Abstract
Motile cilia of multiciliated epithelial cells undergo synchronized beating to produce fluid flow along the luminal surface of various organs. Each motile cilium consists of an axoneme and a basal body (BB), which are linked by a “transition zone” (TZ). The axoneme exhibits a characteristic 9+2 microtubule arrangement important for ciliary motion, but how this microtubule system is generated is not yet fully understood. Here we show that calmodulin-regulated spectrin-associated protein 3 (CAMSAP3), a protein that can stabilize the minus-end of a microtubule, concentrates at multiple sites of the cilium–BB complex, including the upper region of the TZ or the axonemal basal plate (BP) where the central pair of microtubules (CP) initiates. CAMSAP3 dysfunction resulted in loss of the CP and partial distortion of the BP, as well as the failure of multicilia to undergo synchronized beating. These findings suggest that CAMSAP3 plays pivotal roles in the formation or stabilization of the CP by localizing at the basal region of the axoneme and thereby supports the coordinated motion of multicilia in airway epithelial cells.
INTRODUCTION
Multiciliated cells play a central role in generating directional flow of fluids or particles over the surface of various epithelial tissues, such as oviduct epithelium, ependymal cells lining the brain ventricles, and the airway epithelium of the respiratory tract (Brooks and Wallingford, 2014). Motile cilia on these epithelial cells beat synchronously to produce hydrodynamic forces. Failure of cilia to undergo their coordinated movement causes various diseases, including a subgroup of primary ciliary dyskinesia (PCD) (Lee, 2011; Tilley et al., 2015; Damseh et al., 2017).
The individual motile cilium of multiciliated cells consists of a 9+2 microtubule arrangement, that is, nine microtubule doublets and a central pair of single microtubules (CP), which position at the outer and central zones, respectively. This core structure of a cilium, called an axoneme, is linked to the basal body (BB), a form of centriole, from which ciliogenesis begins during development (Dawe et al., 2007a). The connection between the axoneme and the BB is mediated by a zone called the “transition zone” (TZ) (Czarnecki and Shah, 2012; Goncalves and Pelletier, 2017), and the CP sits just above the TZ, although the outer microtubules extend farther downward to join the triplet microtubules of the BB (Greenan et al., 2020). Although extensive studies have been conducted to understand how these ciliary structures form and how motile cilia move in a coordinated manner (Satir and Christensen, 2007), the molecular mechanisms underlying these questions are still largely unresolved.
CP deficiency brings about defective motion of cilia or flagella (Smith and Yang, 2004; Dawe et al., 2007b; Lechtreck et al., 2008; Nozawa et al., 2013; Loreng and Smith, 2017) and is also associated with PCD (Bautista-Harris et al., 2000; Stannard et al., 2004; Burgoyne et al., 2014). For example, removal of the putative serine-threonine kinase fused (Wilson et al., 2009; Nozawa et al., 2013) or radial spoke head proteins such as Rsph1 or Rsph9 (Kott et al., 2013; Zhu et al., 2019) results in partial or complete loss of CP, simultaneously altering the ciliary beat pattern from planar to rotational in motile cilia. CP formation is thought to occur from TZ. Although molecular constituents of the TZ have been analyzed (Czarnecki and Shah, 2012; Dean et al., 2016), how the CP grows out from the TZ in motile cilia, however, remains unknown, particularly because no template for microtubule polymerization is detected at the TZ. In flagella, the CP is anchored to the “basal plate” (BP) at the distal end of the TZ. Recent studies identified a protein named “basalin’ through the analysis of a trypanosome TZ proteome, which is localized at the BP, and its depletion resulted in disruption of the BP as well as collapse of CP (Dean et al., 2016, 2019), suggesting that this structure is important for CP nucleation.
CAMSAP (calmodulin-regulated spectrin-associated protein) and its invertebrate homologues form a protein family, each member of which binds the minus-end of noncentrosomal microtubules (Meng et al., 2008; Baines et al., 2009; Goodwin and Vale, 2010; Tanaka et al., 2012; Hendershott and Vale, 2014; Jiang et al., 2014; King et al., 2014; Richardson et al., 2014). Through this property of CAMSAP, it regulates the dynamics of the minus-ends of microtubules, simultaneously anchoring them to particular subcellular sites (Nashchekin et al., 2016; Toya et al., 2016) and contributes to a variety of cell and tissue morphogenesis (Noordstra et al., 2016; Martin et al., 2018; Takeda et al., 2018; Ko et al., 2019; Kimura et al., 2021; Mitsuhata et al., 2021), including neuritegenesis (Chuang et al., 2014; Yau et al., 2014; Pongrakhananon et al., 2018; Feng et al., 2019; Wang et al., 2019; Chen et al., 2020).
The binding of CAMSAP to microtubules is mediated by its C-terminal CKK domain (Meng et al., 2008; Baines et al., 2009; Hendershott and Vale, 2014; Jiang et al., 2014), and its deletion causes the dysfunction of CAMSAP. For example, CAMSAP3, a member of this molecular family, is concentrated at the apical cortex in intestinal epithelial cells of mice through the action of the CC1 domain, a coiled-coiled region of the protein, tethering noncentrosomal microtubules to these sites, and this results in the assembly of microtubules whose plus ends point basally (Toya et al., 2016). This characteristic microtubule assembly is disrupted in mice bearing a mutant gene encoding a truncated Camsap3 (Camsap3dc/dc mice), in which the CKK domain is deleted, because mRNA transcribed from the mutated gene does not cover the exons that encode the CKK domain (Toya et al., 2016). Camsap3dc/dc mice show other defects, such as cyst formation in proximal renal tubules (Mitsuhata et al., 2021) and narrowing of the lateral ventricles in the brain (Kimura et al., 2021), although the overall brain architecture looks normal. Camsap3dc/dc mice and Camsap3 null mutant mice show similar abnormalities (Pongrakhananon et al., 2018; Mitsuhata et al., 2021), suggesting that CAMSAP3 lacking a CKK domain is fully nonfunctional.
A recent study showed that CAMSAP3 is expressed in nasal multiciliated cells, and its absence causes defects in ciliary motion and polarity (Robinson et al., 2020). Similar observations were also reported using oviduct cells (Usami et al., 2021). However, how CAMSAP3 sustains the structure and function of multicilia remains to be investigated. In the present study, we used airway epithelial cells of adult mice to elucidate the role of CAMSAP3 in coordinated movement of multicilia, finding that CAMSAP3 dysfunction due to CKK domain loss results in a partial disruption of axonemal structures at its proximal end, as well as loss of the CP. Superresolution microscopy revealed that a fraction of CAMSAP3 accumulates around the sites where the CP is supposed to be initiated. Based on these observations, we discuss potential mechanisms by which CAMSAP3 supports CP formation and coordinated motion of multicilia.
RESULTS
Defective ciliary motion in Camsap3-mutated airway cells
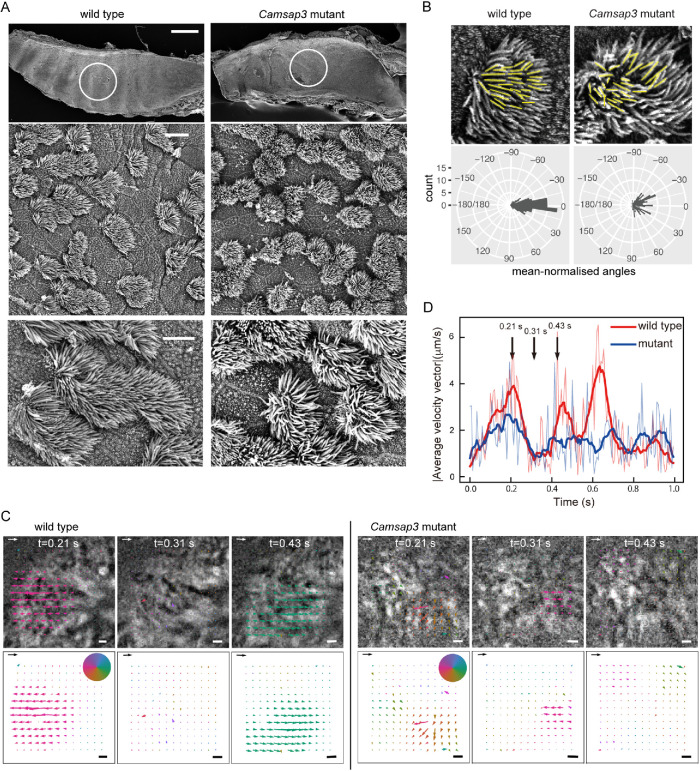
Camsap3dc/dc (Camsap3 mutant) mice frequently produced clicking or chattering sounds (Supplemental Audio S1), prompting us to examine whether the respiratory system has any problem. We observed the luminal surface of trachea collected from postnatal mice using scanning electron-microscopy, finding that multiciliated epithelial cells are normally present in the trachea of the mutant mice, the distribution pattern of which was similar to that observed in wild-type trachea (Figure 1A). In higher magnification views, however, we noted that the alignment of multicilia along the longitudinal axis of trachea, which is generally well ordered in wild-type airway cells, was disorganized to some extent in Camsap3-mutated cells (Figure 1B).
FIGURE 1:
Morphology and movement of motile cilia in airway epithelial cells. (A) Scanning electron microscopical observation of the luminal surface of the trachea. The trachea was isolated from P21 wild-type or Camsap3 mutant male mice. The top row shows a whole-mount trachea, in which the luminal surface is exposed. Part of the encircled region was enlarged in the middle and bottom rows. A representative image of three wild-type and five mutant samples is shown. The oral side is at the right. Scale bars: 500 μm, top; 10 μm, middle; and 5 μm, bottom. (B) Quantitative analysis of ciliary orientation. Cilia were traced with yellow lines for angle measurement, and polar plots for the distribution of ciliary angles were obtained. The ciliary angles were normalized by the average angle in each cell. Angles of 30 cilia in five cells were measured and aggregated for the polar plots (p < 2.2 × 10–16). (C) Quantitative analysis of the collective motion of multicilia, which has been recorded in Videos S1 and S2, for a wild-type and Camsap3-mutated cell, respectively. These videos were used for PIV analysis (see Videos S3 and S4). Three snapshots, chosen from each video, are overlaid with the flow vector field calculated by PIV analysis, and only the vectors, which represent the velocity vectors, are shown at the bottom row. The color of the vectors indicates the angle of the velocity vector, and the colormap is shown as a pie chart. The scale arrow and scale bar on each image are 20 μm/s and 1 μm, respectively. (D) Time-evolution of the magnitude of average velocity vector of ciliary motion plotted for wild-type (red curves) and mutant (blue curves) cells, respectively. The thin curves are the raw data, while the bold curves are the data smoothed by a moving average. The time points shown in B are indicated by black vertical arrows.
Audio S1.
Respiratory sound of a P50 Camsap3 mutant mouse.
To examine whether the motile cilia of the mutant cells have any defects in motility, we isolated trachea and acquired time-lapse images of the cilia on airway epithelial cells. Wild-type cells exhibited a characteristic synchronized beating of multicilia (Supplemental Video S1), whereas those of mutant cells tended to lose such orderly motion, instead displaying aberrant movements individually (Supplemental Video S2). We then analyzed the videos to quantify the pattern of ciliary motion by calculating the flow field of a ciliary carpet using particle image velocimetry (PIV). In PIV analysis, we subdivide each image into smaller areas and then quantify how fast and in which direction each area flows by calculating the cross-correlation between the subdivided areas in subsequent time frames. As a result, velocity is assigned to individual areas, and a flow field is obtained. The flow field for a wild-type cell showed that multiple cilia on the cell oscillated together collectively (Figure 1C and Supplemental Video S3). In contrast, those in a mutant cell were subdivided into several smaller clusters, each of which showed a collective flow in a unique direction (Figure 1C and Supplemental Video S4). Using these data, we quantified the degree of synchrony of ciliary motion by calculating the time-evolution of the average speed of the motion, which is the magnitude of the average velocity over the field of view. Since the individual cilia drive oscillatory motion in the flow field of PIV, if all ciliary motions are synchronized, the magnitude of the average velocity vector is expected to show oscillation. In contrast, if the ciliary motion is all spatially random, the magnitude of the average velocity vector would not oscillate in time. Our analysis showed that the magnitude of the average velocity vector oscillated less prominently in the mutant than in the wild-type cells, indicating that the degree of synchrony of ciliary motion was lower in the mutant cells (Figure 1D).
Movie S1.
Beating multicilia in a wild-type airway epithelial cell. The time-lapse images were acquired at 300 fps and displayed at 31 fps.
Movie S2.
Beating multicilia in a Camsap3-mutated airway epithelial cell. The time-lapse images were acquired at 300 fps and displayed at 31 fps.
Movie S3.
Beating multicilia in a wild-type airway epithelial cell, which is overlaid with a flow vector field calculated by PIV analysis. The flow vector field was also shown separately at the right. The scale arrow and scale bar are 20 μm/s and 1 m, respectively. Part of Video S1 was used for this analysis.
Movie S4.
Beating multicilia in a Camsap3-mutated airway epithelial cell, which is overlaid with a flow vector field calculated by PIV analysis. The flow vector field was also shown separately at the right. The scale arrow and scale bar are 20 μm/s and 1 μm, respectively. Part of Video S2 was used for this analysis.
Loss of the central microtubule pair in the axoneme of Camsap3-mutated motile cilia
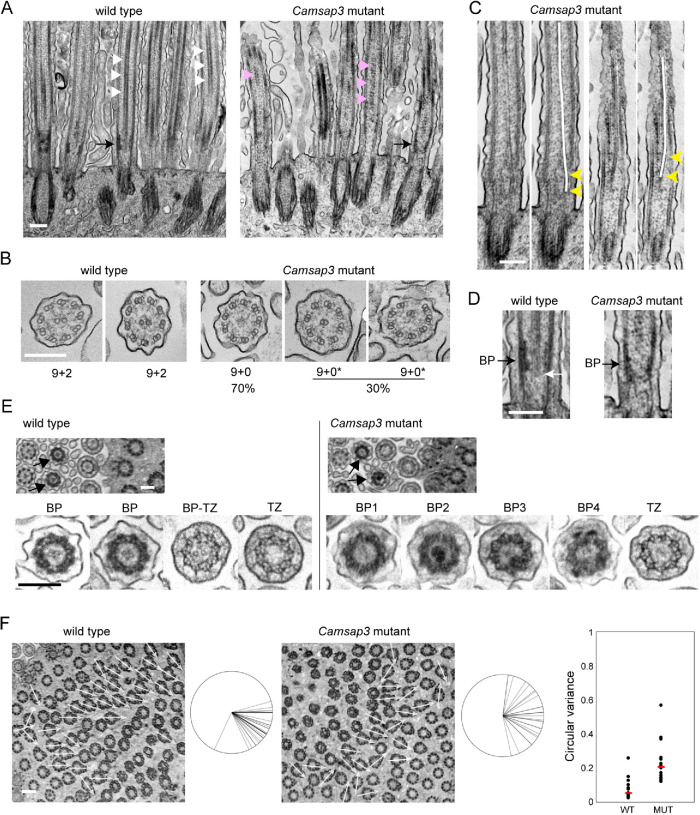
To investigate the structural basis underlying the uncoordinated beating of cilia in Camsap3-mutated cells, we used transmission electron microscopy (TEM). Longitudinal sections along the cilia revealed that the overall morphology of cilia and basal bodies looked normal in the mutants. However, the centrally located electron-dense signals that represent the CP were not clearly detectable in the mutant cells, as they were often discontinuous (Figure 2A). Cross-sections of cilia revealed further details of such abnormalities. While the cilia in wild-type cells exhibit a typical 9 + 2 arrangement of microtubules, any cilium of mutant cells did not have the CP. In these cells, nine outer doublet microtubules were still present in 70% of cilia at the normal positions, but in the rest of them, one of the doublet microtubules was displaced to an inner zone of the axoneme (Figure 2B).
FIGURE 2:
TEM analysis of cilia and basal bodies. (A) Longitudinal section of a multiciliated cell, which yields a longitudinal view of axonemes and basal bodies. White triangles indicate examples of the CP in a wild-type cell; magenta triangles point to intermittent structures at the central position of axoneme in a Camsap3-mutated cell. Black arrows point to the “basal plate” and a corresponding position in the Camsap3 mutant sample. A representative image of more than 50 sections is shown for each genotype. (B) Transverse section of cilia. In the mutant samples, 218 cilia from two independent sections were used for analysis. The mutant axonemes did not contain a CP; instead, some of them (marked as 9+0*) had a doublet microtubule that was mislocalized to central or semicentral positions. (C) Longitudinal section of cilia in Camsap3-mutated cells. A centrally located microtubule structure tilts toward the peripheral zone at basal portions of axoneme, as indicated by yellow arrowheads. Two examples are shown, each of which is duplicated to trace the central microtubule with a white line over the image. (D) Part of the image in A is enlarged. The white arrow points to the proximal end of the CP. BP, BP. (E) Slightly oblique, transverse sections of a multiciliated cell showing cross-sectional views of axoneme and BB at various longitudinal levels, in which arrows point to BPs that are characterized by a dense peripheral ring. Typical examples of ciliary sections at the level of BP and TZ are enlarged at the bottom. The image labeled BP-TZ likely represents a section at the boundary between BP and TZ. In the mutant sections, all of the BPs show some abnormality, including loss or overcondensation of the central structure, and partial deformation of the peripheral ring. Typical examples are shown. More than 20 sections, each of which contains more than 50 cilia, were examined for each genotype. (F) Slightly oblique, transverse section of a multiciliated cell, focusing on the basal foot of basal bodies. The direction of individual basal feet is shown with white arrows on the image, and these are aggregated at the right of each image. The range of orientation of basal feet on the cell varied from 26° to 108° in the wild type, with one exceptional case, and 13° to 172° in the mutant sample. We counted 33 and 26 basal bodies for the wild-type and mutant samples, respectively. The graph shows the variation in basal foot polarity in 13 wild-type or 14 mutant cells, which was quantified using circular variance (CV) that was defined previously (Shi et al., 2014), where cells with lower CV represent uniform BB orientation. Red bar, median. WT, wild type; MUT, Camsap3 mutant. Age of mice used, P126. Thickness of sections, 50 to 70 nm. Scale bars, 200 nm in A–E; 400 nm in F.
To seek the origin of these inner doublet microtubules, we reexamined the longitudinal sections and found that, when centrally located microtubules were detectable, they were tilted along the longitudinal axis, one end of which gets closer to peripheral regions of the axoneme around its basal positions (Figure 2C). These observations suggest the possibility that an outer microtubule doublet was abnormally internalized at upper regions of the axoneme, as observed in certain PCD cases (Burgoyne et al., 2014).
We then focused on the proximal zones of axonemes, where the CP is thought to be nucleated. In longitudinal sections, the CP ended at a zone characterized by the accumulation of electron-dense materials at around the outer microtubule doublets (Figure 2D left, black arrow). As this zone positionally corresponds to the BP or “axonemal plate” that is observed in cilia of unicellular organisms and thought to be the site for CP nucleation (Tucker, 1971; Gilula and Satir, 1972; Dute and Kung, 1978; Hoog et al., 2014; Dean et al., 2016), we also call it BP, although this structure appears to have a cylindrical shape in the case of airway cells. Closer observations showed that the CP began at around the basal end of the BP, but was often sticking out of this BP structure by approximately 20 nm at maximum (n = 25; Figure 2D, white arrow). Axonemes in Camsap3-mutated cells appeared to have a similar zone, except that microtubules were not always detectable in the BP of mutant samples, as seen in upper axonemal zones (Figure 2D right, black arrow).
To confirm these observations, we used transverse sections of an airway cell which are slightly tilted, as these allowed us to observe sequential changes in the inner structures of cilium and BB along the longitudinal axis within a single focal plane. In wild-type samples, we detected a group of ciliary sections, each of which exhibits an electron-dense outer ring with a centrally located CP (Figure 2E, left), just as seen in the BP defined above, leading us to judge that this is a transverse view of the BP. In the cilia of Camsap3-mutated cells, this structure was deformed showing various abnormalities, such as missing CP (Figure 2E, BP1), overly condensed central materials (Figure 2E, BP2), and partial distortion of the outer ring (Figure 2E, BP3). Some of these BPs retained microtubules in the center (Figure 2E, BP4). Thus, not only the CP but also the entire BP architecture appeared abnormal in the mutant.
These sections also allowed us to examine the TZ, which is characterized by the presence of Y-shaped linkers. We did not find any difference in TZ structures between wild-type and mutant cilia, except that a small number of wild-type TZ contained CP (Figure 2E, BP-TZ); this sample probably represents a section at the boundary region of BP and TZ. The structure of the BBs also looked normal in Camsap3-mutated cells, although they tended to lose their rotational polarity in mutant cells, as assessed by measuring the direction of the “basal foot” which normally faces the oral side in wild-type cells (Figure 2F).
Concentration of CAMSAP3 around the proximal ends of cilia
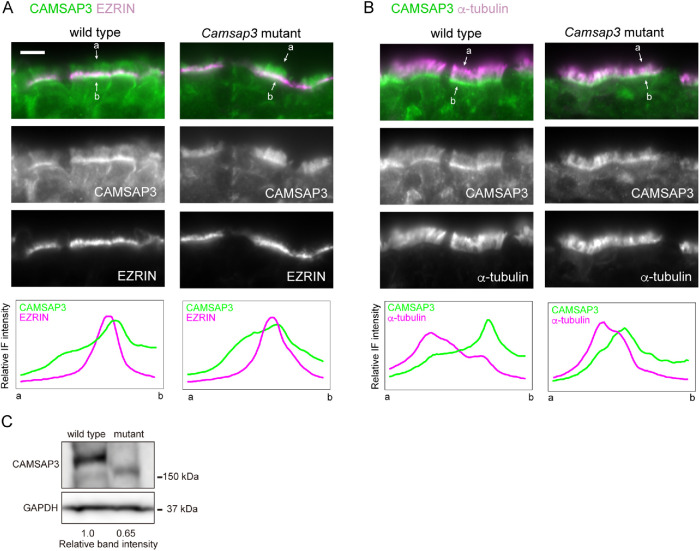
To begin investigating how CAMSAP3 contributes to microtubule assembly in axonemes, we observed its distribution in airway epithelial cells of adult trachea by immunofluorescence (IF) microscopy, using samples fixed with paraformaldehyde (PFA). The results showed that CAMSAP3 was concentrated around the apical plasma membranes (Figure 3A), as assessed by coimmunostaining for EZRIN, which is known to associate with the apical membranes or microvilli (Berryman et al., 1993), as observed in other epithelial cells (Toya et al., 2016; Robinson et al., 2020; Kimura et al., 2021; Usami et al., 2021). This position of CAMSAP3 corresponded to the proximal edge of strong α-tubulin IF signals that should represent ciliary microtubules (Figure 3B). Weak CAMSAP3 IF stains were also detectable along cilia as well as in the cytoplasm. We then examined airway epithelia of Camsap3 mutant mice and found that the mutated CAMSAP3 molecules (CKK domain-truncated CAMSAP3) expressed in the mice also accumulated at the apical zones, overlapping with EZRIN (Figure 3A) or ciliary microtubules at their proximal ends (Figure 3B). Curiously, however, the mutated molecule tended to diffuse into upper portions of cilia, even though its expression level was lower than that of the wild-type one (Figure 3C), suggesting that the truncated CAMSAP3 penetrates into the ciliary body more readily than the wild-type CAMSAP3.
FIGURE 3:
Immunostaining localization of CAMSAP3 in tracheal epithelium. (A, B) Trachea derived from P176 mice were fixed with 2% PFA and processed for coimmunostaining for CAMSAP3 and EZRIN (A) or α-tubulin (B). Images were recorded by conventional IF microscopy. IF signals were scanned along the line (not shown) drawn between the two arrows, across the cilia–BB complex from its apical (a) to basal (b) side. A typical image of three to five sections is shown for each set of immunostaining. Scale bars, 10 μm. (C) Western blots for CAMSAP3 in the trachea isolated from P172 wild-type or Camsap3dc/dc mice.
Localization of CAMSAP3 at three distinct sites
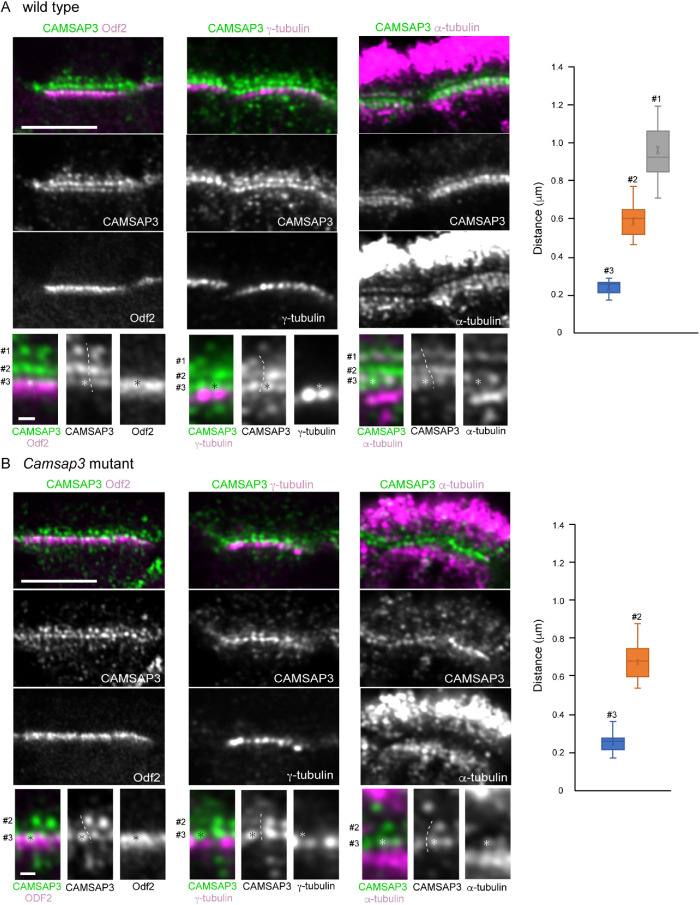
To gain more detailed information on CAMSAP3 localization in the cilia and BBs, we coimmunostained CAMSAP3 and components of the cilium–BB complex, analyzing their IF images by Airyscan confocal laser-scanning microscopy, which can yield around 140 nm lateral resolution. For this experiment, we used methanol for fixation of tissues, since some of the proteins to be examined did not give reproducible IF signals in PFA-fixed specimens. The analysis first revealed that CAMSAP3 IF signals, which are condensed at the apical zone of a cell, were split into at least three rows of CAMSAP3 puncta designated #1, #2, and #3 (Figure 4A). The relative IF intensity of the puncta in one row to another row varied from sample to sample; an example, alternative CAMSAP3 patterns are shown in Supplemental Figure S1. The puncta at the #1 row tended to be variable in morphology, contrasted with a rounded appearance in other rows. Additional puncta were also detectable outside the three rows, but their presence or location was not fixed. Individual CAMSAP3 puncta comprising the three rows were also aligned along the apicobasal axis of the cell, likely due to their association with a single cilium–BB complex at different levels of the apicobasal axis. The distances (mean ± SD) between #1 and #2 and between #2 and #3 along the longitudinal axis were 476 ± 126 nm (n = 30) and 364 ± 47 nm (n = 34), respectively.
FIGURE 4:
Airyscan microscopic analysis of CAMSAP3 distribution in wild-type and Camsap3-mutated multiciliated cells. Longitudinal section of multiciliated cells in wild-type (A) or Camsap3-mutated (B) trachea, which were coimmunostained for the molecules indicated and recorded by Airyscan microscopy. Part of each image was enlarged at the bottom, in which the numbers indicate three different sites of CAMSAP3 puncta. Asterisks mark an example of CAMSAP3 puncta (or their positions) at the #3 site in each image. Dotted lines indicate a putative boundary between the two or three sets of CAMSAP3 puncta aligned along the apicobasal axis. A typical image was selected from 8 to 10 sections of tissues, each of which contain several cells that can be analyzed, for each set of immunostaining. The samples were fixed with methanol. The graphs show the distance of CAMSAP3 puncta at different rows from γ-tubulin puncta. P91, P86,s and P176 mice were used for immunostaining for Odf2, γ-tubulin, and α-tubulin, respectively. Scale bars, 5 and 0.5 μm in the original and enlarged images, respectively.
To determine the structural relationship between these CAMSAP3 puncta and the BB, we coimmunostained for CAMSAP3 and Odf2, which is known to localize at the transition fibers and basal feet (Kunimoto et al., 2012). Odf2 overlapped with the #3 CAMSAP3 puncta at the upper portion of the former, further extending down to CAMSAP3-negative zones (Figure 4A, leftmost). γ-Tubulin, which has been reported to accumulate at the basal foot as well as at additional lower regions of BB in motile cilia (Hagiwara et al., 2000; Clare et al., 2014), was detected below the #3 puncta, occasionally overlapping with faint CAMSAP3 IF signals located underneath the #3 row (Figure 4A, middle). The distances (mean ± SD) between γ-tubulin and the #1, #2 or #3 CAMSAP3 were estimated at 962 ± 130 nm (n = 25), 592 ± 89 nm (n = 18) and 240 ± 37 nm (n = 19), respectively (Figure 4A, graph).
We then detected microtubules by α-tubulin immunostaining (Figure 4A, rightmost). Cytoplasmic microtubules formed meshwork below the #3 CAMSAP3 puncta, and some of them entered into the #3 zone, yielding their overlapping view. On the other hand, ciliary microtubules, which should have a structural continuity to the BB, were not clearly detectable at their proximal regions in our methanol-fixed samples for unknown reasons. Nevertheless, CAMSAP3 puncta at the #1 site exhibited some overlapping with microtubule clusters.
We next analyzed the distribution of the CKK domain-truncated CAMSAP3 by Airyscan microscopy in Camsap3-mutated cells (Figure 4B). As seen for wild-type CAMSAP3, the mutant molecules formed multiple rows. Coimmunostaining for CAMSAP3 and Odf2, γ-tubulin, or α-tubulin showed that their spatial relationships were similar to those found in wild-type cells. However, we could not convincingly detect the #1 row, although CAMSAP3 IF signals were randomly present at the ciliary zone (Figure 4B). We also noted that about half of CAMSAP3 puncta in the #2 row were missing. These suggest that, although the mutated CAMSAP3 still associates with the cilium-BB system at the #3 site, it does not do so at the #1 site, and its binding to the #2 site also became unstable. The distance (mean ± SD) between the #2 and #3 puncta was 335 ± 46 nm (n = 23), and those between γ-tubulin and the #2 or #3 puncta were 674 ± 96 (n = 17) and 250 ± 49 nm (n = 17), respectively (Figure 4B graph).
Localization of CAMSAP3 puncta at the TZ
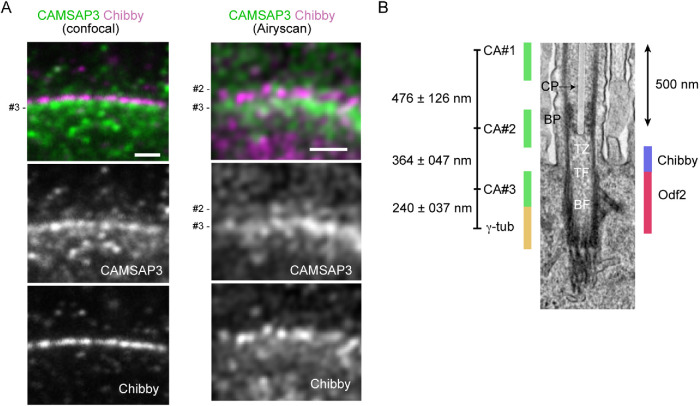
For more precise localization of CAMSAP3 puncta, we coimmunostained wild-type ciliated cells for CAMSAP3 and Chibby, a protein that localizes at a proximal region of TZ, playing a role in ciliogenesis (Enjolras et al., 2012; Burke et al., 2014). Confocal microscopy of longitudinal sections showed that Chibby localizes above the #3 CAMSAP3 puncta (Figure 5A). We then used Airyscan microscopy, confirming that major Chibby puncta are located above the #3 puncta. To relate the distribution of the #2 and #3 CAMSAP3 puncta with the actual structure of the cilia–BB complex, we overlaid their positions on its TEM ultrastructural image, referring to the published location of γ-tubulin (Clare et al., 2014), Odf2 (Kunimoto et al., 2012), and Chibby (Burke et al., 2014) in motile cilia, which were determined at the ultrastructural levels (Figure 5B). This comparison of IF images with the ultrastructure suggests that the #2 CAMSAP3 puncta are localized at a distal portion of TZ or at BP, while the #3 CAMSAP3 puncta, are localized at an upper region of the BB.
FIGURE 5:
CAMASP3 localization in the TZ. (A) Longitudinal section of a wild-type multiciliated cell, coimmunostained for CAMSAP3 and Chibby, and recorded by confocal or Airyscan microscopy. In these samples, the relative IF intensity of #3 CAMSAP3 puncta is high. Trachea were collected from P180 mice and fixed with methanol. Scale bar, 1 μm. (B) A putative map of CAMSAP3 puncta in relation to the ultrastructure of the axoneme, TZ, and BB. The relative position of each CAMSAP3 punctum was determined by measuring the distances between the puncta, and then the position of the entire CAMSAP3 puncta relative to the ultrastructure was adjusted by placing the #3 punctum at the level of the upper half of Odf2 distribution, as observed in Figure 4. The positions of Chibby, Odf2 and γ-tubulin were estimated referring to previous publications, although γ-tubulin localization needs further confirmation. The scale of the ultrastructure image is shown by a 500 nm bar. The CP is overlaid with a semitransparent white color. CP, the central pair of microtubules; BP, basal plate; TZ, transition zone; TF, transition fiber; BF, basal foot; γ-tub, γ-tubulin; CA, CAMSAP3.
Finally, we viewed the overall distribution pattern of basal bodies and microtubules from the top of ciliated cells in which GFP-tagged Centrin 2, a BB protein, is genetically expressed (Higginbotham et al., 2004). Basal bodies exhibited typical horizontal arrays in wild-type cells, and a similar pattern was maintained in Camsap3-mutated cells (Supplemental Figure S2). Microtubules form a horizontal network at the inter-BB spaces (Kunimoto et al., 2012; Tateishi et al., 2013; Tateishi et al., 2017), and we did not find any particular change of this network in Camsap3-mutated cells (Supplemental Figure S2 and Supplemental Videos S5 and S6). Microtubules located below the basal bodies also formed networks, and their pattern was indistinguishable between wild-type and Camsap3-mutated cells. Thus, it seems that Camsap3 mutation did not affect the overall BB assembly, nor did the horizontal networks of microtubules associated with the BBs.
Movie S5.
Animation of sequential optical sections, each of which is 0.2 m thick, of a wild-type multi-ciliated epithelial cell, in which GFP-Centrin (green) and -tubulin (magenta) are visualized by fluorescence signals. The animation view begins around the level where the array of GFP-Centrin is detectable, then shifts toward a more basal view of the cell at a speed of 2 fps. See also Figure 6A.
Movie S6.
Animation of sequential optical sections, each of which is 0.2 m thick, of a Camsap3-mutated multi-ciliated epithelial cell, in which GFP-Centrin (green) and -tubulin (magenta) are visualized by fluorescence signals. The animation view begins at the level where the array of GFP-Centrin is detectable, then shifts toward a more basal view of the cell at a speed of 2 fps. See also Figure 6B.
DISCUSSION
The present study demonstrated that CAMSAP3 concentrates at multiple sites of the cilium–BB complex in tracheal motile cilia in mice, and dysfunction of this molecule results in unsynchronized beating of multicilia, as well as the loss of CP and other structural defects in this complex. Comparison of CAMSAP3 distribution with the ultrastructure of a cilium suggested that the #2 site corresponds to BP or an upper region of TZ, and the #1 and #3 sites are located more distal and proximal than the BP, respectively. A preceding study using nasal epithelial cells also reported that a Camsap3 hypomorphic mutation caused similar functional and structural defects in the multicilia (Robinson et al., 2020). This study, however, detected CAMSAP3 as a single dot located between the axoneme and the BB in P30 mice, contrasted with our observation that the CAMSAP3-bearing region is subdivided into multiple sites. Two possibilities can be considered to explain the difference between the two observations. First, CAMSAP3 may interact with ciliary components in different ways between the nasal and the tracheal multiciliated cells, and second, different sample preparations or microscopic methods can lead to different results. Actually, we noted a variation in the CAMSAP3 distribution pattern. Whichever the case is, the CAMSAP3 dot in P30 nasal cilia may correspond to the #2 or #3 puncta in tracheal cilia, considering their distance from γ-tubulin of the BB. In nasal motile cilia, an additional dot overlapping with γ-tubulin was also observed in P3 mice (Robinson et al., 2020), but we did not examine tracheal motile cilia in such young mice.
The information that CAMSAP3 localizes around at the BP enabled us to discuss its potential role in CP formation on the basis of our knowledge about how CAMSAP3 controls microtubules. That is, it stabilizes the minus-end of a microtubule and simultaneously serves to tether the microtubule filament to particular subcellular sites (Nashchekin et al., 2016; Toya et al., 2016). Here we propose that CAMSAP3 may function to stabilize the CP by binding to the minus-end of microtubules at around the BP, as the minus-ends of axonemal microtubules are oriented basally (Euteneuer and Mcintosh, 1981). Removal of this mechanism would have led to a collapse of the CP. Notably, the CKK-truncated CAMSAP3 expressed in Camsap3dc/dc mice, which are unable to bind the minus-ends of microtubules, were still detected at the #2 site but not always. It is possible that CAMSAP3 can stay at this site without binding microtubules, but its association with a putative binding partner became unstable under this situation, as observed in previous studies (Toya et al., 2016), and consistent with the idea that CAMSAP3 does interact with microtubules at this site. To test these ideas, however, it will be prerequisite to determine whether CAMSAP3 is located inside or outside the axoneme for its interaction with microtubules.
Since BP architecture was also perturbed in Camsap3-mutated airway cells, the presence of CAMSAP3 or CP may also play a role in maintaining this structure. On the other hand, detection of CAMSAP3 puncta at regions more distal than BP (at the #1 site) suggests that CAMSAP3 may interact with microtubules at the axoneme, because its puncta disappeared in Camsap3-mutated cilia. This could be another site for CAMSAP3 to support the CP, but its potential function remains to be further investigated. Mutated CAMSAP3 tended to diffuse into the upper regions of a cilium. This might have occurred due to the release of CAMSAP3 from the #1 or #2 sites.
Loss of CP may explain why the Camsap3-mutated motile cilia showed unsynchronized beating, since CP deficiency due to various mechanisms is known to coincide with disorganized ciliary movement (Wilson et al., 2009; Kott et al., 2013; Nozawa et al., 2013; Zhu et al., 2019). Defective CP may also have been involved in the perturbation of BB polarity observed in the absence of functional CAMSAP3. For example, the aberrant beating of multicilia in Camsap3-mutated cells cannot produce sufficient hydrodynamic forces that are required for the polarized alignment of them (Guirao et al., 2010). On the other hand, we cannot exclude the possibility that CAMSAP3 associated with the cilia–BB complex could have some direct role in determining BB polarity. Defects in this putative mechanism might have perturbed BB polarity and in turn induced uncoordinated beating of cilia.
What is the function of CAMSAP3 at the #3 site that corresponds to the distal part of the BB? It overlaps with the upper component of Odf2 that is known to associate with transition fibers as well as basal feet, removal of which results in loss of BB but without affecting CP (Kunimoto et al., 2012). Despite their overlapping, CAMSAP3 mutants did not mimic what is observed in Odf2 mutants, suggesting that they are not functionally linked to one another. The loss of CAMSAP3 function did not affect the horizontal network of microtubules interspaced between BBs, suggesting that their assembly of microtubes is regulated by factors other than CAMSAP3, such as γ-tubulin, an authentic microtubule nucleator. Thus, the role of CAMSAP3 at the BB remains mysterious. It is noted that the CKK-deficient CAMSAP3 puncta were normally detectable at the #3 site, implying that CAMSAP3 proteins cluster here independently of microtubules.
Curiously, the role of CAMSAP3 in multiciliated cells appears not thoroughly conserved among different organs, contrasted with many other ciliary components whose loss induces PCD. In multiciliated ependymal cells of the brain, CAMSAP3 does not particularly associate with the cilium–BB complex, and their ciliary beating occurs normally in Camsap3-mutant mice (Kimura et al., 2021). In the ependymal cells, CAMSAP3 localizes under the apical plasma membranes and its major role is to regulate the organization of cortical microtubules that are required for establishing a microtubule-dependent signaling system. These observations suggest that CAMSAP3 contributes to multiciliary organization in a tissue-specific manner, although the possibility that other CAMSAP subtypes or unidentified similar proteins have taken over the role of CAMSAP3 in ependymal cells is not excluded. It should also be noted that the CAMSAP family or CKK domain-containing proteins have been detected only in the metazoan (Baines et al., 2009), which implies that their role in the ciliary system is limited to particular animal species. Future studies will be necessary to reveal how CAMSAP3 becomes associated with the cilium–BB complex and supports its structure and function in particular tissues and species.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Mice
A Camsap3 mutant mouse line, Camsap3dc/dc, was described previously (Toya et al., 2016). The transgenic mouse line expressing GFP-Centrin2 (EGFP-CETN2) was a gift from Holden Higginbotham, Brigham Young University (Higginbotham et al., 2004). Female mice of this transgenic line were crossed with Camsap3dc/+ males to generate those expressing GFP-Centrin2 under Camsap3 mutation. ICR mice (Slc; ICR, Japan SLC) were used for immunostaining analysis of Chibby. The age of mice used for the study was recorded for each experiment and described in the figure legends. The experiments using mice were performed in accordance with the protocol(s) approved by the Institutional Animal Care and Use Committee of the RIKEN Center for Biosystems Dynamics Research (RIKEN Kobe Branch), or the Guidelines of Animal Experimentation of National Institutes for Natural Sciences.
Scanning electron microscopy
Mice were anesthetized with isoflurane and dissected to reveal trachea and cut diaphragm. The tracheas were prefixed with a fixative solution (2% PFA and 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4) for 5 min, then removed from mice. The trachea was cut into small pieces, and the samples were immersed in the same fixative solution at room temperature for 2 h. The samples were washed with 0.1 M cacodylate buffer and postfixed in 1% OsO4 in 0.1 M cacodylate buffer on ice for 2 h. Then the samples were washed with distilled water and stained overnight with 0.5% aqueous uranyl acetate at room temperature. The stained samples were dehydrated with ethanol, and the samples were rinsed with distilled water and further stained with 0.5% uranyl acetate solution overnight at room temperature. The samples were rinsed with distilled water and dehydrated through a graded series of ethanol, transferred to isopentyl acetate, and critical point-dried using liquid CO2. The dried specimens were evaporation-coated with Osmium and examined in a JSM-5600LV scanning electron microscope at an accelerating voltage of 10 kV.
TEM
Tracheas were fixed with 2% PFA and 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) at 4°C overnight. After washing five times with 0.1 M cacodylate buffer, the specimens were further fixed with 1% OsO4 in 0.1M cacodylate buffer at 4°C for 1 h and then washed five times with Milli-Q water. The specimens were incubated in 1% uranyl acetate overnight and washed five times with Milli-Q water. Specimens were dehydrated with an ethanol series (20, 50, 70, 90, and 99.5% for 5 min each and twice in 100% for 10 min) and acetone, then followed by infiltration with Epon 812 (TAAB). Polymerization was performed at 60°C for 72 h. Samples were cut with an ultramicrotome (ULTRA CUT UCT, Leica) equipped with a diamond knife (Diatome), doubly stained with uranyl acetate and lead citrate, and examined in a transmission electron microscope (JEM-1400 Plus; JEOL) operated at 100 kV.
IF staining and microscopy
Isolated trachea tissue was fixed in a PEM buffer (PEM: 100 mM PIPES, 5 mM EGTA, and 5 mM MgCl2, adjusted to pH 6.9 with 2 M KOH) containing 2% PFA and 0.1 M sorbitol for 1 h at room temperature, except that the tissue was treated with 4% PFA at 37°C for 1 h and then at 4°C for 24 h for microtubule fixation. Alternatively, trachea tissue was fixed with methanol for 20 to 45 min at –20°C, followed by treatment with acetone for 1 min at –20°C. After fixation, the tissue was rinsed three times with PEM for 10 min. Then, cryoprotection was carried out by incubation with 15% sucrose in PEM overnight at 4°C, followed by incubation with 20% sucrose in PEM for 6 h and 30% sucrose in PEM overnight at 4°C. The tissue was frozen in OCT compound in an embedding mold (Polysciences, Inc.) in liquid nitrogen and stored at –80°C. Frozen samples were sliced at 7 mm thickness with a cryostat (HM500M, MICROM).
After washing twice with phosphate-buffered saline (PBS), the sections were blocked with Blocking One (Nacalai Tesque) for 1 h at room temperature, followed by incubation with primary antibodies in Blocking One (Nacalai Tesque) overnight at 4°C. Sections were washed two times with PBS and incubated for 1 h at RT with corresponding secondary antibodies. Stained sections were washed two times with PBS and mounted with either Fluorsave (Chemicon) or 2,2′‐thiodiethanol (Staudt et al., 2007). For samples used for Airyscan microscopy, antigen retrieval was performed by incubating sections in HistoVT One (Nacalai Tesque) for 3 min at 95°C, prior to the incubation with primary antibodies.
For conventional immunofluorescence microscopy, samples were analyzed by Zeiss Axiopan 2 through Plan-APOCHROMAT 63×/1.4 Oil Dic or Plan-NEOFLUAR 40×/1.3 Oil Dic objectives. For confocal microscopy, a NIKON A1 microscope with Nikon Plan Apo Vc 100×/1.40 oil OFN25 DIC N2 objective lens was used. For superresolution microscopy, stacks of images were taken along the z-axis at optimal intervals using Airyscan (LSM880, Zeiss) with 63×/1.40 NA and 100×/1.46 NA objective lenses. Acquired images were processed using Zen (Zeiss), Adobe Photoshop (Adobe), or ImageJ (Wayne Rasband, National Institutes of Health).
Measurement of distances between CAMSAP3 puncta
To determine the distance between two IF puncta, we measured from the center of one punctum to that of the other punctum using ImageJ. Three to 10 sections were used for each measurement.
Quantification of ciliary orientation in multiciliated epithelial cells
To quantify the alignment of multicilia along the longitudinal axis of trachea, the angles of cilia in SEM images were measured using ImageJ. Briefly, a line was drawn along individual cilia and then the angle of the line was measured. Measured angles of cilia were normalized by the average angle of all the cilia within the cell in order to eliminate a possible difference in the orientation of multiciliated epithelial cells. Polar plots for the distribution of ciliary angles were generated by ggplot2 in R. For the statistical test, we applied the F-test function (var.test) in R.
Live imaging of ciliary motion and quantification of data
Live images of cilia were acquired as described previously (Shinohara et al., 2015). Briefly, tracheas were collected into HEPES-buffered DMEM (pH 7.2) supplemented with 10% fetal bovine serum. Motility of cilia was examined at 25°C with a high-speed CMOS camera (HAS-U2, DITECT) that was connected to a microscope (BX53, Olympus) with a 40× objective lens (UPLFLN40X, Olympus) at a frame rate of 300 frames/s for tracheal cilia. To emphasize the movement of cilia, time-series images were adjusted to gain proper contrast and brightness after subtracting the minimum value through the images from each frame using ImageJ.
The synchrony of the motion of the cilia was quantified by calculating the flow field of the cilia carpet by PIV analysis. We denoised the time-lapse images (128 pix × 128 pix) by a Gaussian filter followed by the enhancement of the contrast using Fiji plugins. Then, the flow field of the time-lapse images was obtained by PIV analysis using a python library (OpenPIV). The interrogation window of PIV was 32 pix, and the overlap between adjacent interrogation areas was 24 pix.
Antibodies
The following primary antibodies were used: rabbit anti-CAMSAP3 (Tanaka et al., 2012, 1: 100), mouse anti-α-tubulin (clone DM1A, Sigma-Aldrich T9026, 1:1000), rabbit monoclonal anti-β-tubulin (clone 9F3, Cell Signaling Technology Cat#2128, 1: 100), mouse anti-γ-tubulin (Sigma-Aldrich T6557, 1:1000), rabbit anti-γ-tubulin (Sigma-Aldrich T3559, 1:500), mouse anti-EZRIN (Abcam 4069, 1:100), rat anti-Odf2 (a gift from Sachiko Tsukita, Osaka University, 1: 2), and mouse monoclonal anti-Chibby (Santa Cruz sc-101551, 1:100). The following secondary antibodies were used: donkey CF-488A anti-rat IgG (Biotium 20027, 1:000) and CF-568 anti-rabbit IgG (Biotium 20698, 1:1000) antibodies; goat Alexa Fluor-488 anti-rat IgG (Invitrogen, A-11006, 1:500), Alexa Fluor-488 anti-rabbit IgG (Invitrogen, A-11034, 1:500), Alexa Fluor-568 anti-mouse IgG (Invitrogen, A-11031, 1:500), Alexa Fluor-594 anti-rabbit IgG (Invitrogen A-11037, 1:500), Alexa Fluor-647 anti-rabbit IgG (Invitrogen, A-21245, 1:500), and Alexa Fluor-647 anti-mouse IgG (Invitrogen, A-21236, 1:1000) antibodies; and goat STAR580 anti-mouse IgG (Abberior 2-0002-005-1, 1:500) and STAR635P anti-rat IgG (Abberior 2-0132-007-5, 1:500). The binding specificity of the anti-CAMSAP3 antibody in the use for IF staining was confirmed by previous studies (Tanaka et al., 2012; Mitsuhata et al., 2021).
Western blot analysis
Trachea tissues were homogenized in TMEN buffer (20 mM Tris-HCl, pH 7.4, 1 mM MgCl2, 2 mM EDTA, and 150 mM NaCl) containing the proteinase inhibitor cocktail cOmplete ultra (Roche, 5892970001) and the phosphatase inhibitor cocktail PhosSTOP (Roche, 4906845001). Then, 2% (wt/vol) SDS solution was added to homogenized trachea tissues. Proteins were separated by 4–20% gradient SDS–PAGE gel (Cosmobio, 414879) and then transferred to PVDF membrane (Millipore, IPVH00010). To detect CAMSAP3 and GAPDH for a loading control, anti-CAMSAP3 rabbit antibody (1:1,000) and anti-GAPDH mouse antibody (MBL, M171-3, 1:1,000) were used as primary antibodies. Quantification of band intensities was done using ImageJ.
Audio recording
The respiratory sound of mice was recorded using the Voice Memos application installed in iPhone XS (Apple).
Supplementary Material
Acknowledgments
We thank Hiroshi Hamada for his support of live imaging experiments and Mitsuru Morimoto for his advice on the study of lung, Sachiko Tsukita for anti-Odf2 antibody, and Ritsu Kamiya for critical reading of the manuscript. This work was supported by the program Grant-in-Aid for Scientific Research (S) (Grant Number 25221104) from the Japan Society for Promotion of Science to M.Ta; the Japan Society for the Promotion of Science, KAKENHI (Grant Numbers 17H03689 and 16H06280) and the Japan Science and Technology Agency, Core Research for Evolutional Science and Technology (JPMJCR1654) to T.F.; the Japan Society for the Promotion of Science, KAKENHI (Grant Number JP20K06645) to M.To; and Daiichi Sankyo Foundation of Life Science to M.S.
Abbreviations used:
- BB
basal body
- BP
basal plate
- CAMSAP
calmodulin-regulated spectrin-associated protein
- CP
central pair of microtubules
- CV
circular variance
- IF
immunofluorescence
- PBS
phosphate-buffered saline
- PCD
primary ciliary dyskinesia
- PFA
paraformaldehyde
- PIV
particle image velocimetry
- TEM
transmission electron microscopy
- TZ
transition zone.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-06-0303).
REFERENCES
- Baines AJ, Bignone PA, King MD, Maggs AM, Bennett PM, Pinder JC, Phillips GW (2009). The CKK domain (DUF1781) binds microtubules and defines the CAMSAP/ssp4 family of animal proteins. Mol Biol Evol 26, 2005–2014. [DOI] [PubMed] [Google Scholar]
- Bautista-Harris G, Julia-Serda G, Rodriguez de Castro F, Santana-Benitez I, Cabrera-Navarro P (2000). Absence of central microtubules and transposition in the ciliary apparatus of three siblings. Respiration 67, 449–452. [DOI] [PubMed] [Google Scholar]
- Berryman M, Franck Z, Bretscher A (1993). Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J Cell Sci 105 (Pt 4), 1025–1043. [DOI] [PubMed] [Google Scholar]
- Brooks ER, Wallingford JB (2014). Multiciliated cells. Curr Biol 24, R973–R982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne T, Lewis A, Dewar A, Luther P, Hogg C, Shoemark A, Dixon M (2014). Characterizing the ultrastructure of primary ciliary dyskinesia transposition defect using electron tomography. Cytoskeleton (Hoboken) 71, 294–301. [DOI] [PubMed] [Google Scholar]
- Burke MC, Li FQ, Cyge B, Arashiro T, Brechbuhl HM, Chen X, Siller SS, Weiss MA, O'Connell CB, Love D, et al. (2014). Chibby promotes ciliary vesicle formation and basal body docking during airway cell differentiation. J Cell Biol 207, 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zheng J, Li X, Zhu L, Shao Z, Yan X, Zhu X (2020). Wdr47 controls neuronal polarization through the Camsap family microtubule minus-end-binding proteins. Cell Rep 31, 107526. [DOI] [PubMed] [Google Scholar]
- Chuang M, Goncharov A, Wang S, Oegema K, Jin Y, Chisholm AD (2014). The microtubule minus-end-binding protein patronin/PTRN-1 is required for axon regeneration in C. elegans. Cell Rep 9, 874–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare DK, Magescas J, Piolot T, Dumoux M, Vesque C, Pichard E, Dang T, Duvauchelle B, Poirier F, Delacour D (2014). Basal foot MTOC organizes pillar MTs required for coordination of beating cilia. Nat Commun 5, 4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki PG, Shah JV (2012). The ciliary transition zone: from morphology and molecules to medicine. Trends Cell Biol 22, 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damseh N, Quercia N, Rumman N, Dell SD, Kim RH (2017). Primary ciliary dyskinesia: mechanisms and management. Appl Clin Genet 10, 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe HR, Farr H, Gull K (2007a). Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J Cell Sci 120, 7–15. [DOI] [PubMed] [Google Scholar]
- Dawe HR, Shaw MK, Farr H, Gull K (2007b). The hydrocephalus inducing gene product, Hydin, positions axonemal central pair microtubules. BMC Biol 5, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean S, Moreira-Leite F, Gull K (2019). Basalin is an evolutionarily unconstrained protein revealed via a conserved role in flagellum basal plate function. Elife 8, e42282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean S, Moreira-Leite F, Varga V, Gull K (2016). Cilium transition zone proteome reveals compartmentalization and differential dynamics of ciliopathy complexes. Proc Natl Acad Sci USA 113, E5135–E5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dute R, Kung C (1978). Ultrastructure of the proximal region of somatic cilia in Paramecium tetraurelia. J Cell Biol 78, 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjolras C, Thomas J, Chhin B, Cortier E, Duteyrat JL, Soulavie F, Kernan MJ, Laurencon A, Durand B (2012). Drosophila chibby is required for basal body formation and ciliogenesis but not for Wg signaling. J Cell Biol 197, 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneuer U, Mcintosh JR (1981). Polarity of some motility-related microtubules. Proc Natl Acad Sci USA 78, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Thyagarajan P, Shorey M, Seebold DY, Weiner AT, Albertson RM, Rao KS, Sagasti A, Goetschius DJ, Rolls MM (2019). Patronin-mediated minus end growth is required for dendritic microtubule polarity. J Cell Biol 218, 2309–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilula NB, Satir P (1972). The ciliary necklace. A ciliary membrane specialization. J Cell Biol 53, 494–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves J, Pelletier L (2017). The ciliary transition zone: finding the pieces and assembling the gate. Mol Cells 40, 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin SS, Vale RD (2010). Patronin regulates the microtubule network by protecting microtubule minus ends. Cell 143, 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenan GA, Vale RD, Agard DA (2020). Electron cryotomography of intact motile cilia defines the basal body to axoneme transition. J Cell Biol 219, e201907060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirao B, Meunier A, Mortaud S, Aguilar A, Corsi JM, Strehl L, Hirota Y, Desoeuvre A, Boutin C, Han YG, et al. (2010). Coupling between hydrodynamic forces and planar cell polarity orients mammalian motile cilia. Nat Cell Biol 12, 341–350. [DOI] [PubMed] [Google Scholar]
- Hagiwara H, Kano A, Aoki T, Ohwada N, Takata K (2000). Localization of gamma-tubulin to the basal foot associated with the basal body extending a cilium. Histochem J 32, 669–671. [DOI] [PubMed] [Google Scholar]
- Hendershott MC, Vale RD (2014). Regulation of microtubule minus-end dynamics by CAMSAPs and Patronin. Proc Natl Acad Sci USA 111, 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham H, Bielas S, Tanaka T, Gleeson JG (2004). Transgenic mouse line with green-fluorescent protein-labeled Centrin 2 allows visualization of the centrosome in living cells. Transgenic Res 13, 155–164. [DOI] [PubMed] [Google Scholar]
- Hoog JL, Lacomble S, O'Toole ET, Hoenger A, McIntosh JR, Gull K (2014). Modes of flagellar assembly in Chlamydomonas reinhardtii and Trypanosoma brucei. Elife 3, e01479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Hua S, Mohan R, Grigoriev I, Yau KW, Liu Q, Katrukha EA, Altelaar AF, Heck AJ, Hoogenraad CC, Akhmanova A (2014). Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev Cell 28, 295–309. [DOI] [PubMed] [Google Scholar]
- Kimura T, Saito H, Kawasaki M, Takeichi M (2021). CAMSAP3 is required for mTORC1-dependent ependymal cell growth and lateral ventricle shaping in mouse brains. Development 148, 195073. [DOI] [PubMed] [Google Scholar]
- King MD, Phillips GW, Bignone PA, Hayes NV, Pinder JC, Baines AJ (2014). A conserved sequence in calmodulin regulated spectrin-associated protein 1 links its interaction with spectrin and calmodulin to neurite outgrowth. J Neurochem 128, 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CS, Tserunyan V, Martin AC (2019). Microtubules promote intercellular contractile force transmission during tissue folding. J Cell Biol 218, 2726–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kott E, Legendre M, Copin B, Papon JF, Dastot-Le Moal F, Montantin G, Duquesnoy P, Piterboth W, Amram D, Bassinet L, et al. (2013). Loss-of-function mutations in RSPH1 cause primary ciliary dyskinesia with central-complex and radial-spoke defects. Am J Hum Genet 93, 561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunimoto K, Yamazaki Y, Nishida T, Shinohara K, Ishikawa H, Hasegawa T, Okanoue T, Hamada H, Noda T, Tamura A, et al. (2012). Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell 148, 189–200. [DOI] [PubMed] [Google Scholar]
- Lechtreck KF, Delmotte P, Robinson ML, Sanderson MJ, Witman GB (2008). Mutations in Hydin impair ciliary motility in mice. J Cell Biol 180, 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L (2011). Mechanisms of mammalian ciliary motility: Insights from primary ciliary dyskinesia genetics. Gene 473, 57–66. [DOI] [PubMed] [Google Scholar]
- Loreng TD, Smith EF (2017). The Central Apparatus of Cilia and Eukaryotic Flagella. Cold Spring Harb Perspect Biol 9, a028118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Veloso A, Wu J, Katrukha EA, Akhmanova A (2018). Control of endothelial cell polarity and sprouting angiogenesis by non-centrosomal microtubules. Elife 7, e33864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W, Mushika Y, Ichii T, Takeichi M (2008). Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell 135, 948–959. [DOI] [PubMed] [Google Scholar]
- Mitsuhata Y, Abe T, Misaki K, Nakajima Y, Kiriya K, Kawasaki M, Kiyonari H, Takeichi M, Toya M, Sato M (2021). Cyst formation in proximal renal tubules caused by dysfunction of the microtubule minus-end regulator CAMSAP3. Sci Rep 11, 5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashchekin D, Fernandes AR, St Johnston D (2016). Patronin/shot cortical foci assemble the noncentrosomal microtubule array that specifies the drosophila anterior-posterior axis. Dev Cell 38, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordstra I, Liu Q, Nijenhuis W, Hua S, Jiang K, Baars M, Remmelzwaal S, Martin M, Kapitein LC, Akhmanova A (2016). Control of apico-basal epithelial polarity by the microtubule minus-end-binding protein CAMSAP3 and spectraplakin ACF7. J Cell Sci 129, 4278–4288. [DOI] [PubMed] [Google Scholar]
- Nozawa YI, Yao E, Lin C, Yang JH, Wilson CW, Gacayan R, Chuang PT (2013). Fused (Stk36) is a ciliary protein required for central pair assembly and motile cilia orientation in the mammalian oviduct. Dev Dyn 242, 1307–1319. [DOI] [PubMed] [Google Scholar]
- Pongrakhananon V, Saito H, Hiver S, Abe T, Shioi G, Meng W, Takeichi M (2018). CAMSAP3 maintains neuronal polarity through regulation of microtubule stability. Proc Natl Acad Sci USA 115, 9750–9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CE, Spilker KA, Cueva JG, Perrino J, Goodman MB, Shen K (2014). PTRN-1, a microtubule minus end-binding CAMSAP homolog, promotes microtubule function in Caenorhabditis elegans neurons. Elife 3, e01498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson AM, Takahashi S, Brotslaw EJ, Ahmad A, Ferrer E, Procissi D, Richter CP, Cheatham MA, Mitchell BJ, Zheng J (2020). CAMSAP3 facilitates basal body polarity and the formation of the central pair of microtubules in motile cilia. Proc Natl Acad Sci USA 117, 13571–13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P, Christensen ST (2007). Overview of structure and function of mammalian cilia. Annu Rev Physiol 69, 377–400. [DOI] [PubMed] [Google Scholar]
- Shi D, Komatsu K, Hirao M, Toyooka Y, Koyama H, Tissir F, Goffinet AM, Uemura T, Fujimori T (2014). Celsr1 is required for the generation of polarity at multiple levels of the mouse oviduct. Development 141, 4558–4568. [DOI] [PubMed] [Google Scholar]
- Shinohara K, Chen D, Nishida T, Misaki K, Yonemura S, Hamada H (2015). Absence of radial spokes in mouse node cilia is required for rotational movement but confers ultrastructural instability as a trade-off. Dev Cell 35, 236–246. [DOI] [PubMed] [Google Scholar]
- Smith EF, Yang P (2004). The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton 57, 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stannard W, Rutman A, Wallis C, O'Callaghan C (2004). Central microtubular agenesis causing primary ciliary dyskinesia. Am J Respir Crit Care Med 169, 634–637. [DOI] [PubMed] [Google Scholar]
- Staudt T, Lang MC, Medda R, Engelhardt J, Hell SW (2007). 2,2'-thiodiethanol: a new water soluble mounting medium for high resolution optical microscopy. Microsc Res Tech 70, 1–9. [DOI] [PubMed] [Google Scholar]
- Takeda M, Sami MM, Wang YC (2018). A homeostatic apical microtubule network shortens cells for epithelial folding via a basal polarity shift. Nat Cell Biol 20, 36–45. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Meng W, Nagae S, Takeichi M (2012). Nezha/CAMSAP3 and CAMSAP2 cooperate in epithelial-specific organization of noncentrosomal microtubules. Proc Natl Acad Sci USA 109, 20029–20034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K, Nishida T, Inoue K, Tsukita S (2017). Three-dimensional organization of layered apical cytoskeletal networks associated with mouse airway tissue development. Sci Rep 7, 43783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K, Yamazaki Y, Nishida T, Watanabe S, Kunimoto K, Ishikawa H, Tsukita S (2013). Two appendages homologous between basal bodies and centrioles are formed using distinct Odf2 domains. J Cell Biol 203, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley AE, Walters MS, Shaykhiev R, Crystal RG (2015). Cilia dysfunction in lung disease. Annu Rev Physiol 77, 379–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toya M, Kobayashi S, Kawasaki M, Shioi G, Kaneko M, Ishiuchi T, Misaki K, Meng W, Takeichi M (2016). CAMSAP3 orients the apical-to-basal polarity of microtubule arrays in epithelial cells. Proc Natl Acad Sci USA 113, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker JB (1971). Development and deployment of cilia, basal bodies, and other microtubular organelles in the cortex of the ciliate Nassula. J Cell Sci 9, 539–567. [DOI] [PubMed] [Google Scholar]
- Usami FM, Arata M, Shi D, Oka S, Higuchi Y, Tissir F, Takeichi M, Fujimori T (2021). Intercellular and intracellular cilia orientation is coordinated by CELSR1 and CAMSAP3 in oviduct multi-ciliated cells. J Cell Sci 134, 257006. [DOI] [PubMed] [Google Scholar]
- Wang Y, Rui M, Tang Q, Bu S, Yu F (2019). Patronin governs minus-end-out orientation of dendritic microtubules to promote dendrite pruning in Drosophila. Elife 8, e39964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CW, Nguyen CT, Chen MH, Yang JH, Gacayan R, Huang J, Chen JN, Chuang PT (2009). Fused has evolved divergent roles in vertebrate Hedgehog signalling and motile ciliogenesis. Nature 459, 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau KW, van Beuningen SF, Cunha-Ferreira I, Cloin BM, van Battum EY, Will L, Schatzle P, Tas RP, van Krugten J, Katrukha EA, et al. (2014). Microtubule minus-end binding protein CAMSAP2 controls axon specification and dendrite development. Neuron 82, 1058–1073. [DOI] [PubMed] [Google Scholar]
- Zhu L, Liu H, Chen Y, Yan X, Zhu X (2019). Rsph9 is critical for ciliary radial spoke assembly and central pair microtubule stability. Biol Cell 111, 29–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.