Abstract
Background
Metabolomic analyses from our group and others have shown that tumors treated with glutamine antagonists (GA) exhibit robust accumulation of formylglycinamide ribonucleotide (FGAR), an intermediate in the de novo purine synthesis pathway. The increase in FGAR is attributed to the inhibition of the enzyme FGAR amidotransferase (FGAR-AT) that catalyzes the ATP-dependent amidation of FGAR to formylglycinamidine ribonucleotide (FGAM). While perturbation of this pathway resulting from GA therapy has long been recognized, no study has reported systematic quantitation and analyses of FGAR in plasma and tumors.
Objective
Herein, we aimed to evaluate the efficacy of our recently discovered tumor-targeted GA prodrug, GA-607 (isopropyl 2-(6-acetamido-2-(adamantane-1-carboxamido)hexanamido)-6-diazo-5-oxohexanoate), and demonstrate its target engagement by quantification of FGAR in plasma and tumors.
Methods
Efficacy and pharmacokinetics of GA-607 were evaluated in a murine EL4 lymphoma model followed by global tumor metabolomic analysis. Liquid chromatography-mass spectrometry (LC-MS) based methods employing the ion-pair chromatography approach were developed and utilized for quantitative FGAR analyses in plasma and tumors.
Results
GA-607 showed preferential tumor distribution and robust single-agent efficacy in a murine EL4 lymphoma model. While several metabolic pathways were perturbed by GA-607 treatment, FGAR showed the highest increase qualitatively. Using our newly developed sensitive and selective LC-MS method, we showed a robust >80- and >10-fold increase in tumor and plasma FGAR levels, respectively, with GA-607 treatment.
Conclusion
These studies describe the importance of FGAR quantification following GA therapy in cancer and underscore its importance as a valuable pharmacodynamic marker in the preclinical and clinical development of GA therapies.
Keywords: Glutamine antagonist, purine synthesis, formylglycinamide ribonucleotide, formylglycinamidine ribonucleotide, biomarker, cancer, LC-MS
1. INTRODUCTION
Purine nucleotide synthesis generates building blocks for DNA, RNA, high-energy intermediates, and signaling molecules in live cells. New purines can be generated by recycling turnover products in salvage pathways or synthesized de novo in a highly conserved ten-step pathway that transforms phosphoribosylpyrophosphate to inosine-5’-monophosphate [1]. Purine synthesis is an essential target in diseases, including viral infections, gout, and cancer [2-6]. Thus, the ability to accurately monitor the effects of purine synthesis blockade is of general interest.
FGAR is an intermediate metabolite at the fourth step of de novo purine synthesis. FGAR is a substrate for the enzyme formylglycinamide ribonucleotide amidotransferase (FGAR-AT, also known as phosphoribosylformylglycinamidine synthase or PFAS). Biochemical studies on prokaryotic homologues provide evidence for FGAR-AT catalyzed hydrolysis of glutamine to ammonia and glutamate; subsequently, ATP activates FGAR for nucleophilic attack by the ammonia generated in the first half of the reaction to produce formylglycinamidine ribonucleotide (FGAM) [7]. Small molecule glutamine antagonists (GAs), including the irreversible inhibitor 6-diazo-5-oxo-L-norleucine (DON), target this conversion of FGAR to FGAM. In mammalian cell extracts enriched for FGAR-AT activity, the inhibitor constant (Ki) for DON was determined to be 1.1 × 10-6 M [8]. Classic studies with the incorporation of 14C-formate into newly synthesized purines in leukemia cells grown in the presence and absence of GA revealed shifts in the HPLC profile of labeled compounds, including significant increases in the peaks corresponding to FGAR when a GA was present [9]. More recently, there has been interest in establishing non-radioactive analytical methods to detect FGAR and other de novo purine synthesis intermediates from human tissue or bio-fluid samples to screen purine synthesis gene products where metabolic causes of disease were not known. To this end, elevation in FGAR levels in human tissue samples was evaluated using an Orbitrap Elite mass spectrometer via MS2 fragmentation analysis followed by confirmation with an FGAR standard synthesized from bacterial recombinant enzymes [10].
Recently, there has been renewed interest in GA as a therapeutic strategy for cancer [11-15]. Blockade of glutamine utilization using a broadly active small molecule GA induces divergent metabolic programs in cancer cells versus effector T-cells, which ultimately results in the ability to overcome tumor immune evasion [13]. Also, myeloid-derived suppressor cells (MDSCs) play important roles in creating an immunosuppressive tumor microenvironment [16] and express enzymes that deplete key nutrients from T-cells. Our previous report revealed that blocking glutamine metabolism markedly inhibits the generation and recruitment of MDSCs [17]. Unfortunately, there are no broadly active GAs available clinically; DON was evaluated in oncology patients several decades ago, but its development was halted due to excessive gastrointestinal toxicity [18]. Our group has recently discovered a tumor-targeted prodrug strategy for DON, designed to mask the active compound with pro-moieties, which are hydrolyzed by tumor-enriched enzymes for bioactivation. One of our lead compounds, termed GA-607 (isopropyl 2-(6-acetamido-2-(adamantane-1-carboxamido)hexanamido)-6-diazo-5-oxohexanoate; comp. 6 in [11]) showed a remarkable 11-fold higher DON exposure to tumor (target tissue) versus GI tissues (toxicity tissue) [11]. However, its in vivo efficacy and target engagement in murine models was not established. Moreover, the effect of GA-607 on the de novo purine synthesis pathway, specifically FGAR, was not determined due to lack of a selective and sensitive bioanalytical method applicable in plasma and tumor samples.
Herein, we report that GA-607 has robust single-agent efficacy in mice bearing EL4 tumors. Using metabolomics, we demonstrate that the purine precursor FGAR as well as other metabolites, are significantly affected by GA in tumors. We developed a novel LC-MS method for the quantitation of FGAR in biological tissues and showed robust accumulation of FGAR in tumors and plasma following GA-607 treatment. This new bioanalytical method could have utility in quantifying FGAR levels as a target engagement biomarker for treatment modalities such as GA-607 that cause inhibition of purine biosynthesis and thus could aid in their clinical development.
2. MATERIALS AND METHODS
2.1. Reagents and Chemicals
GA-607 was synthesized as previously reported [11]. FGAR was provided as a generous gift from Dr. Qi Sun, Laboratory of Organic Chemistry, Jiangxi Science and Technology Normal University (Nanchang, China). Deuterated N-acetyl-L-aspartic acid (NAA-d3; internal standard) was obtained from Canadian Isotopes (Quebec, Canada). LC-MS-grade water, methanol, acetonitrile, and formic acid were obtained from Thermo Fisher Scientific (Waltham, MA, USA). N,N-Dimethylhexylamine (DMHA) was obtained from Sigma Aldrich (St. Louis, MO, USA).
2.2. Evaluation of Efficacy and Pharmacokinetics of GA-607 in Mice Bearing EL4 Flank Tumors
All animal studies were conducted in accordance with the protocols reviewed and approved by the Johns Hopkins Institutional Animal Care and Use Committee in compliance with the Association for Assessment and Accreditation of Laboratory Animal Care International and the Public Health Service Policy on the Humane Care and Use of Laboratory Animals (PHS Policy). Efficacy evaluation was conducted in C57BL/6 CES1-/- mice bearing EL4 lymphoma tumors. Tumors were grafted in mice as described previously with minor modifications [11]. Briefly, mice weighing between 25-30 g and 6-8 weeks of age were maintained on a 12 h light-dark cycle, with access to food and water, ad libitum. EL4 mouse lymphoma cells were obtained as a gift from Dr. Jonathan Powell’s laboratory (Johns Hopkins University, Baltimore, MD). Cells were maintained in RPMI 1640 medium with FBS 10% (v/v), antimycotic/antibiotic 1% (v/v), 2 mM of L-glutamine and 10 mM HEPES in a 5% (v/v) CO2 and 95% (v/v) air incubator.
Upon confluency, mice were injected with EL4 cells (0.3 × 106 cells in 0.2 mL of phosphate-buffered saline, pH 7.4) in one location on the flank. Mice whose tumors reached a mean volume of ~400 mm3 (approximately 7 days post-inoculation) were used for the efficacy study. Mice (n=12/group) were randomized into vehicle-treated or GA-607-treated (3.2 mg/kg of GA-607, equivalent to 1 mg/kg DON) groups, and dosed once daily subcutaneously (SC) in ethanol/tween 80/saline (5:10:85 v/v/v) for 5 consecutive days per week (with a 2-day break in dosing); a regimen which was previously shown to be tolerable and effective with other glutamine antagonists [13]. Tumor volume and body weight were measured and recorded on the days the mice were dosed.
We also assessed the pharmacokinetics of GA-607 and subsequent DON release in a satellite cohort bearing the EL4 tumor. Prior to dosing, the interscapular region of the mice was wiped with alcohol gauze. GA-607 was dissolved immediately (ethanol/tween 80/saline (5:10:85 v/v/v) and was administered to mice as a single SC dose of 3.2 mg/kg (1 mg/kg DON equivalent dose). The mice were euthanized with carbon dioxide at 1 h and 4 h post-drug administration; blood samples (~0.8 mL) were collected in heparinized microtubes by cardiac puncture, and tumors were removed and flash-frozen on dry ice. Blood samples were centrifuged at a temperature of 4°C at 3000 g for 10 min. All samples were kept chilled throughout processing. Plasma samples (0.3 mL) were collected in polypropylene tubes and stored at -80°C until bioanalysis. Flash-frozen tumor samples also were stored at -80°C until bioanalysis.
Bioanalysis of the pharmacokinetic samples was performed as previously described [11]. For quantifying the intact GA-607, standards (0.001-50 nmol/mL), QCs and samples were protein precipitated by adding 5 μL of methanol containing internal standards (losartan: 0.5 μM and 10 μM glutamate-d5) per milligram of tissue, followed by homogenization (tumor tissue) or vortex-mixing (plasma) and then centrifugation at 16,000 g for 5 min at 4°C. Then, 2 μL of the supernatant was injected into the LC-MS/MS system. The [M+H-N2]+ ion transition of GA-607 (m/z 518.142 →153.976, 500.401) and [M+H]+ ion transitions of losartan (IS) (m/z 422.938 → 184.580, 209.275) were used.
For the bioanalysis of DON, the supernatants (100 μL) were transferred to fresh tubes and dried under vacuum at 45°C for 1 h. To each tube, 50 μL of 0.2 M sodium bicarbonate buffer (pH 9.0) and 100 μL of 10 mM dabsyl chloride were added. After vortex-mixing, samples were incubated at 60°C for 15 min to derivatize, followed by centrifugation at 16,000 g for 5 min at 4°C. The supernatants (20 μL) were transferred to a 96-well plate, diluted with 80 μL of water and injected onto LC-MS/MS. Quantification was performed in parallel-reaction monitoring mode as previously described [11, 19].
2.3. Metabolomic Analysis of Mice Following GA-607 Therapy
C57BL/6 CES1-/- mice (n=5/group) bearing flank EL4 tumors were treated with either vehicle or GA-607 (3.2 mg/kg; 1 mg/kg DON equivalent dose); the groups were dosed SC once a day for 4 days until a significant reduction in tumor growth was observed in GA-607 group (tumors still present in sufficient quantity for analyses). One hour after the dose on day 4 mice were euthanized, and tumor tissues were harvested and then flash-frozen in liquid nitrogen. Tumor processing for metabolomics was performed as previously described [15, 17]. Briefly, tumors were homogenized by sonication in 80% cold methanol. Samples were vortexed and stored at -80ºC overnight to precipitate proteins, then centrifuged at 16,000 g for 10 minutes. Supernatants were dried under a steady stream of nitrogen gas and reconstituted in 50% acetonitrile. Samples were analyzed using an Agilent 1290 Infinity Binary UHPLC pump with well-plate autosampler at 4ºC and an Agilent 6520 Q-TOF MS. Chromatography was performed following injection of 4 µL samples over a Zorbax Extend C18 column at 40ºC by gradient elution run from 3% methanol with 5 mmol/L tributylamine to 100% methanol with 5 mmol/L tributylamine over 22 minutes. The MS was equipped with a dual electrospray ion source operated in negative-ion mode. Data were acquired with Agilent MassHunter Acquisition software and processed using MassHunter Qualitative Analysis software and MAVEN [20]. Metabolite identification was performed using mass-to-charge ratio and retention times from known standards or fragmentation analysis. Metabolite peak areas were normalized to extracted tumor weights. Statistical tests for the volcano plot were performed in Excel.
2.4. Ion-exchange LC-MS Method for FGAR Quantification
Various chromatographic conditions were evaluated initially, including hydrophilic interaction liquid chromatography (HILIC), reverse phase (C8, C18, Hypersil BDS) chromatography, and multiple mobile phase combinations for achieving optimal chromatographic separation and sensitivity for FGAR. However, an ion-pairing chromatography approach consisting of DMHA as an aqueous and organic modifier in water and acetonitrile, respectively, and a reverse phase C18 HPLC column gave optimal peak shape and retention. Due to the unavailability of isotopic analogue of FGAR, a stable isotope-labeled deuterated analogue of N-acetyl aspartic acid (NAA-d3) was selected as the internal standard, as under these ion-pair chromatographic conditions, NAA-d3 exhibited similar retention time to FGAR as well as a good chromatographic peak shape and MS signal, and was deemed suitable for quantification of FGAR.
2.4.1. Calibration Standards and Quality Control Samples
The FGAR standard was characterized by NMR and high-resolution mass spectroscopy (supporting Figs. S1-S4). A 100 mM stock solution of FGAR was prepared in water and stored at -20°C. All calibration standards and quality control (QC) samples of FGAR were prepared fresh each day before analysis. The working solutions were prepared by serial dilution in the extraction solvent (10 µM deuterated NAA-d3 in methanol). Calibration standards for FGAR were prepared in plasma (0.03-100 nmol/mL) and tumors (1-1000 nmol/g). Similarly, quality control (QC) samples were prepared independently at three concentrations in plasma (0.08, 2, 80 nmol/mL) and tumor homogenate (20, 200, 2000, nmol/g) covering low, medium and high concentrations levels. Calibration curve fit obtained from peak area ratio (area of analyte/area of internal standard), was assessed by weighted (1/concentration2) quadratic non-linear regression. For tumor analyses, the standard curve was fitted using a blank subtraction method as reported previously [21] to compensate for the presence of endogenous FGAR levels. This was important as it allowed the use of tumor matrix for the calibration while maintaining the recovery and matrix effects between samples and calibration curves. Using this method, LLOQ was determined by t-test and defined as the minimum concentration that resulted in a statistically significant increase above the background [22]. The concentration of each standard was back-calculated from calibration curve parameters, and concentrations in QC, and unknown samples were determined by interpolation. The calibration curve correlation coefficients (R2) ≥0.990 were considered acceptable for all analytical runs. The acceptance criteria for each back-calculated standard concentration was set at ±15% deviation from the nominal value except at the LLOQ, which was set at ±20%.
2.4.2. Chromatographic and Mass Spectrometric Conditions
Samples were analyzed using UltiMate 3000 UHPLC coupled to Q Exactive Focus Orbitrap Mass Spectrometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). The mobile phase used for chromatographic separation consisted of 8 mM DMHA + 0.005% formic acid in water, pH 9 (A), and 8 mM DMHA in acetonitrile (B), delivered at a flow rate of 0.4 mL/min. A gradient LC method (time (min)/%B: 0-0.5/5, 2.5-3.5/95, 3.51-4.50/5) with a short runtime of 4.5 min was developed for the analyses. Separation of analytes was achieved at room temperature using an Agilent Eclipse Plus C18 RRHD 2.1 × 100 mm, 1.8 µm particle size column (Agilent, Santa Clara, CA, USA). Samples were introduced to the interface through a heated ion spray with the capillary temperature setting at 350°C and a spray voltage of 4 kV. Nitrogen was used as the sheath and auxiliary gas set to 30 and 20 arbitrary units, respectively. Samples were subjected to ionization in negative mode and analyzed using the Full MS scan function. Parent ion of FGAR at m/z 313.0442 was used for quantitation and parent ion of NAA-d3 at m/z 177.0596 was used as an internal standard. Data were acquired and quantified with Xcalibur 4.1.31.9.
2.5. Method Validation
The intra-day and inter-day precision and accuracy for QC samples in both plasma and tumor were determined (for inter-day, n=3/day over 2 days), and statistical estimates were tabulated. Extraction efficiency, matrix effect as well as bench-top, freeze-thaw, long-term, and autosampler stability were also evaluated in triplicate at each QC level. Extraction efficiency was calculated by comparing mean peak responses obtained from pre-spiked samples against post-spiked samples. Matrix effect was evaluated from mean peak responses obtained from post-spiked samples compared to neat standards prepared in the extraction solvent. For stability assessment, spiked samples were a) freeze-thawed for three cycles (freeze-thaw stability), b) left at room temperature for 6 h (bench-top stability) or c) frozen at -80°C for 4 weeks before extraction (long-term stability). The autosampler stability was evaluated at 4°C by analyzing QC samples immediately after extraction and after 18 h storage in the autosampler. Regardless of the type of stability experiments conducted, the bias was contained within 85-115% of the nominal values and the precision within ±15% RSD. Assessment of carryover between autosampler injections was made by injecting a double blank sample after the upper limit of quantitation standard in calibration curve (100 nmol/mL in plasma; 10000 nmol/g in tumor). For analyte, carry over in the blank sample following the ULOQ standard should not be greater than 20% of the LLOQ, whereas, for the internal standard, the response in the blank sample should not exceed 5% of the average internal standard response of the calibrators and QCs.
2.6. Quantification of FGAR in Plasma and Tumor Samples
Mice (n=6/group) were randomized into vehicle-treated or GA-607-treated (3.2 mg/kg; 1 mg/kg DON equivalent dose) groups, and were dosed SC once a day for four days. The mice were sacrificed on day 4, and both plasma and tumor samples were collected (1 and 4h post dose) and frozen at -80ºC for FGAR bioanalysis.
Prior to extraction, frozen samples of plasma and tumor were thawed on ice. Sample preparation was performed using a single-step protein precipitation method. For plasma, extraction of analyte was conducted using 20 µL of the sample with 100 µL of extraction solvent (methanol containing 10 µM deuterated N-Acetyl Aspartic acid (NAA-d3) as internal standard), followed by vortex mixing for 30 s and centrifugation at 16,000 g for 5 min at 4°C. The resulting supernatants were transferred to a 96-well plate and analyzed by LC-MS. Similarly, for tumor, ~50 mg of EL4 tumor samples were processed by the addition of 5 μL extraction solvent per each mg of tissue and homogenized with Spex® 2150 stainless steel beads at 1500 rpm for 3 min on Spex® Geno/Grinder® (Spex SamplePrep LLC, Metuchen, NJ, USA). Post homogenization, samples were centrifuged at 16,000 g for 5 min at 4°C, and supernatants were diluted 5-fold in water and analyzed using the LC-MS method described above.
2.7. Statistical Analysis
For biomarker comparisons, groups were statistically compared by one-way ANOVA with Tukey’s post hoc test. The a priori level of significance for all analyses was defined as p < 0.05.
3. RESULTS AND DISCUSSION
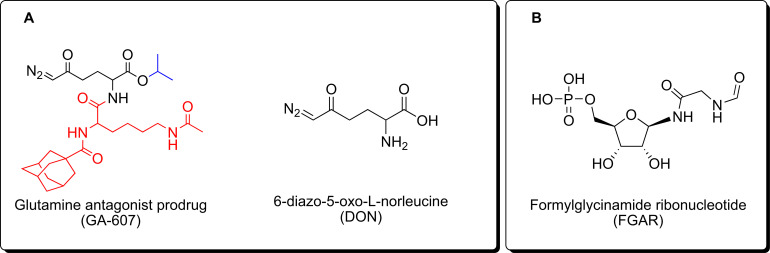
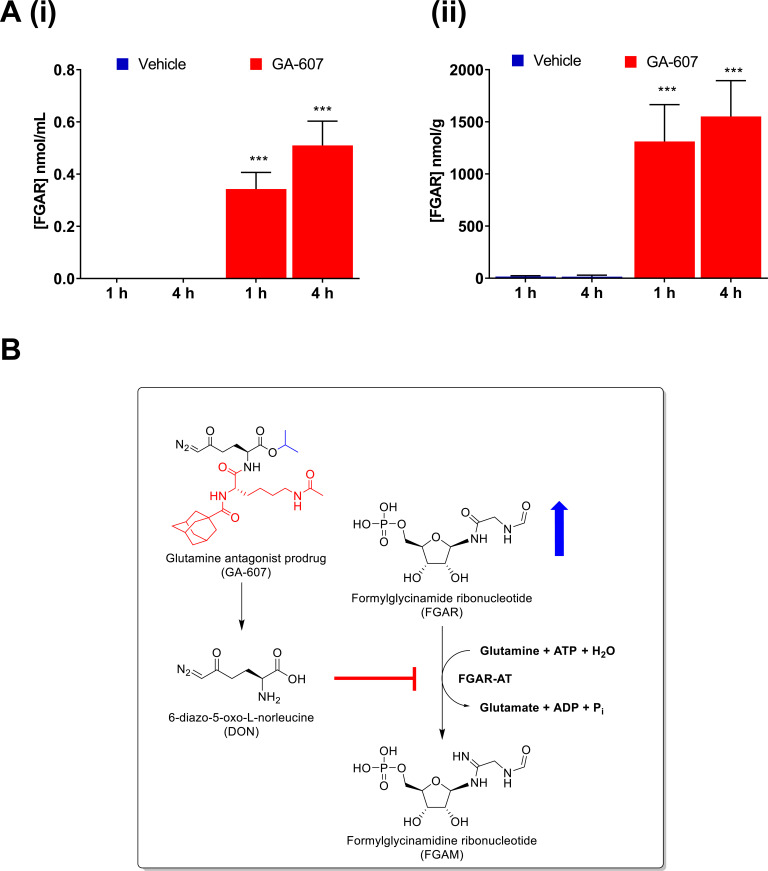
Amongst the GAs reported to-date, DON has been the most extensively studied both preclinically and in clinical trials [23-25]. DON is structurally similar to L-glutamine and broadly inhibits glutamine-utilizing enzymes involved in purine and pyrimidine synthesis, co-enzyme synthesis, amino acid synthesis, and hexosamine production [26]. DON has been evaluated in human studies since the 1950s and was found to elicit a favorable response in cancer patients, however, its efficacy was marred by its toxicity resulting from local glutamine starvation in the gastrointestinal (GI) tract [23, 27-31]. We have previously reported the development of DON prodrugs that preferentially target the tumor environment [11, 13, 32, 33]. One of our lead compounds, GA-607 (Fig. 1A), demonstrated stability in blood and excellent permeability into the tumor where it is selectively transformed to DON. Herein, we first evaluated the efficacy and exposures of GA-607 in a murine EL4 lymphoma model followed by metabolomic analysis to reveal its perturbations of metabolic pathways. Considering the dramatic qualitative increase observed in FGAR (Fig. 1B) levels, we developed a sensitive and robust bioanalytical method to evaluate the target engagement of GA-607 by quantitation of FGAR.
Fig. (1).
Molecular structures of (A) GA-607, DON, and (B) FGAR.
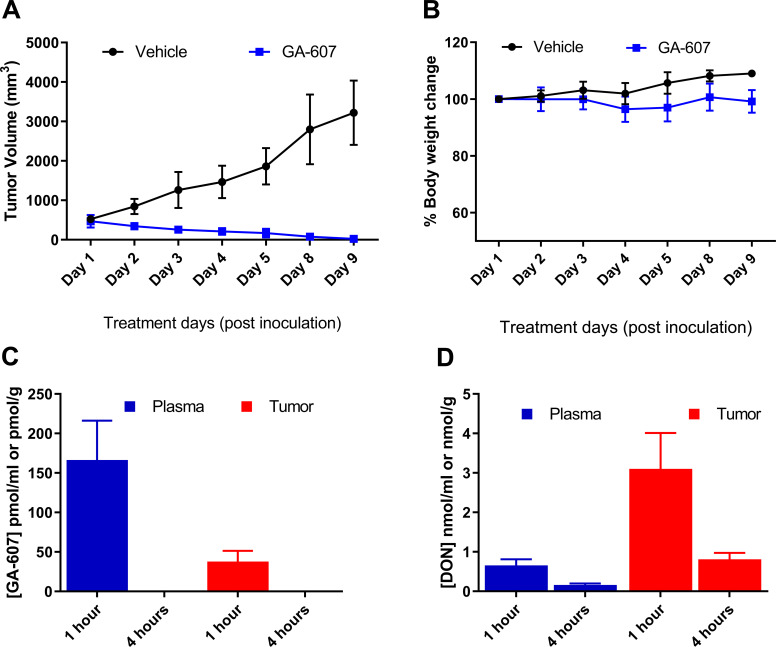
The efficacy of GA-607 was assessed in C57BL/6 CES1-/- mice bearing EL4 lymphoma tumors at 3.2 mg/kg SC (1 mg/kg DON equivalent dose). Our previous published report revealed that this dosing regimen afforded an ideal pharmacokinetic profile [11], releasing efficacious levels of DON within the tumor. As shown in Fig. (2A), the vehicle-treated mice showed continued tumor growth over time, while the mice treated with GA-607 had complete tumor regression. Furthermore, no changes were observed in the body weight (Fig. 2B), general appearance, and behavior, demonstrating the efficacy of GA-607 with good tolerability.
Fig. (2).
Efficacy, tolerability, and pharmacokinetic assessment of GA-607 in EL4 tumor-bearing mice following GA-607 (3.2 mg/kg SC) treatment; dosed 5 consecutive days followed by 2 drug-free days. Tumors volumes and body weights were only measured on the day of dosing. (A) Complete tumor regression was observed following GA-607 administration. (B) No change in body weight was observed following GA-607 administration. (C) GA-607 and (D) GA-607-derived DON levels in plasma and tumors following GA-607 administration. (A higher resolution / color version of this figure is available in the electronic copy of the article).
In addition to efficacy evaluation, drug exposures were also quantified in plasma and tumor tissue for confirmation of tumor distribution. The data obtained from a satellite cohort (Fig. 2C and D) showed preferential DON delivery to the tumor. The intact levels of the GA-607 in plasma were approximately 160 nM at 1-hour post-dose but were below the limit of quantification at 4 hours post-dose (Fig. 2C). The GA-607-derived DON levels in the tumor were approximately 5-fold higher than that of plasma (3.1 µM versus 0.651 µM at 1 h post-dose) (Fig. 2D). A similar 5-fold higher DON tumor exposure was also observed at 4 h (0.75 µM versus 0.16 µM).
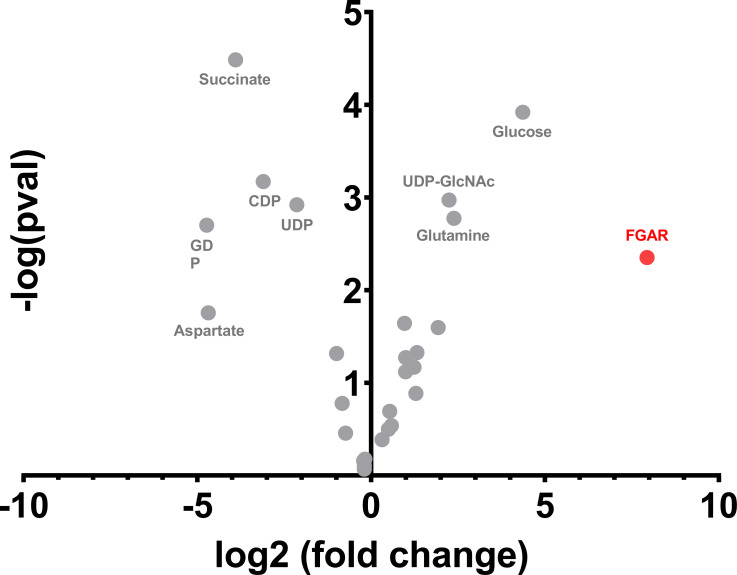
We next conducted global metabolomic analysis in tumors treated with GA-607. Fifty polar metabolites were queried based on mass-to-charge ratio and retention times, and twenty-nine of those were identified among the analyzed samples. Notably, the metabolite with the greatest increase upon GA-607 treatment was FGAR (ratio GA-607/vehicle = 244.96, p-value = 0.0045) (Fig. 3). Other metabolites that demonstrated significant modulation upon GA-607 treatment are also noted in Fig. (3). Consistent with prior studies and the known direct biochemical effect of GA-607, glutamine was also increased (ratio GA-607/vehicle = 5.21, p-value 0.0017). Other significantly modulated metabolites are in pathways containing glutamine amidotransferases including uridine 5′-diphospho-N-acetylglucosamine (UDP-GlcNAc) in hexosamine metabolism, nucleotide diphosphates (guanosine diphosphate (GDP), cytidine diphosphate (CDP), and uridine diphosphate (UDP)) in nucleotide metabolism, and aspartate in amino acid metabolism. Considering the role of FGAR in purine synthesis and its robust modulation upon glutamine antagonism, we focused on development of a sensitive and selective LC-MS assay for FGAR and used it to quantitatively monitor changes in plasma and tumors following GA-607 treatment.
Fig. (3).
Metabolomic analysis of GA-607-versus vehicle-treated EL4 tumors. GA-607 treatment caused an increase in FGAR, glucose, glutamine, and uridine 5′-diphospho-N-acetylglucosamine in the tumor and a decrease in succinate, aspartate, and nucleotide diphosphates. (A higher resolution / color version of this figure is available in the electronic copy of the article).
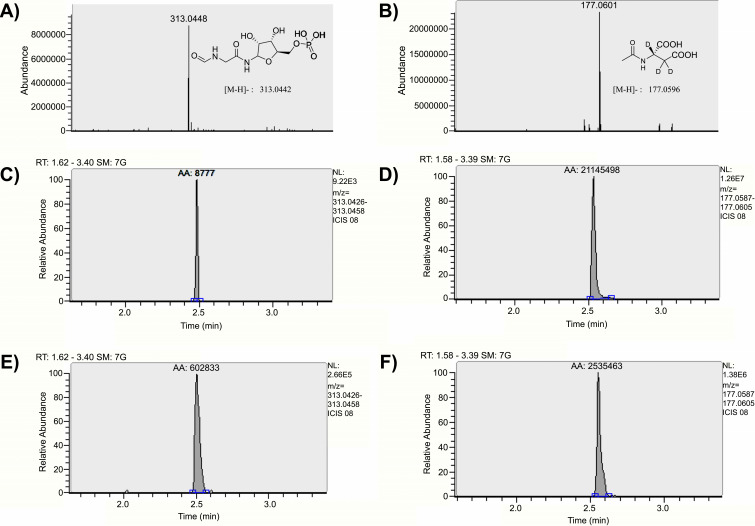
While previous reports of FGAR are qualitative in nature [9, 10], recently Krijt et al., reported an LC-MS/MS-based bioanalytical assay for the quantification of FGAR [34, 35]. However, this method is limited in utility as it has only been tested in urine, dried blood spots and in vitro cultures. The sensitivity of this method has not been evaluated in more complex biomatrices such as plasma and tumor homogenates, as reported here. Furthermore, the method required extensive equilibration time and column regeneration after each run, thus requiring 25 min of runtime for each sample; this severely limits the throughput capacity of the method. We developed a full scan MS analysis using a high-resolution Q Exactive Orbitrap mass spectrometer that provides accurate detection of masses within 2 ppm error with the total runtime of only 4.5 min per sample. The LLOQ for FGAR in plasma was established at 0.03 nmol/mL with an accuracy of 99.0% and precision ≤5.09% (n=6). For determination of LLOQ in tumors we employed a blank subtraction method during data analyses [21]. Tumors have been reported to have baseline levels of endogenous FGAR [36] and simple linear regression without subtraction of endogenous levels would have resulted in erroneous results. Furthermore, this approach also allowed the use of tumor matrix for calibration standards without compromising the recovery or causing differential matrix effects [37-39]; the limitations often encountered when using surrogate matrices. Using this method, the LLOQ for FGAR in the tumor was determined to be 1 nmol/g, as this was the lowest standard concentration that showed statistically significant difference from the background levels (p < 0.05), as determined by t-test [22]. More importantly, the higher LLOQ in tumor versus plasma was not because of the lack of sensitivity of the analytical method, but due to the high endogenous levels of FGAR in tumor. The LLOQ for FGAR in the tumor homogenate exhibited an accuracy and precision of 99.6% and 3.63%, respectively (n=6). Due to a positive parabolic (non-linear) mass spectrometer response observed with increasing concentrations of the FGAR standards across a wide calibration range (0.03-100 nmol/mL in plasma, 1-1000 nmol/g in tumor), a linear regression was unsuitable, and a quadratic fit offered the desired precision and accuracy for the calibration standards [40-42]. Curve fit in plasma and tumor homogenates provided a correlation coefficient of greater than 0.995 ± 0.003 (n=6) and 0.994 ± 0.007 (n=6), respectively. High-resolution mass spectrum and representative chromatographic spectra of FGAR and IS (NAA-d3) in plasma and tumor samples are shown in Fig. (4). Back-calculated concentrations for the calibration standards in plasma and tumor homogenates obtained from regression analysis were within 94.4-105% and 93.0-104%, respectively (Table 1).
Fig. (4).
Representative high-resolution mass spectrum and chromatographic spectra of FGAR and IS. (A) High-resolution full scan mass spectrum of FGAR in negative mode with < 2 ppm error; (B) high-resolution full scan mass spectrum of internal standard (NAA-d3) in negative mode with < 2 ppm error; (C) extracted chromatogram of FGAR spiked in plasma at LLOQ (0.03 nmol/mL); (D) extracted chromatogram for internal standard (NAA-d3) in plasma; (E) extracted chromatogram of FGAR spiked in tumor at LLOQ (1 nmol/g); (F) extracted chromatogram for internal standard (NAA-d3) in EL4 tumor. (A higher resolution / color version of this figure is available in the electronic copy of the article).
Table 1.
Statistical evaluation of calculated concentrations of FGAR obtained from the calibration curves (n=3) prepared in mouse plasma and mouse tumor homogenates.
| Mouse Plasma | Mouse Tumor Homogenate | ||||
|---|---|---|---|---|---|
| Nominal Conc. (nmol/mL) | Accuracy (%) | Precision (% RSD) | Nominal Conc. (nmol/g) | Accuracy (%) | Precision (% RSD) |
| 100 | 94.4 | 5.18 | 10000 | 98.9 | 0.46 |
| 32.0 | 103 | 8.64 | 3000 | 102 | 1.21 |
| 10.0 | 98.9 | 6.76 | 1000 | 102 | 1.51 |
| 3.20 | 105 | 5.31 | 300 | 101 | 1.74 |
| 1.00 | 98.6 | 4.63 | 100 | 100 | 3.27 |
| 0.32 | 97.1 | 10.3 | 30 | 104 | 1.32 |
| 0.10 | 100 | 9.05 | 10 | 95.9 | 1.31 |
| 0.03 | 101 | 7.10 | 3 | 93.0 | 4.83 |
| - | - | - | 1 | 103 | 1.52 |
Extraction efficiency for FGAR quantification was determined by the relative recovery of FGAR extracted from plasma compared to post extraction-spiked samples and ranged from 85% to 99%,
with an average of 89%. Similarly, the extraction efficiency of FGAR from the tumor homogenates ranged from 99% to 104%, with an average of 103%. The results obtained from stability experiments are presented in Tables 2 and 3 for plasma and tumor homogenates, respectively. FGAR was found to be stable at all the tested conditions (freeze-thaw/bench-top/long-term/autosampler) with a <15% deviation from nominal concentrations in both plasma and the tumor homogenates. Matrix effect, as determined by the FGAR peak area between post-extraction spiked and neat standards, ranged from -18% to 6% with an average of -14% in plasma, and ranged from -6% to 17% with an average of 8% in tumor homogenates, and was within the acceptable limits of ± 20% at all tested concentrations. A blank injection following a high standard of 100 nmol/mL in plasma, revealed a minimal carry-over area of 8.97% of the LLOQ standard, which was within the acceptable range. Similarly, carryover in tumor homogenate samples was estimated to be 7.25%. No carry-over was observed for the internal standard. Inter-day accuracy, and precision (%RSD) of QC’s (n=3/day; 2 days) in plasma ranged from 91.0-99.4% and 4.82-13.6%, respectively (Table 4). Likewise, inter-day accuracy and precision (n=3/day; 2 days) for QC’s in the tumor homogenates ranged from 98.1-104% and 4.03-6.03%, respectively (Table 5). These results indicate that the LC-MS method developed is robust, precise, and accurate for the quantitation of FGAR in both plasma and tumor.
Table 2.
Stability of FGAR in mouse plasma after subjecting to 3 freeze-thaw cycles, bench-top (6 h at room temperature), and long-term conditions (-80°C for 4 weeks) before extraction and autosampler (18 h at 4°C) after extraction.
| Nominal Conc. (nmol/mL) | Stability (%) | |||
|---|---|---|---|---|
| Freeze-thaw | Bench-top | Long-term | Autosampler | |
| 80.0 | 107 ± 3.57 | 100 ± 5.32 | 96.5 ± 2.12 | 95.2 ± 2.40 |
| 2.00 | 114 ± 7.84 | 91.9 ± 6.22 | 86.5 ± 6.72 | 96.4 ± 0.591 |
| 0.08 | 112 ± 5.14 | 96.0 ± 12.1 | 92.5 ± 11.2 | 101 ± 8.88 |
Table 3.
Stability of FGAR in mouse tumor homogenates after subjecting to 3 freeze-thaw cycles, bench-top (6 h at room temperature), and long-term conditions (-80°C for 4 weeks) before extraction and autosampler (18 h at 4oC) after extraction.
| Nominal Conc. (nmol/mL) | Stability (%) | |||
|---|---|---|---|---|
| Freeze-Thaw | Bench-Top | Long-Term | Autosampler | |
| 2000 | 104 ± 3.76 | 102 ± 3.17 | 113 ± 2.31 | 103 ± 0.02 |
| 200 | 93.5 ± 3.95 | 106 ± 4.31 | 110 ± 3.85 | 102 ± 0.00 |
| 20 | 104 ± 2.59 | 109 ± 7.93 | 96.2 ± 2.79 | 107 ± 0.06 |
Table 4.
Accuracy and precision in mouse plasma.
| Nominal Conc. (nmol/mL) | Intra-day (n=3) | Inter-day (n=6) | ||
|---|---|---|---|---|
| Accuracy (%) | Precision (% RSD) | Accuracy (%) | Precision (% RSD) | |
| 80.0 | 95.8 | 3.96 | 99.4 | 4.82 |
| 2.00 | 92.0 | 4.19 | 98.9 | 9.39 |
| 0.08 | 111 | 10.6 | 91.0 | 13.6 |
Inter and intra‐day precision and accuracy for FGAR in plasma were determined by analyzing replicates (n=3/day) of spiked samples at 3 different concentration levels over 2 subsequent days. Statistics for inter-day evaluation are generated from n=6 samples.
Table 5.
Accuracy and precision in mouse tumor homogenates.
| Nominal Conc. (nmol/g) | Intra-day (n=3) | Inter-day (n=6) | ||
|---|---|---|---|---|
| Accuracy (%) | Precision (% RSD) | Accuracy (%) | Precision (% RSD) | |
| 2000 | 96.8 | 3.05 | 98.1 | 4.03 |
| 200 | 101 | 3.39 | 104 | 4.11 |
| 20 | 95.2 | 1.69 | 98.2 | 6.03 |
Inter and intra-day precision and accuracy for FGAR in EL4 tumor homogenates were determined by analyzing replicates (n=3/day) of spiked samples at 3 different concentration levels over 2 subsequent days. Statistics for inter-day evaluation are generated from n=6 samples.
FGAR quantification was performed in samples collected after 4 days of GA-607 administration when significant tumor suppression was observed. In the tumors of vehicle-treated mice, an average of 16 and 15 nmol/g FGAR was quantified at 1 h and 4 h post-dose, respectively. In contrast, GA-607 treated animals showed a remarkable 1300 and 1600 nmol/g FGAR at 1 and 4 h post-dose, respectively. These levels represent an 80- to 100-fold increase (Fig. 5A-ii). In contrast, in plasma of GA-607 treated mice, much lower levels were observed (0.3 and 0.5 nmol/mL at 1 and 4 h post-dose, respectively) compared to tumor; however, these levels were still >10-fold higher when compared to vehicle-treated animals, which were generally below the limit of quantification (<0.03 nmol/mL) (Fig. 5A-i). These data support the target engagement of GA-607 to FGAR-AT as presented in Fig. (5B) schematic. Thus, FGAR could be used as a biomarker of the GA-607 effect.
Fig. (5).
FGAR quantification following GA-607 treatment (3.2 mg/kg SC daily for 4 days) in EL4 tumor-bearing mice. (A) (i) FGAR levels in plasma at 1 and 4 hours after GA-607 or vehicle administration; (ii) FGAR levels in tumor at 1 and 4 hours after GA-607 or vehicle administration. Mean ± S.D. ***p > .001 (two-way ANOVA with Tukey’s post hoc test) (B) Schematic representation of the biochemical pathway regulating FGAR metabolism in tumor tissues and the effect of GA therapy. FGAR-AT, an enzyme in de novo purine synthesis catalyzes the conversion of FGAR, ATP, and glutamine to FGAM, ADP, Pi, ammonia, and glutamate, respectively. Upon cleavage in tumors, GA-607 releases DON. The inhibition of FGAR-AT results in elevated FGAR levels. (A higher resolution / color version of this figure is available in the electronic copy of the article).
The FGAR quantification method presented here is a sensitive and selective LC-MS method for direct analysis of FGAR in plasma and tumors. The presented data also makes a strong case for FGAR modulation to corroborate the target engagement of glutamine antagonists. With the prodigious efforts being poured into the discovery and development of glutamine antagonists for the treatment of cancer and other diseases [32, 43-45], development of a biomarker such as FGAR may serve as a valuable tool in both diagnosing dysfunctions in this pathway as well as assessing the target engagement of therapies focused on modulating nucleotide synthesis. Also, while the focus of this report was to demonstrate the efficacy of GA-607 and the identification of FGAR as a target engagement tool, future efforts will be focused on evaluating the effects on downstream metabolites such as FGAM and their quantification. Importantly, although current studies report the evaluation of GA-607 only in EL4 tumor model, we have previously studied our novel GA prodrugs in multiple tumor types (e.g. MC38 colon cancer, CT26 colon cancer, B16 melanoma, 4T1 mammary cancer) with various dosing regimens and treatments when the tumors were small (~30 mm3) as well as large (~300 mm3) and as both single agents and in combination with immunotherapies [13, 15, 17]. Furthermore, our lead glutamine antagonist, DRP-104, is currently in phase I clinical trial (Clinical Trial Identifier: NCT04471415) for advanced solid tumors and has received a fast-track FDA designation for the treatment of advanced non-small-cell lung carcinoma (NSCLC) [46]. These studies underscore the broad applicability of GA therapy to various tumor types and the importance of a biomarker/target engagement tool, such as FGAR that may aid in the further development of these analogs as anticancer agents.
CONCLUSION
The tumor-targeted glutamine antagonist GA-607 was shown, for the first time, to have robust single-agent tumor regression activity in a murine EL4 lymphoma tumor model without any observable signs of toxicity. Metabolomic analyses revealed that GA-607 caused dramatic FGAR accumulation in addition to perturbing other known glutamine-utilizing pathways. A new rapid, sensitive, selective, and reliable LC-MS method was developed and optimized for the quantification of FGAR in plasma and tumor using an N,N-dimethylhexylamine assisted ion-pairing chromatography approach. The method demonstrated good sensitivity, precision, accuracy, and recovery in both plasma and tumor samples. Further, the application of the method to monitor target engagement was demonstrated using GA-607 treatment in EL4 tumored mice. We propose FGAR as a promising pharmacodynamic marker that can quantitatively reveal dysfunction in de novo purine synthesis and validate the target engagement of therapeutic modalities targeting the pathway. Moreover, while studies reported here were limited to only EL4 tumors, the FGAR quantification method described will be broadly applicable to multiple tumor types sensitive to glutamine antagonism in preclinical models and ultimately in clinical studies.
ACKNOWLEDGEMENTS
The authors thank Dr. Jonathan Powell’s Laboratory (Johns Hopkins University, Baltimore, MD) for the generous gift of EL4 mouse lymphoma cells.
LIST OF ABBREVIATIONS
- AUC
Area Under Curve
- CES1-/-
Carboxylesterase 1 Knockout
- DMHA
N,N-dimethylhexylamine
- DON
6-diazo-5-oxo-L-norleucine
- FGAM
Formylglycinamidine Ribonucleotide
- FGAR
Formylglycinamide Ribonucleotide
- FGAR-AT
Formylglycinamide Ribonucleotide Amidotransferase
- GA
Glutamine Antagonist
- HILIC
Hydrophilic Interaction Liquid Chromatography
- LC-MS
Liquid Chromatography-mass Spectrometry
- LLOQ
Lower Limit of Quantitation
- NAA-d3
N-acetyl Aspartate Deuterated
- QC
Quality Control
- SC
Subcutaneous
- UHPLC
Ultra-high Performance Liquid Chromatography
Funding Statement
This research was supported by NIH Grant R01CA193895 (to B.S.S. and R.R.), R01CA229451 (to B.S.S. and R.R.), Bloomberg Kimmel Institute for Cancer Immunotherapy (to B.S.S. and R.R.), LTAUSA18166 by the Ministry of Education, Youth and Sports of the Czech Republic (program INTER-EXCELLENCE to P.M.), and by the Institute of Organic Chemistry and Biochemistry of the Academy of Sciences of the Czech Republic, v.v.i. (RVO 61388963). K.M.L. was supported by a CureSearch Young Investigator Award.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher's website along with the published article.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The protocol of this study was approved from John Hopkins University Animal Care and Use Committee (ACUC), USA (Approval no. MO19M27).
HUMAN AND ANIMAL RIGHTS
No humans were used for studies that are base of this research. Animals studies were carried out under the John Hopkins University Animal Care and Use Committee (ACUC) protocol as stated above.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The datasets generated and analyzed during the current study are available from the corresponding author [RR] on reasonable request.
FUNDING
This research was supported by NIH Grant R01CA193895 (to B.S.S. and R.R.), R01CA229451 (to B.S.S. and R.R.), Bloomberg Kimmel Institute for Cancer Immunotherapy (to B.S.S. and R.R.), LTAUSA18166 by the Ministry of Education, Youth and Sports of the Czech Republic (program INTER-EXCELLENCE to P.M.), and by the Institute of Organic Chemistry and Biochemistry of the Academy of Sciences of the Czech Republic, v.v.i. (RVO 61388963). K.M.L. was supported by a CureSearch Young Investigator Award.
CONFLICT OF INTEREST
The authors declare the following competing financial interest(s): Under a license agreement between Dracen Pharmaceuticals, Inc. and the Johns Hopkins University, Barbara S. Slusher, Rana Rais, and Jesse Alt, are entitled to royalty distributions related to technology used in the research described in this publication. Barbara S. Slusher, Pavel Majer, and Rana Rais, are also co-founders of and hold equity in Dracen Pharmaceuticals, Inc. which is clinically developing GA prodrugs. This arrangement has been reviewed and approved by the Johns Hopkins University in accordance with its conflict of interest policies.
REFERENCES
- 1.Pedley A.M., Benkovic S.J. A new view into the regulation of purine metabolism: The purinosome. Trends Biochem. Sci. 2017;42(2):141–154. doi: 10.1016/j.tibs.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin J., Ren W., Huang X., Deng J., Li T., Yin Y. Potential mechanisms connecting purine metabolism and cancer therapy. Front. Immunol. 2018;9:1697. doi: 10.3389/fimmu.2018.01697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y., Wang W., Xu L., Zhou X., Shokrollahi E., Felczak K., van der Laan L.J., Pankiewicz K.W., Sprengers D., Raat N.J., Metselaar H.J., Peppelenbosch M.P., Pan Q. Cross talk between nucleotide synthesis pathways with cellular immunity in constraining hepatitis E virus replication. Antimicrob. Agents Chemother. 2016;60(5):2834–2848. doi: 10.1128/AAC.02700-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aleo F. The hormonal regulation of purine biosynthesis: basal levels of different hormones in primary gout. 1984. [DOI] [PubMed]
- 5.Hershfield M.S., Seegmiller J.E. Gout and the regulation of purine biosynthesis. Horiz. Biochem. Biophys. 1976;2:134–162. [PubMed] [Google Scholar]
- 6.Grayzel A.I., Seegmiller J.E., Love E. Suppression of uric acid synthesis in the gouty human by the use of 6-diazo-5-oxo-L-norleucine. J. Clin. Invest. 1960;39:447–454. doi: 10.1172/JCI104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoskins A.A., Anand R., Ealick S.E., Stubbe J. The formylglycinamide ribonucleotide amidotransferase complex from Bacillus subtilis: metabolite-mediated complex formation. Biochemistry. 2004;43(32):10314–10327. doi: 10.1021/bi049127h. [DOI] [PubMed] [Google Scholar]
- 8.Levenberg B., Melnick I., Buchanan J.M. Biosynthesis of the purines. XV. The effect of aza-L-serine and 6-diazo-5-oxo-L-norleucine on inosinic acid biosynthesis de novo. J. Biol. Chem. 1957;225(1):163–176. doi: 10.1016/S0021-9258(18)64919-1. [DOI] [PubMed] [Google Scholar]
- 9.Lyons S.D., Sant M.E., Christopherson R.I. Cytotoxic mechanisms of glutamine antagonists in mouse L1210 leukemia. J. Biol. Chem. 1990;265(19):11377–11381. doi: 10.1016/S0021-9258(19)38603-X. [DOI] [PubMed] [Google Scholar]
- 10.Mádrová L., Krijt M., Barešová V., Václavík J., Friedecký D., Dobešová D., Součková O., Škopová V., Adam T., Zikánová M. Mass spectrometric analysis of purine de novo biosynthesis intermediates. PLoS One. 2018;13(12):e0208947. doi: 10.1371/journal.pone.0208947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenora L., Alt J., Dash R.P., Gadiano A.J., Novotná K., Veeravalli V., Lam J., Kirkpatrick Q.R., Lemberg K.M., Majer P., Rais R., Slusher B.S. Tumor-targeted delivery of 6-diazo-5-oxo-l-norleucine (DON) using substituted acetylated lysine prodrugs. J. Med. Chem. 2019;62(7):3524–3538. doi: 10.1021/acs.jmedchem.8b02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S.Z., Poore B., Alt J., Price A., Allen S.J., Hanaford A.R., Kaur H., Orr B.A., Slusher B.S., Eberhart C.G., Raabe E.H., Rubens J.A. Unbiased metabolic profiling predicts sensitivity of high MYC-expressing atypical teratoid/rhabdoid tumors to glutamine inhibition with 6-diazo-5-oxo-l-norleucine. Clin. Cancer Res. 2019;25(19):5925–5936. doi: 10.1158/1078-0432.CCR-19-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leone R.D., Zhao L., Englert J.M., Sun I.M., Oh M.H., Sun I.H., Arwood M.L., Bettencourt I.A., Patel C.H., Wen J., Tam A., Blosser R.L., Prchalova E., Alt J., Rais R., Slusher B.S., Powell J.D. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science. 2019;366(6468):1013–1021. doi: 10.1126/science.aav2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma N.S., Gupta V.K., Garrido V.T., Hadad R., Durden B.C., Kesh K., Giri B., Ferrantella A., Dudeja V., Saluja A., Banerjee S. Targeting tumor-intrinsic hexosamine biosynthesis sensitizes pancreatic cancer to anti-PD1 therapy. J. Clin. Invest. 2020;130(1):451–465. doi: 10.1172/JCI127515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemberg K.M., Zhao L., Wu Y., Veeravalli V., Alt J., Aguilar J.M.H., Dash R.P., Lam J., Tenora L., Rodriguez C., Nedelcovych M.T., Brayton C., Majer P., Blakeley J.O., Rais R., Slusher B.S. The novel glutamine antagonist prodrug JHU395 has antitumor activity in malignant peripheral nerve sheath tumor. Mol. Cancer Ther. 2020;19(2):397–408. doi: 10.1158/1535-7163.MCT-19-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tcyganov E., Mastio J., Chen E., Gabrilovich D.I. Plasticity of myeloid-derived suppressor cells in cancer. Curr. Opin. Immunol. 2018;51:76–82. doi: 10.1016/j.coi.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh M-H., Sun I.H., Zhao L., Leone R.D., Sun I.M., Xu W., Collins S.L., Tam A.J., Blosser R.L., Patel C.H., Englert J.M., Arwood M.L., Wen J., Chan-Li Y., Tenora L., Majer P., Rais R., Slusher B.S., Horton M.R., Powell J.D. Targeting glutamine metabolism enhances tumor-specific immunity by modulating suppressive myeloid cells. J. Clin. Invest. 2020;130(7):3865–3884. doi: 10.1172/JCI131859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cervantes-Madrid D., Romero Y., Dueñas-González A. Reviving lonidamine and 6-diazo-5-oxo-l-norleucine to be used in combination for metabolic cancer therapy. BioMed Res. Int. 2015;2015:690492. doi: 10.1155/2015/690492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alt J., Potter M.C., Rojas C., Slusher B.S. Bioanalysis of 6-diazo-5-oxo-l-norleucine in plasma and brain by ultra-performance liquid chromatography mass spectrometry. Anal. Biochem. 2015;474:28–34. doi: 10.1016/j.ab.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clasquin M.F., Melamud E., Rabinowitz J.D. 2012. [DOI] [PMC free article] [PubMed]
- 21.Thakare R., Chhonker Y.S., Gautam N., Alamoudi J.A., Alnouti Y. Quantitative analysis of endogenous compounds. J. Pharm. Biomed. Anal. 2016;128:426–437. doi: 10.1016/j.jpba.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 22.Strassburg K., Huijbrechts A.M., Kortekaas K.A., Lindeman J.H., Pedersen T.L., Dane A., Berger R., Brenkman A., Hankemeier T., van Duynhoven J., Kalkhoven E., Newman J.W., Vreeken R.J. Quantitative profiling of oxylipins through comprehensive LC-MS/MS analysis: application in cardiac surgery. Anal. Bioanal. Chem. 2012;404(5):1413–1426. doi: 10.1007/s00216-012-6226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magill G.B., Myers W.P., Reilly H.C., Putnam R.C., Magill J.W., Sykes M.P., Escher G.C., Karnofsky D.A., Burchenal J.H. Pharmacological and initial therapeutic observations on 6-diazo-5-oxo-1-norleucine (DON) in human neoplastic disease. Cancer. 1957;10(6):1138–1150. doi: 10.1002/1097-0142(195711/12)10:6<1138:AID-CNCR2820100608>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 24.Earhart R.H., Amato D.J., Chang A.Y., Borden E.C., Shiraki M., Dowd M.E., Comis R.L., Davis T.E., Smith T.J. Phase II trial of 6-diazo-5-oxo-L-norleucine versus aclacinomycin-A in advanced sarcomas and mesotheliomas. Invest. New Drugs. 1990;8(1):113–119. doi: 10.1007/BF00216936. [DOI] [PubMed] [Google Scholar]
- 25.Rahman A., Smith F.P., Luc P.T., Woolley P.V. Phase I study and clinical pharmacology of 6-diazo-5-oxo-L-norleucine (DON). Invest. New Drugs. 1985;3(4):369–374. doi: 10.1007/BF00170760. [DOI] [PubMed] [Google Scholar]
- 26.DeWald H.A., Moore A.M. 6-diazo-5-oxo-l-norleucine, a new tumor-inhibitory substance.1a preparation of l-, d- and dl-forms1b. J. Am. Chem. Soc. 1958;80(15):3941–3945. doi: 10.1021/ja01548a036. [DOI] [Google Scholar]
- 27.Li M.C., Whitmore W.F., Jr, Golbey R., Grabstald H. Effects of combined drug therapy on metastatic cancer of the testis. JAMA. 1960;174:1291–1299. doi: 10.1001/jama.1960.03030100059013. [DOI] [PubMed] [Google Scholar]
- 28.A clinical study of the comparative effect of nitrogen mustard and DON in patients with bronchogenic carcinoma, Hodgkin’s disease, lymphosarcoma, and melanoma. J. Natl. Cancer Inst. 1959;22(2):433–439. doi: 10.1093/jnci/22.2.433. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan M.P., Beatty E.C., Jr, Hyman C.B., Murphy M.L., Pierce M.I., Severo N.C. A comparison of the effectiveness of standard dose 6-mercaptopurine, combination 6-mercaptopurine and DON, and high-loading 6-mercaptopurine therapies in treatment of the acute leukemias of childhood: results of a coperative study. Cancer Chemother. Rep. 1962;18:83–95. [PubMed] [Google Scholar]
- 30.Eagan R.T., Frytak S., Nichols W.C., Creagan E.T., Ingle J.N. Phase II study on DON in patients with previously treated advanced lung cancer. Cancer Treat. Rep. 1982;66(8):1665–1666. [PubMed] [Google Scholar]
- 31.Lemberg K.M., Vornov J.J., Rais R., Slusher B.S. We’re not “don” yet: Optimal dosing and prodrug delivery of 6-diazo-5-oxo-l-norleucine. Mol. Cancer Ther. 2018;17(9):1824–1832. doi: 10.1158/1535-7163.MCT-17-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamashita A.S., da Costa Rosa M., Stumpo V., Rais R., Slusher B.S., Riggins G.J. The glutamine antagonist prodrug JHU-083 slows malignant glioma growth and disrupts mTOR signaling. Neurooncol. Adv. 2020;3(1):a149. doi: 10.1093/noajnl/vdaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanaford A.R., Alt J., Rais R., Wang S.Z., Kaur H., Thorek D.L.J., Eberhart C.G., Slusher B.S., Martin A.M., Raabe E.H. Orally bioavailable glutamine antagonist prodrug JHU-083 penetrates mouse brain and suppresses the growth of MYC-driven medulloblastoma. Transl. Oncol. 2019;12(10):1314–1322. doi: 10.1016/j.tranon.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baresova V., Krijt M., Skopova V., Souckova O., Kmoch S., Zikanova M. CRISPR-Cas9 induced mutations along de novo purine synthesis in HeLa cells result in accumulation of individual enzyme substrates and affect purinosome formation. Mol. Genet. Metab. 2016;119(3):270–277. doi: 10.1016/j.ymgme.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 35.Krijt M., Souckova O., Baresova V., Skopova V., Zikanova M. Metabolic tools for identification of new mutations of enzymes engaged in purine synthesis leading to neurological impairment. Folia Biol. (Praha) 2019;65(3):152–157. doi: 10.14712/fb2019065030152. [DOI] [PubMed] [Google Scholar]
- 36.Bagnara A.S., Letter A.A., Henderson J.F. Multiple mechanisms of regulation of purine biosynthesis de novo in intact tumor cells. Biochim. Biophys. Acta. 1974;374(3):259–270. doi: 10.1016/0005-2787(74)90247-0. [DOI] [PubMed] [Google Scholar]
- 37.Jones B.R., Schultz G.A., Eckstein J.A., Ackermann B.L. Surrogate matrix and surrogate analyte approaches for definitive quantitation of endogenous biomolecules. Bioanalysis. 2012;4(19):2343–2356. doi: 10.4155/bio.12.200. [DOI] [PubMed] [Google Scholar]
- 38.Ho S., Gao H. Surrogate matrix: opportunities and challenges for tissue sample analysis. Bioanalysis. 2015;7(18):2419–2433. doi: 10.4155/bio.15.161. [DOI] [PubMed] [Google Scholar]
- 39.Fan T.W.M., Lorkiewicz P.K., Sellers K., Moseley H.N., Higashi R.M., Lane A.N. Stable isotope-resolved metabolomics and applications for drug development. Pharmacol. Ther. 2012;133(3):366–391. doi: 10.1016/j.pharmthera.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker S.L., Pandey S., Sime F.B., Stuart J., Lipman J., Roberts J.A., Wallis S.C. A validated LC-MS/MS method for the simultaneous quantification of the novel combination antibiotic, ceftolozane-tazobactam, in plasma (total and unbound), CSF, urine and renal replacement therapy effluent: application to pilot pharmacokinetic studies. Clin. Chem. Lab. Med. 2020;59(5):921–933. doi: 10.1515/cclm-2020-1196. [DOI] [PubMed] [Google Scholar]
- 41.Tan A., Awaiye K., Trabelsi F. Impact of calibrator concentrations and their distribution on accuracy of quadratic regression for liquid chromatography-mass spectrometry bioanalysis. Anal. Chim. Acta. 2014;815:33–41. doi: 10.1016/j.aca.2014.01.036. [DOI] [PubMed] [Google Scholar]
- 42.Gu H., Liu G., Wang J., Aubry A.F., Arnold M.E. Selecting the correct weighting factors for linear and quadratic calibration curves with least-squares regression algorithm in bioanalytical LC-MS/MS assays and impacts of using incorrect weighting factors on curve stability, data quality, and assay performance. Anal. Chem. 2014;86(18):8959–8966. doi: 10.1021/ac5018265. [DOI] [PubMed] [Google Scholar]
- 43.Park H.Y., Kim M.J., Lee S., Jin J., Lee S., Kim J.G., Choi Y.K., Park K.G. Inhibitory effect of a glutamine antagonist on proliferation and migration of VSMCs via simultaneous attenuation of glycolysis and oxidative phosphorylation. Int. J. Mol. Sci. 2021;22(11):5602. doi: 10.3390/ijms22115602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pham K., Maxwell M.J., Sweeney H., Alt J., Rais R., Eberhart C.G., Slusher B.S., Raabe E.H. Novel glutamine antagonist JHU395 suppresses MYC-driven medulloblastoma growth and induces apoptosis. J. Neuropathol. Exp. Neurol. 2021;80(4):336–344. doi: 10.1093/jnen/nlab018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemberg K.M. Abstract 1293: A novel protide purine antimetabolite combines with the prodrug glutamine antagonist JHU395 in preclinical models of Ras-driven sarcomas. Cancer Res. 2021;81(13) Suppl.:1293. [Google Scholar]
- 46.Johnson M.L. Phase 1 and phase 2a, first-in-human (FIH) study, of DRP-104, a broad glutamine antagonist, in adult patients with advanced solid tumors. J. Clin. Oncol. 2021;39(15) Suppl.:TPS3149–TPS3149. doi: 10.1200/JCO.2021.39.15_suppl.TPS3149. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher's website along with the published article.
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author [RR] on reasonable request.