Abstract
Here, we report that in T47D breast cancer cells 50 pM progestin is sufficient to activate cell cycle entry and the progesterone gene expression program. At this concentration, equivalent to the progesterone blood levels found around the menopause, progesterone receptor (PR) binds only to 2800 genomic sites, which are accessible to ATAC cleavage prior to hormone exposure. These highly accessible sites (HAs) are surrounded by well-organized nucleosomes and exhibit breast enhancer features, including estrogen receptor alpha (ERα), higher FOXA1 and BRD4 (bromodomain containing 4) occupancy. Although HAs are enriched in RAD21 and CTCF, PR binding is the driving force for the most robust interactions with hormone-regulated genes. HAs show higher frequency of 3D contacts among themselves than with other PR binding sites, indicating colocalization in similar compartments. Gene regulation via HAs is independent of classical coregulators and ATP-activated remodelers, relying mainly on MAP kinase activation that enables PR nuclear engagement. HAs are also preferentially occupied by PR and ERα in breast cancer xenografts derived from MCF-7 cells as well as from patients, indicating their potential usefulness as targets for therapeutic intervention.
INTRODUCTION
Estrogens and progestins exert their effects on breast cancer cells mainly via binding to intracellular receptors, estrogen receptor alpha (ERα) and progesterone receptor (PR), whose binding to genomic hormone response elements (HREs) allows recruitment of chromatin remodeling complexes and transcriptional coregulators (1). In addition, a small proportion of ERα and PR form a complex attached to the cell membrane by palmitoylation (2,3), which is important for the cellular response to hormone by virtue of their crosstalk with SRC/RAS/ERK kinase signaling cascades (4,5). Progestin binding to the membrane-attached PR can activate SRC directly (6) or via the ERα (2,3). Activation of the SRC/RAS/ERK pathway is essential for hormone action on the genome, as ERK phosphorylates the intracellular PR at S294 (pPR) and also activates MSK, resulting in the formation of a ternary complex pPR/ERK/MSK, which is targeted to the genomic HREs, where MSK1 phosphorylates histone H3 at S10 contributing to nucleosome remodeling (7). Activation of the ERK pathway by EGF can mediate the activation of PR through phosphorylation at S294, even in the absence of hormone (8). CDK2 is also activated in response to progestins and can phosphorylate PR at S400 (9), a process that can happen in the absence of hormone (10). CDK2 activated in response to progestin phosphorylates and activates PARP1, which becomes essential for displacement of linker histone H1, the first step in hormone-induced chromatin remodeling (9).
For these and other studies of the effect of progestins in breast cancer cell lines, concentrations ranging between 10 and 100 nM are routinely used. These concentrations are much higher than the levels of progesterone found in women blood around the menopause, when most breast cancers are detected. The median age at breast cancer diagnosis is 61 years, when the progesterone concentrations in blood serum range from 47 to 318 pM (median 127 pM) (11). In addition, for experiments with cell lines synthetic progestins are primarily used, which are poor substrates for metabolic enzymes, leading to increased intracellular availability. Although experiments with high concentrations of progestins have allowed the study of the molecular mechanisms involved in hormonal action, the question remains whether the observed results reflect what would take place at more physiological hormone concentrations.
To address this question, we investigated the initial response of breast cancer cell lines to low concentrations of progestins. Here, we report that breast cancer cells exposed to 50 pM progestin R5020 (promegestone) or progesterone respond with entry in the cell cycle in a way comparable to that of cells exposed to 10 nM progestin. Moreover, while at 10 nM progestin PR binds to >50 000 genomic regions, in cells exposed to 50 pM progestin we detect only 2800 PR-bound genomic regions. These regions are already accessible to cleavage by the ATAC transposase prior to hormone exposure and therefore we named them highly accessible sites (HAs). In hormone-starved cells, HAs exhibit acetylated well-positioned nucleosomes and are enriched in ERα, FOXA1 and BRD4 (bromodomain containing 4). The 3D nuclear architecture analysis shows that HAs tend to contact among themselves more often than expected and interact with PR binding regions observed at higher hormone concentration, suggesting the existence of a functional hierarchy of genomic PR binding regions. We found that prior to hormone exposure T47D cells that express high levels of PR exhibit more open chromatin features over HAs than the same cells partially depleted of PR, or another breast tubular epithelial cell line (MCF-7) with much lower PR levels. In MCF10A, nontumoral cells of the same lineage with undetectable PR expression, chromatin at HAs is poorly open, indicating that a certain level of PR is required for maintaining accessibility of HAs even in the absence of the ligand. Furthermore, we have found that HAs are occupied by ERα and PR in MCF-7 and in patient-derived xenografts (PDXs) at physiological hormone concentrations. Thus, we have unveiled a small subset of regions relevant for the initial response of breast cancer cells to physiological progestin concentrations.
MATERIALS AND METHODS
Cell culture and hormone treatments
T47D breast cancer cells were routinely grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin.
For the experiments, cells were plated in RPMI medium without phenol red supplemented with 10% dextran-coated charcoal-treated FBS (DCC/FBS) and 48 h later medium was replaced by fresh medium without serum. After 24 h in serum-free conditions, cells were incubated with R5020 or progesterone at different concentrations of 0.01, 0.05, 0.5, 0.1, 1 and 10 nM for different times at 37ºC. T47Dwhite cells were routinely grown in RPMI medium without phenol red supplemented with 10% DCC/FBS.
MCF10A cells were routinely grown in DMEM/F-12 medium with 5% horse serum, 20 ng/ml EGF, 100 U/ml penicillin, 100 mg/ml streptomycin, 0.5 mg/ml hydrocortisone, 100 ng/ml cholera toxin and 10 mg/ml insulin.
Specific inhibitors were used along this study: for ERK, PD98059 (50 μM; Calbiochem); for BRD4, JQ1 (500 nM; Sigma, SML0974); for MSK1, H89 (15 μM; InvivoGen, tlrl-h89); for ERα, ICI 182180 (fulvestrant, 10 μM; Tocris, 1047) and tamoxifen (10 μM; Sigma, H7904); for CDK2, CDK2 inhibitor 2 (5 μM; Calbiochem, 21-944); and for p300/CBP, P300i (10 μM; Tocris, 4891). All these inhibitors were added 1 h before hormone induction.
Chromatin immunoprecipitation in cultured cells
Chromatin immunoprecipitation (ChIP) assays were performed as described (12) using anti-PR (H190 SC-7208, Santa Cruz), anti-ERK2(D12) (05-157, Merck), anti-SRC3-(5E11) (#2126, Cell Signaling), anti-p300, clone NM11 (#61401, Active Motif), anti-RNApol II (#2629, Cell Signaling), anti-RAD21 (ab992, Abcam), anti-CTCF (07-729, Merck), anti-H3K27ac (ab4729, Abcam), anti-H3K18ac (#39693, Active Motif) and anti-BRD4 (kind gift from Cheng-Ming Chiang, UT Southwestern Medical Center). Quantification of ChIP was performed by real-time PCR using Roche LightCycler (Roche). The fold enrichment of target sequence in the immunoprecipitated (IP) compared to input (Ref) fractions was calculated using the comparative Ct (the number of cycles required to reach a threshold concentration) method with the equation 2Ct(IP) − Ct(Ref). Each of these values was corrected by the human β-globin gene and referred to as relative abundance over time zero. Primer sequences for HAs, medium accessible sites (MAs) and low accessible sites (LAs) are available in Supplementary Table S1.
Coimmunoprecipitation assay
T47D-MTVL cells treated or not with 50 pM or 10 nM R5020 were lysed, and cell extracts (1 mg protein) were incubated overnight with protein A/G agarose beads previously coupled with 3 μg of the corresponding PR antibody (Santa Cruz, H-190) or an unspecific control antibody. The immunoprecipitated proteins (IPs) were eluted by boiling in SDS sample buffer. Inputs and IPs were analyzed by western blot using PR-, p300-, SRC3- and ERK-specific antibodies.
Cell proliferation
T47D-MTVL cells were cultured as described earlier. Cells (1 × 104) were plated in a 96-well plate in the presence or absence of 0.01, 0.025, 0.050, 0.1, 1 and 10 nM R5020. The cell proliferation ELISA BrdU colorimetric assay (Roche) was performed according to the manufacturer’s instructions. The experiments were performed in quintuplicate.
Flow cytometry
T47D-MTVL cells were plated into duplicate wells of six-well plastic dishes and preincubated as described. After 24 h, 0.01, 0.05, 0.1, 1 and 10 nM R5020 or progesterone was added. Cells were harvested at the start of treatment (control, zero time) and after 24 h of hormone addition. The cell suspension was pelleted, stained with propidium iodide and treated with ribonuclease (RNase). Samples were cooled to 4°C, and 10 000 cells were analyzed on a BD FACS Canto analyzer flow cytometer.
RNA-seq
RNA was extracted from T47D-MTVL cells treated or not for 6 h with 50 pM or 10 nM R5020 and submitted to massive sequencing using the Solexa Genome Analyzer. The protocol followed to analyze the RNA-seq data can be found in the Supplementary Methods section. For RNA-seq experiments, biological duplicates were performed.
ChIP-seq
ChIP-DNA was purified and subjected to deep sequencing using the Solexa Genome Analyzer (Illumina, San Diego, CA). The protocol followed to analyze the ChIP-seq data can be found in Supplementary Methods section. HAs could be identified exclusively using the H190 rabbit antibody from Santa Cruz, which is no longer available; therefore, for PR ChIP-seq experiments, one replicate per point using this antibody followed by deep sequencing with 200 million reads was done. In addition, at least 20 HAs, MAs and LAs were validated by ChIP-qPCR. For T0 and 10 nM R5020 PR ChIP-seq, biological duplicates were done.
ATAC-seq
ATAC experiments were performed as described (13). Extended bioinformatics methods can be found in the Supplementary Data.
Hi-C experiments
Hi-C libraries were generated from T47D cells treated or not with R5020 for 60 min according to the previously published Hi-C protocol with minor adaptations (14). Hi-C libraries were generated independently in both conditions using HindIII and NcoI restriction enzymes. Hi-C libraries were controlled for quality and sequenced on an Illumina Hiseq2000 sequencer. The Illumina Hi-seq paired-end reads were processed by aligning to the reference human genome (GRCh37/hg19) using BWA.
PR HiChIP experiments
HiChIP experiments were performed as described (15) using PR-specific antibody (Santa Cruz, H190). Analysis was performed using dovetail HiChIP pipeline (https://hichip.readthedocs.io/en/latest/).
Rapid immunoprecipitation mass spectrometry of endogenous proteins
Rapid immunoprecipitation mass spectrometry of endogenous protein (RIME) experiments were performed as described (16).
RESULTS
Low concentrations of progestin trigger cell cycle entry, MAP kinase activation and gene regulation
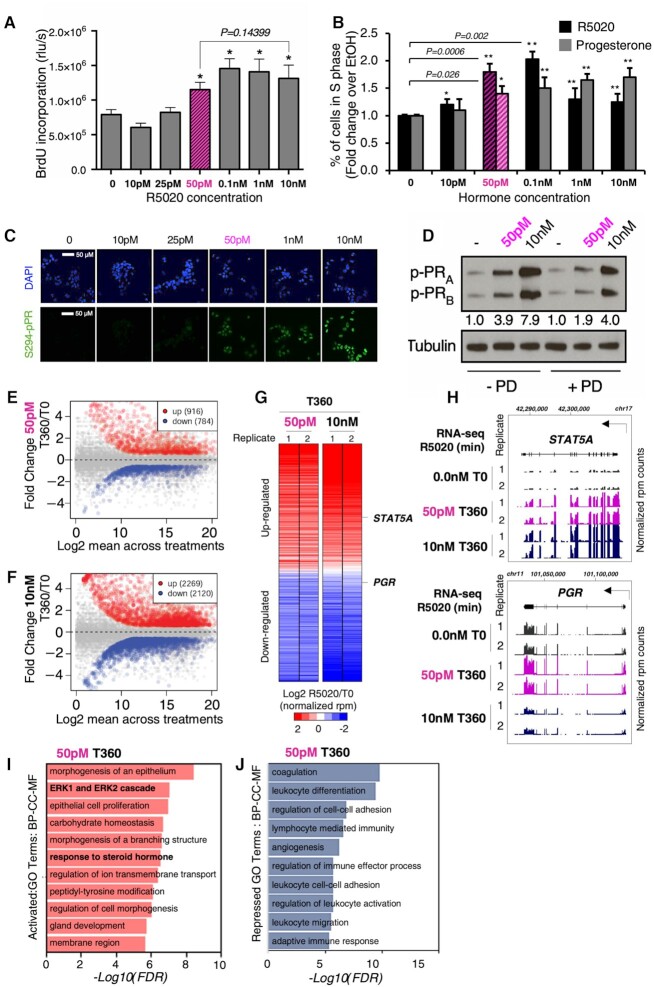
Most of our and other group studies on hormone action in T47D breast cancer cells have been performed using high concentrations (10 nM) of the progestin R5020 (promegestone). Conscious that 10 nM is a very high hormone concentration, we asked what could be the lowest progestin concentration capable of inducing the physiological hormonal response in these cells. To address this point, we carried out a dose–response curve in T47D cells deprived of hormone for 48 h monitoring BrdU incorporation and fluorescence-activated cell sorting (FACS) as indicators of DNA synthesis. By both criteria, we found that, compared to cells exposed just to solvent, 50 pM of R5020 was the lowest concentration able to increase the number of cells entering the cell cycle in a proportion similar to that observed with 10 nM (Figure 1A and B). Thus, a concentration of progestin 200 times lower than routinely used for cell culture was sufficient to induce entry in the cell cycle. Similar results were obtained when progesterone was used instead of R5020 (Figure 1B).
Figure 1.
Effects of various concentrations of R5020 on DNA synthesis, cell cycle, PR phosphorylation and transcriptome. (A) DNA synthesis. T47D-MTVL cells untreated (0) or exposed for 24 h to 10 pM, 25 pM, 50 pM, 100 pM, 1 nM or 10 nM R5020 for 24 h, as indicated. The incorporation of BrdU was measured using an ELISA colorimetric assay (Roche). Bars represent mean ± SD (n = 5). *P < 0.05 using Student’s t‐test. (B) Percentage of cells in S phase. Cells untreated (0) or exposed for 24 h to 10 pM, 25 pM, 50 pM, 100 pM, 1 nM or 10 nM R5020 or progesterone for 24 h, as indicated, were labeled with CD44-APC and CD24-PE antibodies under optimized conditions, and subjected to FACS analysis. Results are represented as mean and SD from three experiments performed in duplicate. *P < 0.05 using Student’s t-test. (C) Phosphorylation of PR at S294. Cells unexposed (T0) or exposed for 30 min to various concentrations of R5020 were subjected to immunofluorescence using a specific antibody against PR phosphorylated at S294 and a secondary antibody labeled with GFP (bottom panel). The top panel shows the same fields with the cells stained with DAPI. (D) Phosphorylation of PR isoforms at S294. Effect of 50 pM and 10 nM progestin on the levels of phosphorylated PR isoforms B and A. Cells pretreated or not with the ERK inhibitor PD98059 (PD) were exposed for 30 min to solvent (−) or to 50 pM or 10 nM R5020 as indicated, lysed and analyzed for the presence of PRS294ph using immunoblotting of cell extracts with specific antibodies and tubulin as a loading control. Scatter plot of gene expression in T47D cells incubated for 360 min (indicated as T360) without or with 50 pM (E) or 10 nM (F). The number of genes up- or downregulated in the T360 condition (−1.5 < FC (fold change) > 1.5; adjusted P-value <0.05) is indicated (see the ‘Materials and Methods’ section for a more detailed description). (G) Heatmaps of differential gene expression for T47D cells exposed to either 50 pM or 10 nM R5020 for 360 min (indicated as T360) compared to time 0 (indicated as T0). Color bar scale stands for log2FC of normalized RNA expression rpm (reads per million base pairs) for the ratio of R5020 versus the T0 condition (R5020/T0) at each indicated concentration (50 pM or 10 nM). (H) Genome browser view of two representative genes from the RNA-seq data from panel (G): STAT5A locus (top panel) and the PGR locus (bottom panel). For both genes, time (360 min, indicated as T360) and concentration of R5020 (50 pM and 10 nM) are indicated on the left. For each experiment, the two biological replicates are also indicated by numbers 1 and 2. Chromosome coordinates are indicated at the top of each gene. Direction of transcription is indicated by the arrow. Plotted are the normalized rpm. WebGestalt gene ontology (GO) analysis (17) of genes upregulated (I) or downregulated (J) in cells exposed to 50 pM R5020 for 360 min (indicated as T360). Reported are the top 10 enriched biological processes, cellular components and molecular functions for the differentially expressed genes with a false discovery rate (FDR) < 10–5 ordered according to −log10(FDR).
Progestins at 10 nM concentration induce signaling via the small proportion of palmitoylated membrane-attached PR that activates the SRC/RAS/ERK pathway and phosphorylation of PR at S294 (3,5). We asked whether 50 pM progestin uses the same pathway. Indeed, 50 pM was the lowest progestin concentration capable of significantly increasing the PRS294ph signal as detected by either immunofluorescence (Figure 1C and Supplementary Figure S1A) or immunoblotting (Supplementary Figure S1B). The 4-fold increase in PRS294ph signal observed at 50 pM progestin was inhibited by the ERK-specific inhibitor PD98059 (Figure 1D). Thus, 50 pM progestin is sufficient to activate the MAP kinase pathway via the membrane-attached PR.
To investigate the effects of low hormone concentrations on gene regulation, we performed RNA-seq analysis in T47D cells untreated or exposed for 6 h (T360), to either 50 pM or 10 nM R5020 (Figure 1E–G). Upon exposure to 50 pM progestin, we found 916 genes upregulated and 784 genes downregulated by the hormone (fold change: 1.5; P < 0.05) (Figure 1E). For the same time of exposure to 10 nM R5020, we found 2269 upregulated and 2120 downregulated genes (Figure 1F). Of these genes, ∼38% and 31.1% were also upregulated and downregulated at 50 pM progestin, respectively (Supplementary Figure S1C). The majority of genes regulated at 50 pM were also regulated at 10 nM (Figure 1G), as exemplified by the STAT5A gene, one of the most upregulated genes at both concentrations (Figure 1H). Progestin-regulated genes were also validated by quantitative PCR (qPCR) after 3 and 6 h of hormone exposure (Supplementary Figure S1D).
Next, we implemented WebGestalt (17) to perform GO analysis of gene expression data. The upregulated gene group at 50 pM progestin showed significant (adjusted P-value <0.001) enrichment for genes associated with regulation of epithelial cell proliferation, ERK1/2 cascade and response to steroid hormones (Figure 1I). Compared to GO terms enriched upon 10 nM progestin exposure (Supplementary Figure S1E), 50 pM progestin seems to act predominantly on genes associated with hormone-related function and the kinase pathway that regulates them. On the other end, downregulated genes at 50 pM were enriched with GO terms associated with cell–cell adhesion and communication (Figure 1J), similarly to the larger group of genes downregulated at 10 nM progestin (Supplementary Figure S1F).
Interestingly, 8.5% of the upregulated genes (82 genes) and 10% of the downregulated genes (107 genes) either are only regulated at 50 pM progestin or showed an opposite tendency at the two progestin concentrations (Supplementary Figure S1G), pointing to distinctive features in gene regulation at low progestin concentration. An interesting case is the PGR gene itself, which was upregulated at 50 pM progestin and downregulated at 10 nM, indicating a positive feedback loop for maintaining hormonal function at low hormone concentration (Figure 1H). The transcription factor RUNX2, whose expression correlates with triple negative breast cancer (18), is also upregulated at 50 pM and inhibited at 10 nM progestin (Supplementary Figure S1H). Conversely, CCAAT/enhancer-binding protein alpha, a transcription factor with antiproliferative function in breast cancer cells (19), and the dual-specificity phosphatase 4 that counteracts the effect of the ERK1/2 and JNK1/2 kinases (20) were both downregulated at 50 pM but activated at 10 nM progestin (Supplementary Figure S1H), underlining the importance of the kinase pathways at low progestin concentration.
Altogether these results clearly emphasize that 50 pM progestin, a more physiological concentration for hormonal studies for breast cancer patients, activates an initial cell proliferation-oriented program largely dependent on kinase signaling.
A small subset of HAs mediates actions of 50 pM progestin
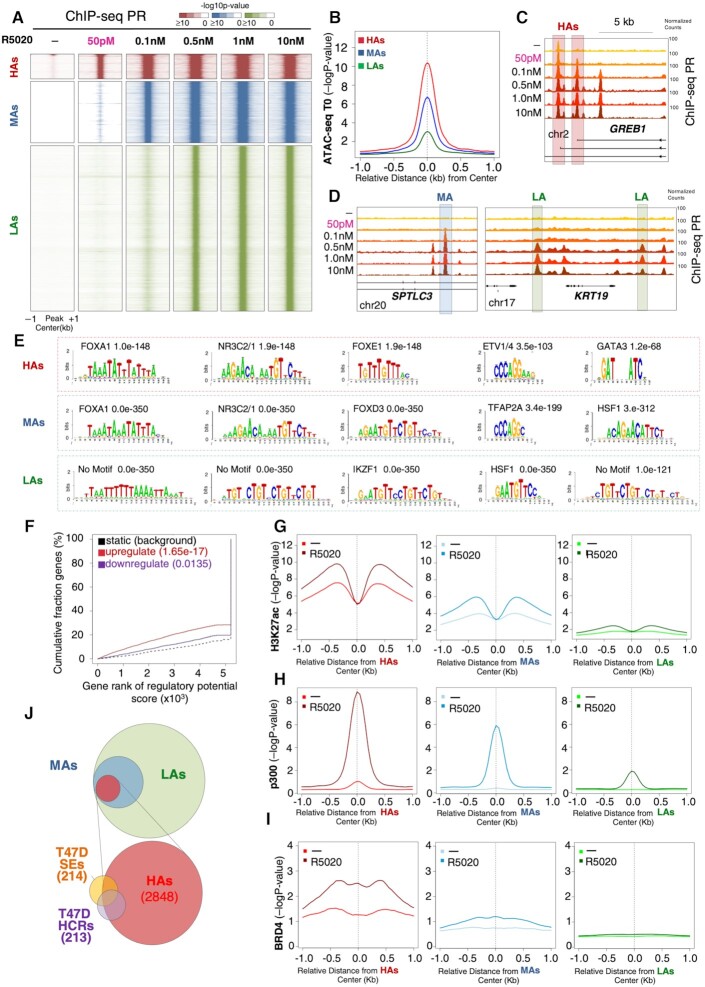
To explore the molecular mechanism behind the biological effects observed at low concentration of hormone, we incubated T47D cells for 30 min with a range of concentrations of R5020 between 50 pM and 10 nM and monitored PR genome occupancy by implementing ChIP-seq with high depth coverage (over 200 million reads per experiment). Apart from a small subset of 474 PR peaks found prior to hormone exposure, the lowest progestin concentration that allowed detection of ligand-dependent PR binding sites (PRBs) was 50 pM. At this concentration, we found 2848 new PRBs, 5.2% of the total peaks observed at 10 nM progestin (Figure 2A and Supplementary Figure S2A). These PRBs were highly accessible to ATAC cleavage before exposure to hormone (Figure 2B) and therefore we called this group HAs. Higher progestin concentration yielded additional PRBs. At 100 pM, we detected a new set of 7820 occupied sites that we called MAs (Figure 2A and B, and Supplementary Figure S2A), and at 0.5 nM and higher progestin we found a different new set of 43 479 occupied sites that we collectively called LAs (Figure 2A and B, and Supplementary Figure S2A). GREB1, a highly progestin upregulated gene, harbors two HAs near its 5′ end, one of which located at the promoter of the shorter transcript is weakly occupied prior to hormone exposure (Figure 2C). One of the introns of the serine palmitoyl transferase long chain base subunit 3 (SPTLC3) gene contains MAs (Figure 2D, left panel), whereas the keratin 19 (KRT19) gene has two different LAs within the surrounding intergenic regions (Figure 2D, right panel). We found that all three classes of PRBs appear mainly within introns and distal intergenic regions (40–50%), as well as near promoters (∼20%) (Supplementary Figure S2B).
Figure 2.
Characterization of PRBs at various progestin concentrations. (A) Heatmap of the dose–response curve of PR binding in T47D‐MTVL cells. ChIP‐seq signal of PR occupancy obtained after 30 min of incubation with 50 pM, 100 pM, 0.5 nM, 1 nM and 10 nM R5020 exposure is represented (window: −1 kb to +1 kb around the PR binding region). Color bar scale with increasing shades of color stands for higher enrichment (−log10 of the Poisson P-value). Based on the analysis of ChIP-seq experiments, sequenced reads were aligned to the human genome, the peaks of enrichment were defined and the P-values of the ChIP reads were calculated in each window (50 bp). Significant peaks were defined as previously shown (58). (B) ATAC-seq results showing chromatin accessibility of the three classes of PRBs prior to hormone exposure (T0) (the y-axis reports −log10 of the Poisson P-value). (C, D) Genome browser images of the ChIP-seq data showing representative genes of the three classes of PRBs: GREB1 (HAs), SPTLC3 (MAs) and KRT19 (LAs). The position of each PRBs is indicated by a gray rectangle. (E) Homer motif analysis of the three classes of PRBs. The most abundant binding motifs identified using Homer motif finding genome analysis tool are shown for HAs, MAs and LAs. The P-values are indicated. Note that although half HRE (TGTTCT) can be inferred from some of the enriched sequences in LAs, by using Homer motif discovery software not known transcription factor binding motif matched the enriched sequence and therefore is indicated as ‘no motif’. (F) Binding and expression target analysis (BETA) of HAs compared to expression data. The red and the purple lines represent the upregulated and downregulated genes after exposure to 50 pM hormone, respectively. The dashed line indicates the nondifferentially expressed genes as background. P-values are indicated and show the significant correlation with genes upregulated upon hormone exposure. The genes are accumulated by the rank on the basis of the regulatory potential score from high to low. The regulatory potential, defined as gene’s likelihood of being regulated by a factor, was estimated for each gene (21). P-values that represent the significance of the up or down group distributions are compared with the non-group by the Kolmogorov–Smirnov test. Average ChIP-seq signal of epigenetic marks at each class of PRBs. The average ChIP-seq signal of H3K27ac (G), p300 (H) and BRD4 (I) at uninduced conditions (T0, lighter color lines) as well as after 30 min exposure to 50 pM (HAs), 0.1 nM (MAs) and 0.5 nM (LAs) R5020 (darker color lines). p300 data were obtained from (38). The graphs are plotted over the summit of centered PRBs (plotted is the −log10 of the Poisson P-value). (J) BEDTools analysis of overlap between HAs and the superenhancers (SEs) (PMID: 30371817) and hormone-controlled regions (HCRs) (30) in T47D‐MTVL breast cancer cells. Overlapping intervals were obtained with BEDTools using the intersectBed function with default parameters (31).
To assure that the results of the RNA-seq experiments at 50 pM progestin are due only to the occupancy of HAs, we performed ChIP experiments for PR at 50 pM for 30 min, 2 h, 4 h and 6 h. As expected, HAs are occupied after 30 min of hormone exposure and the occupancy is maintained for longer times, whereas no occupancy of PR was found at MAs or LAs in the same conditions (Supplementary Figure S2C).
To explore the functional differences between PRBs, we performed motif analysis for all three categories. HAs are enriched in full palindromic HRE GGTACANNNTGTTCT, and in motifs for binding of FOXA1, FOXE1, ETV1/4 and GATA3 (Figure 2E, upper boxed panel). MAs were also enriched in palindromic HREs and motifs for FOXA1, FOXD3, TFAP2A and heat shock factor 1 (HSF1), while LAs were enriched for half palindromic HREs and HSF1 motifs (Figure 2E, middle and lower boxed panels, respectively). Thus, motif analysis suggested that lower concentrations of progestin might favor occupancy by PR of HAs and MAs, as both are enriched in full HREs and FOXA1. In contrast, LAs exhibiting only half HREs and lacking FOXA1 motifs could not support PR recruitment at very low progestin concentration (Figure 2A and E). To explore this, we employed BETA (21) to predict whether PR binding has activating or repressive functions when compared to differential expression of T47D cells upon 6 h exposure to 50 pM progestin. With these conditions, we found that HAs are significantly associated with an activating function (up: P = 1.65 × 10–17 versus down: P = 0.0135 in Figure 2F), also in agreement with a larger number of genes that were activated at 50 pM progestin (Figure 1E). MAs, which appear only with 100 pM, were associated with both gene activation and repression (up: P =2.38 × 10–25 versus down: P =2.74 × 10–7) (Supplementary Figure S2D, left panel), while LAs did not show any strong association with gene expression (up: P =0.0008 versus down: P =0.0038) (Supplementary Figure S2D, right panel) attesting to a less productive binding of the receptor. As for the 474 PRBs detected prior to hormone exposure, their small number does not yield significant motif enrichment.
Thus, our data have identified a minimum set of PRBs engaged at 50 pM progestin that although representing a small fraction of the whole occupied regions at higher hormone concentrations could be sufficient to trigger the observed biological effects.
HAs show chromatin features of active enhancers
We wondered about the reason for the high chromatin accessibility associated with the PRBs obtained in the absence of hormone or at 50 pM progestin concentrations (Figure 2A and B). One possibility was that HAs are localized in nucleosome-free regions (22,23). To test this hypothesis, we took advantage of available MNase-seq data obtained from T47D cells (Supplementary Figure S3A) (24). When we compared all three classes of PRBs, our analysis showed an MNase pattern that supports the existence of an array of about eight to nine well-positioned nucleosomes around HAs with the linker DNA efficiently cleaved by the MNase (Supplementary Figure S3A, left panel). Of note, this pattern turned out to be gradually reduced and restricted in the MAs that exhibit only four to five positioned nucleosomes (Supplementary Figure S3A, middle panel) and in the LAs that exhibits a single strongly positioned nucleosome (Supplementary Figure S3A, right panel).
Given these highly specific nucleosome patterns observed for the three classes of PRBs, we analyzed their epigenetic state by profiling histone H3 lysine 27 and lysine 18 acetylation (H3K27ac and H3K18ac), the lysine acetyltransferase 3B (KAT3B or p300), RNA polymerase II (RNApol II) and BRD4. At uninduced conditions and compared with MAs and LAs, HAs showed significant higher enrichment signal for H3K27ac, H3K18ac, p300, BRD4 (Figure 2G–I) and RNApol II (Supplementary Figure S3B), all features associated with active enhancers (25). Upon exposure to progestin (10 nM), a larger increase in H3K27ac, p300 and BRD4 is detected at HAs and MAs but not at LAs (Figure 2G–I). In addition, H3K18ac, a cancer progression marker associated with transcriptionally active gene promoters (26), was also already high at HAs prior to hormone exposure and increased significantly in the presence of hormone at HAs and MAs but not at LAs (Supplementary Figure S3C, left panel). As histone variant H2A.Z is involved in the activation of neo-enhancers in prostate cancer and participates in the recruitment of FOXA1 to enhancers (27), we asked whether HAs were also enriched in H2A.Z. We found that among all three classes of PRBs, HAs showed the highest level of this histone variant before and after hormone exposure (Supplementary Figure S3D).
These findings suggest that HAs display all features of active enhancers and we speculate whether these PRBs might overlap with the so-called superenhancers (SEs), which are important to control the cell-specific gene expression pattern (28). To date, 214 genomic regions were identified as SEs in T47D breast cancer cells (29) and 213 regions were identified as HCRs, which are frequently bound by unliganded ERα and PR (30). By using BEDTools (31) we found that half of the SEs and 75% of the HCRs contained at least one HA (Figure 2J), supporting the notion that these regions might in fact be key enhancers associated with cellular identity of T47D breast cancer cells and may act as local and global genome organizers.
We conclude that HAs are localized in accessible active chromatin characterized by the presence of a palindromic HRE as well as FOXA1 motifs wrapped around acetylated and H2A.Z-enriched well-positioned nucleosomes (Figure 2G–I, and Supplementary Figures S3A, C and D). In the presence of hormone, PR binding induces further acetylation and BRD4 recruitment to active enhancers (Figure 2I), which overlap with SEs and HCRs.
HAs are hubs of 3D gene interactions
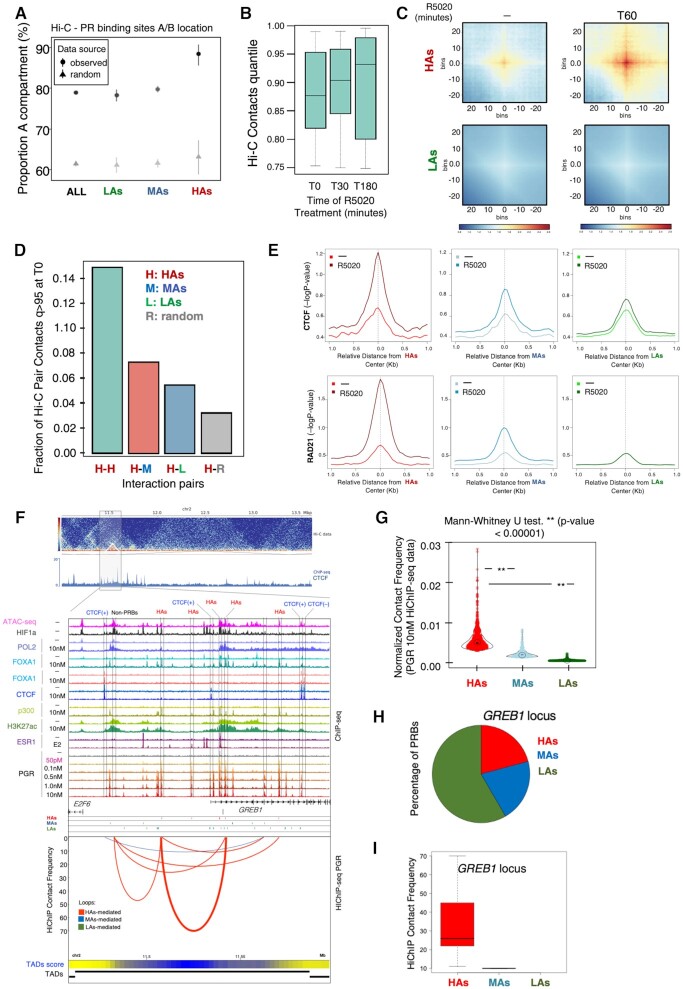
We have previously shown that HCRs establish long-distance inter-TAD (topologically associating domain) interactions between them and organize characteristic looping structures with promoters within their TADs in breast cancer cells expressing PR and ERα (30). Given the observed overlap between HCRs and HAs (Figure 2J), we explored the distribution of the latter into the spatial organization of chromatin in the cell nuclei by performing Hi-C analysis in T47D cells untreated or exposed to progestin. We found that HAs presented 90% of their contacts in compartment A (versus randomly expected 63%), compared with 79% for MAs (versus randomly expected 61%) and 78% for LAs (versus randomly expected 60%) (Figure 3A).
Figure 3.
HAs constitute a hierarchical hub of 3D gene interactions. (A) Distribution of 3D contacts for the different classes of PRBs. Hi-C libraries were generated from T47D‐MTVL cells unexposed or exposed to various concentrations of R5020 for 60 min according to the previously published Hi-C protocol with minor adaptations (14). Distribution of all PRBs, LAs, MAs and HAs, across the A and B compartments was determined from Hi-C assays. (B) Total number of 3D contacts in unexposed cells and in cells exposed to R5020 for 30 or 180 min. The total number of contacts after hormone treatment remained unchanged after 30 and 180 min of exposure to R5020. (C) HiCExplorer aggregate plots. Long-distance interactions among HAs and LAs. Hi-C analysis showed increased contacts among HAs than among LAs, both before and particularly after exposure to R5020 (60 min). The genomic coordinates of the HAs and LAs are centered between half the number of bins and the other half number of bins. Plotted are the submatrices of the aggregated contact frequency for 20 bins (1.5 kb bin size, 35 kb in total) in both upstream and downstream directions. Color bar scale with increasing red shades of color stands for higher contact frequency. (D) Distribution of Hi-C interaction pairs between HAs and all classes of PRBs. High–high, high–medium, high–low and high–random genomic regions in cells exposed for 30 min to 10 nM progestin. (E) Average ChIP-seq signal of CTCF and Rad21 occupancy (upper and lower panels, respectively) in the three classes of PRBs for both unexposed and exposed T47D‐MTVL cells to R5020. The graphs are plotted over the summit of centered PRBs (plotted is the −log10 of the Poisson P-value). (F) Interaction map for PR HiChIP at the GREB1 locus. Top panel: Hi-C data showing calculated TADs. Middle panel: ChIP-seq data for the indicated factor. Bottom panel: interaction frequency map obtained by the loops called on PR HiChIP-seq data. Arcs indicate the interaction between different genomic coordinates and are colored based on their overlap with PRBs (red HAs, blue MAs and green LAs). Thickness of the arc reflects the intensity of the interaction. The exact location of the HAs is indicated by the rectangles. TAD score is also indicated at the bottom as heatmap with blue representing higher score of TAD being present. (G) Violin plots of contact interaction frequency detected by HiChIP-seq of PR in T47D‐MTVL cells unexposed or exposed to R5020. Double asterisk indicates significance of the Mann–Whitney test. (H) Pie chart of the fraction (%) of detected PRBs with the genomic GREB1 locus is shown (right panels). (I) Boxplot showing the median of the contact frequency interactions for the different classes of PRBs indicated as calculated by FitHiChIP (33) at the GREB1 locus.
We therefore hypothesized that HAs might behave as 3D interaction hubs in breast cancer cells. To explore this, we tested the effect of the hormone over the PRB classes in the spatial 3D organization of the genome in T47D cells. After 30 and 180 min of hormone exposure, the total number of contacts remained unchanged compared with the untreated sample (Figure 3B). However, HAs showed a hormone-dependent increase in contacts with themselves compared with LAs (Figure 3C). In fact, HAs tend to interact more frequently with themselves as HCRs do (30), but also interact more frequently with MAs and LAs, as compared with random regions (Figure 3D).
As enhancers possess high frequency of chromatin interactions dictated by the action of CTCF and cohesin (32), both proteins should be detectable at HAs. To explore this, we analyzed the occupancy of the CTCF and the cohesin component, RAD21 at PRBs. Compared with MAs and LAs, the occupancy of CTCF and RAD21 is markedly higher in HAs, especially upon hormone exposure (Figure 3E and F), consistent with a critical role of these factors in mediating chromatin interactions of HAs.
To map interactions that are directly dependent on PR in the presence of hormone, we performed HiChIP experiments of PGR (15) in untreated as well as in T47D cells treated with hormone. Deep sequencing of the HiChIP experiments allowed us to construct normalized interaction maps showing PR-dependent contacts as well as their associated TADs (Supplementary S4A). Under uninduced conditions, although hidden by the overall low signal, genomic compartments can be already distinguished (Supplementary Figure S4A, left T0 panel). However, in the presence of hormone TADs, subdomains and loops are more pronounced and more interactions become evident (Supplementary Figure S4A, right T360 panel), confirming that at global level the HiChIP has captured 3D interactions.
We then moved to call loops in our HiChIP data (33), and found that HAs presented a higher frequency of contacts compared to MAs and LAs, and far from following a random distribution, these interactions occur preferentially between PRBs (Figure 3F and G). In fact, we observed that HAs tended to interact more with each other compared to MAs and LAs (Supplementary Figure S4B), confirming previous Hi-C experiments (Figure 3D). Around the hormone-activated GREB1 gene, the lack of CTCF peaks with convergent orientation could not mediate the formation of loops; however, loops between PRBs and preferentially involving HAs were identified (Figure 3F, H and I). Noticeably, a cluster of HAs can also coordinate looping with non-PRB accessible regions enriched in HIF1a and decorated with H3K27ac as observed in the proximity of the E2F6 gene (Figure 3F).
Another example of how HAs predominantly participate in DNA looping is shown for the SIRT1 regulating lncRNA tumor promoter (SIRLNT) locus (Supplementary Figure S4C–E). We conclude that HAs primarily act as core modules of enhancers that promote interactions in 3D with target genes and mediate the hormonal response of breast cancer cells even in the absence of CTCF-mediated loops.
HA occupancy correlates with PR expression
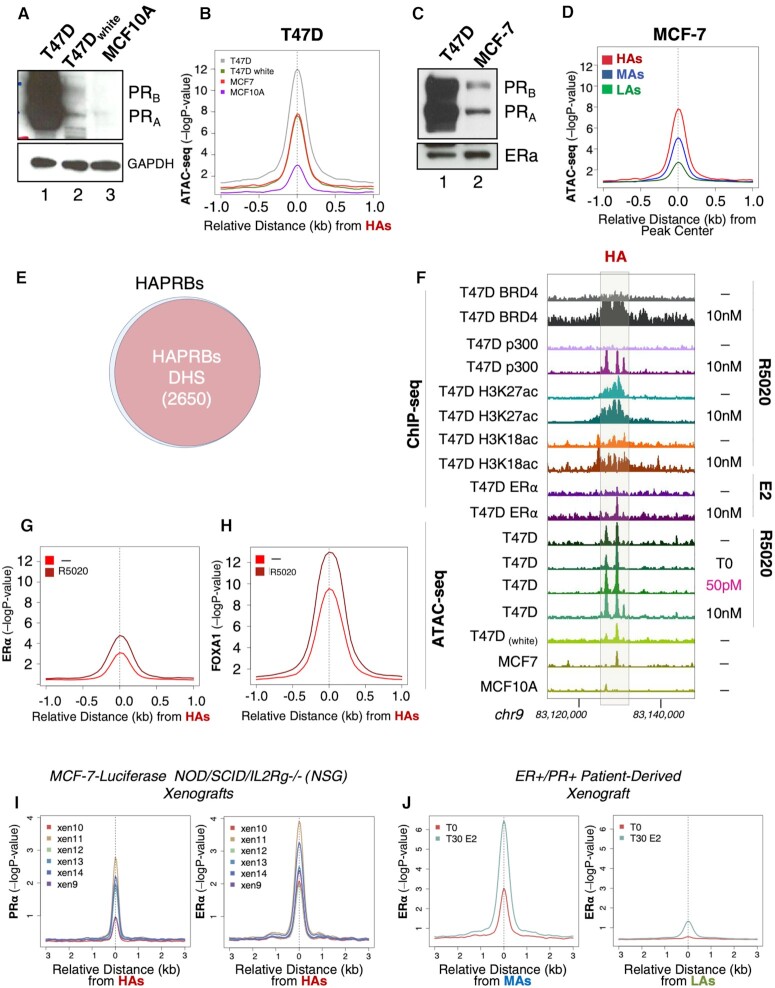
Our results indicate that the regions containing HAs are enhancers ready for PR binding at very low levels of progestin. However, whether PR itself contributes to the accessibility of these genomic regions was not clear. To investigate this issue, we conducted ATAC-seq assays in several cancer cell lines with different endogenous levels of PR and ERα. As previously shown, in T47D (breast cancer, PRhigh), HAs showed the greatest accessibility compared to MAs and LAs, even in the absence of hormone in the media (Figure 2B). Interestingly, when similar experiments were performed in T47D cells that were hormone deprived for several passages (T47Dwhite) and exhibited reduced PR levels (Figure 4A), a remarkable decrease in the accessibility of HAs was observed (Figure 4B and Supplementary Figure S5A). Similar but less significant changes were observed at MAs and LAs (Supplementary Figure S5A and B). In MCF-7 cells, another cancer cell line of the same tubular epithelial type, but with significantly decreased levels of PR (Figure 4C), we found a lower ATAC accessibility of all three classes of PRBs (Figure 4B and D, and Supplementary Figure S5A and B). Lastly, we performed ATAC-seq in the nontumorigenic MCF10A cell line, which does not express PR (Figure 4A), and found a further reduction in the ATAC signal at all PRBs (Figure 4B and Supplementary Figure S5A–C).
Figure 4.
HAs occupancy correlates with PR expression and is detected in physiological breast cancer models. (A) T47D, T47Dwhite and MCF10a cells were lysed and total extracts were analyzed by western blot using PR- and GAPDH (control)-specific antibodies. (B) Sitepro (CEAS package) profile of average ATAC-seq signal in untreated T47D‐MTVL, T47Dwhite, MCF-7 cells (tumoral) and MCF10A (nontumoral) cells. The graphs are plotted over the summit of centered HAs (plotted is the −log10 of the Poisson P-value). (C) T47D and MCF-7 cells were lysed and total extracts were analyzed by western blot using PR- and ERα-specific antibodies. (D) Average ChIP-seq signal for ATAC-seq in untreated MCF-7 cells, around the HAs, MAs and LAs. The graphs are plotted over the summit of centered PRBs of each class (plotted is the −log10 of the Poisson P-value). (E) Venn diagram showing the overlap between HAs and DNase I hypersensitive sites (DHSs) derived from (34). Overlapping intervals were obtained with BEDTools using the intersectBed function with default parameters (31). (F) Genome browser view of ChIP-seq and ATAC-seq data of representative HAs located within the intergenic region of chromosome 9 (in the vicinity of the UGCG locus, not indicated in the figure). The exact location of the HAs is indicated by the gray rectangle. ChIP-ed factor or histone modification is indicated on the left as well as the cell line from which the data have been produced. ATAC-seq data are represented at the bottom for the indicated cell lines. Average ChIP-seq signal for ERα ChIP-seq (G) and FOXA1 ChIP-seq (H) in untreated cells and in cells exposed to estrogen for ERα (G) or progestin (R5020) for FOXA1 (H). The graphs are plotted over a 2 kb window around the summit of centered HAs (plotted is the −log10 of the Poisson P-value). (I) Average profile of PR and ERα occupancy at HAs in six different MCF-7-derived xenografts (38), calculated by centering the position in HAs and plotting the level of occupancy of PR and ERα (left and right panels, respectively) (plotted is the −log10 of the Poisson P-value). (J) Average ChIP-seq signal of ERα occupancy at HAs (left panel) and LAs (right panel) in ER+/PR+ PDXs untreated or treated with estrogen (38), calculated by centering the position of each class of PRBs and plotting the level of occupancy of ERα (plotted is the −log10 of the Poisson P-value).
The difference in ATAC accessibility between T47D, MCF-7 and MCF10A cells was gradually reduced for MAs and LAs compared to HAs (Figure 4B and Supplementary Figure S5A and B). In order to clarify this issue, a quantitative titration with DNase I (0.125–1 unit) was carried out in T47D and MCF10A cell lines (Supplementary Figure S5D and E). Our results showed that while HAs are more open in T47D than in MCF10A, no significant differences were observed in MAs and LAs. Based on these data, it is tempting to speculate that the accessibility of PRBs correlates with PR levels (Figure 4A and B). As an example, we showed the coronin 2B (CORO2B) locus, one of the most upregulated genes by progestin in T47D cells, which harbors HAs at its 3′ end (Supplementary Figure S5F). Our ATAC-seq data with T47Dwhite cells corroborate the notion that receptor levels are indeed correlated with accessibility of HAs (Figure 4B and Supplementary Figure S5G), suggesting a role of PR itself in keeping HAs in an open conformation that could be relevant for sustaining the enhancers responsible for the cell phenotype.
DHSs are in cis genetic markers of regulatory DNA that contain genetic variations associated with diseases and phenotypic traits (34). The recent creation of a census of all human DHSs, each of them accurately associated with a biological phenotype (34), allowed us to verify whether HAs, which show higher accessibility in breast cancer cell lines, could be associated with DHSs linked to cancer phenotype. To investigate this, we overlap our three sets of PRBs with the human DHSs and we found that >93% of HAs are bona fide annotated DHSs (Figure 4E). Moreover, when we analyzed the phenotype associated with the overlapping regions, we found cancer/epithelial terms enriched (Figure 4F). For MAs and LAs, we could detect a reduced percentage of overlap (86% and 79%, respectively) (Supplementary Figure S5H and I, upper panels), but more importantly no breast cancer phenotype was overrepresented in the less accessible PRBs (Supplementary Figure S5H and I, lower panels). Based on these data, we can conclude that HAs are likely breast epithelial DHSs linked to enhancers associated with the establishment of a cancer phenotype in T47D cells. A representative example of such HAs is reported in Figure 4G, where two highly accessible regions are localized within the intergenic region of chromosome 9 close to the UDP-glucose ceramide glucosyltransferase (UGCG) locus associated with prognostic phenotype in breast cancer (35).
To address whether HA occupancy depends on PR level, we checked PR binding at 50 pM R5020 in different breast cancer cell lines: MCF-7, its derived clone MCF-7lowPR (PRlow), T47D and T47Dwhite (PRlow) (see PR levels for each cell line in Supplementary Figure S6A, right). We found that PR recruitment to the HA-associated genes EGF, EGFR, GREB1, TIPARP, STAT5A and BIRC3 was reduced in MCF-7lowPR compared to MCF-7 cells (Supplementary Figure S6A). Similarly, PR binding in T47Dwhite was compromised compared to the wild-type counterpart (Supplementary Figure S6A), confirming the accessibility assays (Figure 4A–D). In T47Dwhite cells, this reduction on PR binding was accompanied by a decrease in the hormonal response of several up- and downregulated genes controlled by HAs compared to T47D cells (Supplementary Figure S6B and C). PR ChIP-seq data obtained in MCF-7 cells, which present lower levels of PR, compared to T47D cells (see Figure 4C), also showed preferential occupation of HAs (Supplementary Figure S6D). Therefore, even if PR levels are low but have reached a given threshold, HAs will be preferentially occupied in the presence of 50 pM hormone. Higher receptor levels will increase HA occupancy rates.
ERα binds to HAs
The ERα actively participates in breast cancer cell proliferation and is the target of the vast majority of endocrine treatments for breast cancer (36). To explore whether ERα participates with PR in shaping the landscape of genomic HAs, we perform ChIP-seq assays of ERα before and after estrogen treatment in T47D cells. We found that compared with MAs and LAs, HAs were enriched in bound ERα both under basal conditions and in the presence of estradiol (Figure 4G and Supplementary Figure S6E). In fact, 32% of the HAs were also bound by ERα in the presence of estradiol and this binding was accompanied by the hormone-dependent recruitment of the pioneer factor forkhead protein FOXA1, a key determinant of ERα function (Figure 4H) (37).
The ERE binding motif is poorly enriched within the ERα-bound HAs (192 regions with EREs, 6.7% of the total), suggesting that ERα binding may be mediated by another transcription factor. One candidate factor is FOXA1 whose motif is highly enriched and presented a strong binding to HAs (compare the P-values on the y-axis in Figure 4H versus Figure 4G and Supplementary Figure S7A). Further analysis of the distance between the FOXA1, PRE and ERE binding motifs in the HAs indicated that the HRE was localized closer to the FOXA1 than to the ERE (median 45 and 90 nt for PRE–FOXA1 and PRE–ERE, respectively) and therefore FOXA1 would also participate in PR binding (Supplementary Figure S7B). Regarding the functional connection between HAs and FOXA1, siRNA-mediated knockdown of FOXA1 compromised hormone-dependent cell proliferation in a wide range of progestin concentrations, including 50 pM (Supplementary Figure S7C). Although FOXA1 depletion was heterogeneous between HAs, hormone-dependent binding of PR was also reduced by FOXA1 knockdown (Supplementary Figure S7D and E). Thus, FOXA1 turned out to be key in HA function.
Relevance of HAs in breast cancer
Our previous observations suggest that HAs may behave as oncogenic enhancers in breast cancer (Figure 4 and Supplementary Figure S5). To test this hypothesis in a more physiological in vivo model, we took advantage of public PR and ERα ChIP-seq data obtained from MCF-7-luciferase xenografts in NOD/SCID/IL2Rg−/− (NSG) mice exposed to slow release of estrogen pellets (38). In this system, HAs were preferentially occupied by PR and ERα in estrogen-treated animals (Figure 4I), while MAs and LAs remained almost unchanged (Supplementary Figure S8A and B).
In order to extend our studies to an even more physiological and clinically relevant system, we tested the genomic distribution of ERα in ER+/PR+ PDX (38) treated with estradiol (Figure 4J). Our results showed that HAs were primarily occupied by ERα compared to LAs, supporting the connection between ERα–PR binding to HAs with breast malignancy in PDXs (Figure 4J).
Receptor binding and cell proliferation at low concentrations of hormone depend on the activation of Erk1/2 signaling pathway
It is widely accepted that PR binding to chromatin in response to 10 nM progestin increases chromatin accessibility through recruitment of chromatin remodelers, which facilitates access of other transcription factors and coregulators as required to favor increasing the rate of transcription (39). We tested whether exposure to picomolar progestin concentration could further increase the already high chromatin accessibility of HAs observed before hormone exposure (Figure 2B). Unexpectedly, the ATAC-seq results showed that chromatin accessibility of HAs was only slightly increased upon progestin exposure, regardless of the concentration (Figure 5A). Notably, HAs that become accessible in the presence of 50 pM turned out to be associated with interesting GO terms such as epithelial cell maturation and development, regulation of Notch and MAPK cascades, and DNA damage (Supplementary Figure S8C).
Figure 5.
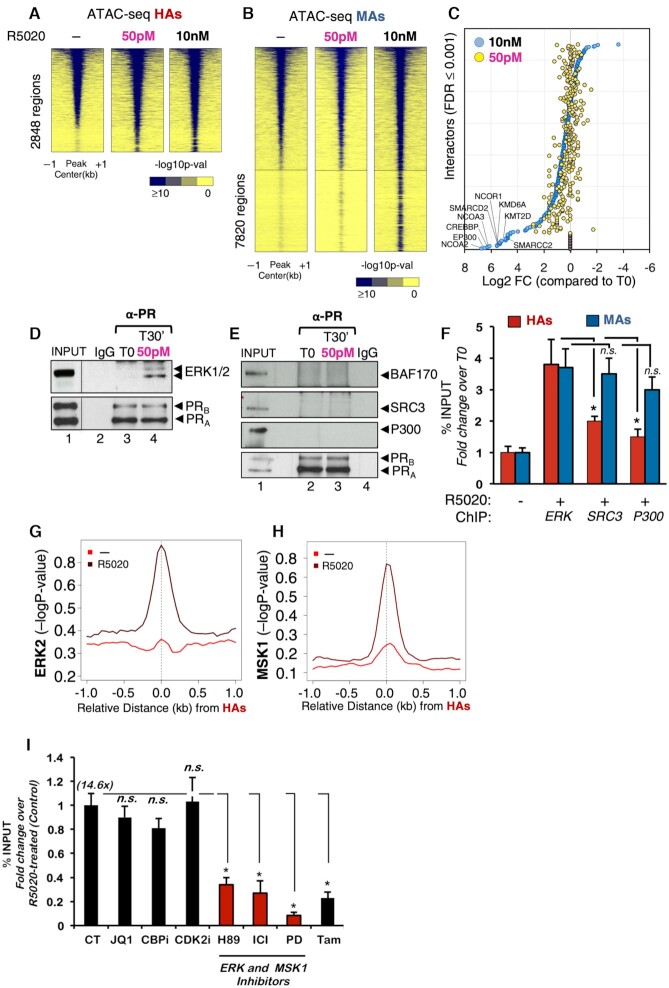
Binding of PR to HAs preferably depends on activation of ERK cell signaling pathway. Heatmap of the average ATAC-seq signal obtained from untreated cells (−) or from cells exposed for 30 min to 50 pM or 10 nM R5020 around the HAs (A) and MAs (B). The graphs are plotted over a 2 kb window around the summit of centered PRBs of each class (plotted is the −log10 of the Poisson P-value). Color bar scale with increasing blue shades of color stands for higher contact ATAC-seq signal. (C) Scatter plot of PR nuclear interactome. RIME assay (59) was performed using PR as bait in cells exposed to low (50 pM) and high (10 nM) progestin concentrations. Interactors were selected based on FDR ≤ 0.001 for two biological replicates. The 10 nM identified interactors (blue circles) were ranked from high log FC (compared to T0 condition). Based on this ranking, the 50 pM interactors were plotted accordingly (yellow circles). SAINT analysis of RIME showed 29 interacting proteins at 50 pM, 225 at both 50 pM and 10 nM, and 123 only at 10 nM R5020. (D, E) T47D-MTVL cells treated or not with 50 pM R5020 were lysed and immunoprecipitated with α-PR-specific antibody or rabbit IgG. The IPs were analyzed by immunoblotting with specific antibodies for ERK1/2 and PR (D) or for BAF170, SRC3, p300 and PR (E). (F) T47D‐MTVL cells untreated or treated for 30 min with 50 pM or 10 nM R5020 were submitted to ChIP assays with α‐ERK, α‐SRC3 and α‐p300 antibodies. Precipitated DNA was analyzed by PCR for the presence of sequences corresponding to several HAs and MAs/LAs (n = 4). Data are represented as mean ± SD from three experiments performed in duplicate. *P < 0.05 using Student’s t‐test. Average ChIP-seq signal of ERK2 (G) and MSK1 (H) in untreated and hormone-treated T47D‐MTVL cells. The graphs are plotted over a 2 kb window around the summit of centered MAs and LAs (plotted is the −log10 of the Poisson P-value). (I) T47D‐MTVL cells untreated (CT) or preincubated for 1 h with JQ1, CBPi, CDK2i, H89, ICI 182780, PD98059 or tamoxifen were untreated or exposed for 30 min to 50 pM R5020 and submitted to ChIP assays with anti‐PR antibody. Data are represented as mean ± SD from three experiments performed in duplicate. *P < 0.05 using Student’s t‐test.
The MAs showed a marked increase in accessibility at 10 nM progestin, while no effect of 50 pM was detected (Figure 5B). These findings suggested that no further chromatin remodeling upon hormone exposure is required for hormonal activation of HAs.
To test whether PR activation of HAs is independent of associated proteins required for chromatin remodeling, we implemented RIME experiments (16) in untreated and R5020-treated T47D cells at 50 pM and 10 nM hormone. Using PR as bait, we identified a set of 225 common nuclear PR interactors, including PARP1, CBX3 (HP1γ), PRKDC and transcription factors such as FOXA1, STAT3 and NR2F2, which were detected at both 50 pM and 10 nM R5020 (Figure 5C). On the other hand, a group of 123 proteins including the already known PR interactors such as NCOA2/3 (SRC2/3), CBP, p300, SMARCD2 (BAF60B), SMARCC2 (BAF170), KDM6A (UTX), KMT2D (MLL2) and the NCOR1/2 (SMRT) were only detected at 10 nM progestin (highlighted in Figure 5C). Co-IP experiments confirmed that PR at 50 pM interacts with ERK1/2 but fails to form a stable complex with BAF170, SRC3 and p300 coregulators (Figure 5D and E).
Next, we tested whether protein kinases were preferentially recruited to HAs, compared to other PR coregulators. By ChIP-qPCR performed at 50 pM progestin, we found ERK1/2 preferentially recruited to HAs compared to the classical chromatin remodelers SRC3 and p300 (Figure 5F, red bars). Conversely, when MAs were tested, a similar recruitment of ERK1/2, SRC3 and p300 was found (Figure 5F, blue bars).
We confirmed by ChIP-seq that in the presence of hormone ERK1/2 as well as its target kinase MSK1 was preferentially recruited to HAs (Figure 5G and H, and Supplementary Figure S9A and B). To test the direct involvement of these kinases in HA function, we monitored PR recruitment to HAs in the presence of the ERK and MSK1 inhibitors, PD98059 and H89, respectively. Upon treatment with these inhibitors, PR recruitment to HAs was greatly reduced (Figure 5I). These differences were not due to the inhibitors affecting PR levels, as they remained unchanged when tested in the same conditions (Supplementary Figure S9C). On the other hand, inhibition of chromatin remodelers such as BRD4 (JQ1), p300 (CBPi) and CDK2 (CDKi) did not affect PR occupancy at HAs (Figure 5I).
Interestingly, inhibition of ERα signaling by the antagonists ICI 182780 or tamoxifen also resulted in decreased PR recruitment to HAs (Figure 5I), highlighting the importance of the PR–ERα crosstalk in HA activation. Moreover, either ERK or MSK1 inhibitors as well ERα antagonists that affect PR binding to HAs also showed significant inhibitory effect in hormone-dependent cell proliferation (Supplementary Figure S9D), supporting the relevance of these genomic regions as potential targets for breast cancer management. Therefore, the receptor activated at low concentration of hormone and mainly associated with kinases is recruited to already accessible HAs and does not require the classical chromatin remodelers indispensable for activation of MAs and LAs.
DISCUSSION
Most of our own knowledge on the molecular mechanism of action of progestins on breast cancer cells was acquired at concentrations around 10 nM, an order of magnitude higher than the progesterone concentration found in the blood of women around the menopause (median 127 pM). Therefore, to what extent the physiological progesterone concentrations can cause similar effects in breast epithelial cells treated with much higher concentrations remained an open question that motivated the experiments presented in this study.
Importantly, progestins promote a single cell cycle followed by proliferation arrest at G1/S, which correlates with a delayed activation of CDK inhibitor p21WAF1 (40). Studies using PR-positive breast cancer cell lines have demonstrated a biphasic cellular response to progestins, with an immediate proliferative burst followed by a sustained growth arrest (41–43). On the other hand, progestins have also been reported as pure antiproliferative agents (44), but the difference with our results relies on the time of induction with the hormone. While we evaluated the initial effect of progestins on cell proliferation at 12–14 h, other studies extend the induction to 48 h, measuring the second antiproliferative phase.
Our unexpected finding is that indeed breast cancer cells exposed to 50 pM progestin respond with very similar biological behavior to that when exposed to 10 nM progestins, although at 50 pM progestin only 2800 sites were found to be occupied by PR instead of >50 000 sites found at 10 nM progestin. This finding highlights that these 2800 sites are pivotal controllers of the hormonal response. Essentially, we find that these sites represent poised breast-specific enhancers that only require progestin activated kinases for PR recruitment and gene regulation even in the absence of extensive chromatin remodeling (Supplementary Figure S10).
The 2800 sites represent HAs activated at picomolar progestin concentrations, which are already accessible even before hormone exposure, and their accessibility does not increase significantly upon exposure to hormone. We provide compelling evidence that they correspond to enhancers associated with cell identity and entry in the cell cycle. We hypothesized that at HAs, but not at MAs or LAs, the preexisting signal of acetylation is followed upon hormone exposure by binding of the epigenetic reader BRD4 that actively participates in RNApol II transcription at promoters and enhancers (45,46) (Supplementary Figure S10).
HAs tend to have more contacts with each other and are enriched in components such as cohesin and CTCF, two factors required for enhancer–promoter loop formation. These sites are frequently occupied by the ERα, which leads us to consider them as hormone-regulated oncogenic enhancers. Interestingly, at picomolar hormone concentration PR nuclear interactors are enriched in kinases, such as ERK1/2 and MSK1, but devoid of some very well known receptor coregulators and chromatin remodelers.
Previous studies with the glucocorticoid receptor have shown that a hormone concentration of 0.5 nM—consistent with the nighttime nadir of human cortisol—regulates the expression of a single gene, the circadian rhythm gene for period 1, supporting that low-dose hormone responses are extraordinarily specific at the genomic scale (47). Our study identifies 1800 genes regulated by 50 pM progestin that are sufficient to control complex biological functions involving hormone-dependent entry in the cell cycle and cell identity. A key concept to stress on this regard is the existence of a restricted number of genomic regions ready to respond to low concentrations of hormone by virtue of their special chromatin accessibility and loaded with cooperating transcription factors prior to hormone exposure. Here, we present evidence that the cellular levels of hormone receptors correlate with the extent of HA accessibility and response to hormone.
Increasing evidence indicates that general and specific transcription factors and their coregulators act as interaction hubs by their ability to form macromolecular condensates enriched in enhancers, silencers and other regulatory sequences, which may facilitate the interaction with gene promoters in a cell-specific manner (48–50). Our results suggest that HAs could form the basis for this kind of hubs in hormone responsive cells, as they tend to interact more frequently between themselves and are enriched in the genome organizers, RAD21 and CTCF. In response to picomolar hormone concentrations, HAs could form macromolecular condensates through the intrinsically disordered domains of pre-bound hormone receptors ERα and PR, protein kinases ERK and MSK, and enhancer markers BRD4 and MED1, all of them factors that will facilitate interactions with the basic transcriptional machinery (51,52). The intrinsically disordered C-terminus of ERα containing activation function 1 has been proved to interact with the P400 complex that deposits H2A.Z, therefore contributing to FoxA1 recruitment (27). We have recently found that 10 pM progestins is sufficient to promote a change in the single molecule trajectories of PR compatible with the formation of small condensates visible in fluorescence microscopy that exhibit properties of liquid droplets (53). Further experiments will be required to demonstrate whether HAs are indeed organized as liquid droplets in cells and how this is regulated by hormone exposure.
Interestingly, HAs are located in chromatin regions with very well organized nucleosomes, indicating an important role of nucleosomal proteins for their function. To maintain this structure, a certain level of PR must be available, suggesting that even in the absence of hormone PR contributes to the competence of the HAs. One possible explanation for the need of nucleosomes is that they are enriched in the histone variant H2A.Z, which weakens the interaction with H3/H4 tetramer making the nucleosomal DNA more accessible for factor binding (54). In fact, we found that H2A.Z turned out to be slightly enriched in HAs compared with MAs and LAs (Supplementary Figure S3D).
A simple model for HA-dependent gene activation in breast cancer cells
We have reported that the genomic and nongenomic pathways converge in the nucleus and more precisely on chromatin during hormonal activation (7). In our model, the hormone-dependent gene activation process consisted of two consecutive cycles, both involving the collaboration between activated kinases, histone modifying enzymes and ATP-dependent chromatin remodelers (55). The first cycle is largely dependent on ERK1/2, MSK1 and CDK2 kinases and concludes with H1 displacement and higher DNA accessibility. In the second cycle, in addition to kinase activation the ATP-dependent remodeling complex BAF actively displaces H2A/B dimers. Here, we have found that the activation of the kinases would constitute the most relevant step from the physiological point of view since it is sufficient to activate the HAs. Hence, we can draw a parallel between rapid nongenomic effects and those found at low concentrations, both activating similar signaling pathways involved in the front line of hormone response.
In fact, we found that activation of the ERK1/2 by EGF while promoting phosphorylation of PR in S294 was per se not sufficient to recruit the receptor to HAs (Supplementary Figure S11A and B). However, EGF did improve PR binding to HAs in the presence of 50 pM progestin, highlighting the connection between these two signaling pathways (Supplementary Figure S11A and B). It is tempting to speculate that binding of 50 pM progestin to membrane-attached PR is sufficient for activation of the ERK/MSK kinases, which in turn will phosphorylate and activate the ligand-free intracellular PR, leading to its translocation into the cell nucleus to bind HAs. Further experiments will be needed to explore this possibility.
Proteasome-dependent PR downregulation was reported in T47D cells treated with progestins (56). Specifically, downregulation of mature PRs occurs by a mechanism in which ligand binding activates PR phosphorylation by MAPKs at a unique serine residue, which then targets the receptors for degradation (56). As the receptor activated at low concentration of hormone is associated with ERK1/2 and recruited in a kinase-dependent manner, to already accessible HAs, we want to address whether PR downregulation was also observed at 50 pM. Our results showed that 50 pM progestin downregulates PR by 30% compared to the 70% depletion detected at 10 nM (Supplementary Figure S11D), which is consistent with the partial activation of the MAPKs observed at low concentration (Figure 1D, lane 2). Conversely, FOXA1 remained unchanged at 50 pM hormone and was slightly decreased at 10 nM (Supplementary Figure S11C and D).
It should be noted that HAs do not correspond to just the strongest PRBs found at 10 nM hormone, but represent a small subset of these sites with specific properties. In fact, the majority of the strongest PRBs exhibited a high nucleosome remodeling index (57), while HAs do not require the chromatin remodeling machineries and are already accessible prior to hormone exposure (Figures 2A and 5A). However, as the HAs are 3D interaction hubs and interact preferentially with other MAs (Figure 3D), they may contribute to the functional relevance of these sites as well.
At low concentration of hormone and in the presence of sufficient amount of PR, the receptor is recruited to HAs. Mechanistically, this process is driven by (i) the accessibility of these sites, (ii) the presence of complete or half HREs and/or FOXA1 DNA binding sequences, and (iii) the ERK1/2 kinases that associate with PR and are involved in the first steps of transcriptional activation.
We found that HA occupancy is conserved from breast cancer cell lines to PDXs. In fact, the percentage of occupancy of the HAs increases as the system becomes more physiological, from cell lines to xenografts and PDXs (data not shown). This supports that HAs would be adapted to respond to physiological concentrations of hormone in the context of tumor tissue to promote its growth.
In our study, we found a correlation between HA occupancy and PDX growth in vivo, but the most convincing proof will be the targeting of HAs and subsequent assessment of tumor growth. These experiments are very challenging due to the abnormal karyotype of tumoral cells that hinders targeting of HAs by CRISPR/Cas9 (see below). Importantly, as the concentration of hormone in PDX models was not measured, the physiological relevance of HAs in breast cancer should be taken with caution and requires further experimentation. In fact, this is a complex issue as the amount of progesterone that is actually acting on the PR in vivo is difficult to estimate since several parameters, including the amount of free hormone (not bound to plasma proteins) and its intracellular metabolization by the enzyme 5-alpha reductase, regulate the effective amount of progesterone in target cells.
Despite the still open questions, our findings unveil that concentrations of progestins comparable to those existing in the blood of women around the menopause are sufficient for the activation of the initial progestin response of cultured breast cancer cells by binding of the receptor to a small subset of ‘special’ PRBs. Future experiments oriented to target the HAs through CRISPR/Cas9-based techniques could allow us to unveil the hierarchy of genomic PRBs. We cannot exclude that PRBs detected at higher hormone concentration—in particular MAs that share some properties with HAs—may be required for sustaining later phases of the hormone response. To explore this possibility, we will need more physiological systems, like cancer spheroids exposed to sustained hormone stimulation, to mimic the tissue situation in vivo.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Luciano Di Croce for advice on the manuscript and Dr Cheng-Ming Chiang (UT Southwestern Medical Center) for providing the BRD4 antibody.
Notes
Present address: Roberto Ferrari, Department of Chemistry, Life Sciences and Environmental Sustainability, University of Parma, Parma, Italy.
Present address: Antonios Lioutas, Genetics Department, Harvard Medical School, Boston, MA, USA.
Contributor Information
Roser Zaurin, Center for Genomic Regulation (CRG), Barcelona, 08003, Spain; Barcelona Institute for Science and Technology (BIST), Barcelona, 08003, Spain.
Roberto Ferrari, Center for Genomic Regulation (CRG), Barcelona, 08003, Spain; Barcelona Institute for Science and Technology (BIST), Barcelona, 08003, Spain.
Ana Silvina Nacht, Center for Genomic Regulation (CRG), Barcelona, 08003, Spain; Barcelona Institute for Science and Technology (BIST), Barcelona, 08003, Spain.
Jose Carbonell, Center for Genomic Regulation (CRG), Barcelona, 08003, Spain; Barcelona Institute for Science and Technology (BIST), Barcelona, 08003, Spain.
Francois Le Dily, Center for Genomic Regulation (CRG), Barcelona, 08003, Spain; Barcelona Institute for Science and Technology (BIST), Barcelona, 08003, Spain.
Jofre Font-Mateu, Center for Genomic Regulation (CRG), Barcelona, 08003, Spain; Barcelona Institute for Science and Technology (BIST), Barcelona, 08003, Spain.
Lara Isabel de Llobet Cucalon, Center for Genomic Regulation (CRG), Barcelona, 08003, Spain; Barcelona Institute for Science and Technology (BIST), Barcelona, 08003, Spain.
Enrique Vidal, Center for Genomic Regulation (CRG), Barcelona, 08003, Spain; Barcelona Institute for Science and Technology (BIST), Barcelona, 08003, Spain.
Antonios Lioutas, Center for Genomic Regulation (CRG), Barcelona, 08003, Spain; Barcelona Institute for Science and Technology (BIST), Barcelona, 08003, Spain.
Miguel Beato, Center for Genomic Regulation (CRG), Barcelona, 08003, Spain; Barcelona Institute for Science and Technology (BIST), Barcelona, 08003, Spain; Universitat Pompeu Fabra (UPF), Barcelona, 08003, Spain.
Guillermo P Vicent, Center for Genomic Regulation (CRG), Barcelona, 08003, Spain; Molecular Biology Institute of Barcelona, Consejo Superior de Investigaciones Científicas (IBMB-CSIC), Barcelona, 08003, Spain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Ciencia e Innovación [PID2019-105173RB-I00 and PID2019-110384GB-C21]; Spanish Ministry of Economy and Competitiveness [SAF2016-75006-P and G62426937]; Consejo Superior de Investigaciones Científicas [201820I131]; Centro de Excelencia Severo Ochoa [SEV-2012-2018]; European Research Council [609989]. Funding for open access charge: Ministerio de Ciencia e Innovación.
Conflict of interest statement. None declared.
REFERENCES
- 1. Beato M., Herrlich P., Schutz G.. Steroid hormone receptors: many actors in search of a plot. Cell. 1995; 83:851–857. [DOI] [PubMed] [Google Scholar]
- 2. Ballare C., Uhrig M., Bechtold T., Sancho E., Di Domenico M., Migliaccio A., Auricchio F., Beato M.. Two domains of the progesterone receptor interact with the estrogen receptor and are required for progesterone activation of the c-Src/Erk pathway in mammalian cells. Mol. Cell. Biol. 2003; 23:1994–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pedram A., Razandi M., Sainson R.C., Kim J.K., Hughes C.C., Levin E.R.. A conserved mechanism for steroid receptor translocation to the plasma membrane. J. Biol. Chem. 2007; 282:22278–22288. [DOI] [PubMed] [Google Scholar]
- 4. Migliaccio A., Di Domenico M., Castoria G., de Falco A., Bontempo P., Nola E., Auricchio F.. Tyrosine kinase/p21ras/MAP-kinase pathway activation by estradiol-receptor complex in MCF-7 cells. EMBO J. 1996; 15:1292–1300. [PMC free article] [PubMed] [Google Scholar]
- 5. Migliaccio A., Piccolo D., Castoria G., Di Domenico M., Bilancio A., Lombardi M., Gong W., Beato M., Auricchio F.. Activation of the Src/p21ras/Erk pathway by progesterone receptor via cross-talk with estrogen receptor. EMBO J. 1998; 17:2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boonyaratanakornkit V., Scott M.P., Ribon V., Sherman L., Anderson S.M., Maller J.L., Miller W.T., Edwards D.P.. Progesterone receptor contains a proline-rich motif that directly interacts with SH3 domains and activates c-Src family tyrosine kinases. Mol. Cell. 2001; 8:269–280. [DOI] [PubMed] [Google Scholar]
- 7. Vicent G.P., Ballare C., Nacht A.S., Clausell J., Subtil-Rodriguez A., Quiles I., Jordan A., Beato M.. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol. Cell. 2006; 24:367–381. [DOI] [PubMed] [Google Scholar]
- 8. Qiu M., Olsen A., Faivre E., Horwitz K.B., Lange C.A.. Mitogen-activated protein kinase regulates nuclear association of human progesterone receptors. Mol. Endocrinol. 2003; 17:628–642. [DOI] [PubMed] [Google Scholar]
- 9. Wright R.H., Castellano G., Bonet J., Le Dily F., Font-Mateu J., Ballare C., Nacht A.S., Soronellas D., Oliva B., Beato M.. CDK2-dependent activation of PARP-1 is required for hormonal gene regulation in breast cancer cells. Genes Dev. 2012; 26:1972–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pierson-Mullany L.K., Lange C.A.. Phosphorylation of progesterone receptor serine 400 mediates ligand-independent transcriptional activity in response to activation of cyclin-dependent protein kinase 2. Mol. Cell. Biol. 2004; 24:10542–10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Missmer S.A., Eliassen A.H., Barbieri R.L., Hankinson S.E.. Endogenous estrogen, androgen, and progesterone concentrations and breast cancer risk among postmenopausal women. J. Natl Cancer Inst. 2004; 96:1856–1865. [DOI] [PubMed] [Google Scholar]
- 12. Strutt H., Paro R.. Mapping DNA target sites of chromatin proteins in vivo by formaldehyde crosslinking. Methods Mol. Biol. 1999; 119:455–467. [DOI] [PubMed] [Google Scholar]
- 13. Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J.. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013; 10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O.et al.. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009; 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mumbach M.R., Rubin A.J., Flynn R.A., Dai C., Khavari P.A., Greenleaf W.J., Chang H.Y.. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods. 2016; 13:919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mohammed H., Taylor C., Brown G.D., Papachristou E.K., Carroll J.S., D’Santos C.S. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat. Protoc. 2016; 11:316–326. [DOI] [PubMed] [Google Scholar]
- 17. Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B.. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019; 47:W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferrari N., McDonald L., Morris J.S., Cameron E.R., Blyth K.. RUNX2 in mammary gland development and breast cancer. J. Cell. Physiol. 2013; 228:1137–1142. [DOI] [PubMed] [Google Scholar]
- 19. Nacht A.S., Ferrari R., Zaurin R., Scabia V., Carbonell-Caballero J., Le Dily F., Quilez J., Leopoldi A., Brisken C., Beato M.et al.. C/EBPalpha mediates the growth inhibitory effect of progestins on breast cancer cells. EMBO J. 2019; 38:e101426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jeffrey K.L., Camps M., Rommel C., Mackay C.R.. Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat. Rev. Drug Discov. 2007; 6:391–403. [DOI] [PubMed] [Google Scholar]
- 21. Wang S., Sun H., Ma J., Zang C., Wang C., Wang J., Tang Q., Meyer C.A., Zhang Y., Liu X.S.. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc. 2013; 8:2502–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boeger H., Griesenbeck J., Strattan J.S., Kornberg R.D.. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell. 2003; 11:1587–1598. [DOI] [PubMed] [Google Scholar]
- 23. Lee C.K., Shibata Y., Rao B., Strahl B.D., Lieb J.D.. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004; 36:900–905. [DOI] [PubMed] [Google Scholar]
- 24. Zhao Y., Wang J., Liang F., Liu Y., Wang Q., Zhang H., Jiang M., Zhang Z., Zhao W., Bao Y.et al.. NucMap: a database of genome-wide nucleosome positioning map across species. Nucleic Acids Res. 2019; 47:D163–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andersson R., Sandelin A.. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 2020; 21:71–87. [DOI] [PubMed] [Google Scholar]
- 26. Halasa M., Wawruszak A., Przybyszewska A., Jaruga A., Guz M., Kalafut J., Stepulak A., Cybulski M.. H3K18Ac as a marker of cancer progression and potential target of anti-cancer therapy. Cells. 2019; 8:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gevry N., Hardy S., Jacques P.E., Laflamme L., Svotelis A., Robert F., Gaudreau L.. Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev. 2009; 23:1522–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiang Y., Qian F., Bai X., Liu Y., Wang Q., Ai B., Han X., Shi S., Zhang J., Li X.et al.. SEdb: a comprehensive human super-enhancer database. Nucleic Acids Res. 2019; 47:D235–D243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Le Dily F., Vidal E., Cuartero Y., Quilez J., Nacht A.S., Vicent G.P., Carbonell-Caballero J., Sharma P., Villanueva-Canas J.L., Ferrari R.et al.. Hormone-control regions mediate steroid receptor-dependent genome organization. Genome Res. 2019; 29:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang J., Li K., Cai W., Liu X., Zhang Y., Orkin S.H., Xu J., Yuan G.C.. Dissecting super-enhancer hierarchy based on chromatin interactions. Nat. Commun. 2018; 9:943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bhattacharyya S., Chandra V., Vijayanand P., Ay F.. Identification of significant chromatin contacts from HiChIP data by FitHiChIP. Nat. Commun. 2019; 10:4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meuleman W., Muratov A., Rynes E., Halow J., Lee K., Bates D., Diegel M., Dunn D., Neri F., Teodosiadis A.et al.. Index and biological spectrum of human DNase I hypersensitive sites. Nature. 2020; 584:244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ruckhaberle E., Karn T., Hanker L., Gatje R., Metzler D., Holtrich U., Kaufmann M., Rody A.. Prognostic relevance of glucosylceramide synthase (GCS) expression in breast cancer. J. Cancer Res. Clin. Oncol. 2009; 135:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lumachi F., Brunello A., Maruzzo M., Basso U., Basso S.M.. Treatment of estrogen receptor-positive breast cancer. Curr. Med. Chem. 2013; 20:596–604. [DOI] [PubMed] [Google Scholar]
- 37. Hurtado A., Holmes K.A., Ross-Innes C.S., Schmidt D., Carroll J.S.. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 2011; 43:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mohammed H., Russell I.A., Stark R., Rueda O.M., Hickey T.E., Tarulli G.A., Serandour A.A., Birrell S.N., Bruna A., Saadi A.et al.. Progesterone receptor modulates ERalpha action in breast cancer. Nature. 2015; 523:313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vicent G.P., Nacht A.S., Zaurin R., Ballare C., Clausell J., Beato M.. Minireview: role of kinases and chromatin remodeling in progesterone signaling to chromatin. Mol. Endocrinol. 2010; 24:2088–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Owen G.I., Richer J.K., Tung L., Takimoto G., Horwitz K.B.. Progesterone regulates transcription of the p21(WAF1) cyclin-dependent kinase inhibitor gene through Sp1 and CBP/p300. J. Biol. Chem. 1998; 273:10696–10701. [DOI] [PubMed] [Google Scholar]
- 41. Groshong S.D., Owen G.I., Grimison B., Schauer I.E., Todd M.C., Langan T.A., Sclafani R.A., Lange C.A., Horwitz K.B.. Biphasic regulation of breast cancer cell growth by progesterone: role of the cyclin-dependent kinase inhibitors, p21 and p27(Kip1). Mol. Endocrinol. 1997; 11:1593–1607. [DOI] [PubMed] [Google Scholar]
- 42. Musgrove E.A., Swarbrick A., Lee C.S., Cornish A.L., Sutherland R.L.. Mechanisms of cyclin-dependent kinase inactivation by progestins. Mol. Cell. Biol. 1998; 18:1812–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Skildum A., Faivre E., Lange C.A.. Progesterone receptors induce cell cycle progression via activation of mitogen-activated protein kinases. Mol. Endocrinol. 2005; 19:327–339. [DOI] [PubMed] [Google Scholar]
- 44. Pang H., Rowan B.G., Al-Dhaheri M., Faber L.E.. Epidermal growth factor suppresses induction by progestin of the adhesion protein desmoplakin in T47D breast cancer cells. Breast Cancer Res. 2004; 6:R239–R245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kanno T., Kanno Y., LeRoy G., Campos E., Sun H.W., Brooks S.R., Vahedi G., Heightman T.D., Garcia B.A., Reinberg D.et al.. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat. Struct. Mol. Biol. 2014; 21:1047–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Loven J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., Young R.A.. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013; 153:320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reddy T.E., Gertz J., Crawford G.E., Garabedian M.J., Myers R.M.. The hypersensitive glucocorticoid response specifically regulates period 1 and expression of circadian genes. Mol. Cell. Biol. 2012; 32:3756–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Beagrie R.A., Scialdone A., Schueler M., Kraemer D.C., Chotalia M., Xie S.Q., Barbieri M., de Santiago I., Lavitas L.M., Branco M.R.et al.. Complex multi-enhancer contacts captured by genome architecture mapping. Nature. 2017; 543:519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Javierre B.M., Burren O.S., Wilder S.P., Kreuzhuber R., Hill S.M., Sewitz S., Cairns J., Wingett S.W., Varnai C., Thiecke M.J.et al.. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016; 167:1369–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stevens T.J., Lando D., Basu S., Atkinson L.P., Cao Y., Lee S.F., Leeb M., Wohlfahrt K.J., Boucher W., O’Shaughnessy-Kirwan A.et al.. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature. 2017; 544:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nair S.J., Yang L., Meluzzi D., Oh S., Yang F., Friedman M.J., Wang S., Suter T., Alshareedah I., Gamliel A.et al.. Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol. 2019; 26:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C.et al.. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018; 361:eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Muñoz-Gil G., Romero-Aristizabal C., Mateos N., de Llobet Cucalon L.I., Beato M., Lewenstein M., Garcia-Parajo M.F., Torreno-Pina J.A. Phase separation of tunable biomolecular condensates predicted by an interacting particle model. 2020; bioRxiv doi:10 September 2020, preprint: not peer reviewed 10.1101/2020.09.09.289876. [DOI]
- 54. Rudnizky S., Bavly A., Malik O., Pnueli L., Melamed P., Kaplan A.. H2A.Z controls the stability and mobility of nucleosomes to regulate expression of the LH genes. Nat. Commun. 2016; 7:12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Beato M., Vicent G.P.. Impact of chromatin structure and dynamics on PR signaling. The initial steps in hormonal gene regulation. Mol. Cell. Endocrinol. 2012; 357:37–42. [DOI] [PubMed] [Google Scholar]
- 56. Lange C.A., Shen T., Horwitz K.B.. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc. Natl Acad. Sci. U.S.A. 2000; 97:1032–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ballare C., Castellano G., Gaveglia L., Althammer S., Gonzalez-Vallinas J., Eyras E., Le Dily F., Zaurin R., Soronellas D., Vicent G.P.et al.. Nucleosome-driven transcription factor binding and gene regulation. Mol. Cell. 2013; 49:67–79. [DOI] [PubMed] [Google Scholar]
- 58. Ferrari R., Su T., Li B., Bonora G., Oberai A., Chan Y., Sasidharan R., Berk A.J., Pellegrini M., Kurdistani S.K. Reorganization of the host epigenome by a viral oncogene. Genome Res. 2012; 22:1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mohammed H., D'Santos C., Serandour A.A., Ali H.R., Brown G.D., Atkins A., Rueda O.M., Holmes K.A., Theodorou V., Robinson J.Let al.. Endogenous purification reveals GREB1 as a key estrogenreceptor regulatory factor. Cell Rep. 2013; 3:342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.