Abstract
In eukaryotes, small RNAs (sRNAs) play critical roles in multiple biological processes. Dicer endonucleases are central to sRNA biogenesis. In plants, DICER-LIKE PROTEIN 3 (DCL3) produces 24-nt small interfering RNAs (siRNAs) that determine the specificity of the RNA-directed DNA methylation (RdDM) pathway. Here, we determined structure of a DCL3-pre-siRNA complex in an active dicing-competent state. The 5’-phosphorylated-A1 of the guide strand and the 1-nt 3’-overhang of the complementary strand are specifically recognized by a positively charged pocket and an aromatic cap, respectively. The 24-nt siRNA length dependence relies on the separation between the 5’-phosphorylated-end of the guide RNA and dual cleavage sites formed by the paired RNaseIII domains. These structural studies, complemented by functional data, reveal insights into the dicing principle for Dicers in general.
One Sentence Summary:
Mechanistic understanding of the terminus-specific, length-dependent, and strand-biased 24-nt siRNA production by Arabidopsis DCL3.
Dicer family RNaseIII enzymes cleave hairpin-shaped pre-microRNA (pre-miRNA) or double-stranded pre-small interfering RNA (pre-siRNA) to generate small RNAs (sRNAs) of defined length (1). After dicing, one sRNA strand is loaded into Argonaute (AGO) proteins, forming the active RNA-induced silencing complex (RISC), which silences target loci (2). Structural and biochemical analysis of Giardia intestinalis, human, and Drosophila Dicers in RNA-free or pre-dicing states have defined overall Dicer topology and function (3–7). However, the structure of Dicer in a cleavage-competent conformation has yet to be determined, limiting current understanding of the dicing mechanism.
In Arabidopsis, there are four DICER LIKE PROTEINS (DCLs). While DCL1 produces 21-nt miRNA (8), DCL2, DCL3, and DCL4 produce 22-nt, 24-nt, and 21-nt siRNAs, respectively (9–12), in which DCL3 functions specifically in the plant-specific RNA-directed DNA methylation (RdDM) pathway. In RdDM, plant-specific RNA polymerase IV (Pol IV) transcribes precursor RNAs, which are converted into dsRNA by RNA DEPENDENT RNA POLYMERASE 2 (RDR2) and diced by DCL3 (13–17). The 24-nt siRNA strand is loaded into AGO4, which interacts with Pol V-transcribed long non-coding RNAs and recruits DOMAINS REARRANGED METHYLASE 2 (DRM2), facilitating DNA methylation (18–21). DCL3 possesses ATP-independent dicing activity and has a substrate preference for pre-siRNAs with a 5’-phosphorylated-A1 on the Pol IV-produced guide strand and a 1-nt 3’-overhangs on the RDR2-produced complementary strand (22–25).
To analyze 24-nt siRNA production by DCL3, we determined the 3.1 Å resolution cryo-EM structure of full length DCL3 in complex with a 40-bp TAS1a derived pre-siRNA with a 5’-phosphorylated-A1 in the guide strand, mimicking the Pol IV strand, and a 1-nt 3’-overhang in the complementary strand, mimicking the RDR2 strand, together with Ca2+ ions mimicking Mg2+ but preventing dicing (Fig. 1A–B and S1 and Tables S1–S2). The helicase domain of DCL3 showed faint density and could not be built into the model (Fig. 1B and S1E). Unlike the several reported Dicer structures in the inactive states (3, 6, 7), our DCL3-RNA structure captured Dicer in a dicing-competent state for the first time. The platform, PAZ, and connector domains form a combined cassette to bind to the RNA duplex end towards the 5’ of the guide strand, and aligns on one side of the RNA with the RNaseIIIa/b domains, forming a continuous positively charged surface to accommodate the RNA (Fig. 1B–D). On the other side, two dsRNA-binding domains (dsRBDs) interact with the RNA backbone and form an enclosed pre-siRNA binding channel, enveloping the bound pre-siRNA (Fig. 1C–D).
Fig. 1. Structure of Arabidopsis DCL3-pre-siRNA complex.
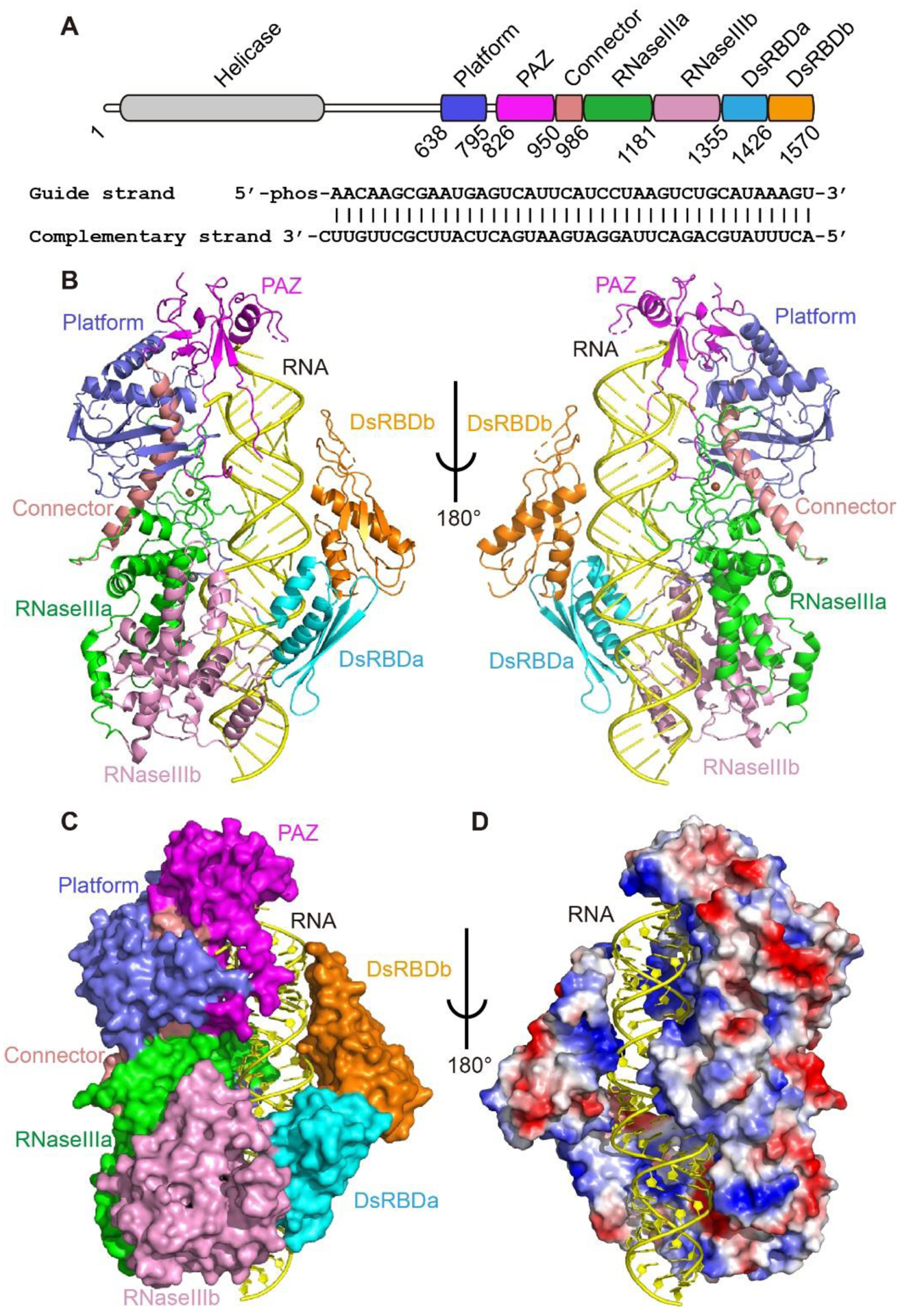
A. Domain architecture of Arabidopsis DCL3 and the sequence of pre-siRNA used for structure determination. B. Overall structure of DCL3-RNA complex in ribbon representation. C. Structure of DCL3 in surface view and RNA in cartoon view, showing that the multiple domains of DCL3 wrapping around the RNA. D. Electrostatic surface view of DLC3 showing the RNA is bound in a positively charged channel. Blue, positively charged region; red, negatively charged region.
35-bp of the pre-siRNA can be traced in the structure and ~33-bp are nearly fully enclosed by DCL3 (Fig. 2A and S2A). The platform, PAZ, and connector domain together accommodate one end of the pre-siRNA (Fig. 2B and S2B), resembling the reported human Dicer platform-PAZ-connector-RNA structure (Fig. S2C) (4). A PAZ domain loop penetrates the RNA duplex, splitting the first base-pair and flipping out the guide strand 5’-phosphorylated-A1 into a platform-PAZ-connector pocket, with the orphaned U1’ retaining its original conformation (Fig. 2B). The 5’-phosphate group inserts into a positively charged pocket formed by surrounding basic residues, including Lys695, Lys903, His906, Arg953, and Lys957, to form extensive electrostatic and hydrogen bonding interactions (Fig. 2C). A sulfate binding pocket observed in the human Dicer platform-PAZ-connector cassette was proposed to be the 5’-phosphate binding site (4, 5). Here, our DCL3-RNA complex structure located the 5’-phosphate binding pocket to similar position (Fig. S2C) and revealed the recognition mechanism, suggesting that Dicer proteins may share the same pocket to recognize the 5’-phosphate group of guide strand RNA.
Fig. 2. The terminus-specific recognition of pre-siRNA.
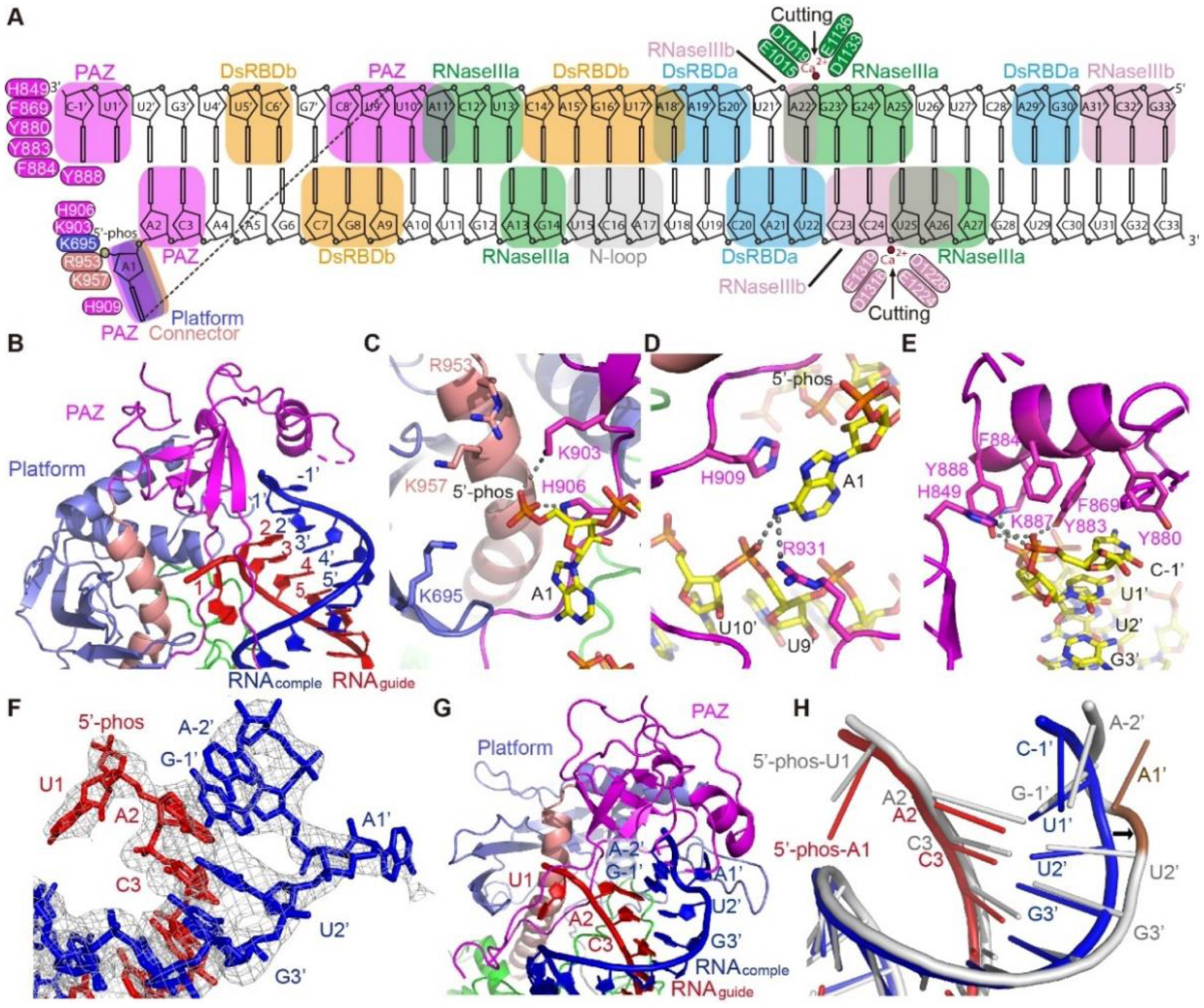
A. Schematic representation of the overall interactions between DCL3 and the pre-siRNA. Key residues are highlighted. The RNA-interacting DCL3 domains are indicated by boxes. B. Overall recognition of the RNA termini by the DCL3 platform-PAZ-connector cassette. The guide strand (RNAguide) and complementary strand (RNAcomple) are highlighted in red and blue, respectively. C. Recognition of the guide strand 5’-phophate group by a positively charged pocket in the platform-PAZ-connector cassette. Interacting residues and hydrogen bonds are highlighted in sticks and dashed-lines, respectively. D. Specific recognition of the A1 base by stacking with DCL3 H909 and hydrogen bonding interactions with DCL3 R931 and the complementary strand. E. An aromatic cap covers and interacts with the last base of the 3’-end of the complementary strand. In addition, the phosphate backbone is recognized by hydrogen bonds. F. Termini of the pre-siRNA with 5’-phosphorylated-U1 on the guide strand and 3’-2-nt overhang on the complementary strand. The cryo-EM map is shown in mesh, showing that both the bases of the U1-A1’ pair were flipped-out. G. Capturing of the RNA termini with 5’-phosphorylated-U1 and 2-nt 3’-overhang by the platform-PAZ-connector cassette. H. Superimposition of the 1-nt 3’-overhang containing RNA duplex (in red and blue) and 2-nt 3’-overhang containing RNA (in silver). The conformational change of RNA is highlighted by the arrow. The flipped out A1’ base allows the G-1’ and A-2’ to fold back and overlap with positions of U1’ and C-1’ of 1-nt-overhang RNA, occupying the same 3’-end position.
The flipped-out A1 base is stabilized through stacking interactions with the PAZ domain residues His909 (Fig. 2D), allowing the A1 base to form hydrogen bonds with both the phosphate group of U9’ on the complementary strand and PAZ domain Arg931 (Fig. 2D), explaining the preference of DCL3 for A1-containing substrate guide strand (22–24, 26). Replacement of A1 with other bases revealed that pyrimidines, such as U and C, are too small to form similar hydrogen-bonding interaction as adenine (Fig. S2D). Although the G base has the potential to form similar hydrogen-bonding interactions (Fig. S2D), the three hydrogen bonds of the G-C pair require more energy to disrupt than the two hydrogen bonds of the A-U pair, making base flipping by DCL3 energetically more favorable for A-U than for G-C. Consistent with this reasoning, U is the second most preferred base at guide strand position 1, with C and G not being preferred (22). In contrast, DCL4 was reported to possess no preference for the 5’-end base of guide strand (22), suggesting that different Dicers may employ different 5’-end base recognition mechanism.
The 3’-end of the complementary strand is recognized by a PAZ domain aromatic cap formed by His849, Phe869, Tyr880, Tyr883, Phe884 and Tyr888, which both encloses and provides stacking opportunities with the 3’-end C-1’ base (Fig. 2E and S2B). Additionally, His849, Tyr883, Lys887, and Tyr888 form salt bridge and hydrogen-bonding interactions with the phosphate groups (Fig. 2E). Thus, recognition of both the 5’-phosphorylated-A1 and 1-nt 3’-overhang of the bound pre-siRNA duplex by the platform-PAZ-connector cassette supports the previous model that Dicer combines the 5’- and 3’-end recognitions as a start to measure the sRNA length (3–5).
DCL3 possesses low but significant activity towards substrates with 5’-phosphorylated-U and/or longer 3’-overhangs (22). To understand this binding tolerance, we determined the cryo-EM structure, at the 3.73 Å resolution, of DCL3 in complex with a 30-bp pre-siRNA having a guide strand 5’-phosphorylated-U1 and a complementary strand 2-nt 3’-overhang (Fig. S3–S4 and Tables S1–2). The structure closely resembles the DCL3-40-bp RNA complex, with an RMSD of 1.9 Å (Fig. S4D), but the faint density corresponding to helicase domain was much weaker and was not built, either (Fig. S3B). The termini of the RNA can be clearly traced with both bases of the U1-A1’ pair flipped-out from the RNA duplex, in contrast to the DCL3-40-bp pre-siRNA complex with only the A1 of guide strand flipped-out (Fig. 2F–H). Recognition of the 5’-phosphorylated-U1 is similar to 5’-phosphorylated-A1, with the major difference being the lack of a hydrogen bond between the U1 base and the complementary strand (Fig. S5A). Superimposition of the dsRNA molecules in the two complexes indicates that the guide strands adopt very similar conformations, while the flip out of the complementary strand A1’ of the 2-nt-overhang RNA distorts the backbone of the RNA (Fig. 2H and S5B). The overhanging G-1’ and A-2’ turn back, superimposing with the U1’ and C-1’ of the 1-nt-overhang RNA in the DCL3-40-bp RNA complex, and are captured by the PAZ domain aromatic cap in a manner similar to the 1-nt-overhang RNA (Fig. 2H and S5C). Because A2-U2’ are paired in both structures, the longer 3’-overhang can loop out from the 1’-position with the last 2-nt of the overhang folding back and being captured by the PAZ aromatic cap, explaining the tolerance mechanism for the longer 3’-overhang.
Similar as previously reported bacterial or yeast RNaseIII homodimer structures (27, 28), the two DCL3 RNaseIII domains adopt a conserved side-by-side arrangement with the two corresponding active sites, marked by Ca2+ ions, located at adjacent positions along the two RNA strands, flanking the minor groove (Fig. 3A). The RNaseIIIa residues Glu1015, Asp1019, Asp1133 and Glu1136 coordinate one Ca2+ ion, mimicking the Mg2+ ion, to participate in the phosphodiester bond cleavage between G23’ and A22’ of the complementary strand (Fig. 3B and S2B), producing a 23-nt-RNA (C-1’-A22’). Similarly, the RNaseIIIb residues Glu1224, Asp1228, Asp1316 and Glu1319 coordinate a second Ca2+ ion to participate in cleavage between C24 and U25 of the guide strand to produce a 24-nt-RNA (A1-C24) (Fig. 3C and S2B). Thus, simultaneous cutting at both strands will produce a dsRNA with newly generated 2-nt 3’-overhang on the guide strand and 1-nt 3’-overhang on the complementary strand (Fig. 2A), consistent with previous biochemical data (22). Consequently, DCL3 mainly produces siRNAs with a 24-nt 5’-phophylated guide strand (Pol IV strand) and a 23-nt 1-nt-overhang complementary strand (RDR2 strand). In planta, AGO4 prefers 24-nt siRNAs possessing 5’-A1 as the guide strand (29, 30), perfectly matching the DCL3 Pol IV strand product, which is thus predominately selected by AGO4 to guide RdDM (23), resulting in strand-biased 24-nt siRNA production (23, 24).
Fig. 3. The active site conformation.
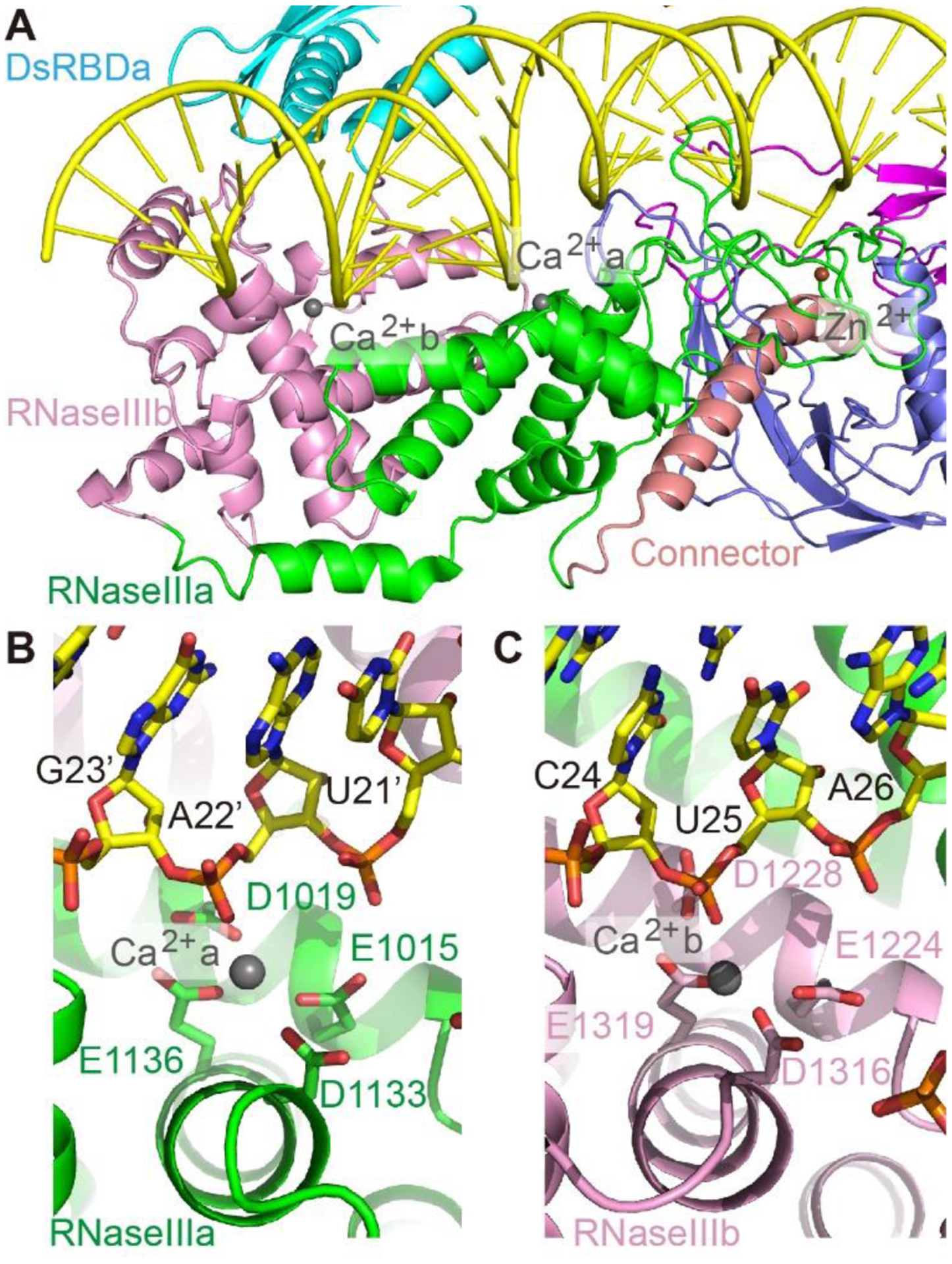
A. Overall structure of the active sites of RNaseIIIa and RNaseIIIb. B-C. Active sites conformation of RNaseIIIa (B) and RNaseIIIb (C).
The two dsRBDs cover the opposite side of pre-siRNA relative to the positions of RNaseIIIa/b (Fig. 1C). The two dsRBDs adopt very similar conformations and their highly positively charged surfaces bind the RNA backbone (Fig. S6A-B). Although both the 5’-half (A2-U18) and 3’-half (C23–C33) of the pre-siRNA adopt A-form conformations, the two segments do not align, because of a conformational distortion in the central region (U19–U22) (Fig. S6C), which can be observed in the DCL3-30-bp RNA complex, too (Fig S6D). This conformational change displaces the 3’-half of the RNA towards the DCL3 active sites (Fig. S6E). In the substrate-loaded complex of yeast RNaseIII Rnt1p-RNA complex structure, while the Rnt1p RNaseIII homodimer is well superimposed with the RNaseIIIa/b of DCL3, the two RNA molecules bounded by Rnt1p homodimer mimic the 5’-half and 3’-half of the DCL3-bounded RNA, respectively, shifting at the same position corresponding to U19 to U22 of DCL3-complexed RNA (Fig. S6E) (27). In contrast, the two dsRBDs of the Rnt1p homodimer occupy different positions other than the dsRBDa/b of DCL3 (Fig. S6E) (27). Together, the similar RNA conformational change upon binding by RNaseIIIs and the different dsRBD binding positions along the RNA suggest a plausible dynamic active dicing model that the RNaseIIIs capture and twist the RNA, while the dsRBDs dynamically bind at different positions during the dicing process. Due to the limited available structures, more Dicer-RNA complex structures in different states are required to shape the precise overall working model for Dicer. .
We performed structure-based mutagenesis studies both in vitro and in vivo. Wild-type (WT) DCL3 cuts a 30-bp pre-siRNA with a 5’-phosphorylated-A1 and a 1-nt 3’-overhang into 24-nt and 23-nt bands efficiently (Fig. 4A–B). Mutations in the 5-phosphate binding pocket (K695A/K957A/K903A), A1 recognition (H909A), and 3’-binding PAZ domain aromatic cap (Δ865–895) all showed decreased activity compared with WT (Fig. 4A and S7A), indicating the role of both the 5’- and 3’-counting in dicing. Both the dsRDBb deletion (Δ1450–1570) and the dsRBDa/b deletion construct (Δ1356–1570) decreased DCL3 activity, suggesting that the dsRBDs contribute to enzymatic processing (Fig. 4A and S7A). An RNaseIIIa active site mutant (E1015A/D1019A/D1133A/E1136A) generated only 24-nt sRNAs and no 23-nt sRNAs (Fig. 4B), whereas, an RNaseIIIb mutant (E1224A/E1228A/D1316/E1319A) generated only 23-nt sRNAs and no 24-nt sRNAs (Fig. 4A), confirming that RNaseIIIa and RNaseIIIb are responsible for complementary strand and guide strand cleavage, respectively.
Fig. 4. Biochemical and in vivo assays.
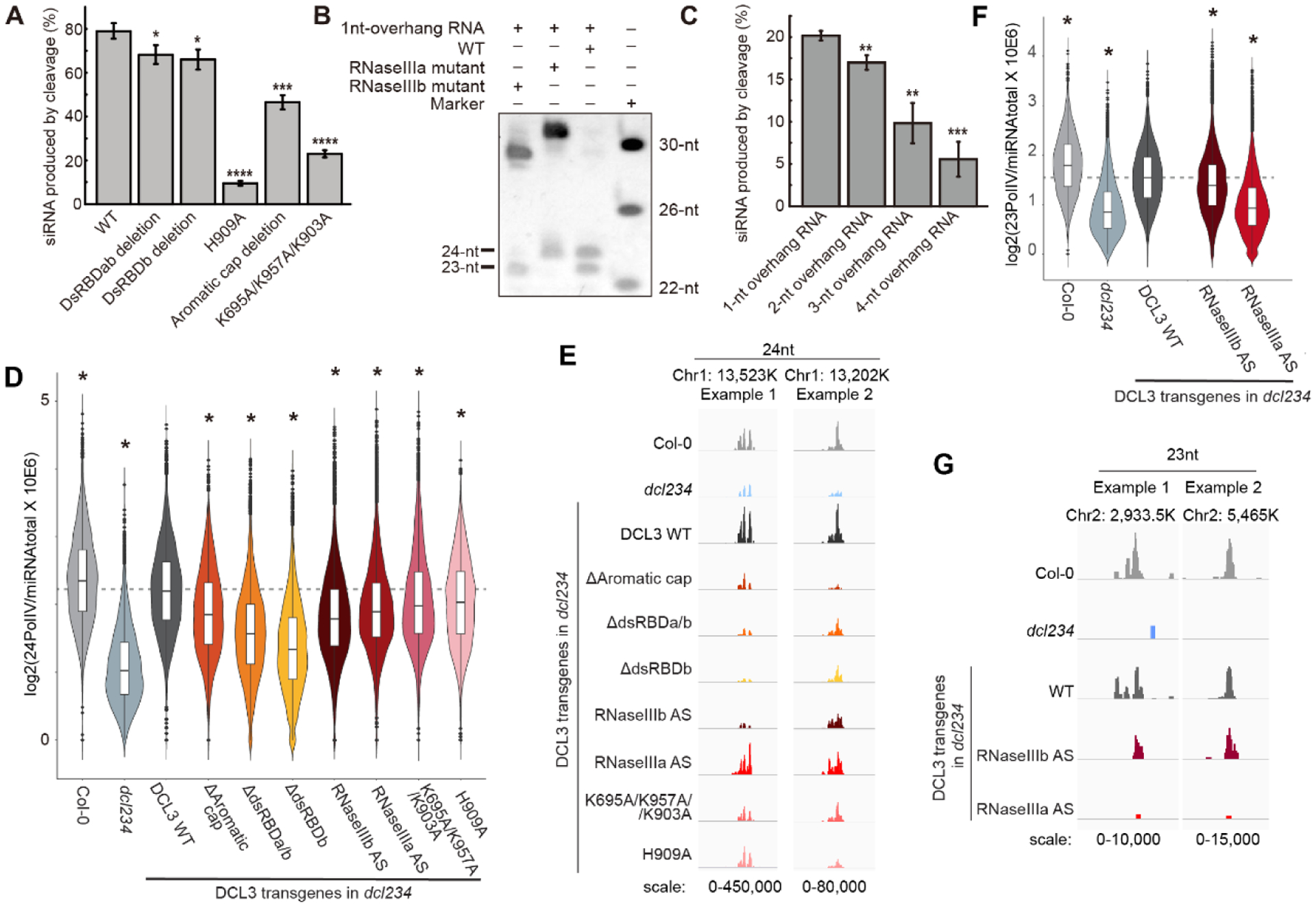
A. In vitro dicing of DCL3 and its mutants. Aromatic cap deletion, Δ865–895; dsRDBb deletion, Δ1450–1570; dsRBDa/b deletion, Δ1356–1570. B. In vitro dicing of DCL3 WT, RNaseIIIa mutant (D1133A/E1136A/E1015A/D1019A), and RNaseIIIb mutants (D1316A/E1319A/E1224A/D1228A). C. DCL3 dicing activity against different RNAs with the same guide strand possessing 5’-phophorylated-A1 and different complementary strands with 3’-overhang ranging from 1-nt to 4-nt. The in vitro assays were performed in a 50 μl reaction system containing 0.003 nmol (A and C) or 0.024 nmol (B) DCL3 or its mutants and 0.01 nmol substrate RNA at 37 °C. The reaction time were set to 5 min (A), 40 min (B), and 2 min (C), respectively. In panels A and C, percentages of the product siRNA were shown as means ± SD (n = 3), two-tailed Student’s t-test was applied, and the conventions used to report the statistical significances of p<0.05, p<0.01, p<0.001, and p<0.0001 were denoted by *, **, ***, and ****, respectively. D and F. Violin plot showing the abundance of 24-nt (D) and 23-nt (F) sRNAs respectively over Pol IV dependent sRNA regions normalized to total miRNAs, n=7,632. Data represent the average of two biological replicates from two independent T1 plants. Two tailed Student’s t-test was used to determine the significance of difference of each indicated genotype compared to the DCL3 WT complementing line. * indicates p-value < 0.01. AS, active site mutant. E and G. Screen shots showing the abundance of 24-nt (E) and 23-nt (G) sRNAs respectively over two example regions.
To further understand the influence of 3’-overhang, we tested RNA substrates with the same 5’-phosphorylated-A1 in the guide strand but with various 3’-overhangs in the complementary strand. Consistent with our structural observations, longer 3’-overhang decreased activity (Fig. 4C and S7B). Moreover, these dsRNAs were cut into one consensus 24-nt band and the other bands ranged from 23–26-nt, depending on overhang lengths (Fig. S7B), suggesting that the 5’-counting predominates over 3’-counting. Thus, the sRNA counting mechanism of DCL3 appears to rely more on measurement of the guide strand RNA length through the recognition of the 5’-phosphate and flipped-out base by the platform-PAZ-connector cassette, and Mg2+ mediated RNA cleavage by the RNaseIIIb domain. In contrast, the Giardia Dicer was reported to strictly obey the 3’-counting rule (3, 5), suggesting that different Dicers possess different counting-end preference.
To assess the in vivo functions of DCL3, wild type pDCL3::DCL3-3xFLAG (hereafter DCL3 WT) or various mutants were transformed into the Arabidopsis dcl2 dcl3 dcl4 (dcl234) triple mutant background, to avoid potential complications caused by the partial functional redundancy with DCL2/4 (9, 16) (Fig. S8). The abundance of 24-nt sRNAs over Pol IV regions (23) was examined in two independent T1 plants of each genotype. DCL3 WT restored the biogenesis of 24-nt sRNAs to near WT levels when transformed into dcl234 (Fig. 4D–E). Consistent with the in vitro studies, the 5’-binding mutants, K695A/K957A/K903A reducing 5’-phosphate binding and H909A reducing 5’-A1 base recognition, and the 3’-binding mutant, Δ865–895 to delete the aromatic cap, all led to significant losses of 24-nt sRNA (Fig. 4D–E). Deletions of dsRBDa/b (Δ1356–1570) or dsRBDb (Δ1450–1570) both failed to restore 24-nt sRNA generation (Fig. 4D–E). The D1316A/E1319A/E1224A/D1228A mutant, which disrupts RNaseIIIb and guide strand processing, led to a significant loss of 24-nt siRNA in vivo (Fig. 4D–E). The D1133A/E1136A/E1015A/D1019A, which affects RNaseIIIa and complementary strand processing, led to a strong loss of 23-nt siRNA in vivo (Fig. 4F–G). Interestingly, we observed a moderate loss of 23-nt sRNAs in RNaseIIIb mutant and a mild loss of 24-nt sRNAs in RNaseIIIa mutant (Fig. 4D–G). These observations likely indicate that the stability of 23- and 24-nt sRNA is dependent on each other. This stability inter-dependence may take place before the 24-nt sRNAs are loaded onto AGO4, or alternatively, the 23-nt passenger strand may facilitate the loading of the 24-nt sRNA onto AGO4 and thus leads to its stabilization. An equally plausible explanation is that cleavage of the two strands are coupled in vivo. Further experimental evidences are required to shed light on the hypotheses.
We further compared DCL3 with human and Giardia Dicers. While the RNaseIIIa/b active sites are conserved, the platform-PAZ-connector cassette of human Dicer is positioned closer to the RNA termini (Fig. S9A), while in Giardia Dicer the platform-PAZ-connector cassette is positioned further away from the RNA termini, allowing the binding of longer RNAs (Fig. S9B), consistent with the human and Giardia Dicers producing slightly shorter (22-nt) or longer (25-nt) sRNAs, respectively, than DCL3 (3, 6). Thus, the sRNA product length of different Dicers likely depends on the relative orientation/positioning between the RNaseIIIa/b active sites and platform-PAZ-connector cassettes. Considering the human and Giardia Dicers structures are not in the dicing-competent conformation, it is also possible that they may take conformational change in active dicing. The future works on more cleavage-competent conformation Dicer structures are required to fully reveal the product length determination mechanism of Dicer proteins.
In conclusion, the DCL3-RNA complex structures in dicing-competent states reveal the molecular basis for the terminus-specificity, accurate length measurement, and the strand-biased 24-nt siRNA production by DCL3. Given the overall shared length measurement and dicing mechanism of Dicers, our structure also provides insight into the general principle of length measurement and asymmetric cutting by Dicers from other species, which may allow us to design engineered Dicers to producing sRNAs with defined terminus preferences and lengths.
Supplementary Material
Acknowledgements:
We thank the staff at SUSTech Cryo-EM Center for assistance during data collection, Dr. Dinshaw Patel for critical reading, and Dr. Guy Riddihough (LSE) for editing.
Funding:
National Key R&D Program (2016YFA0503200), Shenzhen Science and Technology Program (JCYJ20200109110403829 and KQTD20190929173906742), SUSTech (G02226301), Key Laboratory of Molecular Design for Plant Cell Factory of Guangdong Higher Education Institutes (2019KSYS006) to J.D. Guangdong Innovation Research Team Fund (2016ZT06S172) to S.L. NIH to S.E.J. S.E.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Data and materials availability:
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-31963 and EMD-31964. The structures have been deposited in the Protein Data Bank with the accession codes: 7VG2 and 7VG3. Small RNA sequencing data are accessible at NCBI’s Gene Expression Omnibus (GEO) via GEO Series accession number GSE179616.
References and notes:
- 1.Kim VN, Han J, Siomi MC, Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10, 126–139 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Meister G, Argonaute proteins: functional insights and emerging roles. Nat Rev Genet 14, 447–459 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Macrae IJ et al. , Structural basis for double-stranded RNA processing by Dicer. Science 311, 195–198 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Tian Y et al. , A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human Dicer. Mol Cell 53, 606–616 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park JE et al. , Dicer recognizes the 5’ end of RNA for efficient and accurate processing. Nature 475, 201–205 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z et al. , Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate. Cell 173, 1191–1203 e1112 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Sinha NK, Iwasa J, Shen PS, Bass BL, Dicer uses distinct modules for recognizing dsRNA termini. Science 359, 329–334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song X, Li Y, Cao X, Qi Y, MicroRNAs and Their Regulatory Roles in Plant-Environment Interactions. Annu Rev Plant Biol 70, 489–525 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Henderson IR et al. , Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38, 721–725 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Xie Z, Allen E, Wilken A, Carrington JC, DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 102, 12984–12989 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deleris A et al. , Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313, 68–71 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Wu H et al. , Plant 22-nt siRNAs mediate translational repression and stress adaptation. Nature 581, 89–93 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Mosher RA, Schwach F, Studholme D, Baulcombe DC, PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proceedings of the National Academy of Sciences of the United States of America 105, 3145–3150 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ronemus M, Vaughn MW, Martienssen RA, MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by argonaute, dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell 18, 1559–1574 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vazquez F et al. , Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16, 69–79 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Xie Z et al. , Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2, E104 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE, Role of RNA polymerase IV in plant small RNA metabolism. Proceedings of the National Academy of Sciences of the United States of America 104, 4536–4541 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao X, Jacobsen SE, Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12, 1138–1144 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Li CF et al. , An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126, 93–106 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS, RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet 41, 630–634 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zilberman D, Cao X, Jacobsen SE, ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299, 716–719 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Nagano H, Fukudome A, Hiraguri A, Moriyama H, Fukuhara T, Distinct substrate specificities of Arabidopsis DCL3 and DCL4. Nucleic Acids Res 42, 1845–1856 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhai J et al. , A One Precursor One siRNA Model for Pol IV-Dependent siRNA Biogenesis. Cell 163, 445–455 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh J, Mishra V, Wang F, Huang HY, Pikaard CS, Reaction Mechanisms of Pol IV, RDR2, and DCL3 Drive RNA Channeling in the siRNA-Directed DNA Methylation Pathway. Mol Cell 75, 576–589 e575 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi Y, Denli AM, Hannon GJ, Biochemical specialization within Arabidopsis RNA silencing pathways. Molecular Cell 19, 421–428 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Li S et al. , Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res 25, 235–245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song H et al. , The Functional Cycle of Rnt1p: Five Consecutive Steps of Double-Stranded RNA Processing by a Eukaryotic RNase III. Structure 25, 353–363 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gan JH et al. , Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell 124, 355–366 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Mi S et al. , Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell 133, 116–127 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi Y et al. , Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443, 1008–1012 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Mastronarde DN, Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheres SHW, Amyloid structure determination in RELION-3.1. Acta Crystallogr D Struct Biol 76, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zivanov J et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su M, goCTF: Geometrically optimized CTF determination for single-particle cryo-EM. J Struct Biol 205, 22–29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Afonine PV et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersen EF et al. , UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Scheres SHW, Chen S, Prevention of overfitting in cryo-EM structure determination. Nature Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swint-Kruse L, Brown CS, Resmap: automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics 21, 3327–3328 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quinlan AR, Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozomara A et al. , miRBase: from microRNA sequences to function. Nucleic Acids Research 47, D155–D162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan YZ et al. , Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nature Methods 14, 793–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-31963 and EMD-31964. The structures have been deposited in the Protein Data Bank with the accession codes: 7VG2 and 7VG3. Small RNA sequencing data are accessible at NCBI’s Gene Expression Omnibus (GEO) via GEO Series accession number GSE179616.


