Abstract
Rad30 is a member of the newly discovered UmuC/DinB/Rad30 family of DNA polymerases. The N-terminal regions of these proteins are highly homologous, and they contain five conserved motifs, I to V, while their C-terminal regions are quite divergent. We examined the contributions of the C-terminal and N-terminal regions of Rad30 to its activity and biological function. Although deletion of the last 54 amino acids has no effect on DNA polymerase or thymine-thymine (T-T) dimer bypass activity, this C-terminal deletion-containing protein is unable to perform its biological function in vivo. The presence of a bipartite nuclear targeting sequence within this region suggests that at least one function of this portion of Rad30 is nuclear targeting. To identify the active-site residues of Rad30 important for catalysis, we generated mutations of nine acidic residues that are invariant or highly conserved among Rad30 proteins from different eukaryotic species. Mutations of the Asp30 and Glu39 residues present in motif I and of the Asp155 residue present in motif III to alanine completely inactivated the DNA polymerase and T-T dimer bypass activities, and these mutations did not complement the UV sensitivity of the rad30Δ mutation. Mutation of Glu156 in motif III to alanine confers a large reduction in the efficiency of nucleotide incorporation, whereas the remaining five Rad30 mutant proteins retain wild-type levels of DNA polymerase and T-T dimer bypass activities. From these observations, we suggest a role for the Asp30, Glu39, and Asp155 residues in the binding of two metal ions required for the reaction of the incoming deoxynucleoside 5′-triphosphate with the 3′-hydroxyl in the primer terminus, while Glu156 may participate in nucleotide binding.
The RAD30 gene of Saccharomyces cerevisiae functions in error-free bypass of UV-induced DNA lesions (14, 23). RAD30 encodes a DNA polymerase, Polη, that has the ability to efficiently replicate through a cis-syn thymine-thymine (T-T) dimer, and it does so by inserting two A's opposite the two T's of the dimer (12). Inactivation of Polη in humans results in the cancer-prone syndrome, the variant form of xeroderma pigmentosum (11, 22). Cells derived from individuals with the variant form of xeroderma pigmentosum exhibit a deficiency in the replication of UV-damaged DNA (2, 21), and they are hypermutable with UV light (30, 33).
Polη differs from other eukaryotic DNA polymerases in its ability to replicate proficiently through lesions which distort the DNA helix. For example, both the yeast and human Polη enzymes replicate through a cis-syn T-T dimer with the same efficiency and fidelity as they replicate through two undamaged T's (17, 31), and Polη efficiently bypasses an 8-oxoguanine lesion by predominantly inserting a C opposite the lesion (9). Since both of these lesions distort the template strand, the ability of Polη to bypass these and other distorting DNA lesions (8) must derive from an active site that is indifferent to geometric distortions in DNA. In keeping with this idea, Polη is a low-fidelity enzyme, misinserting nucleotides opposite undamaged bases with a frequency of 10−2 to 10−3 (17, 32). The relative insensitivity of Polη to geometric distortions in DNA may derive from a flexible active site able to conform to distortions in Watson-Crick geometry (8, 17, 31, 32).
Polη is a member of the newly discovered Rad30/UmuC/DinB family of DNA polymerases that function in the replication of damaged DNA. In addition to the Escherichia coli UmuC and DinB1 proteins, this family includes eukaryotic Polη proteins (16), human DINB1-encoded Polθ (13), human RAD30B-encoded Polι (15, 29), and the eukaryotic REV1-encoded deoxycytidyl transferase (24). These proteins share extensive sequence homology within their N-terminal regions, and they all contain five conserved motifs (I to V) in this region (14, 16). By contrast, the C-terminal regions of these proteins are rather divergent. Proteins belonging to the Rad30 and DinB subfamilies, however, contain one or two conserved C2H2 or C2HC potential zinc binding motifs within the C-terminus (16).
Structural and mutagenesis studies of a variety of DNA polymerases have indicated that two crucial acidic residues in the polymerase active site bind a pair of divalent metal ions and promote catalysis (see Discussion for details). Although Polη and other members of this family share no sequence homology with other known DNA polymerases, they all possess a set of highly conserved acidic residues in their N termini. In particular, here we identify nine acidic residues that are invariant or conserved in the Rad30 subfamily of proteins and examine the effect of substitution of alanine for these residues on the activity and biological function of Polη. We also examine the contribution of the divergent C-terminal region of Rad30 to its activity and function.
MATERIALS AND METHODS
C-terminal truncations of yeast Rad30 protein.
Five C-terminal truncations of the Rad30 protein were constructed by introducing a stop codon at the desired position by either ligating a linker containing stop codons in all three reading frames, as in the case of the Rad30(1-340) mutant protein retaining the first 340 amino acids, or by using PCR to introduce the TAG stop codon, as for the Rad30(1-398), Rad30(1-452), Rad30(1-513), and Rad30(1-578) truncated proteins, respectively. The five truncated versions of the RAD30 gene were overexpressed in yeast as glutathione S-transferase (GST)-Rad30 fusion proteins under the control of the hybrid GAL PGK promoter in plasmid pHQ241, a 2μm vector with the yeast leu2-d gene as the selectable marker (Table 1). For complementation studies, the five truncated versions were cloned into the low-copy-number CEN/ARS LEU2 vector YCplac111 (6) (Table 1).
TABLE 1.
Plasmids used in this study
| Vector (description) and plasmid | rad30 mutation |
|---|---|
| pHQ241 (GAL PGK GST leu2-d) | |
| pR30.40 | Rad30(1-340) |
| pR30.54 | Rad30(1-398) |
| pR30.55 | Rad30(1-452) |
| pR30.43 | Rad30(1-513) |
| pR30.45 | Rad30(1-578) |
| YCplac111 (CEN/ARS LEU2) | |
| pR30.105 | Rad30(1-340) |
| pR30.56 | Rad30(1-398) |
| pR30.57 | Rad30(1-452) |
| pR30.51 | Rad30(1-513) |
| pR30.50 | Rad30(1-578) |
| pHQ241 (GAL PGK GST leu2-d) | |
| pR30.107 | D30A |
| pR30.115 | E39A |
| pR30.102 | E79A |
| pR30.131 | D155A |
| pR30.132 | E156A |
| pR30.116 | D160A |
| pR30.143 | D228A |
| pR30.146 | D235A |
| pR30.142 | D293A |
| YCplac111 (CEN/ARS LEU2) | |
| pR30.126 | D30A |
| pR30.127 | E39A |
| pR30.113 | E79A |
| pR30.138 | D155A |
| pR30.136 | E156A |
| pR30.120 | D160A |
| pR30.119 | D228A |
| pR30.147 | D235A |
| pR30.145 | D293A |
Point mutations of RAD30.
A series of site-directed mutations of conserved acidic amino acid residues were made in the wild-type RAD30 gene using the QuickChange site-directed mutagenesis kit (Stratagene). The D and E residues that were individually changed to A were D30, E39, D155, E156, D160, D228, D235, and D293. One mutant, E79A, was made using the MORPH-mut S DNA mutagenesis kit (5 Prime→3 Prime, Inc., Boulder, Colo.). DNA fragments containing each mutant rad30 gene were introduced into the GST expression vector pHQ241 and the low-copy-number CEN/ARS LEU2 vector YCplac111 (Table 1).
DNA substrates.
The following two oligodeoxynucleotide substrates were used to assay the biochemical activities of the Rad30 mutant proteins: a 75-nucleotide (nt) template (5′-AGCTA CCATG CCTGC ACGAA GAGTT CGTAT TATGC CTACA CTGGA GTACC GGAGC ATCGT CGTGA CTGGG AAAAC-3′) and a 44-nt primer (5′-GTTTT CCCAG TCACG ACGAT GCTCC GGTAC TCCAG TGTAG GCAT-3′). Another 75-nt template used had the same sequence, except that it had a cis-syn thymine-thymine dimer at the underlined position. The primer was 5′ 32P end labeled by polynucleotide kinase (Boehringer Mannheim), and the primer and template (200 μM) were annealed in 50 mM Tris HCl (pH 7.5)–100 mM NaCl by rapid heating to 95°C and slow cooling to room temperature over several hours.
DNA polymerase assays.
A 5 nM concentration of wild-type and mutant Rad30 proteins, purified from yeast strain BJ5464 as GST fusions as previously described (12), was incubated with the radiolabeled primer-template DNA substrate (10 nM) and all four deoxynucleoside triphosphates (dNTPs; 100 μM each) for 10 min at 25°C in 25 mM Tris HCl (pH 7.5)–5 mM MgCl2–100 μg of bovine serum albumin per ml–10% glycerol. Reactions were stopped by the addition of 10 volumes of formamide loading buffer, and the mixtures were boiled for 2 min and placed on ice. Samples were run on a 10% polyacrylamide sequencing gel containing 5.5 M urea.
UV survival.
Wild-type and mutant Rad30 proteins were tested for biological function in vivo by determining the ability to complement the UV sensitivity of rad5Δ rad30Δ mutant yeast strain YR5-52. Strains harboring the rad30 mutations in low-copy-number CEN/ARS LEU2 vector YCplac111 were grown to late logarithmic or early stationary phase in liquid synthetic complete medium lacking leucine to maintain selection for the plasmid. Cells were washed and diluted in water, and appropriate dilutions were plated on synthetic complete medium lacking leucine for determination of survival following exposure to UV irradiation. After UV irradiation, plates were incubated in the dark at 30°C for 4 days.
Steady-state kinetic analyses.
For steady-state kinetic studies of single-nucleotide incorporation, four 52-nt templates were used (5′-TTCGT ATNAT GCCTA CACTG GAGTA CCGGA GCATC GTCGT GACTG GGAAA AC, where N is G, A, T, or C) and one 44-nt primer was used (5′-GTTTT CCCAG TCACG ACGAT GCTCC GGTAC TCCAG TGTAG GCAT). The wild-type Rad30 protein (1 nM) or the mutant E156A protein (5 nM) was incubated with a 50 nM concentration of the 32P-labeled primer-template DNA substrates in the presence of various concentrations of a single nucleotide for 2, 5, or 15 min in the same buffer used in the DNA polymerase assays. Substrates and products were resolved on a 10% polyacrylamide sequencing gel, and the gel band intensities were determined using a PhosphorImager (Molecular Dynamics). The rate of nucleotide incorporation (amount incorporated per unit of time) was graphed as a function of nucleotide concentration, and the kcat (Vmax per enzyme concentration) and Km parameters were obtained from the best fit to the Michaelis-Menten equation as previously described (3, 7).
RESULTS
Requirement of the Rad30 C terminus for biological function.
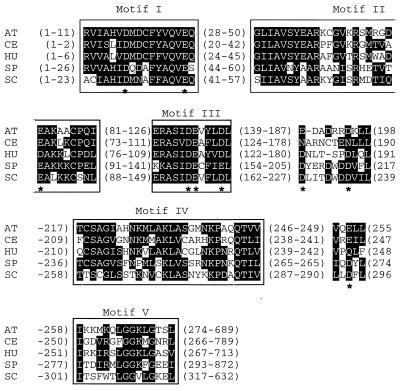
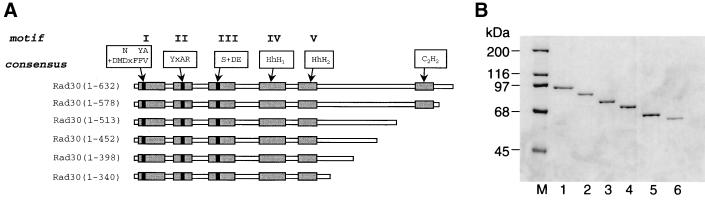
Alignment of Rad30 proteins from different species shows that the N terminus of these proteins is highly conserved and contains five distinctive motifs, I to V (16) (see Fig. 5). The C terminus of the different Rad30 proteins, however is not that well conserved. To examine the function of the C-terminal region of the yeast Rad30 protein, we generated a series of truncated versions of this protein (Fig. 1A). The Rad30(1-578) protein lacks the last 54 amino acids but still retains the C2H2 potential zinc binding motif, which is conserved among the yeast, human, and other Rad30 proteins (16). The Rad30(1-513) protein eliminates the C2H2 motif, and the truncated proteins Rad30(1-452), Rad30(1-398), and Rad30(1-340) removed further amino acid residues from the C terminus. The five C-terminally truncated proteins were purified to near homogeneity using the same purification protocol as was used for wild-type Rad30 protein. The truncated proteins display the expected reduction in molecular size on sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis compared to the wild-type protein (Fig. 1B).
FIG. 5.
Alignment of conserved N termini of Rad30 subfamily DNA polymerases. Sequence alignment of conserved portions of N termini, including motifs I to V of Rad30 proteins from Arabidopsis thaliana (AT), Caenorbabditis elegans (CE), human (HU), Schizosaccharomyces pombe (SP), and S. cerevisiae (SC). Numbers in parenthesis indicate the regions for which amino acid sequences are not shown. Asterisks indicate the highly conserved acidic residues which were individually mutated to alanine in the S. cerevisiae Rad30 protein.
FIG. 1.
C-terminal truncations of the Rad30 protein. (A) Schematic representation of the wild-type and C-terminally truncated Rad30 mutant proteins. Shaded boxes indicate the positions of the conserved motifs (I to V) present in the N terminus and the conserved C2H2 motif located in the C terminus. Motifs I to III are shown with their conserved consensus sequences outlined, where a plus sign indicates a hydrophobic residue (I, L, or V) and the letter x indicates any amino acid. HhH1 and HhH2 in motifs IV and V, respectively, represent the helix-hairpin-helix domains. (B) Analysis of the C-terminally truncated Rad30 proteins by SDS–10% polyacrylamide gel electrophoresis. Each lane has 250 ng of GST-Rad30 protein. Lanes: 1, wild-type Rad30 protein; 2, Rad30(1-578) protein; 3, Rad30(1-513) protein; 4, Rad30(1-452) protein; 5, Rad30(1-398) protein; 6, Rad30(1-340) protein; M, molecular size markers.
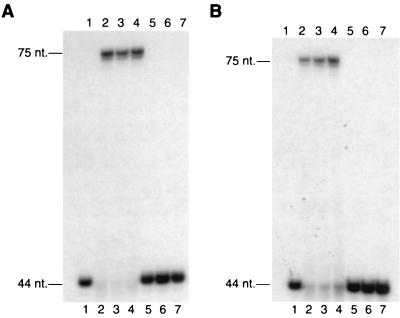
The truncated Rad30 mutant proteins were then examined for DNA polymerase activity and for the ability to replicate through a cis-syn T-T dimer. Proteins (5 nM) were incubated at 25°C for 10 min with either the nondamaged or the damaged DNA substrate (10 nM) and all four dNTPs (100 μM each). Quenched reaction mixtures were subjected to gel electrophoresis and autoradiography to visualize the unextended 44-nt primers and extended full-length 75-nt primers (Fig. 2). The Rad30(1-578) and Rad30(1-513) truncated proteins possessed both DNA polymerase and T-T dimer bypass activities equivalent to those of the wild-type Rad30 protein. By contrast, the Rad30(1-452), Rad30(1-398), and Rad30(1-340) proteins lacked both the DNA polymerase and T-T dimer bypass activities (Fig. 2).
FIG. 2.
DNA polymerase and cis-syn T-T dimer bypass activities of C-terminally truncated Rad30 proteins. (A) DNA polymerase activities of the wild-type and truncated Rad30 mutant proteins were assayed by incubating the proteins (5 nM) for 10 min at 25°C with all four dNTPs (100 μM each) and the nondamaged DNA substrate (10 nM) as described in Materials and Methods, and reactions were stopped and analyzed on a 10% polyacrylamide sequencing gel. Lanes: 1, no protein: 2, wild-type Rad30 protein; 3, Rad30(1-578) protein; 4, Rad30(1-513) protein; 5, Rad30(1-452) protein; 6, Rad30(1-398) protein; 7, Rad30(1-340) protein. (B) T-T dimer bypass activities were assayed as described for the DNA polymerase activities in panel A, except that the DNA substrate used contained a cis-syn T-T dimer in the template strand at nucleotide positions 45 and 46 from the 3′ end.
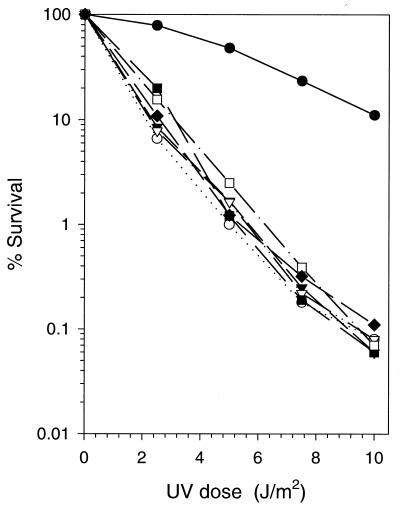
To determine the effect of C-terminal truncations on the in vivo function of Rad30, we cloned the various mutant rad30 genes into a low-copy-number yeast CEN/ARS vector and introduced the plasmid into the rad5Δ rad30Δ double-mutant strain. The RAD5 and RAD30 genes promote replication of UV-damaged DNA via alternate error-free pathways, and as a consequence, the rad5Δ rad30Δ double mutant exhibits a synergistic increase in UV sensitivity compared to the single mutants (14, 23). Also, the frequency of UV-induced mutations is much higher in the rad5Δ rad30Δ double mutant than in the single mutants (14, 23).
As shown in Fig. 3, none of the C-terminally truncated proteins enhanced the UV resistance of the rad5Δ rad30Δ mutant strain whereas expression of the wild-type Rad30 protein in this double mutant led to increased UV resistance, equivalent to that of the rad5Δ strain. Thus, all of these C-terminal truncations inactivate the biological function of Rad30. While this result was expected for the Rad30(1-452), Rad30(1-398), and Rad30(1-340) proteins since they lack the DNA polymerase activity, the inability of the Rad30(1-578) and Rad30(1-513) proteins to complement the UV sensitivity of the rad30Δ mutation was rather surprising, since these C-terminally truncated proteins have proficient DNA synthesis and T-T dimer bypass activities. This observation suggested that the C-terminal portion of the protein, absent in Rad30(1-578), affects the ability of Rad30 to function in vivo.
FIG. 3.
Complementation analysis with C-terminally truncated Rad30 proteins. C-terminal truncations of Rad30 proteins were expressed on a yeast low-copy-number (CEN/ARS) vector in the rad5Δ rad30Δ mutant strain, and their UV sensitivity was compared to that of the rad5Δ rad30Δ mutant strain harboring the wild-type RAD30 gene on a CEN/ARS plasmid. Each data point is the average of three separate experiments. Symbols for the rad5Δ rad30Δ mutant strain carrying the indicated gene on the plasmid: ●, wild-type RAD30; ○, vector only; ▾, Rad30(1-578); ▿, Rad30(1-513); ■, Rad30(1-452); □, Rad30(1-398); ⧫, Rad30(1-340).
A nuclear targeting motif in the C terminus of Rad30.
The requirement of the C-terminal 54 residues of Rad30 for its in vivo function raised the possibility that this region of the protein was important for targeting of the Rad30 protein to the nucleus. Consistent with this idea, we found that residues 601 to 617 of the yeast Rad30 protein contain a bipartite nuclear targeting sequence that has been identified previously in a large number of nuclear proteins (4). In yeast Rad30, this sequence has 3 basic amino acids separated by 10 amino acids and then followed by 2 basic residues (Fig. 4). A similar sequence is also present in the C terminus of the human Rad30 protein, where it contains 2 basic residues separated by 10 amino acids and followed by 5 residues of which 3 are basic (Fig. 4). Such a bipartite motif has been shown to be indispensable for the targeting of a large variety of proteins to the nucleus, and mutagenesis studies have shown that the two clusters of basic amino acids in this sequence are critical for nuclear targeting (4).
FIG. 4.
The nuclear targeting sequences of yeast RAD30- and human RAD30A-encoded DNA Polη are aligned with similar bipartite motifs present in N1, nucleolin, and proteins involved in DNA repair, replication, and transcription. Basic amino acids comprising the two clusters of residues present at the termini of this sequence are in boldface. Each number refers to the position of the first amino acid in the sequence of the protein. H, human; M, mouse; Sc, S. cerevisiae; X, xenopus. This figure is adapted from reference 4.
Acidic residues critical for Rad30 DNA polymerase activity and function.
DNA polymerases require divalent metal ions for their catalytic activity, and extensive structural and mutagenesis studies with a variety of polymerases have indicated a common active-site structure comprised of acidic residues which serve to coordinate a pair of divalent metal ions. Since the Rad30 family of proteins shares no sequence homology with the other prokaryotic or eukaryotic DNA polymerases, in order to identify the acidic residues critical for catalysis, we mutagenized the various invariant and conserved acidic residues present in Rad30 and tested the effects of these mutations on Rad30 activity and function.
As shown in Fig. 5, yeast Rad30 and its counterparts from other species contain nine invariant or highly conserved acidic residues. In yeast Rad30, residues D30 and E39 are in motif I, E79 is in motif II, and D155, E156, and D160 are in motif III while D228 and D235 are located between motifs III and IV and D293 is located between motifs IV and V. Each of these acidic residues in Rad30 was changed to alanine, and the mutant proteins were expressed as GST fusions and purified to near homogeneity from yeast cells using the same procedure as was employed for the wild-type strain. All of the mutant proteins exhibited the same electrophoretic mobility in SDS-polyacrylamide gels as the wild-type protein, and they were expressed to the same degree as the wild-type protein (data not shown).
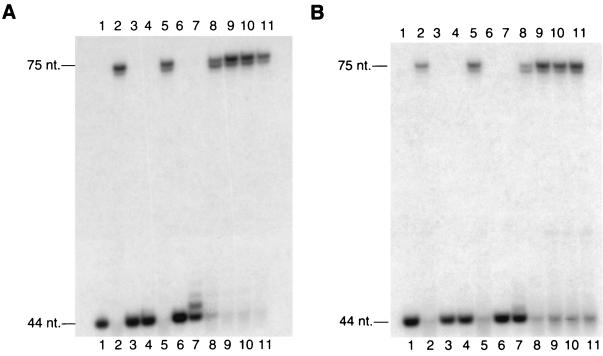
The mutant Rad30 proteins were then assayed for the DNA polymerase and DNA-damage bypass activities. The E79A, D160A, D228A, D235A, and D293A mutant proteins all retained DNA polymerase and T-T dimer bypass activities nearly equivalent to those of the wild-type Rad30 protein (Fig. 6). By contrast, the D30A, E39A, and D155A mutant proteins lacked both of these activities and the E156A mutant protein showed highly inefficient DNA polymerase and dimer bypass activities, extending primers by only a few nucleotides (Fig. 6).
FIG. 6.
DNA polymerase and T-T dimer bypass activities of site-directed Rad30 mutant proteins harboring mutations in the conserved acidic residues. (A) DNA polymerase activities of the wild-type and site-directed Rad30 mutant proteins were assayed by incubating the proteins (5 nM) for 10 min at 25°C with all four dNTPs (100 μM) and the nondamaged DNA substrate (10 nM) as described in Materials and Methods, and reactions were analyzed on a 10% polyacrylamide sequencing gel. Lanes: 1, no protein; 2, wild-type Rad30 protein; 3, D30A mutant protein; 4, E39A mutant protein; 5, E79A mutant protein; 6, D155A mutant protein; 7, E156A mutant protein; 8, D160A mutant protein; 9, D228A mutant protein; 10, D235A mutant protein; 11, D293A mutant protein. (B) T-T dimer bypass activities were assayed as described for the DNA polymerase activities, except that the DNA substrate used contained a cis-syn T-T dimer in the template strand at nucleotide positions 45 and 46 from the 3′ end.
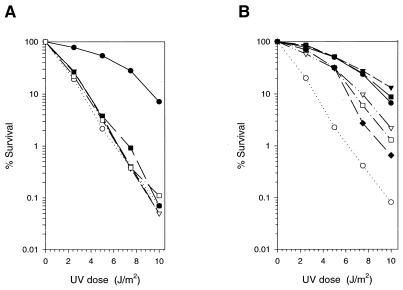
For the in vivo functional analysis of these mutant proteins, the various rad30 mutant genes were cloned into a low-copy-number yeast CEN/ARS vector and the plasmid was introduced into the rad5Δ rad30Δ mutant strain. As shown in Fig. 7A, none of the D30A, E39A, D155A, and E156A mutations, which are all highly defective in the DNA polymerase and dimer bypass activities, could enhance the UV resistance of the rad5Δ rad30Δ strain to the rad5Δ level. Of the mutant proteins which retained full enzymatic activity, the E79A and D228A mutations increased the UV resistance of the rad5Δ rad30Δ strain to the rad5Δ level whereas the D160A, D235A, and D293A mutations complemented the UV sensitivity of the rad5Δ rad30Δ strain to a lesser degree (Fig. 7B).
FIG. 7.
Complementation of UV sensitivity by Rad30 mutant proteins. The various rad30 mutant alleles were cloned into the yeast low-copy-number (CEN/ARS) vector YCplac111. The plasmid was introduced into a rad5Δ rad30Δ mutant strain, and the UV sensitivity of the rad5Δ rad30Δ strain harboring the different rad30 mutations was compared to that of the rad5Δ rad30Δ strain carrying wild-type RAD30 in the same vector, YCplac111. (A) Noncomplementing rad30 mutations. Symbols for the rad5Δ rad30Δ strain carrying the indicated gene on the plasmid: ●, wild-type RAD30; ○, vector only; ▾, D30A; ▿, E39A; ■, D155A; □, E156A. (B) rad30 mutations that complement to various degrees. Symbols for the rad5Δ rad30Δ strain carrying the indicated gene on the plasmid: ●, wild-type RAD30; ○, vector only; ▾, E79A; ▿, D160A; ■, D228A; □, D235A; ⧫, D293A. Each data point in both panels A and B is the average of three separate experiments.
Steady-state kinetic analysis of nucleotide incorporation by the E156A mutant Rad30 protein.
We further characterized the DNA polymerase activity of the E156A mutant Rad30 protein by comparing the steady-state kinetics of single-nucleotide incorporation of the wild-type and E156A mutant Rad30 proteins opposite all four template residues. The rate of single-nucleotide incorporation was measured at various nucleotide concentrations, and the kcat and Km steady-state parameters were determined as described in Materials and Methods. As shown in Table 2, the efficiency (kcat/Km) of correct nucleotide incorporation by the E156A protein was 5,000- to 100,000-fold lower than that of the wild-type protein. This substantial decrease in efficiency resulted primarily from a large increase in the Km for the incoming nucleotide of the E156A mutant protein, which was 200- to 7,000-fold greater than that of the wild-type protein, whereas there was only a modest (10- to 30-fold) decrease in the kcat of the E156A protein relative to the wild-type protein. We also examined the incorporation of incorrect nucleotides by the E156A mutant protein, but because of its low insertion efficiency, we did not detect any misincorporation under these conditions. Thus, the fidelity of nucleotide incorporation by the E156A protein was not significantly disrupted.
TABLE 2.
Steady-state kinetic parameters for nucleotide incorporation by the wild-type Rad30 protein and the E156A mutant Rad30 protein
| Template and protein | dNTP | kcat (min−1) | Km (μM) | kcat/Km ratio (μM−1 min−1) | Relative efficiency |
|---|---|---|---|---|---|
| G | |||||
| Wild type | dCTP | 1 ± 0.09 | 0.06 ± 0.02 | 50 | 1 |
| E156A | dCTP | 0.1 ± 0.006 | 10 ± 2 | 0.01 | 2 × 10−4 |
| A | |||||
| Wild type | dTTP | 1 ± 0.07 | 0.07 ± 0.01 | 20 | 1 |
| E156A | dTTP | 0.03 ± 0.001 | 20 ± 3 | 2 × 10−3 | 1 × 10−4 |
| T | |||||
| Wild type | dATP | 1 ± 0.06 | 0.03 ± 0.01 | 30 | 1 |
| E156A | dATP | 0.07 ± 0.008 | 200 ± 40 | 4 × 10−4 | 1 × 10−5 |
| C | |||||
| Wild type | dGTP | 1 ± 0.07 | 0.02 ± 0.005 | 70 | 1 |
| E156A | dGTP | 0.04 ± 0.001 | 20 ± 2 | 2 × 10−3 | 3 × 10−5 |
In summary, whereas the D30, E39, and D155 residues are essential for Rad30 activity and function, the E156 residue is not essential for activity but it greatly affects the efficiency of nucleotide incorporation. The other conserved acidic residues, E79, D160, D228, D235, and D293, however, are not important for Rad30 polymerase activity. The somewhat lessened ability of some of the mutations in the latter group to complement the UV sensitivity of the rad30Δ mutation may result from structural perturbations which affect the ability of Rad30 to physically interact with other proteins important for its function in vivo.
DISCUSSION
The purpose of this study was to examine the requirement of the C terminus of Rad30 for its function and to identify the acidic residues crucial for Rad30 polymerase activity. Even though the elimination of the last 54 amino acids of Rad30, as in the Rad30(1-578) protein, has no effect on DNA polymerase or T-T dimer bypass activity, this truncation inactivates the biological function of the protein, as judged by the lack of complementation of the UV sensitivity of the rad30Δ mutation. The presence of a bipartite nuclear targeting motif encompassing amino acids 601 to 617 of the yeast Rad30 protein supports an essential role for this C-terminal region in the targeting of this protein to the nucleus.
All proteins belonging to the yeast Rad30 family contain a highly conserved C2H2 sequence motif near the C terminus (16). Deletion of this motif, as in the Rad30(1-513) protein, however, has no discernible effect on DNA synthesis or T-T dimer bypass activity, suggesting that this sequence is dispensable for these Rad30 activities. Further deletion of the C terminus, however, as in the Rad30(1-452) protein, inactivates both the DNA polymerase and dimer bypass activities. Although the first 320 amino acids of Rad30 contain all of conserved motifs I to V and there is no apparent amino acid conservation between residues 452 and 513, the requirement of this C-terminal region for Rad30 activity may reflect a role for this portion in adopting the proper three-dimensional structure of the protein.
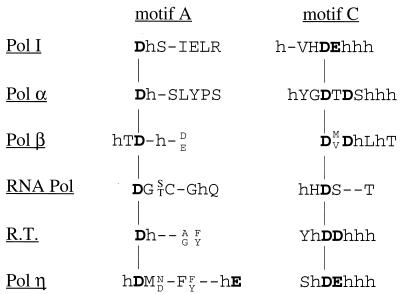
Even though there is little sequence similarity between distantly related DNA and RNA polymerases and reverse transcriptases, they all share two conserved motifs, A and C (18). All polymerases contain an invariant aspartate in motif A, as well as in motif C, and another highly conserved aspartate or glutamate is present in motif C of DNA polymerases but not in DNA-dependent RNA polymerases (Fig. 8). Although in various DNA polymerases, these three acidic residues are in optimal geometric position to promote catalysis by the two-metal-ion mechanism (1, 10, 19, 25), the crystal structure of T7 DNA polymerase complexed with primer-template and a dNTP indicates that the conserved aspartate of motif A and only one of the conserved aspartates present in motif C function in the coordination of divalent metal ions (5). These two aspartates in T7 DNA polymerase correspond to the Asp705 residue of motif A and the Asp882 residue of motif C in the Klenow fragment of E. coli polymerase (Pol I) (Fig. 8 legend). Accordingly, mutagenesis studies with the Klenow fragment show that Asp705 and Asp882 are much more critical for catalysis than Glu883, as mutations of the Asp705 and Asp882 residues to alanine in Klenow result in about 2,500- and 400-fold reductions in kcat, respectively, whereas a mutation of Glu883 to alanine causes only a 25- to 30- fold reduction in kcat (26, 27). The binding of the two metal ions by the polymerase coordinates the interaction of the incoming nucleoside triphosphate with the 3′-hydroxyl on the primer terminus, leading to phosphodiester bond formation (20, 28). In this case, one enzyme-bound metal ion interacts with the 3′-hydroxyl of the primer strand, activating the 3′-hydroxyl oxygen for attack on the α-phosphate of the dNTP while the second metal ion binds to the β- and γ-phosphates of the incoming dNTP and facilitates loss of the pyrophosphate (28). Both metal ions also aid in stabilizing the pentacovalent transition state that occurs during this reaction (28).
FIG. 8.
Alignment of sequences in motifs A and C conserved among polymerases. Motif A of other polymerases resembles motif I of Polη, except that Polη has an additional conserved acidic E residue at position 39 in the yeast protein. Motif C of other DNA polymerases resembles motif III of Polη in that they all contain two conserved acidic residues flanked by hydrophobic residues (h). In Pol I, the D residue in motif A is at position 705 and the D and E residues of motif C occur at positions 882 and 883, respectively. This figure is adapted from Fig. 1 of reference 18. R.T., reverse transcriptase.
Although the Rad30/UmuC/DinB family of proteins shows no sequence similarity to any other DNA polymerases, motifs I and III of these proteins resemble motifs A and C of other polymerases (Fig. 8). The D30 present in motif I of Rad30 resembles the invariant aspartate present in motif A of other polymerases, and the D155 and E156 residues present in motif III of Rad30 resemble the D and D/E residues of motif C in other polymerases. This similarity raised the possibility that of these three acidic residues present in motifs I and III of Rad30, two may be important for catalysis.
We found that the Rad30 protein with the D30A or E39A mutation in motif I had no DNA polymerase or dimer bypass activity and that it also failed to complement the rad30Δ mutation. This suggests that one or both of these residues are involved in the binding of a divalent metal ion. Previously, we simultaneously changed the D155 and E156 residues of Rad30 to alanine, and the resulting Rad30 Ala155-Ala156 mutant protein lacked catalytic activity and was unable to complement the rad30Δ mutation (14). However, it was not possible to determine which amino acid substitution, D155A or E156A, had a more pronounced effect since neither amino acid had been replaced individually. Here we show that the D155A protein completely lacks the enzymatic activity while the E156A protein retains some enzymatic activity. This suggests that D155 is the residue involved in coordination of the metal ion, while E156 may not be directly involved in catalysis.
The E156A mutation causes a substantial increase in the Km for the incoming nucleotide (200- to 7,000-fold) and a modest decrease in the kcat (10- to 30-fold) relative to the wild-type Rad30 protein. The large increase in Km suggests that the E156A mutant protein binds poorly to the incoming dNTP. Thus, Glu156 may directly interact with the incoming nucleotide or the replacement of Glu156 with Ala may alter the active-site conformation such that interactions with the incoming nucleotide are weakened. The modest kcat effect is difficult to interpret, because the kcat likely corresponds to a step following nucleotide incorporation and does not reflect the intrinsic rate of phosphodiester bond formation. This large effect of the E156A mutation on the Km for dNTP differs from the effects reported for the analogous E. coli DNA polymerase I mutation, E883A, as in the polymerase I mutant protein, the Km for the incoming nucleotide increases less than 2-fold, while the kcat decreases 30-fold relative to that of the wild-type protein (26). Thus, whereas substitution of Ala for Glu883 has no effect on nucleotide binding by Pol I, substitution of Ala for Glu156 has a substantial effect on nucleotide binding by Polη.
We have previously suggested that Rad30 has a flexible active site that is more tolerant of DNA distortions and allows it to synthesize DNA opposite a cis-syn T-T dimer and opposite other lesions which distort the DNA helix. It remains unclear how this flexibility in the active site arises. It is possible that the crucial acidic residues involved in metal ion binding are arranged in the active site in Rad30 in a different manner than in DNA polymerases unable to bypass DNA lesions. Although Polη resembles other polymerases in its requirement for the D30 and D155 residues, analogous to the two catalytically essential acidic residues of motifs A and C of other polymerases, it differs from the other polymerases in the additional requirement for E39 in motif I for polymerase activity. Since replacement of any of the acidic residues D30, E39, and D155 with alanine completely inactivates the DNA polymerase activity and the biological function of Rad30, these three acidic residues may all be intimately involved in the coordination of the two metal ions necessary for phosphodiester bond formation. Further, in contrast to the E883 residue of Pol I, the analogous E156 residue of Polη may influence dNTP binding. Thus, the active site of Polη may differ from that of other DNA polymerases in the manner in which acidic residues are used for the binding of metal ions and dNTP.
ACKNOWLEDGMENT
This work was supported by NIH grant GM19261.
REFERENCES
- 1.Beese L S, Derbyshire V, Steitz T A. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 2.Cordeiro-Stone M, Zaritskaya L S, Price L K, Kaufmann W K. Replication fork bypass of a pyrimidine dimer blocking leading strand DNA synthesis. J Biol Chem. 1997;272:13945–13954. doi: 10.1074/jbc.272.21.13945. [DOI] [PubMed] [Google Scholar]
- 3.Creighton S, Bloom L B, Goodman M F. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 4.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 5.Doublie S, Tabor S, Long A M, Richardson C C, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 6.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 7.Goodman M F, Creighton S, Bloom L B, Petruska J. Biochemical basis of DNA replication fidelity. Crit Rev Biochem Mol Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 8.Haracska L, Prakash S, Prakash L. Replication past O6-methylguanine by yeast and human DNA polymerase η. Mol Cell Biol. 2000;20:8001–8007. doi: 10.1128/mcb.20.21.8001-8007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haracska L, Yu S-L, Johnson R E, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 10.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, Hizi A, Hughes S H, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson R E, Kondratick C M, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 12.Johnson R E, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 13.Johnson R E, Prakash S, Prakash L. The human DINB1 gene encodes the DNA polymerase Polθ. Proc Natl Acad Sci USA. 2000;97:3838–3843. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson R E, Prakash S, Prakash L. Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. J Biol Chem. 1999;274:15975–15977. doi: 10.1074/jbc.274.23.15975. [DOI] [PubMed] [Google Scholar]
- 15.Johnson R E, Washington M T, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 16.Johnson R E, Washington M T, Prakash S, Prakash L. Bridging the gap: a family of novel DNA polymerases that replicate faulty DNA. Proc Natl Acad Sci USA. 1999;96:12224–12226. doi: 10.1073/pnas.96.22.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson R E, Washington M T, Prakash S, Prakash L. Fidelity of human DNA polymerase η. J Biol Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 18.Joyce C M, Steitz T A. Function and structure relationships in DNA polymerases. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 19.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel T A, Wilson S H. DNA polymerases on the move. Nat Struct Biol. 1998;5:95–99. doi: 10.1038/nsb0298-95. [DOI] [PubMed] [Google Scholar]
- 21.Lehmann A R, Kirk-Bell S, Arlett C F, Paterson M C, Lohman P H M, de Weerd-Kastelein E A, Bootsma D. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc Natl Acad Sci USA. 1975;72:219–223. doi: 10.1073/pnas.72.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 23.McDonald J P, Levine A S, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson J R, Lawrence C W, Hinkle D C. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 25.Pelletier H, Sawaya M R, Kumar A, Wilson S H, Kraut J. Structure of ternary complexes of rat DNA polymerase β, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 26.Polesky A H, Dahlberg M E, Benkovic S J, Grindley N D F, Joyce C M. Side chains involved in catalysis of the polymerase reaction of DNA polymerase I from Escherichia coli. J Biol Chem. 1992;267:8417–8428. [PubMed] [Google Scholar]
- 27.Polesky A H, Steitz T A, Grindley N D F, Joyce C M. Identification of residues critical for the polymerase activity of the Klenow fragment of DNA polymerase I from Escherichia coli. J Biol Chem. 1990;265:14579–14591. [PubMed] [Google Scholar]
- 28.Steitz T A. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 29.Tissier A, McDonald J P, Frank E G, Woodgate R. polι, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y-C, Maher V M, Mitchell D L, McCormick J J. Evidence from mutation spectra that the UV hypermutability of xeroderma pigmentosum variant cells reflects abnormal, error-prone replication on a template containing photoproducts. Mol Cell Biol. 1993;13:4276–4283. doi: 10.1128/mcb.13.7.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Washington M T, Johnson R E, Prakash S, Prakash L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc Natl Acad Sci USA. 2000;97:3094–3099. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Washington M T, Johnson R E, Prakash S, Prakash L. Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η. J Biol Chem. 1999;274:36835–36838. doi: 10.1074/jbc.274.52.36835. [DOI] [PubMed] [Google Scholar]
- 33.Waters H L, Seetharam S, Seidman M M, Kraemer K H. Ultraviolet hypermutability of a shuttle vector propagated in xeroderma pigmentosum variant cells. J Investig Dermatol. 1993;101:744–748. doi: 10.1111/1523-1747.ep12371686. [DOI] [PubMed] [Google Scholar]