Summary
Because transethnic analysis may facilitate prioritization of causal genetic variants, we performed a genome-wide association study (GWAS) of psoriasis in South Asians (SAS), consisting of 2,590 cases and 1,720 controls. Comparison with our existing European-origin (EUR) GWAS showed that effect sizes of known psoriasis signals were highly correlated in SAS and EUR (Spearman ρ = 0.78; p < 2 × 10−14). Transethnic meta-analysis identified two non-major histocompatibility complex (non-MHC) psoriasis loci (1p36.22 and 1q24.2) not previously identified in EUR, which may have regulatory roles. For these two loci, the transethnic GWAS provided higher genetic resolution and reduced the number of potential causal variants compared to using the EUR sample alone. We then explored multiple strategies to develop reference panels for accurately imputing MHC genotypes in both SAS and EUR populations and conducted a fine mapping of MHC psoriasis associations in SAS and the largest such effort for EUR. HLA-C∗06 was the top-ranking MHC locus in both populations but was even more prominent in SAS based on odds ratio, disease liability, model fit, and predictive power. Transethnic modeling also substantially boosted the probability that the HLA-C∗06 protein variant is causal. Secondary MHC signals included coding variants of HLA-C and HLA-B, but also potential regulatory variants of these two genes as well as HLA-A and several HLA class II genes, with effects on both chromatin accessibility and gene expression. This study highlights the shared genetic basis of psoriasis in SAS and EUR populations and the value of transethnic meta-analysis for discovery and fine mapping of susceptibility loci.
Keywords: genome-wide association study, major histocompatibility complex, human leukocyte antigens, imputation, psoriasis, skin diseases
We combined a GWAS of psoriasis in South Asians with past European studies to identify two novel psoriasis loci and refine Bayesian credible intervals of known loci. Fine-scale mapping of the MHC region in South Asian, European, and transethnic datasets revealed multiple risk loci, including HLA protein-coding and regulatory variants.
Introduction
Psoriasis (MIM: 177900) is a common, chronic, immune-mediated disorder of the skin and joints characterized by cutaneous inflammation, epidermal hyperplasia, and increased risk of arthritis as well as cardiovascular morbidity.1 Substantial evidence indicates that psoriasis is driven by abnormal interactions between the innate and adaptive immune cells, including keratinocytes, neutrophils, macrophages, dendritic cells, and T cells.2,3
Psoriasis affects 0.1% to 6.5% of individuals, depending on ethnicity and geographical location, with higher prevalence recorded at increasing latitudes.4 In European-origin populations, the prevalence of psoriasis is estimated to vary from 2% to 3%,5,6 making psoriasis a favorable target for genome-wide association studies (GWASs). As a result, GWASs to date have identified 87 independent genetic signals for psoriasis at genome-wide significance, with 11 of the 86 shared by European and Chinese populations, 56 established for Europeans only, and 20 for Chinese only.1,7 Secondary psoriasis-association signals (independent of the primary variant) have been reported for at least 11 of the 86 susceptibility regions.1,7,8
With the exception of a relatively small study in Japan,9 most GWASs of psoriasis have been carried out in European-origin and Chinese-origin individuals. In all populations, the strongest psoriasis association signals map to the major histocompatibility complex (MHC), comprising approximately 40% of the detectable heritability of psoriasis.10 Correspondingly, genome-wide significant MHC associations have been reported for Japanese,11 Korean,12 Thai,13 Pakistani,14,15 and Indian16, 17, 18 populations.
Transethnic GWASs provide both advantages and challenges for the study of genetically complex traits, with the advantage of increased sample size being counterbalanced by the potential challenge posed by differences in underlying genetic architecture across populations.19, 20, 21 With these factors in mind, we undertook a GWAS of psoriasis in South Asian populations from India and Pakistan, consisting of 2,590 cases and 1,720 controls. We found that effect sizes of the known psoriasis susceptibility regions were highly correlated in the South Asian (SAS) and European-origin (EUR) datasets, leading us to conduct unconditional and conditional transethnic meta-analyses of psoriasis genetic associations in these two populations. We then investigated whether transethnic analysis could refine Bayesian credible sets for psoriasis loci. Because the MHC carries such a large fraction of the genetic burden for psoriasis,10 much of which appears to map to variation in genes encoding human leukocyte antigens (HLAs) themselves,22 we also developed an improved algorithm for HLA imputation based on SNP2HLA23 and assessed the performance of multiple reference panels derived from EUR and SAS samples. We then built and compared SAS, EUR, and transethnic MHC association models and used multiple bioinformatic tools to explore the potential biological consequences of the many coding and non-coding MHC variants we identified. Finally, we assessed linkage disequilibrium (LD) structure of SAS and EUR to see if it benefited our transethnic analysis.
Subjects and methods
Human subjects
Psoriasis cases were diagnosed by a dermatologist according to established criteria.24 We included individuals ascertained for psoriatic arthritis (PsA [MIM: 6075074]) by a rheumatologist if they manifested joint, skin, scalp, and/or nail lesions consistent with psoriasis. Control individuals were 18 years of age or older, unrelated to affected individuals, and unaffected with psoriasis or PsA. All participating individuals provided written informed consent and were recruited according to the protocols approved by the institutional review boards of each participating institution.
GWAS genotyping
For the SAS GWAS cohort, three batches of genotyping were performed. The first, consisting of 952 cases and 855 unaffected controls after quality control, was typed on an Illumina OmniExpressExome (8v1-1_B) platform. The second and third batches, consisting of a combined total of 1,638 cases and 865 unaffected controls after quality control, were typed on two iterations of the Illumina HumanCoreExome platform (12v1-1_B and 24v1-0_A, respectively). In all, 2,590 cases and 1,720 controls were included after quality control. For quality control, we removed common variants (minor allele frequency [MAF] ≥ 0.05) with a call rate < 95%, rarer variants (MAF < 0.05) with a call rate < 99%, and variants with a Hardy-Weinberg p value in controls < 1 × 10−6. Samples were removed if they had substantial non-South Asian admixture (based on the principal component analysis [PCA] shown in Figure S1), were duplicates or first- or second-degree relatives of other samples (Plink ≥ 0.20), had a genotype call rate < 98%, or had an outlier heterozygosity value (>1.5 × interquartile range above third quartile or below first quartile).
Genotyping, quality control, phasing, and imputation of six EUR-origin GWAS cohorts have previously been described,7 including CASP,25 Kiel,26 Genizon,26 PsA GWAS,27 WTCCC2,28 and Exomechip10 (which contains GWAS content). We also included two datasets based on the Immunochip: PAGE and GAPC.29 Both phase 3 1000 Genomes Project (1KGP)30 and r.1.1 Haplotype Reference Consortium (HRC)31 reference panels were used in imputation, and only well-imputed markers (i.e., imputation quality r2 ≥ 0.7) were used in subsequent analysis; if a marker was well-imputed by both reference panels, the imputed dosage for the panel with the higher imputation quality was used. In all, these datasets included 15,967 cases and 28,194 controls. Finally, we examined all pairwise combinations of the eight EUR cohorts and the two SAS cohorts and removed samples that were duplicates or first- or second-degree relatives with a sample in a different cohort. Characteristics of the 10 studies analyzed for psoriasis associations are described in Table S1.
Genome-wide meta-analysis
We conducted a transethnic meta-analysis using eight EUR and two SAS cohorts, as well as EUR-only and SAS-only meta-analyses. For the EUR-only and transethnic analyses, we included only those markers that were well-imputed in at least half of the studies; for the SAS-only analysis, markers had to be well-imputed in both cohorts. Meta-analysis was carried out using the inverse variance-weighted approach implemented in METAL.32 QQ-plots indicated that the PCA and geographic cohort covariates included in our association models did a good job of controlling for population stratification (Figure S2).
For identifying the Bayesian credible set of markers for each locus, we used association results for genetic variants that were well-imputed in at least half of the cohorts (see Supplemental methods for more details). To compare the number of surrogates for known psoriasis loci, we identified markers in strong LD (r2 ≥ 0.8) with their lead markers, using the EUR and SAS samples from the 1KGP.
Genome-wide conditional analysis
We conducted conditional analysis for each psoriasis-associated locus that achieved genome-wide significance to reveal independent signals in the transethnic meta-analysis. Employing only markers that are well-imputed in all cohorts, we used stepwise conditional analysis to reveal secondary signals within ±500 kb of the psoriasis-associated signals. For each of the secondary signals identified, we computed the 95% credible interval set as described above. For comparison, we took the same marker(s) utilized for each round of the conditional analysis as covariates in a separate analysis using only the EUR cohorts.
HLA genotyping
Eight classical HLA genes—HLA-A (MIM: 142800), HLA-B (MIM: 142830), HLA-C (MIM: 142840), HLA-DPA1 (MIM: 142880), HLA-DPB1 (MIM: 142858), HLA-DQA1 (MIM: 146880), HLA-DQB1 (MIM: 604305), and HLA-DRB1 (MIM: 142857)—were genotyped to 3-field resolution by the Institute of Clinical Molecular Biology at the University of Kiel (IKMB) in Germany. In-solution targeted capturing with an RNA bait panel was designed to accommodate the complete collection of reference sequences for all HLA genes in version 3.09 of the International Immunogenetics information system (IMGT)/HLA database.33 Target DNA was fragmented to 150–300 bp size segments, enriched for HLA gene sequences by hybridization with the bait, and subjected to high-throughput paired-end sequencing with read lengths of 115–125 bp. Reads were aligned against the cDNA collection of the IMGT/HLA database, and the most likely genotype was determined by ranking all possible allele combinations by their harmonic mean for five different quality metrics.
Updated SNP2HLA package
We updated several features of v1.0.3 of SNP2HLA for imputing HLA genotypes.23 Most importantly, we substituted Beagle version 4.134 for the older version 3.0.4 as the imputation engine for both the MakeReference and SNP2HLA scripts, which substantially improved accuracy. The HLA amino acid and SNP sequence dictionaries were updated to a more recent release of the IPD-IMGT/HLA database (r3.30.0, October 2017).35 For the SNP dictionaries we improved handling of polymorphisms for 2-field HLA alleles by employing International Union of Pure and Applied Chemistry (IUPAC) ambiguity codes rather than arbitrary selection of one allele. We also updated the package to accept modern HLA nomenclature.36
Construction of SNP2HLA reference panels
We first built a SNP2HLA reference panel of 397 South Asian individuals. Using a method that maximizes represented genetic diversity,37 288 individuals of Pakistani or Indian ancestry were selected from batch 3 of our South Asian psoriasis GWAS. An additional 192 individuals from north India were obtained from a preliminary version of the IKMB multiethnic HLA reference panel;38 these individuals were originally ascertained for a meta-analysis of inflammatory bowel disease (IBD [MIM: 266600]).39 The 480 selected South Asians were genotyped for eight classical HLA genes; 233 psoriasis GWAS individuals (the University of Michigan [UM] dataset) were successfully genotyped, as were 164 IBD case-control samples, including 141 that were included in the final IKMB panel (IKMB-SAS dataset) and 23 that were not (B.K. Thelma or BKT dataset). We then input into our updated MakeReference script HLA genotypes for the 397 South Asians, along with genotypes for 2,284 SNPs in the classical MHC region common to the microarray used for the UM dataset and the Immunochip platform used for the IKMB-SAS and BKT datasets.
We constructed an additional 18 SNP2HLA reference panels for imputing our South Asian GWAS samples by rebuilding existing HLA panels and by forming various combinations of the UM, IKMB-SAS, and BKT components of the 397-person SAS panel with four other datasets—the non-SAS subset of the IKMB HLA reference panel,38 the European ancestry Type 1 Diabetes Genetics Consortium (T1DGC) SNP2HLA panel,23,40,41 the pan-Asian SNP2HLA panel,42,43 and data from phase 3 of the 1KGP.30,44 HLA genotypes for 1KGP were combined separately with both microarray-based (v1) and sequence-based (v2) MHC genotypes for both the full 1KGP dataset (1KGP-ALL) and its SAS subset (1KGP-SAS), resulting in four versions of 1KGP data used for construction of South Asian panels (1KGP-ALL-v1, 1KGP-SAS-v1, 1KGP-ALL-v2, and 1KGP-SAS-v2). We also built 20 SNP2HLA reference panels for imputation of HLA variants in people of European ancestry. Datasets used for these panels consisted of many of those used for South Asians (T1DGC, UM, BKT, IKMB, 1KGP-ALL-v1, and 1KGP-ALL-v2) as well as the EUR subset of the 1KGP with HLA and either microarray or sequence-based MHC data (1KGP-EUR-v1 and 1KGP-EUR-v2, respectively). Additional details of panel construction are provided in Supplemental methods.
Validation of SNP2HLA reference panels
The performance of each HLA reference panel was assessed by comparing gold-standard HLA genotypes for a population-specific validation set with HLA allele dosages that were imputed by applying the panel with our updated SNP2HLA script to the same validation set. The validation set for South Asians was the sequence-based 2-field genotypes of eight HLA genes for the 397 people of our original SAS panel (UM+BKT+IKMB-SAS datasets). For people of European descent, four validation sets were used, consisting of subsets of four of our psoriasis case-control studies with independent genotyping for five HLA genes (Table S2). Leave-one-out cross-validation was used to compare imputed and genotyped dosages for people shared between a South Asian panel being assessed and the SAS validation set.
Two measures of imputation accuracy were used to compare gold-standard and imputed HLA genotypes. Per-individual accuracy was computed as proposed previously:45
where is the number of 1-field or 2-field alleles for a given HLA gene in the imputation reference panel, is the dosage for genotyped allele for individual , is the dosage for imputed allele for individual normalized to a sum of 2.0, and if > else . The distribution and mean of per-individual accuracy were then examined for all individuals in a validation set. Per-allele accuracy was computed as the squared Pearson correlation of vectors of genotyped and imputed dosages for 1-field or 2-field HLA allele across all individuals in the validation set. Based on these two measures of imputation accuracy, the relative performances of the SNP2HLA reference panels were assessed as described in Supplemental methods.
MHC variant imputation
The best-performing SNP2HLA reference panel for each combination of three groups of HLA genes (HLA-A, -B, -C, -DQB1, -DRB1; HLA-DPA1, -DPB1; HLA-DQA1) and two ancestries (SAS and EUR) was used to impute 1-field, 2-field, amino acid, SNP, and insertion or deletion (indel) alleles of HLA genes into either the two SAS or the eight EUR-ancestry psoriasis case-control studies. Before imputation, genotypes for all SNPs in a 20 Mb region (chr6: 20–40 Mb) encompassing the MHC were extracted from the genome-wide set of quality-controlled microarray genotypes for the target study to be imputed. SNPs from the target study were matched to those in the reference panel based on chromosomal position and allele identities. We then applied our improved SNP2HLA script and updated HLA sequence dictionaries to impute the reference panel variants into the target study, using 35 total iterations of the Beagle phasing algorithm. Imputed variant dosages were extracted for only those HLA genes for which the particular reference panel used provided optimal imputation accuracy.
For each of the 10 case-control studies, we also extracted imputed dosages for all variants in the chr6: 24–36 Mb region from the two genome-wide datasets that were imputed with the 1KGP and HRC reference panels. Biallelic variants were extracted from the 1KGP and HRC datasets as described earlier. Selection and processing of multiallelic variants is described in Supplemental methods.
For each ethnic dataset to be analyzed for association, we merged imputed HLA variant dosages from the three appropriate SNP2HLA panels with MHC variant dosages from the combined 1KGP and HRC panels. For SNPs in HLA genes that were duplicated because they were imputed by both the 1KGP/HRC and SNP2HLA panels, the variant with the lesser mean imputation quality for the studies in the analysis was dropped. We restricted MHC variants used for association analysis to those with a predicted imputation quality (Minimac or Beagle r2) of 0.7 or better for all case-control studies in an analysis.
Association analysis of MHC variants
We tested for association between MHC variants and psoriasis with a logistic regression model. We defined MHC variants to include both biallelic and multiallelic SNPs and indels in the 12 Mb extended MHC region, 1-field and 2-field classical HLA gene alleles, biallelic HLA amino acid polymorphisms for respective residues, and multiallelic HLA amino acid polymorphisms for respective positions. For MHC variants with alleles ( = 2 for biallelic variants and > 2 for multiallelic variants), we included − 1 alleles as independent variables, excluding either the reference genome allele (1KGP/HRC variants) or the most frequent allele (SNP2HLA variants) as a reference, resulting in the following model:
where is the overall intercept, is the additive effect of the dosage of allele for the variant being tested, and is the additive effects of the dosages of optional conditioning variants . is the number of case-control studies, and and are the numbers of study-specific principal components (PCs) and geographic indicator covariates for the study (Table S3 tallies study-specific covariates used for controlling population stratification). Variable is the PC for the study, is the geographic cohort for the study, and is the indicator variable for the study-specific intercept. , , and are the effects of , , and , respectively, and is the error term. Note that the inclusion of study-specific indicator variables assumes fixed effects among the case-control studies. The regression model was fitted using the -glm command of Plink 2.0,46 with the firth-fallback option, which requests a standard logistic regression, followed by Firth regression whenever the logistic regression fails to converge. For each tested MHC variant, an omnibus p value for the association of its − 1 alleles was determined with a multivariate Wald test, which follows a χ2 distribution with − 1 degrees of freedom. For multiallelic variants, in addition to the omnibus test, each of the alleles were tested individually with biallelic Wald tests, including the reference allele. Additional details of association analysis of the MHC region are given in Supplemental methods.
Association model comparison
We used three measures to compare the goodness of fit of pairs of non-nested association regression models: (1) the Akaike Information Criterion (AIC), which was computed by the logistf R package47 using log-likelihoods for models fitted with ordinary logistic regression and penalized log-likelihoods for models fitted with Firth’s bias-reduced logistic regression; (2) the evidence ratio, which quantifies the relative likelihood of one model versus a second and can be computed from the AIC for each model;48 and (3) Tjur’s R2, which quantifies the explanatory power of a logistic model by computing the difference in the means of the model-predicted probabilities of a binary outcome for cases and controls.49 Individual contributions of three groups of regressors (PC and geographic cohort covariates, HLA-C∗06, and all genetic variants other than HLA-C∗06) to the AIC and Tjur’s R2 values of the full model were determined by decomposing the goodness of fit with the Shorrocks-Shapley procedure.50
Bayesian credible sets
Using a Bayesian approach, for association signals in the final regression models we identified the credible set of markers that were 95% likely, based on posterior probability (PP), to contain the causal disease-associated variant. This approach requires specification of a prior distribution for , the effect size of the variant, where is assumed to follow a normal distribution with a prior mean of 0 and a prior variance of .51 For biallelic loci, the original applications of Bayesian credible sets to disease GWASs52,53 used a value of 0.22 for the prior variance, corresponding to a 90% prior probability that lies in the interval [−0.165, 0.165]. Because of the wide range of expected effect sizes for psoriasis-associated MHC loci, for biallelic variants we instead computed a mean Bayes factor for a vector of priors of 0.1, 0.2, 0.4, 0.8, and 1.6, as was first suggested by Wen and Stephens.54 For multiallelic variants, we modified the approach of Wen55 for computing Bayes factors for multiple biallelic SNPs, which uses a -length vector of 0 s for the prior means and a × matrix for the prior variances and cross-covariances for the effect sizes of SNPs in a multiple regression model. We extended this multivariant approach to a single multiallelic variant with alleles in a manner analogous to our association model, namely by decomposing the variant into its biallelic components and dropping the biallelic variant for the most frequent allele (variants imputed with SNP2HLA panels) or the reference genome allele (variants imputed with 1KGP and HRC panels) to avoid complete linear dependency. Priors for the variances of the effect sizes of the remaining biallelic variants were set to one of the five priors used for biallelic variants. Priors for the cross-covariances were determined empirically. For each variant in the final association models for the South Asian and European datasets, we dropped that variant and then repeatedly refitted the regression model for each neighboring (±500 kb) multiallelic variant. The variance-covariance matrices from these refitted regressions were converted to correlation matrices. Most (93.5%) of the resulting 4,861 upper-diagonal cross-correlations were positive due to inherent residual dependencies among the decomposed variants, but the variation in magnitude was large (SD = 0.22). To accommodate this heterogeneity, the first, third, fifth, seventh, and ninth deciles of this set of cross-correlations (0.011, 0.084, 0.177, 0.313, and 0.558), representing the midpoints of each of the five quantiles, were used instead of a single value to convert variances to cross-covariances. We then determined the mean of the approximate Bayes factors computed by formula (11) of Wen55 for each of the 25 combinations of 5 variance priors and 5 cross-correlation priors.
For loci outside the extended MHC region, inclusion into Bayesian credible sets was restricted to biallelic variants present in both the SAS and EUR meta-analyses, well-imputed (r2 ≥ 0.7) for at least half of the participating case-control studies, and within 200 kb of the lead variant for the association signal being analyzed. For both biallelic and multiallelic MHC signals, the window for inclusion was increased to 500 kb to accommodate the unusually long-range LD that characterizes the MHC region. Credible sets for the individual monoethnic and transethnic MHC analyses were also automatically restricted to variants passing the imputation quality threshold of r2 ≥ 0.7 imposed upon the studies contributing to a particular ancestry association model. For comparison of credible sets of the four stepwise-selected variants shared among the different MHC models, credible sets were recomputed for all three ancestry models after further restriction to variants with imputation quality of r2 ≥ 0.7 in all eight European and both South Asian studies.
Multiallelic linkage disequilibrium
We assessed 12 measures of linkage disequilibrium that can handle multiallelic variants. These metrics included five coefficients either reviewed (, , multiallelic ) or proposed (, multiallelic ) by Zhao et al.,56,57 ,58 ,59,60 , and ,60 for a multiallelic variant collapsed down to its most common allele versus the rest,46 ,61, 62, 63 and , a metric we devised equal to the maximum biallelic among all possible pairings of the alleles for two loci. Performance was evaluated empirically for the European and South Asian datasets by computing pairwise LD between all loci in the final full psoriasis association models and their neighboring (±500 kb) variants. Relative magnitudes of the linear (Pearson ) and rank order (Spearman ) correlations between measured LD and the −log10 of the p value for each full-model variant were compared across metrics; equality of slopes for the linear fits of LD versus p value for each full-model variant with its neighboring biallelic versus triallelic versus 4+-allelic variants was also assessed.
Overall, the best-performing measure was , which reduces to the familiar LD coefficient for two biallelic loci. As pointed out by others,60 is also known as Cramer’s statistic,64 which is the statistic for a contingency table relating two categorical variables, normalized to lie in a [0, 1] interval. Although we used as our primary measure of LD between variants, for annotation of psoriasis-associated variants we also used , which was a good performer and is unique among the 12 assessed metrics by being based on differences in the entropy of observed and expected haplotypes rather than differences in their frequencies. Our formulation of multiplies the multiallelic extension63 of the original multilocus statistic61 by two, because as demonstrated by Liu and Lin,62 can attain a maximum value of only , where is the number of loci considered.
Before measurement of LD between pairs of variants, fractional imputed dosages were converted to integer hard calls. For most analyses, individuals with poorer-quality imputed genotypes (>0.25 dosage units from an integer) for either variant of the pair were omitted. For analysis of the correlation of imputed genotype quality with association p value for variants in substantial LD with HLA-C∗06, as well as for determining the number of strong LD proxies for association signals in the full transethnic MHC model, a stricter hard-call threshold of 0.10 dosage units was used instead.
Details of principal components analysis, determination of phenotypic variance explained, MHC variant annotation, and enrichment analysis are presented in Supplemental methods.
Results
Transethnic analysis reveals additional psoriasis loci
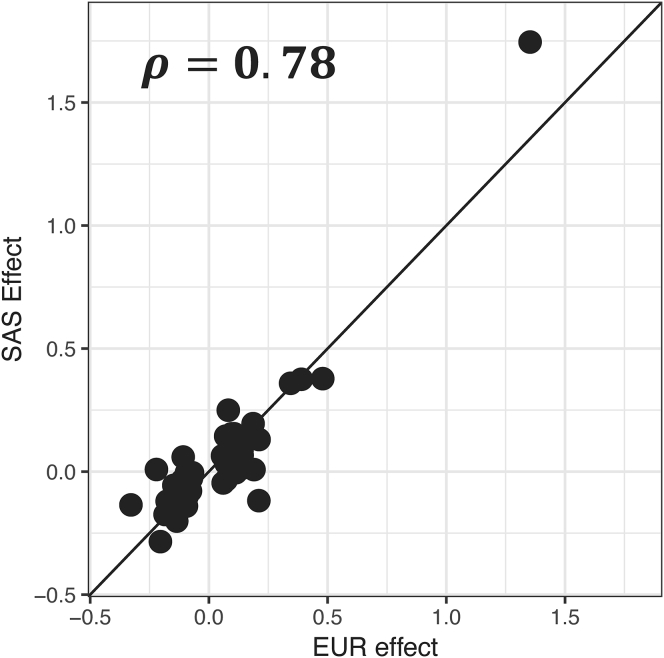
We first compared the European and South Asian GWAS signals, aiming to evaluate shared and unique effects. Despite the lack of power to replicate the established EUR signals in the SAS cohorts using a genome-wide significance threshold, there was a strong correlation between their effect sizes (Figure 1; Spearman’s ρ = 0.78, p < 2 × 10−14), justifying a transethnic analysis of the two populations.
Figure 1.
Correlation of effect sizes of psoriasis-associated loci in the European (x axis) and South Asian (y axis) GWASs
Log(OR) effect sizes are plotted; rho is the Spearman correlation coefficient. The plot includes 66 loci identified by past studies having a genome-wide significant (p ≤ 5 × 10−8) association with psoriasis in European ancestry populations.
For the transethnic, EUR, and SAS meta-analyses, we analyzed 8.95 million, 9.01 million, and 9.20 million well-imputed markers, respectively. The meta-analyses revealed 50, 47, and 3 loci, respectively, for the transethnic, EUR and SAS cohorts, that were associated with psoriasis at a genome-wide level of significance (Figure S3). As shown in Table S4, none of the identified psoriasis loci exhibited significant effect size heterogeneity after correction for multiple testing except the primary MHC locus for the EUR and transethnic meta-analyses, which is likely a consequence of differences across the EUR studies in the proportion of purely cutaneous versus PsA cases.25 The transethnic meta-analysis revealed two psoriasis-associated loci that were previously unreported for either the EUR or SAS populations (Table 1; Figures 2 and S4). Because our signal in 1p36.22 (rs2103876) is close (∼200 kb) to two psoriasis-associated missense variants (rs2274976; rs5063) identified from a Chinese study,65 we investigated pairwise LD among these variants using the EUR samples from the 1KGP. The results indicate D′ > 0.82 and r2 < 0.015 between rs2103876 and both of the missense SNPs, suggesting that they are in LD but exhibit different population allele frequencies: 1KGP risk allele frequencies for rs2103876 are 0.73 (Gujurati Indians in Huston [GIH]), 0.66 (Utah residents with northern and western European ancestry from CEPH collection [CEU]), and 0.67/0.61 (Southern Han Chinese [CHS]/Han Chinese in Beijing [CHB]); for rs2274976 they are 0.78 (GIH), 0.93 (CEU), and 0.9/0.92 (CHS/CHB); and for rs5063 they are 0.86 (GIH), 0.93 (CEU), and 0.91/0.9 (CHS/CHB). Interestingly, the two Chinese missense variations show no evidence of association in our transethnic meta-analysis. However, we did identify potential regulatory roles for each of our two significant non-MHC signals (see Discussion). For the EUR meta-analysis, we also uncovered a genome-wide significant variation (rs77343625; 5:158208927; p = 2.03 × 10−8) >500 kb upstream of the best signal in the IL12B (MIM: 161561) region revealed previously (rs12188300; 5:158829527),29 but this signal is no longer significant after conditioning on the known IL12B signal, indicating that its association results from long-range LD (spreading > 600 kb).
Table 1.
Psoriasis loci established by the transethnic meta-analysis
| Locus | Marker | Location (hg19) | RA/NRA | SAS |
EUR |
Transethnic |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | Direction | ||||
| 1p36.22 | rs2103876 | 1:12053100 | T/C | 1.19 (1.07–1.31) | 7.96E−04 | 1.10 (1.06–1.14) | 1.30E−07 | 1.11 (1.07–1.14) | 1.18E−09 | ++−+++++++ |
| 1q24.2 | rs12046909 | 1:168507463 | C/T | 1.21 (1.09–1.34) | 3.85E−04 | 1.13 (1.08–1.18) | 5.56E−07 | 1.14 (1.09–1.19) | 1.68E−09 | ++++++++++ |
Abbreviations: Direction, the direction of effect of the risk allele for 10 studies: PsA GWAS, CASP GWAS, Kiel GWAS, Genizon GWAS, WTCCC2 GWAS, Exomechip, PAGE, GAPC, and two South Asian GWASs; EUR, European; NRA, non-risk allele; RA, risk allele; SAS, South Asian.
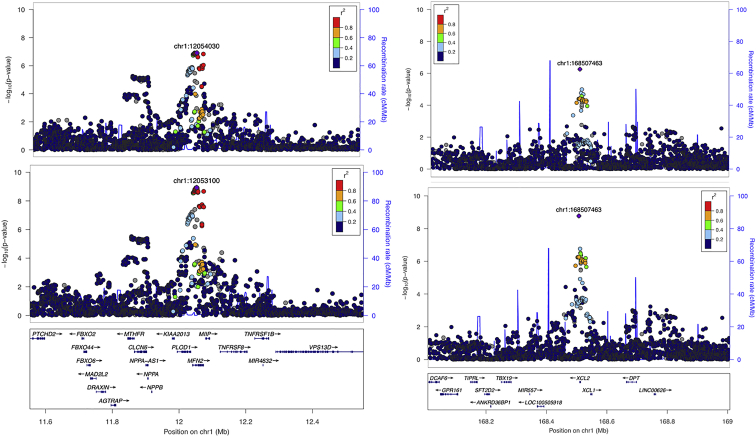
Figure 2.
Association plots for psoriasis loci established by the transethnic meta-analysis
The regional association plots for the 1p36.22 and 1p24.2 loci (left and right pairs of panels) are shown for results from the European and transethnic meta-analyses (top and bottom pairs of panels).
Transethnic Bayesian refinement of primary association signals
To investigate the resolution of localization of causal variants for 65 non-MHC psoriasis loci (63 primary EUR loci from past studies1,7,8 and 2 transethnic loci established by this study), we compared 95% Bayesian credible sets (BCS) for these susceptibility loci in our EUR and transethnic meta-analyses. Among the 41 loci with stronger association signals in the transethnic than EUR meta-analysis, 19 have fewer markers in the 95% BCS for the transethnic model, 14 have the same number of markers, and 8 have more markers (Table S5). Specifically, for the two psoriasis loci established by the transethnic meta-analysis, the number of markers in the 95% BCS drops from 83 to 23 and from 30 to 10 in the transethnic versus EUR-only meta-analyses.
Conditional analysis identifies 12 secondary non-MHC loci
To identify independent signals for known and previously unreported psoriasis loci, we then conducted a transethnic conditional meta-analysis. Analysis was restricted to 46 non-MHC loci achieving genome-wide significant association in the unconditional transethnic analysis and that also had at least one marker well-imputed (r2 ≥ 0.7) for all 10 cohorts, which resulted in an analysis set of 113,745 markers well-imputed in all cohorts mapping to within 500 kb of the lead variant for each of these loci. Altogether, we were able to identify 12 independent signals in nine non-MHC loci (Table S6). Notably, six of the identified independent signals each harbor one genetic variation with a posterior probability ≥ 50% of being causative, with four of the loci (IL23R [MIM: 607562], IFIH1 [MIM: 606951], TRAF3IP2 [MIM: 607043], and NFKBIA [MIM: 164008]) encompassing 10 or fewer variants in their 95% CI set. Overall, all but two of the independent signals harbor fewer variants in the 95% BCS in the transethnic meta-analysis.
Improved imputation of HLA genotypes in both SAS and EUR
Many genetic studies of psoriasis have identified protein and amino acid alleles of HLA genes as potential susceptibility loci.66 The 1KGP and HRC reference panels used to impute genotypes for the GWAS of this study do include some SNPs and indels within HLA genes but no amino acid or classical protein variants. A reference panel explicitly designed for HLA genes is needed for this purpose. We have had good success using the T1DGC reference panel with the SNP2HLA imputation package23 to impute HLA genotypes for people of European ancestry.22 Because no appropriate SNP2HLA panel exists for South Asians, we built a reference panel of 397 individuals of Indian and Pakistani ancestry with our improved version of the SNP2HLA MakeReference script and updated HLA amino acid and DNA sequence dictionaries. This new panel was representative of the population structure of the larger samples of Indians and Pakistanis from which they were drawn, as well as of the various SAS populations represented in the 1KGP (Figure 3). Unfortunately, its imputation accuracy was poor, especially for 2-field alleles that distinguish HLA proteins, where it was under 90% for all but one of eight classical HLA genes (Table S7). As a test, we randomly sampled increasingly large subsets of the T1DGC panel of 5,225 EUR-ancestry individuals, which were then used to build reference panels for HLA imputation of the 397 South Asians. As shown in Figure S5, although the SAS panel did outperform its 397-individual T1DGC counterpart, as the T1DGC subset panels increased in size, mean imputation accuracy increased well beyond that afforded by the SAS panel. We concluded that increasing the sample size of our SAS panel to at least 1,500–2,000 individuals might achieve acceptable imputation accuracies for most HLA genes, with even larger panels needed for imputing HLA-B and HLA-DRB1 genotypes accurately.
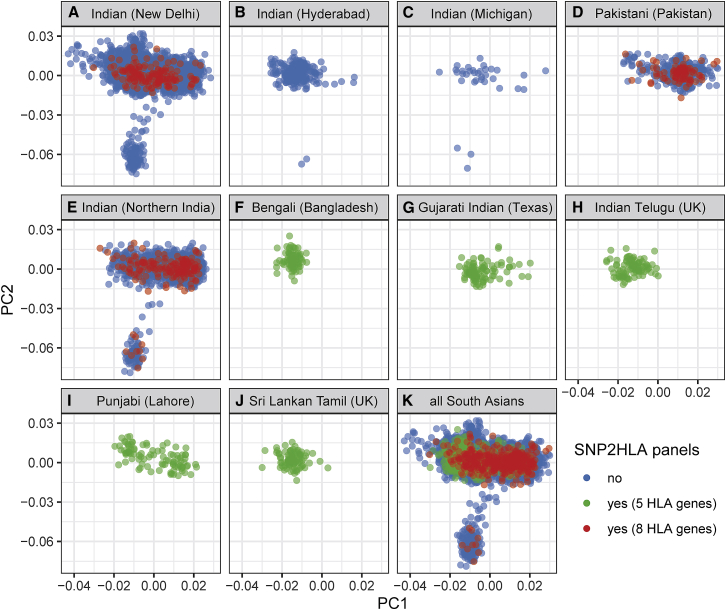
Figure 3.
Population structure of South Asians in the SNP2HLA reference panels
The first two principal components from a PCA of 6,421 individuals of South Asian ancestry are shown. (A–J) Results divided by 10 different source populations: populations in (A)–(D) are from the psoriasis GWAS of this study, the population in (E) is from an inflammatory bowel disease case-control cohort of B.K. Thelma, and populations in (F)–(J) constitute the SAS superpopulation of phase 3 of the 1000 Genomes Project. (K) Results for all populations combined. Points are colored by whether they are part of any of the SNP2HLA panels used for imputation of HLA variants in South Asians; blue, green and red colors indicate whether the individual was not part of any panel, was part of panels used for imputation of 5 HLA genes (A, B, C, DQB1, DRB1), or was part of panels used for imputation of 8 HLA genes (A, B, C, DPA1, DPB1, DQA1, DQB1, DRB1), respectively.
The preferred option of adding more South Asians to our panel was not feasible given the prohibitive cost of HLA genotyping. Past work combining a small population-specific SNP2HLA panel with panels from other populations achieved increased imputation accuracy for the population of interest.42,43 Similarly, large multi-population reference panels have shown good performance when used with other methods of HLA genotype imputation such as HLA∗IMP67 and HIBAG.38 Based on these promising findings, we combined data from several sources (Table S8) to build 19 SNP2HLA reference panels for imputing HLA genotypes in South Asians, varying in size from 397–9,343 people of one to several continental ancestries (Table S9). Each panel was then used as a reference with our updated SNP2HLA script to impute 2-field HLA genotypes for the 397 people of the original SAS panel, using leave-one-out cross-validation to generate imputed dosages for any people shared between the panel and target dataset. For the five HLA genes included in all panels, the multiethnic IKMB+BKT+UM+1KGP-ALL-v2 panel gave the best results (Figures S6 and S7), with mean sample accuracies of 93%–97% (Table S10). Among the 11 panels with genotypes for HLA-DPA1, HLA-DPB1, and HLA-DQA1, the IKMB+BKT+UM panel performed best for HLA-DPA1 and HLA-DPB1 (Figures S8 and S9), and the IKMB+BKT+UM+T1DGC panel performed best for HLA-DQA1 (Figures S10 and S11), with mean accuracies ranging from 93%–94% (Table S10). Imputation of HLA-DQA1 was assessed separately from that for HLA-DPA1 and HLA-DPB1 because of HLA-DQA1 genotyping issues for two of the source datasets (T1DGC and Pan-Asian). As shown in Table S11, the multiethnic IKMB+BKT+UM+1KGP-ALL-v2 reference panel also provided generally excellent imputation results for the global populations of 1KGP, with mean sample accuracies at 2-field HLA resolution of 92%–98% for African, East Asian, EUR, and SAS populations, each constituting 20%–24% of the panel, and 83%–97% accuracy for admixed Americans that constitute only 8% of the panel (Table S8).
We then repeated the procedures used for building South Asian panels to create 20 panels tailored for people of European ancestry (Table S12). Each panel was used to impute 2-field HLA genotypes for four of our psoriasis case-control studies of European ancestry with independent gold-standard HLA genotyping (Table S2). Comparison of imputation accuracy for the five HLA genes in all 20 panels shows that the European-ancestry T1DGC+1KGP-EUR-v2 panel performed best (Figures S12 and S13), with mean sample accuracies of 95%–99% (Table S13). Among the seven panels with HLA-DPA1, HLA-DPB1, and HLA-DQA1 genotypes, the IKMB+BKT panel performed best for imputation of HLA-DQA1 (Figures S14 and S15), achieving a mean sample accuracy of 96% (Table S13). Because we had no independent genotypes for HLA-DPA1 and HLA-DPB1, we assessed panels for imputing these two genes based on imputation accuracies for HLA-A, -B, -C, -DQB1, and -DRB1 as a proxy and found the T1DGC panel performed best (data not shown).
While this study was under review, a large multiethnic HLA reference panel of 21,546 individuals of European, admixed African, East Asian, and Latino ancestry68 became available for use via the Michigan Imputation Server.69 We compared imputation results for our SAS and EUR sample validation sets obtained with this large panel versus what we achieved with our suite of best-performing EUR and SAS HLA panels. As shown in Figure S16, imputation accuracies with our panels equal or exceed those obtained with the new multiethnic panel for both SAS and EUR target samples and both 1-field and 2-field HLA allele resolutions. The gains in accuracy with our panels were generally larger for class II genes, especially for HLA-DQA1, where the conversion of G-group HLA alleles to 2-field alleles that is employed by the newly published panel is most inaccurate.
The three best South Asian reference panels were used with the improved SNP2HLA script to impute HLA genotypes into the two psoriasis case-control studies of SAS ancestry. Similarly, the three best European reference panels were used for imputation into eight case-control studies of EUR ancestry. From the SNP2HLA-imputed genotype datasets, we extracted 1-field, 2-field, amino acid, SNP, and indel variants within those HLA genes for which the panel used was optimal. A comparison of frequencies of imputed 1-field and 2-field HLA alleles reveals many differences between the EUR and SAS populations (Figures S17–S20), which correspond closely to differences in genotyped allele frequencies between these populations published by the National Marrow Donor Program70 (Figures S21 and S22). We expanded our scope beyond classical HLA genes by extracting imputed genotypes for all 1KGP/HRC panel variants in a 12 Mb region (chr6: 24–36) that encompasses the classical MHC region (chr6: 29.64–33.12 Mb), the extended MHC (chr6: 25.73–33.37 Mb) defined by Horton et al.,71 as well as flanking sequence. As shown in Table S14, the density of coding, non-coding, and immune-related genes exceeds the genome-wide average for most segments of the 12 Mb extended MHC, peaking within the classical interval. Frequency distributions of the approximately 288,000 imputed MHC variants, cross-classified by ancestry and MHC region versus reference panel source, MAF and imputation quality, or variant type are shown in Tables S15–S17, respectively.
MHC fine-mapping uncovers multiple independent SAS, EUR, and transethnic loci
Our previous fine-mapping study of the classical MHC22 identified several HLA gene variants that may be driving the multiple independent psoriasis association signals in the region for people of European ancestry. For this study we extended these fine-mapping efforts to South Asians, built upon past work for people of European ancestry with a dataset that includes more individuals and more variants of generally higher imputation quality, and combined our South Asian and European studies to perform a transethnic association analysis.
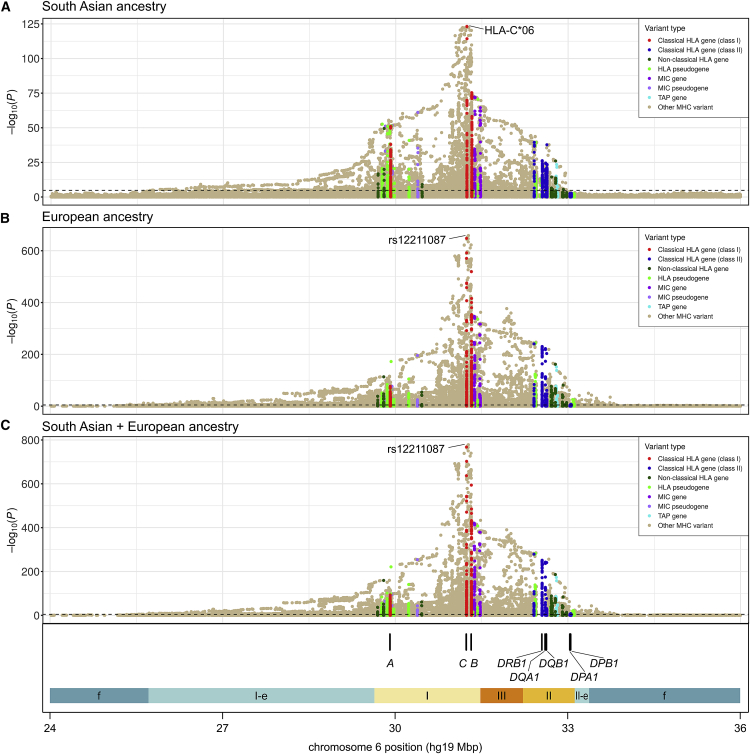
Association analysis of the extended MHC was restricted to variants imputed with good accuracy in all participating studies (Table S18). Tables S19–S21 present frequency distributions of the tested MHC variants, cross-classified by ancestry, MHC region, panel source, MAF, imputation quality, and variant type. Figure 4 plots unconditional association with psoriasis in the extended MHC region for all three ancestry analyses. The most strongly associated variant in the SAS analysis is HLA-C∗06, and the top variant in both the EUR and transethnic analyses is rs12211087, a SNP lying 30 kb upstream of HLA-C that is in nearly perfect LD with HLA-C∗06 ( = 0.986 and 0.990 in SAS and EUR populations, respectively). The unconditional effect size of the lead variant is very large in all cases—odds ratio (OR) (95% confidence interval [CI]) = 5.80 (5.02–6.71), 3.93 (3.75–4.13), 4.09 (3.90–4.28) for SAS, EUR, and transethnic, respectively—but significantly greater for South Asians than Europeans (p = 7.4 × 10−8).
Figure 4.
Plots of unconditional psoriasis association for the MHC and flanking regions
Each circle represents the −log(p) of association of an imputed variant, color-coded based on its membership in various categories of MHC genes, as detailed in the keys. The dashed lines denote thresholds of Bonferroni-corrected significance of 0.05. The locations of the eight HLA genes for which amino acid and protein alleles were imputed are shown at the bottom, along with colored segments denoting the boundaries of the classical MHC region (class I, II, and III), the extended MHC class I (I-e) and II (II-e) regions of Horton et al.,71 and flanking MHC regions (f). We tested association in people of three different ancestries: South Asian (A), European (B), and South Asian and European combined (C).
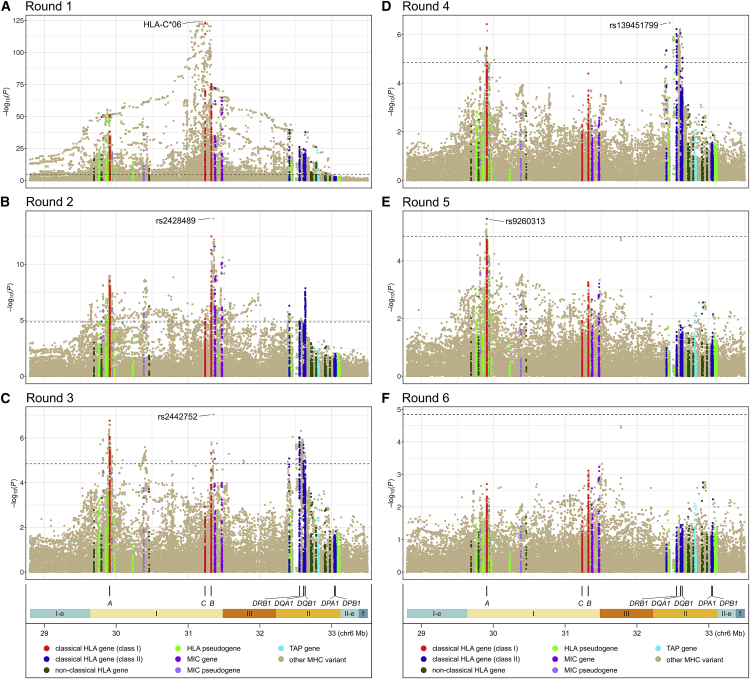
Stepwise analysis identified five independent South Asian MHC psoriasis susceptibility loci (Figures 5, S23, and S24). Pairwise LD values suggested that the five lead variants are mutually independent (Figure S25). Full model effect sizes and association p values indicated that HLA-C∗06 is much more strongly associated with psoriasis in the SAS population than the other four selected variants, contributing 66% to the total variance in disease liability explained by all five loci (Table 2). The second most strongly associated variant is triallelic SNP rs2428489, whose association is mostly driven by its C allele (OR [95% CI] = 1.64 [1.45–1.85], p = 1.8 × 10−15). All five identified variants are situated near one or more classical HLA genes (Figure 5), but the closest genes for three of the variants are not classical HLA genes (Table S22). Furthermore, only one of the variants other than HLA-C∗06 (indel rs139451799) has a plausible protein-changing surrogate (amino acid 13 or 142 of HLA-DRB1) based on two LD measures (Table S22) and comparisons of the magnitude and rank of association p value and Bayesian posterior probability between the variant and its potential surrogate (Tables 2, S22, and S23). Two variants (HLA-C∗06 and rs2442757) have a substantial posterior probability of being causative (0.255 and 0.583, respectively), although the size of the 95% Bayesian credible set for rs2442757 is very large, including 2,868 variants and spanning nearly 1 Mb.
Figure 5.
Plots of stepwise analysis of psoriasis association in the extended MHC region in people of South Asian ancestry
(A)–(F) Association results after each of the six rounds of stepwise regression. Each circle represents the −log(p) of association of an imputed variant, color-coded based on its membership in various categories of MHC genes, as detailed in the key at the bottom. Dashed lines denote thresholds of Bonferroni-corrected significance of 0.05. The locations of the eight HLA genes for which amino acid and protein alleles were imputed are shown at the bottom, along with colored segments denoting the boundaries of the classical MHC region (class I, II, and III), the extended MHC class I (I-e) and II (II-e) regions, and flanking MHC regions (f).
Table 2.
Psoriasis associations from stepwise meta-analysis of the extended MHC region for two studies of South Asian ancestry
| Stepa | Variantb | chr6 positionc | Alleles |
Risk allele frequency |
Association at entry into model |
Association in final full model |
Vg | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Riskd | Nonrisk | Cases | Controls | OR (95% CI) | p value | OR (95% CI) | p value | ||||
| 1 | HLA-C∗06 | 31238192 | C∗06 | other | 0.3361 | 0.1012 | 5.80 (5.02–6.71) | 6.7 × 10−124 | 4.68 (3.99–5.50) | 1.3 × 10−78 | 0.04822 |
| 2 | rs2428489 | 31352972 | C, T | A | NA | NA | NA | 8.2 × 10−15 | NA | 9.6 × 10−16 | 0.01001 |
| C | other | 0.4094 | 0.2654 | 1.41 (1.26–1.58) | 2.1 × 10−9 | 1.64 (1.45–1.85) | 1.8 × 10−15 | NA | |||
| T | other | 0.0840 | 0.1044 | 0.71 (0.87–1.29) | 1.4 × 10−4 | 1.06 (0.87–1.29) | 0.57 | NA | |||
| other | A | 0.4934 | 0.3697 | ref | ref | ref | ref | NA | |||
| 3 | rs2442752 | 31351764 | T | C | 0.6508 | 0.5154 | 1.39 (1.23–1.56) | 8.8 × 10−8 | 1.39 (1.23–1.57) | 8.8 × 10−8 | 0.00576 |
| 4 | rs139451799 | 32454479 | – | other | 0.2295 | 0.2532 | 1.46 (1.26–1.69) | 3.3 × 10−7 | 1.43 (1.23–1.65) | 1.6 × 10−6 | 0.00521 |
| 5 | rs9260313 | 29916885 | T | C | 0.6593 | 0.5341 | 1.29 (1.16–1.44) | 3.4 × 10−6 | 1.29 (1.16–1.44) | 3.4 × 10−6 | 0.00345 |
Abbreviations: chr6, chromosome 6; CI, confidence interval; NA, not applicable; OR, odds ratio; ref, reference; Vg, variance in liability explained by the genetic variant.72
Round of stepwise regression analysis.
Variant notes: variant ID is build 151 dbSNP rsID when applicable; HLA-C∗06 is one biallelic split from a decomposed set of 14 classical 1-field HLA-C alleles; the stepwise-selected variant for triallelic indel rs139451799 is one of its biallelic splits with − versus A+G alleles.
Base pair position in hg19 human reference; for classical HLA proteins the position of the center of the coding unit is given; for indels (all of which are insertions into the reference sequence), the position immediately before the insertion point is given.
Risk allele is based on final full regression model.
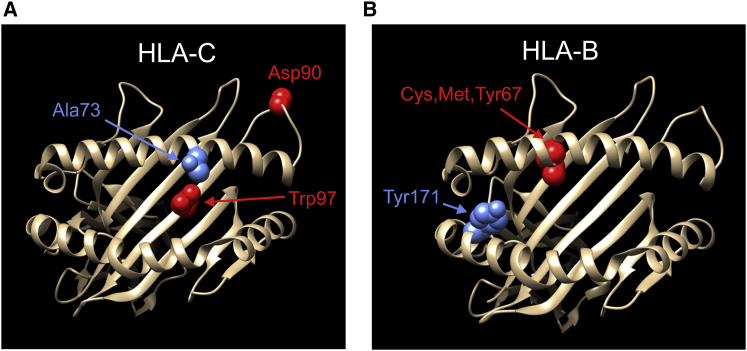
Analysis of the much larger European-ancestry dataset identified 14 independent MHC loci associated with psoriasis (Figures S26–S29). Note that unconditional lead variant rs12211087 (Figure 4) was removed by the backward elimination step after forward selection of HLA-C∗06:02 in the ninth round of the stepwise procedure. Most of the loci in the final EUR regression model are independent of each other, but modest LD is seen between some variants mapping near HLA-B and HLA-C (Figure S30). The strength of association of top-ranking variant HLA-C∗06:02 is not as dominant as seen for South Asians, contributing 47% and 58% of the variance in disease liability explained for all and the top five ranking MHC loci, respectively (Table S24). As shown in Table S25, five of the 14 lead variants alter an HLA protein (HLA-B amino acid positions 67 and 171, HLA-C∗06:02, rs41543814, and HLA-DQA1 Arg52), two (rs137854633 and rs371194629) lie within an HLA gene (HLA-B and HLA-G [MIM: 142871]), and another (rs72866766) is just 468 bp downstream of HLA-B. The location within three-dimensional (3D) ribbon models of five HLA-C and HLA-B amino acids altered by EUR risk variants is illustrated in Figure 6. However, none of the nine non-coding variants have any convincing protein-changing surrogates. Notably, three coding variants (amino acids 67 and 171 of HLA-B, rs41543814) and three non-coding variants (rs1655901, rs72866766, and rs4947340) have strong support for being causative, with posterior probabilities ranging from 0.965–1 (Table S26).
Figure 6.
Protein locations of five HLA amino acid variants with a posterior probability ≥ 0.5 of being causative in at least one of the three full MHC association models (EUR, SAS, transethnic)
Three-dimensional ribbon models for the α chains of HLA-C (A) and HLA-B (B) are based on Protein Data Bank entries 4nt6 and 2bvp, respectively, and were created with UCSF Chimera version 1.15.73 For HLA-C, shown in red is the pairwise combination of Asp90 and Trp97 that makes the HLA-C∗06:02 psoriasis risk allele unique among all 2-field HLA alleles in the minimal allele set constituting a total of 99.9% frequency for the SAS and EUR populations. The Ala73 risk allele of HLA-C, a consequence of the C allele of SNP rs41543814, is shown in blue. For HLA-B, the three residues at position 67 that significantly increase risk for psoriasis in both SAS and EUR are shown in red, and the Tyr171 risk allele is shown in blue.
Fine-mapping of the transethnic dataset revealed 17 independent psoriasis loci in the extended MHC region; their lead variants all lie within the classical MHC (Figures S31–S35). As was true of the two single ancestry analyses, HLA-C∗06 was the top-ranking variant in the full model (OR [95% CI] = 3.18 [2.95–3.43]; p = 3.2 × 10−200), although position 67 of HLA-B was also very strongly associated (multiallelic p = 1.3 × 10−134; OR [95% CI] = 1.98 [1.86–2.10], 1.30 [1.20–1.40], and 1.25 [1.18–1.33] for its cysteine, methionine, and tyrosine residues, respectively). Most of the transethnic signals are independent of each other, with only moderate LD between HLA-C∗06:02 and rs2844626 in South Asians ( = 0.40) and modest LD (0.20 ≤ < 0.40) for seven other pairs of variants in at least one population (Figure S36). LD patterns are broadly similar in EUR and SAS for this set of loci (Figure S37), although there are some substantial differences (e.g., between amino acid 67 of HLA-B and rs2844626 is 0.36 in Europeans and only 0.14 in South Asians). Only two loci in the final transethnic model are protein changing (HLA-C∗06:02 and position 67 of HLA-B), while four others occur in genes: rs1148117870 in the 5′ UTR of HLA-B, and rs1736927, rs112540072, and rs559509014/rs147145279 in introns of HLA-G, TSBP1 [MIM: 618151], and HCP5 [MIM: 604676], respectively (Tables S27 and S28). Of the 15 non-coding variants, only one has a plausible protein-changing surrogate (triallelic indel rs147145279 with surrogate biallelic missense SNP rs41556715 in MICA [MIM: 600169]) based on examination of two LD metrics in both EUR and SAS (Table S28), as well as a comparison of p values and posterior probabilities of the lead variant with possible surrogates (Tables S27–S29). Bayesian posterior probabilities are very high for three of the variants (amino acid 67 of HLA-B, rs1655901, and rs2853998) and exceed 0.50 for three others (HLA-C∗06:02, rs2884626, and rs9271539).
HLA protein-changing variants were highly (11.8-fold) and significantly (p = 4.0 × 10−5) enriched in the final EUR model compared to their proportion among tested classical MHC variants (Table S30). Similar albeit non-significant enrichments of HLA coding variants were observed for the SAS and transethnic models (6.8- and 3.6-fold, respectively). Notably, no such enrichments were observed for protein-changing variants of non-HLA genes. Multiallelic variants were also greatly enriched in all three association models for both the classical and extended MHC regions (Table S30). Compared to the whole genome, both the classical and extended MHC regions show strong enrichment for protein-changing variants and modest enrichment for structural and multiallelic variants (Table S31).
Complete results for the stepwise conditional analysis of SAS, EUR, and transethnic associations in the extended MHC region are provided in Table S32.
SAS and EUR MHC models both similar and dissimilar
We found evidence both for and against the hypothesis that genetic contributions of the MHC region to psoriasis are similar between SAS and EUR.
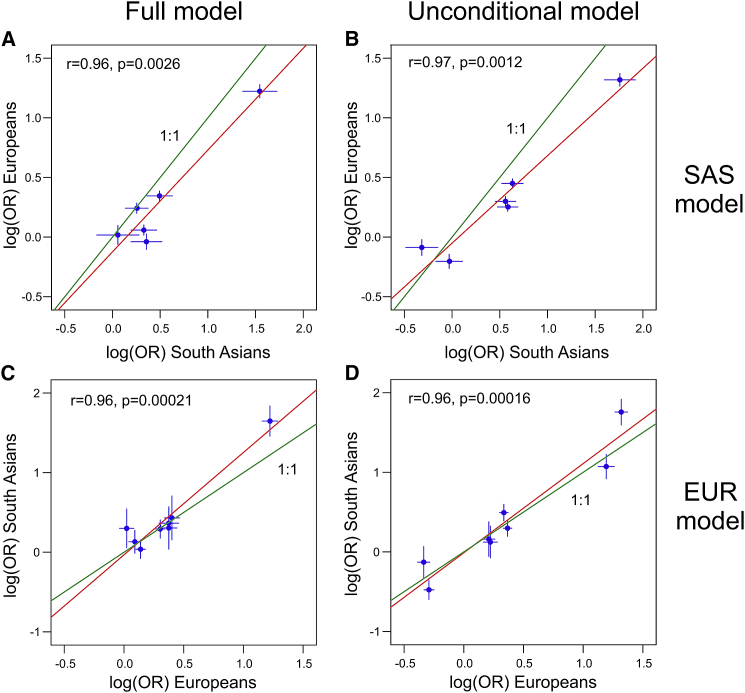
The top signals in the two association models (HLA-C∗06 in SAS and HLA-C∗06:02 in EUR) are essentially identical (LD = 0.9994 and 0.9998 in SAS and EUR, respectively). There is also good correspondence of rs9260303 in the EUR model with rs1655901 in the SAS model; these two SNPs are only 81 bp apart and 3 kb downstream of HLA-A (Tables S22 and S25) and are in substantial LD ( = 0.70 and 0.49 in SAS and EUR, respectively). Furthermore, effect sizes for the five variants in the SAS model are strongly and significantly correlated with their effect sizes when re-estimated for the EUR dataset; this is true both for full model ( = 0.96, p = 0.0026) and unconditional model ( = 0.97, p = 0.0012) coefficients (Figures 7A and 7B). Effect sizes for variants in the EUR model are also significantly correlated with their re-estimated values in the SAS dataset, whether determined for all 14 variants (Figure S38) or for only the top five variants (Figures 7C and 7D). Finally, the explanatory power of the within-population versus cross-population fits of the SAS and EUR models as measured by Tjur’s R2 are similar: R2 = 0.268 for SAS model in SAS versus 0.257 for top 5 of EUR model in SAS, and R2 = 0.304 for top 5 of EUR model in EUR versus 0.296 for SAS in EUR (Table S33).
Figure 7.
Comparison of association effect sizes in Europeans versus South Asians for the top five variants in the South Asian and European regression models for the MHC region
In (A) and (B), the log(OR) of each variant in the South Asian model as estimated in the European dataset is plotted against their estimates in the South Asian dataset. Conversely, in (C) and (D), the log(OR) of each of the top five variants in the European model as estimated in the South Asian dataset is plotted against their estimates in the European dataset. (A) and (C) plot effect sizes for the final regression model containing all variants; (B) and (D) plot unconditional effect sizes for each variant with no other variants in the regression model. Multiallelic variants with m alleles are represented by the set of m − 1 decomposed biallelic variants used for the joint Wald test. The vertical and horizontal bars show the 95% confidence intervals for these estimates in each dataset. Green and red lines depict a 1:1 correspondence and a linear fit, respectively. The Pearson correlation coefficient and its significance are shown in the upper left corner of each plot.
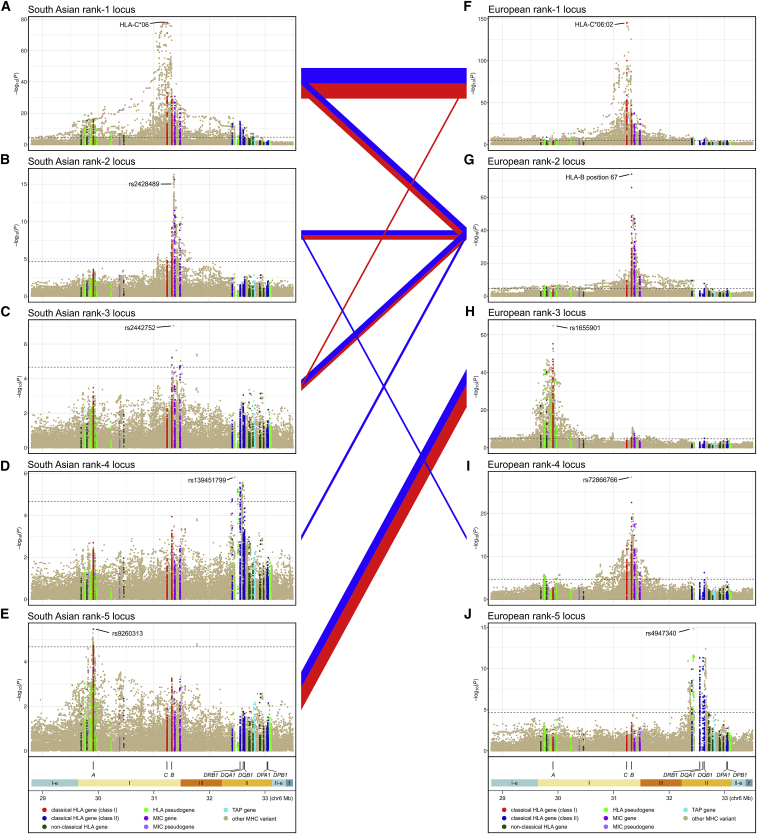
However, there are also differences between the MHC models for SAS and EUR. First, as was true of the unconditional models, the full model effect size for HLA-C∗06 in SAS is significantly greater than that seen for HLA-C∗06:02 in EUR (OR [95% CI] = 4.68 [3.99–5.50] versus 2.80 [2.59–3.03]; p = 1.9 × 10−8). Second, as shown in Figure 8, there is at best only weak correspondence between three of the five SAS loci with any of the top-ranking EUR loci. Finally, examination of goodness-of-fit measures shows much stronger statistical support (ΔAIC > 10 and evidence ratios > 50)48 for within-population MHC models than cross-population models (Table S33).
Figure 8.
Linkage disequilibrium (LD) between the top five MHC association signals in the European and South Asian models
Association plots for the five most significant psoriasis-associated MHC loci in South Asians (A–E) and Europeans (F–J) are shown in decreasing top to bottom order of their significance in the full regression model. LD (Wn2 coefficient) is depicted with line segments connecting pairs of South Asian and European loci; the color of the line indicates the population in which the LD was measured (red, South Asian; blue, European), and the thickness of the line is scaled linearly with the magnitude of LD. Only LD values of Wn2 ≥ 0.1 are shown.
We suspect that the underlying MHC association signals in SAS and EUR are largely the same, but relatively low power (Table S1), coupled with generally lower genotype imputation quality (Table S34), limits accurate identification of lead variants for SAS association signals.
Transethnic MHC model superior to monoethnic models
We used pairwise LD measures to assess the correlation of variants in the monoethnic MHC association models for SAS and EUR with those of the transethnic model. As illustrated in Figure S39, one variant in the SAS model (HLA-C∗06) has a nearly identical surrogate (HLA-C∗06:02) in the transethnic model, and two others exhibit moderately strong LD ( = 0.70 for rs9260313 and rs1655901) or moderate LD ( = 0.51 for rs2428489 and rs9266716) with transethnic variants. A similar proportion of EUR model variants have potential counterparts in the transethnic model (Figure S40): four shared loci (rs1655901 near HLA-A, HLA-C∗06:02, position 67 of HLA-B, and rs147145279), one variant in strong LD ( = 0.81 for rs6935999 and rs28573770), one in moderately strong LD ( = 0.73 for rs371194629 and rs1736927), and three in moderate LD ( = 0.55, 0.43, and 0.43 for rs6935999 with rs28573770, rs1128175 and position 67 of HLA-B, and rs41543814 and rs2844626, respectively). We compared 95% BCS for the four variants in the transethnic model that are shared with either of the two monoethnic models, finding a large increase in posterior probability for HLA-C∗06:02 in the transethnic versus the SAS or EUR models (0.664 versus 0.255 and 0.288) (Tables S23, S26, S29, and S37). The 95% credible sets for position 67 of HLA-B and rs1655901 remained unchanged for the transethnic model compared to the European model (i.e., each variant is the sole member of its set with Bayesian posterior probability = 1.000). In contrast, for the fourth shared variant, rs147145279, the size of the credible set increased and the posterior probability of the lead variant decreased for the transethnic model (Table S35). We also assessed the goodness of fit of the transethnic model to that of the two monoethnic models, finding that predictive performance of the transethnic model, as assessed by Tjur’s R2, was in all cases better than its monoethnic counterparts, but that the increase in performance was slight (Table S36). Decomposing the contribution of different regressors to Tjur’s R2 indicated that variants other than HLA-C∗06 were mostly responsible for the slight edge in goodness of fit of the transethnic models (Table S36).
Regulatory effects of non-coding MHC variants
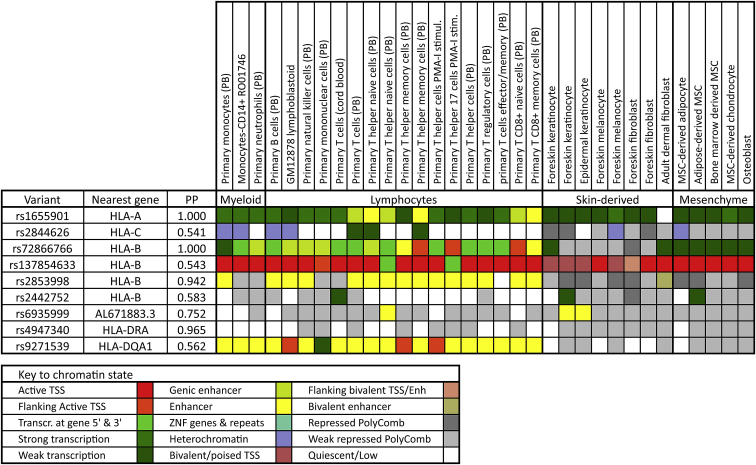
We compiled various potential regulatory features for nine non-coding loci in the final association models that have posterior probabilities exceeding 0.50. These features included cis-expression quantitative trait locus (eQTL) effects (Tables 3 and S37), chromatin state (Figure 9; Table S38), as well as conservation metrics, transcription factor binding data, and scores for the overall likelihood of being regulatory or deleterious (Table S38). Allelic variation at all but one of the nine loci was significantly associated with transcription levels of a specific target gene in a majority of the tested psoriasis-relevant tissues, and there was remarkable consistency across tissues in the direction of the effect of the risk allele on transcription for any given eQTL-gene pair. Nearly all the top target genes for these cis-eQTL effects have a known role in human immunity, including 10 different HLA genes as well as AGER (MIM: 600214), DDX39B (MIM: 142560), HSPA1L (MIM: 140559), MICA, MICB [MIM: 602436), PSMB9 (MIM: 177045), and NCR3 (MIM: 611550). One of the more interesting of these potential regulatory variants is rs137854633, whose SiPhy score indicates significant evolutionary sequence constraint, and which has modestly strong RegulomeDB and CADD scores of being regulatory and deleterious, respectively. The risk alleles of rs137854633 are one base insertions into intron 1 of HLA-B that are only 105 bp from the transcription start site and that are significantly and negatively correlated with HLA-B transcript levels in 29 of 31 tested tissues. The interval containing this variant binds at least 10 different transcription factors in chromatin immunoprecipitation sequencing (ChiP-seq) experiments and is in a region of active or poised transcription in nearly all psoriasis-relevant tissues. Other interesting regulatory candidates include rs2853998 and rs9271539, which lie in an enhancer region in most myeloid and lymphocyte cells but show Polycomb repression in skin-derived and mesenchymal cells. Two other psoriasis risk loci, rs2844626 and rs2442752, are scored by RegulomeDB as likely to affect transcription factor binding and linked to expression of a gene target, which based on cis-eQTL studies appear to be CCHCR1 (MIM: 605310) and HLA-C for the former and HLA-C and HLA-B for the latter.
Table 3.
Summary of top-ranking eGenes for noncoding psoriasis-associated MHC variants with a Bayesian posterior probability > 0.5
| Variant | Ancestry | PP | Nearest gene (position) | Top eGenesa | No. tissuesb | Distance to TSS (kb) | No. tissues with +, − risk allele effectc |
|
|---|---|---|---|---|---|---|---|---|
| Most significant eQTL | Significant eQTL | |||||||
| rs1655901 | EUR | 1.000 | HLA-A (3.1 kb downstream) | HLA-F-AS1 | 46 | 200.0 | 13, 0 | 23, 1 |
| HLA-A | 53 | 6.5 | 11, 0 | 13, 6 | ||||
| HLA-F | 40 | 225.6 | 0, 6 | 0, 15 | ||||
| rs2844626 | SAS+EUR | 0.541 | HLA-C (7.0 kb downstream) | CCHCR1 | 58 | 104.0 | 14, 0 | 29, 6 |
| HLA-C | 58 | 10.3 | 13, 0 | 35, 2 | ||||
| AL645933.2 0.2 | 43 | 133.7 | 0, 16 | 0, 27 | ||||
| rs72866766 | EUR | 1.000 | HLA-B (468 bp downstream) | MICA | 45 | 47.3 | 0, 22 | 0, 29 |
| HLA-C | 58 | 81.3 | 8, 1 | 10, 7 | ||||
| DDX39B | 58 | 188.6 | 5, 0 | 10, 0 | ||||
| rs137854633 | EUR | 0.543 | HLA-B (intron 1) | HLA-B | 31 | 0.1 | 0, 21 | 0, 29 |
| HLA-C | 44 | 85.0 | 0, 10 | 1, 29 | ||||
| MICB | 31 | 137.8 | 3, 0 | 15, 0 | ||||
| rs2853998 | SAS+EUR | 0.942 | HLA-B (2.2 kb upstream) | AL645933.2 | 40 | 36.1 | 0, 11 | 0, 22 |
| HLA-B | 40 | 2.2 | 0, 8 | 0, 35 | ||||
| HLA-C | 53 | 87.3 | 0, 7 | 3, 18 | ||||
| rs2442752 | SAS | 0.583 | AL671883.3 (7.9 kb downstream) | HLA-C | 58 | 111.9 | 11, 8 | 13, 18 |
| HLA-B | 45 | 26.8 | 5, 0 | 23, 0 | ||||
| PSORS1C3 | 48 | 173.1 | 4, 0 | 20, 0 | ||||
| rs6935999 | EUR | 0.752 | HLA-DRA (15 kb upstream) | HSPA1L | 44 | 609.3 | 0, 4 | 0, 9 |
| AGER | 44 | 240.7 | 3, 0 | 6, 0 | ||||
| HLA-DRB1 | 44 | 164.9 | 3, 0 | 5, 1 | ||||
| NCR3 | 37 | 831.2 | 0, 3 | 0, 3 | ||||
| rs4947340 | EUR | 0.965 | HLA-DRA (23 kb upstream) | HLA-DQA2 | 34 | 273.8 | 18, 0 | 33, 0 |
| HLA-DRB1 | 49 | 122.3 | 0, 14 | 0, 48 | ||||
| HLA-DQB2 | 33 | 296.0 | 6, 0 | 30, 1 | ||||
| rs9271539 | SAS+EUR | 0.562 | HLA-DQA1 (15 kb upstream) | HLA-DRB5 | 52 | 92.0 | 28, 0 | 49, 0 |
| HLA-DRB1 | 52 | 32.4 | 12,0 | 22, 4 | ||||
| HLA-DQA1 | 36 | 15.2 | 1, 1 | 8, 5 | ||||
| PSMB9 | 52 | 231.9 | 0, 2 | 0, 24 | ||||
Abbreviations: PP, posterior probability; eGene, gene whose expression is significantly associated with an eQTL; eQTL, expression quantitative trait locus; EUR, European; SAS, South Asian; TSS, transcription start site.
Expressed genes with a TSS within 1 Mb of the variant were ranked by the number of 58 psoriasis-relevant tissues for which the eQTL effects of that variant upon the gene were the most significant. The top three ranking eGenes are shown for each variant (four eGenes shown for two variants because of ties).
Number of 58 tissues from 16 RNA expression studies for which the eQTL effect of the variant on the eGene was assessed. See Table S37 for citations of the 16 expression studies.
Figure 9.
Chromatin states in 33 psoriasis-relevant cell types for psoriasis-associated noncoding variants with a Bayesian posterior probability exceeding 0.50
Chromatin states are derived from the 15-state Roadmap Epigenomics core model based on five chromatin marks.74 Abbreviations: MSC, mesenchymal stem cell; PB, peripheral blood; PMA-I, PMA-ionomycin treatment; PP, posterior probability (Bayesian); TSS, transcription start site.
Reduced extent of LD for SAS versus EUR
We assessed whether there are differences in LD structure between SAS and EUR that may be contributing to the reduction in size of Bayesian credible sets observed for some psoriasis loci in our transethnic models. For non-MHC loci, using EUR and SAS samples of the 1KGP, we found fewer LD surrogates on average for the SAS population—44%, 35%, and 21% of the loci had fewer, equal, or more surrogates in SAS compared to EUR (Figure S41A). We were also able to demonstrate positive albeit nonsignificant correlations between the log2 ratio of the number of LD proxies in EUR versus SAS with both the −log10 (transethnic association p value) (Figure S41B) and with the log2 EUR versus transethnic ratio of the number of markers in the 95% BCS (Figure S41C). Within the MHC region, as shown in Figure S42, the slope of the linear fit for SAS LD values regressed onto EUR LD values was substantially and significantly different from 1.00 (slope = 0.79 and 0.81, with p = 9 × 10−29 and 3 × 10−25 for and LD coefficients, respectively) when considering all 919 unique pairwise combinations of the MHC loci selected by the three stepwise ethnic analyses.
Discussion
We first performed a GWAS of psoriasis in individuals of South Asian descent, followed by a fine-mapping study of MHC psoriasis signals for South Asians and the largest analysis to date of psoriasis MHC signals for people of European ancestry. Finding that effect sizes for known psoriasis loci were strongly correlated in EUR and SAS (ρ = 0.78; p < 2 × 10−14; Figure 1), we conducted a transethnic meta-analysis of non-MHC and MHC signals, identifying two loci that had not previously reached genome-wide significance in EUR populations (Table 1; Figure 2). Inspection of eQTL databases revealed potential regulatory roles for both these signals. For 1p36.22, rs2103876 is a cis-eQTL for multiple genes in blood,75 including MFN2 (MIM: 608507), MIIP (MIM: 608772), MTHFR (MIM: 607093), and CLCN6 (MIM: 602726). MTHFR, which encodes methylenetetrahydrofolate reductase, is the only gene whose expression is positively correlated (p = 4.2 × 10−97) with the psoriasis risk allele (T) and significantly upregulated in psoriatic lesional skin (p = 3.06 × 10−23; fold change [FC] = 1.7).76 Methylenetetrahydrofolate reductase is one of several targets of methotrexate, a highly effective antipsoriatic drug.77 For the 1q24.2 locus, the only significant eQTL target for rs12046909 in whole blood is XCL1 (MIM: 600250),75 whose psoriasis risk allele is negatively associated with XCL1 expression, with no significant eQTL for this genetic variation in other tissues according to GTEx.78 XCL1 encodes a ligand for chemokine receptor XCR1 (MIM: 600552), which is expressed on dendritic cells, and XCL1 is overexpressed in lesional psoriatic versus normal skin (p = 0.011; FC = 1.95).76
Transethnic association studies should improve resolution for fine mapping of genetic variants by leveraging differences in LD architecture among different global populations.30 Our analysis of both non-MHC and MHC psoriasis associations revealed that on average the range of LD with their lead variants was more limited in South Asians than Europeans. The faster decay of LD in SAS versus EUR, which has been reported previously,30 may have contributed to the improved fine-mapping resolution our transethnic models achieved for many associated loci, beyond that afforded by a simple increase in sample size.
Based on their known immunologic functions and large observed effect sizes, HLA protein variants are leading causative candidates for psoriasis.27,79 We improved an existing method to construct high-quality HLA reference panels for South Asians and Europeans and to then impute HLA amino acid, protein, and SNP variants with excellent accuracy. To increase the density and scope of tested MHC variants, we integrated the imputed HLA variants with variants imputed using the 1KGP and HRC reference panels in a 12 Mb interval that encompasses the classical MHC region. For people of EUR ancestry, our MHC fine-mapping analysis surpasses our most recent effort22 in many respects, including effective sample size (39,335 versus 21,137), MHC interval (12 versus 3.7 Mb), total number of tested variants (83,352 versus 8,739), number of tested classical MHC variants (49,407 versus 8,556), and imputed variant quality ( ≥ 0.70 versus no filter). We also included joint association testing of the alleles of multiallelic variants, not only for HLA genes but for the entire 12 Mb extended MHC. To fully incorporate results of multiallelic with biallelic association tests, we assessed 12 metrics for multiallelic LD and utilized the best-performing two, and we also devised a method to compute Bayes factors for multiallelic variants.
Our analysis of the extended MHC region uncovered five independent psoriasis loci in SAS, 14 in EUR, and 17 for the transethnic analysis. Notably, the lead variants for all these risk loci are restricted to the classical MHC. Our efforts to improve HLA variant imputation quality and include multiallelic variants were justified by their significant enrichment in the final regression models. Despite the enrichment of HLA variants, only eight of the 36 identified risk variants change HLA proteins, compared to all six psoriasis loci from our previous MHC fine-mapping study.22 However, four of the non-coding MHC loci lie within HLA protein-coding genes, and 11 are intergenic with an HLA protein-coding gene as their nearest neighbor. Furthermore, based on the LD measure, the best protein-changing surrogates are in HLA genes for 29 of the 36 identified risk loci, even though HLA proteins represent only 19 of 137 coding genes in the classical MHC. Interestingly, the best coding surrogates for five of the remaining seven risk variants are in MICA and TAP2, genes with important immune system functions, whose proteins are either structurally akin to class I HLA molecules (MICA) or involved in translocation of short peptides from cytosol to endoplasmic reticulum for binding to MHC class I proteins (TAP2). Five of the eight HLA coding risk variants have Bayesian posterior probabilities of being causative that exceed 0.50: HLA-C∗06:02 for the transethnic analysis, which differs from other common 2-field HLA-C alleles by amino acid combination Asp90 and Trp97 and by underlying two-SNP haplotype variant rs1131123-T/rs1131118-A, the multiallelic amino acid at position 67 of HLA-B for the European and transethnic analyses, and the essentially biallelic amino acids at positions 171 of HLA-B and 73 of HLA-C for the European analysis. Notably, all these amino acid variants except Asp90 are not only in the antigen-binding groove of the HLA protein (Figure 6) but also interact with bound peptide rather than the T cell receptor.80
Most (28/36) of the identified MHC risk variants are noncoding. Nine of these noncoding variants have posterior probabilities exceeding 0.50, indicating that some of the psoriasis loci in the MHC may have a regulatory function. Most of these nine variants exhibit strong and consistent cis-eQTL effects on target genes with a known function in immunity in a wide variety of psoriasis-relevant tissues. Several of these variants also show evidence of affecting transcription factor binding or correlation with chromatin states indicative of active transcription, enhancers, or regions of Polycomb suppression (Figure 9).
HLA-C∗06 was the top-ranking psoriasis locus for both SAS and EUR, but its strength of association was significantly higher for SAS in both the unconditional and final full regression models. Furthermore, the contribution of HLA-C∗06 to overall psoriasis susceptibility compared to other MHC variants was higher for SAS based on variance in liability explained and goodness-of-fit measures such as AIC and Tjur’s , even when the comparison was restricted to the top five ranking loci in the EUR model. Because HLA-C∗06 is more strongly associated with purely cutaneous psoriasis than with PsA in both EUR-origin individuals27 and our SAS sample (OR [95% CI] = 6.54 [5.50–7.79] versus 5.35 [4.02–7.13]), the stronger HLA-C∗06 association in SAS could be related to the lower prevalence of PsA in SAS (8.7% in the literature81 and 13.0% in our sample) compared to EUR (25%–30%).82 The lower prevalence of PsA in SAS may be a consequence of the substantially lower frequency of several known HLA-B risk alleles for PsA (B∗08, B∗27, B∗38, and B∗39)83,84 in SAS versus EUR that we observed for both our own data (Figures S17B and S19B) and data from the National Marrow Donor Program (NMDP) (Figure S21).
Other than HLA-C∗06, the only other possibly shared locus between the two monoethnic MHC association models is a signal 3 kb downstream of HLA-A. The similarity of effect sizes for the variants in one ethnic model compared to their refitted values for the other ethnic group argues that MHC associations for the two groups may not be as different as the two final regression models seem to indicate but may stem more from a lack of power in the relatively small SAS study to accurately identify signals other than HLA-C∗06. Despite its modest power, combining the SAS dataset with EUR samples in a transethnic analysis did bolster the evidence that the HLA-C∗06 protein variant is causal, increasing its Bayesian posterior probability from 0.255 and 0.288 in the SAS and EUR models to 0.664 in the transethnic model.
Our encouraging findings for the MHC region need to be tempered in light of several limitations. First, many variants in the region were not tested for association. As shown in Table S18, only 42%–52% of the classical MHC variants in the reference panels used for imputation had a high enough imputation quality to qualify for testing, including only 68%–80% and 72%–84% of common (MAF ≥ 0.05) and low-frequency (MAF = 0.01–0.05) variants, respectively. Furthermore, amino acid and full-length protein variants were characterized and tested for only eight of the 137 coding genes in the region (Table S14). The 1KGP and HRC reference panels, which are based on short-read sequences aligned to the reference genome, are deficient in their coverage of indels and larger structural variants in the MHC, as was demonstrated by a Danish study that used a de novo assembly approach.85,86 Second, variation in imputation quality among the tested variants is predicted to affect their power to detect association independent of their true association with disease.87 We demonstrated this for all three of our ancestry datasets, finding a significant positive correlation of full model significance of association with imputation quality for variants in substantial LD with HLA-C∗06 (Figure S43). Third, stepwise regression is unlikely to find the best possible association model, owing to the infeasibility of testing all possible subsets of variants, and to the adverse effects of LD-produced collinearity among genuinely independent susceptibility loci, which can lead to unstable coefficient estimates, inflated standard errors, and a downward bias of their relative importance compared to variants that are uncorrelated with others already in the model.88 We hypothesize that many of the sizeable differences among our three MHC association models arise from these limitations and the modest power of the SAS dataset, rather than from differences in the true but unknown models. However, until this hypothesis can be validated, we believe it safest to treat each model as the best current summary of MHC psoriasis associations for its corresponding population.
To remedy these issues, methods are needed that can address the unique challenges posed by the MHC region. Genotyping every variant in the MHC with close to 100% accuracy is an essential first step, which may require intensive sequence-based approaches with de novo alignment, deep read coverage, very long reads for more accurate haplotype phasing, and multiple libraries with insert sizes ranging from small to very large for proper typing of complex structural variations. Improved association methods are also needed that can better handle large numbers of correlated variants. Notwithstanding these challenges, our results demonstrate the value of genotyping diverse populations both within and beyond the MHC.
Acknowledgments
The authors thank Yang Luo and Soumya Raychaudhuri (Broad Institute of Harvard and MIT) for advice concerning the construction of SNP2HLA reference panels. This work was supported by awards from the National Institutes of Health (R01AR042742, R01AR050511, R01AR054966, R01AR063611, and R01AR065183 to J.T.E.; K01AR072129 to L.C.T.). L.C.T. was also supported by the Dermatology Foundation, the National Psoriasis Foundation, and the Arthritis National Research Foundation. L.C.T., P.E.S., T.T., J.J.V., R.P.N., and J.T.E. are supported by the Dawn and Dudley Holmes Foundation and the Babcock Memorial Trust. J.T.E. is supported by the Ann Arbor Veterans Affairs Hospital. We also wish to acknowledge the Indian Council of Medical Research for support of a foreign collaboration project with the University of Michigan, Ann Arbor, USA funded by National Instititutes of Health, USA (project title Genetic Analysis of Psoriasis and Psoriatic Arthritis in Indians; file no. 50/2/2009-BMS; project code N-1170).
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2021.100069.
Data and code availability
Complete summary statistics for the genome-wide association analyses of the SAS, EUR, and transethnic (SAS and EUR) datasets have been deposited at the NHGRI-EBI GWAS Catalog (accession numbers GCST90019015, GCST90019016, and GCST90019017). Individual-level genotype data for the CASP GWAS, PsA GWAS, and Exomechip case-control studies are available on dbGaP (dbGaP: phs000019.v1.p1, phs000982.v1.p1, and phs001306.v1.p1). Genotypes for the WTCCC2 psoriasis-control study are archived at the European Genome-Phenome Archive (study ID EGAS00000000108) and can be requested by contacting the data access committee at the Wellcome Trust Sanger Institute (datasharing@sanger.ac.uk). Data sharing restrictions do not allow making genotypes available for the remaining six case-control cohorts analyzed by this study (Kiel GWAS, Genizon GWAS, PAGE and GAPC Immunochip studies, batches 1 and 2+3 of SAS GWAS).
Our updated version of the SNP2HLA imputation package is available on GitHub. Five datasets (UM, 1KGP-ALL-v2, T1DGC, BKT, IKMB; see Table S8) were used to build the best-performing MHC reference panels of this study. Our GitHub repository contains three MHC reference panels built by applying our updated methods to the two datasets (UM, 1KGP-ALL-v2) for which genotype data can be freely shared. Genotype data for the T1DGC dataset can requested from the NIDDK Central Repository. Data restrictions preclude sharing any individual-level genotype data for the BKT and IKMB datasets; however, the MHC reference data for the IKMB dataset is available as a HIBAG model for imputing HLA alleles (IKMB models).
Web resources
1000 Genomes, phase 3, Affymetrix 6.0 microarray data, http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/supporting/hd_genotype_chip/
1000 Genomes, phase 3, HLA genotypes, http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/HLA_types/
1000 Genomes, v5 phase3, full integrated variant call set, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/
1000 Genomes, v5 phase3, reduced integrated variant call set (no monomorphic or singleton sites), http://csg.sph.umich.edu/abecasis/MACH/download/1000G.Phase3.v5.html
Beagle 4.1, https://faculty.washington.edu/browning/beagle/b4_1.html
CADD v1.6, https://cadd.gs.washington.edu/
eQTLGen Consortium, https://www.eqtlgen.org/
European Genome-Phenome Archive, https://ega-archive.org/
GTEx Portal, https://www.gtexportal.org/home/
HIBAG model for imputing HLA alleles, https://hibag.s3.amazonaws.com/index.html
IPD-IMGT/HLA database, https://www.ebi.ac.uk/ipd/imgt/hla/
logistf R package, https://cran.r-project.org/web/packages/logistf/index.html
Michigan Imputation Server, https://imputationserver.sph.umich.edu/index.html#!
NHGRI-EBI GWAS Catalog, https://www.ebi.ac.uk/gwas/
OMIM, https://www.omim.org/
Pan-Asian SNP2HLA reference panel, http://software.broadinstitute.org/mpg/snp2hla/
Plink 2.0, https://www.cog-genomics.org/plink/
RegulomeDB 2.0, https://regulomedb.org/regulome-search/
SNP2HLA imputation package, https://github.com/CutaneousBioinf/SNP2HLA_UM
SNP2HLA v1.0.3, http://software.broadinstitute.org/mpg/snp2hla/
T1DGC SNP2HLA reference panel, https://repository.niddk.nih.gov/studies/t1dgc-special/
Supplemental information
References
- 1.Gudjonsson J.E., Elder J.T. In: Kang S., Amagai M., Bruckner A.L., Enk A.H., McMichael A.J., Orringer J.S., editors. McGraw-Hill; New York: 2018. Psoriasis; pp. 457–498. (Dermatology in General Medicine). [Google Scholar]
- 2.Kim J., Krueger J.G. The immunopathogenesis of psoriasis. Dermatol. Clin. 2015;33:13–23. doi: 10.1016/j.det.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Greb J.E., Goldminz A.M., Elder J.T., Lebwohl M.G., Gladman D.D., Wu J.J., Mehta N.N., Finlay A.Y., Gottlieb A.B. Psoriasis. Nat. Rev. Dis. Primers. 2016;2:16082. doi: 10.1038/nrdp.2016.82. [DOI] [PubMed] [Google Scholar]
- 4.Parisi R., Symmons D.P., Griffiths C.E., Ashcroft D.M., Identification and Management of Psoriasis and Associated ComorbidiTy (IMPACT) project team Global epidemiology of psoriasis: a systematic review of incidence and prevalence. J. Invest. Dermatol. 2013;133:377–385. doi: 10.1038/jid.2012.339. [DOI] [PubMed] [Google Scholar]
- 5.Schäfer T. Epidemiology of psoriasis. Review and the German perspective. Dermatology. 2006;212:327–337. doi: 10.1159/000092283. [DOI] [PubMed] [Google Scholar]
- 6.Naldi L. Epidemiology of psoriasis. Curr. Drug Targets Inflamm. Allergy. 2004;3:121–128. doi: 10.2174/1568010043343958. [DOI] [PubMed] [Google Scholar]
- 7.Patrick M.T., Stuart P.E., Raja K., Gudjonsson J.E., Tejasvi T., Yang J., Chandran V., Das S., Callis-Duffin K., Ellinghaus E., et al. Genetic signature to provide robust risk assessment of psoriatic arthritis development in psoriasis patients. Nat. Commun. 2018;9:4178. doi: 10.1038/s41467-018-06672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stuart P.E., Tsoi L.C., Hambro C.A., Elder J.T. In: Gladman D.D., editor. Oxford University Press; New York: 2018. Genetics of Psoriasis; pp. 35–55. (Textbook of Psoriatic Arthritis). [Google Scholar]
- 9.Hirata J., Hirota T., Ozeki T., Kanai M., Sudo T., Tanaka T., Hizawa N., Nakagawa H., Sato S., Mushiroda T., et al. Variants at HLA-A, HLA-C, and HLA-DQB1 Confer Risk of Psoriasis Vulgaris in Japanese. J. Invest. Dermatol. 2018;138:542–548. doi: 10.1016/j.jid.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Tsoi L.C., Stuart P.E., Tian C., Gudjonsson J.E., Das S., Zawistowski M., Ellinghaus E., Barker J.N., Chandran V., Dand N., et al. Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nat. Commun. 2017;8:15382. doi: 10.1038/ncomms15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozawa A., Ohkido M., Inoko H., Ando A., Tsuji K. Specific restriction fragment length polymorphism on the HLA-C region and susceptibility to psoriasis vulgaris. J. Invest. Dermatol. 1988;90:402–405. doi: 10.1111/1523-1747.ep12456500. [DOI] [PubMed] [Google Scholar]
- 12.Kim T.G., Lee H.J., Youn J.I., Kim T.Y., Han H. The association of psoriasis with human leukocyte antigens in Korean population and the influence of age of onset and sex. J. Invest. Dermatol. 2000;114:309–313. doi: 10.1046/j.1523-1747.2000.00863.x. [DOI] [PubMed] [Google Scholar]
- 13.Choonhakarn C., Romphruk A., Puapairoj C., Jirarattanapochai K., Romphruk A., Leelayuwat C. Haplotype associations of the major histocompatibility complex with psoriasis in Northeastern Thais. Int. J. Dermatol. 2002;41:330–334. doi: 10.1046/j.1365-4362.2002.01496.x. [DOI] [PubMed] [Google Scholar]
- 14.Shaiq P.A., Stuart P.E., Latif A., Schmotzer C., Kazmi A.H., Khan M.S., Azam M., Tejasvi T., Voorhees J.J., Raja G.K., et al. Genetic associations of psoriasis in a Pakistani population. Br. J. Dermatol. 2013;169:406–411. doi: 10.1111/bjd.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munir S., ber Rahman S., Rehman S., Saba N., Ahmad W., Nilsson S., Mazhar K., Naluai Å.T. Association analysis of GWAS and candidate gene loci in a Pakistani population with psoriasis. Mol. Immunol. 2015;64:190–194. doi: 10.1016/j.molimm.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 16.Umapathy S., Pawar A., Mitra R., Khuperkar D., Devaraj J.P., Ghosh K., Khopkar U. Hla-a and hla-B alleles associated in psoriasis patients from mumbai, Western India. Indian J. Dermatol. 2011;56:497–500. doi: 10.4103/0019-5154.87128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Indhumathi S., Rajappa M., Chandrashekar L., Ananthanarayanan P.H., Thappa D.M., Negi V.S. The HLA-C∗06 allele as a possible genetic predisposing factor to psoriasis in South Indian Tamils. Arch. Dermatol. Res. 2016;308:193–199. doi: 10.1007/s00403-016-1618-y. [DOI] [PubMed] [Google Scholar]
- 18.Chandra A., Lahiri A., Senapati S., Basu B., Ghosh S., Mukhopadhyay I., Behra A., Sarkar S., Chatterjee G., Chatterjee R. Increased Risk of Psoriasis due to combined effect of HLA-Cw6 and LCE3 risk alleles in Indian population. Sci. Rep. 2016;6:24059. doi: 10.1038/srep24059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marigorta U.M., Navarro A. High trans-ethnic replicability of GWAS results implies common causal variants. PLoS Genet. 2013;9:e1003566. doi: 10.1371/journal.pgen.1003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y.R., Keating B.J. Trans-ethnic genome-wide association studies: advantages and challenges of mapping in diverse populations. Genome Med. 2014;6:91. doi: 10.1186/s13073-014-0091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marigorta U.M., Rodríguez J.A., Gibson G., Navarro A. Replicability and Prediction: Lessons and Challenges from GWAS. Trends Genet. 2018;34:504–517. doi: 10.1016/j.tig.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okada Y., Han B., Tsoi L.C., Stuart P.E., Ellinghaus E., Tejasvi T., Chandran V., Pellett F., Pollock R., Bowcock A.M., et al. Fine mapping major histocompatibility complex associations in psoriasis and its clinical subtypes. Am. J. Hum. Genet. 2014;95:162–172. doi: 10.1016/j.ajhg.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia X., Han B., Onengut-Gumuscu S., Chen W.M., Concannon P.J., Rich S.S., Raychaudhuri S., de Bakker P.I. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nair R.P., Henseler T., Jenisch S., Stuart P., Bichakjian C.K., Lenk W., Westphal E., Guo S.W., Christophers E., Voorhees J.J., Elder J.T. Evidence for two psoriasis susceptibility loci (HLA and 17q) and two novel candidate regions (16q and 20p) by genome-wide scan. Hum. Mol. Genet. 1997;6:1349–1356. doi: 10.1093/hmg/6.8.1349. [DOI] [PubMed] [Google Scholar]
- 25.Nair R.P., Duffin K.C., Helms C., Ding J., Stuart P.E., Goldgar D., Gudjonsson J.E., Li Y., Tejasvi T., Feng B.J., et al. Collaborative Association Study of Psoriasis Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat. Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellinghaus E., Ellinghaus D., Stuart P.E., Nair R.P., Debrus S., Raelson J.V., Belouchi M., Fournier H., Reinhard C., Ding J., et al. Genome-wide association study identifies a psoriasis susceptibility locus at TRAF3IP2. Nat. Genet. 2010;42:991–995. doi: 10.1038/ng.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuart P.E., Nair R.P., Tsoi L.C., Tejasvi T., Das S., Kang H.M., Ellinghaus E., Chandran V., Callis-Duffin K., Ike R., et al. Genome-wide Association Analysis of Psoriatic Arthritis and Cutaneous Psoriasis Reveals Differences in Their Genetic Architecture. Am. J. Hum. Genet. 2015;97:816–836. doi: 10.1016/j.ajhg.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strange A., Capon F., Spencer C.C., Knight J., Weale M.E., Allen M.H., Barton A., Band G., Bellenguez C., Bergboer J.G., et al. Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Control Consortium 2 A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 2010;42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsoi L.C., Spain S.L., Knight J., Ellinghaus E., Stuart P.E., Capon F., Ding J., Li Y., Tejasvi T., Gudjonsson J.E., et al. Collaborative Association Study of Psoriasis (CASP) Genetic Analysis of Psoriasis Consortium. Psoriasis Association Genetics Extension. Wellcome Trust Case Control Consortium 2 Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., et al. Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wittig M., Anmarkrud J.A., Kässens J.C., Koch S., Forster M., Ellinghaus E., Hov J.R., Sauer S., Schimmler M., Ziemann M., et al. Development of a high-resolution NGS-based HLA-typing and analysis pipeline. Nucleic Acids Res. 2015;43:e70. doi: 10.1093/nar/gkv184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Browning B.L., Browning S.R. Genotype Imputation with Millions of Reference Samples. Am. J. Hum. Genet. 2016;98:116–126. doi: 10.1016/j.ajhg.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson J., Barker D.J., Georgiou X., Cooper M.A., Flicek P., Marsh S.G.E. IPD-IMGT/HLA Database. Nucleic Acids Res. 2020;48(D1):D948–D955. doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marsh S.G., Albert E.D., Bodmer W.F., Bontrop R.E., Dupont B., Erlich H.A., Fernández-Viña M., Geraghty D.E., Holdsworth R., Hurley C.K., et al. An update to HLA nomenclature, 2010. Bone Marrow Transplant. 2010;45:846–848. doi: 10.1038/bmt.2010.79. [DOI] [PubMed] [Google Scholar]
- 37.Zhang P., Zhan X., Rosenberg N.A., Zöllner S. Genotype imputation reference panel selection using maximal phylogenetic diversity. Genetics. 2013;195:319–330. doi: 10.1534/genetics.113.154591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Degenhardt F., Wendorff M., Wittig M., Ellinghaus E., Datta L.W., Schembri J., Ng S.C., Rosati E., Hübenthal M., Ellinghaus D., et al. Construction and benchmarking of a multi-ethnic reference panel for the imputation of HLA class I and II alleles. Hum. Mol. Genet. 2019;28:2078–2092. doi: 10.1093/hmg/ddy443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T., et al. International Multiple Sclerosis Genetics Consortium. International IBD Genetics Consortium Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mychaleckyj J.C., Noble J.A., Moonsamy P.V., Carlson J.A., Varney M.D., Post J., Helmberg W., Pierce J.J., Bonella P., Fear A.L., et al. T1DGC HLA genotyping in the international Type 1 Diabetes Genetics Consortium. Clin. Trials. 2010;7(1, Suppl):S75–S87. doi: 10.1177/1740774510373494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onengut-Gumuscu S., Chen W.M., Burren O., Cooper N.J., Quinlan A.R., Mychaleckyj J.C., Farber E., Bonnie J.K., Szpak M., Schofield E., et al. Type 1 Diabetes Genetics Consortium Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pillai N.E., Okada Y., Saw W.Y., Ong R.T., Wang X., Tantoso E., Xu W., Peterson T.A., Bielawny T., Ali M., et al. Predicting HLA alleles from high-resolution SNP data in three Southeast Asian populations. Hum. Mol. Genet. 2014;23:4443–4451. doi: 10.1093/hmg/ddu149. [DOI] [PubMed] [Google Scholar]
- 43.Okada Y., Kim K., Han B., Pillai N.E., Ong R.T., Saw W.Y., Luo M., Jiang L., Yin J., Bang S.Y., et al. Risk for ACPA-positive rheumatoid arthritis is driven by shared HLA amino acid polymorphisms in Asian and European populations. Hum. Mol. Genet. 2014;23:6916–6926. doi: 10.1093/hmg/ddu387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abi-Rached L., Gouret P., Yeh J.H., Di Cristofaro J., Pontarotti P., Picard C., Paganini J. Immune diversity sheds light on missing variation in worldwide genetic diversity panels. PLoS ONE. 2018;13:e0206512. doi: 10.1371/journal.pone.0206512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raychaudhuri S., Sandor C., Stahl E.A., Freudenberg J., Lee H.S., Jia X., Alfredsson L., Padyukov L., Klareskog L., Worthington J., et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heinze G., Schemper M. A solution to the problem of separation in logistic regression. Stat. Med. 2002;21:2409–2419. doi: 10.1002/sim.1047. [DOI] [PubMed] [Google Scholar]
- 48.Burnham K.P., Anderson D.R. Multimodel inference: Understanding AIC and BIC in model selection. Sociol. Methods Res. 2004;33:261–304. [Google Scholar]
- 49.Tjur T. Coefficients of determination in logistic regression models—a new proposal: the coefficient of discrimination. Am. Stat. 2009;63:366–372. [Google Scholar]
- 50.Shorrocks A.F. Decomposition procedures for distributional analysis: a unified framework based on the Shapley value. J. Econ. Inequal. 2013;11:99–126. [Google Scholar]
- 51.Wakefield J. Bayes factors for genome-wide association studies: comparison with P-values. Genet. Epidemiol. 2009;33:79–86. doi: 10.1002/gepi.20359. [DOI] [PubMed] [Google Scholar]
- 52.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maller J.B., McVean G., Byrnes J., Vukcevic D., Palin K., Su Z., Howson J.M., Auton A., Myers S., Morris A., et al. Wellcome Trust Case Control Consortium Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wen X., Stephens M. Bayesian Methods for Genetic Association Analysis with Heterogeneous Subgroups: From Meta-Analyses to Gene-Environment Interactions. Ann. Appl. Stat. 2014;8:176–203. doi: 10.1214/13-AOAS695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wen X. Bayesian model selection in complex linear systems, as illustrated in genetic association studies. Biometrics. 2014;70:73–83. doi: 10.1111/biom.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao H., Nettleton D., Soller M., Dekkers J.C. Evaluation of linkage disequilibrium measures between multi-allelic markers as predictors of linkage disequilibrium between markers and QTL. Genet. Res. 2005;86:77–87. doi: 10.1017/S001667230500769X. [DOI] [PubMed] [Google Scholar]
- 57.Zhao H., Nettleton D., Dekkers J.C. Evaluation of linkage disequilibrium measures between multi-allelic markers as predictors of linkage disequilibrium between single nucleotide polymorphisms. Genet. Res. 2007;89:1–6. doi: 10.1017/S0016672307008634. [DOI] [PubMed] [Google Scholar]
- 58.Hedrick P.W., Thomson G. A two-locus neutrality test: applications to humans, E. coli and lodgepole pine. Genetics. 1986;112:135–156. doi: 10.1093/genetics/112.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamazaki T. The effects of overdominance of linkage in a multilocus system. Genetics. 1977;86:227–236. doi: 10.1093/genetics/86.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomson G., Single R.M. Conditional asymmetric linkage disequilibrium (ALD): extending the biallelic r2 measure. Genetics. 2014;198:321–331. doi: 10.1534/genetics.114.165266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nothnagel M., Fürst R., Rohde K. Entropy as a measure for linkage disequilibrium over multilocus haplotype blocks. Hum. Hered. 2002;54:186–198. doi: 10.1159/000070664. [DOI] [PubMed] [Google Scholar]
- 62.Liu Z., Lin S. Multilocus LD measure and tagging SNP selection with generalized mutual information. Genet. Epidemiol. 2005;29:353–364. doi: 10.1002/gepi.20092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okada Y., Momozawa Y., Ashikawa K., Kanai M., Matsuda K., Kamatani Y., Takahashi A., Kubo M. Construction of a population-specific HLA imputation reference panel and its application to Graves’ disease risk in Japanese. Nat. Genet. 2015;47:798–802. doi: 10.1038/ng.3310. [DOI] [PubMed] [Google Scholar]
- 64.Cramer H. Princeton University Press; Princeton, NJ: 1946. Mathematical Models of Statistics. [Google Scholar]
- 65.Zuo X., Sun L., Yin X., Gao J., Sheng Y., Xu J., Zhang J., He C., Qiu Y., Wen G., et al. Whole-exome SNP array identifies 15 new susceptibility loci for psoriasis. Nat. Commun. 2015;6:6793. doi: 10.1038/ncomms7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Rielly D.D., Jani M., Rahman P., Elder J.T. The Genetics of Psoriasis and Psoriatic Arthritis. J. Rheumatol. Suppl. 2019;95:46–50. doi: 10.3899/jrheum.190119. [DOI] [PubMed] [Google Scholar]
- 67.Motyer A., Vukcevic D., D’ilthey A., Donnelly P., McVean G., Lesie S. Practical use of methods for imputation of HLA alleles from SNP genotype data. bioRxiv. 2016 doi: 10.1101/091009. [DOI] [Google Scholar]
- 68.Luo Y., Kanai M., Choi W., Li X., Sakaue S., Yamamoto K., Ogawa K., Gutierrez-Arcelus M., Gregersen P.K., Stuart P.E., et al. NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium A high-resolution HLA reference panel capturing global population diversity enables multi-ancestry fine-mapping in HIV host response. Nat. Genet. 2021;53:1504–1516. doi: 10.1038/s41588-021-00935-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M., et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gragert L., Madbouly A., Freeman J., Maiers M. Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Hum. Immunol. 2013;74:1313–1320. doi: 10.1016/j.humimm.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 71.Horton R., Wilming L., Rand V., Lovering R.C., Bruford E.A., Khodiyar V.K., Lush M.J., Povey S., Talbot C.C., Jr., Wright M.W., et al. Gene map of the extended human MHC. Nat. Rev. Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 72.So H.C., Gui A.H., Cherny S.S., Sham P.C. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet. Epidemiol. 2011;35:310–317. doi: 10.1002/gepi.20579. [DOI] [PubMed] [Google Scholar]
- 73.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 74.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., et al. Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vosa U., Claringbould A., Westra H.-J., Bonder M.J., Deelen P., Zeng B., Kirsten H., others, Visscher P.M., Scholz M., et al. Unraveling the polygenic architecture of complex traits using blood eQTL meta-analysis. bioRxiv. 2018 doi: 10.1101/447367. [DOI] [Google Scholar]
- 76.Tsoi L.C., Rodriguez E., Degenhardt F., Baurecht H., Wehkamp U., Volks N., Szymczak S., Swindell W.R., Sarkar M.K., Raja K., et al. Atopic Dermatitis Is an IL-13-Dominant Disease with Greater Molecular Heterogeneity Compared to Psoriasis. J. Invest. Dermatol. 2019;139:1480–1489. doi: 10.1016/j.jid.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cronstein B.N., Aune T.M. Methotrexate and its mechanisms of action in inflammatory arthritis. Nat. Rev. Rheumatol. 2020;16:145–154. doi: 10.1038/s41584-020-0373-9. [DOI] [PubMed] [Google Scholar]
- 78.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts. Laboratory, Data Analysis &Coordinating Center (LDACC) NIH program management. Biospecimen collection. Pathology. eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 79.Prinz J.C. Human Leukocyte Antigen-Class I Alleles and the Autoreactive T Cell Response in Psoriasis Pathogenesis. Front. Immunol. 2018;9:954. doi: 10.3389/fimmu.2018.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Deutekom H.W., Keşmir C. Zooming into the binding groove of HLA molecules: which positions and which substitutions change peptide binding most? Immunogenetics. 2015;67:425–436. doi: 10.1007/s00251-015-0849-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar R., Sharma A., Dogra S. Prevalence and clinical patterns of psoriatic arthritis in Indian patients with psoriasis. Indian J. Dermatol. Venereol. Leprol. 2014;80:15–23. doi: 10.4103/0378-6323.125472. [DOI] [PubMed] [Google Scholar]
- 82.Gladman D.D. In: Gordon G.B., Ruderman E., editors. Springer-Verlag; Heidelberg: 2005. Epidemiology; pp. 57–65. (Psoriasis and psoriatic arthrits: An integrated approach). [Google Scholar]
- 83.Eder L., Chandran V., Pellet F., Shanmugarajah S., Rosen C.F., Bull S.B., Gladman D.D. Human leucocyte antigen risk alleles for psoriatic arthritis among patients with psoriasis. Ann. Rheum. Dis. 2011;71:50–55. doi: 10.1136/ard.2011.155044. 21900282. [DOI] [PubMed] [Google Scholar]
- 84.Winchester R., Minevich G., Steshenko V., Kirby B., Kane D., Greenberg D.A., FitzGerald O. HLA associations reveal genetic heterogeneity in psoriatic arthritis and in the psoriasis phenotype. Arthritis Rheum. 2012;64:1134–1144. doi: 10.1002/art.33415. [DOI] [PubMed] [Google Scholar]
- 85.Jensen J.M., Villesen P., Friborg R.M., Mailund T., Besenbacher S., Schierup M.H., Danish Pan-Genome Consortium Assembly and analysis of 100 full MHC haplotypes from the Danish population. Genome Res. 2017;27:1597–1607. doi: 10.1101/gr.218891.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maretty L., Jensen J.M., Petersen B., Sibbesen J.A., Liu S., Villesen P., Skov L., Belling K., Theil Have C., Izarzugaza J.M.G., et al. Sequencing and de novo assembly of 150 genomes from Denmark as a population reference. Nature. 2017;548:87–91. doi: 10.1038/nature23264. [DOI] [PubMed] [Google Scholar]
- 87.Zheng J., Li Y., Abecasis G.R., Scheet P. A comparison of approaches to account for uncertainty in analysis of imputed genotypes. Genet. Epidemiol. 2011;35:102–110. doi: 10.1002/gepi.20552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Harrell F.E. Springer-Verlag; New York: 2001. Regression Modeling Strategies with Applications to Linear Models, Logistic Regression, and Survival Analysis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Complete summary statistics for the genome-wide association analyses of the SAS, EUR, and transethnic (SAS and EUR) datasets have been deposited at the NHGRI-EBI GWAS Catalog (accession numbers GCST90019015, GCST90019016, and GCST90019017). Individual-level genotype data for the CASP GWAS, PsA GWAS, and Exomechip case-control studies are available on dbGaP (dbGaP: phs000019.v1.p1, phs000982.v1.p1, and phs001306.v1.p1). Genotypes for the WTCCC2 psoriasis-control study are archived at the European Genome-Phenome Archive (study ID EGAS00000000108) and can be requested by contacting the data access committee at the Wellcome Trust Sanger Institute (datasharing@sanger.ac.uk). Data sharing restrictions do not allow making genotypes available for the remaining six case-control cohorts analyzed by this study (Kiel GWAS, Genizon GWAS, PAGE and GAPC Immunochip studies, batches 1 and 2+3 of SAS GWAS).
Our updated version of the SNP2HLA imputation package is available on GitHub. Five datasets (UM, 1KGP-ALL-v2, T1DGC, BKT, IKMB; see Table S8) were used to build the best-performing MHC reference panels of this study. Our GitHub repository contains three MHC reference panels built by applying our updated methods to the two datasets (UM, 1KGP-ALL-v2) for which genotype data can be freely shared. Genotype data for the T1DGC dataset can requested from the NIDDK Central Repository. Data restrictions preclude sharing any individual-level genotype data for the BKT and IKMB datasets; however, the MHC reference data for the IKMB dataset is available as a HIBAG model for imputing HLA alleles (IKMB models).