Abstract
Background:
Risk prediction models for postoperative mortality after intraabdominal surgery have typically been developed using preoperative variables. It is unclear if intraoperative data add significant value to these risk prediction models.
Methods:
With IRB approval, an institutional retrospective cohort of intraabdominal surgery patients in the 2005–2015 American College of Surgeons National Surgical Quality Improvement Program was identified. Intraoperative data were obtained from the electronic health record. The primary outcome was 30-day mortality. We evaluated the performance of machine learning algorithms to predict 30-day mortality using (A) baseline variables and (B) baseline + intraoperative variables. Algorithms evaluated were: 1) logistic regression with elastic net selection, 2) random forest (RF), 3) gradient boosting machine (GBM), 4) support vector machine (SVM), and 5) convolutional neural networks (CNN). Model performance was evaluated using the area under the receiver operator characteristic curve (AUC). The sample was randomly divided into a training/testing split with 80%/20% probabilities. Repeated 10-fold cross-validation identified the optimal model hyperparameters in the training dataset for each model, which were then applied to the entire training dataset to train the model. Trained models were applied to the test cohort to evaluate model performance. Statistical significance was evaluated using P<0.05.
Results:
The training and testing cohorts contained 4322 and 1079 patients, respectively, with 62 (1.4%) and 15 (1.4%) experiencing 30-day mortality, respectively. When using only baseline variables to predict mortality, all algorithms except SVM (AUC 0.83 [95% CI 0.69–0.97]) had AUC >0.9: GBM (AUC 0.96 [0.94–1.0]), RF (AUC 0.96 [0.92–1.0]), CNN (AUC 0.96 [0.92–0.99]), and logistic regression (AUC 0.95 [0.91–0.99]). AUC significantly increased with intraoperative variables with CNN (AUC 0.97 [0.96–0.99]; P=0.047 vs. baseline), but there was no improvement with GBM (AUC 0.97 [0.95–0.99]; P=0.3 vs. baseline), RF (AUC 0.96 [0.93–1.0]; P=0.5 vs. baseline), and logistic regression (AUC 0.94 [0.90–0.99]; P=0.6 vs. baseline).
Conclusions:
Postoperative mortality is predicted with excellent discrimination in intraabdominal surgery patients using only preoperative variables in various machine learning algorithms. The addition of intraoperative data to preoperative data also resulted in models with excellent discrimination, but model performance did not improve.
Introduction
Perioperative risk stratification is a critically important aspect of surgical patient management. Many clinical tools have been developed to assess the risk of mortality and other complications following surgery, and risk stratification has typically been performed using preoperative data.1–3 However, perioperative risk stratification should not be thought of as a singular event, but as a dynamic process where risk information is continuously updated as new data become available. The electronic capture of intraoperative management data has facilitated the development of risk prediction tools using the intraoperative period as an additional source of data,4,5 but the specific utility of intraoperative data for mortality risk stratification has not been clearly delineated.
Patient factors are extremely important in assessing postoperative mortality risk, perhaps more so than surgical factors,6 and preoperative models for mortality have excellent performance characteristics.1,2 Although we recently demonstrated that intraoperative data meaningfully contributed to risk prediction models for postoperative acute kidney injury,7 it is not readily apparent if intraoperative data can also improve risk stratification models for mortality as the performance of risk prediction models have varying performance based on the specific complication being assessed.8 Intraoperative data improved prediction models for combined morbidity and mortality in cardiac surgery patients,9 but it is not clear if improvement would be seen for all outcomes if assessed individually. Thus, we questioned whether intraoperative data were necessary to optimize the performance of risk stratification models for postoperative mortality, or if preoperative data were sufficient for this purpose.
We used a cohort of patients at a large academic institution undergoing intraabdominal general surgery to determine whether the addition of intraoperative data to a preoperative prediction model for postoperative mortality improved its performance. Intraabdominal surgery represents a major class of surgical patients with significant risk of morbidity and mortality. We evaluated several machine learning algorithms [1) logistic regression with elastic net selection, 2) random forest, 3) gradient boosting machine, 4) support vector machine, and 5) convolutional neural network] for the prediction of 30-day mortality, comparing the performance of a preoperative model containing only baseline patient data and surgical procedure information to an intraoperative model containing both preoperative and intraoperative data. Our aim is to provide perioperative physicians with important insights on the ways in which intraoperative data can be utilized to stratify surgical patients based on their risk for subsequent mortality.
Methods
Patient selection
This study was approved by the Columbia University Medical Center (CUMC; New York, NY) IRB, including waiver of consent. This is a retrospective study of intraabdominal surgery patients at CUMC participating in the American College of Surgeons National Surgical Quality Improvement Program (ACS NSQIP) from 2005–2015. A different subset of the initial cohort was evaluated for the outcome of acute kidney injury.7 The study was guided by the TRIPOD10 framework. There were 14,606 patients in the ACS NSQIP at CUMC from 2005–15 (Figure 1). Patients with missing or incomplete Anesthesia Information Management Systems (AIMS; CompuRecord, Philips Medical Systems, Eindhoven, The Netherlands) data were excluded. Intraabdominal procedures were identified using the Clinical Classifications Software for Services and Procedures (Agency for Healthcare Research and Quality, Rockville, MD) (Supplemental Table 1). Outpatient procedures were excluded as they have a low risk for mortality. The final cohort included 5401 patients.
Figure 1.
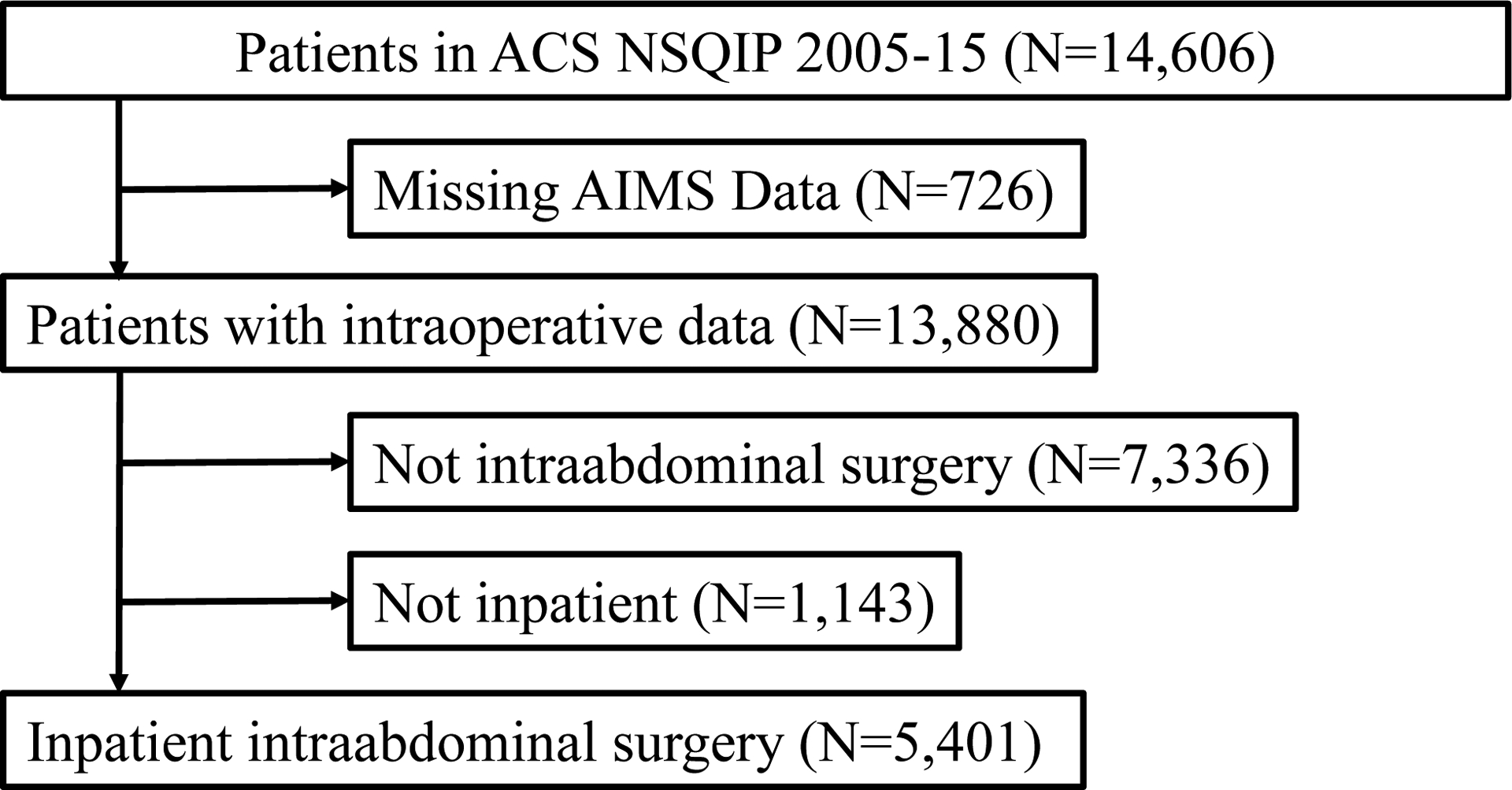
Selection of inpatient intraabdominal surgery procedures, 2005–15. ACS NSQIP, American College of Surgeons National Surgical Quality Improvement Program; AIMS, anesthesia information management system.
Preoperative and intraoperative variables
Variables included in the analyses are listed in Supplemental Tables 2 and 3. Preoperative patient characteristics and demographic data – including comorbidities and laboratory data – were collected from the ACS NSQIP. Missing data were present in preoperative laboratory values and body mass index (BMI; kg/m2) (Supplemental Table 4). No missing data were present in intraoperative variables. Missing laboratory data were imputed using medians within patient groups defined by American Society of Anesthesiologists Physical Status (ASA PS) and emergency status. Missing BMI values were imputed with whole sample average BMI. Intraoperative variables were obtained from the AIMS and reflect routine clinical care. AIMS data were stored in a relational database (Microsoft SQL Server Enterprise 12.0, Redmond, WA) and extracted using structured query language queries.
Intraoperative data were collected as previously described.7 Continuously streamed variables (e.g., blood pressure, anesthetic agent concentration) were captured by the AIMS. Other variables were manually entered by the anesthesia provider (e.g., medications, fluids, etc.). For most medications, the total dose administered to the patient was determined. Exceptions include mannitol (Yes/No), furosemide (Yes/No), ketorolac (Yes/No), and individual antibiotics (Yes/No). We created a variable indicating the number of distinct antibiotics administered, as well as a separate variable indicating the number of distinct antibiotics not including cefazolin, as this was the most commonly used antibiotic. For fluids (both inputs and outputs), total volumes were determined. Blood pressure data (both invasive and non-invasive) were captured in 15-second intervals. If an interval had both invasive and non-invasive measurements, the invasive measurement took precedence. Linear interpolation was used to assign values to intervals without a recorded blood pressure. For this analysis, we measured the duration of time (min) with mean arterial pressure below 55 mm Hg, 60 mm Hg, and 65 mm Hg, and the time-weighted average below the same 3 thresholds (mm Hg × min). For each inhaled anesthetic (sevoflurane, desflurane, isoflurane, and nitrous oxide), two variables were created: 1) a binary variable indicating its use (Yes/No) and 2) the cumulative exposure (%-hours), calculated as the average end-tidal concentration (%) multiplied by the duration of exposure (hours).
Clinical end point
The primary outcome was 30-day mortality, as recorded in the ACS NSQIP dataset.
Statistical Analysis
The analysis plan was approved prior to data analysis. Models were developed in two phases: A) preoperative model incorporating patient characteristics, demographics, and surgical procedure category; and B) intraoperative model incorporating both preoperative and intraoperative variables. The following machine learning algorithms to predict 30-day mortality were evaluated: 1) logistic regression with elastic net selection, 2) random forest (RF), 3) gradient boosting machine (GBM), 4) support vector machine (SVM), and 5) convolutional neural network (CNN). For RF, the Gini index was used as the splitting rule. For GBM, decision trees were used as the weak learners and AdaBoost11 as the loss function. Training data were centered and scaled before fitting the logistic regression, SVM and CNN models. That is, each predictor was subtracted by its mean and divided by its standard deviation.
We used a machine learning framework to tune, train, and test the models to maximize performance while guarding against overfitting.12 The dataset was randomly split by the outcome into training (80%) and testing (20%) datasets. Sampling was done within the levels of the outcome in order to balance the distribution of patients with mortality within the splits. Of the 5401 patients in the sample, 4322 (80.0%) were allocated to the training dataset. Model hyperparameters, which are model parameters not directly learned from the data, were optimized (“tuned”) with repeated 10-fold cross-validation in the training dataset. The optimal hyperparameters, maximizing the cross-validation mean area under the receiver operator characteristic curve (AUCROC), were applied to the entire training dataset to identify model parameters (“training”). The trained models were applied to the testing dataset to evaluate model performance (“testing”), assessed using the AUCROC, area under the precision-recall curve (AUCPR), and calibration plots. The PR curve plots recall (sensitivity) on the x-axis and precision (positive predictive value) on the y-axis to summarize the tradeoff between positive predictive value and sensitivity at different probability thresholds. Point estimates and 95% confidence intervals (CI) for AUCROC and AUCPR were computed with 2000 bootstrap replicates.13 Statistical significance was evaluated using P<0.05.
For interpretable algorithms, variable importance measures (RF and GBM) and odds ratios (logistic regression) evaluated the relative importance of individual variables. For GBM, relative importance measures14 were used, and for RF, Gini impurity15 was used.
Statistical analyses were performed using R Studio (version 1.2.5019; R Studio, Inc., Boston, MA) and R (version 3.6.1; The R Foundation for Statistical Computing, Vienna, Austria). The R package pROC13 calculated AUCROC and compared the AUCROCs of the preoperative and intraoperative models. The R package precrec16 calculated AUCPR. The R package caret17 identified optimal tuning parameters and then fit the final model with the optimal parameters. Logistic regression used the R packages glmnet18 and Matrix.19 Random forest used the R packages ranger20 and e1071.21 GBM used the R package gbm.22 SVM used the R package e1071. Finally, CNN used the R package nnet.23
Results
Preoperative and intraoperative variables among patients with and without 30-day mortality
Of the 4322 patients in the training dataset, 62 (1.4%) died within 30 days of surgery. Many preoperative patient characteristics and comorbidities were significantly different between patients with and without mortality (Table 1). Patients who died were older, more likely to undergo emergent procedures, and had higher rates of risk factors such as diabetes, hypertension, and preoperative sepsis/septic shock. The procedure categories with the highest mortality risk were ‘exploratory laparotomy’ (12%; 16/129), ‘colorectal resection’ (4.3%; 20/469), and ‘excision, lysis peritoneal adhesions’ (3.5%; 2/57). There were no statistically significant differences in preoperative characteristics between the training and testing datasets (data not shown).
Table 1.
Preoperative characteristics and surgical procedure category for intraabdominal surgery patients, by 30-day mortality status.
| Total | Died | Alive | ||
|---|---|---|---|---|
| Variable | 4322 | 62 (1.4%) | 4260 (99%) | P-Value |
| Age (years) | 53 (18) | 72 (14) | 53 (18) | <0.0001 |
| Female | 2517 (58%) | 39 (63%) | 2478 (58%) | 0.5 |
| Race | 0.3 | |||
| White | 1882 (44%) | 23 (37%) | 1859 (44%) | |
| Hispanic | 1358 (31%) | 20 (32%) | 1338 (31%) | |
| Black | 495 (11%) | 11 (18%) | 484 (11%) | |
| Other | 481 (11%) | 8 (13%) | 473 (11%) | |
| Asian | 106 (2%) | 0 (0.0%) | 106 (2.5%) | |
| Emergency | 1130 (26%) | 36 (58%) | 1094 (26%) | <0.0001 |
| Body Mass Index (kg/m2) | 31 (9.9) | 28 (8.1) | 31 (9.9) | 0.03 |
| Diabetes | 759 (18%) | 21 (34%) | 738 (17%) | 0.001 |
| Hypertension | 1872 (43%) | 51 (82%) | 1821 (43%) | <0.0001 |
| Chronic Obstructive Pulmonary Disease | 70 (1.6%) | 7 (11%) | 63 (1.5%) | <0.0001 |
| Current Smoker | 565 (13%) | 12 (19%) | 553 (13%) | 0.2 |
| Dyspnea | 374 (8.7%) | 16 (26%) | 358 (8.4%) | <0.0001 |
| Mechanical Ventilation | 36 (0.8%) | 17 (27%) | 19 (0.4%) | <0.0001 |
| Functionally Dependent | 188 (4.3%) | 31 (50%) | 157 (3.7%) | <0.0001 |
| Ascites | 31 (0.7%) | 6 (10%) | 25 (0.6%) | <0.0001 |
| Estimated Glomerular Filtration Rate (mL/min/1.73 m2) | 87 (30) | 63 (47) | 87 (29) | <0.0001 |
| Preoperative Transfusion | 28 (0.6%) | 7 (11%) | 21 (0.5%) | <0.0001 |
| Bleeding Disorder | 28 (0.6%) | 7 (11%) | 21 (0.5%) | <0.0001 |
| Preoperative Steroid | 28 (0.6%) | 7 (11%) | 21 (0.5%) | <0.0001 |
| Disseminated Cancer | 122 (2.8%) | 9 (15%) | 113 (2.7%) | <0.0001 |
| Wound Infection | 28 (0.6%) | 7 (11%) | 21 (0.5%) | <0.0001 |
| Preoperative Renal Insufficiency/Dialysis | 28 (0.6%) | 7 (11%) | 21 (0.5%) | <0.0001 |
| Preoperative Sepsis/Septic Shock | 252 (5.8%) | 23 (37%) | 229 (5.4%) | <0.0001 |
| Albumin (g/dL) | 4.1 (0.6) | 3.1 (0.8) | 4.1 (0.6) | <0.0001 |
| Alkaline Phosphatase (U/L) | 99 (92) | 126 (126) | 98 (92) | 0.01 |
| Bilirubin (mg/dL) | 0.9 (1.5) | 1.2 (1.0) | 0.9 (1.5) | 0.07 |
| Blood Urea Nitrogen (mg/dL) | 16 (9.3) | 29 (21) | 16 (8.9) | <0.0001 |
| Hematocrit (%) | 39 (5.2) | 33 (7.0) | 39 (5.1) | <0.0001 |
| International Normalized Ratio (Unitless) | 1.1 (0.2) | 1.4 (0.5) | 1.1 (0.2) | <0.0001 |
| Platelets (103/μL) | 266 (94) | 248 (135) | 266 (94) | 0.12 |
| Partial Thromboplastin Time (sec) | 31 (6.5) | 36 (13) | 31 (6.3) | <0.0001 |
| Aspartate Aminotransferase (U/L) | 35 (108) | 169 (747) | 33 (56) | <0.0001 |
| Sodium (mmol/L) | 138 (2.9) | 137 (4.5) | 138 (2.9) | 0.004 |
| White Blood Cells (103/μL) | 9.0 (4.3) | 13 (8.4) | 9.0 (4.2) | <0.0001 |
| Procedure Category | <0.0001 | |||
| Colorectal resection | 469 (11%) | 20 (32%) | 449 (11%) | |
| Exploratory laparotomy | 129 (3.0%) | 16 (26%) | 113 (3%) | |
| Ileostomy and other enterostomy | 450 (10%) | 8 (13%) | 442 (10%) | |
| Other operating room gastrointestinal therapeutic procedures | 449 (10%) | 7 (11%) | 442 (10%) | |
| Small bowel resection | 129 (3.0%) | 3 (4.8%) | 126 (3.0%) | |
| Excision, lysis peritoneal adhesions | 57 (1.3%) | 2 (3.2%) | 55 (1.3%) | |
| Other operating room lower gastrointestinal therapeutic procedures | 166 (3.8%) | 2 (3.2%) | 164 (3.8%) | |
| Cholecystectomy and common duct exploration | 572 (13%) | 2 (3.2%) | 570 (13%) | |
| Appendectomy | 615 (14%) | 1 (1.6%) | 614 (14%) | |
| Gastric bypass and volume reduction | 813 (19%) | 1 (1.6%) | 812 (19%) | |
| Procedures on spleen | 26 (0.6%) | 0 (0.0%) | 26 (0.6%) | |
| Colostomy, temporary and permanent | 18 (0.4%) | 0 (0.0%) | 18 (0.4%) | |
| Gastrectomy, partial and total | 77 (1.8%) | 0 (0.0%) | 77 (1.8%) | |
| Other hernia repair | 352 (8.1%) | 0 (0.0%) | 352 (8.3%) |
For continuous variables, the mean and (standard deviation) are displayed for each group and compared with the t-test. For categorical variables, counts and (%) are displayed for each group and compared with the chi-square test.
There were also many differences in intraoperative variables between patients with and without mortality (Table 2; Supplemental Table 3). Patients who died had longer duration of hypotension, greater estimated blood loss, and received larger doses of vasopressor medications, while also having received larger volumes of fluids such as albumin 5%, red blood cells, colloids, and fresh frozen plasma. There were no statistically significant differences in most intraoperative variables between the training and testing datasets (data not shown). There were statistical differences in cumulative isoflurane exposure and the proportion of patients receiving nitrous oxide (data not shown), but these differences likely had negligible effects on the models.
Table 2.
Intraoperative variables for intraabdominal surgery patients, by 30-day mortality status.
| Total | Died | Alive | ||
|---|---|---|---|---|
| Variable | 4322 | 62 (1.4%) | 4260 (99%) | P-Value |
| Minutes with MAP <55 mm Hg | 2.8 (6.4) | 6.5 (10) | 2.7 (6.3) | <0.0001 |
| Minutes with MAP <60 mm Hg | 7.7 (15) | 16 (22) | 7.6 (14) | <0.0001 |
| Minutes with MAP <65 mm Hg | 18 (27) | 31 (41) | 18 (27) | <0.001 |
| Time Weighted Average with MAP <55 mm Hg (mm Hg*mins) | 14 (35) | 43 (100) | 13 (33) | <0.0001 |
| Time Weighted Average with MAP <60 mm Hg (mm Hg*mins) | 40 (82) | 100 (162) | 39 (80) | <0.0001 |
| Time Weighted Average with MAP <55 mm Hg (mm Hg*mins) | 104 (180) | 221 (305) | 102 (177) | <0.0001 |
| Albumin 5% (mL) | 118 (351) | 298 (510) | 115 (347) | <0.0001 |
| Albumin 25% (mL) | 0.3 (7.3) | 0.0 (0.0) | 0.3 (7.3) | 0.8 |
| Lactated Ringer’s (mL) | 2604 (1731) | 2707 (1992) | 2603 (1727) | 0.6 |
| Other Crystalloid Fluids (mL) | 65 (368) | 411 (990) | 60 (348) | <0.0001 |
| Red Blood Cells (mL) | 52 (276) | 495 (1165) | 45 (235) | <0.0001 |
| Colloids (mL) | 27 (144) | 89 (231) | 26 (143) | <0.001 |
| Fresh Frozen Plasma (mL) | 15 (165) | 294 (1023) | 11 (107) | <0.0001 |
| Platelets (mL) | 2.6 (36) | 32 (121) | 2.2 (33) | <0.0001 |
| Cryoprecipitate (mL) | 0.1 (4.4) | 0.0 (0.0) | 0.1 (4.4) | 0.9 |
| Ascites (mL) | 16 (352) | 56 (287) | 16 (352) | 0.4 |
| Estimated Blood Loss (mL) | 263 (720) | 947 (2958) | 253 (627) | <0.0001 |
| Urine Output (mL) | 287 (388) | 409 (592) | 285 (384) | 0.01 |
| Phenylephrine (mg) | 0.80 (2.4) | 2.4 (4.8) | 0.78 (2.4) | <0.0001 |
| Ephedrine (mg) | 5.9 (12) | 12 (31) | 5.8 (11) | <0.0001 |
| Norepinephrine (mg) | 0.04 (0.4) | 0.94 (2.4) | 0.03 (0.3) | <0.0001 |
| Vasopressin (units) | 0.26 (1.8) | 4.7 (8.2) | 0.20 (1.4) | <0.0001 |
| Epinephrine (mg) | 0.004 (0.1) | 0.202 (1.0) | 0.001 (0.02) | <0.0001 |
| Dopamine (mg) | 0.041 (2) | 2.4 (16) | 0.007 (0.4) | <0.0001 |
| Labetalol (mg) | 2.3 (8.9) | 2.2 (7.1) | 2.3 (9.0) | 0.9 |
| Esmolol (mg) | 2.7 (13) | 8.2 (29) | 2.6 (13) | <0.001 |
| Metoprolol (mg) | 0.28 (1.7) | 0.29 (1.2) | 0.28 (1.7) | 1.0 |
| Hydralazine (mg) | 0.009 (0.2) | 0.081 (0.6) | 0.008 (0.2) | 0.02 |
| Enalaprilat (mg) | 0.001 (0.038) | 0.000 (0.000) | 0.001 (0.038) | 0.9 |
| Clonidine (mg) | 0.0001 (0.003) | 0.0000 (0.0) | 0.0001 (0.003) | 0.9 |
| Nicardipine (mg) | 0.006 (0.085) | 0.005 (0.038) | 0.006 (0.085) | 0.9 |
| Nitroglycerin (mcg) | 0.000 (0.02) | 0.000 (0.000) | 0.0004 (0.02) | 0.9 |
| Mannitol (Yes/No) | 2 (0.05%) | 0 (0%) | 2 (0.05%) | 1.0 |
| Furosemide (Yes/No) | 79 (1.8%) | 8 (13%) | 71 (1.7%) | <0.0001 |
| Ketorolac (Yes/No) | 1088 (25%) | 4 (6.5%) | 1084 (25%) | 0.001 |
| Sevoflurane (Yes/No) | 3632 (84%) | 51 (82%) | 3581 (84%) | 0.8 |
| Sevoflurane (%-Hours) | 3.2 (3.1) | 3.0 (3.7) | 3.2 (3.1) | 0.6 |
| Desflurane (Yes/No) | 1308 (30%) | 10 (16%) | 1298 (30%) | 0.02 |
| Desflurane (%-Hours) | 2.3 (5.6) | 1.0 (4.1) | 2.4 (5.6) | 0.06 |
| Isoflurane (Yes/No) | 691 (16%) | 21 (34%) | 670 (16%) | <0.001 |
| Isoflurane (%-Hours) | 0.5 (1.3) | 0.4 (0.8) | 0.5 (1.3) | 0.7 |
| Nitrous Oxide (Yes/No) | 2403 (56%) | 26 (42%) | 2377 (56%) | 0.04 |
| Nitrous Oxide (%-Hours) | 8.7 (20) | 8.6 (23) | 8.7 (20) | 1.0 |
| Number of distinct antibiotics administered | 0.9 (0.5) | 0.9 (0.7) | 0.9 (0.5) | 0.4 |
| Number of distinct antibiotics administered (not including cefazolin) | 0.6 (0.6) | 0.7 (0.7) | 0.5 (0.6) | 0.01 |
| Propofol (mg) | 218 (231) | 89.3 (131) | 220 (232) | <0.0001 |
| Midazolam (mg) | 1.9 (8.5) | 11 (70) | 1.8 (1.2) | <0.0001 |
| Fentanyl (mg) | 0.3 (0.2) | 0.3 (0.3) | 0.3 (0.2) | 0.6 |
| Morphine (mg) | 1.8 (4.3) | 1.5 (3.5) | 1.8 (4.3) | 0.6 |
| Hydromorphone (mg) | 0.6 (0.9) | 0.4 (0.7) | 0.6 (0.9) | 0.1 |
| Remifentanil (mg) | 0.040 (0.3) | 0.035 (0.3) | 0.040 (0.3) | 0.9 |
| Operative Time (min) | 162 (108) | 177 (139) | 162 (108) | 0.3 |
For continuous variables, the mean and (standard deviation) are displayed for each group and compared with the t-test. For categorical variables, counts and (%) are displayed for each group and compared with the chi-square test.
Comparison of model performance in preoperative and intraoperative machine learning models
When using only preoperative data, the AUCROCs in the test set for GBM, RF, CNN, and logistic elastic net were 0.96 [95% CI 0.94–0.99], 0.96 [95% CI 0.92–1.00], 0.96 [95% CI 0.92–0.99], and 0.95 [95% CI 0.91–0.99], respectively (Table 3; ROC curves in Figure 2). SVM had lower discrimination with an AUCROC of 0.83 [95% CI 0.69–0.97]. When intraoperative data were added to the preoperative model, the AUCROCs in the test set for GBM, RF, CNN, logistic elastic net, and SVM were 0.97 [95% CI 0.95–0.99], 0.97 [95% CI 0.96–0.99], 0.97 [95% CI 0.95–0.99], 0.95 [95% CI 0.91–0.99], and 0.88 [95% CI 0.75–1.00], respectively. The AUCROCs for the preoperative and intraoperative models were significantly different only in the CNN model (P=0.029).
Table 3.
Area under the curve of the receiver operator characteristic curve of preoperative and intraoperative models to predict 30-day mortality in intraabdominal surgery patients.
| Algorithm | Preoperative Model | Intraoperative Model | |||
|---|---|---|---|---|---|
| AUC | 95% CI | AUC | 95% CI | P-value* | |
| Gradient Boosting Machine | 0.96 | [0.94, 0.99] | 0.97 | [0.95, 0.99] | 0.4 |
| Random Forest | 0.96 | [0.92, 1.00] | 0.97 | [0.96, 0.99] | 0.3 |
| Convolutional Neural Network | 0.96 | [0.92, 0.99] | 0.97 | [0.95, 0.99] | 0.029 |
| Logistic Regression | 0.95 | [0.91, 0.99] | 0.95 | [0.91, 0.99] | 0.9 |
| Support Vector Machine | 0.83 | [0.69, 0.97] | 0.88 | [0.75, 1.00] | 0.6 |
AUC, area under the curve of the receiver operator characteristic curve; CI, confidence interval.
AUCs are reported as applied in the test cohort.
Intraoperative model includes both preoperative and intraoperative variables.
P-value compares the preoperative model to the intraoperative model using deLong’s test.
Figure 2.
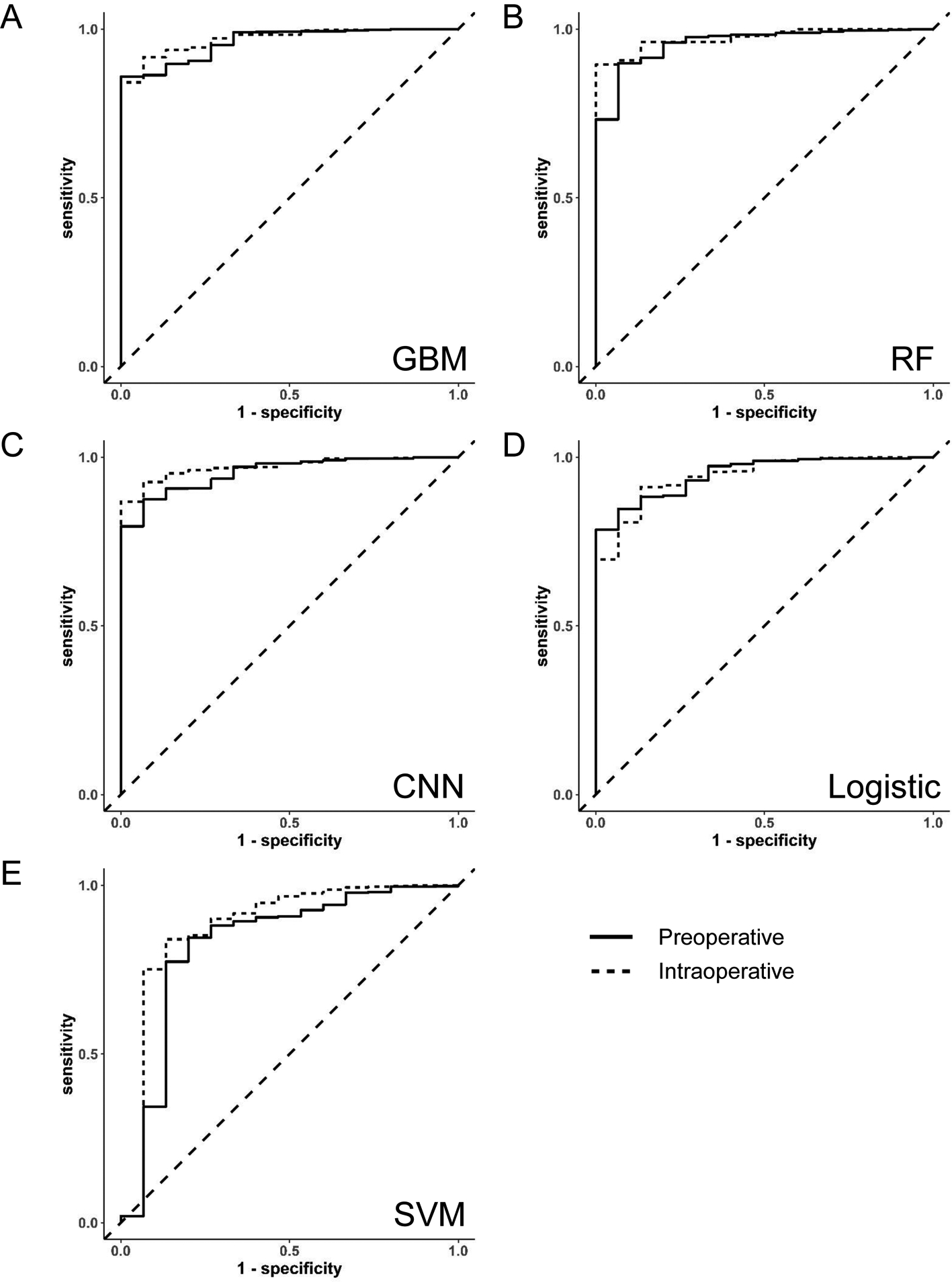
Area under the receiver operator characteristic curve for preoperative and intraoperative machine learning models to predict 30-day mortality in intraabdominal surgery patients. Algorithms evaluated were (A) gradient boosting machine (GBM), (B) random forest (RF), (C) convolutional neural networks (CNN), (D) logistic regression (logistic), and (E) support vector machine (SVM).
Model performance was also assessed with AUCPR. When using only preoperative data, the AUCPRs in the test set for GBM, RF, CNN, logistic elastic net, and SVM were 0.48 [95% CI 0.25–0.70], 0.30 [95% CI 0.16–0.55], 0.30 [95% CI 0.14–0.57], 0.30 [95% CI 0.14–0.55], and 0.12 [95% CI 0.05–0.30], respectively (Supplemental Table 5; PR curves in Supplemental Figure 1). When intraoperative data were added to the preoperative model, the AUCPRs in the test set for GBM, RF, CNN, logistic elastic net, and SVM were 0.50 [95% CI 0.25–0.73], 0.53 [95% CI 0.30–0.75], 0.37 [95% CI 0.18–0.63], 0.43 [95% CI 0.20–0.67], and 0.31 [95% CI 0.10–0.54], respectively. The AUCPRs for the preoperative and intraoperative models were significantly different in the RF (P=0.01) and SVM (P=0.01) models.
Calibration plots are displayed in Figure 3. The logistic regression and SVM models were well-calibrated, while GBM, RF, and CNN had evidence of underestimating or overestimating mortality risk in portions of the calibration plots, particularly in the intraoperative models.
Figure 3.
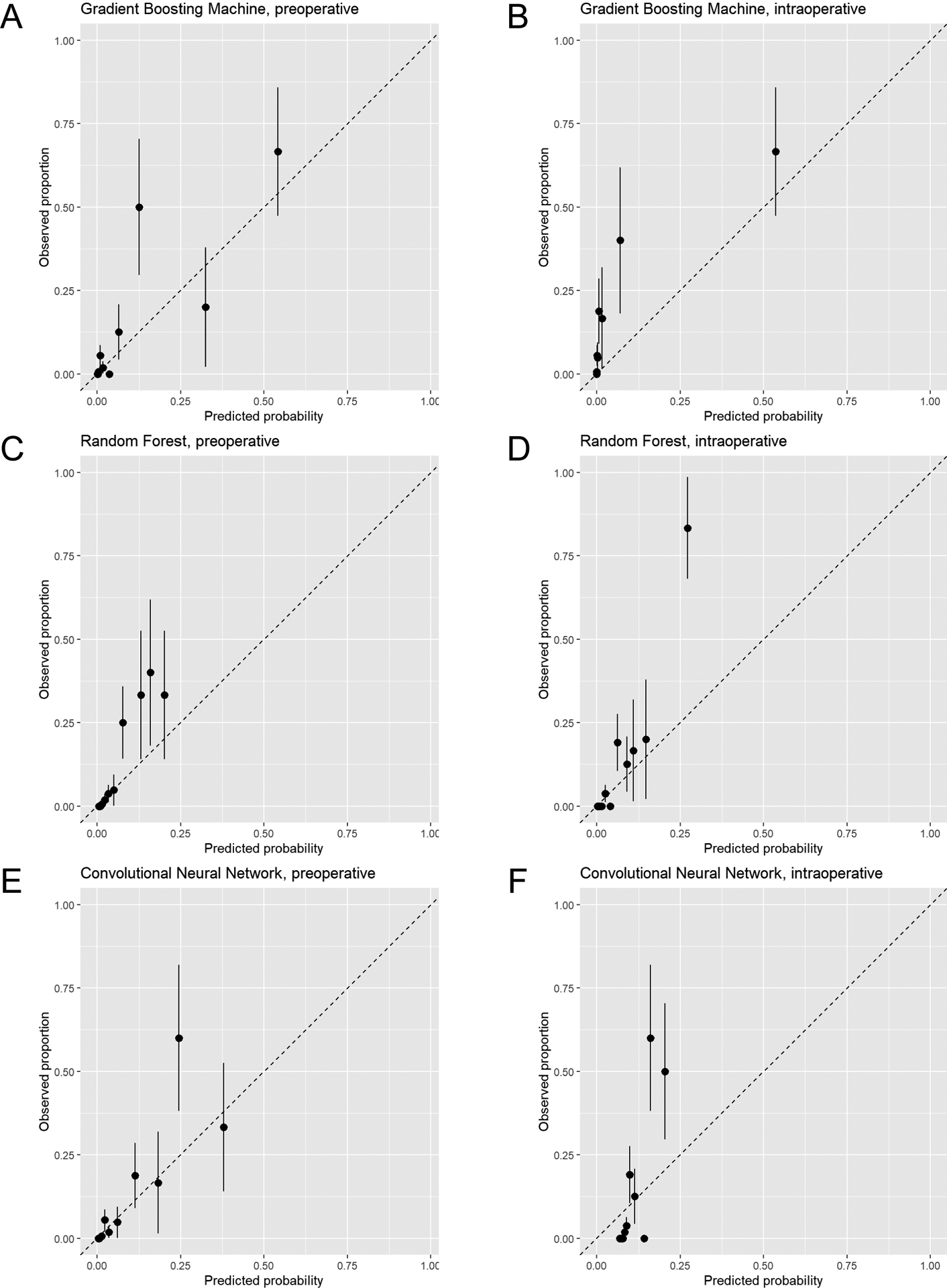
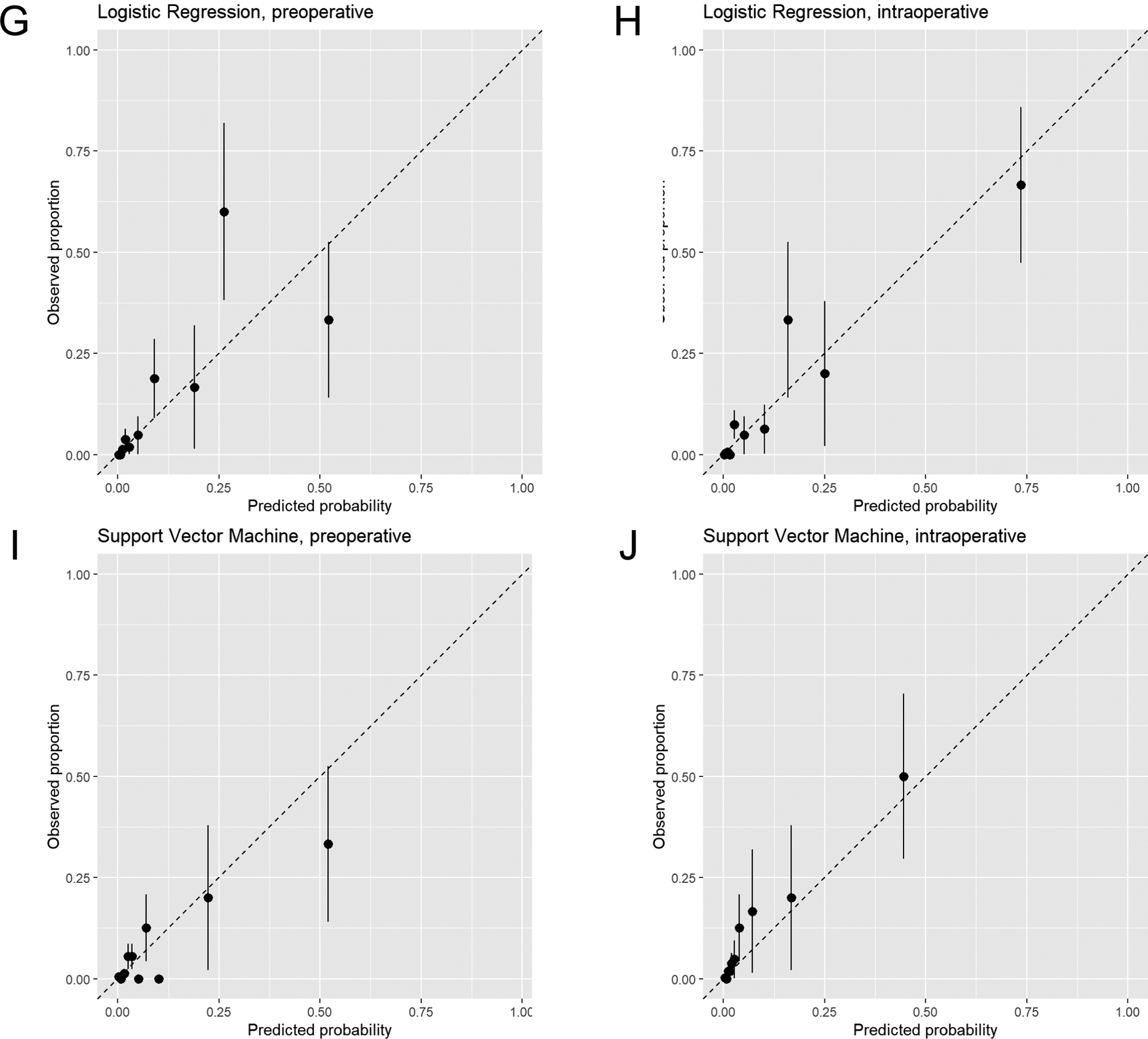
Calibration plots of predicted mortality risk (x-axis) vs. observed mortality (y-axis) in intraabdominal surgery patients. Plots for models using only preoperative data and both preoperative and intraoperative data (intraoperative) are displayed for gradient boosting machine (A and B), random forest (C and D), convolutional neural network (E and F), logistic regression (G and H), and support vector machine (I and J) models. The subjects are divided into 10 groups using the 45th, 70th, 85th, 90th, 95th, 97th, 98.5th, 99th, and 99.5th percentiles of the predicted probability of the fitted model. Groups are divided unequally to account for the right-skewed distribution of predicted probabilities and to focus on patients with high predicted probabilities. Mean predicted probability and 30-day mortality are calculated within each group. All plots reflect calibration in the test dataset.
Variable importance measures in machine learning models
Metrics regarding relative feature importance were available for certain machine learning algorithms (Supplemental Figure 2). Variables with high relative importance included both preoperative characteristics, such as age, functional dependence, and mechanical ventilation, and intraoperative variables such as vasopressor use (e.g., norepinephrine, epinephrine, and vasopressin).
Discussion
We used a machine learning approach to predict 30-day mortality in patients undergoing intraabdominal surgery at a single academic medical center. For 4 out of the 5 machine learning algorithms evaluated, there was excellent performance, as measured by model discrimination, when the models were trained using preoperative patient characteristics and surgical procedure, mimicking the preoperative phase of risk stratification. We then assessed the incremental value of adding intraoperative data to these models and found that while the intraoperative models also had high performance, they did not perform significantly better than models using only preoperative data. Our results suggest that while postoperative mortality after intraabdominal surgery is well-predicted by patients’ underlying medical conditions and intended surgical procedure, the addition of intraoperative data leads to models that have equally high performance.
Anesthesiologists were early proponents of perioperative risk stratification with the development of the ASA PS,24 which has held up over time as a reliable indicator of postoperative morbidity and mortality risk.25 While the ASA PS is somewhat subjective, other risk stratification measures have been developed, such as the Physiological and Operative Severity Score for the enUmeration of Mortality and morbidity (POSSUM)26 that incorporates physiological variables. These measures are evaluated prior to surgery, and preoperative data have been used to develop many well-performing risk stratification models for postoperative mortality.1–3
The advent of AIMS and electronic health records have made the automated capture of data possible, and combined with machine learning approaches, there has been tremendous interest in using these additional data sources to improve perioperative risk stratification. Indeed, machine learning using automated capture of preoperative data outperformed the ASA PS and other traditional risk stratification metrics such as the Charlson comorbidity score.27,28 Automated methods also outperformed the highly-regarded but resource-intensive ACS NSQIP calculator.29,30 It is clear that the availability of electronic patient data has the potential to improve risk stratification in the perioperative period.
The specific benefits of including intraoperative data in mortality prediction models remain unclear. Deep learning using intraoperative features produced well-performing prediction models for postoperative mortality,4,5 but these models did not outperform traditional methods.4,31 Few studies have directly compared preoperative and intraoperative models to assess the incremental value of intraoperative data. Datta et al.32 report that the addition of intraoperative data improved the performance of risk prediction models for in-hospital mortality and other complications, such as mechanical ventilation, neurological complications, cardiovascular complications, and acute kidney injury. We also found that intraoperative data improved models to predict acute kidney injury.7 The value of intraoperative data in predicting surgical outcomes likely varies by the specific outcome being evaluated.
We incorporated a host of intraoperative variables designed to represent various aspects of intraoperative care, such as blood pressure, vasopressor use, fluid management, and anesthetic drug administration. Many studies of the intraoperative period focus on one specific aspect of management (e.g., hypotension)33 but may not adequately account for related aspects that also contribute to adverse outcomes (e.g., vasopressor use). Our models incorporated a wide array of intraoperative variables and machine learning algorithms can account for complex relationships between predictor variables. Many intraoperative variables ranked high in terms of variable importance (Supplemental Figure 2), indicating large effects on model performance, but these analyses are not designed to provide detailed inference on these variables. Future studies may provide a better understanding of the specific relationships between individual variables and mortality through “interpretable” machine learning methodologies.34
Risk prediction for mortality may have different implications than risk prediction for other complications. Surgeons may decline to operate on a patient with a high mortality risk (or patients may refuse surgery),35 but it is not clear if a high predicted risk of other complications, especially potentially reversible complications such as acute kidney injury, would preclude a complicated surgical procedure. For certain conditions, surgeons may choose to operate on a high-risk patient, but perform only a minimal, palliative procedure. Perioperative mortality attributable to anesthesia has dramatically decreased over time,36 and this may be an additional reason why intraoperative data did not improve our models. Our models only include patients who had a surgical procedure, so their utility to assist in the decision to proceed with surgery in a high-risk patient will have to be further evaluated.
There are several differences between our study and others in the literature. We focus on a cohort of intraabdominal surgery patients but many studies include broader categories of surgical procedures,4,5,32 and model performance may be affected by the underlying patient population. Another important aspect of model building using intraoperative data are the specific variables used in the models. For example, intraoperative data include streams of physiological data (e.g., blood pressure every minute), but some degree of data transformation is required (e.g., min, max, mean, standard deviation of blood pressure). The specific variables used vary across studies, and the extent to which these differences account for variation in results is not clear. The type of data may also play a role. For preoperative variables, we used data from the ACS NSQIP which has both advantages and disadvantages compared to automatically captured electronic health record data.37 In addition, machine learning models predominantly use data from a single institution, so institutional differences may account for differences in the results.
While intraoperative data did not improve model performance using our primary metric (AUCROC), there were suggestions of improved performance with AUCPR (Supplemental Table 5). The AUCPR has advantages in evaluating performance in imbalanced datasets.38 However, we are cautious in our interpretation of these results as AUCPR was not our primary metric for assessing model performance. In addition, our models were tuned to optimize AUCROC, and training the models to optimize AUCPR may have produced more optimal results for assessing performance. The AUCROC has consistent interpretations based on its value, but it is difficult to judge model performance based on the AUCPR value alone. The AUCPR of an uninformative model is the baseline event rate, in our case 0.014, and all models had AUCPR values much greater than this baseline rate. As the AUCPR only considers precision (positive predictive value) and recall (sensitivity), true negative results are ignored and a larger sample may be required as a large proportion of patients would be considered true negatives in our analyses.
Our study is subject to certain limitations. The models were developed using data from completed cases, and as such, it is not clear whether there are modifiable factors that can be altered to change the final outcome. However, this study is an initial step towards this goal and future work will delineate the role of modifiable factors, both preoperative and intraoperative, that can improve clinical outcomes after surgery. Machine learning models benefit from larger sample sizes, and it is possible that a larger sample would have altered our results. Imbalanced datasets may affect model performance,39 but we did not specifically perform any data preprocessing, such as oversampling, as our models performed well without this additional step. As an alternative assessment of model performance, we evaluated the precision-recall curve, which may better assess performance in imbalanced datasets.40 In addition, some well-performing models had evidence of miscalibration, particularly in the intraoperative models, and may benefit from recalibration.41
In conclusion, our analyses demonstrate that both preoperative data and intraoperative data can be used to develop well-performing models to predict postoperative mortality in intraabdominal surgery patients. However, in terms of model building, we did not find evidence that intraoperative models outperformed models using only preoperative data. Intraoperative data, nonetheless, has tremendous clinical value but we must determine the specific ways in which this data can be used to improve the care of surgical patients.
Supplementary Material
Key Points Summary.
Question:
Does the addition of intraoperative data to a preoperative model for 30-day mortality after intraabdominal surgery improve the model’s performance?
Findings:
Using several machine learning algorithms, 30-day mortality was predicted with excellent discrimination using only preoperative data, with negligible improvement after adding intraoperative data.
Meaning:
Surgical mortality is well predicted using preoperative patient characteristics, including comorbidities, risk factors, and surgical procedure.
Acknowledgements
The authors acknowledge Mitchell F. Berman, MD and Brian Thumm for their assistance with data from the Anesthesia Information Management System; Nadine M. Thomas for her assistance with local ACS NSQIP data acquisition; and Alla Babina for her assistance with electronic health record data acquisition.
The authors would like to thank Mitchell F. Berman, MD (Columbia University Medical Center) and Brian Thumm (CUMC) for their assistance with data collection from the Anesthesia Information Management System; Nadine M. Thomas (New York Presbyterian Hospital) for her assistance with ACS NSQIP data; and Alla Babina (NYPH) for her assistance with electronic health record data.
Funding:
This work was supported by the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH; Bethesda, MD) through Grant Number KL2 TR001874 (MK) and the National Heart, Lung, and Blood Institute (NHLBI) through grant K23 HL148640 (REF). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Glossary of Terms
- ACS NSQIP
American College of Surgeons National Surgical Quality Improvement Program
- AIMS
Anesthesia Information Management System
- ASA PS
American Society of Anesthesiologists Physical Status
- AUC
area under the curve
- AUCPR
area under the precision-recall curve
- AUCROC
area under the receiver operator characteristic curve
- BMI
body mass index
- CI
confidence interval
- CNN
convolutional neural network
- CUMC
Columbia University Medical Center
- GBM
gradient boosting machine
- RF
random forest
- SVM
support vector machine
Footnotes
Conflicts of Interest: None.
Presented at the American Society of Anesthesiologists’ Annual Meeting (Virtual), October 2020.
References
- 1.Moonesinghe SR, Mythen MG, Das P, Rowan KM, Grocott MP. Risk stratification tools for predicting morbidity and mortality in adult patients undergoing major surgery: qualitative systematic review. Anesthesiology 2013;119:959–81. [DOI] [PubMed] [Google Scholar]
- 2.Le Manach Y, Collins G, Rodseth R et al. Preoperative Score to Predict Postoperative Mortality (POSPOM): Derivation and Validation. Anesthesiology 2016;124:570–9. [DOI] [PubMed] [Google Scholar]
- 3.Kim M, Wall MM, Li G. Applying Latent Class Analysis to Risk Stratification for Perioperative Mortality in Patients Undergoing Intraabdominal General Surgery. Anesth Analg 2016;123:193–205. [DOI] [PubMed] [Google Scholar]
- 4.Lee CK, Hofer I, Gabel E, Baldi P, Cannesson M. Development and Validation of a Deep Neural Network Model for Prediction of Postoperative In-hospital Mortality. Anesthesiology 2018;129:649–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fritz BA, Cui Z, Zhang M et al. Deep-learning model for predicting 30-day postoperative mortality. Br J Anaesth 2019;123:688–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Story DA. Postoperative mortality and complications. Best Pract Res Clin Anaesthesiol 2011;25:319–27. [DOI] [PubMed] [Google Scholar]
- 7.Kim M, Li G, Mohan S et al. Intraoperative Data Enhance the Detection of High-Risk Acute Kidney Injury Patients When Added to a Baseline Prediction Model. Anesth Analg 2021;132:430–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim M, Wall MM, Li G. Risk Stratification for Major Postoperative Complications in Patients Undergoing Intra-abdominal General Surgery Using Latent Class Analysis. Anesth Analg 2018;126:848–57. [DOI] [PubMed] [Google Scholar]
- 9.Durant TJS, Jean RA, Huang C et al. Evaluation of a Risk Stratification Model Using Preoperative and Intraoperative Data for Major Morbidity or Mortality After Cardiac Surgical Treatment. JAMA Netw Open 2020;3:e2028361. [DOI] [PubMed] [Google Scholar]
- 10.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): the TRIPOD Statement. Br J Surg 2015;102:148–58. [DOI] [PubMed] [Google Scholar]
- 11.Freund Y, Schapire RE. A decision-theoretic generalization of on-line learning and an application to boosting Proceedings of the Second European Conference on Computational Learning Theory: Springer-Verlag, 1995:23–37. [Google Scholar]
- 12.Liu Y, Chen PC, Krause J, Peng L. How to Read Articles That Use Machine Learning: Users’ Guides to the Medical Literature. JAMA 2019;322:1806–16. [DOI] [PubMed] [Google Scholar]
- 13.Robin X, Turck N, Hainard A et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman JH. Greedy function approximation: A gradient boosting machine. Ann Stat 2001;29:1189–232. [Google Scholar]
- 15.Nembrini S, Konig IR, Wright MN. The revival of the Gini importance? Bioinformatics 2018;34:3711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito T, Rehmsmeier M. Precrec: fast and accurate precision-recall and ROC curve calculations in R. Bioinformatics 2017;33:145–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn M. Building Predictive Models in R Using the caret Package. 2008 2008;28:26. [Google Scholar]
- 18.Simon N, Friedman JH, Hastie T, Tibshirani R. Regularization Paths for Cox’s Proportional Hazards Model via Coordinate Descent. 2011 2011;39:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bates D, Maechler M. Matrix: Sparse and Dense Matrix Classes and Methods. R package version 1.2-17., 2019. [Google Scholar]
- 20.Wright MN, Ziegler A. ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. 2017 2017;77:17. [Google Scholar]
- 21.Meyer D, Dimitriadou E, Hornik K, Weingessel A, Leisch F. e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien. R package version 1.7-3., 2019. [Google Scholar]
- 22.Greenwell B, Boehmke B, Cunningham J, GBM Developers. gbm: Generalized Boosted Regression Models. R package version 2.1.5., 2019. [Google Scholar]
- 23.Venables WN, Ripley BD, Venables WN. Modern applied statistics with S. 4th ed. New York: Springer, 2002. [Google Scholar]
- 24.Saklad M. Grading of Patients for Surgical Procedures. Anesthesiology 1947;2:281–4. [Google Scholar]
- 25.Hackett NJ, De Oliveira GS, Jain UK, Kim JY. ASA class is a reliable independent predictor of medical complications and mortality following surgery. Int J Surg 2015;18:184–90. [DOI] [PubMed] [Google Scholar]
- 26.Copeland GP, Jones D, Walters M. POSSUM: a scoring system for surgical audit. Br J Surg 1991;78:355–60. [DOI] [PubMed] [Google Scholar]
- 27.Chiew CJ, Liu N, Wong TH, Sim YE, Abdullah HR. Utilizing Machine Learning Methods for Preoperative Prediction of Postsurgical Mortality and Intensive Care Unit Admission. Ann Surg 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill BL, Brown R, Gabel E et al. An automated machine learning-based model predicts postoperative mortality using readily-extractable preoperative electronic health record data. Br J Anaesth 2019;123:877–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corey KM, Kashyap S, Lorenzi E et al. Development and validation of machine learning models to identify high-risk surgical patients using automatically curated electronic health record data (Pythia): A retrospective, single-site study. PLoS Med 2018;15:e1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bilimoria KY, Liu Y, Paruch JL et al. Development and evaluation of the universal ACS NSQIP surgical risk calculator: a decision aid and informed consent tool for patients and surgeons. J Am Coll Surg 2013;217:833–42 e1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cosgriff CV, Celi LA. Deep learning for risk assessment: all about automatic feature extraction. Br J Anaesth 2020;124:131–3. [DOI] [PubMed] [Google Scholar]
- 32.Datta S, Loftus TJ, Ruppert MM et al. Added Value of Intraoperative Data for Predicting Postoperative Complications: The MySurgeryRisk PostOp Extension. J Surg Res 2020;254:350–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salmasi V, Maheshwari K, Yang D et al. Relationship between Intraoperative Hypotension, Defined by Either Reduction from Baseline or Absolute Thresholds, and Acute Kidney and Myocardial Injury after Noncardiac Surgery: A Retrospective Cohort Analysis. Anesthesiology 2017;126:47–65. [DOI] [PubMed] [Google Scholar]
- 34.Lundberg SM, Lee SI. A Unified Approach to Interpreting Model Predictions Advances in Neural Information Processing Systems: Curran Associates., 2017:4765–74. [Google Scholar]
- 35.Dowie J, Wildman M. Choosing the surgical mortality threshold for high risk patients with stage Ia non-small cell lung cancer: insights from decision analysis. Thorax 2002;57:7–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li G, Warner M, Lang BH, Huang L, Sun LS. Epidemiology of anesthesia-related mortality in the United States, 1999–2005. Anesthesiology 2009;110:759–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bensley RP, Yoshida S, Lo RC et al. Accuracy of administrative data versus clinical data to evaluate carotid endarterectomy and carotid stenting. J Vasc Surg 2013;58:412–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One 2015;10:e0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krawczyk B. Learning from imbalanced data: open challenges and future directions. Prog Artif Intell 2016;5:221–32. [Google Scholar]
- 40.Fu GH, Yi LZ, Pan J. Tuning model parameters in class-imbalanced learning with precision-recall curve. Biom J 2019;61:652–64. [DOI] [PubMed] [Google Scholar]
- 41.Fonseca PG, Lopes HD. Calibration of Machine Learning Classifiers for Probability of Default Modelling, 2017:arXiv:1710.08901.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


