Summary
Although some COVID-19 patients maintain SARS-CoV-2-specific serum immunoglobulin G (IgG) for more than 6 months postinfection, others eventually lose IgG levels. We assessed the persistence of SARS-CoV-2-specific B cells in 17 patients, 5 of whom had lost specific IgGs after 5–8 months. Differentiation of blood-derived B cells in vitro revealed persistent SARS-CoV-2-specific IgG B cells in all patients, whereas IgA B cells were maintained in 11. Antibodies derived from cultured B cells blocked binding of viral receptor-binding domain (RBD) to the cellular receptor ACE-2, had neutralizing activity to authentic virus, and recognized the RBD of the variant of concern Alpha similarly to the wild type, whereas reactivity to Beta and Gamma were decreased. Thus, differentiation of memory B cells could be more sensitive for detecting previous infection than measuring serum antibodies. Understanding the persistence of SARS-CoV-2-specific B cells even in the absence of specific serum IgG will help to promote long-term immunity.
Subject areas: Biological sciences, Immunology, Immune response
Graphical Abstract

Highlights
-
•
Memory B cells persist in blood of COVID-19 patients despite loss of specific Ig
-
•
Differentiating B cells in vitro robustly reveals previous SARS-CoV-2 infection
-
•
Ig derived from memory B cells neutralizes SARS-CoV-2 variants
-
•
Persisting memory B cells contribute to protective immunity against SARS-CoV-2
Biological sciences; Immunology; Immune response
Introduction
The development of adaptive immunity to SARS-CoV-2 may provide protection against re-infection and allows the identification of patients who have had a previous infection. Adaptive immunity to SARS-CoV-2 involves antibody (Ab)-producing cells, memory B cells, and several T cell subsets. Analysis of immune responses to different viruses, including other coronaviruses, has shown that the lifespans of the adaptive immune system components vary (Sariol and Perlman, 2020; Sekine et al., 2020).
Details of the kinetics of immune responses to SARS-CoV-2 are beginning to be uncovered (Dan et al., 2021; Quast and Tarlinton, 2021; Rydyznski Moderbacher et al., 2020; Seow et al., 2020; Sette and Crotty, 2021; Sherina et al., 2021; Vabret et al., 2020). Antibody (Ab) responses peak at about 2–3 weeks after infection, at which point the Ab levels decline (Dan et al., 2021; Marot et al., 2021; Perreault et al., 2020). In most individuals, anti-SARS-CoV-2 serum Abs persist for more than 6 months after primary infection, but some patients rapidly lose their specific Abs, especially those that experienced a mild disease course (Dan et al., 2021; Long et al., 2020; Marot et al., 2021; Perreault et al., 2020; Sekine et al., 2020; Seow et al., 2020; Zheng et al., 2021). It has been proposed that, in addition to serum antibody titers, the memory B cell pool should be evaluated to estimate humoral immunity as an indicator of immune protection (Abayasingam et al., 2021; Baumgarth, 2021; Dan et al., 2021; Robbiani et al., 2020).
Initial Ab responses are made by short-lived plasmablasts that develop in extrafollicular sites (Baumgarth, 2021; Baumgarth et al., 2020; Woodruff et al., 2020), and the subsequent development of high-affinity and persistent Abs involves affinity maturation and the expansion of B cells in germinal centers (Gaebler et al., 2021; Robbiani et al., 2020; Sakharkar et al., 2021; Sokal et al., 2021; Weisel and Shlomchik, 2017). Two types of B cells exit in the germinal center: memory B cells and plasmablasts (Baumgarth, 2021; Weisel and Shlomchik, 2017). Many of these plasmablasts are short-lived and die within a few weeks, but some find survival niches in the bone marrow and persist as long-lived plasma cells (Manz et al., 2005). The extent to which these long-lived plasma cells develop differs between different viruses and vaccines (Amanna et al., 2007; Edridge et al., 2020). Different mechanisms regulate the survival of long-lived plasma cells (Amanna and Slifka, 2010; Manz et al., 2005) and memory B cells (Weisel and Shlomchik, 2017).
Recently, SARS-CoV-2 variants of concern (VoCs) have emerged in the United Kingdom (Alpha, B.1.1.7), South Africa (Beta, B.1.351), Brazil (Gamma, P.1.), and India (Delta, B.1.617.2) and elsewhere with multiple substitutions, some of which are in the RBD in the NTD and the receptor-binding motif (RBM) of the RBD (Chen et al., 2021; Hoffmann et al., 2021). These rapidly spreading VoCs are currently causing serious concerns regarding the increased frequency of re-infection, utility of convalescent plasma, and limited vaccine responses (Casadevall et al., 2021; Wibmer et al., 2021).
In this study, we analyzed the persistence of IgA and IgG memory B cells specific for SARS-CoV-2 in COVID-19 patients. We specifically investigated donors who had lost circulating IgG to SARS-CoV-2 and analyzed whether they still harbored specific memory B cells in their blood. To study the patient B cells, we adopted a functional approach, converting blood-derived B cells into Ab-secreting cells in vitro (Pinna et al., 2009; Thaler et al., 2019; Winklmeier et al., 2019). Having identified the SARS-CoV-2-specific memory B cells in the blood, we analyzed whether the secreted Abs have the ability to block binding of the RBD to its cellular receptor ACE-2, show neutralizing activity, and cross-react to the RBDs of VoCs Alpha/B.1.1.7, Beta/B.1.351, and Gamma/P.1. The findings of the study revealed functional properties of persisting memory B cells specific to SARS-CoV-2, which could help to understand and promote protection.
Results
Persistence of IgG memory B cells specific for SARS-CoV-2 in the presence and absence of specific IgG
We analyzed, in parallel, the presence of memory B cells specific for SARS-CoV-2 in blood and specific IgG in serum (Figure 1 shows our approach). Our study included 17 COVID-19 patients who had undergone a mild or asymptomatic disease course (Table 1), and prepandemic blood samples from six HC donors served as the control group. We detected B cells that could be developed into SARS-CoV-2-specific-IgG-secreting plasmablasts in the blood of all COVID-19 patients analyzed. The reactivity to SARS-CoV-2 of these in vitro differentiated plasmablasts and the patient sera from the same blood withdrawal was investigated (Figure 2A). Remarkably, the sera from four COVID-19 patients were negative in the ELISA, and the fifth was borderline. These five donors (HC = 2, MS = 2, SLE = 1; #5, #11, #12, #15, #16) had been seropositive 1–2 months after acute infection (Table 1) but had lost their specific IgG 5–8 months postinfection. Two of these five donors were under immunotherapeutic regimens at the time their blood was sampled for this study (Table 1).
Figure 1.
Experimental scheme
PBMCs from each donor were separated into individual wells and stimulated with the TLR7/8 agonist R848 and IL-2 to differentiate them into Ab-secreting plasmablasts. This was used to compare the serum response to SARS-CoV-2 with that of specific Abs produced in vitro. The frequency of SARS-CoV-2-specific B cells that differentiated into Ab secreting cells was determined. The cross-reactivity to RBDs of emerging variants was tested. The ability of in vitro produced Abs to block the binding of RBD to its receptor ACE-2 and to neutralize infectious virus was determined as outlined.
Table 1.
Characteristics of COVID-19 patients
| ID | Time of infection confirmation | Infection confirmed bya | COVID-19 disease severityb | Age (years) | Gender | Immunotherapy and further diagnosis | Time between infection and first serology (months) | Assay used for first serology test and quantitative resultc | Time between infection and blood sampling for the B cell study (months)d |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 10/2020 | PCR | Mild | 30 | f | None | NA | NA | 1 |
| 2 | 11/2020 | PCR | Mild | 53 | f | None | NA | NA | 1 |
| 3 | 11/2020 | PCR | Mild | 53 | f | None | NA | NA | 1 |
| 4 | 11/2020 | PCR | Mild | 72 | m | None | NA | NA | 1 |
| 5 | 05/2020 | PCR | Asymptomatic | 47 | m | None | 1 | Euroimmun ELISA (0.87) & Roche ECLIA (1.05) | 5 |
| 6 | 04/2020 | PCR | Asymptomatic | 42 | f | Previouslye, MS patient | 6 | Euroimmun ELISA (5,82) | 6 |
| 7 | 05/2020 | Serology | Mild | 24 | m | None | 1 | Roche ECLIA | 6 |
| 8 | 03/2020 | PCR | Mild | 50 | m | None | 2 | Euroimmun ELISA (3,70) | 7 |
| 9 | 03/2020 | Serology | Mild | 27 | m | None | 3 | Euroimmun ELISA (4.60) & Roche ECLIA (152.5) | 7 |
| 10 | 03/2020 | Serology | Mild | 27 | f | None | 3 | Euroimmun ELISA (3.65) & Roche ECLIA (31.05) | 7 |
| 11 | 03/2020 | PCR | Mild | 37 | m | Dimethyl fumarate, MS patient | 2 | Euroimmun ELISA (5,60) | 7 |
| 12 | 03/2020 | PCR | Mild | 46 | f | Canakinumab, Hydroxychloroquine, SLE & SAD | 2 | Euroimmun ELISA (1,20) | 7 |
| 13 | 03/2020 | PCR | Mild | 37 | f | Previouslyf, MS patient | 7 | Roche ECLIA | 7 |
| 14 | 04/2020 | PCR | Mild | 57 | m | None | 2 | Euroimmun ELISA (6.96) & Roche ECLIA (53.34) | 7 |
| 15 | 11/2020 | PCR | Mild | 28 | m | None | 1 | Euroimmun ELISA (1.35) | 7 |
| 16 | 03/2020 | PCR | Mild | 46 | f | Previouslyg, MS patient | 2 | Roche ECLIA | 8 |
| 17 | 03/2020 | PCR | Mild | 45 | f | None | NA | NA | 8 |
Abbreviations: MS, multiple sclerosis; SLE, systemic lupus erythematosus; SAD, suspected autoinflammatory disease.
Three patients did not receive a PCR test due to test shortages at the beginning of the pandemic; however, all three patients displayed symptoms pathognomonic for COVID-19, including fever, respiratory symptoms, and severe anosmia, and tested positive for SARS-CoV-2 antibodies afterward. For these three patients, the month of symptom onset was counted as the time of infection. For all other symptomatic patients, the month of symptom onset and the time of PCR positivity coincided. All study participants tested positive for SARS-CoV-2 antibodies at least once after they were infected.
Mild = no supplementary oxygen needed; no admission to hospital; score <4 according to the WHO clinical progression scale for COVID-19 research (Marshall et al., 2020).
Test results are reported in brackets as OD ratio (ELISA) or cutoff index (ECLIA), respectively.
Donors 5, 11, 12, 15, and 16 had lost SARS-CoV-2-specific IgG at the time of blood sampling for this study.
Status post (s/p) Alemtuzumab (last administration 2017).
s/p Interferon-beta (last administration 2016).
s/p Teriflunomide (last administration 2015).
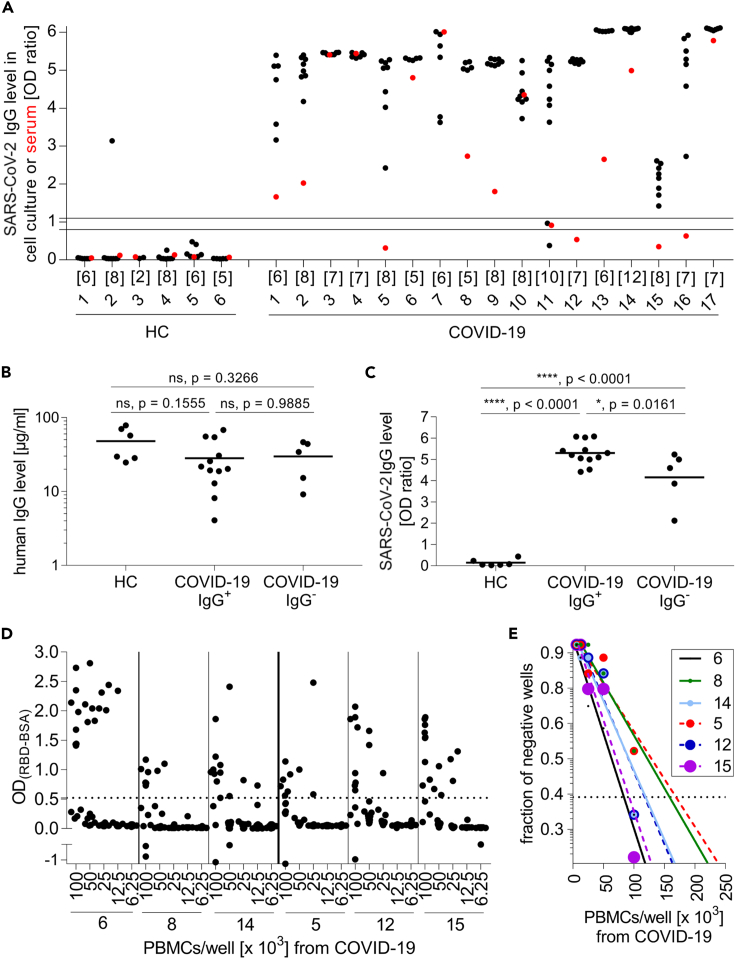
Figure 2.
IgG production by differentiated B cells specific for SARS-CoV-2
(A) PBMCs from healthy controls (HC, left) and COVID-19 patients (right) were differentiated into Ab-secreting cells. The reactivity of IgG against S1 in cell culture supernatants was determined. Each black dot represents one stimulated well. The number of stimulated wells per donor is provided directly under the x axis. The reactivity of the serum in the same ELISA is shown with a red dot. The area between the two horizontal lines were considered to represent the borderline zone of reactivity. COVID-19 patients who were serum-negative for SARS-CoV-2-specific IgG (#5, #11, #12, #15, #16) were designated COVID-19 IgG−, and those who were positive were designated COVID-19 IgG+.
(B and C) Each symbol represents the mean IgG level of all stimulated wells for one donor. Horizontal lines indicate the mean IgG levels of all donors in the respective groups. (B) IgG levels of cell culture supernatants were not significantly different between the groups (one-way ANOVA, Tukey's multiple comparison test; HC = 6, COVID-19 IgG+ = 12, COVID-19 IgG− = 5). (C) Both the COVID-19 IgG− and COVID-19 IgG+ subgroups produced more anti-S1 IgG (one-way ANOVA, Tukey's multiple comparison test; HC = 6, COVID-19 IgG+ = 12, COVID-19 IgG− = 5) than the HC.
(D and E) The raw data for the limiting dilution experiment are shown (D), and the calculation is displayed (E). Continuous lines are shown for donors who still had SARS-CoV-2 IgGs in their serum, and the dotted lines indicate donors who lost their specific serum Abs.
See also Figures S1 and S2.
Based on these findings, we grouped the patients into those with (COVID-19 IgG+) or without (COVID-19 IgG−) serum IgG to SARS-CoV-2. We calculated the mean of the total IgG secreted into the cell culture supernatant and the SARS-CoV-2-specific IgG levels of the samples from each donor and compared the three groups (Figures 2B and 2C). Both the COVID-19 IgG+ and COVID-19 IgG− patients had significantly more SARS-CoV-2-specific B cells in their blood than HC donors (p < 0.0001), even though the amount of total secreted IgG was not significantly different between the groups.
The method of seeding PBMCs into individual wells and testing each well for the development of specific IgG yields a high sensitivity. We noted that all wells containing samples from 15 of the 17 COVID-19 patients were positive for SARS-CoV-2-specific IgG, and for one donor, 6 of the 8 wells were positive, which demonstrated the high frequency of SARS-CoV-2-specific B cells in these patients. We subsequently performed limiting dilution assays on PBMC from six COVID-19 patients (Figures 2D and 2E). We calculated the frequency of specific B cells according to the Poisson distribution and the individual percentage of B cells within the PBMC (between 4% and 15%) and obtained thereby the following rates of B cells that gave rise to Abs to SARS-CoV-2: patient #6 (seropositive): 1:13,000; patient #8 (seropositive): 1:11,000; patient #14 (seropositive): 1:20,000; patient #5 (seronegative): 1:22,000; patient #12 (seronegative) 1:7,000; patient #15 (seronegative) 1:4,000. Thus, in accordance with our calculation of SARS-CoV-2-reactivity in the bulk cultures (Figure 2C), our limiting dilution analysis indicated that the seronegative and seropositive COVID-19 patients had similar frequencies of circulating RBD-specific B cells. The frequencies observed lay within the reactivity ranges previously reported for measles virus and tetanus toxoid (Thaler et al., 2019; Winklmeier et al., 2019). IgG memory B cells constitute about 15% of peripheral B cells, and only 30%–40% of IgG memory B cells are capable of antibody production under these culture conditions (Pinna et al., 2009). We found in 1/35 wells from HCs B cells giving rise to SARS-CoV-2-recognizing Abs, although this HC had no SARS-CoV-2-specific Abs in the serum as expected (Figure 2A). Our assay distinguishes HC from all COVID-19 patients (p < 0.0001) and also from COVID-19 patients who have lost their specific IgG (p < 0.0001) (Figure 2C). The Abs from this single well of one HC did not show neutralizing activity, neither in the ACE-2 binding assay nor in the live cell assay (see below).
As additional readout system to detect SARS-CoV-2 specific B cells, we used an enzyme-linked immune absorbent spot (ELISPOT). This confirmed the presence of SARS-CoV-2-specific B cells in each of the five donors who had lost their SARS-CoV-2 specific IgG (Figure S1).
We also analyzed the presence of anti-SARS-CoV-2 IgA in serum and of specific B cells in blood-secreting IgA (Figures S2A–S2C). The COVID-19 patients had significantly more SARS-CoV-2-specific IgA B cells in their blood than the healthy controls (p = 0.0203, Mann–Whitney U test). We noted that four COVID-19 patients had serum IgA, but no detectable specific IgA B cells. In healthy controls, SARS-CoV-2-specific IgA B cells were seen in 2 out of 6 donors and one was borderline, and no specific serum IgA was detected in 5 of the 6 healthy controls and 1 had borderline levels (Figures S2A–S2C). Although all COVID-19 patients tested had specific IgG B cells in their blood, 12 patients had at least one well with specific IgA B cells. The difference between the occurrences of IgG- and IgA-positive B cells became clearer when we considered the number of positive wells. Specific IgG was detected in 123 out of 125 wells for COVID-19 patients, whereas specific IgA was detected in only 34 wells (p < 0.0001; p values were determined using Fisher's exact test). Therefore, IgA B cells specific for SARS-CoV-2 were detected in the blood of the COVID-19 patients but were less abundant than specific IgG B cells.
Neutralizing activity of immunoglobulins derived from memory B cells
To analyze whether circulating peripheral B cells specific for SARS-CoV-2 can give rise to SARS-CoV-2-neutralizing Abs, we used two readout systems, a surrogate assay to analyze secreted-Ab inhibition of RBD binding to the viral entry receptor ACE-2 (Abe et al., 2020) and a neutralization assay under biosafety level 3 conditions using authentic virus (Figure 1). The significantly higher potency to block binding of RBD to ACE-2 of the COVID-19-patient-derived Abs produced in vitro was evident when compared with the blocking activity of the HC-derived Abs (Figure 3; p = 0.0006, Mann-Whitney U test). Thus, the SARS-CoV-2-specific B cells from COVID-19 patients released substantial amounts of Abs after differentiation into Ab-secreting cells and that were capable of blocking binding of RBD to ACE-2. We also performed a neutralization assay with authentic SARS-CoV-2 including all currently circulating major VoCs (Figure S3). Analyzing patient serum, this neutralization assay essentially confirmed the ELISA results: the group of donors designated COVID-19 IgG− was devoid of neutralizing activity. In contrast, some supernatants from cell culture wells of in vitro differentiated B cells from the same donors showed neutralizing activity (Figure S3).
Figure 3.
Inhibitory activity of Abs after differentiation of memory B cells
PBMCs from healthy controls (HC, left) and COVID-19 patients (right) were differentiated into Ab-secreting cells. The cell culture supernatants (each dot represents an individual well) were added to ELISA plates coated with the RBD. Biotinylated ACE-2 was then added, and its binding was detected with streptavidin–horseradish peroxidase. For calibration, the binding of biotinylated ACE-2 to RBD in the presence of buffer was set as 1. Then, the mean OD of the wells of each donor was calculated to compare the Abs binding to ACE-2 from COVID-19 patients with those from HCs. The Abs from COVID-19 patients reduced ACE-2 binding (p = 0.0006; Mann-Whitney U; HC = 6, COVID-19 = 17).
See also Figure S3.
Cross-reactivity of B cells to variants of concern
We analyzed the cross-reactivity of SARS-CoV-2-specific memory B cells (Figure 4A) against the RBD of three major VoCs in current circulation: Alpha/B.1.1.7, Beta/B.1.351, and Gamma/P.1 (Hoffmann et al., 2021). When we examined the memory B cells from each of the 17 COVID-19 patients, we found that Ab recognition of the RBD of Alpha variant was quantitatively unaltered, whereas recognition of the Beta variant was reduced by approximately 30% (p < 0.0001), and recognition of the Gamma variant was 50% lower than that for the wild type (WT) (p < 0.0001) (Figure 4B). The reactivity pattern SARS-CoV-2 (WT, D614G) = Alpha > Beta > Gamma was seen in the supernatant of the differentiated B cells of each of the COVID-19 patients analyzed.
Figure 4.
B cell reactivity to RBDs of emerging variants
PBMCs from the indicated COVID-19 patients were differentiated into Ab-secreting cells, and the cell culture supernatants were added to ELISA plates coated with the RBDs of wild type (WT, black) and of VoCs Alpha/B.1.1.7 (green), Beta/B.1.351 (orange), or the Gamma/P.1 lineage, also called B.1.1.248 (blue) SARS-CoV-2 variant. (A) From all donors two different cell culture supernatants were examined. Reactivity to the wild type was determined as the delta OD (RBD – BSA) and set as 1, and the relative reactivity to the other RBD variants was calculated and is shown.
(B) The mean reactivity of the tested wells from each donor was determined. Horizontal bars indicate the mean reactivity to the respective RBD variant (one-way ANOVA, Tukey's multiple comparison test; each n = 17).
See also Figure S3.
Discussion
In this study, we robustly detected the persistence of memory B cells specific for SARS-CoV-2 in all COVID-19 patients in our cohort who had undergone mild or asymptomatic acute infections, even if their specific serum IgG had declined to undetectable levels; this has two implications: firstly, the persistence of specific memory B cells, which gave rise to neutralizing Abs and showed differential cross-reactivity to VoCs, helps us to understand and start to develop models for predicting long-term protection against SARS-CoV-2. Secondly, differentiating B cells into Ab-producing cells in vitro could be more sensitive for detecting previous infection than measuring serum levels of SARS-CoV-2 IgG. Starting from differentiation of B cells to Ig-secreting cells, we used four different readout systems, ELISA against spike protein, ELISA against RBDs of VoCs, ACE-2 binding assay, and live-cell neutralization assay. All assays elaborated the presence of functional memory B cells in COVID-19 patients despite loss of serum IgG.
Previous studies have analyzed COVID-19 patients' memory B cells by staining them with labeled antigens (Cagigi et al., 2020; Dan et al., 2021; Gaebler et al., 2021; Hartley et al., 2020; Ogega et al., 2021; Robbiani et al., 2020; Rodda et al., 2021; Vaisman-Mentesh et al., 2020) and using an ELISPOT assay (Ansari et al., 2021; Lyski et al., 2021; Nguyen-Contant et al., 2020; Sherina et al., 2021; Varnaitė et al., 2020). Sorting the SARS-CoV-2-specific B cells and cloning their antigen-receptors provided important insights into clonal turnover and the ongoing somatic hypermutation of SARS-CoV-2-specific B cells associated with antigen persistence (Gaebler et al., 2021; Robbiani et al., 2020). SARS-CoV-2-specific memory B cells from selected patients were analyzed in detail using scRNAseq (Sokal et al., 2021). Our approach, differentiating B cells in vitro to Ab-secreting cells, allowed us to determine the frequency of SARS-CoV-2-specific B cells and their persistence over time. It also allowed us to analyze the function of Ig secreted by memory B cells and their cross-reactivity against VoCs.
We also detected SARS-CoV-2 IgA memory B cells in the blood of the recovered COVID-19 patients, but in contrast to the specific circulating IgG memory B cells, these were only seen in a subset of the cohort and less abundant. Although IgA is best known for its role in the immune response at mucosal sites (Sterlin et al., 2021; Wang et al., 2021), a systemic IgA immune response also occurs that includes the expansion of circulating IgA plasmablasts specific for SARS-CoV-2 (Sterlin et al., 2021). Mucosal IgA secretion continues for longer than the serum IgA response (Sterlin et al., 2021) but was completely lost after 189 days (Sterlin et al., 2021). Circulating IgA declined more rapidly than IgG and decayed by ∼90 days in most COVID-19 cases to levels indistinguishable from controls (Dan et al., 2021). We robustly detected circulating IgG memory B cells more than 6 months after infection. Taken together, the picture is emerging from this study and previous work that when circulating IgG and IgA, mucosal IgA, and circulating IgA memory B cells are gone, circulating IgG memory B cells persist. We found that the Abs produced by these memory B cells had high neutralizing activity, indicating their functional importance upon exposure to the same or mutated SARS-CoV-2.
In general, persisting IgG memory B cells can rapidly differentiate into antibody-secreting cells upon re-exposure (Baumgarth, 2021; Weisel and Shlomchik, 2017). The importance of memory B cells for protection against re-infection is evident from immune responses to other human viruses: protective immunity against hepatitis B was observed despite the loss of Abs (Rosado et al., 2011), and circulating memory B cells to viruses can be sustained for many decades after exposure, well into the 10th decade of life (Yu et al., 2008). In a mouse model of cytomegalovirus, the activation of virus-specific memory B cells to secrete IgG was independent on cognate or bystander T cell help (Hebeis et al., 2004). To provide protection, these memory B cells synergize with SARS-CoV-2-specific long-lived plasma cells that produce the serum IgG and have recently been detected in the bone-marrow (Turner et al., 2021). The persisting memory B cells are expected to be capable of providing relevant functional protection upon subsequent re-exposure with SARS-CoV-2, even if the relevant Abs have vanished.
The global spread of VoCs such as Alpha/B.1.1.7, Beta/B.1.351, and Gamma/P.1 may increase re-infection rates and compromise the success of current vaccines (Lauring and Hodcroft, 2021). We found that the polyclonal memory B cells recognized the RBD of Alpha variant to a similar degree as they did in the WT, whereas reactivity to Beta variant was reduced by 30% and to Gamma variant by as much as 50%. These findings are in accordance with the molecular signatures of these VoCs, as the Alpha VoC contains one mutation within the RBD (N501Y) that results in a higher affinity to the cellular receptor ACE-2, whereas the Beta and Gamma VoCs have three mutations in the RBD (K417N, E484K, N501Y and K417T, E484K, N501Y, respectively) also linked to immune escape (Harvey et al., 2021; Hoffmann et al., 2021). Previously, it was observed that some monoclonal (m)Abs showed reduced neutralizing activity to the Alpha variant; however, the polyclonal Ab response in the patient serum showed little alteration in neutralizing activity (Rees-Spear et al., 2021). An unaltered neutralizing activity to the Alpha variant but reduced reactivity to the Beta variant was reported (Planas et al., 2021). This is exactly in line with our observations for memory B cells from our cohort. We extended our analysis to show that cross-reactivity for the Gamma variant was even lower than for the Beta variant, which agrees with a recent observation that Beta/B.1.351 and Gamma/P.1 can escape neutralization by mAbs and are less efficiently inhibited by sera from BNT162b2-vaccinated individuals (Hoffmann et al., 2021). Although there is consensus that the neutralizing activity of convalescent sera against the Beta VoC is reduced (Wibmer et al., 2021), a stronger reduction of the binding to beta spike was observed in severely affected than in mildly infected patients (Yue et al., 2021). About 90% of the neutralizing activity in sera is directed against the RBD, but other parts, in particular the N-terminal domain of the spike protein, are also targeted by neutralizing Abs. To what extent the reduced humoral activity is associated with an increased susceptibility to these strains is not yet clear. Reactivity of memory B cells against VoCs has also been seen in another recent (Lyski et al., 2021). Interestingly, analysis of the response to other viruses has revealed that the pathogen-specific diversity of memory B cells might be different from long-lived plasma-cell-derived Abs (Lyski et al., 2021; Purtha et al., 2011; Wong et al., 2020). Re-infection with SARS-CoV-2 (Hall et al., 2021) was a possible reason for the resurgence of COVID-19 in Manaus (Sabino et al., 2021). It remains to be analyzed whether patients become re-infected because they have lost a certain type of anti-SARS-CoV-2 immunity or are confronted with a new variant, against which they do not yet have a protective immunity.
Because the consequences of infection with SARS-CoV-2 range from asymptomatic to lethal, accurate confirmation of previous infections is of great epidemiological and prognostic significance. Serological testing has made great advances (Chiereghin et al., 2020; Galipeau et al., 2020; Huang et al., 2020; Zarletti et al., 2020), but the specific IgG response wanes over time, and some donors with previous infections score negative in current serology tests (Abayasingam et al., 2021; Dan et al., 2021). We showed that the donors who lost IgG to SARS-CoV-2 still had specific IgG memory B cells. It has been previously noted that about 5% of infected donors are “nonresponders” who remain seronegative (Marklund et al., 2020; Oved et al., 2020). Using our method, we can analyze whether such donors have memory B cells with potential protective activity.
Our assay, involving the differentiation of B cells into Ab-secreting cells in vitro, identifies circulating memory B cells. We distributed the blood cells into different cell-culture-plate wells and analyzed the wells individually to perform an assay with high sensitivity that allows the identification of even rare autoreactive B cells (Winklmeier et al., 2019). By considering all wells containing samples from each donor, a clear distinction can be made between rare and abundant responses. Thus, the method used allowed us to evaluate whether seronegative individuals have already been infected with SARS-CoV-2. This is of relevance in epidemiology and for optimizing urgently needed immunosuppressive treatments.
We included six prepandemic donors. None of them had anti-SARS-CoV-2 Abs in their serum, in accordance with the very low reactivity in prepandemic donors. From supernatants of the tissue culture wells of these prepandemic donors with B cells differentiated to Ab-secreting cells, 1 out of 35 wells showed a minor reactivity to SARS-CoV-2. This supernatant had no activity to block RBD binding to ACE-2 and no neutralizing activity. A preexisting immunity in serum IgG has been noted in a few cases and was mapped to the S2 subunit of SARS-CoV-2 (Ng et al., 2020); the ELISA we used measures reactivity against S1 subunit. Previously, preexisting humoral immunity had been largely analyzed using serum samples, not by studying memory B cells. Importantly, the fine specificity of memory B cells and plasma cells (responsible for serum IgG) can be different, as in the germinal center memory B cells differentiate from low-affinity precursors, whereas plasma cells differentiate from high-affinity precursors (Viant et al., 2020). A detailed understanding of the presence of preexisting memory B cells with cross-reactivity to SARS-CoV-2 requires larger further studies. Such cross-reactivity could be clinically relevant because a recent infection with endemic human coronaviruses has been reported to be associated with less severe COVID-19 (Sagar et al., 2021).
In this study, we showed that circulating IgG memory B cells specific for SARS-CoV-2 persist in the blood after infection despite the loss of systemic IgG. These persisting B cells can be harnessed to identify a previous infection. Furthermore, these B cells gave rise to neutralizing Abs and showed high cross-reactivity against the emerging Alpha variant, suggesting patients have protection against re-exposure with this variant even after the loss of specific Abs. In contrast to reactivity against the Alpha variant, the reactivity of memory B cells against the Beta variant and, particularly, the Gamma variant was greatly reduced. This warrants further attention and indicates a possible need for follow-up vaccinations covering these mutants (Stamatatos et al., 2021). Thus, our study has added to the efforts to fully uncover the features of SARS-CoV-2-specific memory B cells, which will help us to understand and promote long-term protection.
Limitations of this study
The observation of patients was performed up to 8 months after infection. Longer observation times are necessary to learn more about features of B cell persistence. Two donors, who lost SARS-CoV-2-specific IgG but maintained specific memory B cells received immunosuppressive treatment (canakinumab plus hydroxychloroquine or dimethyl fumarate) at the time of blood sampling for this study. The impact of immunomodulatory therapies on the maintenance of IgG and memory B cells after infection or vaccination has yet to be analyzed in detail. We analyzed the persistence of B cells reactive to the S1 protein, but the maintenance of B cells against other SARS-CoV-2 proteins remains to be analyzed, although the response to the RBD is of paramount importance, as the RBD is targeted by neutralizing Abs (Sette and Crotty, 2021). We analyzed circulating memory B cells but are aware that both systemic and mucosal immunity are relevant for protection. We did not analyze the persistence of B cells in patients very severely affected by COVID-19, but it has been reported that the serum Ig response is even greater and longer-lasting in these patients (Long et al., 2020; Seow et al., 2020). We describe that COVID-19 patients who have seroreverted mount an anti-SARS-CoV-2 IgG response in the supernatant of in vitro differentiated B cells, but we have not phenotyped the original B cells that give rise to these IgGs. We assume that the in vitro produced Abs were derived from memory B cells, based on previous work that analyzed the response of different B cell subsets to various in vitro activators (Pinna et al., 2009). The in vitro differentiation of B cells we apply here could be more sensitive than a serum ELISA to detect a previous infection, but this would require analysis against larger numbers of pre- and postpandemic samples to determine the limit of detection.
STAR★Methods
Key resource table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CD19, PerCP-Cyanine5.5, clone: SJ25C1 | eBioscience | Cat# 45-0198-42; RRID: AB_1518739 |
| Anti-CD19, FITC, clone: HIB19 | Invitrogen | Cat# 11-0199-42; RRID: AB_10669461 |
| Peroxidase AffiniPure F(ab')₂ fragment goat anti-human IgG (H + L), polyclonal | Jackson ImmunoResearch | Cat# 109-036-003; RRID: AB_2337589 |
| Bacterial and virus strains | ||
| SARS-CoV-2 pangolin lineage B.1.177: GISAID EPI ISL 2450298 | This study | N/A |
| SARS-CoV-2 pangolin lineage B.1.1.7: GISAID EPI ISL 2095258 | This study | N/A |
| SARS-CoV-2 pangolin lineage B.1.351: GISAID EPI ISL 1752394 | This study | N/A |
| SARS-CoV-2 pangolin lineage P.1: GISAID EPI ISL 2095178 | This study | N/A |
| SARS-CoV-2 pangolin lineage B.1.617.2: GISAID EPI ISL 2772700 | This study | N/A |
| Lentiviral vector for overexpression of human angiotensin-converting enzyme 2 receptor (hACE2) | (Wratil et al., 2021) | N/A |
| Biological samples | ||
| Human blood samples (serum and PBMCs) | Biobank of the Institute of Clinical Neuroimmunology No. 163-16 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Resiquimod | Sigma-Aldrich | SML0196 |
| Recombinant human IL-2 protein | R&D Systems | 202-IL |
| SARS-CoV-2 RBD (wild type) | Trenzyme | P2020-001 |
| BSA | Sigma-Aldrich | A3059 |
| SARS-CoV-2 RBD (wild type) | ProteoGenix | PX-COV-P046 |
| SARS-CoV-2 RBD (Alpha) | ProteoGenix | PX-COV-P052 |
| SARS-CoV-2 RBD (beta) | ProteoGenix | PX-COV-P053 |
| SARS-CoV-2 RBD (gamma) | ProteoGenix | PX-COV-P054 |
| Angiotensin converting Enzyme-2, ACE2, biotin-tagged, human recombinant | Sigma-Aldrich | SAE0171 |
| Streptavidin-HRP | R&D Systems | 890803 |
| Critical commercial assays | ||
| Human IgG ELISA kit (ALP) | Mabtech | 3850-1AD-6 |
| Human IgA ELISA kit (ALP) | Mabtech | 3860-1AD-6 |
| ELISpot path: Human IgG (SARS-CoV-2, RBD) ALP | Mabtech | 3850-4APW-R1-1 |
| Anti-SARS-CoV-2 ELISA (IgG) | EUROIMMUN | EI 2606-9601 G |
| Anti-SARS-CoV-2 ELISA (IgA) | EUROIMMUN | EI 2606-9601 A |
| CellTiter-Glo 2.0 | Promega | Cat#G9243 |
| Experimental models: Cell lines | ||
| Immortalized cell line: CaCo-2 | American Type Culture Collection | Cat#HTB-37 |
| Immortalized cell line: Vero E6 | American Type Culture Collection | Cat#CRL-1586 |
| Immortalized cell line: MDA-MB-231 | DSMZ-German Collection of Microorganisms and Cell Cultures | Cat#ACC 732 |
| Software and algorithms | ||
| FlowJo version 10.7.1 | BD | N/A |
| Prism 7 and 9.0.2 | GraphPad | https://www.graphpad.com/ |
Resource availability
Lead contact
Resources, reagents and further information requirement should be forwarded to and will be responded by the Lead Contact, Edgar Meinl (edgar.meinl@med.uni-muenchen.de).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Study participants
We analyzed the B-cellular responses to SARS-COV-2 of 17 COVID-19 patients (HC = 12, MS = 4 and SLE = 1) that had a mild or asymptomatic disease course (Table 1). Five of them were identified in a survey of health care workers (Weinberger et al., 2021); we specifically selected patients with a seroreversion. Infection was confirmed by SARS-CoV-2-specific PCR (Muenchhoff et al., 2020) and/or SARS-CoV-2 serology, as indicated in Table 1. Three patients did not receive a PCR test at the beginning of the pandemic but displayed typical symptoms (fever, respiratory symptoms, severe anosmia) and subsequently tested positive for SARS-CoV-2 Abs. All study participants tested positive for SARS-CoV-2 antibodies at least once after they were infected. As a reference, we analyzed pre-pandemic blood samples from six healthy adults (HC), two males and four females, with a mean age of 28 years.
Study approval
The study was approved by the ethical committee of the medical faculty of the LMU Munich. Written informed consent was obtained from each donor prior to their inclusion in the study.
Method details
Differentiation of B cells into Ab-secreting cells and ELISPOT
Our experimental scheme is outlined in Figure 1. PBMCs were obtained by a standard density gradient method using SepMate-50 tubes (STEMCELL Technologies, Vancouver, Canada) and frozen. After thawing, the PBMCs were seeded at 1 × 106 cells into 1 mL of culture medium (RPMI +10% FCS) in 24-well plates and differentiated into Ab-secreting cells using the TLR7/8 ligand resiquimod (2.5 μg/mL; Sigma-Aldrich, St. Louis, MO, USA) and IL-2 (1,000 IU/mL; R&D Systems, Minneapolis, MN, USA) during 11 days of culturing, essentially as previously described (Pinna et al., 2009; Thaler et al., 2019; Winklmeier et al., 2019). This culture system has been tested with different B cell subsets and it was found that resiquimod + IL-2 specifically drives the differentiation of memory B cells into Ab-secreting cells (Pinna et al., 2009). The total IgG and IgA levels of the culture supernatants were measured using human IgG and IgA ELISA development kits (Mabtech, Nacka Strand, Sweden). We considered an IgG concentration of >1 μg/mL to indicate successful differentiation of the B cells and included these wells in the subsequent analysis. To determine the frequency of SARS-CoV-2-specific B cells, 200 μL of PBMCs were distributed as limiting dilutions of between 6.25 × 103 and 6.25 × 105 cells/well in 96-round-bottomed-well plates and stimulated for 11 days culture as described above. The reactivity of the Abs in the supernatants was determined using an RBD ELISA as described below. The antigen-reactive cell frequency was determined according to Poisson distribution as the seeded PBMC count for which 37% of the cultures were negative (Pinna et al., 2009; Thaler et al., 2019; Winklmeier et al., 2019). The total B-cell frequency was measured by flow cytometry using the anti-human CD19-PerCP-Cy5.5 Ab (SJ25C1; eBioscience, San Diego, CA, USA) or the anti-human CD19-FITC Ab (HIB19; Invitrogen/Thermo Fisher Scientific, Waltham, MA, USA), and analysis was performed using FlowJo (10.7.1, BD, Franklin Lakes, NJ, USA).
We also analyzed the presence of SARS-CoV-2 RBD specific B cells by ELISPOT using the kit (3850-4APW-R1-1) from Mabtech. Briefly, PBMCs were activated with resiquimod and IL-2 (Jahnmatz et al., 2013), transferred after four days to wells precoated with anti-human Ig and cultured for one more day. The plate was developed with WASP-tagged RBD and anti-WASP-alkaline phosphatase. To image the ELISPOT, a bright-field microscopy was performed at the Core Facility Bioimaging of the Biomedical Center on a Leica THUNDER Imager 3D Live Cell TIRF inverted microscope, equipped with a motorized stage and a Leica DFC450 camera with 2560 x 1920 pixels. Recordings were done with an N PLAN 5×/0.12 dry objective and 2×2 camera binning (1280 × 960 pixels) resulting in a pixel size of 1.36 μm. Mosaic images of individual wells were recorded and stitched in the LAS X 3.7.4 Navigator tool.
ELISAs to detect specific responses to SARS-COV-2 in serum and cell culture supernatants
SARS-CoV-2-specific IgG and IgA were detected in sera and cell culture supernatants using EUROIMMUN anti-SARS-CoV-2 ELISA kits coated with the S1 domain of the SARS-CoV-2 spike protein (EUROIMMUN, Lübeck, Germany). Serum was diluted 1:101 in sample buffer as indicated in the manufacturer's protocol, and cell culture supernatant was applied undiluted. EUROIMMUN uses a ratio-based analysis for test evaluation and recommends interpreting the results as follows: negative (ratio <0.8), borderline (ratio ≥0.8 to <1.1), positive (ratio ≥1.1). For limiting dilution experiments, Abs against SARS-CoV-2 in the cell culture supernatants were detected using an in-house RBD ELISA. Half-area ELISA plates were coated with 50 μL of RBD (2 μg/mL; P2020-001, Trenzyme, Konstanz, Germany) or bovine serum albumin (BSA, 2 μg/mL; Sigma-Aldrich) overnight at 4°C. The plates were then blocked for 2 h at 37°C with 100 μL of blocking buffer (3% milk in PBS containing 0.05% Tween 20). Undiluted cell culture supernatants (50 μL) were incubated at room temperature for 2 h. Abs were then detected with 50 μL of anti-human IgG horseradish peroxidase (1:5000, 109-036-003, Jackson ImmunoResearch, West Grove, PA, USA) and 50 μL of tetramethylbenzidin (TMB, Sigma-Aldrich) as the substrate. The reaction was stopped by adding 25 μL of 1 M sulfuric acid. The optical density (OD) of the chromogenic reaction was measured at 450 nm, and the plate background was measured at 540 nm. The OD cutoff value for the recognition of RBD was 0.52, which was calculated using the mean +3 SD of the control cell culture supernatants from stimulated wells with 106 PBMCs per 1 mL.
Inhibitory and neutralizing activity of Abs towards SARS-CoV-2
To assess the SARS-CoV-2 inhibitory and neutralizing activity of the Abs, we analyzed whether they blocked the binding of the RBD to ACE-2, the cellular receptor for SARS-CoV-2 according to (Abe et al., 2020) (Figure 1). ELISA plates were coated with RBD and blocked as described above. Then 50 μL of the undiluted cell culture supernatants were added and maintained for 2 h at room temperature, followed by the addition of 50 μL of biotin-tagged ACE-2 (1 μg/mL, SAE0171, Sigma-Aldrich). The binding of ACE-2 was detected by 50 μL of streptavidin conjugated with horseradish peroxidase (1:200, 890803, R&D Systems). ELISAs were developed with TMB as described above, and OD values were normalized to those ACE-2 without cell culture supernatant.
The neutralizing activity of sera and cell culture supernatants was also analyzed by using an S3 neutralization assay. CaCo-2 cells in cell culture medium (Dulbecco's Modified Eagle's Medium containing 2% fetal bovine serum) were challenged for 2 h with clinical isolates of different SARS-CoV-2 variants previously obtained from nasopharyngeal swabs of COVID-19 patients. Subsequently, cell culture medium was exchanged, and three days post infection supernatants were passaged on Vero-E6 cells. After three additional days, cell culture supernatants were harvested and stored at −80°C. Virus stocks were characterized by rRT-PCR. A volume of each stock, which results in a 90% cytopathic effect three days post infection, was incubated for 2 h with the neutralizing samples at different dilutions. Subsequently, 10 μL of the virus-sample mixtures were added to 20 μL MDA-MB-231 cells overexpressing the human angiotensin-converting enzyme 2 receptor (hACE2) cultured in 384-well plates (7,500 cells/well). Three days post infection, 10 μL of CellTiter-Glo 2.0 reagent were added to each well and the luminescence was recorded (0.5 s integration time, no filter). Half-maximal inhibitory concentrations (IC50) for inhibiting virus-mediated cell death were computed in Prism via normalized sigmoidal dose-response curve approximation with variable slopes.
Cross-reactivity to RBD of variants of concern
The cross-reactivities of Abs recognizing SARS-CoV-2 wild type (WT) to RBDs of VoCs Alpha/B.1.1.7 (mutation N501Y), Beta/B.1.351 (mutations K417N, E484K, N501Y) and the Gamma/P.1 lineage (mutations K417T, E484K, N501Y), also called B.1.1.248 (Hoffmann et al., 2021), were determined by ELISA. Half-area ELISA plates were coated with 50 μL of RBD WT (2 μg/mL; PX-COV-P046, ProteoGenix, Schiltigheim, France), RBD Alpha/B.1.1.7 (2 μg/mL; PX-COV-P052, ProteoGenix), RBD Beta/B.1.351 (2 μg/mL; PX-COV-P053, ProteoGenix), RBD Lineage Gamma/P.1 also called B.1.1.248 (2 μg/mL; PX-COV-P054, ProteoGenix), or BSA (2 μg/mL; Sigma-Aldrich) overnight at 4°C. The subsequent procedure was as described for the RBD ELISA.
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism 7 and 9.0.2 (GraphPad Software Inc., La Jolla, CA, USA).
Acknowledgments
This work was supported by the DFG (SFB TR128), the Netzwerk Universitätsmedizin des BMBF, and the MOMENTE program LMU (to SM). The authors would like to express their sincere gratitude to Dr. Andreas Thomae at the Core Facility Bioimaging of the Biomedical Center, LMU Munich, for the assistance in acquiring the ELISPOT images, to Pardis Khosravani at the Core Facility Flow Cytometry of the Biomedical Center, LMU Munich, for the assistance in the flow cytometry analysis, and to PD Dr. Andreas Moosmann at the Helmholtz Zentrum München for providing us with patient samples. The project was funded in part by the BMBF initiative “NaFoUniMedCovid19” (01KX2021, Federal Ministry of Education and Research, Germany), subproject B-FAST, and the ForCOVID research initiative funded by the Free State of Bavaria (both to O.T.K.).
We are grateful to Prof. M. Kerschensteiner for continuous support and Dipl.–Ing. Benjamin Obholzer for image design. The authors want to thank M.Sc. Samantha Ho and Drs. Lisa Ann Gerdes, Reinhard Hohlfeld, Hartmut Wekerle, and Naoto Kawakami for their comments on the manuscript.
Author contributions
SW and EM designed the study and experiments. SW, DT, HR, RG, CS, PRW, and MS conducted experiments and analyzed data. PE provided reagents and analyzed data. KE, OTK, MK, and TK contributed patients’ samples, analyzed data, and edited the manuscript. SW, SM, and EM analyzed data and wrote the manuscript.
Declaration of interests
The authors have no conflicts of interests to declare.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103659.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
References
- Abayasingam A., Balachandran H., Agapiou D., Hammoud M., Rodrigo C., Keoshkerian E., Li H., Brasher N.A., Christ D., Rouet R., et al. Long-term persistence of RBD-positive memory B cells encoding neutralising antibodies in SARS-CoV-2 infection. Cell Rep. Med. 2021;2:100228. doi: 10.1016/j.xcrm.2021.100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe K.T., Li Z., Samson R., Samavarchi-Tehrani P., Valcourt E.J., Wood H., Budylowski P., Dupuis A.P., 2nd, Girardin R.C., Rathod B., et al. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight. 2020;5:142362. doi: 10.1172/jci.insight.142362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanna I.J., Carlson N.E., Slifka M.K. Duration of humoral immunity to common viral and vaccine antigens. N. Engl. J. Med. 2007;357:1903–1915. doi: 10.1056/NEJMoa066092. [DOI] [PubMed] [Google Scholar]
- Amanna I.J., Slifka M.K. Mechanisms that determine plasma cell lifespan and the duration of humoral immunity. Immunol. Rev. 2010;236:125–138. doi: 10.1111/j.1600-065X.2010.00912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A., Arya R., Sachan S., Jha S.N., Kalia A., Lall A., Sette A., Grifoni A., Weiskopf D., Coshic P., et al. Immune memory in mild COVID-19 patients and Unexposed donors reveals persistent T cell responses after SARS-CoV-2 infection. Front. Immunol. 2021;12:636768. doi: 10.3389/fimmu.2021.636768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarth N. The shaping of a B cell pool maximally responsive to infections. Annu. Rev. Immunol. 2021:103–129. doi: 10.1146/annurev-immunol-042718-041238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarth N., Nikolich-Žugich J., Lee F.E., Bhattacharya D. Antibody responses to SARS-CoV-2: let's stick to known knowns. J. Immunol. 2020;205:2342–2350. doi: 10.4049/jimmunol.2000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagigi A., Yu M., Falck-Jones S., Vangeti S., Österberg B., Åhlberg E., Azizmohammadi L., Falck-Jones R., Gubisch P.C., Ödemis M., et al. Airway antibodies wane rapidly after COVID-19 but B cell memory is generated across disease severity. medRxiv. 2020 [Google Scholar]
- Casadevall A., Henderson J.P., Joyner M.J., Pirofski L.A. SARS-CoV-2 variants and convalescent plasma: reality, fallacies, and opportunities. J. Clin. Invest. 2021;131:e148832. doi: 10.1172/jci148832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiereghin A., Zagari R.M., Galli S., Moroni A., Gabrielli L., Venturoli S., Bon I., Rossini G., Saracino I.M., Pavoni M., et al. Recent advances in the evaluation of serological assays for the diagnosis of SARS-CoV-2 infection and COVID-19. Front. Public Health. 2020;8:620222. doi: 10.3389/fpubh.2020.620222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K., Jebbink M.F., Matser A., Kinsella C.M., Rueda P., et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galipeau Y., Greig M., Liu G., Driedger M., Langlois M.A. Humoral responses and serological assays in SARS-CoV-2 infections. Front. Immunol. 2020;11:610688. doi: 10.3389/fimmu.2020.610688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall V.J., Foulkes S., Charlett A., Atti A., Monk E.J.M., Simmons R., Wellington E., Cole M.J., Saei A., Oguti B., et al. SARS-CoV-2 infection rates of antibody-positive compared with antibody-negative health-care workers in England: a large, multicentre, prospective cohort study (SIREN) Lancet. 2021;397:1459–1469. doi: 10.1016/s0140-6736(21)00675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley G.E., Edwards E.S.J., Aui P.M., Varese N., Stojanovic S., McMahon J., Peleg A.Y., Boo I., Drummer H.E., Hogarth P.M., et al. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci. Immunol. 2020;5:eabf8891. doi: 10.1126/sciimmunol.abf8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebeis B.J., Klenovsek K., Rohwer P., Ritter U., Schneider A., Mach M., Winkler T.H. Activation of virus-specific memory B cells in the absence of T cell help. J. Exp. Med. 2004;199:593–602. doi: 10.1084/jem.20030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., Katzelnick L.C., Rattigan S.M., Borgert B.A., Moreno C.A., Solomon B.D., Trimmer-Smith L., et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat. Commun. 2020;11:4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnmatz M., Kesa G., Netterlid E., Buisman A.M., Thorstensson R., Ahlborg N. Optimization of a human IgG B-cell ELISpot assay for the analysis of vaccine-induced B-cell responses. J. Immunol. Methods. 2013;391:50–59. doi: 10.1016/j.jim.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2-what do they mean? JAMA. 2021;325:529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., Hu J.L., Xu W., Zhang Y., Lv F.J., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Lyski Z.L., Brunton A.E., Strnad M.I., Sullivan P.E., Siegel S.A.R., Tafesse F.G., Slifka M.K., Messer W.B. SARS-CoV-2 specific memory B-cells from individuals with diverse disease severities recognize SARS-CoV-2 variants of concern. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiab585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz R.A., Hauser A.E., Hiepe F., Radbruch A. Maintenance of serum antibody levels. Annu.Rev.Immunol. 2005;23:367–386. doi: 10.1146/annurev.immunol.23.021704.115723. [DOI] [PubMed] [Google Scholar]
- Marklund E., Leach S., Axelsson H., Nyström K., Norder H., Bemark M., Angeletti D., Lundgren A., Nilsson S., Andersson L.M., et al. Serum-IgG responses to SARS-CoV-2 after mild and severe COVID-19 infection and analysis of IgG non-responders. PLoS One. 2020;15:e0241104. doi: 10.1371/journal.pone.0241104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marot S., Malet I., Leducq V., Zafilaza K., Sterlin D., Planas D., Gothland A., Jary A., Dorgham K., Bruel T., et al. Rapid decline of neutralizing antibodies against SARS-CoV-2 among infected healthcare workers. Nat. Commun. 2021;12:844. doi: 10.1038/s41467-021-21111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J.C., Murthy S., Diaz J. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect. Dis. 2020;20:e192–e197. doi: 10.1016/s1473-3099(20)30483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenchhoff M., Mairhofer H., Nitschko H., Grzimek-Koschewa N., Hoffmann D., Berger A., Rabenau H., Widera M., Ackermann N., Konrad R., et al. Multicentre comparison of quantitative PCR-based assays to detect SARS-CoV-2, Germany, March 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.Es.2020.25.24.2001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Contant P., Embong A.K., Kanagaiah P., Chaves F.A., Yang H., Branche A.R., Topham D.J., Sangster M.Y. S protein-reactive IgG and memory B cell production after human SARS-CoV-2 infection includes broad reactivity to the S2 subunit. mBio. 2020;11 doi: 10.1128/mBio.01991-20. e01991–01920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogega C.O., Skinner N.E., Blair P.W., Park H.S., Littlefield K., Ganesan A., Dhakal S., Ladiwala P., Antar A.A., Ray S.C., et al. Durable SARS-CoV-2 B cell immunity after mild or severe disease. J. Clin. Invest. 2021;131:e145516. doi: 10.1172/jci145516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oved K., Olmer L., Shemer-Avni Y., Wolf T., Supino-Rosin L., Prajgrod G., Shenhar Y., Payorsky I., Cohen Y., Kohn Y., et al. Multi-center nationwide comparison of seven serology assays reveals a SARS-CoV-2 non-responding seronegative subpopulation. EClinicalMedicine. 2020;29:100651. doi: 10.1016/j.eclinm.2020.100651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault J., Tremblay T., Fournier M.J., Drouin M., Beaudoin-Bussières G., Prévost J., Lewin A., Bégin P., Finzi A., Bazin R. Waning of SARS-CoV-2 RBD antibodies in longitudinal convalescent plasma samples within 4 months after symptom onset. Blood. 2020;136:2588–2591. doi: 10.1182/blood.2020008367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna D., Corti D., Jarrossay D., Sallusto F., Lanzavecchia A. Clonal dissection of the human memory B-cell repertoire following infection and vaccination. Eur. J. Immunol. 2009;39:1260–1270. doi: 10.1002/eji.200839129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., Planchais C., Buchrieser J., Rajah M.M., Bishop E., et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021;27:917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- Purtha W.E., Tedder T.F., Johnson S., Bhattacharya D., Diamond M.S. Memory B cells, but not long-lived plasma cells, possess antigen specificities for viral escape mutants. J. Exp. Med. 2011;208:2599–2606. doi: 10.1084/jem.20110740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast I., Tarlinton D. B cell memory: understanding COVID-19. Immunity. 2021;54:205–210. doi: 10.1016/j.immuni.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees-Spear C., Muir L., Griffith S.A., Heaney J., Aldon Y., Snitselaar J.L., Thomas P., Graham C., Seow J., Lee N., et al. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021;34:108890. doi: 10.1016/j.celrep.2021.108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 2021;184:169–183.e17. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado M.M., Scarsella M., Pandolfi E., Cascioli S., Giorda E., Chionne P., Madonne E., Gesualdo F., Romano M., Ausiello C.M., et al. Switched memory B cells maintain specific memory independently of serum antibodies: the hepatitis B example. Eur. J. Immunol. 2011;41:1800–1808. doi: 10.1002/eji.201041187. [DOI] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino E.C., Buss L.F., Carvalho M.P.S., Prete C.A., Jr., Crispim M.A.E., Fraiji N.A., Pereira R.H.M., Parag K.V., da Silva Peixoto P., Kraemer M.U.G., et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/s0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L.F., Mizgerd J.P. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Invest. 2021;131:e143380. doi: 10.1172/jci143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar M., Rappazzo C.G., Wieland-Alter W.F., Hsieh C.L., Wrapp D., Esterman E.S., Kaku C.I., Wec A.Z., Geoghegan J.C., McLellan J.S., et al. Prolonged evolution of the human B cell response to SARS-CoV-2 infection. Sci. Immunol. 2021;6:eabg6916. doi: 10.1126/sciimmunol.abg6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariol A., Perlman S. Lessons for COVID-19 immunity from other coronavirus infections. Immunity. 2020;53:248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K.J.A., Hemmings O., O'Byrne A., Kouphou N., Galao R.P., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherina N., Piralla A., Du L., Wan H., Kumagai-Braesch M., Andréll J., Braesch-Andersen S., Cassaniti I., Percivalle E., Sarasini A., et al. Persistence of SARS-CoV-2-specific B and T cell responses in convalescent COVID-19 patients 6-8 months after the infection. Med (N Y) 2021;2:281–295.e4. doi: 10.1016/j.medj.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal A., Chappert P., Barba-Spaeth G., Roeser A., Fourati S., Azzaoui I., Vandenberghe A., Fernandez I., Meola A., Bouvier-Alias M., et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell. 2021;184:1201–1213.e14. doi: 10.1016/j.cell.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L., Czartoski J., Wan Y.H., Homad L.J., Rubin V., Glantz H., Neradilek M., Seydoux E., Jennewein M.F., MacCamy A.J., et al. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. 2021:eabg9175. doi: 10.1126/science.abg9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlin D., Mathian A., Miyara M., Mohr A., Anna F., Claër L., Quentric P., Fadlallah J., Devilliers H., Ghillani P., et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021;13:eabd2223. doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler F.S., Thaller A.L., Biljecki M., Schuh E., Winklmeier S., Mahler C.F., Gerhards R., Volk S., Schnorfeil F., Subklewe M., et al. Abundant glutamic acid decarboxylase (GAD)-reactive B cells in gad-antibody-associated neurological disorders. Ann. Neurol. 2019;85:448–454. doi: 10.1002/ana.25414. [DOI] [PubMed] [Google Scholar]
- Turner J.S., Kim W., Kalaidina E., Goss C.W., Rauseo A.M., Schmitz A.J., Hansen L., Haile A., Klebert M.K., Pusic I., et al. SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature. 2021;595:421–425. doi: 10.1038/s41586-021-03647-4. [DOI] [PubMed] [Google Scholar]
- Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman-Mentesh A., Dror Y., Tur-Kaspa R., Markovitch D., Kournos T., Dicker D., Wine Y. SARS-CoV-2 specific memory B cells frequency in recovered patient remains stable while antibodies decay over time. medRxiv. 2020 [Google Scholar]
- Varnaitė R., García M., Glans H., Maleki K.T., Sandberg J.T., Tynell J., Christ W., Lagerqvist N., Asgeirsson H., Ljunggren H.G., et al. Expansion of SARS-CoV-2-specific antibody-secreting cells and generation of neutralizing antibodies in hospitalized COVID-19 patients. J. Immunol. 2020;205:2437–2446. doi: 10.4049/jimmunol.2000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viant C., Weymar G.H.J., Escolano A., Chen S., Hartweger H., Cipolla M., Gazumyan A., Nussenzweig M.C. Antibody affinity shapes the choice between memory and germinal center B cell fates. Cell. 2020;183:1298–1311.e11. doi: 10.1016/j.cell.2020.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Viant C., Gaebler C., Cipolla M., Hoffmann H.H., Oliveira T.Y., Oren D.A., et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger T., Steffen J., Osterman A., Mueller T.T., Muenchhoff M., Wratil P.R., Graf A., Krebs S., Quartucci C., Spaeth P.M., et al. Prospective longitudinal serosurvey of health care workers in the first wave of the SARS-CoV-2 pandemic in a quaternary care hospital in Munich, Germany. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciaa1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisel F., Shlomchik M. Memory B cells of mice and humans. Annu. Rev. Immunol. 2017;35:255–284. doi: 10.1146/annurev-immunol-041015-055531. [DOI] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Winklmeier S., Schlüter M., Spadaro M., Thaler F.S., Vural A., Gerhards R., Macrini C., Mader S., Kurne A., Inan B., et al. Identification of circulating MOG-specific B cells in patients with MOG antibodies. Neurol. Neuroimmunol. Neuroinflamm. 2019;6:625. doi: 10.1212/nxi.0000000000000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R., Belk J.A., Govero J., Uhrlaub J.L., Reinartz D., Zhao H., Errico J.M., D'Souza L., Ripperger T.J., Nikolich-Zugich J., et al. Affinity-restricted memory B cells dominate recall responses to heterologous flaviviruses. Immunity. 2020;53:1078–1094.e7. doi: 10.1016/j.immuni.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff M.C., Ramonell R.P., Nguyen D.C., Cashman K.S., Saini A.S., Haddad N.S., Ley A.M., Kyu S., Howell J.C., Ozturk T., et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wratil P.R., Schmacke N.A., Osterman A., Weinberger T., Rech J., Karakoc B., Zeilberger M., Steffen J., Mueller T.T., Spaeth P.M., et al. In-depth profiling of COVID-19 risk factors and preventive measures in healthcare workers. Infection. 2021:1–14. doi: 10.1007/s15010-021-01672-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Tsibane T., McGraw P.A., House F.S., Keefer C.J., Hicar M.D., Tumpey T.M., Pappas C., Perrone L.A., Martinez O., et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455:532–536. doi: 10.1038/nature07231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue S., Li Z., Lin Y., Yang Y., Yuan M., Pan Z., Hu L., Gao L., Zhou J., Tang J., et al. Sensitivity of SARS-CoV-2 variants to neutralization by convalescent sera and a VH3-30 monoclonal antibody. Front. Immunol. 2021;12:751584. doi: 10.3389/fimmu.2021.751584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarletti G., Tiberi M., De Molfetta V., Bossù M., Toppi E., Bossù P., Scapigliati G. A cell-based ELISA to improve the serological analysis of anti-SARS-CoV-2 IgG. Viruses. 2020;12:1274. doi: 10.3390/v12111274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Zhang Q., Ali A., Li K., Shao N., Zhou X., Ye Z., Chen X., Cao S., Cui J., et al. Sustainability of SARS-CoV-2 induced humoral immune responses in COVID-19 patients from hospitalization to convalescence over six months. Virol. Sin. 2021;36:869–878. doi: 10.1007/s12250-021-00360-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.