Abstract
Mixed lineage leukemia 1 (MLL1) is a histone methyltransferase. Kaposi's sarcoma-associated herpesvirus (KSHV) is a leading cause of malignancy in AIDS. KSHV latently infects tumor cells and its genome is decorated with epigenetic marks. Here, we show that KSHV latency-associated nuclear antigen (LANA) recruits MLL1 to viral DNA where it establishes H3K4me3 modifications at the extensive KSHV terminal repeat elements during primary infection. LANA interacts with MLL1 complex members, including WDR5, integrates into the MLL1 complex, and regulates MLL1 activity. We describe the 1.5-Å crystal structure of N-terminal LANA peptide complexed with MLL1 complex member WDR5, which reveals a potential regulatory mechanism. Disruption of MLL1 expression rendered KSHV latency establishment highly deficient. This deficiency was rescued by MLL1 but not by catalytically inactive MLL1. Therefore, MLL1 is LANA regulable and exerts a central role in virus infection. These results suggest broad potential for MLL1 regulation, including by non-host factors.
INTRODUCTION
Epigenetic H3K4me3 marks are associated with actively transcribed genes and are deposited by COMPASS histone methyltransferase (HMT) complexes. HMT complexes are evolutionarily conserved from yeast to humans, and their methyltransferase activity exerts roles in embryonic stem cell development, hematopoiesis, and neurogenesis (1,2). Mixed lineage leukemia 1 (MLL1) is one of six in a family of HMTs (MLL1−4, Set1A/B) in mammals responsible for catalyzing methylation of histone H3 at lysine 4 through a SET domain. These members have important, non-redundant roles (3). In particular, MLL1 has been intensively studied due to its deregulation in pediatric and adult leukemia (3). Chromosomal translocations involving MLL1 frequently occur in acute myeloid and lymphoid leukemia (1). Mammalian COMPASS complexes are comprised of MLL or SET1, along with WDR5, ASH2L, RBBP5 and DPY30 (4–6).
Kaposi's sarcoma-associated herpesvirus (KSHV) is etiologically linked to Kaposi's sarcoma (KS), the predominant AIDS malignancy, primary effusion lymphoma (PEL) and multicentric Castleman's disease (MCD) (7–10). Upon infection, the KSHV genome is rapidly transported to the nucleus, where it circularizes by fusing at its terminal repeats (TRs) and persists as a multicopy episome in latently infected cells (11). KSHV encodes ∼100 open reading frames (ORFs), although only several viral genes, including the latency-associated nuclear antigen (LANA), are expressed during latency (12). LANA binds KSHV TR DNA to mediate viral episome persistence by tethering viral genomes to host mitotic chromosomes to ensure their segregation to progeny nuclei (13–16).
KSHV enters the cell free of histones and epigenetic modifications. Following infection, epigenetic marks rapidly accumulate on the viral genome to suppress lytic gene expression, while permitting latent gene expression. Broadly distributed repressive modifications, including H3K27me3, H2AK119ub and H3K9me3, deposited by Polycomb Repressive Complex (PRC) 2, PRC1 and IFI16 recruited SUV39H1, respectively, are responsible for silencing lytic genes (17–23), while activating H3K4me3 modifications are deposited at several loci, including the latency promoter (18).
In addition to mediating episome persistence, LANA interacts with chromatin modifiers and histone binding proteins, including KDM3A, SUV39H1, BRD4 and the methyl CpG binding protein MeCP2 and recruits Polycomb repressive complexes to the KSHV genome during de novo infection, indicating a role in epigenetic regulation (19,23–26). Further, LANA is enriched at H3K4me3 peaks at viral and host promoters (27–29). Consistent with its enrichment at these peaks, LANA interacts with SET1, RbBP5, and ASH2L (28). The KSHV TRs comprise ∼20% of the ∼170 kb KSHV genome, are noncoding, and lack promoter activity in latency, yet are highly enriched in H3K4me3 (18,28). Each of the ∼40 TR elements contains three adjacent LANA binding sites (LBSs) (30) suggesting LANA may have a role in modulating TR H3K4me3 levels. The TRs therefore offer a unique opportunity to explore H3K4me3 establishment on naïve DNA, and LANA’s potential role in this process.
Here, we show that LANA recruits MLL1 to KSHV TR DNA and that MLL1 is responsible for a specific temporal pattern of H3K4me3 deposition during primary infection. LANA interacts extensively with MLL1 complex members and regulates MLL1 activity. A crystal structure of N-terminal LANA peptide complexed with complex member WDR5 suggests a mechanism for LANA’s regulatory activity and implies additional, innate host regulation of MLL1. Importantly, knockout of MLL1 results in highly deficient establishment of latent infection, indicating a critical role in KSHV biology.
MATERIALS AND METHODS
Cell lines
BJAB cells were maintained in RPMI medium containing 10% bovine growth serum (BGS; HyClone) and 15 μg/ml gentamicin. KSHV PEL BCBL-1 cells were maintained in RPMI medium containing 10% BGS and 15 μg/ml gentamicin. 293T and iSLK cells were maintained in DMEM containing 10% BGS (Clontech) and 15 μg/mL gentamicin.
Plasmids
pCDNA3.0-Flag-WDR5, -Flag-RbBP5 and -ASH2L (534 amino acid ASH2L isoform) (31) were obtained from Addgene. Full length pcDNA3-MLL-F (32) was obtained from Dr Nancy Zeleznik-Le and generated by Dr S. Korsmeyer and M. Seto. pCDNA3.0-Flag-ASH2L was generated with the Q5 Site-Directed Mutagenesis kit (New England Biolabs) followed by sequencing validation. To enable purification of N-His6-tag recombination proteins, full length WDR5, RbBP5, ASH2L or the MLL1-SET domain (residues 3744-3969) was cloned into pETDuet-1 [Multiple Clone site 1(MCS1)] (Novagen). WDR5 and MLL1-SET (3744–3969) were also cloned into pGEX-6P-1 (Millipore Sigma) to allow expression of GST fusion proteins. pSG5-T7-LANA (33) encodes LANA with an N-terminal T7 epitope tag. Full length MLL1 was cloned to pCMV-3Tag-6 (Agilent Technologies) by an in-fusion recombination-based method (Clontech) through two sequential steps due to the large size of the MLL1 insert. First, the MLL1 gene was PCR amplified (oligonucleotides in Supplementary Table S3) from nucleotide 1 to 8025 to include the AflII restriction site (nt 8010), and inserted into the ClaI and XhoI sites of pCMV-3Tag-6 to construct pCMV-3Tag-6-MLL1-1-8025. This construct was then digested by AflII and XhoI and an independently amplified fragment spanning MLL1-AflII (nt 8010) to the 3’ end of the MLL1 ORF (nt 11910) was inserted, following AflII digestion, to generate full length pCMV-3Tag-6-MLL1, which contains 3X Flag at the N terminus. pCMV-3Tag-6-MLL1 was validated by sequencing. To obtain MLL1 with an alanine substitution at residue 3906, MLL1-N3906A, MLL1-SbfI (nt 10819)-end, was first cloned into pCMV-3Tag-6 using BamHI and XhoI using Clontech In-Fusion cloning. Codon 3906 is just downstream of the SbfI site. The 3906 codon was altered to alanine in pCMV-3Tag-6-MLL1-SbfI (10819)-end using the Q5 Site-Directed Mutagenesis kit (NEB). MLL1-SbfI-N3906A-end was cloned into pCMV-3Tag-6-MLL1 after digestion with SbfI and XhoI to generate full length pCMV-3Tag-6-MLL1/N3906A.
LANA fragments were cloned in pEGFP to generate GFP-LANA 1–32 (34),-LANA 33–331 (35), -LANA 275–777 (36) and -LANA 933–1162 (36). GFP LANA 779–1049 was cloned by Takashi Komatsu. GFP LANA 1031–1119 was cloned by Mary Ballestas, GFP LANA 1031–1065 and GFP LANA 1066–1119 were each cloned using the Q5 Site-Directed Mutagenesis kit (NEB) followed by sequencing validation. GFP LANA 1–32 23CRK→PAA25 was also cloned by Q5 Site-Directed Mutagenesis (New England Biolabs) followed by sequencing validation.
LentiCrispr V2 (37) was obtained from Addgene. Guide RNA sequences, listed in Supplementary Table S3, were designed by CHOPCHOP (38) and were cloned into LentiCrispr V2 as previously described (37). The coding sequences of LANAΔ33–888 (35) were PCR-amplified and inserted into the pMAL vector (NEB) by Murli Narayan or into pETDuet-1[Multiple Clone site 1(MCS1)] (Novagen) to generate 6x His N-terminal tagged fusions, termed pMAL-LANAΔ33–888, or pETDuet-1-LANAΔ33–888, respectively.
Antibodies
Antibodies used included anti-Flag (Sigma, F3165-5MG, clone M2), anti-MLL1C (Bethyl, A300-374A), anti-RbBP5 (Bethyl, A300-109A), anti-ASH2L (Bethyl, A300-112A), anti-WDR5 (Bethyl, A302-430A), anti-GFP (Clontech, 632377, clone JL-8), anti-trimethyl-Histone H3 (Lys4) (EMD Millipore, 07-473), anti-LANA LN53 (Millipore, MABE1109), goat anti-rat IgG (H + L) Alexa Fluor 488 (Invitrogen A11006), goat anti-rabbit IgG (H + L) Alexa Fluor 647 (Invitrogen A21245), human anti -LANA sera adsorbed against uninfected cell extract for western blot or affinity purified against carboxy-terminal LANA for ChIP, Anti-T7 tag® antibody (Abcam, ab9138), and anti-alpha-tubulin (Sigma T9026, cline DM1A).
Virus infections and outgrowth experiments
rKSHV.219 virus was induced from iSLK.219 cells (39). Cells were treated with 1 μg/ml doxycycline, 20 ng/ml TPA and 1mM sodium butyrate for 16 h to induce RTA expression and activate the lytic cycle. The cells were then maintained in DMEM for five additional days. Virus containing supernatant was filtered with a 0.45 μM membrane. Virus was then collected by ultracentrifugation at 25,000g for 4 h and resuspended in medium containing 10% BGS.
For short term experiments, KSHV infection was performed in 12-well plates with cells at ∼80% confluence at a MOI of ∼2–3. Cells were then spinoculated by centrifugation at 2000 × g for 1 h, washed with 1 ml of culture medium, and incubated at 37°C.
For puromycin resistant outgrowth experiments, KSHV infection was performed as above, but with a MOI of 0.1. Forty-eight hours after infection, 2 × 104 cells were seeded in 15 cm dishes containing 20 ml of DMEM with 10% BGS. Twelve hours following seeding, 1 μg/ml puromycin was added to the medium. Puromycin was replenished twice a week in fresh media until individual clones grow out after ∼2 weeks. Cell colonies were stained with crystal violet and counted.
CRSPR knockouts
Gene knockouts were generated by transient transfection of LentiCrispr V2 as previously described (37). Briefly, cells were transfected with gRNA cloned into LentiCrispr V2 by polyethylenimine (PEI) (Sigma). Thirty-six hours post transfection, 1x104 cells were seeded in a 15 cm dish with 25 ml medium containing 2 μg/ml puromycin (InvivoGen) for another 48 h selection to kill untransfected cells. Surviving cells were cultured in medium without drug selection and individual clones selected. Gene knockouts were validated either by immunoblot or sequencing of amplified genomic DNA.
Immunoprecipitation
Cells grown in 6 cm plates were lysed in cell lysis buffer containing 50 mM Tris−HCl pH 7.9, 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM DTT, 4 U/μl micrococcal nuclease (New England Biolabs, Catalog #M0247S), and a complete protease inhibitor tablet (Roche). Antibodies (2 μg anti-LANA or 5 μg anti-MLL1C) conjugated with Dynabeads Protein G (Invitrogen) or anti-Flag M2 (2 μg)-conjugated agarose beads (Sigma) were incubated with whole cell lysates overnight at 4°C. Beads were washed 3–6 times with cell lysis buffer, bound proteins eluted in SDS buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromophenol blue and 125 mM Tris−HCl, pH 6.8), resolved by SDS-PAGE, and detected by western blot. For immunoprecipitations of endogenous proteins, nuclear extract lysate from 20 × 106 cells was used as previously described (40).
ChIP
At different time points post infection, cells from one well of a 12-well plate (∼1 × 106 cells) were cross-linked with 1% formaldehyde for 10 min and the reaction stopped with 125 mM glycine. Cells were resuspended in nuclei lysis buffer (1% SDS, 10 mM EDTA and 50 mM Tris, pH 8.1) and sonicated with a Bioruptor Sonicator (Diagenode). The sonicated cell supernatant was diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X- 100, 1.2 mM EDTA, 16.7 mM Tris−HCl, pH 8.1, 167 mM NaCl). Samples were incubated with the appropriate antibodies [affinity-purified anti-LANA antibody, 2 μg; anti-trimethyl-Histone H3 (Lys4) (Millipore), 1.5 μg; anti-MLL1C (Bethyl), 4 μg; Anti-trimethyl-Histone H3 (Lys27) (Millipore), 2 μg; Anti-trimethyl-Histone H3 (Lys9) (Sigma), 2 μg], or with an equal amount of corresponding IgG control (all ChIP Abs are polyclonal) overnight at 4°C. Protein A or G agarose/salmon sperm DNA beads (Millipore) were added and incubated for 30 min at 4°C with agitation, and immunoprecipitations performed according to the manufacturer's instructions. In brief, immunoprecipitates were washed once, each with low-salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20mM Tris–HCl, pH 8.1, 150 mM NaCl), high-salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, pH 8.1, 500 mM NaCl) and LiCl buffer (0.25 M LiCl, 1% IGEPAL CA630, 1% deoxycholic acid (sodium salt), 1 mM EDTA, 10 mM Tris, pH 8.1), followed by two washes with TE Buffer. DNA was released by reversing crosslinking by heating at 65°C for 4 h and treatment with Proteinase K for 1 h. DNA was purified using a PCR purification kit (Qiagen) and analyzed by quantitative real-time PCR using Power SYBR Green PCR Master Mix (Applied Biosystems) and a QuantStudio 3.0 System (Applied Biosystems) to detect amplified product in accordance with the manufacturer's instructions. Primers used for quantitative PCR reactions were: forward primer 5′-GGGGGACCCCGGGCAGCGAG-3′, reverse primer 5′-GGCTCCCCCAAACAGGCTCA-3′ (flanking KSHV TR nucleotides 677–766). For quantification, 5%, 1%, 0.2% or 0.04% of DNA from KSHV latently infected 293T cells was used to generate a standard curve for quantification comparison, which was performed using QuantStudio 3.0 software.
For assessing LANA-directed TR loading of MLL1 by ChIP assay, 1x107 BJAB cells, or BJAB cells stably expressing LANA (BJAB/LANA), were transfected with 5 μg of p8TR (34) by nucleofection with the Amaxa Nucleofector II, using previously described conditions (40). 5%, 1%, 0.2% or 0.04% of input DNA was used to generate a standard curve for quantification comparison.
Generation of MLL1 expressing cell lines
To stably express MLL1 or MLL1/N3906A in MLL1 KO cells, MLL1 KO clones were transfected with pCMV-3Tag-6-MLL1 or -MLL/N3906A using PEI. Forty-eight hours after transfection, 1 × 105 cells were seeded in 15 cm dishes in DMEM medium. Twenty-four hours later, 100 μg/ml hygromycin was added since the transfected vectors encode for hygromycin resistance. Single colonies that grew out were picked and screened for expression of MLL1 by immunoblot.
Fluorescence microscopy
1 × 105 BCBL1 cells were incubated in a well of a 12-well slide chamber (Ibidi) in 200 μl RPMI with 10% BGS for 24 h at 37°C. Supernatant was removed and cells fixed with 4% paraformaldehyde in PBS for 15 min. After fixation, cells were rinsed ×2 with 1× PBS, and blocked/permeabilized with block/perm buffer (10% BGS, 100 mM glycine, 0.2% Triton-X100 in PBS) with the addition of 5 μg/sample of human IgG. After 30 min, the block/perm buffer was removed and cells were incubated with primary antibodies anti-LANA LN53 (Millipore) and anti-MLL1C (Bethyl), or anti-LANA LN53 and anti-WDR5 (Bethyl) diluted in block/perm buffer at 1:200. After 1h of incubation at room temperature, antibodies were removed, and cells washed ×2 with 1× PBS. Secondary antibodies goat anti rat Alexa Fluor 488 (Invitrogen) and goat anti rabbit Alexa Fluor 647 (Invitrogen)) were added at 1:1000 dilutions, and incubated for 1h at room temperature in dark. Cells were washed ×2 with 1× PBS and coverslips mounted using ProlongGold antifade with DAPI (Life Technologies). Images were captured using a Zeiss LSM 800 confocal microscope and 63× objective. Co-localization was quantified using Zen Blue software and Pearson's correlation coefficient.
Protein expression and purification
pETDuet-1-WDR5, -ASH2L, -RBBP5, -MLL1-SET (residues 3745–3969) or -LANAΔ33–888 were overexpressed as 6× N-terminal histidine fusion proteins in Rosetta cells (Novagen) with 0.1 mM IPTG for 16 h at 18°C. Cells were harvested in buffer containing 50 mM Na2HPO4 pH 8.0, 1.5 M NaCl, 10 mM imidazole, lysed by sonication and clarified by centrifugation and filtration. Proteins were purified by nickel nitrilotriacetic acid chromatography with Protino Ni-NTA Agarose (Macherey-Nagel). Proteins were then eluted with buffer containing 50 mM Na2HPO4 PH 8.0, 300 mM NaCl, 250 mM imidazole followed by dialysis into a final buffer of 20 mM Tris–HCl pH 7.5, 300 mM sodium chloride and 5 mM β-mercaptoethanol (β-ME). pGEX-6p-1-WDR5 and MBP-tagged pMAL-LANA Δ33–888 were overexpressed in Rosetta cells (Novagen) with 0.1 mM IPTG for 16 h at RT. Cells were harvested in phosphate buffered saline for pGEX-6p-1 vectors, or in 20 mM Tris–HCl PH 7.4, 200 mM NaCl, 1 mM EDTA and 10 mM β-ME for pMAL vectors, lysed by sonication, and clarified by centrifugation and filtration. GST-WDR5 was purified by Glutathione Sepharose 4B GST-tagged protein purification resin (GE Life Science) followed by elution with reduced glutathione. MBP-LANA Δ33–888 was purified with Amylose Resin (New England Biolabs) according to the manufacturer's protocol. Purified recombined protein was dialyzed into a final buffer of 20 mM Tris–HCl pH 7.5, 300 mM sodium chloride and 5 mM β-mercaptoethanol (β-ME). Protein concentration was quantified by Bradford assay (Bio-Rad).
For gel-filtration chromatography, LANA (Δ33–888) was cloned into a modified pET28b vector with a 6× His–SUMO tag fused at the N terminus. LANA protein was expressed in Escherichia coli Transetta (DE3) (TransGen Biotech) and purified with Ni-NTA agarose beads (Qiagen) in lysis buffer (50 mM Tris–HCl pH 7.5, 500 mM NaCl, 10% glycerol, 5 mM 2-mercaptoethanol, 1 mM PMSF, 5 mM benzamidine, 1 μg/ml leupeptin and 1 μg/ml pepstatin). The 6× His–SUMO tag was removed by Ulp1 protease digestion. The LANA protein was further purified by HiLoad Superdex 200 gel-filtration chromatography (GE Healthcare) in column buffer A (25 mM Tris–HCl pH 7.5 and 150 mM NaCl). MLL1, WDR5, RBBP5 and ASH2L–DPY30 proteins were purified as described earlier.(41) The purified LANA, MLL1, WDR5, RBBP5 and ASH2L–DPY30 proteins were mixed in a molar ratio of 2.4:2.4:1.2:1.2:1. After incubation on ice for 1 h, the assembled LANA–MLL1 complexes were separated from the free individual components through a Superose 6 gel-filtration chromatography (GE Healthcare) in column buffer A.
For structural studies, the full-length human WDR5 and the N-terminal 24-residue deletion construct WDR5Δ24 were both cloned into pET47b (+) (KanR) plasmid (Novagen), with an N-terminal His-tag, and a 3C protease cleavage site. WDR5 proteins were prepared by plasmid transformation into E. coli BL21 star (DE3) (Invitrogen) with the codon plus plasmid pRARE2 (CamR) (Novagen). WDR5 proteins were overexpressed in ZYP-5052 auto-induction medium supplemented with the corresponding antibiotics. The cells were grown at 37°C until the OD600 reached 0.7 and the temperature was then lowered to 18°C. Cells were harvested 22 h post induction and lysed by sonication in a buffer containing 10 mM PBS pH 7.5, 500 mM NaCl, 5% glycerol, 0.1% CHAPS, 1 mM DTT, supplemented with 5 mM MgCl2, 5 U/ml OmniCleave (Epicentre), 0.25 mg/ml lysozyme and a complete EDTA-free protease inhibitor cocktail (Roche). The lysate was clarified by centrifugation at 19,000 rpm for 45 minutes at 4°C. The supernatant was loaded in a HisTrap FF 5ml column (GE Healthcare) and the protein was eluted with a linear gradient of elution buffer (20 mM HEPES, 500 mM NaCl, 500 mM imidazole, 1 mM DTT, 10% glycerol, pH 7.5). To remove the His-Tag, 3C protease was added in a 1:100 molar ratio to the eluted protein with the addition of 2 mM DTT and 1 mM EDTA. WDR5 was then further purified by size exclusion chromatography (HiLoad 16/600 Superdex 75pg column (GE Healthcare), pre-equilibrated in buffer containing 20 mM HEPES, 150 mM NaCl, 0.1 mM TCEP, pH 7.0 for crystallization experiments or 20 mM Na/K Phosphate, 150 mM NaCl, 0.1 mM TCEP, pH 7.0 for isothermal titration calorimetry analysis (ITC). The peak fraction was collected and concentrated to 10– 15 mg/ml for further use.
In vitro methyltransferase assay
Methyltransferase assays were performed as previously described (42). Briefly, purified, recombinant MLL1-SET was incubated with equimolar amounts of WDR5, RbBP5 and ASH2L in 50 mM Tris pH 8.0, 200 mM NaCl, 3 mM Dithiothreitol (DTT), 5 mM MgCl2, 5% glycerol along with 1 mM histone H3 peptide corresponding to the first N-terminal 21 H3 residues (Millipore). Reactions were initiated by adding 1 μCi of tritiated, S-adenosyl-l-methionine (AdoMet)(PerkinElmer) (0.07 mM tritiated AdoMet) and stopped by spotting reactions onto Whatman P-81 filter papers. Free AdoMet was removed by washing the filter paper four times with 250 ml of 50 mM sodium bicarbonate pH 9.0 and methyltransferase activity quantified by liquid scintillation counting.
GST or MBP fusion protein precipitations
GST or MBP fusion proteins were incubated with purified or in vitro translated protein in binding buffer (50 mM Tris–HCl, pH 7.5, 300 mM NaCl, 0.1% NP-40, 1 mM PMSF) for at least 2 h at 4°C. GST beads (GE Life Sciences) or Amylose Resin (NEB) was added, and the mixture incubated for 1 h with rotation. Beads were then washed three times with binding buffer and bound proteins assessed by SDS–PAGE followed by western blotting or Coomassie blue staining.
Measurement of LANA peptide binding
To understand the energetic contribution made by various contacts between WDR5 and LANA, we performed detailed binding analyses using isothermal titration calorimetry (ITC), using protein preparations of either WDR5 or WDR5Δ24 in the cell. The assays with LANA peptides were performed with wild-type or WDR5Δ24 proteins to assess the roles of N-terminal LANA residues predicted to be essential for peptide binding. Peptides for complex formation were obtained from Genscript, with the exception of WDR510-19, LANA23-32 and LANA21-32 (ProteoGenix) and synthesized with an N-terminal acetyl moiety, and a C-terminal amidylation group.
ITC titrations were performed in a MicroCalTM iTC200 Isothermal Titration Calorimeter (Malvern) at 20°C. WDR5 protein concentration was determined by using the extinction coefficient (69 900 M–1 cm–1) after measuring the absorbance at 280 nm using a NanoDrop spectrophotometer (NanoDrop Technologies). Lyophilized LANA peptides (10 mM) were solubilized in ITC buffer (20 mM Na/K phosphate, 150 mM NaCl, 0.1 mM TCEP, pH 7.0), aliquoted, and stored at –20°C for further use. In each of the experiments, injections of between 1.5 and 2 μl of the titrant were performed at 180 s intervals, with the sample stirred at 800 rpm throughout the experiment. The titration curves were fitted assuming a single-site binding model in the Origin 7.0 software, and included analysis using NITPIC, SEDPHAT and plotted with GUSSI (43). The binding constant (KD) and enthalpy (ΔH) values calculated using SEDPHAT and Origin models were generally in the same range, although in some instances intervention by manual peak integration was required.
Crystallization
Crystallization trials were set up using the vapor diffusion method with the PACT, JCSG and Stura MacroSol screens (Molecular dimensions) at 293 K. The droplets consisted of 100 nl protein solution and 100 nl reservoir solution and were set up in 96-well three-drop Swissci plates sitting drop format and optimized by hanging drop in 24-well XRL Plate (Molecular Dimensions). Crystals grew in 2–3 days and suggested that the protein crystallizes with medium-molecular-weight polyethylene glycols. Diffraction quality crystals were obtained after several rounds of optimization, which included fine screening with PEG 4K, 3350K, 8K and 6K and various buffers at neutral or acidic pH, microseeding, streak-seeding, drop-size and drop-ratio variation. WDR5Δ24 in complex with LANAWIN peptides were prepared by mixing both components at a 1:2 or 1:5 molar ratio.
Crystals appeared after 3–4 days and grew to full size within 1–2 weeks. The WDR5WIN crystals were grown by hanging drop vapor diffusion in the presence of 26% PEG 3350, 100 mM Bis–Tris pH 5.5, 50 mM (NH4)2SO4 at 14.65 mg/ml with 2:1 drop ratio. The WDR5Δ24-LANA23-32 crystals were grown using the same method as described above, with 10 mg/ml with 2-fold peptide molar excess in 1:1 drop ratio in 100 mM Bis–Tris pH 5.5, 50 mM (NH4)2SO4, 25% PEG3350. The crystals used for data collection were stabilized by a 1% increase in PEG3550 to the crystallization buffer and cryo-protected by adding 20% glycerol, before being harvested and flash-cooled in liquid nitrogen.
Crystallographic Data Analysis and Structure determination
The diffraction data were collected on beamline IO4 (WDR5WIN) using X-rays of wavelength 0.9762 Å at Diamond Light Source (Oxfordshire, UK) and ID30B (LANAWIN) using X-rays of wavelength 0.9918 Å at the ESRF (Grenoble, France) Synchrotron sources from crystals cryocooled to 100 K. The WDR5WIN data set was processed by xia2 pipeline (44), automated diffraction data reduction tool which utilizes XDS (45) and the CCP4i2 suite (46,47) for data integration and truncation. The WDR5WIN crystal was indexed in space group P212121, with unit-cell parameters a = 46, b = 57, c = 68.1 Å, α = 90, β = 90, γ = 90°. The structure of the WDR5–WIN complex was determined by molecular replacement with Phaser (48) using the WDR5 structure (PDB entry 4ery) (49) as the search model. Both the WIN motif binding peptide (WDR11–19) and the WDR5 structure were represented by excellent electron density. The structure was refined at 1.3 Å resolution using iterative cycles of manual building in COOT (50) and refinement with REFMAC (51).
The WDR5Δ24-LANA23–32 diffraction data were processed with xia2 (44) using DIALS (52) for indexing, refinement and integration. POINTLESS (53) and AIMLESS (54) were used for scaling and merging of data. The crystal diffracted to 1.25 Å in space group P1 and resulted in 86.4% complete data set to a resolution of 1.50 Å and was indexed in space group P1, with unit-cell parameters a = 51.2, b = 59.7, c = 63.1 Å, α = 80.15, β = 82.98, γ = 89.27°. The Matthews coefficient was consistent with two molecules of WDR5 in the asymmetric unit (VM = 1.94 Å3/Da, solvent content 36.7%). The native Patterson function showed a strong non-origin peak with 20.3% of the origin peak height and located at 0.335, 0.003, 0.001, indicating translational non-crystallographic symmetry (tNCS) with a translation vector of 15.6 Å. The observed translational pseudo-symmetry impacted both the intensity statistics and subsequent analyses, which led to the higher than expected refinement statistics (Supplementary Table S2). The structure of the WDR5Δ24–LANA23-32 complex was determined by molecular replacement with MOLREP (55) using the WDR5WIN structure. The structure was refined at 1.5 Å resolution using iterative cycles of manual building in COOT (50) and refinement with PHENIX (56).
Structural quality was validated by MolProbity and the structure has no Ramachandran outliers, no poor backbone angles or bonds and only 4–5 poor rotamers for the protein side chains (two protomers in the asymmetric unit) in the P1 structure according to the MolProbity server (57). Structure representation was carried out using PyMOL (Schrödinger, LLC) with electrostatic surfaces generated using the APBS plugin (58), the protein structure was prepared using pdb2pqr (59). The coordinates and structure factors have been deposited in the PDB with PDB ID: 7BED for the WDR5WIN structure, and PDB ID: 7BCY for the LANAWIN structure.
Statistical analyses
Statistical analyses were performed using Graphpad 7.
RESULTS
LANA interacts with the MLL1 complex and recruits MLL1 to KSHV DNA
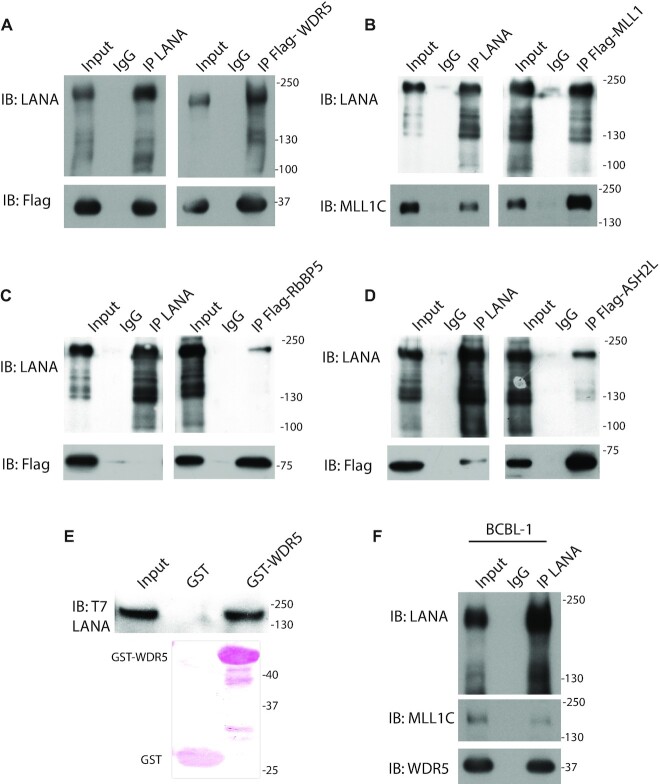
Tandem affinity purification of LANA associated complexes (40) identified core components of the MLL1 histone methyltransferase (HMT) complex including MLL1, WDR5, ASH2L and RBBP5, as potential LANA interacting partners. We further assessed these interactions by co-immunoprecipitation (IP). MLL1, WDR5, or ASH2L, each co-precipitated with LANA after transient co-expression either after IP of LANA or the reciprocal IP (Figure 1A, B, D). However, only weak LANA signal was observed after IP of RbBP5, and no RbBP5 was evident after IP of LANA, suggesting a weak, possibly indirect, interaction between these proteins (Figure 1C). Purified GST-WDR5 also precipitated in vitro translated LANA (Figure 1E).
Figure 1.
LANA interacts with MLL1 complex components. (A–D) Reciprocal co-immunopreciptiations (IPs) of LANA and (A) WDR5, (B) MLL1, (C) RbBP5 or (D) ASH2L from whole cell lysates after expression in 293T cells. WDR5, RbBP5, ASH2L, or MLL1 were immunoprecipitated with anti-Flag, and LANA with anti-LANA antibody, or IgG control used. (E) GST or GST-WDR5 precipitation of in vitro translated full length T7 epitope tagged LANA. Ponceau S stained GST and GST-WDR5 are shown. (F) IP of endogenous LANA from KSHV infected BCBL1 cell nuclear lysate followed by detection of MLL1 or WDR5. (A–E) Molecular weight (kDa) indicated at right.
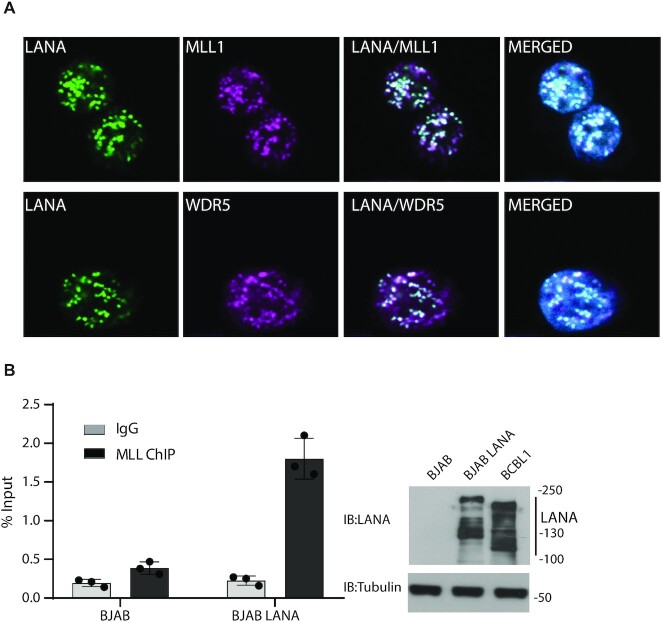
We assessed endogenous interactions between LANA and MLL1 or WDR5 in KSHV naturally infected BCBL1 PEL cells. Both MLL1 and WDR5 co-precipitated with LANA (Figure 1F) from BCBL1 cells. LANA concentrates to dots at sites of KSHV genomes in infected cells (13). Immune fluorescent microscopy demonstrated that LANA (green) colocalized in dots with WDR5 (magenta) or MLL1 (magenta) in BCBL1 cells (overlay generates white) (Figure 2A, Supplementary Figure S1).
Figure 2.
LANA colocalizes with MLL1 and WDR5 in infected cells and recruits MLL1 to TR DNA. (A) Immunofluorescent detection of LANA (green), WDR5 (magenta) or MLL1 (magenta) in BCBL1 primary effusion lymphoma cells. Overlay of green and magenta generates white. Merged images include DAPI (blue). Magnification: 630×. Pearson correlation coefficients are 0.87 for LANA and MLL1, and 0.77 for LANA and WDR5. (B) ChIP for MLL1 binding at TR DNA was performed after transfection of p8TR into uninfected BJAB B lymphoma cells or BJAB cells stably expressing LANA. Bars are averages of three experiments. Individual data points and standard deviation are shown. Immunoblot of LANA or tubulin in BJAB, BJAB/LANA or BCBL1 cells is shown at right. LANA is expressed as multiple bands due to alternative initiation of translation and an alternative poly adenylation signal (85,86). The LANA stably expressed in BJAB cells contains an N-terminal T7 epitope tag.
We asked if LANA can recruit MLL1 to KSHV DNA. Each KSHV TR contains three adjacent LANA binding sites. We performed MLL1 ChIP after transfection of TR DNA into uninfected BJAB B lymphoma cells or BJAB cells stably expressing LANA at physiologic levels. MLL1 association with TR DNA increased in cells expressing LANA (Figure 2B). Together, these data indicate LANA interacts with the MLL1 complex and recruits MLL1 to KSHV TR DNA.
MLL1 deposits H3K4me3 modifications on KSHV TR chromatin
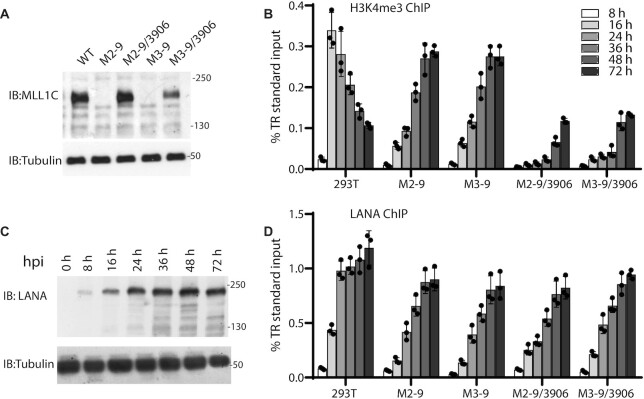
Since high H3K4me3 levels are present at KSHV TR DNA (18,28), we asked if MLL1 is involved in establishing these modifications. We used CRISPR-Cas9 to knock out endogenous MLL1 expression in 293T cells, generating clones M2-9 and M3-9 (Figure 3A, Supplementary Figure S2A). Following KSHV infection in WT cells, H3K4me3 levels peaked at 16 h post-infection (hpi), and then progressively declined (Figure 3B). In contrast, in M2-9 or M3-9 cells, H3K4me3 levels progressively increased through 48–72 hpi. In the absence of MLL1 in these cells, H3K4me3 modifications may be due to LANA recruitment of other MLL/HMT proteins, such as SET1 (28). In fact, in KSHV latently infected MLL1 knockout (KO) cells, LANA retained interaction with endogenous MLL1 complex components WDR5, RBBP5 and ASH2L (Supplementary Figure S2C).
Figure 3.
TR H3K4me3 modification by MLL1 is highly dynamic and varies with LANA occupancy. (A) Immunoblot of MLL1 from 293T, MLL1 KO cell lines M2-9 or M3-9, or MLL1 KO clones expressing MLL1-N3906A. (B) Relative H3K4me3 enrichment levels at KSHV TR DNA as assessed by ChIP in 293T, MLL1 KO, or MLL KO cells expressing MLL1-N3906A at the indicated times following KSHV infection. (C) LANA expression at indicated times following KSHV infection of 293T cells. (D) Relative LANA enrichment levels at the TR as assessed by ChIP in the indicated cell lines at different times post infection (same key for bar patterns as in panel (B).) (B and D) Bars show means from three independent experiments. Individual data points and standard deviation are shown.
We asked if MLL1 catalytic activity is important for the observed rapid, high level, TR H3K4me3 modification. We stably expressed a full length MLL1 catalytic inactive mutant, MLL1-N3906A (60,61), in M2-9 or M3-9 cells (Figure 3A). MLL1-N3906A retained the ability to interact with LANA (Supplementary Figure S2D). Following KSHV infection, H3K4me3 levels progressively increased in M2-9/3906 or M3-9/3906 cells, in a pattern similar to that of M2-9 or M3-9, although H3K4me3 levels were substantially lower (Figure 3B). The lower H3K4me3 levels suggest that the presence of MLL1-N3906A in the complex may inhibit recruitment of other MLL HMT proteins to deposit H3K4me3 onto TR DNA. In contrast, stable expression of WT MLL1 in M2-9 cells resulted in a WT pattern of H3K4me3 deposition (Supplementary Figure S2B). These results indicate that MLL1 is necessary for rapid, high level, H3K4me3 deposition on KSHV TR DNA following infection.
LANA directly interacts with the MLL1 complex and regulates its enzymatic activity
Since LANA interacts with the MLL1 complex and recruits MLL1 to TR DNA, we assessed the relationship between LANA expression and H3K4me3 TR modification. LANA expression gradually increased following infection, peaking at ∼36–48 hpi (Figure 3C). Correspondingly, LANA levels increased over time at TR DNA in 293T, MLL1 KO (M2-9, M3-9), or MLL1-N3906A cells (Figure 3D). Comparison of LANA and H3K4me3 TR levels following infection demonstrated that TR H3K4me3 levels decreased as LANA levels increased, suggesting LANA may inhibit MLL1 activity (Figure 3B, D).
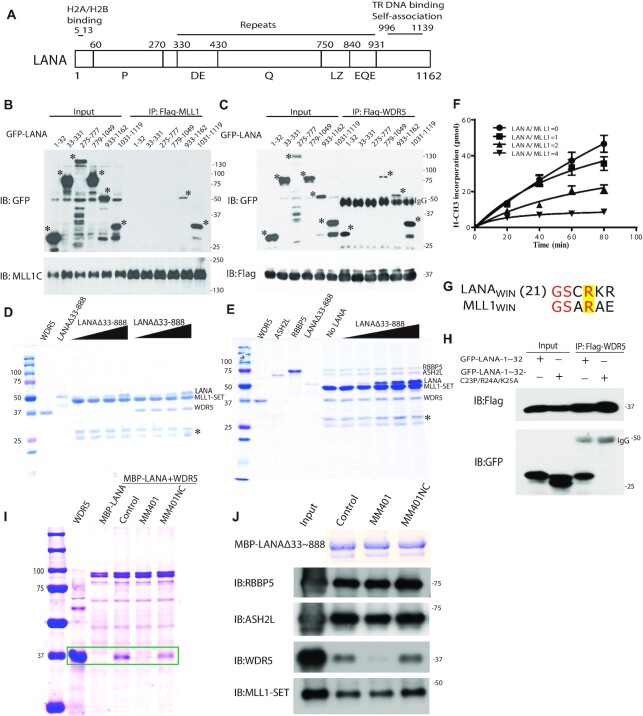
This result led us to pursue the physical interaction of LANA with the MLL1 complex in greater detail. In co-IP experiments following transient expression, MLL1 interacted with C-terminal LANA, while WDR5 interacted with either N- or C-terminal LANA (Figure 4A-C). Although both MLL1 and WDR5 interacted with C-terminal LANA, the binding patterns were different. WDR5 interacted with LANA 779–1049, while MLL1 did not. Further, MLL1 interacted only with LANA 1031–1065, while WDR5 interacted with both LANA 1031–1065 or LANA 1066–1119 (Supplementary Figure S3A, B). Consistent with both these sequences interacting with WDR5, LANA residues 1060–1070 contain a bend, reminiscent of RbBP5’s contact point with WDR5 (62).
Figure 4.
LANA interacts with, and inhibits, the MLL1 complex. (A) Schematic diagram of LANA. LANA proline rich (P), aspartate and glutamate (DE), glutamine (Q), putative leucine zipper (LZ), glutamate and glutamine (EQE) regions. Residues 5–13 bind histones H2A/H2B (34,77). LANA 996 –1139 self-associates to bind terminal repeat (TR) DNA and independently associates with mitotic chromosomes. (B and C) Co-immunoprecipitation (Co-IP) of GFP-LANA regions with (B) MLL1 or (C) WDR5 after expression in 293T cells. IPs were performed using anti-Flag antibody. Asterisks denote GFP-LANA fusions. (D) LANAΔ33–888, or WDR5 (in lanes at right) co-precipitation with GST-MLL1-SET (E) ASH2L, RbBP5, WDR5 and LANAΔ33–888 co-precipitation with GST-MLL1-SET. (D, E) Triangles at top indicate increasing LANAΔ33–888 and asterisk indicates degradation products. (F) Histone methyltransferase activity of the MLL1 complex after incubation with increasing amounts of LANA. Results are the means of three independent experiments. Standard deviation is shown. (G) Sequence alignment of MLL1WIN with LANAWIN starting at LANA residue 21. Yellow highlight indicates the conserved arginine. (H) Co-IP of GFP-LANA-1–32 or GFP LANA-1–32 containing C23P/R24A/K25A substitutions with Flag-WDR5 after co-expression in 293T cells. (I) Co-precipitation of WDR5 with MBP-LANAΔ33–888 in the presence or absence MM401 or enantiomer control. Two left lanes, WDR5 or MBP-LANAΔ33–888 alone. Box encloses WDR5 bands. (J) ASH2L, RbBP5, WDR5, or MLL1-SET co-precipitation with MBP-LANAΔ33–888 after incubation in the presence or absence MM401 or enantiomer control. Proteins were detected by immunoblot except MBP-LANAΔ33–888, which was detected by Coomassie blue. (D, E, I) Proteins were detected with Coomassie blue.
We asked if LANA directly interacts with the MLL1 complex. LANAΔ33–888 contains the LANA N- and C-terminal regions required for interaction with MLL1 or WDR5. Purified GST-MLL1-SET domain co-precipitated purified LANAΔ33–888 and WDR5 (Figure 4D). Since MLL1-SET interacts with WDR5 (63), these proteins may form a ternary complex through multiple interactions. Further, GST-MLL1-SET co-precipitated purified LANAΔ33–888, WDR5, RbBP5, and ASH2L (Figure 4E). Size exclusion chromatography showed LANAΔ33–888 shifted the elution profile of the purified MLL1 complex to an earlier elution volume, and co-eluted with the complex components (Supplementary Figure S3C). These findings indicate LANA directly interacts with MLL1 complex member(s) and integrates into the complex.
We investigated the effect of LANA on MLL1 complex enzymatic activity. The MLL1-SET domain alone exhibited little HMT activity, and the presence of RbBP5 and ASH2L enhanced the activity, consistent with prior data with MLL or SET1 complexes (42). Inclusion of WDR5 was necessary for maximal HMT activity (Supplementary Figure S3D). We next assessed the effect of LANAΔ33–888 on MLL1 complex HMT activity. At an equimolar ratio, LANA exerted little effect on MLL1 catalytic activity (Figure 4F). However, at increasing concentrations, LANA progressively inhibited HMT activity, and activity was undetectable at a 4:1 molar ratio.
This dose-dependent inhibitory effect provided a potential explanation for the dynamic TR H3K4me3 profile early during de novo infection (Figure 3B). At 8 hpi, little LANA is present at the TR (Figure 3D) to recruit MLL1, and therefore, there is little H3K4me3 deposition. By 16 hpi increased LANA levels allow recruitment of MLL1 and high level H3K4me3 deposition. However, as LANA levels subsequently increase at the TR, MLL1 catalytic activity is inhibited, resulting in decreasing levels of H3K4me3 in conjunction with the expected presence of histone demethylase activity such as from KDM1A, KDM1B or KDM2B (64,65). In contrast, in MLL1 KO or MLL1-N3906A cells, the H3K4me3 levels at the TR do not decrease in response to increasing LANA levels (Figure 3B–D), suggesting that the HMT(s) substituting for MLL1 are not inhibited by LANA. MLL1 is unique among MLL proteins in requiring WDR5 for catalytic activity (66), and LANA interacts with WDR5. Therefore, the dose-dependent effect of LANA on the MLL1 complex could indicate a mechanism in which LANA competes with MLL1 for WDR5 binding to inhibit MLL1 activity.
N-terminal LANA contains a WIN motif that binds WDR5
We explored the possibility that LANA might bind WDR5 in a manner similar to that of MLL1. Biochemical and structural studies have demonstrated that the MLL1 WDR5 interaction motif (MLL1WIN) is critical for MLL1 catalytic activity (67,68). The WIN motif is a short 11 amino acid peptide present in all MLL proteins that contains a conserved arginine critical for WDR5 interaction (42). We identified a WIN-like motif in N-terminal LANA that contains the conserved arginine (Figure 4G). Substitution mutations of the arginine along with the preceding and subsequent amino acids eliminated the interaction between N-terminal LANA and WDR5 (Figure 4H). We investigated the interaction of LANAWIN with WDR5 or WDR5Δ24, which lacks the 24 N-terminal WDR5 residues, by isothermal titration calorimetry (ITC) analysis. LANAWIN peptide 16AcAPLTRGSCRKRNRSPERNH232, or similar LANA peptide with N-terminal truncations at residues 19, 21, or 23, bound with affinities ranging from 2.3μM-25μM (Supplementary Figure S4, Supplementary Table S1). Inclusion of LANA residues 28RSPER32 were required for micromolar affinity since 16AcAPLTRGSCRKRNNH227 binding was greatly reduced. Binding was governed by exothermic heat, and entropy driven.
We explored whether LANAWIN occupies the WDR5 WIN binding pocket as expected from its homology with MLLWIN. The small molecule MM-401 occupies the WDR5 WIN binding pocket and thereby inhibits MLL1-WDR5 interaction (66). MM-401, but not its enantiomer, blocked the interaction of WDR5 with LANA (Figure 4I). Since the interaction was blocked despite an independent C-LANA-WDR5 interaction (Figure 4B), C-LANA binding may also be dependent on or may overlap the WDR5 WIN binding pocket. MM-401 also excluded nearly all WDR5 from the LANA/MLL1 complex (Figure 4J) despite WDR5’s additional interaction with RbBP5 (62). These findings demonstrate that LANAWIN binds WDR5 similarly to MLL1WIN and that MM401 excludes WDR5 from the LANA complex.
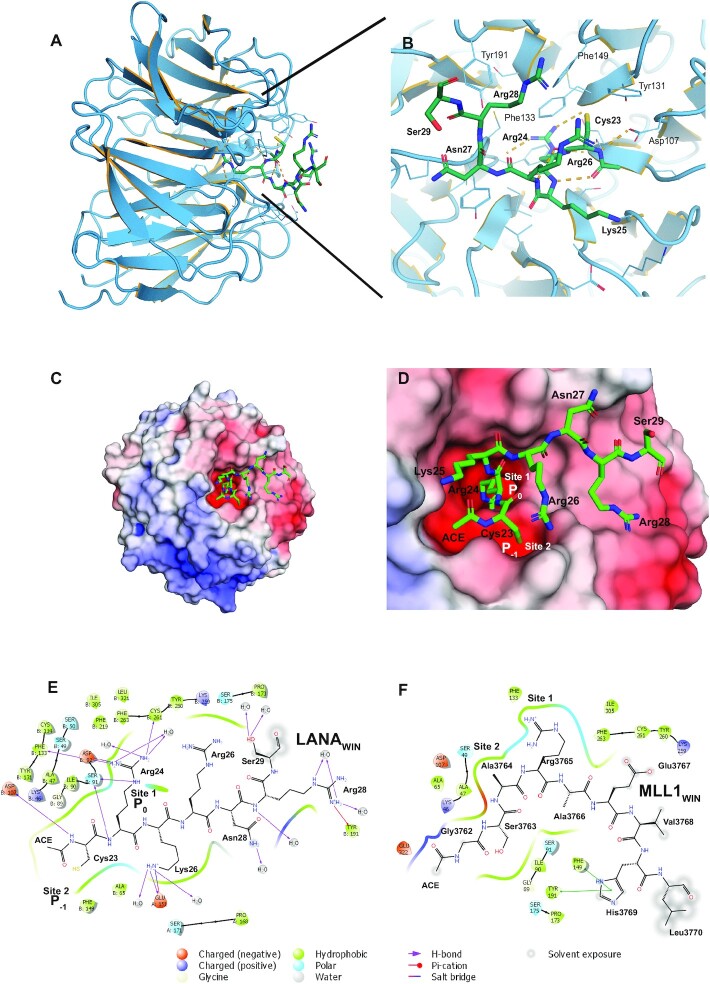
Crystal structure of WDR5 with LANAWIN
We solved the X-ray crystal structure of LANAWIN peptide (23CRKRNRSPER32) complexed with WDR5Δ24 to 1.5 Å resolution (Supplementary Table S2) and built an atomic model for LANA residues 23–29 (Figure 5A–E). Initial attempts to co-crystallize LANAWIN peptide with WDR5 resulted in solution only of the X-ray crystal structure of WDR5 to 1.3 Å resolution. This structure demonstrated WDR5 N-terminal residues (11EAARAQPTP19) at the site of the WIN motif binding pocket (Supplementary Figure S5C). The next visible residue in the crystal structure, Lys27, was too far away (50 Å) from WDR5 Pro19 to be connected by the intervening residues (20SSSATQS26), indicating the WDR5 N-terminal peptide had likely been cleaved to enable binding. Since the WDR5 peptide occupied the pocket, we therefore used WDR5Δ24 for LANA co-crystals. N-terminal LANAWIN binds to WDR5 in a 310-helical conformation with the conserved arginine inserted into the Win motif-binding pocket. N-terminal LANAWIN is well ordered, stabilized by the conserved Arg24 residue that inserts into the central channel of the WDR5 β-propeller, analogous to a pin socket mode of interaction, and forms a large number of interactions through a network of hydrogen bonds and cation-π interactions which provide a major contribution to complex formation (Figure 5A-D). The WDR5 binding cleft contains two adjacent hydrophobic pockets, one (Site 1) that binds LANA Arg24 (P0), and a neighboring pocket (Site 2) that binds Cys23 (P–1), and is selective for small hydrophobic sidechains, such as that of the conserved alanine in the P–1 position that binds similarly in all other WDR5–WIN complexes. The overall structure of WDR5 is maintained when bound with LANAWIN. Comparison of the WDR5-LANAWIN complex with those of WDR5 and MLL1WIN, histone H3WIN, (68–73) or N-terminal WDR5 shows LANA uses the same binding surface for WDR5 interaction (Figure 5E, F, Supplementary Figure S5D–F). LANAWIN binding is most similar to that of H3WIN, with conformational similarity between LANA Lys25 and histone H3 Lys4. LANAWIN can also bind two WDR5 molecules, similar to H3WIN (Supplementary Figure S5A, B). However, LANAWIN differs from other WIN peptides in its use of Cys23 rather than the shorter alanine sidechain at this position. The WDR5 peptidyl-arginine-binding cleft is flexible, allowing structurally divergent conformations of WIN motif C-terminal ends (42). This plasticity is evident as C-terminal LANA peptide shows disorder of both Arg26 and Arg28 although their conformations remain constrained by hydrophobic interactions with Tyr131, Tyr191 and Phe149 of WDR5. The hydrophobic cleft to which LANA binds is negatively charged (Figure 5C, D), providing favorable charge complementarity with the positive electrostatic potential of LANAWIN, and demonstrating N-terminal LANA has evolved for specific WDR5 binding.
Figure 5.
Crystal structure of the LANA WDR5 complex. (A) Overview of LANAWIN (green) bound to WDR5 (blue ribbon). (B) Face on zoomed view of WDR5-binding cleft with LANA carbons green. Key hydrogen bonds are depicted as orange dash lines, solvent molecules and alternate conformations are removed for clarity. (C) Electrostatic surface representation of the ‘top’ face of the WDR5 β-propeller showing the orientation of the LANAWIN motif and (D) a zoomed view of the hydrophobic pockets (‘Site 1, 2’) that recognize both LANAWIN Cys23 and Arg24. Charged surfaces (red, negatively charged; blue, positively charged) were calculated with APBS (87). (E and F) Maestro representations of the ligand interactions of (E) LANAWIN or (F) MLL1WIN at the WDR5 binding site. Arg24 (LANA23-32) is equivalent to Arg3765 (MLL1WIN) in the arginine conserved motif. The WIN motif binding site surface is represented by the contoured line. WDR5 residues are differentiated by colors according to the interaction type as indicated at bottom. The 2D WDR5–ligand interaction diagrams were generated using the Ligand Interaction tool in Maestro (Schrödinger Inc., www.schrodinger.com).
These results suggest a mechanism for LANA inhibition of MLL1 activity. LANAWIN and MLL1WIN both bind the WDR5 WIN pocket. The KD value for MLL1WIN is 1.1 μM (42), indicating MLL1 preferably binds WDR5. At low LANA concentration, the MLL1 complex interacts with WDR5 for optimal catalytic activity. At increasing LANA concentration, competition shifts WDR5 binding from MLL1WIN to LANAWIN, inhibiting MLL1 catalytic activity. However, as LANA also interacts with MLL1-SET (Figure 4A, C, D) through a motif yet to be defined, we cannot exclude the possibility that LANA may directly act at the MLL1 catalytic site.
MLL1 is critical for KSHV latent infection
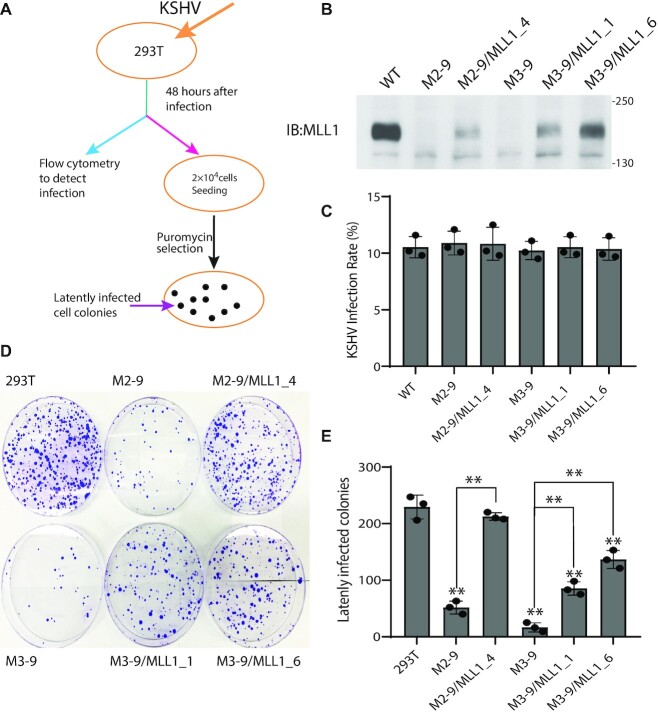
Since LANA functionally interacts with the MLL1 complex, and since MLL1 deposits H3K4me3 marks on KSHV TR chromatin, we asked if MLL1 exerts a role in KSHV latency. We assessed MLL1’s role in latency establishment using a stringent assay requiring stable, long term, latent infection. We infected 293T cells, MLL1 knockout cells M2–9, M3–9 or M3–11 (Figure 6A, 6B, Supplementary Figure S6A), or MLL1 partially deficient cells M2-7 (Supplementary Figure S6A) with KSHV. Forty-eight hours after infection, flow cytometry analysis for GFP expression, constitutively expressed from the recombinant viral genome, demonstrated similar infection rates between the different cell lines (Figure 6C, Supplementary Figure S6B). This result indicates that loss of MLL1 does not alter KSHV infection of cells or trafficking to the nucleus. KSHV latently infects cells as a multicopy episome and selection is required for long term latent infection and maintenance of KSHV episomes ex vivo (13,15,34,74,75). Therefore, KSHV infected cells were placed under puromycin selection, for which the virus encodes resistance. Infection of control 293T cells led to efficient outgrowth of latently infected cell colonies (Figure 6D, E). In contrast, infection of MLL1 knockout cell lines M2-9, M3-9 or M3-11 were highly deficient for outgrowth of latently infected cells (Figure 6D, E; Supplementary Figure S6C, D), averaging at least ∼5-fold fewer drug resistant colonies compared to WT cells. Infection of M2–7 cells, which are partially deficient for MLL1 expression (Supplementary Figure S6A) resulted in outgrowth that was intermediate between WT and MLL1 KO cells (Supplementary Figure S6C, D). Stable expression of MLL1 in MLL1 KO M2-9 or M3-9 cells (Figure 6B; Supplementary Figure S7A) rescued the ability of KSHV to establish long term latency (Figure 6D, E; Supplementary Figure S7B, C). In contrast, stable expression of catalytically inactive MLL1-N3906A in KO M2–9 or M3–9 cells (Supplementary Figure S7A) was highly deficient in its ability to rescue latent infection (Supplementary Figure S7B, C), indicating MLL1 catalytic activity exerts a key role in KSHV infection.
Figure 6.
MLL1 is critical for KSHV to establish latent infection. (A) Experimental design for assessing KSHV infection. 293T cells were infected with a multiplicity of infection of 0.1. (B) Immunoblot for MLL1 in WT 293T, M2-9 or M3-9 MLL1 knockout cells, or after stable expression of full length MLL1 in M2-9 or M3-9 cells. The cross-reactive band migrating below MLL1 indicates similar loading per lane. (C) KSHV infection as detected by flow cytometry for GFP expression at 48 hours post infection. Standard deviation is shown. (D) Crystal violet staining of KSHV latently infected colonies following puromycin resistant outgrowth. (E) Average of colony outgrowth from three independent experiments. Individual data points and standard deviation are shown. **P< 0.01 by the paired sample t-test. Asterisks directly above bars refer to differences from 293T cells.
DISCUSSION
These results demonstrate that MLL1 has a unique role in KSHV infection. LANA recruits MLL1 to KSHV TR DNA and regulates its function, leading to a dynamic H3K4me3 profile during the initial hours following infection. Knockout of MLL1, or loss of MLL1 catalytic activity, led to substantial changes in TR H3K4me3 deposition dynamics. The occurrence of H3K4me3 modifications despite the absence of MLL1 suggests other MLL family members can substitute for MLL1, perhaps within a LANA recruited complex. However, the altered H3K4me3 modification kinetics in the absence of MLL1 indicate other MLL molecules are not functionally interchangeable with MLL1.
This work provides direct evidence for MLL1 regulation by LANA and implies a mechanism through competition for MLLWIN binding to WDR5. MLL1WIN, histone H3WIN, N-terminal WDR5, and LANAWIN all bind to the same WDR5 binding cleft (Figure 5, Supplementary Figure S5) (68–73). MLL1 is unique among the MLL family members in its need for WDR5 binding to stimulate HMT activity. This requirement is likely due to the unique structural organization between MLL1, RBBP5 and WDR5 as compared to that of other MLL family members (41). It has been suggested that competition for binding to WDR5 between MLL1WIN and histone H3WIN (also an MLL1 substrate) may lead to regulation of MLL1 activity (72). Our findings demonstrate that MLL1 methylase activity is regulable by LANA, and imply other WIN motif containing host or pathogen proteins may similarly regulate MLL1. Furthermore, since N-terminal WDR5WIN can occupy the WDR5 pocket, there is potential for regulation by neighboring WDR5 molecules, therefore providing an additional layer of regulatory complexity. These findings suggest competition for MLLWIN binding to WDR5 calibrates MLL1 activity. For instance, increasing or decreasing local LANA concentration could fine tune MLL1 catalytic activity.
LANA’s interaction with MLL1 may occur at the nucleosome. Recent cryo-electron microscopy structures demonstrate the MLL1 complex makes extensive contacts with the nucleosome surface, allowing access to the histone H3 tail in order to deposit methyl marks (41,76). The LANA N-terminus, immediately upstream of LANAWIN, binds the conserved acidic patch at the interface of histones H2A/H2B (77). Superposition of the X-ray crystal structure of N-terminal LANA complexed with the nucleosome (77) with that of the structure of the MLL1 complex and the nucleosome (41) suggests that LANAWIN residues 23–29 are not in close proximity to residues 4–17 binding histones H2A/H2B; therefore, it is unlikely a single LANA molecule simultaneously binds both the WDR5 pocket and histones H2A/H2B. However, LANA dimerizes through its C-terminal domain (78). Therefore, a LANA molecule could bind to the WDR5 pocket while a second LANA molecule within the dimer could interact with the H2A/H2B acidic patch. Since C-LANA interacts with MLL1 or WDR5 (Figure 4), the carboxy-terminal domain of one LANA molecule in a dimer could interact with MLL1, while the other LANA molecule interacts with WDR5. Further, LANA dimers can undergo higher order oligomerization through dimer–dimer interactions (79–82), so additional LANA molecules could potentially interact with an adjacent nucleosome, or with a distinct MLL1 complex.
MLL1 deregulation is linked to leukemia, and this work now links MLL1 to KSHV, a tumor virus. MLL1 deregulation occurs in up to 10% of acute leukemias in all age groups and in ∼75% of acute lymphoblastic leukemia in infants (83). Typically, one MLL1 allele contains a translocation in which one of ∼80 host partners is fused in frame with MLL1, resulting in preservation of N-terminal MLL1 but loss of its C-terminal SET domain (3). Rearranged MLL1 proteins likely promote mislocalization of the fusion partners, leading to expression of genes that promote tumorigenesis (84). It is possible that LANA regulation and/or mislocalization of MLL1 to host genes may promote KSHV tumorigenesis. Alternatively, MLL1 complexes could recruit LANA, which could in turn deregulate genes through decreasing MLL1 activity and/or recruitment of LANA transcriptional regulatory partners. Consistent with these possibilities, ChIP seq analyses demonstrate that LANA often localizes near transcriptional start sites at H3K4me3 peaks (28,29).
This work demonstrates that MLL1 exerts an important role in KSHV latent infection and that its catalytic activity is central to this function. These results suggest that the alterations in TR H3K4me3 levels induced by MLL1 following infection, likely combined with MLL1 activity at additional KSHV genomic sites, are key for latency establishment. In addition, MLL1 may act at host genes that are critical for the promotion and maintenance of latency. Once epigenetic marks are established, the viral genome persists as an episome, and LANA may function with the MLL1 complex to maintain the marks and drive latency, as suggested by LANA’s colocalization with MLL1 and WDR5 in latently infected cells (Figure 2A) in dots at viral genomes. Further, MLL1 deposition of H3K4me3 marks at the TR elements may function in roles other than gene expression regulation. For instance, LANA mediates viral DNA replication of TR DNA and tethers TR DNA to mitotic chromosomes to segregate viral genomes to progeny cell nuclei. It is possible LANA’s MLL1 interaction may affect these functions, which are also essential to latency.
In summary, we show that LANA recruits MLL1 to KSHV DNA where TR H3K4me3 marks are established early following virus infection. LANA regulates MLL1 and the crystal structure of LANAWIN with WDR5 implies a regulatory mechanism and additional host regulation of MLL1. These results suggest local LANA concentration acts to calibrate MLL1 activity and that specific activity levels are key for successful KSHV infection. Strategies that disrupt MLL1 regulation or function may be applicable to prevention or treatment of KSHV malignancy.
DATA AVAILABILITY
Structural data have been deposited at Protein Data Bank (PDB). The accession numbers for the structures are: 7BED, 7BCY. Otherwise, the article includes all data generated/analyzed in the study.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Shaomeng Wang for providing MM401 and enantiomer control, Xiaolong Zhou and Enduo Wang for bacterial expression vectors, Bing Zhu and Charles Richardson for use of their scintillation counter. Mary Ballestas cloned GFP LANA 1031-1119. Murli Narayan cloned pMAL-LANAΔ33-888. We thank Diamond Light Source for beamtime (proposal mx14707), and the staff of beamlines I03, I04 and I04-1 for assistance with crystal testing and data collection. The European Synchrotron Radiation and ALBA Facilities also provided synchrotron radiation access. We thank Montserrat Soler-López for assistance with ESRF beamline ID30B and Roeland Boer for assistance with ALBA beamline BL13-XALOC.
Contributor Information
Min Tan, Departments of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA.
Shijun Li, Departments of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA.
Franceline Juillard, Departments of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA.
Rute Chitas, Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Oeiras 2780-157, Portugal.
Tânia F Custódio, Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Oeiras 2780-157, Portugal.
Han Xue, State Key Laboratory of Molecular Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 200031 Shanghai, China.
Agnieszka Szymula, Departments of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA.
Qiming Sun, Departments of Biochemistry and Cardiology, Second Affiliated Hospital Zhejiang University School of Medicine, Hangzhou, Zhejiang 310058, China.
Bing Liu, Departments of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA.
Ángel L Álvarez, Departments of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA.
She Chen, Proteomics Center, National Institute of Biological Sciences, Beijing 102206, China.
Jing Huang, State Key Laboratory of Oncogenes and Related Genes, Shanghai Institute of Precision Medicine, Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine 200125 Shanghai, China.
J Pedro Simas, Instituto de Medicina Molecular, Avenida Professor Egas Moniz, 1649-028 Lisboa, Portugal; Católica Biomedical Research, Católica Medical School, Universidade Católica Portuguesa, Palma de Cima, 1649-023 Lisboa, Portugal.
Colin E McVey, Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Oeiras 2780-157, Portugal.
Kenneth M Kaye, Departments of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [AI150575, DE025208, CA082036, DE024971 to K.M.K.]; CEM acknowledges FCT Exploratory Grant [IF/01023/2013/CP1173/CT0003]; iNOVA4Health Research Unit [LISBOA-01-0145-FEDER-007344], cofunded by Fundação para a Ciência e Tecnologia/Ministério da Ciência e do Ensino Superior; FEDER under the PT2020 Partnership Agreement; J.P.S. is supported by FCT grants [FPJ000983, FPJ001183]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012; 81:65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Miller T., Krogan N.J., Dover J., Erdjument-Bromage H., Tempst P., Johnston M., Greenblatt J.F., Shilatifard A.. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Piunti A., Shilatifard A.. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016; 352:aad9780. [DOI] [PubMed] [Google Scholar]
- 4. Ansari K.I., Mandal S.S.. Mixed lineage leukemia: roles in gene expression, hormone signaling and mRNA processing. FEBS J. 2010; 277:1790–1804. [DOI] [PubMed] [Google Scholar]
- 5. Hughes C.M., Rozenblatt-Rosen O., Milne T.A., Copeland T.D., Levine S.S., Lee J.C., Hayes D.N., Shanmugam K.S., Bhattacharjee A., Biondi C.A.et al.. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol. Cell. 2004; 13:587–597. [DOI] [PubMed] [Google Scholar]
- 6. Steward M.M., Lee J.S., O’Donovan A., Wyatt M., Bernstein B.E., Shilatifard A.. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 2006; 13:852–854. [DOI] [PubMed] [Google Scholar]
- 7. Chang Y., Cesarman E., Pessin M.S., Lee F., Culpepper J., Knowles D.M., Moore P.S.. Identification of Herpesvirus-like DNA Sequences in AIDS-Associated Kaposi's Sarcoma. Science. 1994; 266:1865–1869. [DOI] [PubMed] [Google Scholar]
- 8. Cesarman E., Chang Y., Moore P.S., Said J.W., Knowles D.M.. Kaposi's Sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. NEJM. 1995; 332:1186–1191. [DOI] [PubMed] [Google Scholar]
- 9. Moore P.S., Chang Y.. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection [see comments]. N. Engl. J. Med. 1995; 332:1181–1185. [DOI] [PubMed] [Google Scholar]
- 10. Soulier J., Grollet L., Oksenhendler E., Cacoub P., Cazals-Hatem D., Babinet P., Agay M.-F., Clauvel J.-P., Raphael M., Degos L.et al.. Kaposi's Sarcoma-Associated Herpesvirus-Like DNA sequences in multicentric castleman's disease. Blood. 1995; 86:1276–1280. [PubMed] [Google Scholar]
- 11. Decker L.L., Shankar P., Khan G., Freeman R.B., Dezube B.J., Lieberman J., Thorley-Lawson D.A.. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J. Exp. Med. 1996; 184:283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Speck S.H., Ganem D.. Viral latency and its regulation: lessons from the gamma-herpesviruses. Cell Host Microbe. 2010; 8:100–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ballestas M.E., Chatis P.A., Kaye K.M.. Efficient persistence of extrachromosomal KSHV DNA mediated by latency- associated nuclear antigen. Science. 1999; 284:641–644. [DOI] [PubMed] [Google Scholar]
- 14. Garber A.C., Hu J., Renne R.. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 2002; 277:27401–27411. [DOI] [PubMed] [Google Scholar]
- 15. Ballestas M.E., Kaye K.M.. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 2001; 75:3250–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cotter M.A. 2nd, Robertson E.S.. The latency-associated nuclear antigen tethers the Kaposi's sarcoma- associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology. 1999; 264:254–264. [DOI] [PubMed] [Google Scholar]
- 17. Toth Z., Brulois K., Lee H.R., Izumiya Y., Tepper C., Kung H.J., Jung J.U.. Biphasic euchromatin-to-heterochromatin transition on the KSHV genome following de novo infection. PLoS Pathog. 2013; 9:e1003813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gunther T., Grundhoff A.. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 2010; 6:e1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Toth Z., Papp B., Brulois K., Choi Y.J., Gao S.J., Jung J.U.. LANA-mediated recruitment of host Polycomb repressive complexes onto the KSHV genome during de novo infection. PLoS Pathog. 2016; 12:e1005878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jha H.C., Lu J., Verma S.C., Banerjee S., Mehta D., Robertson E.S.. Kaposi's sarcoma-associated herpesvirus genome programming during the early stages of primary infection of peripheral blood mononuclear cells. mBio. 2014; 5:e02261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roy A., Ghosh A., Kumar B., Chandran B.. IFI16, a nuclear innate immune DNA sensor, mediates epigenetic silencing of herpesvirus genomes by its association with H3K9 methyltransferases SUV39H1 and GLP. Elife. 2019; 8:e49500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun R., Tan X., Wang X., Wang X., Yang L., Robertson E.S., Lan K.. Epigenetic landscape of kaposi's sarcoma-associated herpesvirus genome in classic kaposi's sarcoma tissues. PLoS Pathog. 2017; 13:e1006167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sakakibara S., Ueda K., Nishimura K., Do E., Ohsaki E., Okuno T., Yamanishi K.. Accumulation of heterochromatin components on the terminal repeat sequence of Kaposi's sarcoma-associated herpesvirus mediated by the latency-associated nuclear antigen. J. Virol. 2004; 78:7299–7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim K.Y., Huerta S.B., Izumiya C., Wang D.H., Martinez A., Shevchenko B., Kung H.J., Campbell M., Izumiya Y.. Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen regulates the KSHV epigenome by association with the histone demethylase KDM3A. J. Virol. 2013; 87:6782–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ottinger M., Christalla T., Nathan K., Brinkmann M.M., Viejo-Borbolla A., Schulz T.F.. Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J. Virol. 2006; 80:10772–10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krithivas A., Fujimuro M., Weidner M., Young D.B., Hayward S.D.. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 2002; 76:11596–11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu F., Tsai K., Chen H.S., Wikramasinghe P., Davuluri R.V., Showe L., Domsic J., Marmorstein R., Lieberman P.M.. Identification of host-chromosome binding sites and candidate gene targets for Kaposi's sarcoma-associated herpesvirus LANA. J. Virol. 2012; 86:5752–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu J., Yang Y., Turner P.C., Jain V., McIntyre L.M., Renne R.. LANA binds to multiple active viral and cellular promoters and associates with the H3K4Methyltransferase hSET1 Complex. PLoS Pathog. 2014; 10:e1004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mercier A., Arias C., Madrid A.S., Holdorf M.M., Ganem D.. Site-Specific association with host and viral chromatin by kaposi's sarcoma-associated herpesvirus LANA and its reversal during lytic reactivation. J. Virol. 2014; 88:6762–6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hellert J., Weidner-Glunde M., Krausze J., Lunsdorf H., Ritter C., Schulz T.F., Luhrs T.. The 3D structure of Kaposi sarcoma herpesvirus LANA C-terminal domain bound to DNA. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:6694–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cho Y.W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G.R., Copeland T.D., Kalkum M.et al.. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 2007; 282:20395–20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xia Z.B., Anderson M., Diaz M.O., Zeleznik-Le N.J.. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:8342–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. De Leon Vazquez E., Carey V.J., Kaye K.M. Identification of kshv lana regions important for episome segregation, replication and persistence. J. Virol. 2013; 87:12270–12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barbera A.J., Ballestas M.E., Kaye K.M.. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence. J. Virol. 2004; 78:294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. De Leon Vazquez E., Kaye K.M.. The internal Kaposi's sarcoma-associated herpesvirus LANA regions exert a critical role on episome persistence. J. Virol. 2011; 85:7622–7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kelley-Clarke B., Ballestas M.E., Komatsu T., Kaye K.M.. Kaposi's sarcoma herpesvirus C-terminal LANA concentrates at pericentromeric and peri-telomeric regions of a subset of mitotic chromosomes. Virology. 2007; 357:149–157. [DOI] [PubMed] [Google Scholar]
- 37. Sanjana N.E., Shalem O., Zhang F.. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014; 11:783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E.. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016; 44:W272–W276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Myoung J., Ganem D.. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods. 2011; 174:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun Q., Tsurimoto T., Juillard F., Li L., Li S., De Leon Vazquez E., Chen S., Kaye K.. Kaposi's sarcoma-associated herpesvirus LANA recruits the DNA polymerase clamp loader to mediate efficient replication and virus persistence. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:11816–11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xue H., Yao T., Cao M., Zhu G., Li Y., Yuan G., Chen Y., Lei M., Huang J.. Structural basis of nucleosome recognition and modification by MLL methyltransferases. Nature. 2019; 573:445–449. [DOI] [PubMed] [Google Scholar]
- 42. Zhang P., Lee H., Brunzelle J.S., Couture J.F.. The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases. Nucleic Acids Res. 2012; 40:4237–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brautigam C.A., Zhao H., Vargas C., Keller S., Schuck P.. Integration and global analysis of isothermal titration calorimetry data for studying macromolecular interactions. Nat. Protoc. 2016; 11:882–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Winter D.C. Commentary: Perspectives on diverticulitis and the art of clinical science - treat the man not the scan. Colorectal Dis. 2010; 12:186–187. [DOI] [PubMed] [Google Scholar]
- 45. Kabsch W. Xds. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Potterton L., Agirre J., Ballard C., Cowtan K., Dodson E., Evans P.R., Jenkins H.T., Keegan R., Krissinel E., Stevenson K.et al.. CCP4i2: the new graphical user interface to the CCP4 program suite. Acta Crystallogr. D Struct. Biol. 2018; 74:68–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A.et al.. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dharmarajan V., Lee J.H., Patel A., Skalnik D.G., Cosgrove M.S.. Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases. J. Biol. Chem. 2012; 287:27275–27289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A.. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Winter G., Waterman D.G., Parkhurst J.M., Brewster A.S., Gildea R.J., Gerstel M., Fuentes-Montero L., Vollmar M., Michels-Clark T., Young I.D.et al.. DIALS: implementation and evaluation of a new integration package. Acta Crystallogr. D Struct. Biol. 2018; 74:85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Evans P. Scaling and assessment of data quality. Acta Crystallogr. D. Biol. Crystallogr. 2006; 62:72–82. [DOI] [PubMed] [Google Scholar]
- 54. Evans P.R., Murshudov G.N.. How good are my data and what is the resolution?. Acta Crystallogr. D. Biol. Crystallogr. 2013; 69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vagin A., Teplyakov A.. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997; 30:1022–1025. [Google Scholar]
- 56. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jurrus E., Engel D., Star K., Monson K., Brandi J., Felberg L.E., Brookes D.H., Wilson L., Chen J., Liles K.et al.. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018; 27:112–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dolinsky T.J., Czodrowski P., Li H., Nielsen J.E., Jensen J.H., Klebe G., Baker N.A.. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007; 35:W522–W525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Patel A., Dharmarajan V., Vought V.E., Cosgrove M.S.. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J. Biol. Chem. 2009; 284:24242–24256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Patel A., Vought V.E., Dharmarajan V., Cosgrove M.S.. A novel non-SET domain multi-subunit methyltransferase required for sequential nucleosomal histone H3 methylation by the mixed lineage leukemia protein-1 (MLL1) core complex. J. Biol. Chem. 2011; 286:3359–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Odho Z., Southall S.M., Wilson J.R.. Characterization of a novel WDR5-binding site that recruits RbBP5 through a conserved motif to enhance methylation of histone H3 lysine 4 by mixed lineage leukemia protein-1. J. Biol. Chem. 2010; 285:32967–32976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rao R.C., Dou Y.. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer. 2015; 15:334–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Naik N.G., Nguyen T.H., Roberts L., Fischer L.T., Glickman K., Golas G., Papp B., Toth Z.. Epigenetic factor siRNA screen during primary KSHV infection identifies novel host restriction factors for the lytic cycle of KSHV. PLoS Pathog. 2020; 16:e1008268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gunther T., Frohlich J., Herrde C., Ohno S., Burkhardt L., Adler H., Grundhoff A.. A comparative epigenome analysis of gammaherpesviruses suggests cis-acting sequence features as critical mediators of rapid polycomb recruitment. PLoS Pathog. 2019; 15:e1007838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cao F., Townsend E.C., Karatas H., Xu J., Li L., Lee S., Liu L., Chen Y., Ouillette P., Zhu J.et al.. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol. Cell. 2014; 53:247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li Y., Han J., Zhang Y., Cao F., Liu Z., Li S., Wu J., Hu C., Wang Y., Shuai J.et al.. Structural basis for activity regulation of MLL family methyltransferases. Nature. 2016; 530:447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Patel A., Vought V.E., Dharmarajan V., Cosgrove M.S.. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J. Biol. Chem. 2008; 283:32162–32175. [DOI] [PubMed] [Google Scholar]
- 69. Couture J.F., Collazo E., Trievel R.C.. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat. Struct. Mol. Biol. 2006; 13:698–703. [DOI] [PubMed] [Google Scholar]
- 70. Schuetz A., Allali-Hassani A., Martin F., Loppnau P., Vedadi M., Bochkarev A., Plotnikov A.N., Arrowsmith C.H., Min J.. Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J. 2006; 25:4245–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ruthenburg A.J., Wang W., Graybosch D.M., Li H., Allis C.D., Patel D.J., Verdine G.L.. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 2006; 13:704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Song J.J., Kingston R.E.. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J. Biol. Chem. 2008; 283:35258–35264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Migliori V., Mapelli M., Guccione E.. On WD40 proteins: propelling our knowledge of transcriptional control?. Epigenetics. 2012; 7:815–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Grundhoff A., Ganem D.. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J. Clin. Invest. 2004; 113:124–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Komatsu T., Ballestas M.E., Barbera A.J., Kelley-Clarke B., Kaye K.M.. KSHV LANA1 binds DNA as an oligomer and residues N-terminal to the oligomerization domain are essential for DNA binding, replication, and episome persistence. Virology. 2004; 319:225–236. [DOI] [PubMed] [Google Scholar]
- 76. Hsu P.L., Shi H., Leonen C., Kang J., Chatterjee C., Zheng N.. Structural Basis of H2B ubiquitination-dependent H3K4 methylation by COMPASS. Mol. Cell. 2019; 76:712–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Barbera A.J., Chodaparambil J.V., Kelley-Clarke B., Joukov V., Walter J.C., Luger K., Kaye K.M.. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006; 311:856–861. [DOI] [PubMed] [Google Scholar]
- 78. Schwam D.R., Luciano R.L., Mahajan S.S., Wong L., Wilson A.C.. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 2000; 74:8532–8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Correia B., Cerqueira S.A., Beauchemin C., Pires de Miranda M., Li S., Ponnusamy R., Rodrigues L., Schneider T.R., Carrondo M.A., Kaye K.M.et al.. Crystal Structure of the Gamma-2 Herpesvirus LANA DNA binding domain identifies charged surface residues which impact viral latency. PLoS Pathog. 2013; 9:e1003673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ponnusamy R., Petoukhov M.V., Correia B., Custodio T.F., Juillard F., Tan M., Pires de Miranda M., Carrondo M.A., Simas J.P., Kaye K.M.et al.. KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA. Nucleic Acids Res. 2015; 43:10039–10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Domsic J.F., Chen H.S., Lu F., Marmorstein R., Lieberman P.M.. Molecular Basis for Oligomeric-DNA binding and episome maintenance by KSHV LANA. PLoS Pathog. 2013; 9:e1003672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hellert J., Weidner-Glunde M., Krausze J., Richter U., Adler H., Fedorov R., Pietrek M., Ruckert J., Ritter C., Schulz T.F.et al.. A structural basis for BRD2/4-Mediated host chromatin interaction and oligomer assembly of kaposi sarcoma-associated herpesvirus and murine gammaherpesvirus LANA proteins. PLoS Pathog. 2013; 9:e1003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Winters A.C., Bernt K.M.. MLL-rearranged leukemias—an update on science and clinical approaches. Front. Pediatr. 2017; 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wilkinson A.W., Gozani O.. Cancer epigenetics: reading the future of leukaemia. Nature. 2017; 543:186–188. [DOI] [PubMed] [Google Scholar]
- 85. Toptan T., Fonseca L., Kwun H.J., Chang Y., Moore P.S.. Complex alternative cytoplasmic protein isoforms of the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 generated through noncanonical translation initiation. J. Virol. 2013; 87:2744–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Canham M., Talbot S.J.. A naturally occurring C-terminal truncated isoform of the latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus does not associate with viral episomal DNA. J. Gen. Virol. 2004; 85:1363–1369. [DOI] [PubMed] [Google Scholar]
- 87. Baker N.A. Poisson-Boltzmann methods for biomolecular electrostatics. Methods Enzymol. 2004; 383:94–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural data have been deposited at Protein Data Bank (PDB). The accession numbers for the structures are: 7BED, 7BCY. Otherwise, the article includes all data generated/analyzed in the study.