microduplication syndrome in monozygotic twins and distal 16p11.2 deletion in their brother. European Journal of Human Genetics, 20(5), 540–546. 10.1038/ejhg.2011.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey, K. E. , Rees, E. , Linden, D. E. , Ripke, S. , Chambert, K. D. , Moran, J. L. , … O'Donovan, M. C. (2016). Common alleles contribute to schizophrenia in CNV carriers. Molecular Psychiatry, 21(8), 1085–1089. 10.1038/mp.2015.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate, D. F. , Dennis, E. L. , Adams, J. T. , Adamson, M. , Belanger, H. G. , Bigler, E. D. , … Wilde, E. A. (2021). Coordinating global multi‐site studies of military‐relevant traumatic brain injury: Opportunities, challenges, and harmonization guidelines. Brain Imaging and Behavior. 10.31234/osf.io/d4qs8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeuw, J. , Brouwer, R. M. , Guimarães, J. , Brandner, P. , Koenis, M. M. G. , Swagerman, S. C. , … Hulshoff Pol, H. E. (2019). Genetic and environmental influences on functional connectivity within and between canonical cortical resting‐state networks throughout adolescent development in boys and girls. NeuroImage, 202, 116073. 10.1016/j.neuroimage.2019.116073 [DOI] [PubMed] [Google Scholar]
- Thompson, P. M. , Jahanshad, N. , Ching, C. R. K. , Salminen, L. E. , Thomopoulos, S. I. , Bright, J. , … Zelman, V. (2020). ENIGMA and global neuroscience: A decade of large‐scale studies of the brain in health and disease across more than 40 countries. Translational Psychiatry, 10(1), 100. 10.1038/s41398-020-0705-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, P. M. , Stein, J. L. , Medland, S. E. , Hibar, D. P. , Vasquez, A. A. , Renteria, M. E. , … Drevets, W. (2014). The ENIGMA consortium: Large‐scale collaborative analyses of neuroimaging and genetic data. Brain Imaging and Behavior, 8(2), 153–182. 10.1007/s11682-013-9,269-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulfarsson, M. O. , Walters, G. B. , Gustafsson, O. , Steinberg, S. , Silva, A. , Doyle, O. M. , … Stefansson, K. (2017). 15q11.2 CNV affects cognitive, structural and functional correlates of dyslexia and dyscalculia. Translational Psychiatry, 7(4), e1109. 10.1038/tp.2017.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer, D. , Sonderby, I. E. , Kaufmann, T. , Walters, G. B. , Abdellaoui, A. , Ames, D. , … Andreassen, O. A. (2020). Association of Copy Number Variation of the 15q11.2 BP1‐BP2 region with cortical and subcortical morphology and cognition. JAMA Psychiatry, 77, 420–430. 10.1001/jamapsychiatry.2019.3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zwaag, B. , Staal, W. G. , Hochstenbach, R. , Poot, M. , Spierenburg, H. A. , de Jonge, M. V. , … Burbach, J. P. (2010). A co‐segregating microduplication of chromosome 15q11.2 pinpoints two risk genes for autism spectrum disorder. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 153b(4), 960–966. 10.1002/ajmg.b.31055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp, T. G. , Hibar, D. P. , Rasmussen, J. M. , Glahn, D. C. , Pearlson, G. D. , Andreassen, O. A. , … Turner, J. A. (2016). Subcortical brain volume abnormalities in 2,028 individuals with schizophrenia and 2,540 healthy controls via the ENIGMA consortium. Molecular Psychiatry, 21(4), 547–553. 10.1038/mp.2015.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp, T. G. M. , Walton, E. , Hibar, D. P. , Schmaal, L. , Jiang, W. , Glahn, D. C. , … Turner, J. A. (2018). Cortical brain abnormalities in 4474 individuals with schizophrenia and 5,098 control subjects via the enhancing Neuro imaging genetics through meta analysis (ENIGMA) consortium. Biological Psychiatry, 84(9), 644–654. 10.1016/j.biopsych.2018.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij, D. , Anagnostou, E. , Arango, C. , Auzias, G. , Behrmann, M. , Busatto, G. F. , … Buitelaar, J. K. (2018). Cortical and subcortical brain Morphometry differences between patients with autism Spectrum disorder and healthy individuals across the lifespan: Results from the ENIGMA ASD working group. The American Journal of Psychiatry, 175(4), 359–369. 10.1176/appi.ajp.2017.17010100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen, J. M. , de Leeuw, N. , Papatsonis, D. N. , Grijseels, E. W. , de Krijger, R. R. , & Wessels, M. W. (2015). Phenotypic variability associated with a large recurrent 1q21.1 microduplication in a three‐generation family. Molecular Syndromology, 6(2), 71–76. 10.1159/000431274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalon Reina, J. , Nir, T. M. , Jahanshad, N. , Kushan, L. , Bearden, C. E. , & Thompson, P. M. (2019). Altered intracellular volume fraction and Neurite dispersion in people with chromosome 22q11.2 copy number variants. Paper presented at the Engineering in Medicine and Biology Conference Berlin, Germany. [Google Scholar]
- Villalon Reina, J. , Nir, T. M. , Kushan, L. , Bearden, C. E. , & Thompson, P. M. (2019). Myelin and G‐ratio imaging in 22q11.2 deletion syndrome: A pilot study. Paper presented at the SFN, Chicago.
- Villalon‐Reina, J. E. , Martinez, K. , Qu, X. , Ching, C. R. K. , Nir, T. M. , Kothapalli, D. , … Bearden, C. E. (2019). Altered white matter microstructure in 22q11.2 deletion syndrome: A multisite diffusion tensor imaging study. Molecular Psychiatry, 25, 2818–2831. 10.1038/s41380-019-0450-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. , Li, M. , Hadley, D. , Liu, R. , Glessner, J. , Grant, S. F. , … Bucan, M. (2007). PennCNV: An integrated hidden Markov model designed for high‐resolution copy number variation detection in whole‐genome SNP genotyping data. Genome Research, 17(11), 1665–1674. 10.1101/g.6861907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Shaw, W. R. , Tsang, H. T. , Reid, E. , & O'Kane, C. J. (2007). Drosophila spichthyin inhibits BMP signaling and regulates synaptic growth and axonal microtubules. Nature Neuroscience, 10(2), 177–185. 10.1038/nn1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warland, A. , Kendall, K. M. , Rees, E. , Kirov, G. , & Caseras, X. (2019). Schizophrenia‐associated genomic copy number variants and subcortical brain volumes in the UKbiobank. Molecular Psychiatry, 25(4), 854–862. 10.1038/s41380-019-0355-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan, C. D. , Altmann, A. , Botia, J. A. , Jahanshad, N. , Hibar, D. P. , Absil, J. , … Sisodiya, S. M. (2018). Structural brain abnormalities in the common epilepsies assessed in a worldwide ENIGMA study. Brain, 141(2), 391–408. 10.1093/brain/awx341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfert, A. B. , Sulovari, A. , Turner, T. N. , Coe, B. P. , & Eichler, E. E. (2017). Recurrent de novo mutations in neurodevelopmental disorders: Properties and clinical implications. Genome Medicine, 9(1), 101. 10.1186/s13073-017-0498-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, Y. J. , Wang, T. , Guadalupe, T. , Nebel, R. A. , Vino, A. , Del Bene, V. A. , … Abrahams, B. S. (2016). A common CYFIP1 variant at the 15q11.2 disease locus is associated with structural variation at the language‐related left Supramarginal Gyrus. PLoS One, 11(6), e0158036. 10.1371/journal.pone.0158036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray, N. R. , Ripke, S. , Mattheisen, M. , Trzaskowski, M. , Byrne, E. M. , Abdellaoui, A. , … Sullivan, P. F. (2018). Genome‐wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nature Genetics, 50(5), 668–681. 10.1038/s41588-018-0090-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


