Abstract
Puumala virus (PUU) nucleocapsid protein (N) was expressed in insect cells by using the Drosophila Expression System (DES; Invitrogen BV, Groningen, The Netherlands). Stable transfectants were established by hygromycin B selection and showed continuous expression of the recombinant protein (DES-PUU-N) for at least 5 months. The antigenic property of DES-PUU-N was shown to be identical to that of native PUU N when examined with a panel of hantavirus-specific monoclonal antibodies. Enzyme-linked immunosorbent assays (ELISAs) for detection of human immunoglobulin M (IgM) and IgG antibodies were established by using DES-PUU-N as antigen and were compared to assays based on native N. The ELISAs were evaluated for patient diagnosis and seroepidemiological purposes with panels of sera collected from patients with hemorrhagic fever with renal syndrome (HFRS) and from healthy blood donors. Equally high sensitivities and specificities for detection of PUU-specific IgM in acute-phase HFRS patient sera were obtained by the ELISA based on DES-PUU-N and the assay based on the native antigen. For detection of PUU-specific IgG, the ELISA based on monoclonal antibody-captured DES-PUU-N antigen showed optimal sensitivity and specificity.
Puumala virus (PUU), a member of the Hantavirus genus, family Bunyaviridae, is a causative agent of hemorrhagic fever with renal syndrome (HFRS) (25). Hantaviruses are negative-stranded RNA viruses with a tripartite genome; the S segment encodes a nucleocapsid protein (N), the M segment encodes two glycoproteins, G1 and G2, and the L segment encodes an RNA polymerase (24, 27). Hantaviruses are transmitted to humans from rodent hosts, probably through inhalation of aerosolized excreta. The bank vole (Clethrionomys glareolus) is the major rodent carrier of PUU (25). HFRS caused by PUU is generally milder than HFRS caused by Dobrava virus (DOB) or Hantaan virus (HTN) and is rarely manifested by hemorrhage. Although the mortality rate from PUU infections is low (<0.2%), the virus causes significant morbidity in northern and eastern Europe; each year western Russia, Finland, Sweden, and Norway report about 5,000, 1,000, 300, and 50 cases, respectively. Sporadic outbreaks are observed in central Europe, and we have recently reported major outbreaks with hundreds of cases in Bosnia and Belgium (7, 17).
The diagnosis of PUU infections requires serological confirmation. Although reverse transcription-PCR has been successfully applied for detection of PUU RNA in a limited number of patient samples (8, 22, 23), the test can be used only during the first week of the disease, and even within this period, the level of viremia seems to be close to the limit of sensitivity (23). Thus, reverse transcription-PCR cannot be recommended as a test that can be used for the routine diagnosis of PUU infections. The N PUU has been demonstrated to be the major antigenic target in the early human antibody response, and high levels of N-specific antibodies are produced during the acute phase of the disease (5, 16, 30). For diagnosis of acute hantavirus infections, assays that measure immunoglobulin M (IgM) antibody levels have been shown to be the most informative; the virus-specific IgM levels rise earlier than the IgG antibody levels, and IgM production is clearly associated with acute infection (1, 12, 16).
The increasing awareness of hantavirus infections in Europe has created an urgent need for rapid and reliable diagnostic assays. As pathogenic hantaviruses require biosafety level 3 facilities for propagation, alternative means of production of viral antigens are preferred. We and others have previously established hantavirus antibody-specific enzyme-linked immunosorbent assays (ELISAs) based on total N or truncated variants expressed in Escherichia coli (3, 5, 11, 12, 33). For the diagnosis of PUU and DOB infections, we recently found that assays based on the total N protein expressed in the baculovirus system had optimal performances (1, 9, 10, 12, 31).
The aim of this study was to produce recombinant PUU N antigen in the Drosophila Expression System (DES; Invitrogen, Groningen, The Netherlands) and to evaluate the suitability of this recombinant protein as a diagnostic antigen.
MATERIALS AND METHODS
Cloning and expression of recombinant protein.
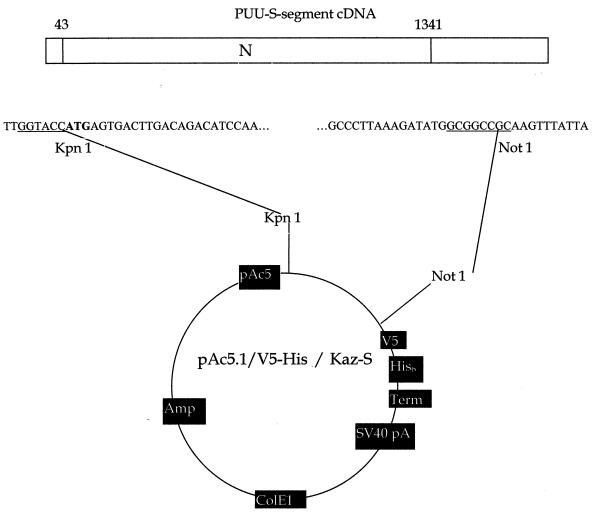
For cloning into the pAc5.1/V5-His vector (DES; Invitrogen) the entire open reading frame (ORF) for the N protein (nucleotides 43 to 1341) was amplified with primers containing KpnI and NotI restriction sites (underlined below; Fig. 1): forward primer, TTG GTA CCA TGA GTG ACT TGA CAG ACA TCC AA; reverse primer, TAA TAA ACT TGC GGC CGC CAT ATC TTT AAG GGC. The cDNA clone of the PUU Kazan strain S segment (14) was used as a template for PCR, according to a standard protocol. The PCR product was cloned into the pAc5.1/V5-His vector and amplified by standard molecular biological techniques and was subsequently prepared for transfection into Schneider (S2) cells (29).
FIG. 1.
Constitution of pAc5.1/V5-His/Kaz-S. The entire ORF for the N protein of PUU (strain Kazan) was amplified and cloned into the KpnI and NotI sites of the pAc5.1/V5-His vector as described in Materials and Methods.
The correct sequence of the recombinant DNA was confirmed by cycle sequencing performed on an FTS-320 Thermal Sequencer by using the PRISM Ready BigDye Terminator Cycle Sequencing kit with AmpliTaq DNA Polymerase FS (Taq-FS; Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). The total reaction volume was 20 μl (8 μl of PRISM premix, 3.2 pmol of sequencing primer, and 500 ng of DNA). The sample was analyzed with an ABI377 sequencer (Perkin-Elmer, Applied Biosystems Division).
Transient transfections of S2 cells were performed according to the manufacturer's instructions (Invitrogen). Briefly, the purified cloned vector (pAc5.1/V5-His/Kaz-S) was transfected by use of calcium phosphate (Invitrogen) into 2 × 106 to 4 × 106 S2 cells/ml in six-well cell culture plates (Costar). The cells were harvested at 48, 72, 96, 120, and 144 h after plasmid transfection.
Cell lines for stable expression of the recombinant protein (DES-PUU-N) were established by selection with hygromycin B according to the manufacturer's instructions (Invitrogen). Briefly, 19 μg of pAc5.1/V5-His/Kaz-S and 1 μg of pCoHYGRO were cotransfected with calcium phosphate into 2 × 106 to 4 × 106 S2 cells/ml in six-well cell culture plates according to the manufacturer's instructions (Invitrogen). After 16 to 24 h of incubation at room temperature, the calcium phosphate was removed by centrifugation. Two days later, 400 μg of hygromycin B per ml was added, as determined by kill-curve titrations (the lowest concentration of hygromycin B which resulted in total cell death after 4 days of incubation), and resistant cells were expanded for 4 weeks. Cells were continuously harvested every 7 days.
Production of recombinant antigen.
For production of DES-PUU-N, transfected cells were cultured in DES Expression media (Invitrogen) supplemented with 10% fetal calf serum, antibiotics, and hygromycin B at room temperature. The cells were collected by centrifugation, and proteinase inhibitors (10 mg of leupeptin per ml, 10 mg of pepstatin A per ml, and 1 mg of aprotinin per ml [Boehringer Mannheim] and 10 mM EDTA) were added. The cell pellets were stored at −20°C until they were used. The cell pellets were dissolved in phosphate-buffered saline (PBS) and sonicated on ice (five times for 5 s each time). To test the antigenicity of the protein, aliquots were tested by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in 4 to 15% polyacrylamide gels, transferred onto nitrocellulose filters, and immunoblotted with a pool of PUU N-reactive monoclonal antibodies (MAbs; 1C12, 4C3, 5B5, and 3G5) as described earlier (15).
Native viral antigen.
Native PUU antigens for ELISA were produced as described previously (15, 16). The antigen used for IgM detection consisted of sonicated extracts of infected Vero E6 cells, and the antigen used for IgG detection consisted of infected cells treated with a detergent-containing buffer.
Serum panels.
Patient sera, previously analyzed in the diagnostic routine at the Swedish Institute for Infectious Disease Control, Stockholm, Sweden, by a μ-capture IgM ELISA based on native viral antigen, as described below, were used to evaluate the DES-PUU-N IgM ELISA. The panel consisted of acute-phase sera from 131 serologically confirmed (PUU IgM-positive) patients, all with clinical symptoms of PUU infection, and 114 serum samples from patients with suspicion of HFRS but with negative serological test results for PUU IgM. In addition, 40 serum samples from patients with other acute viral infections (varicella-zoster virus, measles virus, influenza A virus, and cytomegalovirus) were analyzed.
For evaluation of the IgG assays, a panel of 185 serum samples from apparently healthy individuals (blood donors) in Sweden and Latvia were used.
Buffers for ELISA.
Carbonate buffer (50 mM; pH 9.6) was used for antibody coating in all assays. The blocking step was performed by incubation with 3% bovine serum albumin in PBS for 1 h at 37°C. All sera, conjugates, and the antigens used in the following steps of the ELISAs were diluted in PBS with 0.05% Tween 20 plus 0.5% bovine serum albumin. Tetramethylbenzidine or p-nitrophenylphosphate (pNPP) was used as a substrate, as described by the manufacturer (Sigma). The plates were washed five times in 0.9% NaCl with 0.05% Tween 20 between each step.
MAb detection assay.
A panel of hantavirus-specific MAbs was used to compare the antigenic properties of the recombinant protein with those of the corresponding native antigen. The panel contained the PUU N MAbs 1C12, 4C3, 3G5, 2E12, 5B5, 3H9, 5F4, and 5E1 (15, 19) and the Tula virus (TUL) N MAbs 1C8, 3C11, 7A4, 6A6, and 3D3 (20). The antigenic recognition sites of the MAbs are shown in Table 1. The plates were coated with rabbit anti-PUU diluted 1:300 (17) overnight at 4°C, after which they were blocked prior to use. The antigens were incubated for 1 h at 37°C, followed by incubation of the MAbs at a concentration of 2 μg/ml in duplicate for 1 h at 37°C. Specific antibody binding was detected by incubation of alkaline phosphatase-conjugated anti-mouse IgG (Jackson) diluted 1:1,000 for 1 h at 37°C, followed by incubation of pNPP. Optical densities were measured after 30 min at 405 nm.
TABLE 1.
Antigenic characteristics of DES-PUU-N as analyzed with MAbs
| Virus and epitope (MAb) | Reactivity of the following antigensa:
|
|||||
|---|---|---|---|---|---|---|
| DES-PUU-N | Native PUU N | Bac-PUU-Nb | GST-PUU-Nb | β-Gal-PUU-Nb | PUU rN [poly(His)]c | |
| PUU | ||||||
| N-a (3H9) | + | + | + | + | − | − |
| N-b (5E1) | + | + | + | + | + | + |
| N-c (5B5) | + | + | + | + | + | + |
| N-d (3G5) | + | + | + | + | + | + |
| N-e (5F4) | + | + | + | − | − | − |
| N-f (1C12) | + | + | + | + | + | + |
| N-g (2E12) | + | + | + | + | + | + |
| N-h (4C3) | + | + | + | + | + | + |
| TUL | ||||||
| N-a (3D3) | − | − | − | − | NT | NT |
| N-b (6A6) | − | − | − | − | NT | NT |
| N-c (7A4) | − | − | − | − | NT | NT |
| N-d (3C11) | + | + | + | + | NT | NT |
| N-e (1C8) | + | + | + | + | NT | NT |
FRNT.
The focus-reduction neutralization test (FRNT) was performed as described earlier (17). An 80% reduction in the number of foci compared to the number for the virus control was used as the criterion for virus neutralization titers.
IgM and IgG ELISAs.
PUU IgM μ-capture ELISAs, based on DES-PUU-N or native antigens, were performed essentially as described previously (1, 13). Briefly, microtiter plates were coated with goat anti-human IgM (Cappel, Organon Technica, Turnhout, Belgium), blocked, and incubated with patient or control sera at a 1:200 dilution. Each antigen was added at optimal concentrations, as determined by box titration, followed by the addition of PUU N-specific peroxidase-conjugated bank vole MAb 1C12 (15) and tetramethylbenzidine substrate. To calculate the results, all absorbances were adjusted according to a standard control PUU-positive serum, for which the mean optical density (OD) value for duplicate wells was recalculated to 1.000. Background ODs for the control wells were reduced from the ODs for the wells incubated with antigen. Cutoff values for positive samples were set at an OD of 0.150.
PUU IgG ELISAs based on MAb-captured DES-PUU-N or native antigens were performed essentially as described previously (1, 16). Briefly, microtiter plates were coated with MAb 1C12 (1 μg/ml) and were incubated overnight at 4°C. After blocking, the antigens were incubated, followed by incubation of serum samples (diluted 1:400) in duplicate wells in both antigen-sensitized and control wells. Goat anti-human IgG (γ-chain specific; Sigma) alkaline phosphatase conjugate was added, followed by the addition of pNPP substrate. The results were calculated as described above for the IgM ELISAs except for the use of late-convalescent-phase serum from patients with PUU infection as the standard control and the use of different cutoff values for positive samples, which were set at an OD of 0.100.
RESULTS
Expression of DES-PUU-N.
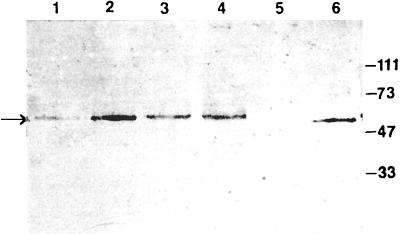
The plasmid pAc5.1/V5-His/Kaz-S, which contains the full-length PUU N ORF correctly inserted, as confirmed by complete nucleotide sequencing, was transfected into S2 cells. To estimate the optimal amount of plasmid DNA required for maximal expression levels, 1, 5, 10, 15, 20, 30, or 40 μg of pAc5.1/V5-His/Kaz-S was transfected to equal numbers of S2 cells. A band of the expected size (approximately 54 kDa) was detected with a pool of PUU-specific MAbs (Fig. 2 and data not shown). The results indicated that 20 μg of the plasmid DNA was the optimal concentration.
FIG. 2.
DES-PUU-N (approximately 54 kDa) immunoblotted with a pool of PUU N-reactive MAbs (MAbs 1C12, 4C3, 5B5, and 3G5) after transient transfection of various amounts of pAc5.1/V5-His/Kaz-S to equal numbers of S2 cells. Lane 1, 10 μg; lane 2, 20 μg; lane 3, 30 μg; lane 4, 40 μg; lane 5, negative control (S2 cells); lane 6, positive control (native PUU N antigen). Molecular masses (in kilodaltons) are indicated to the right.
To estimate the optimal time of cell culture required for maximum expression levels, 20 μg of pAc5.1/V5-His/Kaz-S was transfected to S2 cells, which were harvested 48, 72, 96, 120, and 144 h later. The result indicated that maximum expression of the protein (DES-PUU-N) was obtained at 96 h after the transfection (data not shown).
Antigenic properties of DES-PUU-N.
The hantavirus-specific MAbs reacted with DES-PUU-N in the ELISA with a pattern identical to that seen with native PUU antigen (Table 1). Notably, the two epitopes, N-a and N-e, previously shown to be missing in E. coli-expressed PUU N (4, 18, 31) were clearly recognized.
Establishment of stable transfectants.
The plasmids pAc5.1/V5-His/Kaz-S and pCoHYGRO were cotransfected to 2 × 106 to 4 × 106 S2 cells/ml in 3 ml of medium. After 6 weeks of selection with 400 μg of hygromycin B per ml, cells were continuously analyzed for expression of the recombinant protein at 1.5, 3, 4, and 5 months after the transfection (Table 2). The results indicated a continuous expression for at least 5 months. Frozen stocks of stable DES-PUU-N-expressing cells have been successfully thawed and cultivated after 1 month of storage in liquid nitrogen (Table 2).
TABLE 2.
Continuous expression of DES-PUU-N
| Time after initial transfection | ELISA end-point titera
|
|
|---|---|---|
| IgM/IgG | IgM/IgG after freezing for 1 mob | |
| 6 wk | 80/5120 | NTc/NT |
| 3 mo | 80/1280 | 80/1250 |
| 4 mo | 50/1280 | 60/980 |
| 5 mo | 40/300 | NT/NT |
Sonicated cell extracts were titrated to the end point by the IgM and IgG ELISAs as described in Materials and Methods.
Cells were cultured for 6 weeks, frozen in liquid nitrogen for 1 month, thawed, and subsequently cultured for 2 weeks before analysis as described above.
NT, not tested.
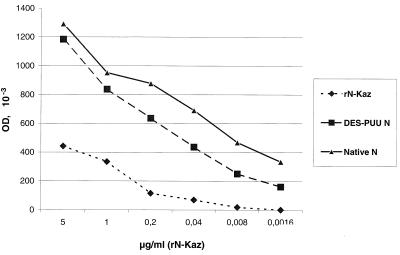
To estimate the amounts of DES-PUU-N expressed by the stable transfectants, crude cell extracts were titrated to the end point, and the results were compared to those for native PUU N and purified recombinant PUU N (rN-Kaz) of known concentration expressed in E. coli (C. de Carvalho Nicacio and Å. Lundkvist, unpublished data) in IgG and IgM ELISAs (Fig. 3 and data not shown). The results indicated that one 225-cm2 flask (inoculated with approximately 3.4 × 107 stable transfectants) yielded 0.6 to 1.8 mg of DES-PUU-N after 1 week of culture, which is equal to the antigen amount required for 80 to 240 IgG ELISA plates or for examination of 1,200 to 3,600 serum samples in duplicate. To obtain similar amounts of native PUU N, harvests from 1.25 to 5 roller bottles (800 cm2) with monolayers of Vero E6 cells inoculated for 14 days were required, indicating an approximately 5- to 20-fold higher efficiency for the DES system.
FIG. 3.
IgG ELISA titration curves. Results for known concentrations of purified rN-Kaz (⧫) were compared to those for dilution series (starting from 1:10 with twofold dilutions) of crude extracts of DES-PUU-N (■; stable transfectants harvested from one 225-cm2 cell culture flask and sonicated in 5 ml) and native PUU N (▴; infected cells from five 800-cm2 roller bottles and sonicated in 5 ml). OD, optical density at 405 nm.
Stability of DES-PUU-N.
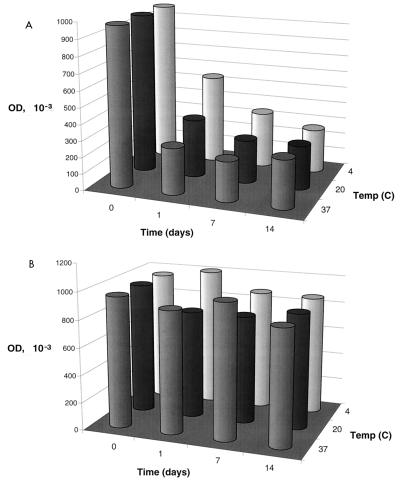
The stability of DES-PUU-N was evaluated by incubating aliquots of sonicated crude cell extracts at 4, 20, and 37°C for 1, 7, and 14 days, respectively. The results revealed a significant decrease in the activity of the antigen over time and at higher temperatures when it was used for the IgM ELISA (Fig. 4A). In contrast, when the antigen was analyzed by the IgG ELISA, almost no difference in activity could be observed (Fig. 4B). The stability of the antigen was further evaluated by repeated freezing-thawing of aliquots stored at −20°C. The results revealed that the antigen was completely stable for up to six cycles of freezing-thawing, irrespective of whether it was used for the IgM or the IgG ELISA (Table 3).
FIG. 4.
DES-PUU-N from sonicated crude cell extracts examined for stability over time and temperature. (A) IgM ELISA. (B) IgG ELISA. OD, optical density at 450 (A) or 405 (B) nm.
TABLE 3.
Stability of DES-PUU-N after repeated freezing-thawing
| No. of cycles | ELISA end-point titersa
|
|
|---|---|---|
| IgM | IgG | |
| 0 | 20 | 320 |
| 1 | 20 | 320 |
| 2 | 20 | 320 |
| 3 | 20 | 320 |
| 4 | 20 | 320 |
| 5 | 20 | 320 |
| 6 | 20 | 320 |
Diluted aliquots of sonicated cell extracts were titrated to the end point by IgM and IgG ELISAs as described in Materials and Methods.
Detection by PUU-specific IgM.
A panel of 131 serum specimens from HFRS patients and from 114 serum specimens from patients with similar clinical symptoms, all previously examined by μ-capture IgM ELISA based on native viral antigen for routine diagnosis, were used for evaluation of the DES-PUU-N-based IgM ELISA (test data are summarized in Table 4). The ELISA showed that optimal specificity and sensitivity were achieved; i.e., all sera reacted identically compared to the reactivities of the sera by the μ-capture ELISA based on the native antigen. Furthermore, none of 40 serum samples from patients with other acute viral infections were reactive.
TABLE 4.
Detection of PUU-specific IgM in acute-phase patient seraa
| Antigen | No. of serum specimens testing positive
|
||
|---|---|---|---|
| HFRS patient sera (n = 131) | Negative control sera Ib(n = 114) | Negative control sera IIc(n = 40) | |
| Native N | 131 | 0 | 0 |
| DES-PUU-N (amino acids 1 to 433) | 131 | 0 | 0 |
Testing was by the μ-capture IgM ELISA.
Negative control sera I, sera from patients with HFRS-like symptoms.
Negative control sera II, sera from patients with other acute viral infections.
Detection by PUU IgG ELISA.
The IgG ELISA based on DES-PUU-N was evaluated with a panel of serum specimens from healthy individuals in Sweden and Latvia (test data are summarized in Table 5). Of 33 PUU-positive serum specimens, as determined by neutralization assay, all were found to be IgG positive by the MAb-antigen-capture ELISAs based on DES-PUU-N or native antigen. All 152 serum samples negative for PUU neutralizing antibodies and negative for PUU IgG when assayed by the ELISA with the native antigen were also negative by the DES-PUU-N IgG ELISA.
TABLE 5.
Detection of PUU-specific IgG in sera from blood donors
| Assay | No. of serum specimens testing positive
|
|
|---|---|---|
| Positive sera (n = 33) | Negative sera (n = 152) | |
| FRNT | 33 | 0 |
| MAb-capture IgG ELISA with native PUU-N | 33 | 0 |
| MAb-capture IgG ELISA with DES-PUU-N (amino acids 1 to 433) | 33 | 0 |
DISCUSSION
Nephropathia epidemica (NE), the form of HFRS caused by PUU which is commonly encountered in northern and central Europe, is a febrile disease associated with acute renal impairment. A rapid and reliable diagnosis is therefore of great importance for differentiation of NE from other acute febrile illnesses in areas where PUU is endemic. Serological assays are needed for diagnosis of hantavirus infection since only two-thirds of PUU-infected patients (22, 23) and approximately 40% of DOB-infected patients (21) are viral RNA positive by current PCR tests and the isolation of hantaviruses from HFRS patients is usually impossible. Due to the high sensitivity combined with the ability for rapid processing of large numbers of samples, ELISA has become the method of choice for the diagnosis of hantavirus infections. Results are obtained within a few hours and do not suffer from ambiguities due to subjective interpretations of immunofluorescence assays. Although the amino-terminal region of PUU N has been shown to constitute a major antigenic target in the human antibody response (4, 6), our data have indicated the presence of immunodominant regions also in the central and the carboxy-terminal parts of the protein, indicating that use of full-length N is essential for proper assay sensitivity (1, 13, 30).
Baculovirus-expressed PUU N antigen has, in contrast to E. coli-expressed PUU N, been found to be antigenically indistinguishable from the native protein (4, 18, 31), and we have to date used this antigen most successfully for diagnosis of PUU infection in patients and seroepidemiological studies (1, 12; K. Brus Sjölander and Å. Lundkvist, unpublished data). However, there are certain drawbacks with the baculovirus system, especially the complicated procedure for antigen production, including repeated titrations of the recombinant virus in plaque assays, the need for specific and separated incubators for handling of the infectious recombinant virus at 27°C, a sensitive target cell line (Sf9) that requires expensive culture media, and the fact that baculovirus kills the target cells in a few days, which makes continuous production of the antigen impossible.
In an attempt to improve and simplify the means of production of a high-quality recombinant antigen for use in the diagnosis of PUU infections, we expressed PUU N in DES. On the basis of our and others' positive experiences with the baculovirus system (1, 10, 12, 26, 28, 31, 32), we selected another insect cell system that has several advantages: (i) the single-step transfection procedure for immediate expression, (ii) the simple and straightforward procedure for the establishment of stable transfectants, and (iii) the easy on-bench handling of the target cells (room temperature, no CO2 requirements, no need for trypsinization).
The antigenic characteristics of DES-PUU-N were found to be indistinguishable from those of native or baculovirus-expressed N when they were analyzed with a panel of MAbs raised against PUU and TUL. Previous data for various E. coli-expressed PUU N proteins have revealed that epitope N-a or N-e, or both, is missing (4, 18, 31). Several assays based on E. coli-expressed N were previously shown to suffer from low sensitivities and specificities, probably due to incorrect folding of the recombinant proteins, i.e., a lack of epitopes important for recognition by human antibodies (1).
The need for purified antigen was efficiently circumvented by the use of the capture format of the ELISAs. The yield of active antigen from crude cell extracts, estimated to be 0.6 to 1.8 mg from 3.4 × 107 cells (similar to the numbers of cells in three 75-cm2 flasks), which was significantly higher than that of native viral antigen, proved the efficiency of the system.
When the stability of the DES-PUU-N antigen was evaluated, the results indicated that the IgM ELISA was more dependent on freshly thawed antigen than the IgG ELISA. Incubation of antigen fractions at different temperatures for 1, 7, and 14 days revealed a clear decrease in IgM ELISA activity over time and with increasing temperature. In contrast, when the same antigen fractions were examined by IgG ELISA, the samples showed almost identical activities independent of time and temperature, indicating that the IgG test is not as dependent as the IgM ELISA on a totally intact antigen. A possible explanation is that the IgM ELISA, in contrast to the IgG ELISA, to some extent requires an aggregated antigen and that the aggregation may be lost over time at higher temperatures. In line with this observation, our previous trials with highly soluble truncated forms of recombinant N in the IgM ELISA were completely unsuccessful, while the same antigen worked well in the IgG ELISA (1). The crude recombinant antigen preparation was, however, found to be stable against repeated freezing-thawing, with no detectable loss of activity after six cycles.
After long-term culture, a decrease in the expression levels over time was revealed by both IgG and IgM ELISA end-point titrations. However, at the termination of the experiment (after 5 months of continuous culture), significant levels of the recombinant antigen were still expressed.
The data concerning IgM assays showed that the IgM μ-capture ELISA based on DES-PUU-N had sensitivity and specificity equal to those of the assay based on native virus antigen. All 131 serum samples from patients with NE and previously confirmed to have acute PUU infection according to the routine diagnosis scored positive, while no nonspecific reactions were seen.
In comparison to the IgG ELISA based on native antigen and the neutralization test results, the IgG ELISA based on crude extracts of DES-PUU-N antigen were found to be equally efficient for the detection of PUU-specific antibodies. Native antigen is difficult to produce due to the hazardous nature of the virus, while DES-PUU-N is easy to produce without the need for biological containment conditions. The higher levels of expression of DES-PUU-N compared to the low levels of antigen obtained when native virus is cultured further points to the advantage of this system. Another advantage of the DES-PUU-N based system, in comparison to E. coli-expressed rN, is that no purification of the antigen seems to be needed.
Although significant serological cross-reactivities are seen among several of the hantaviruses, e.g., within the PUU-Topografov-Khabarovsk-Tula-Prospect Hill hantavirus group, assays based on PUU antigen do not efficiently detect antibodies to more distantly related hantaviruses, such as DOB or HTN. Furthermore, our recent results have proved the need for the homologous DOB antigen for optimal serodiagnosis (2). Therefore, in areas where serologically distinct hantaviruses pathogenic for humans circulate, e.g., in the former Yugoslavia, tests based on at least two antigens are needed for proper diagnosis of patient infections. We are working on the establishment of high-quality serological assays based on recombinant antigens for the diagnosis of DOB infections.
In conclusion, use of DES enabled the straightforward and efficient production of a high-quality recombinant PUU N protein with antigenic characteristics indistinguishable from those of the native antigen. The specificity and sensitivity of the PUU IgM assays based on DES-PUU-N and native N protein were equal. The IgG assay based on MAb-captured DES-PUU-N had results identical to those of an IgG ELISA based on native antigen and the neutralization assay when applied to a panel of serum specimens from healthy individuals.
ACKNOWLEDGMENTS
This work was financially supported by grants from the Swedish Medical Research Council (projects 12177 and 12642) and the Swedish Society of Medicine and by the European Community (contract BMH4-CT97-2499). A.P. was supported by a postdoctoral fellowship from the Karolinska Institute.
REFERENCES
- 1.Brus Sjölander K, Elgh F, Kallio-Kokko H, Vapalahti O, Hägglund M, Palmcrantz V, Juto P, Vaheri A, Niklasson B, Lundkvist Å. Evaluation of serological methods for diagnosis of Puumala hantavirus infection (nephropathia epidemica) J Clin Microbiol. 1997;35:3264–3268. doi: 10.1128/jcm.35.12.3264-3268.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brus Sjölander K, Lundkvist Å. Dobrava virus infection: serological diagnosis and cross-reactions to other hantaviruses. J Virol Methods. 1999;80:137–143. doi: 10.1016/s0166-0934(99)00037-3. [DOI] [PubMed] [Google Scholar]
- 3.Elgh F, Lundkvist Å, Alexeyev O A, Stenlund H, Avsic-Zupanc T, Hjelle B, Lee H W, Smith K J, Vainionpää R, Wiger D, Wadell G, Juto P. Serological diagnosis of hantavirus infection by enzyme-linked immunosorbent assay based on detection of immunoglobulin G and M responses to recombinant nucleocapsid protein of five viral serotypes. J Clin Microbiol. 1997;35:1122–1130. doi: 10.1128/jcm.35.5.1122-1130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elgh F, Lundkvist Å, Alexeyev O A, Wadell G, Juto P. A major antigenic domain for the human humoral response to Puumala virus nucleocapsid protein is located at the amino-terminus. J Virol Methods. 1996;59:161–172. doi: 10.1016/0166-0934(96)02042-3. [DOI] [PubMed] [Google Scholar]
- 5.Elgh F, Wadell G, Juto P. Comparison of the kinetics of Puumala virus specific IgM and IgG antibody responses in nephropathia epidemica as measured by a recombinant antigen-based enzyme-linked immunosorbent assay and an immunofluorescence test. J Med Virol. 1995;45:146–150. doi: 10.1002/jmv.1890450206. [DOI] [PubMed] [Google Scholar]
- 6.Gött P, Zöller L, Darai G, Bautz E K. A major antigenic domain of hantaviruses is located on the aminoproximal site of the viral nucleocapsid protein. Virus Genes. 1997;14:31–40. doi: 10.1023/a:1007983306341. [DOI] [PubMed] [Google Scholar]
- 7.Heyman P, Vervoort T, Colson P, Chu Y K, Avsic-Zupanc T, Lundkvist Å. A major outbreak of hantavirus infection in Belgium in 1995 and 1996. Epidemiol Infect. 1999;122:447–453. doi: 10.1017/s0950268899002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hörling J, Lundkvist Å, Persson K, Mullart M, Czagurova T, Dekonenko A, Tkachenko E, Niklasson B. Detection and subsequent sequencing of Puumala virus from human specimens by polymerase chain reaction. J Clin Microbiol. 1995;33:277–282. doi: 10.1128/jcm.33.2.277-282.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanov A, Vapalahti O, Lankinen H, Tkachenko E, Vaheri A, Niklasson B, Lundkvist Å. Biotin-labeled antigen: a novel approach for detection of Puumala virus-specific IgM. J Virol Methods. 1996;62:87–92. doi: 10.1016/0166-0934(96)02090-3. [DOI] [PubMed] [Google Scholar]
- 10.Kallio-Kokko, H., Å. Lundkvist, A. Plyusnin, T. Avsic-Zupanc, A. Vaheri, and O. Vapalahti. Antigenic properties and diagnostic potential of recombinant Dobrava virus nucleocapsid protein. J. Med. Virol., in press. [PubMed]
- 11.Kallio-Kokko H, Vapalahti O, Hedman K, Brummer-Korvenkontio M, Vaheri A. Puumala virus antibody and immunoglobulin G avidity assays based on a recombinant nucleocapsid antigen. J Clin Microbiol. 1993;31:677–680. doi: 10.1128/jcm.31.3.677-680.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kallio-Kokko H, Vapalahti O, Lundkvist Å, Vaheri A. Evaluation of Puumala virus IgG and IgM enzyme immunoassays based on recombinant baculovirus-expressed nucleocapsid protein for early nephropathia epidemica diagnosis. Clin Diagn Virol. 1998;10:83–90. doi: 10.1016/s0928-0197(97)10019-8. [DOI] [PubMed] [Google Scholar]
- 13.Lundkvist Å, Björsten S, Niklasson B, Ahlborg N. Mapping of B-cell determinants in the nucleocapsid protein of Puumala virus; definition of epitopes specific for acute IgG recognition in humans. Clin Diagn Lab Immunol. 1995;2:82–86. doi: 10.1128/cdli.2.1.82-86.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundkvist Å, Cheng Y, Brus Sjölander K, Niklasson B, Vaheri A, Plyusnin A. Cell culture adaptation of Puumala hantavirus changes the infectivity for its natural reservoir, Clethrionomys glareolus, and leads to accumulation of mutants with altered genomic RNA S segment. J Virol. 1997;71:9515–9523. doi: 10.1128/jvi.71.12.9515-9523.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundkvist Å, Fatouros A, Niklasson B. Antigenic variation of European haemorrhagic fever with renal syndrome virus strains characterized using bank vole monoclonal antibodies. J Gen Virol. 1991;72:2097–2103. doi: 10.1099/0022-1317-72-9-2097. [DOI] [PubMed] [Google Scholar]
- 16.Lundkvist Å, Hörling J, Niklasson B. The humoral response to Puumala virus infection (nephropathia epidemica) investigated by viral protein specific immunoassays. Arch Virol. 1993;130:121–130. doi: 10.1007/BF01319001. [DOI] [PubMed] [Google Scholar]
- 17.Lundkvist Å, Hukic M, Hörling J, Gilljam M, Nichol S, Niklasson S. Puumala and Dobrava viruses cause hemorrhagic fever with renal syndrome in Bosnia-Herzegovina: evidence of highly cross-neutralizing antibody responses in early patient sera. J Med Virol. 1997;53:51–59. [PubMed] [Google Scholar]
- 18.Lundkvist Å, Kallio Kokko H, Sjölander K B, Lankinen H, Niklasson B, Vaheri A, Vapalahti O. Characterization of Puumala virus nucleocapsid protein: identification of B-cell epitopes and domains involved in protective immunity. Virology. 1996;216:397–406. doi: 10.1006/viro.1996.0075. [DOI] [PubMed] [Google Scholar]
- 19.Lundkvist Å, Niklasson B. Bank vole monoclonal antibodies against Puumala virus envelope glycoproteins: identification of epitopes involved in neutralization. Arch Virol. 1992;126:93–105. doi: 10.1007/BF01309687. [DOI] [PubMed] [Google Scholar]
- 20.Lundkvist Å, Vapalahti O, Plyusnin A, Brus Sjölander K, Niklasson B, Vaheri A. Tula hantavirus nucleocapsid protein: characterization of antigenic determinants defined by monoclonal antibodies raised against baculovirus-expressed protein. Virus Res. 1996;45:29–44. doi: 10.1016/0168-1702(96)01360-3. [DOI] [PubMed] [Google Scholar]
- 21.Papa A, Johnson A M, Stockton P C, Bowen M D, Spiropoulou C F, Alexiou-Daniel S, Ksiazek T G, Nichol S T, Antoniadis A. Retrospective serological and genetic study of the distribution of hantaviruses in Greece. J Med Virol. 1998;55:321–327. doi: 10.1002/(sici)1096-9071(199808)55:4<321::aid-jmv11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 22.Plyusnin A, Hörling J, Kanerva M, Mustonen J, Cheng Y, Partanen J, Vapalahti O, Kukkonen S K J, Niemimaa J, Henttonen H, Niklasson B, Lundkvist Å, Vaheri A. Puumala hantavirus genome in patients with nephropathia epidemica: correlation of PCR positivity with HLA haplotype and link to viral sequences in local rodents. J Clin Microbiol. 1997;35:1090–1096. doi: 10.1128/jcm.35.5.1090-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plyusnin A, Mustonen J, Asikainen K, Plyusnina A, Niemimaa J, Henttonen H, Vaheri A. Analysis of Puumala hantavirus genome in patients with nephropathia epidemica and rodent carriers from the sites of infection. J Med Virol. 1999;59:397–405. doi: 10.1002/(sici)1096-9071(199911)59:3<397::aid-jmv21>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 24.Plyusnin A, Vapalahti O, Vaheri A. Hantaviruses: genome structure, expression and evolution. J Gen Virol. 1996;77:2677–2687. doi: 10.1099/0022-1317-77-11-2677. [DOI] [PubMed] [Google Scholar]
- 25.Plyusnin, A., D. Kruger, and Å. Lundkvist. Hantavirus infections in Europe. Adv. Virus Res., in press. [DOI] [PubMed]
- 26.Rossi C A, Schmaljohn C S, Meegan J M, LeDuc J W. Diagnostic potential of a baculovirus-expressed nucleocapsid protein for hantaviruses. Arch. Virol. Suppl. 1. 1990. pp. 19–28. [Google Scholar]
- 27.Schmaljohn C S, Hasty S E, Dalrymple J M, LeDuc J W, Lee H W, von Bonsdorff C-H, Brummer-Korvenkontio M, Vaheri A, Tsai T F, Regnery H L, Goldgaber D, Lee P W. Antigenic and genetic properties of viruses linked to hemorrhagic fever with renal syndrome. Science. 1985;227:1041–1044. doi: 10.1126/science.2858126. [DOI] [PubMed] [Google Scholar]
- 28.Schmaljohn C S, Sugiyama K, Schmaljohn A L, Bishop D H. Baculovirus expression of the small genome segment of Hantaan virus and potential use of the expressed nucleocapsid protein as a diagnostic antigen. J Gen Virol. 1988;69:777–786. doi: 10.1099/0022-1317-69-4-777. [DOI] [PubMed] [Google Scholar]
- 29.Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morph. 1972;27:363–365. [PubMed] [Google Scholar]
- 30.Vapalahti O, Kallio-Kokko H, Närvänen A, Julkunen I, Lundkvist Å, Plyusnin A, Lehväslaiho H, Brummer-Korvenkontio M, Vaheri A, Lankinen H. Human B-cell epitopes of Puumala virus nucleocapsid protein, the major antigen in early serological response. J Med Virol. 1995;46:293–303. doi: 10.1002/jmv.1890460402. [DOI] [PubMed] [Google Scholar]
- 31.Vapalahti O, Lundkvist Å, Kallio-Kokko H, Paukku K, Julkunen I, Lankinen H, Vaheri A. Antigenic properties and diagnostic potential of Puumala virus nucleocapsid protein expressed in insect cells. J Clin Microbiol. 1996;34:119–125. doi: 10.1128/jcm.34.1.119-125.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshimatsu K, Arikawa J, Yoshida R, Lee H, Yoo Y C, Kariwa H, Hashimoto N, Kikinuma M, Nobunaga T, Azuma I. Production of recombinant hantavirus nucleocapsid protein expressed in silkworm larvae and its use as a diagnostic antigen in detecting antibodies in serum from infected rats. Lab Anim Sci. 1995;45:641–646. [PubMed] [Google Scholar]
- 33.Zöller L, Yang S, Gött P, Bautz E K F, Darai G. A novel μ-capture EIA based on recombinant proteins for sensitive and specific diagnosis of hemorrhagic fever with renal syndrome. J Clin Microbiol. 1993;31:1194–1199. doi: 10.1128/jcm.31.5.1194-1199.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]