Abstract
More and more in-depth studies have revealed that the occurrence and development of tumors depend on gene mutation and tumor heterogeneity. The most important manifestation of tumor heterogeneity is the dynamic change of tumor microenvironment (TME) heterogeneity. This depends not only on the tumor cells themselves in the microenvironment where the infiltrating immune cells and matrix together forming an antitumor and/or pro-tumor network. TME has resulted in novel therapeutic interventions as a place beyond tumor beds. The malignant cancer cells, tumor infiltrate immune cells, angiogenic vascular cells, lymphatic endothelial cells, cancer-associated fibroblastic cells, and the released factors including intracellular metabolites, hormonal signals and inflammatory mediators all contribute actively to cancer progression. Protein post-translational modification (PTM) is often regarded as a degradative mechanism in protein destruction or turnover to maintain physiological homeostasis. Advances in quantitative transcriptomics, proteomics, and nuclease-based gene editing are now paving the global ways for exploring PTMs. In this review, we focus on recent developments in the PTM area and speculate on their importance as a critical functional readout for the regulation of TME. A wealth of information has been emerging to prove useful in the search for conventional therapies and the development of global therapeutic strategies.
Subject terms: Cancer microenvironment, Oncogenes
Introduction
Tumor is a complex entity composed of multifactors to control its initiation, progression, and treatment response. The self-sufficient growth signals, the insensitive anti-growth signals, the resistance to apoptosis, the limitless replicative potential, the persistent angiogenesis, the capability for invasion and metastasis, the reprogramed cellular metabolism, and the evading immune destruction are known to be major hallmarks of cancer.1,2 These physiologic changes during tumor development suggest a conceptual rationale for the individualized precise treatment and provide clues for the exploration of new therapy strategies. The overarching task of cancer research is to clarify the functional crosstalk of tumor cells and the surrounding physical and cellular environment, also known as the tumor microenvironment (TME). TME embodies (1) angiogenic vascular cells (AVCs) that contain the tube-forming endothelial cells and multiple angiogenic factors; (2) infiltrating immune cells (IICs) that contain both myeloid- and lymphoid-lineage cells, mitogenic growth mediators, and proteolytic enzymes; (3) cancer-associated fibroblastic cells (CAFs) that have diverse functions in different organ-specific TME.2 The multiple factors in TME and their contributions to cancer hallmarks make TME as a new target for personalized diagnostics and therapeutics.
The abundant vascular system in TME provides nutritional supports for tumor growth and development. IICs including granulocytes, lymphocytes, and macrophages participate in the regulation of local inflammatory response. CAFs help the migration of tumor cells from the primary site to the blood or other metastatic sites and provide channels for angiogenesis of endothelial cells.3,4 The molecular composition and spatial heterogeneity of TME match its functions of promoting and inhibiting cancer, and vary dynamically between different tumor types.4 Conventional radiotherapy and chemotherapy will reshape the heterogeneity of TME and further affect the follow-up treatment response.5,6 The most important regulation pattern of cell–cell interaction in TME is the adaptation and resistance to local immune response, including the dual role of TME in promoting and inhibiting tumors by different lymphocytes crosstalk.7 For example, cytotoxic CD8+ memory T cells infiltrated in TME are supported by interferon (IFN-γ) and interleukin-2 (IL-2) released by CD4+ T-helper cell 1 (Th1), which can kill tumor cells and promote good prognosis by recognizing tumor-specific antigens to activate the immune response.7,8 The Th2 cells can promote B-cell response by producing IL-4, IL-5, and IL-13.9 In contrast, Th17 cells produce IL-17A, IL-17F, IL-21, and IL-22 to promote tumor growth.10 Hypoxic environment in TME can trigger abnormal tumor adaptation and increase its invasiveness and resistance to oxygen-dependent therapy.11–13 Therefore, tumor cells themselves affect TME by releasing cytokines and regulatory small molecules, dynamic changing the dialog with immune cells and local microbiota.14 Genomic and proteomic changes of tumor cells play important roles in the reaction of nutrition, growth, immunity, and mechanical stress in TME. Among them, a variety of protein post-translational modifications regulate the interaction of internal and external signals in tumorigenesis and cancer development. The modification of key signal pathway-related proteins in different cells of TME may have multiple levels of fine regulation patterns, so as to participate in the proliferation, activation, and metabolic reprogramming of immune cells, and then affect the outcome of tumor immunotherapy.
Protein post-translational modifications (PTMs) introduce structural changes in existing proteins to participate in multiple biological processes. Other than the coding diversity of human genes by the specific alternative mRNA splicing, protein PTM on the side chains or backbones promotes the increased complexity from genome level to proteome level, which is the key of proteome diversity. One of the categories of PTM is the covalent conjunction of some chemical groups to protein side chains through enzyme catalyzation, and the other is the cleavage of a protein backbone through proteases or autocatalytic cleavage at a specific peptide.15 PTM enzymes can be divided into three subtypes based on their functional specificity; the “writers” are responsible for adding substrates, the “readers” recognize modified proteins to initiate downstream signaling cascade, and the “erasers” are best known for their role in PTMs removing. One site in the same protein may undergo one or more types of modifications. Similarly, one modulator can perform multiple roles. This crosstalk between PTMs may be positive or negative based on their activation or inhibition function of downstream signals.16 PTMs often modulate electrostatic or structural properties of target proteins to change protein–protein interaction.17 Some modifications provide a scaffold or docking site for the interaction of kinases and their specific substrates.18
With the classification by protein signature, PTM occurs on both histone and nonhistone proteins. Histone modifications are also called histone marks, which play a crucial role in chromatin organization and cellular function such as transcription. These marks are often added on lysine residues with multiple types like acetylation, methylation, propionylation, butyrylation, crotonylation, 2-hydroxyisobutyrylation, malonylation, and succinylation.19 The middle-down proteomics workflow using high-resolution mass spectrometry (MS) is helpful to quantify histone H3 marks.20 Some databases like http://dbPTM.mbc.nctu.edu.tw/ and http://ptmcode.embl.de are developed to validate novel PTM type, predict the potential PTM targets, analysis PTM crosstalk, and assess PTM-associated diseases.21,22
A variety of PTMs have greatly expanded the functional field of proteins, which is particularly important in the immune recognition of tumor therapy. In particular, many peptides presented to T cells through histocompatibility complex are post-translational modified. The regulation of this process may affect the selection of therapeutic targets and the effect of the immune response. In addition to the well-characterized protein PTMs like phosphorylation, methylation, ubiquitination, and sumoylation, other highly prevalent yet less-studied PTMs have attracted considerable attentions in recent years. Although more than 300 PTMs have been identified due to the development of technology, only a few have functional research results at the proteome level.23 Comprehensive and systematic PTM analysis is very challenging. This review summarizes several kinds of these emerging PTMs, including the description of proteins to be modified, the function of PTMs, the enzymes that control them, the technologies to study them and the potential therapy targets. According to different types of modified functional groups, the newly reported PTMs with TME remodeling function are divided into the following types: protein acylation modification, lipid-related protein modification, metabolite-related protein modification, and ubiquitin-like small-molecule protein modification. This review generally described the effect of some PTMs on tumorigenesis and cancer development by affecting the regulatory mechanism of growth and metabolism, immune response, and mechanical stress in tumor microenvironment. Direct targeting tumor microenvironment is the most promising method of tumor therapy, which has great basic and clinical research potential. In-depth analysis of PTM of different target proteins in TME will bring new breakthroughs for effectively relieving immunosuppression and promoting the clinical efficacy and wide application of tumor immunotherapy.
Protein-acylation modification
Acyl CoA compounds are important metabolic intermediates in cells and precursors of many biological macromolecules. With the development of research technology and the deepening of research level, the role of protein acylation modification in the regulation of TME has been gradually explored.
Acetylation
Protein acetylation refers to the transfer of the acetyl group from acetyl coenzyme to lysine under the catalysis of acetyltransferase, which was first reported to occur in the lysine-rich region of the N-terminal of histone.24 Subsequently, the modification-related target proteins, acetyltransferases, deacetylases, and their roles in cell biological regulation were gradually reported. From the perspective of epigenetics, there is a clear causal relationship between histone acetylation and gene transcriptional regulation, including acetylation in nonhistone proteins involved in transcriptional regulation.25 Numerous studies have shown that the protein acetylation at lysine residue is mediated by lysine acetyltransferases (KATs) and deacetylation is mediated by deacetylases (KDACs). Thousands of human histone or nonhistone proteins are characterized to be acetylated through high-resolution MS-based proteomics, in combination with the enrichment of acetylated peptides.26 A large number of studies have gone deeper in the characterization of the protein lysine acetylation.27,28 The intermediate products of cell metabolism dominated by acetyl coenzyme A directly regulate protein acetylation.29 Non-acetyl metabolites can also regulate deacetylases, such as the deacetylation of dinucleotide involved in cell electron transport chain catalyzed by NAD+-assisted sirtuin.30 The details will be described in the following text.
Propionylation
Propionylation is another acetylation-like modification identified by MS, which has been well demonstrated in both prokaryotes and eukaryotes. The bromodomain and plant homeodomain (PHD) finger containing protein 1 (BRPF1) belongs to monocytic leukemic zinc finger (MOZ) histone acetyltransferase (HAT) and recognizes histone acetyl lysine marks to affect gene transcription.31 Lysine acetyltransferase 6A (KAT6A) has essential roles in histone acetylation to regulate chromatin organization and function.32 Recently, BRPF1-KAT6 complexes were identified as propionyltransferases both in vitro and in vivo. As the dual targets of both acetylation and propionylation, histone H3K23 is associated with distinct chromatin impact in response to acetyl-CoA and propionyl-CoA, respectively.33 Acetyltransferases p300 and CBP were found to carry out auto-propionylation. Histone deacetylase sirtuin 1 (SIRT1) is responsible for depropionylation of p53 and p300.34 They may regulate cellular metabolism since the substrate propionyl-CoA is derived from odd-chain fatty acid and amino acid catabolism.35
Butyrylation and isobutyrylation
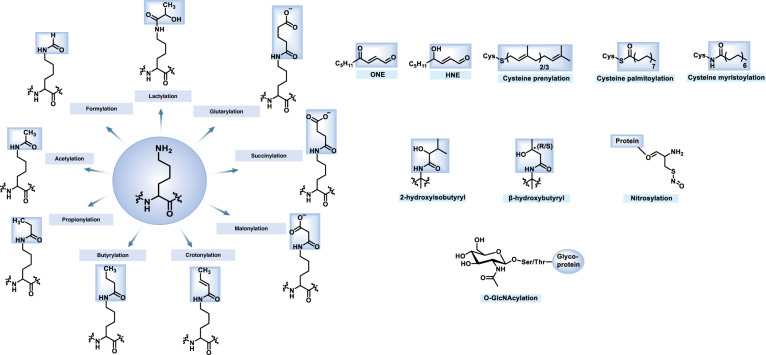
Besides acetyl-CoA and propionyl-CoA, butyryl-CoA also belongs to high-energy molecules to be the modification substrates in lysine side chain formed during the oxidation of fatty acids (Fig. 1). Lysine butyrylation (Kbu) was first discovered by Zhao et al. with synthetic peptides and in vitro enzymatic reactions as a novel lysine modification dependent on n-butyryl-CoA and catalyzed by acetyltransferases.35 Studies demonstrated that p53 can be propionylated and butyrylated in vitro, catalyzed by p300 and CBP.34 This modification can be reversed by the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) with a growth inhibition of neuroblastoma.36 Isobutyryl-CoA is another intermediate derived from branched-chain fatty acid oxidation, leading to lysine isobutyrylation by isobutyryltransferase p300 and HAT1.37,38 Lysine 2-hydroxyisobutyrylation (Khib) in histone H4K8 is abundant and associated with active transcription in male germ cells, opening a new window to study the potential functions of this new modification.39
Fig. 1.
Chemical structures of histone/nonhistone PTMs in this review
Crotonylation
Lysine crotonylation (Kcr) is first identified by Zhao et al. among all core histone proteins including H2A, H2B, H3, H4, and linker histone H1. This modification shows a distinct genomic distribution pattern from acetylation with its specific label property of TSS of active genes.40 The transcriptional coactivator and HAT p300 is reported to have histone crotonyltransferase (HCT) activity. P300-catalyzed histone crotonylation promotes enhanced transcription effect than p300-catalyzed acetylation. In this study, the nucleic/cytosolic acetyl-CoA synthetase enzyme (ACSS2) was implicated to produce intermediate product crotonyl-CoA from the short-chain fatty acid (SCFA) in mammalian cells.41 ACSS2 is overexpressed with the function in acetate uptake and lipid conversion in human cancer. The inhibition of tumor growth and failure of glioma self-renewal were found in ACSS2 deficient mice.42,43 A large number of nonhistone proteins can also be modified by crotonylation in the regulation of subcellular location, cellular composition and function, signal pathways, and biological processes.44 After crotonic acid treatment, serine 46 residue in p53 was identified to be crotonylated other than other protein family members p63 and p73. This directly led to reduced p53 level and consequently abnormality in glycolysis pathway and mitochondrial activity.45 Lysine crotonylation plays an inhibitory role in the development and metastasis of hepatocellular carcinoma.46 Sirtuin family members SIRT1, SIRT2, and SIRT3 are responsible for the removal of histone H3K4 crotonylation in vitro. In addition, SIRT3 selectively targets histone crotonylation and functions as “eraser” to regulate histone crotonylation level and gene expression in living cells.47 Crotonylation is identified preferentially by YEATS (Yaf9, ENL, AF9, Taf14, Sas5) family proteins.48 The co-crystal structure of AF9 YEATS domain in complex with H3K9 acetylation and crotonylation defined a higher affinity of YEATS for crotonyl-lysine, linking histone crotonylation to active transcription.49
β-hydroxybutyrylation and β-hydroxyisobutyrylation
Another kind of modification of histone lysine derived from endogenous short-chain fatty acid metabolism is β-hydroxybutyrylation/β-hydroxyisobutyrylation, also known as 2-hydroxybutyrylation/2-hydroxyisobutyrylation, exists in a variety of bio-fluids in humans. Lysine acetyltransferases p300 is responsible to catalyze both acetylation and 2-hydroxyisobutyration reactions on histones with different activity. A global Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC)-based assay and MS analysis were used to define distinct target protein sequence between p300-mediated acetylation and 2-hydroxyisobutyration. Interesting, most of the β-hydroxyisobutyrylated proteins are glycolytic enzymes, revealing this p300-mediated modification is essential for cellular glucose metabolism regulation.50 Both in vivo and in vitro studies reflect the regulation role of human MYST family acetyltransferase Tip60 in β-hydroxyisobutyrylation at sites H4K8, H4K12, and H4K16. In addition, the removing of β-hydroxyisobutyrylation is mediated by histone deacetylases 2 (HDAC2) and HDAC3.51 However, the Class III sirtuins family member Sirt3 appears to have site-specific activity of β–hydroxybutyrylation in H3K4, K9, K18, K23, K27, and H4K16, which is distinct from the Zn-dependent HDACs.52,53
Malonylation
The property of electron-rich and nucleophilic nature in lysine side chain makes it suitable to undergo covalent post-translational modification, especially bulkier groups carrying negatively charged carboxyl moieties. Lysine malonylation (Kmal) refers to the incorporation of a malonyl group at the ε-amine and this modification was first validated by Zhao et al. in HeLa cells and Escherichia coli (E. coli).54,55 Even the anti-malonylysine antibody is reasonable to detect malonylated proteins, this technology is restricted for tracing the dynamic of malonylation. In another study, an alkyne-functionalized chemical probe was used to profile more than 300 malonylated substrates.56 Abnormal metabolites are associated with many diseases. For example, elevated malonyl-CoA level was found in type 2 diabetic patients as well as type 2 diabetes model db/db mice.57,58 Sirt5 was responsible for demalonylation. Increased protein malonylation was observed in mice lacking Sirt5, which was involved in multiple metabolic networks like glycolysis, gluconeogenesis, urea cycle, and fatty acid β-oxidation.59
Succinylation
Protein succinylation reaction was first identified in bacterial metabolism.60 Thousand of succinylation sites were mapped in diverse organisms including bacteria (E. coli), yeast (S. cerevisiae), human (HeLa) cells, and mouse liver tissue through high-resolution quantitative MS and antibody-based affinity enrichment followed by strong cation exchange (SCX) chromatography.61 Histone succinylation was validated by MS/MS of synthetic peptides, HPLC co-elution, and in vivo isotopic labeling with the enzyme-mediated cofactor succinyl-CoA. This PTM is high conserved from Saccharomyces cerevisiae, Drosophila S2, mouse embryonic fibroblast, and human cells.55 Lysine succinylation may cause the change of charges from 0 to −2, the same effect caused by protein phosphorylation at serine. It is expected that succinylation could have important cellular functions.62 A recent study demonstrated SIRT5 is the major deacylases for lysine desuccinylation.63
Glutarylation
The type of lysine glutarylation (Kglu) was validated as an evolutionarily conserved PTM, in which SIRT5 showed deglutarylation activity both in vitro and in vivo.63,64 Carbamoyl phosphate synthetase 1 (CPS1), the key enzyme important for ammonia-detoxifying urea cycle, was identified to be a glutarylated substrate with inhibit enzyme activity after glutarylation.63,65 Other than MS, the novel bioinformatics tools, the biased support vector machine algorithm, and machine-learning scheme are discovered to predict the potential glutarylation sites and identify physiochemical and sequence-based features.66–70 Glutarylation in mitochondrial proteins suppresses glutamate dehydrogenase (GDH) activity and protein interactions.71
Lipid-related protein modification
Lipids are fundamental structural components of cellular membranes, acting as barriers to separate oneself from the external environment and to divide cells into different functional areas. The interaction of some proteins with specific lipid molecules and the covalent modification of lipid molecules of proteins are two mechanisms of lipid-dependent cell signal transduction in cell. In humans, some proteins undergo more complicate modification such as the addition of lipids to increase plasma membrane docking or regulate protein complex formation. Lipidation modification includes C-terminal glycosylphosphatidylinositol (GPI) anchor, N-terminal myristoylation, S-palmitoylation, and S-prenylation. In this section, we summarize three lipid modification types including myristoylation, palmitoylation, and prenylation.
Myristoylation
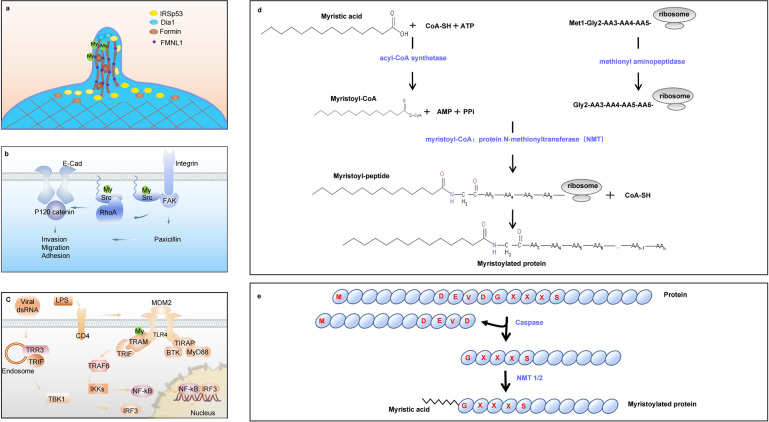
Protein myristoylation refers to the attachment of a 14-carbon fatty acyl group myristic acid to target substrates via an amide bond, which is irreversible and mediated by the eukaryotic enzyme myristoyl-CoA: protein N-myristoyltransferase (NMT).72 Two NMTs are found in human cells (NMT1 and NMT2) according to their protein size.73 This modification exits widely in viral, yeast, plant, and eukaryotic proteins to mediate signaling cascaded.72,74,75 Many enzymes in metabolic pathways undergo this modification, supporting its importance and specific regulatory roles. As myristic acid is a hydrophobic group, the myristoylated proteins are inserted into lipid rafts in the plasma membrane, including endoplasmic reticulum (ER), Golgi apparatus, mitochondria, and nuclear to regulate diverse cellular functions.76 A number of myristoylation sites related to tumors have been identified (Table 1).
Table 1.
Identified protein myristoylation sites related to tumors
| Target | Function in cancer | Ref. |
|---|---|---|
| LAMTOR1 | Inhibits lysosomal degradation. | 588 |
| FMNL1/3 | Controls cytoskeleton remodeling and cellular morphological changes. | 202,465 |
| SFKs | Promotes membrane attachment and Src-family kinase activity. | 256 |
| TRAM | Enhances the membrane attachment for LPS and TRL7 signaling. | 432 |
| Fyn | Promotes subcellular location in immunological synapses and interaction with TCRζ chain. | 459,460 |
| c-Src | Regulates the activity of FAK in cell adhesion and extracellular matrix. | 72,461 |
| Lck | Promotes TCR signal cascade. | 448 |
| eNOS | Regulates enzyme activity and signal transduction. | 470 |
| Fus1 | Facilitates protein docking to the mitochondrial membrane. | 505 |
| BID | Improves its insertion into mitochondria membrane to enhance apoptosis. | 506 |
| PAK2 | Promotes protein location in membrane ruffles and internal membranes. | 507 |
| SAMM50, TOMM40, MIC19, and MIC25 | Promotes mitochondrial localization and membrane binding. | 589 |
| AMPK | Helps the removal of damaged mitochondria by autophagy. | 328 |
| AMPK | Enhances the β oxidation of fatty acids. | 590 |
| gelsolin | Regulate nuclear fragmentation or cytoskeletal remodeling during apoptosis. | 301 |
| DIC2 | ||
| Bap31 | ||
| MACF | ||
| YTHDF2 |
In addition, N-glycine myristoylation-mediated protein–protein and protein-membrane interactions change the subcellular localization of targeted proteins across various cellular processes, suggesting a promising therapy target77 (Fig. 2). A protease from macrophage extracts was reported to cleave myristoylated alanine-rich C kinase substrate (MARCKS), indicating myristoylation can be a substrate for protease.78 The demyristoylation of MARCKS was also observed in cytoplasmic fraction of synaptosomes from the bovine brain with the property of ATP and coenzyme A (CoA) dependence.79–81 Moreover, bio-orthogonal probes and chemical tools, especially the non-radioactive-labeled fatty acids like ω-azido or ω-alkynyl myristate, are effective and easily applicable to identify N-myristoylated proteins.82–85 Recent studies have found the phase separation-related myristoylationon of tumor growth, expanding the mechanism research and treatment direction of lipid-associated protein modification. The oncogenic activity of lysine methyltransferase EZH2 relays on methylation and further phosphorylation of its substrate STAT3.86,87 The hydrophobic effect of EZH2 myristoylation promotes droplets formation to separate from the surrounding solution, which is conducive to the interaction with STAT3 and downstream growth regulation.88 N-Myristoylation Predictive Tools are developed based on the PROSITE motif in substrate proteins89 (http://mendel.imp.univie.ac.at/myristate/), the difference between myristoylated and non-myristoylated proteins90 (http://www.expasy.org/tools/myristoylator/myristoylator.html) and pattern scanning91 (http://www.isv.cnrs-gif.fr/terminator3/index.html).
Fig. 2.
Schematic representation of protein N-myristoylation and function. a The myristoylation of formin family protein FMNL1 leads to membrane association, membrane trafficking and bleb formation. b The N-terminal myristoylation is essential for the membrane attachment and kinase activity of Src. c The myristoylated TRAM tethers it to the membrane, serving as a prerequisite for LPS signaling. d Co-translational N-myristoylation. The initiator methionine is removed by methionine aminopeptidase 2 and myristic acid is transferred to the N-terminal glycine residue by NMT. e Post-translational N-myristoylation. Some proteins are cleaved by a protease to facilitate a myristic acid is covalently attached to glycine residue by NMT
Palmitoylation
Protein palmitoylation, often occurs on cysteine and also known as S-palmitoylation, is the addition of a 16-carbon palmitoyl group to target proteins through thioester bonds. This reaction is reversibly catalyzed by palmitoyl-CoA palmitoyltransferases or non-enzymatic acylation and the converse is depalmitoylase or non-enzymatic hydrolysis.77 The conserved family of protein acyltransferases (PATs) are responsible for protein palmitoylation. They are firstly discovered encoded by the ERF2 and ERF4 genes targeting Ras GTPase.92 In mammalian cells, 23 members of the Golgi-apparatus-specific protein family with the aspartic acid-histidine-histidine-cysteine (DHHC) zinc finger domain are identified to have PAT activity.93,94 In addition, DHHCs generally show substrate specificity and more than one DHHC modify a palmitoylated substrate.94–97 Most mutations of DHHC enzymes are associated with neurodegenerative diseases like Huntington’s, Alzheimer’s, X-linked mental retardation, and schizophrenia, as well as other developmental defects and cancer.77 For example, the depletion of palmitoyltransferase DHHC5 inhibits NSCLC cell growth, colony formation, and cell invasion, indicating the potential of oncogene function of DHHC5.98 Studies provided a dynamic loop between palmitoylation and depalmitoylation, which has been emerging as an important regulation mechanism for cellular homeostasis. Two acyl-protein thioesterase-1 APT1 (also called LYPLA1) and APT2 (also called LYPLA2) catalyze depalmitoylation of palmitoylated proteins anchored in the membrane.99 The radioactive-isotope-labeled palmitic acid and the following immunoprecipitation are effective methods to detect protein palmitoylation.100 Later, the higher sensitive bio-orthogonal probes, which contain a terminal azido or alkynyl group, are developed to obtain precise detection.101 Then a three in vitro steps of acyl-biotin exchange (ABE) assay were developed to detect palmitoylation under any conditions.102
Prenylation (farnesylation + geranylgeranylation)
Protein prenylation refers to the addition of different isoprene unit to C-terminal cysteine residues. The attachment of farnesyl diphosphate (FPP) forms farnesylation and the attachment of geranylgeranyl diphosphate (GGPP) forms geranylgeranylation. These two different sizes of polyisoprenoid groups were identified in Chinese hamster ovary (CHO) cells.103 Furthermore, the geranylgeranyl-modified proteins were discovered in HeLa cells through the analysis of radioactive mevalonic acid-labeled fragments.104 About 2% of total cellular proteins are prenylated in eukaryotic cells with the majority are geranylgeranylated proteins.105 It was reported that protein geranylgeranylation is required for cell-cycle transition from G1 to S phase, but not farnesylation.106
Farnesyltransferase (FT), geranylgeranyltransferase (GGT-1), and Rab Geranylgeranyl transferase (RGGT or GGT-2) are the major protein prenyltransferase.77 The first FT was characterized from rat brain cytosol with the function of transferring 15-carbon farnesyl group farnesyl moiety from farnesyl pyrophosphate to p21RAS cysteine residue.107 In addition, GGT-1 mainly transferred a 20-carbon geranylgeranyl group to substrates and has different preference with FT.108 GGT-2 functions in the linkage of 20-carbon geranylgeranyl groups to cysteine residues in Rab proteins.109 Some website tools are available for protein prenylation prediction such as Prediction Suite (http://mendel.imp.ac.at/PrePS/). Even the thioether bond is stable between farnesyl or geranylgeranyl groups and cysteine, a kind of prenylcysteine lyase located in lysosomal was identified to cleave prenylated proteins.110,111
Metabolism-associated protein modification
Lactylation
According to the restricted oxygen supply in the tumor microenvironment, hypoxia becomes a key hallmark in solid tumors and also promotes tumor progression. Tumor cells have increased glucose uptake and consistent lactate production even in the presence of oxygen (Warburg effect).112 This transition from oxidative phosphorylation (OXPHOS) to an altered glycolysis is regulated by a large number of oncogenes and tumor suppressor genes. Lactate in tumor cells comes from two sources: the pyruvate conversion in the last reaction of glycolytic pathway, and the conversion of phosphoenolpyruvate (PEP) by a tumor-specific isoform of pyruvate kinase (PK) M2.113 Recent data suggest that the lactate may be involved in many biological processes, including epigenetic regulation, intrinsic inflammatory mediation, immunologic escape, tumor cell motility and migration, O2 independent angiogenesis, radioresistance or chemoresistance, acidic or hypoxia tumor microenvironment, and extracellular matrix remodeling.113 The detailed mechanisms of lactate regulation in tumor development and immune response are summarized previously113–119 and we discuss the newly identified protein lactylation in this review. Lysine lactylation was identified by Zhao et al. after the stimulation of lactate with high-performance liquid chromatography (HPLC)-tandem mass spectrometric (MS/MS) analysis.120 Lactic acid from extracellular glucose or hypoxia induced glycolysis promotes histone lactylation and directly regulates gene transcription.120 In addition to the regulation function of lactic acid as a metabolite intra- and extracellular signal molecular on innate and adaptive immunity in the microenvironment,121 protein lactylation plays a role in gene transcription and immune adaptation. The details will be described later.
O-GlcNAcylation
The covalent attachment of glycans to proteins is called protein glycosylation, which is a common contribution to structure and modification diversity.122 Protein GlcNAcylation refers to the addition of single N-acetylglucosamine residue O-linked to the hydroxyl group of serine and threonine residues of multiple nuclear and cytosolic proteins, including transcription factors, tumor suppressors, nuclear pore proteins, kinases, and cytoskeleton proteins. These modified proteins are involved in signal pathway, metabolism, transcriptional, DNA replication, protein trafficking, and other cellular processes.123,124 Multistep radioactivity-based approach and quadrupole time-of-flight (Q-TOF) MS are used to identify the modification sites.125 Since it was first identified on lymphocyte proteins, it was paid more and more attention to support its dynamic regulatory function.123,126 A UDP-N-acetylglucosamine: peptide N-acetylglucosaminyltransferase (O-GlcNAc transferase, OGT) and a neutral and cytoplasmic N-acetyl-β-d-glucosaminidase (O-GlcNAcase, OGA) are responsible for the addition and removal of O-GlcNAc to target proteins, respectively.127,128
Nitrosylation
Nitric oxide (NO) is an important biological second messenger in the regulation of cardiovascular pharmacology, cell migration, inflammatory response, and neurobiology.129 This highly reactive molecule is synthesized by NO synthase (NOS) with l-arginine and molecular oxygen as substrates.130 Protein S-nitrosylation refers to the attachment of an NO group to a reactive cysteine residue, leading to the formation of an S-nitrosoprotein (SNO-protein).131 Dysregulated SNOs are associated with diseases through the influence of nitrosothiol formation, processing, and degradation.132,133 It has become clear that protein S-nitrosylation and denitrosylation play an important role in protein–protein interaction, subcellular localization, and degradation, thus regulating a wide range of cellular processes, including transcription, ion-channel activity, metabolism, cellular redox maintenance, apoptosis, and other kinds of PTM.134
Ubiquitin-like small-molecular protein modification
There exist one kind PTM refers to the conjugation of small peptides by a reversible cycle on the substrates such as the well-known small ubiquitin and ubiquitin-related modifier family in eukaryotic cells. Protein modification by this small-molecule conjugation influence all parts of cellular processes such as mitochondrial function, chromosome remodeling, DNA repair, transcriptional regulation, nuclear transport of intranuclear proteins, heterochromatin construction, plasma membrane association, cytoskeleton dynamics, and protein aggregation.135,136 In addition, humans have a set of ubiquitin-like proteins (UBLs) that covalently modify target proteins.137,138 These UBLs, including ubiquitin-fold modifier 1 (UFM1), Neural precursor cell expressed, developmentally downregulated 8 (Nedd8), small ubiquitin-like modifier 1 (SUMO-1), SUMO-2, SUMO-3, Fau gene-encoded ubiquitin-like protein (FUBI), homologous to ubiquitin 1 (HUB1), interferon-stimulated gene 15 (ISG15), The human leukocyte antigen F-associated transcript 10 (FAT10), ubiquitin-related modifier 1 (URM1), autophagy-related 8 (ATG8), and ATG12, are found in prokaryotes, archaea, yeast, plants, and mammals.139 Moreover, the Ub domain proteins (UDPs) also have a role linking the ubiquitination with the functions of the proteasome. These include the proteasome-associated deubiquitinating enzyme UBP6, Rad23, the DSK2-relative PLIC2/Chap1, BAT3/Chap2, BAG-1, elongin B, and parkin.140 A multistep enzymatic cascade, made up of ATP-dependent activation enzyme E1, binding enzyme E2 and ligase E3, are required in the covalent addition of ubiquitin-like small-molecular protein via the isopeptide bonds between C-terminal diglycine motifs and ε-amino groups corresponding substrates.141 Several E1, dozens of E2, and hundreds of E3 are encoded in the human genome.142–144 As a reversible process, the deubiquitinases (DUBs; also known as deubiquitylating or deubiquitinating enzymes) can remove the UBLs modification from target proteins and break the ubiquitin chains.145,146 These DUBs are divided into six families: ubiquitin-specific proteases (USPs), herpesvirus tegument USPs (htUSPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), the family of Josephins and the JAB1/MPN/MOV34 proteases (JAMMs).139 Many of these enzymes in UBL modification are identified by labeling dependent and independent quantitative proteomics, such as the reported stable isotope labeling by amino acids(SILAC) in cell culture through metabolic incorporation,147,148 isobaric tags for relative and absolute quantification (iTRAQ),149 and the absolute quantification (AQUA) strategy.150,151
UFMylation
UFM1 is a 9.1-kDa protein with a similar structure to ubiquitin.152 It conjugates into target proteins through E1, E2, and E3 enzymes. These refer to the UFM1-activating enzyme (ubiquitin-like modifier-activating enzyme 5; UBA5), the UFM1-conjugating enzyme 1 (UFC1), and the UFM1-specific ligase 1 (UFL1), respectively.153 Protein UFMylation is reversible and conserved among nearly all the eukaryotic organisms, except for yeast and is involved in the homeostasis of the ER stress response, cell differentiation, and NF-κB binding protein-related cell-cycle control.154 UFM1 is cleaved from target proteins by the UFM1-specific proteases (UfSPs).155 Recent studies have demonstrated that UFMylation plays a role in the regulation of ER stress,156 vesicle trafficking,157 cell-autonomous erythroid differentiation,158 hematopoiesis and development,159 β-oxidation of fatty acids,160 and G-protein-coupled receptor (GPCR) biogenesis.161
Neddylation
With the highest amino acid similarity to ubiquitin (identical 58%) of NEDD8 (neural precursor cell-expressed developmentally downregulated 8), protein neddylation is widely studied in cancer. This modification includes the E1 NEDD8-activating enzyme (NAE, composed of NAE1 (APP-BP1) and Uba3 heterodimer), the E2 NEDD8-conjugating enzyme Ubc12 (also known as Ube2m) and Ube2f, and the RING finger domain-containing E3s.162 The Cullin family (Cul1–5, Cul7), PARC, p53/p73/BCA3/VHL, MDM2, and EGFR are substrates of neddylation and functions in the regulation of a set of cellular processes, including cell-cycle inhibition, HIFα degradation, cytokine modulation, oxidative stress-response pathway, DNA replication and nucleotide excision repair, p53 family location and antiapoptotic function of oncogenes.162 In addition to Cullins, the cytoskeleton was another target of Nedd8 conjugation to facilitate the transition from meiosis to mitosis in Caenorhabditis elegans.163 Deneddylation is mediated by the Jab1/MPN domain metalloenzyme (JAMM) motif in zinc metalloprotease COP9 signalosome (CSN) and cysteine protease NEDP1 through conformational changes to promote rapid remodeling of cellular neddylation and deneddylation cycle.164,165 The HEAT repeats protein CAND1 (Cullin-associated and neddylation-dissociate 1) inhibits neddylation through forming a ternary complex with CUL1 and ROC1 and disrupting the linkage between NUB1 (NEDD8 ultimate buster 1) and 26S proteasome and the latter proteasomal-degradation pathway.166–169 Other deubiquitinlase showed deneddylases activity, including ubiquitin-specific peptidase 21 (USP21), deubiquitylating enzyme Ataxin-3 (ATX3), P. falciparum deubiquitinating enzyme PfUCH54, ubiquitin C-terminal hydrolase L1 (UCH-L1), and UCH-L3.162,170,171
The biological effect of PTMs on tumor cells in TME
Chromatin organization and gene transcription
Histones are building blocks of nucleosome, the fundamental part of chromatin. The unstructured N-terminal tail offers a massive opportunity for a large number of PTM. Histone PTM influence various biological processes through two major mechanisms. First, different PTM regulate the interaction between nucleosomes with other nonhistone proteins, leading changes in downstream functions like transcription, replication, and repair. Second, PTM can affect the interaction between nucleosomes with adjacent DNA through the changed properties like net charge, hydrogen bonding, size, or hydrophobicity, thus altering higher-order chromatin structure and DNA-based biological function.172
According to the chemical structures and properties, propionylation (three-carbon molecule: C3) and butyrylation (C4) are the most similar PTMs to acetylation (C2). The site-specific antibodies are effective to characterize propionylation and butyrylation. Histone PTMs at different sites are linked with activation or suppression of transcription through chromatin organization. Lys 23 of histone H3 (H3K23) is detected to be propionylated in the leukemia cell line catalyzed by the histone acetyltransferase p300, promoting an active chromatin structure to regulate transcription even with different dynamics of acetylation.173 Recently, BRPF1 (bromodomain- and PHD finger-containing protein 1) were reported to activate lysine acetyltransferase 6A (KAT6A) and KAT6B, leading to the propionylation of histone H3K23 in vitro and in vivo.33 Lys 14 of histone H3 (H3K14) is considered to be a hallmark of transcriptional activation by the directed recruitment of basal transcription factor TFIID with the demethylation of transcription co-repressor ZMYND8.174,175 A MS analysis identified propionylation and butyrylation in histone H3K14 with a strong linkage with transcriptional activation. Both p300 and the GNAT family HATs, GCN5 and PCAF have acyltransferase activity both in vitro and in vivo.176 The butyrylation activity of p300 was also confirmed in H4K5 and H4K8 and histone butyrylation was observed to directly stimulate gene transcription.177 The isobutyrylation (2-methylpropionylation) is an isomeric structure of butyrylation, taking intermediate product f isobutyryl-CoA generated from the valine metabolism pathway. KAT enzymes, particularly the acetyltransferase p300 and HAT1, are identified to catalyze both lysine propionylation and lysine isobutyrylation to mediate histone H4 isobutyrylation.38 In addition to CBP and p300, another histone acetyltransferases MOF was also showed HCT activity linking with TGFβ-induced transcriptional activation.178 Recently, class I HDACs were demonstrated to possess histone decrotonylation activity in cells. According to the transcription level of six housekeeping genes by qRT-PCR, this histone decrotonylation could repress global transcriptional repression through the disruption of promoter recruitment of crotonylation reader proteins.179 Histone lysine β-hydroxyisobutyrylation is identified at multiple sites in response to β-hydroxybutyrate level with cofactor b-hydroxybutyryl-CoA. Starvation-induced Kbhb at H3K9 may alter chromatin structure to form an active transcription pattern.180 In another study on mice and cells, histone3-lysine9-β-hydroxybutyrylation (H3K9bhb) was induced by fatty acid metabolism-associated β-hydroxybutyrate to promote the expression of growth factor BDNF, thereby regulation depression.181
Histones are identified to be glutarylated in human Hela cells, especially the histone H4 at site Lys91 (H4K91glu). The evolutionarily conserved modification of H4K91glu may inhibit the assembly of H2A-H2B dimers to the H3-H4 tetramer to form histone octamer. The release of H2A-H2B from nucleosome was also observed after glutarylation with a fluorescence resonance energy transfer (FRET) assay.182 Moreover, the mutation of H4K91E, which mimic H4K91glu, resulted in significantly delayed S and G2/M cell cycle, increased sensitivity of DNA damage molecules methyl methanesulfonate (MMS), hydroxyurea (HU), and UV radiation, as well as a less compact chromatin signature.182 Through the screen of all HDACs (HDAC 1–11, sirtuin 1–7) for lysine deglutarylation activity, Sirt5 was found to be a deglutarylase on mitochondrial proteins.63 Even Sirt5 had no function in H4K91 glutarylation, SIRT7 examined the histone deglutarylase activity both in vitro and in cells upon chromatin condensation.182 KAT2A, also known as GCN5, belongs to HAT family. 2-Oxoglutarate dehydrogenase (OGDH) is the E1 subunit of α-ketoglutarate dehydrogenase (α-KGDH) complex, that catalyzes α-ketoglutarate (α-KG) conversion into succinyl-CoA.183 After the interaction between KAT2A and OGDH, the binding affinity of succinyl-CoA was observed, leading to a significant histone H3 succinylation.184 In addition, this interaction also promoted H3K79succ and H4K91glu with the co-factors succinyl-CoA and glutaryl-CoA, respectively. The various function of KAT2A in histone acetyltransferase, succinyltransferase, or glutaryltransferase have highly suggested the tight link between cellular metabolism and epigenetic regulation.182
Histone O-GlcNAcylation regulates chromatin configuration to influence gene expression and DNA repair. Histones and other proteins responsible for DNA assembly are found to be O-GlcNAcylated in vivo, suggesting the role of O-GlcNAcylation as part of the histone code.185 GlcNAcylation of histone H3K4 methyltransferase 5 (MLL5) at Thr440 promote its methylation state, leading to a transcriptionally active chromatin. Histone H2B was reported to serve as an OGT substrate in response to serum glucose stimulation in S112 site. This histone GlcNAcylation may facilitate transcription considering it was located near the transcription start site of many genes.186 Through co-immunoprecipitation and MS analysis, ten-eleven translocation (TET) enzyme TET2 was identified directly interacting with OGT at transcription starting sites (TSS). This interaction not only increases histone O-GlcNAcylation but also regulate gene transcription.187 Moreover, the induction of DNA double-strand breaks (DSBs) increased histone H2B O-GlcNAcylation both in homologous recombination (HR) and non-homologous end-joining (NHEJ) pathway. Functionally, O-GlcNAcylated H2B bind and recruited DNA damage repair protein Nijmegen breakage syndrome 1 (NBS1) and regulated NBS1 foci formation.188
The transcriptional activity of p53 and its homolog TAp73 is negatively regulated by neddylation. The neddylated MDM2 RING finger E3-ubiquitin ligase promotes p53 neddylation to abrogate its transcription function, without a significant influence on MDM2-mediated p53 degradation.189 The p53 family member p73 contains various isoforms and the full length (TAp73) can induce cell-cycle arrest and apoptosis.190 TAp73 was covalently neddylated in an MDM2-dependent manner and deneddylated by NEDP1. TAp73 neddylation promoted its location in the cytoplasm and attenuated transactivation function.191 In addition to MDM2, the E3-ubiquitin ligase F-box only protein 11 (FBXO11), is also responsible for p53 neddylation and suppression of p53 transcriptional activity.192 Ribosomal proteins (RPs) have been identified to interact with MDM2 and negatively regulate p53 under stress signals. Recently, RPL11 are identified to be neddylated in a MDM2-depended manner to avoid destabilization with proteomic approach.193 Moreover, the interaction with MDM2 and the related neddylation of RPL11 triggered a rapid and transient recruitment at promoter sites of p53-regulated genes under actinomycin D (ActD) induced nucleolar stress. This reversed MDM2-mediated p53 transcriptional repression.194 The E2F transcription factor family regulates cell proliferation, cell-cycle transition, differentiation, and apoptosis in response to DNA damage as either activators or repressors.195 Protein interaction and various PTMs regulation are important for E2F1 transcription activity.196 E2F1 was methylated by Set9 at Lys185 to prevent its accumulation at DNA damage sites and downstream gene p73 activation. LSD1 functioned in demethylation of E2F1 to rescue its apoptotic function.197 The phosphorylation of E2F1 was mediated by checkpoint kinase 2 (Chk2) after DNA damage, leading to protein stabilization and proapoptotic transcriptional activation.198 The acetyltransferase p300 is responsible for acetylation and ubiquitination of E2F1.199 E2F1 was later identified to be a new substrate for neddylation, with a negatively regulated transcriptional activity and DNA binding.200
Protein–protein interaction
More and more studies have demonstrated that protein lipidation is essential for protein interactions. The functional role of myristoylation has been reported since many N-myristoylated proteins were key players in various cellular processes. According to the crystal structure of Ca2+-CaM binding with myristoylated peptide derived from N-terminal domain of CAP-23/NAP-22, the function of myristoylation in protein–protein interaction was first defined.201 The inhibition of myristoylation of Formin family proteins FMNL1 and FMNL3 led to cytoskeleton remodeling and later cellular morphological changes.202 Human SOS (hSOS1) is a Ras guanine-nucleotide exchange factor and this activity is highly dependent on protein prenylation.203 The guanine-nucleotide exchange activity appeared on prenylated K-Ras(4B) but not on unprocessed K-Ras(4B) under the stimulation of growth factors.204 The geranylgeranylation of RhoA regulates its interaction with GDP-dissociation inhibitor (GDI) and GDP-dissociation stimulator (GDS).205 The prenylated RhoA also showed a significant interaction with IQ-motif-containing GTPase-activating protein IQGAP1 to regulate breast cancer cell proliferation and migration.206
Recent work revealed protein–protein interaction may be inhibited or activated by O-GlcNAcylation. O-GlcNAcylation modification of transcription factor Sp1 regulated transcription by decreasing its interaction with TATA-binding-protein-associated factor (TAF110) both in vivo and in vitro.207,208 In addition, O-GlcNAcylated serine/threonine-rich region of Sp1 destroys its association with heterotrimeric transcription factor NF-YA.209 The increased O-GlcNAcylation level significantly interrupts the interaction between coactivator Pgc1α and transcription factor PPARγ.210 In order to promote transcription, the N-terminal region of STAT5 was identified to be glycosylated to bind the coactivator of transcription CBP.211 The host cell factor C1 (HCF-1) was reported to recruit OGT to O-GlcNAcylate PGC-1α. This modification increased its interaction with deubiquitinase BAP1 to protect PGC-1α from degradation, thus facilitating downstream transcriptional activation.212 In another example, O-GlcNAcylation of the retinoblastoma-susceptibility gene product (pRB) promoted the binding and inhibition of f transcription factor E2F1.213
Cancer signal cascade
Ketone bodies (β-hydroxybutyrate, acetoacetate, and acetone) serves as a metabolic regulator in glucose utilization, alanine, and non-esterified fatty acid supply, participating in histone PTM in many diseases, including cancer.214,215 β-hydroxybutyrate (BHB) and acetoacetate are the major ketone bodies so that it is important to study BHB-mediated post-translational modification in cancer. The intermediate product β-hydroxybutyrate has been reported to increase “stemness”-associated gene transcription for promoting tumor growth and metastasis.216 The p53 has long been studied in human cancer as a multifunctional transcription factor in the regulation of cell-cycle arrest, proliferation, senescence, aging, apoptosis, autophagy, ferroptosis, cell metabolism, and oxidative balance maintenance.217–224 In addition to histones, the only nonhistone protein with β-hydroxybutyrylation modification is p53. This modification occurs at sites 120, 319, and 370 of p53 lysine after the induction of BHB and is catalyzed by CBP/P300. The acetylation of p53 was reduced after β-hydroxyisobutyrylation, leading to inhibited p53 activation, weakened cell growth arrest, and apoptosis.225 Moreover, β-hydroxybutyration of p53 reduced its acetylation and expression of downstream factors p21 and PUMA, inhibiting cell growth arrest and apoptosis-mediated functions.225
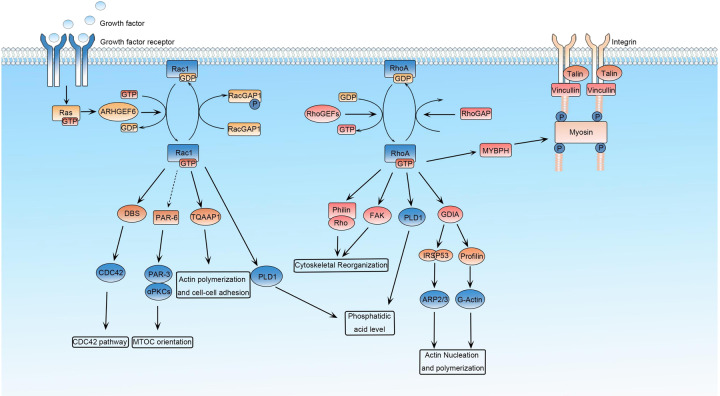
It was found that the lysine at position 222 of lactate dehydrogenase A (LDHA) was recognized and highly succinylated by succinyltransferase CPT1A. The stability of succinylated LDHA was maintained to promote cell growth and invasion by reducing the degradation of ubiquitinated LDHA.226 Glutamine (Gln) can be converted to α- Ketoglutarate enters the tricarboxylic acid (TCA) cycle and plays a key role in synthesizing intermediates of mitochondrial metabolism, maintaining tumor cell growth, and signal regulation.227 The two glutaminase isoforms are liver-type (GLS2) and kidney-type (GLS), in which the site 311 of GLS is activated by succinylation to resist oxidative stress and promote tumor survival by increasing glutamine decomposition and NADPH/glutathione production.228 The growth of renal clear cell carcinoma can be regulated by the interaction between succinate dehydrogenase complex subunit A (SDHA) and SIRT5 and the level of succinylation.229 Many small G-proteins, including Ras, Rho, and Rac families, are prenylated with farnesyl or geranylgeranyl groups to help their membrane location. Several kinds of GTP-binding proteins acting as a molecular switch between active GTP-bound and inactive GDP-bound state are called Rho GTPase. The major function of Rho GTPases is the regulation of actin cytoskeleton and various corresponding cellular processes like cytokinesis, phagocytosis, pinocytosis, cell migration, morphogenesis, and axon guidance.230 In addition, the GTPase also regulate several biological pathways, including transcription factor nuclear factor κB (NF-κB), c-Jun amino-terminal kinases (JNKs) and mitogen-activated protein kinases (MAPKs) pathways, the phagocytic NADPH oxidase complex, G1 cell-cycle progression, the assembly of cadherin containing cell–cell contacts, polarized growth and mitogen-activated protein morphogenesis.231–236 The Rho family members Rac1, RhoA, and CDC42 were reported to be geranylgeranylated and Ras was reported to be farnesylated.237 (Fig. 3) Apoptosis may induce mitochondrial membrane permeabilization (MMP) to release catabolic hydrolases and activators or other proapoptotic molecules, leading to cell death.238,239 The most important regulators of apoptosis are the Bcl-2 protein family, which contains opposite function of antiapoptotic Bcl-2-like proteins (Bcl-2, Bcl-xL, Bcl-w, Mcl-1, and A1/Bfl-1), proapoptotic Bax-like proteins (Bax, Bak, and Bok/Mtd), and proapoptotic BH3-only proteins (Bid, Bim/Bod, Bad, Bmf, Bik/Nbk, Blk, Noxa, Puma/Bbc3, and Hrk/DP5).240,241 High levels of Bcl-2, Bcl-XL, and Mcl-1 were observed in multiple myeloma (MM) and responsible for cell viability and drug resistance.242,243 A post-translational geranylgeranyl lipid modification in C-terminal is essential for anchoring GTPase in the membrane. Other biological functions are found with specific PTM inhibitors. The enzyme 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) inhibitor lovastatin and geranylgeranyltransferase inhibitor (GGTI-298) caused dose-dependent apoptosis in human umbilical vein endothelial cells (HUVECs) through the activation of p53 and other proapoptotic proteins.244 In another study with myeloma cell lines and patient samples, the addition of GGTI-298 induced apoptosis through the downregulation of Mcl-1 protein expression, the disruption of the mitochondrial transmembrane potential, and cytochrome c release.237 The same results were observed in lymphoma cell lines and purified tumor cells from lymphoma patients after the inhibition of prenylation by lovastatin, the farnesyltransferase inhibitor (FTI-277), and GGTI-298.245 In adult T-cell leukemia (ATL), protein geranylgeranylation inhibition suggested anti-proliferative and apoptotic effects.246 Therefore, protein prenylation is a key event in the regulation of cancer cell survival. Ras protein family, including K-Ras, H-Ras, and N-Ras, are activated after GTP binding to regulate signal transduction from the extracellular environment to intracellular hemostasis. Ras activates several downstream signal pathways including Raf-MEK-ERK cascade, PI3K/Akt, and MAPK pathways.247–250 A high frequency of Ras mutation occurs in human cancer with suppressed Ras GTPase activity and constant GTP-binding state.251 This results in permanent signal transduction, leading to tumor cell growth, apoptosis, metabolism, and other biological processes.252 Lipid modifications are commonly occurred in the C-terminal of Ras proteins of yeast and mammalian cells, causing the attachment of Ras proteins with inner surface of the plasma membrane.253 Protein farnesylation occurs on the C-terminal CAAX motif to remove -AAX amino acids and leave C-terminal cysteine (Cys186). Studies found all Ras proteins undergone polyisoprenylation for their membrane association and function activation during human malignancy.254,255
Fig. 3.
Ras is a membrane-associated guanine-nucleotide-binding protein that is normally activated in response to the binding of growth factors. Rac1 is a small G-protein in the Rho family that drives MTOC orientation, actin polymerization, and cell–cell adhesion. Rac1 is activated by ARHGEF6 and repressed by RacGAP in response to upstream regulators such as growth factors. Rho is a member of the Ras superfamily of small GTP-binding proteins that play a central role in diverse biological processes. Rho proteins cycle between an active GTP-bound state and an inactive GDP-bound state, which is controlled by regulatory proteins such as GEFs (guanine exchange factors and GAPs (GTPase-activating proteins). The GTPase RhoA plays a prominent role in regulating the organization of the cytoskeleton by promoting the assembly of focal adhesions and actin stress fibers and by activating FAK. PLD1 catalyzes the hydrolysis of phosphatidylcholine to yield phosphatidic acid and choline. Rho also activates scaffolding proteins such as GDIA and IRSp53 (insulin receptor substrate protein-53). RhoA also binds to Rho/philin and regulate the actin cytoskeleton. The Rho family members Rac1, RhoA, and CDC42 were reported geranylgeranylated, and Ras was reported farnesylated to regulate their cellular functions
The NMT proteins are responsible for protein myristoylation function in various cellular processes in human cancer. The N-terminal myristoylation is essential for the membrane attachment and kinase activity of Src-family protein tyrosine kinases (SFKs, including c-Src, Yes, and Fyn), which transduce signals to control cellular processes such as cell proliferation, adhesion, and motility.256 Early studies identified the positive influence of myristoylation on c-Src kinase activity and cellular transformation in contrast to the case for c-Abl.257,258 The myristoylated Src identified in human colon adenocarcinoma tumor promoted colony formation and cell proliferation.259 The inhibitor of NMT1 block Src myristoylation and cytoplasmic membrane location, as well as the inhibition of cell proliferation, migration, and invasion in prostate cancer cells and the inhibition of tumor growth in vivo.260 In addition to inhibitors, the depletion of NMT isozymes inhibited cell replication, induced apoptosis through regulation BCL family protein level and growth inhibition in vivo.261
The evolutionarily conserved protein p53 is the most intensively studied tumor suppressor in growth arrest and apoptosis and is regulated by a large number of proteins and PTMs. The MDM2 belongs to E3-ubiquitin ligase and promotes p53 ubiquitination and proteasome-dependent degradation.262,263 Since MDM2 was observed to be conjugated to NEDD8, the neddylated p53 was also detected in vitro and in vivo, with the dependence on MDM2-p53 interaction. MDM2-mediated neddylation of p53 suppressed its transcriptional activity.189 The mRNA binding and stabilization protein Hu antigen R (HuR) is another neddylation substrate of MDM2. High expression of HuR was tested in several cancer types to regulate proliferation, differentiation, transformation. In the studies of colon cancer and hepatocellular carcinoma (HCC), MDM2-mediated HuR neddylation at sites K313 and K326 promoted its nuclear localization, avoiding cytoplasmic ubiquitination and degradation.264,265 The higher expression level of neddylation enzymes NAE1 and UBA3 in glioblastoma with poor clinical outcomes indicated neddylation can be considered as a therapeutic target. In glioblastoma cell lines and patient-derived glioblastoma stem cells, the neddylation inhibition by inhibitor MLN4924-induced apoptosis, which was associated with activation of the ERK-MAPK and PI3K-AKT signaling pathways.266
The pathogenic function of NO in tumor depends not only on the direct promotion or inhibition effects of nitrosative and oxidative stress, DNA damage-induced RNS formation, mitochondrial dysfunction, and apoptosis but also on the regulatory effect of protein S-nitrosylation.267 For example, S-nitrosylation may inhibit kinase or phosphatase activity. The apoptosis signal-regulating kinase 1 (ASK1, also MEKK5) is associated with cell apoptosis and mitochondrial damage under cellular stress in p38 kinase-dependent way and Hippo signaling effectors YAP/TAZ mediated cell invasion/migration.268 The interferon-gamma (IFN-γ) induced NO production and S-nitrosylation of ASK1, blocking the interaction of ASK1 with its downstream effectors MKK3 or MKK6.269 Moreover, IFN-γ promoted endogenous S-nitrosylation of JNK activation and disrupted JNK and its substrate c-Jun interaction.270,271 PTEN (phosphatase and tensin homolog) is a tumor suppressor controls a variety of biological processes. PTEN antagonizes the PI3K/Akt signaling pathway and can be regulated by several kinds of PTMs such as ubiquitylation, SUMOylation, neddylation, S-nitrosylation, and phosphorylation.272,273 Cys-83 is the S-nitrosylation site of PTEN under the condition of low NO concentrations and Akt signal is promoted after PTEN S-nitrosylation.274
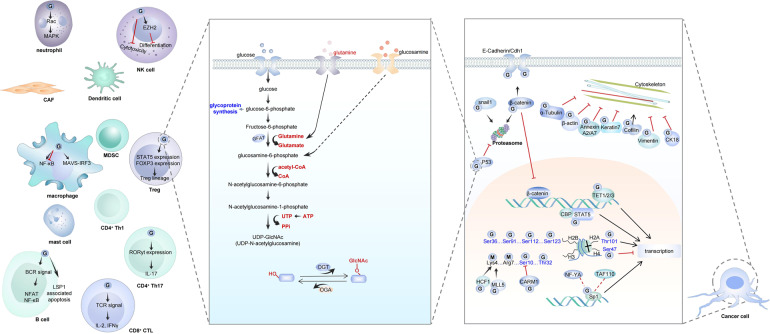
The function of O-GlcNAcylation in human cancer is also confirmed by the studies about O-GlcNAcase. Primary breast tumor tissues showed an increased O-GlcNAcase and lysosomal hexosaminidase activity than the corresponding adjacent normal samples, with a significantly decreased O-GlcNAc mono-glycosylation.275 The enzyme activity of O-GlcNAcase was also increased in thyroid cancers and the modified proteins showed a predominantly nuclear distribution.276 This situation is different in other studies. Researchers identified enhanced GlcNAcylation level in breast tumor as compared to normal samples, and even higher in metastatic lymph nodes. Cell migration and invasion were suppressed after lentiviral-mediated OGT silencing and increased in OGA-specific inhibitors NButGT and PUGNAc treatment.277 In the study of lung and colon cancer, not only O-GlcNAcylation and global O-GlcNAc transferase expression but also the OGT mRNA levels were evaluated in tumor tissues.278 Similarly, a high level of O-GlcNAcylation in target proteins was detected and was associated with the pathogenesis of characterizes chronic lymphocytic leukemia.279 O-GlcNAcylation targets are involved in various biological processes and signal cascades. O-glycosylated carboxy-terminus of p53 increases DNA binding.280 The eukaryotic translation initiation factor 2 (eIF2) is one of the widely studied kinase regulating cell growth, differentiation, metabolism, and stress response, which was reported to be regulated by several PTMs.281 Glycosylation of p67 protein is required to protect the eIF2 alpha-subunit from eIF2 kinase phosphorylation.282 O-GlcNAcylation of mammalian neurofilament-H subunits appears to regulate intermediate filament network formation in large myelinated neurons.283 Considering the central role of O-GlcNAcylation in the regulation of protein interaction and gene transcription, it is reasonable to raise questions on the occurrence and function of O-GlcNAcylation in the cancer signal pathway. Wnt/β-catenin signaling cascade is conserved and has emerged as a fundamental growth and development regulation pathway. Mutation of WNT/β-catenin signaling is frequent in many human cancers.284 Nuclear accumulation of β-catenin enhances the interaction with transcription factors T-cell factor (TCF)/lymphoid enhancer factor (LEF) to activate Wnt target genes. O-GlcNAcylation of β-catenin at Ser23 increased the movement of β-catenin from the nucleus to the plasma membrane, promoted the interaction of β-catenin with E-cadherin, and decreased transcription activity of Wnt signal.285 O-GlcNAcylation of β-catenin at T41 showed a direct competition with phosphorylation and a promotion of α-catenin–β-catenin interaction, which is critical for mucosa integrity.286 FoxM1 is an important transcription factor regulating cell proliferation, differentiation, transformation, cell cycle, and cell invasion in many types of cancer.287–290 Inhibition of the high level of OGT and O-GlcNAc in breast cancer cells reduced expression of FoxM1 and its target proteins, leading to the inhibition of breast cancer phenotypes.291 As one of the most critical transcription regulators in normal and tumor cells, c-Myc was also reported to be modified by O-GlcNAcylation even in the same site of phosphorylation.292,293 PTM of this site may regulate transforming activity and tumorigenicity in lymphomas.294 P53 is a crucial tumor suppressor and its fundamental biological function in tumorigenesis and cancer development is well studied in the regulation of genome instability, transcription, cell-cycle arrest, apoptosis, cellular metabolism as well as autophagy. Researches about the vast regulation network of p53 ranges from gene polymorphisms, transcription, PTM, protein–protein interaction to degradation.295 Besides the regulation by the E3-ligase oncoprotein MDM2 in p53 protein activity and degradation, the cellular function of p53 is also affected by PTM. P53 undergoes O-GlcNAcylation at site Ser149, thus inhibits its phosphorylation at Thr155. This modification abrogates the ubiquitination-dependent proteasomal degradation and p53–MDM2 interactions.296 Kelch ECH-associating protein 1 (Keap1) is a substrate for the Cullin-3 (CUL3)-dependent E3-ubiquitin ligase complex regulating the transcription factor erythroid 2-related factor 2 (Nrf2) to keep intracellular homeostasis under stress condition.297 With the help of a chemical biology approach in MDA-MB-231 cells, O-GlcNAcylation in Keap1 was identified at site Ser104 to promote CUL3 interaction and play a role in ubiquitination and degradation of Nrf2.298
Multiple lipid modifications, including isoprenylation, myristoylation, palmitoylation, and glycosylphosphatidylinositol anchor, may lead to distinct protein subcellular location and various signal transduction.299 The concept of apoptosis, a programmed cell death process, is essential for homeostasis maintenance.300 It was reported that N-terminus myristoylation is due directly to caspase cleavage and autophagy.301 Sirtuins are well known for their lysine deacetylases activity to regulate biological processes. Besides the efficient deacetylase function, SIRT1–3 have been recently identified to remove long-chain fatty acyl groups. Among them, SIRT2 prefers more efficient demyristoylase activity.302 The tumor suppressor SIRT6 is also reported to remove fatty acyl groups from the lysine residues from R-Ras2. Lipid-modified R-Ras2 produces a marked oncogene effect including tumor growth, signal transduction, cytoskeletal dynamics, and PI3K-associated cell proliferation.303
Metabolic regulation
Metabolic reprogramming is a hallmark of cancer cells. A rapid synthesis of structure blocking molecules including nucleotides, proteins, and lipids is necessary to support fast cancer cell growth. Under this condition, cancer cells exhibit increased glucose uptake, as well as fatty acid synthesis and glutamine consumption.304,305 Many metabolic enzymes are reported abnormal in tumorigenesis and cancer development. In order to meet the lipid demand of rapid proliferation, the enzymes related to de novo lipid synthesis in tumor cells are abnormally highly expressed, such as fatty acid synthase (FASN) which catalyzes the synthesis of palmitic acid by acetyl-CoA and malonyl-CoA.306,307 One of the possible mechanisms of upregulated FASN under hypoxia of TME is the activation of Akt-HIF-1 axis and the following induction of transcription factor SREBP-1.308 Multiple acetylation sites were identified on FASN. Acetyltransferase KAT8 mediates ubiquitin–proteasome-related FASN degradation and the deacetylation depends on HDAC3n.309 Therefore, targeting FASN acetylation can be used as a new direction of tumor therapy. Cancer cells expressed a splice isoform of the glycolytic enzyme pyruvate kinase (M2), which was necessary for tumor formation in nude mouse xenografts.310 During Akt-mediated tumorigenesis, the conversion of glucose to lipid depends on the activity of ATP citrate lyase (ACL), which promotes the production of cytosolic acetyl-CoA from mitochondria-derived citrate.311 Conversely, mutations in some oncogenes can affect cancer-associated metabolic reprogramming. Increased expression of the serine/threonine kinase Akt raises glucose consumption to support aerobic glycolysis and tumor survival.312 Increased levels of glycolysis, glutamine metabolism, and nucleotide biosynthesis were also observed after oncogene KEAS expression.313
Multiple CoA generated from the TCA cycle, as well as the metabolism of amino acid and lipids can be the substrates of PTM such as malonyl-CoA, succinyl-CoA, and glutaryl-CoA, highlighting their emerging role in metabolic regulation. Malonyl-CoA is the two-carbon donor in de novo fatty acid biosynthesis and fatty acid elongation, and also the inhibition of carnitine palmitoyltransferase 1 (CPT1) in mitochondrial β-oxidation, hepatic fatty acid synthesis, and ketogenesis.314,315 In mammalian cells, the ACS family member ACSF3 localizes to the mitochondrial matrix and is required for lysine malonylation of mitochondrial proteins.316 Malonyl-CoA can be converted to acetyl-CoA by malonyl-CoA decarboxylase (MCD), which mutations cause an inborn metabolic disorder.317 With affinity enrichment of malonylated peptides, MCD-deficient cells, and SIRT5 KO mice showed increased lysine malonylation, as well as impaired mitochondrial respiration and fatty acid oxidation.318 By the characterization of succinylation proteome, succinylated proteins are discovered significantly enriched in cellular metabolic process, including oxoacid metabolism, oxidation–reduction process, and coenzyme metabolism.319 As previously mentioned, Sirt5 functions in lysine demalonylation and desuccinylation, which has been identified to regulate metabolic proteins. The mitochondrial pyruvate dehydrogenase complex (PDC) catalyzes decarboxylation of pyruvate to acetyl-CoA, linking glycolysis to the TCA cycle.320 In the depletion of SIRT5 in MEF cells, multiple subunits of PDC were hypersuccinylated, suggesting the negative regulation of SIRT5 in PDC activity.319 Succinylation of mitochondrial proteins will affect their normal function and redox state to ensure that cells respond quickly under environmental changes in TME with abnormal metabolic programming.321 Pyruvate kinase PKM2 shows the transcription factor property and enzymatic activity of promoting tumor aerobic glycolysis.322 The activity of PKM2 was enhanced by succinylation at K498. ROS promotes the binding and desuccinylation of PKM2 by SIRT5, reducing NADPH production, so as to achieve growth inhibition.319,323 When the nutritional environment changes such as glucose starvation, the succinylation of PKM2 is involved in regulating its mitochondrial translocation and coordinating cell proliferation or survival.324
Myristoylation and palmitoylation participate in nutrient absorption and cell metabolism by regulating mitochondrial function. AMP-activated protein kinase (AMPK) maintains cellular ATP homeostasis, senses intracellular energy level, and is activated by metabolic stimulation.325,326 Under the starvation condition, cell relies on autophagy to provide energy, and this process is regulated by AMPK.327 Autophagy and impaired mitochondrial clearance in AMPK deficient cells are hindered. Myristoylated AMPK enhances the connection with mitochondria through membrane fusion and maintains the function of AMPK mediated autophagy.328 Fatty acid metabolism is an important step in metabolic reprogramming of tumor cells, which comes from exogenous uptake or de novo synthesis of other metabolic intermediates (glucose or glutamine). Membrane protein CD36 is a fatty acid translocase (FAT) and its palmitoylation can regulate the subcellular distribution and function of proteins.329 The palmitoylation and plasma membrane localization, as well as fatty acid uptake of CD36, require DHHC (Asp–His–His–Cys) motif-containing palmitoyl acyltransferases, DHHC4 and DHHC5.330 Palmitoylation of a coding product of the oncogene KRAS (KRAS4A) determines its interaction with hexokinase 1 (HK-1) and outer mitochondrial membrane (OMM) localization, thereby regulating kinase activity and glycolytic flux.331 Another important oncogene, epidermal growth factor receptor (EGFR), can also be regulated by palmitoylation. The stimulation of palmitic acid de novo synthesis by plasma membrane EGFR (pmEGFR) can promote the palmitoylation of mitochondrial EGFR (mtEGFR), and then promote mitochondrial fusion and tumor cell growth.332 This mechanism extends the understanding of EGFR to promote cancer progression independent of its tyrosine kinase activity. Mevalonate (MVA) is a common building block of many cellular compounds, including the membrane structure and isoprenoid/cholesterol biosynthetic precursor cholesterol,333 the N-linked glycoproteins carrier phosphorylated dolichol,334 the electron transfer lipid in the mitochondrial respiratory chain ubiquinone (UQ, coenzyme Q),335 some tRNAs modification protein substrate isopentenyladenine and the substrates geranylgeranyl pyrophosphate (GGPP) and farnesyl pyrophosphate (FPP) for prenylation.336 HMG-CoA catalyzes the conversion of HMG-CoA to MVA, which is inhibit by lovastatin. Lovastatin is an effective anticancer drug in many types of cancer like acute myelogenous leukemia, medulloblastoma, mesothelioma and neuroblastoma.337–340 The same apoptotic effect of lovastatin was also observed in GGTI-298 and less effective in the FTI-277, indicating the role of prenylation, especially geranylgeranylation, in lovastatin-induced apoptosis in AML.341
In recent years, a series of novel functions of lactate have been discovered such as the fuel for metabolic processes, the promotion role of tumor invasion and metastasis, the function in angiogenesis and especially, the immune suppression in tumor microenvironment. Lactate can be absorbed by cancer cells and transported to mitochondria for oxidation to provide energy. Lactate in tumor microenvironment can inhibit the cytotoxicity of immune cells, suggesting an essential role of lactate in metabolism regulation. Warburg effect in cancer cells explains the lactate formation from glucose. Lactate can also be produced from glutamine metabolism. Glutamine is catalyzed by glutaminase GLS to transform into glutamate, and glutamate dehydrogenase convers glutamate intoαKG to enter TCA cycle.114 In some types of cancers like human pancreatic ductal adenocarcinoma (PDAC), glutamate and oxaloacetate (OAA) are transformed into αKG and aspartate. The aspartate can be converted into oxaloacetate by aspartate transaminase GOT1, followed by the conversion of oxaloacetate to pyruvate.342 Finally, the enzyme LDHA catalyzes the conversion of pyruvate to lactate.114 Lactate transport into cells is mediated by monocarboxylate transporters (MCTs) family proteins MCT1 (also SLC16a1), MCT2 (also SLC16a7), MCT3 (also SLC16a8), and MCT4 (also SLC16a3).343 Since first identified, the lactate-derived lysine lactylation is found associated with cellular metabolism. Glucose can induce lactate production and histone lactylation in a dose-dependent manner and metabolic labeling experiments proved lysine lactylation is endogenously derived from glucose.120 Lactic acid stimulated the increased levels of histone lactation in HK-1 and IDH promoters in NSCLC and breast cancer cells.344 In addition, the transcription of HK-1, G6PD, and PKM was downregulated, while the transcription of SDH, IDH, and HIF1A was upregulated.345 This supports that lactic acid regulates the gene expression of related metabolic enzymes through histone lactylation, so that tumor cells show different metabolic characteristics.
The high-energy nucleotide sugar uridine diphosphate-GlcNAc (UDP-GlcNAc) derived from glucose and other nutrition metabolites is required for protein O-GlcNAcylation.346 The biochemical and phenotypic influence of O-GlcNAcylation is implicated on metabolic disorders and human diseases, particularly cancer. In response to nutrient levels, O-GlcNAcylation was reported to regulate signaling cascade, protein solubility and stability, gene transcription, and genome replication.347 In one study of basal-like breast cancer, a high level of O-GlcNAcylation and α-ketoglutarate-dependent transcription factor HIF-1α, as well as its downstream factor GLUT1, were observed. Mechanistically, changed OGT level regulated HIF-1α proteasomal-dependent degradation, the interaction between HIF-1α and pVHL, ER stress-mediated tumor cell survival, and cell metabolism.348
The newly identified link between neddylation modification with metabolism is studied by neddylation inhibitor MLN4924. As an energy hub in metabolism, mitochondria function, morphology, and fusion-fission dynamics should be fine-tuned.349 Under the condition of oncogene signaling, hypoxia environment, nutrient starvation and anticancer treatment, mitochondria maintain the balance between rounded, fragmented signature with elongated, interconnected shape to regulate its cytoskeletal transport activity and metabolic function. Abnormal mitochondrial regulation contributes to cancer hallmarks, including cellular transformation, cell survival in therapy response, cancer stem cell maintenance, cell differentiation, and migration. Several mammalian mitochondrial proteins undergo PTMs to keep mitochondrial homeostasis.350 Through a global metabolic profiling analysis after the treatment of MLN4924, the nucleotide biosynthesis pathway was disrupted in AML cells.351 A recent study connects altered energy metabolism with protein neddylation. MLN4924 may induce autophagy partially through the inhibition of mTOR activity and induction of ROS stress in liver cancer cells.352 Moreover, mitochondrial fission-to-fusion conversion was induced by MLN4924 treatment in breast cancer cells. The overall metabolism, including carbohydrates, organic acids, amino acids, nucleotides, and lipids were inhibited. As for the altered metabolites, the increased 3-phosphoglyceric acid and pyruvic acid, as well as the decreased succinic acid and fumaric acid implied enhanced glycolysis, reduced mitochondrial functions, increased mtDNA copy number, and mitochondrial respiration (OXPHOS).353 Therefore, the combination strategy of neddylation inhibitor and metabolism regulation agents is the new avenue to enhance anticancer efficiency.
Epithelial–mesenchymal transition (EMT) regulation
Protein post-translational modification participates in all stages of cellular processes (Fig. 4). The interaction between different cells and the attachment of one cell to its extracellular matrix are important for cell movement and tissue structural integrity. As extracellular vehicles (EVs) secreted by tumor and other cells in TME, exosome is involved in intercellular communication and regulate immune response by expressing histocompatibility complex MHC classes I and II.354 Different studies have found that tumor-derived exosomes can promote fibroblast differentiation, contribute to tumor metastasis and participate in TME reprogramming in a TGFβ dependent manner.355,356 A melanoma metastasis model proved that the release of anti-metastatic EVs depends on the acetylation of p53, and BAG6/CBP/P300 is essential in this process.357 The biomechanical properties of tumor cells in TME change with extracellular matrix (ECM) and intracellular cytoskeleton structure to enhance the possibility of invasion.358 The cytoskeleton mainly composed of microtubules and actin not only maintains the basic morphology of cells but also participates in the localization and transportation of organelles.359 Interestingly, the cytoskeleton proteins are often suffered a variety of PTM modifications, including phosphorylation, ubiquitination, palmitoylation, glycosylation, and acetylation.360 Lysine at position 40 of α-tubulin is acetylated by acetyltransferase αTAT1 and this process is reversed by deacetylases HDAC6 and SIRT2.361,362 The high acetylation of α-tubulin is associated with the proliferation and invasive activity of breast cancer and colon cancer, which can be used as a potential marker.363,364 Disruption of microtubule acetylation can cause the increased endoplasmic reticulum stress signal to inhibit tumor progression.365
Fig. 4.
Biological effect of modification on tumor initiation and development. Five groups of PTMs function in cellular biological processes, including altering higher-order chromatin structures to positively or negatively regulate gene transcription, modulating protein attachment on membrane or transcription factors to regulate downstream signal transduction, changing metabolic enzymes and related transcription factors to regulate metabolic reprogramming, as well as remodeling cytoskeleton and cell adhesion system to regulate tumor invasion
The S100 protein family show widely functions in every stage of cancer development by regulating key signal pathways.366 One of the membrane-located S100A10 was reported highly expressed in ovarian cancer, breast cancer, colorectal cancer, renal cell carcinoma, and gastric cancer.367–371 The high level of succinylation of S100A10 at K47 was found in gastric cancer and metastatic lymph node tissue to inhibited ubiquitination-related proteasome degradation. Mechanically, this succinylation is mediated by succinyltransferase CPT1A to promote cell migration and invasion.372
Proteins that participate in cell adhesion system can be divided into three types: cell adhesion molecules (cadherins, integrins, selectins, and immunoglobulins), the highly organized and dynamic three-dimensional (3D) scaffolds ECM (including collagens, fibronectins, laminins, and proteoglycans) and membrane proteins located at cell peripheral, with a linkage function of adhesion systems to the cytoskeleton.373–375 These proteins also regulate signal transduction, membrane trafficking, and cell division.376 Cadherin family members include the epithelia expressed E-cadherin, the neural and mesenchymal tissue expressed N-cadherin, and vascular endothelia-specific VE-cadherin.377 The various cadherin superfamily proteins function as oncogenic or tumor suppressor to affect tumorigenesis and/or tumor progression, making these cadherins potential therapeutic targets for cancer. A current research theme is the various cadherins regulation and cadherin switch during EMT and cancer progression.378 The Rho family of small GTPases, especially Rho, Rac, and Cdc42, were demonstrated to regulate cadherin-mediated adhesion, and thus EMT process.379–382 Ras family also belongs to small GTPases and one of the members Rap1 (with the isoform Rap1A) is a major activator of integrins to regulate cellular functions and cell adhesion.383 Activation of Rap1 promotes prostate cancer survival and metastasis.384 The inhibition of protein geranylgeranylation, but not protein farnesylation, reduced Rap1A and Rab6 geranylgeranylation and prevented prostate cancer metastasis.385 The result shed light on the connection between protein geranylgeranylation with cancer metastasis.
The regulation of O-GlcNAcylation in cancer is also reflected in the EMT process. The snail family member snail1 was reported to induce EMT by directly repressing the transcription of E-cadherin.386 Glycogen synthase kinase 3β (GSK3β) and protein kinase CK1 are responsible for binding and phosphorylating Snail to regulate beta-Trcp-mediated ubiquitination and subcellular localization.387,388 Cancer cells have to break down cell–cell and cell–matrix attachment in EMT, which involves cadherin switch, intrinsic and extrinsic signals, as well as a dramatic reorganization of the cytoskeleton. As a highly dynamic structure regulated by Rho GTPase family, cytoskeleton modulates cell motility and invasion through forming filopodia, lamellipodia, podosomes and invadopodia into the surrounding environment.389 E-cadherin is a single-span transmembrane glycoprotein and a major component of epithelial adheren junctions (AJ) to mediate cell–cell adhesion.390 Under the condition of ER stress-induced apoptosis, E-cadherin and β-catenin were O-glycosylated and led to reduced intercellular adhesion.391 Moreover, the phosphorylation-mediated Snail1 degradation is blocked by O-GlcNAcylation at site Ser112, leading to EMT stimulation through transcriptional suppression of E-cadherin.392
The NAE inhibitor MLN4924 revealed the metastatic regulation of protein neddylation. Liver metastatic veal melanoma (UM) patients showed highly expressed key proteins in the neddylation pathway. The MLN4924 treatment specifically decreased cullin1 neddylation, facilitated the accumulation of CRL substrates, induced mitochondrial damage, triggered intrinsic apoptosis, enhanced DNA double-strand breaks and ROS generation, diminished UM cells migration and invasion, and repressed the cancer stem cells (CSCs) properties through Slug protein degradation. The inhibit tumor outgrowth, disturbed paracrine secretion of VEGF-C, and impaired angiogenesis were observed in NOD-SCID mouse xenograft model.393 The inhibition of the metastatic process by MLN4924 was conformed recently in lung cancer. First, key proteins in neddylation process are overactivated in both lung adenocarcinoma and squamous cell carcinoma. Second, inhibition of neddylation using MLN4924 significantly suppressed cell growth, clone formation, migration, and cell motility in vitro, as well as tumor formation and metastasis in vivo. Lastly, phorbol-12-myristate-13-acetate-induced protein 1 (NOXA)-dependent apoptosis was induced by MLN4924.394 The anti-migration effect by activating E-cadherin and the anti-invasion effect repressing Vimentin were observed after MLN4924 treatment in human clear cell renal carcinoma (ccRCC).395 Mechanistically, MLN4924 inhibit intravascular survival, extravasation, and formation of metastatic colonies to inhibit lung cancer metastasis. At an early stage, MLN4924 downregulated the expression of MMP2, MMP9, and vimentin to disrupt actin cytoskeleton. At a later stage, MLN4924 block neddylation to increase its substrates like p21, p27, and Wee1, leading to cell-cycle arrest and apoptosis.396
PTM-dependent immune surveillance and escape in TME
Acidic environment
Cancer cells export metabolic waste such as lactate into TME to form an acidic environment. The low pH might influence the modification of proteins both in tumor cells, immune cells, fibroblast cells and so on. In addition, low pH is closely corrected with hypoxia and low blood-flow levels, as well as the effectiveness of various therapeutic agents.397
Hypoxia environment
In the inner tumor microenvironment, the concentration of oxygen is much lower and form a condition called hypoxia. The increased oxygen consumption caused by high rate of tumor cell proliferation and decreased oxygen supply caused by compact tumor microvasculature network are main reason of hypoxia formation.398,399 Under conditions of reduced oxygen, hypoxia-inducible factor 1 (HIF-1) is activated, leading to the expression of vascular endothelial growth factor (VEGF) to stimulate angiogenesis.400,401 Up to 95% of FA in tumor cells is de novo synthesis, and the intermediate metabolite palmitic acid is produced from acetyl-CoA, malonyl-CoA, and NADPH through catalysis by FASN.402 In vivo studies found that the angiogenesis of FASN knockout mice was impaired. The depletion of FASN enhanced the accumulation of malonyl-CoA and reduced mTOR activity through mTOR malonylation.403
More and more regulatory functions of HIFα in cancer progression are revealed including the maintenance of cancer stem cells by regulation of Notch pathway,404 the negative regulation of mitochondrial biogenesis and O2 consumption,405 the regulation of glucose metabolism by interacting with pyruvate kinase isoforms PKM2,406 the induction of wild-type EGFR overexpression,407 the suppression of E-cadherin,408 the promotion of metastatic cascade,409,410 and the resistance to radiotherapy and chemotherapy.411,412 Hypoxia-mediated O2 homeostasis plays an important role in the pathogenesis of major causes of mortality, especially in cancer. The expression of some glycolytic enzymes is induced by the transcriptional oncoproteins HIF-1α, reflecting the close correlation of hypoxia microenvironment and cellular metabolism. These enzymes include hexokinase 2 (HK2), glucose-6-phosphate isomerase, 6-phosphofructokinase 1, aldolase, fructose bisphosphate (FBP), triose phosphate isomerase (TPI), phosphoglycerate kinase (PGK1), enolase 1, PKM, and LDHA.114,413–415 In addition, HIF-1 upregulates PD-L1 expression in immune cells like MDSCs and macrophages, constructing an immunosuppressive tumor environment.416–418 Another checkpoint VISTA (V-domain Ig-containing suppressor of T-cell activation) is also overexpressed under hypoxia condition, especially in the immunosuppressive MDSC cells.419 In the phagocytosis function of macrophage, cell surface protein cluster of differentiation 47 (CD47) binds with its ligands, signal regulatory protein α (SIRPα), and thrombospondin-1 (TSP-1), to conduct a “don’t eat me” signal, facilitating tumor cells escape.420,421 Future studies targeting selective inhibitors of hypoxia have a large potential in cancer drug discovery.
Immunosuppression effect
Recently, more and more studies reported the functions of metabolic production and the related protein PTMs in impairing the adaptive immune response or disabling immune surveillance. We discuss the immunosuppression function of several types of PTMs in this review.
Innate immune response and IFN signal
Innate immune response triggered by viral infection may activate transcription factors NF-κB and IRF3 to regulate the expression of type I interferon and interferon-stimulated genes (ISGs).422 After the RNA virus infection, they may replicate to produce double-stranded RNA. Protein RIG-1 and the downstream factor MAVS (mitochondrial antiviral signaling) are involved in recognizing dsRNA and producing interferon to initiate the innate immune response.423,424 The mitochondrial-associated membrane (MAM) links the endoplasmic reticulum to mitochondria, where RIG-1 interacts with MAVS to form an intracellular immune synapse425 (Fig. 5). In a recent study with bone marrow-derived macrophages (BMDMs), protein geranylgeranylation was observed to inhibit RIG-I-like receptor (RLR) mediated innate immune signaling pathways. Moreover, anchoring the Rho GTPase Rac1 into MAM needs protein geranylgeranylation and subsequent palmitoylation. MAM-associated Rac1 promotes MAVS signaling complex formation and then inhibits the interaction between MAVS and the E3 ligase Trim31, facilitating the association of caspase-8 and cFLIPL and MAVS signal termination.426
Fig. 5.
Overview of the hexosamine biosynthetic pathway (HBP) and the regulation of O-GlcNAcylation in TME. Left: O-GlcNAcylation orchestrates immunity. O-GlcNAcylation is increased after neutrophils are activated through the MAPK pathway. In macrophages, O-GlcNAcylation shows both activation and inhibition functions of NF-κB. Upon RNA virus infection, MAVS is modified by OGT to enhance downstream IFN production via IRF3 signaling. O-GlcNAcylation reduces the cytotoxic activity and inhibits NK differentiation by increasing the stability of EZH2. O-GlcNAcylation increases the expression of RORγt and FOXP3 in Th17 and Treg cells. NFAT and NFκB are activated by O-GlcNAcylation in activated B cells and O-GlcNAcylation of LSP1 contributes to B-cell apoptosis. After TCR rearrangement, O-GlcNAcylation promotes positive T-cell development. Middle: nutrient flux modulates protein O-GlcNAcylation. OGT catalyzes the addition of GlcNAc from UDP-GlcNAc to serine and threonine residues, while OGA catalyzes their removal. Right: the function of O-GlcNAcylation in proteasome-associated degradation, cytoskeleton remodeling, and transcriptional regulation
Pathogenic alteration produces host-derived intrinsic stimulation to induce inflammation. The cytokines IL-17A contribute to immune response in cancer and is and is secreted from IL-17A-producing CD4+ T cells (Th17 cells), CD8+ T cells, γδT cells, and natural killer T cells. IL-17A was reported to promote hepatoma stem cells self-renewal and immune escape partly through upregulation of PD-L1.427 Th17-produced IL-17A is induced by IL-1b and IL-23, which is enhanced by lactate.428 Lactate was reported to bind the complex of retinoic acid-inducible gene-I/mitochondrial antiviral-signaling protein (RIG-I/MAVS) transmembrane (TM) domain and inhibit MAVS aggregation, thus suppressing glycolysis-mediated RLR signaling and type I IFN pathway.429
Toll-like receptor (TLR) family has a crucial role in the field of innate immunity and also connects non-specific immunity and specific immunity, even individual TLRs exhibit specific responses.430,431 TLR4 colocalized with TRIF-related adaptor molecule (TRAM) in the plasma membrane and the Golgi apparatus to trigger an intracellular signaling cascade, including the production of IRF3 dependent type I IFN- and NF-κB-dependent pro-inflammatory cytokines. Interestingly, TRAM contains an N-terminal myristoylation sequence, and the myristoylated TRAM tethers it to the membrane, serving as a prerequisite for LPS signaling.432 Moreover, TRAM was reported to function in TLR7 signaling in the detection of single-stranded RNA (ssRNA) derived from RNA viruses. The myriostoylation of TRAM is necessary for TLR7 mediated CCL5, IFN-α, and IFN-β activation.433 These results supported the function of myristoylation in innate immune response mainly by its property in mediating protein location in the cell membrane.
Macrophage
Macrophages are large, mononuclear, highly phagocytic cells derived from monocytes and play a key role in innate immune response and pathogen defense. In addition to phagocytosis, macrophages can secrete a vast array of mediators including enzymes, inflammatory cytokines, growth factors, eicosanoids, and oxidants. The diverse cellular functions are regulated by specific microenvironments and inflammatory mediators.434 The type I cytokines like interferon-γ (IFNγ), tumor necrosis factor-α (TNFα), and pathogen-associated molecular patterns (PAMPs, including lipopolysaccharide (LPS), lipoproteins, dsRNA, lipoteichoic acid) activate macrophage and promote Th1 immune responses, whereas macrophage stimulated by Interleukin (IL)-4, IL-13, IL-1β, IL-10, transforming growth factor-β (TGFβ), and glucocorticoids support Th2-associated function.435 Toll-like receptors (TLRs) recognize microbial components and the best characterized is LPS.436 The interaction between LPS and TLR4 activates the molecules myeloid differentiation factor 88 (MyD88) or TIR-domain-containing adaptor inducing IFN-β associated signal pathways, leading to the phosphorylation and ubiquitin-mediated degradation of IκB proteins and the latter the activation of transcription factor NF-κB.437 The neddylation inhibitor MLN4924 and depletion of Nedd8 or the Nedd8-conjugating enzyme by siRNA are used to demonstrate the regulation of neddylation in macrophage function. The decreased neddylation process disrupted pro-inflammatory cytokines secretion from macrophages through the repression of NF-κB in response to LPS.438 BMDM stimulated by LPS can produce a large amount of malonyl-CoA and cytokines like IL-6 and TNF α. It was further confirmed that the substrate of malonylation induced by LPS was GAPDH at the site of lysine 213, which is essential for cytokine production.439 This confirms the role of PTM regulation targeting GAPDH in the inflammatory response. During LPS stimulation, a select group of macrophage proteins, including a protein kinase C (PKC) substrate, were identified to be myristoylated. This modification targeted protein location to the membrane to interact with PKC.440 Other substrates for PKC in macrophage were myristoylated by IFN-γ, but not IFN-α nor IFN-β, to mediate intracellular signal pathways.441,442 In NMT1+/− mouse model, the depleted colony-forming ability of BMDM was observed, suggesting the role of protein myristoylation in monocytic differentiation.443 Macrophages undergo a metabolic reprogramming from pro-inflammatory “M1” macrophage with high phagocytic and bactericidal potential to “M2” macrophage with anti-parasitic and tissue repair functions.444,445 The low level of arginase-1 (ARG1) produces enhanced nitric oxide synthase (iNOS) to catalyze arginine to citrulline and nitric oxide and then kill pathogens in “M1” macrophage. On the other hand, ARG1 is induced to produce urea, polyamines, and ornithine in “M2” macrophage.446,447 During “M1” macrophage activation, lactate and the corresponding histone lactylation plays a positive role in driving expression of M2-like genes, reflecting a “lactate clock” in gene expression regulation.120
TCR signal
During T-cell development, the suppression of Nmt activity led to impaired TCR signals transmission, decreased T-cell activity, and cytokine secretion after the treatment of CpG and staphylococcal enterotoxin B (SEB) stimulation.448 In addition, lipids modification is common in lipid rafts, the structural components in living cell membrane. Lipid rafts are functional important in early T-cell receptor (TCR) signaling transduction. For example, the aggregation of lipid rafts is involved in TCR activation.449,450 After the presentation of an antigen by antigen-presenting cells (APC), T cells are activated through cell–cell contacts called immunological synapses (IS) or supramolecular activation cluster (SMAC).72,451 The SMAC can be divided into three concentric rings: the central SMAC, the peripheral SMAC, and the distal SMAC. Different SMAC parts enclose various signaling factors to form a stable immunological synapse for T-cell activation452 (Fig. 6). The central SMAC is enriched for TCR, MHC-peptide complexes and signaling molecules like PKC, Src-family kinase Lck and tyrosine kinase Fyn. The peripheral SMAC contains integrin-associated cytoskeletal proteins talin and lymphocyte function-associated antigen 1 (LFA1). The distal SMAC contains CD43, CD44, and CD45.453–456
Fig. 6.
Overview of a mature T-cell synapse. The immunological synapses (IS) is also termed supramolecular activation complexes (SMACs), including the central region of the supramolecular activation complex (c-SMAC), the peripheral ring surrounding the c-SMAC (p-SMAC), the region distal to the synapse outside the p-SMAC (d-SMAC) and the molecules/ligand pairs that are found enriched within. Protein myristoylation makes a significant contribution to IS formation. APC antigen-presenting cell, CTLA4 cytotoxic T-lymphocyte antigen 4, ICAM1 intercellular adhesion molecule 1, LFA1 leukocyte function-associated antigen 1, PI3K phosphatidylinositol 3-kinase, SHP2 SRC homology 2-domain-containing protein tyrosine phosphatase 2, TCR T-cell receptor, ZAP70 ζ-chain-associated protein 70, LCK Src-family kinase
Besides the centrosome translocation to the leading edge mediated by actin and microtubule networks, myristoylation makes a significant contribution to IS formation.72,457 The N-terminal glycine residue (glycine 2) is required for myristylation of tyrosine protein kinase Src family. Mutation of this residue blocked protein stability in the plasma membrane and inhibit TCR-induced tyrosine phosphorylation.458 Another Src-family member Fyn was observed for both N-myristylation and palmitylation, which altered its subcellular localization.459 More importantly, this dual-modification-mediated Fyn location in IS stabilizes the interactions between Fyn and TCRζ chain.460 Focal adhesion kinase (FAK) regulates cell adhesion to the ECM through integrin receptors and participates in integrin-mediated signal transductions, whose activity is influenced by myristoylated c-Src.72,461 N-myristoylation of Lck is indispensable for TCR signaling cascade including CD3ζ, Zap70, and ERK activation, as well as cytokines IFN-γ and IL-2 releasing.448 Cytoskeletal polarization and architecture organization are essential processes in T-cell activation to form F-actin-rich lamellipodium.462 F-actin organization is regulated by Arp2/3 complex to coordinate lamellipodia and filopodia transition in distinct cellular functions.463 The highly conserved Formin family proteins not only functions in filopodium formation, dynamic rearrangement of the actin cytoskeleton, and cell polarity but also is required in cytokinesis and endocytosis.464 The cytoskeletal regulation of Arp2/3 complex and Formins contributes in T-cell activation. Two kinds of Formins, mammalian diaphanous 1 (Dia1) and Formin-like-1 (FMNL1), displayed unique co-localization in the microtubule-organizing center (MTOC) and controlled TCR-mediated centrosome polarization.464 Importantly, N-terminal myristoylation plays essential roles in Formin’s membrane association, membrane trafficking, and bleb formation.202,465 These results support the function of protein myristylation in T-cell antigen receptor signaling.
Dendritic cells
Dendritic cells are efficient stimulators of B and T lymphocytes and mediate both innate and adaptive immune responses.466 DCs undergo maturation process to become highly activated. The metabolic function and/or metabolic byproducts in the tumor microenvironment are increasingly concerned and studied. Among these, the metabolism regulation of immune cells mediates cell differentiation, antigen presentation, ion signaling control, and cytoskeletal remodeling to facilitate downstream responses.467 NO is an essential metabolic regulator in monocyte-derived inflammatory DCs (moDCs) since its production inhibit mitochondrial oxygen consumption to change the OXPHOS and TCA cycle.468 NO synthase (NOS) enzymes include endothelial NOS (eNOS, NOS1) and neuronal NOS (nNOS, NOS3), which were reported to catalyze NO production.469 Based on the subcellular localization analysis of myristoylation-mutant eNOS cDNA, N-terminal myristoylation of the endothelial NO synthase was found to be functionally important in extracellular transport of highly cytotoxic products and signal transduction.470
The neddylation pathway in DCs is associated with its immune regulation function. The release of pro-inflammatory cytokines such as TNF-α and IL-6 by DCs is suppressed by neddylation inhibitor MLN4924 after the stimulation of LPS. The treatment caused a significantly reduced T-cell proliferation and interferon-γ secretion, without any change in apoptosis or cell viability. Mechanically, MLN4924 block the LPS-induced nuclear location of p65 NF-κB in BM-derived DCs (BMDCs) and prevent IκB degradation without affecting the MAPK/ERK pathway.471 IL-12p70 is a key cytokine produced by DCs, which is required to instruct an adaptive Th1 response.472 MLN9424 decreased the secretion of IL-12p70, indicating a restricted capacity of DCs for T-cell activation. Besides downregulated maturation and stimulatory capacity of DCs partially through the mTOR signaling pathway, MLN4924 also promoted caspase-dependent apoptosis of DCs and suppressed T-cell proliferation.473
T-cell function
T-cell activation and the corresponding immune response are along with cytoskeletal rearrangements by segregating engaged TCRs and promoting cytokines delivery.462 During T-cell polarization, MTOC is needed to translocate to IS, allowing T-cell secretion. The myristoylated actin nucleating Formins displayed MTOC-mediated migration without Arp2/3-dependent actin nucleation.474
Other kinds of PTM may affect T-cell activation, especially by metabolism regulation. The switch of T cell from quiescent states (naive and memory T cells) to highly proliferative states (developing thymocytes and effector T cells) is regulated by growth factor signals. One hallmark of this transition in metabolism is the change of ATP production from mitochondrial OXPHOS to high throughput glycolysis.475 The Warburg effect in tumor cells refers to the switch from TCA to aerobic glycolysis, leading to the production of lactate from glucose.112 The mitochondrial membrane-bound enzyme succinate dehydrogenase (SDH) functions in both the TCA cycle and electron transport chain.476 The inhibition of SDH led to decreased proliferation, neutralization of mitochondrial membrane, and impaired T-cell cytokine production, thus affect T-cell biology.477 Studies of Sirt5–SDH axis connect protein PTMs with cell metabolism, as well as T-cell function regulation. Indeed, the depletion of Sirt5 showed a substantial increase in SDH activity and enhanced SDH substrate succinate.319 Cytokines prevent proapoptotic factors release by regulating B-cell lymphoma 2 (BCL-2) activity to maintain mitochondrial integrity. In another way, growth factors promote nutrient uptake through activating protein kinases.478
The dramatically developed field in cancer therapy is immune checkpoint blockade. The immune system recognizes various antigens derived from outside pathogenic infection through TCRs, and is regulated by the immune checkpoint. Immune checkpoints are essential to prevent autoimmunity but often dysregulated in cancer development. The B7:CD28 family regulates co-stimulatory and inhibitory signals to keep a balance of T-cell activation and tolerance.479 The capacity for the selective recognition of antigens derived from any compartments in one cell, the capacity of CD8+ effector T cells (cytotoxic T lymphocytes (CTLs)) to recognize and kill antigen-expressing cells, and the capacity of CD4+ helper T cells to regulate immune responses make T cells a central role in cancer therapeutics.480 Cytotoxic T-lymphocyte-associated antigen 4 (CTLA4 or CD152) and programmed cell death protein 1 (PD-1 or CD279) are the most studied immunoinhibitory receptors. PD-1 ligand 1 (PD-L1, B7-H1 or CD274) and PDL2 (also B7-DC or CD273) are two ligands for PD-1.481,482 Their interaction may cause the inhibition of T-lymphocyte proliferation and cytokine production to suppress the immune response.483 High level of PD-L1 expression was reported in many types of cancer and myeloid cells in the tumor microenvironment.484–489
Studies revealed PD-L1-mediated immunosuppression can be regulated by several PTMs, indicating that except for expression regulation, targeting PTMs of PD-L1 may be a potential strategy to enhance antitumor immune responses (Fig. 7). As a multifunctional serine/threonine kinase, GSK3 regulates a large number of signal pathways through a phosphorylation cascade.490 In a recent study, PD-L1 was observed to undergo N-glycosylated at sites N35, N192, N200, and N219 to facilitate its stabilization. PD-L1 glycosylation antagonized its interaction with GSK3β, preventing GSK3β-mediated PD-L1 destabilization.491 In a study of triple-negative breast cancer, β-1,3-N-acetylglucosaminyltransferase (B3GNT3)-mediated PD-L1 glycosylation increased PD-L1 protein stability and PD-L1/PD-1 interaction. In the co-culture system of tumor cells with PBMC-derived activated T cells, the mutation of PD-L1 glycosylation sites exhibits more sensitivity to T cells.492 A spliced isoform of FK506-binding protein 51 (FKBP51s) catalyzed PD-L1 glycosylation to enhance its expression on the plasma membrane in glioblastoma.493 In CSCs, the EMT process induces N-glycosyltransferase STT3 expression through transcription factor β-catenin and produces a more stable N-glycosylated PD-L1.494 These results suggest that PD-L1 glycosylation is critical for PD-L1-mediated immunosuppression and targeting this PTM is an effective immune checkpoint therapy strategy.
Fig. 7.
The modification of ubiquitination, palmitoylation, and glycosylation of the programmed cell death ligand 1 (PD-L1) protein
Cytotoxic T cells
The inhibitors of farnesyltransferase and geranylgeranylation are reported to regulate immune cell proliferation, differentiation, and function. According to the types of cytokine production and distinct APC populations, the activated CD4+ T cells differentiate into three subtypes. (1) Th1 cells produce IFN-γ and lymphotoxin after the stimulate of cytokines IL-12 and IL-18 to regulate NFAT and NF-κB transcription factors, MAP kinase pathway, as well as the members of the IRF-1 family.495 (2) Th2 cells produce IL-4, IL-5, and IL-13 to regulate the expulsion of parasites and provide B-cell help.495 (3) Stimuli-like microbial factors, adjuvants including LPS or CpG, and β-glucan components in zymosan promote Th17 cells differentiation and production of IL-10, IL-17A, IL-17F, and IL-22 after TGF-β induction to regulate its anti-inflammatory fate in intestinal immune homeostasis maintenance.496,497 The CD4+CD25+Foxp3+ regulatory T cells (nTregs) function in the inhibition of naive T cells differentiate into pathogenic T-helper 1 effectors to suppress autoimmune disease and control a set of physiological and pathological immune responses.498,499 In addition, the depletion of isoprenoid intermediates reduced Ras farnesylation and RhoA geranylgeranylation, preventing Th1-mediated autoimmune disease experimental autoimmune encephalomyelitis (EAE).500 Simvastatin is a semisynthetic compound and cholesterol-lowering drug derived from lovastatin that inhibit HMG-CoA reductase activity. After TGF-β stimulation, simvastatin promotes Foxp3+ CD4+ T-cell differentiation. The same result was observed after GGTI-298 treatment but not FTI-277, suggesting protein geranylgeranylation but not protein farnesylation is responsible for regulating the balance between Th17 cells and Foxp3+ CD4+ T cells.501
Ca2+ channels (CRAC) in the plasma membrane is responsible for Ca2+ entry to mediate biological process as a second messenger in mast and T cells.502 The uptake and release of Ca2+ (also called Ca2+ handling) in mitochondrial regulate energy conversion, metabolism, differentiation, proliferation, gene expression, and immune response. The tumor suppressor Fus1, which is involved in Ca2+-myristoyl switch protein family, has been identified to balance mitochondrial changes during CD4+ T-cell activation.503 In addition, Fus1−/− mouse exhibit abnormal inflammatory cytokine expression, altered T-cell differentiation and changed mitochondrial function.504 More importantly, the regulation function of Fus1 in Ca2+ handling depends on myristoylation. N-myristoylated Fus1 facilitates protein docking to the mitochondrial membrane to ensure mitochondrial Ca2+ handling and its tumor suppressor function.505 Cytotoxic T lymphocytes (CD8+ T cell, CTL) play an essential role in immune defense against intracellular pathogens like viruses, intracellular bacteria, and tumor. The interaction of Fas protein in target cells and Fas ligand (FaL) in CTLs activates caspase cascade in the targeted cells and induces apoptosis. The post-translational myristoylation of protein also regulates CTL function via its role in apoptosis.72 The activated caspase-8 cleaves the cytosolic form of proapoptotic molecule BID, resulting in a truncated fragment that relocates to mitochondria. The early study had identified N-myristoylation of BID after cleavage improved its insertion into mitochondria membrane to enhance cytochrome c release and cell death.506 Subsequently, many other proteins have been identified myristoylated and regulate nuclear fragmentation or cytoskeletal remodeling during apoptosis, these proteins include p21-activated kinase 2 (PAK2), gelsolin, cytoplasmic dynein intermediate chain 2 (DIC2 or CDIC2), B-cell receptor-associated protein 31 (Bap31), microtubule-actin cross-linking factor (MACF), and YTH domain family 2 protein (YTHDF2).301 The myristoylation occurs in caspase-cleaved PAK2 C-terminal kinase fragment to facilitate its location in membrane ruffles and internal membranes, resulting in c-JNK-dependent cell death.507 Targeting myristoylation-related protein subcellular location and cytoskeleton remodeling in apoptosis may lead to advancement in anticancer therapy.
PTMs in PD-L1 and functional regulation
T-cell-based immune response relies on the interaction of TCR on T cells with peptide-major histocompatibility complexes (MHC) on target cells. A series of spatiotemporal control by co-stimulatory or co-inhibitory receptors and their ligands (immune checkpoints) contributes to T-cell activation and function.508 Recently, novel immune checkpoint molecules have been intensively studied except for PD-L1 and CTLA4. These targets contain B- and T-lymphocyte attenuator (BTLA), lymphocyte activation gene-3 (LAG-3), T-cell immunoglobulin and mucin-domain-containing-3 (TIM-3), T-cell immunoglobulin and ITIM domain (TIGIT), and VISTA.509 PD-1 (CD279) is a receptor of the Ig superfamily and can be expressed CD4+ and CD8+ T cells, B cells, macrophages, natural killer T (NKT) cells, and some subset of dendritic cells (DCs) after the immune stimulus.509 Two ligands PD-L1 (CD274, B7-H1) and PDL2 (CD273, B7-DC) bind PD-1 to inhibit T‐cell activation and proliferation, resulting in maintenance of homeostasis and prevention from autoimmunity. PD‐L1 is expressed on several types of cancer as well as on tumor-infiltrating immune cells in the tumor microenvironment. Several signal pathways (MAPK, MEK-ERK pathway and the phosphoinositide 3‐kinase (PI3K)/Akt pathway), the transcription factors (MYC, AP-1, HIF-1, signal transducer, and activator of transcription 3 (STAT3), NF‐ĸB, TGF‐β, GATA‐3, and T‐box transcription factor T‐bet), as well as the activation of KRAS, EGFR, and anaplastic lymphoma kinase (ALK) regulate PD-L1 expression.510,511 PD-1 and PD-L1 interaction not only results in self-tolerance activation but also provides a strategy to escape immune surveillance.512 Considering the central role of PD-L1 in the fundamental biology of immune escape and its predictive value for clinical response, a deep understanding of PD-L1 regulation is need to design effective cancer immune checkpoint therapies. The complex regulatory network of PD-L1 level includes (1) genetic alterations at the PD-L1 locus, (2) inflammatory signals such as pro-inflammatory cytokine IFN-γ and type I interferons (IFN-α and IFN-β), (3) aberrant oncogenic signaling pathways, (4) microRNA-mediated PD-L1 mRNA regulation, and (5) post-translational modulation.511 PD-L1 expression, stability, and the activity to bind PD-1 can be directly regulated by several proteins. With a whole-genome CRISPR/Cas9 deletion library screen, CKLF-like MARVEL transmembrane domain-containing protein 6 (CMTM6) was identified as a major regulator of PD-L1 expression. CMTM6 colocalized with PD-L1 at the plasma membrane and in recycling endosomes to reduce its ubiquitination and prevents PD-L1 from lysosome-mediated degradation. Meanwhile, CMTM6 also fulfills a similar role in the immunological synapse between T cells and cancer cells or APCs.513 Another family member CMTM4 was responsible for the positive regulation of PD-L1 expression in CMTM6 deficient cells.514 The cyclin-dependent kinase 4 (CDK4) phosphorylates SPOP, the E3-ubiquitin ligase of PD-L1, and stabilizes SPOP by dissociating it from the ubiquitin E3-ligase complex APC/CCdh1. The evaluated SPOP promotes ubiquitination-mediated PD-L1 degradation, leading to decreased PD-L1 level.515
In addition to ubiquitination, other protein PTMs of PD-L1 also modulate its immunosuppression activity. The serine/threonine protein kinase GSK3β binds to and phosphorylates PD-L1, leading to ubiquitination-associated PD-L1 degradation. This process was blocked by EGF signal-induced PD-L1 glycosylation.491 Moreover, glycosylation of PD-L1 is necessary for PD-L1 and PD-1 interaction. A type II transmembrane protein B3GNT3 binds to and catalyzes PD-L1 N-linked glycosylation and B3GNT3 depletion inhibit tumor cell growth.492 COP9 signalosome (CSN) specific phosphorylation can target substrates to ubiquitin-dependent degradation.516 One of the family members CSN5 also possesses deubiquitination activity and positively regulates PD-L1. NF-kB induced CSN5 expression, resulting in CSN5-dependent PD-L1 deubiquitination and stabilization.511,517 In the current studies, palmitoylation was discovered on PD-L1 and played an important role in protein stability. PD-L1 expression was decreased after palmitoylation inhibitor treatment. The palmitoylation of PD-L1 also protected cancer cells from T-cell killing and promoted tumor growth. The cysteine-rich zinc finger protein family member ZDHHC9 is responsible for PD-L1 palmitoylation to maintain its stability and cell surface distribution.518 In addition, PD-L1 palmitoylation also blocks its ubiquitination and lysosome-associated degradation, with ZDHHC3 as the main acetyltransferase. Inhibitors targeting PD-L1 palmitoylation were proved to be effective to overcome PD-L1-mediated immune evasion.519 Anti-PD-1/PD-L1 agents and combination therapies with hundreds of chemotherapies, radiotherapy, and anti-angiogenic agents have been tested in active clinical trials and serve as promising attempt in the cancer immunotherapy field.520 Targeting the novel interchange between various PTMs of PD-L1 protein offer novel opportunities to combat immune surveillance and develop more effective drugs and treatments from a new perspective.
Therapy targets
Protein PTMs play key roles in the regulation of cellular hemostasis by targeting both histones and non-histones. Over the past few years, HDAC inhibitors have been discovered and reviewed both as single anticancer agents and in combination with other chemotherapeutic drugs/radiotherapy.521–531 For example, the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA; vorinostat (Zolinza)) may cause cell death in tumor cells and tumor-bearing animals. SAHA has been approved by the US Food and Drug Administration for clinical trials.532 Another potent histone deacetylase inhibitor romidepsin induces cell-cycle arrest or selective apoptosis of malignant cells and has been recently FDA approved in several cancer types, including solid tumors and hematologic malignancies.533,534
Considering the oncogenic function of Ras proteins and their malignant transformation regulation by farnesylation, strategies targeting this PTM are processed to treat cancer a long time ago. Many compounds were identified by random screening to block Ras farnesylation or farnesyl-protein transferase (FPTase) in RAS-transformed cells, human cancer cells, and animal models with no toxicity to normal cells or to normal tissues.535 The anticancer function of farnesyltransferase inhibitors and GGTase I inhibitors on cell-cycle control to induce apoptosis and the related bench-to-bedside translational strategies have been reviewed in detail.536–538 Inhibitors of farnesyltransferase are developed to block farnesyl attachment such as the benzodiazepine peptidomimetics, the CA1A2X peptidomimetics family, and the non-peptide mimetics of CAAX.539–542 The inhibition of isoprenoid biosynthesis and farnesylation reaction are strategies less direct than the inhibition of FPTase to inhibit Ras farnesylation. Targeted screens and rational designs based on substrates are developed to find proper inhibitors.543–546 The CAAX peptidomimetics L-739,749 inhibited anchorage-independent growth in RAS-transformed Ratl cells and was also effective in animal models.547 Other competitive inhibitors targeting farnesyl diphosphate (FPP) were designed and identified for inhibition of Ras processing in NIH3T3 fibroblasts.544,548 K-Ras was reported to show a higher affinity for farnesyltransferase than H-Ras as well as the substrate potential for geranylgeranyltransferase-1. The inhibitor BZA-2B caused the suppression effect of K-RasB both in its farnesylation and geranylgeranylation activity.549 Moreover, the inhibitor L-744,832 was observed repressed tumor growth in both (MMTV)-v-Ha-RAS transgenic mice and MMTV-N-RAS mice.550,551 Several preclinical studies of farnesyltransferase displayed antitumor effect by blocking cell proliferation, inducing apoptosis, increasing reactive oxygen species and causing DNA double-strand breaks with single inhibitors, dual inhibitors, and combination treatments with chemotherapies.552
For the clinical study, an orally bioavailable, nonpeptidomimetic methylquinolone farnesyltransferase inhibitor tipifarnib (Zarnestra, R115777) has exhibited effectiveness in patients with high-risk myelodysplasia, myeloproliferative disorders, and imatinib-resistant chronic myelogenous leukemia.553 In human breast cancer in vivo, tipifarnib inhibits FTase activity and is associated with reduced phospho (p)-STAT3.554 Despite these studies, hundreds of proteins undergo farnesylation, making the mechanisms of drug resistance and several activated pro-survival pathways necessary to be illustrated. In fact, the combination strategy of FTase inhibitors with group I PAK inhibitors is effective in human melanoma, lung, and colon cancer.555 Another farnesyltransferase inhibitor lonafarnib exhibit sufficiently melanoma cell growth inhibition and apoptosis induction after the combination with the pan-RAF inhibitor sorafenib.556 Besides tipifarnib and lonafarnib, other inhibitors including lestaurinib, tandutinib, and PKC 412 have also been designed and tested under early-phase clinical evaluation in acute myeloid leukemia (AML).557 The ongoing clinical trials are listed in Table 2.
Table 2.
The ongoing clinical trials of Farnesyl-transferase inhibitors
| NCT number | Conditions | Drug | Phases |
|---|---|---|---|
| NCT02535650 | Urothelial carcinoma | Tipifarnib | Phase 2 |
| NCT02210858 | Chronic myeloid leukemia | Tipifarnib | Phase 1|Phase 2 |
| NCT00847223 | Mantle cell lymphoma | Tipifarnib | Phase 2 |
| NCT00612651 | Gliosarcoma | Temodar and SCH66336 | Phase 1 |
| NCT00354146 | Leukemia, nonlymphocytic, acute | Tipifarnib (R115777) | Phase 2 |
| NCT00112853 | Leukemia | Tipifarnib|Etoposide | Phase 1 |
| NCT00102635 | Head and neck cancer | Fenretinide (4-HPR) | Drug: SCH66336 | Phase 1 |
| NCT00101153 | Leukemia | Cytarabine|Daunorubicin hydrochloride|Tipifarnib | Phase 1 |
| NCT00096122 | Leukemia | Cytarabine|Idarubicin|Tipifarnib | Phase 1|Phase 2 |
| NCT00093990 | Leukemia | Tipifarnib;Zarnestra; R115777 | Phase 3 |
| NCT00093470 | Leukemia | Tipifarnib | Phase 3 |
| NCT00093418 | Leukemia | Tipifarnib | Phase 2 |
| NCT00004009 | Leukemia | Tipifarnib | Phase 1 |
| NCT00005041 | Lung cancer | Tipifarnib | Phase 2 |
| NCT00012350 | Multiple myeloma | FTI | Phase 2 |
| NCT00083096 | Brain and central nervous system tumors | lonafarnib|Temozolomide | Phase 1 |
| NCT00005989 | Lung cancer | Tipifarnib | Phase 2 |
| NCT00020774 | Liver cancer | Gemcitabine hydrochloride|lonafarnib | Phase 2 |
| NCT00006376 | Bladder cancer|transitional cell cancer of the renal pelvis and ureter | Chemotherapy|Tipifarnib | Phase 2 |
| NCT00006085 | Unspecified adult solid tumor, protocol specific | CP-609,754 | Phase 1 |
| NCT00005848 | Prostate cancer | Chemotherapy|Tipifarnib | Phase 2 |
| NCT00005843 | Pancreatic cancer | Tipifarnib | Phase 2 |
| NCT00077519 | Pancreatic cancer | Tipifarnib|Radiation | Phase 1 |
| NCT00077363 | Recurrent breast cancer|stage iv breast cancer | Tipifarnib|Capecitabine | Phase 2 |
| NCT00076102 | Neurofibromatosis 1|neurofibroma, plexiform | Pirfenidone | Phase 2 |
| NCT00006213 | Leukemia | BMS-214662 | Phase 1 |
| NCT00073450 | Carcinoma, squamous cell|head and neck neoplasms | Farnesyl-protein transferase inhibitor | Phase 2 |
| NCT00070252 | Adult solid neoplasm| breast carcinoma | Capecitabine|Docetaxel|Tipifarnib | Phase 1|Phase 2 |
| NCT00005845 | Leukemia | Tipifarnib | Phase 1 |
| NCT00003707 | Unspecified adult solid tumor, protocol specific | Gemcitabine hydrochloride|Tipifarnib | Phase 1 |
| NCT00022529 | Unspecified adult solid tumor, protocol specific | BMS-214662|Trastuzumab|Pharmacological study | Phase 1 |
| NCT00006242 | Unspecified adult solid tumor, protocol specific | BMS-214662 | Phase 1 |
| NCT00055757 | Stage IIIB non-small cell lung cancer|stage IV non-small cell lung cancer | Tipifarnib|Cisplatin|Gemcitabine hydrochloride | Phase 2 |
| NCT00054470 | Breast cancer | Trastuzumab|Tipifarnib | Phase 2 |
| NCT00045396 | Leukemia | Tipifarnib | Phase 2 |
| NCT00027872 | Leukemia | Tipifarnib | Phase 2 |
| NCT00026104 | Adenocarcinoma of the pancreas| pancreatic cancer | Gemcitabine hydrochloride|Paclitaxel|Tipifarnib|Radiation | Phase 2 |
| NCT00025480 | Lung cancer | Carboplatin|Paclitaxel|Tipifarnib|Radiation | Phase 1 |
| NCT00025038 | Juvenile myelomonocytic leukemia | Tipifarnib|Isotretinoin|Fludarabine phosphate|Cytarabine|Radiation|Cyclophosphamide|Anti-thymocyte globulin | Phase 2 |
| NCT00006351 | Bladder cancer|transitional cell cancer of the renal pelvis and ureter|urethral cancer | Gemcitabine hydrochloride|lonafarnib | Phase 2 |
| NCT00005990 | Unspecified adult solid tumor, protocol specific | Tipifarnib|Topotecan hydrochloride | Phase 1 |
| NCT00005973 | Unspecified adult solid tumor, protocol specific|unspecified childhood solid tumor, protocol specific | BMS-214662|Other: laboratory biomarker analysis | Phase 1 |
| NCT00050986 | Glioblastoma multiforme | Temozolomide|R115777 | Phase 1|Phase 2 |
| NCT00050336 | Carcinoma, non-small-cell lung|metastases, neoplasm | Lonafarnib (SARASAR) | Phase 3 |
| NCT00050141 | Breast cancer | ZARNESTRA, Tipifarnib, R115777 | Phase 2 |
| NCT00048503 | Acute myeloid leukemia | ZARNESTRA, Tipifarnib, R115777 | Phase 2 |
| NCT00040547 | Neoplasms | Farnesyl-Protein Transferase Inhibitor | Phase 1 |
| NCT00040534 | Neoplasms | Farnesyl-Protein Transferase Inhibitor | Phase 1 |
| NCT00038597 | Myelogenous leukemia, chronic | SCH66336 | Phase 2 |
| NCT00038493 | Glioblastoma multiforme | Temozolomide and SCH66336 | Phase 2 |
| NCT00034684 | Leukemia | Farnesyl-protein transferase inhibitor | Phase 1|Phase 2 |
| NCT00021541 | Neurofibroma, plexiform|neurofibromatosis type I | Tipifarnib|Other: placebo | Phase 2 |
Components targeting the ubiquitin–proteasome system is an effective strategy for the treatment of cancer. MLN4924 is a small-molecule inhibitor of NAE screened through a high throughput assay. MLN4924 treatment resulted in S-phase disruption and suppressed tumor growth both in vivo and in vitro.558 Pharmaceutically, MLN4924 forms an irreversible covalent adduct with NEDD8 to disrupt the thioester bond between NEDD8 with NAE and block substrates neddylation.559 The In eukaryotes, ORC (origin recognizing complex) recognizes and binds to replication origins to initiate DNA replication, following recruitment of replication licensing factors Cdt1 and Cdc6, as well as MCM complex to form pre-replicative complex (pre-RC).560–562 The MLN4924 treatment also increased and stabilized Cdt1 protein level, leading to DNA re-replication, the activation of apoptosis, and senescence. Interestingly, compared to wide type cells, p53−/− and p21−/− cells appeared to be more sensitive to MLN4924.563 In addition to S-phase arrest and appearance of re-replicated DNA, MLN4924 was reported to cause other phenotypes including Cullin-RING ligase (CRL) substrates degradation and ATM/ATR kinase-associated single-strand and double-strand breaks.564 Based on these results, MLN4924 has been studied as a promising and effective anticancer strategy. Several clinical trials of MLN4924 are ongoing in anticancer therapy. For the treatment of AML, MLN4924 dramatically disrupted AML cells growth, inhibit clone formation, and decreased Cullin-dependent substrates expression, causing the inhibition of NF-κB transcriptional activity and increased reactive oxygen species (ROS) generation.565 However, there are different biological mechanisms of MLN4924 in distinct genetic subgroups of diffuse large B-cell lymphoma (DLBCL). MLN4924 downregulated NF-κB target genes in activated B-cell-like (ABC) DLBCL and inducted DNA re-replication in germinal-center B-cell-like (GCB) DLBCL.566 The combination strategy of NAE inhibitors showed potential benefit. MLN4924 treatment increased radiosensitivity in pancreatic cancer cells with enhanced G2/M arrest, aneuploidy, and apoptosis. Mechanistically, the inhibition of Cullin neddylation by MLN4924 increased the accumulation of SCF (SKP1, Cullins, and F-box protein) E3 substrates CDT1 and WEE1 after radiation, followed by a remarkable increase in DNA damage and genomic instability.567 Cisplatin is one of the widely used anticancer agents by the formation of inter-strand DNA cross-linkages (ICL) to damage DNA, inhibit DNA synthesis and induce tumor cell death.568 Fanconi anemia (FA) pathway is responsible for resolving ICLs to maintain genomic stability, in which FANCD2 mono-ubiquitination is a central event.569 MLN4924 can suppress ICL-induced FANCD2 mono-ubiquitination, enhancing sensitivity to DNA cross-linking agents.570 In addition, MLN4924 functions not only in the induction of apoptosis to suppress cancer cell growth but also in the induction of an irreversible senescence through persistent accumulation of p21 and sustained activation of DNA damage response.571 In response to the multiunit CRL inactivation, protective autophagy was induced by MLN4924. The corresponding phenotypes of inhibition of the mTOR pathway, accumulation of DEPTOR and HIF-1α, and ROS-induced stress were observed.572,573 More recently, MLN4924 has been shown to suppress cell migration, proliferation, and tube formation in vitro, as well as angiogenesis and tumorigenesis in vivo.574 In conclusion, inhibitors targeting neddylation like MLN4924 may have potential as chemotherapeutics. The clinical trials of MLN4924 are listed in Table S1. However, cancer cells with mutations in the NAE subunit UBA3 have developed MLN4924 resistance and restored cell survival.575,576 Other small-molecule inhibitors are discovered to target E2 conjugating enzyme (UBE2M or UBC12) and DCN1 (DCUN1D1) interaction and DCN1-mediated Cullin neddylation.577,578 The treatment of compound WS-383 inhibited Cul3/1 neddylation, leading to the accumulation of p21, p27, and erythroid 2-related factor 2 (NRF2).579 High-affinity inhibitors DI-404 and DI-591 bind to cellular DCN1 and DCN2 and disrupt the association of DCN1 and UBC12, selectively inhibiting Cullin-3 neddylation and increased NRF2 level.580,581
Given that NMT-mediated protein myristoylation participates widely in signaling processes in many types of cancer, NMT can be considered as a potential target for cancer treatments. The novel ceramidase inhibitors (1 S,2 R)-N-myristoylamino-phenylpropanol-1 (d–e-MAPP) and (1 R,2 R)-N-myristoylamino-4′-nitro-phenylpropandiol-1,3 (B13) was synthesized and examined to block Src N-myristoylation.582 As with NMT1 knockdown, a small-molecule inhibitor B13 blocked NMT1 enzymatic activity and suppressed proliferation, migration, and invasion in vitro and tumor growth in vivo.260 Tris (dibenzylideneacetone) dipalladium (Tris DBA), an organopalladium compound identified in the melanoma study, showed NMT1 inhibition function. This compound was found to have the significant activity to block tumor growth through decreased downstream signaling pathways, including MAPK kinase, PI3K, and Stat-3.583 For the sake of drug resistance of tyrosine kinase inhibitors (TKIs) harboring mutations in the BCR-ABL kinase in chronic myelogenous leukemia (CML), several inhibitors like asciminib (ABL001) and GNF-2 were discovered. Those inhibitors occupied the internal myristate binding site/pocket in the ABL kinase domain to recover the negative regulatory effect of TKIs.584–587 A series of phase I studies are ongoing (ABL001 + Nilotinib, ABL001 + Imatinib, and ABL001 + Dasatinib) in CML or Ph+ ALL patients (https://clinicaltrials.gov/ct2/show/NCT02081378).
Concluding remarks and perspectives
This review highlights the fundamental questions of several highly preserved but less-studied protein PTMs, including the basic signature, the techniques or predictive tools to detect these PTMs, the function and regulation in cancer biology, and suppression effects in the tumor microenvironment. Further characterization of the functional impacts of these novel PTMs on immune regulation may extend our understanding of the multifunctional regulatory network of TME and provide new ideas to improving immunotherapy efficacy.
Given the numerous and diverse links of tumor cell and TME regulation, drugs targeting PTMs have proven rather effective both in preclinical and clinical studies. However, numerous hurdles and uncertainties remain in targeting PTMs into clinical practice. The key challenges include: (1) how does metabolic associated PTMs regulate innate and acquired immunity in cancer progression and treatment; (2) that it is hard to predict a specific modification and its effect in compensatory cellular pathways considering the heterogeneity and plasticity of TME; (3) that it is necessary to identify specific drug targets and biological mechanisms after the treatment of PTM inhibitors; (4) that the effects of endogenous factors, such as patients’ clinical features and microbiota in TME, on the outcomes of treatments remain to be determined; (5) that it is a hot area to construct a preclinical model of TME regulation based on different PTMs and their crosstalk; (6) that an increased understanding of the dominant drivers of PTM regulation is needed to enable reprogramming of TME, which is available for tumor elimination; and (7) that an effective and efficient assessment of PTMs regulation in combination with TME shaping in early-phase clinical studies has the potential to improve the cost-effectiveness of anticancer treatment.
Addressing these challenges will require the combined efforts of basic researchers and clinicians, and the focusing of resources to accelerate understanding of the complex interactions between PTMs regulation and the TME shaping system and the development of improved treatment options for patients with cancer.
Supplementary Information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81873048 to C.X. and 82003114 to W.L.) and the Applied Basic Research Project of Sichuan Science and Technology Department (No. 2020YJ0174 to W.L.).
Author contributions
All authors contributed to writing this paper. All authors have read and approved the article.
Competing interests
The authors declare no competing of interests.
Supplementary information
The online version contains supplementary material available at 10.1038/s41392-021-00825-8.
References
- 1.Corner BM, Cawley PF. Vibrio parahaemolyticus-food poisoning: case report. N. Z. Med. J. 1976;83:155–156. [PubMed] [Google Scholar]
- 2.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.LeBleu VS. Imaging the tumor microenvironment. Cancer J. 2015;21:174–178. doi: 10.1097/PPO.0000000000000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dittmer J, Leyh B. The impact of tumor stroma on drug response in breast cancer. Semin. Cancer Biol. 2015;31:3–15. doi: 10.1016/j.semcancer.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Thompson RF, Maity A. Radiotherapy and the tumor microenvironment: mutual influence and clinical implications. Adv. Exp. Med. Biol. 2014;772:147–165. doi: 10.1007/978-1-4614-5915-6_7. [DOI] [PubMed] [Google Scholar]
- 7.Hivroz C, Chemin K, Tourret M, Bohineust A. Crosstalk between T lymphocytes and dendritic cells. Crit. Rev. Immunol. 2012;32:139–155. doi: 10.1615/critrevimmunol.v32.i2.30. [DOI] [PubMed] [Google Scholar]
- 8.Klebanoff CA, Gattinoni L, Restifo NP. CD8+ T-cell memory in tumor immunology and immunotherapy. Immunol. Rev. 2006;211:214–224. doi: 10.1111/j.0105-2896.2006.00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angell H, Galon J. From the immune contexture to the immunoscore: the role of prognostic and predictive immune markers in cancer. Curr. Opin. Immunol. 2013;25:261–267. doi: 10.1016/j.coi.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Lv L, et al. The accumulation and prognosis value of tumor infiltrating IL-17 producing cells in esophageal squamous cell carcinoma. PLoS ONE. 2011;6:e18219. doi: 10.1371/journal.pone.0018219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hockel M, et al. Intratumoral pO2 predicts survival in advanced cancer of the uterine cervix. Radiother. Oncol. 1993;26:45–50. doi: 10.1016/0167-8140(93)90025-4. [DOI] [PubMed] [Google Scholar]
- 12.Hockel M, et al. Association between tumor hypoxia and malignant progression in advanced cancer of the uterine cervix. Cancer Res. 1996;56:4509–4515. [PubMed] [Google Scholar]
- 13.Vaupel P, Mayer A. Hypoxia in tumors: pathogenesis-related classification, characterization of hypoxia subtypes, and associated biological and clinical implications. Adv. Exp. Med. Biol. 2014;812:19–24. doi: 10.1007/978-1-4939-0620-8_3. [DOI] [PubMed] [Google Scholar]
- 14.Elinav E, Garrett WS, Trinchieri G, Wargo J. The cancer microbiome. Nat. Rev. Cancer. 2019;19:371–376. doi: 10.1038/s41568-019-0155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh CT, Garneau-Tsodikova S, Gatto GJ. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 16.Wu Z, Huang R, Yuan L. Crosstalk of intracellular post-translational modifications in cancer. Arch. Biochem. Biophys. 2019;676:108138. doi: 10.1016/j.abb.2019.108138. [DOI] [PubMed] [Google Scholar]
- 17.Duan G, Walther D. The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput. Biol. 2015;11:e1004049. doi: 10.1371/journal.pcbi.1004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holland PM, Cooper JA. Protein modification: docking sites for kinases. Curr. Biol. 1999;9:R329–R331. doi: 10.1016/s0960-9822(99)80205-x. [DOI] [PubMed] [Google Scholar]
- 19.Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y. SnapShot: histone modifications. Cell. 2014;159:458. doi: 10.1016/j.cell.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwammle V, et al. Systems level analysis of histone H3 post-translational modifications (PTMs) reveals features of PTM crosstalk in chromatin regulation. Mol. Cell. Proteom. 2016;15:2715–2729. doi: 10.1074/mcp.M115.054460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang KY, et al. dbPTM in 2019: exploring disease association and cross-talk of post-translational modifications. Nucleic Acids Res. 2019;47:D298–D308. doi: 10.1093/nar/gky1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minguez P, et al. PTMcode v2: a resource for functional associations of post-translational modifications within and between proteins. Nucleic Acids Res. 2015;43:D494–D502. doi: 10.1093/nar/gku1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y, Jensen ON. Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques. Proteomics. 2009;9:4632–4641. doi: 10.1002/pmic.200900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.PHILLIPS DM. The presence of acetyl groups of histones. Biochem. J. 1963;87:258–263. doi: 10.1042/bj0870258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 26.Gil J, Ramirez-Torres A, Encarnacion-Guevara S. Lysine acetylation and cancer: a proteomics perspective. J. Proteom. 2017;150:297–309. doi: 10.1016/j.jprot.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat. Rev. Drug Discov. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 28.Zhang K, Tian S, Fan E. Protein lysine acetylation analysis: current MS-based proteomic technologies. Analyst. 2013;138:1628–1636. doi: 10.1039/c3an36837h. [DOI] [PubMed] [Google Scholar]
- 29.Park PH, Miller R, Shukla SD. Acetylation of histone H3 at lysine 9 by ethanol in rat hepatocytes. Biochem Biophys. Res. Commun. 2003;306:501–504. doi: 10.1016/s0006-291x(03)01040-4. [DOI] [PubMed] [Google Scholar]
- 30.Avalos JL, Bever KM, Wolberger C. Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol. Cell. 2005;17:855–868. doi: 10.1016/j.molcel.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J, Zhou C, Caflisch A. Structure-based discovery of selective BRPF1 bromodomain inhibitors. Eur. J. Med. Chem. 2018;155:337–352. doi: 10.1016/j.ejmech.2018.05.037. [DOI] [PubMed] [Google Scholar]
- 32.Baell JB, et al. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature. 2018;560:253–257. doi: 10.1038/s41586-018-0387-5. [DOI] [PubMed] [Google Scholar]
- 33.Yan K, et al. Deficient histone H3 propionylation by BRPF1-KAT6 complexes in neurodevelopmental disorders and cancer. Sci. Adv. 2020;6:x21. doi: 10.1126/sciadv.aax0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Z, et al. Molecular characterization of propionyllysines in non-histone proteins. Mol. Cell. Proteom. 2009;8:45–52. doi: 10.1074/mcp.M800224-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteom. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu G, et al. SAHA regulates histone acetylation, butyrylation, and protein expression in neuroblastoma. J. Proteome Res. 2014;13:4211–4219. doi: 10.1021/pr500497e. [DOI] [PubMed] [Google Scholar]
- 37.Bachhawat BK, Coon MJ, Kupiecki FP, Nagle R, Robinson WG. Coenzyme A thiol esters of isobutyric, methacrylic, and beta-hydroxyisobutyric acids as intermediates in the enzymatic degradation of valine. J. Biol. Chem. 1957;224:1–11. [PubMed] [Google Scholar]
- 38.Zhu Z, et al. Identification of lysine isobutyrylation as a new histone modification mark. Nucleic Acids Res. 2021;49:177–189. doi: 10.1093/nar/gkaa1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai L, et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 2014;10:365–370. doi: 10.1038/nchembio.1497. [DOI] [PubMed] [Google Scholar]
- 40.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabari BR, et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell. 2015;58:203–215. doi: 10.1016/j.molcel.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Comerford SA, et al. Acetate dependence of tumors. Cell. 2014;159:1591–1602. doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mashimo T, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159:1603–1614. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu W, et al. Global profiling of crotonylation on non-histone proteins. Cell Res. 2017;27:946–949. doi: 10.1038/cr.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao P, et al. Crotonylation at serine 46 impairs p53 activity. Biochem. Biophys. Res. Commun. 2020;524:730–735. doi: 10.1016/j.bbrc.2020.01.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wan J, Liu H, Ming L. Lysine crotonylation is involved in hepatocellular carcinoma progression. Biomed. Pharmacother. 2019;111:976–982. doi: 10.1016/j.biopha.2018.12.148. [DOI] [PubMed] [Google Scholar]
- 47.Bao, X. et al. Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. eLife. 3, e02999 (2014). [DOI] [PMC free article] [PubMed]
- 48.Zhao D, Li Y, Xiong X, Chen Z, Li H. YEATS domain-A histone acylation reader in health and disease. J. Mol. Biol. 2017;429:1994–2002. doi: 10.1016/j.jmb.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol. Cell. 2016;62:181–193. doi: 10.1016/j.molcel.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang H, et al. p300-mediated lysine 2-hydroxyisobutyrylation regulates glycolysis. Mol. Cell. 2018;70:663–678. doi: 10.1016/j.molcel.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang H, et al. Landscape of the regulatory elements for lysine 2-hydroxyisobutyrylation pathway. Cell Res. 2018;28:111–125. doi: 10.1038/cr.2017.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abmayr SM, Workman JL. Histone lysine de-beta-hydroxybutyrylation by SIRT3. Cell Res. 2019;29:694–695. doi: 10.1038/s41422-019-0211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X, et al. Molecular basis for hierarchical histone de-beta-hydroxybutyrylation by SIRT3. Cell Discov. 2019;5:35. doi: 10.1038/s41421-019-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng C, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteom. 2011;10:M111–M12658. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie Z, et al. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteom. 2012;11:100–107. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bao X, Zhao Q, Yang T, Fung YM, Li XD. A chemical probe for lysine malonylation. Angew. Chem. Int Ed. Engl. 2013;52:4883–4886. doi: 10.1002/anie.201300252. [DOI] [PubMed] [Google Scholar]
- 57.Bandyopadhyay GK, Yu JG, Ofrecio J, Olefsky JM. Increased malonyl-CoA levels in muscle from obese and type 2 diabetic subjects lead to decreased fatty acid oxidation and increased lipogenesis; thiazolidinedione treatment reverses these defects. Diabetes. 2006;55:2277–2285. doi: 10.2337/db06-0062. [DOI] [PubMed] [Google Scholar]
- 58.Du Y, et al. Lysine malonylation is elevated in type 2 diabetic mouse models and enriched in metabolic associated proteins. Mol. Cell. Proteom. 2015;14:227–236. doi: 10.1074/mcp.M114.041947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishida Y, et al. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol. Cell. 2015;59:321–332. doi: 10.1016/j.molcel.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosen R, et al. Probing the active site of homoserine trans-succinylase. FEBS Lett. 2004;577:386–392. doi: 10.1016/j.febslet.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 61.Weinert BT, et al. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Z, et al. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan M, et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hirschey MD, Zhao Y. Metabolic regulation by lysine malonylation, succinylation, and glutarylation. Mol. Cell. Proteom. 2015;14:2308–2315. doi: 10.1074/mcp.R114.046664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao S, et al. Small molecule inhibition of CPS1 activity through an allosteric pocket. Cell Chem. Biol. 2020;27:259–268. doi: 10.1016/j.chembiol.2020.01.009. [DOI] [PubMed] [Google Scholar]
- 66.Ju Z, He JJ. Prediction of lysine glutarylation sites by maximum relevance minimum redundancy feature selection. Anal. Biochem. 2018;550:1–7. doi: 10.1016/j.ab.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 67.Xu Y, Yang Y, Ding J, Li C. iGlu-Lys: a predictor for lysine glutarylation through amino acid pair order features. IEEE Trans. Nanobioscience. 2018;17:394–401. doi: 10.1109/TNB.2018.2848673. [DOI] [PubMed] [Google Scholar]
- 68.Al-Barakati HJ, Saigo H, Newman RH, Kc DB. RF-GlutarySite: a random forest based predictor for glutarylation sites. Mol. Omics. 2019;15:189–204. doi: 10.1039/c9mo00028c. [DOI] [PubMed] [Google Scholar]
- 69.Ju Z, Wang SY. Computational identification of lysine glutarylation sites using positive-unlabeled learning. Curr. Genomics. 2020;21:204–211. doi: 10.2174/1389202921666200511072327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dou L, Li X, Zhang L, Xiang H, Xu L. iGlu_AdaBoost: identification of lysine glutarylation using the AdaBoost classifier. J. Proteome Res. 2021;20:191–201. doi: 10.1021/acs.jproteome.0c00314. [DOI] [PubMed] [Google Scholar]
- 71.Schmiesing J, et al. Disease-linked glutarylation impairs function and interactions of mitochondrial proteins and contributes to mitochondrial heterogeneity. Cell Rep. 2018;24:2946–2956. doi: 10.1016/j.celrep.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 72.Udenwobele DI, et al. Myristoylation: an important protein modification in the immune response. Front. Immunol. 2017;8:751. doi: 10.3389/fimmu.2017.00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giang DK, Cravatt BF. A second mammalian N-myristoyltransferase. J. Biol. Chem. 1998;273:6595–6598. doi: 10.1074/jbc.273.12.6595. [DOI] [PubMed] [Google Scholar]
- 74.Martin DD, Beauchamp E, Berthiaume LG. Post-translational myristoylation: fat matters in cellular life and death. Biochimie. 2011;93:18–31. doi: 10.1016/j.biochi.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 75.Wright MH, Heal WP, Mann DJ, Tate EW. Protein myristoylation in health and disease. J. Chem. Biol. 2010;3:19–35. doi: 10.1007/s12154-009-0032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boutin JA. Myristoylation. Cell. Signal. 1997;9:15–35. doi: 10.1016/s0898-6568(96)00100-3. [DOI] [PubMed] [Google Scholar]
- 77.Jiang H, et al. Protein lipidation: occurrence, mechanisms, biological functions, and enabling technologies. Chem. Rev. 2018;118:919–988. doi: 10.1021/acs.chemrev.6b00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Braun T, McIlhinney RA, Vergeres G. Myristoylation-dependent N-terminal cleavage of the myristoylated alanine-rich C kinase substrate (MARCKS) by cellular extracts. Biochimie. 2000;82:705–715. doi: 10.1016/s0300-9084(00)01154-8. [DOI] [PubMed] [Google Scholar]
- 79.Manenti S, Sorokine O, Van Dorsselaer A, Taniguchi H. Demyristoylation of the major substrate of protein kinase C (MARCKS) by the cytoplasmic fraction of brain synaptosomes. J. Biol. Chem. 1994;269:8309–8313. [PubMed] [Google Scholar]
- 80.Manenti S, Sorokine O, Van Dorsselaer A, Taniguchi H. Demyristoylation of myristoylated alanine-rich C kinase substrate. Biochem Soc. Trans. 1995;23:561–564. doi: 10.1042/bst0230561. [DOI] [PubMed] [Google Scholar]
- 81.Raju RV, Sharma RK. Coenzyme A dependent myristoylation and demyristoylation in the regulation of bovine spleen N-myristoyltransferase. Mol. Cell. Biochem. 1996;158:107–113. doi: 10.1007/BF00225835. [DOI] [PubMed] [Google Scholar]
- 82.Charron G, et al. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J. Am. Chem. Soc. 2009;131:4967–4975. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 83.Hannoush RN, Arenas-Ramirez N. Imaging the lipidome: omega-alkynyl fatty acids for detection and cellular visualization of lipid-modified proteins. ACS Chem. Biol. 2009;4:581–587. doi: 10.1021/cb900085z. [DOI] [PubMed] [Google Scholar]
- 84.Hannoush RN, Sun J. The chemical toolbox for monitoring protein fatty acylation and prenylation. Nat. Chem. Biol. 2010;6:498–506. doi: 10.1038/nchembio.388. [DOI] [PubMed] [Google Scholar]
- 85.Thinon E, et al. Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat. Commun. 2014;5:4919. doi: 10.1038/ncomms5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu K, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim E, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang J, et al. Myristoylation-mediated phase separation of EZH2 compartmentalizes STAT3 to promote lung cancer growth. Cancer Lett. 2021;516:84–98. doi: 10.1016/j.canlet.2021.05.035. [DOI] [PubMed] [Google Scholar]
- 89.Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J. Mol. Biol. 2002;317:541–557. doi: 10.1006/jmbi.2002.5426. [DOI] [PubMed] [Google Scholar]
- 90.Bologna G, Yvon C, Duvaud S, Veuthey AL. N-Terminal myristoylation predictions by ensembles of neural networks. Proteomics. 2004;4:1626–1632. doi: 10.1002/pmic.200300783. [DOI] [PubMed] [Google Scholar]
- 91.Martinez A, et al. Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics. 2008;8:2809–2831. doi: 10.1002/pmic.200701191. [DOI] [PubMed] [Google Scholar]
- 92.Lobo S, Greentree WK, Linder ME, Deschenes RJ. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- 93.Keller CA, et al. The gamma2 subunit of GABA(A) receptors is a substrate for palmitoylation by GODZ. J. Neurosci. 2004;24:5881–5891. doi: 10.1523/JNEUROSCI.1037-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fukata M, Fukata Y, Adesnik H, Nicoll RA, Bredt DS. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44:987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 95.Huang K, et al. Neuronal palmitoyl acyl transferases exhibit distinct substrate specificity. Faseb J. 2009;23:2605–2615. doi: 10.1096/fj.08-127399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Swarthout JT, et al. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J. Biol. Chem. 2005;280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- 97.Baumgart F, Corral-Escariz M, Perez-Gil J, Rodriguez-Crespo I. Palmitoylation of R-Ras by human DHHC19, a palmitoyl transferase with a CaaX box. Biochim Biophys. Acta. 2010;1798:592–604. doi: 10.1016/j.bbamem.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 98.Tian H, et al. Systematic siRNA screen unmasks NSCLC growth dependence by palmitoyltransferase DHHC5. Mol. Cancer Res. 2015;13:784–794. doi: 10.1158/1541-7786.MCR-14-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kong E, et al. Dynamic palmitoylation links cytosol-membrane shuttling of acyl-protein thioesterase-1 and acyl-protein thioesterase-2 with that of proto-oncogene H-ras product and growth-associated protein-43. J. Biol. Chem. 2013;288:9112–9125. doi: 10.1074/jbc.M112.421073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schlesinger MJ, Magee AI, Schmidt MF. Fatty acid acylation of proteins in cultured cells. J. Biol. Chem. 1980;255:10021–10024. [PubMed] [Google Scholar]
- 101.Wang Q, et al. Bioconjugation by copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 102.Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated proteins: purification and identification. Nat. Protoc. 2007;2:1573–1584. doi: 10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- 103.Rilling HC, Breunger E, Epstein WW, Crain PF. Prenylated proteins: the structure of the isoprenoid group. Science. 1990;247:318–320. doi: 10.1126/science.2296720. [DOI] [PubMed] [Google Scholar]
- 104.Farnsworth CC, Gelb MH, Glomset JA. Identification of geranylgeranyl-modified proteins in HeLa cells. Science. 1990;247:320–322. doi: 10.1126/science.2296721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Epstein WW, Lever D, Leining LM, Bruenger E, Rilling HC. Quantitation of prenylcysteines by a selective cleavage reaction. Proc. Natl Acad. Sci. USA. 1991;88:9668–9670. doi: 10.1073/pnas.88.21.9668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vogt A, Qian Y, McGuire TF, Hamilton AD, Sebti SM. Protein geranylgeranylation, not farnesylation, is required for the G1 to S phase transition in mouse fibroblasts. Oncogene. 1996;13:1991–1999. [PubMed] [Google Scholar]
- 107.Reiss Y, Goldstein JL, Seabra MC, Casey PJ, Brown MS. Inhibition of purified p21ras farnesyl:protein transferase by Cys-AAX tetrapeptides. Cell. 1990;62:81–88. doi: 10.1016/0092-8674(90)90242-7. [DOI] [PubMed] [Google Scholar]
- 108.Seabra MC, Reiss Y, Casey PJ, Brown MS, Goldstein JL. Protein farnesyltransferase and geranylgeranyltransferase share a common alpha subunit. Cell. 1991;65:429–434. doi: 10.1016/0092-8674(91)90460-g. [DOI] [PubMed] [Google Scholar]
- 109.Armstrong SA, Seabra MC, Sudhof TC, Goldstein JL, Brown MS. cDNA cloning and expression of the alpha and beta subunits of rat Rab geranylgeranyl transferase. J. Biol. Chem. 1993;268:12221–12229. [PubMed] [Google Scholar]
- 110.Tschantz WR, Zhang L, Casey PJ. Cloning, expression, and cellular localization of a human prenylcysteine lyase. J. Biol. Chem. 1999;274:35802–35808. doi: 10.1074/jbc.274.50.35802. [DOI] [PubMed] [Google Scholar]
- 111.Tschantz WR, Digits JA, Pyun HJ, Coates RM, Casey PJ. Lysosomal prenylcysteine lyase is a FAD-dependent thioether oxidase. J. Biol. Chem. 2001;276:2321–2324. doi: 10.1074/jbc.C000616200. [DOI] [PubMed] [Google Scholar]
- 112.Weinhouse S. On respiratory impairment in cancer cells. Science. 1956;124:267–269. doi: 10.1126/science.124.3215.267. [DOI] [PubMed] [Google Scholar]
- 113.Hirschhaeuser F, Sattler UG, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res. 2011;71:6921–6925. doi: 10.1158/0008-5472.CAN-11-1457. [DOI] [PubMed] [Google Scholar]
- 114.Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J. Clin. Investig. 2013;123:3685–3692. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goodwin ML, Gladden LB, Nijsten MW, Jones KB. Lactate and cancer: revisiting the Warburg effect in an era of lactate shuttling. Front. Nutr. 2014;1:27. doi: 10.3389/fnut.2014.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mu X, et al. Tumor-derived lactate induces M2 macrophage polarization via the activation of the ERK/STAT3 signaling pathway in breast cancer. Cell Cycle. 2018;17:428–438. doi: 10.1080/15384101.2018.1444305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Apicella M, et al. Increased lactate secretion by cancer cells sustains non-cell-autonomous adaptive resistance to MET and EGFR targeted therapies. Cell Metab. 2018;28:848–865. doi: 10.1016/j.cmet.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 118.Goodwin ML, et al. Lactate and cancer: a “lactatic” perspective on spinal tumor metabolism (part 1) Ann. Transl. Med. 2019;7:220. doi: 10.21037/atm.2019.02.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pennington Z, et al. Lactate and cancer: spinal metastases and potential therapeutic targets (part 2) Ann. Transl. Med. 2019;7:221. doi: 10.21037/atm.2019.01.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang D, et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574:575–580. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Haas R, et al. Intermediates of metabolism: from bystanders to signalling molecules. Trends Biochem. Sci. 2016;41:460–471. doi: 10.1016/j.tibs.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 122.Hart GW, Copeland RJ. Glycomics hits the big time. Cell. 2010;143:672–676. doi: 10.1016/j.cell.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wells L, et al. Dynamic O-glycosylation of nuclear and cytosolic proteins: further characterization of the nucleocytoplasmic beta-N-acetylglucosaminidase, O-GlcNAcase. J. Biol. Chem. 2002;277:1755–1761. doi: 10.1074/jbc.m109656200. [DOI] [PubMed] [Google Scholar]
- 124.Slawson C, Hart GW. O-GlcNAc signalling: implications for cancer cell biology. Nat. Rev. Cancer. 2011;11:678–684. doi: 10.1038/nrc3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chalkley RJ, Burlingame AL. Identification of GlcNAcylation sites of peptides and alpha-crystallin using Q-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 2001;12:1106–1113. doi: 10.1016/s1044-0305(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 126.Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 1984;259:3308–3317. [PubMed] [Google Scholar]
- 127.Haltiwanger RS, Holt GD, Hart GW. Enzymatic addition of O-GlcNAc to nuclear and cytoplasmic proteins. Identification of a uridine diphospho-N-acetylglucosamine:peptide beta-N-acetylglucosaminyltransferase. J. Biol. Chem. 1990;265:2563–2568. [PubMed] [Google Scholar]
- 128.Dong DL, Hart GW. Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 1994;269:19321–19330. [PubMed] [Google Scholar]
- 129.Kerwin JJ, Lancaster JJ, Feldman PL. Nitric oxide: a new paradigm for second messengers. J. Med. Chem. 1995;38:4343–4362. doi: 10.1021/jm00022a001. [DOI] [PubMed] [Google Scholar]
- 130.Forstermann U, Sessa WC. Nitric oxide synthases: regulation and function. Eur. Heart J. 2012;33:829–837, 837a. doi: 10.1093/eurheartj/ehr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hess DT, Stamler JS. Regulation by S-nitrosylation of protein post-translational modification. J. Biol. Chem. 2012;287:4411–4418. doi: 10.1074/jbc.R111.285742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol. Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 133.Benhar M, Forrester MT, Stamler JS. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 2009;10:721–732. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- 134.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 135.Dohmen RJ. SUMO protein modification. Biochim Biophys. Acta. 2004;1695:113–131. doi: 10.1016/j.bbamcr.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 136.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 137.Banerjee, S., Kumar, M. & Wiener, R. Decrypting UFMylation: how proteins are modified with UFM1. Biomolecules. 10, 1442 (2020). [DOI] [PMC free article] [PubMed]
- 138.Delgado TC, et al. Neddylation, a novel paradigm in liver cancer. Transl. Gastroenterol. Hepatol. 2018;3:37. doi: 10.21037/tgh.2018.06.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Vertegaal AC. Uncovering ubiquitin and ubiquitin-like signaling networks. Chem. Rev. 2011;111:7923–7940. doi: 10.1021/cr200187e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jentsch S, Pyrowolakis G. Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 2000;10:335–342. doi: 10.1016/s0962-8924(00)01785-2. [DOI] [PubMed] [Google Scholar]
- 141.Eisenberg-Lerner A, Ciechanover A, Merbl Y. Post-translational modification profiling—a novel tool for mapping the protein modification landscape in cancer. Exp. Biol. Med (Maywood). 2016;241:1475–1482. doi: 10.1177/1535370216651732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.van Wijk SJ, Timmers HT. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 2010;24:981–993. doi: 10.1096/fj.09-136259. [DOI] [PubMed] [Google Scholar]
- 144.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 145.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 146.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteom. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 148.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 149.Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteom. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 150.Kirkpatrick DS, Gerber SA, Gygi SP. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods. 2005;35:265–273. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 151.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl Acad. Sci. USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cort JR, Chiang Y, Zheng D, Montelione GT, Kennedy MA. NMR structure of conserved eukaryotic protein ZK652.3 from C. elegans: a ubiquitin-like fold. Proteins. 2002;48:733–736. doi: 10.1002/prot.10197. [DOI] [PubMed] [Google Scholar]
- 153.Wei Y, Xu X. UFMylation: a unique & fashionable modification for life. Genomics Proteom. Bioinforma. 2016;14:140–146. doi: 10.1016/j.gpb.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Daniel J, Liebau E. The ufm1 cascade. Cells. 2014;3:627–638. doi: 10.3390/cells3020627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kang SH, et al. Two novel ubiquitin-fold modifier 1 (Ufm1)-specific proteases, UfSP1 and UfSP2. J. Biol. Chem. 2007;282:5256–5262. doi: 10.1074/jbc.M610590200. [DOI] [PubMed] [Google Scholar]
- 156.Azfer A, Niu J, Rogers LM, Adamski FM, Kolattukudy PE. Activation of endoplasmic reticulum stress response during the development of ischemic heart disease. Am. J. Physiol. Heart Circ. Physiol. 2006;291:H1411–H1420. doi: 10.1152/ajpheart.01378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang Y, Zhang M, Wu J, Lei G, Li H. Transcriptional regulation of the Ufm1 conjugation system in response to disturbance of the endoplasmic reticulum homeostasis and inhibition of vesicle trafficking. PLoS ONE. 2012;7:e48587. doi: 10.1371/journal.pone.0048587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tatsumi K, et al. The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat. Commun. 2011;2:181. doi: 10.1038/ncomms1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang M, et al. RCAD/Ufl1, a Ufm1 E3 ligase, is essential for hematopoietic stem cell function and murine hematopoiesis. Cell Death Differ. 2015;22:1922–1934. doi: 10.1038/cdd.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mor Z, Golan A, Bukovsky I, Caspi E. [Gynecological risk factors for osteoporosis] Harefuah. 1990;119:385–387. [PubMed] [Google Scholar]
- 161.Chen C, Itakura E, Weber KP, Hegde RS, de Bono M. An ER complex of ODR-4 and ODR-8/Ufm1 specific protease 2 promotes GPCR maturation by a Ufm1-independent mechanism. PLoS Genet. 2014;10:e1004082. doi: 10.1371/journal.pgen.1004082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Watson IR, Irwin MS, Ohh M. NEDD8 pathways in cancer, Sine Quibus Non. Cancer Cell. 2011;19:168–176. doi: 10.1016/j.ccr.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 163.Kurz T, et al. Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science. 2002;295:1294–1298. doi: 10.1126/science.1067765. [DOI] [PubMed] [Google Scholar]
- 164.Mosadeghi, R. et al. Structural and kinetic analysis of the COP9-Signalosome activation and the cullin-RING ubiquitin ligase deneddylation cycle. eLife. 5, e12102 (2016). [DOI] [PMC free article] [PubMed]
- 165.Cope GA, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 166.Goldenberg SJ, et al. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 167.Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol. Cell. 2002;10:1511–1518. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- 168.Kamitani T, Kito K, Fukuda-Kamitani T, Yeh ET. Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J. Biol. Chem. 2001;276:46655–46660. doi: 10.1074/jbc.M108636200. [DOI] [PubMed] [Google Scholar]
- 169.Kito K, Yeh ET, Kamitani T. NUB1, a NEDD8-interacting protein, is induced by interferon and down-regulates the NEDD8 expression. J. Biol. Chem. 2001;276:20603–20609. doi: 10.1074/jbc.M100920200. [DOI] [PubMed] [Google Scholar]
- 170.Rabut G, Peter M. Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Xirodimas DP. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc. Trans. 2008;36:802–806. doi: 10.1042/BST0360802. [DOI] [PubMed] [Google Scholar]
- 172.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 173.Liu B, et al. Identification and characterization of propionylation at histone H3 lysine 23 in mammalian cells. J. Biol. Chem. 2009;284:32288–32295. doi: 10.1074/jbc.M109.045856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vermeulen M, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 175.Li N, et al. ZMYND8 reads the dual histone mark H3K4me1-H3K14ac to antagonize the expression of metastasis-linked genes. Mol. Cell. 2016;63:470–484. doi: 10.1016/j.molcel.2016.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kebede AF, et al. Histone propionylation is a mark of active chromatin. Nat. Struct. Mol. Biol. 2017;24:1048–1056. doi: 10.1038/nsmb.3490. [DOI] [PubMed] [Google Scholar]
- 177.Goudarzi A, et al. Dynamic competing histone H4 K5K8 acetylation and butyrylation are hallmarks of highly active gene promoters. Mol. Cell. 2016;62:169–180. doi: 10.1016/j.molcel.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Liu X, et al. MOF as an evolutionarily conserved histone crotonyltransferase and transcriptional activation by histone acetyltransferase-deficient and crotonyltransferase-competent CBP/p300. Cell Discov. 2017;3:17016. doi: 10.1038/celldisc.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wei W, et al. Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017;27:898–915. doi: 10.1038/cr.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xie Z, et al. Metabolic regulation of gene expression by histone lysine beta-hydroxybutyrylation. Mol. Cell. 2016;62:194–206. doi: 10.1016/j.molcel.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen L, Miao Z, Xu X. beta-hydroxybutyrate alleviates depressive behaviors in mice possibly by increasing the histone3-lysine9-beta-hydroxybutyrylation. Biochem. Biophys. Res. Commun. 2017;490:117–122. doi: 10.1016/j.bbrc.2017.05.184. [DOI] [PubMed] [Google Scholar]
- 182.Bao X, et al. Glutarylation of histone H4 Lysine 91 regulates chromatin dynamics. Mol. Cell. 2019;76:660–675. doi: 10.1016/j.molcel.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 183.Szabo P, Cai X, Ali G, Blass JP. Localization of the gene (OGDH) coding for the E1k component of the alpha-ketoglutarate dehydrogenase complex to chromosome 7p13-p11.2. Genomics. 1994;20:324–326. doi: 10.1006/geno.1994.1178. [DOI] [PubMed] [Google Scholar]
- 184.Wang Y, et al. KAT2A coupled with the alpha-KGDH complex acts as a histone H3 succinyltransferase. Nature. 2017;552:273–277. doi: 10.1038/nature25003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl Acad. Sci. USA. 2010;107:19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fujiki R, et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480:557–560. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wang P, et al. OGT mediated histone H2B S112 GlcNAcylation regulates DNA damage response. J. Genet. Genomics. 2015;42:467–475. doi: 10.1016/j.jgg.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 189.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 190.Irwin MS, Miller FD. p73: regulator in cancer and neural development. Cell Death Differ. 2004;11(Suppl 1):S17–S22. doi: 10.1038/sj.cdd.4401452. [DOI] [PubMed] [Google Scholar]
- 191.Watson IR, Blanch A, Lin DC, Ohh M, Irwin MS. Mdm2-mediated NEDD8 modification of TAp73 regulates its transactivation function. J. Biol. Chem. 2006;281:34096–34103. doi: 10.1074/jbc.M603654200. [DOI] [PubMed] [Google Scholar]
- 192.Abida WM, Nikolaev A, Zhao W, Zhang W, Gu W. FBXO11 promotes the Neddylation of p53 and inhibits its transcriptional activity. J. Biol. Chem. 2007;282:1797–1804. doi: 10.1074/jbc.M609001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sundqvist A, Liu G, Mirsaliotis A, Xirodimas DP. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 2009;10:1132–1139. doi: 10.1038/embor.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mahata B, Sundqvist A, Xirodimas DP. Recruitment of RPL11 at promoter sites of p53-regulated genes upon nucleolar stress through NEDD8 and in an Mdm2-dependent manner. Oncogene. 2012;31:3060–3071. doi: 10.1038/onc.2011.482. [DOI] [PubMed] [Google Scholar]
- 195.DeGregori J, Johnson DG. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr. Mol. Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 196.Biswas AK, Johnson DG. Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res. 2012;72:13–17. doi: 10.1158/0008-5472.CAN-11-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kontaki H, Talianidis I. Lysine methylation regulates E2F1-induced cell death. Mol. Cell. 2010;39:152–160. doi: 10.1016/j.molcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 198.Stevens, C., Smith, L. & La Thangue, N. B. Chk2 activates E2F-1 in response to DNA damage. Nat. Cell Biol.5, 401–409 (2003). [DOI] [PubMed]
- 199.Galbiati L, Mendoza-Maldonado R, Gutierrez MI, Giacca M. Regulation of E2F-1 after DNA damage by p300-mediated acetylation and ubiquitination. Cell Cycle. 2005;4:930–939. doi: 10.4161/cc.4.7.1784. [DOI] [PubMed] [Google Scholar]
- 200.Loftus SJ, Liu G, Carr SM, Munro S. & La Thangue, N. B. NEDDylation regulates E2F-1-dependent transcription. EMBO Rep. 2012;13:811–818. doi: 10.1038/embor.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Matsubara M, Nakatsu T, Kato H, Taniguchi H. Crystal structure of a myristoylated CAP-23/NAP-22 N-terminal domain complexed with Ca2+/calmodulin. EMBO J. 2004;23:712–718. doi: 10.1038/sj.emboj.7600093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Moriya K, et al. Protein N-myristoylation is required for cellular morphological changes induced by two formin family proteins, FMNL2 and FMNL3. Biosci. Biotechnol. Biochem. 2012;76:1201–1209. doi: 10.1271/bbb.120069. [DOI] [PubMed] [Google Scholar]
- 203.Pechlivanis M, Ringel R, Popkirova B, Kuhlmann J. Prenylation of Ras facilitates hSOS1-promoted nucleotide exchange, upon Ras binding to the regulatory site. Biochemistry. 2007;46:5341–5348. doi: 10.1021/bi602353k. [DOI] [PubMed] [Google Scholar]
- 204.Porfiri E, Evans T, Chardin P, Hancock JF. Prenylation of Ras proteins is required for efficient hSOS1-promoted guanine nucleotide exchange. J. Biol. Chem. 1994;269:22672–22677. [PubMed] [Google Scholar]
- 205.Hori Y, et al. Post-translational modifications of the C-terminal region of the rho protein are important for its interaction with membranes and the stimulatory and inhibitory GDP/GTP exchange proteins. Oncogene. 1991;6:515–522. [PubMed] [Google Scholar]
- 206.Casteel DE, et al. Rho isoform-specific interaction with IQGAP1 promotes breast cancer cell proliferation and migration. J. Biol. Chem. 2012;287:38367–38378. doi: 10.1074/jbc.M112.377499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Roos MD, Su K, Baker JR, Kudlow JE. O glycosylation of an Sp1-derived peptide blocks known Sp1 protein interactions. Mol. Cell. Biol. 1997;17:6472–6480. doi: 10.1128/mcb.17.11.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yang X, et al. O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc. Natl Acad. Sci. USA. 2001;98:6611–6616. doi: 10.1073/pnas.111099998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lim K, Chang HI. O-GlcNAcylation of Sp1 interrupts Sp1 interaction with NF-Y. Biochem Biophys. Res Commun. 2009;382:593–597. doi: 10.1016/j.bbrc.2009.03.075. [DOI] [PubMed] [Google Scholar]
- 210.Latorre P, Varona L, Burgos C, Carrodeguas JA, Lopez-Buesa P. O-GlcNAcylation mediates the control of cytosolic phosphoenolpyruvate carboxykinase activity via Pgc1alpha. PLoS ONE. 2017;12:e179988. doi: 10.1371/journal.pone.0179988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Gewinner C, et al. The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5. J. Biol. Chem. 2004;279:3563–3572. doi: 10.1074/jbc.M306449200. [DOI] [PubMed] [Google Scholar]
- 212.Ruan HB, et al. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1alpha stability. Cell Metab. 2012;16:226–237. doi: 10.1016/j.cmet.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wells L, Slawson C, Hart GW. The E2F-1 associated retinoblastoma-susceptibility gene product is modified by O-GlcNAc. Amino Acids. 2011;40:877–883. doi: 10.1007/s00726-010-0709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Robinson AM, Williamson DH. Physiological roles of ketone bodies as substrates and signals in mammalian tissues. Physiol. Rev. 1980;60:143–187. doi: 10.1152/physrev.1980.60.1.143. [DOI] [PubMed] [Google Scholar]
- 215.Dabek, A., Wojtala, M., Pirola, L. & Balcerczyk, A. Modulation of cellular biochemistry, epigenetics and metabolomics by ketone bodies. Implications of the ketogenic diet in the physiology of the organism and pathological states. Nutrients12, 788 (2020). [DOI] [PMC free article] [PubMed]
- 216.Martinez-Outschoorn UE, et al. Ketones and lactate increase cancer cell “stemness,” driving recurrence, metastasis and poor clinical outcome in breast cancer: achieving personalized medicine via Metabolo-Genomics. Cell Cycle. 2011;10:1271–1286. doi: 10.4161/cc.10.8.15330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010;2:a1008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 220.Jiang L, Hickman JH, Wang SJ, Gu W. Dynamic roles of p53-mediated metabolic activities in ROS-induced stress responses. Cell Cycle. 2015;14:2881–2885. doi: 10.1080/15384101.2015.1068479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Jiang L, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Feng Z, Lin M, Wu R. The regulation of aging and longevity: a new and complex role of p53. Genes Cancer. 2011;2:443–452. doi: 10.1177/1947601911410223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wang D, et al. Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature. 2016;538:118–122. doi: 10.1038/nature19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Liu K, et al. p53 beta-hydroxybutyrylation attenuates p53 activity. Cell Death Dis. 2019;10:243. doi: 10.1038/s41419-019-1463-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Li X, et al. Lysine-222 succinylation reduces lysosomal degradation of lactate dehydrogenase a and is increased in gastric cancer. J. Exp. Clin. Cancer Res. 2020;39:172. doi: 10.1186/s13046-020-01681-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Michalak KP, Mackowska-Kedziora A, Sobolewski B, Wozniak P. Key roles of glutamine pathways in reprogramming the cancer metabolism. Oxid. Med. Cell. Longev. 2015;2015:964321. doi: 10.1155/2015/964321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Tong Y, et al. SUCLA2-coupled regulation of GLS succinylation and activity counteracts oxidative stress in tumor cells. Mol. Cell. 2021;81:2303–2316. doi: 10.1016/j.molcel.2021.04.002. [DOI] [PubMed] [Google Scholar]
- 229.Ma Y, et al. SIRT5-mediated SDHA desuccinylation promotes clear cell renal cell carcinoma tumorigenesis. Free Radic. Biol. Med. 2019;134:458–467. doi: 10.1016/j.freeradbiomed.2019.01.030. [DOI] [PubMed] [Google Scholar]
- 230.Bishop AL, Hall A. Rho GTPases and their effector proteins. Biochem. J. 2000;348(Pt 2):241–255. [PMC free article] [PubMed] [Google Scholar]
- 231.Perona R, et al. Activation of the nuclear factor-kappaB by Rho, CDC42, and Rac-1 proteins. Genes Dev. 1997;11:463–475. doi: 10.1101/gad.11.4.463. [DOI] [PubMed] [Google Scholar]
- 232.Coso OA, et al. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 233.Abo A, et al. Activation of the NADPH oxidase involves the small GTP-binding protein p21rac1. Nature. 1991;353:668–670. doi: 10.1038/353668a0. [DOI] [PubMed] [Google Scholar]
- 234.Olson MF, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 235.Braga VM. Small GTPases and regulation of cadherin dependent cell-cell adhesion. Mol. Pathol. 1999;52:197–202. doi: 10.1136/mp.52.4.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Johnson DI. Cdc42: An essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol. Biol. Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.van de Donk NW, Kamphuis MM, van Kessel B, Lokhorst HM, Bloem AC. Inhibition of protein geranylgeranylation induces apoptosis in myeloma plasma cells by reducing Mcl-1 protein levels. Blood. 2003;102:3354–3362. doi: 10.1182/blood-2003-03-0970. [DOI] [PubMed] [Google Scholar]
- 238.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 239.Vaux DL. Apoptogenic factors released from mitochondria. Biochim Biophys. Acta. 2011;1813:546–550. doi: 10.1016/j.bbamcr.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 240.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 241.Kvansakul M, et al. Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain-swapped dimer that binds a highly selective subset of BH3-containing death ligands. Cell Death Differ. 2008;15:1564–1571. doi: 10.1038/cdd.2008.83. [DOI] [PubMed] [Google Scholar]
- 242.Zhang B, Gojo I, Fenton RG. Myeloid cell factor-1 is a critical survival factor for multiple myeloma. Blood. 2002;99:1885–1893. doi: 10.1182/blood.v99.6.1885. [DOI] [PubMed] [Google Scholar]
- 243.Gojo I, Zhang B, Fenton RG. The cyclin-dependent kinase inhibitor flavopiridol induces apoptosis in multiple myeloma cells through transcriptional repression and down-regulation of Mcl-1. Clin. Cancer Res. 2002;8:3527–3538. [PubMed] [Google Scholar]
- 244.Li X, et al. Inhibition of protein geranylgeranylation and RhoA/RhoA kinase pathway induces apoptosis in human endothelial cells. J. Biol. Chem. 2002;277:15309–15316. doi: 10.1074/jbc.M201253200. [DOI] [PubMed] [Google Scholar]
- 245.van de Donk NW, et al. Protein geranylgeranylation is critical for the regulation of survival and proliferation of lymphoma tumor cells. Clin. Cancer Res. 2003;9:5735–5748. [PubMed] [Google Scholar]
- 246.Nonaka M, et al. Role for protein geranylgeranylation in adult T-cell leukemia cell survival. Exp. Cell Res. 2009;315:141–150. doi: 10.1016/j.yexcr.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 247.Murugan AK, Munirajan AK, Tsuchida N. Ras oncogenes in oral cancer: the past 20 years. Oral. Oncol. 2012;48:383–392. doi: 10.1016/j.oraloncology.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 248.Stokoe D, Macdonald SG, Cadwallader K, Symons M, Hancock JF. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 249.Schaap D, van der Wal J, Howe LR, Marshall CJ, van Blitterswijk WJ. A dominant-negative mutant of raf blocks mitogen-activated protein kinase activation by growth factors and oncogenic p21ras. J. Biol. Chem. 1993;268:20232–20236. [PubMed] [Google Scholar]
- 250.Rodriguez-Viciana, P., Warne, P. H., Vanhaesebroeck, B., Waterfield, M. D. & Downward, J. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J.15, 2442–2451 (1996). [PMC free article] [PubMed]
- 251.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 252.Zinatizadeh MR, et al. The role and function of Ras-association domain family in cancer: a review. Genes Dis. 2019;6:378–384. doi: 10.1016/j.gendis.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Schafer WR, et al. Genetic and pharmacological suppression of oncogenic mutations in ras genes of yeast and humans. Science. 1989;245:379–385. doi: 10.1126/science.2569235. [DOI] [PubMed] [Google Scholar]
- 254.Hancock JF, Magee AI, Childs JE, Marshall CJ. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 255.Casey PJ, Solski PA, Der CJ, Buss JE. p21ras is modified by a farnesyl isoprenoid. Proc. Natl Acad. Sci. USA. 1989;86:8323–8327. doi: 10.1073/pnas.86.21.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Playford MP, Schaller MD. The interplay between Src and integrins in normal and tumor biology. Oncogene. 2004;23:7928–7946. doi: 10.1038/sj.onc.1208080. [DOI] [PubMed] [Google Scholar]
- 257.Patwardhan P, Resh MD. Myristoylation and membrane binding regulate c-Src stability and kinase activity. Mol. Cell. Biol. 2010;30:4094–4107. doi: 10.1128/MCB.00246-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Kamps MP, Buss JE, Sefton BM. Mutation of NH2-terminal glycine of p60src prevents both myristoylation and morphological transformation. Proc. Natl Acad. Sci. USA. 1985;82:4625–4628. doi: 10.1073/pnas.82.14.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Shoji S, Kurosawa T, Inoue H, Funakoshi T, Kubota Y. Human cellular src gene product: identification of the myristoylated pp60c-src and blockage of its myristoyl acylation with N-fatty acyl compounds resulted in the suppression of colony formation. Biochem. Biophys. Res. Commun. 1990;173:894–901. doi: 10.1016/s0006-291x(05)80870-8. [DOI] [PubMed] [Google Scholar]
- 260.Kim S, et al. Blocking myristoylation of src inhibits its kinase activity and suppresses prostate cancer progression. Cancer Res. 2017;77:6950–6962. doi: 10.1158/0008-5472.CAN-17-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ducker CE, Upson JJ, French KJ, Smith CD. Two N-myristoyltransferase isozymes play unique roles in protein myristoylation, proliferation, and apoptosis. Mol. Cancer Res. 2005;3:463–476. doi: 10.1158/1541-7786.MCR-05-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 263.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 264.Embade N, et al. Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology. 2012;55:1237–1248. doi: 10.1002/hep.24795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Fernandez-Ramos D, Martinez-Chantar ML. NEDDylation in liver cancer: the regulation of the RNA binding protein Hu antigen R. Pancreatology. 2015;15:S49–S54. doi: 10.1016/j.pan.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 266.Han, S. et al. The protein neddylation inhibitor MLN4924 suppresses patient-derived glioblastoma cells via inhibition of ERK and AKT signaling. Cancers11, 1849 (2019). [DOI] [PMC free article] [PubMed]
- 267.Lopez-Sanchez LM, Aranda E, Rodriguez-Ariza A. Nitric oxide and tumor metabolic reprogramming. Biochem. Pharmacol. 2020;176:113769. doi: 10.1016/j.bcp.2019.113769. [DOI] [PubMed] [Google Scholar]
- 268.Han T, et al. ASK1 inhibits proliferation and migration of lung cancer cells via inactivating TAZ. Am. J. Cancer Res. 2020;10:2785–2799. [PMC free article] [PubMed] [Google Scholar]
- 269.Park HS, et al. Inhibition of apoptosis signal-regulating kinase 1 by nitric oxide through a thiol redox mechanism. J. Biol. Chem. 2004;279:7584–7590. doi: 10.1074/jbc.M304183200. [DOI] [PubMed] [Google Scholar]
- 270.Park HS, Huh SH, Kim MS, Lee SH, Choi EJ. Nitric oxide negatively regulates c-Jun N-terminal kinase/stress-activated protein kinase by means of S-nitrosylation. Proc. Natl Acad. Sci. USA. 2000;97:14382–14387. doi: 10.1073/pnas.97.26.14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Park HS, Mo JS, Choi EJ. Nitric oxide inhibits an interaction between JNK1 and c-Jun through nitrosylation. Biochem. Biophys. Res Commun. 2006;351:281–286. doi: 10.1016/j.bbrc.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 272.Lee YR, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat. Rev. Mol. Cell Biol. 2018;19:547–562. doi: 10.1038/s41580-018-0015-0. [DOI] [PubMed] [Google Scholar]
- 273.Xie P, et al. Neddylation of PTEN regulates its nuclear import and promotes tumor development. Cell Res. 2021;31:291–311. doi: 10.1038/s41422-020-00443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Numajiri N, et al. On-off system for PI3-kinase-Akt signaling through S-nitrosylation of phosphatase with sequence homology to tensin (PTEN) Proc. Natl Acad. Sci. USA. 2011;108:10349–10354. doi: 10.1073/pnas.1103503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Slawson C, Pidala J, Potter R. Increased N-acetyl-beta-glucosaminidase activity in primary breast carcinomas corresponds to a decrease in N-acetylglucosamine containing proteins. Biochim Biophys. Acta. 2001;1537:147–157. doi: 10.1016/s0925-4439(01)00067-9. [DOI] [PubMed] [Google Scholar]
- 276.Krzeslak A, Pomorski L, Lipinska A. Elevation of nucleocytoplasmic beta-N-acetylglucosaminidase (O-GlcNAcase) activity in thyroid cancers. Int. J. Mol. Med. 2010;25:643–648. doi: 10.3892/ijmm_00000387. [DOI] [PubMed] [Google Scholar]
- 277.Gu Y, et al. GlcNAcylation plays an essential role in breast cancer metastasis. Cancer Res. 2010;70:6344–6351. doi: 10.1158/0008-5472.CAN-09-1887. [DOI] [PubMed] [Google Scholar]
- 278.Mi W, et al. O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim Biophys. Acta. 2011;1812:514–519. doi: 10.1016/j.bbadis.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 279.Shi Y, et al. Aberrant O-GlcNAcylation characterizes chronic lymphocytic leukemia. Leukemia. 2010;24:1588–1598. doi: 10.1038/leu.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Shaw P, Freeman J, Bovey R, Iggo R. Regulation of specific DNA binding by p53: evidence for a role for O-glycosylation and charged residues at the carboxy-terminus. Oncogene. 1996;12:921–930. [PubMed] [Google Scholar]
- 281.Baird TD, Wek RC. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv. Nutr. 2012;3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Datta B, Ray MK, Chakrabarti D, Wylie DE, Gupta NK. Glycosylation of eukaryotic peptide chain initiation factor 2 (eIF-2)-associated 67-kDa polypeptide (p67) and its possible role in the inhibition of eIF-2 kinase-catalyzed phosphorylation of the eIF-2 alpha-subunit. J. Biol. Chem. 1989;264:20620–20624. [PubMed] [Google Scholar]
- 283.Dong DL, Xu ZS, Hart GW, Cleveland DW. Cytoplasmic O-GlcNAc modification of the head domain and the KSP repeat motif of the neurofilament protein neurofilament-H. J. Biol. Chem. 1996;271:20845–20852. doi: 10.1074/jbc.271.34.20845. [DOI] [PubMed] [Google Scholar]
- 284.Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 285.Ha JR, et al. beta-catenin is O-GlcNAc glycosylated at Serine 23: implications for beta-catenin’s subcellular localization and transactivator function. Exp. Cell Res. 2014;321:153–166. doi: 10.1016/j.yexcr.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 286.Olivier-Van SS, et al. O-GlcNAcylation stabilizes beta-catenin through direct competition with phosphorylation at threonine 41. FASEB J. 2014;28:3325–3338. doi: 10.1096/fj.13-243535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–5189. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- 288.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat. Rev. Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 289.Myatt SS, Lam EW. Targeting FOXM1. Nat. Rev. Cancer. 2008;8:242. doi: 10.1038/nrc2223-c2. [DOI] [PubMed] [Google Scholar]
- 290.Wang, Z., Banerjee, S., Kong, D., Li, Y. & Sarkar, F. H. Retraction: down-regulation of forkhead box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res. 78, 5470 (2018). [DOI] [PubMed]
- 291.Caldwell SA, et al. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene. 2010;29:2831–2842. doi: 10.1038/onc.2010.41. [DOI] [PubMed] [Google Scholar]
- 292.Hart GW, Haltiwanger RS, Holt GD, Kelly WG. Glycosylation in the nucleus and cytoplasm. Annu. Rev. Biochem. 1989;58:841–874. doi: 10.1146/annurev.bi.58.070189.004205. [DOI] [PubMed] [Google Scholar]
- 293.Chou TY, Dang CV, Hart GW. Glycosylation of the c-Myc transactivation domain. Proc. Natl Acad. Sci. USA. 1995;92:4417–4421. doi: 10.1073/pnas.92.10.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Chou TY, Hart GW, Dang CV. c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J. Biol. Chem. 1995;270:18961–18965. doi: 10.1074/jbc.270.32.18961. [DOI] [PubMed] [Google Scholar]
- 295.Brady CA, Attardi LD. p53 at a glance. J. Cell Sci. 2010;123:2527–2532. doi: 10.1242/jcs.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Yang WH, et al. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- 297.Panieri E, Saso L. Inhibition of the NRF2/KEAP1 axis: a promising therapeutic strategy to alter redox balance of cancer cells. Antioxid. Redox Signal. 2021;34:1428–1483. doi: 10.1089/ars.2020.8146. [DOI] [PubMed] [Google Scholar]
- 298.Chen PH, et al. Glycosylation of KEAP1 links nutrient sensing to redox stress signaling. EMBO J. 2017;36:2233–2250. doi: 10.15252/embj.201696113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Johnson DR, Bhatnagar RS, Knoll LJ, Gordon JI. Genetic and biochemical studies of protein N-myristoylation. Annu. Rev. Biochem. 1994;63:869–914. doi: 10.1146/annurev.bi.63.070194.004253. [DOI] [PubMed] [Google Scholar]
- 300.Budd RC. An overview of apoptosis. Coron. Artery Dis. 1997;8:593–597. doi: 10.1097/00019501-199710000-00002. [DOI] [PubMed] [Google Scholar]
- 301.Martin DD, Hayden MR. Post-translational myristoylation at the cross roads of cell death, autophagy and neurodegeneration. Biochem Soc. Trans. 2015;43:229–234. doi: 10.1042/BST20140281. [DOI] [PubMed] [Google Scholar]
- 302.Teng YB, et al. Efficient demyristoylase activity of SIRT2 revealed by kinetic and structural studies. Sci. Rep. 2015;5:8529. doi: 10.1038/srep08529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Zhang, X., Spiegelman, N. A., Nelson, O. D., Jing, H. & Lin, H. SIRT6 regulates Ras-related protein R-Ras2 by lysine defatty-acylation. eLife6, e25158 (2017). [DOI] [PMC free article] [PubMed]
- 304.DeBerardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl Acad. Sci. USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Kuhajda FP. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition. 2000;16:202–208. doi: 10.1016/s0899-9007(99)00266-x. [DOI] [PubMed] [Google Scholar]
- 306.Swinnen JV, et al. Overexpression of fatty acid synthase is an early and common event in the development of prostate cancer. Int. J. Cancer. 2002;98:19–22. doi: 10.1002/ijc.10127. [DOI] [PubMed] [Google Scholar]
- 307.Rashid A, et al. Elevated expression of fatty acid synthase and fatty acid synthetic activity in colorectal neoplasia. Am. J. Pathol. 1997;150:201–208. [PMC free article] [PubMed] [Google Scholar]
- 308.Furuta E, et al. Fatty acid synthase gene is up-regulated by hypoxia via activation of Akt and sterol regulatory element binding protein-1. Cancer Res. 2008;68:1003–1011. doi: 10.1158/0008-5472.CAN-07-2489. [DOI] [PubMed] [Google Scholar]
- 309.Lin HP, et al. Destabilization of fatty acid synthase by acetylation inhibits de novo lipogenesis and tumor cell growth. Cancer Res. 2016;76:6924–6936. doi: 10.1158/0008-5472.CAN-16-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 311.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 312.Elstrom RL, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 313.Gaglio D, et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol. Syst. Biol. 2011;7:523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Kerner J, Minkler PE, Lesnefsky EJ, Hoppel CL. Fatty acid chain elongation in palmitate-perfused working rat heart: mitochondrial acetyl-CoA is the source of two-carbon units for chain elongation. J. Biol. Chem. 2014;289:10223–10234. doi: 10.1074/jbc.M113.524314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Saggerson D. Malonyl-CoA, a key signaling molecule in mammalian cells. Annu. Rev. Nutr. 2008;28:253–272. doi: 10.1146/annurev.nutr.28.061807.155434. [DOI] [PubMed] [Google Scholar]
- 316.Bowman CE, et al. The mammalian malonyl-CoA synthetase ACSF3 is required for mitochondrial protein malonylation and metabolic efficiency. Cell Chem. Biol. 2017;24:673–684. doi: 10.1016/j.chembiol.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.FitzPatrick DR, Hill A, Tolmie JL, Thorburn DR, Christodoulou J. The molecular basis of malonyl-CoA decarboxylase deficiency. Am. J. Hum. Genet. 1999;65:318–326. doi: 10.1086/302492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Colak G, et al. Proteomic and biochemical studies of lysine malonylation suggest its malonic aciduria-associated regulatory role in mitochondrial function and fatty acid oxidation. Mol. Cell. Proteom. 2015;14:3056–3071. doi: 10.1074/mcp.M115.048850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Park J, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Stacpoole PW. Therapeutic targeting of the pyruvate dehydrogenase complex/pyruvate dehydrogenase kinase (PDC/PDK) axis in cancer. J. Natl Cancer Inst. 2017;109:1–14. doi: 10.1093/jnci/djx071. [DOI] [PubMed] [Google Scholar]
- 321.Chen H, et al. Mild metabolic perturbations alter succinylation of mitochondrial proteins. J. Neurosci. Res. 2017;95:2244–2252. doi: 10.1002/jnr.24103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Tamada M, Suematsu M, Saya H. Pyruvate kinase M2: multiple faces for conferring benefits on cancer cells. Clin. Cancer Res. 2012;18:5554–5561. doi: 10.1158/1078-0432.CCR-12-0859. [DOI] [PubMed] [Google Scholar]
- 323.Xiangyun Y, et al. Desuccinylation of pyruvate kinase M2 by SIRT5 contributes to antioxidant response and tumor growth. Oncotarget. 2017;8:6984–6993. doi: 10.18632/oncotarget.14346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Qi H, et al. Succinylation-dependent mitochondrial translocation of PKM2 promotes cell survival in response to nutritional stress. Cell Death Dis. 2019;10:170. doi: 10.1038/s41419-018-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 326.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Liang J, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat. Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 328.Liang J, et al. Myristoylation confers noncanonical AMPK functions in autophagy selectivity and mitochondrial surveillance. Nat. Commun. 2015;6:7926. doi: 10.1038/ncomms8926. [DOI] [PubMed] [Google Scholar]
- 329.Pepino MY, Kuda O, Samovski D, Abumrad NA. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu. Rev. Nutr. 2014;34:281–303. doi: 10.1146/annurev-nutr-071812-161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Wang J, et al. DHHC4 and DHHC5 facilitate fatty acid uptake by palmitoylating and targeting CD36 to the plasma membrane. Cell Rep. 2019;26:209–221. doi: 10.1016/j.celrep.2018.12.022. [DOI] [PubMed] [Google Scholar]
- 331.Amendola CR, et al. KRAS4A directly regulates hexokinase 1. Nature. 2019;576:482–486. doi: 10.1038/s41586-019-1832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Bollu LR, et al. Involvement of de novo synthesized palmitate and mitochondrial EGFR in EGF induced mitochondrial fusion of cancer cells. Cell Cycle. 2014;13:2415–2430. doi: 10.4161/cc.29338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Edwards PA, Ericsson J. Sterols and isoprenoids: signaling molecules derived from the cholesterol biosynthetic pathway. Annu. Rev. Biochem. 1999;68:157–185. doi: 10.1146/annurev.biochem.68.1.157. [DOI] [PubMed] [Google Scholar]
- 334.Kabakoff BD, Doyle JW, Kandutsch AA. Relationships among dolichyl phosphate, glycoprotein synthesis, and cell culture growth. Arch. Biochem. Biophys. 1990;276:382–389. doi: 10.1016/0003-9861(90)90736-i. [DOI] [PubMed] [Google Scholar]
- 335.Wang Y, Hekimi S. The complexity of making ubiquinone. Trends Endocrinol. Metab. 2019;30:929–943. doi: 10.1016/j.tem.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 336.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 337.Dimitroulakos J, et al. Increased sensitivity of acute myeloid leukemias to lovastatin-induced apoptosis: a potential therapeutic approach. Blood. 1999;93:1308–1318. [PubMed] [Google Scholar]
- 338.Macaulay RJ, Wang W, Dimitroulakos J, Becker LE, Yeger H. Lovastatin-induced apoptosis of human medulloblastoma cell lines in vitro. J. Neurooncol. 1999;42:1–11. doi: 10.1023/a:1006164406202. [DOI] [PubMed] [Google Scholar]
- 339.Rubins JB, et al. Lovastatin induces apoptosis in malignant mesothelioma cells. Am. J. Respir. Crit. Care Med. 1998;157:1616–1622. doi: 10.1164/ajrccm.157.5.9709020. [DOI] [PubMed] [Google Scholar]
- 340.Dimitroulakos J, Yeger H. HMG-CoA reductase mediates the biological effects of retinoic acid on human neuroblastoma cells: lovastatin specifically targets P-glycoprotein-expressing cells. Nat. Med. 1996;2:326–333. doi: 10.1038/nm0396-326. [DOI] [PubMed] [Google Scholar]
- 341.Xia Z, et al. Blocking protein geranylgeranylation is essential for lovastatin-induced apoptosis of human acute myeloid leukemia cells. Leukemia. 2001;15:1398–1407. doi: 10.1038/sj.leu.2402196. [DOI] [PubMed] [Google Scholar]
- 342.Son J, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Halestrap AP. The monocarboxylate transporter family-Structure and functional characterization. IUBMB Life. 2012;64:1–9. doi: 10.1002/iub.573. [DOI] [PubMed] [Google Scholar]
- 344.San-Millan I, Julian CG, Matarazzo C, Martinez J, Brooks GA. Is lactate an oncometabolite? Evidence supporting a role for lactate in the regulation of transcriptional activity of cancer-related genes in MCF7 breast cancer cells. Front. Oncol. 2019;9:1536. doi: 10.3389/fonc.2019.01536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Jiang J, et al. Lactate modulates cellular metabolism through histone lactylation-mediated gene expression in non-small cell lung cancer. Front. Oncol. 2021;11:647559. doi: 10.3389/fonc.2021.647559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Bond MR, Hanover JA. A little sugar goes a long way: the cell biology of O-GlcNAc. J. Cell Biol. 2015;208:869–880. doi: 10.1083/jcb.201501101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Bond MR, Hanover JA. O-GlcNAc cycling: a link between metabolism and chronic disease. Annu. Rev. Nutr. 2013;33:205–229. doi: 10.1146/annurev-nutr-071812-161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Ferrer CM, et al. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol. Cell. 2014;54:820–831. doi: 10.1016/j.molcel.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 2014;15:634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Senft D, Ronai ZA. Regulators of mitochondrial dynamics in cancer. Curr. Opin. Cell Biol. 2016;39:43–52. doi: 10.1016/j.ceb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Nawrocki ST, et al. The NEDD8-activating enzyme inhibitor MLN4924 disrupts nucleotide metabolism and augments the efficacy of cytarabine. Clin. Cancer Res. 2015;21:439–447. doi: 10.1158/1078-0432.CCR-14-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Luo Z, et al. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 2012;72:3360–3371. doi: 10.1158/0008-5472.CAN-12-0388. [DOI] [PubMed] [Google Scholar]
- 353.Zhou Q, et al. Inhibiting neddylation modification alters mitochondrial morphology and reprograms energy metabolism in cancer cells. JCI Insight. 2019;4:1–20. doi: 10.1172/jci.insight.121582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Zitvogel L, et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat. Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 355.Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 356.Baglio SR, et al. Blocking tumor-educated MSC paracrine activity halts osteosarcoma progression. Clin. Cancer Res. 2017;23:3721–3733. doi: 10.1158/1078-0432.CCR-16-2726. [DOI] [PubMed] [Google Scholar]
- 357.Schuldner M, et al. Exosome-dependent immune surveillance at the metastatic niche requires BAG6 and CBP/p300-dependent acetylation of p53. Theranostics. 2019;9:6047–6062. doi: 10.7150/thno.36378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Rogers SL, Gelfand VI. Membrane trafficking, organelle transport, and the cytoskeleton. Curr. Opin. Cell Biol. 2000;12:57–62. doi: 10.1016/s0955-0674(99)00057-5. [DOI] [PubMed] [Google Scholar]
- 360.Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat. Rev. Mol. Cell Biol. 2003;4:938–947. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- 361.Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl Acad. Sci. USA. 2010;107:21517–21522. doi: 10.1073/pnas.1013728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Aldana-Masangkay GI, Sakamoto KM. The role of HDAC6 in cancer. J. Biomed. Biotechnol. 2011;2011:875824. doi: 10.1155/2011/875824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Boggs AE, et al. alpha-Tubulin acetylation elevated in metastatic and basal-like breast cancer cells promotes microtentacle formation, adhesion, and invasive migration. Cancer Res. 2015;75:203–215. doi: 10.1158/0008-5472.CAN-13-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Oh S, et al. Genetic disruption of tubulin acetyltransferase, alphaTAT1, inhibits proliferation and invasion of colon cancer cells through decreases in Wnt1/beta-catenin signaling. Biochem Biophys. Res Commun. 2017;482:8–14. doi: 10.1016/j.bbrc.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 365.Ko, P. et al. Microtubule acetylation controls MDA-MB-231 breast cancer cell invasion through the modulation of endoplasmic reticulum stress. Int. J. Mol. Sci. 22, 6018 (2021). [DOI] [PMC free article] [PubMed]
- 366.Chen H, Xu C, Jin Q, Liu Z. S100 protein family in human cancer. Am. J. Cancer Res. 2014;4:89–115. [PMC free article] [PubMed] [Google Scholar]
- 367.Lokman NA, Pyragius CE, Ruszkiewicz A, Oehler MK, Ricciardelli C. Annexin A2 and S100A10 are independent predictors of serous ovarian cancer outcome. Transl. Res. 2016;171:83–95. doi: 10.1016/j.trsl.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 368.Myrvang HK, Guo X, Li C, Dekker LV. Protein interactions between surface annexin A2 and S100A10 mediate adhesion of breast cancer cells to microvascular endothelial cells. FEBS Lett. 2013;587:3210–3215. doi: 10.1016/j.febslet.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 369.Shang J, et al. S100A10 as a novel biomarker in colorectal cancer. Tumour Biol. 2013;34:3785–3790. doi: 10.1007/s13277-013-0962-1. [DOI] [PubMed] [Google Scholar]
- 370.Domoto T, et al. Evaluation of S100A10, annexin II and B-FABP expression as markers for renal cell carcinoma. Cancer Sci. 2007;98:77–82. doi: 10.1111/j.1349-7006.2006.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.El-Rifai W, et al. Gastric cancers overexpress S100A calcium-binding proteins. Cancer Res. 2002;62:6823–6826. [PubMed] [Google Scholar]
- 372.Wang C, et al. CPT1A-mediated succinylation of S100A10 increases human gastric cancer invasion. J. Cell. Mol. Med. 2019;23:293–305. doi: 10.1111/jcmm.13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Janiszewska M, Primi MC, Izard T. Cell adhesion in cancer: beyond the migration of single cells. J. Biol. Chem. 2020;295:2495–2505. doi: 10.1074/jbc.REV119.007759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Theocharis AD, Manou D, Karamanos NK. The extracellular matrix as a multitasking player in disease. FEBS J. 2019;286:2830–2869. doi: 10.1111/febs.14818. [DOI] [PubMed] [Google Scholar]
- 375.Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 376.Whited AM, Johs A. The interactions of peripheral membrane proteins with biological membranes. Chem. Phys. Lipids. 2015;192:51–59. doi: 10.1016/j.chemphyslip.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 377.Ratheesh A, Yap AS. A bigger picture: classical cadherins and the dynamic actin cytoskeleton. Nat. Rev. Mol. Cell Biol. 2012;13:673–679. doi: 10.1038/nrm3431. [DOI] [PubMed] [Google Scholar]
- 378.van Roy F. Beyond E-cadherin: roles of other cadherin superfamily members in cancer. Nat. Rev. Cancer. 2014;14:121–134. doi: 10.1038/nrc3647. [DOI] [PubMed] [Google Scholar]
- 379.Van Aelst L, D’Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 380.Braga VM, Machesky LM, Hall A, Hotchin NA. The small GTPases Rho and Rac are required for the establishment of cadherin-dependent cell-cell contacts. J. Cell Biol. 1997;137:1421–1431. doi: 10.1083/jcb.137.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Hordijk PL, et al. Inhibition of invasion of epithelial cells by Tiam1-Rac signaling. Science. 1997;278:1464–1466. doi: 10.1126/science.278.5342.1464. [DOI] [PubMed] [Google Scholar]
- 382.Takaishi K, Sasaki T, Kotani H, Nishioka H, Takai Y. Regulation of cell-cell adhesion by rac and rho small G proteins in MDCK cells. J. Cell Biol. 1997;139:1047–1059. doi: 10.1083/jcb.139.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Hattori M, Minato N. Rap1 GTPase: functions, regulation, and malignancy. J. Biochem. 2003;134:479–484. doi: 10.1093/jb/mvg180. [DOI] [PubMed] [Google Scholar]
- 384.Bailey CL, Kelly P, Casey PJ. Activation of Rap1 promotes prostate cancer metastasis. Cancer Res. 2009;69:4962–4968. doi: 10.1158/0008-5472.CAN-08-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Reilly JE, Neighbors JD, Hohl RJ. Targeting protein geranylgeranylation slows tumor development in a murine model of prostate cancer metastasis. Cancer Biol. Ther. 2017;18:872–882. doi: 10.1080/15384047.2016.1219817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 387.Zhou BP, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat. Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 388.Xu Y, et al. Role of CK1 in GSK3beta-mediated phosphorylation and degradation of snail. Oncogene. 2010;29:3124–3133. doi: 10.1038/onc.2010.77. [DOI] [PubMed] [Google Scholar]
- 389.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 390.van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell. Mol. Life Sci. 2008;65:3756–3788. doi: 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Zhu W, Leber B, Andrews DW. Cytoplasmic O-glycosylation prevents cell surface transport of E-cadherin during apoptosis. Embo J. 2001;20:5999–6007. doi: 10.1093/emboj/20.21.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Park SY, et al. Snail1 is stabilized by O-GlcNAc modification in hyperglycaemic condition. EMBO J. 2010;29:3787–3796. doi: 10.1038/emboj.2010.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Jin Y, et al. Neddylation blockade diminishes hepatic metastasis by dampening cancer stem-like cells and angiogenesis in uveal melanoma. Clin. Cancer Res. 2018;24:3741–3754. doi: 10.1158/1078-0432.CCR-17-1703. [DOI] [PubMed] [Google Scholar]
- 394.Li L, et al. Overactivated neddylation pathway as a therapeutic target in lung cancer. J. Natl Cancer Inst. 2014;106:u83. doi: 10.1093/jnci/dju083. [DOI] [PubMed] [Google Scholar]
- 395.Tong S, et al. MLN4924 (Pevonedistat), a protein neddylation inhibitor, suppresses proliferation and migration of human clear cell renal cell carcinoma. Sci. Rep. 2017;7:5599. doi: 10.1038/s41598-017-06098-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Jiang Y, et al. Targeting neddylation inhibits intravascular survival and extravasation of cancer cells to prevent lung-cancer metastasis. Cell Biol. Toxicol. 2019;35:233–245. doi: 10.1007/s10565-019-09472-w. [DOI] [PubMed] [Google Scholar]
- 397.Wike-Hooley JL, Haveman J, Reinhold HS. The relevance of tumour pH to the treatment of malignant disease. Radiother. Oncol. 1984;2:343–366. doi: 10.1016/s0167-8140(84)80077-8. [DOI] [PubMed] [Google Scholar]
- 398.Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J. Natl Cancer Inst. 2007;99:1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 399.Jain RK. Normalizing tumor vasculature with anti-angiogenic therapy: a new paradigm for combination therapy. Nat. Med. 2001;7:987–989. doi: 10.1038/nm0901-987. [DOI] [PubMed] [Google Scholar]
- 400.Semenza GL. Oxygen sensing, homeostasis, and disease. N. Engl. J. Med. 2011;365:537–547. doi: 10.1056/NEJMra1011165. [DOI] [PubMed] [Google Scholar]
- 401.Liao D, Johnson RS. Hypoxia: a key regulator of angiogenesis in cancer. Cancer Metastasis Rev. 2007;26:281–290. doi: 10.1007/s10555-007-9066-y. [DOI] [PubMed] [Google Scholar]
- 402.Rohrig F, Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer. 2016;16:732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- 403.Bruning U, et al. Impairment of angiogenesis by fatty acid synthase inhibition involves mTOR malonylation. Cell Metab. 2018;28:866–880. doi: 10.1016/j.cmet.2018.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Wang Y, Liu Y, Malek SN, Zheng P, Liu Y. Targeting HIF1alpha eliminates cancer stem cells in hematological malignancies. Cell. Stem Cell. 2011;8:399–411. doi: 10.1016/j.stem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Zhang H, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 406.Luo W, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Franovic A, et al. Translational up-regulation of the EGFR by tumor hypoxia provides a nonmutational explanation for its overexpression in human cancer. Proc. Natl Acad. Sci. USA. 2007;104:13092–13097. doi: 10.1073/pnas.0702387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Esteban MA, et al. Regulation of E-cadherin expression by VHL and hypoxia-inducible factor. Cancer Res. 2006;66:3567–3575. doi: 10.1158/0008-5472.CAN-05-2670. [DOI] [PubMed] [Google Scholar]
- 409.Sullivan R, Graham CH. Hypoxia-driven selection of the metastatic phenotype. Cancer Metastasis Rev. 2007;26:319–331. doi: 10.1007/s10555-007-9062-2. [DOI] [PubMed] [Google Scholar]
- 410.Krishnamachary B, Semenza GL. Analysis of hypoxia-inducible factor 1alpha expression and its effects on invasion and metastasis. Methods Enzymol. 2007;435:347–354. doi: 10.1016/S0076-6879(07)35017-9. [DOI] [PubMed] [Google Scholar]
- 411.Moeller BJ, Richardson RA, Dewhirst MW. Hypoxia and radiotherapy: opportunities for improved outcomes in cancer treatment. Cancer Metastasis Rev. 2007;26:241–248. doi: 10.1007/s10555-007-9056-0. [DOI] [PubMed] [Google Scholar]
- 412.Rohwer N, Cramer T. Hypoxia-mediated drug resistance: novel insights on the functional interaction of HIFs and cell death pathways. Drug Resist. Updat. 2011;14:191–201. doi: 10.1016/j.drup.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 413.Semenza GL. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J. Appl. Physiol. 2000;88:1474–1480. doi: 10.1152/jappl.2000.88.4.1474. [DOI] [PubMed] [Google Scholar]
- 414.Kim JW, et al. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol. Cell. Biol. 2004;24:5923–5936. doi: 10.1128/MCB.24.13.5923-5936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Shim H, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl Acad. Sci. USA. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Messai Y, et al. Renal cell carcinoma programmed death-ligand 1, a new direct target of hypoxia-inducible factor-2 alpha, is regulated by von Hippel-Lindau gene mutation status. Eur. Urol. 2016;70:623–632. doi: 10.1016/j.eururo.2015.11.029. [DOI] [PubMed] [Google Scholar]
- 417.Noman MZ, et al. PD-L1 is a novel direct target of HIF-1alpha, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J. Exp. Med. 2014;211:781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Noman MZ, Chouaib S. Targeting hypoxia at the forefront of anticancer immune responses. Oncoimmunology. 2014;3:e954463. doi: 10.4161/21624011.2014.954463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Nowak EC, et al. Immunoregulatory functions of VISTA. Immunol. Rev. 2017;276:66–79. doi: 10.1111/imr.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Jaiswal S, et al. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138:271–285. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Willingham SB, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl Acad. Sci. USA. 2012;109:6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 423.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 424.Hou F, et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Horner SM, Liu HM, Park HS, Briley J, Gale MJ. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl Acad. Sci. USA. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Yang S, et al. Control of antiviral innate immune response by protein geranylgeranylation. Sci. Adv. 2019;5:v7999. doi: 10.1126/sciadv.aav7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Wei Y, et al. IL-17A secreted from lymphatic endothelial cells promotes tumorigenesis by upregulation of PD-L1 in hepatoma stem cells. J. Hepatol. 2019;71:1206–1215. doi: 10.1016/j.jhep.2019.08.034. [DOI] [PubMed] [Google Scholar]
- 428.Shime H, et al. Tumor-secreted lactic acid promotes IL-23/IL-17 proinflammatory pathway. J. Immunol. 2008;180:7175–7183. doi: 10.4049/jimmunol.180.11.7175. [DOI] [PubMed] [Google Scholar]
- 429.Zhang W, et al. Lactate is a natural suppressor of RLR signaling by targeting MAVS. Cell. 2019;178:176–189. doi: 10.1016/j.cell.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Medzhitov R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 431.Takeda K, Akira S. TLR signaling pathways. Semin. Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 432.Rowe DC, et al. The myristoylation of TRIF-related adaptor molecule is essential for Toll-like receptor 4 signal transduction. Proc. Natl Acad. Sci. USA. 2006;103:6299–6304. doi: 10.1073/pnas.0510041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Shevlin E, Miggin SM. The TIR-domain containing adaptor TRAM is required for TLR7 mediated RANTES production. PLoS ONE. 2014;9:e107141. doi: 10.1371/journal.pone.0107141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 2019;19:369–382. doi: 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Laskin DL. Macrophages and inflammatory mediators in chemical toxicity: a battle of forces. Chem. Res. Toxicol. 2009;22:1376–1385. doi: 10.1021/tx900086v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Sharif O, Bolshakov VN, Raines S, Newham P, Perkins ND. Transcriptional profiling of the LPS induced NF-kappaB response in macrophages. BMC Immunol. 2007;8:1. doi: 10.1186/1471-2172-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Chang FM, et al. Inhibition of neddylation represses lipopolysaccharide-induced proinflammatory cytokine production in macrophage cells. J. Biol. Chem. 2012;287:35756–35767. doi: 10.1074/jbc.M112.397703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Galvan-Pena S, et al. Malonylation of GAPDH is an inflammatory signal in macrophages. Nat. Commun. 2019;10:338. doi: 10.1038/s41467-018-08187-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Aderem AA. Protein myristoylation as an intermediate step during signal transduction in macrophages: its role in arachidonic acid metabolism and in responses to interferon gamma. J. Cell Sci. Suppl. 1988;9:151–167. doi: 10.1242/jcs.1988.supplement_9.8. [DOI] [PubMed] [Google Scholar]
- 441.Aderem A. The role of myristoylated protein kinase C substrates in intracellular signaling pathways in macrophages. Curr. Top. Microbiol. Immunol. 1992;181:189–207. doi: 10.1007/978-3-642-77377-8_7. [DOI] [PubMed] [Google Scholar]
- 442.Aderem AA, Marratta DE, Cohn ZA. Interferon gamma induces the myristoylation of a 48-kDa protein in macrophages. Proc. Natl Acad. Sci. USA. 1988;85:6310–6313. doi: 10.1073/pnas.85.17.6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Shrivastav A, et al. Requirement of N-myristoyltransferase 1 in the development of monocytic lineage. J. Immunol. 2008;180:1019–1028. doi: 10.4049/jimmunol.180.2.1019. [DOI] [PubMed] [Google Scholar]
- 444.Odegaard JI, Chawla A. Alternative macrophage activation and metabolism. Annu. Rev. Pathol. 2011;6:275–297. doi: 10.1146/annurev-pathol-011110-130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Galvan-Pena S, O’Neill LA. Metabolic reprograming in macrophage polarization. Front. Immunol. 2014;5:420. doi: 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Corraliza IM, Soler G, Eichmann K, Modolell M. Arginase induction by suppressors of nitric oxide synthesis (IL-4, IL-10 and PGE2) in murine bone-marrow-derived macrophages. Biochem. Biophys. Res. Commun. 1995;206:667–673. doi: 10.1006/bbrc.1995.1094. [DOI] [PubMed] [Google Scholar]
- 447.Munder M, Eichmann K, Modolell M. Alternative metabolic states in murine macrophages reflected by the nitric oxide synthase/arginase balance: competitive regulation by CD4+ T cells correlates with Th1/Th2 phenotype. J. Immunol. 1998;160:5347–5354. [PubMed] [Google Scholar]
- 448.Rampoldi F, et al. Immunosuppression and aberrant T cell development in the absence of N-myristoylation. J. Immunol. 2015;195:4228–4243. doi: 10.4049/jimmunol.1500622. [DOI] [PubMed] [Google Scholar]
- 449.He HT, Lellouch A, Marguet D. Lipid rafts and the initiation of T cell receptor signaling. Semin. Immunol. 2005;17:23–33. doi: 10.1016/j.smim.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 450.Janes PW, Ley SC, Magee AI. Aggregation of lipid rafts accompanies signaling via the T cell antigen receptor. J. Cell Biol. 1999;147:447–461. doi: 10.1083/jcb.147.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Rocha-Perugini V, Gordon-Alonso M, Sanchez-Madrid F. Role of drebrin at the immunological synapse. Adv. Exp. Med. Biol. 2017;1006:271–280. doi: 10.1007/978-4-431-56550-5_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Grakoui A, et al. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. [PubMed] [Google Scholar]
- 453.Bromley SK, et al. The immunological synapse. Annu. Rev. Immunol. 2001;19:375–396. doi: 10.1146/annurev.immunol.19.1.375. [DOI] [PubMed] [Google Scholar]
- 454.Davis DM, Dustin ML. What is the importance of the immunological synapse? Trends Immunol. 2004;25:323–327. doi: 10.1016/j.it.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 455.Lee KH, et al. T cell receptor signaling precedes immunological synapse formation. Science. 2002;295:1539–1542. doi: 10.1126/science.1067710. [DOI] [PubMed] [Google Scholar]
- 456.Huppa JB, Davis MM. T-cell-antigen recognition and the immunological synapse. Nat. Rev. Immunol. 2003;3:973–983. doi: 10.1038/nri1245. [DOI] [PubMed] [Google Scholar]
- 457.Ritter AT, Angus KL, Griffiths GM. The role of the cytoskeleton at the immunological synapse. Immunol. Rev. 2013;256:107–117. doi: 10.1111/imr.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Caron L, Abraham N, Pawson T, Veillette A. Structural requirements for enhancement of T-cell responsiveness by the lymphocyte-specific tyrosine protein kinase p56lck. Mol. Cell. Biol. 1992;12:2720–2729. doi: 10.1128/mcb.12.6.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Alland L, Peseckis SM, Atherton RE, Berthiaume L, Resh MD. Dual myristylation and palmitylation of Src family member p59fyn affects subcellular localization. J. Biol. Chem. 1994;269:16701–16705. [PubMed] [Google Scholar]
- 460.Van’T HW, Resh MD. Dual fatty acylation of p59(Fyn) is required for association with the T cell receptor zeta chain through phosphotyrosine-Src homology domain-2 interactions. J. Cell Biol. 1999;145:377–389. doi: 10.1083/jcb.145.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Cohen LA, Guan JL. Mechanisms of focal adhesion kinase regulation. Curr. Cancer Drug Targets. 2005;5:629–643. doi: 10.2174/156800905774932798. [DOI] [PubMed] [Google Scholar]
- 462.Bunnell SC, Kapoor V, Trible RP, Zhang W, Samelson LE. Dynamic actin polymerization drives T cell receptor-induced spreading: a role for the signal transduction adaptor LAT. Immunity. 2001;14:315–329. doi: 10.1016/s1074-7613(01)00112-1. [DOI] [PubMed] [Google Scholar]
- 463.Steffen A, et al. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol. Biol. Cell. 2006;17:2581–2591. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Faix J, Grosse R. Staying in shape with formins. Dev. Cell. 2006;10:693–706. doi: 10.1016/j.devcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 465.Han Y, et al. Formin-like 1 (FMNL1) is regulated by N-terminal myristoylation and induces polarized membrane blebbing. J. Biol. Chem. 2009;284:33409–33417. doi: 10.1074/jbc.M109.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 467.Delmastro-Greenwood MM, Piganelli JD. Changing the energy of an immune response. Am. J. Clin. Exp. Immunol. 2013;2:30–54. [PMC free article] [PubMed] [Google Scholar]
- 468.Everts B, et al. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood. 2012;120:1422–1431. doi: 10.1182/blood-2012-03-419747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Thwe PM, Amiel E. The role of nitric oxide in metabolic regulation of Dendritic cell immune function. Cancer Lett. 2018;412:236–242. doi: 10.1016/j.canlet.2017.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Busconi L, Michel T. Endothelial nitric oxide synthase. N-terminal myristoylation determines subcellular localization. J. Biol. Chem. 1993;268:8410–8413. [PubMed] [Google Scholar]
- 471.Mathewson N, et al. Neddylation plays an important role in the regulation of murine and human dendritic cell function. Blood. 2013;122:2062–2073. doi: 10.1182/blood-2013-02-486373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Keegan C, et al. Mycobacterium tuberculosis transfer RNA induces IL-12p70 via synergistic activation of pattern recognition receptors within a cell network. J. Immunol. 2018;200:3244–3258. doi: 10.4049/jimmunol.1701733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Cheng M, et al. Inhibition of neddylation regulates dendritic cell functions via Deptor accumulation driven mTOR inactivation. Oncotarget. 2016;7:35643–35654. doi: 10.18632/oncotarget.9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Gomez TS, et al. Formins regulate the actin-related protein 2/3 complex-independent polarization of the centrosome to the immunological synapse. Immunity. 2007;26:177–190. doi: 10.1016/j.immuni.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Jones RG, Thompson CB. Revving the engine: signal transduction fuels T cell activation. Immunity. 2007;27:173–178. doi: 10.1016/j.immuni.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 476.Al KF, et al. Unexpected obesity, rather than tumorigenesis, in a conditional mouse model of mitochondrial complex II deficiency. FASEB J. 2021;35:e21227. doi: 10.1096/fj.202002100R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Nastasi C, et al. Inhibition of succinate dehydrogenase activity impairs human T cell activation and function. Sci. Rep. 2021;11:1458. doi: 10.1038/s41598-020-80933-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: energy metabolism and the T-cell response. Nat. Rev. Immunol. 2005;5:844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- 479.Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annu. Rev. Immunol. 2005;23:515–548. doi: 10.1146/annurev.immunol.23.021704.115611. [DOI] [PubMed] [Google Scholar]
- 480.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 1999;5:1365–1369. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 482.Tseng SY, et al. B7-DC, a new dendritic cell molecule with potent costimulatory properties for T cells. J. Exp. Med. 2001;193:839–846. doi: 10.1084/jem.193.7.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Freeman GJ, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Dong H, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 485.Konishi J, et al. B7-H1 expression on non-small cell lung cancer cells and its relationship with tumor-infiltrating lymphocytes and their PD-1 expression. Clin. Cancer Res. 2004;10:5094–5100. doi: 10.1158/1078-0432.CCR-04-0428. [DOI] [PubMed] [Google Scholar]
- 486.Brown JA, et al. Blockade of programmed death-1 ligands on dendritic cells enhances T cell activation and cytokine production. J. Immunol. 2003;170:1257–1266. doi: 10.4049/jimmunol.170.3.1257. [DOI] [PubMed] [Google Scholar]
- 487.Zou W, Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat. Rev. Immunol. 2008;8:467–477. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]
- 488.Curiel TJ, et al. Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity. Nat. Med. 2003;9:562–567. doi: 10.1038/nm863. [DOI] [PubMed] [Google Scholar]
- 489.Liu Y, Zeng B, Zhang Z, Zhang Y, Yang R. B7-H1 on myeloid-derived suppressor cells in immune suppression by a mouse model of ovarian cancer. Clin. Immunol. 2008;129:471–481. doi: 10.1016/j.clim.2008.07.030. [DOI] [PubMed] [Google Scholar]
- 490.Creese I, Feinberg AP, Snyder SH. Butyrophenone influences on the opiate receptor. Eur. J. Pharmacol. 1976;36:231–235. doi: 10.1016/0014-2999(76)90277-6. [DOI] [PubMed] [Google Scholar]
- 491.Li CW, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat. Commun. 2016;7:12632. doi: 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Li CW, et al. Eradication of triple-negative breast cancer cells by targeting glycosylated PD-L1. Cancer Cell. 2018;33:187–201. doi: 10.1016/j.ccell.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.D’Arrigo P, et al. A regulatory role for the co-chaperone FKBP51s in PD-L1 expression in glioma. Oncotarget. 2017;8:68291–68304. doi: 10.18632/oncotarget.19309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Hsu JM, et al. STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat. Commun. 2018;9:1908. doi: 10.1038/s41467-018-04313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Murphy KM, et al. Signaling and transcription in T helper development. Annu. Rev. Immunol. 2000;18:451–494. doi: 10.1146/annurev.immunol.18.1.451. [DOI] [PubMed] [Google Scholar]
- 496.McGeachy MJ, Cua DJ. Th17 cell differentiation: the long and winding road. Immunity. 2008;28:445–453. doi: 10.1016/j.immuni.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 497.Xu H, et al. The induction and function of the anti-inflammatory fate of TH17 cells. Nat. Commun. 2020;11:3334. doi: 10.1038/s41467-020-17097-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Shevach EM, et al. The lifestyle of naturally occurring CD4+ CD25+ Foxp3+ regulatory T cells. Immunol. Rev. 2006;212:60–73. doi: 10.1111/j.0105-2896.2006.00415.x. [DOI] [PubMed] [Google Scholar]
- 499.Sakaguchi S, et al. Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol. Rev. 2006;212:8–27. doi: 10.1111/j.0105-2896.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- 500.Dunn SE, et al. Isoprenoids determine Th1/Th2 fate in pathogenic T cells, providing a mechanism of modulation of autoimmunity by atorvastatin. J. Exp. Med. 2006;203:401–412. doi: 10.1084/jem.20051129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Kagami S, et al. Protein geranylgeranylation regulates the balance between Th17 cells and Foxp3+ regulatory T cells. Int. Immunol. 2009;21:679–689. doi: 10.1093/intimm/dxp037. [DOI] [PubMed] [Google Scholar]
- 502.Di Capite J, Parekh AB. CRAC channels and Ca2+ signaling in mast cells. Immunol. Rev. 2009;231:45–58. doi: 10.1111/j.1600-065X.2009.00808.x. [DOI] [PubMed] [Google Scholar]
- 503.Uzhachenko R, et al. Fus1/Tusc2 is a novel regulator of mitochondrial calcium handling, Ca2+ -coupled mitochondrial processes, and Ca2+ -dependent NFAT and NF-kappaB pathways in CD4+ T cells. Antioxid. Redox Signal. 2014;20:1533–1547. doi: 10.1089/ars.2013.5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Uzhachenko R, et al. Tumour suppressor Fus1 provides a molecular link between inflammatory response and mitochondrial homeostasis. J. Pathol. 2012;227:456–469. doi: 10.1002/path.4039. [DOI] [PubMed] [Google Scholar]
- 505.Uno F, et al. Myristoylation of the fus1 protein is required for tumor suppression in human lung cancer cells. Cancer Res. 2004;64:2969–2976. doi: 10.1158/0008-5472.can-03-3702. [DOI] [PubMed] [Google Scholar]
- 506.Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ. Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science. 2000;290:1761–1765. doi: 10.1126/science.290.5497.1761. [DOI] [PubMed] [Google Scholar]
- 507.Vilas GL, et al. Posttranslational myristoylation of caspase-activated p21-activated protein kinase 2 (PAK2) potentiates late apoptotic events. Proc. Natl Acad. Sci. USA. 2006;103:6542–6547. doi: 10.1073/pnas.0600824103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Wagner M, Jasek M, Karabon L. Immune checkpoint molecules-inherited variations as markers for cancer risk. Front Immunol. 2020;11:606721. doi: 10.3389/fimmu.2020.606721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Qin S, et al. Novel immune checkpoint targets: moving beyond PD-1 and CTLA-4. Mol. Cancer. 2019;18:155. doi: 10.1186/s12943-019-1091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Dermani FK, Samadi P, Rahmani G, Kohlan AK, Najafi R. PD-1/PD-L1 immune checkpoint: potential target for cancer therapy. J. Cell. Physiol. 2019;234:1313–1325. doi: 10.1002/jcp.27172. [DOI] [PubMed] [Google Scholar]
- 511.Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48:434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Seliger, B. Basis of PD1/PD-L1 therapies. J. Clin. Med. 8, 2168 (2019). [DOI] [PMC free article] [PubMed]
- 513.Burr ML, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–105. doi: 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Mezzadra R, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549:106–110. doi: 10.1038/nature23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Zhang J, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Bech-Otschir D, et al. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J. 2001;20:1630–1639. doi: 10.1093/emboj/20.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Lim SO, et al. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30:925–939. doi: 10.1016/j.ccell.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Yang Y, et al. Palmitoylation stabilizes PD-L1 to promote breast tumor growth. Cell Res. 2019;29:83–86. doi: 10.1038/s41422-018-0124-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Yao H, et al. Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat. Biomed. Eng. 2019;3:306–317. doi: 10.1038/s41551-019-0375-6. [DOI] [PubMed] [Google Scholar]
- 520.Tang J, et al. Trial watch: The clinical trial landscape for PD1/PDL1 immune checkpoint inhibitors. Nat. Rev. Drug Discov. 2018;17:854–855. doi: 10.1038/nrd.2018.210. [DOI] [PubMed] [Google Scholar]
- 521.Zhao C, Dong H, Xu Q, Zhang Y. Histone deacetylase (HDAC) inhibitors in cancer: a patent review (2017-present) Expert Opin. Ther. Pat. 2020;30:263–274. doi: 10.1080/13543776.2020.1725470. [DOI] [PubMed] [Google Scholar]
- 522.Zhang Q, Wang S, Chen J, Yu Z. Histone deacetylases (HDACs) guided novel therapies for T-cell lymphomas. Int. J. Med. Sci. 2019;16:424–442. doi: 10.7150/ijms.30154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Marks P, et al. Histone deacetylases and cancer: causes and therapies. Nat. Rev. Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 524.Singh, A. K., Bishayee, A. & Pandey, A. K. Targeting histone deacetylases with natural and synthetic agents: an emerging anticancer strategy. Nutrients10, 731 (2018). [DOI] [PMC free article] [PubMed]
- 525.Lehrmann H, Pritchard LL, Harel-Bellan A. Histone acetyltransferases and deacetylases in the control of cell proliferation and differentiation. Adv. Cancer Res. 2002;86:41–65. doi: 10.1016/s0065-230x(02)86002-x. [DOI] [PubMed] [Google Scholar]
- 526.Marks PA, Dokmanovic M. Histone deacetylase inhibitors: discovery and development as anticancer agents. Expert Opin. Investig. Drugs. 2005;14:1497–1511. doi: 10.1517/13543784.14.12.1497. [DOI] [PubMed] [Google Scholar]
- 527.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. y. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 528.Marks PA, Rifkind RA, Richon VM, Breslow R. Inhibitors of histone deacetylase are potentially effective anticancer agents. Clin. Cancer Res. 2001;7:759–760. [PubMed] [Google Scholar]
- 529.Hess-Stumpp H. Histone deacetylase inhibitors and cancer: from cell biology to the clinic. Eur. J. Cell Biol. 2005;84:109–121. doi: 10.1016/j.ejcb.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 530.Moradei O, Maroun CR, Paquin I, Vaisburg A. Histone deacetylase inhibitors: latest developments, trends and prospects. Curr. Med Chem. Anticancer Agents. 2005;5:529–560. doi: 10.2174/1568011054866946. [DOI] [PubMed] [Google Scholar]
- 531.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 532.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 533.Bertino EM, Otterson GA. Romidepsin: a novel histone deacetylase inhibitor for cancer. Expert Opin. Investig. Drugs. 2011;20:1151–1158. doi: 10.1517/13543784.2011.594437. [DOI] [PubMed] [Google Scholar]
- 534.Harrison SJ, et al. A focus on the preclinical development and clinical status of the histone deacetylase inhibitor, romidepsin (depsipeptide, Istodax((R))) Epigenomics-Uk. 2012;4:571–589. doi: 10.2217/epi.12.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Gibbs JB, Oliff A. The potential of farnesyltransferase inhibitors as cancer chemotherapeutics. Annu Rev. Pharm. Toxicol. 1997;37:143–166. doi: 10.1146/annurev.pharmtox.37.1.143. [DOI] [PubMed] [Google Scholar]
- 536.Sebti SM, Hamilton AD. Farnesyltransferase and geranylgeranyltransferase I inhibitors and cancer therapy: lessons from mechanism and bench-to-bedside translational studies. Oncogene. 2000;19:6584–6593. doi: 10.1038/sj.onc.1204146. [DOI] [PubMed] [Google Scholar]
- 537.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat. Rev. Cancer. 2011;11:775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Appels NM, Beijnen JH, Schellens JH. Development of farnesyl transferase inhibitors: a review. Oncologist. 2005;10:565–578. doi: 10.1634/theoncologist.10-8-565. [DOI] [PubMed] [Google Scholar]
- 539.James GL, et al. Benzodiazepine peptidomimetics: potent inhibitors of Ras farnesylation in animal cells. Science. 1993;260:1937–1942. doi: 10.1126/science.8316834. [DOI] [PubMed] [Google Scholar]
- 540.Qian Y, et al. Design and structural requirements of potent peptidomimetic inhibitors of p21ras farnesyltransferase. J. Biol. Chem. 1994;269:12410–12413. [PubMed] [Google Scholar]
- 541.Nigam M, Seong CM, Qian Y, Hamilton AD, Sebti SM. Potent inhibition of human tumor p21ras farnesyltransferase by A1A2-lacking p21ras CA1A2X peptidomimetics. J. Biol. Chem. 1993;268:20695–20698. [PubMed] [Google Scholar]
- 542.Vogt A, et al. A non-peptide mimetic of Ras-CAAX: selective inhibition of farnesyltransferase and Ras processing. J. Biol. Chem. 1995;270:660–664. doi: 10.1074/jbc.270.2.660. [DOI] [PubMed] [Google Scholar]
- 543.Hara M, et al. Identification of Ras farnesyltransferase inhibitors by microbial screening. Proc. Natl Acad. Sci. USA. 1993;90:2281–2285. doi: 10.1073/pnas.90.6.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Gibbs JB, et al. Selective inhibition of farnesyl-protein transferase blocks ras processing in vivo. J. Biol. Chem. 1993;268:7617–7620. [PubMed] [Google Scholar]
- 545.Garcia AM, Rowell C, Ackermann K, Kowalczyk JJ, Lewis MD. Peptidomimetic inhibitors of Ras farnesylation and function in whole cells. J. Biol. Chem. 1993;268:18415–18418. [PubMed] [Google Scholar]
- 546.Kohl NE, et al. Selective inhibition of ras-dependent transformation by a farnesyltransferase inhibitor. Science. 1993;260:1934–1937. doi: 10.1126/science.8316833. [DOI] [PubMed] [Google Scholar]
- 547.Kohl NE, et al. Development of inhibitors of protein farnesylation as potential chemotherapeutic agents. J. Cell Biochem. Suppy. 1995;22:145–150. doi: 10.1002/jcb.240590819. [DOI] [PubMed] [Google Scholar]
- 548.Scholten JD, et al. Synergy between anions and farnesyldiphosphate competitive inhibitors of farnesyl:protein transferase. J. Biol. Chem. 1997;272:18077–18081. doi: 10.1074/jbc.272.29.18077. [DOI] [PubMed] [Google Scholar]
- 549.James GL, Goldstein JL, Brown MS. Polylysine and CVIM sequences of K-RasB dictate specificity of prenylation and confer resistance to benzodiazepine peptidomimetic in vitro. J. Biol. Chem. 1995;270:6221–6226. doi: 10.1074/jbc.270.11.6221. [DOI] [PubMed] [Google Scholar]
- 550.Omer CA, et al. Mouse mammary tumor virus-Ki-rasB transgenic mice develop mammary carcinomas that can be growth-inhibited by a farnesyl:protein transferase inhibitor. Cancer Res. 2000;60:2680–2688. [PubMed] [Google Scholar]
- 551.Mangues R, et al. Antitumor effect of a farnesyl protein transferase inhibitor in mammary and lymphoid tumors overexpressing N-ras in transgenic mice. Cancer Res. 1998;58:1253–1259. [PubMed] [Google Scholar]
- 552.Basso AD, Kirschmeier P, Bishop WR. Lipid posttranslational modifications. Farnesyl transferase inhibitors. J. Lipid Res. 2006;47:15–31. doi: 10.1194/jlr.R500012-JLR200. [DOI] [PubMed] [Google Scholar]
- 553.Karp JE, Lancet JE. Tipifarnib in the treatment of newly diagnosed acute myelogenous leukemia. Biologics. 2008;2:491–500. doi: 10.2147/btt.s3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Sparano JA, et al. Phase II trial of tipifarnib plus neoadjuvant doxorubicin-cyclophosphamide in patients with clinical stage IIB-IIIC breast cancer. Clin. Cancer Res. 2009;15:2942–2948. doi: 10.1158/1078-0432.CCR-08-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Porcu G, et al. Combined p21-activated kinase and farnesyltransferase inhibitor treatment exhibits enhanced anti-proliferative activity on melanoma, colon and lung cancer cell lines. Mol. Cancer. 2013;12:88. doi: 10.1186/1476-4598-12-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Niessner H, et al. The farnesyl transferase inhibitor lonafarnib inhibits mTOR signaling and enforces sorafenib-induced apoptosis in melanoma cells. J. Investig. Dermatol. 2011;131:468–479. doi: 10.1038/jid.2010.297. [DOI] [PubMed] [Google Scholar]
- 557.Robak T, et al. Novel and emerging drugs for acute myeloid leukemia: pharmacology and therapeutic activity. Curr. Med. Chem. 2011;18:638–666. doi: 10.2174/092986711794480104. [DOI] [PubMed] [Google Scholar]
- 558.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 559.Tanaka T, Nakatani T, Kamitani T. Inhibition of NEDD8-conjugation pathway by novel molecules: potential approaches to anticancer therapy. Mol. Oncol. 2012;6:267–275. doi: 10.1016/j.molonc.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 561.Lei M, Tye BK. Initiating DNA synthesis: from recruiting to activating the MCM complex. J. Cell Sci. 2001;114:1447–1454. doi: 10.1242/jcs.114.8.1447. [DOI] [PubMed] [Google Scholar]
- 562.Kim Y, Kipreos ET. Cdt1 degradation to prevent DNA re-replication: conserved and non-conserved pathways. Cell Div. 2007;2:18. doi: 10.1186/1747-1028-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 2010;70:10310–10320. doi: 10.1158/0008-5472.CAN-10-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Milhollen MA, et al. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res. 2011;71:3042–3051. doi: 10.1158/0008-5472.CAN-10-2122. [DOI] [PubMed] [Google Scholar]
- 565.Swords RT, et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115:3796–3800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- 566.Milhollen MA, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 2010;116:1515–1523. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 567.Wei D, et al. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer Res. 2012;72:282–293. doi: 10.1158/0008-5472.CAN-11-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Basu A, Krishnamurthy S. Cellular responses to Cisplatin-induced DNA damage. J. Nucleic Acids. 2010;2010:1–16. doi: 10.4061/2010/201367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Kee Y, D’Andrea AD. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24:1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Kee Y, et al. Inhibition of the Nedd8 system sensitizes cells to DNA interstrand cross-linking agents. Mol. Cancer Res. 2012;10:369–377. doi: 10.1158/1541-7786.MCR-11-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Jia L, Li H, Sun Y. Induction of p21-dependent senescence by an NAE inhibitor, MLN4924, as a mechanism of growth suppression. Neoplasia. 2011;13:561–569. doi: 10.1593/neo.11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Luo Z, Pan Y, Jeong LS, Liu J, Jia L. Inactivation of the Cullin (CUL)-RING E3 ligase by the NEDD8-activating enzyme inhibitor MLN4924 triggers protective autophagy in cancer cells. Autophagy. 2012;8:1677–1679. doi: 10.4161/auto.21484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Zhao Y, Xiong X, Jia L, Sun Y. Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis. 2012;3:e386. doi: 10.1038/cddis.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Tan M, Li H, Sun Y. Endothelial deletion of Sag/Rbx2/Roc2 E3 ubiquitin ligase causes embryonic lethality and blocks tumor angiogenesis. Oncogene. 2014;33:5211–5220. doi: 10.1038/onc.2013.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Toth JI, Yang L, Dahl R, Petroski MD. A gatekeeper residue for NEDD8-activating enzyme inhibition by MLN4924. Cell Rep. 2012;1:309–316. doi: 10.1016/j.celrep.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Milhollen MA, et al. Treatment-emergent mutations in NAEbeta confer resistance to the NEDD8-activating enzyme inhibitor MLN4924. Cancer Cell. 2012;21:388–401. doi: 10.1016/j.ccr.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 577.Scott DC, et al. Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase. Nat. Chem. Biol. 2017;13:850–857. doi: 10.1038/nchembio.2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Hammill JT, et al. Discovery of an orally bioavailable inhibitor of defective in cullin neddylation 1 (DCN1)-mediated cullin neddylation. J. Med. Chem. 2018;61:2694–2706. doi: 10.1021/acs.jmedchem.7b01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Wang, S. et al. Development of highly potent, selective, and cellular active triazolo[1,5- a]pyrimidine-based inhibitors targeting the DCN1-UBC12 protein-protein interaction. J. Med. Chem. 62, 2772–2797 (2019). [DOI] [PubMed]
- 580.Zhou H, et al. High-affinity peptidomimetic inhibitors of the DCN1-UBC12 protein-protein interaction. J. Med. Chem. 2018;61:1934–1950. doi: 10.1021/acs.jmedchem.7b01455. [DOI] [PubMed] [Google Scholar]
- 581.Zhou H, et al. A potent small-molecule inhibitor of the DCN1-UBC12 interaction that selectively blocks cullin 3 neddylation. Nat. Commun. 2017;8:1150. doi: 10.1038/s41467-017-01243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Bielawska A, et al. Novel analogs of D-e-MAPP and B13. Part 2: signature effects on bioactive sphingolipids. Bioorg. Med. Chem. 2008;16:1032–1045. doi: 10.1016/j.bmc.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Bhandarkar SS, et al. Tris (dibenzylideneacetone) dipalladium, a N-myristoyltransferase-1 inhibitor, is effective against melanoma growth in vitro and in vivo. Clin. Cancer Res. 2008;14:5743–5748. doi: 10.1158/1078-0432.CCR-08-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Schoepfer J, et al. Discovery of asciminib (ABL001), an allosteric inhibitor of the tyrosine kinase activity of BCR-ABL1. J. Med. Chem. 2018;61:8120–8135. doi: 10.1021/acs.jmedchem.8b01040. [DOI] [PubMed] [Google Scholar]
- 585.Radi M, Schenone S, Botta M. Allosteric inhibitors of Bcr-Abl: towards novel myristate-pocket binders. Curr. Pharm. Biotechnol. 2013;14:477–487. doi: 10.2174/138920101405131111103750. [DOI] [PubMed] [Google Scholar]
- 586.Wylie AA, et al. The allosteric inhibitor ABL001 enables dual targeting of BCR-ABL1. Nature. 2017;543:733–737. doi: 10.1038/nature21702. [DOI] [PubMed] [Google Scholar]
- 587.Fabbro D, et al. Inhibitors of the Abl kinase directed at either the ATP- or myristate-binding site. Biochim Biophys. Acta. 2010;1804:454–462. doi: 10.1016/j.bbapap.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 588.Chen YC, et al. N-myristoyltransferase-1 is necessary for lysosomal degradation and mTORC1 activation in cancer cells. Sci. Rep. 2020;10:11952. doi: 10.1038/s41598-020-68615-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Utsumi T, et al. Identification and characterization of protein N-myristoylation occurring on four human mitochondrial proteins, SAMM50, TOMM40, MIC19, and MIC25. PLoS ONE. 2018;13:e206355. doi: 10.1371/journal.pone.0206355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Zhang Q, et al. Metabolic reprogramming of ovarian cancer involves ACSL1-mediated metastasis stimulation through upregulated protein myristoylation. Oncogene. 2021;40:97–111. doi: 10.1038/s41388-020-01516-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.