Abstract
Control of malaria caused by Plasmodium vivax can be improved by the discovery and development of novel drugs against the parasite’s liver stage, which includes relapse-causing hypnozoites. Several recent reports describe breakthroughs in the culture of the P. vivax liver stage in 384-well microtiter plates, with the goal of enabling a hypnozoite-focused drug screen. Herein we describe assay details, protocol developments, and different assay formats to interrogate the chemical sensitivity of the P. vivax liver stage in one such medium-throughput platform. The general assay protocol includes seeding of primary human hepatocytes which are infected with P. vivax sporozoites generated from the feeding of Anopheles dirus mosquitoes on patient isolate bloodmeals. This protocol is unique in that, after source drug plates are supplied, all culture-work steps have been optimized to preclude the need for automated liquid handling, thereby allowing the assay to be performed within resource-limited laboratories in malaria-endemic countries. Throughput is enhanced as complex culture methods, such as extracellular matrix overlays, multiple cell types in co-culture, or hepatic spheroids, are excluded as the workflow consists entirely of routine culture methods for adherent cells. Furthermore, installation of a high-content imager at the study site enables assay data to be read and transmitted with minimal logistical delays. Herein we detail distinct assay improvements which increase data quality, provide a means to limit the confounding effect of hepatic metabolism on assay data, and detect activity of compounds with a slow-clearance phenotype.
Graphical abstract:
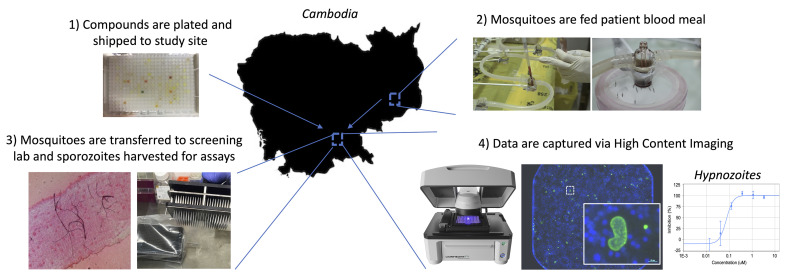
Overview of P. vivax liver stage screening assay performed at the Institute Pasteur of Cambodia.
Keywords: Phenotypic screening, Plasmodium vivax, Hypnozoites, Liver stage assay, Antirelapse, 8-aminoquinolines, Primaquine, Tafenoquine
Background
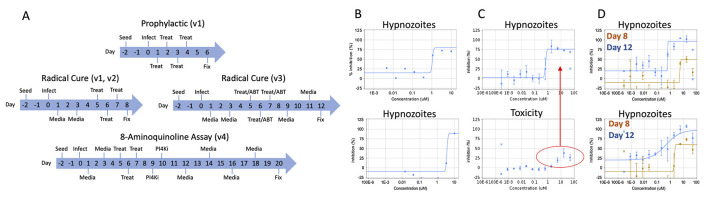
Malaria caused by Plasmodium vivax presents a significant public health burden on endemic countries and can impede developing economies. The Plasmodium parasite exhibits a complex lifecycle, including a rapid population expansion in the Anopheles mosquito vector, in the host liver, and then in the host bloodstream – the stage causing symptomatic malaria. Unique to P. vivax is its ability to produce dormant forms in the liver, termed hypnozoites ( Krotoski et al., 1982 ). When a human is infected with P. vivax from an infectious mosquito bite, hypnozoites persist long after the initial infection is cleared. They can then resume growth and cause a relapsing blood infection. Hypnozoites cannot be killed by most antimalarial drugs, and the only drugs with hypnozonticidal activity are hemolytic ( Chu et al., 2017 ). Until recently, discovery and development of new hypnozonticidal drugs have been slowed by inadequate methods to culture P. vivax liver stage parasites, including hypnozoites, for weeks or even a month in vitro (Figure 1A). This protocol report describes in detail the different assay versions, improvements, and modifications used to generate data for several published and anticipated reports describing P. vivax liver stage compound screening and drug development.
Figure 1. Overview of assay modes, versions, and example data.
A. In prophylactic assays, cultures are treated shortly after sporozoite infection of hepatocytes, in order to assess activity against immature hypnozoites and early schizonts. In radical cure assays, cultures are treated at 5 days post-sporozoite infection to allow for hypnozoites to mature. The v1 and v2 radical cure assays share the same timecourse, but feature different plate maps and normalization protocols. The v3 radical cure assay includes an extended endpoint (12 days post-infection) and addition of 1-aminobenzotriazole (ABT) to the medium on treatment days. The v4 assay includes a further extended endpoint of 20 days post-infection and addition of a PI4K (phosphatidylinositol 4-kinase) inhibitor (“PI4Ki”) to eliminate schizonts present at 9-10 days post-infection. “Seed” indicates hepatocyte seed, “Infect” indicates infection with sporozoites, “Media” indicates media change, “Treat” indicates compound treatment via the pin tool. The endpoint of all assays is fixation (“Fix”) prior to immunofluorescence staining and high content imaging. B. Example data from a v1 radical cure assay for two hit compounds with activity against hypnozoites. C. Example data from a v2 radical cure assay for a compound with nonselective activity against hypnozoites. The highest three doses of compound show a decrease in hepatic nuclei, indicating that cytotoxic conditions (red circle), not hypnozonticidal activity, are killing hypnozoites (top, red arrow) at those concentrations. Bars represent standard deviation (SD). D. Example data from v2 (orange) and v3 (blue) radical cure assays for two developmental compounds with activity against hypnozoites. The v2 curves show partial top plateaus that become complete inhibited with the longer v3 assay. Bars represent SD. Images and data are adapted from Maher et al. (2021) , used with permission. Figure 1A was adapted from Maher et al. (2021) and reproduced under a Creative Commons 4.0 license (http://creativecommons.org/licenses/by/4.0/).
Previously-described platforms to maintain hepatocytes infected with liver stage parasites in vitro include complex culture methodologies such as extracellular matrix overlays ( Dembele et al., 2014 ), micropatterned co-cultures ( March et al., 2013 ; Gural et al., 2018 ), microwell devices ( Maher et al., 2020 ), and hepatic spheroids ( Chua et al., 2019 ). The first report of a medium throughput platform capable of long-term hepatocyte culture and robust Plasmodium infection using standard 384-well microtiter plates was described in 2018 ( Roth et al., 2018 ). Termed the version 1 (v1) assay herein, this format included immunofluorescent (IF) staining ( Schafer et al., 2018 ) and high content imaging (HCI) to quantify growth and persistence of P. vivax liver stages within primary human hepatocytes and was able to produce dose response and single point screening data. However, the v1 assay suffered from poor curve fitting due to the lack of replicate wells for characterizing well-to-well variability (Figure 1B). Nonetheless, this report describes the potent effect of ionophores on hypnozoites, making them useful tool compounds for studying hypnozoite biology and technical positive controls for screening small molecules. The utility of ionophores as control compounds is important because the only drugs that clinically prevent relapse, the 8-aminoquinolines (8AQs) tafenoquine and primaquine, do not exhibit rapid hypnozoite killing in vitro ( Gural et al., 2018 ; Chua et al., 2019 ; Maher et al., 2020 ) and therefore are not ideal positive controls for screening assays. The v1 assay also demonstrated the different chemosensitivity of immature hypnozoites (day 1-4 post-infection) versus mature hypnozoites (by day 5 post-infection) using the phosphatidylinositol 4-kinase inhibitors, underpinning the concept that P. vivax liver stage assays can be performed in a ‘prophylactic’ or ‘radical cure’ mode depending on when treatment is applied ( Zeeman et al., 2016 ; Gural et al., 2018 ; Roth et al., 2018 ) (Figure 1A).
To address the shortcomings of the v1 assay, an improved version 2 (v2) assay was developed ( Maher et al., 2021 ), which includes an expanded dose response (from 8 to 12 points), testing from a higher concentration (from 10 µM to 50 µM), ionophore controls for normalization, and a direct comparison of potency and cytotoxicity to better understand selectivity (Figure 1C). However, two specific deficiencies were noted in the v2 assay. First, when assaying a series of new hypnozonticidal compounds, we frequently obtained a dose response curve with a partial plateau around 50% inhibition. We hypothesized this could be the result of an active compound not having enough time to clear killed hypnozoites from the wells, resulting in dead or dying hypnozoites being aberrantly counted as viable during HCI. By extending the assay 4 days (to 14 days post-seed or 12 days post-infection), we found better clearance of dying hypnozoites that had been treated 7 days earlier (Figure 1D). Second, this platform is unique in that it includes metabolically active hepatocytes, a potential liability when attempting to discover and develop unoptimized small molecules. We performed a series of experiments to characterize the effect of adding 1-aminobenzotriazole (ABT, which has been shown to inhibit cytochrome P450 enzymes) to culture media prior to compound treatment, in an effort to clarify the activity of parent versus possible reactive metabolites against liver stage parasites (Ortiz de Montellano and Mathews, 1981). These improvements led to the version 3 (v3) assay detailed herein (Figure 1D).
The activity of 8AQs against mature hypnozoites has only recently been demonstrated in vitro (Dembélé et al., 2020; Maher et al., 2021 ). In these reports, both primaquine and tafenoquine are made more potent by co-treatment with the blood schizonticide chloroquine, which is currently co-administered with tafenoquine clinically (Llanos- Cuentas et al., 2014 ). We observed non-clearance of 8AQ-treated hypnozoites in our v2 and v3 assay formats; thus, we further extended the assay endpoint (22 days post-seed or 20 days post-infection) in order to enable a longer time for dead hypnozoite clearance, as well as observe schizont formation following quiescence as a hypnozoite ( Maher et al., 2021 ). Termed “reactivation in vitro” (Markus, 2020), this is accomplished by addition of PI4Ki at day 9 post-infection—2 days after test compound treatment—to selectively kill schizonts and not hypnozoites. When 20-day cultures are quantified by HCI, schizonts observed most likely originated from forms that were hypnozoites 10 days sooner, during the selective PI4Ki treatment. These modifications resulted in the version 4 (v4) assay described herein (Figure 1A).
While several reagents and steps are critical to the protocol’s success, perhaps the most fundamentally important reagent is the specific production lot of cryopreserved human hepatocytes. Each production lot exhibits different properties (viability, plateability, purity, etc.) that are, in large part, the result of the process used at the company producing the lot. As a result, lots from some production protocols result in cells that are not supportive of P. vivax and are otherwise difficult or impossible to culture in vitro. We have formed a lot screening and selection process which results in validated lots for P. vivax liver stage studies. The donor selection process should be used first to identify a lot suitable for further assay setup and validation (Table 1).
Table 1. Donor Selection Cascade and Criteria.
| Cascade Step | Summary | Notes |
|---|---|---|
| 1-Preselection |
-Viability > 75% -Cryoplateable -Excess quantity vials available -Pre-tested for microbial contamination -8 × 106 cells/vial -Pure hepatocyte population |
Approximately 40% of harvested human livers are cryoplateable, while 80% of livers contain microbial contamination |
| 2-Donor Screen |
-Obtain 10-30 donor lots -Culture for 3 weeks -Monitor for Stability and Contamination -Infect with sporozoites of same case, quantify relative liver stage infection efficiency |
Approximately 10% of lots tested are noticeably contaminated due to microbial bioburden, while only 5% of lots tested produce robust liver stage parasite numbers |
| 3-Confirmation |
-Buy 10-20 vials of the best lot from step 2 -Perform low-throughput drug assays, look for consistency from thaw to thaw and infection to infection -Run a standard set of compounds to chemically validate the lot |
Reproducibility from vial to vial can be poor, so ensure all vials perform in the same manner. In this stage, 10% of lots failed the reproducibility test |
This protocol includes several technologies or facilities which are often considered ‘infrastructure,’ including an ACL2 insectary, automated liquid handling systems, high-content imaging systems, and drug discovery data management software. It is outside of the scope of this manuscript to provide a full protocol for designing an ACL2 insectary, as well as training scientists how to work in arthropod containment: the knowledge base needed takes months to teach and years to master, but primers are available in the literature (American Committee Of Medical Entomology American Society Of Tropical Medicine And Hygiene, 2019). Likewise, there are many different commercially available systems for automated liquid handling, high content imaging, and drug data management. In this protocol, we describe our use of specific resources (i.e., a Biomek 4000 for automated liquid handling, a Lionheart FX high content imager, and CDD Vault for drug data management) as one example of a complete workflow, but it is outside the scope of this protocol to fully describe the basics of using each. Rather, these systems come with extensive instruction documentation and training modules to enable use. Obtaining patient isolate blood samples infected with P. vivax parasites requires an Institutional Review Board (IRB)-approved Human Subjects Protocol and a medical worker experienced in venipuncture. The elements of a Human Subjects approval are specific to each institution and the governing policies of endemic countries; thus, obtaining Human Subjects approval and demonstrating safe venipuncture are outside the scope of this protocol. Lastly, drug discovery and development is a voluminous field, and a full understanding of this protocol would require a basic understanding of drug discovery and HCI terms and methods.
Materials and Reagents
-
Anopheles dirus rearing and infection
Whatman paper (Whatman®, 3MM CHR paper, catalog number: 3030-917)
Cotton pad
Petri dish (GosselinTM, catalog number: BP93B-101, Diameter: 90 mm, Height: 14.2 mm)
Transfer pipets (COPAN Diagnostics, 3 ml, catalog number: 200CS01)
Transparent plastic cups
Paper cups
Nets
Elastic bands
Glass slides (FisherbrandTM, catalog number: 12-544-2)
Cover slips (MenzelTM, 20 mm × 20 mm, catalog number: BB02000200A153MNZ0)
Heparin tubes (Beckton Dickinson, catalog number: 367886)
Anopheles dirus a mosquito from Pursat Province Cambodia (St Laurent et al., 2015 )
TetraBits Complete fish food (Tetra®)
4-aminobenzoic acid (PABA; Sigma-Aldrich, catalog number: 100536-1KG)
Nutroplex® (Multivitamins, Unilab Inc.)
Mercury dibromofluorescein disodium salt (Mercurochrome, Sigma-Aldrich, catalog number: M7011-25G)
Human AB serum (The Interstate Blood Bank, Inc., USA)
Sucrose (Domino pure cane sugar, UPC 049200045503)
20% sucrose and multivitamins solution, store at 4°C (see Recipes)
-
Dose response drug plate preparation
Small Volume 384 Deep Well Polystyrene MicroplateTM, sterile, (Greiner Bio-One, catalog number: 784261)
Aluminum Foils for PCR and Cold Storage, sterile (VWR®, catalog number: 60941-076)
Filtered Pipette Tips, sterile (Thermo ScientificTM, FinntipTM, catalog number: 94052060)
AP96 P50 Tips, sterile (Beckman Coulter, Biomek®, catalog number: A21582)
DMSO, sterile (Tocris BioscienceTM, catalog number: 3176/100ML)
Test compounds diluted to 50 mM in DMSO, minimum volume of 15 μl each
Nigericin sodium salt (Sigma-Aldrich, catalog number: N7143), diluted to 200 μM in DMSO
-
Hepatocyte seeding and culture
250 ml Vacuum Filtration System (MilliporeSigmaTM, StericupTM, catalog number: S2VPU02RE)
9 × 9 inch Lint-Free Cloth (Electron Microscopy Sciences, catalog number: 100488-446), sterilized by autoclave
Disposable Reagent Reservoir, sterile, 50 ml (Biotang Inc., Accuraci*, catalog number: 90105)
384-well, Collagen Type I-Treated, Flat-Bottom Microplate (Greiner Bio-OneTM, CellCoatTM, catalog number: 781956)
245 mm Square BioAssay Dish (CorningTM, catalog number: 431111)
Cell culture/Petri Dishes (Thermo ScientificTM, NuncTM, catalog number: 150288)
1.5 ml Eppendorf tube (EppendorfTM, catalog number: 022431021)
50 ml Conical Centrifuge tube (CorningTM, Falcon, catalog number: 352070)
Cell culture grade water, sterile (CorningTM, catalog number: 25055CV)
InVitroGROTM CP Medium (BioIVT, catalog number: Z99029)
Penicillin-Streptomycin-Neomycin (PSN) Antibiotic Mixture, 100× (GibcoTM, catalog number: 1560055). Aliquoted and frozen at -20°C
Gentamicin, 10 mg/ml (GibcoTM, catalog number: 1570072)
Trypan Blue cell viability reagent, 0.4% w/v (CorningTM, catalog number: 25900CI)
Complete hepatocyte media (CM), 250 ml (see Recipes)
-
Sporozoite infection
Glass slides (FisherbrandTM, catalog number: 12-544-2), wrapped in a paper towel and autoclaved
Cell culture/Petri Dishes (Thermo ScientificTM, NuncTM, catalog number: 150288)
6-well plate, sterile (CorningTM, CostarTM, catalog number: 3516)
Disposable Pellet Pestel with Tube (FisherbrandTM, catalog number: 12-141-368), unwrapped and sterilized by autoclave
1.5 ml Eppendorf tube (EppendorfTM, catalog number: 022431021), sterilized by autoclave
Spray bottle with 70% ethanol in water
PBS, sterile (GibcoTM, catalog number: 10010023)
Penicillin-Streptomycin-Neomycin (PSN) Antibiotic Mixture, 100× (GibcoTM, catalog number: 1560055), aliquoted and frozen at -20°C
RPMI 1640 Medium, with GlutaMAXTM, without sodium bicarbonate (NaHCO3) (GibcoTM, catalog number: 61870-036)
PSN-PBS mixture for washing mosquitoes, 5 ml (see Recipes)
-
Compound treatment
OmniTrayTM Single-Well Plate (Thermo ScientificTM, NuncTM, catalog number: 264728)
Pin tool Clean Solution (V&P Scientific Inc., catalog number: VP 110)
Cell culture grade water, sterile (CorningTM, catalog number: 25055CV)
DMSO (Fisher BioReagentsTM, catalog number: BP231-100)
70% ethanol in water
Methanol (Fisher, catalog number: S25426)
Lintfree Blot Paper (V&P Scientific Inc., catalog number: VP522100)
1-aminobenzotriazole (Caymen Chemical, catalog number: 15252), diluted to 100 mM in cell culture grade water, sterile (CorningTM, catalog number: 25055CV), filter sterilized, aliquoted, and stored at -20°C (see Recipes)
Complete hepatocyte media (CM) with ABT, 250 ml (see Recipes)
-
Immunofluorescent (IF) detection and high content imaging (HCI)
Aluminum Foils for PCR and Cold Storage, sterile (VWR®, catalog number: 60941-076)
Microplate Sealing Film (AxygenTM, catalog number: PCRSP)
PBS, sterile (GibcoTM, catalog number: 10010023)
Paraformaldehyde, 16% w/v aqueous solution, methanol free (Alfa AesarTM, catalog number: 433689L)
Bovine serum albumin (BSA, Fisher BioReagentsTM, catalog number: BP1600-100)
TritonTM X-100 (Acros OrganicsTM, catalog number: 327372500)
Recombinant mouse anti-UIS4 (Noah Sather, Seattle Children’s Research) ( Schafer et al., 2018 )
IgG (H+L) Highly Cross-Adsorbed Goat anti-Mouse Alexa Fluor®, 488 (InvtrogenTM, catalog number: A11029)
Hoechst 33342 Trihydrochloride, Trihydrate-FluoroPureTM Grade (InvitrogenTM, catalog number: H21492)
4% PFA Fix buffer, 40 ml (see Recipes)
IF stain dilution buffer, 1% w/v BSA, 0.3% v/v Triton X-100, 50 ml (see Recipes)
Primary IF stain (rPvUIS4), 25 ml (see Recipes)
Secondary IF stain (Alexa Fluor 488), 25 ml (see Recipes)
Hoechst stain, 25 ml (see Recipes)
Equipment
-
Anopheles dirus rearing and infection
Dissection forceps (Fine Science Tools Inc., Dumont #5-Fine)
Laboratory trays, polypropylene, autoclavable (Labbox, catalog number: TRYP-001-001. H × L × W: 75 mm × 375 mm × 300 mm & TRYP-002-001 H × L × W: 75 mm × 450 mm × 350 mm)
Centrifuge 5702R (Eppendorf®, catalog number: 5703000322)
Water-jacketed glass mini feeder (Glastechniek Peter Coelen B.V., Netherlands, diameter 15 mm) (Figure 2A and 2B)
Silicone tube for connecting the mini-feeders (Labbox, catalog number: SILT-008-005, diameter inside: 8 mm) (Figure 2A and 2B)
Heating circulator (Huber, reference: KISS 202C)
Custom made rearing cages (30 × 30 × 30 cm and 20 × 20 × 20 cm) covered with nets
Hemotek membrane feeding system (Hemotek®) used with Parafilm membrane (Hemotek, catalog number: SP6W1-1)
InsectaVac vacuum-powered insect aspirator (BioQuip Products, catalog number: 2809B)
Stereomicroscope Leica S6E
Microscope Leica DM1000LED
-
Dose response drug plate preparation
Hemocytometer (SKC, Inc., C-ChipTM, catalog number: DHCN015)
Drug desiccator (Bel-Art, Dry-KeeperTM, catalog number: H420560001/EMD) with DrieriteTM drying agent (Sigma-Aldrich, catalog number: 737828)
F2 16-channel pipette, 1-10 μl (Thermo ScientificTM, FinnpipetteTM, catalog number: 4662080)
F2 16-channel pipette, 5-50 μl (Thermo ScientificTM, FinnpipetteTM, catalog number: 4662090)
Biomek 4000 Automated Liquid Handler (Beckman Coulter) with MP200 8-channel pipette tool, reservoir stand (Beckman Coulter, catalog number: 372795), and 40 ml sterile reservoir (Beckman Coulter, catalog number: 372790)
-
Hepatocyte seeding and culture
Inverted light microscope with 10× objective
37°C cell culture incubator with 5% CO2
Benchtop centrifuge capable of spinning tubes and microtiter plates
Water bath set to 37°C
Aluminum spin holder, custom made via CNC machining (see design in Figure 4C)
-
Sporozoite infection
Hemocytometer (SKC, Inc., C-ChipTM, catalog number: DHCN015)
Inverted light microscope with 10× objective
Tweezers with Straight, Fine Tip, 4.25 inch (Excelta, catalog number: 3C-SA-PI)
Needle Holder for Micro Dissecting Needles (Roboz, catalog number: RS-6063) fitted with a Tungsten Dissecting Needle, 0.25 mm, Ultra Fine (Roboz, catalog number: RS-6064)
Ice bucket with finely crushed ice
-
Compound treatment
Custom made, floating 384-well pin tool, 20 nl slot pin, 0.457 mm diameter, 50.8 mm long, 17 mm exposed pin length (V&P Scientific, Inc., catalog number: FP1S20), kit for manual use (V&P Scientific, Inc., catalog number: BGPK), with docking station (V&P Scientific Inc., catalog number: VP 550A)
Alignment Jig for Registering Floating Pin Multi-Blot Replicators (V&P Scientific, Inc. catalog number: VP 381N), herein called a “Library Copier”
Heavy Duty Blot Station (V&P Scientific Inc., catalog number: VP 540DB)
-
Immunofluorescence detection by High Content Imaging
Lionheart FX Automated Microscope (Biotek®) with FITC and DAPI LED cubes with matching filter sets, a laser autofocus cube, and a 4× objective
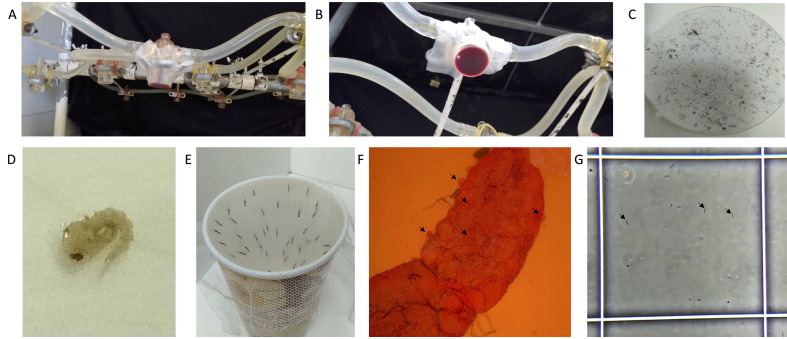
Figure 2. Mosquito rearing and infection.
A. Side view of the apparatus for membrane feeding using a water-jacketed mini-feeder. B. Bottom view of a water-jacketed mini-feeder with a parafilm membrane holding blood. C. Whatman paper with mosquito eggs (a female An. dirus mosquito is included for size context). D. A single An. dirus pupae. E. A paper cup with a mesh lid used to contain females at day 5 post-emergence. F. Mosquito midgut stained with 1% mercurochrome in PBS following dissection at 7-days post-infectious bloodmeal, imaged under magnification with a 10× objective. Arrows show oocysts. G. A hemocytometer with P. vivax sporozoites harvested from An. dirus salivary glands at day 16 post-infectious bloodmeal, imaged under magnification with a 20× objective. The white box is 0.1 mm × 0.1 mm and arrows indicate three example sporozoites.
Figure 4. Overview of salivary gland dissection.
A. Mosquitoes designated for dissection are transferred to plastic cups covered in mesh and moved into the dissection room. B. Mosquito wash buffers are prepared in a 6-well plate on ice. C. Mosquitoes are prepared for dissection by first being sprayed with 70% ethanol and then transferred to the first well of a 6-well plate containing antibiotics (penicillin, streptomycin, and neomycin) in PBS. Individual mosquitoes are then washed in 70% ethanol and rinsed in PBS. E. Washed mosquitoes are placed facing the same direction on a sterile glass slide. Video 4 demonstrates how mosquitoes are washed. F. Slides are viewed under a stereomicroscope. Glands are dissected into an Eppendorf tube with RPMI on ice (blue box at top left). G. A dissector using a dissection needle (right hand) and forceps (left hand). Video 2 demonstrates how salivary glands are dissected.
Software
Gen5 Microplate Reader and Imager Software (Biotek®, www.biotek.com)
CDD Vault (Collaborative Drug Discovery, https://www.collaborativedrug.com/)
Procedure
-
Anopheles dirus rearing
Feed blood to female mosquitoes every 4 days with a Hemotek device placed on top of the cage. Provide females with constant access to 10% sucrose solution. Two-days post-feed, insert a Petri dish containing a humid layer of cotton covered by a disc of Whatman paper into the cage. The next morning, recover the eggs laid on the Whatman paper (Figure 2C). Place the Whatman paper with eggs in a laboratory tray with water.
Upon hatching, feed the larvae every day with ground Koi fish food ad libitium. Change larval water every two days and clean the remaining food every other day. Count larvae at the first transfer to have a density of 150 larvae in the small laboratory trays and 300 larvae in the large laboratory trays.
Collect pupae every morning, using a transfer pipet with the bottom 0.5 cm cut off; this is to ensure a sufficiently large bore opening to avoid damaging the pupae (Figure 2D). Transfer the pupae to transparent plastic cups (maximum 100 pupae/cup). Add water to until 2 cm below the top of the cup. Place the cup in a cage.
For 3 days post-emergence, feed the mosquitoes with 20% sucrose and multivitamins solution. On day 4, change this to 5% sucrose solution. Remove the sugar in the evening of day 4. On day 5, use a vacuum aspirator to transfer females to paper cups covered with nets held by an elastic band (Figure 2D). Keep the cups in the dark. Females are available for an infectious bloodmeal until day 7. On days 5 and 6, if the females have not been used for an infectious bloodmeal, provide 5% sugar solution on a cotton pad for 1 h in the evening.
-
Anopheles dirus infection
Following informed consent, collect isolates of P. vivax-infected blood from symptomatic patients by venipuncture in a heparin tube. Keep blood at 37°C during all manipulations.
Centrifuge the blood at 1,300 × g for 5 min at 37°C.
Remove the patient plasma and replace it by heat-inactivated (56°C for 30 min) naïve human AB serum to avoid transmission-blocking immunity ( Bousema et al., 2012 ). Mix red blood cells and serum. Pipette 450 µl of this mixture into the water-jacketed mini-feeders maintained at 37°C using a pump and water bath (Figure 2A and 2B).
Allow females to engorge for 1 h.
Transfer the females from the cups to the cages. Once in the cage, fed and unfed females can be visually sorted out (using an insect aspirator) by the enlarged darkened abdomen of the blood-fed females. Remove all unfed females. Keep fed females in standard insectary conditions at 26°C and 80% humidity, with 10% sucrose + 0.05% PABA solution until dissection.
On day 6 post-infectious blood meal, dissect the midgut of 10 females and collect midguts in 1% mercurochrome solution under a stereomicroscope (Video 1).
Mount the midguts under cover slips and count the number of oocysts at 10-20× magnification (Figure 2F).
For positive feeds (i.e., females with oocysts on day 6), perform a test salivary gland dissection on 10 females between day 14 and 16 post-feed. Dissect the midguts to assess oocyst opening as well as salivary glands, to determine the sporozoite load (see Procedure E, Figure 2G, and Video 2, Figure 3).
-
Dose response drug plate preparation
The following instructions are for generating a source drug plate containing 14 test compounds in a 12-point dilution series (1:3) from a 50 mM highest dose with duplicate wells per concentration. Map all compounds by row of a 384-well plate, include a DMSO vehicle (negative) control and nigericin (positive) control diluted from 200 μM. Label drug plates as needed (Table S1).
-
Compound providers may send drugs as powder or liquid, either individually vialed or in a microtiter plate. If powder, prepare stock solutions at 10 mM or 50 mM in sterile DMSO; 50 mM stock is ideal to limit the total amount of DMSO in the final working solution and provide enough drug to detect activity, despite the possibility of the compound being inactivated by hepatic metabolism.
Formula weight (FW) is the base and salt (if any) molecular weights together. If the compound has a salt component use FW, not molecular weight of base.
10 mM stock: 100,000× (mg/FW) = μl DMSO to add to make a 10 mM solution.
50 mM stock: 20,000× (mg/FW) = μl DMSO to add to make a 50 mM solution.
For example, primaquine is commercially available as primaquine bisphosphate with a formula weight of 455.34 g/mol. Starting with a vial containing 10 mg of primaquine bisphosphate powder, a 10 mM stock could be made by adding 2,196 µl of DMSO to the vial, or a 50 mM stock could be made by adding 439 µl of DMSO to the vial.
Add 15 µl of 50 mM stock solution to the appropriate row of columns 1 and 13.
Move plates into the hood of the Biomek 4000. Add 10 ml of sterile DMSO to an autoclaved quarter reservoir in the reservoir stand. Using the MP200 head and the dilution series function, mix 5 µl of the source column 5× at 10 µl/s without any air gap or blowout (to reduce bubble formation). Then draw and aspirate 5 µl to the second column and mix 8× as above. Repeat the dilution series until column 12, then stop and replace tips. Apply this pattern to the replicate wells of the dilution series (columns 13-24), and for the alternative rows of compounds that have not yet been diluted by the 8-channel head (Video 3).
Cover diluted plates with a sterile foil seal and spin at 100 × g for 1 min to remove air bubbles (Figure 3A). Plates are stored at -80°C and shipped on dry ice.
-
Hepatocyte seeding and culture
Hepatocyte seeding is performed 2 days before salivary glands are dissected from infected mosquitoes for sporozoite infection.
Prepare for the seed by making 250 ml of complete hepatocyte media (CM, see Recipes below, scale up volume if more media is needed), labeling 384-well assay plates, heating a water bath to 37°C, obtaining a hemocytometer, and transferring necessary reagents to tubes. Speed is important during seeding, so all materials must be ready before thawing.
Using the 16-channel pipettor and media reservoir, add 20 µl of CP medium to each well to pre-wet the assay plate. Ensure liquid is in the bottom of each well (not on the sides); gently tap the assay plate on a hard surface to ensure all liquid rests on the bottom of wells, if needed.
Before proceeding, determine how many cryovials of hepatocytes to thaw. Primary human hepatocytes from BioIVT typically include 6 × 106-10 × 106 live cells per vial and each plate requires 6.25 × 106-8.5 × 106 cells (see section Hepatocyte lot selection above for more information). Then, add (# vials) × 4 ml of CM to a 50 ml conical centrifuge tube.
Add 50 µl of Trypan Blue to an Eppendorf tube.
Following appropriate safety precautions, obtain the cryovials from liquid nitrogen storage. Slightly twist open the cap of each tube to vent possible nitrogen gas inside, thereby preventing possible cryovial explosion. Immediately thaw in the water bath for exactly 120 s, without the vial opening being immersed in the water and without swirling. After 120 s, there will momentarily be an ice sliver inside the vial. Close the caps and quickly rinse vials with 70% ethanol for sterility, then dry the vial with a clean wipe. Move vials into a sterile field (i.e., Class II Biosafety Cabinet-BSC) and remove the vial caps. Using specifically a 1 ml pipette with a 1 ml tip, slowly remove the thawed vial contents and transfer to the 50 ml conical tube with CM. Do not mix the cryovial contents. Once thawed, cryovial contents are collected from all vials, take 1 ml of CM from the 50 ml collection conical and use to rinse and collect all remaining material from the vials (which should now be completely thawed). The 50 ml tube should now have a volume of (# vials) × 5 ml.
-
Swirl the 50 ml conical tube to mix and transfer 50 µl of hepatocytes in CM to the Eppendorf tube with Trypan Blue. Mix slowly thrice, then transfer 10 µl of the mixture to a hemocytometer. Count live cells in all four 4 × 4 quadrants and calculate cell concentration using:
and live cell quantity by using:
Viability should be above 75%. Using CM, adjust concentration to:
For example, assuming a target seed density of 18,000 live cells per well and a thaw with 7.5 × 106 live cells from a single cryovial, adjust concentration to 900 cells per µl using:
– 5000
Mix well using a 10 or 25 ml serological pipette and transfer to a reagent reservoir. Use the 16-channel pipette to transfer 20 µl of cell mixture per well.
Notes:
Critical note 1: Do not use the blowout function on the 16-channel pipette. This will cause bubbles and decrease seed accuracy
Critical note 2: Hepatocytes settle quickly in media. Seed only 1 plate before stopping and re-mixing the hepatocyte suspension using a 10 ml serological pipette and continue with the next plate; mix more often if needed.
To prevent edge-effect due to evaporation, prepare a Bioassay Dish with 4 small Petri dishes with 10 ml water each, as shown in Figure 3B. Place plates in a cell culture incubator at 37°C and 5% CO2. Do not change media before infection with sporozoites.
On the day of infection, place all assay plates to be infected inverted into an aluminum spin holder in the BSC (Figure 3C), then place them in a centrifuge with microtiter plate holders and spin at 40 × g for 1 min with minimum acceleration to remove all media. Transfer the plate and aluminum spin holder to the BSC and clean the top of the plate using a sterile lint-free cloth. Using a media reservoir and 16-channel pipette, add back 20 µl of CM per well. (The additional 20 µl of CM will be added as part of the infection procedure).
-
Sporozoite infection
-
Set up for dissection in a secure Arthropod Containment Facility level 2 (ACL2)
Obtain crushed ice in a benchtop ice bucket.
Add 5 ml of PSN-PBS solution to the top left well of a 6-well plate, 5 ml of 70% ethanol to the middle top well, and 5 ml of PBS to the right top well.
Make up the dissection buffer by adding 10 ml of PBS into a 15 ml conical tube.
Make up the collection buffer by adding 100 µl of RPMI (without NaHCO3) to an Eppendorf tube.
Place the 6-well plate, dissection buffer, and collection buffer on ice.
Obtain the Petri dish and place next to the ice bucket.
Place several puddles of 20 µl dissection buffer on autoclaved glass slides; one puddle will be needed for each mosquito to be dissected. Make only one glass slide at a time.
-
Immobilize mosquitoes by spraying 70% ethanol through the mesh lid, ensure all mosquitoes are immobilized, and remove the lid. Using the forceps, quickly but carefully move all mosquitoes to the Petri dish, and then move all mosquitoes into the PSN-PBS well on ice. Do not let mosquitoes rest in ethanol for more than 2 min as this will dehydrate the mosquitoes and reduce sporozoite viability.
Notes:
Safety note: Make sure all mosquitoes are immobilized with ethanol. Ethanol will not kill the mosquitoes immediately. Regard mosquitoes as infectious vectors until after dissection. Never open a cup outside of an ACL2 room.
Safety note: Obtain all mosquitoes from the cup and account for all mosquitoes before, during, and after dissection.
Safety note: Persons should not enter or exit the ACL2 room when mosquitoes are out of cups.
Safety note: Use lids on the Petri dish and 6-well plate to enclose mosquitoes to further ensure none can escape.
Technical note: Keep mosquitoes on the PSN-PBS buffer on ice to maintain sporozoite viability.
Technical note: Once mosquitoes are sprayed with ethanol, complete dissection steps in less than 60 min; set a timer. Sporozoite viability can be reduced with excessive dissection time.
Using forceps, pick individual mosquitoes by their proboscis and drag each through the middle (70% ethanol) and right (PBS) top wells of the 6 well plate, making sure to let them soak for 1-2 s in each well. Pulling the mosquito through liquid by holding to the proboscis will help dissection greatly, as the legs and wings will be swept back by the forward motion through the liquid. The ethanol step is the most important as an ethanol coating will help discharge static and surface tension on the mosquito surface. Completely dunk the mosquito in the ethanol part of the wash. Be sure to pull the mosquito through the washes in the same direction (i.e., left to right only) to ensure that wings and legs are away from the head. Place each washed mosquito into a separate PBS puddle on the glass slide (Figure 4A-4E and Video 4).
-
(Dissectors may be right or left-handed, the following is for holding the forceps in the right hand and the dissection needle in the left hand). With the proboscis facing right, hold the forceps in the right hand and the needle in the left hand. Bracket the head with the forceps while simultaneously holding the thorax with the needle almost horizontally parallel to the glass slide. Then, simultaneously pull the head to the right while depressing the thorax with the needle. With practice, glands will fall out of the thorax with proper depression and pull pressure – the amount of pressure used for depressing the thorax is key: excess pressure results in a pierced thorax, insufficient pressure results in glands remaining in the thorax when the head is pulled away. While continuing to immobilize the head using the forceps, gather the glands by hooking them with the needles and place them in the collection buffer on ice using a spinning motion, as glands may stick to the needle. Repeat for each mosquito (Video 2).
Notes:
Technical note: Do not rummage around the dissection buffer for missing glands; this increases time per mosquito and increases the chance of microbial contamination downstream.
Technical note: 6 to 10 mosquitoes can be set up per dissection slide. The total number of mosquitoes processed depends on the number of dissectors and the dissection speed. Once the cleanliness of each dissector has been ascertained, salivary glands can be pooled between dissectors.
Centrifuge the collection tube at 1,000 × g for 2 min. Glands will stick to the side of the Eppendorf, and centrifugation is needed to pellet the glands.
Keep the collection buffer with glands on ice and move to a BSC. Use the matching pestle (sterilized by autoclave) to crush the glands by spinning the pestle against the bottom of the Eppendorf tube 10×. Add 100 µl of RPMI (without NaHCO3) to the collection tube to rinse the pestle during removal. Further crush and mix glands and sporozoites by pipetting up and down 100 times using the 100 µl tip.
-
Obtain a new Eppendorf tube and fill with 18 µl of PBS. In this tube, mix 2 µl of crushed glands and sporozoites, making a 1:10 dilution. Transfer 10 µl of the dilution to a hemocytometer and count sporozoites in two 4 × 4 quadrants. Calculate net sporozoites using:
and sporozoite quantity by using:
-
Typically between 15,000 to 20,000 sporozoites are inoculated into each assay well; thus, between 6.4 × 106-8.5 × 106 sporozoites are required per plate. One dissection of 300 mosquitoes typically results in 15 × 106-35 × 106 sporozoites, thus between 2 to 6 plates can be inoculated from a single dissection. Determine how many assay plates can be infected and place:
into a reservoir. Add all sporozoites in the collection buffer, using 200 µl of CM to wash the collection tube. Mix the entire volume at least 10× with a serological pipette to ensure an even sporozoite suspension. Using the 16-channel pipette, add 20 µl of inoculum to each well.
Spin each inoculated assay plate at 200 × g for 5 min and then place the assay plate back into the Bioassay Dish in the incubator. Do not change media later on infection day.
Perform a media change the day after infection, before addition of compounds on each treatment day, and on the days indicated for a specific assay version (Figure 1A). To change media, place a plate inverted into an aluminum spin holder in the BSC (Figure 3C), then place in a centrifuge with microtiter plate holders and spin at 40 × g for 1 min with minimum acceleration to remove all media. Transfer the plate and aluminum spin holder to the BSC and clean the top of the plate using a sterile lint-free cloth. Using a media reservoir and 16-channel pipette, add back 40 µl of CM per well.
-
-
Compound treatment
Thaw drug plates by centrifugation (200 × g for 10 min) to ensure the compound solution is in the bottom of the wells and will not cross-contaminate adjacent wells upon thaw.
All assay versions include a media change prior to treatment. Place the assay plates inverted into an aluminum spin holder in the BSC (Figure 3C), then place in a centrifuge with microtiter plate holders and spin at 40 × g for 1 min with minimum acceleration to remove all media. Transfer the plate and aluminum spin holder to the BSC and clean the top of the plate using a sterile lint-free cloth. Using a media reservoir and 16-channel pipette, add back 40 µl of CM per well (17 ml per plate). For assay versions including an ABT treatment, replace media with ABT media instead of CM.
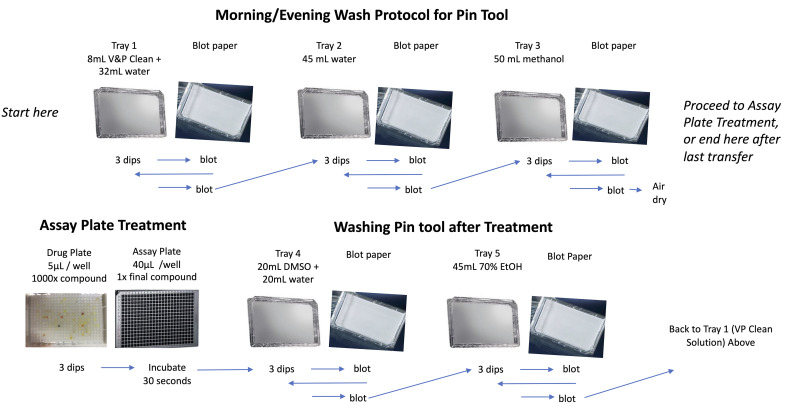
Setup the pin tool items in the BSC. Set out 5 omnitrays and 5 blot trays. Fill the first omnitray with 32 ml of sterile water and 8 ml of V&P clean solution. Fill the second tray with 45 ml of sterile water. Fill the third tray with 50 ml of methanol. Fill the fourth tray with 20 ml of sterile water and 20 ml of DMSO. Fill the fifth tray with 45 ml of 70% ethanol (Figure 5).
Perform the morning pintool wash by dipping pins thrice in the first tray (V&P clean solution), followed by a blot. Repeat the dip and blot. Perform and repeat once the three dip-and-blot wash in the second tray (water) and third tray (methanol). However, after the last methanol dip, place the pin tool in its holder instead of blotting, as this will sterilize the pins (Figure 4 and Video 5).
Set out the drug plate and assay plate, remove lids, and place library copiers on both. Dip the pins thrice in the drug plate and transfer to the assay plate. Let pins rest in the assay plate for 30 s (Figure 5).
If treating multiple plates on the same day, perform the inter-plate pintool wash (Figure 5). Perform and repeat once the three dip-and-blot wash in the fourth tray (water and DMSO), fifth tray (ethanol) first tray (V&P clean solution), second tray (water), and third tray (methanol). Then perform the treatment dip and transfer (Step F5).
After treating all plates, perform the evening pintool wash (Figure 5). Perform and repeat once the three dip-and-blot wash in the fourth tray (water and DMSO), fifth tray (ethanol), first tray (V&P clean solution), second tray (water), and third tray (methanol).
-
Immunofluorescent detection (IF) and High Content Imaging (HCI)
At the endpoint day specific for the assay version being used, remove media by placing assay plates inverted into an aluminum spin holder in the BSC (Figure 3C), then place in a centrifuge with microtiter plate holders and spin at 40 × g for 1 min with minimum acceleration to remove all media. Transfer the plate and aluminum spin holder to the BSC and clean the top of the plate using a sterile lint-free cloth. Use reservoirs and the 16-channel pipette for the following IF steps.
Move plates into a chemical fume hood and add 20 µl of 4% PFA fixing solution per well (8.5 ml per plate), incubate at room temperature for 1 h.
Remove the fix solution and wash twice by addition and removal of 40 µl of PBS per well.
Add 40 µl of primary IF stain (rPvUIS4) to each well (17 ml per plate), cover with a foil seal, and place at 4°C overnight
Remove the primary stain and wash thrice by addition and removal of 40 µl of water per well (net 51 ml per plate). Removal can be done by 16-channel pipette or by flicking the plate into a sink.
Add 40 µl of secondary IF stain (Alexa Fluor 488) to each well (17 ml per plate), cover with a foil seal, and place at 4°C overnight
Remove the secondary stain and wash thrice by addition and removal of 40 µl of water per well (net 51 ml per plate). Removal can be done by 16-channel pipette or by flicking the plate into a sink.
Add 40 µl of Hoechst stain to each well (17 ml per plate) for 1 h at room temperature.
Remove the Hoechst stain and wash twice by addition and removal of 40 µl of water per well (net 34 ml per plate). Removal can be done by 16-channel pipette or by flicking the plate into a sink.
Cover plates with a foil seal and proceed with HCI, or store at 4°C prior to HCI.
Image the plate using a 4×, 10×, or 20× objective. For a Lionheart FX, a 4× objective is used to capture an entire well with 4 fields of view per well. Images are stitched prior to analysis using the standard stitch algorithm available as an image processing module in Gen5 using DAPI as the registration channel.
-
Quantifying High Content Imaging Data
Note: This step is demonstrated using a Lionheart FX with Gen5 analysis software. Similar workflows can be developed on other HCI systems.
Intensity values will vary from run to run; modify the values before performing an analysis.
Pick a well with parasites (i.e., not an effective dose of an active compound) to modify an analysis. Open the analysis setup window.
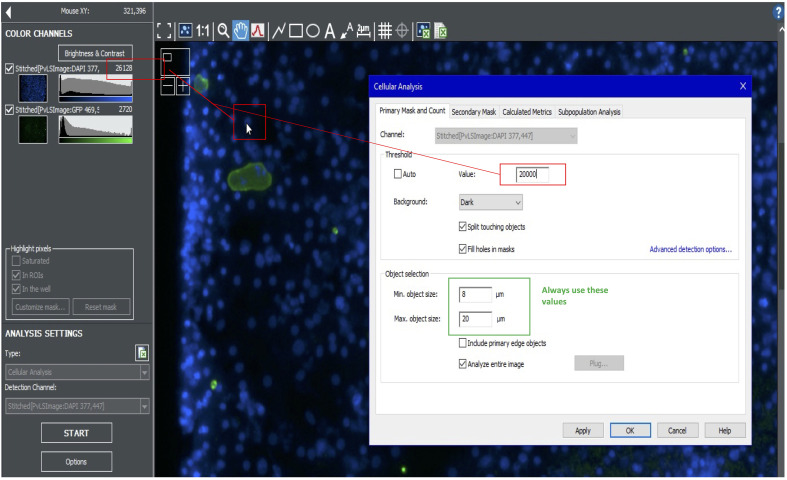
Looking at the blue channel, use the cursor to estimate the average nuclei signal intensity. Also hover over the background to understand background intensity. Take the difference between the nuclei and background intensities and use this number for your primary mask value. For example, if the nuclei intensity is 26,000, and the background is 6,000, use a value of 20,000 for the primary mask. Use a size cutoff of 8-20 µm (Figure 6).
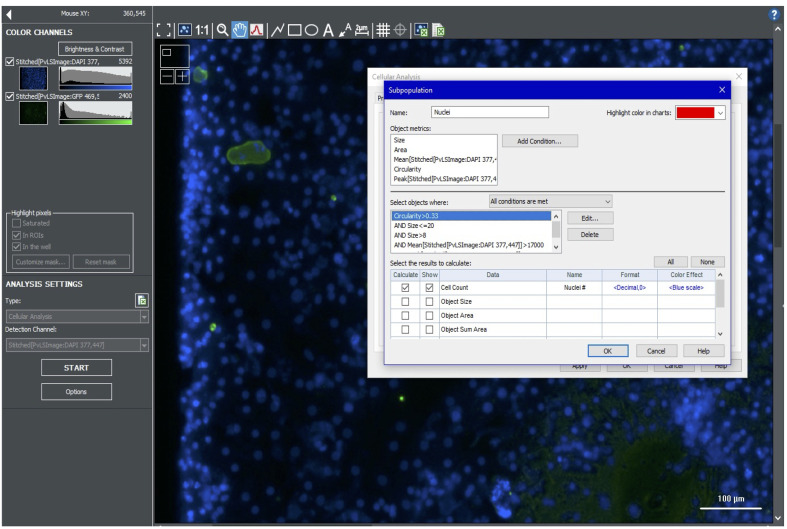
Further refine the primary mask results using a subpopulation analysis. Include a minimum peak value to avoid counting debris; nuclei usually have brighter peak intensity than debris. Other subpopulation parameters include circularity (e.g., >0.33), and mean signal (e.g., >17,000) (Figure 7).
-
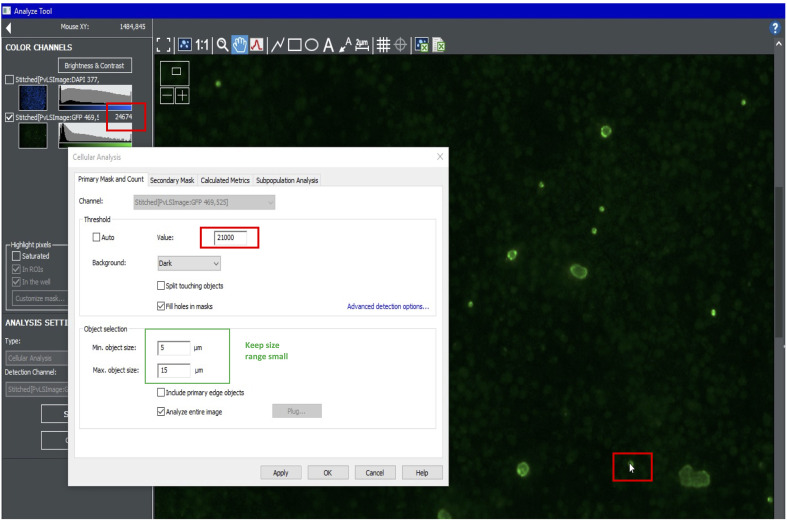
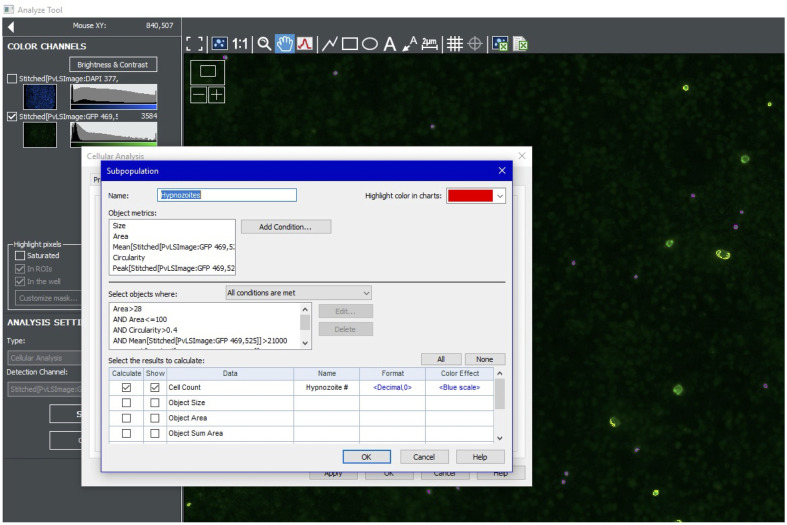
To quantify hypnozoites, turn on the green channel and create a new analysis. Use the cursor to estimate the intensity of the dimmest hypnozoites. For the Primary Mask, use the highest value possible that still highlight all hypnozoites. Use size cutoffs of 5-15 µm (Figure 8).
Further refine the hypnozoite Primary Mask results using a subpopulation analysis. Include a maximum mean value to omit bright artefacts. Other subpopulation parameters include area (e.g., >28 and <100), circularity (e.g., >0.4), mean signal (e.g., >21,000 and <45,000), and peak signal (e.g., >25,000) (Figure 9).
-
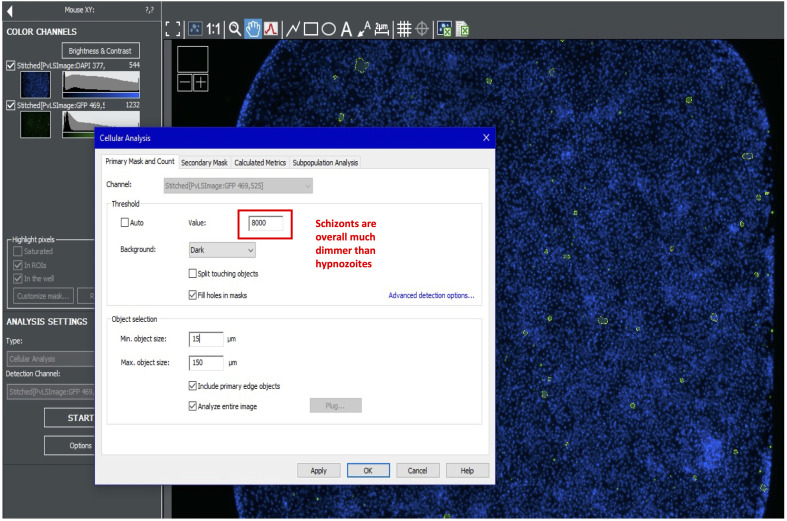
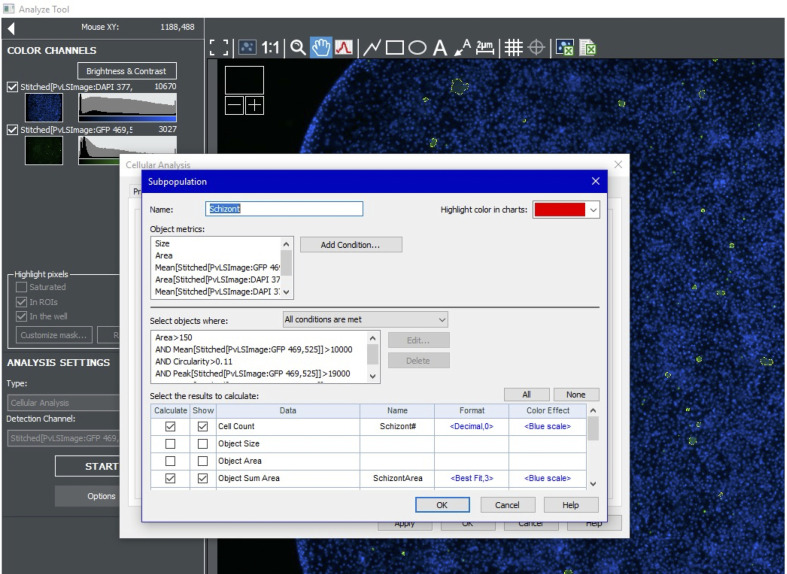
To quantify schizonts, turn on the green channel and create a new analysis. Use the cursor to estimate the intensity of the dimmest schizonts. For the primary mask, use the highest value possible that still highlights all schizonts. Well edges tend to be brighter; avoid highlighting non-specific objects on the well edge. Use size cutoffs of 15-150 µm (Figure 10).
Further refine the schizont primary mask results using a subpopulation analysis. Include a maximum mean value to omit bright artefacts. Other subpopulation parameters include area (e.g., >150 and <10,000), circularity (e.g., >0.11), mean signal (e.g., >10,000 and <30,000), and peak signal (e.g., >19,000) (Figure 11).
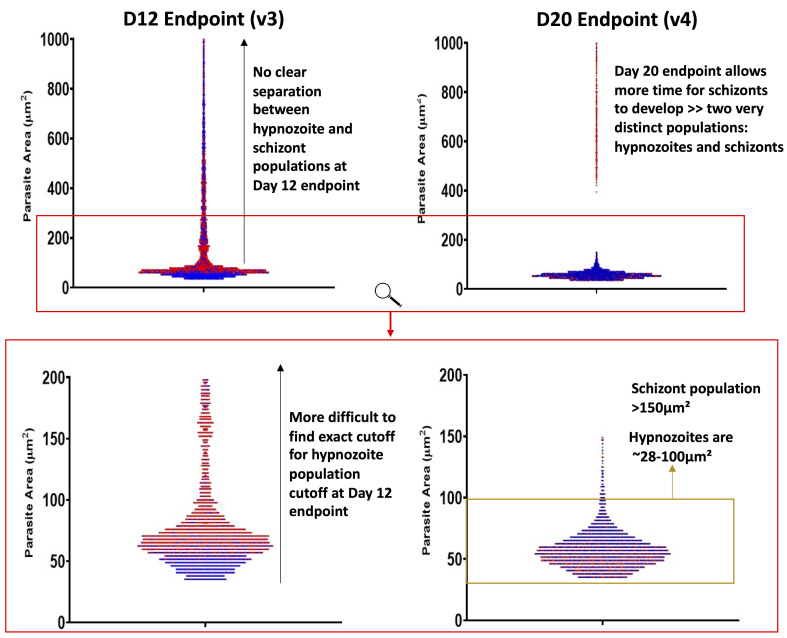
A note regarding the cutoff between hypnozoite and schizont populations: the two populations may not separate entirely in 8 and 12-day experiments (v1, v2, v3), but in our 20-day experiments (v4) we do note hypnozoites are 28-100 µm2 (Figure 12).
Once the analysis is performed, perform an export to obtain the number of nuclei per well, number of hypnozoites per well, number of schizonts for each well, and the net schizont growth (in µm2) per well (see Table S2 as an example). Proceed to data normalization and curve fitting.
Video 1. Occular view of a An. dirus midgut dissection to count oocysts.

Video 2. Occular view of a An. dirus salivary gland dissection to obtain sporozoites.
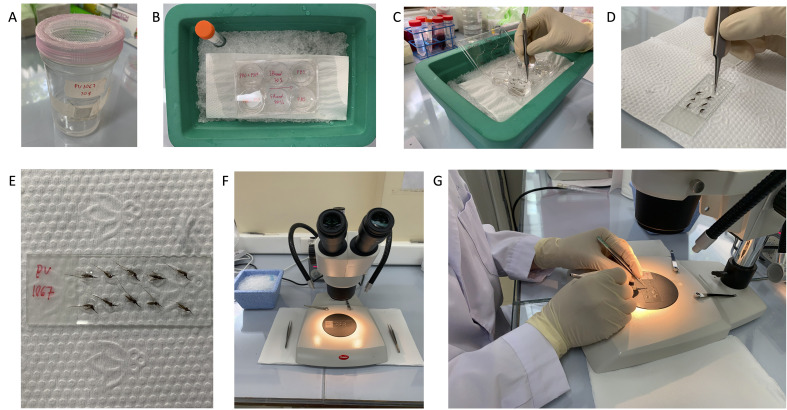
Figure 3. Drug plate preparation, assay plate incubation, and spin holders.
A. An example drug plate following serial dilution using a Biomek 4000. B. Assay plates are stored in assay pans with four small Petri dishes holding water to maintain humidity and prevent edge effects during incubation in a cell culture incubator. The assay pans also help prevent introduction of contaminating microbes when plates are moved between incubators, microscopes, and biosafety cabinets. The white arrow indicates water in a Petri dish, and the yellow arrow indicates an assay plate. C. Design schematics for a custom-fabricated aluminum spin holder for removing spent media, values are mm. The design has been uploaded to the NIH 3D Print Exchange (https://3dprint.nih.gov/discover/3dpx-015808).
Video 3. Making a dilution series in a 384-well microtiter plate with a Biomek 4000.

Video 4. Mosquito wash and placement on slide prior to salivary gland dissection.
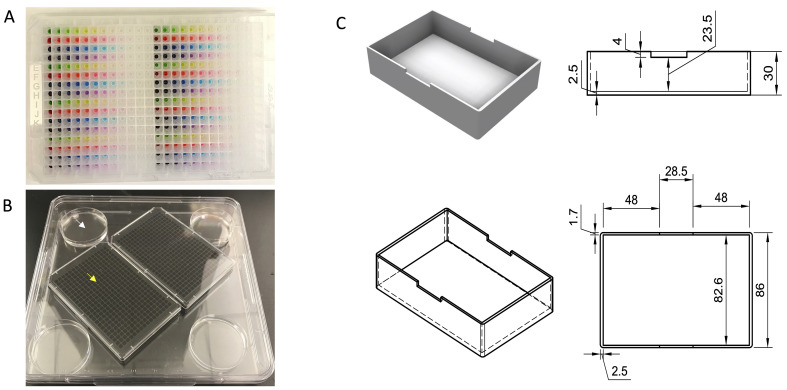
Figure 5. Pin tool wash and compound transfer.
The morning wash steps are used to prepare the pin tool for a treatment. After a treatment, the pin tool is washed and cleaned. After the last plate is treated on a given day, the tool is washed and cleaned again before storage overnight.
Video 5. Method for washing the Pin tool and transferring compounds into assay plates.

Figure 6. Setting the primary mask for hepatic nuclei.
Figure 7. Setting subpopulation parameters for hepatic nuclei.
Figure 8. Setting primary mask for hypnozoites.
Figure 9. Setting subpopulation parameters for hypnozoites.
Figure 10. Setting primary mask for schizonts.
Figure 11. Setting subpopulation parameters for schizonts.
Figure 12. Liver stage parasite size histograms and form classification.
The red box indicates the zoomed area of the top charts. Red and blue dots represent parasites from two different independent experiments. Hypnozoites are 28-100 µm2, but a small number of very large hypnozoites (or, possibly reactivating schizonts) are found between 100-150 µm2.
Data analysis
Technical and independent replication: The original v1 assay included only singleton wells with an 8-point dilution series for dose response assays ( Roth et al., 2018 ). All test compounds were assayed at least once to characterize each as inactive or active. Active compounds were then re-plated and assayed at least once more to obtain a second independent experiment to confirm activity and potency. The v2 and v3 assays include an expanded 12-point dilution series for dose response assays ( Maher et al., 2021 ) and duplicate wells at each concentration, to produce a more accurate dose-response curve. All test compounds were assayed at least once to characterize each as inactive or active. Active compounds were then re-plated and assayed at least once more to obtain a second independent experiment to confirm activity and potency. The v4 assay includes duplicate, triplicate, or quadruplicate wells at each concentration, and at least three independent experiments are performed for each collection of compounds plated together.
Controls: Control wells are used to normalize data. The original v1 was developed before ionophores were found to be active in this assay format; thus, no positive (hypnozoite-killing) control was available. Hypnozoite inhibition was measured as a reduction from negative (DMSO) control wells. Of note, we found using uninfected wells as a positive control (in lieu of an effective treatment for an infected well) artificially increased signal-to-noise as no dead or dying parasites, nor dead parasite debris, which can lead to a high background signal, are produced in an uninfected well. The v2, v3, and v4 assays were developed after monensin was found to kill P. vivax hypnozoites; thus, these assay versions all include an ionophore positive control. Monensin was the first such ionophore used, but nigericin is more potent and consistent regardless of host hepatocyte lot used ( Maher et al., 2021 ). Therefore, inhibition of parasite growth in each well is calculated in CDD Vault by:

Curve fitting and outlier removal: Curve fit settings are managed in CDD Vault’s user interface. Curve fits are constrained to minimum inhibition between -10 to 20%, maximum inhibition ≥50%, and slope ≥0.0, with all curves exhibiting no points with more than 50% inhibition being deemed ‘inactive.’
Outliers are removed when there is an obvious technical error (i.e., edge effect or user error in which hepatocytes or sporozoites are accidentally not added to a column), but because we have noted non-clearance of hypnozoites at cytotoxic concentrations above the effective concentration ( Posfai et al., 2020 ; Maher et al., 2021 ), outliers are also removed in toxic conditions as noted in Figure 13. For nuclei counts as a measure of cytotoxicity, a CC20 is calculated instead of CC50, as most compounds do not exhibit full cytotoxic dose response curves achieving a >50% decrease in hepatic nuclei.
Figure 13. Example dose response curves and toxic point marking.
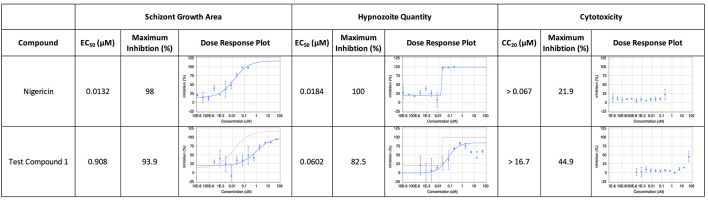
Nigericin is an ionophore control and produces a sigmoidal dose response curve for activity against schizonts and hypnozoites, but not for nuclei. An example test compound produces sigmoidal dose response curves for schizonts and hypnozoites, but is also toxic at the highest 1-2 doses. These points are removed from the hypnozoite dose response curve to enable a better curve fit.
Notes
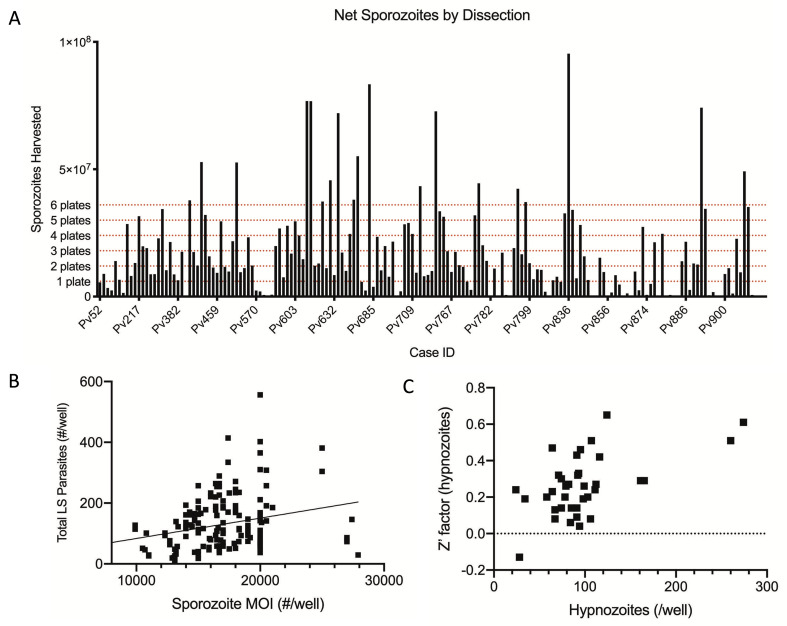
This protocol has been developed to manage variability and unpredictability noted at various points in the assay workflow. For example, no two human hepatocyte donors are the same, and immortalized human hepatocytes do not support Plasmodium infection nearly as well as primary hepatocytes ( Roth et al., 2018 ). Likewise, we expect the exact genetic composition of each P. vivax isolate used to differ; these isolates could consist of a clonal population, a mixed population, or a population consisting of siblings or half-siblings following genetic recombination in the Plasmodium mosquito stages. Furthermore, we have noted different synthetic batches of compounds to exhibit different potencies. Despite these naturally confounding variables, we have tested multiple batches of active compounds against multiple human and parasite backgrounds and found activity to be consistent, with some shift in potency depending on the exact variables mentioned above. Technical variability must also be managed: each mosquito infection produces different numbers of oocysts and sporozoites; thus, a sliding scale of vials to be thawed and assay plates to be infected is used to manage available biomaterials. While we consistently infect wells with 15,000-20,000 sporozoites, there is currently no method to predict how many liver stage parasites will be derived from the inoculum. Figure 14 displays the variability in the number of sporozoites produced from a single dissection and liver stage parasites formed following a specific inoculum.
Figure 14. Demonstrated variability in sporozoite production, inoculum-based infection rate, and well-well infection rate.
A. Example data showing sporozoites harvested from individual dissections of 100-300 mosquitoes. The target number of sporozoites needed to start 1, 2, 3, 4, 5, or 6 assay plates is indicated. B. Example data showing the weak correlation between average number of liver stage (LS) parasites forming following a specific inoculum size (linear regression, Y = 0.006701 * X + 16.54, R2 = 0.07). C. Example data showing the correlation between average number of hypnozoites per well and the Z’ factor for a particular plate. Approximately 50 hypnozoites per well is enough to produce a positive Z’ factor.
Recipes
-
20% sucrose and multivitamins solution (store at 4°C)
10 g sucrose
500 ml purified drinking water
20 drops Nutroplex®
-
Complete hepatocyte media (CM) (250 ml)
250 μl Gentamicin (1:1,000 dilution)
2.5 ml PSN solution (1:100 dilution)
Fill up to 250 ml with CP medium and filter sterilize.
Keep up to 7 days, store at 4°C.
-
ABT stock solution
Make a 100 mM stock by adding 37.3 ml of cell culture grade water to 500 mg 1-aminobenzotriazole.
Filter sterilize, aliquot into sterile Eppendorf tubes, and store at -20°C.
-
Complete hepatocyte media (CM) with ABT (250 ml)
250 μl Gentamicin (1:1,000 dilution)
2.5 ml PSN solution (1:100 dilution)
250 μl 100 mM ABT stock (1:1,000 dilution)
Fill up to 250 ml with CP medium and filter sterilize.
Keep up to 7 days, store at 4°C.
-
PSN-PBS mixture for washing mosquitoes (5 ml)
Add 500 μl of 100× PSN solution to 4.5 ml of PBS (1:10 dilution).
-
4% PFA Fixing buffer (40 ml)
Add 10 ml of 16% paraformaldehyde to 30 ml of PBS (1:4 dilution).
Store at 4°C for up to 1 month.
-
IF stain dilution buffer (1% w/v BSA, 0.3% v/v Triton X-100, 50 ml)
Add 150 μl of Triton X-100 to a 50 ml conical tube.
Add 500 mg BSA to the conical tube.
Fill with PBS to 50 ml and mix by rocking for 1 h.
Store at 4°C for up to 1 month.
-
Primary IF stain (rPvUIS4), 25 ml
Mix 1 μl of rPvUIS4 with 25 ml of IF dilution buffer (1:25,000 dilution)*.
*a 1:10,000 dilution may work better depending on the application
Make fresh when needed.
-
Secondary IF stain (Alexa Fluor 488) (25 ml)
Mix 25 μl of Alexa Fluor 488 with 25 ml of IF dilution buffer (1:1,000 dilution).
Make fresh when needed.
-
Hoechst stain (25 ml)
Mix 25 μl of 10 mg/ml Hoechst 33342 to 25 ml of PBS (1:1,000 dilution, 10 μg/ml final concentration).
Make fresh when needed.
Acknowledgments
We thank the malaria patients of northeastern Cambodia for participation in this study. HCI data from drug studies was produced in part by the Biomedical Microscopy Core at the University of Georgia, supported by the Georgia Research Alliance. Funding was provided by the Bill & Melinda Gates Foundation (OPP1023601 to D.E.K.), and Medicines for Malaria Venture (RD/2017/0042 to B.W. and A.V., RD/16/1082 and RD/15/0022 to S.P.M. and D.E.K.). This protocol reflects contributions from many colleagues, including those authored on our report of the original v1 assay ( Roth et al., 2018 ; Posfai et al., 2020 ) and v2 assay ( Maher et al., 2021 ). Table S1 was adapted from Maher et al. (2021) and reproduced under a Creative Commons 4.0 license (http://creativecommons.org/licenses/by/4.0/).
Competing interests
The authors have no competing interests to declare.
Ethics
The human subjects protocols were approved by the Cambodian National Ethics Committee for Health Research. The protocol conformed to the Helsinki Declaration on ethical principles for medical research involving human subjects (version 2002) and informed written consent was obtained for all volunteers.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Supplementary Data.
References
- 1. American Committee Of Medical Entomology American Society Of Tropical Medicine And Hygiene(2019). Arthropod Containment Guidelines, Version 3.2. Vector Borne Zoonotic Dis 19(3): 152-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bousema T., Dinglasan R. R., Morlais I., Gouagna L. C., van Warmerdam T., Awono-Ambene P. H., Bonnet S., Diallo M., Coulibaly M., Tchuinkam T., et al.(2012). Mosquito feeding assays to determine the infectiousness of naturally infected Plasmodium falciparum gametocyte carriers . PLoS One 7(8): e42821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chu C. S., Bancone G., Moore K. A., Win H. H., Thitipanawan N., Po C., Chowwiwat N., Raksapraidee R., Wilairisak P., Phyo A. P., et al.(2017). Haemolysis in G6PD Heterozygous Females Treated with Primaquine for Plasmodium vivax Malaria: A Nested Cohort in a Trial of Radical Curative Regimens . PLoS Med 14(2): e1002224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chua A. C. Y., Ananthanarayanan A., Ong J. J. Y., Wong J. Y., Yip A., Singh N. H., Qu Y., Dembele L., McMillian M., Ubalee R., et al.(2019). Hepatic spheroids used as an in vitro model to study malaria relapse . Biomaterials 216: 119221. [DOI] [PubMed] [Google Scholar]
- 5. Dembele L., Franetich J. F., Lorthiois A., Gego A., Zeeman A. M., Kocken C. H., Le Grand R., Dereuddre-Bosquet N., van Gemert G. J., Sauerwein R., et al.(2014). Persistence and activation of malaria hypnozoites in long-term primary hepatocyte cultures. Nat Med 20(3): 307-312. [DOI] [PubMed] [Google Scholar]
- 6. Dembélé L., Franetich J. F., Soulard V., Amanzougaghene N., Tajeri S., Bousema T., van Gemert G. J., Le Grand R., Dereuddre-Bosquet N., Baird J. K., et al.(2020). Chloroquine Potentiates Primaquine Activity against Active and Latent Hepatic Plasmodia Ex Vivo: Potentials and Pitfalls . Antimicrob Agents Chemother 65(1): e01416-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gural N., Mancio-Silva L., Miller A. B., Galstian A., Butty V. L., Levine S. S., Patrapuvich R., Desai S. P., Mikolajczak S. A., Kappe S. H. I., et al.(2018). In Vitro Culture, Drug Sensitivity, and Transcriptome of Plasmodium vivax Hypnozoites . Cell Host Microbe 23(3): 395-406 e394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krotoski W. A., Collins W. E., Bray R. S., Garnham P. C., Cogswell F. B., Gwadz R. W., Killick-Kendrick R., Wolf R., Sinden R., Koontz L. C. et al.(1982). Demonstration of hypnozoites in sporozoite-transmitted Plasmodium vivax infection . Am J Trop Med Hyg 31(6): 1291-1293. [DOI] [PubMed] [Google Scholar]
- 9. Llanos-Cuentas A., Lacerda M. V., Rueangweerayut R., Krudsood S., Gupta S. K., Kochar S. K., Arthur P., Chuenchom N., Mohrle J. J., Duparc S., et al.(2014). Tafenoquine plus chloroquine for the treatment and relapse prevention of Plasmodium vivax malaria(DETECTIVE): a multicentre, double-blind, randomised, phase 2b dose-selection study . Lancet 383(9922): 1049-1058. [DOI] [PubMed] [Google Scholar]
- 10. Maher S. P., Conway A. J., Roth A., Adapa S. R., Cualing P., Andolina C., Hsiao J., Turgeon J., Chaumeau V., Johnson M., et al.(2020). An adaptable soft-mold embossing process for fabricating optically-accessible, microfeature-based culture systems and application toward liver stage antimalarial compound testing. Lab Chip 20(6): 1124-1139. [DOI] [PubMed] [Google Scholar]
- 11. Maher S. P., Vantaux A., Chaumeau V., Chua A. C. Y., Cooper C. A., Andolina C., Péneau J., Rouillier M., Rizopoulos Z., Phal S., et al.(2021). Probing the distinct chemosensitivity of Plasmodium vivax liver stage parasites and demonstration of 8-aminoquinoline radical cure activity in vitro . Sci Rep 11: 19905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. March S., Ng S., Velmurugan S., Galstian A., Shan J., Logan D. J., Carpenter A. E., Thomas D., Sim B. K., Mota M. M., et al.(2013). A microscale human liver platform that supports the hepatic stages of Plasmodium falciparum and vivax . Cell Host Microbe 14(1): 104-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Markus M. B.(2020). Transition from Plasmodial Hypnozoite to Schizont Demonstrated. Trends Parasitol 36(5): 407-408. [DOI] [PubMed] [Google Scholar]
- 14. Ortiz de Montellano P. R. and Mathews J. M.(1981). Autocatalytic alkylation of the cytochrome P-450 prosthetic haem group by 1-aminobenzotriazole. Isolation of an NN-bridged benzyne-protoporphyrin IX adduct. Biochem J 195(3): 761-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Posfai D., Maher S. P., Roesch C., Vantaux A., Sylvester K., Peneau J., Popovici J., Kyle D. E., Witkowski B. and Derbyshire E. R.(2020). Plasmodium vivax Liver and Blood Stages Recruit the Druggable Host Membrane Channel Aquaporin-3 . Cell Chem Biol 27(6): 719-727 e715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roth A., Maher S. P., Conway A. J., Ubalee R., Chaumeau V., Andolina C., Kaba S. A., Vantaux A., Bakowski M. A., Thomson-Luque R., et al.(2018). A comprehensive model for assessment of liver stage therapies targeting Plasmodium vivax and Plasmodium falciparum . Nat Commun 9(1): 1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schafer C., Dambrauskas N., Steel R. W., Carbonetti S., Chuenchob V., Flannery E. L., Vigdorovich V., Oliver B. G., Roobsoong W., Maher S. P., et al.(2018). A recombinant antibody against Plasmodium vivax UIS4 for distinguishing replicating from dormant liver stages . Malar J 17(1): 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. St Laurent B., Miller B., Burton T. A., Amaratunga C., Men S., Sovannaroth S., Fay M. P., Miotto O., Gwadz R. W., Anderson J. M. et al.(2015). Artemisinin-resistant Plasmodium falciparum clinical isolates can infect diverse mosquito vectors of Southeast Asia and Africa . Nat Commun 6: 8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeeman A. M., Lakshminarayana S. B., van der Werff N., Klooster E. J., A. Voorberg-van der Wel, Kondreddi R. R., Bodenreider C., Simon O., Sauerwein R., Yeung B. K., et al.(2016). PI4 Kinase Is a Prophylactic but Not Radical Curative Target in Plasmodium vivax-Type Malaria Parasites . Antimicrob Agents Chemother 60(5): 2858-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.